Abstract
The introduction of noncanonical amino acids into proteins and peptides has been of great interest for many years and has facilitated the detailed study of peptide/protein structure and mechanism. In addition to numerous nonproteinogenic α-l-amino acids, bacterial ribosome modification has provided the wherewithal to enable the synthesis of peptides and proteins with a much greater range of structural diversity, as has the use of endogenous bacterial proteins in reconstituted protein synthesizing systems. In a recent report, elongation factor P (EF-P), putatively essential for enabling the incorporation of contiguous proline residues into proteins, was shown to facilitate the introduction of an N-methylated amino acid in addition to proline. This finding prompted us to investigate the properties of this protein factor with a broad variety of structurally diverse amino acid analogues using an optimized suppressor tRNAPro that we designed. While these analogues can generally be incorporated into proteins only in systems containing modified ribosomes specifically selected for their incorporation, we found that EF-P could significantly enhance their incorporation into model protein dihydrofolate reductase using wild-type ribosomes. Plausibly, the increased yields observed in the presence of structurally diverse amino acid analogues may result from the formation of a stabilized ribosomal complex in the presence of EF-P that provides more favorable conditions for peptide bond formation. This finding should enable the facile incorporation of a much broader structural variety of amino acid analogues into proteins and peptides using native ribosomes.
Graphical Abstract
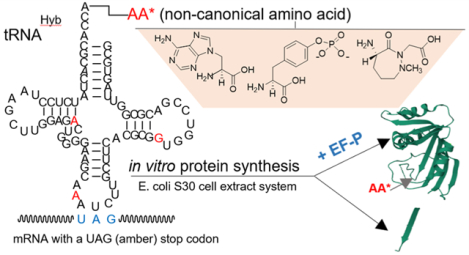
INTRODUCTION
Native proteins are linear polymers built from 20 different genetically encoded amino acids that are linked to each other via peptide bonds. All proteinogenic amino acids possess common structural features, namely, an α-carbon to which an amino group, a carboxyl group, and a side chain with a variable structure and composition are bonded. The single exception is proline, the only amino acid having an α-amino group attached directly to the side chain. Replacement of the primary amino group with a secondary amino group as part of a cyclic pyrrolidine ring structure decreases the efficiency of proline both as an aminoacyl-binding site (A-site) acceptor of the peptidyl moiety during peptide bond formation1 and as a peptidyl-binding site (P-site) peptidyl donor.2–4 As a consequence, polypeptides with a high proline content display poor translational efficiency, especially noticeable in case of di- and triprolyl motifs.5,6 However, the distinctive structure of proline endows exceptional conformational rigidity, and this, in turn, confers a unique role to this cyclic amino acid in defining protein conformation. Proline is commonly found as the first residue of an α-helix; in the edge strands of β-sheets, it aids in the formation of β-turns and introduces kinks into α-helices. Given the structural importance of proline, cells have evolved specialized elongation factors in bacteria (EF-P) and eukaryotes (EF5 and elF-5A), which alleviate proline-induced ribosome stalling.
Bacterial elongation factor P (EF-P) has three β-barrel domains (Figure 1A) and an overall shape reminiscent of the L-shape of tRNA.7,8 The factor binds between the peptidyl (P) and the exit (E) sites of the peptidyl transferase center (PTC) and spans both ribosomal subunits (Figure 1B), as revealed by cocrystallization with the initiator tRNA, mRNA, and 70S ribosome.8,9 EF-P contacts tRNA at three different regions: the acceptor stem, the D-arm, and the anticodon stem-loop. The D-arm appears to be a critical recognition determinant for EF-P, as evidenced by the outcome of the in vitro translation using chimeric and mutagenized tRNA constructs.10 Only the initiator fMet-tRNAfMet and the tRNAPro isoacceptors have D-arms with the characteristics required for interaction with EF-P. Thus, the effect of EF-P is especially pronounced toward translation of mRNAs containing polyproline motifs.4,6 Alleviation of ribosome stalling by EF-P occurs via the organization of a Pro-tRNAPro structure toward a catalytically productive orientation in the PTC.11 The overall stimulatory effect on translation depends on the strength and position of the stalling motif,4,12 physical and chemical properties of the upstream amino acids,13,14 and the rate of translation initiation.15
Figure 1.
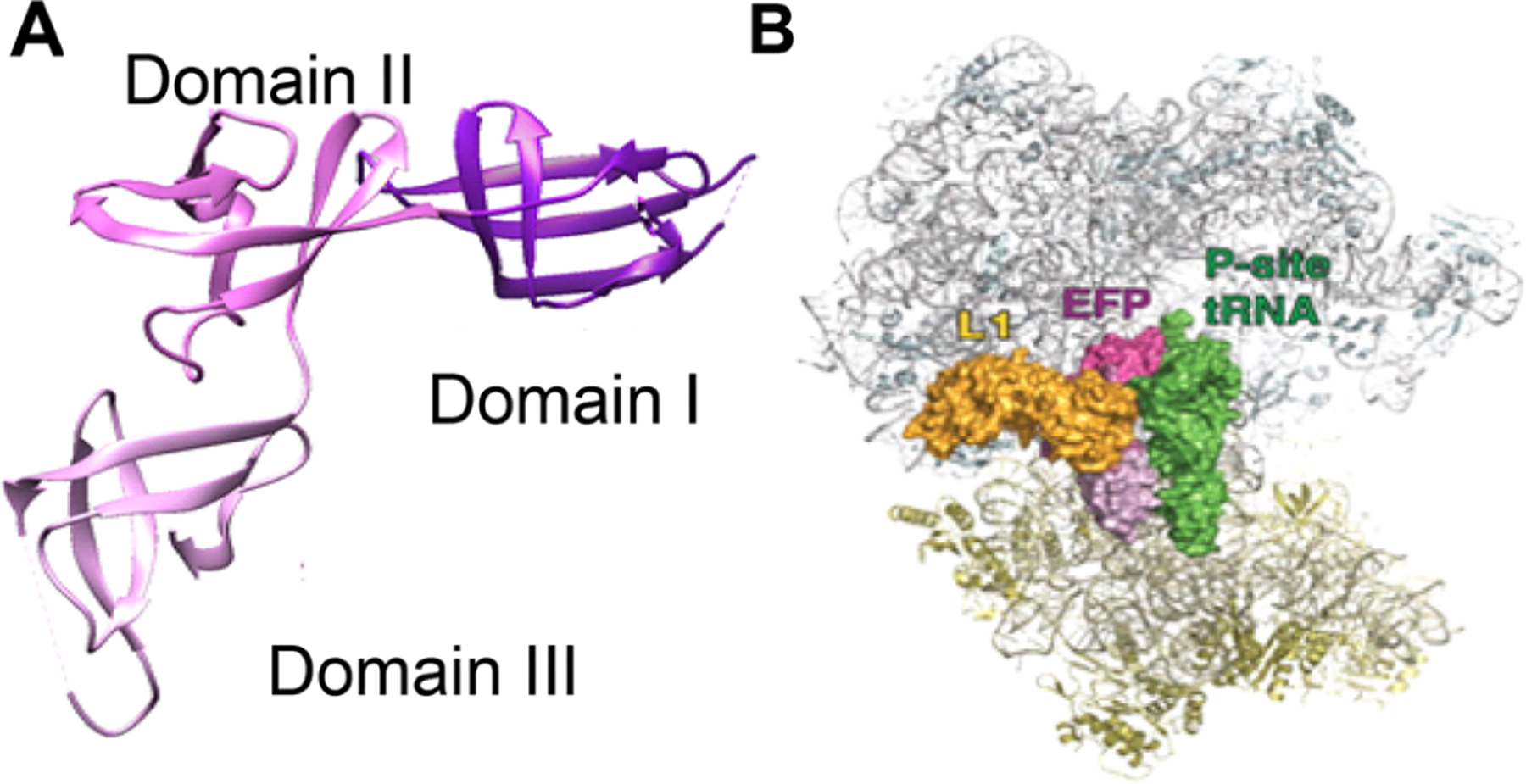
Structure of Clostridium thermocellum EF-P (pdb 1YBY). (A) The three structural domains of EF-P are shown in different shades of magenta. (B) An overview of the binding of EF-P to the 70S ribosome (pdb 2j00 and 2j01; adapted from Blaha, G.; Stanley, R. E.; Steitz, T. A., Science 2009, 325, 966–9709). Reprinted with permission from AAAS. EF-P is in magenta, while the 50S subunit is colored gray, and the 30S subunit is in yellow. Additionally, ribosomal protein L1 is shown in gold, and the P-site tRNA is in green.
EF-P has been reported to become active only following certain post-translational modifications.4,16 In Escherichia coli, the post-translational modifications are β-lysinylation and hydroxylation of Lys34, and they require the consecutive action of three enzymes: lysine 2,3-aminomutase EpmB (YjeK) that converts (S)-α-lysine to (R)-β-lysine, EpmA synthetase (YjeA, a derivative of type II aminoacyl tRNA synthetase) that ligates (R)-β-lysine to the ε-amino group of Lys34,17–20 and EF-P hydroxylase EpmC (YfcM) that hydroxylates β-lysinylated Lys34 at C5(δ).21 In contrast to β-lysinylation, which is obligatory for EF-P activity, hydroxylation, at least in vitro, appears to have only a minor modulating role.4 Some bacteria, such as Pseudomonas aeruginosa, employ EF-P activation through rhamnosylation on Arg32,22 while in eukaryotes, IF-5A, a homologue of EF-P, becomes active following hypusination.23
As reported by Katoh et al.,10 full functionality of post-translationally modified EF-P requires (i) occupancy of the PTC A-site by a tRNA activated with proline or another secondary amino acid and (ii) occupancy of the PTC P-site by tRNAPro activated with peptidyl-proline or at least an activated tRNA having the tRNAPro D-arm sequence. To date, N-methylthreonine has been the only noncanonical amino acid employed to replace the second proline residue in a prolylproline motif in this type of system.10 It may be noted, however, that EF-P has been shown to enhance the expression of peptides containing D-amino acids when also using tRNAs modified in the D-arm and T-stem24 and influences the facility of reaction of structural analogues of proline with puromycin in a ribosomal system.11
The present study further explores the structural boundaries of this system and tests whether EF-P, added exogenously to the E. coli S-30 extract, can enhance positionally predetermined incorporation of unnatural amino acid analogues activated on an optimized suppressor tRNAPro construct during coupled in vitro transcription/translation of a model protein. Using an experimental system in which the codons corresponding to Ala9 and Val10 of E. coli dihydrofolate reductase (DHFR) were altered to encode proline at position 9 and stop codon UAG at position 10, we studied the incorporation of seven different structurally diverse noncanonical amino acids in the presence and absence of modified E. coli EF-P. A majority of these were found to exhibit enhanced DHFR synthesis in the presence of EF-P.
All seven of these noncanonical amino acids have been incorporated into proteins previously in an in vitro S-30 protein synthesizing system, but only one (compound 1, an α-amino acid) was incorporated into protein to a significant extent using wild-type ribosomes; the remainder required modified ribosomes specifically selected for their incorporation. Further investigation using cyclic dipeptide analogue 7 revealed that EF-P produced from a plasmid containing the DNA for EF-P expression, but not for the expression of the modifying enzymes, could also significantly increase their incorporation into DHFR when used at slightly higher concentrations.
RESULTS AND DISCUSSION
To study the effect of EF-P on unnatural amino acid incorporation during in vitro protein translation, a runoff expression vector for E. coli tRNAPro containing a CUA anticodon as a replacement for wild-type CGG anticodon was prepared. In addition, C1:G72 and G2:C71 base pairs in the acceptor stem were interchanged. The modified suppressor tRNA was denoted as tRNAPro1. G72, along with G35 and G36 in the anticodon loop and the discriminator A73, is an element recognizable by prolyl-tRNA synthetase.25,26 In fact, a single substitution at G72 in tRNAPro leads to a large decrease in aminoacylation by the endogenous prolyl-tRNA synthetase.25,27,28 Therefore, the changes introduced were expected to decrease the level of functional interactions with the cognate tRNAPro isoacceptors. The structure of the E. coli suppressor tRNAPro1 is presented in Figure 2A.
Figure 2.
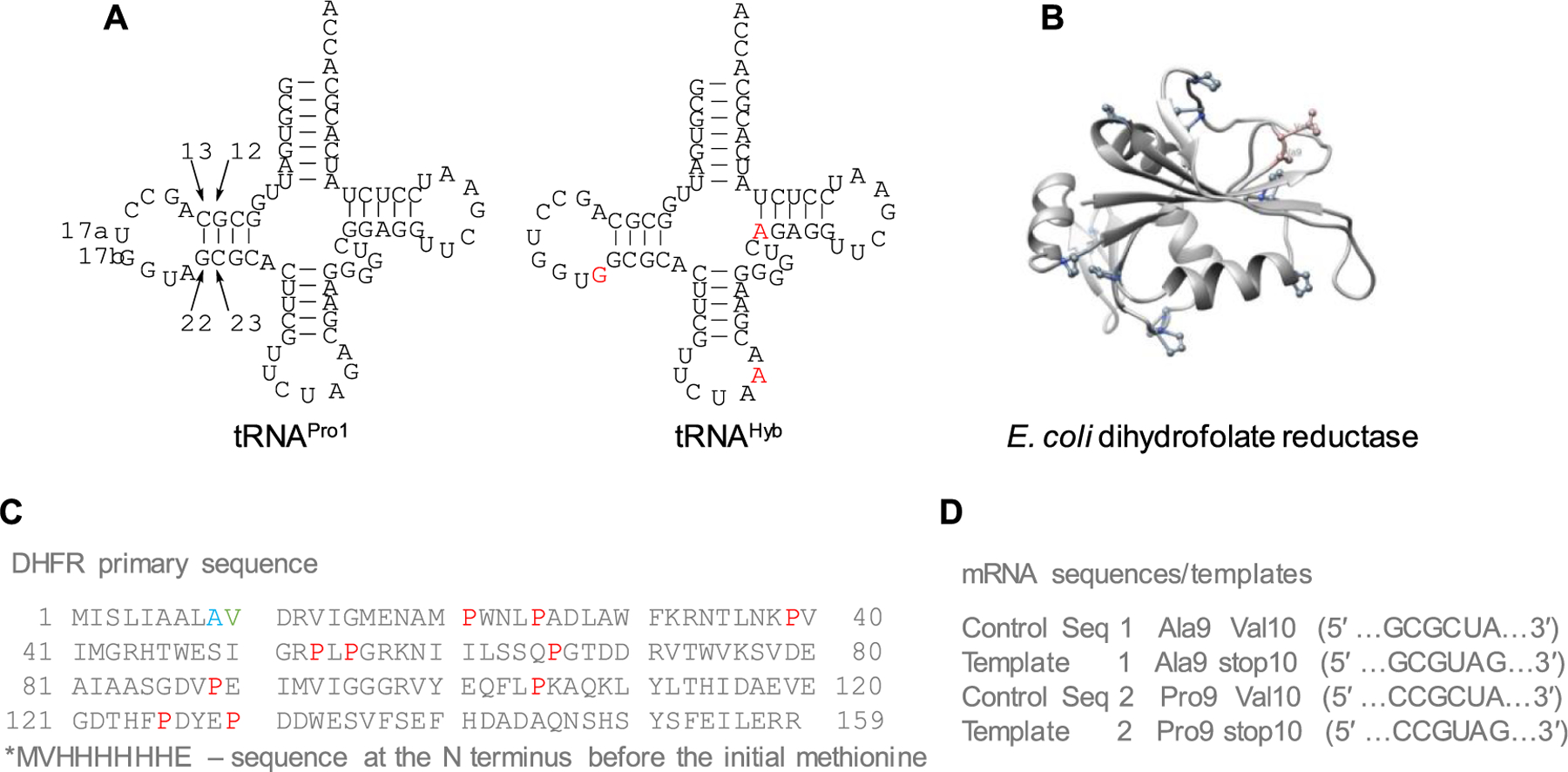
Model system for studying the effect of EF-P on unnatural amino acid incorporation. (A) Secondary structure of E. coli suppressor tRNAPro1 and modified suppressor tRNAHyb. Mutations required to convert tRNAPro1 to tRNAHyb are shown in red. (B) X-ray crystal structure of E. coli DHFR (pdb 5DFR). Key amino acids are shown with their corresponding structures. (C) Primary sequence of E. coli DHFR. Prolines are colored red, mutated Ala9 to Pro9 is colored blue, and mutated Val10 to UAG stop codon is colored green. The star above the initial methionine denotes the position of the His tag. (D) Templates used for coupled in vitro transcription/translation reactions are shown.
Studies have demonstrated that greater cohesion in the anticodon arm of a suppressor tRNA confers increased structural rigidity and higher suppression efficiency.29 The C32UCUAAA38 anticodon loop and a U31–A39 base pair in the anticodon stem are critical in this regard. Among E. coli tRNAs, the G52–C62 and G53–C61 pairs are highly conserved, and the consensus sequence for the most active tRNAs is W49NRGG53-C61CYNW65 (where W is A or T; N is A, T, G, or C; R is A or G; and Y is C or T).30 On the other hand, the stability of the D-arm provided by C13/G22 and G12/C23 pairs along with the 9-nt D-loop appears to be essential.10 Based on that knowledge, a runoff expression vector for another variant of E. coli suppressor tRNAPro1, denoted tRNAHyb, was prepared with three additional point mutations, namely, A21G, G37A, and G49A (Figure 2A).
E. coli DHFR, an enzyme characterized crystallographically (pdb 5DFR) (Figure 2B,C),31 was chosen as a model protein for coupled in vitro transcription/translation studies. DHFR is a small protein (17.96 kDa) that does not contain endogenous diprolyl motifs but has a relatively high proline content (6.3%). The Val10 position was chosen for the site of incorporation of noncanonical amino acids because it is close to the N terminus and is not part of a secondary structure. To this end, the wild-type coding sequence (Figure 2D, control sequence 1) was mutated to generate an amber (UAG) stop codon at position 10 (template 1—DHFR, Ala9stop10). Considering that the effect of EF-P can be more pronounced for the translation of diprolyl motifs,4,5 the preceding Ala9 was additionally replaced with Pro9. This was done by a two-step procedure: the wild-type coding sequence was mutated by a GCG → CCG replacement to afford the Ala9 → Pro9 substitution (control sequence 2). This sequence was further mutated to introduce an amber stop codon in lieu of Val10 (template 2—Pro9stop10) (Figure 2D).
E. coli EF-P, along with three modifying enzymes, was successfully overexpressed from the pET28:EF-P/YjeA/YjeK/YfcM vector (Figure S1A).4,10 Lysine 2,3-aminomutase EpmB (26.3 kDa) migrated separately, while the other three proteins, including EF-P, formed a single band due to the similarity in their molecular masses. However, only EF-P was His-tagged, which allowed its purification by immobilized-metal affinity chromatography (IMAC) (Figure S1B). The N-terminally positioned His-tag was subsequently removed by cleavage with thrombin (Figure S1C).
E. coli suppressors tRNAPro1 and tRNAHyb were initially activated with proteinogenic amino acids. This was accomplished by linearizing the DNA plasmids encoding the two tRNAs without the final CpA sequence. In vitro runoff transcription with T7 RNA polymerase afforded the abbreviated tRNA-COH transcripts. These were ligated to chemically prepared aminoacyl-pdCpA derivatives (Scheme 1).32–35 In addition to proline, phenylalanine and lysine were also employed for suppressor tRNA aminoacylation (Figure S2). Also, phenylalanine and lysine were used to activate yeast suppressor tRNAPhe,33 which we use routinely in our laboratory for in vitro production of modified proteins.35 In vitro DHFR synthesis in the presence of each of the chemically misacylated suppressor tRNAs was performed from template 1 (DHFR, Ala9stop10) and template 2 (DHFR, Pro9stop10), in the absence of exogenous EF-P (Scheme 1, Figure 3). As shown, there was minimal incorporation of any of the amino acids used to activate the E. coli suppressor tRNAPro1. Although some of the recognition elements for E. coli prolyl-tRNA synthetase had been eliminated (Figure 2A), others, including the discriminator A73 base, remained intact. This, and the fact that misacylated E. coli suppressor tRNAPro1 was present in excess and may have undergone partial hydrolysis during the course of in vitro synthesis, could plausibly have resulted in suppressed activation of the cognate tRNAPro isoacceptors by the E. coli prolyl-tRNA synthetase. If actually realized, this could have compromised the translation efficiency. Interestingly, suppressor tRNAHyb did not have such a detrimental effect on DHFR synthesis, likely because of the stimulatory effect of the introduced point mutations (A21G, G37A, and G49A) on suppression efficiency. The effect of template on translation efficiency was negligible when suppressor tRNAHyb was activated with proline but readily apparent when phenylalanine or lysine was used for tRNAHyb activation. Template 2 (DHFR, Pro9stop10) afforded a 3.25- to 3.5-fold greater yield of DHFR in vitro than did template 1 (DHFR, Ala9stop10) (Figure 3). DHFR synthesis was much more effective in control samples when yeast suppressor tRNAPhe activated with phenylalanine was employed, likely because yeast tRNAPhe is hardly recognized (<1%) by the E. coli phenylalanyl-tRNA synthetase.36 In this regard, it should be noted that the combination of template 2 and suppressor tRNAHyb also resulted in significant phenylalanine incorporation, while the alternative use of tRNAPro1 having preserved G37 and G49 (part of the recognition features of tRNAPro for E. coli prolyl-tRNA synthetase26) was highly ineffective (Figure 3B). Other factors may also contribute to the above outcome.
Scheme 1.
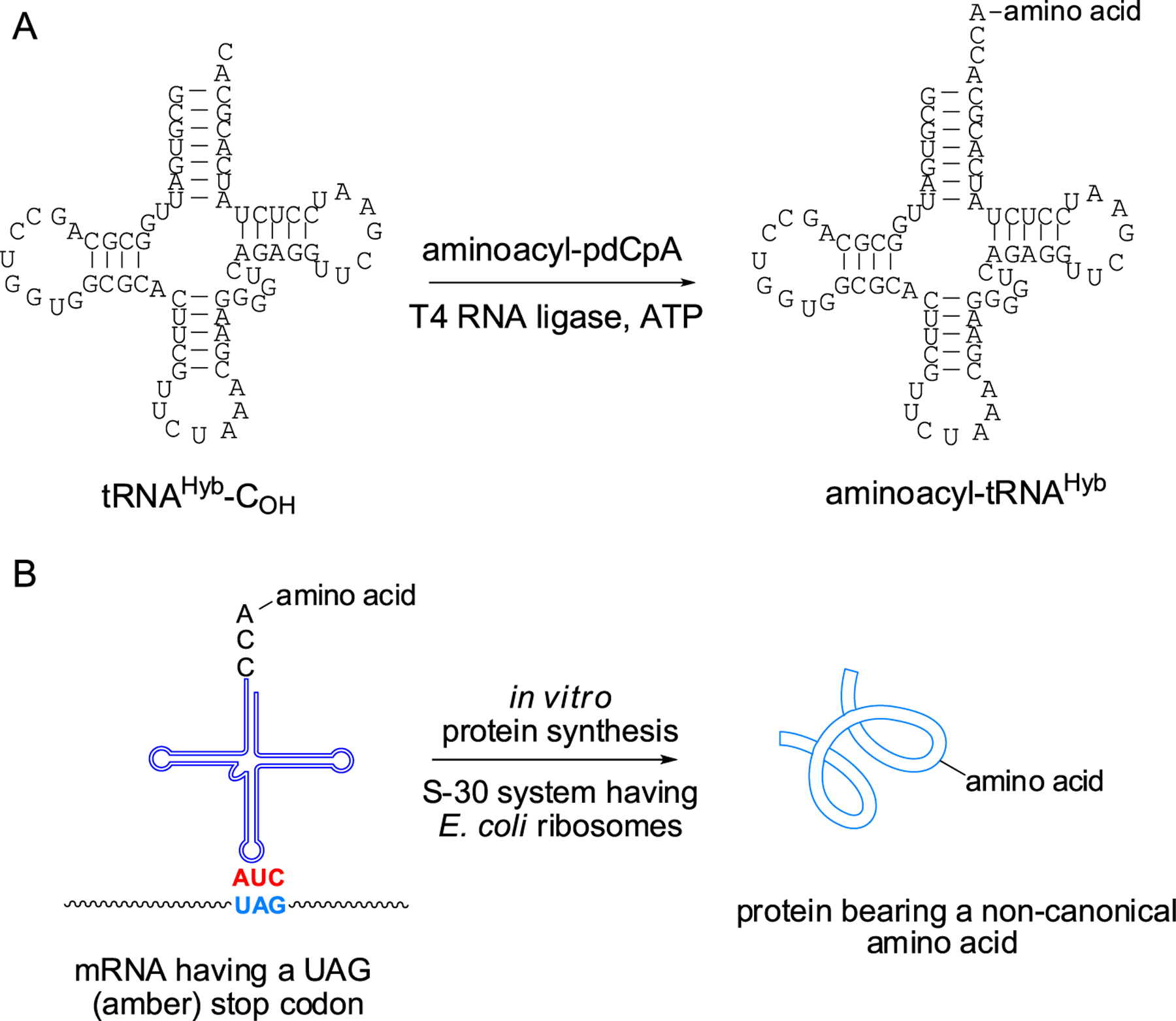
(A) Preparation of a Misacylated Suppressor tRNACUA Transcript by T4 RNA Ligase-Mediated Attachment of an Aminoacylated pdCpA Derivative to an Abbreviated tRNA-COH Lacking the Dinucleotide Sequence Normally Found at the 3′-End of tRNAs. (B) Introduction of a Noncanonical Amino Acid into a Protein Synthesized Ribosomally by Suppression of Nonsense Codon UAG in the mRNA with a Misacylated tRNACUA. All of the Activated tRNAs in This Report Are Amber Suppressor tRNAs
Figure 3.
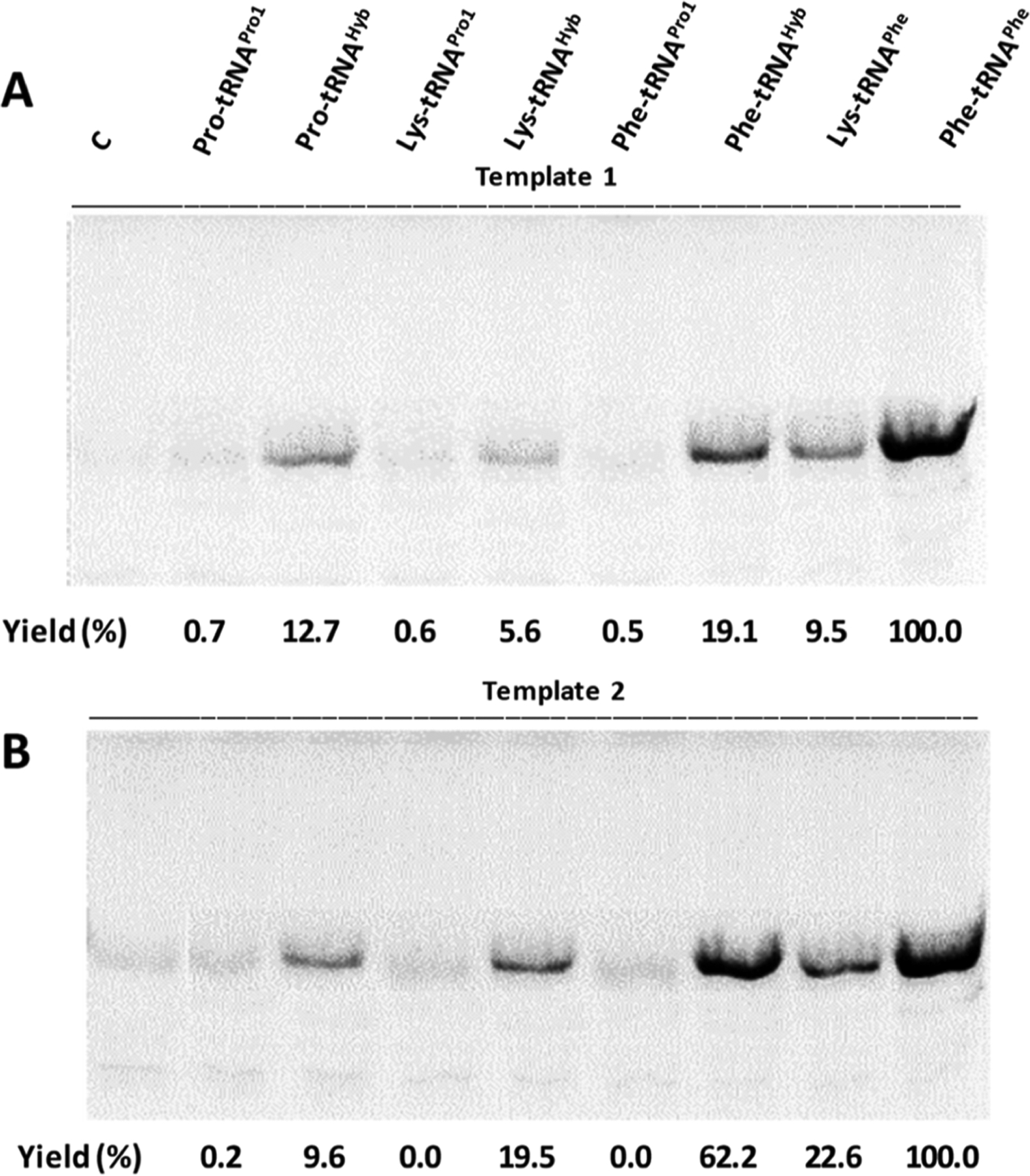
Coupled in vitro transcription/translation guided by (A) template 1 (DHFR, Ala9stop10) or (B) template 2 (DHFR Pro9stop10) in the presence of different activated suppressor tRNAs. Analysis was performed by SDS-PAGE and autoradiography. Yields are expressed as a percentage of those obtained in the presence of yeast suppressor tRNAPhe activated with phenylalanine. A parallel control sample in the absence of any suppressor tRNA (denoted as C) was run for each experiment.
Based on the above results and taking into account that the most essential elements in the D-arm are virtually identical between tRNAPro1 and tRNAHyb, only tRNAHyb was employed in further experiments. Next, the effect of EF-P on the yield of DHFR, translated in the presence of suppressor tRNAHyb aminoacylated with proline (Pro-tRNAHyb) was studied (Figure 4).
Figure 4.
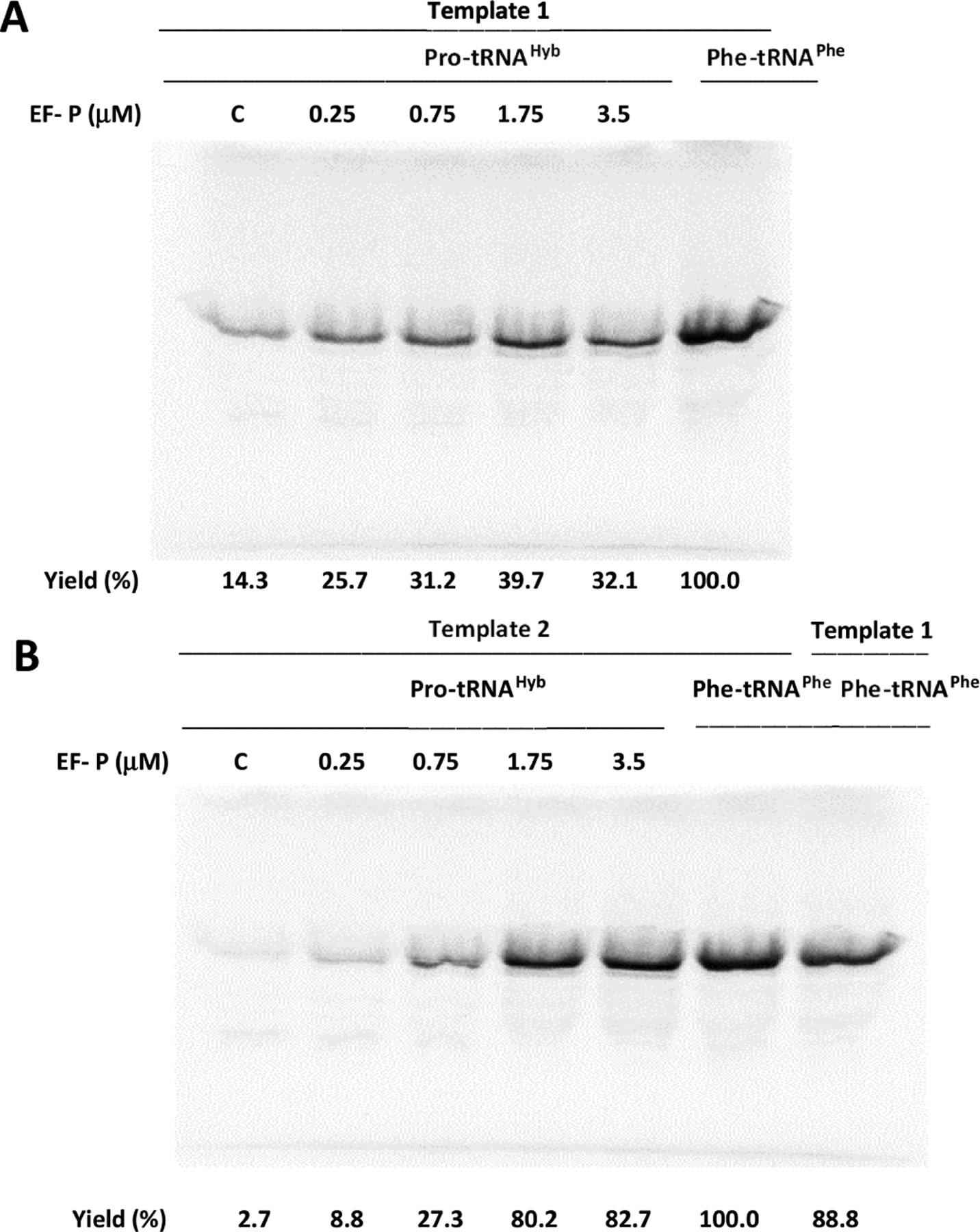
Effect of E. coli EF-P on in vitro translation efficiency from (A) template 1 (DHFR, Ala9stop10) or (B) template 2 (DHFR, Pro9stop10) in the presence of suppressor tRNAHyb acylated with proline (Pro-tRNAHyb). Yields (%) are presented comparatively to the DHFR yield in the presence of yeast suppressor tRNAPhe aminoacylated with phenylalanine (Phe-tRNAPhe) without the addition of exogenous EF-P.
Stepwise increase of exogenous EF-P concentration from 0.25 to 3.5 μM correlated positively with the DHFR production directed by template 1 and template 2, but a better result was realized in the presence of template 2 (Figure 4). For template 1, 1.75 μM EF-P was sufficient to afford a maximum protein yield, ~ 3-fold greater compared to that obtained using the control lacking added EF-P (Figure 4A), while at the same EF-P concentration in the presence of template 2, an ~30-fold increase was observed (Figure 4B).
Comparatively, addition of exogenous EF-P (1.75 μM) had no to little impact on the DHFR yield guided by template 1 in the presence of suppressor tRNAHyb activated with phenylalanine or lysine (Figure 5A). The effects were more pronounced with template 2, with lysine incorporation being affected more than that of phenylalanine (Figure 5B). Possibly, this may simply reflect the greater absolute incorporation of phenylalanine in the absence of EF-P, implying that less improvement is possible via the agency of EF-P. An alternative explanation might involve enhanced binding of ribosome-bound tRNALys by EF-P. EF-P itself contains a modified amino acid with structurally altered side chains that have one or more free amino groups, plausibly able to stabilize the formed ribosomal complex; an analogous function of the lysine ε-amino group esterified to tRNA might produce a similar effect.
Figure 5.
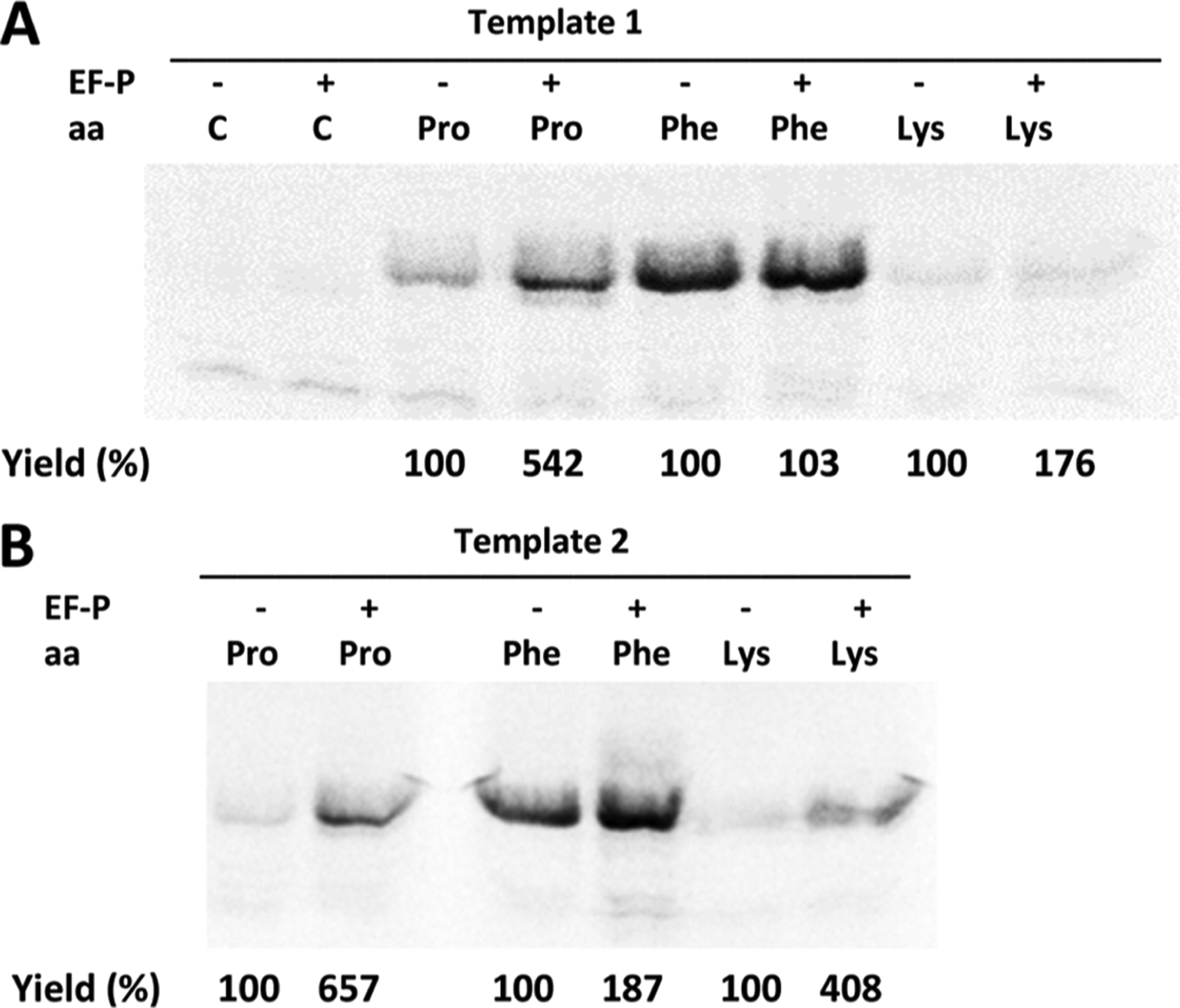
Effect of E. coli EF-P (1.75 μM) on in vitro translation efficiency from (A) template1 (DHFR, Ala9stop10) or (B) template 2 (DHFR, Pro9stop10) in the presence of suppressor tRNAHyb aminoacylated with proline (Pro-tRNAHyb), phenylalanine (Phe-tRNAHyb), or lysine (Lys-tRNAHyb). The outcome of the experiments was monitored by SDS PAGE and autoradiography. Yields (%) are presented comparatively for each amino acid. Control samples (denoted as C) were run in the absence of any suppressor tRNA without (−) or with (+) exogenous EF-P.
However, in accordance with the putative role of EF-P in facilitating the incorporation of contiguous prolines into proteins, the greatest effect of EF-P on nonsense codon suppression was observed for suppressor tRNAHyb activated with proline (Figure 5B).
Although the absolute suppression yields varied somewhat from one experiment to another, these results support the idea that greater EF-P enhancement of nonsense codon suppression may be realized when the PTC P-site is occupied by (peptidyl)prolyl-tRNAPro, consistent with the putative role of EF-P in facilitating translation through contiguous proline codons.37,38 As noted, Katoh et al.10 have shown that it can be beneficial to utilize an A-site tRNA activated with proline, or another amino acid having a secondary α-amine, exemplified by the use of N-methylthreonine.
Because the foregoing data hint at the possible importance of the structure of the aminoacyl moiety of the A-site tRNA in response to EF-P, it seemed of interest to test the effects of EF-P on activated A-site tRNAs bearing unusual noncanonical amino acids. A number of such species, identified in our earlier studies, have shown considerable potential utility as protein constituents, and strategies for facilitating their inclusion into additional proteins would be of substantial interest. Accordingly, several different amino acid analogues (1,39 2,40 3,40 4,41,42 5,43 and 644) were used to activate suppressor tRNAHyb (Figure 6). These activated tRNAs were prepared by T4 RNA ligase-mediated coupling of an in vitro tRNA transcript lacking the 3′-terminal CpA dinucleotide of a mature tRNA with aminoacylated pdCpA transcripts prepared by chemical synthesis (Scheme 1 and Figure S3).32–35
Figure 6.
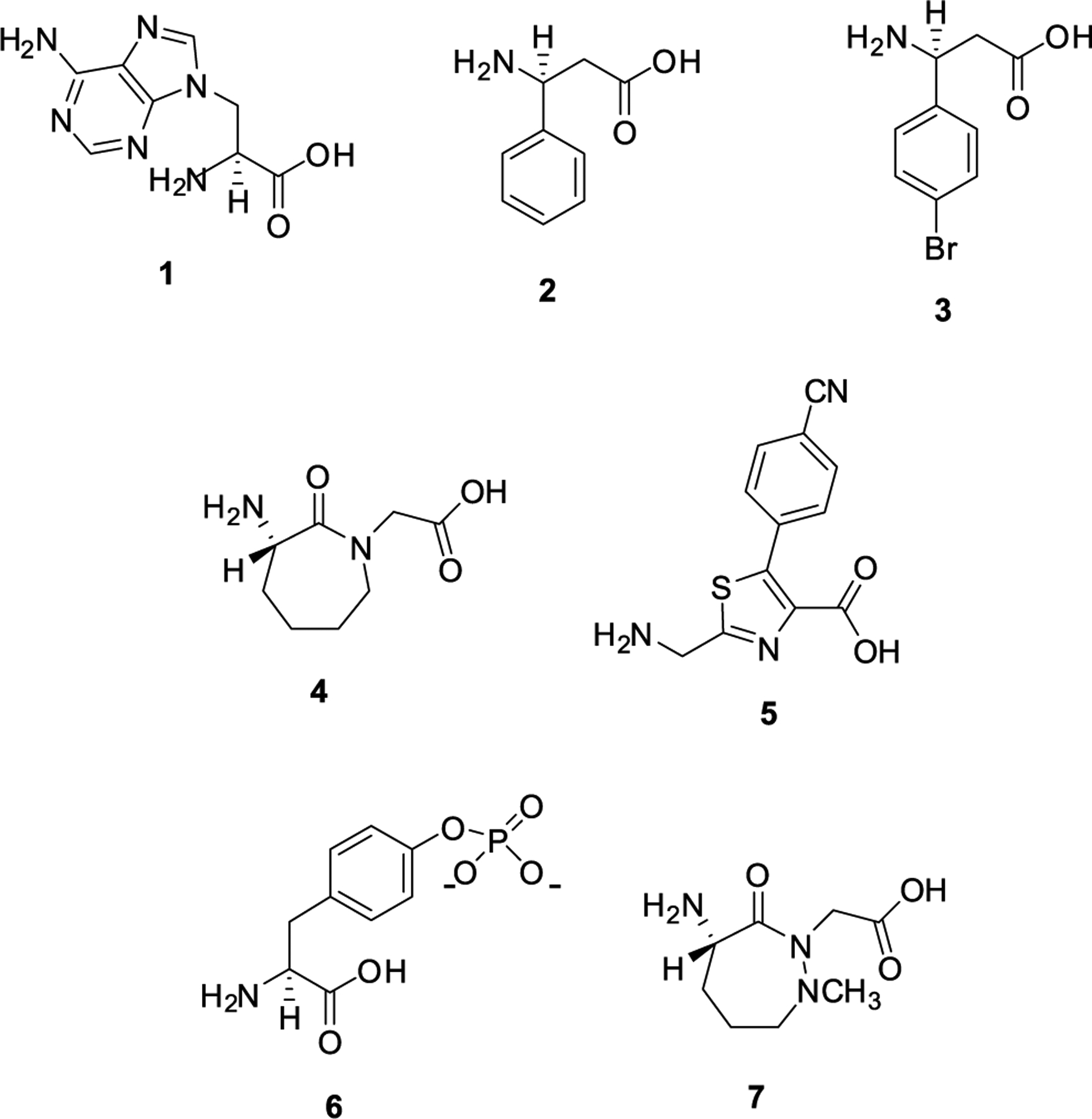
Structures of several noncanonical amino acid analogues used in experiments to determine the effect of EF-P on their incorporation into DHFR: compound 1, an adenine nucleobase amino acid; compound 2, S-β-phenylalanine; compound 3, S-β-p-bromophenylalanine; compound 4, a conformationally constrained dipeptide; compound 5, a fluorescent thiazole dipeptidomimetic; compound 6, S-phosphotyrosine; and compound 7, a conformationally constrained dipeptide.
Nucleobase amino acid 1 and related species have been introduced into key regions of the Rob protein transcription factor45,46 and into an RRM1 binding motif with an affinity for the BCL2 i-motif DNA,47 producing enhanced DNA affinity in each case. β-Amino acids 2 and 3 exemplify novel protein substituents, and a selected example has also been shown to enhance DNA binding.48 Conformationally constrained cyclic dipeptide 4 (and also its congener 7) is reminiscent of cyclic peptide proline and has been shown to be capable of imparting local conformational rigidity to proteins after introduction into a conformationally important protein domain.49 Weakly fluorescent thiazole dipeptidomimetic 5, and several congeners, exhibits dramatic alteration of its photophysical properties when positioned within structured domains of proteins.43,50,51 Finally, by the introduction of phosphotyrosine (pTyr) (6) at a single position of inhibitory protein IκB-α, we have identified a possible new property of this protein in regulating DNA strand exchange by NF-κB.44 Additionally, the introduction of 6 at discrete positions of NF-κB has enabled the regulation of DNA promoter binding and gene transcription/translation in experimental systems.52,53 In the aggregate, these and related amino acid analogues have been incorporated into numerous proteins and peptides, and their incorporation has been studied by multiple analytical techniques including mass spectrometry.
The six misacylated suppressor tRNAsHyb were tested for their ability to incorporate their activated amino acids into position 10 of DHFR when translating templates 1 or 2, with or without added E. coli EF-P (Figures 7 and 8). As shown in Figure 7 for tRNAsHyb activated with amino acids 1, 2, and 3, the results were similar using templates 1 and 2. The presence of EF-P mediated an approximately sevenfold increase in modified DHFR for tRNAHyb activated with nucleobase amino acid 1. The increased incorporation of β-phenylalanine (2) and p-bromo-β-phenylalanine (3) was more modest, but still more than twofold in the presence of EF-P. Only in the case of compound 1 did the absolute amount of modified DHFR formed in the presence of template 2 + EF-P exceed that formed when utilizing template 1 + EF-P. The apparent differences in the extent of the enhancement of protein synthesis may well be related to the earlier finding by Doerfel et al.11 that increased peptide bond formation may be related to conformational adjustment of the aminoacyl-tRNA toward an orientation conducive to peptide bond formation. In the same study, it was also acknowledged that contextual effects have also been noted to be important, such that the structures of flanking amino acids upstream or downstream from the critical amino acid incorporation can affect protein yields. Alternatively, it has been noted in a recent review article that EF-P acts throughout translation elongation, such that other factors may account for the differences in the behavior of individual aminoacyl-tRNAs.38
Figure 7.
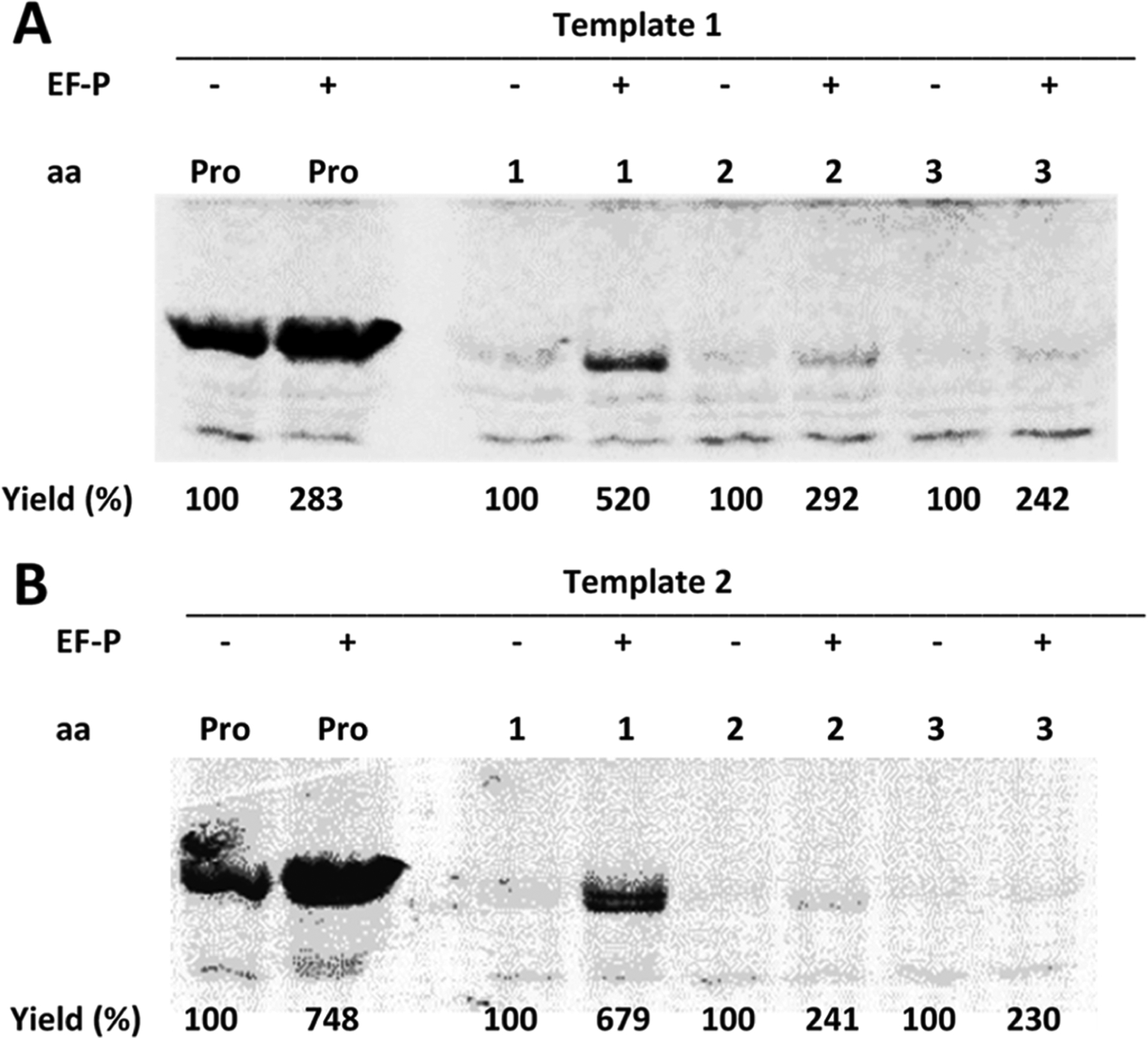
Effect of E. coli EF-P (1.75 μM) on in vitro translation efficiency from (A) template 1 (DHFR, Ala9stop10) or (B) template 2 (DHFR, Pro9stop10) in the presence of suppressor tRNAHyb, activated with proline, or compound 1, 2, or 3. The outcome of the experiment was analyzed by SDS-PAGE and autoradiography. Yields (%) are presented comparatively for each compound.
Figure 8.
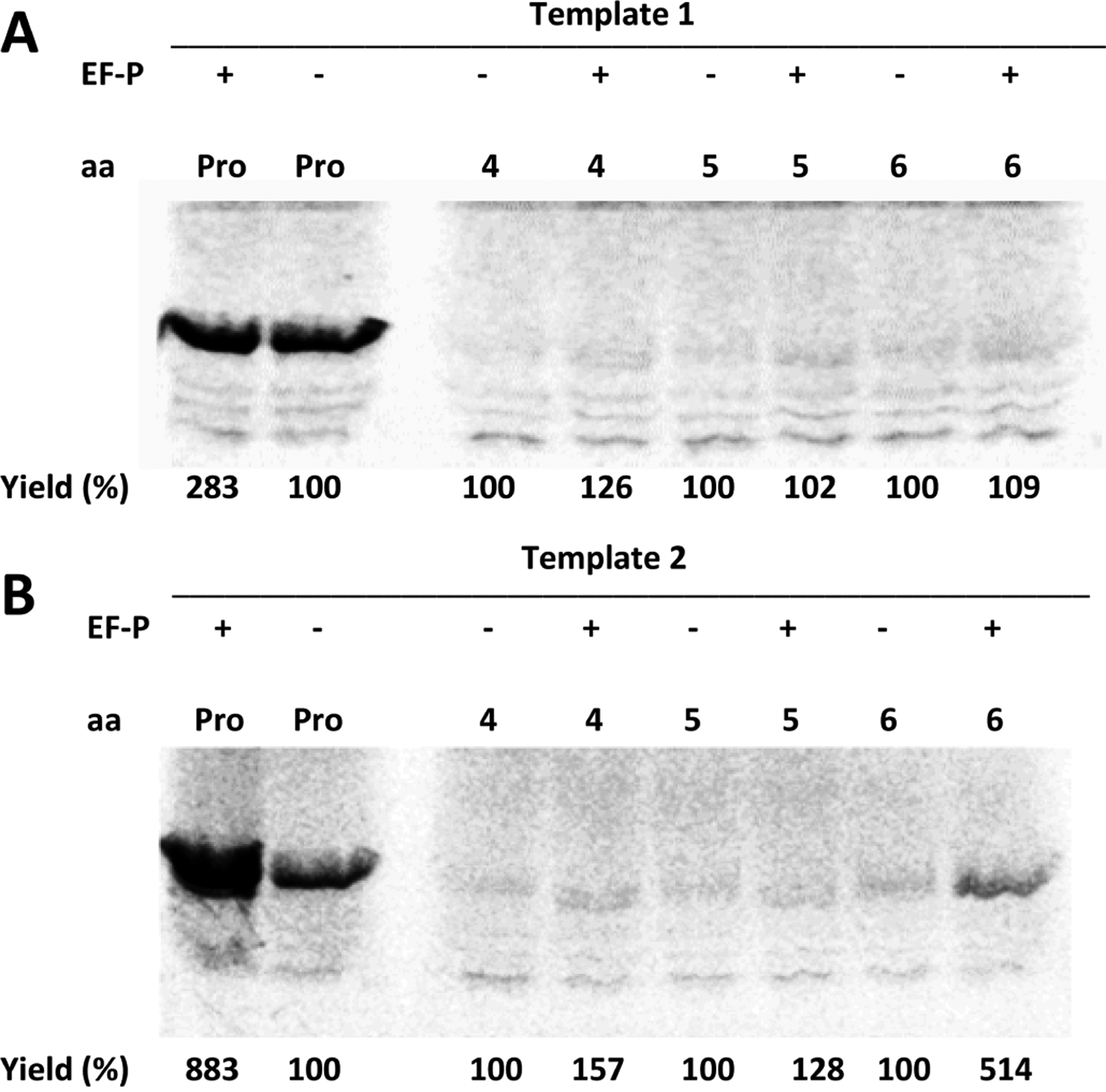
Effect of E. coli EF-P (1.75 μM) on in vitro translation efficiency from (A) template 1 (DHFR, Ala9stop10) or (B) template 2 (DHFR, Pro9stop10) in the presence of suppressor tRNAHyb, activated with proline, or compound 4, 5, or 6. The outcome of the experiment was analyzed by SDS-PAGE and autoradiography. Yields (%) are presented comparatively for each compound.
As shown in Figure 8, when using template 1, the presence of EF-P had a minimal effect on increasing the yields of DHFR containing compounds 4, 5, or 6. In comparison, when using template 2, amino acids 4 and 5 gave measurably enhanced yields of the modified DHFRs, while the greatest enhancement (from 100 → 514%) was observed for phosphotyrosine (6), which is especially notable in that this amino acid was found not to be a substrate for incorporation by wild-type E. coli ribosomes under routine conditions.44
All of the foregoing experiments were carried out utilizing an EF-P preparation that also contained three accessory proteins (YjeA, YjeK, and YfcM), which increased the complexity of EF-P preparation (Figure S1). To determine the extent to which the accessory proteins may be necessary to achieve the results obtained, we also carried out an additional experiment in the presence of a plasmid containing the gene for EF-P, but not for any accessory protein. For this experiment, EF-P was prepared in vivo from a commercially prepared plasmid-borne synthetic gene (Figure S4). DHFR was expressed from template 2 in the presence of suppressor tRNAHyb activated with amino acid 7 (Figure 6) at varying concentrations of either EF-P alone or EF-P expressed in the presence of the three accessory proteins (Figure 9). The reference sample was DHFR prepared in the presence of suppressor tRNAHyb aminoacylated with phenylalanine and gave a yield of DHFR in the absence of added EF-P defined as 100%. As shown, EF-P prepared in the presence of the three modifying enzymes and suppressor tRNAHyb activated with cyclic dipeptide 7 afforded DHFR yields that increased from 33.2 to 41.1% in a concentration-dependent fashion as the EF-P levels were increased from 1 to 4 μM. In comparison, the use of EF-P from a plasmid containing only the gene for EF-P also gave a concentration-dependent increase in DHFR yield but produced less DHFR at every concentration tested (20.1–33.6% at concentrations of EF-P ranging from 1 to 4 μM). Since the translation experiment was carried out in the presence of an S-30 fraction prepared from wild-type E. coli, it seems likely that the extract contained some amount of the EF-P modifying enzymes, and that the yields could have resulted completely or in part from EF-P modified during the DHFR translation experiment.16
Figure 9.
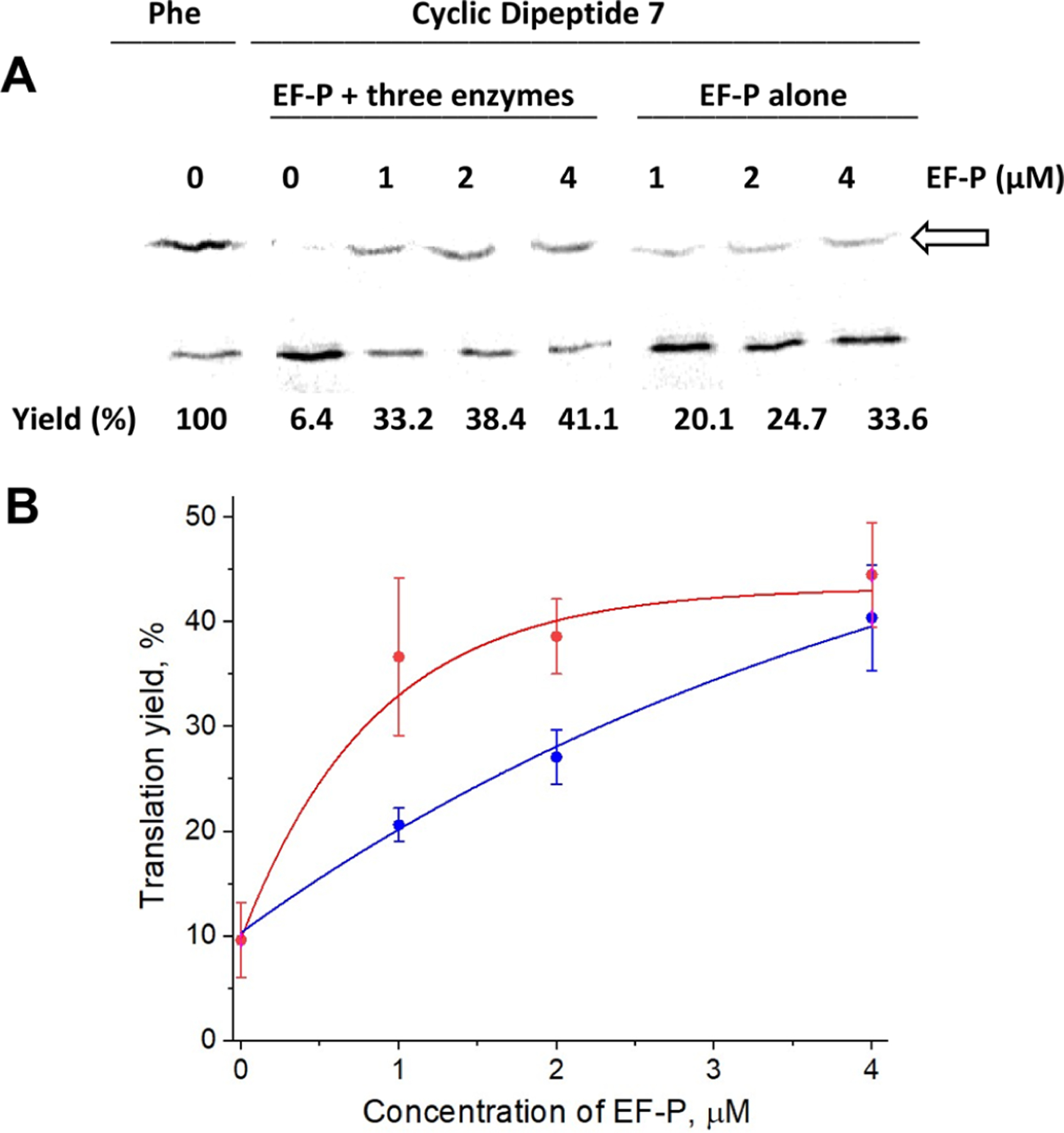
Effect of post-translational modifications of E. coli EF-P on in vitro translation efficiency of DHFR. (A) Template 2 (DHFR, Pro9stop10) was employed in the presence of suppressor tRNAHyb, activated with the cyclic dipeptide 7. Translation was carried out in the presence of 1, 2, or 4 μM modified EF-P (prepared from plasmids pETEF-P/YjeA/YjeK/YfcM) (red curve) and “untreated” EF-P (expressed from a commercial plasmid) (blue curve). The arrow indicates full-length DHFR. The corresponding DHFR yields (%) for modified (B, red curve) and “untreated” EF-P (B, blue curve) are expressed comparatively to the yield of DHFR in the presence of suppressor tRNAHyb activated with phenylalanine.
It should be noted that lack of hydroxylation of β-lysinated Lys34 at C5(δ) seems to have no effect on EF-P activity.4 In contrast, lysylation is an essential modification as it helps to juxtapose the peptide chain into the catalytic core of the peptidyltransferase center and stabilize the terminal 3′-CCA end of the tRNA.20 Lysylated EF-P significantly (12-fold) enhances N-formylmethionyl puromycin synthesis, while unmodified EF-P is still capable of producing an ~3-fold increase,16 which is in the range of the effect the unmodified EF-P exerted on the incorporation of cyclic dipeptide 7 into DHFR (Figure 9).
CONCLUSIONS
A model system was created using in vitro transcription/translation of E. coli DHFR to study the effects of EF-P on the incorporation of a variety of unusual noncanonical amino acids into position 10 of the protein by nonsense codon suppression from a newly designed amber suppressor tRNAHyb. When the tRNAHyb was activated with proline, suppression of the UAG codon at position 10 proceeded with facility and, consistent with previous observations, was enhanced by the post-translationally modified exogenous EF-P and by the presence of a proline codon at position 9 on the template. An analogous observation was made when suppressor tRNAHyb was activated with phenylalanine or lysine. Several noncanonical amino acids of diverse structure, only one of which had previously been found to be incorporated into protein in the presence of wild-type ribosomes, were further tested. While none of the amino acid analogues was readily incorporated into DHFR regardless of the presence or absence of a preceding proline, the addition of fully modified EF-P enhanced the incorporation of all of them into the nascent polypeptide to some extent, and quite noticeably in the cases of amino acid analogues 1, 6, and 7. The greatest increase in the yield of DHFR was noted for adenine nucleobase amino acid 1, the only one of the amino acid analogues tested that could be incorporated into in vitro synthesized proteins using wild-type ribosomes. Enhanced EF-P-mediated incorporation of S-phosphotyrosine (6) adjacent to proline is an interesting finding and might be extended further by the study of additional phosphorylated amino acids. Conformationally constrained dipeptide 7 was also incorporated into DHFR to a significant extent adjacent to proline and in the presence of EF-P. Notably, exogenously added unmodified EF-P also increased the yield of DHFR containing 7, albeit to a slightly lesser extent than fully modified EF-P. Although some portion of the exogenously added EF-P may have been due to post-translational modification caused by the use of an S-30 fraction prepared from E. coli, the result suggests that EF-P could be studied for the enhanced incorporation of nonproteinogenic amino acids into proteins and peptides in a more simplified experimental context.
In the aggregate, two conclusions may reasonably be drawn from the present work, namely, that (i) EF-P is capable of enhancing the incorporation of certain unusual amino acid analogues into proteins and in some cases may be used in lieu of modified ribosomes, and (ii) while fully translationally modified EF-P facilitates the incorporation most effectively, the use of an S-30 system prepared from wild-type E. coli in combination with exogenously supplied unmodified EF-P could also effect the increased incorporation of at least some of them.
Finally, for some of the amino acids whose incorporation was enhanced in the presence of EF-P, the presence of Pro in the protein position preceding the site of modified amino acid incorporation strongly facilitated improved yields. However, this was not uniformly observed (Figure 7), suggesting the possibility of engineering a more global strategy for noncanonical amino acid incorporation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Prof. Marina Rodnina, Max Planck Institute for Multidisciplinary Sciences, Göttingen, Germany, for the plasmid expressing E. coli elongation factor P. This work was supported by Research Grants R01GM103861, R01GM121367, and R35GM140819 from the National Institute of General Medical Sciences, NIH.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.3c07524.
Experimental details, including methods for preparing modified and unmodified elongation factor P and determining the yields of DHFR produced by coupled transcription/translation of a gene for DHFR in the presence of EF-P and suppressor tRNAs misacylated with structurally diverse noncanonical amino acids (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/jacs.3c07524
The authors declare no competing financial interest.
Contributor Information
Sasha M. Daskalova, Biodesign Center for Bioenergetics, and School of Molecular Sciences, Arizona State University, Tempe, Arizona 85287, United States
Larisa M. Dedkova, Biodesign Center for Bioenergetics, and School of Molecular Sciences, Arizona State University, Tempe, Arizona 85287, United States
Rumit Maini, Biodesign Center for Bioenergetics, and School of Molecular Sciences, Arizona State University, Tempe, Arizona 85287, United States.
Poulami Talukder, Biodesign Center for Bioenergetics, and School of Molecular Sciences, Arizona State University, Tempe, Arizona 85287, United States.
Xiaoguang Bai, Biodesign Center for Bioenergetics, and School of Molecular Sciences, Arizona State University, Tempe, Arizona 85287, United States.
Sandipan Roy Chowdhury, Biodesign Center for Bioenergetics, and School of Molecular Sciences, Arizona State University, Tempe, Arizona 85287, United States.
Chao Zhang, Biodesign Center for Bioenergetics, and School of Molecular Sciences, Arizona State University, Tempe, Arizona 85287, United States.
Ryan C. Nangreave, Biodesign Center for Bioenergetics, and School of Molecular Sciences, Arizona State University, Tempe, Arizona 85287, United States
Sidney M. Hecht, Biodesign Center for Bioenergetics, and School of Molecular Sciences, Arizona State University, Tempe, Arizona 85287, United States
REFERENCES
- (1).Pavlov MY; Watts RE; Tan Z; Cornish VW; Ehrenberg M; Forster AC Slow Peptide Bond Formation by Proline and Other N-alkylamino Acids in Translation. Proc. Natl. Acad. Sci. U.S.A 2009, 106, 50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Muto H; Ito K Peptidyl-prolyl-tRNA at the Ribosomal P-site Reacts Poorly with Puromycin. Biochem. Biophys. Res. Commun 2008, 366, 1043–1047. [DOI] [PubMed] [Google Scholar]
- (3).Wohlgemuth I; Brenner S; Beringer M; Rodnina MV Modulation of the Rate of Peptidyl Transfer on the Ribosome by the Nature of Substrates. J. Biol. Chem 2008, 283, 32229–32235. [DOI] [PubMed] [Google Scholar]
- (4).Doerfel LK; Wohlgemuth I; Kothe C; Peske F; Urlaub H; Rodnina MV EF-P is Essential for Rapid Synthesis of Proteins Containing Consecutive Proline Residues. Science 2013, 339, 85–88. [DOI] [PubMed] [Google Scholar]
- (5).Peil L; Starosta AL; Lassak J; Atkinson GC; Virumae K; Spitzer M; Tenson T; Jung K; Remme J; Wilson DN Distinct XPPX Sequence Motifs Induce Ribosome Stalling, Which is Rescued by the Translation Elongation Factor EF-P. Proc. Natl. Acad. Sci. U.S.A 2013, 110, 15265–15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Ude S; Lassak J; Starosta AL; Kraxenberger T; Wilson DN; Jung K Translation Elongation Factor EF-P Alleviates Ribosome Stalling at Polyproline Stretches. Science 2013, 339, 82–85. [DOI] [PubMed] [Google Scholar]
- (7).Hanawa-Suetsugu K; Sekine S; Sakai H; Hori-Takemoto C; Terada T; Unzai S; Tame JR; Kuramitsu S; Shirouzu M; Yokoyama S Crystal Structure of Elongation Factor P from Thermus Thermophilus HB8. Proc. Natl. Acad. Sci. U.S.A 2004, 101, 9595–9600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Choi S; Choe J Crystal Structure of Elongation Factor P from Pseudomonas Aeruginosa at 1.75 Å Resolution. Proteins 2011, 79, 1688–1693. [DOI] [PubMed] [Google Scholar]
- (9).Blaha G; Stanley RE; Steitz TA Formation of the First Peptide Bond: The Structure of EF-P Bound to the 70S Ribosome. Science 2009, 325, 966–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Katoh T; Wohlgemuth I; Nagano M; Rodnina MV; Suga H Essential Structural Elements in tRNAPro for EF-P-Mediated Alleviation of Translation Stalling. Nat. Commun 2016, 7, 11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Doerfel LK; Wohlgemuth I; Kubyshkin V; Starosta AL; Wilson DN; Budisa N; Rodnina MV Entropic Contribution of Elongation Factor P to Proline Positioning at the Catalytic Center of the Ribosome. J. Am. Chem. Soc 2015, 137, 12997–13006. [DOI] [PubMed] [Google Scholar]
- (12).Woolstenhulme CJ; Guydosh NR; Green R; Buskirk AR High-Precision Analysis of Translational Pausing by Ribosome Profiling in Bacteria Lacking EFP. Cell Rep. 2015, 11, 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Elgamal S; Katz A; Hersch SJ; Newsom D; White P; Navarre WW; Ibba M EF-P Dependent Pauses Integrate Proximal and Distal Signals During Translation. PLoS Genet. 2014, 10, No. e1004553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Starosta AL; Lassak J; Peil L; Atkinson GC; Virumae K; Tenson T; Remme J; Jung K; Wilson DN Translational Stalling at Polyproline Stretches is Modulated by the Sequence Context Upstream of the Stall Site. Nucleic Acids Res. 2014, 42, 10711–10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Hersch SJ; Elgamal S; Katz A; Ibba M; Navarre WW Translation Initiation Rate Determines the Impact of Ribosome Stalling on Bacterial Protein Synthesis. J. Biol. Chem 2014, 289, 28160–28171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Park J-H; Johansson HE; Aoki H; Huang BX; Kim H-Y; Ganoza MC; Park MH Post-translational Modification of β-Lysylation is Required for Activity of Escherichia coli Elongation Factor P (EF-P). J. Biol. Chem 2012, 287, 2579–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Behshad E; Ruzicka FJ; Mansoorabadi SO; Chen D; Reed GH; Frey PA Enantiomeric Free Radicals and Enzymatic Control of Stereochemistry in a Radical Mechanism: The Case of Lysine 2,3-Aminomutases. Biochemistry 2006, 45, 12639–12646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Navarre WW; Zou SB; Roy H; Xie JL; Savchenko A; Singer A; Edvokimova E; Prost LR; Kumar R; Ibba M; Fang FC PoxA, YjeK, and Elongation Factor P Coordinately Modulate Virulence and Drug Resistance in Salmonella Enterica. Mol. Cell 2010, 39, 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Sumida T; Yanagisawa T; Ishii R; Yokoyama S Crystallization and Preliminary X-ray Crystallographic Study of GenX, a Lysyl-tRNA Synthetase Paralogue from Escherichia coli, in Complex with Translation Elongation Factor P. Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun 2010, 66, 1115–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Yanagisawa T; Sumida T; Ishii R; Takemoto C; Yokoyama S A Paralog of Lysyl-tRNA Synthetase Aminoacylates a Conserved Lysine Residue in Translation Elongation Factor P. Nat. Struct. Mol. Biol 2010, 17, 1136–1143. [DOI] [PubMed] [Google Scholar]
- (21).Peil L; Starosta AL; Virumae K; Atkinson GC; Tenson T; Remme J; Wilson DN Lys34 of Translation Elongation Factor EF-P is Hydroxylated by YfcM. Nat. Chem. Biol 2012, 8, 695–697. [DOI] [PubMed] [Google Scholar]
- (22).Lassak J; Keilhauer EC; Furst M; Wuichet K; Godeke J; Starosta AL; Chen JM; Søgaard-Andersen L; Rohr J; Wilson DN; Häussler S; Mann M; Jung K Arginine-rhamnosylation as New Strategy to Activate Translation Elongation Factor P. Nat. Chem. Biol 2015, 11, 266–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Cooper HL; Park MH; Folk JE; Safer B; Braverman R Identification of the Hypusine-containing Protein Hy+ as Translation Initiation Factor eIF-4D. Proc. Natl. Acad. Sci. U.S.A 1983, 80, 1854–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Katoh T; Iwane Y; Suga H Logical Engineering of D-arm and T-stem of tRNA that Enhances D-amino Acid Incorporation. Nucleic Acids Res. 2017, 45, 12601–12610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Liu H; Peterson R; Kessler J; Musier-Forsyth K Molecular Recognition of tRNAPro by Escherichia coli Proline tRNA Synthetase In Vitro. Nucleic Acids Res. 1995, 23, 165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Hasegawa T; Yokogawa T Escherichia coli Proline tRNA: Structure and Recognition Sites for Prolyl-tRNA Synthetase. Nucleic Acids Symp. Ser 2000, 44, 7–8. [DOI] [PubMed] [Google Scholar]
- (27).McClain WH; Schneider J; Gabriel K Distinctive Acceptor-End Structure and Other Determinants of Escherichia coli tRNAPro Identity. Nucleic Acids Res. 1994, 22, 522–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).An S; Barany G; Musier-Forsyth K Evolution of Acceptor Stem tRNA Recognition by Class II Prolyl-tRNA Synthetase. Nucleic Acids Res. 2008, 36, 2514–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Yarus M; Cline S; Raftery L; Wier P; Bradley D The Translational Efficiency of tRNA is a Property of the Anticodon Arm. J. Biol. Chem 1986, 261, 10496–10505. [PubMed] [Google Scholar]
- (30).Guo J; Melancçon C; Lee HS; Groff D; Schultz PG Evolution of Amber Suppressor tRNAs for Efficient Bacterial Production of Proteins Containing Nonnatural Amino Acids. Angew. Chem., Int. Ed 2009, 48, 9148–9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Bystroff C; Oatley SJ; Kraut J Crystal Structures of Escherichia coli Dihydrofolate Reductase: The NADP+ Holoenzyme and the Folate·NADP+ Ternary Complex. Substrate Binding and a Model for the Transition State. Biochemistry 1990, 29, 3263–3277. [DOI] [PubMed] [Google Scholar]
- (32).Robertson SA; Noren CJ; Anthony-Cahill J; Griffith MC; Schultz PG The Use of 5′-Phospho-2 deoxyribocytidylylriboadenosine as a Facile Route to Chemical Aminoacylation of tRNA. Nucleic Acids Res. 1989, 17, 9649–9660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Noren CJ; Anthony-Cahill SJ; Noren KA; Griffith MC; Schultz PG; Schultz PG In Vitro Suppression of an Amber Mutation by a Chemically Aminoacylated Transfer RNA Prepared by Runoff Transcription. Nucleic Acids Res. 1990, 18, 83–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Karginov VA; Mamaev SV; An H; Van Cleve MD; Hecht SM; Komatsoulis GA; Abelson JN Probing the Role of an Active Site Aspartic Acid in Dihydrofolate Reductase. J. Am. Chem. Soc 1997, 119, 8166–8176. [Google Scholar]
- (35).Dedkova LM; Hecht SM Expanding the Scope of Protein Synthesis Using Modified Ribosomes. J. Am. Chem. Soc 2019, 141, 6430–6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Kwok Y; Wong JT Evolutionary Relationship Between Halobacterium cutirubrum and Eukaryotes Determined by Use of Aminoacyl-tRNA Synthetases as Phylogenetic Probes. Can. J. Biochem 1980, 58, 213–218. [DOI] [PubMed] [Google Scholar]
- (37).Lassak J; Wilson DN; Jung K Stall No More at Polyproline Stretches with the Translation Elongation Factors EF-P and IF-5A. Mol. Microbiol 2016, 99, 219–235. [DOI] [PubMed] [Google Scholar]
- (38).Hummels KR; Kearns DB Translation Elongation Factor P (EF-P). FEMS Microbiol. Rev 2020, 44, 208–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Talukder P; Dedkova LM; Ellington AD; Yakovchuk P; Lim JEV; Anslyn EV; Hecht SM Synthesis of Alanyl Nucleobase Amino Acids and Their Incorporation Into Proteins. Bioorg. Med. Chem 2016, 24, 4177–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Maini R; Nguyen D; Chen S; Dedkova LM; Roy Chowdhury S; Alcala Torano R; Hecht SM Incorporation of β-Amino Acids into Dihydrofolate Reductase by Ribosomes Having Modifications in the Peptidyltransferase Center. Bioorg. Med. Chem 2013, 21, 1088–1096. [DOI] [PubMed] [Google Scholar]
- (41).Zhang C; Bai X; Dedkova LM; Hecht SM Protein Synthesis with Conformationally Constrained Cyclic Dipeptides. Bioorg. Med. Chem 2020, 28, 115780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Zhang C; Talukder P; Dedkova LM; Hecht SM Facilitated Synthesis of Proteins Containing Modified Dipeptides. Bioorg. Med. Chem 2021, 41, 116210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Roy Chowdhury S; Chauhan PS; Dedkova LM; Bai X; Chen S; Talukder P; Hecht SM Synthesis and Evaluation of a Library of Fluorescent Dipeptidomimetic Analogues as Substrates for Modified Bacterial Ribosomes. Biochemistry 2016, 55, 2427–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Chen S; Maini R; Bai X; Nangreave RC; Dedkova LM; Hecht SM Incorporation of Phosphorylated Tyrosine into Proteins: In Vitro Translation and Study of Phosphorylated IκB-α and its Interaction with NF-κB. J. Am. Chem. Soc 2017, 139, 14098–14108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Zhang C; Chen S; Bai X; Dedkova LM; Hecht SM Alteration of Transcriptional Regulator Rob In Vivo: Enhancement of Promoter DNA Binding and Antibiotic Resistance in the Presence of Nucleobase Amino Acids. Biochemistry 2020, 59, 1217–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Zhang C; Chen S; Dedkova LM; Hecht SM Effects of Nucleobase Amino Acids on the Binding of Rob to its Promoter DNA: Differential Alteration of DNA Affinity and Phenotype. Biochemistry 2020, 59, 2111–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Bai X; Talukder P; Daskalova SM; Roy B; Chen S; Li Z; Dedkova LM; Hecht SM Enhanced Binding Affinity for an i-Motif DNA Substrate Exhibited by a Protein Containing Nucleobase Amino Acids. J. Am. Chem. Soc 2017, 139, 4611–4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Maini R; Roy Chowdhury S; Dedkova LM; Roy B; Daskalova SM; Paul R; Chen S; Hecht SM Protein Synthesis with Ribosomes Selected for the Incorporation of β Amino Acids. Biochemistry 2015, 54, 3694–3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Zhang C; Bai X; Chen S; Dedkova LM; Hecht SM Local Conformational Constraint of Firefly Luciferase Can Affect the Energy of Bioluminescence and Enzyme Stability. CCS Chem. 2022, 4, 1695–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Maini R; Dedkova LM; Paul R; Madathil MM; Roy Chowdhury S; Chen S; Hecht SM Ribosome-Mediated Incorporation of Dipeptides and Dipeptide Analogues Into Proteins In Vitro. J. Am. Chem. Soc 2015, 137, 11206–11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Chen S; Ji X; Gao M; Dedkova LM; Hecht SM In Cellulo Synthesis of Proteins Containing a Fluorescent Oxazole Amino Acid. J. Am. Chem. Soc 2019, 141, 5597–5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Chen S; Ji X; Dedkova LM; Hecht SM Site-Selective Incorporation of Phosphorylated Tyrosine Into the p50 Subunit of NF-κB and Activation of its Downstream Gene CD40. Chem. Commun 2021, 57, 12651–12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Chen S; Ji X; Dedkova LM; Potuganti GR; Hecht SM Site-Selective Tyrosine Phosphorylation in the Activation of the p50 Subunit of NF-κB for DNA Binding and Transcription. ACS Chem. Biol 2023, 18, 59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


