Abstract
Introduction
Artificial intelligence (AI) is expected to impact all facets of inflammatory bowel disease (IBD) management, including disease assessment, treatment decisions, discovery and development of new biomarkers and therapeutics, as well as clinician–patient communication.
Areas covered
This perspective paper provides an overview of the application of AI in the clinical management of IBD through a review of the currently available AI models that could be potential tools for prognosis, shared decision-making, and precision medicine. This overview covers models that measure treatment response based on statistical or machine-learning methods, or a combination of the two. We briefly discuss a computational model that allows integration of immune/biological system knowledge with mathematical modeling and also involves a ‘digital twin’, which allows measurement of temporal trends in mucosal inflammatory activity for predicting treatment response. A viewpoint on AI-enabled wearables and nearables and their use to improve IBD management is also included.
Expert opinion
Although challenges regarding data quality, privacy, and security; ethical concerns; technical limitations; and regulatory barriers remain to be fully addressed, a growing body of evidence suggests a tremendous potential for integration of AI into daily clinical practice to enable precision medicine and shared decision-making.
Keywords: Artificial intelligence, computational model, inflammatory bowel disease, Crohn’s disease, fecal calprotectin, mucosal healing, precision medicine, shared decision-making
ARTICLE HIGHLIGHTS
Advances in artificial intelligence (AI) show promise for improving treatment response prediction, decision-making, and precision medicine in inflammatory bowel disease (IBD).
In particular, AI could improve precision medicine for IBD by enabling identification of disease subtypes, prediction of disease progression and treatment response, selection of personalized treatments, and remote monitoring.
Predictive models can benefit clinicians and patients alike by optimizing shared decision-making processes; patients can also use AI to cope with daily and long-term challenges of the disease.
Beyond patients and practitioners, predictive models may positively impact healthcare structures and payers by enabling effective healthcare-resource utilization.
To increase the accuracy and efficiency of AI models, biomarkers, patient-reported outcomes, and disease scores should be combined within predictive models, and the outputs should be compared with clinical trial data and real-world data for validation.
1. Introduction
Inflammatory bowel disease (IBD) is a chronic, debilitating, inflammatory condition of the gastrointestinal tract, which comprises two main pathologies: ulcerative colitis (UC) and Crohn’s disease (CD) [1]. It is characterized by a lifelong, unpredictable, relapsing-remitting, and destructive course associated with irreversible bowel damage, which causes considerable morbidity and poor quality of life [2]. The pathogenesis is multifactorial, involving loss of tolerance to the commensal gut microbiome, intestinal epithelial barrier dysfunction, and immune dysregulation [3].
IBD diagnosis relies on clinical features, laboratory biomarkers (C-reactive protein [CRP] and fecal calprotectin [FCP]), imaging (endoscopy, magnetic resonance, ultrasonography, nuclear-medicine techniques), and histology [4]. Although treatment algorithms have been developed based on clinical evidence, they do not optimally integrate the heterogeneity in presenting symptoms, treatment response, and long-term clinical outcomes, such as the development of strictures and need for surgery [3]. Therefore, there is an unmet need for precision-medicine strategies to improve diagnostic and therapeutic approaches to IBD [3, 5].
Advancements in artificial intelligence (AI) technologies have transformed the way clinicians and researchers handle, analyze, decode, and interpret large, complex, multifaceted datasets [3]. AI is a multidisciplinary field integrating insights from engineering, computer science, linguistics, and philosophy, wherein systems that could show or mirror human intelligence are comprehended, designed, and developed [3]. Machine learning (ML), a subdiscipline of AI, has significant relevance in the healthcare sector, especially supporting diagnosis and outcome prediction [6].
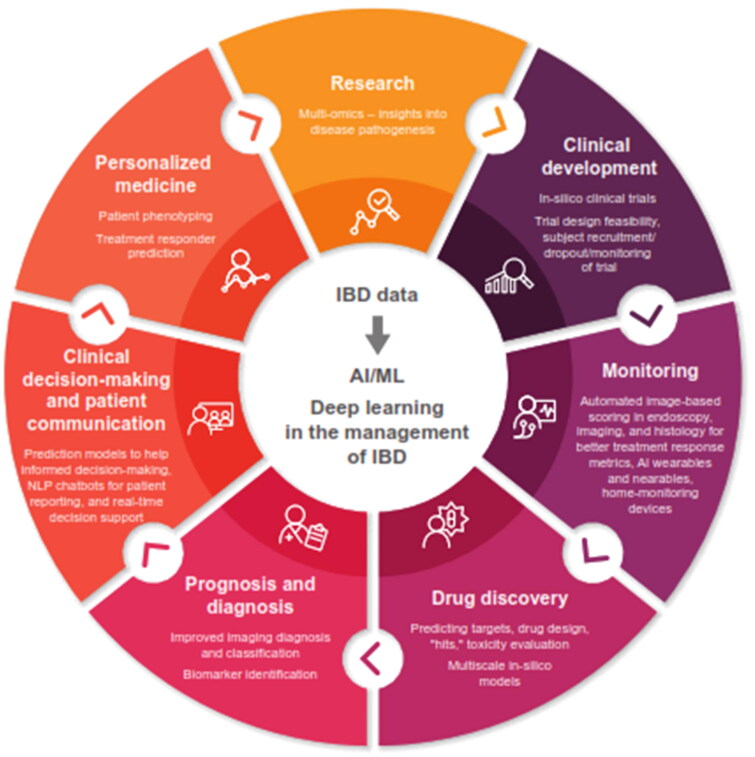
AI has the potential to transform the clinical management of IBD by providing impactful insights through the analysis of large, complex volumes of data [7]. AI could influence almost all domains of IBD management including disease assessment, treatment decisions, discovery, and development of new biomarkers and therapeutics, as well as patient communication (Figure 1).
Figure 1.
Potential use of AI in the management of IBD.
AI, artificial intelligence; IBD, inflammatory bowel disease; ML, machine learning; NLP, natural language processing.
In this perspective article, we briefly outline advances in AI technologies for the diagnosis and endoscopic assessment of IBD. We review various statistical and AI models predicting treatment response and risk of complications in patients with CD, with a deep dive into a multimodal AI model predicting the treatment response to vedolizumab in patients with CD.
CD was chosen as the main focus of the discussion as it is more complex to diagnose and manage than UC. The author’s view on this topic is also presented, along with expert opinion on the impact of AI on prognosis, decision-making, and precision medicine for the management of IBD in general.
2. AI for IBD diagnosis
Most studies evaluating AI for the diagnosis of IBD are based on either ML or convolutional neural networks to analyze genetic/genomic datasets, imaging and endoscopy datasets, and protein expression/proteomics datasets [3].
A major challenge in IBD diagnosis is distinguishing between CD and UC, which is usually done on the basis of clinical features, such as how the inflammation is distributed along the gastrointestinal tract [3]. AI has been used to analyze molecular data to distinguish between CD and UC [8]. Recently, Kraszewski et al. developed a simple diagnostic methodology based only on markers from blood, urine, and stools that could be used by a general practitioner for early diagnosis of IBD [9]. Such a model would be useful for IBD diagnosis in difficult circumstances, like the recent COVID pandemic, during which the gold-standard endoscopic evaluation was difficult to use. Using random forest ML methods, the model could diagnose CD and UC with 97% and 91% mean average precision, respectively, demonstrating the feasibility of IBD diagnosis using noninvasive methods. The UC model included age, gender, and 14 routinely collected laboratory variables (FCP, platelet–large cell ratio, erythrocyte sedimentation rate, creatinine, hemoglobin, mean corpuscular hemoglobin [MCH], low-density lipoprotein cholesterol, peripheral blood erythrocytes, peripheral blood leukocytes, hepatitis B e-antigen, carcinoembryonic antigen, bacteriuria, microscopic stool ova and parasites test, and glucosuria). For diagnosis of CD, the most significant laboratory markers were peripheral blood neutrophils, mean platelet volume, MCH, mean corpuscular hemoglobin concentration, hematocrit, alkaline phosphatase, potassium, total bilirubin, aspartate transaminase, peripheral blood monocytes, erythrocytes, basophils, and erythroblasts [9]. Involvement of genes and their variants in disrupting molecular function are of growing interest for CD diagnosis and AI/ML methods have been used to elucidate such relationships [10, 11].
3. AI for endoscopic assessment
Endoscopic assessment remains the mainstay of objective evaluations in IBD and is a key indicator of therapeutic response, but it is an invasive procedure. Noninvasive prediction models that are based on symptoms or routinely used biomarkers cannot predict endoscopic healing in all patients. Therefore, ileocolonoscopy is the primary approach to assess mucosal disease activity and healing in CD [12]. However, the established endoscopic scoring systems are challenged by recall bias, heterogeneity in clinical presentation, and intra- and inter-observer variability [13]. Application of AI to estimate endoscopic indices/scores has been shown to improve precision and accuracy in quantifying disease severity for both CD and UC [14–20].
Guez et al. developed a multimodal AI model that integrated information from magnetic resonance (MR) enterography and biochemical biomarkers, such as CRP and FCP, to noninvasively assess ileal endoscopic activity in CD [21]. With a better aggregated area under the curve over the folds (0.84 vs 0.8, p < 1e − 9) and median test mean-squared error distribution (7.73 vs 8.8; Wilcoxon test; p < 1e − 5), this model performed better than the current clinical linear models based on the MR index of activity score as well as the ML models created exclusively based on radiological variables or biochemical markers. Use of this model has potential to reduces the number of MR sequences and radiological items that need to be assessed by radiologists to predict ileal endoscopic activity noninvasively. This noninvasive diagnostic method could be an additional advantage for young patients with CD as they need lifelong monitoring.
Iacucci et al. have developed an AI model using white-light endoscopy and virtual chromoendoscopy videos to distinguish histological remission and predict risk of flare. The prediction of histological remission was found to be similar between white-light endoscopy and virtual chromoendoscopy videos, with accuracy ranging between 80% and 85% [22].
4. AI in predicting treatment response in CD
AI has been applied in various fields for predicting treatment response or selecting treatments. For example, it has been used in radiomics to develop a model to predict the sensitivity of tumors to nivolumab, docetaxel, and gefitinib in patients with non-small cell lung cancer [23]. Another example is the development of ML-based ‘personalized antibiograms’. Corbin et al. developed ML models predicting antibiotic-susceptibility patterns, called personalized antibiograms, using electronic health record data. Personalized antibiograms could achieve similar coverage to the clinician benchmark with narrower antibiotics, which could, in turn, improve safety and decrease unnecessary use of broad-spectrum antibiotics [24]. Similarly, considerable research is being devoted to evaluating the use of AI in predicting the treatment response in IBD.
Traditional methods of developing predictive models in IBD are based on statistical regression, which cannot analyze more complex data structures such as repeated measurements. AI can help overcome this limitation [25]. By facilitating reliable prediction of treatment outcomes, AI can inform patient preferences and support shared decision-making, thereby enabling more personalized and cost-effective management of the disease [26]. Nguyen et al. performed a qualitative systematic literature review of studies that compared the performance of ML with traditional statistical models, which were developed based on clinical data routinely collected to predict IBD risk. This review identified 13 studies in which ML-based methods were found to perform better than traditional statistical methods in terms of predicting response to biologics and thiopurines treatment, as well as predicting disease activity and complications over the long term in patients with UC [2]. The performance of some ML models developed for predicting treatment response in patients with CD is summarized in Table 1. Further advancement of the current AI models would make them suitable for use in daily practice, where prediction of treatment response, disease course, or complications can guide clinicians’ treatment decisions.
Table 1.
Performance of AI models developed for patients with CD.
| Study | Objective | AI method used | Performance |
|---|---|---|---|
| Con et al. 2021 [25] | Predicting the response to anti-TNF therapy using conventional vs deep-learning models | Deep learning: feed-forward and recurrent neural network | AuROC and 95% CI:
|
| Waljee et al. 2018 [27] | Predicting the response to vedolizumab treatment | Random forest method | AuROC and 95% CI for corticosteroid-free biologic remission at week 52:
|
| Waljee et al. 2017 [28] | Predicting the response to thiopurine treatment | Random forest method | AuROC and 95% CI for objective remission:
|
| Park et al. 2022 [29] | Predicting the non-durable response to anti-TNF therapy in CD using transcriptome imputed from genotypes | LASSO regression | AuROC (SD) for training and test datasets:
AuROC (SD) for training and test dataset, respectively, for most frequently selected combination of two or three genes for whole-blood expression imputation model:
|
| He et al. 2021 [30] | Predicting response to ustekinumab using gene transcription profiling of patients with CD | Least absolute shrinkage and selection operator regression analysis | AuROC:
|
| Stidham et al. 2021 [7] | Predicting surgical outcomes in US veterans with CD using ML models incorporating routinely collected laboratory studies | LASSO regularized logistic regression | Mean (SD) sensitivity, specificity, AuROC, Brier score, AuROC (random splitting method), and Brier score (random splitting method), for the five models, respectively:
|
| Dong et al. 2019 [31] | Predicting surgery for therapeutic decision-making in Chinese patients with CD | RF, LR, SVM, DT, ANN | Accuracy, precision, true negative rate, and F1 score of the models, respectively:
|
| Venkatapurapu et al. 2022 [32] | Predicting temporal changes in mucosal health using a computational approach integrated with a mechanistic model of CD | A hybrid mechanistic-statistical platform | Overall sensitivity and specificity:
Overall performance of the platform:
|
6-TGN, 6-thioguanine nucleotide; ANN, artificial neural network; AuROC, area under the receiver operator characteristic curve; CD, Crohn’s disease; CI, confidence interval; DT, decision tree; LASSO, least absolute shrinkage and selection operator; LR, logistic regression; ML, machine learning; RF, random forest; SD, standard deviation; SVM, support vector machine; TNF, tumor necrosis factor.
4.1. Prediction of treatment response to anti-tumor necrosis factor (TNF) therapy in CD
In a proof-of-concept study, Con et al. developed deep-learning or artificial neural-network models using the biomarker CRP to predict remission (defined as CRP < 5 mg/L at 12 months) after anti-TNF therapy in CD. The conventional model used baseline data only, while the deep-learning models used baseline and repeated biomarker data. The ML methods showed stronger predictive performance than the conventional statistical model with a significantly higher area under the receiver operator characteristic curve (AuROC; 0.754 [95% CI: 0.674–0.834] vs 0.659 [95% CI: 0.562–0.756]; p = 0.036) [25].
Park et al. developed an ML model using the imputed gene-expression features that could effectively predict non-durable response to anti-TNF agents in patients with CD. The model found that the higher imputed expression levels of the DPY19L3 and GSTT1 genes increased, whereas that of NUCB1 decreased the probability of a non-durable response (Table 1) [29].
4.2. Prediction of treatment response to ustekinumab
He et al. developed an ML model based on the unique expression of four genes (HSD3B1, MUC4, CF1, and CCL11) to predict the response to ustekinumab in patients with CD. The model’s AuROC for the training and testing datasets were 0.746 and 0.734, respectively [30]. This was the first model to build a gene expression-prediction model for response to ustekinumab.
4.3. Prediction of treatment response to thiopurines
Using laboratory values and the age of patients, Waljee et al. developed an ML model to predict objective remission and clinical outcomes with thiopurines in patients with IBD. The AuROC for algorithm-predicted remission in the validation set was 0.79 vs 0.49 for 6-thioguanine nucleotide methods [28].
4.4. Prediction of complications in patients with CD
Siegel et al. developed a CDPATH or PROSPECT model to predict complications (the time from diagnosis to occurrence of an internal penetrating disease, bowel stricture, or non-perianal surgery, such as bowel resection or strictureplasty) in patients with CD [33, 34]. The PROSPECT tool included the following variables: demographic and clinical characteristics, medication exposure, time from diagnosis to complication, NOD2 status, and serologic immune responses. The tool was calibrated and validated in clinical laboratory settings.
4.5. Prediction of risk for surgery
Two ML models were developed for predicting the risk of surgical intervention in patients with CD [31, 35]. Stidham et al. used routinely collected laboratory variables (complete blood counts and metabolic panels) for this. Anti-TNF use was found to be the most impactful predictor; it was associated with a lower risk of surgery within 1 year. In contrast, corticosteroid use was associated an increased risk of surgery. Similarly, high platelet counts and mean cell hemoglobin concentrations, but low levels of albumin and blood urea nitrogen were associated with an increased risk of surgery.
Dong et al. developed ML models predicting the risk of CD-related surgery and complications based on the predictor variables included in a logistic regression model developed by Guizzetti et al. [36], namely, age (at enrollment and diagnosis), gender, smoking status, perianal disease, previous surgical resection for CD, disease location, baseline medication use, abdominal mass and pain, strictures, stool frequency, extraintestinal manifestations, and fistulas. The ML model developed by Dong et al. showed higher accuracy, precision, and F1 score than the statistical model [31].
4.6. Prediction of treatment response to vedolizumab—statistical and single-scale AI modeling
There are two clinical decision-support tools (CDSTs) for predicting treatment response to vedolizumab in patients with CD: one developed by Dulai et al. (2018) and the other developed by Waljee et al. (2018) [27, 37–40].
Dulai et al. used a classic regression approach based on baseline characteristics as predictors for their model. Since vedolizumab exposure is related to its response, higher remission rates can be achieved with dose intensification in patients with low vedolizumab trough concentrations during induction. Moreover, the onset of action of vedolizumab is perceived as being slow. These notions were examined through external validation of the CDST developed by Dulai et al. using three external cohorts [38, 39]. The first external validation was conducted using GEMINI 2 clinical trial data (NCT00783692) to evaluate whether differences in the remission rates are associated with differences in vedolizumab concentrations. The study also assessed whether the CDST can predict variations in the onset of action of vedolizumab. The second external validation was conducted using data from the GETAID prospective study and VICTORY cohorts to evaluate the CDST’s ability to accurately identify patients who may benefit from dose intensification. The CDST’s ability to measure the probability of surgery for CD while on vedolizumab was also evaluated. The tool was further validated to identify patients at risk of higher healthcare-resource utilization, particularly surgery and hospitalization [39]. These validations offer opportunities for therapy optimization, although with some limitations. For example, strong uncertainties remain about the vedolizumab serum cut-off point for clinical remission [41].
Waljee et al. used an ML random forest approach based on baseline characteristics and early treatment outcomes during the vedolizumab induction phase reported from a clinical trial to build their model. The AuROC for corticosteroid-free remission at week 52 considering only baseline data was 0.65 (95% CI: 0.53–0.77), whereas it was 0.75 (95% CI: 0.64–0.86) considering data until week 6. This model could identify patients who were unlikely to attain remission in 6 weeks [27].
5. Deep dive: prediction of treatment response to vedolizumab—multiscale system modeling in immunology
None of the models described in the previous section of this paper can predict temporal changes in treatment outcomes or mucosal health. There is therefore a need for tools that can help bridge the gap between subjective and infrequent objective assessments of disease severity and support decision-making on therapy initiation or continuation.
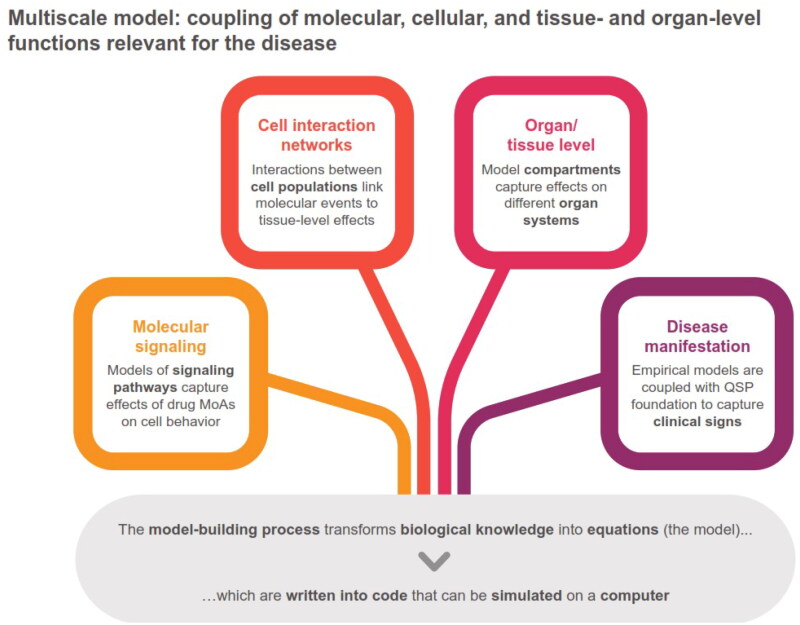
Venkatapurapu et al. tried to address this need by developing a computational platform that can estimate changes in mucosal health and inflammatory activity over a continuous timeline of weeks to years [32]. This model was based on the concept of systems modeling in immunology [32], which involves multiscale modeling integrating molecular, cellular, tissue, and organ-level functions (Figure 2). Such multiscale models are promising in-silico tools to evaluate new therapeutic targets, new biomarkers, and treatment responders and non-responders [42].
Figure 2.
Building the multiscale model platform.
MoA, mode of action; QSP, quantitative systems pharmacology.
5.1. Model description/structure
This multiscale model (Venkatapurapu et al. [32]) integrates the most current knowledge about each immune system with mathematical modeling. The platform has the following three major components: an ML-based responder classifier, a mechanistic model of CD pathophysiology, and a virtual library of patients with CD—termed a ‘digital twin’. Using the decision tree, a response classifier classifies patients qualitatively into having a complete, partial, or no response. The mechanistic model is a mathematical representation of the gut immune system and CD pathophysiological mechanisms in the form of coupled differential equations. This method allows for quick mapping of real patients with their digital twins. The platform can predict endoscopic remission and mucosal healing after vedolizumab treatment with an overall sensitivity of 80% and 75%, and a specificity of 69% and 70%, respectively [32].
5.2. Author’s view on the model’s potential impact on clinical practice
Venkatapurapu et al. referred to STRIDE I (2015) to anchor their model to the therapeutic strategy for CD [32]. However, they seemed to anticipate its evolution (STRIDE II) by integrating biomarkers and endoscopic scores into their platform and considering the VERSIFY study as reference data for training and testing their model [43–45]. The multiscale model has the potential to affect clinical practice in several ways. It was developed and validated to predict the evolution of tissue damage in response to treatment with vedolizumab. Similar training and validation for other biologics will enable forecasting of the effect of treatment on important clinical measures—such as ulcerated area, ulcer size, and overall simple endoscopic score–Crohn’s disease (SES-CD)—as well as various biomarkers. This process could be customized using individual patients’ historical data, rather than relying on a population average, offering an effective CDST for physicians to manage treatment plans and facilitate shared decision-making with patients [32].
Although remarkably innovative and high performing, with inaccurate FCP concentration prediction over time and no integration of the transmural healing aspects within the model, the proposed model does not fully comply with the recent STRIDE II guidelines that recommend selection of therapeutic targets in IBD. Hence, complementary work is needed to optimize the model and its deliverables for more effective utilization in clinical practice. Another limitation is that endoscopic data cannot be used to define transmural damage and extraluminal disease; thus, MR data may be needed to corroborate the model’s predictions. MR, computed tomography, and ultrasonography allow noninvasive measurement of transmural inflammation, strictures, fistulae, abscesses, and proximal small bowel involvement with high sensitivity and specificity [46, 47]. Hence, cross-fertilizing SES-CD with the MR index of activity (MaRIA) score might improve the predictive value of the model, especially for patients with a poor healing response. Thus, the model’s accuracy is currently limited because the SES-CD drives progression-response scenarios and response classifiers, while the FCP and disease location data drive digital-twin assignment.
5.3. Considerations for future development
Certain future standpoints could be considered for advancement of this model. The model should be tested using real-world data either retrospectively or prospectively, including biomarkers such as FCP and CRP, disease scoring such as CDAI or the Lemann index, endoscopic scoring such as SES-CD or modified multiplier SES-CD, and transmural scoring such as MaRIA. The purpose of this testing would be to: (1) verify that the model considers the diversity of disease outcomes through virtual patients; (2) identify which parameters help generate diversity as seen in the real world; (3) adjust the mathematical algorithms; (4) ensure model granularity to generate realistic virtual patients; (5) validate the accuracy of the assumptions made during model development; and (6) reach the relevant magnitude of responses to various treatments. A multicenter and international study on the model’s application in everyday clinical practice is required for it to be valued as an element of precision medicine for IBD in the future.
6. AI apps, chatbots, wearables, and nearables in IBD management
Following the introduction of the STRIDE II guidelines, treatment targets are shifting from symptom-based control to inflammation-based control, which requires monitoring of both symptoms and biomarkers. This evolution has challenged patients as well as providers to find alternative ways for monitoring, communicating, and treatment goal setting. Digital health can help meet this need, and the use of digital health technologies (e.g. tools leveraging smart phones, tablets, internet platforms, and wearables) is growing [48]. A scoping review of digital health apps for the clinical care of patients with IBD identified 11 relevant digital health apps; most of these apps were for obtaining data on patient-reported disease activity, and some were for treatment management [49].
Zand et al. developed a natural language processing (NLP)-based chatbot to categorize electronic messages from patients with IBD into various categories, such as medication queries, laboratory investigation results, and insurance or finance. The appropriateness of this classification was evaluated by three independent physicians. The concordance between the algorithm and physicians was found to be 95% [50].
Jagannath et al. [51] evaluated a wearable sweat sensor that can monitor sweat biomarkers for IBD. Given the role of exercise in IBD management [52], these types of devices could be useful for patients with IBD and become part of routine management for these patients [51].
7. AI in drug discovery for IBD
Evidence shows that AI is being increasingly applied in drug discovery and design, and quantitative structure-activity/property relationship (QSAR/QSPR) modeling presents the state-of-the-art applications in this field [53]. AI can support broad chemical space exploration, fast and easy identification of hit molecules, suggest their route of synthesis, help predict the required chemical structure, and explain drug–target interactions and structure-activity relationship [54]. An AI application ‘Found In Translation (FIT)’ has been developed that uses public gene-expression data to predict novel disease-associated genes. This model can extrapolate the results of a mouse experiment to humans [55]. In addition, the use of virtual patients and in-silico trials may lead to more targeted, safer, and effective treatments and reduce the need for trials and the risk of error [56].
AI-designed drugs for IBD have recently entered phase 1 clinical trials. For example, a protein kinase C (PKC) theta inhibitor was fully designed by AI, and its doses for the first-in-human trial (FIHT) were calculated using ML integrating numerous pharmacological properties [57]. PKC-theta is critical in controlling T-cell functions and drives several autoimmune and inflammatory diseases, including different types of chronic colitis and IBD. The design-to-FIHT duration for such small molecules usually takes 5–10 years, but was about a year in this case [57]. Another example is the development of a barrier-protective therapy for IBD using a Boolean network explorer (a computational platform) by Katkar et al. [58]. This therapy comprises the dual agonism of two nuclear receptors—peroxisome proliferator-activated receptors (PPAR)α and PPARγ—with the aim of altering macrophage processes and mitigate colitis. This therapy could correct gene expression from disease to health [58]. Additionally, Wang et al. developed an ML-based tool (AVA,Dx – Analysis of Variation for Association with Disease) that incorporated exonic variants from whole exome or genome sequencing data to identify IBD pathogenesis pathways [10].
8. Conclusion
A growing body of evidence showcases the tremendous potential for integrating AI into daily clinical practice to enable personalized medicine and shared decision-making in various diseases, including IBD. However, more work is needed to test and validate these ideas and to change treatment practices. Research should consider challenging already established platforms with new omics, molecular, physiological, clinical, imaging, and therapeutic updates.
9. Expert opinion
IBD is a chronic, complex, debilitating disease affecting millions of individuals worldwide. It is difficult to manage and causes significant morbidity and quality-of-life burdens from an individual perspective, and structural and financial concerns from a healthcare perspective. Advances in AI hold promise for improving prediction, fostering shared decision-making between patients and healthcare professionals (HCPs), and precision medicine.
9.1. Impact of AI on prognosis
IBD is a multi-omics disorder involving genomic, transcriptomic, proteomic, and epigenomic variations, as well as environmental contributions [59]. AI could be used to simultaneously analyze different molecules at all these levels, and the results could be integrated into multi-omics models [60]. This approach could provide insights into disease pathogenesis, identify more promising predictive biomarkers, and facilitate early diagnosis and disease management, thereby improving patient outcomes [61]. Such multi-omics projects are underway to investigate IBD heterogeneity and improve precision management [62].
9.2. Impact of AI on clinical decision-making
AI can help patients and clinicians make more informed and shared decisions about treatment options by (1) analyzing patient data, including medical history and progressive treatment outcomes, (2) providing personalized treatment recommendations based on individual characteristics and preferences, and (3) improving treatment acceptance and compliance. This leads to better patient outcomes and, consequently, better financial outcomes for payers. Additionally, based on omics makeup, medical history, and predicted treatment response, AI can identify patterns and biomarkers that indicate the best treatment options for a particular patient by analyzing vast amounts of data from multiple sources.
9.3. Impact of AI on precision medicine
IBD can be effectively treated, but there is no one-size-fits-all treatment approach as the condition has a wide range of phenotypes, disease courses, and outcomes.
It has been shown that single therapeutic agents can reach a therapeutic ceiling with limited remission rates [63]. Given the multiple pathological pathways that drive inflammatory processes in IBD, treatment approaches involving a combination of well-established single agents may be an alternate disease-control strategy [63]. However, there is a need to clarify inflammatory biopathways and omics signatures for each IBD phenotype, to ensure maximal treatment benefit for each patient. Precision medicine supported by AI and ML algorithms can help predict the disease course from the time of diagnosis, thereby supporting the identification of the best treatment approaches to be used for each patient [64].
Predictive ability is a critical factor in precision medicine. ML algorithms are trained to identify the patterns and relationships between clinical and biological variables to predict disease progression, identify patients at risk of complications, and recommend optimal treatment plans. Deep-learning algorithms analyze complex data such as medical images, genetic data, and tissue samples. NLP analyzes unstructured data, such as physician notes and patient reports, to identify key indicators of disease activity and treatment response, providing valuable insights into patient outcomes. Predictive modeling is used to analyze these large datasets of patient information to generate personalized treatment plans that will help optimize treatment approaches.
Thus, AI integrates data from various sources such as electronic medical records, multi-omic and culturomic data, and patient-reported outcomes to create a comprehensive picture of the patient’s health status. A combination of laboratory results, demographic characteristics, and disease location has been shown to be of strong prediction in CD [65]. Integrating genomic and clinical data can help identify the different subtypes of IBD patients [66].
The performance of predictive models is usually assessed by their ability to identify patients at risk, and by how much the predicted risk deviates from the observed risk. However, these statistics do not provide insights into whether the model would provide more benefit than harm if used in clinical practice [66]. A decision-curve analysis to determine the net benefit of a particular model in a particular clinical context can help improve our ability to evaluate performance and clinical utility of these models. The clinical utility would be demonstrated if the model shows a higher net benefit versus alternative ones [67]. Predictive models or CDSTs exist for most of the biologics [25, 29, 30, 38, 40, 68, 69]. Increasing our capability of evaluating the performance of these models would result in improved clinical practice and better outcomes for patients with IBD.
AI can revolutionize daily IBD practices in several ways. First, it supports precision medicine, which allows a more targeted treatment approach. As previously stated, physicians can select the most effective treatment for each individual by analyzing a patient’s omics makeup, medical history, and other factors. This leads to better clinical outcomes and fewer side effects as patients receive the most appropriate treatment for their specific disease phenotype. Second, with precision medicine, physicians can identify patients at a higher risk of developing severe IBD, treatment failure, or loss of response, and they can intervene earlier to prevent disease progression. This would reduce the need for aggressive treatments, surgery, and hospitalization. Precision medicine for monitoring disease progression may include AI-powered remote-monitoring tools that help patients and physicians monitor disease activity and treatment responses in real time, so signs of disease flare-ups can be identified early. Lastly, precision medicine will lead to more efficient use of healthcare resources. Physicians will be able to reduce the number of unnecessary procedures and medications by optimizing treatment selection, leading to cost savings for patients and healthcare systems.
9.4. Challenges to AI adoption in clinical practice
Although AI offers considerable promise for medical diagnostics and clinical management, studies may present with methodological challenges such as small datasets and lack of external validation or comparison with healthcare professionals’ performance using the same sample, as well as limitations with study design, delivery, and reporting [70, 71]. Therefore, there is a need for reporting standards that address specific methodological challenges of AI to enable acceptance and improve perception of this promising technology [71]. Other challenges that currently affect the full adoption of AI in daily clinical practice, include data quality, data privacy and security, ethical concerns, technical limitations, and regulatory barriers [72].
Firstly, AI algorithms require high-quality data for training and validation to generate accurate predictions. However, healthcare data are often incomplete, inconsistent, of poor quality, and lacking standardization. This can limit their accuracy and reliability, leading to inaccurate predictions, difficulties in interpretation, and disparities in care; this would undermine the trust of clinicians in the technology and its adoption in clinical practice. Data from electronic health records are often siloed and not easily accessible. This limits the amount of data available to train algorithms and makes it difficult to develop algorithms with broad applicability [73]. To address these issues of data availability, quality, and completeness, efforts are needed to establish data collection, storage, and sharing standards and to develop technologies that can effectively integrate data from different sources. This can be costly and complex, and HCPs may lack the technical expertise to implement and maintain these systems. Domain expertise may not be available in every healthcare institution. Data scientists and clinicians must collaborate to address this issue and ensure that AI algorithms are clinically relevant and valid.
Secondly, healthcare data are subject to strict privacy and security regulations to protect patient confidentiality, which varies across institutions, countries, and continents. Sharing data between institutions for AI analysis can be challenging because of these regulations, and data breaches can have legal and reputational consequences.
Thirdly, ethical concerns regarding AI application in clinical practice include bias, transparency, and accountability. Physicians may be hesitant to rely on AI predictions if they do not understand how the algorithm arrived at the prediction or if they are concerned about data bias. AI algorithms may produce recommendations that conflict with patients’ preferences or values. To address this issue, guidelines must be developed to ensure that AI can be safely and effectively used to improve patient care. This includes ensuring that patients are adequately informed about AI applications in their care and respecting their rights and preferences [74].
Finally, regulatory approval is required for medical devices that use AI algorithms for clinical decision-making. However, the regulatory process can be lengthy, complex, and costly, and lack of clarity regarding the regulatory framework for AI-based medical devices can hinder their adoption. This limits the ability to develop and deploy AI algorithms rapidly [75, 76].
Addressing these challenges requires collaboration among HCPs, researchers, regulators, and industries. If these challenges are overcome, incorporating AI in precision medicine for IBD is expected to significantly improve standard procedures for disease management over the next 5–10 years.
9.5. Conclusions and future work
There are no definitive endpoints for AI or precision medicine in patients with IBD. AI is an evolving field, and new tools and techniques to improve disease management and patient outcomes are constantly being developed. The potential of AI to contribute to precision medicine continues to grow as our understanding of IBD and the underlying pathomechanism improves. Potential areas of advancement include data source expansion, real-time monitoring, highly specific pan-disease or treatment companion biomarkers, upscaled prediction skills, accelerated identification and development of new treatment targets, and existing drug repositioning.
AI algorithms must be integrated into clinical workflows to ensure they are actionable and can be used in routine clinical practices. One of the challenges in this area is designing interpretable and transparent algorithms so that clinicians can understand how they arrive at a particular recommendation. For this, efforts are needed to develop user-friendly interfaces that can be easily integrated with electronic health record systems.
Further, AI can help optimize clinical trials for new IBD treatments by identifying patients most likely to respond to new therapies. This could accelerate the development and approval of new IBD treatments. The same is true for repositioning existing drugs. Overall, the future of AI application in precision medicine for IBD is very promising, and significant advancements in this field are expected over the next 5 years.
Acknowledgements
Medical writing and editorial assistance were provided by Leena Patel, Cristiana Miglio, and Daria Renshaw from IQVIA, funded by Ferring Pharmaceuticals. Ideation and conception were supported by Yogi Meguro from Sophia University (Tokyo, Japan) and literature searches by Tine Haarmark Nielsen from Ferring Pharmaceuticals (Kastrup, Denmark).
Authors’ contributions
PhP has made substantial contributions to the conception of the work, to the interpretation of data, have drafted the work and substantively revised it, has approved the submitted version, and agrees to be personally accountable for the author’s own contributions and for ensuring that questions related to the accuracy or integrity of any part of the work.
Data sharing
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Disclosure statement
Philippe Pinton is an employee of Ferring Pharmaceuticals and holds stocks of Takeda Pharmaceutical Company Limited.
References
- 1.Lee SH, Kwon JE, Cho M-L.. Immunological pathogenesis of inflammatory bowel disease. Intest Res. 2018;16(1):1–14. doi: 10.5217/ir.2018.16.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nguyen NH, Picetti D, Dulai PS, et al. Machine learning-based prediction models for diagnosis and prognosis in inflammatory bowel diseases: a systematic review. J Crohns Colitis. 2022;16(3):398–413. doi: 10.1093/ecco-jcc/jjab155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gubatan J, Levitte S, Patel A, et al. Artificial intelligence applications in inflammatory bowel disease: emerging technologies and future directions. World J Gastroenterol. 2021;27(17):1920–1935. doi: 10.3748/wjg.v27.i17.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tontini GE, Vecchi M, Pastorelli L, et al. Differential diagnosis in inflammatory bowel disease colitis: state of the art and future perspectives. World J Gastroenterol. 2015;21(1):21–46. doi: 10.3748/wjg.v21.i1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pinton P. Computational models in inflammatory bowel disease. Clin Transl Sci. 2022;15(4):824–830. doi: 10.1111/cts.13228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Javaid M, Haleem A, Singh RP, et al. Significance of machine learning in healthcare: features, pillars and applications. Int J Intell Netw. 2022;3:58–73. doi: 10.1016/j.ijin.2022.05.002. [DOI] [Google Scholar]
- 7.Stidham RW, Takenaka K.. Artificial intelligence for disease assessment in inflammatory bowel disease: how will it change our practice? Gastroenterology. 2022;162(5):1493–1506. doi: 10.1053/j.gastro.2021.12.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smolander J, Dehmer M, Emmert-Streib F.. Comparing deep belief networks with support vector machines for classifying gene expression data from complex disorders. FEBS Open Bio. 2019;9(7):1232–1248. doi: 10.1002/2211-5463.12652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kraszewski S, Szczurek W, Szymczak J, et al. Machine learning prediction model for inflammatory bowel disease based on laboratory markers. Working model in a discovery cohort study. J Clin Med. 2021;10(20):4745. doi: 10.3390/jcm10204745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Miller M, Astrakhan Y, et al. Identifying crohn’s disease signal from variome analysis. Genome Med. 2019;11(1):59. doi: 10.1186/s13073-019-0670-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Lange KM, Moutsianas L, Lee JC, et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet. 2017;49(2):256–261. doi: 10.1038/ng.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brand EC, Elias SG, Minderhoud IM, et al. Systematic review and external validation of prediction models based on symptoms and biomarkers for identifying endoscopic activity in crohn’s disease. Clin Gastroenterol Hepatol. 2020;18(8):1704–1718. doi: 10.1016/j.cgh.2019.12.014. [DOI] [PubMed] [Google Scholar]
- 13.Mosli MH, Feagan BG, Sandborn WJ, et al. Histologic evaluation of ulcerative colitis: a systematic review of disease activity indices. Inflamm Bowel Dis. 2014;20(3):564–575. doi: 10.1097/01.MIB.0000437986.00190.71. [DOI] [PubMed] [Google Scholar]
- 14.Charisis VS, Hadjileontiadis LJ.. Potential of hybrid adaptive filtering in inflammatory lesion detection from capsule endoscopy images. World J Gastroenterol. 2016;22(39):8641–8657. doi: 10.3748/wjg.v22.i39.8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klang E, Barash Y, Margalit RY, et al. Deep learning algorithms for automated detection of crohn’s disease ulcers by video capsule endoscopy. Gastrointest Endosc. 2020;91(3):606–613.e2. doi: 10.1016/j.gie.2019.11.012. [DOI] [PubMed] [Google Scholar]
- 16.Takenaka K, Ohtsuka K, Fujii T, et al. Development and validation of a deep neural network for accurate evaluation of endoscopic images from patients with ulcerative colitis. Gastroenterology. 2020;158(8):2150–2157. doi: 10.1053/j.gastro.2020.02.012. [DOI] [PubMed] [Google Scholar]
- 17.Bossuyt P, Nakase H, Vermeire S, et al. Automatic, computer-aided determination of endoscopic and histological inflammation in patients with mild to moderate ulcerative colitis based on red density. Gut. 2020;69(10):1778–1786. doi: 10.1136/gutjnl-2019-320056. [DOI] [PubMed] [Google Scholar]
- 18.Bhambhvani HP, Zamora A.. Deep learning enabled classification of Mayo endoscopic subscore in patients with ulcerative colitis. Eur J Gastroenterol Hepatol. 2021;33(5):645–649. doi: 10.1097/MEG.0000000000001952. [DOI] [PubMed] [Google Scholar]
- 19.Ozawa T, Ishihara S, Fujishiro M, et al. Novel computer-assisted diagnosis system for endoscopic disease activity in patients with ulcerative colitis. Gastrointest Endosc. 2019;89(2):416–421.e1. doi: 10.1016/j.gie.2018.10.020. [DOI] [PubMed] [Google Scholar]
- 20.Yao H, Najarian K, Gryak J, et al. Fully automated endoscopic disease activity assessment in ulcerative colitis. Gastrointest Endosc. 2021;93(3):728–736.e1. doi: 10.1016/j.gie.2020.08.011. [DOI] [PubMed] [Google Scholar]
- 21.Guez I, Focht G, Greer M-LC, et al. Development of a multimodal machine-learning fusion model to non-invasively assess ileal crohn’s disease endoscopic activity. Comput Methods Programs Biomed. 2022;227:107207. doi: 10.1016/j.cmpb.2022.107207. [DOI] [PubMed] [Google Scholar]
- 22.Iacucci M, Cannatelli R, Parigi TL, et al. A virtual chromoendoscopy artificial intelligence system to detect endoscopic and histologic activity/remission and predict clinical outcomes in ulcerative colitis. Endoscopy. 2023;55(4):332–341. doi: 10.1055/a-1960-3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dercle L, Fronheiser M, Lu L, et al. Identification of non–small cell lung cancer sensitive to systemic cancer therapies using radiomics. Clin Cancer Res. 2020;26(9):2151–2162. doi: 10.1158/1078-0432.CCR-19-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corbin CK, Sung L, Chattopadhyay A, et al. Personalized antibiograms for machine learning driven antibiotic selection. Commun Med (Lond). 2022;2(1):38. doi: 10.1038/s43856-022-00094-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Con D, van Langenberg DR, Vasudevan A.. Deep learning vs conventional learning algorithms for clinical prediction in crohn’s disease: a proof-of-concept study. World J Gastroenterol. 2021;27(38):6476–6488. doi: 10.3748/wjg.v27.i38.6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J, Girard M, Wang S, et al. Using supervised machine learning approach to predict treatment outcomes of vedolizumab in ulcerative colitis patients. J Biopharm Stat. 2022;32(2):330–345. doi: 10.1080/10543406.2021.2009500. [DOI] [PubMed] [Google Scholar]
- 27.Waljee AK, Liu B, Sauder K, et al. Predicting corticosteroid-free biologic remission with vedolizumab in crohn’s disease. Inflamm Bowel Dis. 2018;24(6):1185–1192. doi: 10.1093/ibd/izy031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waljee AK, Sauder K, Patel A, et al. Machine learning algorithms for objective remission and clinical outcomes with thiopurines. J Crohns Colitis. 2017;11(7):801–810. doi: 10.1093/ecco-jcc/jjx014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park SK, Kim YB, Kim S, et al. Development of a machine learning model to predict non-durable response to anti-TNF therapy in crohn’s disease using transcriptome imputed from genotypes. J Pers Med. 2022;12(6):947. doi: 10.3390/jpm12060947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He M, Li C, Tang W, et al. Machine learning gene expression predicting model for ustekinumab response in patients with crohn’s disease. Immun Inflamm Dis. 2021;9(4):1529–1540. doi: 10.1002/iid3.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong Y, Xu L, Fan Y, et al. A novel surgical predictive model for chinese crohn’s disease patients. Medicine (Baltimore). 2019;98(46):e17510. doi: 10.1097/MD.0000000000017510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venkatapurapu SP, Iwakiri R, Udagawa E, et al. A computational platform integrating a mechanistic model of crohn’s disease for predicting temporal progression of mucosal damage and healing. Adv Ther. 2022;39(7):3225–3247. doi: 10.1007/s12325-022-02144-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siegel C, Horton H, Siegel L, et al. A validated web-based tool to display individualised crohn’s disease predicted outcomes based on clinical, serologic and genetic variables. Aliment Pharmacol Ther. 2016;43(2):262–271. doi: 10.1111/apt.13460. [DOI] [PubMed] [Google Scholar]
- 34.Siegel CA, Siegel LS, Dubinsky MC, et al. Performance characteristics of a clinical decision support tool for disease complications in crohn’s disease. Crohns Colitis 360. 2021;3(4):otab074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stidham RW, Liu Y, Enchakalody B, et al. The use of readily available longitudinal data to predict the likelihood of surgery in crohn disease. Inflamm Bowel Dis. 2021;27(8):1328–1334. doi: 10.1093/ibd/izab035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guizzetti L, Zou G, Khanna R, et al. Development of clinical prediction models for surgery and complications in crohn’s disease. J Crohns Colitis. 2018;12(2):167–177. doi: 10.1093/ecco-jcc/jjx130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dulai PS, Boland BS, Singh S, et al. Development and validation of a scoring system to predict outcomes of vedolizumab treatment in patients with crohn’s disease. Gastroenterology. 2018;155(3):687–695. e10. doi: 10.1053/j.gastro.2018.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dulai PS, Amiot A, Peyrin-Biroulet L, et al. A clinical decision support tool may help to optimise vedolizumab therapy in crohn’s disease. Aliment Pharmacol Ther. 2020;51(5):553–564. doi: 10.1111/apt.15609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dulai PS, Wan Y, Huang Z, et al. Probability of response as defined by a clinical decision support tool is associated with lower healthcare resource utilization in vedolizumab-treated patients with crohn’s disease. Crohns Colitis 360. 2022;4(4):otac048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alric H, Amiot A, Kirchgesner J, et al. Vedolizumab clinical decision support tool predicts efficacy of vedolizumab but not ustekinumab in refractory crohn’s disease. Inflamm Bowel Dis. 2022;28(2):218–225. doi: 10.1093/ibd/izab060. [DOI] [PubMed] [Google Scholar]
- 41.Rosario M, Dirks N, Gastonguay M, et al. Population pharmacokinetics–pharmacodynamics of vedolizumab in patients with ulcerative colitis and crohn’s disease. Aliment Pharmacol Ther. 2015;42(2):188–202. doi: 10.1111/apt.13243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Balbas-Martinez V, Ruiz-Cerdá L, Irurzun-Arana I, et al. A systems pharmacology model for inflammatory bowel disease. PLoS One. 2018;13(3):e0192949. doi: 10.1371/journal.pone.0192949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peyrin-Biroulet L, Sandborn W, Sands B, et al. Selecting therapeutic targets in inflammatory bowel disease (STRIDE): determining therapeutic goals for treat-to-target. Am J Gastroenterol. 2015;110(9):1324–1338. doi: 10.1038/ajg.2015.233. [DOI] [PubMed] [Google Scholar]
- 44.Danese S, Sandborn WJ, Colombel J-F, et al. Endoscopic, radiologic, and histologic healing with vedolizumab in patients with active crohn’s disease. Gastroenterology. 2019;157(4):1007–1018. e7. doi: 10.1053/j.gastro.2019.06.038. [DOI] [PubMed] [Google Scholar]
- 45.Turner D, Ricciuto A, Lewis A, et al. STRIDE-II: an update on the selecting therapeutic targets in inflammatory bowel disease (STRIDE) initiative of the international organization for the study of IBD (IOIBD): determining therapeutic goals for treat-to-target strategies in IBD. Gastroenterology. 2021;160(5):1570–1583. doi: 10.1053/j.gastro.2020.12.031. [DOI] [PubMed] [Google Scholar]
- 46.Panés J, Bouzas R, Chaparro M, et al. Systematic review: the use of ultrasonography, computed tomography and magnetic resonance imaging for the diagnosis, assessment of activity and abdominal complications of crohn’s disease. Aliment Pharmacol Ther. 2011;34(2):125–145. doi: 10.1111/j.1365-2036.2011.04710.x. [DOI] [PubMed] [Google Scholar]
- 47.Bruining DH, Zimmermann EM, Loftus Jr EV, et al. Consensus recommendations for evaluation, interpretation, and utilization of computed tomography and magnetic resonance enterography in patients with small bowel crohn’s disease. Gastroenterology. 2018;154(4):1172–1194. doi: 10.1053/j.gastro.2017.11.274. [DOI] [PubMed] [Google Scholar]
- 48.Brooks-Warburton J, Ashton J, Dhar A, et al. Artificial intelligence and inflammatory bowel disease: practicalities and future prospects. Frontline Gastroenterol. 2022;13(4):325–331. doi: 10.1136/flgastro-2021-102003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yin AL, Hachuel D, Pollak JP, et al. Digital health apps in the clinical care of inflammatory bowel disease: scoping review. J Med Internet Res. 2019;21(8):e14630. doi: 10.2196/14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zand A, Sharma A, Stokes Z, et al. An exploration into the use of a chatbot for patients with inflammatory bowel diseases: retrospective cohort study. J Med Internet Res. 2020;22(5):e15589. doi: 10.2196/15589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jagannath B, Muthukumar S, Prasad S.. Wearable sweat sensing device for detection of IBD biomarkers. Inflamm Bowel Dis. 2021;27(Supplement_1):S12–S12. doi: 10.1093/ibd/izaa347.028. [DOI] [Google Scholar]
- 52.Engels M, Cross R, Long M.. Exercise in patients with inflammatory bowel diseases: current perspectives. Clin Exp Gastroenterol. 2018;11:1–11. doi: 10.2147/CEG.S120816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiménez-Luna J, Grisoni F, Weskamp N, et al. Artificial intelligence in drug discovery: recent advances and future perspectives. Expert Opin Drug Discov. 2021;16(9):949–959. doi: 10.1080/17460441.2021.1909567. [DOI] [PubMed] [Google Scholar]
- 54.Paul D, Sanap G, Shenoy S, et al. Artificial intelligence in drug discovery and development. Drug Discov Today. 2021;26(1):80–93. doi: 10.1016/j.drudis.2020.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Normand R, Du W, Briller M, et al. Found in translation: a machine learning model for mouse-to-human inference. Nat Methods. 2018;15(12):1067–1073. doi: 10.1038/s41592-018-0214-9. [DOI] [PubMed] [Google Scholar]
- 56.Sternebring O, Patidar N, Ravi A, et al. PP10-02239. Discrimination of the differential effects of selective suppression of IL-6 trans-signaling by olamkicept in moderate-to-severe ulcerative colitis. United European Gastroenterology Journal. Poster presented at: united European Gastroenterology (UEG) Week 2023; 14–17 October 2023;11:1465–1466. Copenhagen, Denmark. [Google Scholar]
- 57.Exscientia . Exscientia Announces First-in-Human Study for Bristol Myers Squibb In-Licensed PKC Theta Inhibitor, EXS4318. Press release February 2, 2023. Available from: https://investors.exscientia.ai/press-releases/press-release-details/2023/Exscientia-Announces-First-in-Human-Study-for-Bristol-Myers-Squibb-In-Licensed-PKC-Theta-Inhibitor-EXS4318/default.aspx.
- 58.Katkar GD, Sayed IM, Anandachar MS, et al. Artificial intelligence-rationalized balanced PPARα/γ dual agonism resets dysregulated macrophage processes in inflammatory bowel disease. Commun Biol. 2022;5(1):231. doi: 10.1038/s42003-022-03168-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palmieri O, Mazza T, Castellana S, et al. Inflammatory bowel disease meets systems biology: a multi-omics challenge and frontier. OMICS. 2016;20(12):692–698. doi: 10.1089/omi.2016.0147. [DOI] [PubMed] [Google Scholar]
- 60.De Souza HS, Fiocchi C, Iliopoulos D.. The IBD interactome: an integrated view of aetiology, pathogenesis and therapy. Nat Rev Gastroenterol Hepatol. 2017;14(12):739–749. doi: 10.1038/nrgastro.2017.110. [DOI] [PubMed] [Google Scholar]
- 61.Liu X-Y, Tang H, Zhou Q-Y, et al. Advancing the precision management of inflammatory bowel disease in the era of omics approaches and new technology. World J Gastroenterol. 2023;29(2):272–285. doi: 10.3748/wjg.v29.i2.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Verstockt B, Parkes M, Lee JC.. How do we predict a patient’s disease course and whether they will respond to specific treatments? Gastroenterology. 2022;162(5):1383–1395. doi: 10.1053/j.gastro.2021.12.245. [DOI] [PubMed] [Google Scholar]
- 63.Solitano V, Ma C, Hanžel J, et al. Advanced combination treatment with biologic agents and novel small molecule drugs for inflammatory bowel disease. Gastroenterol Hepatol (NY). 2023;19(5):251–263. [PMC free article] [PubMed] [Google Scholar]
- 64.Ashton JJ, Mossotto E, Ennis S, et al. Personalising medicine in inflammatory bowel disease—current and future perspectives. Transl Pediatr. 2019;8(1):56–69. doi: 10.21037/tp.2018.12.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reddy BK, Delen D, Agrawal RK.. Predicting and explaining inflammation in crohn’s disease patients using predictive analytics methods and electronic medical record data. Health Informatics J. 2019;25(4):1201–1218. doi: 10.1177/1460458217751015. [DOI] [PubMed] [Google Scholar]
- 66.Ashton J, Cheng G, Stafford I, et al. DOP64 utilising genomics to predict outcomes and delineate novel subgroups in inflammatory bowel disease. J Crohns Colitis. 2023;17(Supplement_1):i137–i140. doi: 10.1093/ecco-jcc/jjac190.0104. [DOI] [Google Scholar]
- 67.Vickers AJ, Van Calster B, Steyerberg EW.. Net benefit approaches to the evaluation of prediction models, molecular markers, and diagnostic tests. BMJ. 2016;352:i6. doi: 10.1136/bmj.i6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lefevre PL, Dulai PS, Wang Z, et al. A clinical prediction model to determine probability of response to certolizumab pegol for crohn’s disease. BioDrugs. 2022;36(1):85–93. doi: 10.1007/s40259-021-00512-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Park J, Chun J, Yoon H, et al. Feasibility of a clinical decision support tool for ustekinumab to predict clinical remission and relapse in patients with crohn’s disease: a multicenter observational study. Inflamm Bowel Dis. 2023;29(4):548–554. doi: 10.1093/ibd/izac105. [DOI] [PubMed] [Google Scholar]
- 70.Roberts M, Driggs D, Thorpe M, et al. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nat Mach Intell. 2021;3(3):199–217. doi: 10.1038/s42256-021-00307-0. [DOI] [Google Scholar]
- 71.Liu X, Faes L, Kale AU, et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. Lancet Digit Health. 2019;1(6):e271–e297. doi: 10.1016/S2589-7500(19)30123-2. [DOI] [PubMed] [Google Scholar]
- 72.Bajwa J, Munir U, Nori A, et al. Artificial intelligence in healthcare: transforming the practice of medicine. Future Healthc J. 2021;8(2):e188–e194. doi: 10.7861/fhj.2021-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ehsani-Moghaddam B, Martin K, Queenan JA.. Data quality in healthcare: a report of practical experience with the Canadian primary care sentinel surveillance network data. Health Inf Manag. 2021;50(1-2):88–92. doi: 10.1177/1833358319887743. [DOI] [PubMed] [Google Scholar]
- 74.Davenport T, Kalakota R.. The potential for artificial intelligence in healthcare. Future Healthc J. 2019;6(2):94–98. doi: 10.7861/futurehosp.6-2-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fraser AG, Biasin E, Bijnens B, et al. Artificial intelligence in medical device software and high-risk medical devices–a review of definitions, expert recommendations and regulatory initiatives. Expert Rev Med Devices. 2023;20(6):467–491. doi: 10.1080/17434440.2023.2184685. [DOI] [PubMed] [Google Scholar]
- 76.Zanca F, Brusasco C, Pesapane F, et al. Regulatory aspects of the use of artificial intelligence medical software. Semin Radiat Oncol. 2022;32(4):432–441. doi: 10.1016/j.semradonc.2022.06.012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.