Abstract
Myxococcus xanthus DNA segments related to the highly conserved central sequence of ς54 activator proteins have been investigated. A genetic technique designed to inactivate a gene that encodes such an activator by inserting a plasmid-borne internal fragment of the putative gene has been tested. When the internal fragment inserted by homologous recombination into the corresponding chromosomal locus, the expected duplication of the gene was observed by Southern hybridization. The single restriction fragment characteristic of each segment was replaced in the insertion strains by two hybridizing fragments, and one of these fragments hybridized with the kanamycin resistance gene of the plasmid vector. The combined molecular weights of the two fragments from the insertion strains were equal to the molecular weight of the original fragment plus the expected molecular weight contributed by the vector. In the duplication, one copy is expected to have an N-terminal deletion and the other copy is expected to have a C-terminal deletion. In most cases, the net result should be loss of activator function. If an activator is essential for vegetative growth, then it should not be possible to obtain the insertion strain by plasmid integration. Indeed, integrants for three of the segments were not obtained in repeated trials; however, a plausible explanation for these results other than lethality can be offered. Of the seven insertions validated by Southern hybridization, four strains exhibited defects in the development of fruiting bodies. One of these failed to develop in submerged culture, though it developed normally on agar. The other three showed arrested development of fruiting bodies, each at a morphologically different stage of aggregation. One of the mutants may be defective in the reception pathway of A-signal.
Myxococcus xanthus, a gram-negative, rod-shaped, soil bacterium, responds to starvation by differentiating metabolically dormant, spherical myxospores that are resistant to heat and desiccation (for a general review, see reference 9). These spores are contained within a macroscopic structure, the fruiting body, that is formed when the cells glide into and form aggregates of approximately 105 cells. This macroscopic structure is thought to enhance the eventual dispersion of its spores. To achieve its selective advantage, fruiting body development is highly regulated. For example, aggregation and morphogenesis of a fruiting body must precede the physiological and morphological differentiation of cells into the dormant myxospores. Before development, cells assess population density and nutrient conditions (47). Once the process has begun, the population then guides itself through several developmental checkpoints by producing several extracellular signals, the best understood of which are the A-, C-, and E-signals (7, 26, 31, 45). Mutants defective in producing one of these signals fail to develop or sporulate; however, signal-deficient mutants can be induced to sporulate if they are developed in coculture with either a wild-type strain or a mutant from a different signal class (13). A-factor, one of the earliest signals, has been shown to be a mixture of amino acids that must be present at a particular concentration to serve as a quorum sensor (19, 34). The C-signal is a cell surface protein which requires cell alignment for its transmission and helps bring large numbers of cells into late, symmetric aggregates (48).
One genetic route to understanding the mechanisms that regulate development is to find genes that are necessary for, or are regulated by, the developmental process. Two known developmentally regulated genes have promoters that resemble those recognized by the minor sigma factor ς54. Those genes are mbhA (42) and the A-signal-dependent gene termed Ω4521 (24). Keseler and Kaiser have shown that single base changes in six particular residues in the region from −12 to −24 (which distinguishes ς54 from ς70 promoters) ahead of the Ω4521 RNA start site decreased expression of this gene by 90% and that deletions of base −19 decreased expression by 95% (24), as was expected of a ς54 promoter. Attempts to investigate the overall role of ς54 in development by deleting the gene rpoN have been hampered because this gene, which encodes the ς54 subunit in M. xanthus, is essential for viability (25). ς54 promoters are unique in that each requires a specific activator protein in addition to RNA polymerase holoenzyme binding at the promoter, and these activator proteins are often part of sensory circuits. The best-studied examples of such activators are NtrC (NRI), which regulates nitrogen assimilation genes among enteric bacteria, and NifA, which regulates the nitrogen fixation genes in Rhizobium, Klebsiella, and other bacteria (35, 41).
Unable to explore the effects of a deletion of rpoN on development in general, we have turned to its specific activator proteins. The domain structure of the well-documented activator proteins is conserved along genus lines (40) (Fig. 1). The N-terminal domain is usually where the activator protein itself is modified (often by phosphorylation), which can affect the cooperativity of binding to the DNA as well as the ATPase activity of the activator. Promoter binding is directed by the C-terminal domain of the protein, while the central region contains an ATP binding domain, which is also the most highly conserved of the three (40) (Fig. 1).
FIG. 1.
Common structure of ς54 activator proteins. The N-terminal domain is the least conserved among genera and has a length which ranges from 12 to 400 amino acids. Some activators are phosphorylated on an aspartate residue in this domain (P). The C-terminal domain of 65 to 130 amino acids has a helix-turn-helix (HTH) motif characteristic of DNA binding proteins. The central domain of about 240 amino acids is the most highly conserved and has an ATP binding motif. Approximate locations of the PCR primers used to amplify the central domain are indicated by the converging arrows. Consensus data are from reference 40.
A PCR technique that specifically amplifies sequences corresponding to the central, strongly conserved, nucleotide binding domain of ς54 transcriptional activator proteins has been developed by Kaufman and Nixon. Using heterologous primers, they were able to amplify 14 NtrC-like PCR fragments of ∼475 bp each from M. xanthus chromosomal DNA (23). We have used these fragments in a way calculated to cause internal disruptions in their corresponding genes. Each fragment was inserted into a plasmid incapable of replication within M. xanthus, and the resulting plasmids were introduced into M. xanthus. Antibiotic resistance encoded by the plasmid-borne gene should result only by incorporation of that plasmid directly into the gene homologous to the PCR fragment used. The result of the technique should be an altered chromosomal structure resulting from an insertional mutation at the chromosomal locus. We have tested this mutagenesis approach to selectively mutate potential ς54 activator proteins. From 14 individual NtrC-like DNA fragments of 475 bp each, we have attempted disruption of the homologous chromosomal regions. Mutants generated this way were compared with a wild-type strain to assess effects on development. Results of this initial sampling are presented here.
MATERIALS AND METHODS
Media for growth or development.
Myxobacteria were propagated at 32°C in CTT broth (1% Bacto Casitone, 10 mM Tris-HCl [pH 8.0], 8 mM MgSO4, 1 mM KPO4 [pH 7.6]) or CTT agar (CTT broth plus 1.5% Bacto Agar) unless otherwise indicated. CTTYE (CTT plus 0.2% Bacto Yeast Extract) was used after electroporation of M. xanthus. Kanamycin (40 μg/ml) or oxytetracycline (12.5 μg/ml) was added where indicated below. A1 minimal medium has been described previously (3) and was solidified with either 1.5% Bacto Agar or 0.8% agarose. TPM buffer (10 mM Tris-HCl [pH 7.6], 8 mM MgSO4, 1 mM KPO4 [pH 7.6]) and TPM agar (TPM buffer plus 1.5% Bacto Agar) were used to induce development as previously described (28). Briefly, exponential cells were harvested by centrifugation and resuspended in TPM buffer to a concentration of 5 × 109 cells/ml. Ten- or twenty-microliter droplets were then placed on the surfaces of TPM agar plates. Development was also induced in submerged culture (30); cells were developed in 24-well, polystyrene microtiter plates. β-Galactosidase activity was assayed in cells harvested from either submerged culture or TPM agar plates (from five 20-μl droplets) as previously described (21). Escherichia coli cultures were grown in L broth or L agar (44) supplemented with 40 μg of kanamycin per ml where indicated below. Growth, in liquid, of M. xanthus was monitored by measuring turbidity in a Klett-Summerson photoelectric colorimeter equipped with a red filter.
PCR amplification of ς54 transcriptional activator sequences from M. xanthus.
Highly degenerate primers were used to amplify the central domain of ς54 transcriptional activator genes by methods that were previously described (23).
Plasmid and strain construction.
The M. xanthus wild-type strain DK 1622 was used as the starting point for strain construction, so that the products would be isogenic. All the strains and plasmids that were used are listed in Table 1. Thirteen 475-bp (nominal) PCR fragments were ligated into the EcoRI site in pBGS18 (49), generating plasmids pLAG1 through pLAG13. The plasmids were prepared and introduced into E. coli XL1 by either transformation or electroporation according to the methods of Sambrook et al. (44). After propagation in E. coli, these plasmids were extracted and electroporated into M. xanthus as described previously (22). That procedure was modified as follows. Eighty microliters of concentrated M. xanthus cell suspension (10,000 Klett units) was pipetted into a 0.2-cm-gap-size cuvette with 1 to 3 μl of plasmid DNA and electroporated at 1.3 kV, 400 W, and 25 mF, which gave time constants of 8.0 to 9.0 ms. Immediately following electroporation, 1 ml of CTTYE was added to the cuvette; the entire contents were then transferred to a flask containing 1.5 ml of CTTYE, and the mixture was incubated with shaking at 32°C for 5 h before being plated onto selective medium. Colonies were picked, and streaked onto CTT plus kanamycin agar. DNA was prepared from these bacteria (1) and restricted, and the digests were analyzed by Southern hybridization to confirm disruption of the target gene. Southern blots were probed both with the corresponding 475-bp fragment used to produce the gene disruption mutant and with a fragment from the kanamycin resistance gene of pBGS18.
TABLE 1.
Bacterial strains and plasmids
| Strain | Description | Reference or source |
|---|---|---|
| DK 1622 | Wild type | 20 |
| DK 7801 | Incorporation of pLAG 1 | This work |
| DK 7802 | Incorporation of pLAG 4 | This work |
| DK 7803 | Incorporation of pLAG 8 | This work |
| DK 7804 | Incorporation of pLAG 13 | This work |
| DK 7814 | Incorporation of pLAG 11 | This work |
| DK 7823 | Incorporation of pLAG 12 | This work |
| DK 7837 | Incorporation of pLAG 2 | This work |
| DK 3473 | pilR::Tn5 | 54 |
| DK 7808 | DK 7804; Tn5-132lac Ω4521 | This work |
| DK 7809 | Tn5-132lac Ω4521 | This work |
| DK 7824 | Tn5-132lac Ω4455 | This work |
| DK 7825 | Tn5-132lac Ω4469 | This work |
| DK 7826 | Tn5-132lac Ω4411 | This work |
| DK 7840 | DK 7823; Tn5-132lac Ω4521 | This work |
| DK 7842 | DK 7814; Tn5-132lac Ω4414 | This work |
| DK 7853 | asgA 476, Tn5-132 Ω4560 | This work |
| DK 7856 | DK 7837; Tn5-132lac Ω4414 | This work |
| DK 7857 | DK 7823; Tn5-132lac Ω4455 | This work |
| DK 7858 | DK 7823; Tn5-132lac Ω4469 | This work |
| DK 7859 | DK 7823; Tn5-132lac Ω4411 | This work |
| DK 7870 | Tn5-132lac::mbhA | This work |
| DK 7871 | DK 7823; Tn5-132lac::mbhA | This work |
| DK 5208 | Tn5-132::csgA | 27 |
| DK 5511 | Tn5-132lac Ω4414 | 50 |
| DZF 4619 | DZF1, Tn5-132::mbhA | D. Zusman |
| pBGS 18 | Kmr | 49 |
| pLAG 1 | Kmr (pBGS 18) Mxa210 at EcoRI site | This work |
| pLAG 2 | Kmr (pBGS 18) Mxa259 at EcoRI site | This work |
| pLAG 4 | Kmr (pBGS 18) Mxa264 at EcoRI site | This work |
| pLAG 5 | Kmr (pBGS 18) Mxa189 at EcoRI site | This work |
| pLAG 8 | Kmr (pBGS 18) Mxa191 at EcoRI site | This work |
| pLAG 9 | Kmr (pBGS 18) Mxa211 at EcoRI site | This work |
| pLAG 11 | Kmr (pBGS 18) Mxa213 at EcoRI site | This work |
| pLAG 12 | Kmr (pBGS 18) Mxa287 at EcoRI site | This work |
| pLAG 13 | Kmr (pBGS 18) Mxa296 at EcoRI site | This work |
Nutritional requirements.
Strains were inoculated onto A1 minimal agar with kanamycin, grown for 3 to 5 days, and finally transferred to a plate containing A1 agarose plus kanamycin. To assess auxotrophy, the resulting biomass of each disruption strain was compared with that of its nondisrupted parent growing on the same plate.
Sporulation assay.
The efficiency of sporulation was measured as described previously (27) with modification. After development on TPM agar (10-μl droplets of 5 × 109 cells/ml) for 3 days, the cells from five droplets were harvested by scraping them into microcentrifuge tubes containing 1 ml of TPM buffer. The contents of each tube were then treated in a cup horn sonicator (Tekmar) for 5 min at maximum power with ice-water cooling to disrupt fruiting bodies and disperse spores. Residual vegetative cells were killed subsequently by heating the tubes at 50°C for 2 h. After serial dilution and plating of the suspensions, the sporulation efficiency was measured as the number of colonies present relative to the number of cells initially deposited on the TPM agar. To determine whether development could be rescued in a developmentally defective strain by coculture, the activator disruption strains were mixed in a 1:1 ratio with other strains. For these rescue experiments, five 20-μl droplets of the mixed cell suspensions were deposited onto TPM agar and the sporulation efficiency was measured and compared with that of the wild type. The amounts of A- and C-factor produced were determined by the quantitative assessment of how much an asgA or a csgA mutant could be induced to sporulate when it was in coculture with the testing strain compared to the level of sporulation of the wild type under the same conditions.
Introduction of reporter gene fusions into gene disruption mutants.
Myxophage Mx4ts18ts27 hrm (Mx4) (5) was used to transduce Tn5-132lacZ promoter fusions of Ω4521 (from DK 6620) and mbhA (from DZF 4619) into each of the M. xanthus disruption strains. In some cases, the mbhA::Tn5-132lacZ fusion was transduced from DK 7870 into the disruption strains by Mx8clp2 (Mx8) (38). Some developmentally regulated Tn5-132lacZ fusions were transduced into DK 7823 with Mx4 (28). The Tn5-132lacZ fusion Ω4414 was transduced from DK 5511 by Mx8 into DK 7814 and DK 7837. Transductants were selected on CTT plus kanamycin plus oxytetracycline medium, and the structures of the transductants were confirmed by Southern blot analysis (28).
RESULTS
ς54 transcriptional activator sequences from M. xanthus.
With NtrC (NRI) protein as the prototype, the amino acid sequences of members of the family of known transcriptional activators for ς54 promoters have been divided into the three domains shown in Fig. 1. As mentioned above, the N-terminal domain is where the activator protein is often modified and the C-terminal helix-turn-helix domain of the protein is where binding to DNA is mediated. In collaboration with our laboratory, Kaufman and Nixon PCR amplified sequences corresponding to the consensus central ATP binding region from M. xanthus chromosomal DNA. Locations of the PCR primers used by Kaufman and Nixon are indicated in Fig. 1. From M. xanthus they obtained 14 unique NtrC-like fragments from a sample of 60 M13 clones (23). M. xanthus DNA appears to be several times more productive of unique fragments than the three other bacterial species surveyed: Rhizobium meliloti, Bacillus subtilis, and Synechococcus sp. strain PCC7002. The possible significance of this finding is taken up in Discussion.
One of the 14 PCR fragments identified by Kaufman and Nixon, Mxa15, corresponds to a gene, pilR, that had previously been recognized as a ς54 activator in M. xanthus (54). pilR and pilS make up a two-component system that regulates expression of the pilin structural gene pilA. The sequence identified as the promoter of pilA is ς54-like (55). Moreover, M. xanthus pilR is homologous to the pilR gene of Pseudomonas aeruginosa, which encodes a transcriptional activator for a ς54 promoter (15). This finding of an already recognized and sequenced ς54 activator protein within the pool of PCR fragments obtained from the Kaufman and Nixon amplification encouraged pursuit of the others.
Creation of activator-disruption mutant strains.
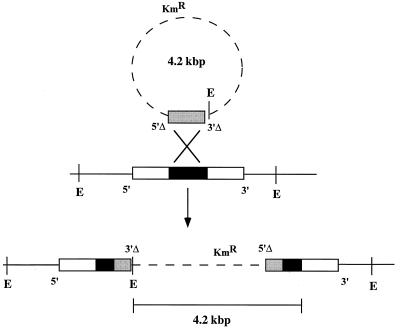
To investigate the physiological functions of putative activators of ς54-dependent genes, each of the PCR-amplified fragments was integrated by homologous recombination into M. xanthus, as diagrammed in Fig. 2. M. xanthus prefers to integrate chimeric plasmids through sequence homology (11). The 475-bp DNA fragments, since they were recovered after PCR amplification with primers that targeted the central domain of the protein, were expected to be completely internal to their corresponding genes (see the PCR primer sites in Fig. 1). As illustrated in Fig. 2, when homologous recombination occurs between an internal fragment and the complete copy that resides in the chromosome, the expected result is two incomplete copies of the gene, one of which, called 5′Δ, is missing sequences upstream of the amplified segment and the other of which, called 3′Δ, is missing sequences downstream. Vector DNA should separate the two incomplete copies from each other; gene function in this case should be totally lost because both copies are defective. The plasmid vector, which bears a gene that encodes kanamycin resistance, is not capable of autonomous replication in M. xanthus. Accordingly, the primary way of obtaining a kanamycin-resistant transformant is by integration, by using the homologous recombination just described, thus creating an insertional mutation that is expected to be null.
FIG. 2.
Gene disruption in M. xanthus by homologous recombination. A plasmid that carries an internal fragment of the ς54 activator target gene (shaded box) recognizes its homolog within the genome (black box). Because the PCR-amplified fragment is completely internal to the central domain (Fig. 1), that fragment can be considered to have the 5′ end of the activator gene (5′Δ) and the 3′ end of the gene (3′Δ) deleted. A single homologous crossover event should lead to a tandem duplication of the internal gene fragment and incorporation of the vector into the chromosomal gene, resulting in a knockout (or null) mutation in that gene. The vector bears the gene that encodes kanamycin resistance. E indicates sites of restriction enzyme cleavage used for Southern blot analysis.
A major technical challenge in generating such gene disruptions has to do with the relatively short length of the internal fragments employed, 475 bp, experience having suggested that fragments less than 700 bp long are incorporated by recombination only rarely into the M. xanthus chromosome. Kmr electroporants were rare, as expected; however, by doubling the ratio of recipient cells to DNA and allowing the electroporated cells to recover from their electric shock in an enriched medium (Casitone supplemented with yeast extract), Kmr electroporants were obtained in almost every experiment.
Kmr insertions in 3 of the 14 potential insertion strains, those of fragments Mxa15, Mxa189, and Mxa211, could not be obtained in several trials each. The reason may be purely technical, given that 475 bp is at the threshold for successful homologous recombination in M. xanthus. Another potential reason for lack of a recombinant is that these sequences may belong to essential genes which cannot be disrupted. The ς54 subunit is essential for viability in M. xanthus (25); therefore, one or more of the potential activators may also be essential.
Some kanamycin-resistant electroporants that did not have a gene disruption (false positives) arose. To discriminate the valid gene disruptions from false positives, DNAs from all Kmr electroporants were prepared and Southern blots were made to reveal their chromosomal compositions.
To detect the false positives, these Southern blots were also probed with a fragment from the kanamycin resistance gene of pBGS18 (the plasmid vector employed). A valid disruption strain, when it was probed with the 475-bp PCR fragment was, according to the procedure outlined in Fig. 2, expected to show two dark bands (one from each copy of the disrupted gene), and one of these bands was expected to hybridize to the kanamycin resistance gene from pBGS 18. One type of false-positive strain, when analyzed in this way, appears identical to the wild type in that it shows one strong band of hybridization to the 475-bp fragment probe but none to the kanamycin resistance gene. Presumably, kanamycin resistance in such false positives results from a chromosomal mutation rather than incorporation of the plasmid. Another type of false-positive strain, which arises from ectopic integration of the plasmid, does not have two new bands hybridizing to the Mxa probe, and pBGS18 does not hybridize to any of the bands to which Mxa hybridizes.
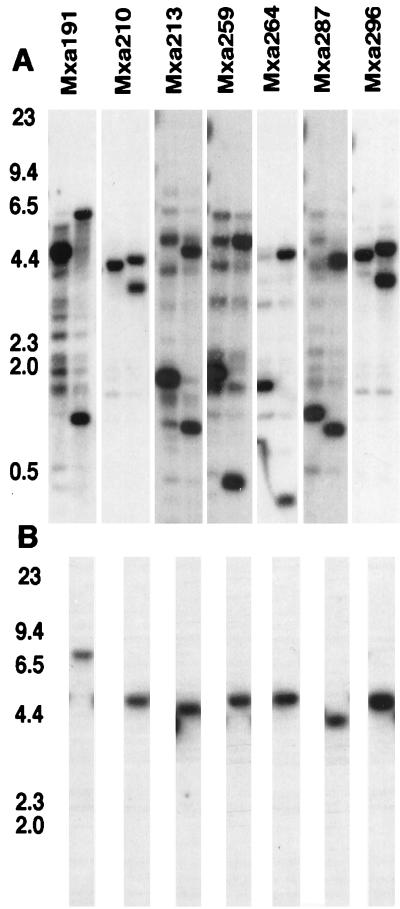
After setting the false positives aside, we made analytical Southern blots of the putative insertion strains, and these are shown in Fig. 3. Seven putative insertion strains showed two new bands in place of one band in the wild type, as was expected for the mechanism shown in Fig. 2. Hybridization of only one of the two new bands to the vector is demonstrated in Fig. 3B. DNA from each strain was also digested with each of several other restriction endonucleases (EcoRI, PstI, and SalI) before being electrophoresed and blotted. Although each enzyme gave a unique set of fragments, the general pattern, namely, two new bands in the putative insertion strain instead of one band as in the wild type, was the same for all four enzymes. For this reason, only the results of the SmaI digestions, which typify the whole set of seven strains, are presented in Fig. 3.
FIG. 3.
Southern blots of the insertion strains. Approximately 1 μg of DNA was loaded in each lane. DNA preparations were restricted with SmaI. SmaI sites are indicated with an E in the diagram of Fig. 2. Probes are listed across the top. The first lane of each blot contains wild-type DNA, and the second lane contains DNA from the insertion mutant. The locations of standard size markers (with sizes in kilobases) are indicated. Each blot has been probed with the PCR fragment used to create that mutant. (B) DNAs from the wild type and each of the mutants were also probed with a portion of the kanamycin resistance gene from pBGS18. Standard size markers of 23, 9.4, 6.5, 4.4, 2.3, and 2.0 kb are shown.
The Kaufman and Nixon technique takes advantage of the fact that the central domain of ς54 activator proteins is conserved. Since 14 of these domains may exist within the M. xanthus genome, Southern blots with any one of the 475-bp fragments to probe a restriction digest of the entire M. xanthus genome may show weak hybridization to numerous bands, most of them only partially homologous. That multiplicity of bands is evident in the Southern blots shown in Fig. 3. Some of the PCR fragments show many partial homologs, Mxa191, for example, whereas others show very few, like Mxa210. Since each Southern blot of Fig. 3 was probed with the particular PCR fragment (Mxa191, etc.) that had given rise to each putative insertion strain, then despite partial homology among the 14 domains, the presence of two new bands in place of one band in the wild-type DNA means that, in each case, insertion had occurred in the completely homologous chromosomal site. If one of the fragments had inserted into one of the partially homologous domains, the wild-type band would still have been present in the digest of the putative insertion strain.
In addition to the replacement pattern, size regularity was expected for insertions created by the mechanism shown in Fig. 2. In reference to the restriction sites shown in Fig. 2, no matter where the insertional recombination occurs within the region of homology, the combined molecular weights of the two new bands in the duplication strain that hybridize to the cloned segment should equal the molecular weight of the band obtained by probing wild-type DNA plus the molecular weight of the vector plasmid. The data of Table 2 reflect results of tests of this prediction in a tabulation of the sum of the sizes of the two major bands in the Southern blots of the insertion strains less the size of the one major band in the wild type. Within the accuracy of molecular weights determined by interpolation from the sizes of the DNA standards, the expected difference—the size of the 4.2-kb vector—was obtained for all of the strains.
TABLE 2.
Size test for insertion by homologous recombination
| Vector or strain | Size difference (kbp)a | Sizes (kbp) of new fragments in the Kmr strainsb |
|---|---|---|
| Vector | 4.2 | NA |
| Mxa191 | 3.6 | 6.7, 1.3 |
| Mxa210 | 3.8 | 4.5, 3.6 |
| Mxa213 | 4.0 | 4.5, 1.4 |
| Mxa259 | 4.4 | 5.3, 1.1 |
| Mxa264 | 3.9 | 4.7, 1.1 |
| Mxa287 | 4.2 | 4.3, 1.5 |
| Mxa296 | 4.1 | 5.0, 3.8 |
Difference = (sum of the sizes of the two new fragments in mutant DNA) − size of the fragment in wild-type DNA. Data were taken from the Southern blots shown in Fig. 3A.
The first-listed size in each entry is the size of the band which hybridizes to pBGS18. NA, not applicable.
According to the Southern blotting data, seven activator disruption strains were successfully constructed by plasmid insertion. In addition, one insertion that we had been unable to obtain by recombination was available in our strain collection as a well-characterized Tn5 insertion into pilR (54). This transposon insertion strain, DK 3473, was used as the Mxa15 activator disruption mutant, bringing the total number of insertion mutants to eight.
Mutant characterization.
The next phase of the investigation sought to find the physiological functions of the putative ς54 activators or, more properly, of their target genes. The strategic reason for placing physiology ahead of gene sequencing was to discover which putative activators, considering their number, have the most relevance to development. Accordingly, growth, swarming motility, sporulation, developmental gene expression, and capacity for fruiting body development were examined for each of the seven disruption strains. Temporarily, these strains are designated by the PCR fragment numbers assigned by Kaufman and Nixon (23), Mxa191 to Mxa296. Functional names will be assigned when the genes to which these fragments correspond have been isolated and characterized.
Growth, colony morphology, and swarming.
The disruption strains were maintained in growth media containing kanamycin to select against loss by excision of the integrated plasmid by homologous recombination between the two copies of the tandemly duplicated gene. For purposes of retaining the duplication, the short length of the duplication and, consequently, less frequent recombination work to advantage in decreasing the frequency of excision. Kanamycin-resistant derivatives of our laboratory strains generally grow more slowly in the presence of this antibiotic than in its absence. DK 1622 Kmr, for example, has a doubling time of 5.5 to 6.5 h in kanamycin medium, 1 or 2 h longer than DK 1622 in drug-free medium. Only one activator-disruption strain showed a significant difference in doubling time from DK 1622 Kmr and all other mutants; Mxa191 doubled after 12 h. All the strains, including Mxa191, were able to grow on the completely defined minimal medium A1 (3), despite their having been selected on the nutritionally rich CTTYE agar, which could have supported growth of many kinds of auxotrophs. The colony morphology of the disruption strains on CTT agar appeared normal (i.e., like that of the reference strain, DK 1622, referred to as the wild type), except for Mxa287, whose colonies were smaller than wild-type colonies, less rough in appearance, and less cohesive as cell masses when they were mechanically disturbed. Their colony morphology resembled that of an asgA mutant, which fails to release A-factor (31). However, the colonies of Mxa287 are not identical to those of an asg mutant, since Mxa287 forms yellow colonies like those of the wild type whereas in general asg mutants form tan colonies (36).
Myxococcus has two supplementary gene systems that control its swarming motility, the A (adventurous) and the S (social) systems (16, 17). Null pilR mutants, which lack the activator denoted Mxa15, lack S motility. They fail to make the pili known to be necessary for the social pattern of swarming, since pilR is the transcriptional activator for the major pilin subunit, pilA (54). The loss of S motility decreases swarming and hence changes the morphology of the colony edge.
Fruiting body development.
Three of the eight disruption mutant strains showed normal fruiting body development during 3 days of incubation on TPM agar (Table 3). However, five were clearly abnormal under laboratory conditions. One of these strains, Mxa296, developed normally on TPM agar but failed to develop in submerged culture. Submerged culture was carried out in the wells of plastic culture dishes, and this type of culture requires that the cells can adhere to and create a biofilm on the polystyrene floor of a well in order to develop fruiting bodies (30). Although the basis for the surface specificity of agar versus that of polystyrene is unknown, the target activator in Mxa296 is clearly necessary for the ability of DK 1622 to initiate development under these particular conditions.
TABLE 3.
Aggregate morphology
| Strain | Terminal morphology and/or nature of aggregationa |
|---|---|
| DK 1622 | Dense, ovoid, uniformly darkened by day 3 |
| Mxa15 | Aggregation delayed 1 to 3 days |
| Mxa191 | Same as for DK 1622 |
| Mxa210 | Same as for DK 1622 |
| Mxa213 | Larger aggregates and irregular darkening |
| Mxa259 | Small, darkened (except for center of aggregate) |
| Mxa264 | Same as for DK 1622 |
| Mxa287 | Flat mat of cells; no darkening |
| Mxa296 | No aggregation in submerged culture; same as for DK 1622 on TPM agar |
Standard conditions were 3 days on TPM agar.
Mxa15 showed delayed development; as pointed out above, this strain has a social motility defect associated with its failure to assemble pili. Since motility in general is necessary for normal fruiting body development (29), and since all S− mutants that fail to assemble pili have been shown to exhibit developmental delay (52), the developmental phenotype of Mxa15 is probably secondary to its motility defect.
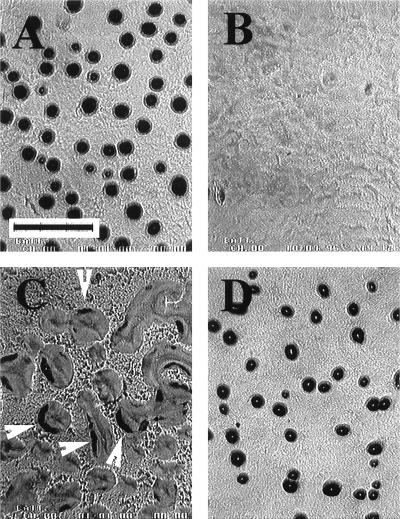
Finally, three strains, Mxa287, Mxa213, and Mxa259, were completely blocked in fruiting body development. All three had normal vegetative swarming patterns, which implies that their failure to completely aggregate is not the consequence of a defect in swarming motility. The three arrested at different stages of aggregation. As illustrated in Fig. 4A, the wild type (DK 1622) formed highly condensed, fully darkened, hemispherical fruiting bodies by 24 h after the initiation of development by starvation. Mxa287 had the earliest apparent point of arrest; it stopped at a stage after formation of linear or branched linear plateau-like elevations that rest on top of an otherwise continuous mat of cells (Fig. 4B). These elevations are low, and it is often difficult to define precise boundaries that separate one elevation from another. In comparing Fig. 4B with 4A, it is evident that these aggregates differ from those of later aggregation stages of normal development in their elevation above the cell mat (lower), distribution (much less regular), and the fraction of the total culture surface they occupy (larger).
FIG. 4.
Development of strains on TPM agar at 24 h. (A) Wild type; (B) Mxa287; (C) Mxa213; (D) Mxa259. Arrowheads indicate incompletely closed structures. The scale bar in panel A measures 0.15 cm, and the scale is the same for all frames.
The second of the three strains whose fruiting body development was blocked, Mxa213, arrested at a more advanced stage in terms of aggregate size, implying the presence of more cells per aggregate (Fig. 4C). Even after 24 h, Mxa213 formed aggregates that, although discrete and generally rounded, were much less condensed than those of the wild type at the same time. The Mxa213 aggregates were loose and only partially darkened, and many had a distorted horseshoe shape, as if they had started to form a loop or ring but failed to close it. These structures had curvy forms which varied widely in size and shape. They had only begun to darken; some were darkened along their peripheries in several discrete segments (Fig. 4C). Others were dark everywhere except at one or both ends.
The third strain, Mxa259, apparently arrested at a stage still later than that of either Mxa287 or Mxa213. Mxa259 cells at their arrest stage (shown in Fig. 4D) were highly condensed, darkened, and closed, compared to those of Mxa213. These aggregates typically had a light center (Fig. 4D), and they appeared to be smaller than wild-type aggregates (compare Fig. 4A and D). On TPM agar their morphology remained the same from day 1 to day 3 or later; they were observed through 5 days. Occasionally, Mxa259 also delayed early aggregation, remaining in a premound state for about a day before proceeding to the condensed stage shown in Fig. 4D.
Sporulation.
The capacity of starvation-induced cultures of each of the three aggregation-defective mutants to sporulate was assessed. Sporulation normally follows aggregation and occurs within the aggregate. The disruption mutants Mxa287, Mxa213, and Mxa259 formed no viable heat- and sonication-resistant spores even after 3 days of development on TPM agar (Table 4). Mxa259 forms darkened aggregates but no viable spores, showing that the darkening of fruiting bodies need not indicate the presence of mature spores. These nonsporulating mutants could also not be induced to form spores by coculturing them with an equal number of wild-type cells. The sporulation level of Mxa287, when it was cocultured with the wild type, remained below 0.1% of wild-type levels (Table 5). It did increase somewhat, but the increase in spore number for Mxa287 was about 3 orders of magnitude less than the rescue seen with signaling mutants such as asg or csg (results shown in Table 5). The failure of wild-type cells to rescue the sporulation defect of Mxa287 and of the other aggregation mutants (data not shown) shows that their primary defects are cell autonomous, not the production of an extracellular factor(s) released by wild-type cells that might be required for sporulation, such as A-factor, C-factor (13), or E-factor (7).
TABLE 4.
Sporulation of aggregation-defective mutants
| Strain | % of cells showing sporulation |
|---|---|
| DK 1622 | 100 |
| Mxa287 | <10−4 (18)a |
| Mxa213 | <10−4 (9)a |
| Mxa259 | <10−4 (9)a |
| DK5208 (csgA) | <10−4 (9)a |
| DK7853 (asgA) | <10−4 (12)a |
No viable spores were detected in this assay; consequently, the frequency is represented by the lower limit of the assay. In parentheses, the number of individual assays performed on the strain is indicated.
TABLE 5.
Sporulation in coculture as a measure of A- or C-factor production
| Strain mixture
|
Sporulation frequency of each strain in the mixturea
|
||
|---|---|---|---|
| Test strain | Coculture strain | Test strain (mean % ± % SD) | Coculture strain (%) |
| Mxa287 | DK 1622 | 0.03 ± 0.02 | 100 |
| DK 5208 (csgA) | DK 1622 | 40 ± 27 | 100 |
| DK 5208 | Mxa287 | 12 ± 8 | <10−4 |
| DK 7853 (asgA) | DK 1622 | 17 ± 11 | 100 |
| DK 7853 | Mxa287 | 25 ± 24 | <10−4 |
Frequency was normalized to that of the wild-type (DK 1622) control in each experiment.
A- and C-factor production.
The aggregation-defective mutant Mxa287 was also specifically tested for its capacity to produce A- and C-factors, because failure to produce either of these factors is known to lead to aggregation arrest and failure to sporulate (32, 48). The ambient levels of A-factor and C-factor were measured by mixing the aggregation mutant with either an A-signal-deficient (asgA) or a C-signal deficient (csgA) mutant, coculturing the mixtures under starvation conditions to induce development, and finally counting the number of spores formed by the asgA and csgA indicator strains, differentially marked by drug resistance. It was evident that the Mxa287 mutant strain made both the A- and C-factors since it induced the asgA and csgA signal-defective mutants to sporulate (Table 5). Mxa287 made roughly wild-type levels of both factors, as judged by the number of viable spores formed. Thus, Mxa287 produces A- and C-factors but fails to respond to the extracellular signals that are contributed by the wild-type during coculture. For these reasons, and others stated below, the putative activator targeted in mutant Mxa287 appears to be part of a signal reception rather than a signal production pathway.
Expression of developmentally regulated reporters.
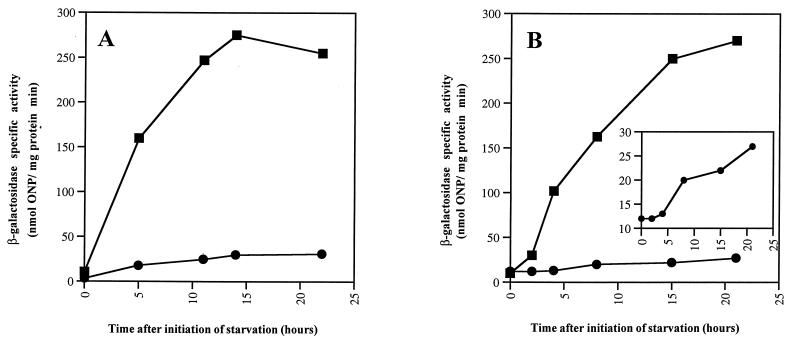
Tn5lacZ transcriptional fusions have been constructed as reporters for two particular ς54-transcribed genes, mbhA and Ω4521 (24, 25, 42). mbhA encodes a myxobacterial hemagglutinin. While these genes are developmentally expressed, neither is essential for development. If the activator proteins sampled in the current collection of disruption mutants include the specific activators for those particular target genes, then those reporters should not be expressed. The mbhA and Ω4521 operon fusions were transduced into all the activator mutants, and the specific activity of β-galactosidase produced from the mbhA or Ω4521 reporter was measured after development had been induced. Activator mutant Mxa287 was unable to express the A-signal-dependent Ω4521 reporter, which is normally expressed at about 2 h into development, the normal time of A-signaling (28). The Mxa296 strain did not express that fusion when it developed in submerged culture but did express Ω4521 when it developed on TPM agar (Fig. 5A). This expression pattern of β-galactosidase from Tn5lacZ Ω4521 parallels the ability of Mxa296 to aggregate on agar and its lack of ability to aggregate in submerged culture. However, Mxa296 was able to express Ω4521 in buffered suspension with either TPM buffer, which is used for TPM agar development, or MC7 buffer, which is used for submerged-culture development. These results also indicate that Mxa296 has a preaggregation defect that is expressed before the strain has developed for 2 to 3 h in submerged culture, the normal time for Ω4521 expression. This defect may be in the recognition of polystyrene as a proper surface for development; alternatively, polystyrene may be inhibitory to development in this mutant.
FIG. 5.
Expression of β-galactosidase from the Tn5lac Ω4521 reporter. (A) Strain Mxa296. The squares indicate β-galactosidase specific activity from cells developed on TPM agar, and the circles indicate β-galactosidase specific activity from cells in submerged culture. (B) Strain Mxa287. The squares reflect β-galactosidase expression in a wild-type background, and the circles show expression in a Mxa287 mutant background. The inset shows Ω4521 expression in the Mxa287 mutant background plotted on a more sensitive scale. ONP, o-nitrophenyl.
Mxa287 failed, by contrast, to express β-galactosidase from the Ω4521 reporter either on TPM agar or in submerged culture; submerged-culture activity is shown in Fig. 5B. An expanded scale (inset of Fig. 5B) does reveal some Ω4521 expression in the Mxa287 mutant background, which reveals the time course typical of asgA mutants (32, 47). In a series of experiments, the residual level of expression ranged from 5 to 10% of that of the wild type. This reporter is generally recognized to be a reliable indicator of the A-signal response pathway (2, 28, 32). A 5 to 10% residual level of Ω4521 expression is also characteristic of asg mutants (32, 47). Thus, the residual level and its apparent time course in the absence of a functional Mxa287 activator imply abnormal A-signaling. Mxa287 insertion strains did express the A-signal-independent transcriptional fusions Ω4411, Ω4469, and Ω4455 at levels within 50% of those in wild-type cells (Fig. 6), which is consistent with an A-signaling defect.
FIG. 6.
Expression of developmentally regulated Tn5lac reporter fusions in Mxa287. The squares represent β-galactosidase specific activity in a wild-type background, and the circles represent β-galactosidase specific activity of the fusions in a Mxa287 mutant background. ONP, o-nitrophenyl.
The mbhA reporter was expressed at roughly normal levels in all the activator disruption strains examined and with roughly wild-type kinetics and timing. One data set for the expression of the mbhA-lacZ fusion in the Mxa287 genetic background is shown in Fig. 6.
Insertions Mxa213 and Mxa259 were both defective in expression of the Ω4414 fusion, which is C-signaling dependent (50). Both expressed the fusion at levels lower than that of the wild type, with Mxa213 at about 8% of the wild-type level and Mxa259 at about 30% of the wild-type level.
DISCUSSION
Endospore formation in B. subtilis has been shown to be regulated by a cascade of different sigma factors, each of which is responsible for the expression of a particular set of genes in the differentiation of the spore from the mother cell (14). At least five sigma factors of the ς70 family have been found in M. xanthus, the deletion of some of which leads to developmental defects (18); however, none of these sigma factors, when they are deleted, cause the extreme developmental defects produced by deletion of ςH, ςF, ςE, ςG, or ςK (all members of the ς70 family), which terminates Bacillus spore formation (14). There is no evidence that these M. xanthus sigma factors are regulated as a cascade.
Sigma factors of the ς54 family are both structurally and functionally different from members of the larger ς70 family (39). ς54 promoters are often used for growth on certain alternate carbon sources among different Pseudomonas spp. (39), on alternate nitrogen sources in some enteric species (35, 41), or for nif genes in some bacteria capable of nitrogen fixation (39, 43). ς54 is cell cycle regulated, but is not vital, in the swarmer cells of Caulobacter crescentus, where expression of several genes of flagellum biosynthesis and assembly are timed by the cell cycle (4).
In contrast to its nonessential role in other bacteria, ς54’s role in the growth of M. xanthus has been found to be essential (25). Although the majority of developmentally regulated promoters recognized to date appear to be recognized by a member of the ς70 family (6), at least three promoters, mbha, pilA (55), and Ω4521, have ς54 hallmarks. Constrained by the requirement of ς54 for growth, to investigate the extent of ς54 use during development, we have created insertion mutations in genes expected to encode ς54 activator proteins. The strategy of Kaufman and Nixon (23) appears to identify multiple ς54 activator proteins in M. xanthus. Starting their search with Rhizobium, they revealed genes for possible activators in Bacillus as well as Myxococcus but none in Synechococcus. With an internal fragment of a presumptive activator gene, the chromosomal gene can be mutated by plasmid insertion. In this way it was possible for us to integrate 7 of 14 Mxa fragments. One other fragment, Mxa15, was represented by a Tn5 insertion in pilR, bringing the number of the 14 fragments that are available for analysis to 8. The remainder of this discussion examines the range of functions suggested in this initial sample of ς54 activators.
The Mxa296 mutant was able to develop fruiting bodies on TPM agar, where it showed a wild-type morphology, but it was unable to develop in submerged culture. Its activator-related defect is early in development, preceding Ω4521 expression at 2 h since the Ω4521 reporter is also not expressed in the insertion mutant in submerged culture but is on agar. Both starvation and A-signaling are required for Ω4521 expression (47). It is therefore possible that either the recognition of starvation or the response to A-signal depends on the Mxa296 activator. However, both starvation recognition and A-signaling can occur in suspension (33) and therefore are not dependent on attachment to a surface. Since the Mxa296 insertion mutant could express the Ω4521 reporter in suspension, it is possible that the polystyrene vessel used for submerged culture inhibited development of this mutant. Whether wild-type cells can adapt to a polystyrene surface while the Mxa296 activator mutant cannot is a possibility that will require further investigation.
One insertion mutant is affected in social gliding motility by the inactivation of pilR, and this mutant has developmental defects as a consequence. ς54 activators often function as response regulators for two-component regulatory systems (35, 46). PilR activates the transcription of pilA, which encodes the major pilin subunit of the polar type IV pili. pilS, which lies immediately upstream of pilR, is homologous to the sensor element of a pilS-pilR two-component system (54). However, pili are found on M. xanthus cells under all laboratory growth conditions so far tested (53), and so the environmental signal that activates the PilR response via PilS has yet to show itself physiologically. The pilR gene could not be disrupted by the insertion of Mxa15 into its target gene in this study. The failure to obtain the Mxa15 insertion mutant is most likely a consequence of the short length of homology of the PCR fragment, since a Tn5 insertion mutant of pilR is viable.
Three other mutants completely arrested aggregation but at distinctly different stages. With the timing of development of the wild type as the reference, Mxa287 halted early (2 to 4 hours), Mxa213 stopped midway in aggregation at about 5 h, and Mxa259 apparently halted near the end of aggregation.
The Mxa287 mutant arrests as an almost featureless mat of cells that resembles the morphology of mutants that do not produce A-factor, such as asgA mutants (31). The Mxa287 mutant also has a sporulation defect which is comparable to that of an asgA mutant. The mutant fails to express the A-signal-dependent Ω4521-lacZ fusion, while it does express the starvation-responsive but A-signal-independent reporters Ω4469, Ω4455, and Ω4411 (Fig. 6). These data indicate that Mxa287 fails to carry out A-signaling. Since, on the one hand, Mxa287 produces wild-type levels of A-factor as judged by its capacity to rescue the sporulation defect of an asgA mutant and, on the other hand, is not rescued by coculture with wild-type cells, the data point to a fault in the A-factor response pathway, as opposed to one in A-factor production.
The failure of this strain to express β-galactosidase from the Ω4521-lacZ promoter fusion, which is known to be driven by a ς54 promoter (24) and possibly to have a binding site for such an activator protein (12), may indicate that the activator lacking in Mxa287 directly controls Ω4521 transcription. As stated above, starvation is a prerequisite for the activation of Ω4521 (47), but Mxa287 is not expected to be a constituent of the starvation recognition circuit since the mutant strain expresses the starvation-dependent, but A-independent, fusion Ω4469 at high levels (Fig. 6). Mxa287 also expresses the developmentally regulated gene fusions Ω4455 and Ω4411 (Fig. 6). These fusions are also considered A-signaling independent (2, 27); the pattern of reporter activity supports the specificity of the Mxa287 activator for A-signaling. In a recent report, Yang and Kaplan (56) cite unpublished experiments which are consistent with Mxa287 being an activator of Ω4521.
That the Ω4521 gene is not essential for development while the Mxa287 mutant fails to develop implies the existence of additional genes that are dependent on the Mxa287 putative activator, at least one of which is necessary for fruiting body development. The identities of these genes remain to be elucidated. The Mxa287 mutant transcribes the mbhA-lacZ promoter fusion at roughly normal levels (Fig. 6), suggesting a different activator for mbhA. It has been reported that asg mutants do not accumulate MbhA protein during development as judged by hemagglutination assays (36), suggesting that mbhA is A-signal dependent. In sum, Mxa287 appears to activate a particular set of A-signal-dependent genes, at least one of which is essential for development.
The mutants Mxa213 and Mxa259 arrest aggregation at stages later than that of the Mxa287 mutant (Fig. 4). Neither of them can sporulate, and neither can be induced to sporulate by exposure to wild-type cells, indicating that their defects are also not in the production of extracellular signals. The failure of Mxa213 and Mxa259 to express the Ω4414-lacZ fusion, which is known to be C-signal dependent (28, 50), may indicate defects in their responses to C-signal.
The activator insertion mutants are expected to be null, because they would produce only a 5′ deletion and a 3′ deletion version of the activator protein. However, the possibility that the 5′ deletion allele retains activity might be considered. Several, but not all, ς54 activator proteins have been found to be active and to become independent of sensory input when they lack only their N-terminal regulatory regions (8, 10, 37). Depending on the extent of 5′ deletion, recombination between the plasmid clone and the M. xanthus activator genes (Fig. 2) might generate a functional central ATP binding domain combined with a functional C-terminal DNA binding domain that would constitute an activator able to function without regulatory control. Regions within the central domain are needed for several functions of the activator protein: cooperative binding of the activator to the DNA, ATP binding, ATP hydrolysis, and binding of the protein to the RNA polymerase holoenzyme (51). The published reports of active 5′ deletion proteins are all of engineered recombinant activator proteins for which care has been taken to retain their entire central domains with all of their functions intact. The PCR primers used by Kaufman and Nixon lie not at the borders of the central domain but fully within it, so it is unlikely that the 5′ deletion recombinant allele retains activity. Nevertheless, as each activator protein is examined, this possibility should be kept in mind.
Fourteen different activator protein sequences were found in the first screening of M. xanthus (23). Eight of these were successfully inactivated in this study. The PCR primers were not designed for the high G+C content (70%) of M. xanthus DNA, and an activator was not found for mbhA, which is thought to have a ς54 promoter (42). It is possible that this first PCR screen has not saturated the genome. In conclusion, the multiplicity and phenotypic range of developmentally defective ς54 activator mutants clearly indicate that these presumed activators, and by implication ς54 itself, play multiple roles in the regulation of development of M. xanthus.
ACKNOWLEDGMENTS
We thank Tracy Nixon and Ilene Kaufman for the generation of the PCR products used in this study. Additionally, we thank the members of the Kaiser laboratory and Ingrid Keseler for insightful discussions and advice. We thank F. J. Murillo and Sydney Kustu for reading the manuscript.
This investigation was supported by U.S. Public Health Service grant GM 23441 to D.K. from the National Institute of General Medical Sciences and postdoctoral fellowship GM 16344 to L.G. from the National Institute of General Medical Sciences, National Institutes of Health.
REFERENCES
- 1.Avery L, Kaiser D. In situ transposon replacement and isolation of a spontaneous tandem genetic duplication. Mol Gen Genet. 1983;191:99–109. doi: 10.1007/BF00330896. [DOI] [PubMed] [Google Scholar]
- 2.Bowden M G, Kaplan H B. The Myxococcus xanthus developmentally expressed asgB-dependent genes can be targets of the A signal-generating or A signal-responding pathway. J Bacteriol. 1996;178:6628–6631. doi: 10.1128/jb.178.22.6628-6631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bretscher A P, Kaiser D. Nutrition of Myxococcus xanthus, a fruiting myxobacterium. J Bacteriol. 1978;133:763–768. doi: 10.1128/jb.133.2.763-768.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brun Y V, Shapiro L. A temporally controlled ς-factor is required for polar morphogenesis and normal cell division in Caulobacter. Genes Dev. 1992;6:2395–2408. doi: 10.1101/gad.6.12a.2395. [DOI] [PubMed] [Google Scholar]
- 5.Campos J M, Geisselsoder J, Zusman D R. Isolation of bacteriophage Mx4, a generalized transducing phage for Myxococcus xanthus. J Mol Biol. 1978;119:167–178. doi: 10.1016/0022-2836(78)90431-x. [DOI] [PubMed] [Google Scholar]
- 6.Downard J, Kroos L. Transcriptional regulation of developmental gene expression in Myxococcus xanthus. In: Dworkin M, Kaiser D, editors. Myxobacteria II. Washington, D.C: American Society for Microbiology; 1993. pp. 183–200. [Google Scholar]
- 7.Downard J S, Ramaswamy S V, Kil K-S. Identification of esg, a genetic locus involved in cell-cell signaling during Myxococcus development. J Bacteriol. 1993;175:7762–7770. doi: 10.1128/jb.175.24.7762-7770.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drummond M H, Contreras A, Mitchenall L A. The function of isolated domains and chimaeric proteins constructed from the transcriptional activators NifA and NtrC of Klebsiella pneumoniae. Mol Microbiol. 1990;4:29–37. doi: 10.1111/j.1365-2958.1990.tb02012.x. [DOI] [PubMed] [Google Scholar]
- 9.Dworkin M. Recent advances in the social and developmental biology of the myxobacteria. Microbiol Rev. 1996;60:70–102. doi: 10.1128/mr.60.1.70-102.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez S, de Lorenzo V, Perez-Martin J. Activation of the transcriptional regulator XylR of Pseudomonas putida by release of repression between functional domains. Mol Microbiol. 1995;16:205–213. doi: 10.1111/j.1365-2958.1995.tb02293.x. [DOI] [PubMed] [Google Scholar]
- 11.Gill R E, Shimkets L J. Genetic approaches for analysis of myxobacterial behavior. In: Dworkin M, Kaiser D, editors. Myxobacteria II. Washington, D.C: American Society for Microbiology; 1993. pp. 129–155. [Google Scholar]
- 12.Gulati P, Xu D, Kaplan H. Identification of the minimum regulatory region of a Myxococcus xanthus A-signal-dedendent developmental gene. J Bacteriol. 1995;177:4645–4651. doi: 10.1128/jb.177.16.4645-4651.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagen D C, Bretscher A P, Kaiser D. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev Biol. 1978;64:284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- 14.Haldenwang W G. The sigma factors of Bacillus subtilis. Microbiol Rev. 1995;59:1–30. doi: 10.1128/mr.59.1.1-30.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hobbs M, Collie E S, Free P D, Livingston S P, Mattick J S. PilS and PilR, a two-component transcriptional regulatory system controlling expression of type 4 fimbriae in Pseudomonas aeruginosa. Mol Microbiol. 1993;7:669–682. doi: 10.1111/j.1365-2958.1993.tb01158.x. [DOI] [PubMed] [Google Scholar]
- 16.Hodgkin J, Kaiser D. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): genes controlling movement of single cells. Mol Gen Genet. 1979;171:167–176. [Google Scholar]
- 17.Hodgkin J, Kaiser D. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): two gene systems control movement. Mol Gen Genet. 1979;171:177–191. [Google Scholar]
- 18.Inouye S, Inouye M. Development-specific gene expression: protein serine/threonine kinases and sigma factors. In: Dworkin M, Kaiser D, editors. Myxobacteria II. Washington, D.C: American Society for Microbiology; 1993. pp. 201–212. [Google Scholar]
- 19.Kaiser D. Bacteria also vote. Science. 1996;272:1598–1599. doi: 10.1126/science.272.5268.1598. [DOI] [PubMed] [Google Scholar]
- 20.Kaiser D. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc Natl Acad Sci USA. 1979;76:5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaplan H B, Kuspa A, Kaiser D. Suppressors that permit A-signal-independent developmental gene expression in Myxococcus xanthus. J Bacteriol. 1991;173:1460–1470. doi: 10.1128/jb.173.4.1460-1470.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kashefi K, Hartzell P L. Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF− defect. Mol Microbiol. 1995;1995:483–494. doi: 10.1111/j.1365-2958.1995.tb02262.x. [DOI] [PubMed] [Google Scholar]
- 23.Kaufman R I, Nixon B T. Use of PCR to isolate genes encoding ς54-dependent activators from diverse bacteria. J Bacteriol. 1996;178:3967–3970. doi: 10.1128/jb.178.13.3967-3970.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keseler I M, Kaiser D. An early A-signal-dependent gene in Myxococcux xanthus has a ς54-like promoter. J Bacteriol. 1995;177:4638–4644. doi: 10.1128/jb.177.16.4638-4644.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keseler I M, Kaiser D. Sigma-54, a vital protein for Myxococcus xanthus. Proc Natl Acad Sci USA. 1997;94:1979–1984. doi: 10.1073/pnas.94.5.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim S K, Kaiser D. C-factor: a cell-cell signaling protein required for fruiting body morphogenesis of M. xanthus. Cell. 1990;61:19–26. doi: 10.1016/0092-8674(90)90211-v. [DOI] [PubMed] [Google Scholar]
- 27.Kroos L, Kaiser D. Expression of many developmentally regulated genes in Myxococcus depends on a sequence of cell interactions. Genes Dev. 1987;1:840–854. doi: 10.1101/gad.1.8.840. [DOI] [PubMed] [Google Scholar]
- 28.Kroos L, Kuspa A, Kaiser D. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev Biol. 1986;117:252–266. doi: 10.1016/0012-1606(86)90368-4. [DOI] [PubMed] [Google Scholar]
- 29.Kroos L P, Hartzell P, Stephens K, Kaiser D. A link between cell movement and gene expression argues that motility is required for cell-cell signalling during fruiting-body development. Genes Dev. 1988;2:1677–1685. doi: 10.1101/gad.2.12a.1677. [DOI] [PubMed] [Google Scholar]
- 30.Kuner J M, Kaiser D. Fruiting body morphogenesis in submerged cultures of Myxococcus xanthus. J Bacteriol. 1982;151:458–461. doi: 10.1128/jb.151.1.458-461.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuspa A, Kaiser D. Genes required for developmental signalling in Myxococcus xanthus: three asg loci. J Bacteriol. 1989;171:2762–2772. doi: 10.1128/jb.171.5.2762-2772.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuspa A, Kroos L, Kaiser D. Intercellular signalling is required for developmental gene expression in Myxococcus xanthus. Dev Biol. 1986;117:267–276. doi: 10.1016/0012-1606(86)90369-6. [DOI] [PubMed] [Google Scholar]
- 33.Kuspa A, Plamann L, Kaiser D. Identification of heat-stable A-factor from Myxococcus xanthus. J Bacteriol. 1992;174:3319–3326. doi: 10.1128/jb.174.10.3319-3326.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuspa A, Plamann L, Kaiser D. A-signalling and the cell density requirement for Myxococcus xanthus development. J Bacteriol. 1992;174:7360–7369. doi: 10.1128/jb.174.22.7360-7369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kustu S, Santero E, Keener J, Popham D, Weiss D. Expression of ς54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol Rev. 1989;53:367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.LaRossa R, Kuner J, Hagen D, Manoil C, Kaiser D. Developmental cell interactions of Myxococcus xanthus: analysis of mutants. J Bacteriol. 1983;153:1394–1404. doi: 10.1128/jb.153.3.1394-1404.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee J H, Scholl D, Nixon B T, Hoover T R. Constitutive ATP hydrolysis and transcription activation by a stable, truncated form of Rhizobium meliloti DctD, a ς54-dependent transcriptional activator. J Biol Chem. 1994;269:20401–20409. [PubMed] [Google Scholar]
- 38.Martin S, Sodergren E, Matsuda T, Kaiser D. Systematic isolation of transducing phages for Myxococcus xanthus. Virology. 1978;88:44–53. doi: 10.1016/0042-6822(78)90108-3. [DOI] [PubMed] [Google Scholar]
- 39.Merrick M J. In a class of its own—the RNA polymerase sigma factor ς54 (ςN) Mol Microbiol. 1993;10:903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 40.Morett E, Segovia L. The ς54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ninfa A J, Atkinson M R, Kamberov E S, Feng J, Ninfa E G. Control of nitrogen assimilation by the NRI-NRII two-component system of enteric bacteria. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: American Society for Microbiology; 1995. pp. 67–88. [Google Scholar]
- 42.Romeo J M, Zusman D R. Transcription of the myxobacterial hemagglutinin gene is mediated by the ς54-like promoter and a cis-acting upstream regulatory region of DNA. J Bacteriol. 1991;173:2969–2976. doi: 10.1128/jb.173.9.2969-2976.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ronson C W, Nixon B T, Albright L M, Ausubel F M. Rhizobium meliloti ntrA (rpoN) gene is required for diverse metabolic functions. J Bacteriol. 1987;169:2424–2430. doi: 10.1128/jb.169.6.2424-2431.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 45.Shimkets L J, Rafiee H. CsgA, an extracellular protein essential for Myxococcus xanthus development. J Bacteriol. 1990;172:5299–5306. doi: 10.1128/jb.172.9.5299-5306.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shingler V. Signal sensing by ς54-dependent regulators: derepression as a control mechanism. Mol Microbiol. 1996;19:409–416. doi: 10.1046/j.1365-2958.1996.388920.x. [DOI] [PubMed] [Google Scholar]
- 47.Singer M, Kaiser D. Ectopic production of guanosine penta- and tetraphosphate can initiate early developmental gene expression in Myxococcus xanthus. Genes Dev. 1995;9:1633–1644. doi: 10.1101/gad.9.13.1633. [DOI] [PubMed] [Google Scholar]
- 48.Sogaard-Andersen L, Slack F J, Kimsey H, Kaiser D. Intercellular C-signalling in Myxococcus xanthus involves a branched signal transduction pathway. Genes Dev. 1996;10:740–754. doi: 10.1101/gad.10.6.740. [DOI] [PubMed] [Google Scholar]
- 49.Spratt B G, Hedge P J, te Heesen S, Edelman A, Broome-Smith J K. Kanamycin-resistant vectors that are analogues of plasmids pUC 8, pUC 9, pEMBL 8, and pEMBL 9. Gene. 1986;41:337–342. doi: 10.1016/0378-1119(86)90117-4. [DOI] [PubMed] [Google Scholar]
- 50.Thony-Meyer L, Kaiser D. devRS, an autoregulated and essential genetic locus for fruiting body development in Myxococcus xanthus. J Bacteriol. 1993;175:7450–7462. doi: 10.1128/jb.175.22.7450-7462.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y-K, Hoover T R. Alterations within the activation domain of the ς54-dependent activator DctD that prevent transcriptional activation. J Bacteriol. 1997;179:5812–5819. doi: 10.1128/jb.179.18.5812-5819.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wee S A. Pilus null mutants show abnormal fruiting body morphogenesis in Myxococcus xanthus. Senior honors thesis. Stanford, Calif: Stanford University; 1996. [Google Scholar]
- 53.Wu, S. S. 1998. Personal communication.
- 54.Wu S S, Kaiser D. Genetic and functional evidence that Type IV pili are required for social gliding motility in Myxococcus xanthus. Mol Microbiol. 1995;18:547–558. doi: 10.1111/j.1365-2958.1995.mmi_18030547.x. [DOI] [PubMed] [Google Scholar]
- 55.Wu S S, Kaiser D. The regulation of expression of the pilA gene in Myxococcus xanthus. J Bacteriol. 1997;179:7748–7758. doi: 10.1128/jb.179.24.7748-7758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang C, Kaplan H B. Myxococcus xanthus sasS encodes a sensor histidine kinase required for early developmental gene expression. J Bacteriol. 1997;179:7759–7767. doi: 10.1128/jb.179.24.7759-7767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]