Graphical abstract
Keywords: Innate immunity, Adaptive immunity, ScRNA-Seq, Across species, Evolution
Highlights
-
•
Macrophages develop evolutionary versatility in expressed genes.
-
•
B cell immunity is relatively conserved among vertebrate species.
-
•
Compensatory TCR cascade components are utilized by different species.
-
•
Mouse species shows the highest similarity in immune transcriptomes to human among multiple vertebrates.
Abstract
Introduction
Both innate and adaptive immune system undergo evolution from low to high vertebrates. Due to the limitation of conventional approaches in identifying broader spectrum of immune cells and molecules from various vertebrates, it remains unclear how immune molecules evolve among vertebrates.
Objectives
Here, we utilized carry out comparative transcriptome analysis in various immune cells across seven vertebrate species.
Methods
Single-cell RNA sequencing (scRNA-seq).
Results
We uncovered both conserved and species-specific profiling of gene expression in innate and adaptive immunity. Macrophages exhibited highly-diversified genes and developed sophisticated molecular signaling networks along with evolution, indicating effective and versatile functions in higher species. In contrast, B cells conservatively evolved with less differentially-expressed genes in analyzed species. Interestingly, T cells represented a dominant immune cell populations in all species and unique T cell populations were identified in zebrafish and pig. We also revealed compensatory TCR cascade components utilized by different species. Inter-species comparison of core gene programs demonstrated mouse species has the highest similarity in immune transcriptomes to human.
Conclusions
Therefore, our comparative study reveals gene transcription characteristics across multiple vertebrate species during the evolution of immune system, providing insights for species-specific immunity as well as the translation of animal studies to human physiology and disease.
Introduction
Over several hundred million years of evolution, the immune system of vertebrates has formed a functional cooperation between innate and adaptive immune cells to orchestrate effective response against invasion of microorganisms, parasites and viruses [1]. Initially, the defense primarily relies on a few sets of pattern-recognition receptors (PRRs) of innate immunity to recognize conserved patterns on different classes of pathogens to trigger an inflammatory response that restricts their invasion [2]. Following an important evolutionary advance, the adaptive immune system constitutively generates a vast repertoire of cells expressing extraordinarily-diversified antigen-receptors to recognize antigenic configurations of specific pathogens, and triggers more antigen-specific and efficient responses [3]. The current understanding in the field indicates that innate immunity may play predominant roles in defending pathogens for lower vertebrates, whereas adaptive immunity in advanced vertebrates gradually develops superior functions due to the highly specific and effective immune response.
The spleen present in all vertebrates is the most ancient and the largest secondary lymphoid organ with a wide range of immunological functions [4], whereas lymph nodes represent a major recent emergence of adaptive immune system unique to mammals and some birds [5], [6]. The basic histological architecture and roles of spleen in maintaining normal physiological levels of platelets [7] and filtering the blood of antigenic particles, aged erythrocytes and apoptotic cells have been conserved during evolution from zebrafish, amphibians, reptiles, birds to mammals [5], [6]. The spleen is composed of a broad spectrum of immune cell types including macrophage, dendritic cell (DC), natural killer cell (NK), neutrophil as well as T cell and B cell etc., which is involved in the initiation and coordination of innate and adaptive immune responses [8], [9]. Thus, the spleen provides a good resource to investigate the evolution of main immune cell populations and their respective subpopulations.
Along with the evolution of immune organs, both innate and adaptive immunity possess a number of genes encoding receptors, transcription factors and other key functional molecules conserved in most vertebrates. The adaptive immune system in both low and high vertebrates generate a diverse repertoire of B and T cell antigen receptors through the rearrangement of immunoglobulin V, D, and J gene fragments mediated by recombination activating genes (RAG) or leucine-rich repeat (LRR) modular units [10]. Several key transcription factors required for B and T cell lineage commitment such as PAX5, E2A, GATA-3, EBF-1, Runx3 and Spi-C, appear to be expressed in both low and high vertebrates [11], [12]. Due to distinct marker expression among different species and lack of certain antibodies especially in lower vertebrates, it is difficult to study immune cells across evolutionary animal phyla. So far, it is still largely unknown that when and how the adaptive immune system across vertebrate species co-evolves in concert with the innate immune system through adapting gene expression.
RNA-seq at single cell level allows for comprehensive identification of cell types, states and gene expression profiles [13], and compared to single-nucleus RNA sequencing (snRNA-seq), scRNA-seq is more preferable for immune cell analysis [14], [15]. Here, we utilized scRNA-seq to dissect the transcriptome of immune cell populations and subpopulations in the spleen across 7 vertebrate species with complete genomic information accessible including Fish (Zebrafish), Frog (Xenopus tropicalis), Mouse (C57BL/6J), Rat (Sprague Dawley), Pig (Bama miniature pig), Monkey (Rhesus macaque) and Human being. Given that the comparisons among species are at molecular levels, most of these species were pooled from multiple individuals for the representative of the species. We compared the immune cell composition and signature genes across species, highlighting both conserved and unique gene expression profiles in major immune cell types T cells, B cells and macrophages. Collectively, these large sets of data provide a comprehensive analysis on gene transcription characteristics across multiple vertebrate species during the evolution of immune system, offering insights into species-specific immunity as well as the translational research from mouse models to human studies.
Materials and methods
Experimental animals and human samples
Spleen samples were collected from 140 zebrafish (1-year-old, male, the Institute of Hydrobiology in Chinese Academy of Sciences), 4 Xenopus tropicalis (2-year-old, male, NASCO), 3 C57BL/6J mice (8-week-old, male, Charles River Laboratories), 3 Rat (6-week-old, male, Charles River Laboratories), 3 Bama miniature pigs (6-month-old male, male, Institute of Zoology in Chinese Academy of Sciences) and 1 Rhesus macaque (10-week-old, male, the Beijing Institute of Xieerxin Biology Resource). The mouse and rat housed in SPF conditions, and zebrafish, Xenopus, pig and Rhesus macaque from clean conditions are included. For human sample, the spleen was obtained from a healthy individual who was admitted to the hospital with a compound injury caused by accident (the First Affiliated Hospital of Xi'an Jiaotong University). The spleen was found to be ruptured during follow-up treatment and was surgically removed. In addition, data of 6 human spleen samples were from published studies. In order to match the human subject, experimental animals at the age equivalent to the adolescent stage of their lifespan were selected. The detailed information of those donors is summarized in the Supplemental Table 1.
Preparation of single cell suspensions and single cell RNA library
The spleen tissues were dissociated into single-cell suspensions with the following procedure: Spleen tissues were processed with the flat end of a syringe in a 100 mm culture dish containing 5 ml cold FACS buffer (2% FBS in PBS), then passed through a 70 μm cell strainer into a 15 ml tube. Cells were centrifuged to remove the supernatant. Cell pellets were treated with 1 ml ACK (Ammonium-Chloride-Potassium) Lysing Buffer to remove the red blood cells. After washing with 10 ml cold FACS buffer, the remaining cells were stained with 7AAD (Part 76332; Lot B226294 Biolegend) for 30 min at 4 °C, then washed and resuspended in cold FACS buffer for flow cytometric sorting using FACS Aria II Cell Sorter (BD Biosciences). By mechanical dissociation and red blood cell lysis, nearly all the single cells released from spleen were immune cells. Further, the live cells were sorted based on 7AAD staining. Sorted 7AAD- cells with a viability higher than 90% were used for 10X genomics scRNA-seq. Approximately 10,000–20,000 cells per sample were loaded into Single Cell A chips (10x Genomics). Single cell RNA libraries were prepared using Chromium Single Cell V2 Reagent Kit according to the manufacturer’s protocol. Libraries were sequenced on the NovaSeq 6000 system with NovaSeq 6000 S4 Reagent Kit (300 cycles).
Raw data processing
Cell Ranger (v3.0.2, https://support.10xgenomics.com/) was used to process scRNA-seq data and generate the matrix data containing gene counts for each cell across species. Briefly, raw base call files from Illumina sequencers were first demultiplexed into FASTQ files with the cellranger mkfastq pipeline. The splicing-aware aligner STAR (https://www.reneshbedre.com/blog/star-aligner) was then used to align FASTQs files to the zebrafish reference genome (GRCz11), the xenopus reference genome (Xenopus_laevis_v2), the mouse reference genome (GRCm38), the rat reference genome (Rnor_6.0), the pig reference genome (Sscrofa11), the rhesus macaque reference genome (Mmul_8), and the human reference genome (GRCh38). The aligned reads were further counted using the cellranger count pipeline.
Identification of homologous genes
First, we defined genes with the same symbol in each species (zebrafish, xenopus, mouse, rat, pig and rhesus macaque) as in human as homologous genes. BLASTP [16] was used to compare proteins from six species (zebrafish, xenopus, mouse, rat, pig and rhesus macaque) with humans to screen for new homologous genes. To avoid evolutionary differences between species, we used different thresholds for each species. First, we sorted the sequence comparison results of the same gene symbol and screened the following four indicators: ppos (Percentage of positive-scoring matches), evalue (Expect value), bitscore (Bit score) and max.cov (https://www.ncbi.nlm.nih.gov/books/NBK279684/). The max.cov is the maximum percentage of the aligned sequence length relative to the subject or query sequence length. Any remaining gene that meets one of the following criteria is defined as a homologous gene: > 0.05 * ppos or < 0.95 * evalue or > 0.95 * bitscore or > 0.95 * max.cov. In addition, since xenopus is tetraploid, we combined the expression levels of the same gene names in the downstream analysis. In addition, since xenopus is an allotetraploid organism that has two copies of each gene, we then combined the expression of two copies for the same gene as the gene expression.
Quality control and data integrating
For each species, the UMI count data were then loaded into R (version 3.6.3) and processed by using the Seurat package (version 3.2.1)[17]. To exclude apoptotic cells and doublets, only cells expressing>200 genes with eligible UMIs per gene (higher than 1.7 and lower than 7) as well as genes expressed in at least 3 cells were retained [18]. Data were scaled to counts per 10,000 and log-normalized. The proportion of mitochondrial genes in 97% of the cells from most of the species was<10%, indicating that the analyzed cells have high quality. To avoid any bias in mitochondria, we removed all mitochondrial genes in the subsequent analyses.
Based on homologous genes determined by BLASTP among 7 species, the canonical correlation analysis (CCA) were applied to correct batch effects by using R package (FindIntegrationAnchors and IntegrateData) with setting dims = 1:40 and anchor.features = 4000. After scaling data with ScaleData, we ran a principal component analysis (PCA) dimensionality reduction (RunPCA) and selected the first 30 PCs to construct a shared nearest neighbor (SNN) graph (FindNeighbors). To visualize the clustering results in 2D space, a t-distributed Stochastic Neighbor Embedding (t-SNE) map was computed using RunTSNE with the first 30 PCs as input features, independent of the clustering step.
Cell type clustering
The cell type clustering consisted of three steps as follows:
Clusters were identified by applying FindClusters with resolution setting to 0.2 and assigned to cell types based on canonical marker genes. Thus, we obtained 7 major cell types, which were annotated as “primary types”.
For T cell, B cell and Macrophages, we extracted the cells of each primary type from the integrated Seurat object and re-ran FindClusters with resolutions setting to 0.3, 0.1, and 0.05, respectively. To visualize these “secondary types” of each primary type in 2D space, the t-SNE coordinates were recalculated as well. We excluded cell types with<100 cells and the smallest subpopulation was Mc-4 with 179 cells.
Differential expression analysis (DEA)
Differential expression analysis was performed between two groups of interest (e.g., T cells and all other cells) within each species by using the “RNA” assay in the integrated Seurat object. The fold change values and P values were calculated by FindMarkers [19] function, with threshold fold change < -1.28 or > 1.28 and P < 0.05. To determine the similarities and differences of different species, the shared and species-specific differentially expressed genes (DEGs) were counted.
Single-cell reference atlases mapping
To valid the representability of our human sample, we applied single-cell reference mapping using our sample as the reference and 6 samples from two other studies as the query [20], [21]. After the same data quality control, total 22,536 cells from those two studies were obtained for further analysis. Marker genes were consistent across reference and query by running FindAllMarkers, and the top 30 principal component analysis (PCA) were selected for cell clustering. Cell types were annotated for query studies according to our cell subsets. Furthermore, we followed the tutorial (https://satijalab.org/seurat/v4.0/reference_mapping.html) to map each donor dataset from the query individually. We used the FindTransferAnchors function with reduction = “pcaproject” and MapQuery function as previously described [22], and setting reduction = “pca” (as the documentation recommends for unimodal analysis).
Correlation and hierarchical clustering
We first calculated the average expression (in non-log space) of selected DEGs such as all DEGs or all up-regulatory DEGs in each species. Spearman correlations were calculated and visualization was done by using ComplexHeatmap (version 2.2.0) [23] in R. In addition, ward.D2 method was applied for hierarchical cluster analysis following Euclidean distance estimated by using the average expression of selected genes between species. For parallel validation analysis of human spleen samples, average expression of all transcriptomes in each cell type was measured. Spearman correlations were calculated and visualization was presented by heatmap between the same cell type from different datasets.
Functional enrichment analysis
Functional enrichment analysis was performed with clusterProfiler (version 3.14.3) [24] and visualized with ggplot2 (version 3.3.2). The annotation Dbi (https://dbi.r-dbi.org/) R package org.Hs.eg.db was used to convert gene symbol to gene id. More specifically, enrichment analysis of Gene Ontology (GO, http://geneontology.org/) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG, https://www.genome.jp/kegg/) pathways were executed by using enrichGO and enrichKEGG. All DEGs were used for GO term/KEGG pathway enrichment, species-shared DEGs were also used to identify immunologic signatures (MSigDB C7 gene sets, https://www.gsea-msigdb.org/gsea/index.jsp). For B cell subset annotation, B cell functional gene sets, including GSE12366, GSE7218, GSE12845, GSE13411, GSE11386, GSE17186 and GSE22886, were collected from C7 collection. Notably, the analyses were only applied for sufficient DEGs (>= 3). If not stated otherwise, P < 0.05 was taken to indicate statistical significance.
Protein–protein interaction analysis
The STRING database [25] was used to predict the regulatory relationship among the top15 markers in each species of Macrophage, T and B cells, and the cytoHubba software of cytoscape [26] was used to map the regulatory network.
Pathway activity score
The metabolic pathway activity score is obtained by using the pathway activity calculation method in the previous studies [27] and defines the pathway activity by using the overall expression level of the gene set in each cluster.
Statistical analysis
Statistical analysis was performed using R (version 3.6.3). Graphs were generated using R package (ggplot2 and Mfuzz [28]) and Cytoscape. The Wilcoxon rank-sum test was used to calculate differential genes between different cell types in the same species, and P < 0.05 was considered significant. Fisher's Exact Test was used for gene function annotation, and Benjamini & Hochberg “BH” method was used to adjust P values.
Results
Characterization of immune cells by scRNA-seq in spleens across species
To explore the gene transcription characteristics across multiple species during immune evolution, we chose 7 species with established genome annotations including Fish (Zebrafish), Frog (Xenopus tropicalis), Mouse (C57BL/6J), Rat (Sprague Dawley), Pig (Bama miniature pig), Monkey (Rhesus macaque) and human being. Two rodents and two primates were included because of their wide application in Biology and Medicine. Spleen was chosen as the source of immune cells since it is the only secondary lymphoid organ present in all vertebrates and also provides a broad spectrum of immune cell types [5].
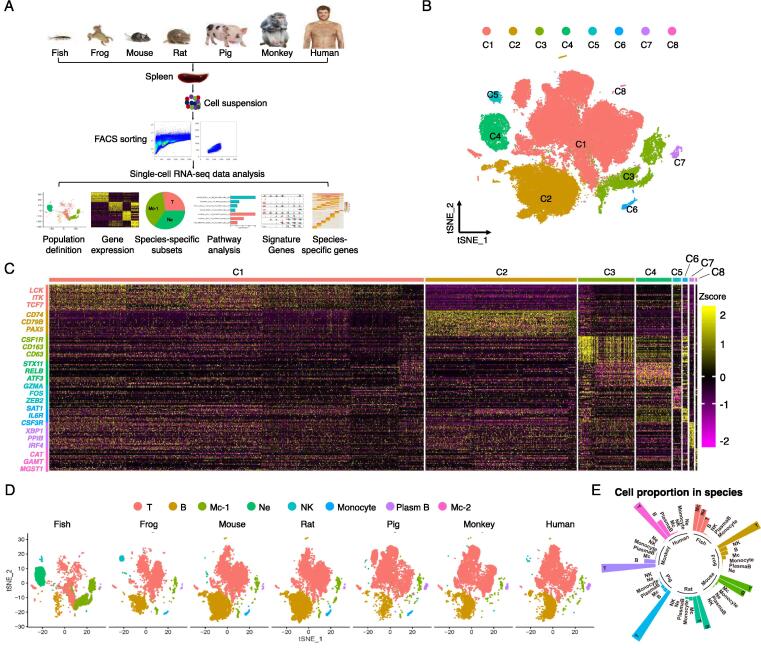
Single-cell sequencing (10x Genomics) was applied to the splenocytes from 7 representative vertebrate species, to assess the changes of immune cell composition and signature gene expression along with evolution (Fig. 1A). After quality control (Methods), a total of 80,315 cells (an average of 10,000 cells for each species) were obtained for subsequent analyses (Fig. S1A). All high-quality cells were divided into 8 main clusters (Fig. 1B and Fig. S1B). With the canonical cell markers, these 8 clusters were assigned to 7 cell types (T cell: CD3E; B cell: CD19; Macrophage1: C1QC; Monocyte: CD33; Neutrophil: ADAM8; NK cell: ITGA2; Macrophage2: CD68; Plasma B cell: IGHG3) (Fig. 1C and Fig. S1C, 1D). More definitively, we defined the cell types as follows: T cell (LCK, ITK, TCF7), B cell (CD74, CD79B, PAX5), Macrophage1 (Mc1: CSF1R, CD163, CD63), Neutrophil (STX11, RELB, ATF3), NK cell (GZMA, FOS, ZEB2), Monocyte (SAT1, IL6R, CSF3R), Macrophage2 (Mc2: XBP1, PPIB, IRF4) and Plasma B cell (CAT, GAMT, MGST1). By analyzing the distribution of each population in all the species (Fig. 1D), we found that T cells, B cells and macrophages were conservatively present with high prevalence in all the species. Of note, T cells and B cells constituted major immune cell populations in all species except for zebrafish, and the fraction of T and B cells showed an increase from bony zebrafish (class Osteichthyes) to xenopus (class Amphibia) to mouse and rat (mammalian rodents), and other mammals (pig and primates). Neutrophils and macrophages are predominant in the zebrafish, while NK cells are mostly present in xenopus (Fig. 1E), indicating zebrafish and xenopus species rely more on the innate immune system to defend against pathogens.
Fig. 1.
Immune cell composition in the spleen across species by scRNA-seq analysis. (A) Flow scheme of the scRNA-seq experiment design and analyses. (B) Clusters of 80,315 cells (all species) in t-SNE space, colors coded by clusters. (C) The heatmap of marker genes in each cluster with important ones labeled on the left. The color bar shows the expression level normalized by the z-score method. (D) The cell clusters (t-SNE plots) and the composition of immune cells in each species. (E) The fraction of major immune cell types in all cells for each species, color coded by species. Mc, Macrophage; Ne, Neutrophil.
To verify the accuracy of homologous genes used to identify cell types across species and avoid the data skewing caused by the integration step, we annotated cell populations using species-specific genomes reference files in zebrafish (Fig. S1E-G) and mouse (Fig. S1H-J), respectively. By comparing the cell types annotated by the zebrafish and human genome according to previously described makers [29], [30], we found that 92.3% were consistent, mainly because some macrophages were annotated as T cells (Fig. S1G). Similarly, by comparing the cell types of the mouse and human genome annotations according to previously described markers [31], we also found that there were some cell annotation errors in mouse, but the consistency was much higher (96.83%) than that in zebrafish (Fig. S1J). Therefore, the use of homologous genes to annotate immune cells across species had high accuracy and authenticity.
Given that only one human spleen was sequenced in this study, we also included public single-cell data of human spleens from two other studies [20], [21], in which one spleen (Spleen_HW) and five spleens (Spleen_ME) were analyzed, respectively. To this end, we applied single-cell reference mapping using our sample as the reference (Spleen_Ref) and others as the query cells [22]. The reference contained total 14,326 cells while the query samples contained 4512 and 18,024 cells. By unbiased analyses of cell-type transcriptomes, our sample and others showed significant similarities among immune cell populations. The major immune cell clusters, including T cells, B cells, plasma cells and macrophages, exhibited well correspondence between the reference and query datasets (Fig. S2A). Remarkably, all major cell subsets in our sample displayed profiles highly similar to that both query samples (Fig. S2B and S2C). All these results demonstrate that our human sample can be considered as a representative human species.
Large evolutionary diversities in macrophages among different species
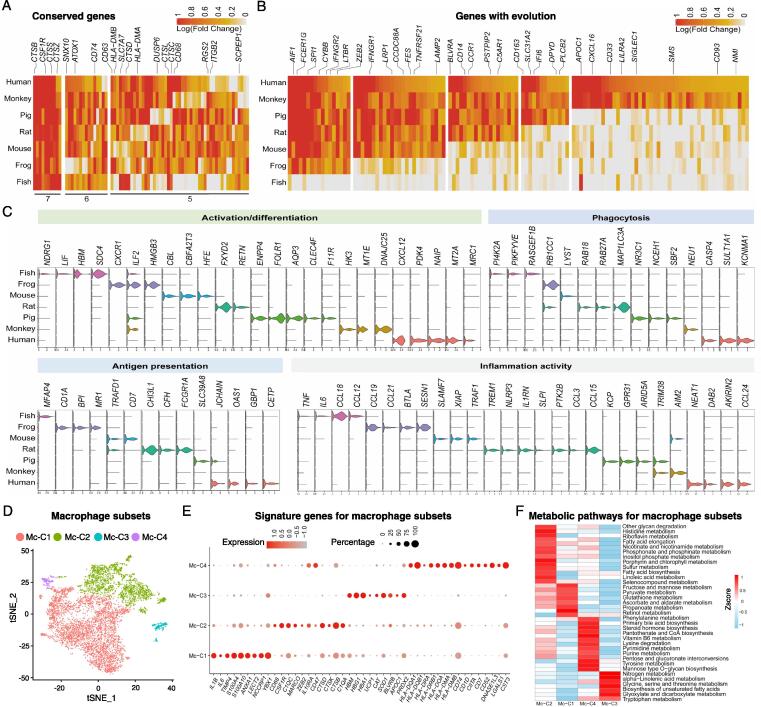
To investigate whether macrophages are conserved in different vertebrate species, we analyzed macrophage genes that were conserved across species (>4 species), and found most of them were associated with lineage identification (e.g., CSF1R, CD63 and CD68), functions of phagocytosis and defenses against infection (e.g., cathepsins CTSB, CTSS, CTSZ, CTSD, CTSL and CTSC, SNX10 and SCPEP1), activation and inflammation (e.g., DUSP6 and RGS2), antigen presentation (e.g., CD74, HLA-DMB and HLA-DMA), as well as migration (e.g., SLC7A7 and ITGB2) (Fig. 2A and Supplemental table 2). Furthermore, protein–protein interaction (PPI) network of DEGs was established and core genes in each species were identified (Fig. S3A). The data showed that CSF1R, as the most important signaling component in regulating macrophage growth and differentiation, was the only hub gene in all 7 species; while other genes, such as CD74, SPL1, AIF1, APP, ITGB2 and TYROBP, involved in its functionality were present in most of the species (>5 species).
Fig. 2.
Macrophages of different species showed great variation. (A, B) Function-related genes in macrophages with conservation (A) and genes emerged expression along with the evolution of species (B) were shown. The numbers in Figure A represent the number of species. (C) Violin plots showing the expression of signature genes related to macrophage activation/differentiation, phagocytosis, antigen presentation and inflammation activity. (D) Composition of macrophage subsets in t-SNE space in each species, color coded by subsets. (E) The expression of marker genes of each macrophage subsets with dot color signifying the relative expression and dot size scaled by the fraction of cells expressing the gene. (F) The heatmap showing the metabolic pathways enriched in each macrophage subsets. The color bar shows the expression level normalized by the z-score method.
Furthermore, we investigated macrophage genes becoming existence or extinction during the evolution (Fig. 2B, S3B and Supplemental table 3, 4). Consistently, genes associated with activation, defenses against infection and phagocytosis/endocytosis functions were conserved across species. Considered as primitive genes, the PRR LY75 and a component of AP1 transcription factor complex JUND were only highly expressed in zebrafish and xenopus, but loss of expression in higher vertebrates (Fig. S3B and Supplemental table 4). However, putative monocyte/macrophage markers CD14, CCR1 and CD163 were not expressed until in mouse macrophages. Interestingly, there were genes with diverse functions appeared even later in the evolution dominantly in rhesus macaque and human, such as CD93 contributing to complement C1q-dependent phagocytosis of apoptotic cells in vivo [32], SIGLEC1 with multifunction in host-pathogen interaction and even in anti-tumor immunity [33], [34], APOC1 involves in lipid metabolism [35], and CXCL16, positively correlated with M2-macrophage attraction and infiltration in injury regeneration and angiogenesis in cancer development [36], [37]. Notably, along with evolutionary progression, large inhibitory receptors were present on macrophages, such as CD33 and LILRA2 [38], [39], suggesting a complex regulatory network in macrophages in higher species (Fig. 2B).
We next found the number of uniquely expressed genes in macrophages by each species was high, indicating a functional diversity in innate immunity (Fig. S3C and Supplemental table 5). We then analyzed the expression of species-specific genes associated with the main functions of macrophages (Fig. 2C). Notably, CXCL12 and MRC1 are uniquely expressed in human macrophages but not in other species. The autocrine CXCL12 production by tumor-associated macrophages (TAMs) modulates the differentiation of macrophages toward a proangiogenic and immunosuppressive phenotype [40]. Additionally, mannose receptor (MRC1/CD206), considered as a typical marker of alternatively activated macrophages (M2), contributes to anti-inflammatory responses, tissue repair, induction of immune tolerance and immune escape mechanism of tumor [41]. Thus, the specific expression of CXCL12 and MRC1 in human macrophages indicated an evolutionarily advanced and sophisticated human immunological function of macrophages. Moreover, phagocytosis is a host defense mechanism that involves endocytosis and degradation of foreign particles, intracellular organelles/vesicles trafficking, fusion and energy consumption. During this process, distinct genes were exploited by macrophages from different species, such as phosphoinositide kinase PI4K2A and PIKFYVE in zebrafish, RB1CC1 in xenopus, LYST in mouse, RAB18, RAB27A and MAP1LC3A in rat and DAB2 in human. Whereas in human, macrophages expressed high levels of KCNMA1 and CASP4. Given the crucial function of K + channels and caspase-mediated pyroptosis in pathogen clearance [42], [43], human macrophages may develop novel strategies for pathogens killing. C–C motif chemokine ligands (CCL), a family of cytokines with chemoattractant activity for most immune cells, exhibited an evolutionary diversity across various species as CCL12 and CCL18 were found highly expressed in zebrafish macrophages, CCL19 and CCL21 in xenopus, CCL3 and CCL15 in rat, and CCL24 in human. All these results demonstrated that certain functional genes may be applied by different species. We also found several species-specific innate receptors and other top 20 genes with unknown functions in macrophages (Fig. S3D). In addition, PRRs were also present in all species albeit with different members in different species (Fig. S3E).
Taking into account of the heterogeneity of macrophages, we further divided them into 4 subtypes based on the gene expression profiles (Fig. 2D). Among those populations, Mc-C1 and Mc-C2 constituted as the major macrophage subtypes across species, suggesting their ancient developmental origin (Fig. 2D). Further signature gene analysis revealed that Mc-C1 highly expressed inflammation-related genes (IL1B, IL6, S100A members and ANXA1), Mc-C2 expressed function regulatory genes (CSF1R, IL10RA, MARCO, CD47 and C1QA), Mc-C3 appeared to be involved in oxidation–reduction reaction by high expression of hemoglobin genes (HBM, HBG1, and HBA1), SOD1 and PRDX2, whereas Mc-C4 represented professional antigen-presenting cells (APCs) as it expressed a number of major histocompatibility complex genes (HLAs), CD74, CD1D and CIITA (Fig. 2E). As metabolic adaptation of macrophages is a key factor in regulating various cellular responses and functions [44], we further analyzed the metabolic characteristics of various macrophage subsets, which also displayed discrete and specific patterns among cell groups (Fig. 2F). These findings shed light on future studies to explore the effects of metabolic reprogramming on macrophage function.
Characterization of B cells in spleens of different species
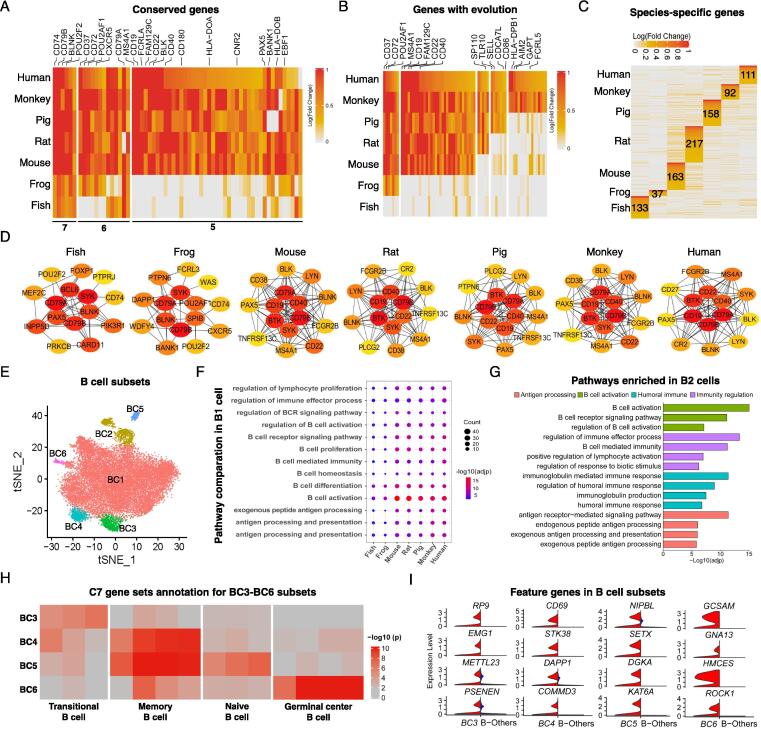
B cells are the major cell population in spleen, and their tissue distribution varies dependent on activation state, indicating divergent expression of receptors and chemotactic molecules [4]. Unlike macrophages, we found that classic B cell markers were conserved across species, including CD79B, BLNK, POU2F2, CD79A, MS4A1, PAX5 and EBF1 (Fig. 3A and Supplemental table 6). Notably, B cell hallmark genes CD19, MS4A1, CD22 and CD40 began to emerge in mouse while some genes are expressed only in rhesus macaque and human, such as AIM2, GAPT and FCRL5 (Fig. 3B and Supplemental table 7). Surprisingly, the number of species-specific genes was much lower in each species compared to that in macrophages (Fig. 3C and Supplemental table 8). The hub genes in PPI network of B cells showed that four key genes (CD79A, CD79B, SYK and BLNK) were present in all species, and six genes (LYN, MS4A1, BLK, CD22, CD19 and CD40) were present in most of the species (>5 species), implicating that CD79A and CD79B may be more accurate to mark B cell lineage, especially for low vertebrates (Fig. 3D). Among species-specific genes expressed in B cells (Fig. S4A), those associated with B cell activation and BCR signaling were LBP, BHLHE40 and RGS13 in zebrafish [45], [46], CLNK and TCIRG1 in xenopus [47], SERINC3 and NFATC3 in mouse, DHRS9, DERL3 and CD302 in pig [48]. Meanwhile, several negative regulatory receptors, such as FCRLB and FCRL3 in zebrafish [49], [50], LAX1 in rat, as well as complement receptor type 1 (CR1, CD35) and FCRL2 in human, have been shown to inhibit B cell activation, differentiation into plasma cells and immunoglobulin production[51], [52]. In addition, zebrafish B cells express high level of Syndecan-4 (SDC4) (Fig. S4A) which is found highly associated with B cell migration in asthma model [53]. Human B cells specifically showed a memory profile with high expression of CD27 [54]. In agreement with the notion that CD4+ helper T (Th) cells drive B cell responses, antibody subclass switching and affinity maturation [55], we have seen unique expression of genes in human B cells involving in cognate T-B cell interaction, such as CD1C, CLECL1 and ALOX5 [56], [57].
Fig. 3.
Characterization of B cells in spleens from different species. (A, B) Function-related genes in B cells with conservation (A) and genes emerged expression along with the evolution of species (B) were shown. The numbers represent the number of species. (C) Heatmap of up-regulated genes that are species-specific in B cells with number labeled. The color indicates the log-scaled fold change of each gene. The numbers represent the number of genes specific to each species. (D) PPI of each species using the top 15 highly expressed genes in B cells. The color represents the degree of association with other genes. (E) The subsets of B cells in t-SNE space in each species, color coded by subsets. (F) B cell functional pathways enriched in each species using up-regulated genes in BC1 comparing to other B cells. The dot color indicates the significance and the dot size is scaled by the number of up-regulated genes in the pathway. (G) Pathways enriched using up-regulated genes in BC2 comparing to other B cells, color coded by the pathway types. (H) C7 gene sets annotation of up-regulated B cell genes in BC3, BC4, BC5 and BC6, color indicates the significance. (I) Feature genes expressed in BC3, BC4, BC5 and BC6 (red) comparing to other cells (blue). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To explore the heterogeneity of B cells, we further divided them into 6 subsets (Fig. 3E). We found that BC1 was the largest population and was present in every species, BC3 and BC4 had higher proportions in mouse and rat, respectively, and BC6 was present in pig and rhesus macaque (Fig. S4B). Correlation analysis of the genes expressed in BC1 showed that BC1 was a relatively conservative subset of B cells (Fig. S4C). Biological process analysis of BC1 subset in all species indicates that similar functional pathways were enriched, namely B cell proliferation and differentiation, B cell activation and BCR signaling pathway (Fig. 3F). In contrast to BC1, the correlation of BC2 subset was relatively variable among species (Fig. S4D). Enrichment analysis of highly expressed genes in BC2 revealed that this subset was a subpopulation of activated B cells with potential immunomodulatory and antigen presentation functions (Fig. 3G). For species-specific B cell subsets, cell annotation of highly expressed genes using C7 gene sets showed that BC3 was a transitional B cell population, BC4 and BC5 were memory/naïve B cells, and BC6 was germinal center (GC) B cells (Fig. 3H and Supplemental table 9). The representative genes of each subset were shown in Fig. 3I.
An important feature of the adaptive immunity is producing antibodies with high specificity and affinity, which is mainly mediated by plasma cells [58]. As shown in Fig. 1D, plasma cells were identified in all species. Biological process analysis verified this cluster by showing that the enriched genes were associated with plasma cells, humoral immune response and immunoglobulin production (Fig. S5A). Immunoglobulin secretion genes (XBP1, ELL2, MZB1 and FKBP11) and B cell surface proteins (CD81 and CD79A) were conserved in plasma cells across species (Fig. S5B). However, plasma cells of each species also showed specifically-expressed genes, of which the pig had the most unique genes (5 2 9) expressed in comparison with other species (Fig. S5C). This finding implied that plasma cells in pig have diverse functions, which may help them to adapt to a wide variety of habitats. We further analyzed top 20 species-specific and up-regulated genes in plasma cells among various species (Fig. S5D and Fig. S5E), and found that human plasma cells expressed a variety of immunoglobulin heavy and light chains, indicating diverse and complex functions in humoral immune responses in human. Together, these data suggest that B cell immunity is relatively conserved among vertebrate species.
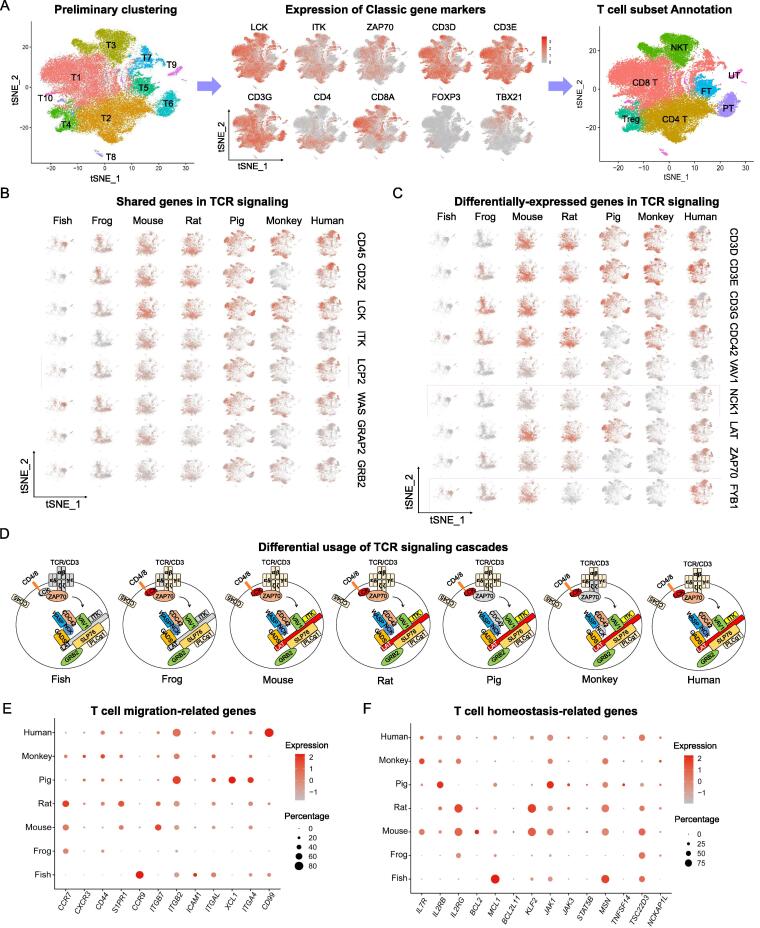
Species-specific profiling of TCR signaling cascades
T cells play a central role in the adaptive immune response fighting against various infectious pathogens. While many studies in the field have been focused on the function of T cells in the spleen, few studies explored the evolution of T cell populations and functions [4]. We first classified 10 T cell subsets expressing hallmark genes (CD3E+CD3G+), then identified CD4+ T (CD4+CD8-), CD8+ T (CD4-CD8+), NKT (TBX21+) and Treg (CD4+FOXP3+) populations. Interestingly, unique T cell subsets were identified in zebrafish and pig annotated as FT and PT, respectively (Fig. 4A). Core genes regulating T cell functions were analyzed in each species (Fig. S6A). TCR signaling, transduced by TCR complex composed of a clonotypic T cell receptor (TCR) α- and β-heterodimer noncovalently associated with multiple signaling subunits, is an essential feature of T cells for antigen recognition, response and lineage differentiation [59]. Core genes in TCR signaling including CD3E, ZAP70, LCK and CD28 were present in all species except for zebrafish, suggesting that zebrafish T cells may not have fully matured TCR functions. In addition, CD45, CD3Z (CD247), LCK, ITK, LCP2, WAS, GRAP2 and GRB2 were detected in T cells from all species (Fig. 4B) while CD3D, CD3E, CD3G, LAT, VAV1, NCK1, CDC42, ZAP70 and FYB1 were partially expressed in T cells from some species (Fig. 4C). For example, ZAP70, a vital mode for controlling TCR downstream signaling, was not expressed in T cells of pig and rhesus macaque. The expression CD3D and LAT was absent in zebrafish and xenopus, while CDC42 and MCK1 were defectively expressed in pig and rhesus macaque, respectively. FMYB1 showed no expression in three species including rat, pig and rhesus macaque. We summarized the differential usage of TCR signaling components in each species in which the expressed molecules were in color while no/low expressed ones were in gray (Fig. 4D). These molecular expression features suggest the compensatory mechanisms from other TCR components utilized in these species.
Fig. 4.
The characteristics of T cells in evolution. (A) Subsets of T cells in t-SNE space, expression of marker genes and the cell types assigned to each subset were shown from left to right. (B, C) Key genes in TCR signaling pathway conservatively (B) or specifically (C) expressed in different species. The color gradient denotes the level of gene expression, red being high whereas gray being low. (D) Differential usage of TCR signaling cascades were summarized in each species. Components expressed in the species were shown in color while no/low expressed ones were in gray. (E, F) The expression of T cell migration-related genes (E) and T cell homeostasis-related genes (F). The dot color indicates the relative gene expression and the dot size is scaled by the percentage of T cells expressing the gene. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Furthermore, we analyzed the distribution of genes related to T cell migration (Fig. 4E) and homeostasis (Fig. 4F) across species. CD99, a cell surface glycoprotein involved in leukocyte migration and adhesion was highly expressed in human T cell [60]. Prominent genes controlling T cell survival and activation, such as IL2RB, BCL2L11, JAK1, JAK3, STAT5B, TNFSF14 (LIGHT), TSC22D3 and NCKAP1L, were highly conserved among various species regarding to both expression prevalence and intensity (Fig. 4F). Nevertheless, species-specific gene patterns were also identified. IL-7 is a well-known vital cytokine for T cell development, naïve T cell survival and memory T cell generation and maintenance [61]. With the observation that IL-7 receptors (IL7R and IL2RG) were enriched in mammals, particularly in mouse, rhesus macaque and human, it is speculated that IL-7 signaling is an advanced product in evolution to regulate T cell function. But T cells in zebrafish specifically expressed high level of MCL1—an anti-apoptotic member of Bcl-2 family—to maintain the survival of CD8+ memory T cells [62], likely compensating the limited expression of other T cell survival signals, e.g., IL7R, IL2R and BCL2. Additionally, rodent (mouse and rat) T cells expressed substantial KLF2 to regulate T cell survival, differentiation, migration and cytokine production [63]. Lastly, species-specific T cell genes were identified but needed a further investigation on their immune functions (Fig. S6B and Supplemental table 10).
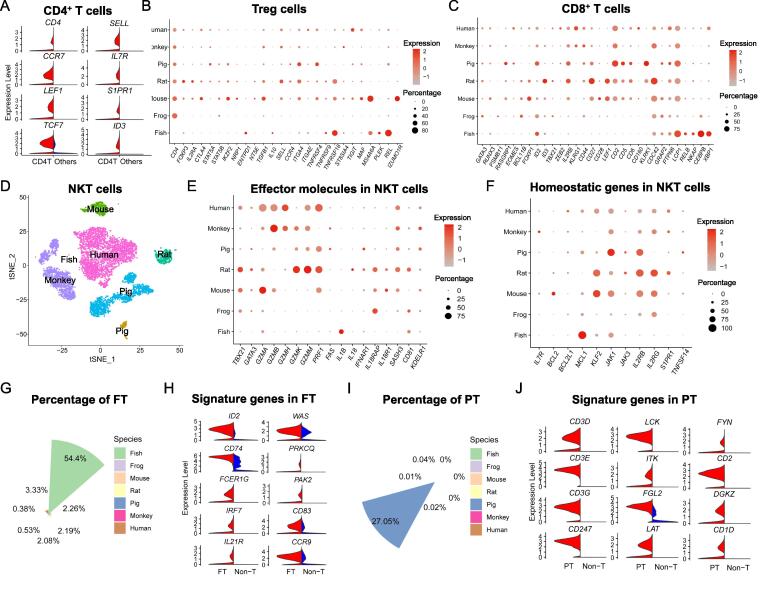
Immune function-related features among T cell subsets across species
T cells can be classified into various subsets based on distinct surface markers, differentiation states and effector molecules. To better understand the divergence and conservation of T cell subsets during evolution, we analyzed signature genes in T cell subsets of CD4+ T, Treg, CD8+ T and NKT cells, which are present in all species. Compared to other immune cells, CD4+ T cells in all species significantly expressed naïve T cell marker genes (CCR7, LEF1, TCF7 and SELL), indicating that most of them were antigen-unexperienced (inactivated) T cells (Fig. 5A). Tregs, a special subset of suppressive CD4+ T cells, are potent mediators of peripheral tolerance to prevent autoimmune reactions [64]. To characterize Treg heterogeneity, we compared Treg functional genes across species (Fig. 5B). Treg program was conserved tremendously in mammals as signature genes were universally distributed among mouse, rat, pig, rhesus macaque and human, such as CD4, FOXP3, IL2RA, CTLA4, STAT5 members, IKZF2, NRP1, NT5E (CD73), TGFB1, IL10, SELL, CCR4, TNFRSF4 (OX40), TNFRSF9 (4-1BB) and TIGIT (Fig. 5B). However, species-specific expression of signature genes was also observed. CD39 (ENTPD1) and CD73 (NT5E) are highly expressed on the surface of Tregs and critical for creating immunosuppressive environment by conversion of ADP/ATP to AMP and AMP to adenosine, respectively [65]. Interestingly, zebrafish Tregs expressed high level of ENTPD1 but not NT5E as in mammals, suggesting suboptimal Treg function in zebrafish. Moreover, Folate receptor 4 (Folr4), encoded by IZUMO1R, a recently discovered Treg marker and maintains Treg cell function [66], appeared to be specifically and highly expressed in mouse.
Fig. 5.
Characterization of immune functions in T cell subsets across species. (A) Up-regulated genes of CD4 T cells (red) compared to other T cells (blue). (B) Dot lot showing the expression of functional genes in Treg cells from different species, in which the dot color indicates the relative gene expression and the dot size is scaled by the percentage of Treg cells expressing the gene. (C) Expression of vital genes associated with CD8+ T cell functions such as activation across species. (D) The fraction of NKT cells in all T cells in each species, color coded by different species. (E, F) Expression of genes related to the effector molecules (E) or homeostasis (F) of NKT cells across species. (G) The fraction of zebrafish-specific T cells (FT) in total T cells in each species, color coded by different species. (H) The expression of innate lymphocyte markers (left chart) and T cell markers (right chart) in FT cells (red) comparing to the non-T cells (blue). (I) The fraction of pig-specific T cells (PT) in all T cells in each species, color coded by different species. (J) Expression of T cell function-related genes in PT cells (red) comparing to the non-T cells (blue). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To reveal the functional features of CD8+ T cells, key transcription factors and surface molecules were analyzed across species (Fig. 5C). Gene profiling in CD8+ T cells was profoundly conserved, particularly those associated with cell survival, differentiation and TCR signaling, such as RUNX3, ID2, TBX21, CDC42, GRAP2, PTPN6 and LCP1. Notably, as the secondary signaling for T cell activation, T cell co-stimulatory receptors (i.e., CD27, CD28, CD2, CD5, CD6 and CD160) exhibited an increasing expression along with the evolutionary path. Lower vertebrate class zebrafish CD8+ T cells expressed specifically high level of transcription factors members CEBPB, XPB1, NF-κB (RELB and NKAP) to regulate T cell functions. In addition, NKT cells play an important role in the protective response against blood-borne infections, and the majority of them patrol around the marginal zone (MZ) and red pulp (RP) of the spleen [67]. Zebrafish and xenopus had very low NKT population, but the proportion of NKT cells was gradually increased from mouse to human (Fig. 5D), implying an evolutionary progression. We then analyzed the key transcription factors and signature molecules of NKT cells across species (Fig. 5E). Interestingly, NKT cells in mammalian species appeared to exert functions mainly via proteases, evidenced by high expression of granzymes (GZMA, GZMB, GZMH, GZMK and GZMM), whereas in low vertebrate classes such as zebrafish and xenopus, cytokines (IL-1, IL-18 and IFN) were likely the major mediators. Furthermore, JAK and IL2 signaling were conserved in NKT cells among species, the anti-apoptotic molecule BCL2 was highly expressed in mouse, while another Bcl-2 family member MCL1 was only detected in zebrafish (Fig. 5F).
Besides, we discovered unique T cell populations in zebrafish and pig species. Although T cell-like activity has long been reported in zebrafish, specific features of T cell types regarding to zebrafish T cell subsets is still lacking [68]. A special T cell population in zebrafish (named as FT) (Fig. 5G) expressed high level of both innate immune molecules and T cell characteristic genes (Fig. 5H). Consistently, GO analysis showed enriched genes in FT were associated with innate immune response, response to interferon, T cell receptor signaling pathway and lymphocyte activation (Fig. S7A and Supplemental table 11). Furthermore, a specific T cell subset (PT) was enriched in pig spleen (Fig. 5I). However, unlike FT, PT already had strong T cell functional characteristics. Violin plots showed CD3 family (CD3D, CD3E, CD3G and CD247) and TCR signaling molecules (LCK, ITK, LAT, FYN and DGKZ) were highly expressed in PT, which indicates that PT represented a group of mature T cell subset (Fig. 5J). Interestingly, PT highly expressed co-receptors (CD2, CD96 and CD84), transcription factors (EOMES and TOX) related to T cell exhaustion, but not GZMK, PRF1 and CXCR3 (Fig. S7B and Supplemental table 12).
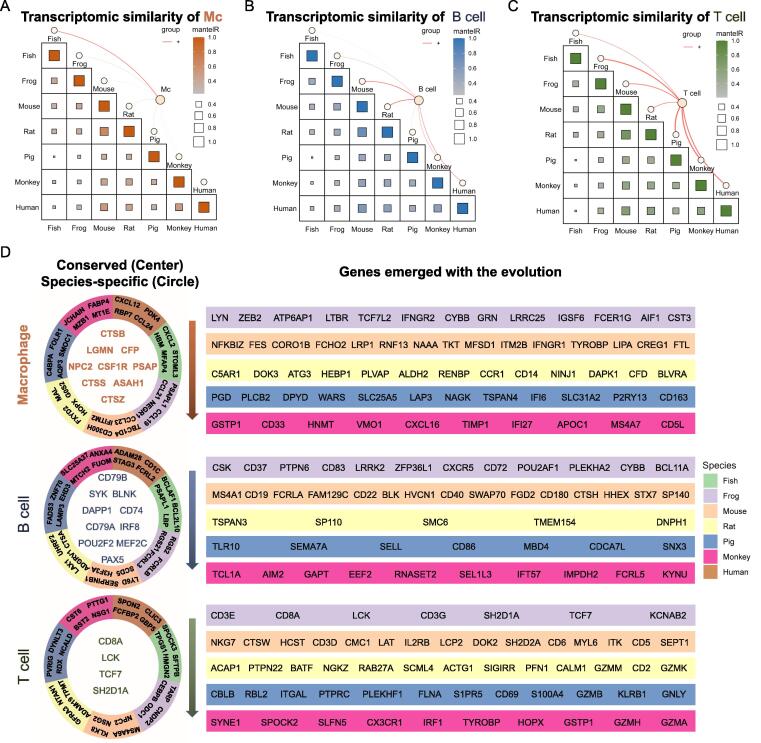
Inter-species comparison identifies core genes program along the evolutionary path
Given that macrophages, B cells and T cells were the major cell populations in the spleen of all species, we further analyzed the signatures of overall genes in these cells across species. From an evolutionary perspective, inter-species comparison of the core gene programs in macrophages (Fig. 6A), B cells (Fig. 6B) and T cells (Fig. 6C) revealed that, rodent mouse had a significant similarity to human, supporting the rationale of using mouse as an appropriate animal model to study human immunity. In addition, as lower vertebrate classes, bony zebrafish had the most similarities to xenopus, followed by mammalian rodents but are phenotypically distant compared to mammalian primates. Of note, T and B cells seemed to display higher similarities than macrophages across species. Additionally, we analyzed key transcription factors essential for the lineage development of macrophages, B cells and T cells (Fig. S8A). Generally, expression of transcription factors in macrophages varies among species more than that in adaptive immune cells. For instance, PPARG, IRF5, IRF8 and STAT1 were highly expressed in macrophages of pig. Transcription factors in B cells had no special patterns across species except that EBF1 was specifically expressed in mouse B cells. TCF7, a transcriptional activator involved in T lymphocyte differentiation, was highly expressed in T cells of xenopus and rat while TCF3 and ID3 were elevated in rat T cells. We further analyzed the gene expression patterns by various cell types across species. In total, we clustered 6 distinct patterns in each cell type which represents gene expression profiling enriched in certain species (Fig. S8B). Among these, clusters 1–6 in macrophages represented genes enriched in zebrafish, zebrafish/xenopus, mouse/rat, mouse/pig, pig and zebrafish/mouse/rhesus macaque. Intriguingly, zebrafish did not show obvious gene enrichment patterns in B and T cells, whereas cluster 6 in T cells exhibited a strikingly high expression in human, indicating those genes play crucial roles in human T cell immunity.
Fig. 6.
Inter-species comparison identifies core gene program across evolutionarily species. (A-C) Inter-species comparison of all expressed genes shows the transcriptomic similarities of macrophages (A), B cells (B) and T cells (C) between different species. The size of the square indicates gene number and the color indicates the degree of similarities. The orange lines represent the cell types in each species and the thickness indicates the cell proportion. (D) Functional genes conserved in all species, with species-specificity and emerged along with the evolution of species were shown for macrophages, B cells and T cells. The conserved genes among all species were marked in the left center with colors showing each cell types. Species-specific genes were located at the left second circle with colors showing each species. Genes emerged with the evolution were marked in the right stripes from xenopus to rhesus macaque. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Finally, we summarized the representative genes conserved in all species, with species-specificity and emerged with species evolution for macrophages, B cells and T cells to track the evolutionary trajectory of those markers (Fig. 6D). In the middle circle, there were 9, 10 and 4 conserved genes existed in all species in macrophages, B cells and T cells, respectively. For instance, CSTB, CSF1R, CTSS and CTSZ in macrophages, CD74, SYK, CD79A and CD79B in B cells, and CD8A, LCK and TCF7 in T cells were primitive genes emerged since early vertebrate. The genes located at the second circle with colors showing different species were species-specific genes. Such as, CXCL12, CCL24, CD1C, STAG3, SPON2 and GBP5, etc. were present only in human, indicating those genes may evolve with unique features. Further investigation for their special functions in human immune system is needed. In addition, genes located at outer circles were shared by multiple species and emerged along with the evolution. Notably, CD8A, LCK and TCF7 were weakly expressed in zebrafish T cells, but much increased in other higher species. Thus, we marked them in both the middle and outer circles (Fig. 6D, right). Taken together, our comparative studies of vertebrate immune systems not only provide critical insights into species-specific innate and adaptive immunity, but also address the uncertainty of the expression patterns of previously proposed immune cell markers across species.
Discussion
Accumulating studies demonstrate that the immune system in higher vertebrate classes has undergone an evolutionary progression from innate immunity as the dominant type of host defense towards tight cooperations between innate and adaptive immunity. Here, we have adopted an integrated and comprehensive transcriptomic analysis of the immune system at the single-cell level to track the molecular and functional features from low to high vertebrates. Generally, the mechanisms underlying innate and adaptive immunity are conserved among vertebrates. Consistently, in this study, we have seen a conserved immune profile throughout 7 species from teleost zebrafish to mammalian human in terms of immune cell populations, signature gene expression, and biological signaling pathways. For instance, the major immune cell types macrophages, T cells and B cells are present in all studied species. Macrophages appear to be functionally conserved in all those taxa, playing major roles in defense against infections, pathogen phagocytosis, cell migration and regulation of pro-inflammatory responses. Core genes involved in T cell lineage, homeostasis/activation and function, as well as B cell hallmarks and immunoglobulin secretion were highly conserved across species.
Although considered primitive during evolution, innate immunity displays less evolutionary conservation compared to adaptive immunity, evidenced by 1) the number of uniquely expressed genes in each species was much higher in innate immune cells than adaptive immune cells. The average number of unique genes from all species in macrophages was 246, whereas there were only 130 and 128 genes in B cells and T cells, respectively. 2) The top 15 hub genes in PPI network of macrophages have changed remarkably from zebrafish to human with only one gene (CFS1R) being present in all 7 species. On the contrary, 4 genes (SYK, CD79A, CD79B, and BLNK) in B cells and 3 genes (LCK, CD3E, and CD8A) in T cells are conserved in all and 6 species (except for zebrafish T cells), respectively. 3) Fewer marker genes were shared across species in innate immune cells. 4) Inter-species comparison of the core gene programs has identified profound similarities between B and T cell population rather than macrophages. 5) Innate immunity in higher organisms (e.g., mammals) has developed sophisticated signaling networks to effectively defend pathogenic infections, trigger pro-inflammatory responses, and coordinate with adaptive immunity. Furthermore, the phagocyte function in innate immunity is evolutionarily conserved to allow for discrimination between self and non-self [69]. Thus, macrophages and genes related to engulf and digest foreign substances (e.g., cathepsins) were conserved in all 7 examined species, indicating those immune components in macrophages are evolutionarily primordial. Both previous reports and our data have shown that in teleost zebrafish, polarized macrophages (e.g., pro-inflammatory and immune-regulatory populations) with distinct effector molecules have evolved to confer host protection and homeostatic regulation [70], [71], highlighting that the divergent pro-inflammatory and homeostatic responses in macrophages have been well established in lower vertebrates. Although the main functions of macrophages are shared across species, featured genes playing these roles are quite different among animal taxon, confirming the evolutionary diversities in macrophages.
The overall fundamental immune elements of adaptive immune system including lymphoid organs, the putative T and B cell populations and genes, TCR repertoires and immunoglobulins (Ig) are highly evolutionarily conserved throughout the jawed vertebrate species [72]. In our study, the B cell populations, including plasma cells, and signature genes associated with B cell markers and functional pathways are conserved in the spleen across all species. Additionally, conserved CD4+ and CD8+ T cell subsets among gnathostome classes have been observed in our study, in agreement with previous reports [73], [74]. Of note, genes in T cells are extremely conserved to be unable to lay out the specific genes emerged along with phylogenetics evolutionary as shown in macrophages and B cells. However, unique T cell populations were identified in zebrafish and pig, and the abundance of individual T cell subsets and expression of key genes vary at different species. TCR signaling cascades are critical for T cell activation, cell survival, proliferation, differentiation, and cytokine production. Importantly, by analyzing the expression of core genes in TCR signaling pathways among various species, we have revealed a conserved yet species-specific profiling of TCR signaling cascades. The presence of molecules in all species (CD45, LCK, LCP2, WAS, etc.) and the absence of molecules in certain species (CD3D, LAT, CDC42, ZAP70, FYB1, etc.) strongly suggest that distinct combinations of TCR components are exploited in a given taxon. The absence of tyrosine kinase ZAP70, an important molecule in initiating TCR signal transduction, in pig and rhesus macaque implicates that compensation from other TCR homologous or non-homologous components may apply in these species.
Intriguingly, immune system in lower vertebrates tends to have mixed innate and adaptive functions. Recent discovery has shown that B cells in all ectotherms tested have potent phagocytic and microbicidal abilities, demonstrating their innate-like nature to act as pivotal APCs in priming adaptive immunity [75], [76]. Similar observations were also reported in amphibians, reptiles and mammals in which one major B cell subgroup B1 (referring to BC-1 in this study) cells exhibited phagocytic and microbicidal abilities [77], [78], [79]. This finding suggests that BC-1 cells represent a more primitive population and have an evolutionary connection with ectotherm B cells [80], [81], which is also supported by our data that BC1 cells and the corresponding enriched genes were present in all species. Moreover, the current study revealed that the unique T cell-like population identified in zebrafish (FT) displayed a strong innate immune signature enriched in lymphocyte activation and response to interferon, and expressed high levels of ID2, IL21R, IRF7, and CCR9, implying it is a group of innate-like lymphocytes as described previously in zebrafish [82], [83], [84], [85]. It will be interesting to further investigate the phenotype and function of this population in future studies. Surprisingly, we have also demonstrated that the immune profiles that resembles human the most is rodents, especially mouse rather than rhesus macaque which is known to share 99 percent identical genome with human [86]. At the genomic level, mouse and human only share approximately 70 percent of the same protein-coding gene sequences, but with varied similarities depending on the tissue, function and transcription of the gene expression [87]. Our data offer profound insights into a conserved immune system shared by mouse and human, supporting the view that mouse is an appropriate model to study human biology and disease related to immune function.
Taken together, our comparative studies of vertebrate immune systems across various species reveal molecular similarity along with species-specific aspects of innate and adaptive immunity. While the current study has provided inspiring insights into the immune evolution, certain limitations must be acknowledged. Some vertebrate classes (e.g., birds) are missing, which may limit the significance of our findings in an evolutionary perspective. Due to the availability of samples in certain species, only male individuals of each species are included in the analysis, which may cause possible bias since sex-based immunological differences exist in males and females [88]. Future studies with female samples may be considered. In addition, samples from different (SPF, clean and conventional) conditions may affect the pattern of gene expression, but the activation markers vary among different species, which may cause complication of analysis. Therefore, the current study focuses on lineage dominant genes for major immune cell populations. Cell populations with low frequency, such as DCs, innate-like lymphocytes or γδ T cells, may fail to be detected due to limited resolution in cell type clustering. Verification of important findings by experiments is encouraged in future works. Collectively, information derived from the current work not only sheds light on the general principles of vertebrate immune systems but also inspires translational research using animal models for studying human physiology and disease.
Code availability
The codes generated in this study are available at the Github repository (https://github.com/Immugent/Single-cell-transcriptomic-atlas-across-species).
Ethics Statement
The animal study was reviewed and approved by the Animal Care Committee of Xi’an Jiaotong University. The human sample was collected approved by the Ethics Committee of Xi’an Jiaotong University.
Funding
This work was supported by grants from the National Key Research and Development Program of China (2021YFA1100702, B.Z.), the Major International (Regional) Joint Research Project (81820108017, B.Z.), the National Natural Science Foundation of China (32170892, B.Z. and 82101826, X.Y.) and the Innovation Capability Support Program of Shaanxi (Program No.: 2021TD-38, B.Z.).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Dr. Guohua Zhang from Beckman Coulter, Prof. Chen Huang and Dr. Xiaofei Wang from Department of Cell Biology and Genetics for flow cytometric technical supports.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2023.02.017.
Contributor Information
Xiaofeng Yang, Email: yangxiaofeng@xjtu.edu.cn.
Lianjun Zhang, Email: zlj@ism.cams.cn.
Baojun Zhang, Email: bj.zhang@mail.xjtu.edu.cn.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Data Availability Statement
Raw data for single cell RNA-seq samples are available in the Gene Expression Omnibus (GEO) database as accession number GSE186158 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE186158).
References
- 1.Iwasaki A., Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327(5963):291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akira S., Uematsu S., Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 3.Cooper M.D., Alder M.N. The evolution of adaptive immune systems. Cell. 2006;124(4):815–822. doi: 10.1016/j.cell.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Lewis S.M., Williams A., Eisenbarth S.C. Structure and function of the immune system in the spleen. Sci Immunol. 2019;4(33) doi: 10.1126/sciimmunol.aau6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boehm T., Hess I., Swann J.B. Evolution of lymphoid tissues. Trends Immunol. 2012;33(6):315–321. doi: 10.1016/j.it.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Brendolan A., et al. Development and function of the mammalian spleen. Bioessays. 2007;29(2):166–177. doi: 10.1002/bies.20528. [DOI] [PubMed] [Google Scholar]
- 7.Abesadze A.I., et al. Role of the spleen in regulating thrombocytopoiesis. Biull Eksp Biol Med. 1978;86(12):718–720. [PubMed] [Google Scholar]
- 8.Morelli A.E., et al. Internalization of circulating apoptotic cells by splenic marginal zone dendritic cells: dependence on complement receptors and effect on cytokine production. Blood. 2003;101(2):611–620. doi: 10.1182/blood-2002-06-1769. [DOI] [PubMed] [Google Scholar]
- 9.Mebius R.E., Kraal G. Structure and function of the spleen. Nat Rev Immunol. 2005;5(8):606–616. doi: 10.1038/nri1669. [DOI] [PubMed] [Google Scholar]
- 10.Pancer Z., Cooper M.D. The evolution of adaptive immunity. Annu Rev Immunol. 2006;24:497–518. doi: 10.1146/annurev.immunol.24.021605.090542. [DOI] [PubMed] [Google Scholar]
- 11.Busslinger M. Transcriptional control of early B cell development. Annu Rev Immunol. 2004;22:55–79. doi: 10.1146/annurev.immunol.22.012703.104807. [DOI] [PubMed] [Google Scholar]
- 12.Anderson M.K., et al. Evolutionary origins of lymphocytes: ensembles of T cell and B cell transcriptional regulators in a cartilaginous fish. J Immunol. 2004;172(10):5851–5860. doi: 10.4049/jimmunol.172.10.5851. [DOI] [PubMed] [Google Scholar]
- 13.Cai S., et al. Integrative single-cell RNA-seq and ATAC-seq analysis of myogenic differentiation in pig. BMC Biol. 2023;21(1):19. doi: 10.1186/s12915-023-01519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrews T.S., et al. Single-cell, single-nucleus, and spatial RNA sequencing of the human liver identifies cholangiocyte and mesenchymal heterogeneity. Hepatol Commun. 2022;6(4):821–840. doi: 10.1002/hep4.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slyper M., et al. A single-cell and single-nucleus RNA-Seq toolbox for fresh and frozen human tumors. Nat Med. 2020;26(5):792–802. doi: 10.1038/s41591-020-0844-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson M., et al. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36(Web Server issue):W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stuart T., et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guilliams M., et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell. 2022;185(2):379–396 e38. doi: 10.1016/j.cell.2021.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butler A., et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He S., et al. Single-cell transcriptome profiling of an adult human cell atlas of 15 major organs. Genome Biol. 2020;21(1):294. doi: 10.1186/s13059-020-02210-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madissoon E., et al. scRNA-seq assessment of the human lung, spleen, and esophagus tissue stability after cold preservation. Genome Biol. 2019;21(1):1. doi: 10.1186/s13059-019-1906-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang J.B., et al. Efficient and precise single-cell reference atlas mapping with Symphony. Nat Commun. 2021;12(1):5890. doi: 10.1038/s41467-021-25957-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 24.Yu G., et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Mering C., et al. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31(1):258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shannon P., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao Z., Dai Z., Locasale J.W. Metabolic landscape of the tumor microenvironment at single cell resolution. Nat Commun. 2019;10(1):3763. doi: 10.1038/s41467-019-11738-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.L. Kumar, E.F. M, Mfuzz: a software package for soft clustering of microarray data. Bioinformation, 2007. 2(1): p. 5-7. [DOI] [PMC free article] [PubMed]
- 29.Tang Q., et al. Dissecting hematopoietic and renal cell heterogeneity in adult zebrafish at single-cell resolution using RNA sequencing. J Exp Med. 2017;214(10):2875–2887. doi: 10.1084/jem.20170976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q., et al. Single-cell RNA-seq landscape midbrain cell responses to red spotted grouper nervous necrosis virus infection. PLoS Pathog. 2021;17(6):e1009665. doi: 10.1371/journal.ppat.1009665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tabula Muris C., et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562(7727):367–372. doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Norsworthy P.J., et al. Murine CD93 (C1qRp) contributes to the removal of apoptotic cells in vivo but is not required for C1q-mediated enhancement of phagocytosis. J Immunol. 2004;172(6):3406–3414. doi: 10.4049/jimmunol.172.6.3406. [DOI] [PubMed] [Google Scholar]
- 33.Hou X., Chen G., Zhao Y. Research progress on CD169-positive macrophages in tumors. Am J Transl Res. 2021;13(8):8589–8597. [PMC free article] [PubMed] [Google Scholar]
- 34.Chang Y.C., Nizet V. Siglecs at the host-pathogen interface. Adv Exp Med Biol. 2020;1204:197–214. doi: 10.1007/978-981-15-1580-4_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li W.H., et al. The apolipoprotein multigene family: biosynthesis, structure, structure-function relationships, and evolution. J Lipid Res. 1988;29(3):245–271. [PubMed] [Google Scholar]
- 36.Kim M.J., et al. CXCL16 positively correlated with M2-macrophage infiltration, enhanced angiogenesis, and poor prognosis in thyroid cancer. Sci Rep. 2019;9(1):13288. doi: 10.1038/s41598-019-49613-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L., et al. Chemokine CXCL16 regulates neutrophil and macrophage infiltration into injured muscle, promoting muscle regeneration. Am J Pathol. 2009;175(6):2518–2527. doi: 10.2353/ajpath.2009.090275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis Marffy A.L., McCarthy A.J. Leukocyte immunoglobulin-like receptors (LILRs) on human neutrophils: modulators of infection and immunity. Front Immunol. 2020;11:857. doi: 10.3389/fimmu.2020.00857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hernandez-Caselles T., et al. CD33 (Siglec-3) inhibitory function: role in the NKG2D/DAP10 activating pathway. J Immunol Res. 2019;2019:6032141. doi: 10.1155/2019/6032141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanchez-Martin L., et al. The chemokine CXCL12 regulates monocyte-macrophage differentiation and RUNX3 expression. Blood. 2011;117(1):88–97. doi: 10.1182/blood-2009-12-258186. [DOI] [PubMed] [Google Scholar]
- 41.Roszer T. Understanding the mysterious M2 macrophage through activation markers and effector mechanisms. Mediators Inflamm. 2015;2015 doi: 10.1155/2015/816460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marie C., et al. A whole-genome RNAi screen uncovers a novel role for human potassium channels in cell killing by the parasite Entamoeba histolytica. Sci Rep. 2015;5:13613. doi: 10.1038/srep13613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Casson C.N., et al. Human caspase-4 mediates noncanonical inflammasome activation against gram-negative bacterial pathogens. PNAS. 2015;112(21):6688–6693. doi: 10.1073/pnas.1421699112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Viola A., et al. The metabolic signature of macrophage responses. Front Immunol. 2019;10:1462. doi: 10.3389/fimmu.2019.01462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kach J., Sethakorn N., Dulin N.O. A finer tuning of G-protein signaling through regulated control of RGS proteins. Am J Phys Heart Circ Phys. 2012;303(1):H19–H35. doi: 10.1152/ajpheart.00764.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cook M.E., et al. Transcription factor Bhlhe40 in immunity and autoimmunity. Trends Immunol. 2020;41(11):1023–1036. doi: 10.1016/j.it.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goitsuka R., et al. MIST functions through distinct domains in immunoreceptor signaling in the presence and absence of LAT. J Biol Chem. 2001;276(38):36043–36050. doi: 10.1074/jbc.M106390200. [DOI] [PubMed] [Google Scholar]
- 48.Lugar P.L., et al. Molecular characterization of circulating plasma cells in patients with active systemic lupus erythematosus. PLoS One. 2012;7(9):e44362. doi: 10.1371/journal.pone.0044362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Masuda K., et al. Defining the immunological phenotype of Fc receptor-like B (FCRLB) deficient mice: confounding role of the inhibitory FcgammaRIIb. Cell Immunol. 2010;266(1):24–31. doi: 10.1016/j.cellimm.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li F.J., et al. FCRL3 promotes TLR9-induced B-cell activation and suppresses plasma cell differentiation. Eur J Immunol. 2013;43(11):2980–2992. doi: 10.1002/eji.201243068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kremlitzka M., et al. Complement receptor type 1 (CR1, CD35) is a potent inhibitor of B-cell functions in rheumatoid arthritis patients. Int Immunol. 2013;25(1):25–33. doi: 10.1093/intimm/dxs090. [DOI] [PubMed] [Google Scholar]
- 52.Jackson T.A., et al. FcR-like 2 inhibition of B cell receptor-mediated activation of B cells. J Immunol. 2010;185(12):7405–7412. doi: 10.4049/jimmunol.1002305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Polte T., et al. Critical role for syndecan-4 in dendritic cell migration during development of allergic airway inflammation. Nat Commun. 2015;6:7554. doi: 10.1038/ncomms8554. [DOI] [PubMed] [Google Scholar]
- 54.Agematsu K. Memory B cells and CD27. Histol Histopathol. 2000;15(2):573–576. doi: 10.14670/HH-15.573. [DOI] [PubMed] [Google Scholar]
- 55.Crotty S. A brief history of T cell help to B cells. Nat Rev Immunol. 2015;15(3):185–189. doi: 10.1038/nri3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Allan L.L., et al. CD1d and CD1c expression in human B cells is regulated by activation and retinoic acid receptor signaling. J Immunol. 2011;186(9):5261–5272. doi: 10.4049/jimmunol.1003615. [DOI] [PubMed] [Google Scholar]
- 57.Nagashima T., et al. Arachidonate 5-lipoxygenase establishes adaptive humoral immunity by controlling primary B cells and their cognate T-cell help. Am J Pathol. 2011;178(1):222–232. doi: 10.1016/j.ajpath.2010.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ionescu L., Urschel S. Memory B cells and long-lived plasma cells. Transplantation. 2019;103(5):890–898. doi: 10.1097/TP.0000000000002594. [DOI] [PubMed] [Google Scholar]
- 59.Gaud G., Lesourne R., Love P.E. Regulatory mechanisms in T cell receptor signalling. Nat Rev Immunol. 2018;18(8):485–497. doi: 10.1038/s41577-018-0020-8. [DOI] [PubMed] [Google Scholar]
- 60.Bixel G., et al. Mouse CD99 participates in T-cell recruitment into inflamed skin. Blood. 2004;104(10):3205–3213. doi: 10.1182/blood-2004-03-1184. [DOI] [PubMed] [Google Scholar]
- 61.Barata J.T., Durum S.K., Seddon B. Flip the coin: IL-7 and IL-7R in health and disease. Nat Immunol. 2019;20(12):1584–1593. doi: 10.1038/s41590-019-0479-x. [DOI] [PubMed] [Google Scholar]
- 62.Gui J., et al. MCL1 enhances the survival of CD8+ memory T Cells after viral infection. J Virol. 2015;89(4):2405–2414. doi: 10.1128/JVI.02480-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee J.Y., et al. The transcription factor KLF2 restrains CD4(+) T follicular helper cell differentiation. Immunity. 2015;42(2):252–264. doi: 10.1016/j.immuni.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Savage P.A., Klawon D.E.J., Miller C.H. Regulatory T cell development. Annu Rev Immunol. 2020;38:421–453. doi: 10.1146/annurev-immunol-100219-020937. [DOI] [PubMed] [Google Scholar]
- 65.Allard B., et al. The ectonucleotidases CD39 and CD73: novel checkpoint inhibitor targets. Immunol Rev. 2017;276(1):121–144. doi: 10.1111/imr.12528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walker L.S. Regulatory T cells: folate receptor 4: a new handle on regulation and memory? Immunol Cell Biol. 2007;85(7):506–507. doi: 10.1038/sj.icb.7100115. [DOI] [PubMed] [Google Scholar]
- 67.Barral P., et al. The location of splenic NKT cells favours their rapid activation by blood-borne antigen. EMBO J. 2012;31(10):2378–2390. doi: 10.1038/emboj.2012.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Laing K.J., Hansen J.D. Fish T cells: recent advances through genomics. Dev Comp Immunol. 2011;35(12):1282–1295. doi: 10.1016/j.dci.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 69.Buchmann K. Evolution of innate immunity: clues from invertebrates via fish to mammals. Front Immunol. 2014;5:459. doi: 10.3389/fimmu.2014.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wiegertjes G.F., et al. Polarization of immune responses in fish: the 'macrophages first' point of view. Mol Immunol. 2016;69:146–156. doi: 10.1016/j.molimm.2015.09.026. [DOI] [PubMed] [Google Scholar]
- 71.Wentzel A.S., et al. Transcriptome sequencing supports a conservation of macrophage polarization in fish. Sci Rep. 2020;10(1):13470. doi: 10.1038/s41598-020-70248-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rauta P.R., Nayak B., Das S. Immune system and immune responses in fish and their role in comparative immunity study: a model for higher organisms. Immunol Lett. 2012;148(1):23–33. doi: 10.1016/j.imlet.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 73.Flajnik M.F. A cold-blooded view of adaptive immunity. Nat Rev Immunol. 2018;18(7):438–453. doi: 10.1038/s41577-018-0003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Toda H., et al. Conservation of characteristics and functions of CD4 positive lymphocytes in a teleost fish. Dev Comp Immunol. 2011;35(6):650–660. doi: 10.1016/j.dci.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 75.Wu L., et al. Recent advances on phagocytic B cells in teleost fish. Front Immunol. 2020;11:824. doi: 10.3389/fimmu.2020.00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhu L.Y., et al. B cells in teleost fish act as pivotal initiating APCs in priming adaptive immunity: an evolutionary perspective on the origin of the B-1 cell subset and B7 molecules. J Immunol. 2014;192(6):2699–2714. doi: 10.4049/jimmunol.1301312. [DOI] [PubMed] [Google Scholar]
- 77.Li J., et al. B lymphocytes from early vertebrates have potent phagocytic and microbicidal abilities. Nat Immunol. 2006;7(10):1116–1124. doi: 10.1038/ni1389. [DOI] [PubMed] [Google Scholar]
- 78.Overland H.S., et al. Phagocytosis by B-cells and neutrophils in Atlantic salmon (Salmo salar L.) and Atlantic cod (Gadus morhua L.) Fish Shellfish Immunol. 2010;28(1):193–204. doi: 10.1016/j.fsi.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 79.Zimmerman L.M., et al. Phagocytic B cells in a reptile. Biol Lett. 2010;6(2):270–273. doi: 10.1098/rsbl.2009.0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Parra D., et al. Pivotal advance: peritoneal cavity B-1 B cells have phagocytic and microbicidal capacities and present phagocytosed antigen to CD4+ T cells. J Leukoc Biol. 2012;91(4):525–536. doi: 10.1189/jlb.0711372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gao J., et al. Novel functions of murine B1 cells: active phagocytic and microbicidal abilities. Eur J Immunol. 2012;42(4):982–992. doi: 10.1002/eji.201141519. [DOI] [PubMed] [Google Scholar]
- 82.He J., et al. IRF-7 Is a critical regulator of Type 2 innate lymphoid cells in allergic airway inflammation. Cell Rep. 2019;29(9):2718–2730 e6. doi: 10.1016/j.celrep.2019.10.077. [DOI] [PubMed] [Google Scholar]
- 83.Poholek C.H., et al. IL-21 controls ILC3 cytokine production and promotes a protective phenotype in a mouse model of colitis. Immunohorizons. 2019;3(6):194–202. doi: 10.4049/immunohorizons.1900005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nechanitzky R., et al. Transcription factor EBF1 is essential for the maintenance of B cell identity and prevention of alternative fates in committed cells. Nat Immunol. 2013;14(8):867–875. doi: 10.1038/ni.2641. [DOI] [PubMed] [Google Scholar]
- 85.Hernandez P.P., et al. Single-cell transcriptional analysis reveals ILC-like cells in zebrafish. Sci Immunol. 2018;3(29) doi: 10.1126/sciimmunol.aau5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chimpanzee S., Analysis C. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437(7055):69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 87.Yue F., et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515(7527):355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Klein S.L., Flanagan K.L. Sex differences in immune responses. Nat Rev Immunol. 2016;16(10):626–638. doi: 10.1038/nri.2016.90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data for single cell RNA-seq samples are available in the Gene Expression Omnibus (GEO) database as accession number GSE186158 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE186158).