Abstract
Transcription-replication conflict is a major cause of replication stress that arises when replication forks collide with the transcription machinery. Replication fork stalling at sites of transcription compromises chromosome replication fidelity and can induce DNA damage with potentially deleterious consequences for genome stability and organismal health. The block to DNA replication by the transcription machinery is complex and can involve stalled or elongating RNA polymerases, promoter-bound transcription factor complexes or DNA topology constraints. In addition, studies over the past two decades have identified co-transcriptional R-loops as a major source for the impairment of DNA replication forks at active genes. However, how R-loops impede DNA replication at the molecular level is incompletely understood. Current evidence suggests that RNA:DNA hybrids, DNA secondary structures, stalled RNA polymerases and condensed chromatin states associated with R-loops contribute to the impediment of fork progression. Moreover, since both R-loops and replication forks are intrinsically asymmetric structures, the outcome of R-loop-replisome collisions is influenced by collision orientation. Collectively, the data suggest that the impact of R-loops on DNA replication is highly dependent on their specific structural composition. Here, we will summarize our current understanding of the molecular basis for R-loop-induced replication fork progression defects.
Keywords: DNA replication, R-loops, transcription, RNA polymerase, genome stability
Introduction
R-loops are three-stranded nucleic acid structures that form when RNA binds to DNA to create a stable RNA:DNA hybrid, leaving a single-stranded DNA (ssDNA) loop exposed. R-loops have been observed in all domains of life and are involved in biological processes, such as transcriptional regulation, immunoglobulin (Ig) class switch recombination (CSR), chromosome segregation, telomere maintenance and DNA replication. However, if occurring in an unscheduled manner, they can compromise genome stability and contribute to human diseases, including cancer, neurodegenerative disorders, and autoimmune diseases (Richard and Manley 2017; Garcia-Muse and Aguilera 2019; Wells et al. 2019; Brickner et al. 2022; Petermann et al. 2022). Understanding the etiology of pathological R-loops and their impact on genome integrity is, therefore, relevant to human health and may ultimately help uncover therapeutic vulnerabilities in associated diseases.
R-loops most commonly form during transcription when the nascent transcript threads back into the DNA template upstream of the RNA polymerase (RNAP). Normally, R-loop levels are tightly controlled by a host of cellular proteins that prevent the formation of R-loops or mediate their resolution. However, when R-loop homeostasis is perturbed, unscheduled R-loops accumulate across the genome, threatening genome stability. One way in which R-loops compromise genome stability is by impeding DNA replication. Since RNAPs and replisomes move processively along DNA, collisions between transcription and replication can occur in a co-directional (CD) or head-on (HO) orientation (Figure 1). In the CD orientation, RNA:DNA hybrids are located on the leading strand template and the displaced ssDNA loop forms the lagging strand template, whereas in the HO orientation, RNA:DNA hybrids occur on the lagging strand and the displaced ssDNA loop forms the leading strand template. Although the genome-destabilizing potential of R-loops is well-documented, the molecular mechanisms involved are incompletely understood. In part, this is due to the fact that cellular R-loops are structurally diverse, featuring distinct sizes, DNA secondary structures and chromatin states that may each individually influence fork progression and are, therefore, difficult to parse in vivo. In addition, common approaches, such as 2D gel-analysis of replication intermediates or DNA fiber labeling to monitor replication fork progression in vivo, are limited in their ability to resolve differential defects in replisome progression, leading and lagging strand synthesis. Recent biochemical studies involving reconstituted E. coli and budding yeast replisomes have begun to address these issues and characterize fork collisions with R-loops at the molecular level. Here, we will review the mechanisms implicated in the block to DNA replication at R-loops.
Figure 1. Co-directional (CD) and head-on (HO) collisions between replisomes and co-transcriptional R-loops in bacteria and eukaryotes.
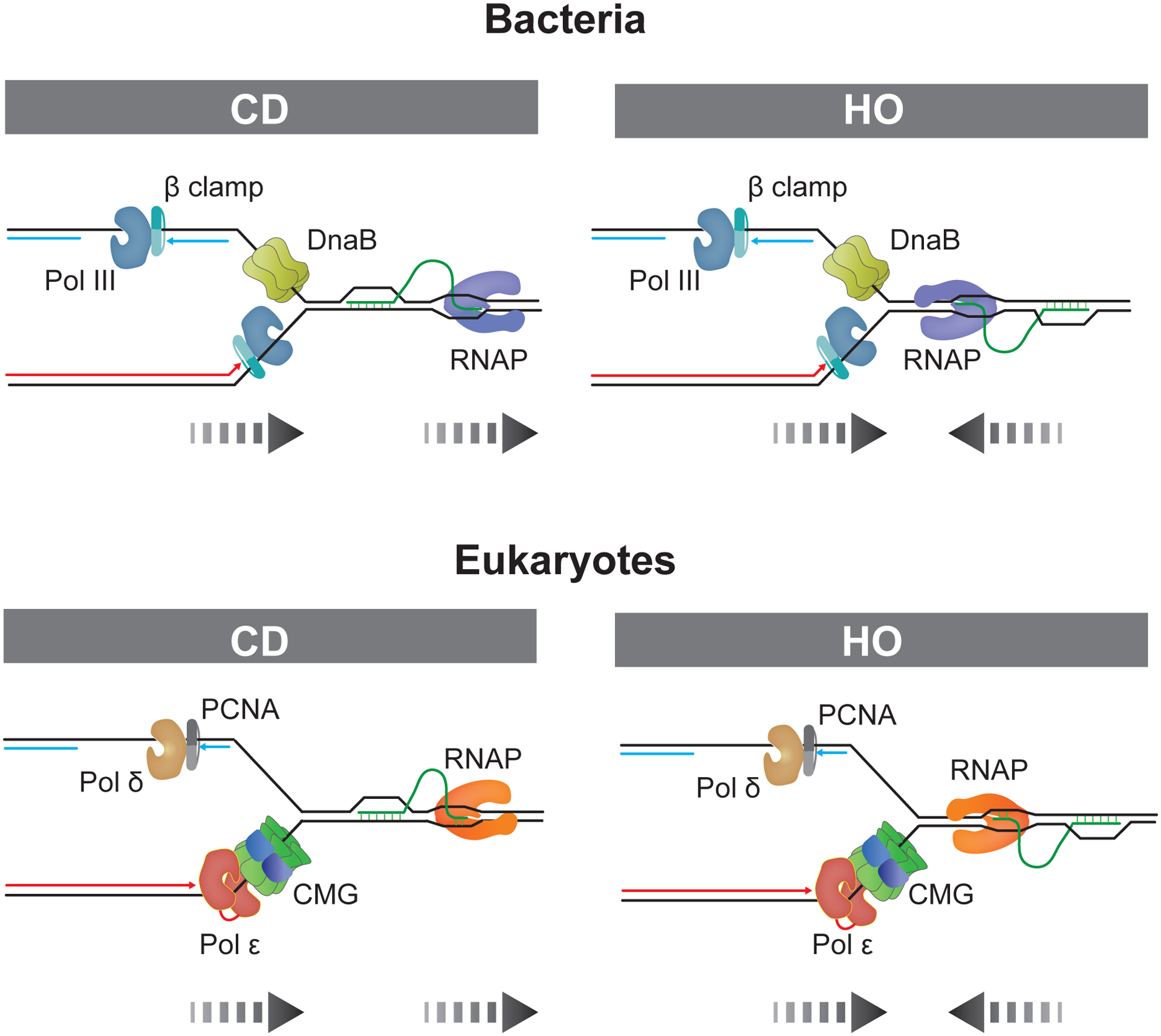
Parental DNA strands are indicated in black, nascent leading and lagging strands are in red and blue, respectively. Dashed arrows represent directions of replication and transcription.
R-loops form co-transcriptionally across the genome
Initially, the ability of RNA molecules to hybridize to dsDNA and form R-loops was demonstrated in vitro and developed as a tool to physically map gene positions in DNA templates by electron microscopy (Thomas et al. 1976). RNA hybridization to dsDNA in these R-loop mapping studies was mediated by reaction conditions that promote the denaturation of dsDNA, such as 70 % formamide and elevated temperature. However, subsequent biochemical studies demonstrated that stable R-loops could also form under native conditions as a product of transcription (Itoh and Tomizawa 1980; Kadesch and Chamberlin 1982; Reaban and Griffin 1990). While R-loop formation in trans, i.e., via invasion of the DNA by an RNA molecule post transcription, has been documented at specific chromosomal loci involved in gene-regulation (Cloutier et al. 2016; Ariel et al. 2020), telomere length control (Feretzaki et al. 2020), or CSR (Ribeiro de Almeida et al. 2018), and either cis or trans mechanisms have been observed in cellular reporter systems (Wahba et al. 2013; Lafuente-Barquero et al. 2020), evidence from genomic R-loop profiling studies suggests that R-loops most commonly form co-transcriptionally in cis.
Methods to map R-loops generally rely on the RNA:DNA hybrid-specific S9.6 monoclonal antibody or catalytically inactive forms of RNase H to isolate RNA:DNA hybrids by affinity purification for high-throughput sequence analyses (Ginno et al. 2012; Chen et al. 2017). Genomic R-loop locations have also been deduced from S9.6 or RNase H binding sites mapped by cleavage under targets and release using nuclease (CUT&RUN) (Yan et al. 2019), cleavage under targets and tagmentation (CUT&Tag) (Wang et al. 2021; Lyu et al. 2022), or RNase H UV crosslinking and analysis of cDNA (H-CRAC) (Aiello et al. 2022). An orthologous approach exploits the sensitivity of exposed cytosine bases in the displaced ssDNA to chemical deamination by bisulfite, which results in cytosine-to-uracil conversions (Yu et al. 2003). R-loops will characteristically exhibit cytosine conversions specifically on one strand, while ssDNA generated by other forms of DNA unwinding will generally expose cytosines on both strands. Bisulfite treatment may also be coupled to RNA:DNA hybrid affinity purification (Dumelie and Jaffrey 2017; Wulfridge and Sarma 2021).
Collectively, the genomic profiles reveal that R-loops are widely distributed, covering 3–10% of eukaryotic genomes (Ginno et al. 2012; Ginno et al. 2013; Sanz et al. 2016; Wahba et al. 2016; Chen et al. 2017; Xu et al. 2017; Sanz and Chedin 2019; Crossley et al. 2020; Promonet et al. 2020; Wang et al. 2021; Lyu et al. 2022). While this represents a population average, recent estimates suggest a steady state level of ~ 300 R-loops per human cell (Crossley et al. 2020). Given an average R-loop half-life of 10–15 minutes, this corresponds to thousands of R-loops being formed and resolved per human cell cycle (Brickner et al. 2022). R-loop-forming regions cover several hundred and up to a few thousand base pairs. However, single molecule analyses of human R-loops using long-range sequencing indicate that R-loop-forming regions often encompass several individual R-loops averaging 200–400 bp in length, with few R-loops exceeding 2,000 bp (Malig et al. 2020). This agrees with a recent electron-microscopic analysis of human cellular R-loops (Stoy et al. 2023).
The genomic R-loop profiles support the co-transcriptional origin of cellular R-loops in multiple ways. First and foremost, a large fraction of genomic R-loops maps to genic regions and features RNA corresponding to the annotated gene transcripts. R-loop levels also often peak at gene boundaries, i.e., transcription start sites (TSSs) and transcription termination sites (TTSs), and promoter-proximal R-loops generally exhibit well-defined 5’ ends that coincide with the TSS (Sanz et al. 2016; Dumelie and Jaffrey 2017; Crossley et al. 2020; Promonet et al. 2020). Moreover, R-loops globally correlate with gene activity, open chromatin, and active promoter and enhancer states (Sanz et al. 2016; Chen et al. 2017; Lyu et al. 2022). Accordingly, R-loops are prevalent at highly expressed loci, such as rRNA and tRNA genes (El Hage et al. 2010; Chan et al. 2014; Wahba et al. 2016; Abraham et al. 2020). Finally, R-loops often extend across exon-intron junctions, indicating that they are formed from unspliced pre-mRNA (Chen et al. 2017; Sanz and Chedin 2019; Malig et al. 2020). Despite the predominant sense-orientation of R-loops across gene bodies, anti-sense R-loops also occur near promoters, where they are involved in transcriptional regulation (Boque-Sastre et al. 2015; Sanz et al. 2016; Dumelie and Jaffrey 2017; Wulfridge and Sarma 2021). Moreover, R-loops can promote antisense transcription, likely through promoter-independent transcription of the displaced ssDNA loop, to control gene-regulation and transcription termination (Skourti-Stathaki et al. 2014; Tan-Wong et al. 2019).
Molecular determinants of co-transcriptional R-loop formation
The co-transcriptional formation of R-loops in cis may be unexpected as RNAPs from all domains of life are designed to prevent extensive hybridization of nascent RNA with template DNA by unpeeling the nascent transcript from the template DNA strand and directing it out of the RNAP core through a specialized RNA exit channel that is distinct from the DNA exit channel (Westover et al. 2004; Vassylyev et al. 2007; Bernecky et al. 2016). Consequently, nascent RNA needs to thread back into the DNA upon exiting the RNAP to form an R-loop. This mechanism is supported by the sensitivity of co-transcriptional R-loops to RNase T1, which specifically degrades single-stranded RNA (Roy et al. 2008). Although the thread-back mechanism is likely the predominant mechanism of co-transcriptional R-loop formation, an alternative mechanism involves the formation of R-loops on the anterior side of RNAPs following RNAP backtracking (Zatreanu et al. 2019). The threading of nascent RNA into DNA depends on the ability of the RNA to pass between separated DNA strands upstream of RNAP and wind around the template DNA strand. Accordingly, R-loop formation may be disfavored at greater distances from the TSS as the ability of the 5’ RNA tail to pass between locally unpaired DNA strands will decrease with the size of the 5’ RNA tail (Castillo-Guzman and Chedin 2021; Belotserkovskii and Hanawalt 2022). Conversely, conditions that aid co-transcriptional R-loop formation (i) increase the conformational flexibility of the nascent RNA emerging from the RNAP, (ii) destabilize the DNA duplex in the wake of elongating RNAP, or (iii) promote the stability of RNA:DNA hybrids relative to their dsDNA counterparts.
The ability of nascent RNA to invade the template DNA is promoted by conditions that prevent the physical sequestration of mRNA chains in co-transcriptional mRNA processing complexes. For example, in bacteria, nascent mRNA is translated by ribosomes that track closely behind RNAP. Consequently, R-loop formation in E. coli is enhanced by the inhibition of co-transcriptional translation (Masse and Drolet 1999). In eukaryotes, pre-mRNA is co-transcriptionally processed into mature mRNA by multi-subunit protein complexes that mediate the capping, splicing, 3’ cleavage and polyadenylation of the nascent transcript, as well as the packaging of the mature mRNA into ribonucleoprotein particles, mRNPs, for export to the cytoplasm (Bentley 2014). Consequently, mutations in eukaryotic mRNA processing factors can increase cellular R-loop levels and R-loop-dependent genome instability. This was first demonstrated for mutations in the Saccharomyces cerevisiae (S. cerevisiae) THO/TREX complex, which licenses mRNAs for nuclear export, and subsequently supported by observations in chicken DT40 cells depleted for the ASF/SF2 splicing factor (Huertas and Aguilera 2003; Li and Manley 2005). Since then, many additional mutations in mRNA biogenesis factors with similar phenotypes have been identified (Garcia-Muse and Aguilera 2019). The observation that spliceosome recruitment in the absence of splicing is sufficient to attenuate R-loop formation supports the notion that physical sequestration of the transcript and not mRNA processing in itself underlies the prevention of co-transcriptional R-loop formation (Bonnet et al. 2017).
As described in the ‘twin supercoil domain’ model, transcription of topologically constrained DNA generates negative DNA supercoils in the wake of RNAP and positive supercoils ahead of it (Liu and Wang 1987). The underwound state of negatively supercoiled DNA may promote R-loop formation by inducing local DNA strand separation. Consistent with this model, negative DNA supercoiling has been linked to R-loop formation in E. coli and promotes co-transcriptional R-loop formation in vitro (Drolet et al. 1994; Stolz et al. 2019). Alternatively, based on the fact that R-loops absorb negative supercoils, it has been suggested that R-loop formation is favored on negatively supercoiled DNA because of the ability of R-loops to convert topologically strained DNA into an energetically more stable relaxed state (Chedin and Benham 2020). Supporting the in vitro data, depletion of DNA topoisomerases that relax DNA supercoils globally drives R-loop formation in bacteria and eukaryotic cells (Masse and Drolet 1999; Tuduri et al. 2009; El Hage et al. 2010).
Finally, R-loop formation is significantly influenced by the DNA sequence at transcribed loci. In general, R-loop formation is promoted by G-richness of the RNA strand due to the exceptional thermodynamic stability of rG:dC base pairs (Stolz et al. 2019). Accordingly, GC-richness coupled to G/C-skew is a prevalent feature of R-loops across the human genome (Ginno et al. 2012; Ginno et al. 2013). This feature was noted early on at Ig switch (S) regions, which form R-loops far more efficiently when transcribed in the physiological orientation, i.e., when producing a G-rich transcript (Reaban et al. 1994; Daniels and Lieber 1995; Shinkura et al. 2003). However, high G-content in the RNA alone was found not to be sufficient for R-loop formation at Ig S regions in vitro. Instead, efficient R-loop formation was specifically promoted by G clustering (Roy et al. 2008). While certain G clusters in the displaced ssDNA of R-loops can form G-quadruplexes (G4s) that stabilize R-loops (Duquette et al. 2004), G clusters at Ig S regions promote R-loop formation even under conditions that disfavor G4s, suggesting that they act instead as nucleation sites for the hybridization of the RNA to DNA (Roy et al. 2008). Once formed, G density in the RNA without clustering is sufficient to stabilize the R-loop (Roy and Lieber 2009). Of note, A/T skew and poly(A) tracts rather than G/C skew appear to be associated with hybrid-prone regions in the budding yeast genome (Wahba et al. 2016).
R-loops induce replication-dependent and -independent genome instability
R-loops threaten genome integrity in multiple ways. One inherently destabilizing feature of R-loops is their partially single-stranded structure, which renders them susceptible to nucleolytic degradation. For example, R-loop junctions are targets for the structure-specific nucleotide excision repair endonucleases XPF and XPG (Tian and Alt 2000; Sollier et al. 2014). Processing of R-loops by XPF and XPG may generate single-stranded DNA breaks (SSBs) or double-stranded DNA breaks (DSBs), depending on which strands are targeted by XPF and XPG, respectively. Alternatively, XPF/XPG-induced SSBs may be converted into DSBs by DNA replication. DSBs may also result from XPF/XPG-induced SSBs when these occur close to SSBs on the opposite strand, as has been suggested for R-loops accumulating at transcription-stalling topoisomerase 1 cleavage complexes (TOP1ccs) that are processed into SSBs by TDP1 (Cristini et al. 2019). Intriguingly, R-loop processing by XPF/XPG can induce an autoimmune response linked to Aicardi-Goutières syndrome, a neuroinflammatory disease, suggesting that R-loop cleavage may induce the release of immunogenic ssDNA or RNA:DNA fragments into the cytoplasm (Lim et al. 2015; Cristini et al. 2022). Consistent with this notion, XPF/XPG activity was recently demonstrated to promote the accumulation of nuclear R-loop-derived RNA:DNA hybrids in the cytoplasm of human cells with defective R-loop homeostasis, activating an innate immune response (Crossley et al. 2023).
DNA breaks at R-loops may also be triggered by the modification or damage of bases exposed in the displaced ssDNA and their subsequent processing by DNA repair enzymes. For example, the base excision repair (BER) machinery has been linked to DNA breaks at R-loops following the deamination of cytosines in the displaced ssDNA by cytosine deaminases, such as mammalian activation-induced deaminase (AID) or yeast Fcy1 (Gomez-Gonzalez and Aguilera 2007; Su and Freudenreich 2017). Uracils resulting from cytosine deamination are excised by uracil DNA glycosylase to generate abasic sites that in turn are processed into SSBs by an apurinic/apyrimidinic (AP) endonuclease. In theory, multiple such ssDNA breaks or nicks may be converted into DSBs if they occur in close proximity and on opposite strands. In addition, R-loops may be processed into DSBs by mismatch repair (MMR) nucleases following cytosine deamination (Su and Freudenreich 2017). The prevalence of such repair-induced DNA breaks at R-loops under physiological conditions remains to be determined, as AID expression is effectively restricted to B cells, where it targets R-loops at Ig S regions to induce CSR. However, AID also targets non-Ig genes in B cells to induce translocations to Ig S regions associated with B cell lymphomas, which was recently shown to be promoted by R-loops induced by the loss of TET family cytosine dioxygenases (Robbiani et al. 2008; Chiarle et al. 2011; Shukla et al. 2022). Whether related cytosine deaminases of the APOBEC family target R-loops in other cell types is not known.
While the presence of ssDNA accounts for some of the instability of R-loops, a major source of R-loop-induced genome instability was revealed by the finding that co-transcriptional R-loops impede normal fork progression. In one study, depletion of Topo I in mammalian cells resulted in transcription-dependent and RNase H1-sensitive DNA breaks and replication defects (Tuduri et al. 2009). Notably, DNA breaks resulting from Topo I deficiency occurred predominantly in S phase, suggesting a causative role for DNA replication in R-loop-mediated genome instability. In another study, co-transcriptional R-loops in E. coli were shown to induce a block to replication fork progression as well as replication-dependent recombination and genome instability, while R-loop induction in HeLa cells by depletion of the splicing factor SRSF1 caused S phase-specific DNA damage and replication impairment (Gan et al. 2011). These findings agreed with prior studies in budding yeast, demonstrating that transcription-associated recombination (TAR) is mediated by replication fork impairment and is dependent on R-loops (Huertas and Aguilera 2003; Prado and Aguilera 2005; Wellinger et al. 2006). Together, these studies established that R-loops could induce genome instability due to replication fork stalling during transcription-replication conflict (TRC).
Co-incidence of replication and transcription can cause transcription-replication conflict
TRC is a major inducer of genome instability in both bacteria and eukaryotes (Lang and Merrikh 2018; Gomez-Gonzalez and Aguilera 2019). In bacteria, transcription-replication collisions are inevitable as these processes are not temporally separated and replication fork progression rates greatly exceed those of RNAP (Mirkin and Mirkin 2005). In contrast, in eukaryotes the spatial and temporal compartmentalization of transcription and replication permits at least partial mitigation of TRC (Lalonde et al. 2021). However, certain long genes associated with common fragile sites (CFSs) require more than one cell cycle to complete transcription and many genes required for cell proliferation are specifically expressed in S phase, limiting the possibility to confine transcription and replication to separate subnuclear compartments (Helmrich et al. 2011). Despite the apparent inevitability of collisions between DNA replication and transcription, eukaryotic replication forks progress at largely uniform rates across most of the genome, indicating that transcription is not a major obstacle for DNA replication under normal conditions (Sekedat et al. 2010; Claussin et al. 2022). Consistent with these observations, a recent study employing a live cell imaging approach in budding yeast to monitor fork progression across a galactose-inducible reporter gene found that transcription did not present a major obstacle to replication forks in wild-type cells (Tsirkas et al. 2022). However, as discussed below, uninterrupted fork progression is highly dependent on ancillary proteins, in particular DNA helicases. Thus, transcription does present a potential barrier to replication forks.
In eukaryotes, DNA helicases implicated in facilitating replisome progression through active genes include Rrm3 and Sen1/senataxin, while DinG, Rep, UvrD and PcrA perform analogous functions in bacteria (Ivessa et al. 2003; Azvolinsky et al. 2009; Guy et al. 2009; Boubakri et al. 2010; Alzu et al. 2012; Merrikh et al. 2015; Osmundson et al. 2017; Tran et al. 2017; Hawkins et al. 2019; Aiello et al. 2022; Claussin et al. 2022). In addition to ancillary helicases, cells employ a variety of additional mechanisms to mitigate transcription-replication collisions. For example, in eukaryotes, transcription-coupled H3K4 methylation (H3K4me) can slow down replication forks to buffer active genes from TRC (Chong et al. 2020). Topoisomerase I has been suggested to buffer gene bodies from TRC by promoting the stable pausing of replication forks at the 3’ end of HO-oriented genes (Promonet et al. 2020). Replisome progression can also be controlled directly to limit TRC. For example, stress-activated protein kinases (SAPKs) can slow down replication forks by phosphorylating Mrc1/Claspin in response to environmental stress and unscheduled transcription (Duch et al. 2013; Duch et al. 2018; Ulsamer et al. 2022). Fork progression is also controlled by the replication checkpoint, which may slow down replication forks in response to TRC by inducing the phosphorylation of Mrc1 and Mcm10 (Seiler et al. 2007; Gomez-Gonzalez et al. 2009; Hamperl et al. 2017; Bacal et al. 2018; Devbhandari and Remus 2020; Promonet et al. 2020; Frattini et al. 2021; McClure and Diffley 2021). Conversely, the replication checkpoint may also down-regulate gene expression to limit TRC under conditions of replication stress (Nguyen et al. 2010).
Collision orientation influences transcription-replication conflict
Since RNAPs encircle both DNA strands during transcription, they may be expected to present an impediment to replication forks irrespective of orientation. However, in principle, replisomes might simply follow behind RNAPs moving in the same direction. While this may happen in eukaryotes, where RNAP and replisomes progress at similar rates, bacterial replisomes advance at a ~ 20x greater rate than RNAP, making CD collisions in bacteria inevitable (Mirkin and Mirkin 2005). Moreover, RNAP progression during transcription is often punctuated by distinct pauses, which increases the potential for CD RNAP-replisome collisions. For example, promoter-proximal RNAP pausing occurs following transcription initiation but prior to elongation, while RNAP pausing is also specifically induced during elongation to coordinate transcription with RNA folding, processing, translation or repair (Mayer et al. 2017). A particularly stable form of RNAP stalling involves RNAP backtracking, which is induced by the misincorporation of rNTPs or when RNAP encounters physical obstacles and involves the disengagement of the RNA 3’ end from the RNAP catalytic site. Consequently, backtracked RNAPs exacerbate replisome-RNAP collisions, causing DSBs specifically during CD collisions in E. coli (Dutta et al. 2011). Similarly, RECQL5, a RECQ family helicase that controls transcription by suppressing RNAP stalling, alleviates TRC in human cells, indicating that stalled RNAPs pose a strong impediment to replication forks also in eukaryotes (Saponaro et al. 2014; Urban et al. 2016).
Although studies in Bacillus subtilis (B. subtilis) demonstrate that CD collisions can challenge uninterrupted fork progression (Merrikh et al. 2011; Mangiameli et al. 2017), HO transcription-replication collisions are generally considered a greater obstacle to replication forks. This had been hypothesized early on based on the observation that genes in the E. coli genome are predominantly co-oriented with replication (Brewer 1988) and was subsequently confirmed experimentally in an electron-microscopic study monitoring fork progression through the highly transcribed rrnB ribosomal RNA operon in E. coli (French 1992). Several later studies in both E. coli and B. subtilis confirmed the increased stalling of replication forks at HO-oriented genes and demonstrated that HO TRC correlates with reduced replication fidelity and genome stability (Mirkin and Mirkin 2005; Wang et al. 2007; Srivatsan et al. 2010; Paul et al. 2013; Sankar et al. 2016; Lang et al. 2017). While several of these studies analyzed TRC at genes featuring RNAP arrays, biochemical studies with reconstituted E. coli or T4 replisomes showed that even single stalled RNAPs could block replisomes in the HO orientation, whereas RNAPs in the CD orientation presented a minor obstacle (Liu et al. 1993; Liu and Alberts 1995; Pomerantz and O’Donnell 2008; Pomerantz and O’Donnell 2010).
Early studies in yeast demonstrated that eukaryotic replication forks are also more sensitive to stalling during HO encounters with highly active genes, such as tRNA genes (Deshpande and Newlon 1996). Although a later genome-wide study found that replication fork stalling at tRNA genes is not strictly dependent on collision orientation, a greater proportion of HO collisions caused fork stalling than CD collisions (Osmundson et al. 2017). HO transcription-replication collisions in yeast were also found to induce significantly higher levels of transcription-associated mutagenesis and recombination than CD collisions (Prado and Aguilera 2005; Kim et al. 2007). Similarly, at yeast ribosomal RNA gene (rDNA) repeats, where polar replication fork barriers (RFBs) ensure the co-orientation of transcription and DNA replication, loss of co-orientation in the absence of functional RFBs resulted in increased recombination (Takeuchi et al. 2003).
Genome-wide replication profiles also reveal a bias towards the co-orientation of transcription and replication in human cells, with replication origins often situated upstream of highly expressed genes and replication termination occurring preferentially downstream of HO-oriented genes (Petryk et al. 2016; Chen et al. 2019). Consequently, perturbation of this organization by oncogene activation or replication stress is linked to genome instability (Barlow et al. 2013; Kotsantis et al. 2016; Stork et al. 2016; Chen et al. 2019). A direct assessment of the importance of collision orientation on TRC was hampered for a long time by the lack of origin specificity in higher eukaryotes. This problem was eventually overcome in human cells with the help of episomes harboring the Epstein-Barr virus replication origin (oriP), which directs the site-specific establishment of unidirectional replication forks (Hamperl et al. 2017). Using this approach, HO transcription-replication collisions were demonstrated to reduce transcription and decrease plasmid stability to a greater degree than CD collisions.
Molecular impediments to replication forks at active genes
Transcription can clearly form an impediment to replication forks that imposes a dependency on ancillary factors and specific genome organization to ensure error-free and complete genome replication. However, the molecular mechanisms underlying TRC remain incompletely understood and may indeed be diverse. Biochemical studies involving reconstituted replisomes suggest that RNAPs sterically block replisome progression (Liu and Alberts 1995; Pomerantz and O’Donnell 2010; Bruning and Marians 2020) (Figure 2a). This is consistent with the observation that mutations that stabilize RNAP II on chromatin exacerbate TRC, while factors that promote the removal of stalled RNAPII attenuate TRC (Felipe-Abrio et al. 2015; Poli et al. 2016; Landsverk et al. 2020; Tsirkas et al. 2022). In addition to RNAP, promoter-bound transcription factor complexes can also create a block to replication forks independently of transcription (Yeung and Smith 2020) (Figure 2b). Moreover, superhelical strain accumulating in the DNA between opposing replisomes and RNAPs or resulting from the physical tethering of active genes to nuclear pore complexes may also stall replisomes (Bermejo et al. 2011; Promonet et al. 2020; Lang and Merrikh 2021; Tsirkas et al. 2022) (Figure 2c). Prolonged stalling of replication forks at DNA supercoils may eventually cause fork collapse due to degradation by structure-specific nucleases, such as Mus81 (Matos et al. 2020; Promonet et al. 2020). Positive torsional strain building up in the DNA between converging RNAPs and replisomes may also be absorbed by reversal of stalled replication forks, generating potentially unstable four-way junctions (Postow et al. 2001). Since fork reversal underpins fork slow-down during replication stress, fork stalling may be an active process mediated by fork remodeling enzymes and not just a consequence of steric inhibition (Vujanovic et al. 2017; Chappidi et al. 2020; Andrs et al. 2023; Stoy et al. 2023) (Figure 2d).
Figure 2: R-loop-independent blocks to replication fork progression during transcription-replication conflict.
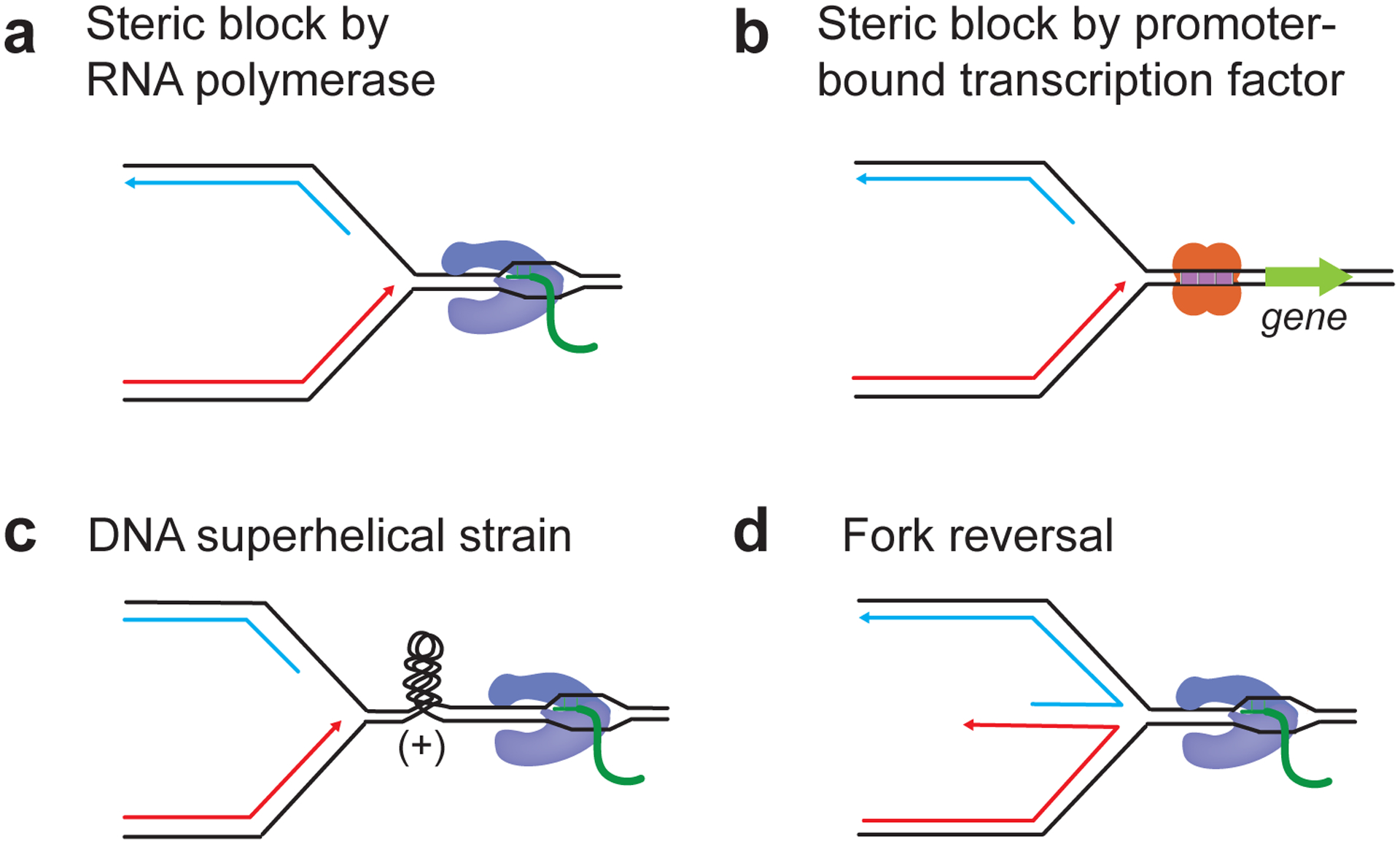
a, Steric inhibition of fork progression by RNA polymerase. b, Steric inhibition of fork progression promoter-bound transcription factor complexes. c, Fork stalling due to positive torsional strain accumulating between converging replication and transcription machineries. d, Fork stalling due to active fork reversal mediated by fork remodeling enzymes.
R-loops exacerbate transcription-replication conflict
Importantly, the observation that RNase H overexpression can rescue replication defects and TRC-associated genome instability identified R-loops as another source for the block to fork progression at active genes. In eukaryotes, this was originally demonstrated in cells harboring mutations in Topo I, the splicing factor SRSF1, or the mRNA export licensing complex THO/TREX (Wellinger et al. 2006; Tuduri et al. 2009; Gan et al. 2011). Since then, many more proteins impacting R-loop homeostasis and R-loop-dependent genome instability have been identified (Garcia-Muse and Aguilera 2019; Brickner et al. 2022). Globally, these can be grouped into four broad functional categories: Factors that (i) prevent the formation of R-loops, (ii) promote the formation of R-loops, (iii) resolve R-loops, or (iv) stabilize R-loops.
Factors preventing the formation of R-loops include proteins involved in mRNA processing and mRNP biogenesis (Garcia-Muse and Aguilera 2019). As described above, mutational inactivation of these complexes may physically expose nascent RNA and thus facilitate RNA:DNA hybridization. Alternatively, the dissociation of pre-mRNA processing factors may also be induced by RNAP pausing at DNA lesions to facilitate RNAP backtracking, with similar effects on R-loop formation (Tresini et al. 2015). Accordingly, proteins preventing extended pausing of RNAP elongation complexes, such as RECQL5, or promoting transcript release from paused RNAP, such as Sen1/senataxin, Xrn2 and BRCA1, also prevent R-loop formation (Mischo et al. 2011; Skourti-Stathaki et al. 2011; Saponaro et al. 2014; Hatchi et al. 2015; Morales et al. 2016; Urban et al. 2016). In bacteria, the physical association of the PcrA helicase with RNAP near the RNA and DNA exit channels may actively suppress R-loops by preventing RNA:DNA hybridization (Urrutia-Irazabal et al. 2021). R-loop formation is also suppressed by proteins inhibiting the formation of ssDNA. In addition to topoisomerases mentioned earlier these include PrimPol, which suppresses ssDNA formation by promoting leading strand restart following replisome uncoupling from leading strand synthesis (Svikovic et al. 2019). R-loop homeostasis in eukaryotes is also greatly influenced by chromatin regulators, such as histone chaperones, chromatin remodelers, and histone modifiers (Bayona-Feliu and Aguilera 2021). Whether these act directly on R-loops or indirectly control chromatin access of R-loop processing factors remains to be determined.
Several proteins, such as Rad51AP1 at telomeres and DSBs (Ouyang et al. 2021; Kaminski et al. 2022; Yadav et al. 2022), PRC2 at polycomb response elements (PREs) (Alecki et al. 2020), or Ddx1 at Ig S regions (Ribeiro de Almeida et al. 2018), drive R-loop formation by facilitating RNA:DNA hybridization. However, these cases occur in non-pathological contexts and may not involve co-transcriptional R-loops. Curiously, although the DHX9 helicase preferentially unwinds RNA:DNA hybrids in vitro and suppresses CPT-induced R-loops in human cells, loss of DHX9 reduces pathological R-loops in cells defective for splicing factors (Chakraborty and Grosse 2011; Chakraborty et al. 2018; Cristini et al. 2018). To reconcile these observations, DHX9 was proposed to resolve RNA secondary structure in nascent RNA, which may normally promote mRNP assembly but promote RNA:DNA hybridization in the absence of splicing factors.
Once formed, R-loops can be resolved by the unwinding or degradation of RNA:DNA hybrids. RNA:DNA hybrid unwinding is catalyzed by RNA/DNA helicases or branchpoint translocases. Examples for the latter include FANCM in eukaryotes and RecG in bacteria, which may extrude RNA strands from hybrids by catalyzing the annealing of the DNA template strands (Hodson et al. 2022). A host of RNA/DNA helicases has been implicated in R-loop removal. In bacteria, these include DinG and UvrD, while a large number of DEAD-box RNA helicases (DDXs), Sen1/senataxin, Aquarius and Sgs1/BLM have been implicated in R-loop suppression in eukaryotes (Boubakri et al. 2010; Garcia-Muse and Aguilera 2019; Bruning and Marians 2021; Brickner et al. 2022). The involvement of multiple helicases in R-loop homeostasis may indicate that they act in a context-specific manner. Such specificity may be established by proteins, such as PARP1, that target helicases to R-loops (Lin et al. 2022; Laspata et al. 2023). Alternatively, helicases may be cell cycle-regulated and directed to select R-loops via physical association with replisomes, such as Sen1 (Alzu et al. 2012; Achar et al. 2020; Appanah et al. 2020; San Martin-Alonso et al. 2021; Zardoni et al. 2021; Aiello et al. 2022).
R-loop resolution by nucleolytic degradation of the RNA strand is predominantly carried out by RNase H enzymes (Hyjek et al. 2019). While E. coli contains a single RNase H enzyme, eukaryotes contain two RNase H enzymes, RNase H1 and RNase H2, which act in a context-specific manner (Zimmer and Koshland 2016; Parajuli et al. 2017; Zhao et al. 2018; Lockhart et al. 2019). In addition to RNase H, RNA exonucleases, such as Xrn2 and the exosome complex, have also been implicated in R-loop resolution (Gavalda et al. 2013; Richard et al. 2013; Morales et al. 2016; Mersaoui et al. 2019). To gain access to their RNA substrate, these nucleases may act coordinately with helicases, such as SETX and DDX5 (Skourti-Stathaki et al. 2011; Richard et al. 2013; Mersaoui et al. 2019). DDX5 has moreover been implicated in an alternative R-loop resolution mechanism involving RNA/DNA decatenation by TOP3B (Saha et al. 2022).
Factors that may influence the stability of RNA:DNA hybrids include RNA:DNA hybrid-binding proteins. One example is yeast Yra1, which binds RNA:DNA hybrids in vitro and can be recruited to chromatin in an R-loop-dependent manner, where it exacerbates TRC-associated genome instability (Garcia-Rubio et al. 2018). Interestingly, Yra1-binding even converts CD-oriented R-loops that do not normally present a threat to genome stability into an obstacle to replication forks, illustrating the potential of R-loop-binding proteins to modulate the block to replication forks at R-loops. In humans, modification of the RNA by N6-methyladenosine (m6A) may either stabilize or destabilize R-loops. In the case of DNA damage-induced pathological R-loops, TonEBP targets the m6A methyltransferase METTL3 to R-loops, which recruits the m6A reader YTHDF2 to induce the degradation of the RNA (Abakir et al. 2020; Kang et al. 2021). Conversely, m6A modification of TERRA by METTL3 stabilizes telomeric R-loops by recruiting the m6A reader YTHDC1 (Chen et al. 2022).
R-loop orientation influences transcription-replication conflict
Consistent with HO transcription-replication collisions generally presenting a more potent block to fork progression than CD collisions, specifically R-loops associated with HO-oriented genes impair fork progression and genome stability in bacteria (Gan et al. 2011; Lang et al. 2017). Moreover, in both human cells and B. subtilis, R-loop levels are by some unknown mechanism aggravated during HO collisions (Hamperl et al. 2017; Lang et al. 2017). While R-loop levels did not correlate with collision orientation in a yeast reporter system, R-loop-mediated genome instability was also here specifically linked to HO collisions (Garcia-Rubio et al. 2018). However, neither R-loop levels nor R-loop-induced DNA damage strongly correlate with collision orientation across the yeast genome, indicating that R-loops can present an impediment to replication in either orientation (Costantino and Koshland 2018; Achar et al. 2020). In agreement with this hypothesis, both CD and HO R-loops induce DNA damage in human cells (Hamperl et al. 2017). Strikingly, the DNA damage in the two orientations is not equivalent as HO R-loops induced an ATR-dependent DNA damage response (DDR), whereas CD R-loops induced an ATM-dependent DDR.
R-loops present multiple distinct impediments to DNA replication
How R-loops block DNA replication at the molecular level is incompletely understood. Fork stalling at R-loops may be an active process involving fork reversal, as inhibition of fork reversal by ZRANB3 depletion suppresses the block to fork progression during R-loop-induced replication stress (Chappidi et al. 2020; Andrs et al. 2023; Stoy et al. 2023). Moreover, since not all replication forks are expected to collide with R-loops at any given time, global fork slowing may also be an indirect consequence of replication checkpoint signaling (Seiler et al. 2007; Mutreja et al. 2018; Promonet et al. 2020). However, several lines of evidence indicate that R-loops also present steric obstacles to replication forks. The mechanistic dissection of these impediments has been hampered by the fact that cellular R-loops are structurally complex and diverse. For example, R-loops exhibit a range of sizes (Malig et al. 2020), associate with many different proteins (Cristini et al. 2018; Wang et al. 2018; Wu et al. 2021; Yan et al. 2022), exhibit distinctive chromatin states (Castellano-Pozo et al. 2013; Skourti-Stathaki et al. 2014; Sanz et al. 2016), and can feature a variety of DNA secondary structures (Duquette et al. 2004; Loomis et al. 2014; Neil et al. 2018; Svikovic et al. 2019), each of which may individually present a block to normal fork progression. Moreover, as both R-loops and replication forks are structurally asymmetric, collision orientation determines which part of an R-loop will be initially encountered by the replicative DNA polymerases or the helicase. In this regard it is important to note that bacterial and eukaryotic replisomes fundamentally differ from each other in that the eukaryotic replicative DNA helicase, CMG (Cdc45-MCM-GINS), encircles the leading strand template, while bacterial replicative DNA helicases, e.g. E. coli DnaB, encircle the lagging strand template. Recent biochemical studies involving reconstituted E. coli and budding yeast replisomes have begun to dissect how RNA:DNA hybrids, DNA secondary structures and stalled RNAPs associated with R-loops differentially impact replisome progression and DNA synthesis on leading and lagging strands (Bruning and Marians 2020; Bruning and Marians 2021; Kumar et al. 2021). These studies also demonstrated that R-loops are not obligatory impediments to fork progression, which agrees with the observations that R-loop orientation influences the outcome of R-loop-replisome collisions and that only a subset of R-loops is associated with DNA damage in eukaryotic cells (Gan et al. 2011; Hamperl et al. 2017; Lang et al. 2017; Costantino and Koshland 2018; Promonet et al. 2020).
The impact of RNA:DNA hybrids on replisome collisions with R-loops
At CD R-loops in eukaryotes, the RNA:DNA hybrid on the leading strand template will be initially encountered by the replicative DNA helicase. CMG can unwind RNA:DNA hybrids, provided the 5’ end of the RNA strand is not hybridized to the DNA, which may explain why replisomes reduce CD R-loop levels in human cells (Hamperl et al. 2017; Kumar et al. 2021) (Figure 3a). CMG can also translocate over RNA:DNA hybrids if the RNA 5’ end is annealed to the DNA (Kumar et al. 2021). This will transfer the RNA:DNA hybrid to the back of the replication fork, where it is encountered by the leading strand polymerase, Pol ε. Since Pol ε lacks strand displacement activity, RNA:DNA hybrids can induce uncoupling of replisome progression from leading strand synthesis (Kumar et al. 2021). Intriguingly, the RNA strand of RNA:DNA hybrids is able to prime leading strand restart in vitro (Kumar et al. 2021) (Figure 3b). Continued replisome progression at CD R-loops thus comes at the cost of retaining RNA in the daughter DNA, which would require post-replicative repair for RNA:DNA hybrid resolution. Interestingly, RNA:DNA hybrid accumulation behind replication forks was recently also observed by electron microscopy in human cells experiencing hormone-induced R-loop-dependent replication stress, raising the possibility that such leading strand restart may occur vivo (Stoy et al. 2023). In addition to RNA:DNA unwinding and bypass, fork stalling also occurs at some CD R-loops in vitro (Kumar et al. 2021). This stalling is prevented by RNase H treatment but exacerbated by pyridostatin, a G4-stabilizing ligand, confirming RNA:DNA hybrids as the cause and implicating G4s in the DNA or RNA strand in the block to replisome progression (Figure 3c). Unlike fork stalling at protein-DNA barriers, fork stalling at CD R-loops is independent of Csm3-Tof1, a subcomplex of the fork protection complex (FPC), indicating that the stall is due to steric hindrance (Kumar et al. 2021).
Figure 3. Consequences of CD R-loop-replisome collisions in eukaryotes.
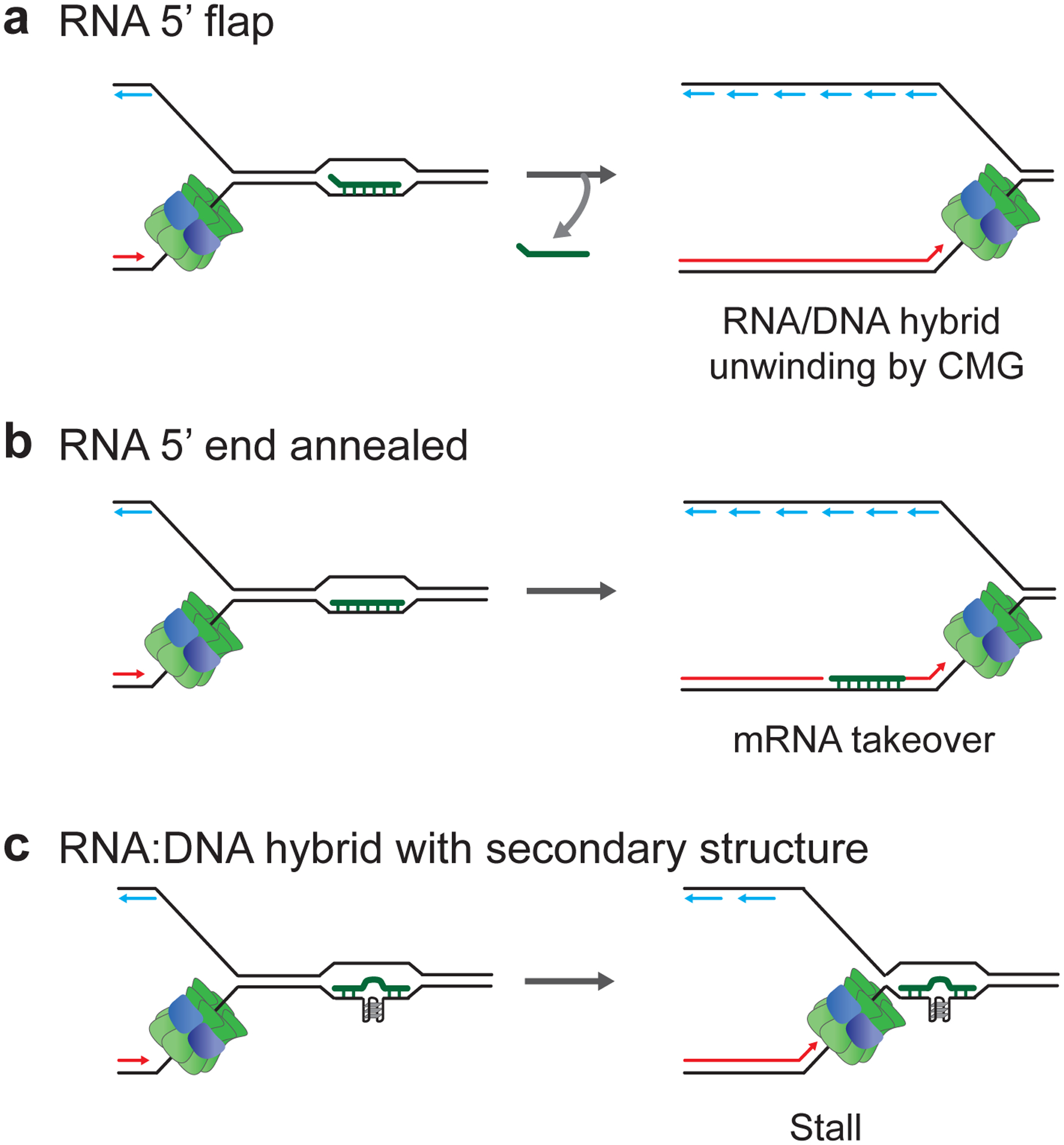
a, CMG unwinding of RNA:DNA hybrids featuring RNA with 5’ flap. b, CMG translocation across RNA:DNA hybrid featuring annealed RNA 5’ end followed by mRNA takeover-mediated leading strand restart. c, Replisome stalling at RNA:DNA hybrids containing secondary structure, such as G4 in DNA or RNA.
In bacteria, RNA:DNA hybrids at CD R-loops are encountered by the leading strand DNA polymerase. In the reconstituted E. coli DNA replication system, short RNA:DNA hybrids of 19 bp length on the leading strand were efficiently unwound, presumably via strand-displacement by DNA Pol III, and thus did not disrupt leading strand synthesis or replisome progression (Bruning and Marians 2020) (Figure 4a). Longer RNA:DNA hybrids of 100 bp length were also readily bypassed by the replisome, but bypass was delayed. This delay was suppressed by UvrD, indicating that RNA:DNA hybrids on the leading strand can interfere with replisome progression despite the fact that the replicative DNA helicase, DnaB, tracks along the lagging strand template. The molecular basis for this is unknown. More importantly, longer RNA:DNA hybrids on the leading strand were retained in the replicated DNA following replisome bypass, resulting in leading strand gaps. Replisome bypass can involve either leading strand skipping, i.e., re-priming of the leading strand by DnaG primase downstream of the RNA:DNA hybrid, or mRNA takeover, i.e., priming of leading strand restart by the RNA of RNA:DNA hybrids (Figure 4b). Replisome skipping is favored at longer RNA:DNA hybrids, while mRNA takeover dominates at shorter RNA:DNA hybrids (Bruning and Marians 2020; Bruning and Marians 2021). Thus, CD R-loops can principally be bypassed by bacterial replisomes in vitro, which is consistent with observations in vivo (Gan et al. 2011; Lang et al. 2017).
Figure 4. Outcomes of CD R-loop-replisome collisions in bacteria.
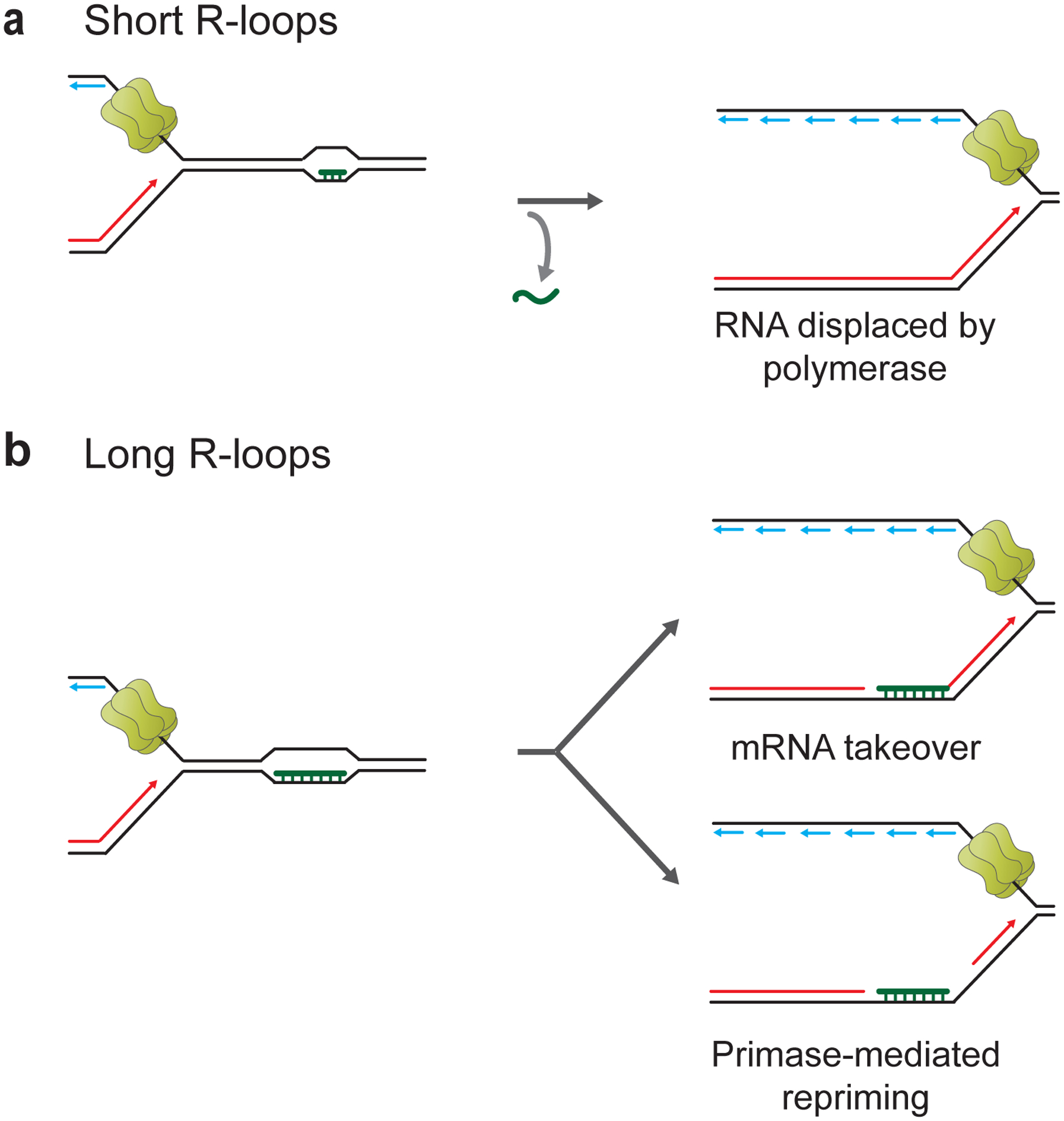
a, CD collisions between replisomes and short naked R-loops allow continued replisome progression and uninterrupted leading strand synthesis. b, Replisome bypass of long naked R-loops or RNAP-associated R-loops can involve mRNA takeover or primase-mediated leading strand restart.
Since CMG encircles the leading strand template, eukaryotic replisomes can generally bypass steric blocks on the lagging strand (Fu et al. 2011; Kose et al. 2019). Accordingly, reconstituted budding yeast replisomes could bypass RNA:DNA hybrids on the lagging strand at HO R-loops in vitro (Kumar et al. 2021). Interestingly, although RNA primers at the 5’ end of Okazaki fragments are efficiently processed by Pol δ and Fen1 during normal lagging strand synthesis, R-loop-associated RNA:DNA hybrids were resistant to such processing and were retained in the newly synthesized lagging strand (Figure 5a). The reasons for this are not clear, but this may provide yet another mechanism for the accumulation of RNA:DNA hybrids behind replication forks in human cells (Stoy et al. 2023).
Figure 5. Consequences of HO R-loop-replisome collisions in eukaryotes and bacteria.
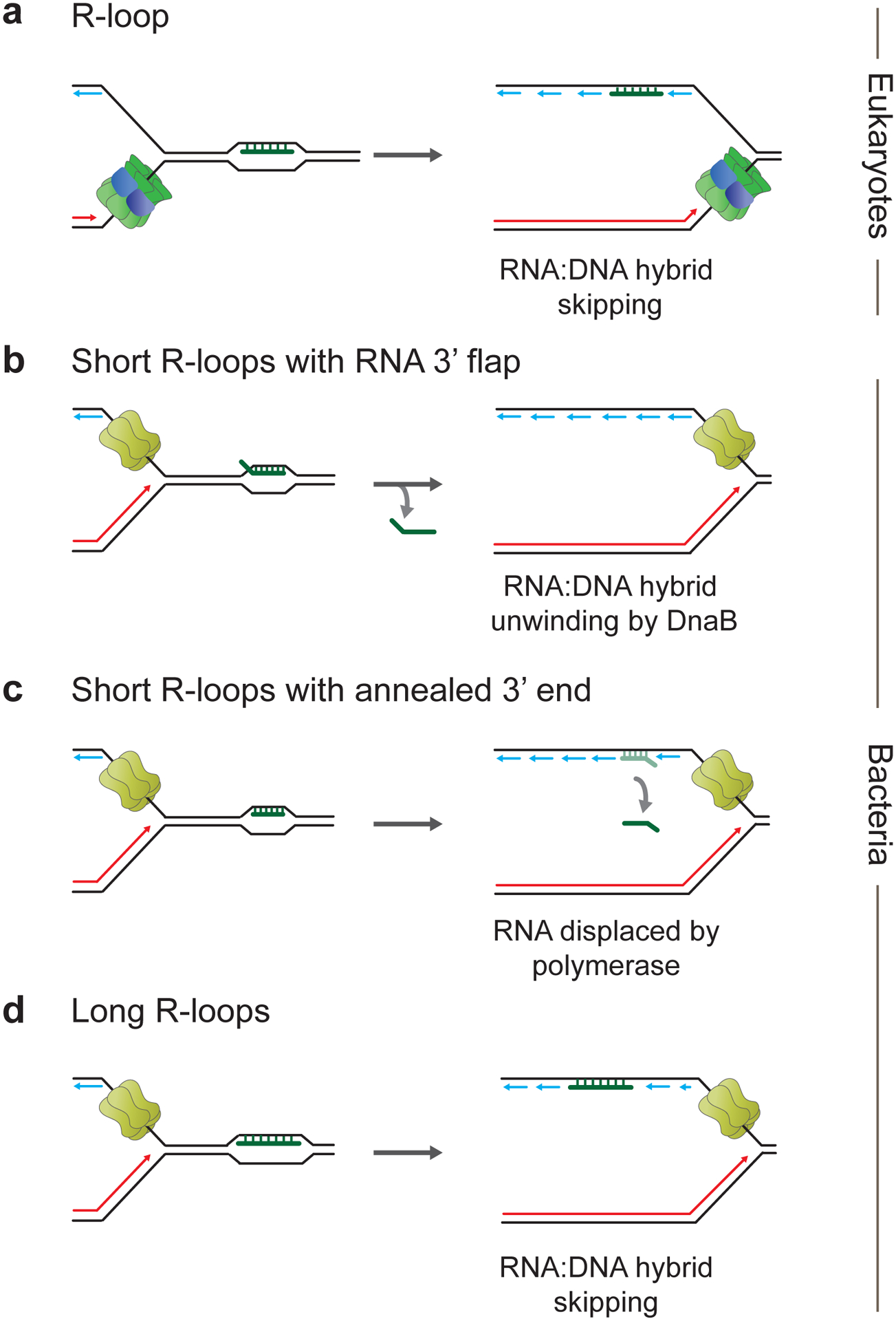
a, Bypass and retention of RNA:DNA hybrid on lagging strand at eukaryotic replication fork. b, Unwinding of RNA:DNA hybrid featuring RNA with unannealed 3’ end by replicative DNA helicase in bacteria. c, Bacterial replisome bypass of short RNA:DNA hybrid featuring RNA with annealed 3’ end followed by RNA displacement by replicative DNA polymerase. d, Bacterial replisome bypass of long RNA:DNA hybrid featuring RNA with annealed 3’ end accompanied by retention of RNA:DNA hybrid in replicated daughter strand.
In the reconstituted E. coli replication system, R-loop-associated RNA:DNA hybrids on the lagging strand that are less than 100 bp in length do not present a significant obstacle to replisome progression and may either get unwound by DnaB (Figure 5b) or strand-displaced by DNA Pol III following DnaB translocation over the RNA:DNA hybrid (Figure 5c) (Bruning and Marians 2020). However, a low level of leading strand blockage was observed at 100 bp long RNA:DNA hybrids, indicative of replisome pausing. Moreover, longer RNA:DNA hybrids remained largely associated with replicated DNA in vitro (Figure 5d), which is consistent with the recent observation of RNA:DNA hybrids behind replication forks following HO R-loop-replisome collisions in B. subtilis cells (Bruning and Marians 2020; Stoy et al. 2023).
Stalled RNAPs exacerbate the R-loop-dependent block to DNA replication
As mentioned above, stalled RNAPs are a major contributor to TRC. Importantly, R-loop formation and RNAP stalling mutually reinforce each other. For example, deregulated promoter-proximal RNAP pausing, RNAP stalling at DNA lesions and uncontrolled RNAP backtracking are potent inducers of R-loops (Sordet et al. 2009; Zhang et al. 2017; Shivji et al. 2018; Herold et al. 2019; Zatreanu et al. 2019). Conversely, R-loops impede RNAP progression (Kireeva et al. 2000; Huertas and Aguilera 2003; Chakraborty et al. 2018). The contribution of stalled RNAPs to the R-loop-dependent block to DNA replication was recently examined in the reconstituted E. coli DNA replication system (Bruning and Marians 2020; Bruning and Marians 2021). RNAPs were stalled by nucleotide restriction and the impact of single stalled RNAPs or arrays of up to three stalled RNAPs at 19–100 bp long R-loops was investigated. Collectively, the data demonstrate that stalled RNAPs exacerbate the impact of R-loops on fork progression and that the impact scales with the number of stalled RNAPs. CD RNAP-R-loop complexes disrupted leading strand synthesis but caused only minor delays in replisome progression, allowing leading strand restart by mRNA takeover downstream of short R-loops associated with a single RNAP (Figure 6a) or by DnaG-catalyzed repriming of the leading strand downstream of larger and more complex CD RNAP-R-loop complexes (Figure 6b). Thus, while a ‘naked’ short R-loop of 19 bp length on the leading strand did not prevent continuous leading strand synthesis, even a single stalled RNAP complex associated with a 19 bp R-loop blocked continuous leading strand synthesis, imposing a requirement for leading strand restart. While the replisome could dislodge CD RNAPs from R-loops, HO RNAP-R-loops presented a strong block to replisomes that had to be cleared by UvrD for uninterrupted fork progression (Figure 6c).
Figure 6. Bacterial replisome collisions with RNAP-associated R-loops.
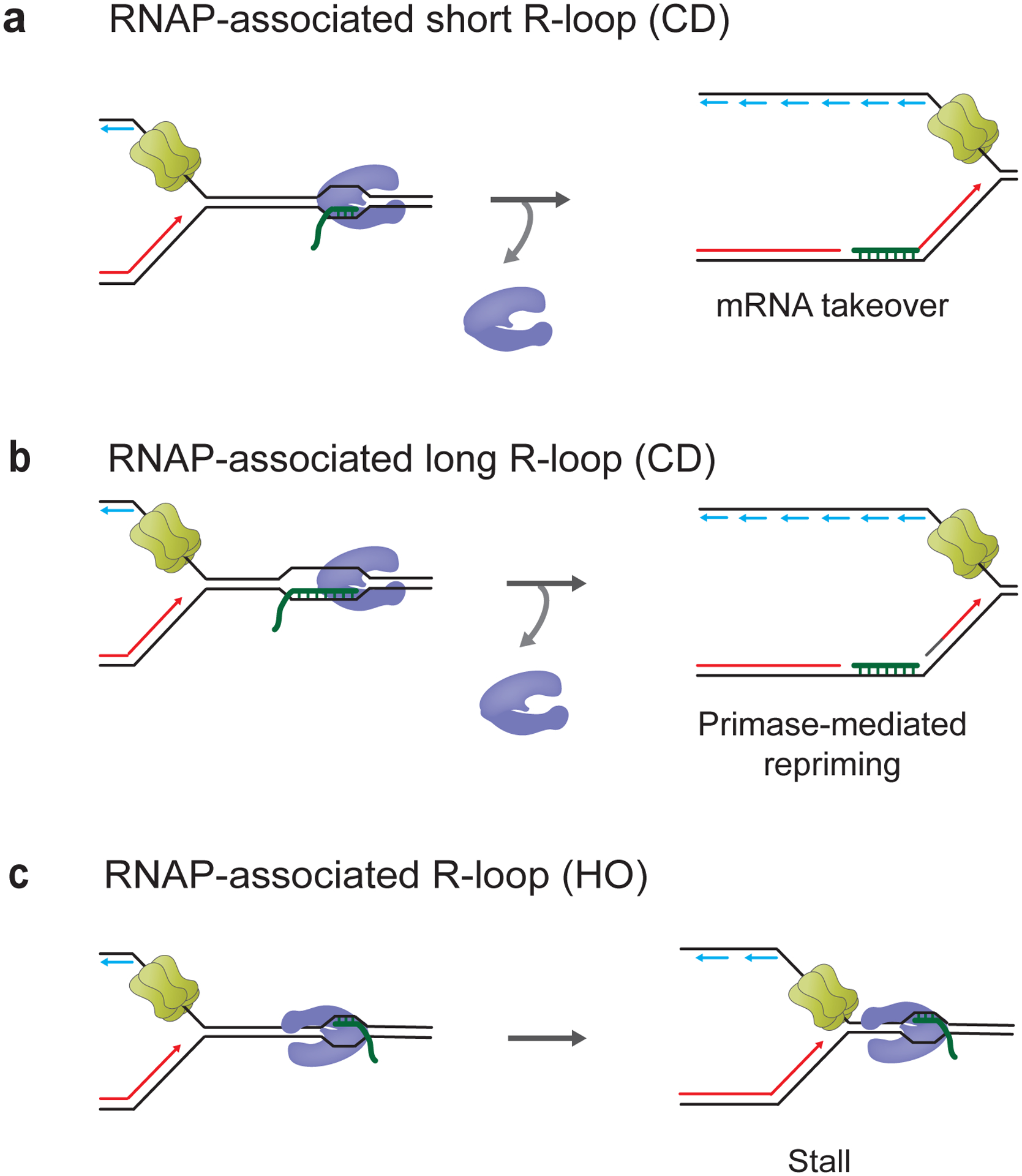
a, Short R-loops associated with RNAP result in mRNA takeover-mediated leading strand restart in CD orientation. b, Long RNAP-associated R-loops cause primase-mediated leading strand restart in CD orientation. c, HO RNA-associated R-loop are strong blocks to fork progression.
DNA secondary structures can inhibit DNA synthesis and replisome progression at R-loops
Non-B-form DNA secondary structures frequently form in repetitive or low-complexity DNA sequences and can impede DNA replication with potentially deleterious consequences for genome stability (Mellor et al. 2022). The folding of many DNA secondary structures, such as hairpins, G-quadruplexes (G4s), H-DNA or i-motif DNA, is facilitated in ssDNA. Consequently, R-loops have an increased potential to form secondary structures on the displaced ssDNA loop. This was first documented at co-transcriptional R-loops featuring G4s on the G-rich displaced strand (Duquette et al. 2004). G4s are compact stacks of G-quartets, each stabilized by Hoogsteen hydrogen-bonded guanines arranged in a planar ring configuration, that form at G-rich repeats composed of at least four tracts of 2–3 Gs (Burge et al. 2006). The characteristic GC-skew associated with many R-loop forming regions thus seems conducive to G4 formation. Indeed, genomic regions exhibiting a high G4 potential, e.g. telomeres, rDNA repeats, and Ig S regions, are well-known to form R-loops and G4 and R-loop positions also correlate globally in mammalian genomes (Yu et al. 2003; El Hage et al. 2010; Chan et al. 2014; Chen et al. 2017; Feretzaki et al. 2020; Lyu et al. 2022). Moreover, R-loops are a source for genome instability induced by chemical G4 ligands in cancer cells, further illustrating the link between R-loops and G4s (De Magis et al. 2019). Importantly, the presence of G4s on the displaced ssDNA loop and an RNA:DNA hybrid on the opposite strand gives rise to potential simultaneous leading and lagging strand obstacles, which complicates the interpretation of which is the primary replication stress-causing element.
G4s have long been recognized to inhibit DNA polymerase progression (Lerner and Sale 2019). In addition, G4s specifically in the leading strand template can inhibit DNA unwinding by CMG (Kumar et al. 2021; Batra et al. 2022). Consequently, G4s especially on the leading strand template are a source for DNA replication stress associated with genetic or epigenetic instability (Sarkies et al. 2010; Lopes et al. 2011; Sato et al. 2021). In contrast, a recent live cell imaging study in budding yeast found that G4 sequences inhibited fork progression specifically when placed on the lagging strand template, indicating that the impact of G4s on DNA replication may be context-dependent (Dahan et al. 2018). Moreover, G4s are structurally diverse and exhibit a range of thermodynamic stabilities that determine their impact on DNA replication (Piazza et al. 2015; Piazza et al. 2017). Accordingly, G4-stabilizing chemical ligands are potent aggravators of G4-dependent replication stress (Piazza et al. 2010; Piazza et al. 2012; Rodriguez et al. 2012). Conversely, many DNA helicases, including WRN, BLM, Sgs1, DDX11, Pif1, FANCJ and RTEL1 in eukaryotes or Rep in bacteria, have been implicated in destabilizing G4s and promoting the replication of G4 DNA (Lerner and Sale 2019).
The impact of G4s on replisome collisions with R-loops was recently investigated using the reconstituted budding yeast DNA replication system (Kumar et al. 2021). In this system, R-loops were formed by transcription of a 1.4 kb region of the mouse Airn gene that was previously shown to be prone to form R-loops (Ginno et al. 2012; Kumar and Remus 2022). This sequence element features a pronounced G/C skew and an array of sequences with G4 potential. Electron-microscopic analysis revealed that R-loops formed at this sequence in vitro were heterogeneous in size and distribution and, therefore, likely featured variable G4 potential on the displaced ssDNA (Kumar et al. 2021). At CD R-loops, when the displaced ssDNA forms the lagging strand template, G4s could be bypassed by the replisome but created gaps in the newly synthesized lagging strand (Figure 7a). In contrast, at R-loops in the HO orientation, i.e., when the displaced ssDNA forms the leading strand template, G4s induced replisome stalling (Figure 7b) or disrupted leading strand synthesis that could be restarted by repriming downstream of the R-loop (Figure 7c). The variability in outcomes is likely derived from the heterogeneity of R-loops. As observed at RNA:DNA hybrids, replisome stalling at G4s was independent of Csm3-Tof1, indicating that G4s present a steric block to the replisome. Moreover, the replication impediments on both the leading and lagging strand were suppressed by the Pif1 helicase, consistent with the ability of Pif1 to promote replication fork progression at G4s in vivo (Ribeyre et al. 2009; Paeschke et al. 2011).
Figure 7: DNA Secondary structures exacerbate consequences of R-loop-replisome collisions in eukaryotes.
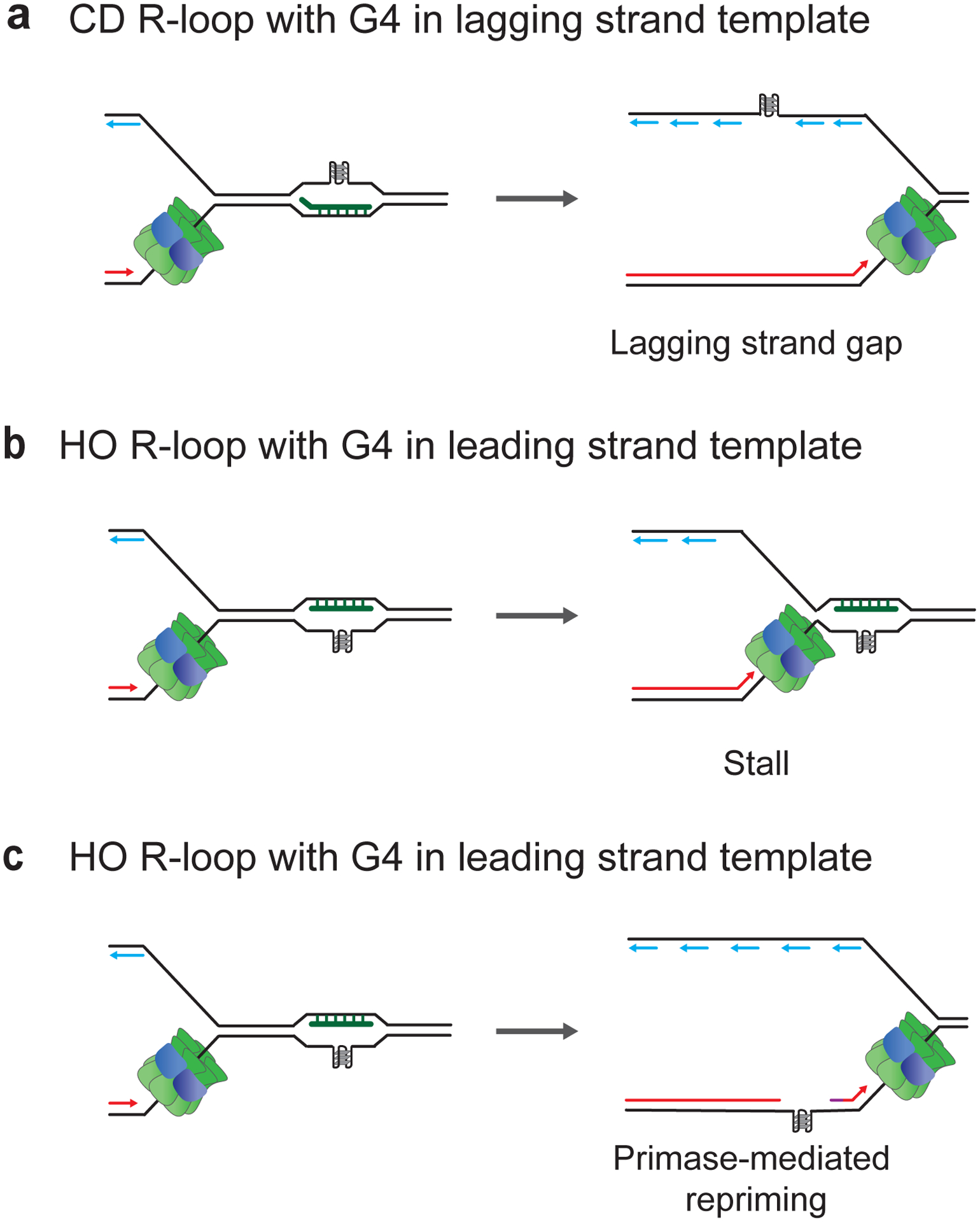
a, G4s in the displaced ssDNA loop on lagging strand cause gaps in lagging strand. b, Replisome stalling at G4s in displaced ssDNA loop on leading strand. c, Replisome bypass and leading strand restart at G4s in displaced ssDNA loop on leading strand.
Other secondary structure-forming sequences associated with R-loops include tri-nucleotide repeats (TNRs) implicated in neurodegenerative diseases (Grabczyk et al. 2007; Lin et al. 2010; Reddy et al. 2011; Groh et al. 2014; Loomis et al. 2014; Su and Freudenreich 2017; Neil et al. 2018; Laverde et al. 2020). The secondary structures formed at TNR-associated R-loops are diverse. For example, CGG repeats associated with fragile X syndrome form simple hairpins on the displaced ssDNA, whereas GAA repeats associated with Friedreich’s ataxia may form H-loops in which the displaced strand engages in triplex formation (H-DNA) (Loomis et al. 2014; Neil et al. 2018). How TNRs affect DNA replication at R-loops is not clear but pathologically long TNRs are known to stall replication forks and induce replication-dependent instability (Krasilnikova and Mirkin 2004; Chandok et al. 2012; Gerhardt et al. 2016; Gellon et al. 2019). One mechanism was suggested by studies in a chicken DT40 cell model, in which even a non-pathological short (GAA)10 repeat could induce replisome uncoupling from leading strand synthesis (Svikovic et al. 2019). Interestingly, this uncoupling was suppressed by RNase H1 overexpression, indicating that R-loop formation rendered the short TNR inhibitory to DNA replication, potentially by forming an H-loop. Moreover, uncoupling was shown to promote further R-loop formation in the absence of PrimPol, indicating that single-stranded DNA generated in the wake of uncoupled replisomes promotes de novo R-loop formation. Genomic R-loop profiles suggest that such uncoupling is suppressed by PrimPol across the genome at sequences exhibiting G4 and H-DNA potential. Collectively, the data thus demonstrate that secondary structures influence the replication-stalling potential of R-loops by modulating R-loop structure and stability and generating strand-specific obstacles to replicative DNA polymerases and helicases.
The chromatin state contributes to the block to replication at R-loops
As mentioned above, R-loops exhibit specific associated proteomes that may influence the fork stalling potential of R-loops, as illustrated by the RNA:DNA hybrid-binding protein Yra1 in yeast, that converts R-loops into orientation-independent replication blocks (Garcia-Rubio et al. 2018) (Figure 8a). In addition, R-loop-forming regions are marked by specific chromatin signatures. Globally, R-loops correlate with sites of increased chromatin accessibility and regions marked by active promoter and enhancer histone modifications, such as H3K4me2, H3K4me3, H3K9ac and H3K27ac, and suppress the establishment of repressed chromatin states by modulating the binding of histone modifying complexes (Chen et al. 2015; Sanz et al. 2016; Chen et al. 2017; Lyu et al. 2022). Moreover, R-loops maintain active gene states by regulating the recruitment of DNA methyltransferases and DNA demethylases to suppress the methylation of CpG island promoters (Ginno et al. 2012; Grunseich et al. 2018; Arab et al. 2019). Conversely, R-loops also contribute to the establishment or maintenance of repressed chromatin states at TTSs, PREs, and TNRs, and associate with phosphorylated H3S10 (H3S10p), a mark of condensed chromatin, which may add another layer of fork stalling potential at R-loops (Castellano-Pozo et al. 2013; Groh et al. 2014; Skourti-Stathaki et al. 2014; Skourti-Stathaki et al. 2019; Alecki et al. 2020) (Figure 8b). Intriguingly, histone mutations that prevent the accumulation of H3S10p suppress R-loop-dependent genome instability in hpr1 and sen1 mutant yeast strains, which has been interpreted to indicate that R-loops inherently do not present an obstacle to fork progression (Garcia-Pichardo et al. 2017). While perhaps difficult to reconcile with histone-independent fork stalling mechanisms implicated in inducing fork stalling at R-loops, this result confirms the potential of specific chromatin states to impede DNA replication at R-loops. This is also supported by the observation that R-loop-dependent TRC is exacerbated in the absence of various histone chaperones and chromatin remodelers, such as FACT, SWI/SNF, and INO80, which promote fork progression through chromatin across the genome (Bayona-Feliu and Aguilera 2021). How H3S10p contributes to R-loop-dependent genome instability, or how chromatin remodelers and histone chaperones facilitate fork progression specifically at R-loops, is mechanistically not understood. Since the three-stranded structure of R-loops is likely refractory to nucleosome assembly, it remains to be determined if and how R-loops and specific chromatin states coincide spatially and temporally. Notably, while condensed chromatin states appear to exacerbate R-loop-dependent TRC in some contexts, they also mitigate TRC by restricting R-loop formation in other contexts (Zeller et al. 2016; Taneja et al. 2017; Almeida et al. 2018). Thus, the detailed mechanisms by which chromatin impacts fork progression at R-loops remain to be clarified.
Figure 8: Effect of chromatin on R-loop-replisome collisions in eukaryotes.
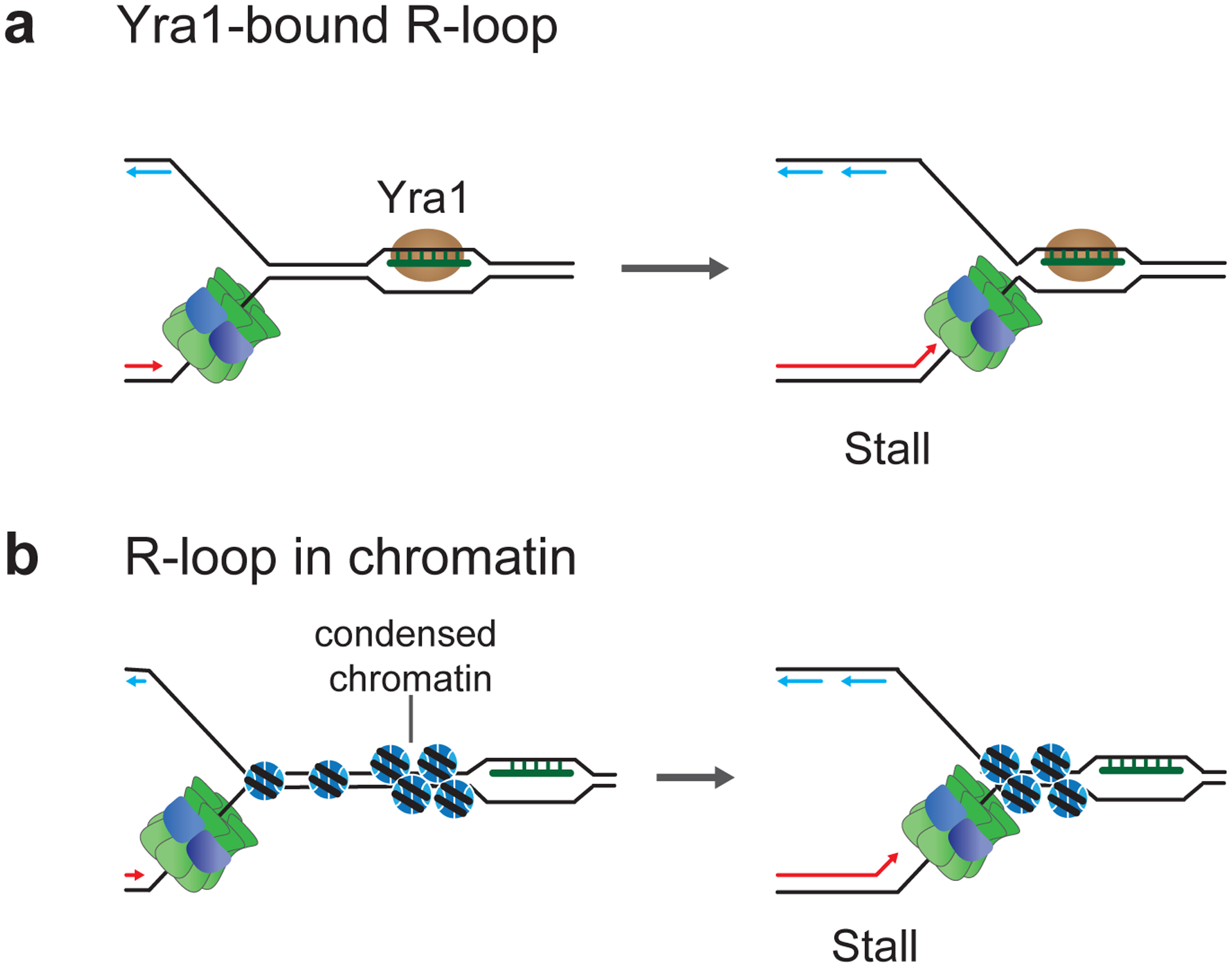
a, Replisome stalling at Yra1-stabilized R-loop. b, Replisome stalling at R-loop-associated condensed chromatin.
Concluding remarks
The studies summarized here demonstrate that the R-loop-dependent block to replication forks is complex and determined by multiple structural features associated with R-loops, including RNA:DNA hybrid length, secondary structure on the displaced ssDNA loop, stalled RNA polymerases, RNA/DNA binding proteins and chromatin structure. This diversity in R-loop structures may in part explain the involvement of a plethora of factors in R-loop removal. Moreover, it suggests that the consequences of R-loop-replisome collisions are unlikely to generalize across the genome but have to be considered in a context-specific manner.
The diversity in R-loop structures implies that the specific mechanisms of fork arrest and restart at R-loops are likely to vary between individual R-loops. For example, in eukaryotes, G4s in the leading strand template can induce the uncoupling of replisome progression from leading strand synthesis, which may potentially promote fork restart via fork reversal upstream of the replisome, as recently observed in the context of other replication stress conditions (Kavlashvili et al. 2023; Liu et al. 2023). In contrast, fork uncoupling would be prevented at obstacles that present a physical block to replisome progression, which may involve either protein-DNA complexes or DNA secondary structures. In this instance, fork restart may require the physical coordination of obstacle resolution by ancillary helicases with replisome progression. How such coordination is achieved is not clear but likely involves differential interactions between ancillary helicases and replisome components as exemplified in eukaryotes by Pif1 and Sen1, which are coupled to replisomes via PCNA or Ctf4/Mrc1, respectively (Dahan et al. 2018; Appanah et al. 2020).
In budding yeast, fork restart at HO R-loops may also occur directly via repriming of the leading strand downstream of G4s, as has been observed in vitro (Kumar et al. 2021). However, analogous leading strand restart by repriming was previously found to be highly inefficient downstream of non-bulky DNA lesions (Taylor and Yeeles 2018), raising the question whether the efficiency of leading strand restart is influenced by the type of obstacle in the DNA. Moreover, in some higher eukaryotes, leading strand restart appears to be promoted by the specialized priming activity of PrimPol (Svikovic et al. 2019). Since PrimPol is not universally conserved in eukaryotes, it may be interesting to determine if similar R-loop structures have distinct consequences on fork progression in different organisms. At CD R-loops, reconstituted budding yeast replisomes have also been observed to restart leading strand synthesis by extending the RNA strand of the RNA:DNA hybrid (Kumar et al. 2021). Such a mechanism involves the translocation of CMG on the RNA:DNA duplex, which may bear the risk of ubiquitin-mediated replisome disassembly in vivo analogous to that occurring on dsDNA (Deegan et al. 2020; Jenkyn-Bedford et al. 2021; Vrtis et al. 2021). Completion of DNA replication in this case may require recombination-based mechanisms or replication rescue by an opposing fork. Future studies involving R-loops with defined structural attributes will be required to dissect the variety of outcomes of R-loop-replisome collisions and to determine the differential involvement of the many factors modulating R-loop stability.
Acknowledgments:
Work in the Remus lab is funded by NIGMS grants R01-GM107239 and R01-GM127428, and NIH-NCI Cancer Center Support Grant P30 CA008748.
Footnotes
Competing interests
The authors declare no competing interests.
Availability of data and materials
Not applicable.
References
- Abakir A, Giles TC, Cristini A, Foster JM, Dai N, Starczak M, Rubio-Roldan A, Li M, Eleftheriou M, Crutchley J et al. 2020. N(6)-methyladenosine regulates the stability of RNA:DNA hybrids in human cells. Nat Genet 52: 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham KJ, Khosraviani N, Chan JNY, Gorthi A, Samman A, Zhao DY, Wang M, Bokros M, Vidya E, Ostrowski LA et al. 2020. Nucleolar RNA polymerase II drives ribosome biogenesis. Nature 585: 298–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achar YJ, Adhil M, Choudhary R, Gilbert N, Foiani M. 2020. Negative supercoil at gene boundaries modulates gene topology. Nature 577: 701–705. [DOI] [PubMed] [Google Scholar]
- Aiello U, Challal D, Wentzinger G, Lengronne A, Appanah R, Pasero P, Palancade B, Libri D. 2022. Sen1 is a key regulator of transcription-driven conflicts. Mol Cell 82: 2952–2966 e2956. [DOI] [PubMed] [Google Scholar]
- Alecki C, Chiwara V, Sanz LA, Grau D, Arias Perez O, Boulier EL, Armache KJ, Chedin F, Francis NJ. 2020. RNA-DNA strand exchange by the Drosophila Polycomb complex PRC2. Nat Commun 11: 1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida R, Fernandez-Justel JM, Santa-Maria C, Cadoret JC, Cano-Aroca L, Lombrana R, Herranz G, Agresti A, Gomez M. 2018. Chromatin conformation regulates the coordination between DNA replication and transcription. Nat Commun 9: 1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzu A, Bermejo R, Begnis M, Lucca C, Piccini D, Carotenuto W, Saponaro M, Brambati A, Cocito A, Foiani M et al. 2012. Senataxin associates with replication forks to protect fork integrity across RNA-polymerase-II-transcribed genes. Cell 151: 835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrs M, Stoy H, Boleslavska B, Chappidi N, Kanagaraj R, Nascakova Z, Menon S, Rao S, Oravetzova A, Dobrovolna J et al. 2023. Excessive reactive oxygen species induce transcription-dependent replication stress. Nat Commun 14: 1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appanah R, Lones EC, Aiello U, Libri D, De Piccoli G. 2020. Sen1 Is Recruited to Replication Forks via Ctf4 and Mrc1 and Promotes Genome Stability. Cell Rep 30: 2094–2105 e2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arab K, Karaulanov E, Musheev M, Trnka P, Schafer A, Grummt I, Niehrs C. 2019. GADD45A binds R-loops and recruits TET1 to CpG island promoters. Nat Genet 51: 217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel F, Lucero L, Christ A, Mammarella MF, Jegu T, Veluchamy A, Mariappan K, Latrasse D, Blein T, Liu C et al. 2020. R-Loop Mediated trans Action of the APOLO Long Noncoding RNA. Mol Cell 77: 1055–1065 e1054. [DOI] [PubMed] [Google Scholar]
- Azvolinsky A, Giresi PG, Lieb JD, Zakian VA. 2009. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol Cell 34: 722–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacal J, Moriel-Carretero M, Pardo B, Barthe A, Sharma S, Chabes A, Lengronne A, Pasero P. 2018. Mrc1 and Rad9 cooperate to regulate initiation and elongation of DNA replication in response to DNA damage. EMBO J 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow JH, Faryabi RB, Callen E, Wong N, Malhowski A, Chen HT, Gutierrez-Cruz G, Sun HW, McKinnon P, Wright G et al. 2013. Identification of early replicating fragile sites that contribute to genome instability. Cell 152: 620–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batra S, Devbhandari S, Remus D. 2022. CMG helicase activity on G4-containing DNA templates. Methods Enzymol 672: 233–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayona-Feliu A, Aguilera A. 2021. The role of chromatin at transcription-replication conflicts as a genome safeguard. Biochem Soc Trans 49: 2727–2736. [DOI] [PubMed] [Google Scholar]
- Belotserkovskii BP, Hanawalt PC. 2022. Topology and kinetics of R-loop formation. Biophys J 121: 3345–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL. 2014. Coupling mRNA processing with transcription in time and space. Nat Rev Genet 15: 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R, Capra T, Jossen R, Colosio A, Frattini C, Carotenuto W, Cocito A, Doksani Y, Klein H, Gomez-Gonzalez B et al. 2011. The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell 146: 233–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernecky C, Herzog F, Baumeister W, Plitzko JM, Cramer P. 2016. Structure of transcribing mammalian RNA polymerase II. Nature 529: 551–554. [DOI] [PubMed] [Google Scholar]
- Bonnet A, Grosso AR, Elkaoutari A, Coleno E, Presle A, Sridhara SC, Janbon G, Geli V, de Almeida SF, Palancade B. 2017. Introns Protect Eukaryotic Genomes from Transcription-Associated Genetic Instability. Mol Cell 67: 608–621 e606. [DOI] [PubMed] [Google Scholar]
- Boque-Sastre R, Soler M, Oliveira-Mateos C, Portela A, Moutinho C, Sayols S, Villanueva A, Esteller M, Guil S. 2015. Head-to-head antisense transcription and R-loop formation promotes transcriptional activation. Proc Natl Acad Sci U S A 112: 5785–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boubakri H, de Septenville AL, Viguera E, Michel B. 2010. The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J 29: 145–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BJ. 1988. When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell 53: 679–686. [DOI] [PubMed] [Google Scholar]
- Brickner JR, Garzon JL, Cimprich KA. 2022. Walking a tightrope: The complex balancing act of R-loops in genome stability. Mol Cell 82: 2267–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruning JG, Marians KJ. 2020. Replisome bypass of transcription complexes and R-loops. Nucleic Acids Res 48: 10353–10367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- -. 2021. Bypass of complex co-directional replication-transcription collisions by replisome skipping. Nucleic Acids Res 49: 9870–9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. 2006. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res 34: 5402–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano-Pozo M, Santos-Pereira JM, Rondon AG, Barroso S, Andujar E, Perez-Alegre M, Garcia-Muse T, Aguilera A. 2013. R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Mol Cell 52: 583–590. [DOI] [PubMed] [Google Scholar]
- Castillo-Guzman D, Chedin F. 2021. Defining R-loop classes and their contributions to genome instability. DNA Repair (Amst) 106: 103182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty P, Grosse F. 2011. Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair (Amst) 10: 654–665. [DOI] [PubMed] [Google Scholar]
- Chakraborty P, Huang JTJ, Hiom K. 2018. DHX9 helicase promotes R-loop formation in cells with impaired RNA splicing. Nat Commun 9: 4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YA, Aristizabal MJ, Lu PY, Luo Z, Hamza A, Kobor MS, Stirling PC, Hieter P. 2014. Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip. PLoS Genet 10: e1004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandok GS, Patel MP, Mirkin SM, Krasilnikova MM. 2012. Effects of Friedreich’s ataxia GAA repeats on DNA replication in mammalian cells. Nucleic Acids Res 40: 3964–3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappidi N, Nascakova Z, Boleslavska B, Zellweger R, Isik E, Andrs M, Menon S, Dobrovolna J, Balbo Pogliano C, Matos J et al. 2020. Fork Cleavage-Religation Cycle and Active Transcription Mediate Replication Restart after Fork Stalling at Co-transcriptional R-Loops. Mol Cell 77: 528–541 e528. [DOI] [PubMed] [Google Scholar]
- Chedin F, Benham CJ. 2020. Emerging roles for R-loop structures in the management of topological stress. J Biol Chem 295: 4684–4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chen JY, Zhang X, Gu Y, Xiao R, Shao C, Tang P, Qian H, Luo D, Li H et al. 2017. R-ChIP Using Inactive RNase H Reveals Dynamic Coupling of R-loops with Transcriptional Pausing at Gene Promoters. Mol Cell 68: 745–757 e745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhang C, Ma W, Huang J, Zhao Y, Liu H. 2022. METTL3-mediated m6A modification stabilizes TERRA and maintains telomere stability. Nucleic Acids Res 50: 11619–11634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PB, Chen HV, Acharya D, Rando OJ, Fazzio TG. 2015. R loops regulate promoter-proximal chromatin architecture and cellular differentiation. Nat Struct Mol Biol 22: 999–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Keegan S, Kahli M, Tonzi P, Fenyo D, Huang TT, Smith DJ. 2019. Transcription shapes DNA replication initiation and termination in human cells. Nat Struct Mol Biol 26: 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarle R, Zhang Y, Frock RL, Lewis SM, Molinie B, Ho YJ, Myers DR, Choi VW, Compagno M, Malkin DJ et al. 2011. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell 147: 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong SY, Cutler S, Lin JJ, Tsai CH, Tsai HK, Biggins S, Tsukiyama T, Lo YC, Kao CF. 2020. H3K4 methylation at active genes mitigates transcription-replication conflicts during replication stress. Nat Commun 11: 809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussin C, Vazquez J, Whitehouse I. 2022. Single-molecule mapping of replisome progression. Mol Cell 82: 1372–1382 e1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier SC, Wang S, Ma WK, Al Husini N, Dhoondia Z, Ansari A, Pascuzzi PE, Tran EJ. 2016. Regulated Formation of lncRNA-DNA Hybrids Enables Faster Transcriptional Induction and Environmental Adaptation. Mol Cell 61: 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino L, Koshland D. 2018. Genome-wide Map of R-Loop-Induced Damage Reveals How a Subset of R-Loops Contributes to Genomic Instability. Mol Cell 71: 487–497 e483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristini A, Groh M, Kristiansen MS, Gromak N. 2018. RNA/DNA Hybrid Interactome Identifies DXH9 as a Molecular Player in Transcriptional Termination and R-Loop-Associated DNA Damage. Cell Rep 23: 1891–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristini A, Ricci G, Britton S, Salimbeni S, Huang SN, Marinello J, Calsou P, Pommier Y, Favre G, Capranico G et al. 2019. Dual Processing of R-Loops and Topoisomerase I Induces Transcription-Dependent DNA Double-Strand Breaks. Cell Rep 28: 3167–3181 e3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristini A, Tellier M, Constantinescu F, Accalai C, Albulescu LO, Heiringhoff R, Bery N, Sordet O, Murphy S, Gromak N. 2022. RNase H2, mutated in Aicardi-Goutieres syndrome, resolves co-transcriptional R-loops to prevent DNA breaks and inflammation. Nat Commun 13: 2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley MP, Bocek MJ, Hamperl S, Swigut T, Cimprich KA. 2020. qDRIP: a method to quantitatively assess RNA-DNA hybrid formation genome-wide. Nucleic Acids Res 48: e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley MP, Song C, Bocek MJ, Choi JH, Kousorous J, Sathirachinda A, Lin C, Brickner JR, Bai G, Lans H et al. 2023. R-loop-derived cytoplasmic RNA-DNA hybrids activate an immune response. Nature 613: 187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan D, Tsirkas I, Dovrat D, Sparks MA, Singh SP, Galletto R, Aharoni A. 2018. Pif1 is essential for efficient replisome progression through lagging strand G-quadruplex DNA secondary structures. Nucleic Acids Res 46: 11847–11857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels GA, Lieber MR. 1995. RNA:DNA complex formation upon transcription of immunoglobulin switch regions: implications for the mechanism and regulation of class switch recombination. Nucleic Acids Res 23: 5006–5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Magis A, Manzo SG, Russo M, Marinello J, Morigi R, Sordet O, Capranico G. 2019. DNA damage and genome instability by G-quadruplex ligands are mediated by R loops in human cancer cells. Proc Natl Acad Sci U S A 116: 816–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deegan TD, Mukherjee PP, Fujisawa R, Polo Rivera C, Labib K. 2020. CMG helicase disassembly is controlled by replication fork DNA, replisome components and a ubiquitin threshold. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande AM, Newlon CS. 1996. DNA replication fork pause sites dependent on transcription. Science 272: 1030–1033. [DOI] [PubMed] [Google Scholar]
- Devbhandari S, Remus D. 2020. Rad53 limits CMG helicase uncoupling from DNA synthesis at replication forks. Nat Struct Mol Biol 27: 461–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drolet M, Bi X, Liu LF. 1994. Hypernegative supercoiling of the DNA template during transcription elongation in vitro. J Biol Chem 269: 2068–2074. [PubMed] [Google Scholar]
- Duch A, Canal B, Barroso SI, Garcia-Rubio M, Seisenbacher G, Aguilera A, de Nadal E, Posas F. 2018. Multiple signaling kinases target Mrc1 to prevent genomic instability triggered by transcription-replication conflicts. Nat Commun 9: 379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duch A, Felipe-Abrio I, Barroso S, Yaakov G, Garcia-Rubio M, Aguilera A, de Nadal E, Posas F. 2013. Coordinated control of replication and transcription by a SAPK protects genomic integrity. Nature 493: 116–119. [DOI] [PubMed] [Google Scholar]
- Dumelie JG, Jaffrey SR. 2017. Defining the location of promoter-associated R-loops at near-nucleotide resolution using bisDRIP-seq. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. 2004. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev 18: 1618–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D, Shatalin K, Epshtein V, Gottesman ME, Nudler E. 2011. Linking RNA polymerase backtracking to genome instability in E. coli. Cell 146: 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hage A, French SL, Beyer AL, Tollervey D. 2010. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev 24: 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felipe-Abrio I, Lafuente-Barquero J, Garcia-Rubio ML, Aguilera A. 2015. RNA polymerase II contributes to preventing transcription-mediated replication fork stalls. EMBO J 34: 236–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feretzaki M, Pospisilova M, Valador Fernandes R, Lunardi T, Krejci L, Lingner J. 2020. RAD51-dependent recruitment of TERRA lncRNA to telomeres through R-loops. Nature 587: 303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frattini C, Promonet A, Alghoul E, Vidal-Eychenie S, Lamarque M, Blanchard MP, Urbach S, Basbous J, Constantinou A. 2021. TopBP1 assembles nuclear condensates to switch on ATR signaling. Mol Cell 81: 1231–1245 e1238. [DOI] [PubMed] [Google Scholar]
- French S 1992. Consequences of replication fork movement through transcription units in vivo. Science 258: 1362–1365. [DOI] [PubMed] [Google Scholar]
- Fu YV, Yardimci H, Long DT, Ho TV, Guainazzi A, Bermudez VP, Hurwitz J, van Oijen A, Scharer OD, Walter JC. 2011. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell 146: 931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan W, Guan Z, Liu J, Gui T, Shen K, Manley JL, Li X. 2011. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev 25: 2041–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Muse T, Aguilera A. 2019. R Loops: From Physiological to Pathological Roles. Cell 179: 604–618. [DOI] [PubMed] [Google Scholar]
- Garcia-Pichardo D, Canas JC, Garcia-Rubio ML, Gomez-Gonzalez B, Rondon AG, Aguilera A. 2017. Histone Mutants Separate R Loop Formation from Genome Instability Induction. Mol Cell 66: 597–609 e595. [DOI] [PubMed] [Google Scholar]
- Garcia-Rubio M, Aguilera P, Lafuente-Barquero J, Ruiz JF, Simon MN, Geli V, Rondon AG, Aguilera A. 2018. Yra1-bound RNA-DNA hybrids cause orientation-independent transcription-replication collisions and telomere instability. Genes Dev 32: 965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavalda S, Gallardo M, Luna R, Aguilera A. 2013. R-loop mediated transcription-associated recombination in trf4Delta mutants reveals new links between RNA surveillance and genome integrity. PLoS One 8: e65541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellon L, Kaushal S, Cebrian J, Lahiri M, Mirkin SM, Freudenreich CH. 2019. Mrc1 and Tof1 prevent fragility and instability at long CAG repeats by their fork stabilizing function. Nucleic Acids Res 47: 794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt J, Bhalla AD, Butler JS, Puckett JW, Dervan PB, Rosenwaks Z, Napierala M. 2016. Stalled DNA Replication Forks at the Endogenous GAA Repeats Drive Repeat Expansion in Friedreich’s Ataxia Cells. Cell Rep 16: 1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginno PA, Lim YW, Lott PL, Korf I, Chedin F. 2013. GC skew at the 5’ and 3’ ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res 23: 1590–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginno PA, Lott PL, Christensen HC, Korf I, Chedin F. 2012. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell 45: 814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gonzalez B, Aguilera A. 2007. Activation-induced cytidine deaminase action is strongly stimulated by mutations of the THO complex. Proc Natl Acad Sci U S A 104: 8409–8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- -. 2019. Transcription-mediated replication hindrance: a major driver of genome instability. Genes Dev 33: 1008–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gonzalez B, Felipe-Abrio I, Aguilera A. 2009. The S-phase checkpoint is required to respond to R-loops accumulated in THO mutants. Mol Cell Biol 29: 5203–5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabczyk E, Mancuso M, Sammarco MC. 2007. A persistent RNA.DNA hybrid formed by transcription of the Friedreich ataxia triplet repeat in live bacteria, and by T7 RNAP in vitro. Nucleic Acids Res 35: 5351–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh M, Lufino MM, Wade-Martins R, Gromak N. 2014. R-loops associated with triplet repeat expansions promote gene silencing in Friedreich ataxia and fragile X syndrome. PLoS Genet 10: e1004318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunseich C, Wang IX, Watts JA, Burdick JT, Guber RD, Zhu Z, Bruzel A, Lanman T, Chen K, Schindler AB et al. 2018. Senataxin Mutation Reveals How R-Loops Promote Transcription by Blocking DNA Methylation at Gene Promoters. Mol Cell 69: 426–437 e427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy CP, Atkinson J, Gupta MK, Mahdi AA, Gwynn EJ, Rudolph CJ, Moon PB, van Knippenberg IC, Cadman CJ, Dillingham MS et al. 2009. Rep provides a second motor at the replisome to promote duplication of protein-bound DNA. Mol Cell 36: 654–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamperl S, Bocek MJ, Saldivar JC, Swigut T, Cimprich KA. 2017. Transcription-Replication Conflict Orientation Modulates R-Loop Levels and Activates Distinct DNA Damage Responses. Cell 170: 774–786 e719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatchi E, Skourti-Stathaki K, Ventz S, Pinello L, Yen A, Kamieniarz-Gdula K, Dimitrov S, Pathania S, McKinney KM, Eaton ML et al. 2015. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol Cell 57: 636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins M, Dimude JU, Howard JAL, Smith AJ, Dillingham MS, Savery NJ, Rudolph CJ, McGlynn P. 2019. Direct removal of RNA polymerase barriers to replication by accessory replicative helicases. Nucleic Acids Res 47: 5100–5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmrich A, Ballarino M, Tora L. 2011. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol Cell 44: 966–977. [DOI] [PubMed] [Google Scholar]
- Herold S, Kalb J, Buchel G, Ade CP, Baluapuri A, Xu J, Koster J, Solvie D, Carstensen A, Klotz C et al. 2019. Recruitment of BRCA1 limits MYCN-driven accumulation of stalled RNA polymerase. Nature 567: 545–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodson C, van Twest S, Dylewska M, O’Rourke JJ, Tan W, Murphy VJ, Walia M, Abbouche L, Nieminuszczy J, Dunn E et al. 2022. Branchpoint translocation by fork remodelers as a general mechanism of R-loop removal. Cell Rep 41: 111749. [DOI] [PubMed] [Google Scholar]
- Huertas P, Aguilera A. 2003. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell 12: 711–721. [DOI] [PubMed] [Google Scholar]
- Hyjek M, Figiel M, Nowotny M. 2019. RNases H: Structure and mechanism. DNA Repair (Amst) 84: 102672. [DOI] [PubMed] [Google Scholar]
- Itoh T, Tomizawa J. 1980. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc Natl Acad Sci U S A 77: 2450–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA. 2003. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol Cell 12: 1525–1536. [DOI] [PubMed] [Google Scholar]
- Jenkyn-Bedford M, Jones ML, Baris Y, Labib KPM, Cannone G, Yeeles JTP, Deegan TD. 2021. A conserved mechanism for regulating replisome disassembly in eukaryotes. Nature 600: 743–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadesch TR, Chamberlin MJ. 1982. Studies of in vitro transcription by calf thymus RNA polymerase II using a novel duplex DNA template. J Biol Chem 257: 5286–5295. [PubMed] [Google Scholar]
- Kaminski N, Wondisford AR, Kwon Y, Lynskey ML, Bhargava R, Barroso-Gonzalez J, Garcia-Exposito L, He B, Xu M, Mellacheruvu D et al. 2022. RAD51AP1 regulates ALT-HDR through chromatin-directed homeostasis of TERRA. Mol Cell 82: 4001–4017 e4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Cheon NY, Park H, Jeong GW, Ye BJ, Yoo EJ, Lee JH, Hur JH, Lee EA, Kim H et al. 2021. TonEBP recognizes R-loops and initiates m6A RNA methylation for R-loop resolution. Nucleic Acids Res 49: 269–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavlashvili T, Liu W, Mohamed TM, Cortez D, Dewar JM. 2023. Replication fork uncoupling causes nascent strand degradation and fork reversal. Nat Struct Mol Biol 30: 115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Abdulovic AL, Gealy R, Lippert MJ, Jinks-Robertson S. 2007. Transcription-associated mutagenesis in yeast is directly proportional to the level of gene expression and influenced by the direction of DNA replication. DNA Repair (Amst) 6: 1285–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireeva ML, Komissarova N, Kashlev M. 2000. Overextended RNA:DNA hybrid as a negative regulator of RNA polymerase II processivity. J Mol Biol 299: 325–335. [DOI] [PubMed] [Google Scholar]
- Kose HB, Larsen NB, Duxin JP, Yardimci H. 2019. Dynamics of the Eukaryotic Replicative Helicase at Lagging-Strand Protein Barriers Support the Steric Exclusion Model. Cell Rep 26: 2113–2125 e2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotsantis P, Silva LM, Irmscher S, Jones RM, Folkes L, Gromak N, Petermann E. 2016. Increased global transcription activity as a mechanism of replication stress in cancer. Nat Commun 7: 13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasilnikova MM, Mirkin SM. 2004. Replication stalling at Friedreich’s ataxia (GAA)n repeats in vivo. Mol Cell Biol 24: 2286–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar C, Batra S, Griffith JD, Remus D. 2021. The interplay of RNA:DNA hybrid structure and G-quadruplexes determines the outcome of R-loop-replisome collisions. Elife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar C, Remus D. 2022. A transcription-based approach to purify R-loop-containing plasmid DNA templates in vitro. STAR Protoc 4: 101937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafuente-Barquero J, Garcia-Rubio ML, Martin-Alonso MS, Gomez-Gonzalez B, Aguilera A. 2020. Harmful DNA:RNA hybrids are formed in cis and in a Rad51-independent manner. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde M, Trauner M, Werner M, Hamperl S. 2021. Consequences and Resolution of Transcription-Replication Conflicts. Life (Basel) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsverk HB, Sandquist LE, Bay LTE, Steurer B, Campsteijn C, Landsverk OJB, Marteijn JA, Petermann E, Trinkle-Mulcahy L, Syljuasen RG. 2020. WDR82/PNUTS-PP1 Prevents Transcription-Replication Conflicts by Promoting RNA Polymerase II Degradation on Chromatin. Cell Rep 33: 108469. [DOI] [PubMed] [Google Scholar]
- Lang KS, Hall AN, Merrikh CN, Ragheb M, Tabakh H, Pollock AJ, Woodward JJ, Dreifus JE, Merrikh H. 2017. Replication-Transcription Conflicts Generate R-Loops that Orchestrate Bacterial Stress Survival and Pathogenesis. Cell 170: 787–799 e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang KS, Merrikh H. 2018. The Clash of Macromolecular Titans: Replication-Transcription Conflicts in Bacteria. Annu Rev Microbiol 72: 71–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- -. 2021. Topological stress is responsible for the detrimental outcomes of head-on replication-transcription conflicts. Cell Rep 34: 108797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laspata N, Kaur P, Mersaoui SY, Muoio D, Liu ZS, Bannister MH, Nguyen HD, Curry C, Pascal JM, Poirier GG et al. 2023. PARP1 associates with R-loops to promote their resolution and genome stability. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laverde EE, Lai Y, Leng F, Balakrishnan L, Freudenreich CH, Liu Y. 2020. R-loops promote trinucleotide repeat deletion through DNA base excision repair enzymatic activities. J Biol Chem 295: 13902–13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner LK, Sale JE. 2019. Replication of G Quadruplex DNA. Genes (Basel) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Manley JL. 2005. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122: 365–378. [DOI] [PubMed] [Google Scholar]
- Lim YW, Sanz LA, Xu X, Hartono SR, Chedin F. 2015. Genome-wide DNA hypomethylation and RNA:DNA hybrid accumulation in Aicardi-Goutieres syndrome. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WL, Chen JK, Wen X, He W, Zarceno GA, Chen Y, Chen S, Paull TT, Liu HW. 2022. DDX18 prevents R-loop-induced DNA damage and genome instability via PARP-1. Cell Rep 40: 111089. [DOI] [PubMed] [Google Scholar]
- Lin Y, Dent SY, Wilson JH, Wells RD, Napierala M. 2010. R loops stimulate genetic instability of CTG.CAG repeats. Proc Natl Acad Sci U S A 107: 692–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Alberts BM. 1995. Head-on collision between a DNA replication apparatus and RNA polymerase transcription complex. Science 267: 1131–1137. [DOI] [PubMed] [Google Scholar]
- Liu B, Wong ML, Tinker RL, Geiduschek EP, Alberts BM. 1993. The DNA replication fork can pass RNA polymerase without displacing the nascent transcript. Nature 366: 33–39. [DOI] [PubMed] [Google Scholar]
- Liu LF, Wang JC. 1987. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A 84: 7024–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Saito Y, Jackson J, Bhowmick R, Kanemaki MT, Vindigni A, Cortez D. 2023. RAD51 bypasses the CMG helicase to promote replication fork reversal. Science 380: 382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart A, Pires VB, Bento F, Kellner V, Luke-Glaser S, Yakoub G, Ulrich HD, Luke B. 2019. RNase H1 and H2 Are Differentially Regulated to Process RNA-DNA Hybrids. Cell Rep 29: 2890–2900 e2895. [DOI] [PubMed] [Google Scholar]
- Loomis EW, Sanz LA, Chedin F, Hagerman PJ. 2014. Transcription-associated R-loop formation across the human FMR1 CGG-repeat region. PLoS Genet 10: e1004294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes J, Piazza A, Bermejo R, Kriegsman B, Colosio A, Teulade-Fichou MP, Foiani M, Nicolas A. 2011. G-quadruplex-induced instability during leading-strand replication. EMBO J 30: 4033–4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu J, Shao R, Kwong Yung PY, Elsasser SJ. 2022. Genome-wide mapping of G-quadruplex structures with CUT&Tag. Nucleic Acids Res 50: e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malig M, Hartono SR, Giafaglione JM, Sanz LA, Chedin F. 2020. Ultra-deep Coverage Single-molecule R-loop Footprinting Reveals Principles of R-loop Formation. J Mol Biol 432: 2271–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiameli SM, Merrikh CN, Wiggins PA, Merrikh H. 2017. Transcription leads to pervasive replisome instability in bacteria. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E, Drolet M. 1999. Escherichia coli DNA topoisomerase I inhibits R-loop formation by relaxing transcription-induced negative supercoiling. J Biol Chem 274: 16659–16664. [DOI] [PubMed] [Google Scholar]
- Matos DA, Zhang JM, Ouyang J, Nguyen HD, Genois MM, Zou L. 2020. ATR Protects the Genome against R Loops through a MUS81-Triggered Feedback Loop. Mol Cell 77: 514–527 e514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, Landry HM, Churchman LS. 2017. Pause & go: from the discovery of RNA polymerase pausing to its functional implications. Curr Opin Cell Biol 46: 72–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure AW, Diffley JF. 2021. Rad53 checkpoint kinase regulation of DNA replication fork rate via Mrc1 phosphorylation. Elife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor C, Perez C, Sale JE. 2022. Creation and resolution of non-B-DNA structural impediments during replication. Crit Rev Biochem Mol Biol 57: 412–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrikh CN, Brewer BJ, Merrikh H. 2015. The B. subtilis Accessory Helicase PcrA Facilitates DNA Replication through Transcription Units. PLoS Genet 11: e1005289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrikh H, Machon C, Grainger WH, Grossman AD, Soultanas P. 2011. Co-directional replication-transcription conflicts lead to replication restart. Nature 470: 554–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersaoui SY, Yu Z, Coulombe Y, Karam M, Busatto FF, Masson JY, Richard S. 2019. Arginine methylation of the DDX5 helicase RGG/RG motif by PRMT5 regulates resolution of RNA:DNA hybrids. EMBO J 38: e100986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin EV, Mirkin SM. 2005. Mechanisms of transcription-replication collisions in bacteria. Mol Cell Biol 25: 888–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischo HE, Gomez-Gonzalez B, Grzechnik P, Rondon AG, Wei W, Steinmetz L, Aguilera A, Proudfoot NJ. 2011. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell 41: 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales JC, Richard P, Patidar PL, Motea EA, Dang TT, Manley JL, Boothman DA. 2016. XRN2 Links Transcription Termination to DNA Damage and Replication Stress. PLoS Genet 12: e1006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutreja K, Krietsch J, Hess J, Ursich S, Berti M, Roessler FK, Zellweger R, Patra M, Gasser G, Lopes M. 2018. ATR-Mediated Global Fork Slowing and Reversal Assist Fork Traverse and Prevent Chromosomal Breakage at DNA Interstrand Cross-Links. Cell Rep 24: 2629–2642 e2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil AJ, Liang MU, Khristich AN, Shah KA, Mirkin SM. 2018. RNA-DNA hybrids promote the expansion of Friedreich’s ataxia (GAA)n repeats via break-induced replication. Nucleic Acids Res 46: 3487–3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VC, Clelland BW, Hockman DJ, Kujat-Choy SL, Mewhort HE, Schultz MC. 2010. Replication stress checkpoint signaling controls tRNA gene transcription. Nat Struct Mol Biol 17: 976–981. [DOI] [PubMed] [Google Scholar]
- Osmundson JS, Kumar J, Yeung R, Smith DJ. 2017. Pif1-family helicases cooperatively suppress widespread replication-fork arrest at tRNA genes. Nat Struct Mol Biol 24: 162–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang J, Yadav T, Zhang JM, Yang H, Rheinbay E, Guo H, Haber DA, Lan L, Zou L. 2021. RNA transcripts stimulate homologous recombination by forming DR-loops. Nature 594: 283–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeschke K, Capra JA, Zakian VA. 2011. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell 145: 678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parajuli S, Teasley DC, Murali B, Jackson J, Vindigni A, Stewart SA. 2017. Human ribonuclease H1 resolves R-loops and thereby enables progression of the DNA replication fork. J Biol Chem 292: 15216–15224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Million-Weaver S, Chattopadhyay S, Sokurenko E, Merrikh H. 2013. Accelerated gene evolution through replication-transcription conflicts. Nature 495: 512–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E, Lan L, Zou L. 2022. Sources, resolution and physiological relevance of R-loops and RNA-DNA hybrids. Nat Rev Mol Cell Biol 23: 521–540. [DOI] [PubMed] [Google Scholar]
- Petryk N, Kahli M, d’Aubenton-Carafa Y, Jaszczyszyn Y, Shen Y, Silvain M, Thermes C, Chen CL, Hyrien O. 2016. Replication landscape of the human genome. Nat Commun 7: 10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza A, Adrian M, Samazan F, Heddi B, Hamon F, Serero A, Lopes J, Teulade-Fichou MP, Phan AT, Nicolas A. 2015. Short loop length and high thermal stability determine genomic instability induced by G-quadruplex-forming minisatellites. EMBO J 34: 1718–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza A, Boule JB, Lopes J, Mingo K, Largy E, Teulade-Fichou MP, Nicolas A. 2010. Genetic instability triggered by G-quadruplex interacting Phen-DC compounds in Saccharomyces cerevisiae. Nucleic Acids Res 38: 4337–4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza A, Cui X, Adrian M, Samazan F, Heddi B, Phan AT, Nicolas AG. 2017. Non-Canonical G-quadruplexes cause the hCEB1 minisatellite instability in Saccharomyces cerevisiae. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza A, Serero A, Boule JB, Legoix-Ne P, Lopes J, Nicolas A. 2012. Stimulation of gross chromosomal rearrangements by the human CEB1 and CEB25 minisatellites in Saccharomyces cerevisiae depends on G-quadruplexes or Cdc13. PLoS Genet 8: e1003033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli J, Gerhold CB, Tosi A, Hustedt N, Seeber A, Sack R, Herzog F, Pasero P, Shimada K, Hopfner KP et al. 2016. Mec1, INO80, and the PAF1 complex cooperate to limit transcription replication conflicts through RNAPII removal during replication stress. Genes Dev 30: 337–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz RT, O’Donnell M. 2008. The replisome uses mRNA as a primer after colliding with RNA polymerase. Nature 456: 762–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- -. 2010. Direct restart of a replication fork stalled by a head-on RNA polymerase. Science 327: 590–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L, Ullsperger C, Keller RW, Bustamante C, Vologodskii AV, Cozzarelli NR. 2001. Positive torsional strain causes the formation of a four-way junction at replication forks. J Biol Chem 276: 2790–2796. [DOI] [PubMed] [Google Scholar]
- Prado F, Aguilera A. 2005. Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J 24: 1267–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promonet A, Padioleau I, Liu Y, Sanz L, Biernacka A, Schmitz AL, Skrzypczak M, Sarrazin A, Mettling C, Rowicka M et al. 2020. Topoisomerase 1 prevents replication stress at R-loop-enriched transcription termination sites. Nat Commun 11: 3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaban ME, Griffin JA. 1990. Induction of RNA-stabilized DNA conformers by transcription of an immunoglobulin switch region. Nature 348: 342–344. [DOI] [PubMed] [Google Scholar]
- Reaban ME, Lebowitz J, Griffin JA. 1994. Transcription induces the formation of a stable RNA.DNA hybrid in the immunoglobulin alpha switch region. J Biol Chem 269: 21850–21857. [PubMed] [Google Scholar]
- Reddy K, Tam M, Bowater RP, Barber M, Tomlinson M, Nichol Edamura K, Wang YH, Pearson CE. 2011. Determinants of R-loop formation at convergent bidirectionally transcribed trinucleotide repeats. Nucleic Acids Res 39: 1749–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro de Almeida C, Dhir S, Dhir A, Moghaddam AE, Sattentau Q, Meinhart A, Proudfoot NJ. 2018. RNA Helicase DDX1 Converts RNA G-Quadruplex Structures into R-Loops to Promote IgH Class Switch Recombination. Mol Cell 70: 650–662 e658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeyre C, Lopes J, Boule JB, Piazza A, Guedin A, Zakian VA, Mergny JL, Nicolas A. 2009. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet 5: e1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Feng S, Manley JL. 2013. A SUMO-dependent interaction between Senataxin and the exosome, disrupted in the neurodegenerative disease AOA2, targets the exosome to sites of transcription-induced DNA damage. Genes Dev 27: 2227–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Manley JL. 2017. R Loops and Links to Human Disease. J Mol Biol 429: 3168–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani DF, Bothmer A, Callen E, Reina-San-Martin B, Dorsett Y, Difilippantonio S, Bolland DJ, Chen HT, Corcoran AE, Nussenzweig A et al. 2008. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell 135: 1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R, Miller KM, Forment JV, Bradshaw CR, Nikan M, Britton S, Oelschlaegel T, Xhemalce B, Balasubramanian S, Jackson SP. 2012. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat Chem Biol 8: 301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D, Lieber MR. 2009. G clustering is important for the initiation of transcription-induced R-loops in vitro, whereas high G density without clustering is sufficient thereafter. Mol Cell Biol 29: 3124–3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D, Yu K, Lieber MR. 2008. Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol Cell Biol 28: 50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Yang X, Huang SN, Agama K, Baechler SA, Sun Y, Zhang H, Saha LK, Su S, Jenkins LM et al. 2022. Resolution of R-loops by topoisomerase III-beta (TOP3B) in coordination with the DEAD-box helicase DDX5. Cell Rep 40: 111067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Martin-Alonso M, Soler-Oliva ME, Garcia-Rubio M, Garcia-Muse T, Aguilera A. 2021. Harmful R-loops are prevented via different cell cycle-specific mechanisms. Nat Commun 12: 4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankar TS, Wastuwidyaningtyas BD, Dong Y, Lewis SA, Wang JD. 2016. The nature of mutations induced by replication-transcription collisions. Nature 535: 178–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz LA, Chedin F. 2019. High-resolution, strand-specific R-loop mapping via S9.6-based DNA-RNA immunoprecipitation and high-throughput sequencing. Nat Protoc 14: 1734–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz LA, Hartono SR, Lim YW, Steyaert S, Rajpurkar A, Ginno PA, Xu X, Chedin F. 2016. Prevalent, Dynamic, and Conserved R-Loop Structures Associate with Specific Epigenomic Signatures in Mammals. Mol Cell 63: 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saponaro M, Kantidakis T, Mitter R, Kelly GP, Heron M, Williams H, Soding J, Stewart A, Svejstrup JQ. 2014. RECQL5 controls transcript elongation and suppresses genome instability associated with transcription stress. Cell 157: 1037–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkies P, Reams C, Simpson LJ, Sale JE. 2010. Epigenetic instability due to defective replication of structured DNA. Mol Cell 40: 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Martin-Pintado N, Post H, Altelaar M, Knipscheer P. 2021. Multistep mechanism of G-quadruplex resolution during DNA replication. Sci Adv 7: eabf8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler JA, Conti C, Syed A, Aladjem MI, Pommier Y. 2007. The intra-S-phase checkpoint affects both DNA replication initiation and elongation: single-cell and -DNA fiber analyses. Mol Cell Biol 27: 5806–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekedat MD, Fenyo D, Rogers RS, Tackett AJ, Aitchison JD, Chait BT. 2010. GINS motion reveals replication fork progression is remarkably uniform throughout the yeast genome. Mol Syst Biol 6: 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkura R, Tian M, Smith M, Chua K, Fujiwara Y, Alt FW. 2003. The influence of transcriptional orientation on endogenous switch region function. Nat Immunol 4: 435–441. [DOI] [PubMed] [Google Scholar]
- Shivji MKK, Renaudin X, Williams CH, Venkitaraman AR. 2018. BRCA2 Regulates Transcription Elongation by RNA Polymerase II to Prevent R-Loop Accumulation. Cell Rep 22: 1031–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla V, Samaniego-Castruita D, Dong Z, Gonzalez-Avalos E, Yan Q, Sarma K, Rao A. 2022. TET deficiency perturbs mature B cell homeostasis and promotes oncogenesis associated with accumulation of G-quadruplex and R-loop structures. Nat Immunol 23: 99–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Kamieniarz-Gdula K, Proudfoot NJ. 2014. R-loops induce repressive chromatin marks over mammalian gene terminators. Nature 516: 436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ, Gromak N. 2011. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell 42: 794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Torlai Triglia E, Warburton M, Voigt P, Bird A, Pombo A. 2019. R-Loops Enhance Polycomb Repression at a Subset of Developmental Regulator Genes. Mol Cell 73: 930–945 e934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollier J, Stork CT, Garcia-Rubio ML, Paulsen RD, Aguilera A, Cimprich KA. 2014. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell 56: 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sordet O, Redon CE, Guirouilh-Barbat J, Smith S, Solier S, Douarre C, Conti C, Nakamura AJ, Das BB, Nicolas E et al. 2009. Ataxia telangiectasia mutated activation by transcription- and topoisomerase I-induced DNA double-strand breaks. EMBO Rep 10: 887–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivatsan A, Tehranchi A, MacAlpine DM, Wang JD. 2010. Co-orientation of replication and transcription preserves genome integrity. PLoS Genet 6: e1000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz R, Sulthana S, Hartono SR, Malig M, Benham CJ, Chedin F. 2019. Interplay between DNA sequence and negative superhelicity drives R-loop structures. Proc Natl Acad Sci U S A 116: 6260–6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork CT, Bocek M, Crossley MP, Sollier J, Sanz LA, Chedin F, Swigut T, Cimprich KA. 2016. Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoy H, Zwicky K, Kuster D, Lang KS, Krietsch J, Crossley MP, Schmid JA, Cimprich KA, Merrikh H, Lopes M. 2023. Direct visualization of transcription-replication conflicts reveals post-replicative DNA:RNA hybrids. Nat Struct Mol Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su XA, Freudenreich CH. 2017. Cytosine deamination and base excision repair cause R-loop-induced CAG repeat fragility and instability in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 114: E8392–E8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svikovic S, Crisp A, Tan-Wong SM, Guilliam TA, Doherty AJ, Proudfoot NJ, Guilbaud G, Sale JE. 2019. R-loop formation during S phase is restricted by PrimPol-mediated repriming. EMBO J 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi Y, Horiuchi T, Kobayashi T. 2003. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev 17: 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan-Wong SM, Dhir S, Proudfoot NJ. 2019. R-Loops Promote Antisense Transcription across the Mammalian Genome. Mol Cell 76: 600–616 e606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja N, Zofall M, Balachandran V, Thillainadesan G, Sugiyama T, Wheeler D, Zhou M, Grewal SI. 2017. SNF2 Family Protein Fft3 Suppresses Nucleosome Turnover to Promote Epigenetic Inheritance and Proper Replication. Mol Cell 66: 50–62 e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MRG, Yeeles JTP. 2018. The Initial Response of a Eukaryotic Replisome to DNA Damage. Mol Cell 70: 1067–1080 e1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M, White RL, Davis RW. 1976. Hybridization of RNA to double-stranded DNA: formation of R-loops. Proc Natl Acad Sci U S A 73: 2294–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Alt FW. 2000. Transcription-induced cleavage of immunoglobulin switch regions by nucleotide excision repair nucleases in vitro. J Biol Chem 275: 24163–24172. [DOI] [PubMed] [Google Scholar]
- Tran PLT, Pohl TJ, Chen CF, Chan A, Pott S, Zakian VA. 2017. PIF1 family DNA helicases suppress R-loop mediated genome instability at tRNA genes. Nat Commun 8: 15025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresini M, Warmerdam DO, Kolovos P, Snijder L, Vrouwe MG, Demmers JA, van IWF, Grosveld FG, Medema RH, Hoeijmakers JH et al. 2015. The core spliceosome as target and effector of non-canonical ATM signalling. Nature 523: 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsirkas I, Dovrat D, Thangaraj M, Brouwer I, Cohen A, Paleiov Z, Meijler MM, Lenstra T, Aharoni A. 2022. Transcription-replication coordination revealed in single live cells. Nucleic Acids Res 50: 2143–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuduri S, Crabbe L, Conti C, Tourriere H, Holtgreve-Grez H, Jauch A, Pantesco V, De Vos J, Thomas A, Theillet C et al. 2009. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol 11: 1315–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulsamer A, Martinez-Limon A, Bader S, Rodriguez-Acebes S, Freire R, Mendez J, de Nadal E, Posas F. 2022. Regulation of Claspin by the p38 stress-activated protein kinase protects cells from DNA damage. Cell Rep 40: 111375. [DOI] [PubMed] [Google Scholar]
- Urban V, Dobrovolna J, Huhn D, Fryzelkova J, Bartek J, Janscak P. 2016. RECQ5 helicase promotes resolution of conflicts between replication and transcription in human cells. J Cell Biol 214: 401–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrutia-Irazabal I, Ault JR, Sobott F, Savery NJ, Dillingham MS. 2021. Analysis of the PcrA-RNA polymerase complex reveals a helicase interaction motif and a role for PcrA/UvrD helicase in the suppression of R-loops. Elife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. 2007. Structural basis for transcription elongation by bacterial RNA polymerase. Nature 448: 157–162. [DOI] [PubMed] [Google Scholar]
- Vrtis KB, Dewar JM, Chistol G, Wu RA, Graham TGW, Walter JC. 2021. Single-strand DNA breaks cause replisome disassembly. Mol Cell 81: 1309–1318 e1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vujanovic M, Krietsch J, Raso MC, Terraneo N, Zellweger R, Schmid JA, Taglialatela A, Huang JW, Holland CL, Zwicky K et al. 2017. Replication Fork Slowing and Reversal upon DNA Damage Require PCNA Polyubiquitination and ZRANB3 DNA Translocase Activity. Mol Cell 67: 882–890 e885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba L, Costantino L, Tan FJ, Zimmer A, Koshland D. 2016. S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev 30: 1327–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba L, Gore SK, Koshland D. 2013. The homologous recombination machinery modulates the formation of RNA-DNA hybrids and associated chromosome instability. Elife 2: e00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IX, Grunseich C, Fox J, Burdick J, Zhu Z, Ravazian N, Hafner M, Cheung VG. 2018. Human proteins that interact with RNA/DNA hybrids. Genome Res 28: 1405–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JD, Berkmen MB, Grossman AD. 2007. Genome-wide coorientation of replication and transcription reduces adverse effects on replication in Bacillus subtilis. Proc Natl Acad Sci U S A 104: 5608–5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Wang H, Li C, Yin Z, Xiao R, Li Q, Xiang Y, Wang W, Huang J, Chen L et al. 2021. Genomic profiling of native R loops with a DNA-RNA hybrid recognition sensor. Sci Adv 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger RE, Prado F, Aguilera A. 2006. Replication fork progression is impaired by transcription in hyperrecombinant yeast cells lacking a functional THO complex. Mol Cell Biol 26: 3327–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells JP, White J, Stirling PC. 2019. R Loops and Their Composite Cancer Connections. Trends Cancer 5: 619–631. [DOI] [PubMed] [Google Scholar]
- Westover KD, Bushnell DA, Kornberg RD. 2004. Structural basis of transcription: separation of RNA from DNA by RNA polymerase II. Science 303: 1014–1016. [DOI] [PubMed] [Google Scholar]
- Wu T, Nance J, Chu F, Fazzio TG. 2021. Characterization of R-Loop-Interacting Proteins in Embryonic Stem Cells Reveals Roles in rRNA Processing and Gene Expression. Mol Cell Proteomics 20: 100142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulfridge P, Sarma K. 2021. A nuclease- and bisulfite-based strategy captures strand-specific R-loops genome-wide. Elife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Xu H, Li K, Fan Y, Liu Y, Yang X, Sun Q. 2017. The R-loop is a common chromatin feature of the Arabidopsis genome. Nat Plants 3: 704–714. [DOI] [PubMed] [Google Scholar]
- Yadav T, Zhang JM, Ouyang J, Leung W, Simoneau A, Zou L. 2022. TERRA and RAD51AP1 promote alternative lengthening of telomeres through an R- to D-loop switch. Mol Cell 82: 3985–4000 e3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Shields EJ, Bonasio R, Sarma K. 2019. Mapping Native R-Loops Genome-wide Using a Targeted Nuclease Approach. Cell Rep 29: 1369–1380 e1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Wulfridge P, Doherty J, Fernandez-Luna JL, Real PJ, Tang HY, Sarma K. 2022. Proximity labeling identifies a repertoire of site-specific R-loop modulators. Nat Commun 13: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung R, Smith DJ. 2020. Determinants of Replication-Fork Pausing at tRNA Genes in Saccharomyces cerevisiae. Genetics 214: 825–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. 2003. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol 4: 442–451. [DOI] [PubMed] [Google Scholar]
- Zardoni L, Nardini E, Brambati A, Lucca C, Choudhary R, Loperfido F, Sabbioneda S, Liberi G. 2021. Elongating RNA polymerase II and RNA:DNA hybrids hinder fork progression and gene expression at sites of head-on replication-transcription collisions. Nucleic Acids Res 49: 12769–12784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatreanu D, Han Z, Mitter R, Tumini E, Williams H, Gregersen L, Dirac-Svejstrup AB, Roma S, Stewart A, Aguilera A et al. 2019. Elongation Factor TFIIS Prevents Transcription Stress and R-Loop Accumulation to Maintain Genome Stability. Mol Cell 76: 57–69 e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller P, Padeken J, van Schendel R, Kalck V, Tijsterman M, Gasser SM. 2016. Histone H3K9 methylation is dispensable for Caenorhabditis elegans development but suppresses RNA:DNA hybrid-associated repeat instability. Nat Genet 48: 1385–1395. [DOI] [PubMed] [Google Scholar]
- Zhang X, Chiang HC, Wang Y, Zhang C, Smith S, Zhao X, Nair SJ, Michalek J, Jatoi I, Lautner M et al. 2017. Attenuation of RNA polymerase II pausing mitigates BRCA1-associated R-loop accumulation and tumorigenesis. Nat Commun 8: 15908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Zhu M, Limbo O, Russell P. 2018. RNase H eliminates R-loops that disrupt DNA replication but is nonessential for efficient DSB repair. EMBO Rep 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer AD, Koshland D. 2016. Differential roles of the RNases H in preventing chromosome instability. Proc Natl Acad Sci U S A 113: 12220–12225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


