Summary
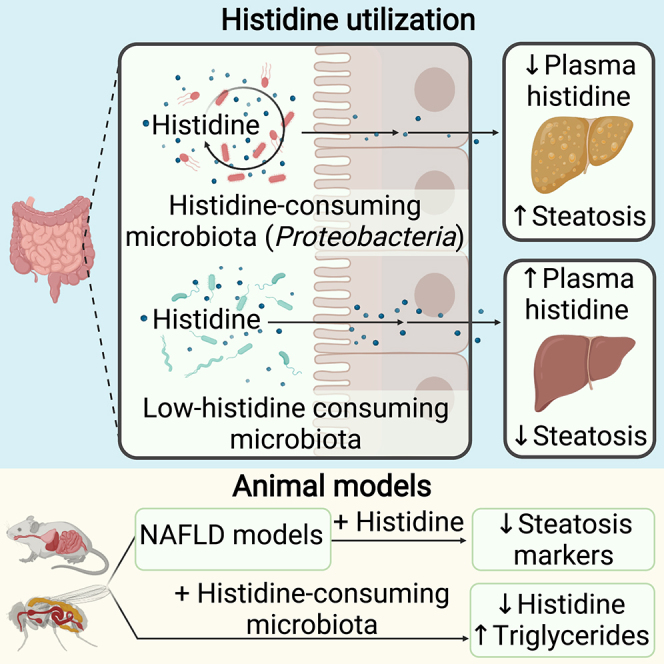
The gut microbiota contributes to the pathophysiology of non-alcoholic fatty liver disease (NAFLD). Histidine is a key energy source for the microbiota, scavenging it from the host. Its role in NAFLD is poorly known. Plasma metabolomics, liver transcriptomics, and fecal metagenomics were performed in three human cohorts coupled with hepatocyte, rodent, and Drosophila models. Machine learning analyses identified plasma histidine as being strongly inversely associated with steatosis and linked to a hepatic transcriptomic signature involved in insulin signaling, inflammation, and trace amine-associated receptor 1. Circulating histidine was inversely associated with Proteobacteria and positively with bacteria lacking the histidine utilization (Hut) system. Histidine supplementation improved NAFLD in different animal models (diet-induced NAFLD in mouse and flies, ob/ob mouse, and ovariectomized rats) and reduced de novo lipogenesis. Fecal microbiota transplantation (FMT) from low-histidine donors and mono-colonization of germ-free flies with Enterobacter cloacae increased triglyceride accumulation and reduced histidine content. The interplay among microbiota, histidine catabolism, and NAFLD opens therapeutic opportunities.
Keywords: amino acids, Hut operon, Proteobacteria, hepatic disease, omics, dysbiosis, NAFLD, histidine
Graphical abstract
Highlights
-
•
Reduced plasma histidine correlates with increased hepatic steatosis in humans
-
•
Histidine alleviates hepatic steatosis in vitro and in vivo in mice, rats, and flies
-
•
Gut microbiota influences NAFLD throughout histidine catabolism
-
•
Drosophila colonization with E. cloacae increases TG and reduces histidine content
Quesada-Vázquez et al. reveal an association between low plasma histidine levels and NAFLD in humans. Histidine supplementation decreased the degree of steatosis in several NAFLD models. Patients with a higher degree of steatosis have increased levels of Proteobacteria and microbial functions involved in histidine catabolism.
Introduction
Non-alcoholic fatty liver disease (NAFLD) is one of the most prevalent metabolic diseases, affecting one-third of the world’s population.1 It is characterized by excessive fat accumulation in the liver, accompanied by low-grade inflammation and insulin resistance, which can lead to fibrosis and, eventually, irreversible cirrhosis or hepatocarcinoma.2 Given the paucity of targeted agents to treat/prevent NAFLD, therapeutic avenues are urgently required.
The liver is a central organ orchestrating amino acid metabolism,3 with these metabolites linked to type 2 diabetes and NAFLD.4,5,6 Different medical conditions, such as obesity, chronic kidney disease, and heart failure, are associated with decreased histidine levels.7,8,9 Histidine has several beneficial effects, including antioxidant, anti-inflammatory, antiglycating, and chelating properties.10 Histidine supplementation has a beneficial effect on ethanol- and acetaminophen-induced mouse models of liver injury.11,12 Results from randomized controlled trials have shown that supplementation with histidine and histidine-containing dipeptides may improve obesity and glycemic control, but the effects on liver function are unclear.13 The only clinical study assessing the effects of histidine supplementation in obese women with metabolic syndrome reported an improvement in insulin resistance through downregulation of inflammation but no changes in serum liver enzymes.14 Therefore, little is known about the associations and underlying mechanisms of histidine in NAFLD. Importantly, the gut microbiota plays a key role in regulating dietary histidine bioavailability15 and is involved in hepatic alterations leading to NAFLD.16 Nevertheless, it is currently unknown whether a connection among the gut microbiota, histidine metabolism, and NAFLD exists.
Thus, we used an integrative systems medicine approach in large, well-characterized NAFLD cohorts, combining plasma metabolomics, fecal metagenomics, hepatic transcriptomics, and fecal microbiota transplantation to assess the relationship between histidine metabolism and NAFLD. Further in vitro human primary hepatocytes and rodent and Drosophila models were used to assess the effects of histidine-related amino acid (HAA) supplementation as a strategy for the treatment of NAFLD.
Results
Circulating histidine levels in different cohorts depending on the degree of steatosis
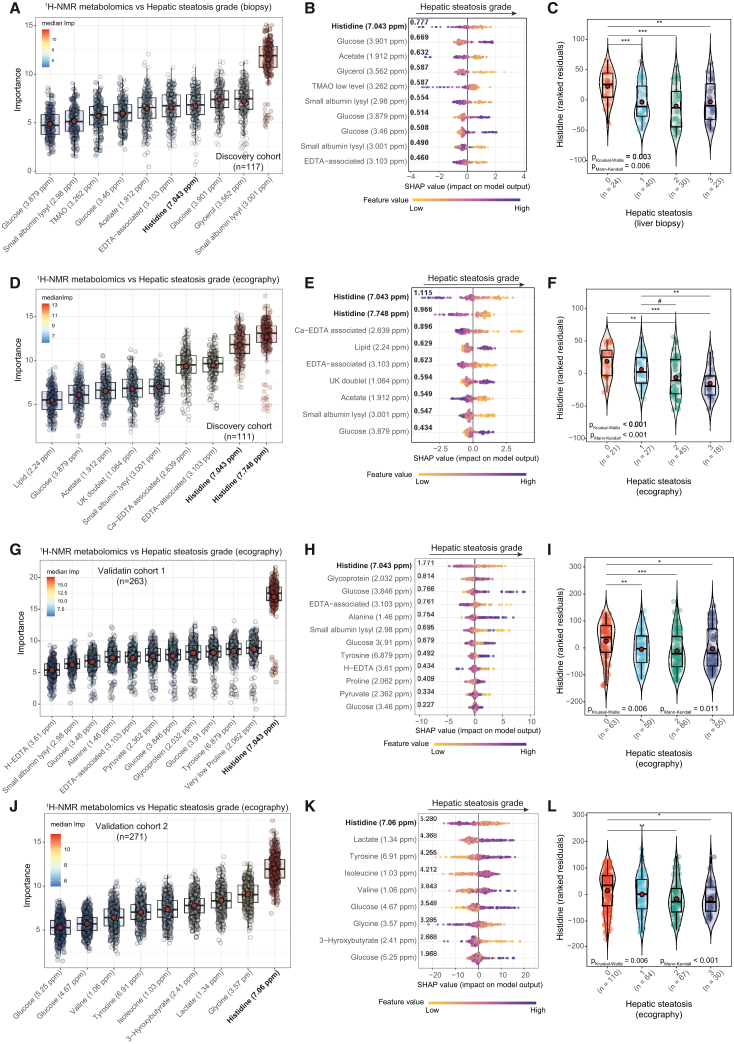
We profiled the plasma metabolome of 117 non-diabetic obese patients from a discovery cohort from Italy and Spain (Table S1) by 1H nuclear magnetic resonance (NMR). Then, we applied a machine learning (ML) variable selection strategy controlling for age, BMI, gender, and country to identify metabolites associated with the liver steatosis grade (pBonferroni < 0.005) as assessed by liver biopsy (Figure 1A; Table S2). We also calculated the exact Shapley additive explanation (SHAP) scores of the selected metabolites to facilitate model interpretation.17 It highlighted histidine as the metabolite with the highest predictive ability (SHAP mean value = 0.777) and negatively associated with the steatosis grade (Figure 1B). Other metabolites negatively associated (significance based on the Boruta algorithm: pBonferroni < 0.005 and sign of the association based on SHAP values) with the steatosis grade included acetate, glycerol, and trimethylamine N-oxide (TMAO), whereas glucose was positively associated (Figures 1A and 1B). NAFLD and diabetes usually coexist and act synergistically to increase the risk of adverse outcomes.18 Although patients in the discovery cohort were non-diabetic, we found that the circulating histidine levels were negatively associated with the fasting glucose levels (Table S3). However, after additionally adjusting the models for glucose concentrations, histidine was still one of the metabolites most associated with the steatosis grade in the discovery cohort (Figure S1A). We also assessed the liver steatosis grade using echography (ultrasound). Our ML approach and the corresponding SHAP values revealed again that histidine had the strongest association (variable importance measure [VIM] = 12.6, pBonferroni < 0.005; absolute SHAP mean value = 1.115) with the ultrasound-based steatosis grade (Figures 1D and 1E; Table S4). Again, we also found negative associations with acetate and positive associations with glucose (Figure 1E).
Figure 1.
Associations of the plasma metabolome with the steatosis grade
(A, D, G, and J) Boxplots of the normalized permutated variable importance measure (VIM) for the metabolites associated with the liver steatosis grade after controlling for age, BMI, gender, and country in the discovery cohorts (n = 117, biopsy; n = 111, echography), validation cohort 1 (n = 263, echography), and validation cohort 2 (n = 271, echography), respectively. Metabolites were identified using the random forest-based ML variable selection algorithm Boruta using 2,000 trees, 500 iterations, and pBonferroni < 0.005.
(B, E, H, and K) SHAP summary plot of the metabolites associated with the liver steatosis grade selected by the Boruta algorithm in the discovery (biopsy and echography), validation 1 (echography), and validation 2 (echography) cohorts, respectively. Each dot represents an individual sample. The x axis represents the SHAP value: the impact of a specific metabolite on the liver steatosis grade prediction of a specific individual. The overall importance for final prediction (average absolute SHAP values) is shown in bold. Colors represent the values of the metabolite normalized concentrations, ranging from yellow (low concentrations of the specific metabolite) to purple (high concentrations of the specific metabolite).
(C, F, I, and L) Violin plots showing the ranked residuals (after adjusting for age, BMI, gender, and country) of plasma histidine levels according to the degree of steatosis assessed by liver biopsy in the discovery cohort (n = 117), liver echography in the discovery cohort (n = 111), liver echography in validation cohort 1 (n = 263), and liver echography in validation cohort 2 (n = 271), respectively. Statistical significance was assessed using both Kruskal-Wallis and Mann-Kendall trend tests, and between-group comparisons were assessed using the Wilcoxon test. #p < 0.1, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
We next sought to validate these associations in a validation cohort of obese patients (BMI > 30 kg/m2) from Italy and Spain (validation cohort 1, n = 263; Table S5) and another validation cohort including obese patients only from Spain (validation cohort 2, n = 271; Table S6) that had measurements of liver steatosis grade by echography. Again, histidine was the metabolite most strongly and negatively associated with the steatosis grade both in validation cohort 1 (VIM = 16.91, pBonferroni < 0.005; absolute SHAP mean value = 1.771) (Figures 1G and 1H; Table S7) and validation cohort 2 (VIM = 12.00, pBonferroni < 0.005; absolute SHAP mean value = 5.280) (Figures 1J and 1K; Table S8) after controlling for age, BMI, gender, and country. The strong associations between histidine and the steatosis degree in the validation cohorts were still significant after additionally controlling for the fasting glucose levels (Figures S1B and S1C). A close look to the histidine levels (adjusting for age, BMI, gender, and country) highlighted that the main difference in histidine levels in all cohorts was between obese patients without (<5%) and with (>5%) steatosis (Figures 1C, 1F, 1I, and 1L).
Microarray analysis of human liver genes associated with circulatory levels of histidine
We next aimed to identify hepatic transcriptomic signatures associated with the plasma histidine levels. We fitted robust linear regression models (controlling for age, BMI, gender, and country) in a subset of patients (n = 88) from the discovery cohort with hepatic transcriptomes to identify significant genes (padj.< 0.1) (Figure 2A; Table S9). Trace amine-associated receptor 1 (TAAR1) was one of the hepatic gene transcripts most significantly associated with plasma histidine levels (log fold change [logFC] = 189.5, padj. = 9.07 × 10−24). Further adjusting the models for the fasting glucose levels did not change the results (Table S10). Including additional potential confounders associated with histidine, such as high-sensitivity C-reactive protein (hs-CRP) and triglyceride (TG) levels did not alter the results either (Table S11). To identify potential mechanisms underlying the effects of histidine on liver steatosis, we integrated gene-gene interaction networks with pathway over-representation analyses on the 93 histidine-associated gene transcripts. On the Reactome database, the most significant cluster included the G alpha (s) (pFDR = 3.6 × 10−6) and the olfactory signaling pathways (pFDR = 8.1 × 10−4) (Figures 2B and 2C; Table S12). Other significant clusters involved the regulation of insulin growth factors (IGFs) (pFDR = 2.63 × 10−5); the activation of fibroblast growth factor receptors (FGFRs), inflammatory regulation pathways, and insulin signaling pathways, such as activation of the phosphatidylinositol 3-kinase (PI3K)-AKT pathway (padj. = 0.0074); and fatty acyl-coenzyme A (CoA) biosynthesis (padj. = 0.035) (Figures 2B and 2C). Significant genes involved in these pathways are shown in Figure 2D. Kyoto Encyclopedia of Genes and Genomes (KEGG)-based analysis also identified fatty acid biosynthesis (padj. = 0.0013), PI3K-AKT signaling (padj. = 0.0076), inflammatory (padj. = 0.0013) and immune regulation (padj. = 0.0045), and olfactory transduction (padj. = 0.024) as relevant pathways linked to circulating histidine levels (Figure S1; Table S13).
Figure 2.
Associations of the liver transcriptome gene expression with the circulating histidine levels
(A) Volcano plot of differentially expressed gene transcripts associated with the plasma histidine levels in the discovery cohort (n = 88), identified using robust linear regression controlling for age, BMI, gender, and country. The log2(FC) associated with a unit change in the plasma histidine levels and the −log10(p value) adjusted for multiple testing are plotted for each transcript.
(B) Dot plot of the results of the Reactome-based over-representation analysis performed on active subnetworks grouped by hierarchical clustering. The x axis represents the fold enrichment defined as the ratio of the frequency of input genes annotated in a pathway to the frequency of all genes annotated to that pathway. The dot size indicates the number of differentially expressed genes in a given pathway. Dots are colored by the −log10(pFDR), with red indicating higher significance.
(C) Enrichment map inter-related significant pathways identified using an active subnetwork-oriented approach. Each color displays a cluster of related pathways using a threshold for kappa statistics of 0.35. The size of the nodes corresponds to its −log10(pFDR). The thickness of the edges between nodes corresponds to the kappa statistic between the two nodes.
(D) Gene concept network depicting significant genes involved in enriched pathways from selected clusters. The dot size of the pathways represents the −log10(pFDR). Pathways with the same color correspond to the same cluster.
To further rule out the contribution of other potential confounders in our findings, we also identified genes associated with hs-CRP, glucose, and TG levels (Table S14. Liver genes associated with hs-CRP in the discovery cohort, controlling for age, BMI, sex, and country (n = 88), related to Figure 2, Table S15. Liver genes associated with glucose levels in the discovery cohort, controlling for age, BMI, sex, and country (n = 88), related to Figure 2, Table S16. Liver genes associated with TG levels in the discovery cohort, controlling for age, BMI, sex, and country (n = 88), related to Figure 2) as well as the related over-represented Reactome pathways (Table S17. Reactome-based over-representation analysis results from significant genes (padj<0.1) associated with hs-CRP, related to Figure 2, Table S18. Reactome-based over-representation analysis results from significant genes (padj<0.1) associated with glucose, related to Figure 2, Table S19. Reactome-based over-representation analysis results from significant genes (padj<0.1) associated with TGs, related to Figure 2). Importantly, the genes associated with these factors were different from those associated with the circulating histidine levels, including TAAR1, which was not associated with any of these confounding factors. In addition, the pathways associated with hs-CRP, glucose, and TGs levels were different from those linked to histidine and involved mainly transporters (Table S17), cell cycle and transcription regulation (Table S18), and collagen formation (Table S19), respectively.
Histidine supplementation modulates NAFLD in human primary hepatocytes
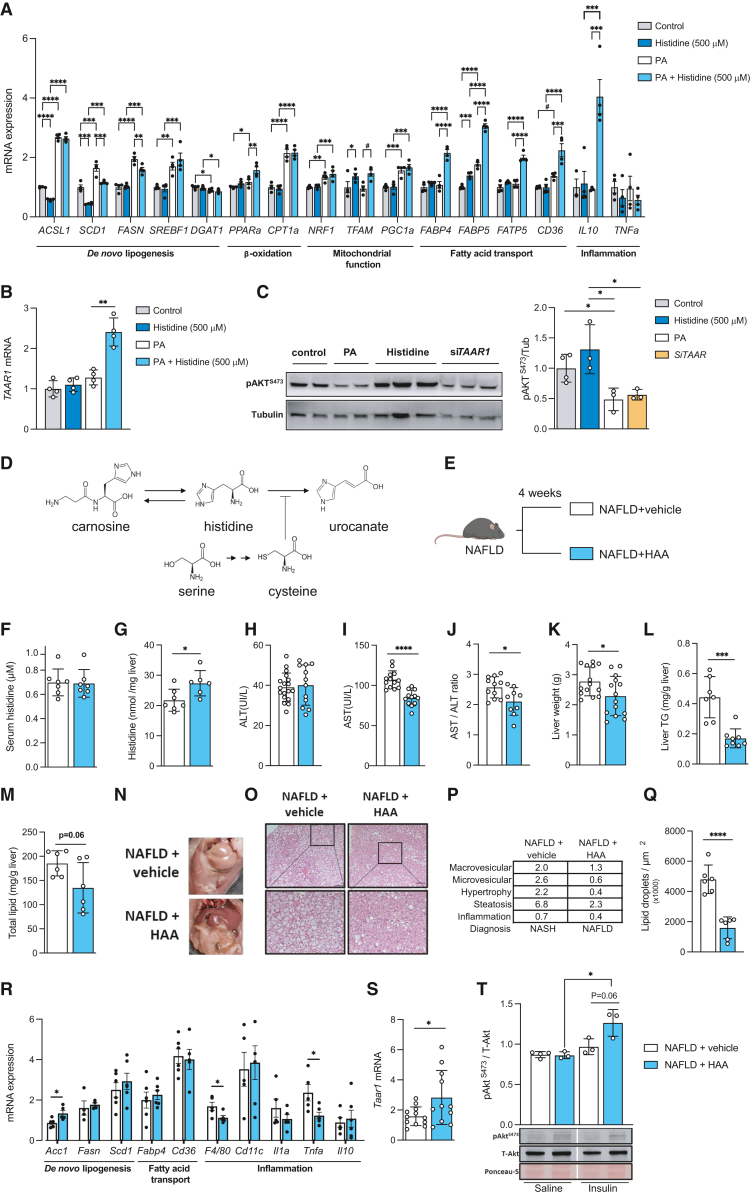
To elucidate the role and underlying mechanisms of histidine in NAFLD, we treated human primary hepatocytes with histidine, palmitic acid (PA; a free fatty acid known to trigger hepatic steatosis in human primary hepatocytes19), or PA and histidine. We then assessed the expression of genes involved in steatosis and de novo hepatic lipogenesis, β-oxidation, mitochondrial function, fatty acid transport, and inflammation. PA induced the expression of lipid metabolism genes (ACSL1, p < 0.0001; SCD1, p = 0.0001; FASN, p < 0.0001; SREBF1, p = 0.003; Figure 3A), whereas co-treatment with PA and histidine led to lower expression of both SCD1 (p = 0.0003) and FASN (p = 0.0034) compared with PA treatment alone, indicating a reduction in de novo hepatic lipogenesis (Figure 3A). We also found an increase in the expression of genes involved in mitochondrial function and fatty acid transport, such as PPARa (p = 0.0026), TFAM (p = 0.014), FABP4 (p < 0.0001), FABP5 (p < 0.0001), FATP5 (p < 0.0001), and CD36 (p = 0.0004) in co-treatment with PA and histidine compared with PA treatment alone (Figure 3A). In addition, co-treatment with PA and histidine promoted a strong upregulation in the expression of the anti-inflammatory cytokine IL10 (p = 0.0004; Figure 3A). Consistent with the results from the human liver microarray, the expression of TAAR1 was strongly induced after co-treatment with PA and histidine (Figure 3B).
Figure 3.
Histidine supplementation modulates NAFLD in human primary hepatocytes and mice
(A) mRNA expression of human primary hepatocyte genes treated with or without histidine (500 μM) and PA, involved in de novo lipogenesis (ACSL1, SCD1, FASN, SREBF1, and DGAT1), β-oxidation (PPARa and CPT1a), mitochondrial function (NRF1, TFAM, and PGC1a), fatty acid transport (FABP4, FABP5, FATP5, and CD36), and inflammation (IL10 and TNFa).
(B) mRNA expression of TAAR1 in primary human hepatocytes treated with or without histidine (500 μM) and PA.
(C) A representative western blot analysis with Akt activation (pAktS473), tubulin levels as housekeeping, and densitometry analysis of phosphorylated AktS473 and tubulin ratio in primary hepatocytes treated with PA or histidine (500 μM) or knockdown of TAAR1 expression.
(D) Schematic of the histidine metabolism pathway.
(E) Schematic of the animal model 1. after induction of NAFLD, animals were treated for 4- weeks with a combination of HAAs (histidine, serine, carnosine and cysteine).
(F–M) Effects of histidine amino acids treatments on the animal model: (F) serum histidine levels; (G) hepatic histidine levels; (H) serum ALT; (I) serum AST; (J) AST/ALT ratio; (K) liver weight; (L) total hepatic TG content; and (M) total hepatic lipid content.
(N) Representative macroscopic appearance of livers.
(O) Hepatic histopathology with hematoxylin and eosin (H&E) staining (magnified area at the bottom). Scale bar, 100 μm).
(P) NAFLD/NASH scoring table. st., steatosis.
(Q) Lipid droplet count.
(R) Hepatic mRNA expression of genes related to de novo lipogenesis (Acc1, Fasn, and Scd1), lipid transport (Cd36 and Fabp4), and inflammation (F4/80, Cd11c, Il1a, Tnfa, and Il10).
(S) Hepatic mRNA expression of Taar1.
(T) Top: representative analysis of Akt activation (pAktS473), total Akt protein levels (T-Akt), and protein loading with Ponceau-S membrane staining. Bottom: densitometry of pAktS473and total Akt ratio.
Data are mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Our pathway enrichment analyses identified PI3K-AKT signaling as one of the pathways most associated with plasma histidine levels. Thus, we next investigated insulin signaling by monitoring AktS473 phosphorylation (Figure 3C). Treatment with PA significantly lowered AktS473 phosphorylation, suggesting that PA reduces the response to insulin, whereas histidine treatment increased phosphorylation (Figure 3C). Remarkably, knockdown of TAAR1 expression with specific small interfering RNA (siRNA) induced a decrease in AktS473 phosphorylation similar to that observed after PA treatment (Figure 3C).
HAA supplementation reduces the main features of NAFLD in mice
To further elucidate the role of histidine in the development of NAFLD, we supplemented a combination of HAAs (histidine, serine, carnosine, and cysteine) to mice fed a high-fructose and high-fat diet (Figures 3D and 3E). We have shown previously that this model induces NAFLD.2 Histidine is an essential amino acid that must be obtained from the diet. Moreover, supplementation of the histidine-containing dipeptide carnosine may increase histidine levels, while cysteine is an inhibitor of histidine ammonia lyase (HAL),20 which catabolizes histidine to urocanic acid, and its levels are regulated by its precursor serine. Although HAA supplementation did not increase serum histidine levels compared with vehicle supplementation (Figure 3F), it increased hepatic histidine levels (p = 0.025; Figure 3G). HAA supplementation had no effect on alanine transaminase (ALT) levels (Figure 3H) but ameliorated several NAFLD-related features, such as aspartate aminotransferase (AST) levels (p < 0.0001; Figure 3I); the AST/ALT ratio (p = 0.018; Figure 3J), which is commonly used to assess liver cell injury;21 the liver weight (Figure 3K); and the hepatic TG levels (Figure 3L). However, it should be acknowledged that AST alone may not be a sufficient indicator of NAFLD. Total hepatic lipid levels also decreased (Figure 3M) in parallel to macroscopic (Figure 3N) and microscopic improvements (Figure 3O). We also observed an amelioration in the histopathological analysis (Figure 3P). This improvement translated from a diagnosis of non-alcoholic steatohepatitis (NASH) in the vehicle animals to a diagnosis of NAFLD in the animals supplemented with HAAs (Figure 3P), together with a reduction in the lipid droplet quantification (p < 0.0001; Figure 3Q).
We next analyzed the liver expression of some genes involved in de novo lipogenesis, lipid transport, and inflammation (Figure 3R). We did not find significant changes in genes involved in lipid transport (Fapb4 and Cd36) or de novo lipid biosynthesis (except for Acc1, p = 0.026) after HAA supplementation. However, it decreased the expression levels of inflammatory markers (F4/80, p = 0.040; Tnfa, p = 0.022) compared with the vehicle (Figure 3R). In line with the human findings, HAA increased the hepatic expression of Taar1 compared with the vehicle group (p = 0.034; Figure 3S). To evaluate the impact of the HAA supplementation on insulin signaling, we performed an intraperitoneal insulin bolus. We observed that NAFLD+HAA mice had a tendency (although not statistically significant, p = 0.06) toward higher pAktS473 phosphorylation after the insulin bolus compared with the NAFLD+vehicle mice, suggesting that HAA supplementation could improve insulin sensitivity (Figure 3T). This result was corroborated by lower fasting glucose (p = 0.011; Figure S3A) and improved responses to glucose tolerance (p = 0.01; Figure S3B) and insulin sensitivity (p = 0.0093; Figure S3C). In addition, HAA supplementation induced a reduction in body weight gain (p = 0.0019; Figure S3D) and adiposity (p = 0.013; Figure S3E).
HAA supplementation protects from hepatic steatosis in a genetic mouse model and a surgical rat model
There are currently no well-established animal models for NAFLD. Therefore, to further validate our findings, we performed additional experiments in two additional murine models of NAFLD: (1) the genetic leptin-deficient ob/ob mouse22 and (2) the surgical ovariectomy (OVX) rat.23 In the ob/ob model, HAA supplementation induced apparent macroscopic (Figure 4A) and microscopic improvements (Figure 4B) with a similar number of lipid droplets (Figure 4C) but with a reduced lipid surface (p = 0.0045; Figure 4D). These features were accompanied by a tendency of reduced total hepatic lipid content (although the results did not reach statistical significance, p = 0.09; Figure 4E) and a reduction in the expression of genes involved in de novo lipogenesis (Acc1, p = 0.0081; Fasn, p = 0.030; Scd1, p = 0.017; Figure 4F). In the OVX model, we found a reduction in the liver weight of HAA-supplemented OVX rats (p = 0.0039; Figure 4G) together with improved microscopic features (Figure 4H), a tendency to reduce the lipid droplets (although not statistically significant, p = 0.07; Figure 4I), and a significant reduction in the lipid surface (p = 0.039; Figure 4J) and total hepatic lipid content (p = 0.031; Figure 4K). In addition, HAA supplementation reduced the expression levels of genes involved in lipid metabolism (Fasn, p = 0.025; Hsl, p = 0.049; Figure 4L) and inflammation (Il1b, p = 0.0361; Figure 4L) compared with supplementation with the vehicle.
Figure 4.
Histidine supplementation alleviates liver steatosis in different animal models
(A‒F) Effects of HAA supplementation in the ob/ob mice model.
(A) Representative macroscopic appearance of livers.
(B) Representative liver micrographs of H&E staining. Scale bar, 100 μm.
(C) Lipid droplet. Count.
(D) Lipid droplet surface field.
(E) Total hepatic lipid content.
(F) Hepatic mRNA expression of genes related to de novo hepatic lipogenesis (Acc1, Fasn, and Scd1), lipid transport (Fabp4), and inflammation (F4/80, Cd11c, Il1a, Tnfa, and Il10).
(G‒L) Effects of HAA supplementation in the OVX rat model.
(G) Liver weight.
(H) Representative liver micrographs of H&E staining. Scale bar, 100 μm.
(I) Lipid droplet count.
(J) Lipid droplet surface field.
(K) Total hepatic lipid content.
(L) Hepatic mRNA expression of genes related to de novo hepatic lipogenesis (Acc1, Fasn, and Scd1), lipolysis (Hsl), lipid transport (Fapt1 and Cd36), and inflammation (Mcp1, Il1b, Tnfa, and Il10).
(M and N) Effects of histidine supplementation (8 g/L) in the NAFLD fly model.
(M) Boxplots represent TG content per fly. TG levels were assessed in 10-day-old flies.
(N) RT-qPCR results of genes related to de novo hepatic lipogenesis (ACC, Desat2, FASN1, SREBP, and mdy), β-oxidation (Eip75B, EcR, whr, and Acoxd57d), lipid transport (Fabp), fatty acid mobilization (Lsd1 and bmm), and insulin receptors (InR and chico).
For all Drosophila experiments, samples contained a pool of 8 flies. Data are mean ± SEM. The p values were determined using Fisher’s least significant difference (LSD) ANOVA test (#p < 0.1, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
Histidine supplementation modulates lipid metabolism in a high-fat diet Drosophila melanogaster model
Finally, we used the alternative model D. melanogaster (fruit flies) to further confirm the role of histidine in the amelioration of NAFLD phenotypes. Drosophila has emerged as a valuable model organism for studying NAFLD. Similar to murine models, the administration of a high-fat diet (HFD) to Drosophila leads to NAFLD-like phenotypes, promoting the accumulation of TGs, stimulation of de novo lipogenesis and fatty acid transport, and a decreased response to insulin.24,25 We found that dietary supplementation of HFD + 8 g/L of histidine significantly reduced the TG accumulation in the flies (p < 0.0001; Figure 4M). At the gene expression level, flies fed an HFD and supplemented with histidine had a significant reduction in the expression of genes related to de novo lipogenesis, such as ACC, Desat2, FASN1, SERPB, and myd (pACC = 0.0007, pDesat2 = 0.0487, pFASN1 < 0.0001, pSERPB = 0.0292, and pmyd = 0.0114). Histidine supplementation also led to a significant increase in expression of Eip75b (pEip75B < 0.0316), the functional homologue of PPARa, and a significant reduction in the expression of whd (pEip75B < 0.0481), the ortholog of human CPT1. It also reduced the expression of genes involved in lipid droplet stabilization (Lsd1, pLsd1 < 0.0001; bmm, pbmm < 0.0001) and had a tendency (although not statistically significant) to downregulate Drosophila insulin receptors (InR, pInR =0.0790), counteracting the effects induced by an HFD (Figure 4N).
Circulating histidine levels are negatively associated with Proteobacteria
We next assessed the potential role of the gut microbiota in modulating plasma histidine levels by performing a shotgun metagenomics analysis of 73 fecal samples of patients from the discovery cohort. We applied the analysis of the composition of the microbiome with bias correction (ANCOM-BC) to identify differential abundant taxa associated with the circulating histidine levels, controlling for age, BMI, gender, and country. Both at the family (Figure 5A; Table S20) and genus (Figure S4A; Table S21) levels, we identified a strong negative association (padj. < 0.1) between histidine levels and members of the phylum Proteobacteria. The inclusion of the fasting glucose levels (Tables S22 and S23) and other potential confounding factors, such as hs-CRP and TGs (Tables S24 and S25), in the models did not alter the results. As we described previously,16 families (Figure 5B; Table S26) and genera (Figure S4B; Table S27) from the phylum Proteobacteria were also strongly associated (padj. < 0.1) with the degree of hepatic steatosis. Circulating histidine levels were also strongly positively associated with Cyanobacteria (logFC = 1.28, padj. = 0.011; Figure 5A), Actinobacteria from the Coriobacteria class (logFC = 3.06, padj. = 0.013; Figures 5A and S4A), and Marinilabiliales (logFC = 1.33, padj. = 0.029; phylum Bacteroidetes) (Figure 5A). At the species level, circulating histidine was positively associated (padj. < 0.1) with bacterial indicators of a healthy gut known to produce anti-inflammatory metabolites, such as species from the genera Faecalibacterium, Bifidobacterium, and Odoribacter26,27(Table S28).
Figure 5.
Associations of plasma histidine and steatosis degree with the gut microbiota and Hut genes
(A and B) Volcano plots of differential bacterial families associated with (A) the circulating histidine levels and (B) hepatic steatosis in the discovery cohort (n = 73), identified using the analysis of microbiomes with bias correction (ANCOM-BC), controlling for age, BMI, gender, and country. The log2(FC) associated with a unit change in the plasma histidine levels and the −log10(p value) adjusted for multiple testing are plotted for each family.
(C) Histidine utilization pathways. The first three pathways appear to be universal. There are two different degradation pathways for formiminoglutamate depending on the genera: hydrolyzation to formamide and glutamate or hydrolyzation to formylglutamate and subsequent hydrolyzation to formate and glutamate.
(D‒G) Violin plots of the centered log ratio-transformed microbial genes involved in histidine utilization (hutH, hutU, hutI, and hutG, respectively) in subjects with a steatosis degree lower or higher than 33%.
(H) Volcano plots of differential bacterial families associated with the hepatic steatosis degree (liver biopsy) in the animal supplementation study for the comparison of NAFLD+vehicle vs. NAFLD+HAA, identified using ANCOM-BC. The log2(FC) and the −log10(p value) adjusted for multiple testing are plotted for each taxon. Significantly different taxa are colored according to phylum.
(I–K) Genus levels of (I) Roseburia and (J) Akkermansia and (K) qPCR of microbial genes expression involved in histidine utilization (hutH and hutG). ∗p < 0.05, ∗∗∗∗p < 0.0001. (K) Data are mean ± SEM.
Histidine is an important carbon and nitrogen source in Proteobacteria28 through its conversion to glutamate. The histidine utilization (hut) operon involves four enzyme-encoding genes: hutH, hutU, hutI, and hutG (Figure 5C).29 Therefore, we next evaluated the associations of these four microbial functions with degree of the steatosis. Remarkably, compared with patients with a steatosis degree of less than 33%, those with greater than 33% of liver steatosis had a higher centered-log ratio (clr)-transformed abundance of microbial genes involved in the first three histidine degradation steps (hutH, p = 0.05; hutU, p = 0.05; hutI, p = 0.07), which appear to be universal and involve the conversion of histidine to urocanate, hydration of urocanate to imidazole propionate, and cleavage of the imidazole ring to give formiminoglutamate (Figures 5D–5G).29
Histidine supplementation ameliorates the gut dysbiosis characteristic of NAFLD
To confirm the direct role of histidine in shaping the composition of the microbiota in NAFLD, we used 16S rRNA gene amplicon sequencing to profile the gut microbiota in the diet-induced NAFLD mouse groups. Consistent with our findings in humans, HAA supplementation resulted in a strong increase in Cyanobacteria (logFC = 2.44, padj. = 0.0005; order Gastranaerophilales) and higher levels of Actinobacteria (logFC = 0.59, padj. = 0.07) and Marinifilaceae (logFC = 1.59, padj. = 0.047) compared with the vehicle-supplemented group (Figure 5H; Table S29). Histidine supplementation also resulted in increased levels of health-promoting bacteria, such as the families Lactobacillaceae (logFC = 2.3, padj. = 0.009) and Christensenellaceae (logFC = 0.51, padj. = 0.08) (Figure 5H). Compared with the vehicle, HAA supplementation in the NAFLD mouse model tended to reduce the levels of the genus Roseburia (although not statistically significant, p = 0.08; Figure 5I) and increase the levels of the genus Akkermansia (Figure 5J), which were decreased with hepatic steatosis in humans (Figure 5B). Remarkably, the expression of bacterial histidine utilization genes (hutH and hutG), which we found to be increased in patients with NAFLD, decreased after HAA supplementation in NAFLD-induced mice compared with their vehicle counterparts (Figure 5K).
Gut microbiota from low-histidine donors promotes hepatic TG accumulation
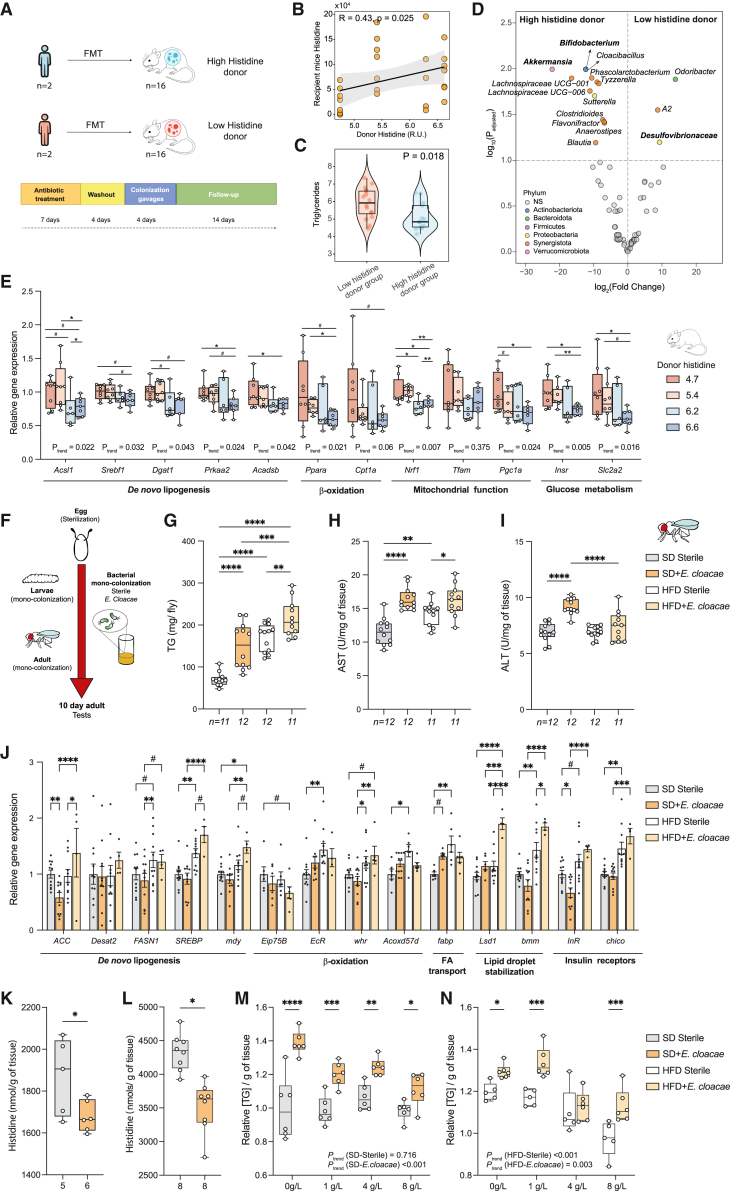
To deepen our insights into the potential role of the microbiota in the progression of NAFLD through alterations in histidine metabolism, we performed an fecal microbiota transplantation (FMT) experiment. We transferred microbiota from donors with histidine levels above the median (high-histidine group, n = 2) and donors with histidine levels below the median (low-histidine group, n = 2) to antibiotic-treated recipient mice (n = 8 mice/donor) (Figure 6A; Table S30). We did not find significant differences in the plasma histidine levels of recipient mice between the FMT groups (p = 0.26; Figure S5A). However, when analyzing the results by donor, we found that mice receiving microbiota from the donor with the lowest histidine levels had lower plasma histidine compared with all other donors (Figure S5B). Therefore, the lack of difference between FMT groups could be due to the fact that, instead of selecting patients with the lowest and highest histidine levels, we selected donors to have a gradual increase in their circulating histidine levels to identify potential dose-response effects. In fact, we found a significant trend toward higher plasma levels in the recipient mice with higher circulating histidine levels of the donor (pMann-Kendall = 0.034) (Figure S5B). Consistently, we also found a positive correlation (R = 0.42, p = 0.025) between the human donor circulating histidine levels and the recipient mouse plasma histidine (Figure 6B). Microbiota transfer from the high-histidine donors also resulted in lower hepatic TGs in recipient mice compared with those receiving a microbiota from the low-histidine donors (p = 0.018; Figure 6C). In addition, we used 16S rRNA gene amplicon sequencing to profile the gut microbiota of the recipient mice (Figure 6D; Table S31). In agreement with our results in humans (Figures 5A and S4A), mice receiving a microbiota from low-histidine donors had higher levels of Desulfovibrionaceae (Figure 6D). Also in line with the findings in humans (Figure 5A; Table S28) and HAA-supplemented mice (Figure 5H; Table S29), mice that received a microbiota from high-histidine donors had higher levels of genera from the phyla Actinobacteria (Bifidobacterium) and Verrucomicrobia (Akkermansia) as well as short-chain fatty-acid (SCFA)-producing genera (Figure 6D). We next predicted the functional potential of the microbiome using PICRUSt2, computed the clr transformation of the knockout (KO), and analyzed the differences in the KO involved in histidine degradation between the two FMT groups. Although not statistically significant, we found a tendency toward lower clr-transformed levels of the KOs involved in the first three histidine degradation steps in mice receiving a microbiota from donors with high histidine levels compared with those receiving a microbiota from donors with low histidine levels (hutH, p = 0.11; hutU, p = 0.12; hutI, p = 0.08; Figures S5C‒S5F), which is similar to our findings in humans (Figures 5D–5G). Finally, we also measured the liver expression of several genes involved in de novo biosynthesis of fatty acids and cholesterol (Acsl1, Srebf1, Dgat1, Prkaa2, and Acadsb), β-oxidation (Ppara and Cpt1a), mitochondrial function (Nrf1, Tfam, and Pgc1a), and glucose metabolism (Insr and Slc2a2). We identified a dose-response effect, with a decrease in the hepatic expression of these genes in the recipient mice with higher histidine levels from the donor (Figure 6E).
Figure 6.
Effects of FMT in mice and mono-colonization of flies with E. cloacae on NAFLD features and histidine
(A) Schematic of FMT from patients with high and low plasma histidine levels to mice (2 human donors per group, 8 mice per human donor).
(B) Spearman correlation between the plasma levels of histidine in the human donor and the recipient mice.
(C) Recipient mouse hepatic TG content in the low- and high-histidine donor groups (Wilcoxon test).
(D) Volcano plot of the recipient mouse microbiota for the comparison between high-histidine donor vs. low-histidine donor groups, identified using ANCOM-BC. The log2(FC) and the –log10(p value) adjusted for multiple testing are plotted for each taxon. Significantly different taxa are colored according to phylum.
(E) Relative gene expression of hepatic genes involved in de novo lipogenesis (Acsl1, Srebf1, Dgat1, Prkaa2, and Acadsb), β-oxidation (Ppara and Cpt1a), mitochondrial function (Nfr1, Tfam, and Pgc1a), and glucose metabolism (Insr and Slc2a2).
(F‒N) Effects of mono-colonization with E. cloacae in germ-free D. melanogaster.
(F) Experimental scheme followed to generate wild-type Drosophila flies under sterile (germ-free) or mono-colonization conditions. After egg sterilization, these were transferred to fly food supplemented E. cloacae or the vehicle for the flies that were left sterile. All tests were performed on day 10 of adulthood.
(G) Boxplots represent TG content per fly.
(H and I) Boxplots represent (H) ALT and (I) AST enzymatic activity in 10-day-old Drosophila flies.
(J) RT-qPCR results. Bars represent relative gene expression of flies fed an SD and HFD and mono-colonized with E. cloacae or left sterile. Error bars represent SEM.
(K and L) Boxplots represent the amount of L-histidine per gram of tissue, measured either by 1H NMR (K) or an ELISA kit (L) of sterile flies or flies mono-colonized with E. cloacae and supplemented with an SD.
(M and N) Boxplots represent TG content per gram of tissue of sterile flies or flies mono-colonized with E. cloacae fed (M) an SD and (N) an HFD and supplemented with 0, 1, 4, or 8 g/L L-histidine.
For all Drosophila experiments, samples contained a pool of 8 flies. Data are mean ± SEM. The p values were determined using Fisher’s LSD ANOVA test (#p < 0.1, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). ptrend was calculated using the non-parametric Mann-Kendall trend test.
Enterobacter cloacae induces an NAFLD-like phenotype through histidine metabolism in D. melanogaster
To further explore the causal role of Proteobacteria in the progression of NAFLD through the catabolism of histidine, we used the model organism D. melanogaster. This organism is easy to maintain under germ-free conditions. Mono-associations between germ-free flies and bacterial spices of interest can then be generated by dietary supplementation to specifically study the effects of a single microorganism on its host (Figure 6F).30 We selected E. cloacae, which is part of the Drosophila microbiome,31 as a representative member of the Proteobacteria, the phylum that had the strongest negative association with plasma histidine levels (Figure 5A) and the strongest positive association with hepatic steatosis (Figure 5B). In addition, E. cloacae has recently shown a causal effect on the induction of NAFLD both in guinea pigs32 and HFD-fed germ-free mice.33 As reported previously,24,25 an HFD induced an NAFLD-like phenotype in flies, characterized by increased TG accumulation (p < 0.0001; Figure 6G) and higher AST activity (p = 0.0012; Figure 6H). Notably, mono-association with E. cloacae worsened this NAFLD-like phenotype when flies were fed either a standard diet (SD) or an HFD. We found a significant increase in TG accumulation in flies mono-associated with E. cloacae compared with sterile flies when these were fed either an SD or HFD (pSD = <0.0001; pHFD = 0.0081; Figure 6G). Similarly, flies mono-colonized with E. cloacae had a significant increase in AST activity when they were fed either an SD or HFD compared with sterile files (pSD < 0.0001; pHFD = 0.028; Figure 6H). Conversely, increased levels of ALT were only found when flies were fed an SD and mono-colonized with E. cloacae (pSD < 0.0001; Figure 6I). In line with these results, HFD-fed flies mono-colonized with E. cloacae had a significant increase in the expression of ACC (pACC = 0.03) involved in de novo lipogenesis compared with HFD sterile flies. We observed the same tendency for SREB (p = 0.08) and myd (p = 0.09), although it was not significant. Although we did not find significative results in genes involved in β-oxidation or insulin receptors (InR and chico, respectively), genes involved in lipid droplet stabilization (Lsd1 and bmm) were significantly upregulated in HFD-fed flies mono-colonized with E. cloacae compared with HFD sterile flies (pLsd1 < 0.0001; pbmm = 0.03; Figure 6J).
We next quantified the histidine per gram of tissue of flies fed an SD diet using both 1H NMR and ELISA. In both cases, we found that the development of an NAFLD-like phenotype due to the mono-association of SD-fed flies with E. cloacae was accompanied by a significant decrease in the flies’ histidine levels per gram of tissue compared with sterile flies (pNMR = 0.0042 [Figure 6K], pELISA = 0.035 [Figure 6L]). Finally, to further validate the role of the gut microbiome in the process of histidine alleviating NAFLD, we explore whether E. cloacae could interfere in the beneficial effects promoted by the dietary supplementation of histidine. Specifically, we measured TG accumulation in flies mono-colonized with E. cloacae and sterile flies fed either an SD or an HFD and supplemented with increasing concentrations of histidine (0, 1, 4 and 8 g/L). In flies fed an SD, the histidine dose had no effect on TG accumulation (ptrend = 0.716), whereas increasing concentrations of histidine progressively decreased TG accumulation in flies mono-colonized with E. cloacae (ptrend <0.001) (Figure 6M). In a situation of an HFD-induced NAFLD phenotype, increasing concentrations of histidine promoted a progressive reduction in TG accumulation both in E. cloacae (ptrend = 0.003) and sterile flies (ptrend <0.001) (Figure 6N). However, this effect was consistently more effective in sterile flies (Figure 6N).
Discussion
Histidine and microbially derived histidine metabolites have been shown to modulate insulin signaling and diabetes.14,34,35 However, little is known about their effects on NAFLD. The current results open insights into the role of histidine catabolism in the pathogenesis of NAFLD. We identified a consistent decrease in circulating histidine levels with increased degree of hepatic steatosis in humans. To validate this association, we assessed whether HAA supplementation ameliorated the features of NAFLD in 3 different rodent models: (1) a mouse model of diet-induced NAFLD, (2) a genetic leptin-deficient mouse model (ob/ob), and (3) a surgical OVX rat model. Previous animal models, which simulated less aggressive hepatic steatosis, indicated only a partial improvement in NALFD features after 4 weeks of histidine supplementation.36,37 Hence, we used a combination of amino acids involved in histidine regulation, aiming to increase histidine levels. We further validated the effects of histidine supplementation in a D. melanogaster model supplemented with an HFD.
HAA treatment in the diet-induced NAFLD mouse model increased hepatic histidine levels in parallel to a significant reduction in NAFLD features. Additional animal models of NAFLD and studies with human primary hepatocytes highlighted the beneficial role of histidine treatment in NAFLD. In all models, we found a consistent reduction in the total lipid content or the TG levels and in the expression of de novo lipogenesis-related genes, whereas the effects on β-oxidation and fatty acid transport was only evident in some models. This suggests that histidine would facilitate the transportation of free fatty acids into the mitochondria to be oxidized, thereby decreasing lipid deposition. Consistent with our findings in human primary hepatocytes, HAA supplementation reduced the hepatic inflammatory status in the diet-induced (decreased gene expression of F4/80 and Tnfα) and OVX (decreased gene expression of IL1b and Tnfa) models of NAFLD. These results agree with those from a study using a histidine decarboxylase (HDC) KO NAFLD mice model, where the lack of HDC activity reduced F4/80 expression in the liver.5 Reduced expression of Tnfα is consistent with observations in a clinical study in obese women supplemented with histidine14 and with a preclinical study where histidine reduced hepatic Tnfα expression.12 We also observed an increased response to insulin after HAA supplementation, which agrees with results from a previous study in obese women where histidine supplementation improved peripheral insulin resistance.14
TAAR1 has recently been proposed as a target for the treatment of type 2 diabetes and obesity.38,39 Notably, TAAR1 activation with a TAAR1 agonist in diet-induced obese mice reduced both postprandial plasma and hepatic TGs compared with vehicle-treated animals.38 Recently, TAAR1 has been shown to be coupled to Gαs signaling pathways in pancreatic β cell lines to improve insulin secretion, β cell function, and proliferation.39 Here, we provide evidence that TAAR1 in the liver and its modulation by histidine may also play a key role in NAFLD. In humans, we found that circulating histidine levels were strongly positively associated with liver expression levels of TAAR1 and supplementation of human primary hepatocytes with histidine led to a strong increase in the expression of TAAR1. Remarkably, silencing of TAAR1 in human primary hepatocytes decreased AKT phosphorylation, suggesting a direct role in reducing the response to insulin, whereas histidine supplementation increased AKT phosphorylation.
Microbiota-host interactions potentially contribute to the development of metabolic diseases, and we have recently unraveled molecular networks linking the gut microbiome to hepatic steatosis.16 Here, we also show that plasma histidine levels are negatively associated with several bacterial families that are also increased in NAFLD, in particular from the phylum Proteobacteria. In particular, the Proteobacteria families Xanthomonadaceae, Rickettsiaceae, and Alteromonadales increased with the degree of hepatic steatosis, which is consistent with findings from previous studies in NASH.40,41,42 At the genus level, Bilophila and Campylobacter have also been described to be increased in NAFLD,43,44 and we found that both genera were associated with the low histidine levels found in NAFLD patients. In addition, both circulating histidine levels in humans and HAA supplementation in mice were associated with increased levels of Actinobacteria, Faecalibacterium, Lactobacillaceae, and Christensenellaceae, which are bacterial families and genera containing several species that have been linked with better metabolic health. In fact, a decrease in the abundance of Faecalibacterium has been found in association with irritable bowel syndrome (IBS), inflammatory bowel disease (IBD), diabetes, and NAFLD.45 Similarly, the Christensenellaceae family has emerged as an important player in human health. Decreased abundance of Christensenellaceae has been associated with obesity, IBD, visceral adipose tissue, and an unhealthier metabolic profile.46,47 HAA supplementation also decreased the levels of Roseburia, which have been associated with NAFLD development in obese individuals,48 and increased the levels of Akkermansia (phylum Verrucomicrobia). Notably, we found decreased levels of Verrucomicrobia with an increase in the degree of steatosis, and Akkermansiaceae administration has been shown to improve several metabolic parameters in an NAFLD animal model.49 Notably, high circulating histidine levels in humans and HAA-supplemented mice are associated with higher levels of Marinifilaceae, which have recently been found to be depleted in patients with liver fibrosis.50
Histidine can be used by many bacteria and is an important carbon, nitrogen, and/or energy source for Proteobacteria. Two recent studies have identified increased abundance of imidazole propionate, a microbially derived histidine catabolite, and hutH, the gene encoding for HAL, which converts histidine to urocanate, in patients with type 2 diabetes.34,35 Therefore, to elucidate a potential role of the gut microbiota in lowering the circulating histidine levels in NAFLD, we analyzed four microbial genes involved in histidine catabolism: hutH, hutU, hutI, and hutG.51 Notably, we found that patients with a higher degree of liver steatosis had higher clr-transformed levels of hutH, hutU, and hutI, suggesting a higher catabolism of histidine by the gut microbiota, which could explain the lower plasma histidine levels found in these patients. Importantly, HAA supplementation reduced the histidine catabolism observed in NAFLD. Thus, NAFLD mice treated with HAA had decreased levels of hutH and hutG compared with NAFLD mouse counterparts. It is worth noting that, although the histidine catabolism pathway is highly conserved among bacteria, it is not universal. Thus, the Hut pathway seems to be absent in Cyanobacteria, Mycoplasma species, and Spirochaete.29 In line with these results, we found that plasma histidine levels were strongly positively associated with Cyanobacteria in humans, whereas Spirochaete had the strongest negative association with the degrees of steatosis. Also consistent with these findings, we found that FMT from donors with low histidine levels resulted in a higher accumulation of hepatic TGs compared with those receiving a microbiota from donors with high histidine levels along with a positive correlation between the histidine levels of the recipient mice and the human donor. Finally, consistent with the role of the Proteobacteria in the development of NAFLD through histidine catabolism, we found that mono-colonization of flies with E. cloacae lead to an accumulation of TG content and AST and ALT activity along with a significant reduction in their histidine content. Importantly, mono-colonization with E. cloacae impaired the progressive reduction in TG accumulation with increasing histidine doses, suggesting that E. cloacae consumes histidine from the medium, preventing, in part, its absorption by the host.27
Limitations of the study
A strength of the present study is the reproducibility of the metabolomic findings related to histidine in three cohorts, three mouse models, and Drosophila. However, it has several limitations. All cohorts were recruited for the FLORINASH study. The validation in a cohort from a different study would provide additional strength to the associations. All subjects had morbid obesity. The results cannot be generalizable to other populations. Although we had a control group comprising obese patients without steatosis, we lack a control group without steatosis and without obesity. Liver transcriptomics and fecal metagenomics data for humans were not available in the validation cohorts. Finally, liver biopsies (the gold standard for identification of NAFLD) were only available for the discovery cohort.
In conclusion, our results show a cross-talk among histidine, hepatic steatosis, and the gut microbiota. We disclosed histidine-linked transcriptomics and metagenomics signatures and uncovered the potential role of the microbiota as a regulator of histidine levels by increased abundance of histidine degradation bacterial species. In addition, the supplementation of histidine may ameliorate hepatic steatosis, inflammation, and insulin resistance, pointing to it being a promising candidate for the treatment of NAFLD.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Phospho-Akt (Ser473) Antibody | Cell Signaling technology, Danvers, MA, USA | Cat#9271; RRID:AB_329825 |
| Secondary antibody anti-rabbit HRP (horseradish peroxidase) | Millipore Sigma, Darmstadt, Germany | Cat#12-348; RRID:AB_390191 |
| Anti-tubulin hFAB rhodamine | BioRad, Hercules, CA, USA | Cat#64225333; RRID:AB_2884950 |
| Akt (pan) (11E7) Rabbit mAb | Cell Signaling technology, Danvers, MA, USA | Cat#4685; RRID:AB_2225340 |
| Phospho-Akt (Ser473) (D9E) XP® Rabbit mAb | Cell Signaling technology, Danvers, MA, USA | Cat#4060; RRID:AB_2315049 |
| β-Actin Antibody (C4) | Santa Cruz Biotechnology, Inc; Dallas, TX, USA | Cat#sc-47778; RRID:AB_626632 |
| IRDye 680RD Goat anti-Rabbit IgG Secondary Antibody | LI-COR Biosciences, Lincoln, NE, USA | Cat#926-68070; RRID:AB_10956588 |
| IRDye® 800CW Goat anti-Rabbit IgG Secondary Antibody | LI-COR Biosciences, Lincoln, NE, USA | Cat#926-32211; RRID:AB_621843 |
| IRDye® 680RD Goat anti-Mouse IgG Secondary Antibody | LI-COR Biosciences, Lincoln, NE, USA | Cat#926-68071; RRID:AB_10956166 |
| Biological samples | ||
| Human body fluids (feces, plasma) | This paper | N/A |
| Human liver biopsy | This paper | N/A |
| Mice body fluids (feces, serum) | This paper | N/A |
| Mice liver | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Methanol LC-MS | Scharlau | Cat#ME03262500 |
| Lysing Matrix E | MP biomedicals | Cat#SKU116914050-CF |
| Acetic acid LC-MS | Scharlau | Cat#AC03470050 |
| Critical commercial assays | ||
| QIAamp DNA Stool Mini kit | QIAGEN | Cat#51504 |
| Bioscientific PCR free library kit | Illumina | N/A |
| Quant-iT dsDNA Assay Kit, high sensitivity kit | Invitrogen | Cat#10164582 |
| Agilent Low Input Quick Amp Labeling Kit | Agilent | Cat#5190-2305 |
| Agilent Gene Expression Hybridization Kit | Agilent | Cat#5188-5242 |
| Mouse Insulin ELISA Kit | Mercodia | Cat#10-1247-01 |
| Glucose Liquid Kit | QCA | Cat#998225 |
| Triglycerides Liquid Kit | QCA | Cat#992330 |
| Total Cholesterol Liquid Kit | QCA | Cat#995282 |
| Nextera XT Index Kit v2 | Illumina | Cat#FC-131-2001 |
| MiSeq® Reagent Kit v3 600 cycle | Illumina | Cat#MS-102-3003 |
| RNeasy Mini Kit | QIAGEN | Cat#74104 |
| High-Capacity RNA-to-cDNA Kit | Applied Biosystems | Cat#4387406 |
| AST and ALT colorimetric assays | QCA | Cat#999500 and 999200 |
| RC/DC Protein Assay | Bio-Rad Laboratories | Cat#5000121 |
| Pierce BCA Protein assay | Thermo Fisher Scientific | Cat#23225 |
| Experimental models: Cell lines | ||
| Primary human hepatocytes | Innoprot, Bizkaia, Spain | Cat#P10651 |
| Experimental models: Organisms/strains | ||
| Mouse C57BL/6J | Envigo+++ | N/A |
| Mouse C57BL/6J | Charles River | N/A |
| Oligonucleotides | ||
| see Table S11 for primers and RNA sequences | N/A | |
| Software and algorithms | ||
| R (version 3.6) | R | https://www.r-projet.org |
| GraphPad Prism 9 software | Graph-Pad Software, La Jolla, CA, USA | https://www.graphpad.com |
| MATLAB (version R20217a) | Mathworks | http://www.mathworks.com/products/matlab.html |
| Statistical Parametric Mapping software (SPM12) | UCL Queen Square Institute of Neurology | https://www.fil.ion.ucl.ac.uk/spm/software/ |
| MassHunter Data Analysis software | Agilent Technologies | https://www.agilent.com/en/product/software-informatics/mass-spectrometry-software |
| Prinseq-lite-0.20.4 | Schmieder and Edwards52 | https://prinseq.sourceforge.net/ |
| FLASh 1.2.11 | Magoč and Salzberg53 | https://ccb.jhu.edu/software/FLASH/ |
| Bowtie2-2.3.4.3 | Langmead and Salzberg54 | https://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| MEGAHIT v1.0 | Li et al.55 | https://github.com/voutcn/megahit |
| Prodigal v2.6.342 | Hyatt et al.56 | https://github.com/hyattpd/Prodigal |
| HMMER | Durbin et al.57 | http://hmmer.org/ |
| Kaiju v1.6.2 | Menzel et al.58 | https://github.com/bioinformatics-centre/kaiju |
| Limma (version 3.30.13) | Smyth59 | https://bioconductor.org/packages/release/bioc/html/limma.html |
| "Pathfinder" R package (version 1.6.4) | Ulgen et al.60 | https://cran.r-project.org/web/packages/pathfindR/index.html |
| “SGoF” R package (version 2.3.3) | Carvajal-Rodríguez et al.61 | https://cran.r-project.org/web/packages/sgof/index.html |
| ARDEP program | Wu et al.62 | http://mem.rcees.ac.cn:8082/ |
| Other | ||
| FastPrep-24TM | MP biomedicals | N/A |
| Bioanalyzer 2100 | Agilent | N/A |
| Hiseq 2500 | Illumina | N/A |
| Qubit 3.0 fluorometer | Thermo Fischer Scientific | N/A |
| High Sensitivity D1000 ScreenTape | Agilent | Cat#5067-5579 |
| Illumina MiSeq | Illumina | N/A |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Jordi Mayneris-Perxachs (jmayneris@idibgi.org).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The data that support the findings of this study are available from the lead contact (jmfreal@idibgi.org) upon reasonable request. The raw metagenomic sequence data for the FLORINASH cohort (with human-associated reads removed) have been deposited under the study accession number PRJEB14215. The raw 16S rRNA gene sequence data associated with the mouse FMT work have been deposited under the study accession number PRJEB24891.
Experimental model and subject details
Clinical study
The discovery cohort included n = 117 well clinically characterized NAFLD patients with morbid obesity aged 22 to 63 years old recruited for the FLORINASH study at the Endocrinology Service of the Hospital Universitari de Girona Dr. Josep Trueta (Girona, Spain) and Policlinico Tor Vergata University of Rome (Rome, Italy).16 Patients in the Discovery cohort underwent surgical intervention during which surgeons performed an intraoperative liver biopsy. The FLORINASH study also included the establishment of two cohorts of clinically well characterized morbidly obese patients to participate in the final validation of markers identified in the discovery cohort. Patients in the validation cohorts did not undergo surgery. Therefore, they did not provide liver biopsies, and NAFLD diagnosis was performed through ultrasound echography. The Validation cohort 1 comprised n = 263 patients with morbid obesity aged 20 to 65 years old at the Endocrinology Service of the Hospital Universitari de Girona Dr Josep Trueta (n = 116) and at the Center for Atherosclerosis of Policlinico Tor Vergata University of Rome (Rome, Italy; n = 147). The Validation cohort 2 comprised n = 271 patients with morbid obesity aged 25 to 66 years old at the Endocrinology Service of the Hospital Universitari de Girona Dr Josep Trueta.
All subjects gave written informed consent, validated and approved by the ethical committee of the Hospital Universitari Dr. Josep Trueta (Comitè d’Ètica d’Investigació Clínica, approval number 2009-046) and Policlinico Tor Vergata University of Rome (Comitato Etico Indipendente, approval number 28-05-2009).
Inclusion criteria: Caucasian origin, stable body weight 3 months before the study, free of any infection 1 month preceding the study and absence of any systemic disease.
Exclusion criteria: presence of liver disease (specifically hepatitis C virus infection and tumoral disease), cancer, iron overload, thyroid dysfunction (by biochemical work-up), >20 g/day of alcohol consumption, hepatitis B, or drug-induced liver injury (determined using a drug questionnaire). Alpha-1 antitrypsin deficiency was excluded by anamnestic data and clinical evidence.
Primary hepatocytes study
Cryopreserved primary human hepatocytes (HHs) were commercially sourced (Innoprot, Bizkaia, Spain) and cultured with hepatocytes medium (Innoprot) supplemented with 5% fetal bovine serum, 1% hepatocytes growth supplement (mixture of growth factors, hormones, and proteins necessary for the culture of primary hepatocytes) and 100 U/mL penicillin and streptomycin (P/S). HHs were grown on fibronectin pre-coated cell dishes at 37°C and 5% CO2 atmosphere.
For histidine supplementation experiments in steatosis, histidine was dissolved in 0.5 M HCl and was used at 500 μM to treat HHs. After 48 h of histidine treatment, cells were treated with PA for 24 h. PA was prepared as follows: 27.84 mg of PA (Sigma, San Luis, MO) was dissolved in 1 mL sterile water to obtain a 100 mM stock solution. Five percent bovine serum albumin (BSA) was prepared in serum-free DMEM and then mixed with PA stock solution for at least 1 h at 40°C to obtain a 5 mM solution. HHs were treated with PA at 200 μM or BSA as the vehicle. All experimental conditions were performed in 4 biological replicates. After treatments, cells were washed with PBS and collected with Qiazol for RNA purification or Protein extraction were performed.
For TAAR1 silencing experiments, 24 h after seeding, HHS were transfected with siRNA against TAAR1 or treated with histidine, for 72 h alone, or in combination with palmitic acid (PA) for 24 h following the siTAAR1/histidine treatment. Briefly, the siRNA (Sigma-Aldrich, St. Louis, MO, USA) against TAAR1 and Lipofectamine RNAiMAX (LifeTechnologies, Darmstadt, Germany) were diluted separately with Opti-MEM I Reduced Serum Medium (Life Technologies, Darmstadt, Germany) and mixed by pipetting afterward. The siRNA-RNAiMAX complexes were left to incubate for 20 min at room temperature and subsequently added on the top of the adherent cells drop-wise. The final concentrations of Lipofectamine RNAiMAX and siRNAs were 1.6 mL/cm2 and 75 nM, respectively, in 24-well cell culture plates, and the final amount of medium per well was 1 mL. Transfection efficiency was assessed by real-time PCR. The siRNAs (Sigma-Aldrich) used were human TAAR1 (SASI_HS01_00134799, with sequences 5′-CAGAAUAUAUCUUAUCGCU[dT][dT]-3′ and 5′-AGCGAUAAGAUAUAUUCUG[dT][dT]-3′).
Animal studies
Animal ethics approvals
Experimental animal procedures for Animal model 1, 2 and 3 were approved by the Animal Ethics Committee of the Technological Unit of Nutrition and Health of Eurecat (Reus, Spain) and the Generalitat de Catalunya (10281 and 11223). Animal model 4 was approved by the local ethical committee (approval number 31–278) of Rangueil University Hospital (Toulouse, France). The experimental protocols followed the “Principles of Laboratory Care” guidelines and were carried out in accordance with the European Communities Council Directive (86/609/EEC).
Animal model 1: Dietary induction of NAFLD in mice
Thirty-two C57BL/6J male mice (Envigo, Sant Feliu de Codines, Barcelona, Spain), 6 weeks old at the beginning of the experiment, were used. Animals were housed in groups (4 mice per cage) under controlled conditions of temperature (22 ± 2°C) and humidity (55 ± 10%), and on a 12-h light/dark cycle with free access to food and water. Mice were left undisturbed to acclimate to the animal facility for one week. NAFLD was induced as previously described.2,63,64 Briefly, after the acclimatisation period, animals were fed with HFHFr diet (HFHC: D12331, Research Diets) supplemented with 23.1 g/L fructose and 18.9 g/L sucrose in the drinking water. Mice were kept on these diets for a period of 20 weeks in ad libitum conditions.
For the last 4 weeks of the experiment (from the 16th to 20th week), NAFLD mice were randomly distributed into two groups: 16 mice were kept under the same fed conditions described before (NAFLD + vehicle), and 16 mice were exposed to histidine-related amino acids treatment (NAFLD + HAA). 20HAA were diluted with drinking water with 23.1 g/L fructose and 18.9 g/L sucrose (vehicle). These specific doses were determined based on previous human clinical supplementation studies65,66,67,68 and a calculation of dose translation from human to animal dosage taking into consideration the body surface area.69 Fresh solutions were freshly prepared three times per week and prepared from stock powders and protected from light. Adiposity index was determined as the sum of the retroperitoneal, mesenteric, epididymal and inguinal white adipose tissues weights. Serum fasting glucose (QCA, Barcelona, Spain) were analyzed by enzymatic colorimetric assays after sacrifice. GTT was performed during the third week of HAA supplementation. Briefly, mice were fasted overnight and after fasted blood glucose levels, were measured, mice were injected i.p. with 1.5 g glucose/kg body weight (Merck KGaA, Darmstadt, Germany), and blood glucose levels were measured every 30, 60, 90, 120, 150, and 180 min, collecting blood from the mice’s tails. For the ITT, during the fourth week of HAA treatment, mice fasted for 6 h and baseline levels of blood glucose were measured using a standard glucometer (LifeScan, Milpitas, CA, USA). Animals were then injected (i.p. 0.375 mU/g of body weight) with insulin (Actrapid Innolet, Novo Nordisk A/S, Bagsvaerd, Denmark), and blood glucose levels were measured as explained in the GTT test process above. Before being euthanized, 10 animals per group were randomly selected to perform an insulin challenge. They were intraperitoneally injected with 1 mU/g of insulin (n = 5 per group) or saline (n = 5 per group) and after 15 min, they were sacrificed.
Animal model 2: Genetic model of NAFLD in mice
Ten 4-weeks old C57BL/6J-ob/ob male mice (genetically obese with leptin deficiency) were obtained from JANVIER LABS. Animals were randomly housed in groups (5 mice per cage) under controlled conditions of temperature (22 ± 2°C) and humidity (55 ± 10%), and on a 12-h light/dark cycle with free access to food and water. Mice were left undisturbed to acclimate to the animal facility for one week. Then, animals were fed and treated for a period of 4 weeks in ad libitum conditions as explained in Animal model 1 and assigned into two experimental groups (ob/ob + vehicle or ob/ob + HAA; n = 5 per group) depending on the treatment received for 4 weeks (vehicle or HAA).
Animal model 3: Surgical model of NAFLD in rats
Ten 24-weeks-old female Sprague-Dawley rats (JANVIER LABS, Saint-Berthevin, France) were bilaterally ovariectomized (OVX) at the JANVIER facilities. The animals were fed a maintenance rat chow diet for 1 week to stabilize after ovariectomy before shipment at 25 weeks of age. After an acclimation period of 1 week at the animal facility of Eurecat, OVX rats were distributed into two experimental groups depending on the treatment received for 8 weeks (OVX + vehicle or OVX + HAA; n = 5 per group). The OVX + HAA was daily supplemented for 8 weeks, which included four amino acids at the following doses: histidine, carnosine and serine at 105 mg/kg; and cysteine at 245 mg/kg. The four bioactive compounds were dissolved together in peach juice. The doses of histidine, carnosine, serine used in mice are equivalent to the daily consumption of 1022 mg of each of these compounds for a 60-kg human. For cysteine, the extrapolated daily intake using the same formula was 2384 mg. For each compound, the dose is considered acceptable, well tolerated and safe in a context of a multi-ingredient supplementation. The OVX + vehicle were also daily supplemented with peach juice (vehicle), for 8 weeks, orally by a syringe of 1 mL in a volume of 0.5–0.6 mL at the same time each day (between 09:00-10:00 a.m.). Four days before the beginning of the treatments, the rats were trained to lick peach juice (0.3 mL) to ensure voluntary consumption.
Animal model 4: Fecal microbiota transplantation in mice
We leveraged data from our previous fecal microbiota transplantation experiment.1 Briefly, fecal samples from low- (n = 2) and high- (n = 2) histidine donors matched for age and BMI were suspended in sterile reduced PBS (N2 gas and thioglycolic acid, Sigma Aldrich, St. Louis, MO). Eight mice (8-week-old C57BL/6 male, Charles River) per patient were treated with an antibiotic mixture for 7 days (neomycin, ampicillin, metronidazole) and after a 4-day washout, mice were administered 20 mg/day of fecal matter for 4 consecutive days. Two weeks later, mice were sacrificed, and liver and plasma were collected and frozen. Experimental groups were randomly allocated.
Drosophila studies
Drosophila melanogaster stocks and maintenance
The Drosophila wild-type strain used in this study was originally obtained from Bestgene. The final stock was created by exchanging the w-allele of the strain that the company regularly uses to inject P-element-based transgenes by a w+ allele.70
Flies were raised and maintained on standard diet (SD) (1L: yeast 23.4 g, yellow cornmeal 44.2 g, sugar 93.5 g, agar 8.5 g, propionic acid 5 mL, 0.1% methylparaben in ethanol 5 mL), high fat diet (HFD) (1L: Coconut oil 150 g, yeast 23.4 g, yellow cornmeal 44.2 g, sugar 93.5 g, agar 8.5 g, propionic acid 5 mL, 0.1% methylparaben in ethanol 5 mL) or HFD supplemented with Histidine 1, 4 or 8 g/L (HFD +, His 1 g/L, His 4 g/L, His 8 g/L). Flies were raised and maintained thought-out their entire development in the corresponding dietary condition, adult flies were transferred to a fresh food vial containing the corresponding diet every 4 days. Fly stocks were maintained at 25°C, in a 12:12 h light-dark cycle.
Method details
Clinical and laboratory parameters
Stool and plasma samples from all subjects were obtained during the week before elective gastric bypass surgery, during which the liver biopsy was sampled. All samples were stored at − 80°C. Liver samples were collected in RNAlater, fragmented, and immediately flash-frozen in liquid nitrogen before storage at − 80°C.
Hepatic steatosis determination and liver histology in the clinical study
For the discovery cohort, liver biopsies were obtained for n = 117 patients who underwent bariatric surgery.16 The investigators were blind to group allocations. Liver biopsies were analyzed by a single pathologist expert in hepatic pathology. For each liver sample, H&E, reticulin and Masson’s trichrome staining were performed. We used a validated histological feature scoring system that addresses the full spectrum of lesions of NAFLD. It includes 4 semi qualitative histological features of steatosis (<5%: 0; 5–33%: 1; 34–66%: 2, >66%: 3), lobular inflammation (No foci: 0; <2 foci per 200x field: 1; 2–4 foci per 200x field: 2; >4 foci x 200x field: 3), hepatocyte ballooning (None: 0; Few balloon cells: 1; Many cells/prominent ballooning: 2), and fibrosis (None: 0; Perisinusoidal: 1; Periportal: 2; Bridging fibrosis: 3; Cirrhosis: 4).71 A NAFLD Activity Score (NAS) validated for use in clinical trials, ranging from 0 to 8, was then calculated as the unweighted sum of steatosis, lobular inflammation, and hepatocellular ballooning scores. NAS of ≥5 is diagnostic of NASH, NAS ≤2 is diagnostic of simple steatosis, and values between are considered indeterminate.72
For the validation cohorts, an ultrasound system with a 3.5 MHz convex transducer (Siemens Acuson S2000, Mochida Siemens Medical System, Tokyo, Japan) was used to scan the liver. Hepatic steatosis was defined as absent (grade 0: <5% steatosis), mild (grade 1: 5–33% steatosis), moderate (grade 2: >33–66% steatosis) or severe (grade 3: >66% steatosis) using the scoring system for NAFLD.71 Images were independently evaluated by two radiologists blinded to clinical and laboratory data.73
Transcriptome and metabolome data analysis in the clinical study
Transcriptomic analyses were performed in a subset of patients from the discovery cohort (n = 88) and have been previously described.16 Briefly, RNA from liver biopsy samples was extracted using standard extraction protocols (TRIzol) by Miltenyi Biotec as previously reported. RNA quality (gel images, RNA integrity number and electropherograms) was assessed using an Agilent 2100 Bioanalyzer platform (Agilent Technologies). An RNA integrity number >6 was considered sufficient for gene expression experiments.74 100 ng of total RNA was used for linear T7-based amplification of RNA for each sample. cRNA was prepared by amplification of the RNA and labeled with Cy3 using the Agilent Low Input Quick Amp Labeling Kit according to the manufacturer’s instructions. The amounts of cRNA and dye that were incorporated were measured by an ND-1000 spectrophotometer (NanoDrop Technologies). Hybridization of the Agilent Whole Human Genome Oligo Microarrays 4 × 44K was done following the Agilent 60-mer oligo microarray processing protocol using the Agilent Gene Expression Hybridization Kit. The fluorescence signals of the hybridized Agilent microarrays were detected using Agilent’s Microarray Scanner after washing with Agilent Gene Expression Wash Buffer twice and with acetonitrile once. Feature intensities were determined using Agilent Feature Extraction Software. Microarray data were processed and normalized using R and the BioConductor package LIMMA (Linear Models for Microarray Data).59 Raw data quality was assessed using pseudoMA and boxplots. A background correction was applied and normalization of the green channel between arrays was done using ‘cyclicloess’ between pairs of arrays. Control and low-expressed probes were removed and only those probes brighter than the negative controls (≥10%) on at least one array were kept. Batch-corrected data was obtained using the remove batch effect based on ‘Batch’.59 Probes with no associated gene ID were removed. Finally, data were averaged based on an association with a particular gene. Raw data quality was assessed using pseudoMA and boxplots. A background correction was applied and normalisation of the green channel between arrays was done using ‘cyclicloess’ between pairs of arrays. Control and low-expressed probes were removed and only those probes brighter than the negative controls (≥10%) on at least one array were kept. Batch-corrected data was obtained using the remove batch effect based on ‘Batch’,59 probes with no associated gene ID were removed. Finally, data were averaged based on an association to a particular gene.
Plasma metabolomics in the clinical study and mice FMT (animal model 4)
The plasma metabolome was determined using NMR. The analyses of the discovery and Validation cohort 1 were performed on a Bruker DRX600 spectrometer equipped with either a 5-mm TXI probe operating at 600.13 MHz or a 5-mm BBI probe operating at 600.44 MHz. The 90° pulse length was determined prior to each run and field frequency was locked using D2O as solvent. Plasma samples were thawed at room temperature and 350 μL aliquots were carefully placed in 5-mm NMR tubes. Then, 150 μL of saline solution (0.9% NaCl prepared with 80:20 H2O/D2O and sodium azide) was added and the mixture was gently vortexed. Spectra were acquired using water suppressed Carr-Purcell-Meiboom-Gill (CPMG) using the Bruker program cpmgpr (recycle delay (RD) −90°-t1-180°-tm-acquire). An RD of 2 s was employed for net magnetization relaxation, during which noise irradiation was applied in order to suppress the large water proton signal. Several loops n = 100 and a spin-echo delay t = 400 μs were used to allow spectral editing through T2 relaxation and therefore attenuation of broad signals. For each sample, 128 scans were recorded in 32K data points with a spectral of 20 ppm.
The analysis of the validation cohort 2 was performed on a Bruker AVANCE II 600 spectrometer fitted with an automatic sample changer and a multinuclear triple resonance (TBI) probe (Bruker Biospin, Germany) at 14.1 T (600.3 MHz). Plasma samples were thawed at room temperature and 400 μL aliquots were combined with 200 μL of saline buffer (9% w/V NaCl, 100% D2O, 10mM TSP). Samples were mixed by vortex and spun for 10 min at 13,000 rpm prior to transferring 550 to a 5 mm NMR tube. Spectra were acquired using water suppressed Carr-Purcell-Meiboom-Gill (CPMG) using the Bruker program cpmgpr (RD-90°t1-180°-tm -acquire free induction decay). The water signal was suppressed by irradiating the RD of 2s with a mixing time (tm) of 10 μs. The acquisition was set to 1.36s and the 90° pulse length of 10.43 μs. Several loops n = 40 and a spin-echo delay t = 400 μs were used to allow spectral editing through T2 relaxation and therefore attenuation of broad signals. For each sample, 256 scans were recorded in 32K data points with a spectral of 20 ppm.
Spectra were manually phased, corrected for baseline distortions and referenced to the center of the α-glucose anomeric doublet (δ 5.23). All spectra were imported to MATLAB and digitized into consecutive integrated spectral regions of equal width. The regions between δ 4.7–4.9 containing the residual water resonance were removed from all spectra in order to minimize the effect of baseline effects caused by imperfect water suppression. Each spectrum was then normalized using a probabilistic quotient normalization algorithm.
Extraction of fecal genomic DNA and whole-genome shotgun sequencing (metagenomics) in the clinical study
Fecal shotgun sequencing data was generated for n = 73 patients from the discovery cohort as previously described.16 Briefly, total DNA was extracted from frozen human stools using the QIAamp DNA Stool Mini kit (Qiagen), with slight modifications by adding a bead-beating step. Quantification of DNA was performed with a Qubit 3.0 fluorometer (Thermo Fisher Scientific, USA), and 3 ng of extracted DNA for each sample was prepared using the Bioscientific PCR free library kit according to the manufacturer’s instructions and sequenced on a Hiseq 2500 (Illumina) with 2 × 150 pair-end chemistry. The obtained input fastq files were decompressed, filtered and 3′ ends-trimmed by quality, using prinseq-lite-0.20.4 program52 and overlapping pairs were joined using FLASH-1.2.11.53 Fastq files were then converted into fast files, and human host reads were removed by mapping the reads against the reference human genome (GRCh38.p11, Dec 2013) using Bowtie 254 with end-to-end and very sensitive options. Next, functional analyses were carried out by assembling the non-host reads into contigs by MEGAHIT v1.1.255 and mapping those reads against the contigs with Bowtie 2. Reads that did not assemble were appended to the contigs. Next, Prodigal v2.6.34256 was used for predicting codifying regions. Functional annotation was carried out with HMMER57 against the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, version 201675 to obtain the functional subcategory, route and annotation of the genes. The filtering of the best annotations and the assignment of the orf annotation to every read were carried out using R 3.1.076 which was also used to count the aligned reads and add the category and its coverage, and finally, build abundance matrices. Taxonomic annotation was implemented with Kaiju v1.6.258 on the human-free reads using a greedy mode. The addition of lineage information, counting of taxa and generation of an abundance matrix for all samples were performed using the package R.77
Preclinical and laboratory parameters (animal model 1)
Serum was obtained by centrifugation and stored at −80°C for further analysis. Serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) were quantified by enzymatic colorimetric assays (QCA, Barcelona, Spain). Fasting glycemia were measured with the Glucose Liquid Kit (QCA, Barcelona, Spain), respectively. Livers were rapidly collected, weighed, and divided into two sections—the lobus hepatis sinister medialis was kept in formalin, and the remaining tissue was frozen in liquid nitrogen and stored at −80°C until further analysis.
Hepatic fat quantification in murine liver (animal model 1, 2, 3)
Hepatic lipids were extracted and quantified following a method previously described.78 Briefly, total lipids were extracted from 80 to 100 mg liver sections by adding 1 mL of hexane/isopropanol (3:2, v/v) and degassing with gas nitrogen. Then, they were left overnight under orbital agitation, at room temperature, and protected from light. After extracting 0.3 mL of Na2SO4 (0.47 M), the organic layer was separated and dried with gas nitrogen. Total lipids were quantified gravimetrically before emulsifying, as described previously.79 Triglycerides and total cholesterol were measured using commercial enzymatic kits (QCA).
Histological evaluation in murine liver (animal model 1, 2, 3)
Liver portions fixed in buffered formalin (4% formaldehyde, 4 gr/L NaH2PO4, 6.5 gr/L Na2HPO4; pH 6.8) were cut at a thickness of 3.5 μm and stained with H&E. Liver images (magnification 40×) were taken with a microscope (ECLIPSE Ti; Nikon, Tokyo, Japan) coupled to a digital sight camera (DS-Ri1, Nikon) and analyzed using ImageJ NDPI software (National Institutes of Health, Bethesda, MD, USA; https://imagej.nih.gov/ij, version 1.52a). To avoid any bias in the analysis, the study had a double-blind design, preventing the reviewers from knowing any data from the mice during the histopathological analysis. A General NAFLD Scoring System was established to diagnose mice with NAFLD/NASH. The key features of NAFLD and NASH were categorized as follows: steatosis was assessed by analysing macrovesicular (0–3) and microvesicular steatosis (0–3) separately, followed by hepatocellular hypertrophy (0–3), which evaluates abnormal cellular enlargement, and finally giving a total score of 9 points of steatosis state. Inflammation was scored by counting cell aggregates (inflammatory foci). The score 0 to 3 depends on the grade of the feature. It is categorized as 0 (<5%), 1 (5–33%), 2 (34–66%) and 3 (>66%), and this scoring is used in each feature of steatosis and then added to the total steatosis score.80 Ballooning was not included in the scoring system, because only quantitative measures were considered for the rodent NAFLD score. It is important to highlight that hypertrophy is not a sign of cellular injury and slightly refers to an anomalous enlargement of the cells without recognizing the source of this enlargement.80
Bacterial genomic DNA isolation, 16S rRNA sequencing and analysis (animal model 1)
Bacterial genomic DNA was isolated from mice fecal samples using the MBP DNA Soil extraction kit. Genomic DNA was normalized to 5 ng/μL with EB (10 mM Tris-HCl) and libraries were performed. Briefly, following a first PCR and clean-up using KAPA Pure Beads (Roche Catalog No. 07983298001) a second PCR master mix was made up using P7 and P5 of Nextera XT Index Kit v2 index primers (Illumina Catalog No. FC-131-2001 to 2004). Following the PCR reaction, the libraries were quantified using the Quant-iT dsDNA Assay Kit, high sensitivity kit (Invitrogen Catalog No. 10164582) and run on a FLUOstar Optima plate reader. Libraries were pooled and run on a High Sensitivity D1000 ScreenTape (Agilent Catalog No. 5067–5579) using the Agilent Tapestation 4200 to calculate the final library pool molarity. The pool was run on an Illumina MiSeq instrument using MiSeq Reagent Kit v3 (600 cycle) (Illumina Catalog FC-102-3003) following the Illumina recommended denaturation and loading recommendations which included a 20% PhiX spike in (PhiX Control v3 Illumina Catalog FC-110-3001). The raw data was analyzed locally on the MiSeq using MiSeq reporter. For the 16S sequence analysis, The LotuS 1.36 used pipeline81,82 in short amplicon mode with default quality filtering. Raw 16S rRNA gene reads were quality filtered to ensure a minimum length of 170 bp, not more than eight homonucleotides, no ambiguous bases, average quality ≥ 27 and an accumulated error below 0.5 and a dereplication filter set to “6:1,3:2”. Cleaned reads were clustered into amplicon sequencing variants (ASVs) using DADA2,83 chimeric ASVs were removed using the DADA2 de novo chimera check. Remainder phiX reads were filtered by mapping ASVs against the phiX reference genome.84 ASV taxonomy was assigned using the LotuS2 LCA algorithms against Silva 138.1 reference database.85 We could assign on average 5500 ± 3108 reads to each sample that were of cyanobacterial origin. Further data analysis was conducted with R statistical language Version 3.00 (The R Foundation, https://www.r-project.org/) as described in Hildebrand et al.,86 employing the rtk software87 or all data normalizations. The 16S raw data can be found at https://1drv.ms/u/s!ApVezdktX3aIlZVJXYWok9s2qhgfvg?e=VDyr2c.
Bacterial genomic DNA isolation, 16S rRNA sequencing and analysis (animal study 4: FMT)
Fecal content was extracted and sequenced by Vaiomer (Vaiomer SAS) as previously described.16 Briefly, total DNA was extracted using the QIAamp DNA Stool Mini Kit (Qiagen) after two mechanical lysis steps in a bead beater. The quality and quantity of extracted DNA were evaluated by gel electrophoresis and NanoDrop 2000 UV spectrophotometer (Thermo Scientific). The V3–V4 hypervariable regions of the 16S rDNA were amplified by two-step PCR using Vaiomer V2 primers and sequenced using MiSeq Reagent Kit v3 (2 × 300-bp paired-end reads; Illumina).
The paired-end reads were filtered and trimmed. Then, amplicon sequence variants (ASVs) were built. Briefly, error rates for each base transition were estimated and dereplication was carried out to combine all identical reads into unique sequences. Then, sequence variants were inferred from dereplication reads and the error estimations. The forward and reverse pairs were merged together (minimum overlapping of 15 bases and maximum of 2 mismatches). Variants reconstructed by combining a left- and right-segment from two more abundant sequences were identified as chimeric and discarded to obtain the ASVs. Then, ASVs were mapped onto the Mus musculus genome (GRCm39 GCF_000001635.27 assembly) using Bowtie2 (version 2.5.0), with a very-sensitive preset. Taxonomy was assigned to each ASV using the Silva database (version 138.1) as reference. In order to identify as many (non-exact matching) species as possible, we mapped the ASVs, with an assigned genus but without species, against the Silva database, using Blastn (version 2.13.0+) with a minimum of 95% of identity and 100% of coverage. For each ASV, the alignment with the highest (best) identity, higher than 97%, with a difference of identity greater than 2 respect to the second-best alignment, was selected. Finally, the species of this selected best alignment were assigned to the corresponding ASV. Functional abundances were predicted for the ASVs with PICRUSt2 (version 2.5.1).88
Primer design for HutH and HutG microbiota functions
Several sequences from prokaryotes species were uploaded from databases to ARDEP program62 in order to design degenerated primers that could amplify in conserved regions for HutH and HutG genes and cover all the species used for the primer design. Based on the k-mer algorithm, primer length is set as k, and all sequences are divided into k-mers. The analysis platform allows for sequence length statistics, length filtering, and redundancy removal to be performed on the sequence database using tools located in the “Sequence Processing” section. After performing sequence database quality control, we could run ARDEP for primer design in the “Primer Processing” section, and we obtained a list of different possibilities of primer sets. The “Covered Taxonomy Calculator “option was used to count the sequence number of taxonomy and functional group covered by primer sets. The basic primer properties were considered (Tm, GC%, primer length) choosing the best option.
Histidine quantification in mice serum samples (animal model 1)
For serum metabolite extraction, 10 μL of the sample was added to a 1.5-mL microcentrifuge tube containing 100 μL of water and 1 μg/mL of Phe-13C as internal standard. Then, samples were incubated in a sample mixer (Thermo Scientific, Barcelona, Spain) for 30 min at 1,200 rpm and 4°C. For protein precipitation, 300 μL of acetonitrile was added to the sample, and the resulting solution was vortexed and incubated in a sample mixer at 1,200 rpm for 30 min at 4°C. Afterward, samples were centrifuged at 13,000 rpm for 15 min at 4°C. The resulting supernatant was transferred into another 1.5-mL tube and incubated overnight at −20°C. The following day, samples were vortexed and centrifuged at 13,000 rpm for 15 min at 4°C. Finally, the supernatant was transferred to vials with glass inserts (Agilent Technologies, Barcelona, Spain) for further analysis after overnight incubation at −20°C. All reagents used were LC-MS grade.
For histidine detection, samples were injected based on the described method.89 Briefly, 4 μL of the extracted sample was injected and chromatographic separation was achieved on a Cogent Diamond Hydride column (15P-2, VWR) equipped with a microfilter (502693, Supelco) with a column temperature of 40°C. The flow rate was 0.4 mL/min for 21 min. Solvent A was composed of water containing 0.1% formic acid (v/v) and solvent B was composed of acetonitrile containing 0.1% formic acid (v/v). The gradient started at 75% of solvent B and was held for 5 min. Then, solvent B was progressively reduced to 60% (in 1 min and held for 1 min), 50% (in 2 min and held for 1 min) and 40% (in 10 min). Finally, solvent B was increased to 100% in 1 min. Post-time was established at 5 min. Electrospray ionization was performed in positive ion mode using N2 at a pressure of 15 psi for the nebulizer with a flow of 11 L/min and a temperature of 300°C, respectively. Data were collected using the MassHunter Data Analysis Software (Agilent Technologies, CA, USA). Samples were decoded and randomized before injection. Metabolite extraction quality controls (plasma samples with internal Phe-13 C) were injected every 10 samples. Peak determination and peak area integration were carried out with MassHunter Quantitative Analyses (Agilent Technologies, CA, USA). Signal normalization was performed using internal standards. Quantification was performed by constructing standard curves. The analysis was performed through liquid chromatography coupled to a hybrid mass spectrometer with electrospray ionization and a triple quadrupole mass analyser. The liquid chromatography system was an ultra-performance liquid chromatography model 1290 coupled to LC-ESI-QqQ-MS/MS model 6420 both from Agilent Technologies (Barcelona, Spain).
Histidine quantification in the liver of mice (animal model 1)
LC-MS-grade acetonitrile (ACN) was purchased from VWR International GmbH (Darmstadt, Germany). Analytical grade formic acid (97%) and hydrochloric acid (37%), LC-MS-grade ammonium formate and the certified reference material amino acids mix (TraceCERT) used to prepare the standard curves were purchased from Merck KGaA (Darmstadt, Germany). The stable isotope-labelled amino acid mix MSK-CAA from Cambridge Isotope Laboratories, Inc., used as an internal standard, was obtained from CK Isotopes Limited (Leicestershire, UK). Sterile 0.45 μm PES filters were purchased from Fisher Scientific UK Ltd (Loughborough, UK).
A 12-point standard curve with concentrations ranging from 0.24 μM to 1 mM was freshly prepared using the commercially available amino acids mix TraceCERT. Approximately 50 mg of liver tissue was homogenized by bead beating using a FastPrep-24 Tissue Homogenizer and 1 mL of 6 N HCl (3 × 30 s at 6.5 m/s cycles, with 5 min on ice in between). The homogenate was placed into a screw cap Pyrex tube and an extra 1mL 6 N HCl was used to ensure all the sample was transferred, making the total volume for hydrolysis 2 mL. To prevent oxidation, the headspace was purged with nitrogen. Samples were then placed into a heat block at 110°C and hydrolyzed overnight for 16h. Once cooled down, hydrolysates were dried at 55°C using a Speedvac centrifugal concentrator, before being reconstituted using 2 mL of H2O, filtered, and stored at −80°C until analysis. Right before being injected, samples were diluted 10X using the LC-MS solvent mix B (90% ACN, 10% H2O, 0.5% FA and 20 mM ammonium formate). Each point of the standard curve and the diluted samples were spiked with the isotopically labeled internal standard mix MSK-CAA-1, to a final 50 μM concentration, before being transferred to LC-MS vials, ready to be analyzed.
Liquid chromatography coupled with mass spectrometry (LC-MS) analysis of amino acids was performed using a ACQUITY UPLC I-Class PLUS (Waters, Wilmslow, UK) coupled to a Xevo TQ-S micro Triple Quadrupole mass spectrometer (Waters, Wilmslow, UK), that was operated in positive electrospray ionization mode (ESI+). Separation was achieved using a Raptor HILIC-Si LC Column (100 × 2.1 mm, 2.7 μm particle size) with a Raptor Polar X EXP guard column cartridge (2.7 μm, 5 mm × 2.1 m), maintained at 35°C (Thames Restek UK Limited, Saunderton, UK). The 13-min gradient recommended by Restek to separate amino acids in human plasma was used,90 but with slight modifications in the mobile phases, injection volume and flow rate. Briefly, mobile phase A consisted of 0.5% formic acid, and 20 mM ammonium formate in water and mobile phase B was 0.5% formic acid, 20 mM ammonium formate, 10% H2O and 90% acetonitrile. The injection volume was 3 μL and the flow rate was set to 400 μL/min. Triplicate injections for each sample were performed. Table S32 lists the exact mass precursor and product ions, retention time (RT), cone voltage and collision energy for each amino acid analyzed. Data was acquired and processed using the MassLynx V4.2 Data and TargetLynx software (Waters, Milford, CT, USA OR Waters, Wilmslow, UK), integrating the area under the peak for each targeted analyte relative to the isotopically labeled internal standard. Processed data were exported to Excel and quantified against the analyzed calibration curves.
RNA extraction and quantitative polymerase chain reaction
Homogenates from livers of Animal model 1–4 were used for total RNA extractions using TriPure reagent (Roche Diagnostic, Sant Cugat del Vallès, Barcelona, Spain) according to the manufacturer’s instructions. RNA homogenates from human primary hepatocytes were extracted and purified using RNeasy Mini Kit (QIAGEN, Gaithersburg, MD). RNA concentration and purity were determined using a nanophotometer (Implen GmbH, München, Germany). RNA was converted to cDNA using the High-Capacity RNA-to-cDNA Kit (Applied Biosystems, Wilmington, DE, USA). The cDNAs were diluted 1:10 before incubation with commercial LightCycler 480 Sybr green I master on a Lightcycler 480 II (Roche Diagnostics GmbH, Manheim, Germany) or TaqMan technology suitable for relative genetic expression quantification. Table S33 shows a list of used primers for this study. 36b4 was used as a housekeeping gene91,92,93 in mice models and Peptidylprolyl isomerase A (PPIA) as housekeeping gen in human primary hepatocytes and OVX rat model.
Protein extraction and Western blot analysis
Human primary hepatocytes were collected by scraping on ice with cell lysis buffer (Cell Signaling Technology). Cell lysates were kept in agitation for 30 min and were centrifuged at 12,000 x g for 10 min at 4°C to remove cell debris. Protein content was determined using the RC/DC Protein Assay (Bio-Rad Laboratories, Hercules, CA, USA). For western blotting, proteins extracts (15 μg) were separated using 4%–20% precast polyacrylamide gel (Bio-Rad) and transferred to polyvinylidene fluoride (PVDF) membranes. After blocking with 5% BSA in TBS-Tween20 (Sigma-Aldrich), membranes were exposed overnight at 4°C to primary antibodies against phospho Akt (pAkt [Ser473], rabbit monoclonal antibody [mAb] #9271) at 1/1,000 dilution (Cell Signaling, Danvers, MA, USA) diluted in 1X TBS containing 0.1% Tween 20, following the recommendations of the manufacturer. After secondary antibody incubation (anti-rabbit HRP), signals were detected using enhanced chemiluminescence horseradish peroxidase substrate (Millipore, Burlington, MA, USA) and analyzed with a luminescent image and signals were quantified by ImageJ software (https://imagej.nih.gov/ij/). Anti-tubulin hFAB rhodamine at 1/1,000 (64225333, BioRad, Hercules, CA, USA) for 2 h exposure was used as control.
In Animal model 1, approximately 20 mg of the liver was homogenized with Tyssuelyser LT (Qiagen, Hilden, Germany) for 50 s in 300 μL lysis buffer (8 mmol/L NaH2PO4, 42 mmol/L Na2HPO4, 1% SDS, 0.1 mol/L NaCl, 0.1% NP40, 1 mmol/L NaF, 10 mmol/L sodium orthovanadate, 2 mmol/L PMSF, and 1% protease inhibitor cocktail 1 (Millipore Sigma, Darmstadt, Germany)). The protein extracts were quantified by the standardized BCA method (Pierce BCA Protein assay kit (Thermo fisher scientific, Waltham, MA, USA). Protein extracts (20–25 μg) were electrophoretically separated on 10% SDS-PAGE and electroblotted to nitrocellulose membranes (Li-cor biosciences, NE, USA).94 Efficient protein transfer was monitored by the Ponceau-S stain. Next, membranes were blocked (5% BSA) at room temperature and probed with specific primary antibodies (diluted 1:1000) overnight at 4°C in 1% BSA: total Akt (4685) (CST, Danvers, MA, USA), phospho-Akt (Ser473) (4060) (CST) and β-Actin (Santa Cruz Biotechnology, Inc; Dallas, TX, USA). Thereafter, infrared fluorescent secondary antibodies anti-rabbit 680, anti-rabbit 800 and anti-mouse 680 (LI-COR Biosciences, Lincoln, NE, USA; 926–68070, 926–32211, and 926–68071, respectively) were used for detection and quantified using ImageJ.95
Drosophila melanogaster studies
mRNA extraction and cDNA synthesis
Ten-day-old wild-type pre-mated females were selected for qRT-PCR. A total of eight flies were collected per sample and transferred to RNAlater solution (Sigma). Total RNA was extracted using the E.Z.N.A Total RNA kit I (Omega Bio-Tek). To avoid amplification from genomic DNA, DNase treatment was performed using the DNA-free Ambion kit and RNA was reverse transcribed into cDNA using the High-Capacity cDNA reverse transcription (Applied Biosystems) according to manufacturers’ procedures.
Quantitative real-time PCR (qRT-PCR)
Quantitative PCRs (qPCRs) were performed using the LightCycler 480 SYBR Green Master (Roche) on a LightCycler 480 II machine (Roche). Primer sequences of the analyzed and the reference genes RNApol2 and 1tub23cf. are provided in Table S33. For each condition, a minimum of 6 biological replicates were analyzed. Differential gene expression was calculated using the 2ΔΔCt method. The average Ct value for each sample was calculated and subtracted from the geometric mean Ct value of the reference genes RNApol2 and 1tub23cf. to calculate the ΔCt value. Fisher’s LSD ANOVA test (GraphPad Prism version 10 for Mac) were employed for calculations of p-values.
Bacterial strains and growth
Enterobacter cloacae (DSM 6234) was obtained from DSMZ, German Collection of Microorganisms and Cell Cultures GmbH (Germany) and was cultured in MRS modified broth96 at 30°C, with agitation, overnight. Mono-associated flies were created inoculating Enterobacter cloacae on top of the sterile diet. 300 μL of culture at an optical density of 0.7 when measuring at 600 nm (OD600) was added into 45 mm diameter vials, with a sterile Whatman filter paper.
Generation of axenic and mono-associated flies
To generate axenic flies, <16 h embryos were collected in apple juice agar plates and sterilized with 50% bleach during 5 min, rinsed twice in sterile water and placed on sterile standard food supplemented with antibiotics (50 μg ampicillin, 50 μg kanamycin, 50 μg tetracycline and 15 μg erythromycin per liter of fly food). Flies were maintained for a minimum of 2 generations in antibiotic media to clear possible Wolbachia infection. Absence of Wolbachia was verified by PCR using Wolbachia-specific primers, as described previously.96 After Wolbachia clearance, conventionally reared stocks were used to regenerate axenic stocks regularly exclusively performing the egg bleaching treatment. For bacterial mono-association, sterilized eggs were transferred to 45 mm vials with sterilized diet inoculated with E. cloacae (see bacterial strains and growth section) and reared until adulthood. The experimental germ-free condition was obtained by inoculating the corresponding amount of sterile MRS to the fly food. Adults were tested for axenicity or the correct bacterial mono-association previous to the experiment plating the fly lysate on different bacterial culture media. All these procedures were performed under the hood, with sterile material and under sterile conditions.
ALT and AST activity
ALT and AST activity were measured using the Alanine Transaminase Colorimetric Assay Kit or Aspartate aminotransferase Colorimetric Assay Kit, both from Cayman Chemicals. Eight females per sample were ground in a tube with 1 mL of cold Tris-HCL pH7.6 and centrifuged at 13,500 rpm (5 min) to extractant the supernatant for three consecutive rounds, to ensure complete clearance of all debris. 10 μL of the sample and positive control were added into a 96-well plate, together with the kit reagents according to manufacturer’s recommendations (final volume per well was 105 μL). Absorbance was measured at 340 nm once every minute for 15 min at 37°C with a microplate reader (Citation 5 imaging reader, BioTek). To determine AST and ALT activity first, the background was subtracted from each well and then the change in absorbance (ΔA340) per minute was plotted to determine the slope at the linear portion of the curve. The reaction rate was determined using the NADH extinction coefficient of 4.11 mM-1 with the following equation ALT or AST activity (U/mL) = [ΔA340/min x 0.105 mL]/[4.11 mM-1 x 0.01 mL] x Sample dilution. One unit is defined as the amount of enzyme that will cause the oxidation of 1.0 μmol of NADH to NAD+ per minute at 37°C.
Triglyceride quantification
The amount of TG was quantified with the Triglyceride Colorimetric Assay Kit (Cayman Chemicals), according to the manufacturer’s protocol. Eight flies were ground in a tube in 300 μL of NP-40 Substitute and supernatant was extracted after centrifugation at 10,000 rpm (5 min) for three consecutive rounds, to ensure complete clearance of debris. Samples were then run through a microplate reader at 535 nm (Citation 5 imaging reader, BioTek). TG levels in flies were calculated by interpolating the OD values to the corresponding standard cube.
Triglyceride quantification in the histidine E. cloacae interaction experiment
The amount of TG was quantified with High Sensitivity Triglyceride Fluorometric Assay Kit (Sigma-Aldrish), according to manufacturer’s protocol. Eight flies were ground in a tube in 7.5 mL of Triglyceride Assay Buffer and supernatant was extracted after centrifugating at 10.000rpm (5 min) for three consecutive rounds, to ensure complete clarence of debris. Samples were then run through a microplate reader at (λex = 535/λem = 587 nm) (Citation 5 imaging reader, BioTek). Relative amounts of TG were calculated respect SD sterile flies TG values.
Histidine quantification
ELISA
Histidine quantification was performed with L-Histidine ELISA from immusmol according to the manufacturer’s protocol. For total histidine quantification, 8 flies were ground in a tube with 500 μL of PBS. After centrifugation at 10,000 rpm for 5 min, supernatant was extracted for three consecutive rounds, to ensure complete clearance of debris. For hemolymph collection 20 adult flies were impaled in the thorax using a 0.2 mm tungsten needle and placed in a 500 Μl tube pierced with a 25 gauge needle and nested inside a 1.5 ML tube for hemolymph collection as described97 and centrifuged at 4 °C at 5,000 g for 5 min 0.5 μL of hemolymph were collected from 20 flies per replicate and volume was brought up to 50 Μl with PBS. Samples were then run through a microplate reader and OD was measured at 450 nm (Citation 5 imaging reader, BioTek), the background of each well was determined at 635 nm. Histidine levels in flies were calculated by interpolating the OD values obtained after subtracting the background to the corresponding standard cube.
NMR
For the quantification of histidine by 1H-NMR, flies were lyophilized overnight and extracted with methanol/water (8:1). Samples were dried with SpeedVact and resuspended in deuterated PBS. Samples were measured and recorded in procno 11. One-dimensional 1H pulse experiments were carried out using the nuclear Overhauser effect spectroscopy (NOESY) presaturation sequence (RD–90°–t1–90°–tm–90° ACQ) to suppress the residual water peak, and the mixing time was set at 100 ms. Solvent presaturation with irradiation power of 125 mW was applied during recycling delay (RD = 5 s) and mixing time. The 90° pulse length was calibrated for each sample and varied from 9.60 to 10.38 μs. The spectral width was 9.6 kHz (16 ppm), and 256 transients were collected into 64 K data points for each 1H spectrum. NMR spectra were recorded at 300 K on an Avance III 600 spectrometer (Bruker, Germany) operating at a proton frequency of 600.20 MHz using a 5 mm PABBO gradient probe. The exponential line broadening applied before Fourier transformation was of 0.3 Hz. The frequency domain spectra were phased, baseline-corrected and referenced to TSP (d = 0 ppm) using TopSpin software (version 4.1.3, Bruker). After pre-processing, histidine specific 1H-NMR region identified in the spectra was integrated using TopSpin software. Histidine concentration was calculated using Eretic signal as reference. Eretic Signal was calibrated using 2 mM sucrose reference sample.
Quantification and statistical analysis
The associations of the plasma histidine levels according to the steatosis grade were calculated using both a Kruskal-Wallis test and a Mann-Kendall trend test controlling for age, BMI, gender, and country. Pairwise comparisons were assessed using the Wilcoxon test. As we adjusted for covariates and these tests are non-parametric, the violin plots were generated with the ranked residuals of the model after adjusting for the selected covariates. Statistical analyses for Animal models and human primary hepatocytes were performed using GraphPad Prism 10 software (Graph-Pad Software, La Jolla, CA, USA). Data are presented as mean ± SEM. Data distribution was analyzed by the Shapiro–Wilk normality test. Differences between the two groups were determined using an unpaired t-test (two-tailed, 95% confidence interval). A p-value below 0.05 was considered statistically significant.
Metabolomic analysis
We used a random-forest based machine learning variable selection strategy using the Boruta algorithm after adjusting for age, BMI, gender, and country, to identify metabolites associated with liver steatosis grade.98 It has been proposed as one of the best performing feature selection methods for high-dimensional omics datasets.99 It comprises four steps: randomization, model building, statistical testing and iteration. For statistical testing, we used a Bonferroni corrected two-tailed binomial test to select relevant features and assess whether these features are important (significantly higher, selected), unimportant (significantly lower, rejected) or tentative (non-significant). The algorithm was run with 400 iterations, a 0.005 cut-off for the Bonferroni adjusted p-values, and 2000 trees.
ML models are referred to as “black-boxes” models due to their nature, complexity, and difficult interpretation. To facilitate model explainability, we used sophisticated tools based on game-theory Shapley values to compute the exact Shapley Additive exPlanations (SHAP) scores and determine the contribution of each selected metabolite for each individual in the model.17 The exact computation of SHAP values guarantees that explanations are always consistent and locally accurate. By contrasting the model prediction with and without the metabolite for each individual, SHAP values establish the significance of a given value in a given metabolite. A given feature may have different SHAP values for different individuals depending on the interactions with other features for each specific individual. The R packages “treeshap” and “SHAPforXGBoost” were used to calculate and plot the SHAP scores.
Transcriptomic analysis
Differential expression gene analyses were performed using the “limma” R package.100 A robust linear regression model adjusted for the age, BMI, gender, and country, was fitted using the “lmFit” function with the option method = “robust”, to limit the influence of outlying samples. Then, an empirical Bayes method was applied to borrow information between genes with the “eBayes” function. p-values were adjusted for multiple comparisons using the Benjamini-Hochberg procedure for False Discovery Rate (pFDR). A cut-off for the pFDR<0.1 was used as a threshold for statistical significance. The functional roles of differentially expressed genes were characterized by integrating the information provided from differential expression analysis, gene-gene interaction networks, and pathway over-representation analysis using the R package “pathfinder” with default parameters.60 Significant genes were initially mapped onto an STRING gene-gene interaction network. Then, active subnetworks of interconnected genes (including genes that are not significant themselves but connect significant genes) in this gene-gene interaction network were identified. Finally, separate pathway over-representation analyses based on the Reactome and KEGG databases were performed for each active subnetwork using the significant genes in each of the active subnetworks and genes in the PIN as the background genes. Pathway statistical significance was set at pFDR<0.05. Significantly enriched pathways were clustered via hierarchical clustering. Genes in each pathway were used to calculate the pairwise kappa statistics, a chance-corrected measure of co-occurrence between pathways. The distance matrix 1- kappa statistic was used for agglomerative hierarchical clustering and a threshold of 0.35 for the kappa statistics was used to define strong relations.
Metagenomic analysis
For metagenomics analyses, we excluded those microbial families, genera, or species having less than 10 counts in 10% of the samples. Differential abundance analyses for taxa associated with the circulating histidine levels, hepatic steatosis, HAA treatment, or the FMT study were performed using the analysis of compositions of microbiomes with bias correction (ANCOM-BC) methodology.101 It considers the bias due to differential sampling fractions across samples by adding a sample-specific offset to a linear regression model, that is estimated from the observed data. The linear regression model in log scale is analogous to log-ratio transformation to consider the compositional nature of metagenomics datasets, while the offset term serves as the bias correction. We adjusted all models in humans for age, gender, BMI, and country. The model for the FMT study was adjusted for the donor’s ferritin levels to control for the potential confounding effect of this parameter on the histidine-derived effects, as ferritin was the only variable that differed between high- and low-histidine donors and we have recently shown that iron influences NAFLD through the gut microbiome.1 p-values were adjusted for multiple comparisons using a Sequential Goodness of Fit61 as implemented in the “SGoF” R package. Unlike FDR methods, which decrease their statistical power as the number of tests increases, SGoF methods increase their power with an increasing number of tests. SGoF has proven to behave particularly better than FDR methods with a high number of tests and low sample size, which is the case of omics large datasets. Statistical significance was set at Padj < 0.1. To assess differences in the microbial Hut genes between patients with a degree of steatosis <33% and a degree of steatosis >33% we performed Mann-Whitney tests to the centered log-ratio transformed counts of the KEGG orthologs (KO) K01479, K01745, K01468, and K01712 corresponding to the hutG, hutH, hutI, and hutU, respectively.
Acknowledgments
We would like to thank Dr. Richard H. Barton for generating the 1H NMR spectra for the discovery cohort and validation cohort 1. This work was funded by ISCIII (PI20/01090) cofunded by ERDF, “A way to make Europe” (to J.M.-P), and ISCIII (CP18/0009) cofunded by ESF, “Investing in your future” (to J.M.-P). This work was also partially funded by Instituto de Salud Carlos III (ISCIII, Madrid, Spain) through the projects PI15/01934, PI18/01022, and PI21/01361 to J.M.F-R, Generalitat de Catalunya 2021-SGR-01263 and 2022-SGR-01556 and funding grant ACCIÓ-Eurecat (PRIV2020-EURHEPAD) (to X.E.), and EU METACARDIS under agreement HEALTH-F4-2012-305312 and MRC MR/M501797/1 (to M.-E.D.). L.H. received an MRC Intermediate Research Fellowship (MR/L01632X/1, UK Med-Bio). N.B. and N.T. were supported by BBSRC through BB/R012504/1, BBS/E/F/000PR10349, BBS/E/F/00044509, and BB/R012490/1. F.H. was supported by ERC-H2020 StG (erc-stg-948219, EPYC) and BBSRC BB/r012490/1 and BBS/e/F/000Pr10355. S.Q.-V. was supported by the Vicente Lopez fellowship (Eurecat). IDIBGI is a CERCA center from the CERCA Program/Generalitat de Catalunya.
Author contributions
S.Q.-V. and A.C.-N. researched the data, performed part of the statistical analyses, and wrote the manuscript. A.C.-N., J.L., and N.O.-C. performed the experiments with human primary hepatocytes. N.T., Y.T., and J.B. performed the histidine supplementation in rodent experiments. A.C.-N. and I.P.-P. performed all experiments in D. melanogaster. F.H. and N.B. performed the 16S analyses and histidine quantification in serum and mouse livers. L.M.-G. and J.C. performed plasma histidine quantification in the FMT study. R.B., M.E.-D., M.F., L.H., J.M.d.B., and A.C. contributed to the discussions and reviewed the manuscript. X.E., J.-M.F.-R., and J.M.-P. carried out the conception and coordination of the study, performed statistical analyses, and wrote the manuscript. All authors approved the final version to be published.
Declaration of interests
The authors declare no competing interests.
Published: December 19, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2023.101341.
Contributor Information
Xavier Escoté, Email: xavier.escote@eurecat.org.
José-Manuel Fernández-Real, Email: jmfreal@idibgi.org.
Jordi Mayneris-Perxachs, Email: jmayneris@idibgi.org.
Supplemental information
Data are mean ± SEM. P values from one-WAY ANOVA and Tuckey’s post hoc test or χ2 test. Different superscript letters indicate statistically significant difference.
The Spearman correlations are also included.
The Spearman correlations are also included.
Data are mean ± SEM. P values from one-WAY ANOVA and Tuckey’s post hoc test or χ2 test. Different superscript letters indicate statistically significant difference.
Data are mean ± SEM. P values from one-WAY ANOVA and Tuckey’s post hoc test or χ2 test. Different superscript letters indicate statistically significant difference.
The Spearman correlations are also included.
The Spearman correlations are also included.
References
- 1.Mayneris-Perxachs J., Cardellini M., Hoyles L., Latorre J., Davato F., Moreno-Navarrete J.M., Arnoriaga-Rodríguez M., Serino M., Abbott J., Barton R.H., et al. Iron status influences non-alcoholic fatty liver disease in obesity through the gut microbiome. Microbiome. 2021;9:104. doi: 10.1186/s40168-021-01052-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quesada-Vázquez S., Colom-Pellicer M., Navarro-Masip È., Aragonès G., del Bas J.M., Caimari A., Escoté X. Supplementation with a Specific Combination of Metabolic Cofactors Ameliorates Non-Alcoholic Fatty Liver Disease and, Hepatic Fibrosis, and Insulin Resistance in Mice. Nutrients. 2021;13:3532. doi: 10.3390/nu13103532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee D.-Y., Kim E.-H. Therapeutic Effects of Amino Acids in Liver Diseases: Current Studies and Future Perspectives. J. Cancer Prev. 2019;24:72–78. doi: 10.15430/JCP.2019.24.2.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kasaoka S., Tsuboyama-Kasaoka N., Kawahara Y., Inoue S., Tsuji M., Ezaki O., Kato H., Tsuchiya T., Okuda H., Nakajima S. Histidine supplementation suppresses food intake and fat accumulation in rats. Nutrition. 2004;20:991–996. doi: 10.1016/j.nut.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Kennedy L., Hargrove L., Demieville J., Bailey J.M., Dar W., Polireddy K., Chen Q., Nevah Rubin M.I., Sybenga A., DeMorrow S., et al. Knockout of l-Histidine Decarboxylase Prevents Cholangiocyte Damage and Hepatic Fibrosis in Mice Subjected to High-Fat Diet Feeding via Disrupted Histamine/Leptin Signaling. Am. J. Pathol. 2018;188:600–615. doi: 10.1016/j.ajpath.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kimura K., Nakamura Y., Inaba Y., Matsumoto M., Kido Y., Asahara S.I., Matsuda T., Watanabe H., Maeda A., Inagaki F., et al. Histidine augments the suppression of hepatic glucose production by central insulin action. Diabetes. 2013;62:2266–2277. doi: 10.2337/db12-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayashi T., Yamashita T., Takahashi T., Tabata T., Watanabe H., Gotoh Y., Shinohara M., Kami K., Tanaka H., Matsumoto K., et al. Uncovering the Role of Gut Microbiota in Amino Acid Metabolic Disturbances in Heart Failure Through Metagenomic Analysis. Front. Cardiovasc. Med. 2021;8:789325. doi: 10.3389/fcvm.2021.789325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niu Y.C., Feng R.N., Hou Y., Li K., Kang Z., Wang J., Sun C.H., Li Y. Histidine and arginine are associated with inflammation and oxidative stress in obese women. Br. J. Nutr. 2012;108:57–61. doi: 10.1017/S0007114511005289. [DOI] [PubMed] [Google Scholar]
- 9.Watanabe M., Suliman M.E., Rashid Qureshi A., Garcia-Lopez E., Bárány P., Heimbürger O., Stenvinkel P., Lindholm B. 2008. Consequences of Low Plasma Histidine in Chronic Kidney Disease Patients: Associations with Inflammation, Oxidative Stress, and Mortality; pp. 1–3. [DOI] [PubMed] [Google Scholar]
- 10.Thalacker-Mercer A.E., Gheller M.E. Benefits and Adverse Effects of Histidine Supplementation. J. Nutr. 2020;150:2588S–2592S. doi: 10.1093/jn/nxaa229. [DOI] [PubMed] [Google Scholar]
- 11.Liu W.h., Liu T.c., Yin M.c. Beneficial effects of histidine and carnosine on ethanol-induced chronic liver injury. Food Chem. Toxicol. 2008;46:1503–1509. doi: 10.1016/j.fct.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 12.Yan S.L., Wu S.T., Yin M.C., Chen H.T., Chen H.C. Protective effects from carnosine and histidine on acetaminophen-induced liver injury. J. Food Sci. 2009;74:H259–H265. doi: 10.1111/j.1750-3841.2009.01330.x. [DOI] [PubMed] [Google Scholar]
- 13.Menon K., Marquina C., Liew D., Mousa A., de Courten B. Histidine-containing dipeptides reduce central obesity and improve glycaemic outcomes: A systematic review and meta-analysis of randomized controlled trials. Obes. Rev. 2020;21:e12975. doi: 10.1111/obr.12975. [DOI] [PubMed] [Google Scholar]
- 14.Feng R.N., Niu Y.C., Sun X.W., Li Q., Zhao C., Wang C., Guo F.C., Sun C.H., Li Y. Histidine supplementation improves insulin resistance through suppressed inflammation in obese women with the metabolic syndrome: A randomised controlled trial. Diabetologia. 2013;56:985–994. doi: 10.1007/s00125-013-2839-7. [DOI] [PubMed] [Google Scholar]
- 15.Portune K.J., Beaumont M., Davila A.M., Tomé D., Blachier F., Sanz Y. Gut microbiota role in dietary protein metabolism and health-related outcomes: The two sides of the coin. Trends Food Sci. Technol. 2016;57:213–232. [Google Scholar]
- 16.Hoyles L., Fernández-Real J.M., Federici M., Serino M., Abbott J., Charpentier J., Heymes C., Luque J.L., Anthony E., Barton R.H., et al. Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat. Med. 2018;24:1070–1080. doi: 10.1038/s41591-018-0061-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lundberg S.M., Erion G., Chen H., DeGrave A., Prutkin J.M., Nair B., Katz R., Himmelfarb J., Bansal N., Lee S.-I. From Local Explanations to Global Understanding with Explainable AI for Trees. Nat. Mach. Intell. 2020;2:56–67. doi: 10.1038/s42256-019-0138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Targher G., Corey K.E., Byrne C.D., Roden M. The complex link between NAFLD and type 2 diabetes mellitus - mechanisms and treatments. Nat. Rev. Gastroenterol. Hepatol. 2021;18:599–612. doi: 10.1038/s41575-021-00448-y. [DOI] [PubMed] [Google Scholar]
- 19.Latorre J., Moreno-Navarrete J.M., Mercader J.M., Sabater M., Rovira Ò., Gironès J., Ricart W., Fernández-Real J.M., Ortega F.J. Decreased lipid metabolism but increased FA biosynthesis are coupled with changes in liver microRNAs in obese subjects with NAFLD. Int. J. Obes. 2017;41:620–630. doi: 10.1038/ijo.2017.21. [DOI] [PubMed] [Google Scholar]
- 20.Weber K., Rétey J. On the Nature of the Irreversible Inhibition of Histidine Ammonia Lyase by Cysteine and Dioxygen. Med. Chem. 1996;4:1001–1006. doi: 10.1016/0968-0896(96)00091-0. [DOI] [PubMed] [Google Scholar]
- 21.Botros M., Sikaris K.A. The De Ritis Ratio: The Test of Time. Clin. Biochem. Rev. 2013;34:117–130. [PMC free article] [PubMed] [Google Scholar]
- 22.Sutter A.G., Palanisamy A.P., Lench J.H., Jessmore A.P., Chavin K.D. Development of steatohepatitis in Ob/Ob mice is dependent on Toll-like receptor 4. Ann. Hepatol. 2015;14:735–743. [PubMed] [Google Scholar]
- 23.Saigo Y., Sasase T., Uno K., Shinozaki Y., Maekawa T., Sano R., Toriniwa Y., Miyajima K., Ohta T. Establishment of a new nonalcoholic steatohepatitis model; Ovariectomy exacerbates nonalcoholic steatohepatitis-like pathology in diabetic rats. J. Pharmacol. Toxicol. Methods. 2022;116 doi: 10.1016/j.vascn.2022.107190. [DOI] [PubMed] [Google Scholar]
- 24.Arquier N., Léopold P. Fly foie gras: modeling fatty liver in Drosophila. Cell Metab. 2007;5:83–85. doi: 10.1016/j.cmet.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 25.Moraes K.C.M., Montagne J. Drosophila melanogaster: A Powerful Tiny Animal Model for the Study of Metabolic Hepatic Diseases. Front. Physiol. 2021;12 doi: 10.3389/fphys.2021.728407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rinninella E., Raoul P., Cintoni M., Franceschi F., Miggiano G.A.D., Gasbarrini A., Mele M.C. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms. 2019;7:14. doi: 10.3390/microorganisms7010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huber-Ruano I., Calvo E., Mayneris-Perxachs J., Rodríguez-Peña M.M., Ceperuelo-Mallafré V., Cedó L., Núñez-Roa C., Miro-Blanch J., Arnoriaga-Rodríguez M., Balvay A., et al. Orally administered Odoribacter laneus improves glucose control and inflammatory profile in obese mice by depleting circulating succinate. Microbiome. 2022;10:135. doi: 10.1186/s40168-022-01306-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wirtz L., Eder M., Brand A.K., Jung H. HutT functions as the major L-histidine transporter in Pseudomonas putida KT2440. FEBS Lett. 2021;595:2113–2126. doi: 10.1002/1873-3468.14159. [DOI] [PubMed] [Google Scholar]
- 29.Bender R.A. Regulation of the Histidine Utilization (Hut) System in Bacteria. Microbiol. Mol. Biol. Rev. 2012;76:565–584. doi: 10.1128/MMBR.00014-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma D., Storelli G., Mitchell M., Leulier F. Studying host-microbiota mutualism in Drosophila: Harnessing the power of gnotobiotic flies. Biomed. J. 2015;38:285–293. doi: 10.4103/2319-4170.158620. [DOI] [PubMed] [Google Scholar]
- 31.Broderick N.A., Lemaitre B. Gut-associated microbes of Drosophila melanogaster. Gut Microb. 2012;3:307–321. doi: 10.4161/gmic.19896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin M., Zheng L., Wei Y., Cheng J., Zhang D., Yan S., Qin H., Wang Q., Ci X., Feng H. Enterobacter cloacae aggravates metabolic disease by inducing inflammation and lipid accumulation. Environ. Toxicol. Pharmacol. 2022;90 doi: 10.1016/j.etap.2022.103819. [DOI] [PubMed] [Google Scholar]
- 33.Fei N., Bruneau A., Zhang X., Wang R., Wang J., Rabot S., Gérard P., Zhao L. Endotoxin Producers Overgrowing in Human Gut Microbiota as the Causative Agents for Nonalcoholic Fatty Liver Disease. mBio. 2020;11 doi: 10.1128/mBio.03263-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koh A., Molinaro A., Ståhlman M., Khan M.T., Schmidt C., Mannerås-Holm L., Wu H., Carreras A., Jeong H., Olofsson L.E., et al. Microbially Produced Imidazole Propionate Impairs Insulin Signaling through mTORC1. Cell. 2018;175:947–961.e17. doi: 10.1016/j.cell.2018.09.055. [DOI] [PubMed] [Google Scholar]
- 35.Molinaro A., Bel Lassen P., Henricsson M., Wu H., Adriouch S., Belda E., Chakaroun R., Nielsen T., Bergh P.O., Rouault C., et al. Imidazole propionate is increased in diabetes and associated with dietary patterns and altered microbial ecology. Nat. Commun. 2020;11:5881. doi: 10.1038/s41467-020-19589-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee Y.T., Hsu C.C., Lin M.H., Liu K.s., Yin M.C. Histidine and carnosine delay diabetic deterioration in mice and protect human low density lipoprotein against oxidation and glycation. Eur. J. Pharmacol. 2005;513:145–150. doi: 10.1016/j.ejphar.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 37.Mong M.C., Chao C.Y., Yin M.C. Histidine and carnosine alleviated hepatic steatosis in mice consumed high saturated fat diet. Eur. J. Pharmacol. 2011;653:82–88. doi: 10.1016/j.ejphar.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 38.Raab S., Wang H., Uhles S., Cole N., Alvarez-Sanchez R., Künnecke B., Ullmer C., Matile H., Bedoucha M., Norcross R.D., et al. Incretin-like effects of small molecule trace amine-associated receptor 1 agonists. Mol. Metab. 2016;5:47–56. doi: 10.1016/j.molmet.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michael E.S., Covic L., Kuliopulos A. Trace amine–associated receptor 1 (TAAR1) promotes anti-diabetic signaling in insulin-secreting cells. J. Biol. Chem. 2019;294:4401–4411. doi: 10.1074/jbc.RA118.005464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quesada-Vázquez S., Aragonès G., del Bas J.M., Escoté X. Diet, Gut Microbiota and Non-Alcoholic Fatty Liver Disease: Three Parts of the Same Axis. Cells. 2020;9:1–17. doi: 10.3390/cells9010176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sookoian S., Salatino A., Castaño G.O., Landa M.S., Fijalkowky C., Garaycoechea M., Pirola C.J. Intrahepatic bacterial metataxonomic signature in non-alcoholic fatty liver disease. Gut. 2020;69:1483–1491. doi: 10.1136/gutjnl-2019-318811. [DOI] [PubMed] [Google Scholar]
- 42.Vallianou N., Christodoulatos G.S., Karampela I., Tsilingiris D., Magkos F., Stratigou T., Kounatidis D., Dalamaga M. Understanding the Role of the Gut Microbiome and Microbial Metabolites in Non-Alcoholic Fatty Liver Disease: Current Evidence and Perspectives. Biomolecules. 2021;12:56. doi: 10.3390/biom12010056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mu H.N., Zhou Q., Yang R.Y., Tang W.Q., Li H.X., Wang S.M., Li J., Chen W.X., Dong J. Caffeic acid prevents non-alcoholic fatty liver disease induced by a high-fat diet through gut microbiota modulation in mice. Food Res. Int. 2021;143:110240. doi: 10.1016/j.foodres.2021.110240. [DOI] [PubMed] [Google Scholar]
- 44.Jian C., Luukkonen P., Sädevirta S., Yki-Järvinen H., Salonen A. Impact of short-term overfeeding of saturated or unsaturated fat or sugars on the gut microbiota in relation to liver fat in obese and overweight adults. Clin. Nutr. 2021;40:207–216. doi: 10.1016/j.clnu.2020.05.008. [DOI] [PubMed] [Google Scholar]
- 45.de Filippis F., Pasolli E., Ercolini D. Newly Explored Faecalibacterium Diversity Is Connected to Age, Lifestyle, Geography, and Disease. Curr. Biol. 2020;30:4932–4943.e4. doi: 10.1016/j.cub.2020.09.063. [DOI] [PubMed] [Google Scholar]
- 46.Waters J.L., Ley R.E. The human gut bacteria Christensenellaceae are widespread, heritable, and associated with health. BMC Biol. 2019;17:83. doi: 10.1186/s12915-019-0699-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tavella T., Rampelli S., Guidarelli G., Bazzocchi A., Gasperini C., Pujos-Guillot E., Comte B., Barone M., Biagi E., Candela M., et al. Elevated gut microbiome abundance of Christensenellaceae, Porphyromonadaceae and Rikenellaceae is associated with reduced visceral adipose tissue and healthier metabolic profile in Italian elderly. Gut Microb. 2021;13:1–19. doi: 10.1080/19490976.2021.1880221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mokhtari Z., Gibson D.L., Hekmatdoost A. Nonalcoholic fatty liver disease, the gut microbiome, and diet. Adv. Nutr. 2017;8:240–252. doi: 10.3945/an.116.013151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Juárez-Fernández M., Porras D., Petrov P., Román-Sagüillo S., García-Mediavilla M.V., Soluyanova P., Martínez-Flórez S., González-Gallego J., Nistal E., Jover R., et al. The Synbiotic Combination of Akkermansia muciniphila and Quercetin Ameliorates Early Obesity and NAFLD through Gut Microbiota Reshaping and Bile Acid Metabolism Modulation. Antioxidants. 2021:10. doi: 10.3390/antiox10122001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwan S.Y., Jiao J., Joon A., Wei P., Petty L.E., Below J.E., Daniel C.R., Wu X., Zhang J., Jenq R.R., et al. Gut microbiome features associated with liver fibrosis in Hispanics, a population at high risk for fatty liver disease. Hepatology. 2022;75:955–967. doi: 10.1002/hep.32197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leyn S.A., Suvorova I.A., Kazakov A.E., Ravcheev D.A., Stepanova V.v., Novichkov P.S., Rodionov D.A. Comparative genomics and evolution of transcriptional regulons in Proteobacteria. Microb. Genom. 2016;2:e000061. doi: 10.1099/mgen.0.000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmieder R., Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Magoč T., Salzberg S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li D., Luo R., Liu C.-M., Leung C.-M., Ting H.-F., Sadakane K., Yamashita H., Lam T.-W. MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods. 2016;102:3–11. doi: 10.1016/j.ymeth.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 56.Hyatt D., Chen G.-L., LoCascio P.F., Land M.L., Larimer F.W., Hauser L.J. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinf. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Durbin R., Eddy S.R., Krogh A., Mitchison G. Cambridge University Press; 1998. Biological Sequence Analysis. [Google Scholar]
- 58.Menzel P., Ng K.L., Krogh A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat. Commun. 2016;7 doi: 10.1038/ncomms11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smyth G.K. In: Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Gentleman R., Carey V.J., Huber W., Irizarry R.A., Dudoit S., editors. Springer; 2005. limma: Linear Models for Microarray Data; pp. 397–420. [Google Scholar]
- 60.Ulgen E., Ozisik O., Sezerman O.U. pathfindR: An R Package for Comprehensive Identification of Enriched Pathways in Omics Data Through Active Subnetworks. Front. Genet. 2019;10:858. doi: 10.3389/fgene.2019.00858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carvajal-Rodríguez A., de Uña-Alvarez J., Rolán-Alvarez E. A new multitest correction (SGoF) that increases its statistical power when increasing the number of tests. BMC Bioinf. 2009;10:209. doi: 10.1186/1471-2105-10-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu Y., Feng K., Wei Z., Wang Z., Deng Y. Ardep, a rapid degenerate primer design pipeline based on k-mers for amplicon microbiome studies. Int. J. Environ. Res. Public Health. 2020;17:5958. doi: 10.3390/ijerph17165958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quesada-Vázquez S., Bone C., Saha S., Triguero I., Colom-Pellicer M., Aragonès G., Hildebrand F., del Bas J.M., Caimari A., Beraza N., Escoté X. Microbiota Dysbiosis and Gut Barrier Dysfunction Associated with Non-Alcoholic Fatty Liver Disease Are Modulated by a Specific Metabolic Cofactors’ Combination. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms232213675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marin V., Rosso N., Dal Ben M., Raseni A., Boschelle M., Degrassi C., Nemeckova I., Nachtigal P., Avellini C., Tiribelli C., Gazzin S. An animal model for the juvenile non-alcoholic fatty liver disease and non-alcoholic steatohepatitis. PLoS One. 2016;11:e0158817. doi: 10.1371/journal.pone.0158817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Du S., Sun S., Liu L., Zhang Q., Guo F., Li C., Feng R., Sun C. Effects of Histidine Supplementation on Global Serum and Urine 1H NMR-based Metabolomics and Serum Amino Acid Profiles in Obese Women from a Randomized Controlled Study. J. Proteome Res. 2017;16:2221–2230. doi: 10.1021/acs.jproteome.7b00030. [DOI] [PubMed] [Google Scholar]
- 66.Dowlati Y., Maheux M., Meyer J.H. The Effect of Oral L-cysteine on Breast Milk and Plasma Cysteine Concentrations. Neuropsychiatr. Dis. Treat. 2020;16:3163–3172. doi: 10.2147/NDT.S255205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fridman V., Suriyanarayanan S., Novak P., David W., Macklin E.A., McKenna-Yasek D., Walsh K., Aziz-Bose R., Oaklander A.L., Brown R., et al. Randomized trial of l-serine in patients with hereditary sensory and autonomic neuropathy type 1. Neurology. 2019;92:E359–E370. doi: 10.1212/WNL.0000000000006811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Houjeghani S., Kheirouri S., Faraji E., Jafarabadi M.A. l-Carnosine supplementation attenuated fasting glucose, triglycerides, advanced glycation end products, and tumor necrosis factor-α levels in patients with type 2 diabetes: a double-blind placebo-controlled randomized clinical trial. Nutr. Res. 2018;49:96–106. doi: 10.1016/j.nutres.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 69.Reagan-Shaw S., Nihal M., Ahmad N. Dose translation from animal to human studies revisited. Faseb. J. 2008;22:659–661. doi: 10.1096/fj.07-9574LSF. [DOI] [PubMed] [Google Scholar]
- 70.Castells-Nobau A., Eidhof I., Fenckova M., Brenman-Suttner D.B., Scheffer-De Gooyert J.M., Christine S., Schellevis R.L., Van Der Laan K., Quentin C., Van Ninhuijs L., et al. Conserved regulation of neurodevelopmental processes and behavior by FoxP in Drosophila. PLoS One. 2019;14:e0211652. doi: 10.1371/journal.pone.0211652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kleiner D.E., Brunt E.M., van Natta M., Behling C., Contos M.J., Cummings O.W., Ferrell L.D., Liu Y.C., Torbenson M.S., Unalp-Arida A., et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 72.Torres D.M., Harrison S.A. Diagnosis and therapy of nonalcoholic steatohepatitis. Gastroenterology. 2008;134:1682–1698. doi: 10.1053/j.gastro.2008.02.077. [DOI] [PubMed] [Google Scholar]
- 73.Strauss S., Gavish E., Gottlieb P., Katsnelson L. Interobserver and intraobserver variability in the sonographic assessment of fatty liver. AJR Am. J. Roentgenol. 2007;189:W320–W323. doi: 10.2214/AJR.07.2123. [DOI] [PubMed] [Google Scholar]
- 74.Fleige S., Pfaffl M.W. RNA integrity and the effect on the real-time qRT-PCR performance. Mol. Aspects Med. 2006;27:126–139. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 75.Kanehisa M., Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arnoriaga-Rodríguez M., Mayneris-Perxachs J., Burokas A., Contreras-Rodríguez O., Blasco G., Coll C., Biarnés C., Miranda-Olivos R., Latorre J., Moreno-Navarrete J.-M., et al. Obesity Impairs Short-Term and Working Memory through Gut Microbial Metabolism of Aromatic Amino Acids. Cell Metab. 2020;32:548–560.e7. doi: 10.1016/j.cmet.2020.09.002. [DOI] [PubMed] [Google Scholar]
- 77.R Development Core Team . 2013. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 78.Caimari A., del Bas J.M., Crescenti A., Arola L. Low doses of grape seed procyanidins reduce adiposity and improve the plasma lipid profile in hamsters. Int. J. Obes. 2013;37:576–583. doi: 10.1038/ijo.2012.75. [DOI] [PubMed] [Google Scholar]
- 79.Rodríguez-Sureda V., Peinado-onsurbe J. A procedure for measuring triacylglyceride and cholesterol content using a small amount of tissue. Anal. Biochem. 2005;343:277–282. doi: 10.1016/j.ab.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 80.Liang W., Menke A.L., Driessen A., Koek G.H., Lindeman J.H. Establishment of a General NAFLD Scoring System for Rodent Models and Comparison to Human Liver Pathology. PLoS One. 2014;9 doi: 10.1371/journal.pone.0115922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hildebrand F., Tadeo R., Voigt A.Y., Bork P., Raes J. LotuS: An efficient and user-friendly OTU processing pipeline. Microbiome. 2014;2:30. doi: 10.1186/2049-2618-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Özkurt E., Fritscher J., Soranzo N., Ng D.Y.K., Davey R.P., Bahram M., Hildebrand F. LotuS2: An ultrafast and highly accurate tool for amplicon sequencing analysis. bioRxiv. 2021 doi: 10.1186/s40168-022-01365-1. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bedarf J.R., Beraza N., Khazneh H., Özkurt E., Baker D., Borger V., Wüllner U., Hildebrand F. Much ado about nothing? Off-target amplification can lead to false-positive bacterial brain microbiome detection in healthy and Parkinson’s disease individuals. Microbiome. 2021;9:75. doi: 10.1186/s40168-021-01012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yilmaz P., Parfrey L.W., Yarza P., Gerken J., Pruesse E., Quast C., Schweer T., Peplies J., Ludwig W., Glöckner F.O. The SILVA and “all-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014;42:D643–D648. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hildebrand F., Moitinho-Silva L., Blasche S., Jahn M.T., Gossmann T.I., Huerta-Cepas J., Hercog R., Luetge M., Bahram M., Pryszlak A., et al. Antibiotics-induced monodominance of a novel gut bacterial order. Gut. 2019;68:1781–1790. doi: 10.1136/gutjnl-2018-317715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Saary P., Forslund K., Bork P., Hildebrand F. RTK: Efficient rarefaction analysis of large datasets. Bioinformatics. 2017;33:2594–2595. doi: 10.1093/bioinformatics/btx206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Douglas G.M., Maffei V.J., Zaneveld J.R., Yurgel S.N., Brown J.R., Taylor C.M., Huttenhower C., Langille M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020;38:685–688. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Puigarnau S., Fernàndez A., Obis E., Jové M., Castañer M., Pamplona R., Portero-Otin M., Camerino O. Metabolomics reveals that fittest trail runners show a better adaptation of bioenergetic pathways. J. Sci. Med. Sport. 2022;25:425–431. doi: 10.1016/j.jsams.2021.12.006. [DOI] [PubMed] [Google Scholar]
- 90.Zhao S., Li H., Han W., Chan W., Li L. Coverage of Chemical-Group-Submetabolome Analysis: Group Classification and Four-Channel Chemical Isotope Labeling LC-MS. Anal. Chem. 2019;91:12108–12115. doi: 10.1021/acs.analchem.9b03431. [DOI] [PubMed] [Google Scholar]
- 91.Yang J., Neira S., Elisa F., Gil-iturbe E., Castilla-madrigal R., Fern M., Mart J.A., Moreno-aliaga M.J. 2021. Effects of Long-Term DHA Supplementation and Physical Aged Female Mice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ballak D.B., van Diepen J.A., Moschen A.R., Jansen H.J., Hijmans A., Groenhof G.J., Leenders F., Bufler P., Boekschoten M.v., Müller M., et al. IL-37 protects against obesity-induced inflammation and insulin resistance. Nat. Commun. 2014;5:4711. doi: 10.1038/ncomms5711. [DOI] [PubMed] [Google Scholar]
- 93.Antraco V.J., Hirata B.K.S., Simão J. de J., Cruz M.M., da Silva V.S., de Sá R.D.C.d.C., Abdala F.M., Armelin-Correa L., Alonso-Vale M.I.C. Omega-3 polyunsaturated fatty acids prevent nonalcoholic steatohepatitis (Nash) and stimulate adipogenesis. Nutrients. 2021;13:1–20. doi: 10.3390/nu13020622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.López-Yoldi M., Fernández-Galilea M., Laiglesia L.M., Larequi E., Prieto J., Martínez J.A., Bustos M., Moreno-Aliaga M.J. Cardiotrophin-1 stimulates lipolysis through the regulation of main adipose tissue lipases. J. Lipid Res. 2014;55:2634–2643. doi: 10.1194/jlr.M055335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Miranda M., Escoté X., Ceperuelo-Mallafré V., Megía A., Caubet E., Näf S., Gómez J.M., González-Clemente J.M., Vicente V., Vendrell J. Relation between human LPIN1, hypoxia and endoplasmic reticulum stress genes in subcutaneous and visceral adipose tissue. Int. J. Obes. 2010;34:679–686. doi: 10.1038/ijo.2009.290. [DOI] [PubMed] [Google Scholar]
- 96.Koyle M.L., Veloz M., Judd A.M., Wong A.C.N., Newell P.D., Douglas A.E., Chaston J.M. Rearing the Fruit Fly Drosophila melanogaster Under Axenic and Gnotobiotic Conditions. J. Vis. Exp. 2016;54219 doi: 10.3791/54219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tuthill B.F., Searcy L.A., Yost R.A., Musselman L.P. Tissue-specific analysis of lipid species in Drosophila during overnutrition by UHPLC-MS/MS and MALDI-MSI. J. Lipid Res. 2020;61:275–290. doi: 10.1194/jlr.RA119000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kursa M.B., Rudnicki W.R. Feature selection with the boruta package. J. Stat. Softw. 2010;36:1–13. [Google Scholar]
- 99.Degenhardt F., Seifert S., Szymczak S. Evaluation of variable selection methods for random forests and omics data sets. Brief. Bioinform. 2019;20:492–503. doi: 10.1093/bib/bbx124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lin H., Peddada S.D. Analysis of compositions of microbiomes with bias correction. Nat. Commun. 2020;11:3514. doi: 10.1038/s41467-020-17041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data are mean ± SEM. P values from one-WAY ANOVA and Tuckey’s post hoc test or χ2 test. Different superscript letters indicate statistically significant difference.
The Spearman correlations are also included.
The Spearman correlations are also included.
Data are mean ± SEM. P values from one-WAY ANOVA and Tuckey’s post hoc test or χ2 test. Different superscript letters indicate statistically significant difference.
Data are mean ± SEM. P values from one-WAY ANOVA and Tuckey’s post hoc test or χ2 test. Different superscript letters indicate statistically significant difference.
The Spearman correlations are also included.
The Spearman correlations are also included.
Data Availability Statement
The data that support the findings of this study are available from the lead contact (jmfreal@idibgi.org) upon reasonable request. The raw metagenomic sequence data for the FLORINASH cohort (with human-associated reads removed) have been deposited under the study accession number PRJEB14215. The raw 16S rRNA gene sequence data associated with the mouse FMT work have been deposited under the study accession number PRJEB24891.