Abstract
Background
Asthma exacerbations reflect disease severity, affect morbidity and mortality, and may lead to declining lung function. Inflammatory endotypes (e.g. T2-high (eosinophilic)) may play a key role in asthma exacerbations. We aimed to assess whether genetic susceptibility underlies asthma exacerbation risk and additionally tested for an interaction between genetic variants and eosinophilia on exacerbation risk.
Methods
UK Biobank data were used to perform a genome-wide association study of individuals with asthma and at least one exacerbation compared to individuals with asthma and no history of exacerbations. Individuals with asthma were identified using self-reported data, hospitalisation data and general practitioner records. Exacerbations were identified as either asthma-related hospitalisation, general practitioner record of asthma exacerbation or an oral corticosteroid burst prescription. A logistic regression model adjusted for age, sex, smoking status and genetic ancestry via principal components was used to assess the association between genetic variants and asthma exacerbations. We sought replication for suggestive associations (p<5×10−6) in the GERA cohort.
Results
In the UK Biobank, we identified 11 604 cases and 37 890 controls. While no variants reached genome-wide significance (p<5×10−8) in the primary analysis, 116 signals were suggestively significant (p<5×10−6). In GERA, two single nucleotide polymorphisms (rs34643691 and rs149721630) replicated (p<0.05), representing signals near the NTRK3 and ABCA13 genes.
Conclusions
Our study has identified reproducible associations with asthma exacerbations in the UK Biobank and GERA cohorts. Confirmation of these findings in different asthma subphenotypes in diverse ancestries and functional investigation will be required to understand their mechanisms of action and potentially inform therapeutic development.
Tweetable abstract
Genetic variations annotated to NTRK3/ABCA13 are associated with asthma exacerbations. The association of an additional locus (CXADRP3/POTEC) shows genetic effects modified by eosinophilia, implicating variants linked with respiratory viruses. https://bit.ly/3QLiZyX
Introduction
Asthma is a heterogeneous chronic respiratory condition, estimated to affect >300 million people worldwide [1], and has a well-established genetic component [2]. The disease features a network of complex inflammatory endotypes, and several clinical phenotypes have been defined based on the onset of asthma, control of symptoms and comorbidity of allergy involved in the underlying pathophysiology [3]. In early attempts to disentangle the molecular pathophysiology of asthma, the disease was broadly divided into two major endotypes: type 2 asthma and non-type 2 asthma [4].
The type 2 inflammatory endotype represents an important clinical challenge, as it is characterised by airway eosinophilia, difficulty to achieve asthma control and a higher frequency of exacerbations [4, 5]. Moreover, a crucial subgroup of individuals with asthma with this endotype are uncontrolled despite treatment with inhaled corticosteroids. This group represents a large proportion of individuals with severe uncontrolled asthma and frequent exacerbations [4]. Type 2 inflammation is driven by increased activity of T-helper cells type 2 (Th2), activated by dendritic cells [4]. These secrete interleukin (IL)-5, IL-4 and IL-13 and other type 2 cytokines, which activate type 2 immunity pathways [6, 7]. Therefore, type 2 asthma is characterised by airway and systemic eosinophilia [4]. Type 2 development is thought to be driven by genetics, as well as epigenetic and environmental factors [4, 8]. Interestingly, Tantisira et al. [9] showed that genetic factors determining susceptibility to asthma differ from those determining its severity. Determining the most important genetic variants affecting asthma severity may be fundamental to understanding drivers of disease activity in asthma [10], and subsequently improve available treatment approaches, either by early prediction of severity, or finding new druggable targets for severe, uncontrolled asthma. Although several studies have attempted to disentangle the underlying genetic variants behind asthma, the power and sample sizes required have necessitated the combination of different potential asthma phenotypes, meaning that these studies may have missed subtype-specific and severity-determining genetic risk factors [2].
Asthma exacerbations are an important cause of asthma morbidity, mortality and healthcare costs [11–15].They are useful in evaluating treatment response and are a marker of asthma control [16]. Moreover, exacerbations may increase the rate of lung function decline, thus representing a clinically important long-term outcome [17, 18]. Asthma exacerbations are known to be affected in part by genetics and epigenetics [2, 19], and are associated with the active inflammatory endotype [5]. Studies investigating asthma exacerbations focused mainly on hospitalisations and emergency department visits, and revealed several genes including IL13, IL4RA, CHI3L1, ORMDL3, CDHR3, CTNNA3, SEMA3D, EXTL2 and PANK1 [9, 20]. However, most studies investigating hospitalisations in asthma focused on childhood asthma, and only included a small number of events or cases [9, 20, 21]. An analysis in the UK Biobank focused on asthma hospitalisations (but not data from primary care records) and implicated genes in the HLA region [22]. In this study, we aimed to evaluate the genetic factors affecting asthma exacerbations in a large, genome-wide study. Additionally, we aimed to investigate whether eosinophilia modifies the effect of variants on exacerbations.
Methods
Study population
This study used a case–control design in two stages. The data source for the primary analysis was the UK Biobank (https://www.ukbiobank.ac.uk), and analysis was performed under approved application 648. The UK Biobank is a population-based study of half a million volunteer participants between the ages of 40 and 69 years, recruited from Great Britain in 2006–2010. In total, 321 057 individuals with genetic data were eligible for inclusion in this analysis. The data source for the replication was the Genetic Epidemiology Research in Aging (GERA) cohort. GERA is a multiethnic cohort of over 110 000 subjects from the Kaiser Permanente Medical Care Plan, Northern California Region (KPNC), Research Program on Genes, Environment, and Health who provided a saliva sample [23]. Ancestry was self-reported, and 80% of subjects were non-Hispanic white. The GERA study was approved by the institutional review boards at KPNC and Brigham and Women's Hospital (2002P000331).
Definition of asthma
In the UK Biobank, asthma was defined as either: self-reported asthma in the touchscreen questionnaire (data field: 6152), an asthma code in general practitioner records (Read v2 and Read v3 codes, full list of codes is provided as supplementary tables S1 and S2; the choice of code was based on Mukherjee et al. [24]), or any hospitalisation event with an asthma ICD10 code as primary or secondary cause for admission (ICD10 codes: J45, J45.0, J45.1, J45.8, J45.9, J46, J46.0). In GERA, over 16 000 asthmatic individuals have genotype data and longitudinal electronic medical records, including detailed diagnosis and medication records. Prescription data were available from outpatient and emergency department visits. Adult asthma cases were defined as patients at least 21 years of age with one or more of the following: physician-diagnosed asthma, self-reported asthma or a report of an asthma exacerbation (i.e. emergency department visit or hospitalisation due to asthma).
Exclusion criteria
In the UK Biobank, individuals were excluded from this study if they had incomplete genotyping data that did not pass quality control (as described in Shrine et al. [25] and Guyatt et al. [26]), non-European ancestry based on K-means clustering after principal component analysis (as described in Shrine et al. [25] and Guyatt et al. [26]), evidence of COPD (defined as either: self-reported COPD in the touchscreen questionnaire; a COPD hospitalisation code (ICD10 codes: J44, J44.0, J44.1, J44.8, J44.9) or a forced expiratory volume in 1 s (FEV1)/forced vital capacity ratio <0.7); evidence of chronic bronchitis and/or emphysema (defined as either: self-reported chronic bronchitis or emphysema in the touchscreen questionnaire or an emphysema hospitalisation code: J43.2, J43.8, J43.9). Related individuals up to second degree relatives were excluded based on kinship coefficients using KING software [27]. In GERA, subjects with COPD, pulmonary embolism, primary pulmonary hypertension, cystic fibrosis, emphysema, chronic bronchitis, bronchiectasis or participants self-reporting a non-European ancestry were excluded.
Definition of asthma exacerbators
Individuals were classified as exacerbators using three different data sources: hospitalisation data, primary care general practitioner records data and primary care prescription data. Hospitalisations were considered the result of an asthma exacerbation if: 1) asthma was listed as a primary cause for hospitalisation; 2) asthma was listed as a secondary cause for hospitalisation with the primary cause being a respiratory infection or condition associated with asthma (codes: J10.0, J10.1, J11.1, J11.8, J20.9, J67.9, J96.0, J96.00, J96.01, J96.09, J96.1, J96.11, J96.19, J96.9, J96.90, J96.91, J96.99, R06.1, R06.2, R06.4, R06.5); 3), asthma was listed a secondary cause and the primary cause was chest pain/dyspnoea (codes: R06.0, R07.0, R07.1, R07.2, R07.3, or R07.4) and the individual had no record of a cardiac condition (all ICD cardiac-related codes I.X). Exacerbations in general practitioner records (available for ∼45% of the UK Biobank population) were identified using read v2 and read v3 codes for exacerbations (codes: H333, H3301, H3311, H33z0, H333z1, XE0YW, Xa1hD, Xafdy, Xafdz, Xafdj, XM0s2, 663d or 8H2P, based on Shah et al. [28]). In the primary care prescription data, oral corticosteroid (OCS) prescriptions were analysed for evidence of OCS bursts [29, 30]. OCS prescriptions were considered as exacerbations if the total dose prescribed was 200–600 mg. Prescriptions <2 weeks apart, <2 weeks after an annual asthma review (to avoid counting rescue packs) or <2 weeks after a general practitioner-recorded exacerbation were excluded. Individuals not meeting any of these definitions were considered controls (non-exacerbators). To account for the potential that prescriptions may have suffered classification bias due to a fraction being prescribed OCS bursts for other conditions, a sensitivity analysis was conducted restricting the definition of exacerbations to those identified using hospitalisation data or primary care general practitioner diagnostic records. In GERA, exacerbations in the GERA cohort were defined as either an emergency department visit or hospitalisation due to asthma (ICD10 codes: J45.XX, J46.XX), or an OCS burst prescription defined as a short course of oral steroids for 3–21 days.
Genotyping
In the UK Biobank, two arrays were used for genotyping: Affymetrix Axiom® UK BiLEVE array and the Affymetrix Axiom® UK Biobank array [31]. The Haplotype Reference Consortium panel was used for imputation. In total, 9 805 379 variants met our quality control criteria: minor allele frequency ≥0.01 and imputation score ≥0.3. GERA sample extraction, genotyping, imputation and QC procedures were previously published [23]. In brief, custom-designed, ethnicity-specific arrays encompassing >650 000 single nucleotide polymorphisms (SNPs) were used for genotyping [23]. The 1000 Genomes phase 3 release panel was used for imputation.
Genome-wide analysis
A logistic regression model was used to determine genome-wide associations between genetic variants and asthma exacerbator status, assuming an additive genetic model. Imputed genotype dosage (effect allele as a continuous variable ranging from 0 to 2, reflecting uncertainty in genotype imputation) was fitted using Plink version 2 [32]. Age, sex, smoking status, the genotyping array and the first 10 principal components (to adjust for population stratification) were included as covariates. From this analysis, we identified independent, associated sentinel variants using clumping in Plink with a p-value threshold of 5×10−6, an r2 (linkage disequilibrium) cut-off of 0.1 and the default distance parameter of 250 kb.
Annotation and functional analysis
A genome-wide association study (GWAS) catalogue was queried for variants associated with asthma at a genome-wide significance level within 1 Mb of the sentinel variant position. Variant annotation and functional mapping were conducted using the Ensembl Variant Effect Predictor (VEP) [33] and SNPnexus [34]. The Kyoto Encyclopedia of Genes and Genomes (KEGG) [35–37] and the Reactome Database [38] were used to investigate the pathways associated with the list of genes annotated to suggestive hits. A lookup of suggestive SNPs (p<10−6) lung cis-eQTL effects was performed using data from the Lung eQTL consortium [39].
Fine-mapping
Fine-mapping was conducted using susieR package version 0.11.42 [40]. All biallelic variants with MAF >0.0005 within a 1 Mb window surrounding the sentinel SNP were included. Summary statistics from logistic regression using Plink were passed to the susie_rss function with a variant correlation matrix generated from hard-calls using the cor function in R. Region plots were generated using LocusZoom [41].
Interaction with eosinophils
Given the potential that genetic variants may have varying effects on exacerbations in different asthma phenotypes, we conducted a secondary analysis to investigate effect modification by eosinophilia (associated with type 2 asthma) on the association between genetic variants and exacerbator status. Eosinophilia was defined as a binary variable using a cut-off of 300 cells·μL−1, collected during any of the three available visits to the UK Biobank assessment centre. Eosinophilia × variant was used as an interaction term, in the following model: Exacerbator(0/1) ∼ SNP + age + sex + smoking + Eosinophilic (0/1) + SNP × Eosinophilic + 10 principal components.
Power analysis
Using Genetic Association Study (GAS) power calculator, detecting variants with a minimum relative risk of 1.3 with 80% power at a 5×10−8 significance level was estimated to require a sample size of at least 1130 cases and 3000 controls for risk alleles with a frequency of at least 10% (assuming frequency of exacerbators 40% among individuals with asthma).
Results
Discovery cohort
In total, 70 918 individuals were selected from the UK Biobank as asthma cases (based on self-reported asthma, hospitalisation with an asthma code or a primary care record of asthma), of which 49 494 individuals met our inclusion criteria for this analysis. The baseline characteristics for all included individuals are shown in table 1. Of included individuals, 11 604 (23.4%) met our criteria for having at least one exacerbation during follow-up (under any of the definitions used) and the remaining individuals (n=37 890) were considered as non-exacerbating controls. Exacerbators were slightly older, included a lower proportion of ever-smokers, had higher levels of eosinophils on average and were less likely to be identified as childhood-onset asthma.
TABLE 1.
Baseline characteristics of cases and controls
| Characteristic | Exacerbators (n=11 604) | Non-exacerbators (n=37 890) |
| Age years, mean±sd | 57±8 | 56±8 |
| Female sex | 7096 (54.5) | 20 542 (54.2) |
| Ever-smoking | 6053 (52.2) | 21 231 (56.0) |
| Eosinophilic (≥0.3 cells·μL−1) | 3117 (26.7) | 8911 (23.5) |
| Childhood-onset asthma (<18 years at age of diagnosis) | 2680 (23.1) | 11 102 (29.3) |
| Primary asthma hospitalisation# | 1670 (14.4) | |
| Secondary hospitalisation# | 717 (6.2) | |
| Chest pain/dyspnoea#,¶ | 1514 (13.0) | |
| Primary care (read codes)# | 1136 (9.8) | |
| Primary care exacerbations (prescriptions)# | 8788 (75.7) |
Data are presented as n (%) unless indicated otherwise. #: some exacerbators met multiple definitions of exacerbations; ¶: secondary asthma hospitalisations with chest pain/dyspnoea as primary hospitalisation cause.
Association signals for asthma exacerbations (discovery)
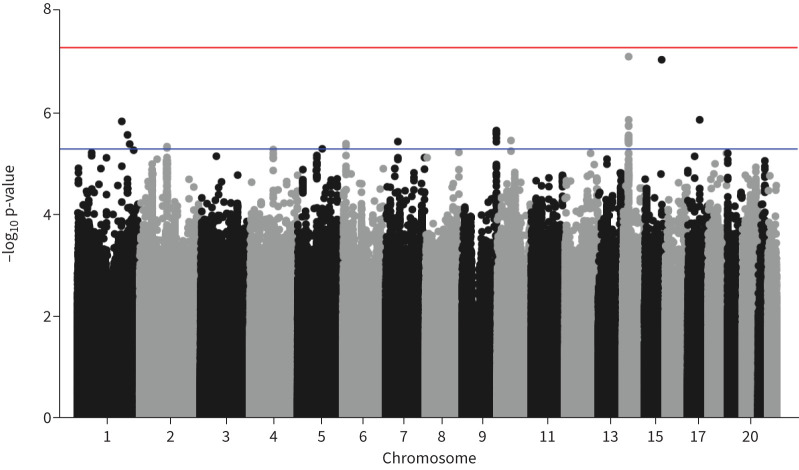
No SNPs were associated with risk of asthma exacerbations at a genome-wide significance threshold (p<5×10−8), but 33 SNPs representing 12 independent loci were significant at a suggestive p-value threshold (p<5×10−6) (figure 1). A quantile–quantile plot is shown in supplementary figure S1. These associations were followed up in a sensitivity analysis where exacerbations were defined as either hospitalisations or general practitioner recorded exacerbations only (therefore excluding prescription data). Two SNPs retained a nominally significant association and direction of effect with exacerbations in the sensitivity analysis.
FIGURE 1.
Manhattan plot of association results for asthma exacerbations in the UK Biobank. The x-axis shows genomic location by chromosome, the y-axis shows the –log10 p-value. The blue line indicates p=5×10−6, and the red line corresponds to p=5×10−8 (commonly known as genome-wide significance level).
Association signals in replication cohort (GERA)
Aiming to replicate our suggestively significant signals from the discovery and to generate more accurate effect estimates for any reproducible loci, all signals showing a suggestive association in the discovery cohort (UK Biobank) were tested for association with exacerbations in an independent study (GERA). In GERA, the analysis included 7927 participants with asthma of European ancestry, of which 40.1% were considered exacerbators. Two variants (rs34643691 and rs149721630) showed consistent direction of effect and nominal significance (p<0.05) for association (table 2) (supplementary file S1).
TABLE 2.
SNPs associated with asthma exacerbations within patients with asthma in both the UK Biobank and GERA
| RsID | Allele | Chr:position | IS | Consequence | Nearest gene | MAF (discovery) | OR (discovery) | p-value (discovery) | OR (replication) | p-value (replication) |
| rs149721630 | C | 7: 48 542 408 | 0.94 | Intronic variant | ABCA13 | 0.012 | 0.70 | 3.57×10–6 | 0.64 | 0.049 |
| rs34643691 | T | 15:88 374 252 | 0.94 | Intergenic variant | NTRK3 | 0.010 | 1.46 | 8.67×10−8 | 1.74 | 0.017 |
SNPs: single nucleotide polymorphisms; GERA: Genetic Epidemiology Research in Aging; IS: imputation info score; MAF: minor allele frequency; IS: imputation info score; OR: odds ratio; RsID: unique SNP identifier.
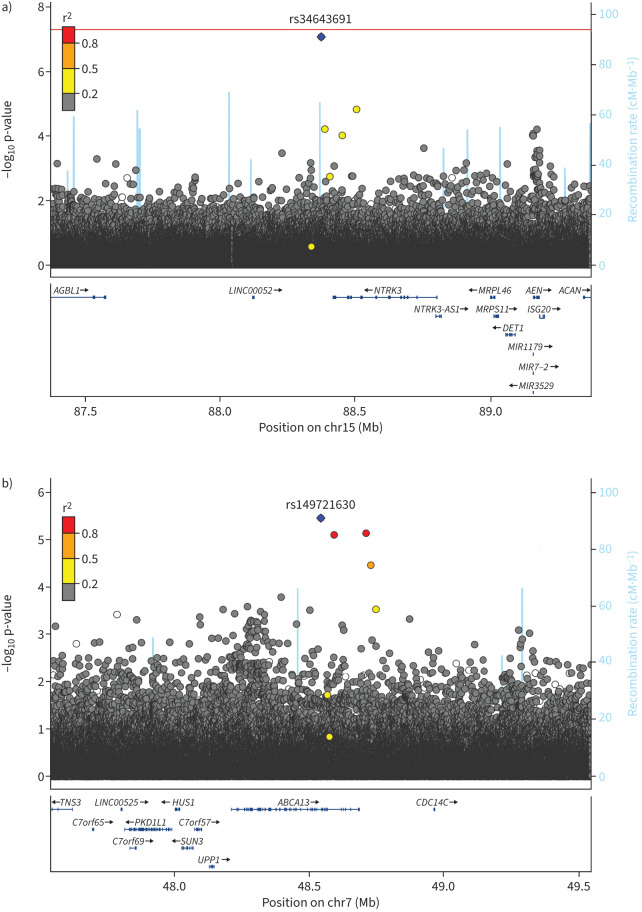
Fine-mapping (UK Biobank)
Figure 2 represents regional association plots for regions (±500 kb) centred at the two replicated SNPs. SNP rs34643691 is in an intergenic region close to Neurotrophic Receptor Tyrosine Kinase 3 (NTRK3) and did not show strong linkage disequilibrium with other SNPs in the region. SNP rs149721630 is an intronic variant within ATP Binding Cassette Subfamily A Member 13 (ABCA13) and is in moderate–high linkage disequilibrium with three other nearby SNPs. Fine-mapping results showed both signals to be the most likely causal variant in their region (supplementary figure S2).
FIGURE 2.
Region plots of replicated signals a) rs34643691 and b) rs149721630 results in discovery. Plot is produced in LocusZoom. Blue diamond: sentinel single nucleotide polymorphism. Genes are shown in the bottom panel in blue, with thicker sections representing exons. Mb: megabase; cM: centimorgan.
SNP by eosinophilia effect on exacerbations
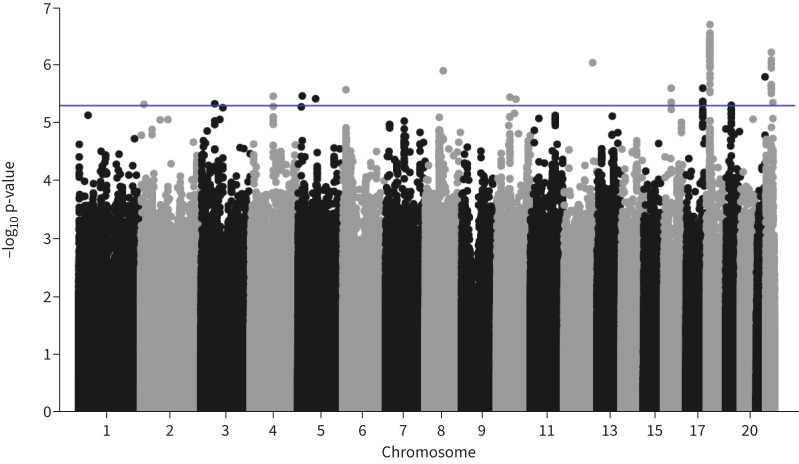
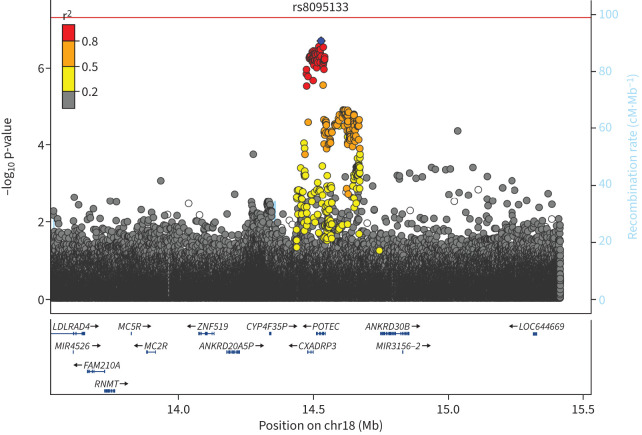
As two genomic loci reproducibly associated with risk of exacerbations in all participants with asthma were identified, we aimed to explore potential endotype-specific effects. To do this, we performed a genome-wide interaction analysis to test for interactions between eosinophilia (indicative of a type 2-high endotype) and variants on exacerbation status. This analysis showed no genome-wide significant associations and 86 signals that were associated with a gene by eosinophilia effect on exacerbations at p<5×10−6 (figure 3, supplementary figure S3). The top hit (rs8095133) comprised a locus on chromosome 18 overlapping two genes (CXADRP3 and POTEC), as shown in the regional association plot (figure 4). The minor allele of the sentinel variant increased exacerbation risk in eosinophilic patients (OR=1.17, p=0.0001) and decreased exacerbation risk in non-eosinophilic patients (OR=0.90, p=4.83×10−5).
FIGURE 3.
Manhattan plot of single nucleotide polymorphisms by eosinophilia interaction results for asthma exacerbations in the UK Biobank. The x-axis shows genomic location by chromosome, the y-axis shows the –log10 p-value. The blue line indicates p=5×10−6.
FIGURE 4.
Genomic region of the chromosome 18 locus showing a significant interaction effect on exacerbation risk. The variants in strong LD are mainly associated with two genes, CXADRP3 and POTEC. The top single nucleotide polymorphism (SNP) (lowest p-value) in the regional analysis is rs8095133(POTEC). Blue diamond: sentinel SNP; red: r2≥0.8; orange: r2 between 0.5 and 0.8; yellow: r2 between 0.2 and 0.5; grey: r2<0.2. Genes are shown in the bottom panel in blue, with thicker sections representing exons. Mb: megabase; cM: centimorgan.
Annotation and functional analyses
Using Haploreg and Ensembl Variant Effect Predictor, we annotated the top hits and identified potential functional consequences of SNPs in both steps of the analyses [33]. GWAS catalogue analyses of the replicated associations as well as the top hit in the interaction analysis did not reveal any previous significant genome-wide associations with asthma. Additionally, the three SNPs did not show an eQTL effect on gene expression levels in the Genotype-Tissue Expression (GTEx) database [42]. Conversely, in the lung eQTL consortium results, the rs149721630 T allele decreased PKD1L1 expression in lung cells (3.085×10−6). Analyses of KEGG and Reactome databases using the annotated genes for the list of suggestive hits, showed no significant associations beyond the false discovery rate, although potential involvement in neutrophil degranulation and immune system pathways was shown (supplementary file 2).
Post hoc power analysis
Aiming to confirm that we had enough power to detect significant associations, we conducted a post hoc power analysis. The number of cases and controls included in our study (11 604 and 37 890, respectively) allows for the detection of variants with a minimum relative risk of 1.2 and an allele frequency of at least 0.05, with >80% power at a 5×10−8 significance level.
Discussion
In this large, genome-wide association study, two novel loci were reproducibly associated with exacerbation risk in individuals with asthma. Exacerbations were identified using three different sources: hospitalisation data, general practitioner records and OCS bursts prescriptions. In an interaction analysis aiming to explore effect modification of eosinophilia on the association between genetic variants and risk of asthma exacerbations, the top locus showed opposite directions of effect on exacerbation risk in eosinophilic versus non-eosinophilic patients.
Two SNPs affected exacerbation status in our primary analysis: rs149721630 and rs34643691. Rs149721630 is an intron variant annotated to the ABCA13 gene, a member of the ATP-binding cassette (ABC) family of transmembrane transporters [43]. ABC is a family of conserved transporters involved in transporting different substrates and consisting of seven subfamilies [43, 44]. Members of the family have been previously associated with several lung conditions [44, 45]. ABCA13 is expressed in the lungs and its expression levels in epithelial cells have been shown to be affected by asthma and smoking [43]. Additionally, decreased expression levels were associated with a trend towards increased asthma severity [43]. Moreover, differential methylation in the region was also associated with rhinovirus-induced wheezing and asthma [46]. Importantly, we have shown that rs149721630 is associated with PKD1L1 expression in lung tissue. PKD1L1 has previously been associated with asthma–COPD overlap in African Americans [47]. It is also a component in cilia motility [48], which affects asthma severity [49]. This suggests that this variant may be associated with asthma exacerbations through an effect on PKD1L1 expression. Finally, ABCA13 is associated with psychiatric disorders, including depression, a condition which shares potential links (including genetic) with asthma [50, 51]. This variant could indicate a potential link between this genetic region and asthma severity through an effect on impaired lipid transport, inflammation or cilia motility [51, 52]. Further investigations may be needed to determine its role in asthma exacerbations. Conversely, rs34643691 lies in an intergenic region, and has not been previously associated to either regulatory elements or phenotypes. The closest gene is NTRK3, previously shown to be involved in the neurotrophin signalling pathway, which may impact phosphatidyl inositol signalling via phosphatidylinositide 3-kinase (PI3K), affecting zileuton (a leukotriene receptor antagonist)-related changes in FEV1 [53].
To investigate the hypothesis that there is effect modification by inflammatory subtype, we conducted an analysis using an interaction term (SNP × eosinophilia). Our interaction analysis identified a suggestive signal on chromosome 18, associated with CXDARP3 and POTEC genes. CXDARP3 is a pseudogene for the CXDAR (Coxsackie Virus And Adenovirus Receptor) gene, and both are expressed in oesophageal mucosal tissues [54]. This indicates a potential link between exacerbations in type 2 asthma and viral respiratory infections. Viral respiratory infections are well known to be one of the most important triggers of asthma exacerbations, and adenoviruses can trigger wheezing and predispose to allergy [55, 56]. Importantly, viral infections and their interaction with allergens not only increase the risk of exacerbations, but also trigger an increased Th2 response [57]. Conversely, POTEC belongs to a multi-gene family encoding Cancer-Testis Antigens and is highly expressed in both ovaries and testes [54, 58]. Importantly, the SNPs identified in the primary analysis were not significant in the interaction analysis, suggesting that the effects of these genetic risk variants are not modified by background eosinophilia.
Our study has several unique strengths: first, we conducted and replicated our GWAS of asthma exacerbations within adults, whereas most previous studies have been conducted in children. Second, we focused on the potential differences between genetic variants affecting the general risk of asthma exacerbations and variants affecting exacerbations modified by type 2 asthma (defined as patients with a blood eosinophil level ≥300 cells·μL−1). This is a unique approach which allows unravelling variants affecting exacerbations in individuals with specific active inflammatory pathways. Finally, we used a broad definition of exacerbations, by including data from various sources in both analysis stages. We included severe exacerbations (hospitalisations) using several definitions, aiming for a wider coverage of hospitalisations associated with asthma. Hospitalisations (in individuals with asthma) recorded as primarily due to influenza, respiratory failure, stridor or wheezing were considered asthma exacerbations. Hospitalisations associated with chest pain or dyspnoea in non-cardiac individuals with asthma were also considered exacerbations. Additionally, we added exacerbations in the community using prescription data and general practitioner records. This broad definition of asthma exacerbations aimed to provide a more accurate picture of asthma exacerbations by including all asthma exacerbators. Using this approach, 23.5% of our asthma population had at least one exacerbation, a figure close to previous estimates indicating the potential usefulness of the broader definition [5]. As most exacerbators were identified based on OCS prescriptions, we carried out sensitivity analyses to show that these variants had consistent magnitude and direction of effect in those identified from diagnostic ICD-10 and primary care read codes alone.
However, our study also had several limitations. First of all, there were no genome-wide significant associations in our primary analysis of exacerbation risk, or in our interaction analysis, within adult asthmatic individuals. This could be in part due to power, especially as most of our top hits had a relatively low minor allele frequency, and heterogeneity of the cases (which were defined using a variety of data sources) [59]. Second, there could be three main criticisms to our definition of asthma exacerbations. 1) We defined all chest pain/dyspnoea in asthmatic non-cardiac patients admitted to a hospital as exacerbations. We aimed to include important signs of exacerbations which would be otherwise missed due to the strict coding system, as these symptoms may be confusing to clinicians on a patient's initial presentation and are common in acute asthma [60]. 2) UK Biobank secondary care data do not include emergency department admissions unless those patients are transferred as inpatients to another department. We will therefore have missed any such exacerbations. 3) We used OCS burst prescriptions to identify asthma exacerbations in the community setting, which could be difficult to fully ascertain as patients may have had additional comorbidities for which they were prescribed OCS at the same dose (also as the available data on the intended duration of the specific prescription were missing). Alternatively, clinicians might have used lower doses intended for exacerbations than the dose threshold we used. Both broad definitions could have misclassified some patients as exacerbators or controls. However, the moderate percentage of exacerbators within our population as well as the results of sensitivity analyses suggest that any potential misclassification had limited effects on our final results. Third, we did not include follow-up time in our model. In the UK Biobank, hospitalisation data were available from 1995 up to 2020, while primary care data (for approximately half the cohort) were available from 1995 until 2016, which could have caused differences in follow-up and therefore the ability to record an exacerbation. Fourth, we did not replicate the interaction analysis assessing the effects of eosinophilia on the association between SNPs and asthma exacerbations, as blood eosinophil levels were not available in the GERA cohort. Fifth, our definition of eosinophilic patients was only based on blood eosinophil levels taken during any of three visits to the UK Biobank assessment centre. Airway eosinophilia could have provided a more accurate definition of eosinophilic patients, but this is less feasible in population-wide studies [61]. Moreover, eosinophilia in participants with less visits to the centres could have been underestimated. Finally, our analysis was restricted to individuals of European ancestry and therefore our results may not be generalisable to other populations.
Understanding the complex factors affecting the severity of asthma is essential to advance the understanding and treatment of asthma. Our study adds to the existing evidence of genetic involvement in the severity of asthma and the presenting clinical phenotype. Further work to translate findings and understand exactly how these variations are contributing towards different inflammatory pathways is required. Additional layers of data (including epigenetic or proteomics data) may be needed to unravel the biologically complex network of interactions, aiming to identify disease markers and/or drug targets.
In conclusion, our GWAS in the UK Biobank identified two variants that are associated with risk of exacerbations, highlighting a potentially important effect for ABCA13 and/or PKD1L1. Additionally, we identified a locus near CXADRP3 and POTEC genes associated with increased exacerbation risk in eosinophilic patients and decreased risk in non-eosinophilic patients. These findings could shed light on important pathways involved in asthma severity.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary figures and tables 00566-2023.SUPPLEMENT (263.3KB, pdf)
Supplementary file 1 00566-2023.SUPPLEMENT (18.4KB, xlsx)
Supplementary file 2 00566-2023.SUPPLEMENT2 (3.3KB, csv)
Asthma readcodes 00566-2023.SUPPLEMENT3 (5.6KB, csv)
Asthma readcodes 2 00566-2023.SUPPLEMENT (3.6KB, csv)
Acknowledgement
We would like to thank the participants in the included studies. Part of the data presented in this manuscript is from the Lung eQTL Consortium. We thank the Publication Committee for granting access to the data, including Wim Timens and Maarten van den Berge from University of Groningen (Groningen, The Netherlands), Don D. Sin and Tillie-Louise Hackett from University of British Columbia (Vancouver, BC, Canada), Ke Hao and David Nickle previously from Merck & Co. Inc., and Yohan Bossé and Philippe Joubert from Laval University (Quebec City, QC, Canada).
Provenance: Submitted article, peer reviewed.
Conflict of interest: M. Tobin received funding from Orion Pharma and GSK, outside the submitted work.
Conflict of interest: L. Lahousse received consulting from AstraZeneca, and honoraria from IPSA vzw and Chiesi, all outside the submitted work; and is a leading member of the European and Belgian Respiratory Societies.
Conflict of interest: A. Edris, K. Voorhies, A.C. Wu, S.M. Lutz, I. Hall, C. Iribarren and K. Fawcett declare no conflict of interest.
Support statement: This work was supported by a Wellcome Trust Institutional Strategic Support Fund (204801/Z/16/Z) and Wellcome Trust Investigator Award (WT202849/Z/16/Z to M. Tobin). M. Tobin holds a National Institute for Health and Care Research Senior Investigator Award. The research was partially supported by the National Institute for Health and Care Research Leicester Biomedical Research Centre; views expressed are those of the authors and not necessarily those of the National Health Service, the National Institute for Health and Care Research or the Department of Health. This work was partly funded by the National Institutes of Health grants R01 HD085993 and NIMH R01MH129337. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. K. Fawcett is supported by an Asthma+Lung Fellowship (AUK-CDA-2019–414). Funding information for this article has been deposited with the Crossref Funder Registry.
Ethics statement: The GERA study was approved by the institutional review boards at KPNC and Brigham and Women's Hospital (2002P000331). UK Biobank analysis was performed under approved application 648.
References
- 1.Boulet LP, Reddel H, Bateman E, et al. The Global Initiative for Asthma (GINA): 25 years later. Eur Respir J 2019; 54: 1900598. doi: 10.1183/13993003.00598-2019 [DOI] [PubMed] [Google Scholar]
- 2.Hernandez-Pacheco N, Pino-Yanes M, Flores C. Genomic predictors of asthma phenotypes and treatment response. Front Pediatr 2019; 7: 6. doi: 10.3389/fped.2019.00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wenzel SE. Emergence of biomolecular pathways to define novel asthma phenotypes. Type-2 immunity and beyond. Am J Respir Cell Mol Biol 2016; 55: 1–4. doi: 10.1165/rcmb.2016-0141PS [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fahy JV. Type 2 inflammation in asthma: present in most, absent in many. Nat Rev Immunol 2015; 15: 57–65. doi: 10.1038/nri3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunican EM, Fahy JV. The role of type 2 inflammation in the pathogenesis of asthma exacerbations. Ann Am Thorac Soc 2015; 12: Suppl. 2, S144–S149. doi: 10.1513/AnnalsATS.201506-377AW [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Georas SN, Rezaee F. Epithelial barrier function: at the front line of asthma immunology and allergic airway inflammation. J Allergy Clin Immunol 2014; 134: 509–520. doi: 10.1016/j.jaci.2014.05.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuruvilla ME, Lee FE, Lee GB. Understanding asthma phenotypes, endotypes, and mechanisms of disease. Clin Rev Allergy Immunol 2019; 56: 219–233. doi: 10.1007/s12016-018-8712-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tyler SR, Bunyavanich S. Leveraging -omics for asthma endotyping. J Allergy Clin Immunol 2019; 144: 13–23. doi: 10.1016/j.jaci.2019.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park HW, Tantisira KG. Genetic signatures of asthma exacerbation. Allergy Asthma Immunol Res 2017; 9: 191–199. doi: 10.4168/aair.2017.9.3.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyers DA, Bleecker ER, Holloway JW, et al. Asthma genetics and personalised medicine. Lancet Respir Med 2014; 2: 405–415. doi: 10.1016/S2213-2600(14)70012-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ivanova JI, Bergman R, Birnbaum HG, et al. Effect of asthma exacerbations on health care costs among asthmatic patients with moderate and severe persistent asthma. J Allergy Clin Immunol 2012; 129: 1229–1235. doi: 10.1016/j.jaci.2012.01.039 [DOI] [PubMed] [Google Scholar]
- 12.Castillo JR, Peters SP, Busse WW. Asthma exacerbations: pathogenesis, prevention, and treatment. J Allergy Clin Immunol Pract 2017; 5: 918–927. doi: 10.1016/j.jaip.2017.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reddel HK, Taylor DR, Bateman ED, et al. An official American Thoracic Society/European Respiratory Society statement: asthma control and exacerbations: standardizing endpoints for clinical asthma trials and clinical practice. Am J Respir Crit Care Med 2009; 180: 59–99. doi: 10.1164/rccm.200801-060ST [DOI] [PubMed] [Google Scholar]
- 14.Jorgensen IM, Jensen VB, Bulow S, et al. Asthma mortality in the Danish child population: risk factors and causes of asthma death. Pediatr Pulmonol 2003; 36: 142–147. doi: 10.1002/ppul.10305 [DOI] [PubMed] [Google Scholar]
- 15.Lloyd A, Price D, Brown R. The impact of asthma exacerbations on health-related quality of life in moderate to severe asthma patients in the UK. Prim Care Respir J 2007; 16: 22–27. doi: 10.3132/pcrj.2007.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuhlbrigge A, Peden D, Apter AJ, et al. Asthma outcomes: exacerbations. J Allergy Clin Immunol 2012; 129: Suppl. 3, S34–S48. doi: 10.1016/j.jaci.2011.12.983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bai TR, Vonk JM, Postma DS, et al. Severe exacerbations predict excess lung function decline in asthma. Eur Respir J 2007; 30: 452–456. doi: 10.1183/09031936.00165106 [DOI] [PubMed] [Google Scholar]
- 18.Rogers AJ, Tantisira KG, Fuhlbrigge AL, et al. Predictors of poor response during asthma therapy differ with definition of outcome. Pharmacogenomics 2009; 10: 1231–1242. doi: 10.2217/pgs.09.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang AL, Gruzieva O, Qiu W, et al. DNA methylation is associated with inhaled corticosteroid response in persistent childhood asthmatics. Clin Exp Allergy 2019; 49: 1225–1234. doi: 10.1111/cea.13447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrera-Luis E, Ortega VE, Ampleford EJ, et al. Multi-ancestry genome-wide association study of asthma exacerbations. Pediatr Allergy Immunol 2022; 33: e13802. doi: 10.1111/pai.13802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Son JH, Park JS, Lee JU, et al. A genome-wide association study on frequent exacerbation of asthma depending on smoking status. Respir Med 2022; 199: 106877. doi: 10.1016/j.rmed.2022.106877 [DOI] [PubMed] [Google Scholar]
- 22.Yan Q, Forno E, Herrera-Luis E, et al. A genome-wide association study of asthma hospitalizations in adults. J Allergy Clin Immunol 2021; 147: 933–940. doi: 10.1016/j.jaci.2020.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kvale MN, Hesselson S, Hoffmann TJ, et al. Genotyping informatics and quality control for 100,000 subjects in the genetic epidemiology research on adult health and aging (GERA) cohort. Genetics 2015; 200: 1051–1060. doi: 10.1534/genetics.115.178905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukherjee M, Gupta R, Farr A, et al. Estimating the incidence, prevalence and true cost of asthma in the UK: secondary analysis of national stand-alone and linked databases in England, Northern Ireland, Scotland and Wales: a study protocol. BMJ Open 2014; 4: e006647. doi: 10.1136/bmjopen-2014-006647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shrine N, Portelli MA, John C, et al. Moderate-to-severe asthma in individuals of European ancestry: a genome-wide association study. Lancet Respir Med 2019; 7: 20–34. doi: 10.1016/S2213-2600(18)30389-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.John C, Guyatt AL, Shrine N, et al. Genetic associations and architecture of asthma–chronic obstructive pulmonary disease overlap. Chest 2022; 161: 1155–1166. [DOI] [PMC free article] [PubMed]
- 27.Manichaikul A, Mychaleckyj JC, Rich SS, et al. Robust relationship inference in genome-wide association studies. Bioinformatics 2010; 26: 2867–2873. doi: 10.1093/bioinformatics/btq559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shah SA, Quint JK, Nwaru BI, et al. Impact of COVID-19 national lockdown on asthma exacerbations: interrupted time-series analysis of English primary care data. Thorax 2021; 76: 860–866. doi: 10.1136/thoraxjnl-2020-216512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGeachie MJ, Wang AL, Lutz SM, et al. Real-life patterns of exacerbations while on inhaled corticosteroids and long-acting beta agonists for asthma over 15 years. J Clin Med 2020; 9: 819. doi: 10.3390/jcm9030819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edris A, de Roos EW, McGeachie MJ, et al. Pharmacogenetics of inhaled corticosteroids and exacerbation risk in adults with asthma. Clin Exp Allergy 2021; 52: 33–45. doi: 10.1111/cea.13829 [DOI] [PubMed] [Google Scholar]
- 31.Wain LV, Shrine N, Miller S, et al. Novel insights into the genetics of smoking behaviour, lung function, and chronic obstructive pulmonary disease (UK BiLEVE): a genetic association study in UK Biobank. Lancet Respir Med 2015; 3: 769–781. doi: 10.1016/S2213-2600(15)00283-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang CC, Chow CC, Tellier LC, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015; 4: 7. doi: 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLaren W, Gil L, Hunt SE, et al. The Ensembl Variant Effect Predictor. Genome Biol 2016; 17: 122. doi: 10.1186/s13059-016-0974-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dayem Ullah AZ, Oscanoa J, Wang J, et al. SNPnexus: assessing the functional relevance of genetic variation to facilitate the promise of precision medicine. Nucleic Acids Res 2018; 46: W109–WW13. doi: 10.1093/nar/gky399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci 2019; 28: 1947–1951. doi: 10.1002/pro.3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanehisa M, Furumichi M, Sato Y, et al. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res 2023; 51: D587–D592. doi: 10.1093/nar/gkac963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000; 28: 27–30. doi: 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gillespie M, Jassal B, Stephan R, et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res 2022; 50: D687–D692. doi: 10.1093/nar/gkab1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hao K, Bosse Y, Nickle DC, et al. Lung eQTLs to help reveal the molecular underpinnings of asthma. PLoS Genet 2012; 8: e1003029. doi: 10.1371/journal.pgen.1003029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang G, Sarkar A, Carbonetto P, et al. A simple new approach to variable selection in regression, with application to genetic fine mapping. J R Stat Soc B 2020; 82: 1273–1300. doi: 10.1111/rssb.12388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 2010; 26: 2336–2337. doi: 10.1093/bioinformatics/btq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013; 45: 580–585. doi: 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aguiar JA, Tamminga A, Lobb B, et al. The impact of cigarette smoke exposure, COPD, or asthma status on ABC transporter gene expression in human airway epithelial cells. Sci Rep 2019; 9: 153. doi: 10.1038/s41598-018-36248-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Deen M, de Vries EG, Timens W, et al. ATP-binding cassette (ABC) transporters in normal and pathological lung. Respir Res 2005; 6: 59. doi: 10.1186/1465-9921-6-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kroner C, Wittmann T, Reu S, et al. Lung disease caused by ABCA3 mutations. Thorax 2017; 72: 213–220. doi: 10.1136/thoraxjnl-2016-208649 [DOI] [PubMed] [Google Scholar]
- 46.Lund RJ, Osmala M, Malonzo M, et al. Atopic asthma after rhinovirus-induced wheezing is associated with DNA methylation change in the SMAD3 gene promoter. Allergy 2018; 73: 1735–1740. doi: 10.1111/all.13473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hardin M, Cho M, McDonald ML, et al. The clinical and genetic features of COPD–asthma overlap syndrome. Eur Respir J 2014; 44: 341–350. doi: 10.1183/09031936.00216013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grimes DT, Keynton JL, Buenavista MT, et al. Genetic analysis reveals a hierarchy of interactions between polycystin-encoding genes and genes controlling cilia function during left-right determination. PLoS Genet 2016; 12: e1006070. doi: 10.1371/journal.pgen.1006070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas B, Rutman A, Hirst RA, et al. Ciliary dysfunction and ultrastructural abnormalities are features of severe asthma. J Allergy Clin Immunol 2010; 126: 722–729.e2. doi: 10.1016/j.jaci.2010.05.046 [DOI] [PubMed] [Google Scholar]
- 50.Zhu Z, Zhu X, Liu CL, et al. Shared genetics of asthma and mental health disorders: a large-scale genome-wide cross-trait analysis. Eur Respir J 2019; 54: 1901507. doi: 10.1183/13993003.01507-2019 [DOI] [PubMed] [Google Scholar]
- 51.Nakato M, Shiranaga N, Tomioka M, et al. ABCA13 dysfunction associated with psychiatric disorders causes impaired cholesterol trafficking. J Biol Chem 2021; 296: 100166. doi: 10.1074/jbc.RA120.015997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chai AB, Ammit AJ, Gelissen IC. Examining the role of ABC lipid transporters in pulmonary lipid homeostasis and inflammation. Respir Res 2017; 18: 41. doi: 10.1186/s12931-017-0526-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dahlin A, Qiu W, Litonjua AA, et al. The phosphatidylinositide 3-kinase (PI3 K) signaling pathway is a determinant of zileuton response in adults with asthma. Pharmacogenomics J 2018; 18: 665–677. doi: 10.1038/s41397-017-0006-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carithers LJ, Ardlie K, Barcus M, et al. A novel approach to high-quality postmortem tissue procurement: the GTEx Project. Biopreserv Biobank 2015; 13: 311–319. doi: 10.1089/bio.2015.0032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jartti T, Bonnelykke K, Elenius V, et al. Role of viruses in asthma. Semin Immunopathol 2020; 42: 61–74. doi: 10.1007/s00281-020-00781-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aydin M, Naumova EA, Paulsen F, et al. House dust mite exposure causes increased susceptibility of nasal epithelial cells to adenovirus infection. Viruses 2020; 12: 1151. doi: 10.3390/v12101151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edwards MR, Strong K, Cameron A, et al. Viral infections in allergy and immunology: how allergic inflammation influences viral infections and illness. J Allergy Clin Immunol 2017; 140: 909–920. doi: 10.1016/j.jaci.2017.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barger CJ, Zhang W, Sharma A, et al. Expression of the POTE gene family in human ovarian cancer. Sci Rep 2018; 8: 17136. doi: 10.1038/s41598-018-35567-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brookes ST, Whitely E, Egger M, et al. Subgroup analyses in randomized trials: risks of subgroup-specific analyses; power and sample size for the interaction test. J Clin Epidemiol 2004; 57: 229–236. doi: 10.1016/j.jclinepi.2003.08.009 [DOI] [PubMed] [Google Scholar]
- 60.Refaat S, Aref H. Acute asthma in emergency department, prevalence of respiratory and non-respiratory symptoms. Egypt J Chest Dis Tuberc 2014; 63: 771–776. doi: 10.1016/j.ejcdt.2014.07.013 [DOI] [Google Scholar]
- 61.Pignatti P, Visca D, Cherubino F, et al. Do blood eosinophils strictly reflect airway inflammation in COPD? Comparison with asthmatic patients. Respir Res 2019; 20: 145. doi: 10.1186/s12931-019-1111-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary figures and tables 00566-2023.SUPPLEMENT (263.3KB, pdf)
Supplementary file 1 00566-2023.SUPPLEMENT (18.4KB, xlsx)
Supplementary file 2 00566-2023.SUPPLEMENT2 (3.3KB, csv)
Asthma readcodes 00566-2023.SUPPLEMENT3 (5.6KB, csv)
Asthma readcodes 2 00566-2023.SUPPLEMENT (3.6KB, csv)