Abstract
The contribution of the microbiota to disease progression and treatment efficacy is often neglected when determining who is at the highest risk of developing gastrointestinal cancers or designing treatment strategies for patients. We reviewed the current literature on the effect of the human microbiota on cancer risk, prognosis, and treatment efficacy. We highlight emerging research that seeks to identify microbial signatures as biomarkers for various gastrointestinal cancers, and discuss how we could harness knowledge of the microbiome to detect, prevent, and treat these cancers. Finally, we outline further research needed in the field of gastrointestinal cancers and the microbiota, and describe the efforts required to increase the accuracy and reproducibility of data linking the microbiome to cancer.
Introduction
Roughly 10–20% of new cancer cases are attributable to carcinogenic infections. This percentage is projected to increase as—through new cancer microbiome research—researchers start to better understand the role of tumour-associated microbes in cancer initiation and progression.1 Cancerous tissue is enriched with specific bacterial communities from the human microbiota, suggesting that such bacteria could be a crucial but often overlooked component of the tumour microenvironment.2–6 Within the tumour microenvironment malignant cells are surrounded by a diverse collection of cell types (figure 1). Cell distribution might be heterogenous across a tumour, whereby different cell types contribute to microniches. The blood vessels integrated within the tumour provide oxygen and nutrients, and the interior of these vessels is lined with endothelial cells. The periphery of the tumour interacts with cancer-associated fibroblasts, which play a role in remodelling the extracellular matrix, a complex network of macromolecules. In addition to varying levels and types of immune infiltrates, tumours harbour an intrinsic microbiota that can also be distributed heterogeneously throughout the tissue or as biofilms on the tissue surface at mucosal sites.
Figure 1:
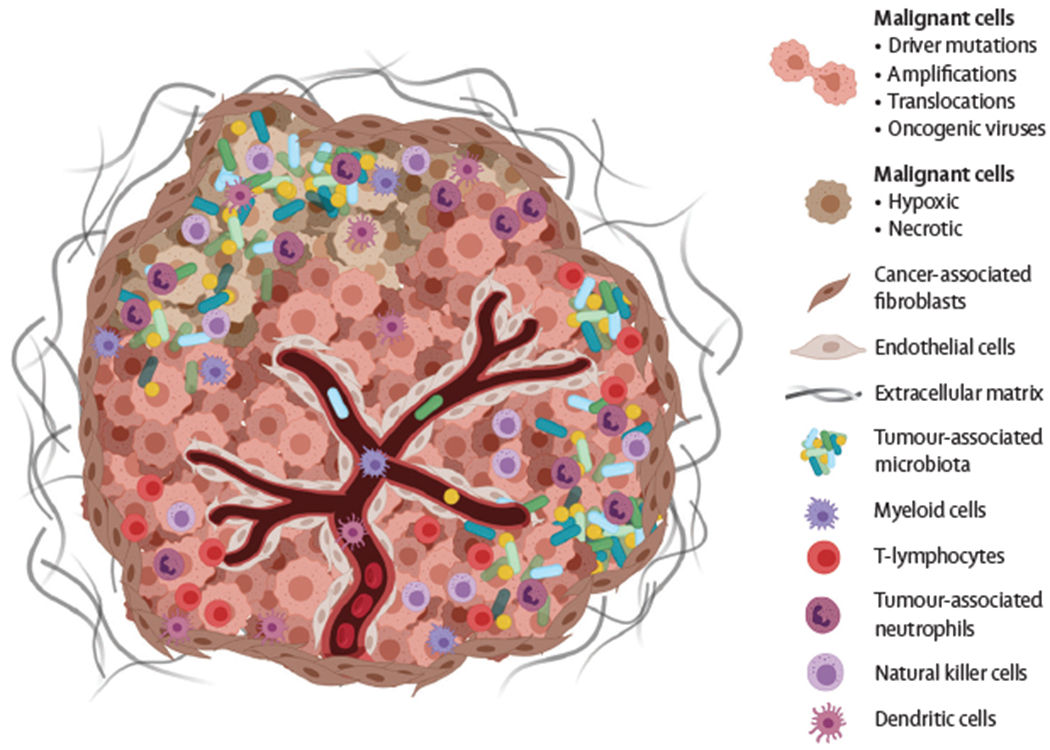
The tumour microenvironment
The human microbiota includes commensal, symbiotic, and pathogenic microorganisms that are found within each body site (eg, skin, oral cavity, and gastrointestinal tract), with the different human—environmental niches harbouring a unique microbial consortium.7 The human body has approximately the same number of human cells as bacterial cells (a ratio of 1:1·3).7 However, the combined genetic material of these bacteria, collectively termed the human microbiome, contains more than 150 times the coding capacity of the human genome.7,8 Most of our bacterial microbiota resides in the gastrointestinal tract, with the highest density in the colorectum, where there can be approximately 1011 bacterial cells per mL of colon content.7 In the colorectum, bacteria are closely associated with the intestinal mucosa and can influence inflammation, immune responses, and the metabolism of drugs or dietary compounds.9,10 The microbiota’s load and composition are influenced by a range of factors including diet, lifestyle, genetics, demographics, and medications.11–13
Since the early 2010s, numerous studies have demonstrated microbial dysbiosis (substantial changes in the diversity and composition of the microbiota) in the gut or specific tissues of people with cancer. At the interface of cancer and microbiome research is an emerging effort to understand the contribution of microbial dysbiosis to cancer initiation, progression, and response to treatment, and its potential for risk prediction. Although this area of cancer research is still very much in its infancy, and many studies to date are correlative, understanding the contribution of the microbiota in human health and disease such as cancer has enormous translational potential. Individuals have the ability to influence the composition of their microbiota, and there is the potential to manipulate it in such a way to mitigate cancer risk, improve cancer treatment efficacy, and replenish protective bacteria lost during dysbiotic states. A deeper understanding of the composition of a healthy and protective microbiota is needed to develop these processes, and to recognise harmful species that exacerbate disease risk.
The microbiota consists of a range of organisms that can impact our gastrointestinal health (bacterial, fungal, viral, and parasitic), however this Review will only focus on the contribution of bacteria to gastrointestinal cancers. In this Review, we highlight key studies that demonstrate the role that bacterial members of the microbiota play in gastrointestinal cancer risk, prognosis, and patient response to treatment. We will also suggest how this field of research could be used to develop bacterial surveillance and modulation treatments, to improve outcomes for patients with cancer.
Microbiome signatures associated with cancer risk and prognosis
Overview
Analyses of tissue samples from a range of gastrointestinal cancers reveal substantial alterations in microbial community structure within tumours and the broader gut microbiome.14 Microbial dysbiosis occurs when there is a change in a microbial community that results in an imbalance of protective and potentially harmful bacteria.15 These alterations often coincide with a loss of microbial diversity, which presents a unique opportunity for otherwise typically rare bacterial community members to thrive and dominate within the tumour tissue or entire gut microbiota.15 Microbial dysbiosis and the growth of specific bacterial species in gastrointestinal cancers has been documented by several studies since 2012. In this section, we will discuss emerging areas of research relating to the assessment of cancer risk and prognosis through analysis of the composition and diversity of microbial communities, or microbial signatures.
Enrichment of specific bacterial species in colorectal cancer
The microbiota of the gastrointestinal tract, and particularly the colorectum, have a well-characterised density and diversity, and so most studies assessing the relationship between microbial dysbiosis and cancer risk or prognosis have focused on colorectal tissue or stool microbiota.
In 2012, it was demonstrated that Fusobacterium nucleatum and other Fusobacterium species are intimately associated with colorectal cancer. This finding was unexpected at the time, because these organisms were commonly viewed as oral pathobionts. Other studies went on to analyse the microbial species composition in over 90 pairs of colorectal tumours and adjacent normal tissue from patients with colorectal cancer.2,3 In addition to the increased prevalence of F nucleatum in colorectal cancer tissue, two such studies showed that this organism was a dominant member of the colorectal cancer microbiota (in some cases reaching 89% of the total bacterial DNA sequence reads2), and exhibited a 79-fold increase in abundance in cancer versus normal tissue.3 A group at the Dana-Farber Cancer Institute analysed two large US prospective studies, the Nurses’ Health Study and the Health Professionals Follow-up Study, and demonstrated that F nucleatum-positivity in colorectal cancer tissue increased along the colon, from the distal to the proximal area. Strikingly, the study found that patients who were harbouring comparatively high levels of F nucleatum in their tumours exhibited poorer survival than those with F nucleatum-negative or F nucleatum-low levels.16,17 Research from our group demonstrated, through microbial culture, microbiome, and imaging analyses, that the association between Fusobacterium and colorectal cancer persists in distant site metastases to the liver.18 Furthermore, many liver metastases share their dominant tumour microbiota with their paired primary cancer.18 Although we have consistent evidence of this bacterium’s enrichment in primary and metastatic colorectal cancer, we do not have a clear picture as to when Fusobacterium species colonise the colorectum and if it occurs prior to tumour formation. A 2013 study reported the detection of Fusobacterium in 48% of adenomas and reported that Fusobacterium was enriched significantly more in the adenoma tissue compared with the surrounding tissue (n=29; p<0·004).19 More recently, in 2020, analyses of adenomas from patients with Lynch syndrome showed that F nucleatum was a minor component of the adenoma microbiota and was not detected in stool of patients with adenomas.20 It has been demonstrated across multiple cohorts that Fusobacterium is detectable in the stool of patients with colorectal cancer,17,21–25 therefore, there is the possibility that faecal microbiome analyses could be used to detect Fusobacterium as a non-invasive screening tool for colorectal cancer. Although Fusobacterium was consistently found in relative abundance in tissue and stools of patients with colorectal cancer, other taxa were found to be substantially—but inconsistently—altered between studies. However, it is possible that such inconsistencies could be due to differences in parameters including geographical location of cohorts, the methods used for analyses, medication such as antibiotic use, and disease severity.
In addition to F nucleatum, two other organisms that are consistently associated with colorectal cancer include enterotoxigenic Bacteroides fragilis (ETBF) and polyketide synthase positive (pks+) Escherichia coli. 26–30 Colibactin is a genotoxin produced by pks+ E coli that induces DNA damage through the alkylation of adenine residues,31–36 and the B fragilis toxin produced by ETBF is a zinc-dependent metalloprotease shown to activate a range of pathways resulting in increased proinflammatory interleukin-8 and c-Myc oncogene expression. A 2018 study demonstrated that both ETBF and pks+ E coli are significantly enriched in the colonic mucosa of patients with familial adenomatous polyposis, a hereditary condition caused by germline mutations in the APC tumour suppressor gene.4 Individuals with this disease develop benign polyps that often progress to colorectal cancer. However, this study demonstrated that ETBF and pks+ E coli colonise, form biofilms, and potentially induce DNA damage in the colonic mucosa of patients with this condition prior to polyp formation. The prevalence of ETBF is also significantly increased in the stool of patients with colorectal cancer, and one study found ETBF in 21 (38%) of 56 patients with colorectal cancer compared with five (12%) in 40 healthy controls (p=0·009).37 A separate study demonstrated significant enrichment of pks+ E coli in mucosa of patients with colorectal cancer, in which 40% of patients with colorectal cancer harboured pks+ E coli compared with 21% of controls.5 This finding suggests that pks+ E coli is associated with colorectal cancer and could affect carcinogenesis.
In a 2020 study, human intestinal organoids were exposed to pks+ E coli through repeated luminal injection followed by whole-genome sequencing, revealing that pks+ E coli induced distinct mutational signatures in human epithelial cells.38 Analysis of genomic data from two independent European cohorts identified a predominance of pks+ E coli mutational signatures in colorectal cancer tissue samples.38 This study provides evidence that a bacterial member of the microbiota can directly contribute to the mutational process; a process that is likely to start early in life. Follow-up studies are warranted, in particular to analyse pks+ mutational signatures in tumours from individuals with familial adenomatous polyposis. Overall, these findings suggest that individuals who harbour specific bacterial strains might possibly be at risk of colorectal cancer with a poor disease prognosis. Later in this Review, we will discuss the potential for targeted microbiome modulation to circumvent such risk and potentially improve prognosis.
It is important to emphasise that, while many studies to date have taken a reductionist approach by focusing on the role of specific bacterial species or strains that are enriched in colorectal cancer tissue, it is also likely that the microbial community as a consortium could contribute to colorectal cancer. For example, there might be bacterial community members that interact synergistically to promote cancer, or alternatively the loss of protective community members could have the same effect. One emerging area of research, related to these hypotheses, is the role of bacterial biofilms in cancer development and prognosis. However, this area of research has been hindered by technical challenges in preserving biofilms in colorectal cancer specimens. For example, formalin fixation of colorectal cancer tissue might strip the biofilm-associated mucus layer from specimens,39,40 and, while Carnoy’s fixative preserves the mucus layer,39 its use can result in lower sensitivity and therefore poorer fluorescent imaging quality of the tissue.40 Current research aims to develop new methods and polymers for preservation of biofilms in tissue samples.40,41
Alterations of microbiome signatures in gastrointestinal cancers
Most gastrointestinal cancers exhibit changes in microbial signatures (a term used to describe the composition and diversity of the microbial community), including in body sites that are considered to have low microbial load. Despite the emerging associations between members of the microbiota and a range of cancers, the bacterium Helicobacter pylori remains the only bacterium considered to be a class I carcinogen by the WHO. Chronic infection by H pylori is one of the greatest risk factors for gastric cancer development.42 Colonisation by H pylori can trigger a proinflammatory cytokine response,43,44 in addition to secretion of virulence factors that are thought to contribute to cancer initiation by inducing DNA damage and increasing cell proliferation and motility.45,46 Even with these potential oncogenic mechanisms, only 1–3% of individuals colonised by H pylori go on to develop gastric cancer, implying the importance of other underlying factors in gastric cancer tumorigenesis.39,40 Once the gastric tumour forms, there is no significant difference in H pylori presence between tumour and adjacent normal tissue.47,48 Several studies have documented broad bacterial dysbiosis in gastric cancer tissue compared with normal tissue, which suggests that bacterial communities might act together to promote gastric cancer progression.49–51 There was, however, little consistency in the enrichment or depletion of specific bacterial taxa between these studies, highlighting the need for larger population-based analyses to overcome individual and geographical microbiota sampling bias.
Distinct microbial signatures have also been reported in gastrointestinal cancers outside of the gastrointestinal tract, including cancers of the pancreas, gallbladder, and bile duct. Although the pancreas was once considered a sterile organ due to the high alkalinity and proteolytic activity of pancreatic juice,52 patients with pancreatic ductal adenocarcinoma exhibit a 1000-fold increase in intratumoural bacteria compared with normal tissue.52,53 Pancreatic tumour tissue harboured a higher relative abundance of Proteobacteria compared with healthy controls, and enrichment of this bacterial phylum was correlated with advanced disease stage.53 There is sparse microbiome data in the literature on gallbladder and biliary tract cancers; however, two meta-analyses have found that asymptomatic carriage of Salmonella typhi and Helicobacter spp is correlated with an elevated risk of gallbladder cancer54 and biliary tract cancer,55 respectively. Microbiome analyses of bile from patients with gallbladder cancer identified the presence of bacteria in 42·9% of samples.56
How such bacterial communities travel to these tissue sites remains an outstanding question. Many gastrointestinal cancers are enriched with bacterial taxa traditionally considered to be oral microbiota, suggesting that these organisms migrate from the mouth to distal sites. In colorectal cancer, it has been suggested that F nucleatum enters the body from the oral cavity and travels to the tumour site through the haematogenous route.57,58 Furthermore, studies of oral samples from colorectal cancer,59 oesophageal,60 and pancreatic cancers61,62 suggest that the oral microbiome composition could be an accurate indicator of cancer risk. Periodontitis and poor oral health are linked to increased risk for multiple gastrointestinal cancers.63–65 While these findings are promising, larger cohort studies are needed.
Blood microbiome signatures as an indicator of cancer risk
The identification of reliable bacterial biomarkers associated with cancer risk, reoccurrence, response to therapy, and patient survival could aid in the stratification of patients for appropriate treatment regimes. A 2020 study demonstrated the potential for blood-based microbial signatures in cancer diagnostics; microbial reads were parsed from published whole-genome and polyA-selected RNA sequencing data from tumour, tumour-adjacent normal tissue, and blood specimens representing 33 cancer types in The Cancer Genome Atlas project (18 116 samples).48 Analyses from this study computationally predicted and subtracted microbial contamination, and detected unique microbial signatures within major cancer types. The study identified microbiome signatures shared between tumour and blood specimens in the project to predict cancers such as pancreatic adenocarcinoma, oesophageal carcinoma, rectum adenocarcinoma, and cholangiocarcinoma. PolyA-selected RNA is not ideal for bacterial analyses because bacterial transcripts lack a polyA tail,66 and so the approach of this study was validated using independent blood-based microbial DNA, derived from plasma, to distinguish signatures from healthy individuals (n=69) and several different cancer types (n=100).48 The use of cell-free DNA for cancer diagnostics is an emerging concept in the field. The collection of this type of DNA is far less invasive than normal gastrointestinal tissue sampling procedures, and it facilitates earlier detection of cancer.67,68 Data supporting the prognostic benefit of blood-based microbial signatures are preliminary but, given the potential for improved early detection and patient outcome, further independent studies to validate these early findings are warranted.
The effect of microbiota compositions on patient prognosis
The presence of specific bacterial populations within patient tumour tissue has been reported to be associated with poor prognosis and outcomes. One US study of 1069 patients found that, during a median follow-up of 10·7 years, the amount of F nucleatum in colorectal carcinoma tissues directly correlated with patient mortality (HR 1·58 in F nucleatum-high cases vs 1·25 in F nucleatum-low cases).17 In this same study, increased loads of F nucleatum were associated with proximal tumour location and high microsatellite instability (MSI-H). These findings were corroborated by another study of F nucleatum burden in colorectal cancer tissues versus adjacent normal tissues (n=190) in which tissues with high F nucleatum load were associated with poorer overall survival (HR 1·68) and MSI-H patient status.
In oesophageal cancer, the presence of either F nucleatum or Porphyromonas gingivalis in tumour tissue has been associated with shorter survival time. In one study, resected oesophageal cancer specimens (n=325) contained significantly greater loads of F nucleatum DNA than did matched normal oesophageal mucosa. F nucleatum was associated with a higher disease stage; stage III cancers showed a F nucleatum positivity rate of 30% versus 15% positivity in stage I cancers. F nucleatum was also associated with shorter survival, with a hazard ratio of 2· 01 (p=0·0068).69 A second study detected P gingivalis in 61% of cancerous tissues, and found that it was undetectable in normal oesophageal mucosa (n=30).70 Even after a relatively short follow-up of 30 months, the mean survival of patients who harboured P gingivalis was 20·1 months versus 25·9 months in the negative group. Both studies show that two predominantly oral microbes, F nucleatum and P gingivalis, are invasive in cancerous tissue and could be risk factors for poor patient prognosis.
By contrast, the presence of other bacterial populations within patient tumour tissue has been reported to be associated with better prognosis and outcomes. A study of tumour microbiota composition in long-term survival (>5 years post-surgery) versus short-term survival (≤5 years post-surgery) in patients with pancreatic ductal adenocarcinoma found significantly increased species diversity in samples of patients with long-term survival compared with those with short-term survival, using stage-matched tissues.71 Patients with pancreatic ductal adenocarcinoma survive for an average of 2·5 years following surgical resection, and only 9% live to 5 years.72 However, a subset of patients survive more than 5 years after diagnosis. Tissue samples of patients with long-term survival had a unique microbial signature that was predictive of long-term survival, and these microbiomes correlated with strong immune infiltration within tumours. Additionally, faecal microbiome transfer from patients who had predicted long-term survival of this disease into a pancreatic mouse model reduced tumour size compared with transfers from patients with predicted short-term survival (p<0·001).71 These findings suggest that gut bacteria of patients with long-term survival pancreatic ductal adenocarcinoma might have a protective effect against tumour proliferation.
When does bacterial dysbiosis begin?
Research is focused on understanding when microbial changes occur during cancer development and whether cancer-associated microbial dysbiosis can be used to assess cancer risk and predict patient prognosis. Two studies catalogued microbial communities within the human gut mucosa at different stages of colorectal cancer tumorigenesis.6,73 These analyses revealed that the abundance of oral microbes increased along the adenoma-to-carcinoma sequence,6,73 with one study validating the metacommunity markers in two additional patient cohorts.6 A separate study characterised the microbiome in early colonic neoplasia through 16S rRNA gene sequencing of aberrant crypt foci and the adjacent normal mucosa within the proximal colon.74 They found that particular microbial signatures within aberrant crypt foci correlated with the presence of polyps and dysbiosis of the colonic mucosa.
A gastric cancer study characterised microbial changes within the gastric mucosa of patients presenting with disease states associated with tumorigenesis.49 In agreement with other studies, this analysis confirmed significant mucosa microbial dysbiosis in patients with gastric cancer.50 Although it is well established that gastric colonisation with H pylori is associated with chronic gastritis and gastric cancer development, H pylori might not be a dominant community member in the gastric mucosa or tumour microbiota of patients once they develop gastric cancer.48,50,75,76 Instead, subsequent inflammation and pH neutralisation within the gastric microenvironment could create a niche for opportunistic microbes to colonise and progress into late-stage disease.
While these studies in colorectal and gastric cancers hint that microbial dysbiosis might occur early in cancer development, these types of investigations are cross-sectional studies that analyse the microbiome at the time of tumour resection. As such, there are no clear data profiling changes in the microbiome during the transition from healthy tissue to malignant lesions.
Direct and indirect effects of the microbiota on cancer treatment efficacy
Overview
Since the early 2010s, there has been a rapidly growing body of research functionally linking the human microbiota to virtually all aspects of human physiology. As a result, researchers have a deeper understanding of how bacterial dysbiosis and alterations in host disease states can contribute to increased severity of human illness. Additionally, this research has revealed that bacteria are in direct contact with the gastrointestinal epithelial layer and are invasive in many gastrointestinal cancer tissues (see table). The proximity of microbial communities, which often surround or exist within neoplastic tissue, to human tumours suggests that bacterial cells are exposed to chemotherapeutic and immunological therapies in real time, alongside local cancer and immune cells. Indeed, it has already been shown that members of the microbiota can metabolise, activate, and inactivate many common drugs,77,78 but their impact on cancer treatment efficacy has received insufficient attention until recently.79,80 The impact of the microbiota on cancer therapies can be separated by their contribution to chemotherapeutic efficacy and modulation of the immune response and, in this section, we will highlight key studies demonstrating both direct and indirect interactions between the microbiota and standard cancer therapeutics that ultimately affect patient treatment response (panel 1).
Table:
Microbial signatures related to gastrointestinal cancer risk and prognosis
| Tissue | Cancer-associated bacteria | |
|---|---|---|
| Oesophagus | Cancerous tissue | Species: Fusobacterium nucleatum,69 Porphyromonas gingivalis73 |
| Stomach | Cancerous tissue | Phylum: Actinobacteria,50
Firmicutes,50
Proteobacteria50 Family: Enterobacteriaceae,50 Pasteurellaceae,51 Xanthomonadaceae,50 Streptococcaceae76 Genus: Achromobacter,50 Bacillus,51 Bifidobacterium,51 Citrobacter,50 Clostridium,50 Fusobacteria,51 Lactococcus,50,51 Leptotrichia,51 Methylobacterium,51 Phyllobacterium,50 Rodococcus,50 Staphylococcus,51 Veillonella 51 Species: Dialister pneumosintes,49,51 Helicobacter pylori,76 Parvimonas micra,49 Peptostreptococcus stomatis,49 Slackia exigua,49 Streptococcus anginosus49 |
| Colon and rectum | Cancerous tissue | Genus: Fusobacterium,6,18
Gemella,6
Granulicatella,6
Parvimonas,6
Peptostreptococcus6 Species: Bacteroides fragilis,6 Escherichia coli,5 F nucleatum2,3,6,19 |
| Colon and rectum | Faeces | Family: Enterococcaceae,23,24
Lachnospiraceae,24,73
Veillonellaceae22 Genus: Alistipes,23,73 Anaerococcus,2 Atopobium,22 Bacteroides,23,24,73 Blautia,23 Campylobacter,23 Clostridium,25 Dorea,23 Escherichia,23 Eubacterium,23 Fastidiosipila,23 Fusobacterium,22–24 Gemella,23 Holdermania,23 Leptotrichia,23 Odoribacter,23,73 Parabacteroides,23,73 Parvimonas,23 Peptostreptococcus,23,25 Phascolarctobacterium,23,24 Porphyromonas,22,24 Pseudomonas, Ruminococcus,23,25 Subdoligranulum23 Species: Alistipes putredinis,73 B fragilis,25 Bilophila wadsworthia,73 E coli,73 Eubacterium spp,25 Clostridium symbiosum,25,73 F nucleatum,25 P stomatis,25 Porphyromonas asaccharolytica,25 Streptococcus salivarius25 |
| Pancreas | Cancerous tissue | Class: Bacteroidea,71
Clostridia,71
Spingobacteriia53 Order: Corynebacteriales,71 Pseudomonadales71 |
| Gallbladder | Bile | Species: Enterobacter spp,56 E coli,56 F nucleatum,56 Salmonella enterica serotype Typhi54 |
| Bile duct | Cancerous tissue | Species: Helicobacter spp55 |
Data are from research studies mentioned in this Review. Only data from patient samples are included (microbial composition from mouse studies were excluded). Not all studies found the same microbial signatures, and they differed in their level of taxonomic resolution.
Panel 1: The effects of microbiota on chemotherapy and the immune response.
Chemotherapy
Directly inactivate or modify drugs (eg, Gammaproteobacteria metabolises gemcitabine81)
Modify tumour cell pathways to promote resistance (eg, F nucleatum induce autophagy82)
Promote chemotherapeutic efficacy (eg, bacteria contribute to the toxic effect of cyclophosphamide,83 and CpG oligonucleotides and oxaliplatin84)
Immune response
Improve efficacy of immune checkpoint inhibitors (eg, anti-PD-1 therapy85–88 and anti-CTLA-4 therapy89)
Aid in anti-cancer immune activation (eg, by stimulating Th1 cells,83 and activating TLR4 and reactive oxygen species84)
Favour immunosupressive environment (eg, bacteria associated with inactivation of natural killer cells,58 reduction in T-cell infiltration,16 and increases in tumour-associated macrophages19)
CpG oligonucleotides=cytosine-phosphorothioate-guanine oligodeoxynucleotides. Th1=T helper type 1. TLR4=Toll-like receptor 4.
Chemotherapy
The human microbiota is metabolically active, while many standard cancer chemotherapeutics are antimetabolites. As such, there is inherent potential for antagonism between the microbiota and chemotherapeutic agents. For example, tumour-associated microbiota might directly affect chemotherapeutic efficacy through chemical or enzymatic modification and inactivation of a drug. One study demonstrated that 86 (76%) of 113 pancreatic cancer tissue samples taken at the time of surgery harboured bacterial communities dominated by Gammaproteobacteria, compared with three (15%) of 20 samples from human pancreas obtained from organ donors (p<0·005).81 The researchers showed via mass spectrometry that Gammaproteobacteria members can metabolise gemcitabine into its inactive form 2′,2′-difluorodeoxyuridine, and confirmed that this reaction contributed to chemoresistance in their mouse model.81 Co-administration of ciprofloxacin, a Gammaproteobacteria-inhibiting antibiotic, along with gemcitabine, prevented chemoresistance in their model. This study suggests that intratumoural bacteria might lower the regional concentration of chemotherapeutics and contribute to chemoresistance.
The gut microbiota can also indirectly augment the activity of several chemotherapeutics by modulating the immune structure of the tumour microenvironment. To date, three different cancer therapeutics have shown reduced efficacy in tumour-bearing mice that lacked a microbiota (either through antibiotic treatment or germ-free derivation): cyclophosphamide, CpG oligonucleotides, and oxaliplatin. Cyclophosphamide is an alkylating agent that stimulates immunogenic cancer cell death through subversion of immunosuppressive T cells and promotes T-helper type 1 and 17 (Th1 and Th17) cell response. Cyclophosphamide increases intestinal permeability, allowing for the translocation of specific microbiota members across the epithelial layer.83 These bacteria are responsible for the stimulation of Th17 cells and Th1 cells in local lymph nodes, resulting in a systemic antitumour response. The bacterial contribution to cyclophosphamide’s toxic effect is lost after treatment with antibiotics, or if the same treatment is performed in germ-free mice. A similar effect was seen in an independent study on the role of the microbiota in CpG oligonucleotide and oxaliplatin treatment efficacy.84 In CpG oligonucleotide and oxaliplatin treatment of tumour-bearing mice, the commensal microbiota triggered TLR4 inflammatory responses and the production of reactive oxygen species by tumour-infiltrating myeloid cells. These responses promoted anti-cancer toxicity. The findings from both studies suggest that there could be unintended risks associated with broad-spectrum antibiotic use during cancer treatment.
Additionally, F nucleatum species were found to indirectly promote resistance to the chemotherapeutics oxaliplatin and fluorouracil through the induction of autophagy.82 One study suggested that F nucleatum activation of TLR4/MYD88 results in downstream inhibition of specific microRNA responsible for the repression of genes that regulate autophagy. Independent validation through multiple cohort analysis determined that F nucleatum abundance in patient tissue strongly correlates with colorectal cancer recurrence and lower survival probability.82 These findings point to the potential benefit of stratifying patients with colorectal cancer based on F nucleatum load, and administering a combined treatment of fluorouracil or oxaliplatin chemotherapy with F nucleatum growth inhibitors. Work from our group demonstrated that F nucleatum and co-occurring anaerobic bacteria persist and remain viable in patient-derived xenografts of colorectal cancer.18 When xenografts from patients with colon cancer are treated with the antibiotic metronidazole, tumour growth, cancer cell proliferation, and tumour Fusobacterium load are all significantly reduced. This finding suggests there could be a role for antibiotics as an adjunct to standard chemotherapy.18 Ideally, a narrow-spectrum inhibitor is required to prevent unwanted antimicrobial activity against beneficial members of the microbiota.
Microbiota and the immune system
Immune checkpoint blockade therapies remove inhibitory signals of T-cell activation and are effective in treating a subset of patients with particular cancer types.91,92 These therapies mainly target PD-1 and CTLA-4 blockades in patients.92,93 Numerous studies, although none so far on gastrointestinal cancers, have established the role of the gut microbiota in promoting the efficacy of anti-PD-1 therapy.85–88 These studies find that the microbiota is required for optimal immune response to treatment, and in particular, the gut commensals Bifidobacterium spp, Collinsella aerofaciens, Enterococcus faecium, Akkermansia muciniphila, and Ruminoccaceae are associated with responder-phenotypes. In anti-CTLA-4 therapy, specific priming of T-cell responses by Bacteroides spp was determined to contribute to antitumor effects.93 By contrast, a separate study found that patients with melanoma with a baseline faecal microbiota that was enriched for Faecalibacterium seemed to have prolonged survival compared with patients with a Bacteroides-enriched microbiota (p=0·051). However, the patient cohort was relatively small and so the research warrants follow-up analyses.94 It is important to note that immune checkpoint inhibitors are not effective treatment for most gastrointestinal cancers, although there has been some success in the treatment of MSI-H tumours.90,91 Therefore, it is critical to understand resistance mechanisms to immunotherapy in these cancers.
Members of the intratumoural microbiota have also been shown to play a role in immunosuppression. For example, F nucleatum was associated with a 7·8-fold increase in tumour-associated macrophages in a colorectal cancer murine model,19 and a lower infiltration of CD3+ T cells in colorectal cancer tissue.16,95 Furthermore, F nucleatum has been shown to hinder components of the immune response by using its Fap2 adhesin to bind inhibitory TIGIT receptors (present on a subset of T cells and on natural killer cells).58 The binding of TIGIT results in the loss of natural killer cell cytotoxicity and T-cell activity, and leads to immune suppression. Conversely, the composition of the extratumoural microbiota has also been shown to affect antitumour immune response. Secondary bile acid metabolism by the gut microbiome results in downstream activation of the natural killer T-cell ligand CXCL16 in liver tissue, promoting antitumour immune responses in liver cancer.96 A 2020 paper revealed that within a variety of tumours, including pancreatic cancer, both cancer and immune cells harboured intracellular bacteria.97 Yet, it is currently unclear how these interactions affect antigen presentation, immune cell infiltration, and immune cell activation.
Harnessing the microbiota in cancer treatment
It is becoming apparent that the structure and function of the human microbiota are intricately associated with gastrointestinal cancer risk, patient prognosis, and treatment outcomes. The evidence evolving from recent literature suggests that a patient’s microbiota should be considered when designing optimal treatment regimens.79,98 In the next decade, research should seek to accurately identify microbial signatures that reproducibly associate with an elevated risk of gastrointestinal cancer, and to develop new technologies to modulate the microbiota and potentiate clinical outcomes for patients. In this section, we propose areas for future studies and discuss current research that supports the benefit microbiota modulation has on patient outcomes (figure 2). We envision that the microbiota could aid in the prediction, prevention, and treatment of gastrointestinal cancers. Prediction could be aided by longitudinal studies monitoring the faecal and oral microbiome, diet, and lifestyle in large, diverse patient cohorts, which could help define reproducible microbiome signatures for patient cancer risk and prognosis. Microbiota modulation could be used to prevent pathogenic or cancer supportive microbial community establishment in patients with an elevated risk of cancer development, in addition to preliminary dietary and lifestyle changes. Lastly, treatment could be positively affected; once a patient is diagnosed with cancer, direct microbiota modification could be used to remove oncogenic and drug metabolising bacteria or deliver bacterial species that improve treatment response. These methods are not mutually exclusive, and a patient could benefit from considering the microbiota at all stages of cancer development.
Figure 2:
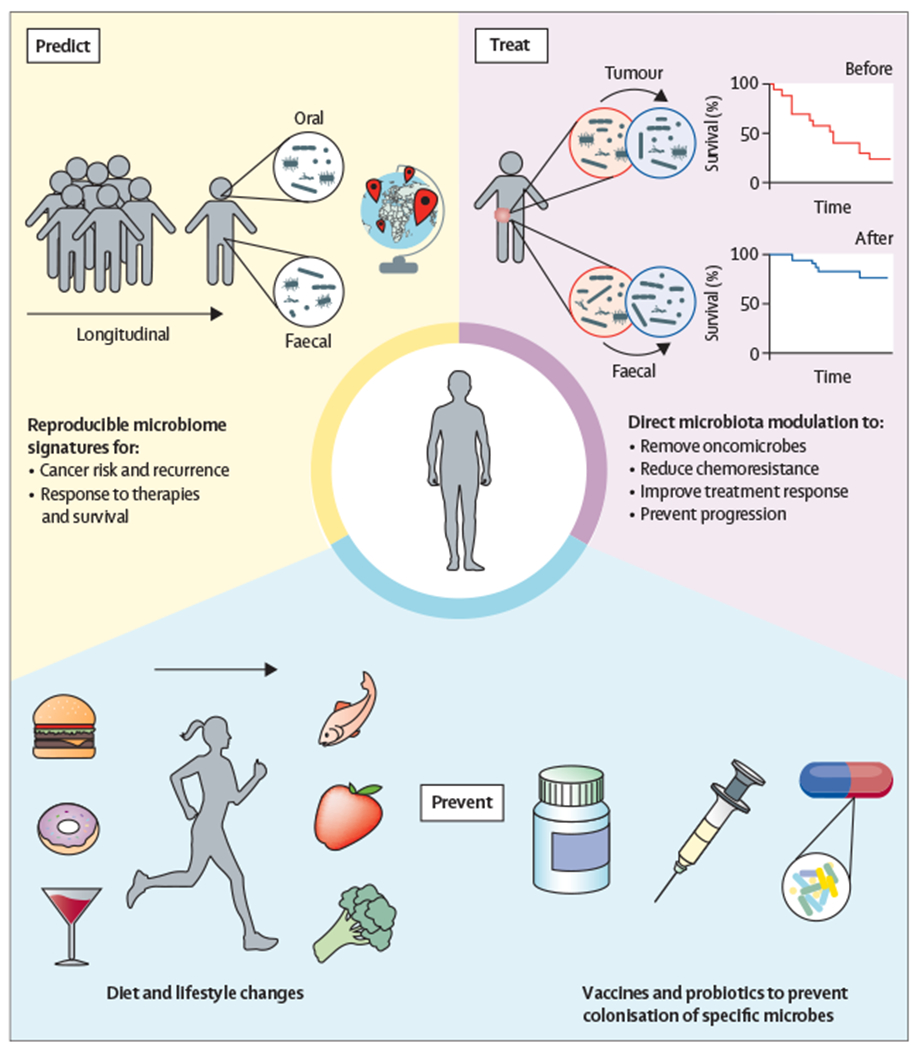
Potential roles for the microbiome in cancer prediction, prevention, and treatment
Microbiota modulation
Prevention
To manipulate the microbiota for cancer prevention, we first need to determine if, and how, the microbiota is contributing to cancer. Most examinations of the microbiota and cancer remain correlative; however, as we build a catalogue of the tumour-associated microbiota, we can move away from relatively straightforward characterisation studies towards functional analyses that demonstrate causation. Detailed elucidation of how microbiota contribute to cancer will reveal viable microbial targets for prophylactic and therapeutic intervention.
Ideally, reduction of microbe-associated risk would occur before the diagnosis of cancer. This reduction could be achieved by altering the composition, and therefore functional capacity, of our microbiota through dietary mitigation and lifestyle changes. Modifications in the diet have been shown to rapidly and reproducibly alter the structure of the gut microbiota.99–101 These studies support a link between animal-based diets with increased bile acids and dietary fat, and microbial dysbiosis leading to systemic inflammation.100,102 In genetically susceptible mice, a high-fat diet has been shown to promote tumour progression and a change in the gut microbiota.103 Tumour growth was attenuated by the addition of butyrate: a short-chain fatty acid derived from bacterial fermentation of complex carbohydrates. Faecal transplant from mice being fed a high-fat diet to mice being fed a normal diet resulted in tumorigenesis; an effect that could be reversed with antibiotic treatment.103 Therefore, it is reasonable to assume that modulation of the gut microbiota via dietary intervention could directly lower the risk of cancer development. The advantage of such approaches is their broad accessibility; however, success relies on patient adherence and it is unclear if dietary changes alone would sufficiently reduce cancer risk.
Vaccination against possibly oncogenic bacteria has the potential to modulate cancer risk permanently, analogous to approaches that circumvent infection with high-risk oncogenic viruses, for example, vaccinating against human papillomavirus to prevent cervical cancer. As previously discussed, the colonic mucosa of patients with familial adenomatous polyposis is enriched for genotoxic pks+ E coli and ETBF; individuals could be stratified by microbiome analyses for pks+ E coli dependent mutational signatures38 and strain positivity for a dual vaccine early in life. However, prophylactic vaccine development is challenging and the process of advancing a candidate through clinical trials can take many years and has a low success rate. For example, H pylori was classified as a class I carcinogen by the WHO in 1994, but despite gastric adenocarcinoma accounting for a third of cancer deaths globally, there is no current vaccine study past a phase 1 clinical trial.104 Substantial innovation is needed to develop effective and safe vaccines for oncogenic bacterial species, as well as studies that assess the physiological consequences of microbial species eradication.
Treatment
The commensal microbiota contributes to human health by providing a protective barrier against invading pathogens, and it is evident from a range of recent studies that particular microbiomes can positively contribute to antitumour immune responses. Therefore, future studies should seek to develop methods for microbial community modulation to potentiate treatment response and improve patient outcomes. Faecal microbiota transplantation has been successful in modulating the microbiota for non-malignant diseases.105 However, these treatments remain investigational, suffer from interindividual donor variation, and bear the inherent risk of pathogen contamination including enteropathogenic E coli and Shiga toxin-producing E coli;106 accordingly, faecal microbiota transplantations are not widely available in standard practice. Instead, the most effective treatment design might be the targeted modulation of specific keystone bacterial species associated with cancer progression and chemoresistance, or alternatively the tailored introduction of a well-defined beneficial bacterial consortium. Both approaches are scalable for widespread application; however, extensive research is needed to define optimal species to target or introduce and the stability of this modulation. Several studies have shed light on the impact of bacterial strain introduction on host immune physiology,107–109 yet few have demonstrated a direct functional capacity to influence cancer.
Promisingly, a recent study identified an 11-strain microbial consortium capable of stimulating a robust IFNγ producing CD8 T-cell response, independent of inflammation, in the gut and other organs.110 Remarkably, while colonisation of this microbial consortium in a syngeneic colon cancer mouse model enhanced the efficacy of anti-PD-1 therapy, it also significantly suppressed tumour growth.110 These 11 strains represent rare and low-abundance species within the human microbiota; therefore, they could become a broadly effective biotherapeutic. The results from this study are encouraging for the potential of microbiome modulation treatment alongside traditional therapy. There is a plethora of microbiota modulation-based clinical trials currently in progress for various cancers.79
Conclusion
Research over the past decade supports a role for bacterial members of the microbiota in gastrointestinal cancer risk, prognosis, and patient response to treatments. Nevertheless, the initial findings that we have highlighted are likely to be just the tip of the iceberg in terms of bacterial organisms’ contributions to cancer initiation and progression. Increased clinical and basic research efforts in this field, serial microbiome sampling of patients, and technological advances to overcome challenges with tissue metagenomics are now needed to delineate host-bacteria functional interactions in the context of cancer. To identify reliable microbial signatures predictive of cancer risk and patient prognosis, or to inform microbiome-based treatment strategies, we need standardised methodologies and uniform study design for longitudinal microbiome sampling across large and diverse patient populations. These approaches are likely to require a concerted effort on behalf of investigators and funding agencies, but they will be crucial to advance our understanding of the microbial landscape of sporadic versus hereditary cancers,111,112 the rise in incidence of early-onset colorectal cancer,113–116 and geographical cancer clusters117–121 (panel 2). We look forward with optimism to the future of this field, as it is evident from current research that there is broad potential for microbiome modulation to mitigate cancer risk and aid in treatment regimens to ultimately improve patient health outcomes.
Panel 2: Outstanding questions and next steps for the future of microbial research.
Longitudinal studies are required to assess cancer risk
To identify the microbial signatures and specimen types that are best suited to assess gastrointestinal cancer risk, faecal and oral specimens from established cohorts are needed to prospectively evaluate the association between the human microbiome and cancer risk. Serial sampling of these cohorts should be used to collect microbiota throughout participants’ lives, and metadata such as lifestyle and dietary information should also be collected. Coordinated global efforts adhering to the open science philosophy would maximise the identification of microbial signatures that are translatable to diverse patient populations and cancer types.
Does the cancer-associated microbiota differ in sporadic versus hereditary cancers?
Most cancers are sporadic in nature, and somatic gene damage can result from many environmental factors including ageing, radiation, chemical exposure, and interactions with bacteria within the microbiota.111,112 By contrast, hereditary cancers arise from pre-existing germline mutations, often in genes that control DNA repair, which predispose the individual to cancer development. Comparative in-depth microbiome profiling of cancerous tissues from both populations could provide valuable insight into bacterial species and pathways associated with cancer risk.
Is the microbiota involved in the increasing incidence of early-onset colorectal cancer?
The rate of early-onset (<50 years) sporadic colorectal cancer is increasing globally. 113–115 Early-onset disease occurs in 11% of colon and 18% of rectal cancers, with a trend toward manifestation in the distal colon.116 Screening for colorectal cancer typically begins after the age of 50 years, and early-onset colorectal cancer is often detected at a more advanced stage. Therefore, it is crucial to understand the risk factors for early-onset colorectal cancer and begin prophylactic screening at a younger age for at-risk groups. The differences between tumour-associated microbiota in patients with early-onset compared with those of a median age (68–72 years old) with the disease remain unexplored. Understanding whether the microbiota is associated with an increased risk of early-onset disease might contribute to the development of prognostic biomarkers, and pave the way for microbiome modulation to reduce risk.
Can geographical cancer clusters be explained by alterations in the microbiota?
Cancer clusters are defined by the US Centers for Disease Control and Prevention as a greater-than-expected number of cancer cases that occur within a group of people in a defined geographic area over a specific period of time. Many gastrointestinal cancers are found in a greater than expected number of cases in specific regions of the world. For example, there is a substantial increase in early-onset colorectal cancer and oesophageal squamous cell carcinoma in sub-Saharan Africa, but the risk factors for the development of these cancers in this population are unclear.117–120 Further research is needed to determine if the microbiome of cancerous tissue correlates with increased cancer incidence in these populations. Confoundingly, the microbiota composition is influenced by genetics, diet, lifestyle, and medical background: all parameters that can greatly differ depending on location, thus making the inclusion of geographical controls of importance. Such studies could be augmented by the application of geographical information systems in cancer surveillance and epidemiology and to uncover hotspots with higher resolution.121
How do we address the issue of reproducibility within this field?
One of the biggest hurdles to the development of reliable microbial biomarkers for cancer is the lack of reproducibility between studies. Numerous variables can affect the quality and composition of microbiome data, including population size, geographical location, patient sample types, methods for sample preparation, controls for contamination, diet, lifestyle, clinicopathological features. A portion of this bias or variability could be reduced by increasing the population size and including multiple cohorts from geographically diverse regions. To control for sample type and preparation, it would be useful to conduct more replication studies to validate promising microbial signatures.
Search strategy and selection criteria.
References for this Review were identified through searches on Google Scholar using the search terms: “microbiota and cancer”, “microbiome and cancer”, “oncogenic bacteria”, “bacteria and cancer treatment”, and “bacteria and immune response to cancer”. Studies on non-gastrointestinal related cancers were excluded unless the research appeared to have translational potential for gastrointestinal cancers. Research articles from Jan 1, 2010 to July 1, 2020, were prioritised unless the seminal research papers on a research topic were published before. Articles were also identified through searches of the authors’ own collection of files on the literature. The final reference list reflects articles that were deemed most relevant and up to date on the topics in this Review. Only articles published in English were reviewed.
Acknowledgments
KDL and SB are supported by the National Cancer Institute of the National Institutes of Health (NIH) R00 CA229984-03 and CDJ is supported by the National Institute of Dental and Craniofacial Research of the NIH and the NIH Office of the Director under NIH Director’s Transformative Research Award R01DE027850.
Declaration of interests
SB is a co-inventor on US Provisional Patent Application number 62/534 672, submitted by the Broad Institute and Dana-Farber Cancer Institute, that covers targeting of Fusobacterium for treatment of colorectal cancer. SB has consulted for X-Biotix Therapeutics and BiomX. SB is on the cancer program scientific advisory board for BiomX. KDL and SB are supported by the National Cancer Institute (NCI) of the National Institutes of Health (NIH) grant number R00 CA229984-03, and CDJ is supported by the National Institute of Dental and Craniofacial Research (NIDCR) of the NIH and the NIH Office of the Director under NIH Director’s Transformative Research Award R01DE027850.
References
- 1.Plummer M, de Martel C, Vignat J, Ferlay J, Bray F, Franceschi S. Global burden of cancers attributable to infections in 2012: a synthetic analysis. Lancet Glob Health 2016; 4: e609–16. [DOI] [PubMed] [Google Scholar]
- 2.Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 2012; 22: 292–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res 2012; 22: 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dejea CM, Fathi P, Craig JM, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 2018; 359: 592–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arthur JC, Perez-Chanona E, Mühlbauer M, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012; 338: 120–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakatsu G, Li X, Zhou H, et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun 2015; 6: 8727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 2016; 14: e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010; 464: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lepage P, Leclerc MC, Joossens M, et al. A metagenomic insight into our gut’s microbiome. Gut 2013; 62: 146–58. [DOI] [PubMed] [Google Scholar]
- 10.Hooper LV. Bacterial contributions to mammalian gut development. Trends Microbiol 2004; 12: 129–34. [DOI] [PubMed] [Google Scholar]
- 11.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 2006; 124: 837–48. [DOI] [PubMed] [Google Scholar]
- 12.Maier L, Pruteanu M, Kuhn M, et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 2018; 555: 623–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodrich JK, Waters JL, Poole AC, et al. Human genetics shape the gut microbiome. Cell 2014; 159: 789–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer 2013; 13: 800–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petersen C, Round JL. Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol 2014; 16: 1024–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mima K, Cao Y, Chan AT, et al. Fusobacterium nucleatum in colorectal carcinoma tissue according to tumor location. Clin Transl Gastroenterol 2016; 7: e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mima K, Nishihara R, Qian ZR, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 2016; 65: 1973–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bullman S, Pedamallu CS, Sicinska E, et al. Analysis of fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017; 358: 1443–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013; 14: 207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan Y, Drew DA, Markowitz A, et al. Structure of the mucosal and stool microbiome in lynch syndrome. Cell Host Microbe 2020; 27: 585–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Y, Weng W, Peng J, et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating Toll-like receptor 4 signaling to nuclear factor-κB, and up-regulating expression of microRNA-21. Gastroenterology 2017; 152: 851–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahn J, Sinha R, Pei Z, et al. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst 2013; 105: 1907–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu N, Yang X, Zhang R, et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol 2013; 66: 462–70. [DOI] [PubMed] [Google Scholar]
- 24.Zackular JP, Rogers MAM, Ruffin MT 4th, Schloss PD. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev Res (Phila) 2014; 7: 1112–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeller G, Tap J, Voigt AY, et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol 2014; 10: 766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boleij A, Hechenbleikner EM, Goodwin AC, et al. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis 2015; 60: 208–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Viljoen KS, Dakshinamurthy A, Goldberg P, Blackburn JM. Quantitative profiling of colorectal cancer-associated bacteria reveals associations between Fusobacterium spp., enterotoxigenic Bacteroides fragilis (ETBF) and clinicopathological features of colorectal cancer. PLoS One 2015; 10: e0119462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe 2014; 15: 317–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swidsinski A, Khilkin M, Kerjaschki D, et al. Association between intraepithelial Escherichia coli and colorectal cancer. Gastroenterology 1998; 115: 281–286. [DOI] [PubMed] [Google Scholar]
- 30.Buc E, Dubois D, Sauvanet P, et al. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS One 2013; 8: e56964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Putze J, Hennequin C, Nougayrède J-P, et al. Genetic structure and distribution of the colibactin genomic island among members of the family Enterobacteriaceae. Infect Immun 2009; 77: 4696–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nougayrède J-P, Homburg S, Taieb F, et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 2006; 313: 848–51. [DOI] [PubMed] [Google Scholar]
- 33.Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrède J-P. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci USA 2010; 107: 11537–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bossuet-Greif N, Vignard J, Taieb F, et al. The Colibactin genotoxin generates DNA interstrand cross-links in infected cells. MBio 2018; 9: e02393–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xue M, Kim CS, Healy AR, et al. Structure elucidation of colibactin and its DNA cross-links. Science 2019; 365: eaax2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson MR, Jiang Y, Villalta PW, et al. The human gut bacterial genotoxin colibactin alkylates DNA. Science 2019; 363: eaar7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toprak NU, Yagci A, Gulluoglu BM, et al. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin Microbiol Infect 2006; 12: 782–86. [DOI] [PubMed] [Google Scholar]
- 38.Pleguezuelos-Manzano C, Puschhof J, Rosendahl Huber A, et al. Mutational signature in colorectal cancer caused by genotoxic pks E. coli. Nature 2020; 580: 269–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johansson MEV, Hansson GC. Preservation of mucus in histological sections, immunostaining of mucins in fixed tissue, and localization of bacteria with FISH. In: McGuckin MA, Thornton DJ, eds. Mucins: Methods and Protocols. Totowa, NJ: Humana Press, 2012: 229–35. [DOI] [PubMed] [Google Scholar]
- 40.Macedonia MC, Drewes JL, Markham NO, et al. Clinically adaptable polymer enables simultaneous spatial analysis of colonic tissues and biofilms. NPJ Biofilms Microbiomes 2020; 6: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dassanayake RP, Falkenberg SM, Stasko JA, Shircliff AL, Lippolis JD, Briggs RE. Identification of a reliable fixative solution to preserve the complex architecture of bacterial biofilms for scanning electron microscopy evaluation. PLoS One 2020; 15: e0233973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nardone G, Compare D. The human gastric microbiota: is it time to rethink the pathogenesis of stomach diseases? United European Gastroenterol J 2015; 3: 255–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brandt S, Kwok T, Hartig R, König W, Backert S. NF-kappaB activation and potentiation of proinflammatory responses by the Helicobacter pylori CagA protein. Proc Natl Acad Sci USA 2005; 102: 9300–05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamb A, Chen L-F. Role of the Helicobacter pylori-induced inflammatory response in the development of gastric cancer. J Cell Biochem 2013; 114: 491–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chaturvedi R, Asim M, Romero-Gallo J, et al. Spermine oxidase mediates the gastric cancer risk associated with Helicobacter pylori CagA. Gastroenterology 2011; 141: 1696–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hatakeyama M Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat Rev Cancer 2004; 4: 688–94. [DOI] [PubMed] [Google Scholar]
- 47.Bass AJ, Thorsson V, Shmulevich I, et al. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014; 513: 202–09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poore GD, Kopylova E, Zhu Q, et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020; 579: 567–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coker OO, Dai Z, Nie Y, et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut 2018; 67: 1024–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, et al. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut 2018; 67: 226–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Castaño-Rodríguez N, Goh K-L, Fock KM, Mitchell HM, Kaakoush NO. Dysbiosis of the microbiome in gastric carcinogenesis. Sci Rep 2017; 7: 15957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei M-Y, Shi S, Liang C, et al. The microbiota and microbiome in pancreatic cancer: more influential than expected. Mol Cancer 2019; 18: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pushalkar S, Hundeyin M, Daley D, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov 2018; 8: 403–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagaraja V, Eslick GD. Systematic review with meta-analysis: the relationship between chronic Salmonella typhi carrier status and gall-bladder cancer. Aliment Pharmacol Ther 2014; 39: 745–50. [DOI] [PubMed] [Google Scholar]
- 55.Zhou D, Wang JD, Weng MZ, et al. Infections of Helicobacter spp. in the biliary system are associated with biliary tract cancer: a meta-analysis. Eur J Gastroenterol Hepatol 2013; 25: 447–54. [DOI] [PubMed] [Google Scholar]
- 56.Tsuchiya Y, Loza E, Villa-Gomez G, et al. Metagenomics of microbial communities in gallbladder bile from patients with gallbladder cancer or cholelithiasis. Asian Pac J Cancer Prev 2018; 19: 961–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abed J, Emgård JEM, Zamir G, et al. Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed gal-GalNAc. Cell Host Microbe 2016; 20: 215–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gur C, Ibrahim Y, Isaacson B, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 2015; 42: 344–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Flemer B, Warren RD, Barrett MP, et al. The oral microbiota in colorectal cancer is distinctive and predictive. Gut 2018; 67: 1454–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peters BA, Wu J, Pei Z, et al. Oral microbiome composition reflects prospective risk for esophageal cancers. Cancer Res 2017; 77: 6777–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Michaud DS, Izard J. Microbiota, oral microbiome, and pancreatic cancer. Cancer J 2014; 20: 203–06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fan X, Alekseyenko AV, Wu J, et al. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut 2018; 67: 120–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nwizu NN, Marshall JR, Moysich K, et al. Periodontal disease and incident cancer risk among postmenopausal women: results from the women’s health initiative observational cohort. Cancer Epidemiol Biomarkers Prev 2017; 26: 1255–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Michaud DS, Lu J, Peacock-Villada AY, et al. Periodontal disease assessed using clinical dental measurements and cancer risk in the ARIC study. J Natl Cancer Inst 2018; 110: 843–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hajishengallis G Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol 2015; 15: 30–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dreyfus M, Régnier P. The poly(A) tail of mRNAs: bodyguard in eukaryotes, scavenger in bacteria. Cell 2002; 111: 611–13. [DOI] [PubMed] [Google Scholar]
- 67.Corcoran RB, Chabner BA. Application of cell-free DNA analysis to cancer treatment. N Engl J Med 2018; 379: 1754–65. [DOI] [PubMed] [Google Scholar]
- 68.Fiala C, Diamandis EP. New approaches for detecting cancer with circulating cell-free DNA. BMC Med 2019; 17: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yamamura K, Baba Y, Nakagawa S, et al. Human microbiome Fusobacterium nucleatum in esophageal cancer tissue is associated with prognosis. Clin Cancer Res 2016; 22: 5574–81. [DOI] [PubMed] [Google Scholar]
- 70.Gao S, Li S, Ma Z, et al. Presence of Porphyromonas gingivalis in esophagus and its association with the clinicopathological characteristics and survival in patients with esophageal cancer. Infect Agent Cancer 2016; 11: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Riquelme E, Zhang Y, Zhang L, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell 2019; 178: 795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018; 68: 7–30. [DOI] [PubMed] [Google Scholar]
- 73.Feng Q, Liang S, Jia H, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun 2015; 6: 6528. [DOI] [PubMed] [Google Scholar]
- 74.Hong B, Ideta T, Lemos BS et al. Characterization of mucosal dysbiosis of early colonic neoplasia. NPJ Precis Oncol 2019; 3: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gao J-J, Zhang Y, Gerhard M, et al. Association between gut microbiota and Helicobacter pylori-related gastric lesions in a high-risk population of gastric cancer. Front Cell Infect Microbiol 2018; 8: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eun CS, Kim BK, Han DS, et al. Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods. Helicobacter 2014; 19: 407–16. [DOI] [PubMed] [Google Scholar]
- 77.Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, Goodman AL. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 2019; 570: 462–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Spanogiannopoulos P, Bess EN, Carmody RN, Turnbaugh PJ. The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism. Nat Rev Microbiol 2016; 14: 273–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Helmink BA, Khan MAW, Hermann A, Gopalakrishnan V, Wargo JA. The microbiome, cancer, and cancer therapy. Nat Med 2019; 25: 377–88. [DOI] [PubMed] [Google Scholar]
- 80.Alexander JL, Wilson ID, Teare J, Marchesi JR, Nicholson JK, Kinross JM. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat Rev Gastroenterol Hepatol 2017; 14: 356–65. [DOI] [PubMed] [Google Scholar]
- 81.Geller LT, Barzily-Rokni M, Danino T, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 2017; 357: 1156–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu T, Guo F, Yu Y, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 2017; 170: 548–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Viaud S, Saccheri F, Mignot G, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013; 342: 971–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Iida N, Dzutsev A, Stewart CA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013; 342: 967–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sivan A, Corrales L, Hubert N, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015; 350: 1084–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018; 359: 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Matson V, Fessler J, Bao R, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018; 359: 104–08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018; 359: 91–97. [DOI] [PubMed] [Google Scholar]
- 89.Vétizou M, Pitt JM, Daillère R, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015; 350: 1079–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kourie HR, Tabchi S, Ghosn M. Checkpoint inhibitors in gastrointestinal cancers: expectations and reality. World J Gastroenterol 2017; 23: 3017–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Miljanic M, Capasso A, Triplett TA, Eckhardt SG, Aung KL. Immune checkpoint blockade in gastrointestinal cancers: the current status and emerging paradigms. J Immunother Precis Oncol 2020; 3: 3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zou W, Wolchok JD, Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci Transl Med 2016; 8: 328rv4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wei SC, Duffy CR, Allison JP. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov 2018; 8: 1069–86. [DOI] [PubMed] [Google Scholar]
- 94.Chaput N, Lepage P, Coutzac C, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol 2017; 28: 1368–79. [DOI] [PubMed] [Google Scholar]
- 95.Serna G, Ruiz-Pace F, Hernando J, et al. Fusobacterium nucleatum persistence and risk of recurrence after preoperative treatment in locally advanced rectal cancer. Ann Oncol 2020; 31: 1366–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ma C, Han M, Heinrich B, et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 2018; 360: eaan5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nejman D, Livyatan I, Fuks G, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 2020; 368: 973–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lam KN, Alexander M, Turnbaugh PJ. Precision medicine goes microscopic: engineering the microbiome to improve drug outcomes. Cell Host Microbe 2019; 26: 22–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Walker AW, Ince J, Duncan SH, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J 2011; 5: 220–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014; 505: 559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cotillard A, Kennedy SP, Kong LC, et al. Dietary intervention impact on gut microbial gene richness. Nature 2013; 500: 585–88. [DOI] [PubMed] [Google Scholar]
- 102.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol 2014; 12: 661–72. [DOI] [PubMed] [Google Scholar]
- 103.Schulz MD, Atay C, Heringer J, et al. High-fat-diet-mediated dysbiosis promotes intestinal carcinogenesis independently of obesity. Nature 2014; 514: 508–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sutton P, Boag JM. Status of vaccine research and development for Helicobacter pylori. Vaccine 2019; 37: 7295–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gough E, Shaikh H, Manges AR. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin Infect Dis 2011; 53: 994–1002. [DOI] [PubMed] [Google Scholar]
- 106.FDA. Fecal microbiota for transplantation: safety alert—risk of serious adverse events likely due to transmission of pathogenic organisms. Mar 12, 2020. https://www.fda.gov/safety/medical-product-safety-information/fecal-microbiota-transplantation-safety-alert-risk-serious-adverse-events-likely-due-transmission (accessed Aug 3, 2020).
- 107.Atarashi K, Tanoue T, Oshima K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013; 500: 232–36. [DOI] [PubMed] [Google Scholar]
- 108.Sefik E, Geva-Zatorsky N, Oh S, et al. Individual intestinal symbionts induce a distinct population of RORã+ regulatory T cells. Science 2015; 349: 993–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tan TG, Sefik E, Geva-Zatorsky N, et al. Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc Natl Acad Sci USA 2016; 113: E8141–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tanoue T, Morita S, Plichta DR, et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature 2019; 565: 600–05. [DOI] [PubMed] [Google Scholar]
- 111.Hope ME, Hold GL, Kain R, El-Omar EM. Sporadic colorectal cancer--role of the commensal microbiota. FEMS Microbiol Lett 2005; 244: 1–7. [DOI] [PubMed] [Google Scholar]
- 112.Martincorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Science 2015; 349: 1483–89. [DOI] [PubMed] [Google Scholar]
- 113.Hofseth LJ, Hebert JR, Chanda A, et al. Early-onset colorectal cancer: initial clues and current views. Nat Rev Gastroenterol Hepatol 2020; 17: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Stigliano V, Sanchez-Mete L, Martayan A, Anti M. Early-onset colorectal cancer: a sporadic or inherited disease? World J Gastroenterol 2014; 20: 12420–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Keum N, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol 2019; 16: 713–32. [DOI] [PubMed] [Google Scholar]
- 116.Ahnen DJ, Wade SW, Jones WF, et al. The increasing incidence of young-onset colorectal cancer: a call to action. Mayo Clin Proc 2014; 89: 216–24. [DOI] [PubMed] [Google Scholar]
- 117.Katsidzira L, Gangaidzo I, Thomson S, Rusakaniko S, Matenga J, Ramesar R. The shifting epidemiology of colorectal cancer in sub-Saharan Africa. Lancet Gastroenterol Hepatol 2017; 2: 377–83. [DOI] [PubMed] [Google Scholar]
- 118.Herman AM, Hawkins AT, Misso K, et al. Colorectal cancer in northern Tanzania: increasing trends and late presentation present major challenges. JCO Glob Oncol 2020; 6: 375–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mmbaga EJ, Deardorff KV, Mushi B, et al. Characteristics of esophageal cancer cases in Tanzania. J Glob Oncol 2018; 4: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Menya D, Maina SK, Kibosia C, et al. Dental fluorosis and oral health in the african esophageal cancer corridor: findings from the Kenya ESCCAPE case-control study and a pan-African perspective. Int J Cancer 2019; 145: 99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sahar L, Foster SL, Sherman RL, et al. GIScience and cancer: State of the art and trends for cancer surveillance and epidemiology. Cancer 2019; 125: 2544–60. [DOI] [PMC free article] [PubMed] [Google Scholar]


