Summary
Background
Since gut microbiome dysbiosis can cause inflammatory disorders by affecting host metabolism, we postulate that the gut microbiome and related metabolites could play a role in hand osteoarthritis. We characterised gut microbiome-related metabolites in people with symptomatic hand osteoarthritis (SHOA) in two independent cohorts.
Methods
Using data collected from a large-sample community-based observational study (discovery cohort), we assessed the relations of the microbial function and plasma key metabolites related to altered microbial function with SHOA. Finally, we verified the relations of plasma metabolites to SHOA in an independent observational study (validation cohort).
Findings
In the discovery cohort (n = 1359), compared to those without SHOA, participants with SHOA had significantly altered microbial functions related to tryptophan metabolism (Q = 0.025). Therefore we measured the plasma tryptophan metabolites and found that participants with SHOA had higher levels of 5-hydroxyindoleacetic acid (odds ratio [OR] = 1.25, 95% confidence interval [CI]: 1.09–1.42) and 5-hydroxytryptophol (OR = 1.13, 95% CI: 1.04–1.23), but lower levels of indole-3-lactic acid (ILA) (OR = 0.85, 95% CI: 0.72–1.00), skatole (OR = 0.93, 95% CI: 0.88–0.99) and 3-hydroxyanthranilic acid (OR = 0.90, 95% CI: 0.85–0.96). Findings from the validation cohort (n = 142) verified that lower levels of ILA were related to SHOA (OR = 0.70, 95% CI: 0.53–0.92).
Interpretation
Alterations of the microbial function of tryptophan biosynthesis and tryptophan metabolites, especially lower levels of ILA, are associated with SHOA. These findings suggest the role of the microbiome and tryptophan metabolites in developing of SHOA and may contribute to future translational opportunities.
Funding
National Key Research and Development Plan and National Natural Science Foundation of China.
Keywords: Hand osteoarthritis, Gut microbiome, Tryptophan metabolites, Population-based study
Research in context.
Evidence before this study
We searched the PubMed database using terms related to “hand osteoarthritis”, “microbiome”, and “metabolites” to identify relevant articles. One previous study found that alteration of gut microbiota composition, analysed using 16S ribosomal RNA (rRNA) sequencing, was associated with symptomatic hand osteoarthritis (SHOA). Another study using an untargeted metabolomics method showed that significant perturbations in microbial tryptophan metabolites of stool samples existed in hand plus knee osteoarthritis cases. However, 16S rRNA sequencing technology is unable to profile the microbial metabolic function directly, and the untargeted metabolomics method has limitations in identifying and quantifying absolute levels of a predefined set of metabolites. To date, few, if any, studies have investigated the alteration of gut microbial function among people with SHOA using metagenome sequencing. Furthermore, the associations of plasma metabolomics modified by altered gut microbiome with SHOA remain unknown.
Added value of this study
Using data from shotgun metagenomic sequencing of stool samples from a large-sample population-based cohort study (i.e., discovery cohort, n = 1359), we found that decreased gut microbial function of tryptophan biosynthesis was associated with SHOA, which might be responsible for the perturbation of plasma tryptophan metabolites, measured by the targeted metabolomics method, in SHOA. Subsequent findings from the independent cohort (i.e., validation cohort, n = 142) verified that alterations of the plasma tryptophan metabolites, especially lower levels of indole-3-lactic acid, were related to SHOA.
Implications of all the available evidence
Our findings provide insights into the role of the gut microbiome and tryptophan metabolites in developing of SHOA and may contribute to translational opportunities.
Introduction
The hand is the second most common site affected by osteoarthritis (OA),1 with the prevalence ranging from 3% to 67% depending on the population and definition of the condition.1,2 Pain, stiffness, and swelling from hand OA can impair the ability to undertake activities of daily living.3 Although the pathogenesis of hand OA remains largely unknown, accumulating evidence suggests that systemic inflammation may play a role in the clinical symptoms and structural change of hand OA.4 Understanding the underlying mechanisms of hand OA could guide the development of preventive and treatment targets for this common disabling condition.5
Gut microbiome dysbiosis can cause inflammatory disorders,6 leading to speculation that the gut microbiome could play a role in the pathogenesis of hand OA. One previous study found that alteration of gut microbiota composition, analysed using 16S ribosomal RNA (rRNA) sequencing, was associated with symptomatic hand OA (SHOA).7 Gut microbiota produces various bioactive metabolites, which can enter the bloodstream of the host, affect host metabolism, and communicate with the host's peripheral organs and tissues.8 However, 16S rRNA sequencing technology is known to be limited by the short read lengths obtained, which cannot directly identify the metabolic functional capabilities of the gut microbiome,9 so the specific microbial function related to SHOA remains undetermined. To date, few, if any, studies have investigated the alteration of gut microbiome among people with SHOA using metagenome sequencing, which could sample all genes and produce detailed metabolic functional profiles of the gut microbiome. Furthermore, the associations of plasma metabolomics modified by altered gut microbiome with SHOA remain unknown.
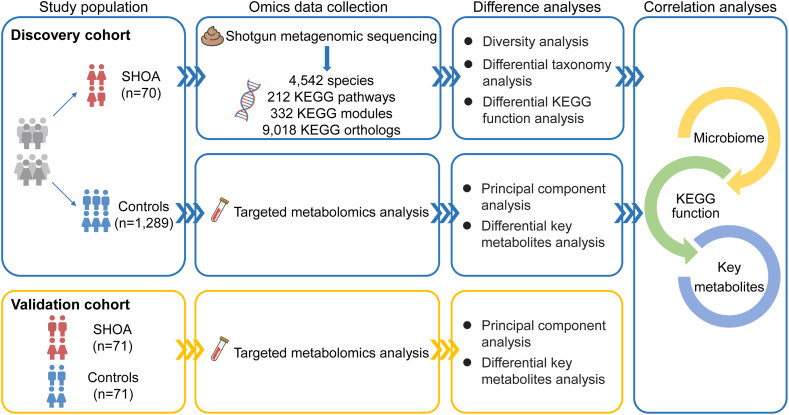
To fill this knowledge gap, we performed shotgun metagenomic sequencing of stool samples for gut microbial function using data from a large-sample community-based observational study (i.e., the discovery cohort).7,10 We performed targeted metabolomics analysis of plasma samples for key metabolites related to altered microbial function in SHOA and compared the key metabolites between participants with and those without SHOA. We also applied a multi-omics analysis approach to examine the associations between SHOA-related microbial function and SHOA-related plasma metabolites. Finally, we verified the relations of plasma key metabolites to SHOA in an independent observational study (validation cohort) (Fig. 1).
Fig. 1.
Summary of the present study. SHOA, symptomatic hand osteoarthritis; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Methods
Study participants
The Xiangya OA (XO) Study (discovery cohort) is a large community-based longitudinal study of the natural history and risk factors of OA in a rural area of China (NCT04033757). Details of the XO Study have been described previously.7,10 Briefly, participants in the XO Study were a randomly selected sample of residents aged 50 years or older from Longshan County in Hunan Province. The XO Study comprises three sub-cohorts, i.e., sub-cohort I (n = 1469), II (n = 1271), and III (n = 1340) initiated in 2015, 2018, and 2019, respectively. Participants from sub-cohort II and sub-cohort III of the XO Study, in which blood and stool samples were collected, were included in the present analysis (n = 2611). Participants were excluded if they did not have the following information: (1) hand posteroanterior radiographic evaluation or hand symptom assessment (n = 86), (2) stool samples for shotgun metagenomic sequencing (n = 567), and (3) plasma metabolites assessment (n = 19). In addition, participants were excluded from the current analyses if they reported antibiotic use within one month before stool sample collection, had a history of inflammatory bowel disease, gastrointestinal tract surgery, cancer, or rheumatoid arthritis, or did not have a sufficient amount of DNA extracted from their stool samples (n = 580). Finally, 1359 participants were included in the discovery cohort in the present analysis.
We then sought to replicate the findings from the XO Study in 71 SHOA patients who were recruited by the Xiangya Hospital of Central South University located in the Changsha City of China, and 71 age (±1 year), sex and BMI (±2 kg/m2) matched controls without SHOA, who were randomly selected from the Xiangya Step Study (validation cohort). The Xiangya Step Study (ChiCTR1800017977) is a population-based longitudinal study initiated in September 2018 in the urban areas of Changsha, China. Participants in the Xiangya Step Study comprised individuals who were between 40 and 79 years, had access to an electronic social network platform to record their daily steps, and came to Xiangya Hospital of Central South University for their annual physical checkup. At each annual physical check-up, participants were given a self-reported questionnaire that queried their socio-demographic and lifestyle factors, and comorbidities. Participants also received physical examinations, including radiographs of multiple joints (i.e., hand, knee, and hip), joint pain assessment, anthropometric measurements, body composition analysis, functional tests, muscle strength measurements of lower-extremity, and blood and urine tests. The sex of each participant was ascertained through self-reported data for both cohorts.
Assessment of SHOA
Participants underwent posteroanterior radiographs of both hands. Radiographs of the second to fifth distal interphalangeal joints, second to fifth proximal interphalangeal joints, first to fifth metacarpophalangeal joints, thumb interphalangeal joints, and thumb base (carpometacarpal) joints of each hand were graded using a modified Kellgren/Lawrence (K/L) scale.11 Radiographic hand OA was defined as a K/L grade of ≥2 in any of the joints listed above. Kappa statistics of intra-rater and inter-rater reliability for the assessment of radiographic hand OA (presence versus absence) were 0.91 (95% CI: 0.83–0.99) and 0.71 (95% CI: 0.45–0.96), respectively, as reported previously.7 The presence of hand symptoms was ascertained by a “yes” response to the question, “On most days, do you have pain, aching, or stiffness in your left/right hand?” A participant was defined as having SHOA if at least one hand had both self-reported symptoms and radiographic OA.12
In the discovery cohort, all participants had hand X-ray assessment. The control group was defined as participants who did not have hand pain or radiographic hand OA (i.e., KL grade <2). In the validation cohort, participants were asked for their hand symptoms first, and those without hand pain or who had hand pain but without radiographic OA were considered the source of the control group.
Stool sample collection and DNA extraction
Stool samples were collected at the recruitment site. The samples were frozen immediately, transported on dry ice within 20 min to the central lab, and stored in −80 °C freezers until further processing. We used 200 mg of stool for DNA exaction using the Magen HiPure Soil DNA Kit (Magen, Guangzhou, China) based on the manufacturer's protocol. Specifically, the stool samples were first homogenised and treated with lysis buffers. Then, a bead-beating process was performed using Tissuelyser-24 L (Jingxin, Shanghai, China) (25 Hz for 5 min) to completely lyse the cells, followed by a 10-min incubation at 65 °C. After the rounds of washing, DNA was eluted using elution buffers. Lastly, the eluted DNA was quantified with a Qubit Fluorometer by using a Qubit dsDNA BR Assay kit, and the quality was checked by running aliquots at 150 V on 1% agarose gel. We retained the integrated and purified DNA samples over 20 Kb and without RNA or protein contamination. In addition, extracted DNA that has a sufficient amount (i.e., ≥1 μg) with a concentration ≥12.5 ng/μL was used for further analysis.
DNA library construction and shotgun metagenomic sequencing
Extracted DNA samples were randomly fragmented by Covaris. Magnetic beads were used to select the fragmented DNA at an average size of 200–400 base pairs. The selected fragments underwent end repair, 3’ adenylation, adapters ligation, PCR amplification, and magnetic-bead purification. The double-stranded PCR products were heat-denatured and circularised by the splint oligo sequence. The single-strand circular DNA was formatted as the final library and qualified. The qualified libraries were sequenced on the MGISEQ-2000 platform (BGI-Shenzhen, China), and paired-end reads of 150-base pair nucleotides were generated.13
To obtain clean data, the original sequencing reads were processed as follows: (1) excluding reads containing 10% uncertain bases (N bases); (2) excluding reads containing adapter sequences (15 bases or more extended sequence aligned to the adapter sequence); (3) excluding reads containing low-quality bases of 20% (bases of Q <20); and (4) a filtering step to remove the sequence of the host genome (SOAP2,14 >90% similarity, a host reference sequence is required). After quality control, high-quality reads were assembled using MEGAHIT.15 For human stool samples, DNA reads were assessed for taxonomics using Kraken2 based on the Unified Human Gastrointestinal Genome (UHGG).16 The alignment results were mapped to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, which provides functional information for the gut microbiome.17
Assessment of plasma metabolites
Blood samples were collected after an overnight fast of 12 h, and all samples were centrifuged at the collection site for 10 min at 3000 RPM using a laboratory centrifuge to separate the plasma. The plasma samples were then stored at −80 °C until further use. After centrifugation, the supernatant was transferred into a new microcentrifuge tube for targeted metabolomics analysis. The concentration of metabolites was calculated according to the calibration curve of the standard substance.
Assessment of plasma tryptophan metabolites
100 μL of plasma sample was extracted with 400 μL acetonitrile: methanol (1:1, v:v) containing isotopically-labelled internal standard mixture. The samples were vortexed for 30 s, and sonicated for 5 min in an ice-water bath, followed by incubation at −40 °C for 1 h. After centrifugation at 12,000 rpm for 15 min at 4 °C, 400 μL supernatant was transferred to an Eppendorf tube and evaporated to dryness under a gentle stream of nitrogen, then reconstituted in 100 μL water containing 0.1% formic acid. After centrifugation at the same condition, the supernatant was transferred into a new microcentrifuge tube for ultra-high performance liquid chromatography-tandem mass spectrometry (UHPLC-MS/MS) analysis. Tryptophan metabolites were measured using an EXIONLC System (Sciex, Framingham, MA, USA) and separated with HSS T3 using a water solution containing 0.1% formic acid and acetonitrile-solution containing 0.1% formic acid in multiple reaction monitoring (MRM) modes. Transitions of each tryptophan metabolite are shown in Supplementary Table S1. SCIEX 6500 QTRAP + triple quadrupole mass spectrometer was applied for assay development in positive and negative modes. MRM data were captured and processed by SCIEX Analyst Work Station Software and Sciex MultiQuant software. Finally, the concentration of tryptophan metabolites was calculated according to the calibration curve of the standard substance.
Statistical analysis
First, we compared the Shannon index (i.e., α-diversity) of microbial genes and species between participants with and those without SHOA from the discovery cohort using the Wilcoxon rank sum test. We assessed the difference in microbiome composition (i.e., β-diversity), measured by Bray–Curtis distance, between the two groups using the permutation multivariate analysis of variance (PERMANOVA) combined with principal coordinates analysis (PCoA) at genes and species levels. We used a 10% prevalence filter to remove extremely rare taxa or microbial function, thus resulting in more robust results.18 For microbial species present in ≥10% of samples, we performed Tweedie compound Poisson generalized linear mixed-effects models (GLMMs) using R package Microbiome Multivariable Associations with Linear Models (MaAsLin2 v.1.7.3)19 to examine the associations of the relative abundance of microbial species with SHOA adjusting for age, sex, BMI, smoking status, alcohol consumption, and frequency of dietary intake of meat/eggs, dairy, and vegetables, where the relative abundance of microbial features was total-sum scaling normalized and log-transformed. The GLMMs enable accurate modelling of non-normally distributed abundance data and correctly account for multiple sources of variation.20 We identified these confounders based on previous publications (Supplementary Figure S1).21, 22, 23 Sensitivity analysis was conducted by further adjusting for medication use and other comorbidities. We also performed the MaAsLin2 to assess the differences in microbial function, represented by KEGG orthologs (KOs), KEGG modules, and KEGG level 3 pathways (present in more than 10% of samples), between participants with and without SHOA.
Second, we compared the compositions of the key plasma metabolites between the two comparison groups using principal component analysis (PCA) without rotation and the PERMANOVA test. The percentages refer to the largest and the second-largest unique proportion of total variance explained by the first and second principal components, respectively. We further evaluated the association of the key plasma metabolite with the prevalence of SHOA using logistic regression adjusting for the aforementioned confounders for the discovery cohort. The linearity assumption underlying the logistic regression model was tested using the Box–Tidwell test.24,25 To assess the robustness of the findings, we performed four sensitivity analyses in the discovery cohort. First, we classified participants with SHOA into two groups, specifically those with only one hand joint with KL grade ≥2, and those with two or more hand joints with KL grade ≥2. We compared the key plasma metabolites among the participants without SHOA, those with one hand joint with SHOA, and those with two or more joints with SHOA using proportional odds logistic regression for related estimates and P for trend. We assessed the proportional odds assumption using the score test.26 Second, among participants without SHOA, we compared key plasma metabolites between participants with asymptomatic hand OA and those without hand OA. Third, we divided the participants into three categories according to the tertiles of key metabolites levels and compared the prevalence of SHOA between the three categories using logistic regression models. Fourth, we assessed whether the associations of key metabolites with SHOA changed from that observed in the primary analysis after adjusting for additional confounders (i.e., medication use and other comorbidities).
Third, we conducted multi-omics analyses to identify the relations of SHOA-related microbial species to the SHOA-related microbial functions (i.e., KEGG modules and KEGG pathways), and the relations of the SHOA-related microbial functions to the SHOA-related metabolites using the partial Spearman's rank correlation test adjusting for the potential confounders. To compute the partial Spearman's rank correlation between X and Y adjusting for Z, the basic approach involves fitting a specified model of X on Z, a specified model of Y on Z, obtaining the residuals from both models and then calculating their Pearson's correlation.27
Finally, we verified the relations of plasma key metabolites to SHOA in the validation cohort. We assessed the associations between the key plasma metabolites and SHOA using the conditional logistic regression model adjusting for smoking status and alcohol consumption.
All statistical analyses were performed with SAS version 9.4 (SAS Institute, Cary, North Carolina, USA) and R version 4.2.0 (R Foundation, Vienna, Austria). P values < 0.05 were considered statistically significant. Multiple testing with the Benjamin & Hochberg False discovery rate method28 for high-throughput sequencing data was applied and corrected P values (Q values) < 0.1 were considered statistically significant.
Ethics
This study was approved by the Research Ethical Committee of Xiangya Hospital, Central South University (approval numbers: 201510506 for the discovery cohort, 201806910 and 202210650 for the validation cohort). All participants provided written informed consent.
Role of funders
No funding bodies played a role in study design, data collection, analyses, interpretation, or writing.
Results
A total of 1359 participants (70 participants with SHOA) were included in the discovery cohort, and 142 participants (71 participants with SHOA) were included in the validation cohort. The basic characteristics of included participants of the discovery cohort and validation cohort are shown in Tables 1 and 2.
Table 1.
Basic characteristics of included participants of the discovery cohort.
| Characteristic | SHOA | Control | P value |
|---|---|---|---|
| N | 70 | 1289 | – |
| Age, median (IQR), years | 72.0 (65.0, 77.0) | 62.0 (55.0, 69.0) | <0.0001 |
| Sex, n (%) | 0.0026 | ||
| Male | 17 (24.3) | 548 (42.5) | |
| Female | 53 (75.7) | 741 (57.5) | |
| BMI, median (IQR), kg/m2 | 23.4 (21.1, 27.6) | 23.6 (21.4, 25.9) | 0.677 |
| Alcohol consumption, n (%) | 0.388 | ||
| None | 37 (54.4) | 616 (48.0) | |
| Past | 9 (13.2) | 145 (11.3) | |
| Current | 22 (32.4) | 523 (40.7) | |
| Smoking status, n (%) | 0.101 | ||
| None | 50 (73.5) | 804 (62.7) | |
| Past | 5 (7.4) | 76 (5.9) | |
| Current | 13 (19.1) | 403 (31.4) | |
| Dietary intake of meat/eggs, median (IQR), times/week | 2.0 (0.6, 4.5) | 2.0 (2.0, 4.5) | 0.274 |
| Dietary intake of dairy products, median (IQR), times/week | 0.0 (0.0, 0.0) | 0.0 (0.0, 0.0) | 0.788 |
| Dietary intake of vegetables, median (IQR), times/week | 14.0 (4.5, 14.0) | 14.0 (4.5, 14.0) | 0.814 |
SHOA, symptomatic hand osteoarthritis; N, number; IQR, interquartile range; BMI, body mass index.
Seven and eight participants with missing values of alcohol consumption status and smoking status, respectively.
Table 2.
Basic characteristics of included participants of the validation cohort.
| Characteristic | SHOA | Control | P value |
|---|---|---|---|
| N | 71 | 71 | – |
| Age, mean (SD), years | 70.6 (8.2) | 70.6 (8.2) | 0.971 |
| Sex, n (%) | 1.000 | ||
| Male | 29 (40.8) | 29 (40.8) | |
| Female | 42 (59.2) | 42 (59.2) | |
| BMI, mean (SD), kg/m2 | 23.8 (3.2) | 23.9 (2.9) | 0.691 |
| Alcohol consumption, n (%) | 0.0005 | ||
| None | 32 (46.4) | 47 (66.2) | |
| Past | 0 (0.0) | 6 (8.5) | |
| Current | 37 (53.6) | 18 (25.3) | |
| Smoking status, n (%) | 0.124 | ||
| None | 44 (63.8) | 52 (73.2) | |
| Past | 7 (10.1) | 10 (14.1) | |
| Current | 18 (26.1) | 9 (12.7) |
SHOA, symptomatic hand osteoarthritis; N, number; SD, standard deviation; BMI, body mass index.
Two participants with missing values of alcohol consumption status and smoking status.
Association of gut microbial function with SHOA
Using the shotgun metagenomic sequencing method, we found that gut microbial α-diversity (i.e., Shannon index) and β-diversity (i.e., Bray–Curtis distance) in either microbial genes (P = 0.008 and 0.045, respectively, PERMANOVA test) or species level (P = 0.009 and 0.039, respectively, PERMANOVA test) differed significantly between participants with and those without SHOA (Supplementary Figure S2a–d). The profile of the gut microbiome was dominated by Prevotella spp, Bacteroides spp, Faecalibacterium prausnitzii, and Escherichia coli at the species level (Supplementary Figure S2e) in both the SHOA and control groups. Compared with those without SHOA, participants with SHOA showed a higher relative abundance of Bilophila wadsworthia (β coefficient: 0.56 [95% CI: 0.24–0.88], Q = 0.025, MaAsLin2), Lactobacillus H mucosae (β coefficient: 2.06 [95% CI: 0.87–3.26], Q = 0.029), Citrobacter B koseri (β coefficient: 2.14 [95% CI: 0.85–3.43], Q = 0.041), and Hungatella hathewayi (β coefficient: 1.53 [95% CI: 0.79–2.26], Q = 0.005) but a lower relative abundance of Roseburia intestinalis (β coefficient: −0.78 [95% CI: −1.29 to −0.26], Q = 0.071), Bacteroides spp (β coefficients = −0.87 [95% CI: −1.43 to −0.31] and −0.48 [95% CI: −0.82 to −0.15], Q = 0.064 and 0.094, respectively), and Haemophilus spp (β coefficients: −1.54 [95% CI: −2.48 to −0.60] to −1.36 [95% CI: −2.29 to −0.43], Q values: 0.086–0.043) compared to the control group (Supplementary Figure S2f and Supplementary Table S2). Sensitivity analysis showed that with further adjustment for medication use and other comorbidities did not change the results materially (Supplementary Table S3).
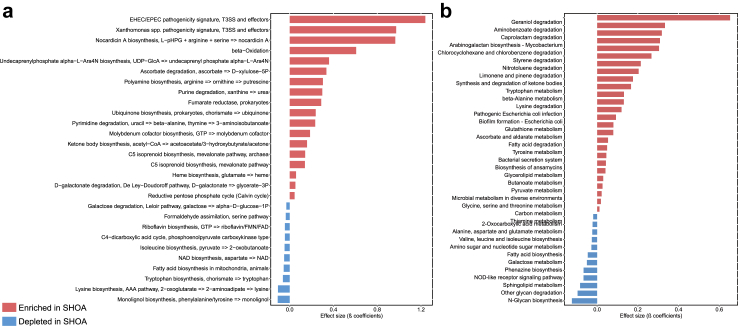
The microbial functions of 296 KOs differed according to the status of SHOA, most of them being engaged in amino acid metabolism, carbohydrate metabolism, metabolism of cofactors and vitamins, and energy metabolism (Supplementary Table S4). Compared to those without SHOA, KEGG modules involving amino acid metabolism (e.g., tryptophan biosynthesis), metabolism of cofactors (e.g., nicotinamide adenine dinucleotide [NAD] biosynthesis) were depleted, while those associated with carbohydrate metabolism (e.g., ascorbate degradation, d-galactonate degradation) and lipid metabolism (e.g., ketone body biosynthesis and beta-oxidation) were enriched in participants with SHOA (Fig. 2a and Supplementary Table S5). In addition, KEGG pathways related to amino acid metabolism of tryptophan, lysine and tyrosine metabolism, carbohydrate metabolism of pyruvate, butanoate, and galactose metabolism, lipid metabolism of fatty acid degradation and biosynthesis were also significantly altered in participants with SHOA (Fig. 2b and Supplementary Table S5).
Fig. 2.
Significant associations of KEGG modules and KEGG pathways with SHOA in the discovery cohort. Significant (Q <0.1, MaAsLin2) associations of KEGG modules (a) and KEGG pathways (b) between participants with SHOA (n = 70) and those without SHOA (n = 1289). β coefficient values shown on the x-axis were generated from the Microbiome Multivariable Associations with Linear Models (MaAsLin2). SHOA, symptomatic hand osteoarthritis; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Association of plasma tryptophan metabolites with SHOA
We searched previous studies evaluating the associations between metabolites and hand OA to help narrow and identify the key metabolites induced by gut microbiome dysbiosis in SHOA. One previous study using untargeted metabolomics analysis for stool samples showed significant perturbations of microbial tryptophan metabolites in hand plus knee OA cases,29 which overlapped with our findings from the microbial function analysis. Thus, we performed a targeted metabolomics analysis of plasma samples for tryptophan metabolites for the participants from the same cohort.
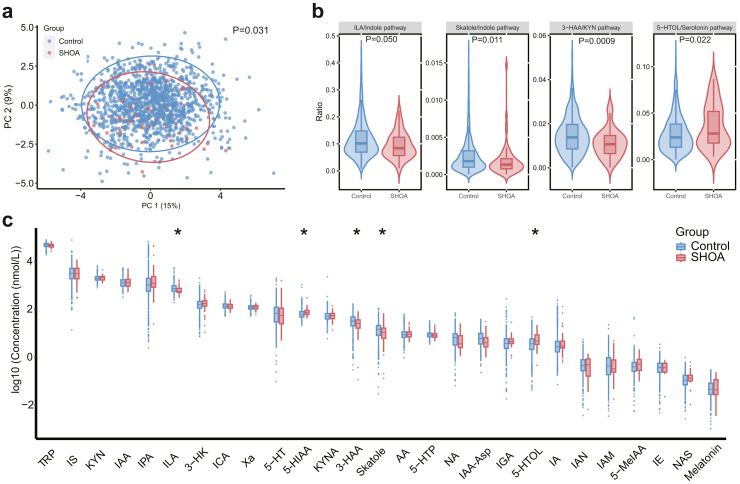
The composition of plasma tryptophan metabolites differed significantly between participants with and those without SHOA in the discovery cohort (P = 0.031, PERMANOVA test, Fig. 3a). According to the previous study,30 tryptophan can be metabolised following three major pathways: the serotonin, the kynurenine (KYN), and the indole. Therefore, we compared the ratios of tryptophan metabolite concentration in their related pathway and the individual tryptophan metabolite level between participants with and those without SHOA. Associations of ratios of tryptophan metabolites concentration in their related pathway with SHOA are shown in Fig. 3b. Specifically, the 5-hydroxytryptophol (5-HTOL)/serotonin pathway ratio was positively associated with SHOA, while the indole-3-lactic acid (ILA)/indole pathway, skatole/indole pathway and 3-hydroxyanthranilic acid (3-HAA)/KYN pathway ratios were inversely associated with SHOA. Higher levels of tryptophan metabolites of 5-hydroxyindoleacetic acid (5-HIAA) and 5-HTOL, but lower levels of ILA, skatole, and 3-HAA were associated with a higher prevalence of SHOA (Fig. 3c). The multivariable-adjusted ORs of SHOA per 0.1 log10 unit increase for 5-HIAA, 5-HTOL, ILA, skatole, and 3-HAA were 1.25 (95% CI: 1.09–1.42), 1.13 (95% CI: 1.04–1.23), 0.85 (95% CI: 0.72–1.00), 0.93 (95% CI: 0.88–0.99) and 0.90 (95% CI: 0.85–0.96), respectively, and the linearity assumption was not violated for the key plasma metabolites. Such differences were more pronounced among participants with more hand joints affected by SHOA (Supplementary Table S6). We divided the participants into three categories according to the tertiles of key metabolites levels and compared the prevalence of SHOA between the three categories (Supplementary Table S7) as well. Specifically, compared with the lowest category of ILA, the multivariable-adjusted ORs of SHOA in the second and third categories of ILA were 0.54 (95% CI: 0.30–0.99) and 0.38 (95% CI: 0.19–0.77), respectively (P for trend = 0.006, logistic regression). Adjusting for additional confounders (i.e., medication use and other comorbidities) showed similar results of the associations of key metabolites with SHOA (Supplementary Table S8). In addition, higher levels of tryptophan metabolites of kynurenic acid, 3-hydroxykynurenine, kynurenine, L-5-hydroxytryptophan, and anthranilic acid, but lower levels of indole-3-acetyl-aspartate and l-tryptophan were associated with asymptomatic hand OA (data not shown).
Fig. 3.
Associations of plasma tryptophan metabolites with SHOA in the discovery cohort. (a) Principal Component Analysis (PCA) plot comparing the composition of plasma tryptophan metabolites between participants with SHOA (n = 70) and those without SHOA (n = 1289) combined with the permutation multivariate analysis of variance (PERMANOVA) test. The null hypothesis tested by PERMANOVA is that, under the assumption of exchangeability of the sample units among the groups, H0: the centroids of the groups, as defined in the space of the chosen resemblance measure, are equivalent for all groups. Significant (∗, P < 0.05, logistic regression) associations of ratios of tryptophan metabolites concentration in their related pathway (b) and individual plasma tryptophan metabolites (c) with SHOA. The upper whisker extends from the hinge to the largest value no further than 1.5 ∗ inter-quartile range (IQR) from the hinge. The lower whisker extends from the hinge to the smallest value at most 1.5 ∗ IQR of the hinge. Data beyond the end of the whiskers are outliers. SHOA, symptomatic hand osteoarthritis; TRP, tryptophan; KYN, kynurenine; ILA, indole-3-lactic acid; 3-HAA, 3-hydroxyanthranilic acid; 5-HIAA, 5-hydroxyindoleacetic acid; 5-HTOL, 5-hydroxytryptophol; 3-HK; 3-hydroxykynurenine; 5-HT, serotonin; 5-HTP, L-5-hydroxytryptophan; 5-MeIAA; 5-methoxy-3-indoleacetic acid; AA, anthranilic acid; IA; indole acrylic acid; IAA, indole-3-acetic acid; IAA-Asp, indole-3-acetyl-aspartate; IAM, indole-3-acetamide; IAN, indole-3-acetonitrile; ICA, indole-3-carboxaldehyde; IE, indole ethanol/tryptophol; IGA, 3-indoleglyoxylic acid; IPA, 3-indolepropionic acid; IS, indoxylsulfate; KYNA, kynurenic acid; NA, nicotinic acid; NAS, N-acetyl-5-hydroxytryptamine; Xa, xanthurenic acid.
Correlation of microbial species to microbial function, and microbial functions to tryptophan metabolites
To identify the possible link between gut microbiome dysbiosis and the alterations of the tryptophan metabolites in SHOA, we assessed the correlations of microbial species to the microbial functions, and microbial functions to the tryptophan metabolites (Supplementary Figures S3 and S4). Specifically, we identified that lower levels of ILA were correlated to fourteen SHOA-related KEGG modules (e.g., decreased KEGG module of tryptophan biosynthesis), which were further associated with eleven classified SHOA-related microbial species (e.g., the lower relative abundance of Bacteroides A mediterraneensis).
Validation of association between tryptophan metabolites and SHOA
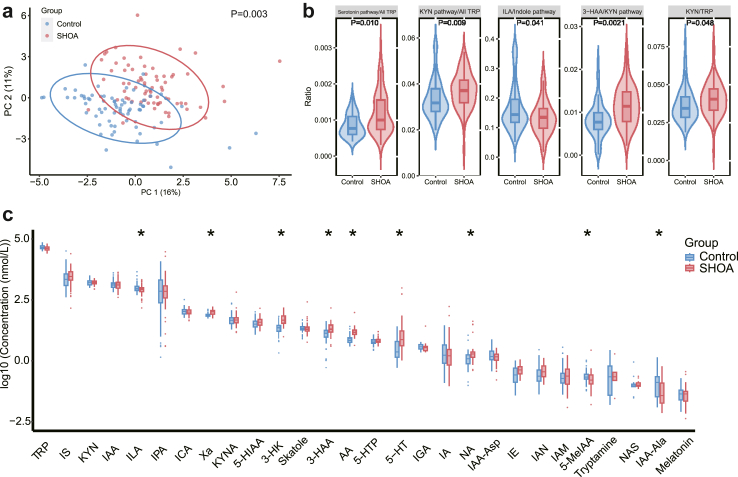
Finally, we verified the associations between plasma tryptophan metabolites and SHOA in the validation cohort. Consistently, there was a significant difference in the composition of plasma tryptophan metabolites between the SHOA group and controls (P = 0.003, PERMANOVA test, Fig. 4a). A total of five ratios of individual metabolite in their related pathways (i.e., ILA/indole pathway, 3-HAA/KYN pathway, KYN pathway/all tryptophan metabolites, serotonin pathway/all tryptophan metabolites and KYN/tryptophan) and nine metabolites (i.e., ILA, 3-HAA, xanthurenic acid, 3-hydroxykynurenine, serotonin, anthranilic acid, nicotinic acid, 5-methoxy-3-indoleacetic acid, and indole-3-acetyl-alanine) differed between the two comparison groups (Fig. 4b–c). Importantly, consistent with the findings in the discovery cohort, we observed a lower ratio of ILA in the indole pathway and a lower level of the ILA in the SHOA group compared with controls. Compared with the lowest category of ILA, ORs were 0.53 [95% CI 0.22–1.27] and 0.38 [95% CI 0.16–0.92] in the second and third categories of ILA, respectively (P for trend = 0.036, logistic regression), with the multivariable-adjusted OR of SHOA per 0.1 log10 unit increase for ILA being 0.70 (95% CI: 0.53–0.92). Further adjustment of medication use and other comorbidities did not change the association of ILA with SHOA materially (OR = 0.69, 95% CI 0.51–0.92 for per 0.1 log10 unit increase of ILA).
Fig. 4.
Associations of plasma tryptophan metabolites with SHOA in the validation cohort. (a) Principal Component Analysis (PCA) plot comparing the composition of plasma tryptophan metabolites between participants with SHOA (n = 71) and those without SHOA (n = 71) combined with the permutation multivariate analysis of variance (PERMANOVA) test. The null hypothesis tested by PERMANOVA is that, under the assumption of exchangeability of the sample units among the groups, H0: the centroids of the groups, as defined in the space of the chosen resemblance measure, are equivalent for all groups. Significant (∗, P < 0.05, logistic regression) associations of ratios of tryptophan metabolites concentration in their related pathway (b) and individual plasma tryptophan metabolites (c) with SHOA adjusting for smoking status and alcohol consumption. The lower and upper hinges of the box plot correspond to the first and third quartiles. The upper whisker extends from the hinge to the largest value no further than 1.5 ∗ inter-quartile range (IQR) from the hinge. The lower whisker extends from the hinge to the smallest value at most 1.5 ∗ IQR of the hinge. Data beyond the end of the whiskers are outliers. SHOA, symptomatic hand osteoarthritis; TRP, tryptophan; KYN, kynurenine; ILA, indole-3-lactic acid; 3-HAA, 3-hydroxyanthranilic acid; 5-HIAA, 5-hydroxyindoleacetic acid; 3-HK; 3-hydroxykynurenine; 5-HT, serotonin; 5-HTP, L-5-hydroxytryptophan; 5-MeIAA; 5-methoxy-3-indoleacetic acid; AA, anthranilic acid; IA; indole acrylic acid; IAA, indole-3-acetic acid; IAA-Asp, indole-3-acetyl-aspartate; IAM, indole-3-acetamide; IAN, indole-3-acetonitrile; ICA, indole-3-carboxaldehyde; IE, indole ethanol/tryptophol; IGA, 3-indoleglyoxylic acid; IPA, 3-indolepropionic acid; IS, indoxylsulfate; KYNA, kynurenic acid; NA, nicotinic acid; NAS, N-acetyl-5-hydroxytryptamine; Xa, xanthurenic acid; IAA-Ala, indole-3-acetyl-alanine.
Discussion
Using data from a large-sample community-based observational study (i.e., the discovery cohort), we found that participants with SHOA had a lower relative abundance of the microbial function of tryptophan biosynthesis. Then we measured the plasma tryptophan metabolites and identified that participants with SHOA had higher levels of 5-HIAA and 5-HTOL, but lower levels of ILA, 3-HAA, and skatole. Moreover, results from multi-omics analysis suggested that the lower level of ILA in SHOA was related to the microbial function of tryptophan biosynthesis, which was further correlated with the microbiome species Bacteroides A mediterraneensis. Finally, findings from the validation cohort verified that alterations of the plasma tryptophan metabolites, especially lower levels of ILA, were related to SHOA.
Several previous studies found that gut microbiome dysbiosis was associated with OA. Results from the Rotterdam study reported a significant difference in the gut microbiome composition between participants with and without OA-related knee pain, and a higher relative abundance of Streptococcus species was associated with increased OA-related knee pain.31 One case–control study identified seven optimal microbial genera biomarkers to distinguish overweight knee OA patients and overweight normal people,32 while another case–control study observed that microbial species of Bifidobacterium longum and Faecalibacterium prausnitzii were decreased, but Clostridium spp. was increased in the OA patients compared with healthy controls.33 Nevertheless, these studies mainly investigated the link between microbiome dysbiosis and knee OA. It is worth noting that knee OA might not necessarily exhibit congruent fundamental mechanisms with hand OA. One previous study reported that gut microbiota composition, profiled by 16S ribosomal RNA gene sequencing, differed according to SHOA status.7 Using an untargeted metabolomics method, previous studies have reported significant perturbations in microbial tryptophan metabolites in people with hand plus knee OA,29 and that plasma l-tryptophan may act as a biomarker for OA.34 However, 16S rRNA sequencing technology cannot differentiate sufficiently the closely related species and directly profile the microbial metabolic function,9 and the untargeted metabolomics method has limitations in identifying a predefined set of metabolites and in quantifying their absolute levels.35 In the present study, using shotgun metagenomic sequencing to profile gut microbial function and targeted metabolomics methods to measure plasma tryptophan metabolites, we have provided evidence that alterations of plasma tryptophan metabolites, which might be induced by microbiome dysbiosis, are associated with SHOA.
Tryptophan metabolites are a crucial category of metabolites engaged in host–microbiota interaction and play a central role in physiology and physiopathology related to systematic inflammation and immune response.30,36,37 Previous studies have shown that the gut microbiota plays a crucial role in the modulation of tryptophan metabolism by directly transforming the tryptophan into several molecules or indirectly affecting the balance among different tryptophan metabolism pathways.30 In addition, alterations in tryptophan metabolism have been reported in several systemic inflammatory diseases.38 For example, indole derivatives (e.g., ILA) produced by tryptophan metabolism benefits intestinal permeability, inflammation regulation, and host immunity by activating aryl hydrocarbon receptors, which are widely expressed in a variety of immune cells.39, 40, 41 Our study found that several tryptophan metabolites are associated with SHOA, and we replicated the inverse association between ILA and SHOA in the validation cohort. This is consistent with the potential role of ILA in antioxidation and anti-inflammation.39,42 Moreover, using multi-omics analysis, we identified the possible link between gut microbiome dysbiosis and tryptophan metabolism alteration in SHOA (e.g., Bacteroides A mediterraneensis to KEGG module of tryptophan biosynthesis, and KEGG module of tryptophan biosynthesis to ILA). We also identified several tryptophan metabolites associated with asymptomatic hand OA, which differed from the SHOA-related tryptophan metabolites. These findings imply that SHOA-related tryptophan metabolites are more involved in pain than in structural changes of hand OA.
Several strengths of our study are worth noting. First, we recruited a large random sample of community-based participants in the discovery cohort. Thus the distribution of the microbiome and the tryptophan metabolites is likely to represent the general population with similar characteristics. Second, the findings of tryptophan metabolites alteration in SHOA in the discovery cohort were verified in the validation cohort, supporting the robustness of the study findings. Third, we performed shotgun metagenomic sequencing for the gut microbiome and the targeted metabolomics analysis for plasma tryptophan metabolites, which allowed us to identify the microbial species and functions, and specific tryptophan metabolites in association with SHOA. Fourth, findings from the multi-omics analyses allowed us to identify the possible link between specific microbial functions and metabolites associated with SHOA. Fifth, we observed more pronounced associations of these SHOA-related tryptophan metabolites with participants with more hand joints affected by SHOA, supporting the robustness of the study findings.
The limitations of the current study should also be acknowledged. First, our study was cross-sectional, so we cannot establish a causal relationship between the gut microbiome, plasma tryptophan metabolites, and the prevalence of SHOA. Thus, we consider our findings from the cross-sectional study as an exploratory analysis. To avoid potential selection bias (e.g., collider bias) when studying the association between each of the microbiome/metabolites and SHOA, we did not adjust for the others (i.e., the remaining microbiome/metabolites). As a result, some of the associations reported in our study may be confounded by the gut microbiome or other plasma metabolites that occurred earlier than the risk factor under the evaluation and were also associated with SHOA. Future prospective cohort studies are needed to establish a temporal relation of the gut microbiome and plasma metabolites to the incident SHOA, and to understand the temporal relationship between the gut microbiome and the key plasma metabolites to obtain a valid effect estimate of each of these factors. Second, several species (e.g., Bacteroides A mediterraneensis) were identified in the possible link between the microbiome and plasma tryptophan metabolites in SHOA. However, other studies have not reported these relations, and their underlying function and mechanisms related to SHOA remain unclear. Third, we were unable to validate our findings in vivo due to the lack of an animal model for hand OA.3 Future studies involving examination of other joint components (such as synovium, subchondral bone, synovial fluid or fat pad) are required to verify these associations and to understand the biological mechanisms involved. Fourth, observational studies cannot rule out unmeasured residual confounding. Fifth, including prevalent cases in the study may cause potential incidence-prevalence bias; further studies limiting the inclusion to incident cases were needed.
Previous studies have shown the impact of the gut microbiome on tryptophan metabolism and tryptophan metabolites, especially ILA, in mitigating systematic inflammation.30,39, 40, 41 In consonance with these antecedent findings, our study provided additional empirical evidence and revealed that alterations of plasma tryptophan metabolites, which might be induced by microbiome dysbiosis, are associated with SHOA. If confirmed by others, these findings hold promise to develop potential therapeutic approaches based on modulating gut microbiome and related metabolites to treat SHOA.
In summary, using data from a population-based observational study (i.e., the discovery cohort), we found that decreased gut microbial function of tryptophan biosynthesis was associated with SHOA, which might be responsible for the perturbation of plasma tryptophan metabolites in SHOA. Subsequently, findings from the independent validation cohort verified that alterations of the plasma tryptophan metabolites, especially lower levels of ILA, are associated with SHOA. Findings from the current exploratory analysis suggest the potential role of the microbiome and tryptophan metabolism in the development of SHOA and may contribute to translational opportunities if they were confirmed by future prospective cohort studies and animal models involving the examination of other joint components.
Contributors
GL and CZ had full access to all of the data in the study, verified the underlying data, and took responsibility for the data's integrity and the data's accuracy. GL and CZ are joint corresponding authors, they are responsible for the decision to submit the manuscript. GL is the lead contact. All authors have read, provided critical feedback on intellectual content, and approved the final manuscript. Concept and design: JW, ZY, JL, YZ, WZ, MD, CZ, GL. Acquisition, analysis, or interpretation of data: JW, ZY, JL, TY, YY, YW, ZW, HL, CL. Drafting of the manuscript: JW, ZY, JL. Critical revision of the manuscript for important intellectual content: JW, ZY, JL, YZ, WZ, MD, TY, YY, HL, YW, ZW, CL, CZ, GL. Obtained funding: GL, CZ, JW. Administrative, technical, or material support: GL, CZ, YZ, WZ, MD. Supervision: GL, CZ, YZ, WZ, MD.
Data sharing statement
The sequencing data have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in the National Genomics Data Center (Nucleic Acids Res 2022), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA012188) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa. Additional data can be obtained from the corresponding authors upon reasonable request.
Declaration of interests
All authors have no competing interests.
Acknowledgements
This work was supported by the National Key Research and Development Plan (GL: 2022YFC3601900, GL: 2022YFC3601901, CZ: 2022YFC2505500), the National Natural Science Foundation of China (GL: 81930071, GL: U21A20352, CZ: 82072502), the Project Program of National Clinical Research Center for Geriatric Disorders (JW: 2021LNJJ06, CZ: 2022LNJJ07), and the Natural Science Foundation of Hunan Province (JW: 2022JJ20100). We are grateful to the Bioinformatics Center, Xiangya Hospital, Central South University for partial support of this work. We appreciate the Shanghai Biotree Biomedical Technology for their service of the assessment of plasma tryptophan metabolites. We thank Abasiama D. Obotiba, Dongxing Xie, Ting Jiang, Xiaoxiao Li, Bei Xu, Jian Tian, Yuqing Wang, Zhenglei Zhu, Ke He, Haochen Wang, Hongyi He, Xin Huang, Ning Wang, Xiang Ding, Bin Zhou, Ruijun Bai, Zhichen Liu, Junyu Zhu, Kun Li, Junyan Liu, Huizhong Long, Xinjia Deng, Wei Li, Yanzhe Liu, Yan Zhou for their contribution to this study.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104892.
Contributor Information
Guanghua Lei, Email: lei_guanghua@csu.edu.cn.
Chao Zeng, Email: zengchao@csu.edu.cn.
Appendix A. Supplementary data
References
- 1.Favero M., Belluzzi E., Ortolan A., et al. Erosive hand osteoarthritis: latest findings and outlook. Nat Rev Rheumatol. 2022;18:171–183. doi: 10.1038/s41584-021-00747-3. [DOI] [PubMed] [Google Scholar]
- 2.Hunter D.J., Bierma-Zeinstra S. Osteoarthritis. Lancet. 2019;393:1745–1759. doi: 10.1016/S0140-6736(19)30417-9. [DOI] [PubMed] [Google Scholar]
- 3.Marshall M., Watt F.E., Vincent T.L., Dziedzic K. Hand osteoarthritis: clinical phenotypes, molecular mechanisms and disease management. Nat Rev Rheumatol. 2018;14:641–656. doi: 10.1038/s41584-018-0095-4. [DOI] [PubMed] [Google Scholar]
- 4.Conaghan P.G., Cook A.D., Hamilton J.A., Tak P.P. Therapeutic options for targeting inflammatory osteoarthritis pain. Nat Rev Rheumatol. 2019;15:355–363. doi: 10.1038/s41584-019-0221-y. [DOI] [PubMed] [Google Scholar]
- 5.Kwok W.Y., Kloppenburg M., Marshall M., et al. Comparison of clinical burden between patients with erosive hand osteoarthritis and inflammatory arthritis in symptomatic community-dwelling adults: the Keele clinical assessment studies. Rheumatology (Oxford) 2013;52:2260–2267. doi: 10.1093/rheumatology/ket267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clemente J.C., Manasson J., Scher J.U. The role of the gut microbiome in systemic inflammatory disease. BMJ. 2018;360:j5145. doi: 10.1136/bmj.j5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei J., Zhang C., Zhang Y., et al. Association between gut microbiota and symptomatic hand osteoarthritis: data from the Xiangya osteoarthritis study. Arthritis Rheumatol. 2021;73:1656–1662. doi: 10.1002/art.41729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tremaroli V., Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 9.Poretsky R., Rodriguez -R.L.M., Luo C., Tsementzi D., Konstantinidis K.T. Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS One. 2014;9 doi: 10.1371/journal.pone.0093827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei J., Zhang Y., Dalbeth N., et al. Association between gut microbiota and elevated serum urate in two independent cohorts. Arthritis Rheumatol. 2022;74:682–691. doi: 10.1002/art.42009. [DOI] [PubMed] [Google Scholar]
- 11.Kellgren J.H., Lawrence J.S. Radiological assessment of osteo-arthrosis. Ann Rheum Dis. 1957;16:494–502. doi: 10.1136/ard.16.4.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snyder E.A., Alvarez C., Golightly Y.M., Renner J.B., Jordan J.M., Nelson A.E. Incidence and progression of hand osteoarthritis in a large community-based cohort: the johnston county osteoarthritis project. Osteoarthritis Cartilage. 2020;28:446–452. doi: 10.1016/j.joca.2020.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foox J., Tighe S.W., Nicolet C.M., et al. Performance assessment of DNA sequencing platforms in the ABRF next-generation sequencing study. Nat Biotechnol. 2021;39:1129–1140. doi: 10.1038/s41587-021-01049-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li R., Yu C., Li Y., et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 15.Li D., Liu C.M., Luo R., Sadakane K., Lam T.W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 16.Almeida A., Nayfach S., Boland M., et al. A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat Biotechnol. 2021;39:105–114. doi: 10.1038/s41587-020-0603-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nearing J.T., Douglas G.M., Hayes M.G., et al. Microbiome differential abundance methods produce different results across 38 datasets. Nat Commun. 2022;13:342. doi: 10.1038/s41467-022-28034-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mallick H., Rahnavard A., Mciver L.J., et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput Biol. 2021;17 doi: 10.1371/journal.pcbi.1009442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolker B.M., Brooks M.E., Clark C.J., et al. Generalized linear mixed models: a practical guide for ecology and evolution. Trends Ecol Evol. 2009;24:127–135. doi: 10.1016/j.tree.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 21.Vujkovic-Cvijin I., Sklar J., Jiang L., Natarajan L., Knight R., Belkaid Y. Host variables confound gut microbiota studies of human disease. Nature. 2020;587:448–454. doi: 10.1038/s41586-020-2881-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dahaghin S., Bierma-Zeinstra S.M., Ginai A.Z., Pols H.A., Hazes J.M., Koes B.W. Prevalence and pattern of radiographic hand osteoarthritis and association with pain and disability (the Rotterdam study) Ann Rheum Dis. 2005;64:682–687. doi: 10.1136/ard.2004.023564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haugen I.K., Englund M., Aliabadi P., et al. Prevalence, incidence and progression of hand osteoarthritis in the general population: the framingham osteoarthritis study. Ann Rheum Dis. 2011;70:1581–1586. doi: 10.1136/ard.2011.150078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gray J.B. Applied regression analysis, linear models, and related methods. Technometrics. 1998;40:156. [Google Scholar]
- 25.Box G.E.P., Tidwell P.W. Transformation of the independent variables. Technometrics. 1962;4:531–550. [Google Scholar]
- 26.Gameroff M.J. Thirtieth Annual SAS Users Group International Conference; 2005. Using the proportional odds model for health-related outcomes : why , when , and how with various SAS ® procedures. [Google Scholar]
- 27.Liu Q., Li C., Wanga V., Shepherd B.E. Covariate-adjusted Spearman's rank correlation with probability-scale residuals. Biometrics. 2018;74:595–605. doi: 10.1111/biom.12812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B. 1995;57:289–300. [Google Scholar]
- 29.Rushing B.R., Mcritchie S., Arbeeva L., et al. Fecal metabolomics reveals products of dysregulated proteolysis and altered microbial metabolism in obesity-related osteoarthritis. Osteoarthritis Cartilage. 2022;30:81–91. doi: 10.1016/j.joca.2021.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agus A., Planchais J., Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe. 2018;23:716–724. doi: 10.1016/j.chom.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 31.Boer C.G., Radjabzadeh D., Medina-Gomez C., et al. Intestinal microbiome composition and its relation to joint pain and inflammation. Nat Commun. 2019;10:4881. doi: 10.1038/s41467-019-12873-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z., Zhu H., Jiang Q., Zhu Y.Z. The gut microbiome as non-invasive biomarkers for identifying overweight people at risk for osteoarthritis. Microb Pathog. 2021;157 doi: 10.1016/j.micpath.2021.104976. [DOI] [PubMed] [Google Scholar]
- 33.Chen J., Wang A., Wang Q. Dysbiosis of the gut microbiome is a risk factor for osteoarthritis in older female adults: a case control study. BMC Bioinf. 2021;22:299. doi: 10.1186/s12859-021-04199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Z., He Z., Kong Y., Liu Z., Gong L. Insight into osteoarthritis through integrative analysis of metabolomics and transcriptomics. Clin Chim Acta. 2020;510:323–329. doi: 10.1016/j.cca.2020.07.010. [DOI] [PubMed] [Google Scholar]
- 35.Chen C.J., Lee D.Y., Yu J., Lin Y.N., Lin T.M. Recent advances in LC-MS-based metabolomics for clinical biomarker discovery. Mass Spectrom Rev. 2022;42(6):2349–2378. doi: 10.1002/mas.21785. [DOI] [PubMed] [Google Scholar]
- 36.Gao J., Xu K., Liu H., et al. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front Cell Infect Microbiol. 2018;8:13. doi: 10.3389/fcimb.2018.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cervenka I., Agudelo L.Z., Ruas J.L. Kynurenines: tryptophan's metabolites in exercise, inflammation, and mental health. Science. 2017;357:f9794. doi: 10.1126/science.aaf9794. [DOI] [PubMed] [Google Scholar]
- 38.Modoux M., Rolhion N., Mani S., Sokol H. Tryptophan metabolism as a pharmacological target. Trends Pharmacol Sci. 2021;42:60–73. doi: 10.1016/j.tips.2020.11.006. [DOI] [PubMed] [Google Scholar]
- 39.Laursen M.F., Sakanaka M., von Burg N., et al. Bifidobacterium species associated with breastfeeding produce aromatic lactic acids in the infant gut. Nat Microbiol. 2021;6:1367–1382. doi: 10.1038/s41564-021-00970-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cervantes-Barragan L., Chai J.N., Tianero M.D., et al. Lactobacillus reuteri induces gut intraepithelial CD4(+)CD8alphaalpha(+) T cells. Science. 2017;357:806–810. doi: 10.1126/science.aah5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jennis M., Cavanaugh C.R., Leo G.C., Mabus J.R., Lenhard J., Hornby P.J. Microbiota-derived tryptophan indoles increase after gastric bypass surgery and reduce intestinal permeability in vitro and in vivo. Neuro Gastroenterol Motil. 2018;30 doi: 10.1111/nmo.13178. [DOI] [PubMed] [Google Scholar]
- 42.Kim J., Balasubramanian I., Bandyopadhyay S., et al. Lactobacillus rhamnosus GG modifies the metabolome of pathobionts in gnotobiotic mice. BMC Microbiol. 2021;21:165. doi: 10.1186/s12866-021-02178-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.