Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is causing the ongoing global pandemic associated with morbidity and mortality in humans. Although disease severity correlates with immune dysregulation, the cellular mechanisms of inflammation and pathogenesis of COVID-19 remain relatively poorly understood. Here, we used mouse-adapted SARS-CoV-2 strain MA10 to investigate the role of adaptive immune cells in disease. We found that while infected wild-type mice lost ~10% weight by 3 to 4 days postinfection, rag−/− mice lacking B and T lymphocytes did not lose weight. Infected lungs at peak weight loss revealed lower pathology scores, fewer neutrophils, and lower interleukin-6 and tumor necrosis factor–α in rag−/− mice. Mice lacking αβ T cells also had less severe weight loss, but adoptive transfer of T and B cells into rag−/− mice did not significantly change the response. Collectively, these findings suggest that while adaptive immune cells are important for clearing SARS-CoV-2 infection, this comes at the expense of increased inflammation and pathology.
The absence of B and T cells protects mice from pathogenesis by mouse-adapted SARS-CoV-2 MA10 despite elevated virus.
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of COVID-19 and is responsible for the ongoing global pandemic that has resulted in more than 0.75 billion cases and nearly 7 million deaths worldwide (1). SARS-CoV-2 infections can result in a wide range of clinical outcomes including severe lethal cases, severe nonlethal cases, mild symptomatic, or asymptomatic infections. Immune cells play an essential role in protecting the host and clearing the virus; however, they can contribute to inflammation resulting in unfavorable clinical outcomes. COVID-19 disease severity has been associated with several factors including immune dysregulation and a cytokine storm (2–5). Whereas much has been uncovered regarding the concerted effort of immune cells during COVID-19 disease, the role and contributions of key immune players remain unclear.
SARS-CoV-2 enters host cells through the human angiotensin converting enzyme-2 (hACE-2) receptor, which limits the use of established animal models for studying immunological factors that influence disease (6). Major mouse models that have shown value in understanding the infection rely on the overexpression (7, 8) or transduction (9) of the hACE-2 receptor and have varying tissue distribution and expression levels of the receptor. While valuable, the use of hACE-2 overexpression and transduction models leads to some caveats in interpreting the host responses. To circumvent this issue, the mouse-adapted virus SARS-CoV-2 MA10 has been developed (10, 11). SARS-CoV-2 MA10 uses the endogenous mouse angiotensin converting enzyme-2 (mACE-2) for infection and recapitulates a mildly symptomatic, nonlethal disease (11). A major advantage of this mouse model is that the receptor is expressed at endogenous levels in the expected tissues similar to the observed expression patterns of hACE-2. Another major advantage of the SARS-CoV-2 MA10 virus is that it allows the use of well-established genetic mouse models to study the role(s) of specific immune cells during viral infection.
Innate immune responses are paramount in mounting an effective response to SARS-CoV-2 infection. Following infection, pattern recognition receptors (PRRs) of innate resident immune cells sense the virus and activate an antiviral state characterized by the expression of type I and III interferon (IFN) and proinflammatory cytokines (4, 12). Elevated expression of chemokines such as CXCL8, CXCL9, and CXCL16 recruits a variety of immune cells, such as neutrophils, cytotoxic T lymphocytes, monocytes, natural killer cells, and others to the site of infection, resulting in increased immunopathology (4, 12). Some of the many cytokines shown to be correlative to human immunopathology are interleukin-6 (IL-6) and tumor necrosis factor–α (TNF-α) (5, 13). Investigation into the causes of immunopathology has shown that patients with severe COVID-19 have altered granulocytes characterized by increased numbers of immature neutrophils and monocytes with an inflammatory signature (3). Blockade of IFN signaling or depletion of the recruited monocytes protects against lethal disease in SARS-CoV-1 infection (12), highlighting the importance of innate immunity in disease progression.
It is well established that humoral and cellular immunity are critical in clearing SARS-CoV-2 infection (9), and while several studies have highlighted the importance of the adaptive immune system in viral clearance, more work is needed to understand its role in virus-induced immunopathology. Here, we explored the role of the adaptive immune cells in the development of immunopathology following SARS-CoV-2 infection. We show that mice lacking adaptive immune cells (rag−/− mice) do not develop clinical signs or weight loss from SARS-CoV-2 MA10 infection, whereas fully immunocompetent wild-type (WT) mice develop disease. While rag−/− mice had little to no lung pathology, virus levels were lower than WT mice early after infection, but not significantly decreased within 2 weeks of infection. Together, these results highlight the importance of the adaptive immune system not only in virus clearance (9), but also in exacerbating immunopathology in SARS-CoV-2 infection.
RESULTS
rag−/− mice do not develop severe disease following SARS-CoV-2 MA10 infection
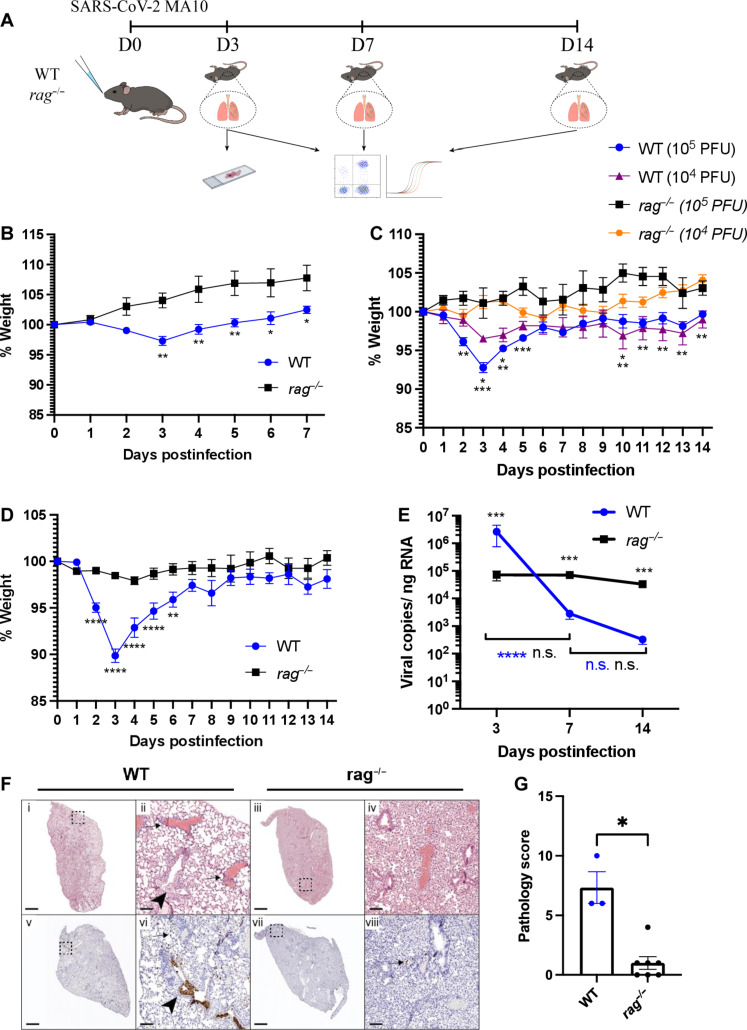
We sought to understand the role of adaptive immune cells in the pathology of SARS-CoV-2 infection using the SARS-CoV-2 MA10 virus because it can infect wild-type mice using the mACE-2 receptor (10, 11). We intranasally infected C57BL/6J WT mice and rag1−/− (rag−/−) mice with SARS-CoV-2 MA10, and weights were monitored daily until animals were euthanized at 3, 7, or 14 days postinfection (dpi) (Fig. 1A). Lungs were collected to measure viral loads using reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR), and sections were imaged after hematoxylin and eosin (H&E) or SARS-CoV-2 nucleoprotein (N) protein immunohistochemistry (IHC) staining to characterize inflammation and viral distribution, respectively. We started by infecting 3-month-old WT and rag−/− mice with 1 × 105 plaque forming units (PFU)/mouse SARS-COV-2 MA10 (Fig. 1B). We observed that infected 3-month-old WT mice lost roughly 3% of body weight at peak weight loss, but, unexpectedly, this was not observed in age-matched rag−/− mice. Next, we evaluated two doses (1 × 104 PFU/mouse and 1 × 105 PFU/mouse) of SARS-CoV-2 MA10 in mice 6 months old or older, which we anticipated would exhibit more severe disease based on prior reports (11). We found that at peak weight loss (3 dpi), 1 × 104 PFU/mouse caused WT mice to lose ~3.5% of their original weight, while rag−/− mice gained ~1% of their original weight (Fig. 1C). With 1 × 105 PFU/mouse, we found that at peak weight loss (3 dpi), WT mice lost roughly 7% of their original weight, while rag−/− mice gained roughly 1% of their original weight. On the basis of these observations, subsequent experiments used mice that were aged 6 months or older with an infectious dose of 1 × 105 PFU/mouse.
Fig. 1. SARS CoV-2 MA10 does not induce weight loss in rag−/− mice.
(A) C57BL/6J wild-type (WT) and rag−/− mice were intranasally infected with 1 × 105 PFU/mouse SARS-CoV-2 MA10 and weighed daily. Animals were euthanized at day 3, 7, or 14 after infection and lungs were collected for histology, qPCR, and flow cytometry analysis. (B) Weight change of 3-month-old WT and rag−/− mice following infection. n = 6 for each strain; the experiment was performed once. Two-way ANOVA with Bonferroni’s multiple comparisons test. (C) Weight change of 6-month-old WT and rag−/− mice following infection with 104 and 105 PFU SARS-CoV-2 MA10. WT: n = 6, rag−/−: n = 3 (104 PFU), WT: n = 7, rag−/−: n = 2 (105 PFU); the experiment was performed once. Two-way ANOVA with Tukey’s multiple comparisons test. (D) Weight change of 6-month-old WT and rag−/− mice following infection with 105 PFU SARS-CoV-2 MA10. WT: n = 19, rag−/−: n = 24; the experiment was performed three times. Two-way ANOVA with Bonferroni’s multiple comparisons test. (E) Lung viral RNA levels at days 3, 7, and 14 after infection in WT and rag−/− mice. Day 3 WT: n = 7, rag−/−: n = 10; day 7 WT: n = 8, rag−/−: n = 7; day 14 WT: n = 4, rag−/−: n = 5; the experiment was performed three times. Two-way ANOVA with Tukey’s multiple comparisons test. (F) H&E staining (top row) of WT and rag−/− mice and immunohistochemistry staining (brown stain, bottom row) against the SARS-CoV-2 MA10 nucleoprotein at day 3 after infection. Experiment was performed once. (G) Histological scoring of WT and rag−/− mice lungs at day 3 after infection. WT: n = 3, rag−/−: n = 7; the experiment was performed once. Mann-Whitney U test.
To further dissect the discrepancy in morbidity, we compared pathology in relation to viral burden in response to an infectious dose of 1 × 105 PFU/mouse. WT mice exhibited weight loss beginning at 2 dpi, with an average weight loss of 10% by 3 dpi, when peak weight loss was observed (Fig. 1D). Four days after infection, the animals started recovering clinically, and by day 7, they returned to their preinfection weight. In sharp contrast, rag−/− mice did not exhibit any significant weight loss through the 14-day experiment. It has been observed that responses to viral infection including SARS-CoV-2 MA10 can differ between mouse strains, with BALB/c mice exhibiting a more severe disease compared to C57BL/6J mice (11). Because rag−/− mice were initially derived from the 129S1/Svlmj and backcrossed to the C57BL/6j background, we investigated whether the phenotype observed in rag−/− mice could be potentially skewed by residual alleles of the 129S1/Svlmj strain. To that end, we compared C57BL/6J mice to 129S1/Svlmj mice following infection with 1 × 105 PFU/mouse SARS-CoV-2 MA10 and found no difference between the two strains (fig. S1A). Adaptive immunity, particularly humoral, has been shown to play a critical role in viral clearance (9), and rag−/− mice lack T and B cells; we investigated the ability of these animals to clear virus following infection. Rag−/− mice had lower viral RNA at 3 dpi compared to WT mice, as measured by RT-qPCR (Fig. 1E). Further, while WT mice were able to significantly reduce viral loads by 14 dpi, the viral loads of the rag−/− mice did not change significantly over 7 or 14 dpi, possibly pointing to a reduced ability of the rag−/− mice to combat infection. To better understand the differences in viral RNA between WT and rag−/− mice (Fig. 1E), we examined mACE-2 expression. Notably, at baseline, WT mice exhibited significantly higher levels of mACE-2 mRNA expression than rag−/− mice (fig. S1G). Following infection, the expression profiles underwent significant shifts, with rag−/− mice consistently displaying higher levels at both 3 and 7 dpi than WT (fig. S1, H and I).
Lung sections were also collected at 3 dpi and stained with H&E and IHC against SARS-CoV-2 N protein. Similar to the findings obtained via RT-qPCR of viral RNA, IHC showed fewer infected cells in the lungs of rag−/− mice compared to WT mice on 3 dpi (Fig. 1F). Histological scoring of lungs of infected WT and rag−/− at 3 dpi showed an increase in perivascular infiltrates in WT mice compared to rag−/− mice, indicating decreased pathology in rag−/− mice (Fig. 1, F and G).
Adenovirus-transduced human ACE-2 (hACE-2) expressing mice clear SARS-CoV-2 WA-1 infection by 7 dpi (9). We thus investigated whether WT and rag−/− mice can clear SARS-CoV-2 MA10 infection. Mice were infected and their weights were monitored up to 30 dpi. Infectious virus was measured by plaque assay and qPCR for viral RNA in the lung (fig. S1, B to D). Consistent with our previous data, weight loss was observed in WT mice but not in rag−/− mice (fig. S1B). Viral RNA was detected in the lungs of both WT and rag−/− mice, albeit at significantly higher levels in the rag−/− mice (fig. S1C). Infectious virus was detected in the lung of rag−/− mice 30 dpi (fig. S1D). Despite the high levels of infectious virus in the lungs of rag−/− mice compared to WT mice, histopathology staining revealed both perivascular and interstitial infiltrates in the lung of both WT and rag−/− mice (fig. S1E). Further, there was a lack of detection of SARS-CoV-2 nucleoprotein by IHC in WT mice compared to the detection of rare positive cells in rag−/− mice (arrows) (fig. S1E). Overall, the lungs of WT mice displayed more pathology than those of rag−/− mice (fig. S1F). Altogether, these data indicate that while rag−/− mice have less viral replication early during infection and do not clear the virus up to 30 dpi, they do not develop the severity of disease and pathology as compared to SARS-CoV-2 MA10–infected WT animals.
rag−/− mice exhibit a dampened inflammatory profile early in the infection
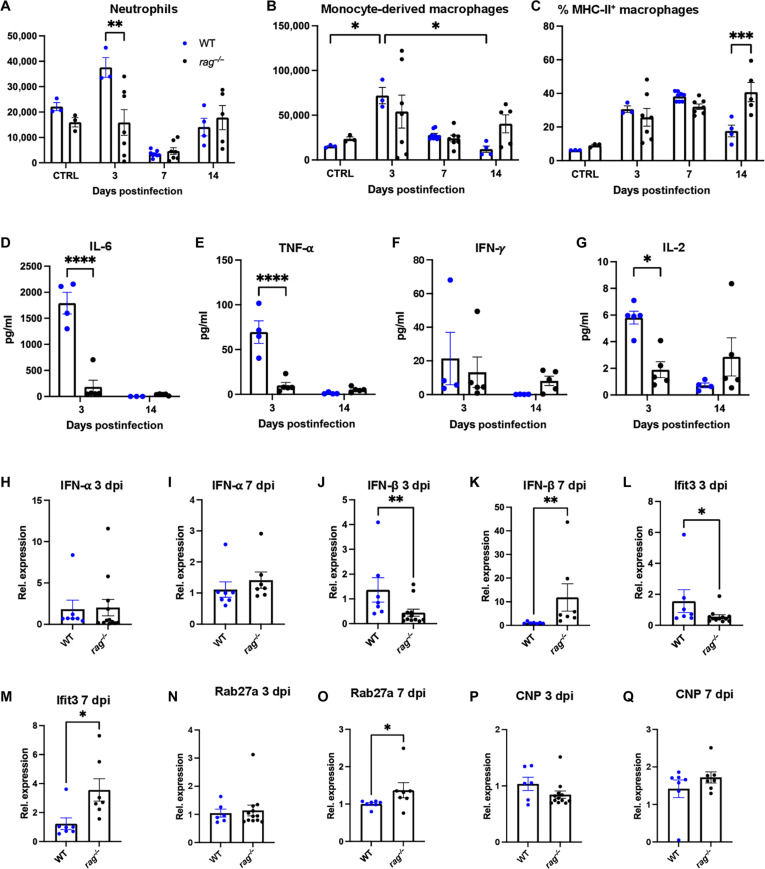
Immune cells such as neutrophils, monocytes, macrophages, and CD4+ and CD8+ T cells work together to kill infected cells, thereby helping to clear the virus (14). Dysregulation of these cells can drive immunopathology, which has been shown to contribute to disease in both fatal and nonfatal cases of SARS-CoV-2 infection (3, 15). To better understand the pathogenesis resulting in the differences in weight loss between WT and rag−/− mice (Fig. 1D), we used flow cytometry to characterize some innate immune cells in animals following infection. At 3 dpi, rag−/− mice had significantly less neutrophils in the lung compared to WT mice (Fig. 2A). At 7 and 14 dpi, the levels of neutrophils in infected WT and rag−/− mice had decreased back to ranges of uninfected control animals. Inflammatory macrophages, which infiltrate the lung as monocytes, have been shown to rapidly increase in numbers following infection, and can be an indicator of potential immunopathology (16). Between rag−/− and WT mice, the number of monocyte-derived macrophages was comparable throughout the experiment (Fig. 2B). When compared to uninfected animals, at 3 dpi WT mice exhibited a significant increase in the number of monocyte-derived macrophages, and then a decrease by 14 dpi, loosely correlating with their relative increase and then their decrease of virus (Figs. 2B and 1E). In contrast, no significant changes in monocyte-derived macrophage numbers were observed in rag−/− mice across the duration of the experiment. Next, we investigated the activation status of the monocyte-derived macrophages by comparing the percent MHC-II+ monocyte-derived macrophages across genotypes. The percentage of activated monocyte-derived macrophages increased at comparable levels in both genotypes at 3 and 7 dpi. However, at 14 dpi, rag−/− mice exhibited significantly more activated monocyte-derived macrophages as compared to WT mice (Fig. 2C), consistent with a higher viral load in rag−/− mice at 14 dpi (Fig. 1E).
Fig. 2. rag−/− mice do not exhibit a heightened proinflammatory cytokine expression at peak weight loss.
(A to C) Flow cytometry analysis of lung cells 3, 7, and 14 dpi. (A) Number of lung neutrophils (CD11b+, MHC-II−, and Ly6G+ cells). (B) Number of monocyte-derived macrophage (CD11b+, CD11c+, and F4/80+ cells). (C) Percentage of MHC-II+ monocyte-derived macrophage (CD11b+, CD11c+, F4/80+, and MHC-II+ cells) in the lungs of wild-type (WT) and rag−/− mice at days 3, 7, and 14 after infection. WT control: n = 3, D3: n = 3, D7: n = 8, D14: n = 3; rag−/− control: n = 3, D3: n = 7, D7: n = 7, D14: n = 5; the experiment was performed once for each time point. Two-way ANOVA with Tukey’s multiple comparisons test. (D to G) Cytokine concentrations from the bronchoalveolar lavage fluid at days 3 and 14 after infection. (D) IL-6, (E) TNF-α, (F) IFN-γ, and (G) IL-2. WT D3: n = 4, D14: n = 3; rag−/− D3: n = 5, D14: n = 5, experiment performed once for each time point. Two-way ANOVA with Sidak’s multiple comparisons test. (H to M) mRNA levels from lung samples of infected animals at days 3 and 7 after infection. (H) Relative expression of IFN-α at 3 dpi WT: n = 7, rag−/−: n = 12. (I) Relative expression of IFN-α at 7 dpi. WT: n = 7, rag−/−: n = 7. (J) Relative expression of IFN-β at 3 dpi. WT: n = 7, rag−/−: n = 12. (K) Relative expression of IFN-β at 7 dpi. WT: n = 7, rag−/−: n = 7. (L) Relative expression of IFIT3 at 3 dpi. WT: n = 7, rag−/−: n = 12. (M) Relative expression of IFIT3 at 7 dpi. WT: n = 7, rag−/−: n = 7. (N) Relative expression of Rab27a 3 dpi. WT: n = 6, rag−/−: n = 12. (O) Relative expression of Rab27a 7 dpi. WT: n = 7, rag−/−: n = 7. (P) Relative expression of CNP 3 dpi. WT: n = 6, rag−/−: n = 12. (Q) Relative expression of CNP 7 dpi. WT: n = 6, rag−/−: n = 12. (H to M) *P < 0.05, **P < 0.01, two-tailed Mann-Whitney test.
In addition to cellular profiles, we sought to investigate the inflammatory cytokine microenvironment that could be causing the differences in morbidity between WT and rag−/− mice. Several studies have shown that cytokine release syndrome (CRS) might be the main contributor of severe cases of SARS-CoV-2, and in some cases, this could result in mortality (5, 17). Multiple cytokines have been implicated in this process for coronaviral infections, including IL-6, TNF-α, and IFN-γ (4, 5, 18). Therefore, we investigated whether these cytokines were differently expressed between WT and rag−/− mice during infection. We collected bronchial alveolar lavage fluid (BALF) from mice at 3 and 14 dpi to assay cytokine levels. Notably, the levels of IL-6 were ~9.7 times lower in rag−/− mice compared to WT mice at 3 dpi (Fig. 2D). By 14 dpi, IL-6 levels had decreased significantly in WT mice and were similar to those of rag−/− mice. Similarly, the levels of TNF-α were seven times lower in rag−/− mice at 3 dpi as compared to those of WT mice, which later decreased and became comparable to rag−/− mice by 14 dpi (Fig. 2E). IFN-γ levels were comparable between rag−/− and WT mice at 3 dpi, and trended higher in rag−/− mice at 14 dpi, albeit not significantly (Fig. 2F). Last, we examined the levels of IL-2, an important cytokine in driving T cell proliferation (19, 20). Rag−/− mice had ~3 times lower levels of BALF IL-2 at 3 dpi, and at 14 dpi, IL-2 levels had significantly decreased in WT mice to levels comparable between WT and rag−/− mice (Fig. 2G).
Considering the critical role of IFN responses, particularly type I IFNs and IFN-stimulated genes (ISGs), in the control of viral infections, we assessed their expression levels at both 3 and 7 dpi within the lung WT and rag−/− mice. We first compared the levels of IFN-α mRNA in WT and rag−/− mice, and they were comparable at 3 and 7 dpi (Fig. 2, H and I). Conversely, we noted a significant increase in IFN-β mRNA levels in WT mice compared to rag−/− mice at 3 dpi (Fig. 2J), which aligns with the higher levels of viral RNA detected in WT mice 3 dpi (Fig. 1E). In contrast, at 7 dpi, we observed a reversal of this trend, with rag−/− mice exhibiting significantly elevated IFN-β levels compared to WT mice (Fig. 2K), and this was consistent with the higher levels of viral RNA detected in rag−/− mice compared to WT mice at 7 dpi (Fig. 1E). Furthermore, we assessed the levels of the ISGs IFN-induced protein with tetratricopeptide repeats 3 (IFIT3), the Ras-related Protein Rab27, and 2′,3′-cyclic nucleotide 3′-phosphodiesterase (CNP). At 3 dpi, WT mice exhibited significantly higher levels of IFIT3 compared to rag−/− mice (Fig. 2L). Conversely, at 7 dpi, rag−/− mice had a substantial increase in IFIT3 expression compared to WT mice (Fig. 2M). Rab27a levels were comparable between WT and rag−/− mice 3 dpi, but were significantly higher in the rag−/− at 7 dpi (Fig. 2, N and O). Comparable levels of CNP were observed in both WT and rag−/− mice at both 3 and 7 dpi (Fig. 2, P and Q). Overall, immunological analysis indicates elevated lung neutrophils and proinflammatory cytokines IL-6, TNF-α, and IL-2 in WT mice compared to rag−/− mice during their peak weight loss after infection. In addition, increased levels of IFN-β and IFIT3 were observed in both WT and rag−/− mice and correlated with high viral RNA in the lung.
tcrα−/− mice lacking αβ T cells display less severe disease compared to WT mice
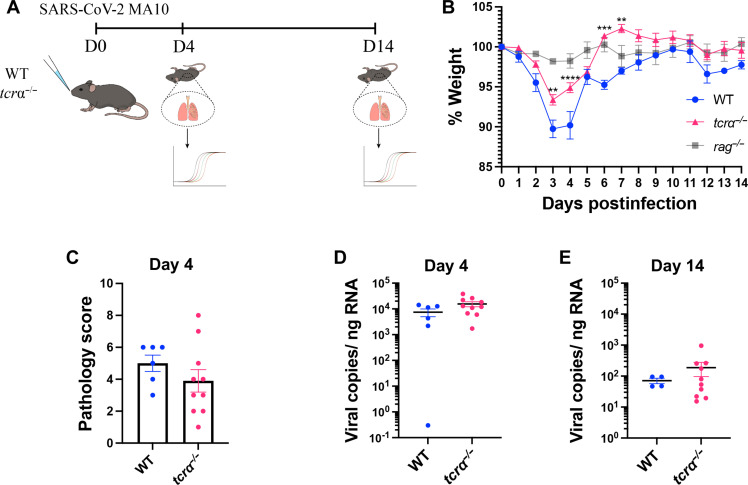
After observing that lymphocytes had a major role in SARS-CoV-2 MA10 pathology of WT mice compared to rag−/− mice, we wanted to isolate potential contributions from key cellular player(s). T helper 1 (TH1) cells and cytotoxic CD8+ T cells are important αβ T cell subsets involved in the response to virus infection that express high levels of the antiviral effector such as IFN-γ, perforin, and granzymes (21–23). We thus chose tcrα−/− mice that genetically lack αβ T cells to investigate their role in SARS-CoV-2 pathology. We infected 6-month-old WT and tcrα−/− mice (24) with SARS-CoV-2 MA10, and subsequently monitored weight loss and collected lungs to analyze pathology and virus load at 4 and 14 dpi (Fig. 3A). Tcrα−/− mice lost some, but not as much weight as WT mice, and a similar trend was observed in 3-month-old mice (fig. S1J). In addition, the tcrα−/− mice recovered their weight faster, such that a significant increase in weight was observed for the tcrα−/− mice at 6 and 7 dpi compared to the WT mice (Fig. 3B). While their weights were different, their lung pathology scores on 4 dpi were not statistically different (Fig. 3C). Similarly, their viral RNA showed similar levels at 4 and 14 dpi (Fig. 3, D and E). Unlike the rag−/− mice, which lacked the ability to reduce the viral RNA levels at 14 dpi (Fig. 1E), the tcrα−/− mice were capable of reducing viral RNA levels equal to WT mice (Fig. 3, D and E). In addition to viral RNA levels, we also assessed whether tcrα−/− mice could clear infection. Thus, we compared infectious virus in the lungs of 3-month-old WT and tcrα−/− mice at 14 dpi. Viral loads were below the limit of detection in 8/10 of the tcrα−/− mice with 2/10 mice testing positive (fig. S1K). Infectious virus was below the limit of detection in 8/8 of the WT mice.
Fig. 3. SARS-CoV-2 MA10 causes a less severe disease in tcrα−/− mice compared to wild-type (WT).
(A) WT and tcrα−/− mice were infected with 1 × 105 PFU/mouse SARS-CoV-2 MA10. On day 4 and 14 after infection, lungs were harvested, and virus RNA was measured by RT-qPCR. (B) WT and tcrα−/− mice weight change following SARS-CoV-2 MA10 infection. WT: n = 10, tcrα−/−: n = 20; the experiment was performed twice. Two-way ANOVA with Sidak’s multiple comparisons test. (Note that the weight loss curve of rag−/− infection from the experiment shown in Fig. 4B was overlaid for comparison purposes only, but not statistical analysis). (C) Pathology scores of WT and tcrα−/− mice lung sections at day 4 after infection (WT: n = 6, tcrα−/−: n = 10); the experiment was performed once. (D) Viral RNA levels on day 4 (WT: n = 6, tcrα−/−: n = 10); the experiment was performed once. (E) Day 14 after infection (WT: n = 4, tcrα−/−: n = 10); experiments were performed once. Two-tailed Mann-Whitney U test.
To gain a better understanding of the role of T cells and potential immunomodulatory regulators in SARS-CoV-2 MA10–induced pathology, we utilized mice lacking the Tec family kinase interleukin 2 inducible kinase (ITK), a crucial signaling kinase downstream of the T cell receptor (TCR), which has altered T cell activation responses (25, 26). To this end, we infected WT, rag−/−, and itk−/− mice with 105 PFU of SARS-CoV-2 MA10 and monitored their weights over a 14-day period. Similar to the observations in tcrα−/− mice, itk−/− mice displayed an intermediate phenotype, suggesting that efficient T cell activation as regulated by ITK plays a role in regulation of SARS-CoV-2 MA10–induced pathology (fig. S1L). In addition, we detected significantly higher levels of viral RNA in the lungs of rag−/− mice compared to both WT and itk−/− mice, but no significant difference was observed between WT and itk−/− mice (fig. S1M). Altogether, these results suggest that αβ T cells are at least partially responsible for immunopathogenesis of this infection, but they do not act alone or are not fully responsible for the full phenotypic difference between WT and rag−/− mice.
Adoptive transfer of splenocytes into rag−/− mice does not significantly affect disease progression
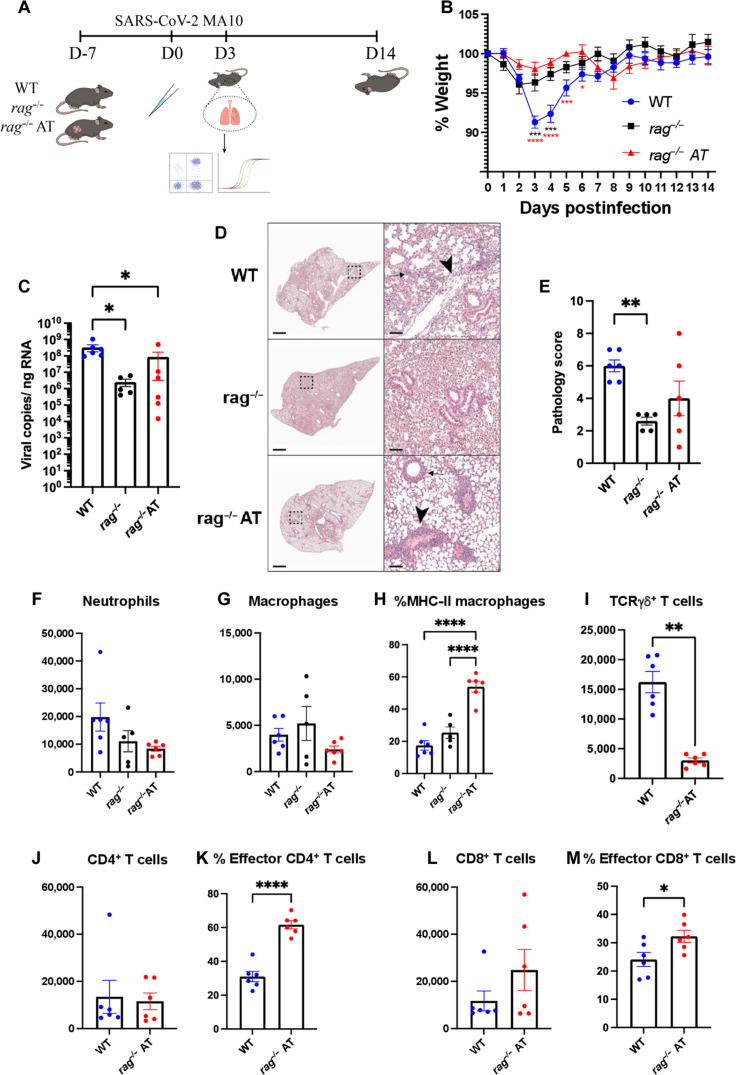
Adaptive immune cells interact with innate immune cells and can influence their differentiation, function, and migration (27, 28). The dysregulation of the innate and adaptive immune systems following SARS-CoV-2 infection has been reported to contribute to immunopathology (29, 30). To investigate whether the introduction of circulating lymphocytes into rag−/− mice could recapitulate the weight loss observed in WT mice following infection, we collected splenocytes from 6-month-old WT mice and adoptively transferred 5 × 107 cells via retroorbital injection 7 days before infection. As controls, WT and rag−/− mice received retroorbital phosphate-buffered saline (PBS) injections before infection. We infected the animals with 1 × 105 PFU/mouse of SARS-CoV-2 MA10 and monitored their weight for 14 days. We then euthanized animals at 3 or 14 dpi (fig. S3A), and analyzed the pathology, immune cells, and viral loads in the lungs (Fig. 4A). Consistent with our previous observations, we found that at 3 dpi, WT mice lost significantly more weight than rag−/− mice following infections. However, there was no difference in weight loss between the rag−/− animals that received adoptively transferred WT splenocytes (rag−/− AT) and the rag−/− animals (Fig. 4B). Furthermore, both rag−/− and rag−/− AT mice had significantly less viral RNA in their lungs at 3 dpi compared to WT mice, and no significant differences were observed between rag−/− and rag−/− AT mice (Fig. 4C). Pathology scores again indicated that rag−/− AT mice did not exhibit the severity of disease as experienced by WT mice (Fig. 4, D and E), although at intermediate levels, the rag−/− AT scores were not significantly different from either of the other two groups. Of note, only WT mice had sloughed or necrotic epithelial cells in bronchioles, while rag−/− and rag−/− AT mice did not (Fig. 4D).
Fig. 4. Adoptive transfer of wild-type (WT) splenocytes into rag−/− mice does not induce weight loss following SARS-CoV-2 MA10 infection.
(A) Phosphate-buffered saline (PBS) or WT splenocytes were adoptively transferred into WT or rag−/− mice (rag−/− AT), respectively, at least 1 day before infection. On day 0, the animals were infected with 1 × 105 PFU/mouse SARS-CoV-2 MA10 and weight was monitored daily. On day 3 after infection, the animals were euthanized, and lungs were collected for flow cytometry, qPCR, and histology. (B) Percent weight change in WT, rag−/−, and rag−/− adoptive transfer (AT) mice following infection. WT: n = 8, rag−/−: n = 4, rag−/− AT: n = 6; experiments were performed four times. Two-way ANOVA with Tukey’s multiple comparisons test. (C) Viral RNA levels from the lungs of infected animals at day 3 after infection; experiments were performed once. One-way ANOVA with Dunn’s multiple comparisons test. (D) H&E staining of lung sections; experiments were performed once. (E) Pathological scoring of the lung sections; experiments were performed once. Two-tailed Mann-Whitney test. (F to M) Flow cytometry of lung cells from day 3 after infection; experiments were performed once. (F) Number of neutrophils (CD11b+ and Ly6Ghigh). (G) Number of monocyte-derived macrophage (CD11b+, Ly6G−, CD11C+, and F4/80+). (H) Percentage of MHC-II+ monocyte-derived macrophages. For (F) to (H), one-way ANOVA with Tukey’s multiple comparisons test. (I) Number of γδ+ T cells. Two-tailed Mann-Whitney test. (J) CD4+ T cells (TCRβ+ and CD4+). (K) Percentage of effector CD4+ T cells (TCRβ+, CD4+, CD62L−, and CD44+). (L) Number of CD8+ T cells (TCRβ+ and CD8+). (M) Percentage of effector CD8+ T cells (TCRβ+, CD8+, CD62L−, and CD44+). For (J) to (M), two-tailed t test. For (C) to (M), WT: n = 6, rag−/−: n = 5, rag−/− AT: n = 6.
Because the adoptive transfer of WT splenocytes into rag−/− mice did not fully recapitulate the disease of WT mice, we investigated the numbers and status of the immune cells at 3 dpi to better understand their behavior following transfer and virus infection. rag−/− and rag−/− AT mice had roughly half as many pulmonary neutrophils compared to WT mice (Fig. 4F), and monocyte-derived macrophage numbers were comparable among all groups (Fig. 4G). Rag−/− AT mice had significantly higher proportion of activated (MHC-II+) monocyte-derived macrophages compared to rag−/− and WT mice, although this parameter was not different between rag−/− and WT mice (Fig. 4H). Because of the lack of weight loss and higher proportions of activated monocyte-derived macrophages in the lungs of rag−/− AT mice, it is possible that activated monocyte-derived macrophages help in virus clearance but do not significantly contribute to immunopathology. Whereas neutrophils and macrophages are present in rag−/− mice, γδ T cells are another important type of innate-like cell that acts quickly in response to mucosal pathogens, which are also absent in rag−/− mice. However, these cells often are long-term resident cells of the lung that establish themselves in their niches early in life to be present for acute responses to pathogens (31, 32). Not unexpectedly, these cells were not restored to WT levels after adoptive transfer plus 3 dpi (Fig. 4I). These resident lymphocytes are an important group of immune cells that have been linked to COVID-19 pathogenesis (33–35), and their absence may shed light on the inability of the adoptive transfer to recapitulate WT disease. γδ T cell numbers were similar to WT mice by 14 dpi (fig. S3B).
In contrast to the γδ T cells, we found that WT and rag−/− AT mice had comparable percentages and numbers of CD4+ and CD8+ T cells within the lungs at 3 dpi (Fig 4, J and L) and at 14 dpi (fig. S3, C and E). Using CD44, a cell adhesion molecule, the expression of which has been shown to identify antigen-experienced T cells (36), we further identified the effector CD4+ and CD8+ T cells. The rag−/− AT mice had a significantly higher percentage of effector CD4+ and CD8+ T cells than WT mice at 3 dpi (Fig. 4, K and M), and similar trends were observed at 14 dpi (fig. S3, D and F). Whether they established an effector phenotype during homeostatic proliferation or during infection, or both, is an area that requires further investigation.
We next investigated the potential role of tissue-resident adaptive immune cells in SARS-CoV-2 MA10–induced pathology. Thus, we blocked B and T cell exit from secondary lymphoid organs using the well-characterized sphingosine-1-phosphate receptor modulator FTY720 (37). WT mice were treated daily with either FTY720 (1 mg/kg) or vehicle control starting on day −2 and infected on day 0 with 1 × 105 PFU/mouse SARS-CoV-2 MA10. To confirm the action of FTY720, analysis of adaptive immune infiltrates in the lungs revealed a significant decrease in αβ+ CD4+ and CD8+ T cells and B cells in FTY720-treated animals compared to vehicle-treated mice (fig. S4, A to D). However, no difference in weight loss was observed between FTY720- and vehicle-treated mice (fig. S4E). To further elucidate the role of circulating T cells in the observed pathology in WT mice, we used T cell–depleting antibodies to deplete circulating T cells (38). WT mice were administered 400 μg of α-CD4 and 400 μg of α-CD8 antibodies every other day, or 800 μg of isotype control, starting from day −2 until 6 dpi. These mice were infected with 105 PFU/mouse of SARS-CoV-2 MA10 on day 0 and subjected to daily monitoring. At 7 dpi, the animals were euthanized, and lung T cells were analyzed via flow cytometry. Comparatively, both the number and frequency of CD4+ and CD8+ T cells in the lungs were markedly lower in mice treated with α-CD4 and α-CD8 antibodies, as opposed to isotype control-treated mice (fig. S4, F to I). However, despite these differences in T cell numbers and frequencies, both the T cell–depleted and isotype control mice exhibited similar weight loss, with T cell–depleted mice displaying a slightly lower average loss compared to the isotype control (fig. S4J). These data suggest that neither the blockade of tissue-infiltrating adaptive immune cells to the site of infections nor the depletion of circulating T cells was sufficient to alter the phenotype observed in WT mice, supporting a role for tissue-resident immune cells in SARS-CoV-2 MA10–induced pathology.
DISCUSSION
The balance between immune defense from pathogens and development of immunopathology is complex and multifaceted (39). Efforts to understand the role of the adaptive immune system in driving disease during viral infection has been difficult because of the importance of the adaptive immunity in controlling virus infection (9, 40, 41). It is well established that immunocompromised animals and humans are at high risk of severe viral infections, such as SARS-CoV-2 (9, 40, 42, 43), and thus, our finding that rag−/− mice lacking B and T cells are protected from viral-induced weight loss and exhibit reduced lung inflammation, while carrying the same or higher levels of virus, was a surprise. These findings suggest that adaptive immune cells play an important role in reducing SARS-CoV-2 levels, but that this benefit may come at the expense of increased inflammation and pathology.
Our findings suggest an important role for the adaptive immune response, particularly likely resident lymphocytes given our results after blocking lymphocyte recirculation and CD4+ and CD8+ T cell depletion, in causing inflammation that contributes to weight loss and lung pathology during SARS-CoV-2 infection. As a trade-off for minimal symptoms during SARS-CoV-2 MA10 infection, rag−/− mice were not able to clear virus, as well as the WT mice (through 30 dpi), supporting other studies reporting the importance of an effective adaptive immune response in virus clearance (9, 44). Despite not decreasing virus levels, rag−/− mice were still able to limit virus replication somewhat, as indicated by the limited virus expansion in the lungs through 30 dpi, likely via innate immune cell responses. Consistent with this proposal that innate cells are able to contribute to limiting infection, Nelson et al. (45) reported that rhesus macaques infected with SARS-CoV-2 exhibit decreased virus loads before the detection of infiltrating antigen-specific T and B cells, thus highlighting a role for innate immunity in contributing to controlling virus levels.
Cytokines are generated by both innate and adaptive immune cells, and have been implicated in both favorable and unfavorable outcomes during virus infection (3, 5, 12, 18, 46). Del Valle et al. (5) found that elevated IL-6 and TNF-α were strong predictors of COVID-19 severity, worse outcomes, and poor survival. IL-6 and TNF-α levels were significantly elevated in the BALF of WT mice compared to rag−/− mice at 3 dpi, coinciding with peak weight loss, making it likely that these cytokines were contributing to the disease. In addition, IL-2 levels were more elevated in WT mice than in rag−/− mice, and this difference could be explained by the lack of IL-2–expressing cells, such as activated T cells, in the rag−/− mice (47, 48). It could be possible that the increased IL-2 levels in WT mice causes pathogenesis by feeding IL-2 receptor–expressing cells, as evidenced by a recent study in which patients with exacerbated SARS-CoV-2 infection had high expression of soluble IL-2 receptor (18). In addition, elevated IFN and ISG gene expression correlated with an increase in viral RNA in the lungs of WT and rag−/− mice.
Multiple studies have thus far highlighted the immune dysregulation in animal models and patients with COVID-19 as a correlate of immune-induced pathology (4, 5, 18). Granulocyte dysregulation has been correlated with exacerbated disease and unfavorable outcomes in SARS-CoV-2 infection (3). We find that rag−/− mice had less lung-infiltrating neutrophils than WT mice at peak weight loss, which warrants further investigation into these cells potentially driving immunopathogenesis and being attractive therapeutic targets. Macrophages and monocytes have been shown to be important in both virus clearance and in resolving inflammation (49–51), but when dysregulated, they can drive inflammation, resulting in tissue damage (52, 53). It has also been shown that macrophages can be infected by SARS-CoV-2 via the FcγR, and this leads to the activation of the inflammasome resulting in pyroptosis (52, 53). In our model, we did not see significant differences in CD11b+ CD11c+ F4/80+ monocyte-derived macrophages at peak weight loss or at disease resolution. However, we did see comparable increases in activated monocyte-derived macrophages in both rag−/− and WT mice at peak weight loss, highlighting the ability of both animal models to respond to infection with acute inflammation.
We found that the adoptive transfer of splenocytes into the rag−/− mice and subsequent infection with SARS-CoV-2 MA10 did not recapitulate the level of disease observed in WT mice. Instead, recipient mice responded similarly to rag−/− mice, or at best had an intermediate phenotype, with significantly less virus than WT mice at 3 dpi. While adoptive transfer experiments can sometimes help to elucidate the role of immune cells, there are some caveats such as the transferred cells may not always completely restore WT immunity. For example, short-term transferred cells are less likely to be able to affect the development or differentiation of innate immune cells, or establish niches as resident cells in peripheral tissues, which can begin during fetal development (54). We also noted that while the T cell compartment in the recipient rag−/− AT and WT mice showed comparable T cell numbers in the lungs at 3 dpi, the rag−/− AT mice had a significantly higher proportion of CD44+ antigen-experienced CD4+ and CD8+ T cells compared to the WT mice. This could be due to the transfer of the cells, as well as the proliferative nature of immune cells entering a lymphopenic environment (55). The absence of significant weight loss in the infected rag−/− AT mice suggests that the antigen-experienced CD4+ and CD8+ T cells observed at 3 dpi are not sufficient to drive immunopathology, although further work is needed to determine the activation kinetics of these cells before 3 dpi. We posit that disease in the WT mice may be driven by the interaction of innate and long-term resident lymphocytes, such as γδ T cells that have had time to establish a particular microenvironment in the lung. Genetic deletion of specific cells or antibody depletion experiments may better probe the role of the adaptive immune system in the pathology of SARS-CoV-2. To this end, we showed that tcrα−/− mice had an intermediate disease phenotype compared to WT and rag−/− mice. While tcrα−/− mice are not able to develop αβ T cells, they do have γδ T cells and B cells (24), which may play roles in the observed weight loss. In addition, it is possible that mechanisms such as antibody-FcγR–mediated infection of macrophages and monocytes, which has been shown to induce inflammation (53, 56), are at play in the tcrα−/− mice, thus driving the intermediate phenotype observed. While αβ T cells do play a partial role in the observed disease, they are not necessary for limiting virus replication, as evidenced by comparable viral RNA levels in WT and tcrα−/− mice at 4 and 14 dpi. Because viral RNA has been shown to persist longer than infectious virus, and C57BL/6J mice can clear SARS-CoV-2 MA10 as early as 5 dpi, it is highly likely that both WT and tcrα−/− mice have successfully cleared the virus by 14 dpi, despite testing positive for viral RNA (11, 57). In addition, we observed no significant difference between 3-month-old WT and tcrα−/− mice in their ability to clear virus, further supporting the notion that αβT cells may not be necessary for SARS-CoV-2 MA10 clearance.
Overall, we have shown that after SARS-CoV-2 MA10 infection, animals lacking an adaptive immune system had lower morbidity than those with a competent adaptive immune system. Transient transfer of immune cells into the immunocompromised animals was not sufficient to induce the pathology observed in WT animals, implicating a role for lung-resident immune cells, including T cells in driving disease. Moreover, rag−/− mice exhibited an attenuated proinflammatory signature compared to WT mice while still limiting virus replication. We present a model that proposes new questions for understanding the role of adaptive immunity in driving COVID-19 pathology, and future studies will seek to uncover the mechanism and pathways by which the adaptive immune cells drive COVID-19 disease. These findings may, in turn, have important implications for other coronavirus-induced infectious diseases.
MATERIALS AND METHODS
Mice and infections
Animal studies were performed following the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Protocols were approved by the Institutional Animal Care and Use Committee at Cornell University. Most of the mice used for the study were on the C57BL/6J background: WT (C57BL/6J) mice, rag1−/− (B6.129S7-Rag1tm1mom/J) mice, tcrα−/− (B6.129S2-Tcratm1mom/J) mice, and itk−/− (B6.129S4-itktm1Litt/PlsJ) mice; one experiment used 129S1/Svlmj mice. All mice were bred and aged in our facility; female and male mice at least 24 weeks old were used for all experiments, unless otherwise noted. Mice were intranasally infected with 1 × 105 PFU/mouse SARS-COV-2 MA-10 under isoflurane anesthesia in an approved animal-BSL-3 facility as previously described (11, 58). Weights were monitored daily, and lung tissue was collected at the indicated time points for downstream analysis. FTY720 (1 mg/kg; Sigma-Aldrich catalog no. SML0700) was prepared per the manufacturer’s recommendations for daily injections.
Virus propagation
Vero E6 cells [African green monkey kidney cells, American Type Culture Collection (ATCC) CRL-1586] were seeded in a T75 flask and infected with 0.1 multiplicity of infection. Infected cells were incubated for 48 hours and supernatant was collected. Supernatants were centrifuged at 1000g for 10 min and virus was transferred into a new tube. The virus was aliquoted and stored and −80°C for titration by plaque assay and mouse infection.
Antibodies and reagents
Antibodies used for flow cytometry are listed in this order: antigen fluorophore (catalog number). CD170 PE (552126), CD117 FITC (553354), and KLRG1 PE-CF594 (565393) were from BD Bioscience. CD16/32 (Fc block 14-0161-85), viability ef506 (65-0866-14), MHC-II AF700 (56-5321-80), CD62L APC (17-0621-83), CD4 AF700 (56-0041-81), CD8 ef450 (48-0081-82), and TCR gamma/delta Super Bright 780 (78-5711-80) were from Invitrogen. CD11b PE-Dazzle594 (101256), CD49b PECy7 (103518), CD11c APC (117310), F4/80 APC-Cy7 (123118), Ly6G Pacific Blue (127612), CD19 FITC (101505), CD127 PerCp-Cy5.5 (135022), TCR-β PE-Cy7 (109222), and CD44 APC-Cy7 (103028) were from BioLegend. Antibodies were used in accordance with the manufacturer’s guidelines or at 1 to 2 μg/ml.
Flow cytometry
Briefly, mice were euthanized with carbon dioxide in accordance with approved protocols. Lungs were collected, digested with Liberase TL and DNase I, and homogenized to create single-cell suspension. Cells were then stained, washed, and fixed in 3% paraformaldehyde (PFA) for 15 min. After fixation, cells were run on an Attune NxT cytometer (Thermo Fisher Scientific). Gating (fig. S2) and downstream analysis was performed using the FlowJo software (BD Biosciences).
Splenocyte isolation and adoptive transfer
Spleens were harvested from WT mice and homogenized to obtain a single-cell suspension. Red blood cells were lysed using ACK buffer, and total splenocytes were counted and adjusted to 5 × 107 cells/200 μl of PBS. Mice were anesthetized and retro-orbitally injected with 5 × 107 splenocytes.
Histology and IHC
Lung tissue was collected, fixed in 10% neutral buffered formalin, and submitted to the Cornell University Animal Health Diagnostic Center for sectioning and staining. Tissue sections (4 μm) were stained with H&E and examined by a board-certified veterinary anatomic pathologist blinded to group categories. Lung pathology was scored based on the percentage of each feature affected across the entire lung section. The features scored were perivascular infiltrates, interstitial infiltrates, presence of cells in alveoli lumina, and type II pneumocyte hyperplasia, the presence of sloughed or necrotic cells in bronchiolar lumina. Each point corresponded to the amount of tissue affected: normal (0); less than 10% (1); between 10 and 25% (2); 26 to 50% (3); and higher than 50% (4). As described previously, IHC was performed on lung tissue with an anti–SARS-CoV-2 nucleoprotein antibody (GeneTex: GTX635679) at a 1:5000 dilution (58).
qPCR and viral RNA quantification
Lung tissues were collected from mice at the indicated endpoints. Tissue samples were dissociated and inactivated in TRIzol, followed by RNA extraction, total RNA quantification, and RT-qPCR using the iTaq Universal Probes One-Step Kit (Bio-Rad) with the following primers and probe from IDT DNA: TGGCCGCAAATTGCACAATT;TGTAGGTCAACCACGTTCCC; /56FAM/CGCATTGGCATGGAAGTCAC/3BHQ_1/ (59). Standard curves of plasmid (IDT DNA: catalog no. 10006625) were run on each assay plate to calculate virus copy number per sample and normalized to total RNA for each sample. The following proprietary primer/probes were purchased from Thermo Fisher Scientific catalog no. 4453320: Cnp (Mm01306641_m1), Rab27a (Mm00469997_m1), mACE2 (Mm01159006_m1), Ifnb1 (Mm00439552_s1), Ifna1 (Mm03030145_gH), Ifit3 (Mm01704846_s1), and Actb (Mm00607939_s1). Relative mRNA quantities were normalized to β actin and presented as 2−ΔΔCt. Plates were run on ViiA7 and analyzed with QuantStudio Real-Time PCR Software (Thermo Fisher Scientific).
Multiplex cytokine assay
Bronchoalveolar lavage washes were performed with 1 ml of 1× PBS immediately after euthanasia, cells were removed via centrifugation, and supernatants were run on the High Sensitivity 5-Plex Mouse ProcartaPlex Panel (ThermoFisher Scientific) per the manufacturer’s instructions, with the addition of a 15-min 3% PFA fixation and wash step after two washes with the Amplification Reagent, before the final Reading Buffer was added. The plate was read on Luminex MagPix and analyzed with xPONENT software (Luminex).
T cell depletion and analysis
α-CD4-clone GK1.5 (catalog no. BE0003-1), α-CD8-clone 2.43 (catalog no. BE0061), and isotype control-clone LTF2 (catalog no. BE0090) depletion antibodies were purchased from Bio X Cell Inc. C57BL6/J WT mice were treated every other day, starting at −2 dpi, with 400 μg each of α-CD4 and α-CD8 antibodies, or 800 μg of isotype control, until 6 dpi. On day 7 after infection, mice were humanely euthanized, and T cells were analyzed by flow cytometry using α-CD4-clone RM4-5 (BD Pharmingen) and α-CD8-clone 53-6.7 (eBioscience) antibodies.
Plaque assay
Lung tissues were collected and homogenized in Dulbecco’s modified Eagle’s medium (DMEM) with 2% fetal bovine serum (FBS). Supernatants were purified by centrifugation and used for plaque assay. Vero E6 cells (African green monkey kidney cells, ATCC CRL-1586) were cultured at a density of 5 × 105 cells per well in 12-well plates and incubated at 37°C before infection the following day. Supernatants were serially diluted in DMEM supplemented with 2% FBS, and cells were infected and incubated for 1 hour at 37°C with gentle shaking every 10 min. Infected cells were overlaid with 1 ml per well of DMEM containing 2% FBS and 0.3% agarose, and cells were incubated at 37°C for 3 days. At 3 dpi, cells were fixed with 4% PFA, stained with 0.5% crystal violet in 30% methanol, and washed three times with water. Plaques were manually counted, and the virus titer was calculated.
Data analysis
Data were analyzed with the statistical test indicated in each figure legend using GraphPad PRISM 9 software. The most common statistical tests used were Mann-Whitney U test or two-way analysis of variance (ANOVA) with the appropriate and indicated posttest. All data are graphed as mean ± SEM. Asterisks indicate statisitcal significance from the listed analysis: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, or no markings for nonsignificant differences.
Acknowledgments
We thank A. Redko and Cornell animal care staff for maintaining mouse colonies; members of the Cornell BSL-3, ABSL-3, and Cornell Biosafety staff including but not limited to P. Jennette, N. Kushner, J. Turse, S. Mattoon, D. Miller, H. B. Roman, B. Singh, D. G. Collin, and T. L. Van Deusen; and members of the Cornell flow Cytometry Core.
Funding: This work was supported by the National Institutes of Health grants AI120701, AI138570, AI129422, and GM125598 to A.A.; the National Institutes of Health grant AI109022; Emergent Ventures at Mercatus Center, George Mason University Fast Grant no. 2103; and the Cornell Seed Grants to H.C.A.
Author contributions: Conceptualization: B.I., J.M.S., H.C.A., and A.A. Methodology: B.I., J.M.S., B.D.R., J.L., and S.D. Investigation: B.I., J.M.S., D.W.B., S.E., I.A.M., M.J., A.C., and R.A.A. Visualization: B.I. and H.B. Supervision: A.A., H.C.A., and G.W. Writing—original draft: B.I., J.M.S., H.C.A., and A.A. Writing—review and editing: D.W.B., S.E., I.A.M., B.D.R., and M.J.
Competing interests: A.A. receives research support from 3M Company. All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S4
REFERENCES AND NOTES
- 1.World-Health-Organization. (https://covid19.who.int/, 2023), vol. 05-01-2023.
- 2.Files J. K., Boppana S., Perez M. D., Sarkar S., Lowman K. E., Qin K., Sterrett S., Carlin E., Bansal A., Sabbaj S., Long D. M., Kutsch O., Kobie J., Goepfert P. A., Erdmann N., Sustained cellular immune dysregulation in individuals recovering from SARS-CoV-2 infection. J. Clin. Investig. 131, 12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schulte-Schrepping J., Reusch N., Paclik D., Bassler K., Schlickeiser S., Zhang B. W., Kramer B., Krammer T., Brumhard S., Bonaguro L., De Domenico E., Wendisch D., Grasshoff M., Kapellos T. S., Beckstette M., Pecht T., Saglam A., Dietrich O., Mei H. E., Schulz A. R., Conrad C., Kunkel D., Vafadarnejad E., Xu C. J., Horne A., Herbert M., Drews A., Thibeault C., Pfeiffer M., Hippenstiel S., Hocke A., Muller-Redetzky H., Heim K. M., Machleidt F., Uhrig A., de Jarcy L. B., Jurgens L., Stegemann M., Glosenkamp C. R., Volk H. D., Goffinet C., Landthaler M., Wyler E., Georg P., Schneider M., Dang-Heine C., Neuwinger N., Kappert K., Tauber R., Corman V., Raabe J., Kaiser K. M., Vinh M. T., Rieke G., Meisel C., Ulas T., Becker M., Geffers R., Witzenrath M., Drosten C., Suttorp N., von Kalle C., Kurth F., Handler K., Schultze J. L., Aschenbrenner A. C., Li Y., Nattermann J., Sawitzki B., Saliba A. E., Sander L. E., Deutsch C.-O. I., Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell 182, 1419–1440 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanco-Melo D., Nilsson-Payant B. E., Liu W. C., Uhl S., Hoagland D., Moller R., Jordan T. X., Oishi K., Panis M., Sachs D., Wang T. T., Schwartz R. E., Lim J. K., Albrecht R. A., tenOever B. R., Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 181, 1036–1045.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Del Valle D. M., Kim-Schulze S., Huang H. H., Beckmann N. D., Nirenberg S., Wang B., Lavin Y., Swartz T. H., Madduri D., Stock A., Marron T. U., Xie H., Patel M., Tuballes K., Van Oekelen O., Rahman A., Kovatch P., Aberg J. A., Schadt E., Jagannath S., Mazumdar M., Charney A. W., Firpo-Betancourt A., Mendu D. R., Jhang J., Reich D., Sigel K., Cordon-Cardo C., Feldmann M., Parekh S., Merad M., Gnjatic S., An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 26, 1636–1643 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou P., Yang X. L., Wang X. G., Hu B., Zhang L., Zhang W., Si H. R., Zhu Y., Li B., Huang C. L., Chen H. D., Chen J., Luo Y., Guo H., Jiang R. D., Liu M. Q., Chen Y., Shen X. R., Wang X., Zheng X. S., Zhao K., Chen Q. J., Deng F., Liu L. L., Yan B., Zhan F. X., Wang Y. Y., Xiao G. F., Shi Z. L., A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arce V. M., Costoya J. A., SARS-CoV-2 infection in K18-ACE2 transgenic mice replicates human pulmonary disease in COVID-19. Cell. Mol. Immunol. 18, 513–514 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winkler E. S., Bailey A. L., Kafai N. M., Nair S., McCune B. T., Yu J. S., Fox J. M., Chen R. T. E., Earnest J. T., Keeler S. P., Ritter J. H., Kang L. I., Dort S., Robichaud A., Head R., Holtzman M. J., Diamond M. S., SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 21, 1327–1335 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Israelow B., Mao T. Y., Klein J., Song E., Menasche B., Omer S. B., Iwasaki A., Adaptive immune determinants of viral clearance and protection in mouse models of SARS-CoV-2. Sci. Immunol. 6, eabl4509 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dinnon K. H., Leist S. R., Schafer A., Edwards C. E., Martinez D. R., Montgomery S. A., West A., Yount B. L., Hou Y. X. J., Adams L. E., Gully K. L., Brown A. J., Huang E., Bryant M. D., Choong I. C., Glenn J. S., Gralinski L. E., Sheahan T. P., Baric R. S., A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 586, 560–566 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leist S. R., Dinnon K. H., Schafer A., Tse L. V., Okuda K., Hou Y. X. J., West A., Edwards C. E., Sanders W., Fritch E. J., Gully K. L., Scobey T., Brown A. J., Sheahan T. P., Moorman N. J., Boucher R. C., Gralinski L. E., Montgomery S. A., Baric R. S., A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell 183, 1070–1085.e12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Channappanavar R., Fehr A. R., Vijay R., Mack M., Zhao J. C., Meyerholz D. K., Perlman S., Dysregulated Type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe 19, 181–193 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montazersaheb S., Khatibi S. M. H., Hejazi M. S., Tarhriz V., Farjami A., Sorbeni F. G., Farahzadi R., Ghasemnejad T., COVID-19 infection: An overview on cytokine storm and related interventions. Virol. J. 19, 92 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guidotti L. G., Chisari F. V., Noncytolytic control of viral infections by the innate and adaptive immune response. Annu. Rev. Immunol. 19, 65–91 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Harrison A. G., Lin T., Wang P. H., Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol. 41, 1100–1115 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Y. G., Fu B. Q., Zheng X. H., Wang D. S., Zhao C. C., Qi Y. J., Sun R., Tian Z. G., Xu X. L., Wei H. M., Pathogenic T-cells and inflammatory monocytes incite inflammatory storms in severe COVID-19 patients. Natl. Sci. Rev. 7, 998–1002 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santa Cruz A., Mendes-Frias A., Oliveira A. I., Dias L., Matos A. R., Carvalho A., Capela C., Pedrosa J., Castro A. G., Silvestre R., Interleukin-6 is a biomarker for the development of fatal severe acute respiratory syndrome coronavirus 2 pneumonia. Front. Immunol. 12, 613422 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H. W., Wang T., Zhang X. Y., Chen H. L., Yu H. J., Zhang X. P., Zhang M. X., Wu S. J., Song J. X., Chen T., Han M. F., Li S. S., Luo X. P., Zhao J. P., Ning Q., Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 130, 2620–2629 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma A., Koka R., Burkett P., Diverse functions of IL-2, IL-15, and IL-7 in lymphoid homeostasis. Annu. Rev. Immunol. 24, 657–679 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Hanke T., Mitnacht R., Boyd R., Hunig T., Induction of interleukin-2 receptor-beta chain expression by self-recognition in the thymus. J. Exp. Med. 180, 1629–1636 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swain S. L., McKinstry K. K., Strutt T. M., Expanding roles for CD4(+) T cells in immunity to viruses. Nat. Rev. Immunol. 12, 136–148 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Battegay M., Moskophidis D., Rahemtulla A., Hengartner H., Mak T. W., Zinkernagel R. M., Enhanced establishment of a virus carrier state in adult CD4+ T-cell-deficient mice. J. Virol. 68, 4700–4704 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huber S. R., van Beek J., de Jonge J., Luytjes W., van Baarle D., T cell responses to viral infections—Opportunities for peptide vaccination. Front. Immunol. 5, 171 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mombaerts P., Clarke A. R., Rudnicki M. A., Iacomini J., Itohara S., Lafaille J. J., Wang L. L., Ichikawa Y., Jaenisch R., Hooper M. L., Tonegawa S., Mutations in T-cell antigen receptor genes α and β block thymocyte development at different stages. Nature 360, 225–231 (1992). [DOI] [PubMed] [Google Scholar]
- 25.Solouki S., August A., Huang W. S., Non-receptor tyrosine kinase signaling in autoimmunity and therapeutic implications. Pharmacol. Ther. 201, 39–50 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaeffer E. M., Debnath J., Yap G., McVicar D., Liao X. C., Littman D. R., Sher A., Varmus H. E., Lenardo M. J., Schwartzberg P. L., Requirement for Tec kinases Rlk and Itk in T cell receptor signaling and immunity. Science 284, 638–641 (1999). [DOI] [PubMed] [Google Scholar]
- 27.You Y., Myers R. C., Freeberg L., Foote J., Kearney J. F., Justement L. B., Carter R. H., Marginal zone B cells regulate antigen capture by marginal zone macrophages. J. Immunol. 186, 2172–2181 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soudja S. M., Chandrabos C., Yakob E., Veenstra M., Palliser D., Lauvau G., Memory-T-cell-derived interferon-gamma instructs potent innate cell activation for protective immunity. Immunity 40, 974–988 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giamarellos-Bourboulis E. J., Netea M. G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., Damoraki G., Gkavogianni T., Adami M. E., Katsaounou P., Ntaganou M., Kyriakopoulou M., Dimopoulos G., Koutsodimitropoulos I., Velissaris D., Koufargyris P., Karageorgos A., Katrini K., Lekakis V., Lupse M., Kotsaki A., Renieris G., Theodoulou D., Panou V., Koukaki E., Koulouris N., Gogos C., Koutsoukou A., Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe 27, 992–1000.e3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lucas C., Wong P., Klein J., Castro T. B. R., Silva J., Sundaram M., Ellingson M. K., Mao T. Y., Oh J. E., Israelow B., Takahashi T., Tokuyama M., Lu P. W., Venkataraman A., Park A., Mohanty S., Wang H. W., Wyllie A. L., Vogels C. B. F., Earnest R., Lapidus S., Ott I. M., Moore A. J., Muenker M. C., Fournier J. B., Campbell M., Odio C. D., Casanovas-Massana A., Herbst R., Shaw A. C., Medzhitov R., Schulz W. L., Grubaugh N. D., Dela Cruz C., Farhadian S., Ko A. I., Omer S. B., Iwasaki A., Yale I. T., Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 584, 463–469 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Massow G., Oh S., Lam A., Gustafsson K., Gamma delta T cells and their involvement in COVID-19 virus infections. Front. Immunol. 12, 741218 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng M., Hu S. L., Lung‐resident γδ T cells and their roles in lung diseases. Immunology 151, 375–384 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lei L., Qian H. B., Yang X. F., Zhang X. Z., Zhang D., Dai T. X., Guo R., Shi L., Cheng Y. B., Zhang B. J., Zhou X. B., Hu J. S., Guo Y. L., The phenotypic changes of γδ T cells in COVID-19 patients. J. Cell. Mol. Med. 24, 11603–11606 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yazdanifar M., Mashkour N., Bertaina A., Making a case for using γδ T cells against SARS-CoV-2. Crit. Rev. Microbiol. 46, 689–702 (2020). [DOI] [PubMed] [Google Scholar]
- 35.M.-S. R. Laetitia Gay, S. Mezouar, M. Richaud, L. Gorvel, E. Foucher, L. Madakamutil, B. La Scola, A. Menard, J. Allardet-Servent, P. Halfon, P. Frohna, C. Cano, J.-L. Mege, D. Olive. Vγ9Vδ2 T cells are potent inhibitors of SARS-CoV-2 replication and exert effector phenotypes in COVID-19 patients. bioRxiv 487518 [Preprint] (2022). 10.1101/2022.04.15.487518. [DOI] [PubMed]
- 36.Schumann J., Stanko K., Schliesser U., Appelt C., Sawitzki B., Differences in CD44 surface expression levels and function discriminates IL-17 and IFN-γ producing helper T cells. PLoS One 10, e0132479 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okuniewska M., Fang V., Baeyens A., Raghavan V., Lee J. Y., Littman D. R., Schwab S. R., SPNS2 enables T cell egress from lymph nodes during an immune response. Cell Rep. 36, 109368 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moynihan K. D., Opel C. F., Szeto G. L., Tzeng A., Zhu E. F., Engreitz J. M., Williams R. T., Rakhra K., Zhang M. H., Rothschilds A. M., Kumari S., Kelly R. L., Kwan B. H., Abraham W., Hu K., Mehta N. K., Kauke M. J., Suh H., Cochran J. R., Lauffenburger D. A., Wittrup K. D., Irvine D. J., Eradication of large established tumors in mice by combination immunotherapy that engages innate and adaptive immune responses. Nat. Med. 22, 1402–1410 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newton A. H., Cardani A., Braciale T. J., The host immune response in respiratory virus infection: Balancing virus clearance and immunopathology. Semin. Immunopathol. 38, 471–482 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mostafa H. H., Vogel P., Srinivasan A., Russell C. J., Non-invasive imaging of sendai virus infection in pharmacologically immunocompromised mice: NK and T cells, but not neutrophils, promote viral clearance after therapy with cyclophosphamide and dexamethasone. PLoS Pathog. 12, e1005875 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Itoh Y., Yoshida R., Shichinohe S., Higuchi M., Ishigaki H., Nakayama M., Pham V. L., Ishida H., Kitano M., Arikata M., Kitagawa N., Mitsuishi Y., Ogasawara K., Tsuchiya H., Hiono T., Okamatsu M., Sakoda Y., Kida H., Ito M., Mai L. Q., Kawaoka Y., Miyamoto H., Ishijima M., Igarashi M., Suzuki Y., Takada A., Protective efficacy of passive immunization with monoclonal antibodies in animal models of H5N1 highly pathogenic avian influenza virus infection. PLoS Pathog. 10, e1004192 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao Y., Chen Y. M., Liu M., Shi S. Z., Tian J. H., Impacts of immunosuppression and immunodeficiency on COVID-19: A systematic review and meta-analysis. J. Infect. 81, e93–e95 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ljungman P., Ward K. N., Crooks B. N. A., Parker A., Martino R., Shaw P. J., Brinch L., Brune M., De La Camara R., Dekker A., Pauksen K., Russell N., Schwarer A. P., Cordonnier C., Respiratory virus infections after stem cell transplantation: A prospective study from the Infectious diseases working party of the European group for blood and marrow transplantation. Bone Marrow Transplant. 28, 479–484 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Grifoni A., Weiskopf D., Ramirez S. I., Mateus J., Dan J. M., Moderbacher C. R., Rawlings S. A., Sutherland A., Premkumar L., Jadi R. S., Marrama D., de Silva A. M., Frazier A., Carlin A. F., Greenbaum J. A., Peters B., Krammer F., Smith D. M., Crotty S., Sette A., Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 181, 1489–1501.e15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nelson C. E., Namasivayam S., Foreman T. W., Kauffman K. D., Sakai S., Dorosky D. E., Lora N. E., Brooks K., Potter E. L., Garza N. L., Lafont B. A. P., Johnson R. F., Roederer M., Sher A., Weiskopf D., Sette A., de Wit E., Hickman H. D., Brenchley J. M., Via L. E., Barber D. L., Imaging N. D. T., Mild SARS-CoV-2 infection in rhesus macaques is associated with viral control prior to antigen-specific T cell responses in tissues. Sci. Immunol. 7, eabo0535 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruan Q. R., Yang K., Wang W. X., Jiang L. Y., Song J. X., Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 46, 846–848 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yui M. A., Hernandez-Hoyos G., Rothenberg E. V., A new regulatory region of the IL-2 locus that confers position-independent transgene expression. J. Immunol. 166, 1730–1739 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Yui M. A., Sharp L. L., Havran W. L., Rothenberg E. V., Preferential activation of an IL-2 regulatory sequence transgene in TCRγδ and NKT cells: Subset-specific differences in IL-2 regulation. J. Immunol. 172, 4691–4699 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Huynh M. L. N., Fadok V. A., Henson P. M., Phosphatidylserine-dependent ingestion of apoptotic cells promotes TGF-β1 secretion and the resolution of inflammation. J. Clin. Investig. 109, 41–50 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fadok V. A., Bratton D. L., Rose D. M., Pearson A., Ezekewitz R. A. B., Henson P. M., A receptor for phosphatidylserine-specific clearance of apoptotic cells. Nature 405, 85–90 (2000). [DOI] [PubMed] [Google Scholar]
- 51.Fadok V. A., Bratton D. L., Konowal A., Freed P. W., Westcott J. Y., Henson P. M., Macrophages that have ingested apoptotic cells in vitro inhibit proinflammatory cytokine production through autocrine/paracrine mechanisms involving TGF-beta, PGE2, and PAF. J. Clin. Investig. 101, 890–898 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sefik E., Qu R. H., Junqueira C., Kaffe E., Mirza H., Zhao J., Brewer J. R., Han A. L., Steach H. R., Israelow B., Blackburn H. N., Velazquez S. E., Chen Y. G., Halene S., Iwasaki A., Meffre E., Nussenzweig M., Lieberman J., Wilen C. B., Kluger Y., Flavell R. A., Inflammasome activation in infected macrophages drives COVID-19 pathology. Nature 606, 585–593 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Junqueira C., Crespo A., Ranjbar S., de Lacerda L. B., Lewandrowski M., Ingber J., Parry B., Ravid S., Clark S., Schrimpf M. R., Ho F. L., Beakes C., Margolin J., Russell N., Kays K., Boucau J., Das Adhikari U., Vora S. M., Leger V., Gehrke L., Henderson L. A., Janssen E., Kwon D., Sander C., Abraham J., Goldberg M. B., Wu H., Mehta G., Bell S., Goldfeld A. E., Filbin M. R., Lieberman J., FcγR-mediated SARS-CoV-2 infection of monocytes activates inflammation. Nature 606, 576–584 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feyaerts D., Urbschat C., Gaudilliere B., Stelzer I. A., Establishment of tissue-resident immune populations in the fetus. Semin. Immunopathol. 44, 747–766 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rocha B., Dautigny N., Pereira P., Peripheral T lymphocytes: Expansion potential and homeostatic regulation of pool sizes and CD4/CD8 ratios in vivo. Eur. J. Immunol. 19, 905–911 (1989). [DOI] [PubMed] [Google Scholar]
- 56.Sefik E., Qu R. H., Junqueira C., Kaffe E., Mirza H., Zhao J., Brewer J. R., Han A. L., Steach H. R., Israelow B., Blackburn H. N., Velazquez S. E., Chen Y. G., Halene S., Iwasaki A., Meffre E., Nussenzweig M., Lieberman J., Wilen C. B., Kluger Y., Flavell R. A., Inflammasome activation in infected macrophages drives COVID-19 pathology. Nature 606, 585–593 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alexandersen S., Chamings A., Bhatta T. R., SARS-CoV-2 genomic and subgenomic RNAs in diagnostic samples are not an indicator of active replication. Nat. Commun. 11, 6059 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shapira T., Monreal I. A., Dion S. P., Buchholz D. W., Imbiakha B., Olmstead A. D., Jager M., Desilets A., Gao G., Martins M., Vandal T., Thompson C. A. H., Chin A., Rees W. D., Steiner T., Nabi I. R., Marsault E., Sahler J., Diel D. G., Van de Walle G. R., August A., Whittaker G. R., Boudreault P. L., Leduc R., Aguilar H. C., Jean F., A TMPRSS2 inhibitor acts as a pan-SARS-CoV-2 prophylactic and therapeutic. Nature 605, 340–348 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Toptan T., Hoehl S., Westhaus S., Bojkova D., Berger A., Rotter B., Hoffmeier K., Cinatl J., Ciesek S., Widera M., Optimized qRT-PCR approach for the detection of intra- and extra-cellular SARS-CoV-2 RNAs. Int. J. Mol. Sci. 21, 4396 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S4