Abstract
Export of oncogenic T-DNA from the phytopathogen Agrobacterium tumefaciens is mediated by the products of the virB operon. It has recently been reported (K. J. Fullner and E. W. Nester, J. Bacteriol. 178:1498–1504, 1996) that DNA transfer does not occur at elevated temperatures; these observations correlate well with much earlier studies on the temperature sensitivity of crown gall tumor development on plants. In testing the hypothesis that this loss of DNA movement reflects a defect in assembly or maintenance of a stable DNA transfer machinery at high temperature, we have found that steady-state levels of VirB10 are sensitive to growth temperature while levels of several other VirB proteins are considerably less affected. This temperature-dependent failure to accumulate VirB10 is exacerbated in an attachment-deficient mutant strain (chvB) which exhibits pleiotropic defects in periplasmic osmoadaption, and virulence of a chvB mutant can be partially restored by lowering the temperature at which the bacteria and the plant tissue are cocultivated. Furthermore, the stability of VirB10 is diminished in cells lacking functional VirB9, but only under conditions of low osmolarity. We propose that newly synthesized VirB10 is inherently labile in the presence of a large osmotic gradient across the inner membrane and is rapidly degraded unless it is stabilized by VirB9-dependent assembly into oligomeric complexes. The possibility that VirB10-containing complexes are not assembled properly at elevated temperatures suggests an explanation for the decades-old observation that tumor formation is exquisitely sensitive to ambient temperature.
The soil bacterium Agrobacterium tumefaciens induces the formation of crown gall tumors on a number of susceptible plants, including most dicotyledenous species. Infection, which occurs at a wound site, requires the presence in the bacterium of a 200-kb tumor-inducing (Ti) plasmid. In a mechanism analogous to conjugal transfer, a single-stranded portion of the Ti plasmid, the T-DNA, is excised and localized to the host plant cell nucleus, where it is stably integrated into the host genome. Expression of genes on the T-DNA encoding biosynthetic enzymes results in the unregulated production of plant growth hormones and uncontrolled plant cell division. Bacterial genes required for transformation include the products of the vir regulon, located on a nontransferred region of the Ti plasmid (for reviews, see references 3, 42, and 61).
Movement of the T-DNA out of the bacterial cell is mediated by the products of the virB operon (for a review, see reference 18). The 11 virB open reading frames exhibit sequence similarity to genes required for the conjugal transfer of plasmids from a number of incompatibility groups (47, 52, 59). Furthermore, the virB genes show considerable conservation in both nucleotide sequence and gene order with the ptl operon, required for the secretion of pertussis toxin from the mammalian pathogen Bordetella pertussis (22, 31, 80). Ten of the 11 virB genes are required for tumorigenesis, although not for the generation of the T strand (8, 9, 24, 26, 37, 64, 78). Almost all of the VirB proteins examined are found in association with the membrane system of A. tumefaciens (8, 20, 25, 33, 34, 44, 60, 63, 68, 73, 74, 79). The recent observation of pili on the surface of A. tumefaciens (38, 46, 51) lends credence to a model in which T-DNA transfer is mediated by a sex pilus similar to those mediating conjugation of a variety of mobilizable plasmids. Further support for the notion that one or more virB gene products are responsible for the elaboration of the pilus comes from the finding that pilus formation requires only virA, virG, virB1 to 11, and one more gene, virD4, which also encodes a membrane-associated protein (58) implicated in T-DNA transfer per se (38, 53).
Several lines of evidence suggest that interactions among VirB proteins are crucial to the assembly of a T-DNA transport apparatus in the bacterial cell membrane. Disulfide bonding between VirB9, located in the periplasm, and VirB7, an outer-membrane-associated lipoprotein (4, 32), stabilizes VirB9, and both VirB7 and VirB9 can also form what are likely to be homomeric complexes (1, 4, 6, 33, 66). Formation of the VirB7-VirB9 dimer appears to play a central role in promoting assembly of the putative VirB-protein transport complex, since both virulence and accumulation of VirB4, VirB5, and VirB8 to -11 are correlated with the modulation of VirB7 levels (33). VirB9 is also required for the formation of VirB10-containing high-molecular-weight aggregates, although VirB9 and VirB10 are not components of the same cross-linkable complexes, and random mutagenesis has been used to identify specific residues in VirB9 that are necessary for VirB10 assembly into complexes (6). VirB11 also appears to interact with VirB9 and VirB10, as evidenced by the recent report that overexpression of virB9, virB10, and/or virB11 (but not virB8) can suppress the avirulent phenotype associated with dominant mutations in virB11 (84). Finally, it has been proposed that VirB9, -10, and -11 may act in a rate-limiting capacity in the assembly of the T-DNA transport machinery; VirB-dependent movement of an RSF1010 derivative into plants (14) suppresses tumor formation by the T-DNA, but overexpression of virB9 to vir11 restores tumorigenicity (77).
It has been known for some time that tumor formation on several host species is optimal at 22°C and does not occur at temperatures above 29°C (62). Loss of virulence at high temperatures is not due to the effects of temperature on the plants (11). Temperature-dependent suppression of tumorigenesis is also not attributable to alterations in the levels of vir gene transcription, which were found to be comparable at 19 and 28°C and only slightly less than the maximal transcriptional induction observed at 22°C (39). Inhibition of vir gene induction does occur at temperatures above 32°C and is likely due to an inactivating conformational change in VirA (43). However, even in a virG mutant strain constitutively expressing the vir genes at 32°C, tumor formation does not occur (43). Using the virB-dependent conjugal transfer of an RSF1010 derivative (7) as a measure of DNA movement across the bacterial membrane, Fullner and Nester recently demonstrated that the avirulence observed at temperatures above 28°C could be attributed to a nonfunctional DNA transfer machinery (39). However, the specific basis for this lack of transfer was not determined.
In A. tumefaciens, growth in hypoosmotic medium results in the accumulation of cyclic β-1,2-glucan in the periplasm (55). Since synthesis of this polysaccharide is reduced in cells growing under conditions of high osmolarity, it is believed that regulated synthesis and export of the glucan to the periplasm provides a mechanism by which the cell can minimize the osmotic gradient across the inner membrane (55). One of the chromosomally encoded gene products required for the periplasmic accumulation of β-1,2-glucan is ChvB, which catalyzes the synthesis of the oligosaccharide from UDP-glucose (85). It has been proposed that the presence of the oligosaccharide in the periplasm may stabilize outer membrane proteins against improper assembly or disassembly (71). By the same token, it seems likely that an excessive osmotic gradient across the inner membrane, such as in a chvB mutant grown in low-osmolarity medium, might well compromise the stability of membrane-associated protein complexes and could contribute to the observed avirulence associated with mutations in chvB.
In this study, we have investigated the role of growth temperature and, indirectly, osmoregulatory mechanisms in the stability of components of the T-DNA transport apparatus. Our findings demonstrate that accumulation of VirB10 is significantly decreased in cells grown at 28°C, compared to that in cells grown at 19°C, and is further diminished in cells lacking a functional ChvB protein. We have also found that VirB9 protects VirB10 from degradation under conditions of osmotic stress. Our data suggest a model in which VirB9-dependent assembly of VirB10 stabilizes the inherently labile protein against turnover. We propose that a lack of VirB10 stabilization at 28°C contributes to the previously documented defect in the T-DNA transfer process at high temperature.
MATERIALS AND METHODS
Bacterial strains and media.
The A. tumefaciens wild-type strain used was A348, which has the C58 chromosomal background and carries the pTiA6NC plasmid (40). The mutant strains used were Ax42 (A348 containing pTiA6 virB9::Tn5virB) (24), 358mx (A348 containing pTiA6 virE::Tn3HoHo1) (67), and A1020, which is A348 chvB::Tn5 (28). Cells were grown on MG/L, and ABIM induction medium was prepared as described previously (34). TYC medium is 0.5% tryptone–0.3% yeast extract supplemented with 7 mM CaCl2 (70).
Chemicals and reagents.
Acetosyringone (AS; 3′,5′-dimethoxy-4′-hydroxyacetophenone) was purchased from Aldrich (Milwaukee, Wis.). Chemical cross-linking agents disuccinimidyl suberate (DSS) and bis(sulfosuccinimidyl suberate) (BS3) were obtained from Pierce (Rockford, Ill.). Timentin was the product of SmithKline Beecham (Philadelphia, Pa.). The ECL Western blotting analysis kit, secondary antibody (donkey anti-rabbit antibody conjugated to horseradish peroxidase), and prestained molecular weight markers were purchased from Amersham (Arlington Heights, Ill.). Acrylamide solutions were obtained from Bio-Rad Laboratories (Hercules, Calif.). Nitrocellulose was the product of Schleicher and Schuell (Keene, N.H.). All other chemicals were the products of Sigma Chemical Co. (St. Louis, Mo.).
Cross-linking and immunoblotting.
Whole cells were treated with either BS3 or DSS as described previously (6), except that 6 ml of cells was pelleted as the starting material. Cross-linking reactions were quenched with 20 mM N-ethylmaleimide in 50 mM Tris-HCl (pH 7.5) and incubated for 5 min at room temperature unless otherwise noted. Where indicated, the washing steps were omitted and the cross-linking reaction was performed on ice. Samples were processed for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) by pelleting the cells, either after the washing and cross-linking steps or directly out of the induction medium, and resuspending them in sample buffer (0.1 M Tris-Cl [pH 6.8], 1.2 M β-mercaptoethanol, 4.3% SDS, 0.13% bromophenol blue, 9.8% glycerol). The volumes of sample buffer were adjusted so that each sample represented an equivalent number of cells per milliliter. Samples were denatured by boiling for 5 min and subjected to electrophoresis on 7 or 10% acrylamide gels (50). Proteins were transferred to nitrocellulose in a Tris-glycine-methanol transfer buffer, using a Hofer (San Francisco, Calif.) transblot apparatus. Nitrocellulose filters were exposed to primary antisera raised in rabbits and secondary donkey anti-rabbit antisera. Detection was performed with an ECL enhanced chemiluminescence kit as described by the manufacturer (Amersham) Antisera recognizing VirB5 (34), VirB8 (34), VirB9 (24), VirB10 (79), and VirB11 (20) have been previously characterized. The more rapidly migrating bands seen in several of the immunoblotting figures (particularly with anti-VirB10 and anti-VirB11) are presumed to be proteolytic products and, at least for VirB10, have been previously reported (79, 84).
Virulence assays.
Leaf square transformation assays were performed on Nicotiana tabacum cv. Havana 425 basically as described previously (6). Bacterial cultures to be tested for virulence were grown at either 19 or 28°C. Where indicated, the bacterial strains were subcultured to ABIM containing 200 μM AS and induced overnight at either 19 or 28°C prior to inoculation. Leaf square explants were infected with a suspension of agrobacteria at an A600 of 0.3 to 0.5. After 2 days of cocultivation at 19 or 28°C on hormone-free Murashige and Skoog (MS) medium (56) supplemented with 100 μM AS, the leaf pieces were washed and then placed on hormone-free MS plates containing 200 μg of timentin per ml to kill the bacteria. After 3 more days of incubation at either 19 or 28°C, the leaf pieces were transferred to 22°C, and they were incubated at that temperature until they were scored.
RESULTS
VirB9 is required for retention of VirB10 in cells washed in buffer.
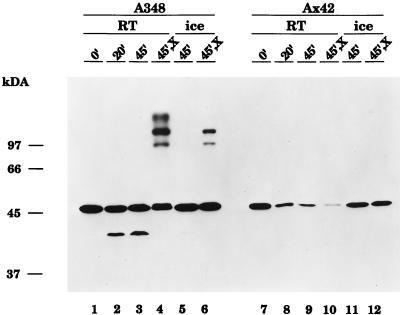
VirB10 exists as a component of several high-molecular-weight complexes that can be visualized by exposing whole cells to chemical cross-linking agents prior to preparation for SDS-PAGE and immunoblotting analysis (79). Beaupre et al. (6) have recently demonstrated that wild-type VirB9 is required for the assembly of VirB10 into cross-linkable aggregates; in strain Ax42, which contains a nonpolar Tn5virB transposon insertion in the virB9 gene (24), no cross-linked complexes are seen. Furthermore, the monomeric form of VirB10 was also almost undetectable in Ax42 cells that had been treated with the cross-linking agent BS3. However, in that study, the cells were washed three times in sodium phosphate buffer (pH 7.6) prior to exposure to BS3 for 30 min at room temperature (6). When we repeated these cross-linking experiments on strain Ax42, omitting the washing steps and incubating with BS3 on ice rather than at room temperature, we obtained a significantly different result (Fig. 1, compare lanes 10 and 12). In particular, the monomeric form of VirB10 (which migrates with an apparent molecular mass of approximately 48 kDa) was far more abundant in cells that had been cross-linked on ice without washing (lane 12); by comparing this sample with a sample pelleted directly from the induction medium and lysed by boiling in SDS-PAGE sample buffer (lane 7), we concluded that in unwashed cells incubated on ice, little loss of VirB10 monomer had occurred. Significantly, however, even under these experimental conditions, at which the monomer levels in strain Ax42 were maintained, no VirB10-containing high-molecular-weight aggregates were observed.
FIG. 1.
Cross-linking conditions influence the stability of VirB10 in Agrobacterium cells lacking wild-type VirB9. Cells of wild-type strain A348 (lanes 1 to 6) or strain Ax42 (virB9::Tn5virB) were induced overnight at 25°C in the presence of 200 μM AS. Cells were pelleted and incubated in 50 mM sodium phosphate buffer, pH 7.6, either at room temperature (RT) (lanes 1 to 4 and 7 to 10) or on ice (lanes 5 to 6 and 11 to 12) for the indicated lengths of time. For the samples in lanes 4, 6, 10, and 12, the cross-linking agent BS3 was included in the incubation at a final concentration of 500 μM (X). At the end of the incubation, cells were quenched with an equal volume of 50 mM Tris-Cl, pH 7.5, and processed for SDS-PAGE and immunoblotting analysis with anti-VirB10 antibodies, as described in Materials and Methods. The migration positions of proteins of known molecular mass (in kilodaltons) are shown at the left.
Further experimentation revealed that incubation for 20 min at room temperature in phosphate buffer, even without washing, was sufficient to cause a loss of VirB10 monomer from Ax42 cells and that depletion of VirB10 monomer was more pronounced when the cells were incubated for longer periods of time at room temperature in the phosphate buffer (Fig. 1, lanes 8 to 10). Curiously, we found that exposure to the cross-linking agent caused a further decrease in the amount of VirB10 monomer compared to samples incubated at room temperature for the same length of time (45 min) under otherwise identical conditions (compare lanes 9 and 10). This observation raised the possibility that some VirB10 in Ax42 cells was indeed aggregated, perhaps into complexes too large to enter the resolving gel. However, neither immunoblotting the stacking gel nor dot blotting the sample revealed any material that cross-reacted with the anti-VirB10 antisera (5a). Accumulation of monomeric VirB10 was not affected when cells were incubated on ice, rather than at room temperature, in phosphate buffer either without (lane 11) or with (lane 12) the cross-linking agent. Furthermore, cells of the wild-type strain A348 subjected to the same incubation conditions, either at room temperature or on ice, did not exhibit any loss of VirB10 monomer (Fig. 1, lanes 1 to 6). From these data, we conclude that VirB10 is rendered more labile in the absence of wild-type VirB9, such that incubation in phosphate buffer at room temperature, but not on ice, leads to substantially diminished steady-state levels of VirB10.
Steady-state levels of VirB10 are greatly diminished in cells grown at 28°C relative to those in cells grown at 19°C.
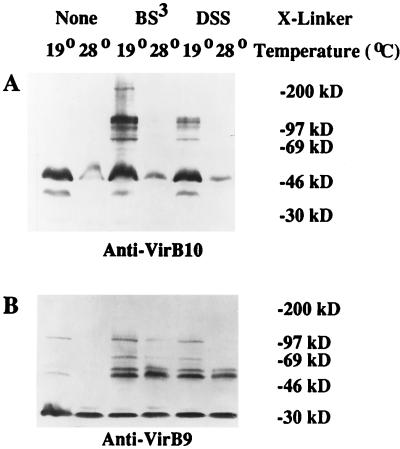
In light of the apparent role of VirB proteins in the formation of a membrane-associated T-DNA transport apparatus, it seems likely that environmental conditions which affect the stability of VirB proteins or impede proper assembly of VirB-containing complexes would result in attenuated virulence. Indeed, experiments by Fullner and Nester (39) demonstrated that conjugal transfer of an RSF1010 derivative via the VirB delivery mechanism occurs at 19°C but not at 28°C; VirB-dependent secretion of the pilin VirB2 (51), and hence formation of pili (38), is also inhibited in cells grown at 28°C. If the defect in DNA transfer at 28°C observed by Fullner and Nester stems from either incomplete assembly or instability of the transport machinery (39), one might predict that the sizes or abundance of VirB9- and/or VirB10-containing complexes identifiable by cross-linking (6, 79) would be altered in cells grown at elevated temperatures. To test this hypothesis, we grew cells of the wild-type strain A348 at either 19 or 28°C and then treated them with one of two related chemical cross-linking agents, BS3 or DSS, before subjecting them to SDS-PAGE and immunoblotting analysis. This experiment revealed that the steady-state levels of VirB10 were much lower in cells grown at 28°C than in cells grown at 19°C (Fig. 2A). Although the cell extracts used in the experiment shown in Fig. 2A were from cells that had been washed in phosphate buffer and incubated at room temperature during the cross-linking step, a significant decrease in the amount of VirB10 was also seen in cells grown at 28°C, pelleted, and lysed directly from induction medium (data not shown) (see Fig. 4A [cf. lanes 1 and 3]). Significantly, growth at 19°C was not sufficient to stabilize the monomeric VirB10 against the loss seen in Ax42 cells washed and incubated at room temperature (Fig. 3A, compare lanes 1 and 3). Thus, steady-state levels of VirB10 are diminished in cells grown at 28°C regardless of whether the cells are washed and incubated at room temperature, and they are decreased in Ax42 cells incubated in phosphate buffer regardless of whether the cells were grown at 19 or 28°C.
FIG. 2.
Effect of growth temperature on the accumulation of VirB9 and VirB10. A. tumefaciens A348 was induced overnight in ABIM containing 200 μM AS at either 19 or 28°C, as indicated. Cells were washed three times in sodium phosphate buffer (pH 7.6), cross-linked with 200 μM BS3 (lanes 3 and 4) or DSS (lanes 5 and 6), and processed for electrophoresis on an SDS–7% (A) or –10% (B) polyacrylamide gel and immunoblotting as described in Materials and Methods. (A) Probed with anti-VirB10; (B) probed with anti-VirB9. The migration positions of proteins of known molecular mass (in kilodaltons) are shown at the right.
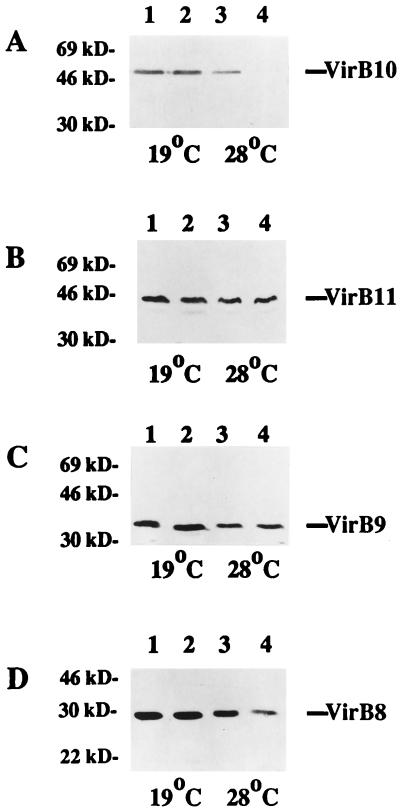
FIG. 4.
Accumulation of VirB proteins in wild-type and chvB mutant cells at 19 and 28°C. A. tumefaciens A348 (wild type) (lanes 1 and 3) and A1020 (chvB::Tn5) (lanes 2 and 4) were induced overnight at either 19 or 28°C, as indicated, in ABIM containing 200 μM AS. Cells were pelleted, and total crude extracts were subjected to electrophoresis on a 10% gel and immunoblot analysis as described in Materials and Methods. (A) Probed with anti-VirB10; (B) probed with anti-VirB11; (C) probed with anti-VirB9; (D) probed with anti-VirB8. The migration positions of proteins of known molecular mass (in kilodaltons) are shown at the left.
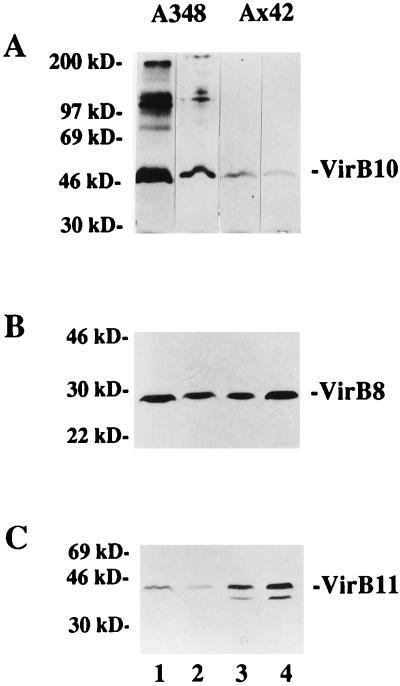
FIG. 3.
Accumulation of VirB proteins at 19 and at 28°C in the presence and absence of functional VirB9. A. tumefaciens A348 (wild type) (lanes 1 and 2) and Ax42 (virB9::Tn5virB) (lanes 3 and 4) were grown and induced in ABIM containing 200 μM AS at either 19°C (odd-numbered lanes) or 28°C (even-numbered lanes). Cells were washed in sodium phosphate buffer and cross-linked with BS3, as described in Materials and Methods, prior to being processed for immunoblotting. (A) Probed with anti-VirB10; (B) probed with anti-VirB8; (C) probed with anti-VirB11. The migration positions of proteins of known molecular mass (in kilodaltons) are shown at the left.
In contrast, accumulation of the monomeric form of VirB9 (Fig. 2B) was relatively unaffected by growth temperature, as was the prominent 60-kDa VirB9-containing aggregate. However, we did observe significantly weaker cross-reactivity in the larger (75- and 130-kDa) VirB9-containing high-molecular-weight species in cells grown at the higher temperature. Thus, although the overall levels of VirB9 are not influenced by temperature, the assembly of VirB9 into large oligomeric complexes does appear to be compromised.
We also examined the effect of growth temperature on the accumulation of other VirB proteins. Cell extracts from wild-type cells grown at either 19 or 28°C were probed with antibodies raised against VirB8 or VirB11. Only small differences in the levels of VirB8 were observed (Fig. 3B). However, cells grown at 28°C consistently accumulated decreased amounts of VirB11 relative to cells grown at 19°C (Fig. 3C). This was true both of cells washed and incubated at room temperature for 30 min and of cells pelleted and resuspended directly in SDS-PAGE sample buffer (data not shown).
Instability of VirB10 at 28°C is exacerbated in a chvB mutant.
Accumulation of cyclic β-1,2 glucan in the periplasm may function to minimize the osmotic gradient across the inner membrane that would otherwise form, particularly in cells grown in a low-osmolarity medium (55). We postulated that an excessive difference in osmolarity across the membrane in a chvB mutant strain might influence the stability of one or more VirB proteins that are localized in the membrane system or periplasm of agrobacteria. To determine whether this was the case, we prepared whole-cell extracts from wild-type and chvB strains grown at 19 or 28°C and subjected them to immunoblotting analysis. As depicted in Fig. 4A, the amount of VirB10 present in the chvB strain A1020 was comparable to the amount in the wild-type strain A348 when the cells were grown at 19°C (lanes 1 and 2). However, when cells were grown at 28°C, a striking decrease in VirB10 levels was seen in chvB cells relative to wild-type cells (Fig. 4A, lanes 3 and 4).
We also compared the amounts of other VirB proteins in chvB and wild-type cells grown at either 19 or 28°C. Levels of VirB11 and VirB9 were indistinguishable in the two strains (Fig. 4B and C), while the amount of VirB8 was moderately diminished in the chvB strain grown at 28°C but not in cells grown at 19°C (Fig. 4D). Taken together, our results indicate that VirB10, which is already less abundant in cells grown at 28°C, is particularly unstable at 28°C in cells exhibiting an apparent defect in osmoadaption; this instability in chvB cells is not a general attribute of all VirB proteins.
Incubation at 19°C restores partial virulence to chvB mutants.
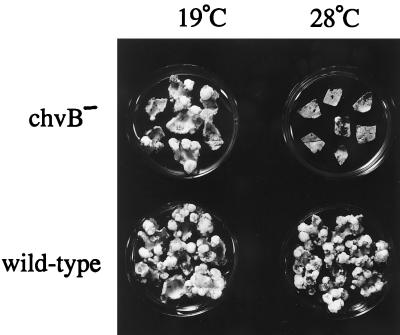
The data described above suggested that chvB mutants, routinely characterized as avirulent in most media, might in fact exhibit some ability to incite tumors if grown and inoculated at 19°C. We tested this hypothesis with tobacco leaf explants and the chvB mutant strain A1020 grown at either 19 or 28°C. Cocultivation was also performed at 19 or 28°C, but all leaf explants were shifted to 22°C 3 days after the elimination of the bacteria. As shown in Fig. 5, a limited number of tumors formed, albeit slowly, on leaf explants infected with strain A1020 at 19°C. In contrast, almost no tumors were seen on explants inoculated with the same strain at 28°C. Pursuant to a previous report (70) suggesting that the virulence of a chvB mutant could be influenced by the osmolarity of the medium in which the cells were grown, we occasionally observed a small enhancement of tumorigenicity when chvB cells were grown at 19°C in medium supplemented with 100 mM NaCl, but in most assays we found no significant effect of inclusion of 100 mM NaCl on the number of tumors incited by strain A1020 (Table 1).
FIG. 5.
Tumor formation by a chvB mutant strain at 19°C but not at 28°C. Tobacco leaf explants were cocultivated at either 19 or 28°C, as described in Materials and Methods, with A. tumefaciens A348 (wild type) or A1020 (chvB::Tn5) which had been grown overnight in TYC at the same temperature. The explants were transferred to selection medium containing hormone-free MS supplemented with 200 μg of timentin per ml and incubated at the same temperature for 3 days and then at 22°C for 21 days.
TABLE 1.
Effect of growth in 100 mM NaCl on tumor formation by a chvB mutant at 19 and 28°C
| Strain | Temp (°C) | Growth mediuma | No. of tumors per leaf pieceb |
|---|---|---|---|
| A1020 | 19 | TYC | 2.9 ± 0.7 |
| 19 | TYC + 100 mM NaCl | 2.4 ± 0.4 | |
| 28 | TYC | 0 ± 0 | |
| 28 | TYC + 100 mM NaCl | 0 ± 0 | |
| A348 | 19 | TYC | 6.2 ± 1.1 |
| 19 | TYC + 100 mM NaCl | 5.3 ± 0.7 | |
| 28 | TYC | 16.9 ± 1.6 | |
| 28 | TYC + 100 mM NaCl | 15.1 ± 1.3 |
Tobacco leaf explants were cocultivated at either 19 or 28°C with A. tumefaciens A348 (wild type) or A1020 (chvB::Tn5) grown in TYC or TYC plus 100 mM NaCl as described in the legend to Fig. 5. Representative leaf pieces (TYC alone) are shown in Fig. 5.
The mean number of tumors ± the standard error was determined 14 days after the leaf pieces were shifted to timentin-containing medium; n = 14 for each sample.
VirB9 stabilizes VirB10 against differences in osmolarity across the inner membrane.
Transcription of the virB operon in response to the inducer AS is dependent on the concerted activities of the phenolic sensor VirA and the transcriptional activator VirG (82). Experiments using bromoacetosyringone, which acts as an inhibitor of AS induction, suggest that vir expression requires continuous stimulation by AS (41). The loss of VirB10 observed when Ax42 cells were washed in sodium phosphate buffer, described above, might therefore be attributable to the removal of AS; if turnover of VirB10 is particularly rapid in the absence of wild-type VirB9, continuous protein synthesis would be required to maintain VirB10 levels, and since prokaryotic regulation is exerted primarily at the level of transcription (49), continuous synthesis might in turn require that levels of the polycistronic virB transcript be continually replenished. On the other hand, the observed instability of VirB10 in a chvB mutant grown at 28°C suggested an alternative explanation, namely, that it is the change in osmotic strength of the medium, rather than the removal of AS, that causes VirB10 to disappear from Ax42 cells upon incubation in phosphate buffer at room temperature.
To distinguish between these two possibilities, we grew both Ax42 and wild-type cells in induction medium (ABIM) containing 200 μM AS, pelleted the cells, and then washed them three times and incubated them for 30 min at room temperature either in sodium phosphate buffer, as before, or in induction medium lacking AS. We reasoned that if removal of AS was sufficient to cause a loss of VirB10, Ax42 cells exposed to either phosphate buffer or ABIM lacking AS should exhibit diminished levels of VirB10; however, if the observed loss was due to an increased osmotic gradient across the inner membrane, only cells shifted from ABIM to phosphate buffer would lose VirB10. The results of this experiment are depicted in Fig. 6. By comparing lanes 3 and 4 or lanes 7 and 8, it is evident that incubation in phosphate buffer, but not ABIM lacking AS, resulted in decreased levels of VirB10; the extent of the loss was comparable in cells grown at either 19 or 28°C. Thus, we conclude that changes in the osmolarity of the medium, rather than a requirement for continuous transcription and translation, can account for the observed loss of VirB10 in Ax42 cells that are washed prior to cross-linking.
FIG. 6.
Comparison of VirB10 levels in a virB9 nonpolar mutant washed in sodium phosphate buffer or in ABIM. A. tumefaciens A348 (wild-type) and Ax42 (virB9::Tn5virB) were induced overnight in ABIM containing 200 μM AS at either 19°C (lanes 1 to 4) or 28°C (lanes 5 to 8). Cells were pelleted and washed three times in either 50 mM sodium phosphate buffer, pH 7.6 (odd-numbered lanes), or ABIM without AS (even-numbered lanes). After incubation for 30 min at room temperature in the same solutions, cells were processed for SDS-PAGE and immunoblotting, using anti-VirB10, as described in Materials and Methods. The migration positions of proteins of known molecular mass (in kilodaltons) are shown at the left.
Turnover of VirB10, VirB11, and VirB8 in the absence of functional VirB9.
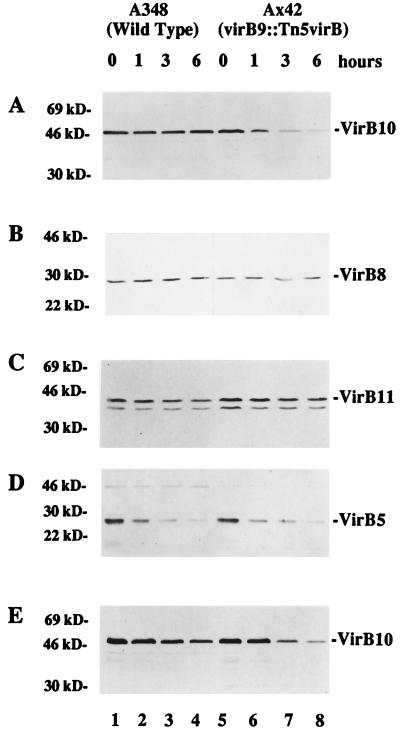
The results described thus far suggest that a large osmotic gradient across the inner membrane can destabilize the VirB10 protein in a strain grown at 28°C or in a strain lacking wild-type VirB9. However, these data do not preclude the possibility that VirB10 has a shorter half-life in Ax42 cells even when they are maintained in ABIM under inducing conditions. To test this more directly, we examined the effect of using chloramphenicol to block protein synthesis in cells growing in induction medium. Strains A348 and Ax42 were induced overnight at 19°C in the presence of 200 μM AS prior to the addition of 100 μg of chloramphenicol/ml, a concentration of antibiotic that we determined to be effective in preventing growth of these strains on both liquid and solid media. Aliquots of cells were removed at 0, 1, 3, and 6 h after the addition of chloramphenicol and processed for SDS-PAGE and immunoblotting analysis. The results depicted in Fig. 7A reveal that as the length of exposure to the protein synthesis inhibitor increased, the amount of VirB10 in strain Ax42 decreased (lanes 5 to 8). In contrast, no such decrease was observed in wild-type cells (Fig. 7A, lanes 1 to 4). Figures 7B and C indicate that unlike VirB10, VirB8 and VirB11 were relatively stable in both A348 and Ax42 cells at 19°C, at least over the 6-h duration of this experiment, and in fact VirB11 appeared to be slightly more stable in Ax42 than in A348. On the other hand, levels of VirB5 diminished rapidly in both A348 and Ax42 cells (Fig. 7D). When this experiment was performed on cells grown at 28°C, some loss of VirB10 was seen even in wild-type cells (Fig. 7E). Likewise, turnover of VirB11 was apparent in wild-type cells at 28°C but not in strain Ax42 (data not shown).
FIG. 7.
Analysis of VirB protein stability in the presence or absence of functional VirB9. A. tumefaciens A348 (wild type) and Ax42 (virB9::Tn5virB) were induced at 19°C (A to D) or 28°C (E) in ABIM containing 200 μM AS to an optical density at 600 nm of approximately 0.5. Chloramphenicol was added to a final concentration of 100 μg/ml, and incubation was continued at the same temperature for the indicated number of hours. Total crude extracts were analyzed by SDS-PAGE and immunoblotting as described in Materials and Methods. (A and E) Probed with anti-VirB10; (B) probed with anti-VirB8; (C) probed with anti-VirB11; (D) probed with anti-VirB5. The migration positions of proteins of known molecular mass (in kilodaltons) are shown at the left.
DISCUSSION
Effect of temperature-dependent VirB10 and VirB11 accumulation on virulence.
Early work on the Agrobacterium-induced formation of crown galls on tomato plants revealed that the efficiency of the infection process was strikingly influenced by temperature. Optimal tumor formation on tomato occurred at 22°C, and the sizes of the tumors were equivalent at 18 and 26°C but decreased as the temperature was raised to 28 to 30°C, above which no tumors were observed (62). Despite the fact that these observations were published more than 7 decades ago, the physiological basis for this temperature dependence has been the subject of relatively few studies in the intervening years. One set of experiments, published in 1950, led to the postulate that it was denaturation of a complex protein structure that was responsible for thermal inactivation of the “tumor inducing principle” (12). In this study, Braun used measurements of the size and weight of tumors and the delay in inception period over a narrow range of temperatures to calculate an energy of inactivation for tumorigenesis. More recently, Fullner and Nester have used the VirB-dependent movement of an RSF1010 derivative between agrobacteria to explore the possibility that the loss of virulence reflects a decrease in the function of the VirB transport machinery at high temperature. In their experiments, these authors found that RSF1010 conjugation was optimal at 19°C and did not occur at temperatures higher than 28°C. Although a decrease in viability of the bacteria in these conjugation assays could account for some of the decrease in transfer frequency between 22 and 25°C, it could not account for the drastic reduction between 25 and 28°C. Additional experiments demonstrated that the genetic requirements for conjugation are identical at 19 and 28°C and that conjugation via virB-independent mechanisms can occur at 28°C, implying that the temperature effect measured was specific to the use of the VirB transport apparatus (39). Interestingly, however, conjugal transfer of the Ti plasmid (which is not mediated by VirB) is also thermosensitive (72). One known consequence of growth at 28°C is a dramatic decrease in the VirB-dependent export of the major pilin VirB2 (51) and hence in the number of pili observed on A. tumefaciens (38). Also apparent are alterations in the number of mating pair aggregates (54) and the morphology of pili (10) elaborated by Escherichia coli carrying certain self-transmissible plasmids when the cells are grown at 37°C rather than at 26 to 30°C. However, these reports leave unresolved the underlying differences in the cell that lead to changes in the abundance or structure of pili at high temperature.
In this report, we have examined the stability of a subset of VirB proteins at permissive and nonpermissive temperatures for virulence. Our data indicate that VirB10, and to a lesser extent VirB11, accumulates to lower levels in cells grown at 28°C than in cells grown at 19°C (Fig. 2A and 3C). Steady-state levels of VirB8 and monomeric VirB9 are relatively unaffected by the growth temperature (Fig. 2B and 3B). (In some previous studies [see, e.g., reference 6], cross-linked versions of VirB10 were detected in cells that were reportedly grown at elevated temperatures, but ours is the first study in which direct comparisons have been made between cells grown at 19 and 28°C, and it seems likely that the growth temperature in the previous reports was less rigorously controlled.) Our findings thus suggest that destabilization of particular VirB proteins may be a key factor in the observed loss of DNA transfer function at high temperature. Both VirB10 and VirB11 have been implicated as essential, and perhaps rate-limiting, components of the T-DNA transfer machinery (77, 84), and VirB10 is known to assemble into multimeric protein complexes (79). We postulate that VirB10 cannot contribute to the formation of a functional transport apparatus because it does not accumulate sufficiently at 28°C or, alternatively, that it is not stable because it is not incorporated into multiprotein transport complexes. Our data do not allow us to discriminate unambiguously between these possibilities, although we favor a model in which assembly of VirB10, rather than its accumulation per se, is the temperature-dependent process (2a). Furthermore, in cells grown at 28°C, VirB9 appears to be stable in its monomeric form and can assemble into its apparent dimeric (60-kDa) form, yet it fails to assemble into the larger-molecular-weight aggregates detectable by chemical cross-linking (Fig. 2B). If DNA transport complex assembly is nucleated by one or two proteins, the loss of those proteins at an elevated temperature could easily destabilize the entire complex (see below). In this context, we note that disruption of energetically favored interactions among VirB proteins, perhaps due to the loss of only a subset of those interacting components, could contribute to the magnitude of the inactivation energy calculated by Braun (12). Finally, although we favor an explanation in which a loss of protein stability accounts for the diminished levels of VirB10 and VirB11, we cannot exclude the possibility that at elevated temperatures, transcription of the virB polycistronic message does not proceed as efficiently all the way to the 3′ end; in the study by Fullner and Nester, transcription was measured from a lacZ fusion located at the very 5′ end of the virB operon (2).
Role of osmoadaption in stability of VirB proteins.
The membrane systems of gram-negative bacteria enclose and segregate two aqueous compartments, the cytoplasm and the periplasm. In a classic study, Stock et al. demonstrated that for E. coli, the osmolarity of periplasm is similar to that of the cytoplasm, approximately 300 mosM for cells grown in standard media (69). It follows that a cell exposed to a hypotonic environment must have a mechanism to maintain a periplasmic osmolarity that is substantially higher than that of the surrounding milieu. For E. coli, it is generally believed that this differential is achieved by the accumulation in the periplasm of membrane-derived oligosaccharides, which with molecular weights of 2,200 to 2,600 are too large to pass through the outer membrane (48). In E. coli, the oligosaccharides that are synthesized in cells shifted to low-osmolarity medium are highly branched β-1,2 glucans that are multiply substituted with anionic residues (48). The negative charges contribute to a Donnan potential of 30 mV (negative inside) across the outer membrane (69). In A. tumefaciens, accumulation of neutral cyclic β-1,2 glucans in the periplasm is similarly osmoregulated, and concentrations of anionic oligosaccharides also rise in cells grown in hypotonic media (55). Periplasmic accumulation of these osmoadaptive cyclic oligosaccharides requires the function of at least two other genes, chvA and exoC, in addition to chvB. The exoC gene encodes the enzyme phosphoglucomutase, which is required for the biosynthesis of UDP-glucose (76). Mutants lacking a functional ChvA protein accumulate β-1,2-glucan in the cytoplasm but do not export it to the periplasm (15).
chvA, chvB, and exoC were first identified as chromosomal loci required for virulence. Originally, mutations in these genes were found to result in defects in the attachment of A. tumefaciens to plant cells (28); chvB mutants exhibit several other pleiotropic phenotypes as well. Subsequent reports have demonstrated that some of the other defects associated with chvB mutations, including poor growth and altered periplasmic protein content, can be suppressed by growing the cells in a high-osmolarity medium (16). chvB mutants are also sensitive to the level of calcium in the growth medium, and Swart et al. showed that defects in motility seen in chvB mutants could be prevented by decreasing the calcium concentration in the growth medium from 7 mM to 0.15 mM (71). However, calcium is required for the activity of the calcium-binding protein rhicadhesin, which mediates the first step in the attachment of A. tumefaciens to plant cells (65). Addition of 100 mM NaCl or 200 mM melezitose to the calcium-containing medium restored rhicadhesin activity, attachment, and virulence to chvB mutants, presumably by preempting the need for cyclic β-1,2-glucan as an osmoregulatory molecule in the periplasm (70). Raising the external osmolarity had a similar effect in Rhizobium meliloti mutants unable to synthesize β-1,2-glucans. In this case, pleiotropic cell surface defects associated with mutations in the ndv genes, the Rhizobium homologs of the chv genes, were rescued by the addition of 100 mM NaCl to the medium (30).
The apparent role of the chvB gene product in osmoadaption has led to the suggestion (71) that periplasmic β-1,2-glucan may interact with the lipid bilayers of both the inner and outer membranes to stabilize them against large differences in osmolarity across the bilayer. Such oligosaccharide effects might in turn modulate assembly of membrane-associated protein complexes, either by contributing directly to the stability of membrane proteins (13) or, potentially, by influencing the fluidity of the membrane (23). In this paper, we report that a chvB mutant exhibits dramatically lowered levels of a specific protein, VirB10, residing in the membrane system of A. tumefaciens. Significantly, the decrease in VirB10 accumulation in the chvB mutant strain was observed in cells grown at 28°C, but not in cells grown at 19°C, and occurred in addition to the temperature-dependent drop in VirB10 pools seen even in wild-type cells (Fig. 4A). These observations suggest that VirB10 may be both inherently labile and fairly sensitive to the existence of an osmotic gradient across the inner membrane. We conclude that in wild-type cells grown at 19°C, VirB10 is apparently stabilized, perhaps by assembly into oligomeric complexes, even in chvB mutant cells, which lack the osmoadaptive β-1,2-glucan in the periplasm. In contrast, when cells are grown at 28°C, pools of VirB10 are smaller and the protein which does accumulate appears not be stabilized against the effects of a hypotonic environment. Attempts to rescue the VirB10 instability phenotype by including 200 mM melezitose or 100 mM NaCl in the growth medium met with limited success; when cells were grown at 28°C in calcium-containing medium plus 200 mM melezitose and then induced in ABIM, the amount of VirB10 in the chvB cells was approximately equal to that in the wild-type cells, but these levels were substantially lower than the VirB10 levels seen in cells grown without melezitose. Inclusion of 100 mM NaCl caused a similar overall reduction in the levels of several VirB proteins (2a), and in fact, the data in Table 1 do not support the claim (70) that addition of 100 mM NaCl to the growth medium can enhance the tumorigenicity of a chvB mutant. However, our finding that virulence can be partially restored to a chvB mutant by growth and cocultivation at 19°C even in the absence of added osmotic support (Table 1; Fig. 5) indicates that one, although certainly not the only, factor contributing to the avirulence of this strain under typical assay conditions may be a lack of VirB10.
VirB9-dependent assembly of VirB10-containing complexes.
It has previously been reported that VirB9 is required for both the stabilization of VirB10 (9, 33) and VirB10 assembly into high-molecular-weight complexes (6). Our results confirm and extend these findings to provide a more nuanced view of the role of VirB9 in promoting the accumulation and proper assembly of VirB10. Beaupre et al. have shown that in strain Ax42, which carries a nonpolar transposon insertion in the virB9 gene (24), no VirB10-containing complexes are detected (6). However, under the conditions used for that cross-linking experiment, the monomeric form of VirB10 was also almost undetectable. This disappearance of VirB10 from strain Ax42 could be the result of enhanced VirB10 degradation, but it is also possible that the absence of VirB9 causes the release of VirB10 from the membrane. Under the cross-linking conditions used by Beaupre et al., we could not detect VirB10 in the culture medium or in the buffer washes (9a), but by omitting the buffer washes and performing the cross-linking on ice, we obtained nearly complete recovery of monomeric VirB10 (Fig. 1). Furthermore, in the present study, we have demonstrated that even under cross-linking conditions in which the monomeric form of VirB10 is present, no cross-linked complexes are seen (Fig. 1). From these data, we conclude that VirB9 facilitates the formation of VirB10-containing aggregates, perhaps by positioning VirB10 so that it can engage in productive interactions, most likely with other VirB10 molecules (6).
Another recent study also suggests that the abundance of VirB9 can influence the conformation of VirB10 in the membrane (84). It may be, for example, that in the absence of VirB9, individual VirB10 molecules are free to diffuse within the plane of the lipid bilayer but that interactions with VirB9 serve to limit movement and thereby promote clustering of VirB10 monomers. In this scenario, periplasmic VirB9 might in turn be immobilized by its interactions with the lipoprotein VirB7, which is anchored in the outer membrane by a fatty acid modification and may therefore serve to tether other components of the transport machinery (4, 32). Our data further indicate that in the absence of interactions with VirB9, VirB10 is rendered more sensitive to the existence of an osmotic gradient across the membrane, such as occurs when cells grown in induction medium are washed in sodium phosphate buffer (Fig. 6). Significantly, accumulation of VirB10 is substantially decreased even in wild-type cells that are not subjected to washing if the cells are grown at 28°C (see, for example, Fig. 4A); conversely, in the absence of VirB9, monomeric VirB10 disappears upon washing in cells grown at both low and high temperature (Fig. 3A), while in a chvB mutant at 28°C, even the presence of VirB9 cannot stabilize VirB10 (Fig. 4A). Taken together, our results suggest a hierarchy of requirements for VirB10 stabilization. In this model, VirB9-dependent assembly of VirB10 into a multiprotein complex stabilizes the inherently labile VirB10, but complex assembly cannot occur at elevated temperatures; thus, in cells grown at 28°C, VirB10 is less abundant even in wild-type cells and its degradation is exacerbated in a cell lacking the osmoadaptive function of the ChvB protein. In contrast, in cells grown at 19°C, VirB10 is insensitive to the absence of the ChvB protein precisely because it is complexed with other components of the transport apparatus.
Berger and Christie (9) and Fernandez et al. (33) have previously published data showing greatly diminished levels of VirB4, VirB8, and VirB11, as well as VirB10, in a strain containing a precise deletion of the virB9 gene. In contrast, our strain Ax42 carries a nonpolar transposon insertion that eliminates only the last 31 codons of virB9 and replaces them with 39 codons from the transposon that happen to be in frame, so that Ax42 expresses, albeit at lowered levels, an almost full-length VirB9. This truncated form of VirB9 is, however, completely nonfunctional in promoting tumorigenesis (24). Interestingly, we observed no decrease in the steady-state levels of VirB8 or VirB11 in strain Ax42 at 19°C (Fig. 7B and C), and the amount of VirB10 was reduced only when Ax42 cells were washed in buffer prior to lysis (Fig. 1 and 6), although the turnover rate for VirB10 in Ax42 appeared to be accelerated compared to that in the wild-type strain (Fig. 7A). These data imply that the truncated version of VirB9 encoded in strain Ax42 may be able to participate in certain stabilizing interactions, despite the fact that it does not appear to interact with VirB7 (6). In fact, we have consistently found significantly more VirB11 in Ax42 than in wild-type cells (see, for example, Fig. 3C), and turnover of VirB11 at temperatures of 28°C or higher is less pronounced in Ax42 than in wild-type strain A348 (data not shown). We cannot rule out the possibility that the proximity of the virB promoter in the Tn5virB transposon within virB9 allows for enhanced expression of virB11 in strain Ax42; however, we do not see a similar enhancement in the VirB11 levels in strain Ax56, which carries the same transposon in virB10 (data not shown). Furthermore, regulated expression of virB7 and virB8 from the lac promoter results in coordinate increases in the levels of VirB7-11, but VirB11 accumulation is actually higher in the absence of induction than in the presence of low concentrations of inducer (33). Perhaps in the absence of any VirB9, VirB11 forms aberrant complexes which stabilize it, while low-level induction of VirB9 results in incorporation of some VirB11 into productive complexes and in turnover of the remaining VirB11. Such a scenario would be consistent with the suggestion that mutant VirB11 molecules can be sequestered by excessive VirB9, VirB10, or VirB11 (84).
Work in several laboratories has led to the proposal that a critical step in the assembly of the T-DNA transport apparatus is the dimerization of VirB9, located in the periplasm, with the lipoprotein VirB7, which is anchored in the outer membrane (84). In this model, additional components are recruited via direct or indirect interactions with this heterodimer (18). An important puzzle that remains to be solved is how associations are forged across the peptidoglycan barrier between the inner and outer membranes (27). This issue is especially trenchant if, as has been proposed, a single multimeric VirB protein complex spans both membranes (18, 83). One attractive model suggests that assembly of the multiprotein DNA transport apparatus may be facilitated by localized lysis of the murein cell wall by VirB1 (5, 57). Furthermore, although the existence of adhesion sites between the inner and outer membranes is highly controversial (27, 29), there are hints from electron microscopic analysis of thin sections that F pili may form at such junctions (35). Fractionation studies suggest that several VirB proteins may partition with both membranes; in many cases, the observed partitioning of a given VirB protein varies from one study to another and may reflect interactions among proteins within a structure that spans the periplasm and which is sheared when the membranes are separated on sucrose gradients (4, 5, 32, 34, 45, 63, 74). One candidate for a peptidoglycan-spanning component of the transport machinery is VirB10. Sequence analysis and studies of virB-phoA fusions indicate that VirB10 contains a single membrane-spanning region with a large periplasmic domain and a small cytoplasmic amino terminus (79). This topology could allow for interactions with both VirB11, located on the inner face of the cytoplasmic membrane, and the periplasmic VirB9. In fact, a small proportion of the VirB10 is found in sucrose gradient fractions with densities intermediate between those of the inner and the outer membranes (34). Second, like its homolog, TraB, which is involved in F pilus assembly, VirB10 has a proline-rich region within the putative periplasmic domain, immediately adjacent to the membrane anchor (36). Although the prolines in VirB10 are not uniformly spaced, it is conceivable that this portion of the periplasmic domain of VirB10 assumes an elongated conformation; it is perhaps worth noting that the VirB10 homolog PtlG contains a similar proline-rich region in which the spacing of the prolines is more regular. Similar proline-rich features in at least two other bacterial proteins, TonB from E. coli and the group C streptococcal M protein, have been proposed to confer on these proteins a rigid, extended shape which allows them to thread through the peptidoglycan layer (81). Third, any model for formation of the VirB pore complex must take into account the observed effect of elevated temperature on T-DNA transport function (39). In this regard, it is noteworthy that of the proteins tested in the present work, it was VirB10 which exhibited the largest decrease in abundance at 28°C.
It is tempting to speculate that VirB10, positioned correctly through its interactions with the VirB9-VirB7 heterodimer, acts to nucleate temperature-dependent assembly of a stable multiprotein complex which mediates T-DNA transfer and that other proteins, such as VirB2 through VirB6, are assembled because of their interactions with VirB10. In fact, a very recent report demonstrates that processed VirB2, the major pilin of the “T pilus,” is exported from A. tumefaciens in a VirB-dependent manner; strikingly, VirB2 accumulates at both 19 and 28°C but appears in the exocellular fraction only at the lower temperature (51). We hypothesize that under conditions (e.g., in a chvB mutant at 28°C) in which the nucleating protein is itself unstable, other interacting species might also be degraded more rapidly. Whether the observed failure of the stable VirB9 monomer to assemble into high-molecular-weight aggregates at 28°C is a cause or a consequence of the loss of VirB10 remains to be determined. Although such a model for assembly of the T-DNA transport apparatus is highly speculative, there is growing evidence to support the notion that certain components may be rate limiting. In particular, Ward et al. showed that T-DNA-mediated virulence is inhibited in A. tumefaciens cells carrying the broad-host-range plasmid RSF1010, which can itself move to the host plant cells in a VirB-dependent manner, but that overexpression of VirB9, VirB10, and VirB11 is sufficient to overcome this oncogenic suppression (77). More intriguing yet, overexpression of either VirB9 or VirB10 alone can suppress the avirulence associated with dominant mutations in VirB11 (84). Finally, the recent discovery in Helicobacter pylori of a cag pathogenicity island consisting of only VirB4, VirB9, VirB10, VirB11, and VirD4 homologs provides support for the notion that these components represent the core structure on which the complete VirB transport machinery is assembled (17, 19, 21, 75).
ACKNOWLEDGMENTS
We are grateful to Andrew Binns, Karl Johnson, Eugene Nester, Lisa Stahl, and Paul Hyman for helpful conversations and support during this project.
This work was supported by grants to L.M.B. and to Andrew N. Binns from the National Science Foundation (MCB9506144 and MCB9513662, respectively) and to J.B. from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Anderson L B, Hertzel A V, Das A. Agrobacterium tumefaciens VirB7 and VirB9 form a disulfide-linked protein complex. Proc Natl Acad Sci USA. 1996;93:8889–8894. doi: 10.1073/pnas.93.17.8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ankenbauer R G, Best E A, Palanca C A, Nester E W. Mutants of the Agrobacterium tumefaciens virA gene exhibiting acetosyringone-independent expression of the vir regulon. Mol Plant-Microbe Interact. 1991;4:400–406. doi: 10.1094/mpmi-4-400. [DOI] [PubMed] [Google Scholar]
- 2a.Banta, L. M. Unpublished data.
- 3.Baron C, Zambryski P C. The plant response in pathogenesis, symbiosis, and wounding: variations on a common theme? Annu Rev Genet. 1995;29:107–129. doi: 10.1146/annurev.ge.29.120195.000543. [DOI] [PubMed] [Google Scholar]
- 4.Baron C, Thorstenson Y R, Zambryski P C. The lipoprotein VirB7 interacts with VirB9 in the membranes of Agrobacterium tumefaciens. J Bacteriol. 1997;179:1211–1218. doi: 10.1128/jb.179.4.1211-1218.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baron C, Llosa M, Zhou S, Zambryski P C. VirB1, a component of the T-complex transfer machinery of Agrobacterium tumefaciens, is processed to a C-terminal secreted product, VirB1*. J Bacteriol. 1997;179:1203–1210. doi: 10.1128/jb.179.4.1203-1210.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Beaupre, C., and J. Bohne. Unpublished data.
- 6.Beaupre C E, Bohne J, Dale E M, Binns A N. Interactions between VirB9 and VirB10 membrane proteins involved in movement of DNA from Agrobacterium tumefaciens into plant cells. J Bacteriol. 1997;179:78–89. doi: 10.1128/jb.179.1.78-89.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beijersbergen A, Dulk-Ras A D, Schilperoort R A, Hooykaas P J J. Conjugative transfer by the virulence system of Agrobacterium tumefaciens. Science. 1992;256:1324–1327. doi: 10.1126/science.256.5061.1324. [DOI] [PubMed] [Google Scholar]
- 8.Berger B R, Christie P J. The Agrobacterium tumefaciens virB4 gene product is an essential virulence protein requiring an intact nucleoside triphosphate-binding domain. J Bacteriol. 1993;175:1723–1734. doi: 10.1128/jb.175.6.1723-1734.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berger B R, Christie P J. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J Bacteriol. 1994;176:3646–3660. doi: 10.1128/jb.176.12.3646-3660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Bohne, J. Unpublished data.
- 10.Bradley D E, Whelan J. Conjugation systems of IncT plasmids. J Gen Microbiol. 1985;131:2665–2671. doi: 10.1099/00221287-131-10-2665. [DOI] [PubMed] [Google Scholar]
- 11.Braun A C. Thermal studies on the factors responsible for tumor initiation in crown gall. Am J Bot. 1947;34:234–240. [PubMed] [Google Scholar]
- 12.Braun A C. Thermal inactivation studies on the tumor-inducing principle in crown gall. Phytopathology. 1950;40:3. [Google Scholar]
- 13.Breedveld M W, Miller K J. Cyclic β-glucans of members of the family Rhizobiaceae. Microbiol Rev. 1994;58:145–161. doi: 10.1128/mr.58.2.145-161.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buchanan-Wollaston V, Passiatore J E, Cannon F. The mob and oriT mobilization functions of a bacterial plasmid promote its transfer to plants. Nature. 1987;328:172–175. [Google Scholar]
- 15.Cangelosi G A, Martinetti G, Leigh J A, Lee C C, Theines C, Nester E W. Role of Agrobacterium tumefaciens ChvA protein in export of β-1,2-glucan. J Bacteriol. 1989;171:1609–1615. doi: 10.1128/jb.171.3.1609-1615.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cangelosi G A, Martinetti G, Nester E W. Osmosensitivity phenotypes of Agrobacterium tumefaciens mutants that lack periplasmic β-1,2-glucan. J Bacteriol. 1990;172:2172–2174. doi: 10.1128/jb.172.4.2172-2174.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Censini S, Lange C, Xiang Z, Crabtree J E, Ghiara P, Bordovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Genetics. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christie P J. Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J Bacteriol. 1997;179:3085–3094. doi: 10.1128/jb.179.10.3085-3094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christie P J. The cag pathogenicity island: mechanistic insights. Trends Microbiol. 1997;5:264–265. doi: 10.1016/S0966-842X(97)88833-6. [DOI] [PubMed] [Google Scholar]
- 20.Christie P J, Ward J E, Gordon M P, Nester E W. A gene required for transfer of T-DNA to plants encodes an ATPase with autophosphorylating activity. Proc Natl Acad Sci USA. 1989;86:9677–9681. doi: 10.1073/pnas.86.24.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Covacci A, Falkow S, Berg D E, Rappuoli R. Did the inheritance of a pathogenicity island modify the virulence of Helicobacter pylori? Trends Microbiol. 1997;5:205–208. doi: 10.1016/S0966-842X(97)01035-4. [DOI] [PubMed] [Google Scholar]
- 22.Covacci A, Rappuoli R. Pertussis toxin export requires accessory genes located downstream from the pertussis toxin operon. Mol Microbiol. 1993;8:429–434. doi: 10.1111/j.1365-2958.1993.tb01587.x. [DOI] [PubMed] [Google Scholar]
- 23.Crowe J H, Whittam M A, Chapman D, Crowe L M. Interactions of phospholipid monolayers with carbohydrates. Biochim Biophys Acta. 1984;769:151–159. doi: 10.1016/0005-2736(84)90018-x. [DOI] [PubMed] [Google Scholar]
- 24.Dale E M, Binns A N, Ward J E., Jr Construction and characterization of Tn5virB, a transposon that generates nonpolar mutations, and its use to define virB8 as an essential virulence gene in Agrobacterium tumefaciens. J Bacteriol. 1993;175:887–891. doi: 10.1128/jb.175.3.887-891.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dang T A T, Christie P J. The VirB4 ATPase of Agrobacterium tumefaciens is a cytoplasmic membrane protein exposed at the periplasmic surface. J Bacteriol. 1997;179:453–462. doi: 10.1128/jb.179.2.453-462.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Vos G, Zambryski P. Expression of Agrobacterium nopaline-specific VirD1, VirD2, and VirC1 proteins and their requirement for T-strand production in E. coli. Mol Plant-Microbe Interact. 1989;2:43–52. doi: 10.1094/mpmi-2-043. [DOI] [PubMed] [Google Scholar]
- 27.Dijkstra A J, Keck W. Peptidoglycan as a barrier to transenvelope transport. J Bacteriol. 1996;178:5555–5562. doi: 10.1128/jb.178.19.5555-5562.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Douglas C J, Staneloni R J, Rubin R A, Nester E W. Identification and genetic analysis of an Agrobacterium tumefaciens chromosomal virulence region. J Bacteriol. 1985;161:850–860. doi: 10.1128/jb.161.3.850-860.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Durrenberger M B, Villiger W, Bachi T. Conjugational junctions: morphology of specific contacts in conjugating Escherichia coli bacteria. J Struct Biol. 1991;107:146–156. doi: 10.1016/1047-8477(91)90018-r. [DOI] [PubMed] [Google Scholar]
- 30.Dylan T, Helinski D R, Ditta G S. Hypoosmotic adaptation in Rhizobium meliloti requires β-(1→2)-glucan. J Bacteriol. 1990;172:1400–1408. doi: 10.1128/jb.172.3.1400-1408.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farizo K M, Cafarella T G, Burns D L. Evidence for a ninth gene, ptlI, in the locus encoding the pertussis toxin secretion system of Bordetella pertussis and formation of a PtlI-PtlF complex. J Biol Chem. 1996;271:31643–31649. doi: 10.1074/jbc.271.49.31643. [DOI] [PubMed] [Google Scholar]
- 32.Fernandez D, Dang T A T, Spudich G M, Zhou X-R, Berger B R, Christie P J. The Agrobacterium tumefaciens virB7 gene product, a proposed component of the T-complex transport apparatus, is a membrane-associated lipoprotein exposed at the periplasmic surface. J Bacteriol. 1996;178:3156–3167. doi: 10.1128/jb.178.11.3156-3167.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernandez D, Spudich G M, Zhou X-R, Christie P J. The Agrobacterium tumefaciens VirB7 lipoprotein is required for stabilization of VirB proteins during assembly of the T-complex transport apparatus. J Bacteriol. 1996;178:3168–3176. doi: 10.1128/jb.178.11.3168-3176.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Finberg K E, Muth T R, Young S P, Maken J B, Heitritter S M, Binns A N, Banta L M. Interactions of VirB9, -10, and -11 with the membrane fraction of Agrobacterium tumefaciens: solubility studies provide evidence for tight associations. J Bacteriol. 1995;177:4881–4889. doi: 10.1128/jb.177.17.4881-4889.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Firth N, Ippen-Ihler K, Skurray R A. Structure and function of the F factor and mechanism of conjugation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C: ASM Press; 1996. pp. 2377–2401. [Google Scholar]
- 36.Frost L S, Ippen-Ihler K, Skurray R A. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol Rev. 1994;58:162–210. doi: 10.1128/mr.58.2.162-210.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fullner K J, Stephens K M, Nester E W. An essential virulence protein of Agrobacterium tumefaciens, VirB4, requires an intact mononucleotide binding domain to function in transfer of T-DNA. Mol Gen Genet. 1994;245:704–715. doi: 10.1007/BF00297277. [DOI] [PubMed] [Google Scholar]
- 38.Fullner K J, Lara J C, Nester E W. Pilus assembly by Agrobacterium T-DNA transfer genes. Science. 1996;273:1107–1109. doi: 10.1126/science.273.5278.1107. [DOI] [PubMed] [Google Scholar]
- 39.Fullner K J, Nester E W. Temperature affects the T-DNA transfer machinery of Agrobacterium tumefaciens. J Bacteriol. 1996;178:1498–1504. doi: 10.1128/jb.178.6.1498-1504.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garfinkel D J, Simpson R B, Ream L W, White F F, Gordon M P, Nester E W. Genetic analysis of crown gall: fine structure map of the T-DNA by site-directed mutagenesis. Cell. 1981;27:143–153. doi: 10.1016/0092-8674(81)90368-8. [DOI] [PubMed] [Google Scholar]
- 41.Hess K M, Dudley M W, Lynn D G, Joerger R D, Binns A N. Mechanism of phenolic activation of Agrobacterium virulence genes: development of a specific inhibitor of bacterial sensor/response systems. Proc Natl Acad Sci USA. 1991;88:7854–7858. doi: 10.1073/pnas.88.17.7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hooykaas P J J, Beijersbergen A G M. The virulence system of Agrobacterium tumefaciens. Annu Rev Phytopathol. 1994;32:157–179. [Google Scholar]
- 43.Jin S, Song Y-N, Deng W-Y, Gordon M P, Nester E W. The regulatory VirA protein of Agrobacterium tumefaciens does not function at elevated temperatures. J Bacteriol. 1993;175:6830–6835. doi: 10.1128/jb.175.21.6830-6835.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones A L, Shirasu K, Kado C I. The product of the virB4 gene of Agrobacterium tumefaciens promotes accumulation of VirB3 protein. J Bacteriol. 1994;176:5255–5261. doi: 10.1128/jb.176.17.5255-5261.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones A L, Lai E-M, Shirasu K, Kado C I. VirB2 is a processed pilin-like protein encoded by the Agrobacterium tumefaciens Ti plasmid. J Bacteriol. 1996;178:5706–5711. doi: 10.1128/jb.178.19.5706-5711.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kado C. Genes of the virB operon are involved in the synthesis of a conjugational pilus-like apparatus required for the transfer of the T-DNA from Agrobacterium tumefaciens to plant cells. In: Furasuki S, Ryer A D, editors. Advances in plant biotechnology. Amsterdam, The Netherlands: Elsevier; 1994. pp. 23–36. [Google Scholar]
- 47.Kado C I. Promiscuous DNA transfer system of Agrobacterium tumefaciens: role of the virB operon in sex pilus assembly and synthesis. Mol Microbiol. 1994;12:17–22. doi: 10.1111/j.1365-2958.1994.tb00990.x. [DOI] [PubMed] [Google Scholar]
- 48.Kennedy E P. Osmotic regulation and the biosynthesis of membrane-derived oligosaccharides in Escherichia coli. Proc Natl Acad Sci USA. 1982;79:1092–1095. doi: 10.1073/pnas.79.4.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kushner S R. Useful host strains and techniques for recombinant DNA experiments. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 2. Washington, D.C: American Society for Microbiology; 1987. pp. 1225–1230. [Google Scholar]
- 50.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 51.Lai E-M, Kado C I. Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens. J Bacteriol. 1998;180:2711–2717. doi: 10.1128/jb.180.10.2711-2717.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lessl M, Balzer D, Pansegrau W, Lanka E. Sequence similarities between the RP4 Tra2 and the Ti VirB region strongly support the conjugation model for T-DNA transfer. J Biol Chem. 1992;267:20471–20480. [PubMed] [Google Scholar]
- 53.Lin T-S, Kado C I. The virD4 gene is required for virulence while virD3 and orf5 are not required for virulence of Agrobacterium tumefaciens. Mol Microbiol. 1993;9:803–812. doi: 10.1111/j.1365-2958.1993.tb01739.x. [DOI] [PubMed] [Google Scholar]
- 54.Maher D, Sherburne R, Taylor D E. H-pilus assembly kinetics determined by electron microscopy. J Bacteriol. 1993;175:2175–2183. doi: 10.1128/jb.175.8.2175-2183.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller K J, Kennedy E P, Reinhold V N. Osmotic adaptation by gram-negative bacteria: possible role for periplasmic oligosaccharides. Science. 1986;231:48–51. doi: 10.1126/science.3941890. [DOI] [PubMed] [Google Scholar]
- 56.Murashige T, Skoog F. A revised medium for rapid growth and bioassay with tobacco tissue culture. Physiol Plant. 1962;15:473–496. [Google Scholar]
- 57.Mushegian A R, Fullner K J, Koonin E V, Nester E W. A family of lysozyme-like virulence factors in bacterial pathogens of plants and animals. Proc Natl Acad Sci USA. 1996;93:7321–7326. doi: 10.1073/pnas.93.14.7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Okamoto S, Toyoda-Yamamoto A, Ito K, Takebe I, Machida Y. Localization and orientation of the VirD4 protein of Agrobacterium tumefaciens in the cell membrane. Mol Gen Genet. 1991;228:24–32. doi: 10.1007/BF00282443. [DOI] [PubMed] [Google Scholar]
- 59.Pohlman R F, Genetti H D, Winans S C. Common ancestry between IncN conjugal transfer genes and macromolecular export systems of plant and animal pathogens. Mol Microbiol. 1994;14:655–668. doi: 10.1111/j.1365-2958.1994.tb01304.x. [DOI] [PubMed] [Google Scholar]
- 60.Rashkova S, Spudich G M, Christie P J. Characterization of membrane and protein interaction determinants of the Agrobacterium tumefaciens VirB11 ATPase. J Bacteriol. 1997;179:583–591. doi: 10.1128/jb.179.3.583-591.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ream W. Agrobacterium tumefaciens and interkingdom genetic exchange. Annu Rev Phytopathol. 1989;27:583–618. doi: 10.1146/annurev.py.27.090189.003055. [DOI] [PubMed] [Google Scholar]
- 62.Riker A J. Studies on the influence of some environmental factors on the development of crown gall. J Agric Res. 1926;32:83–96. [Google Scholar]
- 63.Shirasu K, Kado C I. Membrane location of the Ti plasmid VirB proteins involved in the biosynthesis of a pilin-like conjugative structure on Agrobacterium tumefaciens. FEMS Microbiol Lett. 1993;111:287–294. doi: 10.1111/j.1574-6968.1993.tb06400.x. [DOI] [PubMed] [Google Scholar]
- 64.Shirasu K, Koukolikova-Nicola Z, Hohn B, Kado C I. An inner-membrane-associated virulence protein essential for T-DNA transfer from Agrobacterium tumefaciens to plants exhibits ATPase activity and similarities to conjugative transfer genes. Mol Microbiol. 1994;11:581–588. doi: 10.1111/j.1365-2958.1994.tb00338.x. [DOI] [PubMed] [Google Scholar]
- 65.Smit G, Tubbing D M J, Kijne J W, Lugtenberg B J J. Role of Ca+2 in the activity of ricadhesin from Rhizobium leguminosarum biovar viciae, which mediates the first step in attachment of Rhizobiaceae cells to plant root hair tips. Arch Microbiol. 1991;155:278–283. [Google Scholar]
- 66.Spudich G M, Fernandez D, Zhou X-R, Christie P J. Intermolecular disulfide bonds stabilize VirB7 homodimers and VirB7/VirB9 heterodimers during biogenesis of the Agrobacterium tumefaciens T-complex transport complex. Proc Natl Acad Sci USA. 1996;93:7512–7517. doi: 10.1073/pnas.93.15.7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stachel S E, Nester E W. The genetic and transcriptional organization of the A6 Ti plasmid of Agrobacterium tumefaciens. EMBO J. 1986;5:1445–1454. doi: 10.1002/j.1460-2075.1986.tb04381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stephens K M, Roush C, Nester E. Agrobacterium tumefaciens VirB11 protein requires a consensus nucleotide-binding site for function in virulence. J Bacteriol. 1995;177:27–36. doi: 10.1128/jb.177.1.27-36.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stock J B, Rauch B, Roseman S. Periplasmic space in Salmonella typhimurium and Escherichia coli. J Biol Chem. 1977;252:7850–7861. [PubMed] [Google Scholar]
- 70.Swart S, Lugtenberg B J J, Smit G, Kijne J W. Rhicadhesin-mediated attachment and virulence of an Agrobacterium tumefaciens chvB mutant can be restored by growth in a highly osmotic medium. J Bacteriol. 1994;176:3816–3819. doi: 10.1128/jb.176.12.3816-3819.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Swart S, Logman T J J, Lugtenberg B J J, Smit G, Kijne J W. Several phenotypic changes in the cell envelope of Agrobacterium tumfaciens chvB mutants are prevented by calcium limitation. Arch Microbiol. 1994;161:310–315. [Google Scholar]
- 72.Tempe J, Petit A, Holsters M, van Montagu M, Schell J. Thermosensitive step associated with transfer of the Ti plasmid during conjugation: possible relation to transformation in crown gall. Proc Natl Acad Sci USA. 1977;74:2848–2849. doi: 10.1073/pnas.74.7.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thorstenson Y R, Zambryski P C. The essential virulence protein VirB8 localizes to the inner membrane of Agrobacterium tumefaciens. J Bacteriol. 1994;176:1711–1717. doi: 10.1128/jb.176.6.1711-1717.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thorstenson Y R, Kuldau G A, Zambryski P C. Subcellular localization of seven VirB proteins of Agrobacterium tumefaciens: implications for the formation of a T-DNA transport structure. J Bacteriol. 1993;175:5233–5241. doi: 10.1128/jb.175.16.5233-5241.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tummuru M K R, Sharma S A, Blaser M J. Helicobacter pylori picB, a homologue of the Bordetella pertussis toxin secretion protein, is required for induction of IL-8 in gastric epithelial cells. Mol Microbiol. 1995;18:867–876. doi: 10.1111/j.1365-2958.1995.18050867.x. [DOI] [PubMed] [Google Scholar]
- 76.Uttaro A D, Cangelosi G A, Geremia R A, Nester E W, Ugalde R A. Biochemical characterization of avirulent exoC mutants of Agrobacterium tumefaciens. J Bacteriol. 1990;172:1640–1646. doi: 10.1128/jb.172.3.1640-1646.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ward J E, Dale E M, Binns A N. Activity of the Agrobacterium T-DNA transfer machinery is affected by virB gene products. Proc Natl Acad Sci USA. 1991;88:9350–9354. doi: 10.1073/pnas.88.20.9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ward J E, Dale E M, Christie P J, Nester E W, Binns A N. Complementation analysis of Agrobacterium tumefaciens Ti plasmid virB genes by use of a vir promoter expression vector: virB9, virB10, and virB11 are essential virulence genes. J Bacteriol. 1990;172:5187–5199. doi: 10.1128/jb.172.9.5187-5199.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ward J E, Dale E M, Nester E W, Binns A N. Identification of a VirB10 aggregate in the inner membrane of Agrobacterium tumefaciens. J Bacteriol. 1990;172:5200–5210. doi: 10.1128/jb.172.9.5200-5210.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weiss A A, Johnson F D, Burns D L. Molecular characterization of an operon required for pertussis toxin secretion. Proc Natl Acad Sci USA. 1993;90:2970–2974. doi: 10.1073/pnas.90.7.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Williamson M P. The structure and function of proline-rich regions in proteins. Biochem J. 1994;297:249–260. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Winans S C. Two-way chemical signaling in Agrobacterium-plant interactions. Microbiol Rev. 1992;56:12–31. doi: 10.1128/mr.56.1.12-31.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zambryski P. Chronicles from the Agrobacterium-plant cell DNA transfer story. Ann Rev Plant Physiol Plant Mol Biol. 1992;43:465–490. [Google Scholar]
- 84.Zhou X-R, Christie P J. Suppression of mutant phenotypes of the Agrobacterium tumefaciens VirB11 ATPase by overproduction of VirB proteins. J Bacteriol. 1997;179:5835–5842. doi: 10.1128/jb.179.18.5835-5842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zorreguieta A, Geremia R A, Cavaignac S, Cangelosi G A, Nester E W, Ugalde R A. Identification of the product of an Agrobacterium tumefaciens chromosomal virulence gene. Mol Plant-Microbe Interact. 1988;1:121–127. doi: 10.1094/mpmi-1-121. [DOI] [PubMed] [Google Scholar]