Abstract
Background
Tertiary lymphoid structure (TLS) is a unique organ that carries out tumor cell elimination at tumor sites. It is continuously stimulated by inflammatory tumor signals and has been found to augment immunotherapy response. However, the detailed mechanisms behind it still need to be defined.
Methods
To explore and grasp the whole picture of TLS from a pan-cancer view, we collected nine TLS-related genes from previous studies. We performed a comprehensive analysis of 9637 samples across 33 tumor types accessed from The Cancer Genome Atlas (TCGA) database. EdU, Transwell, and flow cytometry were performed on the feature gene PTGDS in U251 cells. The regulatory role of PTGDS on PD-L1 expression and macrophage polarization was verified.
Results
Alteration analysis showed that mutations of TLS-related genes were widespread and relatively high. Clustering analysis based on the expression of these nine genes obtained two distinct clusters, with high EIF1AY and PTGDS in cluster 2 and better overall survival in cluster 1. To distinguish the two clusters, we utilized six machine learning algorithms and filtrated EIF1AY, PTGDS, SKAP1, and RBP5 as the characteristic genes, among which the former two genes were proved to be hazardous. PTGDS was found to regulate PD-L1 expression and also promoted the proliferation and migration of U251 cells. The knockdown of PTGDS could reduce the migration of macrophages and inhibit the polarization of macrophages into M2-phenotype. In addition, we established a TLS score to demonstrate patients’ TLS activity. The low TLS-score group overlapped with cluster 1 and displayed a better prognosis. Besides, the low TLS-score group was related to better immunotherapy responses. The HE staining of histopathological sections confirmed that the low TLS-score group exhibited higher infiltration of immune cells.
Conclusion
This study reveals broad molecular, tumorigenic, and immunogenic signatures for further functional and therapeutic studies of tertiary lymphoid structure. The TLS score we established effectively predicted immunotherapy response and patients’ survival. Its future application and combination await more research.
Keywords: Tertiary lymphoid structure, Immunotherapy, Machine learning, Pan-cancer, Genomic alteration
1. Introduction
Cancer development is a fierce war between tumor cells and the human immune system. It has long been believed that adaptive immune response against tumor occurs in the secondary lymphoid organs (SLOs) [1]. However, newly published studies revealed further insight into the generation of antitumor response. They found that it could also occur directly at the tumor sites within organized cell aggregate resembling SLOs under the exposure to long-lasting inflammatory signals [2]. This cell aggregate is called tertiary lymphoid structure (TLS).
TLS consists of a CD3+ T cell zone containing dendritic cells (DCs) and fibroblastic reticular cells (FRCs) and a CD20+ B cell zone containing germinal center, plasma cells, and follicular DCs [1]. Being a rich resource of chemokines (including CCL19, CCL21, CXCL13, CCL17, CCL22, and IL-16), TLS could help foster an immunoreactive microenvironment recruiting tumor infiltrating lymphocytes (TILs), including CD4+, CD8+, CD20+ TILs, and plasma cells [[3], [4], [5]]. These compelling compositions facilitate the coordinated actions of CD8+ T cells and B cells, enabling in situ antitumor response and tumor cell elimination [4]. In addition, recent studies demonstrated that TLS could be a prognostic factor for complete pathological response and prolonged disease-free survival in diversified cancer types [3,[6], [7], [8]]. As for immunotherapy which is now considered an unprecedented breakthrough, a high TLS signature was found to predict better response to PD-1 blockade in bladder cancer and melanoma [[9], [10], [11]]. As researchers conclude, there is a disparity between the response rate to immunotherapy and the detected status of conventional biomarkers such as PD-L1 [12], PD-1, and tumor mutation burden (TMB) because one biomarker alone does not predict therapeutic efficacy perfectly [13]. Therefore, the discovery of TLS provides another potential indicator for immunotherapy to be of maximum use.
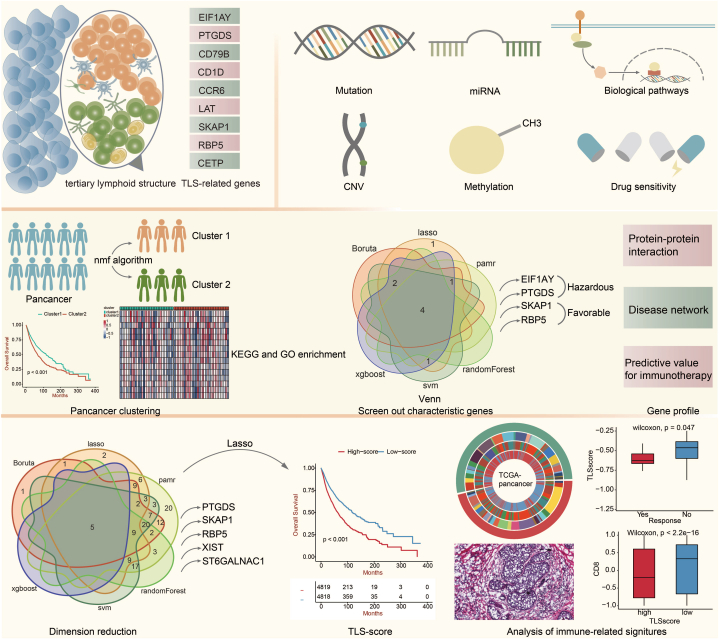
The ubiquitous presence of TLS has been unveiled in cancers at all stages, in primary as well as metastasis [14]. However, its presence varies from tumor to tumor. Regarding its active regulatory role in antitumor immune response, it is crucial to dig into the gene profile, biological function, and predictive value of TLS and understand the whole picture of it in tumors. There are limited studies on the pan-cancer analysis of TLS from all aspects mentioned above. Herein, we conducted an in-depth study based on nine TLS-related genes (EIF1AY, PTGDS, CD79B, CD1D, CCR6, LAT, SKAP1, RBP5, and CETP) [11], ranging from the alteration landscape in pan-tumor to the predictive model of survival, as well as gene function to immunotherapy efficacy (Fig. 1).
Fig. 1.
The flow diagram of this study.
2. Results
2.1. Alteration landscape of TLS-related genes in pan-cancer
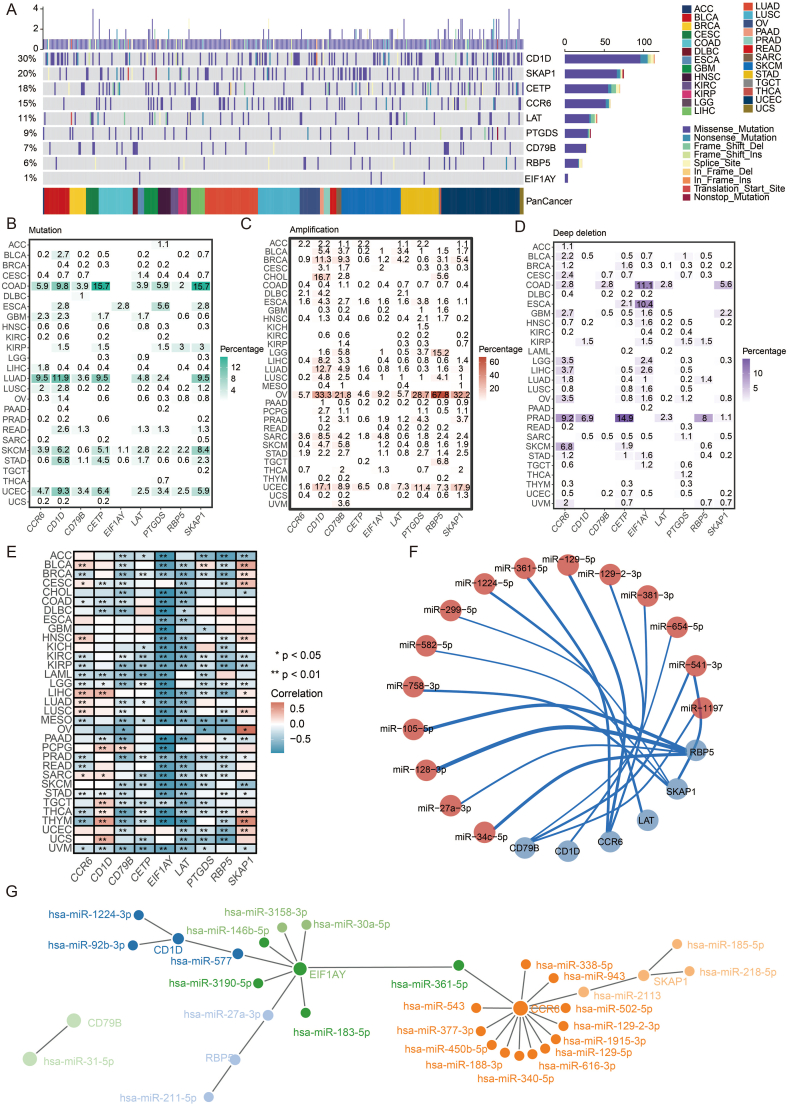
To understand the gene features of TLS-related genes across diversified malignancies, we analyzed 9637 patients across 33 tumors from TCGA and calculated the frequency of alteration, which was defined as mutation and somatic copy number alteration (SCNV) [15]. The overall mutation level of TLS-related genes ranged from 1 % to 30 % (Fig. 2a), which was relatively high and quite widespread in 26 types of tumors. Missense mutation constituted the majority of all mutation types, followed by nonsense mutation, frameshift deletion, and splice site mutation. Among the nine genes, CD1D exhibited the highest mutation level (30 %), most of which came from missense mutation. While EIF1AY showed the lowest mutation level (1 %), with all coming from missense mutation, only occurring in esophageal carcinoma (ESCA), cutaneous skin melanoma (SKCM), and stomach adenocarcinoma (STAD).
Fig. 2.
The mutation landscape and regulation network of TLS genes. a. Landscape of genomic alterations in the TLS genes in cancer. Each row represents a gene, and each column represents a patient. The frequency of alterations in nine genes of TLS is presented. b. Distribution of mutation frequencies over cancer types. c. Distribution of amplification of SCNA frequencies over cancer types. d. Distribution of depletion of SCNA frequencies over cancer types. e. Correlation between methylation and TLS genes over cancer types. f. Circos plot showing the regulation of miRNA on TLS genes. G. Network plot showing the regulation of miRNA on TLS genes.
Across the 26 tumor types, some were observed to have much higher mutation levels than others. Although the percentage was heterogeneous, SKCM and STAD harbored mutations in all nine genes (Fig. 2b). Colon adenocarcinoma (COAD) and uterine corpus endometrial carcinoma (UCEC) presented mutations in eight genes, and lung adenocarcinoma (LUAD) had seven out of nine. On the contrary, adrenocortical carcinoma (ACC), lymphoid neoplasm diffuse large B-cell lymphoma (DLBC), pancreatic adenocarcinoma (PAAD), testicular germ cell tumors (TGCT), and thyroid carcinoma (THCA) barely had mutations in these genes. In addition, a co-mutated pattern of SKAP1, CETP, CD1D, and CCR6 was observed, especially in COAD and LUAD.
Further, we analyzed the SCNV (including amplification and deletion) pattern in 26 tumor types. Ovarian cancer (OV) presented the highest amplification level in all nine genes, among which the amplification frequency of RBP5 was up to 67.8 % (Fig. 2c). The second highest amplification rate was observed in UCEC, which was also a tumor of the female reproductive system. As for deep deletion, prostate adenocarcinoma (PRAD) and COAD showed relatively high frequency (Fig. 2d).
2.2. Regulatory elements, associated biological pathways, and drug sensitivity of TLS-related genes
DNA methylation is a crucial element among the numerous regulatory factors of gene expression. Hypermethylation of the promoter region is usually associated with loss of expression of the target gene [16]. Although in some rare cases, DNA methylation positively correlates with gene expression [17]. Via spearman correlation analysis, methylation of SKAP1 and CD1D was found to correlate positively with gene expression in most tumor types. At the same time, EIF1AY, CD79B, and LAT negatively correlated with gene expression (Fig. 2e). In addition to methylation, miRNA is another key gene expression modulator. To get a comprehensive view of the miRNA regulation network, we screened available databases, and the results are displayed in Fig. 2f and g. The circle's line weight and color depth represent the modulatory intensity.
2.3. Two distinct clusters revealed by the NMF algorithm provided potential prognostic value
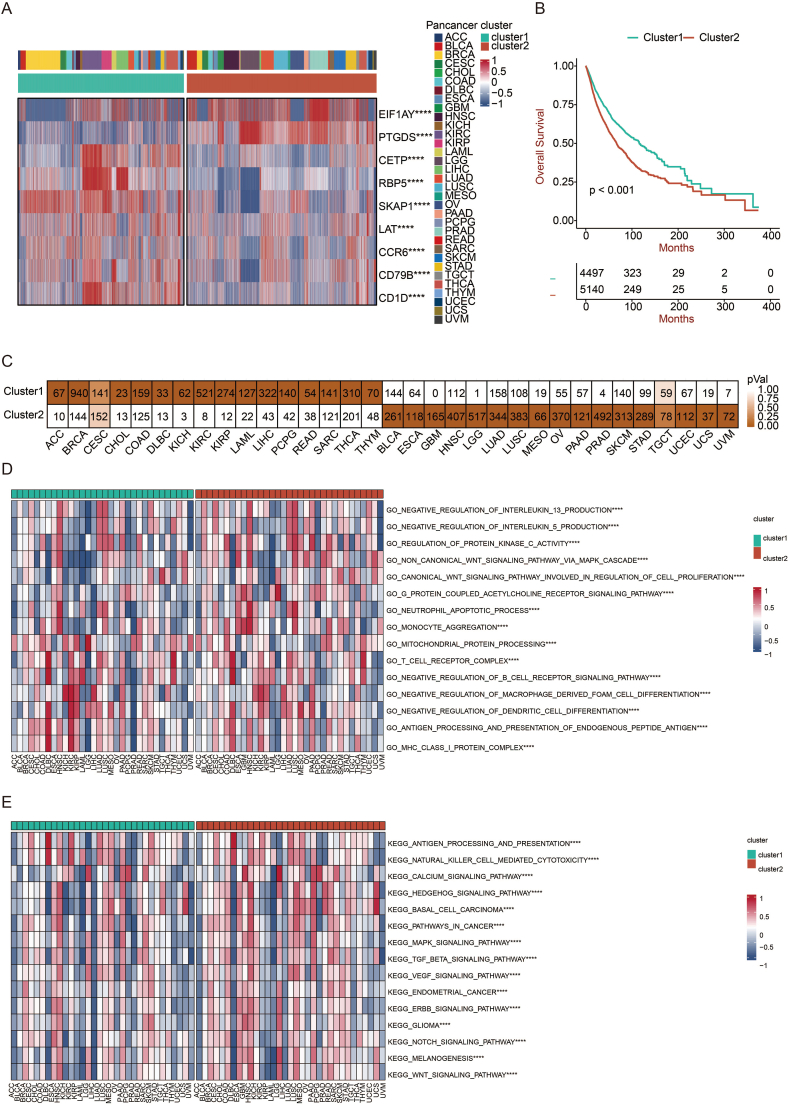
We then carried out a cluster analysis based on TLS-related gene expression via the NMF algorithm in pan-cancer perspective. The tumor was classified into two clusters, with high expression of EIF1AY and PTGDS in cluster 2 and the other seven genes in cluster 1 (Fig. 3a). Moreover, patients in cluster 1 exhibited better overall survival (OS) compared with patients in cluster 2 (p < 0.001) (Fig. 3b). As shown in Fig. 3c, out of 33 tumor types were overrepresented in cluster 1, and 16 tumor types in cluster 2, while cervical squamous cell carcinoma, endocervical adenocarcinoma (CESC) and TGCT were equally abundant in both clusters. The tumor types were uniformly distributed, indicating that this clustering method could distinguish tumors characterized by TLS-related genes.
Fig. 3.
Generation of TLS-related clusters. a. Heatmap showing the distribution of TLS genes in two clusters. b. The survival of patients in cluster 1 was compared with those in cluster 2 using the Kaplan–Meier survival curve in pan-cancer. A log-rank test assessed statistical significance. c. The distribution of two clusters in pan-cancer. d. GSVA results are based on GO terms in two clusters. e. GSVA results are based on KEGG terms in two clusters.
Further, we performed GO and KEGG enrichment analysis and obtained accordant results (Fig. 3d and 3e). Based on available data from GO, antigen processing and presentation, as well as MHC I complex, which are foundations of effective immune response [13], were upregulated in cluster 1. In contrast, the non-canonical WNT signaling pathway was upregulated in cluster 2, associated with neoplastic progression [18]. Upon KEGG analysis, an activated pattern of carcinogenesis pathway was observed in cluster 2. For example, MAPK signaling pathway, ERBB signaling pathway, and WNT signaling pathway were significantly upregulated [19,20].
2.4. Characteristic genes for accurate clustering and their profiles
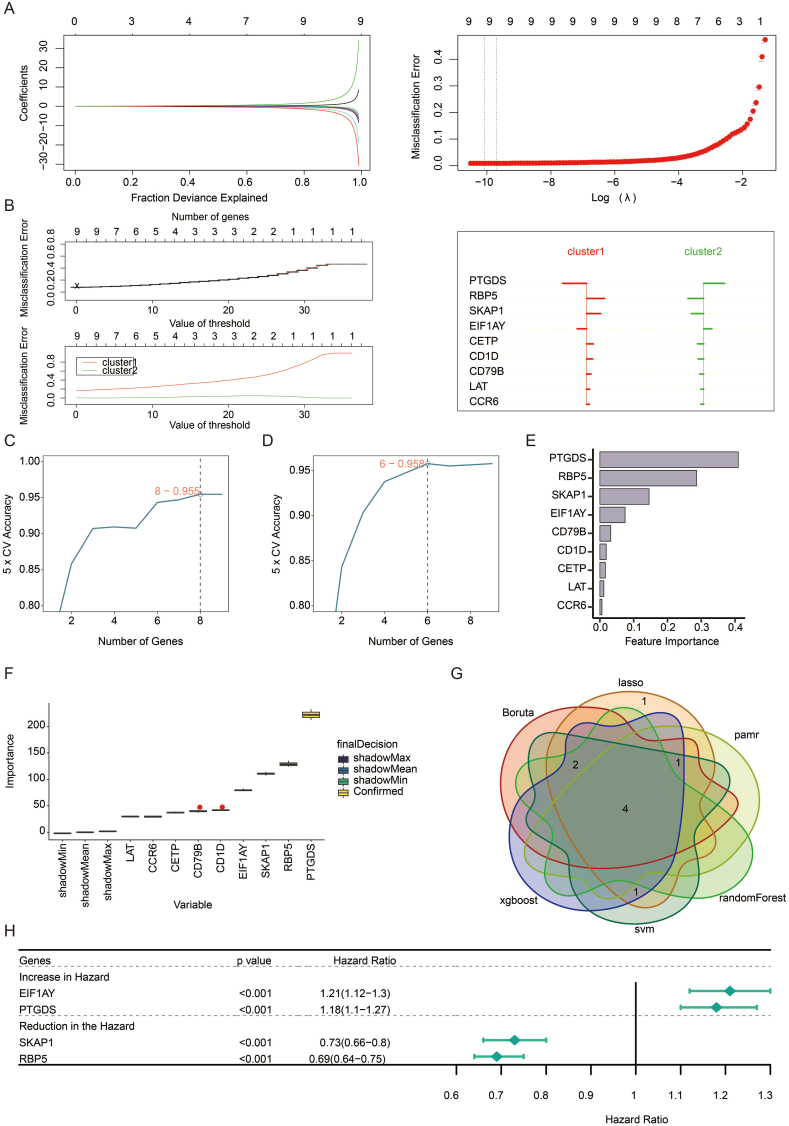
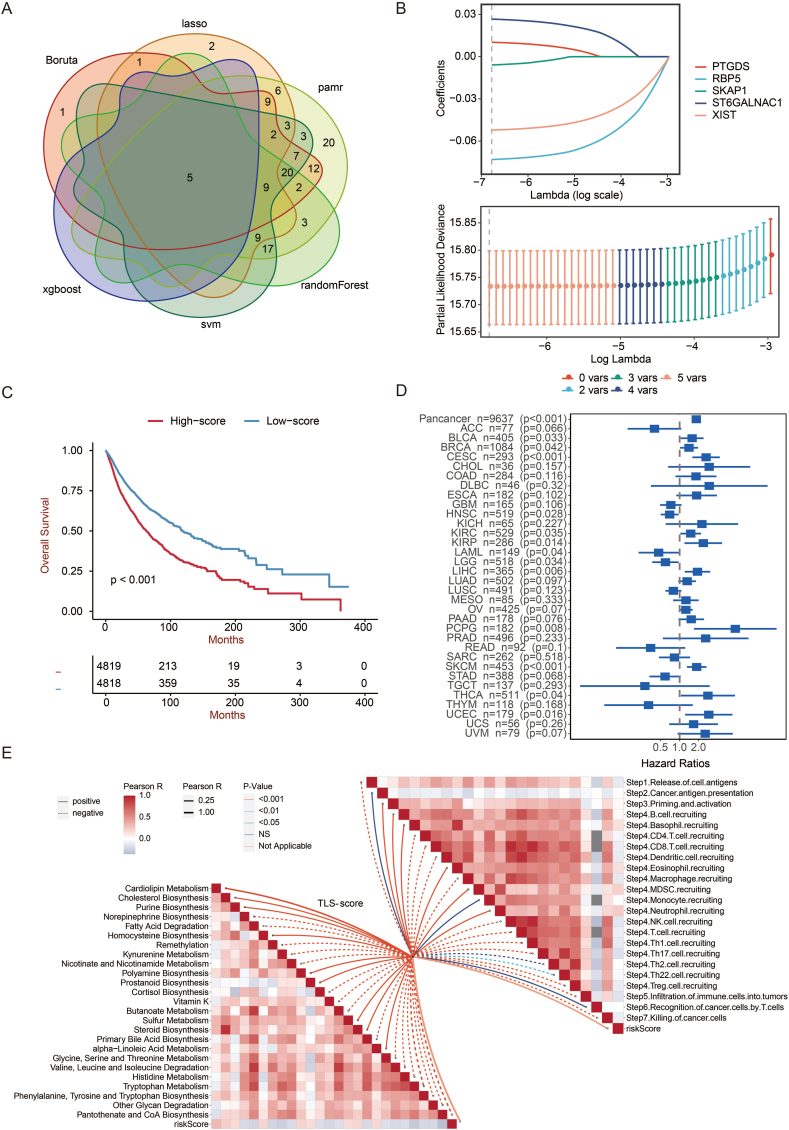
Next, we performed machine learning and prediction on the two populations to screen out the characteristic genes and accurately distinguish the two clusters. As shown in Fig. 4a–f, via LASSO-LR, pamr, SVM, random forest, Xgboost, and Boruta machine learning algorithms, we filtered 9, 9, 8, 6, 9, and 9 genes, respectively. The following Venn diagram displayed an intersection of the six algorithms, including four genes (EIF1AY, PTGDS, SKAP1, RBP5) (Fig. 4g), representing the characteristic genes that best classify the two clusters. By univariate regression analysis, the forest plot displayed an excellent association between these genes and prognosis (Fig. 4h). EIF1AY and PTGDS were both hazardous factors, while SKAP1 and RBP5 were favorable.
Fig. 4.
Excavation of feature genes using machine learning. a. Feature genes between two clusters defined by the LASSO-LR algorithm. b. The pamr algorithm defines feature genes between two clusters. c. The SVM algorithm defines feature genes between two clusters. d. A random forest algorithm defines feature genes between two clusters. e. The Xgboost algorithm defines feature genes between two clusters. f. The Boruta algorithm defines feature genes between two clusters. g. Venn diagram showing the intersected feature genes defined by six machine learning algorithms. h. Forest plot showing the prognostic value of feature genes in pan-cancer.
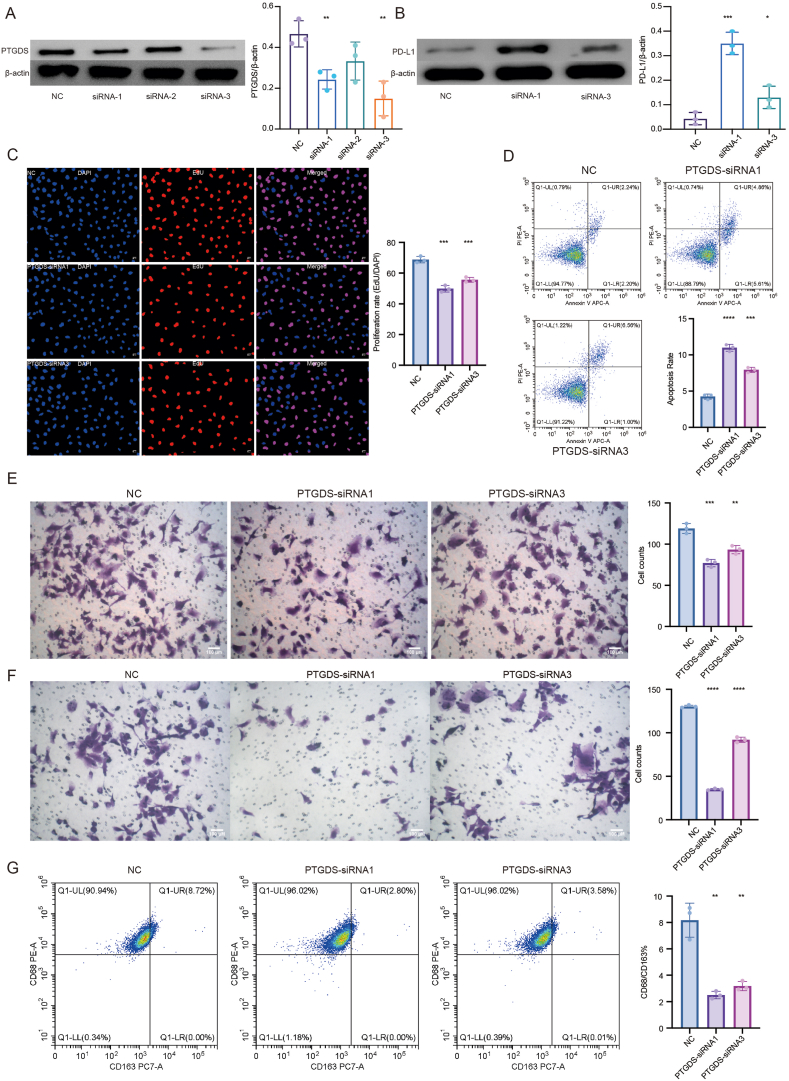
PTGDS has been found to be involved in lipid metabolism, cell self-renewal, tumorigenesis and tumor infiltrating immune cells in multiple cancer types [[21], [22], [23], [24]]. Given the potential value in clinical use as a hazardous factor for prognosis and the relatively adequate studies of PTGDS [25], we performed preliminary experiments to verify the relationship between PTGDS and existing immunotherapy biomarkers. The results showed that the knockdown of PTGDS by siRNA led to increased expression of PD-L1 in U251 cells (Fig. 5aand 5b), indicating the potential regulatory correlation and possible combined therapeutic targets for immunotherapy. Moreover, the knockdown of PTGDS impaired cell proliferation, migration, and invasion and promoted cell apoptosis in U251 cells (Fig. 5c-e), adding to the tumor-killing efficacy of this potential target. We also found that the knockdown of PTGDS could reduce the migration of macrophages and inhibit the polarization of macrophages into M2-phenotype (Fig. 5f and 5g), indicating the potential impact of PTGDS on remodeling tumor immune microenvironment, which is crucial to the formation of TLS and anti-tumor immune response.
Fig. 5.
a and b. Western blots demonstrated an increase of PD-L1 in PTGDS-knockdown U251 cells. The full images of Western blots are provided in the Supplementary files (Figs. S4–7). c. EdU showed a decrease in cell proliferation in PTGDS-knockdown U251 cells. d. Flow cytometry showed an increase of cell apoptosis in PTGDS-knockdown U251 cells. e. Transwell showed a reduction of cell migration and invasion in PTGDS-knockdown U251 cells. f. Transwell showed a reduction of cocultured HMC3 cell migration with PTGDS-knockdown U251 cells. g. Flow cytometry showed a reduction in M2 polarization of cocultured HMC3 cells with PTGDS-knockdown U251 cells.
2.5. Establishment of TLS-score
Based on the two clusters we got, we independently analyzed differential genes and the dimension reduction via the previous six algorithms (Fig. 6a). PTGDS, RBP5, SKAP1, ST6GALNAC1, and XIST were picked out, on the strength of a scoring system we established via the LASSO algorithm. The coefficient of each gene is displayed in Fig. 6b. Next, we divided the patients into a high TLS-score group and a low TLS-score group by the median score. It turned out that the low TLS-score group had a significantly better OS than that of the high TLS-score group (p < 0.001) (Fig. 6c). We further validated the effectiveness of the scoring system in pan-cancer and individual tumors. The forest plot showed that the score was hazardous in most tumor types (Fig. 6d), confirming the result of the Kaplan-Meier curve. As expected, TLS-score negatively correlated with multiple steps in anti-tumor immune response, including the release of cell antigens, recruiting of critical immune cells such as CD4+ T cell, CD8+ T cell, dendritic cell, NK cell, macrophage cell, Th1 cell, and Th17 cell, as well as infiltration of immune cells (Fig. 6e).
Fig. 6.
Generation of TLS score. a. Venn diagram showing the intersected genes after dimension reduction based on six machine learning algorithms. b. Correlation coefficients of intersected genes based on LASSO-LR algorithm for the generation of TLS score. c. The survival of patients with high TLS-scores was compared with those with low TLS-scores using the Kaplan–Meier survival curve in pan-cancer. A log-rank test assessed statistical significance. d. Forest plot showing the prognostic value of TLS score in pan-cancer. e. The butterfly plot shows the correlation between TLS score, metabolism, and cancer immunity cycle.
2.6. TLS-score: an effective predictor for immunotherapy response and prognosis
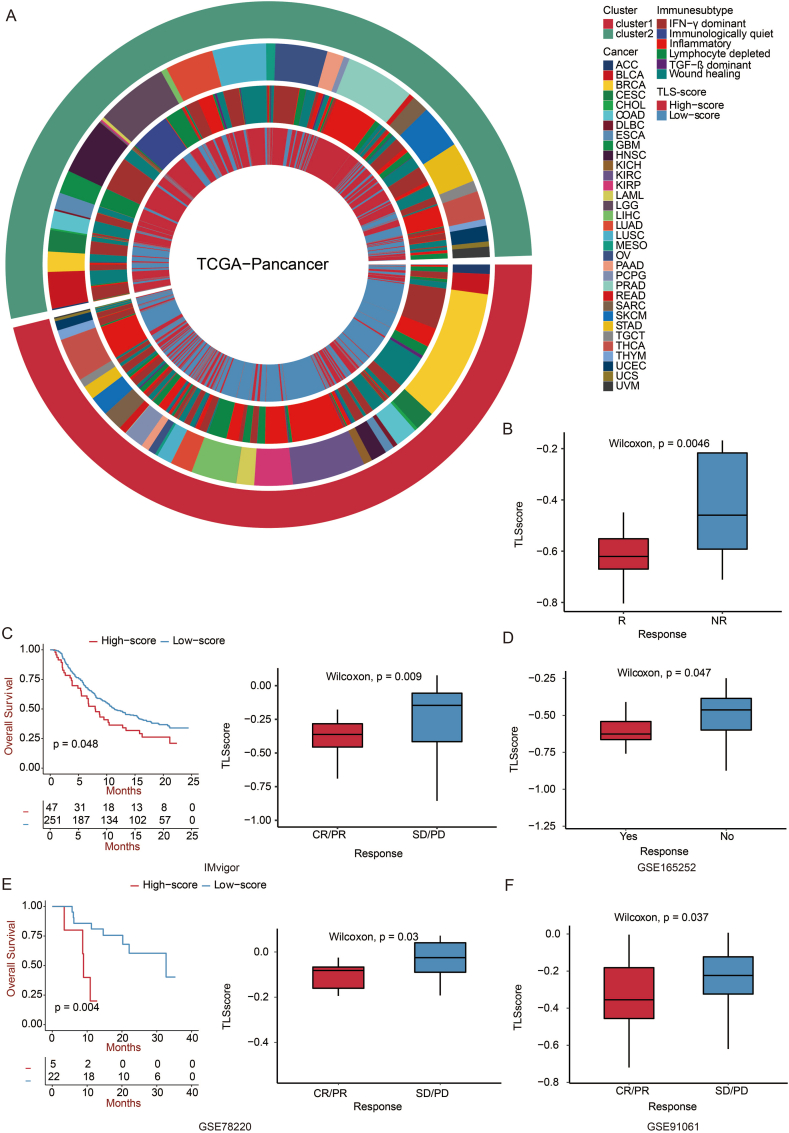
We then calculated the TLS score accordant to the two clusters to verify it. We compared the two score groups and the two clusters in pan-cancer and six immune subtypes [26]. As expected, the low TLS-score group almost all corresponded to cluster 1 and had a better OS (Fig. 7a). Furthermore, cluster 1 exhibited a higher percentage of inflammatory immune subtypes, while the quiet immunological subtype only occurred in cluster 2. According to Thorsson et al. the inflammatory subtype was defined by elevated Th17 and Th1 genes, low to moderate tumor cell proliferation, and the best prognosis. On the contrary, the quiet immunological subtype displayed the lowest lymphocyte infiltration level and the strongest M2 macrophage response [26]. Consistent with the above results, a low TLS score was validated with better OS and superior immunotherapy response in GSE35640, IMvigor, GSE165252, GSE78220, and GSE91061 (Fig. 7b–f). The TLS score was a good predictor for individual tumors for OS (Fig. S1).
Fig. 7.
Prediction of TLS score in immunotherapy response. a. Circos plot showing the distribution of six immune subtypes in two clusters and TLS score groups in pan-cancer. b. Box plot showing the TLS score in patients with or without immunotherapy response in the GSE35640 dataset. The Wilcoxon rank test evaluated statistical importance. c. Overall survival of patients with high TLS-scores was compared with those with low TLS-scores using Kaplan–Meier survival curve in IMvigor210 dataset. A log-rank test assessed statistical significance. Box plot showing the TLS score in patients with complete response/partial response and stable disease/progressive disease. The Wilcoxon rank test assessed statistical significance. d. Box plot showing the TLS score in patients with or without immunotherapy response in the GSE165252 dataset. The Wilcoxon rank test evaluated statistical importance. e. The survival of patients with high TLS-scores was compared with those with low TLS-scores using the Kaplan–Meier survival curve in the GSE78220 dataset. A log-rank test assessed statistical significance. Box plot showing the TLS score in patients with complete response/partial response and stable disease/progressive disease. The Wilcoxon rank test assessed statistical significance. f. Box plot showing the TLS score in patients with complete response/partial response and stable disease/progressive disease in the GSE91061 dataset. The Wilcoxon rank test assessed statistical significance.
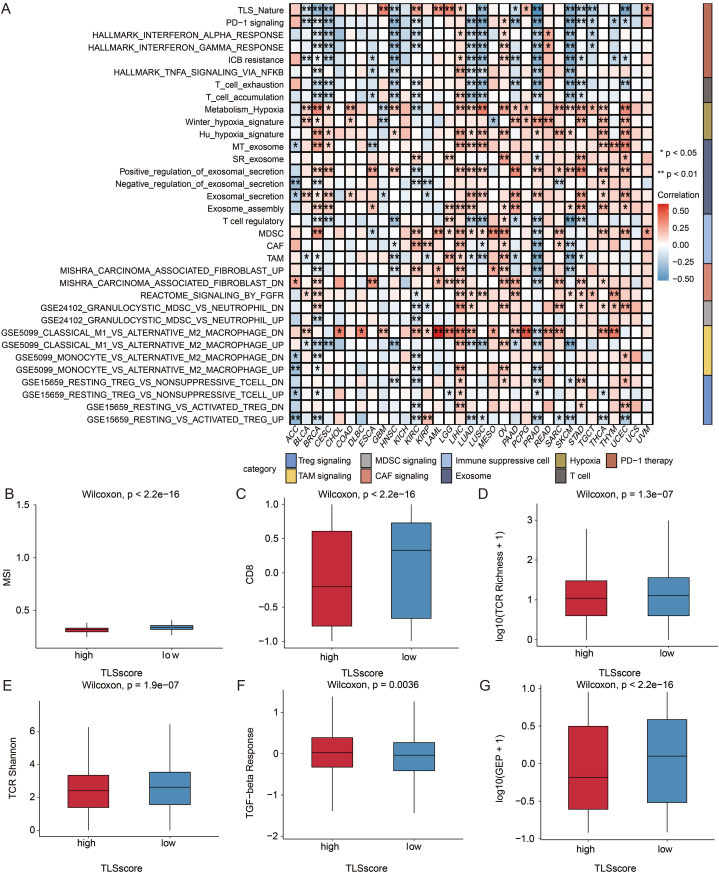
Diving deeper into the mechanisms, we observed a significant positive association between TLS score and hypoxia, exosome, immune suppressive cell, and M2 ratio. In contrast, PD-1 signaling and T-cell signaling were negatively correlated with the TLS score (Fig. 8a). Via ssGSEA immune cell analysis, a trend of negative correlation was also observed in pan-cancer (Fig. S2a). The heatmap of immune checkpoints echoed the above results (Fig. S2b).
Fig. 8.
Immune characteristics of TLS score in pan-cancer. a. Heatmap showing the correlation between selected immune signatures and TLS score. b. Box plot showing the MSI level in patients with high and low TLS scores. The Wilcoxon rank test assessed statistical significance. c. Box plot showing the CD8 level in patients with high and low TLS scores. The Wilcoxon rank test assessed statistical significance. d. Box plot showing the TCR richness level in patients with high and low TLS scores. The Wilcoxon rank test assessed statistical significance. e. Box plot showing the TCR Shannon level in patients with high and low TLS scores. The Wilcoxon rank test assessed statistical significance. f. Box plot showing the TGF-beta level in patients with high and low TLS scores. The Wilcoxon rank test assessed statistical significance. g. Box plot showing the GEP level in patients with high and low TLS scores. The Wilcoxon rank test assessed statistical significance.
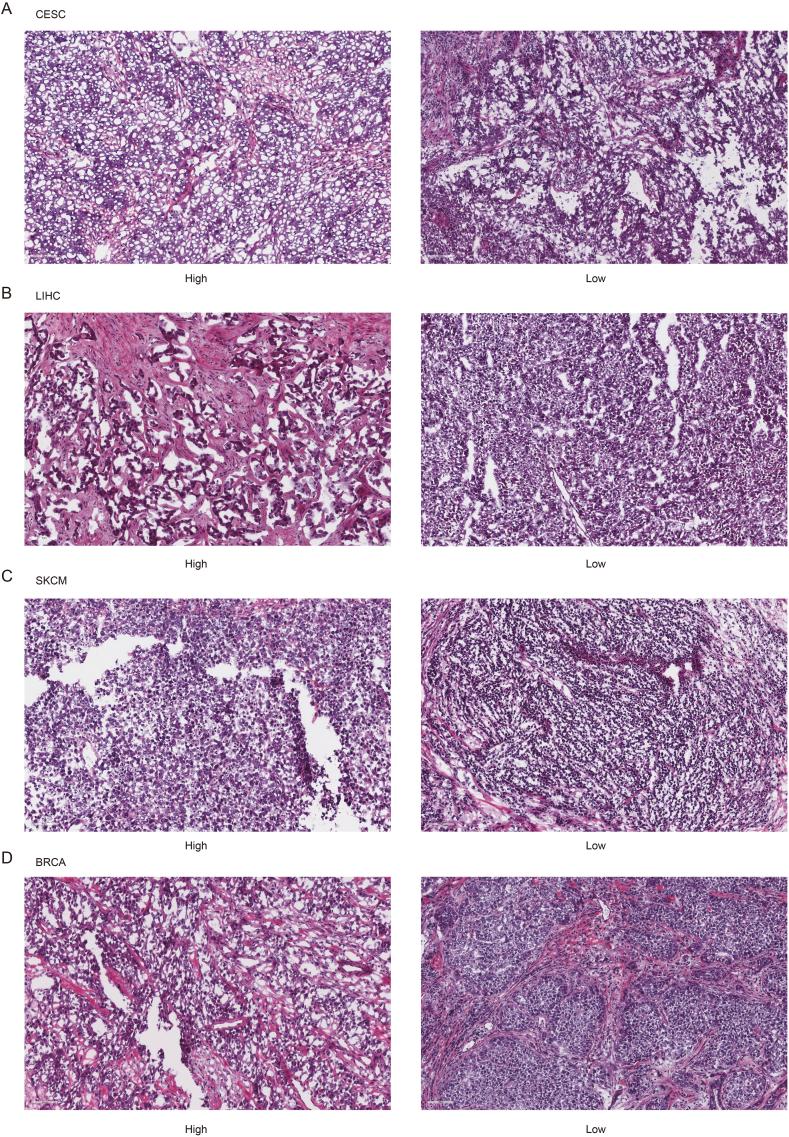
To be more detailed, we compared the high TLS-score and low TLS-score groups with multiple indicators and presented the results in a boxplot. MSI, CD8+ T cell, and TCR richness, landmarks of the immunoreactive tumor microenvironment (TME), were significantly higher in the low TLS-score group (Fig. 8b–e). The HE staining of histopathological sections confirmed that the low TLS-score group exhibited higher infiltration of immune cells (Fig. 9a–d). Transforming growth factor-beta (TGF-beta), which exerted immune suppression and inhibited host immunosurveillance was higher in the high TLS-score group (Fig. 8f) [27]. T cell-inflamed gene expression profile (GEP) was higher in the low TLS-score group, indicating a superior response to PD-1 blockade (Fig. 8g) [28]. Besides, the low TLS-score group exhibited a lower aneuploidy score, altered fraction, number of segments, nonsilent mutation rate, homologous recombination deficiency (HRD), and intratumor heterogeneity (Figs. S3a–f), all of which are clues for genomic instability. As researchers proposed, excessive genomic instability could lead to high intratumor heterogeneity and impair the efficacy of immunotherapy in the end [13]. In addition, the antigen-presenting mechanism (APM) score showed more elevated levels in the low TLS-score group (Fig. S3g). Cancer testis antigen (CTA), a biomarker of tumor proliferation and progression [29], was also lower in the low TLS-score group (Fig. S3h). Moreover, leukocyte fraction and IFNG-related signatures were higher in the low TLS-score group (Fig. S3i-l), suggesting a better footstone for effective immunotherapy.
Fig. 9.
Characteristics of immune infiltration in two TLS score groups. a. Representative pictures of pathological HE staining of two TLS score groups in CESC from TCGA dataset. b. Representative pictures of pathological HE staining of two TLS score groups in LIHC from TCGA dataset. c. Representative pictures of pathological HE staining of two TLS score groups in SKCM from TCGA dataset. d. Representative pictures of pathological HE staining of two TLS score groups in BRCA from TCGA dataset.
3. Discussion
Located in the peripheral region of tumor nests, TLS is sustained by long-lasting tumor-associated inflammation and is privileged to kill tumor cells [30]. It has been demonstrated that TLS could facilitate immunotherapy and promote survival in multiple cancers [11,14,31]. The most important thing right now is to define a group of markers which could best characterize TLS to maximize the value of it as potential biomarker. Machine learning plays a critical role in this part. Gene-based signatures are mathematical models that use various data inputs, such as patient characteristics, genetic information, and clinical data, to predict the outcome or prognosis of a disease. Machine learning algorithms are used to analyze and identify patterns within these data inputs, allowing researchers to develop accurate prognostic models. In this study, to grasp the whole picture of TLS-related genes in cancer, we performed a comprehensive analysis from appearance to essence, including the alteration landscape, the modulators, clustering ability, associated biological pathways, and potential value as biomarkers. For the ultimate goal, we established a TLS score and proved its predictive potency in various ways.
Mutations of TLS-related genes were widespread and exhibited relatively high frequency in cancers. SKCM, STAD, COAD, UCEC, and LUAD had the highest mutation levels. And OV presented the highest amplification level in all nine genes. Interestingly, TLS-related genes displayed a significantly activating effect on ER and PR, which are both hormone receptors, and may be involved in the carcinogenesis of UCEC and OV.
Based on the expression of TLS-related genes, pan-cancer samples fell into two clusters, with high expression of EIF1AY and PTGDS in cluster 2. Patients in cluster 1 exhibited better OS, and correspondingly, antigen processing and MHC I were upregulated in cluster 1, indicating an excellent start to the immune cycle. In contrast, WNT signaling, MAPK signaling, and ERBB signaling were upregulated in cluster 2, which may result in resistance to immunotherapy [32,33]. After LASSO-LR, pamr, SVM, random forest, Xgboost, and Boruta machine learning algorithms [34,35], we screened out EIF1AY, PTGDS, SKAP1, and RBP5 as the characteristic genes to best distinguish the two clusters. Through a series of analyses, EIF1AY and PTGDS were found to be hazardous and more related to the occurrence of diseases. Unlike EIF1AY, which is barely studied in cancers and needs further investigation, PTGDS is a member of the lipocalin superfamily and plays a dual role in prostaglandins metabolism and lipid transport, involved in tumorigenesis of both solid and hematological malignancies [36]. SKAP1 and RBP5 were favorable factors. The former plays a critical role in participating in T cell motility and interactions in lymph nodes [37]. And down-regulation of RBP5 was associated with aggressive features in cancers [38]. Among the four genes, PTGDS regulated the expression of PD-L1 and the proliferation, migration, invasion and apoptosis of tumor cells, and exhibited huge potential in impacting macrophage migration and polarization, showing huge potential for predicting immunotherapy response. Nevertheless, the predictive capacity of these four genes remained unsatisfactory. As many studies have complained, conventional biomarkers like MSI, TMB, and CD274 alone did not predict therapeutic efficacy perfectly [39,40], and combined biomarkers could make up for each other, calling for an effective predictive model using existing biomarkers.
To further explore the association between TLS and immunotherapy, we analyzed the differential genes of the two clusters and established a scoring system through the LASSO-LR algorithm. The TLS score included six genes (PTGDS, RBP5, SKAP1, ST6GALNAC1, and XIST), in addition to the first three genes, high expression of ST6GALNAC1 induced by M2-like macrophages is associated with the onset of colitis-associated colon cancer, and high tumor-initiating, self-renewal, and differentiation abilities [41,42], and XIST mainly functions to modulate cell proliferation and tumor growth [43]. We classified patients into high and low TLS-score groups by the median score. It turned out that the low TLS-score group significantly exhibited a better OS, and almost all overlapped with cluster 1, which verified the efficacy of our clustering and scoring methods. Moreover, after comparing the six immune subtypes demonstrated in the previous study [26], we discovered inflammatory subtype predominated in cluster 1 and the low TLS-score group. This immune subtype was characterized by elevated Th17 and Th1 genes, low to moderate tumor cell proliferation, and displayed the best prognosis. In the end, we found out what a low TLS score really was. It represented a more active immune process, including releasing cell antigens, recruiting immune cells, and upregulating immune infiltration level. It also presented many characteristics beneficial to immunotherapy, such as higher MSI level, CD8+ T cell level, TCR richness, APM score, IFNG level, lower TGF-beta level, intratumor heterogeneity, and CTA score. The characteristic genes of TLS score may impact or reflect multiple levels of the patients’ immune status from a global perspective, such as immunogenic cell death, antigen presentation, and immune cell infiltration, as we have demonstrated PTGDS as a regulator of macrophage recruitment and polarization in this study. The TLS score could serve as a clustering method and distinguish immunoreactive populations from those less likely to benefit from immunotherapy to guide us in making the optimal therapeutic option. Further, previous studies showed that immunotherapy could promote the formation of TLS [14]. Therefore, using TLS score as a longitudinal evaluation could help identify patients who are long-term beneficiaries.
TLS plays a role in orchestrating other lymphoid structures to carry out an anti-tumor immune response. It may be a small part of a larger immune war, but it has the unique advantage of fighting capacity. The TLS score we established effectively predicted immunotherapy response and patients’ survival. Its future application and combination await more research.
4. Materials and methods
4.1. Data collecting and preprocessing
Transcriptomic data (HiSeq Illumina platform) and clinical information of 9637 pan-cancer samples across 33 cancer types were downloaded from The Cancer Genome Atlas (TCGA) dataset (UCSC Xena, https://xenabrowser.net/).
4.2. Genomic alteration analysis
Somatic mutation and copy number alteration data of TCGA pan-cancer samples were collected from Genomic Data Commons. The somatic copy number alteration (SCNA) (amplification and deep deletion) and mutation frequency (truncating and missense) scores of the pan-cancer samples were calculated. Values of the SCNA scores equal to 2 and −2 were referred to as amplification and deep deletion, respectively [44]. The R package ComplexHeatmap (https://github.com/jokergoo/ComplexHeatmap) was used to generate the oncoPrint plot of pan-cancer samples regarding mutation and SCNA of TLS signature genes.
4.3. DNA methylation analysis
DNA methylation data of TCGA pan-cancer samples were collected from Genomic Data Commons. The R package IlluminaHumanMethylation450k.db was used for mapping the methylation array probes to corresponding genes. The median beta values were used for genes mapped with multiple methylation array probes. The median beta value for TLS signature genes in each pan-cancer sample was calculated to evaluate the overall methylation level. The correlation between DNA methylation beta value and mRNA expression for each TLS signature gene was calculated.
4.4. Nonnegative matrix factorization (NMF) clustering for TLS signature genes
TCGA pan-cancer samples with diverse expression patterns of TLS signature genes were classified using the NMF algorithm (https://github.com/renozao/NMF). The optimal number of clusters was determined based on the cophenetic values.
4.5. Estimation of immune infiltration and functional annotation
The relative abundance of immune infiltrating cells was calculated using the xCell algorithm [45] and ssGSEA algorithm. Gene set variation analysis (GSVA) on Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) terms were performed using the R package GSVA [46].
4.6. Machine learning for feature genes
The differentially expressed genes (DEGs) between TLS-related clusters were identified using the R package limma. Machine learning algorithms, including random forest, support vector machines (SVM), and prediction analysis for microarrays (pamr), Xgboost, Boruta, and LASSO-LR, were used for screening out the most potent genes for clustering.
4.7. Cell culture
U251 and HMC3 cells were purchased from iCell (http://www.icellbioscience.com). U251 cells were cultured in Dulbecco's modified eagle medium (DMEM) with 10 % fetal bovine serum (FBS) in the saturated humidity incubator (37 °C, 5 % CO2). HMC3 cells were cultured in 1640 medium with 10 % FBS in the saturated humidity incubator (37 °C, 5 % CO2).
4.8. Cell transfection
PTGDS-siRNA-1 (sense 5′-3′, CCAACUUCCAGCAGGACAAGUTT; antisense 5′-3′, ACUUGUCCUGCUGGAAGUUGGTT), PTGDS-siRNA-2 (sense 5′-3′, CACCUACUCCGUGUCAGUGGUTT; antisense 5′-3′, ACCACUGACACGGAGUAGGUGTT), and PTGDS-siRNA-3 (sense 5′-3′, GAUAAGUGCAUGACGGAACAATT; antisense 5′-3′, UUGUUCCGUCAUGCACUUAUCTT) were used for cell transfection in U251 cells.
4.9. Western blotting
The western blotting assay assessed the expression level of PTGDS, PD-L1, and β-actin. Anti-PD-L1 (Rabbit, 1:2000, Proteintech, China), anti-PTGDS (1:1000, Rabbit, Proteintech, China), and anti-β-actin (Mouse, 1:5000, Proteintech, China) were used as the primary antibody. HRP goat anti-mouse IgG (Mouse, 1:5000, Proteintech, China) and HRP goat anti-rabbit IgG (Rabbit, 1:6000, Proteintech, China) were used as the secondary antibody. ECL development was used for visualization.
4.10. EdU assay
U251 cells transfected with siRNA of PTGDS were collected for EdU assay (NC, PTGDS-siRNA1, PTGDS-siRNA3). Please see the supplementary materials for detailed methods.
4.11. Transwell assay
U251 cells transfected with siRNA of PTGDS were collected for Transwell assay (NC, PTGDS-siRNA1, PTGDS-siRNA3). After the coculture between U251 and HMC3 cells, the cocultured HMC3 cells were collected for Transwell assay (NC, PTGDS-siRNA1, PTGDS-siRNA3). Please see the supplementary materials for detailed methods.
4.12. Apoptosis assay
U251 cells transfected with siRNA of PTGDS were collected for Apoptosis assay (NC, PTGDS-siRNA1, PTGDS-siRNA3). Please see the supplementary materials for detailed methods.
4.13. Flow cytometry
After the coculture between U251 and HMC3 cells, the cocultured HMC3 cells were collected for flow cytometry (NC, PTGDS-siRNA1, PTGDS-siRNA3). Please see the supplementary materials for detailed methods.
4.14. HE staining
Representative pictures of pathological HE staining of two TLS score groups in CESC, LIHC, SKCM, and BRCA samples were collected from TCGA dataset.
4.15. Construction of the risk signature
Machine learning algorithms, including random forest, SVM, pamr, Xgboost, Boruta, and LASSO-LR, were used for dimension reduction, respectively. A risk signature was further developed using LASSO regression analysis based on the intersected genes identified by these machine learning algorithms.
4.16. Prediction of immunotherapy
The GSE35640 (melanoma dataset), IMvigor210 cohort (urothelial carcinoma cohort), GSE91061 (melanoma dataset), GSE165252 (Esophageal adenocarcinoma), and the GSE78220 (melanoma dataset) were collected to predict the immunotherapy response [[47], [48], [49], [50]]. The expression value of raw data from all datasets was transformed to TPM value. TLS signature score was calculated in these cohorts, respectively.
4.17. Annotation from the online portal
The disease network of the feature genes was identified and visualized in the OPENTARGET platform (https://platform.opentargets.org) [51]. The protein interaction of the feature genes was pictured in the STRING database (https://string-db.org).
4.18. Statistical analysis
A univariate Cox proportional hazards regression model was used to assess the prognostic value of the TLS signature score. A two-sided Wilcoxon rank-sum test was used for comparison between the two groups. The Kaplan–Meier method was used to compare survival differences between the two groups. All statistical analyses were performed using the R project (version 3.6.3).
Ethics statement
Review and/or approval by an ethics committee was not needed for this study because there was no patient or animal experiment involved in this study. Informed consent was not required for this study because there were no participants or patients involved in this study.
Consent for publication
Not applicable.
Consent for participate
Written informed consent was obtained from all patients.
Code availability
All statistical analysis was performed on R project 3.6.3.
Funding
The authors acknowledge the financial support from the National Natural Science Foundation of China (NO. 82303035); Hunan Provincial Natural Science Foundation of China (NO. 2021JJ40321); The Science and Technology Innovation Program of Hunan Province (NO. 2020RC2065).
Data availability statement
All data used in this work can be acquired from the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/), the Cancer Genome Atlas (TCGA) datasets (https://xenabrowser.net/), the Chinese Glioma Genome Atlas (CGGA) datasets (http://www.cgga.org.cn/). Data included in article/supp. material/referenced in article.
CRediT authorship contribution statement
Wantao Wu: Writing – review & editing, Writing – original draft, Validation. He Li: Writing – review & editing, Writing – original draft. Zeyu Wang: Data curation. Ziyu Dai: Methodology. Xisong Liang: Software. Peng Luo: Investigation. Kun Liu: Software. Hao Zhang: Formal analysis. Nan Zhang: Methodology. Shuyu Li: Resources, Project administration. Chi Zhang: Supervision, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e23915.
Contributor Information
Shuyu Li, Email: lishuyu@tjh.tjmu.edu.cn.
Chi Zhang, Email: zhangchi25@csu.edu.cn.
Abbreviations
- TLS
Tertiary lymphoid structure
- SLOs
Secondary lymphoid organs
- DCs
Dendritic cells
- FRCs
Fibroblastic reticular cells
- TILs
Tumor-infiltrating lymphocytes
- TMB
Tumor mutation burden
- TCGA
the Cancer Genome Atlas
- SCNA
Somatic copy number alteration
- NMF
Nonnegative Matrix Factorization
- GSVA
Gene set variation analysis
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- DEGs
The differentially expressed genes
- SVM
Support vector machines
- Pamr
Prediction analysis for microarrays
- ESCA
Esophageal carcinoma
- SKCM
Skin cutaneous melanoma
- STAD
Stomach adenocarcinoma
- COAD
Colon adenocarcinoma
- UCEC
Uterine corpus endometrial carcinoma
- LUAD
Lung adenocarcinoma
- ACC
Adrenocortical carcinoma
- DLBC
Lymphoid neoplasm diffuse large B-cell lymphoma
- PAAD
Pancreatic adenocarcinoma
- TGCT
Testicular germ cell tumors
- THCA
Thyroid carcinoma
- OV
Ovarian cancer
- PRAD
Prostate adenocarcinoma
- AR
Androgen receptor
- ER
Estrogen receptor
- EMT
Epithelial-mesenchymal transition
- OS
Overall survival
- CESC
Endocervical adenocarcinoma
- MSI
Microsatellite instability
- IFNG
Interferon-gamma
- TME
Tumor microenvironment
- TGF-beta
Transforming growth factor-beta
- GEP
Gene expression profile
- HRD
Homologous recombination deficiency
- APM
Antigen presenting mechanism
- CTA
Cancer testis antigen
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Sautes-Fridman C., Petitprez F., Calderaro J., Fridman W.H. Tertiary lymphoid structures in the era of cancer immunotherapy. Nat. Rev. Cancer. 2019;19:307–325. doi: 10.1038/s41568-019-0144-6. [DOI] [PubMed] [Google Scholar]
- 2.Drayton D.L., Liao S., Mounzer R.H., Ruddle N.H. Lymphoid organ development: from ontogeny to neogenesis. Nat. Immunol. 2006;7:344–353. doi: 10.1038/ni1330. [DOI] [PubMed] [Google Scholar]
- 3.Zhang W.H., et al. Infiltrating pattern and prognostic value of tertiary lymphoid structures in resected non-functional pancreatic neuroendocrine tumors. J Immunother Cancer. 2020;8 doi: 10.1136/jitc-2020-001188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kroeger D.R., Milne K., Nelson B.H. Tumor-infiltrating plasma cells are associated with tertiary lymphoid structures, cytolytic T-cell responses, and superior prognosis in ovarian cancer. Clin. Cancer Res. 2016;22:3005–3015. doi: 10.1158/1078-0432.CCR-15-2762. [DOI] [PubMed] [Google Scholar]
- 5.de Chaisemartin L., et al. Characterization of chemokines and adhesion molecules associated with T cell presence in tertiary lymphoid structures in human lung cancer. Cancer Res. 2011;71:6391–6399. doi: 10.1158/0008-5472.CAN-11-0952. [DOI] [PubMed] [Google Scholar]
- 6.Song I.H., et al. Predictive value of tertiary lymphoid structures assessed by high endothelial venule counts in the neoadjuvant setting of triple-negative breast cancer. Cancer Res Treat. 2017;49:399–407. doi: 10.4143/crt.2016.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee H.J., et al. Prognostic significance of tumor-infiltrating lymphocytes and the tertiary lymphoid structures in HER2-positive breast cancer treated with adjuvant trastuzumab. Am. J. Clin. Pathol. 2015;144:278–288. doi: 10.1309/AJCPIXUYDVZ0RZ3G. [DOI] [PubMed] [Google Scholar]
- 8.Remark R., et al. Immune contexture and histological response after neoadjuvant chemotherapy predict clinical outcome of lung cancer patients. OncoImmunology. 2016;5 doi: 10.1080/2162402X.2016.1255394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou L., Xu B., Liu Y., Wang Z. Tertiary lymphoid structure signatures are associated with survival and immunotherapy response in muscle-invasive bladder cancer. OncoImmunology. 2021;10 doi: 10.1080/2162402X.2021.1915574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eroglu Z., et al. High response rate to PD-1 blockade in desmoplastic melanomas. Nature. 2018;553:347–350. doi: 10.1038/nature25187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cabrita R., et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature. 2020;577:561–565. doi: 10.1038/s41586-019-1914-8. [DOI] [PubMed] [Google Scholar]
- 12.Zhang H., et al. Regulatory mechanisms of immune checkpoints PD-L1 and CTLA-4 in cancer. J. Exp. Clin. Cancer Res. 2021;40:184. doi: 10.1186/s13046-021-01987-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu W., Liu Y., Zeng S., Han Y., Shen H. Intratumor heterogeneity: the hidden barrier to immunotherapy against MSI tumors from the perspective of IFN-gamma signaling and tumor-infiltrating lymphocytes. J. Hematol. Oncol. 2021;14:160. doi: 10.1186/s13045-021-01166-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helmink B.A., et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature. 2020;577:549–555. doi: 10.1038/s41586-019-1922-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bailey M.H., et al. Comprehensive characterization of cancer driver genes and mutations. Cell. 2018;173:371–385. doi: 10.1016/j.cell.2018.02.060. e318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saghafinia S., Mina M., Riggi N., Hanahan D., Ciriello G. Pan-cancer landscape of aberrant DNA methylation across human tumors. Cell Rep. 2018;25:1066–1080. doi: 10.1016/j.celrep.2018.09.082. e1068. [DOI] [PubMed] [Google Scholar]
- 17.Lee D.D., Komosa M., Nunes N.M., Tabori U. DNA methylation of the TERT promoter and its impact on human cancer. Curr. Opin. Genet. Dev. 2020;60:17–24. doi: 10.1016/j.gde.2020.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Gajos-Michniewicz A., Czyz M. WNT signaling in melanoma. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21144852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumagai S., Koyama S., Nishikawa H. Antitumour immunity regulated by aberrant ERBB family signalling. Nat. Rev. Cancer. 2021;21:181–197. doi: 10.1038/s41568-020-00322-0. [DOI] [PubMed] [Google Scholar]
- 20.Polakis P. Wnt signaling in cancer. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang J., et al. Tumour-infiltrating immune cell-based subtyping and signature gene analysis in breast cancer based on gene expression profiles. J. Cancer. 2020;11:1568–1583. doi: 10.7150/jca.37637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang P., et al. SNX10 and PTGDS are associated with the progression and prognosis of cervical squamous cell carcinoma. BMC Cancer. 2021;21:694. doi: 10.1186/s12885-021-08212-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bie Q., et al. YAP promotes self-renewal of gastric cancer cells by inhibiting expression of L-PTGDS and PTGDR2. Int. J. Clin. Oncol. 2020;25:2055–2065. doi: 10.1007/s10147-020-01771-1. [DOI] [PubMed] [Google Scholar]
- 24.Hu S., et al. Glycoprotein PTGDS promotes tumorigenesis of diffuse large B-cell lymphoma by MYH9-mediated regulation of Wnt-beta-catenin-STAT3 signaling. Cell Death Differ. 2022;29:642–656. doi: 10.1038/s41418-021-00880-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.(!!! INVALID CITATION !!! 21).
- 26.Thorsson V., et al. The immune landscape of cancer. Immunity. 2018;48:812–830. doi: 10.1016/j.immuni.2018.03.023. e814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang L., Pang Y., Moses H.L. TGF-beta and immune cells: an important regulatory axis in the tumor microenvironment and progression. Trends Immunol. 2010;31:220–227. doi: 10.1016/j.it.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erratum for the Research Article "Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy" by. Cristescu R., Mogg R., Ayers M., Albright A., Murphy E., Yearley J., Sher X., Liu X.Q., Lu H., Nebozhyn M., Zhang C., Lunceford J.K., Joe A., Cheng J., Webber A.L., Ibrahim N., Plimack E.R., Ott P.A., Seiwert T.Y., Ribas A., McClanahan T.K., Tomassini J.E., Loboda A., Kaufman D. Science. 2019;363 doi: 10.1126/science.aax1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gordeeva O. Cancer-testis antigens: unique cancer stem cell biomarkers and targets for cancer therapy. Semin. Cancer Biol. 2018;53:75–89. doi: 10.1016/j.semcancer.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 30.Schumacher T.N., Thommen D.S. Tertiary lymphoid structures in cancer. Science. 2022;375 doi: 10.1126/science.abf9419. [DOI] [PubMed] [Google Scholar]
- 31.Calderaro J., et al. Intra-tumoral tertiary lymphoid structures are associated with a low risk of early recurrence of hepatocellular carcinoma. J. Hepatol. 2019;70:58–65. doi: 10.1016/j.jhep.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 32.Spranger S., Bao R., Gajewski T.F. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015;523:231–235. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 33.Grasso C.S., et al. Genetic mechanisms of immune evasion in colorectal cancer. Cancer Discov. 2018;8:730–749. doi: 10.1158/2159-8290.CD-17-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H., et al. Machine learning-based tumor-infiltrating immune cell-associated lncRNAs for predicting prognosis and immunotherapy response in patients with glioblastoma. Briefings Bioinf. 2022;23 doi: 10.1093/bib/bbac386. [DOI] [PubMed] [Google Scholar]
- 35.Zhang N., et al. Machine learning-based identification of tumor-infiltrating immune cell-associated lncRNAs for improving outcomes and immunotherapy responses in patients with low-grade glioma. Theranostics. 2022;12:5931–5948. doi: 10.7150/thno.74281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu S., et al. Glycoprotein PTGDS promotes tumorigenesis of diffuse large B-cell lymphoma by MYH9-mediated regulation of Wnt-beta-catenin-STAT3 signaling. Cell Death Differ. 2021 doi: 10.1038/s41418-021-00880-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raab M., et al. T cell receptor "inside-out" pathway via signaling module SKAP1-RapL regulates T cell motility and interactions in lymph nodes. Immunity. 2010;32:541–556. doi: 10.1016/j.immuni.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ho J.C., et al. Down-regulation of retinol binding protein 5 is associated with aggressive tumor features in hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 2007;133:929–936. doi: 10.1007/s00432-007-0230-0. [DOI] [PubMed] [Google Scholar]
- 39.Chang S.S. Re: atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. J. Urol. 2016;196:1637–1638. doi: 10.1016/j.juro.2016.09.025. [DOI] [PubMed] [Google Scholar]
- 40.Sharma P., et al. Nivolumab in metastatic urothelial carcinoma after platinum therapy (CheckMate 275): a multicentre, single-arm, phase 2 trial. Lancet Oncol. 2017;18:312–322. doi: 10.1016/S1470-2045(17)30065-7. [DOI] [PubMed] [Google Scholar]
- 41.Kvorjak M., et al. Cross-talk between colon cells and macrophages increases ST6GALNAC1 and MUC1-sTn expression in ulcerative colitis and colitis-associated colon cancer. Cancer Immunol. Res. 2020;8:167–178. doi: 10.1158/2326-6066.CIR-19-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang W.Y., et al. Stimulative role of ST6GALNAC1 in proliferation, migration and invasion of ovarian cancer stem cells via the Akt signaling pathway. Cancer Cell Int. 2019;19:86. doi: 10.1186/s12935-019-0780-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu H., Deng H., Zhao Y., Li C., Liang Y. LncRNA XIST/miR-34a axis modulates the cell proliferation and tumor growth of thyroid cancer through MET-PI3K-AKT signaling. J. Exp. Clin. Cancer Res. 2018;37:279. doi: 10.1186/s13046-018-0950-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knijnenburg T.A., et al. Genomic and molecular landscape of DNA damage repair deficiency across the cancer Genome Atlas. Cell Rep. 2018;23:239–254. doi: 10.1016/j.celrep.2018.03.076. e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aran D., Hu Z., Butte A.J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hanzelmann S., Castelo R., Guinney J.G.S.V.A. Gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang S., et al. Immune cell infiltration-based signature for prognosis and immunogenomic analysis in breast cancer. Briefings Bioinf. 2020 doi: 10.1093/bib/bbaa026. [DOI] [PubMed] [Google Scholar]
- 48.Hugo W., et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2017;168:542. doi: 10.1016/j.cell.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 49.van den Ende T., et al. Neoadjuvant chemoradiotherapy combined with atezolizumab for resectable esophageal adenocarcinoma: a single-arm phase II feasibility trial (perfect) Clin. Cancer Res. 2021;27:3351–3359. doi: 10.1158/1078-0432.CCR-20-4443. [DOI] [PubMed] [Google Scholar]
- 50.Riaz N., et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell. 2017;171:934–949. doi: 10.1016/j.cell.2017.09.028. e916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carvalho-Silva D., et al. Open Targets Platform: new developments and updates two years on. Nucleic Acids Res. 2019;47:D1056–D1065. doi: 10.1093/nar/gky1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this work can be acquired from the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/), the Cancer Genome Atlas (TCGA) datasets (https://xenabrowser.net/), the Chinese Glioma Genome Atlas (CGGA) datasets (http://www.cgga.org.cn/). Data included in article/supp. material/referenced in article.