Abstract
Head and neck squamous cell carcinoma (HNSCC) is currently one of the most common malignancies with a poor prognosis worldwide. Meanwhile, small ubiquitin-like modifier (SUMO) specific peptidase 1 (SENP1) was associated with ferroptosis. However, the specific functions and underlying mechanisms of action of SENP1 in ferroptosis and tumor progression of HNSCC remain to be established. The findings of the present study implicated a novel ferroptosis pathway in the initiation and progression of HNSCC, providing new functional targets to guide future therapy. In the present study, The Cancer Genome Atlas database was employed to establish a gene model related to ferroptosis and verified SENP1 as a key gene via transcriptome sequencing. Expression of SENP1 in HNSCC tissue and CAL-27 cells was detected based on reverse transcription-quantitative PCR and western blot analysis. Proliferation and migration abilities of cells were determined using Cell Counting Kit-8, wound healing and Transwell experiments. Expression levels of iron, glutathione (GSH) and lipid peroxidation end-product malondialdehyde (MDA) under conditions of silencing of SENP1 with shRNA lentivirus were assayed. Additionally, the relationship between SENP1 and long-chain acyl-coenzyme A synthase 4 (ACSL4) was validated with the aid of immunoblotting and co-immunoprecipitation (co-IP). Finally, the influence of shSENP1 on the expression of key ferroptosis proteins, glutathione peroxidase 4 (GPX4) and solute carrier family 7 member 11, was evaluated via western blotting. It was revealed that SENP1 was significantly overexpressed in HNSCC and associated with low patient survival. Silencing of SENP1 led to significant suppression of cell proliferation, migration and invasion, increase in the contents of iron ions and MDA and decline in GSH levels in HNSCC cells, thereby enhancing ferroptosis and inhibiting disease progression. Conversely, overexpression of SENP1 suppressed ferroptosis and promoted progression of HNSCC. Co-IP and western blot analyses revealed a SUMOylation link between SENP1 and ACSL4. SENP1 reduced the stability of ACSL4 protein through deSUMOylation, leading to inhibition of ferroptosis. SENP1 silencing further inhibited the expression of the key iron death protein, GPX4, to regulate ferroptosis. Taken together, SENP1 deficiency promoted ferroptosis and inhibited tumor progression through reduction of SUMOylation of ACSL4 in HNSCC. The collective results of the present study supported the utility of SENP1 as an effective predictive biomarker for targeted treatment of HNSCC.
Keywords: SENP1, ACSL4, SUMO, HNSCC, ferroptosis
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer type in the world, with 500,000 new cases diagnosed on average on an annual basis. The low 5-year overall survival rate of ~50% is associated with high rates of metastasis and recurrence (1,2). At the time of diagnosis, >50% of patients are in an advanced stage of the disease (3). The majority of patients are presented with advanced HNSCC despite no clinical history of pre-malignancy (4). Surgery is the most commonly used therapeutic procedure, followed by adjuvant radiation or chemotherapy according to the stage of the disease (5). However, advanced HNSCC is associated with poor prognosis and treatment options are currently limited, highlighting the urgent need for novel therapeutic strategies. Resistance to chemotherapy is a major reason for treatment failure, which is partly mediated by suppression of tumor cell apoptosis. Therefore, elucidation of the mechanisms underlying cancer cell death should provide valuable insights for strategies to develop new therapeutic agents (6).
Ferroptosis, a novel type of programmed cell death characterized by accumulation of free iron and lipid peroxidation, was originally reported by Dixon et al in 2012 (7). This iron-dependent cell death process is caused by the accumulation of reactive oxygen species in cells and is distinct from apoptosis, pyroptosis, autophagy and necroptosis (8). Iron metabolism disorder is a risk factor for cancer demonstrated to facilitate tumor growth. Compared with normal cells, cancer cells show iron addiction, typically manifesting as an excessive reliance on iron for proliferation. In fact, activation of the ferroptosis pathway may underlie resistance to chemotherapy drugs, representing a promising new treatment frontier for cancer (9). A number of ferroptosis-related mRNAs have been identified as biomarkers for the diagnosis, prognosis and treatment of several cancer types. A recent study reported that ferritin heavy chain 1 suppressed proliferation of HNSCC cells by influencing the expression of glutathione (GSH) and Fe2+ and promoting the process of ferroptosis, along with expression of glutathione peroxidase 4 (GPX4) (10). Caveolin-1 is reported to stimulate HNSCC progression by inhibiting ferroptosis (11). However, the potential roles of ferroptosis-related mRNAs in the diagnosis, prognosis and therapy of HNSCC are yet to be clarified.
Small ubiquitin-like modifiers (SUMOs) are highly conserved molecules that couple to lysine residues of target proteins as a post-translational modification process (12). SUMOylation regulates several biological processes, including DNA damage repair, immune response, cancer formation, cell cycle progression and apoptosis. SUMO-specific peptidase 1 (SENP1) is a widely investigated protease that functions in decoupling the SUMO modifier from SUMOylated proteins (13). Various malignancies of the stomach, liver, pancreas, cervix, kidney and breast are associated with abnormal expression and activation of SENP1 (14–17). While earlier findings indicated that SENP1-mediated protein deSUMOylation exerts a protective effect on cardiomyocyte ferroptosis (18), expression of SENP1 and its relationship with ferroptosis in HNSCC are yet to be established.
The acyl CoA synthetase long chain 4 (ACSL4) gene located on human X chromosome encodes an enzyme that preferentially binds 20-carbon polyunsaturated fatty acid (PUFA) substrates, such as linoleic acid (19). ACSL4 plays a key regulatory role in ferroptosis, defined as a process of cell death associated with iron-dependent lipid peroxidation that mainly involves phospholipids containing PUFAs. Silencing of ACSL4 inhibits generation of PUFAs, thus suppressing ferroptosis (20,21). ACSL4 is considered a key disease marker for HNSCC (22). To the best of the authors' knowledge, no studies on the mechanism by which SENP1 regulates ACSL4 and ferroptosis in HNSCC have been documented to date.
In the present study, the prognostic model of HNSCC was created based on expression of ferroptosis-related mRNAs. SENP1 was validated as an important biomarker that inhibits the ferroptosis pathway in HNSCC via deSUMOylation-mediated regulation of ACSL4, in turn, promoting proliferation and invasion of HNSCC cells. The findings of the present study suggested a novel ferroptosis pathway in the initiation and progression of HNSCC, providing new functional targets to guide future therapy.
Materials and methods
Access to The Cancer Genome Atlas (TCGA) public data
RNA-sequencing (RNA-seq) transcriptomic data and the accompanying clinical information for HNSCC and normal control samples were obtained from the Cancer Genome Atlas (TCGA) (https://gdc.cancer.gov/) (23). RNA-seq data were normalized using the Expectation-Maximization method. A total of 502 HNSCC and 44 healthy subjects were subjected to randomized analysis. TCGA transcriptomic data were used to obtain accurate and effective gene expression matrices using Perl language. An expression matrix of mRNAs and lncRNAs was additionally obtained using Perl language. The ‘limma’ package in ‘R’ was utilized to determine differential expression of ferroptosis genes and mRNAs using the following screening conditions: P<0.05, false discovery rate <0.05 and log2 fold change >1. Volcano plots and heatmaps of ferroptosis-related mRNAs were generated. ‘Corfile >0.4, P<0.01’ was set as the screening standard and the relationship between differentially expressed mRNAs and ferroptosis genes determined using R 4.1.3 software (The R Foundation) (24).
Co-expression network
To determine the action sites of key mRNAs, a co-expression network of ferroptosis genes and mRNAs was established, with the aim of screening mRNAs significantly related to ferroptosis-related genes. Screening conditions were ‘P<0.01, coef >0.4’ and the network was established using R software.
Establishment and verification of risk signatures
The differentially expressed mRNA matrix was merged with clinical data downloaded from TCGA and a prognostic model constructed based on differentially expressed mRNAs. Patients with HNSCC were divided into training and test groups. The training group, accounting for 50% of the patients was used to establish a prognostic model, which was validated in the test group. Univariate Cox proportional hazards regression analysis was performed and survival-related mRNAs screened based on P-values <0.05. A least absolute shrinkage and selection operator (LASSO) regression model was generated using the screened mRNAs and 10-fold cross-validation performed with 1,000 cycles to identify the point with the smallest cross-validation error. Significantly expressed mRNAs were identified via LASSO regression and used to generate a Cox model.
Patients and organizations
A total of 15 pairs of tumor tissue (T) and adjacent non-tumor tissue (N) samples were obtained from the Department of Stomatology of the Affiliated Hospital of Xuzhou Medical University from patients with HNSCC without preoperative treatment (Xuzhou, China). Among these 15 patients, there were 9 female patients and 6 male patients, all aged between 40–70 years-old. The tissues were cut and subsequently stored in liquid nitrogen. Samples were independently identified by two pathologists and detailed clinicopathological features were collected. All tissue samples were obtained from July 2020 to June 2022. Informed consent was obtained from each patient prior to any specimen-related studies and the study was approved by the Ethics Committee of the Affiliated Hospital of Xuzhou Medical University (approval no. XYFY2020-KL216-01; Xuzhou, China). Among them, 7 pairs of cancer tissues and 3 cases of adjacent tissues were used for transcriptomics; 15 pairs of cancer tissues and adjacent tissues were applied for reverse transcription-quantitative PCR and western blot experiments.
Transcriptome sequencing
Total RNA was isolated using TRIzol® reagent from cancer tissues of seven patients with HNSCC and three adjacent tissue samples. Detection of the concentration and purity of extracted RNA was performed via spectrophotometry on a Nanodrop 2000 instrument (Agilent Technologies, Inc.); only high-quality RNA sample (OD 260/280=1.8~2.2, OD 260/230 ≥2.0, RIN ≥6.5, 28S:18S ≥1.0, >1 µg) was used to construct sequencing library. Agarose gel electrophoresis was used to determine the integrity of RNA, and RNA integrity number values were established with the Agilent 2100 Bioanalyzer system (Agilent Technologies, Inc.). RNA-seq transcriptome library was prepared following TruSeqTM RNA sample preparation Kit from Illumina, Inc. using 1 µg of total RNA. Shortly, messenger RNA was isolated according to polyA selection method by oligo(dT) beads and then fragmented by fragmentation buffer firstly. Secondly double-stranded cDNA was synthesized using a SuperScript double-stranded cDNA synthesis kit (Invitrogen; Thermo Fisher scientific, Inc.) with random hexamer primers (Illumina, Inc.). Then the synthesized cDNA was subjected to end-repair, phosphorylation and ‘A’ base addition according to Illumina's library construction protocol. Libraries were size selected for cDNA target fragments of 300 bp on 2% Low Range Ultra Agarose followed by PCR amplified using Phusion DNA polymerase (NEB) for 15 PCR cycles. After quantified by TBS380, paired-end RNA-seq sequencing library was sequenced with the Illumina HiSeq xten/NovaSeq 6000 sequencer (2×150 bp read length). according to the manufacturer's protocol. Transcriptomes from all samples were merged to reconstruct a comprehensive transcriptome using perl script. Normalization was performed using R software (v4.1.3) and differentially expressed genes were identified by their P-value and expression fold change.
Cell lines and cell culture
CAL-27 (RRID: CVCL_0030) and HOK (RRID: CVCL_YE19) cells purchased from the Procell Life Science & Technology Co., Ltd. were cultured in DMEM/F12 (1:1) medium (Gibco; Thermo Fisher Scientific, Inc.) containing 10% heat-inactivated FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin and streptomycin. Cells were cultured in a constant temperature incubator under 5% CO2 at 37°C. Erastin (cat. no. HY-15763) was obtained from MedChemExpress.
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
(RT-qPCR) was used to detect Senp1 transcription levels. Total RNA was extracted from cells and tissues using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc). Extracted RNA was reverse-transcribed into cDNA with HiScript® II QRT Super Mix for qPCR (Vazyme Biotech Co., Ltd.) and RT-qPCR was performed using SYBR Green qPCR Master Mix (MedChemExpress) on the Roche Light Cycler 480 II instrument (Roche Diagnostics) using β-actin as an internal standard. Relative gene expression was normalized to β-actin mRNA expression and analyzed using the comparative Cq method (ΔΔCq) (25). The same thermocycling profile conditions were used for all primers sets: 95°C for 10 min, 40 cycles at 95°C for 15 sec, and 60°C for 1 min. The primers used were as follows: β-actin forward, 5′-CTCCATCCTGGCCTCGCTGT-3′ and reverse, 5′-GCTGTCACCTTCACCGTTCC-3′; SENP1 forward, 5′-CCAGCATTTTAACTAACCAGGAAC-3′ and reverse, 5′-GCTAAGTTATCTGGCTGATGTGG-3′; and ACSL4 forward, 5′-CATCCCTGGAGCAGATACTCT-3′ and reverse, 5′-TCACTTAGGATTTCCCTGGTCC-3′.
Western blot analysis
Proteins were extracted from tissues and CAL-27 cells using RIPA lysis buffer (cat. no. 89900; Thermo Fisher Scientific, Inc.) with Halt™ Protease Inhibitor Cocktail (cat. no. 78429; Thermo Fisher Scientific, Inc.). Subsequently, the BCA method was used to quantify the tested protein. Premade adhesive gels (4–20%) were prepared and 30 µg protein samples were loaded per lane, marked on both sides to indicate protein molecular weight and to distinguish left and right; then separated via sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes. Next, membranes were blocked with 5% non-fat milk at room temperature for 2 h, followed by overnight incubation at 4°C with anti-SENP1 (1:1,000; cat. no. 25349-1-AP), anti-GPX4 (1:1,000; cat. no. 67763-1-Ig), anti- solute carrier family 7 member 11 (SLC7A11) (1:1,000; cat. no. 26864-1-AP) and anti-β-actin (1:5,000; cat. no. 20536-1-AP; all from Proteintech Group, Inc.) antibodies. After washing, membranes were incubated with goat anti-mouse (1:5,000; cat. no. ab216776) or goat anti-rabbit secondary antibody (1:5,000; cat. no. ab216773; both from Abcam) for 2 h at room temperature. An enhanced chemiluminescence reagent (Wuhan Servicebio Technology Co., Ltd.) was used to visualize the protein bands. Protein bands were scanned with an Odyssey infrared scanner (LI-COR Biosciences) and band density was analyzed with ImageJ v1.8.0 software (National Institutes of Health).
Cell Counting Kit-8 (CCK-8) assay
The CCK-8 assay (Beyotime Biotechnology Inc.) was employed to detect cell activity. In brief, CAL-27 cells were seeded into a 96-well plate at a density of 1×104 cells/well and cultured for 3 days. Next, cells were transfected with Senp1 short hairpin RNA (shRNA) or overexpression plasmid and survival rates were assessed via CCK-8 colorimetry at 72 h. In brief, CCK-8 was added to each well and was incubated for 1 h at 37°C. CCK-8 solution accounted for ~10% of the volume of the culture medium. OD values were measured at 450 nm using a microplate reader (BioTek Instruments, Inc.).
Wound healing assay
Transfected cells were cultured to ~90% confluence in 12-well plates and a wound area introduced using a sterile 200-µl pipette tip. After washing twice with PBS, cells were cultured in DMEM/F12 without FBS for 48 h. Cell images were obtained using fluorescence microscopy (IX73; Olympus Corporation; scale bars, 100 µm). Wound sizes were measured with ImageJ and calculated as a percentage (from 0 h to 48 h).
Transwell assay
Cells were collected and resuspended in medium (~1×105 cells/ml). An aliquot (200 µl) of the cell suspension was placed in the upper chamber of a Transwell (8-µm pore size; MilliporeSigma), which was inserted into a 24-well plate. After incubation in RPMI-1640 containing 10% (v/v) FBS for 24 h at 37°C, cells on the upper membrane of the insert were removed gently with a cotton swab while those migrating to the bottom surface were fixed with 4% cold methanol for 30 min and stained with 0.1% crystal violet for 1 h. The number of cells on the bottom surface was counted and quantified using fluorescence microscopy (IX73; Olympus Corporation; scale bars, 100 µm).
Lipid peroxidation assay
The TBA method kit (cat. no. A003; Nanjing Jiancheng Bioengineering Institute) was used to measure the concentration of MDA in cells. Cells were mixed well in RIPA lysis buffer. The reagent was added to the centrifuge tube according to the manufacturer's instructions and thoroughly mixed, followed by boiling in a 95°C water bath for 40 min and cooling at room temperature. After centrifugation at 1,600 × g for 10 min at 4°C, absorbance of the supernatant was measured at 532 nm to calculate malondialdehyde (MDA).
Iron assay
An iron assay kit (cat. no. ab83366; Abcam) was employed to evaluate the relative intracellular iron levels. According to the requirements of the reagent supplier, the prepared iron probe, reducing agent, standard diluent (2–10 nmol/well) and test sample (cell lysate) were mixed. Next, 5 µl iron reducing agent was added to each standard well and incubated at 37°C for 30 min, followed by addition of 100 µl iron probe to each well at 37°C and incubation in the dark for 60 min. The sample was immediately evaluated with a colorimetric enzyme marker (593 nm). The total iron content of test samples was determined from the standard curve.
Immunohistochemistry (IHC)
Slices were immunohistochemically stained as experimental procedure. Coverslips seeded with tumor cells or tissue cryo-sections were fixed with 4% paraformaldehyde for 30 min, and then permeabilized with 0.5% Triton X-100 for 10 min. After 2 h of incubation with goat serum (cat. no. 16210064; Thermo Fisher Scientific, Inc.) at 37°C, Consecutively, 4-µm-thick sections were analyzed using primary antibodies against SENP1 (1:1000; cat. no. 25349-1-AP; Proteintech Group, Inc.) at 4°C. Then, sections were incubated with Alexa Fluor (cat. no. R37116; Thermo Fisher Scientific, Inc.) secondary antibody for 1 h at 37°C. Images were obtained using fluorescence microscopy (IX73; Olympus Corporation; scale bars 200 µm).
GSH assay
A GSH colorimetric assay kit (cat. no. CS0260; MilliporeSigma) was used to determine relative GSH levels. This experiment used 5% of 5-thiosalicylic acid solution (SSA), GSH standard solution, working mixture, NADPH solution and test sample (cell extract). SSA (5%) was used as the reagent blank, a total of 10 µl GSH standard solution and 10 µl of test sample were added to a separate well. An aliquot of 150 µl working mixture was added to each well and was incubated at room temperature for 5 min. Finally, a total of 50 µl NADPH solution was added to each well and mixed. After absorbance of each well was determined at 412 nm, the GSH concentration was calculated according to the standard curve.
Lentiviral transfection
To construct stable SENP1-knockdown CAL-27 cell line, shRNA specifically targeting SENP1 was designed by the Thermo Fisher Scientific, Inc. online tool (https://rnaidesigner.thermofisher.com/rnaiexpress/construct.do?pid=−431479708000205999) and cloned into the lentiviral pLVX-sh1 vector. SENP1 overexpressing CAL27 cells were cloned into the lentivirus pLVX–IRES-puro vector. Lentiviruses were synthesized by Genechem Shanghai Co., Ltd. Cells were seeded in cell culture dishes and used to transfect lentiviral with different MOI values (10, 20, 30) at 30% density. On the second day of transfection, cells were cultured in DMEM containing 10% FBS. After 3 days, the transfection effect was observed under fluorescence microscopy (IX73; Olympus Corporation). Then, transfected cells were passaged and cultured with puromycin (5 ng/µl; Shanghai Yeasen Biotechnology Co., Ltd.) used to selected cells for 1 week. Subsequently, stable cell lines were cultured in DMEM containing 10% FBS and puromycin (2 ng/µl). The sequences were as follows: sh-SENP1, 5′-CTCGATGTCTTAGTTCCAGTA-3′; and sh-NC, 5′-GGGACTCGATTCCCACGAATC-3′.
Co-IP
The cells were lysed with 250 µl NP-40 lysis buffer (cat. no. P0013F; Beyotime Institute of Biotechnology). The cell lysates were centrifuged at 4,800 × g for 5 min to remove the cell fragments. The protein stock solution, with anti-SENP1 or ACSL4 antibody bound to 25 µl magnetic beads, was incubated overnight at 4°C. The small pallet was washed three times using the pyrolysis method and sodium dodecyl sulfate samples were boiled for 5 min. SENP1, ACSL4 and SUMO1 levels were analyzed via western blot analysis using anti-SENP1 (1:1,000; cat. no. 25349-1-AP), anti-ACSL4 (1:1,000; cat. no. 22401-1-AP) and anti-SUMO1 (1:1,000; cat. no. 10329-1-AP; all from Proteintech Group, Inc.) antibodies, respectively.
Statistical analysis
All data were analyzed with Graph Pad Prism 7.0 software (GraphPad Software; Dotmatics). Continuous data are presented as the mean ± standard deviation and differences between experimental groups were analyzed using one-way ANOVA or unpaired two-tailed Student's t-test. In addition, to estimate the risk differences between the main outcome groups, a post hoc analysis was conducted using Bonferroni's test following ANOVA. Survival analysis was performed using the Kaplan-Meier method(https://ualcan.path.uab.edu/). The screening of key genes comes from the VENN diagram method (https://bioinfogp.cnb.csic.es/tools/venny/index.html). Differences were considered statistically significant at P<0.05. Experiments were repeated three times and representative experiments are shown.
Results
Expression of ferroptosis-related mRNAs
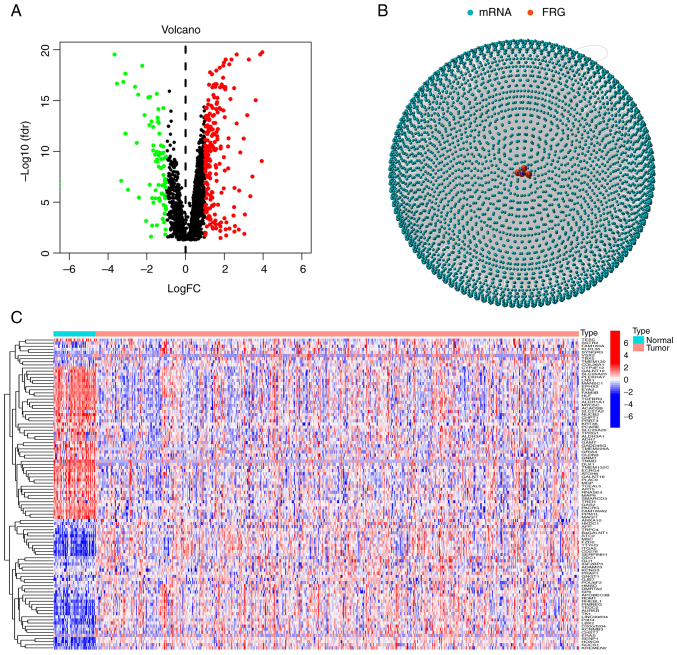
Data from a total of 44 normal and 502 tumor samples were obtained from TCGA database. Based on the co-expression patterns of 40 ferroptosis genes (Table SI) and differentially expressed mRNAs (|log2FC|>1 and P<0.01), a total of 457 ferroptosis-related mRNAs were obtained (correlation coefficients >0.4 and P<0.001). All mRNAs and ferroptosis genes were positively correlated. Volcano plots and heat maps were used to examine expression patterns in normal and tumor groups (Fig. 1A and C) and the relationship between mRNAs and ferroptosis genes was ultimately visualized via network graphs (Fig. 1B).
Figure 1.
Identification of ferroptosis-related mRNAs. (A) Heatmap of 457 ferroptosis-related mRNAs. (B) Volcano plot of 457 ferroptosis-related mRNAs. (C) Network map of different expressive ferroptosis genes and mRNAs. FRG, ferroptosis-related gene.
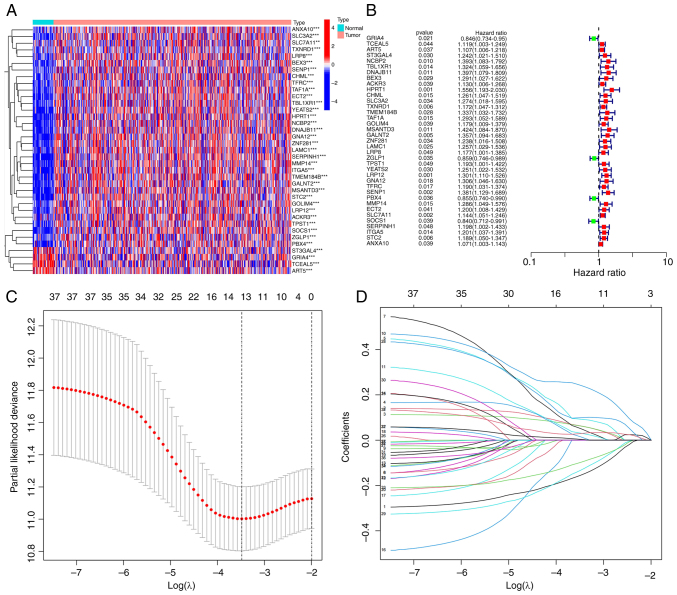
Construction of an mRNA prognostic model
Using univariate Cox proportional hazards regression analysis, 32 significant ferroptosis-related mRNAs among the 457 were identified and a heatmap was generated (Fig. 2A and B). The significant mRNAs obtained via univariate Cox analysis were combined with clinical data to construct LASSO regression curves (Fig. 2C). Cross-validation was performed and the point of the smallest cross-validation error determined. Ultimately, 13 ferroptosis-related mRNAs were screened (Fig. 2D). Following optimization of the Cox model, seven mRNAs (SENP1, PBX4, SOCS1, DNAJB11, ART5, TCEAL5 and GRIA4) were selected. Simultaneous transcriptome analysis of HNSCC and corresponding paracancerous tissues led to the identification of 427 differential genes (199 upregulated and 228 downregulated) (Fig. S1A). When these 427 genes were analyzed with the VENN map with the aforementioned seven genes, a key gene, SENP1, was obtained (Fig. S1B). Thus, SENP1 was selected to start further verification.
Figure 2.
Establishment of a ferroptosis-related mRNA prognostic model in head and neck squamous cell carcinoma. (A) Heatmap of 32 ferroptosis-related mRNAs. (B) A total of 32 ferroptosis-related mRNAs were screened out by univariate Cox regression analysis. (C) Lasso regression analysis screened out 13 mRNAs related to ferroptosis. (D) Lasso regression 10-fold cross-validation to calculate coefficients of mRNAs associated with ferroptosis. LASSO, least absolute shrinkage and selection operator.
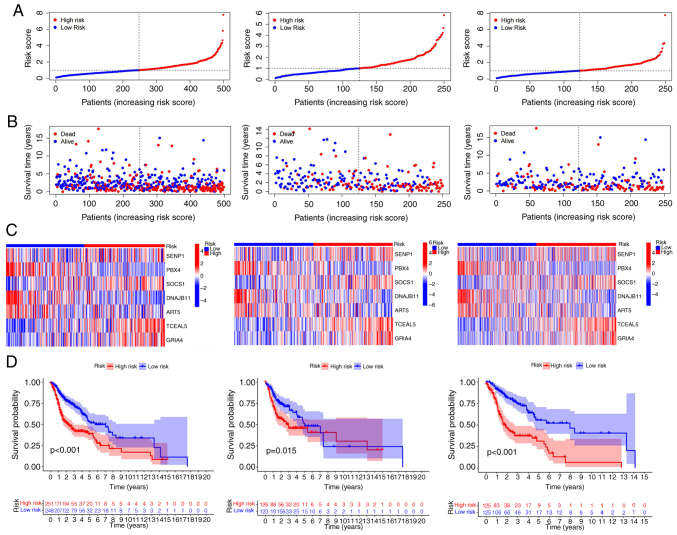
Survival analysis and construction of risk curves
According to the risk score, a Kaplan-Meier curve of overall survival (OS) probability of individuals with ferroptosis-related genes was constructed for the training, test and all groups. The survival rate was significantly lower in high risk than low risk patients for all groups (Fig. 3D). Similar results were obtained for age, sex, stage, type and tumor-node-metastasis (TNM) stage (Fig. S2). Since HNSCC is prone to lymph node metastasis, ‘N’ was specifically taken out as an independent clinical prognostic factor. Effective independent prognostic factors were screened via univariate Cox analysis (screening condition, P<0.05). The risk score of the high-risk group was obviously greater whereas survival time was markedly lower than that of the low-risk group in the training, test and all groups. The differences in expression levels of the seven ferroptosis-related mRNAs in prognostic models of the high risk as well as low risk groups were analyzed using heat maps (Fig. 3A-C).
Figure 3.
Prognostic performance of 7 ferroptosis-related mRNAs in train group, test group and all groups. (A) Display of ferroptosis-related mRNAs prognostic models in train, test and all groups of risk score. (B) State diagram of survival risk in train group, test and all groups. (C) Heat map of ferroptosis-related mRNAs expression levels in high and low risk groups with high expression in red, low expression in blue. (D) Overall survival between high and low risk groups in the train group, test group and all group. All P<0.05.
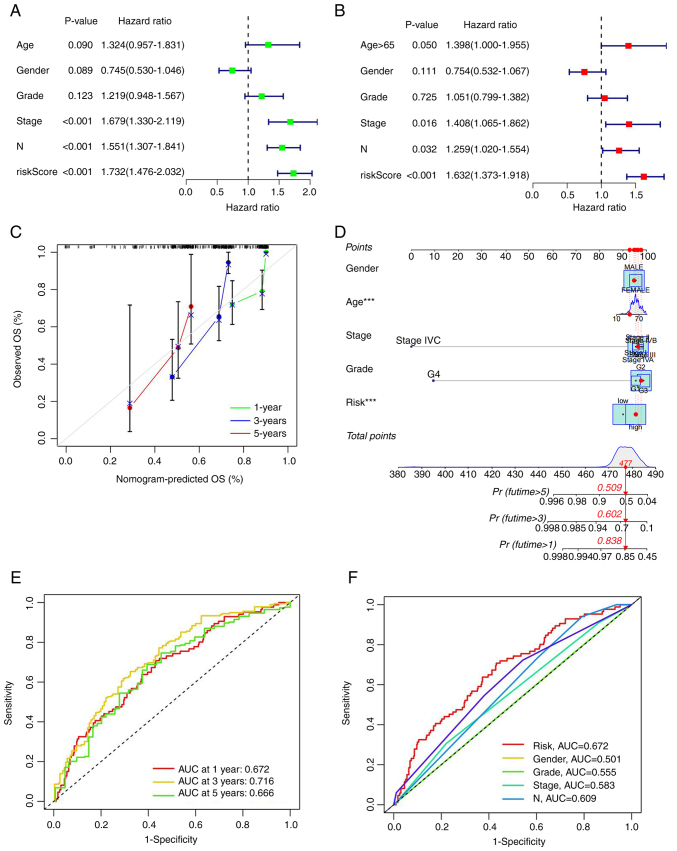
Construction of a nomogram and independent prognostic analysis
As aforementioned, effective independent prognostic factors were screened via univariate Cox analysis (screening condition, P<0.05). The hazard ratio (HR) for the risk score was 1.732 and 95% confidence interval (CI) was 1.476–2.032. ‘Stage’ and ‘N’ were identified as two additional prognostic parameters (Fig. 4A). In multivariate Cox analysis, HR of the risk score was 1.623 with 95% CI of 1.373–1.918. Age, stage and N could be used as additional prognostic parameters (Fig. 4B). A nomogram was generated according to the clinical risk factors identified, specifically, age, sex, stage, grade, TNM stage and comprehensive risk scores were obtained to assess 1-, 3- and 5-year survival rates of patients with HNSCC (Fig. 4C). Calibration curves for 1-, 3- and 5-year survival rates were additionally generated to evaluate the reliability of the nomogram (Fig. 4D). Receiver operation curve (ROC) analysis was utilized to validate the preciseness of the model for predicting patient survival. The predictive values of area under the curve (AUC) were 0.672, 0.716 and 0.666 for 5-, 3- and 1-year survival, respectively (Fig. 4E). All AUC values were >0.65, validating the accuracy of the prognostic model in survival prediction. Moreover, AUCs were larger than those obtained from other clinical traits, suggesting that accuracy of the proposed model was greater than that of other clinical traits (Fig. 4F). The data clearly suggested that the seven mRNA ferroptosis-related models constructed by the authors were closely associated with the pathogenesis of HNSCC.
Figure 4.
Independent prognostic analysis and nomogram plotting. (A) Univariate Cox analysis of clinical traits. (B) Multivariate Cox analysis of clinical traits. (C) Combined risk scores, nomograms of clinical traits anticipate the 5-, 3- and 1-year survival in patients with head and neck squamous cell carcinoma. (D) Calibration curves for nomogram predicting survival. (E) ROC curves of all groups at 5 years, 3 years and 1 year. (F) Comparison of ROC curves of risk groups and clinical traits. ROC, receiver operation curve; AUC, area under the curve; OS, overall survival.
SENP1 is overexpressed in HNSCC
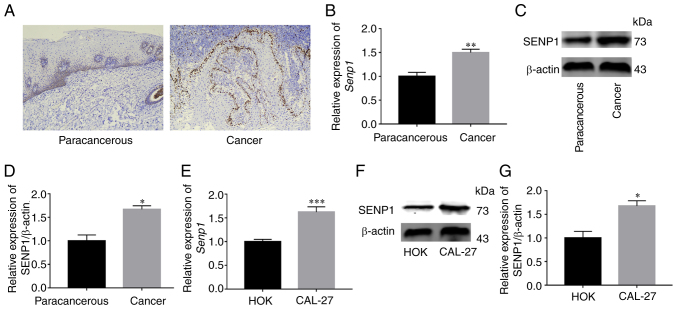
Kaplan-Meier method indicated that high SENP1 expression was generally associated with low survival rates (Fig. S3). SENP1 mRNA that displayed the highest association with ferroptosis was selected for further verification (Fig. S4). Subsequently, the expression of SENP1 in HNSCC tissue was examined via IHC, western blotting and RT-qPCR and was revealed to be significantly higher than that in paracancerous tissue (Fig. 5A-D). Simultaneously, the expression of SENP1 in CAL-27 and HOK was examined via RT-qPCR and western blot analyses. Both sets of experiments revealed higher expression of SENP1 in the CAL-27 cell line (Fig. 5E-G). The results clearly indicated that SENP1 was overexpressed in HNSCC.
Figure 5.
SENP1 is highly expressed in HNSCC tissue and CAL-27 cell line. (A and B) Immunohistochemistry and RT-qPCR indicated high expression of SENP1 in HNSCC tissue. (C and D) Western blotting verified high expression of SENP1 in HNSCC tissue. (E) RT-qPCR indicated high expression of SENP1 in CAL-27 cell line. (F and G) Western blotting verified high expression of SENP1 in CAL-27 cell line. Data are expressed as the mean ± standard error of the mean. *P<0.05, **P<0.01 and ***P<0.001. SENP1, SUMO-specific peptidase 1; HNSCC, head and neck squamous cell carcinoma; RT-qPCR, reverse transcription-quantitative PCR.
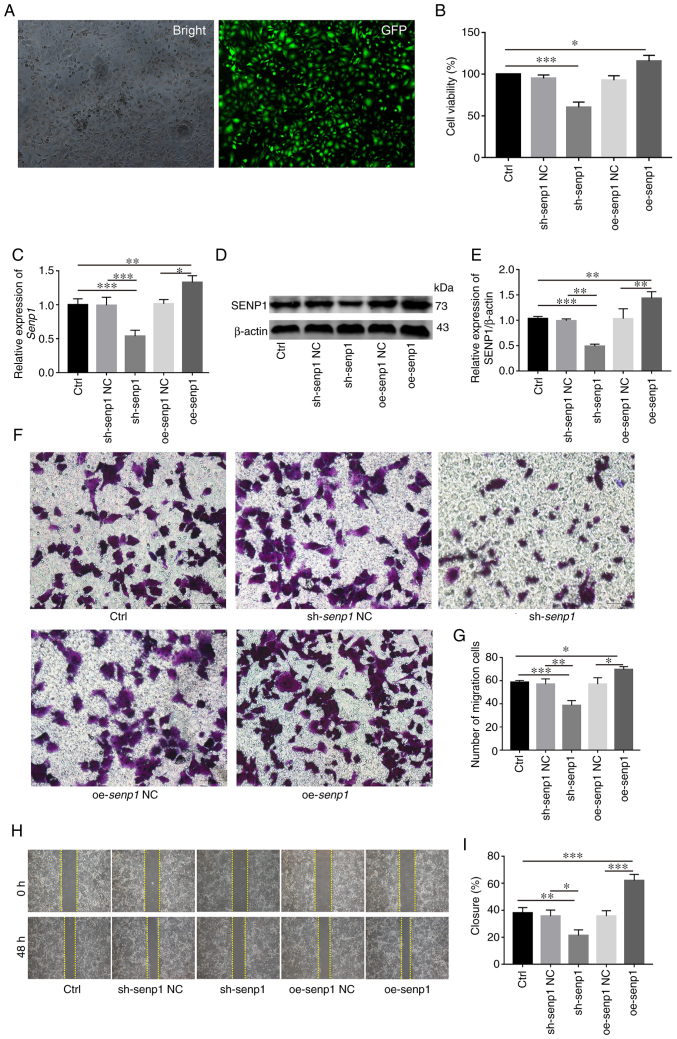
SENP1 accelerates HNSCC cell proliferation and migration
Stable knockout or overexpression cell lines using shRNA and SENP1 overexpression lentivirus, respectively, were first established with a view to investigate the biological role of SENP1 in HNSCC (Fig. 6A). Overexpression and knockdown efficiency of SENP1 was validated at both protein and transcriptional levels (Fig. 6C-E). Data from the CCK-8 assay revealed increased CAL-27 cell proliferation under conditions of overexpression of SENP1 (Fig. 6B). Conversely, knockdown of SENP1 significantly inhibited the viability of CAL-27 cells, suggesting that SENP1 was an important regulator of HNSCC cell proliferation. Transwell assays revealed that compared with the control group, the number of migratory HNSCC cells transfected with sh-SENP1 decreased while upon overexpression of SENP1, the number of migratory HNSCC cells was significantly increased (Fig. 6F and G). The results of the wound healing assay consistently demonstrated that SENP1 promoted cell migration. Compared with the negative control group, the migration ability of oe-SENP1 CAL-27 cells was significantly increased while sh-SENP1 induced a decrease in cell migration ability (Fig. 6H and I). The collective findings indicated the pivotal role of SENP1 in the progression of HNSCC.
Figure 6.
SENP1 promotes the proliferation, migration and invasion of HNSCC in vitro. (A and C) reverse transcription-quantitative PCR revealed transfection efficiency of sh-SENP1 and oe-SENP1. (B) Analysis and evaluation of the impact of knocking down or overexpressing SENP1 on the proliferation ability of HNSCC cells through Cell Counting Kit-8. (D and E) Western blotting verified sh-SENP1l and oe-SENP1 transfection efficiency. (F and G) Transwell assays were conducted to examine the cell migration and invasion ability in HNSCC cells. (H and I) Wound healing assay was employed to detect the cell migration ability of HNSCC cells. Unpaired student's t-test for two-group comparison: *P<0.05, **P<0.01 and ***P<0.001. One-way ANOVA for multi-group comparisons: *P<0.05, **P<0.01 and ***P<0.001. Bonferroni's test for multi-group comparisons: *P<0.05, **P<0.01 and ***P<0.001. SENP1, SUMO-specific peptidase 1; HNSCC, head and neck squamous cell carcinoma; Ctrl, control; shRNA, short hairpin RNA; NC, negative control; oe, overexpressing. Scale bars, 100 µm.
SENP1 is an important regulator of ferroptosis pathway in HNSCC
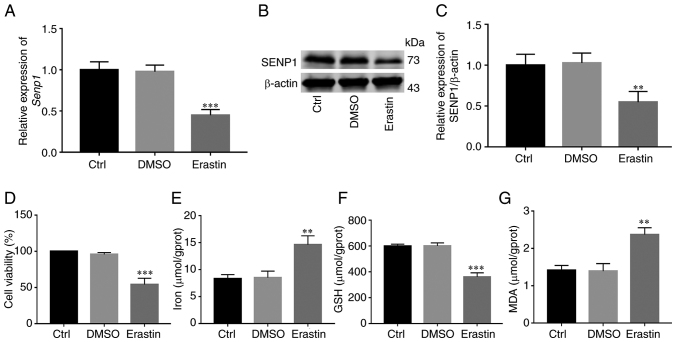
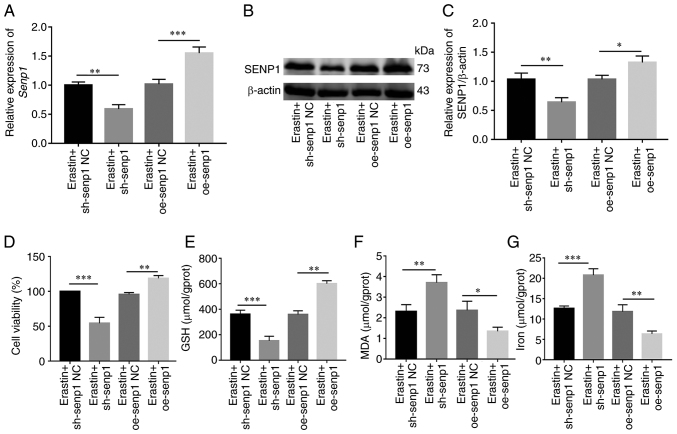
Erastin is a common inducer of ferroptosis. Following activation of ferroptosis of HNSCC cells with erastin, changes in SENP1 expression and its inhibitory mechanisms of action on growth of HNSCC cells were examined. Initial RT-qPCR and western blotting experiments revealed significantly reduced SENP1 expression in cells treated with erastin compared with those treated with DMSO (Fig. 7A-C). To further determine whether inhibition of cell growth was caused by ferroptosis, under conditions of erastin-induced ferroptosis (10 µM, 24 h), the levels of iron death-related indicators (cellular iron, GSH and MDA) in HNSCC cells were detected. Notably, viability of HNSCC cells and levels of GSH were reduced (Fig. 7D and F), whereas levels of iron and MDA, the end product of lipid peroxidation, were significantly increased in cells (Fig. 7E and G). Simultaneously, overexpression and knockdown efficiency of SENP1 at the protein and transcriptional levels were validated under conditions of ferroptosis induction (Fig. 8A-C). Following knockdown of SENP1, compared with the NC group, cell proliferation ability reduced, the end products of cellular iron and lipid peroxidation, MDA, were significantly increased while that of endogenous GSH was significantly decreased (Fig. 8D-G). In addition, the corresponding level of ferroptosis under overexpression of SENP1 was reduced (Fig. 8F). The findings of the aforementioned experiments clearly indicated that SENP1 served as an important regulatory factor of ferroptosis in HSNCC cells.
Figure 7.
Erastin-induced ferroptosis in HNSCC cells and its effect on SENP1. (A) Reverse transcription-quantitative PCR was used to detect the relative expression of SENP1 in HNSCC cells treated with 10 µm erastin for 24 h. (B and C) Western blot analysis detected the relative expression of SENP1 in HNSCC cells treated with 10 µm erastin for 24 h. (D) Cell Counting Kit-8 detected the viability of HNSCC cells after erastin (10 µm, 24 h) treatment. (E) Expression level of iron ions after erastin treatment in HNSCC cells. (F) Expression level of GSH after erastin (10 µm, 24 h) treatment in HNSCC cells. (G) Expression level of MDA after erastin (10 µm, 24 h) treatment in HNSCC cells. **P<0.01 and ***P<0.001. HNSCC, head and neck squamous cell carcinoma; SENP1, SUMO-specific peptidase 1; GSH, glutathione; MDA, malondialdehyde; Ctrl, control.
Figure 8.
SENP1 regulates ferroptosis in HNSCC cells. (A) Reverse transcription-quantitative PCR detected SENP1 expression levels after erastin (10 µm, 24 h) treatment in sh-SENP1 and oe-SENP1. (B and C) Western blotting detected SENP1 expression levels after erastin (10 µm, 24 h) treatment in sh-SENP1 and oe-SENP1. (D-G) After erastin (10 µm, 24 h) treatment in HNSCC cells, SENP1 regulated the (D) cell viability, (E) GSH, (F) MDA and (G) levels of iron. *P<0.05, **P<0.01 and ***P<0.001. SENP1, SUMO-specific peptidase 1; HNSCC, head and neck squamous cell carcinoma; Ctrl, control; shRNA, short hairpin RNA; NC, negative control; oe, overexpressing; GSH, glutathione; MDA, malondialdehyde.
Silencing SENP1 propels ferroptosis by suppressing deSUMOylation of ACSL4 to enhance its protein stability
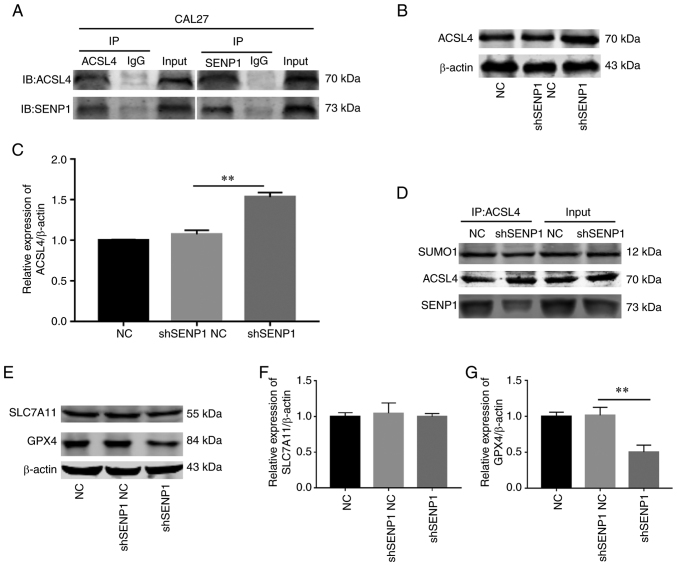
ACSL4 is a key enzyme in the lipid peroxidation procedure that is highly expressed to promote ferroptosis. While previous research has revealed that SENP1 regulates the process of ferroptosis, the association between SENP1 and ACSL4 has not been fully elucidated. It was proposed by the authors that SENP1 reduces the stability of ACSL4 protein via deSUMOylation in HNSCC. Co-IP experiments in CAL-27 cells revealed a significant direct association between SENP1 and ACSL4 (Fig. 9A). Further TCGA and western blot analysis disclosed a negative association between the two molecules (Fig. 9B and C). To further validate this hypothesis, IP experiments on SUMO1 conjugated to ACSL4 in the control and gene knockout groups were conducted. Notably, the SUMO1 level of ACSL4 was decreased in the gene knockout group (Fig. 9D). Furthermore, the impact of SENP1 knockdown on the iron death pathway was evaluated. Protein immunoblotting was employed to determine the levels of key proteins, GPX4 and SLC7A11, of the iron death pathway in samples from each group following SENP1 knockdown, which revealed that the GPX4 level was decreased while that of SLC7A11 remained unchanged (Fig. 9E-G). These data supported a direct correlation between SENP1 and ACSL4. Knockdown of SENP1 may promote SUMO1 conjugation and stability of ACSL4, further consuming GPX4 levels and ultimately, facilitating ferroptosis.
Figure 9.
Interaction between SENP1 and ACSL4 in CAL-27 cells. (A) Co-immunoprecipitation followed by western blot analysis indicated that SENP1 and ACSL4 interacted in HNSCC cells. (B and C) Western blot analysis detected changes in ACSL4 after SENP1 silencing. (D) SUMOylation analysis of ACSL4 in the control group and SENP1 in the silence group. (E and F) Western blot detected ferroptosis-related protein SLC7A11 expression after SENP1 silencing. (E and G) Western blotting detected ferroptosis-related protein GPX4 expression after SENP1 silencing. **P<0.01. SENP1, SUMO-specific peptidase 1; ACSL4, acyl CoA synthetase long chain 4; HNSCC, head and neck squamous cell carcinoma; SLC7A11, solute carrier family 7 member 11; GPX4, glutathione peroxidase 4; NC, negative control; shRNA, short hairpin RNA.
Discussion
Ferroptosis-related mRNAs are associated with progression of HNSCC (26). In the present study, the TCGA database was utilized to explore ferroptosis-related mRNAs in HNSCC. LASSO regression was merged with clinical prognosis data to establish a panel containing seven ferroptosis-related signature mRNAs, which were verified as prognostic markers of HNSCC (Fig. S4). The preciseness of the model was verified according to the AUC values of ROC curves of the risk group. Transcriptome analysis was employed for screening of the key ferroptosis-related SENP1 gene. High expression of SENP1 was associated with poor prognosis and severe malignant progression of HNSCC. Compared with non-neoplastic tissues, expression of SENP1 was elevated in HNSCC, indicating its involvement in tumorigenesis. Notably, silencing of SENP1 had a number of critical effects, including reduced proliferation and migration of HNSCC cells, increased absorption of iron ions, enhanced SUMOylation and stability of ACSL4, suppression of GPX4 activity, and promotion of GSH metabolism, resulting in accumulating cellular lipid peroxidation, and ultimately, ferroptosis. The collective results suggested that SENP1 deficiency promoted ferroptosis and inhibited tumor progression by reducing SUMOylation of ACSL4 in HNSCC.
In recent years, research on ferroptosis was mainly focused on the underlying molecular mechanisms and drug resistance while related protein markers have been relatively overlooked (27,28). To address this gap in knowledge, the present study investigated the relationship between protein biomarkers and ferroptosis in relation to the potential mechanisms underlying HNSCC occurrence. SENP1 monitors cytoactivity by eliminating the conjugation of SUMO proteins from substrate proteins (13). SENP1 targets several SUMO1-modified proteins with effects on cell proliferation, migration and induction of macrophage inflammation and is closely associated with the development of several cancer types (14,29). Previous findings clearly suggested that SENP1 promotes cell migration, invasion and proliferation by regulating DNA methylation of E-cadherin in osteosarcoma (30). Similarly, SENP1 increases androgen sensitivity to promote prostate cancer growth (31) and enhances invasion and metastasis activities of triple-negative breast cancer through elevating CSN5 transcription mediated by GATA1 deSUMOylation (15). Overexpression of SENP1 in HNSCC was originally uncovered via cDNA microarray analysis (32). The results of the present study indicated that SENP1 was an important oncogene significantly upregulated in HNSCC. High expression of SENP1 was positively related to poor survival outcomes and effectively facilitated proliferation and invasion of HNSCC cells.
ACSL4, a lipid droplet-related protein that converts free long-chain fatty acids into fatty acid acyl coenzyme A esters, is involved in both anabolic (fatty acid synthesis and lipogenesis) and catabolic pathways (lipolysis and fatty acid metabolism) of β-oxidation. ACSL4-encoded enzymes have a preferential affinity for 20-carbon PUFA substrates, such as adrenaline (ADA), catalyzing their conversion to AACoA and ADA CoA while inhibiting the process of reducing GSH inhibiting phospholipid synthesis and facilitating lipid peroxidation to promote the ferroptosis process (20). A number of studies suggested that SENP1 is a novel regulator of ferroptosis in cancer cells. Silencing of SENP1 is reported to reduce the stability of A20 to promote ferroptosis and apoptosis in lung cancer cells (33). Upregulation of SENP1 in myocardial cells has also been demonstrated to inhibit erastin-induced ferroptosis (18). However, the relationship between SENP1 and ferroptosis in HNSCC is currently unclear. In the present study, the ferroptosis inducer, erastin, significantly promoted SENP1 expression in HNSCC cells. Low expression of SENP1 in cells promoted ACSL4, thereby leading to GSH-mediated growth of oxidized phospholipids; while promoting the iron death pathway, lipid peroxidation (MDA) and the expression of iron and suppressing the expression of GSH, led to promotion of ferroptosis. However, the scope of the present study was limited to the effect of SENP1 on HNSCC cells and its role in other cancer types requires further exploration.
SENP1 is an important enzyme that induces deSUMOylation (12). Ubiquitination-mediated modification involves degradation of proteasomes, primarily through degrading one or more proteins, to achieve the desired effect. SUMOylation mainly affects the process of protein stability (34,35), with SUMO modification playing a critical role in complex protein regulatory networks. Disruption of SUMOylation can promote the progression of a number of diseases and tumors, highlighting the utility of SUMO as a potential therapeutic target for cancer (36). SENP1 reduces the stability of PTEN proteins through regulating their SUMOylation and ubiquitination. A different study demonstrated that SENP-1 regulated glycolysis in prostate cancer cells by stabilizing HIF-a (37). Moreover, SENP1 can actively regulate the stability and activity of c-Myc in breast cancer through deSUMOylation (38) and reduce the stability of ACSL4 protein by deSUMOylation in hypoxic environments, inhibiting ferroptosis of myocardial cells (33). However, the expression patterns and related mechanisms of associations between SENP1 and ACSL4 in HNSCC have been largely unexplored to date. Data from the present study revealed that SENP1 played an important role in HNSCC progression by modifying ACSL4 through deSUMOylation. SENP1-mediated ASCL4 phosphorylation affected activity, thereby promoting GPX4 expression to inhibit the process of ferroptosis in HNSCC.
In the present study, SENP1 was identified as a novel oncogene with a key role in progression of HNSCC and ferroptosis. SENP1 decreased the stability of ACSL4 protein through deSUMOylation and indirectly inhibited its influence on phospholipid metabolic pathways downstream of ferroptosis. Additionally, SENP1 could serve as an important biomarker for prognosis of patients. However, it must be noted that the present study has a number of limitations that need to be addressed. Additional experiments are necessary to verify SENP1-mediated regulation of SUMOylation of ACSL4. Establishment and characterization of mutant colonies based on predicted SUMO sites of ACSL4 should be performed to confirm the finding that SENP1 regulates the SUMOylation domain of ACSL4 to enhance its stability. Further research is required to facilitate the development of SENP1 as a clinical biomarker of HSNCC and potentially other cancer types.
In conclusion, SENP1 deficiency promoted ferroptosis and inhibited tumor progression through reducing SUMOylation of ACSL4. The collective results of the present study supported the utility of SENP1 as an effective predictive biomarker for targeted treatment of HNSCC.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- HNSCC
head and neck squamous cell carcinoma
- GPX4
glutathione peroxidase 4
- SUMO
small ubiquitin-like modifiers
- SENP1
SUMO-specific peptidase 1
- ACSL4
acyl CoA synthetase long chain 4
- PUFA
polyunsaturated fatty acid
- LASSO
least absolute shrinkage and selection operator
- cDNA
complementary DNA
- CCK-8
Cell Counting Kit-8 assay
- TCGA
The Cancer Genome Atlas
- GSH
glutathione
- TNM
tumor-node-metastasis
- CI
confidence interval
- HR
hazard ratio
- ROC
receiver operation curve
- AUC
area under the curve
- AA
anthotetraenoic acid
- ADA
adrenaline
Funding Statement
The present study was supported by Jiangsu College Natural Science Foundation of China (grant no. 12KJB320015), Xuzhou Science and Technology Planning Project (grant no. KC15SHO15) and Six Talent Peaks Project in Jiangsu (grant no. 2014-wsw-068)
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the Meiji Biotechnology Service Network (https://report.majorbio.com/medical/species_general/task_id/rf7i_7cbvmpp12cp9c3a5fbn4u5). The datasets and The Cancer Genome Atlas (https://gdc.cancer.gov/) repositories. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XX and ZF conceived and designed the study and participated in the processing of the experiments, drafting and revising the first draft. YM conducted data analysis and composition. FD conducted data collection. TG conducted a design for bioinformatics analysis. JZ participated in design of all experiments and wrote the final manuscript determination. All authors read and approved the final manuscript. TG and JZ confirm the authenticity of all the raw data.
Ethics approval and consent to participate
Informed consent was obtained from all the patients and the study was approved by the Ethics Committee of the Affiliated Hospital of Xuzhou Medical University (approval no. XYFY2020-KL216-01; Xuzhou, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Johnson DE, Burtness B, Leemans CR, Lui VWY, Bauman JE, Grandis JR. Head and neck squamous cell carcinoma. Nat Rev Dis Primers. 2020;6:92. doi: 10.1038/s41572-020-00224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Solomon B, Young RJ, Rischin D. Head and neck squamous cell carcinoma: Genomics and emerging biomarkers for immunomodulatory cancer treatments. Semin Cancer Biol. 2018;52:228–240. doi: 10.1016/j.semcancer.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 3.Kitamura N, Sento S, Yoshizawa Y, Sasabe E, Kudo Y, Yamamoto T. Current trends and future prospects of molecular targeted therapy in head and neck squamous cell carcinoma. Int J Mol Sci. 2020;22:240. doi: 10.3390/ijms22010240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Panarese I, Aquino G, Ronchi A, Longo F, Montella M, Cozzolino I, Roccuzzo G, Colella G, Caraglia M, Franco R. Oral and oropharyngeal squamous cell carcinoma: Prognostic and predictive parameters in the etiopathogenetic route. Expert Rev Anticancer Ther. 2019;19:105–119. doi: 10.1080/14737140.2019.1561288. [DOI] [PubMed] [Google Scholar]
- 5.Hanoteau A, Newton JM, Krupar R, Huang C, Liu HC, Gaspero A, Gartrell RD, Saenger YM, Hart TD, Santegoets SJ, et al. Tumor microenvironment modulation enhances immunologic benefit of chemoradiotherapy. J Immunother Cancer. 2019;7:10. doi: 10.1186/s40425-018-0485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Göttgens EL, Ostheimer C, Span PN, Bussink J, Hammond EM. HPV, hypoxia and radiation response in head and neck cancer. Br J Radiol. 2019;92:20180047. doi: 10.1259/bjr.20180047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang C, Liu X, Jin S, Chen Y, Guo R. Ferroptosis in cancer therapy: A novel approach to reversing drug resistance. Mol Cancer. 2022;21:47. doi: 10.1186/s12943-022-01530-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao L, Zhou X, Xie F, Zhang L, Yan H, Huang J, Zhang C, Zhou F, Chen J, Zhang L. Ferroptosis in cancer and cancer immunotherapy. Cancer Commun (Lond) 2022;42:88–116. doi: 10.1002/cac2.12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu L, Li YY, Zhang YC, Wu YX, Guo DD, Long D, Liu ZH. A novel ferroptosis-related gene signature to predict prognosis in patients with head and neck squamous cell carcinoma. Dis Markers. 2021;2021:5759927. doi: 10.1155/2021/5759927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu T, Zhang Z, Pan X, Zhang J, Wang X, Wang M, Li H, Yan M, Chen W. Caveolin-1 promotes cancer progression via inhibiting ferroptosis in head and neck squamous cell carcinoma. J Oral Pathol Med. 2022;51:52–62. doi: 10.1111/jop.13267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yau TY, Sander W, Eidson C, Courey AJ. SUMO interacting motifs: Structure and function. Cells. 2021;10:2825. doi: 10.3390/cells10112825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao X. SUMO-mediated regulation of nuclear functions and signaling processes. Mol Cell. 2018;71:409–418. doi: 10.1016/j.molcel.2018.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Chen L, Li Y, Hou W. SUMO-specific protease 1 inhibitors-A literature and patent overview. Expert Opin Ther Pat. 2022;32:1207–1216. doi: 10.1080/13543776.2022.2165910. [DOI] [PubMed] [Google Scholar]
- 15.Gao Y, Wang R, Liu J, Zhao K, Qian X, He X, Hong L. SENP1 promotes triple-negative breast cancer invasion and metastasis via enhancing CSN5 transcription mediated by GATA1 deSUMOylation. Int J Biol Sci. 2022;18:2186–2201. doi: 10.7150/ijbs.60594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong B, Gao Y, Kang X, Gao H, Zhang J, Guo H, You MJ, Xue W, Cheng J, Huang Y. SENP1 promotes proliferation of clear cell renal cell carcinoma through activation of glycolysis. Oncotarget. 2016;7:80435–80449. doi: 10.18632/oncotarget.12606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun W, Liu X, Yang X, Jing X, Duan C, Yang G, Wu C, Huang H, Luo Q, Xia S, et al. SENP1 regulates the transformation of lung resident mesenchymal stem cells and is associated with idiopathic pulmonary fibrosis progression. Cell Commun Signal. 2022;20:104. doi: 10.1186/s12964-022-00921-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bai YT, Xiao FJ, Wang H, Ge RL, Wang LS. Hypoxia protects H9c2 cells against ferroptosis through SENP1-mediated protein DeSUMOylation. Int J Med Sci. 2021;18:1618–1627. doi: 10.7150/ijms.50804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuwata H, Hara S. Role of acyl-CoA synthetase ACSL4 in arachidonic acid metabolism. Prostaglandins Other Lipid Mediat. 2019;144:106363. doi: 10.1016/j.prostaglandins.2019.106363. [DOI] [PubMed] [Google Scholar]
- 20.Doll S, Proneth B, Tyurina YY, Panzilius E, Kobayashi S, Ingold I, Irmler M, Beckers J, Aichler M, Walch A, et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat Chem Biol. 2017;13:91–98. doi: 10.1038/nchembio.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, Li J, Kang R, Klionsky DJ, Tang D. Ferroptosis: Machinery and regulation. Autophagy. 2021;17:2054–2081. doi: 10.1080/15548627.2020.1810918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quan J, Bode AM, Luo X. ACSL family: The regulatory mechanisms and therapeutic implications in cancer. Eur J Pharmacol. 2021;909:174397. doi: 10.1016/j.ejphar.2021.174397. [DOI] [PubMed] [Google Scholar]
- 23.Blum A, Wang P, Zenklusen JC. SnapShot: TCGA-analyzed tumors. Cell. 2018;173:530. doi: 10.1016/j.cell.2018.03.059. [DOI] [PubMed] [Google Scholar]
- 24.Onofri A, Terzaroli N, Russi L. Linear models for diallel crosses: A review with R functions. Theor Appl Genet. 2021;134:585–601. doi: 10.1007/s00122-020-03716-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.He F, Chen Z, Deng W, Zhan T, Huang X, Zheng Y, Yang H. Development and validation of a novel ferroptosis-related gene signature for predicting prognosis and immune microenvironment in head and neck squamous cell carcinoma. Int Immunopharmacol. 2021;98:107789. doi: 10.1016/j.intimp.2021.107789. [DOI] [PubMed] [Google Scholar]
- 27.Wang W, Zhang J, Wang Y, Xu Y, Zhang S. Identifies microtubule-binding protein CSPP1 as a novel cancer biomarker associated with ferroptosis and tumor microenvironment. Comput Struct Biotechnol J. 2022;20:3322–3335. doi: 10.1016/j.csbj.2022.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu T, Shi L, Yu C, Dong Y, Qiu F, Shen L, Qian Q, Zhou G, Zhu X. Ferroptosis promotes photodynamic therapy: Supramolecular photosensitizer-inducer nanodrug for enhanced cancer treatment. Theranostics. 2019;9:3293–3307. doi: 10.7150/thno.32867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han ZJ, Feng YH, Gu BH, Li YM, Chen H. The post-translational modification, SUMOylation, and cancer (Review) Int J Oncol. 2018;52:1081–1094. doi: 10.3892/ijo.2018.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Liang X, Liang H, Wang B. SENP1/HIF-1α feedback loop modulates hypoxia-induced cell proliferation, invasion, and EMT in human osteosarcoma cells. J Cell Biochem. 2018;119:1819–1826. doi: 10.1002/jcb.26342. [DOI] [PubMed] [Google Scholar]
- 31.Hao L, Dong Y, Zhang JJ, He HG, Chen JG, Zhang SQ, Zhang QJ, Wu W, Han CH, Shi ZD. Melatonin decreases androgen-sensitive prostate cancer growth by suppressing SENP1 expression. Transl Androl Urol. 2022;11:91–103. doi: 10.21037/tau-21-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen X, Ni R, Qian X, Yu C, Wu H, Ni J, Zhu W, Gao X. A verification study on the genes associated with laryngeal squamous cell carcinoma by cDNA microarray. Lin Chuang Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2010;24:411–413. (In Chinese) [PubMed] [Google Scholar]
- 33.Gao C, Xiao F, Zhang L, Sun Y, Wang L, Liu X, Sun H, Xie Z, Liang Y, Xu Q, Wang L. SENP1 inhibition suppresses the growth of lung cancer cells through activation of A20-mediated ferroptosis. Ann Transl Med. 2022;10:224. doi: 10.21037/atm-21-6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vertegaal ACO. Signalling mechanisms and cellular functions of SUMO. Nat Rev Mol Cell Biol. 2022;23:715–731. doi: 10.1038/s41580-022-00500-y. [DOI] [PubMed] [Google Scholar]
- 35.Mevissen TET, Komander D. Mechanisms of deubiquitinase specificity and regulation. Annu Rev Biochem. 2017;86:159–192. doi: 10.1146/annurev-biochem-061516-044916. [DOI] [PubMed] [Google Scholar]
- 36.Kukkula A, Ojala VK, Mendez LM, Sistonen L, Elenius K, Sundvall M. Therapeutic potential of targeting the SUMO pathway in cancer. Cancers (Basel) 2021;13:4402. doi: 10.3390/cancers13174402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bawa-Khalfe T, Yang FM, Ritho J, Lin HK, Cheng J, Yeh ET. SENP1 regulates PTEN stability to dictate prostate cancer development. Oncotarget. 2017;8:17651–17664. doi: 10.18632/oncotarget.13283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun XX, Chen Y, Su Y, Wang X, Chauhan KM, Liang J, Daniel CJ, Sears RC, Dai MS. SUMO protease SENP1 deSUMOylates and stabilizes c-Myc. Proc Natl Acad Sci USA. 2018;115:10983–10988. doi: 10.1073/pnas.1802932115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the Meiji Biotechnology Service Network (https://report.majorbio.com/medical/species_general/task_id/rf7i_7cbvmpp12cp9c3a5fbn4u5). The datasets and The Cancer Genome Atlas (https://gdc.cancer.gov/) repositories. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.