Abstract
Enzymatic methods to quantify deoxyribonucleoside triphosphates have existed for decades. In contrast, no general enzymatic method to quantify ribonucleoside triphosphates (rNTPs), which drive almost all cellular processes and serve as precursors of RNA, exists to date. ATP can be measured with an enzymatic luminometric method employing firefly luciferase, but the quantification of other ribonucleoside mono-, di-, and triphosphates is still a challenge for a non-specialized laboratory and practically impossible without chromatography equipment. To allow feasible quantification of ribonucleoside phosphates in any laboratory with typical molecular biology and biochemistry tools, we developed a robust microplate assay based on real-time detection of the Broccoli RNA aptamer during in vitro transcription. The assay employs the bacteriophage T7 and SP6 RNA polymerases, two oligonucleotide templates encoding the 49-nucleotide Broccoli aptamer, and a high-affinity fluorogenic aptamer-binding dye to quantify each of the four canonical rNTPs. The inclusion of nucleoside mono- and diphosphate kinases in the assay reactions enabled the quantification of the mono- and diphosphate counterparts. The assay is inherently specific and tolerates concentrated tissue and cell extracts. In summary, we describe the first chromatography-free method to quantify ATP, ADP, AMP, GTP, GDP, GMP, UTP, UDP, UMP, CTP, CDP and CMP in biological samples.
Graphical Abstract
Graphical Abstract.
Introduction
Nucleotides are building blocks for nucleic acids, energy carriers in almost all cellular processes, and essential signaling molecules and biosynthetic precursors (1). Information on their cellular concentrations and phosphorylation statuses is increasingly needed in various fields of life sciences. Chromatography-based methods for the quantification of nucleotides have existed for decades (2) and have also more recently undergone modifications to render compatibility with equipment coupled to a mass spectrometer (3–5). In addition, DNA polymerase-based methods for the determination of deoxyribonucleoside triphosphates (dNTPs) have been available since 1969 (6). Nevertheless, reliable nucleotide quantification from complex biological samples is still a challenge, especially for a non-specialized laboratory (7). Recently, we published an improved highly sensitive DNA polymerase-based method for convenient quantification of dNTPs in a high-throughput manner in 384-well microplates (8). This method allows dNTP quantification in any laboratory with a typical qPCR instrument. dNTPs are, however, a minor cellular nucleotide pool needed mainly for DNA repair and replication, and of interest mainly to specialized research niches (9). Ribonucleoside triphosphates (rNTPs) are a several orders of magnitude more abundant class of cellular nucleotides than dNTPs. ATP, the most abundant of them, is easily quantified using a luminometric method employing the firefly luciferase (10). A similarly convenient enzymatic method for the quantification of the other three canonical rNTPs (GTP, UTP and CTP) and the corresponding nucleoside mono- and diphosphates (rNMPs and rNDPs, respectively) is lacking.
Analogous to the DNA polymerase-based methods for dNTPs (8), a similar approach could in principle allow the quantification of rNTPs with RNA polymerases. However, to the best of our knowledge, such a method has never been reported. One obstacle in the development of RNA polymerase-based methods may have been the lack of practical quantitative readout. Unlike DNA polymerases, RNA polymerases undergo cycles of abortive initiation, leading to short (<14 nt) truncated RNA products (11). These short RNA products surpass the number of full-length transcripts when one rNTP is available at a rate-limiting concentration (12). Thus, the total RNA produced or labelled non-limiting nucleotides incorporated are unlikely to serve as sensitive readouts. Since the identification of several fluorogenic dye-binding RNA aptamers (RNA sequences naturally folding into a defined conformation), quantitative in vitro transcription with real-time detection has now, however, become possible (13,14). Here, we utilized the highly sensitive fluorometric detection of the Broccoli aptamer (15) for the development of an RNA polymerase-based method for the quantification of rNTPs and the corresponding mono- and diphosphates.
Materials and methods
Oligonucleotides and other reagents
Integrated DNA Technologies (IDT) (Coralville, Iowa, USA) synthetized all DNA oligonucleotides used in this study (Table 1). The oligonucleotides longer than 50 nt were synthetised using IDT Ultramer technology. Standard desalting was applied to all synthetic oligonucleotides. Double-stranded DNA templates were prepared by heating an equimolar mix of the complementary DNA strands to 95°C and by slowly cooling the mixture to 23–30°C.
Table 1.
DNA templates

|
Supplementary Table S1 lists all essential reagents. For the RNA polymerase-based method, the following molar extinction coefficients (mM−1cm−1) were used to determine the concentrations of the nucleotide standards: 15.4 (259 nm) adenosine, 10 (262 nm) uridine, 8.9 (271 nm) cytidine and 13.7 (252 nm) guanosine. The UV absorbances were measured in Tris-buffered saline at pH 7.6.
Mouse tissues samples
Mice of congenic C57BL/6JCrl background were maintained by the Laboratory Animal Center of the University of Helsinki. Two-month-old male mice were euthanized by cervical dislocation, and the liver tissue was immediately (10–20s) excised and placed in liquid nitrogen and stored at –80°C. For experiments with delayed sample collection, the excised liver was kept at room temperature for the indicated periods.
Cell culture
The mouse hepatocyte line AML12 was maintained in Dulbecco's modified Eagle's medium-F12 mixture (DMEM-F12) with 5 mM glucose and supplemented with 10% fetal bovine serum, 100 U/ml of penicillin, 100 μg/ml of streptomycin,2 mM l-alanyl-l-glutamine, 10 μg/ml insulin, 5.5 μg/ml transferrin and 5 ng/ml selenium. Mouse hepatoma Hepa1–6 cells were maintained in DMEM with 25 mM glucose, 10% fetal bovine serum, penicillin and streptomycin, and 2 mM l-alanyl-l-glutamine. Both cell lines were cultured at 37°C in an atmosphere of 5% CO2 and 95% air. For treatments, the cells were plated in 6-cm plates and grown to 60–80% confluence. Inhibitors targeting mitochondrial ATP production (for details, see figure legends) were added directly to the medium and the cells were incubated for 6 h. To collect the samples, the cells were first rinsed once with cold PBS, followed by the addition of 0.5 ml of ice-cold 80% (v/v) MeOH before the cells were scraped off the plates. The cell suspensions were stored at –80°C until the extraction.
Extraction of nucleotides
Supplementary methods describe an extraction protocol that was employed for the generation of data in Figure 4A and Supplementary Figures S3 and S4. Here, an optimized extraction with a more stringent deproteination is described. Frozen liver samples (18–22 mg) were directly homogenized in 0.5 ml of ice-cold 80% (v/v) MeOH with a battery-operated microtube pestle for 10 s. The homogenization was finalized with a microtip probe sonicator (12 s, amplitude 12%, Branson Digital Sonifier 250). Cultured cells scraped into 80% MeOH were directly sonicated. The samples were kept on dry ice between the homogenization steps. The methanolic extracts were incubated for 3 min at 100°C. Per 0.5 ml of 80% MeOH, 200 μl of chloroform was added to further precipitate macromolecules and to remove nonpolar metabolites. Phase separation was induced by the addition of 200 μl water. The extracts were vortexed for 10 s followed by vigorous shaking for 30 s. After centrifugation (3 min 18 000g at 0°C), the aqueous phase was transferred into a new microtube. MeOH was removed by washing three times with 1.2 ml of diethyl ether. Any remaining layer of diethyl ether was evaporated by a 10-s flow of N2 gas. The traces of the diethyl ether from the aqueous phase were evaporated in a Genevac miVac Duo centrifugal vacuum evaporator for 12 min. The remaining liquid volume after evaporation was estimated by weighing the extracts and assuming the density of 1 mg/μl. Instead of diethyl ether washes, the cell extracts were evaporated to dryness. The extracts were stored at –80°C.
Figure 4.
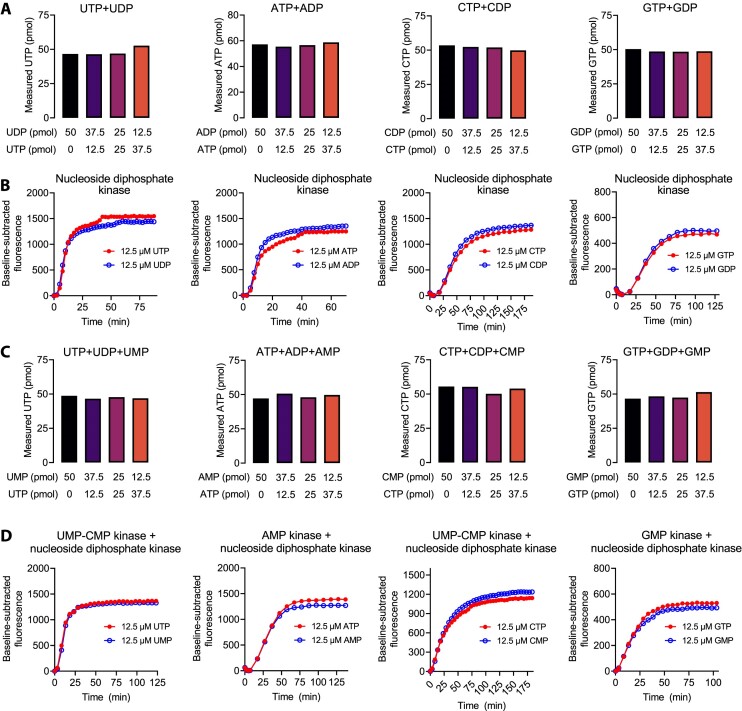
Quantification of rNMPs and rNDPs. (A) Quantification of the indicated rNDP and rNTP combinations by inclusion of nucleoside diphosphate kinase in the assay reactions. (B) Representative fluorescence traces of assay reactions containing nucleoside diphosphate kinase and either rNTP or rNDP as a limiting nucleotide. (C) Quantification of the indicated rNMP and rNTP combinations by inclusion of nucleoside diphosphate kinase and rNMP kinases in the assay reactions. (D) Representative fluorescence traces of assay reactions containing nucleoside diphosphate kinase and rNMP kinase and either an rNTP or rNMP as the limiting nucleotide. Supplementary Figure S2 shows the indispensability of the added kinases for signal generation from rNMPs and rNDPs.
Extraction of total protein fraction
Precipitated proteins from the nucleotide extraction were sedimented by the addition of 800 μl MeOH and centrifugation (3 min 18 000g at 4°C). The protein pellets were dissolved in Laemmli buffer (2% SDS, 1% β-mercaptoethanol, 60 mM Tris–HCl pH 6.8 and 12% glycerol) by sonication and 5 min incubation at 95°C. A modified Bradford reagent containing 2.5 mg/ml α-cyclodextrin to chelate the interfering SDS was employed to measure protein concentrations (16). Bovine serum albumin was used as a reference protein.
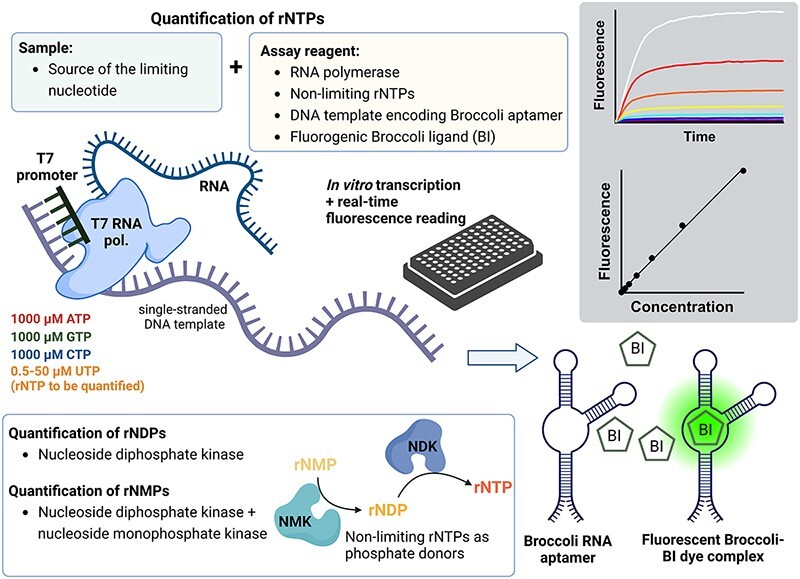
Quantification of rNTPs by in vitro transcription
The assay parameters were varied and optimized throughout this study. The final established assay conditions are described here. T7 RNA polymerase (#M0251, New England BioLabs) and template 3 were the most suitable combination for the quantification of ATP, UTP and CTP. SP6 RNA polymerase (#M0207, New England BioLabs) and template 4 were more optimal for the quantification of GTP. The optimized assay reactions comprised 1x CutSmart buffer (New England BioLabs) (20 mM Tris-acetate pH 7.9, 50 mM potassium acetate, 10 mM Mg-acetate, and 0.1 mg/ml BSA), 2 mM spermidine, 1 mM of each non-limiting rNTP, 1–2 μM limiting rNTP, 5 mM DTT, 20 nM DNA template, 20 μM BI, 0.5 kU/ml RiboLock (EO0382, Thermo Scientific), 0.25 U/ml pyrophosphatase (EF0221, Thermo Scientific), and 1.9 kU/ml T7 RNA polymerase or 0.5 kU/ml SP6 RNA polymerase. The BI stands for Broccoli aptamer ligand (5Z)-3-(1H-Benzimidazol-7-ylmethyl)-5-[(3,5-difluoro-4-hydroxyphenyl)methylene]-3,5-dihydro-2-methyl-4H-imidazol-4-one dihydrochloride) (Cat. No. 7466, Tocris) (17). Some experiments were performed without the ribonuclease inhibitor (Ribolock) as this reagent was found dispensable when adequate measures to prevent ribonuclease contamination were followed. The optimal basal concentration of limiting rNTP was 1 μM for the quantification of ATP, UTP, and CTP, and 2 μM for GTP. Master mixes of the reagents were prepared and mixed with samples in 384-well qPCR plates (#HSP3805, BioRad). The reactions were carried out in volumes of 4–10 μl. In our final established protocol, the reactions comprised one part of sample and one part of 2 × master mix. The reaction set-up was prepared on ice, the plate sealed and briefly centrifuged. The Broccoli aptamer-BI fluorescence was measured upon 470 nm excitation and 510 nm emission in a Synergy H1 plate reader (Biotek) for 2–8 h at 37°C. When accurate temperature control was essential, the plates were read in a Bio-Rad CFX384 qPCR instrument instead. Baseline-subtracted end-point fluorescence values were used to generate the standard curves.
Quantification of rNMPs and rNDPs
The sum of rNDPs + rNTPs of interest was quantified by inclusion of 5 μg/ml of recombinant human nucleoside diphosphate kinase (#NDPK-33, BioNukleo GmbH, Berlin, Germany) in the assay reactions. The sum of target rNMPs + rNDPs + rNTPs were quantified by including the nucleoside diphosphate kinase and 5 μg/ml nucleoside monophosphate kinase in the assay reactions. The following recombinant nucleoside monophosphate kinases produced by BioNukleo were used: AMP kinase (#NMPK-01), GMP kinase (#NMPK-21) and UMP-CMP kinase (#NMPK-22). The concentrations of specific nucleotide species were deduced from three measurements: (i) rNTP, (ii) rNTP + rNDP and (iii) rNTP + rNDP + rNMP.
Luciferin-luciferase assay for ATP
Invitrogen ATP Determination Kit (#A22066) was employed to measure ATP by the firefly luciferase method.
High-performance liquid chromatography (HPLC)
The nucleotides were separated by HPLC run at 1 ml/min using a 150 mm × 4.6 mm SunShell C18-WP HPLC column from ChromaNik Technologies Inc (Osaka, Japan). Based on our previously published protocol for the separation of ADP, rNTPs and dNTPs isocratically on this column (18), where the influence of multiple parameters on nucleotide separation were investigated, we developed a new protocol to also separate rNMPs and the remaining rNDPs. Two main differences were that a phosphate gradient was used to increase the separation of early eluting peaks and that acetonitrile was exchanged for MeOH to enable the separation of GDP and UDP. The mobile phase was a binary mixture of two aqueous solutions, A and B. Solution A contained 5.8% (v/v) MeOH, 0.7 g/l tetrabuytylammonium bromide (TBA-Br) and 46 g/l HPLC-grade KH2PO4 (HiPerSolv Chromanorm grade sold by VWR), and the final solution was adjusted to pH 5.6 withKOH. Solution B had the same concentrations of MeOH and TBA-Br as A but lacked KH2PO4 and did not need pH-adjustment. The nucleotides were analyzed by a linear gradient between 5% A to 85% A over 19 min, before returning to initial conditions over 1 min and equilibrating for 7 min before the loading of the next sample. The HPLC instrument was equipped with a high-pressure mixing chamber with an internal volume of 1740 μl (Dynamic mixer chamber D-14163, Knauer, Berlin, Germany), and a sample loop of 20 μl. Prior to loading, 10 μl of each sample was mixed with 26 μl water and 4 μl 10 × loading buffer. The 10 × loading buffer was prepared by mixing 500 μl 14 mg/ml TBA-Br with 500 μl 23 mg/ml KH2PO4 adjusted to pH 5.6 with KOH. The peak identities were confirmed by their 270 nm/280 nm ratios and with samples spiked with standard solutions. The nucleotides were quantified at 280 nm by comparing the peak heights to a standard nucleotide solution. The use of 280 nm (rather than 260 nm for optimal absorbance of nucleotides) was a way to suppress the effect of IMP, UDP-sugars and AMP on the analyses of GMP, CDP and UDP, respectively.
Protocol details
Step-by-step protocols are provided as supplementary files.
Results
Development of in vitro transcription assay for rNTP quantification
As a starting point we tested an in vitro transcription protocol with fluorescent real-time monitoring by Kartje et al. (13). This assay utilizes T7 RNA polymerase, the Broccoli aptamer and its fluorogenic ligand DFHBI-1T (3,5-difluoro-4hydroxy-benzylidene imidazolinone). While this system functions well for its purpose, characterization and optimization of in vitro transcription for maximum RNA yield, it lacked the sensitivity required for robust quantification of rNTPs (Figure 1A, B, blue lines). To improve the sensitivity, we substituted DFHBI-1T with BI as the Broccoli ligand. BI has a higher affinity for the aptamer and enhanced fluorogenicity in comparison to DFHBI-1T (17). It also promotes the folding and thermal stability of the aptamer. The reaction buffer of the assay described in Kartje et al. includes 5 mM NaCl and 1 mM KCl as enhancers of aptamer folding (13), while the efficient folding of Broccoli aptamers requires ∼50 mM K+ or >200 mM Na+ (19). Therefore, we chose a buffer containing 50 mM potassium acetate. These changes notably increased the aptamer fluorescence (Figure 1A, B). In the initial test runs, we used a rather high template concentration of 1 μM based on previous optimization to maximize the RNA yield when all rNTPs are in excess (13). However, when UTP was at rate-limiting concentration, lowering of the template concentration to below 150 nM increased the RNA yield more than 3-fold (Figure 1C). Any template concentration between 10–150 nM worked well, and 20 nM was selected for further studies. Optimization of the BI concentration led to a further increase in sensitivity (Figure 1D). Together these changes enabled the meaningful quantification of UTP (Figure 1E, F).
Figure 1.
Broccoli aptamer, its fluorogenic ligand BI, and potassium acetate-supplemented buffer enable determination of rNTPs by quantitative in vitro transcription. (A, B) Comparison of Broccoli ligands (DFHBI-1T and BI) and buffer potassium concentration (enhancer of the aptamer folding) on real-time monitoring of in vitro transcription under limiting UTP concentrations with T7 RNA polymerase. (A) Representative fluorescence traces recorded during the in vitro transcription. (B) Standard curves generated from the baseline-subtracted end-point fluorescence values. The symbols and colors represent the same assay conditions as in (A). (C) Optimization of the template concentration. (D) Optimization of BI concentration. (E) Real-time detection of Broccoli-BI fluorescence during the assay reactions with T7 RNA polymerase, and (F) standard curves generated from the end-point fluorescence values. A linear regression line is shown for the linear part of the standard curve. (G) Real-time detection of Broccoli-BI fluorescence during the CTP and GTP assay reactions with SP6 RNA polymerase, and (H) the corresponding standard curves. The initial (0 to 5 min) drop in fluorescence is due to temperature-dependent properties BI (see main text and Supplementary Figure S1I). (I) Assay conditions in graphs A to H. The signal-to-noise ratio was defined as background-subtracted fluorescence divided by the standard deviation of the background (n = 7 replicates).
Next, we tested the same assay conditions for the quantification of the other rNTPs. The quantification of ATP and CTP did not require further optimization (Figure 1E, F). However, the quantification of GTP turned out to be problematic with T7 RNA polymerase as it prefers to initiate transcription with three consecutive guanosines (20) (Table 1 and Figure 1E, F). When GTP is severely limiting, T7 RNA polymerase presumably undergoes excessive cycles of abortive initiation, leading to the consumption of GTP without production of full-length transcripts. To overcome this issue, we tested the T7 class II ϕ2.5 promoter, which enables adenosine-initiating transcripts (21), but without success (Supplementary Figure S1A). SP6 RNA polymerase also prefers to initiate transcription with a guanosine but has less preference for the immediate downstream nucleotide sequence than the T7 RNA polymerase (22). We designed a template for the SP6 RNA polymerase in which a 10-nt pre-aptamer leader sequence contains a single starting guanosine and the next guanosine is at position +11 (Template 2, Table 1). This combination of template and RNA polymerase enabled a useful dynamic range for the quantification of GTP (Figure 1G, H). SP6 RNA polymerase and template 2 also slightly increased the sensitivity for the quantification of CTP (Figure 1H).
Further optimization of the assay
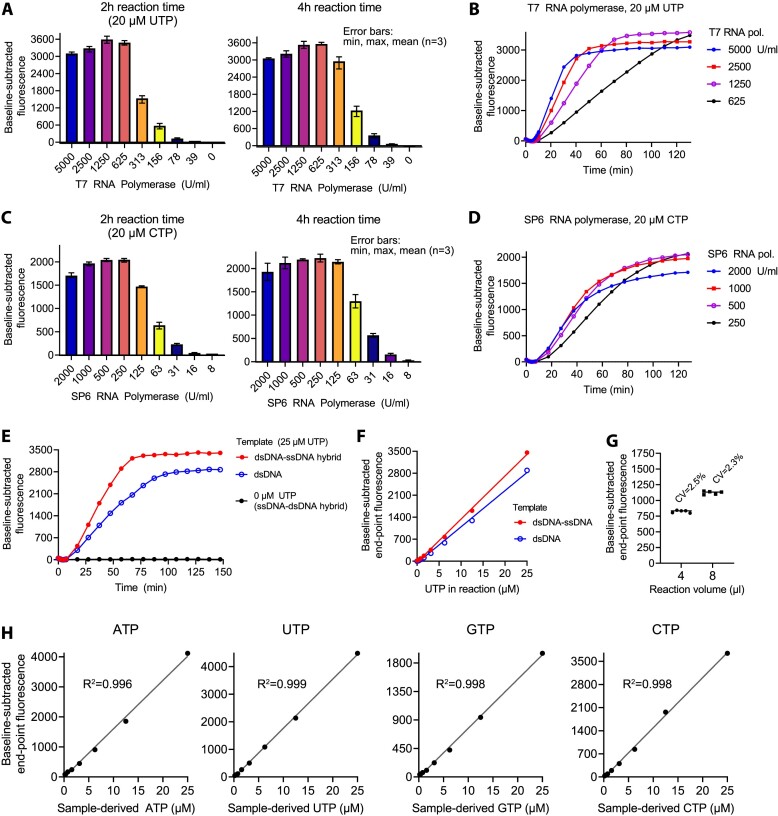
After a successful proof-of-concept, we set out to optimize other assay parameters. The optimal template concentration for T7 RNA polymerase (Figure 1C) proved to be optimal for SP6 RNA polymerase as well (Supplementary Figure S1B). Over 15-fold differences in RNA polymerase concentrations had little effect on the final aptamer yield when the reactions were allowed to progress to completion (Figures 2A–D). Based on maximal end-point fluorescence within 2 h, we selected 1.9 kU/ml as optimal concentration for the T7 RNA polymerase. For the SP6 RNA polymerase, we considered 1 kU/ml and 0.5 kU/ml as optimal concentrations for the quantification of CTP and GTP, respectively.
Figure 2.
Optimization of in vitro transcription for quantification of rNTPs. (A, B) Optimization of T7 RNA polymerase concentration with UTP as the limiting nucleotide. (C, D) Optimization of SP6 RNA polymerase concentration with CTP as the limiting nucleotide. (E, F) Comparison of a double-stranded single-stranded hybrid template and the fully double-stranded equivalent with T7 RNA polymerase. (G) Reaction volume and variation of technical replicates (T7 RNA polymerase and 6.25 μM UTP). CV, coefficient of variation. (H) Linearization of the standard curves by the addition of low basal concentrations of the limiting nucleotide. The basal limiting nucleotide concentrations were 1 μM for the quantification of ATP, UTP, CTP and 2 μM for the quantification of GTP. The standard curves were generated without technical replicates.
In vitro transcription is usually performed at 37–42°C, while the folding of the Broccoli-aptamer is optimal at lower temperatures (19). A decrease of the reaction temperature from 37°C to 30°C slowed down the reaction phase but the end-point fluorescence values were practically identical (Supplementary Figure S1C–H). Cooling down the completed reactions to 23°C marginally increased the aptamer fluorescence (Supplementary Figure S1E, H), indicating close to 100% efficient folding of the Broccoli at 37°C under the given conditions. While manipulating the assay temperature, we observed that the aptamer-independent fluorescence of BI (background) was somewhat sensitive to temperature (Supplementary Figure S1I). When using a qPCR instrument to read the plate, the assay temperature is accurately controlled. However, when using a plate reader, we found it best to determine the baseline fluorescence after a short delay of 4–8 min that is required for signal stabilization.
T7 RNA polymerase requires a double-stranded promoter but efficiently transcribes single-stranded downstream sequences (20). For the experiments above, we used a double-stranded single-stranded hybrid template for the T7 RNA polymerase (Table 1, template 1). A fully double-stranded equivalent proved less optimal (Figures 2E, F). SP6 RNA polymerase has an absolute requirement for fully double-stranded DNA (23). Hence, we did not test hybrid templates with this RNA polymerase.
A T7 RNA polymerase harboring a P266L point mutation shows enhanced promoter clearance and attenuated abortive initiation (24). We tested these potentially beneficial features with UTP as the rNTP to be quantified. The in vitro transcription progressed faster with this mutant than with the wild-type enzyme (Supplementary Figure S1J). However, the sensitivity of the assay slightly decreased (Supplementary Figure S1K).
A 384-well PCR plate allows miniaturization of qPCR reactions to a few microliters. We tested reaction volumes of 4 and 8 μl side by side. The coefficient of variation (CV) between technical replicates was less than 2.6% with both reaction volumes (Figure 2G). The higher reaction volume led to somewhat higher baseline-corrected end-point fluorescence values.
The standard curves for all rNTPs showed wide sensitive linear ranges but clear threshold concentrations below which linear curve fitting did not apply and the resolution of the assay became poor. To eliminate this useless concentration range, we decided to include a low basal concentration (1–2 μM) of the limiting rNTP in the assay reactions. Due to this modification any increase over the basal concentration led to a steep linear increase in the measured signal (Figure 2H). Moreover, the linearization of the standard curve enables easy application of the Standard Addition Method to correct matrix effects if required.
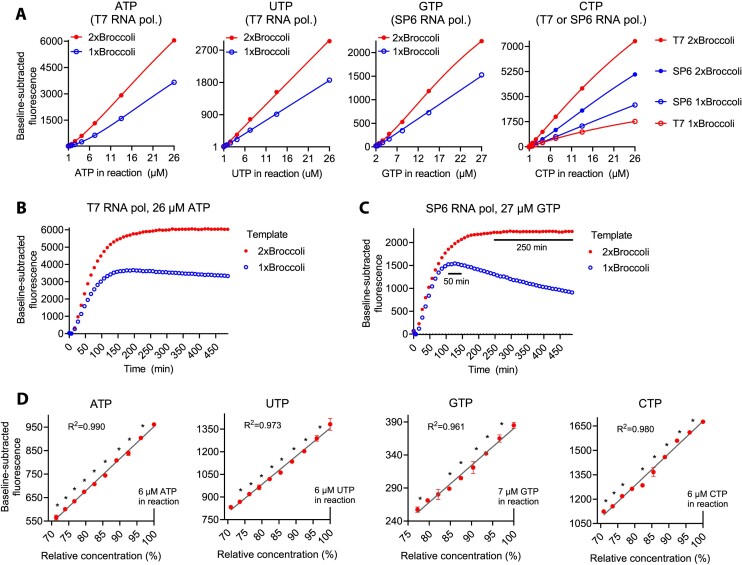
As a potential further signal enhancement, we tested templates encoding dimeric Broccoli (Table 1, template 3 and 4). Instead of concatemeric Broccoli constructs with relative lengthy scaffolds developed for live cell imaging (17), we chose a dimeric Broccoli with a 4 base pair stabilizing stem that prevents misfolding of the aptamer (25). In comparison to the monomeric aptamer, the dimeric counterpart led to approximately 1.6-fold increase in the steepness of ATP, UTP and GTP standard curves (Figure 3A). In the case of CTP quantification, the benefits of dimeric Broccoli were RNA polymerase- and template-dependent. While SP6 RNA polymerase provided higher sensitivity than T7 RNA polymerase with the monomeric Broccoli, the reverse was true with the dimeric Broccoli. With the T7 RNA polymerase the dimeric Broccoli led to over 4-fold increase in the CTP standard curve steepness, making the assay the most sensitive for quantification of CTP (Figure 3A), the least abundant rNTP in many cell types (2). Two drawbacks of the dimeric Broccoli were increased reaction time and necessity to perform non-linear curve fitting for CTP and GTP quantifications (Figures 3A–C). On the other hand, the dimeric Broccoli extended the stable plateau phase of the GTP quantification reactions from ∼50 min to over 4 h (Figure 3C). It is possible that this stabilized aptamer construct is resistant to activation of single-strand RNA/DNA exonuclease activity of some RNA polymerases upon rNTP exhaustion (26).
Figure 3.
Dimeric Broccoli enhances sensitivity and resolution of the rNTP quantification. (A) Signal amplification with stabilized dimeric Broccoli. UTP quantification reactions were performed in a qPCR instrument, and the rest of the reactions in a plate reader. (B) Representative fluorescence traces of ATP quantification reactions with templates encoding monomeric or dimeric Broccoli. (C) Similar experiment as in (B) but with GTP instead of ATP as the limiting nucleotide. Stable plateau phases are marked with black bars for the GTP quantification reactions. (D) Evaluation of assay resolution by quantification of 3–4% differences in limiting rNTP concentration. The symbols represent the mean value of three replicates. The error bars represent SEM. Where error bars are not visible, they are smaller than the symbols. *P< 0.05 for adjacent concentrations (Fisher's LSD, one-sided P value).
Because rNTPs are abundant metabolites and because of the clear threshold concentration (0.2–0.5 μM) of our assay, we did not seek to benchmark the assay based on the traditional lowest limit of detection and quantification. Instead, we concentrated on a more relevant parameter, the quantitative resolution, i.e. the lowest analyte concentration difference the assay can discriminate (27). Using technical triplicates, 10 μl reaction volume and 2× Broccoli templates, we could reliably quantify rNTP differences <4% (Figure 3D), meaning that the resolution was close to pipetting precision.
Modification of the assay for rNMPs and rNDPs
Because most biological processes are sensitive to the energy charge of nucleotides (mainly rNTP-to-rNDP ratio), knowledge on the concentrations of rNMPs and rNDPs can be as important as that of rNTPs. We asked whether we could modify our assay for these nucleotide species by phosphorylating the mono- and diphophates to triphosphates. First, we added a nucleoside diphosphate kinase in the assay reactions and measured the rNTP signal from samples with equal total nucleotide concentration but varying proportions of rNTPs and rNDPs. In this approach, the nucleoside diphosphate kinase utilizes the non-limiting rNTPs as phosphate donors to phosphorylate the target rNDP to the corresponding rNTP, allowing the measurement of the sum of rNDP + rNTP of interest. All rNDP + rNTP mixtures gave essentially the same expected rNTP signal (Figure 4A), implying essentially complete phosphorylation of rNDPs to rNTPs. Monitoring of the transcription kinetics did not reveal differences between rNDP or rNTP samples or their mixtures, indicating almost instant rNDP phosphorylation (Figure 4B). In the absence of nucleoside diphosphate kinase, neither rNDPs nor rNMPs produced a measurable signal (Supplementary Figure S2).
Next, we added the nucleoside diphosphate kinase, and AMP kinase, or GMP kinase, or UMP-CMP kinase in the assay reactions to determine the rNMPs. We repeated the previous experiment but now with varying mixtures of rNMPs and rNTPs. Again, the phosphorylation to rNTPs was almost instant and all mixtures gave essentially the same expected rNTP signal (Figures 4C and D). Both the nucleoside diphosphate kinase and rNMP kinase were required for signal production from rNMPs, verifying that the purified enzymes used in the assay were free from unintended kinase activity (Supplementary Figure S2).
Application of the assay to biological samples
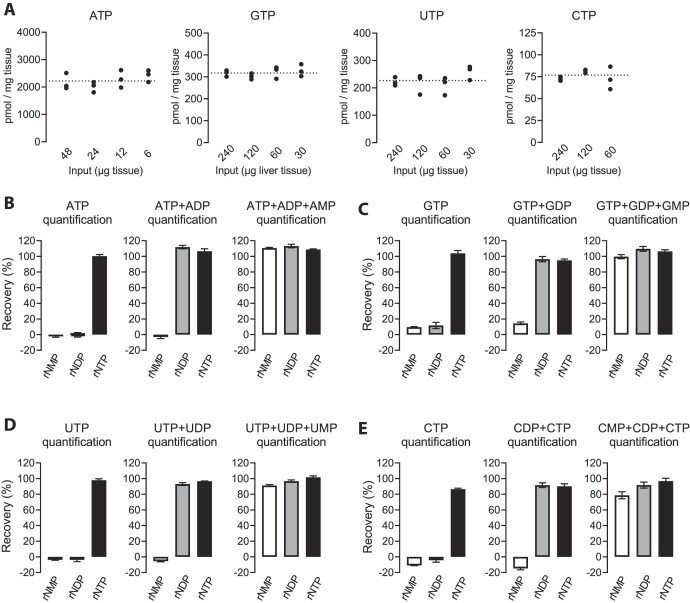
We extracted polar metabolites from mouse liver tissue and made a dilution series of the extracts to assess the applicability of the assay for complex biological samples and proneness to sample-related interference. Each of the four canonical rNTPs were easily quantifiable with our assay (Figure 5A). All dilutions gave essentially the same tissue rNTP concentrations, excluding major sample-related inhibition. The measured liver rNTP concentrations were very close to the values reported for rat liver in the systematic review of Traut el al. (2). As an alternative approach to assess sample-related interference, we spiked known amounts of rNTPs to liver extracts and assessed the in vitro transcription efficiency in comparison to purified rNTPs. The recovery of ATP, GTP and UTP spiked into the extracts was essentially 100% (Figures 5B–D). The recovery of CTP was slightly less (range: 84–96%) (Figure 5E, Supplementary Figure S3). We also assessed liver extracts spiked with rNMPs and rNDPs. We found that extracts that had not undergone a heat denaturation step showed rNMP kinase activity that could phosphorylate at least CMP (Supplementary Figure S3). Slight modifications to the extraction protocol to increase the stringency of extract deproteination, including a heat denaturation step, completely abolished this interference (Figure 5E, Materials and Methods). After these modifications, no unintended nucleotide interconversion took place (Figure 5B–E). With the addition of exogenous nucleotide kinases to quantify rNMPs and rNDPs, the recoveries of these nucleotides spiked to liver extracts were similar to that of rNTPs (Figures 5B–E).
Figure 5.
Quantification of rNTPs from mouse liver. (A) Estimation of sample-related interference by a dilution series. The input refers to the polar metabolite extract worth of initial tissue mass. The reaction volumes were 4 μl (1 μl sample + 3 μl assay reagent). The data points are technical replicates from four pooled liver extracts. (B–E) Recovery of exogenous rNMPs, rNDPs, and rNTPs spiked into liver extracts. The purified nucleotides were added as mixtures comprising 2000 pmol adenosine nucleotides (AMP, ADP or ATP), 200 pmol uridine nucleotides (UMP, UDP or UTP), 200 pmol guanosine nucleotides (GMP, GDP or GTP), and 50 pmol cytidine nucleotides (CMP, CDP or CTP) per mg tissue. Sample inputs (relative to pre-extraction tissue weight) were the following: 11.25 μg for ATP, 112.5 μg for GTP and UTP, and 450 μg for CTP. The reaction volumes were 10 μl (5 μl sample + 5 μl assay reagent). The bar graphs represent mean and SEM from 4 independent liver extracts.
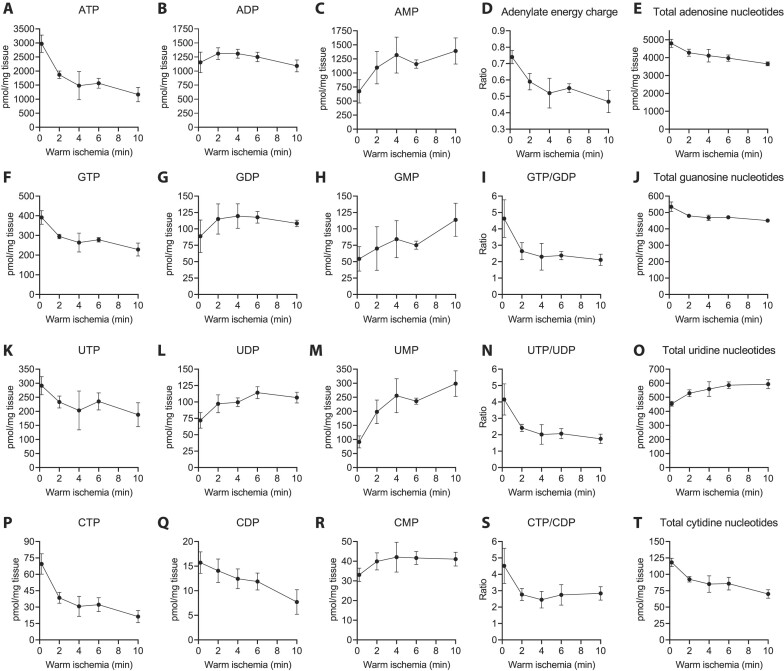
To further test the utility of our assay, we performed a classic warm ischemia experiment to disrupt nucleotide pools ex vivo, exposing subsamples from mouse livers to periods at room temperature (Figure 6 and Supplementary Figure S4). The ATP levels steeply declined for the first 4 min of the ischemia and then briefly stabilized before the decline continued (Figure 6A). After 10 min, the ATP levels were 39% of the initial values. ADP levels were relatively stable (Figure 6B). In contrast, AMP immediately started to accumulate, and its levels mirrored the decline of ATP (Figure 6C). The adenylate energy charge dropped from 0.74 to 0.47 during the follow up of 10 minutes (Figure 6D). At the same time, total adenosine nucleotide content gradually and linearly decreased (Figure 6E).
Figure 6.
Effect of warm ischemia on hepatic ribonucleotide pools. (A–C) Quantification of adenylate nucleotides. (D) Adenylate energy charge defined as (ATP + 0.5ADP)/(ATP + ADP + AMP). (E) Total adenylate nucleotides (ATP + ADP + AMP). (F–J) Quantification of guanosine nucleotides and GTP/GDP ratio. (K–O) Quantification of uridine nucleotides and UTP/UDP ratio. (P–T) Quantification of cytidine nucleotides and CTP/CDP ratio. The data points represent mean and SD from four non-fasted male mice of 2 months of age. Where error bars are not visible, the SD was smaller than the size of the symbol. Supplementary Figure S4 shows data from a similar experiment in which the extracts were prepared as described in the Supplementary methods (no boiling step).
Our assay was also able to quantify all other canonical ribonucleotides and their disturbances during the ischemia. GTP and UTP showed similar patterns of ischemic decline to ATP but with less steep changes (Figures 6F and K). CTP showed the steepest decline among the four rNTPs (Figure 6P). Compared to the rather stable ADP levels (Figure 6B), GDP and UDP accumulated during the ischemia (Figures 6G and L), leading to a clear decrease in rNTP-to-rNDP ratio of the respective nucleotides (Figure 6I, N). In contrast, CDP levels decreased in linear fashion while the CTP-to-CDP ratio still collapsed due to even greater loss of CTP (Figures 6P, Q and S). GMP levels were somewhat variable but showed a clear pattern of accumulation (Figure 6H). The UMP levels showed an even higher relative increase than AMP levels (Figures 6C and M), while the CMP levels showed a small acute increase during the first few minutes and then remained stable (Figure 6R). The size of the total cytidine nucleotide pool decreased linearly and was roughly 50% of the baseline level after 10 min (Figure 6T). The total guanosine pool also decreased linearly but more modestly, while the total uridine nucleotide pool actually increased during the ischemia (Figure 6J, O), potentially due to degradation of nucleotide sugars. Taking into account potential species-specific differences (rat versus mouse), differences between fasted and non-fasted animals, and differing methodology related to sample processing, our results were highly in line with rodent liver reference data (2,28).
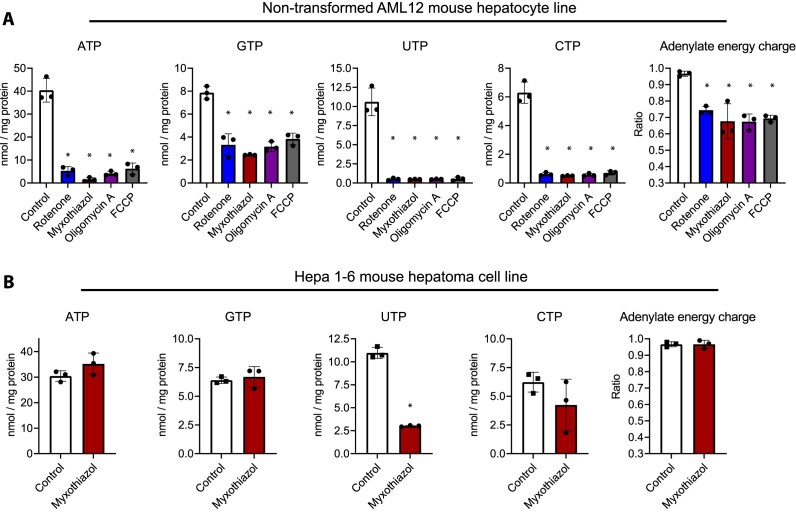
Finally, we assessed the impact of disrupted oxidative phosphorylation (mitochondrial ATP production) on nucleotide levels in two cell lines originating from mouse liver. In the non-transformed AML12 cell line, which derives from hepatocytes of mice overexpressing human transforming growth factor α (29), four inhibitors targeting different sites/processes of the oxidative phosphorylation collapsed the levels of all rNTPs and adenylate energy charge during a 6-h exposure (Figure 7A). This indicated a heavy reliance of these cells on mitochondria for maintenance of nucleotide pools, similarly to observed in primary hepatocytes (30). In contrast, the adenylate energy charge and purine nucleotide concentrations were indifferent to the blockade of oxidative phosphorylation in Hepa1-6 hepatoma cells (Figure 7B). Instead, myxothiazol, which disrupts the oxidative phosphorylation at the level of cytochrome bc1 complex (respiratory complex III), caused a clear UTP depletion in these cells. This is explained by blocked de novo pyrimidine biosynthesis at the level of dihydroorotate dehydrogenase (DHODH), which utilizes ubiquinone as the electron acceptor, the oxidation of which is dependent on complex III (31). Many cancer cells, and apparently Hepa1–6 cells as well, depend on mitochondrial respiration for pyrimidine nucleotide biosynthesis but not necessary for purine nucleotide biosynthesis and ATP generation (32).
Figure 7.
rNTP levels and adenylate energy charge after inhibition of oxidative phosphorylation in cultured mammalian cells. (A) Data from the AML12 mouse hepatocyte line. (B) Data from the Hepa1–6 mouse hepatoma cell line. The inhibitor concentrations were: 100 nM rotenone, 100 nM myxothiazol, 1 μM oligomycin A, and 5 μM FCCP. These inhibitors target mitochondrial NADH dehydrogenase (complex I), cytochrome bc1 complex (complex III), ATP synthase, and mitochondrial membrane potential, respectively. The data points represent individual cell culture dishes and the error bars SD. *, two-sided P value < 0.002 (versus control, one-way ANOVA and Dunnett's test or t-test).
Comparison to luciferin-luciferase ATP assay and HPLC analysis
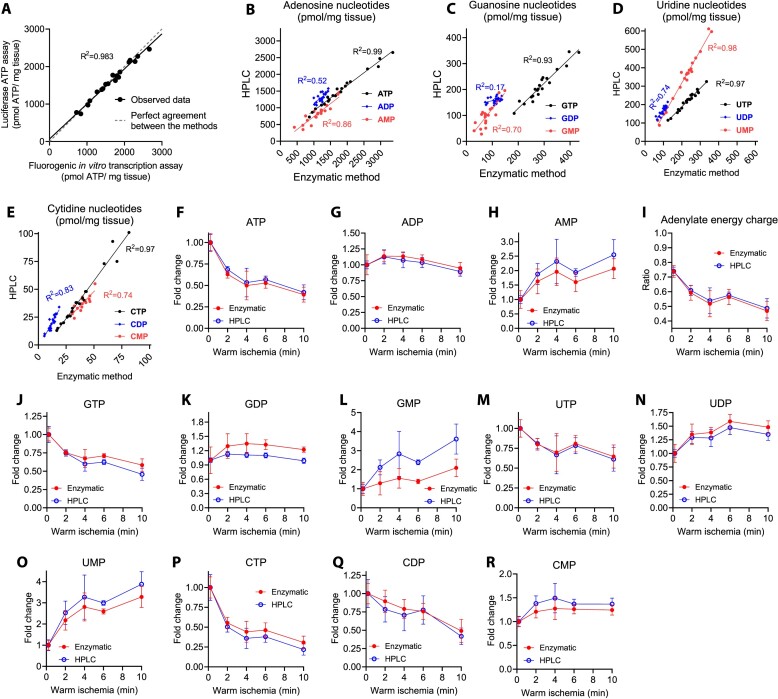
We applied two reference methods to remeasure nucleotides in the ischemic liver samples and compare our RNA polymerase-based method against these. As the first reference method, we employed the luciferin-luciferase enzymatic assay to remeasure ATP in the liver samples. The correlation coefficient (R2) between the methods was 0.983, and the mean difference across the range of 750–2600 pmol ATP/mg liver tissue was 18 pmol (Figures 8A). As the second method, we used a modified variant of a published HPLC protocol (18) for the quantification of the 12 ribonucleotides. The UV-chromatograms showed baseline resolution for 10 nucleotides (Supplementary Figure S5). The GMP and CDP peaks partially overlapped with signals from neighboring peaks. In the case of GMP, the overlapping peak had a retention time indicating that it could be inosine monophosphate (IMP). The rNTP concentrations showed near-perfect correlations between HPLC and the RNA polymerase-based method as judged by the R2 values (Figure 8B–E). However, R2 values depend on both the technical error and the concentration range covered, and it was therefore lower for nucleotides that were not affected as much by the ischemia. ADP and GDP are examples of nucleotides that had too small variation in the experiment to show strong R2 values. Nevertheless, the HPLC analysis reproduced the patterns of ischemic changes in nucleotide levels, regarding rNTPs, rNDP and rNMPs (Figures 8F–R). An exception was GMP levels (Figure 8L), the quantification of which was potentially interfered by ischemic accumulation of IMP in the HPLC analysis (Supplementary Figure S5). Overall, there was a good agreement between the two methods in all cases where the HPLC quantifications were reliable.
Figure 8.
Comparison of the fluorogenic in vitro transcription rNTP assay against luciferin-luciferase ATP assay and HPLC. (A) Correlation between mouse liver ATP levels as determined by the novel RNA polymerase-based method and a luciferin-luciferase ATP assay. The samples derive from the mouse liver ischemia experiment presented in Supplementary Figure S4. (B–E) Correlation plots of mouse liver nucleotide levels determined by the novel enzymatic method and HPLC. In these comparisons, the samples derive from the liver ischemia experiment shown in Figure 6. The same rNTP standards were used in both methods. In the enzymatic method, rNMP and rNDP levels were quantified indirectly (Materials and Methods) using rNTP standards. In HPLC, separate rNMP and rNDP standards were used. (F–R) Relative changes in hepatic nucleotide levels and adenylate energy charge during ischemia as determined by the enzymatic assay (red symbols) and HPLC (blue symbols). The error bars represent SD from four non-fasted male mice of 2 month of age.
Precision of the assay
Tables 2 and 3 show typical CVs for rNTP standards and liver extracts, respectively. Generally, the CV values were well <2.5%.
Table 2.
Intra-assay coefficient of variation (CV) of rNTP standard samples
| rNTP | Input (pmol) | Mean (%) | SD (%) |
|---|---|---|---|
| ATP | 125 | 0.9 | 0.2 |
| ATP | 62.5 | 1.4 | 1.0 |
| ATP | 31.3 | 1.5 | 0.4 |
| ATP | 15.6 | 2.8 | 1.5 |
| ATP | 7.81 | 4.5 | 0.6 |
| GTP | 125 | 1.5 | 0.6 |
| GTP | 62.5 | 2.3 | 1.2 |
| GTP | 31.3 | 1.8 | 1.6 |
| GTP | 15.6 | 1.6 | 0.5 |
| GTP | 7.81 | 5.6 | 2.2 |
| UTP | 125 | 1.4 | 1.1 |
| UTP | 62.5 | 1.8 | 0.6 |
| UTP | 31.3 | 3.5 | 4.0 |
| UTP | 15.6 | 2.4 | 1.5 |
| UTP | 7.81 | 2.4 | 2.5 |
| CTP | 125 | 2.5 | 1.7 |
| CTP | 62.5 | 1.9 | 1.6 |
| CTP | 31.3 | 1.2 | 0.5 |
| CTP | 15.6 | 1.4 | 1.1 |
| CTP | 7.81 | 1.5 | 0.5 |
| CTP | 3.91 | 3.6 | 2.4 |
The mean and SD represent CVs from three independent triplicate measurements. The reaction volumes were 10 μl (5 μl sample and 5 μl assay reagent).
Table 3.
Intra-assay coefficient of variation (CV) of liver extracts
| Nucleotides | Mean (%) | SD (%) |
|---|---|---|
| ATP | 1.7 | 0.9 |
| ATP + ADP | 1.3 | 0.7 |
| ATP + ADP + AMP | 1.3 | 0.5 |
| GTP | 1.9 | 0.7 |
| GTP + GDP | 1.7 | 1.1 |
| GTP + GDP + GMP | 1.2 | 0.8 |
| UTP | 1.5 | 0.9 |
| UTP + UDP | 1.2 | 0.6 |
| UTP + UDP + UMP | 1.4 | 0.8 |
| CTP | 1.7 | 0.9 |
| CTP + CDP | 1.5 | 0.9 |
| CTP + CDP + CMP | 1.5 | 1.0 |
The mean and SD represent CVs from 20 independent triplicate measurements from the liver ischemia time series (Figure 6). The reaction volumes were 10 μl (5 μl sample and 5 μl assay reagent).
Discussion
The bacteriophage T7 and SP6 RNA polymerases are extensively characterized enzymes and vital molecular biology tools. A few studies have also investigated the enzymological properties of these two RNA polymerases under limiting nucleotide concentrations (12,33,34), but the current study demonstrates for the first time their successful employment for the quantification of rNTPs. The assay we describe is simple and straightforward requiring only mixing a detection reagent and a sample in a microplate well. A typical qPCR instrument or plate reader can be used to read the plate and all the reagents are commercially available. The assay shows very low intrinsic variation, meaning that technical replicates are not an absolute requirement for precise rNTP quantification with a good pipetting technique. The modifications enabling the determination of rNMPs and rNDPs, make the assay a highly versatile tool to essentially any laboratory in needs of a convenient method to quantify ribonucleotides.
While methods involving HPLC or capillary electrophoresis exist for quantification of nucleotides (7), many research groups do not have access to the expensive specialized equipment required for these techniques. Moreover, many established chromatography procedures rely on UV absorbance for detection, which works well for abundant late-eluting rNTPs, but which may show interference from unknown overlapping peaks when measuring many of the rNMPs and rNDPs. Mass spectrometry-based detection is less dependent on perfect chromatographic separation of the metabolites as long as there are not too large differences in concentration between the non-resolved components and no overlapping mass fragments, but it is generally not compatible with the mobile phases used in most of the traditional HPLC protocols for nucleotides. Several relatively recent liquid chromatography-mass spectrometry protocols address these challenges but also show that the best practices for nucleotide quantification are still developing (3–5).
We optimized several parameters relevant to the novel assay but found the optimization of the in vitro transcription conditions per se unnecessary because of the extensive literature on the topic (13,35). We, however, do acknowledge the following points as potentially worthy of further assay development. We chose the Broccoli aptamer as the fluorescent reporter. The main rationale for this decision was its short length (49 nt) and it being well characterized (15,19), including for real-time monitoring of in vitro transcription (13). Moreover, the recently identified Broccoli ligand BI makes this aptamer exceptionally stable at optimal in vitro transcription temperatures (17). In comparison to the original and more widely used ligand the DFHBI-1T, BI also induces brighter fluorescence. Numerous other fluorescent aptamer-ligand pairs do, however, exist and may possess potentially beneficial features for the rNTP quantification (recently reviewed in (14)). Because abortive initiation is the main cause of truncated unproductive RNA, it may be possible to increase the sensitivity of the assay by designing templates that encode several concatenated aptamers. A similar approach has facilitated imaging of single transcripts in live cells (17). Our experiments with dimeric Broccoli, nevertheless, suggest that further aptamer concatamerization likely provides only incremental benefits. We settled for two templates that allow a robust quantification of the four canonical rNTPs. Ultimately, the most optimal template likely depends on the rNTP species to be quantified. There is not much room for the manipulation of the promoter or the aptamer sequences, but the pre-aptamer leader sequences are adjustable for minimization of abortive initiation and enhanced promoter clearance to increase the sensitivity of the assay. By employing a suitable set of nucleoside mono- and diphosphate kinases, we extended the assay to cover all canonical ribonucleotide species. In theory, a similar set of kinases targeting the unphosphorylated ribonucleosides (36–38) would enable determination of adenosine, uridine, cytidine, and thymidine as well. One should, however, note that the concentrations of rNMPs and unphosphorylated nucleosides can be very low (even <1% of rNTP concentrations) in healthy tissues and cultured cells, and therefore fall under the limit of quantification. A parallel quantification of dNTPs is also possible with our previously published method that also has similarly convenient real-time fluorometric detection in qPCR plate format (8).
We demonstrated the utility of our method by measuring the ribonucleotides from mouse liver tissue. We excluded major sample-related interference with this tissue and the given extract dilutions. If interference is encountered and absolute quantification is required, our method is amenable to the classic Standard Addition Method as a correction. A more likely source of error is any delay in the quenching of enzymatic activity during sample collection as our ischemia time series and several other studies demonstrate (28,39,40). As the final readout in our method is the increase of fluorescence over the baseline, a modest amount of sample-derived intrinsic fluorescence is not generally a significant source of error. Given the nature of the assay, it can be considered inherently specific as unspecific RNA synthesis is expected to compromise the aptamer structure. Some unnatural RNA-incorporating nucleotide analogues can potentially, however, give signal with our method. More relevantly, however, it is important to follow stringent-enough extraction protocol to avoid sample-derived enzymatic activity and unintended nucleotide interconversion. The nucleotide concentrations in the liver as measured by our method were well within published high-quality reference values (2) and could be reproduced using an HPLC-based method. Moreover, we were able to almost exactly replicate the classic experiment of the effect of ischemia on nucleotide pools (28), validating further the applicability of the assay for determination of not only rNTPs but rNMPs and rNDPs as well.
In addition to being useful for rNTP analyses, our new method will also be useful for the evaluation of the quality of extracts used for the enzymatic determination of dNTPs. Despite the constant development to improve the methodology to quantify dNTPs from cell extracts, large variation between the values from different laboratories has remained. It is difficult to know if this is due to biological differences between cell lines or technical issues such as extraction efficiencies or dephosphorylation of the dNTPs during the process. In this respect, HPLC methods have always had the advantage that they can also measure rNTPs that show much less differences between cell lines and cell cycle stages. The rNTPs can therefore be used as an internal control of the quality of the extract. If the rNTPs are extracted efficiently and not dephosphorylated into rNDPs and rNMPs, it is likely that the dNTPs are not affected either. Our RNA polymerase-dependent method presented here gives the possibility to determine the quality of nucleotide extracts also in laboratories that do not have access to HPLC.
Targeting nucleotide metabolism is increasingly studied as a therapy for cancer, viral infections, and inflammatory diseases (41). Moreover, compromised biosynthesis and maintenance of phosphorylation statuses of nucleotides are proposed primary disease mechanisms in many metabolic diseases such as mitochondrial disorders, yet currently reports of alterations in ribonucleotide levels in these diseases and their model organisms are surprisingly scarce. Given the ubiquitous role of nucleotides in cellular processes, the assay reported here is expected to facilitate research in the various fields of life sciences.
Supplementary Material
Acknowledgements
We thank Vilma Wanne and Diviya Upadhyay for technical assistance, and Nina Sipari and the Viikki metabolomics unit (Helsinki Institute of Life Science) for the efforts to quantify uridine nucleotides by UPLC-MS, and Prof. Vineta Fellman for critically reading the manuscript. The graphical abstract was created with BioRender.com.
Contributor Information
Janne Purhonen, Folkhälsan Research Center, Helsinki 00290, Finland; Stem Cells and Metabolism Research Program, Faculty of Medicine, University of Helsinki, Helsinki 00014, Finland.
Anders Hofer, Department of Medical Biochemistry and Biophysics, Umeå University, Umeå 90187, Sweden.
Jukka Kallijärvi, Folkhälsan Research Center, Helsinki 00290, Finland; Stem Cells and Metabolism Research Program, Faculty of Medicine, University of Helsinki, Helsinki 00014, Finland.
Data availability
The data underlying this article are available in the article and in its online supplementary material. Further data underlying this article will be shared on reasonable request to the corresponding author.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
Samfundet Folkhälsan; Jane and Aatos Erkko Foundation; Medicinska Understödsföreningen Liv och Hälsa and the Swedish Research Council; HiLIFE Laboratory Animal Center Core Facility, University of Helsinki, Finland. Funding for open access charge: Jane and Aatos Erkko foundation, Finland.
Conflict of interest statement. None declared.
References
- 1. Chandel N.S. Nucleotide metabolism. Cold Spring Harb. Perspect. Biol. 2021; 13:a040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Traut T.W. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994; 140:1–22. [DOI] [PubMed] [Google Scholar]
- 3. Zhu B., Wei H., Wang Q., Li F., Dai J., Yan C., Cheng Y.. A simultaneously quantitative method to profiling twenty endogenous nucleosides and nucleotides in cancer cells using UHPLC-MS/MS. Talanta. 2018; 179:615–623. [DOI] [PubMed] [Google Scholar]
- 4. Zborníková E., Knejzlík Z., Hauryliuk V., Krásný L., Rejman D. Analysis of nucleotide pools in bacteria using HPLC-MS in HILIC mode. Talanta. 2019; 205:120161. [DOI] [PubMed] [Google Scholar]
- 5. Kong Z., Jia S., Chabes A.L., Appelblad P., Lundmark R., Moritz T., Chabes A.. Simultaneous determination of ribonucleoside and deoxyribonucleoside triphosphates in biological samples by hydrophilic interaction liquid chromatography coupled with tandem mass spectrometry. Nucleic Acids Res. 2018; 46:e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Solter A.W., Handschumacher R.E.. A rapid quantitative determination of deoxyribonucleoside triphosphates based on the enzymatic synthesis of DNA. Biochim. Biophys. Acta Nucleic Acids Protein Synth. 1969; 174:585–590. [DOI] [PubMed] [Google Scholar]
- 7. Cohen S., Jordheim L.P., Megherbi M., Dumontet C., Guitton J.. Liquid chromatographic methods for the determination of endogenous nucleotides and nucleotide analogs used in cancer therapy: a review. J. Chromatogr. B. 2010; 878:1912–1928. [DOI] [PubMed] [Google Scholar]
- 8. Purhonen J., Banerjee R., McDonald A.E., Fellman V., Kallijärvi J.. A sensitive assay for dNTPs based on long synthetic oligonucleotides, EvaGreen dye and inhibitor-resistant high-fidelity DNA polymerase. Nucleic Acids Res. 2020; 48:e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pai C.-C., Kearsey S.E.. A critical balance: dNTPs and the maintenance of genome stability. Genes (Basel). 2017; 8:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lundin A. Thouand G., Marks R.. Optimization of the firefly luciferase reaction for analytical purposes. Bioluminescence: Fundamentals and Applications in Biotechnology - Vol. 2. 2014; Berlin, Heidelberg: Springer; 31–62.Advances in Biochemical Engineering/Biotechnology. [DOI] [PubMed] [Google Scholar]
- 11. Gong P., Martin C.T.. Mechanism of instability in abortive cycling by T7 RNA polymerase. J. Biol. Chem. 2006; 281:23533–23544. [DOI] [PubMed] [Google Scholar]
- 12. Ling M.-L., Risman S.S., Klement J.F., McGraw N., McAllister W.T.. Abortive initiation by bacteriophage T3 and T7 RNA polymerases under conditions of limiting substrate. Nucleic Acids Res. 1989; 17:1605–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kartje Z.J., Janis H.I., Mukhopadhyay S., Gagnon K.T.. Revisiting T7 RNA polymerase transcription in vitro with the Broccoli RNA aptamer as a simplified real-time fluorescent reporter. J. Biol. Chem. 2021; 296:100175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou H., Zhang S.. Recent development of fluorescent light-up RNA aptamers. Crit. Rev. Anal. Chem. 2022; 52:1644–1661. [DOI] [PubMed] [Google Scholar]
- 15. Filonov G.S., Moon J.D., Svensen N., Jaffrey S.R.. Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J. Am. Chem. Soc. 2014; 136:16299–16308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rabilloud T. Optimization of the cydex blue assay: a one-step colorimetric protein assay using cyclodextrins and compatible with detergents and reducers. PLoS One. 2018; 13:e0195755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li X., Kim H., Litke J.L., Wu J., Jaffrey S.R.. Fluorophore-promoted RNA folding and photostability enables imaging of single broccoli-tagged mRNAs in live mammalian cells. Angew. Chem. Int. Ed. 2020; 59:4511–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ranjbarian F., Sharma S., Falappa G., Taruschio W., Chabes A., Hofer A.. Isocratic HPLC analysis for the simultaneous determination of dNTPs, rNTPs and ADP in biological samples. Nucleic Acids Res. 2022; 50:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ageely E.A., Kartje Z.J., Rohilla K.J., Barkau C.L., Gagnon K.T.. Quadruplex-flanking stem structures modulate the stability and metal ion preferences of RNA mimics of GFP. ACS Chem. Biol. 2016; 11:2398–2406. [DOI] [PubMed] [Google Scholar]
- 20. Milligan J.F., Groebe D.R., Witherell G.W., Uhlenbeck O.C.. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987; 15:8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Coleman T.M., Wang G., Huang F.. Superior 5′ homogeneity of RNA from ATP-initiated transcription under the T7 ϕ2.5 promoter. Nucleic Acids Res. 2004; 32:e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shin I., Kim J., Cantor C.R., Kang C.. Effects of saturation mutagenesis of the phage SP6 promoter on transcription activity, presented by activity logos. Proc. Natl. Acad. Sci. USA. 2000; 97:3890–3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stump W.T., Hall K.B.. SP6 RNA polymerase efficiently synthesizes RNA from short double-stranded DNA templates. Nucleic Acids Res. 1993; 21:5480–5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guillerez J., Lopez P.J., Proux F., Launay H., Dreyfus M.. A mutation in T7 RNA polymerase that facilitates promoter clearance. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:5958–5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alam K.K., Tawiah K.D., Lichte M.F., Porciani D., Burke D.H.. A fluorescent split aptamer for visualizing RNA–RNA assembly in vivo. ACS Synth. Biol. 2017; 6:1710–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sastry S.S., Ross B.M.. Nuclease activity of T7 RNA polymerase and the heterogeneity of transcription elongation complexes. J. Biol. Chem.y. 1997; 272:8644–8652. [DOI] [PubMed] [Google Scholar]
- 27. Wilson B.D., Eisenstein M., Soh H.T.. Comparing assays via the resolution of molecular concentration. Nat. Biomed. Eng. 2022; 6:227–231. [DOI] [PubMed] [Google Scholar]
- 28. Jackson R.C., Boritzki T.J., Morris H.P., Weber G.. Purine and pyrimidine ribonucleotide contents of rat liver and hepatoma 3924A and the effect of ischemia. Life Sci. 1976; 19:1531–1536. [DOI] [PubMed] [Google Scholar]
- 29. Wu J.C., Merlino G., Fausto N.. Establishment and characterization of differentiated, nontransformed hepatocyte cell lines derived from mice transgenic for transforming growth factor alpha. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:674–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matsui Y., Kitade H., Kamiya T., Kanemaki T., Hiramatsu Y., Okumura T., Kamiyama Y.. Adenylate energy charge of rat and human cultured hepatocytes. In Vitro Cell Dev. Biol. Anim. 1994; 30:609–614. [DOI] [PubMed] [Google Scholar]
- 31. Löffler M., Jöckel J., Schuster G.. Dihydroorotat-ubiquinone oxidoreductase links mitochondria in the biosynthesis of pyrimidine nucleotides. Mol. Cell. Biochem. 1997; 174:125–129. [PubMed] [Google Scholar]
- 32. Bajzikova M., Kovarova J., Coelho A.R., Boukalova S., Oh S., Rohlenova K., Svec D., Hubackova S., Endaya B., Judasova K.et al.. Reactivation of dihydroorotate dehydrogenase-driven pyrimidine biosynthesis restores tumor growth of respiration-deficient cancer cells. Cell Metab. 2019; 29:399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Taylor D.R., Mathews M.B.. Transcription by SP6 RNA polymerase exhibits an ATP dependence that is influenced by promoter topology. Nucleic Acids Res. 1993; 21:1927–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Arnold S., Siemann M., Scharnweber K., Werner M., Baumann S., Reuss M.. Kinetic modeling and simulation of in vitro transcription by phage T7 RNA polymerase. Biotechnol. Bioeng. 2001; 72:548–561. [PubMed] [Google Scholar]
- 35. Pokrovskaya I.D., Gurevich V.V.. In vitro transcription: preparative RNA yields in analytical scale reactions. Anal. Biochem. 1994; 220:420–423. [DOI] [PubMed] [Google Scholar]
- 36. Yamada Y., Goto H., Ogasawara N.. Adenosine kinase from human liver. Biochim. Biophys. Acta Enzymol. 1981; 660:36–43. [DOI] [PubMed] [Google Scholar]
- 37. Kawasaki H., Shimaoka M., Usuda Y., Utagawa T.. End-product regulation and kinetic mechanism of guanosine-inosine kinase from Escherichia coli. Biosci. Biotechnol. Biochem. 2000; 64:972–979. [DOI] [PubMed] [Google Scholar]
- 38. Rompay A.R.V., Norda A., Lindén K., Johansson M., Karlsson A.. Phosphorylation of uridine and cytidine nucleoside analogs by two human uridine-cytidine kinases. Mol. Pharmacol. 2001; 59:1181–1186. [DOI] [PubMed] [Google Scholar]
- 39. Overmyer K.A., Thonusin C., Qi N.R., Burant C.F., Evans C.R.. Impact of anesthesia and euthanasia on metabolomics of mammalian tissues: studies in a C57BL/6J mouse model. PLoS One. 2015; 10:e0117232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ryll T., Wagner R.. Improved ion-pair high-performance liquid chromatographic method for the quantification of a wide variety of nucleotides and sugar—Nucleotides in animal cells. J.Chromatogr. B: Biomed. Sci. Appl. 1991; 570:77–88. [DOI] [PubMed] [Google Scholar]
- 41. Ariav Y., Ch’ng J.H., Christofk H.R., Ron-Harel N., Erez A.. Targeting nucleotide metabolism as the nexus of viral infections, cancer, and the immune response. Sci. Adv. 2021; 7:eabg6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material. Further data underlying this article will be shared on reasonable request to the corresponding author.