Summary
The evolution and development of the head have long captivated researchers due to the crucial role of the head as the gateway for sensory stimuli and the intricate structural complexity of the head. Although significant progress has been made in understanding head development in various vertebrate species, our knowledge of early human head ontogeny remains limited. Here, we used advanced whole-mount immunostaining and 3D imaging techniques to generate a comprehensive 3D cellular atlas of human head embryogenesis. We present detailed developmental series of diverse head tissues and cell types, including muscles, vasculature, cartilage, peripheral nerves, and exocrine glands. These datasets, accessible through a dedicated web interface, provide insights into human embryogenesis. We offer perspectives on the branching morphogenesis of human exocrine glands and unknown features of the development of neurovascular and skeletomuscular structures. These insights into human embryology have important implications for understanding craniofacial defects and neurological disorders and advancing diagnostic and therapeutic strategies.
Keywords: Tissue clearing, iDISCO, human embryo, virtual reality, skull, oculomotor system, light-sheet microscopy, vascular
Graphical abstract
Highlights
-
•
3D imaging reveals the developmental sequence of human head organs during gestation
-
•
Salivary gland morphogenesis is asymmetric and shows interindividual variability
-
•
Virtual reality and interactive 3D tools improve our understanding of human development
-
•
Hudeca.com, a web interface and image database to explore and learn human embryology
Using advanced whole-mount immunostaining and 3D imaging, a comprehensive 3D cellular atlas of human head embryogenesis encompassing muscles, vasculature, cartilage, peripheral nerves, and exocrine glands is presented. The datasets are used to provide insights into the branching morphogenesis of exocrine glands and features of neurovascular and skeletomuscular structure development.
Introduction
The appearance of the head has long been recognized as one of the most important and remarkable events in vertebrate evolution.1,2 It is the most intricate structure of the body and acts as the primary gateway for sensory stimuli, including sounds, light, tastes, and odors. The head is protected by muscles and skin and houses the brain within the neurocranium. Additionally, it consists of a diverse range of tissues derived from the three germ layers, precisely organized into endocrine and exocrine glands, sensory organs (such as the eyes/retina, inner ear, taste buds, and olfactory epithelium), and muscles, among others. The prevalence of head malformations in approximately one third of newborns with congenital defects (about 1/700 live births) exemplifies the intricate complexity and precision of cell interactions involved in the development of the human head.3
Neural crest cells (NCCs), in particular, played a crucial role in the emergence of the head2,4 as they contribute significantly to the development of the skull/craniofacial skeleton, cartilage, connective tissue, smooth muscles, and cranial ganglia. NCCs together with cranial neurogenic placodes also influence the differentiation and migration of other head cell types.
The cellular and morphogenetic events accompanying head development have been meticulously described in most vertebrate lineages.5,6,7 These comparative evo-devo studies extend beyond extant vertebrates and encompass fossils, particularly through the analysis of cranial endocasts.8 However, our understanding of the early stages of head development in human embryos remains rudimentary.
Our current knowledge about human head development relies heavily on anatomical data gathered in the first half of the 20th century, using traditional histological methods.9,10 However, only a few studies have utilized cell-specific antibodies to investigate the development of human head organs, resulting in a limited and incomplete understanding of the underlying cellular processes.
Cephalic ontogeny, the development of the head, is a crucial aspect of hominin evolution, reflecting the morphogenic impact of bipedality and encephalization.11 While there is extensive research on brain formation and evolution,12 our understanding of cephalic skeletal development in humans lags behind that of other vertebrates. Current studies on human cephalic skeletal development primarily rely on either histological13 or radiological14,15 techniques, which lack resolution and cell specificity. As a result, we lack a comprehensive and dynamic cellular atlas of human head ontogeny.
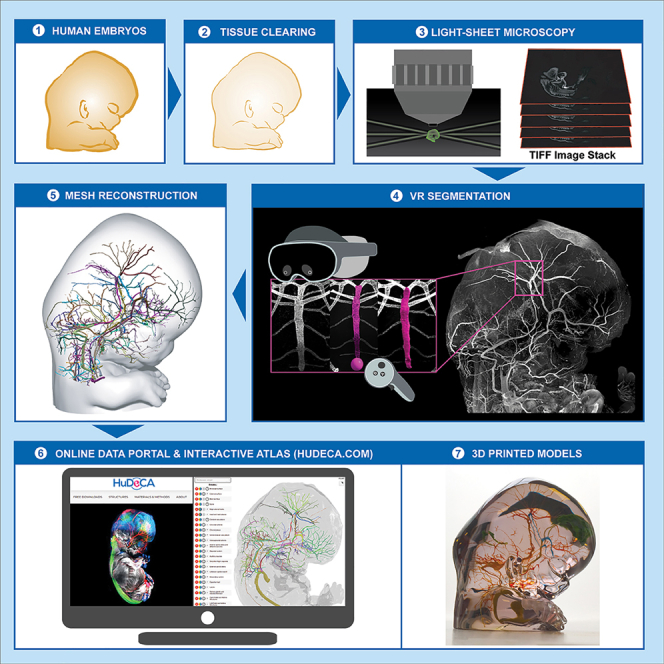
In the past decade, tissue clearing methods have been developed to examine the cellular organization of intact human organs.16 These techniques enable visualization of specific cells through immunohistochemistry and their mapping in 3D using light-sheet fluorescent microscopy (LSFM).17,18 We have applied this combination of whole-mount immunolabeling and 3D imaging to analyze human embryology.19,20 Here, we used this strategy to describe the development of the human head between the fifth and thirteenth post-conceptional weeks (PCW5–PCW13).
Results
To study the development of the human head, we used embryos and fetuses (Table S1A and STAR Methods) from the French Hudeca biobank https://hudeca.genouest.org.
A total 76 specimens (both whole and dissected parts) were used, including 27 embryos (PCW5.5–PCW7.9, of which: 18 males, 4 females, 5 unknown) and 49 fetuses (PCW8.0–PCW13, of which: 20 males, 22 females, 7 unknown). All images are from single samples and are not average reconstructions of multiple samples. The correspondence between samples and figure panels or movies is provided on Table S1D.
All antibodies underwent initial optimization of their dilution using cryostat sections of tissue fragments from human embryos. A total of 70 antibodies were evaluated for this study among which 36 exhibited positive and reproducible staining in 3D (Table S1B). To ensure antibody specificity, we relied on previous studies conducted on mouse embryos and considered the known subcellular locations of recognized epitopes. The brain was not included in our work as it has been extensively studied and building a cell atlas of the developing human brain is beyond the scope of this analysis.
Prior studies combining immunostaining and tissue clearing have relied on a classic two-step method, with a first incubation in the primary antibodies followed by a second step with secondary antibodies conjugated to fluorophores resulting in signal amplification. However, this approach limits the number of antibodies that can be simultaneously employed, as they must be derived from different species to avoid cross-reactivity. This limitation can be overcome by multiplexing approaches using conjugated antibodies, and cyclic-immunostaining procedures have been developed for tissue sections.21
In an effort to expand the range of antibody combinations, we first assessed the efficiency of directly conjugated antibodies for labeling large human embryo and fetal samples and iDISCO/iDISCO+ clearing. Through this approach, we achieved robust passive staining using multiple conjugated antibodies (Table S1B). In addition, we performed on some samples a second round of staining. After the first round of immunostaining, samples were treated with hydrogen peroxide (H2O2), which was previously shown to bleach fluorophores such as Alexa dyes.21 Samples were rehydrated and underwent a second round of immunostaining, with conjugated antibodies, followed by a second round of clearing and imaging.
A total of 76 specimens (whole and dissected parts) were processed and resulted into 400 LSFM scans at multiple magnifications. Up to 4 individual channels were imaged per specimen. This represents more than half a billion individual digital slices (OME.TIFF files) of less than 4 μm, and close to 70 TB of image data.
Development of the head skeleton
Recent studies22 have sparked renewed interest in human cephalic embryology, particularly in understanding the patterning and growth of the basal chondrocranium.23,24,25,26 Although the anatomical details of the human skull base have been documented since the works of His27 and Sutton,28 the origin and assembly of its individual components have remained elusive.29 Histological analysis30 and radio-imaging techniques13 have revealed the sequential formation of the sphenoid bone from the fusion of seven distinct elements. However, the lack of specific markers for mesenchymal, chondrogenic, and osteogenic tissues has hindered the precise mapping of its topography and understanding of its connections with adjacent structures.
Here, we examined chondrogenesis in PCW5.6–PCW11 specimens. To visualize the developing cranium, we used antibodies against Sex-determining region of chromosome Y (SRY)-box-containing gene 9 (Sox9), a transcription factor expressed in cartilage progenitors, and against its downstream target and chondrogenic marker Collagen 2 (Col2), a main component of the cartilage matrix.
First, we established a 3D rendering pipeline using Col2 immunolabeling on a complete PCW7 embryo (Figure 1). The raw signal (Figure 1A) was processed by combining a virtual reality (VR) interface (syGlass) and Imaris software to segment and create mesh structures. Within the head region, we successfully identified and visualized all recognized chondrogenic templates in their respective locations (Figures 1D–1F and S1 and Videos S1A and S1B).
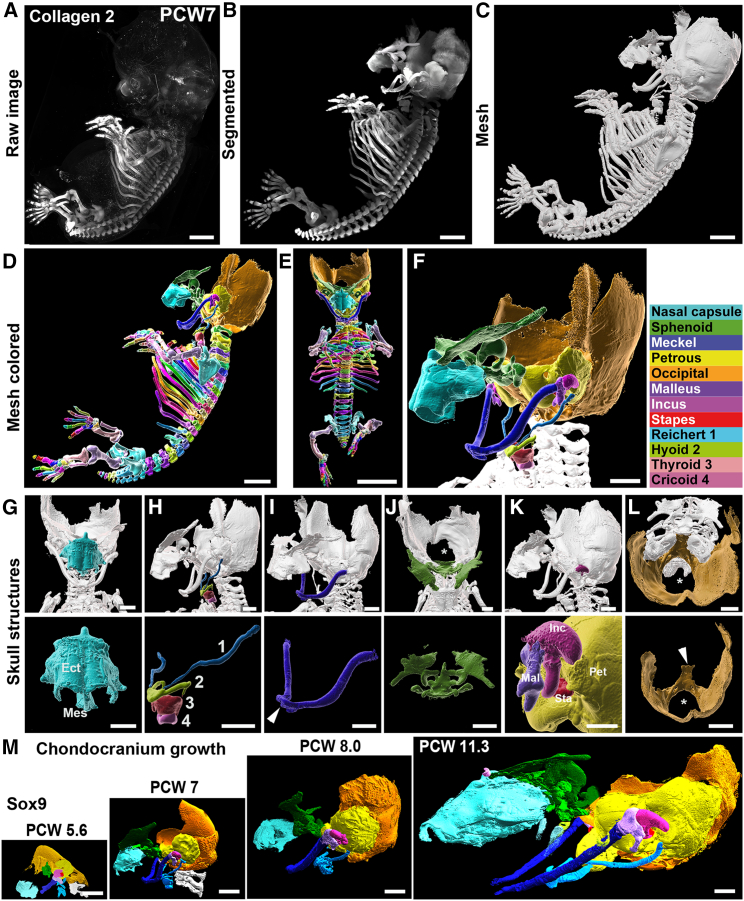
Figure 1.
3D Analysis of the human chondrocranium development
All panels are LSFM images of solvent-cleared embryos and fetus, immunostained with anti-Collagen 2 (A–L) or anti-Sox9 (M) antibodies.
(A–C) Lateral 3D views of a PCW7 embryo illustrating the image processing pipeline. Raw image data (A) are segmented using syGlass (B) to isolate all cartilage templates and a mesh image is built (C) for 3D rendering.
(D and E) (D) All individual skeletal elements have been colored (lateral view). (E) is a frontal view of the same embryo.
(F–M) (F) High-magnification 3D rendering of the cranium with all developing cartilage elements pseudocolored. Names appear on the chart on the right of the image. (G)–(L) are views of each element taken in situ (top panels) or isolated (bottom panels). The nasal capsule (G) assembles the mesethmoid (Mes) and ectethmoid (Ect). The hyoid and the larynx (elements numbered 1–4) are seen in (H). In (I), The Meckel’s cartilages join at the symphysis (arrowhead). (J) Frontal (upper panel) and superior (lower panel) views of the sphenoid. (K) shows the inner ear bones (dark purple, malleus; pink, incus; red, stapes) and their insertion in the petrous (yellow). In (L), the basilar process (arrowhead) and foramen magnum (asterisk) of the chondrogenic ventro-posterior component of the presumptive occipital bone complex are seen. (M) displays the developmental time course of the cranium in human from 5.6 to 11 PCW. All panels are 3D rendering images generated with syGlass from LSFM images of embryos (PCW5.6, PCW7, and PCW8) and fetus (PCW11.3) immunostained with anti-Sox9. Images are presented at the same scale to illustrate the growth of the cranium.
See also Figure S1.
Scale bars: 2 mm in (A)–(D) and (L, top panel), 4 mm in (E), 1 mm in (F)–(K), 3 mm in (L, bottom panel), and 1.5 mm in (M).
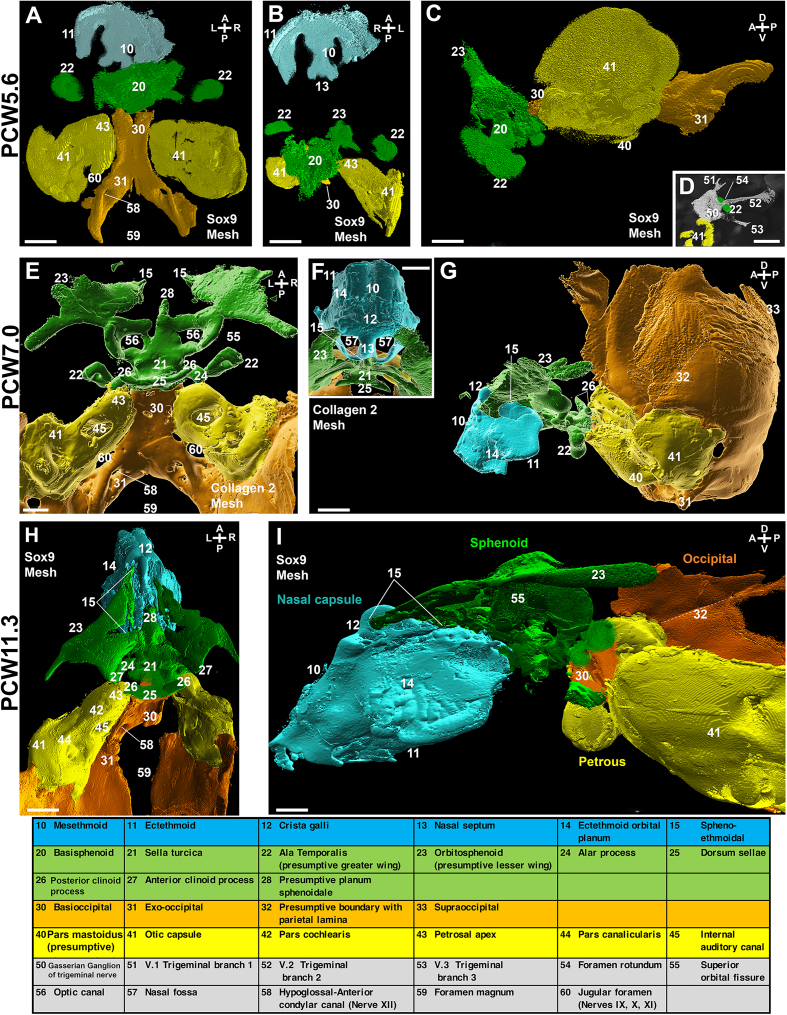
Figure S1.
Sphenoid development in human embryos and fetus, related to Figure 1
(A–I) 3D reconstructions of chondrogenic markers immunolabeling at PCW5.6 (Sox9; A–D), PCW7.0 (Collagen 2; E–G) and PCW11.3 (Sox9; H and I), showing progressive assembly of nasal capsule (blue, 10–15), sphenoidal (green, 20–28), occipital (orange, 30–33), and petrosal (yellow, 40–45) chondrogenic templates of the skull base.
(A–C) The nasal capsule develops remotely, anteriorly from the other three structures, which are already in close proximity at PCW5.6, as seen from superior (A), frontal (B), and lateral (C) views. The isolated sphenoid is a single mass, the basisphenoid (20) flanked by two independent ventro-lateral masses, the ala temporalis (22).
(D) A weak Sox9 labeling delineates the trigeminal nerve, or fifth cranial nerve (CNV) , confirmed on section. This shows the passage of the maxillary (CNV2, 52) trigeminal branch through central holes of the ala temporalis (22), prefiguring the foramen rotundum (54). In contrast, other branches (CNV1, 51, and CNV3, 53, traced from the Gasserian ganglion, 50) run outside of this structure.
(E–G) At PCW7.0 (E, supero-dorsal view; G, lateral view; F, superior close up of the nasal capsule showing the superior surface of the initial basiphenoid (20 in A), which will develop as the main sphenoid body in the adult, splits posteriorly into the sella turcica (21) and anteriorly into the presumptive planum sphenoidale (28). Laterally, the orbitosphenoids (23) that have formed and expanded are connected to the basisphenoid (20 in A), prefiguring the sphenoid lesser wings (23). Likewise, connected to the basisphenoid (20), the alar processes (24) connect to the ala temporalis (22) as the presumptive greater wings. Frontal spheno-ethmoidal cartilage interfaces (15) prefigure the dorsal domain of the orbit. The dorsum sellae (25) rises dorsally, flanked laterally by the posterior clinoid processes (26). A depression of the cartilage plate can be observed as the future hypophyseal fossa or sella turcica (21). The parietal lamina (or plate, delimited by the dotted line) was identified (G and I).
(H and I) Interestingly, the pterygoid plates were not visible (while the pterygoid muscles were present). At PCW11.3 (H, superior view; I, lateral view), two large lateral holes prefigure the superior orbital fissures (55), along with other foramens (45, internal auditory canal; 58, hypoglossal canal; 59, foramen magnum).
The bottom table lists all individual anatomical structures annotated on the figure.
Scale bars: 500 μm in (A)–(D), (F), and (G); 1 mm in (E); and 1.5 mm in (H) and (I).
(A) 3D analysis of the embryonic skeleton of a PCW7 human embryo stained with Col2 antibodies and segmented with syGlass.
(B), 3D visualization of the elements of the cranium in a PCW7 human embryo.
Using Col2, the chondrocranium elements and their interconnected articulations could be visualized. At PCW7, the skull base demonstrated well-organized arrangements centered around the future sphenoid bone (Figure 1J). The sphenoid was observed to connect antero-dorsally with the nasal capsule (Figure 1G), antero-ventrally with the hyoid and Meckel’s cartilage (Figure 1H), laterally with the petrosal and internal ear bones (Figure 1K), and ventro-posteriorly with the chondrogenic components of the presumptive occipital bone (Figure 1L).
Additionally, we examined the growth of the chondrocranium by segmentation of Sox9 immunostaining from PCW5.6 to PCW11.3 (Figure 1M). Notably, the early stages demonstrated significant growth of the presumptive occipital bone compared with other chondrocranium structures. However, from PCW8 to PCW11.3, its growth remained moderate compared with the other structures. Particularly remarkable was the substantial expansion of the sphenoid elements (Figure 1M), encompassing all dimensions and notably affecting the anterior region involved in orbit formation. To understand how the sphenoid elements connect with neighboring structures during skull base assembly, we used both Sox9 and Col2 (Figure S1). At PCW5.6, the segmented Sox9+ staining revealed a large central mass flanked by two smaller pierced masses (Figure S1) corresponding to the basisphenoid (medial) and the ala temporalis (lateral). This finding contradicts a previous report,22 and aligns more closely with a radiologic-based description,13 indicating that the sphenoid initially forms as a composite entity. Interestingly, our reconstruction clearly demonstrates that the basisphenoid is already connected to both the petrosal and ventro-dorsal chondrogenic components of the presumptive occipital bone territory as early as PCW5.6, challenging the notion of a sequential formation. In contrast, the nasal capsule develops as one entity, independently and at a distance from the sphenoidal territory (Figure S1). The rapid differentiation and complexification of the sphenoid elements become evident by PCW7 (Figure 1). The frontal elements adjacent to the nasal capsule have undergone significant expansion (Figure S1), and the establishment of articulations with the future petrous bone (laterally) and basioccipital bone (posteriorly) contributes to the definitive shape of the skull base. This includes the formation of the foramens enabling the passages of neural elements (Figure S1). By PCW11.3, the lesser wings have reached their approximate adult anatomy, while the ala temporalis have yet to fully develop as the greater wings, further underlying the differential growth of chondrocranial structures. At PCW7, Sox9 and Col2 stainings exhibited substantial overlap in all chondrocranium structures. However, on a finer scale, Sox9-positive cells were fewer, less densely distributed, and located more centrally, whereas Col2-expressing cells extended to the periphery and reached more extensive territories, such as the dorsal posterior territory where the occipital, the orbitosphenoidal, and basioccipital bones will form and ossify. These findings suggest that the combined use of both markers captures all chondrogenic elements, including actively proliferating ones like the dorsum sellae, as well as more advanced ones located at the dorsal limits in contact with areas presumed to undergo intramembranous ossification.
Development of head muscles and their innervation
The human head and neck comprise over 75 muscles on each side, responsible for controlling various movements such as eye, lip, and tongue movements, facial expressions, head posture and rotation, speech, chewing, and breathing.31,32 Previous studies on the development of human head muscles have relied on hematoxylin and eosin staining,33 and an earlier investigation of fetal facial muscles suggested that their development is still incomplete at PCW10.34 More recently, a computer-assisted 3D reconstruction of head muscles in a single human embryo reported that at least 30 head and neck muscles were not yet formed by the end of embryogenesis at 8 weeks.35 Unlike trunk muscles, which originate from somites, head muscles have a mixed origin.36 While most of their progenitors arise from the unsegmented presomitic paraxial mesoderm, muscles of the tongue and certain muscles of the pharynx, larynx, and neck come from occipital somites.37,38 Furthermore, the genetic program governing the differentiation of head muscles from cranial mesoderm differs from that of trunk skeletal muscles, which are of somitic origin.39 In our study, we used antibodies against myosin heavy chain (MHC) to identify differentiated muscles (Figure 2A). The use of syGlass facilitated the segmentation of individual muscles throughout the body (Figures 2B–2D and Video S2). Head muscles receive innervation from motor neurons located in the midbrain and hindbrain, specifically in nine motor nuclei (III, IV, V, VI, VII, IX, X, XI, and XII). Visualization of developing motor nerves was achieved using anti-Choline acetyl transferase (ChAT) antibodies.19 Double immunostaining for MHC and ChAT allowed us to track the timing of head muscle innervation.
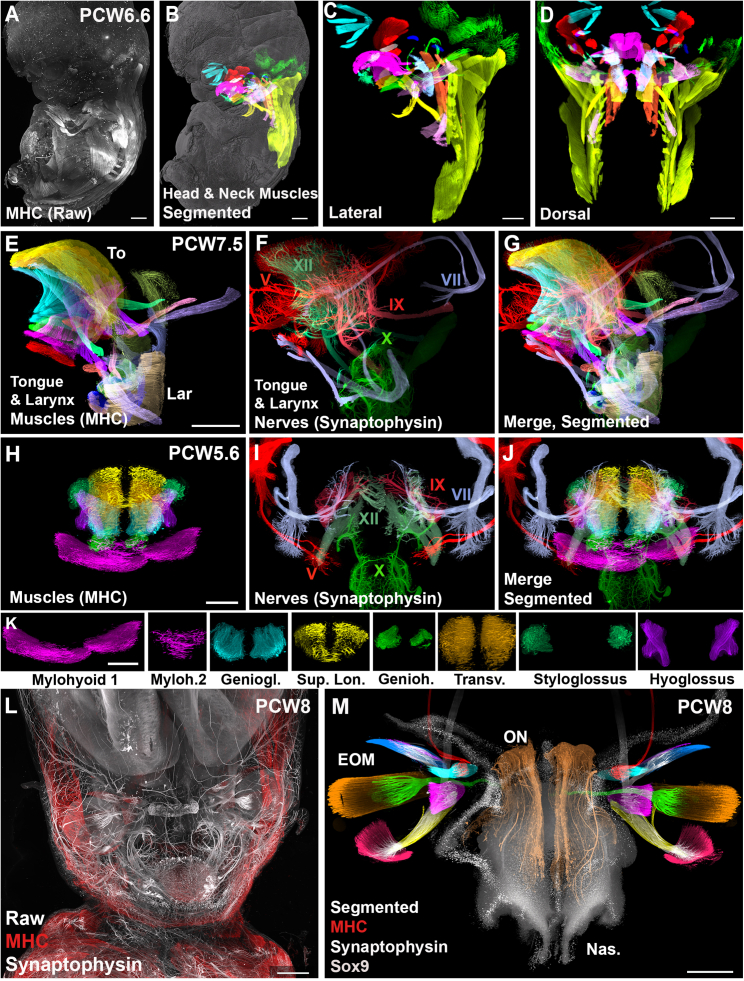
Figure 2.
3D analysis of the development of head and neck muscles in human embryos
All panels are LSFM images of a solvent-cleared embryos immunostained with anti-MHC (A–E, G, H, and J–M), anti-synaptophysin (F, G, I, J, L, and M), and anti-Sox9 (M).
(A) Raw image of a lateral view of the embryo.
(B–D) Lateral (B and C) and dorsal (D) views of the muscles of a PCW6.5 embryo segmented using syGlass. 14 muscle modules are differentially pseudocolored. (B) shows an overlay with the surface shading image (gray).
(E–G) Lateral view of the tongue (To) and larynx (Lar) of a PCW7.5 embryo with all muscles (E) and nerves (F) segmented. (G) Merged image of the muscles and nerves.
(H–K) Frontal view of the tongue of a PCW5.6 embryo with all muscles (H) and nerves (I) segmented. (J) Merged image of the muscles and nerves. (K) shows all individual tongue muscles.
(L and M) Frontal view of the face of a PCW8 embryo with a 3D overlay of muscles (red) and nerves (white) illustrating the complexity of the staining and raw image before segmentation. (M) is an image of the same embryo after segmentation of all extraoculomotor muscles (EOMs; see Figure S4 for the color code), oculomotor nerves, Sox9+ nasal capsule (Nas, gray) and olfactory nerves (ONs, brown).
Abbreviations are as follows: V, trigeminal nerve; VII, facial nerve; IX, glossopharyngeal nerve; X, vagus nerve; XII, hypoglossus nerve; Myloh.2, Mylohyoid 2; Geniogl., genioglossus; Sup. Lon, superior longitudinal; Genioh., geniohyoid; and Transv., transverse.
See also Figures S2 and S3.
Scale bars: 1 mm in (A), (B), (D), (E), and (L); 800 μm in (C) and (M); and 400 μm in (H) and (I); and 300 um in (K).
To facilitate the description of the results, head and neck muscles were split into 14 modules regrouping muscles based on their shared function and/or anatomical locations (Figures 2 and S2 and Table S1C). A total of 45 head tissues from embryos and fetuses were used for this study (Table S1A).
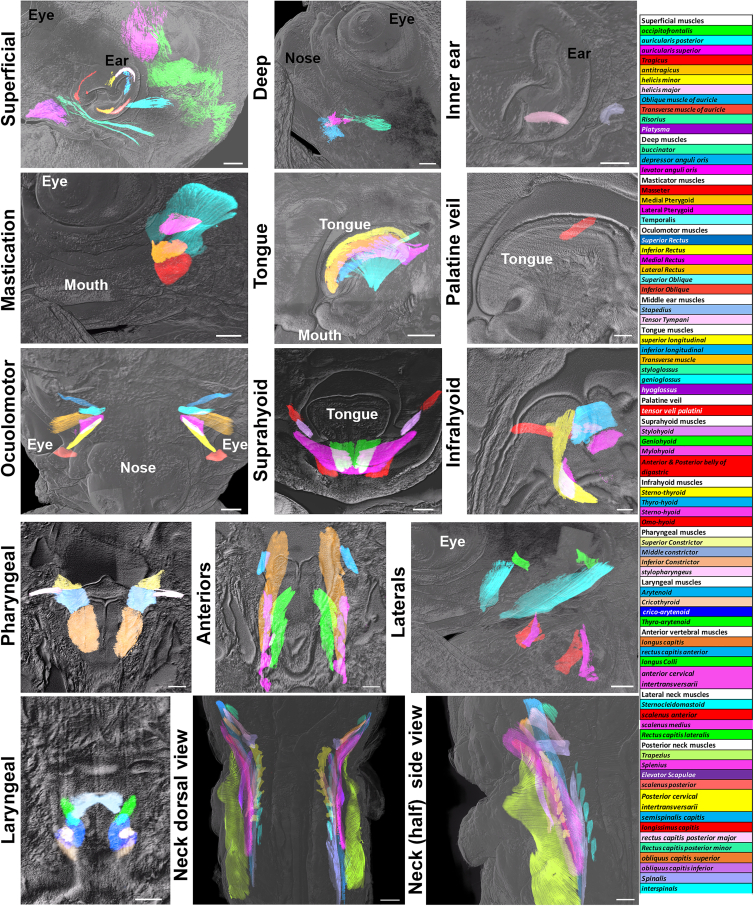
Figure S2.
Organization of head and neck muscles in a PCW6.6 embryo, related to Figure 2
All panels are LSFM images of a solvent-cleared PCW6.6 embryo immunostained with anti-MHC. All head and neck muscles, grouped in 14 anatomical and functional modules are individually segmented. The names of all the modules and muscles (with a color code similar to the corresponding images) are presented on the right side. Two views are shown (bottom panels for the neck muscles).
Scale bars: (panels: superficial, inner ear, mastication, tongue, and laterals) 500 μm, (deep, suprahyoid) 400 μm, (palatine veli) 200 μm, (infrahyoid, pharyngeal, anteriors, and laryngeal) 300 μm, (neck dorsal view) 800 μm, and (neck [half] side view) 700 μm.
We first focused on tongue muscles which derive from the occipital somites. Not much is known about their development. The muscles present in the adult human tongue have been described decades ago,40 but the 3D distribution of their innervation has only been recently studied.41,42,43 Our 3D images of the developing human heads allows very precise insights into the organization of tongue muscles and their relative position. All muscles and nerves could be extracted from samples (n = 19) stained with MHC and ChAT or synaptophysin or β3-tubulin (Figures 2 and S3). This showed that 7 muscles could be seen as early as PCW5.6 (Figures 2H–2K), but that the last two muscles (palatoglossus and pharyngoglossus), still absent at PCW7.5 (Figures 2E–2G) and PCW8 (Figure S3), could be found at PCW11.3 (Table S1C). The arborization of the 5 nerves innervating the tongue and larynx (V, VII, IX, X, and XII) could also be visualized (Figure 2 and Video S3A).
Figure S3.
Description of the muscles of the tongue, suprahyoid, larynx and pharynx in a PCW8 embryo, related to Figure 2 (but the embryo is different)
3D LSFM image of all tongue, suprahyoid, laryngeal, and pharyngeal muscles segmented and pseudocolored in a PCW8 embryo immunostained with anti-MHC, cleared with iDISCO and imaged using LSFM. All muscle names are indicated above each panel.
Scale bars, 500 μm.
(A) 3D visualization of the tongue and larynx muscles and their innervation in a PCW7.5 human embryo, related to Figures 2, S2, and S3.
(B) 3D visualization of extraocular muscles and oculomotor nerves in a PCW8.4 human fetus, related to Figures 3 and S4.
(C) 3D visualization of extraocular muscles, oculomotor nerves and lacrimal gland in a PCW11.3 human fetus, related to Figures 3 and S4.
(D) 3D visualization of the external, middle, and inner ear in a PCW7.5 human embryo, related to Figure S5.
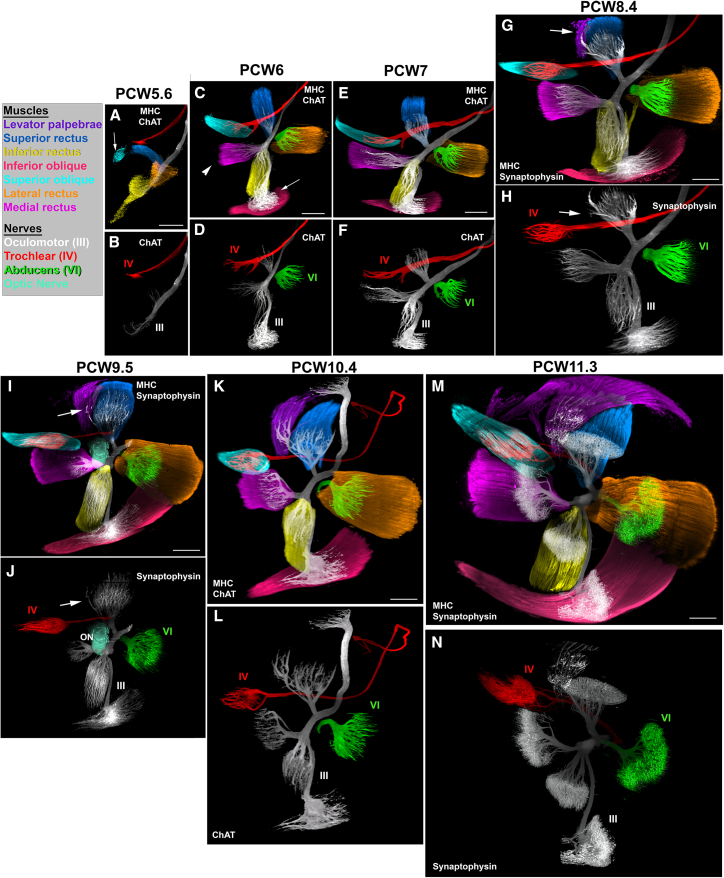
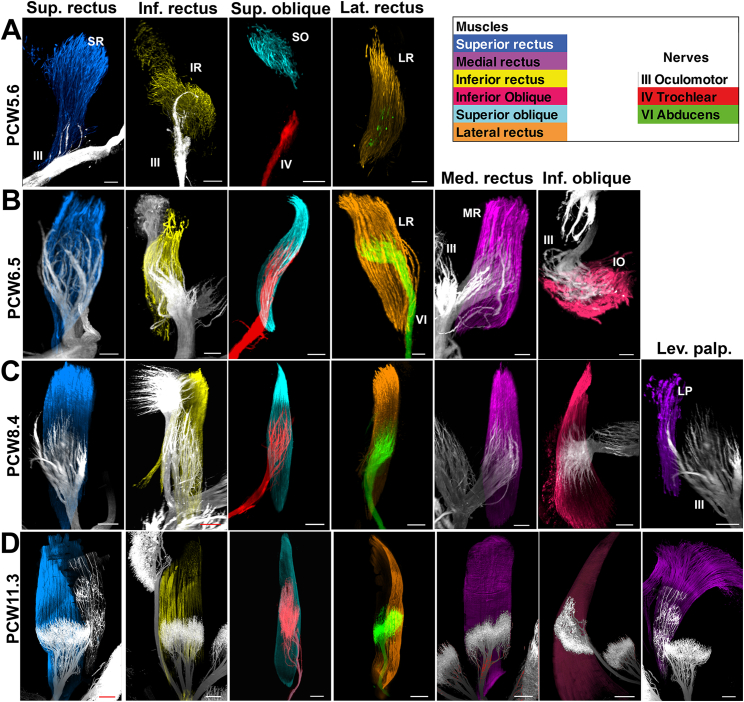
Next, we studied the 6 extraoculomotor muscles (EOMs) and the levator palpebrae, responsible for eye movements and upper eye lid respectively (Figures 2L, 2M, 3, and S4 and Videos S3B and S3C). Their development has been extensively studied in many species,33 but their developmental time course is not fully understood in humans.44,45 Individual muscles and the 3 oculomotor nerves could be easily segmented from raw image datasets (Figures 2M, 3, and S4). In a PCW5.6 embryo, the youngest available, the lateral, superior and inferior recti could be distinguished as well as the primordium of the superior oblique (Figures 3A, 3B, and S4). Double staining with ChAT showed that only the 3 recti were already innervated (although only the terminal part of the abducens nerve was labeled; Figure 3C). By contrast, the trochlear nerve has not yet reached the superior oblique. At PCW6, the muscles have enlarged, and they started to adopt their mature morphologies and positions. A well-defined medial rectus muscle could be seen as well as the inferior oblique (Figure 3C), earlier than previously reported.33 Between PCW6 and PCW7, the six muscles were all innervated including the superior oblique (Figures 3C–3F and S4B). At PCW8.4, the primordium of the levator palpebrae started dissociating from the superior rectus (Figures 3G, 3H, and S4C) and was fully separate from it at PCW10.4 (Figures 3K and 3L). Although ChAT is specific for motor axons, it does not stain motor endplates. We were unable to label endplates in whole mounts using bungarotoxin (data not shown) but found that anti-synaptophysin stains all motor nerves and endplates (Figures 3M, 3N, and S4D). All peripheral nerves, including sensory, could be distinguished from motor nerves. Both were immunoreactive for synaptophysin, but sensory nerves are ChAT- and express the cell-adhesion molecule TAG1/Contactin2 (data not shown). This showed that (as in mature muscles), endplates occupy the center part of each muscle and that nerve arborizations became very dense at PCW11.3 (Figures 3 and S4 and Video S3C).
Figure 3.
Development of the human oculomotor system
All panels are LSFM images of solvent-cleared embryos (A–H) and fetuses (I–N) immunostained with anti-MHC combined with ChAT (A–F, K, and L) or synaptophysin (G–J, M, and N). For each specimen, the upper panel corresponds to the merge image of the motor nerves and oculomotor muscle and the lower panel to the nerves. The inset on the left upper side of the figure provides the color code for muscles and nerves.
(A and B) At PCW5.6, only 4 muscles are visible. All muscles are innervated except the superior oblique (arrow).
(C and D) At PCW6, the 6 extraocular muscles, including the medial rectus (arrowhead) and inferior oblique (arrow) are now present and innervated.
(E and F) This is similar at PCW7.
(G and H) At PCW8.4, the levator palpebrae starts to split from the superior rectus (arrow in G) and receives a small branch coming from the oculomotor nerve (arrow in H). Note the expansion of the inferior oblique.
(I–K) Between PCW9.5 (I and J) and PCW10.4 (K and L), the size of the levator increases and it detaches completely from the superior oblique.
(M and N) At PCW11.3, the terminal branches of the nerves occupy the central part of all muscles and display a large bouquet of synaptophysin+ endplates. Abbreviations are as follows: III, oculomotor nerve; IV: trochlear nerve; and VI, abducens nerve.
See also Figure S4.
Scale bars: 500 μm in (A)–(M).
Figure S4.
Time course of oculomotor nerve development in human embryos, related to Figure 3
All panels are LSFM images of solvent-cleared embryos (A–C) and fetus (D) immunostained with anti-MHC combined with ChAT (A–C) or Synaptophysin (D). (A)–(D) are individual images illustrating the developmental time course of all extraocular muscles and their innervation between PCW5.6 and PCW11.3. The inset on the right gives the color code used for nerve and muscles. Scale bars: all panels are counted from left to right for each row; 100 μm in (A, first and third panels from the left) and (B, fourth and fifth panels); 300 μm in (A, second panel), (C, first, second, sixth, and seventh panels), and (D, first, second, and seventh panels); 200 μm in (A, fourth panel) (B, first and second panels), and (C, third, fourth, and fifth panels); 150 μm in (B, third and sixth panels); and 500 μm in (D, third, fourth, and sixth panels).
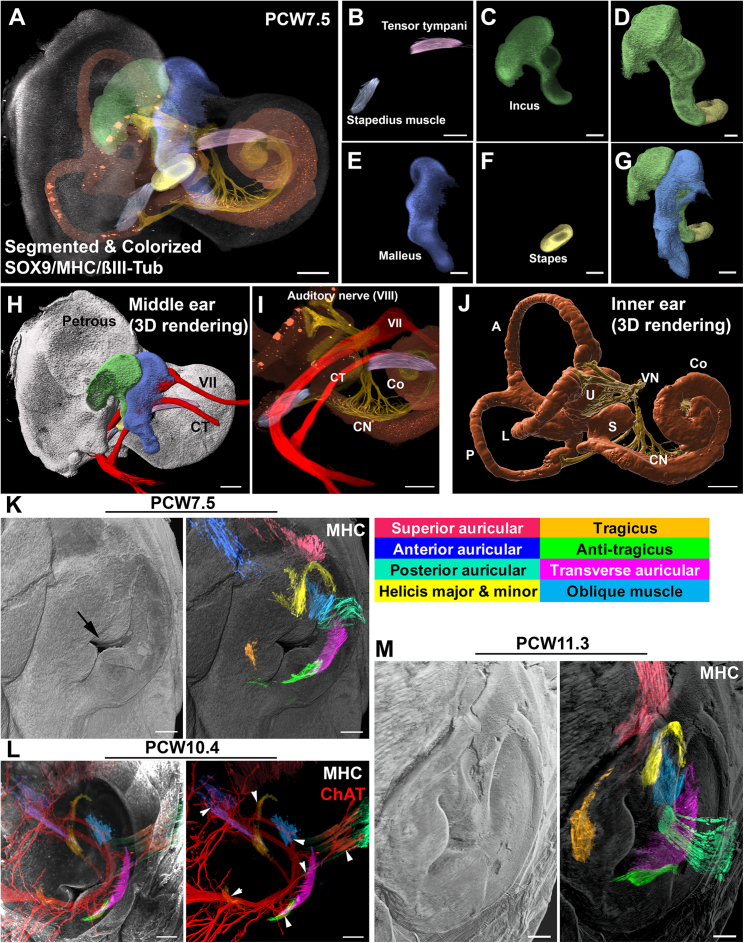
Our data provide a clear view of the organization of the middle and inner ear (Figure S5A and Video S3D), including the stapedius and tensor tympani (Figure S5B), the two smallest muscles in the body, together with the 3 middle ear bones, the incus (Figures S5C and S5D), malleus (Figure S5E), and stapes (Figures S5F and S5G). The cochlea and semicircular canals could be fully segmented just based on background staining, and their innervation observed using anti-βIII-tubulin antibodies (Figures S5H–S5J). Both muscles could be seen as early as PCW6.5 (Table S1C).
Figure S5.
3D organization and ontogenesis of the ear, related to Figure 2
(A–J) 3D LSFM image of the inner and middle ear in a PCW7.5 human embryo stained with MHC, Sox9 and Synaptophysin, cleared and segmented with syGlass. (A) is a merge of all channels, with segmented elements pseudocolored. (B)–(G) shows the two muscles and the three bones of the middle ear, after segmentation (B, C, E, and F) and 3D surface rendering (D and G). (H) is a 3D rendering view showing all elements together: the petrous bone template (gray), the stapedius (light blue) connected to the stapes, the tensor tympani connected to the malleus, the facial nerve (VII), and its chorda tympani (CT) branch. (I) LSFM image showing the relative positions of the two muscles, together with the cochlea (Co), facial nerve (VII), chorda tympani (CT), cochlear nerve (CN), and vestibulocochlear/auditory nerve (VIII). (J) 3D rendering of the cochlea (Co), saccule (S), utricule (U), anterior (A), posterior (P), and lateral (L) semicircular canals together with the vestibular (VN) and cochlear (CN) nerves. The cochlea and canals were segmented based on the background of the Sox9 staining.
(K–M) 3D ontogenesis of external ear muscles in human embryo (K) and fetuses (L and M), immunostained with anti-MHC antibodies (K–M) and ChAT (L). The overlays are surface shading images of the auricle/pinna (gray). Muscles have been segmented using VR and pseudocolored. The color code for muscle identification is on the upper right inset. (K) shows that all individual muscles have started to emerge at PCW7.5. The arrow indicates the developing external auditory meatus. (L) All muscles are innervated by ChAT+ motor nerves (arrowheads show innervating branches). (M) shows the muscles in a PCW11.9 fetus. The size of the muscles has increased, and they now form a ring around the ear.
Scale bars: 500 μm in (A), (B), and (H)–(J); 300 μm in (C) and (E)–(G); 200 μm in (D); and 500 μm in (K)–(M).
Superficial and deep muscles are situated beneath the skin, surrounding areas such as the eyes, ears, mouth, and lips, and play a vital role in facial expressions and chewing. Rather than moving bones, these muscles primarily manipulate the skin or cartilage. They originate from the branchial mesoderm.46 In humans, the ability to move the external ear or auricle is mostly lost, and the auricular muscles are considered vestigial.47 However, at PCW7.5, all three external auricular muscles and six intrinsic muscles were observed (Figure S5K and Table S1C). Through double immunostaining for MHC and ChAT (n = 18), we demonstrated that these nine muscles were innervated (Figure S5L) and therefore likely to be functional during auricle shaping. This supports a role of intrinsic muscles in auricle morphogenesis.48 By PCW11.9, the intrinsic muscles had increased in size, forming a nearly continuous ring within the ear (Figure S5M).
Overall, our data show that most head muscles develop earlier than previously described and are probably all present by the end of the first trimester of gestation.
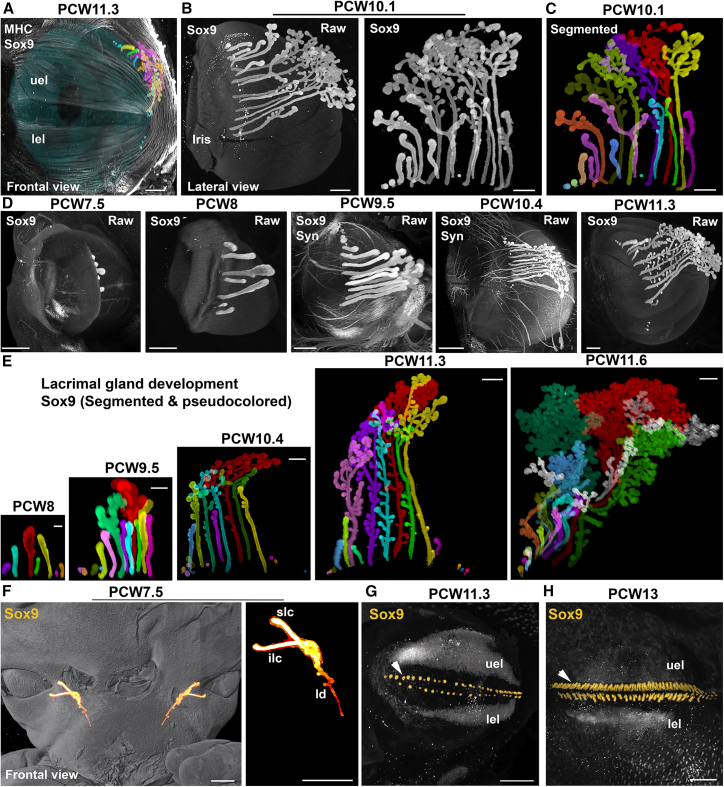
Development of salivary and lacrimal glands in human embryos
Humans have three primary pairs of salivary glands: the parotid, sublingual, and submandibular glands. These glands are connected to the mouth through one or multiple ducts and collectively produce at least one liter of saliva daily. In rodent studies, it has been observed that these glands develop through a process called branching morphogenesis, which involves the successive division and splitting of extending ducts.49 However, the development of salivary glands in humans and the specific details of branching morphogenesis are not well understood. This is primarily due to the difficulty of comprehending the process solely through 2D sections.50 Earlier studies conducted primarily by the Carnegie Institute suggested that gland formation initiates around PCW6.51,52
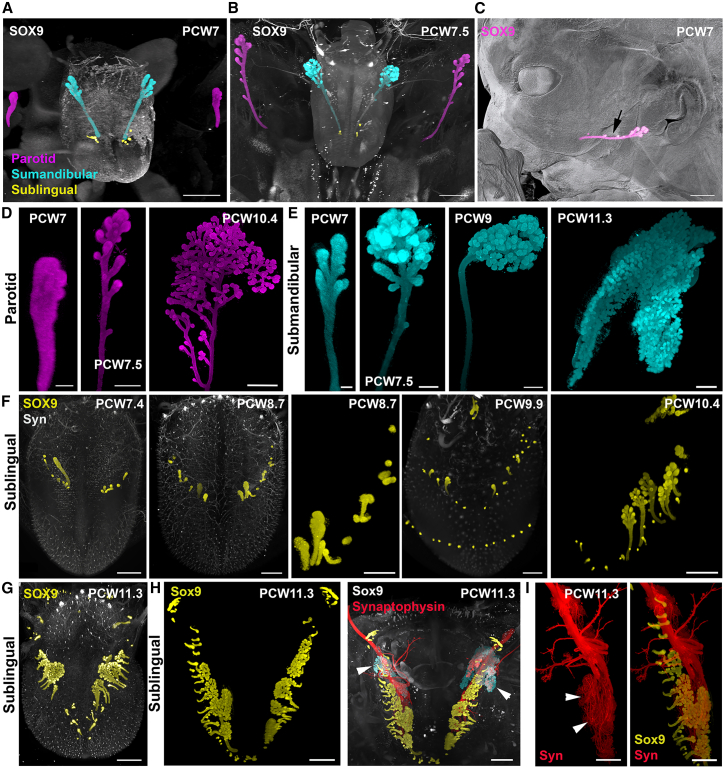
The developing salivary glands were visualized with Sox9 immunolabeling, which is known to be expressed in the embryonic gland epithelium.53 Although Sox9 is present in structures such as the cartilage (Figure 1), Sox9-positive glands could easily be isolated with VR-guided segmentation (Figures 4 and 5 and Video S4A). A total of 38 glands from 20 specimens at PCW6–PCW12 were reconstructed. The first steps of the emergence of the parotid and submaxillary glands could be captured from PCW7–PCW7.5 embryos (Figures 4A and 4B). On each side, each parotid and submaxillary gland extend caudally leaving a single duct behind (Figure 4C) that will give rise to the Stensen and Wharton ducts, respectively. The development of the submaxillaries seemed more advanced, and they branched only at the terminal end (growing tip) whereas for the parotid, a few side branches also formed at a distance from the tip. At later stages, branching was more pronounced and extended to the side branches (Figure 4D). At PCW9, the submaxillary gland had an ovoid shape, but its size significantly increased at PCW11.3 (Figure 4E). The sublingual glands started to bud at PCW7 (Figure 4A) with several individual buds formed on each side of the mouth, under the tongue. After PCW8, some buds started to grow and branch at their tip, but novel and less mature individual buds could be observed (Figure 4F). This was still the case at PCW11 when a large array of more than 20 glands at different stages of complexity was found on each side (Figures 4G and 4H). The branching mode of the sublingual glands appeared very similar to the submaxillary gland, with a pronounced splitting at the tips and not along the basal ducts (Figures 4G–4I). The sublingual and submaxillary glands also received dense innervation from synaptophysin+ axons (Figure 4I).
Figure 4.
Development of human salivary glands
All panels are LSFM images of solvent-cleared embryos (A–F) and fetuses (D, E, and G–I) immunostained with anti-Sox9 (A–I) combined with synaptophysin (F, H, and I). Glands immunolabeled with Sox9 were segmented using VR and pseudocolored.
(A–C) Dorsal view of the mouth and tongue of PCW7 (A) and PCW7.5 (B) embryos where the nascent parotid (magenta), submandibular (cyan), and sublingual (yellow) glands were segmented and pseudocolored. (C) shows a lateral view of the parotid (arrow) overlaid on the 3D rendering image (gray) of the face.
(D and E) Branching morphogenesis of the parotid gland and submandibular glands. A unique duct connects the glands to the mouth.
(F–H) Development of the sublingual glands. The first buds are visible at PCW7 and stay rather short until PCW9 when some sublingual glands start ramifying at their apex (F). (G and H) At PCW11.3, the number of glands has increased on both sides and they are distributed beneath the tongue, along all its length, all individually connected to the mouth floor.
(H and I) Both the submandibular (arrowheads in H) and sublingual (arrowheads in I) are densely innervated by synaptophysin+ axons emanating from the chorda tympani branch of the facial nerve (VII).
Scale bars: all panels are counted from left to right for each row; 700 μm in (A), (B), (D, right), and (I); 1 mm in (C), (G), and (H); 200 µm in (D, left and middlepanels); 100 μm in (E, first panel); 300 μm in (E, second and third panels) and (F, third panel); and 500 μm in (E, fourth panel) and (F, all panels except for the third).
Figure 5.
Development of human lacrimal glands
All panels are LSFM images of solvent-cleared eyes and heads immunostained with anti-Sox9 (A–H) and combined with MHC (A) and synaptophysin (D).
(A) Frontal view of the left eye of a PCW11.3 fetus. The orbiculari oculari muscle is seen as well as the upper (uel) and lower (lel) eye lids. The eye is pseudocolored in blue. The lacrimal gland (arrowheads) and its individual ducts (colored) are seen lying on the superolateral side of the eyeball.
(B–D) Organization the lacrimal gland in a PCW10.1 fetus. (B) shows the raw image obtained with Sox9 staining. In (C), all individual subglands are individually segmented and pseudocolored, the longest one in red. (D) shows the time course of lacrimal gland development, in situ on the eyeballs, between PCW7.5, when only a few buds are observed, and PCW11.3.
(E) All images are at the same scale to illustrate the growth of the lacrimal glands. All subglands have been individually segmented and colored. The color code is conserved, with the longest one in red as a reference (see also Figure S6).
(F) Frontal view of the face and nose/mouth of a PCW7.5 embryo stained with anti-Sox9. Overlays are 3D rendering images of the face (in gray). The superior (slc) and inferior (ilc) lacrimal canaliculi and nasolacrimal duct (ld) have been segmented and pseudocolored in orange.
(G–H) The developing eye lashes are labeled with Sox9 and form 2 arrays lining the edges of the upper (uel) and lower (lel) eyelids at PCW11.3 (arrowheads in G and H). At PCW13 (H), the number of Sox9+ buds has increased and might also include the developing meibomian glands.
Related to Figure S6.
Scale bars: all panels are counted from left to right for each row; 150 μm in (A); 500 μm in (B, left panel), (D), and (E, fourth panel); 400 μm in (B, right panel), (C), and (E, all panels except for the first and fourth); 200 μm in (E, first panel); and 1 mm in (F)–(H).
(A) 3D visualization of the developing exocrine glands in a PCW7.5 human embryo, related to Figures 4 and 5.
(B) 3D visualization of the fetal lacrimal glands in a PCW10.4 human fetus, related to Figures 5 and S7.
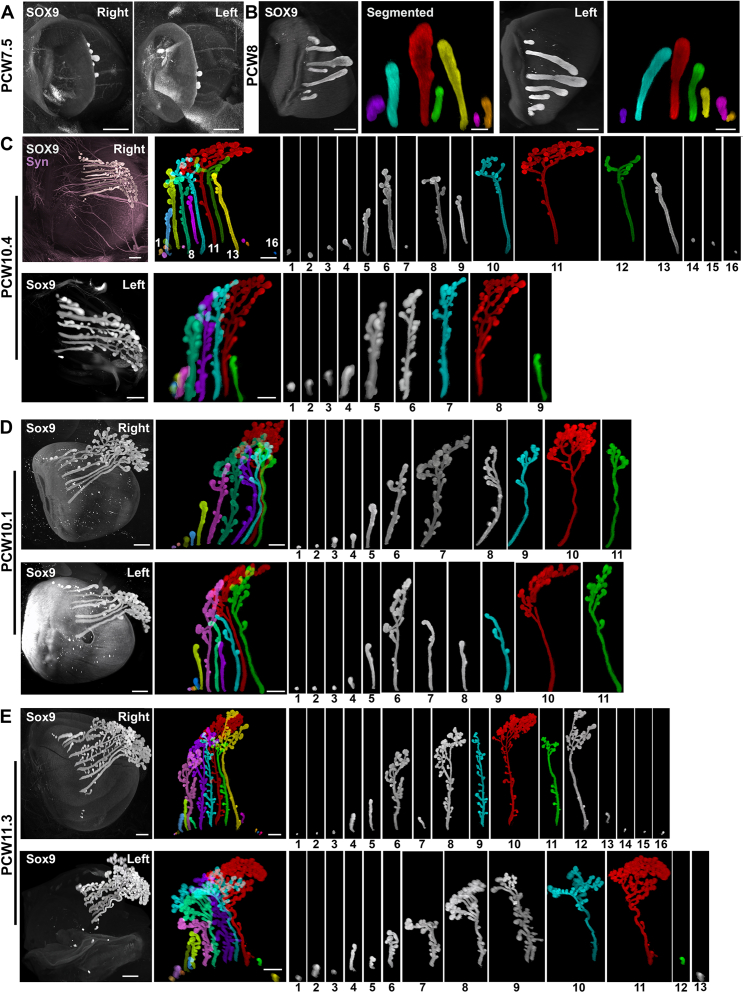
Lacrimal glands produce the tear film that is essential for vision and for cornea homeostasis. As for other head glands, their development was only studied from histological sections.54 In adult they are located in the orbit on the dorsolateral side of the eyes, and this is also the case during development (Figure 5A and Videos S3C and S4B). Using Sox9,55 we could capture all the early phases of lacrimal gland development (n = 30 eyes, from PCW7 to PCW11.8). Immunostaining of PCW10.1 samples (Figures 5B, 5C, and S6C) revealed that the lacrimal glands are composed of multiple individual ducts. These ducts extend caudally along the surface of the eye and develop dense branches in the caudal region. Unlike in mice, where the lacrimal glands form through progressive branching of a single duct, this pattern is different in humans. To better understand the morphology of the intertwined subglands, we used VR to segment and visualize them (Figures 5B, 5C, and S6). Within each eye, the length and complexity of each subgland exhibited significant variability. This variation was observed not only between different eyes but also between the right and left eyes within the same specimen (Figure S6). While one subgland generally appeared more developed than the others, the total number of subglands differed greatly between eyes, even within the same embryo. Additionally, the length and complexity of the glands along the medio-lateral axis also differed. During PCW7, a few Sox9+ buds were observed on each eye (Figure S6A). The number of these buds increased at least until PCW11, with short nascent buds still visible on the medial and lateral sides in proximity to the eyelid (Figures 5D and 5E). Interestingly, the developing nasolacrimal ducts which collect tears and carry them to the nasal cavity, could also be isolated using Sox9 immunostaining (Figure 5F). At PCW11.3, Sox9 was also detected in arrays of small round structures lining the apical edge of the upper and lower eyelids (Figure 5G). At PCW13, their number had significantly increased (Figure 5H), and they were found both on the internal and external side of the eyelids. These structures likely correspond to the nascent eye lash follicles and the first epithelial buds of the meibomian glands.56
Figure S6.
Stochastic development of lacrimal glands in human embryos, related to Figure 6
(A–E) All panels are LSFM images of solvent-cleared embryo (A and B) and fetuses (C–E), immunostained for Sox9 (A–E) and Synaptophysin (Syn, C). In each case, the 2 eyes (right and left) are shown, to illustrate the heterogeneity and asynchrony of lacrimal gland development. A mirror image is presented for the left eye to facilitate the comparison with the right eye.
(A–D) At PCW7 (A) and PCW8 (B) a few buds emerge on both sides. In (B) the longest one is colored in red, the two adjacent ones in green and cyan. The color code is conserved starting from the red/longest duct. Note that from the onset, the number and length of the buds differs between eyes. (C) and (D) shows the right and left eyes of 2 fetuses of similar age (PCW10.4 and PCW10.1 respectively). All subglands have been individually segmented and numbered. The number of glands varies between eyes and between cases. Their respective length is also highly variable with one (in red in all cases) always longer than the others. The relative position, along the superomedial to inferolateral axis, of the longest and most branched subgland (red) also varies between eyes and individuals (position 3/7 in B, 11/16 in right C, 8/9 in left C, 10/11 in D, 10/16 in right E, and 11/13 in left E). Branching mostly occurs at the growing tips, but side branches also form all along the individual ducts.
(E) At PCW11.3, the glands have further developed and have more side branches. Variability in size and branching complexity is still high.
Scale bars: all panels are counted from left to right for each row; 500 μm in (A), (C, first panel first row and second row), (D, first panel first row and second row), and (E, second panel second row); 400 μm in (B, first and third panels), (C, second panel first row), and (D, second panel first row); 200 μm in (B, second and fourth panels); 1 mm in (E, first panel second row).
Development of head and neck arteries
Padget9 based her description of human cerebral vascular development on graphic reconstructions of 22 sectioned embryos of the Carnegie Collection ranging from 3 to 7 weeks (estimated ovulation age) and from 3 (20 somites) to 40 mm crown-rump length (about 10–11 PCW).57 The results of this description have yet to be externally reproduced and most importantly, albeit exquisite in its description, it remains incomplete. In the oldest specimen Padget described, the definitive anatomy of the posterior cerebral artery (PCA) for instance had not yet emerged and the details of its development remain unresolved.9
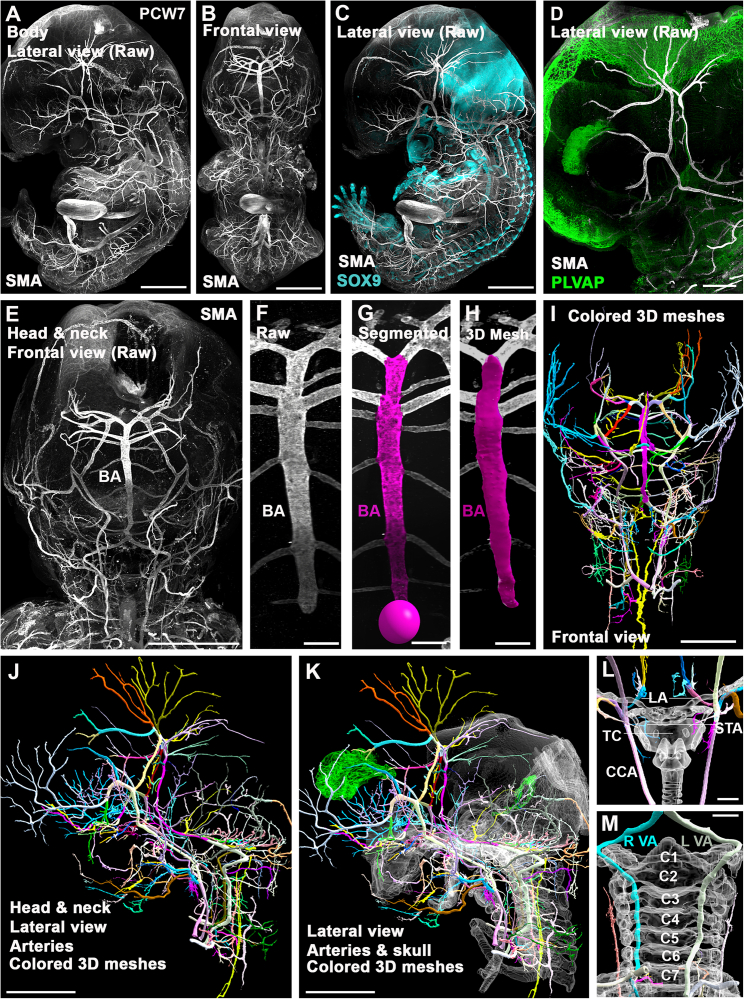
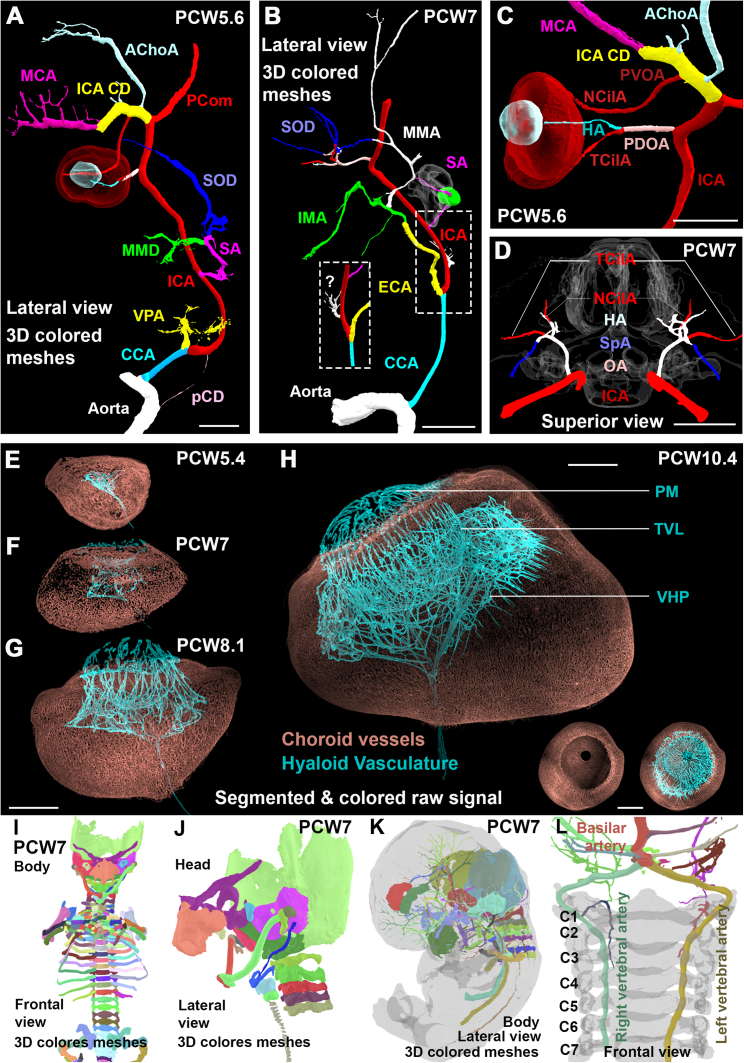
Herein, head and neck arterial 3D organization and development was analyzed in 6 PCW5.6–PCW9 embryos (4 complete embryos and 2 isolated head and neck samples) with immunostaining for smooth muscle actin (SMA; Figures 6A–6E). SMA was chosen as it was previously shown to discriminate arterial vessel walls with high specificity.19 Various other markers were used in the different specimens in conjunction to SMA including different combinations of MHC (data not shown), synaptophysin (Figure 7H), plasmalemma vesicle associated protein PLVAP (Figure 6D), and Sox9 (Figures 6C and 7A), allowing for clear vessel identification in relation to major anatomical landmarks including the developing neuro and splanchnocranium (Figures 6K, 7B, 7C, and 7G), spine (Figure 6M), larynx (Sox9; Figure 6L), choroid plexus (PLVAP; Figures 6D, 6K, 7J, and 7L), muscles (MHC; data not shown) and nerves (synaptophysin; Figure 7H). Embryos showed no sign of congenital anomaly except for one sample (PCW5.6), for which the heart appeared exterior to the thoracic cavity, either related to trauma or a rare congenital malformation termed ectopia cordis.58 Despite this possible congenital defect, the sample was retained for analysis because it was the youngest in the time series, showing a number of developmentally transient arterial structures at an immature stage (Figures S7A and S7D). In 3 of the 6 specimens, arterial wall immunostaining quality was deemed excellent allowing for extensive head and neck arterial segmentation to be performed (Figures 6, 7, and S7A–S7D and Video S5A). In the PCW5.6 (Figures S7A and S7D) and PCW7 (Figures 6E–6K and Video S5A) samples, a total of 95 and 159 head and neck individual arteries and arterial segments where identified and segmented, respectively.
Figure 6.
3D analysis of developing cephalic arteries in human embryos
(A–D) Head and neck vascular segmentation in a PCW7 embryo immunostained for SMA (A, lateral view and B, frontal view), SOX9 (C) and PLVAP (D).
(E–H) Segmentation and mesh generation of the basilar artery (BA).
(I–K) Isolated segmented head and neck arterial vasculature with randomization of individual arterial segment colors is shown in frontal (I) and lateral (J) views. (K) shows head and neck arteries in conjunction with semi-transparent meshes of reference cartilaginous structures (white) and the choroid plexuses (green).
(L) Frontal view showing the right and left superior thyroid arteries (STAs) in relation to the thyroid cartilage (TC). Also showing the common carotid arteries (CCAs) and lingual arteries (LAs).
(M) Frontal view of the right (R VA) and left (L VA) vertebral arteries in relation to semi-transparent cervical vertebra (numbered C1–C7).
Related to Figure S7.
Scale bars: 2.5 mm in (A)–(C), (E), (J), and (K); 1 mm in (D); 500 μm in (F)–(H), (L), and (M); and 2 mm in (I).
Figure 7.
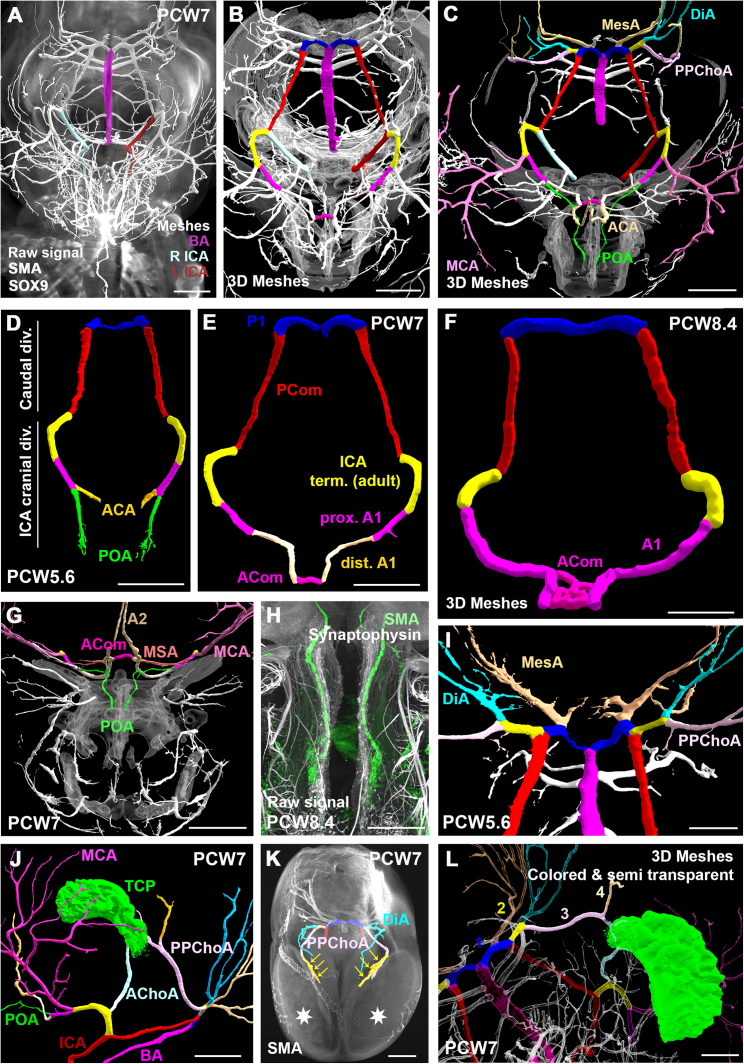
Closure of the arterial circle of Willis and establishment of major cerebral arteries
Superior view of the circle of Willis in a PCW7 embryo (A–C and E).
(A) The circle is fed by the right and left internal carotid arteries (ICAs) and the basilar artery (BA).
(B and E) All segments of the circle of Willis are conspicuous including the caudal division of the ICA, subdivided into the most posterior P1 segment and posterior communicating (PCom) segment, the cranial division of the ICA with the adult ICA terminal segment, proximal A1 (distal to the origin of the middle cerebral artery (MCA) and proximal to the origin of the primitive olfactory artery (POA)), distal A1 and anterior communicating artery (ACom).
(C) Both MCA and post communicating anterior cerebral artery (ACA) are well identified. Posterior branches of the circle of Willis include the mesencephalic arteries (MesAs) as well as the primitive posterior choroidal arteries (PPChoAs) and diencephalic arteries (DiAs) arising from a common trunk (yellow).
(D–F) Anterior closure of the circle of Willis and subsequent growth in PCW5.6, PCW7, and PCW8.4 specimens. The series shows how the anterior cerebral artery first appears as a medial branch of the POA, before overshadowing the latter.
(G and H) The POA is shown to course into the nasal capsule along the olfactory filaments (H). Notice the medial striate arteries (MSAs) or recurrent arteries of Heubner arising from the peri-communicating segment of the ACA.
(I) The stem of the posterior cerebral artery (PCA) is represented by the most distal segment of the ICA caudal division (P1 segment). Notice the asymmetry of the P1 segments in the PCW5.6 embryo, showing early onset of a classical circle of Willis variant.
(J) The telencephalic choroid plexus (TCP) or choroid plexus of the lateral ventricule, is seen to be fed by both the anterior choroidal artery (AChoA) and the PPChoA.
(K) In another late PCW7 specimen, the PPChoAs give rise to prominent branches (yellow arrows) terminating at the medial posterior surface of the cerebral hemispheres (white stars).
(L) The adult posterior cerebral artery is a composite vessel combining the posterior segment of the ICA caudal division (1; blue), the diencephalic-choroid common trunk (2; yellow), the proximal stem of the primitive posterior choroidal artery (3; pink), and post choroidal branches (4; beige) that are shown to vascularize the posterior and medial surface of the developing telencephalic vesicles. The terminal choroidal branches are likely represented in the adult by the lateral posterior choroidal arteries.
Related to Figure S7.
Scale bars: 1 mm in (A)–(H), (J), and (K) and 500 μm in (I) and (L).
Figure S7.
Assembly of the external carotid and ophthalmic arteries and 3D visualization of human embryonic structures with Verge3D, related to Figures 1, 6, and 7
(A) In a PCW5.6 specimen, the left common carotid artery (CCA) primitively divides into an internal carotid artery (ICA) and a ventral pharyngeal artery (VPA), the future stem of the external carotid artery (ECA), not to be confused with the adult ascending pharyngeal artery. Note that the CCA is still connected to the aorta by a putative carotid duct (pCD) remnant. The ICA gives rise to the transient stapedial artery (SA), passing through the stapedial ring obturator foramen (well depicted with SOX9 staining in a PCW7 specimen in B). At PCW5.6 (A), the nascent SA divides into a lower maxillomandibular division (MMD) and an upper supraorbital division (SOD). The ICA (well depicted in C) gives rise to a primitive dorsal ophthalmic artery (PDOA), proximal to the posterior communicating artery (PCom), and a distal primitive ventral ophthalmic artery (PVOA).
(B) Foreshadowing future reconfigurations, at age PCW7, the VPA has anastomosed with the lower division of the stapedial artery, acquiring the internal maxillary artery (IMA) in the process, and the ophthalmic artery has anastomosed with the upper division of the stapedial artery, acquiring its extraocular orbital branches (SOD). The persistent channel between the two (white vessel) is the future middle meningeal artery, connected to the ophthalmic artery by the so-called sphenoidal anastomosis (SpA, well depicted in D). The inset shows an unidentified branch of the ICA, proximal to the origin of the SA, speculatively related to the embryology of the ascending pharyngeal artery and warranting further exploration.
(A and C) Interestingly and contrary to a common misconception, the PVOA is not a branch of the anterior cerebral artery but rather arises from ICA cranial division (ICA CD), opposite to the emergence of the anterior choroidal artery (AChoA) and proximal to the origin of the middle cerebral artery (MCA). The PDOA branches into a temporociliary artery (TCilA) and a hyaloid artery (HA) penetrating the optic nerve. The PVOA terminates as a nasociliary artery (NCilA).
(D) At PCW7 (superior view), the stem of the definitive ophthalmic artery gives rise to both nasociliary and temporociliary arteries as well as the hyaloid artery.
(E–H) PLVAP immuno-staining of four PCW5.4 to PCW10.4 eyes showing the choroid (brown, fed by the ciliary arteries) and hyaloid (blue, fed by the hyaloid artery) vasculatures. The ocular growth series between PCW5.4 and PCW10.4 specimens demonstrates the maturation of the vasa hyaloidea propria (VHP), the tunica vasculosa lentis (TVL), and the pupillary membrane (PM) vessels. Establishment of the PM vascular coverage was the last element of the hyaloid vasculature to appear between PCW7 and PCW8.1.
(I–L) (I and L) Visualization of human embryonic structures, through screenshots of 3D models of two PCW7 human embryo visualized in Verge3D web interface. Structures were immunostained with Collagen 2 (I and J) SMA, PLVAP, and Sox9 (K and L), segmented and 3D meshes were generated. (I) Frontal view of the whole embryo presumptive skeleton. (J) Lateral view of the head and neck presumptive skeleton. (K) shows all segmented structures pseudocolored through the semi-transparent embryo surface. (L) shows the vertebral arteries in relation to the cervical spine (the embryo surface is deleted).
Scale bars: 500 μm in (A), (C), and (E)–(H) and 1 mm in (B), (D), and (L).
(A) 3D Visualization of the head vasculature in a PCW7 human embryo.
(B) Development of the hyaloid vasculature from PCW7 to PCW10.4, related to Figures 7 and S7.
The developmental series captures the sequence of events leading to the construction of the adult external carotid and ophthalmic arteries via the transient stapedial artery (Figures S7A–S7D and Video S5A).9,59 In order to better demonstrate establishment of the ocular vasculature and because distal arterioles were negative for SMA staining, 11 eyes, ranging from PCW5.4 to PCW12.3 were marked for PLVAP and analyzed. This exquisitely demonstrated the distinct choroid and hyaloid vasculatures of the embryonic eye, fed by the ciliary and hyaloid branches of the ophthalmic artery, respectively (Figures S7E–S7H and Video S5B).
Special attention was given to the establishment of cerebral arteries (Figure 7). The data provided resolve the origin of the PCA (Figures 7C and 7I–7L). In the youngest sample (PCW5.6), the caudal division (future posterior communicating artery) of the internal carotid arteries (ICAs), joining bilaterally with the basilar artery cranial tip, gives rise to a primitive posterior choroidal artery, a diencephalic artery, and a mesencephalic artery (Figure 7I). The primitive posterior choroidal and diencephalic arteries arose from a common trunk in all specimens (Figures 7C and 7I). At the earlier stage, both middle cerebral artery and anterior cerebral artery appear as proximal lateral and distal medial branches of the anterior division of the ICA, respectively, overshadowing the primitive olfactory artery, with the anterior communicating complex yet unformed (Figure 7D). In the same specimen, however, the PCA is unidentified. Both primitive anterior and posterior choroidal arteries joined the choroid plexus of the telencephalic vesicles where they anastomose (also seen at PCW7, Figures 7J and 7L). In PCW7 and PCW8.4 samples, the circle of Willis was complete (Figures 7E and 7F) and the primitive posterior choroidal arteries give rise to post choroidal branches arising along the stem of the primitive posterior choroidal artery and coursing along the medial wall of posterior portion of the telencephalon (Figures 7K and 7L), in what is essentially the territory of the adult PCA.
Last, to facilitate the visualization of fully annotated 3D image datasets and their use for outreach and teaching purposes, interactive 3D models of embryonic head arteries and skeleton were created by exporting segmented meshes into Verge3D, a web-based interface and toolkit (Figures S7I–S7L and Videos S6A–S6C; see STAR Methods and supplemental information).
(A) Tutorial for Verge 3D Viewer, illustrating arteries in a PCW7 human embryo, related to Figures 1, 6, and 7.
(B) 3D organization of the circle of Willis in a PCW7 human embryo, related to Figure 7.
(C) 3D organization of the stapedial artery in PCW7 human embryo, related to Figure S7.
(D) Tutorial for the hudeca.com image database, related to all figures.
Discussion
Congenital defects affect more than 3% of births, but despite their prevalence, the molecular mechanisms underlying cell-cell interactions and proper tissue morphogenesis in humans remain largely unknown. Developmental disorders can have lifelong consequences for affected individuals, making it crucial to understand their causes and develop appropriate therapeutic strategies, including in utero interventions. Animal models have provided valuable insights into dysregulated ontogenetic events, but they only partially replicate the human-specific developmental sequences and processes.60,61 This discrepancy has led to a renewed focus on human embryology research and the emergence of human stem cell-derived in vitro models of embryogenesis.62,63,64,65,66 However, the field still lacks precise reference cell atlases of human embryos, and current knowledge is primarily based on outdated studies.
Several recent research initiatives, such as the Human Cell Atlas project, aim to leverage advanced technologies to identify human cell types throughout development and construct high-resolution cellular maps of developing human organs.67,68 Although single-cell RNA sequencing can identify cell types and predict cell trajectories in human embryo organs, it fails to capture and preserve spatial information.69,70,71,72 Commercially available spatial genomic technologies, while capable of analyzing the transcriptome and other molecular information in situ, are currently limited to tissue sections.73,74 However, recent studies suggest that 3D spatial transcriptomics is possible.75,76 Therefore, the combination of whole-mount immunostaining and 3D imaging still represents the best opportunity to construct 3D developmental maps of human embryogenesis.
In this study, we applied this strategy to the developing human head and generated in situ 3D image datasets of various tissues and organs, including muscles, cartilage, and secretory glands. These datasets are accessible through a dedicated web interface. Notably, through image segmentation, we produced high-resolution 3D models of human embryos that can be viewed using multiple platforms, such as virtual reality, interactive web-based interfaces, and even 3D printing. These models also offer teaching tools for medical students.
One limitation of our imaging pipeline is the simultaneous imaging of a limited number of markers (maximum 4). However, our study demonstrates that recombinant conjugated antibodies work effectively with our protocol, allowing the use of human antibodies and the combination of multiple antibodies raised in the same species. This finding supports a previous report in adult mice,77 and our preliminary data hold promise for adapting cyclic-immunostaining methods developed for 2D sections.78 This would enable multiple rounds of staining and clearing, expanding the range of markers that can be utilized in the same human specimen.
Furthermore, our results provide insights into the early stages of secretory gland morphogenesis in the human head. Developmental biologists have extensively studied branching morphogenesis, which explores the regulation of tree-like structures with thousands of branches.79 The intrinsic contribution of a genetically encoded developmental program and the influence of environmental factors (such as morphogens, forces, and cell contacts) have been investigated in rodent models for salivary glands53,80 and lacrimal glands.81,82 However, apart from a century-old study,83 such information is unavailable for human embryos. Our study suggests the existence of a stereotypical growth pattern but also highlights significant individual variability in the process. Particularly, the salivary glands and lacrimal glands consist of a highly heterogeneous number of individual subglands that do not extend and branch at the same rate and pace, resulting in distinct branching complexity among neighboring subglands. Moreover, unlike in mice, where the lacrimal gland possesses a single secretory duct, human lacrimal glands comprise intertwined individual subglands. Investigating their 3D interactions will shed light on how their branching mode compares with rodents, providing essential insights for assessing the developmental relevance of recently developed human lacrimal gland and salivary gland organoids.55,84
By combining whole-mount immunostaining, tissue clearing, and light-sheet imaging, an opportunity arises to revisit and enhance our understanding of the development of human head and neck arteries. This approach minimizes the risk of reconstruction bias and also allows for a more comprehensive description of their development. In adult humans, the PCA gives rise proximally, at the pre-communicating (P1) or post-communicating (P2) segments, to a medial posterior choroidal artery and distally to the lateral posterior choroidal artery, whose relationship to the primitive posterior choroidal artery has been unclear.85,86 Based on the provided data, we can interpret the PCA as primarily representing the trunk of the primitive posterior choroidal artery. As the postero-medial telencephalic branches become dominant, they overshadow the choroidal branches, which subsequently develop into the adult lateral posterior choroidal arteries. This interpretation is consistent with the observation that only the adult lateral posterior choroidal artery anastomoses with the anterior choroidal artery.85,86 These findings align with previous studies in rat embryos.87 Overall, this study provides insights into the embryological origins and development of cerebral arteries, shedding light on the complex composition and relationships of these vascular structures.
Anatomical education, increasingly rely on blended learning approaches, incorporating 3D representations of anatomical specimens and interactive web-based interfaces as valuable learning resources.88,89,90 We firmly believe that the provision of user-friendly web-based interfaces featuring 3D reconstructions of real human embryos can serve as a powerful tool to revitalize human embryology education.
Limitations of the study
Although all specimens were carefully viewed by embryologists for possible anomalies and are considered healthy (to the exception of one embryo with ectopia cordis), we did not perform genetical analysis or karyotyping. Therefore, some of embryos and fetuses studied here could carry mutations and have developmental defects. It will be important to extend this analytical pipeline to pathological embryos with identified genetic defects and also to better take into account possible sex differences. We only provide a rough spatiotemporal 3D template and reference of the development of head organs and tissues, and additional experiments will be needed to fully describe the cellular and molecular mechanisms that shape individual head structures. Cell profiling using transcriptomics and proteomics will have to be performed to identify all cell types in head tissues before we can attempt to precisely map them. This will require to further increase our antibody multiplexing capacities.
This analysis is also limited to specimens at PCW5.6– PCW14, and we are therefore missing the early phases of head assembly (such as NCC migration) as well as the final stages of tissue construction and remodeling. For instance, most of the development of the neurocranium and cranial sutures occurs after PCW14. In addition, the immune system and the central nervous system should be included in future studies. Last, we show that VR-assisted segmentation can handle very large image datasets. However, artificial intelligence (AI) and deep-learning approaches will have to be implemented to facilitate the 3D reconstructions of the samples. This will increase the speed of the analyses, particularly for large fetal organs, and limit the biases inherent to image segmentation performed by humans.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-CD31 | Novus Biologicals | Cat# NB100-2284; RRID:AB_10002513 |
| Armenian Hamster monoclonal anti-CD31 | DSHB | Cat# 2H8; RRID:AB_21610393 |
| Mouse monoclonal anti-CD31 | Cell Signaling | Cat# 3528S; RRID:AB_2160882 |
| Goat polyclonal anti-Choline Acetyltransferase | Millipore | Cat# AB144P; RRID:AB_2079751 |
| Mouse monoclonal anti-Collagen 1 | Abcam | Cat# ab6308; RRID:AB_305411 |
| Rabbit monoclonal anti-Collagen 1 | Abcam | Cat# ab138492; RRID:AB_2861258 |
| Mouse monoclonal anti-Collagen 2 | Abcam | Cat# ab185430; RRID:AB_1603612 |
| Goat polyclonal anti-Lyve1 | R&D systems | Cat# AF2089; RRID:AB_355144 |
| Mouse monoclonal anti-Myosin Heavy Chain | Millipore | Cat# 05-716; RRID:AB_309930 |
| MHC coupled Vio-667 | Miltenyi | N/A (not yet commercialised) |
| Rabbit polyclonal anti-MyH3 | Invitrogen | Cat# PA5-72848; RRID:AB_2718702 |
| Rabbit monoclonal anti-MyoD1 | Abcam | Cat# ab133627; RRID:AB_2890928 |
| Mouse monoclonal anti-Myogenin | Abcam | Cat# ab1835; RRID:AB_302633 |
| Mouse monoclonal anti-Neurofilament-H conjugated to Alexa 488 | Biolegend | Cat# 801612 RRID:AB_2750330 |
| Rabbit monoclonal anti-Osterix | Abcam | Cat# ab209484; RRID:AB_2892207 |
| Rabbit polyclonal anti-Pax7 | Invitrogen | Cat# PA1-117; RRID:AB_2539886 |
| Mouse monoclonal anti-Pax7 | Abcam | Cat# ab218472; RRID:N/A |
| Mouse monoclonal anti-Peripherin | Millipore | Cat# MAB1527; RRID:AB_2284441 |
| Mouse monoclonal anti-PLVAP | Abcam | Cat# ab81719; RRID:AB_1658370 |
| Rabbit monoclonal anti-Prox1 | Abcam | Cat# ab199359; RRID:AB_2868427 |
| Rabbit polyclonal anti-SMA | Abcam | Cat# ab5694; RRID:AB_2223021 |
| Rabbit monoclonal anti-SMA | Abcam | Cat# ab32575; RRID:AB_722538 |
| Rabbit monoclonal anti-SMA conjugated to Alexa488 | Abcam | Cat# ab202295; RRID:AB_2890884 |
| Rabbit monoclonal anti-SMA conjugated to Alexa 555 | Abcam | Cat# ab202509; RRID:AB_2868435 |
| Rabbit monoclonal anti-SMA conjugated to Alexa 594 | Abcam | Cat# ab202510; RRID:N/A |
| Rabbit monoclonal anti-SMA conjugated to Alexa 647 | Abcam | Cat# ab202296; RRID:N/A |
| Goat polyclonal anti-Sox9 | R&D systems | Cat# AF3075; RRID:AB_2194160 |
| Rabbit monoclonal anti-Sox9 conjugated to Alexa647 | Abcam | Cat# ab207677; RRID:N/A |
| Rabbit monoclonal anti-Synaptophysin | Abcam | Cat# ab32127; RRID:AB_2286949 |
| Rabbit monoclonal anti-Synaptophysin conjugated to Alexa 647 | Abcam | Cat# ab196166; RRID:N/A |
| Goat polyclonal anti-Tag1/Contactin2 | R&D Systems | Cat# AF4439; RRID:AB_2044647 |
| Rabbit polyclonal anti-Tenomodulin | Abcam | Cat# ab203676; RRID:AB_2722782 |
| Sheep polyclonal anti-Tyrosine Hydroxylase | Invitrogen | Cat# PA1-4679; RRID:AB_561880 |
| Mouse monoclonal anti-Tubulin beta III | Abcam | Cat# ab78078; RRID:AB_2256751 |
| Rabbit polyclonal anti-Tubulin beta III | Sigma-Aldrich | Cat# T2200; RRID:AB_262133 |
| Donkey Anti-Rabbit IgG H&L (Alexa Fluor 488) | Abcam | Cat# ab150073; RRID:AB_2636877 |
| Donkey Anti-Rabbit IgG H&L (Alexa Fluor 555) | Abcam | Cat# ab150062; RRID:AB_2801638 |
| Donkey Anti-Rabbit IgG H&L (Alexa Fluor 647) | Abcam | Cat# ab150063; RRID:AB_2687541 |
| Donkey Anti-Rabbit IgG H&L (Alexa Fluor 790) | Abcam | Cat# ab186693; RRID:N/A |
| Donkey Anti-Goat IgG H&L (Alexa Fluor 488) | Abcam | Cat# ab150129; RRID:AB_2687506 |
| Donkey Anti-Goat IgG H&L (Cy3) | Abcam | Cat# ab6949; RRID:AB_955018 |
| Donkey Anti-Goat IgG H&L (Alexa Fluor 647) | Abcam | Cat# ab150135; RRID:AB_2687955 |
| Donkey Anti-Goat IgG H&L (Alexa Fluor 750) | Abcam | Cat# ab175745; RRID:AB_2924800 |
| Donkey Anti-Mouse IgG H&L (Alexa Fluor 488) | Abcam | Cat# ab150105; RRID:AB_2732856 |
| Donkey Anti-MouseIgG H&L (Alexa Fluor 555) | Abcam | Cat# ab150110; RRID:AB_2783637 |
| Donkey Anti-Mouse IgG H&L (Alexa Fluor 647) | Abcam | Cat# ab150111; RRID:AB_2890625 |
| Donkey Anti-Mouse IgG H&L (Alexa Fluor 790) | Abcam | Cat# ab186699; RRID:N/A |
| Donkey Anti-Sheep IgG H&L (Alexa Fluor 790) | Jackson ImmunoResearch Labs | Cat# 137327; RRID:N/A |
| Goat Anti-Armenian Hamster IgG H&L (Alexa Fluor 488) | Abcam | Cat# ab173003; RRID:AB_2936402 |
| Biological samples | ||
| Human tissues | INSERM-Hudeca Biobank | this paper |
| Chemicals, peptides, and recombinant proteins | ||
| Phosphate-Buffered Saline | Sigma-Aldrich | Cat# D1408-500ml |
| Helix NP Green | Biolegend | Cat# 425303;RRID:N/A |
| bisBenzimide H (DAPI) | Sigma-Aldrich | Cat# 33258 |
| Agarose | Carl ROTH | Cat# 2267.4 |
| Azide | Sigma-Aldrich | Cat# S2002 |
| Dibenzyl ether (DBE) | Sigma-Aldrich | Cat# 108014 |
| Dichloromethane (DCM) | Sigma-Aldrich | Cat# 270997 |
| Ethylenediaminetetraacetic acid (EDTA) | Sigma-Aldrich | Cat# E7889 |
| Formalin | Sigma-Aldrich | Cat# HT501128 |
| Gelatin | VWR Chemicals | Cat# 24350.262 |
| Hydrogen peroxide solution (H2O2) | Sigma-Aldrich | Cat# 216763 |
| Isopentane | VWR Chemicals | Cat# 24872.298 |
| Methanol (MeOH) | VWR Chemicals | Cat# 20847.360 |
| Mowiol | Sigma-Aldrich | Cat# 81381 |
| Paraformaldehyde | VWR Chemicals | Cat# 28794.295 |
| Polystyrene Weighing dishes | Sigma-Aldrich | Cat# Z154873; Z15488; Z154903 |
| Single Edge Razor Blade | Electron Microscopy Sciences | Cat# 71972 |
| Slides BOND Plus | Leica Biosystems | Cat# S21.2113.A |
| Sucrose | VWR Chemicals | Cat# 27478.296 |
| Syringe filters ROTILABO® MCE, 0,22 μm | Carl ROTH | Cat# KH54.1 |
| TAE Buffer 10X | Invitrogen | Cat# 15558-026 |
| Thimerosal | Sigma-Aldrich | Cat# T8784-5g |
| Thimerosal | Sigma-Aldrich | Cat# T5125 |
| Triton X100 | Sigma-Aldrich | Cat# X100-500ml |
| Tube TPP 15 & 50 mL | Dutscher | Cat# 191115; 191050 |
| Vannas Spring Scissors | Fine Science Tool | Cat# 15000-03 |
| Oligonucleotides | ||
| SRY sense 5′-AGCGATGATTACAGTCCAGC-3′ | Belle et al.19 | N/A |
| SRY antisense 5′-CCTACAGCTTTGTCCAGTGG-3′ | Belle et al.19 | N/A |
| FGF16 sense 5′- CGGGAGGGATACAGGACTAAAC-3′ | Belle et al.19 | N/A |
| FGF16 antisense 5′- CTGTAGGTAGCATCTGTGGC-3′ | Belle et al.19 | N/A |
| Deposited Data | ||
| Raw and analyzed data | This paper | www.hudeca.com |
| Software and algorithms | ||
| Blender (v3.0) | Blender Foundation | https://www.blender.org/ |
| Handbrake (v1.6.1) | Handbrake | https://handbrake.fr/ |
| ImageJ (1.50e, Java 1.8.0_60, 64-bit) | NIH | http://imagej.nih.gov/ij/; RRID:SCR_003070 |
| Imaris x64 software suite (v9 to 10) | Oxford Instruments | https://imaris.oxinst.com/ |
| Imspector software for Blaze Ultramicroscope | Miltenyi | https://www.miltenyibiotec.com/FR-en/products/ultramicroscope-blaze.html |
| iMovie (v10.1.1) | Apple | http://www.apple.com/fr/imovie/ |
| Meshmixer (Version 3.5.474) | Autodesk | https://meshmixer.com/ |
| Meshlab (Version 2021.10) | Cignoni et al.91 | https://www.meshlab.net/# |
| Numpy (Version 1.22.3) | Harris et al.92 | https://numpy.org/ |
| Pandas (Version 1.4.2) | NumFOCUS, Inc. | https://pandas.pydata.org/ |
| Premiere Pro (2019) | Adobe | https://www.adobe.com |
| Pymeshlab (Version 2022.2.post4) | Cignoni et al.91 | https://www.meshlab.net/# |
| Syglass (v1.7.1 & 1.7.2) | IstoVisio, Inc. | https://www.syglass.io/ |
| TIFFFILE (Version 2022.3.25) | 10.5281/zenodo.8357648 | https://github.com/cgohlke/tifffile/ |
| Vedo (Version 2023.4.6) | 10.5281/zenodo.5330735 | https://vedo.embl.es/ |
| Verge3D (v3.9) | Soft8Soft | https://www.soft8soft.com/verge3d/ |
| Wavefront.OBJ (ImageJ plugin) | Virtual Insect Brain (VIB) Protocol | https://imagej.net/formats/wavefront-obj |
| Custom Python code: Mask_labels.ipynb | This paper | https://github.com/Megumi4952/Chedotal_Lab_Head_Development/Mask_labels.ipyn |
| Custom Python code 2: QECD_in_Folder.ipynb | This paper | https://github.com/Megumi4952/Chedotal_Lab_Head_Development/QECD_in_Folder.ipynb |
| Custom Python code 3: Merge_Meshes_for_3DP.ipynb | This paper | https://github.com/Megumi4952/Chedotal_Lab_Head_Development/Merge_Meshes_for_3DP.ipynb |
| Other | ||
| Resource website | Belle et al.19 | https://transparent-human-embryo.com/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Alain Chédotal (alain.chedotal@inserm.fr).
Materials availability
This study did not generate new unique reagents
Data and code availability
-
•
All image data have been deposited at: https://hudeca.com and all original image stacks are accessible to all scientists. Downloaded and exported data is licensed under the Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0).
-
•
All original code has been deposited at GitHub and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental models and study participant details
Human embryos and fetal samples
The human embryos and fetuses included in this study were obtained legally from voluntary abortions, with gestational ages ranging from 5 to 14 weeks post-conception (PCW5-14; Table S1A). Prior to tissue collection, written informed consent was obtained from all donors. All samples were provided by INSERM's HuDeCA Biobank and used in compliance with French regulations. Authorization to use human tissues was granted by the French agency for biomedical research (Agence de la Biomédecine, Saint-Denis La Plaine, France; N° PFS19-012) and the INSERM Ethics Committee (IRB00003888). Embryos (up to PCW8) were classified using Carnegie staging,93 while fetuses were staged based on morphometric correction as described before.94 All specimens were initially selected based on macroscopic morphological criteria, excluding samples with obvious malformations.
The sex of the specimens was determined, whenever possible, by either morphological observations (from PCW8 on) or by PCR genotyping of tissue biopsy. Assignations are reported in Table S1A. From a total of 76 specimens, 36 were males, 23 females and 17 undetermined.
All specimens were photographed and registered from collection to final storage or disposal. The OpenSpecimen platform (www.openspecimen.org) was used to permanently record collection, curation and morphometric annotation. Specimen identifiers described in this work are indexed in accordance to this database (Table S1A).
Method details
Sex determination
In cases where visual assessment of sex was not possible, sex was determined using PCR-based screening. DNA was extracted from tissue biopsies using lysis buffer containing 0,1 mg/ml proteinase K, 5M Sodium Chloride, 20% Sodium dodecyl sulfate, 1M Tris, pH 8.0 solution in water and stored at 54°C overnight. DNA was precipitated with isopropanol (1:1) and re-suspended in RNase/DNase-free water for 3h at 65°C. A PCR was performed in a T100 thermocycler (Biorad) using the following steps: 94°C for 3 min and 35 cycles of 94°C for 1 min; 56°C for 30 s; 72°C for 30 s and 72°C for 5 min. Primer sequences are reported in the key resources table.
Sample Fixation
All samples were fixed in 4% Paraformaldehyde (PFA) in phosphate buffer (0.12M, pH7.4), or 10% formalin, at 4°C for 1-5 days depending upon their size. Specimens were rinsed and cold-stored in fresh 1X Phosphate Buffer Saline containing sodium azide (0.02%) or thimerosal (0.1mg/ml). PBS was changed every 3 months until samples were used. Sampled stored for more than 6 months in PBS were re-fixed overnight in 4% PFA, followed by 3 rinses in 1X PBS, before processing.
Decalcification
Specimens older than PCW9 were decalcified by incubation during 1 week in EDTA 0.5M under agitation at RT. The solution was renewed halfway through the incubation period. The samples were washed twice in PBS 1X during one day.
Bleaching
To remove pigmentation and reduce background related to hematomas, tissue bleaching was carried out.19 Samples were dehydrated for 1hr at RT in ascending concentrations of methanol in H2O (20%, 40%, 60% and 80%).
Next, samples were placed overnight under white light (11W 3000K°) and rolling agitation (IKA, 004011000) with a 6% hydrogen peroxide solution in 100% methanol. The EyeDISCO protocol was used to bleach eye samples.95 A hole was done in the cornea by a needle (30G) and enlarged with scissors. Eyes were incubated for 7 to 10 days in 11% H2O2 on an agitator (IKA, 004011000) and under white light. Next, samples were re-hydrated for 1hr at RT in descending concentrations of methanol (80%,60%,40%,20%) and washed twice and blocked in PBS-Gelatin 0.2% TritonX100 0.5% (PBSGT) solution during 1 day (≤PCW7) to 1 week (≥PCW10).
Antibody screening on cryosections
All antibodies underwent initial testing on cryosections. Embryonic and fetal organs were cryoprotected in a solution of 15% sucrose at 4°C overnight. They were then embedded in a mixture of 7.5% gelatin, 10% sucrose, and 0.12 M phosphate buffer, followed by freezing in isopentane at -55°C and storage at -80°C until needed. Samples were cut into 20μm sections using a cryostat (Leica CM3050) and placed on slides for frozen sections. Cryosections were blocked in PBS-GT (0.2% gelatin, 0.25% Triton-X100 in 1X PBS) for 1 hour and incubated overnight at room temperature (RT) with the primary antibodies diluted in PBS-GT. After 3 washes in PBS, species-specific Alexa-conjugated secondary antibodies were incubated in PBS-GT for 1hr at RT. Sections were counterstained with DAPI and mounted with Mowiol.18 Imaging was conducted using either an epifluorescence microscope (Leica DM6000) or a confocal microscope (Olympus, FV1000).
Whole-mount immunolabelling
For whole-mount immunostaining, samples were transferred to a solution containing the primary antibodies diluted in PBSGT. Volume should be adapted to totally cover the sample. Next, samples were incubated at 37°C with agitation at 20 rpm for 7 (≤PCW8) to 14 days (PCW10-PCW14). This was followed by six washes of 1hour in PBSGT at RT. Next, secondary antibodies were diluted in PBSGT and passed through a 0.22 μm filter. Samples were incubated at 37°C in the secondary antibody solution for 3 (≤PCW8) to 7 (≥PCW10) days and washed six times during 1 hour in PBSGT at RT.
When conjugated antibodies were used in addition to non-conjugated ones, they were added at the same time as the secondary antibodies, if the host species were different from the primary antibodies. Otherwise, conjugated antibodies, diluted in PBSGT, were added after the round of secondary antibodies and incubated for 7 days. Samples were rinsed six times 1 hour in PBSGT at RT.
In the event of a second round of staining, the samples were put back in 100% methanol for 2 times 1 hour and then under white light and rolling agitation in a solution of 6% H2O2 in methanol during 1 day at RT. With this procedure, we could bleach fluorophores in the 555, 647 & 790 nm wavelengths. The samples were re-hydrated in descending concentrations of methanol during 1 hour for each step ((2x)100%,80%,60%,40%20%, PBSGT(2x)) and next incubated with a second mix of antibodies. To avoid cross-reactivity, the host-species of second round antibodies were distinct from those of the first round, unless they were directly conjugated to the fluorophores (Table S1A).
Agarose Embedding
To facilitate the handling and imaging of immune-labelled samples with the light-sheet microscope, tissue samples were embedded prior to clearing in 1.5% agarose. Agarose was dissolved in TAE 1X in an Erlenmeyer flask and heated using a microwave (1000W; reaching boiling point) until a homogeneous solution was obtained. Samples were placed in polystyrene weighing dishes and cooled agarose (about 45°C) was poured over it, avoiding bubbles. Samples were either totally, or partially covered to create a mould. Samples were removed from mould and this latter was cleared separately. Agarose was trimmed as close as possible to the sample with a razor blade. In case of a second round of staining, agarose was not removed and the agarose-embedded tissue was processed.
Sample Clearing
We followed the iDISCO+ protocol.96 After immunolabelling and embedding, samples were placed in TPP tubes (15 or 50ml), dehydrated for 1hour in methanol (20%,40%,60%,80% and 100% (2x)) under rotating agitation (Stuart, SB3). Methanol volumes were equal to about 5 times the sample volume. The samples were next incubated in a solution 67% DCM 33% MeOH overnight followed by 100% DCM during 30 min at RT on a rotator. Lastly, samples were put in 100% DBE during a minimum of 1 day for small samples (≤PCW7) or 1 week with one change after 2 days for larger samples (≥PCW10). The same clearing protocol was used to clear samples reprocessed with a second round of antibodies.
3D Imaging of specimens
Cleared samples were imaged with a Blaze light-sheet microscope (Miltenyi Biotec) equipped with sCMOS camera 5.5MP (2560×2160 pixels) controlled by Imspector Pro 7.5.3 acquisition software (Miltenyi Biotec). The light-sheet, of a 4μm thickness, was generated by lasers at four different wavelengths (488nm, 561nm, 639nm, 785nm). 1X, 4X or 12X objectives with different magnification lenses 0.6X, 1X and 1.66X were used. Samples were supported by a sample holder from Miltenyi and placed in a tank filled with DBE and illuminated by the laser light-sheet from one or both sides depending on the size of the samples. Dynamic horizontal focus, which shifts the focus through the sample while imaging, was used during acquisition to obtain sharper images. Mosaics of 3D image tiles were assembled with an overlap of 10% between the tiles. The images were acquired in a 16 bits TIFF format.
Image analysis
Images were initially processed using Oxford’s Imaris software suite to convert files and stitch mosaics, screen and annotate signal channels and perform initial recording of both 3D and z-Stack staining. Stack images were converted to imaris file (.ims) using ImarisFileConverter. For mosaic reconstruction the.ims file of each tile was imported into ImarisStitcher which performed automatic alignment which we inspected by eye and manually adjusted to create the whole reconstructed.ims file. To isolate a specific structure in Imaris, we used the surface tool with manual selection, and then used the surface to mask the image.
To facilitate the segmentation of anatomical structures, we used a virtual reality software syGlass coupled to an Oculus Quest2 (Meta) headset. Using the ROI tool, each structure was segmented in 3D using a specific channel which could be visualized as a mesh or exported as tiff files. Importantly, all segmentations were checked and validated by at least 3 persons.
To extract the segmentation made in syGlass from the raw data, a Python code was used (see key resources table).
Masks and the raw images were loaded in a Python environment using the tifffile1 library. Next, the features of each labelled mask were obtained using the regionprops.table feature of the sci-kit image library.97 After indexing the features using the pandas dataframe, the raw pixels were extracted with their original characteristics using the numpy.where part of the Numpy4 library as the images are loaded as Numpy arrays by the tifffile library.92 The generated segmented images were saved as Tiff files that can be imported as independent channels in an Imaris dataset, as they share the same dimensionality of the original datasets, allowing a separate pseudo color to be assigned. The resulting images were converted into Imaris 10.0 to be able to visualize a large number of channels together or independently.
Images and Videos were taken by using either the function Camera & Key Frame in syGlass or Snapshot and Animation in Imaris. Adobe Premiere Pro was used to add titles and transitions within the movies.
Raw Image collection as stacks of ome.tiff format files were directly archived to a server (ISILON, Dell, hosted by Genouest), and are available upon request through the Hudeca.com website. This project abides by FAIR principles (Findability, Accessibility, Interoperability, Reusability).
Verge 3D
Meshes created in syGlass were exported as.obj files. To reduce the size of mesh files, meshes were placed in a folder and Python code (https://github.com/Megumi4952/Chedotal_Lab_Head_Development/blob/main/QECD_in_Folder.ipynb) was used to re-mesh each file with Pymeshlab filter “Quadric edge collapse decimation.” An empty Verge3D (Soft8Soft) for Blender app (Version 3.9) was created and meshes were imported into Blender (Version 3.0). Multiple.obj files were imported using the add-on “blender-batch-import-wavefront-obj” (https://github.com/p2or/blender-batch-import-wavefront-obj). Meshes were scaled to 0.001 to bring them into field of view. Meshes were colored by the color picker tool on Base Color under the Principled BSDF shader in the Shading tab. Meshes were joined, and its origins reset to the center. Camera was parented to the center. The default environment.hdf file from Verge3D was used to configure lighting and background. Labels were imported as Empty Plain Axes, and renamed in the Verge3D app manager. Interactions were programmed with Verge3D Puzzles. Applications are uploaded and accessible on the Verge3D network.
For complex files, Verge3D models were built by ChromeLight Studio (Roubaix, France). Verge 3D models are available on the hudeca.com website.
3D Printing
Meshes created in syGlass were exported as.obj files and placed in a folder. Python code (https://github.com/Megumi4952/Chedotal_Lab_Head_Development/blob/main/Merge_Meshes_for_3DP.ipynb) was used to prepare the meshes for 3D printing using the package Pymeshlab.91 Meshes were imported and merged into a single file, smoothed with the filter Laplacian smooth, then simplified to have a maximum of 1,000,000 faces using the filter quadric edge collapse decimation. To combine multiple meshes into one continuous mesh, the file was binarized into an isotropic volume with 7μm voxels using the Python package Vedo.98 The volume was saved as a tiff stack with Python package TIFFFILE99 and opened in FIJI for manual correction. A surface of the volume was reconstructed with the marching cubes algorithm with resample value of 1 using the Wavefront.obj export in FIJI. Finally, the file was imported into Meshmixer for visual inspection, solidified, and exported as an STL.
Informations related to experimental design
Replication
All antibodies used in this study were used on at least 3 distinct specimens to confirm their specificity and reproducibility.
Blinding
All samples and segmentations were analyzed and interpreted by at least three researchers.
Sample-size
Except for the youngest (PCW5) and oldest (PCW13) ages, a minimum of three specimens were processed per stage as reported in Table S1A.
Inclusion-exclusion criteria
Specimens included in this head development study have been screened and included for absence of macroscopic abnormalities. The head of one young specimen (PCW5.6, EH3183) was included here despite an ectopia limited to the thoracic cavity, for which it could not be concluded whether it was a congenital malformation (ectopia cordis) or a surgical trauma.
Quantification and statistical analysis
Morphometric quantifications
To measure objects and assess their length we used a scale bar tool. In Imaris, this scale bar was automatically generated. By default, the scalebar was localized in the frontalmost plane of the virtual 3D frame containing the sample. Since the images are projections of 3D objects, the scale bar set at the frontalmost plane overestimates the dimensions of the most posterior components of the 3D sample. This overestimation is by a factor of 30%. In syGlass, the measurement scale was manually drawn at the level of the structure of interest with the dedicated caliper tool (measuring tool; distance mode). In both Imaris and syGlass, the length of the scale bar was determined by pixel number and size (dependent on microscopic acquisition settings). Pixel sizes range from 0.25 μm to 10.83 μm depending on the lens (1X, 4X or 12X) and magnification (0.6, 1, 1.66 or 2.5). The most frequent acquisition parameters were 4X lens with 0.6 magnification, generating pixels of 2.71 μm. Acquisition parameters are indicated in raw sample file names and metadata, available from our online database. The scale bars were redrawn in Adobe Photoshop (v.2024) for visual homogeneity purposes. The scale bar length was chosen between 100 μm and 3mm in adequation with sample size.
Statistical analysis
All antibody immunofluorescence experiments have been performed at least three times using identical or varying antibody combinations. Antibody immunofluorescent experiment were carried out on tissue samples of at least three human samples. The number of samples processed for each experimental condition is available in the results or in Table S1. All figures and movies, along with their full raw datasets available online, concern unique specimens and are not combinations of several samples. No statistical analysis was performed
Additional resources
We provide 3D embryo models that are accessible via a dedicated web interface. A tutorial is available in Video S6D.
Model 1: Interactive 3D reconstruction of the head arteries (SMA+) of a PCW7 human embryo generated with Verge3D. https://hudeca.com/3D-models/Inserm-foetus-responsive.html
Model 2: Interactive 3D reconstruction of the skeleton (Col2+) of a PCW7 human embryo generated with Verge3D. https://hudeca.com/3D-models/Inserm-foetus2/Inserm-foetus2-responsive.html
Acknowledgments
This work was supported by funding from INSERM HuDeca, INSERM cross-cutting program HuDeCA 2018, the Agence Nationale de la Recherche (ANR), grant ANR-19-CE16-0021-01, LaBex LIFESENSES (ANR-10-LABX-65), HU FOReSIGHT (ANR-18-IAHU-01), and Wellcome Human Developmental Biology Initiative (Wellcome Trust grant 215116/Z/18/Z). We thank the PhD program FIRE and the Graduate School EURIP of Université Paris Cité for financial support to M.I. R.B. was recipient of a fellowship from the “Fondation pour la Recherche Médicale” (FRM). E.S. is recipient of an INSERM-funded Interface Hospital fellowship (CIHU). We thank Dr Stéphane Fouquet and Morgane Belle (Vision Institute Imaging Facility), Michael Morehead, and Nathan Spencer (Istovisio) for technical assistance. This publication is part of the Human Cell Atlas, www.humancellatlas.org/publications. Alain Chédotal is a CIFAR fellow in the McMillan multiscale human program.
Author contributions
A.C. and Y.G. designed the study. A.C., E.S., G.C., R.B., and Y.G. wrote the original manuscript. All authors reviewed and edited the original manuscript before submission. F.Chalmel., M.T., P.G., and S.M.-G. collected, staged, and genotyped the samples. A.F. and R.B. processed the samples. A.A., A.F., E.S., J.B., and R.B. imaged the samples and performed the segmentations. A.C., F.Clarençon., E.S., G.C., R.B., and Y.G. analyzed the data. E.S., M.I., and R.B. generated the Verge3D models. E.H.-G. and M.I. wrote the codes. M.I. generated files for 3D printing.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in their field of research or within their geographical location.
Published: December 8, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2023.11.013.
Contributor Information
Yorick Gitton, Email: yorick.gitton@inserm.fr.
Alain Chédotal, Email: alain.chedotal@inserm.fr.
Supplemental information
A, List and age of the human embryos and fetal samples analyzed. For each sample, sex is indicated and the antibody combinations. B, list of the antibodies used in this study. C, List of head and neck muscles present (green), absent (red) or nascent (orange) between PCW5.6-PCW11.3 (sample ID is indicated). D, listing of the embryo and fetal samples used in each figure and movie.
References
- 1.Darwin C. John Murray; 1859. On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life. [PMC free article] [PubMed] [Google Scholar]
- 2.Gans C., Northcutt R.G. Neural crest and the origin of vertebrates: A new head. Science. 1983;220:268–273. doi: 10.1126/science.220.4594.268. [DOI] [PubMed] [Google Scholar]
- 3.Mai C.T., Isenburg J.L., Canfield M.A., Meyer R.E., Correa A., Alverson C.J., Lupo P.J., Riehle-Colarusso T., Cho S.J., Aggarwal D., et al. National population-based estimates for major birth defects, 2010–2014. Birth Defects Res. 2019;111:1420–1435. doi: 10.1002/bdr2.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martik M.L., Bronner M.E. Riding the crest to get a head: neural crest evolution in vertebrates. Nat. Rev. Neurosci. 2021;22:616–626. doi: 10.1038/s41583-021-00503-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oisi Y., Ota K.G., Kuraku S., Fujimoto S., Kuratani S. Craniofacial development of hagfishes and the evolution of vertebrates. Nature. 2013;493:175–180. doi: 10.1038/nature11794. [DOI] [PubMed] [Google Scholar]
- 6.Dutel H., Galland M., Tafforeau P., Long J.A., Fagan M.J., Janvier P., Herrel A., Santin M.D., Clément G., Herbin M. Neurocranial development of the coelacanth and the evolution of the sarcopterygian head. Nature. 2019;569:556–559. doi: 10.1038/s41586-019-1117-3. [DOI] [PubMed] [Google Scholar]
- 7.Martik M.L., Gandhi S., Uy B.R., Gillis J.A., Green S.A., Simoes-Costa M., Bronner M.E. Evolution of the new head by gradual acquisition of neural crest regulatory circuits. Nature. 2019;574:675–678. doi: 10.1038/s41586-019-1691-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuratani S., Ahlberg P.E. Evolution of the vertebrate neurocranium: problems of the premandibular domain and the origin of the trabecula. Zool. Lett. 2018;4 doi: 10.1186/s40851-017-0083-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Padget D.H. The development of the cranial arteries. Contrib. Embryol. 1948;32:207–261. [Google Scholar]
- 10.Streeter G.L. The development of the cranial and spinal nerves in the occipital region of the human embryo. Am. J. Anat. 1905;4:83–116. [Google Scholar]
- 11.Lesciotto K.M., Richtsmeier J.T. Craniofacial skeletal response to encephalization: how do we know what we think we know? Am. J. Phys. Anthropol. 2019;168:27–46. doi: 10.1002/ajpa.23766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neubauer S., Hublin J.-J., Gunz P. The evolution of modern human brain shape. Sci. Adv. 2018;4 doi: 10.1126/sciadv.aao5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Utsunomiya N., Katsube M., Yamaguchi Y., Yoneyama A., Morimoto N., Yamada S. The first 3D analysis of the sphenoid morphogenesis during the human embryonic period. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-08972-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lhuaire M., Martinez A., Kaplan H., Nuzillard J.M., Renard Y., Tonnelet R., Braun M., Avisse C., Labrousse M. Human developmental anatomy: microscopic magnetic resonance imaging (μMRI) of four human embryos (from Carnegie Stage 10 to 20) Ann. Anat. 2014;196:402–409. doi: 10.1016/j.aanat.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Lombardi S., Scola E., Ippolito D., Zambelli V., Botta G., Cuttin S., Triulzi F., Lombardi C.M. Micro-computed tomography: a new diagnostic tool in postmortem assessment of brain anatomy in small fetuses. Neuroradiology. 2019;61:737–746. doi: 10.1007/s00234-019-02168-2. [DOI] [PubMed] [Google Scholar]
- 16.Ueda H.R., Ertürk A., Chung K., Gradinaru V., Chédotal A., Tomancak P., Keller P.J. Tissue clearing and its applications in neuroscience. Nat. Rev. Neurosci. 2020;21:61–79. doi: 10.1038/s41583-019-0250-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Renier N., Wu Z., Simon D.J., Yang J., Ariel P., Tessier-Lavigne M. iDISCO: A simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 2014;159:896–910. doi: 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 18.Belle M., Godefroy D., Dominici C., Heitz-Marchaland C., Zelina P., Hellal F., Bradke F., Chédotal A. A simple method for 3D analysis of immunolabeled axonal tracts in a transparent nervous system. Cell Rep. 2014;9:1191–1201. doi: 10.1016/j.celrep.2014.10.037. [DOI] [PubMed] [Google Scholar]
- 19.Belle M., Godefroy D., Couly G., Malone S.A.S.A., Collier F., Giacobini P., Chédotal A. Tridimensional visualization and analysis of early human development. Cell. 2017;169:161–173.e12. doi: 10.1016/j.cell.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 20.Casoni F., Malone S.A., Belle M., Luzzati F., Collier F., Allet C., Hrabovszky E., Rasika S., Prevot V., Chédotal A., et al. Development of the neurons controlling fertility in humans: new insights from 3D imaging and transparent fetal brains. Development. 2016;143:3969–3981. doi: 10.1242/dev.139444. [DOI] [PubMed] [Google Scholar]
- 21.Radtke A.J., Kandov E., Lowekamp B., Speranza E., Chu C.J., Gola A., Thakur N., Shih R., Yao L., Yaniv Z.R., et al. IBEX: A versatile multiplex optical imaging approach for deep phenotyping and spatial analysis of cells in complex tissues. Proc. Natl. Acad. Sci. USA. 2020;117:33455–33465. doi: 10.1073/pnas.2018488117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamamoto M., Abe H., Hirouchi H., Sato M., Murakami G., Rodríguez-Vázquez J.F., Abe S. Development of the cartilaginous connecting apparatuses in the fetal sphenoid, with a focus on the alar process. PLoS One. 2021;16 doi: 10.1371/journal.pone.0251068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alatorre Warren J.L., Ponce de León M.S., Hopkins W.D., Zollikofer C.P.E. Evidence for independent brain and neurocranial reorganization during hominin evolution. Proc. Natl. Acad. Sci. USA. 2019;116:22115–22121. doi: 10.1073/pnas.1905071116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lacruz R.S., Stringer C.B., Kimbel W.H., Wood B., Harvati K., O’Higgins P., Bromage T.G., Arsuaga J.-L. The evolutionary history of the human face. Nat. Ecol. Evol. 2019;3:726–736. doi: 10.1038/s41559-019-0865-7. [DOI] [PubMed] [Google Scholar]
- 25.Jeffery N., Spoor F. Ossification and midline shape changes of the human fetal cranial base. Am. J. Phys. Anthropol. 2004;123:78–90. doi: 10.1002/ajpa.10292. [DOI] [PubMed] [Google Scholar]
- 26.Lieberman D.E., McBratney B.M., Krovitz G. The evolution and development of cranial form in Homo sapiens. Proc. Natl. Acad. Sci. USA. 2002;99:1134–1139. doi: 10.1073/pnas.022440799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.His W. [Anatomie Menschlicher Embryonen]. Vogel; 1880. [Google Scholar]
- 28.Sutton J.B. On the development and morphology of the human sphenoid bone. Proc. Zool. Soc. Lond. 1885;53:577–587. [Google Scholar]
- 29.Bastir M., Rosas A., Gunz P., Peña-Melian A., Manzi G., Harvati K., Kruszynski R., Stringer C., Hublin J.J. Evolution of the base of the brain in highly encephalized human species. Nat. Commun. 2011;2 doi: 10.1038/ncomms1593. [DOI] [PubMed] [Google Scholar]
- 30.O’rahilly R., Gardner E. The initial appearance of ossification in staged human embryos. Am. J. Anat. 1972;134:291–301. doi: 10.1002/aja.1001340303. [DOI] [PubMed] [Google Scholar]
- 31.Diogo R., Wood B.A. First Edition. CRC Press; 2012. Comparative Anatomy and Phylogeny of Primate Muscles and Human Evolution. [Google Scholar]
- 32.Cattaneo L., Pavesi G. The facial motor system. Neurosci. Biobehav. Rev. 2014;38:135–159. doi: 10.1016/j.neubiorev.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 33.Gilbert P.W. The origin and development of the human extrinsic ocular muscles. Contrib. Embryol. 1957;36:59–78. [Google Scholar]
- 34.Gasser R.F. The development of the facial muscles in man. Am. J. Anat. 1967;120:357–375. [Google Scholar]
- 35.Warmbrunn M.V., de Bakker B.S., Hagoort J., Alefs-de Bakker P.B., Oostra R.J. Hitherto unknown detailed muscle anatomy in an 8-week-old embryo. J. Anat. 2018;233:243–254. doi: 10.1111/joa.12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Couly G.F., Coltey P.M., Le Douarin N.M. The developmental fate of the cephalic mesoderm in quail-chick chimeras. Development. 1992;114:1–15. doi: 10.1242/dev.114.1.1. [DOI] [PubMed] [Google Scholar]
- 37.Harel I., Tzahor E. In: McLoon K., Andrade F., editors. Springer; 2013. Craniofacial Muscles L. [Google Scholar]
- 38.Couly G.F., Coltey P.M., Le Douarin N.M. The triple origin of skull in higher vertebrates: a study in quail-chick chimeras. Development. 1993;117:409–429. doi: 10.1242/dev.117.2.409. [DOI] [PubMed] [Google Scholar]
- 39.Buckingham M. Skeletal muscle progenitor cells and the role of Pax genes. C. R. Biol. 2007;330:530–533. doi: 10.1016/j.crvi.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 40.Abd-El-Malek S. Observations on the morphology of the human tongue. J. Anat. 1939;73:201–210.3. [PMC free article] [PubMed] [Google Scholar]
- 41.Sanders I., Mu L. A three-dimensional atlas of human tongue muscles. Anat. Rec. (Hoboken) 2013;296:1102–1114. doi: 10.1002/ar.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mu L., Sanders I. Human tongue neuroanatomy: nerve supply and motor endplates. Clin. Anat. 2010;23:777–791. doi: 10.1002/ca.21011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abe S.I., Kikuchi R., Nakao T., Cho B.H., Murakami G., Ide Y. Nerve terminal distribution in the human tongue intrinsic muscles: an immunohistochemical study using midterm fetuses. Clin. Anat. 2012;25:189–197. doi: 10.1002/ca.21201. [DOI] [PubMed] [Google Scholar]
- 44.Tawfik H.A., Dutton J.J. Embryologic and fetal development of the human orbit. Ophthal. Plast. Reconstr. Surg. 2018;34:405–421. doi: 10.1097/IOP.0000000000001172. [DOI] [PubMed] [Google Scholar]
- 45.Comai G.E., Tesařová M., Dupé V., Rhinn M., Vallecillo-García P., da Silva F., Feret B., Exelby K., Dollé P., Carlsson L., et al. Local retinoic acid signaling directs emergence of the extraocular muscle functional unit. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noden D.M., Francis-West P. The differentiation and morphogenesis of craniofacial muscles. Dev. Dyn. 2006;235:1194–1218. doi: 10.1002/dvdy.20697. [DOI] [PubMed] [Google Scholar]
- 47.Strauss D.J., Corona-Strauss F.I., Schroeer A., Flotho P., Hannemann R., Hackley S.A. Vestigial Auriculomotor activity indicates the direction of auditory attention in humans. eLife. 2020;9 doi: 10.7554/eLife.54536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liugan M., Zhang M., Cakmak Y.O. Neuroprosthetics for auricular muscles: neural networks and clinical aspects. Front. Neurol. 2018;8:1–8. doi: 10.3389/fneur.2017.00752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hannezo E., Simons B.D. Multiscale dynamics of branching morphogenesis. Curr. Opin. Cell Biol. 2019;60:99–105. doi: 10.1016/j.ceb.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 50.Quirós-Terrón L., Arráez-Aybar L.A., Murillo-González J., De-la-Cuadra-Blanco C., Martínez-Álvarez M.C., Sanz-Casado J.V., Mérida-Velasco J.R. Initial stages of development of the submandibular gland (human embryos at 5.5-8 weeks of development) J. Anat. 2019;234:700–708. doi: 10.1111/joa.12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keibel F., Mall F.P. Lippincott; 1912. Manual of Human Embryology. [Google Scholar]
- 52.Gasser R.F. The early development of the parotid gland around the facial nerve and its branches in man. Anat. Rec. 1970;167:63–77. doi: 10.1002/ar.1091670107. [DOI] [PubMed] [Google Scholar]
- 53.Chatzeli L., Gaete M., Tucker A.S. Fgf10-Sox9 are essential for establishment of distal progenitor cells during salivary gland development. Development. 2017;144:2294–2305. doi: 10.1242/dev.146019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De la Cuadra-Blanco C., Peces-Peña M.D., Mérida-Velasco J.R. Morphogenesis of the human lacrimal gland. J. Anat. 2003;203:531–536. doi: 10.1046/j.1469-7580.2003.00233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hayashi R., Okubo T., Kudo Y., Ishikawa Y., Imaizumi T., Suzuki K., Shibata S., Katayama T., Park S.J., Young R.D., et al. Generation of 3D lacrimal gland organoids from human pluripotent stem cells. Nature. 2022;605:126–131. doi: 10.1038/s41586-022-04613-4. [DOI] [PubMed] [Google Scholar]
- 56.Duke-Elder S., Cook C. In: Systems of Ophtalmology. Duke-Elder S., editor. Henry Kimpton; 1963. Normal and abnormal development. Part I. Embryology. [Google Scholar]
- 57.Papaioannou G.I., Syngelaki A., Poon L.C.Y., Ross J.A., Nicolaides K.H. Normal ranges of embryonic length, embryonic heart rate, gestational sac diameter and yolk sac diameter at 6–10 weeks. Fetal Diagn. Ther. 2010;28:207–219. doi: 10.1159/000319589. [DOI] [PubMed] [Google Scholar]
- 58.Gabriel A., Donnelly J., Kuc A., Good D., Doros G., Matusz P., Loukas M. Ectopia cordis: A rare congenital anomaly. Clin. Anat. 2014;27:1193–1199. doi: 10.1002/ca.22402. [DOI] [PubMed] [Google Scholar]
- 59.Silbergleit R., Quint D.J., Mehta B.A., Patel S.C., Metes J.J., Noujaim S.E. The persistent stapedial artery. AJNR Am. J. Neurorad. 2000;21:572–577. [PMC free article] [PubMed] [Google Scholar]
- 60.Firmin J., Ecker N., Rivet Danon D., Barraud Lange V., Turlier H., Patrat C., Maître J.-L. 2022. Mechanics of human embryo compaction. [DOI] [PubMed] [Google Scholar]
- 61.So C., Menelaou K., Uraji J., Harasimov K., Steyer A.M., Seres K.B., Bucevičius J., Lukinavičius G., Möbius W., Sibold C., et al. Mechanism of spindle pole organization and instability in human oocytes. Science. 2022;375 doi: 10.1126/science.abj3944. [DOI] [PubMed] [Google Scholar]
- 62.Hyun I., Bredenoord A.L., Briscoe J., Klipstein S., Tan T. Human embryo research beyond the primitive streak. Science. 2021;371:998–1000. doi: 10.1126/science.abf3751. [DOI] [PubMed] [Google Scholar]
- 63.Shen H. Embryo assembly 101. Nature. 2018;559:19–22. doi: 10.1038/d41586-018-05586-z. [DOI] [PubMed] [Google Scholar]
- 64.Matsuda M., Hayashi H., Garcia-Ojalvo J., Yoshioka-Kobayashi K., Kageyama R., Yamanaka Y., Ikeya M., Toguchida J., Alev C., Ebisuya M. Species-specific segmentation clock periods are due to differential biochemical reaction speeds. Science. 2020;369:1450–1455. doi: 10.1126/science.aba7668. [DOI] [PubMed] [Google Scholar]
- 65.Deglincerti A., Croft G.F., Pietila L.N., Zernicka-Goetz M., Siggia E.D., Brivanlou A.H. Self-organization of the in vitro attached human embryo. Nature. 2016;533:251–254. doi: 10.1038/nature17948. [DOI] [PubMed] [Google Scholar]
- 66.Miao Y., Djeffal Y., De Simone A., Zhu K., Lee J.G., Lu Z., Silberfeld A., Rao J., Tarazona O.A., Mongera A., et al. Reconstruction and deconstruction of human somitogenesis in vitro. Nature. 2023;614:500–508. doi: 10.1038/s41586-022-05655-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haniffa M., Taylor D., Linnarsson S., Aronow B.J., Bader G.D., Barker R.A., Camara P.G., Camp J.G., Chédotal A., Copp A., et al. A roadmap for the Human Developmental Cell Atlas. Nature. 2021;597:196–205. doi: 10.1038/s41586-021-03620-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Taylor D.M., Aronow B.J., Tan K., Bernt K., Salomonis N., Greene C.S., Frolova A., Henrickson S.E., Wells A., Pei L., et al. The pediatric cell atlas: defining the growth phase of human development at single-cell resolution. Dev. Cell. 2019;49:10–29. doi: 10.1016/j.devcel.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.He P., Lim K., Sun D., Pett J.P., Jeng Q., Polanski K., Dong Z., Bolt L., Richardson L., Mamanova L., et al. A human fetal lung cell atlas uncovers proximal-distal gradients of differentiation and key regulators of epithelial fates. Cell. 2022;185:4841–4860.e25. doi: 10.1016/j.cell.2022.11.005. [DOI] [PubMed] [Google Scholar]
- 70.Arutyunyan A., Roberts K., Troulé K., Wong F.C.K., Sheridan M.A., Kats I., Garcia-Alonso L., Velten B., Hoo R., Ruiz-Morales E.R., et al. Spatial multiomics map of trophoblast development in early pregnancy. Nature. 2023;616:143–151. doi: 10.1038/s41586-023-05869-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tyser R.C.V., Mahammadov E., Nakanoh S., Vallier L., Scialdone A., Srinivas S. Single-cell transcriptomic characterization of a gastrulating human embryo. Nature. 2021;600:285–289. doi: 10.1038/s41586-021-04158-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Popescu D.M., Botting R.A., Stephenson E., Green K., Webb S., Jardine L., Calderbank E.F., Polanski K., Goh I., Efremova M., et al. Decoding human fetal liver haematopoiesis. Nature. 2019;574:365–371. doi: 10.1038/s41586-019-1652-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Asp M., Giacomello S., Larsson L., Wu C., Fürth D., Qian X., Wärdell E., Custodio J., Reimegård J., Salmén F., et al. A spatiotemporal organ-wide gene expression and cell atlas of the developing human heart. Cell. 2019;179:1647–1660.e19. doi: 10.1016/j.cell.2019.11.025. [DOI] [PubMed] [Google Scholar]
- 74.Sountoulidis A., Marco Salas S., Braun E., Avenel C., Bergenstråhle J., Theelke J., Vicari M., Czarnewski P., Liontos A., Abalo X., et al. A topographic atlas defines developmental origins of cell heterogeneity in the human embryonic lung. Nat. Cell Biol. 2023;25:351–365. doi: 10.1038/s41556-022-01064-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang Y., Eddison M., Fleishman G., Weigert M., Xu S., Wang T., Rokicki K., Goina C., Henry F.E., Lemire A.L., et al. EASI-FISH for thick tissue defines lateral hypothalamus spatio-molecular organization. Cell. 2021;184:6361–6377.e24. doi: 10.1016/j.cell.2021.11.024. [DOI] [PubMed] [Google Scholar]
- 76.Wang X., Allen W.E., Wright M.A., Sylwestrak E.L., Samusik N., Vesuna S., Evans K., Liu C., Ramakrishnan C., Liu J., et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science. 2018;361 doi: 10.1126/science.aat5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mai H., Rong Z., Zhao S., Cai R., Steinke H., Bechmann I., Ertürk A. Scalable tissue labeling and clearing of intact human organs. Nat. Protoc. 2022;17:2188–2215. doi: 10.1038/s41596-022-00712-8. [DOI] [PubMed] [Google Scholar]
- 78.Radtke A.J., Chu C.J., Yaniv Z., Yao L., Marr J., Beuschel R.T., Ichise H., Gola A., Kabat J., Lowekamp B., et al. IBEX: an iterative immunolabeling and chemical bleaching method for high-content imaging of diverse tissues. Nat. Protoc. 2022;17:378–401. doi: 10.1038/s41596-021-00644-9. [DOI] [PubMed] [Google Scholar]
- 79.Hannezo E., Scheele C.L.G.J., Moad M., Drogo N., Heer R., Sampogna R.V., van Rheenen J., Simons B.D. A unifying theory of branching morphogenesis. Cell. 2017;171:242–255.e27. doi: 10.1016/j.cell.2017.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chatzeli L., Teshima T.H.N., Hajihosseini M.K., Gaete M., Proctor G.B., Tucker A.S. Comparing development and regeneration in the submandibular gland highlights distinct mechanisms. J. Anat. 2021;238:1371–1385. doi: 10.1111/joa.13387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kuony A., Michon F. Epithelial markers aSMA, Krt14, and Krt19 unveil elements of murine lacrimal gland morphogenesis and maturation. Front. Physiol. 2017;8 doi: 10.3389/fphys.2017.00739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Miletich I. Molecular regulation of ocular gland development. Semin. Cell Dev. Biol. 2019;91:66–74. doi: 10.1016/j.semcdb.2018.07.023. [DOI] [PubMed] [Google Scholar]
- 83.Speciale-Cirincione Über die Entwicklung der Tränendrüse beim Menschen. Graefes. Arhiv. für Ophthalmologie. 1908;69:193–230. [Google Scholar]
- 84.Yoon Y.J., Kim D., Tak K.Y., Hwang S., Kim J., Sim N.S., Cho J.M., Choi D., Ji Y., Hur J.K., et al. Salivary gland organoid culture maintains distinct glandular properties of murine and human major salivary glands. Nat. Commun. 2022;13 doi: 10.1038/s41467-022-30934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Galloway J.R., Greitz T. The medial and lateral choroid arteries. An anatomic and roentgenographic study. Acta Radiol. 1960;53:353–366. doi: 10.3109/00016926009171684. [DOI] [PubMed] [Google Scholar]
- 86.Fujii K., Lenkey C., Rhoton A.L. Microsurgical anatomy of the choroidal arteries: lateral and third ventricles. J. Neurosurg. 1980;52:165–188. doi: 10.3171/jns.1980.52.2.0165. [DOI] [PubMed] [Google Scholar]
- 87.Moffat D.B. The development of the posterior cerebral artery. J. Anat. 1961;95:485–494. [PMC free article] [PubMed] [Google Scholar]
- 88.Cullinane D.P., Franklin C., Barry D.S. Reviving the anatomic past: breathing new life into historic anatomical teaching tools. J. Anat. 2023;242:701–704. doi: 10.1111/joa.13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Titmus M., Whittaker G., Radunski M., Ellery P., Ir de Oliveira B., Radley H., Helmholz P., Sun Z. A workflow for the creation of photorealistic 3D cadaveric models using photogrammetry. J. Anat. 2023;243:319–333. doi: 10.1111/joa.13872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mai H., Luo J., Hoeher L., Al-Maskari R., Horvath I., Chen Y., Kofler F., Piraud M., Paetzold J.C., Modamio J., et al. Whole-body cellular mapping in mouse using standard IgG antibodies. Nat. Biotechnol. 2023 doi: 10.1038/s41587-023-01846-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cignoni P., Callieri M., Corsini M., Dellepiane M., Ganovelli F., Ranzuglia G. Eurographics Italian Chapter Conference. 2008. Meshlab: an open-source mesh processing tool; pp. 129–136. [Google Scholar]
- 92.Harris C.R., Millman K.J., van der Walt S.J., Gommers R., Virtanen P., Cournapeau D., Wieser E., Taylor J., Berg S., Smith N.J., et al. Array programming with NumPy. Nature. 2020;585:357–362. doi: 10.1038/s41586-020-2649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.O’rahilly R., Müller F. Developmental stages in human embryos: revised and new measurements. Cells Tissues Organs. 2010;192:73–84. doi: 10.1159/000289817. [DOI] [PubMed] [Google Scholar]
- 94.Evtouchenko L., Studer L., Spenger C., Dreher E., Seiler R.W. A mathematical model for the estimation of human embryonic and fetal age. Cell Transplant. 1996;5:453–464. doi: 10.1177/096368979600500404. [DOI] [PubMed] [Google Scholar]
- 95.Vigouroux R.J., Cesar Q., Chédotal A., Nguyen-Ba-Charvet K.T., Chedotal A., Nguyen-Ba-Charvet K.T. Revisiting the role of Dcc in visual system development with a novel eye clearing method. eLife. 2020;9 doi: 10.7554/eLife.51275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Renier N., Adams E.L., Kirst C., Wu Z., Azevedo R., Kohl J., Autry A.E., Kadiri L., Umadevi Venkataraju K., Zhou Y., et al. Mapping of brain activity by automated volume analysis of immediate early Genes. Cell. 2016;165:1789–1802. doi: 10.1016/j.cell.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.van der Walt S., Schönberger J.L., Nunez-Iglesias J., Boulogne F., Warner J.D., Yager N., Gouillart E., Yu T. scikit-image: image processing in Python. PeerJ. 2014;2 doi: 10.7717/peerj.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Musy M., et al. 2021. marcomusy/vedo: 2021.0.5 (v2021.0.5)https://zenodo.org/records/5655358 Zenodo. [Google Scholar]
- 99.Gohlke C. 2023. cgohlke/tifffile: v2023.9.18 (v2023.9.18)https://zenodo.org/records/8115278 Zenodo. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) 3D analysis of the embryonic skeleton of a PCW7 human embryo stained with Col2 antibodies and segmented with syGlass.
(B), 3D visualization of the elements of the cranium in a PCW7 human embryo.
(A) 3D visualization of the tongue and larynx muscles and their innervation in a PCW7.5 human embryo, related to Figures 2, S2, and S3.
(B) 3D visualization of extraocular muscles and oculomotor nerves in a PCW8.4 human fetus, related to Figures 3 and S4.
(C) 3D visualization of extraocular muscles, oculomotor nerves and lacrimal gland in a PCW11.3 human fetus, related to Figures 3 and S4.
(D) 3D visualization of the external, middle, and inner ear in a PCW7.5 human embryo, related to Figure S5.
(A) 3D visualization of the developing exocrine glands in a PCW7.5 human embryo, related to Figures 4 and 5.
(B) 3D visualization of the fetal lacrimal glands in a PCW10.4 human fetus, related to Figures 5 and S7.
(A) 3D Visualization of the head vasculature in a PCW7 human embryo.
(B) Development of the hyaloid vasculature from PCW7 to PCW10.4, related to Figures 7 and S7.
(A) Tutorial for Verge 3D Viewer, illustrating arteries in a PCW7 human embryo, related to Figures 1, 6, and 7.
(B) 3D organization of the circle of Willis in a PCW7 human embryo, related to Figure 7.
(C) 3D organization of the stapedial artery in PCW7 human embryo, related to Figure S7.
(D) Tutorial for the hudeca.com image database, related to all figures.
A, List and age of the human embryos and fetal samples analyzed. For each sample, sex is indicated and the antibody combinations. B, list of the antibodies used in this study. C, List of head and neck muscles present (green), absent (red) or nascent (orange) between PCW5.6-PCW11.3 (sample ID is indicated). D, listing of the embryo and fetal samples used in each figure and movie.
Data Availability Statement
-
•
All image data have been deposited at: https://hudeca.com and all original image stacks are accessible to all scientists. Downloaded and exported data is licensed under the Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0).
-
•
All original code has been deposited at GitHub and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.