Abstract
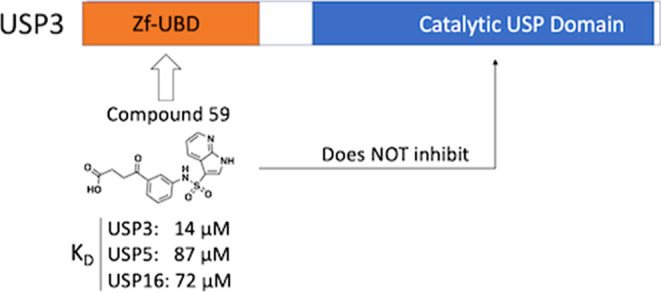
Zinc-finger ubiquitin-binding domains (ZnF-UBDs) are noncatalytic domains mostly found in deubiquitylases (DUBs) such as USP3. They represent an underexplored opportunity for the development of deubiquitylase-targeting chimeras (DUBTACs) to pharmacologically induce the deubiquitylation of target proteins. We previously showed that ZnF-UBDs are ligandable domains. Here, a focused small molecule library screen against a panel of 11 ZnF-UBDs led to the identification of compound 59, a ligand engaging the ZnF-UBD of USP3 with a KD of 14 μM. The compound binds the expected C-terminal ubiquitin binding pocket of USP3 as shown by hydrogen–deuterium exchange mass spectrometry experiments and does not inhibit the cleavage of K48-linked diubiquitin by USP3. As such, this molecule is a chemical starting point toward chemical tools that could be used to interrogate the function of the USP3 Znf-UBD and the consequences of recruiting USP3 to ubiquitylated proteins.
Introduction
Proximity-induced pharmacology is an expanding field in drug discovery in which post-translational modifications can be induced through the recruitment of protein-modifying enzymes to target proteins. Mechanistic proof-of-concept was established with proteolysis targeting chimeras (PROTACs) that induce ubiquitylation and proteasomal degradation of a target protein via the recruitment of an E3 ubiquitin ligase. Conversely, the ability to pharmacologically rescue proteins from proteasomal degradation can allow the investigation of protein function and may have therapeutic benefits in disease settings where a protein is aberrantly degraded. Bifunctional molecules presenting a chemical handle binding a noncatalytic pocket of a deubiquitylase (DUB), an enzyme that removes ubiquitin groups, may enable targeted protein deubiquitylation and subsequent target stabilization. The first reported deubiquitylase-targeting chimera (DUBTAC) recruited OTUB1, a K48 ubiquitin-specific DUB.1 The compound was composed of a covalent ligand binding an allosteric site of OTUB1, linked to a drug that binds a mutant cystic fibrosis transmembrane conductance regulator (CFTR), resulting in stabilized CFTR protein levels in human cystic fibrosis bronchial epithelial cells and restored chloride channel function.1
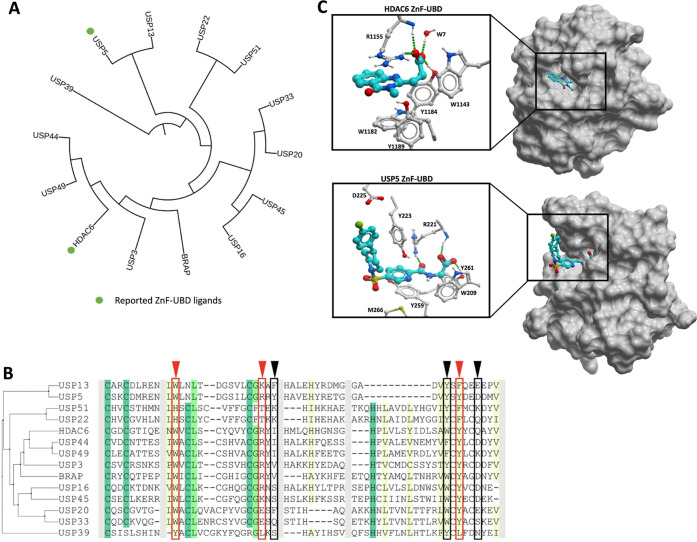
The zinc-finger ubiquitin binding domain (ZnF-UBD) is a noncatalytic structural module found in 12 members of the USP family of deubiquitylases (USP3, USP5, USP13, USP16, USP20, USP22, USP33, USP39, USP44, USP45, USP49, and USP51)2−4 with a unique fold that primarily binds the C-terminal RLRGG motif of ubiquitin (Ub). The ZnF-UBD is also found in the lysine deacetylase HDAC6 and BRAP, a BRCA-1-associated protein and ubiquitin ligase5−8 (Figure 1A). While ZnF-UBDs generally engage ubiquitin (Ub) with a low micromolar binding affinity, this function is lost in some USPs (USP13, USP20, USP22, USP33, USP39, and USP51) due to substitutions of key Ub-coordinating residues3,9−11 (Figure 1B). We previously demonstrated the ligandability of the ZnF-UBD with low micromolar ligands for the ZnF-UBD of USP5 and a nM ligand for HDAC612−16 (Figure 1A,C). The function of ZnF-UBDs and their role in the catalytic activity of DUBs are poorly understood. We have shown that ligands targeting the ZnF-UBD of USP5 inhibit its catalytic activity12,13 and therefore cannot be used as chemical handles for the development of USP5-recruiting DUBTACs, but the same may not be true for small molecules targeting the ZnF-UBD of other USPs.
Figure 1.
Human ZnF-UBDs: (A) (Left) Dendrogram of full-length human proteins that contain a ZnF-UBD. The dendrogram was created by first doing a multiple sequence alignment using Clustal Omega,17 followed by tree generation with iTol.18 Reported ZnF-UBD ligands are highlighted with green circles. (B) Sequence alignment of human ZnF-UBDs. Conserved residues are indicated with shading. USP5 residues that interact with Ub are indicated with arrowheads and boxes (red: expected to be essential for Ub binding; black: not essential). (C) Co-crystal structures of HDAC6 and USP5 ZnF-UBD in complex with ligands (PDB: 6CED, 7MS7).
Here, we report the discovery and characterization of USP3 ZnF-UBD ligands. Screening of a small, focused chemical library against a panel of ZnF-UBDs led to the discovery of a moderately selective USP3 ligand that does not inhibit the catalytic function of USP3. Our work provides a chemical starting point for the development of more potent USP3 ZnF-UBD ligands that can serve as tools to study the function of the USP3 ZnF-UBD or bifunctional molecules to recruit USP3 to ubiquitylated proteins.
Results
Discovery of a Ligand Binding the ZnF-UBD of USP3
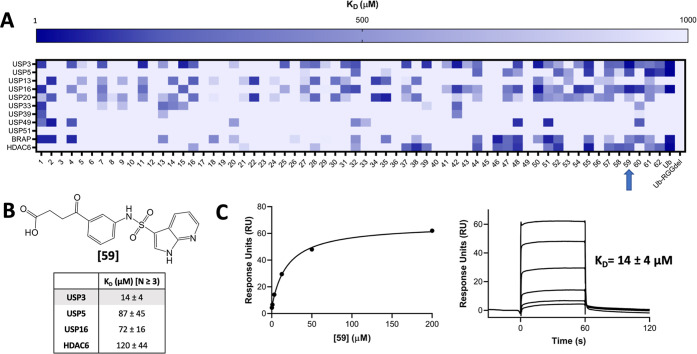
Following our observation that an aliphatic chain ending with a carboxylic acid mimicking the endogenous substrate of the ZnF-UBD of HDAC6 and USP5 was essential for binding (Figure 1C), we compiled a focused library of 670 molecules bearing this moiety from our in-house collection to target the Ub-binding ZnF-UBDs of USP3, USP5, USP13, USP16, USP20, USP22, USP33, USP39, USP49, USP51, BRAP and HDAC6. Compounds were grouped into 62 clusters with ICM (Molsoft) based on their chemical similarity, and the binding of 62 representative molecules to our panel of 11 ZnF-UBDs was measured in a surface plasmon resonance (SPR) assay. Ub and a C-terminal RGG-deleted Ub construct were used as positive and negative controls, respectively (Figure 2A, Tables S1 and S2). As expected, no binding to Ub was observed for USP13, USP20, USP33, USP39, and USP51, but low micromolar binding was observed for USP3, USP5, USP16, BRAP, and HDAC6. No binding to RGG-deleted Ub was observed by any ZnF-UBD protein. Interestingly, no Ub binding was observed for USP49, despite the conservation of Ub-binding residues.
Figure 2.
Small molecule screen identifies ZnF-UBD hits. (A) Heat map showing binding of 64 ligands to 11 ZnF-UBDs by SPR. A 4-fold 6-point dilution series beginning at 200 μM was used for KD determination with N = 1. (B) Summary of binding data for compound 59 by SPR, N ≥ 3. (C) Representative SPR binding curve from steady-state fit analysis and sensorgrams for USP3 ZnF-UBD and 59. A KD of 14 ± 4 μM was obtained from the average of seven independent measurements.
Many ligands from the screen bound USP3 but few showed selectivity for USP3 over other ZnF-UBDs. Compound 59 was identified and confirmed as a promising USP3 hit and was more than 5-times selective for USP3 over USP5, USP16, and HDAC6 ZnF-UBDs (KD: USP3 = 14 ± 4 μM; USP5 = 87 ± 45 μM; USP16 = 72 ± 16 μM; HDAC6 = 120 ± 44 μM) (Figure 2B,C and Table S2). Although more selective compounds were identified, including 25 and 39, their affinity for USP3 was substantially weaker (KD: 191 and 102 μM respectively) and they were not pursued within the confines of this project. We note that 59 contains a pyrrolopyridine moiety which is a known hinge binder for ATP-competitive kinase inhibitors. In a profiling experiment, 59 inhibited only one of the 58 kinases screened (44% inhibition of Aurora kinase A at 10 μM. Table S3). This confirms that the compound is selective overall and that kinase inhibition should be monitored in future hit optimization.
Compound 59 Does Not Perturb the Enzymatic Activity of USP3
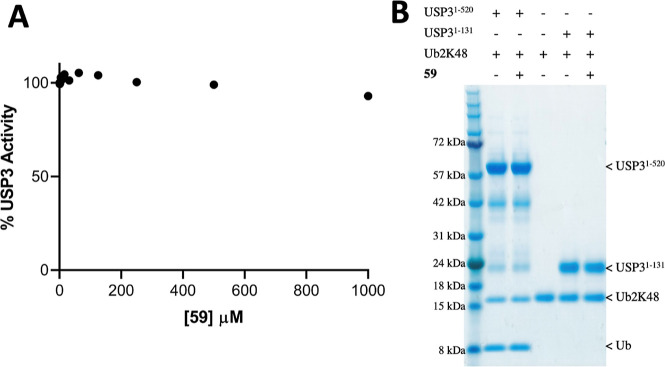
We next used a fluorogenic ubiquitin rhodamine assay to test whether 59 inhibited USP3 DUB catalytic activity.19 We found that the enzyme remains fully active even in the presence of a 1 mM compound (Figure 3A). This is in contrast with USP5, where ligands targeting the ZnF-UBD reduce enzymatic activity to a basal level.13 To verify that binding of the C-terminal Ub tail to the ZnF-UBD of USP3 was not necessary for the cleavage of endogenous substrates, we used a gel-based assay to monitor the cleavage of a K48-linked diubiquitin substrate in the absence and presence of 59. Again, we saw no sign of inhibition of deubiquitylase activity, suggesting that USP3 ZnF-UBD targeting ligands could be used as chemical starting points for USP3-recruiting DUBTACs (Figure 3B). For both assays, no positive control catalytic inhibitor could be included, as no USP3 inhibitors have been described to date in the literature.
Figure 3.
59 does not inhibit USP3 catalytic activity. (A) UbRho110 assay (N = 2). Fluorescence signal is normalized to the control [no compound, dimethyl sulfoxide (DMSO) only]. (B) Cleavage of Ub2K48 in the absence or presence of 59 (70 μM final concentration).
Compound 59 Binds the C-Terminal Ubiquitin Binding Site of USP3
The structure of USP3 ZnF-UBD is not available from the PDB and, despite extensive crystallization screening efforts, we were unable to solve it by X-ray crystallography, in either its apo form or in complex with Ub or 59. To identify the binding site of 59 and characterize the binding event’s corresponding allosteric effects, hydrogen–deuterium exchange mass spectrometry (HDX-MS) was used.20,21 A sequence coverage of over 90% was obtained for USP3 ZnF-UBD with a peptide per amino acid redundancy of 5.24 (Table S3 and Figure S1).
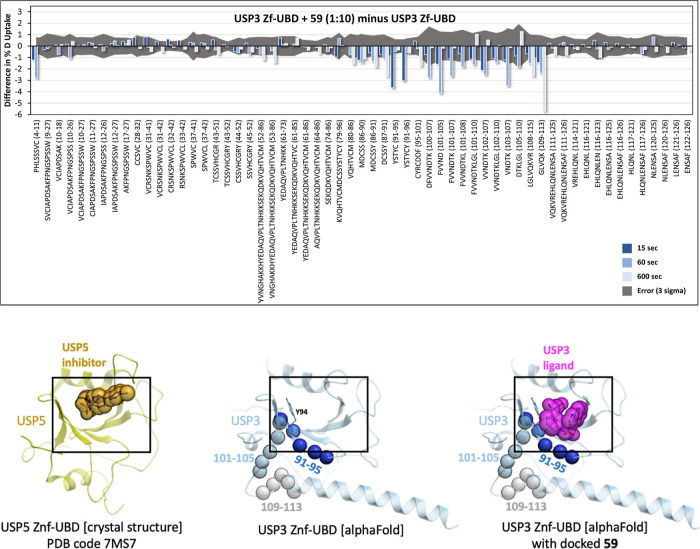
HDX-MS analysis was performed for USP3 ZnF-UBD alone and in complex with 59 (1:10 and 1:20) for 15, 60, and 600 s at 20 °C (Figures 4 and S1 and S2). Given the 14 μM equilibrium dissociation constant (KD), the bound fractions in the 1:10 and 1:20 states upon D2O buffer addition were expected to be 40.1 and 58.1%, respectively. The HDX data described here are differential (deuterium uptake of bound state minus unbound state), in technical triplicate (n = 3), and both the 1:10 and 1:20 ligand ratios show consistent results. To be considered statistically significant, differences must have exceeded triple the propagated standard error. Complexation resulted in significant attenuation of deuterium uptake rates in 20 of 59 peptides, all localized to the same region (Figure 4). At the earliest time point (15 s), a high-intensity, negative uptake difference was observed at residues 91–96 (YSTYCY), with the magnitude of the difference decreasing rapidly over the next two time points (1 and 10 min). Significant uptake differences appear for peptides 5–10 (HLSSSVC) and 102–107 (VVNDTK) at the 1 min time point. Finally, residues 110–113 (LVQK) exhibit a slowly evolving decrease deuterium uptake, whose magnitude becomes the dominant difference signal at the longest measured time point (10 min).
Figure 4.
Mapping of the ligand binding site by HDX-MS. Top: the difference in deuterium fractional uptake (%) between USP3 ZnF-UBD + 59 (1:10) and USP3 ZnF-UBD is plotted as a function of each time point per peptide. The time points are 15 s (dark blue), 60 s (light blue), and 600 s (light grey). The error (3 times the propagated error) is shown as a dark gray area. If the bars exceed this gray area, they are statistically significant. Bars pointing down indicate a decrease in the level of deuterium uptake in the presence of the ligand. Bottom: HDX signals observed at 15, 60, and 600 s (same color-coding as above) are highlighted as spheres on an AlphaFold prediction of the USP3 ZnF-UBD structure (center). A crystal structure of the homologous USP5 ZnF-UBD bound to a ligand (PDB ID: 7MS7) (left) and a model of 59 docked to the USP3 ZnF-UBD AlphaFold model prediction (right) are shown as references.
Together, these data support a direct binding event at residues 91–95 (as discussed below), which superimpose with the binding pocket of an analogous compound cocrystallized in complex with the ZnF-UBD of USP5 (Figure 4).13 A predicted model of 59 bound to this site shows that the compound is expected to occlude residues 91–95 from the solvent, in agreement with the decrease in deuterium exchange observed immediately at this site upon treatment with 59 (Figure 4). We also note that most side chains lining the binding pocket delineated by our docking model and confirmed by HDX are not conserved across ZnF-UBDs and some are unique to USP3, supporting the notion that this site can be targeted by USP3-selective ligands (Figure S3).
Discussion
USP3 is a nuclear DUB that primarily deubiquitylates and stabilizes targets for maintenance of genome stability, regulation of cell proliferation, DNA damage response, and the innate immune response.22−27 There are currently no selective USP3 inhibitors due to the high conservation of the catalytic USP domain. The ZnF-UBD of USP3 is required for its interaction with and deubiquitylation of targets such as histone H2A and RIG-I.22,25
Here, we identified a ligand of USP3 ZnF-UBD that does not inhibit the enzymatic activity of full-length USP3. We were unable to solve the crystal structure of the USP3 in complex with 59 but used HDX-MS to map its binding site. While our HDX data indicate a direct binding event at a site known in the context of USP5 to recognize the C-terminal extremity of Ub,2 and to be exploited by chemical analogs of 59,13 we are also observing deuterium exchange at an adjacent site (Figure 4). Attenuated deuterium uptake kinetics in the presence of a ligand can arise from “direct” effects (i.e., new intermolecular hydrogen bonding or solvent exclusion resulting from direct contact between the protein and the ligand) or “induced” effects (i.e., increased intramolecular hydrogen-bond stability upon binding, resulting in reduced sampling of a broader conformational ensemble accessible to the native, unbound protein).20,21,28 HDX uptake kinetics can therefore provide insight on whether an observed decrease in deuterium uptake in a particular region is a direct or allosteric effect of ligand binding.29 Briefly, this is because direct effects are a function of the rate of binding equilibration (i.e., kon + koff), which in this case will cause an “early” time of maximum difference in the HDX data that rapidly diminishes due to the high koff of 59 (as observed in the SPR sensorgrams, see Figure 2C). Indeed, the HDX data indicate that 59 makes direct contact at residues 91–95, which uniquely exhibits the expected direct effect difference kinetics profile. Notably, this agrees exactly with a critical π-stacking in the docked model between the acetophenone group of 59 and Y94 (Figure 4), corresponding to the π-stacking of the crystallized USP5 ligand with Y259. All other significant decreases in uptake, corresponding to residues 102–107, and 110–113, are near the predicted binding pocket, but exhibit slower uptake kinetics, indicative of induced (orthosteric) effects. All HDX differences observed localize to one region of USP3, highlighting the same pocket verified in an analogous structure, PDB 7MS7. Here, the USP5 ZnF-UBD is bound to compound N-{5-[4-(4-chlorophenyl)piperidine-1-sulfonyl]pyridine-2-carbonyl}glycine (ZQ1), which shares a similar chemical scaffold to 59.13 In the USP5 structure, there is a close π-stacking interaction between the pyridine moieties of ZQ1 and Y259, which is analogous to the HDX-supported 59/USP3 Y94 interaction discussed above.
Compound 59 binds USP3 with a KD of around 14 μM, which would not be sufficient for a traditional occupancy-based competitor. Proximity-inducing pharmacological agents such as PROTACs bind at the interface of proteins and exploit, rather than compete against protein–protein interactions.30−32 As such, they can sometimes be derived from chemical handles with a relatively low affinity. As an example, Apcin, a ligand binding the E3 ligase CDC20 with a KD of 10 μM, was successfully used as a chemical handle for the development of a CDC20-recruiting PROTAC.33
Interestingly, USP3 is not inhibited by 59, although we previously found that ligands targeting the same domain of USP5 do inhibit its catalytic function.13 This suggests distinct structure–function relationships of Znf-UBD domains across the USP family and supports the continued development of ligands for these domains to further interrogate this biology.
In conclusion, 59 may be a good chemical starting point for the development of protein–protein interaction inhibitors that would antagonize the binding of USP3 to some of its endogenous targets, given that they occupy the same pocket as the C-terminal diglycine motif. Conversely, 59 or more potent compounds could serve as chemical handles for the development of bifunctional molecules that recruit USP3 to other proteins, which may lead to their deubiquitylation. While our docked model of 59 (Figure 4) and previously published ligands targeting other ZnF-UBD proteins12−16 (Figure 1) indicate that the carboxylic moiety of the compound is deeply buried in the binding pocket, the pyrrolopyridine ring is predicted to be partially solvent exposed and may serve as an anchor point for linker attachment for the development of proximity pharmacology agents. In addition to 59, the focused chemical library screen also identified hits against other ZnF-UBDs, such as USP16 and BRAP (50 and 46, respectively) which require further validation, but may be promising starting points for hit expansion for these other disease-relevant protein targets.34,35
Materials and Methods
Cloning, Protein Expression, and Purification
cDNAs encoding Ub1–76, Ub1–73, USP5171–290, and HDAC61109–1215 were cloned as previously described.12−15 DNA encoding USP31–131, USP1625–185, USP201–141, USP3329–134, USP3984–194, USP491–115, USP51176–305, and BRAP304–390 were subcloned into a modified vector encoding an N-terminal AviTag for biotinylation and a C-terminal His6 tag (p28BIOH-LIC), while USP13183–307 was subcloned into a vector with an N-terminal His-tag and TEV protease cleavage site, and a C-terminal biotinylation sequence (pNICBIO2). USP31–520 was subcloned into a pFBOH-LIC vector and overexpressed in Sf9 cells, where cultures were grown in HyQ SFX Insect Serum Free Medium (Fisher Scientific) to a density of 4 × 106 cells/mL and infected with 10 mL of P3 viral stock media per 1 L of cell culture. Cell culture medium was collected after 4 days of incubation in a shaker at 27 °C. Proteins were purified as before;12−15 briefly, all proteins were purified by metal affinity chromatography, gel filtration, and ion exchange chromatography. The final concentration of purified proteins was measured by the UV absorbance at 280 nm. Protein identity was confirmed by mass spectrometry, and purity was assessed by SDS-PAGE.
Surface Plasmon Resonance Assay
Studies were performed with a Biacore T200 (GE Health Sciences) at 20 °C. Approximately, 3000–6000 response units (RU) of biotinylated ZnF-UBDs were captured to flow cells of a streptavidin-conjugated SA chip per the manufacturer’s protocol, and an empty flow cell was used for reference subtraction. Serial dilutions were prepared in 20 mM N-(2-hydroxyethyl)piperazine-N′-ethanesulfonic acid (HEPES) at pH 7.4, 150 mM NaCl, 1 mM TCEP, 0.005% Tween-20 (v/v), and 1% DMSO (v/v). KD determination experiments were performed using multicycle kinetics with 60 s contact time, and 60 s dissociation time at a flow rate of 30 μL/min at 20 °C. KD values were calculated using steady-state affinity fitting with the Biacore T200 evaluation software (GE Health Sciences).
Ubiquitin-Rhodamine 110 Assay
Experiments were performed in a total volume of 60 μL in 384-well black polypropylene microplates (Grenier). Fluorescence was measured using a Biotek Synergy H1 microplate reader (Biotek) at excitation and emission wavelengths of 485 and 528 nm, respectively. Ligands were prepared in 20 mM Tris pH 7.5, 125 mM NaCl, 1 mM DTT, 0.01% TX-100 (v/v), and 1% DMSO (v/v) for a 2-fold dilution titration series. 500 nM USP31–520 and 200 nM ubiquitin-rhodamine 110 (UBPBio) were added to each well. Following a 1 min centrifugation at 250g, fluorescence readings were immediately taken for 10 min. The data were analyzed using GraphPad Prism 8.2.0.
Ub2K48 Cleavage Assay
70 pmoles each of USP3 full-length (USP31–520) or USP3 ZnF-UBD (USP31–131) were incubated in 10 mM HEPES pH7.5, 150 mM NaCl, 3 mM EDTA, 0.05% Tween-20, 1 mM TCEP with or without compound 59 at 10-fold molar excess, for 1 h at room temperature. 60 pmoles of Ub2K48 (Boston Biochem) were added to all reactions, bringing the final volume to 10 μL and reactions were incubated for 1 h at room temperature. Reactions were stopped by the addition of SDS-PAGE loading dye to a final concentration of 1× and incubation at 95 °C for 5 min. Reactions were analyzed by Coomassie-stained SDS-PAGE using NuPAGE 4–12% Bis-Tris gel Invitrogen).
Kinase Profiling
Kinase profiling was conducted at Eurofins, with a radiometric activity assay in the presence of 10 μM of 59.
Hydrogen–Deuterium Exchange Mass Spectrometry
To begin, three samples were prepared and incubated for 30 min at 0 °C: 7.5 μM USP3 ZnF-UBD (apo), 7.5 μM USP3 ZnF-UBD and 75 μM 59 (1:10 complex), and 7.5 μM USP3 ZnF-UBD and 150 μM 59 (1:20 complex). All samples were then held at 0 °C prior to mixing 8 μL of the sample with 52 μL of buffered D2O (10 mM phosphate buffer pD 7.5, 150 mM NaCl) to yield a final D2O concentration of 87%. The HDX reaction was allowed to take place for 15, 60, or 600 s at 20 °C. Then, 50 μL of the HDX reaction was quenched for 1 min at 0 °C with 50 μL Quench buffer (100 mM phosphate buffer, pH 2.5) to stop the HDX reaction. Next, 50 μL (25 pmol) of the quenched sample was loaded onto a mixed Nepenthensin-2 Pepsin (1:1) column (Affipro) and desalted (ACQUITY UPLC BEH C18 VanGuard Precolumn, 130 Å, 1.7 μm, 2.1 mm × 5 mm, Waters) using Mobile Phase A (0.1% formic acid in water) at 200 μL/min for 3 min. The peptides were then reverse-phase separated (ACQUITY UPLC BEH C18 Column, 130 Å, 1.7 μm, 2.1 mm × 50 mm, Waters) at 40 μL/min using a gradient from 5 to 35% Mobile Phase B (0.1% formic acid in acetonitrile). Eluted peptides were electrosprayed into the Select Series Cyclic IMS (Q-IMS-TOF, Waters) while Leu-Enk was used for lockspray. Fragmentation was conducted using collision-induced dissociation in the HDMSe mode with a collisional energy ramp from 20 to 29 V (in the transfer cell). To obtain undeuterated peptides, the above steps were followed except in place of deuteration buffer, and an equilibration buffer was used (10 mM Phosphate Buffer pH 7.5, 150 mM NaCl). HDX was automated using the ACQUITY UPLC M-Class System with HDX Technology (Waters) and PAL3 liquid handling (CTC Analytics AG). Peptide identification was conducted using ProteinLynx Global Server 3.0.3 (PLGS, Waters). HDX analysis and visualization were conducted using DynamX 3.0 (Waters) and PyMOL 2.5.0.
Molecular Modeling
The model of USP3 predicted by AlphaFold was downloaded from https://alphafold.ebi.ac.uk/ and superimposed to the crystal structure of USP5 ZnF-UBD in a complex with a chemical analog of 59. Compound 59 was then docked to the corresponding pocket using a grid representation of the receptor with ICM (Molsoft, San Diego). The system was further relaxed with 100 ns molecular dynamics (MD) simulations with ICM using OpenMM.36 Six independent simulations did not all converge to the same pose, reflecting the challenge of docking to the AlphaFold structures. The binding pose shown in Figure 4 was from one of the most stable simulations (Figure S4) and is provided as an example. All binding poses were at the same site and occluded residues 91–95.
Chemistry
The purity of compound 59 was confirmed to be greater than 95% by LCMS.
Acknowledgments
The Structural Genomics Consortium is a registered charity (no: 1097737) that receives funds from Bayer AG, Boehringer Ingelheim, Bristol Myers Squibb, Genentech, Genome Canada through Ontario Genomics Institute [OGI-196], EU/EFPIA/OICR/McGill/KTH/Diamond Innovative Medicines Initiative 2 Joint Undertaking [EUbOPEN grant 875510], Janssen, Merck KGaA (aka EMD in Canada and US), Pfizer and Takeda. We thank Dr. Vijayaratnam Santhakumar for his advice and Dr. Radek Laufer at OICR for providing kinase profiling data from Eurofins.
Glossary
Abbreviations
- BRAP
BRCA-1-associated protein
- DMSO
dimethyl sulfoxide
- DUB
deubiquitylase
- DUBTAC
deubiquitylase targeting chimera
- HDAC6
histone deacetylase 6
- HDX
hydrogen–deuterium exchange
- KD
dissociation constant
- K48
lysine 48
- MD
molecular dynamics
- PROTAC
proteolysis targeting chimera
- MS
mass spectrometry
- SD
standard deviation
- SPR
surface plasmon resonance
- Ub
ubiquitin
- USP
ubiquitin-specific protease
- ZnF-UBD
ubiquitin-specific processing protease zinc-finger
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c07070.
Table S1: compounds smiles strings and catalog numbers, Table S2: matrix Screen results, Table S3: kinase profiling; Table S4: PLGS filtering parameters, Figure S1: HDX sequence coverage of USP31–131, Figure S2: differential HDX heatmaps, Figure S3: diversity of sidechains lining the binding pocket; Figure S4: ligand RMSD over 100 ns MD simulation (PDF)
Author Contributions
The manuscript was written by M.K.M., E.W., D.J.W, and M.S. All authors have given approval to the final version of the manuscript. M.K.M. expressed and purified proteins, designed and optimized biophysical assays and tested compounds., B.W conducted analytical chemistry. E.W conducted and D.J.W. supervised HDX experiments. H.K. conducted molecular modeling and simulations. A.B. dissolved, plated, and managed compounds. R.J.H. and M.S. advised throughout the project.
M.S. gratefully acknowledges support from NSERC [grant RGPIN-2019-04416] and MITACS Accelerate (IT13051). M.K.M. acknowledges CQDM (Quantum Leap-176) and the Peterborough K.M. Hunter Charitable Foundation for financial support.
The authors declare no competing financial interest.
Supplementary Material
References
- Henning N. J.; Boike L.; Spradlin J. N.; Ward C. C.; Liu G.; Zhang E.; Belcher B. P.; Brittain S. M.; Hesse M. J.; Dovala D.; McGregor L. M.; Valdez Misiolek R.; Plasschaert L. W.; Rowlands D. J.; Wang F.; Frank A. O.; Fuller D.; Estes A. R.; Randal K. L.; Panidapu A.; McKenna J. M.; Tallarico J. A.; Schirle M.; Nomura D. K. Deubiquitinase-Targeting Chimeras for Targeted Protein Stabilization. Nat. Chem. Biol. 2022, 18 (4), 412–421. 10.1038/s41589-022-00971-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu F. E.; Horton J. R.; Mullally J. E.; Heroux A.; Cheng X.; Wilkinson K. D. The Ubiquitin Binding Domain ZnF UBP Recognizes the C-Terminal Diglycine Motif of Unanchored Ubiquitin. Cell 2006, 124, 1197–1208. 10.1016/j.cell.2006.02.038. [DOI] [PubMed] [Google Scholar]
- Bonnet J.; Romier C.; Tora L.; Devys D. Zinc-Finger UBPs: Regulators of Deubiquitylation. Trends Biochem. Sci. 2008, 33 (8), 369–375. 10.1016/j.tibs.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Pai M. T.; Tzeng S. R.; Kovacs J. J.; Keaton M. A.; Li S. S. C.; Yao T. P.; Zhou P. Solution Structure of the Ubp-M BUZ Domain, a Highly Specific Protein Module That Recognizes the C-Terminal Tail of Free Ubiquitin. J. Mol. Biol. 2007, 370, 290–302. 10.1016/j.jmb.2007.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheny S. A.; Chen C.; Kortum R. L.; Razidlo G. L.; Lewis R. E.; White M. A. Ras Regulates Assembly of Mitogenic Signalling Complexes through the Effector Protein IMP. Nature 2004, 427, 256–260. 10.1038/nature02237. [DOI] [PubMed] [Google Scholar]
- Hook S. S.; Orian A.; Cowley S. M.; Eisenman R. N. Histone Deacetylase 6 Binds Polyubiquitin through Its Zinc Finger (PAZ Domain) and Copurifies with Deubiquitinating Enzymes. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 13425–13430. 10.1073/pnas.172511699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L.; Schubert H. L.; Hill C. P. Ubiquitin-Binding Domains. Nat. Rev. Mol. Cell Biol. 2005, 6, 610–621. 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- Boyault C.; Gilquin B.; Zhang Y.; Rybin V.; Garman E.; Meyer-Klaucke W.; Matthias P.; Müller C. W.; Khochbin S. HDAC6-P97/VCP Controlled Polyubiquitin Chain Turnover. EMBO J. 2006, 25 (14), 3357–3366. 10.1038/sj.emboj.7601210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. H.; Zhou C. J.; Zhou Z. R.; Song A. X.; Hu H. Y. Domain Analysis Reveals That a Deubiquitinating Enzyme Usp13 Performs Non-Activating Catalysis for Lys63-Linked Polyubiquitin. PLoS One 2011, 6, e29362 10.1371/journal.pone.0029362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Ding Y.; Zhou C.; Wen Y.; Zhang N. Structural and Functional Studies of USP20 ZnF-UBP Domain by NMR. Protein Sci. 2019, 28 (9), 1606–1619. 10.1002/pro.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen M. D.; Bycroft M. The Solution Structure of the ZnF UBP Domain of USP33/VDU1. Protein Sci. 2007, 16 (9), 2072–2075. 10.1110/ps.072967807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M. K.; Franzoni I.; De Freitas R. F.; Tempel W.; Houliston S.; Smith L.; Vedadi M.; Arrowsmith C. H.; Harding R. J.; Schapira M. Discovery of Small Molecule Antagonists of the USP5 Zinc Finger Ubiquitin-Binding Domain. J. Med. Chem. 2019, 62, 10144–10155. 10.1021/acs.jmedchem.9b00988. [DOI] [PubMed] [Google Scholar]
- Mann M. K.; Zepeda-Velázquez C. A.; González-Álvarez H.; Dong A.; Kiyota T.; Aman A. M.; Loppnau P.; Li Y.; Wilson B.; Arrowsmith C. H.; Al-Awar R.; Harding R. J.; Schapira M. Structure-Activity Relationship of USP5 Inhibitors. J. Med. Chem. 2021, 64 (20), 15017–15036. 10.1021/acs.jmedchem.1c00889. [DOI] [PubMed] [Google Scholar]
- Harding R. J.; Ferreira De Freitas R.; Collins P.; Franzoni I.; Ravichandran M.; Ouyang H.; Juarez-Ornelas K. A.; Lautens M.; Schapira M.; Von Delft F.; Santhakumar V.; Arrowsmith C. H. Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin. J. Med. Chem. 2017, 60, 9090–9096. 10.1021/acs.jmedchem.7b00933. [DOI] [PubMed] [Google Scholar]
- Ferreira De Freitas R.; Harding R. J.; Franzoni I.; Ravichandran M.; Mann M. K.; Ouyang H.; Lautens M.; Santhakumar V.; Arrowsmith C. H.; Schapira M. Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors. J. Med. Chem. 2018, 61, 4517–4527. 10.1021/acs.jmedchem.8b00258. [DOI] [PubMed] [Google Scholar]
- Harding R. J.; Franzoni I.; Mann M. K.; Szewczyk M. M.; Mirabi B.; Ferreira de Freitas R.; Owens D. D. G.; Ackloo S.; Scheremetjew A.; Juarez-Ornelas K. A.; Sanichar R.; Baker R. J.; Dank C.; Brown P. J.; Barsyte-Lovejoy D.; Santhakumar V.; Schapira M.; Lautens M.; Arrowsmith C. H. Discovery and Characterization of a Chemical Probe Targeting the Zinc-Finger Ubiquitin-Binding Domain of HDAC6. J. Med. Chem. 2023, 66 (15), 10273–10288. 10.1021/acs.jmedchem.3c00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F.; Wilm A.; Dineen D.; Gibson T. J.; Karplus K.; Li W.; Lopez R.; McWilliam H.; Remmert M.; Söding J.; Thompson J. D.; Higgins D. G. Fast, Scalable Generation of High-Quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2011, 7 (1), 539. 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]; School of Medicine and Medical Science, UCD Conway Institute of Biomolecular and Biomedical Research, University College Dublin, Dublin, Ireland
- Letunic I.; Bork P. Interactive Tree Of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49 (W1), W293–W296. 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varca A. C.; Casalena D.; Auld D.; Buhrlage S. J. Identification of Deubiquitinase Inhibitors via High-Throughput Screening Using a Fluorogenic Ubiquitin-Rhodamine Assay. STAR Protoc. 2021, 2 (4), 100896. 10.1016/j.xpro.2021.100896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James E. I.; Murphree T. A.; Vorauer C.; Engen J. R.; Guttman M. Advances in Hydrogen/Deuterium Exchange Mass Spectrometry and the Pursuit of Challenging Biological Systems. Chem. Rev. 2022, 122 (8), 7562–7623. 10.1021/acs.chemrev.1c00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oganesyan I.; Lento C.; Wilson D. J. Contemporary Hydrogen Deuterium Exchange Mass Spectrometry. Methods 2018, 144, 27–42. 10.1016/j.ymeth.2018.04.023. [DOI] [PubMed] [Google Scholar]
- Nicassio F.; Corrado N.; Vissers J. H. A.; Areces L. B.; Bergink S.; Marteijn J. A.; Geverts B.; Houtsmuller A. B.; Vermeulen W.; Di Fiore P. P.; Citterio E. Human USP3 Is a Chromatin Modifier Required for S Phase Progression and Genome Stability. Curr. Biol. 2007, 17, 1972–1977. 10.1016/j.cub.2007.10.034. [DOI] [PubMed] [Google Scholar]
- Liang R.-P.; Zhang X.-X.; Zhao J.; Zhu R.-T.; Wang W.-J.; Lu Q.-W.; Sun Y.-L. Ubiquitin-Specific Protease 3 Facilitates Cell Proliferation by Deubiquitinating Pyruvate Kinase L/R in Gallbladder Cancer. Lab. Invest. 2022, 102 (12), 1367–1376. 10.1038/s41374-022-00836-1. [DOI] [PubMed] [Google Scholar]
- Zhuang W.; Zhang L.; Zheng Y.; Liu B.; Ma C.; Zhao W.; Liu S.; Liu F.; Gao C. USP3 Deubiquitinates and Stabilizes the Adapter Protein ASC to Regulate Inflammasome Activation. Cell. Mol. Immunol. 2022, 19 (10), 1141–1152. 10.1038/s41423-022-00917-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J.; Song Y.; Li Y.; Zhu Q.; Tan P.; Qin Y.; Wang H. Y.; Wang R.-F. USP3 Inhibits Type I Interferon Signaling by Deubiquitinating RIG-I-like Receptors. Cell Res. 2014, 24 (4), 400–416. 10.1038/cr.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N.; Zhu Q.; Wani G.; He J.; Wang Q.; Wani A. A. USP3 Counteracts RNF168 via Deubiquitinating H2A and ΓH2AX at Lysine 13 and 15. Cell Cycle 2014, 13 (1), 106–114. 10.4161/cc.26814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.; Qin J.; Li F.; Yang C.; Li Z.; Zhou Z.; Zhang H.; Li Y.; Wang X.; Liu R.; Tao Q.; Chen W.; Chen C. USP3 Promotes Breast Cancer Cell Proliferation by Deubiquitinating KLF5. J. Biol. Chem. 2019, 294 (47), 17837–17847. 10.1074/jbc.RA119.009102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narang D.; Lento C.; J Wilson D. HDX-MS: An Analytical Tool to Capture Protein Motion in Action. Biomedicines 2020, 8 (7), 224. 10.3390/biomedicines8070224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen T. J. D.; Gårdsvoll H.; Danø K.; Roepstorff P.; Ploug M. Dynamics of Urokinase Receptor Interaction with Peptide Antagonists Studied by Amide Hydrogen Exchange and Mass Spectrometry. Biochemistry 2004, 43 (47), 15044–15057. 10.1021/bi048706j. [DOI] [PubMed] [Google Scholar]
- Békés M.; Langley D. R.; Crews C. M. PROTAC Targeted Protein Degraders: The Past Is Prologue. Nat. Rev. Drug Discov. 2022, 21 (3), 181–200. 10.1038/s41573-021-00371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirnomas D.; Hornberger K. R.; Crews C. M. Protein Degraders Enter the Clinic - a New Approach to Cancer Therapy. Nat. Rev. Clin. Oncol. 2023, 20 (4), 265–278. 10.1038/s41571-023-00736-3. [DOI] [PubMed] [Google Scholar]
- Schapira M.; Calabrese M. F.; Bullock A. N.; Crews C. M. Targeted Protein Degradation: Expanding the Toolbox. Nat. Rev. Drug Discov. 2019, 18 (12), 949–963. 10.1038/s41573-019-0047-y. [DOI] [PubMed] [Google Scholar]
- Chi J. J.; Li H.; Zhou Z.; Izquierdo-Ferrer J.; Xue Y.; Wavelet C. M.; Schiltz G. E.; Zhang B.; Cristofanilli M.; Lu X.; Bahar I.; Wan Y. A Novel Strategy to Block Mitotic Progression for Targeted Therapy. EBioMedicine 2019, 49, 40–54. 10.1016/j.ebiom.2019.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinitz F.; Chen E. Y.; Nicolis di Robilant B.; Chuluun B.; Antony J.; Jones R. C.; Gubbi N.; Lee K.; Ho W. H. D.; Kolluru S. S.; Qian D.; Adorno M.; Piltti K.; Anderson A.; Monje M.; Heller H. C.; Quake S. R.; Clarke M. F. Inhibiting USP16 Rescues Stem Cell Aging and Memory in an Alzheimer’s Model. Elife 2022, 11, e66037 10.7554/eLife.66037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priest C.; Nagari R. T.; Bideyan L.; Lee S. D.; Nguyen A.; Xiao X.; Tontonoz P. Brap Regulates Liver Morphology and Hepatocyte Turnover via Modulation of the Hippo Pathway. Proc. Natl. Acad. Sci. U.S.A. 2022, 119 (18), e2201859119 10.1073/pnas.2201859119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman P.; Swails J.; Chodera J. D.; McGibbon R. T.; Zhao Y.; Beauchamp K. A.; Wang L.-P.; Simmonett A. C.; Harrigan M. P.; Stern C. D.; Wiewiora R. P.; Brooks B. R.; Pande V. S. OpenMM 7: Rapid Development of High Performance Algorithms for Molecular Dynamics. PLoS Comput. Biol. 2017, 13 (7), e1005659 10.1371/journal.pcbi.1005659. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.