Abstract
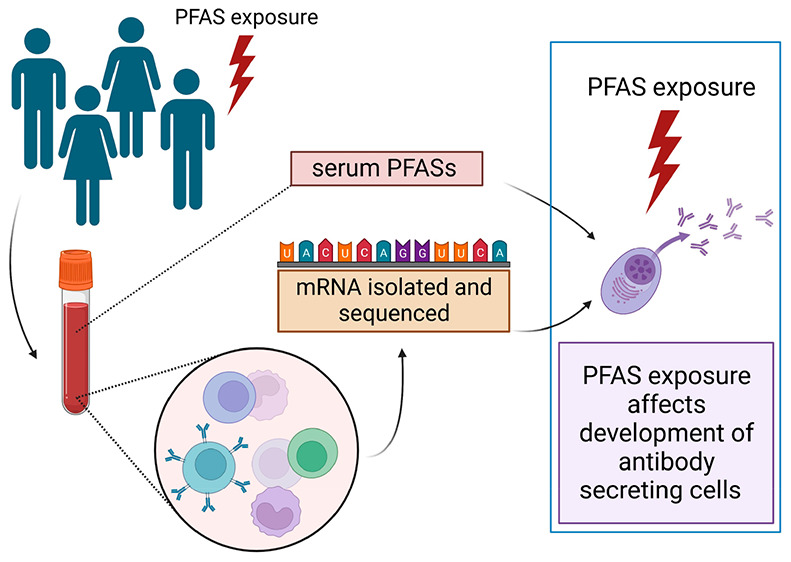
While the immunomodulation effects of per- and polyfluoroalkyl substances (PFASs) are described on the level of clinical signs in epidemiological studies (e.g., suppressed antibody response after vaccination), the underlying mechanism has still not been fully elucidated. To reveal mechanisms of PFAS exposure on immunity, we investigated the genome-wide transcriptomic changes of peripheral blood mononuclear cells (PBMCs) responding to PFAS exposure (specifically, exposure to PFPA, PFOA, PFNA, PFDA, PFUnDA, PFHxS, and PFOS). Blood samples and the chemical load in the blood were analyzed under the cross-sectional CELSPAC: Young Adults study. The overall aim of the study was to identify sensitive gene sets and cellular pathways conserved for multiple PFAS chemicals. Transcriptome networks related to adaptive immunity were perturbed by multiple PFAS exposure (i.e., blood levels of at least four PFASs). Specifically, processes tightly connected with late B cell development, such as B cell receptor signaling, germinal center reactions, and plasma cell development, were shown to be affected. Our comprehensive transcriptome analysis identified the disruption of B cell development, specifically the impact on the maturation of antibody-secreting cells, as a potential mechanism underlying PFAS immunotoxicity.
Keywords: Perfluoroalkyl substances, gene expression, peripheral blood mononuclear cells, adult cohort, transcriptomics, immunotoxicity, B cell, plasma cell
Short abstract
The disruption of B cell development by PFASs can compromise adaptive immunity, making organisms more vulnerable to stressors and pathogens and potentially and ultimately reducing the fitness of the population.
1. Introduction
Per- and polyfluoroalkyl substances (PFASs) are emerging environmental contaminants that have been used since the 1940s. PFASs, due to their surfactant properties and chemical stability, have found many applications in industry as well as in the consumer sector.1 Due to their high stability, PFASs persist in the environment and thus can be found in water, soil, and air. Therefore, people and other living organisms are continuously exposed to these chemicals.2 Alarmingly, PFASs have been found in human matrices with high frequency. The United States of America (U.S.A.) and European epidemiological and biomonitoring studies report that PFASs are present in human blood, often with a detection frequency above 90%.3,4
PFAS exposure has been associated with adverse health outcomes, such as liver damage, endocrine disruption, liver and testicular cancer, and immune disruption.5 Suppressed antibody response after vaccination is one of the frequently described effects of the immune disruption associated with PFASs.6,7 Further, PFAS exposure has been associated with an increased risk of infectious diseases, the prevalence of asthma, and altered immunological responses in allergies.7 Taken together, it is evident that PFASs are immunomodulatory stressors; however, the mechanism of action has still not been fully elucidated, specifically in humans.
A number of epidemiological studies have indicated that PFASs interfere with antibody production. Experimental studies reviewed recently by Ehrlich et al. in 2023 suggest the involvement of nuclear receptors, such as NF-κB and PPARs, and/or calcium signaling.8 Data from both epidemiological and toxicological studies are valuable for determining adverse outcome pathways (AOPs), i.e., the set of casually linked events leading from the initial molecular event to the apical health effect. Especially, the identification and quantification of biomarkers of effect provide valuable data for building AOPs.9 By implementing omics and advanced bioinformatics, biomarkers of effect on biological levels such as the genome, transcriptome, proteome, and metabolome can be revealed. These omics technologies are useful for characterizing the effect of PFASs on human health and, most importantly, revealing an early event that may lead to adverse health effects.10,11 Genome-wide transcriptomic analysis of immune blood cells, for example, can uncover valuable information about complex immune signaling. This technique allows researchers to analyze the complete set of ribonucleic acid (RNA) transcripts present in cells, providing valuable insights into the gene expression patterns underlying immune responses. Genome-wide transcriptomic studies are prevalently performed on samples from in vitro studies and animal in vivo studies. Transcriptomic analyses of samples from epidemiological studies are scarce despite their outputs being valuable, as they can demonstrate the transcriptomic activity of cells in living human organisms. By employing transcriptomic techniques, we can quantify the specific transcripts and molecular pathways involved in immunity, offering a comprehensive view of the immune system’s dynamics.12 This approach enables the characterization of immune cells and their interactions in response to various stimuli, including pathogens, toxins, and (most importantly) environmental factors, and therefore provides evidence about potential biomarkers of effect.
However, a biomarker of effect that can be identified in a study focusing on a particular outcome does not have to be considered the only one because chemical compounds can have multiple modes of action and mechanisms to affect human health. Nevertheless, these single studies that identify biomarkers are important, as they can be later compared to other similar studies in a focused meta-analysis and the most relevant mechanism can be identified. Further, because PFASs are a group of compounds that are structurally alike, we hypothesize that there could be a conserved mechanism of action for multiple PFASs, which can be elucidated by capturing the biomarker of effect.
Therefore, the objective of this study was to identify a transcriptomic response that is conserved for multiple PFASs, including the seven most abundant ones: perfluoropentanoate (PFPA), perfluorooctanoate (PFOA), perfluorononanoate (PFNA), perfluorodecanoate (PFDA), perfluoroundecanoate (PFUnDA), pefluorohexanesulfonate (PFHxS), and perfluorooctanesulfonate (PFOS). To reach this aim, the gene expression profiles within human immune cells in relation to PFAS blood levels from a cross-sectional Czech adult cohort study were researched. Using this approach, we aimed to uncover the molecular responses underlying PFAS-associated immunomodulation in humans. Through the utilization of transcriptomics, our research endeavors shed light on the specific gene expression patterns, molecular pathways, and regulatory mechanisms involved in the immune system’s response to PFAS exposure.
2. Materials and Methods
2.1. Study Population
We employed data from the cross-sectional Central European Longitudinal Studies of Parents and Children: Young Adults (CELSPAC: YA) study, which is an ongoing follow-up re-examination of the Czech part of the ELSPAC birth cohort (European Longitudinal Study of Pregnancy and Childhood) that was initiated in 1991–1992 in the Czech Republic. Detailed information about the ELSPAC-CZ study is provided in ref (13). The CELSPAC: YA study collected a broad spectrum of data, including lifestyle and health questionnaires, blood and urine samples, and chemical analysis of blood. We examined CELSPAC: YA participants that had all of the input data available for the analysis (PFAS blood levels, transcriptomic profile, and questionnaire data), i.e., 288 participants, which included a comparable number of men (n = 143) and women (n = 145). The participants that were examined were around 27 years old (geometric mean = 27, minimum (min) = 20, and maximum (max) = 37) and were generally of normal weight (with a median body mass index (BMI) of 23.5). They prevalently had a university education (75%), were nonsmokers (69%), and a comparable number of them rarely (55%) or often (45%) consumed alcohol. The general characteristics of the cohort are summarized in Table S1 in the Supporting Information (SI). For more details about the cohort study and the collected data, see our previous work.14 The CELSPAC: YA study was approved by the ELSPAC Ethics Committee (ref no. ELSPAC/EK/2/2019, dated March 13, 2019).
2.2. Analysis of PFAS in Blood Samples
Blood samples were processed and stored in a −80 °C freezer within 4 h after collection. PFAS serum levels were measured by high-pressure liquid chromatography coupled with tandem mass spectrometry (HPLC-MS/MS). The detailed analytical procedure has already been published in our previous work.14 In brief, 12 different PFASs were analyzed in serum samples: PFPA (CAS 2706-90-3), PFOA (CAS 335-67-1), PFNA (CAS 375-95-1), PFDA (CAS 335-76-2), PFUnDA (CAS 2058-94-8), PFHxS (CAS 355-46-4), PFOS (CAS 1763-23-1), perfluorohexanoate (PFHxA, CAS 307-24-4), perfluoroheptanoate (PFHpA, CAS 375-85-9), perfluorododecanoate (PFDoDA, CAS 307-55-1), perfluorobutanesulfonate (PFBS, CAS 375-73-5), and perfluoroheptanesulfonate (PFHpS, CAS 375-92-8). Nevertheless, only PFPA, PFOA, PFNA, PFDA, PFUnDA, PFHxS, and PFOS were included in the investigation as the most abundant chemicals, as at least 97% of their values were above the limit of detection (LOD). The serum concentrations of all 12 PFASs, together with their detection frequencies, are given in the SI, Table S2. A correlation matrix of the seven studied PFASs is depicted in Figure S1.
2.3. Peripheral Blood Mononuclear Cell (PBMC) Extraction and RNA Isolation
After the blood collection from the participants, the whole blood samples (9 mL) were immediately centrifuged, and the buffy coat fraction (i.e., white blood cell fraction) was separated by Ficoll-Paque to isolate the peripheral blood mononuclear cells (PBMCs). The PBMC fraction was suspended in RNAprotect Cell Reagent and frozen (−80 °C) in 300 μL aliquots containing ∼13 million cells until use for analysis (not longer than 3 years). The RNA was then extracted from the PBMCs in the RNAprotect Cell Reagent with the Zymo Research Quick-RNA Whole Blood (R1201) extraction kit according to the manufacturer’s instructions. Quality parameters such as the concentration, purity (NanoDrop, Thermo Fisher Scientific), and integrity (5200 Fragment Analyzer system, Agilent) of the extracted RNA were determined. For library preparation and sequencing, 1 μg of high-quality RNA per sample was used. The mean RNA integrity number (RIN) for the samples was 9.0 (min–max: 7.3–10.0).
2.4. Library Preparation and Sequencing
Genome-wide analysis of gene expression was conducted using a next-generation sequencing (NGS) platform with the QuantSeq library preparation step. cDNA libraries for each sample (RNA) were generated from 1 μg of the total RNA using the QuantSeq 3′ mRNA-Seq library prep kit for Illumina (Lexogen) following the manufacturer’s instructions. QuantSeq generates highly strand-specific NGS libraries close to the 3′ end of poly-A RNA.15 Standard external barcodes were ligated to allow multiplex sequencing. After PCR amplification, the libraries were size-selected with Agencourt AMPure XP magnetic beads (Beckman Coulter). The libraries were quantified by Qubit (Life Technologies), and their size (∼250 bp) was determined by using an Agilent 2100 Bioanalyzer. The libraries were sequenced (Illumina NovaSeq platform) and quality checked (110 bp single read) to obtain a minimum of 20–25 million reads per sample. Further, the NGS data were demultiplexed. The quality of the samples was continuously checked using FastQC (0.11.5), Qualimap (11_12–16) and MultiQC (1.8). All of the reads were trimmed, and bad-quality reads were removed using BBMap (38.42). Mapping reads were done by STAR (2.7.7a) using a GRCH38 human reference. Deduplication of the samples was done using umi_tools (1.0.0). Transcript features were counted by using htseq-count (0.11.1) and mmquant (1.3). Samtools (1.9) was used to manipulate the sequencing files.
2.5. Statistical Analysis
Data were processed
in R programming software (version 4.2.2).16 Exposure data below the LOD or between the LOD and the limit of
quantitation (LOQ) were imputed using LOD/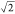 and LOQ/
and LOQ/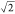 , respectively.
Genes with at least 5 CPM
(counts per million) in at least 20% of the samples were kept to analyze.
Data were normalized using TMM (trimmed mean of M values) normalization and were transformed to a continuous log2
scale using limma voom.17,18 The influence of the
batches (i.e., the batches for RNA extraction and library preparation)
was checked by principal component analysis (PCA) plots and by the
correlation of principal components with potential confounders. Surrogate
variable analysis was performed on the data, and the first 10 surrogate
variables were used to adjust the unknown cell blood composition.19−21 Gene expression associated with individual PFASs was identified
using the limma lmfit model, and p values were corrected
for multiple testing using the Benjamini–Hochberg false discovery
rate (FDR).22 The model was adjusted for
biological, socioeconomic, and technical covariates (sex, age, BMI,
education, smoking status, alcohol consumption, and library preparation
batch). Genes were annotated using GeneCards.23 The genes whose expression was associated with four or more PFASs
were used for subsequent enrichment analysis to uncover conserved
PFAS effects on immunity.
, respectively.
Genes with at least 5 CPM
(counts per million) in at least 20% of the samples were kept to analyze.
Data were normalized using TMM (trimmed mean of M values) normalization and were transformed to a continuous log2
scale using limma voom.17,18 The influence of the
batches (i.e., the batches for RNA extraction and library preparation)
was checked by principal component analysis (PCA) plots and by the
correlation of principal components with potential confounders. Surrogate
variable analysis was performed on the data, and the first 10 surrogate
variables were used to adjust the unknown cell blood composition.19−21 Gene expression associated with individual PFASs was identified
using the limma lmfit model, and p values were corrected
for multiple testing using the Benjamini–Hochberg false discovery
rate (FDR).22 The model was adjusted for
biological, socioeconomic, and technical covariates (sex, age, BMI,
education, smoking status, alcohol consumption, and library preparation
batch). Genes were annotated using GeneCards.23 The genes whose expression was associated with four or more PFASs
were used for subsequent enrichment analysis to uncover conserved
PFAS effects on immunity.
2.6. Enrichment Analysis
Gene Set Enrichment Analysis (GSEA) was conducted using Pathway Studio (version 12.0). Gene sets were permutated 1000 times using the Kolmogorov–Smirnov classic approach as an enrichment algorithm. To broaden the analysis, all pathways were expanded to include cell processes and functional classes in target gene seeds. The enrichment p value cutoff was set at p < 0.05. Subnetwork Enrichment Analysis (SNEA) was also performed as previously described.24 The enrichment p value for the gene seeds was set at p < 0.05.
3. Results and Discussion
3.1. Gene Expression Significantly Associated with Multiple PFAS Exposure
Our analysis identified 166 genes that were significantly (p < 0.05) associated with exposure to at least four of the seven examined PFASs (PFPA, PFOA, PFNA, PFDA, PFUnDA, PFHxS, and PFOS; Figure S2). However, no genes were significantly associated when the more stringent statistical analysis (FDR correction) was applied. The analysis did not identify a single gene associated with all seven PFASs. However, 10 out of the 166 genes were associated with the levels of six PFASs, and 44 genes were associated with five PFASs. It is worth noting that, apart from PFPA, all of the other PFASs (PFOA, PFNA, PFDA, PFUnDA, PFHxS, and PFOS) demonstrated a congruent direction of effect for individual genes, either the downregulation or the upregulation of gene expression (Figure S2). This trend is visible when looking at the same color in each row in Figure S2: red indicates the upregulation of gene expression, and blue indicates the downregulation of gene expression. This overall trend revealed a contrasting relationship between PFPA exposure and gene expression compared to the other PFASs, as the associations exhibit opposite directions. Further, compared to the other PFASs, PFPA had only a few statistically significant (p < 0.05) associations (Table S3).
PFPA stands out among other PFAS compounds due to its shorter perfluorinated carbon chain comprising only four fluorinated carbons and due to it being the only short-chain PFAS. In contrast to the other long-chain PFAS compounds analyzed in this study, this disparity in carbon chain length grants PFPA distinctive chemical properties and attributes.25 Consequently, this variation could potentially result in distinct biological effects. Chain-length-dependent biological activity has been shown in in vivo studies (e.g., mice, rats, and marine mussel models).26−28 In a study with mice, Lee and Kim suggested that the length of the perfluorinated chain determines the effect, as they observed increased NF-κB activity in the case of longer PFASs (C10 and C11) and no NF-κB activity in the case of shorter PFASs (C7 and C9). Similarly, Stevenson et al. observed a chain-length-specific interaction between PFASs and efflux transporters, a multixenobiotic resistance mechanism that triggered the exporting chemicals from cells.27 Further, a study with rats showed distinct toxicokinetic properties for PFASs with different carbon chain lengths; specifically, there was a high clearance rate for PFASs with shorter carbon chain lengths.28 Different chemical–physical properties, i.e., differences in lipophobic/hydrophobic perfluoroalkyl tails, can thus trigger both the fate of the chemicals in the organisms and the biological activity. It is important to note that specific studies and research are needed to comprehensively evaluate and determine the precise biological effects of PFPA compared to those of other PFAS compounds.
3.2. Pathways Enriched by Genes Associated with Multiple PFAS Exposure Identified by SNEA
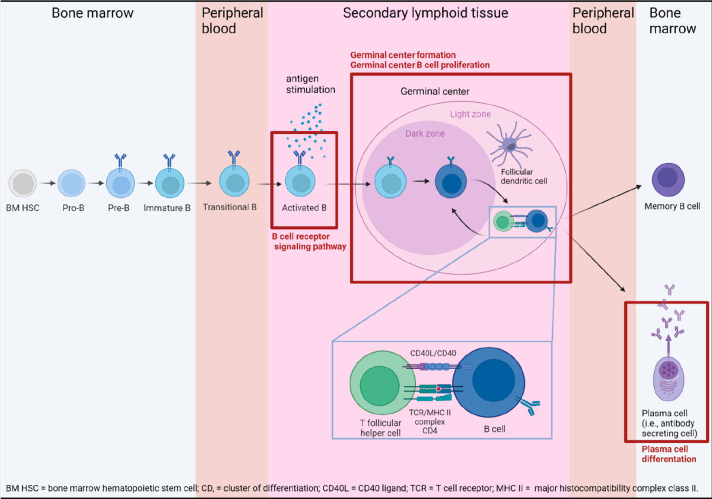
Four cell processes were identified by SNEA as being enriched (Table 1 and Figure 1): germinal center B cell differentiation, germinal center formation, the B cell receptor signaling pathway, and plasma cell differentiation. These enriched processes suggest that PFASs target B cell development, especially late B cell development (impaired processes are visualized in Figure 1).
Table 1. All Statistically Significant Cell Processes Identified by SNEA of Deregulated Genes for Multiple PFAS Exposure (i.e., at Least Four PFASs).
| cell process | overlapping entities | p value |
|---|---|---|
| germinal center B cell differentiation | TCF3; CD19; EBF1; KDM1A; SPIB | 0.008 |
| germinal center formation | SCD; TCF3; POU2F2; IL17RA; CD19; EBF1; KDM1A; SPIB; NLRP3 | 0.008 |
| B cell receptor signaling pathway | TCF3; POU2F2; RASGRP3; SLA; CD19; EBF1; SPIB; LAX1; IGHM; TCF4; FCRLA | 0.011 |
| plasma cell differentiation | TCF3; POU2F2; CDKN2C; CD19; EBF1; SPIB; TCF4 | 0.038 |
Figure 1.
B cell development. Processes in red frames were enriched by genes associated with multiple PFAS exposure (i.e., with at least four PFASs; see Table 1). B cells develop in bone marrow, where their B cell receptors (BCRs) undergo somatic recombination, resulting in high variability in the BCR specificity (for a particular antigen) of individual B cells. These immature B cells enter the bloodstream and migrate to secondary lymphoid organs (e.g., the spleen and lymphoid nodes), where they become activated and create germinal centers. In a germinal center, a B cell undergoes somatic hypermutation, selection, and class-switch recombination, and it becomes either a memory cell or a plasma cell, whereas the latter is crucial for antibody production.
Genes overlapping within significant cell processes (i.e., those listed in Table 1) are shown in Table 2. These include the genes coding transcription factors E2A, E2-2, EBF1, OCT-2, and SPI-B, further the transmembrane molecule CD19, and the histone demethylase LSD1, all of which are significantly involved in B cell development, including germinal center (GC) reactions. GCs play a crucial role in the formation and maturation of plasma cells, which are a type of B cells that are responsible for producing antibodies.
Table 2. Key Transcripts of Genes and Respective Proteins Associated with Multiple PFAS Exposure Involved in Enriched Cell Processesa.
| Gene name | Coded protein | PFDA | PFHxS | PFNA | PFOA | PFOS | PFPA | PFUnDA |
|---|---|---|---|---|---|---|---|---|
| CD19 | CD19 | −0.037 | −0.059* | −0.068* | −0.052* | −0.071* | 0.060* | −0.019 |
| EBF1 | EBF1 | −0.070* | −0.102* | −0.082* | −0.029 | −0.097* | 0.035 | −0.063* |
| KDM1A | LSD1 | −0.075* | −0.077* | −0.070* | −0.039 | −0.071* | 0.015 | −0.049* |
| POU2F2 | OCT-2 | −0.029* | −0.029* | −0.030* | −0.007 | −0.029* | −0.003 | −0.010 |
| SPIB | SPI-B | −0.030 | −0.074* | −0.058* | −0.014 | −0.070* | 0.060* | −0.060* |
| TCF3 | E2A | −0.019 | −0.028* | −0.026* | −0.038* | −0.036* | 0.010 | −0.013 |
| TCF4 | E2-2 | −0.038 | −0.036 | −0.053* | −0.036 | −0.071* | 0.062* | −0.046* |
Results are expressed as β coefficients produced by linear regression. Statistically significant results (p < 0.05) are marked in bold with *. PFPA = perfluoropentanoate, PFOA = perfluorooctanoate, PFNA = perfluorononanoate, PFDA = perfluorodecanoate, PFUnDA = perfluoroundecanoate, PFHxS = pefluorohexane sulfonate, and PFOS = perfluorooctane sulfonate.
The transcription factors E2A (encoded by the TCF3 gene) and E2-2 (encoded by the TCF4 gene) belong to a group of E-proteins, and together, they cooperate to regulate B cell immunity, especially late B cell development. Both play a crucial role in controlling GC and plasma cell development. While E2A has been shown to be a dominant E-protein in GC B cell differentiation, E2-2 plays a dominant role in plasma cell development.29
Another transcription factor, which often cooperates with E2A, is early B cell factor EBF1. As its name indicates, EBF1 plays a key role in the early stages of B cell development, specifically in the pro-B cell stage. Additionally, EBF1 is known to play an important role in GC formation.30,31 Specifically, EBF1 has been shown to interact with PAX5, one of the crucial players in B cell differentiation, and together, they regulate the transcription of many genes during B cell development. The cooperation of these two transcription factors allows the expression of molecules such as CD19 and CD79b, which are important players in B cell signaling.32
CD19, besides being a unique characterization surface marker of B cells, has an indispensable functional role. CD19 modulates both BCR-dependent and BCR-independent signaling.33 Specifically, CD19 often functions with CD21 (which is activated by binding of the antigen-C3d complex), CD81 (TAPA-1), and CD225 to comprise a multimolecular complex that can transduce signals in both a BCR-dependent and BCR-independent fashion. CD19 is a transmembrane protein transducing signal to downstream protein kinases such as Lyn, Fyn (Src family), Abl, Btk, PI3K, and Ras family kinases.33,34 Further, CD19 is also required for optimal MHC class II-mediated signaling through Akt kinase.35
OCT-2, encoded by the POU2F2 gene, is a transcription factor that is indispensable for GC formation. OCT-2 action is accompanied by the OBF1 coactivator, which stabilizes the binding of OCT-2 to chromatin.36,37 Interestingly, these two factors (OCT2 and OBF1) are essential for the proliferation and survival of diffuse large B cell lymphoma.37,38
SPI-B is a transcription factor that represses B cell differentiation.39 However, SPI-B together with PU.1 (encoded by the SPI1 gene) is essential for signaling through BCR and through receptors for CD40L, BAFF, and TLR ligands.40
LSD1, encoded by KDM1A, is a histone demethylase that can regulate gene expression. It interacts with Blimp-1, which is an essential transcription factor for plasma cell differentiation.41 On the basis of LSD1-deficient mice, LSD1 has been shown to be a crucial epigenetic modifier in plasma cell development by regulating chromatin accessibility.41
Overall, SNEA revealed four statistically significant cell processes that were all related to B cells, specifically to GC reactions. Further, genes abundantly present in these cell processes were negatively associated with PFAS exposure except for PFPA. Such a negative association between the genes and PFAS exposure indicates the downregulation of these genes (Table 2) and their respective cell processes (Table 1). This finding is in line with epidemiological studies connecting PFAS exposure to decreased antibody production, as GCs and plasma cells are crucial for antibody response.
3.3. Pathways Enriched by Genes Associated with Multiple PFAS Exposure Identified by GSEA
In total, 126 entities were significantly enriched by genes associated with multiple PFAS exposure by GSEA (Table S4). GSEA provides detailed information about enriched entities on levels such as biomarkers, signal processing, biological processes, and diseases. The majority of the enriched entities were related to immunity terms, but there were also non-immune terms and general biological processes. Enriched immune-related entities included terms related to both innate and adaptive immunity. A prevalent motif within adaptive immunity terms is B cell activation and development, which corresponds to the results produced by SNEA (Table 2). The identified B cell-related entities were clustered into four main domains: B cell receptor signaling, TLR signaling, T cell–B cell interaction, and E2A signaling (Table 3).
Table 3. B Cell-Related Enriched Processes, Biomarkers, and Diseases Associated with Multiple PFAS Exposures Identified by GSEA.
| cluster | name | overlapping genes | p value | hit type |
|---|---|---|---|---|
| B cell receptor signaling | CD72 → AP-1 expression targets | MAP2K4; E2F5; CDCA4; MAP2K3 | 0.0007 | biomarkers |
| B cell receptor → NFATC signaling | CD19; IGHM | 0.0175 | signal processing | |
| B cell receptor → NF-κB signaling | CD19; IGHM | 0.0295 | signal processing | |
| B cell receptor → AP-1 signaling | CD19; IGHM | 0.0449 | signal processing | |
| TLR signaling | TLR4 → AP-1 expression targets | LDLR; MAP2K3; MAP2K4 | 0.0177 | biomarkers |
| TLR → AP-1 signaling | MAP2K4; MAP2K3 | 0.0191 | signal processing | |
| T cell–B cell interaction | T cell-dependent B cell activation | RASGRP3; MAP2K3; IGHM | 0.0277 | biological process |
| MHC2-mediated antigen presentation | HLA-DMA; IFI30 | 0.0313 | biological process | |
| E2A signaling | NOTCH → TCF3 signaling | MAP2K4; MAP2K3; TCF3 | 0.0002 | signal processing |
| Hodgkin and Reed–Sternberg cell reprogramming | POU2F2; EBF1; TCF3 | 0.0014 | disease |
B cell receptor (BCR) signaling is crucial for B cell activation and, therefore, for the subsequent creation of GCs. BCR signaling is very complex, as BCR cooperates with many other receptors and has several downstream intracellular signaling pathways42 (Table 3, B cell receptor signaling cluster). Specifically, BCR can activate NFAT signaling downstream.43 Even though NFAT signaling was discovered in T cells, it was later revealed that this signaling pathway is essential for normal B cell homeostasis and differentiation.44 Further, BCR signaling can lead to the activation of the transcription factors NF-κB and AP-1, which are highly involved in regulating immune response.43,45 Moreover, B cells at all stages (except plasma cells) express the CD72 molecule, which is a coreceptor that regulates BCR signaling.46 In addition to BCRs, B cells express toll-like receptors (TLRs), which play an important role in B cell activation. TLR signaling can therefore enhance the signal for B cell activation, and it is important for B cell differentiation in plasma cells.47
When a GC is formed, T follicular helper cells play an important role in the coactivation of B cells, which is crucial for their future differentiation into plasma cells (Table 3, T cell–B cell interaction cluster). This coactivation is ensured by a T cell receptor recognizing the antigen on the MHC class-II molecule expressed by a B cell, and further by the CD40L of the T cell binding to the CD40 receptor of the B cell (Figure 1).42,48−50
As stated above, the TCF3 gene codes the transcription factor E2A, which is essential for GC B cell differentiation (Table 3, E2A signaling cluster). Nevertheless, the E2A function is regulated by other players, one of these being Notch proteins.51 Notch proteins regulate whole B cell development, including plasma cell differentiation.52 The importance of E2A in B cell development is also demonstrated by the fact that the disruption of the E2A function contributes to the progression of Hodgkin lymphoma.53,54 Hodgkin lymphoma is a B cell-derived cancer and is characterized by the presence of Reed–Sternberg cells and Hodgkin cells.55
The overall GSEA results indicate that multiple PFAS exposure affects pathways and genes of adaptive immunity; however, general entities (pathways and biomarkers) that play a role in the entire immune system, including innate immunity, were also identified. Within entities of adaptive immunity, processes involved in B cell activation, development, and differentiation were abundantly included, which is in line with the results of SNEA. However, the results of GSEA indicate that PFAS exposure may also affect other processes within the entire immune system, demonstrating the complexity of PFAS immunomodulation.
3.4. Possible Underlying Mechanism of PFAS-Induced Immunomodulation
Taken together, the results of the current study indicate that multiple PFAS exposure influences the immune system in the phase of late B cell development, specifically B cell activation, GC reactions, and plasma cell development. As GCs and plasma cells are essential for antibody production, the findings of the current study are in line with suppressed antibody responses after vaccination associated with PFAS exposure, which has been largely reported in the literature.6,56−60 Similarly, a recent in vivo study on mice showed a decreased level of antibodies accompanied by a decreased number of splenic B cells, including plasma cells, due to PFOA exposure.61 Further, the affected development of plasma cells, i.e., antibody-secreting cells, that was observed in our study could be the underlying mechanism for the altered prevalence of allergic diseases, as the IgE antibodies are the main effector agent in allergic diseases.62 Interestingly, PFAS exposure was associated with both the increased63,64 and decreased14,65 prevalence of allergic diseases. This evidence indicates that the effects of PFAS exposure are complex and complicated, especially when we are assessing exposure to a mixture of them, as different PFASs can act differently, as was also shown in our study with the example of PFPA.
Despite the fact that transcriptomic data from epidemiological studies are highly valuable, there is a scarce number of them. Similar to our study, the effect of PFAS exposure on adaptive immunity, specifically the deregulation of T cell signaling, was observed in a Norwegian BraMat human cohort study investigating transcriptomic profiles in neonatal cord blood.66 Although both studies focused on immunotoxicity, their design and samples differed. Because the Norwegian study focused on the genes common for PFAS exposure and anti-rubella antibody levels at 3 years of age, the genes common for PFAS exposure, and a number of common cold episodes until 3 years of age, the results are not directly comparable to the those of this current study. Interestingly, a large cross-species transcriptomics analysis, which included human samples, identified the neutrophil tertiary granule mechanism as being strongly conserved among species and proposed it as a potential mechanism underlying PFAS immunotoxicity. Neutrophils are the most abundant cells of innate immunity and are evolutionarily more conserved through different species compared to B cell signaling and other adaptive immunity processes. A strong pattern related to neutrophils was not shown in our study, as our analysis was based on the transcriptome of PBMCs that do not contain neutrophils. Even though a PBMC sample does not contain all of the immune cells in the same ratio as in a living organism (innate immune cells in particular are not represented in full numbers), PBMCs are considered a valuable matrix for transcriptomic analysis.67
The detailed mechanism behind PFAS immunotoxicity is still not fully understood. We described for the first time the alteration in B cell development as a potential mode of action on human samples. Among others, we identified NF-κB and NFAT signaling, which have already been described as being associated with PFAS exposure.8 In both human and animal studies, the association of PFAS exposure with a disbalance of Th1/Th2 cytokines has been observed.8 However, the evidence of this imbalance is inconsistent between published studies and is thus inconclusive. Nevertheless, a disbalance of Th1/Th2 cytokines could further affect B cell development in stages such as B cell activation, GC formation, antibody-secreting plasma cell development, or memory B cells differentiation.
In conclusion, the utilization of genome-wide sequencing in this study revealed that BCR signaling, GC reactions, and the development of plasma cells could be behind the potential mechanisms underlying PFAS immunotoxicity. Future research should be conducted on B cell development and germinal centers as the target for PFASs to verify the results of the current study, preferably using controlled toxicological experiments. A further relevant avenue that needs to be explored is determining the long-term effects of PFAS exposure on immune function and potential immunological memory and/or examining the broader consequences of PFAS-induced immunotoxicity on the overall immune response, including the susceptibility to infections, allergies, autoimmune diseases, and impaired vaccine responses. We believe that such further investigations may enhance our understanding of immunotoxicity not only for currently used PFASs but also prospectively for their chemically similar alternatives.
Acknowledgments
The authors thank RECETOX Research Infrastructure (LM2023069) financed by the Ministry of Education, Youth and Sports, and the Operational Programme Research, Development and Education (the CETOCOEN EXCELLENCE Project CZ.02.1.01/0.0/0.0/17_043/0009632 and Cetocoen Plus CZ.02.1.01/0.0/0.0/15_003/0000469) for supportive background. The laboratory part of this work was supported by the Project BBMRI.cz (LM2023033). This work was supported by the European Union’s Horizon 2020 research and innovation program under Grant 857560. This publication reflects only the authors’ view, and the European Commission is not responsible for any use that may be made of the information it contains. Figures were created using the paid version of Biorender.com.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.est.3c05109.
Population characteristics, PFAS serum levels, a correlation matrix of the PFASs, counts of associated genes, genes associated with PFAS exposure, and enriched entities identified by GSEA (PDF)
Author Contributions
B.R.: conceptualization, methodology, investigation, formal analysis, writing—original draft, and writing—review and editing. V.T.: conceptualization, supervision, and writing—review and editing. H.V.: methodology, investigation, formal analysis, and writing—review and editing;. C.J.M.: formal analysis and writing—review and editing. P.P.: conceptualization, coordination, and writing—review and editing. M.Z.: conceptualization, coordination, and writing—review and editing. J.K.: conceptualization, coordination, resources, and writing—review and editing. L.B.: conceptualization, supervision, and writing—review and editing. O.A.: conceptualization, methodology, supervision, and writing—review and editing.
The authors declare no competing financial interest.
Special Issue
Published as part of Environmental Science & Technologyvirtual special issue “The Exposome and Human Health”.
Supplementary Material
References
- Dean W. S.; Adejumo H. A.; Caiati A.; Garay P. M.; Harmata A. S.; Li L.; Rodriguez E. E.; Sundar S. A Framework for Regulation of New and Existing PFAS by EPA. Journal of Science Policy & Governance 2020, 16 (1), 14. [Google Scholar]
- De Silva A. O.; Armitage J. M.; Bruton T. A.; Dassuncao C.; Heiger-Bernays W.; Hu X. C.; Kärrman A.; Kelly B.; Ng C.; Robuck A.; Sun M.; Webster T. F.; Sunderland E. M. PFAS Exposure Pathways for Humans and Wildlife: A Synthesis of Current Knowledge and Key Gaps in Understanding. Environ. Toxicol. Chem. 2021, 40 (3), 631–657. 10.1002/etc.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calafat A. M.; Wong L.-Y.; Kuklenyik Z.; Reidy J. A.; Needham L. L. Polyfluoroalkyl Chemicals in the U.S. Population: Data from the National Health and Nutrition Examination Survey (NHANES) 2003–2004 and Comparisons with NHANES 1999–2000. Environ. Health Perspect. 2007, 115 (11), 1596–1602. 10.1289/ehp.10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EU HBM Dashboard. HBM4EU. https://www.hbm4eu.eu/what-we-do/european-hbm-platform/eu-hbm-dashboard/ (accessed 2023-01-09).
- Fenton S. E.; Ducatman A.; Boobis A.; DeWitt J. C.; Lau C.; Ng C.; Smith J. S.; Roberts S. M. Per- and Polyfluoroalkyl Substance Toxicity and Human Health Review: Current State of Knowledge and Strategies for Informing Future Research. Environ. Toxicol. Chem. 2021, 40 (3), 606–630. 10.1002/etc.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Xue L.; Deji Z.; Wang X.; Liu P.; Lu J.; Zhou R.; Huang Z. Effects of Exposure to Per- and Polyfluoroalkyl Substances on Vaccine Antibodies: A Systematic Review and Meta-Analysis Based on Epidemiological Studies. Environ. Pollut. 2022, 306, 119442. 10.1016/j.envpol.2022.119442. [DOI] [PubMed] [Google Scholar]
- von Holst H.; Nayak P.; Dembek Z.; Buehler S.; Echeverria D.; Fallacara D.; John L. Perfluoroalkyl Substances Exposure and Immunity, Allergic Response, Infection, and Asthma in Children: Review of Epidemiologic Studies. Heliyon 2021, 7 (10), e08160 10.1016/j.heliyon.2021.e08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich V.; Bil W.; Vandebriel R.; Granum B.; Luijten M.; Lindeman B.; Grandjean P.; Kaiser A.-M.; Hauzenberger I.; Hartmann C.; Gundacker C.; Uhl M. Consideration of Pathways for Immunotoxicity of Per- and Polyfluoroalkyl Substances (PFAS). Environ. Health 2023, 22 (1), 19. 10.1186/s12940-022-00958-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Carrillo A.; Mustieles V.; Salamanca-Fernández E.; Olivas-Martínez A.; Suárez B.; Bajard L.; Baken K.; Blaha L.; Bonefeld-Jørgensen E. C.; Couderq S.; D’Cruz S. C.; Fini J.-B.; Govarts E.; Gundacker C.; Hernández A. F.; Lacasaña M.; Laguzzi F.; Linderman B.; Long M.; Louro H.; Neophytou C.; Oberemn A.; Remy S.; Rosenmai A. K.; Saber A. T.; Schoeters G.; Silva M. J.; Smagulova F.; Uhl M.; Vinggaard A. M.; Vogel U.; Wielsøe M.; Olea N.; Fernández M. F. Implementation of Effect Biomarkers in Human Biomonitoring Studies: A Systematic Approach Synergizing Toxicological and Epidemiological Knowledge. Int. J. Hyg. Environ. Health 2023, 249, 114140. 10.1016/j.ijheh.2023.114140. [DOI] [PubMed] [Google Scholar]
- Yao X.; Cao D.; Wang F.; Zhang W.; Ma C.; Song M. An Overview of Omics Approaches to Characterize the Effect of Perfluoroalkyl Substances in Environmental Health. TrAC Trends Anal. Chem. 2019, 121, 115367. 10.1016/j.trac.2018.12.021. [DOI] [Google Scholar]
- Beale D. J.; Sinclair G. M.; Shah R.; Paten A. M.; Kumar A.; Long S. M.; Vardy S.; Jones O. A. H. A Review of Omics-Based PFAS Exposure Studies Reveals Common Biochemical Response Pathways. Sci. Total Environ. 2022, 845, 157255. 10.1016/j.scitotenv.2022.157255. [DOI] [PubMed] [Google Scholar]
- Chaussabel D. Assessment of Immune Status Using Blood Transcriptomics and Potential Implications for Global Health. Semin. Immunol. 2015, 27 (1), 58–66. 10.1016/j.smim.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Piler P.; Kandrnal V.; Kukla L.; Andrýsková L.; Švancara J.; Jarkovský J.; Dušek L.; Pikhart H.; Bobák M.; Klánová J. Cohort Profile: The European Longitudinal Study of Pregnancy and Childhood (ELSPAC) in the Czech Republic. Int. J. Epidemiol. 2017, 46 (5), 1379. 10.1093/ije/dyw091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudzanova B.; Vlaanderen J.; Kalina J.; Piler P.; Zvonar M.; Klanova J.; Blaha L.; Adamovsky O. Impact of PFAS Exposure on Prevalence of Immune-Mediated Diseases in Adults in the Czech Republic. Environ. Res. 2023, 229, 115969. 10.1016/j.envres.2023.115969. [DOI] [PubMed] [Google Scholar]
- Moll P.; Ante M.; Seitz A.; Reda T. QuantSeq 3′ mRNA Sequencing for RNA Quantification. Nat. Methods 2014, 11 (12), i–iii. 10.1038/nmeth.f.376. [DOI] [Google Scholar]
- R Core Team . R: A Language and Environment for Statistical Computing. 2022. https://www.r-project.org/ (accessed 2022-11-11).
- Smyth G. K.limma: Linear Models for Microarray Data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Gentleman R., Carey V. J., Huber W., Irizarry R. A., Dudoit S., Eds.; Springer, New York, New York, U.S., 2005; p 397–420. 10.1007/0-387-29362-0_23. [DOI] [Google Scholar]
- Law C. W.; Chen Y.; Shi W.; Smyth G. K. Voom: Precision Weights Unlock Linear Model Analysis Tools for RNA-Seq Read Counts. Genome Biol. 2014, 15 (2), R29. 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J. T.; Storey J. D. Capturing Heterogeneity in Gene Expression Studies by Surrogate Variable Analysis. PLoS Genet. 2007, 3 (9), e161. 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff A. E.Isva: Independent Surrogate Variable Analysis. 2017. https://cran.r-project.org/web/packages/isva/index.html (accessed 2023-06-15).
- Chen J.; Behnam E.. SmartSVA: Fast and Robust Surrogate Variable Analysis. 2017. https://cran.r-project.org/web/packages/SmartSVA/index.html (accessed 2023-06-15).
- Benjamini Y.; Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57 (1), 289–300. 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- Stelzer G.; Rosen N.; Plaschkes I.; Zimmerman S.; Twik M.; Fishilevich S.; Stein T. I.; Nudel R.; Lieder I.; Mazor Y.; Kaplan S.; Dahary D.; Warshawsky D.; Guan-Golan Y.; Kohn A.; Rappaport N.; Safran M.; Lancet D. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinforma. 2016, 54 (1), 1. 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- Langlois V. S.; Martyniuk C. J. Genome Wide Analysis of Silurana (Xenopus) Tropicalis Development Reveals Dynamic Expression Using Network Enrichment Analysis. Mech. Dev. 2013, 130 (4), 304–322. 10.1016/j.mod.2012.12.002. [DOI] [PubMed] [Google Scholar]
- Dhore R.; Murthy G. S. Per/Polyfluoroalkyl Substances Production, Applications and Environmental Impacts. Bioresour. Technol. 2021, 341, 125808. 10.1016/j.biortech.2021.125808. [DOI] [PubMed] [Google Scholar]
- Lee J.-K.; Kim S.-H. Correlation between Mast Cell-Mediated Allergic Inflammation and Length of Perfluorinated Compounds. J. Toxicol. Environ. Health A 2018, 81 (9), 302–313. 10.1080/15287394.2018.1440188. [DOI] [PubMed] [Google Scholar]
- Stevenson C. N.; MacManus-Spencer L. A.; Luckenbach T.; Luthy R. G.; Epel D. New Perspectives on Perfluorochemical Ecotoxicology: Inhibition and Induction of an Efflux Transporter in the Marine Mussel, Mytilus Californianus. Environ. Sci. Technol. 2006, 40 (17), 5580–5585. 10.1021/es0602593. [DOI] [PubMed] [Google Scholar]
- Ohmori K.; Kudo N.; Katayama K.; Kawashima Y. Comparison of the Toxicokinetics between Perfluorocarboxylic Acids with Different Carbon Chain Length. Toxicology 2003, 184 (2), 135–140. 10.1016/S0300-483X(02)00573-5. [DOI] [PubMed] [Google Scholar]
- Wöhner M.; Tagoh H.; Bilic I.; Jaritz M.; Poliakova D. K.; Fischer M.; Busslinger M. Molecular Functions of the Transcription Factors E2A and E2–2 in Controlling Germinal Center B Cell and Plasma Cell Development. J. Exp. Med. 2016, 213 (7), 1201–1221. 10.1084/jem.20152002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Györy I.; Boller S.; Nechanitzky R.; Mandel E.; Pott S.; Liu E.; Grosschedl R. Transcription Factor Ebf1 Regulates Differentiation Stage-Specific Signaling, Proliferation, and Survival of B Cells. Genes Dev. 2012, 26 (7), 668–682. 10.1101/gad.187328.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilagos B.; Hoffmann M.; Souabni A.; Sun Q.; Werner B.; Medvedovic J.; Bilic I.; Minnich M.; Axelsson E.; Jaritz M.; Busslinger M. Essential Role of EBF1 in the Generation and Function of Distinct Mature B Cell Types. J. Exp. Med. 2012, 209 (4), 775–792. 10.1084/jem.20112422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullerwell C. E.; Robichaud P. P.; Deprez P. M. L.; Joy A. P.; Wajnberg G.; D’Souza D.; Chacko S.; Fournier S.; Crapoulet N.; Barnett D. A.; Lewis S. M.; Ouellette R. J. EBF1 Drives Hallmark B Cell Gene Expression by Enabling the Interaction of PAX5 with the MLL H3K4Methyltransferase Complex. Sci. Rep. 2021, 11 (1), 1537. 10.1038/s41598-021-81000-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K.; Wei G.; Liu D. CD19: A Biomarker for B Cell Development, Lymphoma Diagnosis and Therapy. Exp. Hematol. Oncol. 2012, 1 (1), 36. 10.1186/2162-3619-1-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Burg M.; van Zelm M. C.; Driessen G. J. A.; van Dongen J. J. M. New Frontiers of Primary Antibody Deficiencies. Cell. Mol. Life Sci. 2012, 69 (1), 59–73. 10.1007/s00018-011-0836-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills D. M.; Stolpa J. C.; Cambier J. C.. Modulation of MHC Class II Signal Transduction by CD19. In Mechanisms of Lymphocyte Activation and Immune Regulation XI; Gupta S., Alt F., Cooper M., Melchers F., Rajewsky K., Eds.; Springer, Boston, Massachusetts, U.S., 2007; p 139–148. 10.1007/0-387-46530-8_12. [DOI] [PubMed] [Google Scholar]
- Song S.; Cao C.; Choukrallah M.-A.; Tang F.; Christofori G.; Kohler H.; Wu F.; Fodor B. D.; Frederiksen M.; Willis S. N.; Jackson J. T.; Nutt S. L.; Dirnhofer S.; Stadler M. B.; Matthias P. OBF1 and Oct Factors Control the Germinal Center Transcriptional Program. Blood 2021, 137 (21), 2920–2934. 10.1182/blood.2020010175. [DOI] [PubMed] [Google Scholar]
- Küppers R. OBF1 and OCT1/2 Regulate the Germinal Center B-Cell Program. Blood 2021, 137 (21), 2862–2863. 10.1182/blood.2021010689. [DOI] [PubMed] [Google Scholar]
- Hodson D. J.; Shaffer A. L.; Xiao W.; Wright G. W.; Schmitz R.; Phelan J. D.; Yang Y.; Webster D. E.; Rui L.; Kohlhammer H.; Nakagawa M.; Waldmann T. A.; Staudt L. M. Regulation of Normal B-Cell Differentiation and Malignant B-Cell Survival by OCT2. Proc. Natl. Acad. Sci. U.S.A. 2016, 113 (14), E2039–E2046. 10.1073/pnas.1600557113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidlin H.; Diehl S. A.; Nagasawa M.; Scheeren F. A.; Schotte R.; Uittenbogaart C. H.; Spits H.; Blom B. Spi-B Inhibits Human Plasma Cell Differentiation by Repressing BLIMP1 and XBP-1 Expression. Blood 2008, 112 (5), 1804–1812. 10.1182/blood-2008-01-136440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis S. N.; Tellier J.; Liao Y.; Trezise S.; Light A.; O’Donnell K.; Garrett-Sinha L. A.; Shi W.; Tarlinton D. M.; Nutt S. L. Environmental Sensing by Mature B Cells Is Controlled by the Transcription Factors PU.1 and SpiB. Nat. Commun. 2017, 8 (1), 1426. 10.1038/s41467-017-01605-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines R. R.; Barwick B. G.; Scharer C. D.; Majumder P.; Randall T. D.; Boss J. M. The Histone Demethylase LSD1 Regulates B Cell Proliferation and Plasmablast Differentiation. J. Immunol. 2018, 201 (9), 2799–2811. 10.4049/jimmunol.1800952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper K.; Grimbacher B.; Eibel H. B-Cell Biology and Development. J. Allergy Clin. Immunol. 2013, 131 (4), 959–971. 10.1016/j.jaci.2013.01.046. [DOI] [PubMed] [Google Scholar]
- de Gorter D. J. J.; Vos J. C. M.; Pals S. T.; Spaargaren M. The B Cell Antigen Receptor Controls AP-1 and NFAT Activity through Ras-Mediated Activation of Ral1. J. Immunol. 2007, 178 (3), 1405–1414. 10.4049/jimmunol.178.3.1405. [DOI] [PubMed] [Google Scholar]
- Peng S. L.; Gerth A. J.; Ranger A. M.; Glimcher L. H. NFATc1 and NFATc2 Together Control Both T and B Cell Activation and Differentiation. Immunity 2001, 14 (1), 13–20. 10.1016/S1074-7613(01)00085-1. [DOI] [PubMed] [Google Scholar]
- Weil R.; Israël A. T-Cell-Receptor- and B-Cell-Receptor-Mediated Activation of NF-κB in Lymphocytes. Curr. Opin. Immunol. 2004, 16 (3), 374–381. 10.1016/j.coi.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Wu H.-J.; Bondada S. Positive and Negative Roles of CD72 in B Cell Function. Immunol. Res. 2002, 25 (2), 155–166. 10.1385/IR:25:2:155. [DOI] [PubMed] [Google Scholar]
- Hua Z.; Hou B. TLR Signaling in B-Cell Development and Activation. Cell. Mol. Immunol. 2013, 10 (2), 103–106. 10.1038/cmi.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon V.; Wolf H. M.; Sasgary M.; Litzman J.; Samstag A.; Hauber I.; Lokaj J.; Eibl M. M. Defective Integration of Activating Signals Derived from the T Cell Receptor (TCR) and Costimulatory Molecules in Both CD4+ and CD8+ T Lymphocytes of Common Variable Immunodeficiency (CVID) Patients. Clin. Exp. Immunol. 2007, 110 (2), 174–181. 10.1111/j.1365-2249.1997.tb08314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon V.; Eggenbauer H.; Wolf H. M.; Fischer M. B.; Litzman J.; Lokaj J.; Eibl M. M. Antigen Presentation by Common Variable Immunodeficiency (CVID) B Cells and Monocytes Is Unimpaired. Clin. Exp. Immunol. 2003, 108 (1), 1–8. 10.1046/j.1365-2249.1997.d01-989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chovancova Z.; Vlkova M.; Litzman J.; Lokaj J.; Thon V. Antibody Forming Cells and Plasmablasts in Peripheral Blood in CVID Patients after Vaccination. Vaccine 2011, 29 (24), 4142–4150. 10.1016/j.vaccine.2011.03.087. [DOI] [PubMed] [Google Scholar]
- Nie L.; Xu M.; Vladimirova A.; Sun X.-H. Notch-Induced E2A Ubiquitination and Degradation Are Controlled by MAP Kinase Activities. EMBO J. 2003, 22 (21), 5780–5792. 10.1093/emboj/cdg567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garis M.; Garrett-Sinha L. A. Notch Signaling in B Cell Immune Responses. Front. Immunol. 2021, 11, 609324. 10.3389/fimmu.2020.609324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozen W.; Timofeeva M. N.; Li D.; Diepstra A.; Hazelett D.; Delahaye-Sourdeix M.; Edlund C. K.; Franke L.; Rostgaard K.; Van Den Berg D. J.; Cortessis V. K.; Smedby K. E.; Glaser S. L.; Westra H.-J.; Robison L. L.; Mack T. M.; Ghesquieres H.; Hwang A. E.; Nieters A.; de Sanjose S.; Lightfoot T.; Becker N.; Maynadie M.; Foretova L.; Roman E.; Benavente Y.; Rand K. A.; Nathwani B. N.; Glimelius B.; Staines A.; Boffetta P.; Link B. K.; Kiemeney L.; Ansell S. M.; Bhatia S.; Strong L. C.; Galan P.; Vatten L.; Habermann T. M.; Duell E. J.; Lake A.; Veenstra R. N.; Visser L.; Liu Y.; Urayama K. Y.; Montgomery D.; Gaborieau V.; Weiss L. M.; Byrnes G.; Lathrop M.; Cocco P.; Best T.; Skol A. D.; Adami H.-O.; Melbye M.; Cerhan J. R.; Gallagher A.; Taylor G. M.; Slager S. L.; Brennan P.; Coetzee G. A.; Conti D. V.; Onel K.; Jarrett R. F.; Hjalgrim H.; van den Berg A.; McKay J. D. A Meta-Analysis of Hodgkin Lymphoma Reveals 19p13.3 TCF3 as a Novel Susceptibility Locus. Nat. Commun. 2014, 5 (1), 3856. 10.1038/ncomms4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan H.; Xie L.; Wirth T.; Ushmorov A. Repression of TCF3/E2A Contributes to Hodgkin Lymphomagenesis. Oncotarget 2016, 7 (24), 36854–36864. 10.18632/oncotarget.9210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weniger M. A.; Küppers R. Molecular Biology of Hodgkin Lymphoma. Leukemia 2021, 35 (4), 968–981. 10.1038/s41375-021-01204-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham K.; Mielke H.; Fromme H.; Völkel W.; Menzel J.; Peiser M.; Zepp F.; Willich S. N.; Weikert C. Internal Exposure to Perfluoroalkyl Substances (PFASs) and Biological Markers in 101 Healthy 1-Year-Old Children: Associations between Levels of Perfluorooctanoic Acid (PFOA) and Vaccine Response. Arch. Toxicol. 2020, 94 (6), 2131–2147. 10.1007/s00204-020-02715-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean P.; Heilmann C.; Weihe P.; Nielsen F.; Mogensen U. B.; Budtz-Jørgensen E. Serum Vaccine Antibody Concentrations in Adolescents Exposed to Perfluorinated Compounds. Environ. Health Perspect. 2017, 125 (7), 077018. 10.1289/EHP275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielsen K.; Shamim Z.; Ryder L. P.; Nielsen F.; Grandjean P.; Budtz-Jørgensen E.; Heilmann C. Antibody Response to Booster Vaccination with Tetanus and Diphtheria in Adults Exposed to Perfluorinated Alkylates. J. Immunotoxicol. 2016, 13 (2), 270–273. 10.3109/1547691X.2015.1067259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looker C.; Luster M. I.; Calafat A. M.; Johnson V. J.; Burleson G. R.; Burleson F. G.; Fletcher T. Influenza Vaccine Response in Adults Exposed to Perfluorooctanoate and Perfluorooctanesulfonate. Toxicol. Sci. Off. J. Soc. Toxicol. 2014, 138 (1), 76–88. 10.1093/toxsci/kft269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermann C. A. G.; Jensen K. J.; Nielsen F.; Budtz-Jørgensen E.; van der Klis F.; Benn C. S.; Grandjean P.; Fisker A. B. Serum Perfluoroalkyl Substances, Vaccine Responses, and Morbidity in a Cohort of Guinea-Bissau Children. Environ. Health Perspect. 2020, 128 (8), 87002. 10.1289/EHP6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor K. D.; Woodlief T. L.; Ahmed A.; Hu Q.; Duncker P. C.; DeWitt J. C. Quantifying the Impact of PFOA Exposure on B-Cell Development and Antibody Production. Toxicol. Sci. 2023, 194 (1), 101–108. 10.1093/toxsci/kfad043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averbeck M.; Gebhardt C.; Emmrich F.; Treudler R.; Simon J. C. Immunologic Principles of Allergic Disease. JDDG J. Dtsch. Dermatol. Ges. 2007, 5 (11), 1015–1027. 10.1111/j.1610-0387.2007.06538.x. [DOI] [PubMed] [Google Scholar]
- Buser M. C.; Scinicariello F. Perfluoroalkyl Substances and Food Allergies in Adolescents. Environ. Int. 2016, 88, 74–79. 10.1016/j.envint.2015.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang I.-J.; Hsieh W.-S.; Chen C.-Y.; Fletcher T.; Lien G.-W.; Chiang H.-L.; Chiang C.-F.; Wu T.-N.; Chen P.-C. The Effect of Prenatal Perfluorinated Chemicals Exposures on Pediatric Atopy. Environ. Res. 2011, 111 (6), 785–791. 10.1016/j.envres.2011.04.006. [DOI] [PubMed] [Google Scholar]
- Lowe A. J.; Dharmage S. C.; Abramson M. J.; Vijayasarathy S.; Erbas B.; Mueller J. F.; Lodge C. J. Cord-Serum per- and Poly-Fluoroalkyl Substances and Atopy and Eczema at 12-Months. Allergy 2019, 74 (4), 812–815. 10.1111/all.13669. [DOI] [PubMed] [Google Scholar]
- Pennings J. L. A.; Jennen D. G. J.; Nygaard U. C.; Namork E.; Haug L. S.; van Loveren H.; Granum B. Cord Blood Gene Expression Supports That Prenatal Exposure to Perfluoroalkyl Substances Causes Depressed Immune Functionality in Early Childhood. J. Immunotoxicol. 2016, 13 (2), 173–180. 10.3109/1547691X.2015.1029147. [DOI] [PubMed] [Google Scholar]
- Reynés B.; Priego T.; Cifre M.; Oliver P.; Palou A. Peripheral Blood Cells, a Transcriptomic Tool in Nutrigenomic and Obesity Studies: Current State of the Art. Compr. Rev. Food Sci. Food Saf. 2018, 17 (4), 1006–1020. 10.1111/1541-4337.12363. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.