Abstract
Fargesin, a bioactive lignan derived from Flos Magnoliae, possesses anti-inflammatory, anti-oxidative, anti-melanogenic, and anti-apoptotic effects. This study compared the metabolic profiles of fargesin in human, dog, monkey, mouse, and rat hepatocytes using liquid chromatography-high resolution mass spectrometry. In addition, we investigated the human cytochrome P450 (CYP), UDP-glucuronosyltransferase (UGT), and sulfotransferase (SULT) enzymes responsible for fargesin metabolism. The hepatic extraction ratio of fargesin among the five species ranged from 0.59 to 0.78, suggesting that it undergoes a moderate-to-extensive degree of hepatic metabolism. During metabolism, fargesin generates three phase 1 metabolites, including fargesin catechol (M1) and O-desmethylfargesin (M2 and M3), and 11 phase 2 metabolites, including O-methyl-M1 (M4 and M5) via catechol O-methyltransferase (COMT), glucuronides of M1, M2, M4, and M5, and sulfates of M1–M5. The production of M1 from fargesin via O-demethylenation is catalyzed by CYP2C9, CYP3A4, CYP2C19, and CYP2C8 enzymes, whereas the formation of M2 and M3 (O-desmethylfargesin) is catalyzed by CYP2C9, CYP2B6, CYP2C19, CYP3A4, CYP1A2, and CYP2D6 enzymes. M4 is metabolized to M4 glucuronide by UGT1A3, UGT1A8, UGT1A10, UGT2B15, and UGT2B17 enzymes, whereas M4 sulfate is generated by multiple SULT enzymes. Fargesin is extensively metabolized in human hepatocytes by CYP, COMT, UGT, and SULT enzymes. These findings help to elucidate the pharmacokinetics and drug interactions of fargesin.
Keywords: Fargesin, Hepatocytes, In vitro metabolism, Cytochrome P450, UDP-glucuronosyltransferase
Introduction
Fargesin, 5-[(3S,3aR,6R,6aR)-6-(3,4-dimethoxyphenyl)-1,3,3a,4,6,6a-hexahydrofuro[3,4-c]furan-3-yl]-1,3-benzodioxole is a bioactive lignan derived from Flos Magnoliae [1]. It has anti-allergic effects [2], inhibits inducible nitric oxide synthase [3, 4], and improves osteoarthritis, chemical-induced bowel disease, and atherosclerosis via anti-inflammatory effects [5–9]. Furthermore, it promotes reverse cholesterol transport [9] and has in vivo anti-hypertensive effects via attenuation of oxidative stress and apoptosis [10, 11]. It also improves lipid and glucose metabolism [12, 13] and inhibits colon cancer cell growth via suppression of the CDK2/cyclin E signaling pathway [14] and melanin synthesis [15].
During the development of a new drug, drug candidates are selected based on efficacy and pharmacokinetic attributes, such as clearance, oral bioavailability, and metabolic profile [16]. During the preclinical development stage, the metabolism and pharmacokinetic properties of drug candidates must be characterized to predict the drug exposure levels and safety in humans. Therefore, it is necessary to characterize the metabolic stability and metabolic pathways of drugs in human and animal hepatocytes, the gold standard model for in vitro metabolism studies. Furthermore, the identification of drug-metabolizing enzymes, such as cytochrome P450 (CYP), UDP-glucuronosyltransferase (UGT), and sulfotransferase (SULT), which are responsible for the metabolism of candidate drugs, can predict pharmacokinetics and drug interactions [16, 17].
Few studies have evaluated the pharmacokinetics of fargesin in rats and mice [18, 19]. In one such study on male ICR mice, intravenous or oral administration of fargesin at 1, 2, and 4 mg/kg led to high clearance (53.2–55.5 mL/min/kg), short half-life (84.7–140.0 min), low absolute bioavailability (4.1–9.6%), no urinary recovery, and negligible cumulative fecal recovery for 48 h (< 0.089% of the dose), suggesting that the high clearance of fargesin is attributable to its metabolism [18]. However, no studies have evaluated the metabolism of fargesin in experimental animals and humans.
In two previous studies, fargesin inhibited CYP2C9-mediated diclofenac hydroxylation (Ki: 16.3 µM), UGT1A1-mediated SN-38 glucuronidation (Ki: 25.3 µM), and UGT1A3-mediated chenodeoxycholic acid 24-acyl-glucuronidation (Ki: 24.5 µM) in human liver microsomes [20, 21]. Furthermore, it showed time-dependent inhibition of CYP2C8-catalyzed amodiaquine N-deethylation (Ki: 10.7 µM and kinact: 0.082 min−1), CYP2C19-catalyzed [S]-mephenytoin hydroxylation (Ki: 3.7 µM and kinact: 0.102 min−1), and CYP3A4-catalyzed midazolam hydroxylation (Ki: 23.0 µM and kinact: 0.050 min−1) in human liver microsomes [20].
The present study explored the in vitro metabolic pathways of fargesin in hepatocytes from humans and four animal models using liquid chromatography-high resolution mass spectrometry (LC-HRMS), and characterized the human drug-metabolizing enzymes involved in its metabolism to predict pharmacokinetics and drug interactions.
Materials and methods
Materials and reagents
Fargesin (purity, 98.0%), sylvatesmin (purity, 98.54%, M4), and magnolin (purity, 98.9%; internal standard) were obtained from Tokyo Chemical Industry (Tokyo, Japan), Toronto Research Chemicals (Toronto, Canada), and PhytoLab GmbH & Co. (Vestenbergsgreuth, Germany), respectively. Alamethicin, reduced nicotinamide adenine dinucleotide phosphate (NADPH), 3-phosphoadenosine-5-phosphosulfate (PAPS), S-adenosylmethionine (SAM), and uridine 5′-diphosphoglucuronic acid (UDPGA) were obtained from Sigma-Aldrich Co. (St. Louis, MO, USA). Pooled human liver microsomes, pooled human liver S9 fraction, cryopreserved dog, monkey, mouse, and rat hepatocytes, human cDNA-expressed CYP enzymes (CYP1A2, 2A6, 2B6, 2C8, 2C9, 2C19, 2D6, 2E1, 3A4, and 3A5) and UGT enzymes (UGT 1A1, 1A3, 1A4, 1A6, 1A7, 1A8, 1A9, 1A10, 2B4, 2B7, 2B10, 2B15, and 2B17) were obtained from Corning Life Sciences (Woburn, MA, USA). Human cDNA-expressed SULT enzymes (SULT 1A1*1, 1A1*2, 1A2, 1A3, 1B1, 1C2, 1C4, 1E1, and 2A1) were obtained from Cyprotex Ltd. (Scotland, UK). LiverPool™ pooled human hepatocytes (50-donor mixed gender), INVITROGRO HT medium, and INVITROGRO KHB were obtained from BioIVT (Westbury, NY, USA). Cryopreserved hepatocyte recovery medium for cryopreserved dog, monkey, mouse, and rat hepatocytes was obtained from Invitrogen (Waltham, MA, USA). Methanol and water (HPLC grade) were obtained from Thermo Fischer Scientific (Fair Lawn, NJ, USA). All other chemicals were of the highest quality available.
Metabolic stability of fargesin in human and animal hepatocytes
To evaluate the metabolic stability of fargesin in human, dog, monkey, mouse, and rat hepatocytes, pooled cryopreserved hepatocytes were carefully thawed in recovery medium and resuspended in incubation medium (Krebs–Henseleit buffer for mouse, rat, and human hepatocytes; William’s E media for dog and monkey hepatocytes) to achieve a final density of 5 × 105 cells/mL. Then, 60 µL hepatocyte suspension and an equal volume of 1 μM fargesin in the incubation medium were mixed in 96-well plates and incubated in triplicate for 0, 15, 30, 45, 60, 90, and 120 min at 37 °C in a CO2 incubator. Next, 120 μL ice-cold magnolin (internal standard) in methanol was added to each well and the mixtures were sonicated for 5 min, followed by centrifugation at 13,000 rpm for 10 min at 4 °C. An aliquot of each supernatant was transferred to an autosampler vial, and 5 µL supernatant was analyzed via LC-HRMS, as described previously [18].
The depletion rate constant (k) was calculated from the slope of the natural logarithm plots of the percentage of fargesin remaining at each time point compared to 0 min. In vitro parameters of metabolic stability, such as half-life (t1/2), intrinsic clearance (Clint), and hepatic clearance (Clhep), and hepatic extraction ratio of fargesin in human, dog, monkey, mouse, and rat hepatocytes [22] were calculated using the following equations:
Here, A has values of 87.5, 40, 32, 32, and 25.7; B has values of 135, 117, 215, 120, and 139; Qh (representing hepatic blood flow) has values of 90, 55.2, 30.9, 43.4, and 20.7 mL/min/kg for mouse, rat, dog, monkey, and human, respectively [23]. Hepatic extraction ratio values of ≤ 0.25, 0.25–0.75, and ≥ 0.75 were regarded as low, moderate, and high, respectively [22].
Metabolite profiling of fargesin in hepatocytes
To identify in vitro metabolites of fargesin, 60 µL aliquots of dog, monkey, mouse, rat, or human hepatocyte suspensions (5 × 105 cells/mL) and an equal volume of 10 μM fargesin in the incubation medium were mixed in 96-well plates and incubated for 2 h in a CO2 incubator at 37 °C. 120 μL ice-cold methanol was added to each well and the mixtures were sonicated for 5 min, followed by centrifugation at 13,000 rpm for 10 min at 4 ℃. The supernatants were evaporated to dryness using a vacuum concentrator. The residues were dissolved in 100 μL 40% methanol and 5 μL aliquots were injected into the LC-HRMS system.
To identify the glucuronidation of phase 1 metabolites, M1–M3, 100 μL reaction mixture containing 50 mM potassium phosphate buffer (pH 7.4), 10 mM magnesium chloride, human liver microsomes (20 μg protein), 1 mM NADPH, 2 mM UDPGA, and 10 μM fargesin was used. The mixture was incubated at 37 °C for 30 min. The reactions were quenched by adding 200 μL methanol. After centrifugation, the supernatants were evaporated to dryness using a vacuum concentrator. The residues were dissolved in 100 μL 40% methanol and 5 μL aliquots were injected into the LC-HRMS system.
To identify fargesin and its metabolites, we used a Q-Exactive Orbitrap mass spectrometer (Thermo Fisher Scientific) coupled with Nexera X2 UPLC (Shimadzu, Kyoto, Japan). Fargesin and its metabolites were separated on a Halo C18 column (2.1 × 100 mm, 2.7 μm; Advanced Material Technology, Wilmington, DE, USA) via gradient elution using 5% (v/v) methanol in 10 mM ammonium formate (mobile phase A) and 95% methanol (mobile phase B) at a flow rate of 0.25 mL/min: 40% mobile phase B for 0.5 min, 40 to 60% mobile phase B over 29.5 min, 60 to 90% mobile phase B over 0.5 min, 90% mobile phase B for 3 min, 90 to 40% mobile phase B over 0.5 min, and 40% mobile phase B for 3.5 min. The column and autosampler temperatures were 40 °C and 4 °C, respectively. Accurate mass values of fargesin and its metabolites were measured via positive electrospray ionization, with the following electrospray source settings: capillary temperature, 250 °C; aux gas heater temperature, 200 °C; spray voltage, 3.8 kV; nitrogen sheath gas, 40 arbitrary units; and auxiliary gas, 10 arbitrary units. Full scan MS1 with the data-dependent MS2 acquisition mode was used to obtain MS scan data ranging from m/z 100 to 1000 with resolution of 70,000. Nitrogen gas was used for higher-energy collision dissociation at an energy of 15 eV to obtain product ion spectra of fargesin and its metabolites. Data were processed using Xcalibur software version 2.2 (Thermo Fisher Scientific). The drug structures were analyzed using Mass Frontier software (version 6.0; HighChem Ltd., Bratislava, Slovakia).
Screening of CYP enzymes involved in the phase 1 metabolism of fargesin
Incubation mixtures (95 μL) containing fargesin (final concentration: 5 μM), 10 mM magnesium chloride, and 10 human cDNA-expressed CYP enzymes (CYP1A2, CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP2E1, CYP3A4, and CYP3A5; 4 pmol) in 50 mM potassium phosphate buffer (pH 7.4) were mixed with 5 μL 1 mM NADPH and incubated at 37 °C for 30 min in a shaking water bath. The reactions were terminated by adding 100 μL ice-cold magnolin (100 ng/mL) in methanol. The samples were centrifuged at 13,500 rpm for 10 min at 4 °C and 5 μL supernatants were analyzed via LC-HRMS, as described above.
In the CYP screening experiments, the levels of fargesin and its metabolites, M1–M3 were quantified based on the parallel reaction monitoring (PRM) transitions using m/z 388.17547 → 135.04407 for fargesin, m/z 376.17547 → 123.04408 for M1, m/z 374.15982 → 137.05974 for M2 and M3. The concentrations of M1–M3 metabolites were determined using the standard curve of fargesin in the absence of authentic standards.
The relative contributions of CYP isoforms to the formation of M1–M3 metabolites from fargesin in human liver microsomes were determined using where RAF is the relative activity factor that encompasses the hepatic abundance of each CYP isoform and the differences in activity per unit enzyme between human cDNA-expressed CYPs and human liver microsomal CYPs [24].
Screening of UGT enzymes involved in M4 glucuronidation
A mixture (95 μL) containing M4 (final concentration: 10 μM), alamethacin, and 13 human cDNA-expressed UGT enzymes (UGTs 1A1, 1A3, 1A4, 1A6, 1A7, 1A8, 1A9, 1A10, 2B4, 2B7, 2B10, 2B15, and 2B17; 10 μg protein) in 50 mM Tris buffer (pH 7.4) was mixed with 5 μL UDPGA and incubated at 37 °C for 30 min in a shaking water bath. The reactions were stopped by adding 200 μL ice-cold magnolin (100 ng/mL) in methanol. The samples were centrifuged at 13,500 rpm for 10 min at 4 °C and 5 μL supernatant was analyzed via LC-HRMS, as described above.
The levels of M4 and its metabolite M4 glucuronide (M4-G) were quantified based on PRM transitions using m/z 390.1907 → 137.05971 for M4 and m/z 566.22375 → 355.15320 for M4-G. As no authentic standard of the M4-G metabolite was available, the metabolite was quantified based on the standard curve of M4 (sylvatesmin).
Screening of SULT enzymes involved in M4 sulfation
A mixture (95 μL) containing M4 (final concentration: 10 μM) and human liver S9 fraction or 10 human cDNA-expressed SULT enzymes (SULTs 1A1*1, 1A1*2, 1A2, 1A3, 1B1, 1C2, 1C4, 1E1, and 2A1; 0.25 μg protein) in 50 mM phosphate buffer (pH 7.4) were mixed with 5 μL 0.4 mM PAPS and incubated at 37 °C for 30 min. The reactions were terminated by adding 200 μL ice-cold magnolin (100 ng/mL) in methanol. The samples were centrifuged at 13,500 rpm for 10 min at 4 °C and 5 μL supernatant was analyzed via LC-HRMS, as described above.
The levels of M4 and its metabolite M4 sulfate (M4-S) were quantified based on PRM transitions using m/z 390.1907 → 137.05971 for M4 and m/z 470.14750 → 315.05255 for M4-S. As no authentic standard of the M4-S metabolite was available, its levels were quantified on the basis of the standard curve of M4.
Results
Metabolic stability of fargesin in hepatocytes
The in vitro parameters of metabolic stability (e.g., t1/2, CLint, CLhep, and hepatic extraction ratio) of fargesin obtained after incubation with human, dog, monkey, mouse, and rat hepatocytes are shown in Table 1. The t1/2 values of fargesin in human, dog, monkey, mouse, and rat hepatocytes were 68.3, 204.0, 46.2, 130.6, and 104.2 min, respectively. Using a well-stirred model, the CLint values were 72.5, 46.7, 115.2, 125.4, and 62.3 mL/min/kg, whereas the CLhep values were 16.1, 18.6, 31.5, 52.4, and 29.3 mL/min/kg, respectively (Table 1). The hepatic extraction ratios were 0.78, 0.60, 0.73, 0.58, and 0.53, respectively, indicating that it is highly metabolized (> 0.75) in humans but is moderately metabolized (0.25–0.75) in the others [22].
Table 1.
Half-life (t1/2), intrinsic clearance (CLint), hepatic clearance (CLhep), and hepatic extraction ratio values of fargesin obtained by incubation with human, dog, monkey, mouse, and rat hepatocytes at 37 °C in a CO2 incubator
| Parameters | Human | Dog | Monkey | Mouse | Rat |
|---|---|---|---|---|---|
| t1/2 (min) | 68.3 | 204.0 | 46.2 | 130.6 | 104.2 |
| CLint (mL/min/kg) | 72.5 | 46.7 | 115.2 | 125.4 | 62.3 |
| CLhep (mL/min/kg) | 16.1 | 18.6 | 31.5 | 52.4 | 29.3 |
| Hepatic extraction ratio | 0.78 | 0.60 | 0.73 | 0.58 | 0.53 |
Metabolic profiles of fargesin in hepatocytes
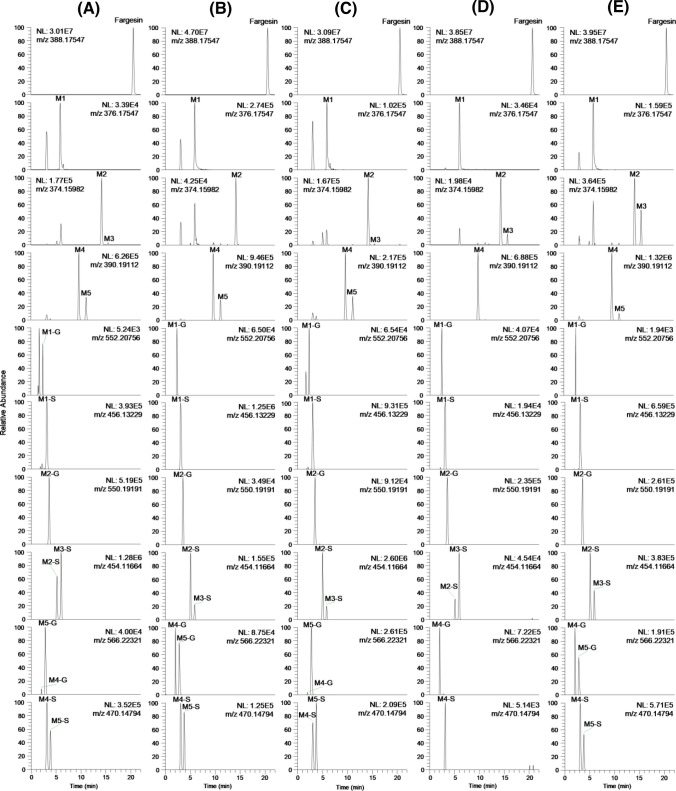
Incubation of fargesin with each type of hepatocyte resulted in the formation of 14 metabolites. The extracted ion chromatograms of fargesin, three phase 1 metabolites (M1–M3), and 11 phase 2 metabolites are shown in Fig. 1. The retention time, elemental composition, observed molecular ion, mass error, and product ions of fargesin and its 14 metabolites are presented in Table 2. M1–M4, M1-G, M1-S, M2-G, M2-S, M3-S, M4-G, and M4-S were formed in all species. M5, M5-G, and M5-S were formed from fargesin in all species except mice.
Fig. 1.
Extracted ion chromatograms of fargesin and its possible 14 metabolites obtained following incubation of 10 μM fargesin with a human, b dog, c monkey, d mouse, and e rat hepatocytes at 37 ℃ for 2 h
Table 2.
Retention times (tR), molecular formulae, observed molecular ions, mass errors, product ions, and biotransformation of fargesin and its metabolites identified after incubation of 10 μM fargesin with human, dog, monkey, mouse, and rat hepatocytes at 37 °C
| Metabolite | tR (min) | Molecular formula | Detected [M + NH4]+ (m/z) | Mass error (ppm) | Product ions (m/z) | Biotransformation |
|---|---|---|---|---|---|---|
| Fargesin | 20.56 | C21H22O6 | 388.17548 | 0.03 | 135.04388, 151.07513, 203.07022, 233.08055, 249.11177, 353.13803 | |
| M1 | 5.87 | C20H22O6 | 376.17593 | 1.22 | 123.04385, 151.07544, 191.06989, 221.08063, 249.11093, 341.13812 | O-demethylenation |
| M1-G | 2.35 | C26H30O12 | 552.20776 | 0.36 | 191.06966, 221.08070, 249.11020, 341.13760, 517.16846 | O-demethylenation and glucuronidation |
| M1-S | 3.14 | C20H22O9S | 456.13129 | −2.19 | 123.04344, 249.11234, 271.02643, 301.03720, 341.13879, 376.17484, 421.09473 | O-demethylenation and sulfation |
| M2 | 14.18 | C20H20O6 | 374.15967 | −0.40 | 135.04411, 137.05966, 203.07039, 233.08035, 235.09657, 339.12103 | O-demethylation |
| M3 | 15.46 | C20H20O6 | 374.15976 | −0.16 | 135.04417, 137.05978, 203.07045, 233.08090, 235.09656, 339.12311 | O-demethylation |
| M2-G | 3.55 | C26H28O12 | 550.19257 | 1.20 | 203.06995, 233.08055, 235.09608, 339.12250 | O-demethylation and glucuronidation |
| M2-S | 5.20 | C20H20O9S | 454.11606 | −1.28 | 135.04382, 233.08055, 315.05307, 339.12241, 374.15915, 419.07907 | O-demethylation and sulfation |
| M3-S | 6.04 | C20H20O9S | 454.11685 | 0.46 | 135.04407, 233.08089, 315.05330, 339.12283, 374.15967, 419.07959 | O-demethylation and sulfation |
| M4 | 9.61 | C21H24O6 | 390.19040 | −1.85 | 137.05954, 151.07513, 205.08569, 235.09628, 249.11174, 355.15353 | O-demethylenation and O-methylation |
| M5 | 11.08 | C21H24O6 | 390.19116 | 0.10 | 137.05988, 151.07544, 205.08586, 235.09680, 249.11200, 355.15384 | O-demethylenation and O-methylation |
| M4-G | 2.06 | C27H32O12 | 566.22186 | −2.38 | 235.09625, 249.11206, 355.15320 | O-demethylenation, O-methylation, and glucuronidation |
| M5-G | 2.87 | C27H32O12 | 566.22375 | 0.95 | 235.09702, 249.11147, 355.15448 | O-demethylenation, O-methylation, and glucuronidation |
| M4-S | 3.12 | C21H24O9S | 470.14792 | −0.04 | 235.09683, 249.11256, 315.05347, 355.15262, 390.19131, 435.11053 | O-demethylenation, O-methylation, and sulfation |
| M5-S | 3.87 | C21H24O9S | 470.14795 | 0.02 | 235.09674, 249.11244, 315.05365, 355.15466, 390.18954, 435.11108 | O-demethylenation, O-methylation, and sulfation |
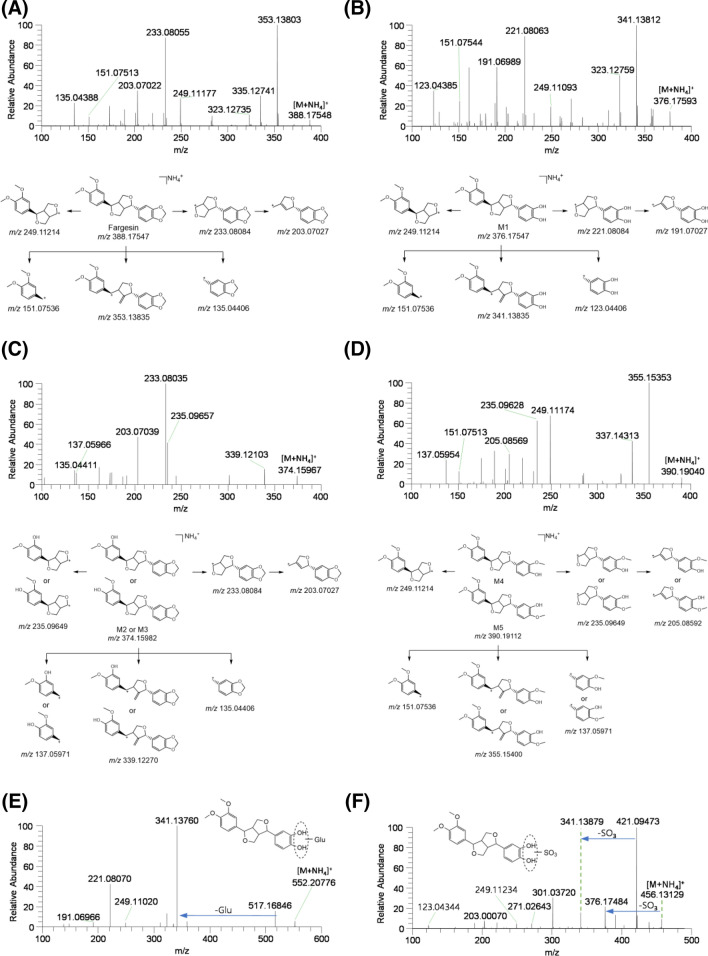
Fargesin exhibited an ammonium adduct ([M + NH4]+) peak at m/z 388.17548 as molecular ion in MS spectrum. The characteristic product ions at m/z 353.13803 (loss of water and NH3 from [M + NH4]+ ion), m/z 249.11177 (4-(3,4-dimethoxyphenyl)tetrahydro-1H,3H-furo[3,4-c]furan-1-ylium ion), m/z 233.08055 (4-(benzo[d][1,3]dioxol-5-yl)tetrahydro-1H,3H-furo[3,4-c]furan-1-ylium ion), m/z 203.07022 (5-(benzo[d][1,3]dioxol-5-yl)-2,5-dihydrofuran-3-yl)methylium ion), m/z 151.07513 ((3,4-dimethoxyphenyl)methylium ion), and m/z 135.04388 (benzo[1.3]dioxol-5-ylmethylium ion) were produced within MS/MS spectrum of fargesin ([M + NH4]+; Fig. 2a).
Fig. 2.
MS/MS spectra and fragmentation of a fargesin and its possible metabolites, b M1, c M2 and M3, d M4 and M5, e M1 glucuronide (M1-G), and f M1 sulfate (M1-S)
M1 exhibited an [M + NH4]+ ion at m/z 376.17593 with 12 atomic mass units (amu) less than that of the [M + NH4]+ ion of fargesin. The MS/MS spectrum of M1 included product ions at m/z 341.13812 (loss of water and NH3 from [M + NH4]+ ion), m/z 249.11093, m/z 221.08063 (4-(3,4-dihydroxyphenyl)tetrahydro-1H,3H-furo[3,4-c]furan-1-ylium), m/z 191.06989 ((5-(3,4-dihydroxyphenyl)-2,5-dihydrofuran-3-yl)methylium ion), m/z 151.07544, and m/z 123.04385 ((3,4-dihydroxyphenyl)methylium ion) (Fig. 2b), indicating that M1 was produced by demethylenation of benzo[1,3]dioxole to pyrocatechol. As a result, M1 was presumed to be catechol metabolite of fargesin.
M2 and M3 exhibited an [M + NH4]+ ion at m/z 374.15967 with 14 amu less than that of the [M + NH4]+ ion of fargesin, indicating O-demethylation of fargesin. The MS/MS spectra of M2 and M3 generated the product ions at m/z 339.12103 (loss of water and NH3 from [M + NH4]+ ion), m/z 235.09657 (4-(3-hydroxy-4-methoxyphenyl)tetrahydro-1H,3H-furo[3,4-c]furan-1-ylium or 4-(4-hydroxy-3-methoxyphenyl)tetrahydro-1H,3H-furo[3,4-c]furan-1-ylium ion), m/z 233.08035, m/z 203.07039, m/z 137.05966 ((3-hydroxy-4-methoxyphenyl)methylium or (4-methoxy-3-hydroxyphenyl)methylium ion), and m/z 135.04411 (Fig. 2c). M2 and M3 were presumed to be 3-O-desmethyl- and 4-O-desmethyl-fargesin, whereas the accurate position of O-demethylation at the 3,4-dimethoxybenzyl moiety of M2 and M3 could not be ascertained due to a lack of authentic standards.
M4 and M5 exhibited an [M + NH4]+ ion at m/z 390.19040, with 14 amu more than that of the [M + NH4]+ ion of M1, indicating O-methylation of M1. The MS/MS spectra of M4 and M5 generated the product ions at m/z 355.15353 (loss of water and NH3 from [M + NH4]+ ion), m/z 249.11174, m/z 235.09628 (4-(4-hydroxy-3-methoxyphenyl)tetrahydro-1H,3H-furo[3,4-c]furan-1-ylium and 4-(3-hydroxy-4-methoxyphenyl)tetrahydro-1H,3H-furo[3,4-c]furan-1-ylium ion for M4 and M5, respectively), m/z 205.08569 ((5-(4-hydroxy-3-methoxyphenyl)-2,5-dihydrofuran-3-yl)methylium or (5-(3-hydroxy-4-methoxyphenyl)-2,5-dihydrofuran-3-yl)methylium ion), m/z 151.07513, and m/z 137.05954 ((4-hydroxy-3-methoxyphenyl)methylium or (3-hydroxy-4-methoxyphenyl)methylium ion) (Fig. 2d). M4 was confirmed as sylvatesmin on the basis of the retention time and MS/MS spectrum of the authentic standard; therefore, M5 was identified as 4-O-methyl-M1. Incubation of fargesin with the human liver S9 fraction in the presence of NADPH and SAM as cofactors at 37 °C for 30 min resulted in the formation of M1–M5, suggesting that M4 and M5 were 3- and 4-O-methyl-M1 formed from fargesin by CYP and catechol O-methyltransferase (COMT).
M1-G exhibited an [M + NH4]+ ion at m/z 552.20776, with 176 amu higher than that of the [M + NH4]+ ion of M1, and generated the product ions at m/z 517.16846 (loss of water and NH3 from [M + NH4]+ ion), m/z 341.13760 (loss of glucuronic acid from m/z 517.16846 ion), m/z 249.11020, m/z 221.08070, and m/z 191.06966 (Fig. 2e). M1-G was presumed to be M1 glucuronide, although the accurate position of glucuronidation at the benzene 1,2-diol moiety of M1 could not be determined due to a lack of an authentic standard.
M1-S exhibited an [M + NH4]+ ion at m/z 456.13129, with 80 amu higher than that of the [M + NH4]+ ion of M1, and generated the product ions at m/z 421.09473 (loss of water and NH3 from [M + NH4]+ ion), m/z 376.17484 (loss of sulfate group from [M + NH4]+ ion), m/z 341.13879 (loss of sulfate group from m/z 421.09473 ion), and m/z 123.04344 (Fig. 2f). M1-S was presumed to be M1 sulfate, although the accurate position of sulfation at benzene 1,2-diol moiety of M1 could not be determined due to a lack of an authentic standard.
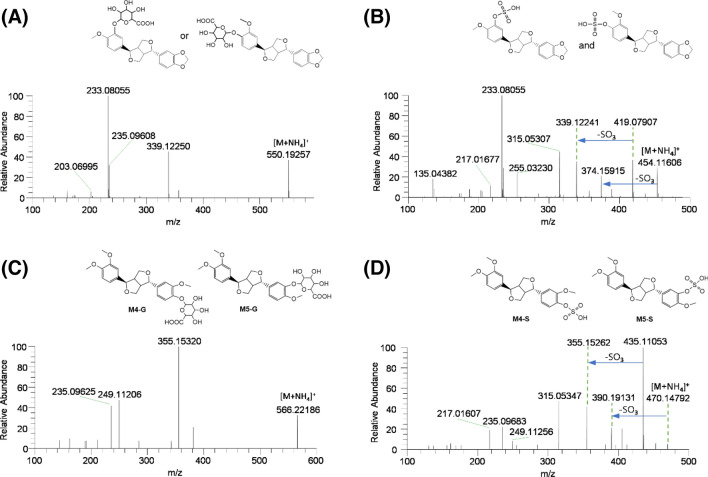
M2-G exhibited an [M + NH4]+ ion at m/z 550.19257, with 176 amu higher than that of the [M + NH4]+ ion of O-desmethylfargesin (M2 and M3), indicating the glucuronidation of O-desmethylfargesin. The MS/MS spectrum of M2-G included the product ions at m/z 339.12250, m/z 235.09608, m/z 233.08055, and m/z 203.06995 (Fig. 3a). Compared to the incubation of fargesin in the presence of NADPH, incubation of fargesin with human liver microsomes in the presence of NADPH and UDPGA resulted in the formation of O-desmethylfargesin glucuronide and a decrease in M2 concentration, without a change in M3 concentration. These results indicate that M2 was further metabolized to M2 glucuronide; therefore, M2-G is presumed to be M2 glucuronide.
Fig. 3.
MS/MS spectra of possible fargesin metabolites: a M2 glucuronide (M2-G), b M2 sulfate (M2-S) and M3 sulfate (M3-S), c M4 glucuronide (M4-G) and M5 glucuronide (M5-G), and d M4 sulfate (M4-S) and M5 sulfate (M5-S)
M2-S and M3-S exhibited an [M + NH4]+ ion at m/z 454.11606, with 80 amu higher than that of the [M + NH4]+ ion of M2 and M3, and generated product ions at m/z 419.07907 (loss of water and NH3 from [M + NH4]+ ion), m/z 374.15915 (loss of sulfate group from [M + NH4]+ ion), m/z 339.12241 (loss of sulfate group from m/z 419.07907 ion), m/z 315.05307 (loss of 1,3-benzodioxole moiety and NH3 from [M + NH4]+ ion), m/z 233.08055, and m/z 135.04382 (Fig. 3b). M2-S and M3-S were presumed to be M2 sulfate and M3 sulfate, respectively; however, the position of sulfation could not be accurately determined due to a lack of authentic standard.
M4-G and M5-G exhibited an [M + NH4]+ ion at m/z 566.22186, with 176 amu higher than that of the [M + NH4]+ ion of M4 and M5, indicating the glucuronidation of M4 (sylvatesmin) and M5. Their MS/MS spectra showed product ions at m/z 355.15320 (loss of glucuronosyl group, water, and NH3 from [M + NH4]+ ion), m/z 249.11206, and m/z 235.09625 (Fig. 3c). Based on the retention time and MS/MS spectrum of M4 glucuronide (M4-G) formed after the incubation of sylvatesmin (M4) with human liver microsomes and UDPGA at 37 °C for 30 min, M4-G was identified as sylvatesmin (M4) glucuronide. M5-G was characterized as M5 glucuronide.
M4-S and M5-S exhibited an [M + NH4]+ ion at m/z 470.14792, with 80 amu more than that of the [M + NH4]+ ion of M4 and M5, and generated product ions at m/z 435.11053 (loss of water and NH3 from [M + NH4]+ ion), m/z 390.19131 (loss of sulfate moiety from [M + NH4]+ ion), m/z 355.15262 (loss of sulfate moiety from m/z 435.11053 ion), m/z 315.05347 (loss of 3,4-dimethoxyphenyl moiety from molecular ion), m/z 249.11256, and m/z 235.09683 (loss of sulfate moiety from m/z 315.05262) (Fig. 3d), indicating the sulfation of M4 and M5. Based on the retention time and MS/MS spectrum of M4 sulfate (M4-S) formed after incubation of sylvatesmin (M4) with human liver S9 fractions and PAPS at 37 °C for 30 min, M4-S was identified as sylvatesmin (M4) sulfate. M5-S was characterized as M5 sulfate.
Screening of CYP, UGT, and SULT enzymes involved in fargesin metabolism
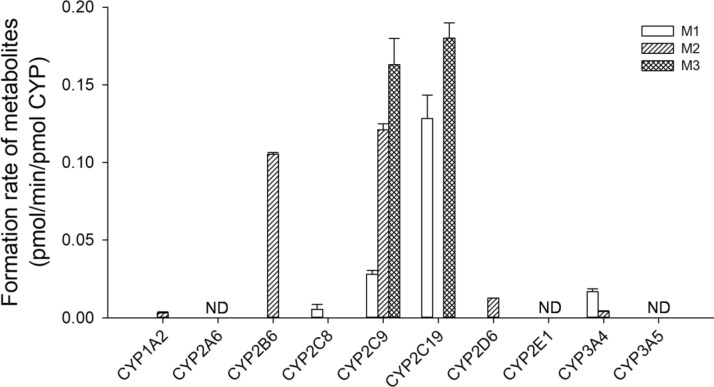
To characterize the CYP enzymes involved in fargesin metabolism, the formation rates of phase I metabolites, such as M1–M3, from 5 μM fargesin were evaluated by incubation with 10 human cDNA-expressed CYPs (1A2, 2A6, 2B6, 2C8, 2C9, 2C19, 2D6, 2E1, 3A4, and 3A5) and NADPH at 37 °C for 30 min. CYP1A2, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, and CYP3A4 enzymes were involved in fargesin metabolism, whereas CYP2A6, CYP2E1, and CYP3A5 enzymes were not involved (Fig. 4). The relative contributions of CYP2C8, CYP2C9, CYP2C19, and CYP3A4 to M1 formation from fargesin were 1.5%, 84.2%, 5.3%, and 9.0%, respectively. The relative contributions of CYP1A2, CYP2B6, CYP2C9, CYP2D6, and CYP3A4 to M2 formation from fargesin were 0.2%, 31.8%, 67.4%, 0.1%, and 0.4%, respectively, whereas M3 formation from fargesin was catalyzed by CYP2C9 and CYP2C19 based on their relative contributions (98.5% and 1.5%, respectively). These results indicate that CYP2C9 enzyme plays a major role in the formation of M1–M3 from fargesin, with minor contributions from CYP1A2, CYP2B6, CYP2C8, CYP2C19, CYP2D6, and CYP3A4.
Fig. 4.
Formation rates of M1–M3 following incubation of 5 μM fargesin with human cDNA-expressed CYP enzymes (1A2, 2A6, 2B6, 2C8, 2C9, 2C19, 2D6, 2E1, 3A4, and 3A5). Data points represent the mean ± standard deviation (n = 3). ND not detected (lower limit of quantification: 0.002 pmol/min/pmol CYP)
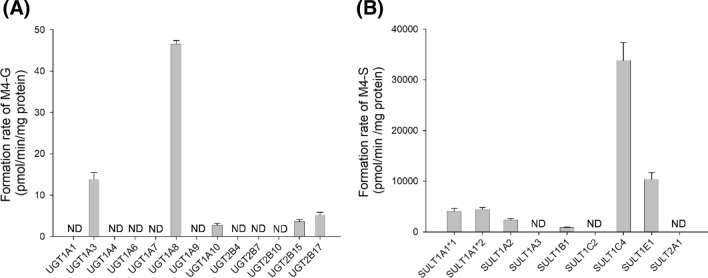
To characterize the UGT enzymes involved in the metabolism of M4 to M4 glucuronide (M4-G), the formation rates of M4-G from 10 μM sylvatesmin (M4) were evaluated by incubation with 13 human cDNA-expressed UGTs (1A1, 1A3, 1A4, 1A6, 1A7, 1A8, 1A9, 1A10, 2B4, 2B7, 2B10, 2B15, and 2B17) and UDPGA at 37 °C for 30 min. UGT1A3, UGT1A8, UGT1A10, UGT2B15, and UGT2B17 were involved in the formation of M4-G from M4 (Fig. 5a).
Fig. 5.
Formation rates of a M4 glucuronide (M4-G) and b M4 sulfate (M4-S) following incubation of 10 μM sylvatesmin (M4) with human cDNA-expressed UGT enzymes (1A1, 1A3, 1A4, 1A6, 1A7, 1A8, 1A9, 1A10, 2B4, 2B7, 2B10, 2B15, and 2B17) and SULT enzymes (1A1*1, 1A1*2, 1A2, 1A3, 1B1, 1C2, 1E1, and 2A1), respectively. Data points represent the mean ± standard deviation (n = 3). ND not detected (lower limit of quantification: 0.5 pmol/min/mg protein)
To characterize the SULT enzymes involved in the metabolism of M4 to M4 sulfate (M4-S), the formation rates of M4-S from 10 μM sylvatesmin (M4) were evaluated by incubation with human cDNA-expressed SULTs (1A1*1, 1A1*2, 1A2, 1A3, 1B1, 1C2, 1E1, and 2A1) and PAPS at 37 °C for 30 min. SULT1A1*1, SULT1A1*2, SULT1A2, SULT1B1, SULT1C4, and SULT1E1 enzymes were responsible for the formation of M4-S from M4 (Fig. 5b).
Discussion
Our data indicate that fargesin is extensively metabolized in humans but moderately metabolized in dogs, monkeys, mice, and rats. The CLhep value (52.4 mL/min/kg) in mouse hepatocytes was comparable to the systemic clearance value (53.2–55.5 mL/min/kg) following intravenous injection of fargesin in male ICR mice [18], suggesting that hepatic metabolism is the major clearance pathway of fargesin in experimental animals and humans.
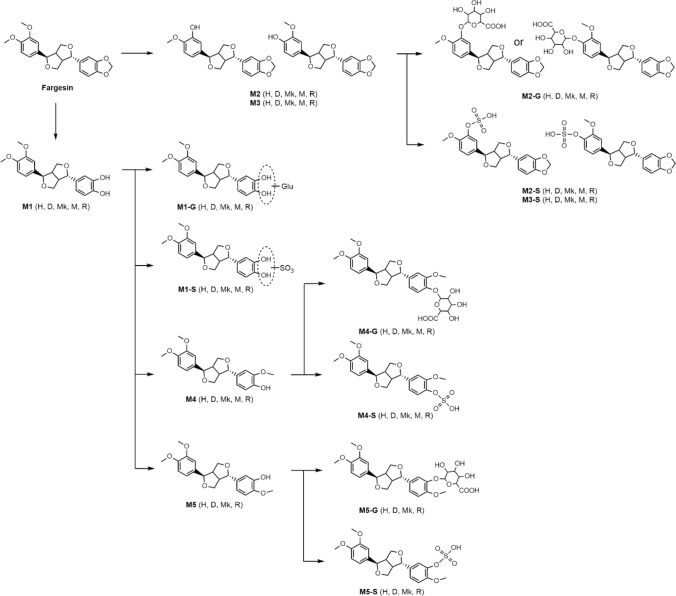
The incubation of fargesin with human, dog, monkey, mouse, and rat hepatocytes resulted in the formation of three phase 1 metabolites and 11 phase 2 metabolites via O-methylation, glucuronidation, and sulfation of three phase 1 metabolites (Fig. 1; Table 2). The potential in vitro metabolic pathways of fargesin in each species are summarized in Fig. 6.
Fig. 6.
Potential metabolic pathways of fargesin in human (H), dog (D), monkey (Mk), mouse (M), and rat (R) hepatocytes. SO3: sulfate; Glu: glucuronosyl
The metabolism of fargesin to fargesin catechol (M1) is a major metabolic pathway in the hepatocytes of all five species. This is analogous to the CYP-mediated O-demethylenation of 1,3-benzodioxole compounds such as myristicin [25–27], irisflorenin [28], sesamin [29–31], piperine [32], dihydromethysticin [33], tetrahydropiperine [34], deoxypodophyllotoxin [35], and podophyllotoxin [36] to each catechol metabolite, which are further metabolized to phase 2 metabolites by glucuronidation, sulfation, O-methylation, or combination. In our study, fargesin catechol (M1) was further metabolized to M1 glucuronide (M1-G), M1 sulfate (M1-S), and O-methyl-M1 (M4 and M5), which were further metabolized to their glucuronides (M4-G and M5-G) and sulfates (M4-S and M5-S) in human, dog, monkey, and rat hepatocytes (Fig. 6).
1,3-Benzodioxle compounds can form a reactive metabolite intermediate at methylene group and inhibit CYPs [37]. Sesamin exhibited the mechanism-based inhibition of CYP2C9, which plays a major role in the oxidation of sesamin to sesamin monocatechol [29, 31]. The oxidation of myristicin to myristicin catechol is catalyzed by CYP3A4 and CYP1A2 [26]. Myristicin is a mechanism-based inhibitor of CYP1A2 via the formation of myristicin catechol [27]. Fargesin exhibited competitive inhibition of CYP2C9 activity and time-dependent inhibition of CYP2C8, CYP2C19, and CYP3A4 activities in human liver microsomes, whereas the other tetrahydrofuranoid lignans without a 1,3-benzodioxole group, such as epimagnolin A, eudesmin, magnolin, and dimethyllirioresinol, did not inhibit CYP activities [20]. The metabolism of magnolin to the two major metabolites, 4′-O-desmethylmagnolin and 4′′-O-desmethylmagnolin, is catalyzed by CYP2C9, CYP3A4, CP2C8, and CYP2C19 enzymes [38]. Therefore, fargesin inhibits CYP2C8, CYP2C9, CYP2C19, and CYP3A4 activities through the oxidation of 1,3-benzodioxole group to M1.
Incubation of fargesin with human liver S9 fractions in the presence of SAM, a cofactor of COMT, and NADPH resulted in the formation of M4 and M5 and a decrease in M1 compared to incubation in the absence of SAM, supporting that M4 (sylvatesmin) and M5 might be formed from M1 via COMT-mediated O-methylation. COMT promotes meta-O-methylation over para-O-methylation of the catechol moiety and the COMT activity in rats and dogs is higher than that in humans, mice, and monkeys [39, 40], suggesting that the peak area of M4 produced by fargesin was larger than that of M5 in human, rat, dog, and monkey hepatocytes. Furthermore, M5 was not detected in mouse hepatocytes (Fig. 1).
Gastrointestine-specific UGT1A8 and UGT1A10 as well as UGT1A3, UGT2B15, and UGT2B17 are involved in the glucuronidation of M4 to M4-G (Fig. 5a), suggesting that M4-G can be produced from fargesin in the intestine as well as liver. The formation of M4-S from M4 was catalyzed by SULT1A1*1, SULT1A1*2, SULT1A2, SULT1B1, SULT1C4, and SULT1E1 enzymes (Fig. 5b).
In conclusion, fargesin metabolism produced 14 metabolites in human, rat, mouse, monkey, and dog hepatocytes. These included three phase 1 metabolites; fargesin catechol (M1) by the CYP2C8, CYP2C9, CYP2C19, and CYP3A4 enzymes; and O-desmethylfargesin (M2, M3) by the CYP1A2, CYP2B6, CYP2C9, CYP2C19, CYP2D6, and CYP3A4 enzymes; as well as 11 phase 2 metabolites, such as O-methyl-M1 (M4 and M5) by COMT, M1-G, M2-G, M4-G, and M5-G by UGTs, and M1-S, M2-S, M3-S, M4-S, and M5-S by SULTs.
Funding
This work was supported by a grant from the National Research Foundation of Korea (NRF) funded by the Korea government (MSIT) (NRF-2023R1A2C2007632) and the Research Fund, 2021 of The Catholic University of Korea.
Data availability
Data are available upon request.
Declarations
Conflict of interests
The authors have no conflicts of interest to declare.
Footnotes
Min Seo Lee and Eun Jeong Park have contributed equally to this work.
References
- 1.Shen Y, Li CG, Zhou SF, Pang EC, Story DF, Xue CC. Chemistry and bioactivity of Flos Magnoliae, a Chinese herb for rhinitis and sinusitis. Curr Med Chem. 2008;15:1616–1627. doi: 10.2174/092986708784911515. [DOI] [PubMed] [Google Scholar]
- 2.Hong PTL, Kim HJ, Kim WK, Nam JH. Flos magnoliae constituent fargesin has an anti-allergic effect via ORAI1 channel inhibition. Korean J Physiol Pharmacol. 2021;25:251–258. doi: 10.4196/kjpp.2021.25.3.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baek JA, Lee YD, Lee CB, Go HK, Kim JP, Seo JJ, Rhee YK, Kim AM, Na DJ. Extracts of Magnoliae flos inhibit inducible nitric oxide synthase via ERK in human respiratory epithelial cells. Nitric Oxide. 2009;20:122–128. doi: 10.1016/j.niox.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Kim JS, Kim JY, Lee HJ, Lim HJ, Lee DY, Kim DH, Ryu JH. Suppression of inducible nitric oxide synthase expression by furfuran lignans from flower buds of Magnolia fargesii in BV-2 microglial cells. Phytother Res. 2010;24:748–753. doi: 10.1002/ptr.3028. [DOI] [PubMed] [Google Scholar]
- 5.Pham TH, Kim MS, Le MQ, Song YS, Bak Y, Ryu HW, Oh SR, Yoon DY. Fargesin exerts anti-inflammatory effects in THP-1 monocytes by suppressing PKC-dependent AP-1 and NF-ĸB signaling. Phytomedicine. 2017;24:96–103. doi: 10.1016/j.phymed.2016.11.014. [DOI] [PubMed] [Google Scholar]
- 6.Yue B, Ren YJ, Zhang JJ, Luo XP, Yu ZL, Ren GY, Sun AN, Deng C, Wang ZT, Dou W. Anti-inflammatory effects of fargesin on chemically induced inflammatory bowel disease in mice. Molecules. 2018;23:1380. doi: 10.3390/molecules23061380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu J, Zhang H, Pan J, Hu Z, Liu L, Liu Y, Yu X, Bai X, Cai D, Zhang H. Fargesin ameliorates osteoarthritis via macrophage reprogramming by downregulating MAPK and NF-κB pathways. Arthritis Res Ther. 2021;23:142. doi: 10.1186/s13075-021-02512-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jun AY, Kim HJ, Park KK, Son KH, Lee DH, Woo MH, Chung WY. Tetrahydrofurofuran-type lignans inhibit breast cancer-mediated bone destruction by blocking the vicious cycle between cancer cells, osteoblasts and osteoclasts. Invest New Drugs. 2014;32:1–13. doi: 10.1007/s10637-013-9969-0. [DOI] [PubMed] [Google Scholar]
- 9.Wang G, Gao J, He LH, Yu XH, Zhao ZW, Zou J, Wen FJ, Zhou L, Wan XJ, Tang CK. Fargesin alleviates atherosclerosis by promoting reverse cholesterol transport and reducing inflammatory response. Biochim Biophys Acta Mol Cell Biol Lipids. 2020;1865:158633. doi: 10.1016/j.bbalip.2020.158633. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Cheng Y, Xue H, Yue Y, Zhang W, Li X. Fargesin as a potential β1 adrenergic receptor antagonist protects the hearts against ischemia/reperfusion injury in rats via attenuating oxidative stress and apoptosis. Fitoterapia. 2015;105:16–25. doi: 10.1016/j.fitote.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 11.Sha S, Xu D, Wang Y, Zhao W, Li X. Antihypertensive effects of fargesin in vitro and in vivo via attenuating oxidative stress and promoting nitric oxide release. Can J Physiol Pharmacol. 2016;94:900–906. doi: 10.1139/cjpp-2015-0615. [DOI] [PubMed] [Google Scholar]
- 12.Lee YS, Cha BY, Choi SS, Harada Y, Choi BK, Yonezawa T, Teruya T, Nagai K, Woo JT. Fargesin improves lipid and glucose metabolism in 3T3-L1 adipocytes and high-fat diet-induced obese mice. BioFactors. 2012;38:300–308. doi: 10.1002/biof.1022. [DOI] [PubMed] [Google Scholar]
- 13.Choi SS, Cha BY, Choi BK, Lee YS, Yonezawa T, Teruya T, Nagai K, Woo JT. Fargesin, a component of Flos Magnoliae, stimulates glucose uptake in L6 myotubes. J Nat Med. 2013;67:320–326. doi: 10.1007/s11418-012-0685-4. [DOI] [PubMed] [Google Scholar]
- 14.Lee GE, Lee CJ, An HJ, Kang HC, Lee HS, Lee JY, Oh SR, Cho SJ, Kim DJ, Cho YY. Fargesin inhibits EGF-induced cell transformation and colon cancer cell growth by suppression of CDK2/Cyclin E signaling pathway. Int J Mol Sci. 2021;22:2073. doi: 10.3390/ijms22042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu T, Chai B, Shi Y, Dang Y, Ye X. Fargesin inhibits melanin synthesis in murine malignant and immortalized melanocytes by regulating PKA/CREB and P38/MAPK signaling pathways. J Dermatol Sci. 2019;94:213–219. doi: 10.1016/j.jdermsci.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Foti RS, Dalvie DK. Cytochrome P450 and non-cytochrome P450 oxidative metabolism: contributions to the pharmacokinetics, safety, and efficacy of xenobiotics. Drug Metab Dispos. 2016;44:1229–1245. doi: 10.1124/dmd.116.071753. [DOI] [PubMed] [Google Scholar]
- 17.Hwang DK, Kim JH, Shin Y, Choi WG, Kim S, Cho YY, Lee JY, Kang HC, Lee HS. Identification of catalposide metabolites in human liver and intestinal preparations and characterization of the relevant sulfotransferase, UDP-glucuronosyltransferase, and carboxylesterase enzymes. Pharmaceutics. 2019;11:355. doi: 10.3390/pharmaceutics11070355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee MS, Lim CH, Bang YY, Lee HS. Quantification of fargesin in mouse plasma using liquid chromatography-high resolution mass spectrometry: application to pharmacokinetics of fargesin in mice. Mass Spectro Lett. 2022;13:20–25. doi: 10.5478/MSL.2022.13.1.20. [DOI] [Google Scholar]
- 19.Zhang Z, Liu X, Cao W, Xiao X, Qu J, Zhao M, Li X. Pharmacokinetics and tissue distribution of fargesin after oral administration in rats by high performance liquid chromatography. Curr Pharm Anal. 2016;12:379. doi: 10.2174/1573412912666151211191600. [DOI] [Google Scholar]
- 20.Kim JH, Kwon SS, Jeong H, Lee HS. Inhibitory effects of dimethyllirioresinol, epimagnolin A, eudesmin, fargesin, and magnolin on cytochrome P450 enzyme activities in human liver microsomes. Int J Mol Sci. 2017;18:952. doi: 10.3390/ijms18050952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park R, Park EJ, Cho YY, Lee JY, Kang HC, Song IS, Lee HS. Tetrahydrofurofuranoid lignans, eudesmin, fargesin, epimagnolin A, magnolin, and yangambin inhibit UDP-glucuronosyltransferase 1A1 and 1A3 activities in human liver microsomes. Pharmaceutics. 2021;13:187. doi: 10.3390/pharmaceutics13020187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim JH, Kim DK, Choi WG, Ji HY, Choi JS, Song IS, Lee S, Lee HS. In vitro metabolism of DWP16001, a novel sodium-glucose cotransporter 2 inhibitor, in human and animal hepatocytes. Pharmaceutics. 2020;12:865. doi: 10.3390/pharmaceutics12090865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davies B, Morris T. Physiological parameters in laboratory animals and humans. Pharm Res. 1993;10:1093–1095. doi: 10.1023/a:1018943613122. [DOI] [PubMed] [Google Scholar]
- 24.Kong TY, Kim JH, Choi WG, Lee JY, Kim HS, Kim JY, In MK, Lee HS. Metabolic characterization of (1-(5-fluoropentyl)-1H-indol-3-yl)(4-methyl-1-naphthalenyl)-methanone (MAM-2201) using human liver microsomes and cDNA-overexpressed cytochrome P450 enzymes. Anal Bioanal Chem. 2017;409:1667–1680. doi: 10.1007/s00216-016-0113-9. [DOI] [PubMed] [Google Scholar]
- 25.Lee HS, Jeong TC, Kim JH. In vitro and in vivo metabolism of myristicin in the rat. J Chromatogr B Biomed Sci Appl. 1998;705:367–372. doi: 10.1016/s0378-4347(97)00531-8. [DOI] [PubMed] [Google Scholar]
- 26.Yun CH, Lee HS, Lee HY, Yim SK, Kim KH, Kim E, Yea SS, Guengerich FP. Roles of human liver cytochrome P450 3A4 and 1A2 enzymes in the oxidation of myristicin. Toxicol Lett. 2003;137:143–150. doi: 10.1016/s0378-4274(02)00397-1. [DOI] [PubMed] [Google Scholar]
- 27.Yang AH, He X, Chen JX, He LN, Jin CH, Wang LL, Zhang FL, An LJ. Identification and characterization of reactive metabolites in myristicin-mediated mechanism-based inhibition of CYP1A2. Chem Biol Interact. 2015;237:133–140. doi: 10.1016/j.cbi.2015.06.018. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, Qiao GX, Zhao GF, Zhao SF. Characterization of the metabolites of irisflorentin by using ultra-high performance liquid chromatography combined with quadrupole/orbitrap tandem mass spectrometry. J Pharm Biomed Anal. 2021;203:114222. doi: 10.1016/j.jpba.2021.114222. [DOI] [PubMed] [Google Scholar]
- 29.Yasuda K, Ikushiro S, Kamakura M, Ohta M, Sakaki T. Metabolism of sesamin by cytochrome P450 in human liver microsomes. Drug Metab Dispos. 2010;38:2117–2123. doi: 10.1124/dmd.110.035659. [DOI] [PubMed] [Google Scholar]
- 30.Yasuda K, Ikushiro S, Kamakura M, Munetsuna E, Ohta M, Sakaki T. Sequential metabolism of sesamin by cytochrome P450 and UDP-glucuronosyltransferase in human liver. Drug Metab Dispos. 2011;39:1538–1545. doi: 10.1124/dmd.111.039875. [DOI] [PubMed] [Google Scholar]
- 31.Yasuda K, Sakaki T. How is sesamin metabolised in the human liver to show its biological effects? Expert Opin Drug Metab Toxicol. 2012;8:93–102. doi: 10.1517/17425255.2012.637917. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Li M, Wang Z, Wen M, Tang J. Identification of the metabolites of piperine via hepatocyte incubation and liquid chromatography combined with diode-array detection and high-resolution mass spectrometry. Rapid Commun Mass Spectrom. 2020;34:e8947. doi: 10.1002/rcm.8947. [DOI] [PubMed] [Google Scholar]
- 33.Cheng C, Zhao S, Gu YL, Pang J, Zhao Y. Characterization and identification of the metabolites of dihydromethysticin by ultra-high-performance liquid chromatography orbitrap high-resolution mass spectrometry. J Sep Sci. 2022;45:2914–2923. doi: 10.1002/jssc.202200250. [DOI] [PubMed] [Google Scholar]
- 34.Chen X, Li Y. Identification of the stable and reactive metabolites of tetrahydropiperine using ultrahigh-performance liquid chromatography combined with diode-array detection and high-resolution mass spectrometry. Rapid Commun Mass Spectrom. 2021;35:e8975. doi: 10.1002/rcm.8975. [DOI] [PubMed] [Google Scholar]
- 35.Xie Q, Chen Y, Liu F, Zhong Z, Zhao K, Ling Z, Wang F, Tang X, Wang Z, Liu L, Liu X. Interspecies differences in metabolism of deoxypodophyllotoxin in hepatic microsomes from human, monkey, rat, mouse and dog. Drug Metab Pharmacokinet. 2016;31:314–322. doi: 10.1016/j.dmpk.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 36.Sun D, Gao X, Wang Q, Krausz KW, Fang Z, Zhang Y, Xie C, Gonzalez FJ. Metabolic map of the antiviral drug podophyllotoxin provides insights into hepatotoxicity. Xenobiotica. 2021;51:1047–1059. doi: 10.1080/00498254.2021.1961920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simonneaux G, Le Maux P. Carbene complexes of heme proteins and iron porphyrin models. Top Organomet Chem. 2006;17:83–122. doi: 10.1007/3418_006. [DOI] [Google Scholar]
- 38.Kim DK, Liu K-H, Jeong JH, Ji HY, Oh SR, Lee H-K, Lee HS. In vitro metabolism of magnolin and characterization of cytochrome P450 enzymes responsible for its metabolism in human liver microsomes. Xenobiotica. 2011;41:358–371. doi: 10.3109/00498254.2010.549968. [DOI] [PubMed] [Google Scholar]
- 39.Xia Y, Pang H, Dou T, Wang P, Ge G. Interspecies comparison in the COMT-mediated methylation of 3-BTD. RSC Adv. 2018;8:16278–16284. doi: 10.1039/c8ra01938j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jalkanen A, Lassheikki V, Torsti T, Gharib E, Lehtonen M, Juvonen RO. Tissue and interspecies comparison of catechol-O-methyltransferase mediated catalysis of 6-O-methylation of esculetin to scopoletin and its inhibition by entacapone and tolcapone. Xenobiotica. 2021;51:268–278. doi: 10.1080/00498254.2020.1853850. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available upon request.