Abstract
Long non-coding RNAs (lncRNAs) play an important role in gene regulation and contribute to tumorigenesis. While pan-cancer studies of lncRNA expression have been performed for adult malignancies, the lncRNA landscape across pediatric cancers remains largely uncharted. Here, we curated RNA sequencing data for 1,044 pediatric leukemia and extra-cranial solid tumors and integrated paired tumor whole genome sequencing and epigenetic data in relevant cell line models to explore lncRNA expression, regulation, and association with cancer. A total of 2,657 lncRNAs were robustly expressed across six pediatric cancers, including 1,142 exhibiting histotype-elevated expression. DNA copy number alterations contributed to lncRNA dysregulation at a proportion comparable to protein coding genes. Application of a multi-dimensional framework to identify and prioritize lncRNAs impacting gene networks revealed that lncRNAs dysregulated in pediatric cancer are associated with proliferation, metabolism, and DNA damage hallmarks. Analysis of upstream regulation via cell-type specific transcription factors further implicated distinct histotype-elevated and developmental lncRNAs. Integration of these analyses prioritized lncRNAs for experimental validation, and silencing of TBX2-AS1, the top-prioritized neuroblastoma-specific lncRNA, resulted in significant growth inhibition of neuroblastoma cells, confirming the computational predictions. Taken together, these data provide a comprehensive characterization of lncRNA regulation and function in pediatric cancers and pave the way for future mechanistic studies.
Keywords: pediatric cancer, long non-coding RNAs, neuroblastoma, gene regulation
Introduction
Long non-coding RNAs (lncRNAs) are transcribed RNA molecules greater than 200 nucleotides in length that do not code for proteins. These molecules account for 70% of the expressed human transcriptome and provide a key aspect of gene regulation[1, 2]. Compared to protein coding genes (PCGs), lncRNAs typically have fewer exons, weaker conservation, and lower abundance[1]. Despite this, lncRNAs have been shown to play significant roles in both transcriptional and post-transcriptional gene regulation[3]. LncRNAs perform these roles by physically interacting with a variety of substrates, including proteins (transcription co-factors), RNAs (microRNA sponges), and DNA (chromatin interaction scaffolds). While the mechanisms and function for the majority of lncRNAs remain unknown[2], those that have been experimentally characterized are involved in a variety of cellular processes, including gene silencing (ANRIL)[4], modulation of chromatin architecture (Xist)[5], and pre-mRNA processing (MALAT1)[6]. LncRNAs are also important in development. For example, the H19 lncRNA is involved in imprinting[7], while the well-conserved TUNA lncRNA controls stem cell pluripotency and lineage differentiation[8].
Dysregulation of lncRNA expression has been widely observed in cancer[1, 9, 10] and studies have shown that lncRNAs play important roles in tumor initiation and progression[11]. LncRNAs can function as tumor suppressors, such as the PANDA lncRNA which regulates DNA damage response in diffuse large B-cell lymphoma[12]; however, many more lncRNAs appear to be oncogenes. Examples include the HOTAIR and PVT1 lncRNAs which promote proliferation in various cancers through tissue specific mechanisms[13, 14]. Pan-cancer analyses of lncRNA expression in adult malignancies have uncovered many cancer-associated lncRNAs[1, 9–11, 15, 16]. Identification of functional lncRNAs amongst the large set of cancer-associated lncRNAs, however, remains challenging[9, 17]. Current methods to identify putative functional lncRNAs involve identifying lncRNA-specific genetic aberrations [9, 10] or using lncRNA expression to predict overall patient survival[10]. To more systematically address how lncRNAs drive the pathogenesis of cancer, recent computational methods seek to assign function to these molecules based on predicted target genes and regulatory network models. These methods have been applied to adult malignancies and allow for more focused hypotheses to be tested[15, 16].
LncRNA studies in pediatric cancers have limited their focused to single histotypes, specifically neuroblastoma, leukemias, and brain cancer[18–23]. CASC15 and NBAT-1 are a sense-antisense lncRNA pair that map to a NBL susceptibility locus identified by genome-wide association study[24, 25]. Both lncRNAs are downregulated in high-risk NBL tumors and have been shown to be involved in cell proliferation and differentiation[18, 24]. In pediatric T-ALL, the NOTCH-regulated lncRNA, LUNAR1, promotes T-ALL cell growth by sustaining IGF1 signaling[21]. To date, it is unknown whether lncRNAs function as common drivers across multiple pediatric cancers, or if instead, the majority of lncRNAs influence oncogenesis in a histotype-specific manner. Furthermore, given that pediatric cancers typically arise from primitive embryonic and mesodermal cells, rather than adult epithelial cells, it is unclear whether adult cancer lncRNA drivers will also be implicated in childhood cancer.
Here, we perform a pan-pediatric cancer study of lncRNAs across 1,044 pediatric leukemias and extra-cranial solid tumors. We present the landscape of lncRNA expression across these childhood cancers and perform integrative multi-omic analyses to assess tissue elevated expression, regulation, and putative function. To validate our approach, we show that silencing of the top-prioritized NBL-specific lncRNA, TBX2-AS1, impairs NBL cell growth in human-derived NBL cell line models.
Materials and Methods
RNA-seq data processing.
A comprehensive RNA-seq analysis pipeline was used on all samples (Supplementary Table S1, Supplementary Fig. S1). First, FASTQC (RRID:SCR_014583) was run on all samples and any samples that had a Phred score < 30 for more than 25% of read bases were removed. Samples were then aligned using STAR_2.4.2a (RRID:SCR_004463) [26] with the following parameters: “STAR --runMode alignReads --runThreadN 10 --twopassMode Basic --twopass1readsN −1 --chimSegmentMin 15 --chimOutType WithinBAM –genomeDir X--genomeFastaFiles ucsc.hg19.fa --readFilesIn fasta1 fasta2 --readFilesCommand zcat --outSAMtype BAM SortedByCoordinate --outFileNamePrefix X --outSAMstrandField intronMotif --quantMode TranscriptomeSAM GeneCounts --sjdbGTFfile gencode.v19.annotation.gtf --sjdbOverhang X.” To assess the quality of the aligned RNA-seq data we ran MultiQC [27] (RRID:SCR_014982), and removed samples with < 70% uniquely mapped reads and < 10 million mapped reads. Details of RNA-seq data read quantification can be found in Supplementary Methods.
Tissue specific gene expression.
The tau score, a measure of the tissue specific expression of a gene was calculated as described by Yanai et. al[28]. The formula for the score is listed below. xi is defined as the mean expression of a gene in a particular cancer and n is the total number of cancers considered, in this case n = 6.
CNV and structural variant detection.
Copy number calls were made by Complete Genomics (CGI) from WGS for NBL, WT, AML, and B-ALL. We used CGI files “somaticCnvDetailsDiploidBeta” containing ploidy estimates and tumor/blood coverage along 2kb bins across the genome. To create segmentation files, we used custom scripts to reformat CGI coverage data to meet requirements of the “copynumber” R bioconductor package (RRID:SCR_006442) as previously described[29]. Segmentation files were visualized using the R package svpluscnv [30]. We then ran GISTIC2.0 (RRID:SCR_000151), using segmentation data as inputs using parameters: “GISTIC2 -v 30 -refgene hg19 -genegistic 1 -smallmem 1 -broad 1 -twoside 1 -brlen 0.98 -conf 0.90 -armpeel 1 -savegene 1 -gcm extreme -js 2 -rx 0”. To determine genes impacted by copy number we intersected CNV regions listed in the “all_lesions.conf_90.txt” file from GISTIC output with gene positions. We used section 1 from the “all_lesions.conf_90.txt” file to assign a binary descriptor to each gene as either being not amplified or deleted (CNV-no) if the sample had actual copy gain 0 for the region containing the gene. We assigned CNV-yes if the region containing the gene was amplified or deleted, which included samples with actual copy gain 1 or 2, where 1 indicates low level copy number aberration (exceeds low threshold of copy number: 1: 0.1<t< 0.9) and 2 indicates a high level of copy number aberration, CNV exceeds high threshold (t>0.9) according to GISTIC.
Structural variants were identified from WGS as previously described[29]. To obtain a high confidence set of junctions, filtering was applied to obtain the highConfidenceSomaticAllJunctionsBeta:
DiscordantMatePairAlignments ≥ 10 (10 or more discordant mate pairs in cluster
JunctionSequenceResolve = Y (local de novo assembly is successful)
Exclude interchromosomal junction if present in any genomes in baseline samples (FrequencyInBaseline > 0)
Exclude the junction if overlap with known underrepresented repeats (KnownUnderrepresentedRepeat = Y): ALR/Alpha, GAATGn, HSATII, LSU_rRNA_Hsa, and RSU_rRNA_Hsa
Exclude the junction if the length of either of the side sections is less than 70 base pairs.
Further filtering of these high confidence structural variants included removing rare/common germline variants that passed the CGI filters. We used the Database of Genomic Variants (DGV v. 2016–05-15, GRCh37 RRID:SCR_007000) to remove SVs that had at least 50% reciprocal overlap with DGV annotated common events and were type matched.
Identification of gene regulatory networks using the lncMod framework.
We developed custom Python 2.7 (RRID:SCR_008394) scripts to implement the general framework of the lncMod method [31]. We first identified transcription factor target gene regulation specific to each cancer by performing motif analysis upstream of all candidate target genes. We then delineated genes (TF, target genes, or lncRNAs) that had high expression variance (IQR > 1.5). For each differentially expressed lncRNA, we sorted cancer samples from low to high lncRNA expression. We then determined the correlation (Spearman’s rho) between the expression of all transcription factor and target gene pairs for the 25% of samples with the lowest lncRNA expression and separately for the 25% of samples with the highest expression for the given lncRNA [15, 16, 31]. Significant lncRNA modulators were identified based on a significance difference between the TF-target gene correlation in the low vs high lncRNA expression groups, which was assessed using the rewiring score. Permutation analysis of randomized lncRNA TF-target gene triplets was performed to determine the significance of the rewire score in the context of multiple testing hypothesis. Further details about implementation of the lncMod framework can be found in the Supplementary Methods.
Promoter-focused Capture C data generation.
High resolution promoter-focused Capture C was performed in the neuroblastoma cell line, NB1643 (RRID:CVCL_5627), (untreated) in triplicate. Cell fixation, 3C library generation, capture C, and sequencing was performed as previously described [32–35]. For each replicate, 107 fixed cells were centrifuged to cell pellets and split to 6 tubes for a pre-digestion incubation with 0.3%SDS, 1x NEB DpnII restriction buffer, and dH2O for 1hr at 37ºC shaking at 1,000rpm. A 1.7% solution of Triton X-100 was added to each tube and shaking was continued for another hour.10 ul of DpnII (NEB, 50 U/μL) was added to each sample tube and continued shaking for 2 days. 100uL Digestion reaction was then removed and set aside for digestion efficiency QC. The remaining samples were heat inactivated incubated at 1000 rpm in a MultiTherm for 20 min, at 65°C to inactivate the DpnII, and cooled on ice for 20 additional minutes. Digested samples were ligated with 8 uL of T4 DNA ligase (HC ThermoFisher, 30 U/μL) and 1X ligase buffer at 1,000 rpm overnight at 16°C. The ligated samples were then de-crosslinked overnight at 65°C with Proteinase K (20 mg/mL, Denville Scientific) along with pre-digestion and digestion control. Both controls and ligated samples were incubated for 30 min at 37°C with RNase A (Millipore), followed by phenol/chloroform extraction, ethanol precipitation at −20°C, then the 3C libraries were centrifuged at 3000 rpm for 45 min at 4°C to pellet the samples. The pellets of 3C libraries and controls were resuspended in 300uL and 20μL dH2O, respectively, and stored at −20°C. Sample concentrations were measured by Qubit. Digestion and ligation efficiencies were assessed by gel electrophoresis on a 0.9% agarose gel and by quantitative PCR (SYBR green, Thermo Fisher).
Isolated DNA from 3C libraries was quantified using a Qubit fluorometer (Life technologies), and 10 μg of each library was sheared in dH2O using a QSonica Q800R to an average fragment size of 350bp.QSonica settings used were 60% amplitude, 30s on, 30s off, 2 min intervals, for a total of 5 intervals at 4 °C. After shearing, DNA was purified using AMPureXP beads (Agencourt). DNA size was assessed on a Bioanalyzer 2100 using a DNA 1000 Chip (Agilent) and DNA concentration was checked via Qubit. SureSelect XT library prep kits (Agilent) were used to repair DNA ends and for adaptor ligation following the manufacturer protocol. Excess adaptors were removed using AMPureXP beads. Size and concentration were checked again by Bioanalyzer 2100 using a DNA 1000 Chip and by Qubit fluorometer before hybridization. One microgram of adaptor-ligated library was used as input for the SureSelect XT capture kit using manufacturer protocol and custom-designed 41K promoter Capture-C probe set. The quantity and quality of the captured libraries were assessed by Bioanalyzer using a high sensitivity DNA Chip and by Qubit fluorometer. SureSelect XT libraries were then paired-end sequenced on Illumina NovaSeq 6000 platform (51bp read length) at the Center for Spatial and Functional Genomics at CHOP.
Cell lines and reagents.
NBL cell lines were obtained from the American Type Tissue Culture Collection (ATCC) and grown in RPM1–1640 with HEPES, L-glutamine and phenol red, supplemented with 10% FBS, 1% L-glutamine in an incubator at 37°C with 5% CO2. All cell lines used in experiments for this study had a passage less than 15. Cell line identity was confirmed biennially through genotyping and confirmation of STR (short tandem repeat) profiles, while routine testing for Mycoplasma contamination was confirmed to be negative.
siRNA and growth assays.
The NBL cell lines, NLF (RRID:CVCL_E217) and SKNSH (RRID:CVCL_0531), were plated in a 96-well RTCES microelectronic sensor array (ACEA Biosciences, San Diego, CA, USA). Cell density measurements were made every hour and were normalized to 24 hours post-plating (at transfection time). We used siRNAs to knockdown the expression of genes in NLF and SKNSH. The siRNAs utilized included a non-targeting negative control siRNA (Silencer™ Select Negative Control siRNA, cat #4390843), TBX2 Silencer™ siRNA (assay ID 115748), TBX2-AS1 Silencer™ Select siRNA (assay ID n514841) referred to as siTBX2-AS1 and siTBX2-AS1-A, TBX2-AS1 Silencer™ Select siRNA (assay ID n550888) referred to as siTBX2-AS1-B, TBX2-AS1 Silencer™ Select siRNA (assay ID S197244) referred to as siTBX2-AS1-C, TBX2-AS1 Silencer™ Select siRNA (assay ID n543080) referred to as siTBX2-AS1-D, and SMARTpool: ON-TARGETplus PLK1 siRNA (cat # L-003290–00-0010). Transfection of cells was done using the DharmaFECT 1 transfection reagent (cat # T-2001–02). siRNA at a concentration of 50nM and 2% (NLF) and 2–4% (SKNSH) DharmaFECT was added to RPMI medium without 10% FBS or any antibiotic separately and then incubated at room temperature for 5 minutes. The siRNA medium was then added to the DharmaFECT and incubated for another 20 minutes to form a complex. This solution was then mixed with our normal growth media and applied to cells 24 hours after they had been initially plated. All experiments were repeated in triplicate, with technical replicates (n=3) being averaged per biological replicate.
Real time quantitative PCR.
Total RNA was extracted from NBL cells using miRNeasy kit (Qiagen) and the provided protocol for animal cells. The concentration of RNA was determined with the Nanodrop (Thermo Scientific). cDNA synthesis was performed using the SuperScript™ First-Strand Synthesis System for RT-PCR using the SuperScript™ reverse transcriptase (Invitrogen). 5–20ng of cDNA were mixed with the TaqMan Universal PCR Master Mix (Thermo Fisher Scientific) and TaqMan probes/primers for either TBX2-AS1 (Hs00417285_m1) or the house keeping gene, HPRT1 (Hs02800695_m1). Gene expression from these reactions were measured using RT-qPCR and TBX2-AS1 expression was normalized to HPRT1 expression.
Protein isolation and western blotting.
Whole cell lysates were made using denaturing lysis buffer containing protease/phosphatase inhibitors (Cell Signaling Technology 5872). Cells were kept on ice and lysed for 30 min. Samples were then sonicated for 5 sec and spun at max speed in a microcentrifuge for 15 min at 4°C, after which supernatant was collected into a clean tube. Protein was quantified using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific 23227) and 30 μg protein was then loaded on 4–12% Tris-Glycine gels, transferred to PVDF membrane, and probed with antibodies in 5% milk in TBST. Primary antibodies used include: Actin Beta (Santa Cruz Biotechnology sc-47778, RRID:AB_626632) used at 1:2500 and TBX2 (Santa Cruz Biotechnology sc-514291, RRID:AB_2941848) used at 1:250. The secondary antibody used was Goat anti-mouse HRP (Thermo Fisher 31430, RRID:AB_228307) at 1:25,000. Blots were developed using SuperSignal West Femto Maximium Sensitivity Substrate (Thermo Fisher Scientific 23227) on the Chemidoc Imaging System (Biorad).
Data Availability
All TARGET RNA and DNA-sequencing data analyzed in this study are available through the database of Genotypes and Phenotypes (dbGaP; https://www.ncbi.nlm.nih.gov/gap/) under study-id phs000218 and accession number phs000467. GMKF RNA-sequencing data are available through dbGAP study accession phs001436.v1.p1. Neuroblastoma cell line RNA-sequencing data analyzed in this study are available through GEO at accessions GSE89413. NBL histone ChIP-seq and transcription factor ChIP-seq data used in this study are both available through GEO at accessions: GSE138315 and GSE94822, respectively. RNA-sequencing generated for NLF cells treated with siRNAs in this study is available through GEO at accession: GSE238166. Code used to generate figures and analyze data is available at: https://github.com/diskin-lab-chop/PanTARGET_lncRNA_Study. All other raw data generated in this study are available upon request from the corresponding author.
Results
Identification of robustly expressed lncRNAs across pediatric cancers
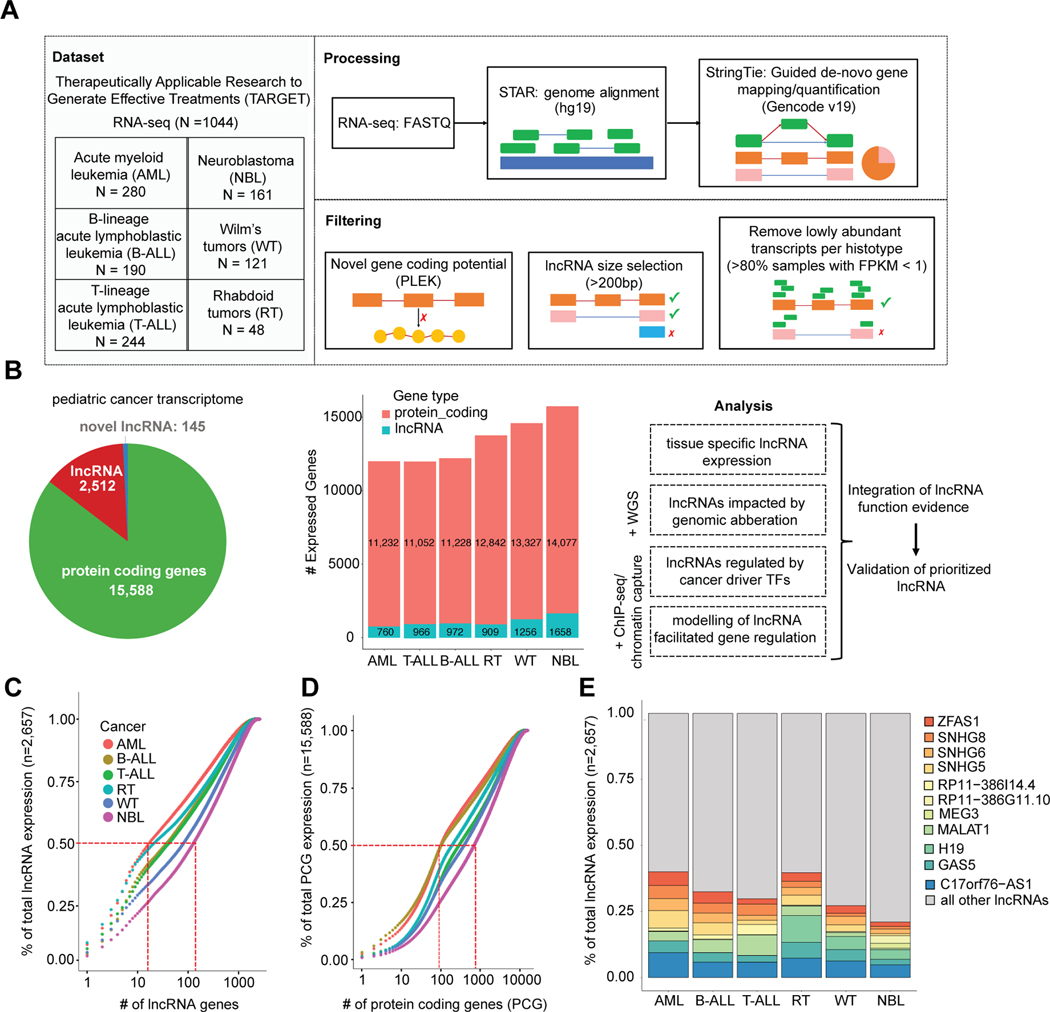
To define the repertoire of highly expressed lncRNAs in childhood cancers, we analyzed RNA-sequencing data from six distinct pediatric cancer histotypes profiled through the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) project (https://www.cancer.gov/ccg/research/genome-sequencing/target) (Supplementary Table S1). This curated set of 1,044 leukemia and solid tumor samples includes 280 acute myeloid leukemia (AML), 190 B-lymphoblastic leukemias (B-ALL), 244 T-lymphoblastic leukemias (T-ALL), 121 Wilms tumors (WT), 48 extracranial rhabdoid tumors (RT), and 161 neuroblastomas (NBL) (Fig. 1A). To identify novel cancer-associated lncRNAs, we performed guided de novo transcriptome assembly using StringTie v1.3.3[36] with the GENCODE v19 database [37] as a gene annotation reference (Supplementary Fig. S1A). Expressed gene sequences that did not match exons and transcript structures of any known gene in the GENCODE v19 or RefSeq v74 databases were considered putative novel genes (Supplementary Fig. S1A–B). Of these novel genes, we identified candidate lncRNAs by using the PLEK v1 algorithm[38] to assess non-coding potential, and then additionally filtered hits by transcript length, exon read coverage, and genomic location (Fig. 1A, Supplementary Fig. S1A). As validation of our lncRNA discovery pipeline, we observed that 36% (87 of 242) of identified novel lncRNAs not annotated in Gencode v19 (hg19) were indeed annotated in the more recent Gencode v29 (hg38) genome build (Supplementary Table S2). To ensure that we selected robustly expressed genes in the setting of cancer heterogeneity and sequencing variability, we applied a conservative expression cutoff of Fragments Per Kilobase of transcript per Million mapped reads (FPKM) >1 in at least 20% of samples for each cancer. Across all cancers there were 15,588 PCGs, 2,512 known lncRNAs, and 145 novel lncRNAs expressed, though the total number of expressed genes varied per cancer (Fig. 1B, Supplementary Table S3). Principal component analysis (PCA) of lncRNA gene expression showed that blood (AML, B-ALL, T-ALL) and solid (NBL, WT, RT) cancers form two distinct groups. Moreover, individual cancer histotypes clustered more closely using lncRNA expression than combined PCG and lncRNA expression (Supplementary Fig. S2A–C), consistent with the known tissue specific nature of lncRNA expression and function[1].
Figure 1: Pan-pediatric cancer transcriptome characterization.
(A) Overview of pan-pediatric cancer RNA-seq dataset and schematic of data processing and filtering. Reads from RNA-seq fastq files were aligned using the STAR algorithm and then gene transcripts were mapped in a guided de novo manner and quantified via the StringTie algorithm. Genes were considered novel if they did not have transcript exon structures matching genes in the GENCODE v19 or RefSeq v74 databases. Novel genes were assigned as lncRNAs based on length >200bp and non-coding potential calculated using the PLEK algorithm. Transcripts with low expression (FPKM <1 in >80% samples) were not considered for further analysis. (B) Pie graph showing the quantity of robustly expressed protein coding genes, GENCODE/RefSeq annotated lncRNAs, and novel lncRNAs. The number of genes expressed per cancer is also shown. Adjoining schematic gives overview of additional data types that were integrated with transcriptome data: WGS, ChIP-seq, and chromatin capture. Listed are the analyses used to elucidate lncRNAs with functional roles in pediatric cancer. (C) Cumulative expression plots comparing the number of lncRNAs and (D) protein coding genes, respectively, that constitute the total sum of gene expression (FPKM) per pediatric cancer. (E) Percentage of total lncRNA expression (FPKM) accounted for by the union of top five expressed lncRNAs per cancer (total 11 lncRNAs).
Overall, lncRNAs had lower average expression compared to PCGs resulting in fewer highly expressed lncRNAs (Supplementary Fig. S2D). Between 10–100 (3.7%) lncRNAs accounted for 50% of the total sum of lncRNA expression (Fig. 1C). In contrast, between 100–1000 (6.4%) PCGs accounted for 50% of the total sum of PCG expression (Fig. 1D). We examined the union of the top five most highly expressed lncRNAs across pediatric cancers (total 11 lncRNAs). Some of these lncRNAs had higher expression in the blood cancers (MALAT1 and RP11–386I14.4), in the solid cancers (H19), or in only one cancer, such as MEG3 and RP11–386G11.10 in NBL (Fig. 1E). Five of these highly expressed lncRNAs were among the top 10 lncRNAs expressed across normal tissues in the Genotype-Tissue Expression (GTEx) project [39]. Specifically, C17orf76-AS1 (LRRC75A-AS1), MALAT1, GAS5, SNHG6, SNHG8 were expressed ubiquitously in 30 of the 49 GTEx tissues (Supplementary Table S4).
Tissue-elevated lncRNA expression distinguishes pediatric cancers
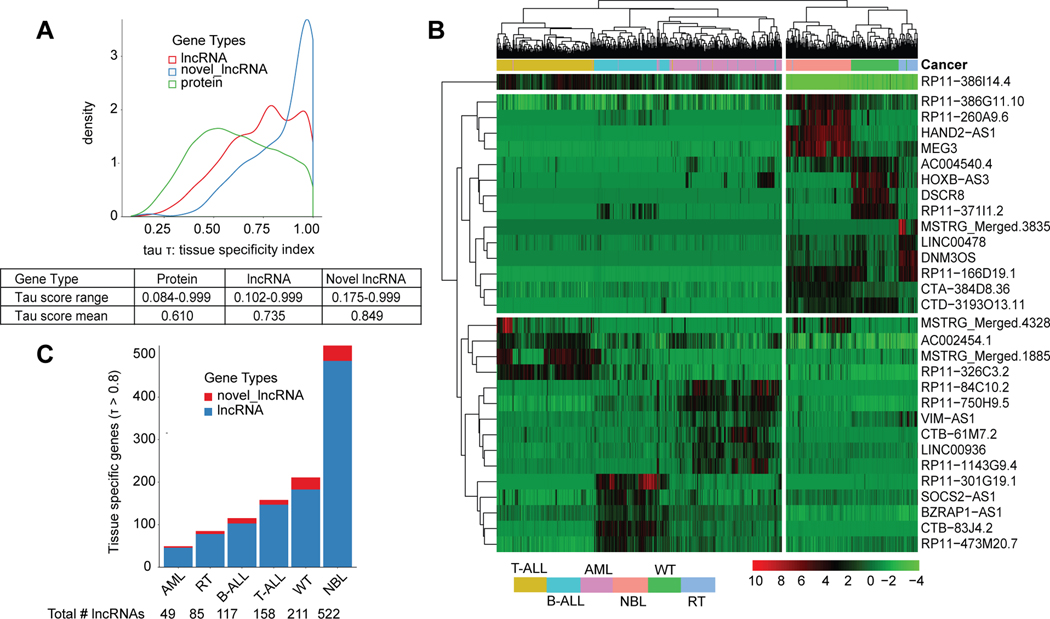
To evaluate more formally the relative expression of lncRNAs, we annotated all genes with a tissue specificity index (tau score)[28, 40]. The established tau score ranges from 0 (ubiquitous expression) to 1 (tissue-elevated). As an example, the highly expressed lncRNA C17orf76-AS1 yielded a tau score of 0.296 in this study, indicating ubiquitous expression (Supplementary Fig. S2E). In contrast, the highly expressed MEG3 lncRNA, which is known to have tissue-elevated expression in NBL[19, 41], yielded a tau score of 0.986 (Supplementary Fig. S2F). Overall, we observed that lncRNAs yielded a higher tau score range and mean, and thus greater tissue specific expression than PCGs (t-test p-value=1.62×10−42). Novel lncRNAs had the greatest tissue specific expression (t-test: vs proteins- p-value=1.62×10−42, vs known lncRNAs- p-value = 3.39×10−13) (Fig. 2A). A tau score threshold of 0.8 has been suggested to distinguish tissue specific genes[40], and using this cutoff we identified 1,142 (42%) tissue elevated (TE) lncRNAs (Fig. S2B, Supplementary Table S5). To assess how well TE lncRNAs distinguish cancers, we performed clustering based on the top five highest expressed TE lncRNAs per cancer (30 total). The expression of just these lncRNAs was sufficient to cluster samples of the same cancer type (Fig. 2B). Furthermore, the blood and solid cancers separately clustered together with little expression overlap observed between the two groups across the 30 genes (Fig. 2B). Finally, we identified a similar proportion of TE lncRNAs (38%, n = 1624) across 12 adult cancers from The Cancer Genome Atlas (TCGA) and observed that adult cancer tissue types were also well distinguished based on the expression of the top 5 most TS lncRNAs (Supplementary Fig. S2G–H).
Figure 2: LncRNAs exhibit tissue specific expression that can distinguish cancers.
(A) Tissue specificity index (tau score) which ranges from 0 (ubiquitously expressed) to 1 (tissue specific) is plotted for genes across three gene types: protein coding genes, lncRNAs, and novel lncRNAs. Table shows the tau score range and mean per gene type. (B) Heatmap showing the hierarchically clustered gene expression for the top five most tissue specific lncRNAs per cancer, ranked by highest tau score. Samples from each cancer cluster together based on expression of these genes alone. (C) Number of tissue specific known and novel lncRNAs in each cancer as defined by tissue specific gene threshold: tau score > 0.8.
Notably, NBL tumors expressed 2.5x more TE lncRNAs (n=522) than the cancer with the next highest: WT (TE lncRNAs: n=211), and 10x more than AML, which had the least number of TE lncRNAs (n=49) (Fig. 2C). To validate NBL’s striking quantity of TE lncRNAs, we first assessed whether immune and stromal cell infiltration[42] could be contributing to the variety of lncRNAs expressed. We ran the ESTIMATE algorithm as previously described[42] to determine levels of immune and stromal cell presence in each tumor sample using expression data. We then re-calculated each cancer’s tau score, restricting our analysis to NBL samples with either 80% or 90% tumor purity. In both cases, we found that NBL still had the greatest number of TE lncRNAs (n =588, NBL 90% purity) compared to other cancers (Supplementary Table S6). NBL was the only cancer studied that used an un-stranded RNA-seq protocol (Supplementary Table S1). To assess whether this explained the high number of TE lncRNAs, we first compared lncRNA quantification between 14 samples in our cohort that were later sequenced using stranded RNA-seq as part of the Gabriela Miller Kids First (GMKF) cohort. The median expression correlation across all lncRNAs and the antisense lncRNAs subset was r=0.691 and r=0.688, respectively. We then validated the TE lncRNAs in NBL using the full GMKF cohort (n=223 tumors) and observed that 48% of expressed lncRNAs were tissue elevated, an increase from the 31% observed in the TARGET cohort (Supplementary Table S6). These results confirm lncRNA abundance in NBL and demonstrate that the tau score robustly identifies TE lncRNAs across varying datasets.
Somatic DNA copy number alterations impact lncRNA expression
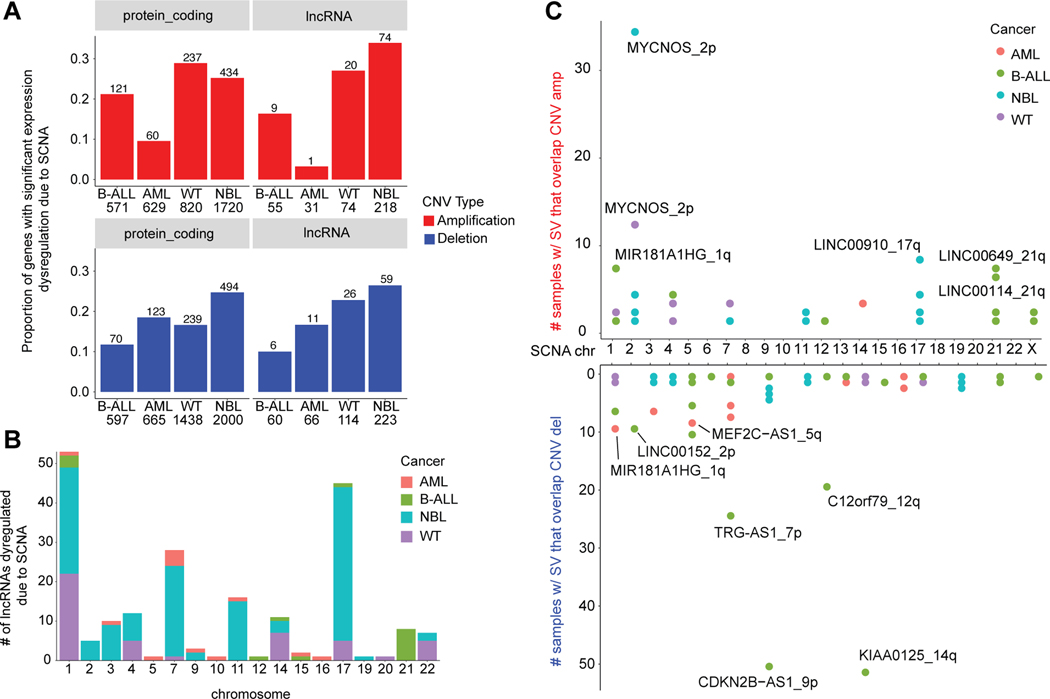
Many pediatric cancers are marked by a lower somatic single nucleotide variant (SNV) and insertion-deletion (indel) burden than observed in adult cancers[42]. Instead, large chromosomal events, such as somatic copy number aberrations (SCNAs) and other structural variants (SVs) have been shown to dysregulate protein coding driver genes[29, 42]. The extent to which large chromosomal alterations impact lncRNAs in pediatric cancers remains unknown. We thus sought to identify SCNAs and SVs using whole genome sequencing (WGS) data from the TARGET project available for NBL (n=146), B-ALL (n=302), AML (n=297), and WT (n=81). We observed that NBL had the greatest frequency of copy number events (Supplementary Fig. S3A) and highest correlation between number of lncRNAs in CNV regions and expressed lncRNAs per chromosome (Pearson’s r=0.556). The GISTIC v2 algorithm[43] was applied to detect regions of recurrent SCNA (q-value < 0.25). We identified 673 expressed lncRNAs overlapping 176 significant SCNA regions across the cancers (Supplementary Table S7). WGS samples with matched RNA-sequencing were then used to compare lncRNA expression in samples with or without an SCNA event and determine significant differential expression (DE) (Supplementary Table S8). Across all cancers, between 10–30% of expressed genes overlapping SCNA regions showed significant differential expression based on SCNA, a proportion that was similar for both PCGs and lncRNAs (Fig. 3A). Altogether, there were 198 (29%) unique lncRNAs with significant DE due to SCNA (Supplementary Fig. S3B). The majority of the significantly dysregulated lncRNAs were identified in the two cancers with the greatest overall number of expressed lncRNAs, NBL and WT, and mapped to regions with highly recurrent SCNAs in those cancers (chromosomes 1, 7, 11, and 17) (Fig. 3B).
Figure 3: A similar proportion of lncRNAs and protein coding genes are dysregulated due to SCNA.
(A) The proportion of protein coding and lncRNA genes that have significant differential expression due SCNA, separated by copy number type (amplification or deletion). The number of genes found in SCNA loci is shown per cancer. Genes were evaluated to have differential expression due to copy number using the Wilcoxon rank sum test (p-value < 0.05) and log |fold change| > 1.5), comparing samples with no SCNA to samples with low/high SCNA as defined by GISTIC scores. (B) The number of differentially expressed lncRNAs per chromosome and per cancer, distinguished by color. Chromosomes 1 and 17 had the most dysregulated lncRNAs associating with the greater frequency of SCNA on these chromosomes across cancers. (C) Number of samples with structural variant breakpoints in or near (+/− 2.5kb) lncRNAs and that are also located in copy number regions, stratified by amplification or deletion status of the locus.
While SCNAs can cause the dysregulation of lncRNA expression based on gene dosage, structural variant (SV) breakpoints within a lncRNA could cause loss or gain of function[29, 42]. We utilized WGS data to identify lncRNAs disrupted by SV breakpoints using a previously described combination approach involving copy number read-depth and discordant junction approach[29]. There were 650 unique expressed lncRNA genes disrupted by SVs, 89% of which were found in only one sample (Supplementary Fig. S4A). We observed 212 SV-impacted lncRNA genes located at SCNA regions (Fig. 3C), and 65% of lncRNAs genes disrupted by SV breakpoints in at least five samples that overlapped an SCNA regions (Supplementary Fig. S4B, Supplementary Table S9). Indeed, the top-ranked SV-impacted lncRNA in both NBL and WT, MYCNOS, associates with the disease-driving chr2p24 amplification[44, 45] (Supplementary Fig. S4C–D). In B-ALL, the SV-impacted lncRNAs: KIAA0125 and CDKN2B-AS1 (ANRIL) associate with the well-studied IGH translocation and CDKN2A/B deletion locus (Supplementary Fig. S4E)[46]. The top-ranked SV-impacted lncRNA in AML, MIR181A1HG (MONC), associates with a recurrent SCNA deletion on 1q and is mildly up-regulated in the AML dataset (p = 0.061, Supplementary Fig. S4F). MIR181A1HG (MONC) was described previously as an oncogene in acute megakaryoblastic leukemia[47]. Finally, we observed 30 lncRNAs with pan-cancer (n>3) expression and SV breakpoints (Supplementary Fig. S4G). The most number of breakpoints across unique samples was observed in LINC00910, which was shown previously to be essential for cell growth in the K562 cell line[48].
Characterization of transcriptional network perturbation mediated by dysregulated lncRNAs
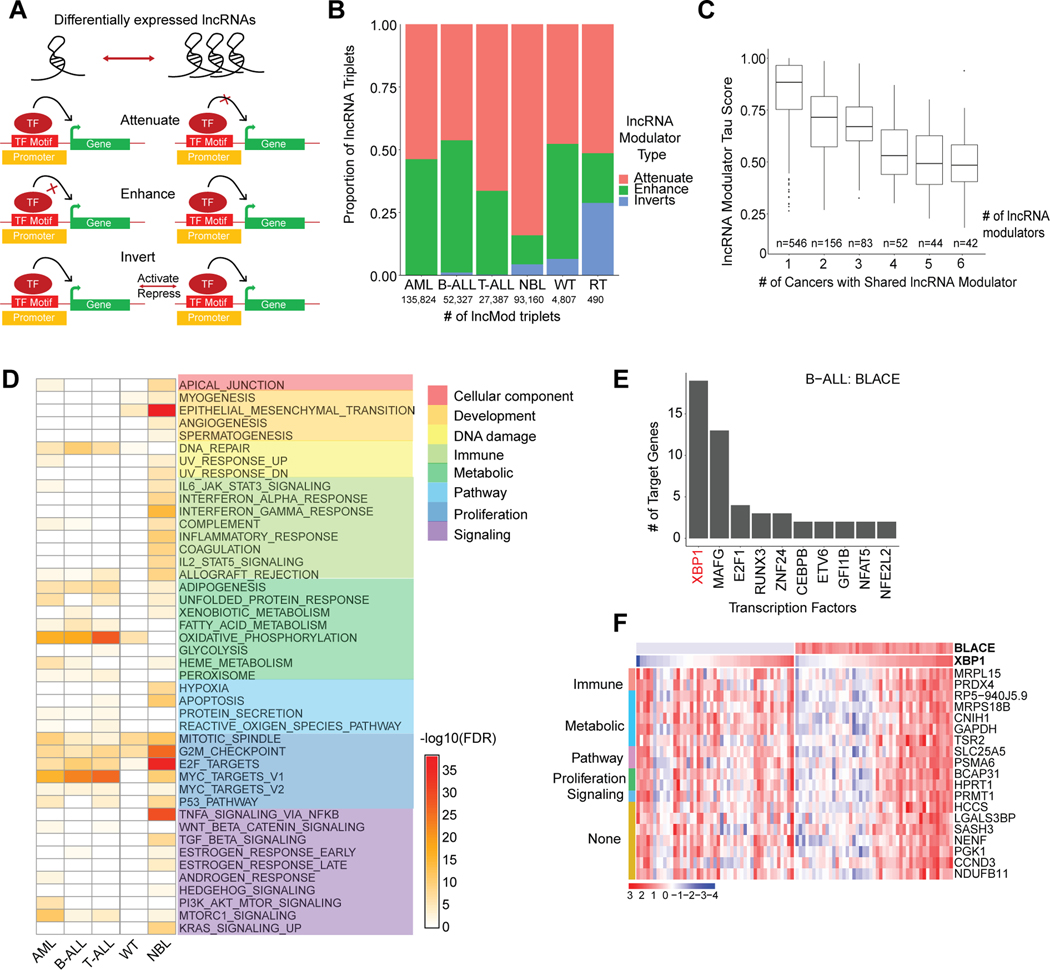
To determine how lncRNAs may drive pediatric cancers, we computationally predicted the downstream impact of lncRNAs on gene regulation. We focused on identifying lncRNAs that mediate transcriptional regulation by modulating TF activity (lncRNA modulators)[49–51]. We wrote custom scripts implementing the lncMod computational framework[31] to first identify DE-lncRNAs, and then to assess their impact on correlated expression between a TF and its target genes[15, 31] (Fig. 4A). Across all cancers studied, we identified 313,370 unique, dysregulated lncMod triplets (lncRNA-TF-target gene), representing 0.02–0.2% of possible triplets, which have significant correlation differences between a TF and target gene upon lncRNA expression dysregulation (Supplementary Table S10–S11, Supplementary Fig. S5A). This proportion was consistent with previous findings from the lncMap study in adult cancers[15], although more triplets were identified in datasets with greater sample size (Supplementary Table S10–S11). We observed that the majority of lncRNA modulators are either intergenic (39%) or antisense (38%) lncRNAs, with only 15% of the antisense lncRNAs having a correlation of r>0.6 with their sense-protein coding gene. The majority of lncRNA modulators appear to function in trans, with only 5% of lncRNA-TF pairs belonging to the same chromosome. LncRNA modulators were categorized into one of three categories based on their impact on TF-target gene correlation; either the correlation was enhanced, attenuated, or inverted (Fig. 4A–B). LncRNA modulators have context specific function such that for different TF-target gene pairs they could exert different types of regulation (Supplementary Fig. S5B). The majority of lncRNA modulators appeared to be active in only one cancer, with only 15% (138 of 923 lncRNAs) having pan-cancer activity (n>3) (Fig. 4C).
Figure 4: lncRNA modulators impact transcriptional networks involving proliferation.
(A) Schematic that shows the three ways (attenuate, enhance, or invert) in which differentially expressed lncRNA modulators can impact transcription factor and target gene relationships. lncRNA modulators are associated with a TF-target gene pair based on a significant difference between TF-target gene expression correlation in samples with low lncRNA expression (lowest quartile) vs samples with high lncRNA expression (highest quartile). (B) The proportion of lncRNA modulator types associated with significantly dysregulated lncRNA modulator- TF-target gene (lncMod) triplets. The number of significantly dysregulated lncMod triplets is listed per cancer. (C) Number of lncRNA modulators genes that are common in lncMod triplets across cancers. Common lncRNA modulator genes tend to have a lower tau score compared to lncRNA modulators only associated with one cancer. (D) Gene set enrichment using the MSigDB Hallmark gene set, of target genes associated with lncRNA modulators in each cancer (Fisher’s exact test, FDR < 0.1). (E) Transcription factors associated with the B-ALL expression specific lncRNA, BLACE, ranked based on number of regulated target genes. (F) Expression heatmap of BLACE and the target genes of the XBP1 transcription factor, grouped by associated hallmark gene set, in samples within the bottom and top quartiles of BLACE expression in B-ALL.
To determine the biological impact of lncRNA modulators, we identified lncRNAs whose target genes were enriched in MSigDB’s Hallmark Gene Sets (HMS)[52] (Fisher’s exact test, FDR < 0.1). Across most cancers, lncRNA modulator target genes had significant enrichment in the proliferation, metabolism, and DNA damage hallmark categories (FDR range: 0.1 to 2.24×10−36; Fig. 4D). Overall, the top-enriched hallmark pathways closely mirrored those found for lncRNA modulators in adult cancers[16]. Consistent with its role in development and as an oncogene in certain cancers [17], the top-enriched hallmarks for the H19 lncRNA, dysregulated in NBL, were the EMT (development) and G2M-checkpoint (proliferation) hallmarks (Supplementary Fig. S5C). The blood cancers exhibited strong enrichment of lncRNA modulators regulating MYC targets, which has a well-established role in leukemias[53]. Furthermore, in AML, we observed that gene targets of the myeloid-specific lncRNA, HOTAIRM1, were most enriched for proliferation hallmarks (Supplementary Fig. S5D), consistent with this lncRNA’s known role in proliferation as an oncogene in adult AML[54].
Finally, we sought to determine potential lncRNA mechanism by identifying recurring patterns of regulation amongst lncMod triplets. To this end, we nominated candidate lncRNA-TF associations by ranking TFs based on the number of target genes regulated by each given TF (Supplementary Table S12). As proof-of-concept, we were able to detect known lncRNA-TF associations such as GAS5 with E2F4[55] (RNA-protein), and SNHG1 with TP53[56] (RNA-RNA) amongst lncMod triplets in our study (Supplementary Fig. S5E–F). A notable example from the hundreds of novel associations identified is between the B-ALL specific lncRNA, BLACE (B-cell acute lymphoblastic leukemia expressed, tau score: 0.999) and its top associated TF, XBP1, which has known roles in pre-B-ALL cell proliferation and tumorigenesis[57] (Fig. 4E–F). These predictions of lncRNA transcriptional networks provide focused avenues to elucidate the mechanisms through which lncRNAs can drive pediatric cancers.
lncRNA expression distinguishes cancer cell lineages in neuroblastoma
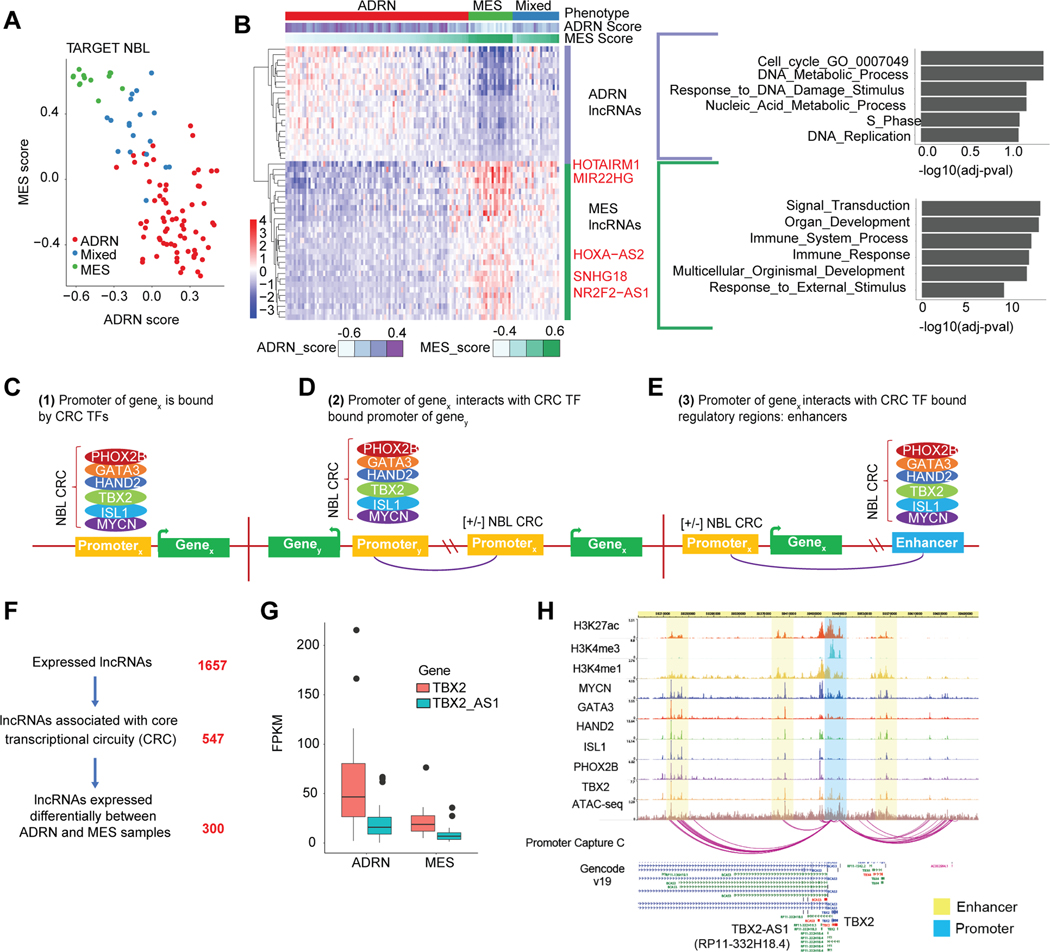
Pediatric cancers arise in the context of normal human development where cells do not differentiate as they should, resulting in malignant cell transformation[58]. Some tumors are comprised of heterogenous cells that resemble distinct differentiation lineages with distinct transcriptomic states due to specialized super enhancer transcription factor networks, [59]. We sought to discover lncRNAs associated with these varying cell lineages as they may contribute to pediatric cancer etiology. We used NBL as a model given its heterogeneity and two confirmed tumor cell states: the undifferentiated mesenchymal (MES) cells and the committed adrenergic (ADRN) cells, which can interconvert[60]. Given that NBL precursor cells, the neural crest cells, have been shown to have a more MES gene expression signature[59, 60], we hypothesized that lncRNAs correlated with an MES signature may play a role in NBL development. Using the gene set variation analysis (GSVA) method[61] we assigned for each NBL Stage 4 sample (n=130), both a MES and ADRN score. Using hierarchical clustering (Supplementary Fig. S6A) we categorized samples based on their primary gene expression phenotype as ADRN, MES, or mixed (Fig. 5A). We next correlated the MES and ADRN score with lncRNA expression across NBL samples. We observed 29 lncRNAs associated with MES samples and 21 lncRNAs associated with ADRN samples (Fig. 5B) (Spearman’s |rho| >0.6, adj. p-value < 0.01). A guilt-by-association analysis[62] analysis was performed to determine the potential functional pathway for these lncRNAs based on the pathway of their correlated protein coding genes. Gene set enrichment was performed using the gene ontology (GO) biological processes gene set. Intriguingly, the ADRN group of lncRNAs showed enrichment for DNA replication and cell cycle associated gene sets, whereas the MES lncRNAs were associated with organ development and immune response (Fig. 5B). The ADRN cell state is known to be dependent on MYCN [63], which may drive enrichment for cell-cycle associated lncRNAs. Indeed, we observed that ADRN samples had greater expression of MYCN compared to MES samples, despite including a lower proportion with MYCN amplification (Supplementary Fig. S6B). Additionally, the enriched immune response gene sets in the MES tumors are consistent with two recent, complementary findings demonstrating the immunogenicity of MES subtype [64, 65]. These pathway results were validated in an independent analysis of the GMKF NBL cohort restricted to Stage 4 samples (n=67) (Supplementary Fig. S6C). Across both TARGET and GMKF cohorts we observed 13 lncRNAs strongly associated with MES samples (Supplementary Fig. S6D), which warrant further study for their potential role in NBL development.
Figure 5: Identification of lncRNAs associated with distinct neuroblastoma cell states.
(A) The MES and ADRN signature score for TARGET NBL samples, with each sample labeled with either ADRN, Mixed, or MES phenotype based on clustering analysis. (B) Heatmap of the expression of lncRNAs that have significant correlation with either the MES or ADRN score (|r| >0.6, p-value < 0.01). lncRNAs were correlated with protein coding genes on the same chromosome and subsequent gene set enrichment analysis was performed for MES and ADRN protein coding genes separately. (C) Schematic of how ADRN associated CRC regulated genes are identified using ChIP-seq and chromatin interaction data. We identified lncRNAs based on three types of regulation. 1) CRC transcription factors binding directly at the promoter of the lncRNA. (D) 2) CRC TFs bind an enhancer region that interacts with a lncRNA promoter. (E) 3) CRC TFs bind the promoter of a different gene, and this promoter interacts with a lncRNA promoter. CRC TF binding was identified from ChIP-seq data, while enhancer-promoter and promoter-promoter interactions were identified from chromatin capture data. (F) Filtering of lncRNAs expressed in NBL based on CRC TF regulation and differential expression based on sample phenotypes (ADRN or MES). (G) Expression of TBX2 and TBX2-AS1 stratified by NBL sample phenotype (ADRN or MES). (H) ChIP-seq tracks for histone marks and CRC transcription factors in the NBL cell line: BE(2)C, and promoter capture C chromatin interactions in NBL cell line: NB1643, at the TBX2/TBX2-AS1 locus.
Identification of potential cancer driver lncRNAs via integration of epigenetic data
While majority of our NBL cohort has an ADRN phenotype we observed heterogeneity in lncRNA expression across ADRN samples. To better define ADRN associated lncRNAs, we integrated information about transcription factors within the ADRN core transcriptional circuitry (CRC). This set of co-bound and auto-regulated TF’s (MYCN, PHOX2B, HAND2, GATA3, ISL1, and TBX2[59, 63]) has been found to regulate many key neuroblastoma driver genes such as ALK and altogether drives cell identity. To distinguish potential lncRNA driver genes in NBL, we used epigenetic data to identify lncRNAs regulated by the CRC TFs. CRC-driven gene regulation can occur both by direct binding of TFs to the promoter of the gene of interest (Fig. 5C) or via long-range chromatin interactions and distal binding to other promoters (Fig. 5D) and enhancer regions (Fig. 5E) [59, 63, 66, 67]. CRC-bound regulatory loci were identified from publicly available ChIP-seq data for all ADRN TFs across two MYCN-amplified NBL cell lines: SKNBE(2)C and KELLY[63]. To comprehensively identify both short- and long- range CRC gene regulation, we generated high-resolution (i.e. using 4-cutter restriction enzyme DpnII) genome-wide promoter-focused Capture C[32] in the NBL cell line NB1643. After pinpointing gene promoters interacting with CRC TF bound regulatory loci (promoters or enhancers) (Fig. 5C–E), we identified 547 lncRNA genes associated with the NBL CRC (Fig. 5F, Supplementary Table S13), with only 249 (45%) of these lncRNA genes being bound by CRC TFs within their promoter regions. We further distinguished 300 ADRN lncRNAs based on differential expression (DE) between ADRN and MES samples (Fig. 5F, Supplementary Table S14), but note that 28% of these genes are also in NBL-associated copy number altered regions. For example, in the 17q amplified region, the TBX2-AS1 lncRNA is highly correlated to the CRC TF: TBX2 (Pearson’s r=0.77) and both are up-regulated in ADRN samples (Fig. 5G). CRC binding is observed at both the shared promoter region of TBX2 and TBX2-AS1 and at an interacting distal enhancer (Fig. 5H). TBX2 was recently shown to be involved in NBL cell proliferation[68] but the role of TBX2-AS1 in NBL is unknown.
Integrative multi-omic analysis prioritizes TBX2-AS1 as a candidate functional lncRNA in NBL
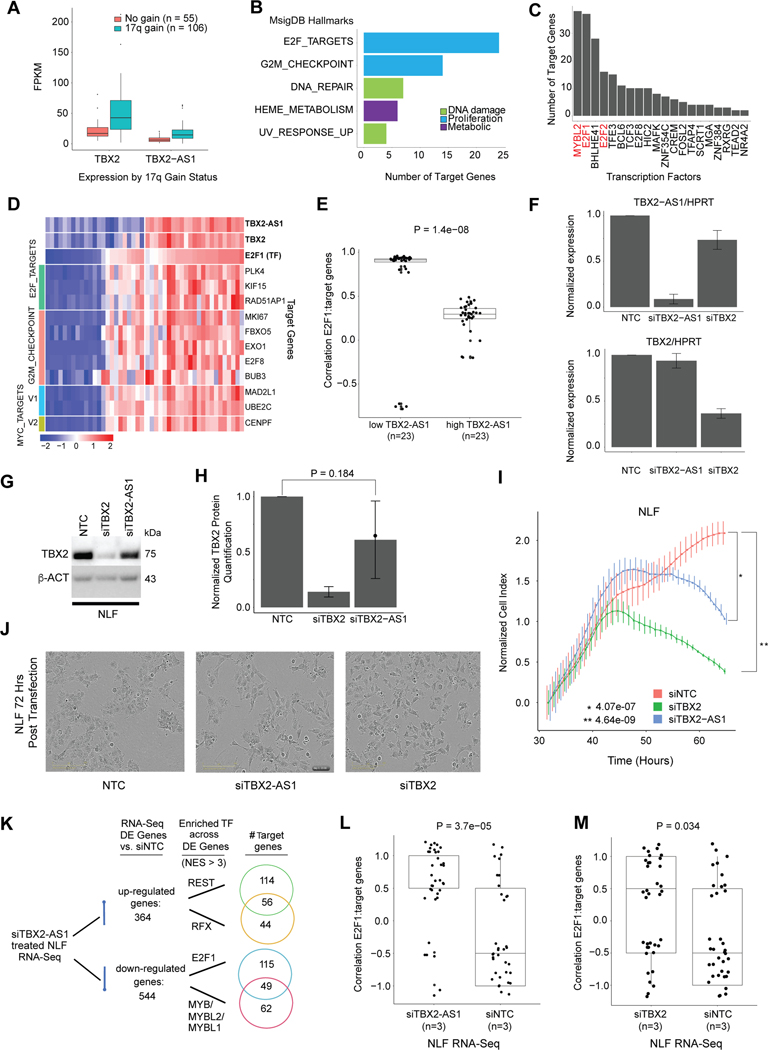
To obtain a comprehensive prioritization of candidate functional lncRNAs for each cancer histotype, we integrated information for (1) tissue elevated expression, (2) dysregulation due to DNA copy number aberration, (3) regulation by CRC TFs, and (4) significance in regulatory modulation (Supplementary Table S15). We further investigated NBL lncRNAs that had concurrent annotation as tissue elevated, lncRNA modulator, and as CRC regulated (Supplementary Table S16). The top ranked lncRNA in NBL was MEG3, which has a known role in both NBL and other cancers[41]. Given that 36% of tissue-elevated lncRNAs and 36% of NBL lncRNA modulators are CRC regulated, the next notable prioritized lncRNA was TBX2-AS1 which is co-regulated by a CRC TF, TBX2 (tau score: TBX2- 0.807, TBX2-AS1- 0.86; Supplementary Fig. S7A). We first confirmed that TBX2 and antisense lncRNA TBX2-AS1 had comparable expression in the stranded GMKF NBL cohort (Supplementary Fig. S7B). We next looked at copy number association and observed that TBX2-AS1 is up-regulated due to chromosome 17q gain (Fig. 6A) as has previously been shown for TBX2 [68]. TBX2 has been shown to drive NBL proliferation via the FOXM1/E2F1 gene regulatory network and we hypothesized that TBX2-AS1 may play a similar role given the predictions from our lncMod analysis indicating that TBX2-AS1 impacts E2F targets and G2M checkpoint genes (Fig. 6B). Furthermore, the TFs primarily impacted by TBX2 knockdown, MYBL2 and E2F1, were found to have the most target genes predicted to be regulated by TBX2-AS1 (Fig 6C). Evidence for this association was further supported by the correlation (Spearman’s rho > 0.4) between TBX2-AS1 and TBX2’s target TFs, including: FOXM1, E2F1, and MYBL2 (Supplementary Fig. S7C). We observed that samples with lower TBX2-AS1 expression had significantly greater correlation between E2F1 and its lncMod predicted target genes (Wilcoxon p-value=1.4×10−8, than samples with high TBX2-AS1 expression, suggesting a role in regulation of E2F1 targeting (Fig. 6D–E). While the strong correlation between TBX2-AS1 and TBX2 may confound our predictions, a previous study showed positionally conserved lncRNAs[54], including TBX2-AS1, often regulate their neighboring developmental TFs (TBX2) and can play roles in genome organization and cancer[54]. Based on the promising in silico evidence, we prioritized TBX2-AS1 for experimental study.
Figure 6: TBX2-AS1 influences NBL cell proliferation and E2F1-target gene expression.
(A)Expression of TBX2 and TBX2-AS1 in NBL tumor samples with and without 17q gain. (B) The top MSigDB Hallmarks enriched across targets genes (p-value < 0.01) regulated by TBX2-AS1 as predicted from lncMod analysis. (C) The transcription factors with most target genes regulated by TBX2-AS1 as predicted from lncMod analysis. (D) Expression of gene targets of the E2F1 transcription factor that are enriched for proliferation hallmarks, in samples with low and high TBX2 and TBX2-AS1 expression. TBX2 expression is highly correlated with that of TBX2-AS1 (Pearson’s r=0.77). (E) Expression correlation between E2F1 and its lncMod predicted target genes (n=36) in TARGET NBL Stage 4 non MYCN amplified samples with the lowest 25% versus highest 25% quartile of TBX2-AS1 expression. (F) siRNA knockdown efficiency of TBX2-AS1 and TBX2 in the NBL cell line, NLF. (G) Western blot analysis of TBX2 in siTBX2 and siTBX2-AS1 treated NLF cell line (representative blot shown). (H) Quantification of TBX2 protein expression from three Western blots of independent knockdown experiments. (I) Representative image of cell growth (as measured by RT-Ces assay) of the NBL cell lines, NLF. Cell index is normalized to time point when siRNA reagent is added at 24 hours post cell plating. (J) Images of NLF cells after siTBX2-AS1 and siTBX2 show morphology changes. (K) Results from iRegulon analysis for genes that are up- or down-regulated upon siTBX2-AS1 treatment in NLF. Number of genes shown in Venn diagram with evidence of motif or ChIP-seq binding of the listed transcription factors. (L) Expression correlation between E2F1 and its lncMod predicted target genes (n=36) identified using RNA-sequencing expression profiling from the NLF cell line treated with either siNTC or siTBX2-AS1. (M) Expression correlation between E2F1 and its lncMod predicted target genes (n=36) identified using RNA-sequencing expression profiling from the NLF cell line treated with either siNTC or siTBX2.
Silencing of TBX2-AS1 inhibits neuroblastoma cell growth and validates lncMod pathway prediction
We assessed the role of TBX2-AS1 using human-derived NBL cell line models. First, we evaluated TBX2-AS1 expression across 38 NBL cell lines using RNA-seq[69] followed by validation of eight cell lines using RT-qPCR (Supplementary Figs. S7D–E). We selected NLF and SKNSH models for further study based on their high TBX2-AS1 expression and varying expression levels of TBX2. To ensure lncRNA specific silencing in the context of co-regulation at the genomic locus of TBX2 and TBX2-AS1, we chose to use small interfering RNA (siRNA) to knockdown TBX2-AS1 and TBX2. There was a 91% and 27% reduction of TBX2-AS1 expression in NLF treated with siTBX2-AS1 and siTBX2, respectively (Fig. 6F). Though TBX2 was slightly down-regulated in the siTBX2-AS1 treated cells, this change was not significant at the RNA or protein level (Fig. 6F–H), while siTBX2 treated cells had 63% reduced TBX2 expression. Given the known role of TBX2 in NBL cell proliferation[68], we measured cell growth of siTBX2-AS1 treated NLF cells to determine if TBX2-AS1 functions similarly. When the non-targeting control (siNTC) treated cells reached confluence, NLF cell growth index was reduced 42%.6 and 64.4% in the siTBX2-AS1 and siTBX2 treated cells, respectively (n=3, p-value < 0.01) (Fig. 6I). Knockdown of TBX2-AS1 with three additional unique siRNAs each reduced NLF cell growth (p <0.05) and did not impact TBX2 expression (Supplementary Fig. S7F–H). Live cell imaging using the IncuCyte revealed changes in cell morphology for siTBX2-AS1 and siTBX2 treated NLF cells, featuring an appearance of disrupted cell to cell adhesion and elongated cell body (Fig. 6J), suggestive of a neuronal differentiation phenotype. We repeated this knockdown study in the SKNSH cell line and observed similar trends in expression changes and growth reduction (Supplementary Fig. S7I–N).
RNA-sequencing following TBX2-AS1 silencing validates E2F1-target gene correlation
To identify pathways impacted by TBX2-AS1 and TBX2 knockdown, we performed total RNA sequencing in triplicate of NLF cells following siRNA treatment. We observed that the expression of TBX2 was slightly, but not-significantly, decreased upon TBX2-AS1 knockdown, confirming our qPCR and Western blot results (Supplementary Fig. S8A–B). TBX2-AS1 expression did not significantly change in the siTBX2 condition, confirming previous studies [68]. From the expression profiling we identified differentially expressed (DE) genes between control (siNTC) and siTBX2-AS1 (n=908) and control versus siTBX2 treated cells (n=569) (Supplementary Fig. S8C–D). In the siTBX2-AS1 condition, we observed that TBX2-AS1 was in the 99th percentile of all DE genes (log-fold change > 1.5, adj p-value < 0.1), ranking 11 out of 908 based on fold change, while TBX2 ranked in the 77th percentile of DE genes in the siTBX2 condition (Supplementary Fig. S8E–F). While most of the DE genes were unique to each condition, the log fold change between the 130 DE genes common to both knockdown conditions were highly correlated (r=0.877, Supplementary Fig. S8G). Gene set enrichment analysis (GSEA) of the 364 significantly up-regulated genes (log-fold change > 1.5, adj p-value < 0.1), associated with siTBX2-AS1, revealed enrichment (FDR < 0.1) for hallmarks associated with inflammation including: TNFA signaling and interferon gamma response (Supplementary Table S17). Across the 544 siTBX2-AS1 down-regulated genes, the E2F target genes hallmark was the only significantly enriched gene set. GSEA of the upregulated genes in the siTBX2 condition were associated with proliferation such as MYC targets and TP53 pathway, while downregulated genes were enriched for inflammation associated interferon alpha and gamma response gene sets (Supplementary Table S17). To determine whether differentially expressed genes shared common regulation, we used the iRegulon program[70] to search upstream of genes for TF motifs and TF ChIP-seq tracks from ENCODE. Using a normalized enrichment score (NES) of at least 3, we observed motif enrichment for the neuronal differentiation repressor REST and the RFX family of transcription factors in 59% of siTBX2-AS1 up-regulated genes (Fig. 6K). In 42% of downregulated genes, the top enriched TFs were MYBL2 and E2F1, corroborating earlier GSEA results. Of the E2F1 and MYBL2 target genes, 46% were also TBX2/neuroblastoma CRC target genes.
While growth assays confirmed our lncMod pathway prediction that TBX2-AS1 impacts NBL proliferation, gene expression profiling revealed a significant increase (Wilcoxon, p-value=3.7×10−5) in the correlation between E2F1 and predicted target genes associated with TBX2-AS1 knockdown, reflective of the association we observed in-vivo between E2F1 and its target genes in patients with lower TBX2-AS1 expression (Fig. 6L). E2F1-target gene correlation modestly changed upon TBX2 knockdown (Fig. 6M). These data thus demonstrate the utility of our integrative lncRNA characterization and prioritization approach for future validation experiments across all cancers considered in this study. Furthermore, we uncovered a functional role for TBX2-AS1 in NBL proliferation impacting target genes of TBX2, E2F, MYBL2, and REST.
Discussion
LncRNAs have emerged as important regulators of gene expression and their dysregulation can impact key cancer pathways and drive tumorigenesis[1, 2]. Despite this, relatively few lncRNAs have been experimentally characterized. Tools such as LncSpA have emerged to query lncRNA expression in normal and cancerous tissues, including a subset of pediatric cancer samples from TARGET[71]. However, the functional mechanisms of these expressed lncRNAs remain unknown. In this study, we explored lncRNA expression, cancer association, and regulatory networks across 1,044 pediatric leukemias and solid tumors, representing six different cancer types. The careful curation, quality control, and breadth of samples included allowed for robust identification of tissue-elevated lncRNAs. Furthermore, systems modelling identified expression patterns for both up- and downstream lncRNA gene regulation. Altogether, we provide multi-dimensional insight into the predicted biological and functional relevance of lncRNAs by integrating WGS, ChIP-seq, chromatin capture, and predictions of transcriptional networks.
Analysis of the lncRNA landscape across pediatric cancers revealed the histotype and context-specific nature of lncRNAs. We report a total of 2,657 robustly expressed lncRNAs across the six cancer types studied. This number is notably smaller than reports from previous pan-cancer studies in both adult and childhood malignancies[9, 11, 71], due to the smaller number of cancer types studied here and conservative expression threshold applied. However, similar to findings in adult cancers, 43% (1,142/ 2,657) of expressed lncRNAs exhibited tissue-elevated (TE) expression across pediatric cancers. Indeed, lncRNAs had significantly greater tissue specificity than protein coding genes, making them more ideal candidates as biomarkers. Currently there is one lncRNA, PCA3, that is FDA-approved as a biomarker for prostate cancer[72], while multiple trials investigating ncRNAs in cancer prognostics are underway[73]. In this study, the top five most tissue-elevated lncRNAs per cancer were sufficient to differentiate each cancer histotype. Furthermore, we identify lncRNAs specific to distinct cell lineages within NBL, suggesting there is potential for lncRNAs to be used as highly sensitive markers to differentiate cancer subtypes more accurately.
Typically, investigation of lncRNA dysregulation involves comparing lncRNA expression between cancer and normal control samples and is an analysis that amply yields adult cancer associated lncRNAs[9]. However, the lack of normal expression controls for many pediatric cancers[42, 71] is a major complication in defining pediatric cancer-associated lncRNAs. To overcome this, we leveraged information about how pediatric cancers are epigenetically regulated. In particular, NBL is composed of two cell lineages representing different development stages and each with distinct super-enhancer transcription factor networks. Given the tie between organogenesis and tumorigenesis in pediatric cancer[58], we hypothesized that lncRNAs associated with these cell states may also be involved in NBL development. Through correlation and pathway analyses, we discovered that lncRNAs associated with the mesenchymal cell lineage had enrichment for organogenesis gene sets, while adrenergic-associated lncRNAs were predicted to be involved in proliferation based on enrichment for DNA replication and cell cycle gene sets. The majority of NBL samples have cells with an adrenergic gene expression signature, which could suggest that ADRN lncRNAs are major drivers of disease and thus potential therapeutic targets. To better identify these ADRN lncRNAs, we integrated ChIP-sequencing of core regulatory (CRC) transcription factors for ADRN cells with our expression data to identify cancer driver lncRNAs. CRC TFs bind to cell-type-specific enhancers and regulate the expression of cell-type-specific genes. By delineating enhancer associated gene regulation we were able to prioritize lncRNAs likely to be important for cancer cell identity based on CRC TF regulation. CRC TFs have been well defined for NBL[63, 67]; however, the fact that they largely bind enhancer regions necessitated that we also use chromatin interaction data to accurately determine the regulated genes. Incorporation of these datasets allowed us to identify 2-fold more CRC regulated lncRNAs in NBL as compared to using just ChIP-seq data alone. Similar analyses of lncRNAs regulated by the TAL1 CRC has previously revealed novel T-ALL associated genes[20]. An important next step in distinguishing cancer-specific lncRNAs will be the application of this novel analysis to a broader set of pediatric cancers.
While upstream regulation can help nominate cancer-associated lncRNAs, an understanding of lncRNA mechanism of action in a histology-specific manner is also crucial. However, prediction of lncRNA function is limited given that very few lncRNA mechanisms have been fully established for any cell type and lncRNAs lack conserved sequence and structure[74]. Many studies instead use correlated protein coding gene expression as a proxy to define lncRNA pathways, but this approach often results in many false positives and does not provide mechanistic insight[71, 74]. To address this, we used the lncMod method[15, 31] to model the functional mechanism of dysregulated lncRNAs by examining correlated changes in transcription factor to target gene regulation. We used motif presence and regression analysis to identify TF-target gene relationships, though future studies will be strengthened by incorporating TF ChIP-seq data, when it becomes more widely available for pediatric cancers. Nevertheless, we were able to successfully associate lncRNAs to TFs with known interactions, such as SNHG1 with TP53[56], while also providing a prioritized list of novel associations that serve as a starting point for targeted experimental studies such as RIP/MS and ChiRP-seq[75]. While our lncMod analysis was focused on transcriptional regulation, the addition of microRNA binding and RNA-binding protein data, as utilized in adult cancers[16], is an important next step in understanding how lncRNAs impact post-transcriptional regulation in pediatric cancers. The recent lncRNAfunc knowledgebase, a curated list of lncRNA functional mechanisms in TCGA samples from adults, may provide complementary information to our lncMod analysis by further enhancing our understanding of how pediatric lncRNAs regulate their target genes[76].
In this study we delineated high confidence lncRNA expression across pediatric cancers within the restrictions set by the sequencing depth and RNA-seq type available per cancer dataset. We required RNA-seq samples included in our study to have at least 10 million reads and read length of at least 75 bp. We used the StringTie method for expression quantification, which more conservatively assigns reads associated with lncRNAs, especially those that overlap protein coding genes, resulting in slightly lower but likely more accurate expression values. Given that all samples in this study were poly-A selected, with the exception of the T-ALL cohort, our analyses were restricted to poly-A tailed lncRNAs, which tend to be processed similarly to protein coding mRNAs and have more stable transcripts[77]. Future studies involving total RNA-seq, greater sequencing depth, and longer read sizes could capture a larger diversity of expressed lncRNAs by accounting for non-polyadenylated genes and identify scarcer or temporally expressed lncRNAs. Nevertheless, given that our study focuses on highly expressed lncRNAs there is reduced potential for transcriptional noise to confound our functional predictions[78]. In addition to having a limited number of RNA matched WGS samples, the Complete Genomics short read technology limits the detection of structural variants based on size as previously described[29, 42]. The use of long-read sequencing and greater sequencing depth in future studies will enable more accurate copy number and structural variant detection in pediatric cancers.
Finally, multi-dimensional integration of our computational predictions resulted in the nomination of functionally relevant lncRNAs in each pediatric cancer. We annotated tissue specificity, copy number, pathway, and likely targets for these lncRNAs, providing a solid foundation for mechanistic studies. As proof-of-principle we demonstrate that the top-prioritized tissue-specific and CRC-regulated lncRNA, TBX2-AS1, impacts NBL cell growth, validating our in-vivo pathway predictions. Through in-vitro transcriptomic profiling, following TBX2-AS1 knockdown, we were able to show that the correlation of E2F1 and its target genes significantly increases, consistent with our predictions using in-vivo data. We did not observe a significant difference in cells treated with siTBX2, which may suggest that E2F1 regulation is driven by TBX2-AS1. This was further supported by GSEA of siTBX2 vs siTBX2-AS1 treated cells, which revealed differentially enriched pathways. siTBX2 cells were associated with changes in expression of MYC targets, consistent with previous findings, while siTBX2-AS1 associated with changes in E2F1 target gene expression.
Investigating the mechanism through which TBX2-AS1 impacts gene regulation is an important future direction, for which we have derived several potential hypotheses from our studies. First, we observed that both E2F1 target genes and CRC target genes (including those of TBX2) are impacted by TBX2-AS1 knockdown. However, the observation that TBX2 expression was non-significantly down-regulated in siTBX2-AS1 treated cells suggests that TBX2-AS1 function is not entirely TBX2 dependent. Instead, one possibility is that TBX2-AS1 binds directly to TBX2 and/or E2F1 in order to guide them to gene targets. Another possibility is that of a regulatory cascade in which TBX2-AS1 regulates TBX2 which in turn regulates E2F1. Application of techniques, such as ChIRP-seq, that can uncover lncRNA- DNA and lncRNA- protein binding partners, could illuminate the functional mechanism of TBX2-AS1 in the future. TBX2-AS1 was also previously shown to be among a group of lncRNAs which are positionally conserved and near developmental associated TFs[54]. This group of lncRNAs and their neighboring TFs, typically have tissue specific expression, can be involved in cancer development, and affect each other’s expression[54], all of which we observed for TBX2 and TBX2-AS1. In combination these genes may contribute in tandem to the proliferative state of NBL cells and have potential as novel therapeutic targets.
Altogether, this study provides a comprehensive characterization of the most highly expressed lncRNAs across six high-risk pediatric cancers and serves as a rich resource for future mechanistic studies; these data may aid in the selection of cancer biomarkers and candidate therapeutic lncRNA targets.
Supplementary Material
Significance.
Comprehensive characterization of lncRNAs in pediatric cancer leads to the identification of highly expressed lncRNAs across childhood cancers, annotation of lncRNAs showing histotype-specific elevated expression, and prediction of lncRNA gene regulatory networks.
Acknowledgments:
This work was supported in part by NIH grants R01-CA124709 (S.J.D.), R01-CA237562 (S.J.D.), R03-CA230366 (S.J.D.), X01-HL136997 (J.M.M.), and T32-HG46-18 (A.M.). This project was also funded in part by a supplement to the Children’s Oncology Group Chair’s grant CA098543 and with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E to S.J.D and Complete Genomics. Promoter Capture C studies were partially funded by the Center for Spatial and Functional Genomics (A.D.W and S.FA.G) at CHOP. S.FA.G. was supported by the Daniel B. Burke Endowed and Chair for Diabetes Research and R01 HG010067. We thank Dr. Daniela S. Gerhard for her leadership and steadfast commitment to childhood cancer research, including the TARGET project. Dr. Gerhard collaborated on this study and provided important feedback on the initial manuscript draft prior to her passing on June 25, 2021. We also thank Dr. Jo Lynne Rokita and Dr. Amber Weiner for insightful discussions which contributed to this project.
Footnotes
Conflicts of interest statement:
The authors declare no potential conflicts of interest.
References
- 1.Iyer MK, et al. , The landscape of long noncoding RNAs in the human transcriptome. Nature genetics, 2015. 47: p. 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gil N. and Ulitsky I, Regulation of gene expression by cis-acting long non-coding RNAs. Nat Rev Genet, 2020. 21(2): p. 102–117. [DOI] [PubMed] [Google Scholar]
- 3.Dykes IM and Emanueli C, Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genomics Proteomics Bioinformatics, 2017. 15(3): p. 177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kotake Y, et al. , Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene, 2011. 30(16): p. 1956–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engreitz JM, et al. , The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science, 2013. 341(6147): p. 1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tripathi V, et al. , The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell, 2010. 39(6): p. 925–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monnier P, et al. , H19 lncRNA controls gene expression of the Imprinted Gene Network by recruiting MBD1. Proc Natl Acad Sci U S A, 2013. 110(51): p. 20693–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin N, et al. , An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell, 2014. 53(6): p. 1005–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan X, et al. , Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell, 2015. 28(4): p. 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du Z, et al. , Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol, 2013. 20(7): p. 908–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lanzós A, et al. , Discovery of Cancer Driver Long Noncoding RNAs across 1112 Tumour Genomes: New Candidates and Distinguishing Features. Scientific Reports, 2017. 7: p. 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, et al. , Discovery and validation of the tumor-suppressive function of long noncoding RNA PANDA in human diffuse large B-cell lymphoma through the inactivation of MAPK/ERK signaling pathway. Oncotarget, 2017. 8(42): p. 72182–72196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hajjari M. and Salavaty A, HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med, 2015. 12(1): p. 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Onagoruwa OT, et al. , Oncogenic Role of PVT1 and Therapeutic Implications. Front Oncol, 2020. 10: p. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, et al. , LncMAP: Pan-cancer Atlas of long noncoding RNA-mediated transcriptional network perturbations. Nucleic Acids Research, 2018. 46: p. 1113–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiu HS, et al. , Pan-Cancer Analysis of lncRNA Regulation Supports Their Targeting of Cancer Genes in Each Tumor Context. Cell Rep, 2018. 23(1): p. 297–312 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huarte M, The emerging role of lncRNAs in cancer. Nature medicine, 2015. 21: p. 1253–61. [DOI] [PubMed] [Google Scholar]
- 18.Mondal T, et al. , Sense-Antisense lncRNA Pair Encoded by Locus 6p22.3 Determines Neuroblastoma Susceptibility via the USP36-CHD7-SOX9 Regulatory Axis. Cancer Cell, 2018. 33: p. 417–434.e7. [DOI] [PubMed] [Google Scholar]
- 19.Rombaut D, et al. , Integrative analysis identifies lincRNAs up- and downstream of neuroblastoma driver genes. Sci Rep, 2019. 9(1): p. 5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ngoc PCT, et al. , Identification of novel lncRNAs regulated by the TAL1 complex in T-cell acute lymphoblastic leukemia. Leukemia, 2018. 32(10): p. 2138–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trimarchi T, et al. , Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell, 2014. 158(3): p. 593–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vanhooren J, et al. , Deciphering the Non-Coding RNA Landscape of Pediatric Acute Myeloid Leukemia. Cancers (Basel), 2022. 14(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kesherwani V, et al. , Long non-coding RNA profiling of pediatric Medulloblastoma. BMC Med Genomics, 2020. 13(1): p. 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russell MR, et al. , CASC15-S is a tumor suppressor lncRNA at the 6p22 neuroblastoma susceptibility locus. Cancer Res, 2016. 75: p. 3155–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDaniel LD, et al. , Common variants upstream of MLF1 at 3q25 and within CPZ at 4p16 associated with neuroblastoma. PLoS Genet, 2017. 13(5): p. e1006787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dobin A, et al. , STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 2013. 29(1): p. 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ewels P, et al. , MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics, 2016. 32(19): p. 3047–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yanai I, et al. , Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics, 2005. 21(5): p. 650–9. [DOI] [PubMed] [Google Scholar]
- 29.Lopez G, et al. , Somatic structural variation targets neurodevelopmental genes and identifies SHANK2 as a tumor suppressor in neuroblastoma. Genome Res, 2020. 30(9): p. 1228–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopez G, et al. , svpluscnv: analysis and visualization of complex structural variation data. Bioinformatics, 2021. 37(13): p. 1912–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, et al. , Identification and characterization of lncRNA mediated transcriptional dysregulation dictates lncRNA roles in glioblastoma. Oncotarget, 2016. 7: p. 45027–45041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chesi A, et al. , Genome-scale Capture C promoter interactions implicate effector genes at GWAS loci for bone mineral density. Nat Commun, 2019. 10(1): p. 1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su C, et al. , Mapping effector genes at lupus GWAS loci using promoter Capture-C in follicular helper T cells. Nat Commun, 2020. 11(1): p. 3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pahl MC, et al. , Implicating effector genes at COVID-19 GWAS loci using promoter-focused Capture-C in disease-relevant immune cell types. Genome Biol, 2022. 23(1): p. 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palermo J, et al. , Variant-to-gene mapping followed by cross-species genetic screening identifies GPI-anchor biosynthesis as a regulator of sleep. Sci Adv, 2023. 9(1): p. eabq0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pertea M, et al. , StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature biotechnology, 2015. 33: p. 290–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frankish A, et al. , GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res, 2019. 47(D1): p. D766–D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li A, Zhang J, and Zhou Z, PLEK: a tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinformatics, 2014. 15: p. 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Consortium GT, Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science, 2015. 348(6235): p. 648–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kryuchkova-Mostacci N. and Robinson-Rechavi M, A benchmark of gene expression tissue-specificity metrics. Brief Bioinform, 2017. 18(2): p. 205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong K, Tang W, and Dong R, MEG3, HCN3 and linc01105 influence proliferation and apoptosis of neuroblastoma cells via HIF-1 alpha and p53 pathway. Pediatric Blood and Cancer, 2016. 63: p. S194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma X, et al. , Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature, 2018. 555(7696): p. 371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mermel CH, et al. , GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol, 2011. 12(4): p. R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pugh TJ, et al. , The genetic landscape of high-risk neuroblastoma. Nat Genet, 2013. 45(3): p. 279–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gadd S, et al. , A Children’s Oncology Group and TARGET initiative exploring the genetic landscape of Wilms tumor. Nat Genet, 2017. 49(10): p. 1487–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harvey RC, et al. , Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood, 2010. 116(23): p. 4874–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emmrich S, et al. , LincRNAs MONC and MIR100HG act as oncogenes in acute megakaryoblastic leukemia. Mol Cancer, 2014. 13: p. 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, et al. , Genome-wide screening for functional long noncoding RNAs in human cells by Cas9 targeting of splice sites. Nat Biotechnol, 2018. [DOI] [PubMed] [Google Scholar]
- 49.Zhao X, et al. , CTCF cooperates with noncoding RNA MYCNOS to promote neuroblastoma progression through facilitating MYCN expression. Oncogene, 2015: p. 1–12. [DOI] [PubMed] [Google Scholar]
- 50.Ng SY, et al. , The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Molecular Cell, 2013. 51: p. 349–359. [DOI] [PubMed] [Google Scholar]
- 51.Cho SW, et al. , Promoter of lncRNA Gene PVT1 Is a Tumor-Suppressor DNA Boundary Element. Cell, 2018. 173(6): p. 1398–1412 e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liberzon A, et al. , The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst, 2015. 1(6): p. 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ahmadi SE, et al. , MYC: a multipurpose oncogene with prognostic and therapeutic implications in blood malignancies. J Hematol Oncol, 2021. 14(1): p. 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amaral PP, et al. , Genomic positional conservation identifies topological anchor point RNAs linked to developmental loci. Genome Biol, 2018. 19(1): p. 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang M, et al. , Long noncoding RNA GAS5 promotes bladder cancer cells apoptosis through inhibiting EZH2 transcription. Cell Death Dis, 2018. 9(2): p. 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao Y, et al. , Long non-coding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) promote cell proliferation in colorectal cancer by affecting P53. Eur Rev Med Pharmacol Sci, 2018. 22(4): p. 976–984. [DOI] [PubMed] [Google Scholar]
- 57.Kharabi Masouleh B, et al. , Mechanistic rationale for targeting the unfolded protein response in pre-B acute lymphoblastic leukemia. Proc Natl Acad Sci U S A, 2014. 111(21): p. E2219–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Federico S, Brennan R, and Dyer MA, Childhood cancer and developmental biology a crucial partnership. Curr Top Dev Biol, 2011. 94: p. 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boeva V, et al. , Heterogeneity of neuroblastoma cell identity defined by transcriptional circuitries. Nat Genet, 2017. 49(9): p. 1408–1413. [DOI] [PubMed] [Google Scholar]
- 60.van Groningen T, et al. , Neuroblastoma is composed of two super-enhancer-associated differentiation states. Nat Genet, 2017. 49(8): p. 1261–1266. [DOI] [PubMed] [Google Scholar]
- 61.Hanzelmann S, Castelo R, and Guinney J, GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics, 2013. 14: p. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Signal B, Gloss BS, and Dinger ME, Computational Approaches for Functional Prediction and Characterisation of Long Noncoding RNAs. Trends in Genetics, 2016. 32: p. 620–637. [DOI] [PubMed] [Google Scholar]
- 63.Durbin AD, et al. , Selective gene dependencies in MYCN-amplified neuroblastoma include the core transcriptional regulatory circuitry. Nat Genet, 2018. 50(9): p. 1240–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sengupta S, et al. , Mesenchymal and adrenergic cell lineage states in neuroblastoma possess distinct immunogenic phenotypes. Nat Cancer, 2022. 3(10): p. 1228–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wolpaw AJ, et al. , Epigenetic state determines inflammatory sensing in neuroblastoma. Proc Natl Acad Sci U S A, 2022. 119(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mansour MR, et al. , An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science, 2014. 346: p. 1373–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sanda T, et al. , Core transcriptional regulatory circuit controlled by the TAL1 complex in human T cell acute lymphoblastic leukemia. Cancer Cell, 2012. 22(2): p. 209–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Decaesteker B, et al. , TBX2 is a neuroblastoma core regulatory circuitry component enhancing MYCN/FOXM1 reactivation of DREAM targets. Nat Commun, 2018. 9(1): p. 4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Harenza JL, et al. , Transcriptomic profiling of 39 commonly-used neuroblastoma cell lines. Sci Data, 2017. 4: p. 170033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Janky R, et al. , iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput Biol, 2014. 10(7): p. e1003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lv D, et al. , LncSpA: LncRNA Spatial Atlas of Expression across Normal and Cancer Tissues. Cancer Res, 2020. 80(10): p. 2067–2071. [DOI] [PubMed] [Google Scholar]
- 72.Lemos AEG, et al. , The long non-coding RNA PCA3: an update of its functions and clinical applications as a biomarker in prostate cancer. Oncotarget, 2019. 10(61): p. 6589–6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Slack FJ and Chinnaiyan AM, The Role of Non-coding RNAs in Oncology. Cell, 2019. 179(5): p. 1033–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang X. and Ho TT, Computational Analysis of lncRNA Function in Cancer. Methods Mol Biol, 2019. 1878: p. 139–155. [DOI] [PubMed] [Google Scholar]
- 75.Chu C, Spitale RC, and Chang HY, Technologies to probe functions and mechanisms of long noncoding RNAs. Nature Structural & Molecular Biology, 2015. 22: p. 29–35. [DOI] [PubMed] [Google Scholar]
- 76.Yang M, et al. , lncRNAfunc: a knowledgebase of lncRNA function in human cancer. Nucleic Acids Res, 2022. 50(D1): p. D1295–D1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Clark MB, et al. , Genome-wide analysis of long noncoding RNA stability. Genome Res, 2012. 22(5): p. 885–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hon CC, et al. , An atlas of human long non-coding RNAs with accurate 5′ ends. Nature, 2017. 543: p. 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All TARGET RNA and DNA-sequencing data analyzed in this study are available through the database of Genotypes and Phenotypes (dbGaP; https://www.ncbi.nlm.nih.gov/gap/) under study-id phs000218 and accession number phs000467. GMKF RNA-sequencing data are available through dbGAP study accession phs001436.v1.p1. Neuroblastoma cell line RNA-sequencing data analyzed in this study are available through GEO at accessions GSE89413. NBL histone ChIP-seq and transcription factor ChIP-seq data used in this study are both available through GEO at accessions: GSE138315 and GSE94822, respectively. RNA-sequencing generated for NLF cells treated with siRNAs in this study is available through GEO at accession: GSE238166. Code used to generate figures and analyze data is available at: https://github.com/diskin-lab-chop/PanTARGET_lncRNA_Study. All other raw data generated in this study are available upon request from the corresponding author.