Abstract
The human leukocyte antigen (HLA) locus plays a critical role in complex traits spanning autoimmune and infectious diseases, transplantation and cancer. While coding variation in HLA genes has been extensively documented, regulatory genetic variation modulating HLA expression levels has not been comprehensively investigated. Here we mapped expression quantitative trait loci (eQTLs) for classical HLA genes across 1,073 individuals and 1,131,414 single cells from three tissues. To mitigate technical confounding, we developed scHLApers, a pipeline to accurately quantify single-cell HLA expression using personalized reference genomes. We identified cell-type-specific cis-eQTLs for every classical HLA gene. Modeling eQTLs at single-cell resolution revealed that many eQTL effects are dynamic across cell states even within a cell type. HLA-DQ genes exhibit particularly cell-state-dependent effects within myeloid, B and T cells. For example, a T cell HLA-DQA1 eQTL (rs3104371) is strongest in cytotoxic cells. Dynamic HLA regulation may underlie important interindividual variability in immune responses.
The human leukocyte antigen (HLA) genes, located within the major histocompatibility complex (MHC) region on chromosome 6, are central to the immune response. Classical HLA class I and II molecules trigger adaptive immunity by presenting antigens to CD8+ and CD4+ T cells, respectively. Positive and balancing selection has made the coding sequences of these genes among the most polymorphic in the genome1. The HLA locus has the greatest number of associations with immune-mediated diseases and typically has larger effect sizes than all other loci combined1-4. For example, the HLA-C*06:02 allele is the major genetic risk factor for psoriasis5, and HLA-DRB1 alleles modulate risk for rheumatoid arthritis (RA)6 and multiple sclerosis7. HLA genes also play key roles in cancer by presenting neoantigens and in transplantation, where mismatched HLA alleles can result in rejection.
The regulatory mechanisms governing HLA genes are not yet well understood. Previous studies have focused on coding variation altering HLA protein structure, which may affect antigen binding6,8,9 or restrict the T cell receptor repertoire10-12. However, mounting evidence indicates that noncoding HLA regulatory variation can influence disease13-15. Higher HLA-C expression was found to control HIV infection but increase Crohn’s disease risk13. Investigators have argued that risk alleles for systemic lupus erythematosus and vitiligo lie within regulatory regions that increase class II expression in myeloid cells14,15. Understanding the role of noncoding HLA variation in disease requires defining the genetic variation regulating HLA gene expression. Previous bulk RNA-sequencing (RNA-seq) studies have identified expression quantitative trait loci (eQTLs) for HLA genes in homogeneous cell lines16,17. However, HLA gene regulation may be context dependent, varying across cell types or finer-grained cell states within a cell type. For example, we previously demonstrated that allele-specific expression of HLA class II changes dynamically in activated memory CD4+ T cells in vitro18. Single-cell RNA-seq (scRNA-seq) may offer a more comprehensive understanding of HLA expression and its regulation by assaying cell states in vivo and mapping context-dependent eQTLs19-21.
Because HLA genes are highly polymorphic, standard short-read sequencing pipelines that align reads to a single reference genome are biased when quantifying HLA expression22,23. Reads can fail to align if an individual’s allele is dissimilar from the reference allele, resulting in unmapped reads, or reads can ‘multi-map’ to multiple HLA genes due to sequence similarity between genes24. This bias confounds eQTL analysis, making it difficult to distinguish genuine genetic associations with HLA expression from inaccurate read alignment. In bulk data, personalized reference genomes accounting for individuals’ HLA genotypes have been used to overcome this bias16,17,25,26. In this Analysis, we developed a personalized pipeline (scHLApers; Fig. 1c) extending this approach to single-cell data. We integrated four datasets (Fig. 1a) to explore how genetic regulation of classical HLA class I (HLA-A, HLA-B and HLA-C) and class II (HLA-DPA1, HLA-DPB1, HLA-DQA1, HLA-DQB1 and HLA-DRB1) gene expression varies dynamically across diverse immune cell states (Fig. 1d), offering new insights into complex diseases.
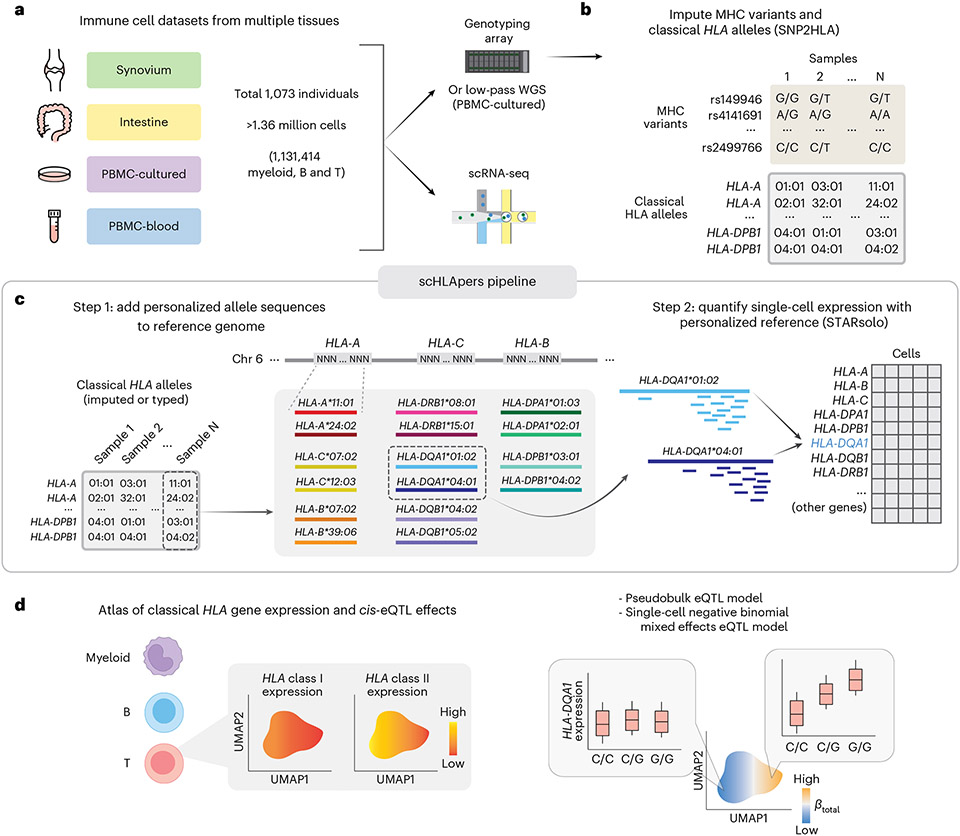
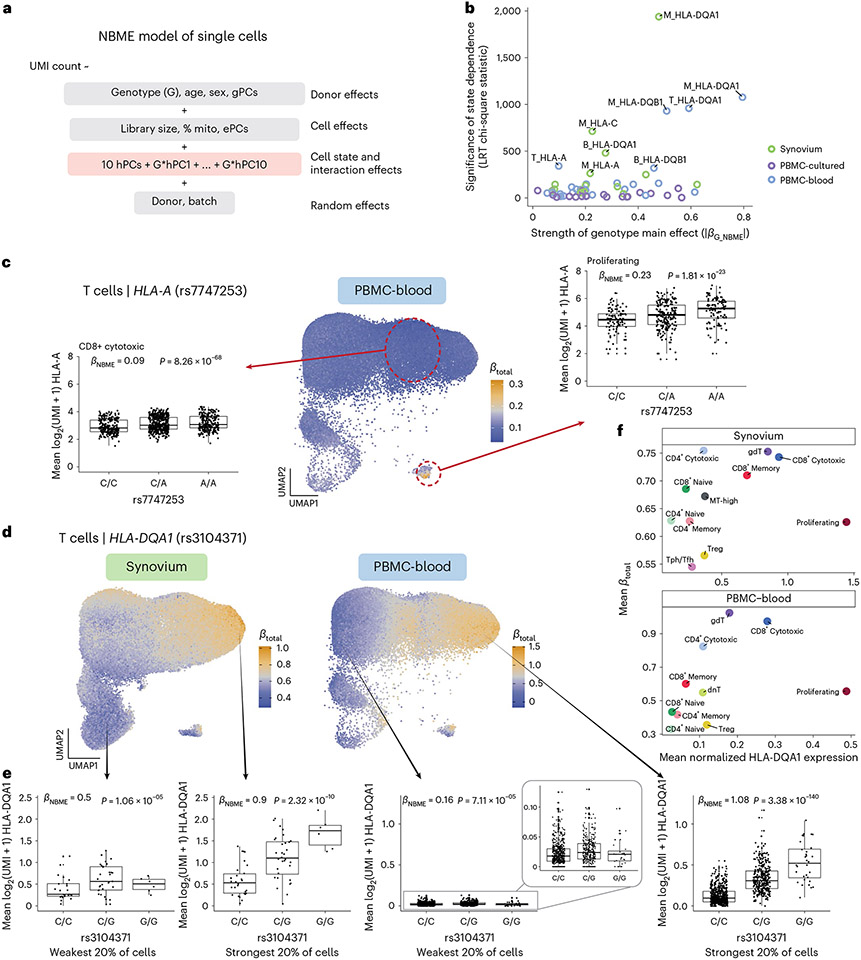
Fig. 1 ∣. Overview of study and scHLApers pipeline.
a, We used four datasets with genotype and scRNA-seq data: synovium (n = 69 individuals, m = 275,323 cells), intestine (n = 22, m = 137,321), PBMC-cultured (n = 73, m = 188,507), and PBMC-blood (n = 909, m = 765,079). b, Using the genotype data, we imputed SNPs within the MHC and one- and two-field classical HLA alleles. c, Schematic of scHLApers pipeline, where scRNA-seq reads are aligned to a personalized reference for each individual based on classical HLA alleles. In the example, an individual is heterozygous for all eight HLA genes, so 16 additional contigs are added to the reference. Original reference gene sequences are masked with Ns. scHLApers outputs a whole-transcriptome counts matrix with improved HLA gene estimates. Both alleles contribute to count estimation for each gene. d, We generated an atlas of HLA expression across all cell types (left) and mapped eQTLs for HLA genes in myeloid, B and T cells. Schematic of example dynamic eQTL (right), where eQTL strength (slope, ) changes across T cell states.
Results
Quantifying single-cell HLA expression with scHLApers
We developed scHLApers, a pipeline that accurately quantifies single-cell HLA expression using a personalized reference (Fig. 1c, Methods and Supplementary Note 1). First, scHLApers uses an individual’s unique classical HLA alleles (Fig. 1b) to add the personalized genomic sequences for each two-field allele from the Immuno Polymorphism Database-ImMunoGeneTics/HLA (IPD-IMGT/HLA) database27 to the standard reference genome in place of the original HLA gene sequences. scHLApers then uses STARsolo28 to quantify whole-transcriptome expression in single-cells with multimapping.
Four cohorts with genotype and scRNA-seq data
To study immune cell states from diverse tissues and biological conditions, including some from disease conditions, we used four scRNA-seq datasets with paired genotype data (Fig. 1a, Supplementary Table 1 and Supplementary Fig. 1). After quality control (QC) (Methods and Supplementary Table 2), the combined dataset of 1,073 individuals comprised synovial joint biopsies from an RA cohort29 (synovium, n = 69 individuals), intestinal biopsies from an ulcerative colitis (UC) cohort30 (intestine, n = 22), peripheral blood mononuclear cells (PBMCs) from healthy males cultured in vitro with influenza A virus and control conditions31 (PBMC-cultured, n = 73), and PBMCs from a large Australian cohort32 (PBMC-blood, n = 909).
Imputing HLA alleles and MHC variants
Using SNP2HLA with our group’s multi-ancestry HLA reference panel24,33,34 (Methods, Fig. 1b and Supplementary Fig. 2), we inferred a common set of 12,050 variants in the MHC with imputation dosage R2 > 0.8 and minor allele frequency (MAF) >1% in each cohort. These included 11,938 single nucleotide polymorphisms (SNPs) and 112 one- and two-field alleles for classical HLA genes (Fig. 2a and Supplementary Table 3). We used the two-field alleles to quantify expression with scHLApers (Fig. 1c), and we used both types of variation as input for downstream eQTL analysis (Fig. 1d).
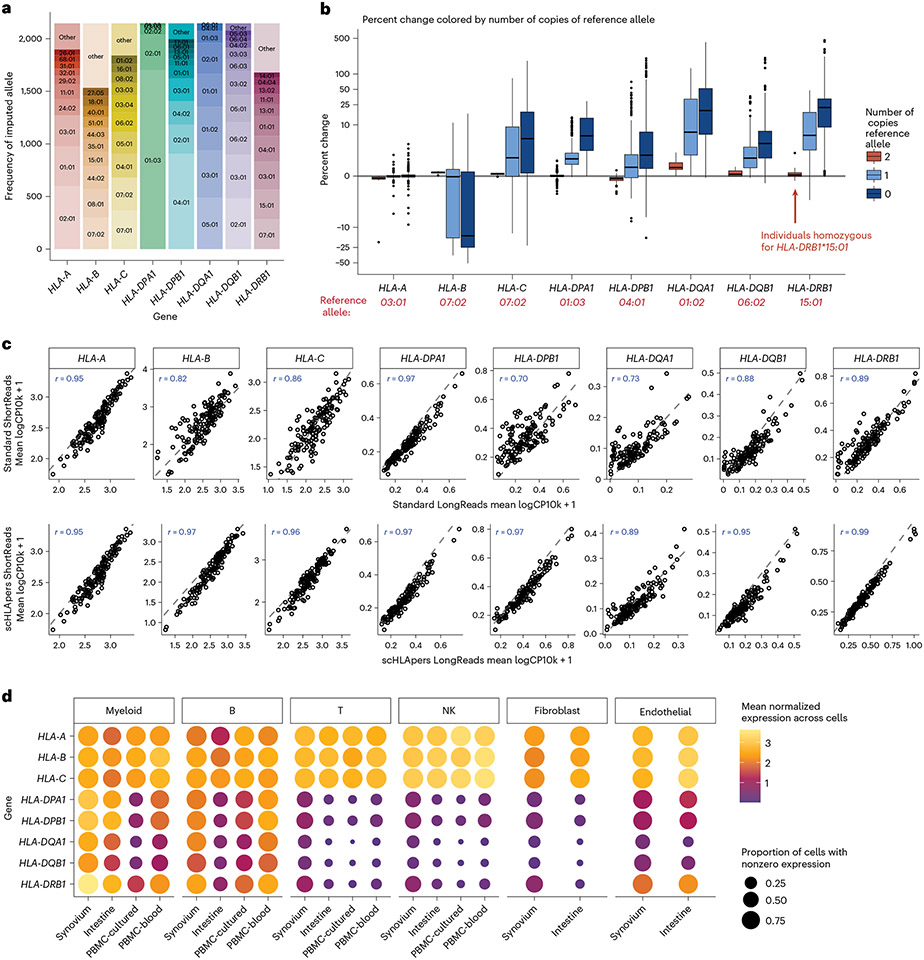
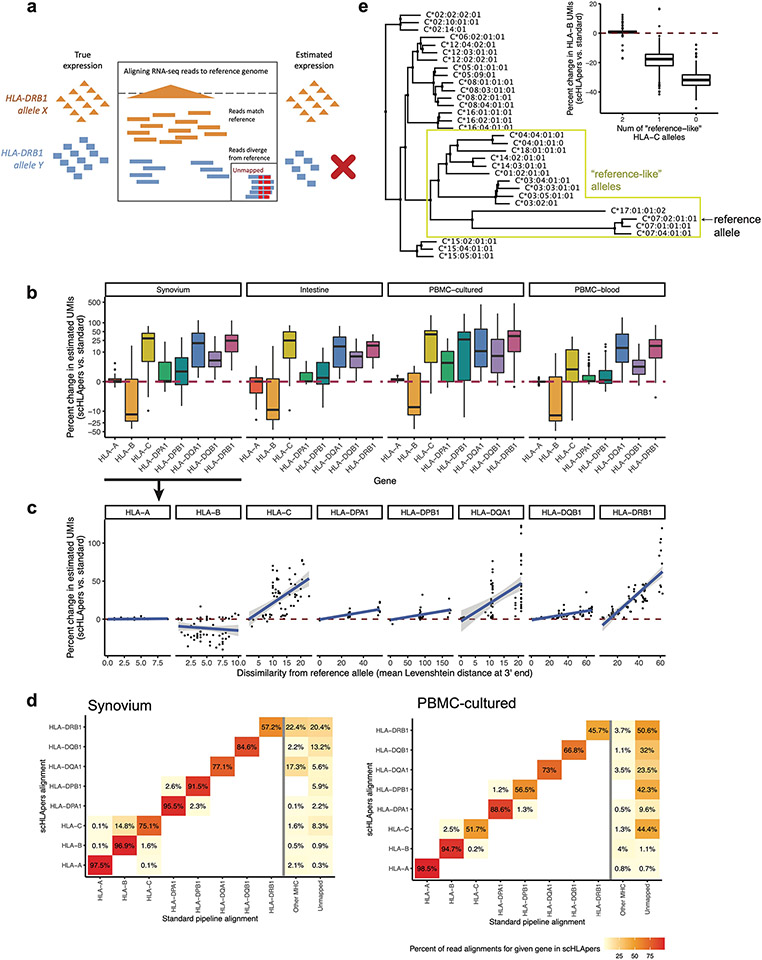
Fig. 2 ∣. Quantifying single-cell HLA expression using scHLApers.
a, Frequency of each imputed two-field HLA allele across all cohorts. Most common alleles (up to ten) labeled for each gene, with other alleles grouped into ‘other’. Alleles with frequency of less than five are not labeled. b, Boxplot (each observation is one sample) showing percentage change (y axis) in the estimated UMIs for each HLA gene (x axis) summed across all cells after quantification with scHLApers (compared to a pipeline using the standard reference genome) in the combined dataset (n = 1,073 individuals), colored by number of copies of the reference allele (labeled in red below for each gene). Gray horizontal line denotes no change. Boxplot center line represents median, lower/upper box limits represent 25/75% quantiles, whiskers extend to box limit ± 1.5× interquartile range, and outlying points are plotted individually. c, Comparing the estimated HLA expression as measured using shorter reads (84 bp, y axis) versus longer reads (289 bp, x axis) in the standard pipeline (top row) compared to scHLApers (bottom row). Each dot shows the mean log(CP10k + 1)-normalized expression across cells for one sample in PBMC-cultured (n = 146 samples from 73 individuals). r is Pearson correlation; dashed gray line is identity line. d, HLA expression in different cell types across cohorts: myeloid (m = 145,090 cells), B (m = 180,935), T (m = 805,389), NK (m = 125,865), fibroblasts (m = 82,651) and endothelial (m = 26,300). Dot size indicates proportion of cells with nonzero expression; color indicates log(CP10k + 1)-normalized expression (mean across cells).
Assessing the performance of scHLApers
We assessed the performance of scHLApers compared to a pipeline without personalization, that is, using the standard GRCh38 reference genome (Methods and Extended Data Fig. 1). We expected estimated HLA gene expression to generally increase with scHLApers since it rescues previously unmapped reads. For each individual, we calculated the percentage change in the total unique molecular identifier (UMI) count for each HLA gene across all cells after personalization. Personalization indeed generally led to higher estimated expression (Fig. 2b), with concordant trends across cohorts (Extended Data Fig. 1b). We reasoned that if scHLApers aligns reads more appropriately, then personalization should have larger effects for individuals whose alleles diverge more from reference genome alleles. Encouragingly, for individuals homozygous for the reference allele for a given gene (for example, HLA-DRB1*15:01), the scHLApers estimate highly coincided with the standard pipeline’s estimate; in contrast, greater dosage of non-reference alleles led to greater changes in estimated expression after personalization (Fig. 2b). To further quantify this, we compared the percentage change in estimated expression per individual to their alleles’ sequence dissimilarity to the reference (based on Levenshtein distance, Methods). For all genes except HLA-B, individuals with alleles more different from the reference tended to show a greater increase in expression after personalization (Extended Data Fig. 1c). The genes whose expression increased the most per individual were HLA-DRB1 (mean +29% change, 25th to 75th percentile (+10% to 38% change) in synovium), HLA-DQA1 (+29% (+3% to 44%)), HLA-C (+26% (+5% to 44%)), and HLA-DQB1 (+7% (+3% to 10%)), consistent with prior findings in bulk RNA-seq17. Expression of HLA-DPB1, HLA-DPA1 and HLA-A also increased but to a lesser extent (Supplementary Table 4). Unexpectedly, we observed an overall decrease in HLA-B counts across all cohorts (Extended Data Fig. 1b). After detailed investigation, we determined this was not a mishandling of reads by scHLApers, but rather was explained by scHLApers improving the assignments of reads from HLA-B to HLA-C (Supplementary Note 1). For individuals with both HLA-C alleles similar to the reference allele (HLA-C*07:02), HLA-B was less affected by personalization (Extended Data Fig. 1e). In contrast, for individuals with at least one non-reference-like HLA-C allele (that is, different from HLA-C*07:02), more reads aligning to HLA-B in the standard pipeline aligned better to HLA-C in scHLApers, leading to appropriately decreased HLA-B counts observed after personalization.
To assess if scHLApers improved the consistency of expression quantification, we leveraged the fact that each PBMC-cultured library was sequenced using two read lengths (289 bp and 84bp). We reasoned that a standard pipeline might lead to inconsistent quantification between the longer and shorter read versions of the dataset due to different types of mapping biases for different read lengths. In contrast, personalization should result in consistent quantification of each HLA gene between the two versions. Indeed, personalization increased the correlation between the estimated expression in shorter- and longer-read data for all genes across samples (Fig. 2c; HLA-B Spearman r = 0.97 scHLApers versus 0.82 standard; HLA-C r = 0.96 versus 0.86; HLA-DPB1 r = 0.97 versus 0.70). Together, our results demonstrate that aligning reads to a personalized reference improves precision in quantifying single-cell HLA expression.
While all four datasets were sequenced using 10x Genomics (10x) 3′ protocols, we also applied scHLApers to a separate dataset of synovium samples with matched 10x 5′ data (n = 9 individuals, 26,638 cells)35. We found that scHLApers led to a greater increase in HLA-A and HLA-B counts after personalization in 5′ data compared to 3′ data, due to increased dissimilarity from the reference allele on the 5′ end of the genes compared to the 3′ end (Supplementary Note 1, Supplementary Table 5 and Supplementary Fig. 3).
HLA gene expression across major cell types
After removing low-quality cells (Supplementary Table 2, Supplementary Note 2 and Supplementary Fig. 4a-c), we grouped cells from the four datasets into six major cell types (Methods and Supplementary Table 6) to investigate cell-type-specific HLA expression using scHLApers. These include four immune cell types from all cohorts: 145,090 myeloid cells (monocytes, macrophages and dendritic cells (DCs)), 180,935 B cells (including plasma cells), 805,389 T cells and 125,865 natural killer (NK) cells. It also includes stromal cells from the two solid tissue datasets: 82,651 fibroblasts and 26,300 endothelial cells. We examined HLA gene expression patterns across cell types. As expected, we found that all cell types highly express HLA class I genes across tissues, consistent with ubiquitous presentation of self-peptides, whereas class II expression varied (Fig. 2d). Specifically, myeloid cells and B cells expressed the highest levels of class II, consistent with their role as professional antigen-presenting cells. Interestingly, all other cell types, such as T cells, also express class II genes, albeit at lower levels. Human T cells have been previously observed to express HLA class II upon activation18,36-38, though its function is not well understood39-41.
Multi-cohort analysis identifies HLA regulatory variants
To identify eQTLs for classical HLA genes, we tested the 12,050 MHC-wide variants (Fig. 3a and Supplementary Table 7) for association with the expression of each HLA gene in myeloid, B and T cells. We chose these three cell types because they are well represented in all datasets and have known roles in antigen presentation (myeloid and B) or prior evidence for state-dependent HLA regulation (T)18. For each cell type and individual, we aggregated single-cell expression profiles into a single ‘pseudobulk’ measurement (Methods and Supplementary Fig. 4d,e). We used linear regression and analyzed all four cohorts together, controlling for covariates and testing 289,200 pairs of variants and HLA genes (Methods, Supplementary Fig. 5 and Supplementary Data 1).
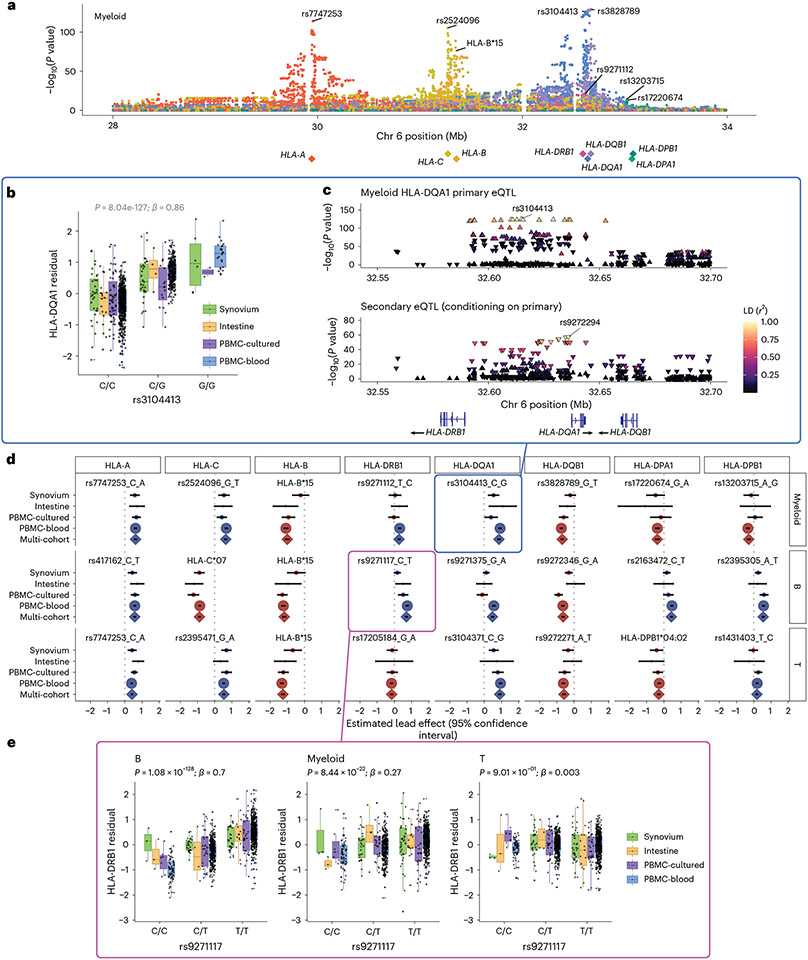
Fig. 3 ∣. eQTLs for classical HLA genes from pseudobulk analysis.
a, Manhattan plot showing the significance (y axis) of association between tested MHC variants (x axis) and expression of each HLA gene (color) in myeloid cells from the multi-cohort model. Most significant (lead) eQTLs are labeled. Diamonds indicate TSS of each gene. b, Boxplot showing an example lead eQTL (rs3104413). Increased dosage of the G allele (x axis) associates with higher HLA-DQA1 expression in myeloid cells (y axis: units are the residual of inverse normal transformed mean log(CP10k + 1)-normalized expression across cells after regressing out covariates), n = 1,025 individuals total (synovium n = 69, intestine n = 22, PBMC-cultured n = 73, PBMC-blood n = 861), plotted by dataset (color). All lead eQTLs shown in Supplementary Fig. 6c, Locus zoom plot for the primary (rs3104413) and secondary (rs9272294) eQTLs for HLA-DQA1 in myeloid cells. Significance of association (y axis) is shown for nearby variants on chromosome 6 (x axis); color denotes LD (r2 with lead eQTL in multi-ancestry HLA reference). Triangles point upwards for a positive (downwards for negative) effect on expression. Gene bodies and direction of transcription (arrows) for HLA-DRB1, HLA-DQA1 and HLA-DQB1 are underneath. d, Grid showing lead eQTLs for each HLA gene (columns) in each cell type (rows: myeloid cells n = 1,025, B cells n = 1,069, and T cells n = 1,072 individuals total; for dataset breakdown, see Supplementary Table 2). Each element of the grid includes a forest plot with the estimated lead effect size (x axis) and 95% confidence interval (mean ± 1.96 standard error) of the estimate from the multi-cohort analysis (diamond) and the same variant-gene pair tested for an association within each cohort separately (dots above). Size of the dots/diamond indicates cohort size; color indicates sign of the ALT allele’s effect on expression (blue for positive, red for negative). The eQTLs boxed in blue and magenta are highlighted in b and c and in e, respectively. e, Example of a cell-type-dependent eQTL (rs9271117) that was the lead eQTL for HLA-DRB1 and strongest in B cells. Boxplots are formatted analogously to b and show the eQTL’s effect for all three cell types separately. In a–c and e, nominal Wald P values are derived from linear regression (two-sided test).
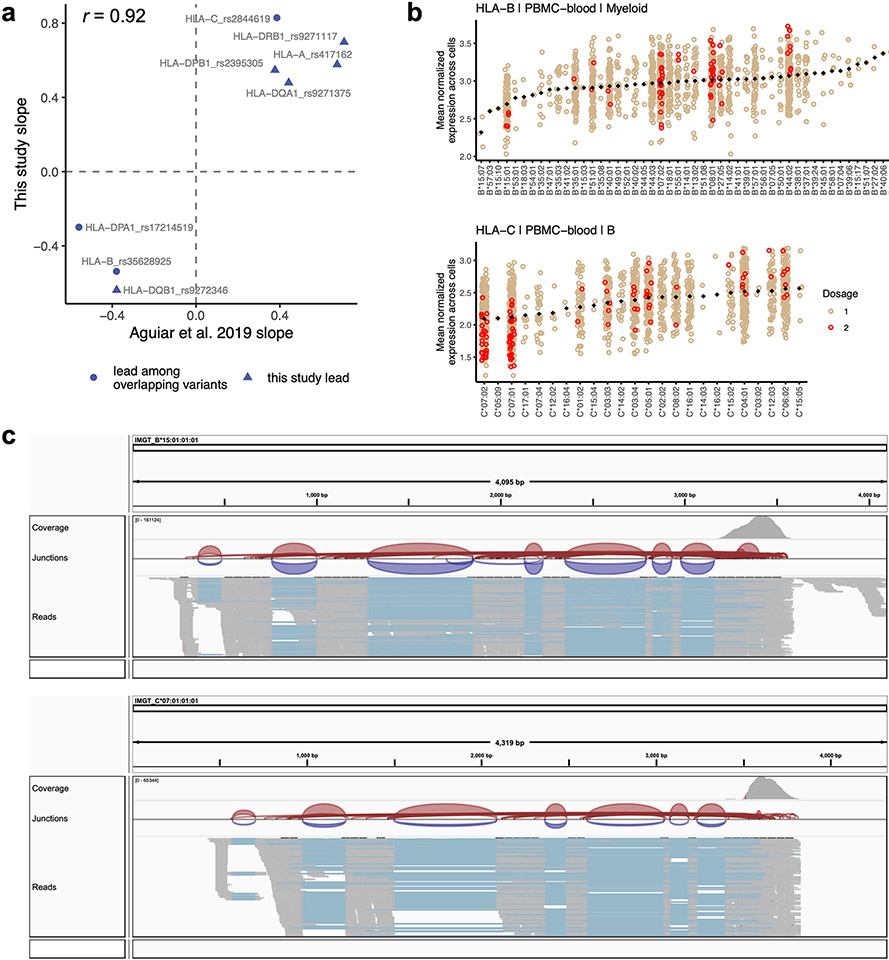
We detected an eQTL for every HLA gene in every cell type (P values <4 × 10−9; Fig. 3b-e, Supplementary Fig. 6 and Supplementary Table 8). Calculating the effect size of each lead eQTL in each cohort separately, we observed 91.7% (88/96) mean directional concordance across cohorts (Fig. 3d and Supplementary Table 9), suggesting consistent effects across datasets. The B cell results were highly concordant with a previous study on HLA eQTLs17, which used bulk RNA-seq data from lymphoblastoid cell lines and found that all eight variants included in both studies showed consistent directions of effect (Pearson r = 0.92, Extended Data Fig. 2a).
Most lead variants (19/24) were individual SNPs within the MHC. For example, rs3104413, the lead variant for HLA-DQA1 in myeloid cells, is located between HLA-DRB1 and HLA-DQA1 (P = 8.04 × 10−127; Fig. 3b,c). This SNP commonly co-occurs with the classical HLA-DQA1*03:01 allele (87.5% of DQA1*03:01 haplotypes are in phase with the G allele of rs3104413; Supplementary Table 10). The HLA-DQA1*03:01 allele is part of the DQ8 haplotype, which is associated with type 1 diabetes and celiac disease42.
Some lead eQTLs were individual one- or two-field HLA alleles. For example, HLA-B*15 was the lead eQTL for HLA-B in all three cell types (P < 3 × 10−81) and associated with lower expression of HLA-B (Extended Data Fig. 2b,c). A recent study using a new capture RNA-seq method also found that HLA-B*15 alleles were among the lowest expressed in bulk PBMCs, consistent with our observations43. HLA-C*07 was the most significant variant for HLA-C in B cells (P = 2.87 × 10−210; Supplementary Fig. 6b and Extended Data Fig. 2b), reflecting reduced expression of HLA-C*07 alleles relative to other HLA-C alleles. This finding could not be explained by read alignment bias (Extended Data Fig. 2c) and is supported by previous work showing that HLA-C*07 alleles contain a 3′ untranslated region microRNA binding site that reduces HLA-C expression44,45. Interestingly, the HLA-C*06:02 and HLA-C*12:03 alleles, major risk factors for psoriasis5, were associated with higher HLA-C expression in all three cell types (P < 8 × 10−40 and 3 × 10−8, respectively; Supplementary Data 1). The increased expression of these HLA-C alleles may contribute to psoriasis disease risk46.
scHLApers improves eQTL estimates
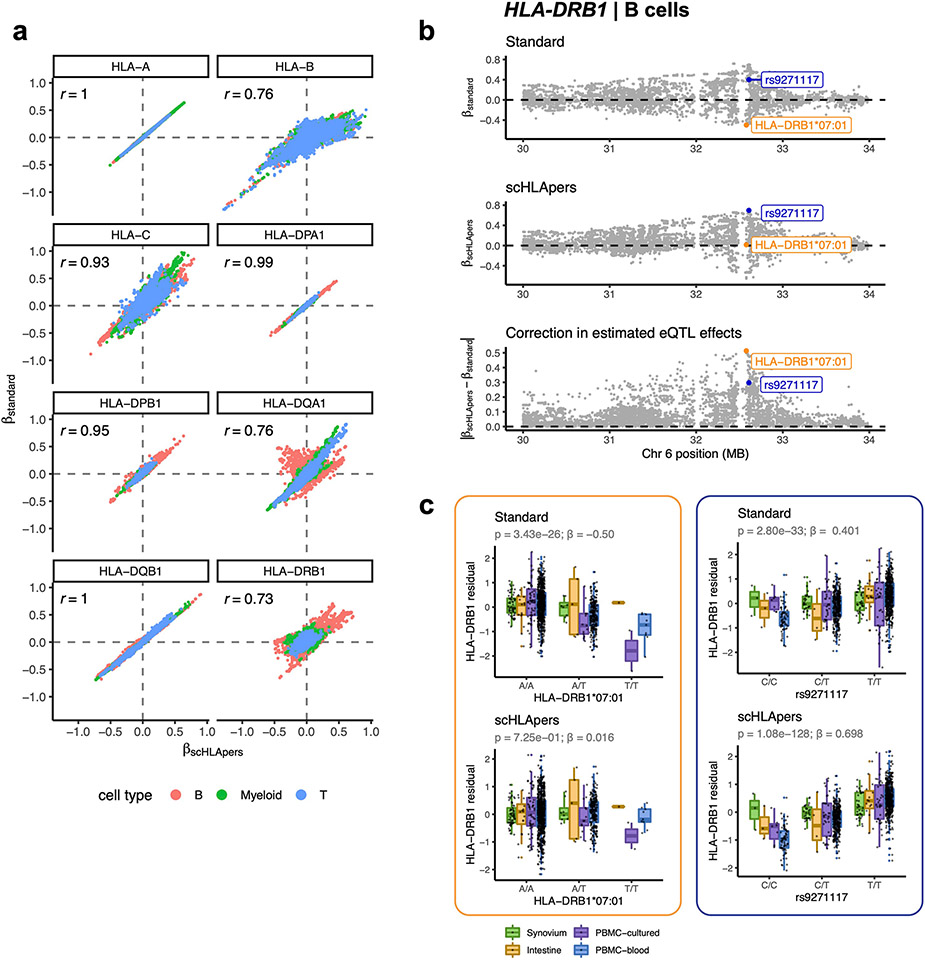
We compared the eQTL effect sizes estimated using expression values from scHLApers versus the standard pipeline. For genes whose expression were most affected by personalization, eQTL estimates were meaningfully impacted (Pearson r = 0.73 for HLA-DRB1, 0.76 for HLA-DQA1, 0.76 for HLA-B, 0.93 for HLA-C; Extended Data Fig. 3a). These improved eQTL estimates probably reflect the reduction of spurious eQTL signals caused by reference bias. For example, using the standard pipeline, the two-field allele HLA-DRB1*07:01 was significantly associated with HLA-DRB1 expression in B cells (β = −0.50, P = 3.43 × 10−26). However, with scHLApers, the effect was corrected away (β = 0.02, P = 0.73) (Extended Data Fig. 3b,c). In contrast, the lead HLA-DRB1 eQTL for scHLApers (rs9271117) was significant in both pipelines (Extended Data Fig. 3b,c).
HLA eQTLs are cell type dependent
We next explored whether HLA eQTLs are cell type dependent, as reported for other genes32,47. To test this, we used a mixed-effects model including an interaction term for cell type with genotype (Methods). Almost all (22/24) eQTLs exhibited statistically significant cell-type dependency (interaction P < 2.08 × 10−3 = 0.05/24 tests), and several showed dramatic effects (Supplementary Table 11). The strongest example was the lead eQTL for HLA-DRB1 in B cells (rs9271117, β = 0.7, P = 1.08 × 10−128), which was ~3-fold weaker in myeloid cells (β = 0.27, P = 8.44 × 10−22) and altogether absent in T cells (P = 0.90) (Fig. 3e). Similarly, eQTLs for HLA-DPA1 and HLA-DPB1 (rs2163472 and rs2395305) exhibited much stronger regulatory effects in B cells compared to myeloid and T cells (Supplementary Fig. 8a,b, β = 0.43 in B versus 0.04 and 0.08 in myeloid and T; β = 0.55 versus 0.07 and 0.12, respectively). These results highlight the importance of considering cell type when studying the genetic basis of HLA expression.
Conditional analysis identifies multiple eQTLs per gene
We used conditional analysis to identify additional regulatory variants beyond the primary eQTL (Supplementary Data 2). For example, after controlling for the effect of rs3104413, a secondary independent variant (rs9272294, linkage disequilibrium (LD) r2 = 0.04 with rs3104413) located ~1.4 kb upstream of HLA-DQA1 was also associated with HLA-DQA1 expression in myeloid cells (P = 3.06 × 10−58; Fig. 3c). We repeated this process to identify up to three additional independent eQTLs (P < 5 × 10−8) for each gene in each cell type (Supplementary Fig. 7). HLA-B, HLA-C and HLA-DQB1 exhibited the most independent signals (three or more eQTLs per cell type). Most associations (76% = 44/58) were unique to a gene and cell type (r2 < 0.8 with all other lead variants; Supplementary Fig. 8c), but some were shared. For example, the primary eQTLs for HLA-DPA1 and HLA-DPB1 in B cells (rs2163472 and rs2395305, respectively) were tightly linked to each other (r2 = 1.0) and to the secondary signal for HLA-DPB1 in T cells (rs4435981, r2 = 0.99). Additionally, the primary eQTLs for HLA-DQA1 in myeloid and T cells (rs3104413 and rs3104371) were linked (r2 = 0.86), and the secondary signals shared the same lead variant (rs9272294).
HLA genes exhibit cell-state-dependent expression
We next investigated whether HLA expression varies across cell states. Here, ‘cell state’ refers to finer-grained transcriptional phenotypes of cells within a major cell type. While there are multiple ways to represent cell state, we used harmonized expression principal components (hPCs) as latent variables capturing the main axes of transcriptional variation among the cells corrected for technical covariates. We integrated the single cells from all four datasets into a unified, continuous, low-dimensional embedding space for each cell type (myeloid, B or T) (Fig. 4a-c). This integration was accomplished by applying PC analysis to the two tissue datasets and removing batch and dataset-specific effects using Harmony48, then projecting the cells from the two PBMC datasets onto the same hPC axes using Symphony49 (Methods and Supplementary Fig. 9). The resulting hPC space appropriately captured transcriptional variation as reflected by the cell state annotations from the original studies (Fig. 4d and Supplementary Fig. 13), but does not rely on a specific clustering resolution.
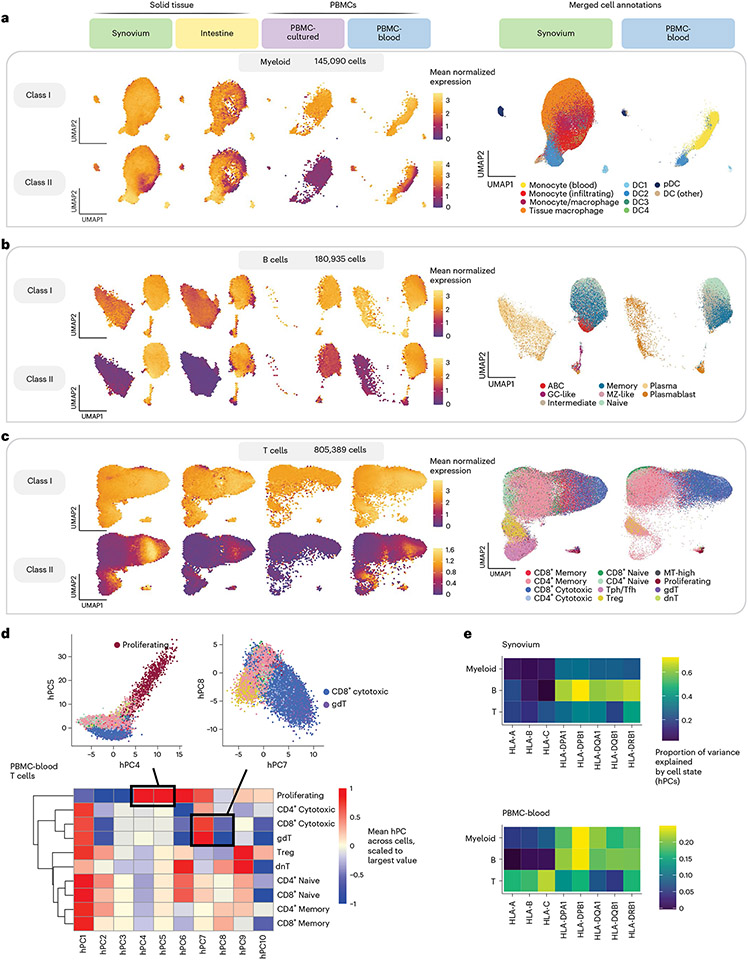
Fig. 4 ∣. Integrating single cells into a unified cell state embedding across datasets.
a–c, UMAP of cells generated using tissue-defined embedding (top ten hPCs from synovium and intestine), with PBMC datasets projected into the same space. The plot is divided into three sections: myeloid cells (a), B cells (b) and T cells (c). Left: class I and II HLA expression across cells across datasets. Cells are binned into hexagons to avoid overplotting (50 bins per horizontal and vertical UMAP directions) and colored by mean log(CP10k + 1)-normalized expression of class I/II genes per bin (for example, for class I, mean of HLA-A, HLA-B and HLA-C). Right: cell state annotations (color) for a representative PBMC (PBMC-blood) and solid tissue (synovium) dataset from merging annotations from each dataset to a shared set of labels. d, Heatmap showing mean value for each hPC (color) across cells for each discrete cell annotation within T cells in PBMC-blood. Values are scaled relative to the most extreme value across cell states. Black boxes and inset figures above show examples of how hPCs are linked to original cell state labels: proliferating cells (high in hPC4 and hPC5) and CD8+ cytotoxic and γδ (gdT) cells (high in hPC7, low in hPC8). e, Estimated proportion of variance in UMIs explained by cell state hPCs (color) across HLA genes and cell types.
The shared single-cell embedding allowed us to compare HLA expression patterns across fine-grained transcriptional states. Both class I and II expression varied widely across cell states within a given cell type (Fig. 4a-c and Supplementary Figs. 10-12). By quantifying the variance explained by cell state for each gene (Methods), we found that cell state generally explained a greater proportion of variance in class II expression (mean 30%, 25th to 75th percentile (17–37%) across all cohorts) compared to class I (mean 19% (8–34%)) (Fig. 4e and Supplementary Table 12). The abundance of certain cell states differed considerably between blood and tissues. For example, tissue macrophages and infiltrating monocytes were absent or at low abundance in PBMCs. However, HLA expression patterns were generally similar in cell states shared across tissues, suggesting that cell state rather than tissue context was driving expression. For example, conventional DC1 and DC2 cells expressed the highest levels of class II among myeloid cells in both blood and tissue (Fig. 4a). Among B cells (Fig. 4b), class II expression was lower in plasma cells than in B cells, reflecting the downregulation of class II in the transition to plasma cells50,51. Among T cells, proliferating and CD8+ cytotoxic cells expressed the highest levels of class II (Fig. 4c).
Modeling dynamic eQTLs at single-cell resolution
Single-cell-resolution eQTL models19,20,52,53, which model expression in individual cells, can identify dynamic eQTLs–regulatory effects that change as cells transition across continuous cell states. Dynamic effects can be masked in pseudobulk analysis and may reflect cell-state-specific transcription factors binding to specific regulatory elements.
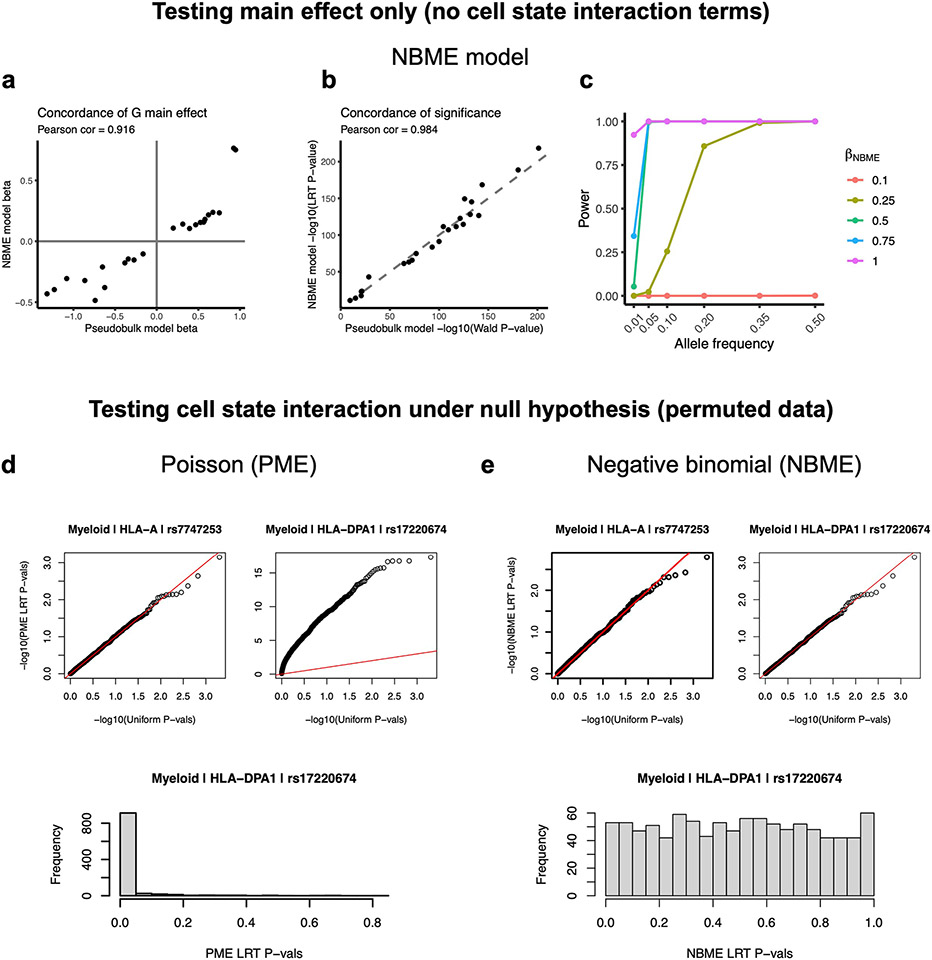
To investigate whether HLA eQTLs are dynamic, we used a single-cell negative binomial mixed-effects (NBME) model (Methods). Briefly, we modeled the UMI count of each gene as a function of genotype and its interaction with cell state, accounting for sample-level covariates (age, sex and ancestry), cell-level fixed effects (library size, percentage mitochondrial UMIs, and expression principal components (PCs)), and random effects for donor and batch (Fig. 5a). The NBME model showed high concordance with the pseudobulk model when testing for eQTL main effect size and significance (Pearson r = 0.916 for effect, 0.984 for significance; Extended Data Fig. 4a,b). By simulating single-cell datasets across a range of allele frequencies with different eQTL effect sizes (Methods), we determined that the NBME model has adequate power to detect eQTLs for our application (Extended Data Fig. 4c). We then used the top ten hPCs for each major cell type (Methods) as a continuous multivariate representation of cell state when modeling eQTLs and tested for cell-state interactions (G × hPC) within each dataset using the same cell-state definitions across datasets. We tested the lead eQTLs identified by our pseudobulk analysis, comprising 58 variant-gene pairs with robust genotype main effects and excluding the Intestine dataset due to its small sample size (Methods). We confirmed that the model has well calibrated type I error when testing for cell-state interactions (Extended Data Fig. 4d,e).
Fig. 5 ∣. Identifying dynamic eQTLs by modeling single cells.
a, NBME model of single cells used to identify cell-state-dependent regulatory effects. Pink box highlights terms for cell state (ten hPCs per cell type) and their interaction with genotype. b, Testing lead eQTLs identified in multi-cohort pseudobulk analysis for cell-state dependence using the NBME model in each dataset (color) in myeloid (‘M’), B and T cells. Magnitude of genotype main effect (x axis) versus the significance of cell-state interaction (y axis), measured using chi-square (χ2) statistic from LRT comparing full model (a) to null model without G × hPC interactions. c, Dynamic HLA-A eQTL (rs7747253) in T cells (n = 909 individuals, m = 538,579 cells in PBMC-blood). UMAP shows T cells colored by estimated eQTL strength . Boxplots for the eQTL effect are shown for two annotated cell states (CD8+ cytotoxic and proliferating, outlined in red circles), showing mean log2(UMI + 1) of HLA-A across all cells in the cell state per individual by genotype. and P values are derived from fitting the NBME model without cell-state interaction terms on the discrete cell populations and comparing to a null model without genotype using an LRT (n = 908 individuals, m = 96,516 cells for CD8+ cytotoxic; n = 409, m = 739 for proliferating). Boxplot center line represents median, lower/upper box limits represent 25/75% quantiles, whiskers extend to box limit ± 1.5× interquartile range, and outlying points are plotted individually. d–f, Dynamic HLA-DQA1 eQTL (rs3104371) in T cells (n = 68 individuals, m = 82,423 cells in synovium; n = 909, m = 538,579 in PBMC-blood). UMAP (d) colored by eQTL strength , from blue (weakest) to orange (strongest). Boxplots (e) showing the eQTL effects in cells from the top and bottom quintile of , showing mean log2(UMI + 1) per individual (y axis) by genotype. Labeled and P value are derived from fitting the NBME model without cell-state interaction terms on the cells from the discrete quintile and comparing to a null model without genotype using an LRT. Boxplot elements defined as in c. Scatterplot (f) showing the mean (y axis) compared to the mean log(CP10k + 1)-normalized expression of HLA-DQA1 (x axis) across annotated cell states (color). LRT, likelihood ratio test (one-sided).
We observed that most eQTLs (78% = 45/58) showed statistically significant cell-state dependence (interaction P < 8.6 × 10−4 = 0.05/58 tests; Supplementary Table 13). Indeed, every HLA gene tested was dynamic in at least one cell type, and HLA-DQA1, HLA-DQB1, HLA-C and HLA-A were the most state dependent (Supplementary Table 14). Most interaction effects were modest relative to the main genotype effect (Supplementary Table 13). Interestingly, the PBMC-cultured dataset exhibited much less significant cell-state interactions overall (Fig. 5b), despite being similar in size to the synovium dataset. This is possibly due to cell state differences in cultured cells compared to cells collected in vivo.
Comparing dynamic effects across cell states
We next assessed the strength of dynamic regulatory effects in relation to annotated cell states. For each eQTL, we calculated each cell’s estimated total eQTL effect size from the genotype main effect and interaction effects weighted by the cell’s position along each hPC (Methods)19. This allowed us to compare the eQTL’s strength across cell states. For example, in PBMC-blood T cells, the effect of the HLA-A eQTL (rs7747253, interaction P = 4.9 × 10−68) was strongest in proliferating cells (mean for proliferating versus 0.10 for other T cells; Fig. 5c), suggesting the variant plays a more substantial role in regulating HLA-A expression during T cell proliferation than at rest. This eQTL was also cell state dependent in myeloid cells (Supplementary Fig. 14a-d).
We explored whether cell-state-interacting eQTLs may contribute to interactions with contextual factors that have been tested in bulk-level analyses47,54-56, including age, sex and interferon response. Our findings indicate that if an eQTL interacts with cell states whose abundance changes with a sample-level factor, the factor can show an interaction in bulk; however, single-cell interaction testing is better powered (Supplementary Note 3, Supplementary Table 15 and Supplementary Fig. 15).
We observed the most significant cell-state interaction effects for HLA-DQ genes (Fig. 5b), specifically HLA-DQA1 in T cells (interaction P = 2.9 × 10−200 in PBMC-blood) and HLA-DQA1 and HLA-DQB1 in myeloid cells (interaction P < 1× 10−195 in both synovium and PBMC-blood). In T cells (Fig. 5d-f), the HLA-DQA1 eQTL (rs3104371) had the strongest effects in gamma-delta (γδ), cytotoxic CD8+ and cytotoxic CD4+ T cells, a finding that replicated in synovium (Fig. 5f). All three of these cell states exhibit cytotoxic activity. Our results indicate that HLA-DQA1 expression is under dynamic genetic regulation in T cells, and further studies to clarify its functional role are warranted.
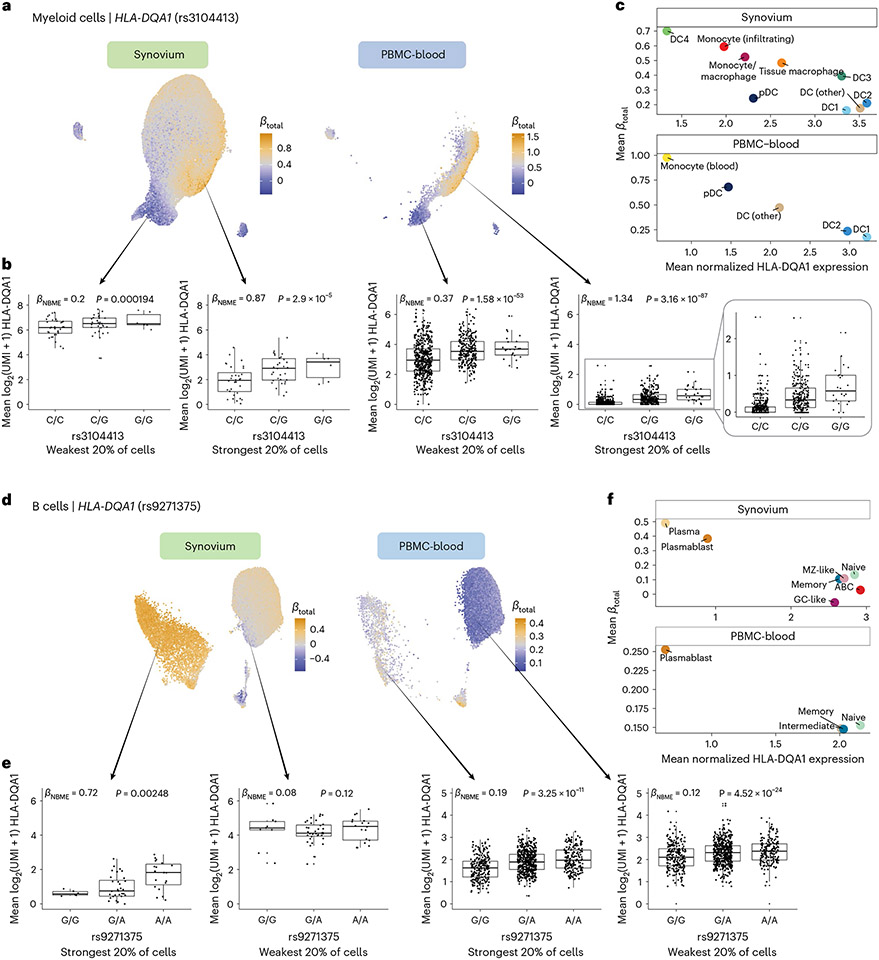
In myeloid cells, PBMC-blood and synovium showed similar patterns of regulation for the HLA-DQA1 eQTL (rs3104413; Fig. 6a-c). The strongest effects were observed in a subpopulation of monocytes in PBMC-blood and infiltrating monocytes and DC4 cells (which are similar to CD16+ monocytes57) in synovium (Fig. 6c), suggesting that the underlying regulatory mechanisms governing the dynamic eQTL are active in both blood and synovium. The estimated values were robust to whether the embedding was defined using the tissue datasets or PBMC-blood dataset alone (Pearson r across cells, 0.896; Supplementary Fig. 14e-g). In contrast to the T cell HLA-DQA1 example, the eQTL strength was negatively correlated with the expression of the gene. That is, the expression of HLA-DQA1 is highest in conventional DC1 and DC2 cells, but the eQTL is weakest in those states (Fig. 6c). HLA-DQB1 also showed similar patterns of eQTL strength as HLA-DQA1 in PBMC-blood (r across cells, 0.953), suggesting that HLA-DQ genes are coordinately regulated.
Fig. 6 ∣. Dynamic HLA-DQ eQTLs in myeloid and B cells.
a–c, Dynamic HLA-DQA1 eQTL (rs3104413) in myeloid cells (n = 69 individuals, m = 66,789 cells in synovium; n = 861, m = 40,568 in PBMC-blood). UMAP (a) of cells for tissue-defined embedding, colored by , from blue (weakest) to orange (strongest). Boxplot (b) showing the eQTL effect across individuals in the bottom and top quintiles of estimated . Labeled and P value are from fitting the NBME model without cell-state interaction terms on the cells from the discrete quintile and comparing to a null model without genotype using an LRT. Mean log2(UMI + 1) across cells per individual (y axis) by each genotype. Boxplot center line represents median, lower/upper box limits represent 25/75% quantiles, whiskers extend to box limit ± 1.5× interquartile range, and outlying points are plotted individually. Scatterplot (c) showing the mean estimated (y axis) compared to the mean log(CP10k + 1)-normalized expression of HLA-DQA1 (x axis) across annotated cell states (color). d–f, Dynamic HLA-DQA1 eQTL in B cells (n = 65 individuals, m = 25,917 cells in synovium; n = 909 individuals, m = 80,784 in PBMC-blood). d–f are analogous to a–c, respectively. LRT, likelihood ratio test (one-sided).
In B cells, the HLA-DQA1 and HLA-DQB1 eQTLs (rs9271375 and rs927346) were also state dependent (interaction P < 2× 10−9 in synovium and PBMC-blood), with plasma cells and plasmablasts exhibiting the strongest effects (Fig. 6d-f). Interestingly, the overall trend in B cells was similar to myeloid cells (and opposite of T cells) in that cell states with higher HLA-DQ expression (pre-activated B cells and conventional DCs, respectively) had weaker eQTL effects. In contrast, states with lower expression (plasma cells and monocytes) had stronger effects. A potential explanation is that cells critical for antigen presentation, such as DCs and pre-activated B cells58,59, have mechanisms to maintain high HLA-DQ expression to ensure proper function, such that genetic effects contribute less to expression differences. Meanwhile, cell states with lower expression may have evolved greater genetic diversity in their antigen presentation capabilities, leading to diversity in immune responses across individuals.
Discussion
This study demonstrates highly variable cell-type and cell-state-specific expression and genetic regulation of HLA genes. By integrating four diverse datasets from multiple tissues capturing a broad set of cell states and contexts, we found that classical HLA gene expression is under cis-regulation. Class II genes show particularly variable strengths of genetic regulation depending on cellular context. At the cell-type level, B cells display much stronger regulatory effects for HLA-DRB1, HLA-DPA1 and HLA-DPB1 than myeloid and T cells (Fig. 3e and Supplementary Fig. 8a,b). Single-cell resolution eQTL modeling revealed that many eQTLs are cell state dependent, especially for HLA-DQ genes (Figs. 5 and 6). We previously showed that HLA-DQ exhibits state-dependent regulation in CD4+ T cells ex vivo18. Here, we demonstrated that HLA-DQ is dynamically regulated in multiple cell types across tissues in vivo.
Variation in the HLA is hypothesized to have evolved to confer selective advantages in immune response to pathogens60, maternal–fetal tolerance61 and susceptibility to autoimmune diseases62, depending on environmental contexts. Coding variation in HLA genes affects the quality of presented antigens by determining which peptide sequences are presented, and population diversity enables collective responsiveness to diverse pathogens. Concurrently, HLA regulatory variation may affect the quantity of antigen presentation, leading to different thresholds of immune responsiveness. It has been shown that the expression levels of HLA-C alleles can affect immunogenicity in unrelated donor hematopoietic cell transplantation63, and HLA downregulation in tumors may affect response to immune checkpoint inhibitors64,65. The presence of multiple independent regulatory effects at each HLA gene and cell-type and cell-state-specific effects suggests that regulatory variation may have been selected to ensure diverse immune responses within a population.
There are several limitations of this study. First, our reference-based HLA imputation may have missed ultra-rare alleles. Long-read sequencing or sequence-based typing with polymerase chain reaction could eventually improve the detection of all possible noncoding HLA variants66,67. Second, we were not able to fine-map the eQTLs to precise causal variants because of the high degree of linkage disequilibrium (LD) in the MHC region. Functional work evaluating candidate causal variation may ultimately define causal variation. Finally, we did not perform colocalization with genome-wide association study associations for several reasons. Standard tools (for example, coloc68) that assume a single causal variant are not appropriate within the HLA locus because genome-wide association study signal may jointly arise from both coding and regulatory variation, rather than acting exclusively through gene expression. Moreover, although colocalization can be paired with conditional analyses or fine-mapping approaches69 to test multiple independent effects in a region, the extensive LD poses a challenge. Colocalization analyses within the HLA have not been systematically evaluated for accuracy and replication and warrant future investigation.
Future data generation efforts that increase the size and ancestral diversity of genotyped single-cell cohorts will continue to improve our understanding of state-dependent and population-specific regulatory effects and aid in fine-mapping efforts70.
Methods
Quantifying single-cell HLA expression with scHLApers
We developed the scHLApers (single-cell HLA expression using a personalized reference) pipeline to accurately quantify classical HLA expression in scRNA-seq data. As input, the pipeline takes in scRNA-seq read-level data (FASTQ or BAM) and HLA allele calls. If sequence-based typing is unavailable, HLA alleles can be imputed using genotyping data (see ‘HLA imputation’ section). A personalized reference is created for each individual by adding personalized HLA allele sequences as extra contigs to the reference and masking the original reference HLA gene sequences. The output is a whole-transcriptome counts matrix with improved HLA expression estimates. The code and tutorials to run scHLApers are available at ref. 71 (v1.0 used for this study).
Preparing the HLA allelic sequence database.
scHLApers requires a database of genomic HLA allele sequences. To prepare this, we downloaded the IPD-IMGT/HLA database72 (v3.47.0). The database contains sequence alignment files for full-length genomic sequences (that is, four-field resolution, ending in ‘gen.txt’) and nucleotide coding sequences (that is, two- and three-field resolution, ending in ‘nuc.txt’). We filled in any incomplete genomic sequences with bases from the most similar complete allele using the hla_compile_index function from the ‘hlaseqlib’ R package (v0.0.3)73. Coding allele sequences with no corresponding genomic sequence were substituted with the genomic sequence of the most similar allele with a genomic sequence based on the Hamming distance of coding sequences. For HLA-A, HLA-DQA1, HLA-DQB1, HLA-DPA1 and HLA-DPB1, we padded the 5′ and 3′ ends of the allelic sequences from IPD-IMGT/HLA with extra bases from the GRCh38 reference to ensure that they did not have any missing sequence content compared to the reference sequences. The reference gene boundaries were defined by the Gencode v38 annotation file.
Creating personalized reference genome and annotation files.
scHLApers creates a personalized reference genome (FASTA) and annotation file (GTF) for each individual. Based on the HLA allele calls, scHLApers creates a FASTA file for each individual with their genomic allelic sequences from the allelic sequence database. Each allele is included as a separate contig, with the allele name as the identifier. If multiple four-field versions exist for a given two-field allele, the corresponding XX:XX:01:01 allele sequence is chosen. The original reference classical HLA gene sequences are masked with ‘NNN…’ to prevent reads from aligning to them. The personalized allelic sequences are then concatenated with the masked GRCh38 reference genome to produce the personalized reference.
In the personalized annotation file (GTF), all entries corresponding to the classical HLA genes are removed from the original Gencode v38 annotation file. New entries are added for each personalized allele with the ‘seqname’ column labeled as the allele name (matching the identifier in the personalized reference FASTA file), the ‘feature name’ as ‘exon’ to enable read alignments to the entire sequence, the ‘start’ and ‘end’ positions as ‘1’ and the length of the sequence, respectively, and the strand as ‘+’ since all sequences in the database are defined as the forward strand. The ‘attribute’ column is labeled with ‘transcript_id’ as the allele name (for example, IMGT_A*01:01:01:01) and ‘gene_id’ and ‘gene_name’ as the gene name (for example, IMGT_A), allowing alignments to either allele of the gene to contribute to its total UMI count.
Quantifying single-cell expression.
Using the personalized genome and annotations, scHLApers performs single-cell read alignment and expression quantification using STARsolo28 (v2.7.10a). STARsolo performs barcode correction, UMI collapsing and optimal distribution of multimapping reads (that is, reads mapping to either overlapping genes or multiple paralogous genes at separate loci), which are typically discarded in standard pipelines. We chose STARsolo over pseudoalignment-to-transcriptome methods because it can identify splice junctions de novo, which is useful because the transcript isoform usage for each HLA allele is not readily available. The personalized genome index is generated using STARsolo –runMode genomeGenerate, and read alignment is performed with –runMode alignReads. The user specifies the appropriate UMI length (–soloUMIlen), cell barcode whitelist file (–soloCBwhitelist), and assay type (–soloType CB_UMI_Simple for droplet-based data). Additionally, scHLApers counts all reads overlapping gene’s introns and exons (–soloFeatures GeneFull_Ex50pAS) and optimally distributes multimapping reads using an expectation-maximization algorithm (–soloMultiMappers EM). The parameters –soloCBmatchWLtype 1MM_Nbase_pseudocounts, –soloUMIfiltering MultiGeneUMI_CR and –soloUMIdedup 1MM_CR are used to match CellRanger results. Users can output a coordinate-sorted BAM file to view individual read alignments (–outSAMtype BAM SortedByCoordinate and –outSAMunmapped Within).
Cohorts with paired single-cell transcriptomics and genotype data
We obtained data from four existing studies with scRNA-seq and genotype data from the same individuals (Supplementary Table 1). These include (1) synovial biopsies from patients with RA and from osteoarthritis controls (synovium, n = 69 individuals after sample QC)29, (2) intestinal biopsies from patients with UC and from healthy controls (intestine, n = 22)30, (3) PBMCs from healthy males that were treated in vitro with both influenza A virus and mock conditions (PBMC-cultured, n = 73)31, and (4) PBMCs collected from a large population cohort (PBMC-blood, n = 909)32. For details regarding the collection of these cohorts and determination of the number of samples per cohort included in this study, see Supplementary Note 4.
QC of genotyping data
All cohorts were genotyped using genotyping arrays, except for PBMC-cultured, which used low-pass whole-genome sequencing (WGS) (Supplementary Table 1). We processed the genotyping data and performed QC using PLINK v1.90, as described in Supplementary Note 4 following the tutorial at ref. 74 (ref. 24). Genome-wide variants were used to calculate PCs to control for genetic ancestry in eQTL analysis, and variants in the extended MHC (defined here as chr 6: 28000000–34000000) were used for HLA imputation.
HLA imputation
HLA imputation with SNP2HLA.
We used SNP2HLA75 to perform HLA imputation using version 2 of our group’s multi-ethnic reference panel described in Sakaue et al.24,33,34. We performed imputation on the full genotyping datasets (that is, not limited to samples with paired scRNA-seq), then subset the imputed VCF file to the samples with scRNA-seq. Two types of genetic variation were imputed: SNPs within the MHC (n = 14,691) and classical HLA alleles at one- and two-field resolution for HLA-A, HLA-B, HLA-C, HLA-DPA1, HLA-DPB1, HLA-DQA1, HLA-DQB1 and HLA-DRB1 (n = 570). In SNP2HLA output, reference (REF) and alternative (ALT) values for classical alleles are set to ‘A’ and ‘T’ denoting absence and presence of the allele, respectively.
The two-field HLA alleles were used in scHLApers to make personalized references. SNP2HLA outputs an individual’s imputed dosage (0–2) and inferred genotype (GT: 0∣0, 0∣1, 1∣0 or 1∣1) for every HLA allele in the reference panel. Note that, for a subset of individuals, we could not confidently call two-field alleles for one or more HLA genes, and the dosage was split across multiple alleles (<0.5 for any given allele). We excluded these individuals (9 synovium, 3 intestine, 15 PBMC-cultured and 60 PBMC-blood individuals, representing <8% of total samples) to avoid introducing a technical batch effect. All downstream analyses included 1,073 individuals for whom we could confidently impute phased alleles for every HLA gene (GT: 0∣1 and 1∣0 for two alleles or GT: 1∣1 for one allele).
QC of imputed MHC variants.
We performed QC on the imputed MHC-wide variants using custom R scripts and the ’vcfR’ (v1.12.0) package. Because the HLA reference uses hg19 coordinates, we first lifted over the imputed variants to GRCh38 using CrossMap (v0.6.1) and chain file76. Then, we subset to the relevant samples and calculated the MAF within the subset. We retained variants with imputation dosage R2 (DR2, the estimated squared correlation between the estimated allele dose and the true allele dose) >0.8 and MAF >0.01 in each cohort. For the intestine cohort, which was genotyped on two different arrays, we first filtered by DR2 within each array then merged them by the intersecting variants before filtering by MAF >0.01 across the merged cohort. We took the intersection of variants across all four cohorts passing our QC thresholds to arrive at a final set of 12,050 variants for eQTL testing (Supplementary Fig. 2): 112 one- and two-field HLA alleles and 11,938 intergenic variants.
Assessing the performance of scHLApers
Applying scHLApers to all four cohorts.
We applied scHLApers to quantify single-cell expression for all four datasets. As a comparison, we also ran a standard pipeline that used STARsolo with the same parameters as scHLApers but with the original GRCh38 reference (with no personalization) and discarding multimapping reads. For both versions, we generated BAM files containing unmapped reads (samtools view -b -f 4) and reads aligning to the MHC and personalized contigs using samtools (v1.4.1). We removed empty droplets and low-quality cells by filtering the count matrices by cell barcodes (see ‘Processing single-cell expression data’ section).
For the read length concordance analysis, the PBMC-cultured dataset contained reads of two different lengths (84 and 289 bp). We generated long- and short-read versions of the dataset by creating separate BAM files by sequence length and running scHLApers on longer and shorter reads separately. To visually inspect read alignments and coverage across the personalized allelic contigs in scHLApers, we used Integrative Genomics Viewer (IGV v2.11.2).
Comparing percent change to dissimilarity from the reference alleles.
We assessed how expression estimates (summed UMI counts across all cells for a sample) changed from a standard pipeline (sp_exp) to the scHLApers pipeline (pers_exp) (equation (1)) with respect to the dissimilarity between the reference allele and personalized alleles.
| (1) |
Dissimilarity was defined as the Levenshtein distance between the genomic GRCh38 allele and personalized allele sequences, calculated using the stringdist function in the ‘stringdist’ (v0.9.8) R package. Since all datasets used 10x 3′ assays, the read coverage was predominantly at the 3′ end of the gene (Supplementary Fig. 3). Hence, distances were calculated at the 3′ end using sequence segments of 500 bp (HLA-A, HLA-B, HLA-C and HLA-DRB1), 1,000 bp (HLA-DQA1 and HLA-DPA1), 1,500 bp (HLA-DQB1) or 2,500 bp (HLA-DPB1), encompassing the region where reads accumulated. For individuals heterozygous for a gene, we took the mean of the two distances. The GRCh38 reference allele sequences are listed in Darby et al.77 (A*03:01, B*07:02, C*07:02, DQA1*01:02, DQB1*06:02, DRB1*15:01, DPA1*01:03 and DPB1*04:01). We confirmed these by performing a multiple sequence alignment between the IPD-IMGT/HLA allelic sequences and the reference sequence using the msaClustalW function from the ‘msa’ (v1.22.0) R package.
Application of scHLApers to 5′-based data.
We applied scHLApers to a separate dataset from a subset of synovium individuals with matching 10x 5′ data (n = 9 individuals, 26,638 cells)35. To compare the dissimilarity of HLA class I alleles to the reference alleles at the 5′ end (500-bp region), we calculated Levenshtein distance at the 5′ end of the multiple sequence alignment, as described for 3′ data above.
Investigating read mapping between HLA-B and HLA-C.
To quantify the rescuing of unmapped reads and identify reads ‘jumping’ between different genes, we tracked where reads aligned in scHLApers versus the standard pipeline. We analyzed the BAM files output from both pipelines using a custom R script and the scanbam function in ‘Rsamtools’ (v2.6.0). A given read can align to the classical HLA genes (that is, personalized contigs for scHLApers or gene regions defined by Gencode v38 for the standard pipeline), another location in the MHC outside of classical HLA genes, another location outside of the MHC, or be unmapped. We used the multiple sequence alignment for HLA-C to generate a phylogenetic tree of HLA-C allele sequences using the ‘Neighbor Joining’ option in Jalview (v2.11.0). By grouping the HLA-C alleles by similarity to the reference allele (C*07:02) based on the tree, we could observe the relationship between the dosage of ‘reference-like’ HLA-C alleles and the change in HLA-B counts.
Processing single-cell expression data
QC of single-cell data.
For synovium, intestine and PBMC-cultured datasets, we subset the count matrix output from scHLApers to the cells passing QC in the original studies (that is, barcodes present in published cell metadata). For the PBMC-blood dataset, we started from the original cells but performed additional filtering steps to remove suspected doublets (Supplementary Note 2). Then, we performed uniform cell-level QC procedures on all cohorts, removing cells with <500 genes and >20% mitochondrial counts.
Defining major cell types and merged cell annotations.
We defined a common set of six major cell types across the four datasets–myeloid (monocytes, macrophages and DCs), B (including plasma), T, NK, fibroblast and endothelial–by aggregating fine-grained cell annotations. For synovium, intestine and PBMC-cultured, these fine-grained annotations came from the originally published cell annotations. For PBMC-blood, we used the Seurat Azimuth PBMC CITE-seq reference78 to transfer labels to the cells following the more stringent doublet removal (Supplementary Note 2). We removed cells from the following annotations that did not fall under our major cell type categories of interest: ‘Mu-0: Mural’ and ‘T-21: Innate-like’ cells in synovium; ‘Glia’, ‘CD69− Mast’, ‘CD69+ Mast’ and ‘Pericytes’ for intestine; ‘NKT’ and ‘neutrophils’ for intestine; and ‘HSPC’, ‘Platelet’, ‘Doublet’, ‘Eryth’ and ‘MAIT’ for PBMC-blood. The final cell numbers can be found in Supplementary Table 2. We generated cell-type-specific count matrices for downstream analyses, removing cells from individuals with fewer than five cells of the cell type. To obtain a version of finer-grained cell annotations to aid in the interpretation of cell embeddings, we manually merged the fine-grained cell annotations for myeloid, B and T cells in synovium and PBMC-blood datasets to a shared set of common cell state annotations (for example, PBMC-blood ‘CD4 CTL’ and ‘CD4 TEM’ and synovium ‘T-12: CD4+ GNLY+’ were merged into ‘CD4+ Cytotoxic’; Supplementary Table 6).
Pseudobulk eQTL analysis
Generation of pseudobulk profiles.
For each cell type (myeloid, B and T), we generated ‘pseudobulk’ versions for each dataset. First, we performed library size normalization using log(CP10k + 1) within each cell, then aggregated all cells per sample by taking the mean normalized expression of each gene to obtain a samples-by-genes matrix79. We excluded individuals with fewer than five cells of the cell type. We performed rank-based inverse normal transformation for each gene, including genes with nonzero expression in greater than half of the samples.
Multi-cohort eQTL model.
To control for genetic ancestry, we used PLINK (v1.90) to calculate genotype PCs (gPCs) using 66,827 shared genome-wide variants across all four datasets. For PC analysis, we included all individuals from the full array cohorts passing QC (including those without paired scRNA-seq data, Supplementary Fig. 1f). To infer hidden determinants of gene expression variation, we ran probabilistic estimation of expression residuals (PEER)80 on each pseudobulk expression matrix for each dataset and cell type separately, using the ‘peer’ R package (v1.0). We used different numbers of PEER factors for each dataset to account for the varying number of individuals in each cohort ( for synovium, 2 for intestine, 7 for PBMC-cultured and 20 for PBMC-blood; Supplementary Fig. 5a). We generated covariate-corrected expression residuals, accounting for sex, age, ancestry (five gPCs), 10x chemistry (for intestine) and PEER factors.
To identify eQTLs for each classical HLA gene, we incorporated all four datasets into a single model (‘multi-cohort model’) to boost power. We combined the expression residuals from all datasets together for each cell type (Supplementary Fig. 5b). For PBMC-cultured, which included both influenza-stimulated and noninfected cells for each sample, we included only the noninfected cells in the analysis. We tested each of the 12,050 MHC-wide variants for association with residualized expression using linear regression (equation (2)), controlling for the dataset to account for systematic differences across cohorts. This provided a pooled estimate for each eQTL effect across datasets. For lead eQTLs in the multi-cohort model, we also ran the model in each dataset separately (without the dataset term) to compare the concordance across datasets. We also ran the same model using the HLA expression estimates from the standard pipeline to compare to the scElLApers results.
| (2) |
Comparison to Aguiar et al. bulk eQTL study.
We compared the lead eQTL effects identified in this study to a bulk RNA-seq study by Aguiar et al.17 on HLA eQTLs in lymphoblastoid cell lines (LCLs). We obtained eQTL summary statistics from the original authors and limited the comparison to B cells in this study as they are most biologically similar to LCLs. Because some variants tested in this study were not tested in Aguiar et al., we restricted the comparison to the lead variants among those tested in both.
Grouping classical HLA alleles by lead eQTL variants.
To determine how classical one- and two-field HLA alleles track with lead eQTL variants, we compared the co-occurrence between eQTL variants and HLA alleles for the associated gene. To calculate co-occurrence (, ranging from 0 to 1), we used the multi-ethnic HLA reference panel dataset from HLA imputation24. Because the reference dataset is phased, we could calculate the proportion of reference haplotypes (n = 20,349 samples × 2 chromosomes = 40,698 haplotypes) containing the ALT allele of each lead eQTL using a custom R script (equation (3)).
| (3) |
Cell-type interaction analysis.
To determine whether lead eQTLs are cell type dependent, we modeled the residualized expression from all three cell types together using a linear mixed-effects model, adding a fixed effect for cell type (myeloid, B or T), an interaction term between variant and cell type (G × cell_type), and a random effect for donor to account for the non-independent sampling of cell types from the same donor (equation (4)). To ascertain the significance of the cell type dependency, we compared the full model to a null model without the interaction term using a likelihood ratio test (LRT) (lrtest function from ‘lmtest’ v0.9-39R package).
| (4) |
Conditional analysis.
To identify additional eQTLs independent from the lead eQTL, we performed up to three additional rounds of conditional analysis for each gene and cell type using the multi-cohort model, conditioning on the lead eQTL(s) from the previous round(s). We terminated early if the lead eQTL did not reach a significance of P < 5 × 10−8. We used PLINK (v1.90) (−ld) to calculate LD r2 values between every pair of lead eQTLs across cell types and rounds of conditional analysis using the multi-ethnic HLA reference panel.
Visualizations.
To generate boxplots of pseudobulk eQTL effects, we used the expression residuals and regressed out the effect of dataset (not already corrected during PEER). For the Manhattan plots, because each gene has multiple potential transcription start sites (TSS) depending on the transcript, we selected the transcript with the midpoint chromosomal start position across transcripts. LD r2 values for the locus zoom plot were calculated using PLINK (v.1.90) and the multi-ethnic HLA reference panel. For generating figures, we used R packages ‘ggrastr’ (v1.0.1), ‘ggrepel’ (v0.9.1), ‘patchwork’ (v1.1.1) and ggplot2’ (v3.3.5).
Creating a single-cell atlas of HLA expression
Mapping cells into a shared embedding.
To create low-dimensional cell state embeddings of single cells across datasets, we first integrated the two tissue datasets (synovium and intestine). For each cell type (myeloid, B and T), we concatenated the counts matrices from both datasets and filtered to the union of the top 1,500 variable genes per dataset calculated using the variance stabilizing transform (vst) method, excluding cell cycle genes (Seurat v4.1.0s.genes and g2m.genes), mitochondrial (MT-) and ribosomal (RPL-, RPS-) genes. We scaled the variable genes across all cells using R package ‘singlecellmethods’ (v0.1.0), calculated the top ten PCs (using the ‘irlba’ v2.3.5 R package), then removed sample and dataset-specific effects using Harmony48 (v0.1.0) (parameters: , nclust 50 and sigma 0.2), resulting in a ten-dimensional ‘Harmonized PC’ (hPC) embedding. We visualized the embedding in 2D using uniform manifold approximation and projection (UMAP), calculated with the umap function in the ‘uwot’ (v0.1.11) R package, with n_neighbors = 30 and min_dist = 0.2. We then projected the two PBMC datasets into the same tissue-defined embedding using Symphony49 (v0.1.0) to align analogous cell states across tissues. For PBMC-cultured, we included cells from both influenza-stimulated and noninfected samples. Symphony mapping was performed one query dataset at a time, correcting for ‘sample’ effects in the query.
As an alternative approach, we also explored de novo integration of all four datasets together. We used the top 1,500 variable genes per dataset (top 1,000 for T cells) and Harmony integration with , and (batch defined as the sample for Synovium, 10x chemistry for intestine, and experimental batch for PBMC datasets). However, the tissue-defined embeddings produced a cleaner visual separation of cell states, particularly for myeloid cells (Supplementary Fig. 9) and were therefore used for downstream analysis.
Quantifying proportion of expression variance explained by cell state.
To estimate the percent of variance in HLA expression explained by cell state, we fit an NBME model of the UMI count of each HLA gene across cells in each cell type. We included donor-level fixed effects for age, sex and ancestry (five gPCs), cell-level fixed effects for scaled log(total UMI count), scaled percent mitochondrial UMIs, and cell state (ten hPCs), and random effects for donor (and experimental batch for PBMC datasets). The NBME models (including all other versions described in subsequent sections) were fit using the glmer.nb function from the ‘lme4’ (v1.1-28) R package with options nAGQ = 0 and ‘nloptwrap’ optimizer. We used the r.squaredGLMM function from the ‘MuMIn’ (v.1.43.17) R package81 to estimate the marginal R2 using the ‘delta’ method for the full model (equation (5)) as well as a model without cell state terms. The difference between the R2 values between the two models was used to estimate the proportion of variance explained by cell state.
| (5) |
Defining a cell embedding using PBMC-blood alone.
We also defined an alternative cell state embedding for each cell type using cells from PBMC-blood alone. To do this, we used the same dimensionality reduction pipeline described above for the tissue-defined embedding, except we used the top 2,000 variable genes across PBMC-blood for each cell type and corrected for experimental batch with Harmony .
Single-cell eQTL analysis
We used a single-cell NBME eQTL model to test HLA eQTLs for cell-state dependency. The model is adapted from the Poisson mixed-effects (PME) model recently described by our group19. We used NBME in this study because we found that the LRT P values from the PME model exhibited inflation when testing for cell-state interactions (Extended Data Fig. 4d; see ‘Evaluating model calibration for testing cell-state interaction’ section), probably because HLA genes exhibit greater overdispersion than other genes, whereas NBME was well calibrated. We first used an NBME model without cell state to define the set of variant-gene pairs with robust genotype main effects within each dataset. We then used an NBME model with cell state to test for dynamic effects. We excluded the Intestine dataset due to small sample size (n = 22).
Testing for genotype effect using NBME model without cell state.
Using the lead eQTL variants identified in the pseudobulk multi-cohort model above (8 genes × 3 cell types = 24 variants), we tested each eQTL using a single-cell NBME model (equation (6)) to assess the genotype effect. We modeled the per-cell UMI count of each HLA gene in each major cell type and dataset separately (24 variants × 3 datasets = 72 variant-gene pairs to test). We included the same donor and cell-level fixed and random effects as in equation (5), except without cell state terms (hPCs) and adding additional terms for donor genotype (G) and five expression PCs (ePCs), which are calculated on each dataset separately to account for technical effects (akin to PEER factors in pseudobulk). We determined the significance of the genotype effect by comparing to a null model without genotype using an LRT with 1 degree of freedom.
| (6) |
We compared the genotype main effect size and significance from the NBME model (equation (6)) to the pseudobulk eQTL model using the PBMC-blood dataset. Significance was represented by LRT P values in the NBME model and Wald P values in the pseudobulk linear model (run on PBMC-blood separately).
To define variants with robust main effects to test for cell-state interaction, we included only variant-gene pairs within a cell type and dataset with a significant genotype main effect (LRT P value <0.05), resulting in a total of 58 variant-gene pairs.
Power analysis for NBME model.
We estimated the power to detect a spectrum of effect sizes across a range of allele frequencies using our NBME model (methods detailed in Supplementary Note 4).
Testing for cell-state interaction using NBME model.
To test the 58 variant-gene pairs for dynamic regulatory effects, we modeled the eQTLs at single-cell resolution using an NBME model (equation (7)). While the model can use any cell state variable (for example, clusters and pseudotime trajectory), we reasoned that hPCs would provide a principled and unbiased way to define continuous cell states. We include the same donor and cell-level fixed and random effects as in equation (6), with the addition of cell state (hPC1-10 from the tissue-defined Symphony embeddings) and genotype interaction with cell state . To assess whether the eQTL is cell state dependent, we compared the full model (equation (7)) to a null model without interaction terms using an LRT with 10 degrees of freedom.
| (7) |
Evaluating model calibration for testing cell-state interaction.
We analyzed the calibration of the NBME model when testing for interaction between genotype and cell state. Using the PBMC-blood cells and embedding defined in PBMC-blood alone, we permuted cell state (ten hPCs as a block) across all cells, then ran the NBME model for each variant-gene pair (equation (7)) and assessed its significance using LRT, which should yield uniform P values if the model is well calibrated. We repeated this process for 1,000 permutations and compared the results to the equivalent analysis performed with a PME model (glmer function from ‘lme4’ R package with family = ‘poisson’).
Comparing eQTL strength across cell states.
For a given eQTL, we combined the genotype main effect with the interaction effects of each hPC (estimated in equation (7)), weighted by each cell’s position along each hPC (for example, ) to score each cell on the basis of its estimated total eQTL effect size (equation (8)). This allowed us to compare the strength of the eQTL across cell states by plotting the estimated of each cell in UMAP coordinates and comparing the mean across cell state annotations.
| (8) |
By binning cells by five quantiles of estimated , we calculated the main genotype effect in each quantile separately using equation (6), determining significance by LRT comparing to a null model without the genotype term. For the T cell HLA-A dynamic eQTL, the dynamic effect was very specific to proliferating cells. Hence, for visualization, we did not bin the cells by five quantiles based on hPCs because proliferating cells were rare (n = 739 cells) and instead calculated the main genotype effect in proliferating cells and CD8+ Cytotoxic cells (n = 96,516) for comparison.
To compare the , estimates derived from the tissue-defined embedding to those from the embedding defined using PBMC-blood alone for the myeloid HLA-DQA1 eQTL (rs3104413), we ran the same NBME cell-state interaction model (equation (7)) except using ten hPCs defined in PBMC-blood (see ‘Defining a cell embedding using PBMC-blood alone’ section). We calculated the Pearson correlation between the estimates produced by the two embeddings. We also tested for eQTL interactions with contextual factors (age, sex and interferon response) as described in Supplementary Note 4.
Extended Data
Extended Data Fig. 1 ∣. Correcting HLA expression estimation bias with scHLApers.
a, Schematic showing how high HLA gene polymorphism leads to bias in read alignment to a single reference genome. Consider two hypothetical individuals who are either homozygous for HLA-DRB1 allele X (orange) or allele Y (blue), where the reference allele is X. Reads from X will align perfectly to the reference, leading to accurate HLA-DRB1 quantification. However, for Y, reads will fail to align to the reference due to discordant sequence content, leading to unmapped reads and underestimation of expression. b, Percentage change in expression (total UMIs for HLA gene per individual, y-axis) across cohorts (synovium, n = 69 individuals; intestine, n = 22; PBMC-cultured, n = 73; PBMC-blood, n = 909). c, Percentage change in estimated expression (total UMIs for HLA gene per individual, y-axis) in synovium (n = 69) as a function of the mean (between the individual’s two alleles) Levenshtein distance relative to the GRCh38 reference allele at the 3’ end of each gene (x-axis). For b and c, dashed horizontal red lines denote no change. Fitted linear regression line (blue) shown with 95% confidence region. d, Heatmap showing the alignment of reads to each gene in scHLApers (rows) versus where the same read aligned (‘came from’) in the standard pipeline (columns) for synovium (top) and PBMC-cultured (bottom). Columns include HLA genes, other regions in the extended MHC, or unmapped reads. Rows sum to 100%, and a darker color indicates that more of the reads aligning to a given gene in scHLApers came from the corresponding location in the standard pipeline. e, Phylogenetic tree derived from a multiple sequence alignment of HLA-C allelic genomic sequences. The reference allele is C*07:02. Yellow box shows alleles similar to the reference (‘reference-like’). Boxplot on right shows the change in HLA-B estimated UMI counts summed across cells from each sample (y-axis) compared to the genotype for HLA-C in terms of dosage of ‘reference-like’ alleles (x-axis), across n = 1,073 individuals from all cohorts. For b and e, boxplot center line represents median, lower/upper box limits represent 25/75% quantiles, whiskers extend to box limit ±1.5 × IQR, and outlying points are plotted individually.
Extended Data Fig. 2 ∣. Concordance of eQTLs with bulk RNA-seq, differential allelic expression, and read alignment visualization.
a, Concordance between the effect sizes of lead HLA eQTLs identified in the multi-cohort pseudobulk model for B cells (this study, y-axis) and the same variant’s effect in LCLs identified through bulk RNA-seq eQTL analysis (Aguiar et al., x-axis). Because not all lead variants in this study were directly comparable due to different sets of tested variants, we tested the concordance of the most significant variant present in both datasets (triangles indicate that the exact lead variant in this study was also tested in Aguiar et al., whereas circles indicate ‘substitute’ lead variants was used for comparison). b, HLA-B expression in myeloid cells (top, n = 861 individuals) and HLA-C expression in B cells (bottom, n = 909), showing mean log(CP10k + 1)-normalized expression (y-axis) across cells for each individual in PBMC-blood by allele (x-axis). Each individual’s expression value is plotted once if they are homozygous (red) and twice if heterozygous (tan) for each allele (imputed dosage is rounded to the nearest integer). The black diamonds show the mean value for each allele (used to order the x-axis). c, Integrative Genomics Viewer (IGV) screenshots showing read alignments for alleles HLA-B*15:01 and HLA-C*07:01, associated with lower expression of the respective genes, for a representative individual in synovium.
Extended Data Fig. 3 ∣. Personalization improves eQTL effect size estimates.
a, Comparison of eQTL effect size estimates calculated using expression quantified by scHLApers (x-axis) vs. standard pipeline (y-axis). Each dot represents one of 12,045 MHC-wide genetic variants tested using the pseudobulk eQTL model per cell type (color). Pearson correlation is labeled for each gene. b, Example of eQTL effect correction through the use of corrected expression estimates, shown for HLA-DRB1 in B cells. eQTL effect sizes (y-axis) estimated for MHC variants along Chr. 6 (x-axis), shown for standard pipeline (top), scHLApers pipeline (middle), and the magnitude of difference between the betas from the two pipelines (bottom). The variant with the largest correction in estimated eQTL effect (HLA-DRB1*07:01) is labeled in orange, and the lead variant in the scHLApers pipeline (rs9271117) is labeled in blue. c, Boxplots visualizing the eQTL effects across individuals for HLA-DRB1*07:01 (left) and rs9271117 (right) using HLA-DRB1 expression estimates from the standard (top) vs. scHLApers (bottom) pipelines. Increased dosage of the ALT allele (x-axis) vs. HLA-DRB1 expression in B cells (y-axis: units are residual of inverse normal transformed mean log(CP10k + 1)-normalized expression across cells after regressing out covariates), across n = 1,069 individuals total (synovium, n = 65; intestine, n = 22; PBMC-cultured, n = 73; PBMC-blood, n = 909), plotted by dataset (color). For HLA-DRB1*07:01, ‘A’ denotes absence of the allele, and ‘T’ denotes presence (rather than REF/ALT nucleotides). Nominal Wald P-values are derived from linear regression (two-sided test).
Extended Data Fig. 4 ∣. Testing single-cell NBME model for concordance with pseudobulk and for calibration for genotype-cell-state interactions.
a-e, The models in a-c test genotype main effects, whereas d and e test genotype-cell-state interaction. a,b, Concordance of genotype main effect estimates (a) and significance of genotype main effect (b) between the NBME model (y-axis) and the pseudobulk model for the PBMC-blood dataset (x-axis) across all cell types and classical HLA genes. c, Power of the NBME single-cell eQTL model to detect regulatory effects across allele frequencies. The proportion of simulations where the null hypothesis was appropriately rejected at α = 5 × 10−8 (y-axis) in the presence of a simulated eQTL effect across 1000 simulations. Simulations were run across a range of eQTL allele frequencies (x-axis) and effect sizes (colors) using the PBMC-blood myeloid data and HLA-DQA1 expression. d,e, We permuted cell state (10 hPCs as a block) for 1,000 tests and obtained interaction P-values from a one-sided likelihood ratio test (LRT) comparing to the null model without G×hPC interaction terms. Q-Q plots showing statistical calibration (compared to uniform P-values) for PME model (d) versus NBME model (e) when testing for cell state interactions for representative class I (HLA-A) and class II (HLA-DPA1) genes in myeloid cells in PBMC-blood. The red line is the identity line. The histograms below show distributions of LRT P-values for HLA-DPA1.
Supplementary Material
Acknowledgements
We thank A. Dobin, H. Randolph, H. Lau, C. Stevens and members of the Raychaudhuri Lab, in particular A. Gupta and Y. Baglaenko, for their helpful input and discussions. This work was funded by the National Institutes of Health grants T32GM007753 and T32GM144273 (J.B.K., L.R. and K.A.L.), F30AI172238 (J.B.K.), T32HG002295 (A.Z.S. and L.R.), T32AR007530 (A.N.), F30AI157385 (L.R.), R01AR063759 (S.R.), U01HG012009 (S.R.) and UC2AR081023 (S.R.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This project also received funding from the MGH Center for the Study of Inflammatory Bowel Disease grant DK-43351 (R.J.X.), a fellowship from the Fok Foundation (J.E.P.), the Arthritis National Research Foundation (M.G.-A.), Gilead Sciences Research Scholar grant (M.G.-A.), Lupus Research Alliance (M.G.-A.) and a Kennedy Trust KTRR Senior Research Fellowship (KENN202109) (Y.L).
Accelerating Medicines Partnership Program: Rheumatoid Arthritis and Systemic Lupus Erythematosus (AMP RA/SLE) Network
Jennifer Albrecht22, William Apruzzese5, Nirmal Banda24, Jennifer L. Barnas22, Joan M. Bathon25, Ami Ben-Artzi26, Brendan F. Boyce27, David L. Boyle28, S. Louis Bridges Jr.20,21, Vivian P. Bykerk20,21, Debbie Campbell22, Hayley L. Carr28,29, Arnold Ceponis30, Adam Chicoine5, Andrew Cordle31, Michelle Curtis1,2,3,4,5, Kevin D. Deane24, Edward DiCarlo32, Patrick Dunn33,34, Andrew Filer28,29,35, Gary S. Firestein30, Lindsy Forbess28, Laura Geraldino-Pardilla25, Susan M. Goodman20,21, Ellen M. Gravallese5, Peter K. Gregersen36, Joel M. Guthridge37, V. Michael Holers24, Diane Horowitz36, Laura B. Hughes38, Kazuyoshi Ishigaki1,2,3,4,5,39, Lionel B. Ivashkiv20,21, Judith A. James37, Gregory Keras5, Ilya Korsunsky1,2,3,4,5, Amit Lakhanpal20,21, James A. Lederer40, Myles Lewis41,42, Zhihan J. Li5, Yuhong Li5, Katherine P. Liao3,5, Arthur M. Mandelin II43, Ian Mantel20,21, Kathryne E. Marks5, Mark Maybury28, Andrew McDavid44, Mandy J. McGeachy45, Joseph Mears1,2,3,4,5, Nida Meednu22, Nghia Millard1,2,3,4,5, Larry W. Moreland24,45, Saba Nayar28,29,35, Alessandra Nerviani41,42, Dana E. Orange20,46, Harris Perlman43, Costantino Pitzalis41,42,47, Javier Rangel-Moreno22, Karim Raza28,29, Yakir Reshef1,2,3,4,5, Christopher Ritchlin22, Felice Rivellese41,42, William H. Robinson48, Ilfita Sahbudin28, Anvita Singaraju20,21, Jennifer A. Seifert24, Kamil Slowikowski3,4,49,50, Melanie H. Smith20, Darren Tabechian22, Dagmar Scheel-Toellner28,29, Paul J. Utz48, Gerald F. M. Watts5, Kevin Wei5, Kathryn Weinand1,2,3,4,5, Dana Weisenfeld5, Michael H. Weisman26,48, Aaron Wyse31, Qian Xiao1,2,3,4,5 & Zhu Zhu5
24Division of Rheumatology, University of Colorado School of Medicine, Aurora, CO, USA. 25Division of Rheumatology, Columbia University College of Physicians and Surgeons, New York, NY, USA. 26Division of Rheumatology, Cedars-Sinai Medical Center, Los Angeles, CA, USA. 27Department of Pathology and Laboratory Medicine, University of Rochester Medical Center, Rochester, NY, USA. 28Rheumatology Research Group, Institute for Inflammation and Ageing, University of Birmingham, Birmingham, UK. 29NIHR Birmingham Biomedical Research Center and Clinical Research Facility, University of Birmingham, Queen Elizabeth Hospital, Birmingham, UK. 30Division of Rheumatology, Allergy and Immunology, University of California, San Diego, La Jolla, CA, USA. 31Department of Radiology, University of Pittsburgh Medical Center, Pittsburgh, PA, USA. 32Department of Pathology and Laboratory Medicine, Hospital for Special Surgery, New York, NY, USA. 33Division of Allergy, Immunology, and Transplantation, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, USA. 34Northrop Grumman Health Solutions, Rockville, MD, USA. 35Birmingham Tissue Analytics, Institute of Translational Medicine, University of Birmingham, Birmingham, UK. 36Feinstein Institute for Medical Research, Northwell Health, Manhasset, New York, NY, USA. 37Department of Arthritis & Clinical Immunology, Oklahoma Medical Research Foundation, Oklahoma City, OK, USA. 38Division of Clinical Immunology and Rheumatology, Department of Medicine, University of Alabama at Birmingham, Birmingham, AL, USA. 39Laboratory for Human Immunogenetics, RIKEN Center for Integrative Medical Sciences, Yokohama, Japan. 40Department of Surgery, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA, USA. 41Centre for Experimental Medicine & Rheumatology, William Harvey Research Institute, Queen Mary University of London, London, UK. 42Barts Health NHS Trust, Barts Biomedical Research Centre, National Institute for Health and Care Research, London, UK. 43Division of Rheumatology, Department of Medicine, Northwestern University Feinberg School of Medicine, Chicago, IL, USA. 44Department of Biostatistics and Computational Biology, University of Rochester School of Medicine and Dentistry, Rochester, NY, USA. 45Division of Rheumatology and Clinical Immunology, University of Pittsburgh School of Medicine, Pittsburgh, PA, USA. 46Laboratory of Molecular Neuro-Oncology, The Rockefeller University, New York, NY, USA. 47Department of Biomedical Sciences, Humanitas University and Humanitas Research Hospital, Milan, Italy. 48Division of Immunology and Rheumatology, Institute for Immunity, Transplantation and Infection, Stanford University School of Medicine, Stanford, CA, USA. 49Center for Immunology and Inflammatory Diseases, Department of Medicine, Massachusetts General Hospital, Boston, MA, USA. 50MGH Cancer Center, Boston, MA, USA.
Footnotes
Competing interests
J.B.K. is a consultant to Aditum Bio. R.J.X. is co-founder of Jnana Therapeutics and Celsius Therapeutics, scientific advisory board member at Nestlé, and board director at MoonLake Immunotherapeutics; these organizations had no roles in this study. M.B.B. is a consultant to GSK, 4FO Ventures, Third Rock Ventures and consultant and founder of Mestag Therapeutics. S.R. is a scientific advisor to Pfizer, Janssen and Sonoma Biotherapeutics, a founder of Mestag Therapeutics, and a consultant for AbbVie, Sanofi, Biogen and Nimbus Therapeutics. The remaining authors declare no competing interests.
Extended data is available for this paper at https://doi.org/10.1038/s41588-023-01586-6.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41588-023-01586-6.
A list of authors and their affiliations appears at the end of the paper.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41588-023-01586-6.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The GRCh38 reference genome (primary assembly) and Gencode v38 annotation file can be downloaded at https://www.gencodegenes.org/human/release_38.html. For the synovium dataset, the single-cell expression data are available on Synapse at https://doi.org/10.7303/syn52297840. Genotype data are available on the Arthritis and Autoimmune and Related Diseases Knowledge Portal (ARK Portal, https://arkportal.synapse.org/Explore/Datasets/DetailsPage?id=syn52297840). For intestine, the raw scRNA-seq data (bam files) was obtained from the Broad Data Use Oversight System (DUOS) (dataset name: Ulcerative_Colitis_in_Colon_Regev_Xavier); the genotype data are available on dbGaP (phs001642). For PBMC-cultured, the raw scRNA-seq data (FASTQ files) was obtained from GEO (PRJNA682434), and the imputed low-pass WGS data is publicly available at SRA (PRJNA736483) and Zenodo (https://doi.org/10.5281/zenodo.4273999). For PBMC-blood (OneK1K cohort), both the raw scRNA-seq data (bam files) and genotyping data are publicly available on GEO (GSE196830). The reprocessed versions of all scRNA-seq count matrices from this study after realignment with scHLApers are publicly available on Figshare (https://doi.org/10.6084/m9.figshare.24311335).
Code availability
Code and tutorials to run the scHLApers pipeline (v1.0) are available on GitHub (https://github.com/immunogenomics/scHLApers) and Zenodo (https://doi.org/10.5281/zenodo.10003910). Scripts for reproducing analyses in the manuscript are also available on GitHub (https://github.com/immunogenomics/hla2023) and Zenodo (https://doi.org/10.5281/zenodo.10003911).
References
- 1.Lenz TL, Spirin V, Jordan DM & Sunyaev SR Excess of deleterious mutations around HLA genes reveals evolutionary cost of balancing selection. Mol. Biol. Evol 33, 2555–2564 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dendrou CA, Petersen J, Rossjohn J & Fugger L HLA variation and disease. Nat. Rev. Immunol 18, 325–339 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Matzaraki V, Kumar V, Wijmenga C & Zhernakova A The MHC locus and genetic susceptibility to autoimmune and infectious diseases. Genome Biol. 18, 76 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trowsdale J & Knight JC Major histocompatibility complex genomics and human disease. Annu. Rev. Genomics Hum. Genet 14, 301–323 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okada Y. et al. Fine mapping major histocompatibility complex associations in psoriasis and its clinical subtypes. Am. J. Hum. Genet 95, 162–172 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raychaudhuri S. et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat. Genet 44, 291–296 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hollenbach JA & Oksenberg JR The immunogenetics of multiple sclerosis: a comprehensive review. J. Autoimmun 64, 13–25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vader W. et al. The HLA-DQ2 gene dose effect in celiac disease is directly related to the magnitude and breadth of gluten-specific T cell responses. Proc. Natl Acad. Sci. USA 100, 12390–12395 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu X. et al. Additive and interaction effects at three amino acid positions in HLA-DQ and HLA-DR molecules drive type 1 diabetes risk. Nat. Genet 47, 898–905 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishigaki K. et al. HLA autoimmune risk alleles restrict the hypervariable region of T cell receptors. Nat. Genet 54,393–402 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharon E. et al. Genetic variation in MHC proteins is associated with T cell receptor expression biases. Nat. Genet 48, 995–1002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Broughton SE et al. Biased T cell receptor usage directed against human leukocyte antigen DQ8-restricted gliadin peptides is associated with celiac disease. Immunity 37, 611–621 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Apps R. et al. Influence of HLA-C expression level on HIV control. Science 340, 87–91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cavalli G. et al. MHC class II super-enhancer increases surface expression of HLA-DR and HLA-DQ and affects cytokine production in autoimmune vitiligo. Proc. Natl Acad. Sci. USA 113, 1363–1368 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raj P. et al. Regulatory polymorphisms modulate the expression of HLA class II molecules and promote autoimmunity. eLife 5, e12089 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D’Antonio M. et al. Systematic genetic analysis of the MHC region reveals mechanistic underpinnings of HLA type associations with disease. eLife 8, e48476 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aguiar VRC, César J, Delaneau O, Dermitzakis ET & Meyer D Expression estimation and eQTL mapping for HLA genes with a personalized pipeline. PLoS Genet. 15, e1008091 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutierrez-Arcelus M. et al. Allele-specific expression changes dynamically during T cell activation in HLA and other autoimmune loci. Nat. Genet 52, 247–253 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nathan A. et al. Single-cell eQTL models reveal dynamic T cell state dependence of disease loci. Nature 606, 120–128 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cuomo ASE et al. CellRegMap: a statistical framework for mapping context-specific regulatory variants using scRNA-seq. Mol. Syst. Biol 18, e10663 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmiedel BJ et al. Single-cell eQTL analysis of activated T cell subsets reveals activation and cell type–dependent effects of disease-risk variants. Sci. Immunol 7, eabm2508 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer D, Aguiar VRC, Bitarello BD, Brandt DYC & Nunes K A genomic perspective on HLA evolution. Immunogenetics 70, 5–27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brandt DYC et al. Mapping bias overestimates reference allele frequencies at the HLA genes in the 1000 Genomes Project Phase I data. G3 5, 931–941 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakaue S. et al. A statistical genetics guide to identifying HLA alleles driving complex disease. Nat. Protoc 18, 2625–2641 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aguiar VRC, Masotti C, Camargo AA & Meyer D HLApers: HLA typing and quantification of expression with personalized index. Methods Mol. Biol 2120, 101–112 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Bettens F. et al. Regulation of HLA class I expression by non-coding gene variations. PLoS Genet. 18, e1010212 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson J. et al. IPD-IMGT/HLA database. Nucleic Acids Res. 48, D948–D955 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaminow B, Yunusov D & Dobin A STARsolo: accurate, fast and versatile mapping/quantification of single-cell and single-nucleus RNA-seq data. Preprint at bioRxiv 10.1101/2021.05.05.442755 (2021). [DOI] [Google Scholar]
- 29.Zhang F et al. Deconstruction of rheumatoid arthritis synovium defines inflammatory subtypes. Nature 10.1038/s41586-023-06708-y (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smillie CS et al. Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell 178, 714–730.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Randolph HE et al. Genetic ancestry effects on the response to viral infection are pervasive but cell type specific. Science 374, 1127–1133 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yazar S. et al. Single-cell eQTL mapping identifies cell type-specific genetic control of autoimmune disease. Science 376, eabf3041 (2022). [DOI] [PubMed] [Google Scholar]
- 33.Jia X. et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS ONE 8, e64683 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo Y. et al. A high-resolution HLA reference panel capturing global population diversity enables multi-ancestry fine-mapping in HIV host response. Nat. Genet 53, 1504–1516 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dunlap G. et al. Clonal associations of lymphocyte subsets and functional states revealed by single cell antigen receptor profiling of T and B cells in rheumatoid arthritis synovium. Preprint at bioRxiv 10.1101/2023.03.18.533282 (2023). [DOI] [Google Scholar]
- 36.Wang Z. et al. Clonally diverse CD38+HLA-DR+CD8+ T cells persist during fatal H7N9 disease. Nat. Commun 9, 824 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tippalagama R. et al. HLA-DR marks recently divided antigen-specific effector CD4 T cells in active tuberculosis patients. J. Immunol 207, 523–533 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soskic B. et al. Immune disease risk variants regulate gene expression dynamics during CD4+ T cell activation. Nat. Genet 54, 817–826 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holling TM, Schooten E & van Den Elsen PJ Function and regulation of MHC class II molecules in T-lymphocytes: of mice and men. Hum. Immunol 65, 282–290 (2004). [DOI] [PubMed] [Google Scholar]
- 40.LaSalle JM, Tolentino PJ, Freeman GJ, Nadler LM & Hafler DA Early signaling defects in human T cells anergized by T cell presentation of autoantigen. J. Exp. Med 176, 177–186 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lanzavecchia A, Roosnek E, Gregory T, Berman P & Abrignani S T cells can present antigens such as HIV gp120 targeted to their own surface molecules. Nature 334, 530–532 (1988). [DOI] [PubMed] [Google Scholar]
- 42.Hagopian W. et al. Co-occurrence of type 1 diabetes and celiac disease autoimmunity. Pediatrics 140, e20171305 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamamoto F. et al. Capturing differential allele-level expression and genotypes of all classical HLA loci and haplotypes by a new capture RNA-seq method. Front. Immunol 11, 941 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaur G. et al. Structural and regulatory diversity shape HLA-C protein expression levels. Nat. Commun 8, 15924 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kulkarni S. et al. Genetic interplay between HLA-C and MIR148A in HIV control and Crohn disease. Proc. Natl Acad. Sci. USA 110, 20705–20710 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chandran V. et al. Killer-cell immunoglobulin-like receptor gene polymorphisms and susceptibility to psoriatic arthritis. Rheumatology 53, 233–239 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Ota M. et al. Dynamic landscape of immune cell-specific gene regulation in immune-mediated diseases. Cell 184, 3006–3021. e17 (2021). [DOI] [PubMed] [Google Scholar]
- 48.Korsunsky I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kang JB et al. Efficient and precise single-cell reference atlas mapping with Symphony. Nat. Commun 12, 5890 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilkinson ST et al. Partial plasma cell differentiation as a mechanism of lost major histocompatibility complex class II expression in diffuse large B-cell lymphoma. Blood 119, 1459–1467 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoon HS et al. ZBTB32 is an early repressor of the CIITA and MHC class II gene expression during B cell differentiation to plasma cells. J. Immunol 189, 2393–2403 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumasaka N. et al. Mapping interindividual dynamics of innate immune response at single-cell resolution. Nat. Genet 55, 1066–1075 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kang JB, Raveane A, Nathan A, Soranzo N & Raychaudhuri S Methods and insights from single-cell expression quantitative trait loci. Annu. Rev. Genomics Hum. Genet 24, 277–303 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yao C. et al. Sex- and age-interacting eQTLs in human complex diseases. Hum. Mol. Genet 23, 1947–1956 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davenport EE et al. Discovering in vivo cytokine-eQTL interactions from a lupus clinical trial. Genome Biol. 19, 168 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhernakova DV et al. Identification of context-dependent expression quantitative trait loci in whole blood. Nat. Genet 49, 139–145 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Calzetti F. et al. Human dendritic cell subset 4 (DC4) correlates to a subset of CD14dim/−CD16++ monocytes. J. Allergy Clin. Immunol 141, 2276–2279.e3 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Janeway CA, Travers P, Walport M & Shlomchik MJ Immunobiology (CRC Press, 2001). [Google Scholar]
- 59.Kambayashi T & Laufer TM Atypical MHC class II-expressing antigen-presenting cells: can anything replace a dendritic cell? Nat. Rev. Immunol 14, 719–730 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Prugnolle F. et al. Pathogen-driven selection and worldwide HLA class I diversity. Curr. Biol 15, 1022–1027 (2005). [DOI] [PubMed] [Google Scholar]
- 61.Yeung H-Y & Dendrou CA Pregnancy immunogenetics and genomics: implications for pregnancy-related complications and autoimmune disease. Annu. Rev. Genomics Hum. Genet 20, 73–97 (2019). [DOI] [PubMed] [Google Scholar]
- 62.Barreiro LB & Quintana-Murci L From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat. Rev. Genet 11, 17–30 (2010). [DOI] [PubMed] [Google Scholar]
- 63.Petersdorf EW et al. HLA-C expression levels define permissible mismatches in hematopoietic cell transplantation. Blood 124, 3996–4003 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chowell D. et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 359, 582–587 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Naranbhai V. et al. HLA-A*03 and response to immune checkpoint blockade in cancer: an epidemiological biomarker study. Lancet Oncol. 23, 172–184 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matern BM et al. Long-read nanopore sequencing validated for human leukocyte antigen class I typing in routine diagnostics. J. Mol. Diagn 22, 912–919 (2020). [DOI] [PubMed] [Google Scholar]
- 67.Liu C. et al. High-resolution HLA typing by long reads from the R10.3 Oxford nanopore flow cells. Hum. Immunol 82, 288–295 (2021). [DOI] [PubMed] [Google Scholar]
- 68.Giambartolomei C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wallace C. A more accurate method for colocalisation analysis allowing for multiple causal variants. PLoS Genet. 17, e1009440 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van der Wijst M. et al. The single-cell eQTLGen consortium. eLife 9, e52155 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.scHLApers. GitHub. https://github.com/immunogenomics/scHLApers (2023). [Google Scholar]
- 72.IMGTHLA. GitHub. https://github.com/ANHIG/IMGTHLA (2023). [Google Scholar]
- 73.hlaseqlib. GitHub. https://github.com/genevol-usp/hlaseqlib (2022). [Google Scholar]
- 74.tutorial_HLAQCImputation.ipynb. GitHub. https://github.com/immunogenomics/HLA_analyses_tutorial/blob/main/tutorial_HLAQCImputation.ipynb (2023). [Google Scholar]
- 75.SNP2HLA.py. GitHub. https://github.com/immunogenomics/HLA_analyses_tutorial/blob/main/scripts/SNP2HLA.py (2023). [Google Scholar]
- 76.Chain file for hg19 to hg38 liftover. UCSC. http://hgdownload.soe.ucsc.edu/goldenPath/hg19/liftOver/hg19ToHg38.over.chain.gz (2013). [Google Scholar]
- 77.Darby CA, Stubbington MJT, Marks PJ, Martínez Barrio Á & Fiddes IT scHLAcount: allele-specific HLA expression from single-cell gene expression data. Bioinformatics 36, 3905–3906 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Azimuth. HuBMAP Consortium. https://app.azimuth.hubmapconsortium.org/app/human-pbmc (2020). [Google Scholar]
- 79.Cuomo ASE et al. Optimizing expression quantitative trait locus mapping workflows for single-cell studies. Genome Biol. 22, 188 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stegle O, Parts L, Piipari M, Winn J & Durbin R Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat. Protoc 7, 500–507 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nakagawa S, Johnson PCD & Schielzeth H The coefficient of determination R2 and intra-class correlation coefficient from generalized linear mixed-effects models revisited and expanded. J. R. Soc. Interface 14, 20170213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The GRCh38 reference genome (primary assembly) and Gencode v38 annotation file can be downloaded at https://www.gencodegenes.org/human/release_38.html. For the synovium dataset, the single-cell expression data are available on Synapse at https://doi.org/10.7303/syn52297840. Genotype data are available on the Arthritis and Autoimmune and Related Diseases Knowledge Portal (ARK Portal, https://arkportal.synapse.org/Explore/Datasets/DetailsPage?id=syn52297840). For intestine, the raw scRNA-seq data (bam files) was obtained from the Broad Data Use Oversight System (DUOS) (dataset name: Ulcerative_Colitis_in_Colon_Regev_Xavier); the genotype data are available on dbGaP (phs001642). For PBMC-cultured, the raw scRNA-seq data (FASTQ files) was obtained from GEO (PRJNA682434), and the imputed low-pass WGS data is publicly available at SRA (PRJNA736483) and Zenodo (https://doi.org/10.5281/zenodo.4273999). For PBMC-blood (OneK1K cohort), both the raw scRNA-seq data (bam files) and genotyping data are publicly available on GEO (GSE196830). The reprocessed versions of all scRNA-seq count matrices from this study after realignment with scHLApers are publicly available on Figshare (https://doi.org/10.6084/m9.figshare.24311335).
Code and tutorials to run the scHLApers pipeline (v1.0) are available on GitHub (https://github.com/immunogenomics/scHLApers) and Zenodo (https://doi.org/10.5281/zenodo.10003910). Scripts for reproducing analyses in the manuscript are also available on GitHub (https://github.com/immunogenomics/hla2023) and Zenodo (https://doi.org/10.5281/zenodo.10003911).