1. Introduction
Use of antipsychotic drugs in patients with schizophrenia (SCZ) is standard treatment and often long-term. However repeated treatment failure with inadequate improvement in symptoms occurs in approximately 30% of patients treated with antipsychotics. This clinical picture is termed treatment-resistant schizophrenia (TRS)(Meltzer, 1997). The biological mechanisms underlying TRS remain unclear, although disease heterogeneity, pharmacokinetics and/or genetic diversity, are likely of major relevance(Kinon, 2018). Initial genetic studies attempting to better understand the molecular mechanisms of TRS produced conflicting results, due in part to differences in the patient groups studied as well as the use of the candidate gene approach(Gillespie et al., 2017; Lally et al., 2016). Most recently, a genome-wide association study (GWAS) of TRS confirmed that it is polygenic in nature (estimated SNP-based heritability of 1–6%), despite not identifying any genome-wide significant loci(Pardinas et al., 2022). Obtaining a better understanding of the underlying mechanisms and early identification of TRS remain critical priorities in SCZ research.
Clozapine (CLZ) is superior to other antipsychotics in terms of clinical effect and is the only licensed treatment for TRS(Huhn et al., 2019; Kumra et al., 2008). Despite this, the mechanism of action underlying the superior efficacy of CLZ remains unknown(Pardinas et al., 2021). Despite being effective in ~60% of TRS cases however, treatment may be accompanied by severe adverse effects such as agranulocytosis, seizures and myocarditis, limiting its use as a first line SCZ drug(Gurrera et al., 2022; Kumra et al., 2008). Treatment-induced weight gain and accompanying dyslipidemia and glucose dysregulation are also major adverse effects which may limit adherence and success of antipsychotic treatment(Huhn et al., 2019; Leucht et al., 2013; Raben et al., 2017). A recent review investigated the effects of 18 different antipsychotics on metabolic function and identified large heterogeneity in their effect on body weight, body mass index (BMI), cholesterol and glucose concentrations, with CLZ associated with the largest degree of metabolic dysfunction(Pillinger et al., 2020). Improvements in symptom severity were associated with increases in body weight, BMI, low-density lipoprotein (LDL) and total cholesterol, highlighting that the most efficacious treatment options, such as CLZ, also result in the greatest metabolic dysfunction(Pillinger et al., 2020). Moreover, higher BMI is commonly observed in TRS patients taking CLZ when compared to patients with SCZ responding to antipsychotics other than CLZ(Garriga et al., 2022; Pillinger et al., 2020). Findings from a clinical trial indicated that for olanzapine and CLZ, therapeutic response was closely related to gain in weight and BMI (Czobor et al., 2002). The relationship between antipsychotic treatment and metabolic dysfunction is well described(Huhn et al., 2019; Leucht et al., 2013; Raben et al., 2017), and patients with the best effect of olanzapine or CLZ on symptom levels seem to have the largest increase in BMI (Czobor et al., 2002). Still it remains unknown whether metabolic disturbances are associated with antipsychotic efficacy due to overlapping molecular mechanisms(Pillinger et al., 2020), or if these observations are due to other factors such as increased compliance and off-target effects.
Given our limited understanding of the underlying genetics of TRS based on the conventional GWAS approach(Pardinas et al., 2022), additional statical approaches may be employed to boost statistical power for genetic discovery. The conditional false discovery rate (cFDR) framework increases discovery and improves polygenic prediction in underpowered GWAS by leveraging genetic overlap with a second, more powerful GWAS(Andreassen et al., 2013; Andreassen et al., 2014; Smeland et al., 2020; van der Meer et al., 2022). In addition, the cFDR approach is agnostic to effect direction, and so is able to leverage shared genetic variants regardless of effect direction or genome-wide genetic correlation (Smeland et al., 2020). Given evidence that treatment response is associated with weight gain and treatment resistance is associated with a lack of weight gain, we hypothesized that TRS and measures of metabolic function (such as BMI) may have a shared genetic basis. In contrast to the TRS GWAS which included only 10,501 TRS cases(Pardinas et al., 2022), the latest GWAS of BMI includes almost 800,000 individuals and identified approximately 100 genome-wide significant loci(Yengo et al., 2018). Therefore, we sought to utilize the cFDR framework to increase power for genetic discovery and improve polygenic prediction of TRS, by conditioning on BMI.
2. Methods
2.1. GWAS Sample Description
Publicly available GWAS summary statistics for TRS, BMI and SCZ were obtained. The TRS GWAS sample comprised 10,501 TRS and 20,325 non-TRS patients(Pardinas et al., 2022), while the latest GWAS of BMI included 795,640 individuals(Yengo et al., 2018). Moreover, a second GWAS of BMI in 28,681 children at eight years old was also obtained for replication analysis(Helgeland et al., 2022). The SCZ GWAS sample comprised 53,386 cases and 77,258 controls(Trubetskoy et al., 2022) and was only used in the polygenic score analysis as a comparator to show the specificity of the TRS GWAS in predicting TRS outcome. The SCZ GWAS was not used for cFDR analysis. All GWAS data used in these analyses were from individuals of European ancestry. An overview of the study design and samples is shown in Figure 1. The Norwegian Institutional Review Board for the South-East Norway Region has evaluated the current protocol and found that no additional institutional review board approval was needed because no individual data were used. More detailed descriptions are available in the Supplementary methods and original publications(Helgeland et al., 2022; Pardinas et al., 2022; Trubetskoy et al., 2022; Yengo et al., 2018).
Figure 1.
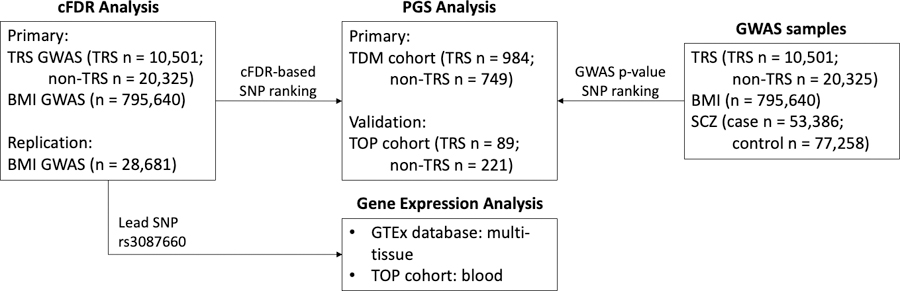
An overview of the study design and the included samples. (A) The conditional false discovery rate (cFDR) approach was used to improve discovery of genetic variants associated with treatment resistant schizophrenia (TRS) after conditioning on body-mass index (BMI). (B) Polygenic score (PGS) analyses were conducted to determine the variance in TRS explained by common genetic variants. PGSs were constructed from GWAS summary statistics of TRS, BMI and SCZ (i), as well as the re-ranked TRS data after conditioning on BMI (ii). (C) The lead SNP rs3087660, associated with TRS after conditioning on BMI, was mapped to genes using the Open Targets platform. The identified gene-variant relationships were further queried across numerous expression quantitative trait loci databases. Lasty, the identified eQTLs were investigated in the TOP TRS cohort.
2.2. Conditional/Conjunctional False Discovery Rate
We conducted cFDR analysis, conditioning genetic associations with TRS on BMI, using default settings (Supplementary Methods). cFDR leverages statistical pleiotropy, the presence of SNP-associations with both traits (Watanabe et al., 2019), to identify variants more likely to be true associations despite p-values below the genome-wide significance threshold. In short, the cFDR procedure re-ranks the test statistics in a primary phenotype (TRS) conditional on the associations in a secondary phenotype (BMI). Hence, TRS variants jointly associated with BMI will attain lower cFDR estimates. An FDR level of 0.01 was set for cFDR, corresponding to 1 false positive per 100 reported associations. More details are provided in the Supplementary Methods, original publications (Andreassen et al., 2013; Andreassen et al., 2014) and subsequent review (Smeland et al., 2020).
2.3. Locus Definitions and Annotations
Genetic loci were defined based on association summary statistics produced with cFDR following the protocol implemented in the Functional Mapping and Annotation (FUMA) online platform with default parameters(Watanabe et al., 2017). Briefly, independent significant genetic variants were identified as variants with cFDR<0.01 and linkage disequilibrium (LD) r2<0.6 with each other. A subset of these independent significant variants with LD r2<0.1 were then selected as lead variants. Moreover, for each independent significant variant all candidate variants were identified as variants with cFDR<0.1 and LD r2≥0.6. For a given lead variant the borders of the genomic locus were defined as minimum/maximum positional coordinates over all corresponding candidate variants. Finally, loci were merged if they were separated by less than 250kb.
Genes were mapped to each identified locus using the Open Targets Genetics platform (https://genetics.opentargets.org/)(Ghoussaini et al., 2021). This platform aggregates human GWAS and functional genomics data including gene expression, protein abundance, chromatin interaction and conformation data from a wide range of cell types and tissues to make robust connections between GWAS-associated loci, variants and likely causal genes. To assign likely causal genes for a given variant, a disease-agnostic Variant to Gene (V2G) analysis pipeline provides a single aggregated V2G score for each variant-gene prediction. For each locus we considered the top 3 genes with the highest V2G score. Finally, we queried SNPs for known expression quantitative trait loci (eQTLs) across multiple tissues using the GTEx portal (GTEx v8)(Consortium, 2015), in peripheral blood using the eQTLGen(Vosa et al., 2021) and in different brain tissues using the BRAINEAC portal(Hernandez et al., 2012). We used the LocusCompare online tool to visualize the colocaliztion of SNPs in the chr15:66,624,854–66,797,492 cFDR locus and tissue-specific MAP2K1 gene expression from the GTEx database (Liu et al., 2019).
2.4. Polygenic Score Analysis and SNP Annotations
Polygenic scores (PGSs) were constructed using PRSice-2(Choi and O’Reilly, 2019) from summary statistics from TRS(Pardinas et al., 2022), BMI(Yengo et al., 2018) and SCZ(Trubetskoy et al., 2022). Using the pleioPGS approach(van der Meer et al., 2022), we leveraged our multivariate analysis by comparing standard GWAS-ranked lead SNPs with cFDR-based ranking, using the same weights derived from the original TRS GWAS. We calculated the scores for specific numbers of lead SNPs rather than for significance thresholds to allow for direct comparison between the approaches, at equal numbers of SNPs in each set. We compared the top 100 (approximate number of SNPs p<1E-5 in the original TRS GWAS) to 105,000 SNPs (number of independent SNPs p<1 in the original TRS GWAS). Sex, age and 10 principal components were included as covariates. Functional consequences of the SNPs contributing to the PGSs explaining the most variability in TRS status were annotated using FUMA(Watanabe et al., 2017).
The initial PGS target sample (TDM cohort) included patients phenotyped with TRS (n=984) or non-TRS (n=749) from the therapeutic drug monitoring (TDM) service at Center for Psychopharmacology in Diakonhjemmet Hospital, Oslo, Norway during the period January 2005 and August 2020 (Figure 1, Supplementary Table 1). The TRS cohort was defined based on the use/prescription of CLZ, the main drug indicated in TRS(FDA, 2014), verified by detectable serum concentration of CLZ and/or available CLZ dose information in TDM registries. All non-TRS patients had only used non-CLZ antipsychotic drug(s) and had never been prescribed CLZ as confirmed by their longitudinal TDM profiles. In Norway, TDM analyses are used as a tool to aid personalized treatment and clinical follow-up in psychiatry. Routine TDM analyses of CLZ is performed due its extensive pharmacokinetic variability and narrow therapeutic range (Hiemke et al., 2018). The analyses guide physicians to optimize dosage for prevention of concentration-dependent severe side effects (e.g., tonic-clonic seizures, sedation) while ensuring adherence to treatment and optimal clinical effect in relation to the therapeutic concentration reference range. The validation PGS target sample (TOP cohort) included 310 patients with SCZ with reported antipsychotic treatment history and RNA microarray data from peripheral blood (Figure 1, Supplementary Methods, Supplementary Table 1). The recruitment procedure and clinical evaluation for this study sample are described in detail in previous reports(Simonsen et al., 2011; Szabo et al., 2022; Werner et al., 2020). Briefly, patients were defined as being TRS (n=89) based on two or more failed trials of antipsychotic treatment, each of at least six weeks duration and with therapeutic dosage. At least one of the antipsychotics had to be a second-generation antipsychotic. All other patients with SCZ (n=221) were considered non-TRS. All individuals in both the TDM and TOP cohorts were of European ancestry.
All participants gave written informed consent and the study was approved by the Norwegian Regional Committee for Medical Research Ethics and the Norwegian Data Inspectorate. All procedures and methods were carried out in accordance with relevant guidelines and regulations.
2.5. Gene Expression Analysis
The TOP cohort was used to assess differences in MAP2K1 mRNA levels that were investigated between the TRS and non-TRS patient groups, between rs3087660 genotype groups, and between rs3087660 genotype groups within the TRS and non-TRS patient groups (Figure 1). Details of the microarray analysis and subsequent quality control are provided in the Supplementary Methods. Analyses were performed using linear regression, adjusting for age and sex. These analyses were considered validation of the eQTL analyses described above and so a p-value threshold of p<0.05 was applied.
2.6. Statistical Analysis
Statistical analyses were performed in R using the base statistical package(R Core Team, 2020). Welch two sample t-tests and chi-squared tests were used to compare baseline charactersistcs (age, sex, BMI and proportion of TRS patients) between the TDM and TOP cohorts.
3. Results
3.1. Genetic Overlap and Discovery
Genome-wide genetic correlation (rg=0.048) was calculated between TRS and BMI using linkage disequilibrium score regression (Bulik-Sullivan et al., 2015). The conditional quantile-quantile (QQ) plots suggest the presence of enrichment for TRS given BMI (Figure 2A), shown by the incremental incidence of association with TRS (leftward deflection) as a function of the significance of association with BMI. We leveraged this cross-phenotype polygenic enrichment using cFDR analyses and re-ranked TRS SNPs conditionally on their association with BMI. At cFDR < 0.01 we identified 2 loci (rs7560232 chr2:207,121,329–207,207,137 and rs3087660 chr15:66,624,854–66,797,492) associated with TRS after conditioning on BMI (Figure 2B, Table 1, Supplementary Table 2). Investigation of these loci in the GWAS catalogue(Buniello et al., 2019) showed that the locus on chromosome 2 has been previously associated with changes in volume of the cerebellar vermal lobules(Zhao et al., 2019), lower self-rated health(Harris et al., 2017) and age-related hearing impairment(Praveen et al., 2022), while the locus on chromosome 15 is only previously associated with BMI(Zhu et al., 2018). The top three genes with the highest V2G score for the locus on chromosome 2 include ZDBF2, CMKLR2 and ADAM23, and for the locus on chromosome 15 include SNAPC5, TIPIN and MAP2K1 (Figure 2B, Table 1). As a replication analysis, we similarly re-ranked TRS SNPs conditionally on their association with BMI in an independent BMI GWAS in children at eight years old(Helgeland et al., 2022) (Supplementary Figure 1 and Supplementary Table 3) and confirmed the association of the locus on chr15 (cFDR = 9.8E-03; rs11631065 chr15:66,603,388–66,797,492) with TRS. We did not observe consistent enrichment for BMI given TRS (Supplementary Figure 2), and so the reverse cFDR analysis was not pursued.
Figure 2.
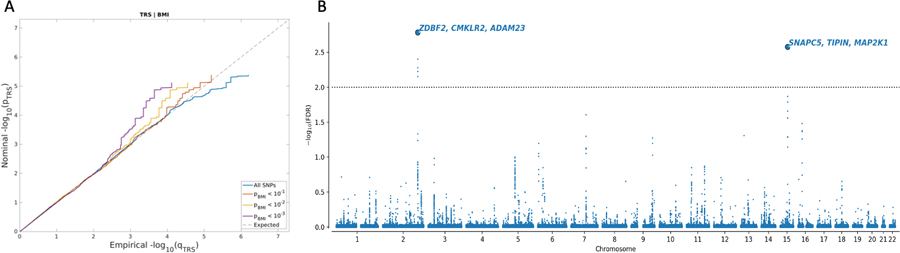
(A) Conditional QQ plots showing cross-phenotype polygenic enrichment between treatment resistant schizophrenia (TRS) and body-mass index (BMI). Plotted are the nominal vs empirical −log10 p values (corrected for inflation) for TRS, below the standard genome-wide association study threshold of p < 5.0 × 10−8, as a function of significance of association with BMI at the levels of p ≤ 0.10, p ≤ 0.01, and p ≤ 0.001. The dashed lines indicate the null hypothesis. (B) Common genetic variants associated with TRS after conditioning on BMI at conditional False Discovery Rate (cFDR) < 0.01. Manhattan plot showing the −log10 transformed cFDR values for each SNP on the y-axis and chromosomal positions along the x-axis. The dotted horizontal line represents the threshold for significant shared associations (cFDR < 0.01). Independent lead SNPs are encircled in black and are annotated to the top three genes with highest variant-to-gene scores. Further details for these loci are provided in Supplementary Table 2.
Table 1.
Novel loci for treatment resistant schizophrenia (TRS) after conditioning on body-mass index (BMI)
| Chr | Range | Lead SNP | A1/A2# | Mapped Genes$ | Functional category | p-value TRS | OR TRS | Rank TRS | p-value BMI | Beta BMI | cFDR |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 207,121,329 – 207,207,137 | rs7560232 | G/A | ZDBF2, CMKLR2, ADAM23 | Intergenic | 1.9E-06 | 0.898 | 4th | 1.1E-09 | 0.011 | 1.6E-03 |
| 15* | 66,624,854 – 66,797,492 | rs3087660 | G/A | SNAPC5, TIPIN, MAP2K1 | 5’ UTR (ZWILCH), Intronic (RPL4) | 3.1E-06 | 0.899 | 13th | 1.3E-10 | 0.011 | 2.7E-03 |
Lead SNPs in independent genomic loci associated with TRS after conditioning on BMI at cFDR < 0.01, after merging regions <250 kb apart into a single locus. The table presents chromosomal position (Chr), mapped genes and functional category, as well as p-values and effect sizes (odds ratios (OR) and betas) from the original summary statistics on TRS(Pardinas et al., 2022) and BMI(Yengo et al., 2018). The effect sizes are given with reference to allele 2 (A1). For more details and a list of all candidate variants in these loci, see Supplementary Table 2.
A1 is effect allele. Rank TRS: p-value based rank of the SNP in the original TRS GWAS.
Locus replicated with analysis using independent BMI GWAS data, see Supplementary Table 3.
Genes listed here are the top 3 genes with the highest variant to gene score in the Open Targets Genetics platform (Ghoussaini et al., 2021).
Additional assessment of the variant-gene relationships in the GTEx database showed significant associations between the lead SNP rs3087660 on chromosome 15 and MAP2K1 gene expression in the cerebellum (p=6.6E-5) and adipose subcutaneous tissues (p=7.8E-7) (Supplementary Figures 3 and 4). Similarly, the rs3087660 G allele was associated with increased MAP2K1 expression in peripheral blood in the eQTLGen database (p=1.5E-11). Although not significant, a similar pattern is also observed in the cerebellum in the BRAINEAC database (p=0.15) (Supplementary Figure 5). LocusCompare results suggest that the rs3087660 variant is the most likely eQTL for MAP2K1 in the cerebellum, while other variants in the locus are stronger eQTLs for MAP2K1 in adipose subcutaneous tissue, and whole blood (Supplementary Figure 6). We further assessed the associations between rs3087660 genotype and MAP2K1 gene expression in the TOP cohort of 310 patients with SCZ, including 89 TRS and 221 non-TRS patients. MAP2K1 gene expression was significantly (p=0.010) lower in the TRS group compared to the non-TRS group (Supplementary Figure 7A and Supplementary Table 4). Like our observations in the GTEx database, greater MAP2K1 gene expression was observed in patients with the rs3087660 GG genotype when compared to those with the GA and AA genotypes, although these differences between genotype groups were not significant (Supplementary Figure 7B and Supplementary Table 4). Although not significant, when MAP2K1 gene expression was compared between rs3087660 genotype groups, in TRS and non-TRS patients separately, the expected dose effect was greater in the non-TRS group than in the TRS group (Supplementary Figure 7C and Supplementary Table 4).
3.2. Polygenic Prediction
We compared the top 100–105,000 SNPs using original TRS GWAS p-value ranking and cFDR-based ranking (Supplementary Figure 8), hypothesizing that the boosted power from our conditional analysis would select more informative variants than standard GWAS, resulting in improved PGS performance. The greatest variance explained by any of the tested PGS was captured within the first 500 SNPs (Figure 3). The maximum variance explained by the original TRS PGS (liability r2 = 4.96%, p = 0.013) was achieved using the top 500 independent SNPs. Interestingly, cFDR-based ranking of TRS conditioned on BMI achieved a similar explained variance (liability r2 = 4.77%, p = 0.017) using only the top 100 independent SNPs, while the maximum variance explained (liability r2 = 5.62%, p = 0.011) required only the top 300 independent SNPs and outperformed the original TRS PGS by 1.13 fold. PGSs for BMI and SCZ were also plotted for comparison.
Figure 3.
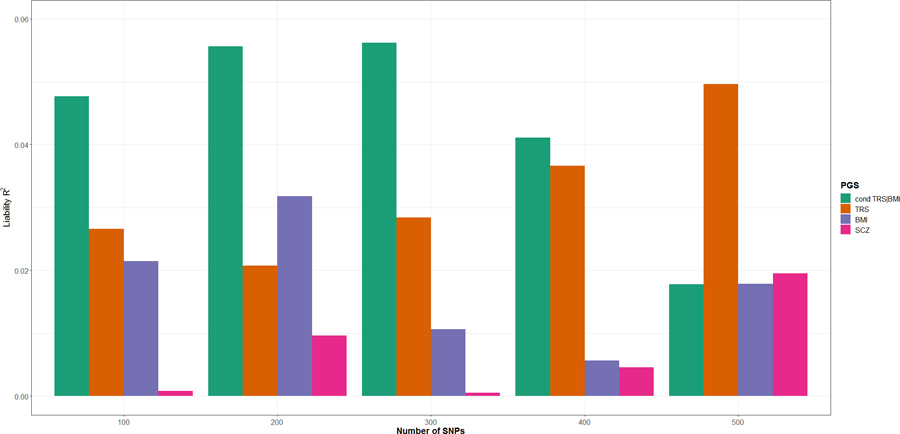
Explained variance (on the liability scale) of treatment resistant schizophrenia (TRS, orange), body-mass index (BMI, purple), TRS after conditioning on BMI (cond TRS|BMI, green) and schizophrenia (SCZ, pink) polygenic scores (PGS) (Liability r2, y-axis) for top 100– 500 independent variants from the TRS GWAS summary statistics. The results of explained variance for all SNP ranges test (100 – 105,000 SNPs) are presented in Supplementary Figure 6.
Of the top 100 independent SNPs after cFDR-based ranking of TRS conditioned on BMI, only 29 were included in the top 100 independent SNPs from the original TRS GWAS, and 46 were included in the top 500 independent SNPs from the original TRS GWAS (Supplementary Table 5). Similarly, when considering the top 300 independent SNPs after cFDR-based ranking of TRS conditioned on BMI (which explained the most variance; Figure 3), only 42 were included in the top 100 independent SNPs from the original TRS GWAS, and 96 were included in the top 500 independent SNPs from the original TRS GWAS (Supplementary Table 5).
Assessment of the functional consequences of the SNPs included in the PGSs showed enrichment of intronic SNPs (enrichment = 1.29, p = 3.74E-130) and depletion of intergenic (enrichment = 0.846, p = 8.94E-60) and ncRNA intronic (enrichment = 0.828, p = 6.86E-13) SNPs in the cFDR-based PGS relative to the reference. In contrast, intergenic SNPs (enrichment = 1.06, p = 9.62E-11) and ncRNA intronic (enrichment = 1.09, p = 2.41E-4) SNPs were enriched, while intronic SNPs were depleted (enrichment = 0.908, p = 3.93E-14) in the original TRS PGS relative to the reference panel (Supplementary Figure 9).
We sought to validate these results in the smaller TOP cohort of SCZ patients used to investigate the differential gene expression of MAP2K1 as described above. Although the greatest maximum variance explained was observed for the original TRS PGS (liability r2 = 0.70%, p = 0.205, 700 SNPs) and not with the cFDR-based ranking of TRS conditioned on BMI (liability r2 = 0.47%, p = 0.299, 600 SNPs), we did observe greater explained variance for the cFDR-based ranking for all SNP thresholds below 700 SNPs (Supplementary Figure 10).
Patients in the TDM cohort (mean age = 49.04 ± 16.13) were significantly older (p<2.2E-16) when compared to those in the TOP cohort (mean age = 30.42 ± 9.36), although no differences were identified between TRS and non-TRS patients in either cohort (Supplementary Table 1). Moreover, the proportion of TRS patients was significantly greater (p<2.2E-16) in the TDM cohort (n = 984/1733) than in the TOP cohort (n = 98/310) (Supplementary Table 1).
4. Discussion
Here we applied the cFDR method to boost discovery of genetic variants associated with TRS, and identified two novel loci associated with TRS after conditioning on BMI. These results provide evidence for a genetic overlap between TRS and BMI. Identified specific loci may provide insights into the shared genetic mechanisms influencing TRS and BMI and inform on their biological underpinnings. In addition, we showed that the polygenic prediction of TRS is improved by leveraging the identified genetic overlap with BMI.
Using the cFDR approach, we identified two novel loci (rs7560232 on chromosome 2 and rs3087660 on chromosome 15) associated with TRS after conditioning on BMI, one (rs3087660) of which replicated using an independent BMI GWAS(Helgeland et al., 2022). The top three genes mapped to this locus (rs3087660) include SNAPC5, TIPIN and MAP2K1. Of these, MAP2K1 is of particular interest since it forms part of the MAPK/ERK pathway activated by antipsychotics(de Bartolomeis et al., 2022). Activation of this pathway results in the phosphorylation of proteins involved in several biological processes including, transcriptional and translational regulation, cellular excitability, dendritic organization, long-term potentiation and depression, neuronal survival, synaptogenesis and neurotransmitter release(Engel et al., 2009), and ERK activation specifically contributes to synaptic plasticity and connectivity(Harrison and Weinberger, 2005; Konradi and Heckers, 2001). The MAP2K1 gene was recently shown to be downregulated in peripheral blood of SCZ patients prior to treatment, when compared to healthy controls, and upregulated after treatment with antipsychotics(Wang et al., 2022). Moreover, a strong positive correlation between demethylation of the MAP2K1 gene promoter region and lifetime antipsychotic use was identified in frontal cortex postmortem tissue samples collected from patients with SCZ(Mill et al., 2008). Investigation of the rs3087660-MAP2K1 relationship in the GTEx database suggests that this variant may influence MAP2K1 gene expression, particularly in the cerebellum and adipose subcutaneous tissues where the rs3087660 G allele increases expression. Comparable findings were also confirmed in peripheral blood and the cerebellum in the eQTLGen and BRAINEAC databases, respectively. Further evaluation of this relationship in our naturalistic TOP cohort of TRS and non-TRS SCZ patients showed a similar pattern in peripheral blood samples, where patients with the GG genotype had greater MAP2K1 gene expression when compared to patients with the GA or AA genotypes. Moreover, stratifying this cohort by TRS status suggests that this effect may be greater in non-TRS patients than in TRS patients. Our cFDR findings show that the rs3087660 G allele is associated with reduced risk of TRS. It is tempting to speculate that the increase in MAP2K1 activity expected after treatment with conventional antipsychotics may be facilitated by the presence of the rs3087660 G in non-TRS patients, and that MAP2K1 activity is reduced in TRS patients regardless of rs3087660 genotype. Interestingly, although CLZ is also shown to increase MAP2K1 activity (Browning et al., 2005), the mechanism is distinct from other antipsychotics(Pereira et al., 2012). Thus, response to CLZ in TRS patients may, in part, be due to the specific and unique manner in which CLZ influences MAP2K1 gene expression. These results provide a novel target mechanism that should be explored to better understand the mechanism of action underlying the superior efficacy of CLZ. Less is known about how the two other genes mapped to rs3087660, SNAPC5 and TIPIN, may relate to TRS.
Genes mapped to the other identified locus (rs7560232) include ZDBF2, CMKLR2 and ADAM23. The ZDBF2 gene is mostly expressed in the nucleus accumbens(Puighermanal et al., 2020), and has been shown to be downregulated in SCZ(Glatt et al., 2011; Stock et al., 2021), but it is not known how ZDBF2 expression relates to TRS. A recent study has shown reduced appetite and consequent reduced body weight gain after birth in ZDBF2 knock-out mice providing some evidence for how this locus relates to BMI (Glaser et al., 2022). Moreover, this locus has previously been implicated in other complex health-related traits including volume of the cerebellar vermal lobules(Zhao et al., 2019), lower self-rated health(Harris et al., 2017) and age-related hearing impairment(Praveen et al., 2022). Interestingly, the effects of both lead SNPs (rs3087660 and rs7560232), as well as all other candidate SNPs within these loci in LD with the lead SNPs, are in line with the observed relationship between antipsychotic treatment and metabolic dysfunction(Huhn et al., 2019; Leucht et al., 2013; Raben et al., 2017), that these variants reduce risk of TRS while increasing risk of higher BMI. Moreover, the receptor encoded by the CMKLR2 gene, mostly expressed in the central nervous system (CNS)(Helfer and Wu, 2018), has been shown to bind chemerin(De Henau et al., 2016; Helfer and Wu, 2018), an adipokine involved in inflammation, cell expansion, and angiogenesis in adipose tissue(Helfer and Wu, 2018). Chemerin activates hypothalamic stem cells to increase food intake, and has been positively associated with BMI, obesity, and metabolic disorders. Like CLZ, CMKLR2 activates the ERK1/2-MAPK pathways, which has been related to the superior efficacy of CLZ compared to other antipsychotics(Aringhieri et al., 2017). The other gene mapped to rs7560232, ADAM23, is widely expressed in the CNS and has also been involved in both immune(Elizondo et al., 2016) and adipocytes functions(Kim et al., 2012). Taken together, these findings suggest that similar underlying biological processes contribute to antipsychotic response and metabolic dysfunction, such as increased BMI, and supports the concept that metabolic disturbances may be a component underlying antipsychotic drug efficacy and not the result of off-target actions(Pillinger et al., 2020). This hypothesis, which needs to be investigated in more detail in future studies, is in line with the fact that CLZ exhibits the best clinical response in TRS but is also associated with the highest BMI increase(Pillinger et al., 2020).
By leveraging the identified genetic overlap between TRS and BMI, and re-ranking genetic variants according to the TRS|BMI cFDR values, we also show a 1.13 fold improvement in TRS polygenic prediction in the TDM cohort. This pleioPGS approach outperformed the standard GWAS-based ranking, utilizing less SNPs in the PGS, despite using the same SNP weightings. We hypothesized that improved PGS performance would be due to the inclusion of more informative SNPs, based on cFDR re-ranking, in the PGS model. This is supported by the successive inclusion of more highly ranked cFDR SNPs in the PGS constructed from the original TRS GWAS, and corresponding increase in explained variance observed, as more SNPs are added to the PGS model. Annotation of the variants contributing to the PGSs with greatest variance explained showed enrichment of intronic SNPs and depletion of intergenic and ncRNA intronic SNPs within those contributing to the cFDR PGS, while SNPs contributing to the original TRS PGS showed the opposite pattern with enrichment of intergenic and ncRNA intronic SNPs and depletion of intronic SNPs. Recent evidence suggests that intronic regions are enriched for regulatory elements of expression of genes involved in tissue-specific functions, while housekeeping genes are more often controlled by intergenic enhancers(Borsari et al., 2021). This suggests that the pleioPGS method may be prioritizing tissue-specific regulatory variants more likely to be involved in TRS aetiology and therefore more predictive of the outcome.
To validate these findings, we repeated this PGS analysis in the independent TOP cohort. We observed the expected greater variance explained by the TRS|BMI PGS compared to the standard TRS PGS for all SNP thresholds below 700 SNPs, however the greatest variance explained was by the standard TRS PGS at 700 SNPs. Variance explained by PGS in this validation cohort (maximum variance explained = 0.70%) was also considerably less than for the initial test cohort (maximum variance explained = 5.62%). This may be due to the smaller sample size of the TOP cohort, as well as, the difference in TRS definitions used. The TDM cohort TRS definition was based on CLZ use, matching the definition used in the TRS GWAS(Pardinas et al., 2022), while for the TOP cohort the TRS definition was based on two or more failed trials of antipsychotic treatment. Thus, the phenotype for the TDM cohort better matches the discovery GWAS from which PGS SNP weights were assigned.
The results of the cFDR analysis highlight how data from small GWAS might still be used to understand genetic architecture and overlap between traits. However, despite identifying novel loci for TRS, larger GWAS of TRS are still required to better understand the underlying genetic etiology. One limitation that the cFDR method inherits from the GWASs it draws upon is that it is agnostic regarding the specific causal variants underlying the overlapping genomic loci. These overlapping loci could result from both shared or separate causal variants, or “mediated pleiotropy”, where one phenotype is causative of the other (Solovieff et al., 2013). Another limitation to the cFDR approach is that the cross-trait enrichment both reflects the extent of polygenic overlap between the phenotypes and the power of the two GWASs analyzed and therefore, cross-trait enrichment will be harder to detect if one or both investigated GWASs are inadequately powered (Smeland et al., 2020). Moreover, the FDR values generated from the cFDR method cannot be used in a number of post-GWAS analyses which require p-values as input, such as MAGMA (de Leeuw et al., 2015) and colocalization (Giambartolomei et al., 2014; Hormozdiari et al., 2016). In addition, the predictive ability of the PGS remains far from being clinically relevant, and larger GWAS of TRS will also improve SNP weights for polygenic prediction. Moreover, these analyses were limited to European-ancestry participants due to available data and differences in linkage disequilibrium between ancestral groups. Larger, more diverse TRS samples, as well as new trans-ancestral methods and analyses are required to increase the generalizability of these findings. Finally, although other measures, such as waist-to-hip ratio, adiposity and body fat distribution may be considered more optimal markers of metabolic function (Okorodudu et al., 2010), BMI was investigated due to the power of the available GWASs for these traits. Thus, the shared genetic architecture of TRS and other measures of metabolic function should be investigated when more well powered data become available.
In conclusion, we identified two novel loci associated with TRS and showed improved prediction of TRS, by leveraging genetic overlap between TRS and BMI. In addition, we identified a novel target mechanism that should be explored to better understand the mechanism of action underlying the superior efficacy of CLZ. These findings confirm that shared genetic mechanisms influence both TRS and BMI and provide new insights into the biological underpinnings of metabolic dysfunction and response to antipsychotic treatment.
Supplementary Material
Acknowledgements
We thank the research participants, employees and researchers of the PGC, GIANT and TOP for making this research possible. This work was partly performed on the TSD (Tjeneste for Sensitive Data) facilities, owned by the University of Oslo, operated and developed by the TSD service group at the University of Oslo, IT-Department (USIT). Computations were also performed on resources provided by UNINETT Sigma2—the National Infrastructure for High Performance Computing and Data Storage in Norway. We gratefully acknowledge support from the American National Institutes of Health (R01MH124875, NS057198, EB00790), the Research Council of Norway (RCN) (248980, 296030, 229129, 213837, 324252, 300309, 273291, 223273), the South-East Norway Regional Health Authority (2022-073), and KG Jebsen Stiftelsen (SKGJ-MED-021). This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 847776 and 964874.
Footnotes
Conflict of Interest
Dr. Andreassen reported grants from Stiftelsen Kristian Gerhard Jebsen, South-East Regional Health Authority, Research Council of Norway, and European Union’s Horizon 2020 during the conduct of the study; personal fees from HealthLytix (stock options), Lundbeck (speaker’s honorarium), and Sunovion (speaker’s honorarium) outside the submitted work; and had a pending patent for systems and methods for identifying polymorphisms. Dr. Dale reported grants from the National Institutes of Health outside the submitted work; had a patent for US7324842 licensed to Siemens Healthineers; is a founder of and holds equity in Cortechs Labs and serves on its scientific advisory board; is a member of the scientific advisory board of Human Longevity; a member of the scientific advisory board of Healthlytix; and receives funding through a research agreement with GE Healthcare. No other disclosures were reported.
Code Availability
Code for cFDR and pleioFDR are publicly available at https://github.com/precimed/pleiofdr and https://github.com/norment/open-science/tree/main/2021_VanderMeer_medRxiv_pleioPGS, respectively.
References
- Andreassen OA, Djurovic S, Thompson WK, Schork AJ, Kendler KS, O’Donovan MC, Rujescu D, Werge T, van de Bunt M, Morris AP, McCarthy MI, International Consortium for Blood Pressure, G., Diabetes Genetics, R., Meta-analysis, C., Psychiatric Genomics Consortium Schizophrenia Working, G., Roddey JC, McEvoy LK, Desikan RS, Dale AM, 2013. Improved detection of common variants associated with schizophrenia by leveraging pleiotropy with cardiovascular-disease risk factors. Am J Hum Genet 92 (2), 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassen OA, Thompson WK, Dale AM, 2014. Boosting the power of schizophrenia genetics by leveraging new statistical tools. Schizophr Bull 40 (1), 13–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aringhieri S, Kolachalam S, Gerace C, Carli M, Verdesca V, Brunacci MG, Rossi C, Ippolito C, Solini A, Corsini GU, Scarselli M, 2017. Clozapine as the most efficacious antipsychotic for activating ERK 1/2 kinases: Role of 5-HT(2A) receptor agonism. Eur Neuropsychopharmacol 27 (4), 383–398. [DOI] [PubMed] [Google Scholar]
- Borsari B, Villegas-Miron P, Perez-Lluch S, Turpin I, Laayouni H, Segarra-Casas A, Bertranpetit J, Guigo R, Acosta S, 2021. Enhancers with tissue-specific activity are enriched in intronic regions. Genome Res 31 (8), 1325–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning JL, Patel T, Brandt PC, Young KA, Holcomb LA, Hicks PB, 2005. Clozapine and the mitogen-activated protein kinase signal transduction pathway: implications for antipsychotic actions. Biol Psychiatry 57 (6), 617–623. [DOI] [PubMed] [Google Scholar]
- Bulik-Sullivan BK, Loh P-R, Finucane H, Daly MJ, Patterson N, Price AL, Neale BM, 2015. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nature Genetics 47, 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, McMahon A, Morales J, Mountjoy E, Sollis E, Suveges D, Vrousgou O, Whetzel PL, Amode R, Guillen JA, Riat HS, Trevanion SJ, Hall P, Junkins H, Flicek P, Burdett T, Hindorff LA, Cunningham F, Parkinson H, 2019. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 47 (D1), D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SW, O’Reilly PF, 2019. PRSice-2: Polygenic Risk Score software for biobank-scale data. Gigascience 8 (7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium G, 2015. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348 (6235), 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czobor P, Volavka J, Sheitman B, Lindenmayer JP, Citrome L, McEvoy J, Cooper TB, Chakos M, Lieberman JA, 2002. Antipsychotic-induced weight gain and therapeutic response: a differential association. J Clin Psychopharmacol 22 (3), 244–251. [DOI] [PubMed] [Google Scholar]
- de Bartolomeis A, Barone A, Begni V, Riva MA, 2022. Present and future antipsychotic drugs: A systematic review of the putative mechanisms of action for efficacy and a critical appraisal under a translational perspective. Pharmacol Res 176, 106078. [DOI] [PubMed] [Google Scholar]
- De Henau O, Degroot GN, Imbault V, Robert V, De Poorter C, McHeik S, Gales C, Parmentier M, Springael JY, 2016. Signaling Properties of Chemerin Receptors CMKLR1, GPR1 and CCRL2. PLoS One 11 (10), e0164179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw CA, Mooij JM, Heskes T, Posthuma D, 2015. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 11 (4), e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elizondo DM, Andargie TE, Marshall KM, Zariwala AM, Lipscomb MW, 2016. Dendritic cell expression of ADAM23 governs T cell proliferation and cytokine production through the alpha(v)beta(3) integrin receptor. J Leukoc Biol 100 (5), 855–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel SR, Creson TK, Hao Y, Shen Y, Maeng S, Nekrasova T, Landreth GE, Manji HK, Chen G, 2009. The extracellular signal-regulated kinase pathway contributes to the control of behavioral excitement. Mol Psychiatry 14 (4), 448–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA, 2014. U.S. Food and Drug Administration, Clozaril, label approval, NDA no. 019758
- Garriga M, Mallorqui A, Bernad S, Ruiz-Cortes V, Oliveira C, Amoretti S, Mezquida G, Bioque M, Molina O, Gomez-Ramiro M, Vieta E, Bernardo M, Parellada E, Garcia-Rizo C, 2022. Antipsychotic-Associated Weight Gain and Clinical Improvement Under Clozapine Treatment. J Clin Psychopharmacol 42 (1), 75–80. [DOI] [PubMed] [Google Scholar]
- Ghoussaini M, Mountjoy E, Carmona M, Peat G, Schmidt EM, Hercules A, Fumis L, Miranda A, Carvalho-Silva D, Buniello A, Burdett T, Hayhurst J, Baker J, Ferrer J, Gonzalez-Uriarte A, Jupp S, Karim MA, Koscielny G, Machlitt-Northen S, Malangone C, Pendlington ZM, Roncaglia P, Suveges D, Wright D, Vrousgou O, Papa E, Parkinson H, MacArthur JAL, Todd JA, Barrett JC, Schwartzentruber J, Hulcoop DG, Ochoa D, McDonagh EM, Dunham I, 2021. Open Targets Genetics: systematic identification of trait-associated genes using large-scale genetics and functional genomics. Nucleic Acids Res 49 (D1), D1311–D1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, Plagnol V, 2014. Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics. PLoS genetics 10 (5), e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie AL, Samanaite R, Mill J, Egerton A, MacCabe JH, 2017. Is treatment-resistant schizophrenia categorically distinct from treatment-responsive schizophrenia? a systematic review. BMC Psychiatry 17 (1), 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser J, Iranzo J, Borensztein M, Marinucci M, Gualtieri A, Jouhanneau C, Teissandier A, Gaston-Massuet C, Bourc’his D, 2022. The imprinted Zdbf2 gene finely tunes control of feeding and growth in neonates. Elife 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatt SJ, Stone WS, Nossova N, Liew CC, Seidman LJ, Tsuang MT, 2011. Similarities and differences in peripheral blood gene-expression signatures of individuals with schizophrenia and their first-degree biological relatives. Am J Med Genet B Neuropsychiatr Genet 156B (8), 869–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurrera RJ, Gearin PF, Love J, Li KJ, Xu A, Donaghey FH, Gerace MR, 2022. Recognition and management of clozapine adverse effects: A systematic review and qualitative synthesis. Acta Psychiatr Scand 145 (5), 423–441. [DOI] [PubMed] [Google Scholar]
- Harris SE, Hagenaars SP, Davies G, David Hill W, Liewald DCM, Ritchie SJ, Marioni RE, Metastroke Consortium IC f.B.P.G.-W.A.S., International Consortium for Blood Pressure Genome-Wide Association, S., Aging, C.C., Longevity, G., Group, C.C.C., Sudlow CLM, Wardlaw JM, McIntosh AM, Gale CR, Deary IJ, 2017. Molecular genetic contributions to self-rated health. Int J Epidemiol 46 (3), 994–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PJ, Weinberger DR, 2005. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry 10 (1), 40–68; image 45. [DOI] [PubMed] [Google Scholar]
- Helfer G, Wu QF, 2018. Chemerin: a multifaceted adipokine involved in metabolic disorders. J Endocrinol 238 (2), R79–R94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgeland O, Vaudel M, Sole-Navais P, Flatley C, Juodakis J, Bacelis J, Koloen IL, Knudsen GP, Johansson BB, Magnus P, Kjennerud TR, Juliusson PB, Stoltenberg C, Holmen OL, Andreassen OA, Jacobsson B, Njolstad PR, Johansson S, 2022. Characterization of the genetic architecture of infant and early childhood body mass index. Nat Metab 4 (3), 344–358. [DOI] [PubMed] [Google Scholar]
- Hernandez DG, Nalls MA, Moore M, Chong S, Dillman A, Trabzuni D, Gibbs JR, Ryten M, Arepalli S, Weale ME, Zonderman AB, Troncoso J, O’Brien R, Walker R, Smith C, Bandinelli S, Traynor BJ, Hardy J, Singleton AB, Cookson MR, 2012. Integration of GWAS SNPs and tissue specific expression profiling reveal discrete eQTLs for human traits in blood and brain. Neurobiol Dis 47 (1), 20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiemke C, Bergemann N, Clement HW, Conca A, Deckert J, Domschke K, Eckermann G, Egberts K, Gerlach M, Greiner C, Grunder G, Haen E, Havemann-Reinecke U, Hefner G, Helmer R, Janssen G, Jaquenoud E, Laux G, Messer T, Mossner R, Muller MJ, Paulzen M, Pfuhlmann B, Riederer P, Saria A, Schoppek B, Schoretsanitis G, Schwarz M, Gracia MS, Stegmann B, Steimer W, Stingl JC, Uhr M, Ulrich S, Unterecker S, Waschgler R, Zernig G, Zurek G, Baumann P, 2018. Consensus Guidelines for Therapeutic Drug Monitoring in Neuropsychopharmacology: Update 2017. Pharmacopsychiatry 51 (1–02), 9–62. [DOI] [PubMed] [Google Scholar]
- Hormozdiari F, van de Bunt M, Segre AV, Li X, Joo JWJ, Bilow M, Sul JH, Sankararaman S, Pasaniuc B, Eskin E, 2016. Colocalization of GWAS and eQTL Signals Detects Target Genes. Am J Hum Genet 99 (6), 1245–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huhn M, Nikolakopoulou A, Schneider-Thoma J, Krause M, Samara M, Peter N, Arndt T, Backers L, Rothe P, Cipriani A, Davis J, Salanti G, Leucht S, 2019. Comparative efficacy and tolerability of 32 oral antipsychotics for the acute treatment of adults with multi-episode schizophrenia: a systematic review and network meta-analysis. Lancet 394 (10202), 939–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HA, Park WJ, Jeong HS, Lee HE, Lee SH, Kwon NS, Baek KJ, Kim DS, Yun HY, 2012. Leucine-rich glioma inactivated 3 regulates adipogenesis through ADAM23. Biochim Biophys Acta 1821 (6), 914–922. [DOI] [PubMed] [Google Scholar]
- Kinon BJ, 2018. The Group of Treatment Resistant Schizophrenias. Heterogeneity in Treatment Resistant Schizophrenia (TRS). Front Psychiatry 9, 757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konradi C, Heckers S, 2001. Antipsychotic drugs and neuroplasticity: insights into the treatment and neurobiology of schizophrenia. Biol Psychiatry 50 (10), 729–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumra S, Kranzler H, Gerbino-Rosen G, Kester HM, De Thomas C, Kafantaris V, Correll CU, Kane JM, 2008. Clozapine and “high-dose” olanzapine in refractory early-onset schizophrenia: a 12-week randomized and double-blind comparison. Biol Psychiatry 63 (5), 524–529. [DOI] [PubMed] [Google Scholar]
- Lally J, Gaughran F, Timms P, Curran SR, 2016. Treatment-resistant schizophrenia: current insights on the pharmacogenomics of antipsychotics. Pharmgenomics Pers Med 9, 117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leucht S, Cipriani A, Spineli L, Mavridis D, Orey D, Richter F, Samara M, Barbui C, Engel RR, Geddes JR, Kissling W, Stapf MP, Lassig B, Salanti G, Davis JM, 2013. Comparative efficacy and tolerability of 15 antipsychotic drugs in schizophrenia: a multiple-treatments meta-analysis. Lancet 382 (9896), 951–962. [DOI] [PubMed] [Google Scholar]
- Liu B, Gloudemans MJ, Rao AS, Ingelsson E, Montgomery SB, 2019. Abundant associations with gene expression complicate GWAS follow-up. Nat Genet 51 (5), 768–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meltzer HY, 1997. Treatment-resistant schizophrenia--the role of clozapine. Curr Med Res Opin 14 (1), 1–20. [DOI] [PubMed] [Google Scholar]
- Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, Jia P, Assadzadeh A, Flanagan J, Schumacher A, Wang SC, Petronis A, 2008. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet 82 (3), 696–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okorodudu DO, Jumean MF, Montori VM, Romero-Corral A, Somers VK, Erwin PJ, Lopez-Jimenez F, 2010. Diagnostic performance of body mass index to identify obesity as defined by body adiposity: a systematic review and meta-analysis. Int J Obes (Lond) 34 (5), 791–799. [DOI] [PubMed] [Google Scholar]
- Pardinas AF, Owen MJ, Walters JTR, 2021. Pharmacogenomics: A road ahead for precision medicine in psychiatry. Neuron 109 (24), 3914–3929. [DOI] [PubMed] [Google Scholar]
- Pardinas AF, Smart SE, Willcocks IR, Holmans PA, Dennison CA, Lynham AJ, Legge SE, Baune BT, Bigdeli TB, Cairns MJ, Corvin A, Fanous AH, Frank J, Kelly B, McQuillin A, Melle I, Mortensen PB, Mowry BJ, Pato CN, Periyasamy S, Rietschel M, Rujescu D, Simonsen C, St Clair D, Tooney P, Wu JQ, Andreassen OA, Kowalec K, Sullivan PF, Murray RM, Owen MJ, MacCabe JH, O’Donovan MC, Walters JTR, Genetics Workstream of the Schizophrenia Treatment, R., Therapeutic Advances, C., the Schizophrenia Working Group of the Psychiatric Genomics, C., Ajnakina O, Alameda L, Barnes TRE, Berardi D, Bonora E, Camporesi S, Cleusix M, Conus P, Crespo-Facorro B, D’Andrea G, Demjaha A, Do KQ, Doody GA, Eap CB, Ferchiou A, Di Forti M, Guidi L, Homman L, Jenni R, Joyce EM, Kassoumeri L, Khadimallah I, Lastrina O, Muratori R, Noyan H, O’Neill FA, Pignon B, Restellini R, Richard JR, Schurhoff F, Spaniel F, Szoke A, Tarricone I, Tortelli A, Ucok A, Vazquez-Bourgon J, 2022. Interaction Testing and Polygenic Risk Scoring to Estimate the Association of Common Genetic Variants With Treatment Resistance in Schizophrenia. JAMA Psychiatry 79 (3), 260–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira A, Sugiharto-Winarno A, Zhang B, Malcolm P, Fink G, Sundram S, 2012. Clozapine induction of ERK1/2 cell signalling via the EGF receptor in mouse prefrontal cortex and striatum is distinct from other antipsychotic drugs. Int J Neuropsychopharmacol 15 (8), 1149–1160. [DOI] [PubMed] [Google Scholar]
- Pillinger T, McCutcheon RA, Vano L, Mizuno Y, Arumuham A, Hindley G, Beck K, Natesan S, Efthimiou O, Cipriani A, Howes OD, 2020. Comparative effects of 18 antipsychotics on metabolic function in patients with schizophrenia, predictors of metabolic dysregulation, and association with psychopathology: a systematic review and network meta-analysis. Lancet Psychiatry 7 (1), 64–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praveen K, Dobbyn L, Gurski L, Ayer AH, Staples J, Mishra S, Bai Y, Kaufman A, Moscati A, Benner C, Chen E, Chen S, Popov A, Smith J, collaboration G-RD, Regeneron Genetics C, Decibel R.c., Melander O, Jones MB, Marchini J, Balasubramanian S, Zambrowicz B, Drummond MC, Baras A, Abecasis GR, Ferreira MA, Stahl EA, Coppola G, 2022. Population-scale analysis of common and rare genetic variation associated with hearing loss in adults. Commun Biol 5 (1), 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puighermanal E, Castell L, Esteve-Codina A, Melser S, Kaganovsky K, Zussy C, Boubaker-Vitre J, Gut M, Rialle S, Kellendonk C, Sanz E, Quintana A, Marsicano G, Martin M, Rubinstein M, Girault JA, Ding JB, Valjent E, 2020. Functional and molecular heterogeneity of D2R neurons along dorsal ventral axis in the striatum. Nat Commun 11 (1), 1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team, 2020. R: A language and environment for statistical computing R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Raben AT, Marshe VS, Chintoh A, Gorbovskaya I, Muller DJ, Hahn MK, 2017. The Complex Relationship between Antipsychotic-Induced Weight Gain and Therapeutic Benefits: A Systematic Review and Implications for Treatment. Front Neurosci 11, 741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen C, Sundet K, Vaskinn A, Birkenaes AB, Engh JA, Færden A, Jónsdóttir H, Ringen PA, Opjordsmoen S, Melle I, Friis S, Andreassen OA, 2011. Neurocognitive dysfunction in bipolar and schizophrenia spectrum disorders depends on history of psychosis rather than diagnostic group. Schizophr Bull 37 (1), 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeland OB, Frei O, Shadrin A, O’Connell K, Fan CC, Bahrami S, Holland D, Djurovic S, Thompson WK, Dale AM, Andreassen OA, 2020. Discovery of shared genomic loci using the conditional false discovery rate approach. Hum Genet 139 (1), 85–94. [DOI] [PubMed] [Google Scholar]
- Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW, 2013. Pleiotropy in complex traits: challenges and strategies. Nat Rev Genet 14 (7), 483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock R, Jeckel P, Kraushaar U, Wust R, Fallgatter A, Volkmer H, 2021. The potential of induced pluripotent stem cells for discriminating neurodevelopmental disorders. Stem Cells Transl Med 10 (1), 50–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo A, O’Connell KS, Ueland T, Sheikh MA, Agartz I, Andreou D, Aukrust P, Boye B, Boen E, Drange OK, Elvsashagen T, Engh JA, Hope S, Collier Hoegh M, Joa I, Johnsen E, Kroken RA, Vik Lagerberg T, Lekva T, Malt UF, Melle I, Morken G, Naerland T, Steen VM, Sorensen K, Wedervang-Resell K, Auten Weibell M, Westlye LT, Steen NE, Andreassen O, Djurovic S, 2022. Increased circulating IL-18 levels in severe mental disorders indicate systemic inflammasome activation. Brain Behav Immun 99, 299–306. [DOI] [PubMed] [Google Scholar]
- Trubetskoy V, Pardinas AF, Qi T, Panagiotaropoulou G, Awasthi S, Bigdeli TB, Bryois J, Chen CY, Dennison CA, Hall LS, Lam M, Watanabe K, Frei O, Ge T, Harwood JC, Koopmans F, Magnusson S, Richards AL, Sidorenko J, Wu Y, Zeng J, Grove J, Kim M, Li Z, Voloudakis G, Zhang W, Adams M, Agartz I, Atkinson EG, Agerbo E, Al Eissa M, Albus M, Alexander M, Alizadeh BZ, Alptekin K, Als TD, Amin F, Arolt V, Arrojo M, Athanasiu L, Azevedo MH, Bacanu SA, Bass NJ, Begemann M, Belliveau RA, Bene J, Benyamin B, Bergen SE, Blasi G, Bobes J, Bonassi S, Braun A, Bressan RA, Bromet EJ, Bruggeman R, Buckley PF, Buckner RL, Bybjerg-Grauholm J, Cahn W, Cairns MJ, Calkins ME, Carr VJ, Castle D, Catts SV, Chambert KD, Chan RCK, Chaumette B, Cheng W, Cheung EFC, Chong SA, Cohen D, Consoli A, Cordeiro Q, Costas J, Curtis C, Davidson M, Davis KL, de Haan L, Degenhardt F, DeLisi LE, Demontis D, Dickerson F, Dikeos D, Dinan T, Djurovic S, Duan J, Ducci G, Dudbridge F, Eriksson JG, Fananas L, Faraone SV, Fiorentino A, Forstner A, Frank J, Freimer NB, Fromer M, Frustaci A, Gadelha A, Genovese G, Gershon ES, Giannitelli M, Giegling I, Giusti-Rodriguez P, Godard S, Goldstein JI, Gonzalez Penas J, Gonzalez-Pinto A, Gopal S, Gratten J, Green MF, Greenwood TA, Guillin O, Guloksuz S, Gur RE, Gur RC, Gutierrez B, Hahn E, Hakonarson H, Haroutunian V, Hartmann AM, Harvey C, Hayward C, Henskens FA, Herms S, Hoffmann P, Howrigan DP, Ikeda M, Iyegbe C, Joa I, Julia A, Kahler AK, Kam-Thong T, Kamatani Y, Karachanak-Yankova S, Kebir O, Keller MC, Kelly BJ, Khrunin A, Kim SW, Klovins J, Kondratiev N, Konte B, Kraft J, Kubo M, Kucinskas V, Kucinskiene ZA, Kusumawardhani A, Kuzelova-Ptackova H, Landi S, Lazzeroni LC, Lee PH, Legge SE, Lehrer DS, Lencer R, Lerer B, Li M, Lieberman J, Light GA, Limborska S, Liu CM, Lonnqvist J, Loughland CM, Lubinski J, Luykx JJ, Lynham A, Macek M Jr., Mackinnon A, Magnusson PKE, Maher BS, Maier W, Malaspina D, Mallet J, Marder SR, Marsal S, Martin AR, Martorell L, Mattheisen M, McCarley RW, McDonald C, McGrath JJ, Medeiros H, Meier S, Melegh B, Melle I, Mesholam-Gately RI, Metspalu A, Michie PT, Milani L, Milanova V, Mitjans M, Molden E, Molina E, Molto MD, Mondelli V, Moreno C, Morley CP, Muntane G, Murphy KC, Myin-Germeys I, Nenadic I, Nestadt G, Nikitina-Zake L, Noto C, Nuechterlein KH, O’Brien NL, O’Neill FA, Oh SY, Olincy A, Ota VK, Pantelis C, Papadimitriou GN, Parellada M, Paunio T, Pellegrino R, Periyasamy S, Perkins DO, Pfuhlmann B, Pietilainen O, Pimm J, Porteous D, Powell J, Quattrone D, Quested D, Radant AD, Rampino A, Rapaport MH, Rautanen A, Reichenberg A, Roe C, Roffman JL, Roth J, Rothermundt M, Rutten BPF, Saker-Delye S, Salomaa V, Sanjuan J, Santoro ML, Savitz A, Schall U, Scott RJ, Seidman LJ, Sharp SI, Shi J, Siever LJ, Sigurdsson E, Sim K, Skarabis N, Slominsky P, So HC, Sobell JL, Soderman E, Stain HJ, Steen NE, Steixner-Kumar AA, Stogmann E, Stone WS, Straub RE, Streit F, Strengman E, Stroup TS, Subramaniam M, Sugar CA, Suvisaari J, Svrakic DM, Swerdlow NR, Szatkiewicz JP, Ta TMT, Takahashi A, Terao C, Thibaut F, Toncheva D, Tooney PA, Torretta S, Tosato S, Tura GB, Turetsky BI, Ucok A, Vaaler A, van Amelsvoort T, van Winkel R, Veijola J, Waddington J, Walter H, Waterreus A, Webb BT, Weiser M, Williams NM, Witt SH, Wormley BK, Wu JQ, Xu Z, Yolken R, Zai CC, Zhou W, Zhu F, Zimprich F, Atbasoglu EC, Ayub M, Benner C, Bertolino A, Black DW, Bray NJ, Breen G, Buccola NG, Byerley WF, Chen WJ, Cloninger CR, Crespo-Facorro B, Donohoe G, Freedman R, Galletly C, Gandal MJ, Gennarelli M, Hougaard DM, Hwu HG, Jablensky AV, McCarroll SA, Moran JL, Mors O, Mortensen PB, Muller-Myhsok B, Neil AL, Nordentoft M, Pato MT, Petryshen TL, Pirinen M, Pulver AE, Schulze TG, Silverman JM, Smoller JW, Stahl EA, Tsuang DW, Vilella E, Wang SH, Xu S, Indonesia Schizophrenia C, PsychEncode, Psychosis Endophenotypes International, C., Syn, G.O.C., Adolfsson R, Arango C, Baune BT, Belangero SI, Borglum AD, Braff D, Bramon E, Buxbaum JD, Campion D, Cervilla JA, Cichon S, Collier DA, Corvin A, Curtis D, Forti MD, Domenici E, Ehrenreich H, Escott-Price V, Esko T, Fanous AH, Gareeva A, Gawlik M, Gejman PV, Gill M, Glatt SJ, Golimbet V, Hong KS, Hultman CM, Hyman SE, Iwata N, Jonsson EG, Kahn RS, Kennedy JL, Khusnutdinova E, Kirov G, Knowles JA, Krebs MO, Laurent-Levinson C, Lee J, Lencz T, Levinson DF, Li QS, Liu J, Malhotra AK, Malhotra D, McIntosh A, McQuillin A, Menezes PR, Morgan VA, Morris DW, Mowry BJ, Murray RM, Nimgaonkar V, Nothen MM, Ophoff RA, Paciga SA, Palotie A, Pato CN, Qin S, Rietschel M, Riley BP, Rivera M, Rujescu D, Saka MC, Sanders AR, Schwab SG, Serretti A, Sham PC, Shi Y, St Clair D, Stefansson H, Stefansson K, Tsuang MT, van Os J, Vawter MP, Weinberger DR, Werge T, Wildenauer DB, Yu X, Yue W, Holmans PA, Pocklington AJ, Roussos P, Vassos E, Verhage M, Visscher PM, Yang J, Posthuma D, Andreassen OA, Kendler KS, Owen MJ, Wray NR, Daly MJ, Huang H, Neale BM, Sullivan PF, Ripke S, Walters JTR, O’Donovan MC, Schizophrenia Working Group of the Psychiatric Genomics, C., 2022. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 604 (7906), 502–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer D, Shadrin AA, O’Connell K, Bettella F, Djurovic S, Wolfers T, Alnaes D, Agartz I, Smeland OB, Melle I, Sanchez JM, Linden DEJ, Dale AM, Westlye LT, Andreassen OA, Frei O, Kaufmann T, 2022. Boosting Schizophrenia Genetics by Utilizing Genetic Overlap With Brain Morphology. Biol Psychiatry [DOI] [PubMed] [Google Scholar]
- Vosa U, Claringbould A, Westra HJ, Bonder MJ, Deelen P, Zeng B, Kirsten H, Saha A, Kreuzhuber R, Yazar S, Brugge H, Oelen R, de Vries DH, van der Wijst MGP, Kasela S, Pervjakova N, Alves I, Fave MJ, Agbessi M, Christiansen MW, Jansen R, Seppala I, Tong L, Teumer A, Schramm K, Hemani G, Verlouw J, Yaghootkar H, Sonmez Flitman R, Brown A, Kukushkina V, Kalnapenkis A, Rueger S, Porcu E, Kronberg J, Kettunen J, Lee B, Zhang F, Qi T, Hernandez JA, Arindrarto W, Beutner F, Consortium B, i, Q.T.L.C., Dmitrieva J, Elansary M, Fairfax BP, Georges M, Heijmans BT, Hewitt AW, Kahonen M, Kim Y, Knight JC, Kovacs P, Krohn K, Li S, Loeffler M, Marigorta UM, Mei H, Momozawa Y, Muller-Nurasyid M, Nauck M, Nivard MG, Penninx B, Pritchard JK, Raitakari OT, Rotzschke O, Slagboom EP, Stehouwer CDA, Stumvoll M, Sullivan P, t Hoen PAC, Thiery J, Tonjes A, van Dongen J, van Iterson M, Veldink JH, Volker U, Warmerdam R, Wijmenga C, Swertz M, Andiappan A, Montgomery GW, Ripatti S, Perola M, Kutalik Z, Dermitzakis E, Bergmann S, Frayling T, van Meurs J, Prokisch H, Ahsan H, Pierce BL, Lehtimaki T, Boomsma DI, Psaty BM, Gharib SA, Awadalla P, Milani L, Ouwehand WH, Downes K, Stegle O, Battle A, Visscher PM, Yang J, Scholz M, Powell J, Gibson G, Esko T, Franke L, 2021. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat Genet 53 (9), 1300–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Su P, Yang J, Xu L, Yuan A, Li C, Zhang T, Dong F, Zhou J, Samsom J, Wong AHC, Liu F, 2022. The D2R-DISC1 protein complex and associated proteins are altered in schizophrenia and normalized with antipsychotic treatment. J Psychiatry Neurosci 47 (2), E134–E147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Stringer S, Frei O, Umicevic Mirkov M, de Leeuw C, Polderman TJC, van der Sluis S, Andreassen OA, Neale BM, Posthuma D, 2019. A global overview of pleiotropy and genetic architecture in complex traits. Nat Genet 51 (9), 1339–1348. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Taskesen E, van Bochoven A, Posthuma D, 2017. Functional mapping and annotation of genetic associations with FUMA. Nat Commun 8 (1), 1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner MCF, Wirgenes KV, Haram M, Bettella F, Lunding SH, Rodevand L, Hjell G, Agartz I, Djurovic S, Melle I, Andreassen OA, Steen NE, 2020. Indicated association between polygenic risk score and treatment-resistance in a naturalistic sample of patients with schizophrenia spectrum disorders. Schizophr Res 218, 55–62. [DOI] [PubMed] [Google Scholar]
- Yengo L, Sidorenko J, Kemper KE, Zheng Z, Wood AR, Weedon MN, Frayling TM, Hirschhorn J, Yang J, Visscher PM, Consortium G, 2018. Meta-analysis of genome-wide association studies for height and body mass index in approximately 700000 individuals of European ancestry. Hum Mol Genet 27 (20), 3641–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Luo T, Li T, Li Y, Zhang J, Shan Y, Wang X, Yang L, Zhou F, Zhu Z, Alzheimer’s Disease Neuroimaging, I., Pediatric Imaging, Genetics N, Zhu H, 2019. Genome-wide association analysis of 19,629 individuals identifies variants influencing regional brain volumes and refines their genetic co-architecture with cognitive and mental health traits. Nat Genet 51 (11), 1637–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Zheng Z, Zhang F, Wu Y, Trzaskowski M, Maier R, Robinson MR, McGrath JJ, Visscher PM, Wray NR, Yang J, 2018. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Commun 9 (1), 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


