Abstract
White matter hyperintensities (WMHs) are well‐established markers of cerebral small vessel disease, and are associated with an increased risk of stroke, dementia, and mortality. Although their prevalence increases with age, small and punctate WMHs have been reported with surprisingly high frequency even in young, neurologically asymptomatic adults. However, most automated methods to segment WMH published to date are not optimized for detecting small and sparse WMH. Here we present the SHIVA‐WMH tool, a deep‐learning (DL)‐based automatic WMH segmentation tool that has been trained with manual segmentations of WMH in a wide range of WMH severity. We show that it is able to detect WMH with high efficiency in subjects with only small punctate WMH as well as in subjects with large WMHs (i.e., with confluency) in evaluation datasets from three distinct databases: magnetic resonance imaging‐Share consisting of young university students, MICCAI 2017 WMH challenge dataset consisting of older patients from memory clinics, and UK Biobank with community‐dwelling middle‐aged and older adults. Across these three cohorts with a wide‐ranging WMH load, our tool achieved voxel‐level and individual lesion cluster‐level Dice scores of 0.66 and 0.71, respectively, which were higher than for three reference tools tested: the lesion prediction algorithm implemented in the lesion segmentation toolbox (LPA: Schmidt), PGS tool, a DL‐based algorithm and the current winner of the MICCAI 2017 WMH challenge (Park et al.), and HyperMapper tool (Mojiri Forooshani et al.), another DL‐based method with high reported performance in subjects with mild WMH burden. Our tool is publicly and openly available to the research community to facilitate investigations of WMH across a wide range of severity in other cohorts, and to contribute to our understanding of the emergence and progression of WMH.
Keywords: automatic segmentation, cerebral small vessel disease, deep‐learning, magnetic resonance imaging, white matter hyperintensities
We propose a novel 3D Unet‐based model, SHIVA‐white matter hyperintensity (WMH) detector, with much improved detection of small WMH across subjects with a wide range of WMH burden compared to existing methods. It can be a valuable tool for characterizing much understudied, early WMH found even in young adults.
1. INTRODUCTION
Cerebral small vessel disease (cSVD) represents a spectrum of pathological processes affecting small vessels of the brain. It is a leading vascular cause of dementia and accounts for up to 25% of strokes (Cannistraro et al., 2019; Wardlaw et al., 2013). Most often, cSVD is covert and can be detected on brain imaging of individuals without clinical manifestation of stroke. White matter hyperintensity (WMH) is one of the most well‐established imaging markers of cSVD, and is characterized by heightened signal intensity on T2‐weighted‐fluid‐attenuated inversion recovery (FLAIR) sequences of magnetic resonance imaging (MRI). WMH of presumed vascular origin is highly prevalent in neurologically asymptomatic older individuals, and is associated with an increased risk of stroke, cognitive decline, dementia, and mortality (Debette et al., 2019; Debette & Markus, 2010; Wardlaw et al., 2021). So far, the exact etiology and pathogenesis of these age‐related WMHs remain elusive, though they are known to be associated with common cardiovascular risk factors, including smoking and hypertension (Moroni et al., 2018). Regardless, they represent an important preclinical biomarker of cSVD that should trigger preventive interventions to reduce the risk of stroke and dementia and can be used as a surrogate endpoint in clinical trials (Wardlaw et al., 2021).
Although their prevalence increases with age, small and punctate WMHs have been reported with surprisingly high frequency even in young adults under 40 years of age (Keřkovský et al., 2019; Wadhwa et al., 2019; Wang et al., 2019; Williamson et al., 2018). If they represent early forms of covert cSVD, it is crucial to investigate their emergence and progression in order to study their pathophysiological correlates as well as their association with genetic, environmental, and behavioral risk factors. Clinically, the presence and severity of WMH on MRI are most commonly assessed with visual rating scales, such as the Fazekas (Fazekas et al., 1987) or more anatomically detailed Scheltens Scale (Scheltens et al., 1993) and the Age‐Related White Matter Changes (ARWMC) scale (Wahlund et al., 2001). They are all point scales expressing the severity of WMH, with grade 0 indicating no lesion and increasing grade indicating the size and/or number of the lesion, as well as the degree of confluency. While they can be effective for differentiating the most severe cases of WMH from milder cases, these clinical scales are inadequate for the characterization of subjects with the early stages of cSVD, who would at most receive a grade of 1. They also provide very limited information on the spatial extent and distribution of WMH, only distinguishing between WMH found in the periventricular region from deep white matter (for Fazekas) or different lobes in each hemisphere (for ARWMC). It is therefore essential to have automated methods to segment WMH in order to quantify and provide precise spatial information of any lesions in cohorts across the disease spectrum.
While numerous WMH segmentation tools and algorithms exist, including an increasing number of deep‐learning (DL) based methods in recent years, there is a lack of automated methods that have been validated in populations with low prevalence and small overall lesion load, hampering the detailed characterization of WMH in young subjects. Indeed, most methods published to date are optimized for detection in older subjects (Guerrero et al., 2018; Li et al., 2018, 2022; Sundaresan et al., 2021; Umapathy et al., 2021) or patients with multiple sclerosis (MS; reviewed in Zeng et al., 2020) who typically manifest a higher load of WMH with sharper boundaries and large confluent lesions. In these populations, the advantage of more advanced DL‐based methods over more traditional signal‐processing and machine‐learning‐based methods is reported to be minimal (Balakrishnan et al., 2021). The true advantage of these advanced methods may be more evident for the segmentation of WMH in subjects with relatively mild lesion load (<5 ml), as recently suggested (Khademi et al., 2021; Li et al., 2022; Rachmadi et al., 2018). However, even those studies evaluating their method in subjects with mild WMH burden with DL‐based (Khademi et al., 2021; Rachmadi et al., 2018) or other approaches (Ong et al., 2022; Rachmadi et al., 2020) primarily use databases with 2D FLAIR acquisition with the slice thickness ranging from 3 to 5 mm, which precludes detection of small WMH not on the plane of acquisition. To our knowledge, no study has explicitly optimized the segmentation performance in high‐quality 3D FLAIR scans from healthy young‐ to middle‐aged adults with very mild WMH burden.
Among the DL‐based approaches, Unet‐based architecture (Ronneberger et al., 2015) has been by far the most popular and successful, with the top two winning methods in the MICCAI 2017 WMH segmentation challenge (MWC: Kuijf et al., 2019) using variations of Unet architecture (Li et al., 2018; Park et al., 2021). Designed for biomedical image segmentation tasks that require pixel‐by‐pixel classifications, Unet offers an elegant architecture that allows efficient learning from limited sources of training images (Ronneberger et al., 2015). Unlike many publicized neural networks for image classification that are typically trained on a large annotated dataset (e.g., ImageNet dataset with ~1.2 million training images), Ronneberger et al. (2015) demonstrated a successful segmentation of neuronal structure by their Unet model trained with only 30 training dataset of electron microscopy images. The model uses successive contracting layers of fully convolutional network, followed by the upsampling operators that are more or less symmetric to the contracting path, resulting in the characteristic “u‐shape” architecture (and hence their name). The skip connections that connect feature maps of the contracting path to the expanding path help preserve spatial information and allow the model to learn fine‐grained detail with the full spatial context of the input image. Li et al. (2018) and Park et al. (2021) both used the Unet architecture with 2D input, partly because the imaging resolution along z‐direction of the MWC training dataset is rather poor and vary from one data source to the other.
In the present study, we describe the development of a 3D Unet‐based tool we call “SHIVA‐WMH” detector, optimized to detect a full range of WMH severity, including very mild cases that can be observed in young subjects from general population. Our tool was developed in the context of the SHIVA project (https://rhu-shiva.com/), whose aim is to prevent cognitive decline and dementia through a better understanding of cSVD, and is based on the architecture of “SHIVA‐perivascular spaces (PVS)” detector we described previously (Boutinaud et al., 2021: https://github.com/pboutinaud/SHIVA_PVS) that segments PVS, another marker of covert cSVD (Wardlaw et al., 2013; Yu et al., 2022). We took advantage of the 1 mm isotropic 3D FLAIR and T1‐weighted anatomical scans of the unique, large neuroimaging database of French university students called MRi‐Share (Tsuchida et al., 2021). The whole‐brain manual segmentation of WMH in subsample of MRi‐Share subjects were combined with the publicly available MRI from the MWC training data (Kuijf et al., 2019) to cover the full range of age‐related WMH severity when tuning and evaluating our model. We aimed to create a ready‐to‐use tool that can chart the emergence and progression of WMH across the adult lifespan in multicohort studies with varying age ranges. Our SHIVA‐WMH detector tool with pretrained models is made openly available at (https://github.com/pboutinaud/SHIVA_WMH) to other researchers to encourage replication and further research into the earliest forms of WMH.
2. MATERIALS AND METHODS
2.1. Overview
The present paper first describes the development of SHIVA‐WMH detector, trained with manually traced WMH from 90 subjects (40 MRi‐Share and 50 MWC) and synthetic WMH from 360 other subjects (all from MRi‐Share). A separate set of 10 subjects each from manually traced MRi‐Share and MWC, as well as 11 subjects from the UK Biobank data (Alfaro‐Almagro et al., 2018), served as an independent, held‐out evaluation set (total of 31 subjects). The latter represented a sample from a cohort never seen during training and constituted an important test of generalizability of the tool performance.
We then describe the evaluation of our tool against three reference WMH segmentation tools that could be applied out‐of‐the‐box (i.e., without retraining or fine‐tuning) in the held‐out test set: (1) lesion prediction algorithm implemented in the lesion segmentation toolbox (LPA‐LST; Schmidt, 2017a; Schmidt, 2017b), a clinical reference tool based on the conventional signal‐processing, (2) PGS (Park et al., 2021), a state‐of‐the‐art, ensemble 2D Unet‐based model that is currently the winner of the MWC, and (3) HyperMapper (HPM; Mojiri Forooshani et al., 2022), another state‐of‐the‐art 3D Unet‐based tool with promising results in subjects with mild WMH burden. We present the performance metrics both at the voxel‐ and individual lesion cluster‐level for each tool, and provide cohort‐by‐cohort analysis for performance comparison.
Finally, we stress the importance of early identification and characterization of the small WMH we attempted to optimize with our tool by comparing microstructural properties inside and outside the manually traced WMH in MRi‐Share subjects using the multishell diffusion‐weighted imaging (DWI) available for this dataset.
2.2. Participants and MRI data description
Table 1 summarizes the key acquisition parameters for the T1‐weighted (T1w) and FLAIR images, sample sizes for manually traced WMH, and the range of total lesion load for the three datasets used in the present work, acquired across five different scanners.
TABLE 1.
Summary of key acquisition parameters and the range of manually traced WMH lesion load in the three cohorts.
| Dataset | Institute | Scanner | T1‐weighted voxel size (TR/TE/TI) | FLAIR voxel size (TR/TE/TI) | Train | Test | WMH load range in ml |
|---|---|---|---|---|---|---|---|
| MRi‐Share | Bordeaux University | 3 T Siemens Prisma | 1.00 × 1.00 × 1.00 mm3 (2000/2.0/880 ms) | 1.00 × 1.00 × 1.00 mm3 (5000/394/1800 ms) | 40 | 10 | 0–1.8 |
| MWC | UMC Utrecht | 3 T Philips Achieva | 1.00 × 1.00 × 1.00 mm3 (7.9/4.5/—ms) | 0.96 × 0.95 × 3.00 mm3 (11,000/125/2800 ms) | 17 | 3 | 0.8–75.0 |
| VU Amsterdam | 3 T GE Signa HDxt | 0.94 × 0.94 × 1.00 mm3 (7.8/3.0/−−ms) | 0.98 × 0.98 × .20 mm3 (8000/126/2340 ms) | 17 | 3 | ||
| NUHS Singapore | 3 T Siemens TrioTim | 1.00 × 1.00 × 1.00 mm3 (2300/1.9/900 ms) | 1.00 × 1.00 × 3.00 mm3 (9000/82/2500 ms) | 16 | 4 | ||
| UKB | UKB Imaging Centre | 3 T Siemens Skyra | 1.00 × 1.00 × 1.00 mm3 (2000/2.01/800 ms) | 1.05 × 1.00 × 1.00 mm3 (5000/395/1800 ms) | — | 11 | 0.1–22.5 |
| Total | 90 | 31 |
Abbreviations: MWC, MICCAI 2017 WMH segmentation challenge dataset; NUHS, the National University Health System; TE, echo time; TI, inversion time; TR, repetition time; UKB, UK Biobank; UMC, the University Medical Center; VU, Vrije Universiteit University Medical Centre.
2.2.1. MRi‐Share
The MRi‐Share database is a subcomponent of a larger, prospective cohort study on French university students' health, called iShare (internet‐based Student Health Research enterprise, www.i-share.fr). The detailed study and MRI acquisition protocol have been described in Tsuchida et al. (2021). For the training and evaluation of the SHIVA‐WMH detector, we used the MRI data and manually traced WMH from the same subsample of 50 subjects described in Boutinaud et al. (2021), drawn from the total sample of 1867 MRi‐Share subjects (mean age 22.1 years, range 18–35, 72% female). We also used the T1w and FLAIR data from additional 360 subjects, selected from the 1817 subjects without any manual tracing of WMH, to enhance the training dataset, as described in Section 2.4.3.
All participants were imaged between 2015 and 2018 on a 3 T Siemens Prisma MRI scanner (Siemens Healthcare, Erlangen, Germany) with a 64‐channel head coil at Bordeaux University, in a single MRI session lasting for ~45 min. It included a 3D T1w magnetization‐prepared rapid gradient‐echo as well as a 3D SPACE FLAIR sequence, both with 1.0 mm isotropic resolution (Table 1). Diffusion‐weighted MRI (DWI) data were acquired using a multishell multiband x3 sequence with 100 noncollinear diffusion gradient directions (b0/d8 each for anterior‐to‐posterior and posterior‐to‐anterior phase encoding; b300/d8; b1000/d32; b2000/d60) with the following parameters: TR/TE = 3540/75 ms; FOV = 118 × 118 mm2; 84 slices; 1.7 mm isotropic resolution.
2.2.2. MICCAI 2017 WMH challenge dataset (MWC)
We supplemented the training and evaluation datasets with the MRI and manual tracing of WMH from 60 subjects provided as training datasets in the MICCAI 2017 WMH segmentation challenge (http://wmh.isi.uu.nl/; Kuijf et al., 2019). The set of 3D T1w and 2D or 3D multi‐slice FLAIR images were acquired at three different institutes: the University Medical Center (UMC) Utrecht, Vrije Universiteit University Medical Centre (VU) Amsterdam, and the National University Health System (NUHS) in Singapore (20 subjects per site; Table 1). The 3D FLAIR images had been resampled into the transversal direction with a slice thickness of 3 mm by the MWC organizers to make them similar to other 2D FLAIR images in the dataset and to save time for manual annotation. All subjects were recruited at the memory clinics on each site as part of larger cohort studies (UMC Utrecht and VU Amsterdam data from a cohort of 861 subjects, mean age 67.7 years and 46.3% female (Boomsma et al., 2017); NUHS Singapore data from a cohort of 238 subjects, mean age 72.5 years, range 50–95 years, 51% female (van Veluw et al., 2015)), and were selected randomly by the MWC organizers.
2.2.3. UK Biobank
UK Biobank is the largest cohort study with brain MRI measurements from approximately 50,000 middle‐aged and older adults (as of 2022), recruited from communities across the United Kingdom (Miller et al., 2016). All brain imaging data were acquired at one of the three dedicated imaging centers equipped with identical 3 T Siemens Skyra scanners (Siemens Healthcare, Erlangen, Germany) with the standard Siemens 32‐channel head coil (Alfaro‐Almagro et al., 2018). For the present work, we use the raw T1w and FLAIR images acquired using similar sequences as those for MRi‐Share (Table 1).
2.3. Manual tracing of WMH
2.3.1. MRi‐Share
Subjects for manual tracing of WMH were selected based on the visual inspection of a neuroradiologist (BM) who reviewed the raw T1w and FLAIR images of the entire dataset, to cover varying degrees of both WMH and visible PVS, from no detectable WMH or PVS to many visible WMH (>10) and/or PVS. A trained investigator (AT) then performed voxelwise manual segmentation of each WMH on the raw FLAIR images using Medical Image Processing, Analysis and Visualization (MIPAV) software (v 7.4.0).
Specifically, WMH was segmented on each axial slice of the FLAIR image, viewed along with coronal and sagittal views using the 3D view setting of the MIPAV to check the 3D shape and extent of hyperintense signals. Any punctate region of increased intensity within the white matter, as well as hyperintense rims around the ventricles that were thicker than 2 mm, were segmented. This included punctate hyperintense regions sometimes found around the PVS visible on T1w images. For each hyperintense region found, the “paint grow” tool of the MIPAV was applied to automatically paint every neighboring voxels that have a higher intensity than and within a 3 mm distance from the selected voxel. Following the initial segmentation of the first 10 subjects, they were reviewed and modified by a second expert (LL). Any discordance between the two raters was then reviewed together to reach a consensus. Subsequently, the remaining 40 MRI datasets were manually segmented by the first expert only.
2.3.2. MICCAI 2017 WMH segmentation challenge
We used the publicly available manual tracing of WMH for the 60 MWC training dataset. The details of the procedure are described in Kuijf et al. (2019). Briefly, manual tracing of WMH, as well as any other pathologies, was performed by two expert raters by consensus, in which the tracing performed by the first rater was reviewed by the second rater and corrected by the first rater.
2.3.3. UK Biobank
A small sample of 11 subjects was selected to cover a range of estimated WMH load (0.7–16 ml, representing values in the first and last decile of the entire dataset) from a pool of 13,554 subjects (available at the time of the selection) with “usable” quality T1w and FLAIR images as well as the WMH load estimated by the Brain Intensity Abnormality Classification Algorithm tool (Griffanti et al., 2016). The same rater who performed the manual tracing of WMH for the MRi‐Share database (AT) manually traced WMH on the raw FLAIR images using the 3D Slicer tool (version 4.11.20210226: https://www.slicer.org), using the same criteria applied during the segmentation of WMH for MRi‐Share. With the 3D Slicer tool, the “Threshold” effect in the Segment Editor module was used to specify an intensity range that preliminarily isolated visible hyperintensities from the surrounding white matter in any given location. As in MRi‐Share, each axial slice was reviewed slice by slice, together with coronal and sagittal views to check the entire 3D extent of each lesion, and the “Paint” tool that painted regions with the specified intensity range was used to segment individual hyperintensities deemed as lesion, in each plane of the 3D view.
2.4. SHIVA‐WMH detector
2.4.1. Preprocessing
In order to prepare the input image arrays for the SHIVA‐WMH detector, the following preprocessing steps were performed on the T1w, FLAIR, and manually traced WMH masks:
Reorientation to match either LAS or RAS (left or right/anterior/superior) orientation using fslreorient2std tool from the FSL.
For the MWC dataset, images were resampled to 1 mm isotropic using flirt from the FSL (Jenkinson et al., 2002), with—applyisoxfm and—noresampleblur options.
Linear coregistration of FLAIR to T1w image was performed with flirt (Jenkinson et al., 2002) for MRi‐Share and UKB datasets, and the generated transformation matrices were applied to the WMH masks to bring them to the reference T1w images. This step was skipped for the MWC dataset, which had already been aligned by the organizers.
A brain mask created based on the individual T1w image was used to obtain a bounding box around the brain (centered on the mass center of the brain mask), and to crop all images to a uniform dimension of (160 × 216 × 176 voxels).
Voxel intensity values inside the brain mask were linearly rescaled to values between 0 and 1 by setting the 99th percentile value as the maximum and setting any higher intensity values as 1.
2.4.2. SHIVA‐WMH architecture and implementation
Our model is based on the previously published PVS detector (Boutinaud et al., 2021) with an Unet‐like architecture of Ronneberger et al. (2015). We made the following modifications to improve performance or to adapt it for the specific task of WMH segmentation:
For the primary multichannel model, the input layer was modified to accept multimodal input of T1w and FLAIR images.
Pretraining with auto‐encoder was not performed since the larger training set with multimodal input in the present work allowed relatively fast training without the pretraining with auto‐encoder.
The architecture of the network was modified to have an increased number of initial kernels (feature maps, nKinit), from 8 to 10, and the multiplication factor (mF) applied to the number of kernels at the first convolution layer of each stage (or depth) after the first was slightly reduced from 2 to 1.8.
The dropout rate applied at each stage (after the max pooling for encoding or after the last convolution for decoding blocks) was increased from 0.1 to 0.5, except in the first encoding block, which had a reduced dropout rate of 0.05 to increase the rate of retention, reflecting the general recommendation for optimal dropout rate across a wide range of networks and tasks (Srivastava et al., 2014).
Convolution blocks now use a Swish activation function (Equation (1)) rather than the rectified linear unit (ReLU; Glorot & Bengio, 2010) used previously, since it has shown an advantage over ReLU on deeper models across a number of challenging datasets (Ramachandran et al., 2017).
| (1) |
Figure 1 provides a schematic overview of the resulting architecture of the SHIVA‐WMH model. The modified model has slightly less trainable parameters (40 million rather than 44 million in the Boutinaud et al.'s study) and also extracts a higher number of features at higher resolution upper stages and less at deeper levels, which we found to be advantageous for the detection of small lesions like PVS and small WMH.
FIGURE 1.
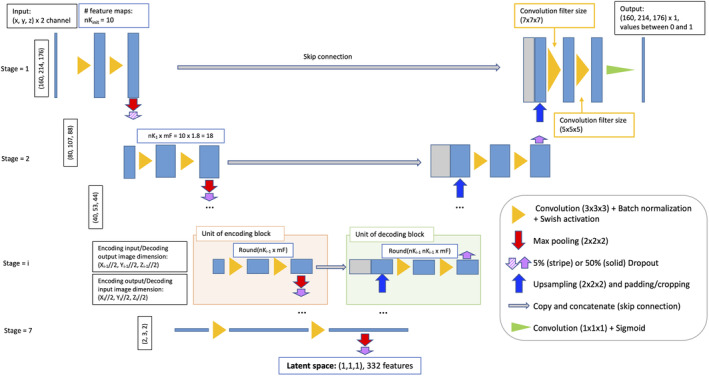
Schematic overview of the SHIVA‐white matter hyperintensity (WMH) detector network architecture. The figure illustrates the 3D Unet architecture used for the SHIVA‐WMH detector. Each blue box corresponds to the multichannel feature maps, with the number of features at each stage indicated in the white box with blue outline. The gray boxes are copied and concatenated feature maps from the encoding path to the decoding path. The arrows and triangles stand for different operations as indicated inside right legend.
In addition to the network architecture modifications, we performed the following data augmentations to the training dataset to increase the model robustness: (i) flipping on the midsagittal plane, (ii) voxel translations (up to plus or minus five voxels in each orthogonal axis), and (iii) nonlinear voxel intensity value transformation using a Bézier curve, in the similar fashion as in Zhou et al. (2021), with two endpoints set to [0, 0] and [1, 1] and two control points within this range generated randomly. The probability of each type of augmentation for an image at a training epoch was set to 0.5, 0.9, and 0.9, respectively. In particular, the nonlinear voxel intensity value transformation was found to significantly improve the generalizability of the detector when predicting lesions in unseen datasets (unpublished observation from the PVS detector).
We implemented the network in Python 3.7, using Tensorflow 2.7 with Keras backend, scikit‐learn (1.0.1), and scikit‐image (0.18.3). The network was trained on a computer (Ubuntu 22.04) with a Xeon ES2640, 40 cores, 256 GB RAM, and a Tesla V100 GPU with 32 GB RAM; inferences can be done on any GPU compatible with the Tensorflow version with at least 8 GB RAM. We used a fivefold cross‐validation scheme to train the network, stratified on WMH voxel load and cohort. We used the Adam optimizer with the default parameters of beta1 = .9, beta2 = .999, epsilon = 1e–7, and used a cyclical learning rate with exponential decay, with the initial and maximum learning rates set to 1e‐6 and 0.001, respectively. As in the Boutinaud et al.'s study, we used a Dice loss function (Equation (2)) as a loss function:
| (2) |
where y true and y pred represent the image arrays for the ground truth (manually traced WMH or synthetic WMH labels in the case of enhanced training set: see Section 2.4.3) and predicted WMH, respectively, and (y true × y pred ) is an element‐wise multiplication of the two arrays, representing the intersection between the two images. We used the smoothing constant 𝜖 of 1e‐6 to prevent the division by 0. We used a batch size of 4 for each training fold to fit in the available GPU memory. The output maps from each fold, valued between 0 and 1, were averaged to create the final WMH prediction map, also valued between 0 and 1.
2.4.3. Training and enhancement
We initially trained our model with T1w and FLAIR images from 40 MRi‐Share and 50 MWC subjects with manually traced WMH (Table 1), using the fivefold cross validation scheme, in which 20% of the training data served as validation data in each fold. We call this model our “base” model. However, due to the large imbalance in the total number of voxels labeled as WMH in the two cohorts (only ~8 K voxels of WMH collectively in MRi‐Share compared to ~777 K voxels in MWC training data; also see Figure 2), the Dice loss function would inevitably bias the optimization toward models that can detect larger WMH lesions. In order to gauge how much the presence of MWC training data degrades the performance of our model in MRi‐Share subjects, we trained another model with only MRi‐Share training data, which we call “MRi‐Share‐specific” model. As shown in the Supplemental Figure 1, the comparison of segmentation accuracy in the MRi‐Share subjects from the validation sets in each training fold indicated lower accuracy of the “base” model relative to “MRi‐Share‐specific” model, as expected.
FIGURE 2.
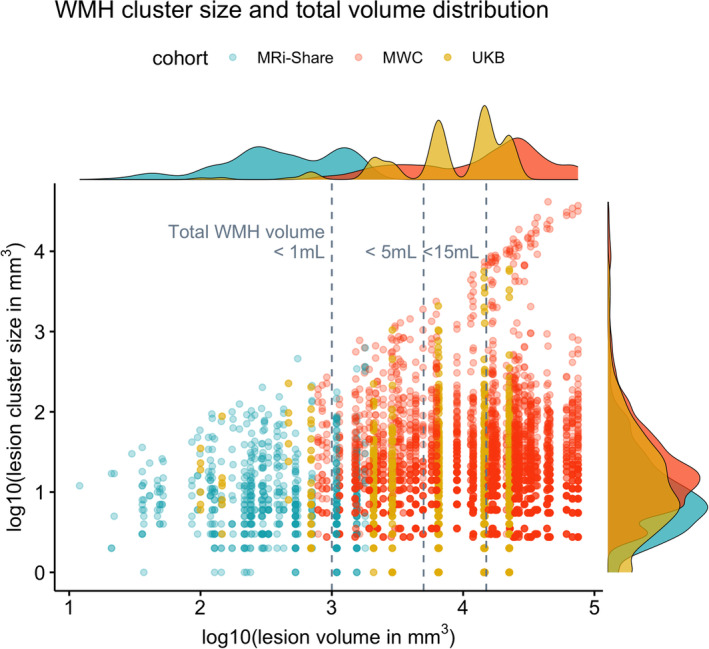
Distribution of total lesion load and individual distributions of lesion‐cluster sizes in each subject with the manually delineated WMH in the three datasets. The x‐axis plots the log10‐transformed total WMH volume (in mm3) against the y‐axis showing the distribution of the log10‐transformed individual lesion cluster size (in mm3) in each subject, based on the manually traced WMH (i.e., each dot along the given x‐value representing the lesion clusters in a single subject). Colors indicate subjects from each cohort (MRi‐Share in turquoise, MWC in orange, and UKB in gold colors). Each subject is assigned one of five shapes randomly to separate data points coming from subjects with very close overall lesion volumes. Histograms on the margins show the distribution of the total lesion volume (top) or the lesion cluster size (right) separately for each cohort.
To overcome the problem of imbalances, we took the advantage of the large pool of still unannotated MRi‐Share dataset to generate pseudo WMH labels using the specialized “MRi‐Share‐specific” model that could segment small WMH with relatively high accuracy and use them to enhance the original training data composed of manually traced WMH labels. This is akin to “semisupervised learning with pseudo annotations” reviewed by Tajbakhsh et al. (2020) as one promising approach to tackle the problem of scarce annotated training data common in medical image segmentation tasks (Tajbakhsh et al., 2020). In this approach, pseudo annotations are first assigned to unlabeled data, then a new segmentation model is trained with combined data that include both the labeled and pseudo labeled data. We used this approach more specifically to ameliorate the voxel imbalance between the examples of small WMH primarily found in MRi‐Share and those of large WMH found in MWC data. We first obtained the predictions of WMH from the “MRi‐Share‐specific” model in 100 randomly selected MRi‐Share subjects out of 1817 subjects without the manual tracing of WMH. We filtered out those with very low or high predicted load of WMH by removing subjects in the top and bottom 5 percentile of the total estimated WMH volume. This is because subjects at the bottom 5 percentile would scarcely add examples of small WMH that can be learned by the model, and conversely, we did not need to add examples of subjects with the highest load, which is already provided by the manually traced MWC training data. We then used the pseudo WMH labels in the remaining 90 subjects to supplement the original training data from 90 subjects with manual tracing (“enh90” model, for enhancement with 90 predicted data).
While the “base” model used the Glorot uniform initializer to initialize its weights, “enh90” model used the weights of the “base” model as the initial weights to speed up the training. Since it showed some indication of improvement over the “base” model in the validation sets of both MRi‐Share and MWC training data (Supplemental Figure 1), we repeated the process with an additional enhancement using a new set of pseudo WMH labels from 270 MRi‐Share subjects generated similarly from the “MRi‐Share‐specific” model to train another model (“enh270” model). As its initial weights, we used the weights of “enh90” model, thus resulting in a model that was cumulatively trained with 360 synthetic WMH labels. This model showed further improvement in the MRi‐Share validation sets and similar performance in the MWC. We repeated the process with increasing numbers of enhancements (300 and 400 additional training labels), each using the weights of the preceding model as the initial weights. However, since there were no signs of further improvement in the MRi‐Share validation sets after the “enh270” model (Supplemental Figure 1), we selected this model as the optimal detector; it will be referred to in the following as the SHIVA‐WMH detector.
2.4.4. FLAIR‐only version
We initially focused on developing a model that uses both T1w and FLAIR as inputs, following an earlier observation of the superior performance of multimodal over FLAIR‐only models. However, to formally compare the impact of having only FLAIR as an input and to potentially allow quantification of WMH in datasets with only FLAIR images, we created the modification of the SHIVA‐WMH detector with FLAIR only input. The training process of the FLAIR‐only version and its performance comparison with the multimodal version is described in the Supplemental Material.
2.5. Performance evaluation
We evaluated the performance of the SHIVA‐WMH detector and existing methods in the held‐out test dataset from the three cohorts, including 10 unseen data each from MRi‐Share and MWC and 11 data from UKB. The UKB test data represent a dataset coming outside of cohorts used in training (Table 1), with the levels of WMH severity being the intermediate between those of MRi‐Share and MWC.
2.5.1. Evaluation metrics
We focused on the metrics that quantify the spatial similarity of the ground truth (manually traced WMH of the held‐out test set) and predicted WMH both at the voxel‐ or individual lesion cluster‐level. Specifically, we counted the number of true positives (TP), false negatives (FN), and false positives (FP) voxel‐by‐voxel or at the level of individual lesion clusters (each individual lesion cluster defined as a 3D connected component using voxel connectivity of 26) to compute the following performance metrics.
Voxel‐level or cluster‐level true positive rate (VL‐ or CL‐TPR): It is measured as the number of TP voxels or lesion clusters divided by the number of ground truth voxels or clusters (i.e., TP + FN), and is equivalent to sensitivity or recall.
Voxel‐level or cluster‐level positive predictive value (VL‐ or CL‐PPV): It is measured as the number of TP voxels or clusters divided by the number of predicted WMH voxels or clusters (i.e., TP + FP), and is equivalent to precision.
- Voxel‐level or cluster‐level Dice coefficient (VL‐ or CL‐Dice): It is the harmonic mean of the TPR and the PPV, or, equivalently, it can be expressed as:
Note that the term F1 score is used sometimes to refer to lesion cluster‐based metric (CL‐Dice in our terminology) to distinguish from the VL‐Dice, which is often referred as Dice score or Dice similarity coefficient (e.g., Kuijf et al., 2019), even though mathematically the Dice and F1 scores are equivalent (Reinke et al., 2021). While both VL‐ and CL‐metrics are overlap‐based measures that quantify the spatial similarity between the predicted and ground truth label as a reference, CL‐metrics emphasize the detection quality over segmentation quality: in other words, CL‐metrics quantify the ability to identify every lesion cluster, rather than the ability to precisely segment any given lesion cluster, and thus more relevant for small WMH lesions where detection of every lesion cluster may be more pertinent than the precise delineation of lesion boundaries (Park et al., 2018). In contrast, VL‐metrics are biased toward larger lesions in that large lesion clusters with more voxel‐count influence them disproportionately than lesion clusters with very small voxel‐count. In other words, when there is mixture of small and large lesions, any given segmentation tool can achieve high VL‐metrics without detecting any of the small lesions if the collective voxel counts of the small lesions are smaller than those of the large lesions (Reinke et al., 2021).
We also report the modified Hausdorff distance (95th percentile: HD95) as a measure of the accuracy of the segmentation boundaries. It is defined as the 95th percentile of the symmetric surface distances (Hausdorff distance) of two binary images, with lower values indicating shorter overall distances between the two images. We used the implementation of HD95 computation from the MedPy package (version 0.4.0).
2.5.2. Metric comparisons with existing methods
We performed sets of paired t tests with subject as the within‐factor that compared each performance metric of the SHIVA‐WMH against the LST‐LPA, PGS, and HPM, separately for each of the three cohorts in the held‐out test set to allow evaluation of performance in the cohorts with very different demographic and WMH lesion characteristics. Note that PGS performance could not be compared for the MWC test subjects, since they were part of the training data used for this tool. Each comparison was Bonferroni corrected for the number of comparisons made for the given cohort (three for MRi‐Share and UKB, two for MWC). All paired t tests were performed in R, version 4.2.2 (R Core Team, 2018), and visualized using ggpubr package (Kassambara, 2022). Summary tables were generated using gt package (Iannone et al., 2020).
For all metric computations, prediction maps were thresholded at 0.5 to make a fair comparison with the PGS tool, whose output prediction maps were already thresholded at this value. For all other tools (SHIVA‐WMH, LST‐LPA, HPM), lower thresholds improved the VL‐ and CL‐Dice scores slightly (thresholds that resulted in the highest average VL‐ and CL‐Dice scores were 0.2 for SHIVA‐WMH, 0.1 for LST‐LPA and HPM), but the overall patterns remained essentially the same (not shown).
2.6. Comparison algorithms
2.6.1. Lesion segmentation toolbox–LPA
The LST‐LPA is an open‐source MATLAB tool that uses only an FLAIR image as an input to segment WMH without requiring any optimizations or retraining (Schmidt, 2017b). It is based on a conventional signal processing method with binary classification using a logistic regression model that has been trained with 53 MS patients with severe lesion patterns. While it was originally developed to detect WMH in MS patients (Schmidt et al., 2012), it has been applied in the context of age‐related WMH and widely used (Garnier‐Crussard et al., 2020; Ribaldi et al., 2021; Vanderbecq et al., 2020). Being part of the toolbox for the SPM software, it is fully automated and simple to use, and is often selected as a reference tool for the new WMH detection method development (Balakrishnan et al., 2021). Since it comes with a built‐in preprocessing pipeline that includes intensity normalization, we used the raw FLAIR images from the test dataset as the input to obtain the predicted maps of WMH.
2.6.2. PGS
The PGS tool is a 2D Unet‐style model with a multi‐scale highlight foreground method to augment the influence of small lesions or voxels lying in the lesion boundaries, and is currently the best‐ranking method that has been submitted to the MICCAI 2017 WMH challenge (Park et al., 2021). The pretrained model submitted to the challenge is available as the Docker‐contained code from the challenge website (https://wmh.isi.uu.nl/results/pgs/). It has been trained on the 2D axial slices of T1w and FLAIR images from the same MWC dataset used in the present study. Because our MWC test dataset is part of the training dataset in the challenge (and therefore in the PGS model), evaluation of this tool in the MWC test dataset was not performed. Since this tool had been trained on the bias‐field corrected data prepared by the challenge organizers (using SPM12), we used the bias‐field corrected T1w and FLAIR images of the MRi‐Share (based on SPM12, as described in Tsuchida et al., 2021) and UKB datasets (based on FSL FAST, as described in Alfaro‐Almagro et al., 2018) as inputs for this tool.
2.6.3. HyperMapper
The HPM tool is a 3D Unet model with Monte Carlo dropout layers incorporated in the encoding layers, and has been trained with T1w and FLAIR images of 432 subjects from multicenter studies that represented various diagnostic groups including cognitively normal, cerebrovascular disease, Parkinson's disease, Alzheimer's disease, and frontal temporal dementia (Mojiri Forooshani et al., 2022). The authors have reported high accuracy on a large and diverse test dataset representing 158 subjects from five different studies, including one unseen dataset. Of particular interest from our perspective, it also reported very high accuracy on the subsample of 50 test subjects with mild WMH cases with the average of 2 ml of lesions. Unlike the PGS tool and many other recently published methods (including all other tools submitted to the MICCAI 2017 WMH challenge), the HPM did not use the MWC dataset to train the models. As such, its performance on the MWC test dataset could be evaluated against the SHIVA‐WMH. The pretrained HPM tool is publicly available from https://github.com/AICONSlab/HyperMapp3r. We used the Docker‐contained image included in the repository to perform predictions on the test datasets. Since this tool also has been trained on the bias‐field‐corrected training data, we used the bias‐field corrected T1w and FLAIR images for all test datasets.
2.7. Comparison of microstructural properties inside WMH to normal‐appearing white matter in MRi‐Share
The processing pipeline of DWI data to obtain diffusion tensor imaging (DTI: Basser et al., 1994) and neurite orientation dispersion and density imaging (NODDI: Zhang et al., 2012) metrics in MRi‐Share has been described in detail in Tsuchida et al. (2021). Briefly, DWI data were preprocessed with the Eddy tool from FMRIB Software Library (FSL, version 5.0.10: https://fsl.fmrib.ox.ac.uk/fsl) to correct for susceptibility and eddy‐current distortion, then denoised using the nonlocal means filter (Coupe et al., 2008; Coupe et al., 2011) using nlmeans tool implemented in the Dipy package (0.12.0: Garyfallidis et al., 2014). The DTI model was fit using the Dipy package, while the NODDI model was fit using the AMICO tool (Daducci et al., 2015).
The resulting scalar images of DTI and NODDI metrics were coregistered to the native T1w image space using antsRegistrationSyNQuick script of Advanced Normalization Tools (ANTs, version 2.1: http://stnava.github.io/ANTs/) package. For the present work, we focused on the neurite density index (NDI) from NODDI, fractional anisotropy (FA), and mean diffusivity (MD) from DTI metrics, which all have been shown to be altered in WMH in MS (Alotaibi et al., 2021) or in older subjects (Muñoz Maniega et al., 2015; Riphagen et al., 2018).
To compare NDI, FA, and MD values inside WMH and normal‐appearing white matter (NAWM) in the 50 MRi‐Share subjects with the manual tracing of WMH, manually traced WMH masks in the native FLAIR space were coregistered linearly to the T1w image by applying the transformation matrix generated by the coregistration of FLAIR to T1w with flirt tool from the FSL (Jenkinson et al., 2002). The mask of NAWM was generated by combining both the manually traced WMH and PVS masks, then subtracting this from the cerebral white matter labels generated by the Freesurfer (v6.0: http://surfer.nmr.mgh.harvard.edu/) in the native T1w space. To remove any partial volume effects near the border of lesion or other tissue types (gray matter and cerebrospinal fluid), the mask of cerebral white matter was eroded once using fslmaths tool from the FSL. To avoid the partial volume effects of the cerebrospinal fluid in the WMH mask, periventricular lesion clusters within a 2 mm distance of individual ventricle maps (generated with Freesurfer v6.0) were removed from the analysis. Then, 46 out of 50 subjects had non‐empty WMH masks and could be included in the analysis. Mean values of NDI, FA, and MD inside the resulting WMH and NAWM masks were computed and compared by performing a within‐subject t test for each metric.
All paired t‐tests were performed in R, version 4.2.2 (R Core Team, 2018), and visualized using ggpubr package (Kassambara, 2022).
3. RESULTS
3.1. Characterization of manually traced WMH in the three cohorts
The sample of subjects with manual segmentations of WMH in the present study comprised 50 young adults (18–35 years of age) from the MRi‐Share study, 11 middle‐aged to older UKB participants (>40 years of age), and 60 memory clinic patients (approximately >50 years of age) from the MWC. Figure 2 shows the distribution of total WMH lesion volume and individual lesion cluster sizes of each participant (with the x‐axis effectively ordering every subject according to the total lesion volume) in each of the three cohorts with the manual tracing of the WMH, in order to appreciate the range of cluster sizes observed in individuals with varying load of WMH. Not surprisingly, young subjects of the MRi‐Share with very mild lesion load only have small WMH clusters, and the maximum size of individual lesion increases with the total lesion volume, with very large lesions in older MWC subjects with the most severe cases of WMH, likely representing the confluence of deep and periventricular WMH. However, it should be noted that subjects with high WMH load also have many relatively small lesion clusters, which underscores the importance of accurately segmenting small lesions for comprehensive characterization of WMH. Figure 2 also demonstrates the vastly different scales of the overall amount of WMH lesions in the three cohorts in this study: without the log10‐transformation of the scales, the total WMH volumes for the MRi‐Share subjects would cluster around 0, since most subjects have less than 1 ml of WMH in total. Representing middle‐aged and older subjects, the 11 subjects from the UKB show levels of WMH load intermediate between those of the MRi‐Share and MWC subjects.
3.2. Detection of WMH with SHIVA‐WMH detector across a wide range of lesion loads
We used the manually traced lesions from both MRi‐Share and MWC, and enhanced training data from additional MRi‐Share participants to train the SHIVA‐WMH detector. We then evaluated the performance of our detector against three reference methods (LST‐LPA, PGS, and HPM), in the held‐out evaluation test subjects comprised of 10 MRi‐Share, 11 UKB, and 10 MWC subjects. Table 2 shows the summary of performance metrics for each method across all 31 test subjects, except for PGS, which was not evaluated for MWC test subjects. Overall, they indicate the superior segmentation accuracy of the SHIVA‐WMH detector over the three reference methods (LST‐LPA, PGS, and HPM), with higher sensitivity (TPR) and precision (PPV) both at the voxel‐ and lesion cluster‐level than any of the reference methods, resulting in the significantly higher VL‐ and CL‐Dice scores (difference of 0.34, 0.27, and 0.24 in VL‐Dice, all paired t tests p < .001, and 0.50, 0.31, and 0.41 in CL‐Dice, all p < .0001, against LST‐LPA, PGS, and HPM, respectively). The correlation between the log‐transformed volume of manually traced lesion and segmented WMH across the test set subjects was also the highest for SHIVA‐WMH compared to the three reference methods (Figure 3). Although FLAIR‐only version of SHIVA‐WMH had slightly worse performance than the primary multimodal input version, it still had nominally better VL‐Dice scores (difference of 0.27, 0.20, and 0.17 with p < .001, NS, 0.5 against LST‐LPA, PGS, and HPM, respectively) and significantly better CL‐Dice scores against all three reference methods tested (difference of 0.50, 0.31, and 0.41 with all p < .0001 for CL‐Dice; Supplemental Table 1).
TABLE 2.
Comparison of SHIVA‐WMH against three reference methods across the 31 test subjects for each performance metric.
| Mean (SD) | |||||||
|---|---|---|---|---|---|---|---|
| VL‐TPR | VL‐PPV | VL‐Dice | CL‐TPR | CL‐PPV | CL‐Dice | HD95 | |
| All (N = 31 a ) | |||||||
| SHIVA | 0.63 (0.20) | 0.76 (0.18) | 0.66 (0.16) | 0.66 (0.16) | 0.83 (0.17) | 0.71 (0.13) | 2.82 (3.21) |
| LST‐LPA | 0.30**** (0.32) | 0.48** (0.36) | 0.32**** (0.29) | 0.20**** (0.20) | 0.37**** (0.31) | 0.21**** (0.17) | 4.55 (3.93) |
| PGS | 0.45* (0.18) | 0.41**** (0.32) | 0.39*** (0.26) | 0.62 (0.17) | 0.34**** (0.28) | 0.40**** (0.26) | 3.83 (8.48) |
| HPM | 0.34**** (0.24) | 0.64 (0.38) | 0.42*** (0.29) | 0.25**** (0.19) | 0.58** (0.33) | 0.30**** (0.17) | 3.45 (3.24) |
Note: Mean and standard deviations (SD) of each metric across all the test subjects are shown for SHIVA‐WMH and the three reference methods (LST‐LPA, PGS, HPM). For each metric, best scores are indicated in bold. Asterisk indicates the degree of statistical significance for each paired t test comparing SHIVA‐WMH against each of the reference methods.
Comparison with PGS was performed in 21 test subjects that excluded subjects from MWC.
p < .0001.
.0001 ⩽ p < .001.
.001 ⩽ p < .01.
.01 ⩽ p < .05.
FIGURE 3.
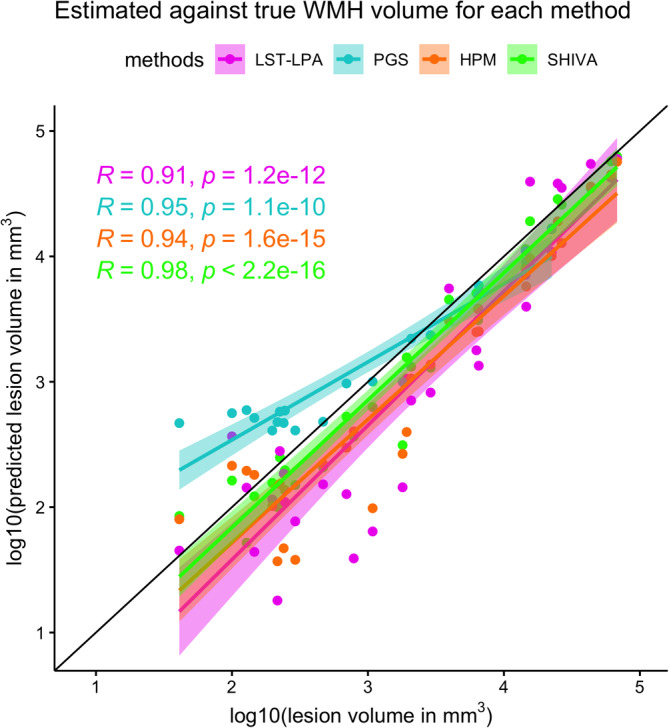
Correlations between total white matter hyperintensity (WMH) volume of the manually delineated lesions and WMH segmented by each method across the test‐set subjects. The x‐axis plots the log10‐transformed total WMH volume (in mm3) based on the manually traced WMH against the y‐axis showing also the log10‐transformed total volume (in mm3) of WMH segmented by each method. Each dot along the given x‐value represents a single subject, with different colors indicating WMH volume estimates based on each method (LST‐LPA in pink, PGS in turquoise, HPM in orange, and SHIVA‐WMH in green). Regression lines with confidence intervals are also shown for each method in respective colors, as well as Pearson's correlation coefficients (R) between the log10‐transformed volumes and associated p values.
Because the evaluation test set comprised subjects with very different demographics and WMH burden, we performed more detailed analysis by comparing the performance metrics separately for each of the three cohorts. Figure 4 graphically illustrates quantitative comparisons of the two main metrics of interest, VL‐ and CL‐Dice scores, in each cohort. Supplemental Figure 2 shows similar cohort‐specific comparisons for FLAIR‐only version of SHIVA‐WMH. Table 3 provides the same summary as in Table 2, but separately for each cohort. Qualitative comparisons of example segmentations from each method are shown in Figure 5, separately for representative test set subjects with either only small WMH lesions (mild) or with large confluent lesions (severe) from each cohort (except for MRi‐Share, in which none of the subjects had severe WMH). Finally, supplemental Figure 3 plots individual VL and CL‐Dice scores as a function of maximum size of WMH cluster in each subject to further illustrate the performance differences of each tool across subjects with a range of WMH burden.
FIGURE 4.
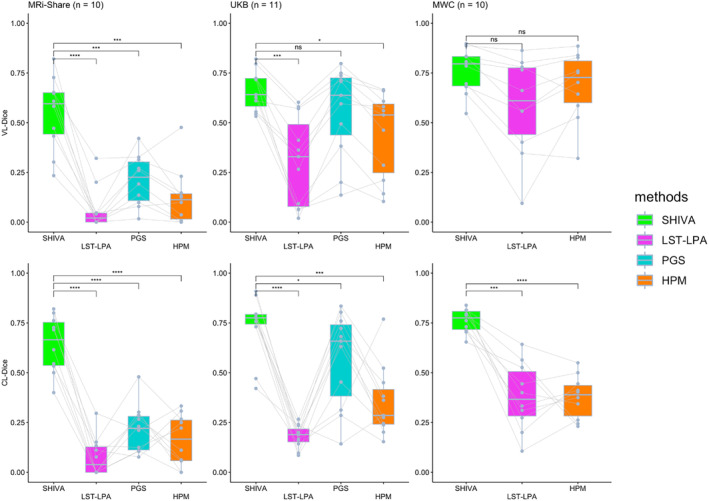
Voxel (VL‐) and cluster‐level (CL‐) Dice scores of SHIVA‐white matter hyperintensity (WMH) compared with lesion prediction algorithm implemented in the lesion segmentation toolbox (LST‐LPA), PGS, and HyperMapper (HPM) tools in the test‐set subjects in each cohort. Comparisons of VL‐Dice (top row) and CL‐Dice (bottom row) scores between SHIVA‐WMH against the reference tools (LST‐LPA, PGS, and HPM) are shown, separately for MRi‐Share (n = 10), UKB (n = 11), and MWC (n = 10) test subjects. Asterisk indicates the degree of statistical significance for each paired t test comparing SHIVA‐WMH against each of the reference methods: ****p < .0001, ***.0001 ⩽ p < .001, **.001 ⩽ p < .01, *.01 ⩽ p < .05, ns p ⩾ .05.
TABLE 3.
Summary of performance metric comparisons between SHIVA‐WMH against the three reference methods separately for each test cohort.
| Mean (SD) | |||||||
|---|---|---|---|---|---|---|---|
| VL‐TPR | VL‐PPV | VL‐Dice | CL‐TPR | CL‐PPV | CL‐Dice | HD95 | |
| MRi‐Share (n = 10) | |||||||
| SHIVA | 0.55 (0.25) | 0.66 (0.22) | 0.55 (0.19) | 0.52 (0.18) | 0.91 (0.11) | 0.64 (0.14) | 3.25 (4.46) |
| LST‐LPA | 0.06*** (0.11) | 0.15*** (0.19) | 0.07**** (0.11) | 0.10*** (0.14) | 0.08**** (0.11) | 0.08**** (0.10) | 7.82 (7.53) |
| PGS | 0.32** (0.17) | 0.19**** (0.14) | 0.21*** (0.13) | 0.50 (0.15) | 0.15**** (0.11) | 0.22**** (0.12) | 2.13 (1.21) |
| HPM | 0.10** (0.12) | 0.32* (0.37) | 0.12*** (0.14) | 0.22* (0.26) | 0.29*** (0.33) | 0.16**** (0.13) | 5.74 (5.56) |
| UKB (n = 11) | |||||||
| SHIVA | 0.58 (0.12) | 0.83 (0.16) | 0.66 (0.10) | 0.72 (0.09) | 0.78 (0.22) | 0.73 (0.15) | 2.87 (3.16) |
| LST‐LPA | 0.22*** (0.16) | 0.57* (0.37) | 0.31*** (0.22) | 0.16**** (0.09) | 0.37*** (0.25) | 0.18**** (0.06) | 3.92 (1.61) |
| PGS | 0.57 (0.10) | 0.60* (0.31) | 0.56 (0.23) | 0.73 (0.11) | 0.52** (0.27) | 0.57* (0.23) | 5.38 (11.70) |
| HPM | 0.34** (0.15) | 0.66 (0.36) | 0.44* (0.21) | 0.28*** (0.20) | 0.64 (0.28) | 0.35*** (0.18) | 2.94 (0.87) |
| MWC (n = 10) | |||||||
| SHIVA | 0.76 (0.15) | 0.78 (0.12) | 0.76 (0.11) | 0.74 (0.09) | 0.81 (0.13) | 0.76 (0.06) | 2.31 (1.72) |
| LST‐LPA | 0.63 (0.32) | 0.71 (0.23) | 0.58 (0.24) | 0.34** (0.25) | 0.66 (0.23) | 0.38*** (0.17) | 3.62 (2.73) |
| HPM | 0.57 (0.19) | 0.93 * (0.06) | 0.68 (0.17) | 0.25**** (0.09) | 0.80 (0.15) | 0.38**** (0.11) | 2.19 (1.01) |
Note: Mean and standard deviations (SD) of each metric in each test cohort are shown for SHIVA‐WMH and the three reference methods (LST‐LPA, PGS, HPM). For each metric in each cohort, best scores are indicated in bold. Asterisk indicates the degree of statistical significance for each paired t test comparing SHIVA‐WMH against each of the reference methods.
p < .0001.
.0001 ⩽ p < .001.
.001 ⩽ p < .01.
.01 ⩽ p < .05.
FIGURE 5.
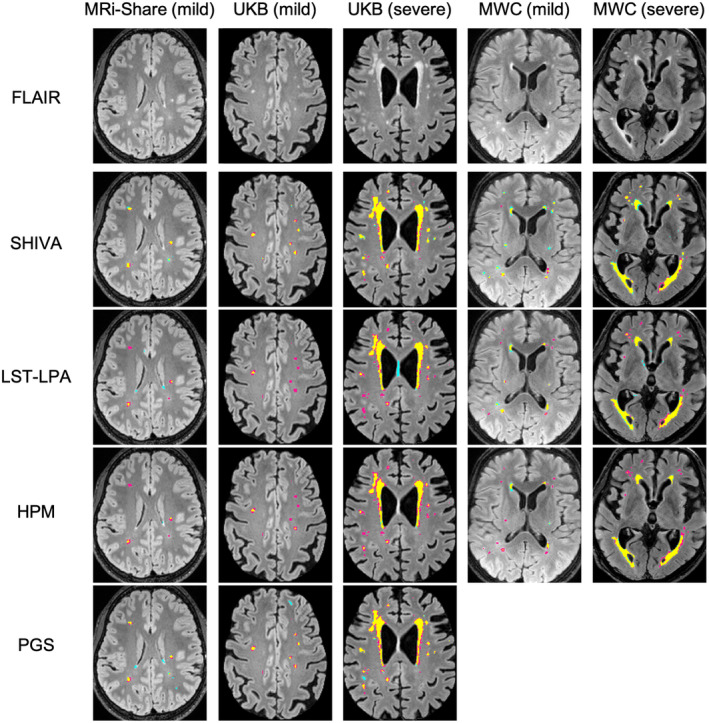
Examples of segmentation output by SHIVA‐white matter hyperintensity (WMH) detector, lesion prediction algorithm implemented in the lesion segmentation toolbox (LST‐LPA), PGS, and HyperMapper (HPM) tools. Examples of WMH segmentations by SHIVA‐WMH, LST‐LPA, PGS, and HPM are shown separately for representative subject(s) in each cohort with either mild (<5 ml) or severe (>5 ml) WMH load. The top row shows the selected axial slices from each subject, and second to last rows show the segmentation results of each tool, with yellow, pink, and cyan colors indicating true positive, false negative, and false positive voxels, respectively.
They indicate the clear advantage of the SHIVA‐WMH detector in the MRi‐Share test dataset, both at the voxel‐ and lesion cluster‐level: Our tool shows both higher TPR and PPV than any of the three reference methods, resulting in the significantly higher VL‐ and CL‐Dice scores (difference of 0.48, 0.34, and 0.43 in VL‐Dice, with all paired t tests p < .001, and 0.56, 0.42, and 0.48 in CL‐Dice, with all p < .0001 against LST‐LPA, PGS, and HPM, respectively; Figure 4 and Table 3). FLAIR‐only version shows a similar pattern, albeit with slightly lower VL‐ and CL‐Dice scores compared to the multimodal version and weaker significance when compared against other reference methods (Supplemental Figure 2).
In the MWC test dataset, both multimodal and FLAIR‐only versions of SHIVA‐WMH have significantly higher CL‐Dice (difference of 0.38 with p < .001 against both LST‐LPA and HPM) and numerically better VL‐Dice scores than the LST‐LPA or HPM, suggesting that our tool is detecting small lesion clusters better than these tools. While HPM had significantly better VL‐PPV than SHIVA‐WMH, it came at the cost of lower VL‐TPR. In this cohort, we did not perform direct comparison with PGS, since the model had been trained with the subjects we used as the evaluation test set in the present work.
Finally, in the unseen cohort of UKB, with the intermediate level of the overall lesion severity compared to the other two cohorts, multimodal SHIVA‐WMH is clearly superior to the LST‐LPA and HPM, with significantly better VL‐ and CL‐Dice scores (difference of 0.35 and 0.22 in VL‐Dice, with p < .01 and 0.05, and CL‐Dice difference of 0.55 and 0.38, with both p < .001 against LST‐LPA and HPM, respectively). It also shows a significantly better CL‐Dice than the PGS (difference of 0.16, p < .05), in particular showing better VL‐ and CL‐PPV (Figure 4 and Table 3; also see Figure 5 for an example of FP in PGS). FLAIR‐only version has a similar CL‐Dice score as the multimodal version, but shows a slightly worse VL‐Dice score in this cohort (Supplemental Figure 2).
The summary of subject‐by‐subject VL‐ and CL‐Dice scores across the range of WMH lesion sizes complements these findings in the cohort‐specific analyses: Overall, when subjects are burdened with large WMH lesions, as in the case for most MWC subjects, VL‐Dice scores are not very different across tools, but the advantage of SHIVA‐WMH is evident in subjects with only small WMH lesions (Supplemental Figure 3). At the level of lesion‐clusters, SHIVA‐WMH consistently shows higher CL‐Dice scores than the reference tools across the entire range of WMH burden (Supplemental Figure 3). It indicates the superior detection of individual lesion clusters by the SHIVA‐WMH compared to other tools even in subjects with the high WMH burden, and also highlights the dangers of focusing solely on VL‐Dice scores when assessing prediction quality in the presence of mixture of small and large lesions.
3.3. Comparison of microstructural properties inside WMH to NAWM in young subjects of MRi‐Share
Despite their relative sparsity and small sizes, within‐subject comparisons of white matter properties inside and outside the manually traced lesions revealed significant differences in NDI, FA, and MD values in the WMH found in MRi‐Share participants: compared to NAWM, WMH showed a decreased NDI (mean difference [95% confidence intervals] = −0.179 [−0.145, −0.213], paired t test p < .0001) and FA (−0.125 [−0.104, −0.144], paired t test p < .0001) values, and an elevated MD values (+2.2 × 10−4 [1.88 × 10−4, 2.57 × 10−4] mm2/s, paired t test p < .0001) (Figure 6). The changes in NDI, FA, and MD values were visible at the level of individual lesion clusters in some cases, as in the example shown in Figure 3.
FIGURE 6.
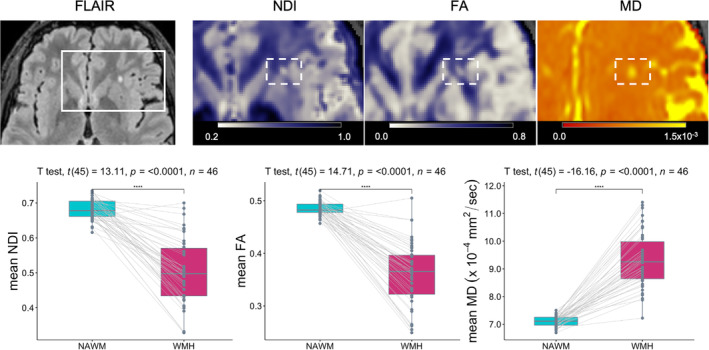
White matter microstructural properties of small white matter hyperintensity (WMH) in magnetic resonance imaging (MRi)‐Share. The top row shows an example axial slice of an FLAIR image of an MRi‐Share subject showing an isolated punctate WMH, together with blown‐up images of NDI, FA, and MD maps in the same slice to highlight the decreased NDI and FA values and increased MD values within the WMH (in the dotted rectangles). The bottom row shows the within‐subject comparisons of mean NDI, FA, or MD values inside the NAWM and WMH in 46 subjects with at least one WMH after removing WMH too close to ventricles to avoid partial volume effects from the cerebrospinal fluid in the ventricles.
4. DISCUSSION
Long dismissed as the “normal” radiological finding in the aging brain, WMH is now firmly established as the most common marker of covert cSVD (Debette & Markus, 2010; Wardlaw et al., 2015). In the present work, we leveraged the high‐quality research scans from the MRi‐Share study (Tsuchida et al., 2021) to demonstrate that small punctate WMH can already be observed in young adults in their twenties. We described the SHIVA‐WMH detector, trained with both the MRi‐Share and publicly available MWC dataset from older subjects (Kuijf et al., 2019), with a specific aim to optimize WMH detection across a wider range of WMH burdens than existing methods that are ready to be used out‐of‐the‐box (i.e., without retraining). We demonstrated the superior performance of our tool relative to three reference methods in accurately segmenting WMH in the test dataset composed of three databases representing young, middle‐aged, and older individuals with varying degrees of WMH burden. When performance was compared using Dice score, both at the voxel‐by‐voxel‐ level (VL‐Dice) and at the level of each lesion cluster (CL‐Dice), to evaluate the similarity between the manually traced and predicted WMH segmentations, our tool had the highest average scores across the test dataset, with the overall VL‐Dice and CL‐Dice scores of 0.66 and 0.71, respectively. Detailed evaluation in each cohort separately indicated that the SHIVA‐WMH detector achieved significantly higher CL‐Dice scores than all three reference methods across the three cohorts, indicating the highest detection accuracy at the lesion cluster‐level. It also had significantly higher VL‐Dice score in the MRi‐Share test set, and significantly or nominally higher Dice scores in both UKB and WMC test datasets than the reference methods, attesting to its segmentation accuracy as well. In WMC, VL‐ and CL‐Dice scores attained by SHIVA‐WMH (mean of 0.76 for both VL‐ and CL‐Dice) approached or were on a par with those reported for two trained human observers who annotated WMH in WMC training data, whose labels were compared against the consensus labels created by two experts (mean of 0.77/0.79 for VL‐Dice and 0.74/0.76 for CL‐Dice for each of the two observer) (Kuijf et al., 2019). It can use either multimodal inputs of coregistered T1w and FLAIR images or FLAIR image only, although some performance metrics drop slightly for FLAIR‐only version. To our knowledge, this is the first automated WMH detection tool that incorporated 3D FLAIR data from young, neurologically asymptomatic adults to train and validate the method in subjects with very mild WMH burden (<2 ml). Finally, we demonstrated the relevancy of identifying small punctate WMH in otherwise healthy individuals by comparing DWI‐based metrics inside manually traced WMH and NAWM: despite their small size and sparsity, WMH in the MRi‐Share subjects exhibited signs of compromised white matter integrity, with lower FA and NDI and elevated MD inside WMH relative to NAWM. It highlights the value of having an automated tool like our SHIVA‐WMH for further investigation and characterization of WMH as they emerge and progress into more severe forms in large‐scale population‐based studies.
Prior imaging studies have shown that there are significant increases in MD and decreases in FA in WMH compared to NAWM in community‐dwelling elderly (Muñoz Maniega et al., 2015; Riphagen et al., 2018; Wardlaw et al., 2015). While few studies have characterized WMH in younger adults, one study that examined the diffusion properties of relatively mild WMH (total lesion volume <6 ml) in 3D FLAIR scans of neurologically asymptomatic subjects aged between 21 and 60 also reported similar changes, with a significant MD increase and nonsignificant FA decrease in WMH compared to NAWM (Keřkovský et al., 2019). We extend these earlier observations by demonstrating that the same changes in DTI metrics can already be detected in even milder WMH in young university students. Furthermore, we showed that NDI derived from the NODDI model is sensitive to the microstructural changes in the WMH found in these young subjects, potentially giving more specific insight into the early pathophysiology than DTI metrics alone. It suggests that increased water diffusivity and decreased directionality of diffusion (indicated by MD and FA, respectively) may be driven at least partially by lower myelination or axon density, as indicated by NDI. Although NDI is known to be lower in WMH found in patients with MS (Alotaibi et al., 2021; Mustafi et al., 2019), to our knowledge, this is the first to demonstrate the sensitivity of NDI to WMH in asymptomatic young subjects. Our finding is consistent with early neuropathological work on punctate WMH indicating the reduced myelin content with neuropil atrophy in perivascular tissues in the deep white matter (Fazekas et al., 1998), and indicates that subtle microstructural changes are already detectable with MRI even in young subjects with very low overall WMH load.
Although the present work does not address the functional significance of the small amount of WMH in these young subjects, recent work has highlighted the associations between higher WMH and poorer executive task performance even in young adults aged between 20 and 40 who exhibited a similar degree of WMH burden as MRi‐Share subjects in our study (Garnier‐Crussard et al., 2020). Further, there is evidence that the amount of WMH found in young adults is associated with several modifiable cardiovascular risk factors, such as body mass index, physical activity, smoking, and alcohol consumption (Williamson et al., 2018). Together, it underscores the importance of accurately charting the early emergence and progression of WMH for a better understanding of its pathophysiology, its genetic and environmental determinants, and ultimately for early intervention.
To this end, we combined the WMH labels of the MRi‐Share with the publicly available MWC dataset to develop the SHIVA‐WMH detector, based on our prior work that used the 3D Unet‐based architecture to detect PVS, another marker of covert cSVD (Boutinaud et al., 2021). Even though, there has been an increasing number of studies applying Unet‐based models for WMH detection, it has not been used to push the limit of early detection in young, neurologically asymptomatic cohorts. Further, with few exceptions (Tran et al., 2022; Umapathy et al., 2021), most work on automatic segmentation methods for age‐related WMH so far has focused on developing and evaluating their tools on more conventional 2D FLAIR images with thick slices (typically 3 to 5 mm). However, there is an inherent limitation in accurately quantifying the amount of small WMH with 2D FLAIR acquisitions, since small lesions out of the plane of acquisition cannot be detected. More modern 3D FLAIR acquisitions with isotropic resolution are known to achieve a better signal/contrast‐to‐noise ratio and allow greater sensitivity to WMH lesions than 2D acquisitions (Bink et al., 2006; Chagla et al., 2008). They are also increasingly available in large neuroimaging databases in population‐based studies (e.g., UKB; Alfaro‐Almagro et al., 2018; ADNI‐3; Gunter et al., 2017; Rhineland study; Lohner et al., 2022). Yet, several recent works on age‐related WMH detection methods that explicitly evaluated their methods in participants with mild WMH burden (<5 ml) had not included 3D FLAIR datasets for training or evaluation (Khademi et al., 2021; Mojiri Forooshani et al., 2022; Ong et al., 2022; Rachmadi et al., 2018). Our work is unique in taking advantage of the 1 mm isotropic 3D FLAIR scans from young adults and the detailed delineation of every visible WMH in every slice to train the 3D Unet‐based model, with the objective to apply our tool in other cohorts with similar acquisitions to accurately describe WMH burden across the adult lifespan.
We evaluated the performance of the SHIVA‐WMH detector in the 10 subjects each from the MRi‐Share and MWC datasets that had been set aside (i.e., not seen during training), as well as in the 11 additional subjects from the UKB dataset, representing adults from the general population in the age range and with the level of WMH burden in between MRi‐Share and MWC. As the reference tools for comparison, we selected one conventional signal intensity‐based method (LST‐LPA) and two existing Unet‐based methods (PGS and HPM) that could be used out‐of‐the‐box (i.e., with pretrained models available for Unet‐based methods). The LST‐LPA is based on the logistic regression model that uses intensity and location information of FLAIR images to classify WMH. It was originally developed to segment WMH in MS patients (Schmidt, 2017a), but has been applied widely to quantify age‐related WMH as well (Ribaldi et al., 2021) and has also been demonstrated to show robust performance across diverse datasets with age‐related WMH (Heinen et al., 2019; Vanderbecq et al., 2020). It is also by far the most common algorithm used as a reference method in studies proposing new WMH detection methods (29 out of 37 studies reviewed in Balakrishnan et al., 2021). The PGS is a 2D Unet‐based method and the current winner of the MWC 2017 challenge (https://wmh.isi.uu.nl/results/), with the VL‐ and CL‐Dice scores of 0.81 and 0.79, respectively, in the held‐out test dataset of the challenge (Park et al., 2021). It combines the Unet architecture with what the authors call a multi‐scale highlighting foregrounds approach, in which the ground truth labels are downsampled at multiple scales to allow loss minimization at each layer of the Unet. This approach has the effect of emphasizing the contributions of small lesions and the voxels in the lesion boundaries during the network training, and as a result should improve accurate detection of WMH voxels with high uncertainty due to partial volume effect, including small lesions. Another recently published Unet‐based model, HPM uses 3D input like our SHIVA‐WMH detector. It was chosen based on the high reported performance in cases of very mild WMH burden in a multi‐cohort dataset of 50 subjects with mean WMH volume of ~2 ml, with VL‐ and CL‐Dice scores of 0.84 and 0.72, respectively (Mojiri Forooshani et al., 2022). These scores are the highest among several recent works that explicitly evaluated their methods in participants with mild WMH burden of less than 5 ml (Khademi et al., 2021; Mojiri Forooshani et al., 2022; Ong et al., 2022; Rachmadi et al., 2018). For the purpose of comparison with our tool, it also had the advantage of being trained with multisite imaging datasets that did not include the MWC dataset, allowing for a fair comparison of performance with our tool in this cohort, unlike the PGS, whose training data included MWC testing data set aside in the present study.
Of the three reference methods, LST‐LPA had the lowest overall Dice scores, and performed progressively worse in the cohorts with lower overall WMH burden. It had comparable Dice scores as SHIVA‐WMH in the MWC test set, but only at the voxel‐level. Lesion‐wise CL‐Dice was significantly worse than SHIVA‐WMH even in this test cohort with the largest overall WMH burden. It suggests that while it is able to detect relatively large WMH in subjects with moderate‐ to high‐lesion burden, small lesions found in these subjects are missed by LST‐LPA. This observation is consistent with other studies demonstrating the superior performance of Unet‐based methods over LST‐LPA primarily in subjects with lower overall lesion burden (Khademi et al., 2021; Li et al., 2022). Among the three reference methods, PGS had the highest sensitivity in the MRi‐Share and UKB test datasets, with comparable VL‐ and CL‐TPR in UKB and lesion‐wise CL‐TPR in MRi‐Share as SHIVA‐WMH, attesting to the stated advantage of their approach. However, the relatively high sensitivity came at the cost of low precision in both test datasets, resulting in the significantly lower VL‐ and CL‐Dice scores in MRi‐Share and CL‐Dice score in UKB compared to SHIVA‐WMH. The lower precision likely results from the 2D input they use for their Unet model, since islands of cortical ribbons on some axial slices are difficult to distinguish from WMH without the 3D context. The underperformance of HPM was somewhat surprising, given their high reported performance in subjects with mild WMH burden and the fact that it has been trained with a large and diverse dataset representing 432 individuals from 4 multicenter studies, using the 3D input for their Unet model as in SHIVA‐WMH. Although speculative, we suspect that the reason may be the nature of the training dataset they used: All their training and testing data were from 2D FLAIR with 3 mm slice thickness, which may have limited the advantage of full 3D model. Further, in order to prepare the large number of reference WMH labels to train their model, they used a semiautomated pipeline that generated intensity‐based segmentations. Although these labels were then reviewed and manually edited by trained human annotators, it is generally more difficult and time consuming to add lesions missed by the automated method than rejecting FPs during such manual editing, which can result in more conservative labels missing small lesions not detected by the semiautomated method. In contrast, we used the high‐quality, high‐resolution manual labels for the MRi‐Share training dataset consisting of 40 subjects to train a model specific to this cohort, then used the predictions generated by the model in unannotated MRi‐Share subjects to iteratively train our model until optimal performance was reached in the manually traced MRi‐Share and MWC validation sets. Such an approach has been recently suggested as one of the effective solutions for enhancing limited high‐quality annotations for training DL‐based models (Tajbakhsh et al., 2020), here applied specifically to enhance performance for more difficult cases of subjects with very low WMH burden.
The superior performance of the SHIVA‐WMH detector relative to other reference methods in the UKB test set is worth being emphasized, as this test set represents data coming from an unseen cohort, and thus constitutes an important test case for the transferability of our detector to cohorts not seen during the training. Further, the UKB dataset has been acquired with a modern scanner with high‐quality 3D FLAIR acquisitions similar to MRi‐Share, and represents one of the intended target populations to apply our tool in future studies to characterize the full extent of WMH in neurologically asymptomatic adults. Relative to SHIVA‐WMH, only PGS showed comparable sensitivity to WMH found in UKB, but at the cost of lower precision, resulting in the lower and more variable Dice scores in this cohort than our tool. It suggests that our tool can predict variable burden of WMH in this cohort more accurately and consistently than the reference tools tested here.
It should be noted that we compared the performance of SHIVA‐WMH against PGS and HPM without retraining the latter two methods with the same training data used by SHIVA‐WMH, since our aim was to evaluate the direct applicability of these publicly available tools, rather than to test the ability of different Unet‐based models to learn new dataset. Although beyond the scope of this study, it is possible that innovations in the model architectures, such as the multiscale highlighting foregrounds approach in PGS, can further improve small lesion detection when used in combination with the high‐resolution training data we used. Also, the present work focused on WMH detection in asymptomatic or presymptomatic adults, with the assumption that WMH found in these adults are primarily early stages of age‐related WMH of presumed vascular origin, thus combining MRi‐Share and MWC dataset for training our model. However, in reality, WMH found in MRi‐Share may represent mixed pathology, with lesions in some subjects caused by pre‐ or sub‐clinical inflammatory conditions (Hosseiny et al., 2020). While the model learning the WMH in a given training data is agnostic about their etiology, it can learn any global or local spatial and intensity features of the lesions present in the training dataset. To the extent that there are etiology‐specific patterns in WMH appearance and spatial distributions, it is possible that SHIVA‐WMH has learnt predominant WMH patterns in the specific training dataset we used, and may be less sensitive to lesions found in other conditions we did not explicitly focus on, such as MS (training dataset for SHIVA‐WMH did not contain any incidental MS subjects). Beyond the sample‐specific characteristics of WMH patterns, any other sample‐specific image characteristics (scanner‐ and/or center‐specific characteristics as a result of specific acquisition parameters and protocols, etc.) may also influence and potentially bias the trained model. Diverse sources of training data that represent data from four different cohort studies, each acquired using a 3 T scanner at different institutes, safeguard against such biases. Yet, it is possible that our model may have over‐learned any idiosyncratic image features of MRi‐Share, given the enhancement procedure that increased the proportion of MRi‐Share data in the model training. However, the superior performance of our tool against other reference tools in the UKB cohort that had not been seen during the training indicates more benefits of our enhancement procedure than any detrimental effects of over‐learning and the resulting loss of generalizability. Even so, we plan to continuously improve our tool by fine‐tuning our model using any new data available for training and evaluation. For example, we are in the process of including the publicly available WMH labels from MS and other clinical populations to improve the robustness of our detector across different datasets and etiology.
4.1. Conclusion
To summarize, we presented the SHIVA‐WMH detector, a 3D‐Unet based model trained with both MRi‐Share and MWC dataset with the specific aim to improve detection of small WMH in asymptomatic or pre‐symptomatic adults in population‐based studies. Our tool outperformed both a classic WMH segmentation tool (LST‐LPA) and existing state‐of‐the‐art Unet‐based tools (PGS and HPM) in segmenting small WMH in non‐clinical, community‐dwelling adults represented by MRi‐Share and UKB. Our tool can effectively segment WMH across a wider range of WMH burden than existing methods, and thus can be a valuable tool for studies aiming to characterize the emergence and progression of WMH lesions. Such studies are essential for understanding the pathophysiology and early‐life factors associated with the most common etiology of WMH in the population, namely cSVD. Our demonstration of altered diffusion properties of small WMH in MRi‐Share, bearing the hallmark of compromised microstructural integrity similar to those found in WMH of older subjects or MS patients, also underscores the importance of early detection and intervention. To encourage more research on the early detection and characterization of WMH, we make the SHIVA‐WMH detector freely and openly available at (https://github.com/pboutinaud/SHIVA_WMH), including the current version and any future upgrades.
AUTHOR CONTRIBUTIONS
Ami Tsuchida: Conceptualization, formal analysis, investigation, data curation, writing‐original draft, visualization. Philippe Boutinaud: Conceptualization, methodology, software, writing‐review and editing. Violaine Verrecchia: Data curation. Christophe Tzourio: Funding acquisition, writing‐review and editing. Stéphanie Debette: Funding acquisition, project administration, writing‐review and editing. Marc Joliot: Conceptualization, supervision, project administration, writing‐review and editing.
FUNDING INFORMATION
This work has been supported by a grant overseen by the French National Research Agency (ANR) as part of the “Investissements d'Avenir” Program ANR‐18‐RHUS‐002. This work was supported by a grant from the French National Research Agency (ANR‐16‐LCV2‐0006‐01, LABCOM Ginesislab). The i‐Share study has received funding from the ANR (Agence Nationale de la Recherche) via the “Investissements d'Avenir” Program (grant ANR‐10‐COHO‐05). The MRi‐Share Cohort was supported by grant ANR‐10‐LABX‐57 and supplementary funding was received from the Conseil Régional of Nouvelle Aquitaine (ref. 4370420). The work was also supported by the “France Investissements d'Avenir” Program (ANR‐10‐IDEX‐03‐0). We thank the Precision and Global Vascular Brain Health Institute (VBHI) funded by France 2030 IHU3 initiative.
CONFLICT OF INTEREST
The authors have nothing to declare.
Supporting information
DATA S1 Supplementary Information.
ACKNOWLEDGMENTS
The authors like to acknowledge Prof Bernard Mazoyer for the initial conception of the project to characterize early signs of white matter anomalies in the young subjects of MRi‐Share and his reviewing of raw T1w and FLAIR images in this database to select 50 subjects with ranging amount of both WMH and PVS. The authors would also like to acknowledge Iana Astafeva from the GIN and Pierre Yves Hervé from Fealinx for participating to the software developments, data handling and computations. The authors are also indebted to the following individuals for their invaluable contribution to the MRi‐Share project: Serge Anandra, Amandine André, Gregory Beaudet, Christophe Bernard, Bruno Brochet, Aurore Capelli, Claire Cardona, Arnaud Chaussé, Christophe Delalande, Vincent Durand, Louise Knafo, Morgane Lachaize, Alexandre Laurent, Hugues Loiseau, Elena Milesi, Marie Mougin, Maylis Melin, Guy Perchey, Clothilde Pollet, Thomas Tourdias, Cécile Marchal, Guillaume Penchet, Cécile Dulau, Igor Sibon, Sabrina Debruxelle, Sophie Auriacombe, Caroline Roussillon, Nicolas Vinuesa, and the i‐Share “relay” students. The authors also like to express our gratitude to Paul Matthews (Imperial College, London, UK) and to the personnel of the UK‐Biobank imaging center at Stockport (UK) for their help while designing the MRi‐Share image acquisition protocol, and to Maxime Descoteaux (Sherbrooke University, Canada) for his help in implementing the DWI processing and QC pipelines. Finally, the authors would like to express their gratitude to the 1,870 students of the Bordeaux University who gave their consent to participate in MRi‐Share.
Tsuchida, A. , Boutinaud, P. , Verrecchia, V. , Tzourio, C. , Debette, S. , & Joliot, M. (2024). Early detection of white matter hyperintensities using SHIVA‐WMH detector. Human Brain Mapping, 45(1), e26548. 10.1002/hbm.26548
DATA AVAILABILITY STATEMENT
MRi‐Share data used in this study cannot be shared through a public repository due to French regulations regarding sharing of the medical imaging data. However, de‐identified data can be requested to the i‐Share Scientific Collaborations Coordinator (ilaria.montagni@u-bordeaux.fr) with a letter of intent (explaining the rationale and objectives of the research proposal), and a brief summary of the planned means and options for funding. MICCAI 2017 WMH segmentation challenge dataset used in the present work is freely and publicly available at the challenge homepage (https://wmh.isi.uu.nl/data/). This work also used the neuroimaging dataset obtained from the UK Biobank Resource (application number 18359 and 94113). Source codes for the statistical analysis presented in the manuscript are available on GitHub (https://github.com/atsuch/SHIVA-WMHpaper). SHIVA‐WMH detector presented in this work is also publicly available at https://github.com/pboutinaud/SHIVA_WMH.
REFERENCES
- Alfaro‐Almagro, F. , Jenkinson, M. , Bangerter, N. K. , Andersson, J. L. R. , Griffanti, L. , Douaud, G. , Sotiropoulos, S. N. , Jbabdi, S. , Hernandez‐Fernandez, M. , Vallee, E. , Vidaurre, D. , Webster, M. , McCarthy, P. , Rorden, C. , Daducci, A. , Alexander, D. C. , Zhang, H. , Dragonu, I. , Matthews, P. M. , … Smith, S. M. (2018). Image processing and quality control for the first 10,000 brain imaging datasets from UK Biobank. Neuroimage, 166, 400–424. 10.1016/j.neuroimage.2017.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alotaibi, A. , Podlasek, A. , AlTokhis, A. , Aldhebaib, A. , Dineen, R. A. , & Constantinescu, C. S. (2021). Investigating microstructural changes in white matter in multiple sclerosis: A systematic review and meta‐analysis of neurite orientation dispersion and density imaging. Brain Sciences, 11(9), 1151. 10.3390/brainsci11091151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan, R. , Valdés Hernández, M. D. C. , & Farrall, A. J. (2021). Automatic segmentation of white matter hyperintensities from brain magnetic resonance images in the era of deep learning and big data—A systematic review. Computerized Medical Imaging and Graphics, 88(101), 867. 10.1016/j.compmedimag.2021.101867 [DOI] [PubMed] [Google Scholar]
- Basser, P. J. , Mattiello, J. , & LeBihan, D. (1994). MR diffusion tensor spectroscopy and imaging. Biophysical Journal, 66, 259–267. 10.1016/S0006-3495(94)80775-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bink, A. , Schmitt, M. , Gaa, J. , Mugler, J. P. , Lanfermann, H. , & Zanella, F. E. (2006). Detection of lesions in multiple sclerosis by 2D FLAIR and single‐slab 3D FLAIR sequences at 3.0 T: Initial results. European Radiology, 16, 1104–1110. 10.1007/s00330-005-0107-z [DOI] [PubMed] [Google Scholar]
- Boomsma, J. M. F. , Exalto, L. G. , Barkhof, F. , van den Berg, E. , de Bresser, J. , Heinen, R. , Koek, H. L. , Prins, N. D. , Scheltens, P. , Weinstein, H. C. , van der Flier, W. M. , & Biessels, G. J. (2017). Vascular cognitive impairment in a memory clinic population: Rationale and design of the “Utrecht‐Amsterdam clinical features and prognosis in vascular cognitive impairment” (TRACE‐VCI) study. JMIR Research Protocols, 6, e60. 10.2196/resprot.6864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutinaud, P. , Tsuchida, A. , Laurent, A. , Adonias, F. , Hanifehlou, Z. , Nozais, V. , Verrecchia, V. , Lampe, L. , Zhang, J. , Zhu, Y.‐C. , Tzourio, C. , Mazoyer, B. , & Joliot, M. (2021). 3D segmentation of perivascular spaces on T1‐weighted 3 tesla MR images with a convolutional autoencoder and a U‐shaped neural network. Frontiers in Neuroinformatics, 15(641), 600. 10.3389/fninf.2021.641600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannistraro, R. J. , Badi, M. , Eidelman, B. H. , Dickson, D. W. , Middlebrooks, E. H. , & Meschia, J. F. (2019). CNS small vessel disease: A clinical review. Neurology, 92, 1146–1156. 10.1212/WNL.0000000000007654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagla, G. H. , Busse, R. F. , Sydnor, R. , Rowley, H. A. , & Turski, P. A. (2008). Three‐dimensional fluid attenuated inversion recovery imaging with isotropic resolution and nonselective adiabatic inversion provides improved three‐dimensional visualization and cerebrospinal fluid suppression compared to two‐dimensional flair at 3 tesla. Investigative Radiology, 43, 547–551. 10.1097/RLI.0b013e3181814d28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coupe, P. , Manjon, J. , Robles, M. , & Collins, L. D. (2011). Adaptive multiresolution non‐local means filter for 3D MR image denoising. IET Image Processing, 6, 558. [Google Scholar]
- Coupe, P. , Yger, P. , Prima, S. , Hellier, P. , Kervrann, C. , & Barillot, C. (2008). An optimized blockwise nonlocal means denoising filter for 3‐D magnetic resonance images. IEEE Transactions on Medical Imaging, 27, 425–441. 10.1109/TMI.2007.906087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daducci, A. , Canales‐Rodríguez, E. J. , Zhang, H. , Dyrby, T. B. , Alexander, D. C. , & Thiran, J.‐P. (2015). Accelerated microstructure imaging via convex optimization (AMICO) from diffusion MRI data. Neuroimage, 105, 32–44. 10.1016/j.neuroimage.2014.10.026 [DOI] [PubMed] [Google Scholar]
- Debette, S. , & Markus, H. S. (2010). The clinical importance of white matter hyperintensities on brain magnetic resonance imaging: Systematic review and meta‐analysis. BMJ, 341, c3666. 10.1136/bmj.c3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debette, S. , Schilling, S. , Duperron, M.‐G. , Larsson, S. C. , & Markus, H. S. (2019). Clinical significance of magnetic resonance imaging markers of vascular brain injury: A systematic review and meta‐analysis. JAMA Neurology, 76, 81–94. 10.1001/jamaneurol.2018.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazekas, F. , Chawluk, J. B. , Alavi, A. , Hurtig, H. I. , & Zimmerman, R. A. (1987). MR signal abnormalities at 1.5 T in Alzheimer's dementia and normal aging. American Journal of Roentgenology, 149, 351–356. 10.2214/ajr.149.2.351 [DOI] [PubMed] [Google Scholar]
- Fazekas, F. , Schmidt, R. , & Scheltens, P. (1998). Pathophysiologic mechanisms in the development of age‐related white matter changes of the brain. Dementia and Geriatric Cognitive Disorders, 9(Suppl 1), 2–5. 10.1159/000051182 [DOI] [PubMed] [Google Scholar]
- Garnier‐Crussard, A. , Bougacha, S. , Wirth, M. , André, C. , Delarue, M. , Landeau, B. , Mézenge, F. , Kuhn, E. , Gonneaud, J. , Chocat, A. , Quillard, A. , Ferrand‐Devouge, E. , de La Sayette, V. , Vivien, D. , Krolak‐Salmon, P. , & Chételat, G. (2020). White matter hyperintensities across the adult lifespan: Relation to age, Aβ load, and cognition. Alzheimer's Research & Therapy, 12, 127. 10.1186/s13195-020-00669-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garyfallidis, E. , Brett, M. , Amirbekian, B. , Rokem, A. , van der Walt, S. , Descoteaux, M. , Nimmo‐Smith, I. , & Dipy Contributors . (2014). Dipy, a library for the analysis of diffusion MRI data. Frontiers in Neuroinformatics, 8, 8. 10.3389/fninf.2014.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glorot, X. , & Bengio, Y. (2010). Understanding the difficulty of training deep feedforward neural networks, in: Teh, Y. W., Titterington, M. (Eds.), Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics, Proceedings of Machine Learning Research. Presented at the International Conference on Artificial Intelligence and Statistics, PMLR, Chia Laguna Resort, Sardinia, Italy, pp. 249–256.
- Griffanti, L. , Zamboni, G. , Khan, A. , Li, L. , Bonifacio, G. , Sundaresan, V. , Schulz, U. G. , Kuker, W. , Battaglini, M. , Rothwell, P. M. , & Jenkinson, M. (2016). BIANCA (brain intensity abnormality classification algorithm): A new tool for automated segmentation of white matter hyperintensities. Neuroimage, 141, 191–205. 10.1016/j.neuroimage.2016.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero, R. , Qin, C. , Oktay, O. , Bowles, C. , Chen, L. , Joules, R. , Wolz, R. , Valdés‐Hernández, M. C. , Dickie, D. A. , Wardlaw, J. , & Rueckert, D. (2018). White matter hyperintensity and stroke lesion segmentation and differentiation using convolutional neural networks. NeuroImage: Clinical, 17, 918–934. 10.1016/j.nicl.2017.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunter, J. L. , Borowski, B. J. , Thostenson, K. , Arani, A. , Reid, R. I. , Cash, D. M. , Thomas, D. L. , Zhang, H. , DeCarli, C. S. , Fox, N. C. , Thompson, P. M. , Tosun, D. , Weiner, M. , & Jack, C. R. (2017). ADNI‐3 MRI PROTOCOL. Alzheimer's & Dementia, 13, P104–P105. 10.1016/j.jalz.2017.06.2411 [DOI] [Google Scholar]
- Heinen, R. , Steenwijk, M. D. , Barkhof, F. , Biesbroek, J. M. , van der Flier, W. M. , Kuijf, H. J. , Prins, N. D. , Vrenken, H. , Biessels, G. J. , de Bresser, J. , & TRACE‐VCI study group . (2019). Performance of five automated white matter hyperintensity segmentation methods in a multicenter dataset. Scientific Reports, 9(16), 742. 10.1038/s41598-019-52966-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseiny, M. , Newsome, S. D. , & Yousem, D. M. (2020). Radiologically isolated syndrome: A review for neuroradiologists. American Journal of Neuroradiology, 41, 1542–1549. 10.3174/ajnr.A6649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannone, R. , Cheng, J. , & Schloerke, B. (2020). gt: Easily create presentation‐ready display tables.
- Jenkinson, M. , Bannister, P. , Brady, M. , & Smith, S. (2002). Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage, 17, 825–841. 10.1016/s1053-8119(02)91132-8 [DOI] [PubMed] [Google Scholar]
- Kassambara, A. (2022). ggpubr: “ggplot2” based publication ready plots.
- Keřkovský, M. , Stulík, J. , Dostál, M. , Kuhn, M. , Lošák, J. , Praksová, P. , Hulová, M. , Bednařík, J. , Šprláková‐Puková, A. , & Mechl, M. (2019). Structural and functional MRI correlates of T2 hyperintensities of brain white matter in young neurologically asymptomatic adults. European Radiology, 29, 7027–7036. 10.1007/s00330-019-06268-8 [DOI] [PubMed] [Google Scholar]
- Khademi, A. , Gibicar, A. , Arezza, G. , DiGregorio, J. , Tyrrell, P. N. , & Moody, A. R. (2021). Segmentation of white matter lesions in multicentre FLAIR MRI. Neuroimage: Reports, 1, 100044. 10.1016/j.ynirp.2021.100044 [DOI] [Google Scholar]
- Kuijf, H. J. , Biesbroek, J. M. , De Bresser, J. , Heinen, R. , Andermatt, S. , Bento, M. , Berseth, M. , Belyaev, M. , Cardoso, M. J. , Casamitjana, A. , Collins, D. L. , Dadar, M. , Georgiou, A. , Ghafoorian, M. , Jin, D. , Khademi, A. , Knight, J. , Li, H. , Llado, X. , … Biessels, G. J. (2019). Standardized assessment of automatic segmentation of white matter hyperintensities and results of the WMH segmentation challenge. IEEE Transactions on Medical Imaging, 38, 2556–2568. 10.1109/TMI.2019.2905770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Jiang, G. , Zhang, J. , Wang, R. , Wang, Z. , Zheng, W.‐S. , & Menze, B. (2018). Fully convolutional network ensembles for white matter hyperintensities segmentation in MR images. Neuroimage, 183, 650–665. 10.1016/j.neuroimage.2018.07.005 [DOI] [PubMed] [Google Scholar]
- Li, X. , Zhao, Y. , Jiang, J. , Cheng, J. , Zhu, W. , Wu, Z. , Jing, J. , Zhang, Z. , Wen, W. , Sachdev, P. S. , Wang, Y. , Liu, T. , & Li, Z. (2022). White matter hyperintensities segmentation using an ensemble of neural networks. Human Brain Mapping, 43, 929–939. 10.1002/hbm.25695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohner, V. , Enkirch, S. J. , Hattingen, E. , Stöcker, T. , & Breteler, M. M. B. (2022). Safety of tattoos, permanent make‐up, and medical implants in population‐based 3 T magnetic resonance brain imaging: The Rhineland study. Frontiers in Neurology, 13(795), 573. 10.3389/fneur.2022.795573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, K. L. , Alfaro‐Almagro, F. , Bangerter, N. K. , Thomas, D. L. , Yacoub, E. , Xu, J. , Bartsch, A. J. , Jbabdi, S. , Sotiropoulos, S. N. , Andersson, J. L. R. , Griffanti, L. , Douaud, G. , Okell, T. W. , Weale, P. , Dragonu, I. , Garratt, S. , Hudson, S. , Collins, R. , Jenkinson, M. , … Smith, S. M. (2016). Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nature Neuroscience, 19, 1523–1536. 10.1038/nn.4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojiri Forooshani, P. , Biparva, M. , Ntiri, E. E. , Ramirez, J. , Boone, L. , Holmes, M. F. , Adamo, S. , Gao, F. , Ozzoude, M. , Scott, C. J. M. , Dowlatshahi, D. , Lawrence‐Dewar, J. M. , Kwan, D. , Lang, A. E. , Marcotte, K. , Leonard, C. , Rochon, E. , Heyn, C. , Bartha, R. , … Goubran, M. (2022). Deep Bayesian networks for uncertainty estimation and adversarial resistance of white matter hyperintensity segmentation. Human Brain Mapping, 43, 2089–2108. 10.1002/hbm.25784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroni, F. , Ammirati, E. , Rocca, M. A. , Filippi, M. , Magnoni, M. , & Camici, P. G. (2018). Cardiovascular disease and brain health: Focus on white matter hyperintensities. International Journal of Cardiology. Heart & Vasculature, 19, 63–69. 10.1016/j.ijcha.2018.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafi, S. M. , Harezlak, J. , Kodiweera, C. , Randolph, J. S. , Ford, J. C. , Wishart, H. A. , & Wu, Y.‐C. (2019). Detecting white matter alterations in multiple sclerosis using advanced diffusion magnetic resonance imaging. Neural Regeneration Research, 14, 114–123. 10.4103/1673-5374.243716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz Maniega, S. , Valdés Hernández, M. C. , Clayden, J. D. , Royle, N. A. , Murray, C. , Morris, Z. , Aribisala, B. S. , Gow, A. J. , Starr, J. M. , Bastin, M. E. , Deary, I. J. , & Wardlaw, J. M. (2015). White matter hyperintensities and normal‐appearing white matter integrity in the aging brain. Neurobiology of Aging, 36, 909–918. 10.1016/j.neurobiolaging.2014.07.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong, K. , Young, D. M. , Sulaiman, S. , Shamsuddin, S. M. , Mohd Zain, N. R. , Hashim, H. , Yuen, K. , Sanders, S. J. , Yu, W. , & Hang, S. (2022). Detection of subtle white matter lesions in MRI through texture feature extraction and boundary delineation using an embedded clustering strategy. Scientific Reports, 12, 4433. 10.1038/s41598-022-07843-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, B.‐Y. , Lee, M. J. , Lee, S.‐H. , Cha, J. , Chung, C.‐S. , Kim, S. T. , Park, H. (2018). DEWS (deep white matter hyperintensity segmentation framework): A fully automated pipeline for detecting small deep white matter hyperintensities in migraineurs. Neuroimage Clin, 18, 638–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, G. , Hong, J. , Duffy, B. A. , Lee, J.‐M. , & Kim, H. (2021). White matter hyperintensities segmentation using the ensemble U‐net with multi‐scale highlighting foregrounds. Neuroimage, 237(118), 140. 10.1016/j.neuroimage.2021.118140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . (2018). R: A language and environment for statistical computing. R Core Team. [Google Scholar]
- Rachmadi, M. F. , Valdés‐Hernández, M. D. C. , Agan, M. L. F. , Di Perri, C. , Komura, T. , & Alzheimer's Disease Neuroimaging Initiative . (2018). Segmentation of white matter hyperintensities using convolutional neural networks with global spatial information in routine clinical brain MRI with none or mild vascular pathology. Computerized Medical Imaging and Graphics, 66, 28–43. 10.1016/j.compmedimag.2018.02.002 [DOI] [PubMed] [Google Scholar]
- Rachmadi, M. F. , Valdés‐Hernández, M. D. C. , Li, H. , Guerrero, R. , Meijboom, R. , Wiseman, S. , Waldman, A. , Zhang, J. , Rueckert, D. , Wardlaw, J. , & Komura, T. (2020). Limited one‐time sampling irregularity map (LOTS‐IM) for automatic unsupervised assessment of white matter hyperintensities and multiple sclerosis lesions in structural brain magnetic resonance images. Computerized Medical Imaging and Graphics, 79(101), 685. 10.1016/j.compmedimag.2019.101685 [DOI] [PubMed] [Google Scholar]
- Ramachandran, P. , Zoph, B. , & Le, Q. V. (2017). Searching for activation functions. arXiv. 10.48550/arxiv.1710.05941 [DOI]
- Reinke, A. , Tizabi, M. D. , Sudre, C. H. , Eisenmann, M. , Rädsch, T. , Baumgartner, M. , Acion, L. , Antonelli, M. , Arbel, T. , Bakas, S. , Bankhead, P. , Benis, A. , Cardoso, M. J. , Cheplygina, V. , Cimini, B. , Collins, G. S. , Farahani, K. , Glocker, B. , Godau, P. , … Maier‐Hein, L. (2021). Common limitations of image processing metrics: A picture story. arXiv. 10.48550/arxiv.2104.05642 [DOI]
- Ribaldi, F. , Altomare, D. , Jovicich, J. , Ferrari, C. , Picco, A. , Pizzini, F. B. , Soricelli, A. , Mega, A. , Ferretti, A. , Drevelegas, A. , Bosch, B. , Müller, B. W. , Marra, C. , Cavaliere, C. , Bartrés‐Faz, D. , Nobili, F. , Alessandrini, F. , Barkhof, F. , Gros‐Dagnac, H. , … Marizzoni, M. (2021). Accuracy and reproducibility of automated white matter hyperintensities segmentation with lesion segmentation tool: A European multi‐site 3 T study. Magnetic Resonance Imaging, 76, 108–115. 10.1016/j.mri.2020.11.008 [DOI] [PubMed] [Google Scholar]
- Riphagen, J. M. , Gronenschild, E. H. B. M. , Salat, D. H. , Freeze, W. M. , Ivanov, D. , Clerx, L. , Verhey, F. R. J. , Aalten, P. , & Jacobs, H. I. L. (2018). Shades of white: Diffusion properties of T1‐ and FLAIR‐defined white matter signal abnormalities differ in stages from cognitively normal to dementia. Neurobiology of Aging, 68, 48–58. 10.1016/j.neurobiolaging.2018.03.029 [DOI] [PubMed] [Google Scholar]
- Ronneberger, O. , Fischer, P. , & Brox, T. (2015). U‐net: Convolutional networks for biomedical image segmentation. In Navab N., Hornegger J., Wells W. M., & Frangi A. F. (Eds.), Medical image computing and computer‐assisted intervention (MICCAI), Lecture notes in computer science (pp. 234–241). Springer International Publishing. 10.1007/978-3-319-24,574-4_28 [DOI] [Google Scholar]
- Scheltens, P. , Barkhof, F. , Leys, D. , Pruvo, J. P. , Nauta, J. J. , Vermersch, P. , Steinling, M. , & Valk, J. (1993). A semiquantative rating scale for the assessment of signal hyperintensities on magnetic resonance imaging. Journal of the Neurological Sciences, 114, 7–12. 10.1016/0022-510x(93)90041-v [DOI] [PubMed] [Google Scholar]
- Schmidt, P. (2017a). Bayesian inference for structured additive regression models for large‐scale problems with applications to medical imaging (Doctoral dissertation).
- Schmidt, P. (2017b). Bayesian inference for structured additive regression models for large‐scale problems with applications to medical imaging. Ludwig‐Maximilians‐Universität München. 10.5282/edoc.20373 [DOI] [Google Scholar]
- Schmidt, P. , Gaser, C. , Arsic, M. , Buck, D. , Förschler, A. , Berthele, A. , Hoshi, M. , Ilg, R. , Schmid, V. J. , Zimmer, C. , Hemmer, B. , & Mühlau, M. (2012). An automated tool for detection of FLAIR‐hyperintense white‐matter lesions in multiple sclerosis. Neuroimage, 59, 3774–3783. 10.1016/j.neuroimage.2011.11.032 [DOI] [PubMed] [Google Scholar]
- Srivastava, N. , Hinton, G. , Krizhevsky, A. , Sutskever, I. , & Salakhutdinov, R. (2014). Dropout: A simple way to prevent neural networks from overfitting. Journal of Machine Learning Research, 15, 1929–1958. [Google Scholar]
- Sundaresan, V. , Zamboni, G. , Rothwell, P. M. , Jenkinson, M. , & Griffanti, L. (2021). Triplanar ensemble U‐net model for white matter hyperintensities segmentation on MR images. Medical Image Analysis, 73(102), 184. 10.1016/j.media.2021.102184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajbakhsh, N. , Jeyaseelan, L. , Li, Q. , Chiang, J. N. , Wu, Z. , & Ding, X. (2020). Embracing imperfect datasets: A review of deep learning solutions for medical image segmentation. Medical Image Analysis, 63(101), 693. 10.1016/j.media.2020.101693 [DOI] [PubMed] [Google Scholar]
- Tran, P. , Thoprakarn, U. , Gourieux, E. , Dos Santos, C. L. , Cavedo, E. , Guizard, N. , Cotton, F. , Krolak‐Salmon, P. , Delmaire, C. , Heidelberg, D. , Pyatigorskaya, N. , Ströer, S. , Dormont, D. , Martini, J.‐B. , Chupin, M. , Alzheimer's Disease Neuroimaging Initiatives , & for the Frontotemporal Lobar Degeneration Neuroimaging Initiative . (2022). Automatic segmentation of white matter hyperintensities: Validation and comparison with state‐of‐the‐art methods on both multiple sclerosis and elderly subjects. NeuroImage: Clinical, 33(102), 940. 10.1016/j.nicl.2022.102940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida, A. , Laurent, A. , Crivello, F. , Petit, L. , Joliot, M. , Pepe, A. , Beguedou, N. , Gueye, M.‐F. , Verrecchia, V. , Nozais, V. , Zago, L. , Mellet, E. , Debette, S. , Tzourio, C. , & Mazoyer, B. (2021). The MRi‐Share database: Brain imaging in a cross‐sectional cohort of 1870 university students. Brain Structure and Function, 226, 2057–2085. 10.1007/s00429-021-02334-4 [DOI] [PubMed] [Google Scholar]
- Umapathy, L. , Perez‐Carrillo, G. G. , Keerthivasan, M. B. , Rosado‐Toro, J. A. , Altbach, M. I. , Winegar, B. , Weinkauf, C. , Bilgin, A. , & Alzheimer's Disease Neuroimaging Initiative . (2021). A stacked generalization of 3D orthogonal deep learning convolutional neural networks for improved detection of white matter hyperintensities in 3D FLAIR images. American Journal of Neuroradiology, 42, 639–647. 10.3174/ajnr.A6970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veluw, S. J. , Hilal, S. , Kuijf, H. J. , Ikram, M. K. , Xin, X. , Yeow, T. B. , Venketasubramanian, N. , Biessels, G. J. , & Chen, C. (2015). Cortical microinfarcts on 3 T MRI: Clinical correlates in memory‐clinic patients. Alzheimer's & Dementia, 11, 1500–1509. 10.1016/j.jalz.2014.12.010 [DOI] [PubMed] [Google Scholar]
- Vanderbecq, Q. , Xu, E. , Ströer, S. , Couvy‐Duchesne, B. , Diaz Melo, M. , Dormont, D. , Colliot, O. , & Alzheimer's Disease Neuroimaging Initiative . (2020). Comparison and validation of seven white matter hyperintensities segmentation software in elderly patients. NeuroImage: Clinical, 27(102), 357. 10.1016/j.nicl.2020.102357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhwa, R. , Wen, W. , Frankland, A. , Leung, V. , Sinbandhit, C. , Stuart, A. , Dawes, L. , Hadzi‐Pavlovic, D. , Levy, F. , Lenrootl, R. , Mitchell, P. B. , & Roberts, G. (2019). White matter hyperintensities in young individuals with bipolar disorder or at high genetic risk. Journal of Affective Disorders, 245, 228–236. 10.1016/j.jad.2018.10.368 [DOI] [PubMed] [Google Scholar]
- Wahlund, L. O. , Barkhof, F. , Fazekas, F. , Bronge, L. , Augustin, M. , Sjögren, M. , Wallin, A. , Ader, H. , Leys, D. , Pantoni, L. , Pasquier, F. , Erkinjuntti, T. , Scheltens, P. , & European Task Force on Age‐Related White Matter Changes . (2001). A new rating scale for age‐related white matter changes applicable to MRI and CT. Stroke, 32, 1318–1322. 10.1161/01.STR.32.6.1318 [DOI] [PubMed] [Google Scholar]
- Wang, M.‐L. , Zhang, X.‐X. , Yu, M.‐M. , Li, W.‐B. , & Li, Y.‐H. (2019). Prevalence of white matter hyperintensity in young clinical patients. American Journal of Roentgenology, 213, 667–671. 10.2214/AJR.18.20888 [DOI] [PubMed] [Google Scholar]
- Wardlaw, J. M. , Debette, S. , Jokinen, H. , De Leeuw, F.‐E. , Pantoni, L. , Chabriat, H. , Staals, J. , Doubal, F. , Rudilosso, S. , Eppinger, S. , Schilling, S. , Ornello, R. , Enzinger, C. , Cordonnier, C. , Taylor‐Rowan, M. , & Lindgren, A. G. (2021). ESO guideline on covert cerebral small vessel disease. European Stroke Journal, 6, CXI–CLXII. 10.1177/23969873211012132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardlaw, J. M. , Smith, C. , & Dichgans, M. (2013). Mechanisms of sporadic cerebral small vessel disease: Insights from neuroimaging. Lancet Neurology, 12, 483–497. 10.1016/S1474-4422(13)70060-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardlaw, J. M. , Valdés Hernández, M. C. , & Muñoz‐Maniega, S. (2015). What are white matter hyperintensities made of? Relevance to vascular cognitive impairment. Journal of the American Heart Association, 4(6), e001140. 10.1161/JAHA.114.001140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson, W. , Lewandowski, A. J. , Forkert, N. D. , Griffanti, L. , Okell, T. W. , Betts, J. , Boardman, H. , Siepmann, T. , McKean, D. , Huckstep, O. , Francis, J. M. , Neubauer, S. , Phellan, R. , Jenkinson, M. , Doherty, A. , Dawes, H. , Frangou, E. , Malamateniou, C. , Foster, C. , & Leeson, P. (2018). Association of cardiovascular risk factors with MRI indices of cerebrovascular structure and function and white matter hyperintensities in young adults. JAMA, 320, 665–673. 10.1001/jama.2018.11498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, L. , Hu, X. , Li, H. , & Zhao, Y. (2022). Perivascular spaces, glymphatic system and MR. Frontiers in Neurology, 13(844), 938. 10.3389/fneur.2022.844938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, C. , Gu, L. , Liu, Z. , & Zhao, S. (2020). Review of deep learning approaches for the segmentation of multiple sclerosis lesions on brain MRI. Frontiers in Neuroinformatics, 14, 610967. 10.3389/fninf.2020.610967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Schneider, T. , Wheeler‐Kingshott, C. A. , & Alexander, D. C. (2012). NODDI: Practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage, 61, 1000–1016. 10.1016/j.neuroimage.2012.03.072 [DOI] [PubMed] [Google Scholar]
- Zhou, Z. , Sodha, V. , Pang, J. , Gotway, M. B. , & Liang, J. (2021). Models genesis. Medical Image Analysis, 67(101), 840. 10.1016/j.media.2020.101840 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DATA S1 Supplementary Information.
Data Availability Statement
MRi‐Share data used in this study cannot be shared through a public repository due to French regulations regarding sharing of the medical imaging data. However, de‐identified data can be requested to the i‐Share Scientific Collaborations Coordinator (ilaria.montagni@u-bordeaux.fr) with a letter of intent (explaining the rationale and objectives of the research proposal), and a brief summary of the planned means and options for funding. MICCAI 2017 WMH segmentation challenge dataset used in the present work is freely and publicly available at the challenge homepage (https://wmh.isi.uu.nl/data/). This work also used the neuroimaging dataset obtained from the UK Biobank Resource (application number 18359 and 94113). Source codes for the statistical analysis presented in the manuscript are available on GitHub (https://github.com/atsuch/SHIVA-WMHpaper). SHIVA‐WMH detector presented in this work is also publicly available at https://github.com/pboutinaud/SHIVA_WMH.


