Abstract
Purpose
Vascular endothelial growth factor tyrosine kinase inhibitors (TKIs) have been the standard of care for advanced and metastatic clear cell renal cell carcinoma (ccRCC). However, the therapeutic effect of TKI monotherapy remains unsatisfactory given the high rates of acquired resistance to TKI therapy despite favorable initial tumor response.
Materials and Methods
To define the TKI-resistance mechanism and identify new therapeutic target for TKI-resistant ccRCC, an integrative differential gene expression analysis was performed using acquired resistant cohort and a public dataset. Sunitinib-resistant RCC cell lines were established and used to test their malignant behaviors of TKI resistance through in vitro and in vivo studies. Immunohistochemistry was conducted to compare expression between the tumor and normal kidney and verify expression of pathway-related proteins.
Results
Integrated differential gene expression analysis revealed increased interferon-induced transmembrane protein 3 (IFITM3) expression in post-TKI samples. IFITM3 expression was increased in ccRCC compared with the normal kidney. TKI-resistant RCC cells showed high expression of IFITM3 compared with TKI-sensitive cells and displayed aggressive biologic features such as higher proliferative ability, clonogenic survival, migration, and invasion while being treated with sunitinib. These aggressive features were suppressed by the inhibition of IFITM3 expression and promoted by IFITM3 overexpression, and these findings were confirmed in a xenograft model. IFITM3-mediated TKI resistance was associated with the activation of TRAF6 and MAPK/AP-1 pathways.
Conclusions
These results demonstrate IFITM3-mediated activation of the TRAF6/MAPK/AP-1 pathways as a mechanism of acquired TKI resistance, and suggest IFITM3 as a new target for TKI-resistant ccRCC.
Keywords: Carcinoma, renal cell; Drug resistance; IFITM3 protein, human; Mitogen-activated protein kinase 1; TNF receptor-associated factor 6
Graphical Abstract
INTRODUCTION
Vascular endothelial growth factor receptor (VEGFR)-targeted therapy, employing tyrosine kinase inhibitors (TKIs) and anti-VEGF antibodies, has been widely used as a first- or second-line treatment option in metastatic renal cell carcinoma (mRCC), particularly in the clear cell subtype. However, the therapeutic effect of TKI monotherapy appears unsatisfactory because intrinsic resistance is common, and nearly all patients eventually develop acquired resistance to TKIs. With the recent introduction of immune checkpoint inhibitors (ICIs) and the development of combination strategies using ICIs and TKI, the management of mRCC has undergone a paradigm shift. Nevertheless, single-agent TKI treatments such as sunitinib are still used, especially for patients who wish to avoid the potential toxicities of ICI-based regimens, whose access to the combination regimens is limited, or who require next-line treatment after tumor progression. Therefore, the mechanism of TKI resistance must be defined to overcome drug resistance and develop an effective treatment approach for TKI monotherapy and combination therapy.
Our previous studies investigated the differential gene expression of RCC tumor samples harvested before TKI treatment and immediately after disease progression despite treatment [1,2]. Interferon-induced transmembrane protein 3 (IFITM3) was one of commonly upregulated genes in the acquired TKI resistance [2]. IFITM3 has been actively studied for its anti-inflammatory action and antiviral effects [3,4]. Although less studied in neoplastic disease, previous reports showed that IFITM3 expression was increased in malignant tumors including the colon, breast, and prostate with poor clinical outcomes [5,6,7,8,9]. Since IFITM3 was under-evaluated in cancer therapeutics, it was selected for further analysis in this study to find the clinical significance and underlying mechanisms of dysregulation of IFITM3 expression in TKI-resistant clear cell renal cell carcinoma (ccRCC).
TNF receptor-associated factor 6 (TRAF6) is a cytoplasmic adaptor protein that plays an important role in signal transduction from various immune receptors. Upon stimulation, TRAF6 activates mitogen-activated protein kinase 1 (MAPK)/activator protein 1 (AP-1) pathways and induces the expression of genes associated with innate immunity, inflammation, and cell survival [10,11]. Overexpression of TRAF6 is associated with oncogenesis and is involved in tumor cell proliferation, survival, apoptosis, and invasion. TRAF6 is also upregulated in various malignant tumors, including pancreatic, colorectal, breast, and ovary, with poor survival outcomes [12].
MATERIALS AND METHODS
1. Patients
This retrospective study was approved by the Asan Meical Center Institutional Review Board (2012-0788) with a waiver of patient consent. The acquired resistant cohort of 10 ccRCC patients with paired tumor samples harvested at pre- and post-TKI treatment periods has been described previously [1,2]. The intrinsic resistance cohort comprised 89 advanced ccRCC cases, for which formalin-fixed paraffin-embedded tumor blocks were available for IFITM3 immunostaining from the previous study [2]. For the immunohistochemical comparison of gene expression between normal kidneys and tumors, 14 cases of surgically resected localized ccRCC were included.
Response to VEGFR-TKI was assessed according to the Revised Response Evaluation Criteria in Solid Tumors guidelines (version 1.1). Patients with complete response, partial response, and stable disease for at least 24 weeks were defined as the clinical benefit group. In contrast, those with progressive or stable disease for less than 24 weeks were defined as the clinical non-benefit group [13].
To verify the clinical significance of candidate gene expression on intrinsic TKI resistance, we searched for gene expression studies on patients with RCC treated with TKI and available TKI responsiveness. We found that Beuselinck et al. [14] generated transcriptome data of 53 patients with ccRCC treated with first-line sunitinib, and their data were retrieved for analysis.
2. Differential gene expression and integrated DEG analysis
The methods for gene expression profiling and integrated DEG analysis have been described previously [1,2]. Briefly, the 20 matched pairs of pre- and post-TKI treatment tumor samples from the 10 patients were used for expression profiling of the acquired resistance cohort. Their differential gene expression between paired pre- and post-TKI tumor samples was analyzed with the empirical Bayes moderated paired t-test using R package limma.
To overcome the limitation of the small number (10) of cases in our acquired resistance cohort, we used a public dataset (GSE76068) generated from a ccRCC patient-derived xenograft (PDX) model [15]. We also used GEPIA, which provides gene expression analysis based on tumor and normal samples from the TCGA and the GTEx databases. Functional transcriptome analyses were performed using Ingenuity Pathway Analysis (IPA) and MetaCore (Clarivate Analytics) software with default settings.
3. Cell culture and establishment of TKI-resistant cell lines
The human RCC cell lines 769-P, 786-O, ACHN, Caki-1, and A704 were purchased from the American Type Culture Collection.
To define the sensitivity of the RCC cells to sunitinib (Sutent), a dose-response study was first performed by treating the cells with various concentrations of sunitinib from 0.1 to 50 µM for 72 hours. Then a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay was performed, and various levels of inhibitory concentration (IC) values were estimated (Supplementary Table 1). Next, to develop sunitinib-resistant renal cell carcinoma cell lines designated as suR, the RCC cells were treated with sunitinib for 72 hours. After 24 hours of stabilization in the culture medium without sunitinib, cells were re-treated with sunitinib at a higher concentration by either long-term exposure to sunitinib at a concentration of 2.5 µM for 6 months (ACHN/suR) or stepwise increase of sunitinib concentration from IC10 to the final concentration up to 17 µM for 5–6 months (Caki-1/suR, 769-P/suR, 786-O/suR, and A704/suR).
4. Cell viability assay and clonogenic survival assay
The MTT assay was used for the cell viability assessment. For the clonogenic survival assay, cells were diluted into a cell density of 500 cells/mL and plated in triplicate into six-well plates. After 7–11 days in culture, the colonies were examined and subjected to photographs. Each experiment was performed in triplicate and repeated at least three times [16].
5. RNA interference and mimic expression
For the RNAi-mediated IFITM3 knock-down assay, shRNAs designed for IFITM3 were cloned into the pLKO.1-CMV-Neo vector (SHCLND, TRCN0000118024; Sigma-Aldrich). Among the three candidate sequences, the shRNA4 sequence, 5′-CCTCATGACCATTCTGCTCAT-3′, showed the best performance and was used for further experiments (Supplementary Fig. 1). The lentivirus containing the IFITM3 shRNA was delivered into 786-O/suR as previously described [17]. For ectopic overexpression of IFITM3 proteins, a lentivirus containing the pCMV6-Entry vector cloned with IFITM3 ORF construct was purchased (RC201635, OriGene). The recombinant pseudo-lentiviral particles were concentrated using the Lenti-X Concentrator kit (Clontech), then infected into 786-O/P using 6 µg/mL polybrene (Invitrogen).
6. Western blotting and immunoprecipitation pull-down assay
For western blotting, total protein was extracted from the cultured cells using RIPA lysis and extraction buffer (Thermo Fisher Scientific) and separated on a 12% SDS–PAGE polyacrylamide gel.
The primary antibodies used in this study were IFITM3 (1:1,000 dilution, ab109429, Abcam), TRAF6 (1:100, sc-8409, Santa Cruz Biotechnology), IRAK1 (1:200, ab238, Abcam), p-ERK (1:800, #4376, Cell Signaling Technology), c-Fos (1:20,#4384, Cell Signaling Technology), c-Myc (1:100, 395R16, Cell Marque), and survivin (1:50, ab76424, Abcam). β-actin served as the internal control.
For the immunoprecipitation (IP) assay, the protein extracts from cultured cells were prepared using IP lysis buffer (50 mM Tris–Cl [pH 7.4], 0.5% NP-40, 150 mM NaCl, 1.5 mM MgCl2, 2 mM DTT, 2 mM EGTA) supplemented with protease/phosphatase inhibitor mixtures (Roche), then centrifuged (12,000 g for 10 minutes at 4℃). The IP assay was performed as previously described [18].
7. Scratch assay, Matrigel invasion assay and soft agar colony formation assay
The scratch assay and Matrigel invasion assay were performed as previously described [1,2]. Anchorage-independent growth was examined by performing soft-agar assays using Agarose (A9539, Sigma-Aldrich). Cells (300 cells/well) were plated in triplicates in 6-well plates. After 7–14 days, colonies were stained with Crystal Violet (C1035, Biosesang), counted, and subjected to photographs. Each experiment was repeated at least three times.
8. Immunohistochemistry
Immunohistochemical staining was performed using an automated staining system (BenchMark XT, Ventana Medical Systems) with the abovementioned antibodies. All immunostained slides were evaluated by H-score assessment semi-quantitatively by two independent pathologists (S.U.J. and Y.M.C.), whom were blinded to clinical and pathological information [19].
9. Xenograft model of renal cell carcinoma
The animal study was approved by the Institutional Animal Care and Use Committee of Asan Institute for Life Sciences, Asan Medical Center, Seoul, Republic of Korea, which abides by the Institute of Laboratory Animal Resources guide (approval number: 2021-12-134). Male SID (NSGA mouse; Scid mediated-total Immune Deficient mouse) mice were supplied by Orient. After expansion in vitro, 1×107 cells were inoculated subcutaneously into the dorsum of the right thigh of 6-week-old mice. Ten mice were used in each replicate. Sunitinib (20 mg/kg) was administered by gavage every 3 days beginning on the 31st day after transplantation [20]. Tumors were measured with calipers twice weekly after transplantation. Mice were euthanized when a significant loss of body weight (more than 20%) was noted, according to the Laboratory Animal Resources guide. The xenograft tumors were dissected, fixed in 10% buffered formalin, embedded in paraffin, and cut into 4 µm thick sections for pathologic examination.
10. Statistical analysis
A Student’s t-test for continuous variables, expressed as mean±standard error, was used to compare statistical significance in the intrinsic resistance cohort. The Kaplan–Meier method and the log-rank test were used to evaluate differences in survival outcomes. The threshold for statistical significance was a p-value of less than 0.05.
RESULTS
1. Upregulation of IFITM3 in ccRCC compared with normal kidney
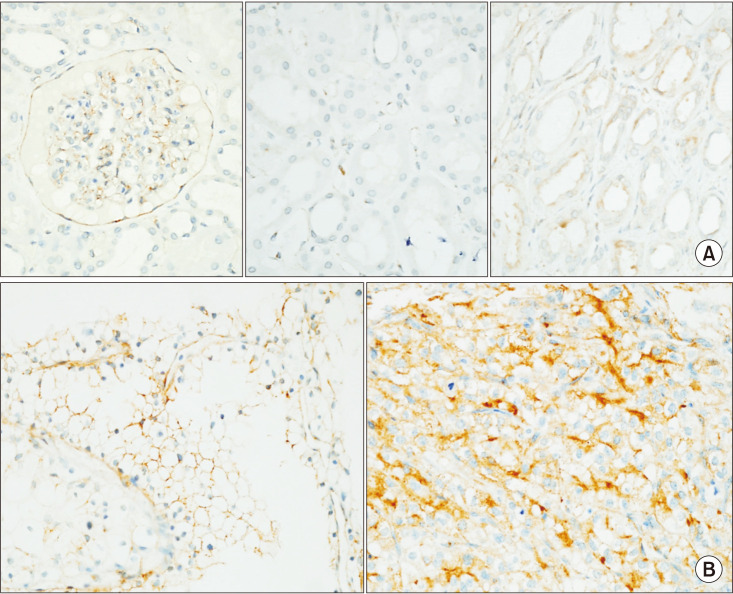
In the normal kidney, IFITM3 was expressed in glomerular and interstitial vascular endothelial cells and glomerular parietal epithelial cells as a cytoplasmic pattern. IFITM3 expression was inconspicuous in tubular epithelial cells, including proximal convoluted tubules, from which ccRCC was proposed to arise. In contrast, damaged or atrophic tubules expressed IFITM3 (Fig. 1A).
Fig. 1. Interferon-induced transmembrane protein 3 (IFITM3) expression in the normal kidney and clear cell renal cell carcinoma (ccRCC). (A) In normal renal parenchyma, IFITM3 was immunoreactive in glomerular parietal epithelial cells, capillaries (left), and damaged or atrophic tubules (right). IFITM3 expression was inconspicuous in tubular epithelial cells (middle). (B) In ccRCC, IFITM3 was immunoreactive as a membranous pattern in low-grade tumors (left), whereas cytoplasmic patterns are observed in high-grade tumors (right) (magnification, ×400).
In ccRCC, IFITM3 was expressed in tumor cells with a tendency for membranous pattern in low-grade tumors, whereas membranous and cytoplasmic patterns were observed in high-grade tumors (Fig. 1B). The higher expression of IFITM3 in RCC was also confirmed in the public database GEPIA (Supplementary Fig. 2) [21].
2. Role of IFITM3 on aggressive behavior of sunitinib-resistant RCC cells
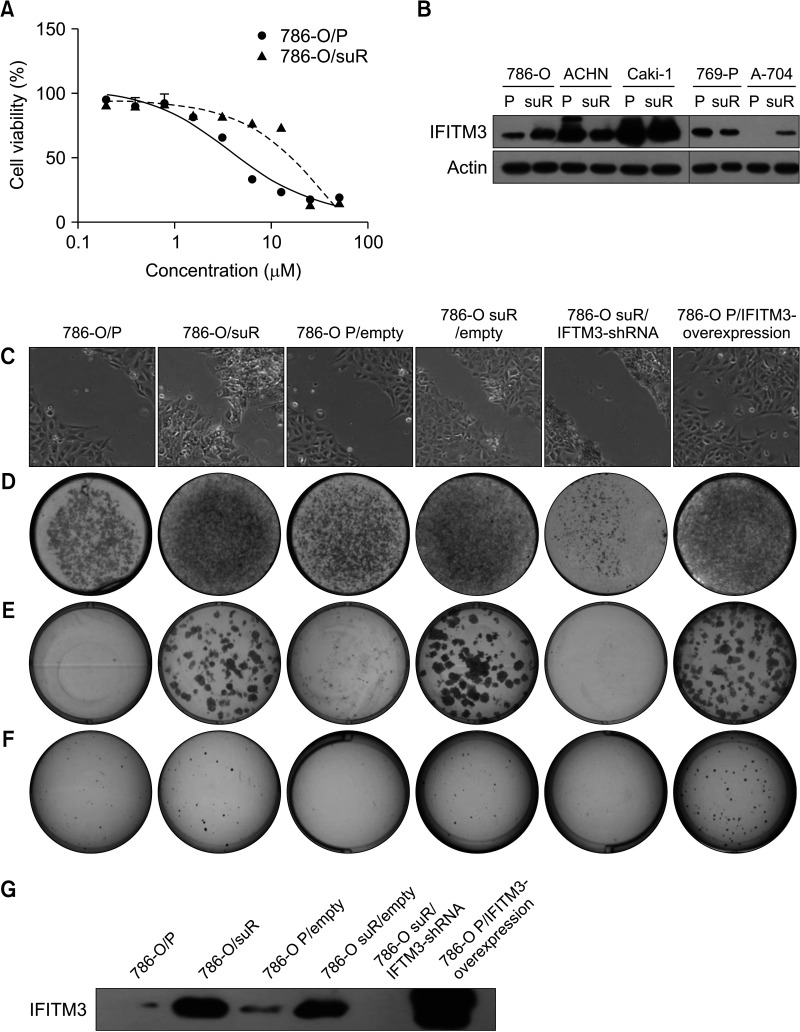
The sunitinib-resistant renal cell carcinoma cell lines showed higher cell viability at various concentrations of sunitinib compared with the parental cell lines, confirming their TKI resistance (Fig. 2A and Supplementary Fig. 3). Among the five suR RCC cells, 786-O/suR and A704/suR showed increased expression of IFITM3 compared with their parental cells (Fig. 2B). Based on this finding and the slow growth rate of A704/suR, 786-O/parental cells (786-O/P) and 786-O/suR were selected for further experiments.
Fig. 2. Generation of sunitinib-resistant renal cell carcinoma (RCC) cell lines and role of interferon-induced transmembrane protein 3 (IFITM3) on aggressive behavior of sunitinib-resistant RCC cells. (A) Sunitinib-resistant RCC cell lines showed higher cell viability at various concentrations of sunitinib compared with the parental cell lines of five sunitinib-resistant RCC cell lines (786-O/suR and 786-O/P shown). Although both 786-O/suR and A-704/suR showed increased IFITM3 protein expression compared with the parental cell lines (B), 786-O/suR was finally selected for further analysis due to the slow proliferative rate of A704/suR. The migration (C), invasion (D), and proliferative capacities (E, F) of 786-O/P, 786-O/suR, 786-O P/empty, 786-O suR/empty, 786-O/suR with inhibition of IFITM3 and 786-O/P with overexpression of IFITM3 were determined by the scratch test assay, the Matrigel invasion assay and the soft agar colony formation assay, respectively. (G) The expression of IFITM3 was inhibited by shRNA transfection into 786-O/suR or increased by IFITM3 mimic transfection into 786-O/P.
To assess the role of IFITM3 on the aggressive behavior of RCC during TKI treatment, the following experiments were performed using 786-O/P and 786-O/suR subjected to sunitinib at 2.0 µM. 786-O/suR migrated rapidly into the cleft in the scratch assay and showed higher invasion activity with more cells invading the Matrigel membrane (Fig. 2C, D). In addition to the high proliferation (Fig. 2A), 786-O/suR showed higher clonogenic survival with larger numbers and sizes of colonies in the colony-forming assay compared with 786-O/P (Fig. 2E). Furthermore, 786-O/suR showed an increase in anchorage-independent growth compared with 786-O/P (Fig. 2F). Taken together, these results suggest that 786-O/suR has aggressive cancer cell properties, similar to patients with TKI-resistant RCC.
To evaluate whether IFITM3 could regulate the aggressive behavior of TKI-resistant RCC, the aforementioned experiments were conducted while the expression of IFITM3 was inhibited by shRNA transfection into 786-O/suR or increased by IFITM3 mimic transfection into 786-O/P (Fig. 2G). Inhibition of IFITM3 expression resulted in the suppression of proliferation, clonogenic survival, migration, and invasion of 786-O/suR. In contrast, these aggressive behaviors increased as a result of the IFITM3 overexpression in 786-O/P (Fig. 2C-F).
3. Role of IFITM3 in tumor growth in TKI-resistant RCC in vivo
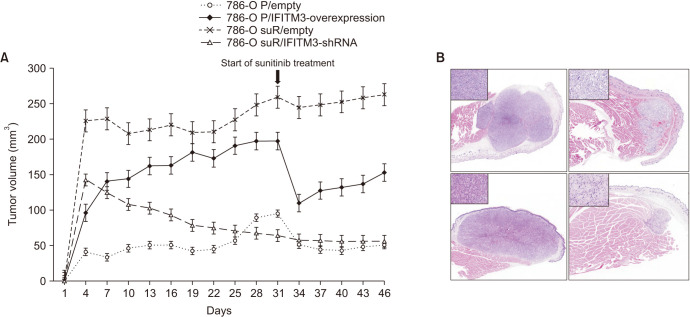
To examine the in vivo relevance of the above results, the tumorigenicity of IFITM3-overexpressing 786-O/P and IFITM3-inhibited 786-O/suR along with 786-O/P and 786-O/suR was examined in a xenograft model (Fig. 3). 786-O/suR were grown as larger tumors compared with 786-O/P throughout the experiments. IFITM3-overexpressing 786-O/P also exhibited significantly higher tumor growth than 786-O/P cells, whereas IFITM3-inhibited 786-O/suR showed significantly smaller growth than 786-O/suR.
Fig. 3. (A) The tumorigenicity of 786-O P/empty, 786-O suR/empty, 786-O P/IFITM3-overexpression, and 786-O suR/IFITM3-shRNA was evaluated in a xenograft model. Sunitinib was administered 31 days after transplantation. (B) Representative images 786-O P/IFITM3-overexpression (left upper), 786-O P/empty, 786-O suR/IFITM3-shRNA, and 786-O suR/empty were presented clockwise. Representative images of the high-power field were presented as inset. IFITM3, interferon-induced transmembrane protein 3 (magnification, ×40; ×400).
When sunitinib was administered on the 31st day after the transplantation, the tumor size of IFITM3-overexpressing 786-O/P decreased in the xenografts. However, we observed a marked rebound in tumor growth, restoring a mean of 66% (range, 55%–77%) of tumor volume before the sunitinib treatment in 12 days, whereas 786-O/P maintained reduced tumor volume (Fig. 3A). 786-O/suR was barely affected by the sunitinib administration, but IFITM3-inhibited 786-O/suR remained small, similar to those of 786-O/P after the sunitinib treatment.
On microscopic examination, transfected cells of the xenografts of parental and TKI-resistant 786-O showed similar histologic features: nests of eosinophilic to clear high-grade cells with a retained vascular network and variable amounts of sarcomatoid change and necrosis (Fig. 3B). These findings suggest that the aggressive behavior of TKI-resistant RCC could be regulated by IFITM3 expression in vivo.
4. IFITM3-mediated activation of TRAF6/MAPK/AP-1 signaling pathway in TKI-resistant RCC cells
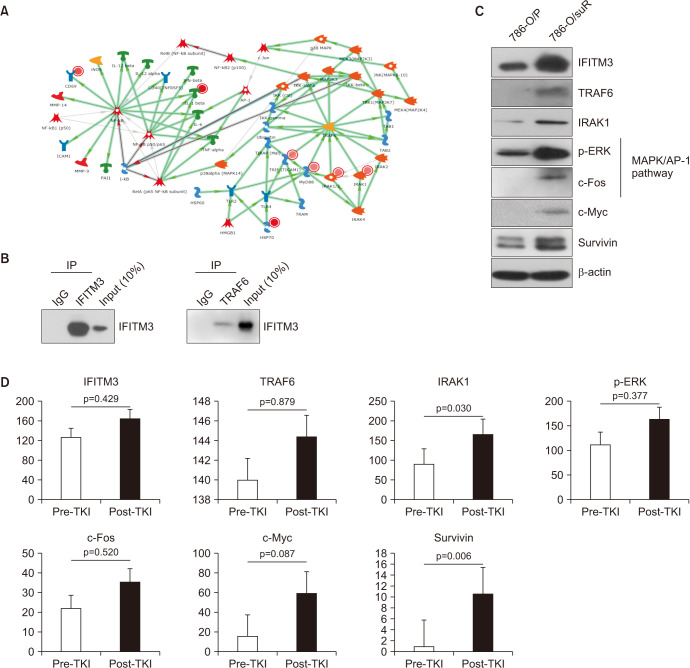
We performed MetaCore gene network analysis using the transcriptome profile of the 10 paired pre- and post-TKI tumor tissues to get a mechanistic insight. The MetaCore analysis revealed that several TRAF6-related genes were up-regulated in post-TKI RCC (Fig. 4A). We speculated that there may be a direct protein interaction between IFITM3 and TRAF6. IP pull-down assay demonstrated that IFITM3 physically interacted with TRAF6 in 786-O/suR cells (Fig. 4B).
Fig. 4. Interferon-induced transmembrane protein 3 (IFITM3)-mediated TRAF6/MAPK/AP-1 signaling pathway activation in tyrosine kinase inhibitor (TKI)-resistant renal cell carcinoma (RCC) cells and the 10 paired clear cell RCC tissues. (A) The results of MetaCore analysis using the transcriptome profile of the 10 paired pre- and post-TKI tumor tissues revealed that several TRAF6-related genes were up-regulated in post-TKI RCC. (B) An immunoprecipitation (IP) pull-down assay was performed to examine the interaction between IFITM3 and TRAF6 proteins in 786-O/suR cells. (C) The expression of IFITM3 with genes related to TRAF6/MAPK/AP-1 signaling pathways (TRAF6, p-ERK, and c-Fos), IRAK1, c-Myc, and survivin were upregulated in 786-O/suR compared with 786-O/P. (D) All antibodies increased in post-TKI tissues compared with pre-TKI tissues with statistical significance in IRAK1 and survivin.
TRAF6 functions as a signal transducer and activates the MAPK pathway. To validate the association of IFITM3-mediated TKI resistance with the signaling pathway, we examined TRAF6 expression levels in 786-O cells. 786-O/suR showed upregulation of TRAF6 compared with 786-O/P (Fig. 4C). Accordingly, IRAK1, genes related to MAPK/AP-1 signaling pathways (p-ERK and c-Fos), c-Myc, and survivin were upregulated or activated in 786-O/suR compared with 786-O/P (Fig. 4C).
5. Verification of the correlation between expression of IFITM3 and TRAF6/MAPK/AP-1 pathways in human TKI-resistant RCC tissue
To verify the IFITM3-mediated activation of TRAF6/MAPK/AP-1 signaling pathway at the protein level in human RCC, immunohistochemical staining was performed on the 10 paired ccRCC tissues, among which 1 case could not be examined due to the lack of post-TKI RCC tissue available. TRAF6 and IRAK1 showed membranous/cytoplasmic patterns, p-ERK showed nuclear and cytoplasmic/membranous patterns, and c-Fos, c-Myc, and survivin showed nuclear patterns (Supplementary Fig. 4). The expression of all relevant proteins increased in post-TKI tissues compared with pre-TKI tissues with statistical significance in IRAK1 and survivin (Fig. 4D).
6. No correlation between IFITM3 expression and intrinsic TKI resistance
We next speculated that IFITM3 also plays a role in intrinsic TKI resistance. IFITM3 immunohistochemistry was conducted to evaluate its expression and its association with TKI response in a separate intrinsic resistance cohort of 89 patients.
IFITM3 was expressed variably from negative to positive in tumor cells (mean H-score 184.0). IFITM3 expression slightly increased in the non-responder group (53 cases, mean H-score 187.7) compared with the responder group (36 cases, mean H-score 178.6); however, the difference did not meet the statistical significance (p=0.694). There was no prognostic significance of IFITM3 expression in overall and progression-free survival (Supplementary Fig. 5A). The lack of predictive and prognostic significance was confirmed through the analysis using previously reported transcriptomic data from 53 patients (Supplementary Fig. 5B).
DISCUSSION
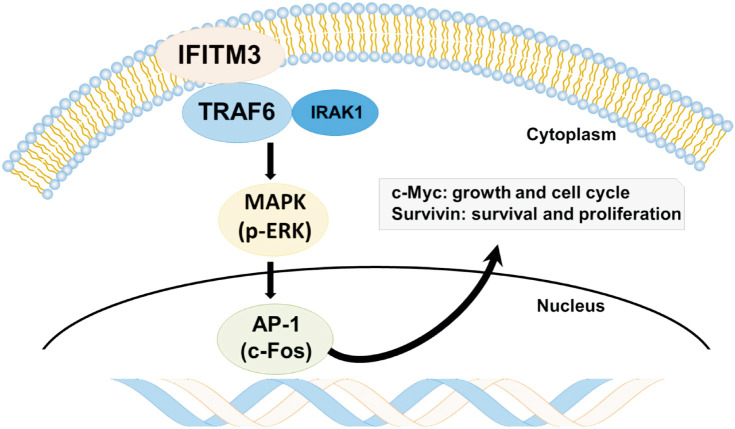
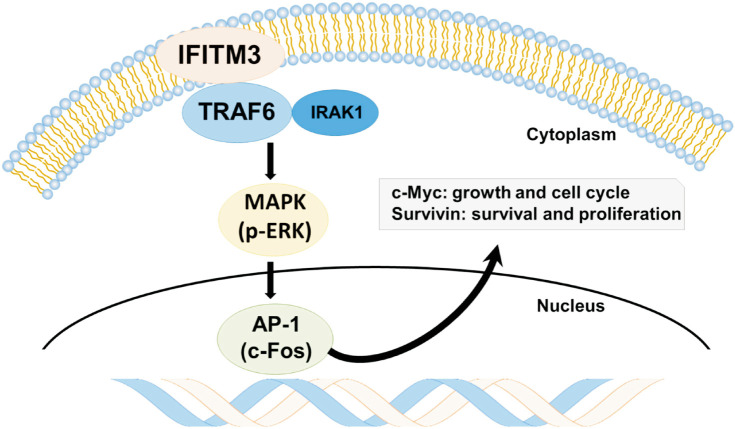
This study demonstrates that IFITM3 expression was increased not only in ccRCC compared with normal kidneys but also in ccRCC with acquired TKI resistance compared with TKI-naive ccRCC. We also observed that IFITM3 expression was responsible for aggressive behaviors of TKI-resistant ccRCC cells, mediated through TRAF6/MAPK/AP-1 pathways using in vitro and in vivo studies. These results suggest IFITM3-mediated activation of TRAF6/MAPK/AP-1 pathways as a mechanism of TKI resistance, and IFITM3 as a new therapeutic target for TKI-resistant ccRCC. As summarized in Fig. 5, ccRCC initially responds to sunitinib, but IFITM3 overexpression is eventually induced as speculated below. The binding of IFITM3 to TRAF6 results in the activation of TRAF6, which induces oncogenic signaling pathways, including MAPK/AP-1 pathways, and accelerates tumor growth and migration despite treatment with TKIs.
Fig. 5. The interferon-induced transmembrane protein 3 (IFITM3)-induced acquired tyrosine kinase inhibitor (TKI) resistance mechanism represented in a schematic diagram. In patients resistant to sunitinib, overexpression of IFITM3 is observed. The binding of IFITM3 to TRAF6 leads to the activation of TRAF6 associated with IRAK1, which induces oncogenic signaling pathways, including MAPK/AP-1 pathways. As a result, clear cell renal cell carcinoma (ccRCC) can accelerate tumor growth, survival, and proliferation despite TKI treatment.
IFITM3, also known as fragilis or I-8U, is a 15-kD transmembrane protein and belongs to the IFITM family. IFITM3 is induced by interferon and plays a critical role in antiviral activity by interfering with the fusion of the viral envelope and the endosome membrane during virus entry into host cells. In addition, IFITM3 has been implicated in human disease, such as acute lung injury, leukemia, and malignant solid tumors [3,22]. IFITM3 is expressed in a variety of normal tissues, with particularly high expression in female genital tracts such as the fallopian tube, uterine cervix and endometrium, lung, and adipose tissue [23]. Although IFITM3 expression levels differed across various types of tumors, this study showed its increased expression in most tumors compared with corresponding normal tissues [23]. The overexpression of IFITM3 has been reported to enhance cancer properties such as proliferation, stemness, replicative immortality, migration, and invasion, similar to this study [3]. Furthermore, reports have also suggested that Bcr-Abl TKI imatinib resistant leukemia cells extensively release IFITM3-containing exosomes, increasing the survival of imatinib sensitive cells treated with imatinib [24].
The overexpression of IFITM3 has been largely suggested as an indirect effect of inflammatory/immune reaction and epithelial–mesenchymal transition (EMT). In ccRCC, the VEGFR-targeted TKI therapy inhibits multiple receptor tyrosine kinases important for tumor angiogenesis, and results in marked atrophy of capillary sinus surrounding tumor cells. Consequently, the lack of or limited blood supply leads to various degenerative changes in tumor cells, such as nuclear pyknosis, degeneration, and necrosis, which may trigger an inflammatory reaction and increase of IFITM3 expression [25]. Given that IFITM3 expression was higher in TKI-resistant ccRCC, an alternative mechanism such as EMT may also play a role in the overexpression of IFTIM3. We have previously reported that the expression of EMT-related genes such as SNAI2, TWIST, CD44, and CLDN1 was upregulated in TKI-resistant ccRCC tumors in this acquired resistance cohort [1]. Emerging evidence also suggests that the expression of IFITM3 is regulated by multiple signaling pathways, including interferon-mediated JAK/STAT pathway, transforming growth factor-β-mediated activation of SMAD proteins and MAPK/PI3 pathways, and Wnt/β-catenin pathway [8,26,27].
Our study has some limitations. First, it is a retrospective single-center study. Although we showed IFITM3 and TRAF6 expression was increased in TKI-resistant ccRCC by comparing matched ccRCC tissues harvested at pre- and post-TKI treatment periods, only a small number of patients (10 patients) could be analyzed. It has been difficult to procure post-treated tumor tissues within 1-month post-clinical recognition of tumor progression. Once the diagnosis of ccRCC is confirmed during the pretreatment period, treatment responsiveness is usually determined clinically by imaging studies. To overcome this limitation, we performed an integrated analysis using a public dataset. While we showed that IFITM3 increased in a subset of TKI-resistant ccRCC patients via the TRAF6/MAPK/AP-1 pathway, we cannot exclude the possibility of other mechanisms being involved. Therefore, conducting prospective multi-center studies with a large number of matched pre- and post-TKI treatment tumor tissues is necessary to confirm the study’s results and identify potential alternative mechanisms. With the recent introduction of combination therapy using ICIs and TKIs, examining whether the IFITM3-mediated activation of TRAF6/MAPK/AP-1 pathways also plays a role in acquired resistant RCC after the combination therapy would be interesting.
CONCLUSIONS
IFITM3 expression was increased in ccRCC than normal kidneys and in ccRCC with acquired TKI resistance than TKI-naive ccRCC. IFITM3-mediated activation of the TRAF6/MAPK/AP-1 pathways is a mechanism of acquired TKI resistance in ccRCC. IFITM3 may serve as a new potential therapeutic target for TKI-resistant ccRCC.
Footnotes
CONFLICTS OF INTEREST: The authors have nothing to disclose.
FUNDING: This work is funded by the Ministry of Science, ICT and Future Planning (2019R1A2C1088246) and a grant (2022IL0018-1) from the Asan Institute for Life Sciences, Asan Medical Center, Seoul, Korea.
- Research conception and design: Yong Mee Cho.
- Data acquisition: Yong Mee Cho, Hee Sang Hwang, and Heounjeong Go.
- Statistical analysis: Se Un Jeong.
- Data analysis and interpretation: Hee Sang Hwang, Hyein Ju, Dong-Myung Shin, Heounjeong Go, Chang Ohk Sung, and Gowun Jeong.
- Drafting of the manuscript: Yong Mee Cho and Se Un Jeong.
- Critical revision of the manuscript: Yong Mee Cho and Se Un Jeong.
- Obtaining funding: Yong Mee Cho.
- Cell line, immunoprecipitation, and animal experiments: Ja-Min Park and Sun Young Yoon.
- Supervision: Yong Mee Cho and Jae-Lyun Lee.
- Approval of the final manuscript: all authors.
SUPPLEMENTARY MATERIALS
Supplementary materials can be found via https://doi.org/10.4111/icu.20230294.
Interferon-induced transmembrane protein 3 (IFITM3) shRNA sequence. Among the three candidate sequences, the shRNA4 showed the best performance and was used for further experiments.
Increased expression of IFITM3 (interferon-induced transmembrane protein 3) in renal cell carcinoma (RCC) compared to the normal kidney in a public database GEPIA.
The sunitinib-resistant renal cell carcinoma cell lines including Caki-1 (A), 769-P (B), ACHN (C), and A-704 (D) showed higher cell viability at various concentrations of sunitinib compared to the parental cell lines, confirming their tyrosine kinase inhibitor (TKI) resistance.
The representative expression of TRAF6, IRAK1, p-ERK, Survivin, c-Myc, and c-Fos (clockwise). TRAF6 and IRAK1 showed membranous/cytoplasmic pattern while p-ERK showed mixed membranous/cytoplasmic and nuclear pattern. Survivin, c-Myc, and c-Fos showed nuclear pattern (magnification, ×400).
No correlation of interferon-induced transmembrane protein 3 (IFITM3) expression in intrinsic tyrosine kinase inhibitor (TKI) resistance. There was no prognostic significance of IFITM3 expression in the overall and progression-free survivals of 89 cases of advanced clear cell renal cell carcinoma (ccRCC) cohort (A), and in recurrence and progression-free survival of previously reported 53 ccRCC patients treated with sunitinib (B).
IC values by sunitinib in various renal cell carcinoma cell lines
References
- 1.Hwang HS, Go H, Park JM, Yoon SY, Lee JL, Jeong SU, et al. Epithelial-mesenchymal transition as a mechanism of resistance to tyrosine kinase inhibitors in clear cell renal cell carcinoma. Lab Invest. 2019;99:659–670. doi: 10.1038/s41374-019-0188-y. [DOI] [PubMed] [Google Scholar]
- 2.Hwang HS, Park YY, Shin SJ, Go H, Park JM, Yoon SY, et al. Involvement of the TNF-α pathway in TKI resistance and suggestion of TNFR1 as a predictive biomarker for TKI responsiveness in clear cell renal cell carcinoma. J Korean Med Sci. 2020;35:e31. doi: 10.3346/jkms.2020.35.e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rajapaksa US, Jin C, Dong T. Malignancy and IFITM3: friend or foe? Front Oncol. 2020;10:593245. doi: 10.3389/fonc.2020.593245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lange UC, Saitou M, Western PS, Barton SC, Surani MA. The fragilis interferon-inducible gene family of transmembrane proteins is associated with germ cell specification in mice. BMC Dev Biol. 2003;3:1. doi: 10.1186/1471-213X-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fan J, Peng Z, Zhou C, Qiu G, Tang H, Sun Y, et al. Gene-expression profiling in Chinese patients with colon cancer by coupling experimental and bioinformatic genomewide gene-expression analyses: identification and validation of IFITM3 as a biomarker of early colon carcinogenesis. Cancer. 2008;113:266–275. doi: 10.1002/cncr.23551. [DOI] [PubMed] [Google Scholar]
- 6.Yang M, Gao H, Chen P, Jia J, Wu S. Knockdown of interferon-induced transmembrane protein 3 expression suppresses breast cancer cell growth and colony formation and affects the cell cycle. Oncol Rep. 2013;30:171–178. doi: 10.3892/or.2013.2428. [DOI] [PubMed] [Google Scholar]
- 7.Liu X, Chen L, Fan Y, Hong Y, Yang X, Li Y, et al. IFITM3 promotes bone metastasis of prostate cancer cells by mediating activation of the TGF-β signaling pathway. Cell Death Dis. 2019;10:517. doi: 10.1038/s41419-019-1750-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Min J, Feng Q, Liao W, Liang Y, Gong C, Li E, et al. IFITM3 promotes hepatocellular carcinoma invasion and metastasis by regulating MMP9 through p38/MAPK signaling. FEBS Open Bio. 2018;8:1299–1311. doi: 10.1002/2211-5463.12479. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Lee J, Robinson ME, Ma N, Artadji D, Ahmed MA, Xiao G, et al. IFITM3 functions as a PIP3 scaffold to amplify PI3K signalling in B cells. Nature. 2020;588:491–497. doi: 10.1038/s41586-020-2884-6. Erratum in: Nature 2021;592:E3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi JH, Sun SC. Tumor Necrosis factor receptor-associated factor regulation of nuclear factor κB and mitogen-activated protein kinase pathways. Front Immunol. 2018;9:1849. doi: 10.3389/fimmu.2018.01849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanos T, Marinissen MJ, Leskow FC, Hochbaum D, Martinetto H, Gutkind JS, et al. Phosphorylation of c-Fos by members of the p38 MAPK family. Role in the AP-1 response to UV light. J Biol Chem. 2005;280:18842–18852. doi: 10.1074/jbc.M500620200. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Liu N, Tang L, Yan B, Chen X, Zhang J, et al. The relationship between TRAF6 and tumors. Cancer Cell Int. 2020;20:429. doi: 10.1186/s12935-020-01517-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Leo A, Jerusalem G, Petruzelka L, Torres R, Bondarenko IN, Khasanov R, et al. Results of the CONFIRM phase III trial comparing fulvestrant 250 mg with fulvestrant 500 mg in postmenopausal women with estrogen receptor-positive advanced breast cancer. J Clin Oncol. 2010;28:4594–4600. doi: 10.1200/JCO.2010.28.8415. Erratum in: J Clin Oncol 2011;29:2293. [DOI] [PubMed] [Google Scholar]
- 14.Beuselinck B, Job S, Becht E, Karadimou A, Verkarre V, Couchy G, et al. Molecular subtypes of clear cell renal cell carcinoma are associated with sunitinib response in the metastatic setting. Clin Cancer Res. 2015;21:1329–1339. doi: 10.1158/1078-0432.CCR-14-1128. [DOI] [PubMed] [Google Scholar]
- 15.Diaz-Montero CM, Mao FJ, Barnard J, Parker Y, Zamanian-Daryoush M, Pink JJ, et al. MEK inhibition abrogates sunitinib resistance in a renal cell carcinoma patient-derived xenograft model. Br J Cancer. 2016;115:920–928. doi: 10.1038/bjc.2016.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeong SU, Hwang HS, Park JM, Yoon SY, Shin SJ, Go H, et al. PD-L1 upregulation by the mTOR pathway in VEGFR-TKI-resistant metastatic clear cell renal cell carcinoma. Cancer Res Treat. 2023;55:231–244. doi: 10.4143/crt.2021.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heo J, Lee J, Nam YJ, Kim Y, Yun H, Lee S, et al. The CDK1/TFCP2L1/ID2 cascade offers a novel combination therapy strategy in a preclinical model of bladder cancer. Exp Mol Med. 2022;54:801–811. doi: 10.1038/s12276-022-00786-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heo J, Lim J, Lee S, Jeong J, Kang H, Kim Y, et al. Sirt1 regulates DNA methylation and differentiation potential of embryonic stem cells by antagonizing Dnmt3l. Cell Rep. 2017;18:1930–1945. doi: 10.1016/j.celrep.2017.01.074. [DOI] [PubMed] [Google Scholar]
- 19.Chiu K, Lee L, Cheung S, Churg AM. Glypican-1 immunohistochemistry does not separate mesothelioma from pulmonary adenocarcinoma. Mod Pathol. 2018;31:1400–1403. doi: 10.1038/s41379-018-0066-y. [DOI] [PubMed] [Google Scholar]
- 20.Zhou L, Liu XD, Sun M, Zhang X, German P, Bai S, et al. Targeting MET and AXL overcomes resistance to sunitinib therapy in renal cell carcinoma. Oncogene. 2016;35:2687–2697. doi: 10.1038/onc.2015.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang LQ, Adyshev DM, Singleton P, Li H, Cepeda J, Huang SY, et al. Interactions between PBEF and oxidative stress proteins--a potential new mechanism underlying PBEF in the pathogenesis of acute lung injury. FEBS Lett. 2008;582:1802–1808. doi: 10.1016/j.febslet.2008.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu P, Zhang Y, Zhang S, Peng C, Yang W, Li X, et al. Integrative overview of IFITMs family based on Bioinformatics analysis. Intractable Rare Dis Res. 2021;10:165–172. doi: 10.5582/irdr.2021.01041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hrdinova T, Toman O, Dresler J, Klimentova J, Salovska B, Pajer P, et al. Exosomes released by imatinib-resistant K562 cells contain specific membrane markers, IFITM3, CD146 and CD36 and increase the survival of imatinib-sensitive cells in the presence of imatinib. Int J Oncol. 2021;58:238–250. doi: 10.3892/ijo.2020.5163. [DOI] [PubMed] [Google Scholar]
- 25.Kondo T, Hashimoto Y, Kobayashi H, Iizuka J, Nishikawa T, Nakano M, et al. Presurgical targeted therapy with tyrosine kinase inhibitors for advanced renal cell carcinoma: clinical results and histopathological therapeutic effects. Jpn J Clin Oncol. 2010;40:1173–1179. doi: 10.1093/jjco/hyq150. [DOI] [PubMed] [Google Scholar]
- 26.Schoenherr C, Byron A, Sandilands E, Paliashvili K, Baillie GS, Garcia-Munoz A, et al. Ambra1 spatially regulates Src activity and Src/FAK-mediated cancer cell invasion via trafficking networks. Elife. 2017;6:e23172. doi: 10.7554/eLife.23172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao Y, Li J, Qiao N, Meng Q, Zhang M, Wang X, et al. Adrenomedullin blockade suppresses sunitinib-resistant renal cell carcinoma growth by targeting the ERK/MAPK pathway. Oncotarget. 2016;7:63374–63387. doi: 10.18632/oncotarget.11463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Interferon-induced transmembrane protein 3 (IFITM3) shRNA sequence. Among the three candidate sequences, the shRNA4 showed the best performance and was used for further experiments.
Increased expression of IFITM3 (interferon-induced transmembrane protein 3) in renal cell carcinoma (RCC) compared to the normal kidney in a public database GEPIA.
The sunitinib-resistant renal cell carcinoma cell lines including Caki-1 (A), 769-P (B), ACHN (C), and A-704 (D) showed higher cell viability at various concentrations of sunitinib compared to the parental cell lines, confirming their tyrosine kinase inhibitor (TKI) resistance.
The representative expression of TRAF6, IRAK1, p-ERK, Survivin, c-Myc, and c-Fos (clockwise). TRAF6 and IRAK1 showed membranous/cytoplasmic pattern while p-ERK showed mixed membranous/cytoplasmic and nuclear pattern. Survivin, c-Myc, and c-Fos showed nuclear pattern (magnification, ×400).
No correlation of interferon-induced transmembrane protein 3 (IFITM3) expression in intrinsic tyrosine kinase inhibitor (TKI) resistance. There was no prognostic significance of IFITM3 expression in the overall and progression-free survivals of 89 cases of advanced clear cell renal cell carcinoma (ccRCC) cohort (A), and in recurrence and progression-free survival of previously reported 53 ccRCC patients treated with sunitinib (B).
IC values by sunitinib in various renal cell carcinoma cell lines