Abstract
Rosa (Rosaceae) is an important ornamental and medicinal plant genus worldwide, with several species being cultivated in China. Members of Sporocadaceae (pestalotioid fungi) are globally distributed and include endophytes, saprobes but also plant pathogens, infecting a broad range of host plants on which they can cause important plant diseases. Although several Sporocadaceae species were recorded to inhabit Rosa spp., the taxa occurring on Rosa remain largely unresolved. In this study, a total of 295 diseased samples were collected from branches, fruits, leaves and spines of eight Rosa species (R. chinensis, R. helenae, R. laevigata, R. multiflora, R. omeiensis, R. rugosa, R. spinosissima and R. xanthina) in Gansu, Henan, Hunan, Qinghai, Shaanxi Provinces and the Ningxia Autonomous Region of China. Subsequently 126 strains were obtained and identified based on comparisons of DNA sequence data. Based on these results 15 species residing in six genera of Sporocadaceae were delineated, including four known species (Pestalotiopsis chamaeropis, Pes. rhodomyrtus, Sporocadus sorbi and Spo. trimorphus) and 11 new species described here as Monochaetia rosarum, Neopestalotiopsis concentrica, N. subepidermalis, Pestalotiopsis tumida, Seimatosporium centrale, Seim. gracile, Seim. nonappendiculatum, Seim. parvum, Seiridium rosae, Sporocadus brevis, and Spo. spiniger. This study also represents the first report of Pes. chamaeropis, Pes. rhodomyrtus and Spo. sorbi on Rosa. The overall data revealed that Pestalotiopsis was the most prevalent genus, followed by Seimatosporium, while Pes. chamaeropis and Pes. rhodomyrtus were the two most prevalent species. Analysis of Sporocadaceae abundance on Rosa species and plant organs revealed that spines of R. chinensis had the highest species diversity.
Citation: Peng C, Crous PW, Jiang N, et al. 2022. Diversity of Sporocadaceae (pestalotioid fungi) from Rosa in China. Persoonia 49: 201–260. https://doi.org/10.3767/persoonia.2022.49.07.
Keywords: Amphisphaeriales, Ascomycota, new taxa, phylogeny, taxonomy
INTRODUCTION
Sporocadaceae (Xylariales, Sordariomycetes) is a well-known fungal family containing pestalotioid fungi. Traditionally, pestalotioid fungi are circumscribed as a group of coelomycetous fungi having fusoid or nearly fusoid, multi-septate conidia, with appendages at one or both ends (Nag Raj 1993, Maharachchikumbura et al. 2014, Liu et al. 2019).
Pestalotioid fungi were previously classified in Amphisphaeriaceae, Amphisphaeriales (Eriksson 1986, Samuels et al. 1987). Subsequently, several studies suggested that Amphisphaeriales should not be accepted due to the lack of stable phylogenetic support, and hence it was treated as synonym of Xylariales (Eriksson 1987, Kang et al. 1999, Smith et al. 2003). Later, Senanayake et al. (2015) revised Xylariomycetidae and transferred several important genera of pestalotioid fungi from Amphisphaeriaceae to three new families, Bartaliniaceae, Discosiaceae and Pestalotiopsidaceae. Genera such as Bartalinia and Broomella were transferred to Bartaliniaceae, Discosia and Seimatosporium to Discosiaceae, and Neopestalotiopsis, Pestalotiopsis, Pseudopestalotiopsis, Monochaetia and Seiridium to Pestalotiopsidaceae. Crous et al. (2015a) introduced a new family Robillardaceae to accommodate Robillarda. Subsequently, Jaklitsch et al. (2016) grouped the pestalotioid fungi into a single family and revived the older family name Sporocadaceae. Therefore, Bartaliniaceae, Discosiaceae, Pestalotiopsidaceae and Robillardaceae became synonyms of Sporocadaceae. These families were classified in Amphisphaeriales which was resurrected instead of Xylariales (Senanayake et al. 2015). Recently, several studies treated Amphisphaeriales as a distinct order (Senanayake et al. 2015, Samarakoon 2016, Hongsanan et al. 2017, Wijayawardene et al. 2020) . Based on multi-locus phylogenetic analyses with morphological characters, Liu et al. (2019) confirmed the natural taxonomic status of Sporocadaceae, which currently contains 33 genera (Liu et al. 2019, Wijayawardene et al. 2020).
Sporocadaceae contains many important plant pathogens associated with diseases on a wide range of plant hosts worldwide (Maharachchikumbura et al. 2014, Liu et al. 2019, Norphanphoun et al. 2019). Within the family, pestalotiopsis-like taxa (Neopestalotiopsis, Pestalotiopsis and Pseudopestalotiopsis) are the group that has received the most attention (Maharachchikumbura et al. 2014, Wang et al. 2019b, Gualberto et al. 2021) . For example, N. mangiferae and N. palmarum cause leaf diseases on a variety of cash crops in Brazil, South Africa and India, weakening tree vigour, and even reducing yield in severe cases (Spaulding 1949, Mendes et al. 1998, Crous et al. 2000). Pestalotiopsis pini is an emerging pathogen causing shoot blight and stem necrosis on Pinus pinea (Silva et al. 2020), while in Australia, Pes. telopeae causes a serious leaf spot disease of Telopea spp. (Maharachchikumbura et al. 2014). Furthermore, pestalotiopsis-like fungi are widespread, occur on many hosts in Proteaceae, and are generally regarded to be saprobic or weakly pathogenic (Crous et al. 2013). Neopestalotiopsis protearum was recorded as causing leaf spots and blight on several Protea and Leucospermum hosts in Zimbabwe (Swart et al. 1999, Crous et al. 2011b). This species is also reported from Australia and Proteaceae in the Western Cape Province of South Africa (Crous et al. 2013). Neopestalotiopsis protearum is probably only a problem of commercial importance in summer rainfall areas and it has been intercepted at quarantine inspection points (Taylor 2000). Pestalotiopsis montellicoides was isolated from Protea cynaroides leaves from South Africa (Mordue 1986), and a Pestalotiopsis sp. (asexual Pestalosphaeria leucospermi), was described from living leaves of a Leucospermum sp. in New Zealand (Samuels et al. 1987). In Portugal and the Canary Islands, a species of Pestalotiopsis is commonly associated with tip blight and leaf spot symptoms on Protea, Leucospermum and Leucadendron species, although pathogenicity studies have not yet been conducted (Crous et al. 2013). Members of Pseudopestalotiopsis are cosmopolitan in distribution and have often been regarded as leaves spot pathogens occurring on a broad host range, e.g., Pse. elaeidis and Pse. theae cause foliar diseases in more than 60 hosts around the world in tropical and subtropical areas (Maharachchikumbura et al. 2014, Liu et al. 2019). In addition to pestalotiopsis-like species, the diseases caused by other groups of Sporocadaceae cannot be underestimated. Cypress canker is caused by several species of Seiridium (Bonthond et al. 2018), Allelochaeta is an important foliar pathogen of eucalypts (Crous et al. 2019b), and some species of Distononappendiculata and Truncatella cause diseases on a wide range of hosts (Crous et al. 2011a, 2013, Liu et al. 2019).
Sporocadaceae has an extremely rich species diversity in China. The investigation of the biodiversity of plant-associated pestalotioid fungi in China date back as far as 1886, when Patouillard collected and described many species from Yunnan (Patouillard 1886). With subsequent research, a total of 310 species belonging to 22 genera were reported in China, inhabiting many hosts, especially in Juglandaceae, Myrtaceae, Pinaceae, Podocarpaceae, Rhododendronaceae, Rosaceae, Theaceae and Vitaceae (Chen 2003, Ge et al. 2009, Liu et al. 2019). Most of the previous studies on Sporocadaceae in China focused on Pestalotiopsis. Previous investigations on Pestalotiopsis in China were summarised by Tai (1979), in which 38 species from 52 plant hosts were listed. A wider survey included 153 species obtained from at least 406 plant species, 67 of which are endemic to China (Ge et al. 2009). Hitherto 203 species have been reported in China, accounting for more than 65 % of the total records of this family in China. However, the distribution of 11 genera in China is still unknown, i.e., Ciliochorella, Clypeosphaeria, Diploceras, Disaeta, Distononappendiculata, Heterotruncatella, Hyalotiella, Morinia, Nonappendiculata, Parabartalinia and Xenseimatosporium. Furthermore, many species of Sporocadaceae can also cause serious plant diseases in China. Pseudopestalotiopsis camelliae-sinensis is responsible for grey blight of tea plants and causes serious losses in some tea-growing regions of China (Wang et al. 2019b), while Pes. apiculata causes severe top blight of cedar seedlings (Ge et al. 2009). Monochaetia kansensis and M. monochaeta cause leaf spots on a variety of Quercus and Castanea plants (Teng 1996, Chen et al. 2002, Chen 2003), and Truncatella laurocerasi causes grey blight and leaf spot on Eriobotrya in China (Tai 1979). Considering the importance of pestalotioid fungi, it is necessary to clarify the species diversity and distribution of Sporocadaceae in China in a modern taxonomic framework.
Rosa (Rosaceae) is widely distributed in tropical to cold temperate regions of the Northern Hemisphere, including approximately 200 species (Bruneau et al. 2007, Fougère-Danezan et al. 2015). China is the main distribution area of Rosa plants globally (Wu et al. 2003). There are currently 95 Rosa spp. in China (65 of which are endemic), accounting for about 41 % of the global total (Jin 2020). Rosa species are widely cultivated and are of immense economic importance in China (Liu 2016). As important ornamental plants, Rosa spp. play a key role in Chinese landscaping (Zhang et al. 2009). Furthermore, Rosa species are important raw materials for the spice and food industry, and a rose industry has been established in many parts of China, generating huge income for the local economy (Wang 2021). Most Rosa spp. can be used in traditional Chinese medicines, having great nutritional and medicinal value (Wang 2021). In addition to these, Rosa spp. are important resource species for ecological and vegetation restoration, having great ecological value in China, because many rose species have strong resistance to stress and can survive in harsh environments (Liu 2016).
Many fungal taxa such as Botryosphaeria dothidea, Botrytis cinerea, Chaetomella raphigera, Colletotrichum siamense, Cytospora spp., Diplocarpon rosae, Elsinoe rosarum and Lasiodiplodia theobromae have in the past been identified as the causal agents of various diseases of Rosa spp. in China, and severely limited their production. (Zhang et al. 2014, Bagsic et al. 2016, Chen et al. 2016, Debener 2019, Feng et al. 2019 Jia et al. 2019, Munoz et al. 2019, Pan et al. 2020). Members of Sporocadaceae have also been reported to cause diseases on Rosa spp. Examples include cankers caused by N. rosicola, dieback caused by Ciliochorella mangiferae and Robillarda sessilis, stem lesions caused by N. rosae and R. sessilis, and leaf spots caused by Diploceras discosioides, Discosia artocreas and Truncatella angustata (Weiss 1950, Mathur 1979, Peregrine & Ahmad 1982, Eken et al. 2009, Maharachchikumbura et al. 2014, Jiang et al. 2018). Furthermore, Rosa has proven to represent a rich niche of undescribed species of Sporocadaceae, with many remaining poorly identified, as the generic concepts have been in flux until recently (Liu et al. 2019). Therefore, conducting detailed surveys of pestalotioid fungi from Rosa spp. in China was necessary. The aims of the present study were thus to identify these fungi based on phylogenetic analyses and morphological comparisons, describe the species new to science, and gain a better understanding of the diversity and prevalence of Sporocadaceae associated with Rosa spp. in China.
MATERIALS AND METHODS
Sampling and isolation
A total of 295 Rosa samples (branches, fruits, leaves and spines) showing disease symptoms (Fig. 1) were collected from five provinces (Gansu, Henan, Hunan, Qinghai and Shaanxi) and the Ningxia Autonomous Region of China, which are the main production areas of Rosa plants in China. The Rosa species sampled include R. chinensis, R. helenae, R. laevigata, R. multiflora, R. omeiensis, R. rugosa, R. spinosissima and R. xanthina.
Fig. 1.

Disease symptoms on Rosa associated with infection by Sporocadaceae. a–c. Leaves spots on Rosa rugosa caused by Pestalotiopsis rhodomyrtus; d–e. lesion developing on the fruits of Rosa laevigata infected by Seimatosporium nonappendiculatum; f. dying bush; g–h. dieback on (g) Rosa rugosa and (h) Rosa xanthina caused by Seiridium rosae and Sporocadus sorbi; i–j. sporocarps of Pestalotiopsis chamaeropis and Seimatosporium centrale on the spines of (i) Rosa rugosa and (j) Rosa chinensis.
A total of 126 strains were obtained by removing the spore mass from conidiomata and generating single spore colonies, or plating superficially sterilised diseased tissue on potato dextrose agar (PDA, 20 % diced potatoes, 2 % agar and 2 % glucose) and incubating Petri dishes at 25 °C in the dark for 2–3 d. When colonies just formed, they were hyphal-tipped and transferred to fresh PDA Petri dishes (Crous et al. 2019a). Type specimens of new species from this study were deposited in the Museum of the Beijing Forestry University (BJFC), and ex-type living cultures were deposited in the China Forestry Culture Collection Centre (CFCC), Beijing, China.
Morphological analyses
Cultures were incubated on PDA at 25 °C in a 12 h day/night regime (Crous et al. 2019a). After 15 d, colony diameters were measured and colony colours were rated according to Rayner (1970). Slide preparations were mounted in lactic acid or water, from colonies sporulating on PDA, autoclaved pine needles on 2 % tap water agar (Smith et al. 1996), and incubated at 25 °C under continuous nuv-light to promote sporulation. Sections through stromata were made by hand. Observations were made with a Leica DM 2500 dissecting microscope (Wetzlar, Germany), and with a Nikon Eclipse 80i compound microscope using differential interference contrast (DIC) illumination and images recorded on a Nis DS-Ri2 camera with the Nikon NisElements F4.30.01 software. Conidial length was measured from the base of the basal cell to the base of the apical appendage, and conidial width was measured at the widest point of the conidium (Bonthond et al. 2018). Taxonomic novelties were deposited in MycoBank (Crous et al. 2004).
DNA extraction, PCR amplification and sequencing
Total genomic DNA was extracted from axenic cultures using a modified cetyltrimethylammonium bromide (CTAB) protocol (Doyle & Doyle 1990). DNA products were stored at -20 °C. The extracted DNA was used as template for the polymerase chain reaction (PCR). PCR reaction primers (forward and reverse) of each fungal genus are found in Table 1. PCR parameters were initiated with 95 °C for 5 min, followed by 34 cycles of denaturation at 95 °C for 30 s, annealing at a suitable temperature for 30 s (56 °C for ITS and LSU, 52 °C for TEF, 52 °C for RPB2 and 60 °C for TUB), and extension at 72 °C for 30 s, and terminated with a final elongation step at 72 °C for 10 min. The final PCR products were examined by electrophoresis in 2 % agarose gels. The amplified PCR products were sent to a commercial sequencing provider (Tsingke Biotechnology Co. Ltd, Beijing, China).
Table 1.
PCR reaction primers (forward and reverse) for amplification of gene loci of each fungal genus.
| Genus | Loci used for amplification | References | ||||
|---|---|---|---|---|---|---|
| ITS | LSU | RPB2 | TEF | TUB | ||
| Monochaetia | ITS1/ITS4 | LROR/LR5 | RPB2-5F2/fRPB2-7cR | Liu et al. (2019), Jiang et al. (2021) | ||
| Neopestalotiopsis | ITS1/ITS4 | LROR/LR5 | RPB2-5F2/fRPB2-7cR | EF1-728F/EF-2 | T1/Bt2b | Liu et al. (2019), Norphanphoun et al. (2019), Jiang et al. (2021) |
| Pestalotiopsis | ITS1/ITS4 | LROR/LR5 | RPB2-5F2/fRPB2-7cR | EF1-728F/EF-2 | T1/Bt2b | Liu et al. (2019), Norphanphoun et al. (2019), Jiang et al. (2021) |
| Seimatosporium | ITS1/ITS4 | LROR/LR5 | RPB2-5F2/fRPB2-7cR | Goonasekara et al. (2016), Wijayawardene et al. (2016a), Liu et al. (2019) | ||
| Seiridium | ITS1/ITS4 | LROR/LR5 | RPB2-5F2/fRPB2-7cR | EF1-728F/EF-2 | T1/Bt2b | Jiang et al. (2019), Liu et al. (2019), Marin-Felix et al. (2019) |
| Sporocadus | ITS1/ITS4 | LROR/LR5 | RPB2-5F2/fRPB2-7cR | EF1-728F/EF-2 | T1/Bt2b | Liu et al. (2019) |
Phylogenetic analyses
All nucleotide sequences generated from different primer pairs in this study were deposited in GenBank (Table 2). Sequences were BLASTn searched in NCBI to obtain the related sequences from recent publications and were analysed (Table 3). Sequences were aligned in MAFFT v. 7 at the web server (http://mafft.cbrc.jp/alignment/server) (Katoh & Standley 2013, Katoh et al. 2019) and manually adjusted in MEGA v. 6 (Tamura et al. 2013).
Table 2.
Isolates sequenced and used for phylogenetic analyses in the current study.
| Species1 | Culture no. | Status2 | Host | Tissues | Origin | GenBank accession no. | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| ITS | LSU | RPB2 | TEF | TUB | ||||||
| Monochaetia rosarum | CFCC55172 = ROC 099 | T | Rosa chinensis | branches | Nanyang, Henan | MZ292088 | OK560346 | OL742111 | OL814484 | OM103654 |
| CFCC55173 = ROC 100 | Rosa chinensis | branches | Nanyang, Henan | MZ292089 | OK560347 | OL742112 | OL814485 | OM103655 | ||
| ROC 098 | Rosa chinensis | branches | Nanyang, Henan | MZ292090 | OK560348 | OL742113 | OL814486 | OM103656 | ||
| ROC 101 | Rosa chinensis | branches | Nanyang, Henan | MZ292091 | OK560349 | OL742114 | OL814487 | OM103657 | ||
| Neopestalotiopsis concentrica | CFCC 55162 = ROC 53 | T | Rosa rugosa | spines | Xinyang, Henan | OK560707 | OK560440 | OL742115 | OM622433 | OM117698 |
| CFCC 55163 = ROC 64 | Rosa chinensis | spines | Xinyang, Henan | OK560708 | OK560441 | OL742116 | OM622434 | OM117699 | ||
| ROC 135 | Rosa chinensis | spines | Xinyang, Henan | OK560709 | OK560442 | OL742117 | OM622435 | OM117700 | ||
| ROC 136 | Rosa chinensis | spines | Xinyang, Henan | OK560710 | OK560443 | OL742118 | OM622436 | OM117701 | ||
| ROC 137 | Rosa chinensis | spines | Nanyang, Henan | OK560711 | OK560444 | OL742119 | OM622437 | OM117702 | ||
| ROC 138 | Rosa chinensis | spines | Nanyang, Henan | OK560712 | OK560445 | OL742120 | OM622438 | OM117703 | ||
| ROC 140 | Rosa chinensis | spines | Nanyang, Henan | OK560713 | OK560446 | OL742121 | OM622439 | OM117704 | ||
| ROC 141 | Rosa chinensis | spines | Nanyang, Henan | OK560714 | OK560447 | OL742122 | OM622440 | OM117705 | ||
| ROC 142 | Rosa chinensis | spines | Nanyang, Henan | OK560715 | OK560448 | OL742123 | OM622441 | OM117706 | ||
| N. subepidermalis | CFCC 55160 = ROC 161 | T | Rosa rugosa | spines | Xinyang, Henan | OK560699 | OK560432 | – | OM622425 | OM117690 |
| ROC 162 | Rosa rugosa | spines | Xinyang, Henan | OK560700 | OK560433 | – | OM622426 | OM117691 | ||
| CFCC 55161 = ROC 169 | Rosa chinensis | spines | Xinyang, Henan | OK560701 | OK560434 | – | OM622427 | OM117692 | ||
| ROC 170 | Rosa chinensis | spines | Xinyang, Henan | OK560702 | OK560435 | – | OM622428 | OM117693 | ||
| ROC 171 | Rosa chinensis | branches | Xinyang, Henan | OK560703 | OK560436 | – | OM622429 | OM117694 | ||
| ROC 172 | Rosa chinensis | branches | Xinyang, Henan | OK560704 | OK560437 | – | OM622430 | OM117695 | ||
| ROC 173 | Rosa chinensis | branches | Xinyang, Henan | OK560705 | OK560438 | – | OM622431 | OM117696 | ||
| ROC 174 | Rosa chinensis | spines | Xinyang, Henan | OK560706 | OK560439 | – | OM622432 | OM117697 | ||
| Pestalotiopsis chamaeropis | CFCC 55156 = ROC 23-1 | Rosa chinensis | spines | Tianshui, Gansu | OK560574 | OK560306 | OL742124 | OL814488 | OM158138 | |
| ROC 270 | Rosa chinensis | spines | Tianshui, Gansu | OK560575 | OK560307 | OL742125 | OL814489 | OM158139 | ||
| ROC 272 | Rosa chinensis | spines | Tianshui, Gansu | OK560576 | OK560308 | OL742126 | OL814490 | OM158140 | ||
| ROC 273 | Rosa chinensis | spines | Tianshui, Gansu | OK560577 | OK560309 | OL742127 | OL814491 | OM158141 | ||
| ROC 275 | Rosa chinensis | spines | Tianshui, Gansu | OK560578 | OK560310 | OL742128 | OL814492 | OM158142 | ||
| CFCC 55157 = ROC 23-2 | Rosa chinensis | spines | Tianshui, Gansu | OK560579 | OK560311 | OL742129 | OL814493 | OM158143 | ||
| ROC 276 | Rosa chinensis | spines | Tianshui, Gansu | OK560580 | OK560312 | OL742130 | OL814494 | OM158144 | ||
| ROC 278 | Rosa chinensis | spines | Tianshui, Gansu | OK560581 | OK560313 | OL742131 | OL814495 | OM158145 | ||
| ROC 279 | Rosa chinensis | spines | Tianshui, Gansu | OK560582 | OK560314 | OL742132 | OL814496 | OM158146 | ||
| ROC 280 | Rosa chinensis | spines | Tianshui, Gansu | OK560583 | OK560315 | OL742133 | OL814497 | OM158147 | ||
| ROC 281 | Rosa chinensis | spines | Tianshui, Gansu | OK560584 | OK560316 | OL742134 | OL814498 | OM158148 | ||
| ROC 282 | Rosa chinensis | spines | Tianshui, Gansu | OK560585 | OK560317 | OL742135 | OL814499 | OM158149 | ||
| ROC 283 | Rosa chinensis | spines | Tianshui, Gansu | OK560586 | OK560318 | OL742136 | OL814500 | OM158150 | ||
| ROC 286 | Rosa chinensis | spines | Tianshui, Gansu | OK560587 | OK560319 | OL742137 | OL814501 | OM158151 | ||
| ROC 289 | Rosa chinensis | spines | Tianshui, Gansu | OK560588 | OK560320 | OL742138 | OL814502 | OM158152 | ||
| ROC 290 | Rosa chinensis | spines | Tianshui, Gansu | OK560589 | OK560321 | OL742139 | OL814503 | OM158153 | ||
| ROC 292 | Rosa chinensis | spines | Tianshui, Gansu | OK560590 | OK560322 | OL742140 | OL814504 | OM158154 | ||
| ROC 293 | Rosa chinensis | spines | Tianshui, Gansu | OK560591 | OK560323 | OL742141 | OL814505 | OM158155 | ||
| Pes. rhodomyrtus | ROC 056 | Rosa rugosa | leaves | Nanyang, Henan | OK560592 | – | – | OL814506 | OM158156 | |
| ROC 057 | Rosa rugosa | leaves | Nanyang, Henan | OK560593 | – | – | OL814507 | OM158157 | ||
| ROC 058 | Rosa rugosa | leaves | Nanyang, Henan | OK560594 | – | – | OL814508 | OM158158 | ||
| ROC 059 | Rosa rugosa | leaves | Nanyang, Henan | OK560595 | – | – | OL814509 | OM158159 | ||
| ROC 060 | Rosa rugosa | leaves | Nanyang, Henan | OK560596 | – | – | OL814510 | OM158160 | ||
| ROC 061 | Rosa rugosa | leaves | Nanyang, Henan | OK560597 | – | – | OL814511 | OM158161 | ||
| ROC 062 | Rosa rugosa | leaves | Nanyang, Henan | OK560598 | – | – | OL814512 | OM158162 | ||
| ROC 303 | Rosa multiflora | spines | Gannan, Gansu | OK560599 | – | – | OL814513 | OM158163 | ||
| ROC 304 | Rosa multiflora | spines | Gannan, Gansu | OK560600 | – | – | OL814514 | OM158164 | ||
| ROC 305 | Rosa multiflora | spines | Gannan, Gansu | OK560601 | – | – | OL814515 | OM158165 | ||
| ROC 306 | Rosa multiflora | spines | Gannan, Gansu | OK560602 | – | – | OL814516 | OM158166 | ||
| ROC 307 | Rosa multiflora | spines | Gannan, Gansu | OK560603 | – | – | OL814517 | OM158167 | ||
| ROC 309 | Rosa multiflora | spines | Gannan, Gansu | OK560604 | – | – | OL814518 | OM158168 | ||
| ROC 311 | Rosa multiflora | spines | Gannan, Gansu | OK560605 | – | – | OL814519 | OM158169 | ||
| Pes. rhodomyrtus (cont.) | ROC 356 | Rosa chinensis | branches | Changsha, Hunan | OK560606 | – | – | OL814520 | OM158170 | |
| ROC 357 | Rosa chinensis | branches | Changsha, Hunan | OK560607 | – | – | OL814521 | OM158171 | ||
| ROC 358 | Rosa chinensis | branches | Changsha, Hunan | OK560608 | – | – | OL814522 | OM158172 | ||
| ROC 359 | Rosa chinensis | branches | Changsha, Hunan | OK560609 | – | – | OL814523 | OM158173 | ||
| Pes. tumida | CFCC 55158 = ROC 110 | T | Rosa chinensis | spines | Tianshui, Gansu | OK560610 | OK560324 | OL742142 | OL814524 | OM158174 |
| ROC 109 | Rosa chinensis | spines | Tianshui, Gansu | OK560611 | OK560325 | OL742143 | OL814525 | OM158175 | ||
| ROC 108 | Rosa chinensis | spines | Tianshui, Gansu | OK560612 | OK560326 | OL742144 | OL814526 | OM158176 | ||
| CFCC 55159 = ROC 234 | Rosa chinensis | branches | Tianshui, Gansu | OK560613 | OK560327 | OL742145 | OL814527 | OM158177 | ||
| ROC 235 | Rosa chinensis | branches | Tianshui, Gansu | OK560614 | OK560328 | OL742146 | OL814528 | OM158178 | ||
| ROC 236 | Rosa chinensis | branches | Tianshui, Gansu | OK560615 | OK560329 | OL742147 | OL814529 | OM158179 | ||
| ROC 237 | Rosa chinensis | spines | Tianshui, Gansu | OK560616 | OK560330 | OL742148 | OL814530 | OM158180 | ||
| ROC 238 | Rosa chinensis | spines | Tianshui, Gansu | OK560617 | OK560331 | OL742149 | OL814531 | OM158181 | ||
| ROC 240 | Rosa chinensis | branches | Tianshui, Gansu | OK560618 | OK560332 | OL742150 | OL814532 | OM158182 | ||
| Seimatosporium centrale | CFCC 55166 = ROC 003 | T | Rosa chinensis | spines | Tianshui, Gansu | OK560629 | OK560399 | ON055447 | OM986918 | OM301641 |
| ROC 001 | Rosa chinensis | spines | Tianshui, Gansu | OK560630 | OK560400 | ON055448 | OM986919 | OM301642 | ||
| ROC 002 | Rosa chinensis | spines | Tianshui, Gansu | OK560631 | OK560401 | ON055449 | OM986920 | OM301643 | ||
| CFCC 55169 =ROC 014 | Rosa chinensis | spines | Baoji, Shaanxi | OK560632 | OK560402 | ON055450 | OM986921 | OM301644 | ||
| ROC 015 | Rosa chinensis | spines | Baoji, Shaanxi | OK560633 | OK560403 | ON055451 | OM986922 | OM301645 | ||
| ROC 016 | Rosa chinensis | spines | Baoji, Shaanxi | OK560634 | OK560404 | ON055452 | OM986923 | OM301646 | ||
| ROC 145 | Rosa chinensis | spines | Tianshui, Gansu | OK560635 | OK560405 | ON055453 | OM986924 | OM301647 | ||
| ROC 146 | Rosa chinensis | spines | Tianshui, Gansu | OK560636 | OK560406 | ON055454 | OM986925 | OM301648 | ||
| ROC 147 | Rosa chinensis | spines | Tianshui, Gansu | OK560637 | OK560407 | ON055455 | OM986926 | OM301649 | ||
| Seim, gracile | CFCC 55167 = ROC 004 | T | Rosa xanthina | spines | Tianshui, Gansu | OK560638 | OK560408 | ON055456 | OM986927 | OM301650 |
| ROC 005 | Rosa xanthina | spines | Tianshui, Gansu | OK560639 | OK560409 | ON055457 | OM986928 | OM301651 | ||
| ROC 006 | Rosa xanthina | spines | Tianshui, Gansu | OK560640 | OK560410 | ON055458 | OM986929 | OM301652 | ||
| ROC 007 | Rosa xanthina | spines | Tianshui, Gansu | OK560641 | OK560411 | ON055459 | OM986930 | OM301653 | ||
| ROC 008 | Rosa xanthina | spines | Tianshui, Gansu | OK560642 | OK560412 | ON055460 | OM986931 | OM301654 | ||
| ROC 009 | Rosa xanthina | spines | Tianshui, Gansu | OK560643 | OK560413 | ON055461 | OM986932 | OM301655 | ||
| ROC 010 | Rosa xanthina | spines | Tianshui, Gansu | OK560644 | OK560414 | ON055462 | OM986933 | OM301656 | ||
| ROC 011 | Rosa xanthina | spines | Tianshui, Gansu | OK560645 | OK560415 | ON055463 | OM986934 | OM301657 | ||
| ROC 012 | Rosa xanthina | spines | Tianshui, Gansu | OK560646 | OK560416 | ON055464 | OM986935 | OM301658 | ||
| Seim, nonappendiculatum | CFCC 55168 = ROC 377 | T | Rosa laevigata | fruits | Guyuan, Ningxia | OK560657 | OK560427 | ON055475 | OM986946 | OM301669 |
| ROC 378 | Rosa laevigata | fruits | Guyuan, Ningxia | OK560658 | OK560428 | ON055476 | OM986947 | OM301670 | ||
| ROC 379 | Rosa laevigata | fruits | Guyuan, Ningxia | OK560659 | OK560429 | ON055477 | OM986948 | OM301671 | ||
| ROC 380 | Rosa laevigata | fruits | Guyuan, Ningxia | OK560660 | OK560430 | ON055478 | OM986949 | OM301672 | ||
| ROC 381 | Rosa laevigata | fruits | Guyuan, Ningxia | OK560661 | OK560431 | ON055479 | OM986950 | OM301673 | ||
| Seim, parvum | CFCC 55164 = ROC 038 | T | Rosa spinosissima | spines | Huangnan, Qinghai | OK560647 | OK560417 | ON055465 | OM986936 | OM301659 |
| ROC 039 | Rosa spinosissima | spines | Huangnan, Qinghai | OK560648 | OK560418 | ON055466 | OM986937 | OM301660 | ||
| ROC 040 | Rosa spinosissima | spines | Huangnan, Qinghai | OK560649 | OK560419 | ON055467 | OM986938 | OM301661 | ||
| ROC 041 | Rosa spinosissima | spines | Huangnan, Qinghai | OK560650 | OK560420 | ON055468 | OM986939 | OM301662 | ||
| ROC 042 | Rosa spinosissima | spines | Huangnan, Qinghai | OK560651 | OK560421 | ON055469 | OM986940 | OM301663 | ||
| ROC 043 | Rosa spinosissima | spines | Huangnan, Qinghai | OK560652 | OK560422 | ON055470 | OM986941 | OM301664 | ||
| CFCC 55165 =ROC 017 | Rosa helenae | spines | Huangnan, Qinghai | OK560653 | OK560423 | ON055471 | OM986942 | OM301665 | ||
| ROC 018 | Rosa helenae | spines | Huangnan, Qinghai | OK560654 | OK560424 | ON055472 | OM986943 | OM301666 | ||
| ROC 019 | Rosa helenae | spines | Huangnan, Qinghai | OK560655 | OK560425 | ON055473 | OM986944 | OM301667 | ||
| ROC 020 | Rosa helenae | spines | Huangnan, Qinghai | OK560656 | OK560426 | ON055474 | OM986945 | OM301668 | ||
| Seiridium rosae | CFCC 55174 = ROC 208 | T | Rosa rugosa | branches | Guyuan, Ningxia | OK560681 | OK560394 | OL742151 | OL814533 | OM313314 |
| ROC 209 | Rosa rugosa | branches | Guyuan, Ningxia | OK560682 | OK560395 | OL742152 | OL814534 | OM313315 | ||
| CFCC 55175 = ROC 267 | Rosa rugosa | branches | Guyuan, Ningxia | OK560683 | OK560396 | OL742153 | OL814535 | OM313316 | ||
| ROC 268 | Rosa rugosa | branches | Guyuan, Ningxia | OK560684 | OK560397 | OL742154 | OL814536 | OM313317 | ||
| Sporocadus brevis | CFCC 55170 = ROC 091 | T | Rosa spinosissima | spines | Gannan, Gansu | OK655780 | OK560371 | OL742155 | OL814537 | OM401659 |
| ROC 092 | Rosa spinosissima | spines | Gannan, Gansu | OK655781 | OK560372 | OL742156 | OL814538 | OM401660 | ||
| ROC 093 | Rosa spinosissima | spines | Gannan, Gansu | OK655782 | OK560373 | OL742157 | OL814539 | OM401661 | ||
| Sporocadus brevis (cont.) | ROC 094 | Rosa spinosissima | spines | Gannan, Gansu | OK655783 | OK560374 | OL742158 | OL814540 | OM401662 | |
| ROC 095 | Rosa spinosissima | spines | Gannan, Gansu | OK655784 | OK560375 | OL742159 | OL814541 | OM401663 | ||
| Spo. sorbi | ROC 105 | Rosa xanthina | branches | Lanzhou, Gansu | OK655785 | OK560376 | OL742160 | OL814542 | OM401664 | |
| ROC 102 | Rosa xanthina | branches | Lanzhou, Gansu | OK655786 | OK560377 | OL742161 | OL814543 | OM401665 | ||
| ROC 103 | Rosa xanthina | branches | Lanzhou, Gansu | OK655787 | OK560378 | OL742162 | OL814544 | OM401666 | ||
| ROC 159 | Rosa xanthina | spines | Ganan, Gansu | OK655788 | OK560379 | OL742163 | OL814545 | OM401667 | ||
| ROC 160 | Rosa xanthina | spines | Ganan, Gansu | OK655789 | OK560380 | OL742164 | OL814546 | OM401668 | ||
| ROC 161 | Rosa xanthina | spines | Ganan, Gansu | OK655790 | OK560381 | OL742165 | OL814547 | OM401669 | ||
| Spo. spiniger | ROC 119 | T | Rosa omeiensis | spines | Lanzhou, Gansu | OK655791 | OK560382 | OL742166 | OL814548 | OM401670 |
| ROC 120 | Rosa omeiensis | spines | Lanzhou, Gansu | OK655792 | OK560383 | OL742167 | OL814549 | OM401671 | ||
| ROC 121 | Rosa omeiensis | spines | Lanzhou, Gansu | OK655793 | OK560384 | OL742168 | OL814550 | OM401672 | ||
| ROC 122 | Rosa omeiensis | spines | Lanzhou, Gansu | OK655794 | OK560385 | OL742169 | OL814551 | OM401673 | ||
| ROC 123 | Rosa omeiensis | spines | Lanzhou, Gansu | OK655795 | OK560386 | OL742170 | OL814552 | OM401674 | ||
| ROC 124 | Rosa omeiensis | spines | Lanzhou, Gansu | OK655796 | OK560387 | OL742171 | OL814553 | OM401675 | ||
| ROC 125 | Rosa omeiensis | spines | Lanzhou, Gansu | OK655797 | OK560388 | OL742172 | OL814554 | OM401676 | ||
| Spo. trimorphus | CFCC 55171 = ROC 112 | Rosa xanthina | branches | Lanzhou, Gansu | OK655798 | OK560389 | OL742173 | OL814555 | OM401677 | |
| ROC 113 | Rosa xanthina | branches | Lanzhou, Gansu | OK655799 | OK560390 | OL742174 | OL814556 | OM401678 | ||
| ROC 114 | Rosa xanthina | branches | Lanzhou, Gansu | OK655800 | OK560391 | OL742175 | OL814557 | OM401679 | ||
| ROC 115 | Rosa xanthina | branches | Lanzhou, Gansu | OK655801 | OK560392 | OL742176 | OL814558 | OM401680 | ||
| ROC 116 | Rosa xanthina | branches | Lanzhou, Gansu | OK655802 | OK560393 | OL742177 | OL814559 | OM401681 | ||
1 Newly described taxa and deposited sequences are in bold.
2 T: ex-type.
Table 3.
Isolates from previous studies used in the phylogenetic analyses in the current study.
| Species | Strain number1 | Status2 | Country | Substrate | GenBank accession no. | References | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| ITS | LSU | RPB2 | TEF | TUB | ||||||
| Allelochaeta acuta | CPC 16629 | Australia | Eucalyptus dives | MH554086 | MH554297 | MH555000 | – | – | Crous et al. (2018) | |
| All. elegans | CBS 187.81 | ET | Australia | Melaleuca lanceolata | MH554014 | MH554234 | MH554927 | – | – | Crous et al. (2018) |
| All. falcata | CPC 13580 | Australia | Eucalyptus alligatrix | MH554073 | MH554284 | MH554985 | – | – | Crous et al. (2018) | |
| CBS 131117 | ET | Australia | Eucalyptus alligatrix | MH553999 | MH554217 | MH554907 | – | – | Crous et al. (2018) | |
| All. fusispora | CPC 17616 | Australia | Eucalyptus sp. | MH554094 | MH554304 | MH555008 | – | – | Crous et al. (2018) | |
| CBS 810.73 | IT | Australia | Eucalyptus polyanthemos | MH554067 | MH554279 | MH554980 | – | – | Crous et al. (2018) | |
| All. kriegeriana | CBS 188.81 | Australia | Callistemon sieberi | MH554015 | MH554235 | MH554928 | – | – | Crous et al. (2018) | |
| All. neoacuta | CBS 115131 | T | South Africa | Eucalyptus smithii | JN871200 | JN871209 | MH554998 | – | – | Crous et al. (2018) |
| CBS 110733 | South Africa | Eucalyptus smithii | JN871201 | JN871210 | MH554999 | – | – | Crous et al. (2018) | ||
| All. neodilophospora | CPC 17161 | T | Australia | Callistemon pinifolius | MH554090 | MH554300 | MH555004 | – | – | Crous et al. (2018) |
| All. neoorbicularis | CPC 13581 | Australia | Eucalyptus regnans | MH554074 | MH554285 | MH554986 | – | – | Crous et al. (2018) | |
| All. obliquae | CPC 20191 | T | Australia | Eucalyptus obliqua | MH554105 | MH554315 | MH555018 | – | – | Crous et al. (2018) |
| All. orbicularis | CBS 131118 | ET | Australia | Corymbia henryi | MH554000 | MH554218 | MH554908 | – | – | Crous et al. (2018) |
| All. paraelegans | CBS 150.71 | T | Australia | Melaleuca ericifolia | MH554007 | MH554228 | MH554923 | – | – | Crous et al. (2018) |
| All. pseudowalkeri | CPC 17043 | T | Australia | Eucalyptus sp. | MH554089 | MH554299 | MH555003 | – | – | Crous et al. (2018) |
| All. sparsifoliae | CPC 14529 | Australia | Eucalyptus sparsifolia | MH554083 | MH554294 | MH554995 | – | – | Crous et al. (2018) | |
| CPC 14502 | T | Australia | Eucalyptus sparsifolia | MH554082 | MH554293 | MH554994 | – | – | Crous et al. (2018) | |
| Bartalinia bella | CBS 125525 | South Africa | Maytenus abbottii | GU291796 | MH554214 | MH554904 | – | – | Liu et al. (2019) | |
| CBS 464.61 | T | Brazil | Air | MH554051 | MH554264 | MH554964 | – | – | Liu et al. (2019) | |
| Bar. pini | CBS 143891 | T | Uganda | Pinus patula | MH554125 | MH554330 | MH555033 | – | – | Liu et al. (2019) |
| CBS 144141 | USA | Acacia koa | MH554170 | MH554364 | MH555067 | – | – | Liu et al. (2019) | ||
| Bar. robillardoides | CBS 122705 | ET | Italy | Leptoglossus occidentalis | LT853104 | KJ710438 | LT853152 | – | – | Liu et al. (2019) |
| CBS 122615 | South Africa | Cupressus lusitanica | MH553989 | MH554207 | MH554897 | – | – | Liu et al. (2019) | ||
| Beltrania pseudorhombica | CPC 23656 | China | Pinus tabulaeformis | MH554124 | KJ869215 | MH555032 | – | – | Crous et al. (2014a), | |
| Liu et al. (2019) | ||||||||||
| Bel. rhombica | CBS 123.58 | T | Mozambique | Sand near mangrove swamp | MH553990 | MH554209 | MH554899 | – | – | Liu et al. (2019) |
| Broomella vitalbae | HPC 1154 | – | – | MH554173 | MH554367 | – | – | – | Liu et al. (2019) | |
| MFLUCC 13-0798 | ET | Italy | Clematis vitalba | NR 153610 | KP757749 | – | – | – | Liu et al. (2019) | |
| Ciliochorella phanericola | MFLUCC 12-0310 | Thailand | Dead leaves | KF827444 | KF827445 | KF827479 | – | – | Hyde et al. (2016) | |
| MFLUCC 14-0984 | T | Thailand | Phanera purpurea | KX789680 | KX789681 | – | – | – | Hyde et al. (2016) | |
| Clypeosphaeria uniseptata | CBS 114967 | Hong Kong, China | Wood | MH553979 | MH554197 | MH554878 | – | – | Liu et al. (2019) | |
| Diploceras hypericinum | CBS 109058 | New Zealand | Hypericum sp. | MH553955 | MH554178 | MH554852 | – | – | Liu et al. (2019) | |
| CBS 492.97 | Netherlands | Hypericum perforatum | MH554054 | MH554267 | MH554967 | – | – | Liu et al. (2019) | ||
| CBS 197.36 | Switzerland | Hypericum sp. | MH554017 | MH554237 | MH554930 | – | – | Liu et al. (2019) | ||
| CBS 143885 | ET | Netherlands | Hypericum perforatum | MH554108 | MH554316 | MH555019 | – | – | Liu et al. (2019) | |
| Disaeta arbuti | CBS 143903 | Australia | Acacia pycnantha | MH554148 | MH554346 | MH555050 | – | – | Liu et al. (2019) | |
| Discosia artocreas | CBS 124848 | ET | Germany | Fagus sylvatica | MH553994 | MH554213 | MH554903 | – | – | Liu et al. (2019) |
| Dis. brasiliensis | NTCL095 | Thailand | Dead leaf | KF827433 | KF827437 | KF827474 | – | – | Liu et al. (2019) | |
| NTCL097-2 | Thailand | Dead leaf | KF827434 | KF827438 | KF827475 | – | – | Liu et al. (2019) | ||
| NTCL094-2 | Thailand | Dead leaf | KF827432 | KF827436 | KF827473 | – | – | Liu et al. (2019) | ||
| Discosia sp. 6 | CBS 241.66 | South Africa | Acacia karroo | MH554022 | MH554244 | MH554933 | – | – | Liu et al. (2019) | |
| Discosia sp. 7 | CBS 684.70 | Netherlands | Aesculus hippocastanu | MH554064 | MH554277 | MH554978 | – | – | Liu et al. (2019) | |
| Distononappendiculata banksiae | CPC 17658 | Australia | Banksia marginata | MH554097 | MH554307 | MH555011 | – | – | Liu et al. (2019) | |
| CBS 131308 | T | Australia | Banksia marginata | JQ044422 | JQ044442 | MH554909 | – | – | Liu et al. (2019) | |
| CPC 20185 | Australia | Banksia marginata | MH554104 | MH554314 | MH555017 | – | – | Liu et al. (2019) | ||
| CBS 143906 | Australia | Banksia marginata | MH554158 | MH554354 | MH555057 | – | – | Liu et al. (2019) | ||
| Dist. casuarinae | CBS 143884 | T | Australia | Casuarina sp. | MH554093 | MH554303 | MH555007 | – | – | Liu et al. (2019) |
| Dist. verruca ta | CBS 144032 | T | Australia | Banksia repens | MH554163 | MH554359 | MH555062 | – | – | Liu et al. (2019) |
| Diversimediispora humicola | CBS 302.86 | T | USA | Soil | MH554028 | MH554247 | MH554941 | – | – | Liu et al. (2019) |
| Heterotruncatella lutea | CBS 349.73 | IT | Australia | Acacia pycnantha | LT853099 | DQ414533 | LT853146 | – | – | Liu et al. (2019) |
| Het. proteicola | CBS 123029 | South Africa | Protea acaulis | MH553993 | MH554212 | MH554902 | – | – | Liu et al. (2019) | |
| Het. restionacearum | CBS 119210 | South Africa | Ischyrolepis cf. gaudichaudiana | DQ278915 | DQ278929 | MH554892 | – | – | Liu et al. (2019) | |
| CBS 118150 | South Africa | Restio filiformis | DQ278914 | MH554203 | MH554889 | – | – | Liu et al. (2019) | ||
| Het. spadicea | CBS 118148 | South Africa | Rhodocoma capensis | DQ278913 | DQ278928 | MH554888 | – | – | Liu et al. (2019) | |
| CBS 118144 | South Africa | Ischyrolepis sp. | DQ278921 | DQ278926 | MH554886 | – | – | Liu et al. (2019) | ||
| Hyalotiella spartii | MFLUCC 13-0397 | T | Italy | Spartium junceum | KP757756 | KP757752 | – | – | – | Li et al. (2015) |
| Hyalotiella transvalensis | CBS 303.65 | T | South Africa | Soil | MH554029 | MH554248 | MH554942 | – | – | Liu et al. (2019) |
| Hymenopleella austroafricana | CBS 143886 | T | South Africa | Gleditsia triacanthos | MH554115 | MH554320 | MH555023 | – | – | Liu et al. (2019) |
| CBS 144027 | Zambia | Combretum hereroense | MH554119 | MH554324 | MH555027 | – | – | Liu et al. (2019) | ||
| CBS 144026 | South Africa | Bridelia mollis | MH554117 | MH554322 | MH555025 | – | – | Liu et al. (2019) | ||
| Hym. endophytica | EMLAS5-1 | T | Korea | Abies firma | KX216520 | KX216518 | – | – | – | Liu et al. (2019) |
| Hym. hippophaëicola | CBS 113687 | Sweden | Hippophae rhamnoides | MH553969 | MH554188 | MH554863 | – | – | Jaklitsch et al. (2016) | |
| CBS 140410 | ET | Austria | Hippophae rhamnoides | KT949901 | MH554224 | MH554919 | – | – | Jaklitsch et al. (2016) | |
| Hym. lakefuxianensis | HKUCC 7303 | T | China | Submerged wood | – | AF452047 | – | – | – | Liu et al. (2019) |
| Hym. polyseptata | CBS 143887 | T | South Africa | Combretum sp. | MH554116 | MH554321 | MH555024 | – | – | Liu et al. (2019) |
| Hym. subcylindrica | CBS 164.77 | India | Cocos nucifera | MH554009 | MH554230 | MH554925 | – | – | Liu et al. (2019) | |
| CBS 647.74 | T | India | Gypsophilla seeds | MH554062 | MH554275 | MH554976 | – | – | Liu et al. (2019) | |
| Immersidiscosia eucalypti | MAFF 242781 | Japan | Unknown dead leaves | AB594793 | AB593725 | – | – | – | Tanaka et al. (2011) | |
| NBRC 104197 | Japan | Ardisia japonica | AB594792 | AB593724 | – | – | – | Tanaka et al. (2011) | ||
| Lepteutypa fuckelii | CBS 140409 | NT | Belgium | Tilia cordata | NR 154123 | KT949902 | MH554918 | – | – | Liu et al. (2019) |
| Lep. sambuci | CBS 131707 | T | UK | Sambucus nigra | NR 154124 | MH554219 | MH554911 | – | – | Liu et al. (2019) |
| Microdochium lycopodinum | CBS 125585 | T | Austria | Lycopodium annotinum | KP859016 | KP858952 | KP859125 | – | – | Hernandez-Restrepo et al. (2016), |
| Liu et al. (2019) | ||||||||||
| Mic. phragmitis | CBS 285.71 | ET | Poland | Puccinia teleutosorus | KP859013 | KP858949 | KP859122 | – | – | Liu et al. (2019) |
| Mic. seminicola | CBS 139951 | T | Switzerland | Maize kernels | NR 155375 | KP858974 | KP859147 | – | – | Hernandez-Restrepo et al. (2016), |
| Liu et al. (2019) | ||||||||||
| Monochaetia camelliae | PSH2000I-151 | China | Camellia hongkongensis | AY682948 | – | – | – | – | Liu et al. (2019) | |
| PSH2000I-146 | China | Camellia pitardii | AY682947 | – | – | – | – | Liu et al. (2019) | ||
| Μ. castaneae | CFCC 54354 = SM9-1 | T | China | Castanea mollissima | MW166222 | – | – | – | – | Jiang et al. (2021) |
| SM9-2 | China | Castanea mollissima | MW166223 | – | – | – | – | Jiang et al. (2021) | ||
| Μ. dimorphospora | NBRC 9980 | Japan | Castanea pubinervis | LC146750 | – | – | – | – | De Silva et al. (2018) | |
| NNIBRFG396 | Korea | Fresh water | MT271967 | – | – | – | – | De Silva et al. (2018) | ||
| Μ. ilicis | CBS 101009 | Japan | Air | MH553953 | MH554176 | MH554849 | – | – | Liu et al. (2019) | |
| KUMCC 15-0520 | T | China | llex sp. | KX984153 | – | – | – | – | Liu et al. (2019) | |
| Μ. junipericola | CBS:143391 | Germany | Juniperus communis | MH107900 | – | – | – | – | Liu et al. (2019) | |
| Μ. kansensis | ZJLQ468 | China | Pseudotaxus chienii | KC345692 | – | – | – | – | De Silva et al. (2018) | |
| PSHI2004Endo1030 | China | Cyclobalaopsis sp. | DQ534044 | – | – | – | – | De Silva et al. (2018) | ||
| PSHI2004Endo1031 | China | Quercus aliena | DQ534045 | – | – | – | – | De Silva et al. (2018) | ||
| Μ. mochaeta | CBS 315.54 | England | Quercus sp. | MH554030 | MH554249 | MH554943 | – | – | Liu et al. (2019) | |
| CBS 658.95 | Netherlands | Quercus robur | MH554063 | MH554276 | MH554977 | – | – | Liu et al. (2019) | ||
| CBS 115004 | Netherlands | Quercus robur | AY853243 | MH554198 | MH554879 | – | – | Liu et al. (2019) | ||
| CBS 546.80 | Netherlands | Culture contaminant | MH554056 | MH554270 | MH554969 | – | – | Liu et al. (2019) | ||
| CBS 199.82 | ET | Italy | Quercus pubescens | MH554018 | MH554238 | MH554931 | – | – | Liu et al. (2019) | |
| M18 | Italy | Unknown plant | JX262802 | – | – | – | – | Liu et al. (2019) | ||
| Μ. quercus | CBS 144034 = CPC 29514 | T | Mexico | Quercus eduardi | MH554171 | MH554365 | MH555068 | – | – | De Silva et al. (2018) |
| CBS 144034 = CPC 29515 | Mexico | Quercus eduardi | NR_161110 | – | – | – | – | De Silva et al. (2018) | ||
| Μ. schimae | SAUCC212201 | T | China | Schima superba | MZ577565 | – | – | OK104874 | OK104867 | Zhang et al. (2022) |
| SAUCC212202 | China | Schima superba | MZ577566 | – | – | OK104875 | OK104868 | Zhang et al. (2022) | ||
| SAUCC212203 | China | Schima superba | MZ577567 | – | – | OK104876 | OK104869 | Zhang et al. (2022) | ||
| Μ. sinensis | HKAS 10065 | China | Quercus sp. | NR_161064 | – | – | – | – | De Silva et al. (2019) | |
| KUMCC 15-0517 | China | Quercus sp. | MH 115996 | – | – | – | – | De Silva et al. (2020) | ||
| Morinia acaciae | CBS 100230 | New Zealand | Prunus salicina cv. ‘Omega’ | MH553950 | MH554174 | MH554847 | – | – | Liu et al. (2019) | |
| CBS 137994 | T | France | Acacia melanoxylon | MH554002 | MH554221 | MH554914 | – | – | Liu et al. (2019) | |
| Mor. crini | CBS 143888 | T | South Africa | Crinum bulbispermum | MH554118 | MH554323 | MH555026 | – | – | Liu et al. (2019) |
| Mor. longiappendiculata | CBS 117603 | T | Spain | Calluna vulgaris | AY929324 | MH554202 | MH554885 | – | – | Collado et al. (2006), |
| Liu et al. (2019) | ||||||||||
| Mor. pestalozzioides | F090354 | ET | Spain | Sedum sediforme | AY929325 | – | – | – | – | Liu et al. (2019) |
| Neopestalotiopsis acrostichi | MFLUCC 17-1754 | T | Thailand | Acrostichum aureum | MK764272 | – | – | MK764316 | MK764338 | Norphanphoun et al. (2019) |
| MFLUCC 17-1755 | Thailand | Acrostichum aureum | MK764273 | – | – | MK764317 | MK764339 | Norphanphoun et al. (2019) | ||
| N. alpapicalis | MFLUCC 17-2544 | T | Thailand | Rhizophora mucronata | MK357772 | – | – | MK463547 | MK463545 | Kumar et al. (2019) |
| MFLUCC 17-2545 | Thailand | Rhizophora apiculata | MK357773 | – | – | MK463548 | MK463546 | Kumar et al. (2019) | ||
| N. aotearoa | CBS 367.54 | T | New Zealand | Canvas | KM 199369 | – | – | KM 199526 | KM 199454 | Maharachchikumbura et al. (2014) |
| N. asiatica | MFLUCC 12-0286 | T | China | Leaves | JX398983 | – | – | JX399049 | JX399018 | Maharachchikumbura et al. (2014) |
| N. australis | CBS 114159 | T | Australia | Telopea sp. | KM 199348 | – | – | KM 199537 | KM 199432 | Maharachchikumbura et al. (2014) |
| N. brachiata | MFLUCC 17-1555 | T | Thailand | Rhizophora apiculata | MK764274 | – | – | MK764318 | MK764340 | Norphanphoun et al. (2019) |
| N. brasiliensis | COAD 2166 | T | Brazil | Psidium guajava | MG686469 | – | – | MG692402 | MG692400 | Bezerra et al. (2018) |
| N. camelliae-oleiferae | CSUFTCC81 | T | China | Camellia oleifera | OK493585 | – | – | OK507955 | OK562360 | Li etal. (2021) |
| N. cavernicola | KUMCC 20-0269 | T | China | Cave rock surface | MW545802 | – | – | MW550735 | MW557596 | Liu et al. (2021) |
| N. chiangmaiensis | MFLUCC 18-0113 | Thailand | Dead leaves | – | – | – | MH388404 | MH412725 | Tibpromma et al. (2018) | |
| N. chrysea | MFLUCC 12-0261 | T | China | Pandanus sp. | JX398985 | – | – | JX399051 | JX399020 | Maharachchikumbura et al. (2014) |
| N. clavispora | MFLUCC 12-0281 | T | China | Magnolia sp. | JX398979 | – | – | JX399045 | JX399014 | Maharachchikumbura et al. (2014) |
| N. cocoes | MFLUCC 15-0152 | Thailand | Cocos nucifera | KX789687 | – | – | KX789689 | – | Hyde et al. (2016) | |
| N. coffeae-arabicae | HGUP4019 | T | China | Coffea arabica | KF412649 | – | – | KF412646 | KF412643 | Song et al. (2013) |
| N. cubana | CBS 600.96 | T | Cuba | Leaf litter | KM 199347 | KM116253 | MH554973 | KM 199521 | KM 199438 | Maharachchikumbura et al. (2014) |
| N. dendrobii | MFLUCC 14-0106 | Thailand | Dendrobium cariniferum | MK993571 | – | – | MK975829 | MK975835 | Ma et al. (2019) | |
| N. drenthii | BRIP 72264a | T | Australia | Macadamia integrifolia | MZ303787 | – | – | MZ344172 | MZ312680 | Prasannath et al. (2021) |
| N. egyptiaca | CBS 140162 | T | Egypt | Mangifera indica | KP943747 | – | – | KP943748 | KP943746 | Crous et al. (2015b) |
| N. ellipsospora | CBS 115113 | T | China | Dead plant material | KM 199343 | – | – | KM 199544 | KM 199450 | Maharachchikumbura et al. (2014) |
| MFLUCC 12-0283 | China | Dead plant material | JX398980 | – | – | JX399047 | JX399016 | Maharachchikumbura et al. (2014) | ||
| N. eucalypticola | CBS 264.37 | T | – | Eucalyptus globulus | KM 199376 | KM116256 | MH554935 | KM199551 | KM199431 | Maharachchikumbura et al. (2014) |
| N. eucalyptorum | CBS 147684 | T | Portugal | Eucalyptus globulus | MW794108 | – | – | MW805397 | MW802841 | Diogo et al. (2021) |
| N. foedans | CGMCC 3.9123 | T | China | Mangrove plant | JX398987 | – | – | JX399053 | JX399022 | Maharachchikumbura et al. (2014) |
| N. formicarum | CBS 362.72 | T | Cuba | Plant debris | MH860500 | – | – | KM199517 | KM 199455 | Maharachchikumbura et al. (2014) |
| N. guajavae | FMBCC 11.1 | T | Pakistan | Psidium guajava | MF783085 | – | – | MH460868 | MH460871 | Ul Haq et al. (2021) |
| N. guajavicola | FMBCC 11.4 | T | Pakistan | Psidium guajava | MH209245 | – | – | MH460870 | MH460873 | Ul Haq et al. (2021) |
| N. hadrolaeliae | COAD 2637 | T | Brazil | Hadrolaelia jongheana | MK454709 | – | – | MK465122 | MK465120 | Freitas et al. (2019) |
| N. haikouensis | SAUCC212271 | T | China | Ilex chinensis | OK087294 | – | – | OK104877 | OK104870 | Zhang et al. (2022) |
| SAUCC212272 | China | Ilex chinensis | OK087295 | – | – | OK104878 | OK104871 | Zhang et al. (2022) | ||
| N. hispanica | CBS 147686 | T | Portugal | Eucalyptus globulus | MW794107 | – | – | MW805399 | MW802840 | Diogo et al. (2021) |
| N. honoluluana | CBS 114495 | T | USA | Telopea sp. | KM 199364 | – | – | KM 199548 | KM199461 | Maharachchikumbura et al. (2014) |
| N. hydeana | MFLUCC 20-0132 | T | Thailand | Artocarpus heterophyllus | MW266069 | – | – | MW251129 | MW251119 | Huanluek et al. (2021) |
| N. iberica | CBS 147688 | T | Portugal | Eucalyptus globulus | MW794111 | – | – | MW805402 | MW802844 | Diogo et al. (2021) |
| N. iraniensis | CBS 137768 | Iran | Fragaria x ananassa | KM074045 | – | – | KM074051 | KM074056 | Norphanphoun et al. (2019) | |
| N. javaensis | CBS 257.31 | T | Indonesia | Cocos nucifera | KM 199357 | – | – | KM 199543 | KM 199437 | Maharachchikumbura et al. (2014) |
| N. longiappendiculata | CBS 147690 | T | Portugal | Eucalyptus globulus | MW794112 | – | – | MW805404 | MW802845 | Diogo et al. (2021) |
| N. lusitanica | CBS 147692 | T | Portugal | Eucalyptus globulus | MW794110 | – | – | MW805406 | MW802843 | Diogo et al. (2021) |
| N. macadamiae | BRIP 63737c | T | New South Wales | Macadamia integrifolia | KX186604 | – | – | KX186627 | KX186654 | Akinsanmi et al. (2017) |
| N. maddoxii | BRIP 72266a | T | Australia | Macadamia integrifolia | MZ303782 | – | – | MZ344167 | MZ312675 | Prasannath et al. (2021) |
| N. magna | MFLUCC 12-0652 | T | France | Pteridium sp. | KF582795 | – | – | KF582791 | KF582793 | Maharachchikumbura et al. (2014) |
| N. mesopotamica | CBS 336.86 | T | Iraq | Pinus brutia | KM 199362 | KM116271 | MH554944 | KM 199555 | KM199441 | Maharachchikumbura et al. (2014) |
| N. musae | MFLU 16-1279 | T | Thailand | Musa sp. | KX789683 | – | – | KX789685 | KX789686 | Hyde et al. (2016) |
| N. natalensis | CBS 138.41 | T | South Africa | Acacia mollissima | KM199377 | – | – | KM 199552 | KM 199466 | Maharachchikumbura et al. (2014) |
| N. nebuloides | BRIP 66617 | T | Australia | Sporobolus elongatus | MK966338 | – | – | MK977633 | MK977632 | Crous et al. (2020) |
| N. olumideae | BRIP 72273a | T | Australia | Macadamia integrifolia | MZ303790 | – | – | MZ344175 | MZ312683 | Prasannath et al. (2021) |
| N. palmarum | PSHI2004Endo458 | Cuba | Zalacca wallichiana | DQ813426 | – | – | – | DQ787836 | Liu et al. (2010) | |
| PSHI2004Endo454 | Cuba | Roystonea regia | DQ813427 | – | – | – | DQ787837 | Liu et al. (2010) | ||
| N. pandanicola | KUMCC 17-0175 | – | Pandanus sp. | – | – | – | MH388389 | MH412720 | Tibpromma et al. (2018) | |
| N. pemambucana | URM 7148-01 | T | Brazil | Vismia guianensis | KJ792466 | – | – | KU306739 | – | Silvério et al. (2016) |
| N. perukae | FMBCC 11.3 | T | Pakistan | Psidium guajava | MH209077 | – | – | MH523647 | MH460876 | Ul Haq et al. (2021) |
| N. petila | MFLUCC 17-1738 | T | Thailand | Rhizophora mucronata | MK764275 | – | – | MK764319 | MK764341 | Norphanphoun et al. (2019) |
| MFLUCC 17-1737 | Thailand | Rhizophora mucronata | MK764276 | – | – | MK764320 | MK764342 | Norphanphoun et al. (2019) | ||
| N. phangngaensis | MFLUCC 18-0119 | – | Pandanus sp. | MH388354 | – | – | MH388390 | MH412721 | Tibpromma et al. (2018) | |
| N. piceana | CBS 394.48 | T | UK | Picea sp. | KM 199368 | – | – | KM199527 | KM 199453 | Maharachchikumbura et al. (2014) |
| N. protearum | CBS 114178 | T | Zimbabwe | Leucospermum cuneiforme | JN712498 | JN712564 | MH554873 | LT853201 | KM 199463 | Maharachchikumbura et al. (2014) |
| N. psidii | FMBCC 11.2 | T | Pakistan | Psidium guajava | MF783082 | – | – | MH460874 | MH477870 | Ul Haq et al. (2021) |
| N. rhapidis | GUCC 21501 | T | China | Rhododendron simsii | MW931620 | – | – | MW980442 | MW980441 | Yang et al. (2021) |
| N. rhizophorae | MFLUCC 17-1551 | T | Thailand | Rhizophora mucronata | MK764277 | – | – | MK764321 | MK764343 | Norphanphoun et al. (2019) |
| MFLUCC 17-1550 | Thailand | Rhizophora mucronata | MK764278 | – | – | MK764322 | MK764344 | Norphanphoun et al. (2019) | ||
| N. rhododendri | GUCC 21504 | T | China | Rhododendron simsii | MW979577 | – | – | MW980444 | MW980443 | Yang et al. (2021) |
| N. rosae | CBS 101057 | T | New Zealand | Rosa sp. | KM 199359 | KM 116245 | MH554850 | KM 199524 | KM 199430 | Maharachchikumbura et al. (2014) |
| CBS 124745 | USA | Paeonia suffruticosa | KM 199360 | – | – | KM 199524 | KM 199430 | Maharachchikumbura et al. (2014) | ||
| CRM-FRC | Mexico | Fragaria x ananassa | MN385718 | – | – | MN268532 | MN268529 | Rebollar-Alviter et al. (2020) | ||
| AC50 | Italy | Persea americana | ON117810 | – | – | ON107276 | ON209165 | Alberto et al. (2022) | ||
| N. rosicola | CFCC 51992 | T | China | Rosa chinensis | KY885239 | – | – | KY885243 | KY885245 | Jiang et al. (2018) |
| CFCC 51993 | China | Rosa chinensis | KY885240 | – | – | KY885244 | KY885246 | Jiang et al. (2018) | ||
| N. samaragenensis | MFLUCC 12-0233 | T | Thailand | Syzygium samarangense | KM 199365 | – | – | KM 199556 | KM 199447 | Norphanphoun et al. (2019) |
| N. saprophytica | MFLUCC 12-0282 | T | China | Magnolia sp. | KY606286 | – | – | JX399048 | JX399017 | Maharachchikumbura et al. (2014) |
| N. scalabiensis | CAA1029 | T | Portugal | Vaccinium corymbosum | MW969748 | MW959100 | MW934611 | Santos et al. (2022) | ||
| N. sichuanensis | CFCC 54338 = SM15-1 | T | China | Castanea mollissima | MW166231 | – | – | MW199750 | MW218524 | Jiang et al. (2021) |
| SM15-1C | China | Castanea mollissima | MW166232 | – | – | MW199751 | MW218525 | Jiang et al. (2021) | ||
| N. siciliana | AC46 | Italy | Persea americana | ON117813 | – | – | ON107273 | ON209162 | Alberto et al. (2022) | |
| AC48 | Italy | Persea americana | ON117812 | – | – | ON107274 | ON209163 | Alberto et al. (2022) | ||
| AC49 | Italy | Persea americana | ON117811 | – | – | ON107275 | ON209164 | Alberto et al. (2022) | ||
| N. sonneratae | MFLUCC 17-1744 | T | Thailand | Sonneronata alba | MK764279 | – | – | MK764323 | MK764345 | Norphanphoun et al. (2019) |
| MFLUCC 17-1745 | Thailand | Sonneronata alba | MK764280 | – | – | MK764324 | MK764346 | Norphanphoun et al. (2019) | ||
| Neopestalotiopsis sp.1 | VRes4 | Colombia | Scab disease of Guava | KR493566 | – | – | KR493638 | – | Kumar et al. (2019) | |
| Neopestalotiopsis sp.2 | BPca2 | Colombia | Scab disease of Guava | KR493559 | – | – | KR493627 | KR493666 | Kumar et al. (2019) | |
| Neopestalotiopsis sp.3 | VrleP | Colombia | Scab disease of Guava | KR493520 | – | – | KR493660 | KR493719 | Kumar et al. (2019) | |
| Neopestalotiopsis sp.4 | VTman4 | Colombia | Scab disease of Guava | KR493554 | – | – | KR493611 | KR493724 | Kumar et al. (2019) | |
| Neopestalotiopsis sp.5 | BVayr1 | Colombia | Scab disease of Guava | KR493545 | – | – | KR493629 | KR493739 | Kumar et al. (2019) | |
| Neopestalotiopsis sp.6 | BRIP 63740a | Australia | Dry flower | KX186617 | – | – | KX186628 | KX186656 | Kumar et al. (2019) | |
| Neopestalotiopsis sp.7 | BRIP 63745a | Australia | Dry flower | KX186614 | – | – | KX186633 | KX186660 | Kumar et al. (2019) | |
| Neopestalotiopsis sp.8 | CBS 119.75 | India | Achras sapota | KM 199356 | – | – | KM199531 | KM 199439 | Kumar et al. (2019) | |
| Neopestalotiopsis sp.9 | CM M1363 | Brazil | Opuntia stricta | KY549599 | – | – | KY549596 | KY549634 | Kumar et al. (2019) | |
| Neopestalotiopsis sp.10 | LC6489 | China | Camellia sp. | KX895020 | – | – | KX895239 | KX895353 | Kumar et al. (2019) | |
| Neopestalotiopsis sp.11 | MFLUCC 12-0614 | Midi-pyrénées | Unidentified host | KX816919 | – | – | KX816889 | KX816947 | Kumar et al. (2019) | |
| Neopestalotiopsis sp.12 | MMf0011 | Japan | Lilium speciosum | LC184188 | – | – | LC184190 | LC184189 | Kumar et al. (2019) | |
| Neopestalotiopsis sp.13 | SC2A3 | China | Tea plant | KU252210 | – | – | KU252390 | KU252477 | Kumar et al. (2019) | |
| Neopestalotiopsis sp.14 | CBS 164.42 | France | Sand dune | KM 199367 | – | – | KM 199520 | KM 199434 | Kumar et al. (2019) | |
| Neopestalotiopsis sp.15 | CBS 266.80 | India | Vitis vinifera | KM 199352 | – | – | KM 199532 | – | Kumar et al. (2019) | |
| Neopestalotiopsis sp.16 | CBS 360.61 | Guinea | Cinchona sp. | KM 199346 | – | – | KM 199522 | KM 199440 | Kumar et al. (2019) | |
| Neopestalotiopsis sp.17 | CBS 361.61 | Netherlands | Cissus sp. | KM 199355 | – | – | KM 199549 | KM 199460 | Kumar et al. (2019) | |
| N. steyaertii | IMI 192475 | Australia | Eucalyptus viminalis | KF582796 | – | – | KF582792 | KF582794 | Maharachchikumbura et al. (2014) | |
| N. surinamensis | CBS 450.74 | T | Suriname | Soil | KM 199351 | KM 116258 | MH554962 | KM199518 | KM 199465 | Maharachchikumbura et al. (2014) |
| N. thailandica | MFLUCC 17-1730 | T | Thailand | Rhizophora mucronata | MK764281 | – | – | MK764325 | MK764347 | Norphanphoun et al. (2019) |
| N. umbrinospora | MFLUCC 12-0285 | T | China | Dead leaves | JX398984 | – | – | JX399050 | JX399019 | Norphanphoun et al. (2019) |
| N. vaccinii | CAA1059 | T | Portugal | Vaccinium corymbosum | MW969747 | – | – | MW959099 | MW934610 | Santos et al. (2022) |
| N. vacciniicola | CAA1055 | T | Portugal | Vaccinium corymbosum | MW969751 | – | – | MW959103 | MW934614 | Santos et al. (2022) |
| N. versicolor | CBS 303.49 | China | Myrica rubra | MH856535 | – | – | – | – | Vu et al. (2019) | |
| N. vheenae | BRIP 72293a | T | Australia | Macadamia integrifolia | MZ303792 | – | – | MZ344177 | MZ312685 | Prasannath et al. (2021) |
| N. vitis | MFLUCC 15-1265 | China | Vitis vinifera | KU140694 | – | – | KU140676 | KU140685 | Jayawardena et al. (2016) | |
| N. zakeelii | BRIP 72282a | T | Australia | Macadamia integrifolia | MZ303789 | – | – | MZ344174 | MZ312682 | Prasannath et al. (2021) |
| N. zimbabwana | CBS H-21769 | Zimbabwe | Leucospermum cunciforme | – | JX556249 | MH554855 | KM 199545 | KM 199456 | Maharachchikumbura et al. (2014) | |
| Nonappendiculata quercina | CBS 116061 | T | Italy | Quercus suber | MH553982 | MH554199 | MH554882 | – | – | Liu et al. (2019) |
| CBS 270.82 | Italy | Quercus pubescens | MH554025 | MH554246 | MH554937 | – | – | Liu et al. (2019) | ||
| Parabartalinia lateralis | CBS 399.71 | T | South Africa | Acacia karroo | MH554043 | MH554256 | MH554954 | – | – | Liu et al. (2019) |
| Pestalotiopsis abietis | CFCC 53011 | T | China | Abies fargesii | MK397013 | – | – | MK622277 | MK622280 | Gu et al. (2021) |
| CFCC 53012 | China | Abies fargesii | MK397014 | – | – | MK622278 | MK622281 | Gu et al. (2021) | ||
| CFCC 53013 | China | Abies fargesii | MK397015 | – | – | MK622279 | MK622282 | Gu et al. (2021) | ||
| Pes. adusta | ICMP 6088 | T | Fiji | Refrigerator door PVC gasket | JX399006 | – | – | JX399070 | JX399037 | Norphanphoun et al. (2019) |
| CBS 263.33 | Netherlands | Rhododendron ponticum | KM199316 | – | – | KM 199489 | KM199414 | Norphanphoun et al. (2019) | ||
| Pes. aggestorum | LC6301 | T | China | Camellia sinensis | KX895015 | – | – | KX895234 | KX895348 | Liu et al. (2017) |
| Pes. anacardiacearum | IFRDCC 2397 | T | China | Mangifera indica | KC247154 | – | – | KC247156 | KC247155 | Maharachchikumbura et al. (2013b) |
| Pes. arceuthobii | CBS 433.65 | USA | Arceuthobium campylopodum | MH554046 | – | – | MH554481 | MH554722 | Maharachchikumbura et al. (2014) | |
| CBS 434.65 | T | USA | Arceuthobium campylopodum | KM 199341 | – | – | KM199516 | KM199427 | Maharachchikumbura et al. (2014) | |
| Pes. arengae | CBS 331.92 | T | Singapore | Arenga undulatifolia | KM 199340 | – | – | KM199515 | KM 199426 | Maharachchikumbura et al. (2014) |
| Pes. australasiae | CBS 114126 | T | New Zealand | Knightia sp. | KM 199297 | KM116218 | MH554867 | KM 199499 | KM 199409 | Maharachchikumbura et al. (2014) |
| CBS 114141 | Australia | Protea cv. ’Pink Ice’ | KM 199298 | – | – | KM199501 | KM199410 | Maharachchikumbura et al. (2014) | ||
| Pes. australis | CBS 114193 | T | Australia | Grevillea sp. | KM 199332 | KM116197 | MH554875 | KM 199475 | KM 199383 | Maharachchikumbura et al. (2014) |
| CBS 119350 | South Africa | Brabejum stellatifolium | KM 199333 | – | – | KM 199476 | KM 199384 | Maharachchikumbura et al. (2014) | ||
| MEAN 1096 = CPC 36750 = | Portugal | Pinus pinea | MT374679 | – | – | MT374692 | MT374704 | Silva et al. (2020) | ||
| CBS 146843 | ||||||||||
| MEAN 1109 | Portugal | Pinus pinea | MT374683 | – | – | – | MT374708 | Silva et al. (2020) | ||
| MEAN 1110 | Portugal | Pinus pinea | MT374684 | – | – | MT374696 | MT374709 | Silva et al. (2020) | ||
| MEAN 1111 | Portugal | Pinus pinea | MT374685 | – | – | MT374697 | MT374710 | Silva et al. (2020) | ||
| MEAN 1112 | Portugal | Pinus pinea | MT374686 | – | – | MT374698 | MT374711 | Silva et al. (2020) | ||
| Pes. biciliata | CBS 124463 | T | Slovakia | Platanus x hispanica | KM 199308 | – | – | KM 199505 | KM 199399 | Maharachchikumbura et al. (2014) |
| CBS 236.38 | Italy | Paeonia sp. | KM 199309 | – | – | KM 199506 | KM199401 | Maharachchikumbura et al. (2014) | ||
| MEAN 1168 | Portugal | Pinus pinea | MT374690 | – | – | MT374702 | MT374715 | Maharachchikumbura et al. (2014) | ||
| Pes. brachiata | LC2988 | T | China | Camellia sp. | KX894933 | – | – | KX895150 | KX895265 | Maharachchikumbura et al. (2014) |
| Pes. brassicae | CBS 170.26 | T | New Zealand | Brassica napus | KM 199379 | – | – | KM 199558 | – | Maharachchikumbura et al. (2014) |
| Pes. camelliae | CBS 443.62 | Turkey | Camellia sinensis | KM 199336 | – | – | KM199512 | KM 199424 | Zhang et al. (2012b) | |
| MFLUCC 12-0277 | T | China | Camellia japonica | JX399010 | – | – | JX399074 | JX399041 | Zhang et al. (2012b) | |
| Pes. camelliae-oleiferae | CSUFTCC08 | T | China | Camellia oleifera | OK493593 | OK507963 | OK562368 | Li et al. (2021) | ||
| CSUFTCC09 | China | Camellia oleifera | OK493594 | OK507964 | OK562369 | Li et al. (2021) | ||||
| CSUFTCC10 | China | Camellia oleifera | OK493595 | OK507965 | OK562370 | Li et al. (2021) | ||||
| Pes. chamaeropis | CBS 113607 | – | – | KM 199325 | – | – | KM 199472 | KM 199390 | Maharachchikumbura et al. (2014) | |
| CBS 186.71 | T | Italy | Chamaerops humilis | KM 199326 | – | – | KM 199473 | KM199391 | Maharachchikumbura et al. (2014) | |
| Pes. chiaroscuro | BRIP 72970 | T | Australia | Sporobolus natalensis | OK422510 | – | – | – | – | Crous et al. (2022) |
| Pes. clavata | MFLUCC 12-0268 | T | China | Buxus sp. | JX398990 | – | – | JX399056 | JX399025 | Maharachchikumbura et al. (2012) |
| Pes. colombiensis | CBS 118553 | T | Colombia | Eucalyptus eurograndis | KM 199307 | – | – | KM 199488 | KM 199421 | Maharachchikumbura et al. (2014) |
| Pes. digitalis | ICMP 5434 | T | New Zealand | Digitalis purpurea | KP781879 | – | – | – | KP781883 | Liu et al. (2015) |
| Pes. diploclisiae | CBS 115587 | T | Hong Kong, China | Diploclisia glaucescens | KM 199320 | – | – | KM 199486 | KM199419 | Maharachchikumbura et al. (2014) |
| Pes. disseminata | CBS 118552 | New Zealand | Eucalyptus botryoides | MH553986 | – | – | MH554410 | MH554652 | Maharachchikumbura et al. (2014) | |
| CBS 143904 | New Zealand | Persea americana | MH554152 | – | – | MH554587 | MH554825 | Maharachchikumbura et al. (2014) | ||
| MEAN 1165 | Portugal | Pinus pinea, blighted shoot | MT374687 | – | – | MT374699 | MT374712 | Silva et al. (2020) | ||
| MEAN 1166 | Portugal | Pinus pinea, blighted shoot | MT374688 | – | – | MT374700 | MT374713 | Silva et al. (2020) | ||
| Pes. distincta | LC3232 | T | China | Camellia sinensis | KX894961 | – | – | KX895178 | KX895293 | Silva et al. (2020) |
| LC8184 | China | Camellia sinensis | KY464138 | – | – | KY464148 | KY464158 | Silva et al. (2020) | ||
| Pes. diversiseta | MFLUCC 12-0287 | T | China | Rhododendron sp. | JX399009 | – | – | JX399073 | JX399040 | Maharachchikumbura et al. (2012) |
| Pes. doitungensis | MFLUCC 14-0115 | T | Thailand | Dendrobium sp. | MK993574 | – | – | MK975832 | MK975837 | Ma et al. (2019) |
| Pes. dracaenicola | MFLUCC 18-0913 | T | Thailand | Dracaena sp. | MN962731 | – | – | MN962732 | MN962733 | Chaiwan et al. (2020) |
| Pes. dracontomelonis | MFLUCC 10-0149 | T | Thailand | Dracontomelon dao | KP781877 | – | – | KP781880 | – | Liu et al. (2015) |
| Pes. ericacearum | IFRDCC 2439 | T | China | Rhododendron delavayi | KC537807 | – | – | KC537814 | KC537821 | Zhang et al. (2013b) |
| Pes. etonensis | BRIP 66615 | T | Australia | Sporobolus jacquemontii | MK966339 | – | – | MK977635 | MK977634 | Crous et al. (2020) |
| Pes. formosana | NTUCC 17-009 | T | Taiwan,China | On dead grass | MH809381 | – | – | MH809389 | MH809385 | Ariyawansa & Hyde (2018) |
| Pes. furcata | MFLUCC 12-0054 | T | Thailand | Camellia sinensis | JQ683724 | – | – | JQ683740 | JQ683708 | Maharachchikumbura et al. (2013a) |
| Pes. gaultheriae | IFRD 411-014 | T | China | Gaultheria forrestii | KC537805 | – | – | KC537812 | KC537819 | Zhang et al. (2013) |
| Pes. gibbosa | NOF3175 | T | Canada | Gaultheria shallon | LC311589 | – | – | LC311591 | LC311590 | Watanabe et al. (2018) |
| Pes. grevilleae | CBS 114127 | T | Australia | Grevillea sp. | KM 199300 | KM116212 | MH554868 | KM 199504 | KM 199407 | Maharachchikumbura et al. (2014) |
| Pes. hawaiiensis | CBS 114491 | T | USA | Leucospermum cv. ’Coral’ | KM 199339 | – | – | KM199514 | KM 199428 | Maharachchikumbura et al. (2014) |
| Pes. hispanica | CBS 115.391 | T | Spain | Protea cv. ’Susara’ | MH553981 | – | – | MH554399 | MH554640 | Liu etal. (2019) |
| Pes. hollandica | CBS 265.33 | T | Netherlands | Sciadopitys verticillata | KM 199328 | KM 116228 | MH554936 | KM199481 | KM 199388 | Maharachchikumbura et al. (2014) |
| MEAN 1091 = CPC 36745 = | Portugal | Pinus pinea, blighted shoot | MT374678 | – | – | MT374691 | MT374703 | Maharachchikumbura et al. (2014) | ||
| CBS 146839 | ||||||||||
| Pes. humicola | CBS 115450 | T | Hong Kong, China | Ilex cinerea | KM199319 | KM 116208 | MH554881 | KM 199487 | KM199418 | Maharachchikumbura et al. (2014) |
| CBS 336.97 | Papua New Guinea | Soil | KM 199317 | – | – | KM 199484 | KM 199420 | Maharachchikumbura et al. (2014) | ||
| Pes. hunanensis | CSUFTCC15 | T | China | Camellia oleifera | OK493599 | – | – | OK507969 | OK562374 | Li et al. (2021) |
| CSUFTCC18 | China | Camellia oleifera | OK493600 | – | – | OK507970 | OK562375 | Li et al. (2021) | ||
| CSUFTCC19 | China | Camellia oleifera | OK493601 | – | – | OK507971 | OK562376 | Li et al. (2021) | ||
| Pes. hydei | MFLUCC 20-0135 | T | Thailand | Litsea petiolata | NR_172003 | – | – | MW251113 | MW251112 | Huanluek et al. (2021) |
| Pes. inflexa | MFLUCC 12-0270 | T | China | Unidentified tree | JX399008 | – | – | JX399072 | JX399039 | Maharachchikumbura et al. (2012) |
| Pes. intermedia | MFLUCC 12-0259 | T | China | Unidentified tree | JX398993 | – | – | JX399059 | JX399028 | Maharachchikumbura et al. (2012) |
| Pes. italiana | MFLUCC 12-0657 | T | Italy | Cupressus glabra | KP781878 | – | – | KP781881 | KP781882 | Liu et al. (2015) |
| Pes. jesteri | CBS 109350 | T | Papua New Guinea | Fragraea bodenii | KM 199380 | – | – | KM 199554 | KM 199468 | Strobel et al. (2000) |
| Pes. jiangxiensis | LC4399 | T | China | Camellia sp. | KX895009 | – | – | KX895227 | KX895341 | Liu et al. (2017) |
| Pes. jinchanghens | LC6636 | T | China | Camellia sinensis | KX895028 | – | – | KX895247 | KX895361 | Liu et al. (2017) |
| Pes. kandelicola | NCYUCC 19-0355 | Taiwan,China | Kandelia candel | MT560722 | – | – | MT563101 | MT563099 | Hyde et al. (2020) | |
| NCYUCC 19-0354 | Taiwan,China | Kandelia candel | MT560723 | – | – | MT563102 | MT563100 | Hyde et al. (2020) | ||
| Pes. kenyana | CBS 442.67 | T | Kenya | Coffea sp. | KM 199302 | KM 116234 | MH554958 | KM 199502 | KM 199395 | Maharachchikumbura et al. (2014) |
| Pes. knightiae | CBS 114138 | T | New Zealand | Knightia sp. | KM199310 | KM116227 | MH554870 | KM 199497 | KM 199408 | Maharachchikumbura et al. (2014) |
| Pes. leucadendri | CBS 121417 | T | South Africa | Leucadendron sp. | MH553987 | – | – | MH554412 | MH554654 | Liu et al. (2019) |
| Pes. licualicola | HGUP 4057 | T | China | Licuala grandis | KC492509 | – | – | KC481684 | KC481683 | Geng et al. (2013) |
| Pes. linearis | MFLUCC 12-0271 | T | China | Trachelospermum sp. | JX398992 | – | – | JX399058 | JX399027 | Maharachchikumbura et al. (2012) |
| Pes. longiappendiculata | LC3013 | T | China | Camellia sinensis | KX894939 | – | – | KX895156 | KX895271 | Liu et al. (2017) |
| Pes. lushanensis | LC4344 | T | China | Camellia sp. | KX895005 | – | – | KX895223 | KX895337 | Liu et al. (2017) |
| Pes. macadamiae | BRIP 63738b | T | Australia | Macadamia integrifolia | KX186588 | – | – | KX186621 | KX186680 | Akinsanmi et al. (2017) |
| Pes. malayana | CBS 102220 | T | Malaysia | Macaranga triloba | KM 199306 | – | – | KM 199482 | KM199411 | Maharachchikumbura et al. (2014) |
| Pes. monochaeta | CBS 144.97 | T | Netherlands | Quercus robur | KM 199327 | – | – | KM 199479 | KM 199386 | Maharachchikumbura et al. (2014) |
| Pes. nanjingensis | CSUFTCC16 | T | China | Camellia oleifera | OK493602 | – | – | OK507972 | OK562377 | Li et al. (2021) |
| CSUFTCC20 | China | Camellia oleifera | OK493603 | – | – | OK507973 | OK562378 | Li et al. (2021) | ||
| CSUFTCC04 | China | Camellia oleifera | OK493604 | – | – | OK507974 | OK562379 | Li et al. (2021) | ||
| Pes. nanningensis | CSUFTCC10 | T | China | Camellia oleifera | OK562371 | – | – | OK507966 | OK562371 | Li et al. (2021) |
| CSUFTCC11 | China | Camellia oleifera | OK562372 | – | – | OK507967 | OK562372 | Li et al. (2021) | ||
| CSUFTCC12 | China | Camellia oleifera | OK562373 | – | – | OK507968 | OK562373 | Li et al. (2021) | ||
| Pes. neolitseae | NTUCC 17-011 | T | Taiwan,China | Neolitsea villosa | MH809383 | – | – | MH809391 | MH809387 | Ariyawansa & Hyde (2018) |
| Pes. novae-hollandiae | CBS 130973 | T | Australia | Banksia grandis | KM 199337 | – | – | KM199511 | KM 199425 | Maharachchikumbura et al. (2014) |
| Pes. oryzae | CBS 353.69 | T | Denmark | Oryza sativa | KM 199299 | KM116221 | MH554947 | KM 199496 | KM 199398 | Maharachchikumbura et al. (2014) |
| Pes. pallidotheae | MAFF 240993 | T | Japan | Pieris japonica | NR111022 | – | – | LC311585 | LC311584 | Watanabe et al. (2010) |
| Pes. pandanicola | MFLUCC 16-0255 | T | Thailand | Pandanus sp. | MH388361 | – | – | MH388396 | MH412723 | Li etal. (2021) |
| Pes. papuana | CBS 331.96 | T | Papua New Guinea | Soil | KM 199321 | – | – | KM199491 | KM199413 | Maharachchikumbura et al. (2014) |
| Pes. parva | CBS 114972 | Hong Kong, China | Leaf | MH553980 | – | – | MH554397 | MH704625 | Maharachchikumbura et al. (2014) | |
| CBS 278.35 | T | – | Leucothoe fontanesiana | KM199313 | KM 116205 | MH554939 | KM 199509 | KM 199405 | Maharachchikumbura et al. (2014) | |
| Pes. photinicola | GZCC 16-0028 | T | China | Photinia serrulata | KY092404 | – | – | KY047662 | KY047663 | Chen etal. (2017) |
| Pes. pini | MEAN 1092 = CPC 36746 = | Portugal | Pinus pinea | MT374680 | – | – | MT374693 | MT374705 | Silva et al. (2020) | |
| CBS 146840 | ||||||||||
| MEAN 1094 = CPC 36748 = | T | Portugal | Pinus pinea | MT374681 | – | – | MT374694 | MT374706 | Silva et al. (2020) | |
| CBS 146841 | ||||||||||
| MEAN 1095 = CPC 36749 = | Portugal | Pinus pinea | MT374682 | – | – | MT374695 | MT374707 | Silva et al. (2020) | ||
| CBS 146842 | ||||||||||
| MEAN 1167 | Portugal | Pinus pinaster | MT374689 | – | – | MT374701 | MT374714 | Silva et al. (2020) | ||
| Pes. pinicola | KUMCC 19-0183 | T | China | Pinus armandii | MN412636 | – | – | MN417509 | MN417507 | Tibpromma et al. (2019) |
| Pes. portugallica | CBS 684.85 | New Zealand | Camellia japonica | MH554065 | – | – | MH554501 | MH554741 | Maharachchikumbura et al. (2014) | |
| CBS 393.48 | T | Portugal | – | KM 199335 | KM 116233 | MH554951 | KM199510 | KM 199422 | Maharachchikumbura et al. (2014) | |
| Pes. rhizophorae | MFLUCC 17-0416 | T | Thailand | Rhizophora apiculata | MK764283 | – | – | MK764327 | MK764349 | Norphanphoun et al. (2019) |
| Pes. rhododendri | IFRDCC 2399 | T | China | Rhododendron sinogrande | KC537804 | – | – | KC537811 | KC537818 | Norphanphoun et al. (2019) |
| CBS 144024 | Zimbabwe | Pinus sp. | MH554109 | – | – | MH554543 | MH554782 | Norphanphoun et al. (2019) | ||
| Pes. rhodomyrtus | HGUP 4230 | T | China | Rhodomyrtus tomentosa | KF412648 | – | – | KF412645 | KF412642 | Norphanphoun et al. (2019) |
| LC3413 | China | Camellia sinensis | KX894981 | – | – | KX895198 | KX895313 | Norphanphoun et al. (2019) | ||
| Pes. rosea | MFLUCC 12-0258 | T | China | Pinus sp. | JX399005 | – | – | JX399069 | JX399036 | Maharachchikumbura et al. (2012) |
| Pes. scoparia | CBS 176.25 | T | – | Chamaecyparis sp. | KM 199330 | – | – | KM 199478 | KM 199393 | Maharachchikumbura et al. (2014) |
| Pes. sequoiae | MFLUCC 13-0399 | T | Italy | Sequoia sempervirens | KX572339 | – | – | – | – | Maharachchikumbura et al. (2014) |
| Pes. shorea | MFLUCC 12-0314 | T | Thailand | Shorea obtusa | KJ503811 | – | – | KJ503817 | KJ503814 | Song et al. (2013) |
| Pestalotiopsis 7 FL 2019 | CBS 110326 | USA | Pinus sp. | MH553957 | – | – | MH554375 | MH554616 | Silva et al. (2020) | |
| Pestalotiopsis 7 FL 2019 | CBS 127.80 | Chile | Pinus radiata | MH553995 | – | – | MH554422 | MH554664 | Silva et al. (2020) | |
| Pes. spathulata | CBS 356.86 | T | Chile | Guevina avellana | KM 199338 | – | – | KM199513 | KM 199423 | Maharachchikumbura et al. (2014) |
| Pes. spathuliappendiculata | CBS 144035 | T | Australia | Phoenix canariensis | MH554172 | MH554366 | – | MH554607 | MH554845 | Liu et al. (2019) |
| Pes. telopeae | CBS 114137 | Australia | Protea cv. ’Pink Ice’ | KM 199301 | – | – | KM 199559 | KM 199469 | Maharachchikumbura et al. (2014) | |
| CBS 114161 | T | Australia | Telopea sp. | KM 199296 | – | – | KM 199500 | KM 199403 | Maharachchikumbura et al. (2014) | |
| Pes. terricola | CBS 141.69 | T | Pacific Islands | Soil | MH554004 | – | – | MH554438 | MH554680 | Maharachchikumbura et al. (2014) |
| Pes. thailandica | MFLUCC 17-1616 | T | Thailand | Rhizophora apiculata | MK764285 | – | – | MK764329 | MK764351 | Maharachchikumbura et al. (2014) |
| Pes. trachycarpicola | IFRDCC 2440 | T | China | Trachycarpus fortunei | JQ845947 | – | – | JQ845946 | JQ845945 | Zhang et al. (2012a) |
| Pes. unicolor | MFLUCC 12-0275 | China | Unidentified tree | JX398998 | – | – | JX399063 | JX399029 | Maharachchikumbura et al. (2012) | |
| MFLUCC 12-0276 | T | China | Rhododendron sp. | JX398999 | – | – | – | JX399030 | Maharachchikumbura et al. (2012) | |
| Pes. verruculosa | MFLUCC 12-0274 | T | China | Rhododendron sp. | JX398996 | – | – | JX399061 | – | Maharachchikumbura et al. (2012) |
| Pes. cf. verruculosa | CBS 365.54 | Netherlands | Chamaecyparis lawsoniana | MH554037 | – | – | MH554472 | MH554713 | Maharachchikumbura et al. (2012) | |
| Pes. yanglingensis | LC3412 | China | Camellia sinensis | KX894980 | – | – | KX895197 | KX895312 | Liu et al. (2017) | |
| LC4553 | T | China | Camellia sinensis | KX895012 | – | – | KX895231 | KX895345 | Liu et al. (2017) | |
| Pes. yunnanensis | HMAS 96359 | T | China | Podocarpus macrophyllus | AY373375 | – | – | – | – | Wei et al. (2013) |
| Phlogicylindrium eucalypti | CBS 120080 | T | Australia | Eucalyptus globulus | NR_132813 | DQ923534 | MH554893 | – | – | Liu et al. (2019) |
| Phlogicylindrium eucalyptorum | CBS 120221 | T | Australia | Eucalyptus globus | EU040223 | MH554204 | MH554894 | – | – | Liu et al. (2019) |
| Phlogicylindrium uniforme | CBS 131312 | T | Australia | Eucalyptus globus | JQ044426 | JQ044445 | MH554910 | – | – | Crous et al. (2011a) |
| Pseudopestalotiopsis cocos | CBS 272.29 | T | Indonesia | Cocos nucifera | KM 199378 | KM 116276 | MH554938 | – | – | Maharachchikumbura et al. (2014) |
| Pse. elaeidis | CBS 413.62 | IT | Nigeria | Elaeis guineensis | MH554044 | MH554257 | MH554955 | – | – | Liu et al. (2019) |
| Pse. indica | CBS 459.78 | T | India | Rosa sinensis | KM 199381 | MH554263 | MH554963 | – | – | Maharachchikumbura et al. (2014) |
| Pseudosarcostroma osyridicola | CBS 103.76 | T | France | Osyris alba | MH553954 | MH554177 | MH554851 | – | – | Liu et al. (2019) |
| Robillarda africana | CBS 122.75 | T | South Africa | – | KR873253 | KR873281 | MH554896 | – | – | Crous et al. (2015a) |
| Rob. australiana | CBS 143882 | T | Australia | – | MH554091 | MH554301 | MH555005 | – | – | Liu et al. (2019) |
| Rob. roystoneae | CBS 115445 | T | Hong Kong, China | Roystonea regia | KR873254 | KR873282 | MH554880 | – | – | Crous et al. (2015a) |
| Rob. sessilis | CBS 114312 | ET | Germany | Dust | KR873256 | KR873284 | MH554877 | – | – | Liu et al. (2019) |
| Rob. terrae | CBS 587.71 | T | India | Soil | KJ710484 | KJ710459 | MH554971 | – | – | Crous et al. (2015a) |
| Sarcostroma australiense | CBS 144160 | T | Australia | Daviesia latifolia | MH554138 | MH554340 | MH555044 | – | – | Liu et al. (2019) |
| Sar. diversiseptatum | CBS 189.81 | T | Australia | Correa reflexa | MH554016 | MH554236 | MH554929 | – | – | Liu et al. (2019) |
| Sar. grevilleae | CBS 143418 | Australia | Grevillea sp. | MH554006 | MH554227 | MH554922 | – | – | Liu et al. (2019) | |
| Sar. leucospermi | CBS 111290 | T | South Africa | Leucospermum cv. ‘High Gold’ | MH554081 | MH554292 | MH554993 | – | – | Liu et al. (2019) |
| CBS 111309 | South Africa | Leucospermum cv. ‘High Gold’ | MH554079 | MH554290 | MH554991 | – | – | Liu et al. (2019) | ||
| Sar. longiappendiculatum | CBS 111308 | South Africa | Leucospermum cv. ‘High Gold’ | MH554080 | MH554291 | MH554992 | – | – | Liu et al. (2019) | |
| Sar. paragrevilleae | CBS 114142 | T | Australia | Grevillea sp. | MH553974 | MH554193 | MH554871 | – | – | Liu et al. (2019) |
| Sar. proteae | CBS 113610 | T | Australia | Protea magnifica | MH553968 | MH554187 | MH554862 | – | – | Liu et al. (2019) |
| CBS 114189 | Australia | Protea magnifica | MH553976 | MH554195 | MH554874 | – | – | Liu et al. (2019) | ||
| Sar. restionis | CBS 118153 | South Africa | Ischyrolepis cf. sieben | DQ278923 | DQ278925 | MH554890 | – | – | Liu et al. (2019) | |
| CBS 282.65 | UK | Pteridium aquilinum | AB594804 | AB593736 | MH554940 | – | – | Liu et al. (2019) | ||
| CBS 118154 | T | South Africa | Restio filiformis | DQ278922 | DQ278924 | MH554891 | – | – | Liu et al. (2019) | |
| CBS 111311 | New Zealand | – | MH553958 | MH554180 | MH554854 | – | – | Liu et al. (2019) | ||
| CBS 114130 | South Africa | Leucospermum sp. | MH553973 | MH554192 | MH554869 | – | – | Liu et al. (2019) | ||
| Seimatosporium botan | HMUC-sei-302PD | Chile | Vitis vinifera | JN088482 | – | – | – | – | Hatakeyama & Harada (2004) | |
| NBRC 104200 = H4619 | T | Japan | Paeonia suffruticosa | AB594799 | AB593731 | – | – | LC047770 | Hatakeyama & Harada (2004) | |
| Seim, discosioides | NBRC 104201 | Canada | Punica granatum | AB594800 | AB593732 | – | – | Tanaka et al. (2011) | ||
| Seim, elegans | NBRC 32674 | Japan | Melaleuca ericifolia | AB594801 | AB593733 | – | – | – | Wijayawardene et al. (2016a) | |
| Seim, eucalypti | CPC 156 | South Africa | Eucalyptus smithii | JN871200 | JN871209 | – | – | – | Wijayawardene et al. (2016a) | |
| Seim, falcatum | CPC 13578 | Australia | Eucalyptus sp. | JN871204 | JN871213 | – | – | – | Wijayawardene et al. (2016a) | |
| Seim, ficeae | MFLUCC 15-0519 | T | Italy | Rubus sp. | KR092800 | KR920686 | – | – | – | Wijayawardene et al. (2016a) |
| Seim, foliicola | NBRC 32676 | Japan | Juniperus phoenicea | AB594802 | AB593734 | – | – | – | Wijayawardene et al. (2016a) | |
| Seim, germanicum | CBS 437.87 | T | Germany | – | MH554047 | MH554259 | MH554957 | MH554482 | MH554723 | Liu et al. (2019) |
| Seim, glandigenum | NBRC 32677 | Japan | Fagus sylvatica | AB594803 | AB593735 | – | – | – | Liu et al. (2019) | |
| Seim, grevilleae | ICMP 10981 | South Africa | Protea sp. | AF405304 | AF382372 | – | – | – | Wijayawardene et al. (2016a) | |
| Seim, hakeae | NBRC 32678 | Japan | Pteridium aquilinum | AB594804 | AB593736 | – | – | – | Wijayawardene et al. (2016a) | |
| Seim, hypericinum | NBRC 32647 | Japan | Hypericum sp. | AB594805 | AB593737 | – | – | – | Wijayawardene et al. (2016a) | |
| Seim, luteosporum | Napa754 | USA | Prunus persica | KY706283 | KY706308 | – | KY706333 | KY706258 | Liu et al. (2019) | |
| CBS 142599 | T | USA | Vitis vinifera | KY706284 | KY706309 | – | KY706334 | KY706259 | Liu et al. (2019) | |
| Seim, mariae | NBRC 32681 | Japan | Correa reflexa | AB594807 | AB593740 | – | – | – | Wijayawardene et al. (2016a) | |
| Seim, marivanicum | IRAN 2310CT = CBS 143781 | T | Iran | Vitis vinifera | MW361952 | MW361960 | – | MW375358 | MW375352 | Moghadam et al. (2022) |
| IRAN 2300C = CBS 143780 | Iran | Vitis vinifera | MW361951 | MW361959 | – | MW375357 | MW375351 | Moghadam et al. (2022) | ||
| Seim, obtusum | CPC 12935 | T | Australia | Corymbia henryi | JN871206 | JN871215 | – | – | – | Barber et al. (2011) |
| Seim, parasiticum | NBRC 32682 | Japan | Physocarpus amurensis | AB594808 | AB593741 | – | – | – | Wijayawardene et al. (2016a) | |
| Seim, pezizoides | 71TB | Unknown | Unknown | KF573991 | – | – | – | – | Lawrence et al. (2018) | |
| Seim, physocarpi | CBS 139968 = | T | Russia | Physocarpus opulifolius | KT198722 | KT198723 | MH554917 | MH554434 | MH554676 | Liu et al. (2019) |
| MFLUCC 14-0625 | ||||||||||
| CBS 789.68 = NBRC 32682 | Netherlands | Physocarpus amurensis | MH554066 | MH554278 | MH554979 | MH554502 | MH554742 | Liu et al. (2019) | ||
| Seim, pistaciae | CBS 138865 = CPC 24455 | T | Iran | Pistacia vera | KP004463 | KP004491 | MH554915 | MH554432 | MH554674 | Crous et al. (2014b) |
| CPC 24457 | Iran | Pistacia vera | MH554126 | MH554331 | MH555035 | MH554561 | MH554799 | Crous et al. (2014b) | ||
| Seim, pseudorosae | MFLUCC 14-0468 | T | Italy | Rosa villosa | – | KU359035 | – | – | – | Li et al. (2016) |
| Seim, quercinum | MFLUCC 14-1198 | T | Germany | Quercus robur | KU974965 | KU974964 | – | – | – | Goonasekara et al. (2016) |
| Seim, restionis | CBS 118154 | South Africa | Restio filiformis | DQ278922 | DQ278924 | – | – | – | Lawrence et al. (2018) | |
| Seim, rhombisporium | MFLUCC 15-0727 | T | Italy | Vaccinium myrtillus | KR092792 | KR092780 | – | – | – | Wijayawardene et al. (2016a) |
| Seim, rosae | CBS 139823 = MFLUCC 14-0621 | ET | Russia | Rosa kalmiussica | KT198726 | KT198727 | LT853153 | LT853203 | LT853253 | Liu et al. (2019) |
| Seim, soli | CBS 941.69 | T | Denmark | Soil | MH554071 | MH554282 | MH554983 | MH554507 | – | Liu et al. (2019) |
| Seim, vitifusiforme | CBS 142600 | T | USA | Vitis vinifera | KY706296 | KY706321 | – | KY706346 | KY706271 | Lawrence et al. (2018) |
| Napa751 | USA | Vitis vinifera | KY706289 | KY706314 | – | KY706339 | KY706264 | Lawrence et al. (2018) | ||
| Seim, vitis | MFLUCC 14-0051 | T | Italy | Vitis vinifera | KR920363 | KR920362 | – | – | – | Senanayake et al. (2015) |
| Seim, vitis-viniferae | CBS 123004 | T | Spain | Vitis vinifera | MH553992 | MH554211 | MH554901 | MH554418 | MH554660 | Liu et al. (2019) |
| CBS 116499 | Iran | Vitis vinifera | MH553984 | MH554201 | MH554884 | MH554402 | MH554643 | Liu et al. (2019) | ||
| Seim, walken | CPC 17644 | Australia | Eucalyptus sp. | JN871207 | JN871216 | – | – | – | Barber et al. (2011) | |
| Seiridium aquaticum | MFLUCC 17-0474 | T | China | Decaying wood | MK828605 | – | – | – | – | Luo et al. (2019) |
| S-136 | China | Decaying wood | MK828606 | – | – | – | – | Luo et al. (2019) | ||
| Seir. camelliae | MFLUCC 12-0647 | T | China | Camellia reticula | JQ683725 | – | – | JQ683741 | JQ683709 | Maharachchikumbura et al. (2015) |
| Seir. cancrinum | CBS 226.55 = IMI 052256 | T | Kenya | Cupressus macrocarpa | LT853089 | MH554241 | LT853137 | LT853186 | LT853236 | Bonthond et al. (2018) |
| CBS 907.85 | South Africa | Cupressus lustinaca | LT853090 | – | LT853138 | LT853187 | LT853237 | Bonthond et al. (2018) | ||
| Seir. Cardinale | CBS 909.85 | T | South Africa | Cupressus lustinaca | LT853064 | – | LT853113 | LT853161 | LT853211 | Marin-Felix et al. (2019) |
| CBS 910.85 | South Africa | Cupressus sempervirens | LT853065 | – | LT853114 | LT853162 | LT853212 | Marin-Felix et al. (2019) | ||
| CBS 523.82 | New Zealand | Cupressocyparis sp. | LT853062 | – | LT853111 | LT853159 | LT853209 | Marin-Felix et al. (2019) | ||
| Seir. ceratosporum | PHSI2001Pathcw07 | China | Vitis vinifera | AY687314 | – | – | – | – | Jiang et al. (2019) | |
| Seir. chinense | CFCC 53031 | T | China | Trachycarpus fortunei | MK353158 | – | MK351796 | MK351799 | MK351802 | Jiang et al. (2019) |
| CFCC 53032 | China | Trachycarpus fortunei | MK353159 | – | MK351797 | MK351800 | MK351803 | Jiang et al. (2019) | ||
| CFCC 53033 | China | Trachycarpus fortunei | MK353160 | – | MK351798 | MK351801 | MK351804 | Jiang et al. (2019) | ||
| Seir. cupressi | CBS 122616 | Greece | Cupressus sp. | LT853082 | – | LT853130 | LT853179 | LT853229 | Jiang et al. (2019) | |
| CBS 224.55 = IMI 052254 | ET | Kenya | Cupressus macrocarpa | LT853083 | – | LT853131 | LT853180 | LT853230 | Jiang et al. (2019) | |
| Seir. eucalypti | CBS 343.97 | ET | Australia | Eucalyptus delegatensis | LT853099 | MH554251 | LT853146 | LT853196 | LT853246 | Jiang et al. (2019) |
| Seir. kartense | CBS 142629 = CPC 20183 | T | Australia | Eucalyptus cladocalyx | LT853100 | – | LT853147 | LT853197 | LT853247 | Bonthond et al. (2018) |
| Seir. kenyanium | CBS 228.55 = IMI 052257 | T | Kenya | Juniperus procera | LT853098 | MH554242 | LT853145 | LT853195 | LT853245 | Jiang et al. (2019) |
| Seir. marginatum | CBS 140403 | ET | France | Rosa canina | KT949914 | MH554223 | LT853149 | LT853199 | LT853249 | Jaklitsch et al. (2016) |
| Seir. neocupressi | CBS 142625 = CPC 23786 | T | Italy | Cupressus sempervirens | LT853079 | MH554329 | LT853127 | LT853176 | LT853226 | Bonthond et al. (2018) |
| CBS 142626 | Italy | Cupressus sempervirens | LT853080 | – | LT853128 | LT853177 | LT853227 | Bonthond et al. (2018) | ||
| Seir. papillatum | CBS 340.97 = VPRI 20827 | T | Australia | Eucalyptus delegatensis | LT853102 | DQ414531 | LT853150 | LT853200 | LT853250 | Jiang et al. (2019) |
| Seir. pezizoides | CBS 145115 | Italy | Vitis vinifera | MK079342 | – | MK058475 | MK058480 | MK058485 | Marin-Felix et al. (2019) | |
| Seir. phylicae | CBS 133587 = CPC 19964 | T | Tristan da Cunha | Phylica arborea | LT853091 | – | LT853139 | LT853188 | – | Crous et al. (2012) |
| CPC 19962 | Tristan da Cunha | Phylica arborea | LT853092 | – | LT853140 | LT853189 | – | Crous et al. (2012) | ||
| Seir. podocarpi | CBS 137995 | T | South Africa | Podocarpus latifolus | LT853101 | – | LT853148 | LT853198 | LT853248 | Crous et al. (2014a) |
| Seir. pseudocardinale | MFLUCC 13-0525 | Italy | Cupressus arizonica | KU848210 | – | – | – | – | Wijayawardene et al. (2016b) | |
| CBS 122613 = CMW 1648 | Portugal | Cupressus sp. | LT853096 | MH554206 | LT853143 | LT853193 | LT853243 | Wijayawardene et al. (2016b) | ||
| Seir. rosarum | MFLUCC 17-0654 | T | Italy | Rosa sp. | MG828961 | – | – | – | – | Wanasinghe et al. (2018) |
| Seir. spyridicola | CBS 142628 | T | Australia | Spyridium globosum | LT853095 | – | LT853142 | LT853192 | LT853242 | Bonthond et al. (2018) |
| Seir. unicorne | CBS 120306 | South Africa | Cupressus sempervirens | LT853087 | – | LT853135 | LT853184 | LT853234 | Bonthond et al. (2018) | |
| CBS 538.82 = NBRC 32684 | T | New Zealand | Cryptomeria japonica | LT853088 | MH554269 | LT853136 | LT853185 | LT853235 | Bonthond et al. (2018) | |
| Seir. venetum | MFLU 14-0265 | T | Italy | Cornus mas | KT438836 | – | – | – | KT438837 | Maharachchikumbura et al. (2015) |
| Sporocadus biseptatus | CBS 110324 = MYC 754 | T | – | – | MH553956 | MH554179 | MH554853 | MH554374 | MH554615 | Liu et al. (2019) |
| Spo. cornicola | CBS 143889 = CPC 23235 | Germany | Cornus sanguinea | MH554121 | MH554326 | MH555029 | MH554555 | MH554794 | Liu et al. (2019) | |
| MFLUCC 14-0448 | T | Italy | Cornus sanguinea | KU974967 | – | – | – | – | Liu et al. (2019) | |
| Spo. cornii | MFLUCC 14-0467 | T | Italy | Cornus sp. | KT162918 | KR559739 | – | – | – | Moghadam et al. (2022) |
| Spo. cotini | CBS 139966 = | T | Russia | Cotinus coggygria | MH554003 | MH554222 | MH554916 | MH554433 | MH554675 | Liu et al. (2019) |
| MFLUCC 14-0623 | ||||||||||
| Spo. incanus | CBS 123003 | T | Spain | Prunus dulcis | MH553991 | MH554210 | MH554900 | MH554417 | MH554659 | Liu et al. (2019) |
| Spo. italicus | MFLUCC 14-1196 | T | Italy | Crategus sp. | MF614829 | MF614829 | – | – | – | Moghadam et al. (2022) |
| Spo. kurdistanicus | IRAN 2356C = CBS 143778 | T | Iran | Vitis vinifera | MW361950 | MW361958 | – | MW375356 | MW375350 | Moghadam et al. (2022) |
| IRAN 2355C | Iran | Vitis vinifera | MW361949 | – | – | – | – | Moghadam et al. (2022) | ||
| IRAN 2354C | Iran | Vitis vinifera | MW361948 | MW361957 | – | MW375355 | MW375349 | Moghadam et al. (2022) | ||
| IRAN 2313C = CBS 143777 | Iran | Vitis vinifera | MW361947 | MW361956 | – | MW375354 | MW375348 | Moghadam et al. (2022) | ||
| Spo. lichenicola | CBS 354.90 = NBRC 32677 | Germany | Fagus sylvatica | MH554035 | MH554252 | MH554948 | MH554470 | MH554711 | Liu et al. (2019) | |
| CPC 24528 | Germany | Juniperus communis | MH554127 | MH554332 | MH555036 | MH554562 | MH554800 | Liu et al. (2019) | ||
| NBRC 32625 = IMI 079706 | ET | UK | Rosa canina | MH883643 | MH883646 | MH883647 | MH883644 | MH883645 | Liu et al. (2019) | |
| Spo. mali | CBS 446.70 | T | Netherlands | Malus sylvestris | MH554049 | MH554261 | MH554960 | MH554484 | MH554725 | Liu et al. (2019) |
| Spo. microcyclus | CBS 424.95 | T | Germany | Sorbus aria | MH554045 | MH554258 | MH554956 | MH554480 | MH554721 | Liu et al. (2019) |
| CBS 887.68 = NBRC 32680 | Netherlands | Ribes sp. | MH554068 | MH554280 | MH554981 | MH554504 | MH554744 | Liu et al. (2019) | ||
| Spo. multiseptatus | CBS 143899 = CPC 26606 | T | Serbia | Viburnum sp. | MH554141 | MH554343 | MH555047 | MH554576 | MH554814 | Liu et al. (2019) |
| Spo. pseudocorni | MFLU 13-0529 | T | Italy | Cornus sp. | – | KU359033 | – | – | – | Moghadam et al. (2022) |
| Spo. rosarum | CBS 113832 = UPSC2172 | Sweden | Rosa canina | MH553970 | MH554189 | MH554864 | MH554388 | MH554629 | Liu et al. (2019) | |
| MFLUCC 15-0563 | T | Italy | Rosa canina | MG828960 | MG829071 | – | – | – | Liu et al. (2019) | |
| MFLUCC 14-0466 | T | Italy | Rosa canina | KT284775 | KT281912 | – | – | – | Liu et al. (2019) | |
| Spo. rosigena | CBS 116498 | Iran | Vitis vinifera | MH553983 | MH554200 | MH554883 | MH554401 | MH554642 | Liu et al. (2019) | |
| CBS 129166 = MSCL 860 | Latvia | Rhododendron | MH553996 | MH554215 | MH554905 | MH554423 | MH554665 | Liu et al. (2019) | ||
| CBS 182.50 | Netherlands | Pyrus communis | MH554013 | MH554233 | MH554926 | MH554447 | MH554689 | Liu et al. (2019) | ||
| CBS 250.49 | Netherlands | Rubus fruticosus | MH554023 | MH554245 | MH554934 | MH554457 | MH554699 | Liu et al. (2019) | ||
| CBS 466.96 | Netherlands | Rubus sp. | MH554052 | MH554265 | MH554965 | MH554487 | MH554728 | Liu et al. (2019) | ||
| MFLU 16-0239 | T | Italy | Rosa canina | MG828958 | MG829069 | – | – | – | Liu et al. (2019) | |
| Spo. rotundatus | CBS 616.83 | T | Canada | Arceuthobium pussilum | MH554060 | MH554273 | MH554974 | MH554496 | MH554737 | Liu et al. (2019) |
| Spo. sorbi | CBS 160.25 | – | – | MH554008 | MH554229 | MH554924 | MH554442 | MH554684 | Liu et al. (2019) | |
| MFLUCC 14-0469 | T | Italy | Sorbus torminalis | KT284774 | KT281911 | – | – | – | Liu et al. (2019) | |
| Spotocadus sp. | CBS 506.71 | Italy | Euphorbia sp. | MH554055 | MH554268 | MH554968 | MH554490 | MH554731 | Liu et al. (2019) | |
| Spo. trimorphus | CBS 114203 = UPSC2430 | T | Sweden | Rosa canina | MH553977 | MH554196 | MH554876 | MH554395 | MH554636 | Liu et al. (2019) |
| Strickeria kochii | CBS 140411 | ET | Austria | Robinia pseudoacacia | NR_154423 | KT949918 | MH554920 | – | – | Liu et al. (2019) |
| Synnemapestaloides juniperi | CBS 477.77 | T | France | Juniperus phoenicea | MH554053 | MH554266 | MH554966 | – | – | Liu et al. (2019) |
| Syn. rhododendri | MAFF 239201 | T | Japan | Rhododendron brachycarpum | LC047753 | LC047744 | – | – | – | Liu et al. (2019) |
| MAFF 243052 | Japan | Rhododendron brachycarpum | LC047757 | LC047748 | – | – | – | Liu et al. (2019) | ||
| Truncatella angustata | CBS 398.71 | Turkey | Soil | MH554042 | MH554255 | MH554953 | – | – | Liu et al. (2019) | |
| CBS 231.77 | Turkey | Gossypium sp. | MH554021 | MH554243 | MH554932 | – | – | Liu et al. (2019) | ||
| CPC 21354 | France | Vitis vinifera cv. ‘Prunelard’ | MH554111 | MH554317 | MH555020 | – | – | Liu et al. (2019) | ||
| CBS 642.97 | Switzerland | Heterodera carotae | MH554061 | MH554274 | MH554975 | – | – | Liu et al. (2019) | ||
| CBS 393.80 | Chile | Gevuina avellana | MH554041 | MH554254 | MH554952 | – | – | Liu et al. (2019) | ||
| CBS 938.70 | Netherlands | Prunus laurocerasus | MH554070 | MH554281 | MH554982 | – | – | Liu et al. (2019) | ||
| CBS 356.33 | – | Prunus sp. | MH554036 | MH554253 | MH554949 | – | – | Liu et al. (2019) | ||
| CBS 144025 | NT | France | Vitis vinifera cv. ‘Prunelard | MH554112 | MH554318 | MH555021 | – | – | Liu et al. (2019) | |
| CBS 338.32 | Netherlands | Lupinus sp. | MH554033 | MH554250 | MH554945 | – | – | Liu et al. (2019) | ||
| CBS 165.25 | – | Prunus armeniaca | MH554010 | MH554231 | – | – | – | Liu et al. (2019) | ||
| undetermined sp. 1 | CBS 113991 | Sweden | Salix caprea | MH553971 | MH554190 | MH554865 | – | – | Liu et al. (2019) | |
| undetermined sp. 2 | CBS 387.77 | Finland | Skin of man | MH554040 | KM 116277 | MH554950 | – | – | Liu et al. (2019) | |
| Xenoseimatosporium quercinum | MFLUCC 14-1198 | T | Germany | Quercus robur | NR_155804 | NG_059681 | – | – | – | Liu et al. (2019) |
| CBS 129171 | Rhododendron sp. | MH553997 | MH554216 | MH554906 | – | – | Liu et al. (2029) | |||
1 BRIP: Queensland Plant Pathology Herbarium, Australia; CBS: Culture collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; CFCC: China Forestry Culture Collection Center, China; CMW: Culture Collection of the Forestry and Agricultural Biotechnology Institute (FABI), University of Pretoria, Pretoria, South Africa; CPC: Culture collection of Pedro Crous, housed at the Westerdijk Institute; HGUP: Plant Pathology Herbarium of Guizhou University; HHUF: herbarium of Hirosaki University; HKUCC: The University of Hong Kong Culture Collection; HPC: Herbarium of Pedro Crous, housed at the Westerdijk Institute; ICMP: International Collection of Micro-organisms from Plants, Landcare Research, Private Bag 92170, Auckland, New Zealand; IFRDCC: International Fungal Research and Development Culture Collection; IMI: International Mycological Institute, CABI-Bioscience, Egham, Bakeham Lane, United Kingdom; LC: working collection of Lei Cai, housed at the Institute of Microbiology, Chinese Academy of Sciences, Beijing, China; MAFF: Ministry of Agriculture, Forestry and Fisheries, Tsukuba, Ibaraki, Japan; MEAN: culture collection of INIAV Institute, Oeiras, Portugal; MFLUCC: Mae Fah Luang University Culture Collection; NOF: The Fungus Culture Collection of the Northern Forestry Centre, Alberta, Canada; MSCL: Microbial Strain Collection of Latvia; NBRC: Biological Resource Center; NTUCC: National Taiwan University Culture Collection, Taiwan; UPSC: Uppsala University Culture Collection of Fungi, Sweden; VPRI: Victorian Plant Disease Herbarium, Australia.
2 Status: status of the strains. ET: ex-epitype; IT: ex-isotype; NT: ex-neotype; R: reference strain; ST: ex-syntype; T: ex-type.
3 MFLUCC 15-0563: Type of Seimatosporium rosigenum, MFLUCC 14-0466: Type of Seimatosporium pseudorosarum.
Seven phylogenetic analyses were conducted based on both individual and combined loci for one family (Sporocadaceae) and six genera (Monochaetia, Neopestalotiopsis, Pestalotiopsis, Seimatosporium, Seiridium and Sporocadus). Maximum Parsimony (MP) was used to construct phylogenies using PAUP v. 4.0b10 (Swofford 2003), Maximum Likelihood (ML) was executed in RAxML (Dean et al. 2014) implemented in raxmlGUI v. 1.5b1 (Silvestro & Michalak 2012) and MrBayes v. 3.1.2 was used for Bayesian analyses (BA) (Huelsenbeck & Ronquist 2001).
Trees were visualised with FigTree v. 1.4.0 (http://tree.bio.ed.ac.uksoftware/figtree/), and additional layout was done with Adobe Illustrator CS v. 5. Maximum-likelihood bootstrap values (MLBP) and Maximum-parsimony bootstrap values (MPBP) equal or greater than 50 % are provided for each tree. Bayesian posterior probabilities (BYPP) > 0.90 are indicated as thickened lines.
Prevalence
To determine the prevalence of Sporocadaceae genera and species on Rosa spp. and plant parts (branches, fruits, leaves or spines), the Isolation Rate (RI) was calculated for each species with the formula: RI % = (NS / NI) × 100, where NS was the number of isolates from the same genera or species, and NI was the total number of isolates from each Rosa sp. or plant part. The overall RI was calculated using the NI value equal to the total number of isolates obtained from Rosa plants (Vieira et al. 2014, Fu et al. 2019, Guo et al. 2020).
RESULTS
Phylogenetic analyses
The concatenated DNA sequence dataset (ITS, LSU and RPB2) was used to infer delimitation at the family level. The concatenated alignment had a total length of 2419 characters including alignment gaps (659 for ITS, 890 for LSU and 832 for RPB2). Of these, 1 401 characters were constant, 144 variable characters were parsimony-uninformative and 874 characters were parsimony informative. The ML search resolved a best tree with an InL of -36418.696700. The BA lasted for 1 855 000 generations and the 50 % consensus tree and posterior probabilities were calculated from 2 784 trees from two runs. The ML tree confirmed the same tree topology and the clades as those presented in the Bayesian phylogeny (Fig. 2).
Fig. 2.



Phylogenetic tree of Sporocadaceae (50 % majority rule consensus) based on maximum likelihood (ML) analysis of the combined LSU, ITS and RPB2 sequence alignment. Nodes are labelled with bootstrap values from RAxML/Bayesian posterior probabilities. Nodes receiving below 50 bootstrap values and 0.5 probability values are not labelled. The scale bar represents the expected number of changes per site. Genera in this study are delimited in coloured boxes and isolates collected in this study are in bold. Ex-type strains are represented in bold.
Following alignment of Monochaetia, the ITS sequence data with a total of 599 characters including gaps, of which 496 characters were constant, 15 variable characters were parsimony uninformative, and 88 characters were variable and parsimony informative. The parsimony analysis resulted in 1 000 equally parsimonious trees, one of which is in Fig. 3 (CI = 0.780, RI = 0.887, RC = 0.692, HI = 0.220). The topology of the phylogenetic trees generated from the MP, ML and BA methods were congruent.
Fig. 3.

Phylogenetic tree of Monochaetia resulting from maximum likelihood (ML) analysis of the ITS sequence alignment. Nodes are labelled with bootstrap values from RAxML/Parsimony bootstrap/Bayesian posterior probabilities values. Nodes receiving below 50 bootstrap values and 0.5 probability values are not labelled. The scale bar represents the expected number of changes per site. Ex-type strains are represented in bold; strains obtained in the current study in blue. The flower symbol represents species that have been recorded from Rosa.
In the phylogenetic tree constructed for Neopestalotiopsis and Pestalotiopsis, sequences of the combined ITS, TEF and TUB were aligned. In Neopestalotiopsis, the alignment comprises 2 263 characters including alignment gaps after alignment (580 for ITS, 592 for TEF and 787 for TUB). Of these, 1647 characters were constant, 383 variable characters were parsimonyuninformative and 233 characters were parsimony informative. The MP analysis resulted in a single equally most parsimonious tree (CI = 0.672, RI = 0.659, RC = 0.442, HI = 0.328). In Pestalotiopsis, the alignment comprises 2 183 characters including alignment gaps after alignment (590 for ITS, 670 for TEF and 909 for TUB), of which 1165 characters were constant, 384 variable characters were parsimony-uninformative and 634 characters were parsimony informative. The MP analysis resulted in a single equally most parsimonious tree (CI = 0.565, RI = 0.845, RC = 0.478, HI = 0.435). MP was similar to the topology from ML and BA (Fig. 4, 5).
Fig. 4.


The best Maximum Likelihood tree from the multi-gene alignment (ITS, TUB and TEF) for the Neopestalotiopsis. Nodes are labelled with bootstrap values from RAxML/Parsimony bootstrap/Bayesian posterior probabilities values. Nodes receiving below 50 bootstrap values and 0.5 probability values are not labelled. The scale bar represents the expected number of changes per site. Ex-type strains are represented in bold; strains obtained in the current study in blue. The flower symbol represents species that have been recorded from Rosa.
Fig. 5.


The best Maximum Likelihood tree from the multi-gene alignment (ITS, TUB and TEF) for the Pestalotiopsis. Nodes are labelled with bootstrap values from RAxML/Parsimony bootstrap/Bayesian posterior probabilities values. Nodes receiving below 50 bootstrap values and 0.5 probability values are not labelled. The scale bar represents the expected number of changes per site. Ex-type strains are represented in bold; strains obtained in the current study in blue. The flower symbol represents species that have been recorded from Rosa.
In Seimatosporium, the combined ITS + LSU dataset had an aligned length of 1495 characters in the dataset (601 for ITS and 887 for LSU), of which 1 181 characters are constant, 118 are variable and parsimony-uninformative, and 196 are parsimony informative. Maximum Parsimony analysis yielded 750 equally parsimonious trees (CI = 0.674, RI = 0.738, RC = 0.497, HI = 0.326), and a strict consensus tree is shown in Fig. 6.
Fig. 6.

The best Maximum Likelihood tree from the multi-gene alignment (ITS and LSU) for the Seimatosporium. Nodes are labelled with bootstrap values from RAxML/Parsimony bootstrap/Bayesian posterior probabilities values. Nodes receiving below 50 bootstrap values and 0.5 probability values are not labelled. The scale bar represents the expected number of changes per site. Ex-type strains are represented in bold; strains obtained in the current study in blue. The flower symbol represents species that have been recorded from Rosa.
In Seiridium, the combined ITS, RPB2, TEF and TUB dataset consists of 2 897 characters including alignment gaps (598 for ITS, 818 for RPB2, 606 for TEF and 829 for TUB), of which 1 637 are constant, 454 are variable parsimony uninformative characters and 806 are parsimony-informative characters. The MP analysis resulted in a single equally most parsimonious tree (CI = 0.664, RI = 0.689, RC = 0.457, HI = 0.336) (Fig. 7).
Fig. 7.

The best Maximum Likelihood tree from the multi-gene alignment (ITS, RPB2, TEF and TUB) for the Seiridium. Nodes are labelled with bootstrap values from RAxML/Parsimony bootstrap/Bayesian posterior probabilities values. Nodes receiving below 50 bootstrap values and 0.5 probability values are not labelled. The scale bar represents the expected number of changes per site. Ex-type strains are represented in bold; strains obtained in the current study in blue. The flower symbol represents species that have been recorded from Rosa.
In Sporocadus, the combined sequences of ITS, LSU, RPB2, TEF and TUB were aligned. The combined data comprises 3619 characters including alignment gaps (566 for ITS, 900 for LSU, 869 for RPB2, 496 for TEF and 760 for TUB). Of these, 2 671 characters were constant, 263 variable characters were parsimony-uninformative and 685 characters were parsimony informative. The MP analysis resulted in a single equally most parsimonious tree (CI = 0.657, RI = 0.808, RC = 0.531, HI = 0.343) (Fig. 8).
Fig. 8.

The best Maximum Likelihood tree from the multi-gene alignment (ITS, LSU, RPB2, TEF and TUB) for the Sporocadus. Nodes are labelled with bootstrap values from RAxML/Parsimony bootstrap/Bayesian posterior probabilities values. Nodes receiving below 50 bootstrap values and 0.5 probability values are not labelled. The scale bar represents the expected number of changes per site. Ex-type strains are represented in bold; strains obtained in the current study in blue. The flower symbol represents species that have been recorded from Rosa.
All the MP analyses resulted in a tree with the same topology and terminal clades as the ML and BA trees. The new species from the present study appeared in distinct clades with high bootstrap support.
Taxonomy
Based on the morphology and multi-locus phylogeny, 126 isolates were assigned to 15 species belonging to six genera in Sporocadaceae, including 11 species newly described below.
Monochaetia rosarum C. Peng & C.M. Tian, sp. nov. — MycoBank MB 843811; Fig. 9
Fig. 9.

Monochaetia rosarum (BJFC-S1877, holotype). a. Disease symptoms; b–c. habit of conidiomata on branch; d. colonies on PDA at 3 d (left) and 15 d (right); e. longitudinal section through conidioma; f. conidiomata on PDA; g–i. conidiogenous cells with attached conidia; k. conidia. — Scale bars: b–c, e = 50 μm; f = 200 μm; g–k = 10 μm.
Etymology. The species epithet reflects the name of the host plant genus Rosa.
Typus. China, Henan Province, Nanyang City, Neixiang county, Baotianman Nature Reserve, N33°29'29" E111°55'50", alt. 1311 m, on branches of R. chinensis, 7 Aug. 2020, C.M. Tian, Y.M. Liang & C. Peng (holotype BJFC-S1877, ex-type culture CFCC 55172 = ROC 099).
Symptoms appeared as circular to irregular, red or dark brown and raised, dehiscent lesions on twigs and branches (Fig. 9b–c). Sexual morph not observed. Asexual morph: Acervular conidiomata visible on the host, globose or irregular, superficial to semiimmersed, scattered or aggregated, black or red, 76–118 μm diam. Conidiophores septate and branched, hyaline, smooth-walled or verruculose. Conidiogenous cells annellidic, discrete or integrated, cylindrical, subcylindrical, hyaline or pale brown, smooth, 6.5–14(–15) × (1–)1.5–2 μm (av. = 10.7 ± 2.84 × 1.7 ± 0.26 μm). Conidia fusoid, straight or slightly curved, 4-septate, wall smooth, not constricted at the septa, (17.5–)18–20(–21) × 5–6(–7) μm (av. = 19.5 ± 0.95 × 5.4 ± 0.37 μm); basal cell obconic with or without truncate base, thin-walled, hyaline, 2.5–3(–3.5) μm (av. = 2.6 ± 0.27 μm) long; three median cells, doliiform or subcylindrical, mid-brown, thick-walled, the first median cell from base (3–)3.5–5 μm (av. = 4.2 ± 0.42 μm) long, the second cell (2.5–)3–3.5(–4.5) μm (av. = 3.4 ± 0.27 μm) long, the third cell (3–)3.5–4(–5) μm (av. = 3.7 ± 0.36 μm) long, together (10.5–)11.5–12.5(–13) μm (av. = 12.1 ± 0.47 μm) long; apical cell conic with an acute apex, thin-walled, hyaline, 2–2.5(–3) μm (av. = 2.4 ± 0.31 μm) long; apical appendage single, tubular, attenuated, unbranched, variously bent; (8–)9–12(–12.5) μm (av. = 10.8 ± 1.36 μm) long; basal appendage single, unbranched, centric, 4.5–6(–6.5) μm (av. = 5.5 ± 0.48 μm); mean conidium length/width ratio = 3.25 : 1.
Culture characteristics — On PDA, colonies slowly growing, up to 10 mm diam after 3 d and reaching 38–40 mm after 15 d, white to pale grey with a uniform texture, with an irregular margin, lacking aerial mycelium, becoming greyish and yellowish after 30 d, reverse yellowish brown. Conidiomata globose, distributed irregularly on the medium surface, exuding globose, dark brown to black conidial masses.
Additional materials examined. China, Henan Province, Nanyang City, Neixiang county, Baotianman Nature Reserve, N33°29'68" E111°55'16", alt. 1329 m, on branches of R. chinensis, 7 Aug. 2020, C.M. Tian, Y.M. Liang & C. Peng (BJFC-S1878, cultures CFCC 55173 = ROC 100, ROC 98, ROC 101).
Notes — Monochaetia rosarum forms an independent clade and is phylogenetically distinct from M. kansensis (Fig. 3). Monochaetia rosarum can be distinguished from M. kansensis in ITS loci by 13 bp differences (from 561 characters, with 97.6 % sequence identity, including 3 bp gaps). In addition, M. rosarum differs from M. kansensis in producing thinner conidia (5–6 μmvs 6–8 μm). Furthermore, the apical and basal appendages of M. rosarum are significantly shorter than those of M. kansensis (9–12 μm vs 15–26 μm and 4.5–6 μm vs 10–38 μm).
Four Monochaetia species have been reported from Rosa spp., i.e., M. concentrica, M. rosae-caninae, M. seiridioides and M. turgida (Guba 1961, Tai 1979, Nag Raj 1993, Chen 2003). These four species are easily distinguishable from our new species based on conidial dimensions. Monochaetia concentrica and M. seiridioides have longer conidia than M. rosarum (20–26 μm vs 18–20 μm and 26–28 μm vs 18–20 μm). Monochaetia turgida has wider conidia than M. rosarum (7.6–8.3 μm vs 5–6 μm).
Although M. rosarum and M. rosae-caninae have similar conidial width (5–6 μm vs 5–7 μm), the conidia of M. rosarum is significantly longer than those of M. rosae-caninae (18–20 μm vs 16–18 μm). In addition, M. rosarum is similar to M. camelliae in conidial dimensions (18–20 × 5–6 μm vs 18–20 × 4–7 μm), the latter having been previously reported from Camellia, Eucalyptus and Peltophorum. However, based on the combined gene phylogenetic analysis, M. rosarum is separated from M. camelliae and the apical appendages of M. rosarum are shorter than those of M. camelliae (9–12 μm vs 12–14 μm).
Key to Monochaetia species on Rosa spp.
1. Conidial length more than 20 μm 2 . . . . . . . . . . . . . . . . . . . 2
1. Conidial length less than 20 μm 3 . . . . . . . . . . . . . . . . . . . . 3
2. Conidia 20–26 × 6.5–8.5 μm, apical appendage 15 μm long . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . M. concentrica
2. Conidia 26–28 × 6–7 μm, apical appendage 12 μm long . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .M. seiridioides
3. Conidial width more than 8 μm . . . . . . . . . . . . . M. turgida
3. Conidial width less than 8 μm . . . . . . . . . . . . . . . . . . . . . 4
4. Conidia 16–18 × 5–7 μm . . . . . . . . . . . .M. rosae-caninae
4. Conidia 18–20 × 5–6 μm . . . . . . . . . . . . . . . .M. rosarum
Neopestalotiopsis concentrica C. Peng & C.M. Tian, sp. nov. — MycoBank MB 843813; Fig. 10
Fig. 10.

Neopestalotiopsis concentrica (BJFC-S1882, holotype). a–b. Appearance of conidiomata on host substrate; c. conidiomata on PDA; d. colonies on PDA at 3 d (left) and 15 d (right); e–l. conidiogenous cells with attached conidia; m. conidia. — Scale bars: c = 200 μm; e–l = 10 μm.
Etymology. Name refers to the concentric circles formed by colonies on PDA.
Typus. China, Henan Province, Xinyang City, Jigong Mountain, N31°49'3" E114°4'33", alt. 717 m, on spines of R. rugosa, 6 Aug. 2020, C. Peng & S. Jia (holotype BJFC-S1882, ex-type culture CFCC 55162 = ROC 053).
Sexual morph not observed. Asexual morph: Acervular conidiomata globose or irregular, superficial to semi-immersed, scattered or aggregated, visible as black acervuli on the host, 93–117 μm diam. Conidiophores often reduced to conidiogenous cells. Conidiogenous cells discrete, ampulliform or irregular, variable in size, (3–)3.5–14.5(–15) × 3–5(–5.5) μm (av. = 7.8 ± 4.96 × 4.0 ± 1.21 μm), hyaline to subhyaline. Conidia fusoid, ellipsoid, straight to slightly curved, 4-septate, 14–18.5(–19) × 4.5–5(–6) μm (av. = 15.7 ± 1.06 × 4.9 ± 0.69 μm); basal cell conic to obconic with a truncate base, hyaline and thin-walled, 3–3.5(–4.0) μm (av. = 4.3 ± 0.21 μm) long; three median cells doliiform, (9–)9.5–10.5(–11) μm (av. = 10.0 ± 0.96 μm) long, wall rugose, versicoloured, septa darker than the rest of the cell, second cell from the base pale brown, 3–3.5 μm (av. = 3.3 ± 0.28 μm) long; third cell honey brown, 3–4 μm (av. = 3.74 ± 0.44 μm) long; fourth cell brown, 3–3.5 μm (av. = 3.1 ± 0.31 μm) long; apical cell (2.5–)3–3.5(–4) μm (av. = 3.4 ± 0.86 μm) long, hyaline, cylindrical, thinand smooth-walled; with 2–3 tubular apical appendages (mostly three), branched or unbranched, filiform, 19–26(–26.5) μm (av. = 22.6 ± 2.14 μm) long; basal appendage single, tubular, unbranched, centric, (3–)3.5–5.5(–6.0) μm (av. = 4.1 ± 0.95 μm) long.
Culture characteristics — Colony on PDA with flattened mycelium, white, smoke grey in the centre, reverse with smoke grey pigments formed a concentric ring pattern, reaching at 58–62 mm diam in 15 d at 28 °C. Conidiomata globose, irregularly distributed on the medium surface, exuding globose, dark brown to black conidial masses.
Additional materials examined. China, Henan Province, Xinyang City, Jigong Mountain, N31°49'1" E114°4'12", alt. 723 m, on spines of R. chinensis, 6 Aug. 2020, C. Peng & S. Jia (BJFC-S1883, living culture CFCC 55163 = ROC 064, ROC 135, ROC 136); Henan Province, Nanyang City, Xinyang City, Shihe District, N31°49'8" E114°3'58", alt. 727 m, on spines of R. chinensis, 6 Aug. 2020, C. Peng & S. Jia (BJFC-S1884, cultures ROC 137, ROC 138, ROC 140, ROC 141, ROC 142).
Notes — This species is most closely related to N. ellipsospora, N. rhapidis and N. samarangensis (Fig. 4), but distinguished from N. ellipsospora by 15 bp difference in the concatenated alignment (two in the ITS region (482/484, 99.5 % with no gaps), nine TEF (531/540, 98.3 % with four gaps) and four TUB (436/440, 99.0 % with four gaps)), from N. rhapidis by 13 bp difference (six in the ITS region (507/513, 98.8 % with no gaps), three TEF (499/502, 99.4 % with three gaps) and four TUB (743/747, 99.4 % with no gaps)) and from N. samarangensis by 12 bp difference (three in the ITS region (481/484, 99.3 % with two gaps), three TEF (537/540, 99.4 % with three gaps) and six TUB (434/440, 98.6 % with no gaps)). Moreover, N. concentrica differs from these two species in morphology, namely by having longer apical appendages than N. ellipsospora (19–26 μm vs 5–12 μm), N. rhapidis (19–26 μm vs 11–16 μm) and N. samarangensis (19–26 μm vs 12–18 μm). However, its conidia are shorter than those of N. ellipsospora (14–18.5 μm vs 19–25 μm), N. rhapidis (14–18.5 μm vs 22–25.5 μm) and N. samarangensis (14–18.5 μm vs 18–21 μm).
Neopestalotiopsis concentrica is phylogenetically and morphologically distinct from N. clavispora, N. palmarum, N. rosae, N. rosicola and N. versicolor, which were previously also reported from Rosa (Liu et al. 2010, Feng et al. 2014, Maharachchikumbura et al. 2014, Jiang et al. 2018, Vu et al. 2019). Its conidia are smaller than N. clavispora (14–18.5 × 4.5–5 μm vs 20–24 × 6.5–8.5 μm), N. palmarum (14–18.5 × 4.5–5 μm vs 19–23.6 × 5.6–6.6 μm), N. rosae (14–18.5 × 4.5–5 μm vs 22–37 × 7.5–9.5 μm), N. rosicola (14–18.5 × 4.5–5 μm vs 20.2–25.5 × 5.5–8 μm) and N. versicolor (14–18.5 × 4.5–5 μm vs 21.2–28.3 × 7.1–8.3 μm).
Neopestalotiopsis subepidermalis C. Peng & C.M. Tian, sp. nov. — MycoBank MB 843812; Fig. 11
Fig. 11.
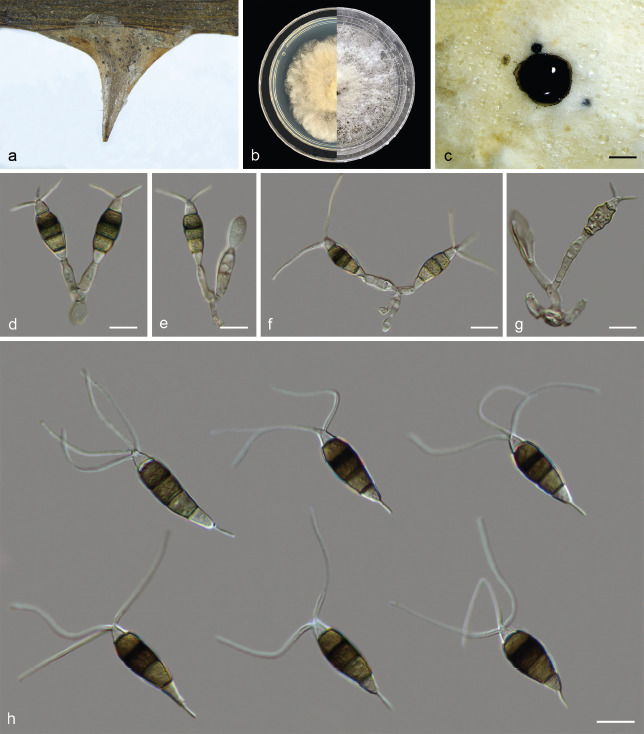
Neopestalotiopsis subepidermalis (BJFC-S1879, holotype). a. Appearance of conidiomata on host substrate; b. colonies on PDA at 3 d (left) and 15 d (right); c. conidiomata on PDA; d–g. conidiogenous cells with attached conidia; h. conidia. — Scale bars: c = 200 μm; d–h = 10 μm.
Etymology. Referring to the conidioma occurring under the epidermis of spines, not breaking through the epidermis.
Typus. China, Henan Province, Xinyang City, Jigong Mountain, N31°49'15" E114°3'31", alt. 163.4 m, on spines of R. rugosa, 5 Aug. 2020, C. Peng & S. Jia (holotype BJFC-S1879, ex-type culture CFCC 55160 = ROC 161, ROC 162).
Sexual morph not observed. Asexual morph: Acervular conidiomata globose or irregular, superficial to semi-immersed, scattered or aggregated, visible as black acervuli on the host, 80–122 μm diam. Conidiophores septate, branched, subcylindrical, hyaline to subhyaline, often reduced to conidiogenous cells. Conidiogenous cells discrete, collarette present and not flared, cylindrical, hyaline to subhyaline, (5.5–)6–18(–18.5) Sexual morph not observed. 3–5 μm (av. = 11.2 ± 2.90 × 3.8 ± 0.60 μm). Conidia fusoid, ellipsoid, straight to slightly curved, 4-septate, (19.5–)20–25(–26) × 7.5–9(–9.5) μm (av. = 22.6 ± 2.10 × 8.4 ± 0.95 μm); basal cell conic to obconic with a truncate base, hyaline and thin-walled, 4.0–5.5 μm (av. = 4.6 ± 0.64 μm) long; three median cells doliiform, 14–16 μm (av. = 15.0 ± 1.06 μm) long, wall rugose, versicoloured, septa darker than the rest of the cell, second cell from the base pale brown, (4.5–)5–6 μm (av. = 5.7 ± 0.44 μm) long; third cell honey brown, (4.5–)5–6 μm (av. = 5.4 ± 0.36 μm) long; fourth cell brown, (3.5–)4–5 μm (av. = 4.5 ± 0.72 μm) long; apical cell 3–3.5(–4) μm (av. = 3.2 ± 0.27 μm) long, hyaline, cylindrical, thinand smooth-walled; with 2–4 tubular apical appendages, branched or unbranched, filiform, (26.5–) 27–32.5(–33.5) μm (av. = 29.2 ± 1.79 μm) long; basal appendage single, tubular, unbranched, centric, (6.5–)7–7.5(–8) μm (av. = 7.3 ± 0.58 μm) long.
Culture characteristics — Colonies on PDA with aerial mycelium white, fluffy, reverse yellowish pigment accumulation in the centre, surrounded by amber, pure white at the colony margin, reaching at 58–60 mm diam in 15 d at 28 °C. Conidiomata globose, irregularly distributed on the medium surface, exuding globose, dark brown to black conidial masses.
Additional materials examined. China, Henan Province, Xinyang City, Jigong Mounta, N31°49'20" E114°32'6", alt. 220.1 m , on spines of R. chinensis, 5 Aug. 2020, C. Peng & S. Jia (BJFC-S1880, cultures CFCC 55161 = ROC 169, ROC 170); Henan Province, Xinyang City, Shihe District, N31°49'23" E114°3'24", alt. 199.6 m, on branches and spines of R. chinensis, 5 Aug. 2020, C. Peng & S. Jia (BJFC-S1881, cultures ROC 171, ROC 172, ROC 173, ROC 174).
Notes — The eight strains of N. subepidermalis in the present study form a well-supported independent clade distinct from known Neopestalotiopsis species (ML/MP/BI = 87/84/0.98). Neopestalotiopsis subepidermalis is most closely related to N. camelliae-oleiferae, N. mesopotamica, N. rosae and N. vacciniicola (Fig. 4), but differs from them in concatenated alignment. A comparison of the ITS region showed one nucleotide difference (453/454, 99.7 % with two gaps) with N. camelliaeoleiferae, three nucleotide differences (526/529, 99.4 % with a single gap) with N. mesopotamica, two nucleotide differences (524/526, 99.6 % with no gaps) with N. rosae, one nucleotide differences (519/520, 99.8 % with a single gap) with N. vacciniicola. Comparison of the TEF region revealed 15 bp differences (520/535, 97.1 % with no gaps) with N. camelliae-oleiferae, three bp differences (469/472, 99.5 % with no gaps) with N. mesopotamica, two bp differences (479/481, 99.5 % with no gaps) with N. rosae, 13 bp differences (462/475, 97.2 % with no gaps) with N. vacciniicola. Comparison of the TUB region revealed two bp differences (400/402, 99.5 % with no gaps) with N. camelliae-oleiferae, eight bp differences (723/731, 98.9 % with no gaps) with N. mesopotamica, 11 bp differences (736/747, 98.5 % with no gaps) with N. rosae, seven bp differences (412/419, 98.3 % with no gaps) with N. vacciniicola. Neopestalotiopsis rosae was also recorded from Rosa, but the morphological characteristics of N. rosae, in which the apical appendages do not arise from the apical crest are distinct from N. subepidermalis and other taxa in this genus (Maharachchikumbura et al. 2014). In addition, N. subepidermalis differs from the three other species in the dimensions of its conidia and apical appendages: the conidia of N. subepidermalis are shorter than N. mesopotamica (20–25 μm vs 26–32 μm), but longer than those of N. vacciniicola (20–25 μm vs 14.5–15.2 μm). Furthermore, the apical appendages of N. subepidermalis are significantly longer than those of N. camelliae-oleiferae (27–32.5 μm vs 15.5–18.5 μm) and N. vacciniicola (27–32.5 μm vs 4.3–24.3 μm).
Based on the phylogeny, N. subepidermalis is distinct from N. clavispora, N. palmarum, N. rosicola and N. versicolor, the four Neopestalotiopsis species associated with Rosa besides N. rosae (Liu et al. 2010, Feng et al. 2014, Jiang et al. 2018, Vu et al. 2019) (Fig. 4). Furthermore, N. subepidermalis differs from N. clavispora and N. versicolor in having the longer basal appendage (7–7.5 μm vs 3–5.5 μm and 7–7.5 μm vs 2.4–6.8 μm), while N. subepidermalis has longer apical appendages than N. rosicola and N. palmarum (27–32.5 μm vs 17–22.8 μm and 27–32.5 μm vs 11.8–18.9 μm).
Key to Neopestalotiopsis species on Rosa spp.
1. Conidial width less than 6 μm . . . . . . . . . . . . . . . . . . . . . . 2
1. Conidial width more than 6 μm . . . . . . . . . . . . . . . . . . . . . 3
2. Conidia 14–18.5 × 4.5–5 μm, apical appendage 19–26 μm long . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . N. concentrica
2. Conidia 19–23.6 × 5.6–6.6 μm, apical appendage 11.8–18.9 μm long . . . . . . . . . . . . . . . . . . . . . . . . . . . . . N. palmarum
3. Basal appendages less than 5.5 μm . . . . . . . . . . . . . . . . . 4
3. Basal appendages more than 5.5 μm . . . . . . . . . . . . . . . . 5
4. Conidia 20–24 × 6–8 μm, apical appendages 22–32 μm . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . N. clavispora
4. Conidia 21.2–28.3 × 7.1–8.3 μm, apical appendage 18.9–30.7 μm long . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . N. versicolor
5. Apical appendages do not arise from the apical crest . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . N. rosae
5. Apical appendages arise from the apical crest . . . . . . . . . 6
6. Conidia 20.2–25.5 × 5.5–8.0 μm, apical appendages 17.0–22.8 μm . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . N. rosicola
6. Conidia 20–25 × 7.5–9 μm, apical appendages 27–32.5 μm . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . N. subepidermalis
Pestalotiopsis chamaeropis Maharachch. et al., Stud. Mycol. 79: 158. 2014 — Fig. 12
Fig. 12.

Pestalotiopsis chamaeropis (BJFC-S1885). a. Appearance of conidiomata on host substrate; b. colonies on PDA at 3 d (left) and 15 d (right); c. conidiomata on PDA; d–i. conidiogenous cells with attached conidia; j. conidia. — Scale bars: c = 200 μm; d–i = 10 μm.
Sexual morph not observed. Asexual morph: Acervular conidiomata globose or irregular, superficial to semi-immersed, scattered or aggregated, visible as black acervuli on the host. Conidiophores septate, branched, subcylindrical, hyaline, verruculose. Conidiogenous cells discrete, cylindrical, hyaline, smooth-walled, collarette present and not flared, with prominent periclinal thickening, (5.5–)7–8.5(–10) × 2–4(–4.5) μm (av. = 7.6 ± 1.32 × 3.4 ± 0.66 μm). Conidia fusoid, ellipsoid, straight to slightly curved, 4-septate, (25–)26–27.5(–28) × 7–8.5(–9) μm (av. = 27 ± 0.54 × 7.9 ± 0.56 μm); basal cell obconic with a truncate base, hyaline, thin-walled, 4.5–5.5(–6) μm (av. = 5.1 ± O. 36 μm) long; three median cells doliiform to subcylindrical, (15–)16.5–17(–18) μm (av. = 16.7 ± 0.52 μm) long, wall verruculose, concolourous, but occasionally the two upper median cells are slightly darker than the lower median cell, brown, second cell from the base 3.5–6(–6.5) μm (av. = 5.2 ± 0.81 μm) long, third cell (3–)5–6(–6.5) μm (av. = 5.4 ± 0.35 μm) long, fourth cell (3.5–)5.5–6(–6.5) μm (av. = 5.7 ± 0.42 μm) long; apical cell 3.5–5(–6) μm (av. = 4.4 ± 0.57 μm) long, hyaline, subcylindrical, thinand smooth walled; with 2–3 tubular apical appendages (mostly three), arising from the apical crest, unbranched, filiform, variable in size, 6–24(–24.5) μm (av. = 16.4 ± 3.66 μm) long; basal appendage single, tubular, unbranched, centric, 5.5–8(–8.5) μm (av. = 6.7 ± 1.07 μm) long.
Culture characteristics — Colony on PDA with fluffy mycelium, panniform, aerial mycelium white, reverse white or light yellow, being yellow at the centre and white at the edge. Colony 60–66 mm diam in 15 d at 28 °C. Conidiomata globose, irregularly distributed on the medium surface, exuding globose, dark brown to black conidial masses.
Materials examined. China, Gansu Province, Tianshui City, Maiji District, Maiji Mountain, N34°20'35" E106°0'22", alt. 1509 m, on spines of R. chinensis, 18 Aug. 2020, C. Peng & S. Jia (BJFC-S1885, cultures CFCC 55156 = ROC 23-1, ROC 270, ROC 272, ROC 273, ROC 275); Gansu Province, Tianshui City, Maiji District, Maiji Mountain, N34°20'52" E106°0'16", alt. 1508 m, on spines of R. chinensis, 18 Aug. 2020, C. Peng & S. Jia (BJFC-S1886, cultures CFCC 55157 = ROC 23-2, ROC 276, ROC 278, ROC 279, ROC 280); Gansu Province, Tianshui City, Maiji District, Maiji Mountain, N34°20'17" E106°0'36", alt. 1500 m, on spines of R. chinensis, 19 Aug. 2020, C. Peng & S. Jia (BJFC-S1887, cultures ROC 281, ROC 282, ROC 283, ROC 286); ibid. (cultures ROC 289, ROC 290, ROC 292, ROC 293).
Notes — Pestalotiopsis chamaeropis was first described from leaves of Chamaerops humilis in Italy and subsequently reported on a wide range of hosts (e.g., Camellia sinensis, Pieris japonica, Prostanthera rotundifolia, Vandopsis spp. and Vitis vinifera) (Maharachchikumbura et al. 2014, Moslemi & Taylor 2015, Liu et al. 2017, Ran et al. 2017, Jayawardena et al. 2018, Nozawa et al. 2019, Wang et al. 2019b). In this study, 18 isolates clustered together with the ex-type culture of Pes. chamaeropis (CBS 186.71) in the multi-locus phylogenetic tree (Fig. 5). This is the first report of this fungus on R. chinensis. Compared with the description of the ex-type isolate CBS 186.71, CFCC 55156 has smaller conidiogenous cells (7–8.5 × 2–4 μm vs 20–50 × 2–5 μm).
Pestalotiopsis rhodomyrtus Y Song et al., Phytotaxa 126: 27. 2013 — Fig. 13
Fig. 13.

Pestalotiopsis rhodomyrtus (BJFC-S1888). a. Disease symptoms; b–c. appearance of conidiomata on host substrate; d. conidiomata on PDA; d–h. conidiogenous cells with attached conidia; i. conidia. — Scale bars: d = 200 μm; e–i = 10 μm.
Sexual morph not observed. Asexual morph: Acervular conidiomata globose or irregular. superficial to semi-immersed, scattered or aggregated, visible as black acervuli on the host, 86–99 μm. Conidiophores branched, subcylindrical, hyaline. Conidiogenous cells discrete, cylindrical or ampulliform, hyaline, smooth-walled, collarette present and not flared, with prominent periclinal thickening, (5–)6.5–10(–11.5) × (1.5–)2–3 μm (av. = 8.4 ± 1.35 × 2.7 ± 0.67 μm). Conidia fusoid, ellipsoid, straight to slightly curved, 4-septate, (17.5–)19–26 × (5–) 5.5–6.5(–7) μm (av. = 22.7 ± 1.21 × 5.9 ± 0.52 μm); basal cell obconic with a truncate base, hyaline, minutely verruculose and thin-walled, 3–5(–5.5) μm (av. = 4.2 ± 0.52 μm) long; three median cells doliiform to subcylindrical, (11–)12–18.5(–19.5) μm (av. = 14.9 ± 1.46 μm) long, wall verruculose, concolourous, pale brown, second cell from the base (3–)4.5–5.5 μm (av. = 4.8 ± 0.36 μm) long, third cell (3–)3.5–5(–5.5) μm (av. = 4.6 ± 0.54 μm), fourth cell 4–5.5(–6) μm (av. = 4.9 ± 0.38 μm); apical cell (3–)3.5–5.5 μm (av. = 4.4 ± 0.85 μm) long, hyaline, cylindrical to subcylindrical, thin and smooth walled; with 2–3 tubular apical appendages (mostly three), arising from the apical crest, unbranched, filiform, flexuous, (9–)11.5–20(–21.5) μm (av. = 15.7 ± 1.58 μm) long; one basal appendage, tubular, centric appendage tubular, branched or unbranched, (5.5–)6–7.5 μm (av. = 6.9 ± 1.74 μm) long.
Culture characteristics — Colonies on PDA flat with entire margin, aerial mycelium white, cottony, fluffy; reverse white in the centre and faint yellow margin, colony diam 59–63 mm diam in 15 d. Conidia in mass black. Conidiomata globose, irregularly distributed on the medium surface, exuding globose, black conidial masses.
Materials examined. China, Henan Province, Nanyang City, Neixiang county, Mahuang Village, N33°20'14" E114°52'1", alt. 266 m, on leaves of R. rugosa, 7 Aug. 2020, Y.M. Liang & C. Peng (BJFC-S1888, cultures ROC 056, ROC 057, ROC 058); ibid. (cultures ROC 059, ROC 060, ROC 061, ROC 062); Gansu Province, Gannan Tibetan Autonomous Prefecture, Lintan County, Yeliguan, N34°53'48" E103°35'2", alt. 2740 m, on spines of R. multiflora, 18 Aug. 2020, C. Peng & S. Jia (BJFC-S1889, cultures ROC 303, ROC 304, ROC 305, ROC 306); ibid. (cultures ROC 307, ROC 309, ROC 311); Hunan Province, Changsha City, Changsha County, N28°58'52" E113°34'38", alt. 61 m, on branches of R. chinensis, 10 Nov. 2020, C.M. Tian & N. Jiang (BJFC-S1890, cultures ROC 356, ROC 357, ROC 358, ROC 359).
Notes — Pestalotiopsis rhodomyrtus was initially described from Rhodomyrtus tomentosa in Guangxi Province, China (Song et al. 2013). In addition, this species was discovered on Camellia sinensis (Liu et al. 2017, Wang et al. 2019a). In this study, 18 isolates were identified as belonging to this species (Fig. 5) and this is the first report of this fungus on the host genus Rosa.
Compared with the description of the ex-type isolate HGUP 4230, isolate ROC 056 has longer apical and basal appendages (11.5–20 μm vs 7.5–14.9 μm and 6–7.5 μm vs 2.8–4.9 μm).
Pestalotiopsis tumida C. Peng & C.M. Tian, sp. nov. — MycoBank MB 843814; Fig. 14
Fig. 14.

Pestalotiopsis tumida (BJFC-S1891 holotype). a. Disease symptoms; b. appearance of conidiomata on host substrate; c–d. conidiomata on PDA; e. colonies on PDA at 3 d (left) and 15 d (right); f. conidiomata on Rosa chinensis; g–k. conidiogenous cells with attached conidia; l. conidia. — Scale bars: c–d = 200 μm; f–l = 10 μm.
Etymology. Name refers to the basal appendage that is occasionally swollen at the tip.
Typus. China, Gansu Province, Tianshui City, Maiji District, Maiji Mounta, N34°20'32" E106°0'29", alt. 1601 m, on spines of R. chinensis, 15 Aug. 2020, C. Peng & S. Jia (holotype BJFC-S1891, ex-type cultures CFCC 55158 = ROC 110, ROC 108, ROC 109).
Sexual morph not observed. Asexual morph: Acervular conidiomata globose or irregular. superficial to semi-immersed, scattered or aggregated, visible as black acervuli on the host. Conidiophores branched, subcylindrical, hyaline. Conidiogenous cells discrete, cylindrical or ampulliform, hyaline, smooth-walled, collarette present and not flared, with prominent periclinal thickening, (7–)8–11.5(–12) × (1.5–)2–4.5 μm (av. = 10.4 ± 2.23 × 2.9 ± 0.83 μm). Conidia fusoid, ellipsoid, straight to slightly curved, 4-septate, (19–)19.5–23.5(–24) × 6.5–7.5 μm (av. = 21.4 ± 1.33 × 6.9 ± 0.58 μm); basal cell obconic with a truncate base, hyaline, minutely verruculose and thin-walled, (2.5–)3–4.5 μm (av. = 3.9 ± 0.58 μm) long; three median cells doliiform to subcylindrical, (14–)14.5–18(–18.5) μm (av. = 16.6 ± 1.68 μm) long, wall verruculose, concolourous, light brown, second cell from the base (3.5–)4–5 μm (av. = 4.6 ± 0.36 μm) long, third cell (4–)4.5–6.5 μm (av. = 5.3 ± 0.68 μm), fourth cell (4.5–)5.5 μm (av. = 5.1 ± 0.34 μm); apical cell 4.5–6 μm (av. = 5.1 ± 0.49 μm) long, hyaline, cylindrical to subcylindrical, thinand smooth walled; with 2–3 tubular apical appendages (mostly three), arising from the apical crest, unbranched, filiform, flexuous, (10–)10.5–15.5(–16) μm (av. = 13.1 ± 2.07 μm) long; 1–2 basal appendages, tubular, centric appendage tubular, branched or unbranched, occasionally swollen at the tip, (6.5–)7–19(–19.5) μm (av. = 10.9 ± 3.44 μm) long, excentric appendage tubular, 6.5–7.5(–8) μm (av. = 7.3 ± 0.26 μm) long.
Culture characteristics — Colonies on PDA with aerial mycelium white, fluffy, reverse yellow pigment accumulation in the centre, surrounded by amber, pure white at the colony margin. Colony 56–60 mm diam in 15 d at 28 °C. Conidiomata globose, irregularly distributed on the medium surface, exuding globose, black conidial masses.
Additional materials examined. China, Gansu Province, Tianshui City, Maiji District, Maiji Mounta, N34°20'36" E106°0'23", alt. 1557 m, on branches and spines of R. chinensis, 15 Aug. 2020, C. Peng & S. Jia (BJFC-S1892, cultures CFCC 55159 = ROC 234, ROC 235, ROC 236, ROC 237); ibid. (cultures ROC 238, ROC 240).
Notes — Pestalotiopsis tumida formed a distinct clade (ML/ MP/BI = 100/100/1) in the multi-locus analyses and is sister to Pes. intermedia and Pes. linearis. Pestalotiopsis tumida (Fig. 5) can be distinguished from Pes. intermedia and Pes. linearis in ITS (two different unique fixed alleles in Pes. intermedia (531/533, 99.5 % with a single gap) and two in Pes. linearis (536/538, 99.5 % with no gaps)), TEF loci (six different unique fixed alleles in Pes. intermedia (526/532, 98.6 % with no gaps) and four in Pes. linearis (528/532, 99.2 % with no gaps)) and TUB loci (10 different unique fixed alleles in Pes. intermedia (443/553, 97.7 % with four gaps) and 18 in Pes. linearis (415/433, 95.7 % with three gaps)). Moreover, the conidial morphology of Pes. tumida is significantly different from these two species. Pestalotiopsis tumida differs from Pes. intermedia and Pes. linearis by shorter conidia (Pes. tumida: 19.5–23.5 μm vs Pes. intermedia: 24–28 μm, Pes. linearis: 24–33 μm). In addition, it is different from Pes. linearis in bearing shorter apical appendages (10.5–15.5 μm vs 24–33 μm) and longer basal appendages (7–19 μm vs 4–7 μm).
There are 11 species of Pestalotiopsis that have been recorded from Rosa, namely Pes. adusta, Pes. algeriensis, Pes. aquatica, Pes. lespedezae, Pes. longisetula, Pes. macrochaeta, Pes. maculans, Pes. oleandri, Pes. populi-nigrae, Pes. rosae and Pes. suffocata (Riley 1960, Guba 1961, Mathur 1979, Rai 1990, Zhu et al. 1991, Nag Raj 1993, Mendes et al. 1998, Sameva 2004, Wei et al. 2005, Kobayashi 2007, Ge et al. 2009). Pestalotiopsis tumida can be easily distinguished from Pes. adusta, Pes. algeriensis, Pes. aquatica, Pes. longisetula and Pes. oleandri by the colour of its median conidial cells. The median conidial cells of these five species are versicolourous while the three median conidial cells of Pes. tumida are concolourous. Pestalotiopsis tumida can be distinguished from other Rosa associated species by the size of its conidia and length of appendages (Table 4). Furthermore, the basal appendage of Pes. tumida is occasionally swollen at the tip, a unique morphological character that distinguishes it from other species that have been recorded from Rosa.
Table 4.
Synopsis of Pestalotiopsis species with concolourous median conidium cells from Rosa.
| Species | Conidia length (μm) | Conidia width (μm) | Middle cells length (μm) | Length of apical appendages (μm) | Length of basal appendages (μm) | Host | Country | References |
|---|---|---|---|---|---|---|---|---|
| Pestalotiopsis lespedezae | 20.2–25 | 7.5–8.8 | 13.8–17.5 | 15–27.5 | 5–6.3 | Rosa canina | India | Mathur (1979) |
| Pes. macrochaeta | 24.8–33 | 6.6–7.6 | 13.0–21.2 | 15.3–34.2 | 7.1–11.8 | Rosa sp. | Brazil | Mendes al. (1998) |
| Pes. maculans | 19–27.5 | 6–8 | 10–15.5 | 16–24 | 4.2–5 | Rosa sp. | Bulgaria | Sameva ((2004) |
| Pes. populi-nigrae | 21.2–28.3 | 7.1–8.3 | 14.2–16.5 | 21.2–31.9 | 2.4–7.1 | Rosa hybrida | Japan | Kobayashi (2007) |
| Pes. rosae | 20.5–28.8 | 5–6.3 | 11.8–16.3 | 10–25 | 4–5.3 | Rosa sp. | China | Ge et al. (2009) |
| Pes. suffocata | 20.8–28.6 | 5.8–7.1 | 13.6–18.8 | 15.6–35.1 | 1.3–3.5 | Rosa sp. | China | Ge et al. (2009) |
| Pes. tumida | 19.5–23.5 | 6.5–7.5 | 14.5–18 | 10.5–15.5 | 7–19 | Rosa chinensis | China | Present study |
* Newly described taxon is in bold.
Key to Pestalotiopsis species on Rosa spp.
1. Median conidial cells versicolourous . . . . . . . . . . . . . . . 2
1. Median conidial cells concolourous . . . . . . . . . . . . . . . . 6
2. Length of apical appendages less than 12 μm . . . . . . . . 3
2. Length of apical appendages more than 12 μm . . . . . . 5
3. Basal appendages less than 3.5 μm . . . . . . . . . . . . . . . 4
3. Basal appendages more than 3.5 μm . . Pes. algeriensis
4. Conidia not or slightly constricted at the septa, 16–22 × 5–7 μm, apical appendages 5–12 μm .... Pes. adusta
4. Conidia constricted at the septa, 18.9–28.3 × 4.7–7.1 μm, apical appendages 16.5–28.3 μm . . . . . . . Pes. oleandri
5. Conidia 20.6–28.2 × 7.2–8.2 μm, apical appendages 12.8–23 μm, basal appendages 2–4 μm . . . . . . Pes. aquatica
5. Conidia 20.6–25.7 × 6.4–7.7 μm, apical appendages 20.6–36 μm, basal appendages 5.1–7.7 μm . Pes. longisetula
6. Basal appendages less than 5 μm . . . . . . . . . . . . . . . . 7
6. Basal appendages more than 5 μm . . . . . . . . . . . . . . . . 9
7. Conidia 3–7.514.94-septate . . . . . . . . . . . . . . . . . . . . Pes. rosae
7. Conidia 4-septate . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 8
8. Conidia 21.8–28.3 × 6.6–8.3 μm, apical appendages 21.2–31.9 μm, basal appendages 1.5–3 μm Pes. populi-nigrae
8. Conidia 19–27.5 × 6–8 μm, apical appendages 16–24 μm, basal appendages 4.2–5 μm . . . . . Pes. maculans
9. Conidial length less than 26 μm . . . . . . . . . . . . . . . . . 10
9. Conidial length more than 26 μm . . . . . . . . . . . . . . . . . 12
10. Basal appendages occasionally swollen at the tip . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .Pes. tumida
10. Basal appendages not occasionally swollen at the tip 11
11. Conidia 19.7–26.3 × 4.9–6.7 μm, apical appendages 7.5–14.9 μm, basal appendages 2.8–4.9 μm . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Pes. rhodomyrtus
11. Conidia 20–25 × 7.5–8.5 μm, apical appendages 15–22.5 μm, basal appendages 3.7–7.5 μm Pes. lespedezae
12. Basal appendages less than 5.5 μm .... Pes. suffocata
12. Basal appendages more than 5.5 μm . . . . . . . . . . . . . . 13
13. On PDA, reverse of colonies cinnamon-rufous, conidia 24.8–33 × 6.6–7.6 μm, apical appendages 15.3–34.2 μm, basal appendages 7.1–11.8 μm . . . . . . . . . Pes. macrochaeta
13. On PDA, reverse of colonies white, conidia 26–27.5 × 7–8.5 μm, apical appendages 6–24 μm, basal appendages 5.5–8 μm . . . . . . . . . . . . . . . . . . . . . . Pes. chamaeropis
Seimatosporium centrale C. Peng & C.M. Tian, sp. nov. — MycoBank MB 843816; Fig. 15
Fig. 15.

Seimatosporium centrale (BJFC-S1893, holotype). a–b. Disease symptoms; c. appearance of conidiomata on host substrate; d. conidiomata on PDA; e. colonies on PDA at 3 d (left) and 15 d (right); f. longitudinal section through conidiomata; g–k. conidiogenous cells with attached conidia; l. conidia. — Scale bars: c–d = 200 μm; f = 20 μm; g–l = 10 μm.
Etymology. Name refers to conidiomata produced in the centre of the colonies on PDA.
Typus. China, Gansu Province, Tianshui City, Maiji District, Dongcha Town, N34°19'34" E106°34'27", alt. 1120 m, on spines of R. chinensis, 16 July 2019, C.M. Tian, Y.M. Liang & C. Peng (holotype BJFC-S1893, ex-type cultures CFCC 55166 = ROC 003, ROC 001, ROC 002).
Sexual morph not observed. Asexual morph: Acervular conidiomata solitary to gregarious, immersed to semi-immersed, unilocular, subglobose, visible as black acervuli on the spines of R. chinensis. Conidiophores branched at the base, hyaline. Conidiogenous cells discrete or integrated, cylindrical, subcylindrical or irregular, hyaline, smooth, thick-walled, (6–)7.5–21.5(–23.5) × (2–)2.5–3(–3.5) μm (av. = 14.4 ± 3.23 × 2.7 ± 0.35 μm). Conidia straight or slightly curved, fusoid, cylindrical or subcylindrical, concolourous, 3-septate, mostly constricted at the septa, transverse septa fairly thick, smooth-walled, (18.5–) 19.5–23.5(–24) × (5–)5.5–7 μm (av. = 20.9 ± 1.58 × 6.1 ± 0.52 μm); basal cell obconic with a truncate base, hyaline to pale brown, (4–)4.5–5.5(–6) μm (av. = 5.0 ± 0.64 μm) long; middle cells cylindrical, pale brown, second cell from base (4–)4.5–5.5 μm (av. = 5.0 ± 0.39 μm), third cell from base (4.5–)5–6(–6.5) μm (av. = 5.4 ± 0.58 μm), together (9.5–)10–11.5(–12) μm (av. = 10.7 ± 0.88 μm) long; apical cell conical, pale brown, (3.5–)4–5 μm (av. = 4.2 ± 0.32 μm); apical appendage single, unbranched, centric, occasionally excentric, 3.5–5.5(–7) μm (av. = 4.9 ± 0.67 μm) long; basal appendage single, unbranched, excentric, (3–)4.5–5(–6) μm (av. = 5.0 ± 0.27 μm) long; mean conidium length/width ratio = 3.40 : 1.
Culture characteristics — Colonies on PDA with aerial mycelium white, dense, reverse with tawny pigment accumulation, being darker at the centre and paler at the edge. Colony 56–58 mm diam in 15 d at 28 °C. Conidiomata are observed around 25 d and are mostly produced in the centre of the colony. Conidiomata globose, exuding black conidial masses.
Additional materials examined. China, Shaanxi Province, Baoji City, Fengxian County, Xinjia Mountain, N34°12'45" E106°36'25", alt. 1609 m, on spines of R. chinensis, 16 July 2019, C.M. Tian, Y.M. Liang & C. Peng (BJFC-S1894, cultures CFCC 55169 = ROC 014, ROC 015, ROC 016); Gansu Province, Tianshui City, Maiji District, Maiji Mountain, N34°20'44" E106°0'22", alt. 1601 m, on spines of R. chinensis, 15 Aug. 2020, C. Peng & S. Jia (BJFC-S1895, cultures ROC 145, ROC 146, ROC 147).
Notes — The nine isolates studied form a well-supported independent clade distinct from known Seimatosporium species (ML/MP/BI = 98/96/1). Seimatosporium centrale is most closely related to Seim. botan, Seim. discosioides, Seim. gracile sp. nov. and Seim. nonappendiculatum sp. nov. (Fig. 6). Among them, Seim. discosioides as pathogens occurring on some Rosaceae hosts, especially on Rosa spp. (Shoemaker 1964, Nag Raj 1993). A pairwise comparison of the ITS sequence showed seven nucleotide differences out of 557 nucleotides (98.7 % similarity, including two gaps) between Seim. centrale and Seim. discosioides, while LSU showed six nucleotide differences out of 794 nucleotides (99.2 % similarity, without gaps). Meanwhile, Seim. discosioides morphologically differs from our new species and other Seimatosporium species associated with Rosa by the absent of appendages. Seimatosporium centrale differs from Seim. botan in morphology, namely having wider conidia (5.5–7 μm vs 4–5 μm) and their mean conidium length/ width ratio is quite distinct (3.4 : 1 in Seim. centrale vs 4.6 : 1 in Seim. botan). In addition, Seim. botan has two types of conidia (with only basal appendage and both apical and basal appendages) while Seim. centrale has only one conidial type (both apical and basal appendages). Furthermore, Seim. centrale can be distinguished from Seim. gracile by its extremely high mean conidium length/width ratio (3.40 : 1 vs 6.19 : 1) and from Seim. nonappendiculatum by the colour of its basal cell (pale brown vs hyaline).
Besides Seim. discosioides, four species of Seimatosporium have been recorded from Rosa, namely Seim. caninum, Seim. pseudorosae, Seim. salicinum and Seim. rosae (Nag Raj 1993, Kobayashi 2007, Mulenko et al. 2008, Norphanphoun et al. 2015, Hyde et al. 2016). Seimatosporium centrale has larger conidia than Seim. pseudorosae and Seim. rosae (19.5–23.5 × 5.5–7 μm vs 12–17.5 × 3–6 μm and 19.5–23.5 × 5.5–7 μm vs 10–16 × 3–4.5 μm). However, the apical appendages of Seim. centrale are noticeably shorter than those of the two species (3.5–5.5 μm vs 8–25 μm and 3.5–5.5 μm vs 1.5–15 μm). Seimatosporium caninum and Seim. salicinum can be distinguished from Seim. centrale based on the number of conidial septa. While Seim. centrale has 3-septate conidia, Seim. salicinum has 2–3-septate and Seim. caninum only has 2-septate conidia (Wanasinghe et al. 2018).
Seimatosporium gracile C. Peng & C.M. Tian, sp. nov. — MycoBank MB 843818; Fig. 16
Fig. 16.

Seimatosporium gracile (BJFC-S1896, holotype). a. Appearance of conidiomata on host substrate; b. colonies on PDA at 3 d (left) and 15 d (right); c. conidiomata formed on pine needles; d–k. conidiogenous cells with attached conidia; l. conidia. — Scale bars: c = 200 μm; d–l = 10 μm.
Etymology. From Latin ‘gracile’ means slender/slim, which refers to the shape of the conidia.
Typus. China, Gansu Province, Tianshui City, Qingshui county, Shidong Mountain, N34°41'4" E106°21'34", alt. 1697 m, on spines of R. xanthina, 17 July 2019, C.M. Tian, Y.M. Liang & C. Peng (holotype BJFC-S1896, ex-type cultures CFCC 55167 = ROC 004, ROC 005, ROC 006).
Sexual morph not observed. Asexual morph: Acervular conidiomata solitary to gregarious, immersed when immature, becoming erumpent at maturity, subglobose, up to 95–337 μm diam, 56–104 μm high, visible as black conidiomata on the spines. Conidiophores irregularly branched, mostly branched at the base, 1–3-septate, hyaline. Conidiogenous cells discrete or integrated, ampulliform, cylindrical or subcylindrical, hyaline, smooth, thick-walled, variable in size, (7–)8.5–21.5(–23) × 1.52(–3) μm (av. = 15.6 ± 3.23 × 1.6 ± 0.18 μm). Conidia straight or slightly curved, cymbiform or fusoid, concolourous, 3-septate, barely constricted at the septa, transverse septa dark brown and fairly thick, smooth-walled, (13.5–)14.5–19.5(–20) × 2.5–3(–3.5) μm (av. = 16.7 ± 1.82 × 2.7 ± 0.25 μm); basal cell obconic with or without a truncate base, pale brown to hyaline, (2.5–)3–4(–4.5) μm (av. = 3.6 ± 0.47 μm) long; middle cells cylindrical, pale brown, second cell from base (3–)3.5–4.5 μm (av. = 3.9 ± 0.37 μm), third cell from base 3.5–4(–4.5) μm (av. = 3.8 ± 0.27 μm), together 7–8(–9) μm (av. = 7.7 ± 0.43 μm) long; apical cell conical, pale brown, (2.5–)3–3.5(–4.5) μm (av. = 3.2 ± 0.28 μm); apical appendage single, unbranched, centric, (3–)4–8(–8.5) μm (av. = 6.3 ± 1.58 μm) long; basal appendage single, unbranched, excentric, 4.5–8.5(–9) μm (av. = 6.1 ± 1.22 μm) long; mean conidium length/width ratio = 6.19 : 1.
Culture characteristics — Colonies on PDA flat with irregular margin, colony yellow or pale yellow in the centre with fluffy aerial mycelia and pale white margin; reverse tawny pigment accumulation formed in the shape of a concentric ring pattern. Colony 56–58 mm diam in 15 d at 28 °C. Conidiomata sparse, concentrically and irregularly distributed on the medium surface.
Additional materials examined. China, Gansu Province, Tianshui City, Qingshui county, Shidong mountain, N34°41'4" E106°21'35", alt. 1699 m, on spines of R. xanthina, 17 July 2019, C.M. Tian, Y.M. Liang & C. Peng (BJFC-S1897, cultures ROC 007, ROC 008, ROC 009); ibid. (cultures ROC 010, ROC 011, ROC 012).
Notes — Seimatosporium gracile forms an independent clade (ML/MP/BI = 100/100/1) and is phylogenetically distinct from Seim. nonappendiculatum (described below) (Fig. 6). A comparison of the ITS region showed 0.35 % differences (two bp difference of 566 bp, with one single gap), 0.23 % bp differences (2 bp difference of 846 bp, with no gaps) in the LSU region, 2.1 % bp differences (7 bp difference of 326 bp, with no gaps) in TEF, 2.4 % bp differences (11 bp difference of 464 bp, with no gaps) in TUB and 0.38 % bp differences (4 bp difference of 1051 bp, with no gaps) in RPB2, which is evidence for new species rank. Moreover, the two species are morphologically distinct. Seimatosporium gracile differs from Seim. nonappendiculatum in the colour of its basal cell and presence or absence of appendages. While Seim. gracile has a pale brown basal cell with both apical and basal appendages, Seim. nonappen diculatum has a hyaline basal cell with no appendages or only basal appendage. Furthermore, the typical characteristic of Seim. gracile is that the conidia are slender and has a markedly higher mean conidial length/width ratios (6.19 : 1) than other species that have been recorded from Rosa in this genus.
Seimatosporium nonappendiculatum C. Peng & C.M. Tian, sp. nov. — MycoBank MB 843820; Fig. 17
Fig. 17.

Seimatosporium nonappendiculatum (BJFC-S1901, holotype). a–b. Disease symptoms; c. appearance of conidiomata on host substrate; d. conidiomata formed on PDA; e. longitudinal section through conidiomata; f–m. conidiogenous cells with attached conidia; n. conidia. — Scale bars: d = 200 μm; e = 20 μm; f–n = 10 μm.
Etymology. Referring to conidia lacking an apical appendage.
Typus. China, Ningxia Hui Autonomous Region, Guyuan City, Jingyuan County, N35°23'53" E106°22'33", alt. 1786 m, on fruits of R. laevigata, 29 Aug. 2020, C. Peng (holotype BJFC-S1901, ex-type cultures CFCC 55168 = ROC 377, ROC 378).
Symptoms are marked by slightly collapsed lesions, with regular margin, black dark brown, round spots on fruits (Fig. 19a–c). Sexual morph not observed. Asexual morph: Acervular conidiomata solitary to gregarious, subglobose, globose or irregular, up to 287–436 μm diam, 135–160 μm high. Conidiophores cylindrical, branched at the base, 1–3-septate, hyaline, smooth-walled. Conidiogenous cells discrete or integrated, ampulliform, cylindrical or subcylindrical, hyaline, smooth, thick-walled, variable in size, (5–)6.5–13.5(–16) × (1.5–)2–2.5(–3) μm (av. = 9.9 ± 2.21 × 2.3 ± 0.25 μm). Conidia straight or curved, fusoid or pyriform, 3-septate, distoseptate or euseptate, with or without constrictions at the septa, transverse septa dark brown and fairly thick, smooth-walled, (17–)18–23.5(–24) × (5–)5.5–7 μm (av. = 20.3 ± 1.47 × 6.4 ± 0.52 μm); basal cell obconic with a truncate base, hyaline or almost hyaline, (3–)3.5–4.5 μm (av. = 3.9 ± 0.39 μm) long; middle cells cylindrical, pale to medium brown, second cell from base 3–5(–5.5) μm (av. = 4.3 ± 0.32 μm), third cell from base (3.5–)4–4.5(–5) μm (av. = 4.3 ± 0.44 μm), together (7–)8–10(–11) μm (av. = 8.7 ± 0.32 μm) long; apical cell obtuse or conical, pale brown, (3–)4.5–5.5 μm (av. = 4.8 ± 0.31 μm); apical appendage absent; basal appendage lacking or, when present, single, unbranched, excentric, 1.5–4 μm (av. = 2.9 ± 0.93 μm) long; mean conidium length/ width ratio = 3.17 : 1.
Fig. 19.

Asexual morph of Seimatosporium parvum (BJFC-S1898, holotype). a. Appearance of conidiomata on host substrate; b. appearance of conidiomata on host substrate; c. conidiomata formed on pine needles; d. colonies on PDA at 3 d (left) and 15 d (right); e–l. conidiogenous cells with attached conidia; m. conidia. — Scale bars: c = 200 μm; e–m = 10 μm.
Culture characteristics — Colonies on PDA flat with entire margin, colony yellow or pale yellow in the centre with fluffy aerial mycelia and pale white margin; reverse with yellow pigment. Colony 50–52 mm diam in 15 d at 25 °C. Conidiomata sparse, concentrically and irregularly distributed on the medium surface.
Additional material examined. China, Ningxia Hui Autonomous Region, Guyuan City, Jingyuan County, N35°23'53" E106°22'36", alt. 1788 m, on fruits of R. laevigata, 29 Aug. 2020, C. Peng (BJFC-S1902, cultures ROC 379, ROC 380, ROC 381).
Notes — Seimatosporium nonappendiculatum forms an independent clade (ML/MP/BI = 89/93/1) and is phylogenetically distinct from Seim. gracile (Fig. 6). The differences between Seim. nonappendiculatum and Seim. gracile have been mentioned above (see Seim. gracile).
Seimatosporium nonappendiculatum is characterised by its fusoid to pyriform, 3-septate conidia with a hyaline basal cell, lacking an apical appendage or with an excentric basal appendage. These morphological characteristics can easily distinguish Seim. nonappendiculatum from other Seimatosporium species previously reported from Rosa. Seimatosporium botan which was collected from Paeonia suffruticosa in Japan also has two types of conidia (Hatakeyama & Harada 2004). Seimatosporium botan can produce conidia with only one basal appendage which morphologically resembles our new Seimatosporium species, but Seim. botan has longer basal appendages (4–8 μm vs 1.5–4 μm) and larger mean conidium length/width ratio (4.6 : 1 vs 3.17 : 1). The other conidial type of Seim. botan bears appendages at both ends, which is very different from the conidia of Seim. nonappendiculatum. Besides Seim. botan, Seim. grammitum, Seim. hebeiense and Seim. hysterioides produce conidia with only a basal appendage (Shoemaker 1964, Hatakeyama & Harada 2004). Seimatosporium nonappendiculatum can be distinguished from these three species by its larger conidia (18–23.5 × 5.5–7 μm vs 12–18.5 × 3.5–5.5 μm, 18–23.5 × 5.5–7 μm vs 13.5–17.6 × 4.5–6.5 μm and 18–23.5 × 5.5–7 μm vs 12–18 × 5.5–6.5 μm). Seimatosporium pseudoglandigenum produces conidia without appendages which morphologically resemble the other conidial type of Seim. nonappendiculatum (Wijayawardene et al. 2016a). However, the conidia of Seim. pseudoglandigenum only has one conidial type. Although Seim. nonappendiculatum is similar to Seim. pseudoglandigenum in conidial size (18–23.5 × 5.5–7 μm vs 15–23 × 5–8 μm), Seim. pseudoglandigenum can be distinguished from our new species based on the colour of its apical conidial cells. While Seim. nonappendiculatum has pale brown apical cells and the apical cell is heterochromatic to the basal cells, Seim. pseudoglandigenum has hyaline apical cells and the apical cell is concolourous to the basal cells (Wijayawardene et al. 2016a).
Seimatosporiumparvum C. Peng & C.M. Tian, sp. nov. — MycoBank MB 843819; Fig. 18, 19
Fig. 18.

Sexual morph of Seimatosporiumparvum (BJFC-S1898, holotype). a. Disease symptoms; b. appearance of ascomata on host substrate; c–d. longitudinal section through ascomata; e. asci and pseudoparaphyses; f–g. asci; h–i. ascospores. — Scale bars: c–d = 50 μm; f–k = 10 μm.
Etymology. From Latin parva ‘small’, referring to smaller conidia.
Typus. China, Qinghai Province, Huangnan Tibetan Autonomous Prefecture, Zeku County, N37°18'51" E101°56'34", alt. 2890 m, on spines of R. spinosissima, 12 July 2019, C.M. Tian, Y.M. Liang & C. Peng (holotype BJFC-S1898, ex-type cultures CFCC 55164 = ROC 038, ROC 039, ROC 040).
Sexual morph: Ascomata 52–155 μm high × 102–254 μm diam, black, immersed, solitary or gregarious, fully or partly erumpent, globose, uniloculate. Ostiole 6.5–16.5 μm, single, dark grey to black. Peridium 22–68 μm wide, comprising 2 layers, outer most layer heavily pigmented, thick-walled, comprising dark brown cells of textura angularis, inner layer composed of hyaline, flattened cells of textura angularis. Pseudoparaphyses numerous, 1.6–2.7 μm wide, filamentous, branched. Asci (86.5–) 91.599(–102.5) × (9–)10–11(–12.5) μm (av. = 96.7 ± 4.13 × 10.6 ± 1.16 μm), 8-spored, bitunicate, fissitunicate, clavate to cylindrical. Ascospores (21–)23.5–29(–30.5) × (3–)4.5–6(–7) μm (av. = 26.4 ± 2.21 × 5.4 ± 1.01 μm), overlapping biseriate, fusiform with narrow ends, mostly curved, 4–5-septate, constricted at the central septum, cells above central septum swollen, (3.5–)4.5–5.5(–6) × (4–)4.5–6 μm (av. = 5.1 ± 0.37 × 5.5 ± 1.02 μm). Asexual morph: Acervuli conidiomata solitary to gregarious, immersed when immature, becoming erumpent at maturity, unilocular, subglobose, up to 133–262 μm diam, 80–121 μm high, visible as black acervuli on the spines of R. spinosissima. Conidiophores cylindrical, branched at the base, smooth. Conidiogenous cells discrete or integrated, ampulliform, cylindrical or subcylindrical, hyaline, smooth, thick-walled, variable in size, (7–)9.5–24.5(–26.5) × (1–)1.5–2(–2.5) μm (av. = 16.5 ± 3.23 × 1.7 ± 0.97 μm). Conidia straight or slightly curved, cylindrical or subcylindrical, concolourous, 3-septate, without constrictions at the septa, transverse septa dark brown and fairly thick, smooth-walled, (10–)11–13.5(–14) × (2–)2.5–3.5(–4) μm (av. = 12.3 ± 0.51 × 2.9 ± 0.25 μm); basal cell obconic with a truncate base, pale brown to brown, (2–)2.5–3.5(–4) μm (av. = 2.6 ± 0.32 μm) long; middle cells cylindrical, pale brown to medium brown, second cell from base 2–3(–3.5) μm (av. = 2.7 ± 0.27 μm), third cell from base 2–3(–4) μm (av. = 2.4 ± 0.36 μm), together (5.5–)6–7(–8) μm (av. = 6.4 ± 0.41 μm) long; apical cell obtuse or conical, pale brown, (2–) 2.5–3.5 μm (av. = 3.1 ± 0.31 μm); apical appendage single, unbranched, centric, (17–)19.5–22.5(–23) μm (av. = 21.3 ± 2.13 μm) long; basal appendage single, unbranched, excentric, (18–)19.5–28.5(–30) μm (av. = 25.8 ± 2.32 μm) long; mean conidium length/width ratio = 3.95 : 1.
Culture characteristics — Colonies on PDA flat with entire margin, colony yellow or pale yellow in the centre with fluffy aerial mycelia and pale white margin; reverse with yellow pigment. Colony 54–58 mm diam in 15 d at 28 °C. Conidiomata sparse, concentrically and irregularly distributed on the medium surface.
Additional materials examined. China, Qinghai Province, Huangnan Tibetan Autonomous Prefecture, Zeku County, N37°18'60" E101°56'43", alt. 2899 m, on spines of R. spinosissima, 12 July 2019, C.M. Tian, Y.M. Liang & C. Peng (BJFC-S1899, cultures ROC 041, ROC 042, ROC 043); ibid., N37°18'56" E101°56'09", alt. 2910 m, on branches of R. helenae, 13 July 2019, C.M. Tian, Y.M. Liang & C. Peng (BJFC-S1900, cultures CFCC 55165 = ROC 017, ROC 018, ROC 019, ROC 020).
Notes — Seimatosporium parvum is most closely related to Seim. pseudorosae which has also been reported from Rosa (Hyde et al. 2016) (Fig. 6). Pairwise comparison of the LSU sequence data reveals 7 bp difference from 668 (98.7 %, without gap region) between this species and Seim. pseudorosae.
Moreover, morphological differences between Seim. parvum and Seim. pseudorosae are obvious, namely Seim. parvum have smaller conidia (11–13.5 × 2.5–3.5 μm vs 12–17.5 × 4–6 μm). Furthermore, the basal appendages of Seim. parvum are longer than those of Seim. pseudorosae (19.5–28.5 μm vs 6–15 μm).
In addition to Seim. pseudorosae, Seim. parvum is separated from Seim. rosae which has also been reported from Rosa based on phylogenetic analysis (Norphanphoun et al. 2015) (Fig. 6). Furthermore, Seim. parvum differs from other Seimatosporium species associated with Rosa in producing thinner conidia (Seim. parvum: 2.5–3.5 μm vs Seim. salicinum: 4–6 μm and Seim. rosae: 3–4.5 μm) and having longer apical appendage (Seim. parvum: 19.5–22.5 μm vs Seim. salicinum: 9–14 μm and Seim. rosae: 5–10 μm). Although Seim. caninum is similar to Seim. parvum in conidial dimensions (9.5–12 × 4–5 μm vs 11–13.5 × 2.5–3.5 μm), the conidia of this species only have two septa and lack appendages, which make Seim. caninum easily distinguishable from other species of Seimatosporium (Nag Raj 1993).
Key to Seimatosporium species on Rosa spp.
1. Conidia lacking appendages . . . . . . . . . . . . . . . . . . . . . . . 2
1. Conidia with appendages at both ends . . . . . . . . . . . . . . . 3
2. Conidia 2-septate . . . . . . . . . . . . . . . . . . . . Seim caninum
2. Conidia 3-septate . . . . . . . . . . . . . . . . . Seim. discosioides
3. Appendages less than 8 μm . . . . . . . . . . . . . . . . . . . . . . . 4
3. Appendages more than 8 μm . . . . . . . . . . . . . . . . . . . . . 7
4. Conidia with appendages at both ends . . . . . . . . . . . . . . . 5
4. Conidia without apical appendage, with or without basal appendage . . . . . . . . . . . . . . . . Seim. nonappendiculatum
5. Conidial length more than 15 μm . . . . . . . . . . . . . . . . . . . 6
5. Conidial length less than 15 μm . . . . . . . . . . . Seim. rosae
6. Mean conidium length/width ratio = 3.40 : 1, conidia 19.5–23.5 × 5.5–7 μm . . . . . . . . . . . . . . . . . . . . . Seim. centrale
6. Mean conidium length/width ratio = 6.19 : 1, conidia 14.5–19.5 × 2.5–3 μm . . . . . . . . . . . . . . . . . . . . . . Seim. gracile
7. Mean conidium length/width ratio less than 6 . . . . . . . . . 8
7. Mean conidium length/width ratio more than 6 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Seim. parvum
8. Conidia 3-septate, 12–17.5 × 3–6 μm Seim. pseudorosae
8. Conidia 2–3-septate, 11–17 × 4–6 μm . . . Seim. salicinum
Seiridium rosae C. Peng & C.M. Tian, sp. nov. — MycoBank MB 843821; Fig. 20
Fig. 20.

Seiridium rosae (BJFC-S1903, holotype). a–b. Disease symptoms c. appearance of conidiomata on host substrate; d. conidiomata on PDA; e. colonies on PDA at 3 d (left) and 15 d (right); f–g. conidiomata on Rosa rugosa; h–n. conidiogenous cells with attached conidia; o. conidia. — Scale bars: c–d = 200 μm; h–o = 10 μm.
Etymology. The epithet reflects the name of the host plant genus Rosa.
Typus. China, Ningxia Hui Autonomous Region, Guyuan City, Jingyuan County, N35°18'52" E106°21'53", alt. 2157 m, on branches of R. rugosa, 27 Aug. 2020, C.M. Tian, Y.M. Liang & C. Peng (holotype BJFC-S1903, ex-type cultures CFCC55174 = ROC 208, ROC 209).
Symptoms appeared as elongate and ovoid, red or dark red, raised, dehiscent lesions on twigs or branches (Fig. 20a–c). Sexual morph not observed. Asexual morph: Acervular conidiomata irregularly scattered over the surface, solitary to gregarious, immersed when immature, erumpent from tissue at maturity, subglobose to globose and occasionally irregular. Conidiophores septate, cylindrical, irregularly branched, mostly branched at the base. Conidiogenous cells discrete, hyaline, subcylindrical to cylindrical, smooth-and thin-walled, the size varies tremendously, (8–)12–64.5(–70) × (1–)1.5–2(–3) μm (av. = 27.8 ± 15.43 × 1.7 ± 0.16 μm). Conidia fusoid, straight or slightly curved, 5-septate, not striate, distoseptate without pores, bearing one or two basal and one or more apical appendages, (29–)31–35(–36.5) × (7–)8–9.5 μm (av. = 32.6 ± 0.83 × 8.8 ± 0.47 μm); basal cell obconic, hyaline, smooth-walled, with marginal frill, (3–)3.5–4.5 μm (av. = 4.1 ± 0.63 μm); four median cells brown, smooth-walled, cylindrical to doliiform; second cell from base (4–)5.5–6.5(–7) μm (av. = 6.1 ± 0.21 μm); third cell (3–)4–5.5(–6) μm (av. = 4.9 ± 0.52 μm); fourth cell (3–) 3.5–5.5(–6.5) μm (av. = 4.7 ± 0.68 μm), fifth cell (4–)6–7.5 μm (av. = 6.5 ± 0.48 μm), together (18–)20.5–24.5(–26) μm (av. = 22.8 ± 2.65 μm); apical cell conical, hyaline, smooth-walled, (3–)3.5–5.5(–7) μm (av. = 4.1 ± 1.05 μm) long; apical appendages single or multiple, centric, branched or unbranched, occasionally swollen at the tip, (27–)28.5–44.5(–46) μm (av. = 35.3 ± 4.68 μm); 1–2 basal appendages (mostly two), cylindrical, centric, occasionally excentric, unbranched or branched, (2–)3–4.5 μm (av. = 3.7 ± 0.55 μm).
Culture characteristics — Colony on PDA with flattened mycelium, white, smoke grey in the centre, reverse with pale yellow pigments formed in concentric pattern. Colony 36–40 mm diam in 15 d at 25 °C, with concentrically distributed conidiomata.
Additional materials examined. China, Ningxia Hui Autonomous Region, Guyuan City, Jingyuan County, N35°18’52” E106°21’55”, alt. 2158 m, on branches of R. rugosa, 27 Aug. 2020, C.M. Tian, Y.M. Liang & C. Peng (cultures CFCC 55175 = ROC 267, ROC 268).
Notes — Seiridium rosae clustered in a well-supported independent clade (ML/MP/BI = 100/100/1) closely related to Seir. aquaticum and Seir. venetum (Fig. 7). Seiridium rosae can be distinguished from Seir. venetum in ITS (19 bp difference from 463 characters, with 95.8 % similarity, including 28 gaps) and TUB (7 bp difference from 365 characters, with 98.1 % similarity, including no gaps) sequences. Pairwise comparison of the ITS sequence data reveals 49 bp difference from 526 (90.6 % similarity, including 10 gaps) between our new species and the Seir. aquaticum. Seiridium rosae is distinct from Seir. aquaticum by its branched appendages, while Seir. aquaticum has conidia bearing single, short and unbranched appendage (Luo et al. 2019). Additionally, Seir. rosae has thinner conidia (8–9.5 μm vs 12–14 μm) and longer basal appendages (the basal appendages of Seir. aquaticum reduced to marginal frills) than Seir. aquaticum (Luo et al. 2019). Seiridium venetum can also produce conidia with branched apical appendages (Nag Raj 1985), but this species can be distinguished from Seir. rosae based on conidial dimensions (31–35 × 8–9.5 μm vs 20–30 × 6.5–8.5 μm). Seiridium rosae morphologically resembles Seir. pezizoides (basionym: Pestalotia pezizoides) which was isolated from Vitis sp. in Italy and the USA having 5-distoseptate conidia with one or more, branched or unbranched apical appendages (Nag Raj 1985). However, Seir. rosae differs from Seir. pezizoides in having longer apical appendages (28.5–44.5 μm vs 8.5–27 μm) and shorter basal appendages (3–4.5 μm vs 5.5–14 μm). Besides, the conidia of Seir. rosae mostly bear two basal appendages while Seir. pezizoides only has one basal appendage (Nag Raj 1985, Marin-Felix et al. 2019). Seiridium rosae is also morphologically distinct from other Seridium species associated with Rosa (i.e., Seir. marginatum and Seir. rosarum) (Jaklitsch et al. 2016, Wanasinghe et al. 2018). Seiridium rosae differs from Seir. marginatum in having smaller conidia (31–35 × 8–9.5 μm vs 38–42 × 8.8–10.2 μm), and differs from Seir. rosarum in having larger conidia (31–35 × 8–9.5 μm vs 22–28 × 7–9 μm) and longer conidiogenous cell (12–64.5 μm vs 5–20 μm) (Jaklitsch et al. 2016, Wanasinghe et al. 2018).
Key to Seiridium species on Rosa spp.
1. Conidia with one or more, branched or unbranched apical appendages . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Seir. rosae
1. Conidia with one unbranched apical appendage . . . . . . . 2
2. Conidia 38.2–42 × 8.8–10.2 μm, appendages up to 52 μm long . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Seir. marginatum
2. Conidia 22–28 × 7–9 μm, appendages up to 12 μm long . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Seir. rosarum
Sporocadus brevis C. Peng & C.M. Tian, sp. nov. — Myco-Bank MB 843822; Fig. 21
Fig. 21.

Sporocadus brevis (BJFC-S1904, holotype). a. Disease symptoms; b–c. appearance of conidiomata on host substrate; d. conidiomata formed on pine needles; e. longitudinal section through conidiomata; f. colonies on PDA at 3 d (left) and 15 d (right); g–m. conidiogenous cells with attached conidia; n. conidia. — Scale bars: c–d = 200 μm; e = 50 μm; g–n = 10 μm.
Etymology. The Latin ‘brevis’ meaning short, which refers to the conidial size.
Typus. China, Gansu Province, Gannan Tibetan Autonomous Prefecture, Lintan County, Yeliguan, N34°53'43" E103°35'2", alt. 2808 m, on spines of R. spinosissima, 21 Aug. 2020, C. Peng & S. Jia (holotype BJFC-S1904, ex-type cultures CFCC 55170 = ROC 091, ROC 092, ROC 093).
Sexual morph not observed. Asexual morph: Acervular conidiomata solitary to gregarious, mostly irregular or subglobose, erumpent through the surface of bark when mature, up to 187–251 μm diam, visible as black acervuli on the spines. Conidiophores septate, branched at base, smooth, hyaline, mostly reduced to conidiogenous cells. Conidiogenous cells discrete or integrated, lageniform, filiform or ampulliform, sometimes irregular, hyaline, smooth, without annellations, (2.5–)3.5–19(–21) × 1–2(–2.5) μm (av. = 8.2 ± 3.62 × 1.6 ± 0.24 μm). Conidia straight or slightly curved, fusoid or obovoid, pale brown, mostly 2-septate, rarely 3-septate, septa fairly thick-walled and darker than the rest of the cell, wall smooth and occasionally slightly constricted at the septa, (9–)10–12(–12.5) × (6–)6.5–7(–7.5) μm (av. = 11.3 ± 0.74 × 6.4 ± 0.22 μm), lacking appendages; basal cell obconic with or without a truncate base, hyaline to pale brown, thick-walled, shorter than other cells, 1.5–3.5(–4) μm (av. = 2.4 ± 0.47 μm) long; median cells doliiform, each (2–)3–4.5(–5) μm (av. = 3.7 ± 0.43 μm) long, pale to mid-brown; apical cell (2.5–)3–5(–5.5) μm (av. = 4.2 ± 0.68 μm), short conic with a wide round apex, concolourous with the median cell in 2-septate conidia paler than median cells in 3-septate conidia; mean conidium length/width ratio = 1.76 : 1.
Culture characteristics — Cultures on PDA with aerial mycelium white, fluffy, reverse with a mottled tawny pigment. Colony 54–56 mm diam in 15 d at 28 °C. Conidiomata sparse and distributed irregularly on the medium surface.
Additional material examined. China, Gansu Province, Gannan Tibetan Autonomous Prefecture, Lintan County, Yeliguan, N34°53'48" E103°35'2", alt. 2806 m, on spines of R. spinosissima, 21 Aug. 2020, C. Peng & S. Jia (BJFC-S1905, cultures ROC 094, ROC 095).
Notes — Sporocadus brevis is introduced based on multigene phylogenetic analysis, with five isolates clustering separately in a well-supported clade (ML/MP/BI = 100/100/1) (Fig. 8). Sporocadus brevis is most closely related to Spo. trimorphus, which was isolated from R. canina in Sweden (Liu et al. 2019), but distinguished based on ITS, RPB2, TEF and TUB loci from Spo. trimorphus by 34 bp in the concatenated alignment, in which 2 bp are distinct in the ITS region (from 506 characters, with 99.6 % sequence identity, including 1 gap), 5 bp in the RPB2 region (from 832 characters, with 99.3 % sequence identity, no gaps), 11 bp in the TEF region (from 468 characters, with 97.6 % sequence identity, including 1 gap) and 16 bp in the TUB region (from 702 characters, with 97.7 % sequence identity, including 1 gap). Morphologically, Spo. trimorphus can produce conidia with appendages but the conidia of Spo. brevis lack appendages (Liu et al. 2019). Moreover, conidia of Spo. brevis are wider than those of Spo. trimorphus (6.5–7 μm vs 3–4.5 μm).
In addition to Sporocadus trimorphus, Spo. lichenicola, Spo. rosarum and Spo. rosigena are also associated with Rosa spp. (Wanasinghe et al. 2018, Liu et al. 2019). Sporocadus brevis can be easily distinguished from these species by the conidial length (10–12 μm in Spo. brevis vs 18–25 μm in Spo. lichenicola) and width (6.5–7 μm in Spo. brevis vs 4–6 μm in Spo. rosarum, 3.5–6.5 μm in Spo. rosigena). Furthermore, the mean conidium length/width ratio of Spo. brevis is smaller than these four species (Spo. brevis: 1.76 : 1 vs Spo. lichenicola: 3 : 1, Spo. rosarum: 2.24 : 1, Spo. rosigena: 2.4 : 1 and Spo. trimorphus: 3.4 : 1).
Sporocadus sorbi (Wijayaw. et al.) F. Liu et al., Stud. Mycol. 92: 404. 2019 — Fig. 22
Fig. 22.

Sporocadus sorbi (BJFC-S1906). a–b. Appearance of conidiomata on host substrate; c. conidiomata on PDA; d–g. conidiogenous cells with attached conidia; h. conidia. — Scale bars: c = 200 μm; d–h = 10 μm.
Sexual morph not observed. Asexual morph: Acervular conidiomata solitary to gregarious, mostly subglobose, immersed to semi-immersed, up to 390–420 μm diam, 121–130 μm high, visible as black or dark red acervuli on the stems. Conidiophores septate, mostly branched at base, hyaline, smooth.
Conidiogenous cells discrete or integrated, sub-cylindrical or lageniform, variable in size, hyaline, smooth, with annellations, (7–)12–43.5(–48) × 1.5–2(–2.5) μm (av. = 17.2 ± 9.78 × 1.9 ± 0.34 μm). Conidia fusoid or obovoid, straight, 3-septate, transverse septa brown to dark brown, slightly constricted at the septa, wall smooth, (9.5–)11–15.5(–17) × (3–)3.5–5.5(–6) μm (av. = 13.7 ± 1.54 × 4.3 ± 0.21 μm), lacking appendages; basal cell obconic with or without a truncate base, hyaline to pale grey, 2–3(–3.5) μm (av. = 2.7 ± 0.38 μm) long; median cells mostly two, cylindrical, fairly thick-walled and pale brown, each 3–4.5(–5) μm (av. = 4.1 ± 0.54 μm) long; apical cell conic, hyaline or concolourous with median cells, 2–3(–4) μm (av. = 2.6 ± 0.32 μm) long; mean conidium length/width ratio = 3.18 : 1.
Culture characteristics — Colony on PDA with fluffy aerial mycelium, panniform, aerial mycelium white to pale brown, reverse umber coloured, being darker at the centre and paler at the edge. Colony 36–42 mm diam in 15 d at 25 °C. Conidio-mata black, distributed circularly at the colony margin on the medium surface.
Materials examined. China, Gansu Province, Lanzhou City, Yuzhong County, Xinglong Mountain, N35°48'2" E104°3'57", alt. 2160 m, on branches of R. xanthina, 26 Aug. 2020, C. Peng (BJFC-S1906, cultures ROC 102, ROC 103, ROC 105); Gansu Province, Gannan Tibetan Autonomous Prefecture, Lintan County, Yeliguan, N34°53'45" E103°35'2", alt. 2806 m, on spines of R. xanthina, 21 Aug. 2020, C. Peng & S. Jia (BJFC-S1907, cultures ROC 159, ROC 160, ROC 161).
Notes — Sporocadus sorbi was first reported from a dead leaf of Sorbus torminalis in Italy (Lawrence et al. 2018). In this study, six isolates of Sporocadus were identified as belonging to this species, and this is the first report of this fungus on Rosa plants, and in China.
Conidia of the ex-type (MFLUCC 14-0469) of Spo. sorbi are slightly wider than those of isolate ROC 101 (5.5–7.5 μm vs 3.5–5.5 μm), and it has shorter conidiogenous cells (8–20 μm vs 12–43.5 μm).
Sporocadus spiniger C. Peng & C.M. Tian, sp. nov. — MycoBank MB 843823; Fig. 23
Fig. 23.

Sporocadus spiniger (BJFC-S1908, holotype). a. Disease symptoms; b. colonies on PDA at 3 d (left) and 15 d (right); c–d. appearance of conidiomata on host substrate; e. conidiomata formed on pine needles; f–h, j–m. conidiogenous cells with attached conidia; i. longitudinal section through conidiomata; n. conidia. — Scale bars: e = 200 μm; f–h, j–m, n = 10 μm; i = 50 μm.
Etymology. Referring to the fact that this strain was isolated from spines.
Typus. China, Gansu Province, Lanzhou City, Yongdeng County, Dayou Town, N36°44'20" E102°49'27", alt. 2743 m, on spines of R. omeiensis, 25 Oct. 2020, C. Peng (holotype BJFC-S1908, ex-type cultures ROC 119, ROC 120, ROC 121).
Sexual morph not observed. Asexual morph: Acervular conidiomata solitary to gregarious, mostly irregular, occasionally subglobose, superficial to semi-immersed, up to 154–196 μm diam, 61–95 μm high, visible as black acervuli on the spines. Conidiophores septate, mostly branched at base, hyaline, smooth. Conidiogenous cells discrete, filiform or ampulliform, sometimes cylindrical and subcylindrical, hyaline, smooth, without annellations, (12–)14.5–30.5(–34) × 1–1.5 μm (av. = 23.8 ± 4.27 × 1.2 ± 0.18 μm). Conidia straight or slightly curved, clavate or subcylindrical, with round ends, pale brown, (2–)3-septate, septa darker than the rest of cell, wall smooth and slightly constricted at the septa, (12.5–)14.5–16.5(–17.5) × (4–)4.5–5(–5.5) μm (av. = 15.8 ± 0.54 × 4.9 ± 0.31 μm), lacking appendages; basal cell obconic with round base, hyaline to pale brown, paler than other cells, thick-walled, (2–)2.5–3.5(–4) μm (av. = 2.8 ± 0.21 μm) long; median cells doliiform, the second cell from base is slightly longer than the third cell in 3-septate conidia, 3.5–4.5 μm (av. = 4.2 ± 2.17 μm) vs 2.5–3 μm (av. = 2.7 ± 0.16 μm), pale to mid-brown; median cell 4.9–5.0 μm (av. = 5.0 ± 0.07 μm), pale brown in 2-septate conidia; apical cell in 2-septate conidia significantly longer than apical cell in 3-septate conidia, (5.5–)6–6.5 μm (av. = 6.2 ± 0.09 μm) vs (2–)2.5–3.5 μm (av. = 2.7 ± 0.31 μm), short conic with a wide round apex, concolourous with the median cells; mean conidium length/width ratio = 3.22 : 1.
Culture characteristics — Cultures on PDA with white aerial mycelium and regular margin, fluffy, reverse yellowish brown. Colony 48–52 mm diam in 15 d at 25 °C. Acervuli black, distributed irregularly at the colony margin on the medium surface.
Additional material examined. China, Lanzhou City, Yongdeng County, Dayou Town, N36°44'17" E102°49'25", alt. 2745 m, on spines of R. omeiensis, 25 Oct. 2020, C. Peng (BJFC-S1909, cultures ROC 122, ROC 123, ROC 124, ROC 125).
Notes — Based on the multi-locus phylogenetic analysis, the seven isolates cluster separately in a well-supported clade (ML/ MP/BI = 100/100/1) (Fig. 8). Sporocadus spiniger is most closely related to Spo. brevis and Spo. trimorphus, differentiated from them in ITS (three different unique fixed alleles in Spo. brevis (521/524, 99.4 % with no gaps) and three in Spo. trimorphus (503/506, 99.4 % with no gaps)), LSU loci (four bp in Spo. brevis (884/888, 99.5 % with a single gap) and eight bp in Spo. trimorphus (876/884, 99.0 % with no gaps)), RPB2 loci (35 bp in Spo. brevis (812/847, 95.8 % with no gaps) and 39 bp in Spo. trimorphus (793/832, 95.3 % with no gaps)), TEF loci (40 bp in Spo. brevis (433/473, 91.5 % with 11 gaps) and 45 bp in Spo. trimorphus (427/472, 91.5 % with 11 gaps)) and TUB loci (45 bp in Spo. brevis (663/708, 93.6 % with 10 gaps) and 41 bp in Spo. trimorphus (667/708, 94.2 % with 10 gaps)). Moreover, Spo. spiniger differs from Spo. brevis in having longer conidia (14.5–16.5 μm vs 10–12 μm) and conidiogenous cells (14.5–30.5 μm vs 3.5–19 μm). The conidia are also wider than those of Spo. trimorphus (4.5–5 μm vs 3–4.5 μm).
Other species of Sporocadus that have been recorded from Rosa can be distinguished from this new species based on the number of septa and presence or absence of appendages. While Spo. spiniger has mostly 3-septate conidia without appendages, Spo. lichenicola has 4–5-septate conidia and Spo. rosarum has conidia with one apical appendage (Liu et al. 2019). Sporocadus rosigena mostly has 3-septate conidia without appendages that are morphologically similar to those of Spo. spiniger (Liu et al. 2019), but these two species have a distinctly different mean conidium length/width ratio (Spo. spiniger: 3.22 : 1 vs Spo. rosigena: 2.4 : 1).
Sporocadus trimorphus F. Liu et al., Stud. Mycol. 92: 406. 2018 — Fig. 24
Fig. 24.

Sporocadus trimorphus (BJFC-S1910). a. Disease symptoms; b. colonies on PDA at 3 d (left) and 15 d (right); c–d. appearance of conidiomata on host substrate; e. conidiomata on PDA; f. longitudinal section through conidiomata; g–k. conidiogenous cells with attached conidia; l. conidia. — Scale bars: e = 200 μm; f = 20 μm; g–l = 10 μm.
Sexual morph not observed. Asexual morph: Acervular stromata ostiolate, immersed or semi-immersed in plant tissues, slightly to strongly erumpent through the bark surface, sometimes delimited by a black or dark red marginal line, up to 207–269 μm diam, 82–144 μm high. Conidiophores septate, mostly branched at base, hyaline, smooth. Conidiogenous cells discrete or integrated, sub-cylindrical, lageniform or ampulliform, hyaline, smooth, with annellations, (11.5–)13–34(–38.5) × 1.5–2.5 μm (av. = 24.5 ± 6.43 × 2.0 ± 0.31 μm). Conidia fusoid or obovoid, straight, 3-septate, transverse septa fairly thick, wall smooth, (11–)12–16 × 4.5–5.5 μm (av. = 14.1 ± 1.22 × 4.8 ± 0.14 μm), bearing appendages; basal cell obconic with or without a truncate base, hyaline to pale brown, 2–3 μm (av. = 2.7 ± 0.41 μm) long; median cells mostly two, cylindrical, fairly thick-walled and pale brown, each (3–)3.5–4.5(–5) μm (av. = 4.1 ± 0.88 μm) long; apical cell conic, hyaline or concolourous with median cells, (1.5–)2–3.5(–4.5) μm (av. = 2.7 ± 0.28 μm) long; apical appendage lacking or, when present, single, unbranched, attenuated, tubular or flexuous, (6.5–)8–13.5 μm (av. = 11.3 ± 1.39 μm) long; basal appendage lacking or, when present, unbranched, tubular or flexuous, centric or excentric, (7–)9–12(–13.5) μm (av. = 10.4 ± 1.27 μm) long; mean conidium length/width ratio = 2.94 : 1.
Culture characteristics — On PDA, colonies initially white, irregular, lacking aerial mycelium, fast growing, reaching up to 60–62 mm diam in 15 d. Colonies pale white to pale grey after 15 d, lacking aerial mycelium. Conidiomata distributed sparsely over the medium surface.
Material examined. China, Gansu Province, Lanzhou City, Yuzhong County, Xinglong Mountain, N35°47'37" E104°3'53", alt. 2168 m, on branches of R. xanthina, 25 Aug. 2020, C. Peng (BJFC-S1910, cultures CFCC 55171 = ROC 112, ROC 113, ROC 114, ROC 115, ROC 116).
Notes — Sporocadus trimorphus was first described from R. canina in Sweden (Liu et al. 2019). In this study, five isolates were identified as belonging to this species and this species is also reported for the first time from China. Sporocadus trimorphus is characterised by three conidial types, i.e., non-appendaged, either apical or basal appendaged, and both apical and basal appendaged conidia (Liu et al. 2019).
Compared with the description of the ex-type isolate CBS 114203, isolate CFCC 55171 has shorter apical appendages (8–13.5 μm vs 2–20 μm), slightly wider conidia (4.5–5.5 μm vs 3–4.5 μm) and longer conidiogenous cells (13–34 μm vs 4.5–14 μm).
Key to Sporocadus species on Rosa spp.
1. Conidia with appendages . . . . . . . . . . . . . . . . . . . . . . . . . 2
1. Conidia lacking appendages . . . . . . . . . . . . . . . . . . . . . . . 6
2. Conidia 3–5-septate . . . . . . . . . . . . . . . . . Spo. lichenicola
2. Conidia 3-septate . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 3
3. Mean conidium length/width ratio less than 3 . . . . . . . . . 4
3. Mean conidium length/width ratio more than 3 . . . . . . . . 5
4. Conidia 10–12 × 6.5–7 μm . . . . . . . . . . . . . . . . Spo. brevis
4. Conidia 10–15 × 3.5–6.5 μm . . . . . . . . . . . . Spo. rosigena
5. Conidia 11–15.5 × 3.5–5.5 μm, conidiogenous cells 11.0–28.5 × 1.0–1.7 μm . . . . . . . . . . . . . . . . . . . . . . . . Spo. sorbi
5. Conidia 14.5–16.5 × 4.5–5 μm, conidiogenous cells 14.5–30.5 × 1.0–1.5 μm . . . . . . . . . . . . . . . . . . . . . Spo. spiniger
6. Conidia of two types, i.e., non-appendaged or only basal appendaged . . . . . . . . . . . . . . . . . . . . . . . . . . . Spo. rosarum
6. Conidia of three types, i.e., non-appendaged, either apical or basal appendaged, or both apical and basal appendaged conidia . . . . . . . . . . . . . . . . . . . . . . . . . . . Spo. trimorphous
Prevalence
Pestalotiopsis was the most prevalent genus (45 isolates, 35.2 % of the total isolates from Rosa), followed by Seimatosporium (33 isolates, 26.2 %) and Sporocadus (23 isolates, 18.3 %). The isolation rate of the other genera varied from 3.1 % to 13.5 % (Fig. 25). Pestalotiopsis was also the most abundant genus in all tissues investigated and had the highest isolation rate from all tissues. Seiridium was the genus with the lowest richness among all tissues, isolated only from branches (Fig. 26).
Fig. 25.

Overall isolation rate (%) of Sporocadaceae genera and species.
Fig. 26.

Isolation rate (%) of Sporocadaceae genera from each Rosa organs.
Fifteen species present in six genera (Monochaetia, Neopestalotiopsis, Pestalotiopsis, Seimatosporium, Seiridium and Sporocadus) were identified. Pestalotiopsis chamaeropis and Pes. rhodomyrtus (18 isolates, 14 % of the isolates from Rosa) were the two most prevalent species, followed by Seim. parvum (10 isolates, 8 %). Neopestalotiopsis concentrica, Seim. centrale and Seim. gracile have the same isolation rate of 7 %. In the current study, N. concentrica, N. subepidermalis, Pes. rhodomyrtus and Seim. parvum were isolated from spines/branches of two and three species of Rosa. Each of the remaining 11 species of Sporocadaceae was only isolated from one Rosa species (Fig. 29).
Fig. 29.

Isolation rate (%) of Sporocadaceae species from each Rosa sp.
The isolation rate of Seimatosporium in fruits and Pestalotiopsis in leaves was 100 %. These data reveal low generic and species diversity in these plant organs. In contrast, the branches had the highest genus diversity among all organs in this study, except for Seimatosporium, the other five genera were isolated from branches. Although the generic diversity of spines was slightly lower than that of branches, the number of strains isolated from the spines and the species diversity of spines was the highest of all tissues and was much higher than that of any other tissues (Fig. 26, 27).
Analysis of the abundance of Sporocadaceae species on Rosa species revealed seven species from R. chinensis, four from R. rugosa, three from R. xanthina and two from R. spinosissima, respectively (Fig. 29), with only one species on the remaining Rosa species, namely R. multiflora (Pes. rhodomyrtus), R. helenae (Seim. parvum), R. laevigata (Seim. nonappendiculatum) and R. omeiensis (Spo. spiniger) (Fig. 29). These findings might be due to the smaller sampling size of R. multiflora, R. helenae, R. laevigata and R. omeiensis obtained, since symptomatic branches were far less commonly observed than for the other Rosa species.
DISCUSSION
In this study, many Rosa specimens were collected from six major rose production areas (Gansu, Henan, Hunan, Ningxia, Qinghai and Shaanxi) in China, and 126 pestalotioid strains were obtained elsewhere. Multi-locus phylogenetic and morphological analyses revealed 15 species belonging to six genera in Sporocadaceae, namely Monochaetia, Neopestalotiopsis, Pestalotiopsis, Seimatosporium, Seiridium and Sporocadus.
Seimatosporium species have been extensively studied on several hosts (Tanaka et al. 2011, Liu et al. 2019), but not yet on Rosa spp. Up to now, five species of Seimatosporium have been recorded from Rosa, namely Seim. caninum on R. canina, Seim. pseudorosae on R. villosa, Seim. salicinum on R. multiflora and R. spinosissima, Seim. rosae on R. canina, R. kalmiussica, R. mucosa and R. pomifera, Seim. discosioides on R. blanda, R. heliophila, R. moschata, R. multiflora and R. rugosa (Weiss 1950, Nag Raj 1993, Cho & Shin 2004, Kobayashi 2007, Li et al. 2016, Crous et al. 2018, Liu et al. 2019). Among them, Seim. salicinum has been recorded in China (Tai 1979). In this study, a total of four new Seimatosporium species were introduced from Rosa in China. Seimatosporium is the genus which has the most species found and has a relatively high isolation rate (26.2 %) (Fig. 25). Seimatosporium species from Rosa spp. in previous and present studies show quite a difference based on morphology and DNA sequence data. Seimatosporium rosae is distinguished from the four new species in this study based on DNA phylogenetic data (Fig. 6) and the number of conidial septa (3–6-septate vs 3-septate). Although Seim. pseudorosae is closely related to Seim. parvum in this study (Fig. 6), these two species can be clearly distinguished by the size of their conidia and the length of their appendages. There are no sequence data available for Seim. caninum and Seim. salicinum for comparison, but the morphological characteristics of Seim. caninum (appendages lacking, 2-septate conidia) clearly distinguish it from other Seimatosporium species (Nag Raj 1993). Seimatosporium salicinum also produces 2-septate conidia (Sutton 1975, Nag Raj 1993), and this morphological characteristic distinguishes it from our four new species. Except for Seim. salicinum (the main hosts are Salix spp. and Rosaceae plants) (Nag Raj 1993, Tanaka et al. 2011), the other species from Rosa spp. are rarely recorded on other hosts (Nag Raj 1993, Mulenko et al. 2008, Li et al. 2016, Bonthond et al. 2018). Currently, seven Seimatosporium species have been reported from China besides our new species and Seim. salicinum. However, none of them occur on Rosa spp., i.e., Seim. botan, Seim. grammitum and Seim. hebeiensis on Paeonia suffruticosa, Seim. lonicerae on Pinus tabulaeformis, Seim. rhododendri on Rhododendron aureum, Seim. piceae on Picea jezoensis and Seim. maria (Tai 1979, Wang 1985, Nag Raj 1993, Guo 2004, Duan et al. 2011). Among these species, Seim. botan can produce conidia with only basal appendages (Hatakeyama & Harada 2004), which is similar to Seim. nonappendiculatum, to which it formed a sister clade. Seimatosporium grammitum and Seim. hebeiensis produce conidia lacking appendages and are therefore similar to Seim. nonappendiculatum. However, they differ significantly in their conidial dimensions. Having conidia with only basal appendages or without appendages, makes Seim. nonappendiculatum unique in the Sporocadaceae, and phylogenetically it clustered closer Sporocadus (conidia lacking appendages) than other Seimatosporium species (Fig. 2). The most typical morphological characteristics of Seim. piceae is that the length of the appendages is longer than the conidia itself (Wang 1985), which is most similar to the new species Seim. parvum (Fig. 6), but the conidia and appendages of Seim. parvum are significantly shorter than Seim. picea (14.3–15 × 4.21–4.34 μm vs 18–23 × 5–7 μm). In addition, the conidial dimensions of the other two new species, Seim. centrale and Seim. gracile, are significantly different from those species currently recorded in China.
Sporocadus is the type of Sporocadaceae and generally characterised by non-appendaged, 3-septate conidia (Liu et al. 2019). Sporocadus has long been assumed to be a synonym of Seimatosporium (Sutton 1975), but was confirmed to be a separate genus based on multigene phylogeny and conidial morphology (Brockmann 1976, Nag Raj 1993, Liu et al. 2019, Wijayawardene et al. 2020). However, in this study, although ITS-Blast is useful to locate species at the generic and family level, relatively weak in distinguishing Seimatosporium from Sporocadus, and even less so at the species level. Therefore, analysing these two genera at the family level is necessary to classify species into the correct genus (Fig. 2). Corresponding to the taxonomic classification determined by multi-locus phylogenetic analyses (Fig. 2), most Sporocadus species also exhibit characteristic morphological characters, including their non-appendaged and 3-septate conidia. In previous studies, most of these features have been used to delimit this genus (Brockmann 1976, Nag Raj 1993). It is worth to note that the presence or absence of conidial appendages is not a stable taxonomic character for the genus (Liu et al. 2019). For example, the conidia of Seim. discosioides and Seim. germanicum are also non-appendaged (Shoemaker 1964, Liu et al. 2019). Liu et al. (2019) introduced Spo. trimorphus and the conidia of this species have three types, i.e., non-appendaged which is the typical morphological characteristics of this genus; both apical and basal appendaged conidia which is the typical characteristics of Seimatosporium and either apical or basal appendaged conidia. This was also the first time that species with appendaged conidia were observed in Sporocadus, which renders morphology-based identifications more difficult, and underlines the necessity of molecular data for accurate identification. This study also found Spo. trimorphus on R. xanthina, which is also a first report from China. The other important morphological characteristic of this genus is 3-septate conidia (Nag Raj 1993). However, the number of conidial septa is far too unstable a feature to determine generic delineations. Liu et al. (2019) were the first to suggest that Sporocadus species could produce 2-septate conidia and introduced five species with this conidial type, namely Spo. biseptatus, Spo. incanus, Spo. rosarum, Spo. rosigena and Spo. rotundatus. Although the newly described species Spo. brevis can also produce conidia with two septa, this species is phylogenetically distinct from these five species (Fig. 8) and has significantly wider conidia (Table 5). To our knowledge, this study represents the first resolution of Sporocadus species in China based on multilocus sequence data. A total of four species of Sporocadus were identified in this study, and the isolation rate is similar to that of Seimatosporium (Fig. 25). Besides Spo. brevis and Spo. trimorphus, this study also described a new species Spo. spiniger, and represents the first report of Spo. sorbi on Rosa globally. Currently, only one Sporocadus species has been reported from China, namely Spo. lichenicola, the type species of the genus (Tai 1979). The hosts of this species include many Rosa plants and can cause leaf spot or brown spot of R. xanthina in China (Tai 1979, Wu 1992, Chen 2003). Based on multigene phylogenetic analyses, Spo. lichenicola is also separated from the two new species (Fig. 8). Moreover, the conidia of Spo. lichenicola are 4–5-septate, which is also an important morphological characteristic distinguishing it from other species in Sporocadus (Norphanphoun et al. 2015). In addition to Spo. lichenicola, relatively little has been recorded about Sporocadus species associated with Rosa. Liu et al. (2019) transferred four Seimatosporium species associated with Rosa to Sporocadus based on molecular systematics and morphological characteristics, namely Spo. rosarum (synonyms: Seim. rosarum, Seim. pseudorosarum and Seim. rosigenum) and Spo. rosigena (basionym: Seim. rosicola). Sporocadus rosarum forms a large sister clade with the two new species and Spo. trimorphus (Fig. 8), but the conidia of Spo. rosarum have an apical appendage, which makes it easy to distinguish them from other species of Sporocadus (Wanasinghe et al. 2018, Liu et al. 2019). Sporocadus rosigena is separated from the two new species in this study based on DNA phylogeny (Fig. 8) and there are also differences in conidial dimensions. It is remarkable that Seim. caninum isolated from R. chinensis has the typical morphological characteristics of Sporocadus, that is, with conidia lacking appendages (Sutton 1975). The feature of 2-septate conidia distinguishes Seim. caninum from other species in Seimatosporium and most of the members in Sporocadus. As described above, six Sporocadus species (including the new species described here) can produce 2-septate conidia similar to those of Seim. caninum, but the conidia of Seim. caninum are smaller than those of other species (Table 5). Therefore, since related sequence data of Seim. caninum are unavailable, further research is needed to resolve which genus it belongs to.
Table 5.
Synopsis of Seimatosporium caninum and species with 2-septate conidia in Sporocadus.
| Species | Length of conidia (μm) | Width of conidia (μm) | Host | Country | References |
|---|---|---|---|---|---|
| Seimatosporium caninum | 9.5–12 | 4.5–5.5 | Quercus incana | India | Sutton (1975) |
| Sporocadus biseptatus | 12.5–19.5 | 4.5–9 | Unknown | Unknown | Liu et al. (2019) |
| Spo. brevis | 10–12 | 6.5–7 | Rosa spinosissima | China | Present study |
| Spo. incanus | 11.5–20 | 4.5–6.5 | Prunus dulcis | Spain | Liu et al. (2019) |
| Spo. rosarum | 9.0–14.0 | 4.0–6.0 | Rosa canina | Europe | Liu et al. (2019) |
| Spo. rosigena | 10.0–15.0 | 3.5–6.5 | Vitis vinifera | Iran | Liu et al. (2019) |
| Rosa canina | Italy | Liu et al. (2019) | |||
| Rh ododendron sp. | Latvia | Liu et al. (2019) | |||
| Rubus fruticosus | Netherlands | Liu et al. (2019) | |||
| Spo. rotundatus | 9.0–16.5 | 4.5–7.5 | Arceuthobium pussilum | Canada | Liu et al. (2019) |
* Newly described taxon is in bold.
Pestalotiopsis is the genus with the highest isolation rate in this study, namely 29.6 % (Fig. 25), which is basically in line with expectations, because Pestalotiopsis, as an important plant pathogen, has many records and can parasitise more than 50 plant families in China (Tai 1979, Chen 2003, Ge et al. 2009). Cash crops such as palm, eucalypts, guava and tea trees have suffered serious damage due to species of Pestalotiopsis (Ge et al. 2009, Maharachchikumbura et al. 2013a, Wang et al. 2019a, b). This genus also has many records on Rosa plants. In China, five species of Pestalotiopsis have been recorded from Rosa, namely Pes. rosae and Pes. suffocata that are parasitic on R. chinensis, Pes. oleandri that is parasitic on R. laevigata, Pes. longisetula that is parasitic on R. henryi and Pes. aquatica that is saprophytic on R. chinensis (Zhu et al. 1991, Wei et al. 2005, Ge et al. 2009). Beyond that, there are six species of Pestalotiopsis recorded from Rosa in different countries (Riley 1960, Guba 1961, Mathur 1979, Rai 1990, Nag Raj 1993, Mendes et al. 1998, Sameva 2004, Kobayashi 2007). In this study, Pes. chamaeropis and Pes. rhodomyrtus are newly reported from Rosa. For the newly described species, Pes. tumida isolated from R. chinensis, morphological differences distinguish this species and other known species recorded on Rosa, especially in terms of the conidial dimensions and length of the appendages, as well as shape of basal appendages. The results of our study suggest that the species diversity of Pestalotiopsis on Rosa in China may be higher than what was previously expected.
Neopestalotiopsis was separated from Pestalotiopsis based on its versicolourous median conidium cells and indistinct conidiophores (Maharachchikumbura et al. 2014, Liu et al. 2017, 2019). In this study, two new species of Neopestalotiopsis were introduced from Rosa based on the phylogenetic analyses and morphological characteristics, namely N. concentrica and N. subepidermalis. Five Neopestalotiopsis species associated with Rosa have been recorded, namely N. clavispora, N. palmarum, N. rosae, N. rosicola and N. versicolor (Liu et al. 2010, Feng et al. 2014, Maharachchikumbura et al. 2014, Jiang et al. 2018, Vu et al. 2019). For these five species, N. clavispora, N. palmarum, N. rosicola and N. versicolor have been reported in China and cause leaf blotch, stem canker of R. chinensis and leaf spot disease of many other hosts (Ge et al. 2009, Feng et al. 2014, Jiang et al. 2018), but these four species are phylogenetically separated from our new species (Fig. 4) and there are also differences in their conidial morphology. Neopestalotiopsis rosae is morphologically quite distinct from other taxa in the genus (has appendages which do not arise from the apical crest, but at different regions in the upper half of the apical cell; Maharachchikumbura et al. 2014). With the increase of collections and DNA data, N. clavispora, N. palmarum, N. rosae and N. versicolor appear to be widely distributed in different countries where they cause severe diseases on different hosts, e.g., N. rosae causes dieback, crown rot, fruit rot and root rot of many economically important plants including Eucalyptus, Fragaria, Paeonia and Vitis (Maharachchikumbura et al. 2014, Rebollar-Alviter et al. 2020, Santos et al. 2020, Cosseboom & Hu 2021), and N. clavispora can infect more than 50 plant species belonging to 27 families in China and about 17 other countries, resulting in severe leaf diseases (Qiu et al. 2020). For this reason, the two new species isolated from Rosa may present dangerous pathogens for other hosts, and further work is needed to better understand the role and distribution of these two new Neopestalotiopsis species. For other pestalotiopsis-like species associated with Rosa, as mentioned above, some Pestalotiopsis spp. on Rosa (Pes. adusta, Pes. algeriensis, Pes. aquatica, Pes. longisetula and Pes. oleandri) have the typical morphological characteristics of Neopestalotiopsis, namely versicolourous median conidium cells (Guba 1961, Mordue & Holliday 1971, Zhu et al. 1991, Wei et al. 2005, Ge et al. 2009, Cardoso et al. 2017). However, due to the lack of related DNA sequence data, their taxonomy still needs to be resolved. However, our two new Neopestalotiopsis species can be distinguished from these five Pestalotiopsis species based on their conidial and appendage morphology (Table 6). This study also encountered the same problem as previous studies on Neopetalotiopsis compared to other genera, namely the short branch length and low support rate of the current phylogenetic tree based on ITS, TEF and TUB (Liu et al. 2017, Norphanphoun et al. 2019). More gene regions will have to be introduced in future studies to provide better support for species in the genus.
Table 6.
Synopsis of two new Neopestalotiopsis species and species with versicolourous median conidium cells in Pestalotiopsis.
| Species | Length of conidia (μm) | Width of conidia (μm) | Length of apical appendages (μm) | Length of basal appendages (μm) | Host | Country | References |
|---|---|---|---|---|---|---|---|
| Neopestalotiopsis concentrica | 14–18.5 | 4.5–5 | 19–26 | 3.5–5.5 | Rosa rugosa | China | Present study |
| N. subepidermalis | 20–25 | 7.5–9 | 27–32.5 | 7–7.5 | Rosa rugosa | China | Present study |
| Pestalotiopsis adusta | 16–22.4 | 4.7–6.6 | 5 –12 | 3.5 | Rosa indica | India | Rai (1990) |
| Pes. algeriensis | 20.5–31.3 | 6.3–8.0 | 13.3–17.8 | 3–6.3 | Rosa sempervirens | Algeria | Nag Raj (1993) |
| Pes. aquatica | 20.6–28.2 | 7.2–8.2 | 12.8–23 | 2–4 | Rosa sp. | China | Ge et al. (2009) |
| Pes. longisetula | 20.6–25.7 | 6.4–7.7 | 20.6–36 | 5.1–7.7 | Rosa henryi | China | Ge et al. (2009) |
| Rosa sp. | China | Ge et al. (2009) | |||||
| Pes. oleandri | 15–22.6 | 5–7.5 | 14–16.5 | 6.1–11.5 | Rosa laevigata | China | Ge et al. (2009) |
| Rosa sp. | China | Zhu et al. (1991) |
* Newly described taxa are in bold.
Many species of Pestalotiopsis and Neopestalotiopsis have overlapping morphological characteristics, and it is difficult to identify them solely based on morphology (Maharachchikumbura et al. 2014, Liu et al. 2017). Some morphological characteristics are unstable and vary with host range, culture and other environmental conditions (Maharachchikumbura et al. 2011, Norphanphoun et al. 2019). But at the same time, conidial morphology can still provide a good reference for species distinction of pestalotioid fungi (Crous et al. 2012). Therefore, the combination of morphology and molecular systematics based on sequence data has become an important means to solve the classification of pestalotiopsis-like and support the introduction of new species (Crous et al. 2012, Maharachchikumbura et al. 2014, Liu et al. 2019).
Monochaetia has undergone a lot of changes since it was introduced, and many species have been transferred to other genera, such as Diploceras, Monochaetinula, Sarcostroma, Seimatosporium and Seiridium (De Silva et al. 2018, Liu et al. 2019). However, species of Monochaetia are morphologically conserved at present and the typical morphological characteristic of this genus is that conidia have a single centric apical and basal appendage (if present) (De Silva et al. 2018, Liu et al. 2019). Previous reports on the hosts of Monochaetia were mainly concentrated on Fagaceae (especially Castanea and Quercus spp.) (De Silva et al. 2018, Liu et al. 2019). In this study one new species, M. rosarum, was isolated from Rosa and can be distinguished from other phylogenetically allied species based on conidial dimensions. In addition, there are currently four species of Monochaetia that have been recorded from Rosa, namely M. concentrica, M. rosae-caninae, M. seiridioides and M. turgida (Guba 1961, Tai 1979, Chen 2003). Except for M. rosae-caninae, three other species have also been recorded in China (Tai 1979, Chen 2003). The species associated with Rosa have relatively wide host ranges mainly concentrated in the Rosaceae, excluding the Fagaceae (Weiss 1950, Sutton 1980, Nag Raj 1993), and the choice of host may also be affected by the geographical environment. For example, M. turgida, has been recorded on Rosa spp. in India, on Crataegus spp. in the USA, and on Pyrus spp in China (Mathur 1979, Tai 1979, Nag Raj 1993). Besides the three species on Rosa mentioned above (M. concentrica, M. seiridioides and M. turgida), 16 species of Monochaetia have been recorded from China (Table 7). Among them, Chen et al. (2002) introduced seven Monochaetia species based on morphology and host, i.e., M. caryotae on Caryota ochlandra, M. cycadis on Cycas revoluta, M. elaeocarpi on Elaeocarpus serratus, M. garciniae on Garcinia tinctoria, M. hirta on Castanopsis fabri, M. sabinae on Sabina chinensis and M. salaccae on Salacca secanda, and all these species are endemic to China. Although sequence data for these species are not available and reliance only on morphology and host data in Monochaetia taxonomy is far from perfect, the ongoing discovery of this genus from other hosts except for Fagaceae in China will deepen our understanding of the species in this genus. It is worth mentioning that Zhao & Li (1994) have described a species M. nodosporella, the hosts of which include also Castanopsis spp. Characteristics of this species are quite different from morphological characters of Monochaetia, including 4-celled distoseptate conidia with two olivaceous-brown median cells, lacking appendages. Considering the limited sampling and the lack of sequence data, the taxonomy of M. nodosporella remains unresolved and will be treated once DNA sequence data have been obtained. Aside from the species associated with Rosa and the Chinese endemic species mentioned above, the remaining species recorded in China have a relatively small range of host plants mainly on Castanea and Quercus, and are geographically widespread (Tai 1979, Zhao & Li 1994, Chen et al. 2002). For example, M. monochaeta and M. kansensis have also been recorded in many countries besides China, but the hosts are basically Quercus and Castanea (Ellis & Everhart 1893, Weiss 1950, Guba 1961, Nag Raj 1993, Cho & Shin 2004, Kobayashi 2007). Therefore, for the new species M. rosarum described in this study, many specimens still need to be collected to improve our understanding of its host range and distribution.
Table 7.
Monochaetia species that have been recorded in China in previous studies
| Species | Host | Location | Collector | References |
|---|---|---|---|---|
| Monochaetia caryotae | Caryota ochlandra | Guangxi, China | Y.X. Chen & G. Wei | Chen et al. (2002) |
| M. castaneae | Castanea mollissima | Sichuan, China | N. Jiang | Jiang et al. (2021) |
| M. celaeocarpi | Elaeocarpus serratus | Hainan, China | Z.W. Wang & G. Wei | Chen et al. (2002) |
| M. concentrica | Rosa xanthina | Jilin, China | P.K. Qi | Tai (1979) |
| M. cycadis | Cycas revoluta | Guangxi, China | G. Wei | Chen et al. (2002) |
| M. diospyri | Diospyros lotus | – | – | Chen et al. (2002) |
| M. garciniae | Garcinia tinctoria | Hainan, China | Z.W. Wang & G. Wei | Chen et al. (2002) |
| M. hirta | Castanopsis fabri | Hainan, China | Z.W. Wang & G. Wei | Chen et al. (2002) |
| M. kansensis | Castanea mollissima | – | – | Chen et al. (2002) |
| Quercus dentata | Henan, China | M.Q, Wang | Tai (1979) | |
| Q. dentata | Gansu, China | Y.R. Meng | Meng (2003) | |
| M. monochaeta | Q. dentata | Gansu, China | Y.R. Meng | Meng et al. (2003) |
| Q. variabilis | Jiangsu, China | – | Teng (1963) | |
| Q. variabilis | Jiangsu, China | Q.Y. Shen | Tai (1979) | |
| M. nattrassii | Camellia sinensis | Hong Kong | – | Sutton et al. (1980) |
| M. nodosporella | Castanopsis delavayi | Yunnan, China | Zhao & Li | Zhao & Li (1994) |
| M. sabinae | Sabina chinensis | Guangxi, China | G. Wei | Chen et al. (2002) |
| M. saccardoi | Q. variabilis | Jiangsu, China | – | Teng (1963) |
| M. salaccae | Salacca secunda | Yunnan, China | G. Wei | Chen et al. (2002) |
| M. seiridioides | R. xanthina | Henan, China | M.Q, Wang | Tai (1979) |
| M. turgida | Pyrus communis | Guangdong, China | Z. Tu | Tai (1979) |
Seiridium generally produces 5-septate conidia with a single centric apical and single excentric basal appendage (Bonthond et al. 2018, Liu et al. 2019). Nees (1817) established Seiridium based on Seir. marginatum which was collected from rose stems in Germany, and this is also the earliest record of pestalotioid fungi on the Rosa plants. Presently, besides the type species Seir. marginatum, only one Seiridium species has been recorded from Rosa, namely Seir. rosarum, which is distributed in Europe and Australia (Sutton 1980, Nag Raj 1993, Jaklitsch et al. 2016). In this study, a new species Seir. rosae was introduced from R. multiflora in China. The newly described species can be distinguished from most Seiridium species by its numerous appendages, which are branched, while in Seiridium appendages are fewer and generally unbranched (Maharachchikumbura et al. 2014). However, conidial appendages are branched, which is not unique to Seir. rosae in Seiridium and is also typical for Seir. indicum, Seir. pezizoides and Seir. venetum (Pavgi & Singh 1970, Maharachchikumbura et al. 2015, Marin-Felix et al. 2019). Seiridium rosae is easily distinguishable from these three species based on conidial morphology (Table 8). It is worth mentioning that Seiridium is the most important pathogen group in Sporocadaceae (Bonthond et al. 2018). Seiridium is the main pathogen of cypress canker and causes huge economic losses worldwide (Boesewinkel 1983, Graniti 1986, 1993, 1998, Barnes et al. 2001, Tsopelas et al. 2007). There are currently nine species of Seiridium recorded in China, with diverse hosts (Table 9). However, thus far China still has no record of Seiridium spp. on Cupressaceae.
Table 8.
Synopsis of species with branched appendages in Seiridium.
| Species | Length of conidia (μm) | Width of conidia (μm) | Length of apical appendages (μm) | Length of basal appendages (μm) | Host | Country | References |
|---|---|---|---|---|---|---|---|
| Seiridium indicum | 22–30.8 | 7.7–8.8 | 4.4–11 | 1–1.8 | Spirea micrantha | India | Nag Raj (1993) |
| Seir. pezizoides | 28–33.5 | 7–8 | 8.5–27 | 5.5–14 | Vitis vinifera | Italy | Marin-Felix et al. (2019) |
| Seir. rosae | 31–35 | 8–9.5 | 28.5–44.5 | 3–4.5 | Rosa rugosa | China | Present study |
| Seir. venetum | 20–30 | 6.5–8.5 | 10–35 | 2–5 | Cornus sanguinea | Italy | Nag Raj (1993) |
* Newly described taxa is in bold.
Table 9.
Seiridium species that have been recorded in China in previous studies.
| Species | Host | Location | Collector | References |
|---|---|---|---|---|
| Seiridium camelliae | Camellia reticulata | Yunnan, China | Y.M. Zhang | Maharachchikumbura et al. (2015) |
| Seir. ceratosporum | Vitis vinifera | Yunnan, China | – | Bonthond et al. (2018) |
| Seir. chinense | Trachycarpus fortunei | Shaanxi, China | N. Jiang & C.M. Tian | Jiang et al. (2018) |
| Seir. cupressi | Vitis sp. | Zhejiang, China | – | Teng (1996) |
| Seir. eriobotryae | Eriobotrya japonica | Guangxi, China | G. Wei | Chen et al. (2002) |
| Seir. manilkarae | Manilkara zapota | Yunnan, China | G. Wei | Chen et al. (2002) |
| Seir. pezizoides | Camellia oleifera | Hunan, China | J.X. Yu | Yu et al. (2018) |
| Seir. unicorne | Eriobotrya japonica | Jiangsu, China | F.L. Tai | Tai (1979) |
| Malus mandshurica | Jilin, China | Miura | Tai (1979) | |
| M. prunifolia | Jilin, China | P.K. Qi | Tai (1979) | |
| M. pumila | Yunnan, China | L.D. Lin | Tai (1979) | |
| Pyrus ussuriensis | Jilin, China | P.K. Qi | Tai (1979) | |
| Sorbus alnifolia | Jilin, China | Miura | Tai (1979) | |
| V. vinifera | Jiangsu, China | L. Ling | Tai (1979) | |
| Seir. venetum | Cornus sp. | Yunnan, China | – | Tai (1979) |
In this study, the species diversity of Sporocadaceae on the stems and spines of Rosa was significantly higher than that on the leaves and fruits. Nevertheless, a new species Seim. nonappendiculatum was isolated from the fruits of Rosa laevigata and Pes. rhodomyrtus, that was first reported on Rosa, was isolated from leaves of Rosa rugosa. Therefore, the leaves and fruits of Rosa plants might harbour many more species of pestalotioid fungi yet unknown to science. This study represents the first systematic investigation, morphological and molecular characterisation of Sporocadaceae on Rosa. The findings of this study reveal taxonomic, morphological and biological diversity of Sporocadaceae associated with different Rosa spp. in China. Other than expanding our knowledge of the genetic diversity and hosts of Sporocadaceae on Rosa, it provides crucially important information to understand the ecology of the Sporocadaceae associated with Rosa. Further study is needed to test the pathogenicity of these species and understand their biological and epidemiological characteristics to contribute towards better disease management.
Fig. 27.

Isolation rate (%) of Sporocadaceae species from each Rosa organs.
Fig. 28.

Isolation rate (%) of Sporocadaceae genera from each Rosa sp.
Acknowledgements
This study is financed by National Natural Science Foundation of China (Project No.: 31670647).
Declaration on conflict of interest
The authors declare that there is no conflict of interest.
Supplementary material
Phylogenetic tree of Monochaetia resulting from maximum likelihood (ML) analysis of the ITS sequence alignment. Nodes are labelled with bootstrap values from RAxML/Parsimony bootstrap/Bayesian posterior probabilities values. Nodes receiving below 50 bootstrap values and 0.5 probability values are not labelled. The scale bar represents the expected number of changes per site. Isolates collected in this study are in bold and blue.
Phylogenetic trees based on maximum likelihood (ML) analyses for species in Neopestalotiopsis. a. ITS region; b. TEF gene region; c. TUB gene region. Nodes are labelled with bootstrap values from RAxML/Parsimony bootstrap/Bayesian posterior probabilities values. Nodes receiving below 50 bootstrap values and 0.5 probability values are not labelled. The scale bar represents the expected number of changes per site. Isolates collected in this study are in bold and blue.
Phylogenetic trees based on maximum likelihood (ML) analyses for species in Pestalotiopsis. a. ITS region; b. TEF gene region; c. TUB gene region. Nodes are labelled with bootstrap values from RAxML/Parsimony bootstrap/Bayesian posterior probabilities values. Nodes receiving below 50 bootstrap values and 0.5 probability values are not labelled. The scale bar represents the expected number of changes per site. Isolates collected in this study are in bold and blue.
Phylogenetic trees based on maximum likelihood (ML) analyses for species in Seimatosporium. a. ITS region; b. LSU gene region. Nodes are labelled with bootstrap values from RAxML/Parsimony bootstrap/Bayesian posterior probabilities values. Nodes receiving below 50 bootstrap values and 0.5 probability values are not labelled. The scale bar represents the expected number of changes per site. Isolates collected in this study are in bold and blue.
Phylogenetic trees based on maximum likelihood (ML) analyses for species in Seiridium. a. ITS region; b. RPB2 gene region; c. TEF gene region; d. TUB gene region. Nodes are labelled with bootstrap values from RaxML/Parsimony bootstrap/Bayesian posterior probabilities values. Nodes receiving below 50 bootstrap values and 0.5 probability values are not labelled. The scale bar represents the expected number of changes per site. Isolates collected in this study are in bold and blue.
Phylogenetic trees based on maximum likelihood (ML) analyses for species in Sporocadus. a. ITS region; b. LSU gene region; c. RPB2 gene region; d. TEF gene region; e. TUB gene region. Nodes are labelled with bootstrap values from RAxML/Parsimony bootstrap/Bayesian posterior probabilities values. Nodes receiving below 50 bootstrap values and 0.5 probability values are not labelled. The scale bar represents the expected number of changes per site. Isolates collected in this study are in bold and blue.
REFERENCES
- Akinsanmi OA, Nisa S, Jeff-Ego OS, et al. 2017. Dry flower disease of macadamia in Australia caused by Neopestalotiopsis macadamiae sp. nov. and Pestalotiopsis macadamiae sp. nov. Plant Disease 101: 45–53. [DOI] [PubMed] [Google Scholar]
- Alberto F, Giorgio G, Dalia A. 2022. Neopestalotiopsis siciliana sp. nov. and N. rosae causing stem lesion and dieback on avocado plants in Italy. Journal of Fungi 8: 562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariyawansa HA, Hyde KD. 2018. Additions to Pestalotiopsis in Taiwan. Mycosphere 9: 999–1013. [Google Scholar]
- Bagsic I, Linde M, Debener T. 2016. Genetic diversity and pathogenicity of Sphaceloma rosarum (teleomorph Elsinoë rosarum) causing spot anthracnose on roses. Plant Pathology 65: 978–986. [Google Scholar]
- Barber PA, Crous PW, Groenewald JZ, et al. 2011. Reassessing Vermisporium (Amphisphaeriaceae), a genus of foliar pathogens of eucalypts. Persoonia 27: 90–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes I, Roux J, Wingfield MJ, et al. 2001. Characterization of Seiridium spp. associated with cypress canker based on ß-tubulin and histone sequences. Plant Disease 85: 317–321. [DOI] [PubMed] [Google Scholar]
- Bezerra JDP, Machado AR, Firmino AL, et al. 2018. Mycological diversity description I. Acta Botanica Brasilica 32: 656–666. [Google Scholar]
- Boesewinkel H. 1983. New records of the three fungi causing cypress canker in New Zealand, Seiridium cupressi (Guba) comb. nov. and S. cardinale on Cupressocyparis and S. unicorne on Cryptomeria and Cupressus. Transactions of the British Mycological Society 80: 544–547. [Google Scholar]
- Bonthond G, Sandoval-Denis M, Groenewald JZ, et al. 2018. Seiridium (Sporocadaceae): an important genus of plant pathogenic fungi. Persoonia 40: 96–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmann I. 1976. Untersuchungen über die Gattung Discostroma Clements (Ascomycetes). Sydowia 28: 275–338. [Google Scholar]
- Bruneau A, Starr JR, Joly S. 2007. Phylogenetic relationships in the genus Rosa: new evidence from chloroplast DNA sequences and an appraisal of current knowledge. Systematic Botany 32: 366–378. [Google Scholar]
- Cardoso JK, Nanami DS, Zanutto CA, et al. 2017. Description and identification of two new diseases of guariroba palm (Syagrus oleraceae) in Brazil. Journal of Phytopathology 165: 610–619. [Google Scholar]
- Chaiwan N, Wanasinghe DN, Mapook A, et al. 2020. Novel species of Pestalotiopsis fungi on Dracaena from Thailand. Mycology 11: 306–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MM. 2003. Forest fungi phytogeography: Forest fungi phytogeography of China, North America, and Siberia and international quarantine of tree pathogens. Pacific Mushroom Research and Education Center Sacramento USA. [Google Scholar]
- Chen SF, Li GQ, Liu JQ, et al. 2016. Characteristics of Lasiodiplodia theobromae from Rosa rugosa in South China. Crop Protection 79: 51–55. [Google Scholar]
- Chen YX, Wai G, Chen WP. 2002. New species of Monochaetia and Seiridium in China. Mycosystema 21: 503–510. [Google Scholar]
- Chen YY, Maharachchikumbura SSN, Liu JK. 2017. Fungi from Asian Karst formations I. Pestalotiopsis photinicola sp. nov., causing leaf spots of Photinia serrulata. Mycosphere 8: 103–110. [Google Scholar]
- Cho WD, Shin HD. 2004. List of plant diseases in Korea. Korean Society of Plant Pathology, Korea. [Google Scholar]
- Collado J, Platas G, Bills GF, et al. 2006. Studies on Morinia: Recognition of Morinia longiappendiculata sp. nov. as a new endophytic fungus, and a new circumscription of Morinia pestalozzioides. Mycologia 98: 616–627. [DOI] [PubMed] [Google Scholar]
- Cosseboom SD, Hu M. 2021. Diversity, pathogenicity, and fungicide sensitivity of fungal species associated with late-season rots of wine grape in the Mid-Atlantic United States. Plant Disease 105: 3101–3110. [DOI] [PubMed] [Google Scholar]
- Crous PW, Boers J, Holdom D, et al. 2022. Fungal Planet description sheets: 1383–1435. Persoonia 48: 261–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Carris LM, Giraldo A, et al. 2015a. The genera of fungi-fixing the application of the type species of generic names-G 2: Allantophomopsis, Latorua, Macrodiplodiopsis, Macrohilum, Milospium, Protostegia, Pyricularia, Robillarda, Rotula, Septoriella, Torula, and Wojnowicia. IMA Fungus 6: 163–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Denman S, Taylor JE, et al. 2013. Cultivation and diseases of Proteaceae: Leucadendron, Leucospermum and Protea. CBS Biodiversity Series 13. CBS-KNAW Fungal Biodiversity Centre. [Google Scholar]
- Crous PW, Gams W, Stalpers JA, et al. 2004. MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22. [Google Scholar]
- Crous PW, Liu F, Cai L, et al. 2018. Allelochaeta (Sporocadaceae): pigmentation lost and gained. Fungal Systematics and Evolution 2: 273–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Phillips A, Baxter A. 2000. Phytopathogenic fungi from South Africa: 73–75. Department of Plant Pathology Press, University of Stellenbosch, South Africa. [Google Scholar]
- Crous PW, Shivas RG, Quaedvlieg W, et al. 2014a. Fungal Planet description sheets: 214–280. Persoonia 32: 184–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Summerell BA, Shivas RG, et al. 2011a. Fungal Planet description sheets: 92–106. Persoonia 27: 130–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Summerell BA, Swart L, et al. 2011b. Fungal pathogens of Proteaceae. Persoonia 27: 20–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Verkley G, Christensen M, et al. 2012. How important are conidial appendages? Persoonia 28: 126–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Verkley G, Groenewald J, et al. 2019a. Fungal Biodiversity. Westerdijk Laboratory Manual Series no. 1. Utrecht, Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands. [Google Scholar]
- Crous PW, Wingfield MJ, Cheewangkoon R, et al. 2019b. Foliar pathogens of eucalypts. Studies in Mycology 94: 125–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Chooi YH, et al. 2020. Fungal Planet description sheets: 1042–1111. Persoonia 44: 301–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Le Roux JJ, et al. 2015b. Fungal Planet description sheets: 371–399. Persoonia 35: 264–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Schumacher RK, et al. 2014b. Fungal Planet description sheets: 281–319. Persoonia 33: 212–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva NI, Maharachchikumbura SSN, Bhat DJ, et al. 2018. Monochaetia sinensis sp. nov. from Yunnan Province in China. Phytotaxa 375: 059–069. [Google Scholar]
- Dean RA, Lichens-Park A, Kole C. 2014. Genomics of plant-associated fungi: monocot pathogens. Berlin, Heidelberg, Germany. [Google Scholar]
- Debener T. 2019. The Beast and the Beauty: What do we know about Black Spot in roses? Critical Reviews in Plant Sciences 38: 313–326. [Google Scholar]
- Diogo E, Gonçalves CI, Silva AC, et al. 2021. Five new species of Neopestalotiopsis associated with diseased Eucalyptus spp. in Portugal. Mycological Progress 20: 1441–1456. [Google Scholar]
- Doyle JJ, Doyle JL. 1990. Isolation of plant DNA from fresh tissue. Focus 12: 39–40. [Google Scholar]
- Duan YB, Yu ZZ, Kang YB. 2011. First report of leaf spot disease of peony caused by Seimatosporium botan in China. Plant Disease 95: 226. [DOI] [PubMed] [Google Scholar]
- Eken C, Spanbayev A, Tulegenova Z, et al. 2009. First report of Truncatella angustata causing leaf spot on Rosa canina in Kazakhstan. Australasian Plant Disease Notes 4: 44–45. [Google Scholar]
- Ellis JB, Everhart BM. 1893. New species of fungi from various localities. Proceedings of the Academy of Natural Sciences of Philadelphia 45: 440–466. [Google Scholar]
- Eriksson O. 1986. Notes on ascomycete systematics, Nos. 1–224. Systema Ascomycetum 5: 113–174. [Google Scholar]
- Eriksson O. 1987. Notes on ascomycete systematics. Nos. 225–463. Systema Ascomycetum 9: 11–165. [Google Scholar]
- Feng F, Zhou G, Li H. 2019. First report of Colletotrichum siamense causing anthracnose on Rosa chinensis in China. Plant Disease 103: 1422–1422. [Google Scholar]
- Feng Y, Liu B, Sun B. 2014. First report of leaf blotch caused by Pestalotiopsis clavispora on Rosa chinensis in China. Plant Disease 98: 1009–1009. [DOI] [PubMed] [Google Scholar]
- Fougère-Danezan M, Joly S, Bruneau A, et al. 2015. Phylogeny and biogeography of wild roses with specific attention to polyploids. Annals of Botany 115: 275–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas EFS, Da Silva M, Barros MVP, et al. 2019. Neopestalotiopsis hadrolaeliae sp. nov., a new endophytic species from the roots of the endangered orchid Hadrolaelia jongheana in Brazil. Phytotaxa 416: 211–220. [Google Scholar]
- Fu M, Crous PW, Bai Q, et al. 2019. Colletotrichum species associated with anthracnose of Pyrus spp. in China. Persoonia 42: 1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge QX, Chen YX, Xu T. 2009. Flora Fungorum Sinicorum. Vol. 38. Pestalotiopsis. Science Press. Beijing, China. [In Chinese.] [Google Scholar]
- Geng K, Zhang B, Song Y, et al. 2013. A new species of Pestalotiopsis from leaf spots of Licuala grandis from Hainan, China. Phytotaxa 88: 49–54. [Google Scholar]
- Goonasekara ID, Maharachchikumbura SSN, Wijayawardene NN, et al. 2016. Seimatosporium quercina sp. nov. (Discosiaceae) on Quercus robur from Germany. Phytotaxa 255: 240–280. [Google Scholar]
- Graniti A. 1986. Seiridium cardinale and other cypress cankers 1. EPPO Bulletin 16: 479–486. [Google Scholar]
- Graniti A. 1993. Seiridium blight of cypress-another ecological disaster? Plant Disease 77: 1–6. [Google Scholar]
- Graniti A. 1998. Cypress canker: a pandemic in progress. Annual Review of Phytopathology 36: 91–114. [DOI] [PubMed] [Google Scholar]
- Gu M, Hu D, Han B, et al. 2021. Pestalotiopsis abietis sp. nov. from Abies fargesii in China. Phytotaxa 509: 93–105. [Google Scholar]
- Gualberto GF, Catarino ADM, Sousa TF, et al. 2021. Pseudopestalotiopsis gilvanii sp. nov. and Neopestalotiopsis formicarum leaves spot pathogens from guarana plant: a new threat to global tropical hosts. Phytotaxa 489: 121–139. [Google Scholar]
- Guba EF. 1961. Monograph of Monochaetia and Pestalotia. Harvard University Press, Cambridge, Massachusetts, USA. [Google Scholar]
- Guo LD. 2004. Endophytic Fungi II. New records from pine in China. Mycosystema 23: 24–27. [Google Scholar]
- Guo Y, Crous PW, Bai Q, et al. 2020. High diversity of Diaporthe species associated with pear shoot canker in China. Persoonia 77: 132–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama S, Harada Y. 2004. A new species of Discostroma and its anamorph Seimatosporium with two morphological types of conidia, isolated from the stems of Paeonia suffruticosa. Mycoscience 45: 106–111. [Google Scholar]
- Hernández-Restrepo M, Groenewald JZ, Crous PW, et al. 2016. Taxonomic and phylogenetic re-evaluation of Microdochium, Monographella and Idriella. Persoonia 36: 57–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongsanan S, Maharachchikumbura SSN, Hyde KD, et al. 2017. An updated phylogeny of Sordariomycetes based on phylogenetic and molecular clock evidence. Fungal Diversity 84: 25–41. [Google Scholar]
- Huanluek N, Jayawardena RS, Maharachchikumbura SSN, et al. 2021. Additions to pestalotioid fungi in Thailand: Neopestalotiopsis hydeana sp. nov. and Pestalotiopsis hydei sp. nov. Phytotaxa 479: 23–43. [Google Scholar]
- Huelsenbeck JP, Ronquist F. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Hyde KD, Hongsanan S, Jeewon R, et al. 2016. Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 80: 1–270. [Google Scholar]
- Hyde KD, Jeewon R, Chen YJ, et al. 2020. The numbers of fungi: is the descriptive curve flattening. Fungal Diversity 103: 219–271. [Google Scholar]
- Jaklitsch WM, Gardiennet A, Voglmayr H. 2016. Resolution of morphology-based taxonomic delusions: Acrocordiella, Basiseptospora, Blogiascospora, Clypeosphaeria, Hymenopleella, Lepteutypa, Pseudapiospora, Requienella, Seiridium and Strickeria. Persoonia 37: 82–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayawardena R, Hyde KD, Chethana K, et al. 2018. Mycosphere notes 102–168: Saprotrophic fungi on Vitis in China, Italy, Russia and Thailand. Mycosphere 9: 1–114. [Google Scholar]
- Jayawardena RS, Liu M, Maharachchikumbura SSN. 2016. Neopestalotiopsis vitis sp. nov. causing grapevine leaf spot in China. Phytotaxa 258: 63–74. [Google Scholar]
- Jia J, Li X, Zhang W, et al. 2019. First report of Botryosphaeria dothidea associated with stem canker on Rosa chinensis in China. Plant Disease 103: 3280. [Google Scholar]
- Jiang N, Bonthond G, Fan XL, et al. 2018. Neopestalotiopsis rosicola sp. nov. causing stem canker of Rosa chinensis in China. Mycotaxon 133: 271–283. [Google Scholar]
- Jiang N, Fan XL, Tian CM. 2021. Identification and characterization of feafinhabiting fungi from Castanea plantations in China. Journal of Fungi 7: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Liang YM, Tian CM. 2019. Morphological and phylogenic evidences reveal a new Seiridium species in China. Phytotaxa 418: 287–293. [Google Scholar]
- Jin J. 2020. Investigation and application of Rosa resources in Guizhou, China. Seed 39: 61–65. [In Chinese.] [Google Scholar]
- Kang JC, Hyde KD, Kong RY. 1999. Studies on Amphisphaeriales: the Amphisphaeriaceae (sensu stricto). Mycological Research 103: 53–64. [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20: 1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T. 2007. Index of fungi inhabiting woody plants in Japan. Zenkoku Noson Kyoiku Kyokai, Tokyo, Japan. [Google Scholar]
- Kumar V, Cheewangkoon R, Gentekaki E. 2019. Neopestalotiopsis alpapicalis sp. nov. a new endophyte from tropical mangrove trees in Krabi Province (Thailand). Phytotaxa 393: 251–262. [Google Scholar]
- Lawrence DP, Travadon R, Baumgartner K. 2018. Novel Seimatosporium species from grapevine in northern California and their interactions with fungal pathogens involved in the trunk-disease complex. Plant Disease 102: 1081–1092. [DOI] [PubMed] [Google Scholar]
- Li GJ, Hyde KD, Zhao RL, et al. 2016. Fungal diversity notes 253–366: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 78: 1–237. [Google Scholar]
- Li L, Yang Q, Li H. 2021. Morphology, phylogeny, and pathogenicity of pestalotioid species on Camellia oleifera in China. Journal of Fungi 7: 1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WJ, Maharachchikumbura SSN, Li QR. 2015. Epitypification of Broomella vitalbae and introduction of a novel species of Hyalotiella. Cryptogamie, Mycologie 36: 93–108. [Google Scholar]
- Liu AR, Chen SC, Wu SY, et al. 2010. Cultural studies coupled with DNA based sequence analyses and its implication on pigmentation as a phylogenetic marker in Pestalotiopsis taxonomy. Molecular Phylogenetics and Evolution 57: 528–535. [DOI] [PubMed] [Google Scholar]
- Liu F, Bonthond G, Groenewald JZ, et al. 2019. Sporocadaceae, a family of coelomycetous fungi with appendage-bearing conidia. Studies in Mycology 92: 287–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Hou L, Raza M, et al. 2017. Pestalotiopsis and allied genera from Camellia, with description of 11 new species from China. Scientific Reports 7: 866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LL. 2016. Review on the research and utilization of the genus Rosa in China. Agricultural Science and Technology: 1–5.
- Liu JK, Hyde KD, Jones EB, et al. 2015. Fungal diversity notes 1–110: taxonomic and phylogenetic contributions to fungal species. Fungal Diversity 72: 1–197. [Google Scholar]
- Liu X, Tibpromma S, Zhang F, et al. 2021. Neopestalotiopsis cavernicola sp. nov. from Gem Cave in Yunnan Province, China. Phytotaxa 512: 1–27. [Google Scholar]
- Luo ZL, Hyde KD, Liu J, et al. 2019. Freshwater Sordariomycetes. Fungal Diversity 99: 451–660. [Google Scholar]
- Ma XY, Maharachchikumbura SSN, Chen BW, et al. 2019. Endophytic pestalotiod taxa in Dendrobium orchids. Phytotaxa 419: 268–286. [Google Scholar]
- Maharachchikumbura SSN, Camporesi E, Liu ZY, et al. 2015. Seiridium venetum redescribed, and S. camelliae, a new species from Camellia reticulata in China. Mycological Progress 14: 85. [Google Scholar]
- Maharachchikumbura SSN, Chukeatirote E, Guo LD, et al. 2013a. Pestalotiopsis species associated with Camellia sinensis (tea). Mycotaxon 123: 47–61. [Google Scholar]
- Maharachchikumbura SSN, Guo LD, Cai L, et al. 2012. A multi-locus backbone tree for Pestalotiopsis, with a polyphasic characterization of 14 new species. Fungal Diversity 56: 95–129. [Google Scholar]
- Maharachchikumbura SSN, Guo LD, Chukeatirote E, et al. 2011. Pestalotiopsis-morphology, phylogeny, biochemistry and diversity. Fungal Diversity 50: 167–187. [Google Scholar]
- Maharachchikumbura SSN, Hyde KD, Groenewald JZ, et al. 2014. Pestalotiopsis revisited. Studies in Mycology 79: 121–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharachchikumburaa SSN, Zhang YM, Wang Y, et al. 2013b. Pestalotiopsis anacardiacearum sp. nov. Amphisphaeriaceae has an intricate relationship with Penicillaria jocosatrix, the mango tip borer. Phytotaxa 99: 49–57. [Google Scholar]
- Marin-Felix Y, Hernandez-Restrepo M, Iturrieta-Gonzalez I, et al. 2019. Genera of phytopathogenic fungi: GOPHY 3. Studies in Mycology 94: 1–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur RS. 1979. The Coelomycetes of India. Bishen Singh Mahendra Pal Singh. [Google Scholar]
- Mendes M, Da Silva V, Dianese J. 1998. Fungos em Plants no Brasil. Embrapa-SPI/Embrapa-Cenargen, Brasilia. [Google Scholar]
- Meng YR. 2003. Diseases of economic plants in Gansu province. Gansu Science and Technology Press. Lanzhou, China. [In Chinese.] [Google Scholar]
- Moghadam JN, Khaledi E, Abdollahzadeh J, et al. 2022. Seimatosporium marivanicum, Sporocadus kurdistanicus, and Xenoseimatosporium kurdistanicum: three new pestalotioid species associated with grapevine trunk diseases from the Kurdistan Province, Iran. Mycological Progress 21: 427–446. [Google Scholar]
- Mordue J. 1986. Another unusual species of Pestalotiopsis: P. montellicoides sp. nov. Transactions of the British Mycological Society 86: 665–668. [Google Scholar]
- Mordue J, Holliday P. 1971. Pestalotiopsis palmarum. [Descriptions of Fungi and Bacteria]. IMI Descriptions of Fungi and Bacteria No. 319.
- Moslemi A, Taylor PW. 2015. Pestalotiopsis chamaeropis causing leaf spot disease of round leaf mint-bush (Prostanthera rotundifolia) in Australia. Australasian Plant Disease Notes 10: 1–5. [Google Scholar]
- Mułenko W, Majewski T, Ruszkiewicz-Michalska M. 2008. A preliminary checklist of micromycetes in Poland. W. Szafer Institute of Botany, Polish Academy of Sciences, Poland. [Google Scholar]
- Munoz M, Faust JE, Schnabel G. 2019. Characterization of Botrytis cinerea from commercial cut flower roses. Plant Disease 103: 1577–1583. [DOI] [PubMed] [Google Scholar]
- Nag Raj T. 1985. Redisposals and redescriptions in the Monochaetia-Seiridium, Pestalotia-Pestalotiopsis complexes. I: The correct name for the type species of Pestalotiopsis. II: Pestalotiopsis besseyii (Guba) comb. nov. and Pestalosphaeria varia sp. nov. III: Monochaetia ilicina (Sacc.) comb. nov. IV: On Monochaetia miersi. Mycotaxon 22: 43–75. [Google Scholar]
- Nag Raj T. 1993. Coelomycetous anamorphs with appendage-bearing conidia. Mycologue Publications, Waterloo, Canada. [Google Scholar]
- Nees C. 1817. Das system der Pilze und Schwämme. Stahelsche Buchhandlung, Würzburg, Germany. [Google Scholar]
- Norphanphoun C, Jayawardena RS, Chen Y, et al. 2019. Morphological and phylogenetic characterization of novel pestalotioid species associated with mangroves in Thailand. Mycosphere 10: 531–578. [Google Scholar]
- Norphanphoun C, Maharachchikumbura SSN, Daranagama A, et al. 2015. Towards a backbone tree for Seimatosporium, with S. physocarpi sp. nov. Mycosphere 6: 385–400. [Google Scholar]
- Nozawa S, Seto Y, Watanabe K. 2019. First report of leaf blight caused by Pestalotiopsis chamaeropis and Neopestalotiopsis sp. in Japanese andromeda. Journal of General Plant Pathology 85: 449–452. [Google Scholar]
- Pan M, Zhu H, Bonthond G, et al. 2020. High diversity of Cytospora associated with canker and dieback of Rosaceae in China, with 10 new species described. Frontiers in Plant Science 11: 690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patouillard N. 1886. Quelques champignons de la Chine, récoltés par M. l’abbé Delavay dans la province du Yunnan. Revue Mycologique 8: 179–182. [Google Scholar]
- Pavgi M, Singh U. 1970. Parasitic fungi from North India. IX. Sydowia 24: 113–119. [Google Scholar]
- Peregrine WTH, Ahmad KB. 1982. Brunei: A first annotated list of plant diseases and associated organisms. Phytopathological Papers 27: 1–87. [Google Scholar]
- Prasannath K, Shivas RG, Galea VJ, et al. 2021. Neopestalotiopsis species associated with flower diseases of Macadamia integrifolia in Australia. Journal of Fungi 7: 771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu F, Xu G, Zheng FQ, et al. 2020. First report of Neopestalotiopsis clavispora causing leaf spot on macadamia (Macadamia integrifolia) in China. Plant Disease 104: 288–289. [Google Scholar]
- Rai M. 1990. New records of fungi from India. Indian Journal of Mycology and Plant Pathology 20: 199–201. [Google Scholar]
- Ran S, Maharachchikumbura SSN, Ren Y, et al. 2017. Two new records in Pestalotiopsidaceae associated with Orchidaceae disease in Guangxi Province, China. Mycosphere 8: 121–130. [Google Scholar]
- Rayner RW. 1970. A mycological colour chart. CMI and British Mycological Society, Kew, Surrey, UK. [Google Scholar]
- Rebollar-Alviter A, Silva-Rojas HV, Fuentes-Aragón D, et al. 2020. An emerging strawberry fungal disease associated with root rot, crown rot and leaf spot caused by Neopestalotiopsis rosae in Mexico. Plant Disease 104: 2054–2059. [DOI] [PubMed] [Google Scholar]
- Riley EA. 1960. A revised list of plant diseases in Tanganyika Territory.
- Samarakoon M. 2016. Evolution of Xylariomycetidae (Ascomycota: Sordariomycetes). Mycosphere 7: 1746–1761. [Google Scholar]
- Sameva EF. 2004. New records of anamorphic fungi from Bulgaria. Micologia Balcanica 1: 55–57. [Google Scholar]
- Samuels G, Müller E, Petrini O. 1987. Studies in the Amphisphaeriaceae (sensu lato). III: New species of Monographella and Pestalosphaeria, and two new genera. Mycotaxon 28: 473–499. [Google Scholar]
- Santos GS, Mafia RG, Aguiar AM, et al. 2020. Stem rot of eucalyptus cuttings caused by Neopestalotiopsis spp. in Brazil. Journal of Phytopathology 168: 311–321. [Google Scholar]
- Santos J, Hilário S, Pinto G, et al. 2022. Diversity and pathogenicity of pestalotioid fungi associated with blueberry plants in Portugal, with description of three novel species of Neopestalotiopsis. European Journal of Plant Pathology 162: 539–555. [Google Scholar]
- Senanayake IC, Maharachchikumbura SSN, Hyde KD, et al. 2015. Towards unraveling relationships in Xylariomycetidae (Sordariomycetes). Fungal Diversity 73: 73–144. [Google Scholar]
- Shoemaker R. 1964. Seimatosporium (= Cryptostictis) parasites of Rosa, Vitis, and Cornus. Canadian Journal of Botany 42: 411–421. [Google Scholar]
- Silva AC, Diogo E, Henriques J, et al. 2020. Pestalotiopsis pini sp. nov., an emerging pathogen on stone pine (Pinus pinea L.). Forests 11: 805. [Google Scholar]
- Silvério ML, Cavalcanti MA, Silva GA. 2016. A new epifoliar species of Neo-pestalotiopsis from Brazil. Agrotropica 28: 151–158. [Google Scholar]
- Silvestro D, Michalak I. 2012. raxmlGUI: a graphical front-end for RAxML. Organisms Diversity & Evolution 12: 335–337. [Google Scholar]
- Smith GJ, Liew EC, Hyde KD. 2003. The Xylariales: a monophyletic order containing 7 families. Fungal Diversity 13: 185–218. [Google Scholar]
- Smith H, Wingfield M, Coutinho T, et al. 1996. Sphaeropsis sapinea and Botryosphaeria dothidea endophytic in Pinus spp. and Eucalyptus spp. in South Africa. South African Journal of Botany 62: 86–88. [Google Scholar]
- Song Y, Geng K, Hyde KD, et al. 2013. Two new species of Pestalotiopsis from Southern China. Phytotaxa 126: 22–32. [Google Scholar]
- Spaulding P. 1949. Foreign diseases of forest trees of the world: an annotated list. US Department of Agriculture, USA.
- Sutton BC. 1975. Coelomycetes. V. Coryneum. Mycological Papers 138: 1–224. Commonwealth Mycological Institute, Kew. [Google Scholar]
- Sutton BC. 1980. The coelomycetes. Fungi imperfecti with pycnidia, acervuli and stromata. Commonwealth Mycological Institute, Kew. [Google Scholar]
- Swart L, Taylor J, Crous PW, et al. 1999. Pestalotiopsis leaf spot disease of Proteaceae in Zimbabwe. South African Journal of Botany 65: 239–242. [Google Scholar]
- Swofford DL. 2003. PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods) version 4.0b10. Sinauer Associates, Sunderland, MA, USA. [Google Scholar]
- Tai FL. 1979. Sylloge Fungorum Sinicorum. Science Press, Academia Sinica. Beijing, China. [In Chinese.] [Google Scholar]
- Tamura K, Stecher G, Peterson D, et al. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Endo M, Hirayama K, et al. 2011. Phylogeny of Discosia and Seimatosporium, and introduction of Adisciso and Immersidiscosia genera nova. Persoonia 26: 85–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. 2000. Proteaceae pathogens: the significance of their distribution in relation to recent changes in phytosanitary regulations. Acta Horticulturae 545: 253–264. [Google Scholar]
- Teng SC. 1963. Fungi of China. Science Press. Beijing, China. [In Chinese.] [Google Scholar]
- Teng SC. 1996. Fungi of China. Ithaca, Mycotaxon, Ltd., NY, USA. [Google Scholar]
- Tibpromma S, Hyde KD, McKenzie EH, et al. 2018. Fungal diversity notes 840–928: micro-fungi associated with Pandanaceae. Fungal Diversity 93: 1–160. [Google Scholar]
- Tibpromma S, Mortimer PE, Karunarathna SC, et al. 2019. Morphology and multi-gene phylogeny reveal Pestalotiopsis pinicola sp. nov. and a new host record of Cladosporium anthropophilum from edible pine (Pinus armandii) seeds in Yunnan province, China. Pathogens 8: 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsopelas P, Barnes I, Wingfield M, et al. 2007. Seiridium cardinale on Juniperus species in Greece. Forest Pathology 37: 338–347. [Google Scholar]
- Ul Haq I, Ijaz S, Khan NA, et al. 2021. Genealogical concordance of phylogenetic species recognition-based delimitation of Neopestalotiopsis species associated with leaf spots and fruit canker disease affected guava plants. Pakistan Journal of Agricultural Sciences 58: 1301–1313. [Google Scholar]
- Vieira WA, Michereff SJ, De Morais MA, et al. 2014. Endophytic species of Colletotrichum associated with mango in northeastern Brazil. Fungal Diversity 67: 181–202. [Google Scholar]
- Vu D, Groenewald M, De Vries M, et al. 2019. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Studies in Mycology 92: 135–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanasinghe DN, Phukhamsakda C, Hyde KD, et al. 2018. Fungal diversity notes 709–839: taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Diversity 89: 1–236. [Google Scholar]
- Wang G. 1985. New species and records of fungi from Changbai mountain, China (I). Bulletin of Botanical Research 5: 137–139. [In Chinese.] [Google Scholar]
- Wang S. 2021. Relationships between species richness patterns of Rosa L. and environmental factors in China. Acta Ecologica Sinica 42: 1–8. [Google Scholar]
- Wang S, Mi X, Wu Z, et al. 2019a. Characterization and pathogenicity of pestalotiopsis-like species associated with gray blight disease on Camellia sinensis in Anhui province, China. Plant Disease 103: 2786–2797. [DOI] [PubMed] [Google Scholar]
- Wang Y, Xiong F, Lu Q, et al. 2019b. Diversity of pestalotiopsis-like species causing Gray Blight Disease of tea plants (Camellia sinensis) in China, including two novel Pestalotiopsis species, and analysis of their pathogenicity. Plant Disease 103: 2548–2558. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Motohashi K, Ono Y. 2010. Description of Pestalotiopsis pallidotheae: a new species from Japan. Mycoscience 51: 182–188. [Google Scholar]
- Watanabe K, Nozawa S, Hsiang T. 2018. The cup fungus Pestalopezia brunneopruinosa is Pestalotiopsis gibbosa and belongs to Sordariomycetes. PloS one 13: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss F. 1950. Index of plant diseases in the United States. Parts I, II, III. U.S. Department of Agrculture, Plant Disease Survey Special Publication. [Google Scholar]
- Wei JG, Phan CK, Wang L, et al. 2013. Pestalotiopsis yunnanensis sp. nov., an endophyte from Podocarpus macrophyllus (Podocarpaceae) based on morphology and ITS sequence data. Mycological Progress 12: 563–568. [Google Scholar]
- Wei JG, Xu T, Guo LD, et al. 2005. Endophytic Pestalotiopsis species from southern China. Mycosystema 24: 481–493. [Google Scholar]
- Wijayawardene NN, Goonasekara I, Camporesi E, et al. 2016a. Two new Seimatosporium species from Italy. Mycosphere 7: 204–213. [Google Scholar]
- Wijayawardene NN, Hyde KD, Al-Ani LKT, et al. 2020. Outline of fungi and fungus-like taxa. Mycosphere 11: 1060–1456. [Google Scholar]
- Wijayawardene NN, Hyde KD, Wanasinghe DN, et al. 2016b. Taxonomy and phylogeny of dematiaceous coelomycetes. Fungal Diversity 77: 1–316. [Google Scholar]
- Wu W. 1992. Two new Seimatosporium species from China. Journal of Hebei Academy of Sciences 2: 61–64. [Google Scholar]
- Wu ZY, Raven PH, Hong DY. (eds). 2003. Flora of China. Vol. 9 (Pittosporaceae through Connaraceae). Science Press, Beijing, and Missouri Botanical Garden Press, St. Louis. [Google Scholar]
- Yang Q, Zeng XY, Yuan J, et al. 2021. Two new species of Neopestalotiopsis from southern China. Biodiversity Data Journal 9: e70446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Wu Y, He Z, et al. 2018. Diversity and antifungal activity of endophytic fungi associated with Camellia oleifera. Mycobiology 46: 85–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Li J, Wu H, et al. 2014. First report of Chaetomella raphigera causing leaf spot on Rosa chinensis in China. Plant Disease 98: 569–569. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Jiang Y, Zu W. 2009. Resources of the genus Rosa in China and their application prospects in gardens. Seed 28: 1–9. [In Chinese.] [Google Scholar]
- Zhang YM, Maharachchikumbura SSN, Mckenzie EH. 2012a. A novel species of Pestalotiopsis causing leaf spots of Trachycarpus fortunei. Cryptogamie, Mycologie 33: 311–318. [Google Scholar]
- Zhang YM, Maharachchikumbura SSN, Tian Q. 2013. Pestalotiopsis species on ornamental plants in Yunnan Province, China. Sydowia 65: 113–128. [Google Scholar]
- Zhang YM, Maharachchikumbura SSN, Wei JG. 2012b. Pestalotiopsis camelliae, a new species associated with grey blight of Camellia japonica in China. Sydowia 64: 335–344. [Google Scholar]
- Zhang ZX, Liu RY, Liu SB, et al. 2022. Morphological and phylogenetic analyses reveal two new species of Sporocadaceae from Hainan, China. MycoKeys 88: 171–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Li N. 1994. A new species of Monochaetia (Coelomycetes) from China. Mycotaxon 52: 187–191. [Google Scholar]
- Zhu PL, Ge QX, Xu T. 1991. Seven new combinations of Pestalotiopsis from China. Acta Mycologica Sinica 10: 273–279. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic tree of Monochaetia resulting from maximum likelihood (ML) analysis of the ITS sequence alignment. Nodes are labelled with bootstrap values from RAxML/Parsimony bootstrap/Bayesian posterior probabilities values. Nodes receiving below 50 bootstrap values and 0.5 probability values are not labelled. The scale bar represents the expected number of changes per site. Isolates collected in this study are in bold and blue.
Phylogenetic trees based on maximum likelihood (ML) analyses for species in Neopestalotiopsis. a. ITS region; b. TEF gene region; c. TUB gene region. Nodes are labelled with bootstrap values from RAxML/Parsimony bootstrap/Bayesian posterior probabilities values. Nodes receiving below 50 bootstrap values and 0.5 probability values are not labelled. The scale bar represents the expected number of changes per site. Isolates collected in this study are in bold and blue.
Phylogenetic trees based on maximum likelihood (ML) analyses for species in Pestalotiopsis. a. ITS region; b. TEF gene region; c. TUB gene region. Nodes are labelled with bootstrap values from RAxML/Parsimony bootstrap/Bayesian posterior probabilities values. Nodes receiving below 50 bootstrap values and 0.5 probability values are not labelled. The scale bar represents the expected number of changes per site. Isolates collected in this study are in bold and blue.
Phylogenetic trees based on maximum likelihood (ML) analyses for species in Seimatosporium. a. ITS region; b. LSU gene region. Nodes are labelled with bootstrap values from RAxML/Parsimony bootstrap/Bayesian posterior probabilities values. Nodes receiving below 50 bootstrap values and 0.5 probability values are not labelled. The scale bar represents the expected number of changes per site. Isolates collected in this study are in bold and blue.
Phylogenetic trees based on maximum likelihood (ML) analyses for species in Seiridium. a. ITS region; b. RPB2 gene region; c. TEF gene region; d. TUB gene region. Nodes are labelled with bootstrap values from RaxML/Parsimony bootstrap/Bayesian posterior probabilities values. Nodes receiving below 50 bootstrap values and 0.5 probability values are not labelled. The scale bar represents the expected number of changes per site. Isolates collected in this study are in bold and blue.
Phylogenetic trees based on maximum likelihood (ML) analyses for species in Sporocadus. a. ITS region; b. LSU gene region; c. RPB2 gene region; d. TEF gene region; e. TUB gene region. Nodes are labelled with bootstrap values from RAxML/Parsimony bootstrap/Bayesian posterior probabilities values. Nodes receiving below 50 bootstrap values and 0.5 probability values are not labelled. The scale bar represents the expected number of changes per site. Isolates collected in this study are in bold and blue.


