Abstract
Clostridioides difficile is the leading cause of antibiotic-associated diarrhea worldwide with significant morbidity and mortality. This organism is naturally resistant to several beta-lactam antibiotics that inhibit the polymerization of peptidoglycan, an essential component of the bacteria cell envelope. Previous work has revealed that C. difficile peptidoglycan has an unusual composition. It mostly contains 3-3 cross-links, catalyzed by enzymes called L,D-transpeptidases (Ldts) that are poorly inhibited by beta-lactams. It was therefore hypothesized that peptidoglycan polymerization by these enzymes could underpin antibiotic resistance. Here, we investigated the catalytic activity of the three canonical Ldts encoded by C. difficile (LdtCd1, LdtCd2, and LdtCd3) in vitro and explored their contribution to growth and antibiotic resistance. We show that two of these enzymes catalyze the formation of novel types of peptidoglycan cross-links using meso-diaminopimelic acid both as a donor and an acceptor, also observed in peptidoglycan sacculi. We demonstrate that the simultaneous deletion of these three genes only has a minor impact on both peptidoglycan structure and resistance to beta-lactams. This unexpected result therefore implies that the formation of 3-3 peptidoglycan cross-links in C. difficile is catalyzed by as yet unidentified noncanonical Ldt enzymes.
Keywords: Clostridioides difficile; peptidoglycan; L,D-transpeptidase; cross-link; bacterial cell envelope; antibiotics; beta-lactams
Clostridioides difficile is a spore-forming Gram-positive obligate anaerobe that can cause hospital-associated diarrhea worldwide, representing increasing healthcare resource and economic burden (1). Although C. difficile has been recognized as a major cause of healthcare-associated infections since the 1970s, the more recent increase in morbidity and mortality is linked to the emergence of virulent epidemic strains including those belonging to ribotype 027 (2). C. difficile infections are underpinned by the natural resistance of this organism to several antibiotics including broad-spectrum beta-lactams such as cephalosporins. The dysbiosis caused by an antibiotic treatment creates an environment conducive to the germination of C. difficile spores and the production of virulence factors including toxins and several surface proteins (3).
The resistance of C. difficile to beta-lactams is poorly understood. These antibiotics covalently bind to D,D-transpeptidases (also known as penicillin-binding proteins) and irreversibly inhibit the enzymatic activity of these enzymes (4). In most bacteria, inhibition of D,D-transpeptidation disrupts the polymerization of peptidoglycan, the major and essential component of the bacterial cell wall and prevents bacterial growth (5). The peptidoglycan of C. difficile has an unusual composition. It is mostly polymerized by a class of enzymes called L,D transpeptidases (Ldts) (6). Unlike D,D-transpeptidases, which form bonds between the amino acids in positions 3 and 4 of peptidoglycan peptide stems (4-3 cross-links), Ldts form bonds between two amino acids in positions 3 (3-3 cross-links). The activity of Ldts involves a catalytic mechanism distinct from the mechanism of D,D-transpeptidases, and Ldts are not inhibited by beta-lactams, with the exception of penems and carbapenems (7). The C. difficile genome encodes three enzymes called LdtCd1, LdtCd2, and LdtCd3, which contain a canonical Ldt domain (YkuD). The contribution of these three enzymes to the peptidoglycan structure was investigated in strain 630 (6). Despite attempts to generate a triple knockout strain, only genes encoding LdtCd1 and LdtCd2 could be inactivated simultaneously (6) so it was suggested that 3-3 cross-links were required for viability. Analysis of the peptidoglycan structure in the double mutant strain revealed a limited decrease of 3-3 cross-links (6). The mutant remained able to perform 3-3 cross-links in the presence of ampicillin, suggesting that C. difficile Ldts were insensitive to this antibiotic (6). In vitro experiments revealed that these enzymes display distinct enzymatic activities and inhibition by beta-lactams (8). All enzymes were reported to have L,D carboxypeptidase activity, but L,D-transpeptidation and exchange of the amino acid in position 4 could only be detected for LdtCd2 and LdtCd3. Interestingly, LdtCd3 could not be acylated by any of the beta-lactams tested. The acylation efficacy of LdtCd1 and LdtCd2 by penicillin and cephalosporin antibiotics was much lower than the acylation by carbapenems, and the hydrolysis of these antibiotics was more efficient. It was therefore concluded that LdtCd activity could only be inhibited by carbapenems (8).
Outstanding questions remain on the individual role of LdtCd enzymes in peptidoglycan polymerization, the essentiality of the L,D-transpeptidation pathway in C. difficile and its contribution to antibiotic resistance. In this work, we further investigate the enzymatic activity of C. difficile Ldts, both in vitro and during vegetative growth. We show that LdtCd2 and LdtCd3 display novel enzymatic activities and that the genes encoding the three canonical Ldts can be deleted simultaneously. High-resolution structure of the wildtype and triple mutant peptidoglycan only revealed a minor impact on muropeptide composition, and no change in resistance to beta-lactams could be detected in the mutant strain. This work therefore provides new insights into the catalytic activities of Ldts and implies that the existence of another unidentified type of enzyme(s) that does not contain a canonical YkuD domain is able to catalyze the formation of 3-3 cross-links in C. difficile.
Results
In vitro assays with recombinant LdtCd1, LdtCd2, and LdtCd3 reveal distinct activities and a novel type of L,D-transpeptidation
We sought to investigate the activities of the three LdtCd enzymes to identify their specific roles in peptidoglycan remodeling. The recombinant enzymes were purified (Fig. S1) to test their enzymatic activities using four types of purified substrates: (i) a disaccharide-tetrapeptide alone (GlcNAc-MurNAc-L-Ala-D-isoGlu-meso-DAP-D-Ala; gm-AEJA, where “J” represents meso-diaminopimelic acid [DAP]) to test L,D-carboxypeptidase and L,D-transpeptidase activities (Fig. 1A); (ii) the same disaccharide-tetrapeptide (gm-AEJA) in the presence of D-methionine to test fourth amino acid exchange (Fig. 1B); (iii) a 4-3 cross-linked dimer ((GlcNAc-MurNAc-L-Ala-D-isoGlu-meso-DAP-D-Ala)2; gm-AEJA=gm-AEJA, where “=” represents a peptidoglycan cross-link) (Fig. 1C); and (iv) a 3-3 cross-linked dimer (GlcN-MurNAc-L-Ala-D-isoGlu-meso-DAP-D-Ala-GlcN-MurNAc-L-Ala-D-isoGlu-meso-DAP (g(-Ac)m-AEJA=g(-Ac)m-AEJA) to test endopeptidase activities ((Fig. 1D). The monomer and 4-3 cross-linked dimer were purified from the Escherichia coli Δ6ldt strain and therefore contain fully acetylated sugars (9). The 3-3 cross-linked dimer was purified from C. difficile and therefore contained deacetylated GlcNAc (GlcN).
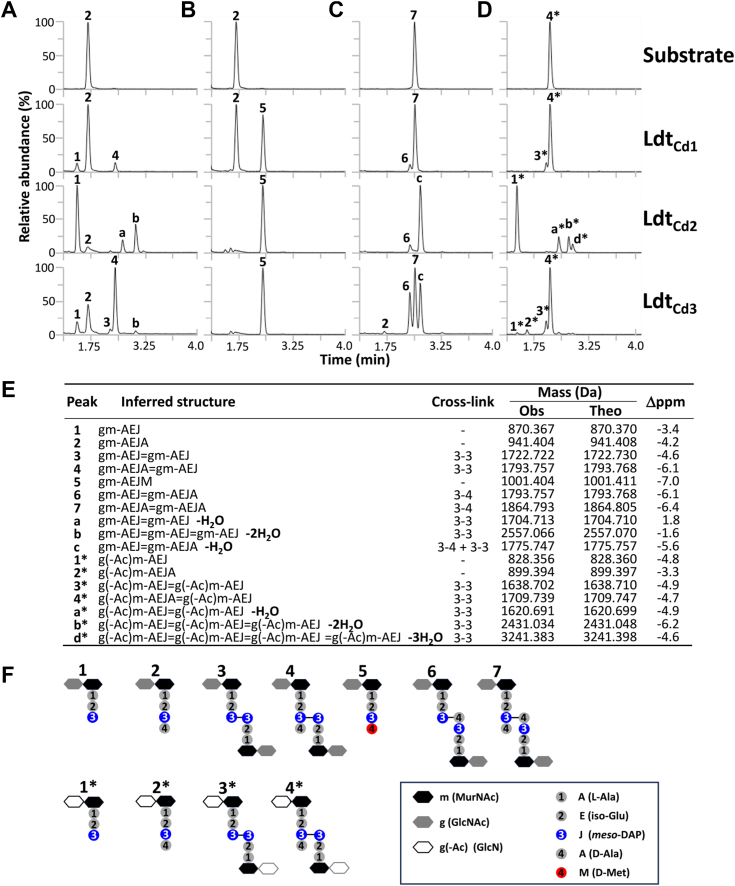
Figure 1.
HPLC-MS chromatograms of Ldt in vitro assays. Recombinant enzymes were incubated in the presence of a disaccharide-tetrapeptide substrate to test carboxypeptidase and transpeptidase activity and exchange (A and B, respectively). Endopeptidase activity was tested using either a 4-3 cross-linked or a 3-3 cross-linked dimer (C and D, respectively). The inferred structures from LC-MS analysis (E) as well as expected structures (F) are described. All traces correspond to Total Ion Chromatograms (LC-MS data) corresponding to average intensity values from three independent experiments. The difference between observed and theoretical masses (Δppm) was calculated ((Theoretical mass – Observed mass)/Theoretical mass∗10E6).
LdtCd recombinant enzymes were active against all substrates tested and revealed distinct preferential activities. LdtCd1 displayed a low carboxypeptidase and transpeptidase activity and only converted half of the substrate during the exchange reaction. No endopeptidase activity was detected with any of the dimers.
LdtCd2 had a preferential carboxypeptidase activity on the gm-AEJA substrate that was mostly converted into a disaccharide-tripeptide (gm-AEJ) and transformed all the monomer into gm-AEJM. A very weak carboxypeptidase activity was detected with the 4-3 dimer while all the 3-3 dimers were completely cleaved, releasing disaccharide-tripeptides (g(-Ac)m-AEJ) as the most abundant products. LdtCd2 was only active on 3-3 cross-linked dimers. Interestingly, several multimers matching the expected mass for dimers, trimers, and tetramers lacking a molecule of water were detected (labeled as a, b, c, a∗, b∗ and d∗; Fig. 1E). These were further analyzed by NMR.
LdtCd3 had the highest L,D-transpeptidase activity of all enzymes, and the 3-3 cross-linked dimer was the most abundant product generated from the gm-AEJA substrate. This enzyme also displayed some carboxypeptidase activity, using the monomer or both dimers and converted all the monomer into gm-AEJM. Surprisingly, the carboxypeptidase activity of LdtCd3 was higher on the 4-3 dimer than on the 3-3 dimer. LdtCd3 also produced two transpeptidation products matching the mass of a 3-3 dimer lacking a molecule of water (peak b, also detected with LdtCd2) and the mass of a 4-3 dimer lacking a molecule of water (peak c). The structures of all expected and previously described muropeptides produced by LdtCd1, LdtCd2, and LdtCd3 are described in Figure 1E.
Tandem mass spectrometry and NMR analyses of LdtCd2 and LdtCd3 transpeptidation products reveal a novel type of peptidoglycan cross-links
The muropeptide contained in peak a (Fig. 1) was analyzed by tandem mass spectrometry (MS/MS). The Fragmentation spectrum confirmed the inferred structure for a dimer with doubly cross-linked meso-DAP residues used both as an acceptor and as a donor group (Fig. 2A; see ions labeled). Several signature ions were found, including a doubly cross-linked DAP–DAP fragment.
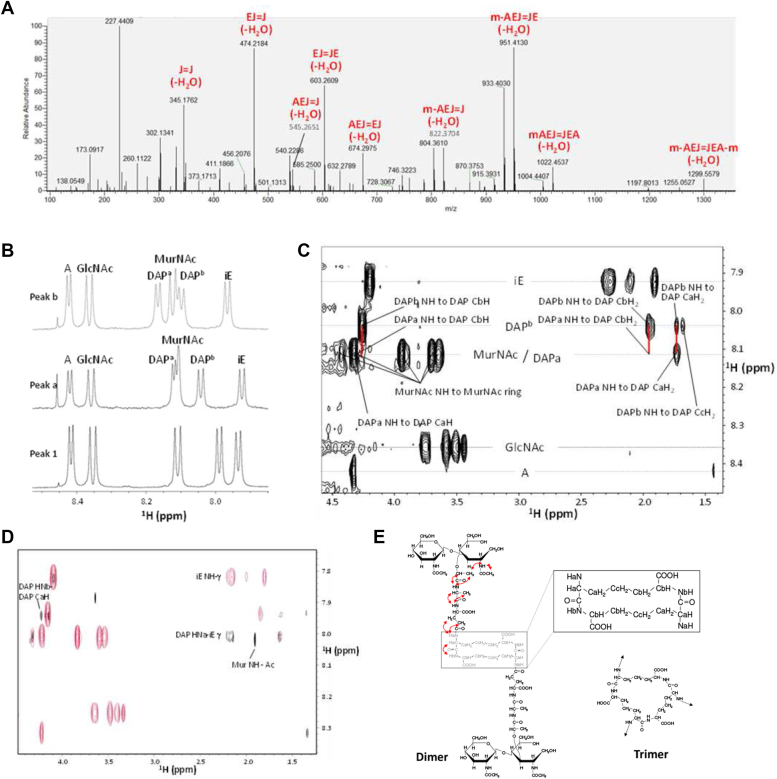
Figure 2.
MS/MS and NMR analysis of unusual muropeptides a and b.A, MS/MS analysis of the ion corresponding to peak a (m/z = 1705.7211). Nine fragment ions corresponding to peptides with doubly bonded m-DAP residues are indicated. B, 1D NMR spectra of peptidoglycan fragments identified during in vitro assays (see Fig. 1): peak 1 (gm-AEJ) was used as a control; peak a corresponds to a dimer (gm-AEJ=gm-AEJ) with an unusual cross-link; peak b corresponds to a trimer (gm-AEJ=gm-AEJ=gm-AEJ) with an unusual cross-link The identity of each amide proton is indicated on the spectra. C, part of the TOCSY spectrum of the muropeptide in peak a, showing connectivities between amide protons and side chains. The signals linked by red lines are the connectivities for the DAPa and DAPb amides, showing that they connect to identical side-chain frequencies and are therefore part of the same spin system. D, TOCSY (red) and NOESY (black) spectra of the dimer. Significant peaks are marked. E, structures of (left) dimer (with NOEs indicated) and (right) trimer. Only the central part of the trimer is shown, with arrows indicating where the AEJ chains are attached. A, alanine; DAPa, meso-diaminopimelic acid backbone (directly bonded to the isoglutamate); DAPb, meso-diaminopimelic acid side chain; GlcNAc, N-acetylglucosamine; iE, isoglutamate; MurNAc, N-acetylmuramic acid.
Peptidoglycan fragments in peaks 1, a, and b (Fig. 1E) were purified and further analyzed by NMR. One-dimensional (1D) NMR spectra of the peptidoglycan fragments demonstrated a high purity for each. The monomer gm-AEJ (peak 1) used as a control (Fig. 2B, bottom panel) had all the amide signals expected, namely, two sugar N-acetyl signals, and one signal each for the Ala, DAP, and isoGlu (iE) residues. There is only one signal for DAP because one amine forms an amide with iE, while the other is a free amino group and therefore exchanges too fast with water to be visible. All the other signals are as expected, including the presence of two N-acetyl methyl singlets from the two sugars and two methyl doublets from Ala and the lactyl group on MurNAc.
The dimer corresponding to peak a has a remarkably simple NMR spectrum (Fig. 2B, middle panel). The amide region contains only six amide doublets, two N-acetyl methyl singlets, and two methyl doublets, as seen in the monomer (Fig. S2). This simplicity very strongly suggests a symmetrical dimer, and the similarity of the amide chemical shifts between the monomer and dimer implies a similar covalent structure.
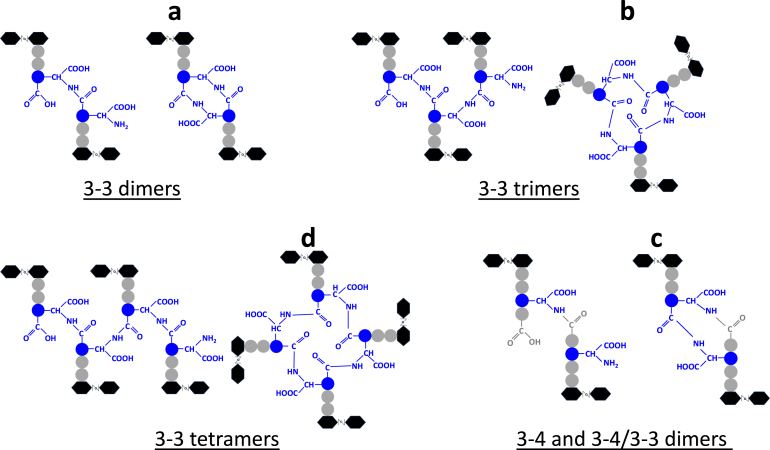
There are two amide signals from the diaminopimelate, visible in the total correlation spectroscopy (TOCSY) spectrum because they belong to the same spin system (Fig. 2C), indicating that both amines in the diaminopimelate take part in amide bonds. Chemical shift assignments for the dimer are listed in Table S1. In the nuclear Overhauser enhancement spectroscopy (NOESY) spectrum, there are the expected sequential NOEs present between NHi and protons in residue (i-1), as indicated in Figure 2D. Crucially, these include NOEs between DAP NHa and iE CγH2, and the other “sequential” NOE of DAP NHb to DAP CaH (Fig. 2, D and E). Similarly, the spectrum of trimer b (Fig. 2B, top panel) is also very similar. The chemical shifts remain very similar to the monomer and dimer, and again there is only one set of signals, indicating a symmetrical trimer. The NMR spectra are thus fully consistent with the structures described in Figure 3, and the simplicity of the spectrum means that no unsymmetrical structure is possible. Based on our NMR data, we conclude that the muropeptides in peaks a, b, c, and d all correspond to multiply cross-linked structures (Figs. 2 and 3).
Figure 3.
Unusual peptidoglycan cross-links catalyzed by LdtCd2and LdtCd3. Based on NMR data, the structure of muropeptides corresponding to peaks a, b, c, and d are shown next to the structure of the canonical 3-3 or 4-3 dimers, 3-3 trimer, and 3-3 tetramer. Dimer c contains both types of cross-links (3-3 and 4-3), resulting from D,D- and L,D-transpeptidation. Muropeptides a∗, b∗, d∗ display the same cross-links as a, b, d but contain GlcN instead of GlcNAc.
TraDIS data mining and gene deletion reveal that the three canonical C. difficile Ldts are nonessential
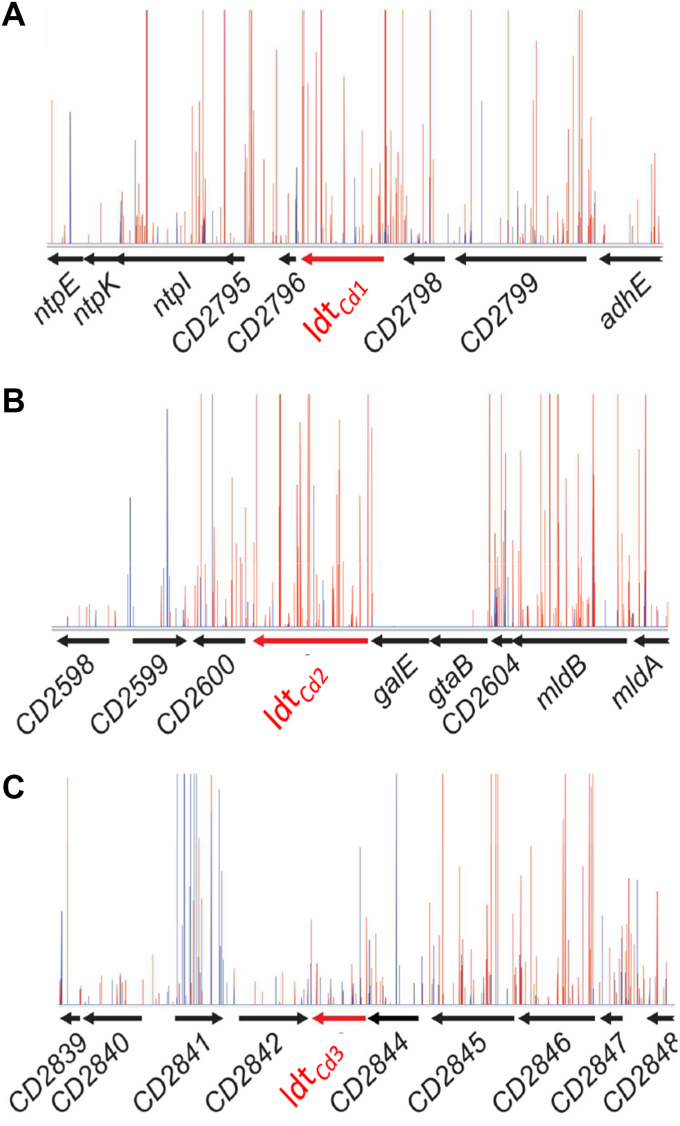
Previous attempts to build a mutant with deletions in all three genes encoding the canonical Ldts were unsuccessful (6), suggesting either that one of them is essential or that 3-3 cross-linking is essential. We took advantage of transposon-directed insertion site sequencing (TraDIS) data previously published in a study that identified essential genes in C. difficile R20291 (10). Using the number of transposon insertions in each ldtCd gene as a proxy to determine essentiality, we concluded that none of these genes was essential (Fig. 4), leaving the possibility that the combined deletion of ldtCd1, ldtCd2, and ldtCd3 could be non viable. To test this hypothesis, we sought to generate a series of in-frame deletions in ldtCd1, ldtCd2, and ldtCd3. All genes were deleted individually or simultaneously. All the combinations of deletions, including the triple deletion mutant, could be obtained, showing that these genes are not required for viability. Since this result was unexpected, we performed whole genome sequencing on the mutants and confirmed the deletion of the ldt genes in the strains sequenced. Single nucleotide polymorphism analysis identified a unique single mutation in the triple deletion mutant (File S1 and Fig. S3). The mutation (T>TA) occurred at position 581480 in the intergenic mutation between CD0482 and glsA, two genes with no known link to peptidoglycan polymerization, encoding a putative phosphoribulokinase/uridine kinase and a putative glutaminase, respectively. This result therefore suggests that the deletion of the ldtCd genes does not lead to genetic mutations likely to compensate for the lack of L,D-transpeptidase activity.
Figure 4.
TraDIS analysis of the ldtCd loci. The number of transposition events in ldtCd1 (A), ldtCd2 (B), and ldtCd3 (C) are shown as histograms depicting the localization (x-axis) and the frequency (y-axis) of transposon insertion sites (in red are antisense insertions; in blue are sense insertions).
High-resolution structure of the wildtype and triple mutant peptidoglycans
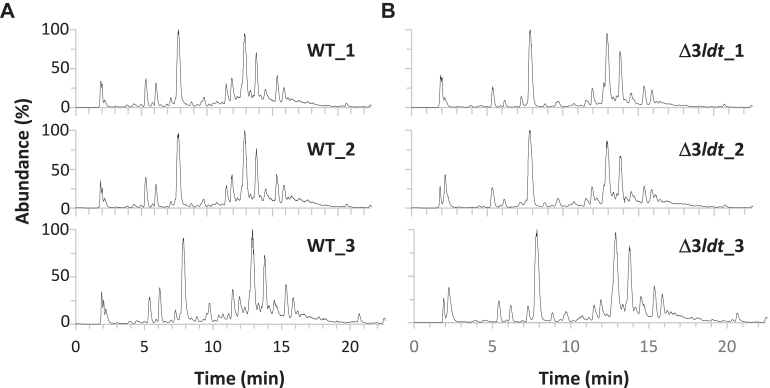
Peptidoglycan was extracted from vegetative cells in stationary phase, and soluble fragments released after mutanolysin digestion were analyzed by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS). Surprisingly, the chromatograms of the WT and triple mutant were virtually identical, indicating a very minor contribution of the three Ldts to peptidoglycan structure (Fig. 5).
Figure 5.
HPLC-MS chromatogram of C. difficile reduced disaccharide-peptides. Each total ion chromatogram corresponds to a biological replicate of strain R20291 (WT) (A) and its isogenic derivative with in-frame deletions in genes ldtCd1, ldtCd2, and ldtCd3 (Δ3ldt) (B).
To investigate subtle differences associated with the simultaneous deletion of the three ldtCd genes, we performed a high-resolution analysis of the LC-MS/MS datasets using the Byos (Protein Metrics by Dotmatics) and PGFinder software (11). A bespoke search strategy was designed (Fig. S4). A first search was performed to identify the monomer search space using the Byonic module from the proprietary software Byos. Thirty-four disaccharide-peptides with a fragmentation showing more than half of the expected b and y ions were identified (Fig. S5). These included 24 deacetylated monomers containing di-, tri-, tetra-, and pentapeptide stems (g(-Ac)m-AE, g(-Ac)m-AEJ, g(-Ac)m-AEJX, and g(-Ac)m-AEJAX) and 10 fully acetylated monomers (gm-AE, gm-AEJ, gm-AEJX, and gm-AEJAX), where X can be any amino acid. A database called DB_0 made of these 34 monomers was used to perform a PGFinder search (step 2 in Fig. S4) to identify the most abundant monomers. Thirteen disaccharide-peptides accounting for more than 98% of the monomers identified were selected to create a second database (DB_1) containing dimers resulting from 3-3 and 4-3 cross-linking. A third PGFinder search (step 3 in Fig. S4) was performed to identify the most abundant dimers and generate the next database containing monomers, dimers, and trimers (DB_2). The next search with PGFinder and DB_2 (step 4 in Fig. S4) identified the most abundant trimers. A final database called DB_3 was created using all the information from sequential searches; it contained all MS/MS-checked monomers, 26 dimers, 16 trimers, and all AnhydroMurNAc derivatives of the 10 most abundant mono-, di-, and trimers, as well as 4 unusually cross-linked structures identified during in vitro assays.
The result of the PGFinder search using DB_3 and biological replicates from the WT and triple mutant is described in Table 1. The search strategy described here combining both LC-MS and LC-MS/MS analysis allowed us to identify 97 muropeptides, which is an unprecedentedly detailed analysis. The comparison between the two strains revealed a remarkable similarity between the two peptidoglycan compositions (Table 2). No significant difference was found when comparing the proportion of monomers, dimers, trimers, or glycan chain length. Cross-linking index as well as the proportion of 3-3 cross-links was also similar. The only difference found was a significant decrease in the exchange reaction (23.4 ± 0.7% in the WT and 16.9 ± 1.4% in the triple mutant). Overall, our analysis therefore demonstrated that the three canonical L,D-transpeptidases LdtCd1, LdtCd2, and LdtCd3 only contribute marginally to remodel the peptidoglycan of C. difficile vegetative cells.
Table 1.
Muropeptide analysis of C. difficile WT and triple ldtCd1, ldtCd2, ldtCd3 mutant (Δ3ldt)
| Muropeptidea | Ldt activity/crosslinkb | Abundance (%)c |
RT (min) | Averaged Δppm | |
|---|---|---|---|---|---|
| WT | Δ3ldt | ||||
| gm(-Ac)-AEJA|1 | N/A | 21.594% ± 1.050% | 28.192% ± 4.671% | 7.90 ± 0.02 | 2.71 |
| gm(-Ac)-AEJ|1 | Carboxypeptidase | 9.002% ± 1.348% | 8.235% ± 1.026% | 5.39 ± 0.03 | 2.85 |
| gm(-Ac)-AEJG|1 | Exchange | 9.814% ± 0.907% | 4.696% ± 1.177% | 6.16 ± 0.02 | 2.55 |
| gm(-Ac)-AEJF|1 | Exchange | 1.820% ± 0.806% | 2.083% ± 0.886% | 20.72 ± 0.03 | 2.14 |
| gm(-Ac)-AE|1 | N/A | 3.174% ± 0.185% | 2.297% ± 0.520% | 7.74 ± 0.05 | 2.31 |
| gm-AEJA|1 | N/A | 0.512% ± 0.061% | 0.716% ± 0.211% | 8.84 ± 0.02 | 1.50 |
| gm(-Ac)-AEJV|1 | Exchange | 0.944% ± 0.111% | 0.570% ± 0.113% | 13.83 ± 0.02 | 1.71 |
| gm(-Ac)-AEJI|1 | Exchange | 0.294% ± 0.076% | 0.389% ± 0.131% | 18.20 ± 0.04 | 0.66 |
| gm(-Ac)-AEJY|1 | Exchange | 0.172% | 0.328% 0.072% | 15.82 ± 0.04 | 0.99 |
| gm(-Ac)-AEJAA|1 | N/A | 0.244% ± 0.007% | 0.311% ± 0.053% | 9.63 ± 0.08 | 1.68 |
| gm-AEJA (Anh)|1 | N/A | 0.168% ± 0.010% | 0.270% ± 0.085% | 7.97 ± 0.11 | 2.24 |
| gm(-Ac)-AEJS|1 | Exchange | 0.302% ± 0.036% | 0.234% ± 0.039% | 5.52 ± 0.02 | 2.04 |
| gm-AEJ|1 | N/A | 0.197% ± 0.025% | 0.207% ± 0.130% | 9.41 ± 4.13 | 1.89 |
| gm(-Ac)-AEJK|1 | Exchange | 0.324% ± 0.086% | 0.184% ± 0.053% | 4.55 ± 0.06 | 2.08 |
| gm(-Ac)-AEJQ|1, gm(-Ac)-AEJAG|1 | Exchange | 0.107% ± 0.011% | 0.095% ± 0.022% | 8.41 ± 1.39 | 0.86 |
| gm(-Ac)-AEJA (Anh)|1 | N/A | 0.033% ± 0.002% | 0.084% ± 0.010% | 11.35 ± 0.02 | 0.31 |
| gm-AEJG|1 | Exchange | 0.115% ± 0.026% | 0.071% ± 0.010% | 7.74 ± 0.75 | 1.08 |
| gm-AE|1 | N/A | 0.081% ± 0.016% | 0.062% ± 0.019% | 8.65 ± 0.03 | 0.41 |
| gm(-Ac)-AEJAT|1 | N/A | 0.177% ± 0.021% | 0.096% ± 0.022% | 6.63 ± 0.04 | 0.61 |
| gm(-Ac)-AEJAF|1 | N/A | 0.042% ± 0.015% | 0.055% ± 0.018% | 24.36 ± 0.02 | 1.30 |
| gm-AEJI|1 | Exchange | 0.015% ± 0.009% | 0.042% ± 0.028% | 16.08 ± 4.43 | 0.88 |
| gm(-Ac)-AEJN|1 | Exchange | 0.235% ± 0.065% | 0.062% ± 0.004% | 5.05 ± 0.03 | 1.49 |
| gm(-Ac)-AEJH|1 | Exchange | 0.104% ± 0.080% | ND | 5.19 ± 0.20 | 2.01 |
| gm(-Ac)-AEJE|1 | Exchange | 0.062% ± 0.010% | 0.041% ± 0.003% | 7.51 ± 0.03 | 1.37 |
| gm-AEJF|1 | Exchange | 0.019% ± 0.013% | 0.020% ± 0.014% | 21.95 ± 0.01 | 1.35 |
| gm-AEJK|1, gm(-Ac)-AEJAV|1 (mixture) | Exchange | 0.000% ± 0.000% | 0.000% ± 0.000% | 6.69 ± 3.18 | 1.88 |
| gm(-Ac)-AEJAI|1 | N/A | 0.021% ± 0.003% | 0.023% ± 0.003% | 19.36 ± 0.02 | 0.76 |
| gm(-Ac)-AEJAK|1 | N/A | 0.019% ± 0.001% | 0.024% ± 0.006% | 7.22 ± 0.12 | 1.79 |
| gm(-Ac)-AEJD|1 | N/A | 0.125% ± 0.027% | 0.024% ± 0.004% | 6.27 ± 0.04 | 2.03 |
| gm-AEJH|1 | Exchange | 0.014% ± 0.007% | ND | 6.40 ± 0.02 | 0.74 |
| gm(-Ac)-AEJT|1 | Exchange | 0.025% ± 0.002% | 0.020% ± 0.002% | 6.49 ± 0.14 | 1.08 |
| gm(-Ac)-AE (Anh)|1 | N/A | 0.029% ± 0.004% | 0.015% ± 0.004% | 12.00 ± 0.01 | 1.58 |
| gm-AEJAA|1 | N/A | 0.010% ± 0.001% | 0.010% ± 0.000% | 10.79 ± 1.54 | 1.10 |
| gm(-Ac)-AEJ (Anh)|1 | N/A | 0.025% ± 0.005% | 0.013% ± 0.003% | 8.56 ± 0.01 | 0.58 |
| gm(-Ac)-AEJAY|1 | N/A | 0.005% | 0.005% ± 0.004% | 17.18 ± 0.03 | 0.22 |
| gm(-Ac)-AEJG (Anh)|1 | Exchange | 0.015% ± 0.004% | 0.006% ± 0.003% | 9.31 ± 0.10 | 0.46 |
| gm(-Ac)-AEJAA (Anh)|1 | Exchange | ND | 0.001% ± 0.000% | 9.49 ± 0.04 | 1.74 |
| gm(-Ac)-AEJY (Anh)|1 | Exchange | ND | 0.000% | 24.42 ± 0.03 | 1.39 |
| gm-AEJAF|1 | N/A | 0.001% ± 0.001% | 0.001% ± 0.001% | 24.07 ± 0.01 | 1.04 |
| gm(-Ac)-AEJA=gm(-Ac)-AEJ|2 | 3-3 | 16.323% ± 0.074% | 19.316% ± 1.031% | 13.09 ± 0.08 | 1.31 |
| gm(-Ac)-AEJA=gm(-Ac)-AEJA|2 | 3-4 | 6.806% ± 1.727% | 8.963% ± 1.708% | 13.93 ± 0.11 | 1.20 |
| gm(-Ac)-AEJ=gm(-Ac)-AEJ|2 | 3-3 | 5.903% ± 1.156% | 5.446% ± 0.636% | 11.99 ± 0.03 | 1.57 |
| gm(-Ac)-AEJG=gm(-Ac)-AEJ|2 | 3-3 | 5.741% ± 0.730% | 2.688% ± 0.881% | 11.54 ± 0.04 | 1.39 |
| gm-AEJA=gm(-Ac)-AEJ|2 | 3-3 | 1.238% ± 0.141% | 1.677% ± 0.398% | 13.80 ± 0.03 | 1.77 |
| gm(-Ac)-AEJF=gm(-Ac)-AEJ|2 | 3-3 | 0.682% ± 0.213% | 0.719% ± 0.283% | 22.56 ± 0.01 | 1.85 |
| gm-AEJA=gm(-Ac)-AEJA|2 | 3-4 | 0.560% ± 0.052% | 0.713% ± 0.151% | 14.78 ± 0.08 | 0.91 |
| gm(-Ac)-AEJG=gm(-Ac)-AEJA|2 | 3-4 | 1.112% ± 0.202% | 0.467% ± 0.114% | 12.40 ± 0.02 | 0.68 |
| gm-AEJA=gm(-Ac)-AEJ (Anh)|2 | 3-3 | 0.244% ± 0.036% | 0.374% ± 0.131% | 19.22 ± 0.10 | 0.83 |
| gm(-Ac)-AEJV=gm(-Ac)-AEJ|2 | 3-3 | 0.308% ± 0.047% | 0.205% ± 0.057% | 17.77 ± 0.02 | 0.57 |
| gm(-Ac)-AEJA=gm(-Ac)-AEJ (Anh)|2 | 3-3 | 0.337% ± 0.040% | 0.244% ± 0.023% | 15.68 ± 0.32 | 1.47 |
| gm(-Ac)-AEJI=gm(-Ac)-AEJ|2 | 3-3 | 0.138% ± 0.055% | 0.176% ± 0.073% | 20.98 ± 0.02 | 0.88 |
| gm-AEJ=gm(-Ac)-AEJ|2 | 3-3 | 0.250% ± 0.011% | 0.250% ± 0.092% | 12.69 ± 0.03 | 1.26 |
| gm(-Ac)-AEJA=gm(-Ac)-AEJA (Anh)|2 | 3-4 | 0.113% ± 0.007% | 0.176% ± 0.011% | 16.99 ± 0.02 | 0.34 |
| gm(-Ac)-AEJF=gm(-Ac)-AEJA|2 | 3-4 | 0.161% ± 0.061% | 0.130% ± 0.043% | 23.14 ± 0.02 | 1.43 |
| gm(-Ac)-AEJY=gm(-Ac)-AEJ|2 | 3-3 | 0.063% ± 0.084% | 0.107% ± 0.034% | 18.74 ± 1.18 | 0.37 |
| gm(-Ac)-AEJS=gm(-Ac)-AEJ|2 | 3-3 | 0.207% ± 0.005% | 0.133% ± 0.014% | 11.41 ± 0.14 | 0.63 |
| gm-AEJA=gm(-Ac)-AEJA (Anh)|2 | 3-4 | 0.072% ± 0.019% | 0.125% ± 0.012% | 14.02 ± 0.30 | 9.40 |
| gm(-Ac)-AEJK=gm(-Ac)-AEJ|2 | 3-3 | 0.154% ± 0.033% | 0.080% ± 0.022% | 10.20 ± 0.02 | 0.27 |
| gm(-Ac)-AEJI=gm(-Ac)-AEJA|2 | 3-4 | 0.037% ± 0.013% | 0.042% ± 0.016% | 21.78 ± 0.02 | 0.42 |
| gm(-Ac)-AEJV=gm(-Ac)-AEJA|2 | 3-4 | 0.067% ± 0.006% | 0.038% ± 0.011% | 18.60 ± 0.02 | 0.35 |
| gm(-Ac)-AEJS=gm(-Ac)-AEJA|2 | 3-4 | 0.053% ± 0.006% | 0.046% ± 0.008% | 12.09 ± 0.02 | 1.10 |
| gm(-Ac)-AEJAA=gm(-Ac)-AEJA|2 | 3-4 | 0.013% ± 0.006% | 0.027% ± 0.002% | 13.96 ± 0.51 | 0.48 |
| gm(-Ac)-AEJ=gm(-Ac)-AEJ (Anh)|2 | 3-3 | 0.110% ± 0.021% | 0.011% | 14.84 ± 0.22 | 0.30 |
| gm(-Ac)-AEJN=gm(-Ac)-AEJ|2 | 3-3 | 0.094% ± 0.025% | 0.028% ± 0.005% | 10.88 ± 0.02 | 0.64 |
| gm(-Ac)-AEJK=gm(-Ac)-AEJA|2 | 3-4 | 0.044% ± 0.008% | 0.027% ± 0.002% | 10.97 ± 0.02 | 0.56 |
| gm(-Ac)-AEJY=gm(-Ac)-AEJA|2 | 3-4 | 0.013% ± 0.000% | 0.021% ± 0.005% | 18.92 ± 0.02 | 0.31 |
| gm(-Ac)-AEJA=gm(-Ac)-AEJ (-H2O)|2 | 3-3∗ | 0.138% ± 0.000% | 0.016% ± 0.005% | 14.67 ± 0.54 | 0.55 |
| gm(-Ac)-AEJN=gm(-Ac)-AEJA|2 | 3-4 | 0.021% ± 0.006% | 0.009% ± 0.001% | 11.62 ± 0.02 | 1.08 |
| gm(-Ac)-AEJG=gm(-Ac)-AEJA (Anh)|2 | 3-4 | 0.004% | 0.000% | 15.38 ± 0.00 | 2.26 |
| gm(-Ac)-AEJF=gm(-Ac)-AEJ (Anh)|2 | 3-3 | 0.010% ± 0.002% | 0.014% ± 0.014% | 20.96 ± 4.59 | 5.28 |
| gm(-Ac)-AEJG=gm(-Ac)-AEJ (Anh)|2 | 3-3 | 0.126% ± 0.017% | 0.001% | 15.56 ± 2.77 | 0.79 |
| gm(-Ac)-AEJI=gm(-Ac)-AEJ (Anh)|2 | 3-3 | 0.002% ± 0.001% | 0.001% ± 0.000% | 23.32 ± 0.03 | 1.04 |
| gm(-Ac)-AEJV=gm(-Ac)-AEJ (Anh)|2 | 3-3 | 0.015% ± 0.011% | 0.001% | 19.18 ± 3.13 | 2.32 |
| gm(-Ac)-AEJA=gm(-Ac)-AEJ=gm(-Ac)-AEJ|3 | 3-3 | 3.428% ± 0.114% | 3.041% ± 0.233% | 15.39 ± 0.04 | 1.42 |
| gm(-Ac)-AEJA=gm(-Ac)-AEJA=gm(-Ac)-AEJ|3 | 3-3, 3-4 | 1.823% ± 0.087% | 2.273% ± 0.327% | 15.98 ± 0.02 | 1.09 |
| gm-AEJA=gm(-Ac)-AEJA=gm(-Ac)-AEJ|3 | 3-3, 3-4 | 0.344% ± 0.047% | 0.446% ± 0.133% | 16.76 ± 0.02 | 0.27 |
| gm-AEJA=gm(-Ac)-AEJ=gm(-Ac)-AEJ|3 | 3-3 | 0.505% ± 0.052% | 0.449% ± 0.135% | 16.19 ± 0.02 | 0.48 |
| gm(-Ac)-AEJA=gm(-Ac)-AEJA=gm(-Ac)-AEJA|3 | 3-4 | 0.348% ± 0.040% | 0.468% ± 0.060% | 16.55 ± 0.03 | 0.30 |
| gm-AEJA=gm(-Ac)-AEJ=gm(-Ac)-AEJ (Anh)|3 | 3-3 | 0.259% ± 0.026% | 0.342% ± 0.073% | 20.34 ± 0.03 | 1.20 |
| gm(-Ac)-AEJ=gm(-Ac)-AEJ=gm(-Ac)-AEJ|3 | 3-3 | 0.638% ± 0.165% | 0.395% ± 0.046% | 14.46 ± 0.02 | 0.48 |
| gm(-Ac)-AEJG=gm(-Ac)-AEJ=gm(-Ac)-AEJ|3 | 3-3 | 0.565% ± 0.090% | 0.194% ± 0.068% | 14.13 ± 0.02 | 0.23 |
| gm-AEJA=gm(-Ac)-AEJA=gm(-Ac)-AEJA|3 | 3-4 | 0.095% ± 0.015% | 0.142% ± 0.032% | 17.30 ± 0.02 | 0.31 |
| gm(-Ac)-AEJG=gm(-Ac)-AEJA=gm(-Ac)-AEJ|3 | 3-3, 3-4 | 0.253% ± 0.052% | 0.115% ± 0.038% | 14.69 ± 0.01 | 0.31 |
| gm-AEJA=gm(-Ac)-AEJA=gm(-Ac)-AEJ (Anh)|3 | 3-3, 3-4 | 0.097% ± 0.008% | 0.136% ± 0.017% | 20.18 ± 1.58 | 2.32 |
| gm(-Ac)-AEJF=gm(-Ac)-AEJ=gm(-Ac)-AEJ|3 | 3-3 | 0.061% ± 0.018% | 0.041% ± 0.019% | 23.03 ± 0.01 | 1.05 |
| gm(-Ac)-AEJA=gm(-Ac)-AEJ=gm(-Ac)-AEJ (Anh)|3 | 3-3 | 0.174% ± 0.019% | 0.049% ± 0.008% | 17.55 ± 0.16 | 0.30 |
| gm(-Ac)-AEJA=gm(-Ac)-AEJA=gm(-Ac)-AEJ (Anh)|3 | 3-3, 3-4 | 0.062% ± 0.007% | 0.051% ± 0.005% | 18.40 ± 0.09 | 0.40 |
| gm(-Ac)-AEJF=gm(-Ac)-AEJ=gm(-Ac)-AEJA|3 | 3-3, 3-4 | 0.036% ± 0.013% | 0.026% ± 0.010% | 23.39 ± 0.02 | 0.40 |
| gm(-Ac)-AEJG=gm(-Ac)-AEJA=gm(-Ac)-AEJA|3 | 3-4 | 0.047% ± 0.012% | 0.021% ± 0.008% | 15.23 ± 0.01 | 0.59 |
| gm(-Ac)-AEJG=gm(-Ac)-AEJ=gm(-Ac)-AEJ (Anh)|3 | 3-3 | 0.047% ± 0.007% | ND | 16.24 ± 0.02 | 0.22 |
| gm(-Ac)-AEJ=gm(-Ac)-AEJ=gm(-Ac)-AEJ (-H2O)|3 | 3-3∗ | 0.056% ± 0.015% | ND | 16.98 ± 0.03 | 1.18 |
| gm(-Ac)-AEJA=gm(-Ac)-AEJA=gm(-Ac)-AEJA (Anh)|3 | 3-4 | 0.013% ± 0.001% | 0.017% ± 0.002% | 19.02 ± 0.04 | 0.40 |
| gm(-Ac)-AEJG=gm(-Ac)-AEJ=gm(-Ac)-AEJA (Anh)|3 | 3-3, 3-4 | 0.011% ± 0.001% | ND | 16.82 ± 0.12 | 0.25 |
| gm(-Ac)-AEJA=gm(-Ac)-AEJ=gm(-Ac)-AEJ (-H2O)|3 | 3-3 | 0.002% | ND | 16.83 ± 0.00 | 2.64 |
| gm-AEJA=gm(-Ac)-AEJA=gm(-Ac)-AEJA (Anh)|3 | 3-4 | 0.004% ± 0.006% | 0.016% ± 0.007% | 17.35 ± 1.74 | 4.89 |
| gm(-Ac)-AEJ=gm(-Ac)-AEJ=gm(-Ac)-AEJ (Anh)|3 | 3-3 | 0.083% ± 0.006% | 0.005% ± 0.003% | 16.68 ± 0.28 | 1.56 |
| gm(-Ac)-AEJF=gm(-Ac)-AEJ=gm(-Ac)-AEJ (Anh)|3 | 3-3 | 0.002% ± 0.000% | 0.005% ± 0.005% | 23.51 ± 3.39 | 3.09 |
| gm(-Ac)-AEJ=gm(-Ac)-AEJ=gm(-Ac)-AEJ (Anh)|3 | 3-3 | 0.083% ± 0.006% | 0.005% ± 0.003% | 16.68 ± 0.28 | 1.56 |
| gm(-Ac)-AEJF=gm(-Ac)-AEJ=gm(-Ac)-AEJ (Anh)|3 | 3-3 | 0.002% ± 0.000% | 0.005% ± 0.005% | 23.51 ± 3.39 | 3.09 |
Abbreviation: RT, retention time.
When multiple dimer structures are possible, the most likely structure is proposed based on the abundance of acceptor stems.
N/A, not applicable; 3-3∗ crosslinks correspond to L,D-transpeptidation products doubly crosslinked.
ND, not detected; no standard deviation is provided when muropeptides were only identified in a single replicate.
Absolute mass difference between observed and theoretical mass in parts per million (ppm).
Table 2.
Summary of WT and 3Δldt PG properties
| Muropeptides/properties | WT | Δ3ldt |
|---|---|---|
| Monomers | 49.93% ± 2.82% | 49.96% ± 3.13% |
| Dimers | 41.12% ± 2.15% | 41.94% ± 2.19% |
| Trimers | 8.95% ± 1.00% | 8.09% ± 1.04% |
| Cross-linking index | 23.51% ± 1.40% | 23.64% ± 1.44% |
| Glycan chain length | 96.6 ± 1.8 | 87.3 ± 18.7 |
| gm-AEJX | 23.39% ± 0.70% | 16.86% ± 1.41% |
| 3-3 cross-links | 81.33% ± 2.54% | 78.35% ± 2.68% |
Comparative phenomics of the parental R20291 strain and its isogenic Δ3ldt derivative
A comprehensive set of experiments were carried out to compare the phenotype of the R20291 strain and the triple ldtCd mutant. As expected, based on the results from peptidoglycan analysis, no significant differences were observed between the two strains in cell size (Fig. S6), sporulation (Fig. S7), or toxin release (Fig. S8). Minimum inhibitory concentrations (MICs) for several beta-lactams were also tested for all the mutants generated in this study and did not reveal any difference in the resistance against any of these antibiotics (Table S2).
Discussion
Recombinant L,D-transpeptidases represent a class of enzymes that are amenable to study in vitro since they can use soluble peptidoglycan fragments as a substrate. In vitro assays with distinct peptidoglycan fragments purified from intact sacculi were used to explore the catalytic activities of the three C. difficile Ldts. Our comparative analysis based on an LC-MS/MS assay with several substrates provided information about the preferential activity of each Ldt. Previous studies reported that LdtCd1 only displayed carboxypeptidase activity (8). Our data confirmed this result and also revealed that it can also perform transpeptidation and exchange reactions, even though this enzyme was poorly active on all substrates tested. LdtCd2 was able to perform all reactions but preferentially acted as a carboxypeptidase. Remarkably, LdtCd2 was the only enzyme with endopeptidase activity, exclusively using 3-3 dimers as a substrate. Unlike other endopeptidases that cleave 3-3 cross-links (MepA, MepM, and MepK), LdtCd2 has a strict substrate specificity for 3-3 cross-links since no activity against 4-3 cross-links could be detected (12, 13) (Table S3). LdtCd2 activity therefore appears to be unique since it is the first enzyme described that is only active on 3-3 cross-links.
LdtCd3 displayed the highest transpeptidase/exchange activity and a relatively weak carboxypeptidase and endopeptidase activity, preferentially against 3-3 cross-linked dimers. Unlike LdtCd2, LdtCd3 could cleave 4-3 dimers with low efficiency.
Besides the exhaustive description of expected LdtCd activities, our in vitro assays also revealed that LdtCd2 and LdtCd3 can generate a novel type of peptidoglycan cross-links. These result from double transpeptidation reactions that use meso-DAP both as a donor and an acceptor group. Interestingly, we identified double cross-linked dimers containing either two 3-3 cross-links or a mixture of 4-3 and 3-3 cross-links. In hindsight, this result is not entirely surprising since the catalytic reaction leading to this type of bond is the same as the reaction leading to the formation of “normal” 3-3 bonds. Double cross-links can be detected in the peptidoglycan from C. difficile as well as in the peptidoglycan from Gram-negative organisms producing Ldts (E. coli, Rhizobium leguminosarum, and Brucella abortus; S. Mesnage, unpublished). Interestingly, double 4-3 cross-links resulting from D,D-transpeptidation have also been described in Staphylococcus aureus (14). The physiological role of these peptidoglycan cross-links remains unknown and awaits further studies.
Based on the impact of L,D-transpeptidation on antibiotic resistance in Enterococcus faecium, it is tempting to assume that L,D-transpeptidation in C. difficile could underpin beta-lactam resistance. This remains an open question since we were unable to generate a mutant devoid of 3-3 cross-links. The mutant harboring deletions in the genes encoding the three canonical Ldts still contained 78% of 3-3 cross-links, indicating that this organism encodes (an)other enzyme(s) that does not contain a YkuD domain but is (are) able to make 3-3 cross-links. Our findings are surprising and somewhat contrasting with a previous study, where the combined deletion of ldtCd1 and ldtCd2 led to a decrease in the cross-linking index (18.2% as compared with 33.8% for the WT strain). This discrepancy is difficult to explain but could be attributed either to the different strain analyzed (C. difficile 630 versus R20291) or to the different strategies followed for peptidoglycan analysis. The work by Peltier et al. involved offline analysis of individual fractions from a single replicate and a quantification of muropeptides based on UV whilst our analysis involved LC-MS analysis (online) of biological triplicates and a quantification using ion intensity. Based on the remarkably similar peptidoglycan structure of the WT and mutant strain described here (Tables 1 and 2), we are confident that the three canonical LdtCd enzymes have a minor contribution to the formation of 3-3 cross-links and only contribute significantly to incorporate noncanonical amino acids in the position 4 of peptide stems.
The interaction of recombinant Ldts with beta-lactams has been extensively studied in vitro, and the data reported support the idea that these enzymes play a role in resistance to these antibiotics. Ldts are acylated by beta-lactams, but enzyme inactivation only occurs in the presence of carbapenems and penems. Other beta-lactams such as cephems (cephalosporin) are poor inhibitors since acylation is slow and the thioester bond formed in the enzyme–antibiotic adduct is prone to hydrolysis (15). This has been shown for model organisms including E. faecium (15), Mycobacterium tuberculosis (16), and C. difficile Ldts (8) and is true for most Ldts studied to date despite some exceptions for M. tuberculosis LdtMt5 (16), Acinetobacter baumanii LdtAb (17), and C. difficile LdtCd3 (8), which are not inhibited by carbapenems. The contribution of Ldts to beta-lactam resistance has been documented in E. faecium (18), M. tuberculosis (19), Mycobacterium smegmatis (20), and A. baumanii (17). In C. difficile, the inactivation of two of the three Ldts did not lead to a change in beta-lactam resistance (6). Our data revealed that the inactivation of all canonical C. difficile ldts has no impact on beta-lactam resistance, as expected, based on the results from peptidoglycan analysis.
The identification of alternative (noncanonical) Ldt(s) encoded by C. difficile is therefore required to investigate (i) whether L,D-transpeptidation is essential in this organism and (ii) whether this mode of peptidoglycan polymerization underpins beta-lactam resistance.
Experimental procedures
Bacterial strains, plasmids, oligonucleotides, and growth conditions
Bacterial strains, plasmids, and oligonucleotides are described in Table S4. C. difficile R20291 (ribotype 027) and isogenic derivatives were grown on BHI agar plates or in TY broth. During selection of mutants, strains were grown on C. difficile minimal medium (21) supplemented with 5-fluorocytosine (50 μg/ml) when required. Cultures were incubated at 37 °C in an anaerobic cabinet under an atmosphere containing 80% nitrogen, 10% hydrogen, and 10% carbon dioxide. E. coli was grown on Luria Bertani (LB) agar plates or in LB broth at 37 °C. When needed, thiamphenicol was added (30 μg/ml).
Construction of C. difficile deletion mutants
C. difficile mutant strains were constructed by homologous recombination. Briefly, 1.2 kb upstream and downstream of the region to be deleted was synthesized as a single DNA fragment (Genewiz) and cloned between BamHI and SacI sites in pJAK112, yielding pNG007 (ldtCd1 deletion), pNG008 (ldtCd2 deletion), and pNG009 (ldtCd3 deletion). Plasmids were introduced into C. difficile strain R20291 by conjugation (22), and allelic exchange was carried out as described (10).
Determination of minimum inhibitory concentrations
MICs were determined according to an agar dilution method using Wilkins Chalgren agar and as recommended by the Clinical and Laboratory Standards Institute guidelines. C. difficile isolates were cultured on fresh blood agar plates, prior to inoculation of single colonies into prereduced Schaedler Anaerobic Broths and anaerobic culture for 24 h. Cultures were diluted in prereduced phosphate-buffered saline to achieve a 1 McFarland standard equivalent, and 105 colony-forming units were spotted on Wilkins Chalgren agar containing doubling antibiotic dilutions and non-antibiotic-containing controls. Agar plates containing amoxicillin clavulanate were prepared with a fixed concentration of 2 mg/l clavulanate, and those containing piperacillin tazobactam were prepared with a fixed concentration of 4 mg/l tazobactam, as recommended by European Committee on Antimicrobial Susceptibility Testing guidelines. Agar plates were incubated anaerobically for 48 h before reading. The MIC was defined as the lowest concentration of antibiotic completely preventing growth, significantly reducing it to a haze or one to three discrete colonies.
Chromosomal DNA extraction, sequencing, and genome analysis
Genomic DNA was purified using phenol–chloroform extraction as described (10), and whole genome sequencing was performed by MicrobesNG using their standard Illumina service. Sequence analysis was performed using a custom script, as described (23). In brief, reads were aligned to the C. difficile R20291 reference (accession number: FN545816) using BWA-mem (v0.7.17) and sorted using SAMtools (v1.43) (24). PCR duplicates were removed via Picard (v2.25.2) (http://broadinstitute.github.io/picard/). SAMtools (v1.43) mpileup was used to generate the mpileup prerequisite for Varscan. Varscan (v2.4.3-1) (25) was then used to call variants using parameters previously described (23), and snpEff (v5.0) (26) was used to annotate variants. Variants that co-occurred in the WT were removed to generate a list of mutations unique to mutant strains. Mutations were visualized on the genome using a previously published custom script in RStudio (v4.1.0) using the Plotrix package (27).
TraDIS analysis
The construction of the transposon library, the sequencing of insertion sites, and the mapping to their corresponding reference sequences were described (10). Visualization of insertion sites was done using the Artemis genome browser (28).
Peptidoglycan extraction
C. difficile strains were grown overnight in 10 ml of TY broth from a single colony. The starter cultures were used to inoculate 100 ml TY medium (1/100 dilution). After 48 h at 37 °C, cells were spun, supernatant was discarded, and cell pellet was snap frozen in liquid nitrogen; the cell pellet was then resuspended in 20 ml of boiling MilliQ water (MQ) before the addition of 5 ml warm 20% (w/v) SDS (4% SDS final concentration). After 30 min at 100 °C, the cells were allowed to cool down to room temperature. Insoluble cell walls were pelleted at 45,000g for 20 min and washed 5 times using warm MQ water. Proteins covalently bound to peptidoglycan were removed by pronase treatment (final concentration of 2 mg/ml, 4 h at 60 °C). Protease-treated cell walls were washed 6 times with 30 ml of MQ water before covalently bound polymers were removed by incubation in 1 M HCl for 5 h at 37 °C. Insoluble pure peptidoglycan was washed 6 times with MQ water, snap frozen in liquid nitrogen, freeze-dried and resuspended at a final concentration of 10 mg/ml.
LdtCd production and purification
The plasmids for protein production were designed as described (8). LdtCd1 and LdtCd3 were expressed as full-length His-tagged proteins. LdtCd2 could not be produced as a stable full-length protein, so the catalytic domain was purified. Recombinant LdtCd were produced in E. coli BL21(DE3) grown in LB broth. One-liter cultures were inoculated at an OD600nm of 0.05, and protein expression was induced with 1 mM isopropyl ß-D-1-thiogalactopyranoside when the cultures reached an OD600nm of 0.7. They were then cooled down to 20 °C and incubated for 16 h at this temperature. Cells were harvested, resuspended in a buffer containing 50 mM Tris-HCl (pH8.0) + 500 mM NaCl, and mechanically broken using a French press (2 passages at 1250 psi). Cell debris were removed by centrifuging the crude cell extract at 45,000g for 30 min at 4 °C. The entire soluble fraction was loaded on a 5-ml HiTrap column equilibrated in buffer A at a flow rate of 5 ml/min. Elution was performed using a 10 column volume gradient to 250 mM imidazole in buffer A. Fractions containing the LdtCd proteins were pooled, concentrated to 2 mg/ml and further purified by gel filtration chromatography using a Hiload 16/600 superdex 75 column equilibrated in 50 mM Tris-HCl pH 8.5 + 250 mM NaCl). LdtCd proteins were concentrated on an Amicon centrifugal filter to a final concentration of 2 mg/ml and stored at −80 °C until further use.
Purification of substrates for in vitro assays
Peptidoglycan fragments used as substrates were purified from E. coli or C. difficile sacculi digested with mutanolysin and reduced with sodium borohydride. Digestion products were separated by reversed-phase HPLC using a Hypersil column (4.6 mm × 250 mm, 5 μm particle size) using a water–acetonitrile–0.1% (v/v) formic acid gradient. Fractions containing the muropeptides of interest were freeze-dried and quantified by NMR using trimethylsilyl propionate as a standard.
In vitro Ldt assays
Each in vitro assay was carried out in triplicate, and average chromatograms are shown in Figure 1. Each reaction was carried out in a phosphate saline buffer (pH 8.0) in a final volume of 50 μl and contained 100 μM substrate and 10 μM enzyme. For exchange reactions, D-methionine was added at a concentration of 1 mM. Reactions were incubated at 37 °C for 4 h.
Preparation of soluble muropeptides for peptidoglycan structural analysis
Purified peptidoglycan (1 mg) was digested overnight in 50 mM phosphate buffer (pH 5.5) supplemented with 200 U of mutanolysin (Sigma) in a final volume of 125 μl. Following heat inactivation of mutanolysin (5 min at 100 °C), soluble disaccharide peptides were mixed with an equal volume of 250 mM borate buffer (pH 9.25) and reduced with 0.2% (w/v) sodium borohydride. After 20 min at room temperature, the pH was adjusted to 4.5 to 5.5 using phosphoric acid.
Ultrahigh-Performance chromatography coupled to tandem mass spectrometry
An Ultimate 3000 UHPLC (Dionex/Thermo Fisher Scientific) system coupled with a high-resolution Q Exactive Focus mass spectrometer (Thermo Fisher Scientific) was used for LC-MS analysis. Muropeptides were separated using a C18 analytical column (Hypersil Gold aQ, 1.9-μm particles, 150 mm × 2.1 mm; Thermo Fisher Scientific) at a temperature of 50 °C for peptidoglycan analysis or on a smaller C18 column for in vitro assays (Hypersil Gold aQ, 1.9-μm particles, 50 mm × 2.1 mm; Thermo Fisher Scientific). For peptidoglycan analysis, muropeptide elution was performed at 0.25 ml/min by applying a mixture of solvent A (water, 0.1% [v/v] formic acid) and solvent B (acetonitrile, 0.1% [v/v] formic acid). Liquid chromatography conditions were 0 to 12.5% B for 25 min increasing to 20% B for 10 min. After 5 min at 95%, the column was re-equilibrated for 10 min with 100% buffer A. For in vitro assays, a flow rate of 0.4 ml/min was used. PG fragments were eluted with a 5-min gradient to 15% B followed by 2 min at 95% B. The column was re-equilibrated for 6 min with 100% buffer A.
The Orbitrap Exploris 240 was operated under electrospray ionization (H-ESI high flow)-positive mode, full scan (m/z 150–2250) at resolution 120,000 (full width at half maximum) at m/z 200, with normalized AGC Target 100%, and automated maximum ion injection time. Data-dependent MS/MS were acquired on a “Top 5” data-dependent mode using the following parameters: resolution 30,000; AGC 100%, automated injection time, with normalized collision energy 25%.
Nuclear magnetic resonance
Purified peptidoglycan fragments were dissolved in 90% H2O/10% D2O. They were analyzed by NMR at 298 K on a Bruker DRX-600 equipped with a cryoprobe. TOCSY spectra were acquired using a 60-ms spin-lock with a field strength of 10 kHz. NOESY spectra used a 200-ms mixing time. All data were analyzed using Topspin 4.0.5.
Analysis of PG structure
LC-MS datasets were deconvoluted with the Byos software v3.11 (Protein Metrics). Sequential searches were carried out with PGFinder v1.0.3, with default settings (10 ppm tolerance, 0.5 min cleanup window) following the strategy described in Fig. S4. Data from individual matching output was consolidated as previously described to calculate average intensities, retention times, observed monoisotopic masses, and ppm differences. The output from individual searches and consolidated data are described in File S2). Cross-linking index and glycan chain length were determined as previously (29). The cross-linking index is defined as 0.5 ∗ (% of dimers) + 0.66 ∗ (% of trimers); glycan chain length was inferred from the abundance of anhydroMurNAc groups, which are found at the end of glycan chains. It is defined as 1/(% of AnhydroMurNAc monomers + 0.5 ∗ (% of AnhydroMurNAc dimers) + 0.33 ∗ (% of AnhydroMurNAc trimers).
Flow cytometry
Cells corresponding to biological replicates were grown overnight, diluted 1:100 into fresh broth (OD600 ∼ 0.02), and grown to mid-exponential phase (OD600 ∼ 0.5). Bacteria were diluted 1:100 in filtered phosphate-buffered saline and analyzed by flow cytometry using Millipore Guava Easy Cyte system. Light scatter data were obtained with logarithmic amplifiers for 2500 events. Forward scattered and side-scattered light values were compared using Student t test with Welch’s correction using GraphPad Prism.
Sporulation
Sporulation efficiency was assessed as described (30). Briefly, stationary phase cultures of C. difficile were incubated anaerobically for 5 days and the total and heat-resistant (spore) colony-forming units (65 °C for 30 min) were determined every 24 h. Strains were assayed in technical triplicate and the data presented as the mean and standard deviation.
Toxin release assays
Toxin production was detected in whole cell lysates or concentrated culture supernatants by Western blot. For both fractions, material corresponding to the equivalent of 20 ml of culture at OD600nm = 1 was loaded onto a 6% SDS PAGE, transferred on a polyvinylidene fluoride membrane, and probed with a mouse monoclonal antibody (MA1-7413, Thermo Fisher) against toxin B at a 1/1000 dilution. A secondary rabbit anti-mouse antibody coupled to horseradish peroxidase (#31450, Thermo Fisher) was used at a 1/10,000 dilution. Blots were revealed by chemiluminescence using a BioRad chemidoc system.
Data availability
LC-MS/MS datasets have been deposited in the GLYCOPOST repository (GPST000371). NMR assignments have been deposited in the Biological Magnetic Resonance Data Bank (52169). Sequencing data were deposited in the NCBI Sequence Read Archive (SRA) under Bioproject ID PRJNA1026070.
Supporting information
This article contains supporting information (Figs. S1–S8, Tables S1–S4 and Files S1 and S2).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors would like to thank Nick W. Turner (University of Sheffield) for insightful suggestions to draw unusual peptidoglycan fragments.
Author contributions
S. M. conceptualization; N. F. G. and S. M. data curation; S. M. formal analysis; S. M., M. H. W., R. P. F., and A. L. L. funding acquisition; N. F. G., D. G., M. G. A.-Z., M. P. W., C. A. E., W. D. S., J. E. B., and J. F. investigation; N. F. G., C. A. E., M. J. D., and R. P. F. methodology; S. M. and G. L. D. project administration; S. M., R. P. F., and G. L. D. supervision; N. F. G. and S. M. validation; M. G. A.-Z., M. P. W., and S. M. visualization; S.M. writing – original draft; N. F. G., D. G., M. G. A.-Z., M. P. W., C. A. E., J. E. B., J. F., G. L. D., A. L. L., R. P. F., and S. M. writing – review and editing.
Funding and additional information
This work was supported by a Medical Research Council grant to S. M., R. F. P., M. H. W., and A. L. L. (MR/S009272/1). C. A. E. and M. J. D. received support from the Biotechnology and Biological Science Research Council (BB/M012166/1). M. G. A.-Z. is funded by a Mexican government PhD scholarship (CONAHCYT, 2021-000007-01EXTF-00221); J. E. B. is funded by a studentship from the MRC Discovery Medicine North (DiMeN) Doctoral Training Partnership (MR/R015902/1).
Reviewed by members of the JBC Editorial Board. Edited by Chris Whitfield
Supporting information
References
- 1.Wingen-Heimann S.M., Davies K., Viprey V.F., Davis G., Wilcox M.H., Vehreschild M., et al. Clostridioides difficile infection (CDI): a pan-European multi-center cost and resource utilization study, results from the Combatting Bacterial Resistance in Europe CDI (COMBACTE-CDI) Clin. Microbiol. Infect. 2023;29:651.e1–651.e8. doi: 10.1016/j.cmi.2022.12.019. [DOI] [PubMed] [Google Scholar]
- 2.Lessa F.C., Gould C.V., McDonald L.C. Current status of Clostridium difficile infection epidemiology. Clin. Infect. Dis. 2012;55:S65–S70. doi: 10.1093/cid/cis319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buddle J.E., Fagan R.P. Pathogenicity and virulence of Clostridioides difficile. Virulence. 2023;14 doi: 10.1080/21505594.2022.2150452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frere J.M. Mechanism of action of beta-lactam antibiotics at the molecular level. Biochem. Pharmacol. 1977;26:2203–2210. doi: 10.1016/0006-2952(77)90280-5. [DOI] [PubMed] [Google Scholar]
- 5.Cho H., Uehara T., Bernhardt T.G. Beta-lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell. 2014;159:1300–1311. doi: 10.1016/j.cell.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peltier J., Courtin P., El Meouche I., Lemee L., Chapot-Chartier M.P., Pons J.L. Clostridium difficile has an original peptidoglycan structure with a high level of N-acetylglucosamine deacetylation and mainly 3-3 cross-links. J. Biol. Chem. 2011;286:29053–29062. doi: 10.1074/jbc.M111.259150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aliashkevich A., Cava F. L,D-transpeptidases: the great unknown among the peptidoglycan cross-linkers. FEBS J. 2021;289:4718–4730. doi: 10.1111/febs.16066. [DOI] [PubMed] [Google Scholar]
- 8.Sutterlin L., Edoo Z., Hugonnet J.E., Mainardi J.L., Arthur M. Peptidoglycan cross-linking activity of L,D-Transpeptidases from Clostridium difficile and inactivation of these enzymes by beta-lactams. Antimicrob. Agents Chemother. 2018;62 doi: 10.1128/AAC.01607-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.More N., Martorana A.M., Biboy J., Otten C., Winkle M., Serrano C.K.G., et al. Peptidoglycan remodeling enables Escherichia coli to survive severe outer membrane assembly defect. mBio. 2019;10 doi: 10.1128/mBio.02729-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dembek M., Barquist L., Boinett C.J., Cain A.K., Mayho M., Lawley T.D., et al. High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. mBio. 2015;6 doi: 10.1128/mBio.02383-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel A.V., Turner R.D., Rifflet A., Acosta-Martin A.E., Nichols A., Awad M.M., et al. PGFinder, a novel analysis pipeline for the consistent, reproducible and high-resolution structural analysis of bacterial peptidoglycans. eLife. 2021;10:e70597. doi: 10.7554/eLife.70597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voedts H., Dorchene D., Lodge A., Vollmer W., Arthur M., Hugonnet J.E. Role of endopeptidases in peptidoglycan synthesis mediated by alternative cross-linking enzymes in Escherichia coli. EMBO J. 2021;40 doi: 10.15252/embj.2021108126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chodisetti P.K., Reddy M. Peptidoglycan hydrolase of an unusual cross-link cleavage specificity contributes to bacterial cell wall synthesis. Proc. Natl. Acad. Sci. U. S. A. 2019;116:7825–7830. doi: 10.1073/pnas.1816893116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boneca I.G., Xu N., Gage D.A., de Jonge B.L., Tomasz A. Structural characterization of an abnormally cross-linked muropeptide dimer that is accumulated in the peptidoglycan of methicillin- and cefotaxime-resistant mutants of Staphylococcus aureus. J. Biol. Chem. 1997;272:29053–29059. doi: 10.1074/jbc.272.46.29053. [DOI] [PubMed] [Google Scholar]
- 15.Triboulet S., Dubee V., Lecoq L., Bougault C., Mainardi J.L., Rice L.B., et al. Kinetic features of L,D-transpeptidase inactivation critical for beta-lactam antibacterial activity. PLoS One. 2013;8 doi: 10.1371/journal.pone.0067831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cordillot M., Dubee V., Triboulet S., Dubost L., Marie A., Hugonnet J.E., et al. In vitro cross-linking of Mycobacterium tuberculosis peptidoglycan by L,D-transpeptidases and inactivation of these enzymes by carbapenems. Antimicrob. Agents Chemother. 2013;57:5940–5945. doi: 10.1128/AAC.01663-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toth M., Stewart N.K., Smith C.A., Lee M., Vakulenko S.B. The l,d-transpeptidase ldt(Ab) from Acinetobacter baumannii is poorly inhibited by carbapenems and has a unique structural architecture. ACS Infect. Dis. 2022;8:1948–1961. doi: 10.1021/acsinfecdis.2c00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mainardi J.L., Legrand R., Arthur M., Schoot B., van Heijenoort J., Gutmann L. Novel mechanism of beta-lactam resistance due to bypass of DD-transpeptidation in Enterococcus faecium. J. Biol. Chem. 2000;275:16490–16496. doi: 10.1074/jbc.M909877199. [DOI] [PubMed] [Google Scholar]
- 19.Gupta R., Lavollay M., Mainardi J.L., Arthur M., Bishai W.R., Lamichhane G. The Mycobacterium tuberculosis protein LdtMt2 is a nonclassical transpeptidase required for virulence and resistance to amoxicillin. Nat. Med. 2010;16:466–469. doi: 10.1038/nm.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baranowski C., Welsh M.A., Sham L.T., Eskandarian H.A., Lim H.C., Kieser K.J., et al. Maturing Mycobacterium smegmatis peptidoglycan requires non-canonical crosslinks to maintain shape. eLife. 2018;7 doi: 10.7554/eLife.37516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karasawa T., Ikoma S., Yamakawa K., Nakamura S. A defined growth medium for Clostridium difficile. Microbiology (Reading) 1995;141:371–375. doi: 10.1099/13500872-141-2-371. [DOI] [PubMed] [Google Scholar]
- 22.Kirk J.A., Fagan R.P. Heat shock increases conjugation efficiency in Clostridium difficile. Anaerobe. 2016;42:1–5. doi: 10.1016/j.anaerobe.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buddle J.E., Wright R.C.T., Turner C.E., Chaudhuri R.R., Brockhurst M.A., Fagan R.P. Multiple evolutionary pathways lead to vancomycin resistance in Clostridioides difficile. bioRxiv. 2023 doi: 10.1101/2023.1109.1115.557922. [preprint] [DOI] [Google Scholar]
- 24.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koboldt D.C., Chen K., Wylie T., Larson D.E., McLellan M.D., Mardis E.R., et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25:2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., Wang L., et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrison E., Guymer D., Spiers A.J., Paterson S., Brockhurst M.A. Parallel compensatory evolution stabilizes plasmids across the parasitism-mutualism continuum. Curr. Biol. 2015;25:2034–2039. doi: 10.1016/j.cub.2015.06.024. [DOI] [PubMed] [Google Scholar]
- 28.Carver T., Harris S.R., Berriman M., Parkhill J., McQuillan J.A. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics. 2012;28:464–469. doi: 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glauner B. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal. Biochem. 1988;172:451–464. doi: 10.1016/0003-2697(88)90468-x. [DOI] [PubMed] [Google Scholar]
- 30.Kirk J.A., Gebhart D., Buckley A.M., Lok S., Scholl D., Douce G.R., et al. New class of precision antimicrobials redefines role of Clostridium difficile S-layer in virulence and viability. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aah6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
LC-MS/MS datasets have been deposited in the GLYCOPOST repository (GPST000371). NMR assignments have been deposited in the Biological Magnetic Resonance Data Bank (52169). Sequencing data were deposited in the NCBI Sequence Read Archive (SRA) under Bioproject ID PRJNA1026070.