Abstract
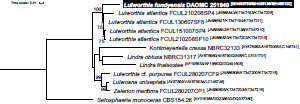
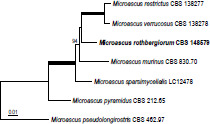
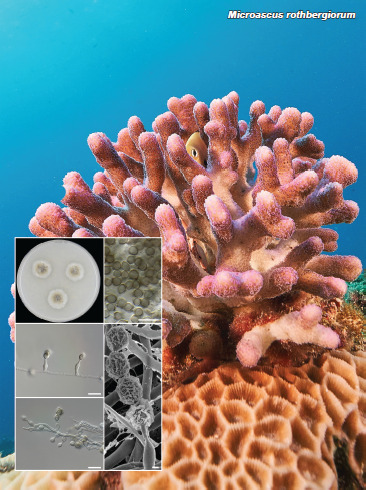
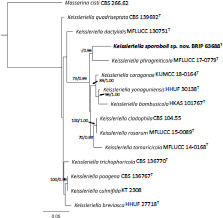
Novel species of fungi described in this study include those from various countries as follows: Australia, Agaricus albofoetidus, Agaricus aureoelephanti and Agaricus parviumbrus on soil, Fusarium ramsdenii from stem cankers of Araucaria cunninghamii, Keissleriella sporoboli from stem of Sporobolus natalensis, Leptosphaerulina queenslandica and Pestalotiopsis chiaroscuro from leaves of Sporobolus natalensis, Serendipita petricolae as endophyte from roots of Eriochilus petricola, Stagonospora tauntonensis from stem of Sporobolus natalensis, Teratosphaeria carnegiei from leaves of Eucalyptus grandis × E. camaldulensis and Wongia ficherai from roots of Eragrostis curvula. Canada, Lulworthia fundyensis from intertidal wood and Newbrunswickomyces abietophilus (incl. Newbrunswickomyces gen. nov.) on buds of Abies balsamea. Czech Republic, Geosmithia funiculosa from a bark beetle gallery on Ulmus minor and Neoherpotrichiella juglandicola (incl. Neoherpotrichiella gen. nov.) from wood of Juglans regia. France, Aspergillus rouenensis and Neoacrodontium gallica (incl. Neoacrodontium gen. nov.) from bore dust of Xestobium rufovillosum feeding on Quercus wood, Endoradiciella communis (incl. Endoradiciella gen. nov.) endophytic in roots of Microthlaspi perfoliatum and Entoloma simulans on soil. India, Amanita konajensis on soil and Keithomyces indicus from soil. Israel, Microascus rothbergiorum from Stylophora pistillata. Italy, Calonarius ligusticus on soil. Netherlands, Appendopyricularia juncicola (incl. Appendopyricularia gen. nov.), Eriospora juncicola and Tetraploa juncicola on dead culms of Juncus effusus, Gonatophragmium physciae on Physcia caesia and Paracosmospora physciae (incl. Paracosmospora gen. nov.) on Physcia tenella, Myrmecridium phragmitigenum on dead culm of Phragmites australis, Neochalara lolae on stems of Pteridium aquilinum, Niesslia nieuwwulvenica on dead culm of undetermined Poaceae, Nothodevriesia narthecii (incl. Nothodevriesia gen. nov.) on dead leaves of Narthecium ossifragum and Parastenospora pini (incl. Parastenospora gen. nov.) on dead twigs of Pinus sylvestris. Norway, Verticillium bjoernoeyanum from sand grains attached to a piece of driftwood on a sandy beach. Portugal, Collybiopsis cimrmanii on the base of living Quercus ilex and amongst dead leaves of Laurus and herbs. South Africa, Paraproliferophorum hyphaenes (incl. Paraproliferophorum gen. nov.) on living leaves of Hyphaene sp. and Saccothecium widdringtoniae on twigs of Widdringtonia wallichii. Spain, Cortinarius dryosalor on soil, Cyphellophora endoradicis endophytic in roots of Microthlaspi perfoliatum, Geoglossum laurisilvae on soil, Leptographium gemmatum from fluvial sediments, Physalacria auricularioides from a dead twig of Castanea sativa, Terfezia bertae and Tuber davidlopezii in soil. Sweden, Alpova larskersii, Inocybe alpestris and Inocybe boreogodeyi on soil. Thailand, Russula banwatchanensis, Russula purpureoviridis and Russula lilacina on soil. Ukraine, Nectriella adonidis on overwintered stems of Adonis vernalis. USA, Microcyclus jacquiniae from living leaves of Jacquinia keyensis and Penicillium neoherquei from a minute mushroom sporocarp. Morphological and culture characteristics are supported by DNA barcodes.
Citation: Crous PW, Boers J, Holdom D, et al. 2022. Fungal Planet description sheets: 1383–1435. Persoonia 48: 261–371. https://doi.org/10.3767/persoonia.2022.48.08.
Keywords: ITS nrDNA barcodes, LSU, new taxa, systematics
Acknowledgments
Teresa Lebel and colleagues acknowledge the Royal Botanic Gardens Victoria and the Friends of the Royal Botanic Gardens Victoria for funding and support for Northern Territory field and laboratory work; Northern Territory Herbarium staff for space and laboratory access and help with collection curation; State Herbarium of South Australia for support of the undergraduate intern J. Broadbridge. This research was also supported through funding from Australian Biological Resources Study grant (4-EHOJ161) to the State Herbarium of South Australia. Ellen Larsson acknowledges the Swedish Taxonomy Initiative, SLU, Artdatabanken, Uppsala (dha.2019.4.3-13) and the curators of herbaria O, S, UME and UPS. Fernando Esteve-Raventós acknowledges the FEDER/Ministerio de Ciencia, Innovación y Universidades – Agencia Estatal de Investigación, Spain (Project CGL2017-86540-P). Jordi Vila (Institut d’Estudis Catalans) is thanked for sending us material and a photograph of Inocybe alpestris. The studies of Hana Ševčíková and Vladimír Antonín was facilitated via institutional support of the long-term conceptual development of research institutions for the Moravian Museum provided by the Czech Ministry of Culture (ref. MK000094862). François Armada and colleagues acknowledge Clotilde & Didier Borgarino and Patrice Tanchaud for collections, and Francisco Donaire (JA-CUSSTA), as fungarium curator. Gregorio Delgado is grateful to William Colbert and Kamash Pillai (Eurofins EMLab P&K) for provision of laboratory facilities. Jose G. Maciá-Vicente acknowledges support from the German Research Foundation under grant MA7171/1-1, and from the Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz (LOEWE) of the state of Hesse within the framework of the Cluster for Integrative Fungal Research (IPF). Kandikere R. Sridhar and colleagues are thankful to Prof. Sajeewa S.N. Maharachchikumbura, University of Electronic Science and Technology of China, P.R. China for facilitating phylogenetic tree construction and interpretation. Kai Reschke and colleagues thank Machiel E. Noordeloos for extensive support with their Entoloma research. The work of Bálint Dima was supported by the ELTE Thematic Excellence Programme 2020 (TKP2020-IKA-05), financed by the National Research, Development and Innovation Office of Hungary. Antonio Mateos and colleagues thank Amalio Gutiérrez for his collaboration and cooperation in the field collections in Extremadura. Ajay C. Lagashetti and colleagues thank the Director, MACS-Agharkar Research Institute, Pune, for providing necessary facilities, the CSIR, New Delhi for a Senior Research Fellowship (SRF) and S.P. Pune University for granting permission to register for a PhD degree. The study of Daniel Torres-Garcia and colleagues was partially supported by the Spanish Ministerio de Economía, Industria y Competitividad (grant CGL2017-88094-P). Lukasz Istel and co-authors thank Jan Dijksterhuis for the examination of Microascus rothbergiorum using Scanning Electron Microscopy. Oded Yarden and Nofar Lifshitz were funded by the Israel Science Foundation. The research of Milan Spetik and co-authors was supported by project No. IGA-ZF/2021-ST2003. Saúl De la Peña-Lastra and colleagues thank the Atlantic Islands National Maritime-Terrestrial Park authorities and guards, especially Víctor Rivas Iglesias, and also Cristóbal Burgos Morillo for his generous help in processing the images. Tracey Steinrucken and colleagues were supported by AgriFutures Australia (Rural Industries Research and Development Corporation), through funding from the Australian Government Department of Agriculture, Water and the Environment, as part of its Rural Research and Development for Profit program (PRJ-010527). Janneke Aylward and colleagues are grateful for funding received from the Department of Science and Innovation (DSI)-National Research Foundation (NRF) Centre of Excellence in Plant Health Biotechnology (CPHB), South Africa and the DST-NRF SARChI chair in Fungal Genomics. Julio Cabero acknowledges Berta Martín from the regional administration of Zamora (Spain) for supporting this work, as well as Hipólito Hernandez, environmental agent in the area of Sanabria, for sharing his knowledge about local flora and ecology. Asunción Morte is grateful to Fundación Séneca- Agencia de Ciencia y Tecnología de la Región de Murcia (20866/PI/18) for financial support. Umpawa Pinruan and colleagues were financially supported by the Platform Technology Management Section, National Center for Genetic Engineering and Biotechnology (BIOTEC), Project Grant No. P19-50231. Konstanze Bensch (Westerdijk Fungal Biodiversity Institute, Utrecht) is thanked for correcting the spelling of various Latin epithets.
REFERENCES
- Abdelrazek S, Choudhari S, Thimmapuram J, et al. 2020. Changes in the core endophytic mycobiome of carrot taproots in response to crop management and genotype. Scientific Reports 10: 13685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agerer R. 1980. Contribution to neotropical cyphellaceous fungi. 11. Deigloria gen. nov. (Physalacriaceae). Mycotaxon 12: 185–200. [Google Scholar]
- Al Subeh ZY, Raja HA, Burdette JE, et al. 2021. Three diketomorpholines from a Penicillium sp. (strain G1071). Phytochemistry 189: 112830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfieri SA, Jr, Samuels GJ. 1979. Nectriella pironii and its kutilakesa-like anamorph, a parasite of ornamental shrubs. Mycologia 71: 1178–1185. [Google Scholar]
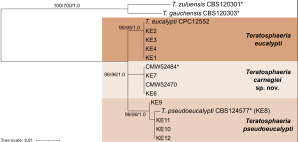
- Andjic V, Pegg GS, Carnegie AJ, et al. 2010. Teratosphaeria pseudoeucalypti, new cryptic species responsible for leaf blight of Eucalyptus in subtropical and tropical Australia. Plant Pathology 59: 900–912. [Google Scholar]
- Antonín V, Noordeloos M. 2010. A monograph of marasmioid and collybioid fungi in Europe. IHW Verlag. [Google Scholar]
- Arauzo S, Iglesias P. 2014. La familia Geoglossaceae ss. str. en la península Ibérica y la Macaronesia. Errotari 11: 166–259. [Google Scholar]
- Arya CP, Manoj Kumar A, Pradeep CK, et al. 2022. Agaricus brunneodiscus, a new species of Agaricus sect. Rarolentes from India. Phytotaxa 533: 181–193. [Google Scholar]
- Arzanlou M, Groenewald JZ, Gams W, et al. 2007. Phylogenetic and morphotaxonomic revision of Ramichloridium and allied genera. Studies in Mycology 58: 57–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylward J, Havenga M, Dreyer LL, et al. 2021. Genetic diversity of Teratosphaeria pseudoeucalypti in Eucalyptus plantations in Australia and Uruguay. Australasian Plant Pathology 50: 639–649. [Google Scholar]
- Azevedo E, Barata M, Marques MI, et al. 2017. Lulworthia atlantica: a new species supported by molecular phylogeny and morphological analysis. Mycologia 109: 287–295. [DOI] [PubMed] [Google Scholar]
- Azuddin NF, Mohd MH, Rosely NFN, et al. 2021. Molecular phylogeny of endophytic fungi from Rattan (Calamus castaneus Griff.) spines and their antagonistic activities against plant pathogenic fungi. Journal of Fungi 7: 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandini D, Oertel B, Schüssler C, et al. 2020. Noch mehr Risspilze: Fünfzehn neue und zwei wenig bekannte Arten der Gattung Inocybe. Mycologia Bavarica 20: 13–101. [Google Scholar]
- Bao D-F, Hyde KD, McKenzie EHC, et al. 2021. Biodiversity of lignicolous freshwater hyphomycetes from China and Thailand and description of sixteen species. Journal of Fungi 7: 669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr ME. 1972. Preliminary studies on the Dothideales in temperate North America. Contributions from the University of Michigan Herbarium 9: 523–638. [Google Scholar]
- Bashir H, Chen J, Jabeen S, et al. 2021. An overview of Agaricus section Hondenses and Agaricus section Xanthodermatei with description of eight new species from Pakistan. Scientific Reports 11: 12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell J, Yokoya K, Kendon JP, et al. 2020. Diversity of root-associated culturable fungi of Cephalanthera rubra (Orchidaceae) in relation to soil characteristics. PeerJ 8: e8695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellanger J-M, Moreau P-A, Corriol G, et al. 2015. Plunging hands into the mushroom jar: a phylogenetic framework for Lyophyllaceae (Agaricales, Basidiomycota). Genetica 143: 169–194. [DOI] [PubMed] [Google Scholar]
- Berger F, Braun U, Heuchert B. 2015. Gonatophragmium lichenophilum sp. nov. – a new lichenicolous hyphomycete from Austria. Mycobiota 5: 7–13. [Google Scholar]
- Berthier J. 1985. Les Physalacriaceae du Globe. Biblioteca Mycologica 98: 1–128. [Google Scholar]
- Bidaud A. 2011. Cortinaires rares ou nouveaux de la région Rhône-Alpes (France). Journal des J.E.C. 13: 4–24. [Google Scholar]
- Bonito G, Hameed K, Ventura R, et al. 2016. Isolating a functionally relevant guild of fungi from the root microbiome of Populus. Fungal Ecology 22: 35–42. [Google Scholar]
- Brandrud TE, Lindström H, Marklund H, et al. 1992. Cortinarius: Flora Photographica. Vol. II (Swedish version). Cortinarius HB, Sweden. [Google Scholar]
- Braun U, Crous PW, Nakashima C. 2015. Cercosporoid fungi (Mycosphaerellaceae) 4. Species on dicots (Acanthaceae to Amaranthaceae). IMA Fungus 6: 373–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun U, Nakashima C, Crous PW. 2013. Cercosporoid fungi (Mycosphaerellaceae) 1. Species on other fungi, Pteridophyta and Gymnospermae. IMA Fungus 4: 265–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess TI, Wingfield MJ. 2017. Pathogens on the move: A 100-year global experiment with planted eucalypts. Bioscience 67: 14–25. [Google Scholar]
- Cannon P, Carmarán C, Romero A. 1995. Studies on biotrophic fungi from Argentina: Microcyclus porlieriae, with a key to South American species of Microcyclus. Mycological Research 99: 353–356. [Google Scholar]
- Ceruti A, Fontana A, Nosenzo C. 2003. Le specie europee del genere Tuber: una revisione storica. Vol. 37. Turin, Italy, Museo Regionale di Scienze Naturali. [Google Scholar]
- Chang R, Cao W, Wang Y, et al. 2022. Melanodevriesia, a new genus of endolichenic oleaginous black yeast recovered from the Inner Mongolia Region of China. Fungal Systematics and Evolution 9: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Callac P, Parra LA, et al. 2017. Study in Agaricus subgenus Minores and allied clades reveals a new American subgenus and contrasting phylogenetic patterns in Europe and Greater Mekong Subregion. Persoonia 38: 170–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Parra L, Kesel A, et al. 2016. Inter- and intra-specific diversity in Agaricus endoxanthus and allied species reveals a new taxon, A. punjabensis. Phytotaxa 252: 1–16. [Google Scholar]
- Cochard H, Réaudin D. 2019. Physalacria stilboidea (Cooke) Sacc., espèce exotique nouvelle pour la France. Bulletin Mycologique et Botanique Dauphiné-Savoie 235: 11–16. [Google Scholar]
- Cooke MC. 1889. New Australian fungi. Grevillea 18: 1–8. [Google Scholar]
- Corner EJH. 1950. A monograph of Clavaria and allied genera. Annals of Botany Memoirs 1: 1–740. [Google Scholar]
- Cripps CL, Larsson E, Vauras J. 2020. Nodulose-spored Inocybe species from the Rocky Mountain alpine zone: molecularly linked to European type specimens. Mycologia 112: 133–153. [DOI] [PubMed] [Google Scholar]
- Crous PW, Cowan DA, Maggs-Colling G, et al. 2020a. Fungal Planet description sheets: 1112–1181. Persoonia 45: 251–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Cowan DA, Maggs-Colling G, et al. 2021a. Fungal Planet description sheets: 1182–1283. Persoonia 46: 313–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Groenewald JZ. 2011. Why everlastings don’t last. Persoonia 26: 70–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Hernández-Restrepo M, Schumacher RK, et al. 2021b. New and interesting fungi 4. Fungal Systematics and Evolution 7: 255–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Osieck ER, Jurjević Ž, et al. 2021c. Fungal Planet description sheets: 1284–1382. Persoonia 47: 178–374. [DOI] [PubMed] [Google Scholar]
- Crous PW, Schumacher RK, Akulov A, et al. 2019. New and interesting fungi. 2. Fungal Systematics and Evolution 3: 57–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Shivas RG, Quaedvlieg W, et al. 2014. Fungal Planet description sheets: 214–280. Persoonia 32: 184–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Summerell BA, Shivas RG, et al. 2011. Fungal Planet description sheets: 92–106. Persoonia 27: 130–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI, et al. 2018. Fungal Planet description sheets: 716–784. Persoonia 40: 239–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Chooi Y-H, et al. 2020b. Fungal Planet description sheets: 1042–1111. Persoonia 44: 301–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Richardson DM, et al. 2016. Fungal Planet description sheets: 400–468. Persoonia 36: 316–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Schumacher RK, et al. 2020c. New and interesting fungi 3. Fungal Systematics and Evolution 6: 157–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Hora Júnior BT, De Macedo DM, Barreto RW, et al. 2014. Erasing the past: a new identity for the Damoclean pathogen causing South American leaf blight of rubber. PLoS ONE 9: e104750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Beer ZW, Wingfield MJ. 2013. Emerging lineages in the Ophiostomatales. In: Seifert KA, De Beer ZW, Wingfield MJ. (eds), The Ophiostomatoid fungi: expanding frontiers: 21–46. CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands. [Google Scholar]
- De Hoog GS. 1972. The genera Beauveria, Isaria, Tritirachium and Acrodontium gen. nov. Studies in Mycology 1: 1–41. [Google Scholar]
- Dereeper A, Guignon V, Blanc G, et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Research 1: 36 (Web Server issue): W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardin DE. 1987. New and noteworthy marasmioid fungi from California. Mycologia 79: 123–134. [Google Scholar]
- Desjardin DE, Perry BA. 2017. The gymnopoid fungi (Basidiomycota, Agari-cales) from the Republic of São Tomé and Príncipe, West Africa. Mycosphere 8: 1317–1391. [Google Scholar]
- Devadatha B, Calabon MS, Abeywickrama PD, et al. 2020. Molecular data reveals a new holomorphic marine fungus, Halobyssothecium estuariae, and the asexual morph of Keissleriella phragmiticola. Mycology 11:167–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta AK, Wilson AW, Antonín V, et al. 2015. Taxonomic and phylogenetic study on gymnopoid fungi from Eastern India I. Mycological Progress 14: 79. [Google Scholar]
- Eberhart J, Trappe J, Piña Páez C, et al. 2020. Tuber luomae, a new spiny-spored truffle species from the Pacific Northwest, USA. Fungal Systematics and Evolution 6: 299–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MB. 1949. Tetraploa. Transactions British Mycological Society 32: 246–251. [Google Scholar]
- Esteve-Raventós F, Bandini D, Oertel B, et al. 2018. Advances in the knowledge of the Inocybe mixtilis group (Inocybaceae, Agaricomycetes), through molecular and morphological studies. Persoonia 41: 213–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
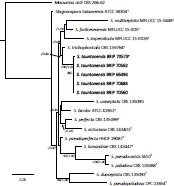
- Fritsche Y, Lopes ME, Selosse M-A, et al. 2021. Serendipita restingae sp. nov. (Sebacinales); an orchid mycorrhizal agaricomycete with wide host range. Mycorrhiza 30: 1–15. [DOI] [PubMed] [Google Scholar]
- Gams W, Philippi S. 1992. A study of Cyathicula strobilina and its Chalara anamorph in vitro. Persoonia 14: 547–552. [Google Scholar]
- Gams W, Stielow B, Gräfenhan T. et al. 2019. The ascomycete genus Niesslia and associated monocillium-like anamorphs. Mycological Progress 18: 5–76. [Google Scholar]
- Garnica S, Schön ME, Abarenkov K, et al. 2016. Determining threshold values for barcoding fungi: lessons from Cortinarius (Basidiomycota), a highly diverse and widespread ectomycorrhizal genus. FEMS Microbiology Ecology 92: fiw045. [DOI] [PubMed] [Google Scholar]
- Geiser DM, Ivery MLL, Hakiza G, et al. 2005. Gibberella xylarioides (anamorph: Fusarium xylarioides), a causative agent of coffee wilt disease in Africa, is a previously unrecognized member of the G. fujikuroi species complex. Mycologia 97: 191–201. [DOI] [PubMed] [Google Scholar]
- Gerlach W. 1977. Fusarium robustum spec. nov., der Erreger einer Stammfäule an Araucaria angustifolia (Bertol.) O. Kuntze in Argentinien? Journal of Phytopathology 88: 29–37. [Google Scholar]
- Gerlach W, Nirenberg H. 1982. The genus Fusarium – a pictorial atlas. Mitteilungen aus der Biologischen Bundesanstalt für Land- und Forstwirtschaft Berlin-Dahlem 209: 1–406. [Google Scholar]
- Glynou K, Ali T, Buch A-K, et al. 2016. The local environment determines the assembly of root endophytic fungi at a continental scale. Environmental Microbiology 18: 2418–2434. [DOI] [PubMed] [Google Scholar]
- Greif MD, Gibas CFC, Currah RS. 2006. Leptographium piriforme sp. nov., from a taxonomically diverse collection of arthropods collected in an aspen-dominated forest in western Canada. Mycologia 98: 771–780. [DOI] [PubMed] [Google Scholar]
- Gui Y, Zhu GS, Callac P, et al. 2015. Agaricus section Arvenses: three new species in highland subtropical Southwest China. Fungal Biology 119: 79–94. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, et al. 2010. New algorithms and methods to estimate maximum likelihood phylogenies: Assessing the performance of PhyML 3.0. Systematic Biology 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Havenga M, Wingfield BD, Wingfield MJ, et al. 2020. Diagnostic markers for Teratosphaeria destructans and closely related species. Forest Pathology 50: e12645. [Google Scholar]
- Hayward J, Tourtellot SG, Horton TR. 2014. A revision of the Alpova diplophloeus complex in North America. Mycologia 106: 846–855. [DOI] [PubMed] [Google Scholar]
- Hernández-Restrepo M, Giraldo A, Van Doorn R, et al. 2020. The genera of fungi - G6: Arthrographis, Kramasamuha, Melnikomyces, Thysanorea, and Verruconis. Fungal Systematics and Evolution 6: 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera CS, Rossman AY, Samuels GJ, et al. 2013. Pseudocosmospora, a new genus to accommodate Cosmospora vilior and related species. Mycologia 105: 1287–1305. [DOI] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, Von Haeseler A, et al. 2018. UFBoot2: Improving the ultrafast bootstrap approximation. Molecular Biology and Evolution 35: 518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horak E, Desjardin DE. 1994. Reduced marasmioid and mycenoid Agarics from Australasia. Australian Systematic Botany 7: 153–170. [Google Scholar]
- Hou LW, Groenewald JZ, Pfenning LH, et al. 2020. The phoma-like dilemma. Studies in Mycology 96: 309–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J, Kocsubé S, Visagie CM, et al. 2020. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Studies in Mycology 95: 5–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson O, Buchholz M, Doyle V, et al. 2019. Multilocus phylogeny of Acrospermaceae: New epibiotic species and placement of Gonatophragmium, Pseudovirgaria, and Phaeodactylium anamorphs. Mycologia 111: 1041–1055. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Hyde KD, Hongsanan S, Jeewon R, et al. 2016. Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 80: 1–270. [Google Scholar]
- Inderbitzin P, Bostock RM, Davis RM. et al. 2011. Phylogenetics and taxonomy of the fungal vascular wilt pathogen Verticillium, with the descriptions of five new species. PLoS ONE 6: e28341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob M, Bhat DJ. 2000. Two new endophytic fungi from India. Cryptogamie, Mycologie 21: 81–88. [Google Scholar]
- Jacobsson S, Larsson E. 2018. Inocybe (Fr.) Fr. In: Knudsen H, Vesterholt J. (eds), Funga Nordica. Agaricoid, boletoid, cyphelloid and gasteroid genera. Second Edition: 981–1021. Nordsvamp, Copenhagen. [Google Scholar]
- Jankowiak R, Kolařík M. 2010. Leptographium piriforme – first record for Europe and of potential pathogenicity. Biologia 65: 754–757. [Google Scholar]
- Jayawardena RS, Hyde KD, McKenzie EH, et al. 2019. One stop shop III: taxonomic update with molecular phylogeny for important phytopathogenic genera: 51–75 (2019). Fungal Diversity 98: 77–160. [Google Scholar]
- Jones EBG, Pang K-L, Abdel-Wahab MA, et al. 2019. An online resource for marine fungi. Fungal Diversity 96: 347–433. [Google Scholar]
- Jones EBG, Suetrong S, Sakayaroj J, et al. 2015. Classification of marine Ascomycota, Basidiomycota, Blastocladiomycota and Chytridiomycota. Fungal Diversity 73: 1–72. [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, et al. 2017. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature Methods 14: 587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamara AM, El-Lakany MH, Badran OA, et al. 1981. Seed pathology of Araucaria spp. 1. A survey of seed-borne fungi associated with four Araucaria spp. Australian Forest Research 11: 269–274. [Google Scholar]
- Karunarathna A, Działak P, Jayawardena RS, et al. 2021. A novel addition to the Pezizellaceae (Rhytismatales, Ascomycota). Phytotaxa 480: 251–261. [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software version 7 : improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrigan RW. 2016. Agaricus of North America. Memoirs of The New York Botanical Garden Volume 114. NYBG Press. [Google Scholar]
- Kers LE. 1981. Några anmärkningsvärda fynd av hypogeiska svampar i Sverige. Svensk Botanisk Tidskrift 75: 129–140. [Google Scholar]
- Kers LE. 1983. Några svenska fynd av hypogeiska svampar. Svensk Botanisk Tidskrift 77: 259–268. [Google Scholar]
- Kers LE. 1986. Några norska fynd av hypogeer. Agarica 7: 30–48. [Google Scholar]
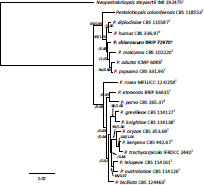
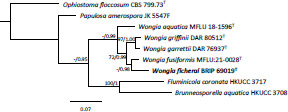
- Khemmuk W, Geering ADW, Shivas RG. 2016. Wongia gen. nov. (Papulosaceae, Sordariomycetes), a new generic name for two root-infecting fungi from Australia. IMA Fungus 7: 247–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaubauf S, Tharreau D, Fournier E, et al. 2014. Resolving the polyphyletic nature of Pyricularia (Pyriculariaceae). Studies in Mycology 79: 85–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp DG, Imrefi I, Boldpurev E, et al. 2019. Root-colonizing endophytic fungi of the dominant grass Stipa krylovii from a Mongolian Steppe grassland. Frontiers in Microbiology 10: 2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolařík M, Hulcr J, Tisserat N, et al. 2017. Geosmithia associated with bark beetles and woodborers in the western USA: taxonomic diversity and vector specificity. Mycologia 109: 185–199. [DOI] [PubMed] [Google Scholar]
- Kolařík M, Jankowiak R. 2013. Vector affinity and diversity of Geosmithia fungi living on subcortical insects inhabiting Pinaceae species in Central and Northeastern Europe. Microbial Ecology 66: 682–700. [DOI] [PubMed] [Google Scholar]
- Kolařík M, Kostovčík M, Pažoutová S. 2007. Host range and diversity of the genus Geosmithia (Ascomycota: Hypocreales) living in association with bark beetles in the Mediterranean area. Mycological Research 111: 1298–1310. [DOI] [PubMed] [Google Scholar]
- Kolařík M, Kubátová A, Pažoutová S. 2004. Morphological and molecular characterisation of Geosmithia putterillii, G. pallida comb. nov. and G. flava sp. nov., associated with subcorticolous insects. Mycological Research 108: 1053–1069. [PubMed] [Google Scholar]
- Kornerup A, Wanscher JH. 1978. Methuen handbook of colour. 3rd ed. Eyre Methuen, London. [Google Scholar]
- Kornerup A, Wanscher JH. 1981. Taschenlexikon der Farben. Muster-Schmidt Verlag, Göttingen. [Google Scholar]
- Kovăcs G, Calonge FD, Martín MP. 2011. The diversity of Terfezia desert truffles: new species and a highly variable species complex with intrasporocarpic nrDNA ITS heterogeneity. Mycologia 103: 841–853. [DOI] [PubMed] [Google Scholar]
- Kozlov AM, Darriba D, Flouri T, et al. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35: 4453–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, et al. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Molecular Biology and Evolution 35: 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Molecular Biology and Evolution 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyper TW. 1986. A revision of the genus Inocybe in Europe. I. Subgenus Inosperma and the smooth-spored species of subgenus Inocybe. Persoonia Supplement 3(1): 1–247. [Google Scholar]
- Laessøe T, Spooner BM. 1993. New British records. 103. Physalacria cryptomeriae Berthier & Rogerson. 104. Physalacria stilboidea (Cooke) Sacc. Mycologist 7: 162–163. [Google Scholar]
- Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Research 47: W256–W259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Deng WQ, Li TH, et al. 2018. Endophytic fungal communities associated with leaves, stems and roots of four medicinal plants in South China. Studies in Fungi 3: 126–140. [Google Scholar]
- Li X, Li W, Chu L, et al. 2016. Diversity and heavy metal tolerance of endophytic fungi from Dysphania ambrosioides, a hyperaccumulator from Pb–Zn contaminated soils. Journal of Plant Interactions 11: 186–192. [Google Scholar]
- Liu H, Li T, Ding Y, et al. 2017. Dark septate endophytes colonizing the roots of ‘non-mycorrhizal’ plants in a mine tailing pond and in a relatively undisturbed environment, Southwest China. Journal of Plant Interactions 12: 264–271. [Google Scholar]
- Liu JK, Hyde KD, Gareth EBG, et al. 2015. Fungal diversity notes 1–110: taxonomic and phylogenetic contributions to fungal species. Fungal Diversity 72: 1–197. [Google Scholar]
- Loizides M, Kyriakou T, Tziakouris A. 2011. Edible & toxic Fungi of Cyprus. Published by the authors. [Google Scholar]
- Madrid H, Hernández M, Gené J, et al. 2016. New and interesting chaetothyrialean fungi from Spain. Mycological Progress 15: 1179–1201. [Google Scholar]
- Maharachchikumbura SSN, Guo LD, Chukeatirote E, et al. 2011. Pestalotiopsis-morphology, phylogeny, biochemistry and diversity. Fungal Diversity 50: 167–187. [Google Scholar]
- Maharachchikumbura SSN, Hyde KD, Groenewald JZ, et al. 2014. Pestalotiopsis revisited. Studies in Mycology 79: 121–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlpine D. 1902. Fungus diseases of stone-fruit trees in Australia and their treatment. Melbourne, Victorian Department of Agriculture. [Google Scholar]
- Mello A, Vizzini A, Longato S, et al. 2000. Tuber borchii versus Tuber maculatum: neotype studies and DNA analyses. Mycologia 92: 326–331. [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), 14 Nov. 2010, New Orleans: 1–8. [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, et al. 2020. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution 37: 1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
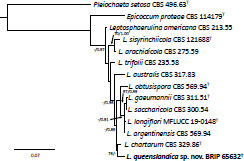
- Mitkowski NA, Browning M. 2004. Leptosphaerulina australis associated with intensively managed stands of Poa annua and Agrostis palustris. Canadian Journal of Plant Pathology 26: 193–198. [Google Scholar]
- Mongkolsamrit S, Khonsanit A, Thanakitpipattana D, et al. 2020. Revisiting Metarhizium and the description of new species from Thailand. Studies in Mycology 95: 171–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monkai J, Liu J-K, Boonmee S, et al. 2013. Planistromellaceae (Botryosphaeriales). Cryptogamie, Mycologie 34: 45–77. [Google Scholar]
- Moreau PA, Rochet J, Richard F, et al. 2011. Taxonomy of Alnus-associated hypogeous species of Alpova and Melanogaster (Basidiomycota, Paxillaceae) in Europe. Cryptogamie, Mycologie 32: 33–62. [Google Scholar]
- Münzmay T, Saar G, Schmidt-Stohn G, et al. 2009. Cortinarius laberiae Münzmay, B. Oertel & Saar nov. spec. und zwei weitere, wenig bekannte Arten aus der Gattung Cortinarius, Untergattung Phlegmacium in Europa. Journées Européenes du Cortinaire 11: 32–40. [Google Scholar]
- Nag Raj TR, Kendrick WB. 1975. A monograph of Chalara and allied genera. Wilfred Laurier University Press, Waterloo, Ontario, Canada. [Google Scholar]
- Nguyen L-T, Schmidt HA, Von Haeseler A, et al. 2015. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Molecular Biology and Evolution 32: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordeloos ME. 1980. Entoloma subgenus Nolanea in the Netherlands and adjacent regions with a reconnaissance of its remaining taxa in Europe. Persoonia 10: 427–534. [Google Scholar]
- Noordeloos ME, Hausknecht A. 1998. Rezente Rötlingsfunde aus Österreich und Italien. Österreichische Zeitschrift für Pilzkunde 7: 227–261. [Google Scholar]
- Noordeloos ME, Vila J, Jordal JB, et al. 2022. Contributions to the revision of the genus Entoloma (Basidiomycota, Agaricales) in Europe: six new species from subgenus Cyanula and typification of E. incarnatofuscescens. Fungal Systematics and Evolution 9: 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oktalira FT, May TW, Dearnaley JDW, et al. 2021. Seven new Serendipita species associated with Australian terrestrial orchids. Mycologia 113: 968–987. [DOI] [PubMed] [Google Scholar]
- Pegler DN. 1977. A preliminary Agaric flora of East Africa. Kew Bulletin Additional Series 6: 1–615. [Google Scholar]
- Pepori AL, Kolařík M, Bettini PP, et al. 2015. Morphological and molecular characterisation of Geosmithia species on European elms. Fungal Biology 119: 1063–1074. [DOI] [PubMed] [Google Scholar]
- Petersen RH, Hughes KW. 2021. Collybiopsis and its type species, Co. ramealis. Mycotaxon 136: 263–349. [Google Scholar]
- Phookamsak R, Hyde KD, Jeewon R, et al. 2019. Fungal diversity notes 929–1035: taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Diversity 95: 1–273. [Google Scholar]
- Pitt JI. 1980. The genus Penicillium and its teleomorphic states Eupenicillium and Talaromyces. London, Academic Press. [Google Scholar]
- Quaedvlieg W, Binder M, Groenewald JZ, et al. 2014. Introducing the Consolidated Species Concept to resolve species in the Teratosphaeriaceae. Persoonia 33: 1–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quin J, Yang ZL. 2016. Three new species of Physalacria from China, with a key to the Asian taxa. Mycologia 108: 215–226. [DOI] [PubMed] [Google Scholar]
- Radic T, Likar M, Hancevic K, et al. 2021. Root-associated community composition and co-occurrence patterns of fungi in wild grapevine. Fungal Ecology 50: 101034. [Google Scholar]
- Ramirez C. 1982. Manual and atlas of the Penicillia. Amsterdam-New York-Oxford, Elsevier Biomedical Press. [Google Scholar]
- Raper KB, Thom C. 1949. A manual of the Penicillia. Baltimore, Waverly Press, INC for The Williams & Wilkins Company. [Google Scholar]
- Redhead SA. 1979. Physalacria subpeltata sp. nov. from Hawaii. Mycotaxon 10: 46–48. [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Van der Mark P, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman AY, Samuels GJ, Rogerson CT, et al. 1999. Genera of Bionectriaceae, Hypocreaceae and Nectriaceae (Hypocreales, Ascomycetes). Studies in Mycology 42: 1–248. [Google Scholar]
- Roux C. 1986. Leptosphaerulina chartarum sp.nov., the teleomorph of Pitho-myces chartarum. Transactions of the British Mycological Society 86: 319–323. [Google Scholar]
- Sandoval-Denis S, Gené J, Sutton DA, et al. 2016. Redefining Microascus, Scopulariopsis and allied genera. Studies in Mycology 36: 1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santana-Ortiz B, Chen J, Parra L, et al. 2021. The genus Agaricus in the Caribbean II. Refined phylogeny of Agaricus subg. Spissicaules with description of two new sections and eight new species. Mycological Progress 20: 381–411. [Google Scholar]
- Seifert KA, Nickerson NL, Corlett M, et al. 2004. Devriesia, a new hyphomycete genus to accommodate heat-resistant, cladosporium-like fungi. Canadian Journal of Botany 82: 914–926. [Google Scholar]
- Shemesh H, Boaz BE, Millar CI, et al. 2020. Symbiotic interactions above treeline of long-lived pines: Mycorrhizal advantage of limber pine (Pinus flexilis) over Great Basin bristlecone pine (Pinus longaeva) at the seedling stage. Journal of Ecology 108: 908–916. [Google Scholar]
- Singer R. 1969. Mycoflora australis. Beihefte zur Nova Hedwigia 29: 1–405. [Google Scholar]
- Singer R. 1973. The genera Marasmiellus, Crepidotus, and Simocybe in the Neotropics. Beihefte zur Nova Hedwigia 44: 1–517. [Google Scholar]
- Singer R. 1976. Marasmieae (Basidiomycetes-Tricholomataceae). Flora Neo-tropica Monograph 17: 1–347. [Google Scholar]
- Singer R. 1986. The Agaricales in modern taxonomy. 4th edn. Koeltz Scientific Books, Kijnigstein, Germany. [Google Scholar]
- Singer R, Digilio APL. 1951. Pródromo de la Flora Agaricina Argentina. Lilloa 25: 5–461. [Google Scholar]
- Soop K, Dima B, Cooper JA, et al. 2019. A phylogenetic approach to a global supraspecific taxonomy of Cortinarius (Agaricales) with an emphasis on the southern mycota. Persoonia 42: 261–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: A toll for phylogenetic analysis and post-analysis of large phylogenenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology 57: 758–771. [DOI] [PubMed] [Google Scholar]
- Stangl J. 1989. Die Gattung Inocybe in Bayern. Hoppea 46: 5–388. [Google Scholar]
- Strzałka B, Jankowiak R, Bilański P, et al. 2020. Two new species of Ophiostomatales (Sordariomycetes) associated with the bark beetle Dryocoetes alni from Poland. MycoKeys 68: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strzałka B, Kolařík M, Jankowiak R. 2021. Geosmithia associated with hardwood-infesting bark and ambrosia beetles, with the description of three new species from Poland. Antonie van Leeuwenhoek 114: 169–194. [DOI] [PubMed] [Google Scholar]
- Sutton BC. 1980. The Coelomycetes. Fungi Imperfecti with Pycnidia, Acervuli and Stromata. CMI, Kew, UK. [Google Scholar]
- Swofford DL. 2003. PAUP* 4.0b10. Phylogenetic Analysis Using Parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland, MA, USA. [Google Scholar]
- Takahashi H. 2000. Two new species of Marasmiellus from eastern Honshu, Japan. Mycoscience 41: 467–472. [Google Scholar]
- Tamura K, Stecher G, Kumar S. 2021. MEGA 11: Molecular Evolutionary Genetics Analysis Version 11. Molecular Biology and Evolution 38: 3022–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, et al. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Hirayama K, Yonezawa H, et al. 2009. Molecular taxonomy of bambusicolous fungi: Tetraplosphaeriaceae, a new pleosporalean family with tetraploa-like anamorphs. Studies in Mycology 64: 175–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennakoon DS, Thambugala KM, De Silva ED, et al. 2019. Leaf litter saprobic Didymellaceae (Dothideomycetes): Leptosphaerulina longiflori sp., nov. and Didymella sinensis, a new record from Roystonea regia. Asian Journal of Mycology 2: 87–100. [Google Scholar]
- Thambugala KM, Wanasinghe DN, Phillips AJL, et al. 2017. Mycosphere notes 1–50: Grass (Poaceae) inhabiting Dothideomycetes. Mycosphere 8: 697–796. [Google Scholar]
- Thongklang N, Nawaz R, Khalid AN, et al. 2014. Morphological and molecular characterization of three Agaricus species from tropical Asia (Pakistan, Thailand) reveals a new group in section Xanthodermatei. Mycologia 106: 1220–1232. [DOI] [PubMed] [Google Scholar]
- Tulasne LR, Tulasne C. 1851. Fungi Hypogaei. In: Klincksieck F. (ed.), Histoire et Monographie des Champignons Hypogés. Paris, France. [Google Scholar]
- Vauras J, Larsson E. 2016. Inocybe caprimulgi and I. lacunarum, two new nodulose-spored species from Fennoscandia. Karstenia 55: 1–18. [Google Scholar]
- Videira S, Groenewald JZ, Nakashima C, et al. 2017. Mycosphaerellaceae – chaos or clarity? Studies in Mycology 87: 257–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Videira SIR, Groenewald JZ, Braun U, et al. 2016. All that glitters is not Ramularia. Studies in Mycology 83: 49–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila J, Carbó J, Caballero F, et al. 2013. A first approach to the study of the genus Entoloma subgenus Nolanea sensu lato using molecular and morphological data. Fungi non Delineati 66: 3–62. [Google Scholar]
- Visagie CM, Goodwell M, Nkwe DO. 2021. Aspergillus diversity from the Gcwihaba Cave in Botswana and description of one new species. Fungal Systematics and Evolution 8: 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J, Sutton BC, Pascoe IG. 1992. Phaeoseptoria eucalypti and similar fungi on Eucalyptus, with description of Kirramyces gen. nov. (Coelomycetes). Mycological Research 96: 911–924. [Google Scholar]
- Walz A, De Hoog GS. 1987. A new species of Cyphellophora. Antonie van Leeuwenhoek 53: 143–146. [DOI] [PubMed] [Google Scholar]
- Wanasinghe DN, Phukhamsakda C, Hyde KD, et al. 2018. Fungal diversity notes 709–839: taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Diversity 89: 1–236. [Google Scholar]
- Wang XC, Chen K, Zeng ZQ, et al. 2017. Phylogeny and morphological analyses of Penicillium section Sclerotiora (Fungi) lead to the discovery of five new species. Scientific Reports 7: 8233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warcup JH, Talbot PHB. 1967. Perfect states of Rhizoctonias associated with orchids. New Phytologist 66: 631–641. [Google Scholar]
- Weiss M, Waller F, Zuccaro A, et al. 2016. Sebacinales – one thousand and one interactions with land plants. New Phytologist 211: 20–40. [DOI] [PubMed] [Google Scholar]
- Wijayawardene NN, Hyde KD, Wanasinghe DN, et al. 2016. Taxonomy and phylogeny of dematiaceous coelomycetes. Fungal Diversity 77: 1–316. [Google Scholar]
- Wijayawardene NN, Phillips AJL, Tibpromma S, et al. 2021. Looking for the undiscovered asexual taxa; case studies from lesser studied life modes and habitats. Mycosphere 12: 1290–1333. [Google Scholar]
- Wong PTW, Dong C, Stirling AM, et al. 2012. Two new Magnaporthe species pathogenic to warm-season turfgrassses in Australia. Australasian Plant Pathology 41: 321–329. [Google Scholar]
- Woudenberg JHC, Meijer M, Houbraken J, et al. 2017. Scopulariopsis and scopulariopsis-like species from indoor environments. Studies in Mycology 88: 1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaguchi T, Miyadoh S, Udagawa S. 1993. Chromocleista, a new cleistothecial genus with a Geosmithia anamorph. Transactions of the Mycological Society of Japan 34: 101–108. [Google Scholar]
- Yen LTH, Dung NL, Van Hop D, et al. 2012. Condylospora vietnamensis, a new Ingoldian hyphomycete isolated from fallen leaves in Vietnam. Mycoscience 53: 326–329. [Google Scholar]
- Yin M, Wingfield MJ, Zhou X, et al. 2019. Taxonomy and phylogeny of the Leptographium olivaceum complex (Ophiostomatales, Ascomycota), including descriptions of six new species from China and Europe. MycoKeys 60: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoya K, Postel S, Fang R, et al. 2017. Endophytic fungal diversity of Fragaria vesca, a crop wild relative of strawberry, along environmental gradients within a small geographical area. PeerJ 5: e2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Gao F, Jakovlić I, et al. 2020. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular Ecology Resources 20: 348–355. [DOI] [PubMed] [Google Scholar]
- Zhang X, Li Y, Si H, et al. 2022. Geosmithia species associated with bark beetles from China, with the description of eleven new species. Frontiers in Microbiology 124. 10.3389/fmicb.2022.820402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z-F, Zhou S-Y, Eurwilaichitr L, et al. 2021. Culturable mycobiota from Karst caves in China II, with descriptions of 33 new species. Fungal Diversity 106: 29–136. [Google Scholar]
- Zhou J-L, Su S-Y, Su H, et al. 2016. A description of eleven new species of Agaricus sections Xanthodermatei and Hondenses collected from Tibet and the surrounding areas. Phytotaxa 257: 99–121. [Google Scholar]