Abstract
Predatory bacteriophages have evolved a vast array of depolymerases for bacteria capture and deprotection. These depolymerases are enzymes responsible for degrading diverse bacterial surface carbohydrates. They are exploited as antibiofilm agents and antimicrobial adjuvants while rarely inducing bacterial resistance, making them an invaluable asset in the era of antibiotic resistance. Numerous depolymerases have been investigated preclinically, with evidence indicating that depolymerases with appropriate dose regimens can safely and effectively combat different multidrug-resistant pathogens in animal infection models. Additionally, some formulation approaches have been developed for improved stability and activity of depolymerases. However, depolymerase formulation is limited to liquid dosage form and remains in its infancy, posing a significant hurdle to their clinical translation, compounded by challenges in their applicability and manufacturing. Future development must address these obstacles for clinical utility. Here, after unravelling the history, diversity, and therapeutic use of depolymerases, we summarized the preclinical efficacy and existing formulation findings of recombinant depolymerases. Finally, the challenges and perspectives of depolymerases as therapeutics for humans were assessed to provide insights for their further development.
Key words: Phage-derived polysaccharide depolymerases, Bacteriophage proteins, Bacterial capsules, Polysaccharide capsules, Antibiotic resistance, Biofilms, Antibacterial therapy, Antimicrobial adjuvants, Combination treatment
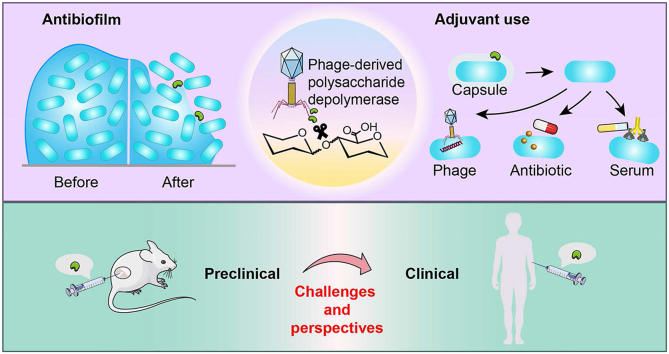
Graphical abstract
Phage-derived polysaccharide depolymerases are useful as antibiofilm agents and antimicrobial adjuvants. Their preclinical efficacy was reviewed, and their challenges and prospects for clinical translation were discussed.
1. Introduction
The overuse and misuse of antibiotics have promoted bacterial resistance to antibiotics, resulting in the prevalence of superbugs and thereby causing more difficult-to-treat infectious diseases1. The situation is worsening by the increasing challenges in discovering novel antibiotics and the rapidly evolved resistance rate to the newly marketed antibiotics2. Therefore, developing novel alternatives to combat multidrug-resistant (MDR) bacteria with a low risk of bacterial resistance development is highly desirable.
Bacteriophage (phage) therapy, developed before the discovery of penicillin by using bacterial viruses to treat bacterial infections, is re-emerging as a promising alternative to conventional antibiotics in the fight against MDR bacteria3. Phages possess a co-evolved mechanism enabling them to overcome bacterial resistance4,5. One typical bacterial resistance mechanism involves the secretion of slime layers (biofilms) and cell-surface capsules, which mask the primary receptors and hinder phage adsorption to host bacteria6, 7, 8, 9. Fortunately, some phages have adapted to such resistance by encoding depolymerases to degrade the polysaccharide components of capsules and biofilm slime4. This exemplifies the remarkable advantage of phage therapy over conventional antibiotics in combating MDR bacteria. However, the activity of whole phage particles is compromised by the high-frequency and rapid emergence of phage resistance10. In contrast, free depolymerases, different from whole phage particles, barely promote resistant mutations as they do not directly kill bacteria11.
Recently, depolymerases have been identified as novel alternative antimicrobials against MDR bacteria and biofilm-related infections. These enzymes act on the bacterial surface decorated polysaccharides, including capsular polysaccharides (CPS), exopolysaccharides (EPS), and lipopolysaccharides (LPS), to unmask the binding sites of different antimicrobial agents. Aside from facilitating phage diffusion and adsorption, depolymerases can sensitize bacteria to antibiotic treatments and boost the innate immune responses (i.e., increasing serum complement deposition and promoting phagocytosis), thereby significantly enhancing the antibacterial efficacy12,13. Many recombinant depolymerases have been investigated to treat different MDR bacterial infections in vivo. Moreover, the development of formulations for the efficient delivery of these proteinaceous agents has been budding. These endeavors promote our understanding of the challenges and prospects associated with translating depolymerases into antibacterial therapeutics.
In this review, we start with overviewing the development history of depolymerases, followed by unravelling the diversity of depolymerases in terms of their structure, mechanisms of action, and substrate specificity. We then survey the therapeutic use of depolymerases in antibiofilm, assisting phage therapy, antibiotic treatment and immune attack, and suppressing resistance development. We focused on summarizing the therapeutic efficacy of diverse recombinant depolymerases against different MDR pathogens in preclinical animal models, shedding light on the existing formulation findings, and finally assessing the challenges and perspectives of translating them into therapeutics for humans.
2. History of depolymerases
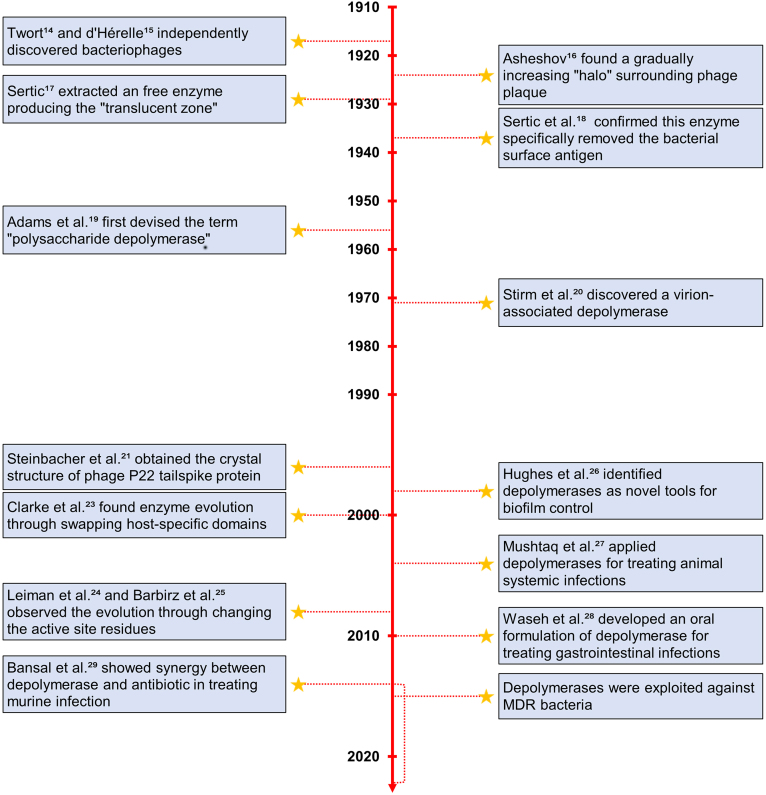
The major events in the development history of depolymerases were summarized (Fig. 1). Phages were discovered independently by Twort in 191514 and d'Hérelle in 191715. Certain phages produced plaques surrounded by a gradually increasing “translucent halo”16. In 1929, Sertic extracted an enzyme that reproduced the halo on the bacterial culture, first revealing the mechanism of halo production17. Later in 1937, the enzyme was demonstrated to specifically remove the bacterial surface antigen and increase the sensitivity of bacterium to phages without causing bacterial cell death18. Up until 1956, Adams et al.19 isolated a new enzyme capable of hydrolyzing bacterial capsular polysaccharides and devised the term “polysaccharide depolymerase”. This fostered the discovery of various kinds of depolymerases since the 1960s.
Figure 1.
A timeline of depolymerase discovery and development (by publication year).
Different from the before-found depolymerases that were in free form, Stirm et al.20 discovered a phage particle carried endoglycosidases that depolymerized capsular polysaccharides in 1971, providing early evidence of depolymerases as a structural part of phage. The following studies started to explore the structural features of depolymerases. In 1996, Steinbacher et al.21 crystalized phage P22 tailspike protein (TSP) with rhamnosidase activity for bacterial O-antigens. This research provided the structural basis of the interaction between TSP and its specific polysaccharide substrate. TSP was demonstrated to form elongated homotrimers with a β-helical central domain responsible for substrate recognition and binding. Since then, the identical architecture was confirmed in many other TSPs22. Evolutionary changes were observed between these proteins through two approaches: (1) swapping host-specific domains23 and (2) changing the active site residues24,25. Entering the 21st century, Clarke et al.23 initially proposed the evolutionary process of phage K5A to phage K1E using the first strategy, which involved the acquisition of the K1 endosialidase and the loss of the K5 lyase. A few years later, Leiman et al.24 and Barbirz et al.25 observed the evolution in the TSPs of phage P22, Sf6 and HK620 by the second strategy. These three TSPs exhibited similar structures in the same robust β-helical fold but possessed changes in their active site residues.
The applications of depolymerases were extensively explored. In 1998, Hughes et al.26 discussed the potential of depolymerases in degrading the extracellular polysaccharide matrix of biofilms, offering insights into a new tool for biofilm control. In 2004, Mushtaq et al.27 investigated the therapeutic efficacy of depolymerases in preventing and curing systemic infection. A significant reduction in mortality was observed after intraperitoneal administration with 20 μg of depolymerase. This might be ascribed to the removal of the bacterial surface barrier, allowing the complement system to kill bacteria. On this basis, the application of depolymerases for the treatment of systemic infections was opened up. In 2010, the first oral formulation of depolymerase was developed for preventing bacterial colonization of the gut28. In 2014, Bansal et al.29 conducted a pioneer study investigating the combined effect of depolymerase and gentamicin in treating systemic and lung infections. The combination treatment significantly reduced bacterial load compared to the single treatments. The synergy between depolymerases and antibiotics offered a promising avenue to overcome the growing antimicrobial resistance problem. Nowadays, many studies put efforts into applying depolymerases as antibiotic alternatives to combat MDR bacteria11,30,31.
3. Diversity of depolymerases
3.1. Structure
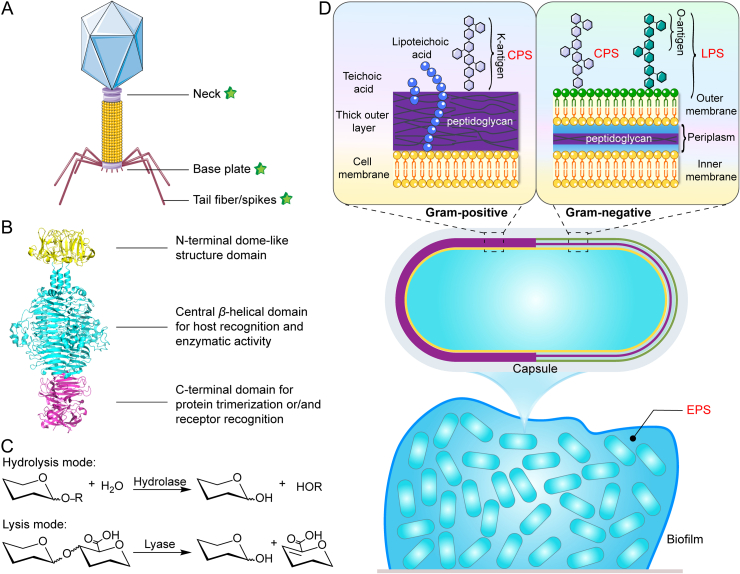
Phage–bacteria evolutionary machinery causes the alteration of bacterial carbohydrate structures and diversifies the active depolymerases. Many known depolymerases share common structural features despite a high diversity in their enzymatic specificity forced by the variable carbohydrate structures of bacteria32. Previously, 160 putative depolymerases in 143 phages infecting 24 genera of bacteria have been overviewed33. Depolymerases are structural components of the phage adsorption apparatus that mostly appear as tail fibers or tail spikes attached to the base plate, but also with few exceptions on the neck (Fig. 2A). Another present form of depolymerases is soluble proteins distant from the phage structure34. Predominantly, depolymerases locate within receptor binding proteins (RBPs) to help initiation of phage infection by recognizing and degrading bacterial capsules as the first step35. At the structural level, RBPs with depolymerase activity typically have a modular architecture consisting of three domains: a conserved N-terminal domain, a variable central domain and a C-terminal domain (Fig. 2B). Specifically, the N-terminal domain forms a dome-like structure responsible for anchoring the RBP to the tail structure of the phage particle. The central domain forms an elongated parallel β-helical homotrimer structure with a specific catalytic pocket that contributes to host recognition and enzymatic activity. This highly interwoven complex structure also determines its high protein stability in harsh environments. The C-terminal domain is generally involved in receptor binding and may contain a chaperone to assist proper protein folding and functionality22,36, 37, 38. This modular architecture of RBPs allows for swift modification of the host specificity through two different strategies: (i) swapping the host-specific central domain via intense horizontal gene transfer and (ii) changing the polysaccharide-depolymerizing domains via the accumulation of mutations (vertical gene transfer)22,37,38.
Figure 2.
Structure, mechanisms of action, and substrate specificity of depolymerases. (A) Distribution of depolymerase through the structure of bacteriophages. (B) The modular structure of the model tail spike (PDB ID 2XC1) of phage P22, consists of the N-terminal domain (yellow), central domain (cyan), and C-terminal domain (magentas). (C) General hydrolysis and lysis reaction mechanisms for depolymerases catalyzing polysaccharide degradation. (D) Varied targets of depolymerases in the architecture of bacteria and biofilms, including capsular polysaccharides (CPS), exopolysaccharides (EPS), and lipopolysaccharides (LPS).
3.2. Mechanism of action
Mechanistically, depolymerases cleave the O-glycosidic bonds of bacterial polysaccharides to obtain oligosaccharide products either in a hydrolysis or lysis mode (Fig. 2C). Such modes of action accordingly divide the depolymerases into two main classes: hydrolases and lyases. The diversity of depolymerases in terms of the two mechanisms of action has been summarized by Pires et al33. Hydrolases including sialidases, levanases, xylosidases, dextranases, hyaluronidases, and peptidases use a water molecule for cleavage. In contrast, lyases including hyaluronate, alginate, pectin/pectate lyases and K5 lyases, cleave a glycosidic bond by β-elimination with the introduction of a new double bond without involving a water molecule4,34,39.
3.3. Substrate specificity
The substrate specificity of depolymerases also account for their diversity. The structures and types of polysaccharide substrates produced by bacteria is diverse (Fig. 2D). Specifically, the Gram-negative bacteria can produce LPS as the outer membrane-embedded components with the O-antigens at the end and have thin peptidoglycan (PG) layers in the periplasmic space. The Gram-positive bacteria lack the LPS-embedded outer membranes, producing instead a much thicker PG layer. Both Gram-negative and Gram-positive bacteria can produce CPS as the outermost enveloped layers and secrete EPS as the biofilm constitutes32. In a broad sense, the phage-encoded peptidoglycan hydrolases, called endolysins, are also depolymerases that refer to any generic enzymes degrading polymers. However, distinguished from endolysins, depolymerases commonly refer to enzymes targeting CPS (also called K-antigen), EPS, and LPS (carrying the O-antigen). The surface CPS/LPS are enormously variable among bacteria and can be changed or modified by gene mutations, deletions or insertion in the carbohydrate biosynthesis loci32. The diversities in the sugar composition and structure of O- or K-antigen distinguish immunologically distinct strains by the corresponding O- or K-serotyping schemes40. Correlatively, depolymerases with a high serotype specificity also exhibit a high diversity to target different O- or K-types of bacteria. The diversity of depolymerases targeting CPS/EPS/LPS in terms of substrate specificity has previously been overviewed by Knecht et al4.
4. Potential applications of depolymerases
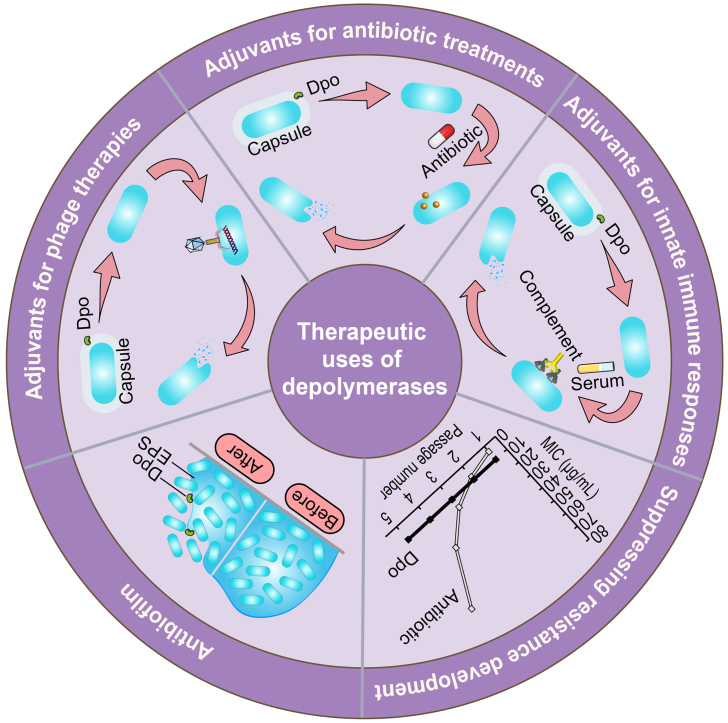
Depolymerases can be exploited as antibacterial therapeutics in two forms: phage-expressing enzymes and recombinant proteins; and both forms of proteins can be genetically engineered or mixed as cocktails to expand their host ranges41. Applications of phage and phage-derived proteins have been extensively reviewed34,42,43. Depolymerases can degrade CPS/EPS/LPS in the biofilm and bacterial surface barrier, but they do not kill bacteria. Therefore, they are expected to have great therapeutic potential as antibiofilm agents and antimicrobial adjuvants without inducing resistance development. Specifically, they can be exploited in the below five main strategies (Fig. 3).
Figure 3.
Schematic diagram of the therapeutic uses of depolymerases (Dpo).
4.1. Antibiofilm
Over 80% of bacterial infections are associated with biofilms and resistant to conventional antibiotic medications with a 1000-fold decrease in susceptibility44,45. The structural and functional integrity of bacterial biofilms and their virulence are mainly affected by the extracellular polymeric substances, accounting over 90% of the biofilms8,46. Many studies have demonstrated that depolymerases are effective in both preventing and eradicating biofilms, with combination treatments and cocktail therapies further enhancing the antibiofilm efficacy11,47, 48, 49, 50, 51, 52, 53, 54.
Gutiérrez et al.48 first identified a putative EPS depolymerase in a temperate phage vB_SepiS-phiIPLA7 infecting a host strain Staphylococcus epidermidis F12. This study confirmed the ability of depolymerase-bearing phage to mediate biofilm degradation with the viability of the host bacteria in the biofilm readily reduced and only 5% of the total counts survived after 3 h. In their subsequent study49, the gene encoding the depolymerase was cloned and the recombinant protein named Dpo7 was overexpressed and purified. Dpo7 was demonstrated to significantly reduce the biomass and adhered cells in the preformed biofilm in a dose-dependent but time-independent mode. Also, it was highly capable of preventing the formation of host bacterial biofilms. This study also found that Dpo7 can disperse biofilms generated by some other strains of S. epidermidis and Staphylococcus aureus. However, total removal of preformed biofilm and complete inhibition of biofilm formation by Dpo7 was not observed. Hopefully, combination therapy using depolymerases together with antibiotics or other antibacterial agents has been proved to enhance antibiofilm effectiveness. In another study, the effects of a depolymerase, Dpo71, in combination with colistin against Acinetobacter baumannii biofilm formation were tested11. The combination treatment significantly enhanced the antibiofilm efficiency compared with single-agent treatments. Similar results were also achieved in other combination therapy studies, such as investigating the efficacy of an endolysin LysK and a depolymerase DA7 against both the static and dynamic Staphylococcal biofilms50; testing the activity of phage-borne depolymerase and chloride dioxide (ClO2) against Klebsiella biofilm51; and evaluating the synergistic effect of honey and phage-bearing depolymerase against Escherichia coli biofilm52. Moreover, a cocktail therapy of three depolymerase-containing phages was preliminarily reported to eradicate Enterobacter cloacae biofilms completely53.
Topka-Bielecka et al.12 have comprehensively reviewed the application of depolymerases against biofilms. However, the antibiofilm activity is mostly confirmed under in vitro conditions. Such conditions lack the in vivo harsh environment and the response of immune system, thereby cannot wholly reflect the actual effects against biofilm infections. The in vivo antibiofilm efficacy of depolymerases has not yet been studied, which is worth to be considered in further research.
4.2. Adjuvants for phage therapies
For phages evolved with polysaccharide depolymerization activities, the phage receptor molecules at the bacterial surface can be exposed through degrading the carbohydrate layers, facilitating phage adsorption to allow genome injection, replication and finally disintegration of host bacterial cells43. However, the chance of isolating a natural phage that is simultaneously susceptible to the targeted bacteria and capable of expressing a relevant EPS-degrading enzyme is, to our surprise, not particularly high55. Therefore, for phages without polysaccharide depolymerizing domains, combined therapy with biofilm-susceptible depolymerases or engineering the phage with depolymerase activity are two possible approaches to improve their efficacy in removing biofilms and host pathogens54,56, 57, 58.
Hughes et al.57 first elucidated the role of polysaccharide depolymerases in phage attacking its host bacteria within biofilms. They proposed that the susceptibility of bacteria to phage and the ability of the phage-borne depolymerase to degrade EPS collaboratively affected the fate of biofilms. An efficient disruption of the matrix barrier by depolymerase followed by sufficient phage infection could ultimately undermine the whole biofilm structure. In contrast, an inefficient depolymerase may only allow phage infection against bacteria on the surface of biofilms. Hanlon et al.58 have also proved that phage diffusion through Pseudomonas aeruginosa biofilm may be facilitated by a reduction in EPS viscosity caused by enzymatic degradation. Rather than trying to select a combination or isolate a natural phage, Lu et al.54 proposed a modular design strategy of engineering the non-enzymatic T7 phage to express the most effective EPS-degrading enzymes (Dispersin B) specific to adhesins in the biofilms formed by different bacterial species. The engineered enzymatic phage was more efficacious at removing bacterial biofilms and reducing bacterial cell counts than the non-enzymatic phage. Moreover, such EPS-degrading enzyme rendered the phage a much wider range of biofilm-targeting abilities and thus could be applicable to a greater number of infections.
4.3. Adjuvants for antibiotic treatments
The polysaccharide components protect bacteria from antimicrobials46. The removal of bacterial CPS/LPS/EPS by depolymerases is supposed to enhance the entry of antimicrobials into bacterial cells. Combination treatments using antimicrobials and depolymerases have been reported to be promising in controlling bacterial infections, especially those caused by MDR pathogens11,59,60.
The synergistic effects of depolymerase and gentamicin against Klebsiella pneumoniae biofilm was investigated59. Depolymerases were reported to effectively disperse the biofilm matrix to facilitate the penetration of gentamicin, resulting in a significant reduction of bacterial counts compared with monotherapies. A similar synergy was also observed in the combination treatments of the Dep42 depolymerase and polymyxin against K. pneumoniae biofilms60, as well as the Dpo71 and colistin combination against A. baumannii biofilms11. The authors11 investigated the mechanisms responsible for the adjunctive effect of Dpo71 on colistin, showing that the bacterial decapsulation by depolymerase promoted the interaction between colistin and the bacteria membrane, and hence their entry.
4.4. Adjuvants for innate immune responses
The polysaccharide components also shield bacteria from immune attack (e.g., complement killing and phagocytosis)46. Applying depolymerases in the presence of human serum or macrophages has been reported to yield a significant antibacterial performance31,61, 62, 63.
The influence of two capsular depolymerases, KP32gp37 and KP32gp38, on the sensitivity of K. pneumoniae to complement-mediated killing and phagocytosis was evaluated61. Capsule degradation with a serotype-specific depolymerase produced a time-dependent decline in bacterial survival mediated by complement killing. The decapsulated strains were prone to phagocytosis with a two-fold increased uptake and microphage killing. Another research31 on a K64-specific capsule depolymerase (K64dep) against K. pneumoniae also confirmed the sensitized serum and neutrophil killing of enzyme-treated bacteria. Although depolymerases lack intrinsic antibacterial activity, they render the pathogens deprotected and fully susceptible to innate immune responses in a serum/phagocyte concentration-dependent manner.
4.5. Suppressing resistance development
Due to the selective pressure of phage therapy and the natural mechanism of bacteria-virus coevolution, the emergence of phage-resistant bacteria is unavoidable42. In contrast to using a whole phage, recombinant depolymerases rarely promote bacterial mutations and thus are recommended for therapeutic applications to reduce the resistance risk11,63,64. Another approach is applying combination treatment with antimicrobials harnessing different mechanisms so that the bacteria are less likely to develop resistance to multiple drugs simultaneously65, 66, 67.
The resistance development of host bacteria upon exposure to a depolymerase (DpoMK34) and its parent phage PMK34 was evaluated63. The emergence of phage-resistant mutants was observed, while DpoMK34-insensitive mutants were not yielded after repeated challenges. The resistance development was also not easily detected in the K2 depolymerase-challenged cultures64 and Dpo71-challenged bacteria11. In the combined therapy approach, the unwanted emergence of phage-resistant variants can also be delayed or reduced through using an effective phage cocktail or phage/depolymerase combination66. Majkowska-Skrobek and co-workers revealed that the phage cocktails consisting of capsule-specific (KP34) and capsule-independent phages (KP15 and KP27) could produce an effective treatment limiting the development of phage resistance in K. pneumoniae population67. The reduced bacterial resistance was also achieved in the combined treatment with KP34p57, KP15 and KP27 after replacing phage KP34 with its produced capsule depolymerase (KP34p57)66.
5. Therapeutic efficacy of depolymerases in vivo
After confirming the therapeutic ability of depolymerases in vitro, it is essential to prove that they are also active in vivo. Here, we will only focus on work applying recombinant depolymerases in the free form, not those used as an integral part of phage. To date, the in vivo evaluation of depolymerase efficacy has been limited to animal tests68. This section will summarize the preclinical findings of diverse reported recombinant depolymerases against several MDR pathogens in animal infection models (Table 1)11,27, 28, 29, 30, 31,64,69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83. In summary, depolymerases with proper dose regimens were safe and effective as antimicrobials in vivo.
Table 1.
Summary of preclinical efficacy of phage-derived depolymerases.
| Depolymerase name | Animal infection model | Dose regimen | Result | Ref. |
|---|---|---|---|---|
| K. pneumoniae | ||||
| K2 depolymerase | Normal mouse pneumonia and septicemia models | A single dose of 50 μg enzyme or in combination with 1.5 mg/kg gentamicin given 24 h (pneumonia model) or 6 h (septicemia model) post-infection via i.p. injection | The bacteria count, inflammatory cytokine (IL-1ß, IL-10 and TNFα) levels and neutrophil infiltration were significantly reduced in both infection models. Combination treatment caused more bacteria reduction and cytokine downregulation than sole depolymerase treatment. | 29 |
| K64dep | Immunocompromised mouse bacteremia model | A single dose of 18.75 or 150 μg enzyme given 1 or 8 h post–challenge via i.p. injection, respectively | The survival rate was 100% and 12.5% when treatment was given at 1 and 8 h post-infection, respectively. Significant antibacterial effectiveness was yielded by earlier treatment. No toxicity and pathological changes in liver, kidney, and spleen were observed. |
31 |
| K1-ORF34 | Immunocompetent and immunocompromised mouse invasive infection | A single dose of 25 μg enzyme given 30 min post-infection via i.p. injection | The survival rate was 100% in treating immunocompetent mice infected with 3.3 × 103 CFUs bacteria and immunocompromised mice infected with 1.6 × 103 CFUs bacteria. This indicated the antibacterial effects of depolymerases to both models. | 69 |
| Dep_kpv74 | Normal mouse hip infection model | A single dose of 10, 20, or 40 μg enzyme given 0.5 h post-infection via i.p. injection A single dose of 40 μg enzyme given 0.5, 3, or 24 h post-infection via i.p. injection |
The survival rate was 90% (40 μg, 0.5 h group), 80% (10 and 20 μg, 0.5 h groups) and 60% (40 μg, 3 and 24 h groups). A higher dose and earlier treatment can lead to improved treatment outcomes. | 70 |
| P560dep | Normal mouse bacteremia model | A single dose of 50 μg enzyme given 1 h prior to bacterial infection or 1 h post-infection via i.p. injection | 100% and 90% of mice were protected from death in the prevention and treatment groups, respectively, indicating significant prevention effect and antibacterial effect. | 71 |
| Dp42 | Normal mouse bacteremia model | A single dose of 50 μg enzyme given 6 h prior to bacterial infection or 0.5 h post-infection via i.p. injection | 100% of mice were protected from death, indicating significant prevention effect and antibacterial effect. Bacterial load in the mouse organs were significantly reduced. |
72 |
| depoKP36 | Galleria mellonella larvae infection model | A single dose of 2.8 μg depoKP36 given 5 min post-infection, or bacteria pretreated with 280 μg/mL enzyme for 2 h before inoculation via injection into the last proleg | The survival rate at 24, 48, 72 h was 40%, 30%, 20% in the post-infection treatment group and 77%, 47%, 43% in the pre-infection treatment group, respectively. Pretreatment of bacteria with depolymerase showed an increased survival rate compared to post-infection treatment. | 73 |
| Dep_kpv79 Dep_kpv767 | Normal mouse sepsis and hip infection model | A single dose of 50 μg enzyme given 0.5 h post-infection via i.p. injection | 80%–100% of animals were protected against death. | 74 |
| Dep622 DepS8 | G. mellonella larvae infection model | A single dose of 2 μg Dep622 or DepS8 simultaneously given at the same time of bacteria inoculation via injection into the hemocoel | ∼90% and ∼70% of larvae inoculated with 3 × 105 and 3 × 106 CFU bacteria survived within 5 days, respectively. The antibacterial efficacy was negatively correlated to the severity of infections. | 75 |
| A. baumannii | ||||
| Dpo71 | G. mellonella larvae infection model | A single dose of 5 μg Dpo71 or in combination with 1 μg colistin given 0.5 h post-infection via injection into the last right proleg | 40% and 80% of infected worms were rescued from the Dpo71-alone treatment and combination treatment, respectively. The antibacterial effect was significantly improved in combination treatment compared with the single treatments. | 11 |
| Tailspike protein of φAB6 phage | Zebrafish infection model | A single dose of 20 μg enzyme given 0.5 h post-infection via injection into cloaca | 80% of zebrafish survived within 4 days. No toxicity was observed. |
30 |
| K2 depolymerase | G. mellonella larvae infection model and immunocompromised mouse sepsis model | A single dose of 0.25, 0.5, and 3 μg per larva given 30 min post-infection, or bacteria pretreatment for 2 h before inoculation via injection into the penultimate right proleg A single dose of 50 μg enzyme per mouse given 1 h post-infection via i.p. injection |
The larvae survival rate was significantly increased in a dose-dependent and dosing time-dependent manner. Treated with 0.25, 0.5, and 3 μg enzyme, 53%, 69% and 88% of larvae survived in the pretreatment group, and 15%, 56% and 70% of larvae survived in the post-infection treatment group. 60% of enzyme-treated mice survived. The inflammatory factors (TNF-α and IL-6) were significantly reduced. |
64 |
| Dpo48 | G. mellonella larvae infection model, normal and immunocompromised mouse sepsis model | A single dose of 5 μg Dpo48 given 5 min post-infection, or bacteria pretreated with 50 μg/mL enzyme for 1 h before inoculation via injection into the last proleg A single dose of 50 μg enzyme per mice given 2 h post-infection via i.p. injection |
93% and 26% of larvae survived in the pretreatment group and treatment group within 3 days, respectively. Pretreatment of bacteria with depolymerase showed an increased survival rate compared to post-infection treatment. 100% of mice were protected from death. No significant differences in blood biochemical indicators and histopathological changes in the liver, spleen, lung and kidney were observed. |
76 |
| Dp49 | Normal mouse bacteremia infection | A single dose of 50 μg enzyme given 0.5 h post-infection via i.p. injection | 100% of mice were protected from death within 96 h. The bacterial loads from organs such as liver, spleen, and lungs all significantly decreased. |
77 |
| E. coli | ||||
| EndoE (K1E) | Neonatal rat gastrointestinal colonization-induced bacteremia model | 20 μg enzyme given by i.p. injection on Days 1, 2, 3, 4, and 5 post-infection; A single dose of 20 μg enzyme given on Days 1 or 3 post-infection via i.p. injection; A single dose of 0.125–20 μg enzyme given on Day 1 post-infection via i.p. injection | EndoE prevented the death and invasion of bacteria from the gastrointestinal tract into the bloodstream from the first dose. A single dose of depolymerase was sufficient to produce a therapeutic effect comparable to that of multiple doses. Single-dose treatment on Day 1 was more effective than that on Day 3. Earlier treatment with depolymerase yielded a better antibacterial efficacy. The minimal dose of 0.25 μg prevented the death of at least 80% of rats. |
27,78 |
| K1F, K1H, K5, K30 gp41, K30 gp42 depolymerases | Normal and immunocompromised mouse systemic infection model | Normal: 0, 2, 5, or 20 μg enzyme given within 0.5 h post-infection via injection into the right thigh; 20 μg enzyme given 8 h post-infection via injection into contralateral thigh; 20 μg K1E, 2 μg K1F and 2 μg K1H given immediately via i.p. and i.m. injection Immunocompromised: 20 μg enzyme given immediately or 8 h post-infection via i.m injection |
For normal mice, they were rescued in a dose-dependent manner. Delayed treatment with K1F, K1H and K5 enzymes resulted in 50%–60% survival compared to 90%–100% in immediate treatment. K1E significantly improved survival rate via i.p. injection compared with i.m. injection; K1F enzyme showed the opposite pattern; K1H yielded similarly low rescue rates for both routes. No toxicity was observed. For immunocompetent mice, immediate treatment was superior to delayed treatment for all enzymes. |
79,80 |
| Dep6 | Normal mouse systemic infection model | A single dose of 30 μg Dep6 given 3 h prior to infection, simultaneously with bacteria, and 3 h post–challenge | The survival rates were 33%, 83%, 100% in the post-infection treatment group, simultaneous treatment group and pretreatment group, respectively, indicating a dosing time-dependent manner. No toxic effects were observed. |
81 |
| P. aeruginosa | ||||
| LKA1gp49 | G. mellonella larvae infection model | A single dose of 5 or 50 μg/mL given 15 min post-infection, or bacteria pretreated with 5 μg/mL enzyme for 1 h before inoculation via injection into the hindmost proleg | 20% of larvae survived after 72 h in the treatment group, independent of LKA1gp49 concentration. 35% of larvae survived after 72 h in the pretreatment group. Pretreatment of bacteria with depolymerase showed an increased survival rate compared to post-infection treatment. | 82 |
| S. enterica | ||||
| P22sTsp | Chicken gastrointestinal tract infection model | 3 doses of 30 mg P22sTsp given 1, 18 and 42 h (protocol 1) or 18, 42 and 66 h (protocol 2) via oral garage. | A significant reduction of Salmonella in the cecum, liver and spleen in Protocol 1 but not in Protocol 2. Significant antibacterial effectiveness was yielded by earlier treatment. | 28 |
| S. aureus | ||||
| DA7 | Murine model of fracture-related infections | A single dose of 50 μg equimolar enzybiotics (M23, GH15, DA7) or in combination with 200 μg gentamicin via irrigation and 110 mg/kg vancomycin via s.c. | Enzybiotics alone did not significantly reduce overall CFU levels in soft tissue and implant but did so in combination with classical antibiotics. The antibacterial effect was significantly improved in combination treatment compared to the single treatments. | 83 |
5.1. Animal infection models
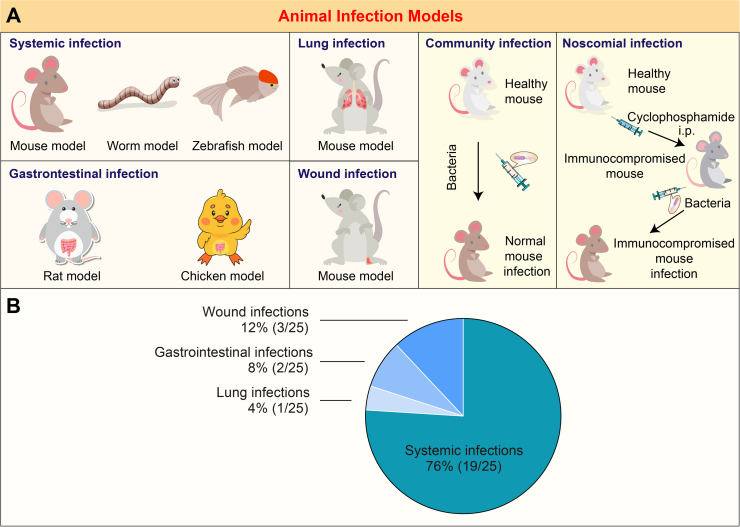
Currently, many depolymerases have been identified against some MDR bacteria, including the Gram-negative bacteria (K. pneumoniae, A. baumannii, E. coli, P. aeruginosa and Salmonella enterica) and the Gram-positive bacteria (S. aureus), that are listed as priority pathogens in urgent need of new control by the World Health Organization84. Because of bacterial surface carbohydrate layers, these virulent and resistant pathogens always cause severe infectious diseases, such as bloodstream infections, pneumonia, wound infections, and gastrointestinal infections, in hospitalized environments or communities13,41,85. Different animal models have been established to mimic these infections (Table 1 and Fig. 4A). Most studies targeted systemic infections, mainly based on murine models and few on Galleria mellonella and Zebrafish models. The murine animals were also used to investigate the efficacy of depolymerases against lung infections29 and fracture-related infections83. Other animals, such as rats and chickens, were employed to assess the therapeutic efficacy of depolymerases against intestinal infections27,28,78. While the community infection models can be established by direct injection of bacteria in healthy animals, the approach to develop nosocomial infection models was achieved by treating healthy mammalian animals with an immunosuppressive agent, like cyclophosphamide, before bacterial inoculation31,64,69,76,80. The immunocompromised animals afforded a greater range of useful inoculum sizes, which also increased the latitude of experimental designs80. However, monotherapy of depolymerases was mainly limited to systemic infections due to their dependency on serum-mediated bacteria killing. It is worth noting that few studies focus on local infections, which suggests more efforts to extend depolymerase applicability (Fig. 4B).
Figure 4.
Different animal infection models and the proportion of their associated studies. (A) Schematic diagram of various animal infection models. (B) Pie chart depicting the fraction of four different infection models reported in 22 references summarized in Table 1. Literature involving two types of models was counted twice. The total counts were 25.
5.2. Dose regimen
The administration route, dose, timing and frequency are critical factors affecting the success of depolymerase therapy86. First, the administration routes of depolymerase varied depending on the targeted site of action and the properties of enzymes. Parenteral administration, including intraperitoneal (i.p.), intravenous (i.v.), subcutaneous (s.c.), and intramuscular (i.m.), was used for systemic infections, while oral delivery was used for gastrointestinal infections28, and topical delivery was applied directly to the infected area83. However, it is noted that different administration routes of a specific enzyme can result in different bioavailability and therefore affect the therapeutic efficacy80. Second, the enzyme dose was determined by the type of animal models, the activity of the specific enzyme used, and the severity of infections. These were evidenced by varied enzyme doses used in different animal models64,76, different effective doses of three K1 capsular polysaccharide depolymerases targeting the same bacterial infection model79, and a larger dose of K64dep used for late-stage infections31. A higher dose may result in more significant degradation of targeted polysaccharides, leading to improved treatment outcomes64,70,79. Third, determined by the changes in the levels of targeted polysaccharides, the dosing time played a pivotal role in the treatment outcomes. Administering depolymerases as a pre-treatment before infection initiation or as an immediate/early treatment after infection establishment was demonstrated to yield a more significant therapeutic effectiveness27,28,31,70,76,80, 81, 82. Last, a single dose of depolymerase was sufficient to produce a therapeutic effect comparable to that of multiple doses27. In a few cases, continuous or regularly scheduled dosing may be necessary to maintain therapeutic enzyme levels28. In summary, the optimal dose regimen must be carefully determined on a case-by-case basis.
5.3. Effectiveness
Based on the development of appropriate animal infection models and dose regimens, the prevention, anti-virulence and anti-bacterial effects of depolymerases have been demonstrated by the improved survival rates and decreased bacterial counts. For instance, a prophylactic administration of depolymerase before the bacterial challenge was demonstrated to prevent infectious disease and rescue 100% of infected animals from death71,72,81. Before inoculation, pretreatment of bacteria with depolymerase also showed an increased survival rate64,73,76,82. Depolymerases removed surface carbohydrate layers and generated less virulent pathogens, thereby reducing the pathogenicity and host damage. Moreover, post-infection treatment with depolymerases improved survival, especially immediate treatment. Due to the exposure of pathogens to the host immune system by depolymerase, bacteria were vulnerable to serum and neutrophil killing, resulting in a decreased bacterial load72,77. Depolymerase monotherapy has been seen to be sufficient to overcome systemic infections. However, it must be noted that different depolymerizing proteins displayed various degrees of antimicrobial activity. For example, when the bacteria causing the same lethality to the untreated groups, K2 depolymerase showed a lower efficacy in rescuing infected mice compared to Dpo48 given at the same dose64,76. The discrepancy in efficacy might be attributed to their differences in enzymatic activities, synergism with other antibacterial factors, and variations in bacterial cell characteristics.
A strategy for improving the efficacies of recombinant depolymerases is to combine them with other antimicrobials, which may achieve a synergistic effect against the target bacteria and represent a promising treatment against local infections. Compared to antimicrobials used alone, the combination treatments of K2 depolymerase and gentamicin29, enzybiotics and gentamicin/vancomycin83 significantly reduced the bacterial counts for lung infections and fracture-related infections, respectively. This can be attributed to the dispersal of polysaccharide matrix mediated by depolymerases, leading to improved susceptibility of bacteria towards otherwise ineffective concentration of antibiotics. The synergistic combination could kill bacteria with different mechanisms of action and reduce the doses of individual antimicrobial, thus reducing the risk of developing bacterial resistance and possible side effects65.
5.4. Safety
The safety of depolymerases was evaluated by monitoring their impact on inflammatory response and toxicity in animal models. The level of inflammatory cytokines was elevated in response to bacterial challenge but reduced after depolymerase treatments29,64. This indicated the ability of depolymerases to control the immune response by reducing virulence and tissue damage caused by pathogens. Although proteinaceous depolymerases systemically administered at a single dose of 50 μg per mouse have the potential to trigger an immune response and generate neutralizing antibodies, no apparent adverse effects or reduction in the antibacterial activity were observed29. However, further research is still needed to investigate the risk of immunogenicity associated with higher doses. Acute toxicity of depolymerases was also assessed by examining blood biochemical markers, histopathological damage, survival rate, body weight and abnormal behaviour in normal mice or cyclophosphamide-treated mice. No significant differences were observed between the treated and control groups with all mice surviving and appearing healthy without any abnormal behaviour30,31,76,79. Notably, even treatment with a high dose of enzyme (up to 150 μg) showed no toxic side effects31. While the results are promising in the reported studies, the safety and efficacy of depolymerase therapy still require further evaluation in larger animal models and clinical trials.
6. Formulation of depolymerase therapeutics
Formulation strategies play a crucial role in the ultimate usefulness of enzyme products that may otherwise be limited by harsh in vivo environments. Factors such as pH, temperature, and chemical presence impact enzyme stability and activity, requiring consideration during the formulation process. In the current practice, depolymerases used for therapeutic purposes are predominantly prepared as liquid-based formulations for parenteral injections in most cases, and non-invasive oral and topical delivery in other cases (Table 1). This section summarizes the existing findings in optimizing the delivery performance of depolymerase-based liquid formulations in terms of improving stability and activity.
The effects of long-term storage, pH, temperature, buffer composition, and protein interactions on the activity of depolymerases were investigated. Chen et al.11 found that storing Dpo71 dissolved in phosphate-buffered saline at 4 °C was stable for up to 6 months without significant activity loss, suggesting the potential of developing depolymerases as commercially viable products. Many depolymerases were observed to retain activity across a broad range of pH and temperature conditions due to their trimeric structure80,82,83,87. Kassa et al.88 found that the addition of metal ions such as Na+, K+, Ca2+, Mg2+, Cu2+, and Al3+ to the buffer system could enhance the depolymerase activity, while Zn2+, Mn2+, and Fe2+ inhibited the activity. The presence of surfactants [Triton X-100, Tween-20, Tween-80, and sodium dodecyl sulfate (SDS)], a reductant (2-mercaptoethanol), and a protease inhibitor [ethylenediaminetetraacetic acid (EDTA)] at 100 mmol/L concentration did not affect the depolymerase activity, whereas another protease inhibitor [phenylmethanesulfonyl fluoride (PMSF)] at 1 mmol/L concentration inhibited the enzyme activity up to 38%. Majkowska-Skrobek et al.61 showed that KP32gp37 was resistant to SDS detergent and trypsin protease, but KP32gp38 was not. These findings suggested the need for formulation strategies to avoid the denaturation of some depolymerases in body fluid. For example, Waseh et al.28 reported that P22sTsp was resistant to trypsin but sensitive to chymotrypsin and pepsin digestion. To diminish the possible adverse effects of these gastrointestinal proteases in vivo, 10% bovine serum albumin (BSA) was added to the P22sTsp formulation for protective purposes. Another study pioneeringly immobilized a depolymerase, alginate lyase, in the hyaluronic acid-cholesterol nanohydrogels, which almost fully preserved the enzymatic activity89. However, studies on formulating depolymerases are still scarce and further research would be needed for a more promising antimicrobial efficacy.
7. Challenges of developing depolymerases into antimicrobial drugs
7.1. Applicability issues
A major challenge in the drug development of depolymerases is their narrow host spectrum90. One depolymerase is effective only against particular bacterial strains, requiring precise diagnostic tests to identify the bacterial strain causing the infection before initiating therapy12. Failure to recognize the causative bacterial strain can render the depolymerase treatment ineffective, thus limiting its broad clinical utility. To address this challenge, combination therapy41, cocktail therapy53, and genetic engineering54 approaches can be employed to broaden the range of bacterial strains targeted and expand the applicability of depolymerases. Nevertheless, more research is needed to validate the effectiveness and safety of these approaches used in vivo. Despite their limitations, the high specificity of depolymerases also offers advantages by protecting the normal human microbiota, reducing the risk of unintentional harm to the host91. Therefore, balancing the potential limitations and benefits of the narrow host range is necessary for the successful development and clinical application of depolymerase therapeutics.
7.2. Manufacturing issues
The large-scale production of depolymerases for widespread use needs to overcome several issues regarding expression platforms, purification degree, safety concerns, and production costs92,93. The scale-up production requires the efficient harvesting of the desired enzyme from the expression platform, such as E. coli, methylotrophic yeast, or filamentous fungi. However, the most efficient platform needs to be selected for each individual depolymerase. The subsequent purification process implicates the safety of the final product. To date, depolymerases used for therapeutic purposes are primarily administrated parenterally. The risk of inadequate purification and contamination during this process would increase immunogenicity and allergenicity, resulting in patient harm. Therefore, the proteins must be highly purified under strict operations. This calls for multiple separation, filtration, and sterilization steps in conformity with Good Manufacturing Practices (GMPs)94. To meet the requirements, companies must invest in production lines. Nevertheless, efforts are needed to reduce the total production costs to make the final product of depolymerases economically viable.
7.3. Formulation issues
One main challenge in formulating depolymerases is ensuring their physiochemical stability during storage and delivery to the patient95. It is essential to characterize the properties of each depolymerase before identifying the suitable excipients. For example, for proteins that can readily aggregate and be vulnerable to chemical degradation, the addition of stabilizing excipients, such as surfactants and protease inhibitors can form a protective barrier around the protein, reducing its exposure to environmental stressors28,88. However, the current formulations of depolymerases are limited to liquid dosage form and scarcely reported. To attain adequate shelf-life, stable in vivo transport and efficient delivery to the target site, further development of formulation approaches is still needed. The formulation strategies might include (1) the lyophilization of depolymerases for long-term storage stability, (2) the design of nano-based delivery systems that can shield depolymerase from harsh conditions with improved bioavailability and enhanced target specificity, and (3) the preparation of topical formulations for direct delivery of depolymerase to localized infections96. In summary, the optimal formulations compatible with the administration route for steadily transport and enhanced delivery to the target site must be carefully determined for each depolymerase.
8. Concluding remarks and future perspectives
In the era of antibiotic resistance, phage-derived polysaccharide depolymerases have emerged as alternative antimicrobials. Their variety in structure, mechanisms of action, and substrate specificity have revealed the diversity of depolymerases and expanded our understanding of their applications. A noteworthy finding is that susceptible bacterial strains of a specific depolymerase might share the same serotype73, suggesting the potential utility of depolymerases as a probe for rapid and flexible bacterial typing. Although the phage-typing method has been employed for subdividing serotypes of bacteria97, it is limited due to phage-type alterations following lysogenic conversion98. Surprisingly, phage depolymerase-based typing in this context remains unexplored, presenting an intriguing avenue to harness depolymerases for diagnostic purposes aside from therapeutic applications.
In vitro studies have shed light on the therapeutic uses of depolymerases as antibiofilm agents. However, it is worth noting that current antibiofilm studies are primarily conducted under in vitro conditions, which may not fully reflect the actual biofilm infections99. To gain a more comprehensive understanding of the efficacy of depolymerases, it is essential to characterize their performance in actual infection settings by direct sampling from infection sites100. Advancements in scientific techniques, such as RNA sequencing, metabolomics, immunohistochemistry, and microscopy imaging, allow for the characterization of infectious microenvironments and validation of the actual effects of depolymerases in vivo. By leveraging these state-of-the-art methods, we can pave the way for applying depolymerases in effectively combating biofilm-related infections.
Furthermore, the adjuvant properties of depolymerases, which enhance the efficacy of conventional antimicrobials while minimizing the emergence of resistance, present a promising approach to fighting against MDR bacteria. Preclinical in vivo studies have demonstrated the safety and effectiveness of depolymerases against various MDR bacterial infections. However, the therapeutic application of depolymerases has not yet entered the clinical stage, and formulations are in their infancy. More efforts are needed to gather additional preclinical data regarding their pharmacokinetics, potential toxicity, and resistance selection to proceed to clinical treatments. In addition, several gaps need to be filled to successfully develop depolymerase therapeutics, including (1) exploring the innovative approaches to expand the applicability of depolymerases; (2) establishing production specifications to ensure the quality and safety of the final product; and (3) formulating depolymerases into various delivery systems to ensure their stability during storage and transport, and efficient delivery to the target site.
To fully realize the therapeutic potential of depolymerases, cutting-edge research directions will focus on combination therapy, cocktail therapy, and genetic engineering in the future. Synergistic combinations of depolymerases and other antimicrobials capable of efficiently eradicating MDR bacteria and biofilms have to be explored. On the other hand, implementing cocktail therapy and genetic engineering strategies to broaden the host range of depolymerases are also worth future study. Owing to the recent advancements in structural bioinformatics and computational modeling, rational design and optimization of the enzymatic activity of depolymerases can be facilitated22. Ultimately, these innovative approaches have the potential to revolutionize the field of depolymerase-based therapeutics, reinforcing their combat against MDR bacterial infections and biofilm-related diseases. Despite the challenges in developing depolymerases into antimicrobial drugs, we firmly believe these obstacles will be overcome, resulting in significant societal benefits.
Acknowledgments
This work was supported by the University Grants Committee, Hong Kong SAR Government (No. 14112921, China). The support of HKPFS from the University Grants Committee to Honglan Wang was greatly acknowledged.
Author contributions
Honglan Wang conceptualized the article, performed the literature search, and wrote the manuscript. Yannan Liu and Changqing Bai reviewed the manuscript. Sharon Shui Yee Leung conceptualized the article, supervision, and revised the manuscript. All of the authors have read and approved the final manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
References
- 1.Christaki E., Marcou M., Tofarides A. Antimicrobial resistance in bacteria: mechanisms, evolution, and persistence. J Mol Evol. 2020;88:26–40. doi: 10.1007/s00239-019-09914-3. [DOI] [PubMed] [Google Scholar]
- 2.Baker S.J., Payne D.J., Rappuoli R., De Gregorio E. Technologies to address antimicrobial resistance. Proc Natl Acad Sci U S A. 2018;115:12887–12895. doi: 10.1073/pnas.1717160115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mousavi S.M., Babakhani S., Moradi L., Karami S., Shahbandeh M., Mirshekar M., et al. Bacteriophage as a novel therapeutic weapon for killing colistin-resistant multi-drug-resistant and extensively drug-resistant Gram-negative bacteria. Curr Microbiol. 2021;78:4023–4036. doi: 10.1007/s00284-021-02662-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knecht L.E., Veljkovic M., Fieseler L. Diversity and function of phage encoded depolymerases. Front Microbiol. 2020;10:2949. doi: 10.3389/fmicb.2019.02949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koskella B., Brockhurst M.A. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol Rev. 2014;38:916–931. doi: 10.1111/1574-6976.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abarquero D., Renes E., Fresno J.M., Tornadijo M.E. Study of exopolysaccharides from lactic acid bacteria and their industrial applications: a review. Int J Food Sci Technol. 2021;57:16–26. [Google Scholar]
- 7.Hoiby N. A short history of microbial biofilms and biofilm infections. APMIS. 2017;125:272–275. doi: 10.1111/apm.12686. [DOI] [PubMed] [Google Scholar]
- 8.Flemming H.C., Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 9.Labrie S.J., Samson J.E., Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 10.Oechslin F. Resistance development to bacteriophages occurring during bacteriophage therapy. Viruses. 2018;10:351. doi: 10.3390/v10070351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X., Liu M., Zhang P., Xu M., Yuan W., Bian L., et al. Phage-derived depolymerase as an antibiotic adjuvant against multidrug-resistant Acinetobacter baumannii. Front Microbiol. 2022;13 doi: 10.3389/fmicb.2022.845500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Topka-Bielecka G., Dydecka A., Necel A., Bloch S., Nejman-Falenczyk B., Wegrzyn G., et al. Bacteriophage-derived depolymerases against bacterial biofilm. Antibiotics. 2021;10:175. doi: 10.3390/antibiotics10020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liston S.D., McMahon S.A., Le Bas A., Suits M.D.L., Naismith J.H., Whitfield C. Periplasmic depolymerase provides insight into ABC transporter-dependent secretion of bacterial capsular polysaccharides. Proc Natl Acad Sci U S A. 2018;115:E4870–E4879. doi: 10.1073/pnas.1801336115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Twort F.W. An investigation on the nature of ultra-microscopic viruses. Lancet. 1915;186:1241–1243. doi: 10.1017/s0022172400043606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.d'Herelle F. An invisible microbe that is antagonistic to the dysentery bacillus. C R Acad Sci. 1917;165:373–375. [Google Scholar]
- 16.Asheshov I.N. Experimental studies on the bacteriophage. J Infect Dis. 1924;34:536–548. [Google Scholar]
- 17.Sertic V. Origine de la lysine d'une race du bacteriophage. Compt Rend Soc Biol. 1929;100:477–479. [Google Scholar]
- 18.Sertic V., Boulgaxov N. Sur la sensibilite d'une souche d'Escherichia coli au bactdriophage, en relation avec les caracteres antig6niques. Compt Rend Soc Biol. 1937;126:734–736. [Google Scholar]
- 19.Adams M.H., Park B.H. An enzyme produced by a phage-host cell system: II. the properties of the polysaccharide depolymerase. Virology. 1956;2:719–736. doi: 10.1016/0042-6822(56)90054-x. [DOI] [PubMed] [Google Scholar]
- 20.Stirm S., Bessler W., Fehmel F., Freund-Mölbert E. Bacteriophage particles with endo-glycosidase activity. J Virol. 1971;8:343–346. doi: 10.1128/jvi.8.3.343-346.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steinbacher S., Baxa U., Miller S., Weintraub A., Seckler R., Huber R. Crystal structure of phage P22 tailspike protein complexed with Salmonella sp. O-antigen receptors. Proc Natl Acad Sci U S A. 1996;93:10584–10588. doi: 10.1073/pnas.93.20.10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Latka A., Leiman P.G., Drulis-Kawa Z., Briers Y. Modeling the architecture of depolymerase-containing receptor binding proteins in Klebsiella phages. Front Microbiol. 2019;10:2649. doi: 10.3389/fmicb.2019.02649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clarke B.R., Esumeh F., Roberts I.S. Cloning, expression, and purification of the K5 capsular polysaccharide lyase (KflA) from coliphage K5A: evidence for two distinct K5 lyase enzymes. J Bacteriol. 2000;182:3761–3766. doi: 10.1128/jb.182.13.3761-3766.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leiman P.G., Molineux I.J. Evolution of a new enzyme activity from the same motif fold. Mol Microbiol. 2008;69:287–290. doi: 10.1111/j.1365-2958.2008.06241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barbirz S., Muller J.J., Uetrecht C., Clark A.J., Heinemann U., Seckler R. Crystal structure of Escherichia coli phage HK620 tailspike: podoviral tailspike endoglycosidase modules are evolutionarily related. Mol Microbiol. 2008;69:303–316. doi: 10.1111/j.1365-2958.2008.06311.x. [DOI] [PubMed] [Google Scholar]
- 26.Hughes K., Sutherland I., Clark J., Jones M. Bacteriophage and associated polysaccharide depolymerases—novel tools for study of bacterial biofilms. J Appl Microbiol. 1998;85:583–590. doi: 10.1046/j.1365-2672.1998.853541.x. [DOI] [PubMed] [Google Scholar]
- 27.Mushtaq N., Redpath M.B., Luzio J.P., Taylor P.W. Prevention and cure of systemic Escherichia coli K1 infection by modification of the bacterial phenotype. Antimicrob Agents Chemother. 2004;48:1503–1508. doi: 10.1128/AAC.48.5.1503-1508.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waseh S., Hanifi-Moghaddam P., Coleman R., Masotti M., Ryan S., Foss M., et al. Orally administered P22 phage tailspike protein reduces salmonella colonization in chickens: prospects of a novel therapy against bacterial infections. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bansal S., Harjai K., Chhibber S. Depolymerase improves gentamicin efficacy during Klebsiella pneumoniae induced murine infection. BMC Infect Dis. 2014;14:456. doi: 10.1186/1471-2334-14-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shahed-Al-Mahmud M., Roy R., Sugiokto F.G., Islam M.N., Lin M.D., Lin L.C., et al. Phage phiAB6-borne depolymerase combats Acinetobacter baumannii biofilm formation and infection. Antibiotics. 2021;10:279. doi: 10.3390/antibiotics10030279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan Y.J., Lin T.L., Lin Y.T., Su P.A., Chen C.T., Hsieh P.F., et al. Identification of capsular types in carbapenem-resistant Klebsiella pneumoniae strains by wzc sequencing and implications for capsule depolymerase treatment. Antimicrob Agents Chemother. 2015;59:1038–1047. doi: 10.1128/AAC.03560-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mostowy R.J., Holt K.E. Diversity-generating machines: genetics of bacterial sugar-coating. Trends Microbiol. 2018;26:1008–1021. doi: 10.1016/j.tim.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pires D.P., Oliveira H., Melo L.D., Sillankorva S., Azeredo J. Bacteriophage-encoded depolymerases: their diversity and biotechnological applications. Appl Microbiol Biotechnol. 2016;100:2141–2151. doi: 10.1007/s00253-015-7247-0. [DOI] [PubMed] [Google Scholar]
- 34.Drulis-Kawa Z., Majkowska-Skrobek G., Maciejewska B. Bacteriophages and phage-derived proteins—application approaches. Curr Med Chem. 2015;22:1757–1773. doi: 10.2174/0929867322666150209152851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nobrega F.L., Vlot M., de Jonge P.A., Dreesens L.L., Beaumont H.J.E., Lavigne R., et al. Targeting mechanisms of tailed bacteriophages. Nat Rev Microbiol. 2018;16:760–773. doi: 10.1038/s41579-018-0070-8. [DOI] [PubMed] [Google Scholar]
- 36.Squeglia F., Maciejewska B., Latka A., Ruggiero A., Briers Y., Drulis-Kawa Z., et al. Structural and functional studies of a Klebsiella phage capsule depolymerase tailspike: mechanistic insights into capsular degradation. Structure. 2020;28:613–624.e4. doi: 10.1016/j.str.2020.04.015. [DOI] [PubMed] [Google Scholar]
- 37.Latka A., Maciejewska B., Majkowska-Skrobek G., Briers Y., Drulis-Kawa Z. Bacteriophage-encoded virion-associated enzymes to overcome the carbohydrate barriers during the infection process. Appl Microbiol Biotechnol. 2017;101:3103–3119. doi: 10.1007/s00253-017-8224-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Latka A., Lemire S., Grimon D., Dams D., Maciejewska B., Lu T., et al. Engineering the modular receptor-binding proteins of Klebsiella phages switches their capsule serotype specificity. mBio. 2021;12 doi: 10.1128/mBio.00455-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan J., Mao J., Xie J. Bacteriophage polysaccharide depolymerases and biomedical applications. BioDrugs. 2014;28:265–274. doi: 10.1007/s40259-013-0081-y. [DOI] [PubMed] [Google Scholar]
- 40.Patro L.P.P., Sudhakar K.U., Rathinavelan T.K.-P.A.M. A unified platform to distinguish Klebsiella species K- and O-antigen types, model antigen structures and identify hypervirulent strains. Sci Rep. 2020;10 doi: 10.1038/s41598-020-73360-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oliveira H., Drulis-Kawa Z., Azeredo J. Exploiting phage-derived carbohydrate depolymerases for combating infectious diseases. Trends Microbiol. 2022;30:707–709. doi: 10.1016/j.tim.2022.05.002. [DOI] [PubMed] [Google Scholar]
- 42.Reuter M., Kruger D.H. Approaches to optimize therapeutic bacteriophage and bacteriophage-derived products to combat bacterial infections. Virus Gene. 2020;56:136–149. doi: 10.1007/s11262-020-01735-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Drulis-Kawa Z., Majkowska-Skrobek G., Maciejewska B., Delattre A.S., Lavigne R. Learning from bacteriophages—advantages and limitations of phage and phage-encoded protein applications. Curr Protein Pept Sci. 2012;13:699–722. doi: 10.2174/138920312804871193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li X.H., Lee J.H. Antibiofilm agents: a new perspective for antimicrobial strategy. J Microbiol. 2017;55:753–766. doi: 10.1007/s12275-017-7274-x. [DOI] [PubMed] [Google Scholar]
- 45.Taraszkiewicz A., Fila G., Grinholc M., Nakonieczna J. Innovative strategies to overcome biofilm resistance. BioMed Res Int. 2013;2013 doi: 10.1155/2013/150653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Limoli D.H., Jones C.J., Wozniak D.J. Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol Spectr. 2015;3:10–1128. doi: 10.1128/microbiolspec.MB-0011-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hernandez-Morales A.C., Lessor L.L., Wood T.L., Migl D., Mijalis E.M., Cahill J., et al. Genomic and biochemical characterization of Acinetobacter podophage Petty reveals a novel lysis mechanism and tail-associated depolymerase activity. J Virol. 2018;92 doi: 10.1128/JVI.01064-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gutiérrez D., Martínez B., Rodríguez A., García P. Genomic characterization of two Staphylococcus epidermidis bacteriophages with anti-biofilm potential. BMC Genom. 2012;13:228. doi: 10.1186/1471-2164-13-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gutiérrez D., Briers Y., Rodríguez-Rubio L., Martínez B., Rodríguez A., Lavigne R., et al. Role of the pre-neck appendage protein (Dpo7) from phage vB_SepiS-phiIPLA7 as an anti-biofilm agent in Staphylococcal species. Front Microbiol. 2015;6:1315. doi: 10.3389/fmicb.2015.01315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Olsen N.M.C., Thiran E., Hasler T., Vanzieleghem T., Belibasakis G.N., Mahillon J., et al. Synergistic removal of static and dynamic Staphylococcus aureus biofilms by combined treatment with a bacteriophage endolysin and a polysaccharide depolymerase. Viruses. 2018;10:438. doi: 10.3390/v10080438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chai Z., Wang J., Tao S., Mou H. Application of bacteriophage-borne enzyme combined with chlorine dioxide on controlling bacterial biofilm. LWT--Food Sci Technol. 2014;59:1159–1165. [Google Scholar]
- 52.Oliveira A., Ribeiro H.G., Silva A.C., Silva M.D., Sousa J.C., Rodrigues C.F., et al. Synergistic antimicrobial interaction between honey and phage against Escherichia coli biofilms. Front Microbiol. 2017;8:2407. doi: 10.3389/fmicb.2017.02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tait K., Skillman L.C., Sutherland I.W. The efficacy of bacteriophage as a method of biofilm eradication. Biofouling. 2002;18:305–311. [Google Scholar]
- 54.Lu T.K., Collins J.J. Dispersing biofilms with engineered enzymatic bacteriophage. Proc Natl Acad Sci U S A. 2007;104:11197–11202. doi: 10.1073/pnas.0704624104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Projan S. Phage-inspired antibiotics?. Nat Biotechnol. 2004;22:167–168. doi: 10.1038/nbt0204-167. [DOI] [PubMed] [Google Scholar]
- 56.Pires D.P., Melo L., Vilas Boas D., Sillankorva S., Azeredo J. Phage therapy as an alternative or complementary strategy to prevent and control biofilm-related infections. Curr Opin Microbiol. 2017;39:48–56. doi: 10.1016/j.mib.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 57.Hughes K.A., Sutherland I.W., Jones M.V. Biofilm susceptibility to bacteriophage attack: the role of phage-borne polysaccharide depolymerase. Microbiology (Read) 1998;144:3039–3047. doi: 10.1099/00221287-144-11-3039. [DOI] [PubMed] [Google Scholar]
- 58.Hanlon G.W., Denyer S.P., Olliff C.J., Ibrahim L.J. Reduction in exopolysaccharide viscosity as an aid to bacteriophage penetration through Pseudomonas aeruginosa biofilms. Appl Environ Microbiol. 2001;67:2746–2753. doi: 10.1128/AEM.67.6.2746-2753.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bansal S., Harjai K., Chhibber S. Aeromonas punctata derived depolymerase improves susceptibility of Klebsiella pneumoniae biofilm to gentamicin. BMC Microbiol. 2015;15:119. doi: 10.1186/s12866-015-0455-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu Y., Wang R., Xu M., Liu Y., Zhu X., Qiu J., et al. A novel polysaccharide depolymerase encoded by the phage SH-KP152226 confers specific activity against multidrug-resistant Klebsiella pneumoniae via biofilm degradation. Front Microbiol. 2019;10:2768. doi: 10.3389/fmicb.2019.02768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Majkowska-Skrobek G., Latka A., Berisio R., Squeglia F., Maciejewska B., Briers Y., et al. Phage-borne depolymerases decrease Klebsiella pneumoniae resistance to innate defense mechanisms. Front Microbiol. 2018;9:2517. doi: 10.3389/fmicb.2018.02517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oliveira H., Pinto G., Mendes B., Dias O., Hendrix H., Akturk E., et al. A tailspike with exopolysaccharide depolymerase activity from a new Providencia stuartii phage makes multidrug-resistant bacteria susceptible to serum-mediated killing. Appl Environ Microbiol. 2020;86 doi: 10.1128/AEM.00073-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abdelkader K., Gutierrez D., Latka A., Boeckaerts D., Drulis-Kawa Z., Criel B., et al. The specific capsule depolymerase of phage PMK34 sensitizes Acinetobacter baumannii to serum killing. Antibiotics. 2022;11:677. doi: 10.3390/antibiotics11050677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oliveira H., Mendes A., Fraga A.G., Ferreira A., Pimenta A.I., Mil-Homens D., et al. K2 Capsule depolymerase is highly stable, is refractory to resistance, and protects larvae and mice from Acinetobacter baumannii sepsis. Appl Environ Microbiol. 2019;85 doi: 10.1128/AEM.00934-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marimani M. In: Combination therapy against multidrug resistance. Wani M.Y., Ahmad A., editors. Academic Press; Cambridge MA: 2020. Combination therapy against multidrug resistance; pp. 39–64. [Google Scholar]
- 66.Smug B.J., Majkowska-Skrobek G., Drulis-Kawa Z. PhREEPred: phage resistance emergence prediction web tool to foresee encapsulated bacterial escape from phage cocktail treatment. J Mol Biol. 2022;434 doi: 10.1016/j.jmb.2022.167670. [DOI] [PubMed] [Google Scholar]
- 67.Majkowska-Skrobek G., Markwitz P., Sosnowska E., Lood C., Lavigne R., Drulis-Kawa Z. The evolutionary trade-offs in phage-resistant Klebsiella pneumoniae entail cross-phage sensitization and loss of multidrug resistance. Environ Microbiol. 2021;23:7723–7740. doi: 10.1111/1462-2920.15476. [DOI] [PubMed] [Google Scholar]
- 68.Danis-Wlodarczyk K.M., Wozniak D.J., Abedon S.T. Treating bacterial infections with bacteriophage-based enzybiotics: in vitro, in vivo and clinical application. Antibiotics. 2021;10:1497. doi: 10.3390/antibiotics10121497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin T.L., Hsieh P.F., Huang Y.T., Lee W.C., Tsai Y.T., Su P.A., et al. Isolation of a bacteriophage and its depolymerase specific for K1 capsule of Klebsiella pneumoniae: implication in typing and treatment. J Infect Dis. 2014;210:1734–1744. doi: 10.1093/infdis/jiu332. [DOI] [PubMed] [Google Scholar]
- 70.Volozhantsev N.V., Borzilov A.I., Shpirt A.M., Krasilnikova V.M., Verevkin V.V., Denisenko E.A., et al. Comparison of the therapeutic potential of bacteriophage KpV74 and phage-derived depolymerase (beta-glucosidase) against Klebsiella pneumoniae capsular type K2. Virus Res. 2022;322 doi: 10.1016/j.virusres.2022.198951. [DOI] [PubMed] [Google Scholar]
- 71.Li M., Wang H., Chen L., Guo G., Li P., Ma J., et al. Identification of a phage-derived depolymerase specific for KL47 capsule of Klebsiella pneumoniae and its therapeutic potential in mice. Virol Sin. 2022;37:538–546. doi: 10.1016/j.virs.2022.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang C., Li P., Niu W., Yuan X., Liu H., Huang Y., et al. Protective and therapeutic application of the depolymerase derived from a novel KN1 genotype of Klebsiella pneumoniae bacteriophage in mice. Res Microbiol. 2019;170:156–164. doi: 10.1016/j.resmic.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 73.Majkowska-Skrobek G., Latka A., Berisio R., Maciejewska B., Squeglia F., Romano M., et al. Capsule-targeting depolymerase, derived from Klebsiella KP36 phage, as a tool for the development of anti-virulent strategy. Viruses. 2016;8:324. doi: 10.3390/v8120324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Volozhantsev N.V., Shpirt A.M., Borzilov A.I., Komisarova E.V., Krasilnikova V.M., Shashkov A.S., et al. Characterization and therapeutic potential of bacteriophage-encoded polysaccharide depolymerases with beta galactosidase activity against Klebsiella pneumoniae K57 capsular type. Antibiotics. 2020;9:732. doi: 10.3390/antibiotics9110732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gorodnichev R.B., Volozhantsev N.V., Krasilnikova V.M., Bodoev I.N., Kornienko M.A., Kuptsov N.S., et al. Novel Klebsiella pneumoniae K23-specific bacteriophages from different families: similarity of depolymerases and their therapeutic potential. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.669618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu Y., Leung S.S.Y., Guo Y., Zhao L., Jiang N., Mi L., et al. The capsule depolymerase Dpo48 rescues Galleria mellonella and mice from Acinetobacter baumannii systemic infections. Front Microbiol. 2019;10:545. doi: 10.3389/fmicb.2019.00545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang C., Li P., Zhu Y., Huang Y., Gao M., Yuan X., et al. Identification of a novel Acinetobacter baumannii phage-derived depolymerase and its therapeutic application in mice. Front Microbiol. 2020;11:1407. doi: 10.3389/fmicb.2020.01407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mushtaq N., Redpath M.B., Luzio J.P., Taylor P.W. Treatment of experimental Escherichia coli infection with recombinant bacteriophage-derived capsule depolymerase. J Antimicrob Chemother. 2005;56:160–165. doi: 10.1093/jac/dki177. [DOI] [PubMed] [Google Scholar]
- 79.Lin H., Paff M.L., Molineux I.J., Bull J.J. Therapeutic application of phage capsule depolymerases against K1, K5, and K30 capsulated E. coli in mice. Front Microbiol. 2017;8:2257. doi: 10.3389/fmicb.2017.02257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lin H., Paff M.L., Molineux I.J., Bull J.J. Antibiotic therapy using phage depolymerases: robustness across a range of conditions. Viruses. 2018;10:622. doi: 10.3390/v10110622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen Y., Li X., Wang S., Guan L., Li X., Hu D., et al. A novel tail-associated O91-specific polysaccharide depolymerase from a podophage reveals lytic efficacy of Shiga toxin-producing Escherichia coli. Appl Environ Microbiol. 2020;86 doi: 10.1128/AEM.00145-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Olszak T., Shneider M.M., Latka A., Maciejewska B., Browning C., Sycheva L.V., et al. The O-specific polysaccharide lyase from the phage LKA1 tailspike reduces Pseudomonas virulence. Sci Rep. 2017;7 doi: 10.1038/s41598-017-16411-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sumrall E.T., Hofstee M.I., Arens D., Rohrig C., Baertl S., Gehweiler D., et al. An enzybiotic regimen for the treatment of methicillin-resistant Staphylococcus aureus orthopaedic device-related infection. Antibiotics. 2021;10:1186. doi: 10.3390/antibiotics10101186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.M Campos J.C., Antunes L.C., Ferreira R.B. Global priority pathogens: virulence, antimicrobial resistance and prospective treatment options. Future Microbiol. 2020;15:649–677. doi: 10.2217/fmb-2019-0333. [DOI] [PubMed] [Google Scholar]
- 85.Ma Y.X., Wang C.Y., Li Y.Y., Li J., Wan Q.Q., Chen J.H., et al. Considerations and caveats in combating ESKAPE pathogens against nosocomial infections. Adv Sci. 2019;7 doi: 10.1002/advs.201901872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ryan E.M., Gorman S.P., Donnelly R.F., Gilmore B.F. Recent advances in bacteriophage therapy: how delivery routes, formulation, concentration and timing influence the success of phage therapy. J Pharm Pharmacol. 2011;63:1253–1264. doi: 10.1111/j.2042-7158.2011.01324.x. [DOI] [PubMed] [Google Scholar]
- 87.Liu Y., Mi Z., Mi L., Huang Y., Li P., Liu H., et al. Identification and characterization of capsule depolymerase Dpo48 from Acinetobacter baumannii phage IME200. PeerJ. 2019;7 doi: 10.7717/peerj.6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kassa T., Chhibber S. Thermal treatment of the bacteriophage lysate of Klebsiella pneumoniae B5055 as a step for the purification of capsular depolymerase enzyme. J Virol Methods. 2012;179:135–141. doi: 10.1016/j.jviromet.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 89.Montanari E., Di Meo C., Sennato S., Francioso A., Marinelli A.L., Ranzo F., et al. Hyaluronan-cholesterol nanohydrogels: characterisation and effectiveness in carrying alginate lyase. N Biotech. 2017;37:80–89. doi: 10.1016/j.nbt.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 90.Dunstan R.A., Bamert R.S., Belousoff M.J., Short F.L., Barlow C.K., Pickard D.J., et al. Mechanistic insights into the capsule-targeting depolymerase from a Klebsiella pneumoniae bacteriophage. Microbiol Spectr. 2021;9 doi: 10.1128/spectrum.01023-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Libertucci J., Young V.B. The role of the microbiota in infectious diseases. Nat Microbiol. 2019;4:35–45. doi: 10.1038/s41564-018-0278-4. [DOI] [PubMed] [Google Scholar]
- 92.Gutierrez D., Fernandez L., Rodriguez A., Garcia P. Are phage lytic proteins the secret weapon to kill Staphylococcus aureus?. mBio. 2018;9 doi: 10.1128/mBio.01923-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Structural Genomics Consortium, China Structural Genomics Consortium, Northeast Structural Genomics Consortium, Graslund S, Nordlund P, Weigelt J, et al. Protein production and purification. Nat Methods. 2008;5:135–146. doi: 10.1038/nmeth.f.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Klimyuk V., Pogue G., Herz S., Butler J., Haydon H. Production of recombinant antigens and antibodies in Nicotiana benthamiana using 'magnifection' technology: GMP-compliant facilities for small- and large-scale manufacturing. Curr Top Microbiol Immunol. 2014;375:127–154. doi: 10.1007/82_2012_212. [DOI] [PubMed] [Google Scholar]
- 95.Frokjaer S., Otzen D.E. Protein drug stability: a formulation challenge. Nat Rev Drug Discov. 2005;4:298–306. doi: 10.1038/nrd1695. [DOI] [PubMed] [Google Scholar]
- 96.Mitragotri S., Burke P.A., Langer R. Overcoming the challenges in administering biopharmaceuticals: formulation and delivery strategies. Nat Rev Drug Discov. 2014;13:655–672. doi: 10.1038/nrd4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dijkshoorn L., Towner K. In: New approaches for the generation and analysis of microbial typing data. Lenie Dijkshoorn K.J., Towner M.S., editors. Elsevier Science B.V.; Amsterdam: 2001. An introduction to the generation and analysis of microbial typing data; pp. 1–30. [Google Scholar]
- 98.Federici S., Nobs S.P., Elinav E. Phages and their potential to modulate the microbiome and immunity. Cell Mol Immunol. 2021;18:889–904. doi: 10.1038/s41423-020-00532-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cornforth D.M., Dees J.L., Ibberson C.B., Huse H.K., Mathiesen I.H., Kirketerp-Moller K., et al. Pseudomonas aeruginosa transcriptome during human infection. Proc Natl Acad Sci U S A. 2018;115:E5125–E5134. doi: 10.1073/pnas.1717525115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bjarnsholt T., Whiteley M., Rumbaugh K.P., Stewart P.S., Jensen P.Ø., Frimodt-Møller N. The importance of understanding the infectious microenvironment. Lancet Infect Dis. 2022;22:e88–e92. doi: 10.1016/S1473-3099(21)00122-5. [DOI] [PMC free article] [PubMed] [Google Scholar]