Abstract
The trillions of microorganisms in the human intestine are important regulators of health, and disruptions in the gut microbial communities can cause disease. The gut, liver, and immune system have a symbiotic relationship with these microorganisms. Environmental factors, such as high-fat diets and alcohol consumption, can disrupt and alter microbial communities. This dysbiosis can lead to dysfunction of the intestinal barrier, translocation of microbial components to liver, and development or progression of liver disease. Changes in metabolites produced by gut microorganisms can also contribute to liver disease. In this Review, we discuss the importance of the gut microbiota in maintenance of health and the alterations in microbial mediators that contribute to liver disease. We present strategies for modulation of the intestinal microbiota and/or their metabolites as potential treatments for liver disease.
Table of contents
In this Review, Hsu and Schnabl examine the role of the gut microbiota in liver and intestine function, how gut microorganisms contribute to liver diseases, and explore different microbiota-based strategies for the treatment of liver disease.
Introduction
The gut and liver interact via several pathways. One way is their anatomical association — venous blood from the small and large intestine drains into the portal vein, which provides 75% of the liver’s blood supply. These organs also interact via the microbiota — the liver is the first organ to encounter enterally absorbed nutrients and microbial metabolites. The liver, in turn, regulates the gut by secreting bile containing bile acids, immunoglobulin A, and antimicrobial molecules directly into the small intestine, which completes the cycle by modulating the microbiota (FIG. 1).
Fig. 1. The gut–liver axis.
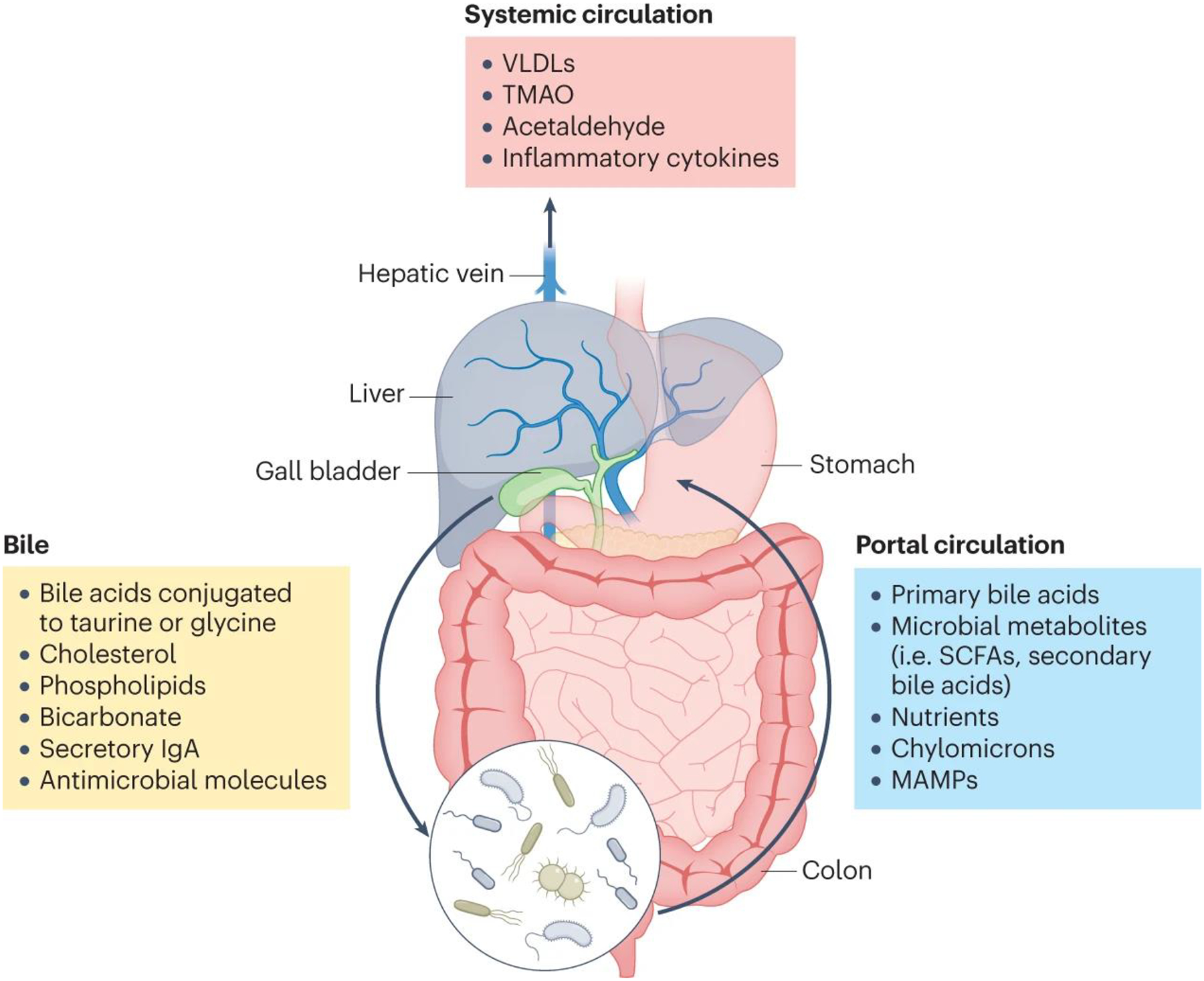
The liver produces bile, which is composed of cholesterol, phospholipids, proteins, bicarbonate, and bile acids that have been conjugated by the liver to taurine or glycine (yellow box). Bile is directed into the proximal small intestine via the bile ducts, where it aids in lipid digestion and absorption. In the colon, gut bacteria convert primary bile acids into secondary bile acids via dihydroxylation, and eventually both primary and secondary bile acids are reabsorbed back into the liver via the portal circulation (blue box). The portal circulation also carries absorbed nutrients, lipids, microbial products such as lipopolisaccharide, and microbial metabolites such as short-chain fatty acids (SCFAs) back to the liver. Toxic metabolites such as acetaldehyde and inflammatory cytokines produced by the liver then continue into the systemic circulation (red box). IgA, immunoglobulin A; SCFAs, small-chain fatty acids; MAMPs, microbe-associated molecular patterns; VLDLs, very low-density lipoproteins; TMAO, trimethylamine N-oxide.
An intact intestinal barrier is important for absorption of nutrients, but also keeps trillions of microorganisms contained in the lumen, to prevent them from spreading systemically. Microbial products and metabolites help maintain the gut barrier and liver health1. The disruption of intestinal microbial communities (dysbiosis), due to dietary or environmental changes such as high caloric intake or alcohol consumption, is a risk factor for liver disease. Although mechanisms of pathogenesis differ among hepatic diseases, intestinal dysbiosis has been associated with metabolic liver diseases such as alcohol-associated liver disease and non-alcoholic fatty liver disease (NAFLD). Dysbiosis of the microbiota has been shown to modulate liver diseases — liver disease development can be transmitted with fecal microbiota in preclinical models2,3, and modest improvement in metabolic and liver pathologies are seen after limited follow-up in patients following fecal microbiota transplantation from healthy donors4. These observations indicate the importance of the state of the intestinal microbiota in liver function. In this Review, we explore how the gut, liver, and intestinal microbiota interact under healthy conditions, how a dysbiotic microbiota affects functions of the intestine and the liver, and potential interventional approaches to restore the intestinal ecosystem.
The gut microbiota and the gut–liver axis in health
The human gastrointestinal tract harbors over 100 trillion microorganisms including bacteria, fungi, viruses, and archaea that make up the gut microbiota5–7. Though the gut microbiota is often referred to as a uniform entity, there is considerable compositional variation between the different sections of the gastrointestinal tract from the mouth to the colon due to environmental differences such as pH and oxygen availability. For example, the proportion of obligate anaerobes increases from the duodenum (proximal small intestine) to the ileum (distal small intestine) and colon due to decreased oxygen availability, and the microbial density is highest in the colon due to the longer transit time, which allows for more proliferation8. Most of our understanding of the human gut microbiome is derived from the fecal microbiome, which cannot be extrapolated to the entire gastrointestinal tract. The phyla Bacteroidetes and Firmicutes predominate in the gastrointestinal tract, representing 90% of the gut microbiota9,10. At a genus level, the most abundant genera are Bacteroides, Alistipes, Parabacteroides, and Prevotella within the Bacteroidetes phyla, and Faecalibacterium, Lachnospiraceae, and Roseburia within the Firmicutes phylum. The Actinobacteria phylum is proportionally less abundant and mainly represented by the Bifidobacterium genus.
The gut microbiome is highly dynamic, with significant interindividual as well as intraindividual variation across time due to influences not limited to diet, activity, medications, and circadian rhythm. In a deeply symbiotic relationship, the gut microbiota carries out critical functions for their host, such as assisting with efficient nutrient extraction, modulating hormonal pathways via microbial-derived metabolites, metabolizing drugs and xenobiotics, and priming host immunity11,12. For example, intestinal microorganisms degrade fibers into short-chain fatty acids (SCFA) such as acetate, butyrate, and propionate, which provide an important energy source for enterocytes in the colon and stimulate the antimicrobial activity of macrophages13 and regulatory T cells (TReg cells)14. Meanwhile, the gut maintains a nutrient-rich, anaerobic luminal environment by consuming oxygen in the mitochondria of colonocytes to maintain an oxygen partial pressure of less than 7.6 mmHg15, and by producing inducible nitric oxide and nicotinamide adenine dinucleotide phosphate (NADPH) oxidase 1 in the ileum16.
The gut is also responsible for preventing the translocation of microorganisms and microbial antigens to the extraintestinal space, and this is regulated by several important barrier systems (FIG. 2). The mucus layer separates luminal bacteria from the underlying intestinal epithelial cells (IEC). It consists of mucins rich in glycoproteins and its composition is dictated by the gut microbiota17. The mucus layer is thickest in the colon and is divided into an inner and outer mucus layer18. Whereas the outer mucus layer contains bacteria, the inner mucus layer is largely devoid of bacteria. This is facilitated by secretion of antimicrobial molecules by intestinal epithelial cells and Paneth cells19. Plasma cells in the lamina propria produce immunoglobulin A (IgA), which is secreted as secretory IgA into the intestinal lumen20. IgA is essential for protection of the mucosal surface by binding and neutralizing harmful pathobionts and pathogens20. Besides secreting important antimicrobial defense molecules into the intestinal lumen, epithelial cells are also an important physical barrier. Epithelial cells are tightly bound together by adherens and tight junctions, which restrict the passage of viable bacteria or fungi, but allow paracellular passage of small molecules, such as SCFAs21.
Fig. 2. Barrier systems against translocation of microorganisms.
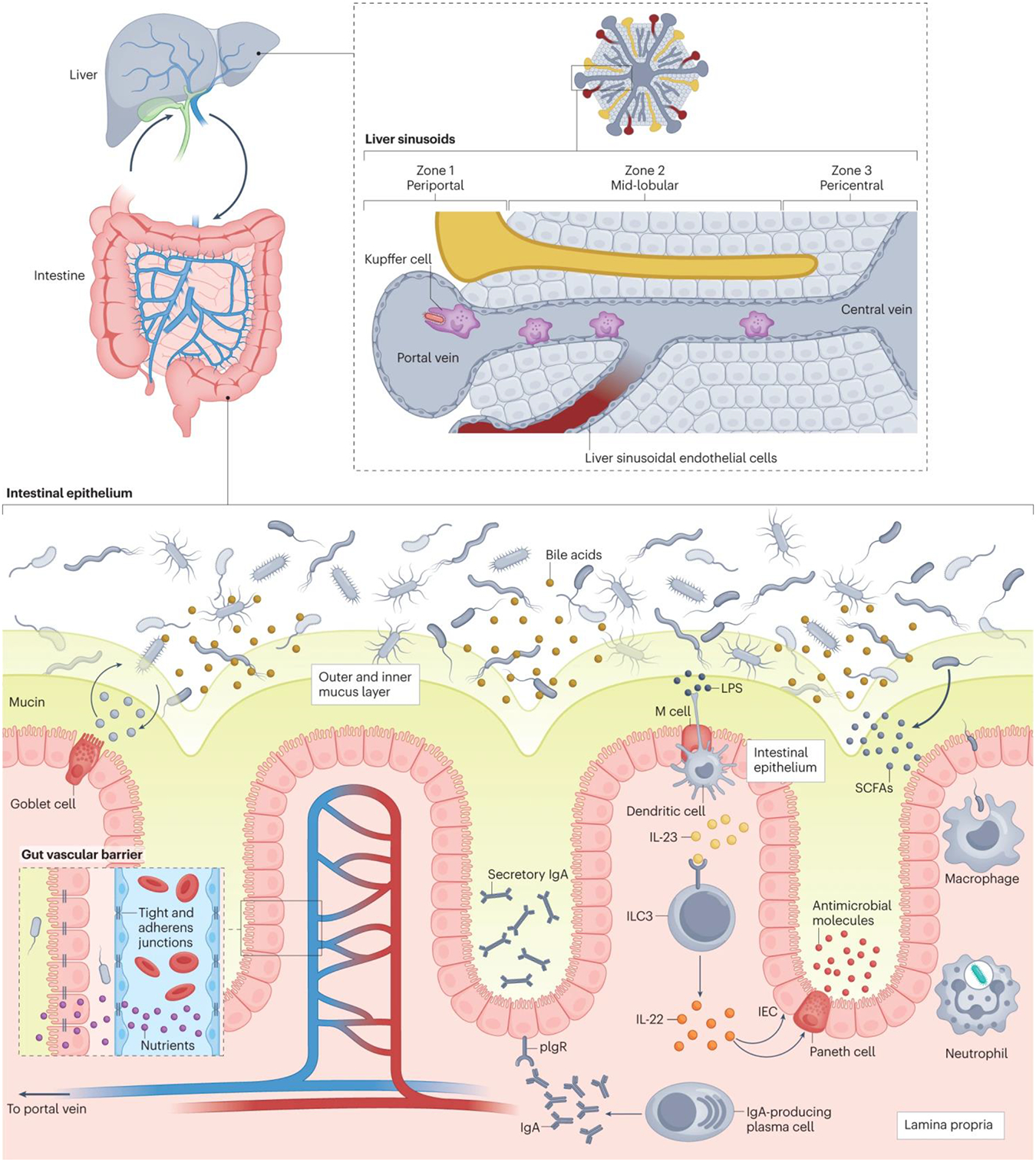
There are multiple layers of defense that prevent translocation of gut bacteria into the systemic circulation. The intestinal epithelium is the first interface, as it separates the bacteria in the gut lumen from the lamina propria below. A mucus layer (green), composed of mucins produced by goblet cells, separates the bacteria from the underlying intestinal epithelial cells. In the lamina propria (beige), whole bacteria that cross the intestinal epithelium are phagocytosed by macrophages and neutrophils. Plasma cells produce immunoglobulin A (IgA), which binds to pIgR, a receptor on enterocytes, for transport across the epithelium and secretion into the mucus layer, where this secretory IgA binds and neutralizes harmful pathobionts and pathogens. M cells in the intestinal epithelium relay microbial antigens to dendritic cells, which activate Type 3 innate lymphoid cells (ILC3) (via interleukin 23 (IL-23)) to produce interleukin 22 (IL-22). IL-22 ultimately signals Paneth cells and intestinal epithelial cells (IEC) to secrete antimicrobial molecules into the mucus layer. Short-chain fatty acids (SCFAs), produced by bacterial microbial degradation of fiber, are an energy source for enterocytes and enhance the antimicrobial activity of macrophages. Another physical barrier against bacterial translocation is the gut vascular barrier (inset box). Tight junctions and adherens junctions on endothelial cells regulate the permeability of the gut vascular barrier, allowing passage of nutrients and small molecules into the enteral circulation but restricting the passage of viable bacteria and microbial antigens. Finally, gut microorganisms that do translocate into the enteral circulation will face the liver barrier via the portal vein. In the liver, blood from the portal vein flows into the central vein via sinusoids (grey), which are lined with liver sinusoidal endothelial cells that release chemokines in response to microorganisms and microbial products. This creates a chemokine gradient that recruits Kupffer cells to the periportal zone, where they can neutralize translocated microorganims. LPS, lipopolysaccharide.
The next barrier comprises the immunologic defense systems in the intraepithelial compartment, lamina propria, and mesenteric lymph nodes. Under healthy conditions, the epithelial barrier allows microbial antigens and stimuli to pass into the lamina propria, where many different immune cells can respond. These interactions are critical for maintaining gut homeostasis and immune responses. For example, lamina propria M-cells and dendritic cells sample luminal and lamina propria microbial antigens and present them to naïve T cells in mesenteric lymph nodes to trigger an adaptive immune response and induce CD4+ Treg cells, which are crucial for developing tolerance and preventing chronic inflammation. Primed T-helper 17 (Th17) cells control the mucosal interface and prevent expansion of luminal fungi and other microorganisms22. Different T cell subsets and innate lymphoid cells secrete cytokines rapidly upon a microbial trigger. Type 3 innate lymphoid cells (ILC3) are activated by microbial metabolites including indoles as microbial-derived tryptophan metabolites, to produce interleukin 22 (IL-22). IL-22 signals via its receptor on intestinal epithelial cells to induce expression of antimicrobial genes and increase cell proliferation. Finally, neutrophils and macrophages in the lamina propria phagocyte viable bacteria that breach the mucus and epithelial cell layer.
Most venous blood from the small and large intestine drains into the portal vein. The gut vascular barrier, comprised of tight and adherens junction proteins on endothelial cells, prevents the translocation of viable bacteria from the lamina propria into the portal circulation. Portal vein blood contains enterally absorbed nutrients such as carbohydrates, lipids, and amino acids, which are taken up and metabolized or stored in hepatocytes, as well as microbial-derived metabolites. The liver is the first major organ to face these microbial-derived metabolites, which are important for the immune and metabolic functions of the liver. For example, commensal-derived D-lactic acid promotes killing of circulating pathogens by Kupffer cells, the resident macrophages in the liver23. Bacterial sphingolipids from the gut symbiont Bacteroides thetaiotaomicron transfer to the liver and reduce lipid accumulation by increasing beta-oxidation in mice24. Lipopolysaccharides (LPS) and other microbial products are important to support immune tolerance and xenobiotic metabolism locally. Complete absence of microbiota in germ-free mice renders the liver more susceptible to toxins1. Kupffer cells are an important protective barrier, as they phagocytose translocated microbial products and viable bacteria. Sinusoidal endothelial cells sense microorganisms and signal for Kupffer cells and natural killer T (NKT) cells to localize to the periportal area, as the entry point of invading microorganisms25. This immune zonation in the liver protects against translocating gut bacteria25. IgA-producing plasma cells are primed in the intestine, migrate to the liver, and secrete IgA26.
Gut–liver interactions are often considered a one-way path from the gut to the liver, but the liver also directly communicates with the gut via the biliary system. Hepatocytes synthesize and secrete bile, a solution rich in IgA, bicarbonate, antimicrobial molecules, and bile acids. Bile acids have multiple functions, such as facilitating the digestion and absorption of lipids and lipid-soluble vitamins and shape the gut microbial composition. Bile acids exert bacteriostatic effects both directly via their detergent properties and indirectly via activation of the bile acid receptor farnesoid X receptor (FXR) to stimulate the production of antimicrobial molecules in intestinal epithelial cells27 and maintain the gut vascular barrier28. In rats, interruption of bile flow to the gut by bile duct ligation or choledochovesical fistula (diversion of bile flow from the bile duct directly into the urinary bladder) resulted in an increase in the cecal Gram-negative aerobic population and increased bacterial translocation29. In healthy human volunteers, suppression of endogenous bile acid synthesis by obeticholic acid, a bile acid analog and FXR agonist, led to proliferation of small-intestinal Gram-positive bacteria30. Conversely, gut microorganisms can also shape bile acid supply and composition by deconjugating and dehydroxylating liver-derived primary bile acids into secondary bile acids, which are not as efficiently reabsorbed in the ileum31. In the ileum, bile acid activation of the FXR target gene fibroblast growth factor (FGF) 15 in mice and FGF19 in humans not only suppresses hepatic synthesis of bile acids in a self-regulating feedback loop32, but also mediates metabolic effects such as regulation of insulin sensitivity and stimulation of hepatic protein and glycogen synthesis33.
A third route of communication between the gut and the liver is the systemic circulation, through which metabolites, products, or inflammatory mediators derived from the liver reach the gut via the arterial blood supply. Taken together, the gut and liver have a multidirectional interaction that is vital for both organs.
The gut microbiota and the gut–liver axis in liver disease
Non-alcoholic fatty liver disease
Non-alcoholic fatty liver disease (NAFLD) encompasses a spectrum of liver pathologies, including fatty liver, which progresses in a minority of patients to nonalcoholic steatohepatitis (NASH) with fibrosis34 (BOX 1). Patients with NAFLD present with changes in the intestinal microbiota composition (FIG. 3), though taxonomic differences specific to NAFLD are occasionally conflicting and difficult to parse from those caused by or resulting from comorbid conditions of obesity, diet, and metabolic syndrome. For example, bacterial dysbiosis is different in patients with NAFLD who are lean versus obese. A study of patients with NAFLD and without obesity in Asia found decreased bacterial diversity, reduced proportions of Ruminococcaceae and increased proportions of Veillonellaceae, and these changes were associated with fibrosis severity35. Similarly, patients with NASH or liver fibrosis in stages F2–F4 and without obesity showed a distinct fecal fungal microbiome (mycobiome) signature36. The degree of liver fibrosis also affects the microbiome composition, as fecal proportions of Ruminococcus obeum CAG:39 and E. rectale were significantly lower in patients with advanced fibrosis than mild or moderate NAFLD37. Patients with NASH with advanced fibrosis also had a decrease in intestinal viral diversity and a reduction in the proportion of bacteriophages (phages) compared with other intestinal viruses38. Specific viral taxa, such as several Lactococcus phages, were less frequently present in patients with more severe disease, though whether this is caused by gut bacterial dysbiosis or a result of dysbiosis is unknown38.
BOX 1. Liver disease overview.
Liver diseases have different etiologies, including viral (hepatitis A, B, or C viruses), autoimmune (acute autoimmune hepatitis, primary biliary cirrhosis, primary sclerosing cholangitis), metabolic (nonalcoholic fatty liver disease), and toxic causes (alcohol, acetaminophen). Over time, all forms of chronic liver injury progress from fibrosis (hepatocyte death and scarring of the liver) to cirrhosis (advanced fibrosis and liver failure). Liver fibrosis and cirrhosis are clinically diagnosed by liver stiffness measurement, using elastography (transient elastography or magnetic resonance elastography) or characteristic imaging and serum laboratory analyses. Patients with cirrhosis are susceptible to decompensation events secondary to increased portal pressures such as the development of ascites, esophageal variceal bleeding, hepatic encephalopathy, or hepatocellular carcinoma, which are associated with large decreases in patients’ prognoses and quality of life.
Clinically, the gut microbiota is intimately linked with the pathogenesis of increased portal pressures and every aspect of cirrhotic decompensation. For example, one serious complication of ascites associated with significant morbidity and mortality is spontaneous bacterial peritonitis, caused by translocation of gut bacteria into the sterile peritoneal space. For patients with cirrhosis who develop variceal bleeding, prophylactic antibiotics are standard of care, and 30% to 48% of patients who did not receive antibiotics developed bacterial infections149,150. Patients with cirrhosis may also accumulate nitrogenous waste products, such as ammonia, in the systemic circulation, leading to hepatic encephalopathy. The mainstays of hepatic encephalopathy therapy are lactulose, a disaccharide that acts as a laxative and has been shown to enrich for beneficial gut microorganisms that reduce ammonia production and acidify the colonic contents, allowing for excretion of ammonia. Hepatic encephalopathy is also treated with rifaximin, a non-absorbable broad-spectrum antibiotic151.
Fig. 3. Gut microbiomes differ with etiology of liver disease.
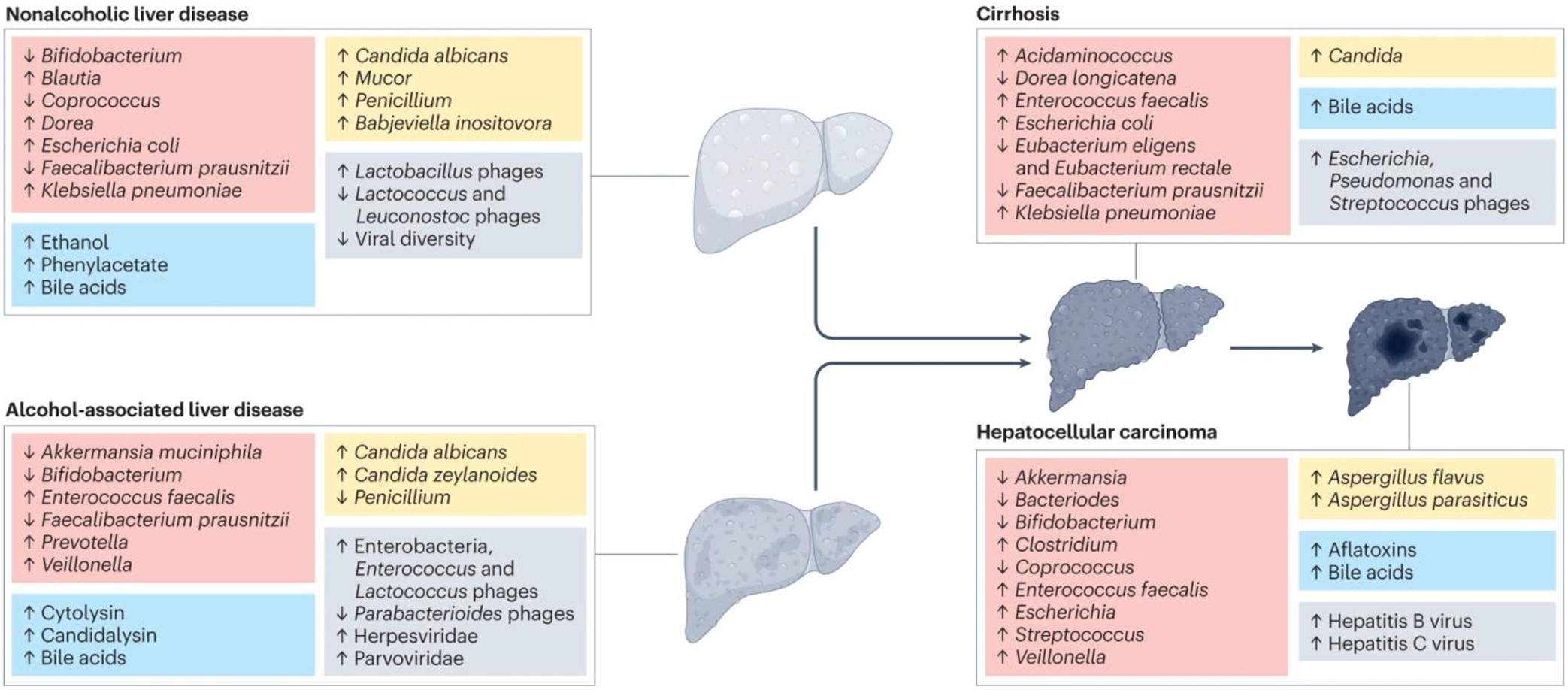
Non-alcoholic liver disease and alcohol-associated liver disease are associated with different changes in the composition of bacteria, fungi, viruses, and metabolites in the gut. Liver disease progression to cirrhosis and subsequent hepatocarcinogenesis (development of hepatocellular carcinoma) are associated with further changes in the gut microbiome and metabolites. The red boxes represent bacteria, blue boxes metabolites, yellow boxes fungi, and grey boxes viruses.
Gut dysbiosis induces intestinal inflammation, which contributes to intestinal barrier dysfunction and translocation of microbe-associated molecular patterns (MAMPs), such as LPS, to the liver and systemic circulation39. This pathophysiology was seen in a high fat diet mouse model of NASH which demonstrated dysbiosis and increased epithelial and gut vascular permeability, suggesting a possible mechanism of NASH pathogenesis28, and mouse models of diet-induced steatohepatitis demonstrate positive correlation between levels of serum LPS and liver injury40. However, only about one half of patients with NASH have increased intestinal permeability41.
Dysbiosis is also associated with functional metabolic consequences. Anaerobic bacteria ferment carbohydrates to produce ethanol, which is increased in patients with NASH under conditions that are incompletely understood. Individuals with NAFLD and NASH exhibited higher ethanol concentrations in their portal vein blood compared with individuals without hepatic steatosis42, and similar results were also seen in the peripheral blood of pediatric NAFLD patients43. In the most extreme case, the microbiota of patients with auto-brewery syndrome produce enough ethanol to make them drunk and incapacitated44. Hepatocytes metabolize ethanol into acetate and eventually triglycerides, resulting in steatosis. This is accompanied by oxidative stress resulting in death of hepatocytes and progression of liver disease44. The prevalence of patients who develop liver disease via this mechanism is not known, but it is likely that ethanol-producing bacteria contribute to liver disease in only a subgroup of NAFLD patients.
Other microbial metabolites such as phenylactetate and N,N,N-trimethyl-5-aminovaleric acid (TMAVA) have also been implicated in NAFLD pathogenesis. Serum phenylacetate was significantly associated with steatosis in women with obesity, and mice fed phenylacetate demonstrated increased hepatic triglyceride accumulation45. Plasma levels of TMAVA, metabolized from trimethyllysin by intestinal Enterococcus faecalis and Pseudomonas aeruginosa, were increased in patients with hepatic steatosis, and mice given TMAVA on a high fat diet demonstrated reduced carnitine synthesis and fatty acid oxidation and increased hepatic steatosis46. Conversely, gut microorganism-derived tryptophan metabolites such as indole-3-acetate and indole-3-propionate have been shown to suppress inflammation and attenuate hepatic steatosis in NAFLD or NASH47,48. Tryptophan metabolites and their effects on the immune system in NAFLD have been thoroughly reviewed49.
Patients with NAFLD or NASH also have higher serum levels of total bile acids compared with healthy controls, but a relative increase of secondary bile acids compared to primary bile acids along with a higher abundance of taurine and glycine-metabolizing gut bacteria50,51. These changes in the composition of the bile acid profile results in reduced FXR activity in enterocytes and a dysfunctional negative-feedback loop, via FGF15 or FGF19. Despite increased systemic levels of bile acid, de novo hepatic synthesis of bile acids is also increased in patients with NASH52. Interestingly, the role of FXR signaling in NAFLD pathogenesis is controversial. While one study demonstrated that high-fat diet-induced hepatic steatosis was reduced with intestinal FXR stimulation in mice53, another showed that it was reduced in intestine-specific Fxr-knockout mice54. Because of the bidirectional relationship of bile acids in the gut–liver axis, it remains to be shown what is a cause and what is a consequence. Nevertheless, bile acids control immune system functions through their interaction between host and bacteria. Several secondary bile acids induce Treg cells, either directly or by stimulating dendritic cells through FXR55–57. It is not known whether this mechanism is involved in modulating liver disease. In addition, bile acids are amphipathic detergents that can directly cause hepatocyte damage — especially in patients with increased bile acid levels due to cholestatic conditions (reduction in bile secretion and flow)58. Finally, while much of our understanding of bile acids in health and disease stems from rodent studies, it is important to consider some significant differences in bile acid composition between humans and mice59. For example, mice have more abundant hydrophilic bile acid species and 6-hydroxylated bile acids, which are quite rare in humans.
Studies of fecal microbiota transfer in preclinical models indicate that progression, and in some cases the onset, of hepatic steatosis and steatohepatitis is affected by the gut microbiota. Antibiotics and antifungals reduce diet-induced steatohepatitis in mice36,39. Furthermore, early-life exposure to microbiota from wild mice confers resistance to high-fat diet-induced obesity and hepatic steatosis60. It is not clear how much of the clinical NAFLD phenotype in patients is driven by dysbiosis and its functional consequences. Gut dysbiosis can be considered a risk factor in a subset of patients and might be an additional hit that causes progression of disease in patients who otherwise would not progress to more advanced disease. It will be important to better phenotype patients with NAFLD and identify which patients might benefit from a microbiome-centered therapy. Such a personalized medicine approach is likely to achieve better treatment outcomes than the current one-size-fits-all clinical trials.
Alcohol-associated liver disease
Hepatic steatosis will develop in most patients with alcohol use disorder, but less than 50% will progress to more advanced disease with fibrosis, cirrhosis, or alcohol-associated hepatitis (BOX 1). Patients with alcohol-associated liver disease have a dysbiotic gut microbiota that is generally characterized by decreased bacterial diversity, increased proportions of pathobionts such as enterococci61, and reductions in beneficial bacteria such as Akkermansia muciniphila62 (FIG. 3). Stool samples from patients with alcohol-associated liver disease have decreased fungal diversity and increased Candida albicans63,64. One study of stool samples from patients with alcohol use disorder found higher abundance of Enterobacteria and Lactococcus phages in samples from patients with more progressive liver disease65, and a higher abundance of Lactococcus phages was also seen in NAFLD patients who consumed low levels of alcohol66. Another study found that Escherichia, Enterobacteria, and Enterococcus phages were overrepresented in stool samples from patients with alcohol-associated hepatitis, whereas Parabacteroides phages were underrepresented67. Viral diversity and the proportion of mammalian viruses are increased in fecal samples from patients with alcohol-associated hepatitis.
Similar to NAFLD, an altered gut microbiota is considered a risk factor for progression of liver disease, but it is not clear if it is a causative factor. Increased intestinal permeability is present in less than half of patients with alcohol use disorder and early liver disease68. MAMPs such as LPS translocate to the liver and cause progression of liver disease via activation of pathogen recognition receptors such as toll like receptor 4 (TLR4) in the liver69. This more traditional view of pathogenesis has dominated the field for many years. However, we now know that increased permeability is only one piece of the puzzle. MAMP signaling via their receptors alone is necessary, but not sufficient to cause alcohol-associated liver disease70.
Several dysregulated microbial metabolites might also contribute to gut barrier dysfunction. SCFAs are decreased in the stool of patients with alcohol-associated liver disease71, which is associated with gut barrier dysfunction72, though a limitation to the measurement of fecal SCFAs is their rapid metabolism by gut microbiota or absorption by colonocytes limiting the remaining measurable SCFAs pool in feces73. Microbial metabolites of tryptophan metabolism such as indoles are decreased in fecal samples of patients with alcohol-associated hepatitis74. Indoles are aryl hydrocarbon receptor ligands and stimulate IL-22 in ILC3 cells, which induces antimicrobial proteins including regenerating islet derived factor 3 (REG3)74. These molecules maintain an inner mucus layer devoid of bacteria to reduce bacterial translocation. Once this protective barrier is disrupted during chronic ethanol administration, translocation of viable bacteria occurs even during early liver disease in mice70.
Bile acid metabolism is also dysregulated in patients with alcohol-associated liver disease. This is characterized by increased total systemic bile acids and suppressed hepatic synthesis of bile acids58. Intestinal inflammation can be reduced, gut barrier function restored, and ethanol-induced liver disease decreased following intestine-restricted FXR stimulation in mice75.
Plasma levels of trimethylamine (TMA), a microbial product of choline metabolism, were elevated in patients with alcohol-associated hepatitis, whereas TMA n-oxide (TMAO), a co-metabolite generated in hepatocytes from TMA and linked to cardiovascular disease, was decreased in patients with alcohol-associated hepatitis. Small molecule inhibition of gut microbial CutC and CutD activity, which is involved in TMA synthesis, reduced ethanol-induced liver disease in mice76. Further studies are required to test whether TMA affects the gut barrier or exacerbates disease at the level of the liver.
Exotoxins are actively secreted microbial toxins that target other bacteria or host cells. Pore-forming toxins often consist of multiple subunits that oligomerize and form pores in cell membranes. Cytolysin is a two-subunit exotoxin secreted by E. faecalis77. Approximately one third of patients with a severe form of alcohol-associated liver disease, alcohol-associated hepatitis, are colonized by cytolysin-positive E. faecalis, which is associated with high mortality61. Cytolysin appears to be specific for alcohol-associated liver disease, as the prevalence for cytolysin positivity was low in patients with NAFLD and there was no correlation with disease severity78. Cytolysin promotes ethanol-induced liver disease in preclinical models and causes direct hepatocyte death, presumably via direct pore formation in the cell membrane61. It is feasible that within-host evolution of cytolysin-positive E. faecalis allows colonization of the mucosal niche and facilitate bacterial translocation, as was shown for the pathobiont Enterococcus gallinarium79.
Candidalysin is another pore-forming, cytolytic peptide secreted by C. albicans, and it is encoded by hyphae-associated gene ECE180. Patients with alcohol-associated hepatitis and colonized with ECE1-positive C. albicans had increased mortality63. Candidalysin exacerbates ethanol-induced liver disease and damages hepatocytes in vitro63. Candidalysin-induced Th17 response81 and intracellular activation of the Nlrp3 inflammasome in macrophages and dendritic cells82 might further enhance tissue damage. Cytolysin and candidalysin are not associated with increased epithelial cell damage or gut barrier dysfunction in liver disease models. It is unknown how the peptide toxins get to the liver—whether by direct translocation of the encoding viable microorganisms to the liver or in synergy with other microbial virulence factors produced and released in the intestinal lumen.
Taken together, several microbiota-related pathways contribute to alcohol-associated liver disease, including gut barrier dysfunction.
Cirrhosis
Cirrhosis, characterized by a decrease in functional hepatocytes and an increase in fibrotic tissue, is the end stage of chronic liver disease and can have multiple etiologies (BOX 1). A common consequence of cirrhosis is decreased bile flow (cholestasis) due to remodeling and scarring of liver tissue. These changes, along with decreased gut motility83, lead to the small intestinal bacterial overgrowth84 and intestinal dysbiosis seen commonly in cirrhosis. In addition to decreased bacterial diversity, proportional increases in Escherichia coli, Acidaminococcus sp. D21, and Klebsiella pneumoniae; and decreases in Eubacterium eligens, Eubacterium rectale, Dorea longicatena, and Faecalibacterium prausnitzii were found in patients with cirrhosis56 (FIG. 3). Patients with cirrhosis also present differences in the fecal virome. One study showed that Bullavirinae, Felixounavirus, Streptococcus, Escherichia, and Pseudomonas phages were positively linked with model for end-stage liver disease (MELD) score, whereas Faecalibacterium phages were negatively linked with MELD score85. Differences in intestinal fungal populations were found to be predictive of hospitalization in patients with cirrhosis; a lower ratio of the bacterial phylum Bacteroidetes to the fungal phylum Ascomycota was associated with lower likelihood of 90-day hospitalization86.
Most patients with cirrhosis have increased intestinal permeability, which allows MAMPs to translocate to the circulation and contribute to an increase in systemic inflammation87. Once disease progresses, patients with decompensated cirrhosis have translocation of viable bacteria, causing infections such as bacterial peritonitis. Cirrhosis-associated immune dysfunction, with a decrease in phagocytic activity of bacteria by neutrophils, monocytes, and macrophages, contributes to increased infections in patients with decompensated cirrhosis88. A vicious cycle that includes worsening gut dysbiosis, increased gut permeability, systemic inflammation, and deteriorating liver disease with less bile flow, exacerbates progression of disease.
Dysbiosis changes the metabolism of intestinal bile acids, with less conversion of primary to secondary bile acids. Patients with advanced cirrhosis had the lowest amount of total and secondary bile acids in stool, compared with controls or patients with early cirrhosis89. Patients with cirrhosis had increased levels, in particular, of conjugated serum bile acids89. Total and conjugated serum bile acids correlated with liver disease severity in patients with alcohol-associated hepatitis, most of which had underlying cirrhosis58. Changes in luminal bile acids and in FXR activity are connected to gut barrier dysfunction and bacterial translocation. Oral treatment with FXR agonists reduced translocation of viable bacteria from the intestine to the liver in mice with cirrhosis by increasing expression of tight-junction proteins in epithelial cells and restoring the gut vascular barrier90.
Hepatocellular carcinoma
Hepatocellular carcinoma (HCC) is a feared complication of advanced liver disease that develops in about one third of patients with compensated cirrhosis (BOX 1). By contrast, few patients develop HCC in the absence of advanced fibrosis or cirrhosis. The main risk factors for HCC are chronic hepatitis B or C virus infection, heavy alcohol use, diabetes, and more recently described, NAFLD91.
A few studies have compared the gut microbiomes of patients with cirrhosis, with and without HCC. Fecal microbial diversity is decreased in patients with cirrhosis compared with healthy controls, but then diversity increases with progression of cirrhosis to HCC92. Other microbiota changes include decreased Bifidobacterium and Akkermansia species and increased Enterococcus species92–94 in fecal samples from patients with HCC compared to patients with cirrhosis without HCC (FIG. 3). Further, butyrate-producing bacteria such as the genera Ruminococcus, Oscillibacter, Faecalibacterium, Clostridium IV and Coprococcus were decreased in samples from patients with early-stage HCC, whereas LPS-producers such as Klebsiella and Haemophilus were increased compared with controls92.
Gut microbiota are believed to contribute to the pathogenesis of HCC. MYC-transgenic mice, which are predisposed to development of spontaneous HCC, were found to have fewer and smaller HCCs when they were given antibiotics95. This study further demonstrated that primary bile acids increased accumulation of hepatic NKT cells, whereas secondary bile acids reversed this; therefore, depletion of secondary bile acids by antibiotics led to NKT cell accumulation and antitumor activity in the liver. One specific secondary bile acid, deoxycholic acid, was found to be more abundant in individuals with obesity; blocking production of deoxycholic acid with antibiotics prevented HCC development in obese mice96.
In addition to modulating immune reactions in the liver, secondary bile acids may also affect gut permeability. One proposed mechanism is via the proteolytic enzyme gelatinase E (GelE), secreted by E. faecalis. GelE induces loss of barrier function in mice with colitis97. Patients with chronic liver disease have increased colonization by GelE-positive E. faecalis, through altered deoxycholic acid production. GelE-positive E. faecalis increase gut permeability, and their translocation to the liver promotes liver carcinogenesis98. GelE-positive E. faecalis therefore have synergistic detrimental effects in intestine and liver.
Previous studies have linked gut permeability-induced activation of inflammatory signaling via toll-like receptors (TLRs), gut translocation of MAMPs, and development of liver disease and HCC. Rats with diethylnitrosamine-induced HCC have increased serum levels of LPS, and administration of antibiotics decreased tumor size and number99. The same study showed that TLR4-knockout mice given injections of diethylnitrosamine have significantly decreased tumor burdens compared with wild-type mice. Gut sterilization significantly reduced tumor burden in mice given diethylnitrosamine plus carbon tetrachloride to induce HCC. However prolonged administration of low-dose LPS after gut sterilization significantly increased HCC development100. In humans, higher circulating levels of serum anti-LPS and anti-flagellin immunoglobulins, consistent with chronic exposure to bacterial products LPS and flagellin, was significantly associated with increased risk of HCC101.
Translating microbiota research into novel therapies for liver disease
Lifestyle modifications such as alcohol cessation and weight loss remain the cornerstones of treatment for alcohol-associated and non-alcoholic liver disease, respectively, and gut microbiota play a key role. The gut microbiota is critical for nutrient absorption – germ-free mice have lower body fat than conventionally raised mice despite consuming more calories102, and obesity is transmissible via colonization of germ-free mice with gut microbiota from obese mice103. Similarly, alcohol use behavior may be modulated by gut microbiota. Several bacteria are capable of producing neurotransmitters such as γ-aminobutyric acid, serotonin, and dopamine104,105 and gut microbiota influence blood-brain barrier permeability105. Given the scope of these topics, this section will not cover the exciting preclinical work focused on diet and lifestyle-based therapies for liver disease.
Here, we will review emerging therapies that manipulate the gut–liver interaction for treatment of liver disease. Small clinical trials of untargeted therapies such as fecal microbiota transplantation (FMT), probiotics, and antibiotics (Table 1) have had encouraging outcomes. Preclinical studies of engineered bacteria or phages have revealed novel therapeutic approaches. We discuss the results from studies of untargeted therapies and provide a detailed summary of recent randomized clinical trials (Table 1) and a comprehensive overview of targeted therapies (FIG. 4).
Table 1.
Clinical trials of microbiome-based therapies for liver diseases
| Reference | Intervention | Subjects | Results and conclusions |
|---|---|---|---|
| Fecal microbiota transplantation | |||
| 128 | Oral probiotics (Bifidobacterium and Lactobacillus acidophilus) versus heterologous donor FMT via colonoscopy | 75 NAFLD patients: 28 in probiotic arm, 47 in FMT arm. Of the 47 FMT patients, 15 were lean and 32 had obesity | FMT was associated with improvement in hepatic steatosis as measured by Fibroscan compared to oral probiotics and more improvement in lean NAFLD |
| 129 | Standard of care versus FMT via nasojejunal tube | 33 patients with severe alcoholic hepatitis and acute-on-chronic liver failure; 20 received standard of care, 13 received FMT | Survival at 28 and 90 days; hepatic encephalopathy and ascites resolution was significantly better in the FMT group. |
| 106 | Placebo versus FMT enema from a donor enriched in Lachnospiraceae and Ruminococcaceae | 20 men with AUD-related cirrhosis, randomized 1:1 | FMT is safe and associated with short-term reductions in alcohol craving and consumption, with favorable microbial changes, as well as reduction in alcohol use disorder-related events over 6 months |
| 130 | Allogenic or autologous FMT delivered by upper endoscopy to distal duodenum | 21 patients with NAFLD, randomized 3:1 to allogenic vs autologous FMT | No significant changes in insulin resistance or hepatic steatosis in patients who received the allogenic or autologous FMT. Patients with elevated small intestinal permeability at baseline who received allogenic FMT had a significant reduction after 6 weeks |
| 131 | Three FMT via colonoscopy, each 8 weeks apart, from autologous vs lean, vegan donors | 21 overweight patients with metabolic syndrome and hepatic steatosis, randomized 1:1 | Allogenic FMT from lean, vegan donors showed a trend towards decrease in necro-inflammation score compared with autologous FMT, though study was underpowered for significance |
| 111 | Placebo versus 15 FMT oral capsules from a single donor enriched in Lachnospiraceae and Ruminococcaceae | 20 patients with cirrhosis and recurrent hepatic encephalopathy, randomized 1:1 | Oral FMT capsules were safe and well-tolerated in patient with cirrhosis and recurrent hepatic encephalopathy, and associated with improved duodenal mucosal diversity, dysbiosis, and duodenal anti-microbial peptide expression |
| 132 | Standard of care (no treatment) versus 5 days of antibiotics followed by FMT enema from a single donor enriched in Lachnospiraceae and Ruminococcaceae | 20 cirrhotic patients with recurrent hepatic encephalopathy | FMT treatment is safe and associated with reduction in hospitalizations due to liver-related and portal hypertensive complications and improvement in cognition |
| Probiotics | |||
| 133 | Lactulose, rifaximin, or probiotic Escherichia coli Nissle (EcN) 1917 strain for 1 month | 45 patients with cirrhosis and hepatic encephalopathy, randomized 1:1:1 | Patients given rifaximin or the EcN probiotic had more significant reduction in serum ammonia and proinflammatory cytokines, normalization of fecal Bifidobacteria and Lactobacilli abundance, and improved cognition compared with lactulose treatment |
| 134 | Rosuvastatin only versus rosuvastatin plus Clostridium butyricum capsules for 6 months | 96 patients with NAFLD, randomized 1:1 | Probiotic treatment was associated with significantly lower serum total cholesterol, triglycerides, free fatty acids, total bilirubin, direct bilirubin, ALT, AST, and inflammatory markers compared with controls |
| 135 | Placebo versus probiotics sachet containing six Lactobacillus and Bifidobacterium species at a concentration of 30 billion CFU for 6 months | 39 patients with NAFLD, randomized 1:1 | No significant changes in hepatic steatosis, fibrosis, or liver-specific lab parameters between the two treatments. |
| 136 | Placebo versus VSL#3 probiotic supplementation for 10 weeks | 35 patients with NAFLD, randomized 1:1 | VSL#3 did not significantly improve markers of cardiovascular risk or liver injury in patients with NAFLD. |
| 137 | Probiotic capsule + placebo of prebiotic versus oligofructose + placebo of probiotic vs placebo of probiotic + placebo of prebiotic | 111 patients with NAFLD, randomized 1:1:1 | Probiotic supplementation was associated with decreases in triglyceride, ALT, AST, GGT, and alkaline phosphatase compared with placebo |
| 138 | Placebo versus symbiotic capsules (fructo-oligosaccharides plus Bifidobacterium animalis lactis BB-12) for 10–14 months | 104 patients with NAFLD, randomized 1:1:1 | 1 year of administration of a synbiotic combination (probiotic and prebiotic) altered the fecal microbiome but did not reduce liver fat content or markers of liver fibrosis in patients with NAFLD |
| 139 | Placebo versus probiotic mixture (Lactobacillus acidophilus, L. rhamnosus, L. paracasei, Pediococcus pentosaceus, Bifidobacterium lactis, and B. breve) for 12 weeks | 68 patients with NAFLD and obesity, randomized 1:1 | 12 weeks of probiotic treatment significantly reduced hepatic fat and BMI in NAFLD patients with obesity |
| 140 | Placebo versus probiotic mixture (14 probiotic bacteria genera Bifidobacterium, Lactobacillus, Lactococcus, Propionibacterium) for 8 weeks | 58 patients with NAFLD and type 2 diabetes, randomized 1:1 | Probiotic treatment was associated with reduction in fatty liver index, AST, and GGT, but not liver stiffness, compared with placebo |
| 141 | Placebo versus probiotic mixture (Lactobacilli, Bifidobacteria, and Streptococcus thermophilus) for 12 weeks | 75 patients with NASH were randomized 1:1 | Probiotic treatment was associated with significant reduction in serum ALT and cholesterol, liver stiffness, and BMI as compared with placebo |
| 142 | Placebo versus probiotic mixture (Lactobacillus acidophilus, Bifidobacterium lactis, Bifidobacterium bifidum, and Lactobacillus rhamnosus) for 12 weeks | 64children with obesity and sonographic NAFLD, randomized 1:1 | Probiotic treatment was associated with more normal liver sonography and significant decrease in serum ALT, AST, cholesterol, triglycerides, and LDL as compared with placebo |
| Bile acids and metabolites | |||
| 143 | Placebo versus 50 mg or 80 mg of MET409 (a non-bile acid FXR agonist) for 12 weeks | 58 patients with NASH, randomized 1:1:1 | MET409 lowered liver fat content, but was associated with decrease in HDL and increase in LDL compared with placebo |
| 144 | Placebo versus aldafermin (an engineered analog of FGF19) for 24 weeks | 78 patients with NASH, randomized 1:2 | Aldafermin was associated with a significant reduction in absolute liver fat content and serum ALT and AST compared with placebo |
| 145 | Placebo versus 5, 10, or 20 mg volixibat (an inhibitor of the apical sodium-dependent bile acid transporter) for 48 weeks | 197 patients with NASH without cirrhosis, randomized 1:1:1:1 | Volixibat had no effect on steatosis, serum level of ALT, or liver histology in adults with NASH |
| 146 | Inulin control versus inulin-propionate ester for 42 days | 18 patients with NAFLD, randomized 1:1 | Intrahepatocellular lipid was significantly increased in the inulin-control group but not observed in the inulin-propionate ester group. |
| 147 | Placebo versus 500 mg or 1500 mg norursodeoxycholic acid capsules for 12 weeks | 198 patients with NAFLD and increased serum ALT, randomized 1:1:1 | 1500 mg norursodeoxycholic acid resulted in a significant reduction of serum ALT within 12 weeks of treatment compared with placebo |
| 148 | Placebo versus 3 mg or 6 mg of subcutaneous NGM282 for 12 weeks | 82 patients with biopsy-confirmed NASH, randomized 1:1:1 | NGM282 was associated with significant reduction in liver fat content compared with placebo |
FMT, fecal microbiota transplantation; NAFLD, nonalcoholic fatty liver disease; AUD, alcohol use disorder; EcN, Escherichia coli Nissle 1917; CFU, colony-forming unit; ALT, alanine aminotransferase; AST, aspartate aminotransferase; GGT, γ-glutamyltransferase; BMI, body-mass index; NASH, nonalcoholic steatohepatitis; LDL, low-density lipoprotein; HDL, high-density lipoprotein; FXR, farnesoid X receptor.
Fig. 4. Targeted approaches for treatment of liver disease.
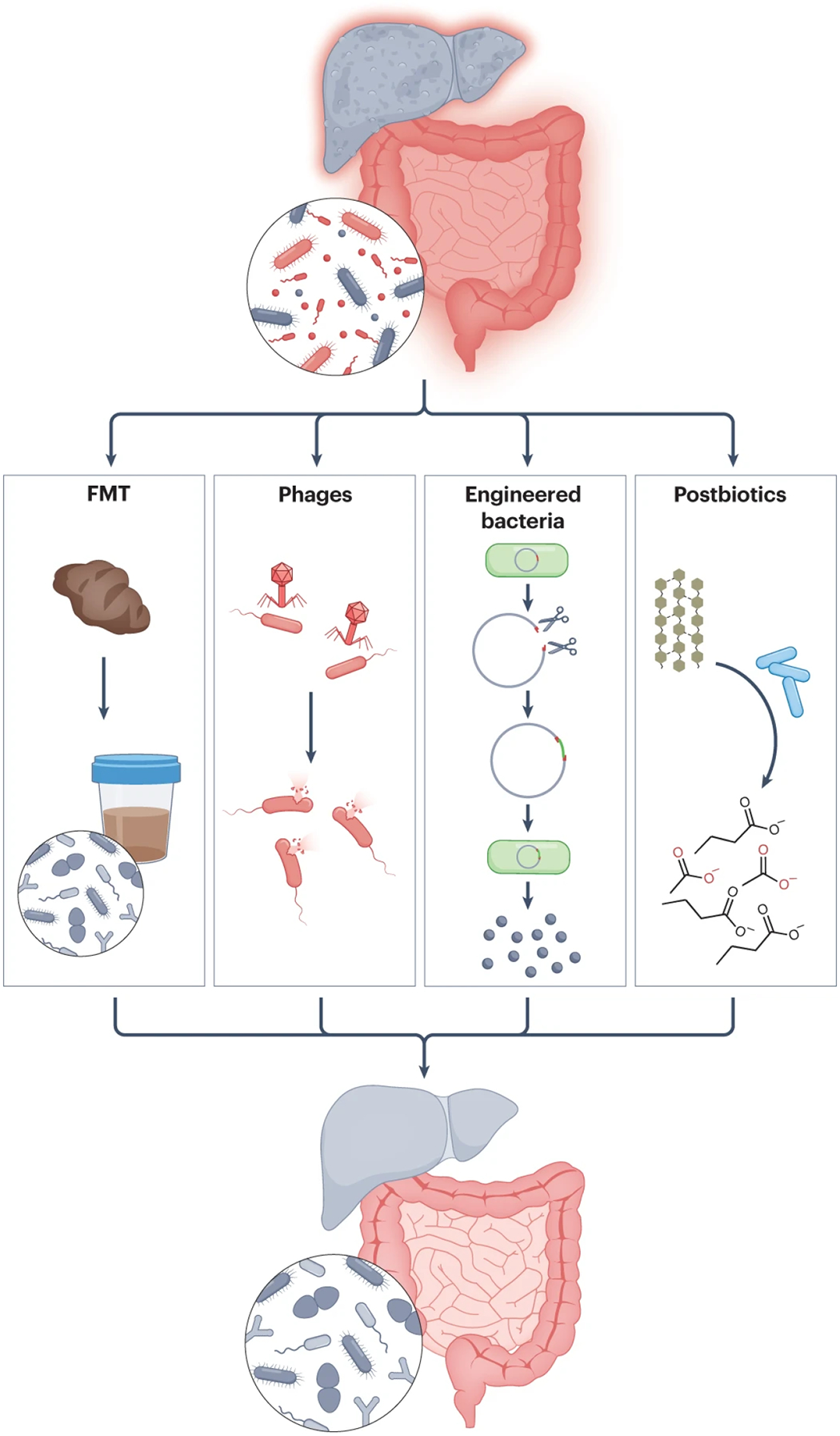
Fecal microbiota transplantation (FMT) involves transplantation of the entire gut microbial community. More targeted treatment approaches include use of bacteriophages (phages) to target specific bacterial strains that produce toxic metabolites, genetically engineered bacteria to produce beneficial metabolites, and supplementation of beneficial microbially produced metabolites or postbiotics.
Fecal microbiota transplantation
Fecal microbiota transplantation, which introduces stool from a healthy donor into a patient’s gastrointestinal tract, is a Food and Drug Administration-approved therapy for recurrent Clostridioides difficile infection and has had promising results in clinical trials for NAFLD, alcohol-associated liver disease, alcohol-associated hepatitis, and hepatic encephalopathy (Table 1). Although the exact mechanisms of FMT are unclear, it has been shown to restore gut barrier function and increases synthesis of SCFA106,107. However, there are still several outstanding issues surrounding broader implementation of FMT in the treatment of chronic liver diseases. For example, the route and frequency of fecal microbiota administration varies among clinical trials, and includes via oral capsules, nasogastric or nasojejunal tubes, upper endoscopy, colonoscopy, or rectal enemas, performed once in some studies, or through several daily or monthly instillations in others (Table 1)108. While there are no head-to-head clinical trials comparing efficacy and engraftment achieved by the different routes and frequencies for FMT, one FMT trial in patients with type 2 diabetes mellitus and obesity showed improved microbial engraftment with repeated FMTs and association with improved liver stiffness109. A recent study performed metagenomic analysis of the stool microbiomes of 226 triads of donors, pre-FMT recipients, and post-FMT recipients to compare methods of FMT in a systematic meta-analysis of 24 studies that used FMT in different clinical settings110. This study found that strain engraftment rate positively correlated with clinical response to FMT, and antibiotic treatment prior to FMT greatly improved strain engraftment. Additionally, the route of FMT delivery was most significantly associated with engraftment rate, with mixed route of FMT administration (combining upper and lower gastrointestinal tract administration) yielding the highest engraftment. Bacteroidetes and Actinobacteria species demonstrated higher average strain engraftment rates than Firmicutes and Proteobacteria species, and Gram-positive bacteria were less likely to have high rates of engraftment compared with more resistant Gram-negative species. These findings emphasize the importance of characterizing and possibly even standardizing FMT donor stool. Some trials attempt to provide more targeted FMT therapy by selecting a donor sample enriched in Lachnospiraceae and Ruminococcaceae106,111, but concern regarding transfer of multidrug-resistant bacteria after FMT still exists112. SER-109, an oral therapeutic composed of live purified Firmicutes bacterial spores, circumvents this concern and effectively reduced risk of C. difficile infection recurrence in patients with history of recurrent C. difficile infection113. Similar purified bacterial formulations may be developed in the future for the treatment of liver diseases.
Phages
Phages are the natural predators of bacteria and make up the majority of the intestinal virome114. Recent metagenomic studies demonstrated changes in intestinal phage composition in patients with liver disease65,67. There have been preclinical studies of the ability of phages to selectively target pathogenic bacterial species and their effects in models of liver disease. Colonization of gnotobiotic mice with fecal microbiota from patients with alcohol-associated hepatitis containing cytolysin-positive E. faecalis resulted in exacerbation of liver disease; administration of phages that target cytolytic E. faecalis reversed liver disease progression61. Steatohepatitis develops in healthy mice transplanted with fecal microbiota from gnotobiotic mice that had received fecal microbiota from a patient with NASH; but pre-treatment with phages that selectively target an ethanol-producing strain of Klebsiella pneumoniae prevented the development of diet-induced steatohepatitis44. Although the target selectivity of phages can be a therapeutic advantage, it also hinders widespread clinical use, because only patients with specific bacterial strains in the intestinal microbiota are likely to respond. The development of phage cocktails or genetic engineering of a phage species to target multiple receptors might extend its host range for broader therapeutic applications.
Engineered bacteria
Probiotics (live bacteria thought to promote health) have been known to have beneficial effects for decades. Some clinical trials of probiotics, mostly from the Lactobacillus and Bifidobacterium genera, in patients with liver diseases such as NAFLD, alcohol-associated liver disease, or hepatic encephalopathy, have had moderate success while others have had equivocal results (Table 1). Due to the wide variability of probiotic formulations and their untargeted nature, results from clinical trials have been difficult to replicate.
The idea of using genetically engineered bacteria as targeted therapies has gained traction. For example, expression of IL-22, a cytokine that regulates the production of REG3G lectins to mediate an intestinal immune response against pathogens, is lower in ethanol-induced liver disease. One study showed that mice fed Lactobacillus reuteri engineered to produce IL-22 along with an ethanol diet developed less severe liver damage, inflammation, and bacterial translocation to the liver as compared with mice fed a wild-type strain of L. reuteri74. In another study, oral administration of a genetically engineered E. coli strain (SYNB1020) that metabolized ammonia and converted it to L-arginine in the gut to mice with thioacetamide-induced liver injury lowered the systemic levels of ammonia115. Unfortunately, a phase Ib/IIa randomized controlled trial in patients with cirrhosis and elevated blood ammonia levels did not show a change in systemic ammonia or other endpoints, compared with placebo (ClinicalTrials.gov Identifier: NCT03447730)116. Nevertheless, the concept of using genetically engineered bacteria to treat liver diseases remains attractive and may be facilitated by the development of methods for genetically engineering native host bacteria as a chassis for transgene delivery117. In this study, authors engineered the native host E. coli to express bile salt hydrolase and demonstrated stable engraftment and improved insulin sensitivity and glucose tolerance in treated mice.
Prebiotics and postbiotics
An alternative to viable microorganisms is the use of prebiotics, non-digestible dietary substances, or postbiotics, inactivated microbial cells or cell components118 that confer health benefits. For example, the prebiotic lactulose, a non-absorbable disaccharide, is a clinical staple for the treatment of hepatic encephalopathy (BOX1). Preclinical trials suggest that supplementation with prebiotics such as inulin, fructooligosaccharides, and pectin may attenuate hepatic steatosis and liver injury in diet- and ethanol-induced liver injury models, respectively119–121. In addition to modifying the composition and function of gut microbiota, prebiotics are fermented by the gut microorganisms into SCFA122. In a mouse model of ethanol-induced liver injury, supplementation of the SCFA butyrate in the form of tributyrin protected mice from disrupted intestinal permeability and liver injury117. However, the benefits of SCFA supplementation for liver disease are controversial. One study demonstrated that increased SCFA production in response to supplementation of the soluble fiber inulin in dysbiotic TLR5-deficient mice that were fed a high-fat diet led to development of HCC123. There are no clinical trials to date testing the therapeutic efficacy of SCFA in patients with liver disease.
Postbiotics previously encompassed a broader definition118, but some preclinical and clinical trials of postbiotics under the new stricter definition do show promise. A small trial of 32 insulin-resistant volunteers with obesity or who are overweight showed that oral supplementation of pasteurized Akkermansia muciniphila improved metabolic parameters compared with control124. Preclinical trials using exopolysaccharide derived from lactic acid bacteria or postbiotics prepared from Lactobacillus paracasei demonstrated benefits in insulin sensitivity and NAFLD125,126. Further research is needed to understand what specific postbiotic components are most likely to benefit patients with liver disease and in what context.
Implications and future directions
The composition of the intestinal microbiota affects all aspects of gut–liver interactions, from the intestinal epithelium, to the gut vascular barrier, to the liver sinusoids, and back to the gut via the biliary tree and the systemic circulation. In a healthy state, the gut microbiota is contained within the gut lumen, but its metabolites act locally and systemically to maintain homeostasis of the gut–liver axis. Disruption of the homeostasis at any of these interfaces can contribute to liver disease. Hence, therapeutic strategies for liver diseases that modulate the gut microbiota, such as fine-tuning of fecal microbiota transplantation, or tailored approaches that specifically target the mechanisms by which dysbiosis leads to liver injury, are being developed.
An important aspect to the success of therapeutics that aim to target the gut–liver axis is the ability to personalize the intervention to specific patients within a disease population. One relatively unexplored avenue of research into more individualized microbiome risk factors for liver disease is to study the interplay between individuals’ genetics and their gut microbiome. For example, variants in the patatin-like phospholipase 3 (PNPLA3), transmembrane 6 superfamily member 2 (TM6SF2), and membrane bound O-acyltransferase domain-containing 7 (MBOAT7) genes are some of the strongest genetic contributors to NAFLD and alcohol-associated liver disease127 and their effects on the microbiome needs further study.
Another avenue that will open new opportunities for both patient research and diagnostic testing is the development of better technologies to measure microbial communities and metabolites in vivo. Currently, our standard methods for investigating patients’ gut microbial communities or gut microbial metabolite concentrations are from fecal samples, which limits investigation of the whole gastrointestinal tract, or from endoscopic evaluation, which requires an invasive procedure. Development of non-invasive methods to detect concentrations of microbial metabolites in vivo will allow researchers to understand where in the gastrointestinal tract these metabolites are being produced and allow for increased ease of diagnostic testing in patients.
Finally, development of more precise therapeutic options will improve both the safety and efficacy of gut microbiota-modulating treatments. Preclinical studies have evaluated new precision approaches, such as the use of bacteriophages or engineered bacteria, but these results have yet to be replicated in humans. If they are found to be safe and effective in clinical trials, these therapies have the potential of transforming the way liver disease is managed in clinical practice, including more precise interventions with fewer side effects. Safety remains a concern for translating these therapies into humans. The durability of treatment has been another concern, though more widespread use of genetic engineering of native (host) bacteria may facilitate stable engraftment and durable results. Although preclinical studies have provided valuable proof of concept, translating their findings into humans is critical, as rodent microbiomes, even those of gnotobiotic mice implanted with human microbiota, cannot fully recapitulate the intricacies of the human gut–liver axis. Nevertheless, our growing understanding of the gut–liver axis confirms that the intestinal microbiota is a compelling target for novel liver disease therapies.
BOX 2. Microbial metabolites and molecules involved in bidirectional communication between gut and liver.
Ethanol.
Ethanol can be produced by the gut microbiota via saccharolytic fermentation and has been implicated in liver disease progression in a subset of patients with nonalcoholic steatohepatitis who were found to have increased abundance of ethanol-producing bacteria in their stool and increased concentrations of ethanol in their systemic circulation42–44. Ethanol and its metabolite acetaldehyde cause hepatic damage, directly and indirectly, via disruptions in gut barrier function.
Bile acids.
Primary bile acids are produced from cholesterol in the liver, conjugated with glycine or taurine, and released into the intestine, where they are deconjugated and dehydroxylated by gut microbiota to secondary bile acids. Primary and secondary bile acids are reabsorbed and returned to the liver via the portal vein. In addition to aiding nutrient absorption, bile acids also serve as signaling molecules, by binding to cellular receptors such as FXR to regulate bile acid synthesis, lipid metabolism, and hepatic gluconeogenesis via fibroblast growth factor 15 (FGF15) and FGF1933.
Short chain fatty acids (SCFA).
SCFA such as acetate, propionate, and butyrate are synthesized via anaerobic fermentation of non-digestible proteins and fibers by the gut microbiota and delivered to the liver via the portal circulation. In addition to serving as an energy source, SCFA also act on signaling pathways to affect hepatic metabolism. Known prolific butyrate producers belong to the Lachnospiriaceae and Ruminococcacecae families and include Eubacterium rectale, Faecalibacterium prausnitzii, and several Roseburia and Anaerostipes. species152.
Indoles and tryptophan metabolites.
Indole is produced from L-tryptophan by gut microbial enzymes and decreases gut inflammation, prevents gut barrier dysfunction, and reduces lipopolysaccharide-induced hepatic inflammation.
Trimethylamine (TMA) and trimethylamine N-oxide (TMAO).
Choline is converted to TMA by gut microbiota and can then be oxidized to TMAO by hepatic monooxygenases153. Serum levels of TMAO have been positively correlated with severity of hepatic steatosis. TMAO is thought to increase the risk for cardiovascular disease by promoting endothelial dysfunction and thrombus formation, reducing glucose tolerance, and increased foam cell production154,155.
Microbiota-derived hepatotoxins.
Bacterial lipopolysaccharides, found in the outer membrane of Gram-negative bacteria, activate microbe-associated molecular pattern receptors such as toll-like receptors. Microorganism-derived exotoxins such as cytolysin from E. faecalis and candidalysin from C. albicans have hepatoxic effects, forming pores in cell membranes.
Acknowledgments
C.L.H. is supported by a Pilot and Feasibility Award from the Southern California Research Center for ALPD and Cirrhosis P50 AA011999 and by T32 DK007202. The review was supported by services provided by NIH center P30 DK120515.
Footnotes
Third party rights statement
This review does not include any display items from previously published or copyrighted material.
Competing interests
B.S. has been consulting for Ambys Medicines, Ferring Research Institute, Gelesis, HOST Therabiomics, Intercept Pharmaceuticals, Mabwell Therapeutics, Patara Pharmaceuticals and Takeda. B.S. is founder of Nterica Bio. UC San Diego has filed several patents with B.S. as inventor. B.S.’s institution UC San Diego has received research support from Artizan Biosciences, Axial Biotherapeutics, BiomX, CymaBay Therapeutics, NGM Biopharmaceuticals, Prodigy Biotech and Synlogic Operating Company. C.H. declares no competing interests.
References
- 1.Mazagova M et al. Commensal microbiota is hepatoprotective and prevents liver fibrosis in mice. FASEB J 29, 1043–1055, doi:fj.14–259515 [pii] 10.1096/fj.14-259515 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henao-Mejia J et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 482, 179–185, doi: 10.1038/nature10809 nature10809 [pii] (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]; First paper to report transmissibility of experimental NASH into mice by fecal microbiota transplantion.
- 3.Llopis M et al. Intestinal microbiota contributes to individual susceptibility to alcoholic liver disease. Gut 65, 830–839, doi:gutjnl-2015–310585 [pii] 10.1136/gutjnl-2015-310585 (2016). [DOI] [PubMed] [Google Scholar]; First paper to report transmissibility of experimental ALD in gnotobiotic mice.
- 4.Philips CA, Ahamed R, Rajesh S, Abduljaleel JKP & Augustine P Long-term Outcomes of Stool Transplant in Alcohol-associated Hepatitis-Analysis of Clinical Outcomes, Relapse, Gut Microbiota and Comparisons with Standard Care. J Clin Exp Hepatol 12, 1124–1132, doi: 10.1016/j.jceh.2022.01.001 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qin J et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65, doi: 10.1038/nature08821 nature08821 [pii] (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Minot S et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res 21, 1616–1625, doi: 10.1101/gr.122705.111 gr.122705.111 [pii] (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suhr MJ, Banjara N & Hallen-Adams HE Sequence-based methods for detecting and evaluating the human gut mycobiome. Lett Appl Microbiol 62, 209–215, doi: 10.1111/lam.12539 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Sender R, Fuchs S & Milo R Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol 14, e1002533, doi: 10.1371/journal.pbio.1002533 e1002533 PBIOLOGY-D-16–00889 [pii] (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eckburg PB et al. Diversity of the human intestinal microbial flora. Science 308, 1635–1638, doi:1110591 [pii] 10.1126/science.1110591 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arumugam M et al. Enterotypes of the human gut microbiome. Nature 473, 174–180, doi: 10.1038/nature09944 nature09944 [pii] (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan Y & Pedersen O Gut microbiota in human metabolic health and disease. Nat Rev Microbiol 19, 55–71, doi: 10.1038/s41579-020-0433-9 10.1038/s41579-020-0433-9 [pii] (2021). [DOI] [PubMed] [Google Scholar]
- 12.Hooper LV, Midtvedt T & Gordon JI How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr 22, 283–307, doi:011602.092259 [pii] 10.1146/annurev.nutr.22.011602.092259 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Schulthess J et al. The Short Chain Fatty Acid Butyrate Imprints an Antimicrobial Program in Macrophages. Immunity 50, 432–445 e437, doi: 10.1016/j.immuni.2018.12.018 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arpaia N et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455, doi: 10.1038/nature12726 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Litvak Y, Byndloss MX & Baumler AJ Colonocyte metabolism shapes the gut microbiota. Science 362, doi: 10.1126/science.aat9076 eaat9076 362/6418/eaat9076 [pii] (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JY, Tsolis RM & Baumler AJ The microbiome and gut homeostasis. Science 377, eabp9960, doi: 10.1126/science.abp9960 (2022). [DOI] [PubMed] [Google Scholar]
- 17.Jakobsson HE et al. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep 16, 164–177, doi: 10.15252/embr.201439263 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johansson ME et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A 105, 15064–15069, doi:0803124105 [pii] 10.1073/pnas.0803124105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates two distinct mucus layers coating the colonic epithelium, both comprised in large part by the mucin Muc2, that separate bacteria from the colon epithelia.
- 19.Vaishnava S et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 334, 255–258, doi:334/6053/255 [pii] 10.1126/science.1209791 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bunker JJ & Bendelac A IgA Responses to Microbiota. Immunity 49, 211–224, doi: 10.1016/j.immuni.2018.08.011 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turner JR Intestinal mucosal barrier function in health and disease. Nat Rev Immunol 9, 799–809, doi:nri2653 [pii] 10.1038/nri2653 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Bacher P et al. Human Anti-fungal Th17 Immunity and Pathology Rely on Cross-Reactivity against Candida albicans. Cell 176, 1340–1355 e1315, doi:S0092–8674(19)30104–7 [pii] 10.1016/j.cell.2019.01.041 (2019). [DOI] [PubMed] [Google Scholar]
- 23.McDonald B et al. Programing of an Intravascular Immune Firewall by the Gut Microbiota Protects against Pathogen Dissemination during Infection. Cell Host Microbe 28, 660–668 e664, doi: 10.1016/j.chom.2020.07.014 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Le HH, Lee MT, Besler KR & Johnson EL Host hepatic metabolism is modulated by gut microbiota-derived sphingolipids. Cell Host Microbe 30, 798–808 e797, doi: 10.1016/j.chom.2022.05.002 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gola A et al. Commensal-driven immune zonation of the liver promotes host defence. Nature 589, 131–136, doi: 10.1038/s41586-020-2977-2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moro-Sibilot L et al. Mouse and Human Liver Contain Immunoglobulin A-Secreting Cells Originating From Peyer’s Patches and Directed Against Intestinal Antigens. Gastroenterology, doi:S0016–5085(16)34318–9 [pii] 10.1053/j.gastro.2016.04.014 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Inagaki T et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci U S A 103, 3920–3925, doi:0509592103 [pii] 10.1073/pnas.0509592103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study elucidated the role of farnesoid X receptor in maintaining the intestinal barrier and preventing bacterial overgrowth and translocation.
- 28.Mouries J et al. Microbiota-driven gut vascular barrier disruption is a prerequisite for non-alcoholic steatohepatitis development. J Hepatol 71, 1216–1228, doi: 10.1016/j.jhep.2019.08.005 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clements WD et al. Role of the gut in the pathophysiology of extrahepatic biliary obstruction. Gut 39, 587–593, doi: 10.1136/gut.39.4.587 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedman ES et al. FXR-Dependent Modulation of the Human Small Intestinal Microbiome by the Bile Acid Derivative Obeticholic Acid. Gastroenterology 155, 1741–1752 e1745, doi:S0016–5085(18)34887-X [pii] 10.1053/j.gastro.2018.08.022 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuchs CD & Trauner M Role of bile acids and their receptors in gastrointestinal and hepatic pathophysiology. Nat Rev Gastroenterol Hepatol 19, 432–450, doi: 10.1038/s41575-021-00566-7 10.1038/s41575-021-00566-7 [pii] (2022). [DOI] [PubMed] [Google Scholar]
- 32.Inagaki T et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab 2, 217–225, doi:S1550–4131(05)00261–5 [pii] 10.1016/j.cmet.2005.09.001 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Kliewer SA & Mangelsdorf DJ Bile Acids as Hormones: The FXR-FGF15/19 Pathway. Dig Dis 33, 327–331, doi: 10.1159/000371670 000371670 [pii] (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Powell EE, Wong VW & Rinella M Non-alcoholic fatty liver disease. Lancet 397, 2212–2224, doi: 10.1016/S0140-6736(20)32511-3 (2021). [DOI] [PubMed] [Google Scholar]
- 35.Lee G et al. Distinct signatures of gut microbiome and metabolites associated with significant fibrosis in non-obese NAFLD. Nat Commun 11, 4982, doi: 10.1038/s41467-020-18754-5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Demir M et al. The fecal mycobiome in non-alcoholic fatty liver disease. J Hepatol 76, 788–799, doi:S0168–8278(21)02238–8 [pii] 10.1016/j.jhep.2021.11.029 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loomba R et al. Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metab 25, 1054–1062 e1055, doi:S1550–4131(17)30206–1 [pii] 10.1016/j.cmet.2017.04.001 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lang S et al. Intestinal Virome Signature Associated With Severity of Nonalcoholic Fatty Liver Disease. Gastroenterology 159, 1839–1852, doi:S0016–5085(20)34923–4 [pii] 10.1053/j.gastro.2020.07.005 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; First study to interrogate the composition of the intestinal virome and its association with severity of NAFLD and fibrosis.
- 39.Rahman K et al. Loss of Junctional Adhesion Molecule A Promotes Severe Steatohepatitis in Mice on a Diet High in Saturated Fat, Fructose, and Cholesterol. Gastroenterology 151, 733–746 e712, doi: 10.1053/j.gastro.2016.06.022 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsu CL et al. Differences in Bacterial Translocation and Liver Injury in Ethanol Versus Diet-Induced Liver Disease. Dig Dis Sci, doi: 10.1007/s10620-023-07860-1 10.1007/s10620-023-07860-1 [pii] (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luther J et al. Hepatic Injury in Nonalcoholic Steatohepatitis Contributes to Altered Intestinal Permeability. Cell Mol Gastroenterol Hepatol 1, 222–232, doi: 10.1016/j.jcmgh.2015.01.001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meijnikman AS et al. Microbiome-derived ethanol in nonalcoholic fatty liver disease. Nat Med 28, 2100–2106, doi: 10.1038/s41591-022-02016-6 10.1038/s41591-022-02016-6 [pii] (2022). [DOI] [PubMed] [Google Scholar]; This is the first study to measure portal vein ethanol concentrations in patients with NAFLD and demonstrate higher portal vein ethanol concentrations in patients post-prandially and with more advanced liver disease.
- 43.Zhu L et al. Characterization of the gut microbiome in non-alcoholic steatohepatitis (NASH) patients: A connection between endogenous alcohol and NASH. Hepatology 57, 601–609, doi: 10.1002/hep.26093 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Yuan J et al. Fatty Liver Disease Caused by High-Alcohol-Producing Klebsiella pneumoniae. Cell Metab 30, 675–688 e677, doi:S1550–4131(19)30447–4 [pii] 10.1016/j.cmet.2019.08.018 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Hoyles L et al. Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat Med 24, 1070–1080, doi: 10.1038/s41591-018-0061-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao M et al. TMAVA, a Metabolite of Intestinal Microbes, Is Increased in Plasma From Patients With Liver Steatosis, Inhibits gamma-Butyrobetaine Hydroxylase, and Exacerbates Fatty Liver in Mice. Gastroenterology 158, 2266–2281 e2227, doi:S0016–5085(20)30244–4 [pii] 10.1053/j.gastro.2020.02.033 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Beaumont M et al. The gut microbiota metabolite indole alleviates liver inflammation in mice. FASEB J 32, fj201800544, doi: 10.1096/fj.201800544 FJ_201800544 [pii] (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma L et al. Indole Alleviates Diet-Induced Hepatic Steatosis and Inflammation in a Manner Involving Myeloid Cell 6-Phosphofructo-2-Kinase/Fructose-2,6-Biphosphatase 3. Hepatology 72, 1191–1203, doi: 10.1002/hep.31115 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teunis C, Nieuwdorp M & Hanssen N Interactions between Tryptophan Metabolism, the Gut Microbiome and the Immune System as Potential Drivers of Non-Alcoholic Fatty Liver Disease (NAFLD) and Metabolic Diseases. Metabolites 12, doi: 10.3390/metabo12060514 514 metabo12060514 [pii] metabolites-12–00514 [pii] (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiao N et al. Suppressed hepatic bile acid signalling despite elevated production of primary and secondary bile acids in NAFLD. Gut 67, 1881–1891, doi: 10.1136/gutjnl-2017-314307 gutjnl-2017–314307 [pii] (2018). [DOI] [PubMed] [Google Scholar]
- 51.Ferslew BC et al. Altered Bile Acid Metabolome in Patients with Nonalcoholic Steatohepatitis. Dig Dis Sci 60, 3318–3328, doi: 10.1007/s10620-015-3776-8 10.1007/s10620-015-3776-8 [pii] (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mouzaki M et al. Bile Acids and Dysbiosis in Non-Alcoholic Fatty Liver Disease. PLoS One 11, e0151829, doi: 10.1371/journal.pone.0151829 PONE-D-15–55006 [pii] (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fang S et al. Intestinal FXR agonism promotes adipose tissue browning and reduces obesity and insulin resistance. Nat Med 21, 159–165, doi: 10.1038/nm.3760 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang C et al. Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. J Clin Invest 125, 386–402, doi:76738 [pii] 10.1172/JCI76738 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Campbell C et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature 581, 475–479, doi: 10.1038/s41586-020-2193-0 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song X et al. Microbial bile acid metabolites modulate gut RORgamma(+) regulatory T cell homeostasis. Nature 577, 410–415, doi: 10.1038/s41586-019-1865-0 10.1038/s41586-019-1865-0 [pii] (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hang S et al. Bile acid metabolites control TH17 and Treg cell differentiation. Nature 576, 143–148, doi: 10.1038/s41586-019-1785-z (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identified bile acid metabolites capable of regulating T cell differentiation, reducing T helper cells expressing IL-17a and increasing regulatory T cells.
- 58.Brandl K et al. Dysregulation of serum bile acids and FGF19 in alcoholic hepatitis. J Hepatol 69, 396–405, doi:S0168–8278(18)31973–1 [pii] 10.1016/j.jhep.2018.03.031 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li J & Dawson PA Animal models to study bile acid metabolism. Biochim Biophys Acta Mol Basis Dis 1865, 895–911, doi:S0925–4439(18)30181–9 [pii] 10.1016/j.bbadis.2018.05.011 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hild B et al. Neonatal exposure to a wild-derived microbiome protects mice against diet-induced obesity. Nat Metab 3, 1042–1057, doi: 10.1038/s42255-021-00439-y (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Duan Y et al. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature 575, 505–511, doi: 10.1038/s41586-019-1742-x 10.1038/s41586-019-1742-x [pii] (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; First preclinical trial demonstrating presence of cytolysin, an exotoxin secreted by Enterococcus faecalis, is associated with severity of alcoholic hepatitis and employing phage-based therapy in a murine model of a non-infectious disease.
- 62.Grander C et al. Recovery of ethanol-induced Akkermansia muciniphila depletion ameliorates alcoholic liver disease. Gut 67, 891–901, doi: 10.1136/gutjnl-2016-313432 gutjnl-2016–313432 [pii] (2018). [DOI] [PubMed] [Google Scholar]
- 63.Chu H et al. The Candida albicans exotoxin candidalysin promotes alcohol-associated liver disease. J Hepatol 72, 391–400, doi:S0168–8278(19)30599–9 [pii] 10.1016/j.jhep.2019.09.029 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lang S et al. Intestinal Fungal Dysbiosis and Systemic Immune Response to Fungi in Patients With Alcoholic Hepatitis. Hepatology 71, 522–538, doi: 10.1002/hep.30832 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; First study demonstrating association of anti-fungal antibodies with mortality in patients with alcoholic hepatitis.
- 65.Hsu CL et al. Intestinal virome in patients with alcohol use disorder and after abstinence. Hepatol Commun 6, 2058–2069, doi: 10.1002/hep4.1947 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hsu CL et al. Any alcohol use in NAFLD patients is associated with significant changes to the intestinal virome. Hepatology, doi: 10.1097/HEP.0000000000000238 01515467-990000000-00266 [pii] (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jiang L et al. Intestinal Virome in Patients With Alcoholic Hepatitis. Hepatology 72, 2182–2196, doi: 10.1002/hep.31459 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leclercq S et al. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc Natl Acad Sci U S A 111, E4485–4493, doi: 10.1073/pnas.1415174111 1415174111 [pii] (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]; First study demonstrating association of gut permeability and gut-microbiota composition with addiction severity in patients with alcohol use disorder.
- 69.Lang S & Schnabl B Microbiota and Fatty Liver Disease-the Known, the Unknown, and the Future. Cell Host Microbe 28, 233–244, doi:S1931–3128(20)30403–0 [pii] 10.1016/j.chom.2020.07.007 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang L et al. Intestinal REG3 Lectins Protect against Alcoholic Steatohepatitis by Reducing Mucosa-Associated Microbiota and Preventing Bacterial Translocation. Cell Host Microbe 19, 227–239, doi:S1931–3128(16)30002–6 [pii] 10.1016/j.chom.2016.01.003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates that intestinal antimicrobial molecules such as REG3 lectins protect the host from bacterial translocation to extra-intestinal tissues caused by chronic ethanol exposure.
- 71.Couch RD et al. Alcohol induced alterations to the human fecal VOC metabolome. PLoS One 10, e0119362, doi: 10.1371/journal.pone.0119362 PONE-D-14–39664 [pii] (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cresci GA et al. Prophylactic tributyrin treatment mitigates chronic-binge alcohol-induced intestinal barrier and liver injury. J Gastroenterol Hepatol, doi: 10.1111/jgh.13731 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Morrison DJ & Preston T Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 7, 189–200, doi: 10.1080/19490976.2015.1134082 1134082 [pii] (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hendrikx T et al. Bacteria engineered to produce IL-22 in intestine induce expression of REG3G to reduce ethanol-induced liver disease in mice. Gut 68, 1504–1515, doi: 10.1136/gutjnl-2018-317232 gutjnl-2018–317232 gutjnl-2018–317232 [pii] (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hartmann P et al. Modulation of the intestinal bile acid/farnesoid X receptor/fibroblast growth factor 15 axis improves alcoholic liver disease in mice. Hepatology 67, 2150–2166, doi: 10.1002/hep.29676 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Helsley RN et al. Gut microbial trimethylamine is elevated in alcohol-associated hepatitis and contributes to ethanol-induced liver injury in mice. Elife 11, doi: 10.7554/eLife.76554 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Haas W, Shepard BD & Gilmore MS Two-component regulator of Enterococcus faecalis cytolysin responds to quorum-sensing autoinduction. Nature 415, 84–87, doi: 10.1038/415084a (2002). [DOI] [PubMed] [Google Scholar]
- 78.Lang S, Demir M, Duan Y, Martin A & Schnabl B Cytolysin-positive Enterococcus faecalis is not increased in patients with non-alcoholic steatohepatitis. Liver Int 40, 860–865, doi: 10.1111/liv.14377 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang Y et al. Within-host evolution of a gut pathobiont facilitates liver translocation. Nature 607, 563–570, doi: 10.1038/s41586-022-04949-x (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Richardson JP et al. Candidalysins Are a New Family of Cytolytic Fungal Peptide Toxins. mBio, e0351021, doi: 10.1128/mbio.03510-21 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Verma AH et al. Oral epithelial cells orchestrate innate type 17 responses to Candida albicans through the virulence factor candidalysin. Sci Immunol 2, doi: 10.1126/sciimmunol.aam8834 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kasper L et al. The fungal peptide toxin Candidalysin activates the NLRP3 inflammasome and causes cytolysis in mononuclear phagocytes. Nat Commun 9, 4260, doi: 10.1038/s41467-018-06607-1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chang CS, Chen GH, Lien HC & Yeh HZ Small intestine dysmotility and bacterial overgrowth in cirrhotic patients with spontaneous bacterial peritonitis. Hepatology 28, 1187–1190, doi:S0270913998004339 [pii] 10.1002/hep.510280504 (1998). [DOI] [PubMed] [Google Scholar]
- 84.Bauer TM et al. Small intestinal bacterial overgrowth in human cirrhosis is associated with systemic endotoxemia. Am J Gastroenterol 97, 2364–2370, doi: 10.1111/j.1572-0241.2002.05791.x (2002). [DOI] [PubMed] [Google Scholar]
- 85.Bajaj JS et al. Interaction of bacterial metagenome and virome in patients with cirrhosis and hepatic encephalopathy. Gut 70, 1162–1173, doi: 10.1136/gutjnl-2020-322470 (2021). [DOI] [PubMed] [Google Scholar]
- 86.Bajaj JS et al. Fungal dysbiosis in cirrhosis. Gut 67, 1146–1154, doi: 10.1136/gutjnl-2016-313170 (2018). [DOI] [PubMed] [Google Scholar]
- 87.Hasa E, Hartmann P & Schnabl B Liver cirrhosis and immune dysfunction. Int Immunol 34, 455–466, doi: 10.1093/intimm/dxac030 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Trebicka J, Macnaughtan J, Schnabl B, Shawcross DL & Bajaj JS The microbiota in cirrhosis and its role in hepatic decompensation. J Hepatol 75 Suppl 1, S67–S81, doi: 10.1016/j.jhep.2020.11.013 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kakiyama G et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol 58, 949–955, doi: 10.1016/j.jhep.2013.01.003 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sorribas M et al. FXR modulates the gut-vascular barrier by regulating the entry sites for bacterial translocation in experimental cirrhosis. J Hepatol 71, 1126–1140, doi: 10.1016/j.jhep.2019.06.017 (2019). [DOI] [PubMed] [Google Scholar]
- 91.Kulik L & El-Serag HB Epidemiology and Management of Hepatocellular Carcinoma. Gastroenterology 156, 477–491 e471, doi: 10.1053/j.gastro.2018.08.065 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ren Z et al. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut 68, 1014–1023, doi: 10.1136/gutjnl-2017-315084 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ponziani FR et al. Hepatocellular Carcinoma Is Associated With Gut Microbiota Profile and Inflammation in Nonalcoholic Fatty Liver Disease. Hepatology 69, 107–120, doi: 10.1002/hep.30036 (2019). [DOI] [PubMed] [Google Scholar]
- 94.Zheng R et al. Liver cirrhosis contributes to the disorder of gut microbiota in patients with hepatocellular carcinoma. Cancer Med 9, 4232–4250, doi: 10.1002/cam4.3045 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ma C et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 360, doi: 10.1126/science.aan5931 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yoshimoto S et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499, 97–101, doi: 10.1038/nature12347 (2013). [DOI] [PubMed] [Google Scholar]
- 97.Steck N et al. Enterococcus faecalis metalloprotease compromises epithelial barrier and contributes to intestinal inflammation. Gastroenterology 141, 959–971, doi: 10.1053/j.gastro.2011.05.035 (2011). [DOI] [PubMed] [Google Scholar]
- 98.Iida N et al. Chronic liver disease enables gut Enterococcus faecalis colonization to promote liver carcinogenesis. Nat Cancer 2, 1039–1054, doi: 10.1038/s43018-021-00251-3 (2021). [DOI] [PubMed] [Google Scholar]
- 99.Yu LX et al. Endotoxin accumulation prevents carcinogen-induced apoptosis and promotes liver tumorigenesis in rodents. Hepatology 52, 1322–1333, doi: 10.1002/hep.23845 (2010). [DOI] [PubMed] [Google Scholar]
- 100.Dapito DH et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell 21, 504–516, doi: 10.1016/j.ccr.2012.02.007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fedirko V et al. Exposure to bacterial products lipopolysaccharide and flagellin and hepatocellular carcinoma: a nested case-control study. BMC Med 15, 72, doi: 10.1186/s12916-017-0830-8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Backhed F et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A 101, 15718–15723, doi:0407076101 [pii] 010115718 [pii] 10.1073/pnas.0407076101 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Turnbaugh PJ et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031, doi:nature05414 [pii] 10.1038/nature05414 (2006). [DOI] [PubMed] [Google Scholar]; First study to demonstrate that the microbiome of individuals with obesity have an increased capacity for energy harvest from the diet and obesity was transmissible to germ-free mice with transplantation of this microbiota.
- 104.Strandwitz P Neurotransmitter modulation by the gut microbiota. Brain Res 1693, 128–133, doi:S0006–8993(18)30150–1 [pii] 10.1016/j.brainres.2018.03.015 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Braniste V et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci Transl Med 6, 263ra158, doi: 10.1126/scitranslmed.3009759 6/263/263ra158 [pii] (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bajaj JS et al. A Randomized Clinical Trial of Fecal Microbiota Transplant for Alcohol Use Disorder. Hepatology 73, 1688–1700, doi: 10.1002/hep.31496 (2021). [DOI] [PubMed] [Google Scholar]; First human study to demonstrate improvement in alcohol craving symptoms in patients with alcohol-associated cirrhosis and alcohol use disorder after FMT.
- 107.Bajaj JS et al. Antibiotic-Associated Disruption of Microbiota Composition and Function in Cirrhosis Is Restored by Fecal Transplant. Hepatology 68, 1549–1558, doi: 10.1002/hep.30037 (2018). [DOI] [PubMed] [Google Scholar]
- 108.Gulati M, Singh SK, Corrie L, Kaur IP & Chandwani L Delivery routes for faecal microbiota transplants: Available, anticipated and aspired. Pharmacol Res 159, 104954, doi:S1043–6618(20)31262–7 [pii] 10.1016/j.phrs.2020.104954 (2020). [DOI] [PubMed] [Google Scholar]
- 109.Ng SC et al. Microbiota engraftment after faecal microbiota transplantation in obese subjects with type 2 diabetes: a 24-week, double-blind, randomised controlled trial. Gut 71, 716–723, doi: 10.1136/gutjnl-2020-323617 (2022). [DOI] [PubMed] [Google Scholar]
- 110.Ianiro G et al. Variability of strain engraftment and predictability of microbiome composition after fecal microbiota transplantation across different diseases. Nat Med 28, 1913–1923, doi: 10.1038/s41591-022-01964-3 10.1038/s41591-022-01964-3 [pii] 1964 [pii] (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bajaj JS et al. Fecal Microbial Transplant Capsules Are Safe in Hepatic Encephalopathy: A Phase 1, Randomized, Placebo-Controlled Trial. Hepatology 70, 1690–1703, doi: 10.1002/hep.30690 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.DeFilipp Z et al. Drug-Resistant E. coli Bacteremia Transmitted by Fecal Microbiota Transplant. N Engl J Med 381, 2043–2050, doi: 10.1056/NEJMoa1910437 (2019). [DOI] [PubMed] [Google Scholar]
- 113.Feuerstadt P et al. SER-109, an Oral Microbiome Therapy for Recurrent Clostridioides difficile Infection. N Engl J Med 386, 220–229, doi: 10.1056/NEJMoa2106516 (2022). [DOI] [PubMed] [Google Scholar]
- 114.Hsu CL, Duan Y, Fouts DE & Schnabl B Intestinal virome and therapeutic potential of bacteriophages in liver disease. J Hepatol 75, 1465–1475, doi:S0168–8278(21)01997–8 [pii] 10.1016/j.jhep.2021.08.003 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kurtz CB et al. An engineered E. coli Nissle improves hyperammonemia and survival in mice and shows dose-dependent exposure in healthy humans. Sci Transl Med 11, doi: 10.1126/scitranslmed.aau7975 (2019). [DOI] [PubMed] [Google Scholar]
- 116.US National Library of Medicine. ClinicalTrials.gov, h. c. g. c. s. N. [DOI] [PubMed]
- 117.Russell BJ et al. Intestinal transgene delivery with native E. coli chassis allows persistent physiological changes. Cell 185, 3263–3277 e3215, doi: 10.1016/j.cell.2022.06.050 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Salminen S et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat Rev Gastroenterol Hepatol 18, 649–667, doi: 10.1038/s41575-021-00440-6 10.1038/s41575-021-00440-6 [pii] 440 [pii] (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yang Z et al. Inulin intervention attenuates hepatic steatosis in rats via modulating gut microbiota and maintaining intestinal barrier function. Food Res Int 163, 112309, doi:S0963–9969(22)01367–9 [pii] 10.1016/j.foodres.2022.112309 (2023). [DOI] [PubMed] [Google Scholar]
- 120.Huang X et al. Fructooligosaccharides attenuate non-alcoholic fatty liver disease by remodeling gut microbiota and association with lipid metabolism. Biomed Pharmacother 159, 114300, doi:S0753–3322(23)00088–4 [pii] 10.1016/j.biopha.2023.114300 (2023). [DOI] [PubMed] [Google Scholar]
- 121.Wrzosek L et al. Microbiota tryptophan metabolism induces aryl hydrocarbon receptor activation and improves alcohol-induced liver injury. Gut 70, 1299–1308, doi: 10.1136/gutjnl-2020-321565 gutjnl-2020–321565 [pii] (2021). [DOI] [PubMed] [Google Scholar]
- 122.Pohl K, Moodley P & Dhanda A The effect of increasing intestinal short-chain fatty acid concentration on gut permeability and liver injury in the context of liver disease: A systematic review. J Gastroenterol Hepatol 37, 1498–1506, doi: 10.1111/jgh.15899 JGH15899 [pii] (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Singh V et al. Dysregulated Microbial Fermentation of Soluble Fiber Induces Cholestatic Liver Cancer. Cell 175, 679–694 e622, doi: 10.1016/j.cell.2018.09.004 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Depommier C et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat Med 25, 1096–1103, doi: 10.1038/s41591-019-0495-2 10.1038/s41591-019-0495-2 [pii] (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhou Y, Cui Y & Qu X Exopolysaccharides of lactic acid bacteria: Structure, bioactivity and associations: A review. Carbohydr Polym 207, 317–332, doi:S0144–8617(18)31426–7 [pii] 10.1016/j.carbpol.2018.11.093 (2019). [DOI] [PubMed] [Google Scholar]
- 126.Pan Z et al. Postbiotics Prepared Using Lactobacillus paracasei CCFM1224 Prevent Nonalcoholic Fatty Liver Disease by Modulating the Gut Microbiota and Liver Metabolism. Int J Mol Sci 23, doi: 10.3390/ijms232113522 13522 ijms232113522 [pii] ijms-23–13522 [pii] (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Anstee QM, Seth D & Day CP Genetic Factors That Affect Risk of Alcoholic and Nonalcoholic Fatty Liver Disease. Gastroenterology 150, 1728–1744 e1727, doi:S0016–5085(16)00138–4 [pii] 10.1053/j.gastro.2016.01.037 (2016). [DOI] [PubMed] [Google Scholar]
- 128.Xue L, Deng Z, Luo W, He X & Chen Y Effect of Fecal Microbiota Transplantation on Non-Alcoholic Fatty Liver Disease: A Randomized Clinical Trial. Front Cell Infect Microbiol 12, 759306, doi: 10.3389/fcimb.2022.759306 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sharma A et al. Fecal microbiota transplantation in alcohol-associated acute-on-chronic liver failure: an open-label clinical trial. Hepatol Int 16, 433–446, doi: 10.1007/s12072-022-10312-z (2022). [DOI] [PubMed] [Google Scholar]
- 130.Craven L et al. Allogenic Fecal Microbiota Transplantation in Patients With Nonalcoholic Fatty Liver Disease Improves Abnormal Small Intestinal Permeability: A Randomized Control Trial. Am J Gastroenterol 115, 1055–1065, doi: 10.14309/ajg.0000000000000661 (2020). [DOI] [PubMed] [Google Scholar]
- 131.Witjes JJ et al. Donor Fecal Microbiota Transplantation Alters Gut Microbiota and Metabolites in Obese Individuals With Steatohepatitis. Hepatol Commun 4, 1578–1590, doi: 10.1002/hep4.1601 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bajaj JS et al. Fecal microbiota transplant from a rational stool donor improves hepatic encephalopathy: A randomized clinical trial. Hepatology 66, 1727–1738, doi: 10.1002/hep.29306 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; First human randomized clinical trial using FMT for the treatment of hepatic encephalopathy in patients with cirrhosis showing reduction in hospitalizations due to liver-related complications.
- 133.Manzhalii E et al. Effect of a specific Escherichia coli Nissle 1917 strain on minimal/mild hepatic encephalopathy treatment. World J Hepatol 14, 634–646, doi: 10.4254/wjh.v14.i3.634 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhu W, Yan M, Cao H, Zhou J & Xu Z Effects of Clostridium butyricum Capsules Combined with Rosuvastatin on Intestinal Flora, Lipid Metabolism, Liver Function and Inflammation in NAFLD Patients. Cell Mol Biol (Noisy-le-grand) 68, 64–69, doi: 10.14715/cmb/2022.68.2.10 (2022). [DOI] [PubMed] [Google Scholar]
- 135.Mohamad Nor MH et al. The Effect of Probiotics (MCP((R)) BCMC((R)) Strains) on Hepatic Steatosis, Small Intestinal Mucosal Immune Function, and Intestinal Barrier in Patients with Non-Alcoholic Fatty Liver Disease. Nutrients 13, doi: 10.3390/nu13093192 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chong PL, Laight D, Aspinall RJ, Higginson A & Cummings MH A randomised placebo controlled trial of VSL#3((R)) probiotic on biomarkers of cardiovascular risk and liver injury in non-alcoholic fatty liver disease. BMC Gastroenterol 21, 144, doi: 10.1186/s12876-021-01660-5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Behrouz V, Aryaeian N, Zahedi MJ & Jazayeri S Effects of probiotic and prebiotic supplementation on metabolic parameters, liver aminotransferases, and systemic inflammation in nonalcoholic fatty liver disease: A randomized clinical trial. J Food Sci 85, 3611–3617, doi: 10.1111/1750-3841.15367 (2020). [DOI] [PubMed] [Google Scholar]
- 138.Scorletti E et al. Synbiotics Alter Fecal Microbiomes, But Not Liver Fat or Fibrosis, in a Randomized Trial of Patients With Nonalcoholic Fatty Liver Disease. Gastroenterology 158, 1597–1610 e1597, doi: 10.1053/j.gastro.2020.01.031 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ahn SB et al. Randomized, Double-blind, Placebo-controlled Study of a Multispecies Probiotic Mixture in Nonalcoholic Fatty Liver Disease. Sci Rep 9, 5688, doi: 10.1038/s41598-019-42059-3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kobyliak N et al. A Multi-strain Probiotic Reduces the Fatty Liver Index, Cytokines and Aminotransferase levels in NAFLD Patients: Evidence from a Randomized Clinical Trial. J Gastrointestin Liver Dis 27, 41–49, doi: 10.15403/jgld.2014.1121.271.kby (2018). [DOI] [PubMed] [Google Scholar]
- 141.Manzhalii E, Virchenko O, Falalyeyeva T, Beregova T & Stremmel W Treatment efficacy of a probiotic preparation for non-alcoholic steatohepatitis: A pilot trial. J Dig Dis 18, 698–703, doi: 10.1111/1751-2980.12561 (2017). [DOI] [PubMed] [Google Scholar]
- 142.Famouri F, Shariat Z, Hashemipour M, Keikha M & Kelishadi R Effects of Probiotics on Nonalcoholic Fatty Liver Disease in Obese Children and Adolescents. J Pediatr Gastroenterol Nutr 64, 413–417, doi: 10.1097/MPG.0000000000001422 (2017). [DOI] [PubMed] [Google Scholar]
- 143.Harrison SA et al. A structurally optimized FXR agonist, MET409, reduced liver fat content over 12 weeks in patients with non-alcoholic steatohepatitis. J Hepatol 75, 25–33, doi: 10.1016/j.jhep.2021.01.047 (2021). [DOI] [PubMed] [Google Scholar]
- 144.Harrison SA et al. Efficacy and Safety of Aldafermin, an Engineered FGF19 Analog, in a Randomized, Double-Blind, Placebo-Controlled Trial of Patients With Nonalcoholic Steatohepatitis. Gastroenterology 160, 219–231 e211, doi: 10.1053/j.gastro.2020.08.004 (2021). [DOI] [PubMed] [Google Scholar]; First randomized clinical trial showing that an engineered FGF19 analog could improve hepatic steatosis and liver injury in patients with NASH.
- 145.Newsome PN et al. Volixibat in adults with non-alcoholic steatohepatitis: 24-week interim analysis from a randomized, phase II study. J Hepatol 73, 231–240, doi: 10.1016/j.jhep.2020.03.024 (2020). [DOI] [PubMed] [Google Scholar]
- 146.Chambers ES et al. The effects of dietary supplementation with inulin and inulin-propionate ester on hepatic steatosis in adults with non-alcoholic fatty liver disease. Diabetes Obes Metab 21, 372–376, doi: 10.1111/dom.13500 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Traussnigg S et al. Norursodeoxycholic acid versus placebo in the treatment of non-alcoholic fatty liver disease: a double-blind, randomised, placebo-controlled, phase 2 dose-finding trial. Lancet Gastroenterol Hepatol 4, 781–793, doi: 10.1016/S2468-1253(19)30184-0 (2019). [DOI] [PubMed] [Google Scholar]; First randomized clinical trial showing that supplementation of a modified secondary bile acid in patients with NAFLD improved hepatic injury.
- 148.Harrison SA et al. NGM282 for treatment of non-alcoholic steatohepatitis: a multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet 391, 1174–1185, doi: 10.1016/S0140-6736(18)30474-4 (2018). [DOI] [PubMed] [Google Scholar]
- 149.Bernard B et al. Antibiotic prophylaxis for the prevention of bacterial infections in cirrhotic patients with gastrointestinal bleeding: a meta-analysis. Hepatology 29, 1655–1661, doi: 10.1002/hep.510290608 (1999). [DOI] [PubMed] [Google Scholar]
- 150.Moon AM, Dominitz JA, Ioannou GN, Lowy E & Beste LA Use of Antibiotics Among Patients With Cirrhosis and Upper Gastrointestinal Bleeding Is Associated With Reduced Mortality. Clin Gastroenterol Hepatol 14, 1629–1637 e1621, doi: 10.1016/j.cgh.2016.05.040 (2016). [DOI] [PubMed] [Google Scholar]
- 151.Gluud LL, Vilstrup H & Morgan MY Non-absorbable disaccharides versus placebo/no intervention and lactulose versus lactitol for the prevention and treatment of hepatic encephalopathy in people with cirrhosis. Cochrane Database Syst Rev, CD003044, doi: 10.1002/14651858.CD003044.pub4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Baxter NT et al. Dynamics of Human Gut Microbiota and Short-Chain Fatty Acids in Response to Dietary Interventions with Three Fermentable Fibers. mBio 10, doi: 10.1128/mBio.02566-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wang Z et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63, doi: 10.1038/nature09922 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tang WH & Hazen SL Microbiome, trimethylamine N-oxide, and cardiometabolic disease. Transl Res 179, 108–115, doi: 10.1016/j.trsl.2016.07.007 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Witkowski M, Weeks TL & Hazen SL Gut Microbiota and Cardiovascular Disease. Circ Res 127, 553–570, doi: 10.1161/CIRCRESAHA.120.316242 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]


