Summary
Background
The occult lymph node metastasis (LNM) of laryngeal squamous cell carcinoma (LSCC) affects the treatment and prognosis of patients. This study aimed to comprehensively compare the performance of the three-dimensional and two-dimensional deep learning models, radiomics model, and the fusion models for predicting occult LNM in LSCC.
Methods
In this retrospective diagnostic study, a total of 553 patients with clinical N0 stage LSCC, who underwent surgical treatment without distant metastasis and multiple primary cancers, were consecutively enrolled from four Chinese medical centres between January 01, 2016 and December 30, 2020. The participant data were manually retrieved from medical records, imaging databases, and pathology reports. The study cohort was divided into a training set (n = 300), an internal test set (n = 89), and two external test sets (n = 120 and 44, respectively). The three-dimensional deep learning (3D DL), two-dimensional deep learning (2D DL), and radiomics model were developed using CT images of the primary tumor. The clinical model was constructed based on clinical and radiological features. Two fusion strategies were utilized to develop the fusion model: the feature-based DLRad_FB model and the decision-based DLRad_DB model. The discriminative ability and correlation of 3D DL, 2D DL and radiomics features were analysed comprehensively. The performances of the predictive models were evaluated based on the pathological diagnosis.
Findings
The 3D DL features had superior discriminative ability and lower internal redundancy compared to 2D DL and radiomics features. The DLRad_DB model achieved the highest AUC (0.89–0.90) among all the study sets, significantly outperforming the clinical model (AUC = 0.73–0.78, P = 0.0001–0.042, Delong test). Compared to the DLRad_DB model, the AUC values for the DLRad_FB, 3D DL, 2D DL, and radiomics models were 0.82–0.84 (P = 0.025–0.46), 0.86–0.89 (P = 0.75–0.97), 0.83–0.86 (P = 0.029–0.66), and 0.79–0.82 (P = 0.0072–0.10), respectively in the study sets. Additionally, the DLRad_DB model exhibited the best sensitivity (82–88%) and specificity (79–85%) in the test sets.
Interpretation
The decision-based fusion model DLRad_DB, which combines 3D DL, 2D DL, radiomics, and clinical data, can be utilized to predict occult LNM in LSCC. This has the potential to minimize unnecessary lymph node dissection and prophylactic radiotherapy in patients with cN0 disease.
Funding
National Natural Science Foundation of China, Natural Science Foundation of Shandong Province.
Keywords: Deep learning, Artificial intelligence, Radiomics, Laryngeal cancer, Occult lymph node metastasis
Research in context.
Evidence before this study
We searched PubMed on August 6, 2023 using the term (“Laryngeal cancer” OR “Laryngeal squamous cell carcinoma”) AND (“occult lymph node metastasis” OR “lymph node metastasis”) AND (“Deep learning” OR “Convolutional neural network” OR “Artificial intelligence” OR “Radiomics”), without language restriction. We did not find any deep learning-based models. There is only two single-centre study using radiomics-based methods to predict lymph node metastasis (LNM) in laryngeal cancer (not occult LNM). We performed an updated search of PubMed on September 6, 2023 using the term (“Head Neck Cancer” OR “HNSCC”) AND (“occult lymph node metastasis” OR “lymph node metastasis”) AND (“Deep learning” OR “Convolutional neural network” OR “Artificial intelligence” OR “Radiomics”), without language restriction. We have identified 18 studies that utilized artificial intelligence (AI) methods based on MRI, CT, or PET for predicting LNM in oral cancer or head and neck squamous cell carcinoma (HNSCC). Among them, 6 studies reported on the prediction of occult LNM using radiomics in patients with oral cancer. These studies were all conducted in single-centre settings, with the largest sample size being 313 cases. There were no reports on using deep learning for predicting occult LNM in HNSCC.
Added value of this study
This study conducted a comparative analysis of three-dimensional and two-dimensional deep-learning, radiomics, clinical, and fusion models for predicting occult LNM in patients with laryngeal squamous cell carcinoma (LSCC). The study was conducted in multi-centre cohorts and excluded cases with obvious LNM features on radiology and physical examination. The findings suggested that the proposed DLRad_DB fusion model was a reasonable approach for predicting occult LNM in LSCC. Additionally, the performance of the multidomain fusion model could be influenced by the fusion strategy. The early fusion strategy may be susceptible to the correlation redundancy among features.
Implications of all the available evidence
Our study demonstrated that the proposed DLRad_DB model enabled early prediction of occult LNM in LSCC using CT images. This provides a foundation for clinical decision-making, minimizing unnecessary surgical interventions or radiotherapy. Additionally, the DLRad_DB model may have the potential to be applied to other HNSCC cancers.
Introduction
Laryngeal squamous cell carcinoma (LSCC) is the most prevalent type of cancer in the upper aerodigestive tract.1 In the United States, an estimated 12,380 new cases of laryngeal cancer are projected for 2023, with 3820 deaths expected from laryngeal cancer.2 Occult lymph node metastasis (LNM) refers to the presence of cancer cells in the lymph nodes that cannot be detected through routine imaging techniques or physical examination. These metastases are typically microscopic and do not cause visible enlargement or abnormalities. In early-stage LSCC (T1-2), occult LNM was estimated to occur in approximately 13–16% of cases. The rate of occult LNM was higher in advanced-stage LSCC (T3-4), ranging from about 20 to 25%.3,4 The presence of cervical LNM had been associated with a 50% reduction in overall survival, and regional lymph node recurrence was considered the main cause of treatment failure in LSCC.5,6 As a result, clinicians often perform prophylactic elective neck dissection (END) and radiotherapy in patients with cN0 LSCC, except for cases of T1-2 glottic larynx cancers where the risk of LNM is relatively low. However, it has been reported that prophylactic END or radiotherapy did not significantly improve survival compared to an observation strategy for early-stage LSCC.7 Moreover, these treatment approaches might lead to complications that can impair the quality of life.8 Therefore, accurate prediction of occult LNM preoperatively is crucial for the precise diagnosis and intervention of LSCC.
Various approaches have been proposed to predict occult LNM in LSCC, including clinical models, molecular markers, and sentinel lymph node biopsy.9, 10, 11 However, their performances and practicalities have been less satisfactory. In recent years, radiomics and deep learning (DL) techniques have demonstrated remarkable capabilities in disease diagnosis, molecular typing, and predicting therapeutic responses.12, 13, 14 Zhang et al. conducted a study using radiomics and DL methods to analyse CT of 276 patients with cervical enlarged lymph nodes. The study achieved a high predicted area under the curve (AUC) of 0.92 for metastatic lymph nodes.15 While studies demonstrated impressive performances, they primarily focused on analyzing lymph node images that typically displayed structurally abnormal characteristics in cases of metastasis.16 Few studies have focused on the challenge of predicting pathological LNM that cannot be visualized through imaging techniques. Previous studies have shown a correlation between radiological characteristics of primary tumors and LNM.17,18 Building on this knowledge, we hypothesized that artificial intelligence (AI) image analysis of primary sites may be a reasonable method for predicting occult LNM.
Medical images, such as CT and MRI, exist in three-dimensional (3D) spaces, and using a 2D method to address 3D tasks may result in the loss of crucial structural information. Training a 3D DCNN often requires larger datasets compared to a 2D DCNN, which poses a challenge for medical research due to limited data availability.19 Additionally, deep learning features may exhibit sensitivity to global translation, rotation, and scaling of images, while radiomics features do not possess such susceptibility.20 Consequently, the 3D/2D deep learning features and radiomics features may be complementary. Recent studies have shown that the multidomain fusion model of radiomics and deep learning outperforms individual models.21 Despite the promising perspective shown by the multidomain fusion approach, previous studies have mostly relied on a single fusion strategy or either 3D or 2D deep learning techniques.22,23 The combination of 2D and 3D deep learning and the impact of different fusion strategies on model performance have not been explored explicitly.
Collectively, our research aimed to compare the roles of 3D and 2D deep learning, radiomics, and fusion models in predicting occult LNM in LSCC. We employed two model fusion strategies: feature-based early fusion and decision-based late fusion. Furthermore, we investigated the intrinsic connections and distinctions among 3D/2D deep learning and radiomics features, which may help explain the performance differences between model fusion strategies.
Methods
Study cohort
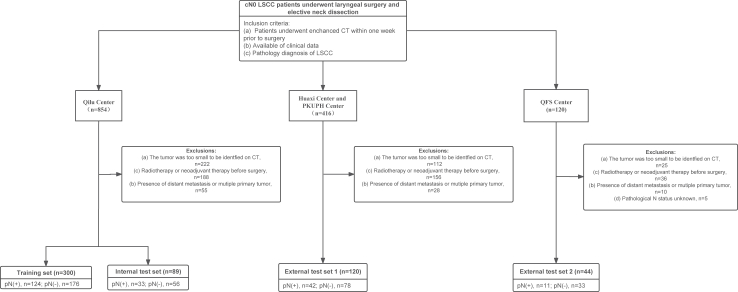
A total of 533 patients with cN0 LSCC from four Chinese hospitals were retrospectively collected (Fig. 1). The training set included 300 patients who were consecutively treated at Qilu Hospital of Shandong University (known as Qilu) from January 1 2016 to April 30, 2019. The internal test set included 89 patients who were consecutively treated at Qilu from May 1, 2019 to December 30, 2020. The external test set 1 included 120 patients who were consecutively treated at West China Hospital of Sichuan University (known as Huaxi, n = 62) and Peking University People’s Hospital (known as PKUPH, n = 58) from January 1, 2019 to December 30, 2020. The external test set 2 included 44 patients who were consecutively treated at the Shandong Provincial Qianfoshan Hospital (known as QFS) from January 1, 2019 to December 30, 2020. The inclusion criteria were as follows: (1) Clinical examination, B-ultrasonography, and CT/MRI confirmed lymph node stage cN0; (2) Pathological verification of laryngeal squamous cell carcinoma; (3) Enhanced CT examination performed within one week prior to surgery; (4) Initial treatment involved surgery without neoadjuvant chemoradiotherapy; (5) No evidence of distant metastasis or multiple primary cancers. The study followed the STARD reporting guideline. Detailed information about treatment protocols and clinical information were described in the Supplementary Methods. The overall workflow of this study is illustrated in Fig. 2.
Fig. 1.
Flowchart diagram shows the patient selection process from four medical centres. LSCC indicates laryngeal squamous cell carcinoma; PKUPH, Peking University People’s Hospital; QFS, Shandong Provincial Qianfoshan Hospital; pN (+), pathological lymph node positive; pN (−), pathological lymph node negative; and cN0, clinical N0 stage.
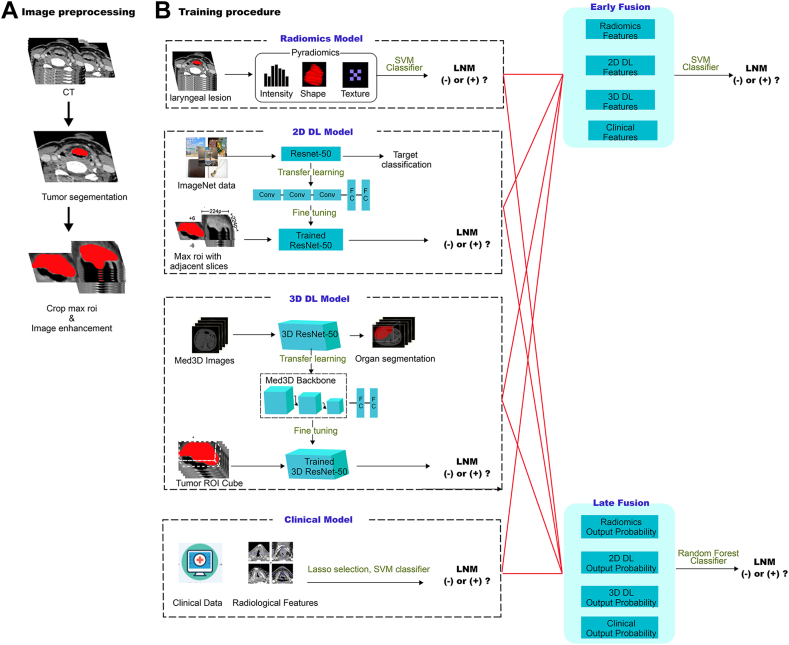
Fig. 2.
Workflow diagram for the development of the predictive models. Tumor segmentation and region of interest (ROI) delineation are performed by experienced radiologists. The Radiomics model is developed using PyRadiomics. For the two-dimensional deep learning (2D DL) model, the pre-trained ImageNet ResNet50 is fine-tuned based on our training data. The tumor's maximal ROI cross-section and six adjacent CT slices above and below it, are cropped as the input for ResNet50. The patient-level probability is calculated by averaging the probabilities of all CT slices belonging to one patient. For the three-dimensional deep learning (3D DL) model, the pre-trained 3D ResNet50 backbone is retained and transferred to optimize parameters in our training data. The clinical data and radiological features are used to construct the clinical model. For the early fusion model, the extracted features from four basic models are combined to train an SVM classifier. For the late fusion model, the output probabilities from four basic models are used to develop a stacking model with a random forest classifier.
Our study was approved by the ethics committee of Qilu Hospital, Shandong University (KYLL-2020(KS)-320). Since it was a retrospective study, informed consent was waived.
CT examination and image preprocessing
All patients included in the study underwent preoperative CT scans using a multi-slice CT system while in the supine position. The CT scans were performed from the skull base to the sternoclavicular joint. During the scanning process, patients were instructed to maintain calm respiration and avoid swallowing to minimize motion artifacts. Intravenous contrast-enhanced images were acquired with a delay of 60–65 s after the injection of the contrast agent. To achieve clear visualization of tissues in the head and neck region, the CT images were standardized using a window width of 350 and a window level of 50. The Hounsfield Unit (HU) values were adjusted to range from −125 to 225. Bi-cubic spline interpolation was used to resample the CT images to a voxel size of 1 mm∗1 mm∗1 mm.
Two experienced radiologists, in a blinded manner, utilized ITK-Snap software (v.3.6.0) to independently delineate tumor boundaries, creating regions of interest (ROI). Radiologists could refer to images from flexible nasopharyngoscopy to determine the boundaries of a tumor. These ROIs encompassed the primary tumor areas as well as areas susceptible to infiltration and inflammation. To assess the reliability and consistency of the ROI delineations, two months later, all patients' ROIs were re-annotated, and this data was used for intraclass correlation coefficient (ICC) analysis.
Radiomics feature extraction
In our study, we utilized PyRadiomics to extract the radiomics features.24 A total of 1834 radiomics features were extracted, including first-order features, shape-based features, and texture features. The detailed parameters regarding radiomics feature extraction are described in the Supplementary Methods and the PyRadiomics website (https://pyradiomics.readthedocs.io/en/latest/). The configuration file for feature extraction is provided in the Supplementary File.
Feature selection and model construction
The radiomics features were standardized using z-score normalization. To handle strong correlations between features (Spearman correlation coefficient ≥ 0.9), we employed a greedy recursive feature deletion strategy for feature filtering. This strategy entails iteratively removing the feature with the highest redundancy within the current feature set, until the current set no longer contains features with a correlation coefficient greater than 0.9. Next, features with high stability, as determined by intraobserver and interobserver ICC values exceeding 0.75, were also preserved.
To further refine the features, the multivariate least absolute shrinkage and selection operator (LASSO) regression was employed. The SVM (support vector machine) algorithm, known for its ability to identify optimal hyperplanes for distinguishing between different targets, has been used in many studies.25 We trained the SVM classifier to construct the predictive models. The five-fold cross-validation strategy was utilized in selecting the optimal model configuration and hyperparameter settings (Supplementary Methods). The SVM classifier received the training dataset as input, where each sample consisted of a set of features and a corresponding label indicating the presence of LNM. The output was a trained classifier model for predicting patients in the test set. The model output represented the probability (ranging from 0 to 1) of LNM in the tested patient.
Clinical model construction
The clinical characteristics and radiological features in Table 1 were used to construct the clinical model. These features were standardized using z-score normalization. Next, feature selection was performed using LASSO regression, followed by training an SVM classifier for classification prediction. Detailed procedures were described in the above section of "Feature Selection and Model Construction".
Table 1.
Baseline characteristics of study sets.a
| Characteristics | Training set (n = 300) |
Internal test set (n = 89) |
External test set 1 (n = 120) |
External test set 2 (n = 44) |
P valuec |
|---|---|---|---|---|---|
| Age (years)b | 62 (56–68) | 62 (55–67) | 62 (55–66) | 62 (57–68) | 0.82 |
| Sex | 0.063 | ||||
| Female | 11 (4) | 5 (6) | 1 (1) | 4 (9) | |
| Male | 289 (96) | 84 (94) | 119 (99) | 40 (91) | |
| Primary site | <0.0001 | ||||
| Glottic | 44 (15) | 11 (12) | 36 (30) | 17 (39) | |
| Supraglottic | 256 (85) | 78 (88) | 84 (70) | 27 (61) | |
| T stage | <0.0001 | ||||
| T1 | 96 (32) | 31 (35) | 33 (28) | 2 (5) | |
| T2 | 117 (39) | 34 (38) | 52 (43) | 6 (13) | |
| T3 | 67 (22) | 19 (21) | 29 (24) | 15 (34) | |
| T4 | 20 (7) | 6 (6) | 6 (5) | 21 (48) | |
| Differentiation status | 0.082 | ||||
| Well | 84 (28) | 33 (37) | 36 (30) | 7 (16) | |
| Moderate | 179 (60) | 46 (52) | 61 (51) | 28 (64) | |
| Poor or undifferentiated | 37 (12) | 10 (11) | 23 (19) | 9 (20) | |
| Radiological features-paraglottic/preepiglottic space invasion | 0.072 | ||||
| None | 259 (86) | 67 (75) | 96 (80) | 37 (84) | |
| Yes | 41 (14) | 22 (25) | 24 (20) | 7 (16) | |
| Radiological features-cartilage invasion | 0.47 | ||||
| None | 292 (97) | 85 (96) | 108 (90) | 42 (95) | |
| Yes | 8 (3) | 4 (4) | 12 (10) | 2 (5) | |
| Radiological features-maximum tumor diameter (cm)b | 3.2 (0.7–5.7) | 3.6 (1.2–6.6) | 3.7 (1.3–6.5) | 4.6 (0.5–8.1) | 0.22 |
| Radiological features-enhancement pattern | 0.55 | ||||
| Homogenous enhancement | 231 (77) | 72 (81) | 90 (75) | 32 (73) | |
| Inhomogeneous enhancement | 69 (23) | 17 (19) | 30 (25) | 12 (27) | |
| Pathological LNM | 124 (41) | 33 (37) | 42 (35) | 11 (25) | 0.17 |
Abbreviation: LNM, lymph node metastasis.
Unless otherwise indicated, data are numbers of patients, and data in parentheses are percentage.
Data are means, with IQRs in parentheses.
P value was calculated by Chi-square test or t-test.
2D DL model development and feature extraction
In 2D DL, a bounding box referred to the bounding rectangle of the largest tumor cross-section. It was used to crop the maximum tumor ROI and its adjacent six slices. Consequently, each patient comprised the maximum ROI of the primary tumor, as well as up to six adjacent patches above and below it. For small tumors, the number of adjacent CT slices may be fewer than six. After the cropping process, the images were resized to a standardized 256 × 256 pixel size using the linear interpolation method. Data enhancement strategies were employed, including random horizontal and vertical flips, and random cropping to a size of 224 × 224 pixels. The input image size was set to 224 × 224 pixels. The ResNet50 network was pre-trained using the ImageNet dataset, and transfer learning was subsequently performed on the training set. ImageNet is a large-scale image database that contains millions of labeled images across thousands of categories. ImageNet-based transfer learning has been used in many medical studies.14,26 We employed a global fine-tuning strategy to update the parameters, thereby adapting the ResNet50 for prediction of LNM.
Multiple patches from one patient were assigned identical labels indicating the presence of LNM. The images of primary tumors along with labels were used as input for the 2D DCNN. During the training process, the ResNet50 parameters were updated iteratively using backpropagation, and the cross-entropy loss function was employed, considering the output probability and pathology tags. The learning rate was set to 1 × 10−4, and the Adam optimizer was used to update the parameters. A batch size of 64 was utilized, and L2 regularization and early stop strategies were implemented to prevent overfitting (Supplementary Methods). The trained 2D DCNN could be used to predict the probability of LNM for each CT patch. The patient-level probability was then obtained by averaging the probabilities of all CT patches from one patient.27
After completing the training of ResNet50, we utilized the ResNet50 to extract 2048 deep learning features of each patch from the penultimate average pooling layer in ResNet50. The features were then compressed to a set of 256 features using PCA.
3D DL model development and feature extraction
Recognizing the limitations of 2D DCNNs in capturing the full extent of 3D structures, 3D ResNet was developed as an improvement over ResNet. 3D ResNet leverages automatic parameter learning to extract contextual features while preserving stereoscopic information.28
To address the scarcity of pretrained models on medical 3D images, the Med3D dataset, a multi-modal, multi-organ medical image segmentation dataset, was introduced.29 In our study, we employed the 3D ResNet50 pretrained on Med3D as our backbone. We replaced the decoder layers with fully connected layers.30 All layers in the 3D ResNet50 backbone were initialized using pre-trained weights and fine-tuned using our training set.
In 3D DL, the bounding box referred to the bounding cube of the tumor ROI. All ROI cubes were resized to a dimension of 96∗96∗96 using linear interpolation. Additionally, data augmentation techniques such as X, Y, and Z axis inversion strategies were employed to enhance the dataset. The 3D images of primary tumor along with their corresponding labels were then utilized as input for the 3D DCNN. The network parameters were updated through backpropagation and optimized using the Adam optimizer. We conducted 300 epochs of training, with a learning rate set to 0.02. Finally, the LNM of a patient could be predicted by the trained 3D DCNN.
To obtain the 3D DL features of an individual patient, a total of 2048 features of the tumor image were extracted from the penultimate averaging pooling layer of the 3D ResNet50 model. The features were then compressed to a set of 256 features using PCA.
Construction of the fusion model
The current study employed two fusion strategies to establish the fusion model.31
Feature-level fusion, also known as early fusion, involves connecting all features from different modalities into a single feature vector. The radiomics features of the primary tumor were extracted using PyRadiomics, while the 2D/3D DL features were obtained through DCNNs, as described above. These features, along with clinical and radiological characteristics, were standardized using z-score normalization. Subsequently, Spearman correlation analysis, ICC analysis, and LASSO analysis were performed to select the features. Finally, the SVM classifier was trained to construct the feature-based fusion model, known as DLRad_FB. Detailed methods are described in the above section “Feature selection and model construction”.
Decision-level fusion, also known as late fusion, combines the output probabilities of different models. We employed the stacking ensemble learning strategy to integrate the output probabilities from the four basic models.32 We evaluated the performance of three stacking models–Random Forest, Support Vector Machine, and Gradient Boosting Decision Tree–on the training set. After comparing their results (data not shown), we chose to use Random Forest classifier for further analysis. The optimal hyperparameters of the classifier were determined through a five-fold cross-validation method applied to the training set. Finally, the trained best stacking model “DLRad_DB” was evaluated in both the internal and external test sets.
Statistical analysis
Chi-square test or Fischer test was used to compare between categorical variables, Mann–Whitney U test or independent T-test was used for continuous variables. Receiver operating characteristic (ROC) curves and AUC, accuracy, sensitivity, and specificity were used to assess the performance of the predictive model. To determine the 95% Confidence Interval (CI) for the AUC, we employed the ci.auc function from the pROC R package. The Delong test was used to compare AUCs. P < 0.05 was considered statistically significant. All statistical analyses were performed using the R language (version 3.4.3) and the scikit-learn package (version: 0.18) in Python 3.70.
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. DPL, JL, and LHZ had access to the dataset and had the final responsibility for the decision to submit for publication.
Results
Patient characteristics
This study analysed a total of 553 patients (mean age, 61 years ±8 [SD]; 532 male) with cN0 LSCC, including 300 patients in the training set, 89 patients in the internal test set, 120 patients in the external test set 1, and 44 patients in the external test set 2. Regarding the 2D DL process, this study produced 7026 CT slices, with 3817 from the training set, 1150 from the internal test set, 1512 from the external test set 1, and 547 from the external test set 2. Detailed information is summarized in Table 1. Compared to the training set (glottic 15%, 44/300; supraglottic 85%, 256/300) and internal test set (glottic 12%, 11/89; supraglottic 88%, 78/89), both the external test set 1 (glottic 30%, 36/120; supraglottic 70%, 84/120) and external test set 2 (glottic 39%, 17/44; supraglottic 61%, 27/44) had a greater proportion of patients with glottic cancer (P < 0.05, pairwise comparison of Chi-square test). Additionally, the external test set 2 had a higher percentage of advanced T3-4 tumors (82%, 36/44) compared to the training set (29%, 87/300), internal test set (27%, 25/89), and external test set 1 (29%, 35/120, P < 0.05 after pairwise comparison of Chi-square test). There were no significant differences in the rate of occult LNM across all the study sets (P = 0.17).
In the training and internal test sets from Qilu centre, a total of 7284 lymph nodes were collected from 389 patients, revealing 416 lymph nodes to be pathologically positive for metastasis. The overall rate of occult LNM was 40.4% (157 of 389), with 98.1% (154 of 157) of these cases exhibiting ipsilateral or bilateral level II metastasis (Supplementary Table S1), suggesting that level II was the primary site for LSCC metastasis. In the external test set 1, there was 35% (42 of 120) of patients with occult lymph node metastasis, with 100% (42 of 42) of these cases exhibiting ipsilateral or bilateral level II metastasis. The rate of occult LNM was 25% (11 of 44) in the external test set 2. The information of metastatic region was not available from the external test set 2.
Comprehensive analysis of radiomics, 2D and 3D deep learning features
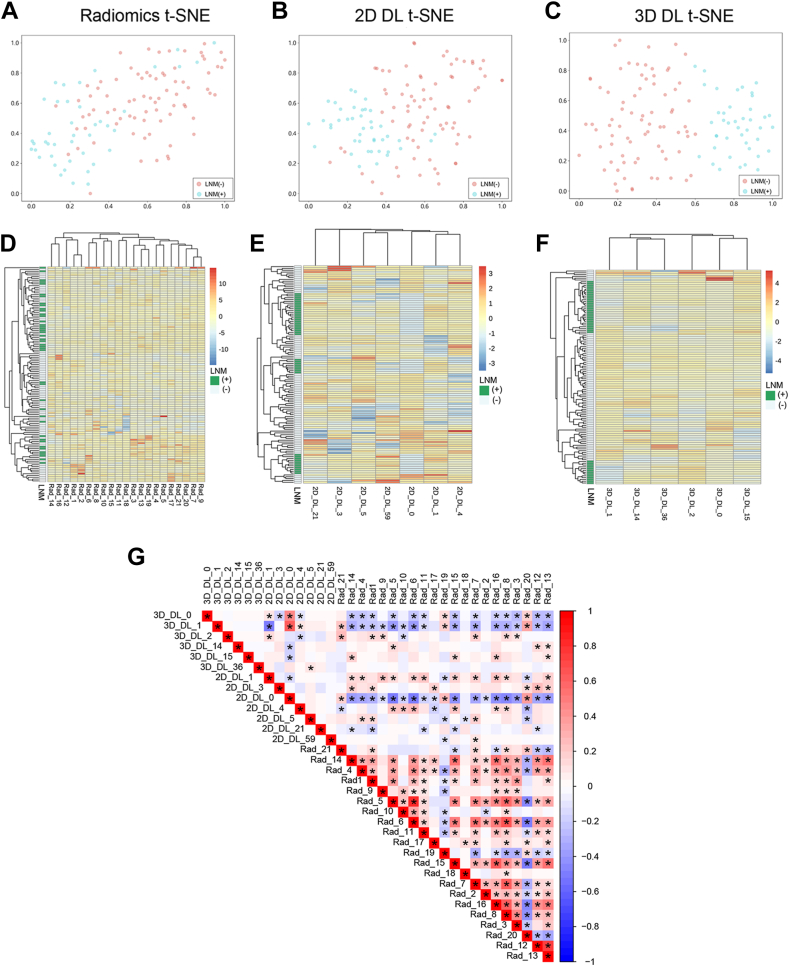
After conducting LASSO feature selection, we identified 21 key features out of 1834 radiomics features. Additionally, seven 2D DL features and six 3D DL features were found to have significantly high weights, and were considered as the top features (Supplementary Figure S1). T-SNE dimension reduction was used for visualization of these features. As shown in Fig. 3A–C and Supplementary Figure S2, compared with radiomics and 2D DL features, 3D DL features had better discriminative capabilities. The hierarchical clustering heatmap indicated that 3D DL features were more effective in distinguishing different LNM statuses (Fig. 3D–F and Supplementary Figure S3). The Spearman correlation analysis revealed a high degree of redundancy among the radiomics features (Fig. 3G), while there was practically no intrinsic correlation among the 3D DL features. Radiomics features exhibited partial correlations with both the 2D and 3D DL features; a few correlations were also observed between the latter two.
Fig. 3.
Discrimination ability and correlation analysis of radiomics and deep learning (DL) features in the external test set 1. T-distributed stochastic neighbour embedding (t-SNE) visualizations for the radiomics features (A), two-dimensional (2D) DL features (B), and three-dimensional (3D) DL features (C) in the external test set 1. Each dot represents a patient. Blue dots indicate patients with lymph node metastasis (LNM), and red dots indicate patients without LNM. Hierarchical clustering heatmap for key radiomics features (D), top 2D DL features (E), and top 3D DL features (F) in the external test set 1. The x-axis represents the IDs of radiomics and DL features, and the y-axis represents 120 patients. Patients belong to the same cluster (adjacent rows) share similar features in the Euclidean space. The status of LNM is displayed on the white-green bar located on the left side next to the y-axis. Supplementary Table S2 provides the descriptions of the key radiomics features. (G) The correlation heatmap of key radiomics features, top 2D and 3D DL features in external test set 1, where the color represents the magnitude of the spearmen correlation coefficient, and the asterisks indicate P < 0.05. Red indicates radiomics.
Performance analysis of the clinical, radiomics, 2D/3D deep learning models and the fusion models
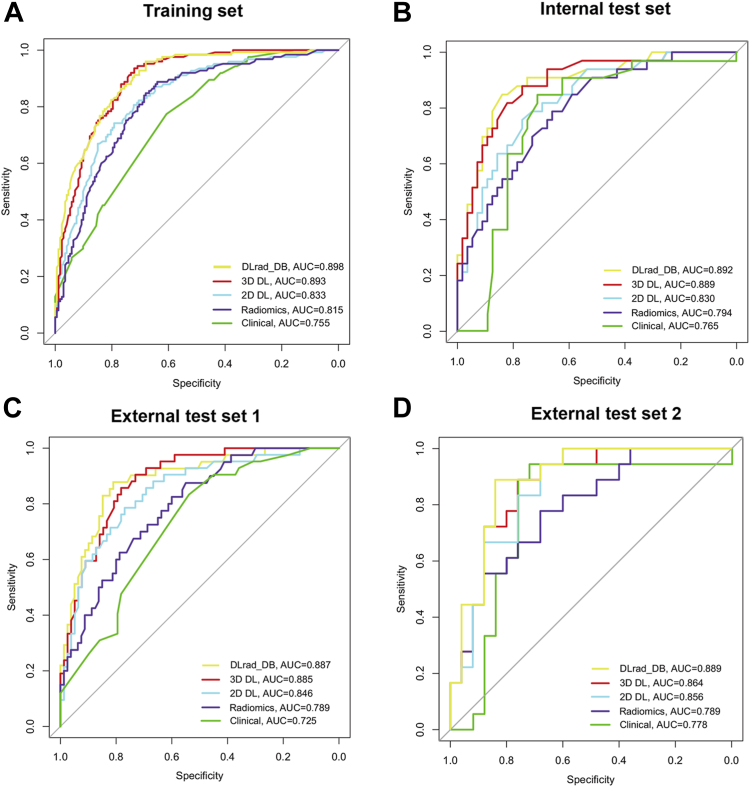
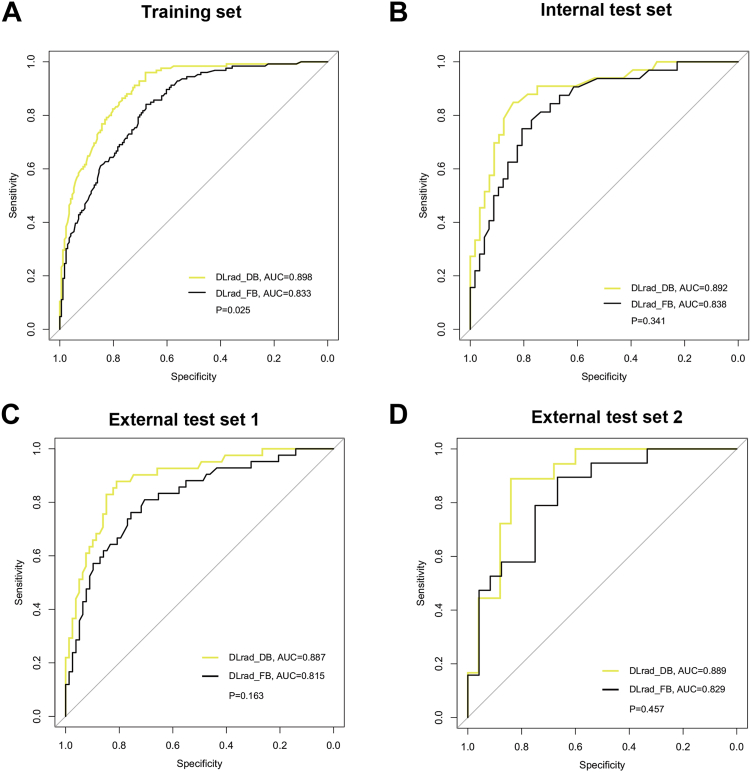
The diagnostic indicators of the predictive models in study sets are described in Table 2. The DLRad_DB model achieved the highest AUC (0.89–0.90) among all the study sets, significantly outperforming the clinical model (AUC = 0.73–0.78, P = 0.0001–0.042) (Fig. 4). Compared with the DLRad_DB model, the AUCs for the DLRad_FB, 3D DL, 2D DL, and radiomics models were 0.82–0.84 (P = 0.025–0.46), 0.86–0.89 (P = 0.75–0.97), 0.83–0.86 (P = 0.029–0.66), and 0.79–0.82 (P = 0.0072–0.10), respectively in the study sets. Furthermore, the 3D DL model demonstrated a satisfactory AUC of 0.89 in the training set, surpassing that of the 2D DL (AUC = 0.83, P = 0.042), radiomics (AUC = 0.82, P = 0.013), and clinical models (AUC = 0.76, P < 0.0001). In the test sets, no significant differences were observed in AUC between the 3D DL model (AUC = 0.86–0.89) and both the 2D DL model (AUC = 0.83–0.86, P = 0.30–0.91) and the radiomics model (AUC = 0.79, P = 0.062–0.40). The feature-based fusion model DLRad_FB did not demonstrate superiority in the study sets (Fig. 5). The performance of DLRad_FB (AUC = 0.83) in the training set was inferior to that of the 3D DL model (AUC = 0.89, P = 0.039) and the DLRad_DB model (AUC = 0.90, P = 0.025). In the test sets, there was no significant difference in performance between the DLRad_FB model (AUC = 0.82–0.84) and either the 3D DL model (AUC = 0.86–0.89, P = 0.17–0.67) or the DLRad_DB model (AUC = 0.89, P = 0.16–0.46). The calibration curves of the DLRad_DB model exhibited good consistency across a wider range of probabilities in the study sets (Supplementary Figure S4A–D). DCA curves demonstrated that the DLRad_DB model provided greater net benefit (Supplementary Figure S4E).
Table 2.
Performances of the predictive models in the training and test sets.a
| Model and metric | AUCb | Accuracy (%) | Sensitivity (%) | Specificity (%) | P valuec | P valuec | P valuec | P valuec |
|---|---|---|---|---|---|---|---|---|
| Training set (n = 300) | ||||||||
| DLRad_DB | 0.90 (0.86–0.93) | 82 (245/300) | 86 (107/124) | 78 (138/176) | Reference | 0.025 | 0.86 | <0.0001 |
| DLRad_FB | 0.83 (0.79–0.89) | 75 (225/300) | 82 (102/124) | 70 (123/176) | 0.025 | Reference | 0.039 | 0.028 |
| 3D DL | 0.89 (0.86–0.93) | 80 (239/300) | 82 (102/124) | 78 (137/176) | 0.86 | 0.039 | Reference | <0.0001 |
| 2D DL | 0.83 (0.79–0.88) | 77 (231/300) | 74 (92/124) | 79 (139/176) | 0.029 | 0.99 | 0.042 | 0.031 |
| Radiomics | 0.82 (0.77–0.86) | 75 (224/300) | 73 (91/124) | 76 (133/176) | 0.0072 | 0.61 | 0.013 | 0.10 |
| Clinical | 0.76 (0.70–0.81) | 63 (190/300) | 67 (83/124) | 61 (107/176) | <0.0001 | 0.028 | <0.0001 | Reference |
| Internal test set (n = 89) | ||||||||
| DLRad_DB | 0.89 (0.82–0.96) | 82 (73/89) | 88 (29/33) | 79 (44/56) | Reference | 0.34 | 0.97 | 0.042 |
| DLRad_FB | 0.84 (0.75–0.92) | 78 (69/89) | 79 (26/33) | 77 (43/56) | 0.34 | Reference | 0.36 | 0.29 |
| 3D DL | 0.89 (0.82–0.96) | 80 (71/89) | 85 (28/33) | 77 (43/56) | 0.97 | 0.36 | Reference | 0.043 |
| 2D DL | 0.83 (0.75–0.92) | 76 (68/89) | 76 (25/33) | 77 (43/56) | 0.28 | 0.91 | 0.30 | 0.35 |
| Radiomics | 0.79 (0.70–0.89) | 70 (62/89) | 73 (24/33) | 68 (38/56) | 0.10 | 0.50 | 0.11 | 0.69 |
| Clinical | 0.77 (0.66–0.85) | 67 (60/89) | 70 (23/33) | 66 (37/56) | 0.042 | 0.29 | 0.043 | Reference |
| External test set 1 (n = 120) | ||||||||
| DLRad_DB | 0.89 (0.83–0.95) | 83 (100/120) | 88 (37/42) | 81 (63/78) | Reference | 0.16 | 0.95 | 0.0042 |
| DLRad_FB | 0.82 (0.73–0.90) | 76 (91/120) | 74 (31/42) | 77 (60/78) | 0.16 | Reference | 0.17 | 0.15 |
| 3D DL | 0.89 (0.83–0.94) | 81 (97/120) | 83 (35/42) | 79 (62/78) | 0.95 | 0.17 | Reference | 0.0041 |
| 2D DL | 0.85 (0.77–0.92) | 78 (93/120) | 76 (32/42) | 78 (61/78) | 0.39 | 0.58 | 0.41 | 0.04 |
| Radiomics | 0.79 (0.71–0.87) | 70 (84/120) | 74 (31/42) | 68 (53/78) | 0.062 | 0.66 | 0.062 | 0.31 |
| Clinical | 0.73 (0.64–0.82) | 67 (80/120) | 69 (29/42) | 65 (51/78) | 0.0042 | 0.15 | 0.0041 | Reference |
| External test set 2 (n = 44) | ||||||||
| DLRad_DB | 0.89 (0.81–0.98) | 84 (37/44) | 82 (9/11) | 85 (28/33) | Reference | 0.46 | 0.75 | 0.042 |
| DLRad_FB | 0.83 (0.71–0.95) | 73 (32/44) | 64 (7/11) | 76 (25/33) | 0.46 | Reference | 0.67 | 0.62 |
| 3D DL | 0.86 (0.76–0.97) | 77 (34/44) | 82 (9/11) | 76 (25/33) | 0.75 | 0.67 | Reference | 0.40 |
| 2D DL | 0.86 (0.74–0.97) | 75 (33/44) | 73 (8/11) | 76 (25/33) | 0.66 | 0.75 | 0.91 | 0.43 |
| Radiomics | 0.79 (0.66–0.88) | 68 (30/44) | 73 (8/11) | 67 (22/33) | 0.083 | 0.67 | 0.40 | 0.83 |
| Clinical | 0.78 (0.66–0.85) | 73 (32/44) | 64 (7/11) | 76 (25/33) | 0.042 | 0.62 | 0.40 | Reference |
Abbreviations: AUC, area under the curve; DL, deep learning; 3D, three-dimensional; 2D, two-dimensional.
Bold text indicates that the P-value is less than 0.05.
Unless otherwise specified, data are percentages, with proportions of patients (numerator/denominator) in parentheses.
Data in parentheses are 95% CIs.
P value was calculated by the Delong test.
Fig. 4.
Performances for occult lymph node metastasis (LNM) prediction. The receiver operating characteristic (ROC) curves of the DLRad_DB model, three-dimensional (3D) deep learning (DL) model, two-dimensional (2D) DL model, radiomics model, and clinical model in the training set (A), internal test set (B), external test set 1 (C), and external test set 2 (D). AUC indicates area under the curve.
Fig. 5.
Performances for occult lymph node metastasis (LNM) prediction. The receiver operator characteristic (ROC) curves of the DLRad_DB model and the DLRad_FB model in the training set (A), internal test set (B), external test set 1 (C), and external test set 2 (D). AUC indicates area under the curve. P was calculated through the Delong test.
The optimal cutoff value of the predictive model was determined based on the maximum Youden index calculated in the training set. The optimal cutoff values were 0.555, 0.612, 0.512, 0.572, 0.645, and 0.672 for the DLRad_DB, DLRad_FB, 3D DL, 2D DL, radiomics, and clinical models, respectively. Based on the optimal cutoff values, the DLRad_DB exhibited the best sensitivity (82–88%) and specificity (79–85%) in the test sets (Table 2).
Stratification analysis of the model performances
It has been reported that the invasion depth and primary site of LSCC were related to LNM. We further performed stratification analysis of the performance of the predictive models. Overall, the T stage (T1-2 vs T3-4, Supplementary Figure S5) and primary tumor site (glottic vs supraglottic, Supplementary Figure S6) did not significantly affect performances of the predictive models. The radiomics, 2D DL, 3D DL, and DLRad_DB models exhibited stable performances among different clinical settings.
Discussion
Currently, there is no ideal method to predict the presence of occult LNM in LSCC.9,10 In this study, we developed radiomics, 2D and 3D DL models based on the CT images of primary tumor, alongside a clinical model, for the prediction of occult LNM. Two fusion strategies were employed to construct the multidomain fusion model: the feature-based DLRad_FB model and the decision-based DLRad_DB model. The results showed that the DLRad_DB model exhibited the best performance, with an AUC of 0.89 in test sets. The feature-based analysis indicated that 3D DL features exhibited the lowest internal redundancy and possessed superior discriminative ability. On the other hand, radiomics features demonstrated significant intrinsic redundancy and displayed multiple correlations with both 2D and 3D DL features.
The assessment of occult LNM in head and neck squamous cell carcinoma (HNSCC) has attracted much attention for a long time. Meler et al. examined the transcriptional expression of SEMA3F and NRP2 in patients with cN0 HNSCC, and found that the expressions of SEMA3F and NRP2 were independent risk factors for occult LNM.33 PET-CT can provide valuable information about the morphology and metabolic activity of lymph nodes, aiding in the detection of LNM. PET-CT has been shown to have a high negative predictive value ranging from 87% to 95%.34,35 However, PET sensitivity was compromised when it came to detecting sub-centimeter or micrometastasis LNM.36 Additionally, PET false positives were common due to the presence of inflammatory and infectious lymph nodes, which could interfere with accurate interpretation of PET scans.37
Using deep learning and radiomics to predict LNM is promising. This approach primarily relied on analyzing images of lymph nodes or primary tumors.15,16 In reality, radiological examinations often failed to show pathological lymph nodes measuring less than 3 mm in size.38 Additionally, the segmentation of lymph nodes is a time-intensive task that heavily relies on the expertise of radiologists. These limitations hinder the practical implementation of AI models that rely on lymph node imaging for accurate metastasis prediction. Yuan and Ren et al. utilized radiomics features derived from the primary tumor to predict occult LNM in tongue cancer. Their methods yielded promising results, with accuracies ranging from 0.74 to 0.82.39,40 Currently, there is a lack of studies utilizing deep learning techniques to predict occult LNM of HNSCC. In this study, cases with obvious signs of LNM on radiological and physical examination were excluded. Our findings indicated that the artificial intelligence model based on the primary tumor was a reasonable approach for predicting occult LNM in LSCC. In fact, occult LNM of LSCC was closely related to the primary tumor site and volume.41 Our clinical model also showed a relatively high AUC value of 0.73–0.78 in test sets.
3D CNNs require a larger dataset to effectively learn complex spatial features. Insufficient data can lead to suboptimal performance or overfitting.19 Kim et al. constructed a 2D Desnet model (they called 2.5D Desnet) by analyzing the axial, coronal and sagittal sections of pulmonary CT.42 The performance of the model for predicting lung cancer surpassed that of the 3D Desnet model. Chen et al. reported the advantages of 3D transfer learning.29 By utilizing the 3D ResNet backbone pretrained on Med3D, they achieved significant improvements in both speed and accuracy compared to de novo training. In the present study, we employed the Med3D database to pretrain 3D Resnet50. The 3D DL features demonstrated superior discriminative ability in comparison to 2D DL and radiomics features. In the training set, the 3D DL model also outperformed the 2D DL and radiomics models. In this study, the radiomics features exhibited significant internal correlations, whereas the 3D DL features showed low internal correlations. This observation could also be attributed to the utilization of PCA during the selection process of 3D DL features, which effectively reduced data dimensions and generated uncorrelated features.
A recent systematic review compared the performance of the radiomics model, DL model, and multidomain fusion model in medical researches.43 The findings revealed that the fusion model outperformed in 63% of the examined studies, underperformed in 25% of the studies, and had comparable performance in 13% of the studies. Our results suggested that the performance of the fusion model could be impacted by the fusion strategies. The majority of biomedical researches employed the early fusion strategy.44 However, the early fusion approach does not necessarily constitute the best practice. Huang et al. utilized a CT-based CNN and clinical data to detect pulmonary embolism, and compared seven fusion architectures.45 They found that the late fusion strategy yielded the most optimal performance, which was similar with our results. In this study, the correlation between deep learning features and radiomics features may lead to estimate instability, potentially compromising the performance of the feature-based DLRad_FB model. Therefore, when selecting a fusion approach for a specific medical issue, it is not advisable to solely rely on experience; conducting preliminary experiments for multiple approaches and selecting the most optimal one is recommended.
Our study presented a noninvasive preoperative approach utilizing an AI-based model to predict occult LNM in laryngeal cancer, thereby providing diagnostic assistance and facilitating informed treatment decisions. For high-risk patients predicted by the model, END or elective neck irradiation (ENI) was recommended, while minimizing the potential for surgical trauma and complications in low-risk patients. The AI model can also assist physicians in post-treatment surveillance, facilitating prompt identification of potential metastatic lesions and enabling timely implementation of appropriate therapeutic interventions. Furthermore, our model has the potential to be used for continuous learning and optimizing parameters in other types of HNSCC.46 A recent phase II clinical study predicted the metastasis of lymph nodes in HNSCC based on an AI algorithm, and strictly treated the affected and suspicious lymph nodes with intensive-modulated radiation therapy instead of ENI. They found that the AI-guided radiotherapy was oncological reasonable.47 This study offered valuable insights into the validation of AI-assisted diagnosis for LNM and the improvement of treatment strategies. However, the AI algorithm utilized in this single-centre study (n = 68) was based on lymph node images, thus failing to address the challenge of occult LNM. Consequently, in order to ascertain whether AI-driven diagnosis of occult LNM can effectively translate into therapeutic survival benefits, it is imperative to conduct additional prospective multicentre intervention trials that yield more compelling evidence.
It is important to consider the limitations of our study when interpreting the results. Firstly, this was a retrospective analysis, and prospective multicentre cases are needed to confirm our findings. Secondly, our study had a relatively high proportion of patients with T3-4 diseases. However, the prediction of occult LNM holds greater significance in early-stage LSCC. This is because END is more controversial in early-stage cases, whereas it is considered the standard treatment for advanced laryngeal cancer. Due to the small size of the primary tumor in early-stage LSCC and the limited number of cases with LNM labels, it was challenging to train an effective predictive model. Although our study included T3-4 LSCC, which may potentially impact the model’s performance in the T1-2 stages, our hierarchical analysis based on T stage did not support this claim. The models demonstrated comparable performance in both T1-2 and T3-4 stages. Additionally, the inclusion of additional T3-4 LSCC cases could potentially improve the generalizability of our predictive model in other types of HNSCC, assuming these cancers share similar imaging characteristics.
Another limitation of our model was that our model was unable to locate the region of occult LNM. However, our data indicated that the ipsilateral cervical level II was the most common site of LSCC metastasis (98.1%), which was consistent with previous studies (75%–95%).6,48 Therefore, for patients with high risk of LNM predicted by the model, we recommend performing a prophylactic END involving minimal ipsilateral level II-III.
In conclusion, the proposed multidomain fusion model DLRad_DB could be used for predicting occult LNM in LSCC based on pretreatment CT images. Although additional evidence is required before implementing the DLRad_DB model in a clinical setting, our study facilitates accurate preoperative diagnosis and informed treatment decision-making for LSCC.
Contributors
DPL, JL, and LHZ have accessed and verified the underlying data, and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Concept and design: DPL, JL, and LHZ.
Acquisition, analysis, or interpretation of data: WLW, HL, ZYZ, CYX, DMW, and WML.
Drafting of the manuscript: WLW.
Critical revision of the manuscript for important intellectual content: YQ and WML.
Statistical analysis: WLW.
Supervision: DPL, JL, and LHZ.
Data sharing statement
The code utilized in the study is accessible online (https://github.com/headneckwwl/occult-LNM). The data used in this research are not publicly available due to ethical considerations regarding the inclusion of facial information of patients. However, interested parties may be able to obtain access to the data through the necessary institutional approvals and signed agreements for data usage.
Declaration of interests
All authors declare no competing interests.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (82071918 to Dapeng Lei) and Natural Science Foundation of Shandong Province (ZR2023QH515 to Wenlun Wang). We would like to express our gratitude to OnekeyAI company for their technical assistance.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.eclinm.2023.102385.
Contributor Information
Lihong Zhang, Email: lihongzhang@vip.sina.com.
Jun Liu, Email: hxheadneckjunl@163.com.
Dapeng Lei, Email: leidapeng@sdu.edu.cn.
Appendix A. Supplementary data
References
- 1.Steuer C.E., El-Deiry M., Parks J.R., Higgins K.A., Saba N.F. An update on larynx cancer. CA Cancer J Clin. 2017;67(1):31–50. doi: 10.3322/caac.21386. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Wagle N.S., Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73(1):17–48. doi: 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- 3.Sharbel D.D., Abkemeier M., Groves M.W., Albergotti W.G., Byrd J.K., Reyes-Gelves C. Occult metastasis in laryngeal squamous cell carcinoma: a systematic review and meta-analysis. Ann Otol Rhinol Laryngol. 2021;130(1):67–77. doi: 10.1177/0003489420937744. [DOI] [PubMed] [Google Scholar]
- 4.Saito Y., Yoshimoto S., Kitayama M., et al. Elective neck dissection for T3-T4N0 laryngeal carcinoma: evidence from Japan's National Head and Neck Cancer Registry. Int J Clin Oncol. 2023;28(2):209–220. doi: 10.1007/s10147-022-02275-w. [DOI] [PubMed] [Google Scholar]
- 5.Petrarolha S., Dedivitis R., Matos L., Ramos D., Kulcsar M. Lymph node density as a predictive factor for worse outcomes in laryngeal cancer. Eur Arch Otorhinolaryngol. 2020;277(3):833–840. doi: 10.1007/s00405-020-05789-z. [DOI] [PubMed] [Google Scholar]
- 6.Wang S.X., Ning W.J., Zhang X.W., Tang P.Z., Li Z.J., Liu W.S. Predictors of occult lymph node metastasis and prognosis in patients with cN0 T1-T2 supraglottic laryngeal carcinoma: a retrospective study. ORL J Otorhinolaryngol Relat Spec. 2019;81(5–6):317–326. doi: 10.1159/000503007. [DOI] [PubMed] [Google Scholar]
- 7.Patel T.R., Eggerstedt M., Toor J., et al. Occult lymph node metastasis in early-stage glottic cancer in the national cancer database. Laryngoscope. 2021;131(4):E1139–E1146. doi: 10.1002/lary.28995. [DOI] [PubMed] [Google Scholar]
- 8.Deganello A., Gitti G., Meccariello G., Parrinello G., Mannelli G., Gallo O. Effectiveness and pitfalls of elective neck dissection in N0 laryngeal cancer. Acta Otorhinolaryngol Ital. 2011;31(4):216–221. [PMC free article] [PubMed] [Google Scholar]
- 9.Ma H., Lian M., Feng L., et al. Factors contributing to lymph node occult metastasis in supraglottic laryngeal carcinoma cT2-T4 N0M0 and metastasis predictive equation. Chin J Cancer Res. 2014;26(6):685–691. doi: 10.3978/j.issn.1000-9604.2014.12.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J., Zhang F., Hua M., Song X., Liu S., Dong Z. Prognostic value of lymphatic vessel density in oral squamous cell carcinoma. Life Sci. 2021;265 doi: 10.1016/j.lfs.2020.118746. [DOI] [PubMed] [Google Scholar]
- 11.Flach G.B., Bloemena E., van Schie A., et al. Sentinel node identification in laryngeal cancer: feasible in primary cancer with previously untreated neck. Oral Oncol. 2013;49(2):165–168. doi: 10.1016/j.oraloncology.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Wang H., Wang L., Lee E.H., et al. Decoding COVID-19 pneumonia: comparison of deep learning and radiomics CT image signatures. Eur J Nucl Med Mol Imaging. 2021;48(5):1478–1486. doi: 10.1007/s00259-020-05075-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le N.Q.K., Kha Q.H., Nguyen V.H., Chen Y.C., Cheng S.J., Chen C.Y. Machine learning-based radiomics signatures for EGFR and KRAS mutations prediction in non-small-cell lung cancer. Int J Mol Sci. 2021;22(17):9254. doi: 10.3390/ijms22179254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomita H., Kobayashi T., Takaya E., et al. Deep learning approach of diffusion-weighted imaging as an outcome predictor in laryngeal and hypopharyngeal cancer patients with radiotherapy-related curative treatment: a preliminary study. Eur Radiol. 2022;32(8):5353–5361. doi: 10.1007/s00330-022-08630-9. [DOI] [PubMed] [Google Scholar]
- 15.Zhang W., Peng J., Zhao S., et al. Deep learning combined with radiomics for the classification of enlarged cervical lymph nodes. J Cancer Res Clin Oncol. 2022;148(10):2773–2780. doi: 10.1007/s00432-022-04047-5. [DOI] [PubMed] [Google Scholar]
- 16.Chen L., Zhou Z., Sher D., et al. Combining many-objective radiomics and 3D convolutional neural network through evidential reasoning to predict lymph node metastasis in head and neck cancer. Phys Med Biol. 2019;64(7) doi: 10.1088/1361-6560/ab083a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang F., Tan R., Feng K., et al. Magnetic resonance imaging-based radiomics features associated with depth of invasion predicted lymph node metastasis and prognosis in tongue cancer. J Magn Reson Imaging. 2022;56(1):196–209. doi: 10.1002/jmri.28019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forghani R., Chatterjee A., Reinhold C., et al. Head and neck squamous cell carcinoma: prediction of cervical lymph node metastasis by dual-energy CT texture analysis with machine learning. Eur Radiol. 2019;29(11):6172–6181. doi: 10.1007/s00330-019-06159-y. [DOI] [PubMed] [Google Scholar]
- 19.Singh S.P., Wang L., Gupta S., Goli H., Padmanabhan P., Gulyás B. 3D deep learning on medical images: a review. Sensors. 2020;20(18):5097. doi: 10.3390/s20185097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gong Y., Wang L., Guo R., Lazebnik S. Springer International Publishing; Cham: 2014. Multi-scale orderless pooling of deep convolutional activation features; pp. 392–407. [Google Scholar]
- 21.Pease M., Arefan D., Barber J., et al. Outcome prediction in patients with severe traumatic brain injury using deep learning from head CT scans. Radiology. 2022;304(2):385–394. doi: 10.1148/radiol.212181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nie D., Lu J., Zhang H., et al. Multi-channel 3D deep feature learning for survival time prediction of brain tumor patients using multi-modal neuroimages. Sci Rep. 2019;9(1):1103. doi: 10.1038/s41598-018-37387-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y., Lobo-Mueller E.M., Karanicolas P., Gallinger S., Haider M.A., Khalvati F. Improving prognostic performance in resectable pancreatic ductal adenocarcinoma using radiomics and deep learning features fusion in CT images. Sci Rep. 2021;11(1):1378. doi: 10.1038/s41598-021-80998-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Griethuysen J.J.M., Fedorov A., Parmar C., et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(21):e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukherjee S., Patra A., Khasawneh H., et al. Radiomics-based machine-learning models can detect pancreatic cancer on prediagnostic computed tomography scans at a substantial lead time before clinical diagnosis. Gastroenterology. 2022;163(5):1435–1446.e3. doi: 10.1053/j.gastro.2022.06.066. [DOI] [PubMed] [Google Scholar]
- 26.Lang D.M., Peeken J.C., Combs S.E., Wilkens J.J., Bartzsch S. Deep learning based HPV status prediction for oropharyngeal cancer patients. Cancers. 2021;13(4):786. doi: 10.3390/cancers13040786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X., Yang W., Weinreb J., et al. Searching for prostate cancer by fully automated magnetic resonance imaging classification: deep learning versus non-deep learning. Sci Rep. 2017;7(1) doi: 10.1038/s41598-017-15720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hara K., Kataoka H., Satoh Y. 2018 IEEE/CVF conference on computer vision and pattern recognition; 2018 18-23 June 2018. 2018. Can spatiotemporal 3D CNNs retrace the history of 2D CNNs and ImageNet? pp. 6546–6555. [Google Scholar]
- 29.Chen S., Ma K., Zheng Y. Med3D: transfer learning for 3D medical image analysis. ArXiv. 2019 abs/1904.00625. [Google Scholar]
- 30.He K., Zhang X., Ren S., Sun J. Spatial pyramid pooling in deep convolutional networks for visual recognition. IEEE Trans Pattern Anal Mach Intell. 2015;37(9):1904–1916. doi: 10.1109/TPAMI.2015.2389824. [DOI] [PubMed] [Google Scholar]
- 31.Barata C., Emre Celebi M., Marques J.S. Melanoma detection algorithm based on feature fusion. Annu Int Conf IEEE Eng Med Biol Soc. 2015;2015:2653–2656. doi: 10.1109/EMBC.2015.7318937. [DOI] [PubMed] [Google Scholar]
- 32.Zhao S., Wang J., Jin C., et al. Stacking ensemble learning-based [(18)F]FDG PET radiomics for outcome prediction in diffuse large B-cell lymphoma. J Nucl Med. 2023;64(10):1603–1609. doi: 10.2967/jnumed.122.265244. [DOI] [PubMed] [Google Scholar]
- 33.Meler-Claramonte C., Avilés-Jurado F.X., Vilaseca I., et al. Semaphorin-3F/Neuropilin-2 transcriptional expression as a predictive biomarker of occult lymph node metastases in HNSCC. Cancers. 2022;14(9):2259. doi: 10.3390/cancers14092259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowe V.J., Duan F., Subramaniam R.M., et al. Multicenter trial of [(18)F]fluorodeoxyglucose positron emission tomography/computed tomography staging of head and neck cancer and negative predictive value and surgical impact in the N0 neck: results from ACRIN 6685. J Clin Oncol. 2019;37(20):1704–1712. doi: 10.1200/JCO.18.01182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao G., Sun J., Ba K., Zhang Y. Significance of PET-CT for detecting occult lymph node metastasis and affecting prognosis in early-stage tongue squamous cell carcinoma. Front Oncol. 2020;10:386. doi: 10.3389/fonc.2020.00386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim S.J., Pak K., Kim K. Diagnostic accuracy of F-18 FDG PET or PET/CT for detection of lymph node metastasis in clinically node negative head and neck cancer patients; a systematic review and meta-analysis. Am J Otolaryngol. 2019;40(2):297–305. doi: 10.1016/j.amjoto.2018.10.013. [DOI] [PubMed] [Google Scholar]
- 37.Zhang H., Seikaly H., Biron V.L., Jeffery C.C. Utility of PET-CT in detecting nodal metastasis in cN0 early stage oral cavity squamous cell carcinoma. Oral Oncol. 2018;80:89–92. doi: 10.1016/j.oraloncology.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 38.Laimer J., Lauinger A., Steinmassl O., et al. Cervical lymph node metastases in oral squamous cell carcinoma-how much imaging do we need? Diagnostics. 2020;10(4):199. doi: 10.3390/diagnostics10040199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan Y., Ren J., Tao X. Machine learning-based MRI texture analysis to predict occult lymph node metastasis in early-stage oral tongue squamous cell carcinoma. Eur Radiol. 2021;31(9):6429–6437. doi: 10.1007/s00330-021-07731-1. [DOI] [PubMed] [Google Scholar]
- 40.Ren J., Yuan Y., Tao X. Histogram analysis of diffusion-weighted imaging and dynamic contrast-enhanced MRI for predicting occult lymph node metastasis in early-stage oral tongue squamous cell carcinoma. Eur Radiol. 2022;32(4):2739–2747. doi: 10.1007/s00330-021-08310-0. [DOI] [PubMed] [Google Scholar]
- 41.Sanabria A., Shah J.P., Medina J.E., et al. Incidence of occult lymph node metastasis in primary larynx squamous cell carcinoma, by subsite, T classification and neck level: a systematic review. Cancers. 2020;12(4):1059. doi: 10.3390/cancers12041059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim H., Lee D., Cho W.S., et al. CT-based deep learning model to differentiate invasive pulmonary adenocarcinomas appearing as subsolid nodules among surgical candidates: comparison of the diagnostic performance with a size-based logistic model and radiologists. Eur Radiol. 2020;30(6):3295–3305. doi: 10.1007/s00330-019-06628-4. [DOI] [PubMed] [Google Scholar]
- 43.Demircioğlu A. Are deep models in radiomics performing better than generic models? A systematic review. Eur Radiol Exp. 2023;7(1):11. doi: 10.1186/s41747-023-00325-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mohsen F., Ali H., El Hajj N., Shah Z. Artificial intelligence-based methods for fusion of electronic health records and imaging data. Sci Rep. 2022;12(1) doi: 10.1038/s41598-022-22514-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang S.C., Pareek A., Zamanian R., Banerjee I., Lungren M.P. Multimodal fusion with deep neural networks for leveraging CT imaging and electronic health record: a case-study in pulmonary embolism detection. Sci Rep. 2020;10(1) doi: 10.1038/s41598-020-78888-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bándi P., Balkenhol M., van Dijk M., et al. Continual learning strategies for cancer-independent detection of lymph node metastases. Med Image Anal. 2023;85 doi: 10.1016/j.media.2023.102755. [DOI] [PubMed] [Google Scholar]
- 47.Sher D.J., Moon D.H., Vo D., et al. Efficacy and quality-of-life following involved nodal radiotherapy for head and neck squamous cell carcinoma: the INRT-AIR phase II clinical trial. Clin Cancer Res. 2023;29(17):3284–3291. doi: 10.1158/1078-0432.CCR-23-0334. [DOI] [PubMed] [Google Scholar]
- 48.Djordjevic V., Bukurov B., Arsovic N., et al. Prospective case-control study of efficacy of bilateral selective neck dissection in primary surgical treatment of supraglottic laryngeal cancers with clinically negative cervical findings (N0) Clin Otolaryngol. 2016;41(6):634–639. doi: 10.1111/coa.12570. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.