Abstract
Background
Neutrophil extracellular traps (NETs) have repeatedly been related to COVID-19 severity and mortality. However, there is no consensus on their quantification, and there are scarce data on their evolution during the disease. We studied circulating NET markers in patients with COVID-19 throughout their hospitalization.
Methods
We prospectively included 93 patients (201 blood samples), evaluating the disease severity in 3 evolutionary phases (viral, early, and late inflammation). Of these, 72 had 180 samples in various phases. We also evaluated 55 controls with similar age, sex and comorbidities. We measured 4 NET markers in serum: cfDNA, CitH3, and MPO-DNA and NE-DNA complexes; as well as neutrophil-related cytokines IL-8 and G-CSF.
Results
The COVID-19 group had higher CitH3 (28.29 vs 20.29 pg/mL, p = 0.022), and cfDNA, MPO-DNA, and NE-DNA (7.87 vs 2.56 ng/mL; 0.80 vs 0.52 and 1.04 vs 0.72, respectively, p < 0.001 for all) than the controls throughout hospitalisation. cfDNA was the only NET marker clearly related to severity, and it remained higher in non-survivors during the 3 phases. Only cfDNA was an independent risk factor for mortality and need for intensive care. Neutrophil count, IL-8, and G-CSF were significantly related to severity. MPO-DNA and NE-DNA showed significant correlations (r: 0.483, p < 0.001), including all 3 phases and across all severity grades, and they only remained significantly higher on days 10–16 of evolution in those who died. Correlations among the other NET markers were lower than expected.
Conclusions
The circulating biomarkers of NETs were present in patients with COVID-19 throughout hospitalization. cfDNA was associated with severity and mortality, but the three other markers showed little or no association with these outcomes. Neutrophil activity and neutrophil count were also associated with severity. MPO-DNA and NE-DNA better reflected NET formation. cfDNA appeared to be more associated with overall tissue damage; previous widespread use of this marker could have overestimated the relationship between NETs and severity. Currently, there are limitations to accurate NET markers measurement that make it difficult to assess its true role in COVID-19 pathogenesis.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12931-023-02650-9.
Keywords: COVID-19, Neutrophil extracellular traps, NET markers, Longitudinal study, Severity, Mortality
Summary at a Glance
Circulating markers evidenced NETs formation by Covid-19 throughout hospitalization and in all grades of severity, but showed limited relationship with severity and mortality, and different degrees of specificity, making it difficult to assess their true role in the disease outcomes.
Background
Elevated neutrophil levels are early indicators of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and severe disease [1–5]. Their effector functions are multiple, including enzyme degranulation, pathogen destruction by reactive oxygen species (ROS), phagocytosis, and neutrophil extracellular traps (NETs). NETs are networks of chromatin, intranuclear proteins (histones), and granule enzymes present in extracellular medium after neutrophilic destruction, to further control bacterial, fungal, viral, or parasitic infection [6]. This cell death mechanism, or NETosis, is distinct from apoptosis and necrosis, with an alternative mechanism, exocytosis of granules and nuclear material, termed vital or non-lytic NETosis [7, 8]. Peptidylarginine deiminase (PAD4) activation results in histone citrullination [9], although it is not known whether citrullination is a cause or a consequence of histone externalization. All these products, in cases of excessive production and/or reduced clearance [10, 11], can cause significant tissue damage. NETosis triggers are multiple, including virus-damaged epithelial cells, activated platelets and endothelial cells, and inflammatory cytokines, including interleukin (IL)-1, IL-8, and granulocyte colony-stimulating factor (GCSF) [2]. During the COVID-19 pandemic, certain NET components, such as cell-free DNA (cfDNA) and extracellular histones, have been shown to be damage-associated molecular patterns (DAMPs) from damaged cells, which regardless of their origin have been involved in its pathogenesis [12]. The presence of potential circulating NET markers [2, 4, 5, 12–14], in respiratory samples [13, 15] or in tissues [5, 15–18] has been associated with severity and mortality.
NET quantification typically assesses the simultaneous presence of its components as biomarkers, including cfDNA, histones (H3) or citrullinated histones (CitH3), myeloperoxidase (MPO) isolated or in complexes with DNA (MPO-DNA), neutrophil elastase (NE), and NE-DNA complexes. Those considered more specific are those less likely to originate from activation and death of neutrophils other than NETs, such as from other cells or damaged tissues, or incidentally from molecular interactions in plasma. The choice of markers is a potential source of contradictory conclusions, and few studies exist in longitudinal cohorts to improve knowledge of their predictive power. After having demonstrated the usefulness of cfDNA as a prognostic marker in patients hospitalised for COVID-19 [19], we aimed to check whether these elevated figures were related to NETs, studying the most specific NET markers longitudinally and throughout the hospitalization of our patient group.
Since the beginning of the pandemic, the disease progression time has been considered important for prognostic and therapeutic measures. From the onset of symptoms, there is an initial viral replication and elimination phase lasting approximately 7 days, followed by a decrease in viral presence and increased inflammation (early inflammation phase). In the case of progression, there is disbalance of the immune response by day 16 and later (late inflammation phase), and potentially life-threatening pulmonary and systemic alterations can occur [20]. Our objective was to study the evolution of circulating markers of neutrophilic activation and NETs, including those considered to have high specificity [21], throughout the hospitalization of patients with Covid-19 and their relationship with severity and mortality.
Our hypotheses:
NETs markers are increased in patients with COVID-19 throughout their hospital evolution.
These markers are associated with severity or mortality at some point in the evolution of hospitalised patients with COVID-19.
Methods
Patients and controls
Patients with COVID-19 hospitalised in April and May 2020 were prospectively enrolled. Exclusion criteria were refusal to sign informed consent, concomitant infection during hospitalization, and immunosuppression (transplant recipients, hematologic malignancies, chemotherapy, or prednisone equivalent ≥ 20 mg/day). The patients were grouped according to severity, based on the Chinese Center for Disease Control and Prevention (CCDC) classification [22] and the World Health Organization Ordinal Scale (WHO OS) [23] as follows: moderately ill (MI), severely ill (SI), and critically ill (CI). According to the day of symptom onset, 3 phases were considered: viral (1–9 days from symptom onset), early inflammation (10–16 days from symptom onset), and late inflammation (> 16 days from symptom onset).
We selected 55 non-hospitalised controls without any infection, of similar age and chronic pathologies as the patients, all of whom underwent routine laboratory tests. All met the same exclusion criteria as the patient group. To compare the three dependent groups (severity groups or the three disease progression phases), we would need a total sample size of 100 (33 by group).
Blood samples
The samples were obtained from blood requested for healthcare reasons. Patients with at least 2 blood samples in 2 consecutive phases were included in the longitudinal study. We measured neutrophil count, cytokines related to neutrophil activation and recruitment (IL-8, G-CSF), and 4 NET markers: cfDNA, CitH3, and MPO-DNA and NE-DNA complexes.
IL-8 and G-CSF
IL-8 and G-CSF in serum were quantified employing a bead-based multiplex immunoassay for R&D (LXSAHM-07) and a LABSCAN 100 (Luminex corporation). Data were analysed with Xponent software.
NE-DNA and MPO-DNA complexes
Serum samples were analysed by enzyme-linked immunosorbent assay (ELISA) of NE-DNA and MPO-DNA complexes, employing anti-NE and anti-MPO antibodies (07–496-I and MABS461 from Sigma Aldrich) and peroxidase conjugate anti-DNA antibody (Cell Death Detection ELISA Kit 154467500 Roche) as previously described [24].
Citrullinated histone H3
Serum samples were analysed employing a citrullinated histone H3 (Clone 11D3) ELISA kit (Cayman 501620) for quantification according to the manufacturer’s instructions, and 450-nm ODs were read in a Sinergy HT reader (Biotek).
Cell-free DNA
DNA was purified following the manufacturer’s procedure (ChargeSwitch gDNA 1 mL Serum Kit), adjusting the reactant quantity to the serum sample volume (100 μL). PicoGreen reagent was added in a 1:1 ratio to the purified sample (previously diluted to 1/5 in TE buffer), and the mix was incubated 5 min at room temperature in the dark (Quant-iT PicoGreen dsDNA Assay Kit, Invitrogen P7589). The Synergy HT reader (Biotek) was used to read the sample’s fluorescence (excitation 485 nm, emission 528 nm), and cfDNA concentrations were calculated with the standard provided in the kit.
Statistical analysis
According to normal criteria (Shapiro–Wilk test), data were presented as mean (standard deviation) or median (interquartile range) and analyzed with the Mann–Whitney U, Kruskal–Wallis, Student’s, or analysis of variance test, as appropriate. Correlations between biomarkers and clinical variables were analysed with Spearman’s rho correlation. Receiver operating characteristic (ROC) analysis was performed to determine the predictive power of biomarkers for various outcomes. A Wilcoxon signed-rank test was performed for the longitudinal study.
Logistic regression was used for specific outcomes, adjusting for age > 60 years, male sex, hypertension, obesity, diabetes, and corticosteroid therapy. The variables selected for the multivariate model were age, sex, and those with a p-value less than 0.05 in the bivariate analysis. All statistical analyses were performed with R version 4.0.5 (R Core Team 2021), and P-values < 0.05 were considered significant. The study was approved (03/04/2020) by IRB Aragón (Aragon Research Ethics Committee [CEICA]) (07/2020).
Results
Patients and controls
We collected 201 samples from 93 patients, 72 of whom provided samples (180) from at least 2 disease progression stages. Of the 180 samples from the 72 patients, 87 corresponded to the viral phase, 53 to the early inflammatory phase and 40 to the late inflammatory phase. Thirty-two patients had three or more samples, and eight patients had samples from all three disease progression phases. The median number of days from symptoms onset for the viral, early inflammatory, and late inflammatory phase samples were 6 (5–8), 12 (11–14), and 20 (18–24), respectively. Twelve patients required admission to the intensive care unit (ICU); 19 died. There were no statistically significant differences in the patients’ (n = 93) and controls’ (n = 55) demographics (age and sex) or comorbidities (hypertension; diabetes; obesity; respiratory, cardiac, renal, or hepatic disease; dementia; dyslipidemia) (p > 0.05 for all) (Additional file 1: Table S1). At admission, 56 of the study 93 patients had moderate disease, 29 had severe disease, and 8 were initially classified as critical. Table 1 presents the patients’ information according to severity and samples at each stage.
Table 1.
Number of patients in each severity group and number of samples of the patients followed longitudinally, according to time from symptom onset
| WHO OS scale | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | Total |
|---|---|---|---|---|---|---|---|---|---|
| Patients (n) | 0 | 0 | 7 | 41 | 20 | 1 | 5 | 19 | 93 |
| CCDC scale | Mild-moderate | Severe | Critical | Total | |||||
|---|---|---|---|---|---|---|---|---|---|
| Patients (n) | 51 | 15 | 27 | 93 | |||||
| Illness onset | Viral phase | Early inflammation | Late inflammation | Total | |||||
|---|---|---|---|---|---|---|---|---|---|
| Samples (n) from 93 patients | 97 | 59 | 45 | 93 | |||||
| Samples (n) from 72 patients | 87 | 53 | 40 | ||||||
CCDC scale Chinese Center for Disease Control and Prevention classification, WHO OS World Health Organization Ordinal Scale
NETs markers in patients and controls
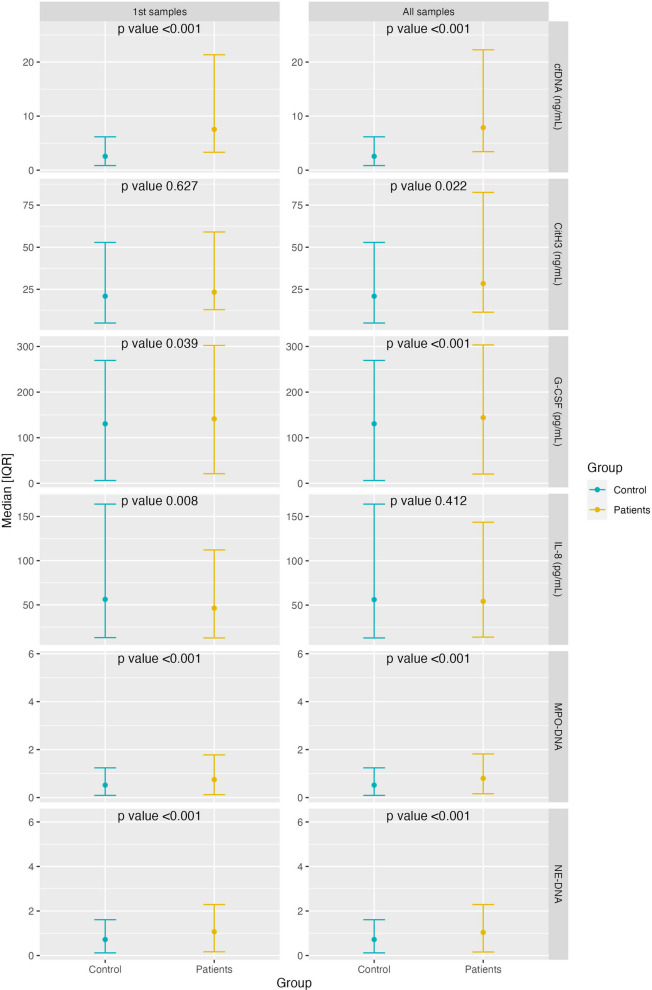
On admission, we observed elevated levels compared with controls in 3 of the 4 NET markers studied: cfDNA and MPO-DNA and NE-DNA complexes (p < 0.001 for all) (Fig. 1). The same occurred in the first 9 days of disease evolution (viral phase), and from days 10–16 (early inflammation phase), in which we also observed elevated CitH3 values (p = 0.003). These differences were maintained from day 17 (late inflammation phase) (p < 0.001 for all 4 markers) (Table 2). Even the MI patients (according to WHO OS) presented increased values in the 4 NET markers: MPO-DNA (p < 0.001), NE-DNA (p < 0.001), CitH3 (p = 0.05), and cfDNA (p < 0.001). In the SI and CI patients together, all 4 NETs markers remained significantly elevated relative to controls (p = 0.041 for CitH3 and p < 0.001 for the others).
Fig. 1.
Comparison of biomarkers between patients and controls for all samples and for only the first sample after admission. Median comparison was performed using Wilcoxon signed-rank test
Table 2.
Comparison of biomarkers between patients and controls during the three disease progression phases
| Viral phase (1–9 days) |
Patients N = 93 |
Controls N = 55 |
p-value |
|---|---|---|---|
| IL-8 (pg/mL) | 50.31 [34.62; 69.05] | 56.21 [43.26; 107.75] | 0.025 |
| G-CSF (pg/mL) | 138.42 [120.43; 159.45] | 130.47 [124.32; 138.97] | 0.032 |
| MPO-DNA | 0.75 [0.63; 0.95] | 0.52 [0.43; 0.72] | < 0.001 |
| NE-DNA | 1.06 [0.87; 1.25] | 0.72 [0.60; 0.89] | < 0.001 |
| cfDNA (ng/mL) | 6.67 [4.04; 13.69] | 2.56 [1.71; 3.61] | < 0.001 |
| CitH3 (ng/mL) | 23.35 [11.32; 34.99] | 20.91 [15.92; 31.94] | 0.599 |
| Early inflammation (10–16 days) |
Patients N = 59 |
Controls N = 55 |
p-value |
|---|---|---|---|
| IL-8 (pg/mL) | 55.02 [42.51; 88.89] | 56.21 [43.26; 107.75] | 0.725 |
| G-CSF (pg/mL) | 146.39 [129.14; 162.05] | 130.47 [124.32; 138.97] | 0.001 |
| MPO-DNA | 0.90 [0.64; 1.10] | 0.52 [0.43; 0.72] | < 0.001 |
| NE-DNA | 1.03 [0.87; 1.25] | 0.72 [0.60; 0.89] | < 0.001 |
| cfDNA (ng/mL) | 7.51 [5.04; 11.66] | 2.56 [1.71; 3.61] | < 0.001 |
| CitH3 (ng/mL) | 35.89 [21.49; 68.04] | 20.91 [15.92; 31.94] | 0.003 |
| Late inflammation (> 16 days) | Patients N = 45 |
Controls N = 55 |
p-value |
|---|---|---|---|
| IL-8 (pg/mL) | 78.18 [50.36; 111.21] | 56.21 [43.26; 107.75] | 0.093 |
| G-CSF (pg/mL) | 149.48 [132.43;157.64] | 130.47 [124.32; 138.97] | < 0.001 |
| MPO-DNA | 0.87 [0.74; 1.05] | 0.52 [0.43; 0.72] | < 0.001 |
| NE-DNA | 1.00 [0.88; 1.32] | 0.72 [0.60; 0.89] | < 0.001 |
| cfDNA (ng/mL) | 11.24 [5.77; 17.77] | 2.56 [1.71;3.61] | < 0.001 |
| CitH3 (ng/mL) | 54.54 [28.72; 89.53] | 20.91 [15.92; 31.94] | < 0.001 |
NET markers, neutrophil count, IL-8, G-CSF, and severity and mortality
Among the 4 NETs markers, we only found significant cfDNA increases in MI vs SI and CI, but not in CitH3, MPO-DNA, or NE-DNA complexes. The same occurred comparing MI and SI vs CI (CCDC) (Table 3). Of the 4 NET markers, only cfDNA showed significant increases in CI vs non-CI (CCDC and WHO OS scales), in the first sample and in the 3 phases of the study. Neutrophil count, IL-8, and G-CSF were significantly related to severity (Table 3). cfDNA was also the single NET marker with higher levels in non-survivors compared with survivors, including in the total sample (p < 0.001), at admission, and in the 3 phases of the disease. The same occurred with neutrophil count. MPO-DNA and NE-DNA showed higher values in non-survivors, but only in the early inflammation phase (Table 4). The non-survivors showed elevated neutrophils, IL-8, and cfDNA in the first sample after admission. There were no differences in CitH3 levels between non-survivors and survivors at any time. In our multivariate model, only cfDNA and obesity were independent risk factors for mortality and need for ICU (Table 5, and Additional file 2: Table S2). The area under the curve (AUC) ROC for critical status and mortality was clearly higher for cfDNA than for the other parameters (Table 6). Analysis examining the relationship between NETs markers and sex, age, and obesity only revealed a statistically significant association between cfDNA and sex (p = 0.002), with higher levels observed in males (14.7 ng/mL) compared to females (7.5 ng/mL). However, this association was not observed in the control group (p = 0.317) (Additional file 3: Table S3). Sex-biased studies were performed for the different outcomes and the same results were obtained regardless of sex (Additional file 4: Table S4).
Table 3.
Comparisons of biomarkers according to severity (CCDC scale and WHO OS)
| Severe and critical | Moderate | p-value | |
|---|---|---|---|
| CCDC scale | |||
| Neutrophils/mm3 | 6700 [4575; 10725] | 4600 [3500; 6400] | < 0.001 |
| IL-8 (pg/mL) | 58.22 [46.90; 97.08] | 49.89 [35.59; 80.94] | 0.015 |
| G-CSF (pg/mL) | 148.16 [126.10; 163.97] | 142.01 [122.64; 156.55] | 0.162 |
| MPO-DNA | 0.84 [0.64; 1.03] | 0.78 [0.63; 0.99] | 0.563 |
| NE-DNA | 0.98 [0.86; 1.29] | 1.06 [0.91; 1.24] | 0.394 |
| cfDNA (ng/mL) | 9.44 [6.15; 20.19] | 6.21 [3.46; 10.44] | < 0.001 |
| CitH3 (ng/mL) | 28.28 [16.19; 57.92] | 28.43 [17.93; 51.13] | 0.670 |
| WHO OS | |||
| Neutrophils/mm3 | 6600 [4500; 10775] | 4600 [3550; 6350] | < 0.001 |
| IL-8 (pg/mL) | 58.80 [46.72; 102.29] | 49.15 [36.85; 72.20] | 0.007 |
| G-CSF (pg/mL) | 150.75 [131.66; 168.10] | 136.76 [120.48; 153.66] | 0.002 |
| MPO-DNA | 0.80 [0.64; 1.02] | 0.80 [0.64; 1.00] | 0.873 |
| NE-DNA | 1.04 [0.87; 1.31] | 1.05 [0.89; 1.24] | 0.945 |
| cfDNA (ng/mL) | 9.08 [5.54; 19.38] | 6.88 [3.61; 10.73] | 0.001 |
| CitH3 (ng/mL) | 31.79 [15.08; 57.53] | 26.80 [18.37; 49.48] | 0.388 |
| Critical | Moderate and severe | p-value | |
|---|---|---|---|
| CCDC scale | |||
| Neutrophils/mm3 | 8250 [5400; 12100] | 4900 [3700; 6600] | < 0.001 |
| IL-8 (pg/mL) | 68.60 [47.40; 94.41] | 52.57 [37.56; 82.75] | 0.058 |
| G-CSF (pg/mL) | 152.80 [131.66; 177.42] | 140.17 [122.64; 156.55] | 0.009 |
| MPO-DNA | 0.84 [0.68; 1.01] | 0.78 [0.63; 1.01] | 0.470 |
| NE-DNA | 0.99 [0.86; 1.35] | 1.05 [0.89; 1.24] | 0.669 |
| cfDNA (ng/mL) | 14.11 [7.13; 22.67] | 7.01 [4.01; 10.93] | < 0.001 |
| CitH3 (ng/mL) | 27.67 [12.88; 53.68] | 30.58 [19.08; 54.70] | 0.324 |
Table 4.
Comparisons of median biomarker levels between survivors and non-survivors
| A | |||
|---|---|---|---|
| Total samples (n = 201) |
Survivors (n = 158) | Non-survivors (n = 43) | p-value |
| Neutrophils/mm3 | 5100 [3700; 6700] | 8400 [6050; 13600] | < 0.001 |
| IL-8 (pg/mL) | 52.82 [37.84; 83.46] | 69.12 [49.89; 104.92] | 0.043 |
| G-CSF (pg/mL) | 142.57 [122.64; 157.28] | 149.55 [130.26; 165.07] | 0.090 |
| MPO-DNA | 0.80 [0.63; 1.00] | 0.80 [0.71; 1.02] | 0.517 |
| NE-DNA | 1.04 [0.87; 1.23] | 1.05 [0.90; 1.48] | 0.439 |
| cfDNA (ng/mL) | 7.06 [4.12; 11.40] | 14.98 [7.42; 25.74] | < 0.001 |
| CitH3 (ng/mL) | 28.29 [18.10; 53.97] | 29.29 [12.34; 58.58] | 0.745 |
| 1st sample (n = 93) | Survivors (n = 74) | Non-survivors (n = 19) | p-value |
|---|---|---|---|
| Neutrophils/mm3 | 5050 [3725; 6550] | 8400 [4950; 11750] | 0.009 |
| IL-8 (pg/mL) | 42.11 [33.18; 56.48] | 61.83 [52.88; 89.06] | 0.016 |
| G-CSF (pg/mL) | 134.22 [118.12; 153.99] | 160.09 [139.92; 184.10] | 0.012 |
| MPO-DNA | 0.75 [0.63; 1.01] | 0.77 [0.71; 1.01] | 0.508 |
| NE-DNA | 1.07 [0.88; 1.19] | 1.02 [0.94; 1.57] | 0.559 |
| cfDNA (ng/mL) | 6.87 [4.01; 10.81] | 14.98 [7.73; 22.51] | 0.002 |
| CitH3 (ng/mL) | 24.51 [12.61; 34.99] | 14.80 [6.70; 29.79] | 0.129 |
| B | |||
|---|---|---|---|
| Viral phase (1–9 days) |
Survivors N =78 |
Non-survivors N =19 |
p-value |
| Neutrophils/mm3 | 4500 [3150; 6600] | 7000 [4600; 10550] | 0.011 |
| IL-8 (pg/mL) | 49.03 [34.24; 61.20] | 54.29 [42.30; 74.82] | 0.269 |
| G-CSF (pg/mL) | 136.45 [118.12; 153.91] | 155.74 [128.91; 168.02] | 0.069 |
| MPO-DNA | 0.72 [0.63; 0.95] | 0.75 [0.65; 0.90] | 0.988 |
| NE-DNA | 1.07 [0.91; 1.23] | 0.96 [0.78; 1.34] | 0.342 |
| cfDNA (ng/mL) | 6.11 [3.48; 11.45] | 7.87 [5.08; 22.21] | 0.037 |
| CitH3 (ng/mL) | 24.51 [13.76; 34.99] | 16.19 [7.76; 33.13] | 0.295 |
| Early inflammation (10 –16 days) |
Survivors N = 48 |
Non-survivors N = 11 |
p-value |
|---|---|---|---|
| Neutrophils/mm3 | 5600 [4275; 7000] | 11400 [7650; 14450] | < 0.001 |
| IL-8 (pg/mL) | 51.93 [40.97; 81.89] | 77.53 [60.24; 110.09] | 0.052 |
| G-CSF (pg/mL) | 146.64 [129.14; 159.15] | 142.57 [125.89; 175.26] | 0.731 |
| MPO-DNA | 0.85 [0.61; 1.03] | 1.61 [0.90; 2.87] | 0.026 |
| NE-DNA | 0.99 [0.86; 1.14] | 1.37 [1.10; 1.68] | 0.016 |
| cfDNA (ng/mL) | 7.11 [4.46; 9.75] | 12.59 [7.88; 26.36] | 0.008 |
| CitH3 (ng/mL) | 35.89 [21.92; 65.96] | 34.49 [17.23; 67.53] | 1.000 |
| Late inflammation (>16 days) | Survivors N = 32 |
Non-survivors N = 13 |
p-value |
|---|---|---|---|
| Neutrophils/mm3 | 5250 [4075; 6625] | 8400.00 [6500.00; 16400.00] | 0.001 |
| IL-8 (pg/mL) | 79.03 [49.63; 110.03] | 75.08 [51.72; 107.04] | 0.966 |
| G-CSF (pg/mL) | 149.55 [135.01; 156.55] | 149.42 [131.66; 161.75] | 0.920 |
| MPO-DNA | 0.87 [0.77; 1.05] | 0.78 [0.69; 1.02] | 0.466 |
| NE-DNA | 0.99 [0.88; 1.33] | 1.06 [0.90; 1.11] | 0.548 |
| cfDNA (ng/mL) | 8.35 [4.91; 12.94] | 24.62 [19.21; 39.06] | < 0.001 |
| CitH3 (ng/mL) | 54.13 [29.46; 79.69] | 73.28 [34.28; 102.09] | 0.520 |
(MPO-DNA and NE-DNA complex, cfDNA and CitH3) in all samples (n = 201)A: total samples and first sample; B: throughout the 3 evolutionary phases
Table 5.
Multivariate model for ICU need
| ICU admittance | Yes | No | OR univariate | OR multivariate |
|---|---|---|---|---|
|
Age > 60 years n (%) |
51 (85.0) | 9 (15.0) | 1.76 (0.48–8.41, p = 0.421) | 0.40 (0.01–9.74, p = 0.599) |
|
Male sex n (%) |
34 (85.0) | 8 (19.0) | 2.76 (0.80–11.06, p = 0.119) | 1.84 (0.08–63.05, p = 0.695) |
|
Hypertension n (%) |
40 (88.9) | 5 (11.1) | 0.73 (0.20–2.48, p = 0.619) | – |
|
Obesity n (%) |
7 (53.8) | 6 (46.2) | 10.57 (2.69–43.67, p = 0.001) | 46.21 (3.76–2138.71, p = 0.011) |
| Corticosteroid therapy n (%) | 36 (76.6) | 11 (23.4) | 13.75 (2.50–257.24, p = 0.014 | 19.62 (1.07–2584.67, p = 0.114) |
|
Diabetes n (%) |
25 (89.3) | 3 (10.7) | 0.75 (0.16–2.75, p = 0.680) | – |
|
IL-8 Mean (SD) |
71.3 (97.6) | 67.9 (32.7) | 1.00 (0.99–1.01, p = 0.909) | – |
|
G-CSF Mean (SD) |
199.5 (324.7) | 170.9 (41.8) | 1.00 (0.99–1.00, p = 0.775) | – |
|
cfDNA Mean (SD) |
8.7 (6.5) | 23.7 (17.4) | 1.14 (1.06–1.25, p = 0.001) | 1.22 (1.07–1.53, p = 0.025) |
|
MPO-DNA Mean (SD) |
0.9 (0.6) | 1.1 (0.8) | 1.57 (0.61–3.46, p = 0.272) | – |
|
NE-DNA Mean (SD) |
1.1 (0.4) | 1.4 (0.7) | 2.49 (0.78–7.95, p = 0.100) | – |
|
CitH3 Mean (SD) |
30.1 (25.8) | 18.8 (15.3) | 0.97 (0.92–1.00, p = 0.174) | – |
Bold highlights parameters with significant differences (p < 0.05)
Table 6.
AUC ROC of NET markers for severity and mortality
| Marker | AUC | Threshold | Specificity | Sensitivity |
|---|---|---|---|---|
| AUC ROC for critical status | ||||
| cfDNA | 0.716 (0.63–0.8) | 14.10 | 0.509 | 0.818 |
| MPO-DNA | 0.535 (0.44–0.63) | 0.71 | 0.740 | 0.379 |
| NE-DNA | 0.520 (0.42–0.62) | 0.94 | 0.431 | 0.691 |
| CitH3 | 0.546 (0.45–0.65) | 17.14 | 0.396 | 0.803 |
| AUC ROC for mortality | ||||
| cfDNA | 0.748 (0.66–0.84) | 20.17 | 0.947 | 0.450 |
| MPO-DNA | 0.465 (0.36–0.57) | 0.75 | 0.579 | 0.459 |
| NE-DNA | 0.540 (0.43–0.65) | 1.53 | 0.899 | 0.256 |
| CitH3 | 0.483 (0.37–0.6) | 73.18 | 0.880 | 0.250 |
Evolution of NET markers throughout the disease course
In the longitudinal study, the MPO-DNA and NE-DNA complexes remained elevated and stable during the 3 phases of the disease. cfDNA increased, especially in the late inflammation phase (p = 0.043) compared with the viral phase; levels of CitH3 progressively increased throughout the 3 phases (p < 0.001), doubling between the first and last sample obtained (p < 0.001) (Table 7).
Table 7.
Comparison of biomarkers among the three disease progression phases
| Viral phase (1) N = 87 |
Early inflammation (2) N = 53 |
Late inflammation (3) N = 40 |
p-value | p-value 1 vs 2 | p-value 1 vs 3 | p-value 2 vs 3 | |
|---|---|---|---|---|---|---|---|
| Neutrophils/mm3 | 5100 [3350; 6950] | 6000 [4400; 8700] | 6050 [4100; 8225] | 0.069 | 0.119 | 0.119 | 0.874 |
| IL-8 (pg/mL) | 52.01 [34.96; 68.72] | 55.52 [45.64; 91.01] | 77.32 [50.36; 106.06] | 0.002 | 0.048 | 0.002 | 0.108 |
| G-CSF (pg/mL) | 139.08 [120.98; 160.64] | 147.85 [129.84; 162.19] | 148.79 [132.43; 157.64] | 0.478 | 0.488 | 0.488 | 0.964 |
| MPO-DNA | 0.75 [0.62; 0.94] | 0.87 [0.63; 1.03] | 0.87 [0.72; 1.03] | 0.092 | 0.211 | 0.112 | 0.833 |
| NE-DNA | 1.05 [0.88; 1.25] | 1.03 [0.86; 1.25] | 0.97 [0.88; 1.27] | 0.909 | 0.868 | 0.868 | 0.868 |
| cfDNA (ng/mL) | 6.40 [3.61; 13.99] | 7.87 [5.34; 14.65] | 10.53 [5.56; 17.30] | 0.041 | 0.200 | 0.043 | 0.200 |
| CitH3 (ng/mL) | 23.64 [12.73; 35.10] | 35.89 [21.92; 61.36] | 54.54 [28.72; 91.81] | < 0.001 | 0.001 | < 0.001 | 0.043 |
Median comparison was performed using Wilcoxon signed-rank test
Correlations
There were significant correlations between MPO-DNA and NE-DNA complexes (r = 0.483, p < 0.001), and less correlation between cfDNA and CitH3 (r = 0.277, p < 0.001) and between cfDNA and NE-DNA (r = 0.168, p = 0.022) (Table 8). We found no other correlations among the 4 NET markers. There were significant correlations between MPO-DNA and NE-DNA (r = 0.793, p < 0.001 in MI; r = 0.472, p < 0.001 in SI; and r = 0.627, p < 0.001 in CI [WHO OS]), which remained significant from the sample at hospital arrival (r = 0.586, p < 0.001) and during the 3 phases (r = 0.705, p < 0.001 in viral; r = 0.581, p < 0.001 in early inflammation; and r = 0.476, p = 0.002 in late inflammation). We found no correlation between CitH3 and MPO-DNA or NE-DNA. There were less strong, but significant, correlations between cfDNA and NE-DNA in CI (r = 0.297, p = 0.043) and between cfDNA and CitH3 in CI (r = 0.371, p = 0.009) and SI (r = 0.289, p = 0.004) (WHO OS). In the total sample, cfDNA was the only NET marker that correlated with typical clinical severity factors, such as lymphopenia, neutrophil/lymphocyte ratio, C-reactive protein, lactate dehydrogenase levels, and baseline SaO2 (See Additional file 5: Table S5).
Table 8.
Correlations between 4 circulating NET biomarkers
| Total samples (n = 201) | r | p |
|---|---|---|
| cfDNA vs MPO-DNA | 0.095 | 0.203 |
| cfDNA vs NE-DNA | 0.168 | 0.022 |
| cfDNA vs CitH3 | 0.277 | < 0.001 |
| MPO-DNA vs NE-DNA | 0.483 | < 0.001 |
| MPO-DNA vs CitH3 | 0.019 | 0.797 |
| NE-DNA vs CitH3 | − 0.010 | 0.891 |
Discussion
The most important results of our study included a clear increase in circulating NET markers in patients with COVID-19 compared with controls, confirming NET formation in hospitalised patients with COVID-19, from admission throughout hospitalization, including less severe cases. cfDNA was the NET marker most strongly associated with severity and mortality, from the first sample and throughout the disease. It was an independent risk factor and showed the best AUC ROC for both outcomes. However, we found less association than expected between the other 3 NET markers and poor outcomes: only increased MPO-DNA and NE-DNA complexes during the second week (days 10–16 from symptom onset) in non-surviving patients. We observed a significant correlation between markers considered highly specific for NETosis: MPO-DNA and NE-DNA complexes, during the entire evolution and at all severity levels, were much higher than that of the other markers among each other. Throughout the evolution, neutrophil count increased with severity and mortality, and IL-8 increased with severity.
The presence of NET biomarkers in patients with COVID-19 has generated interest [2, 4, 5, 12, 18, 25]; most studies have found increased markers in these patients with respect to healthy controls [2, 5, 12, 13, 26]. Increased NET formation from ex vivo neutrophils of patients with COVID-19 [27] and impaired NET degradation in those with COVID-19 have been reported [5, 25].
In our study, every NETosis marker remained elevated from admission (CitH3 from day 10 after the onset of the disease), during the entire hospitalization and at the 3 levels of severity. This, and the high MPO-DNA and NE-DNA correlations in the 3 levels of severity and during the entire evolution, offer little doubt as to the existence of NETosis in hospitalised patients with COVID-19, including in less severe patients. Interestingly, the delayed onset and increased CitH3 suggest that citrullination is a consequence rather than a cause of histone externalization, which is a matter of debate [9]. Other studies have concluded that circulating NET marker levels are related to severity and mortality in COVID-19, and that NETs could play an important role in pathogenesis [2, 4, 5, 12, 14, 15, 26, 28–30], including their identification in fatal disease tissues [5, 30] and the observation of small pulmonary vessel occlusion by NET aggregates [5, 16]. This NETs–severity relationship is the dominant opinion today [11].
However, contradictory results have been observed for the relationship between circulating NET markers and COVID-19 severity [5, 31, 32], viral load [13, 30], or associated obstructive vascular phenomena [5, 13, 16]. Some reasons could include the non-standardized choice of NET markers, which varies between studies (including some less specific ones); an apparent discrepancy between the markers in tissues and in peripheral blood [13]), as well as between serum or plasma [33]; levels can vary significantly with underlying chronic diseases [5]; and some appear to be associated with severity more than others [5]. Furthermore, plasma levels of markers considered specific, such as MPO-DNA appear to be less sensitive in discriminating severity than other NETs measurement methods, such as NETosis induction in plasma samples, as has been recently reported in patients with COVID-19 [32]. In addition, the heterogeneity of the populations, the short half-life of NET markers [34], and the variety of enzymatic pathways triggering NETs [13] can modify the markers and lead to divergent results.
In addition to cfDNA, included in most of the previous studies, we chose 3 NET markers considered the most specific: CitH3, and MPO-DNA and NE-DNA complexes [21]. It is difficult to distinguish NETosis markers from those of neutrophil activation or other types of cell death [35, 36]. cfDNA or nucleosomes (DNA formations with histones or nuclear proteins, such as H3) can also originate from necrotic processes. MPO and NE frequently originate from neutrophil degranulation; however, their binding to DNA (NE-DNA and MPO-DNA complexes used in our study) likely makes them the most specific for NETs, because they are less likely to be formed incidentally by molecular interactions in plasma [21, 35]. Citrullinated histones, such as CitH3, although considered specific, represent only one NETosis pathway (via PAD4) [37]; their levels may be similar in children with COVID-19 as in healthy children [38], and can vary depending on stimulus [39]. A recent study found a much stronger relationship between COVID-19 severity (and long COVID) and increased NETosis-induction capacity measured ex vivo, than that of circulating NET-specific marker levels (MPO-DNA complex) [32].
The lack of correlations observed between some markers considered specific, such as CitH3 with MPO-DNA and NE-DNA, has been reported in COVID-19 between CitH3 and MPO-DNA [2], and between CitH4-DNA and NE-DNA, which showed opposite trends in COVID-19 severity assessment [5]. In another study, CitH3-NE complexes did not differ from those of controls [31]. The simultaneous involvement of alternative non-PDA4 pathway-dependent NETosis in COVID-19 [2, 40] and the existence of sequestered NET fragments in damaged organs could explain the discrepancies between measurements of circulating citrullinated histones and other markers and with those of tissues [31].
Also, neutrophils can generate markers such as cfDNA and CitH3 by mechanisms other than NETosis [2, 12]. Therefore, the use of citrullinated histones (CitH3) as specific NET markers, despite frequent employment, is controversial [35, 37]. Lastly, markers with the potential to detect NETs have not been designed, and it is unclear whether NET markers are drivers of disease severity, a simple consequence of acute inflammation [2], or both.
The clear association of cfDNA, neutrophil counts, and IL-8 with severity and mortality, and the (much poorer) relationship of the more specific markers of NETosis suggest an important role for neutrophils in severe COVID-19 cases [30, 41]; however, it is difficult to establish a clear relationship between circulating NET markers and these outcomes. The strong correlation of the 2 markers considered the more specific for NETs, MPO-DNA and NE-DNA, in all severity groups and throughout hospitalization, and the very limited correlation of the remaining biomarkers with each other, suggest that MPO-DNA and NE-DNA are the best circulating NETosis markers. Overall, the results of our study suggest, like others [2, 12, 42, 43], that cfDNA is not a marker with high specificity for NETosis, because it is also released by various hematopoietic cells and from a wide range of tissues after cell destruction, as has been demonstrated in COVID-19 [19, 34, 42]. Also, cfDNA is a DAMP, capable of amplifying the inflammatory response [44, 45] via toll-like receptor 9 (TLR-9) [44]. NET formation is likely better reflected by more neutrophil-specific markers, such as MPO-DNA and NE-DNA, whereas cfDNA better reflects cellular damage in a broader sense, correlating with disease severity parameters and better predicting disease outcomes [2, 12]. The widespread inclusion of cfDNA in previous studies could have overestimated the role of NETs in COVID-19 severity. The relationship detected between cfDNA and sex (p = 0.002) only in the COVID group (not in the controls) supports that males have a higher immune response and tissue damage than females, as has been previously reported [19].
On days 10–16, when viral control appears to be achieved in those with a good prognosis, the significantly higher blood levels of MPO-DNA and NE-DNA complexes in those who died suggest increased NET production, concurrent with a lack of viral clearance in the second week of evolution [46], associated with poor prognoses [47]. Our results suggest that NET status varies at different time points in the evolution of COVID-19; that an increase in NET release during the second week of disease progression, probably associated with a higher viral load, can contribute to poorer outcomes; and that NET detection during this period could predict its evolution. An association between NETs and COVID-19 severity could encourage new treatment approaches, such as DNases [25], recombinant human DNase-I [48], IL-8 receptor antagonists [30], certain monoclonal antibodies, PAD4 or nicotinamide adenine dinucleotide phosphate (NADPH) inhibitors [49], Fostamatinib [50], Resolvin T-series [51], and current and novel histone inhibitors [36].
Our study has limitations. Our cohort was small, from a single hospital. We were unable to measure viral load and thus demonstrate a parallel association of increased and sustained viral load and NET formation on days 10–16 in patients with poor prognosis. Quantification problems for NETosis and cytokines have been mentioned. The strengths of our study include that there is scarce information on NETosis markers on patients with COVID-19 progression. As far as we know, ours is the first study systematically including 3 evolutive phases, including the NET biomarker most used in previous studies and 3 markers of those considered more specific combined. Our controls were not healthy, as in most studies, but had comorbidities similar to those of our patients, which gives greater value to comparisons of their parameters with those of the patients.
Conclusions
Our study has demonstrated the presence of increased markers of both circulating NETs and neutrophil activation and recruitment at all COVID-19 severity levels, from admission throughout the evolution of hospitalised patients, but suggests differences in specificity and quality of some NETosis markers. Although previous studies have reported that these marker levels could be related to disease severity, at present there is no evidence that SARS-CoV-2 can directly induce NET production, and more information is needed on the possible mechanism by which NETs would promote COVID-19 progression [2, 29]. Our results contradict in some ways some of the other previous results. However, in our previous study with the same samples, AUCs for other markers well known for their association with outcomes in COVID-19, such as IL-6, were validated [19]. It seems clear that, with our current limitations in accurately measuring NETs by circulating markers, it is difficult to reach definitive conclusions about the proper role of NETs in the pathogenesis of COVID-19.
Supplementary Information
Additional file 1: Table S1. Demographics and comorbidities of the patients and controls
Additional file 2: Table S2: Detailed view of the multivariate model for severity, mortality, and need for ICU.
Additional file 3: Table S3. Relationship between NET markers and sex, age, and obesity separately in patients and controls.
Additional file 4: Table S4. Sex-biased analysis of NET markers and mortality in total samples.
Additional file 5: Table S5. Correlations between usual clinical parameters of severity, neutrophil counts, neutrophil associated cytokines (IL-8 and G-CSF), and NET markers
Acknowledgements
We thank Mar Gonzalez Cantalejo, head of the library of the Miguel Servet University Hospital and all its staff for their invaluable help in the availability of data and materials.
Abbreviations
- CCDC score
Chinese Center for Disease Control and Prevention classification
- cfDNA
Cell-free DNA
- CI
Critically ill
- CitH3
Citrullinated histone 3
- CitH3-NE
Citrullinated histone 3-neutrophil elastase complexes
- CitH4-DNA
Citrullinated histone 4-DNA complexes
- DAMPs
Damage-associated molecular patterns
- GCSF
Granulocyte colony-stimulating factor
- H3
Histone 3
- IL-1
Interleukin 1
- IL-8
Interleukin 8
- MI
Moderately ill
- MPO-DNA
Myeloperoxidase-DNA complexes
- NADPH
Nicotinamide adenine dinucleotide phosphate
- NE-DNA
Neutrophilic elastase-DNA complexes
- NETs
Neutrophil extracellular traps
- PAD4
Peptidylarginine deiminase
- ROS
Reactive oxygen species
- SI
Severely ill
- TLR-9
Toll like receptor 9
- WHO OS
World Health Organization ordinal scale
Author contributions
Conception and design: SB, ABL, CDD. Acquisition of data: LLV, CDD, LT, PRG, ATE, RL, CA, CB. Analysis and interpretation of data: JG, AC, BJ, ABL, SB, CDD. Writing of the manuscript: SB, ABL, CDD. Drafting or revising of the article: SB, ABL, CDD. Final approval of the manuscript: SB, ABL, LLV, CDD, LT, PRG, RL, CA, JG, AC, BJ, CB, ATE.
Funding
This work was supported with funds from the Pneumoaragon Foundation of the Aragonese Society of Pneumology (SADAR).
Availability of data and materials
The datasets supporting the conclusions of this article are available in the Zenodo repository, in https://zenodo.org/record/8205103 [52].
Declarations
Ethics approval and consent to participate
The study was approved (01/04/2020) by IRB Aragón (Aragon Research Ethics Committee [CEICA]) (Record No. 07/2020). All patients and controls signed the informed consent form to participate in the study. Our research was conducted in accordance with the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors report no conflicts of interest related to the submitted work.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pedersen SF, Ho YC. SARS-CoV-2: a storm is raging. J Clin Invest. 2020;130(5):2202–2205. doi: 10.1172/JCI137647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zuo Y, Yalavarthi S, Shi H, et al. Neutrophil extracellular traps in COVID-19. JCI Insight. 2020;5(11):e138999. doi: 10.1172/jci.insight.138999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fox SE, Akmatbekov A, Harbert JL, Li G, Brown JQ, Vander Heide RS. Pulmonary and cardiac pathology in COVID-19: the first autopsy series from New Orleans. Lancet Respir Med. 2020;8:681–686. doi: 10.1016/S2213-2600(20)30243-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes BJ, Adrover JM, Baxter-Stoltzfus A, et al. Targeting potential drivers of COVID-19: neutrophil extracellular traps. J Exp Med. 2020;217:e20200652. doi: 10.1084/jem.20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carmona-Rivera C, Zhang Y, Dobbs K, et al. Multicenter analysis of neutrophil extracellular trap dysregulation in adult and pediatric COVID-19. JCI Insight. 2022;7(16):e160332. doi: 10.1172/jci.insight.160332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brinkmann V, Reichard U, Goosmann C, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303(5663):1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 7.Papayannopoulos V. Neutrophil extracellular traps in immunity and disease. Nat Rev Immunol. 2018;18(2):134–147. doi: 10.1038/nri.2017.105. [DOI] [PubMed] [Google Scholar]
- 8.Johansson C, Kirsebom FCM. Neutrophils in respiratory viral infections. Mucosal Immunol. 2021;14(4):815–827. doi: 10.1038/s41385-021-00397-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nathan C. Neutrophils and COVID-19: Nots, NETs, and knots. J Exp Med. 2020;217(9):e20201439. doi: 10.1084/jem.20201439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ackermann M, Anders HJ, Bilyy R, et al. Patients with COVID-19: in the dark-NETs of neutrophils. Cell Death Differ. 2021;28(11):3125–3139. doi: 10.1038/s41418-021-00805-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jo A, Kim DW. Neutrophil extracellular traps in airway diseases: pathological roles and therapeutic implications. Int J Mol Sci. 2023;24(5):5034. doi: 10.3390/ijms24055034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huckriede J, Anderberg SB, Morales A, et al. Evolution of NETosis markers and DAMPs have prognostic value in critically ill COVID-19 patients. Sci Rep. 2021;11(1):15701. doi: 10.1038/s41598-021-95209-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ouwendijk WJD, Raadsen MP, van Kampen JJA, et al. High levels of neutrophil extracellular traps persist in the lower respiratory tract of critically ill patients with coronavirus disease 2019. J Infect Dis. 2021;223(9):1512–1521. doi: 10.1093/infdis/jiab050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borges L, Pithon-Curi TC, Curi R, Hatanaka E. COVID-19 and neutrophils: the relationship between hyperinflammation and neutrophil extracellular traps. Mediators Inflamm. 2020;2020:8829674. doi: 10.1155/2020/8829674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Veras FP, Pontelli MC, Silva CM, et al. SARS-CoV-2-triggered neutrophil extracellular traps mediate COVID-19 pathology. J Exp Med. 2020;217(12):e20201129. doi: 10.1084/jem.20201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Middleton EA, He XY, Denorme F, et al. Neutrophil extracellular traps contribute to immunothrombosis in COVID-19 acute respiratory distress syndrome. Blood. 2020;136:1169–1179. doi: 10.1182/blood.2020007008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Radermecker C, Detrembleur N, Guiot J, et al. Neutrophil extracellular traps infiltrate the lung airway, interstitial, and vascular compartments in severe COVID-19. J Exp Med. 2020;217(12):e20201012. doi: 10.1084/jem.20201012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rowley AH. Understanding SARS-CoV-2-related multisystem inflammatory syndrome in children. Nat Rev Immunol. 2020;20(8):453–454. doi: 10.1038/s41577-020-0367-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bello S, Lasierra AB, López-Vergara L, et al. IL-6 and cfDNA monitoring throughout COVID-19 hospitalization are accurate markers of its outcomes. Respir Res. 2023;24(1):125. doi: 10.1186/s12931-023-02426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Durán-Méndez A, Aguilar-Arroyo AD, Vivanco-Gómez E, et al. Tocilizumab reduces COVID-19 mortality and pathology in a dose and timing-dependent fashion: a multi-centric study. Sci Rep. 2021;11(1):19728. doi: 10.1038/s41598-021-99291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayden H, Ibrahim N, Klopf J, et al. ELISA detection of MPO-DNA complexes in human plasma is error-prone and yields limited information on neutrophil extracellular traps formed in vivo. PLoS ONE. 2021;16(4):e0250265. doi: 10.1371/journal.pone.0250265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in china: summary of a report of 72314 cases from the Chinese Center for disease control and prevention. JAMA. 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 23.Marshall JC, Murthy S, Diaz J, et al. A minimal common outcome measure set for COVID-19 clinical research. Lancet Infect Dis. 2020;20(8):e192–e197. doi: 10.1016/S1473-3099(20)30483-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kano H, Huq MA, Tsuda M, Noguchi H, Takeyama N. Sandwich ELISA for circulating myeloperoxidase- and neutrophil elastase-DNA complexes released from neutrophil extracellular traps. Adv Tech Biol Med. 2016;5:196. doi: 10.4172/2379-1764.1000196. [DOI] [Google Scholar]
- 25.Veras FP, Gomes GF, Silva BMS, et al. Targeting neutrophils extracellular traps (NETs) reduces multiple organ injury in a COVID-19 mouse model. Respir Res. 2023;24(1):66. doi: 10.1186/s12931-023-02336-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Li Q, Yin Y, et al. Excessive neutrophils and neutrophil extracellular traps in COVID-19. Front Immunol. 2020;11:2063. doi: 10.3389/fimmu.2020.02063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masso-Silva JA, Moshensky A, Lam MTY, et al. Increased peripheral blood neutrophil activation phenotypes and neutrophil extracellular trap formation in critically ill coronavirus disease 2019 (COVID-19) patients: a case series and review of the literature. Clin Infect Dis. 2022;74(3):479–489. doi: 10.1093/cid/ciab437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silvestre-Roig C, Braster Q, Wichapong K, et al. Externalized histone H4 orchestrates chronic inflammation by inducing lytic cell death. Nature. 2019;569(7755):236–240. doi: 10.1038/s41586-019-1167-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao X, Zhou L, Kou Y, Kou J. Activated neutrophils in the initiation and progression of COVID-19: hyperinflammation and immunothrombosis in COVID-19. Am J Transl Res. 2022;14(3):1454–1468. [PMC free article] [PubMed] [Google Scholar]
- 30.Melero I, Villalba-Esparza M, Recalde-Zamacona B, et al. Neutrophil extracellular traps, local IL-8 expression, and cytotoxic T-lymphocyte response in the lungs of patients with fatal COVID-19. Chest. 2022;162(5):1006–1016. doi: 10.1016/j.chest.2022.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kinnare N, Hook JS, Patel PA, Monson NL, Moreland JG. Neutrophil extracellular trap formation potential correlates with lung disease severity in COVID-19 patients. Inflammation. 2022;45(2):800–811. doi: 10.1007/s10753-021-01585-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krinsky N, Sizikov S, Nissim S, et al. NETosis induction reflects COVID-19 severity and Long COVID: insights from a two-center patient cohort study in Israel. J Thromb Haemost. 2023 doi: 10.1016/j.jtha.2023.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trigg RM, Martinson LJ, Parpart-Li S, Shaw JA. Factors that influence quality and yield of circulating-free DNA: a systematic review of the methodology literature. Heliyon. 2018;4(7):e00699. doi: 10.1016/j.heliyon.2018.e00699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Z, Zhang H, Qu M, et al. Review: the emerging role of neutrophil extracellular traps in sepsis and sepsis-associated thrombosis. Front Cell Infect Microbiol. 2021;11:653228. doi: 10.3389/fcimb.2021.653228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Buhr N, von Köckritz-Blickwede M. Detection, visualization, and quantification of neutrophil extracellular traps (NETs) and NET markers. Methods Mol Biol. 2020;2087:425–442. doi: 10.1007/978-1-0716-0154-9_25. [DOI] [PubMed] [Google Scholar]
- 36.de Vries F, Huckriede J, Wichapong K, Reutelingsperger C, Nicolaes GAF. The role of extracellular histones in COVID-19. J Intern Med. 2023;293(3):275–292. doi: 10.1111/joim.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kenny EF, Herzig A, Kruger R, et al. Diverse stimuli engage different neutrophil extracellular trap pathways. Elife. 2017;6:e24437. doi: 10.7554/eLife.24437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seery V, Raiden SC, Algieri SC, et al. Blood neutrophils from children with COVID-19 exhibit both inflammatory and anti-inflammatory markers. EBioMedicine. 2021;67:103357. doi: 10.1016/j.ebiom.2021.103357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jorch SK, Kubes P. An emerging role for neutrophil extracellular traps in noninfectious disease. Nat Med. 2017;23(3):279–287. doi: 10.1038/nm.4294. [DOI] [PubMed] [Google Scholar]
- 40.Boeltz S, Amini P, Anders H-J, et al. To NET or not to NET: Current opinions and state of the science regarding the formation of neutrophil extracellular traps. Cell Death Differ. 2019;26(3):395–408. doi: 10.1038/s41418-018-0261-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cesta MC, Zippoli M, Marsiglia C, et al. Neutrophil activation and neutrophil extracellular traps (NETs) in COVID-19 ARDS and immunothrombosis. Eur J Immunol. 2023;53(1):e2250010. doi: 10.1002/eji.202250010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andargie TE, Tsuji N, Seifuddin F, et al. Cell-free DNA maps COVID-19 tissue injury and risk of death and can cause tissue injury. JCI Insight. 2021;6(7):e147610. doi: 10.1172/jci.insight.147610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hammad R, Eldosoky MAER, Fouad SH, et al. Circulating cell-free DNA, peripheral lymphocyte subsets alterations and neutrophil lymphocyte ratio in assessment of COVID-19 severity. Innate Immun. 2021;27(3):240–250. doi: 10.1177/1753425921995577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsuji N, Tsuji T, Ohashi N, et al. Role of mitochondrial DNA in septic AKI via toll-like receptor 9. J Am Soc Nephrol. 2016;27(7):2009–2020. doi: 10.1681/ASN.2015040376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hauser C, Otterbein L. Danger signals from mitochondrial DAMPS in trauma and post-injury sepsis. Eur J Trauma Emerg Surg. 2018;44(3):317–324. doi: 10.1007/s00068-018-0963-2. [DOI] [PubMed] [Google Scholar]
- 46.Munker D, Osterman A, Stubbe H, et al. Dynamics of SARS-CoV-2 shedding in the respiratory tract depends on the severity of disease in COVID-19 patients. Eur Respir J. 2021;58(1):2002724. doi: 10.1183/13993003.02724-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Gu X, Li H, et al. Risk factors of viral RNAaemia and its association with clinical prognosis among patients with severe COVID-19. Chest. 2021;159(4):1382–1386. doi: 10.1016/j.chest.2020.11.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jarrahi A, Khodadadi H, Moore NS, Lu Y, Awad ME, Salles EL, Vaibhav K, Baban B, Dhandapani KM. Recombinant human DNase-I improves acute respiratory distress syndrome via neutrophil extracellular trap degradation. J Thromb Haemost. 2023;21(9):2473–2484. doi: 10.1016/j.jtha.2023.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiménez-Alcázar M, Rangaswamy C, Panda R, et al. Host DNases prevent vascular occlusion by neutrophil extracellular traps. Science. 2017;358(6367):1202–1206. doi: 10.1126/science.aam8897. [DOI] [PubMed] [Google Scholar]
- 50.Strich JR, Ramos-Benitez MJ, Randazzo D, et al. Fostamatinib inhibits neutrophils extracellular traps induced by COVID-19 patient plasma: a potential therapeutic. J Infect Dis. 2021;223(6):981–984. doi: 10.1093/infdis/jiaa789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chiang N, Sakuma M, Rodriguez AR, Spur BW, Irimia D, Serhan CN. Resolvin T-series reduce neutrophil extracellular traps. Blood. 2022;139(8):1222–1233. doi: 10.1182/blood.2021013422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Diego C, Lasierra AB, López-Vergara L, Torralba L, Ruiz de Gopegui P, Lahoz R, Abadía C, Godino J, Cebollada A, Jimeno B, Bello C, Tejada A, Bello S. What is the actual relationship between neutrophil extracellular traps and COVID-19 severity? A longitudinal study. 2023. https://zenodo.org/record/8205103 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Demographics and comorbidities of the patients and controls
Additional file 2: Table S2: Detailed view of the multivariate model for severity, mortality, and need for ICU.
Additional file 3: Table S3. Relationship between NET markers and sex, age, and obesity separately in patients and controls.
Additional file 4: Table S4. Sex-biased analysis of NET markers and mortality in total samples.
Additional file 5: Table S5. Correlations between usual clinical parameters of severity, neutrophil counts, neutrophil associated cytokines (IL-8 and G-CSF), and NET markers
Data Availability Statement
The datasets supporting the conclusions of this article are available in the Zenodo repository, in https://zenodo.org/record/8205103 [52].