Abstract
Ancient bony fishes had heterocercal tails, like modern sharks and sturgeons, with asymmetric caudal fins and a vertebral column extending into an elongated upper lobe. Teleost fishes, in contrast, developed a homocercal tail characterized by two separate equal-sized fin lobes and the body axis not extending into the caudal fin. A similar heterocercal-to-homocercal transition occurs during teleost ontogeny, although the underlying genetic and developmental mechanisms for either transition remain unresolved. Here, we investigated the role of hox13 genes in caudal fin formation as these genes control posterior identity in animals. Analysis of expression profiles of zebrafish hox13 paralogs and phenotypes of CRISPR/Cas9-induced mutants showed that double hoxb13a and hoxc13a mutants fail to form a caudal fin. Furthermore, single mutants display heterocercal-like morphologies not seen since Mesozoic fossil teleosteomorphs. Relaxation of functional constraints after the teleost genome duplication may have allowed hox13 duplicates to neo- or subfunctionalize, ultimately contributing to the evolution of a homocercal tail in teleost fishes.
hoxb13a and hoxc13a control caudal fin patterning, with mutant phenotypes revealing evolutionary transitions of teleost tails.
INTRODUCTION
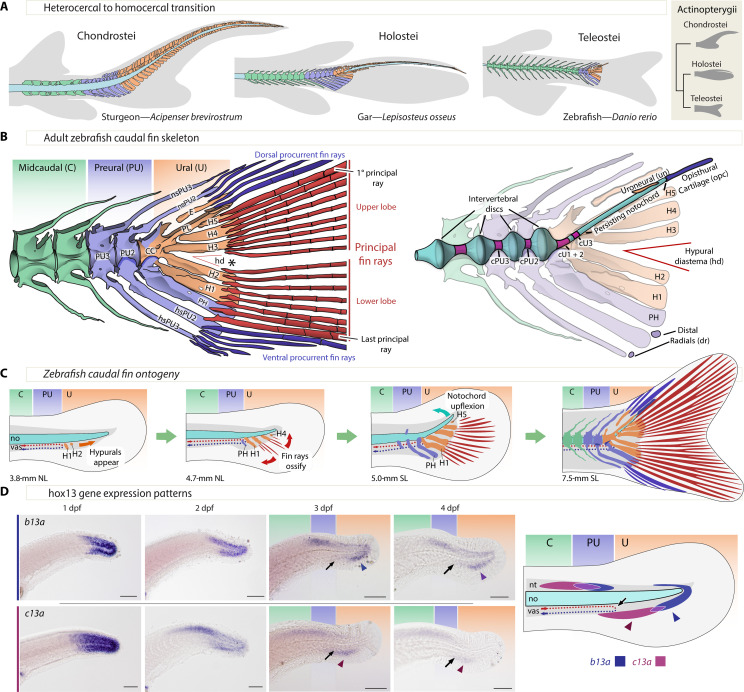
A pivotal event in teleost fish evolution was the transition from a heterocercal to a homocercal tail, which restructured the morphology of the posterior body axis (1–6), enhancing propulsion and maneuverability. Heterocercal tails, like those of modern sturgeons, have a larger upper lobe containing the vertebral column and a continuous set of bony fin rays. Homocercal tails have, instead, a short upward-bent column that ends at the base of the fin and two sets of caudal rays separated into discrete upper and lower lobes of roughly equal size (Fig. 1, A to D) (3, 4, 6–8). Early naturalists identified a similar morphological transformation during the ontogeny of modern teleosts, which initially develop a heterocercal-like tail (Fig. 1C) that then acquires the homocercal shape of adult tails (1–3, 6, 9). Thus, the main difference between heterocercal and homocercal tails in development and evolution lies in the organization and extension of the posterior end of the body axis (Fig. 1, A to C).
Fig. 1. Zebrafish caudal anatomy, development, evolution, and b13a and c13a expression.
(A) Representative caudal fin morphologies in actinopterygians, featuring gray-shaded body contour and fin rays, with midcaudal, preural, and ural elements in green, blue, and orange, respectively. (B) Left: Adult zebrafish caudal fin skeleton, highlighting the three regions of the caudal fin and the separation of upper and lower principal rays (asterisk, *). Right: Description of internal elements within the caudal fin skeleton, including the notochord. (C) Developmental relocation of the zebrafish caudal fin elements following bending of the notochord and the appearance of principal fin rays. (D) In situ hybridizations showing b13a and c13a transcripts from 1 to 4 days postfertilization (dpf) and schematic representation of expression domains of both genes at 4 dpf. Black arrows indicate the end of the caudal artery at the PU-U boundary, while blue and red arrowheads mark the ventral expression domains of b13a and c13a, respectively. Scale bars, 0.1 mm. CC, compound centrum; cPU2 and cPU3, preural chordacentra 2 and 3; cU1 to cU3, ural chordacentra 1 to 3; dr, distal radial; E, epural; H1 to H5, hypurals 1 to 5; hd, hypural diastema; hsPU2 and hsPU3, haemal spines of the preural centra 2 and 3; nsPU2 and sPU3, neural spines of the preural centra 2 and 3; NL, notochordal length; no, notochord; opc, opisthural cartilage; PH, parhypural; PL, pleurostyle; PU2 and PU3, preural centra 2 and 3; SL, standard length; un, uroneural; vas, vasculature.
The heterocercal-to-homocercal transition entailed a significant reduction in the number of preural and ural vertebrae supporting an increased number of bony fin rays (Fig. 1A) (4, 5, 7). The preural region encompasses a transitional region between midcaudal and ural vertebrae, from the most anterior vertebrae supporting rays up to the final vertebra bearing a haemal arch (Fig. 1, A and B). Posterior to the preural lies the ural region, characterized by an upward-bent body axis that ventrally supports hypurals (H in Fig. 1B) (4, 5, 7). This upwardly bent body axis is achieved by a dorsoventral (DV) symmetry break during ontogeny that relocates the posterior hypurals dorsally (Fig. 1C). The external DV symmetry of the homocercal tail is then achieved by fin rays developing symmetrically in pairs above and below the hypural diastema, located between the central hypurals 2 and 3 (Fig. 1B). Fin rays originating posterior to the diastema contribute to the upper lobe, while those anterior to it form the lower lobe (Fig. 1C) (4, 6, 10, 11).
Considering these transformations in body axis elements, posterior Hox genes likely regulate homocercal fin patterning and evolution. Hox genes encode homeodomain-containing transcription factors organized into four clusters in tetrapods (12, 13). Their anteroposterior expression during embryogenesis generally correlates with their genomic organization (14–17). Hox13 paralogs are expressed most caudally and are present in all four vertebrate clusters, suggesting preserved nonredundant functions. In chicken and mouse, Hoxb13 and Hoxc13 affect body axis elongation and patterning. Gain-of-function mutants of both genes display reduced tail elongation (18) and fewer caudal vertebrae (19, 20). Conversely, Hoxb13 loss-of-function mutant mice exhibit extra caudal vertebrae (21), and Hoxc13 null mutant mice undergo vertebral identity changes (22). Teleosts, though, experienced a whole genome duplication initially resulting in eight hox clusters (23–25), with six of these clusters containing a hox13 paralog. All six hox13 paralogs are expressed in the zebrafish tail bud (26–28), with hoxa13b (henceforth, a13b) and d13a required for tail bud elongation (28) and a13a, a13b, c13a, and c13b involved in caudal fin ray regeneration (29, 30). No studies, however, have explored hox13 paralog contribution to caudal patterning in fish.
In this work, we tested the hypothesis that members of the hox13 gene set directly regulate the formation of the caudal fin and that they are linked to the evolution of the homocercal tail. We analyzed gene expression profiles for all six zebrafish hox13 paralogs and selected two candidate genes that were then mutated using CRISPR-Cas9. Our results show that hoxb13a and hoxc13a direct caudal fin patterning and that phenotypic transformations elicited in homozygous mutants recapitulate several evolutionary transformations that characterize the heterocercal-to-homocercal transition.
RESULTS
hoxb13a and hoxc13a are candidate regulators of caudal fin patterning
We first determined the expression patterns of all six zebrafish hox13 paralogs during the first four days postfertilization (dpf) (Fig. 1D, figs. S1 and S2, and Supplementary Text). By 2 dpf, distinct expression patterns were observed for each of the hox13 genes in various organs (figs. S1 and S2). Notably, b13a and c13a exhibited strong labeling in the tail, persisting until at least 4 dpf (Fig. 1D), suggesting their involvement in caudal fin patterning, which becomes evident around 3.8-mm notochordal length (NL), between 10 and 12 dpf (Fig. 1C). In contrast, a13a, a13b, and d13a displayed stronger labeling in the pectoral fin buds than in the tail (figs. S1 and S2) and have been shown by gene expression and phenotypes of deletion mutants to play a role in patterning vertebrate paired appendages (31–33) but not caudal fins (34). Considering that hox genes controlling caudal fin patterning should be widely conserved in teleosts, we found that only three hox13 genes (a13b, b13a, and c13a) were conserved in annotated genome assemblies of all major teleost lineages (fig. S3 and table S1). Thus, the intersection of expression patterns and evolutionary conservation identified b13a and c13a as the best candidate genes for caudal fin patterning.
To delimit the anteroposterior hox13 gene expression domains, we used adult morphological landmarks in 3- and 4-dpf zebrafish larvae. The DV U-turn of the caudal vasculature defines the preural-to-ural boundary (black arrow, Fig. 1D) (4, 5), while the last two or three myomeres delimit the preural region, based on the number of preural vertebrae in the adult (Fig. 1B). The ventral expression of c13a localizes to the preural-to-ural boundary, while its dorsal domain extends further anteriorly (Fig. 1D). In contrast, the ventral domain of b13a marks the ural region, while its dorsal domain marks the preural region (Fig. 1D). The complementarity of these two gene expression patterns is notable because, as the notochord bends dorsally at about 4.5 mm standard length (SL) (Fig. 1C) (35), the ventral posterior b13a domain corresponds to the upper lobe of the caudal fin, while the ventral anterior c13a domain corresponds to the lower lobe, with their overlap aligning with the hypural diastema.
hoxb13a and hoxc13a genes regulate caudal fin length and symmetry
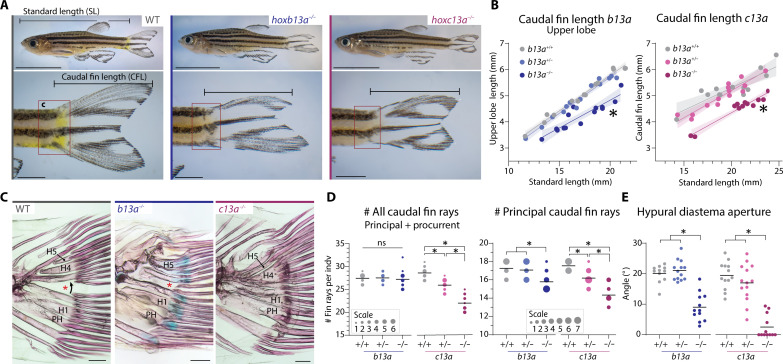
Using CRISPR-Cas9 mutagenesis, we generated b13a and c13a loss-of-function mutant zebrafish with frameshift mutations upstream of the homeodomain leading to premature stop codons (fig. S4, A and B). These mutations were inherited in Mendelian ratios (fig. S4C). Homozygous mutants for b13a and c13a were viable and displayed distinct phenotypic changes in the caudal fin compared to their heterozygous and wild-type (WT) siblings (Fig. 2A). Homozygous b13a−/− mutants showed a shorter upper lobe, resulting in an asymmetric caudal fin (Fig. 2, A and B, and fig. S5A), while homozygous c13a−/− mutants had symmetric but shorter and narrower fins in the DV axis (Fig. 2, A and B).
Fig. 2. Caudal fin phenotypes in b13a and c13a zebrafish mutants.
(A) Lateral views of WT and homozygous b13a and c13a mutants (10 mm scale bar for whole fish; 2 mm for caudal fin close-up). (B) Upper lobe length relative to SL in WT, heterozygous, and homozygous mutant siblings (n = b13a+/+:14, b13a+/−:14, b13a−/−:14, c13a+/+:13, c13a+/−:14, and c13a−/−:12). Analysis of covariance indicated a significant reduction in upper lobe length in both homozygous mutants [b13a: F2,38 = 88.0, P < 0.0001; c13a: F2,35 = 59.86, P < 0.0001]. Shaded area represents the 95% confidence interval around the regression line. (C) Skeletal staining showing caudal ray bases, hypurals, and the hypural diastema (red asterisk). Scale bars, 200 μm. (D) Caudal and principal ray number in both mutants (n = b13a+/+:12, b13a+/−:14, b13a−/−:14, c13a+/+:13, c13a+/−:15, and c13a−/−:12). Analysis of variance (ANOVA) revealed a significant reduction in total fin ray number for c13a [F2,37 = 67.02, P < 0.0001], but not for b13a mutants [F2,37 = 0.2223, P = 0.8017]. Multiple comparisons indicated a c13a gene-dosage effect (P < 0.0001 for all comparisons). Both mutants reduced principal ray number [b13a: F2,37 = 10,11, P = 0.0003; c13a: F2,37 = 36.93, P < 0.0001] with only b13a−/− differing from its siblings [P = 0.0021 versus b13a+/− and P = 0.0008 versus b13a+/+], while c13a demonstrating the same gene-dosage effect (P < 0.0001 for all comparisons). (E) Hypural diastema aperture angle in both mutants (n = b13a+/+:12, b13a+/−:14, b13a−/−:14, c13a+/+:13, c13a+/−:14, and c13a−/−:12). ANOVA revealed a significant reduction in diastema aperture for both genes [b13a: F2,33 = 39.89, P < 0.0001; c13a: F2,36 = 42.28, P < 0.0001]. Multiple comparisons showed this effect in homozygous mutants only (P < 0.0001 for b13a−/− and c13a−/−). For (D) and (E), horizontal lines represent the mean; circle size is proportional to fish scored. See Fig. 1 for anatomical terminology.
We tested whether morphological changes in mutant fins were associated with caudal ray number. Caudal fin rays are of two types. Principal rays (labeled in red, Fig. 1, B and C) are the central and longest rays, including all bifurcated rays and the first nonbifurcated ray of each fin lobe. Procurrent rays (dark blue) consist of two series of smaller, nonbifurcated rays anterior to the principal rays (36). The c13a−/− mutants had significantly fewer caudal rays than their WT siblings in both caudal fin lobes, losing a total of three principal and three procurrent rays (Fig. 2D and fig. S5, C and D). Heterozygous c13a+/− siblings displayed an intermediate phenotype, indicating a partially dominant mutant allele. In contrast, b13a−/− mutants retained the same total caudal ray number as WT but lost one or two principal rays in the upper lobe, compensated by additional procurrent rays (Fig. 2D and fig. S5, B to D). Both mutants lacked separation between caudal rays of the upper and lower lobes and had a reduced aperture of the hypural diastema (red asterisk, Fig. 2, C and E), particularly evident in homozygous c13a−/− mutants (Fig. 2, C and E). In addition, in most homozygous single mutants for each gene, the uppermost principal ray originated above the notochord, unlike in WTs where all principal rays are hypaxial (fig. S5, E and F). Approximately half of c13a−/− mutants had a short upper principal ray that did not extend along the entire leading margin of the fin (fig. S5, E and F), implying that this phenotype had reduced penetrance in c13a single mutant homozygotes.
To verify the specificity of b13a and c13a phenotypes, we examined all other fins and found that the number of dorsal, anal, pectoral, and pelvic fin rays and endoskeletal radials remained unaltered (fig. S6). This finding indicates that both b13a and c13a genes specifically regulate caudal fin shape, length, and number of rays and promote the separation between the upper and lower fin lobes and do not affect the fins regulated by a13a, a13b, and d13a (31–33).
hoxb13a and hoxc13a genes regulate the number and identity of posterior vertebrae
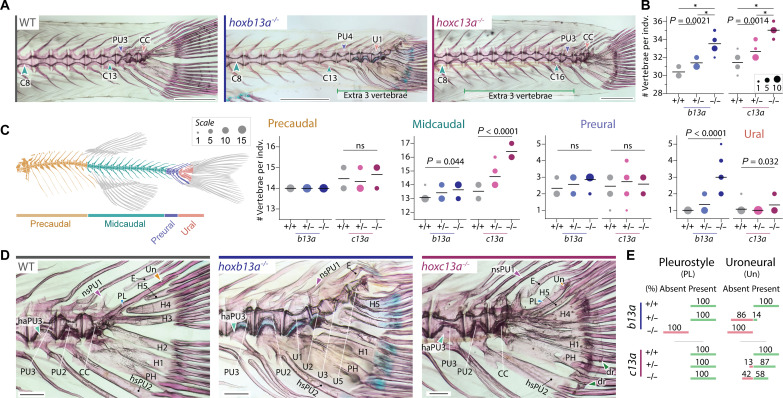
Examination of the axial skeleton revealed that both homozygous single mutants had approximately three extra vertebrae compared to WT siblings, while heterozygous siblings had one additional vertebra, again indicating partially dominant phenotypes (Fig. 3, A and B). Analysis of the different vertebral regions along the body axis (Fig. 3C) (7, 37) showed that c13a mutants had additional midcaudal vertebrae, while b13a mutants primarily had extra ural vertebrae (Fig. 3C), consistent with their respective expression domains. The extra midcaudal vertebrae in c13a mutants were positioned between the anal fin and the preural vertebrae, resembling other midcaudal vertebrae (Fig. 3A). Conversely, the extra ural vertebrae in b13a mutants extended to the posterior end of the notochord (Fig. 3D and red arrowheads in fig. S7A), a region lacking centra that maintains a persistent notochord in WTs (Figs. 1B and 3D and fig. S7A). In addition, b13a−/− mutants either lacked or had a significantly reduced opisthural cartilage, a specialized structure derived from the notochordal sheath (38) that covers the posterior end of the notochord (yellow arrowheads, fig. S7A) (7). In both mutants, but particularly marked in larger b13a−/− specimens, the notochord terminated near the base of hypural 5 (H5) instead of reaching the posterior end of H5 (black versus red arrowheads, fig. S7A). Together, these findings demonstrate a positional correlation between the expression domains of c13a and b13a (Fig. 1D) and the changes in vertebral number observed in the mutants (Fig. 3, A and C).
Fig. 3. Vertebral phenotypes of b13a and c13a zebrafish mutants.
(A) Cleared and stained skeletal preparations of WT and homozygous b13a and c13a mutants showing variation in tail vertebrae number. Scale bars, 1 mm. For (B), (C), and (E), n = b13a+/+:12, b13a+/−:14, b13a−/−:14, c13a+/+:13, c13a+/−:15, and c13a−/−:12. (B) Total number of vertebrae in both mutants. ANOVA revealed a significant increase in vertebrae number [b13a: F2,37 = 78.28, P < 0.0001; c13a: F2,37 = 63.49, P < 0.0001] and multiple comparisons showed a gene-dosage effect for both genes (*P < 0.0001). (C) Vertebral number per body region in both mutants. Poisson regressions revealed significant increases for both mutants in midcaudal [b13a: χ2(2) = 6.23, P = 0.044; c13a: χ2(2) = 30.003, P < 0.0001] and ural vertebrae [b13a: χ2(2) = 41.22, P < 0.0001; c13a: χ2(2) = 6.88, P = 0.032]. Other comparisons were not significant (ns) [precaudal | c13a: χ2(2) = 1.54, P = 0.46; preural | b13a: χ2(2) = 3.08, P = 0.21; preural | c13a: χ2(2) = 0.32, P = 0.85). Post hoc tests were not applied due to the minimal variance or absence of variance in several observations, rendering those tests invalid. The line is at the mean and circle size is proportional to the number of individuals. (D) Caudal fin endoskeleton of WT and homozygous b13a and c13a mutants. The pleurostyle (blue arrowhead) and uroneural (orange arrowhead) are absent in b13a−/− mutants. An elongated neural spine of the preural centrum 1 (purple arrowhead) is present in both mutants. Extra distal radials (green arrowheads) are present in c13a−/− mutants. A cartilaginous articulation of the haemal arch of the preural centrum 3 with its centrum (light blue arrowhead) is present in the b13a−/− mutant, as revealed by Alcian blue staining. Scale bars, 200 μm. (E) Frequencies of pleurostyle and uroneural loss in b13a and c13a mutants. See Fig. 1 for anatomical terminology.
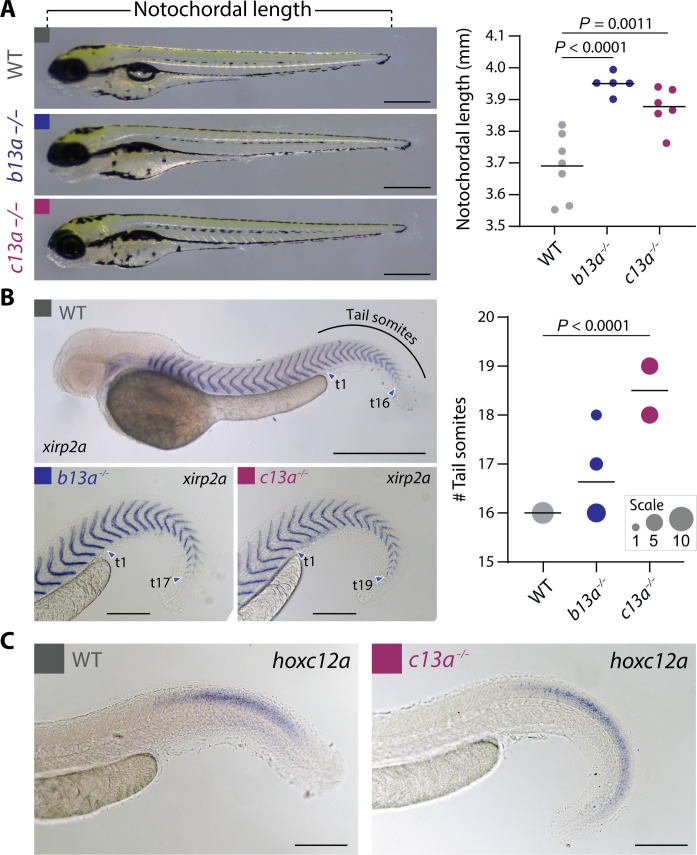
To investigate the developmental origins of these segmental identity changes, we measured larval NL (Fig. 4A) and somite number in b13a−/− and c13a−/− mutants using the myotome boundary marker xirp2a (Fig. 4B) (39). Both mutants showed increased body length and somite number compared to WT animals (Fig. 4, A and B). The number of extra somites in c13a−/− mutant larvae matched the additional caudal vertebrae in mutant adults (Fig. 3, A to D). However, in b13a−/− mutants, the extra ural vertebrae in adults did not correspond to additional somites in larvae (Fig. 3, A to D), consistent with ural vertebrae forming independently of somites (40). In addition, in c13a−/− mutants, we examined the expression domain of hoxc12a, the gene preceding c13a in the hoxca cluster. In the absence of a functional c13a gene, c12a expression extended posteriorly into the elongated portion of the tail (Fig. 4C), consistent with the expected sequential repression of hox genes by their neighboring 3′ gene (41). These findings highlight the role of b13a and c13a genes in establishing posterior segmental identity and repressing body axis elongation.
Fig. 4. Length and somite number in mutant b13a−/− and c13a−/− larvae.
(A) NL of 3-dpf WT, b13a−/−, and c13a−/− mutant larvae. n = WT:7, b13a−/−:5, and c13a−/−:6. ANOVA revealed a significant change in larval length [F2,15 = 18.59, P < 0.0001]. Multiple comparisons indicated significantly longer b13a (P < 0.0001) and c13a (P = 0.0011) larvae compared with WTs. Scale bars, 0.5 mm. (B) Tail somite number in 2-dpf WT, b13a−/−, and c13a−/− mutant larvae, using in situ hybridization with the myotome boundary marker xirp2a. (n = WT:8, b13a−/−:11, and c13a−/−:10). Poisson regression revealed significant increases in somite number for both mutants [χ2(2) = 33.19, P < 0.0001]. Post hoc tests were not applied due to the absence of variance in WTs, rendering those tests invalid. Circle size is proportional to fish scored (0.5 mm scale bar for WT, 0.2 mm scale bars for b13a−/− and c13a−/− mutants). (C) In situ hybridization against hoxc12a in 2-dpf WT and c13a−/− mutant embryos. Scale bars, 0.2 mm. For (A) and (B), graph horizontal lines represent the mean.
hoxb13a and hoxc13a mutants display non-overlapping caudal fin phenotypes
Next, we examined skeletal changes in the caudal fin complex of hox13 mutants (Fig. 3, D and E). Notably, b13a−/− mutants lacked both the uroneural and pleurostyle (Fig. 3, D and E), which are modified neural arches covering the lateral surface of the posterior-most vertebrae and notochord in the ural region and are characteristic of homocercal tails (Fig. 1C) (4, 7, 40). Uroneural loss was also observed in most heterozygous b13a+/− and half of the homozygous c13a−/− individuals (Fig. 3E). In addition, b13a−/− mutants displayed an elongated preural neural spine on top of the last centrum and a cartilaginous articulation between preural centrum 3 and its haemal arch, which is fused in WTs (Fig. 3D and fig. S7B). In contrast, c13a−/− mutants developed extra distal radials at the ends of the parhypural and hypurals 1 and 2 (green arrowheads, Fig. 3D and fig. S7, B and E), with some radials ossified in specimens over 20-mm SL, resembling the supernumerary pectoral fin distal radials observed in zebrafish hoxa13 mutants (34). The epural number, however, showed no change (fig. S7C), and the dorsal hypural number (H3 to H5) was only slightly decreased in b13a−/− mutants (fig. S7D). These findings revealed gene-specific alterations in the caudal endoskeleton, including, in b13a−/− mutants, the absence of pleurostyle and uroneural along with modified preural arches and spines, and, in c13a−/− mutants, the presence of extra distal radials.
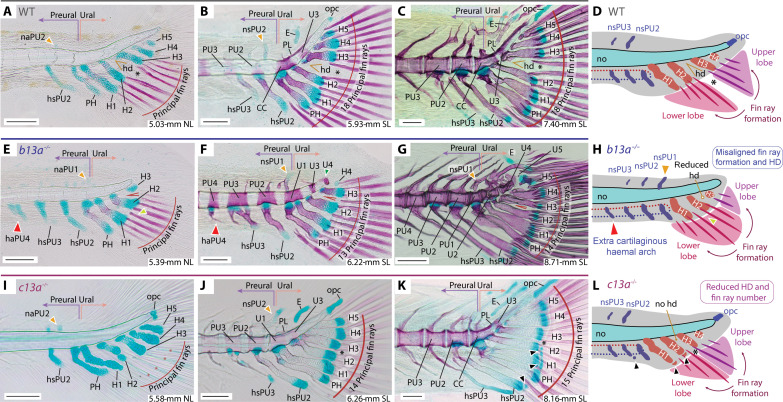
Ontogenetic analysis of b13a−/− mutants further revealed noteworthy differences with WT fish (Fig. 5, A to D versus E to H), including (i) the development of hypurals 1 to 5 closer to the notochord’s posterior end (Fig. 5, E to H), accompanied by an additional pair of cartilaginous preural neural and haemal arches between the midcaudal and preural regions (red arrowhead, Fig. 5, E to G). (ii) The first neural arch to develop formed dorsal to the parhypural (orange arrowhead, Fig. 5, E and F), instead of dorsal to the preural centrum 2 as in WTs (Fig. 5, A and B). (iii) The first principal fin rays appeared between hypurals 1 and 2 (yellow arrowhead, Fig. 5, E and H), rather than aligned with the hypural diastema between hypurals 2 and 3 (Fig. 5, A and D). (iv) The upward bending of the notochord was less pronounced than in WTs (Fig. 5, A to C versus E to G). (v) Hourglass-shaped vertebral centra with neural arches extended to the notochord’s posterior tip (green arrowhead, Fig. 5, F and G). The observed changes in developing b13a mutants correlated with adult phenotypes, suggesting that b13a (Fig. 5H) (i) inhibits preural element formation; (ii) suppresses vertebral centra formation, resulting in a persistent notochord; (iii) induces modification of posterior ural neural arches into a pleurostyle and uroneural; (iv) drives opisthural cartilage formation at the notochord’s posterior end; and (v) aligns the hypural diastema and first developing principal rays.
Fig. 5. Ontogenetic series of cleared and stained skeletal preparations for b13a−/− and c13a−/− mutants.
The figure displays the development of the caudal fin in zebrafish WT, b13a−/−, and c13a−/− mutants, focusing on the formation of fin rays and posterior vertebrae. (A) 5.03-mm NL WT specimen. (B) 5.93-mm SL WT specimen. (C) 7.40-mm SL WT specimen. (D) Diagram showing normal zebrafish caudal fin development, fin ray appearance is aligned with the hypural diastema. (E) 5.38-mm NL b13a−/− mutant specimen. (F) 6.22-mm SL b13a−/− mutant specimen. (G) 8.71-mm SL b13a−/− mutant specimen. (H) Diagram showing the b13a−/− mutant caudal fin development, with a reduced hypural diastema and the appearance of fin rays not aligned with the position of the diastema. The notochord is abbreviated, with no opisthural cartilage, the base of hypurals is closer to the notochord posterior end, and an additional preural element is present anteriorly. (I) 5.58-mm NL c13a−/− mutant specimen. (J) 6.26-mm SL c13a−/− mutant specimen. (K) 8.16-mm SL c13a−/− mutant specimen. Black arrowheads, extra distal radials. (L) Diagram showing the c13a−/− mutant caudal fin development, lacking a hypural diastema, with fewer fin rays forming, more spaced between them, and additional radials at the distal ends of the hypurals 1 and 2. Red arrowheads, extra cartilaginous haemal arch or spine; yellow arrowheads, first neural arch/spine that develops; black asterisk, separation between upper and lower caudal fin lobes. See Fig. 1 for anatomical terminology. Scale bars, 0.1 mm.
Ontogenetic analysis of c13a−/− mutants also revealed notable differences with WTs (Fig. 5, A to D versus I to L). Specifically, c13a−/− mutants lacked a hypural diastema from early on (Fig. 5, A to C versus I to K). While the first pair of rays developed between hypurals 2 and 3 as in WTs (Fig. 5I), mutants exhibited a continuous array of caudal rays, contrasting with the separate anterior and posterior clusters seen in WTs (Fig. 5A). In addition, c13a−/− juveniles formed distal radials at the ends of the parhypural and hypurals 1 and 2 (black arrowhead, Fig. 5K), absent in WTs (Fig. 5C). These observations suggest that c13a (Fig. 5L) (i) promotes the formation of caudal fin rays and their separation into upper and lower clusters, (ii) induces hypural diastema formation, and (iii) inhibits distal radial development.
hoxb13a−/−; hoxc13a−/− double mutants lack a caudal fin
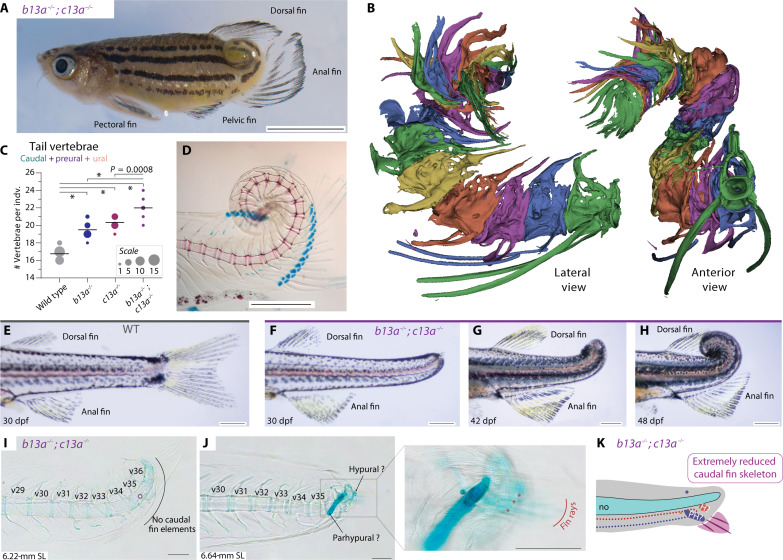
b13a−/−; c13a−/− double mutants exhibited severe tail abnormalities, including dorsal coiling of the tail from the anterior end of the anal fin and complete absence of the caudal fin (Fig. 6A); hence, the anal fin became positioned caudally and assumed the functional role of a caudal fin in swimming due to tail distortion (Fig. 6A and movie S1). Microcomputed tomography (micro-CT) imaging and conventional skeletal staining revealed malformed, fused, and atrophied skeletal elements in the tail of adult double mutants (Fig. 6, B and D; fig. S8, A and B, and movie S2). Furthermore, double mutants had an average of 22 tail vertebrae, exceeding the counts of single mutants by two or three and WTs by five (Fig. 6C). Notably, the dorsal, anal, pectoral, and pelvic fins showed no defects (fig. S6). Therefore, b13a and c13a act additively in arresting body axis elongation and are redundant but necessary to promote the formation of a caudal fin.
Fig. 6. b13a−/−; c13a−/− double mutants have coiled tails that lack a caudal fin.
(A) Adult double mutant zebrafish showing a coiled tail and posterior repositioning of the anal fin. Scale bar, 5 mm. (B) Segmented micro-CT images of the double mutant tail with each vertebra colored differently. The posterior vertebrae are abbreviated and highly fused. (C) Tail vertebral number in double mutants, single mutants, and WTs. ANOVA revealed a significant increase in tail vertebrae [F3,53 = 101.91, P < 0.0001]. Multiple comparisons indicated that, except for the b13a-c13a pair, all comparisons were statistically significant, indicating that both genes act additively contributing to tail vertebrae number. *P < 0.0001. The line indicates the mean and circle size is proportional to the number of individuals. (D) Cleared and stained skeletal preparation of an adult double mutant tail, revealing a spiral tail without caudal fin rays at the tip. Scale bar, 1 mm. (E) Lateral view of a WT tail. (F to H) Ontogenetic sequence of tail transformations in b13a−/−; c13a−/− double mutants. Scale bars, 1 mm. (I) A cleared and stained skeletal preparation of 6.22-mm NL double mutant larva reveals the absence of any cartilaginous or bony element unique to the caudal fin, including no hypural or fin rays. (J) 6.64-mm NL double mutant larva shows two cartilaginous elements and three rudimentary fin rays. (K) Diagram of the same specimen depicting the presence of few fin rays and abbreviated caudal fin, consisting of a single hypural and a single preural element. Scale bars, 0.1 mm [(I) to (K)]. For anatomical terminology, see Fig. 1.
To understand the origin of this phenotype, we examined the development of double mutants (Fig. 6, F to K). Before fin fold resorption, at around 30 dpf, the tail’s posterior end was still mostly straight but longer than in WTs (Fig. 6, E and F). Subsequently, the tail began coiling dorsally in a spiral manner (Fig. 6, G and H). Notably, double mutant larvae did not develop caudal fin skeletal elements (Fig. 6I), although one of five larvae analyzed formed three rudimentary fin rays at the end of an elongated cartilaginous element potentially corresponding to the parhypural due to the presence of a haemal arch (Fig. 6, J and K). Thus, in comparison to single mutants, b13a−/−; c13a−/− double mutants lost most, if not all, preural and ural elements of the caudal fin (Figs. 6K and 7A).
Fig. 7. b13a−/− and c13a−/− zebrafish mutants recapitulate the heterocercal-to-homocercal transition.
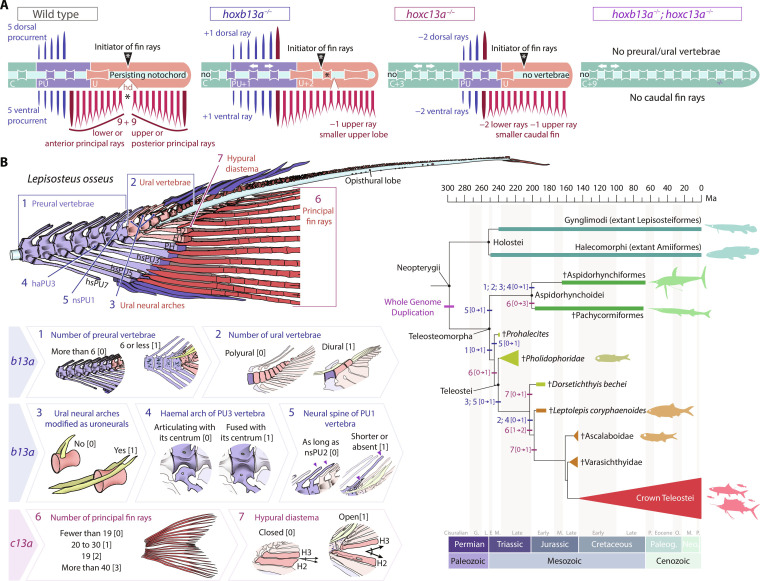
(A) Linearized diagram illustrating the phenotypic differences in a single and double b13a−/− and c13a−/− mutants compared to a WT tail. White arrows indicate expanded body regions. “Initiator of fin rays” indicates the position where caudal rays appear during ontogeny. The black asterisk represents the separation between upper and lower lobe caudal fin ray clusters. (B) Diagram of the caudal fin skeleton of the longnose gar Lepisosteus osseus, highlighting the main morphological characters reversed in b13a−/− and c13a−/− mutants (numbered from 1 to 7). States of the seven characters numbered from [0] to [3]. These characters are mapped on a simplified phylogeny of stem teleosts (44–46), with Holostei as an outgroup. Most characters experienced high homoplasy and underwent modifications during the evolution of basal teleosts in the Triassic during the heterocercal-to-homocercal transition. See Fig. 1 for anatomical terminology.
A homeotic transformation underlies the heterocercal-to-homocercal transition
Intriguingly, zebrafish b13a−/− and c13a−/− single mutants exhibit similarities to the heterocercal tails of primitive teleosteomorphs, including the absence of uroneurals, additional preural and ural centra, a reduced number of principal rays, and the lack of the hypural diastema (6, 8, 42, 43). Thus, to critically compare b13a−/− and c13a−/− phenotypes to ancestral caudal fin morphologies, we extracted a subset of caudal fin characters that were significantly modified in our mutants from morphological matrices of stem teleost phylogenies (Supplementary Text and table S2) (44–46) and coded mutant phenotypes as character states (Fig. 7B). The polarity of changes for each modified character was subsequently mapped back into a simplified phylogeny of stem teleost lineages, with holosteans as an outgroup (Fig. 7B) (44–46).
We identified seven modified characters traceable to the fossil record. The primitive condition, as illustrated in the living holostean Lepisosteus osseus (Fig. 7B), included more than six preural vertebrae (Char. 1 [0]); a polyural skeleton with over three ural vertebrae (Char. 2 [0]); ural vertebrae with neural arch instead of uroneurals (Char. 3 [0]); an autogenous haemal arch of preural centrum 3 (Char. 4 [0]); neural spine of preural centrum 1 as long as the preceding spine (Char. 5 [0]); fewer than 19 principal fin rays (Char. 6 [0]); and a closed hypural diastema (Char. 7 [0]) (table S2). The b13a−/− mutants reversed characters 2 to 5 (Fig. 3, C to E, and fig. S7B) and partially reversed characters 1, 6, and 7 (Figs. 2, D and E, and 3C), while c13a−/− mutants reversed characters 6 and 7 (Fig. 2, D and E) and partially reversed character 3 (Fig. 3E). In addition, b13a−/− mutants exhibited alterations untraceable to the fossil record, including opisthural cartilage absence (fig. S7B), and misalignment between first caudal fin rays forming and hypural diastema (Fig. 5H), which are however characters seen in living holosteans (4, 6, 10, 11). Overall, both mutants showed partial or complete reversals to ancestral heterocercal caudal fin morphologies.
The described characters emerged among the earliest diverging stem teleosts, exhibiting homoplasy across clades. Mesozoic †Pachycormiformes and †Aspidorhynchiformes showed tail modifications convergent with teleost characters 1 to 6 (Fig. 7B). †Pachycormiformes increased principal ray number to more than 40 (Char. 6 [3]), while †Aspidorhynchiformes acquired characters attributed to b13a (Chars. 1 to 4, [1]), including the convergent modification of ural neural arches into uroneurals (4, 7). In the lineage leading to modern teleosts (†Prohalecites plus Teleostei), all seven characters were gradually acquired until the establishment of the homocercal tail in the early Jurassic †Leptolepis coryphaenoides (Fig. 7B) (8). Subsequently, the teleost caudal skeleton stabilized with fewer than six preurals (Char. 1 [1]), two ural vertebrae (Char. 2 [1]), uroneurals (Char. 3 [1]), fused haemal arch with the preural centrum 3 (Char. 4 [1]), short neural spine of preural centrum 1 (Char. 5 [1]), 19 principal rays (Char. 6 [2]), and an open hypural diastema (Char. 7 [1]). Thus, the altered characters in b13a−/− and c13a−/− mutants stabilized concurrently with the development of the homocercal tail after an initial phase of morphological diversification in Teleosteomorpha.
DISCUSSION
Homeotic patterning of the zebrafish tail
We describe homeotic transformations in the zebrafish tail caused by mutations in two posterior hox genes. Vertebrate Hox13 paralogs, along with Hox9 to Hox12 paralogs, are orthologs of the Drosophila posterior Hox gene Abd-B (15, 47), which determines posterior segment identity (48). Gain-of-function mutations in Drosophila Hox genes tend to transform body segments into more posterior ones, such as in Antennapedia (49, 50). Conversely, loss-of-function mutations result in anteriorization, with segments assuming a more anterior fate (48). This phenomenon, known as posterior prevalence (41), arises because Hox genes expressed in more posterior domains (and more 5′ in the cluster) have phenotypic precedence over Hox genes expressed in more anterior domains (and more 3′ in the cluster). Posterior prevalence can explain the phenotypes described here. For instance, b13a−/− mutants exhibit preural-like vertebrae in the ural region, which normally lack centra but retain the notochord (Fig. 7A). Mutants for c13a were also anteriorized, because segments that would typically form preurals instead develop midcaudal vertebrae, accompanied by expanded expression of the adjacent gene c12a (Fig. 4C). Therefore, both b13a−/− and c13a−/− zebrafish mutants exhibit anteriorization, as do Drosophila mutants lacking the Hox13 ortholog Abd-B, and are, therefore, homeotic mutants.
Zebrafish b13a−/− and c13a−/− mutant phenotypes resemble mouse Hoxb13−/− and Hoxc13−/− mutants, which exhibit tail anteriorization: Hoxb13−/− and Hoxc13−/− mutant mice have dorsal root ganglia and vertebral lateral processes in more posterior tail segments, respectively (21, 22). Furthermore, expression patterns of Hoxb13 and Hoxc13 in chicken (18) show the same posterior-anterior relationship observed in zebrafish (Fig. 1D). Similar anteriorization phenotypes and expression patterns across zebrafish, mouse, and chicken support a general function of Hox13 paralogs in specifying vertebrate tail identity. Furthermore, posterior Hox genes not only determine segmental identity but also regulate body axis elongation. Hox13 genes inhibit somitogenesis and axis elongation by affecting mesoderm ingression (18), the Lin28/let-7 pathway (51), and growth differentiation factor signaling (19). In mice, gain of function of either Hoxb13 or Hoxc13 truncates the body axis (20), while Hoxb13 loss of function increases the number of vertebrae (21). Accordingly, our results show that b13a and c13a loss of function in zebrafish increases the number of somites (Fig. 4B) and vertebrae (Fig. 3A), indicating a conserved role in arresting body axis elongation.
The zebrafish tail elongates as a result of posterior tail bud progenitors converging to the midline, where they intercalate and compact (52, 53) in the absence of cell proliferation (54, 55), thereby lengthening the tail without an increase in volume (52, 56). This suggests that forming additional somites would deplete the posterior mesodermal progenitor population, responsible for migrating distally and giving rise to caudal fin rays (57). Consequently, the elongated tails with extra somites observed in our single b13a and c13a mutants (Figs. 3, A to C, and 4B) explain the smaller caudal fin size with fewer rays (Fig. 2, A to D) due to the reduced number of remaining fin ray–forming posterior progenitors. Thus, hox13 mutants would anteriorize the zebrafish body axis by shifting posterior mesodermal progenitor identity from fin ray precursors to somite precursors, explaining the observed gene-dosage effect of both single mutants, resulting in fewer rays and additional vertebrae compared with WT (Figs. 2D and 3B). With c13a expressed more anteriorly than b13a at 1 dpf (Fig. 1D), mutants would repattern caudal fin ray progenitors toward the more anterior midcaudal vertebrae, while posteriorly expressed b13a (Fig. 1D) would shift upper lobe fin ray identity toward the more posterior ural vertebrae.
The b13a−/−; c13a−/− double mutant zebrafish exhibit severe anteriorization, lacking caudal fin rays and endoskeletal fin elements (Fig. 6), implying partial redundancy of the two paralogs in specifying a caudal fin. As hypothesized above, depletion of the fin ray progenitor population due to those cells contributing to a longer axis instead would explain the complete loss of the caudal fin. Intriguingly, the spiraling tail and posterior repositioning of the anal fin in double mutants (Fig. 6) mimic the coiled tail of seahorses, which swim using their dorsal fin in the absence of a caudal fin (58), despite retaining copies of both paralogs (fig. S3). A schematic representation of a straightened double mutant tail (Fig. 7A) reveals a tetrapod-like structure with neural and haemal arches, indicating conservation of axial tail patterning across bony vertebrates. The caudal fin is ancestrally shared by ray- and lobe-finned fishes (59) and so is the broad functional conservation of Hoxb13 and Hoxc13 paralogs (fig. S3), suggesting that the independent losses of the caudal fin in tetrapods and certain teleost lineages, like seahorses or sunfishes, may have involved the same developmental mechanism proposed for b13a−/−; c13a−/− double mutants, by shifting posterior identity from forming caudal fin rays toward an increase in somite number and a lengthening of the tail.
Evolution of Hox13 gene functions
The b13a−/−; c13a−/− zebrafish double mutants displayed normal patterning of dorsal, anal, pectoral, and pelvic fins despite its severe tail defects. In contrast, a13a−/−; a13b−/− double mutant zebrafish, reciprocally, had impaired or missing dorsal, anal, pectoral, and pelvic fins while having a normal caudal fin (34). Furthermore, the caudal fin is clearly distinguishable from other fins because of (i) its direct anchoring to the body axis through posterior vertebrae, (ii) an absence of a zone of polarizing activity (ZPA) for establishing its anteroposterior axis (60, 61), and (iii) the lack of nested Hoxd9–13 expression (62, 63). Because the caudal fin likely evolved first among vertebrate fins (59), a single proto-Hox13 gene may have initially specified caudal fin formation linked to posterior identity. Following the two rounds of whole genome duplication at the base of vertebrate lineage (13), the fin-forming function of Hox13 genes may have been repurposed from the axis to other appendages (62, 64). In this scenario, Hoxb13 and Hoxc13 retained the ancestral caudal fin patterning function, while Hoxa13 and Hoxd13 underwent neofunctionalization and subfunctionalization (65) to pattern the dorsal, anal, and paired fins during vertebrate evolution.
This study has established previously unforeseen correlations between successive evolutionary transformations leading to the homocercal tail and the phenotypic changes observed in hox13 mutants. While mammals and nonteleost ray-finned fishes have a single copy of each Hox13 gene, teleosts experienced a whole genome duplication (TGD), doubling their number (fig. S3) (23, 25, 66). The TGD likely occurred at the base of Teleosteomorpha (Fig. 7B) (24) and provided opportunities for neofunctionalization and subfunctionalization events (65) without pervasive pleiotropic consequences (67, 68) that may have facilitated parallel evolution (69, 70). As expected for this model, the stepwise emergence of the homocercal caudal fin followed the TGD associated with substantial early homoplasy (Fig. 7B). Thus, the TGD likely enabled relaxation of constraints that allowed the evolution of original functions for genes such as b13a and c13a, contributing, for instance, to the appearance of uroneurals and a prominent hypural diastema. Moreover, considering that in heterocercal tails, the body axis extends along the caudal fin upper lobe (Fig. 1A), spatiotemporal shifts in hox13 gene expression might have led to an earlier arrest of axis extension and increased caudal ray number, at the expense of posterior vertebrae number, allowing the evolution of a homocercal caudal fin.
Hox13 genes as a basis for teleost caudal fin morphological diversification
The phenotype of b13a−/− mutants reflects evolutionary changes in the homocercal tail. In primitive heterocercal tails in holosteans and early diverging teleosteomorphs, the notochord extends far beyond the hypurals (Figs. 1A and 7B) (3, 71, 72). The evolution of the homocercal tail involved reduction and internalization of this notochordal extension, resulting in an enclosed notochord ending in an opisthural cartilage over the last series of hypurals in stem teleost lineages (Fig. 1, B and D) (4, 38, 40, 73, 74). Many teleost lineages with derived morphologies, such as mormyrids, toadfishes, and atherinomorphs (75–77), further specialized by ossifying the notochord’s posterior end and losing the opisthural cartilage (1–3). Zebrafish b13a−/− mutants exhibited notochord ossification up to its posterior end and the absence of an opisthural cartilage (fig. S6A), resembling this later stage of homocercal evolution. Thus, these transformational series could be attributed to homeotic changes associated with the b13a gene during teleost evolution.
The c13a−/− mutants also provide insights into the regulation of caudal fin ray number and lobe separation. In heterocercal tails, principal ray number matches the number of supporting haemal spines and hypurals (4). Early diverging teleosteomorphs convergently increased principal fin ray number (Fig. 7B), resulting in DV mirror-image symmetry of 19 principal fin rays reflected around an open hypural diastema after the acquisition of a homocercal tail (Fig. 7B) (4, 10). This pattern has been retained in early diverging crown teleosts, such as elopiforms, clupeiforms, and salmoniforms (4, 7, 73, 74). However, in lineages with derived morphologies, such as flatfishes, cobitids, and cyprinodontids (77–79), the caudal fin shares characteristics similar to c13a−/− mutants, including reduced fin ray number and a closed hypural diastema, resulting in a continuous array of fin rays (Fig. 3D). Thus, regulation of c13a likely played a role not only in the evolution of a symmetric homocercal tail but also in the series of caudal fin transformations that convergently occurred during the diversification of teleosts.
Our findings provide a robust developmental genetic explanation for the evolution of a major vertebrate innovation: the transition of the caudal fin from a heterocercal to a homocercal form. These findings link the segmental identity of the posterior end of the teleost body to two hox13 paralogs, hoxb13a and hoxc13a, which pattern the oldest vertebrate appendage, the caudal fin. The origin of these genes after two successive rounds of vertebrate whole genome duplications followed by a teleost-specific genome duplication may have released genetic constraints, permitting the complex series of morphological transformations that resulted in the highly derived homocercal caudal fin, which may have provided advantages in mobility that contributed to teleost fishes radiating during the Mesozoic and coming to dominate extant ichthyofauna. Alterations to the regulation of b13a and c13a expression might have further provided a substrate for the vast and beautiful morphological diversity of teleost caudal fins.
MATERIALS AND METHODS
Zebrafish husbandry and experimental conditions
Zebrafish embryos were naturally spawned and collected in the zebrafish facility at the Facultad de Ciencias, Universidad de Chile and then raised in petri dishes at a constant temperature of 28°C in E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.3 mM MgSO4, and 0.1% methylene blue, buffered at pH 7.0), according to standard procedures (80). Larvae and juveniles were maintained at a density of 20 fish/liter under a 14:10 light-dark cycle. Animals were fed two to three times a day with dry food particles (Gemma, Skretting, Norway). Water conditions were maintained at 28°C within a pH range of 7 to 7.3 and a conductivity of 600 to 800 μS. Adult fish were housed at a density of four to eight fish/liter and fed twice a day. All fish manipulations were performed under anesthesia or after euthanasia using MS-222 (Tricaine), the animal procedures adhered to the guidelines and received approval from the Institutional Animal Use and Care Committee of the University of Chile, certificate number 18141-FCS-UCH.
Staging of zebrafish embryos, juveniles, and adults
We identified zebrafish embryonic stages until 4 dpf according to previous descriptions (81). For postembryonic stages, we relied on external anatomy and length measurements, using NL for preflexion stages or SL for postflexion stages. In addition, we compared the sequence of ossification in the axial and caudal fin skeleton with previous studies (7, 37, 82, 83).
In situ hybridization
Specific primers were designed to target and amplify the cDNA sequences of hoxa13a, hoxb13a, hoxc13a, hoxc13b, and hoxc12a using National Center for Biotechnology Information (NCBI) primer designing tool (table S3). Recombinant DNA plasmids were generated for each of these genes by cloning the polymerase chain reaction (PCR) product into the dual promoter pCRII-TOPO vector, following instructions provided with the TOPO TA Cloning kit. Plasmids were then purified using an E.Z.N.A. Plasmid Miniprep kit II. The direction of insertion into the TOPO TA vector was confirmed through restriction enzyme digestion assays. For in vitro transcription of digoxigenin-labeled RNA, SP6, T3, or T7 RNA polymerases were used (84), along with a DIG labeling mix (Roche) followed by a deoxyribonuclease step and ethanol precipitation. The labeled in situ RNA probes were resuspended in nuclease-free water and stored at −20°C. Plasmids for hoxa13b and hoxd13a in situ probes were provided by T. Schilling (85), while the plasmid for xirp2a in situ probe was a gift from S. Schulte-Merker (39).
We followed a modified version of the in situ hybridization protocol described by Thisse and Thisse (86). The modifications are as follows: For fixation, we used a 4% paraformaldehyde solution with 1% dimethyl sulfoxide (DMSO) for 1 to 3 hours at room temperature. To achieve dehydration, we directly transferred embryos from PBST (1× PBS, 1% Tween 20) into 100% methanol. Bleaching was performed using a solution of 3% H2O2 and 0.5% (w/v) KOH immediately after rehydration. The digestion step involved treating embryos with proteinase K (PK) solution in 1% DMSO adjusted to 10 μg/ml PK for 10 min for 1-dpf embryos, 20 μg/ml PK for 20 min for 2-dpf embryos, and 20 μg/ml PK for 30 min for 3- and 4-dpf embryos. For prehybridization, we used a solution without ribosomal RNA and heparin (50% formamide, 5× SSC, 0.2% Tween 20, and 0.0092 M citric acid). The in situ hybridization probes were used at a concentration of 0.55 ng/μl, with the hybridization solution supplemented with 5% dextran sulfate. The anti-DIG antibody was used at a concentration of 1:3000, with the blocking buffer containing 1% DMSO. Last, before transfer into glycerol, embryos were dehydrated in methanol to enhance contrast.
Genome editing
Genome editing was performed using the CRISPR-Cas9 system (87–89). Two single guide RNAs (sgRNAs) per gene were designed, targeting the first exon of hoxb13a and hoxc13a genes. CRISPRscan (table S3) (90) was used for sgRNA design, selecting guides with the lowest off-target cutting frequency determination scores. The selected sgRNAs targeted the following sequences: hoxb13a, 5′ ACCGTACGGGATGGGACCTG 3′; 5′ GAGCCATGGACAAGGCTAGA 3′; hoxc13a, 5′ GAGCAGCGCTTTGCCCACAG 3′; 5′ GAAGCGCTAGATGACGTCTG 3′. The sgRNAs were synthetized using a PCR-based strategy (91) and transcribed using the MEGAscript T7 Transcription Kit. Cas9 mRNA was transcribed from the pCS2 + hspCas9 plasmid (92) using the mMESSAGE mMACHINE SP6 Transcription kit.
One-cell stage zebrafish embryos were injected with a final concentration of Cas9 mRNA (400 ng/μl) and sgRNA (100 ng/μl). The efficiency of each sgRNA was assessed using a heteroduplex shift assay (93) with specific primers (table S3). The injected F0 generation of CRISPR-Cas9 was raised and outcrossed to WTs. F1 progeny was genotyped by Sanger sequencing to identify transmitted mutations. Frameshift mutations resulting in an early stop codon in the first exon of the hoxb13a and hoxc13a genes were selected. Heterozygous F1 fish carrying the same mutation were incrossed to generate the F2 generation. The F2 generation was raised together in the same rack until adulthood and then was genotyped and categorized into WT, heterozygous, or homozygous mutants for subsequent morphological comparisons. This experimental setup minimized the likelihood that morphological differences among them result from difference other than the selected mutation.
Genotyping
Genotyping was performed using a heteroduplex mobility shift assay (93) (fig. S4C) to identify mutations using genomic DNA provided by clipping the anal fin of adult fish. Tissue samples were incubated in a 50 mM NaOH solution and heated at 98°C for 15 min. Mechanical homogenization was conducted using pipette tips, and samples were buffered with 100 mM tris (pH 8.0). PCR primers were designed to amplify a 300-bp product encompassing the targeted site of CRISPR-Cas9 mutagenesis (table S3). PCR amplification was performed using SapphireAmp Fast PCR Master Mix directly from unidentified DNA samples. The resulting PCR products were loaded onto acrylamide gels [15% (w/v) acrylamide:bisacrilamide 29:1 solution, 1× tris-borate EDTA (TBE), 0.167% ammonium persulfate, and 0.001% tetramethylethylenediamine (TEMED)] in a vertical electrophoresis chamber (Bio-Rad) filled with 1× TBE (89 mM tris, 89 mM boric acid, and 2 mM EDTA). The gels were then subjected to electrophoresis at 180 V for 90 min.
The presence of both a mutant and a WT allele in the same sample, indicating heterozygosity, was visualized by a mobility shift and observed as a delay in DNA migration on the acrylamide gel (red arrowhead, fig. S4C). To distinguish homozygous mutants from WTs, high-quality genomic DNA was extracted from hoxb13a and hoxc13a mutants by anal fin clipping and lysis buffer incubation [10 mM tris (pH 7.5), 10 mM EDT (pH 8), 200 mM NaCl, 0.1% IGEPAL CA-630, and PK (200 μg/ml)] for 2 hours at 50°C, followed by phenol:chloroform extraction and ethanol precipitation. The purified genomic DNA of homozygous mutants was combined with each sample to perform a mobility shift assay. When a WT sample was combined with a homozygous mutant allele, it generated a mobility shift similar to a heterozygous genotype (blue asterisks, samples 5 and 6, fig. S4D). Conversely, combining mutant DNA with a mutant sample did not result in a delay in electrophoretic mobility (red asterisk, sample 7, fig. S4C). By using both approaches, we could unambiguously identify all homozygous and heterozygous genotypes.
Genomic identification of hox13 orthologs
Annotations of hox13 genes were obtained from high-quality genome assemblies in the NCBI database. Each Hox cluster was analyzed individually to determine the presence or absence of hox13 genes. Taxon sampling included all teleost fish families available in the database, ensuring representation of at least one member from each family. This sampling strategy covered three elopomorphs, three osteoglossomorphs, three clupeomorphs, and several ostariophysan and euteleosteomorph families.
Clearing and staining
Clearing and staining procedures were conducted using alizarin red and Alcian blue dyes. For juveniles and adults, a standard acid protocol was followed (40). In contrast, an acid-free protocol was used for larvae (94).
Image acquisition and processing
Photographs of in situ hybridization as well as cleared and stained specimens were obtained using a transmitted light Olympus BX51 and Zeiss Axiolab 5 microscopes, using 4×, 10×, and 20× objectives. Micro-CT images were acquired using a Skyscan 1272 scanner with imaging parameters set at 80 kV and 125 μA, achieving a resolution of 5 μm. The image acquisition process involved capturing images at intervals of 0.2° around a complete 360° rotation, with each image exposed for 2 s. Images were analyzed and segmented using Slicer 3D.
Multiple images were stacked and merged using Photoshop auto-align and auto-blend tools, and background was adjusted using the fill tool in Photoshop. Image size and resolution were adjusted in Photoshop to 300 dots per inch. Brightness, contrast, white balance, and tone processing were performed in Adobe Lightroom Classic. Final figures and diagrams were assembled in Adobe Illustrator.
Quantification and statistical analyses
NL was measured for preflexion larvae, extending from the tip of the snout to the posterior tip of the notochord. SL was used for postflexion larvae, juveniles, and adults, measured from the tip of the snout to the posterior margin of the hypural plate. Caudal fin length was measured from the posterior margin of the hypurals to the most distal tip of the upper or lower caudal fin lobes. To assess differences in caudal fin size between different genotypes, length measurements were logarithmically transformed to accommodate for scale variations (95), followed by an analysis of covariance, using a model II linear regression. Statistical analyses for fin ray counts involved conducting two-tailed analysis of variance (ANOVA) test followed by Tukey’s multiple comparisons test. Brown-Forsythe tests were conducted to assess equal variances. Statistical analyses for the number of vertebrae, somites, hypurals, epurals, caudal fin distal radials, and principal and procurrent rays involved fitting Poisson generalized linear models (GLM) in R software. For each comparison, the minimum or maximum value was established as zero, thereby preserving count differences as positive integers relative to this reference point. Analyses of deviance were performed using the ANOVA function of the “car” R package, calculating a likelihood ratio chi-square for Poisson GLM regressions. When variance exceeded zero, multiple comparisons were conducted using the “emmeans” R package. Residual diagnostics, including equal variance assessments, were performed using the “DHARMa” R package.
Acknowledgments
We thank F. Espinoza for management in the Allende Lab; Y. Segovia, F. Benassi, P. Vargas, and D. Henríquez for their assistance with fish husbandry; J. Botelho (PUC) for help with micro-CT and discussions; and F. Gajardo, C. Vásquez, A. Vargas, Á. Glavic, V. Muñoz (U. de Chile), and S. Marcellini (U. de Concepcion) for comments and discussions. We thank T. Schilling and S. Schultze-Merker for in situ probes against hoxa13b, hoxd13a, and xirp2a.
Funding: This work was supported by ANID grant Fondecyt Postdoctorado (Chile): 3210275 (N.C), ANID Millennium Institute Center for Genome Regulation: ICN2021_044 (Chile) (M.L.A.), ANID grant Fondecyt regular (Chile): 1180606 (M.LA), ANID grant Doctorado Nacional 2015 (Chile): 21150789 (N.C), NSF grant (USA): 043162 (G.A.), and NIH grant R01 (USA): OD011116 (J.H.P.)
Author contributions: Conceptualization: N.C., G.A., M.L.A., T.D., J.H.P., and S.M.-S. Methodology: N.C., G.A., and M.L.A. Formal analysis: N.C. Investigation: N.C., G.A., and S.M.-S. Resources: M.L.A. and N.C. Visualization: N.C., G.A., S.M.-S., and M.L.A. Funding acquisition: M.L.A., N.C., and J.H.P. Project administration: N.C. and M.L.A. Supervision: M.L.A., N.C., and T.D. Validation: N.C., M.L.A., and T.D. Data curation: N.C. and M.L.A. Writing—original draft: N.C., G.A., and M.L.A. Writing—review and editing: N.C., G.A., J.H.P., T.D., M.L.A., and S.M.-S.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S8
Tables S1 to S3
Legends for movies S1 and S2
References
Other Supplementary Material for this manuscript includes the following:
Movies S1 and S2
REFERENCES AND NOTES
- 1.Heckel J. J., Über das Wirbelsäulen-Ende bei Ganoiden und Teleostiern. Sber. Akad. Wiss. Wien 5, 143–147 (1850). [Google Scholar]
- 2.A. Kölliker, Über das Ende der Wirbelsäule der Ganoiden und einiger Teleostier. (von Willhelm Engelmann, 1860).
- 3.J. A. Ryder, "On the origin of heterocercy and the evolution of the fins and fin rays of fishes," Rep. US Comm. Fish and Fisheries. (US Government Printing Office, 1884), pp. 981–1107.
- 4.Schultze H.-P., Arratia G., The composition of the caudal skeleton of teleosts (Actinopterygil: Osteichthyes). Zool. J. Linn. Soc. 97, 189–231 (1989). [Google Scholar]
- 5.Nybelin O., Zur Morphologie und Terminologie des Schwanzskelettes der Actinopterygier. Ark. Zool. 15, 485–516 (1963). [Google Scholar]
- 6.B. D. Metscher, P. E. Ahlberg, "Origin of the teleost tail: Phylogenetic frameworks for developmental studies" in Major Events in Early Vertebrate Evolution, P. E. Ahlberg, Ed. (Taylor & Francis, 2001), chap. 19, pp. 333–349.
- 7.H.-P. Schultze, G. Arratia, "The caudal skeleton of basal teleosts, its conventions, and some of its major evolutionary novelties in a temporal dimension" in Mesozoic Fishes 5 – Global Diversity and Evolution, G. Arratia, H.-P. Schultze, M. V. H. Wilson, Eds. (Verlag Dr. Friedrich Pfeil, 2013), pp. 187–246.
- 8.Arratia G., Complexities of early Teleostei and the evolution of particular morphological structures through time. Copeia 103, 999–1025 (2015). [Google Scholar]
- 9.Huxley T. H., Original communications: Observations on the development of some parts of the skeleton of fishes. J. Cell Sci. 1, 33–46 (1859). [Google Scholar]
- 10.Desvignes T., Carey A., Postlethwait J. H., Evolution of caudal fin ray development and caudal fin hypural diastema complex in spotted gar, teleosts, and other neopterygian fishes. Dev. Dyn. 247, 832–853 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desvignes T., Robbins A. E., Carey A. Z., Bailon-Zambrano R., Nichols J. T., Postlethwait J. H., Stankunas K., Coordinated patterning of zebrafish caudal fin symmetry by a central and two peripheral organizers. Dev. Dyn. 251, 1306–1321 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakatani Y., Shingate P., Ravi V., Pillai N. E., Prasad A., McLysaght A., Venkatesh B., Reconstruction of proto-vertebrate, proto-cyclostome and proto-gnathostome genomes provides new insights into early vertebrate evolution. Nat. Commun. 12, 4489 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simakov O., Marletaz F., Yue J. X., O'Connell B., Jenkins J., Brandt A., Calef R., Tung C. H., Huang T. K., Schmutz J., Satoh N., Yu J. K., Putnam N. H., Green R. E., Rokhsar D. S., Deeply conserved synteny resolves early events in vertebrate evolution. Nat. Ecol. Evol. 4, 820–830 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis E. B., A gene complex controlling segmentation in Drosophila. Nature 276, 565–570 (1978). [DOI] [PubMed] [Google Scholar]
- 15.Duboule D., Dollé P., The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO J. 8, 1497–1505 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaunt S. J., Krumlauf R., Duboule D., Mouse homeo-genes within a subfamily, Hox-1.4, −2.6 and −5.1, display similar anteroposterior domains of expression in the embryo, but show stage- and tissue-dependent differences in their regulation. Development 107, 131–141 (1989). [DOI] [PubMed] [Google Scholar]
- 17.Graham A., Papalopulu N., Krumlauf R., The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell 57, 367–378 (1989). [DOI] [PubMed] [Google Scholar]
- 18.Denans N., Iimura T., Pourquié O., Hox genes control vertebrate body elongation by collinear Wnt repression. eLife 4, e04379 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aires R., de Lemos L., Novoa A., Jurberg A. D., Mascrez B., Duboule D., Mallo M., Tail Bud progenitor activity relies on a network comprising Gdf11, Lin28, and Hox13 genes. Dev. Cell 48, 383–395.e8 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Young T., Rowland J. E., van de Ven C., Bialecka M., Novoa A., Carapuco M., van Nes J., de Graaff W., Duluc I., Freund J. N., Beck F., Mallo M., Deschamps J., Cdx and Hox genes differentially regulate posterior axial growth in mammalian embryos. Dev. Cell 17, 516–526 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Economides K. D., Zeltser L., Capecchi M. R., Hoxb13 mutations cause overgrowth of caudal spinal cordand tail vertebrae. Dev. Biol. 256, 317–330 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Godwin A. R., Capecchi M. R., Hoxc13 mutant mice lack external hair. Genes Dev. 12, 11–20 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amores A., Force A., Yan Y. L., Joly L., Amemiya C., Fritz A., Ho R. K., Langeland J., Prince V., Wang Y. L., Westerfield M., Ekker M., Postlethwait J. H., Zebrafish hox Clusters and vertebrate genome evolution. Science 282, 1711–1714 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Davesne D., Friedman M., Schmitt A. D., Fernandez V., Carnevale G., Ahlberg P. E., Sanchez S., Benson R. B. J., Fossilized cell structures identify an ancient origin for the teleost whole-genome duplication. Proc. Natl. Acad. Sci. U.S.A. 118, e2101780118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor J. S., Braasch I., Frickey T., Meyer A., Van de Peer Y., Genome duplication, a trait shared by 22000 species of ray-finned fish. Genome Res. 13, 382–390 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thummel R., Li L., Tanase C., Sarras M. P. Jr., Godwin A. R., Differences in expression pattern and function between zebrafish hoxc13 orthologs: Recruitment of Hoxc13b into an early embryonic role. Dev. Biol. 274, 318–333 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Corredor-Adámez M., Welten M., Spaink H., Jeffery J., Schoon R., De Bakker M., Bagowski C., Meijer A., Verbeek F., Richardson M., Genomic annotation and transcriptome analysis of the zebrafish (Danio rerio) hoxcomplex with description of a novel member, hoxb13a. Evol. Dev. 7, 362–375 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Ye Z., Kimelman D., Hox13 genes are required for mesoderm formation and axis elongation during early zebrafish development. Development 147, dev185298 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Géraudie J., Borday Birraux V., Posterior hoxa genes expression during zebrafish bony fin ray development and regeneration suggests their involvement in scleroblast differentiation. Dev. Genes Evol. 213, 182–186 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Thummel R., Ju M., Sarras M. P. Jr., Godwin A. R., Both Hoxc13 orthologs are functionally important for zebrafish tail fin regeneration. Dev. Genes Evol. 217, 413–420 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Fromental-Ramain C., Warot X., Messadecq N., LeMeur M., Dollé P., Chambon P., Hoxa-13 and Hoxd-13 play a crucial role in the patterning of the limb autopod. Development 122, 2997–3011 (1996). [DOI] [PubMed] [Google Scholar]
- 32.Ahn D., Ho R. K., Tri-phasic expression of posterior Hox genes during development of pectoral fins in zebrafish: Implications for the evolution of vertebrate paired appendages. Dev. Biol. 322, 220–233 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Freitas R., Gomez-Marin C., Wilson J. M., Casares F., Gomez-Skarmeta J. L., Hoxd13 contribution to the evolution of vertebrate appendages. Dev. Cell 23, 1219–1229 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Nakamura T., Gehrke A. R., Lemberg J., Szymaszek J., Shubin N. H., Digits and fin rays share common developmental histories. Nature 537, 225–228 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parichy D. M., Elizondo M. R., Mills M. G., Gordon T. N., Engeszer R. E., Normal table of postembryonic zebrafish development: Staging by externally visible anatomy of the living fish. Dev. Dyn. 238, 2975–3015 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.G. Arratia, Actinopterygian postcranial skeleton with special reference to the diversity of fin ray elements, and the problem of identifying homologies in Mesozoic Fishes 4 – Homology and Phylogeny, G. Arratia, H.-P. Schultze, M. V. H. Wilson, Eds. (Verlag Dr. Friedrich Pfeil, 2008), pp. 49–101.
- 37.Bird N. C., Mabee P. M., Developmental morphology of the axial skeleton of the zebrafish, Danio rerio (Ostariophysi: Cyprinidae). Dev. Dyn. 228, 337–357 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Kryvi H., Nordvik K., Fjelldal P. G., Eilertsen M., Helvik J. V., Storen E. N., Long J. H. Jr., Heads and tails: The notochord develops differently in the cranium and caudal fin of Atlantic Salmon (Salmo salar, L.). Anat. Rec. 304, 1629–1649 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lleras Forero L., Narayanan R., Huitema L. F., VanBergen M., Apschner A., Peterson-Maduro J., Logister I., Valentin G., Morelli L. G., Oates A. C., Schulte-Merker S., Segmentation of the zebrafish axial skeleton relies on notochord sheath cells and not on the segmentation clock. eLife 7, e33843 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cumplido N., Allende M. L., Arratia G., From Devo to Evo: Patterning, fusion and evolution of the zebrafish terminal vertebra. Front. Zool. 17, 18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duboule D., Morata G., Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet. 10, 358–364 (1994). [DOI] [PubMed] [Google Scholar]
- 42.Patterson C., The caudal skeleton in lower Liassic pholidophorid fishes. Bull.Br.Mus.Nat.Hist.Geol. 16, 201–239 (1968). [Google Scholar]
- 43.G. Arratia, "The caudal skeleton of Jurassic teleosts; a phylogenetic analysis" in Early vertebrates and related problems in evolutionary biology, M. Chang, H. Liu, G.-R. Zhang, Eds. (Science Press, 1991), pp. 249–340. [Google Scholar]
- 44.Arratia G., New Triassic teleosts (Actinopterygii, Teleosteomorpha) from northern Italy and their phylogenetic relationships among the most basal teleosts. J. Vertebr. Paleontol. 37, e1312690 (2017). [Google Scholar]
- 45.Arratia G., Morphology, taxonomy, and phylogeny of Triassic pholidophorid fishes (Actinopterygii, Teleostei). J. Vertebr. Paleontol. 33, 1–138 (2013). [Google Scholar]
- 46.Arratia G., Schultze H. P., Tischlinger H., On a remarkable new species of Tharsis, a Late Jurassic teleostean fish from southern Germany: Its morphology and phylogenetic relationships. Foss. Rec. 22, 1–23 (2019). [Google Scholar]
- 47.McGinnis W., Garber R. L., Wirz J., Kuroiwa A., Gehring W. J., A homologous protein-coding sequence in Drosophila homeotic genes and its conservation in other metazoans. Cell 37, 403–408 (1984). [DOI] [PubMed] [Google Scholar]
- 48.Lewis E. B., The bithorax complex: The first fifty years. Int. J. Dev. Biol. 42, 403–415 (1998). [PubMed] [Google Scholar]
- 49.Postlethwait J. H., Schneiderman H. A., A clonal analysis of determination in Antennapedia a homoeotic mutant of Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 64, 176–183 (1969). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Postlethwait J. H., Schneiderman H. A., Pattern formation and determination in the antenna of the homoeotic mutant Antennapedia of Drosophila melanogaster. Dev. Biol. 25, 606–640 (1971). [DOI] [PubMed] [Google Scholar]
- 51.Sato T., Kataoka K., Ito Y., Yokoyama S., Inui M., Mori M., Takahashi S., Akita K., Takada S., Ueno-Kudoh H., Asahara H., Lin28a/let-7 pathway modulates the Hox code via Polycomb regulation during axial patterning in vertebrates. eLife 9, e53608 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomson L., Muresan L., Steventon B., The zebrafish presomitic mesoderm elongates through compaction-extension. Cells Dev. 168, 203748 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mongera A., Rowghanian P., Gustafson H. J., Shelton E., Kealhofer D. A., Carn E. K., Serwane F., Lucio A. A., Giammona J., Campàs O., A fluid-to-solid jamming transition underlies vertebrate body axis elongation. Nature 561, 401–405 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang L., Kendrick C., Julich D., Holley S. A., Cell cycle progression is required for zebrafish somite morphogenesis but not segmentation clock function. Development 135, 2065–2070 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bouldin C. M., Snelson C. D., Farr G. H., Kimelman D., Restricted expression of cdc25a in the tailbud is essential for formation of the zebrafish posterior body. Genes Dev. 28, 384–395 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steventon B., Duarte F., Lagadec R., Mazan S., Nicolas J. F., Hirsinger E., Species-specific contribution of volumetric growth and tissue convergence to posterior body elongation in vertebrates. Development 143, 1732–1741 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Lee R. T., Thiery J. P., Carney T. J., Dermal fin rays and scales derive from mesoderm, not neural crest. Curr. Biol. 23, R336–R337 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Neutens C., Adriaens D., Christiaens J., De Kegel B., Dierick M., Boistel R., Van Hoorebeke L., Grasping convergent evolution in syngnathids: A unique tale of tails. J. Anat. 224, 710–723 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Larouche O., Zelditch M. L., Cloutier R., A critical appraisal of appendage disparity and homology in fishes. Fish Fish. 20, 1138–1175 (2019). [Google Scholar]
- 60.Hadzhiev Y., Lele Z., Schindler S., Wilson S. W., Ahlberg P., Strahle U., Muller F., Hedgehog signaling patterns the outgrowth of unpaired skeletal appendages in zebrafish. BMC Dev. Biol. 7, 75 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Letelier J., de la Calle-Mustienes E., Pieretti J., Naranjo S., Maeso I., Nakamura T., Pascual-Anaya J., Shubin N. H., Schneider I., Martinez-Morales J. R., Gomez-Skarmeta J. L., A conserved Shh cis-regulatory module highlights a common developmental origin of unpaired and paired fins. Nat. Genet. 50, 504–509 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Freitas R., Zhang G., Cohn M. J., Evidence that mechanisms of fin development evolved in the midline of early vertebrates. Nature 442, 1033–1037 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Freitas R., Zhang G. J., Cohn M. J., Biphasic Hoxd gene expression in shark paired fins reveals an ancient origin of the distal limb domain. PLOS ONE 2, e754 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Minelli A., The origin and evolution of appendages. Int. J. Dev. Biol. 47, 573–581 (2003). [PubMed] [Google Scholar]
- 65.Force A., Lynch M., Pickett F. B., Amores A., Yan Y. L., Postlethwait J., Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Postlethwait J. H., Woods I. G., Ngo-Hazelett P., Yan Y. L., Kelly P. D., Chu F., Huang H., Hill-Force A., Talbot W. S., Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 10, 1890–1902 (2000). [DOI] [PubMed] [Google Scholar]
- 67.Hittinger C. T., Carroll S. B., Gene duplication and the adaptive evolution of a classic genetic switch. Nature 449, 677–681 (2007). [DOI] [PubMed] [Google Scholar]
- 68.Stern D. L., The genetic causes of convergent evolution. Nat. Rev. Genet. 14, 751–764 (2013). [DOI] [PubMed] [Google Scholar]
- 69.Chen L., DeVries A. L., Cheng C. H., Convergent evolution of antifreeze glycoproteins in Antarctic notothenioid fish and Arctic cod. Proc. Natl. Acad. Sci. U.S.A. 94, 3817–3822 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stern D. L., Orgogozo V., The loci of evolution: How predictable is genetic evolution? Evolution 62, 2155–2177 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grande L., Bemis W. E., A comprehensive phylogenetic study of amiid fishes (Amiidae) based on comparative skeletal anatomy. an empirical search for interconnected patterns of natural history. J. Vertebr. Paleontol. 18, 1–696 (1998). [Google Scholar]
- 72.Desvignes T., Carey A., Braasch I., Enright T., Postlethwait J. H., Skeletal development in the heterocercal caudal fin of spotted gar (Lepisosteus oculatus) and other lepisosteiformes. Dev. Dyn. 247, 724–740 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schultze H. P., Arratia G., Reevaluation of the caudal skeleton of some actinopterygian fishes: II. Hiodon, Elops, and Albula. J. Morphol. 195, 257–303 (1988). [DOI] [PubMed] [Google Scholar]
- 74.Arratia G., Schultze H.-P., Reevaluation of the caudal skeleton of certain actinopterygian fishes: III. Salmonidae. Homologization of caudal skeletal structures. J. Morphol. 214, 187–249 (1992). [DOI] [PubMed] [Google Scholar]
- 75.E. J. Hilton, R. Britz, "The caudal skeleton of osteoglossomorph fishes, revisited: Comparisons, homologies, and characters" in Origin and Phylogenetic Interrelationships of Teleosts, J. S. Nelson, H.-P. Schultze, M. V. H. Wilson, Eds. (Verlag Dr. Friedrich Pfeil, 2010), pp. 219–237. [Google Scholar]
- 76.Vaz D. F., Hilton E. J., The caudal skeleton of batrachoidiformes (Teleostei: Percomorphacea): A study of morphological diversity, intraspecific variation, and phylogenetic inferences. Zool. J. Linn. Soc. 189, 228–286 (2020). [Google Scholar]
- 77.Thieme P., Warth P., Moritz T., Development of the caudal-fin skeleton reveals multiple convergent fusions within Atherinomorpha. Front. Zool. 18, 20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gosline W. A., Some osteological features of modern lower teleostean fishes. Smithson. Misc. Collect. Washingt. 142, 1–42 (1961). [Google Scholar]
- 79.Hoshino K., Homologies of the caudal fin rays of Pleuronectiformes (Teleostei). Ichthyol. Res. 48, 231–246 (2001). [Google Scholar]
- 80.M. Westerfield, The zebrafish book: A guide for the laboratory use of zebrafish (Danio rerio). (Univ. of Oregon Press, ed. 4th, 2000). [Google Scholar]
- 81.Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F., Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310 (1995). [DOI] [PubMed] [Google Scholar]
- 82.Wiley E. O., Fuiten A. M., Doosey M. H., Lohman B. K., Merkes C., Azuma M., The caudal skeleton of the zebrafish, Danio rerio, from a phylogenetic perspective: A polyural interpretation of homologous structures. Copeia 103, 740–750 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bensimon-Brito A., Cancela M. L., Huysseune A., Witten P. E., The zebrafish (Danio rerio) caudal complex - a model to study vertebral body fusion. J. Appl. Ichthyol. 26, 235–238 (2010). [Google Scholar]
- 84.H. Acloque, D. G. Wilkinson, M. A. Nieto, "Chapter 9 In Situ Hybridization Analysis of Chick Embryos in Whole-Mount and Tissue Sections" in Avian Embryology, 2nd Edition. (2008), pp. 169–185. [DOI] [PubMed] [Google Scholar]
- 85.Muto A., Ikeda S., Lopez-Burks M. E., Kikuchi Y., Calof A. L., Lander A. D., Schilling T. F., Nipbl and mediator cooperatively regulate gene expression to control limb development. PLOS Genet. 10, e1004671 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thisse C., Thisse B., High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69 (2008). [DOI] [PubMed] [Google Scholar]
- 87.Hruscha A., Krawitz P., Rechenberg A., Heinrich V., Hecht J., Haass C., Schmid B., Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development 140, 4982–4987 (2013). [DOI] [PubMed] [Google Scholar]
- 88.Hwang W. Y., Fu Y., Reyon D., Maeder M. L., Tsai S. Q., Sander J. D., Peterson R. T., Yeh J. R., Joung J. K., Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31, 227–229 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jao L. E., Wente S. R., Chen W., Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. U.S.A. 110, 13904–13909 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moreno-Mateos M. A., Vejnar C. E., Beaudoin J.-D., Fernandez J. P., Mis E. K., Khokha M. K., Giraldez A. J., CRISPRscan: Designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods 12, 982–988 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nakayama T., Blitz I. L., Fish M. B., Odeleye A. O., Manohar S., Cho K. W., Grainger R. M., Cas9-based genome editing in Xenopus tropicalis. Method. Enzymol. 546, 355–375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ansai S., Kinoshita M., Targeted mutagenesis using CRISPR/Cas system in medaka. Biol. Open 3, 362–371 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sorlien E. L., Witucki M. A., Ogas J., Efficient production and identification of CRISPR/Cas9-generated gene knockouts in the model system Danio rerio. J. Vis. Exp., e56969 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Walker M. B., Kimmel C. B., A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech. Histochem. 82, 23–28 (2007). [DOI] [PubMed] [Google Scholar]
- 95.Bryant E. H., On use of logarithms to accommodate scale. Syst. Zool. 35, 552–559 (1986). [Google Scholar]
- 96.Sordino P., Duboule D., Kondo T., Zebrafish Hoxa and Evx-2 genes: Cloning, developmental expression and implications for the functional evolution of posterior Hox genes. Mech. Dev. 59, 165–175 (1996). [DOI] [PubMed] [Google Scholar]
- 97.Thisse C., Thisse B., High throughput expression analysis of ZF-models consortium clones. ZFIN Direct Data Submission., (2005). [Google Scholar]
- 98.C. Patterson, "The contribution of paleontology to teleostean phylogeny" in Major Patterns in Vertebrate Evolution, M. K. Hecht, P. C. Goody, B. M. Hecht, Eds. (Plenum, 1977), vol. 14, pp. 579–643. [Google Scholar]
- 99.Brito P. M., Révision des Aspidorhynchidae (Pisces, Actinopterygii) du Mésozoïque: Ostéologie, relations phylogénétiques, données environnementales et biogéographiques. Geodiversitas 19, 681–772 (1997). [Google Scholar]
- 100.Schultze H.-P., Arratia G., Reevaluation of the caudal skeleton of actinopterygian fishes: I. Lepisosteus and Amia. J. Morphol. 190, 215–241 (1986). [DOI] [PubMed] [Google Scholar]
- 101.G. Arratia, "The monophyly of Teleostei and stem-group teleosts" in Mesozoic Fishes 2 – Systematics and Fossil Record, G. Arratia, H.-P. Schultze, Eds. (Dr. Friedrich Pfeil, München, 1999), vol. 2, pp. 265–334. [Google Scholar]
- 102.Arratia G., Basal teleosts and teleostean phylogeny. Paleo Ichth. 7, 5–168 (1997). [Google Scholar]
- 103.Arratia G., Remarkable teleostean fishes from the Late Jurassic of southern Germany and their phylogenetic relationships. Mitt. Mus. Nat. Kd., Berl. Geowiss. Reihe 3, 137–179 (2000). [Google Scholar]
- 104.Patterson C., Rosen D. E., Review of ichthyodectiform and other Mesozoic teleost fishes, and the theory and practice of classifying fossils. Bull. Am. Mus. Nat. Hist. 158, 81–172 (1977). [Google Scholar]
- 105.Betancur R. R., Wiley E. O., Arratia G., Acero A., Bailly N., Miya M., Lecointre G., Orti G., Phylogenetic classification of bony fishes. BMC Evol. Biol. 17, 162 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Parey E., Louis A., Montfort J., Bouchez O., Roques C., Iampietro C., Lluch J., Castinel A., Donnadieu C., Desvignes T., Floi Bucao C., Jouanno E., Wen M., Mejri S., Dirks R., Jansen H., Henkel C., Chen W. J., Zahm M., Cabau C., Klopp C., Thompson A. W., Robinson-Rechavi M., Braasch I., Lecointre G., Bobe J., Postlethwait J. H., Berthelot C., Crollius H. R., Guiguen Y., Genome structures resolve the early diversification of teleost fishes. Science 379, 572–575 (2023). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S8
Tables S1 to S3
Legends for movies S1 and S2
References
Movies S1 and S2