Abstract
Legionella pneumophila replicates within a specialized phagosome in cultured cells, a function necessary for its pathogenicity. The replicative phagosome lacks membrane marker proteins, such as the glycoprotein LAMP-1, that are indicators of the normal endocytic pathway. We describe the isolation of several Legionella genes essential for intracellular growth and evasion of the endocytic pathway, using a genetic and cell biological approach. We screened 4,960 ethyl methanesulfonate-mutagenized colonies for defects in intracellular growth and trafficking to the replicative phagosome. Six mutant strains of L. pneumophila that had severe intracellular growth defects in mouse bone marrow-derived macrophages were identified. All six mutants were found in phagosomes that colocalized with LAMP-1, indicating defects in intracellular trafficking. The growth defects of two of these strains were complemented by molecular clones from a bank constructed from a wild-type L. pneumophila strain. The inserts from these clones are located in a region of the chromosome contiguous with several other genes essential for intracellular growth. Three mutants could be complemented by single open reading frames placed in trans, one mutant by a gene termed dotH and two additional mutants by a gene termed dotO. A deletion mutation was created in a third gene, dotI, which is located directly upstream of dotH. The ΔdotI strain was also defective for intracellular growth in macrophages, and this defect was complemented by a single open reading frame in trans. Based on sequence analysis and structural predictions, possible roles of dotH, dotI, and dotO in intracellular growth are discussed.
Legionella pneumophila is a gram-negative, facultative intracellular bacterium and the causative agent of Legionnaires’ pneumonia. The organism is able to infect and survive inside human monocytes and macrophages (29), in addition to growing within freshwater amoebae, which are thought to represent its natural reservoir (13, 46). Replication within macrophages appears critical for disease, as mutants defective for intracellular growth in vitro are unable to cause disease in animal models (9).
Legionnaires’ pneumonia results when L. pneumophila is inhaled as an aerosol by a susceptible individual. Once inside the lung, these bacteria are phagocytosed by resident alveolar macrophages (48), where they establish a specialized compartment for intracellular growth called a replicative phagosome (19). The nature of this replicative niche has been described in detail (19), although few molecular details are known. Initially, L. pneumophila is engulfed by the macrophage (20). Following its engulfment, the bacterium is found within a phagosome bounded by a single membrane inside the eukaryotic cell (18, 20). Phagosomes carrying virulent L. pneumophila are significantly less acidic than those bearing nonpathogenic bacteria (21). In addition, fusion of these phagosomes with the lysosomal compartment does not occur, as the normal endocytic pathway is subverted (10, 19). Recent data suggest that L. pneumophila may alter the maturation of its phagosome before fusion with late endosomes, thus preventing the acquisition of late endosomal and lysosomal markers CD63, LAMP-1, LAMP-2, and cathepsin D (10, 32). This specialized phagosome containing the organism becomes associated sequentially with small vesicles, mitochondria, and rough endoplasmic reticulum, forming a compartment in which the organism replicates to large numbers (19, 40). The ultrastructure of this compartment bears striking similarity to that of autophagous vacuoles (40). The molecular mechanism for formation of this replicative phagosome is unknown.
Several bacterial gene products that are essential for intracellular growth of L. pneumophila have been described. A number of these genes are located in a contiguous region, the dotA (defective in organelle trafficking) and icmWXYZ genes (2, 4). The dotA gene product is a large inner membrane protein of 1,048 amino acids with eight transmembrane domains (2, 33). The predicted topology of DotA relative to the membrane is similar to MalF, an essential component of an ABC-type transport system in Escherichia coli (33). DotA may play a similar role, possibly transporting a substance(s) necessary for intracellular growth and evasion of the endocytic pathway across the bacterial membranes. The icm gene cluster, located adjacent to the dotA gene (4, 25), may also be involved in this function. At least one gene product in this region is likely to be transported across the inner membrane, based on sequence analysis (4). A second cluster of three genes essential for intracellular growth, located approximately 10 kb from dotA, has been identified recently. This locus includes the dotB gene (36, 43), which encodes a predicted protein similar to a family of nucleotide binding proteins involved in the transport of macromolecules across bacterial membranes (16).
It has been shown recently that contact of L. pneumophila with macrophages and erythrocytes at high multiplicities of infection results in pore formation, causing cellular lysis (22, 23). Cytotoxicity is not seen at low multiplicities of infection, suggesting that the eukaryotic cell is able to withstand the insertion of a small number of pores in its plasma membrane, or else the pore is blocked. This observed cytotoxicity is dependent on bacterial proteins required for intracellular growth, such as DotA (23). A bacterial membrane transport complex may be involved in the formation of a pore in eukaryotic cell membranes as an essential step in establishing a replicative phagosome.
This work was initiated to identify and characterize factors of L. pneumophila important for intracellular growth and for targeting to the replicative phagosome. To this end, we describe the characterization of six mutants that were isolated based on defective intracellular growth and an inability to bypass the normal endocytic pathway. This resulted in the identification of three linked genes necessary for intracellular growth and targeting of L. pneumophila.
MATERIALS AND METHODS
Bacterial strains and media.
All L. pneumophila derivatives are from Lp01 (hsdR rpsL), a virulent L. pneumophila Philadelphia-1, serogroup 1 strain that grows intracellularly (2) (Table 1). L. pneumophila strains were routinely cultured either on buffered charcoal-yeast extract agar (BCYE) (12) or in Aces-buffered yeast extract broth (AYE) (15). Mutant L. pneumophila strains were tested for salt resistance by titering on BCYE plates containing 0.65% NaCl (6, 17). Casamino Acids medium (CAA) was used to test mutant L. pneumophila strains for thymine, tryptophan, and nucleoside auxotrophies (28). For L. pneumophila, antibiotics were used at the following concentrations: kanamycin, 20 μg/ml; streptomycin, 50 μg/ml; rifampin, 5 μg/ml; and trimethoprim, 50 μg/ml.
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Relevant genotype | Reference or source |
|---|---|---|
| L. pneumophilaa | ||
| Lp01 | Philadelphia-1 rpsL hsdR | 2 |
| Lp02 | Lp01 thyA | 2 |
| HL019 | Lp01/pMS8 | This work |
| HL900 | Lp01 dotG? | This work |
| HL1000 | Lp01 dotO2 | This work |
| HL1005 | Lp01 dotO2/pMS8 | This work |
| HL1009 | Lp01 dotO2/p1415 | This work |
| HL1011 | Lp01 dotO2/p1A | This work |
| HL1012 | Lp01 dotO2/p2A | This work |
| HL1300 | Lp01 Dot− | This work |
| HL1400 | Lp01 dotO1 | This work |
| HL1405 | HL1400/pMS8 | This work |
| HL1419 | HL1400/p1415 | This work |
| HL1421 | Lp01 dotO1/p1A | This work |
| HL1422 | Lp01 dotO1/p2A | This work |
| HL1600 | Lp01 Dot− | This work |
| HL1700 | Lp01 dotH1 | This work |
| HL1705 | HL1700/pMS8 | This work |
| HL1719 | HL1700/p1713 | This work |
| HL1725 | HL1700/pH3 | This work |
| HL056 | Lp01 dotIΔ1 | This work |
| HL057 | HL056/pMS8 | This work |
| HL059 | HL056/pI1 | This work |
| E. coli | ||
| DH5α | supE44 ΔlacU169 (Φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 49 |
| DH5α (λpir) | DH5α (λpir) tet::Mu recA | 24 |
| XL1-Blue | F′::Tn10 proA+B+ lacIq Δ(lacZ)M15/recA1 endA1 gyrA96 (Nalr) thi hsdR17 (rK− mK+) supE44 relA1 lac | 5 |
| MT607 | recA56 pro-82 thi-1 hsdR17 supE44, RK600 | 14 |
| Plasmids | ||
| pMS4 | oriRSF1010 RP4 mob kan | 39 |
| pMS8 | pMS4 sacB | 37 |
| p1415 | pMS8 proP+ dotO+ | This work |
| p1A | pMS8 proP′ | This work |
| p2A | pMS8 dotO+ | This work |
| p1713 | pMS8 dotH+ dotI+ | This work |
| pH3 | pMS8 dotH+ | This work |
| pI1 | pMS8 dotI+ | This work |
| pSR47 | oriTRP4 oriR6K kan | 27 |
| pSR47S | pSR47 sacB | 45a |
| pHJK | pSR47s dotH+ dotJ+ dotK+ | This work |
All were derived from Philadelphia-1, serogroup 1 strain Lp01 (CDC, Atlanta, Ga.).
Initial isolation of plasmids was in E. coli DH5α or XL1-Blue. E. coli MT607 bearing RK600 (Tra+) was used for mobilizing plasmids into L. pneumophila by triparental conjugation as previously described, using either E. coli DH5α or XL1-Blue as the donor strain (39). For the propagation of suicide plasmids requiring R6K π protein, E. coli DH5α (λpir) (24) was used. Antibiotics were used at the following concentrations with E. coli: kanamycin, 40 μg/ml; and ampicillin, 150 μg/ml.
Cell culture.
Bone marrow-derived macrophages from female A/J mice were prepared as described previously (40). After culturing in L-cell conditioned medium, the macrophages were replated for use by lifting cells in phosphate-buffered saline (PBS) on ice for 5 to 10 min, harvesting cells by centrifugation, and resuspending cells in RPMI 1640 containing 10% fetal bovine serum. Macrophages were used for quick-screen poke plaque assays, immunofluorescence microscopy, and growth curve assays. Macrophage-like U937 cells (American Type Culture Collection) were cultured as described previously (2) and differentiated by treatment with phorbol 12-myristate 13-acetate (Sigma).
Mutagenesis.
Liquid cultures (50 ml) of virulent L. pneumophila strain Lp01 (wild type) were grown to mid-logarithmic phase in AYE medium at 37°C, and bacteria were collected by centrifugation at 2,500 rpm for 10 min (Beckman centrifuge, JA-14 rotor). Bacteria were washed two times in PBS at 37°C and collected by centrifugation at 5,000 rpm for 5 min. Washed bacteria were resuspended in PBS at 37°C and were mutagenized by adding ethyl methanesulfonate (EMS; Eastman Kodak, Rochester, N.Y.) to a 2% (vol/vol) final concentration and aerating for 30 min at 37°C. Mutagenesis was stopped by the addition of 5 volumes of fresh AYE, and bacteria were collected by centrifugation at 5,000 rpm for 5 min. Mutagenized bacteria were resuspended in fresh AYE and divided into pools. These pools were permitted to recover and grow with aeration for 20 h at 37°C. Each pool was titered for viability (approximately 50%) and resistance to rifampin (approximately 80- to 100-fold increase relative to cultures incubated in absence of EMS). Mutagenized pools were collected by centrifugation and frozen in glycerol at −80°C until screening.
Screen for mutants defective for intracellular growth.
Mutagenized bacteria were screened for defects in intracellular growth by using the quick-screen poke plaque assay previously described (2), with minor alterations. Briefly, monolayers of A/J mouse bone marrow macrophages were plated at 2 × 106 to 3 × 106 cells/well in six-well tissue culture dishes. Monolayers were overlaid with a solution of molten 0.7% Noble agar prepared in 0.8% RPMI with 20% fetal bovine serum. Colonies isolated on BCYE plates from the mutagenized pools were poked into the monolayer. Infected monolayers were incubated at 37°C in 5% CO2 for 3 days, overlaid again with a solution of molten 0.9% Noble agar in RPMI with 0.01% neutral red, and incubated for 1 to 3 h at 37°C. Stained monolayers were then inspected for the presence of plaques at the sites of bacterial inoculation. Bacterial isolates that failed to form plaques on macrophage monolayers were retained and retested by using this assay. Intracellular growth defects of mutagenized strains that failed to form plaques in two quick-screen poke plaque assays were assayed by examining intracellular growth yield (38).
To determine intracellular growth yield, bone marrow-derived macrophages were plated at a density of 2.5 × 105 cells/well in 24-well tissue culture dishes. When U937 cells were used for this type of assay, they were plated at a density of 106 cells/well in 24-well tissue culture dishes. Bacterial strains were grown in patches on BCYE plates for 48 h at 37°C, resuspended in PBS, diluted in RPMI, and used to infect monolayers at approximately 105 bacteria/well. Two hours after infection, medium on infected monolayers was replaced. The monolayers were lysed in sterile H2O at this time point (t0), and viable counts were determined to quantitate cell-associated bacteria. A series of identically infected monolayers was maintained at 37°C in a humidified incubator in 5% CO2 for up to 3 days. Supernatants containing bacteria were pooled with lysates from the corresponding infected monolayers and titered for bacterial CFU on BCYE at the times indicated.
Immunofluorescence microscopy.
To assay intracellular trafficking of L. pneumophila derivatives, bone marrow-derived macrophages were plated on 12-mm-diameter glass coverslips in 24-well tissue culture dishes at a density of 105 cells/well. Monolayers were incubated with 106 bacteria/well with L. pneumophila cultured on BCYE for 48 h. Formalin killing of Lp01 was done as previously described (18). After 2 h at 37°C, supernatants were removed and the monolayers were fixed in periodate-lysine-paraformaldehyde (26) containing 5% sucrose for 20 min at room temperature. Fixed monolayers were washed three times in PBS and stored at 4°C until staining.
To stain samples, wells were successively incubated three times for 5 min in blocking buffer (PBS containing 2% goat serum) at room temperature. All antibody probing steps were for 1 h at 37°C in a humidified incubator. After blocking, samples were stained with anti-L. pneumophila polyclonal rabbit serum (produced by K. Berger) diluted 1:10,000 in blocking buffer to identify extracellular bacteria. Samples were washed three times for 5 min with blocking buffer, stained with Cascade blue-conjugated goat anti-rabbit immunoglobulin G (IgG; Molecular Probes, Eugene, Oreg.) diluted 1:500 in blocking buffer, and incubated as described above. Samples were washed three times in PBS for 5 min and then permeabilized in −20°C methanol for 10 s. After incubating three times for 5 min with blocking buffer, samples were stained with anti-L. pneumophila rabbit serum diluted 1:10,000 in blocking buffer to identify both intracellular and extracellular bacteria. Following three 5-min washes with blocking buffer, samples were stained with anti-LAMP-1 rat monoclonal antibody IB4 diluted 1:100 in blocking solution (obtained from the Developmental Studies Hybridoma Bank of the Department of Pharmacology and Molecular Sciences, Johns Hopkins University School of Medicine, Baltimore, Md., and the Department of Biology, University of Iowa, Iowa City) (39). After washing three times for 5 min in blocking buffer, samples were stained simultaneously with rhodamine-conjugated goat anti-rabbit IgG (Molecular Probes) and fluorescein isothiocyanate-conjugated goat anti-rat IgG. Samples were placed in mounting medium (90% glycerol containing 1 mg of phenylenediamine per ml in PBS [pH 9.0]) and visualized by fluorescence microscopy (Zeiss Axioskop).
Complementation of mutants defective for intracellular growth.
An L. pneumophila genomic library cloned in pMS8, containing average inserts of 5 kb in size, was introduced into intracellular growth-defective strains HL1400 and HL1700 by conjugation (39). Plasmid pMS8 is a derivative of plasmid pMS4 (39) bearing the sacB gene in the SalI site (37). The Lp02 (Lp02 is a derivative of Lp01 which is a thymine auxotroph) genomic library used in this study was made by cloning Sau3A fragments from a genomic partial digest into a unique BamHI site in pMS8, created by site-directed mutagenesis of a unique EcoRI site in the sacB gene (37). This strategy was used to provide a selection against vector alone. The library was harbored in XL1-Blue (Table 1). Transconjugants were pooled and frozen at −80°C until use. After thawing, pools were enriched for strains that were able to grow intracellularly by incubating these pools with phorbol ester-treated U937 cells. In six-well tissue culture dishes, approximately 8 × 105 to 1 × 106 bacteria were added to 1 × 107 to 2.5 × 107 U937 cells/well and incubated for 3 to 4 days. At this point, monolayers were lysed and pooled with culture supernatants, and bacteria were centrifuged at 2,500 rpm for 10 min. Recovered bacteria were resuspended in fresh tissue culture medium and introduced onto a second set of U937 cell monolayers for 3 to 4 days. After this time, infected U937 cell monolayers were lysed in H2O, lysates were plated on BCYE containing kanamycin, and colonies on plates from each well were pooled and stored at −80°C. Approximately 7 × 104 to 1 × 105 colonies were recovered per well.
Enriched pools were screened for strains able to grow intracellularly, using single plaques on U937 cells, as previously described (2). For each complemented strain, eight plaques were picked and streaked on BCYE medium containing kanamycin, and four bacterial colonies were selected from each plaque for quantitation of intracellular growth, using U937 cells. Since all 32 strains selected in this fashion for each mutant grew intracellularly, plasmids were isolated from one strain from each plaque (eight plaques). Plasmids were compared by restriction mapping.
DNA manipulations.
Endonuclease digestions, ligations, and DNA purification were performed by following standard protocols (34). Plasmid DNA was prepared from L. pneumophila and E. coli by nonalkaline lysis followed by precipitation with hexadecyltrimethylammonium bromide (11) and ethanol precipitation.
PCRs were in 100-μl reaction volumes, using Vent polymerase in reaction buffer (New England Biolabs), deoxynucleoside triphosphates (200 μM each; Pharmacia), and 50 mM MgCl2 (New England Biolabs). Circular plasmid DNA and genomic DNA isolated from single colonies were used as templates (3).
DNA sequence analysis.
DNA sequences of the inserts of complementing clones p1415 and p1713 were determined by dideoxynucleotide sequencing (Howard Hughes Medical Institute sequencing facility, Harvard Medical School). Inserts of complementing clones were subcloned into pBluescript SK, and both strands were sequenced by using either standard pBluescript SK primers and/or primers located on the sequenced fragments. MacVector 5.0 and AssemblyLIGN were used for sequence analysis and assembly. Predicted protein sequences were compared to protein sequences in the National Center for Biotechnology Information database, using BLASTP analysis.
Plasmid constructions.
Plasmids bearing dotH or dotI were constructed by using fragments generated by PCR and ligated into the BamHI site of pMS8. Plasmid pH3 contained the complete dotH open reading frame, generated by PCR using primers HAp18 (3′ end of dotH; 5′-CGGGATCCCTTTTTTGCTCGCCATTTGC-3′) and HAp19 (5′ end of dotH; 5′-CGGGATCCTTCACAATTTGTTGTTGGAC-3′), each of which contains a BamHI cleavage site. Annealing temperatures used were 44°C for 3 cycles and 58°C for 11 subsequent cycles. Plasmid pI1 contains the dotI open reading frame and was generated by PCR using primers HAp20 (3′ end of dotI; 5′-CGGGATCCGCCTATCACCAAACAATATT-3′) and HAp21 (5′ end of dotI; 5′-CGGGATCCCCGCAATAATTTTTAGAGGA-3′), which also contain BamHI cleavage sites. Annealing temperatures used were 44°C for 2 cycles and 56°C for 11 subsequent cycles. All fragments were digested with BamHI, purified by agarose gel electrophoresis, and ligated to BamHI-cut pMS8.
Plasmids p1A and p2A were generated by cloning KpnI subfragments of L. pneumophila genomic DNA from p1415 back into a unique KpnI site in pMS8. p1A contains an approximately 3-kb KpnI fragment of p1415 containing two incomplete open reading frames including a gene homologous to the citA gene of E. coli (35). p2A contains an approximately 4-kb KpnI fragment of p1415 on which the only complete open reading frame is dotO.
Suicide plasmid pSR47s is a derivative of pSR47 (oriTRP4 oriR6K kan) (27) bearing the sacB gene in the SalI site. This plasmid bears selectable and counterselectable markers that allow both integration and excision to be selected. Plasmid pHJK is a derivative of plasmid pSR47s harboring dotH, dotJ, and dotK, used for making an in-frame deletion within the dotI gene. PCR was used to generate fragments flanking the dotI gene by using the following primers and restriction sites. A 991-bp fragment spanning a region upstream of dotI and ending at the 5′ end of the dotI gene was generated with the primers HAp38 (5′-ATTTGCGGCCGCGGGGATAACAGGTGAGATCACTTCG-3′, containing a NotI site) and HAp37 (5′-GCTCTAGATAACGCCAAAATGACTTTGCGTTGAC-3′, containing an XbaI site). A 1,170-bp fragment beginning near the 3′ end of the dotI gene and extending downstream was generated by using the primers HAp36 (5′-GGACTAGTTCGCCTAGAGGGATAGGTATTTCAC-3′, containing an SpeI site) and HAp35 (5′-ACGCGTCGACTTGCTTATAACCCTTCTACCTTGAGTTGC-3′, containing a SalI site). The annealing temperatures used for both reactions were 54°C for 3 cycles and 60°C for an additional 17 cycles. PCR products were purified by using Qiaquick spin columns (Qiagen), digested with the appropriate restriction enzymes, and purified by agarose gel electrophoresis using low-melting-point agarose. The downstream 1,161-bp fragment was ligated to pSR47s digested with SalI and SpeI and transformed into DH5α (λpir). The resulting plasmid was cut with XbaI and NotI and ligated to the upstream 991-bp fragment cleaved with XbaI and NotI to create plasmid pHJK.
Nucleotide sequence accession number.
The sequence shown in Fig. 4 has been assigned GenBank accession no. AF026534.
FIG. 4.
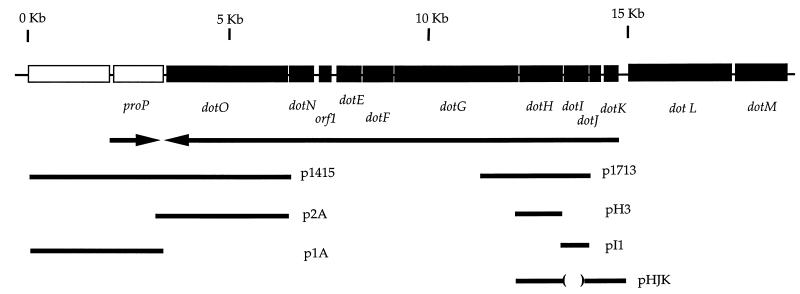
Physical map of new loci in L. pneumophila essential for intracellular growth. Arrows illustrate the direction of transcription and do not imply operon structure. Inserts of plasmids isolated in the complementation test from L. pneumophila genomic library, as well as inserts from plasmids containing open reading frames generated by PCR or subcloning, are shown.
RESULTS
Isolation of intracellular growth mutants.
Two enrichments for L. pneumophila mutants defective for intracellular growth had been performed previously in this laboratory (2, 39). The primary bias of these procedures was that they required the desired strains to survive for many hours within a macrophage or a macrophage-like cell line. Furthermore, a large number of the survivors from these enrichments that showed defects in intracellular trafficking had mutations in the dotA gene. To overcome these problems, we used a general screen for mutants that did not require survival within macrophages. The ability of intracellular growth mutants to target to the replicative phagosome was characterized by fluorescence microscopy after the isolation of candidate mutants.
Approximately 4,960 EMS-mutagenized bacterial strains from 36 different pools were tested for the ability to form plaques on bone marrow macrophage monolayers. Of these, 36 strains were unable to form plaques in two consecutive poke plaque assays. Growth curve assays showed that 17 of these strains had decreased intracellular growth rates. Six strains (from five different pools) which showed no change in yield of viable counts after 72 h in mouse bone marrow macrophages (data not shown) also had intracellular growth defects in culture with macrophage-like U937 cells (Table 2). By comparison, wild-type L. pneumophila (Lp01) increased intracellularly in viable counts by 3 orders of magnitude over 3 days of culture in these cells (Table 2). The remaining 11 strains showed lower levels of intracellular growth in bone marrow-derived macrophages and U937 cells (approximately 10- to 700-fold less than Lp01) than wild-type L. pneumophila (data not shown) but were not studied further. The six strains with the most severe intracellular growth and trafficking defects were chosen for further analysis.
TABLE 2.
Phenotypes of intracellular growth mutants of L. pneumophila
| Strain | Pool no. | Fold growth in U937 cellsa | Total errorb | Colocalization with LAMP-1c | Growth on:
|
|
|---|---|---|---|---|---|---|
| CAA agar | 0.65% NaCld | |||||
| Lp01 | 1.7 × 103 | 4.9 × 102 | − | + | − | |
| HL900 | 24 | 0.56 | 0.59 | + | + | + |
| HL1000 | 24 | 0.68 | 0.68 | + | + | + |
| HL1300 | 34 | 0.66 | 0.13 | + | + | + |
| HL1400 | 35 | 0.72 | 0.19 | + | + | + |
| HL1600 | 40 | 0.34 | 0.16 | + | + | + |
| HL1700 | 1 | 1.1 | 0.29 | + | + | + |
Phorbol ester-treated U937 cell monolayers were incubated with 105 CFU of L. pneumophila derivatives. Viable counts were determined after 2 h (t0) and again at 72 h as described (Materials and Methods). Total growth was calculated as total mean CFU at 72 h/total mean CFU at t0. Values shown are calculated from averages of triplicate samples from a typical experiment. A value of 1 represents equivalent numbers of bacteria at t0 and 72 h.
Total growth [(standard error for t0) + (standard error for 72 h)].
Measured as described (Materials and Methods).
+, approximately 100- to 1,000-fold increase in plating efficiency compared to Lp01.
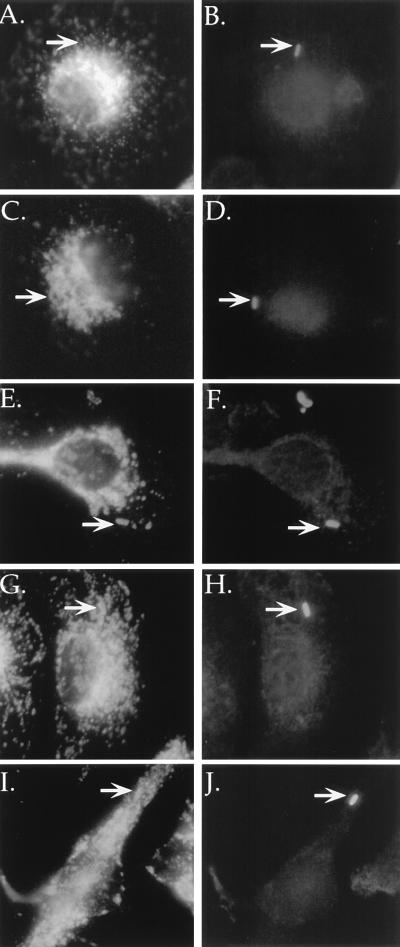
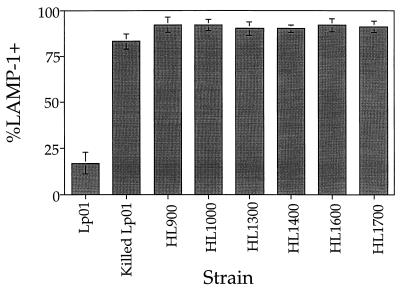
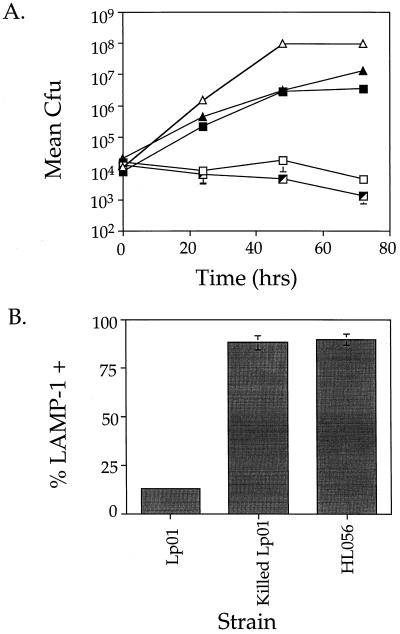
Mutants defective for intracellular growth were examined for aberrant intracellular trafficking in macrophages. The ability of these mutants to evade the endocytic pathway was evaluated by using the late endosomal and lysosomal marker LAMP-1 (8). Bone marrow macrophages were infected with either mutant or growth-competent L. pneumophila for 2 h and observed by fluorescence microscopy to determine if intracellular bacteria colocalized with the late endosomal and lysosomal marker LAMP-1 (Fig. 1). Virulent organisms largely evaded colocalization with LAMP-1 (Fig. 1A), whereas mutants colocalized with LAMP-1. For the parental L. pneumophila strain, 17% ± 6% (Fig. 2, Lp01) of the intracellular bacteria colocalized with LAMP-1. For the mutants studied here, approximately 90% of the intracellular bacteria colocalized with this marker (Fig. 2). The high frequency with which the mutants colocalized with LAMP-1 was similar to the frequency with which formalin-killed Lp01 colocalized with this marker (Fig. 2, Killed Lp01). These results indicate that the mutants isolated in this study were unable to evade the endocytic pathway.
FIG. 1.
Colocalization of intracellular growth mutants with late endosomal, lysosomal marker LAMP-1 in mouse bone marrow-derived macrophages by immunofluorescence microscopy. Macrophages were infected with wild-type or mutant strains of L. pneumophila for 2 h, fixed, and stained for LAMP-1 colocalization (A, C, E, G, and I) and intracellular versus extracellular bacteria (B, D, F, H, and J). Neighboring panels show LAMP-1 staining (left) and corresponding intracellular bacteria (right). (A and B) Lp01 (dot+); (C and D) formalin-killed Lp01 (dot+); (E and F) HL1400 (dotO); (G and H) HL1700 (dotH); (I and J) HL056 (dotI).
FIG. 2.
Intracellular growth mutants of L. pneumophila colocalize with LAMP-1. For each sample, mouse bone marrow macrophages were incubated with mutant or wild-type strains for 2 h, fixed, stained for intracellular versus extracellular bacteria and LAMP-1 colocalization, and examined. Data were collected from 100 intracellular bacteria in total. Percent LAMP-1 positive was calculated by dividing the number of intracellular rod-shaped bacteria colocalizing with LAMP-1 by the total number of intracellular rod-shaped bacteria scored. Values shown are averages of duplicate samples from two identical experiments (four samples in total) and their standard deviations.
All six mutant strains shared several other phenotypic traits. CAA agar can be used to test for thymidine, tryptophan, and nucleoside auxotrophies (28). All mutant strains isolated here grew as well as wild-type L. pneumophila on CAA agar (Table 2). In addition, it has been shown previously that the growth of virulent L. pneumophila is inhibited by 0.65% NaCl (6, 17). While the basis of this phenomenon is unknown, this property has been used to select for avirulent mutants (6, 17, 43). All of the intracellular growth mutants described in this study had an approximately 100- to 1,000-fold-higher plating efficiency on BCYE containing 0.65% NaCl than the parental Lp01 (Table 2).
Complementation of intracellular growth mutants.
The six mutants which were most defective for intracellular growth and failed to evade the endocytic pathway were chosen for further analysis. All of these mutants were tested for complementation by dotA, icmXWYZ, and dotB, genes previously shown to be essential for intracellular growth (2, 4, 43). Plasmids bearing these genes were unable to restore the ability of these six mutants to grow intracellularly (data not shown).
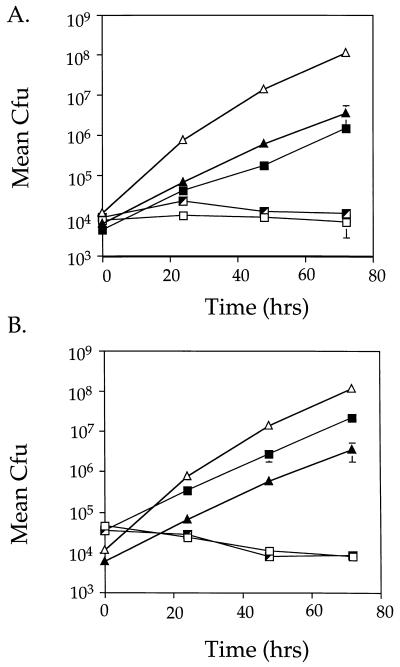
Of the six intracellular growth mutants isolated in this study, HL1400 and HL1700 were chosen to identify DNA fragments capable of complementing intracellular growth defects. These two mutants were isolated from different mutagenized pools and were therefore not siblings. A plasmid gene bank containing inserts from a virulent L. pneumophila strain was introduced into each mutant, and strains able to grow within phorbol ester-treated U937 cells were isolated after two cycles of enrichment. For each mutant, the strains that survived the enrichment had plasmids with identical restriction patterns (data not shown). Plasmids isolated from complemented HL1400 strains and complemented HL1700 strains did not share common restriction patterns or fragments, however, and thus represented two distinct molecular clones.
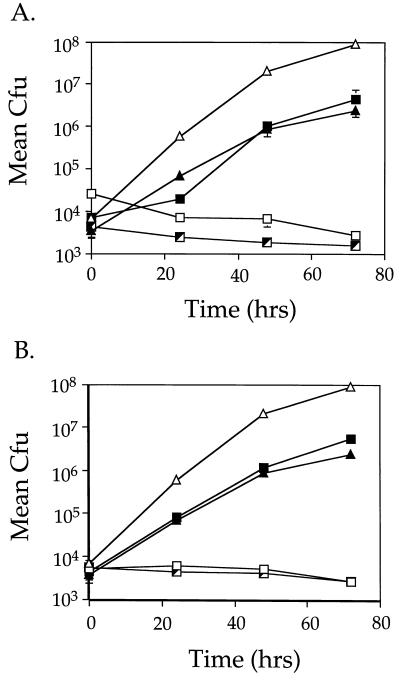
To ensure that restoration of intracellular growth was linked to the two plasmids, each was reintroduced into a fresh background and the resultant strains were tested for intracellular growth. Plasmid p1713 restored intracellular growth to mutant HL1700, and plasmid p1415 restored intracellular growth to mutant strain HL1400 (Fig. 3). The growth rate for each complemented strain was almost indistinguishable from that of Lp01 bearing the cloning vector, pMS8 (Fig. 3A [compare HL019 to HL1719]; Fig. 3B [compare HL019 to HL1419]). In addition, both plasmids were tested for the ability to complement the remaining four mutants isolated in this study. p1415 complemented one additional mutant strain, HL1000, and the other three mutants were not complemented by either plasmid (data not shown).
FIG. 3.
Complementation of intracellular growth defects of L. pneumophila mutants HL1700 and HL1400. Growth was monitored for 72 h to measure the ability of p1713 and p1415 to allow mutants HL1700 (A) and HL1400 (B) to grow intracellularly in phorbol ester-treated U937 cells. Data points and error bars represent the mean CFU of triplicate samples from a typical experiment (performed at least twice) and their standard deviations. (A) Lp01 (dot+; ▵), HL019 (dot+, pMS8; ▴), HL1700 (dotH; □), HL1705 (dotH, pMS8; ┘) and HL1719 (dotH, p1713; ▪). (B) Lp01 (dot+; ▵), HL019 (dot+, pMS8; ▴), HL1400 (dotO; □), HL1405 (dotO, pMS8; ┘), and HL1419 (dotO, p1415; ▪).
Sequence analysis.
Complete double-stranded sequence was obtained for the region spanning the inserts of p1415 and p1713. The chromosomal regions of these plasmids were found to be linked to each other, separated by approximately 5 kb on the L. pneumophila chromosome. Since the area between these two fragments is also required for L. pneumophila virulence (44), the nucleotide sequence of the entire region was determined. The region contains 14 open reading frames, 12 of which are translated in the same orientation, and spans approximately 20 kb on the L. pneumophila chromosome. A physical map for this region, showing the locations of the inserts of complementing plasmids, is shown in Fig. 4.
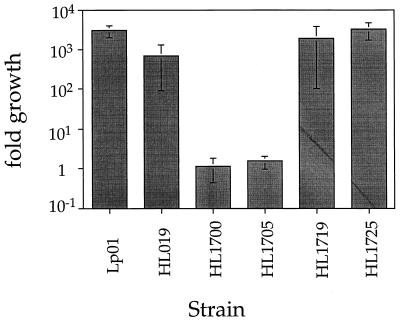
The chromosomal fragment in p1713 contained two complete open reading frames, of 636 (upstream) and 1,080 (downstream) bp oriented in the same direction (Fig. 4). To determine which open reading frame was responsible for complementing the intracellular growth defect of HL1700, DNA fragments spanning each of these open reading frames were generated separately by PCR, and plasmids harboring these fragments were tested for complementing ability. The 1,080-bp open reading frame alone restored the ability of HL1700 to grow intracellularly (Fig. 5; compare HL1705 to HL1725). This gene was named dotH in accordance with the defect in bypassing the endocytic pathway. The mutant strain containing the dotH gene on a plasmid grew as well as the wild-type organism harboring the vector alone (Fig. 5; compare HL019 to HL1725), while the 636-bp gene was unable to restore intracellular growth ability to this strain (data not shown).
FIG. 5.
dotH complements the intracellular growth defect of mutant HL1700 in U937 cells. L. pneumophila strains harboring plasmids were tested for growth in phorbol ester-treated U937 cells. Mean CFU were calculated as the average of duplicate samples from two identical experiments. Values shown represent fold growth, which was determined by dividing mean CFU at 72 h by the mean CFU at t0. Error bars represent the total error, which is equal to the total growth multiplied by the combination of the fractional errors for each time point. Strains: Lp01 (dot+); HL019 (dot+, pMS8); HL1700 (dotH); HL1705 (dotH, pMS8); HL1719 (dotH, p1713); HL1725 (dotH, pH3).
The insert of plasmid p1415 also contained two complete open reading frames oriented in opposite directions (Fig. 4). One of these showed strong sequence homology to the E. coli gene citA (35). The other open reading frame was approximately 3 kb in size and did not show homology with any previously identified proteins. To show which gene complemented the intracellular growth defect of HL1400, plasmids (Fig. 4, p1A and p2A) harboring fragments of p1415 were generated and tested for complementation. Plasmid p2A, bearing a 4-kb KpnI subfragment of p1415, contains the 3-kb open reading frame as the only intact open reading frame. p2A was able to complement the intracellular growth defect of HL1400 (Fig. 6A; compare HL019 to HL1422). This gene, designated dotO, also restored intracellular growth ability to HL1000 (Fig. 6B; compare HL019 to HL1012).
FIG. 6.
Mutants HL1400 and HL1000 are complemented for intracellular growth by the dotO gene in trans. Growth was measured for 72 h to examine the ability of p2A (dotO+) to allow HL1400 (A) and HL1000 (B) to grow intracellularly. Data points and error bars represent the mean CFU of triplicate samples from a typical experiment (performed at least twice) and their standard deviations. (A) Lp01 (dot+; ▵), HL019 (dot+, pMS8; ▴), HL1400 (dotO1; □), HL1405 (dotO1, pMS8; ┘), and HL1422 (dotO1, p2A; ▪). (B) Lp01 (dot+; ▵), HL019 (dot+, pMS8; ▴), HL1000 (dotO2; □), HL1005 (dotO2, pMS8; ┘), and HL1012 (dotO2, p2A; ▪).
dotI is essential for intracellular growth and is required for targeting.
To determine whether the 636-bp open reading frame (dotI) adjacent to and upstream of dotH on p1713 was involved in intracellular growth, we constructed a chromosomal in-frame deletion and tested the deletion mutant for intracellular growth. The ΔdotI strain was unable to grow intracellularly (Fig. 7A, HL056), while dotI, in trans, complemented this defect (Fig. 7A, HL059) and grew nearly as well as wild-type L. pneumophila harboring vector alone (Fig. 7A, HL019).
FIG. 7.
(A) ΔdotI mutation causes defective intracellular growth in phorbol ester-treated U937 cells over 72 h, and this defect is complemented by dotI in trans. Growth of L. pneumophila was monitored for 72 h. Values and error bars represent the average of triplicate samples from a typical experiment (performed at least twice) and their standard deviations. Strains: Lp01 (dot+; ▵), HL019 (dot+, pMS8; ▴), HL056 (dotI; □), HL057 (dotI, pMS8; ┘), and HL059 (dotI, pI1; ▪). (B) ΔdotI mutants colocalize with LAMP-1. Mouse bone marrow-derived macrophages were incubated with mutant or virulent strains for 2 h, fixed, and stained for intracellular versus extracellular bacteria and LAMP-1 colocalization. Data were collected from 100 intracellular bacteria. Percent LAMP-1 positive was calculated by dividing the number of intracellular rod-shaped bacteria colocalizing with LAMP-1 by the total number of intracellular rod-shaped bacteria scored. Values shown are the averages of duplicate samples from two identical experiments (four samples in total) and their standard deviations.
This ΔdotI mutant was also tested for the ability to target properly within bone marrow-derived macrophages. In these experiments, ΔdotI mutants failed to evade the endocytic pathway, colocalizing with LAMP-1 as frequently as killed L. pneumophila (Fig. 1I and J; Fig. 7B [compare Killed Lp01 to HL056]).
Hydrophilicity analysis and structural predictions.
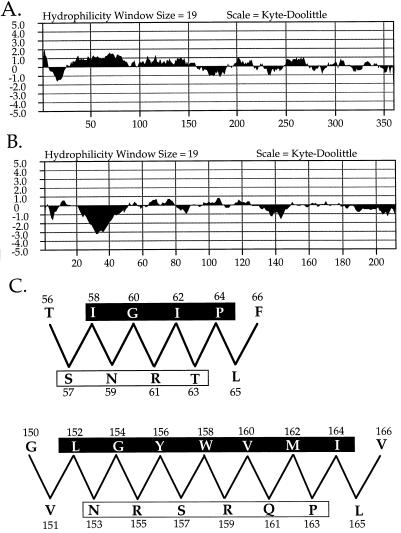
The nucleotide sequence of the dotH gene predicts a protein of 360 amino acids which does not show similarity with any previously described proteins. Kyte-Doolittle hydrophilicity analysis of DotH (Fig. 8A), using a window size of 19, predicts a classical N-terminal secretion signal sequence. The dotI nucleotide sequence predicts a protein of 212 amino acids. DotI is similar (P = 10−9) to the product, of unknown function, of orf3, located on the IncM plasmid R446. DotI hydrophilicity analysis (Fig. 8B) predicts a single transmembrane domain near the N-terminal end of the protein. In addition, amphiphilicity analysis predicts a strongly amphipathic β-sheet structure between amino acids 152 and 164 and an additional amphipathic β-sheet between amino acids 57 and 64 (Fig. 8C). Finally, the dotO gene predicts a protein of 1,010 amino acids. DotO has no homologs in GenBank, and its hydrophilicity analysis shows no strong transmembrane prediction (data not shown).
FIG. 8.
Hydrophilicity analyses and model of predicted amphiphilic β-sheet of proteins shown to be essential for intracellular growth of L. pneumophila. (A) Hydrophilicity analysis of DotH predicts an N-terminal secretion signal sequence. (B) Hydrophilicity analysis of DotI predicts a membrane-spanning domain between amino acid residues 20 and 50. (C) Model of amphipathic β-sheets in DotI between amino acids 57 and 64 and amino acids 152 and 164. Hydrophobic faces of the predicted β-sheets are shown in a shaded box, while the polar faces are shown in an unshaded box.
DISCUSSION
The goal of this study was to isolate genes of L. pneumophila important for intracellular growth and proper trafficking, to better understand these processes at the molecular level. Screening of EMS-mutagenized colonies for the ability to form plaques on bone marrow-derived macrophages yielded 36 strains which failed to plaque. Of these 36 strains, 17 showed reduced intracellular growth to various degrees compared to wild-type L. pneumophila. The remaining 19 strains grew as well as wild-type L. pneumophila in growth assays and were not studied further. This group may have been less cytotoxic than the parental strain or may reflect the frequency of false positives inherent in this method.
Of the 17 strains which showed reduced intracellular growth ability, 6 strains had very severe growth defects and were targeted to a late endosomal or lysosomal compartment, in contrast to virulent L. pneumophila. All six mutants showed intracellular growth and targeting phenotypes identical to those of dotA mutants, although none of these mutants were complemented by dotA in trans (2, 33).
Complementation analysis using two mutants isolated in this study, HL1700 and HL1400, allowed us to identify two genes of L. pneumophila, dotH and dotO, that restored the mutants’ ability to grow intracellularly. The dotO gene placed in trans also complemented a third mutant isolated in this study, HL1000. Finally, an additional gene, dotI, located upstream of dotH, was also found to be essential for intracellular growth.
Of the three remaining mutants, the mutation responsible for the intracellular growth defect in one, HL900, is probably linked to this region as well. Western analysis using antisera recognizing DotG showed that this mutant produces truncated DotG (1), making it likely that this lesion is responsible for the intracellular growth and trafficking defects of this strain. The two remaining mutants, HL1300 and HL1600, have not been tested for complementation using an L. pneumophila genomic library.
DotH, DotI, and DotO have few notable features. DotH, which contains a secretion signal sequence, is likely to be located outside the bacterial cytoplasm, while the location of DotO cannot be predicted from primary sequence information alone. DotI contains two notable structural features: a transmembrane domain and two amphipathic β-sheet regions. The amphipathic regions are intriguing because some secreted pore-forming toxins, such as Staphylococcus aureus α-hemolysin, and bacterial membrane porins exist in which amphipathic β-sheets are responsible for forming the pore structure (30, 31, 42). The most recently described example is protective antigen of anthrax toxin, monomers of which contain an amphipathic β-hairpin. When the monomers heptamerize and insert into a eukaryotic membrane, an antiparallel β-barrel structure is formed, which is the pore (31). Whether the amphipathic regions in DotI form a membrane-spanning pore structure, and whether this structure is necessary for intracellular growth and proper evasion of the endocytic pathway, remains to be determined.
The dotI gene is similar to orf3 of the IncM plasmid R446, located in a region near two genes (imlA and imlB) involved in regulation of conjugal pilus formation (41). Although the significance of this similarity for DotI function is unknown, it has been shown that other members of this new dot region show homology to genes involved in conjugation (44). In addition, structural similarities between conjugal transfer systems and export systems for virulence determinants in pathogenic bacteria have been described for several microorganisms, including Agrobacterium tumefaciens, Bordetella pertussis, and Helicobacter pylori (7, 47).
Recently, it has been shown that L. pneumophila is cytotoxic to macrophages at high multiplicities of infection and causes contact dependent hemolysis of erythrocytes (22, 23). This activity is most likely due to the ability of the bacterium to induce pore formation in eukaryotic membranes. Genes critical for intracellular growth and evasion of the endocytic pathway, including dotH, dotI, and dotO, are essential for cytotoxic activity (23). The inability of many of the dot mutants tested to demonstrate cytotoxicity may indicate that their products perform common or closely related functions in the assembly or insertion of the pore itself.
Although the proximity of this new dot region to the dotA-icm region is unknown, the products of the genes in both regions may participate in a common function (2, 4, 25, 43). Many of the predicted products of these loci have homologs that are components of large multisubunit complexes involved in transport of macromolecules across bacterial membranes (33, 43). Mutations in both regions result in failure to grow intracellularly and failure to evade the endocytic pathway indicated by colocalization with LAMP-1 (2, 32). The previously isolated mutant 25D also fails to grow intracellularly and fails to prevent phagolysosome fusion (17). The high degree of similarity in intracellular growth and targeting defects between dotA, icmWXYZ mutants, and the dot mutants described in this report is suggestive of shared or closely related functions.
The broad mutant isolation procedure used in this study did not screen sufficient numbers of mutants to make this screen saturating. This fact may explain why no additional dotA mutations were isolated in this study. In addition, this is also a potential explanation for the fact that no mutants that lost significant viability in macrophage culture were isolated. Two mutants have been isolated in this laboratory that are killed in macrophage culture (39, 45). One of these strains has a mutation in a gene involved in cell wall biosynthesis as well as a second mutation in a region previously shown to be important for intracellular growth (icmWXYZ) (45). This raises the possibility that L. pneumophila shows some inherent resistance to damage, allowing persistence when improperly targeted within the macrophage, at least in bone marrow-derived macrophages and U937 cells. Multiple mutations may be required to debilitate L. pneumophila sufficiently to make it susceptible to macrophage killing. The gene affected in the second mutant (39) remains to be identified.
We avoided using enrichment procedures, such as the thymineless death enrichment strategy previously utilized in mutant isolation procedures in this laboratory (2, 39). This enrichment protocol prevented the isolation of mutants which lost viability in macrophage culture, a potentially interesting class of mutants which our screening procedure should be able to isolate. In addition, many of the mutants isolated by using thymineless death enrichment had mutations in a single locus, dotA. The reasons for this strong bias toward dotA are unclear.
In summary, we have identified three new genes that are part of a 20-kb region of the L. pneumophila chromosome in which genes essential for evasion of the endocytic pathway and subsequent intracellular growth are encoded. We predict that the Dot proteins encoded in this region, including DotH, DotI, and DotO, are components of a multisubunit membrane complex that plays an essential role in the establishment and maintenance of intracellular growth. Future work will focus on providing evidence for this hypothesis and for elucidating the role of these proteins in proper targeting to the replicative phagosome and intracellular growth.
ACKNOWLEDGMENTS
We thank Michele Swanson for the generous gift of her L. pneumophila genomic library. In addition, we thank Michele Swanson and Dorothy Fallows for critical reading of the manuscript.
This work was supported by the Howard Hughes Medical Institute. J.P.V. was supported by a postdoctoral fellowship from the Medical Foundation. H.L.A. was supported by NIH training grants 5T32 A107422-04 and 5T32 A107422-5.
ADDENDUM IN PROOF
Genes dotL and dotM have been described previously by G. Segal and H. A. Shuman (Infect. Immun. 65:5057–5066, 1997) as icmO and icmP, respectively (EMBL accession no. Y12705).
REFERENCES
- 1.Andrews, H. L., J. P. Vogel, and R. R. Isberg. Unpublished data.
- 2.Berger K H, Isberg R R. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 3.Berger K H, Merriam J J, Isberg R R. Altered intracellular targeting properties associated with mutations in the Legionella pneumophila dotA gene. Mol Microbiol. 1994;14:809–822. doi: 10.1111/j.1365-2958.1994.tb01317.x. [DOI] [PubMed] [Google Scholar]
- 4.Brand B C, Sadosky A B, Shuman H A. The Legionella pneumophila icm locus: a set of genes required for intracellular multiplication in human macrophages. Mol Microbiol. 1994;14:797–808. doi: 10.1111/j.1365-2958.1994.tb01316.x. [DOI] [PubMed] [Google Scholar]
- 5.Bullock W O, Fernandez J M, Short J M. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques. 1987;5:376–378. [Google Scholar]
- 6.Catrenich C E, Johnson W. Characterization of the selective inhibition of growth of virulent Legionella pneumophila by supplemented Mueller-Hinton medium. Infect Immun. 1989;57:1862–1864. doi: 10.1128/iai.57.6.1862-1864.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Censini S, Lange C, Xiang Z, Crabtree J E, Ghiara P, Borodovski M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J W, Cha Y, Yuksel K U, Gracy R W, August J T. Isolation and sequencing of a cDNA clone encoding lysosomal membrane glycoprotein mouse LAMP-1. J Biol Chem. 1988;263:8754–8758. [PubMed] [Google Scholar]
- 9.Cianciotto N, Eisenstein B I, Engleberg N C. Genetics and molecular pathogenesis of Legionella pneumophila, an intracellular parasite of macrophages. Mol Biol Med. 1989;6:409–424. [PubMed] [Google Scholar]
- 10.Clemens D L, Horwitz M A. Characterization of the Mycobacterium tuberculosis phagosome and evidence that phagosomal maturation is inhibited. J Exp Med. 1995;181:257–270. doi: 10.1084/jem.181.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Del Sal G, Manfioletti G, Schneider C. The CTAB-DNA precipitation method: a common mini-scale preparation of template DNA from phagemids, phages, or plasmids suitable for sequencing. BioTechniques. 1989;7:514–519. [PubMed] [Google Scholar]
- 12.Feeley J C, Gibson R J, Gorman G W, Langford N C, Rasheed J K, Makel D C, Blaine W B. Charcoal yeast extract agar: primary isolation medium for Legionella pneumophila. J Clin Microbiol. 1979;10:437–441. doi: 10.1128/jcm.10.4.437-441.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fields B S, Sanden G N, Barbaree J M, Morrill W E, Wadowsky R M, White E H, Feeley J C. Intracellular multiplication of Legionella pneumophila in amoebae isolated from hospital hot water tanks. Curr Microbiol. 1989;18:131–137. [Google Scholar]
- 14.Finan T M, Kunkel B, De Vos G, Signer E R. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J Bacteriol. 1986;167:66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gabay J E, Horwitz M A. Isolation and characterization of the cytoplasmic and outer membranes of the Legionnaires’ disease bacterium (Legionella pneumophila) J Exp Med. 1985;161:409–422. doi: 10.1084/jem.161.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hobbs M, Mattick J S. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol Microbiol. 1993;10:233–243. doi: 10.1111/j.1365-2958.1993.tb01949.x. [DOI] [PubMed] [Google Scholar]
- 17.Horwitz M A. Characterization of avirulent mutant Legionella pneumophila that survive but do not multiply within human monocytes. J Exp Med. 1987;166:1310–1328. doi: 10.1084/jem.166.5.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horwitz M A. Formation of a novel phagosome by the Legionnaires’ disease bacterium (Legionella pneumophila) in human monocytes. J Exp Med. 1983;158:1319–1331. doi: 10.1084/jem.158.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horwitz M A. The Legionnaires’ disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J Exp Med. 1983;158:2108–2126. doi: 10.1084/jem.158.6.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horwitz M A. Phagocytosis of the Legionnaires’ disease bacterium (Legionella pneumophila) occurs by a novel mechanism: engulfment within a pseudopod coil. Cell. 1984;36:27–33. doi: 10.1016/0092-8674(84)90070-9. [DOI] [PubMed] [Google Scholar]
- 21.Horwitz M A, Maxfield F R. Legionella pneumophila inhibits acidification of its phagosome in human monocytes. J Cell Biol. 1984;99:1936–1943. doi: 10.1083/jcb.99.6.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Husmann L K, Johnson W. Cytotoxicity of extracellular Legionella pneumophila. Infect Immun. 1994;62:2111–2114. doi: 10.1128/iai.62.5.2111-2114.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirby, J. E., J. P. Vogel, H. L. Andrews, and R. R. Isberg. Evidence for pore forming ability by Legionella pneumophila. Mol. Microbiol., in press. [DOI] [PubMed]
- 24.Kolter R, Inuzuka M, Helinski D R. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell. 1978;15:1199–1208. doi: 10.1016/0092-8674(78)90046-6. [DOI] [PubMed] [Google Scholar]
- 25.Marra A, Blander S J, Horwitz M A, Shuman H A. Identification of Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proc Natl Acad Sci USA. 1992;89:9607–9611. doi: 10.1073/pnas.89.20.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLean I W, Nakane P K. Periodate-lysine-paraformaldehyde fixative: a new fixative for immunoelectron microscopy. J Histochem Cytochem. 1974;22:1077–1083. doi: 10.1177/22.12.1077. [DOI] [PubMed] [Google Scholar]
- 27.Merriam J J, Mathur R, Maxfield-Boumil R, Isberg R R. Analysis of the Legionella pneumophila fliI gene: intracellular growth of a defined mutant defective for flagellum biosynthesis. Infect Immun. 1997;65:2497–2501. doi: 10.1128/iai.65.6.2497-2501.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mintz C S, Chen J, Shuman H A. Isolation and characterization of auxotrophic mutants of Legionella pneumophila that fail to multiply in human monocytes. Infect Immun. 1988;56:1449–1455. doi: 10.1128/iai.56.6.1449-1455.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nash T W, Libby D M, Horwitz M A. Interaction between the Legionnaires’ disease bacterium (Legionella pneumophila) and human alveolar macrophages. J Clin Invest. 1984;74:771–782. doi: 10.1172/JCI111493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikaido H. Porins and specific diffusion channels in bacterial outer membranes. J Biol Chem. 1994;269:3905–3908. [PubMed] [Google Scholar]
- 31.Petosa C, Collier R J, Klimpel K R, Leppla S H, Liddington R C. Crystal structure of the anthrax toxin protective antigen. Nature. 1997;385:833–838. doi: 10.1038/385833a0. [DOI] [PubMed] [Google Scholar]
- 32.Roy, C. R., K. H. Berger, and R. R. Isberg. Legionella pneumophila DotA protein is required for early phagosome trafficking decisions that occur within minutes of bacterial uptake. Submitted for publication. [DOI] [PubMed]
- 33.Roy C R, Isberg R R. Topology of Legionella pneumophila DotA: an inner membrane protein required for replication in macrophages. Infect Immun. 1997;65:571–578. doi: 10.1128/iai.65.2.571-578.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Sasatsu M, Misra T K, Chu L, Laddaga R, Silver S. Cloning and DNA sequence of a plasmid-determined citrate utilization system in Escherichia coli. J Bacteriol. 1985;164:983–993. doi: 10.1128/jb.164.3.983-993.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shuman, H. Personal communication.
- 37.Swanson, M. S. Unpublished data.
- 38.Swanson M S, Isberg R R. Analysis of intracellular fate of Legionella pneumophila mutants. Ann N Y Acad Sci. 1996;797:8–18. doi: 10.1111/j.1749-6632.1996.tb52944.x. [DOI] [PubMed] [Google Scholar]
- 39.Swanson M S, Isberg R R. Identification of Legionella pneumophila mutants that have aberrant intracellular fates. Infect Immun. 1996;64:2585–2594. doi: 10.1128/iai.64.7.2585-2594.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swanson M S, Isberg R R. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect Immun. 1995;63:3609–3620. doi: 10.1128/iai.63.9.3609-3620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tietze E, Tschape H. Temperature-dependent expression of conjugation pili by IncM plasmid-harbouring bacteria: identification of plasmid-encoded regulatory functions. J Basic Microbiol. 1994;34:105–116. doi: 10.1002/jobm.3620340206. [DOI] [PubMed] [Google Scholar]
- 42.Vecsey-Semjen B, Lesieur C, Moellby R, Gisou van der Goot F. Conformational changes due to membrane binding and channel formation by staphylococcal α-toxin. J Biol Chem. 1997;272:5709–5717. doi: 10.1074/jbc.272.9.5709. [DOI] [PubMed] [Google Scholar]
- 43.Vogel J P, Roy C, Isberg R R. Use of salt to isolate Legionella pneumophila mutants unable to replicate in macrophages. Ann N Y Acad Sci. 1996;797:271–272. doi: 10.1111/j.1749-6632.1996.tb52975.x. [DOI] [PubMed] [Google Scholar]
- 44.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. Conjugal transfer by the virulence system of Legionella pneumophila. Science, in press. [DOI] [PubMed]
- 45.Vogel, J. P., and R. R. Isberg. Unpublished data.
- 45a.Vogel, J. P., et al. Unpublished data.
- 46.Wadowsky R M, Butler L J, K. C M, Verma S M, Paul M A, Fields B S, Keleti G, Sykora J L, Yee R B. Growth-supporting activity for Legionella pneumophila in tap water cultures and implication of hartmannellid amoebae as growth factors. Appl Environ Microbiol. 1988;54:2677–2682. doi: 10.1128/aem.54.11.2677-2682.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winans S C, Burns D L, Christie P J. Adaptation of a conjugal transfer system for the export of pathogenic macromolecules. Trends Microbiol. 1996;6:64–68. doi: 10.1016/0966-842X(96)81513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winn W C, Myerowitz R L. The pathology of the Legionella pneumonias. Hum Pathol. 1981;12:401–422. doi: 10.1016/s0046-8177(81)80021-4. [DOI] [PubMed] [Google Scholar]
- 49.Woodcock D M, Crowther P J, Doherty J, Jefferson S, DeCruz E, Noyer-Weidner M, Smith S S, Michael M Z, Graham M W. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 1989;17:3469–3478. doi: 10.1093/nar/17.9.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]