Significance
Activity in the developing brain affects the fate of multipotent neural progenitor cells, for instance, whether they continue dividing or differentiate into neurons. We conducted an unbiased screen to identify candidate mechanisms that influence neural progenitor cell fate by analyzing differentially expressed transcriptomes from neural progenitor cells and newly differentiated neurons in Xenopus tadpoles following exposure to a visual experience regime known to affect neurogenesis. We identified BRCA1 and ELK-1 as members of a differentially expressed network of transcriptional regulators. Longitudinal in vivo time-lapse imaging indicates that BRCA1 and ELK1 regulate neural progenitor cell fate and that the effects of visual experience on cell fate decisions require BRCA1 and ELK-1. This study expands our understanding of the mechanisms governing brain development.
Keywords: neural progenitor cell, differential expression, Xenopus, transcription, BRCA1
Abstract
In developing Xenopus tadpoles, the optic tectum begins to receive patterned visual input while visuomotor circuits are still undergoing neurogenesis and circuit assembly. This visual input regulates neural progenitor cell fate decisions such that maintaining tadpoles in the dark increases proliferation, expanding the progenitor pool, while visual stimulation promotes neuronal differentiation. To identify regulators of activity-dependent neural progenitor cell fate, we profiled the transcriptomes of proliferating neural progenitor cells and newly differentiated neurons using RNA-Seq. We used advanced bioinformatic analysis of 1,130 differentially expressed transcripts to identify six differentially regulated transcriptional regulators, including Breast Cancer 1 (BRCA1) and the ETS-family transcription factor, ELK-1, which are predicted to regulate the majority of the other differentially expressed transcripts. BRCA1 is known for its role in cancers, but relatively little is known about its potential role in regulating neural progenitor cell fate. ELK-1 is a multifunctional transcription factor which regulates immediate early gene expression. We investigated the potential functions of BRCA1 and ELK-1 in activity-regulated neurogenesis in the tadpole visual system using in vivo time-lapse imaging to monitor the fate of GFP-expressing SOX2+ neural progenitor cells in the optic tectum. Our longitudinal in vivo imaging analysis showed that knockdown of either BRCA1 or ELK-1 altered the fates of neural progenitor cells and furthermore that the effects of visual experience on neurogenesis depend on BRCA1 and ELK-1 expression. These studies provide insight into the potential mechanisms by which neural activity affects neural progenitor cell fate.
Neurogenesis is the collective process of cell proliferation, differentiation, migration, and survival, which lead to the generation of functional neurons. Dysregulation of neural progenitor cell (NPC) fate decisions affecting neurogenesis results in abnormal brain development (1–3). Insight into the regulation of cell proliferation, neuronal differentiation, and apoptosis will advance understanding brain development.
Neuronal circuit activity regulates multiple aspects of brain development (4, 5). Xenopus laevis tadpoles are an excellent system to investigate how sensory experience modulates neural development because tadpoles receive and respond to patterned visual stimuli while neurogenesis and circuit assembly are occurring. In tadpoles, visual stimulation increases integration of newly generated neurons into the tectal circuit by regulating neuronal structural development, synaptic connectivity, and biophysical properties that affect neuronal firing (6–8). Visual experience also regulates neural progenitor proliferation, cell fate, and neuronal differentiation. For instance, exposing tadpoles to dark continuously over 2 d, instead of the normal 12-h light/12-h dark cycle, significantly increased NPC proliferation, expanding the progenitor pool, whereas exposing animals to visual stimulation for 24 h decreased NPC proliferation and increased neuronal differentiation (9, 10). These studies indicate that visual experience conditions that increase or decrease tectal circuit activity lead to different NPC fates; however, little is known about the molecular mechanisms that regulate these activity-induced cell fate decisions.
Differential transcriptome analysis has been a productive approach to generate hypotheses and identify molecular mechanisms involved in cell fate regulation in a variety of experimental systems, including Xenopus (11–16). Here, we leverage prior studies in Xenopus tadpoles to investigate activity-dependent regulation of NPC fate in vivo. We used transcriptomic profiling of birthdated cohorts of NPCs and newly differentiated neurons with RNA-seq to identify differentially expressed (DE) transcripts. Datamining our DE dataset revealed cellular mechanisms underlying NPC fate decisions and neuronal differentiation and identified BRCA1 and ELK-1 as candidate molecular regulators of NPC fate. In vivo time-lapse imaging combined with knock down strategies indicated that BRCA1 and ELK-1 regulate NPC fate and that the effects of visual experience on NPC fate depend on ELK-1 and BRCA1 expression.
Results
Differential Expression Analysis of Transcripts Expressed by Neural Progenitor Cells and Immature Neurons.
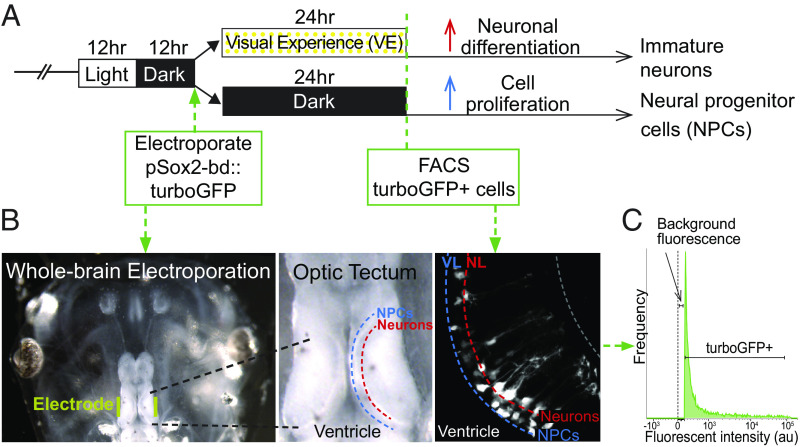
We used RNA-seq to profile transcripts in NPCs and their neuronal progeny isolated from animals exposed to different visual experience regimes (Fig. 1A). To isolate enriched populations of NPCs and immature neurons, we expressed turbo-GFP (tGFP) in NPCs in vivo by electroporating the optic tectum with pSOX2-bd::tGFP, a plasmid that drives tGFP expression upon binding of endogenous SOX2 (10, 17). The use of pSOX2-bd::tGFP provides many benefits for this experiment (10): 1) it only labels cells that express SOX2, resulting in tGFP expression specifically in NPCs, birthdating the SOX2+ NPCs and their progeny in the tectum; 2) it uses Gal4-UAS to amplify tGFP expression; 3) the rapid maturation kinetics of tGFP, together with Gal4-UAS amplified gene expression, enabled us to label and isolate SOX2+ NPCs and their progeny within 1 d of electroporation; 4) tGFP is stable for days; and 5) it labels SOX2+ cells even under conditions that might alter the generation of additional neural progenitors (SI Appendix, Fig. S1). After exposing electroporated animals to dark or visual stimulation (Fig. 1 A and B), we dissected the midbrains and isolated ~40,000 tGFP+ cells by FACS (Fig. 1C and SI Appendix, Methods).
Fig. 1.
Isolation of neural progenitor cells and immature neurons from the optic tectum. (A) Visual experience paradigm used to enrich for NPCs and immature neurons. Animals were reared in 12 h light/12 h dark until stage 46 when the midbrain was electroporated with pSOX2-bd::tGFP plasmid. After electroporation, animals were randomly divided into two groups, one exposed to enhanced visual experience (VE) for 24 h. (B) Whole brain electroporation labels NPCs and neurons in the optic tectum. Left: Image of the head of the tadpole with electrodes (yellow) on each side of the optic tectum. Middle: Dorsal view of the optic tectum. The ventricle, NPC layer, and neuronal layer are labeled. Right: in vivo 2-photon image of tGFP-labeled NPCs and neurons in the ventricular layer (VL) and neuronal layer (NL). (C) Fluorescence histogram indicates the gate setting of the fluorescence-activated cell sorting (FACS) to isolate tGFP+ cells. Control is set to the background fluorescence from nonelectroporated midbrain cells.
We identified 1,130 transcripts that were differentially expressed between NPCs and immature neurons using DESeq2 (SI Appendix, Figs. S2 and S3 and Table S1 and Dataset S2). neurod1, wnt1, fgf2, vegfa, nfkb1, and smad9 were enriched in immature neurons, and elk-1, e4f1, sstr4, bmp4, jak2, and nr2f5 were enriched in NPCs (SI Appendix, Table S2) validating enrichment of NPCs and immature neurons.
Bioinformatic Analysis of Differentially Expressed Genes Identified Functional Categories and Interaction Networks.
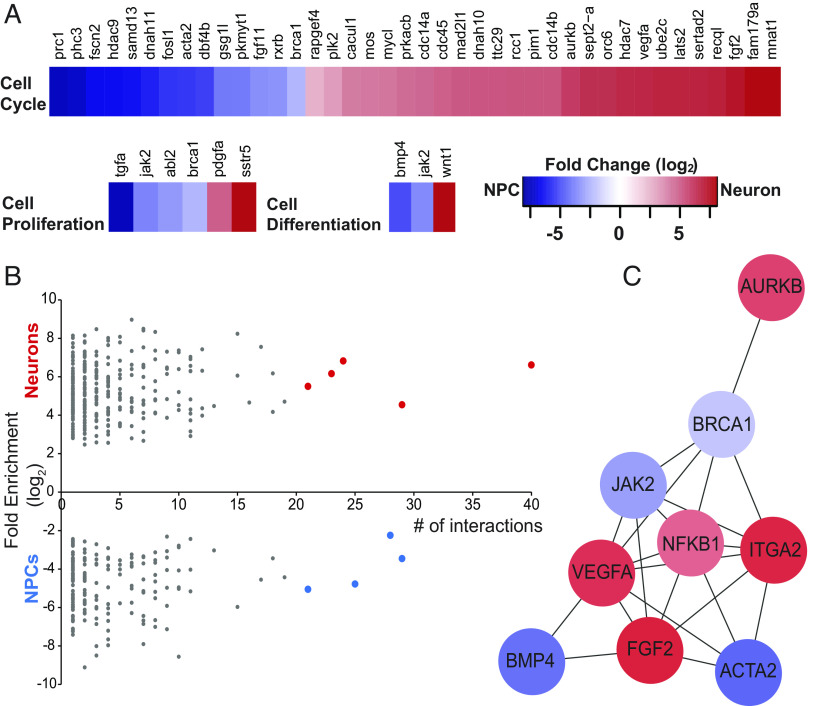
To identify molecular and cellular processes affecting NPC proliferation and neuronal differentiation governed by DE transcripts, we categorized these transcripts using the PANTHER (Protein Analysis Through Evolutionary Relationships) classification system (SI Appendix, Figs. S4 and S5 and Dataset S2). PANTHER classified 367 transcripts which clustered catalytic activity (220 genes), DNA binding (98), receptor-mediated signaling (146), and structural proteins (89) (SI Appendix, Fig. S4 and Dataset S2). Three GO slim biological processes, cell proliferation, cell differentiation, and cell cycle, were enriched in our DE dataset (Fig. 2A and Dataset S2). This analysis identified DE transcripts known to affect cell fate changes and those that are less well known for a role in neurogenesis, which may be subject of further investigation.
Fig. 2.
Bioinformatic analysis of differentially expressed transcripts identifies functional categories and interaction networks. (A) Fold change of transcript expression between NPCs and immature neurons in GO biological processes: cell proliferation, cell differentiation, and cell cycle. (B) Protein interaction network analysis of DE transcripts, arranged by number of binding partners (interactions) in NCPs (blue) and neurons (red). (C) Network analysis of the nine transcripts highlighted in (B) with the most binding partners, blue indicates genes up-regulated in neural progenitor cells and red indicates increased expression in immature neurons. Fold change scale applies to (A) and (C).
We used STRING and Cytoscape to identify protein–protein interaction networks among DE transcripts. STRING identified 458 proteins with one or more interaction partners (Fig. 2 and Dataset S3). Ranking protein importance within network nodes based on degree centrality and closeness centrality (18) indicates that proteins with more than 20 binding partners are likely to play an important role in neurogenesis. Nine proteins had 20 or more interaction partners, and 4 of these, ACTA2, BMP4, JAK2, and BRCA1, were enriched in NPCs, while the other 5, ITGA2, VEGFA, FGF2, AURKB, and NFKB1, were enriched in immature neurons (Fig. 2B). Furthermore, network analysis indicated that these nine proteins interact with each other (Fig. 2C), supporting the idea of protein network-based regulation of fundamental neurodevelopment events such as cell fate.
Differentially Expressed Master Regulator Network.
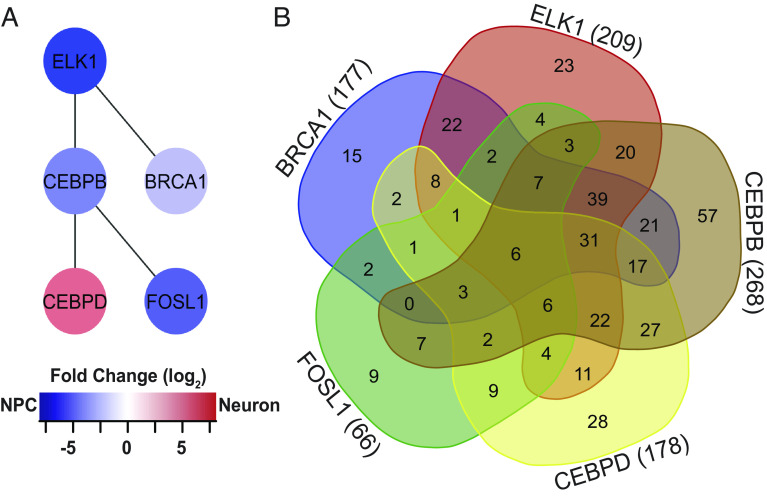
We were interested in determining whether upstream DE transcriptional regulators could operate as master regulators, controlling the differential expression of other transcripts in NPCs and immature neurons. We identified candidate master regulators based on two criteria: the number of their targeted genes and the number of interactions they have with other transcriptional regulators, assuming that a candidate with more protein–protein interactions can indirectly regulate more transcripts. After testing this strategy by mining the ENCODE database (SI Appendix, Fig. S6), we tested our DE dataset and identified six candidates that regulated other DE transcripts: BRCA1, ELK-1, CEBPB, CEBPD, FOSL1, and BRF1. Using protein network analyses, we were surprised to find that five of these DE upstream transcriptional regulators, excluding BRF1 (transcription factor IIIB 90 kDa subunit), interact with each other (Fig. 3A). This analysis suggests that differential transcript expression in NPCs and newly differentiated neurons may be governed by a synergistic network of these five transcriptional regulators. A Venn diagram of the genes targeted by CEBPB (268), ELK-1 (209), CEBPD (178), BRCA1 (177), and FOSL1 (66) (Fig. 3B and SI Appendix, Table S3) identified potential coregulated transcripts, including a diverse range of cellular processes that affect progenitor cell and neuronal functions, including cytoskeletal regulation, apoptosis, TGFβ signaling, and transcriptional regulation, consistent with the idea that spatiotemporal dynamics of transcriptional regulation may provide an important element of control (19).
Fig. 3.
Network of differentially expressed transcriptional regulators. (A) ELK1, BRCA1, CEBPB, CEBPD, and FOSL1 form a network of DE transcription factors that regulates other DE transcription factors. Color refers to fold change enrichment between NPCs (blue) and neurons (red). (B) 5-way Venn diagram showing the numbers of transcripts targeted by the 5 DE transcription factors in (B). For specific targets, see SI Appendix, Table S3.
Of these five networked transcriptional regulators, we selected BRCA1 and ELK-1 for further investigation as potential master regulators in NPC fate in the developing brain, based on their enriched expression in NPCs (Fig. 2), the observation that BRCA1 and ELK-1 interact with each other in a network of candidate master regulators and regulate a total of 270 target genes (Fig. 3 and SI Appendix, Table S2) and the relative paucity of information about their roles in NPCs (20–23).
Visual Experience Alters BRCA1 and ELK-1 Protein Expression in Neural Progenitor Cells.
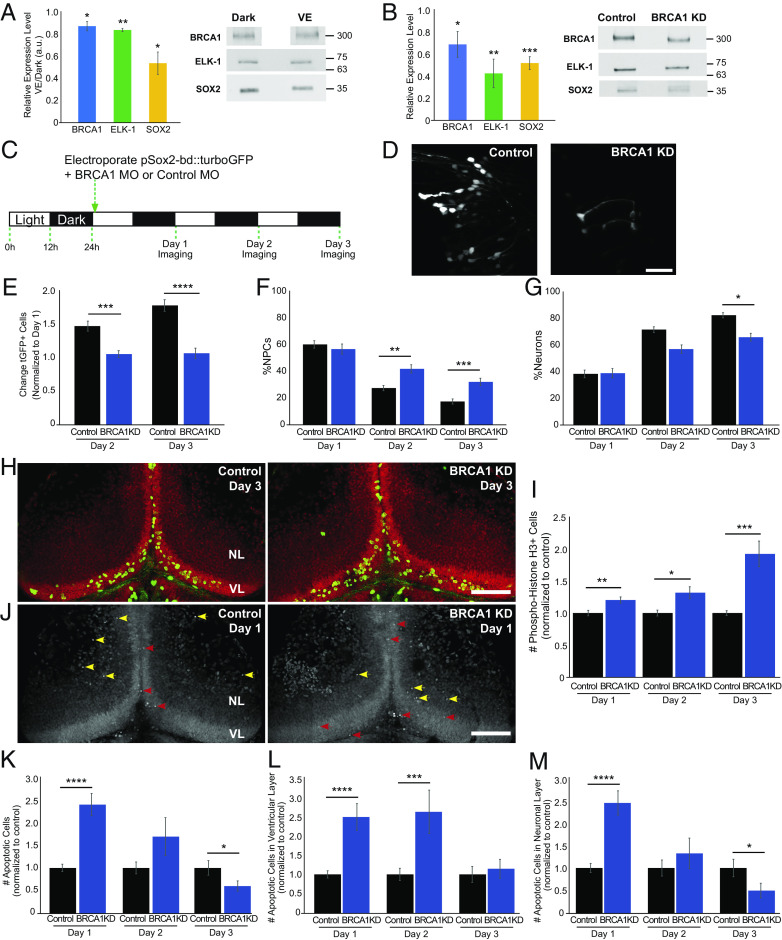
To address the potential roles of BRCA1 and ELK-1 in NPC fate under different visual experience conditions, we first examined whether exposing animals to dark affected SOX2, BRCA1, and ELK-1 expression compared to enhanced visual experience (VE). Western blots of the midbrain indicate that exposure to dark significantly increased SOX2, BRCA1, and ELK-1 compared to VE (Fig. 4A). These results are consistent with our previous findings that dark exposure increases SOX2+ NPC proliferation (9, 10) and the higher brca1 and elk-1 transcript expression in NPCs (Figs. 2 A and C and 3A and SI Appendix, Table S2).
Fig. 4.
BRCA1 regulates neural progenitor cell fate. (A) BRCA1 (blue), ELK-1 (green), and SOX2 (yellow) expression are increased in animals maintained in the dark compared to enhanced visual experience (VE). Quantitation, normalized to Ponceau staining (Left). n: BRCA1 (5); ELK-1 (4), SOX2 (4). (B) BRCA1 knockdown lowers BRCA1 (blue, n = 5), ELK-1 (green, n = 6), and SOX2 (yellow, n = 6) protein expression. Midbrains were dissected for western blots 2 d after electroporating morpholinos. (C) Diagram of in vivo imaging protocol. Animals were maintained in a 12 h light/12 h dark cycle. After coelectroporation with tGFP and brca1 or control morpholinos, animals were imaged for 3 consecutive days. (D) Representative in vivo images of tGFP+ tectal cells from control morpholino and BRCA1 KD animals. (Scale bar, 50 µm.) (E) BRCA1 KD significantly reduced tGFP+ cell numbers compared to control morpholino-treated animals on days 2 and 3. n = 45 to 46 animals per condition. (F and G) Of the cells shown in (E), the percentage of NPCs significantly increased (F) and neurons decreased (G) with BRCA1 KD compared to control morpholino treatment. (H) Z-projection images showing pH3 immunolabeling (green) and SYTOX nuclear labeling (red) on day 3 in the ventricular layer (VL) and neuronal layer (NL). (Scale bar: 100 µm.) (I) BRCA1 KD significantly increased the total number of pH3+ cells in the tectum compared to control morpholino-treated animals over 3 d. n = 37 to 48 animals per group/timepoint. (J) Confocal Z-projection images of SYTOX-labeled apoptotic nuclei in NPCs in the ventricular layer on day 1 (VL, red arrowheads) and apoptotic neurons in the neuronal layers (NL, yellow arrowheads). (Scale bar: 100 µm.) (K) BRCA1 KD significantly affected the total number of apoptotic cells compared to control morpholino-treated animals. n = 34 to 38 animals per group/timepoint. (L and M) Of the cells shown in (K), BRCA1 KD significantly increased apoptosis in NPCs on days 1 and 2 (L) and in neurons on day 1 compared to control morpholino-treated animals. (M) *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; two-tailed Student’s t test in (A); one-tailed Student’s t test in (B); Mann–Whitney U test in (E–G), (I), and (K–M).
BRCA1 Regulates the Fate of Neural Progenitor Cells.
To examine the function of BRCA1 in NPCs, we knocked down BRCA1 using translation-blocking antisense morpholinos. Two days after electroporating brca1 or control morpholinos, BRCA1 was reduced, and both ELK-1 and SOX2 protein were decreased (Fig. 4B), suggesting that either BRCA1 directly regulates these proteins or that BRCA1 is required to maintain SOX2- and ELK1-expressing NPC numbers.
We then used in vivo time-lapse imaging to visualize the effect of BRCA1 KD on NPCs in the optic tectum. Following the protocol in Fig. 4C, we imaged tGFP+ tectal cells over 3 d, classified them as NPCs or neurons (10), and determined the changes in tGFP+ cell types in BRCA1 KD and control conditions (Fig. 4D). In control animals, tGFP+ cell numbers increased over 3 d (Fig. 4E), as reported (10, 17). BRCA1 KD blocked the normal increase in tGFP+ cells, resulting in a constant number of tGFP+ cells over the imaging period (Fig. 4E), consistent with the decrease in SOX2 in western blots (Fig. 4B). In addition, BRCA1 KD increased the proportion of tGFP+ NPCs (Fig. 4F) and decreased the proportion of tGFP+ neurons (Fig. 4G) compared to control. These data indicate that BRCA1 KD alters the fate of NPCs and suggest that BRCA1 may affect both NPC proliferation and their differentiation into neurons.
To examine whether BRCA1 KD blocked the increase in the total tGFP+ cells by blocking cell division, we immunolabeled cells for the cell division marker phospho-histone H3 (pH3) (Fig. 4H, green cells), a SOX2-independent measure of proliferation. BRCA1 KD increased the number of pH3+ cells in the optic tectum compared to controls, indicating that BRCA1 KD did not block NPC proliferation, but instead, increased NPC proliferation (Fig. 4I), consistent with the increased proportion of NPCs seen with BRCA1 KD (Fig. 4F).
To investigate whether BRCA1 KD increases apoptosis as well as proliferation, thereby maintaining a constant number of tGFP+ cells over the 3-d period, we labeled apoptotic cells with SYTOX nuclear dye (Fig. 4J). BRCA1 KD transiently increased apoptosis in the optic tectum on days 1 and 2, then decreased apoptosis on day 3 compared to controls (Fig. 4K). BRCA1 KD transiently increased apoptosis in NPCs at day 1 and day 2 and in neurons at day 1 (Fig. 4 L and M). These data indicate that BRCA1 KD increases NPC proliferation and transiently increases apoptosis in NPCs and neurons. Combined with the observations that BRCA1 KD blocks the normal increase in SOX2+ progeny, these data suggest that BRCA1 KD biases NPCs to undergo symmetric divisions, generating NPC daughter cells, many of which become apoptotic along with some neurons.
ELK-1 Regulates the Fate of Neural Progenitor Cells.
We examined the function of ELK-1 in NPCs by knocking down ELK-1 with translation-blocking morpholinos (Fig. 5A, green). ELK-1 KD also decreased SOX2 compared to control morpholinos (Fig. 5A, yellow). Following the protocol in Fig. 4C, we found that ELK-1 KD resulted in a delayed block in the normal increase in total tGFP+ cells. From day 1 to day 2, tGFP+ cell numbers increased to a comparable extent in ELK-1 KD and control animals. In contrast to the continued increase in cell numbers in controls, (Fig. 5C, black bars), ELK-1 KD tGFP+ cell numbers failed to increase (Fig. 5C, green bars). Furthermore, ELK-1 KD significantly increased tGFP+ NPCs and decreased tGFP+ neurons compared to control (Fig. 5 D and E). ELK-1 KD transiently increased pH3+ cells at day 1 compared to control (Fig. 5 F and G), while ELK-1 KD increased SYTOX+ cells in the optic tectum (Fig. 5 H and I), possibly explaining why the day 1–day 2 increase in total tGFP+ cell number is the same between ELK-1 KD and controls (Fig. 5C). Specifically, ELK-1 KD increased apoptosis in NPCs over all 3 d of the experiment (Fig. 5J), and transiently increased apoptosis in neurons at day 1 compared to control (Fig. 5K). These data suggest that ELK-1 KD increased the proportion of NPCs in the total tGFP+ pool. Underlying this increase in the proportion of NPCs was a transient increase in NPC proliferation combined with increased apoptosis in NPCs over 3 d and a transient increase in apoptosis in neurons. Together, these studies suggest that ELK-1 KD drives NPCs to undergo symmetric divisions, expanding the progenitor pool, while also increasing NPC apoptosis. The experiments described above suggest that BRCA1 and ELK-1 regulate the fate of NPCs in the developing optic tectum.
Fig. 5.
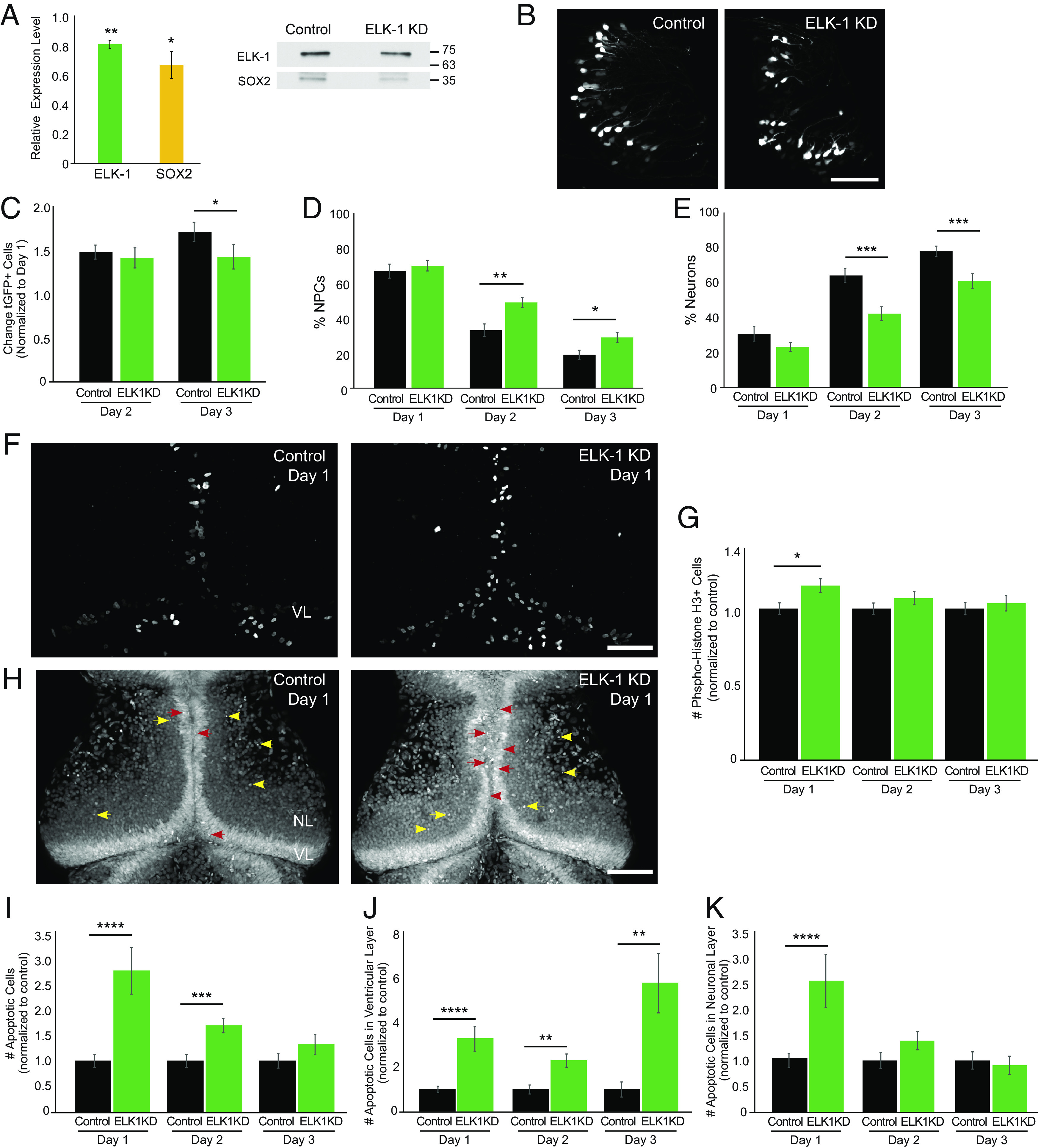
ELK-1 regulates neural progenitor cell fate. (A) elk-1 morpholino reduces ELK-1 (green, n = 3) and SOX2 (yellow, n = 3) compared to control morpholino-treated animals. Midbrains were dissected for western blots 2 d after electroporating morpholinos. Blots were normalized to Ponceau staining for quantification. (B) Representative in vivo images of tGFP+ cells in the optic tectum from control morpholino-treated animals and ELK-1 KD animals. The experimental protocol is shown in Fig. 4C. (Scale bar: 100 µm.) (C) ELK-1 KD significantly reduces the total number of tGFP+ progeny number on day 3 compared to control morpholino-treated animals. n = 12 animals per condition. (D and E) ELK-1 KD significantly increases the percentage of NPCs (D) and decreases neurons (E) in total eGFP+ cells compared to control morpholino-treated animals. (F) Confocal Z-projection of phospho-Histone H3 (pH3) immunolabeling in control morpholino-treated animals and ELK-1 KD tecta. (Scale bar: 100 µm.) (G) ELK-1 KD increases pH3+ cells on day 1 compared to control morpholino-treated animals. n = 40 to 52 animals per group/timepoint. (H) Confocal Z-projection images of SYTOX+ apoptotic nuclei in NPCs in the ventricular layer (VL) on day 1 (red arrowheads) and apoptotic neurons in the neuronal layers (NL, yellow arrowheads). (Scale bar: 100 µm.) (I) ELK-1 KD significantly increases the total number of apoptotic cells on days 1 and 2 compared to control morpholino-treated animals. n = 39 to 51 animals per group/timepoint. (J and K) ELK-1 KD significantly increased apoptosis in NPCs across all days tested (J) and in neurons on day 1 (K) compared to control morpholino-treated animals. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; one-tailed Student’s t test in (A); Mann–Whitney U test in (C–E), (G), and (I–K).
Visual Experience Effects on Neural Progenitor Cell Fate Depend on BRCA1.
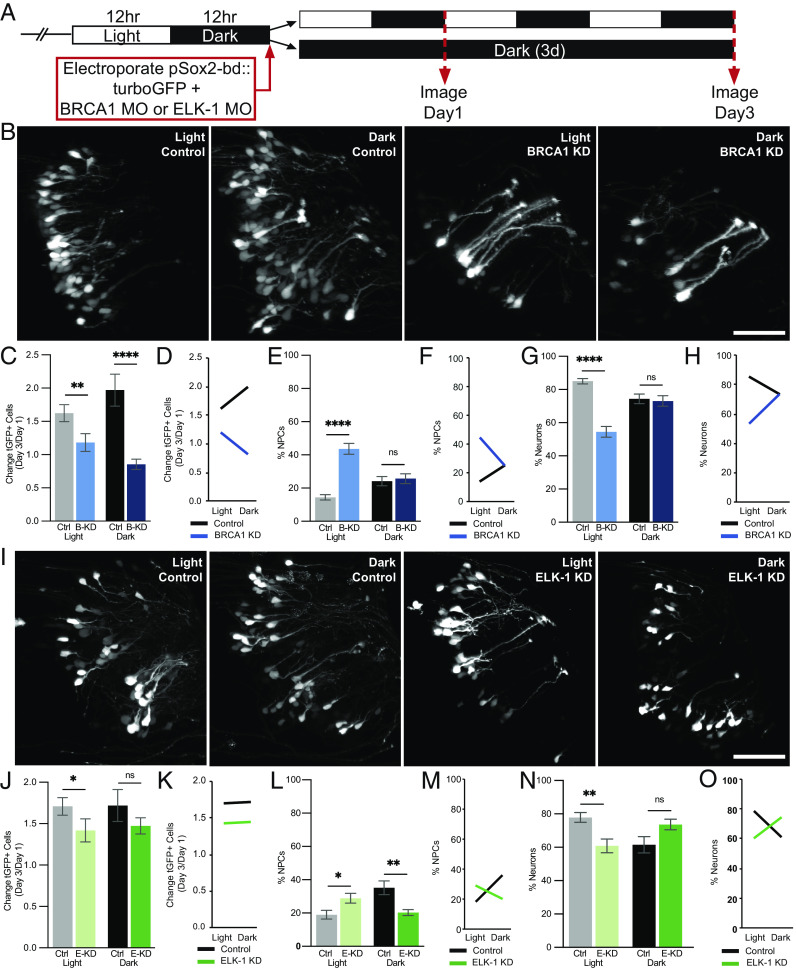
Data described above indicate that BRCA1 is enriched in NPCs (Figs. 2 and 3), that BRCA1 limits NPC proliferation in animals maintained in the 12 h light/12 h dark condition, and that BRCA1 and SOX2 expression increase in the optic tectum when animals are exposed to dark (Fig. 4). To investigate whether the effects of visual experience on NPC fate require BRCA1 expression, we collected in vivo time-lapse images from BRCA1 KD and control animals that were exposed to light/dark conditions or maintained in the dark (Fig. 6 A and B and SI Appendix, Methods for details). Plotting the normalized change in total tGFP+ cell numbers between days 1 and 3 showed that BRCA1 KD decreased tGFP+ cell numbers in animals exposed to the light/dark condition (Fig. 6C, gray vs. light blue bars), reproducing data shown in Fig. 4E. In addition, BRCA1 KD blocked the dark-induced increase in tGFP+ cell numbers (Fig. 6C, black vs. dark blue bars). To further dissect how visual experience and BRCA1 protein expression are involved in regulating the fate of tGFP+ NPCs, we conducted a two-way ANOVA statistical analysis which reveals a statistically significant interaction between visual experience and BRCA1 protein expression on tGFP+ cell number. The factorial experimental design we employed enabled us to perform such analysis. The profile plot shown in Fig. 6D illustrates the relationship between the visual experience conditions and BRCA1 protein expression. In control animals (Fig. 6D, black line), the fold change in tGFP+ cells increased in the dark compared to light/dark conditions. By contrast, when the BRCA1 KD animals were maintained in dark over the 3-d experiment (Fig. 6D, blue line), tGFP+ cell numbers decreased compared to control animals. These data indicate that effects of visual experience on tGFP+ cell numbers depend on BRCA1 protein expression.
Fig. 6.
BRCA1 and ELK-1 mediate visual experience–dependent effects on neural progenitor cell fate. (A) Diagram of treatment and imaging protocol. (B–H) BRCA1 mediates the effects of visual stimulation on NPC fate. (B) Representative images of tGFP+ cells collected on day 3 from animals exposed to 12 h light/12 h dark (light) or continuous dark (dark) with BRCA1 KD or control morpholinos. (Scale bar: 50 µm.) (C–H) Quantitative analysis of imaging data. (C) BRCA1 KD (B-KD) blocked the normal increase in GFP+ cells over the 3-d imaging period in animals exposed to 12 h light/12 h dark (light: gray bar vs. light blue bar) or dark (dark: black bar vs. dark blue bar) compared to control morpholino-treated animals. n = 17 to 24 animals per condition/timepoint. (D) Profile plot of data in (C) demonstrating that the effect of visual experience on tGFP+ cell number depends on BRCA1. (E and G) BRCA KD significantly increased NPCs in animals exposed to light [(E) light: gray bar vs. light blue bar] and decreased neurons [(G) light: gray bar vs. light blue bar] compared to control morpholino-treated animals. BRCA1 KD did not affect NPCs (E) or neurons (G) in animals exposed to dark (black vs. dark blue bars). (F and H) Profile plots of data in (E) and (G) demonstrating that the effect of visual experience on NPC fate depends on BRCA1 expression. (I–O) ELK-1 mediates the effects of visual stimulation on NPC fate. (I) Representative images of tGFP+ cells collected on day 3 in animals exposed to light or dark with ELK-1 KD or control morpholinos. (Scale bar: 50 µm.) (J–O) Quantitative analysis of in vivo imaging data. (J) ELK-1 KD (E-KD) blocked the normal increase in GFP+ cells in animals exposed to light (light: gray vs. light green bars) but not dark (black vs. dark green bars) compared to control morpholino-treated animals. n = 11 to 14 animals per condition/timepoint. (K) Profile plot of factorial comparison of data in (I), demonstrates that the effect of visual experience on the total number of tGFP+ cells is not dependent on ELK-1. (L) ELK-1 KD significantly increased NPCs under light conditions (gray vs. light green bars) and significantly decreased neural progenitors under dark conditions (black vs. dark green bars) compared to control morpholino-treated animals. (N) ELK-1 KD significantly decreased neurons in animals exposed to light (gray vs. light green bars) but did not significantly affect neurons in animals in dark (black vs. dark green bars) compared to control morpholino-treated animals. (M and O) Profile plots of data in (L) and (N) demonstrate that the effect of visual experience conditions on the fate of NPCs and neurons depends on ELK-1 expression. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; Mann–Whitney U test in (C), (E), (G), (J), (L), and (N). Two-way ANOVA analysis was used in (D), (F), (H), (K), (M), and (O).
We next examined whether the influence of visual experience on NPCs is affected by BRCA1. Visual experience provided in the light/dark condition significantly increases NPCs with BRCA1 KD (Fig. 4F) and this is independently reproduced in Fig. 6E (gray vs. light blue bars), suggesting that BRCA1 normally limits the generation of NPCs under control 12 h light/12 h dark conditions. In contrast, BRCA1 KD does not change the proportion of NPCs in animals maintained in the dark (Fig. 6E, black vs. dark blue bars), suggesting that dark-induced increase in proliferation [previously reported (9, 10) and shown in Fig. 6C, gray and black bars] is not sensitive to decreasing BRCA1. Two-way ANOVA analysis shows a statistically significant interaction between visual experience and BRCA1 on the proportion of NPCs in the tGFP+ cells (Fig. 6F). Analysis of the proportion of neurons in the tGFP+ cell population supports a similar conclusion. In animals exposed to light/dark conditions, BRCA1 KD significantly decreases neurons compared to controls (Fig. 6G, gray vs. light blue bars) and there is a significant interaction between the visual experience conditions and BRCA1 on the proportion of neurons (Fig. 6H), indicating that BRCA1 affects neuron number under light/dark conditions. However, as predicted from the NPC data in Fig. 6E, the proportions of neurons are similar in BRCA1 KD and control animals maintained in the dark (Fig. 6G, black vs. dark blue bars). These data suggest that BRCA1 is involved in NPC fate in response to light, that the dark-induced increase in proliferation (Fig. 6C, gray and black bars) is not sensitive to decreasing BRCA1 and that the increase in neuron numbers in light/dark conditions requires BRCA1. Together, these data indicate that the effects of light/dark exposure on NPC fate depend on BRCA1.
Visual Experience Effects on Neural Progenitor Cell Fate Are Mediated by ELK-1.
Thus far, our data indicate that ELK-1 is more highly expressed in NPCs than neurons (Fig. 3), that exposing animals to dark increases ELK-1 (Fig. 4), and that ELK-1 limits neurogenesis by affecting both NPC proliferation and apoptosis (Fig. 5) in animals maintained in the light/dark condition. To further understand the mechanisms by which visual experience conditions affect NPC fate, we tested whether the effects of visual experience on NPC fate are mediated by ELK-1 using the protocol in Fig. 6A. Representative images of tGFP+ cells are shown for the control morpholino-treated animals and ELK-1 KD animals after 3 d of light/dark or dark conditions (Fig. 6I). ELK-1 KD decreased the number of tGFP+ cells in animals maintained in 12 h light/12 h dark conditions, independently reproducing data in Fig. 5, but ELK-1 KD did not significantly change tGFP+ cell numbers in animals exposed to dark (Fig. 6J). Furthermore, two-way ANOVA statistical analysis of ELK-1 expression and visual experience conditions suggests that these two factors do not interact to regulate the number of tGFP+ cells, indicated by the two parallel lines in the profile plot (Fig. 6K).
We next examined whether the effect of dark exposure on increasing NPCs depends on ELK-1. Under the 12 h light/dark visual experience condition, ELK-1 KD increased NPCs compared to controls (Fig. 6L, gray vs. light green bars), independently replicating data in Fig. 5D. Furthermore, ELK-1 KD blocked the dark-induced increase in NPCs (Fig. 6L, black vs. dark green bars). These data indicate that ELK-1 normally limits the generation of NPCs under control visual experience conditions and that ELK-1 is required for the dark-induced expansion of the neural progenitor pool. We find a statistically significant interaction between the visual experience condition and ELK-1 expression on the proportion of NPCs in the tGFP+ cell population based on the two-way ANOVA analysis (Fig. 6M). These data indicate that ELK-1 may be involved in NPC fate in both light/dark and continuous dark conditions. ELK-1 KD significantly decreased neurons in light/dark conditions, compared to control morpholinos (Fig. 6N, gray vs. light green bars) and there was a significant interaction between the visual experience condition and ELK-1 expression on neurons (Fig. 6O). However, ELK-1 KD did not significantly affect the proportion of neurons generated under dark conditions (Fig. 6N, black vs. dark green bars), suggesting that ELK-1 is required specifically to mediate the effects of dark exposure on expanding the NPC pool and the effects of light in promoting neuronal differentiation. Together, these data demonstrate that the effects of visual experience conditions on NPC fate depend on ELK-1.
Discussion
This study profiled the transcriptomes of activity-induced proliferating NPCs and newly differentiated immature neurons in X. laevis tadpole brains using RNA-seq. Neuronal activity regulates the fate of NPCs in the CNS (4, 5, 9, 10, 17, 24). Here, we exposed tadpoles to dark or 12 h light/12 h dark for 24 h to bias the in vivo fate of the tGFP-birthdated cohort of SOX2+ optic tectal NPCs toward cell proliferation or differentiation, respectivel, y and used RNA-Seq of FAC sorted tGFP+ cells to identify molecular signatures of activity-dependent effects on neural progenitor cell fate. This experimental design enabled us to detect differences in the transcriptomes of NPCs and newly differentiated neurons induced within 24 h in the unperturbed neurogenic niche in the intact developing brain. Our transcriptome analysis identified 1,130 DE transcripts between NPCs and neurons. Data-mining and bioinformatic analyses revealed an overview of the potential roles of the DE transcripts in neurogenesis and identified a network of DE upstream transcriptional regulators, including BRCA1 and ELK-1, which is predicted to regulate the expression of other DE transcripts in NPCs and neurons during in vivo sensory experience-driven brain development. In vivo time-lapse two-photon imaging showed that BRCA1 and ELK-1 affect neurogenesis. Furthermore, BRCA1 and ELK-1 mediate sensory experience–dependent effects on NPC fate. Together these studies provide a resource for transcriptomic profiles of enriched populations of NPCs and immature CNS neurons from X. laevis, increase our understanding of cellular and molecular mechanisms underlying experience-dependent NPC fate decisions, and suggest roles for BRCA1 and ELK-1 in experience-dependent neurogenesis in the developing vertebrate brain.
Bioinformatic Analysis Identified Functional Categories and Networks of Differentially Expressed Genes.
We mined several databases to reveal the differences in transcriptomes between NPCs and immature neurons and the potential implications these differences represent. Recognizing that we are not analyzing pure populations of single neural cell types, our bioinformatic approaches emphasized network interactions, which weigh multiple components with known interactions or operating within known signaling pathways. PANTHER’s functional categorization of the DE transcripts identified molecular components and signaling pathways induced in response to activity and extracellular signaling events that affect cell proliferation or cell cycle exit and neuronal differentiation. Prominent categories of the DE transcripts include proteases and transcription factors which regulate progenitor cell fate in other systems, validating our approach. Other DE transcripts that are less well known with respect to NPC fate regulation may reveal additional mechanisms involved in this context (SI Appendix, Figs. S5 and S6).
Cell proliferation, cell differentiation, and cell cycle were identified by PANTHER as prominent functional categories represented in the DE transcripts. These processes are most relevant to maintaining NPC self-renewability and neuronal differentiation. Among the DE transcripts in our dataset that are included in these processes, brca1, tgfa, and jak2, which are enriched in NPCs, are known to promote cell proliferation (25, 26), while sstr5, which is decreased in NPCs, inhibits cell proliferation (27). pdgfa, in addition to its role promoting cell proliferation, regulates cell migration (28), consistent with our observation that pdgfa expression was increased in immature neurons. Together, this analysis identifies neurodevelopmental cellular processes that are associated with the DE transcripts and importantly generates hypotheses implicating less well-known candidates in neurodevelopmental processes.
Protein–Protein Interaction Networks Identify Key Players in Neurodevelopment among the Differentially Expressed Transcripts.
Further analysis of the DE transcripts identified a network of highly connected hub proteins that interact with each other: ACTA2, BMP4, JAK2, and BRCA1, which were enriched in NPCs, and ITGA2, VEGFA, FGF2, AURKB, and NFKB1, which were enriched in immature neurons (Fig. 2 A–C and SI Appendix, Fig. S5). Focusing on the NPCs, brca1 (breast cancer 1) and jak2 (Janus kinase 2, a nonreceptor tyrosine kinase) are enriched in NPCs, and their proteins each have 28 and 29 interaction partners in this network, respectively. BRCA1 can regulate the expression and modulate the activity of JAK2 (29). JAK2 activates the STAT signaling cascade, and stat2 itself is enriched in NPCs. The JAK/STAT signaling cascade modulates proliferation of NPCs (30, 31). bmp4 (bone morphogenetic protein 4), also enriched in NPCs, has 25 interactions in this network and maintains self-renewal of mouse embryonic stem cells (32). In summary, proteins generated from the DE transcripts occupy important positions within network nodes regulating cell proliferation, differentiation, and survival, with distinct functions in regulating neurogenesis. Proteins generated from DE transcripts are themselves well-connected in a network of other proteins derived from DE transcripts, consistent with coordinated transcriptional control generating functional protein interaction networks.
Bioinformatic Identification of a Transcriptional Regulatory Network that May Mediate Activity-Dependent Control of Neural Progenitor Cell Fate.
A major interest in our bioinformatic analysis is to identify DE master transcriptional regulators that could function in a network to regulate other DE transcripts in NPCs and immature neurons in response to different visual stimulation conditions. Using a dual criteria strategy to search for transcription factor master regulators based on 1) capacity to regulate the majority of DE transcripts in our dataset, and 2) large number of interaction partners, successfully identified five candidate master regulators that each have multiple interaction partners and together form a network: CEBPD, FOSL1, CEBPB, ELK-1, and BRCA1. Are these candidates likely to mediate sensory experience–dependent effects on transcript expression? Are they likely to regulate NPC proliferation and neuronal differentiation? Activity-dependent regulation of cell proliferation and neuronal differentiation is mediated by intercellular signaling between neurons and NPCs that initiates ERK/MAPK signaling (5, 33). ERK/MAPK signaling induces transcription of immediate early genes in NPCs, which in turn can induce expression of diverse genes (34). This network is well positioned to transduce activity-triggered intercellular signals to affect NPC fate. FOSL1, CEBPB, ELK-1, and BRCA1 are enriched in NPCs and interact with each other. FOSL1 (fos-like antigen 1) binds c-Jun to form the activator protein 1 (AP-1) transcription complex and promotes cell cycle progression (35). CEBPB [CCAAT/enhancer binding protein (C/EBP), beta] promotes self-renewal and proliferation of NPCs and survival of new-born neurons (36) and synergizes with ELK-1 (37).
BRCA1 is a multifunctional protein, widely known as a tumor suppressor, with roles in genome stability, checkpoint control, replication fork stability, and DNA double-strand break repair via homologous recombination (26, 38). In mice, BRCA1 null is embryonic lethal (39, 40), but mice with spatial and temporal control of BRCA1 loss of function have neurodevelopmental defects, specifically related to BRCA1’s role in neurogenesis (20, 41). BRCA1’s function in NPCs may be related to its function repairing double-stranded DNA damage because their high rate of proliferation makes them prone to DNA double strand breaks (39); however, BRCA1 is also a component of core transcriptional machinery, where it can act as a transcriptional activator or repressor, depending on its interaction partners (42). BRCA1 is an upstream regulator of elk-1, and ELK-1 interacts with BRCA1 to augment BRCA1’s growth suppressive function in cancer cells (43, 44). In addition, BRCA1 can regulate expression and modulate activity of JAK2 (29), which we identified as a DE transcript with one of the highest number of protein–protein interactions, and can reportedly induce cell proliferation by activating promoters of c-fos and c-myc, which itself was differentially expressed and has a high number of protein–protein interactions.
ELK-1 is expressed in SOX2+ NPCs and neurons throughout development and in adult animals (45). ELK-1 is phosphorylated by MAPKs, including ERK, resulting in translocation of pELK-1 to the nucleus and induced transcription of diverse target genes. The specificity of ELK-1-regulated transcriptional responses is likely due to spatiotemporal control of recruitment of specific coactivators, such as CREB binding protein, p300, and serum response factor, resulting in diverse downstream outcomes concerning pluripotency, apoptosis, proliferation, and survival in neural progenitors or synaptic plasticity in neurons (21). Nuclear translocation of pELK-1 in SOX2+ neural progenitors induced transcription of immediate early genes (IEGs), including egr1 (aka zif266) and c-fos, as well as other targets pertaining to proliferation and pluripotency, such as sox2, oct4, and nanog (45). This convergence of extracellular signaling to IEGs and sox2 suggests a mechanism for extracellular activating signals to regulate NPC fate, consistent with studies demonstrating a role for neuronal activity in neurogenesis (4, 5).
CEBPD, [CCAAT/enhancer binding protein (C/EBP), delta] inhibits cell proliferation by down-regulating c-Myc and cyclins and increasing expression of differentiation-related genes (36). This suggests that CEBPD may inhibit cell proliferation and drive neuronal differentiation, consistent with the increased expression we observed in immature neurons. In combination with the enriched expression of neuroD1 and fgf2, that are known to induce terminal neuronal differentiation (46), visual experience-induced increased CEBPD expression in tadpoles may promote NPCs to exit the cell cycle and differentiate into neurons. Together, analysis of the DE transcriptomes identified factors and mechanisms that may regulate NPC proliferation and neuronal differentiation in the developing brain in response to activity-driven cues.
BRCA1 and ELK-1 Are Required for Visual Experience–Dependent Regulation of Neural Progenitor Cell Fate.
Our bioinformatic analysis indicated that BRCA1 and ELK-1 are members of a network of DE transcriptional regulators which may in turn target a large proportion of the transcripts that are differentially expressed between neural progenitors and neurons in our study. This suggested that different visual experience conditions, light or dark, lead to sequentially amplifying effects on differential transcript expression in NPCs and neurons. Indeed, our in vivo imaging data indicated that BRCA1 and ELK-1 are required for neurogenesis and furthermore that effects of visual experience conditions on expansion of the progenitor pool and neuronal differentiation are mediated by BRCA1 and ELK-1.
We targeted BRCA1 and ELK1 KD to the optic tectum of stage 46 tadpoles to avoid early lethality and large-scale neurodevelopmental defects (20, 39–41) and to assess direct effects of BRCA1 or ELK1 manipulation in animals under different visual experience conditions. BRCA1 KD decreased ELK-1 and SOX2 in midbrain lysates, consistent with the increased apoptosis in NPCs. The overall decrease in tGFP+ cell numbers with BRCA1 KD seen with in vivo imaging followed a rapid increase in apoptosis, principally in NPCs, consistent with BRCA1’s role in homologous recombination-mediated DNA repair in proliferative cells in the rodent brain (20, 39, 41). The increased apoptosis seen with BRCA1 KD changes the proportion of NPCs and neurons in the in vivo imaging data. In control animals, the proportion of NPCs decreases as the proportion of neurons increases reciprocally, but apoptosis in NPCs blocked the normal increase in neuronal differentiation, reducing the proportion of neurons in the tGFP+ population and paradoxically increasing the proportion of neural progenitors. It is interesting that we observe an increase in pH3 labeling in response to BRCA1 KD, suggesting that BRCA1 KD does not interfere with S-phase DNA replication.
Visual experience–dependent effects on neural progenitor fate are mediated by BRCA1 and ELK-1. BRCA1 KD experiments suggest that the visual experience–dependent increase in cell numbers requires BRCA1. BRCA1 KD increased the proportion of NPCs and decreased the proportion of neurons in the tGFP+ population in response to visual stimulation provided in the 12 h light/12 h dark condition. This altered cell fate suggests that BRCA1 is required for the visual stimulation–induced differentiation of neurons such that with decreased BRCA1, NPCs fail to differentiate and their relative numbers increase. The influence of visual stimulation conditions on the proportions of NPCs and neurons is significantly altered by ELK-1 KD, suggesting that ELK-1 is required for both the increased NPC proliferation in the dark and the visual stimulation-induced neuronal differentiation. These effects of ELK-1 on proliferation and differentiation are likely mediated by different ELK-1 targets and signaling pathways. Together these data indicate that BRCA1 and ELK-1 regulate neural progenitor cell fate, through both shared and diverse molecular and cellular pathways.
The interplay between BRCA1 and ELK-1 is particularly interesting in light of their shared roles in NPC fate in response to visual activity. Animals exposed to visual stimulation have decreased expression of BRCA1, ELK-1, and SOX2 compared to animals exposed to dark. Furthermore, BRCA1 KD decreases ELK-1 expression, consistent with other studies indicating that BRCA1 negatively regulates elk-1 transcription (43). In addition, BRCA1 and ELK-1 interact and this interaction enhances BRCA1 function, suggesting that ELK-1 may function downstream of BRCA1 (44). Decreased levels of BRCA1 and ELK-1 in the tectum increased NPC apoptosis, consistent with the observation that ELK-1 KD decreases expression of “stemness” genes, related to self-renewal and differentiation, such as sox2, oct4, and nanog (22). Although we have not examined whether BRCA1 and ELK-1 are coexpressed in Xenopus optic tectal NPCs, these studies suggest that BRCA1 KD and ELK-1 KD drive NPCs to expand the progenitor pool through symmetric divisions, while also increasing NPC apoptosis.
We propose that BRCA1 and ELK-1 are master transcriptional regulators of activity-regulated changes in neurogenesis. This idea is supported by the bioinformatic analysis that they potentially regulate 270 differentially expressed transcripts in our dataset and that they have the capacity to operate in a network with the three other differentially expressed transcriptional regulators, CEBPB, CEBPD, and FOSL1, to target a total of 409 transcripts and by experimental evidence for their roles in visual experience–dependent modulation of NPC fate.
Materials and Methods
Animals.
Albino X. laevis tadpoles of both sexes were reared under a 12-h light/12-h dark cycle, anesthetized before all procedures, and killed as described (10), and See SI Appendix, Methods. All animal protocols were approved by the Institutional Animal Care and Use Committee of Scripps Research (approval # 08-0083-3).
Isolation of Enriched Neural Progenitor Cells and Immature Neurons.
Cell samples were collected from tadpole midbrain as described briefly here and in detail in SI Appendix, Methods. The brains of anesthetized stage 46 tadpoles were electroporated with pSOX2-bd::tGFP (10) (SI Appendix, Methods and Fig. S1). Animals were exposed to visual stimulation, to enrich for immature neurons, or dark for 24 h to enrich for NPCs, and midbrains from ~100 animals per condition were dissected and dissociated into single cells. tGFP+ cells were isolated using fluorescence-activated cell sorting (FACS; FACSAria II, BD Biosciences, USA; RRID:SCR_018934).
RNA-Seq of Neural Progenitor Cells and Immature Neurons.
Total RNA from samples collected above was extracted using the mirVana kit (Life Technologies), followed by DNase treatment and clean-up using the RNeasy mini kit (Qiagen, USA). Samples with RNA integrity number (RIN) >8 were used for analysis. Three biological replicates were analyzed for each condition. NextGen sequencing was done at the Sequencing Core at the Scripps Research Institute (La Jolla) using the HiSeq2000 platform (Illumina; RRID:SCR_020132) and read quality was analyzed (SI Appendix, Table S1 and Figs. S2 and S3). We used the DE analysis package, DESeq2 (RRID:SCR_015687), and R for graphics (v3.1.2; cran.r-project.org; RRID:SCR_001905) through Bioconductor (RRID:SCR_006442). See SI Appendix, Methods.
Bioinformatic Analysis.
Bioinformatic analysis was conducted using STRING (v10; RRID:SCR_005223), Cytoscape (v3.2.1; RRID:SCR_015784) (https://www.cytoscape.org/), ClueGO (v2.1.7; RRID:SCR_005748), PANTHER (RRID: SCR_004869), ENCODE (RRID:SCR_015482), and Cufflinks suite (v2.2.1; RRID:SCR_014597) (SI Appendix, Methods).
In Vivo Time-Lapse Imaging.
The brains of anesthetized late-stage 46 tadpoles were coelectroporated with pSOX2-bd::tGFP and antisense translation-blocking morpholino oligonucleotides tagged with lissamine fluorophores (47) targeted against brca1, elk-1, or a control sequence (GeneTools). Tadpoles were imaged with a 20× (Olympus XLUMPlanFL 0.95 NA) water immersion lens on a custom-built two-photon microscope. All samples were imaged in parallel using identical image acquisition parameters. See SI Appendix, Methods.
Immunohistochemistry.
Anesthetized animals were fixed in 4% paraformaldehyde (Electron Microscopy Sciences) in phosphate-buffered saline (pH 7.4). Dissected brains were incubated in blocking solution, followed by anti-pH3 antibody and Alexa488 donkey anti-mouse secondary antibody or Sytox Orange (SytoxO). Brains were mounted in 6 M urea in 50% glycerol for imaging (Nikon C2, 20× Plan Apo lens with 0.75 NA). ImageJ Cell Count plugin was used for analysis. See SI Appendix, Methods.
Western Blots.
Experimental and control samples were prepared and processed in parallel. For ELK-1 (Abcam #ab188316; RRID:AB_2890919) and SOX2 (Cell Signaling Technology #3579S; RRID:AB_2195767) antibodies, tissues were homogenized in RIPA buffer. For BRCA1 antibody labeling (SCBT #SC-646; RRID:AB_630945), tissue was prepared as described in SI Appendix, Methods. Antibodies were detected by goat anti-mouse/rabbit HRP-conjugated secondaries (BioRad) followed by ECL (Pierce, Thermo Fisher Scientific, 32209). Quantification was performed using densitometry (ImageJ), using different exposures to avoid saturation and normalized to Ponceau S staining (SI Appendix, Methods).
Statistical Tests.
The nonparametric Mann–Whitney U test and one-tailed or two-tailed Student’s t test were used for comparisons of two groups using Prism 9 statistics software (Graphpad Prism; RRID:SCR_002798). For unbalanced two-way ANOVA analysis, car package in R was used (RRID:SCR_001905).
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Acknowledgments
This work was supported by the California Institute of Regenerative Medicine Training Grant #01165 to The Scripps Research Institute (L-C.H.), Dart Neuroscience, the NIH (NS076006, NS114975, EY011261, EY027437, and EY031597), and an endowment from the Hahn Foundation to H.T.C. We thank members of the Cline lab for helpful comments and discussion and both the Fluorescence Activated Cell Sorting Core and the Next Generation Sequencing Core at The Scripps Research Institute.
Author contributions
L.-C.H., C.R.M., and H.T.C. designed research; L.-C.H., C.R.M., and H.-Y.H. performed research; L.-C.H., C.R.M., H.-Y.H., and A.C.T. analyzed data; and L.-C.H., C.R.M., and H.T.C. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: J.L., Pontificia Universidad Católica de Chile; and A.K.S., University of Houston.
Data, Materials, and Software Availability
All raw X. laevis data are available on GEO as GSE184315 (48) and on Xenbase. pSox2-bd::FP plasmid is available from Addgene, plasmid #34703. Morpholino sequences are provided in the accompanying reagent list. The DE read data are provided in Dataset S1. The Panther analysis, STRING analysis, and CytoScape data are provided in Datasets S2–S4, respectively.
Supporting Information
References
- 1.Pramparo T., et al. , Cell cycle networks link gene expression dysregulation, mutation, and brain maldevelopment in autistic toddlers. Mol. Syst. Biol. 11, 841 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gotz M., Nakafuku M., Petrik D., Neurogenesis in the developing and adult brain-similarities and key differences. Cold Spring Harb. Perspect. Biol. 8, a018853 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chenn A., Walsh C. A., Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science 297, 365–369 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Pan Y., Monje M., Activity shapes neural circuit form and function: A historical perspective. J. Neurosci. 40, 944–954 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirischuk S., et al. , Modulation of neocortical development by early neuronal activity: Physiology and pathophysiology. Front Cell Neurosci. 11, 379 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sin W. C., Haas K., Ruthazer E. S., Cline H. T., Dendrite growth increased by visual activity requires NMDA receptor and Rho GTPases. Nature 419, 475–480 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Aizenman C. D., Cline H. T., Enhanced visual activity in vivo forms nascent synapses in the developing retinotectal projection. J. Neurophysiol. 97, 2949–2957 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Gambrill A. C., Faulkner R. L., McKeown C. R., Cline H. T., Enhanced visual experience rehabilitates the injured brain in Xenopus tadpoles in an NMDAR-dependent manner. J. Neurophysiol. 121, 306–320 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma P., Cline H. T., Visual activity regulates neural progenitor cells in developing xenopus CNS through musashi1. Neuron 68, 442–455 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bestman J. E., Lee-Osbourne J., Cline H. T., In vivo time-lapse imaging of cell proliferation and differentiation in the optic tectum of Xenopus laevis tadpoles. J. Comp. Neurol. 520, 401–433 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Azim K., et al. , Transcriptional hallmarks of heterogeneous neural stem cell niches of the subventricular zone. Stem Cells 33, 2232–2242 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Berger C., et al. , FACS purification and transcriptome analysis of drosophila neural stem cells reveals a role for Klumpfuss in self-renewal. Cell Rep. 2, 407–418 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee-Liu D., et al. , Genome-wide expression profile of the response to spinal cord injury in Xenopus laevis reveals extensive differences between regenerative and non-regenerative stages. Neural Dev. 9, 12 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raj S., Sifuentes C. J., Kyono Y., Denver R. J., Metamorphic gene regulation programs in Xenopus tropicalis tadpole brain. PLoS One 18, e0287858 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ta A. C., et al. , Temporal and spatial transcriptomic dynamics across brain development in Xenopus laevis tadpoles. G3 (Bethesda) 12, jkab387 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cordero-Veliz C., Larrain J., Faunes F., Transcriptome analysis of the response to thyroid hormone in Xenopus neural stem and progenitor cells. Dev. Dyn. 252, 294–304 (2023). [DOI] [PubMed] [Google Scholar]
- 17.Bestman J. E., Huang L. C., Lee-Osbourne J., Cheung P., Cline H. T., An in vivo screen to identify candidate neurogenic genes in the developing Xenopus visual system. Dev. Biol. 408, 269–291 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang S., Zhao J., Multi-attribute intergrated measurement of node importance in complex networks. Chaos 25, 113105 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Yosef N., Regev A., Impulse control: Temporal dynamics in gene transcription. Cell 144, 886–896 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pao G. M., et al. , Role of BRCA1 in brain development. Proc. Natl. Acad. Sci. U.S.A. 111, E1240–E1248 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Besnard A., Galan-Rodriguez B., Vanhoutte P., Caboche J., Elk-1 a transcription factor with multiple facets in the brain. Front. Neurosci. 5, 35 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sogut M. S., et al. , ETS-domain transcription factor Elk-1 regulates stemness genes in brain tumors and CD133+ braintumor-initiating cells. J. Pers. Med. 11, 125 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noro T., et al. , Elk-1 regulates retinal ganglion cell axon regeneration after injury. Sci. Rep. 12, 17446 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall Z. J., Tropepe V., Movement maintains forebrain neurogenesis via peripheral neural feedback in larval zebrafish. Elife 7, e31045 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim Y. H., et al. , Differential regulation of proliferation and differentiation in neural precursor cells by the Jak pathway. Stem Cells 28, 1816–1828 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Prakash R., Zhang Y., Feng W., Jasin M., Homologous recombination and human health: The roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb. Perspect Biol. 7, a016600 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barbieri F., et al. , Somatostatin receptors 1, 2, and 5 cooperate in the somatostatin inhibition of C6 glioma cell proliferation in vitro via a phosphotyrosine phosphatase-eta-dependent inhibition of extracellularly regulated kinase-1/2. Endocrinology 149, 4736–4746 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Nagel M., Tahinci E., Symes K., Winklbauer R., Guidance of mesoderm cell migration in the Xenopus gastrula requires PDGF signaling. Development 131, 2727–2736 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Gao B., et al. , Constitutive activation of JAK-STAT3 signaling by BRCA1 in human prostate cancer cells. FEBS Lett. 488, 179–184 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Kang M. K., Kang S. K., Interleukin-6 induces proliferation in adult spinal cord-derived neural progenitors via the JAK2/STAT3 pathway with EGF-induced MAPK phosphorylation. Cell Prolif. 41, 377–392 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chung H., Li E., Kim Y., Kim S., Park S., Multiple signaling pathways mediate ghrelin-induced proliferation of hippocampal neural stem cells. J. Endocrinol. 218, 49–59 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Zhang J., et al. , BMP induces cochlin expression to facilitate self-renewal and suppress neural differentiation of mouse embryonic stem cells. J. Biol. Chem. 288, 8053–8060 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma D. K., et al. , Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 323, 1074–1077 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yap E. L., Greenberg M. E., Activity-regulated transcription: Bridging the gap between neural activity and behavior. Neuron 100, 330–348 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hess J., Angel P., Schorpp-Kistner M., AP-1 subunits: Quarrel and harmony among siblings. J. Cell Sci. 117, 5965–5973 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Pulido-Salgado M., Vidal-Taboada J. M., Saura J., C/EBPbeta and C/EBPdelta transcription factors: Basic biology and roles in the CNS. Prog. Neurobiol. 132, 1–33 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Hanlon M., Bundy L. M., Sealy L., C/EBP beta and Elk-1 synergistically transactivate the c-fos serum response element. BMC Cell Biol. 1, 2 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Densham R. M., Morris J. R., The BRCA1 ubiquitin ligase function sets a new trend for remodelling in DNA repair. Nucleus 8, 116–125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orii K. E., Lee Y., Kondo N., McKinnon P. J., Selective utilization of nonhomologous end-joining and homologous recombination DNA repair pathways during nervous system development. Proc. Natl. Acad. Sci. U.S.A. 103, 10017–10022 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gowen L. C., Johnson B. L., Latour A. M., Sulik K. K., Koller B. H., Brca1 deficiency results in early embryonic lethality characterized by neuroepithelial abnormalities. Nat. Genet. 12, 191–194 (1996). [DOI] [PubMed] [Google Scholar]
- 41.Pulvers J. N., Huttner W. B., Brca1 is required for embryonic development of the mouse cerebral cortex to normal size by preventing apoptosis of early neural progenitors. Development 136, 1859–1868 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Mullan P. B., Quinn J. E., Harkin D. P., The role of BRCA1 in transcriptional regulation and cell cycle control. Oncogene 25, 5854–5863 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Maniccia A. W., et al. , Mitochondrial localization, ELK-1 transcriptional regulation and growth inhibitory functions of BRCA1, BRCA1a, and BRCA1b proteins. J. Cell Physiol. 219, 634–641 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chai Y., et al. , c-Fos oncogene regulator Elk-1 interacts with BRCA1 splice variants BRCA1a/1b and enhances BRCA1a/1b-mediated growth suppression in breast cancer cells. Oncogene 20, 1357–1367 (2001). [DOI] [PubMed] [Google Scholar]
- 45.Wells T., Rough K., Carter D. A., Transcription mapping of embryonic rat brain reveals EGR-1 induction in SOX2 neural progenitor cells. Front Mol. Neurosci. 4, 6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Louvi A., Artavanis-Tsakonas S., Notch signalling in vertebrate neural development. Nat. Rev. Neurosci. 7, 93–102 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Bestman J. E., Cline H. T., Morpholino studies in Xenopus brain development. Methods Mol. Biol. 2047, 377–395 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Chien Huang L., et al. , BRCA1 and ELK-1 regulate neural progenitor cell fate in the optic tectum in response to visual experience in xenopus laevis tadpoles3. NIH GEO4. https://www.ncbi.nlm.nih.gov/geo/GSE184315/5. Deposited 16 September 2021. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Data Availability Statement
All raw X. laevis data are available on GEO as GSE184315 (48) and on Xenbase. pSox2-bd::FP plasmid is available from Addgene, plasmid #34703. Morpholino sequences are provided in the accompanying reagent list. The DE read data are provided in Dataset S1. The Panther analysis, STRING analysis, and CytoScape data are provided in Datasets S2–S4, respectively.