Summary
Bulk-tissue molecular quantitative trait loci (QTLs) have been the starting point for interpreting disease-associated variants, and context-specific QTLs show particular relevance for disease. Here, we present the results of mapping interaction QTLs (iQTLs) for cell type, age, and other phenotypic variables in multi-omic, longitudinal data from the blood of individuals of diverse ancestries. By modeling the interaction between genotype and estimated cell-type proportions, we demonstrate that cell-type iQTLs could be considered as proxies for cell-type-specific QTL effects, particularly for the most abundant cell type in the tissue. The interpretation of age iQTLs, however, warrants caution because the moderation effect of age on the genotype and molecular phenotype association could be mediated by changes in cell-type composition. Finally, we show that cell-type iQTLs contribute to cell-type-specific enrichment of diseases that, in combination with additional functional data, could guide future functional studies. Overall, this study highlights the use of iQTLs to gain insights into the context specificity of regulatory effects.
Keywords: gene expression, DNA methylation, interaction QTL, cell-type composition, gene-environment interaction
To identify regulatory variants with plasticity in effect size, we performed interaction molecular quantitative trait loci (iQTL) mapping with cell-type abundance, age, sex, and smoking as the environmental factors. Our results highlight the usefulness of iQTLs for gaining insights into the context specificity of regulatory effects.
Introduction
Bulk-tissue molecular quantitative trait loci (molQTLs) have been valuable in highlighting potential target genes and gene regulatory mechanisms of disease-associated genetic variants.1,2,3 However, context-specific regulatory variants, such as cell-type-specific or response QTLs, exhibit particular relevance for disease when compared to standard molQTLs from steady-state tissues.4 Mapping cell-type interaction expression QTLs by modeling the interaction effect between the genotype of an SNP and computationally inferred cell-type estimates has been shown to aid in the discovery of cell-type-specific effects of expression QTLs.5,6,7 Pinpointing the true mediating cell type with this approach might still be challenging because of the properties of the interaction model and correlations between cell-type proportions. Thus, rigorous interpretation of cell-type interaction molecular quantitative trait loci (iQTLs) is important for inferring insights about the true cell-type specificity of these effects.
The etiology of most complex diseases is recognized to be influenced both by genetic and environmental factors and their interactions.8 Detecting gene-environment (G × E) interactions in genome-wide association studies (GWASs) has proven difficult as a result of small effect sizes and computational challenges.9,10 Mapping interaction molQTLs for physiological environments, such as age, sex, smoking, or inflammation, offers an opportunity to identify G × E interactions at the molecular level with improved statistical power attributed to stronger effects of regulatory variants. Recently, a transcription-based framework has shown the potential to link genes with genetic variant-age interactions to age-associated diseases,11 suggesting the benefits of focusing on regulatory variants to study their complex interplay with other factors contributing jointly to variability in traits and diseases.
To comprehensively assess the utility of interaction molQTLs, we performed cell-type interaction molQTL (iQTL) mapping from gene expression (RNA-seq) and DNA methylation (EPIC array) in 1,319 participants of diverse ancestries as part of the Trans-Omics for Precision Medicine (TOPMed) program Multi-Ethnic Study of Atherosclerosis (MESA) multi-omics pilot with data from two time points (exam 1 and exam 5, 10 years apart) (Figures 1A and S1A). This longitudinal design enabled us to assess the robustness of cell-type iQTLs. Additionally, we characterize the sharing, replication, and functional enrichment of cell-type iQTLs with respect to their direction of effect. MESA phenotyping data allows us to map age, sex, and smoking iQTLs and study the mediation by cell-type iQTLs. Finally, we highlight the informativeness of cell-type iQTLs for proposing cell-type-specific mechanisms underlying diseases.
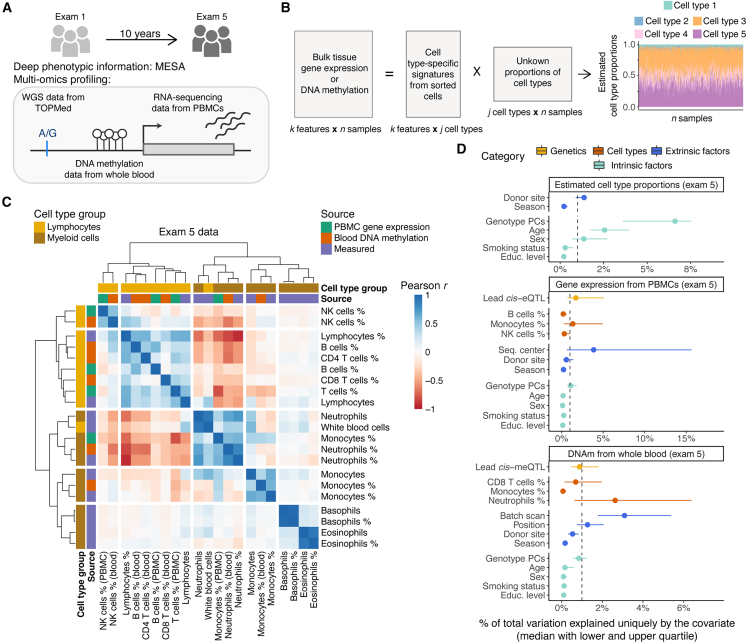
Figure 1.
Study design and overview of the estimated cell-type proportions
(A) Illustration of the study design and data types profiled for 1,319 individuals.
(B) Graphical illustration of cell-type deconvolution.
(C) Correlation of cell-type proportions. Exam 5 data from three sources were used: data estimated with the CIBERSORT method from PBMC gene expression, data estimated with the Houseman method from whole-blood DNA methylation, and cell counts measured by flow cytometry.
(D) Sources of variability in estimated cell-type proportions with CIBERSORT and the Houseman method, gene expression from PBMCs, and DNA methylation from whole-blood according to exam 5 data. The median of the total explained variation is calculated across all the tested cell types, genes, and CpG sites. A gray dashed line denotes 1% of the total explained variance. Error bars denote the lower and upper quartile of the total explained variation.
Material and methods
MESA multi-omics pilot
MESA is a prospective cohort study with the goal of identifying the progression of subclinical atherosclerosis.12 MESA recruited 6,814 participants, ages 45–84 years and free of clinical cardiovascular disease, at six field centers from 2000–2002. MESA included multiple race and ethnic groups (38% non-Hispanic white, 28% African American, 22% Hispanic, and 12% Asian Americans). It comprises 53% females and includes 49% ever-smokers (18% current). All MESA participants provided written informed consent, and the study was approved by the institutional review boards of collaborating institutions.
The MESA multi-omics pilot data include 30× whole-genome sequencing (WGS) of ∼4,600 individuals through the Trans-Omics for Precision Medicine (TOPMed) project,13 wherein ∼1,000 participant samples were collected at two time points (exam 1 and exam 5, ten years apart). Whole-blood and/or cell types (peripheral blood mononuclear cells (PBMCs), monocytes, and T cells) were assayed for transcriptome (RNA-seq), Illumina EPIC methylomics data, plasma targeted and untargeted metabolomics data, and plasma proteomics data. The MESA multi-omics pilot biospecimen collection, molecular phenotype data production, and quality control (QC) are described in detail in the supplemental material and methods.
Here, we included data from 1,319 participants (701 women and 618 men; an average age of 60.4 in exam 1 and 69.7 in exam 5) of the MESA multi-omics pilot in the analyses. Specifically, we analyzed PBMC gene expression data for 19,699 genes from exam 1 (n = 931) and exam 5 (n = 864), and whole-blood DNA methylation (DNAm) data for 747,868 CpG sites from exam 1 (n = 900; 740,291 CpG sites passed QC) and/or exam 5 (n = 899; 747,771 CpG sites passed QC), together with genotype data from TOPMed Freeze 8.
Cell-type deconvolution
We estimated the cell-type composition of PBMC expression and whole-blood DNAm by applying two widely used methods: CIBERSORT14 and the Houseman method,15 respectively.
We employed the R implementation of CIBERSORT with default settings and utilized the LM22 gene signature matrix provided with the software. CIBERSORT was run on the TPM gene expression matrix containing the 2,648 analysis freeze samples, which included samples that passed QC and also samples that came from related individuals but were left out in interaction cis-eQTL (ieQTL) mapping. Specifically, the analysis freeze contained 972 samples from PBMC exam 1, 916 samples from PBMC exam 5, 375 samples from sorted CD19+ monocytes, and 385 samples from sorted CD4+ T cells. For duplicate gene symbols, the gene with the highest mean expression across all samples was retained, and others were removed from the input. The proportions of cell-type subcategories were summed, resulting in proportions for the following broad cell-type categories: B cells, dendritic cells, eosinophils, macrophages, mast cells, monocytes, natural killer (NK) cells, neutrophils, plasma cells, and T cells.
We used the Houseman method implemented in the meffil R package,16 along with the whole-blood reference from Reinius et al.,17,18 and used the meffil.qc function with the "blood gse35069 complete" reference applied to the DNAm IDAT files. Importantly, in meffil each sample is individually normalized to the cell-type reference dataset so that dependence between other samples and cell-type composition estimates is avoided.
For downstream analysis of cell-type estimates, for each cell type we excluded data points that were more than ±3 standard deviations (SD) from the mean.
Variability in cell-type composition, gene expression, and DNAm
To estimate the unique contribution of different traits to variation in estimated cell-type proportions, gene expression, and DNAm, we used fixed-effects linear models with no interactions as follows:
where (1) cis genetics include lead cis-molQTLs mapped in MESA (supplemental material and methods); (2) cell-type composition includes centered and scaled, moderately correlated estimated cell-type proportions (pairwise |r | < 0.6); (3) extrinsic technical and/or biological factors include batch variables, donor site, and season; and (4) intrinsic biological variables include centered and scaled genotype principal components (PCs), which are moderately correlated in MESA (pairwise |r | < 0.6), from TOPMed Freeze 8 and centered and scaled age, sex, smoking status, and educational attainment as a proxy for socioeconomic status. We applied inverse normal transformation on the response variable (cell-type proportions, gene expression levels of autosomal genes, and DNA methylation levels of 100,000 randomly selected autosomal CpG sites). Of note, to avoid ties, we added random noise from a normal distribution N(0, 10−16) to cell-type proportions before applying inverse normal transformation. More specifically, we used the type II test for computation of sums of squares (SS) to assess the significance of the main effects19 by using the car R package. To calculate the proportion of variation uniquely explained by a given trait, we used the eta-squared metric, which involves dividing the SS of each term by the total SS.
Association between estimated cell-type proportions and different traits
We measured the effect of various traits on estimated cell-type proportions by using a linear model. First, we applied inverse normal transformation on the estimated cell-type proportions to justify the assumptions of linear regression. To avoid ties, we added random noise from a normal distribution N(0, 10−16) before the transformation. Second, we leveraged the rich phenotype data available in MESA. We selected traits from 11 different categories, defined as baseline covariates (including age, sex, and genotype PCs from TOPMed Freeze 8), anthropometric characteristics, smoking habits, alcohol consumption, physical activity, atherosclerosis, blood pressure, inflammation, kidney function, lipids, and lung function. We applied a log transformation with a pseudo-count of 1 to the molecular traits. We exclude data points that were >|3| SD from the mean and scaled numeric variables by dividing by two times their SD. This transformation ensured that the resulting coefficients were comparable for both untransformed binary traits and numeric traits.20 Third, we fit linear regression with a cell-type proportion as the response variable and a trait as the explanatory variable and adjusted for age, sex, self-reported race or ethnicity, educational attainment, site, and season of sample collection. If genotype PCs were the traits of interest, then self-reported race or ethnicity was excluded from the list of covariates. To adjust for multiple testing, we applied Bonferroni correction and considered associations to be significant if p value/(# of traits groups × # cell type groups) < 0.05, where the count of cell-type groups is equal to 5, corresponding to B cells; T cells, CD4 T cells, or CD8 T cells; NK cells; monocytes; and neutrophils.
Mapping of interaction QTLs (iQTLs)
Interaction QTLs (iQTLs), which serve as a proxy for cell-type-specific QTLs, are molQTLs, whose effect is dependent on the cell-type abundance.7 Importantly, the iQTL model is not suitable for detecting molQTLs that are present only in one cell type and the molecular phenotype is expressed only in that particular cell type (Note S1). Although the total expression of a molecular phenotype is correlated with the proportion of the given cell type in the tissue sample, there is no interaction between the genotype and cell-type abundance because the molQTL effect stays constant across the range of the given cell-type abundance in the tissue sample.
We mapped iQTLs by using TensorQTL.21 Namely, we fit a linear regression model Y ∼ G + E+ G × E + C, where Y is the molecular phenotype (gene expression or DNAm; inverse normal-transformed), G is the genotype of the genetic variant with MAF > 0.01 in the MESA multi-omics pilot data, E is the environmental variable (estimated cell-type proportions, age, sex, smoking phenotype; mean-centered), G × E is the interaction effect between the genotype and environmental variable, and C represents additional covariates that correspond to 11 genotype PCs from TOPMed Freeze 8, sex, and probabilistic estimation of expression residuals (PEER22) factors to account for unobserved confounders in the molecular data. The estimation of PEER factors is described in the supplemental material and methods. We applied inverse normal transformation to the molecular phenotypes to minimize outlier effects and to satisfy the assumptions of the linear regression model. Importantly, because transformed molecular phenotypes are the response variables in the iQTL model, interaction effect size might lack biological interpretation, as previously noted for cis-regulatory effect size in eQTL studies.23 Besides the lack of biological interpretability, as a result of a non-linear transformation (such as log transformation or inverse normal transformation) of the response variable, a significant interaction effect on the transformed scale in an additive model is multiplicative on the original scale. The absence of an interaction effect on the transformed scale does not automatically imply the absence of an interaction on the original scale. However, ieQTL validation using allele-specific expression (ASE) data of eQTL heterozygotes assured that ieQTLs are not statistical artifacts of the linear model with an interaction term.7 We mean-centered environmental variables by subtracting the sample mean from every value of a variable to better interpret the main effects. Smoking phenotypes that were considered as environmental variables included current smoking (a binary variable), smoking status (a numeric variable whereby current smokers are coded as 2, former smokers as 1, and never smokers as 0), and cotinine levels (inverse normal transformed with random noise added from a normal distribution N(0, 10−16) before the transformation so that ties were avoided).
As for regular QTL mapping in the MESA multi-omics data (supplemental material and methods), iQTLs were tested for variants ±1 Mb of the gene’s transcription start site (TSS) or ±500 kb of the CpG site. To avoid potential outlier effects in cell-type iQTLs, we only included variants with MAF > 0.05 in the samples belonging to the top and bottom halves of the distribution of estimated cell-type proportions in the analyses. For age, sex, and smoking iQTLs, we applied a more stringent MAF filter to the top and bottom halves of interaction values (MAF interaction > 0.1).
To identify genes with significant ieQTLs (ieGenes) or CpG sites with significant imeQTLs (imeSites), we corrected the top nominal p values for each molecular phenotype for multiple testing at the phenotype level by using eigenMT24 and across molecular phenotypes by using the Benjamini-Hochberg procedure to control the false-discovery rate (FDR). As the significance threshold, we used FDR < 0.05 for cell-type iQTLs and FDR < 0.25 for trait iQTLs. We further combined significant iQTLs across exams by selecting the molecular phenotype-variant pair with the lower interaction p value.
We note that cell-type iQTLs could be confounded by factors that affect both the cell-type abundance and the molQTL effect size. However, correcting cell-type abundances for these factors and using residualized cell-type proportions in the iQTL model can reduce study power in most typical scenarios (Note S2).
We noticed a considerably lower number of monocyte imeQTLs than other cell-type imeQTLs, which is probably attributable to the lower variance in monocyte estimates (SD = 0.02, SD > 0.03 for other cell-type estimates). Thus, we only show data related to monocyte imeQTLs in the supplemental figures and tables.
Direction of iQTL effect
For the continuous environmental variable used for testing the interaction effect with a genotype, we grouped the direction of the iQTL effect into three categories: (1) positive (increasing)—the QTL effect size is positively correlated (increasing) with the environmental variable, (2) negative (decreasing)—the QTL effect size is negatively correlated (decreasing) with the environmental variable, and (3) uncertain. Assignment of iQTLs into these three categories was based on the estimates from the linear model. iQTLs with a nominally non-significant genotype main effect (pG > 0.05) were assigned to the “uncertain” group. For clarification, with mean-centered environmental variables, the genotype main effect corresponds to the QTL effect when the environmental variable is 0. Thus, the genotype effect crosses in the middle when the environmental variable is plotted against the molecular phenotype and coloring data points according to the genotype of the iQTL variant. iQTLs with a nominally significant genotype main effect (pG < 0.05) were assigned to the “positive” or “negative” group if the product of the genotype main effect and interaction effect ( × ) was greater or smaller than 0, respectively.
For binary environmental variables, we fitted QTL models separately for both groups. We assigned iQTLs to one of the four categories: (1) no effect in one—nominally non-significant genotype effect in one of the groups, (2) magnitude difference—nominally significant genotype effects with the same sign of the estimate in both of the groups, (3) opposite effect—nominally significant genotype effect with the opposite sign of the estimate in both of the groups, and (4) uncertain—nominally non-significant genotype effect in both of the groups.
Sentinel CpG sites for imeQTLs
Bisulfite DNA sequencing indicates that significant correlation in DNAm between CpG sites (co-methylation) has been observed for short distances up to 1 kb and decreases to baseline after 2 kb.25 To investigate the extent of co-methylation in the EPIC array, we calculated pairwise Pearson correlation coefficients between CpG sites within 500 kb on chromosome 22. We used inverse normal-transformed DNAm data from exam 5 as an example. We observed that the degree of co-methylation dropped rapidly within 500 bp and stayed on average around 0.19 after 1 kb. Of note, a similar observation of shorter distances for stronger co-methylation has been previously made on the basis of the Illumina 450K array.26 Because of this, for the imeQTLs, we defined sentinel CpG sites to be used in all the downstream analyses by keeping the CpG site-variant pair with the most significant interaction p value in a 2 kb window (±1 kb from the most associated CpG site).
Reproducibility of iQTLs
We leveraged data from two time points in MESA to estimate the reproducibility of iQTLs by treating one of the time points as the discovery set and the other as the validation set. We calculated the proportion of true positives27 (π1) on the basis of the interaction p value observed in the validation data by using the qvalue R package. This metric was used if more than 20 phenotype-variant pairs were found in the validation set (in the pi0est() function, where we set lambda = 0.5 if more than 20 phenotype-variant pairs were found or lambda = 0.85 if more than 100 phenotype-variant pairs were found). Additionally, we calculated the fraction of phenotype-variant pairs showing at least nominal significance of the interaction effect in the validation set.
Sharing of cell-type iQTLs
We estimated sharing among the same type of cell-type iQTLs by using the π1 statistic as in the reproducibility of iQTLs analysis.
To estimate sharing between cell-type ieQTLs and sentinel cell-type imeQTLs (FDR < 0.05 in exam 1 or exam 5), we focused on ieQTLs that are in LD (r2 ≥ 0.5 within 1 Mb) with imeQTLs, and vice versa. First, we calculated LD between the cell-type iQTLs by using MESA multi-omics pilot data. Second, for a given query and validation set, we calculated what proportion of variants from the query set was in LD with the variants from the validation set. For this, we set the denominator to the minimum number of variants from the query and validation sets; this number was termed the normalized overlap. Third, to estimate the significance of sharing between a cell-type ieQTL (query set) and a cell-type imeQTL (validation set), or vice versa, we asked whether the query set with positive direction is more likely to overlap with the validation set with positive direction than with the validation set with negative direction. For this, we calculated the odds ratio (OR) as the ratio of the odds of the two aforementioned events. To estimate the OR if any cell is equal to zero in the 2 × 2 table, we applied the Haldane-Anscombe correction28 by adding a fixed value of 0.5 to all cells.
Sharing of cell-type iQTLs across populations
To assess the sharing of cell-type iQTLs across self-reported race or ethnicity groups in MESA, we leveraged eQTL data from purified cell types from MESA. Namely, expression data from monocytes and T cells were available for a subset of individuals from exam 5 (n = 355 and n = 362, respectively). We chose monocytes as the cell type of interest for this analysis because of the high quality of the data. There was more variability among estimated cell-type proportions from T cell data. eQTL mapping in monocytes was done according to the standard pipeline (supplemental material and methods). Monocyte eQTLs were fine mapped via SuSiE29 to 95% credible sets of putative causal variants across all the individuals and by self-reported race or ethnicity groups. Then, we calculated the maximum LD between the monocyte ieQTLs and fine-mapped variants from the credible set across all individuals with expression data from monocytes or by self-reported race or ethnicity. For comparison between self-reported race or ethnicity groups, we focused on whether the ieGene has been fine-mapped to putative causal variants in monocytes and, if so, whether the maximum LD is above or below a specified threshold.
Replication of cell-type ieQTLs in the eQTL Catalogue
We performed replication analysis of cell-type ieQTLs in 45 eQTL datasets from purified blood cell types (with and without stimulation) from the eQTL Catalogue.30 For studies based on microarray technology, if multiple probes per gene existed, we chose the one with the lowest eQTL p value.
We estimated replication by using three different metrics: (1) the proportion of true positives27 (π1) determined with the qvalue R package if more than 20 or more than 100 gene-variant pairs were found in the replication data when lambda = 0.5 or lambda = 0.85 in the pi0est() function, respectively; (2) effect size quantified as the absolute value of the median of the genotype effect in the replication data; and (3) concordance in allelic direction, defined as the proportion of gene-variant pairs having the same directionality of the genotype effect in the replication data and genotype main effect in the cell-type iQTL data.
Functional enrichment analysis
For functional enrichment analysis, we used the registry of candidate cis-regulatory elements (cCREs) produced by the ENCODE consortium.31 The registry V2 consisted of 926,535 human cCREs covering 839 cell and tissue types. We downloaded 61 files representing unique samples with cCREs from various blood cell types, corresponding to 19 unique blood cell types. To maximize data about cCREs available per cell type, we combined data across different samples per cell type. For example, for a H3K27ac-high feature, we required that all samples with H3K27ac data available have an indication of high H3K27ac signal.
To test for the significance of overlap between cell-type iQTLs and cCREs, we used the Genomic Annotation Shifter32 (GoShifter) method. GoShifter tests for enrichment by locally shifting annotations within the boundaries of associated loci. To generate a null distribution, we repeated the shifting process 10,000 times. As input, we only used independent (sentinel) cell-type iQTLs that had FDR < 0.05 in exam 1 or exam 5 and that had positive or negative direction, and we provided a list of their LD proxies. To ensure independence of cell-type iQTLs, we performed LD pruning with PLINK33 at an r2 threshold of 0.1 in a window consisting of 1,000 variants sliding by one variant at a time. LD proxies were defined as variants with r2 ≥ 0.8 within 100 kb of the cell-type iQTLs. LD was calculated on the basis of the unrelated 1,319 individuals from the MESA multi-omics pilot.
To quantify the observed enrichment, we used the delta-overlap parameter. Delta overlap is defined as the difference between the observed proportion of loci overlapping a cCRE and the mean of the proportion of loci overlapping the cCRE under the null distribution. Thus, larger delta-overlap values show stronger enrichment. To estimate the significance of the enrichment, we calculated one-sided permutation p value as the proportion of permuted loci overlapping a cCRE is equal to or greater than the observed overlap (adding a pseudo-count of 1 to numerator and denominator). To account for multiple testing, we applied the Bonferroni correction method and accounted for the number of target cell types with the given cCRE data available, the number of cell types tested for interaction effect, and the number of groups of direction of effect. This was applied separately for each of the tested cCRE and cell-type iQTL combinations.
Colocalization analysis of cell-type iQTLs
To investigate whether cell-type iQTLs provide insights into cell-type-specific mechanisms of disease, we performed colocalization analysis with cell-type iQTLs with positive or negative direction and selected diseases and traits. We focused on seven immunological diseases (asthma,34 hay fever,34 Crohn disease,35 inflammatory bowel disease,35 rheumatoid arthritis,36 systemic lupus erythematosus,37 and ulcerative colitis35) and three metabolic traits (HDL cholesterol,38 LDL cholesterol,38 and triglycerides38). We used the GWAS summary statistics harmonized and imputed by the GTEx consortium.39 For this analysis, we used autosomal cell-type ieQTLs with FDR < 0.25 in exam 1 or exam 5 and autosomal sentinel cell-type imeQTLs with FDR < 0.05 in exam 1 or exam 5, with either a positive or negative direction of effect.
We performed colocalization analysis with coloc40 by using the coloc R package and assuming one causal variant. Coloc was run on a 400 kb region centered on each cell-type iQTL (±200 kb from the iQTL) that had at least one GWAS variant with p value <10−5 within 100 kb of the iQTL. Priors were set as follows: p1 = 10−4, p2 = 10−4, p3 = 5 × 10−6, as suggested.41 As input for cell type iQTL data, we used regression beta and the variance of beta, and for GWAS data, we used the p values. We excluded loci, where the molecular phenotype (TSS of a gene or CpG site) fell into the MHC region, due to complicated LD patterns in this region. Posterior probability for colocalization (PP4) > 0.5 was used as evidence for colocalization. For visualization of colocalized loci, we used locuszoom-like figures with LD calculated based on MESA individuals used for iQTL mapping.
Next, we tested whether we observed more colocalized loci for cell type iQTLs with a positive direction and a given disease/trait compared to height.34 Height was used as a comparison to account for the enrichment of regulatory variants among trait-associated variants. We calculated the odds ratio (OR) as the ratio of the odds of cell type iQTL colocalizing with a trait of interest to the odds of cell type iQTL colocalizing with height. To estimate the OR if any cell is equal to zero in the 2x2 table, we applied the Haldane-Anscombe correction28 by adding a fixed value of 0.5 to all cells. To test the significance of the OR, we required that at least 10 loci be tested for colocalization with the trait of interest. Bonferroni correction was applied separately for cell type ieQTLs and cell type imeQTLs to account for the number of cell type iQTL and disease/trait pairs used in enrichment testing.
Mediated moderation
We hypothesized that trait iQTLs may be mediated by G × cell-type effects. First, we assessed whether we observed enrichment of cell-type iQTL effects among our trait iQTLs (age, sex, and smoking iQTLs). For this, we evaluated the interaction effect between the genotype of the iQTL variant and cell-type proportions of the trait iQTLs. Enrichment was estimated from the inflation marker lambda (λ), which is calculated as the ratio of the median observed χ2 test statistic to the median expected χ2 test statistic under the null hypothesis.
Second, to formally assess mediation, we formulated the mediated moderation model,42 where the effect of a moderator (W, e.g., age) on the association between the independent variable (G, e.g., genotype) and dependent variable (Y, e.g., molecular phenotype) is transmitted through a mediator (M, e.g., cell-type proportion). Structural equation modeling (SEM) has been proposed for the analysis of mediated moderation.42 Because the mediated moderation effect is described by the path XW → XM → Y, we observed in a simulation analysis that we could obtain similar results by using mediation analysis techniques instead of SEM. Thus, we applied mediation analysis by using the mediation R package43 for added flexibility to account for additional covariates. More precisely, we defined the mediator and outcome models as follows:
where C is the covariate matrix, including 11 genotype PCs from TOPMed, sex, and PEER factors. G, M, and W were mean-centered for the mediation analysis.
We estimated the significance of the average causal mediation effect (ACME), average direct effect (ADE), total effect, and proportion of mediated effect by bootstrapping with k = 1,000 Monte Carlo draws and calculated 95% confidence intervals by using the bias-corrected and accelerated (BCa) method. A p value for ACME < 0.05 was used as an indicator of support for mediation.
Cell-type composition probes
To evaluate whether imeSites are likely to be associated with cell-type composition, we used the “Cell Composition Association Table” from the FlowSorted.Blood.450k R package.44 This table summarizes the association, determined by ANOVA, between each autosomal probe that is included in the Illumina 450k array and does not contain annotated SNPs and blood cell composition.
Results
Cell-type composition of blood tissue
We used two methods to characterize the cellular composition of peripheral blood mononuclear cells (PBMCs) from RNA-seq and whole blood from DNA methylation (DNAm) data in MESA—CIBERSORT14 and the Houseman method,15 respectively. These deconvolution methods leverage external purified leukocyte data to infer the proportions of white blood cells (WBCs) in heterogeneous tissue samples by modeling bulk tissue data as the sum of weighted cell-type-specific expression or DNAm signatures (Figure 1B).
Neutrophils were the most abundant cell type in whole-blood samples, as expected, but were depleted in PBMC samples, where monocytes and T cells constituted a majority of the cell populations (Figure S1B). We observed a moderate correlation between the CIBERSORT and Houseman estimates for the same cell type (Pearson correlation 0.42 < r < 0.57 in exam 5 data for B cell, NK cell, and T cell comparisons, Figure S2). Furthermore, clustering of the cell-type abundances showed good concordance between the estimated proportions from different molecular datasets and measured cell-type estimates available for a subset of individuals at the exam 5 time point (Figure 1C). However, more rare cell types, such as eosinophils, were not estimated as accurately as more abundant cell types (Figure S3). Of note, for more abundant cell types, correlation coefficients were similarly high across the different self-reported race and ethnicity groups (Figure S4). Also, there were significant differences by exam in cell-type composition, reflecting the impact of a 10-year difference between the exams (Figure S5).
Next, we sought to identify factors that account for variability in cell-type composition. Genotype PCs that reflect genome-wide genetic effects and population structure explained the highest median proportion of variance (∼7%), followed by age, sex and donor site (Figure 1D). For a more detailed quantification of the unique contributions to total variation in gene expression and DNAm, we studied four categories of factors: (1) cis genetics—lead cis-molQTLs mapped in MESA (supplemental material and methods), (2) cell-type composition—estimated cell-type proportions, (3) extrinsic technical and/or biological factors—batch variables, donor site, and season, and (4) intrinsic biological variables—genotype PCs, age, sex, smoking status, and educational attainment as a proxy for socioeconomic status. The total amount of variability explained by all considered factors varied greatly: it ranged from ∼5% to 90% per gene or CpG site (medians ranged from 40% to 20%, respectively, Figure S6). Both in gene expression and DNAm data, the largest fraction of inter-sample variation was accounted for by batch variables, estimated cell-type proportions, and lead cis-molQTLs after other variables were controlled for (Figure 1D). Although the median contribution of intrinsic biological variables was lower than that of other categories, the loci where a large proportion of variation was explained by age or smoking status highlighted known molecular biomarkers for aging (e.g., CD248,45 ELOVL2, and FHL246), or smoking (e.g., AHRR47,48 and GPR1549) (Figure S6). Discovery of age-related differences might be confounded, however, by relative changes in cell-type composition because of the impact of age on cell-type proportions.44 This issue is generalizable to any outcome of interest that correlates with cell-type composition. This highlights the importance of accounting for cell-type composition as one of the largest sources of variability in studies analyzing gene expression or DNAm.
Cell-type interaction expression QTLs and interaction methylation QTLs in blood
Variability in cell-type composition can be exploited to identify cell-type interaction QTLs,7,50,51 where the effect size of the regulatory variants increases (positive direction of effect) or decreases (negative direction of effect) depending on cell-type abundance; cell-type interaction QTLs can thus serve as proxies for cell-type-specific QTLs (Figure 2A). Applying this framework, we identified cell-type interaction cis-eQTLs (ieQTLs) for 2,130 genes (out of 19,699, ±1Mb of the TSS) and cell-type interaction cis-meQTLs (imeQTLs) for 22,141 CpG sites (out of 747,868, ± 500 kb of the CpG site) at at least one of the time points with false-discovery rate (FDR) < 0.05 across ancestries (Figure 2B). Given the correlation in DNAm between proximal CpG sites,25,26 we defined 20,099 sentinel CpG sites for imeQTLs to represent independent loci by keeping the most significant association in a 2 kb window; these were used for further analyses as described below.
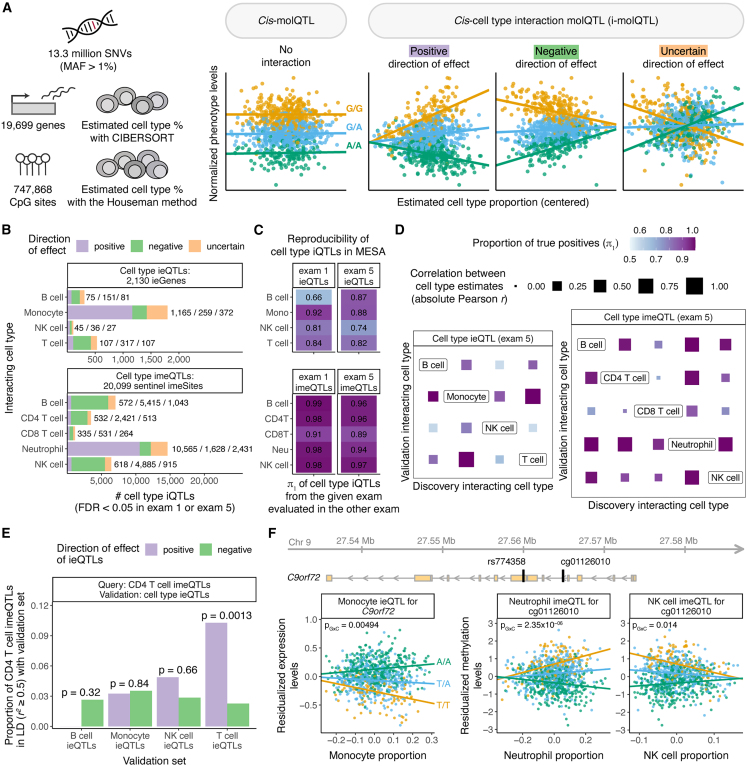
Figure 2.
Discovery of cell-type ieQTLs and imeQTLs
(A) Illustration of the approach used for mapping cell-type interaction molQTLs in MESA.
(B) Number of significant cell-type ieQTLs and imeQTLs combined across exams (FDR < 0.05 in exam 1 or exam 5 data) stratified by direction of the iQTL effect.
(C) Reproducibility of cell-type iQTLs with a positive or negative direction of effect; one of the exams was used for discovery and the other for validation, and vice versa. The proportion of true positives (π1 statistic) is used as a measure of reproducibility.
(D) Sharing among cell-type ieQTLs and cell-type imeQTLs with a positive or negative direction of effect on the basis of exam 5 data is quantified as the proportion of true positives (π1). The size of the square represents the correlation between the two estimated cell-type proportions measured via the absolute value of the Pearson correlation coefficient (r).
(E) Sharing between CD4 T cell imeQTLs (query set) and cell-type ieQTLs (validation set) combined across exams is quantified as the proportion of CD4 T cell imeQTLs that have a positive direction of effect and are in LD (r2 ≥ 0.5) with ieQTLs that have either positive or negative direction of effect from the given validation set. The p value shows the significance of the odds that CD4 T cell imeQTL with a positive direction of effect overlaps with a cell-type ieQTL with a positive direction of effect as compared to the odds that a CD4 T cell imeQTL with a positive direction of effect overlaps with a cell-type ieQTL with a negative direction of effect.
(F) Example of a cell-type iQTL (rs774358) affecting both the expression levels of a gene (C9orf72) and a nearby CpG site (cg01126010). The p value of the interaction effect from the linear model fitted with TensorQTL is shown.
Discovery of both cell-type ieQTLs and imeQTLs was dominated by the most abundant cell type, as previously observed;51 the majority of these aforementioned iQTLs had a positive direction of effect (Figure 2B). A relatively small percentage of all significant cell-type iQTLs (on average, 16.8% across cell-type iQTLs and exams) belonged to the “uncertain” group enriched for variants with lower minor-allele frequency (MAF) and higher association p values of the interaction effect, indicative of likely false-positive results7 (Figure S7). Using one of the time points for discovery and the other for validation, we observed high reproducibility rates for all cell-type iQTLs with either positive or negative direction of effect as an internal quality measure (mean π1 of 0.84 and 0.96 for cell-type ieQTLs and imeQTLs, respectively, Figure 2C). Cell-type iQTLs with uncertain direction had considerably lower nominal reproducibility rates (Figure S8) and were excluded from subsequent analyses.
The MESA cohort design allowed us to investigate population-specific effects of cell-type iQTLs. By comparing allele-frequency estimates for lead monocyte ieQTLs with positive direction across self-reported race and ethnicity groups, we observed that 0%–14% of ieQTLs did not meet the MAF > 0.01 criteria in one of the specific populations (Figure S9A). To study whether the likely causal variants are the same across populations, we leveraged the fine-mapped eQTL data by self-reported race or ethnicity from purified monocytes from MESA exam 5 (supplemental material and methods). First, we observed that 66.5%–74.8% of the monocyte ieGenes with positive direction of effect were fine mapped to likely causal eQTLs in monocytes; there was an overlap of 883 (93.1%) ieGenes fine mapped in at least two self-reported race or ethnicity groups (Figure S9B). Second, we calculated LD between the lead ieQTL and fine-mapped variants by self-reported race or ethnicity groups. Although there were considerable group-to-group differences between the fraction of ieGenes with a positive direction of effect and lead ieQTLs in strong LD (r2 > 0.5) with fine-mapped eQTLs, these differences were less pronounced when a more lenient r2 threshold was used (Figure S9C). This is consistent with the plausible scenario that cell-type ieQTLs are largely shared across major ancestral groups when differences in LD and allele frequency are taken into account, as shown for eQTLs.1
Sharing between cell-type ieQTLs and imeQTLs
Next, we sought to analyze the extent of sharing between cell-type ieQTLs and imeQTLs. We noticed that the iQTLs for highly abundant cell types—monocyte ieQTLs and neutrophil imeQTLs—with a negative direction of effect can often be found as an iQTL for another cell-type with a positive direction of effect, and vice versa (Figure S10). In general, the high degree of sharing among cell-type ieQTLs and imeQTLs reflected the magnitude of (anti)correlation between estimated cell-type proportions (Figure 2D), suggesting that cell-type iQTLs with specific genetic effects in one (or more) cell types often manifest in other (anti)correlated cell types. We also discovered indications of the same cell-type iQTL affecting both expression levels of a gene and DNA methylation levels of a nearby CpG site (Figure S11; Table S1). For example, CD4+ T cell imeQTLs with a positive direction of effect overlapped significantly more often with T cell ieQTLs with a positive direction of effect (Figure 2E, p = 0.0013 as compared to T cell ieQTLs with a negative direction of effect). Across 500 unique gene-CpG site pairs associated with the same iQTL (or lead iQTLs in strong LD, r2 > 0.5), where both the ieQTL and imeQTL effect were positive, we observed a discordant genotype main effect for the majority of the pairs (64.4%), indicative of a mostly negative correlation between gene expression and DNAm, as described before for methylation-expression associations (eQTMs).52
An example of a shared cell-type iQTL is rs774358, a variant associated with the expression of C9orf72 and with DNAm of the nearby CpG site cg01126010; the molQTL effect increases with monocyte and neutrophil abundance, respectively (Figure 2F). rs774358 is also an NK cell imeQTL with a negative direction of effect, possibly as a result of a negative correlation between the proportion of neutrophils and that of NK cells in blood. Expansion of a GGGGCC hexanucleotide repeat in C9orf72 (MIM: 105550) is one of the genetic hallmarks of amyotrophic lateral sclerosis (ALS). The expression of C9orf72 is highest in myeloid cells,53,54 indicative of myeloid-cell-specific molQTLs captured by our iQTL approach.
Cell-type specificity of cell-type iQTLs
To analyze specificity of cell-type ieQTLs by comparing their effects in purified cell types, we leveraged data from the eQTL Catalogue.30 This resource includes 45 eQTL datasets from various blood cell types with and without stimulation from the lymphocyte and myeloid lineage. We observed, in general, high replication rates for the cell-type ieQTLs with a positive direction of effect in eQTL data from the corresponding cell (sub)type (max π1 > 0.8 except for B cell ieQTLs, Figure S12); these high replication rates further manifested as higher median effect size and concordant allelic direction (Figure 3A; Table S2). For instance, monocyte ieQTLs with a positive direction of effect replicated well in eQTL data from steady-state monocytes as compared to stimulated monocytes, reflecting the need to map response QTLs to discover novel genes with molQTL specific to a cell state. We observed the highest replication rates for T cell ieQTLs with a positive direction of effect in different CD4 memory T cell subsets, most likely reflecting the shift from naive to memory T cells with age55 in the elderly study subjects from MESA. Importantly, the broad replication patterns matched the corresponding cell type for ieQTLs with a positive but not a negative direction, and replication in other cell types mirrored the sharing of ieQTLs and (anti)correlation between cell-type proportions (Figure S12).
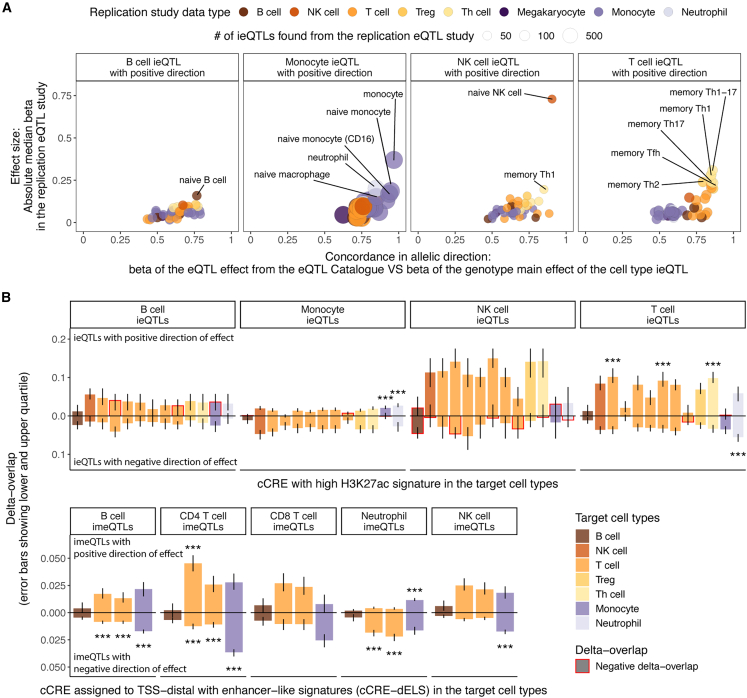
Figure 3.
Replication and functional enrichment analysis of cell-type iQTLs
(A) Replication of ieQTLs with a positive direction of effect in eQTL datasets from purified cell types from the eQTL Catalogue was based on effect size in eQTL data and allelic concordance. Highlighted are up to five datasets with absolute median effect size (beta) > 0.15 in the eQTL dataset and the proportion of QTLs with the same allelic direction >0.75 for B cell ieQTLs or >0.8 for other cell-type ieQTLs. Numerical results for all reference cell types are reported in Table S2.
(B) Functional enrichment analysis performed with GoShifter shows the delta overlap, which is the difference between the observed proportion of loci overlapping a cCRE and the null for cell-type ieQTLs (upper panel) overlapping cCRE with high H3K27ac and for cell-type imeQTLs (lower panel) overlapping cCRE-dELSs. Negative delta overlap denotes that a smaller proportion of iQTL variants overlap with a cCRE than in the null distribution. Error bars denote the lower and upper quartile of the delta overlap. ∗∗∗Significant association (adjusted p < 0.05) after correction for the number of target cell types with cCRE data, the number of cell types tested for interaction effect, and the number of groups of direction of effect. Numerical results for all reference cell types are reported in Table S3.
Cis-eQTLs and cis-meQTLs have been shown to be enriched in functional elements of the genome.1,52 We analyzed the candidate cis-regulatory elements (cCREs) from various blood cell types produced by the ENCODE project.31 After accounting for local genomic structure with GoShifter,32 we observed highly cell-type-specific enrichments of cell-type iQTLs with a positive direction of effect in distal enhancer-like signatures (cCRE-dELS) and enhancer-associated H3K27ac marks (Figures 3B and S13; Table S3), consistent with the tissue-specific nature of enhancers.56,57 As an example, monocyte ieQTLs were characterized by high H3K27ac in monocytes and neutrophils (the cells of the myeloid phagocyte system58), and T cell ieQTLs and CD4+ T cell imeQTLs were enriched in T cell subtypes. When focusing on promoter-like signatures (cCRE-PLS), we observed evidence for enrichment of the best powered cell-type iQTLs, monocyte ieQTLs and neutrophil imeQTLs with a positive direction of effect, in all the five assayed cell types (Figure S13). cCRE-PLS was also a highly shared feature, in contrast to cCRE-dELS; 64.6% of cCRE-PLSs were present in all blood cell types, whereas 60.9% of cCRE-dELSs were found only in one of the assayed blood cell types.
As exemplified by the results, cell-type iQTLs can capture cell-type-specific effects rather than overall cell-type dependence with good resolution. The interpretation of cell-type iQTLs, however, requires consideration of the direction of effect, correlation between cell types, and the quality of the deconvolution. Together, these results support mapping cell-type iQTLs as proxies for cell-type-specific QTL effects, particularly for the most abundant cell type in the tissue.
Environmental modifiers of the molQTL effect
Next, we leveraged the variation in age, sex, and three smoking phenotypes to quantify the impact of the selected higher-order phenotypes as modifiers of cis-QTL effects, i.e., to discover trait iQTLs, where the regulatory variant has a context-specific effect. Compared with cell-type iQTLs, trait iQTLs were less abundant. Using a relaxed FDR < 0.25, we identified 277 genes with age, smoking, or sex interaction eQTLs and identified 2,397 CpG sites with age, smoking, or sex interaction meQTLs (Figure 4A). Reproducibility rates between exams added confidence to the robustness of these trait iQTLs (Figure S14), given that independent replication data are scarce. As an example, we discovered an AHRR eQTL that was significant only in current smokers (Figure 4B). Hypomethylation of AHRR is one of the most replicated biomarkers for active smoking,59 and coordinated changes in both DNAm and gene expression across several tissues have been reported.48
Figure 4.
Trait iQTLs and mediated moderation
(A) Number of significant trait ieQTLs and imeQTLs in exam 1 and exam 5 (FDR < 0.25) by direction of the iQTL effect. For numeric traits, direction of the iQTL effect is defined as (1) positive—genotype effect size increases depending on the trait or (2) negative—genotype effect size decreases depending on the trait. For binary traits, the direction of the effect is defined as (1) no effect in one—nominally non-significant genotype effect in one of the groups, (2) magnitude difference—nominally significant genotype effect in both groups with the same sign of the estimate, or (3) opposite effect—nominally significant genotype effect in both groups with the opposite sign of the estimate.
(B) Example of smoking-current ieQTL for AHRR (upper plot) and age imeQTL for cg06953865 (lower plot); the p value of the interaction effect from the linear model fitted with TensorQTL is shown.
(C) Inflation of G × monocyte effect among age ieQTLs and G × neutrophil effect among age imeQTLs in exam 5 data by direction of age iQTL effect. λ is the inflation factor.
(D) Schema of the mediated-moderation approach, where the moderation effect of age on the genotype to DNAm association is mediated by changes in neutrophil proportions. The mediated-moderation effect is described by the G × age → G × neutrophil → DNAm path. p value histogram of the average causal mediation effect (ACME) of the G × neutrophil effect mediating the G × age effect on DNAm for 32 age imeQTLs with a positive or negative direction of effect.
As observed for cell-type iQTLs, a significant G × E term from the interaction model is not specific to the environment tested and might capture effects related to factors correlated with the environment. Across different traits available in MESA, age, sex and smoking are the main non-genetic factors associated with cell-type composition (Figures 1D and S15), similar to previous findings.60 Indeed, we observed a strong enrichment of age iQTLs with a positive or negative direction of effect as cell-type iQTLs when compared to age iQTLs with an uncertain direction of effect as a background (λ = 19.89 vs. 1.56 and 17.0 vs. 1.83 for the G × monocyte and G × neutrophil effect in exam 5, respectively, Figures 4C, S16A, and S16F), suggesting that some of the age iQTLs might be mediated by cell-type iQTLs. Although some sex and smoking iQTLs were very strong cell-type iQTLs, the evidence for global inflation was weaker (with median λ = 2.64 and 1.52 for G × monocyte and G × neutrophil interactions in exam 5, respectively, Figure S16). This is in line with the finding that the effects of age in DNAm were largely mediated by changes in immune cell proportions, whereas the effects of sex were typically independent of cellular composition.61 However, because our cell-type iQTL mapping is dominated by the most abundant cell type, we might be underpowered to detect global inflation of interaction with rarer immune cell types in blood.
To formally test for the effect of age iQTLs mediated by cell-type iQTLs, we adapted the concept of mediated moderation,42 where the effect of a moderator (age) on the association between genotype and molecular phenotype is transmitted through a mediator (cell-type proportion) (Figures S17A and S17B). We evaluated this hypothesis by using neutrophil proportion as the mediator for age imeQTLs in exam 5 because the observed inflation of the G × neutrophil effect was the strongest. As a basis for mediated moderation, neutrophil proportion was positively correlated with age (r = 0.14, p = 4.81 × 10−5, Figure S17C), in line with a reported continuous increase of the percentage of neutrophils with age.62 As a result, we observed support for the notion that the G × neutrophil effect mediated the G × age effect on DNAm for 43.8% (14/32) of age imeQTLs with a positive or negative direction of effect (p value of average causal mediation effect (ACME) < 0.05, Figure 4D, Table S4); on average, 15.5% of the total effect was explained by the mediator (Figure S17D). Of note, the mediation signal was driven primarily by age imeQTLs with a positive direction of effect, where 50% (13/26) showed nominal support for mediation. Interestingly, 71.4% (5 out of 7 CpG sites also present on the 450K array) of the imeSites with support for mediation have been identified as CpG sites associated with blood cell composition44 (Figure S18). As an example, rs7258891 is an age imeQTL for cg06953865 (Figure 4B), where we observed strong evidence for mediation (ACME p < 0.001). This variant has mainly been associated with various cell-count phenotypes, including neutrophil percentage,63 and the CpG site exhibits different average DNAm across cell types (p = 1.33 × 10−7), suggesting that the effect of age on the meQTL is most likely mediated by changes in cell-type composition.
Together, these results suggest that cell-type composition changes might confound trait iQTLs by mediating the moderation effect of a trait on genotype and molecular phenotype association, as previously observed for differential expression and differential methylation analysis.64,65 Thus, an apparent age iQTL effect may arise when a certain cell-type proportion varies with age and the regulatory variant has a cell-type-specific effect on a molecular phenotype. This warrants caution in interpreting G × E effects on a molecular level.
Cell-type iQTLs contribute to immune-mediated inflammatory diseases
Genetic regulatory effects can aid in elucidating the tissue specificity of heritable traits and diseases.66 Given the observed cell-type-specific nature of cell-type iQTLs with a positive direction of effect, we analyzed whether cell-type iQTLs provide insights into cell-type-specific mechanisms of diseases. We performed colocalization analysis with coloc40 of cell-type iQTLs (FDR <0.25 for ieQTLs and FDR <0.05 for imeQTLs) and a selection of immune diseases and cardiometabolic traits (Figure S19; Table S5). To account for widespread enrichment of QTLs among trait-associated variants,38 we compared the results of colocalization analysis to the number of cell-type iQTLs colocalizing with height, Our data confirmed several previously observed cell-type-specific enrichments for traits and diseases (Figure 5A): monocytes with lipid traits,18 B cells with systemic lupus erythematosus,67 and many different immune cell types, including NK cells, T cells, and B cells with inflammatory bowel disease.68 Given the varying number of cell-type iQTLs with a positive direction of effect, we had greater statistical power to detect significant associations involving cell-type imeQTLs, particularly neutrophil imeQTLs. Emerging evidence also suggests the contribution of neutrophils in the pathogenesis of autoimmune and inflammatory diseases.69,70
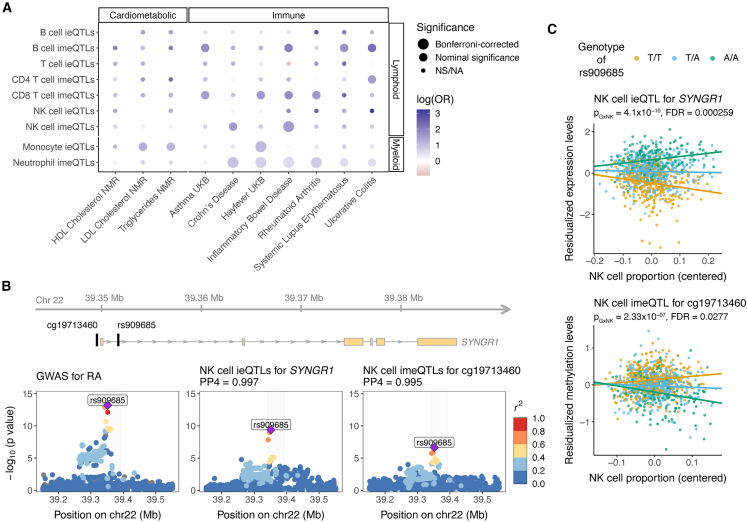
Figure 5.
Cell-type interaction QTLs and relevance for diseases
(A) Relevance of cell-type ieQTLs (FDR < 0.25) and cell-type imeQTLs (FDR < 0.05) for selected cardiometabolic and immune diseases compared to height. For each of the cell-type iQTLs, we calculated the odds ratio (OR) as the ratio of the odds that an iQTL would colocalize with a cardiometabolic or immune disease to the odds that an iQTL would colocalize with height. For testing the significance of the OR, at least 10 loci had to be tested for colocalization; otherwise, the significance is noted as NA (not available). Bonferroni correction was applied separately for cell-type ieQTLs and cell-type imeQTLs. NS: not significant.
(B) Colocalization between GWAS for RA by Okada et al.,36 NK-cell ieQTLs for SYNGR1, and imeQTLs for a nearby CpG site cg19713460, shown as regional association plots. The highlighted region is depicted at the top and shows the location of rs909685, the lead GWAS variant for RA, and the CpG site relative to SYNGR1.
(C) Association plot for the NK cell ieQTL for SYNGR1 and the NK-cell imeQTL for cg19713460. The p value of the interaction effect from the linear model fitted with TensorQTL is shown. Dots are colored on the basis of the genotype of rs909685. Data in (B) and (C) are from exam 1, where we observed the lowest interaction p values.
In addition to being useful for studying disease-specific enrichment, cell-type iQTLs can help researchers to understand the cell-type-specific mechanism of a disease-associated variant. For instance, the A allele of rs909685 (T/A), located in the intron of the synaptogyrin-1 (SYNGR1) gene, has been shown to increase the susceptibility to rheumatoid arthritis (RA) for individuals of European, Asian, and African ancestries.36,71,72 In our data, rs909685 was associated with SYNGR1 expression; the effect size increased (positive direction of effect) with NK cell and T cell proportions and decreased (negative direction of effect) with monocyte proportion (Figures 5C and S20B). rs909685 was also associated with the methylation levels of cg19713460, located in the promoter region of SYNGR1 (400 bp from the TSS); the effect size increased with NK cell proportion (Figure 5C). Of note, the A allele of rs909685 was associated with higher expression levels of SYNGR1 and lower methylation levels of cg19713460 (Figures 5C and S20B). For the cell-type iQTLs, we observed very strong evidence for colocalization with the RA GWAS signal (PP4 > 0.99) (Figures 5B and S20A). Interestingly, rs909685 falls into the cCRE that is characterized by high DNase and H3K27ac in NK cells, CD8+ T cells, and B cells (Figure S20C). Furthermore, the SYNGR1 knockdown lowered the release of pro-inflammatory cytokines or chemokines (e.g., IFN-γ, TNF, and RANTES) by activated NK cells, suggesting a functional role of SYNGR1 in NK cells.73 Together, these data suggest that rs909685 influences susceptibility to RA via NK-cell-specific action, as captured by our cell-type iQTLs integrated with functional annotation data. As a likely mechanism, a causal chain in which methylation of the promoter of SYNGR1 leads to an effect on mRNA expression and then a subsequent effect on RA risk has been proposed.74 This example highlights the usefulness of incorporating cell-type iQTLs and functional data into investigations of cell type-specific mechanisms of disease-associated variants.
Discussion
We performed interaction QTL mapping with cell-type abundance, age, sex, and smoking as the environmental factors to identify regulatory variants with plasticity in effect size rather than constant molecular effects. Although a sample size of ∼900 individuals of multi-ethnic background at two time points was sufficient for mapping cell-type iQTLs for a large number of genes and CpG sites, discovery of molQTLs interacting with higher-order physiological traits was limited. Given the unique aspects of our study design, we were able to assess the reproducibility of the iQTLs between time points to demonstrate the robustness of the results and highlight the sharing between cell-type ieQTLs and imeQTLs; this sharing was characterized mostly by a negative correlation between gene expression and DNAm and a discordant genotype main effect. Importantly, the interpretation of cell-type iQTLs depends on several factors—direction of effect, correlation between cell types within the tissue, and resolution of the cell-type deconvolution. Our results suggest that the biologically most informative results are obtained for molQTLs when the iQTL effect size is increasing (positive direction) with the most abundant cell type in the tissue.
Even though cell-type iQTLs cannot be considered cell-type-specific per se, cell-type iQTLs with a positive direction of effect replicate well in eQTL datasets from purified cell types and show enrichment in cCREs from the interacting (or similar) cell type. We demonstrated this concept in whole blood, which had the necessary cell-type-specific eQTL replication data. Our results show promise for interaction QTL approaches for identification of cell-type-specific QTLs in other tissues where single-cell or cell-type-specific data are not available or easily acquired. Moreover, cell-type iQTLs combined with functional annotations of the genome can help prioritize cell types for functional follow-up studies.
Importantly, iQTLs present molQTLs, where the molQTL effect is dependent on the environmental variable. More specifically, for estimation of iQTLs, the transformed gene expression levels are modeled as a function of a genetic variant, environmental variable, and an interaction between the two variables. This approach is not suitable for detecting molQTLs that are present only in one cell type. For example, negative binomial regression that allows modeling the dependence of RNA-seq transcripts on cell-type proportions and the genotype of a genetic variant would be an alternative approach that might overcome this. In addition to using molQTL data from purified cell types, the forthcoming population-scale single-cell QTL studies, such as that undertaken by the single-cell eQTLGen consortium,75 would allow additional validation of iQTLs.
molQTLs with G × E interactions at the molecular level hold promise for guiding the discovery of G × E interactions in complex diseases.76,77,78,79,80,81 These loci might mark the genetic component of inter-individual variation in response to different environments or physiological states, including disease, thus contributing to phenotypic variation in humans. Our results with age imeQTLs, however, suggest that cell-type composition changes could partly mediate the moderation effect of age. Similar observations have previously been made for sex-biased cis-eQTLs,82 yet the confounding effect that cell-type composition has on molQTL effect-size variation has not been appreciated to the same extent as in differential expression and methylation studies, particularly in epigenome-wide association studies.44,65
On the basis of our results, we propose that mediation by cell-type composition is the primary starting hypothesis for molQTLs with G × E effects, and this should be explicitly ruled out before other molecular moderation mechanisms are postulated. We further hypothesize that in cases where trait iQTL and GWAS signal colocalize, only molQTLs with G × E not mediated by cell types would have a G × E interaction at the GWAS level—whereas molQTLs with support for mediation most likely are subject to confounding. Future studies with larger sample sizes will be needed for the proper evaluation of this hypothesis.
Overall, the integration of genomic data with functional multi-omic data in large and diverse longitudinal cohorts offers an opportunity to map genetic effects on molecular traits and to study its complex interplay with other environmental factors. Our study shows the value of mapping interaction QTLs as a feasible computational approach that can provide insights into the context specificity of regulatory effects.
Data and code availability
MESA WGS data are part of the NHLBI Trans-Omics for Precision Medicine (TOPMed) Whole-Genome Sequencing Program and are available through dbGaP (dbGaP: phs001416.v1.p1). MESA molecular data are also part of this program (dbGaP: phs001416.v3.p1). Comprehensive phenotypic data for MESA study participants are available through dbGaP as well (dbGaP: phs000209.v13.p3). The full summary statistics of cell-type interaction molQTLs are available on request to the corresponding authors without restrictions (∼11 GB per cell-type ieQTL and ∼200 GB per cell-type imeQTL, including summary statistics for both exams). The significant cell-type and trait interaction molQTLs are available at Figshare (https://figshare.com/projects/Interaction_molecular_QTL_mapping_discovers_cellular_and_environmental_modifiers_of_genetic_regulatory_effects/184462). Code for mapping iQTLs with TensorQTL is available at Github (https://github.com/broadinstitute/tensorqtl).
Acknowledgments
We thank all members of the Lappalainen laboratory for valuable discussions and support. We also thank Paul J. Hoffman, Grant T. Hiura, Kristina L. Buschur, Sailalitha Bollepalli, and Inga-Maria Launonen for their input and advice on earlier versions of this manuscript. S.K., R.G.B., and T.L. were supported by a grant from the National Heart, Lung, and Blood Institute (NHLBI) of the National Institutes of Health (NIH) (grant R01HL142028). S.K.H. is supported by Marie-Skłodowska Curie fellowship H2020 grant 706636, Helmholtz Young Investigator grant VH-NG-1620, and Deutsche Forschungsgemeinschaft Emmy Noether Programme grant KI 2091/2-1. B.C.B. is supported by the NIH’s National Human Genome Research Institute (NHGRI) (grant K99HG012373). T.L. is supported by the National Institute of Mental Healthof the NIH (grant R01MH106842), NHGRI grant UM1HG008901,and the NIH’s National Institute of General Medical Sciences (grant R01GM122924). Whole-genome sequencing (WGS) for the Trans-Omics in Precision Medicine (TOPMed) program was supported by the NHLBI. The MESA projects are conducted and supported by NHLBI in collaboration with MESA investigators. Support for the Multi-Ethnic Study of Atherosclerosis (MESA) projects are conducted and supported by NHLBI in collaboration with MESA investigators. Additional acknowledgments and funding information are available in the supplemental acknowledgments.
Author contributions
S.K. and T.L. designed the study. S.K. performed the analyses. F.A., S.K.H., B.C.B., and D.C.N. contributed to the analyses of the data. J.I.R., S.S.R., R.P.T., P.D., Y.L., K.D.T., W.C.J., D.V.D.B., S.G., N.G., J.D.S., and T.W.B. were involved in the acquisition and processing of data. T.L., R.G.B., S.S.R., A.M., and K.G.A. supervised the work. S.K. and T.L. wrote the first draft. F.A., S.S.R., A.M., S.K.H., and B.C.B. contributed to the editing of the manuscript. All authors approved the final version of the manuscript.
Declaration of interests
F.A. is an employee of Illumina, Inc. and an inventor on a patent application related to TensorQTL. T.L. advises Variant Bio, Goldfinch Bio, GlaxoSmithKline, and Pfizer and has equity in Variant Bio.
Published: January 4, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2023.11.013.
Contributor Information
Silva Kasela, Email: skasela@nygenome.org.
Tuuli Lappalainen, Email: tlappalainen@nygenome.org.
Web resources
CIBERSORT, https://rdrr.io/github/IOBR/IOBR/src/R/CIBERSORT.R
TensorQTL, https://github.com/broadinstitute/tensorqtl
GoShifter, https://github.com/immunogenomics/goshifter
mediation, https://github.com/kosukeimai/mediation
FlowSorted.Blood.450k, https://bioconductor.org/packages/FlowSorted.Blood.450k/
Supplemental information
References
- 1.GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Võsa U., Claringbould A., Westra H.-J., Bonder M.J., Deelen P., Zeng B., Kirsten H., Saha A., Kreuzhuber R., Yazar S., et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat. Genet. 2021;53:1300–1310. doi: 10.1038/s41588-021-00913-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Min J.L., Hemani G., Hannon E., Dekkers K.F., Castillo-Fernandez J., Luijk R., Carnero-Montoro E., Lawson D.J., Burrows K., Suderman M., et al. Genomic and phenotypic insights from an atlas of genetic effects on DNA methylation. Nat. Genet. 2021;53:1311–1321. doi: 10.1038/s41588-021-00923-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Umans B.D., Battle A., Gilad Y. Where Are the Disease-Associated eQTLs? Trends Genet. 2021;37:109–124. doi: 10.1016/j.tig.2020.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Westra H.-J., Arends D., Esko T., Peters M.J., Schurmann C., Schramm K., Kettunen J., Yaghootkar H., Fairfax B.P., Andiappan A.K., et al. Cell Specific eQTL Analysis without Sorting Cells. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aguirre-Gamboa R., de Klein N., di Tommaso J., Claringbould A., van der Wijst M.G., de Vries D., Brugge H., Oelen R., Võsa U., Zorro M.M., et al. Deconvolution of bulk blood eQTL effects into immune cell subpopulations. BMC Bioinf. 2020;21:243. doi: 10.1186/s12859-020-03576-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim-Hellmuth S., Aguet F., Oliva M., Muñoz-Aguirre M., Kasela S., Wucher V., Castel S.E., Hamel A.R., Viñuela A., Roberts A.L., et al. Cell type-specific genetic regulation of gene expression across human tissues. Science. 2020;369 doi: 10.1126/science.aaz8528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hunter D.J. Gene-environment interactions in human diseases. Nat. Rev. Genet. 2005;6:287–298. doi: 10.1038/nrg1578. [DOI] [PubMed] [Google Scholar]
- 9.McAllister K., Mechanic L.E., Amos C., Aschard H., Blair I.A., Chatterjee N., Conti D., Gauderman W.J., Hsu L., Hutter C.M., et al. Current Challenges and New Opportunities for Gene-Environment Interaction Studies of Complex Diseases. Am. J. Epidemiol. 2017;186:753–761. doi: 10.1093/aje/kwx227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han S.S., Chatterjee N. Review of Statistical Methods for Gene-Environment Interaction Analysis. Curr. Epidemiol. Rep. 2018;5:39–45. [Google Scholar]
- 11.Wang K., Basu M., Malin J., Hannenhalli S. A transcription-centric model of SNP-age interaction. PLoS Genet. 2021;17 doi: 10.1371/journal.pgen.1009427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bild D.E., Bluemke D.A., Burke G.L., Detrano R., Diez Roux A.V., Folsom A.R., Greenland P., Jacob D.R., Kronmal R., Liu K., et al. Multi-Ethnic Study of Atherosclerosis: objectives and design. Am. J. Epidemiol. 2002;156:871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 13.Taliun D., Harris D.N., Kessler M.D., Carlson J., Szpiech Z.A., Torres R., Taliun S.A.G., Corvelo A., Gogarten S.M., Kang H.M., et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–299. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., Hoang C.D., Diehn M., Alizadeh A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houseman E.A., Accomando W.P., Koestler D.C., Christensen B.C., Marsit C.J., Nelson H.H., Wiencke J.K., Kelsey K.T. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinf. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Min J.L., Hemani G., Davey Smith G., Relton C., Suderman M. Meffil: efficient normalization and analysis of very large DNA methylation datasets. Bioinforma. Oxf. Engl. 2018;34:3983–3989. doi: 10.1093/bioinformatics/bty476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reinius L.E., Acevedo N., Joerink M., Pershagen G., Dahlén S.E., Greco D., Söderhäll C., Scheynius A., Kere J. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One. 2012;7 doi: 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raj T., Rothamel K., Mostafavi S., Ye C., Lee M.N., Replogle J.M., Feng T., Lee M., Asinovski N., Frohlich I., et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science. 2014;344:519–523. doi: 10.1126/science.1249547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langsrud Ø. ANOVA for unbalanced data: Use Type II instead of Type III sums of squares. Stat. Comput. 2003;13:163–167. [Google Scholar]
- 20.Gelman A. Scaling regression inputs by dividing by two standard deviations. Stat. Med. 2008;27:2865–2873. doi: 10.1002/sim.3107. [DOI] [PubMed] [Google Scholar]
- 21.Taylor-Weiner A., Aguet F., Haradhvala N.J., Gosai S., Anand S., Kim J., Ardlie K., Van Allen E.M., Getz G. Scaling computational genomics to millions of individuals with GPUs. Genome Biol. 2019;20:228. doi: 10.1186/s13059-019-1836-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stegle O., Parts L., Durbin R., Winn J. A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLoS Comput. Biol. 2010;6 doi: 10.1371/journal.pcbi.1000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohammadi P., Castel S.E., Brown A.A., Lappalainen T. Quantifying the regulatory effect size of cis-acting genetic variation using allelic fold change. Genome Res. 2017;27:1872–1884. doi: 10.1101/gr.216747.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis J.R., Fresard L., Knowles D.A., Pala M., Bustamante C.D., Battle A., Montgomery S.B. An Efficient Multiple-Testing Adjustment for eQTL Studies that Accounts for Linkage Disequilibrium between Variants. Am. J. Hum. Genet. 2016;98:216–224. doi: 10.1016/j.ajhg.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eckhardt F., Lewin J., Cortese R., Rakyan V.K., Attwood J., Burger M., Burton J., Cox T.V., Davies R., Down T.A., et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat. Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang W., Spector T.D., Deloukas P., Bell J.T., Engelhardt B.E. Predicting genome-wide DNA methylation using methylation marks, genomic position, and DNA regulatory elements. Genome Biol. 2015;16:14. doi: 10.1186/s13059-015-0581-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawson R. Small Sample Confidence Intervals for the Odds Ratio. Commun. Stat. - Simul. Comput. 2004;33:1095–1113. [Google Scholar]
- 29.Wang G., Sarkar A., Carbonetto P., Stephens M. A simple new approach to variable selection in regression, with application to genetic fine mapping. J. R. Stat. Soc. Series B Stat. Methodol. 2020;82:1273–1300. doi: 10.1111/rssb.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kerimov N., Hayhurst J.D., Peikova K., Manning J.R., Walter P., Kolberg L., Samoviča M., Sakthivel M.P., Kuzmin I., Trevanion S.J., et al. A compendium of uniformly processed human gene expression and splicing quantitative trait loci. Nat. Genet. 2021;53:1290–1299. doi: 10.1038/s41588-021-00924-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.ENCODE Project Consortium. Moore J.E., Purcaro M.J., Pratt H.E., Epstein C.B., Shoresh N., Adrian J., Kawli T., Davis C.A., Dobin A., et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature. 2020;583:699–710. doi: 10.1038/s41586-020-2493-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trynka G., Westra H.-J., Slowikowski K., Hu X., Xu H., Stranger B.E., Klein R.J., Han B., Raychaudhuri S. Disentangling the Effects of Colocalizing Genomic Annotations to Functionally Prioritize Non-coding Variants within Complex-Trait Loci. Am. J. Hum. Genet. 2015;97:139–152. doi: 10.1016/j.ajhg.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J., et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu J.Z., van Sommeren S., Huang H., Ng S.C., Alberts R., Takahashi A., Ripke S., Lee J.C., Jostins L., Shah T., et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okada Y., Wu D., Trynka G., Raj T., Terao C., Ikari K., Kochi Y., Ohmura K., Suzuki A., Yoshida S., et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bentham J., Morris D.L., Graham D.S.C., Pinder C.L., Tombleson P., Behrens T.W., Martín J., Fairfax B.P., Knight J.C., Chen L., et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat. Genet. 2015;47:1457–1464. doi: 10.1038/ng.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kettunen J., Demirkan A., Würtz P., Draisma H.H.M., Haller T., Rawal R., Vaarhorst A., Kangas A.J., Lyytikäinen L.P., Pirinen M., et al. Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat. Commun. 2016;7 doi: 10.1038/ncomms11122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barbeira A.N., Bonazzola R., Gamazon E.R., Liang Y., Park Y., Kim-Hellmuth S., Wang G., Jiang Z., Zhou D., Hormozdiari F., et al. Exploiting the GTEx resources to decipher the mechanisms at GWAS loci. Genome Biol. 2021;22:49. doi: 10.1186/s13059-020-02252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giambartolomei C., Vukcevic D., Schadt E.E., Franke L., Hingorani A.D., Wallace C., Plagnol V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wallace C. Eliciting priors and relaxing the single causal variant assumption in colocalisation analyses. PLoS Genet. 2020;16 doi: 10.1371/journal.pgen.1008720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwan J.L.Y., Chan W. Variable system: An alternative approach for the analysis of mediated moderation. Psychol. Methods. 2018;23:262–277. doi: 10.1037/met0000160. [DOI] [PubMed] [Google Scholar]
- 43.Tingley D., Yamamoto T., Hirose K., Keele L., Imai K. mediation: R Package for Causal Mediation Analysis. J. Stat. Softw. 2014;59:1–38. [Google Scholar]
- 44.Jaffe A.E., Irizarry R.A. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 2014;15:R31. doi: 10.1186/gb-2014-15-2-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harries L.W., Hernandez D., Henley W., Wood A.R., Holly A.C., Bradley-Smith R.M., Yaghootkar H., Dutta A., Murray A., Frayling T.M., et al. Human aging is characterized by focused changes in gene expression and deregulation of alternative splicing. Aging Cell. 2011;10:868–878. doi: 10.1111/j.1474-9726.2011.00726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garagnani P., Bacalini M.G., Pirazzini C., Gori D., Giuliani C., Mari D., Di Blasio A.M., Gentilini D., Vitale G., Collino S., et al. Methylation of ELOVL2 gene as a new epigenetic marker of age. Aging Cell. 2012;11:1132–1134. doi: 10.1111/acel.12005. [DOI] [PubMed] [Google Scholar]
- 47.Bojesen S.E., Timpson N., Relton C., Davey Smith G., Nordestgaard B.G. AHRR (cg05575921) hypomethylation marks smoking behaviour, morbidity and mortality. Thorax. 2017;72:646–653. doi: 10.1136/thoraxjnl-2016-208789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsai P.-C., Glastonbury C.A., Eliot M.N., Bollepalli S., Yet I., Castillo-Fernandez J.E., Carnero-Montoro E., Hardiman T., Martin T.C., Vickers A., et al. Smoking induces coordinated DNA methylation and gene expression changes in adipose tissue with consequences for metabolic health. Clin. Epigenetics. 2018;10:126. doi: 10.1186/s13148-018-0558-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huan T., Joehanes R., Schurmann C., Schramm K., Pilling L.C., Peters M.J., Mägi R., DeMeo D., O’Connor G.T., Ferrucci L., et al. A whole-blood transcriptome meta-analysis identifies gene expression signatures of cigarette smoking. Hum. Mol. Genet. 2016;25:4611–4623. doi: 10.1093/hmg/ddw288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hawe J.S., Wilson R., Schmid K.T., Zhou L., Lakshmanan L.N., Lehne B.C., Kühnel B., Scott W.R., Wielscher M., Yew Y.W., et al. Genetic variation influencing DNA methylation provides insights into molecular mechanisms regulating genomic function. Nat. Genet. 2022;54:18–29. doi: 10.1038/s41588-021-00969-x. [DOI] [PubMed] [Google Scholar]
- 51.de Klein N., Tsai E.A., Vochteloo M., Baird D., Huang Y., Chen C.-Y., van Dam S., Oelen R., Deelen P., Bakker O.B., et al. Brain expression quantitative trait locus and network analyses reveal downstream effects and putative drivers for brain-related diseases. Nat. Genet. 2023;55:377–388. doi: 10.1038/s41588-023-01300-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonder M.J., Luijk R., Zhernakova D.V., Moed M., Deelen P., Vermaat M., van Iterson M., van Dijk F., van Galen M., Bot J., et al. Disease variants alter transcription factor levels and methylation of their binding sites. Nat. Genet. 2017;49:131–138. doi: 10.1038/ng.3721. [DOI] [PubMed] [Google Scholar]
- 53.Rizzu P., Blauwendraat C., Heetveld S., Lynes E.M., Castillo-Lizardo M., Dhingra A., Pyz E., Hobert M., Synofzik M., Simón-Sánchez J., et al. C9orf72 is differentially expressed in the central nervous system and myeloid cells and consistently reduced in C9orf72, MAPT and GRN mutation carriers. Acta Neuropathol. Commun. 2016;4:37. doi: 10.1186/s40478-016-0306-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O’Rourke J.G., Bogdanik L., Yáñez A., Lall D., Wolf A.J., Muhammad A.K.M.G., Ho R., Carmona S., Vit J.P., Zarrow J., et al. C9orf72 is required for proper macrophage and microglial function in mice. Science. 2016;351:1324–1329. doi: 10.1126/science.aaf1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Surh C.D., Sprent J. Homeostasis of naive and memory T cells. Immunity. 2008;29:848–862. doi: 10.1016/j.immuni.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 56.Roadmap Epigenomics Consortium. Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hill M.S., Vande Zande P., Wittkopp P.J. Molecular and evolutionary processes generating variation in gene expression. Nat. Rev. Genet. 2021;22:203–215. doi: 10.1038/s41576-020-00304-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Silva M.T., Correia-Neves M. Neutrophils and macrophages: the main partners of phagocyte cell systems. Front. Immunol. 2012;3:174. doi: 10.3389/fimmu.2012.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao X., Jia M., Zhang Y., Breitling L.P., Brenner H. DNA methylation changes of whole blood cells in response to active smoking exposure in adults: a systematic review of DNA methylation studies. Clin. Epigenetics. 2015;7:113. doi: 10.1186/s13148-015-0148-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Patin E., Hasan M., Bergstedt J., Rouilly V., Libri V., Urrutia A., Alanio C., Scepanovic P., Hammer C., Jönsson F., et al. Natural variation in the parameters of innate immune cells is preferentially driven by genetic factors. Nat. Immunol. 2018;19:302–314. doi: 10.1038/s41590-018-0049-7. [DOI] [PubMed] [Google Scholar]
- 61.Bergstedt J., Azzou S.A.K., Tsuo K., Jaquaniello A., Urrutia A., Rotival M., Lin D.T.S., MacIsaac J.L., Kobor M.S., Albert M.L., et al. The immune factors driving DNA methylation variation in human blood. Nat. Commun. 2022;13:5895. doi: 10.1038/s41467-022-33511-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Valiathan R., Ashman M., Asthana D. Effects of Ageing on the Immune System: Infants to Elderly. Scand. J. Immunol. 2016;83:255–266. doi: 10.1111/sji.12413. [DOI] [PubMed] [Google Scholar]
- 63.Vuckovic D., Bao E.L., Akbari P., Lareau C.A., Mousas A., Jiang T., Chen M.-H., Raffield L.M., Tardaguila M., Huffman J.E., et al. The Polygenic and Monogenic Basis of Blood Traits and Diseases. Cell. 2020;182:1214–1231.e11. doi: 10.1016/j.cell.2020.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shen-Orr S.S., Gaujoux R. Computational deconvolution: extracting cell type-specific information from heterogeneous samples. Curr. Opin. Immunol. 2013;25:571–578. doi: 10.1016/j.coi.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Teschendorff A.E., Relton C.L. Statistical and integrative system-level analysis of DNA methylation data. Nat. Rev. Genet. 2018;19:129–147. doi: 10.1038/nrg.2017.86. [DOI] [PubMed] [Google Scholar]
- 66.Hekselman I., Yeger-Lotem E. Mechanisms of tissue and cell-type specificity in heritable traits and diseases. Nat. Rev. Genet. 2020;21:137–150. doi: 10.1038/s41576-019-0200-9. [DOI] [PubMed] [Google Scholar]
- 67.Soskic B., Cano-Gamez E., Smyth D.J., Rowan W.C., Nakic N., Esparza-Gordillo J., Bossini-Castillo L., Tough D.F., Larminie C.G.C., Bronson P.G., et al. Chromatin activity at GWAS loci identifies T cell states driving complex immune diseases. Nat. Genet. 2019;51:1486–1493. doi: 10.1038/s41588-019-0493-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Finucane H.K., Reshef Y.A., Anttila V., Slowikowski K., Gusev A., Byrnes A., Gazal S., Loh P.-R., Lareau C., Shoresh N., et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet. 2018;50:621–629. doi: 10.1038/s41588-018-0081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mantovani A., Cassatella M.A., Costantini C., Jaillon S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat. Rev. Immunol. 2011;11:519–531. doi: 10.1038/nri3024. [DOI] [PubMed] [Google Scholar]
- 70.Németh T., Sperandio M., Mócsai A. Neutrophils as emerging therapeutic targets. Nat. Rev. Drug Discov. 2020;19:253–275. doi: 10.1038/s41573-019-0054-z. [DOI] [PubMed] [Google Scholar]
- 71.Kim K., Bang S.-Y., Lee H.-S., Cho S.-K., Choi C.-B., Sung Y.-K., Kim T.-H., Jun J.-B., Yoo D.H., Kang Y.M., et al. High-density genotyping of immune loci in Koreans and Europeans identifies eight new rheumatoid arthritis risk loci. Ann. Rheum. Dis. 2015;74:e13. doi: 10.1136/annrheumdis-2013-204749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Laufer V.A., Tiwari H.K., Reynolds R.J., Danila M.I., Wang J., Edberg J.C., Kimberly R.P., Kottyan L.C., Harley J.B., Mikuls T.R., et al. Genetic influences on susceptibility to rheumatoid arthritis in African-Americans. Hum. Mol. Genet. 2019;28:858–874. doi: 10.1093/hmg/ddy395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schmiedel B.J., Singh D., Madrigal A., Valdovino-Gonzalez A.G., White B.M., Zapardiel-Gonzalo J., Ha B., Altay G., Greenbaum J.A., McVicker G., et al. Impact of Genetic Polymorphisms on Human Immune Cell Gene Expression. Cell. 2018;175:1701–1715.e16. doi: 10.1016/j.cell.2018.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mo X.-B., Zhang Y.-H., Lei S.-F. Integrative analysis identifies potential causal methylation-mRNA regulation chains for rheumatoid arthritis. Mol. Immunol. 2021;131:89–96. doi: 10.1016/j.molimm.2020.12.021. [DOI] [PubMed] [Google Scholar]
- 75.van der Wijst M., de Vries D.H., Groot H.E., Trynka G., Hon C.C., Bonder M.J., Stegle O., Nawijn M.C., Idaghdour Y., van der Harst P., et al. The single-cell eQTLGen consortium. Elife. 2020;9 doi: 10.7554/eLife.52155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yao C., Joehanes R., Johnson A.D., Huan T., Esko T., Ying S., Freedman J.E., Murabito J., Lunetta K.L., Metspalu A., et al. Sex- and age-interacting eQTLs in human complex diseases. Hum. Mol. Genet. 2014;23:1947–1956. doi: 10.1093/hmg/ddt582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Knowles D.A., Davis J.R., Edgington H., Raj A., Favé M.J., Zhu X., Potash J.B., Weissman M.M., Shi J., Levinson D.F., et al. Allele-specific expression reveals interactions between genetic variation and environment. Nat. Methods. 2017;14:699–702. doi: 10.1038/nmeth.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim-Hellmuth S., Bechheim M., Pütz B., Mohammadi P., Nédélec Y., Giangreco N., Becker J., Kaiser V., Fricker N., Beier E., et al. Genetic regulatory effects modified by immune activation contribute to autoimmune disease associations. Nat. Commun. 2017;8:266. doi: 10.1038/s41467-017-00366-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Findley A.S., Monziani A., Richards A.L., Rhodes K., Ward M.C., Kalita C.A., Alazizi A., Pazokitoroudi A., Sankararaman S., Wen X., et al. Functional dynamic genetic effects on gene regulation are specific to particular cell types and environmental conditions. Elife. 2021;10 doi: 10.7554/eLife.67077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Meng W., Zhu Z., Jiang X., Too C.L., Uebe S., Jagodic M., Kockum I., Murad S., Ferrucci L., Alfredsson L., et al. DNA methylation mediates genotype and smoking interaction in the development of anti-citrullinated peptide antibody-positive rheumatoid arthritis. Arthritis Res. Ther. 2017;19:71. doi: 10.1186/s13075-017-1276-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Teh A.L., Pan H., Chen L., Ong M.-L., Dogra S., Wong J., MacIsaac J.L., Mah S.M., McEwen L.M., Saw S.-M., et al. The effect of genotype and in utero environment on interindividual variation in neonate DNA methylomes. Genome Res. 2014;24:1064–1074. doi: 10.1101/gr.171439.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Oliva M., Muñoz-Aguirre M., Kim-Hellmuth S., Wucher V., Gewirtz A.D.H., Cotter D.J., Parsana P., Kasela S., Balliu B., Viñuela A., et al. The impact of sex on gene expression across human tissues. Science. 2020;369 doi: 10.1126/science.aba3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MESA WGS data are part of the NHLBI Trans-Omics for Precision Medicine (TOPMed) Whole-Genome Sequencing Program and are available through dbGaP (dbGaP: phs001416.v1.p1). MESA molecular data are also part of this program (dbGaP: phs001416.v3.p1). Comprehensive phenotypic data for MESA study participants are available through dbGaP as well (dbGaP: phs000209.v13.p3). The full summary statistics of cell-type interaction molQTLs are available on request to the corresponding authors without restrictions (∼11 GB per cell-type ieQTL and ∼200 GB per cell-type imeQTL, including summary statistics for both exams). The significant cell-type and trait interaction molQTLs are available at Figshare (https://figshare.com/projects/Interaction_molecular_QTL_mapping_discovers_cellular_and_environmental_modifiers_of_genetic_regulatory_effects/184462). Code for mapping iQTLs with TensorQTL is available at Github (https://github.com/broadinstitute/tensorqtl).