Abstract
Pathogen recognition triggers energy-intensive defense systems. Although successful defense should depend on energy availability, how metabolic information is communicated to defense remains unclear. We show that sugar, especially glucose-6-phosphate (G6P), is critical in coordinating defense in Arabidopsis. Under sugar-sufficient conditions, phosphorylation levels of calcium-dependent protein kinase 5 (CPK5) are elevated by G6P-mediated suppression of protein phosphatases, enhancing defense responses before pathogen invasion. Subsequently, recognition of bacterial flagellin activates sugar transporters, leading to increased cellular G6P, which elicits CPK5-independent signaling promoting synthesis of the phytohormone salicylic acid (SA) for antibacterial defense. In contrast, while perception of fungal chitin does not promote sugar influx or SA accumulation, chitin-induced synthesis of the antifungal compound camalexin requires basal sugar influx activity. By monitoring sugar levels, plants determine defense levels and execute appropriate outputs against bacterial and fungal pathogens. Together, our findings provide a comprehensive view of the roles of sugar in defense.
In plants, the types and amplitudes of defense outputs against bacterial and fungal pathogens depend on cellular sugars.
INTRODUCTION
In plants, perception of pathogen-associated molecular patterns (PAMPs) on the plasma membrane triggers a cascade of defense responses, including extensive transcriptional reprogramming and de novo synthesis of metabolites such as phytoalexins and phytohormones (1). Despite the energy-intensive nature of defense signaling, the molecular link between defense signaling and cellular metabolic status remains largely unknown. For example, defense signaling should be suppressed under low nutritional conditions and enhanced under sufficient nutritional conditions. Of primary metabolites, sugar is known to affect various signal pathways not only as an energy precursor and a carbon skeleton but also as a signal molecule (2). Sugar has been proposed to stimulate plant defense, by a mechanism whose molecular basis has remained unclear for more than two decades (3). In addition, exogenous application of sugars to plants is reported to enhance defense responses against bacterial and fungal pathogens (4, 5). Glucose (Glc) is recognized by hexokinase 1 (HXK1) (6), a dual-function enzyme with both catalytic and sensory activity for Glc, in the model plant Arabidopsis thaliana. Sensing of Glc via HXK1 triggers down-regulation of photosynthetic genes (6). On the other hand, Glc catalysis mediated by HXK1 induces defense signaling. Overexpression of not only Arabidopsis HXK1 but also yeast HXK2 elevated expression of defense-responsive genes (7). Furthermore, cell death induced by loss of the myo-inositol 1-phosphatase synthase 1 gene was attenuated in the absence of HXK1’s enzymatic activity (8). Therefore, a Glc-derived metabolite(s) catalyzed by HXK, rather than Glc per se, is thought to stimulate defense signaling (3).
Cellular Ca2+ concentration fluctuates in response to various environmental cues, including pathogen recognition. In response to PAMPs, calcium-dependent protein kinases (CPKs/CDPKs) were rapidly activated by an increase in cytosolic Ca2+, regulating the expression of defense-related genes. Among Arabidopsis CPKs, CPK4/5/6/11, belonging to subgroup I, are especially involved in defense signaling (9). For instance, during fungal invasion, CPK5 and CPK6 mediate the synthesis of the antifungal compound camalexin by phosphorylating the transcription factor WRKY33 (10, 11). In addition, CPK5 interacts with the atypical nucleotide binding domain and leucine-rich repeat protein TN2, activating TN2-mediated defense responses when EXO70B1, a subunit of the exocyst complex, is perturbed (12).
In this study, we focused on how cellular sugar conditions are communicated to plant defense signaling. We showed that Glc-6-phosphate (G6P), not Glc, affects multiple signal branches that orchestrate defense outputs, using genetic and pharmacological analyses. CPK5 stimulates the expression of sugar-responsive genes, and its activity is suppressed by clade A protein phosphatase 2Cs (PP2Cs). We found that G6P inhibits their phosphatase activities, leading to de-suppression of CPK5 activity. Therefore, defense responses are enhanced in advance of subsequent pathogen recognition under sugar-sufficient conditions. In addition, after perception of bacterial and fungal PAMPs, sugar influx transporters regulate cellular G6P amounts, which elicits CPK5-independent signal pathways that cooperatively or antagonistically act on CPK5-mediated signaling for executing appropriate defense outputs against bacterial and fungal pathogens. Thus, this work provides a comprehensive view of the roles of sugar in coordinating plant defense signaling.
RESULTS
Sugar activates expression of defense-related genes via HXK-mediated Glc phosphorylation
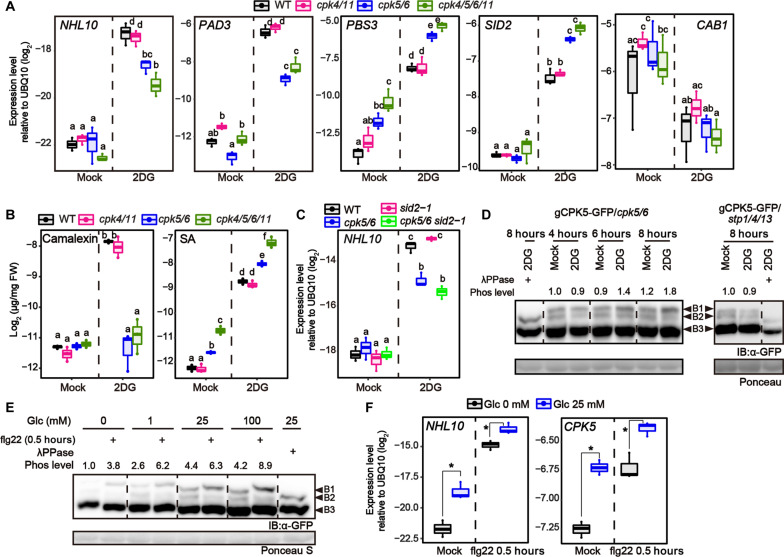
First, to obtain an overview of how sugar stimulates defense signaling in plants, we studied the effect of sucrose (Suc) application on gene expression by RNA sequencing (RNA-seq) analysis. We found that application of Suc to Arabidopsis seedlings induced defense-related genes (fig. S1, A and B, cluster III). In addition, we showed that Glc application also induced expression of the defense marker gene NDR1/HIN1-LIKE 10 (NHL10) and the camalexin synthetic gene PHYTOALEXIN DEFICIENT 3 (PAD3) (fig. S1, C and D), showing that sugar stimulates defense signaling in Arabidopsis. On the other hand, expression of the photosynthesis-related gene CHLOROPHYLL A/B BINDING PROTEIN 1 (CAB1) was repressed (fig. S1, C and D), which confirmed the activation of sugar signaling under these experimental conditions (above 25 mM Glc or Suc). We next investigated whether sugar also promotes gene expression during pattern-triggered immunity (PTI). Seedlings were treated with flg22 peptide, a derivative of bacterial flagellin, for 8 hours with or without Glc, Suc, or mannitol (Mtl) (Fig. 1A). Flg22-triggered expression of NHL10 and PAD3 was higher in the presence of Glc or Suc but unaffected by Mtl (Fig. 1B). These data suggested that the recognition of metabolizable sugars and/or sugar metabolism affects defense signaling, and this effect is not due to alteration of osmotic pressure. Because Suc effects were sometimes stronger than Glc effects as for PAD3 expression (Fig. 1B), we next asked whether Glc and Suc differentially activate defense signaling. In Arabidopsis, two cytoplasmic HXKs, HXK1 and HXK2 (6), phosphorylate Glc but not Suc. To explore the involvement of HXKs in Glc- and/or Suc-mediated stimulation of defense signaling, we established hxk1 hxk2 (hxk1/2) mutants (fig. S2, E and F). The hxk2 mutant was presumed to express HXK2 with a C-terminal 13–amino acid deletion (fig. S2C). The activity of HXK2ΔC13 was significantly weaker than that of HXK2 in vitro (fig. S2D). Therefore, the hxk2 mutant was used in this study although HXK2 activity was not completely absent. We found that both Glc- and Suc-induced expression of NHL10 and PAD3 were reduced in hxk1/2 plants (Fig. 1C), suggesting that Glc, hydrolyzed from Suc, stimulates their expression under Suc conditions. Suc accumulates to high levels in the cytoplasm of plant cells, while Glc is mainly compartmentalized in the vacuole (13). Therefore, Suc may stimulate gene expression more easily than Glc, although both sugars act through HXKs.
Fig. 1. Sugar promotes plant defense signaling.
(A) A schematic description of experimental procedures for flg22 application in the presence of glucose (Glc), sucrose (Suc), or mannitol (Mtl) (i) or 2DG treatment (ii). (B and C) Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis of defense-related gene expression in Arabidopsis seedlings exposed to mock or 1 μM flg22 for 8 hours in the presence of 25 mM Glc, 25 mM Suc, or 25 mM Mtl (n = 3, biologically independent samples). (D) qRT-PCR analysis of defense-related gene expression in Arabidopsis seedlings exposed to 25 mM Glc, 25 mM 3-O-methyl-d-Glc (3OMG), or 0.5 mM 2-deoxy-d-Glc (2DG) for 24 hours (n = 3, biologically independent samples). (E) Growth of Pst DC3000 syringe-inoculated into rosette leaves of 4-week-old Arabidopsis. Bacteria were inoculated 1 day after 0.5 mM 2DG treatment. Bacterial titers were determined 3 days after inoculation (n = 4, biologically independent samples). (F) Quantification of camalexin and SA 24 hours after 0.5 mM 2DG treatment (n = 3, biologically independent samples). FW indicates fresh weight. Statistically significant differences among samples were determined using the two-tailed Welch’s t test (P < 0.05) (E) or one-way analysis of variance (ANOVA) with multiple comparison tests (Tukey HSD) and are represented by asterisks or different letters (P < 0.05). Experiments were repeated at least three times, and similar results were obtained.
Next, to explore the detailed mechanisms by which Glc activates defense signaling, we applied two non-metabolizable Glc analogs, 3-O-methyl-d-Glc and 2-deoxy-d-Glc (2DG), to seedlings for 24 hours. Although both Glc analogs are imported into the cytoplasm via transporters (14), only 2DG is phosphorylated by HXK (2). NHL10 expression was significantly induced only by 2DG application (Fig. 1D), to a level lower than that induced by flg22 (fig. S1E). In addition, 2DG treatment enhanced resistance to the pathogenic bacterium Pseudomonas syringae pv. tomato (Pst) DC3000 in 4-week-old plants (Fig. 1E), indicating that 2DG induces defense signaling in both seedlings and mature plants. Furthermore, RNA-seq analyses revealed that defense-related genes, including flg22-responsive genes, were overrepresented among 2DG-responsive genes in seedlings (fig. S1, A and B). Not only camalexin synthetic genes such as PAD3 but also SA-synthetic genes such as AVRPPHB SUSCEPTIBLE 3 (PBS3) and SALICYLIC ACID INDUCTION DEFICIENT 2 (SID2) were strongly induced in response to 2DG (figs. S1A and S3A). Consistently, camalexin and SA accumulated 24 hours after 2DG application (Fig. 1F). We confirmed that PBS3 expression also responded to Glc and Suc. While Glc or Suc alone did not activate PBS3 expression, they promoted flg22-triggered PBS3 expression (Fig. 1B). Note that flg22-triggered SA accumulation also increased in the presence of Glc and Suc, while the application of Suc, but not Glc, induced SA synthesis even without flg22 treatment (fig. S1F). Camalexin accumulation also showed similar patterns (fig. S1F).
2DG is a more potent inducer of many defense-related genes than Glc (Fig. 1D). However, simultaneous application of excess Glc suppressed 2DG-induced expression to the levels under Glc conditions (Fig. 2A), indicating competitive effects between 2DG and Glc on cellular activities such as transport and metabolism. Because 2DG is not further catabolized in glycolysis after HXK-mediated phosphorylation, 2DG-6-phosphate (2DG6P) accumulates following 2DG application (Fig. 2B). However, we showed that cellular 2DG and 2DG6P decreased markedly following simultaneous application of excess amounts of Glc (Fig. 2B). Therefore, the amounts of cellular 2DG and/or 2DG6P appeared to correlate with the amplitude of defense signaling in response to 2DG. 2DG and 2DG6P act as inhibitors of HXK and phosphoglucose isomerase, respectively (15, 16), thereby causing inhibition of glycolysis. We therefore deduced that glycolysis-derived energy is dispensable for 2DG-induced defense stimulation. In addition, we concluded that energy depletion caused by 2DG treatment does not lead to defense stimulation either, because the mitochondrial electron transport inhibitor antimycin A, the mitochondria uncoupler 2,4-dinitrophenol, and the photosynthesis inhibitor 3-(3,4-dichlorophenyl)-1,1-dimethylurea did not induce camalexin or SA (fig. S1G).
Fig. 2. HXK-mediated Glc phosphorylation is required for sugar-induced defense signaling.
(A and B) qRT-PCR analysis of the expression of defense-related genes (A) and quantification of metabolites (B) in Arabidopsis seedlings exposed to 1 mM 2DG and/or 25 mM Glc for 24 hours (n = 3, biologically independent samples). 2DG + Glc indicates simultaneous application of 1 mM 2DG and 25 mM Glc. (C and D) qRT-PCR analysis of defense-related gene expression in Arabidopsis seedlings exposed to mock or 0.5 mM 2DG for 8 hours (n = 3, biologically independent samples). (E) qRT-PCR analysis of defense-related gene expression in Arabidopsis seedlings exposed to 0.5 mM 2DG for 8 hours (n = 3, biologically independent samples). (F) Quantification of 2DG6P and 2DG after 0.5 mM 2DG treatment (n = 3, biologically independent samples). Statistically significant differences among samples were determined using one-way ANOVA with multiple comparison tests [Tukey HSD or Dunnett’s test between wild-type (WT) and other genotypes (E)] and are represented by asterisks or different letters (P < 0.05). Experiments were repeated at least three times, and similar results were obtained.
The results described above suggested contributions of sugar transporters and HXK to 2DG-induced defense signaling. We focused on 8 hours after 2DG application, as, at this time point, 2DG responsiveness of most genes was observable (fig. S3A) and the secondary effect of SA accumulation was minimized because SA had not yet accumulated (fig. S3B). Of 14 SUGAR TRANSPORT PROTEIN (STP) genes that mainly transport monosaccharides in Arabidopsis, STP1, STP4, and STP13 are the most abundantly expressed (17). Therefore, we established stp1 stp4 stp13 (stp1/4/13) triple mutants by introducing a 17–base pair (bp) deletion in the STP4 locus into stp1/13 plants using CRISPR-Cas9 (fig. S4, A to F). As expected, stp1/4/13 plants exhibited a significant reduction in 2DG influx activity (fig. S4D). We found that stp1/4/13 plants displayed no induction of NHL10 or PBS3 in response to 2DG (Fig. 2C). Moreover, hxk1/2 plants also showed reduced expression of NHL10 and PBS3 in response to 2DG (Fig. 2D). 2DG-triggered induction was still detectable in hxk1/2 plants, likely due to residual HXK2 activity (fig. S2D). Introduction of an HXK1 cDNA complemented the reductions in 2DG-induced defense gene expression and 2DG6P production in hxk1/2 plants (Fig. 2E and fig. S2H). On the other hand, introduction of HXK1(G104D) (6), which acts only as a Glc sensor and lacks catalytic activity, did not complement these phenotypes (Fig. 2E and fig. S2H). Thus, HXK-mediated 2DG phosphorylation is required for 2DG-induced defense signaling. However, we noticed that 2DG-induced SID2 expression appeared to be restored by HXK1(G104D) (fig. S2G), implying that HXK1’s sensor activity also contributes to 2DG-induced defense signaling. We found that 2DG6P, but not 2DG, significantly decreased in hxk1/2 plants; stp1/4/13 plants showed a reduction of both 2DG and 2DG6P (Fig. 2F). Therefore, 2DG6P levels correlated with expression levels of defense-related genes in hxk1/2 plants in response to 2DG. Together, these data suggested that 2DG6P, which HXK1/2 generate, primarily mediates defense signaling after 2DG application.
Sugar stimulates the protein and phosphorylation levels of CPK5
Calcium-dependent protein kinases, particularly CPK4/5/6/11 belonging to subgroup I, activate NHL10 expression in response to flg22 (9); moreover, CPK5/6 induce PAD3 during fungal infection (10, 11). We found that loss of CPK5/6 attenuated the induction of NHL10 and PAD3 in response to 2DG, while 2DG-induced expression of PBS3 and SID2 was enhanced in cpk5/6 plants (Fig. 3A). Consistently, accumulation of camalexin was almost abolished but that of SA was further increased in cpk5/6 plants in response to 2DG (Fig. 3B). CPK4/11 partially contributes to suppression of SA biosynthesis because cpk4/5/6/11 plants showed higher SA accumulation than cpk5/6 plants in response to 2DG, although 2DG-induced SA accumulation did not differ between wild-type (WT) and cpk4/11 plants (Fig. 3B). Introduction of a C-terminally green fluorescent protein (GFP)–fused CPK5 genomic fragment containing its promoter region (gCPK5-GFP) fully complemented 2DG-induced gene expression in cpk5/6 plants (fig. S5A). Therefore, CPK5/6, especially CPK5, affect 2DG-induced signaling by positively and negatively regulating the expression of NHL10/PAD3 and PBS3/SID2, respectively, downstream of HXK1/2. Previously, plasma membrane-localized tobacco CDPKs, similar to Arabidopsis CPK5, were reported to be stimulated by application of Glc or Suc (18). This is consistent with our data showing a relationship between sugar and CPKs. In addition, 2DG-induced CAB1 attenuation was not affected by loss of CPK5/6, suggesting the presence of a CPK5/6-independent sugar signaling pathway(s) in Arabidopsis (Fig. 3A).
Fig. 3. The protein and phosphorylation levels of CPK5 are regulated by sugar conditions.
(A and C) qRT-PCR analysis of defense-related gene expression in Arabidopsis seedlings exposed to mock or 0.5 mM 2DG for 8 hours (n = 3, biologically independent samples). (B) Quantification of camalexin and SA 24 hours after mock or 0.5 mM 2DG treatment (n = 3, biologically independent samples). (D and E) Immunoblot analysis of CPK5-GFP in Arabidopsis seedlings after 0.5 mM 2DG application (D) or 1 μM flg22 application (E) (Mn2+-Phos-tag SDS–polyacrylamide gel electrophoresis). Phospho levels were calculated as the ratios of intensities of B1 and B3. The phospho levels of mock 4 hours (D) and mock in the absence of Glc (E) were set to 1.0. IB denotes immunoblotting with anti-GFP antibodies. Ponceau S–stained membranes are shown below. (F) qRT-PCR analysis of defense-related gene expression in Arabidopsis seedlings exposed to mock or flg22 for 0.5 hours in the absence/presence of 25 mM Glc (n = 3, biologically independent samples). Statistically significant differences among samples were determined using the two-tailed Welch’s t test (P < 0.05) (F) or one-way ANOVA with multiple comparison tests (Tukey HSD) and are represented by asterisks or different letters (P < 0.05). Experiments were repeated at least three times, and similar results were obtained.
NHL10 expression was similar in cpk5/6 and cpk5/6/sid2 plants (Fig. 3C and fig. S5B), indicating that enhanced SA levels in cpk5/6 plants did not reduce 2DG-induced NHL10 expression. CPK5/6 phosphorylate the transcription factors WRKY33 (group I) and WRKY8/28/48 (group IIc), transducing signals to downstream pathways (10, 19). PAD3 expression in response to 2DG was reduced in wrky33 plants but not wrky8 plants (fig. S5C), suggesting that WRKY33 acts as a downstream factor of CPK5/6 in 2DG-induced signaling.
We next asked whether 2DG application affects CPK5 status. An immunoblot showed three bands of CPK5-GFP (Fig. 3D). Of these, the uppermost band (B1) was abolished by treatment with lambda protein phosphatase (λPPase) (Fig. 3D), indicating that B1 represents the major phosphorylated form of CPK5-GFP. CPKs are rapidly activated upon PAMP perception following Ca2+ influx (9). We found that flg22 application increased the intensity of B1 and reduced that of the middle band (B2) but did not affect the lowest band (B3) of CPK5-GFP after transient elevation of the cytosolic Ca2+ concentration (fig. S6A). These data indicated that the B2 form of CPK5-GFP becomes phosphorylated and shifts to the B1 form in response to flg22. We concluded that the B3 form is an alternative splicing variant because it was only faintly observable in plants expressing CPK5(cDNA)-GFP from the constitutive 35S promoter (fig. S6B). We evaluated the dynamics of CPK5 phosphorylation by calculating the B1/B3 ratio of CPK5-GFP. The B2 intensity was sometimes too weak to use for normalization, so instead we used the stable signal of B3 as an indicator of CPK5 protein abundance. The CPK5 phosphorylation level (B1/B3 ratio) increased under 2DG treatment (Fig. 3D), but there was no such increase in stp1/4/13 plants, indicating that 2DG uptake leads to CPK5 phosphorylation.
Next, we investigated the phosphorylation status of CPK5-GFP in the presence of Glc. Consistent with the 2DG experiments, phosphorylation levels of CPK5-GFP increased under higher Glc conditions (Fig. 3E). The protein abundance of CPK5-GFP was elevated in a Glc-dependent manner (Fig. 3E). We found that CPK5 expression was induced by Glc application (Fig. 3F), likely contributing to an increase in CPK5 protein amounts. On the other hand, flg22-triggered CPK5 phosphorylation occurred even in the absence of Glc (Fig. 3E), suggesting that Glc is dispensable for flg22-triggered CPK5 phosphorylation. However, a Glc-dependent increase in phosphorylation levels before flg22 application ensured higher flg22-triggered phosphorylation levels of CPK5-GFP (Fig. 3E). NHL10 expression 0.5 hours after flg22 application appeared to correlate with protein and phosphorylation levels of CPK5-GFP (Fig. 3F). Together, these data suggested that Glc modulates CPK5 activity for defense gene expression by affecting its protein and phosphorylation levels.
Dephosphorylation of CPK5 by clade A PP2Cs is suppressed by G6P
PAMP treatment induced a transient, strong elevation of cytosolic Ca2+ (Fig. 4A), leading to CPK5 phosphorylation (Fig. 3E). On the other hand, despite inducing CPK5 phosphorylation, 2DG treatment did not trigger discernible elevation of cytosolic Ca2+ (Fig. 4A). This prompted us to explore Ca2+-independent CPK5 regulatory mechanisms. CPK6 and CPK11 were previously reported to bind to clade A PP2Cs (20, 21). We asked whether CPK5 also interacts with clade A PP2Cs in yeast two-hybrid and split-luciferase assays. Of the clade A PP2Cs that we tested, we found that ABI1 and PP2CA strongly, and HAI1 weakly, bound to CPK5, whereas HAB1 did not (Fig. 4B and fig. S6C). On the other hand, AP2C1 (clade B) and PP2C6-6 (clade E) did not bind to CPK5 (fig. S6D), suggesting that CPK5 interacted specifically with clade A PP2Cs. Consistent with the protein interaction data, in vitro CPK5 phosphorylation levels, detected by anti-phosphothreonine antibody, were massively reduced by ABI1 and PP2CA but not by HAB1 (fig. S6E). Furthermore, CPK5 trans-phosphorylation activity on an N-terminal fragment of RBOHD (RBOHD-NT) (22) and a WRKY33 wbox1 fragment (10) was weakened in the presence of ABI1 (Fig. 4C and fig. S6F). Consistent with previous reports (20, 23), in vitro trans-phosphorylation activity of the protein kinase OPEN STOMATA 1 (OST1) was also suppressed in the presence of ABI1 (fig. S6G). When we added ABI1 to the reaction after stopping CPK5 activity using the kinase inhibitor staurosporine, the phosphorylation levels of CPK5 substrates were not reduced (Fig. 4C and fig. S6F). Therefore, we can exclude the possibility that ABI1 directly dephosphorylated CPK5 substrates. This ABI1 effect on CPK5 activity was alleviated in the presence of Ca2+ (Fig. 4C), indicating that Ca2+-activated CPK5 can overcome ABI1-mediated suppression. We inferred from these data that clade A PP2Cs suppress CPK5 activity, especially before PAMP-triggered Ca2+ influx.
Fig. 4. G6P suppresses dephosphorylation of CPK5 by clade A PP2Cs.
(A) Cytosolic Ca2+ concentrations were measured via aequorin-mediated luminescence in response to PAMPs (left) or 2DG (right) within 15 min (top) or 8 hour (bottom) (means ± SE, n = 8, biologically independent samples). (B) Split-luciferase assay for protein interactions between ABI1 and CPK5 in N. benthamiana leaves. Agrobacterium strains harboring indicated constructs were co-infiltrated into N. benthamiana. Luminescence was detected after infiltrating luciferin into leaves. (C) Autoradiograph of RBOHD-NT phosphorylation by CPK5 in vitro for 30 min with [γ-32P] adenosine triphosphate (ATP) (top images). Parentheses indicate addition 15 min after the reaction was started. Coomassie Brilliant Blue (CBB)–stained gels are shown below. (D) ABI1 dephosphorylate CPK5 in vitro. Phosphorylation of CPK5 was detected by anti-phosphothreonine (pThr) antibody. Sugars (10 mM) were applied. IB denotes immunoblotting. Fru-6P, Man-6P, GlcN-6P, Sor-6P, Glc-1P, Gal1P, and uridine diphosphate (UDP)–Glc indicate fructose-6-phosphate, mannose-6-phosphate, glucosamine-6-phosphate, sorbitol-6-phosphate, glucose-1-phosphate, galactose-6-phosphate, and UDP-glucose, respectively. (E and F) in vitro phosphatase activity of ABI1. Sugars (10 mM) were applied. (G) In vitro protein interaction between CPK5 and ABI1. Sugars (10 mM) were applied. IB denotes immunoblotting with anti-MBP or anti-GST antibodies. (H) Quantification of camalexin and SA 24 hours after mock or 0.5 mM 2DG treatment (n = 3, biologically independent samples). Statistically significant differences among samples were determined using one-way ANOVA with multiple comparison tests (Tukey HSD) and are represented by different letters (P < 0.05). Experiments were repeated at least three times, and similar results were obtained.
We next asked whether sugar affects ABI1-mediated CPK5 regulation. Because we found that 2DG6P is involved in 2DG-induced defense signaling, we investigated whether 2DG6P and G6P, which is the natural counterpart of 2DG6P, affect ABI1-mediated CPK5 dephosphorylation. We demonstrated here that application of 2DG6P and G6P, but not 2DG or Glc, reduced ABI1-mediated CPK5 dephosphorylation in vitro (Fig. 4D). We found that G6P suppressed the in vitro protein phosphatase activities of ABI1 and HAB1 (Fig. 4, E and F, and fig. S6I), while the activity of PP2C6-6 (clade E) was not affected (Fig. 4F). In addition, λPPase-mediated CPK5 dephosphorylation was not reduced by G6P (fig. S6H), implying that G6P specifically influences the activity of clade A PP2Cs. Furthermore, we revealed that the application of 2DG6P and G6P, but not 2DG or Glc, dissociated protein interactions between CPK5 and ABI1 (Fig. 4G).
Other hexose-phosphates including hexose-1-phosphates, but not the sugar nucleotide uridine diphosphate–Glc, also inhibited ABI1-mediated CPK5 dephosphorylation, whereas Glc-1-phosphate (G1P) had a weaker effect than G6P (Fig. 4D and fig. S6J). In contrast, sorbitol-6-phosphate did not affect ABI1 activity (Fig. 4D), suggesting that the phosphate group did not directly inhibit ABI1 activity. G6P concentration was previously reported to be around 3 to 5 mM in the cytoplasm of plant cells (13). G6P at this concentration may not be enough to suppress PP2C activity; G6P above 10 mM inhibited ABI1 activity in vitro (fig. S6I). We speculate that combinations of G6P and other sugar phosphates may lead to suppression of PP2C activity in plant cells under sugar-sufficient conditions. The combination of 5 mM G6P and 5 mM fructose-6-phosphate suppressed ABI1-mediated CPK5 dephosphorylation, although either hexose-phosphate of 5 mM alone did not (fig. S6K).
We next investigated the relationship between CPK5/6 and ABI1/2 in plants. While camalexin synthesis in response to 2DG was similar in WT and abi1/2 plants, 2DG-induced SA was reduced in abi1/2 plants, compared with WT plants (Fig. 4H). On the other hand, SA accumulation in response to 2DG was elevated in abi1/2/cpk5/6 plants to a similar extent as in cpk5/6 plants (Fig. 4H), implying that enhanced CPK5/6 activity caused a reduction in 2DG-induced SA synthesis in abi1/2 plants.
Sugar influx contributes to PTI
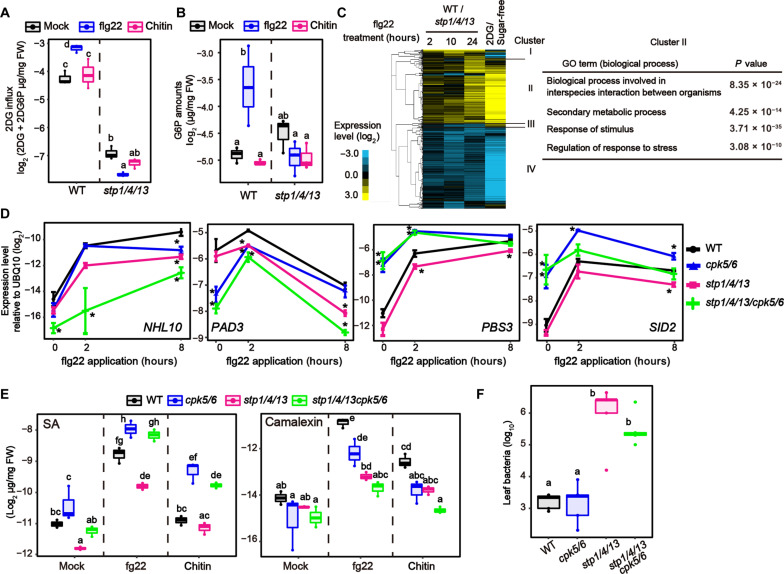
We have reported that elevated sugar influx activity in response to flg22 interrupts sugar acquisition by pathogens (24). Given that, while apoplasmic sugar amounts decrease, cytoplasmic sugar should concomitantly increase in the cytoplasm during flg22-triggered immunity. We found that G6P amounts were elevated in response to flg22 under 25 mM Suc conditions, dependent on the activation of sugar transporter STPs (Fig. 5, A and B). Flg22-triggered expression of defense-related genes decreased in hxk1/2 plants in the presence of 25 mM Glc or Suc (Fig. 1C). Flg22-triggered synthesis of SA and camalexin was also reduced in hxk1/2 plants under these sugar conditions (fig. S7A), suggesting that G6P, which HXK1/2 generate, affects flg22-triggered defense responses. Together, we hypothesized that STP-mediated sugar influx contributes to flg22-triggered signaling. We showed that sugar influx activity was enhanced 8 and 24 hours, but not 3 hours, after flg22 application (fig. S7B). Therefore, we focused on the contribution of STP-mediated sugar influx to flg22-triggered signaling during these later time points. To this end, we treated plants with flg22 under 25 mM Suc conditions. Whereas possible differences in cellular sugar amounts before PAMP recognition would cause different CPK5 phosphorylation status between WT and stp1/4/13 plants, this effect can be minimized under sugar-sufficient conditions. Notably, RNA-seq analyses revealed that transcriptional profiles in response to flg22 in the presence of 25 mM Suc were substantially altered in stp1/4/13 plants compared with WT plants (Fig. 5C and fig. S7C). As expected, these differences in transcriptional profiles increased at the later stage of flg22-triggered signaling (Fig. 5C), correlating with the timing of enhancement of sugar uptake activity in response to flg22 (fig. S7B). Of particular interest was the observation that STP-dependent genes largely overlapped with 2DG-responsive genes during flg22-triggered defense (Fig. 5C). This suggests that sugar signaling activated by STP-mediated sugar influx stimulates defense signaling in response to flg22. We also found that STP-mediated sugar influx was stimulated in response to another bacterial PAMP, elf18 peptide that is a fragment of bacterial elongation factor Tu (fig. S8A). Furthermore, STP-mediated sugar influx also contributed to elf18-triggered defense signaling (fig. S8, B and C). Consistently, stp1/4/13 plants showed higher susceptibility to Pst DC3000 (ΔavrPtoΔavrPtoB), a less virulent strain of Pst DC3000 (Fig. 5F).
Fig. 5. Sugar influx contributes to pattern-triggered signaling.
(A) 2DG influx activity in Arabidopsis seedlings 8 hours after 1 μM flg22 or chitin (250 μg/ml) treatment (n = 3, biologically independent samples). 2DG influx activity was measured by calculating the mixed amounts of 2DG and 2DG6P 1 hour after 0.5 mM 2DG application. (B) Quantification of G6P in Arabidopsis seedlings 8 hours after 1 μM flg22 or chitin (250 μg/ml) treatment under 25 mM Suc conditions (n = 3, biologically independent samples). (C) Heatmaps displaying expression patterns of genes that show significant expression changes in stp1/4/13 plants, relative to WT plants, in response to 1 μM flg22 in the presence of 25 mM Suc (q < 0.01 and|log2FC| > 1). The log2 fold changes relative to WT were subjected to hierarchical clustering analysis (left). Overrepresented Gene Ontology (GO) terms in cluster II are presented (right). (D) qRT-PCR analysis of defense-related gene expression in Arabidopsis seedlings exposed to 1 μM flg22 under 25 mM Suc conditions (n = 3, biologically independent samples). WT data are shown in fig. S7D. (E) Quantification of SA and camalexin in Arabidopsis seedlings 8 hours after 1 μM flg22 treatment or chitin (250 μg/ml) treatment under 25 mM Suc conditions (n = 3, biologically independent samples). (F) Growth of Pst DC3000(ΔavrPtoΔavrPtoB) spray-inoculated onto rosette leaves of 4-week-old Arabidopsis. Bacterial titers were determined 3 days after inoculation (n = 5, biologically independent samples). Statistically significant differences among samples were determined using one-way ANOVA with multiple comparison tests [Tukey HSD or Dunnett’s test between WT and other genotypes at each time point (D)] and are represented by asterisks or different letters (P < 0.05). Experiments were repeated at least three times, and similar results were obtained.
We found that, in the presence of 25 mM Suc, loss of CPK5/6 reduced flg22-triggered expression of NHL10 and PAD3 to a similar extent as loss of STP1/4/13 (Fig. 5D). In addition, flg22-triggered camalexin synthesis was reduced in cpk5/6 and stp1/4/13 plants (Fig. 5E). We expected that STP-mediated sugar influx would contribute to flg22-triggered defense gene expression by enhancing CPK5/6 activity. However, CPK5 phosphorylation levels were not altered between WT and stp1/4/13 plants 8 hours after flg22 application (fig. S7E), at which time G6P accumulates via sugar influx (Fig. 5, A and B). We showed that Ca2+-activated CPK5 overcomes ABI1-mediated dephosphorylation (Fig. 3C). Although flg22-triggered Ca2+-influx has already returned to baseline by 8 hours after flg22 application (Fig. 4A), CPK5, once activated by Ca2+, may not be affected by G6P for some time. Next, to dissect the relationship between CPK5/6-mediated and STP-mediated signaling during PTI, we established stp1/4/13/cpk5/6 quintuple mutants (fig. S9A). These stp1/4/13/cpk5/6 plants showed almost no response to 2DG, like stp1/4/13 plants (fig. S9B). Thus, stp1/4/13 mutations are epistatic to cpk5/6 mutations with respect to 2DG response because 2DG influx is essential to trigger 2DG-induced signaling. On the other hand, in response to flg22, stp1/4/13/cpk5/6 plants showed an additive phenotype relative to cpk5/6 and stp1/4/13 plants. stp1/4/13/cpk5/6 plants exhibited a further reduction in flg22-triggered expression of NHL10 and PAD3, compared with cpk5/6 and stp1/4/13 plants (Fig. 5D). These additive effects suggested that STP-mediated sugar influx stimulates CPK5/6-independent signaling, which promotes flg22-triggered expression of NHL10 and PAD3 together with Ca2+-activated CPK5/6-mediated signaling.
Expression of PBS3 and SID2 in response to flg22 were also reduced in stp1/4/13 plants (Fig. 5D). Consistently, flg22-induced SA amounts were less induced in stp1/4/13 plants (Fig. 5E), indicating that sugar influx activates the SA biosynthesis pathway in response to flg22. On the other hand, we found that SA-synthetic gene expression and SA accumulation were highly induced in cpk5/6 plants in response to flg22 as well as 2DG (Fig. 5, D and E), although CPK5 was previously reported to promote SA synthesis during effector-triggered immunity (12, 19). We confirmed that enhanced SA amounts in cpk5/6 plants were also detected in mature leaves after flg22 application or inoculation of Pst DC3000 (fig. S10A). In addition, although cpk5/6 plants were less susceptible to spray-inoculated Pst DC3000 than WT plants, addition to cpk5/6 plants of a sid2 mutation causing SA deficiency resulted in higher susceptibility to Pst DC3000 to cpk5/6 plants (fig. S10B), suggesting that high SA levels elevated antibacterial resistance in cpk5/6 plants under our experimental conditions. We demonstrated that cpk5/6 mutations were epistatic to stp1/4/13 mutations with respect to flg22-triggered SA synthesis. In response to flg22, stp1/4/13/cpk5/6 plants showed high SA accumulation similar to cpk5/6 plants, whereas SA levels were reduced in stp1/4/13 plants (Fig. 5E). These results revealed that CPK5/6-mediated SA suppression becomes dominant in stp1/4/13 plants and suggested that STP-mediated sugar signaling promotes SA synthesis by attenuating this CPK5/6 effect in WT plants. However, we think that STP-mediated sugar influx does not lead to inhibition of CPK5/6 activity, because flg22-triggered SA synthesis was not altered between WT and cpk5/6 plants under sugar-deficient conditions (fig. S10C), indicating that loss of CPK5/6 activity was not a direct trigger for SA synthesis. Therefore, we concluded that STP-mediated sugar influx enhances SA-synthetic signaling that acts antagonistically on CPK5/6-mediated SA suppression during flg22-triggered immunity. On the other hand, elevated susceptibility to Pst DC3000 (ΔavrPtoΔavrPtoB) in stp1/4/13 plants was not significantly reduced in stp1/4/13/cpk5/6 plants despite higher SA accumulation (Fig. 5F). Because an increase in apoplasmic sugar amounts by loss of STPs elevates bacterial virulence (24), it could counteract SA-mediated immunity from loss of CPK5/6.
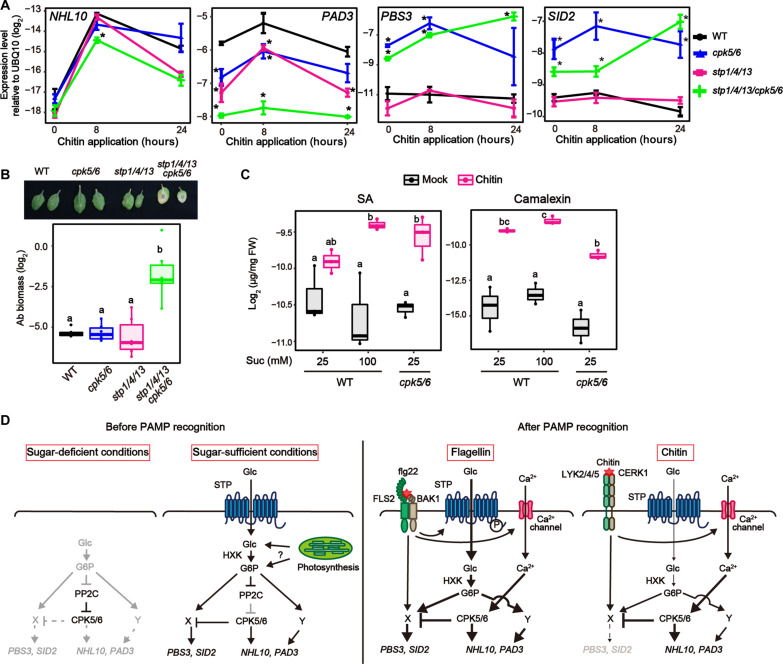
We found that, unlike flg22, the fungal cell wall component chitin did not stimulate sugar influx activity or increase G6P amounts (Fig. 5, A and B). STP13, a gene responsible for flg22-induced sugar influx activity (24), was less induced in response to chitin than in response to flg22 (fig. S12, A and B). Moreover, CERK1, the co-receptor for chitin receptors, did not phosphorylate STP13 T485 (24), which is the important residue for elevation of sugar influx activity (fig. S12C), although BAK1, the co-receptor for the flagellin receptor, did in vitro (24). Therefore, we wondered whether sugar influx activity does not contribute to chitin-triggered signaling. However, chitin-triggered PAD3 expression and camalexin synthesis were reduced in stp1/4/13 plants (Figs. 5E and 6A), indicating that basal sugar influx activity mediated by STPs is required for chitin-triggered signaling. On the other hand, chitin-triggered NHL10 expression was not reduced, while flg22-triggered expression was reduced, in stp1/4/13 plants (Fig. 6A), suggesting that the contribution of STP-mediated sugar influx to chitin-triggered signaling is lower than to flg22-triggered signaling. STP-mediated sugar influx led to activation of CPK5-independent signaling pathways in response to chitin as well as flg22. PAD3 expression and camalexin accumulation were further reduced in stp1/4/13/cpk5/6 plants in response to chitin, compared to cpk5/6 and stp1/4/13 plants (Figs. 5E and 6A). Furthermore, antifungal defense against the necrotrophic fungus Alternaria brassicicola was massively reduced in stp1/4/13/cpk5/6 plants (Fig. 6B). Together, these data show that STP-mediated sugar influx contributes to defense against both bacterial and fungal pathogens, although it affects these PAMP receptor signaling processes differentially.
Fig. 6. Cellular sugar level is key to coordinating defense outputs during PTI.
(A) qRT-PCR analysis of defense-related gene expression in Arabidopsis seedlings exposed to chitin (250 μg/ml) under 25 mM Suc conditions (n = 3, biologically independent samples). WT data are shown in fig. S11. (B) Biomass of A. brassicicola on detached rosette leaves of 4-week-old Arabidopsis after 5 days (n = 4, biologically independent samples). (C) Quantification of SA and camalexin in Arabidopsis seedlings 8 hours after chitin (250 μg/ml) treatment under 25 mM or 100 mM Suc conditions (n = 3, biologically independent samples). (D) A model proposed in this study. An increase in cellular G6P, possibly through sugar influx and photosynthesis, results in suppression of clade A PP2Cs to enhance CPK5/6 activities. During PTI, sugar influx triggers CPK5-independent signaling. One pathway (Y) additively activates NHL10/PAD3 with CPK5/6. The other pathway (X) is antagonistic to CPK5/6 that suppress PBS3/SID2. Sugar influx activity is enhanced in response to flg22, leading to SA accumulation that promotes antibacterial defense. On the other hand, chitin recognition only promotes camalexin synthesis, because sugar influx activity is not affected. Statistically significant differences among samples were determined using one-way ANOVA with multiple comparison tests [Tukey HSD or Dunnett’s test between WT and other genotypes at each time point (A)] and are represented by asterisks or different letters (P < 0.05). Experiments were repeated at least three times, and similar results were obtained.
Cellular sugar amounts determine SA synthesis during PTI
In contrast to flg22 application (Fig. 5, D and E, and fig. S7D), we found that chitin application did not induce SA-synthetic gene expression or SA synthesis under our experimental conditions (25 mM Suc) (Figs. 5E and 6A and fig. S11). Previous studies also reported that chitin treatment did not induce expression of the SA marker gene PR1 (25). However, chitin treatment induced SA accumulation in cpk5/6 plants (Figs. 5E and 6C). This result suggests that chitin signaling can stimulate SA-synthetic signaling but is too weak to overcome CPK5/6-mediated suppression. We noticed that it resembles flg22-triggered SA accumulation patterns in stp1/4/13 and stp1/4/13/cpk5/6 plants. Therefore, unincreased cytoplasmic sugar levels (Fig. 5B), due to unenhanced sugar influx activity (Fig. 5A), may cause undetectable chitin-triggered SA synthesis. To investigate this hypothesis, we treated plants with chitin under higher sugar conditions (100 mM Suc) to elevate basal cellular sugar amounts. Notably, chitin-triggered SA accumulation was observable even in WT plants under these conditions (Fig. 6C). Together, these data suggested that cellular sugar levels are key factors for SA synthesis during PTI.
DISCUSSION
In this study, we provide evidence that cellular sugar level is a critical factor regulating defense outputs. We found that G6P contributes to defense activation in Arabidopsis. Likewise, G6P has been reported to stimulate sugar signaling in animals, for instance, by promoting the nuclear localization of the transcription factor MondoA (26). Because Glc is rapidly phosphorylated by HXK in the cytoplasm, Glc amounts are limited in the cytoplasm; Glc is mainly compartmentalized into the vacuole in plant cells (13). Therefore, cytoplasmic G6P sensory mechanisms may have evolved in various organisms. However, the mode of action of these G6P sensors has been elusive. We propose a mechanism by which G6P suppresses the activity of clade A PP2Cs and/or dissociates the PP2Cs from CPK5, leading to an increase in CPK5 phosphorylation levels. This G6P-mediated CPK5 regulation contributes to enhancement of defense levels before pathogen recognition (Fig. 6D). We showed that other hexose-phosphates, including G1P, also suppresses ABI1-mediated CPK5 dephosphorylation (Fig. 4D). However, G1P effects were weaker than G6P effects (fig. S6J). To understand these differences, the molecular mechanisms of hexose-phosphate recognition by ABI1 need to be uncovered. In this study, we investigated the roles of hexose-phosphates in sugar signaling by using 2DG. We found that the effects of 2DG treatment were stronger than those of Glc treatment (Fig. 1D) and 2DG treatments altered the expression of genes that are not responsive to flg22 (fig. S1A). It is possible that some effects of 2DG treatment resulted from pleiotropic effects of non-metabolizable 2DG6P, which accumulated in the cytoplasm. However, in addition to gene expression data, we provided other evidence that not only 2DG but also Glc treatment induced CPK5 phosphorylation in plants and that both 2DG6P and G6P suppress ABI1-mediated CPK5 dephosphorylation in vitro.
After recognition of PAMPs, STP-mediated sugar influx activates CPK5-independent signaling. We showed that at least two CPK5-independent pathways are stimulated by sugar influx during PTI. One stimulates NHL10 and PAD3, like the CPK5/6 pathway (Fig. 6D). The other acts antagonistically on the CPK5/6 pathway that represses PBS3 and SID2 (Fig. 6D). The later signal pathway is required to synthesize SA during PTI. The sugar sensor SnRK1 becomes active under sugar-starved conditions, prioritizing the catabolic rather than the anabolic pathway. It was reported that SnRK1 overexpression led to suppression of Glc-induced PR1 (27). Because G6P is a candidate molecule for suppression of SnRK1 activity (28), enhanced G6P levels via STPs could inhibit SnRK1 activity for SA synthesis in response to flg22.
Because most phytopathogenic bacteria are hemi-biotrophic (29), SA signaling would be effective to suppress their growth during biotrophic phases. On the other hand, SA signaling can have negative effects on antifungal defense, especially against necrotrophic fungal pathogens (30) although the growth of A. brassicicola did not increase in cpk5/6 plants in which SA accumulated to high levels (Fig. 6B). Our study demonstrated that flg22-triggered signaling forces sugar transporters to elevate cellular sugar amounts, activating SA synthesis. In contrast, basal sugar amounts determined SA synthesis during chitin-triggered signaling. Chitin signaling increased SA synthesis only in the presence of high levels of sugar. It was reported that infection by biotrophic fungi led to an elevation of cellular sugar levels in infected plant tissues (31); biotrophic fungi are thought to strategically acquire sugars from host cells by manipulating host sugar transporters (32, 33). Therefore, it seems reasonable that chitin-triggered SA synthesis occurs dependent on cellular sugar levels. Together, we showed that flg22 and chitin receptor signaling differentially affect sugar influx activity, promoting appropriate defense outputs against pathogens with different sensitivities to SA-mediated defense. However, sugar influx activity was reported to be enhanced during infection by the necrotrophic fungus Botrytis cinerea (34), suggesting that fungal PAMPs other than chitin could activate sugar influx activity. We found that elf18-triggered signaling also stimulate STP-mediated sugar influx (fig. S8). Because BAK1 is also a co-receptor of the elf18 receptor EFR, BAK1-mediated signaling may promote sugar influx, possibly by phosphorylating STP13. For instance, RLP23 that contributes to defense against B. cinerea forms complexes with BAK1 (35, 36), thereby possibly enhancing STP-mediated sugar influx during B. cinerea infection. Other kinases, such as receptor-like cytoplasmic kinases, might also contribute to regulating sugar influx activity during fungal infection.
We showed qualitative differences between flg22-triggered and chitin-triggered defense outputs in a manner that depends on cellular sugar amounts. Although recognition of each PAMP immediately induces similar sets of defense-related genes (37), transcriptional profiles triggered by each PAMP during later phases were reported to be different (38). We expected that sugar influx might cause these differences during the later PTI phases. The contribution of STPs became larger during the late phase of flg22-triggered signaling, apparently correlating with the timing of enhancement of STP-mediated sugar influx activity (fig. S7B). Because nutrient competition occurs at the interface of host-pathogen interactions, we previously described sugar uptake by STPs as contributing to interruption of pathogen sugar acquisition (24). We now highlight it again as an important plant defense system that not only suppresses the pathogen but also coordinates the plant defense. Together, we provide a comprehensive view of how sugar contributes to defense signaling in plants. Because defense signaling is energy-intensive, estimating sugar availability by monitoring cellular G6P amounts may allow fine-tuning of defense outputs. In nature, G6P signaling may interact with other processes that affect defense signaling. Environmental cues such as light, humidity, and temperature dynamically influence cellular sugar levels (39, 40). These environments were also reported to affect plant defense signaling. For example, dark-induced sugar-starvation elevated susceptibility to pathogens (41). In addition, cold and drought conditions, which increase cellular sugar amounts in plants, induced PR1 expression (42, 43). It will be interesting to analyze whether sugar plays a role in molecular integration of environmental conditions into defense signaling.
MATERIALS AND METHODS
Plant materials and growth conditions
The Columbia-0 (Col-0) ecotype of A. thaliana was used as WT. Col-0 mutants stp1 (SALK_048848) (17), stp13 (SALK_045494) (17), cpk4 (SALK_000685) (44), cpk5 (SAIL_657_C06) (19), cpk6 (SALK_025460) (19), cpk11 (SALK_054495) (9), wrky33-1 (SALK_006603) (45), wrky8-1 (SALK_107668C) (19), hxk1 (WiscDsLoxHs044_02E), hxk2 (WiscDsLox289_292O3), sid2-1 (46), pad3-1 (47), abi1-2 (SALK_072009) (48) and abi2-2 (SALK_015166) (48) were used. To establish the stp1/4/13 mutant, a 17-bp deletion in the STP4 locus was introduced into stp1 stp13 plants using CRISPR-Cas9. To establish stp4, stp1 stp4, and stp4 stp13 plants, stp1/4/13 plants were crossed with Col-0 plants, and the desired genotypes were detected from the F2 progeny. pMAQ2 (49) plants were used to measure [Ca2+]cyt. Plants were grown on soil or 0.5× Murashige and Skoog (MS) agar medium [0.5× MS salt (Wako), 0.5× Gamborg’s vitamins (Sigma-Aldrich), 25 mM Suc, MES (0.5 g/liter) (pH 5.7), and 0.8% agar] at 22°C under 10-hour light/14-hour dark or 16-hour light/8-hour dark conditions, respectively.
Plasmid construction
The 35S GFP NosT fragment was amplified from pRI 35S-GFP (24) and inserted into the Hind III/Eco RI sites of pCAMBIA1300. This plasmid was named pCAMBIA 35S-GFP. To establish transgenic Arabidopsis, a genomic fragment including the upstream and coding regions of CPK5 or STP13 was inserted into the Hind III/Sal I sites of pCAMBIA 35S-GFP, or a CPK5 cDNA fragment was inserted into the Xba I/Sal I site of pCAMBIA 35S-GFP. A cDNA fragment of HXK1 or HXK1(G104D) was inserted into the Xba I/Sal I sites of pCAMBIA 35S-GFP. For obtaining recombinant proteins, cDNA fragments of CPK5 or OST1 were inserted into the Bam HI/Hind III sites of pET-28a(+). cDNA fragments of ABI1, HAB1, ABF1, HXK2, or HXK2ΔC13 were inserted into the Bam HI/Sal I sites of pGEX-6P-1. cDNA fragments of PP2CA, N-terminal truncated RBOHD, and WRKY33 wbox1 were inserted into the Eco RI/Sal I site of pMAL-c2. For yeast two-hybrid assays, cDNA fragments of ABI1, HAB1, PP2CA, or HAI1 were inserted into the Nde I/Sal I sites of pGADT7.cDNA fragments of full-length CPK5, N-terminal truncated CPK5(D221A), or C-terminal truncated CPK5 were inserted into the Nde I/Eco RI sites of pGADT7. For split-luciferase assays, a CPK5 cDNA fragment was inserted into the Bam HI/Sal I sites of pCAMBIA nLuc. cDNA fragments of ABI1, AP2C1, or PP2C6-6 were inserted into the Bam HI/Sal I sites of pCAMBIA cLuc. For CRISPR-Cas9, a guide RNA (gRNA) sequence of STP4 was inserted into the Aar I sites of pKIR1.1 (50). All cloning reactions except pKIR1.1 were performed using SLiCE (51) as described previously. For pKIR1.1, a gRNA fragment was ligated using Ligation high Ver. 2 (TOYOBO). Primers used for plasmid construction are listed in table S1.
Pathogen inoculation
A. brassicicola (NBRC 31226) was cultured on potato-dextrose agar medium and incubated at 25°C. Single 10-μl drops of spore suspension (5 × 105 spores/ml) were applied to detached 4-week-old Arabidopsis leaves. Inoculated leaves were then incubated on wet paper in petri dishes for 5 days. Six leaf discs from six independent inoculated leaves were pooled into a tube for genomic DNA extraction. The biomass of A. brassicicola on inoculated leaves was assessed by measuring DNA amounts of the fungus (ABU03393) and Arabidopsis (At5g26751) (52). Strains of P. syringae pv. tomato (Pst) DC3000 and Pst DC3000(ΔavrPtoΔavrPtoB) were cultured in NYGA medium [Bacto Peptone (5 g/liter), yeast extract (3 g/liter), and glycerol (20 ml/liter)] with rifampicin (50 μg/ml) at 30°C overnight. The cells were washed twice and resuspended in 10 mM MgCl2. These bacteria [optical density at 600 nm (OD600) = 0.02] in 0.02% Silwet L-77 were sprayed onto 4-week-old plants, which were then placed under plastic covers for 2 (Pst DC3000) or 3 days [Pst DC3000(ΔavrPtoΔavrPtoB)]. Alternatively, Pst DC3000 (OD600 = 0.0002) was syringe-infiltrated into leaves of 4-week-old plants 1 day after 0.5 mM 2DG infiltration, which were then placed under plastic covers for 3 days. Four leaf discs obtained from four independent leaves were pooled. Serially diluted suspensions in 10 mM MgCl2 were plated on NYGA agar medium with rifampicin and incubated at 30°C for 2 days to enumerate colonies.
In vitro kinase assay
The in vitro kinase assay followed a previously described method (24) with some modifications. Recombinant proteins were purified from Escherichia coli BL21 (DE3) with TALON Metal Affinity Resin (Takara), Pierce Glutathione Agarose (Thermo Fisher Scientific), or Amylose Resin [New England Biolabs (NEB)]. Kinases (CPK5, 300 ng; and OST1, 100 ng), their substrates (2000 ng), and phosphatases (1000 ng) were incubated in kinase reaction buffer [100 mM tris-HCl (pH 8.0), 10 mM MgCl2, 2 mM EGTA, and 1 mM dithiothreitol (DTT)] in the absence or presence of 1.5 mM CaCl2 with 100 μM non-radiolabeled adenosine triphosphate (ATP) containing 370 kBq of [γ-32P]ATP (PerkinElmer; NEG002A) for 30 min at 25°C. The buffer Ca2+ concentration was estimated at approximately 120 μM by MaxChelator (Ca/Mg/ATP/EGTA Calculator 2.2b) (https://somapp.ucdmc.ucdavis.edu/pharmacology/bers/maxchelator/). To investigate whether clade A PP2Cs dephosphorylate kinase substrates, kinase reactions were stopped by addition of 100 μM staurosporine (Wako) for 15 min. Then, clade A PP2Cs were added to the reactions and incubated for a further 15 min. The reactions were lastly stopped by the addition of SDS sample buffer and incubated at 95°C for 5 min. The proteins were separated in 10% polyacrylamide gels. The gels were dried after visualization with Coomassie Brilliant Blue (CBB). Radioactivity in the dried gel was detected by FLA5000 (Fujifilm).
Quantitative reverse transcription polymerase chain reaction analysis
Arabidopsis seedlings were grown on agar medium containing 25 mM Suc for 6 days and transferred into 2 ml of liquid medium (25 mM Suc) in 24-well plates to acclimate plants to liquid medium. The next day, after the medium was replaced with fresh liquid medium with 25 mM Suc for PAMP treatment or without sugars for 2DG treatment, 1 μM flg22 peptide (QRLSTGSRINSAKDDAAGLQIA), 1 μM elf18 peptide (Ac-SKEKFERTKPHVNVGTIG), chitin (250 μg/ml), or 0.5 mM 2DG were applied. Chitin solution was prepared by repeatedly grinding crab or shrimp shell powder (Sigma-Aldrich, C9752) in water with a pestle and heating to 60°C. Undissolved particles were removed by centrifugation and filtration. Or, seedlings were grown on agar medium containing 25 mM Suc for 6 days and transferred into 2 ml of liquid medium without sugars in 24-well plates to reset cellular sugar conditions. The next day, after changing to fresh medium with the indicated sugars, plants were further incubated for 24 hours. Harvested plant samples were frozen and ground in liquid N2. Total RNA was isolated using Sepasol RNA I Super G reagent (Nacalai Tesque), followed by treatment with RQ1 ribonuclease (RNase)–Free deoxyribonuclease (DNase) (Promega), and reverse-transcribed using the ReverTra Ace qPCR RT Master Mix with gDNA remover (TOYOBO) following the manufacturer’s instructions. Quantitative polymerase chain reaction (qPCR) was performed with GoTaq qPCR Master Mix (Promega) on a LightCycler 96 (Roche). The expression levels of genes of interest were normalized relative to those of a reference gene, UBQ10. Relative expression (log2) was calculated by subtracting Ct values of genes of interest from those of UBQ10.
Metabolite quantification
For camalexin and salicylic acid (SA) measurement, frozen and ground samples were homogenized with 500 μl of 80% methanol with 3-indolepropionic acid (TCI, I0032) and d4-SA (Toronto Research, S088127) (0.5 μg each) as internal controls for camalexin and SA, respectively. Mixtures were incubated for 30 min at 65°C. After centrifugation, 400 μl of the supernatant was transferred to a new tube. Metabolites were repeatedly extracted from the pellets with 80% methanol without internal controls. For sugar measurement, frozen and ground samples were homogenized with 1 ml of extraction buffer (methanol:water:chloroform, 5:2:2) with 10 μg of ribitol (Wako) as an internal control. Mixtures were incubated for 30 min at 37°C. After centrifugation, 900 μl of the supernatant was transferred to a new tube, and 400 μl of water was added. After centrifugation, the upper phase was retrieved. These samples were evaporated using a spin dryer at 50°C and subsequently freeze-dried. Samples were sonicated in 30 μl of methoxyamine (Sigma-Aldrich) [20 mg/ml dissolved in pyridine (Wako)] and incubated for 90 min at 30°C. Subsequently, 30 μl of MSTFA + 1% TMCS (Thermo Fisher Scientific) was added, and incubation was continued for 30 min at 37°C. After centrifugation, the supernatants were subjected to gas chromatography–mass spectrometry analysis. Each sample (1 μl) was separated on a gas chromatograph (7820A, Agilent Technologies) combined with a mass spectrometric detector (5977B, Agilent Technologies). For quantitative determination of metabolites, peaks that originated from selected ion chromatograms (camalexin 272, SA 267, 3-indolepropionic acid 333, d4-SA 271, Glc 319, G6P 387, 2DG 319, 2DG6P 387, and ribitol 319) were used.
Detection of CPK5 phosphorylation
Arabidopsis seedlings were grown on agar medium containing 25 mM Suc for 6 days and then transferred into 2 ml of liquid medium with 25 mM Suc in 24-well plate. The next day, the medium was replaced with fresh liquid medium with 25 mM Suc for flg22 treatment or without sugars for 2DG treatment. Plants were treated with 1 μM flg22 peptide or 0.5 mM 2DG. In addition, for Glc treatment, seedlings were grown on agar medium containing 25 mM Suc for 6 days and transferred into 2 ml of liquid medium without sugars i n 24-well plates to reset cellular sugar conditions. The next day, the medium was replaced with fresh medium containing the indicated concentrations of Glc. Plants were incubated for 24 hours and then treated with 1 μM flg22. After freezing samples and grinding them in liquid N2, proteins were extracted with lysis buffer [50 mM tris-HCl (pH 7.5), 300 mM NaCl, 15% glycerol, 1% Triton X-100, and 2 mM DTT]. Lambda protein phosphatase (BioAcademia) treatment was for 30 min at 30°C. Proteins were separated in 8% polyacrylamide gels containing 50 μM Phos-tag Acrylamide (Wako) and 100 μM MnCl2. CPK5-GFP was detected with anti-GFP antibody (B-2, Santa Cruz Biotechnology). Band intensity was measured using ImageJ. For detecting in vitro CPK5 phosphorylation status, HisT7-CPK5 (250 ng) and glutathione S-transferase (GST)–ABI1 (500 ng) were incubated with reaction buffer [100 mM tris-HCl (pH 8.0), 10 mM MgCl2, 2 mM EGTA, 1 mM DTT, 1 mM ATP, 1 mM MnCl2, and protease inhibitor cocktail (EDTA-free) (Nacalai)] in the absence or presence of the indicated sugars for 45 min at 30°C. The reactions were stopped by addition of SDS sample buffer and incubated at 95°C for 5 min. The proteins were separated in 10% polyacrylamide gels. CPK5 phosphorylation status was detected using anti-phosphothreonine antibody (9381S, Cell Signaling Technology). The membranes were stained with CBB.
Hexokinase activity assay
Hexokinase activity was measured using an enzyme-linked assay following a previously described method (53) with some modifications. GST-fused recombinant hexokinases were purified from E. coli BL21 (DE3) with Pierce Glutathione Agarose (Thermo Fisher Scientific) and incubated in reaction buffer [50 mM Hepes-NaOH (pH 7.5), 2 mM MgCl2, 1 mM EDTA, 9 mM KCl, 1 mM nicotinamide adenine dinucleotide (NAD), 1 mM ATP, 2 mM Glc, and 0.8 U of NAD-dependent G6P dehydrogenase (TOYOBO, G6D-321)] at room temperature. Absorbance at 340 nm was monitored continuously every 5 s with a spectrophotometer (MULTISKAN FC; Thermo Fisher Scientific) to detect the reduced form of NAD (oxidized form) production.
Protein interaction assay
Yeast two-hybrid assays were performed using strain AH109 (Takara). Vectors pGADT7 and pGBKT7 were sequentially transformed into this strain. Transformants were incubated on medium without histidine, leucine, and tryptophan to detect protein interactions. In addition, 1 mM 3-AT was optionally added to medium to investigate strong protein interactions. For split-luciferase assays, Agrobacterium strains (GV3101) transformed with nLuc or cLuc plasmids in infiltration buffer [10 mM MES-KOH (pH 5.5) and 10 mM MgCl2] were simultaneously infiltrated into Nicotiana benthamiana. Leaves. After 2 days, 10 mM luciferin (P1041, Promega) in infiltration buffer was infiltrated into the Agrobacterium-infected leaves. Luminescence was detected using LAS4000 (Cytiva). Luminescence data were colored using ImageJ. For pull-down assays, maltose-binding protein (MBP)–CPK5 (5000 ng) and GST or GST-ABI1 (1000 ng) were added into 100 μl of reaction buffer [100 mM Hepes (pH 7.5), 10 mM MgCl2, 2 mM EGTA, and 2 mM DTT] in the presence/absence of the indicated sugars (10 mM). After incubation for 30 min at 25°C, 900 μl of interaction buffer [50 mM tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 2 mM DTT, 15% glycerol, and 1% Triton X-100] was added to the reactions with MagneGST Glutathione Particles (Promega) in the presence/absence of the indicated sugars (10 mM). After mixing the tubes at 4°C for 1 hour and 15 min, the beads were washed with 1 ml of interaction buffer three times. SDS sample buffers were added, and the tubes were incubated at 95°C for 5 min. The proteins were separated in 10% polyacrylamide gels. MBP-tagged and GST-tagged proteins were detected using anti-MBP (E8032S, NEB) and anti-GST antibody (60-021, BioAcademia), respectively.
Aequorin luminescence measurement
Six-day-old Arabidopsis seedlings expressing p35S-apoaequorin (pMAQ2) grown on MS agar medium with 25 mM Suc were transferred into 200 μl of liquid medium with 20 μM coelenterazine h (Wako) in 96-well plates. The next day, after transfer to fresh medium with 20 μM coelenterazine h, luminescence was recorded using a luminometer (LUMINOSKAN; Thermo Fisher Scientific).
Measurement of 2DG influx activity
Glc influx activity into Arabidopsis seedlings was analyzed by measuring 2DG influx activity. While Glc is immediately metabolized after absorption into cells, 2DG is not further catabolized after HXK-mediated phosphorylation. We therefore estimated Glc influx activity by measuring the accumulation of 2DG and 2DG6P. Arabidopsis seedlings were grown on agar medium containing 25 mM Suc for 6 days and then transferred into 2 ml of liquid medium with 25 mM Suc in a 24-well plate. The next day, the medium was replaced with fresh liquid medium containing 25 mM Suc. Plants were treated with 1 μM flg22 peptide or chitin (250 μg/ml). After 8 hours, the medium was replaced with fresh liquid medium without Suc. Plants were harvested 1 hour after 0.5 mM 2DG treatment. Accumulation of 2DG and 2DG6P was measured using the metabolite analysis described above.
RNA-seq and data analysis
Total RNAs were extracted using Sepazol-RNA I Super G (Nacalai Tesque), followed by treatment with RQ1 RNase-Free DNase (Promega). These DNase-treated RNAs (500 ng) were used for library preparation using BrAD-seq to create strand-specific 3′ digital gene expression libraries (54). The libraries were sequenced on a HiSeq X Ten platform at Macrogen (Tokyo, Japan), producing 150-bp paired-end reads. Because the quality of reverse reads was poor due to the poly(A) sequence, only forward reads were used for the analysis. The first eight bases and adaptors were trimmed and quality filtering was performed by fastp (version 0.19.7) (55). The trimmed and quality-filtered reads were mapped to the Arabidopsis genome (TAIR10) using STAR (version 2.6.1b) (56) and transformed to a count per gene per library using featureCounts (version 1.6.0) (57). Statistical analysis of the RNA-seq data was performed in the R environment (version 3.5.3). Because BrAD-seq involves poly(A) enrichment, mitochondrial and chloroplast genes were excluded. Genes with mean read counts of fewer than 10 per library were considered to be expressed at low levels and were excluded from the analysis. The resulting count data were subjected to trimmed-mean of M-value normalization using the function calcNormFactors in the package edgeR, followed by log transformation by the function voom in the package limma to yield log2 counts per million. To each gene, a linear model was fit by the function lmFit in the limma package with the following terms: Sg = Gg + Rr + ɛg, where S is log2 expression value, G is genotype, R is biological replicate and ɛ is residual; or Sgt = GTgt + Rr + ɛgt, where S is log2 expression value, GT is genotype:treatment interaction, R is biological replicate, and ɛ is residual. For variance shrinkage in the calculation of P values, the eBayes function in the limma package was used; the resulting P values were then corrected for multiple hypothesis testing by calculating Storey’s q values using the function qvalue in the package qvalue. To extract genes with significant expression changes, cutoffs of q < 0.01 and |log2FC| > 1 were applied. To create heatmaps, average linkage hierarchical clustering with uncentered Pearson correlation as a distance measure was carried out using CLUSTER (58), followed by visualization using TREEVIEW (58).
Data availability
The RNA-seq data used in this study were deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (accession no. GSE169473). Details of the parameters used for quality filtering, mapping, and counting are available in the Gene Expression Omnibus submission.
Statistics
All data displayed in this study were analyzed using R. No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those generally used in the field. For plant studies, plants were rotated several times in growth chambers to minimize allocation effects. Samples were not blinded for allocation or data analyses. In box plots, center lines represent the medians, box edges delimit lower and upper quartiles, and whiskers show the highest and lowest data points.
Acknowledgments
We thank R. Matsumoto (Tokushima University) for technical assistance; Y. Takano (Kyoto University) for providing reagents; and the Advanced Radiation Research, Education, and Management Center (Tokushima University) for radioisotope experiments.
Funding: This work was supported by JST PRESTO (JPMJPR17Q9, K.Y.; and JPMJPR17Q6, A.M.), JSPS KAKENHI (20K21280 and 21H02157, K.Y.), the Sumitomo Foundation (K.Y.), and the Mitsubishi Foundation (K.Y.).
Author contributions: Conceptualization: K.Y. Methodology: K.Y. and A.M. Investigation: K.Y. and A.M. Visualization: K.Y. and A.M. Writing—original draft: K.Y. Writing—review and editing: K.Y. and A.M.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Sequencing data are available in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database (accession no. GSE169473).
Supplementary Materials
This PDF file includes:
Figs. S1 to S12
Table S1
REFERENCES AND NOTES
- 1.Couto D., Zipfel C., Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16, 537–552 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Jang J.-C., Sheen J., Sugar sensing in higher plants. Plant Cell 6, 1665–1679 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sheen J., Zhou L., Jang J.-C., Sugars as signaling molecules. Curr. Opin. Plant Biol. 2, 410–418 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Gómez-Ariza J., Campo S., Rufat M., Estopà M., Messeguer J., Segundo B. S., Coca M., Sucrose-mediated priming of plant defense responses and broad-spectrum disease resistance by overexpression of the maize pathogenesis-related PRms protein in rice plants. MPMI 20, 832–842 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Tsutsui T., Nakano A., Ueda T., The plant-specific RAB5 GTPase ARA6 is required for starch and sugar homeostasis in Arabidopsis thaliana. Plant and Cell Physiology 56, 1073–1083 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Moore B., Zhou L., Rolland F., Hall Q., Cheng W.-H., Liu Y.-X., Hwang I., Jones T., Sheen J., Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300, 332–336 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Xiao W., Sheen J., Jang J.-C., The role of hexokinase in plant sugar signal transduction and growth and development. Plant Mol. Biol. 44, 451–461 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Bruggeman Q., Prunier F., Mazubert C., de Bont L., Garmier M., Lugan R., Benhamed M., Bergounioux C., Raynaud C., Delarue M., Involvement of Arabidopsis Hexokinase1 in cell death mediated byMyo-Inositol accumulation. Plant Cell 27, 1801–1814 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boudsocq M., Willmann M. R., McCormack M., Lee H., Shan L., He P., Bush J., Cheng S.-H., Sheen J., Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464, 418–422 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou J., Wang X., He Y., Sang T., Wang P., Dai S., Zhang S., Meng X., Differential phosphorylation of the transcription factor WRKY33 by the protein kinases CPK5/CPK6 and MPK3/MPK6 cooperatively regulates camalexin biosynthesis in arabidopsis. Plant Cell 32, 2621–2638 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang L., Zhang Y., Guan R., Li S., Xu X., Zhang S., Xu J., Co-regulation of indole glucosinolates and camalexin biosynthesis by CPK5/CPK6 and MPK3/MPK6 signaling pathways. J. Integr. Plant Biol. 62, 1780–1796 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu N., Hake K., Wang W., Zhao T., Romeis T., Tang D., CALCIUM-DEPENDENT PROTEIN KINASE5 associates with the truncated NLR protein TIR-NBS2 to contribute to exo70B1-mediated immunity. Plant Cell 29, 746–759 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leidreiter K., Kruse A., Heineke D., Robinson D. G., Heldt H.-W., Subcellular volumes and metabolite concentrations in potato (Solanum tuberosum cv. Désirée) Leaves†. Botanica Acta 108, 439–444 (1995). [Google Scholar]
- 14.Boorer K. J., Loo D. D., Wright E. M., Steady-state and presteady-state kinetics of the H+/hexose cotransporter (STP1) from Arabidopsis thaliana expressed in Xenopus oocytes. J. Biol. Chem. 269, 20417–20424 (1994). [PubMed] [Google Scholar]
- 15.Sols A., Crane R. K., Substrate specificity of brain hexokinase. J. Biol. Chem. 210, 581–595 (1954). [PubMed] [Google Scholar]
- 16.Wick A. N., Drury D. R., Nakada H. I., Wolfe J. B., Britton B., Grabowski R., Localization of the primary metabolic block producED BY 2-deoxyglucose. J. Biol. Chem. 224, 963–969 (1957). [PubMed] [Google Scholar]
- 17.Yamada K., Kanai M., Osakabe Y., Ohiraki H., Shinozaki K., Yamaguchi-Shinozaki K., Monosaccharide absorption activity of arabidopsis roots depends on expression profiles of transporter genes under high salinity conditions. J. Biol. Chem. 286, 43577–43586 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohto M., Nakamura K., Sugar-induced increase of calcium-dependent protein kinases associated with the plasma membrane in leaf tissues of tobacco. Plant Physiol. 109, 973–981 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao X., Chen X., Lin W., Chen S., Lu D., Niu Y., Li L., Cheng C., McCormack M., Sheen J., Shan L., He P., Bifurcation of Arabidopsis NLR immune signaling via Ca2+-dependent protein kinases. PLOS Pathog. 9, e1003127 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lynch T., Erickson B. J., Finkelstein R. R., Direct interactions of ABA-insensitive(ABI)-clade protein phosphatase(PP)2Cs with calcium-dependent protein kinases and ABA response element-binding bZIPs may contribute to turning off ABA response. Plant Mol. Biol. 80, 647–658 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Brandt B., Munemasa S., Wang C., Nguyen D., Yong T., Yang P. G., Poretsky E., Belknap T. F., Waadt R., Alemán F., Schroeder J. I., Calcium specificity signaling mechanisms in abscisic acid signal transduction in Arabidopsis guard cells. eLife 4, e03599 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kadota Y., Sklenar J., Derbyshire P., Stransfeld L., Asai S., Ntoukakis V., Jones J. D., Shirasu K., Menke F., Jones A., Zipfel C., Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 54, 43–55 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Geiger D., Scherzer S., Mumm P., Marten I., Ache P., Matschi S., Liese A., Wellmann C., Al-Rasheid K. A. S., Grill E., Romeis T., Hedrich R., Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc. Natl. Acad. Sci. 107, 8023–8028 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamada K., Saijo Y., Nakagami H., Takano Y., Regulation of sugar transporter activity for antibacterial defense in Arabidopsis. Science 354, 1427–1430 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Gust A. A., Biswas R., Lenz H. D., Rauhut T., Ranf S., Kemmerling B., Götz F., Glawischnig E., Lee J., Felix G., Nürnberger T., Bacteria-derived peptidoglycans constitute pathogen-associated molecular patterns triggering innate immunity in arabidopsis. J. Biol. Chem. 282, 32338–32348 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Stoltzman C. A., Peterson C. W., Breen K. T., Muoio D. M., Billin A. N., Ayer D. E., Glucose sensing by MondoA:Mlx complexes: A role for hexokinases and direct regulation of thioredoxin-interacting protein expression. Proc. Natl. Acad. Sci. U.S.A. 105, 6912–6917 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jossier M., Bouly J.-P., Meimoun P., Arjmand A., Lessard P., Hawley S., Grahame Hardie D., Thomas M., SnRK1 (SNF1-related kinase 1) has a central role in sugar and ABA signalling in Arabidopsis thaliana. Plant J. 59, 316–328 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Toroser D., Plaut Z., Huber S. C., Regulation of a plant SNF1-related protein kinase by glucose-6-phosphate. Plant Physiol. 123, 403–412 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kraepiel Y., Barny M., Gram-negative phytopathogenic bacteria, all hemibiotrophs after all? Mol. Plant Pathol. 17, 313–316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flors V., Ton J., Van Doorn R., Jakab G., García-Agustín P., Mauch-Mani B., Interplay between JA, SA and ABA signalling during basal and induced resistance against Pseudomonas syringae and Alternaria brassicicola. Plant J. 54, 81–92 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Chang Q., Liu J., Wang Q., Han L., Liu J., Li M., Huang L., Yang J., Kang Z., The effect of Puccinia striiformis f. sp. tritici on the levels of water-soluble carbohydrates and the photosynthetic rate in wheat leaves. Physiol. Mol. Plant Pathol. 84, 131–137 (2013). [Google Scholar]
- 32.Sutton P. N., Gilbert M. J., Williams L. E., Hall J. L., Powdery mildew infection of wheat leaves changes host solute transport and invertase activity. Physiol. Plant. 129, 787–795 (2007). [Google Scholar]
- 33.Liu J., Liu M., Tan L., Huai B., Ma X., Pan Q., Zheng P., Wen Y., Zhang Q., Zhao Q., Kang Z., Xiao S., AtSTP8, an endoplasmic reticulum-localised monosaccharide transporter from Arabidopsis, is recruited to the extrahaustorial membrane during powdery mildew infection. New Phytol. 230, 2404–2419 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Veillet F., Gaillard C., Lemonnier P., Coutos-Thévenot P., La Camera S., The molecular dialogue between Arabidopsis thaliana and the necrotrophic fungus Botrytis cinerea leads to major changes in host carbon metabolism. Sci. Rep. 7, 17121 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Albert I., Böhm H., Albert M., Feiler C. E., Imkampe J., Wallmeroth N., Brancato C., Raaymakers T. M., Oome S., Zhang H., Krol E., Grefen C., Gust A. A., Chai J., Hedrich R., Van den Ackerveken G., Nürnberger T., An RLP23–SOBIR1–BAK1 complex mediates NLP-triggered immunity. Nature Plants 1, 1–9 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Ono E., Mise K., Takano Y., RLP23 is required for Arabidopsis immunity against the grey mould pathogen Botrytis cinerea. Sci. Rep. 10, 13798 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bjornson M., Pimprikar P., Nürnberger T., Zipfel C., The transcriptional landscape of Arabidopsis thaliana pattern-triggered immunity. Nat. Plants 7, 579–586 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim Y., Tsuda K., Igarashi D., Hillmer R. A., Sakakibara H., Myers C. L., Katagiri F., Mechanisms underlying robustness and tunability in a plant immune signaling network. Cell Host Microbe 15, 84–94 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gibon Y., Bläsing O. E., Palacios-Rojas N., Pankovic D., Hendriks J. H. M., Fisahn J., Höhne M., Günther M., Stitt M., Adjustment of diurnal starch turnover to short days: Depletion of sugar during the night leads to a temporary inhibition of carbohydrate utilization, accumulation of sugars and post-translational activation of ADP-glucose pyrophosphorylase in the following light period. Plant J. 39, 847–862 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Maruyama K., Takeda M., Kidokoro S., Yamada K., Sakuma Y., Urano K., Fujita M., Yoshiwara K., Matsukura S., Morishita Y., Sasaki R., Suzuki H., Saito K., Shibata D., Shinozaki K., Yamaguchi-Shinozaki K., Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol. 150, 1972–1980 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Engelsdorf T., Horst R. J., Pröls R., Pröschel M., Dietz F., Hückelhoven R., Voll L. M., Reduced carbohydrate availability enhances the susceptibility of arabidopsis toward Colletotrichum higginsianum. Plant Physiol. 162, 225–238 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim Y. S., An C., Park S., Gilmour S. J., Wang L., Renna L., Brandizzi F., Grumet R., Thomashow M. F., CAMTA-mediated regulation of salicylic acid immunity pathway genes in arabidopsis exposed to low temperature and pathogen infection. Plant Cell 29, 2465–2477 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu W.-X., Zhang F.-C., Zhang W.-Z., Song L.-F., Wu W.-H., Chen Y.-F., Arabidopsis Di19 functions as a transcription factor and modulates PR1, PR2, and PR5 expression in response to drought stress. Mol. Plant 6, 1487–1502 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Hubbard K. E., Siegel R. S., Valerio G., Brandt B., Schroeder J. I., Abscisic acid and CO2 signalling via calcium sensitivity priming in guard cells, new CDPK mutant phenotypes and a method for improved resolution of stomatal stimulus-response analyses. Ann. Bot. 109, 5–17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng Z., Qamar S. A., Chen Z., Mengiste T., Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48, 592–605 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Wildermuth M. C., Dewdney J., Wu G., Ausubel F. M., Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565 (2001). [DOI] [PubMed] [Google Scholar]
- 47.Glazebrook J., Ausubel F. M., Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc. Natl. Acad. Sci. 91, 8955–8959 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rubio S., Rodrigues A., Saez A., Dizon M. B., Galle A., Kim T.-H., Santiago J., Flexas J., Schroeder J. I., Rodriguez P. L., Triple Loss of Function of Protein Phosphatases Type 2C Leads to Partial Constitutive Response to Endogenous Abscisic Acid. Plant Physiology 150, 1345–1355 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Knight M. R., Campbell A. K., Smith S. M., Trewavas A. J., Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature 352, 524–526 (1991). [DOI] [PubMed] [Google Scholar]
- 50.Tsutsui H., Higashiyama T., pKAMA-ITACHI vectors for highly efficient CRISPR/Cas9-mediated gene knockout in Arabidopsis thaliana. Plant Cell Physiol. 58, 46–56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okegawa Y., Motohashi K., A simple and ultra-low cost homemade seamless ligation cloning extract (SLiCE) as an alternative to a commercially available seamless DNA cloning kit. Biochem. Biophys. Rep. 4, 148–151 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gachon C., Saindrenan P., Real-time PCR monitoring of fungal development in Arabidopsis thaliana infected by Alternaria brassicicola and Botrytis cinerea. Plant Physiol. Biochem. 42, 367–371 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Feng J., Zhao S., Chen X., Wang W., Dong W., Chen J., Shen J.-R., Liu L., Kuang T., Biochemical and structural study of Arabidopsis hexokinase 1. Acta Crystallogr. D Biol. Crystallogr. 71, 367–375 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Townsley B. T., Covington M. F., Ichihashi Y., Zumstein K., Sinha N. R., BrAD-seq: Breath Adapter Directional sequencing: A streamlined, ultra-simple and fast library preparation protocol for strand specific mRNA library construction. Front. Plant Sci. 6, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen S., Zhou Y., Chen Y., Gu J., Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Eisen M. B., Spellman P. T., Brown P. O., Botstein D., Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. 95, 14863–14868 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S12
Table S1
Data Availability Statement
The RNA-seq data used in this study were deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (accession no. GSE169473). Details of the parameters used for quality filtering, mapping, and counting are available in the Gene Expression Omnibus submission.