Abstract
The smooth and precise transition from totipotency to pluripotency is a key process in embryonic development, generating pluripotent stem cells capable of forming all cell types. While endogenous retroviruses (ERVs) are essential for early development, their precise roles in this transition remains mysterious. Using cutting-edge genetic and biochemical techniques in mice, we identify MERVL-gag, a retroviral protein, as a crucial modulator of pluripotent factors OCT4 and SOX2 during lineage specification. MERVL-gag tightly operates with URI, a prefoldin protein that concurs with pluripotency bias in mouse blastomeres, and which is indeed required for totipotency-to-pluripotency transition. Accordingly, URI loss promotes a stable totipotent-like state and embryo arrest at 2C stage. Mechanistically, URI binds and shields OCT4 and SOX2 from proteasome degradation, while MERVL-gag displaces URI from pluripotent factor interaction, causing their degradation. Our findings reveal the symbiotic coevolution of ERVs with their host cells to ensure the smooth and timely progression of early embryo development.
Endogenous retroviral protein intricately guides pluripotent transition in mouse embryos, engaging a symbiotic dance with URI.
INTRODUCTION
The transition of a fertilized oocyte into a totipotent zygote marks the beginning of embryonic development. This transition relies on extensive replacement of the maternal transcriptome by the zygotic genome activation, together with well-orchestrated epigenetic reprogramming of both parental nuclei (1–3). The totipotent zygote generates the entire living organism by later segregating the two first cell lineages to establish the pluripotent inner cell mass (ICM), as well as the surrounding outer trophectoderm (TE) layer that nourishes and sustains development. Within the ICM, a second lineage specification takes place at the late preimplantation stage to form both the pluripotent epiblast (EPI) from which the embryo proper derives as well as the primitive endoderm (PrE) that will give rise to the extraembryonic endoderm layers of the yolk sac. TE, EPI, and PrE cells constitute the blastocyst, a structure formed at the last preimplantation stage.
The octamer-binding 4 (OCT4) and the sex-determining region Y box 2 (SOX2) transcriptional factors are indispensable to the foundation of pluripotency in the early embryo, and for the complete segregation of the EPI and TE lineages between eight-cell (8C) stage to the early blastocyst before implantation (4). However, in the early 2C embryo, which is considered to have two equally totipotent blastomeres, differences in the developmental potential and pluripotency lineage contribution between the two blastomeres have been reported (5–7). OCT4 and SOX2 activity in the early embryo blastomeres is known to predict cell lineage contribution. Specifically, long stable binding of OCT4 and SOX2 to DNA, as well as their heterogeneous target gene expression patterns reportedly bias cell fate in the 4C mouse embryo (7–9). Furthermore, their DNA binding depends on histone H3 arginine 26 (H3R26) methylation levels (8, 9), which are controlled by the coactivator-associated arginine methyltransferase 1 (CARM1), which is heterogeneously expressed at the first steps of murine development (7, 10, 11). However, the mechanisms that control the stability of the pluripotency factors that are indispensable for cell fate decisions remain to be elucidated. Approximatively, 10% of the mammalian genome is composed of nontransposable endogenous retroviruses (ERVs), remnants of ancient viral infections that have become integrated into the genome of their hosts, and erroneously considered to be nonfunctional “junk” DNA. ERVs become active in a specific pattern during embryo development, potentially regulating chromatin organization and transcription (12–14). The murine class III ERV with leucine transfer RNA primer binding site (MERVL) is specifically expressed during the zygotic genome activation at the 2C stage (12, 15), is one of the first transcripts expressed in the totipotent embryo, and reportedly acts as a proximal promoter to drive genes related to totipotency (12). Accordingly, activation of MERVL in pluripotent stem cells can induce a 2C-like cell state (14, 16, 17). Moreover, while derepression of ERVs can be lethal at various embryonic stages (18–21), depletion of MERVL specifically causes lineage segregation failure in early mouse embryo (22, 23). MERVL also encodes the group-specific antigen (gag) protein, known as MERVL-gag which forms virus-like particles (15); however, the mechanism by which ERVs control cell fate in the early embryo at the transition from totipotency to pluripotency requires further investigation. Understanding this mechanism is crucial, as the stability of pluripotency factors is indispensable for cell fate decisions during the timely progression of embryonic development.
The unconventional prefoldin RPB5 interactor (URI) is an atypically large member of the unconventional prefoldin complex (24, 25). Although the function of the URI prefoldin-like complex remains largely unknown, prefoldins have proven to be essential for the health and integrity of cells, protecting against unfolded protein aggregation (25). URI has been initially identified as a downstream component of the nutrient and growth factor signalling cascade and has been shown to coordinate nutrient availability with gene expression (24, 26). Moreover, findings indicate that overexpressing URI in mice facilitates oncogenic activity (27–29), while its genetic ablation in adult mice leads to organ failure (28, 30). Maintaining homeostatic URI levels is therefore essential to preserve organ homeostasis during adulthood, and hence, URI might have a critical role during mouse embryo development. This is supported by studies conducted in Caenorhabditis elegans and Drosophila showing that deletion of prefoldin results in embryonic lethality (31, 32). However, it remains unknown whether URI is required for early embryonic development and how it interacts with key players in the first lineage specifications during preimplantation mouse embryos.
RESULTS
URI concurs with blastomere pluripotency bias in the early mouse embryo
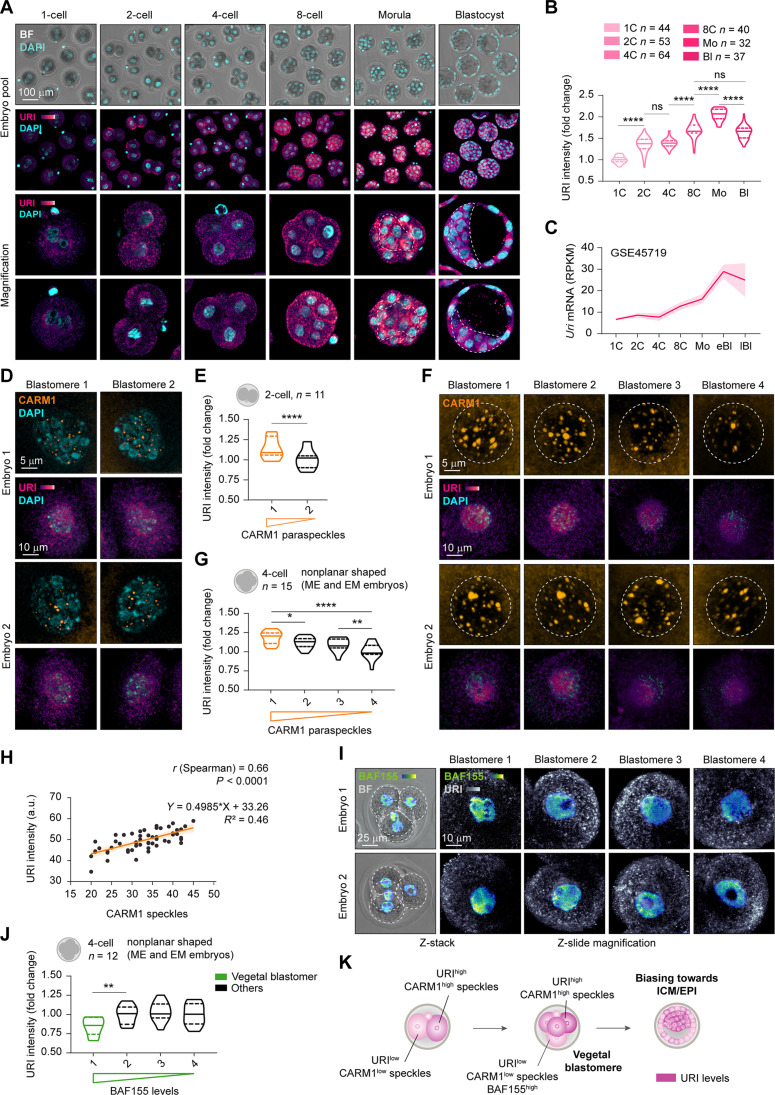
To study the role of URI in early embryonic development, we examined its expression along a preimplanted mouse embryo. We found that its protein and mRNA levels were gradually increased until the final blastocyst stage (Fig. 1, A to C, and table S1). Immunofluorescence assays demonstrated that URI was heterogeneously expressed between the 2C blastomeres, with a modest increased expression in the high CARM1 blastomere, reported to contribute predominantly to the embryonic axis (mainly the ICM and polar TE) (Fig. 1, D and E, and fig. S1, A to C) (7, 10, 11). At this stage, highly variable genes known to contribute to cell-to-cell variation (33) were detected in 2C embryos by single-cell RNA sequencing (scRNA-seq), and the results showed no differences in Uri mRNA levels, either by gene dispersion analysis or by hierarchical clustering of blastomeres (fig. S1, D to G, and table S1).
Fig. 1. URI concurs with blastomere pluripotency bias in the early mouse embryo.
(A) Immunofluorescence (IF) of URI in murine preimplanted embryos at different time points of embryonic development. Bottom represents magnifications for single embryo; dotted lines indicate ICM compartment. Scale bar, 100 μm. BF, bright-field. (B) URI intensity across early murine development from (A). One-way analysis of variance (ANOVA; Tukey post hoc test); ****P < 0.0001; ns, nonsignificant. (C) Uri expression along murine preimplantation development from normalized RNA-seq dataset analysis. Data are represented as mean of pooled embryos ±95% confidence intervals. (D) IF of URI and CARM1 speckles in mouse 2C embryos. Scale bars, 10 and 5 μm. (E) URI intensity in high versus low CARM1 speckles in 2C blastomeres from (D). Paired t test; ****P < 0.0001. (F) IF of URI and CARM1 nuclear speckles in 4C embryos. Scale bars, 10 and 5 μm. (G) URI intensity in high versus low CARM1 speckles number from (F). Matched one-way ANOVA (Tukey correction); *P < 0.05, **P < 0.01, and ****P < 0.0001. (H) Linear regression and correlation analysis of URI and CARM1 speckles number in 4C embryos from (F). a.u., arbitrary units. (I) IF of URI and BAF155 in mouse 4C blastomeres. Scale bars, 25 and 10 μm. (J) URI intensity in high versus low BAF155 blastomeres from (I). Matched one-way ANOVA (Geisser-Greenhouse correction and Holm-Sidak post hoc test); **P < 0.01. (K) Scheme summarizing URI expression among blastomeres at early developmental stages. Total number of embryos is referred in each panel. Repository accession number for sequencing dataset analysis are indicated in respective panel and compiled in table S1. Mo, Morula embryo; eBl, early blastocyst embryo; lBl, late blastocyst embryo.
The 4C embryo gathers greater blastomeric differences at transcriptomic levels, which are responsible for the greater lineage-dependent bias and developmental potency compared to 2C embryos (7, 34). As seen in 2C embryos, coimmunofluorescence analysis indicated that URI remained heterogeneously expressed among blastomeres; its expression was low in the BAF155high vegetal blastomere, which mostly contributes to the TE lineage (35), whereas it concurred with high CARM1 levels (Fig. 1, F to J, and fig. S1, H to J). Moreover, analysis of scRNA-seq datasets conducted in 4C embryos and combined with single-cell gene set enrichment analysis (GSEA) revealed that Uri was among the most variable genes and was possibly targeted by OCT4 and SOX2 as its levels correlated with a highly variable OCT4 and SOX2 target signature (fig. S1, K to Q, and table S1). Accordingly, chromatin immunoprecipitation sequencing analysis (ChIP-seq) performed in pluripotent mouse embryonic stem cells (mESCs) indicated that Uri expression was regulated by OCT4 and SOX2 via binding to an active superenhancer region, which are known to be key nodes for cell identity and tightly regulated during cell state transitions (fig. S1, R and S, and table S1) (36). Therefore, as reported for OCT4 and SOX2 and their target genes (7–9), URI expression might concur with pluripotency bias in mouse blastomeres (Fig. 1K).
Increased URI levels correlate with pluripotent identity
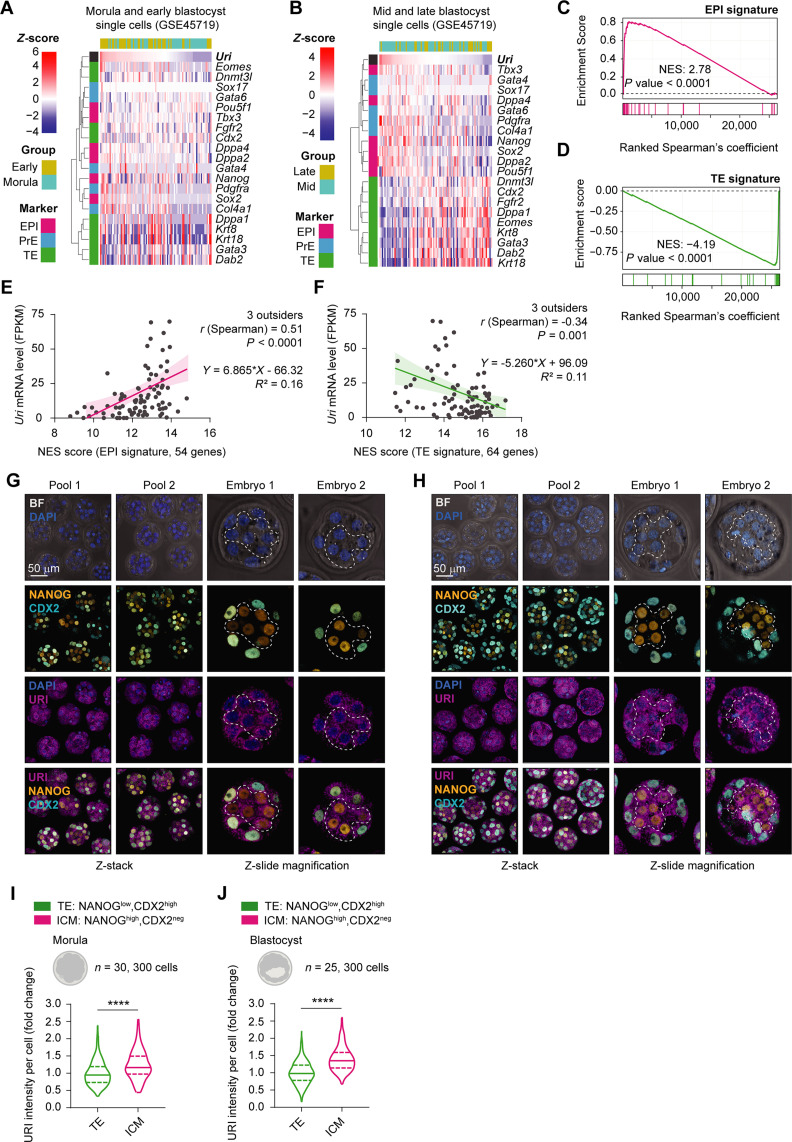
Although URI levels were found to continuously increase across preimplantation stage, in parallel, a heterogenous URI pattern was observed, correlating with pluripotency biases in the 2C and 4C embryos. We therefore sought to establish whether this correlation was maintained across the preimplantation process by analysing publicly available RNA-seq datasets across morulas and blastocysts. The results indicated that Uri expression was positively associated with the pluripotent EPI lineages and negatively associated with the TE signature in mid and late blastocysts (Fig. 2, A to F, and table S1). In addition, coimmunofluorescence analysis in the morula and blastocyst stages demonstrated that URI expression was slightly higher in the NANOG-positive EPI/ICM compartment but lower in the CDX2-positive TE (Fig. 2, G to J). Subsequently, the results of assay for transposase-accessible chromatin (ATAC-seq), deoxyribonuclease (DNase) I hypersensitive sites sequencing (DNase-seq), and H3K4me3 ChIP-seq conducted in preimplanted embryos confirmed that the Uri enhancer region was accessible at the onset of pluripotency, shortly after the 4C embryo stage (fig. S2, A to E, and table S1). In line with these findings, ChIP-seq analysis of mESCs and trophoblast stem cells (TSCs) revealed decreased accessibility and activity in the Uri enhancer locus (where binding of essential transcription factors was decreased), clarifying the differences in URI level expression between EPI and TE cells (fig. S2, F to L, and table S1). Therefore, high URI expression correlates with pluripotent identity during embryonic development.
Fig. 2. High URI expression is restricted to the EPI compartment.
(A) Heatmap showing Uri-ranked mRNA expression of EPI (fuchsia), PrE (blue), and TE (green) marker genes in morula and early blastocysts from scRNA-seq datasets. (B) Heat map showing Uri-ranked mRNA expression of EPI, PrE, and TE marker genes in mid and late blastocyst single cells from referenced RNA-seq datasets. (C and D) GSEA showing EPI (C) or TE (D) gene signature distribution in ranked gene correlation with Uri expression from (A and B, respectively). (E and F) Linear regression analysis of normalized Uri mRNA levels and normalized enrichment score (NES) of GSEA for EPI (C) or TE signature (D). (G and H) IF of URI, NANOG, and CDX2 in pooled morula (G) or blastocyst embryos (H). Magnification pictures for single embryos are shown. Dashed lines delimit ICM compartment. Scale bar, 50 μm. (I and J) URI intensity in ICM and TE compartment from morula (I) or blastocyst (J) embryos from (G and H, respectively). t test; ****P < 0.0001. Total number of embryos is referred in each panel. Repository accession number for sequencing dataset analysis are indicated in respective panel and compiled in table S1. Gene signature lists are arranged in table S2.
Genetic URI ablation impairs postimplantation development
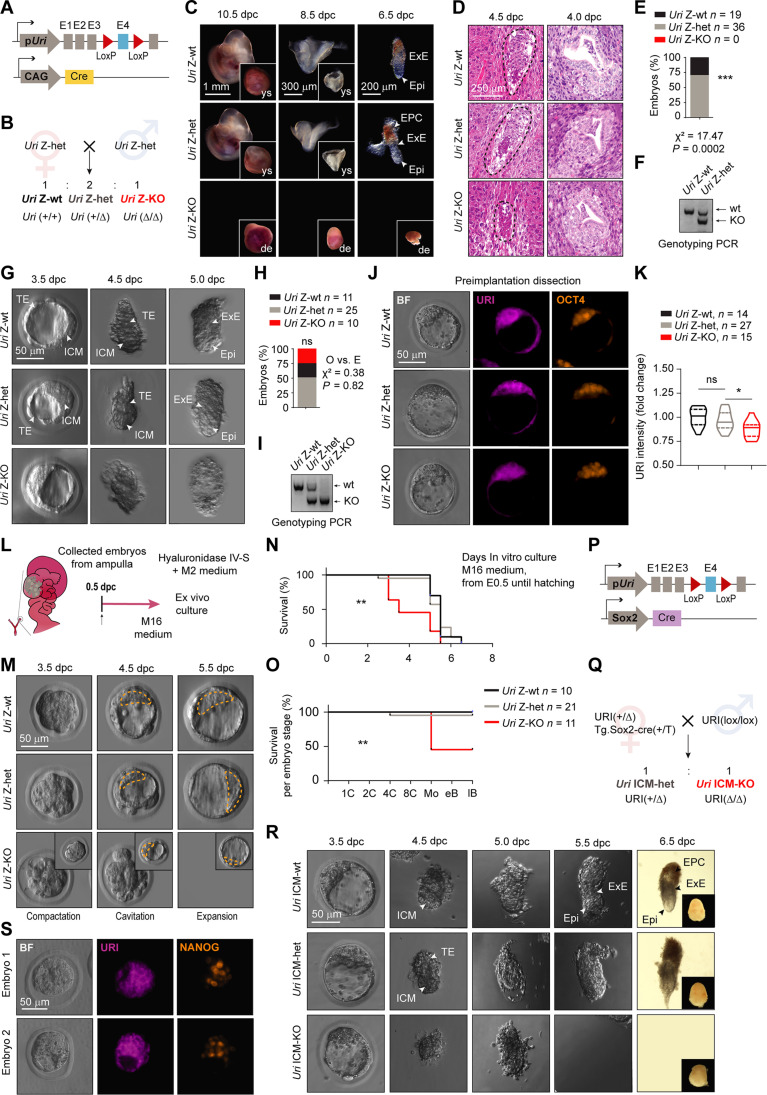
The above data suggest that URI is required for pluripotency onset to guide the totipotent-to-pluripotent transition during preimplantation mouse embryo development. Thus, to explore the function of URI in vivo in preimplanted embryos, we generated Uri zygotic-knockout (Uri Z-KO) embryos by crossing Tg.CAG-Cre mice, carrying Cre recombinase under the control of the hybrid CMV enhancer chicken β-actin promoter (CAG) element (37), with conditional Uri knockout (URI lox) (30) mice (Fig. 3, A and B). Embryos with heterozygous URI ablation (Uri Z-het) developed in line with their littermate wild-type embryos (Uri Z-wt), while Uri Z-KO embryos displayed an unexpected late phenotype, with a defective implantation process leading to early embryonic death around 5.0 days after conception (Fig. 3, C to I). Immunofluorescence revealed residual URI levels in Uri Z-KO blastocysts (Fig. 3, J and K), most likely due to the contribution and compensation of maternal Uri mRNA from the oocyte. Uri Z-KO embryos reached a well-formed and integrous blastocyst in vivo with the expected Mendelian proportion but showed lower developmental efficiency when cultured ex vivo (Fig. 3, L to O).
Fig. 3. URI Z-KO embryos undergo lethality at implantation stage.
(A) Scheme of Uri Z-KO embryo. (B) Breeding strategy to generate Uri zygotic wild-type (Z-wt), heterozygous (Z-het), and knockout (Z-KO) embryos. (C) Postimplanted embryos at different days postconception (dpc) from (B). Arrows indicate EPI, ectoplacental cone (EPC) and extraembryonic ectoderm (ExE). Deciduation (de) and yolk sac (ys) pictures are shown. Scale bars, 1 mm (10.5 dpc), 300 μm (8.5 dpc), and 200 μm (6.5 dpc). (D) H&E staining of embryos at the time of implantation. Scale bars, 100 and 500 μm. (E) Percentage of embryos found in (C). Chi-squared test applied; ***P < 0.0001. (F) Genotyping of embryos found in (C). (G) Peri-implanted embryos at different timepoints from (B). TE, ExE, and ICM are pointed with white arrows. Scale bar, 50 μm. (H) Number of embryos found in (A). χ2 test applied for the expected versus observed events; ***P < 0.0001. (I) Genotyping of embryos found in (G). (J) IF of URI and OCT4 in blastocysts. Scale bar, 50 μm. (K) URI intensity in embryos from (J). One-way ANOVA test (Tukey correction); *P < 0.05; ns, nonsignificant. (L) Scheme for ex vivo embryo culture. (M) Ex vivo cultured embryos from (L). Scale bar, 50 μm. (N) Survival of embryos during ex vivo culturing overtime from (M). Zona pellucida hatching was also considered a death event. Mantel-Cox test; **P < 0.01. (O) Survival plot of developmentally normal embryos per stage from (M). Mantel-Cox test; **P < 0.01. (P) Scheme of Uri ICM-KO embryo. (Q) Breeding strategy to generate Uri ICM wild-type (ICM-wt), heterozygous (ICM-het), and knockout (ICM-KO) embryos. (R) Dissected embryos from (Q). Scale bar, 50 μm. (S) IF of URI and NANOG in the last pre-implantation blastocyst state (3.5 dpc) in different embryos from (Q). Scale bar, 50 μm.
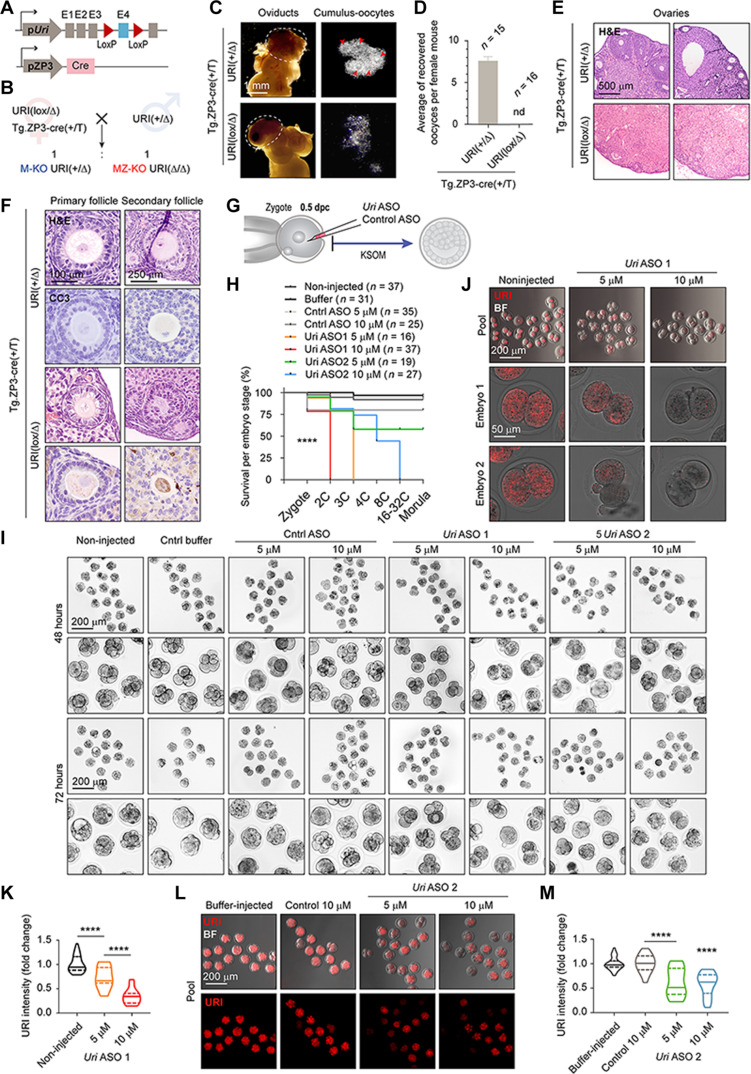
To confirm that postimplantation lethality of Uri Z-KO embryos was due to a dysfunctional ICM/EPI and not a defective TE compartment, we generated Uri ICM-KO embryos by crossing Sox2-Cre mice (38) with URI lox mice (Fig. 3, P and Q). The Sox2-Cre mouse displays a sex-determined phenotype where Cre recombinase is expressed in the female germline and exhibits early activity regardless of the embryo genotype, thereby generating an EPI compartment-derived knockout (39). As observed for Uri Z-KO mice, Uri ICM-KO embryos had normal preimplantation development but failed to progress to the peri-implantation stage (Fig. 3, R and S), indicating that URI ablation specifically in the ICM is sufficient to impair postimplantation development, likely due to a compromised pluripotent compartment.
Therefore, to determine whether maternal Uri mRNA phenotypically compensates for Uri zygotic ablation, we crossed oocyte-specific ZP3-Cre mice (40) with URI lox mice to generate both Uri maternal-zygotic knockout (MZ-KO) and maternal knockout (M-KO) embryos (Fig. 4, A and B). Unfortunately, both Uri MZ-KO and M-KO embryos could not be generated (Fig. 4, C and D), apparently due to an impairment in oocyte development and maturation in the primary and secondary ovary follicles (Fig. 4, E and F). Notably, similar to the Uri Z-KO embryo phenotype, embryos with zygotic ablation of OCT4 (Pou5f1) (41) or SOX2 (Sox2) (42) had a defective ICM, with compromised development during implantation. Moreover, targeting both maternal and zygotic (MZ-KD) mRNA of OCT4 (43, 44) or SOX2 (45) arrested embryonic development at the 4C or 8C stage. While OCT4 MZ-KO embryos reached blastocyst stage when knocked out using the genetic model (46), these discrepancies might result from adaptative mechanism during oocyte maturation following gene depletion.
Fig. 4. URI is required for embryo development beyond totipotent 2C.
(A) Scheme of Uri maternal and zygotic ablation by crossing the oocyte-specific ZP3-Cre mice with the Uri lox mice. Uri maternal-zygotic knockout (MZ-KO) and maternal knockout (M-KO) embryos are generated. (B) Breeding strategy to generate Uri MZ-KO and M-KO embryos. (C) Representative BF images of ovaries and oviducts from referred genotype mice. Dashed lines depict ovaries. Arrowheads point zygotes. Scale bar, 1 mm. (D) Number of zygotes dissected from the ampulla region in the oviducts from panel (C). (E) Histological analysis by hematoxylin and eosin staining (H&E) of ovaries in maternal URI-depleted oocyte from (B). Scale bar, 500 μm. (F) Immunohistochemistry of cleaved caspase-3 (CC3) and histological analysis by H&E in primary and secondary ovary follicles from (E). Scale bars, 100 and 250 μm. (G) Scheme of zygote microinjection for targeting maternal and zygotic Uri mRNA with antisense-oligonucleotide (ASO). (H) Survival of embryos per preimplantation stage 72 hours after microinjection from (G). Mantel-Cox test; ****P < 0.0001. (I) Phase-contrast images for noninjected or buffer-microinjected, control ASO-microinjected (both 5 and 10 μM), and Uri ASO-microinjected (5 μM and 10 μM) embryos after 48 and 72 hours. Scale bars, 200 μm. Higher-magnification images are shown next to each condition. (J) IF of URI in Uri ASO1-microinjected embryos 72 hours after microinjection compared to noninjected 2C embryos. BF images are merged. Scale bars, 200 and 50 μm. (K) URI intensity in noninjected and Uri ASO1-microinjected embryos normalized to fold change. One-way ANOVA test (Tukey correction); ****P < 0.0001. (L) IF of URI in Uri ASO2-microinjected embryos 72 hours after microinjection compared to buffer and 10 μM control-microinjected embryos. BF images are merged. Scale bars, 200 μm. M, URI intensity in Uri ASO2-microinjected embryos normalized to fold change. One-way ANOVA test (Tukey correction); ****P < 0.0001. Total number of embryos is referred in each panel.
EPI cells from the ICM functionally retain self-renewal ability and can be isolated and cultured in vitro in the presence of the cytokine leukemia inhibitory factor (LIF) alone (47, 48), or for better efficiency, with the addition of two inhibitors targeting the kinases MEK and GSK3, in a process termed “2i” addition (fig. S3A) (49). As expected, mESCs could be derived neither from Uri Z-KO (fig. S3, B to E) nor from Uri ICM-KO embryos (fig. S3, F to I), suggesting that URI loss compromises the pluripotent potential of EPI cells.
URI is required for embryo development beyond totipotency
As the presence of the reminiscent maternal URI in the ICM compartment of Uri Z-KO and Uri ICM-KO blastocysts might have obscured an earlier phenotype in both Uri KO embryos, we generated maternal-zygotic Uri knockdown (Uri MZ-KD) embryos by injecting antisense-oligonucleotide (ASO) against Uri in early zygotes (Fig. 4G). The results showed that efficient URI depletion dose-dependently blocked the development of mouse embryos at the 2C (10 μM) and 3C-4C stages (5 μM) (Fig. 4, H to K). Partial URI down-regulation by the microinjection of a second Uri ASO (Fig. 4, L and M), resulted in embryos with an inefficient compaction process during the morula stage. Therefore, URI loss might prevent early totipotency-to-pluripotency transition and impede normal lineage segregation at later stages.
URI is essential for pluripotency maintenance
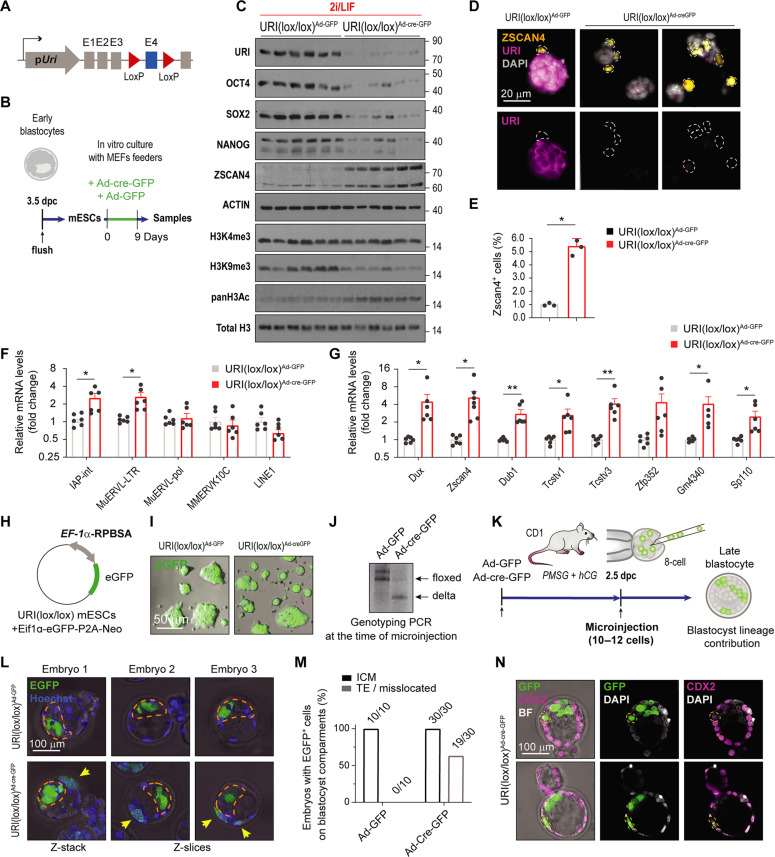
To further explore the consequences of URI loss in the preimplantation embryo and because mESCs cannot be rederived in vitro from URI-depleted embryos, mESCs were established from conditional URI-floxed embryos, cultured in the presence of 2i/LIF, and infected with adenovirus expressing either enhanced green fluorescent protein (EGFP) alone (AdV-EGFP) or in combination with Cre recombinase (AdV-Cre-EGFP) (Fig. 5, A and B). AdV-Cre–mediated URI ablation resulted in a marked reduction in the protein levels of the core pluripotency factors OCT4, SOX2, and NANOG (Fig. 5C), while the mRNA levels remained unaffected (fig. S4A). To confirm these results, we generated inducible URI knockout mESCs by crossing the hUBC-CreERT2 line (50) with URI lox mice (30) (fig. S4B). Isolated mESCs were cultured in the presence of 2i/LIF and treated with 4-hydroxytamoxifen to deplete URI (fig. S4C). Accordingly, URI loss was accompanied by down-regulation of OCT4, SOX2, and NANOG (fig. S4D), suggesting that URI is required for the maintenance of pluripotency factors and that its loss may compromise the mESC developmental potential.
Fig. 5. URI is essential for pluripotency maintenance.
(A) Scheme of conditional knockout URI lox mice. (B) Scheme of mESCs derivation from URI knockout mice treated with adenoviruses expressing either Cre recombinase combined with an enhanced green fluorescence protein EGFP (AdV-Cre-EGFP) or EGFP alone (AdV-EGFP) as control and cultured in presence of 2i/LIF. (C) Western blot (WB) of mESCs treated either with AdV-EGFP or AdV-Cre-EGFP as reported in (B). (D) IF of mESCs for URI and the totipotent-like marker ZSCAN4 after treatment with either Ad-EGFP or AdV-Cre-EGFP as reported in (B). Dashed outlines denote totipotent-like cells. Scale bar, 20 μm. (E) Abundance of the ZSCAN4 population in mESCs treated with either AdV-EGFP or AdV-Cre-EGFP as reported in from (D). t test; *P < 0.05. (F) qRT-PCR of transposable elements in mESCs treated either with AdV-EGFP or AdV-Cre-EGFP. Multi-t test; *P < 0.05. (G) qRT-PCR of totipotent-like genes in mESCs treated either with AdV-EGFP or AdV-Cre-EGFP. Multi-t test; *P < 0.05 and **P < 0.01. (H) Plasmid construct for permanent labeling of mESCs with EGFP. (I) BF merged images for endogenous EGFP signal in mESCs stably electroporated with plasmid from (H). Scale bar, 50 μm. (J) DNA electrophoresis of genotyping PCR for postinfected mESCs at the time of microinjection from (I). (K) Scheme of microinjection and tracking of labeled mESCs in 8C embryos. (L) Representative phase-contrast images for microinjected embryos as described in (K) showing endogenous EGFP signal for Z-stacked projection or single Z-slides in different embryos. Dashed orange outline denotes ICM compartment; yellow arrowheads point integrated cells outside the ICM in the TE compartment. Scale bar, 100 μm. (M) Frequency of microinjected mESCs in different compartment location from (L). (N) IF of microinjected embryos at blastocyst stage for GFP and CDX2 marker. Yellow dashed outlines identify integrated cells in TE. Scale bar, 100 μm.
Immediately after blastocyst implantation, EPI cells polarize to establish the basal-apical axis. Proteins of the cell polarity PAR complex (e.g., PAR6B) form the apical domain de novo, which involves coating with adherent junction and anti-adhesive glycoproteins such as the podocalyxin-like protein (PODXL). Shortly after, the cells alter their shape to form a rosette-like structure [embryonic day 4.75 (E4.75) to E5.0] with a small luminal space in the center of the rosette or proamniotic cavity (E5.0 to E5.25) (4, 51). To determine whether URI loss can affect the features of the progressing EPI cells, pluripotent mESCs were cultured on Matrigel containing N2B27, and 2is were withdrawn to mimic the changes in the EPI compartment that occur during blastocyst implantation (fig. S4, E and F). Immunofluorescence of γ-TUBULIN, required for microtubule nucleation from centrosomes to the cortical actin-rich domains PAR6B, PODXL, and E-cadherin revealed defective cell polarization and failure of URI-depleted cells to develop into epithelial rosette-like structures (fig. S4, G to J). Therefore, URI depletion compromises the development potential of pluripotent EPI cells, most likely by down-regulating OCT4 and SOX2 (52).
URI loss stochastically reactivates the totipotent-like state
As the pluripotent factors are reportedly down-regulated in 2C-like cells at the protein but not at the transcriptional levels (12), as similarly observed in URI-depleted mESCs, we sought to determine whether the 2C-like state was reinforced in the absence of URI. Remarkably, URI depleted-mESCs showed increased expression of the well-known 2C-like cell marker ZSCAN4 (Fig. 5C), with a fivefold increase in the ZSCAN4-positive mESC population (Fig. 5, D and E). Totipotent 2C-like cells are reportedly characterized by the reactivation of some transposable elements, specifically from the class I endogenous retroviral long terminal repeats (LTRs), which serve as proximal gene promoters (12, 17). The 2C-like state also concurs with well-known epigenomic changes such as global genome hypomethylation and histone hyperacetylation (12, 53, 54). Accordingly, transcriptional expression of transposable elements and totipotent-like markers was found to be significantly increased in URI-depleted mESCs (Fig. 5, F and G). In addition, URI depletion in mESCs reduced histone 3 lysine-9 trimethylation (H3K9me3), a repressive transcriptional epigenetic mark implicated in reawakening of ERV LTRs (12, 53, 54) (Fig. 5C). Notably, H3K4me3, which is known to be positively correlated with active promoter state and transcription (53), remained unchanged (Fig. 5C). However, increased histone pan-acetylation levels, which are necessary for chromatin remodeling in the 2C-like state transition, were detected in URI-depleted mESCs (Fig. 5C).
As totipotent 2C-like cells contribute to both embryonic and extra-embryonic tissues (12, 17), we next investigated whether URI-depleted mESCs could colonize the TE compartment when injected in preimplanted mouse embryos. mESCs derived from URI-floxed mice were stably transfected and labelled with a construct containing EGFP under the control of the synthetic RPBSA promoter (Fig. 5, H and I). EGFP-positive mESCs were depleted of URI using AdV-Cre, before being microinjected at the early 8C stage to track their contribution to blastocyst chimeras (Fig. 5, J and K). URI depletion resulted in the relocation of a large proportion of heterochromatic mESCs into the extraembryonic TE compartment (Fig. 5, L and M). Notably, no TE-integrated URI-depleted mESCs stained positive for CDX2 (Fig. 5N), meaning that either CDX2 is dispensable for mESC differentiation into TE once OCT4 is repressed (55) or that SOX2 plays an important role in TE specification in murine embryos (45), possibly conferring incomplete TE self-renewal potential. Accordingly, we cannot disregard the potential role of URI in TE lineage specification, considering its expression in this compartment. Together, these data indicate that URI depletion facilitates the pluripotency-to-totipotency transition in mESCs.
URI loss is a hallmark of 2C-like cells
We next examined whether URI down-regulation could be a hallmark of 2C-like cells. A marginal proportion of normal pluripotent mESCs (<1%) cycles in and out of a transient 2C-like state simultaneously under standard mESC culture conditions (12, 17). As shown by coimmunofluorescence assays performed in wild-type mESCs, the expression of ZSCAN4 or MERVL-gag correlated with little or no URI expression (fig. S5, A to D). In agreement with these findings, analysis of publicly available scRNA-seq data obtained from different populations of pluripotent and totipotent-like mESCs revealed that Uri expression was low for Zscan4 and MERVL markers (fig. S5, E and F, and table S1), indicating that the 2C-like markers negatively correlate with Uri levels. To assess those results at the protein level, we electroporated mESCs with well-known 2C-like reporters Zscan4-mEmerald (56) or MERVL-LTR-tdTomato (12) constructs. As a result, decreased URI levels were detected in both types of reporter-transfected totipotent-like cells (fig. S5, G to J).
In addition, ChIP-seq analysis performed in pluripotent mESCs (Zscan4−; ERVL−LTR− cells) and compared to two states of 2C-like cells (Zscan4+; MERVL−LTR− cells and Zscan4+; MERVL−LTR+ cells) revealed that the H3K4me3 levels and chromatin accessibility were decreased at the Uri promoter in both totipotent-like states (fig. S5, K and L, and table S1), indicating that Uri transcription is reduced in 2C-like cells.
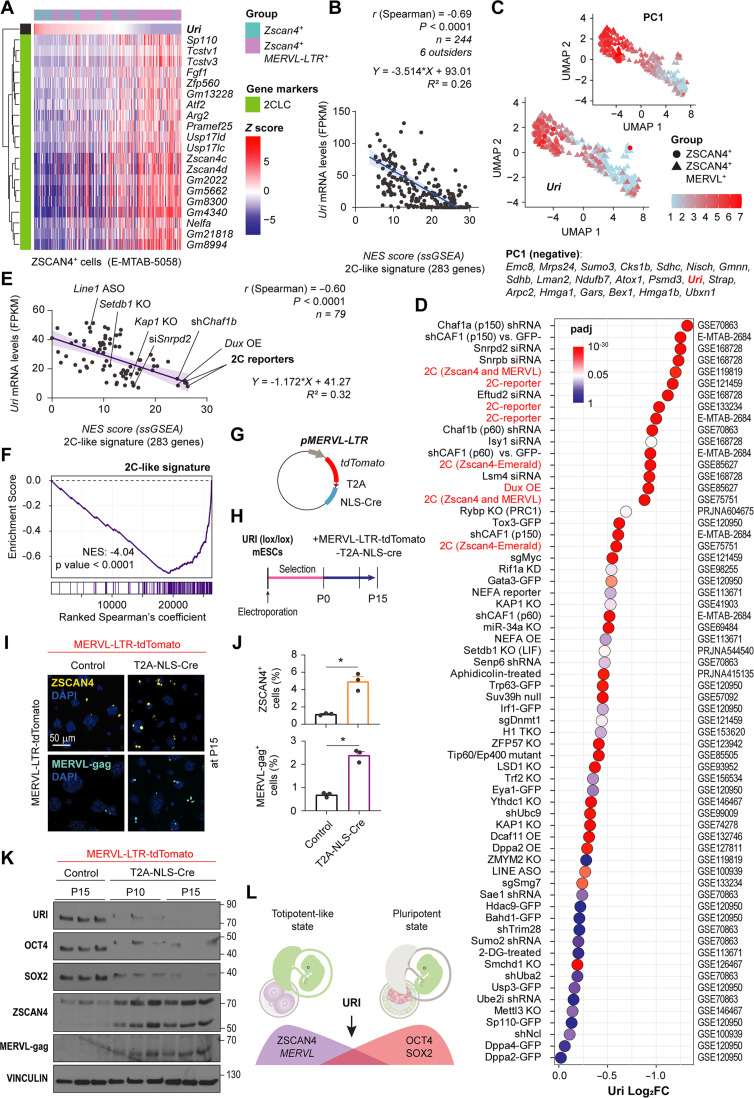
Accordingly, analysis of RNA-seq data obtained from different mouse totipotent 2C-like cells (Zscan4+; MERVL−LTR+ cells and Zscan4+ cells) showed that the stronger the 2C-like state signature, the lower the Uri expression (Fig. 6, A and B, and table S1). We also observed a negative correlation between Uri and MERVL repeats expression in mESCs (fig. S5, M to O). Principal components analysis was used to compute the detected genes in the different totipotent 2C-like cells and unbiasedly identified Uri as a top candidate gene, the levels of which could differentiate the transcriptomic state of 2C-like cells (Fig. 6C and table S1). Analysis of 79 published RNA-seq datasets of totipotent 2C-like cells from synthetic (knockout-knockdown and overexpressing) and reporter models confirmed that Uri expression was significantly down-regulated in the majority of these models. Uri expression was also negatively correlated with the signature of 2C-like cells across all models, such that the lower the Uri expression, the stronger the totipotent-like signature (Fig. 6, D to F, and table S1). Together, these data indicated that URI low is a molecular hallmark of totipotent 2C-like cells.
Fig. 6. Uri low marks 2C-like state.
(A) Heat map showing Uri-ranked mRNA expression of totipotent-like marker genes from scRNA-seq datasets. (B) Linear regression and correlation analysis between normalized Uri mRNA levels and NES of GSEA for 2C-like signature. Data are depicted as mean ± 95% confidence intervals. (C) UMAP plot depicting Zscan4+ and Zscan4+; MERVL-LTR+ scRNA-seq. Color lookup table renders normalized Uri expression. Uri is algorithmically identified as part of the PC1, also displayed. (D) Dot plot for differential expression analysis of Uri across multiple 2C-like cells, ERV activating, or sorted reporter models. Color depicts significance from P-adjusted (q) values white centered at 0.05 value. (E) Linear regression and correlation analysis showing the negative correlation between normalized Uri mRNA levels and NES of GSEA with 2C-like signature for totipotent-like models in (D). Data are depicted as mean ± 95% confidence intervals. (F) GSEA for 2C-like signature for ranked gene correlation with Uri across models from (D). (G) Plasmid construct for depleting URI in 2C-like cells under the MERVL-LTR promoter. (H) Timeline for URI depletion in totipotent-like cells by electroporating plasmid from (G). (I) IF of the totipotent-like marker ZSCAN4 or MERVL-gag in long term culture 2C-like dependent URI-depleted mESCs from (H). Scale bar, 50 μm. (J) Abundance of 2C-like cells after 15 passages from panel (I). t test analysis; *P < 0.05. (K) WB of post-electroporated mESCs at indicated number of passages as reported in (H). (L) Scheme representing the pluripotent state with URI expression opposed to the totipotent-like state. Repository accession number for sequencing dataset analysis are indicated in respective panel and compiled in table S1. Gene signature lists are arranged in table S2.
Next, to determine whether the loss of URI irreversibly blocks the transition to pluripotency from the totipotent-like state, two reporters of 2C-like cells were generated (Zscan4c-mEmerald-Cre or MERVL-LTR-tdTomato-Cre constructs) and electroporated in conditional URI-depleted mESCs. Following URI loss, a greater percentage of 2C-like cells arose when compared to the control, which were stably maintained over time with several passages (Fig. 6, G to K, and fig. S5, P to T). In addition, URI loss in mESCs impairs the pluripotency acquisition from a totipotent-like state, as shown by the absence of core pluripotent factors (Fig. 6S and fig. S5T). These results support the transition arrest of embryos from 2C or 4C to further progression following the loss of URI, as reported above (Fig. 6L).
URI is necessary to protect core pluripotent factors from degradation
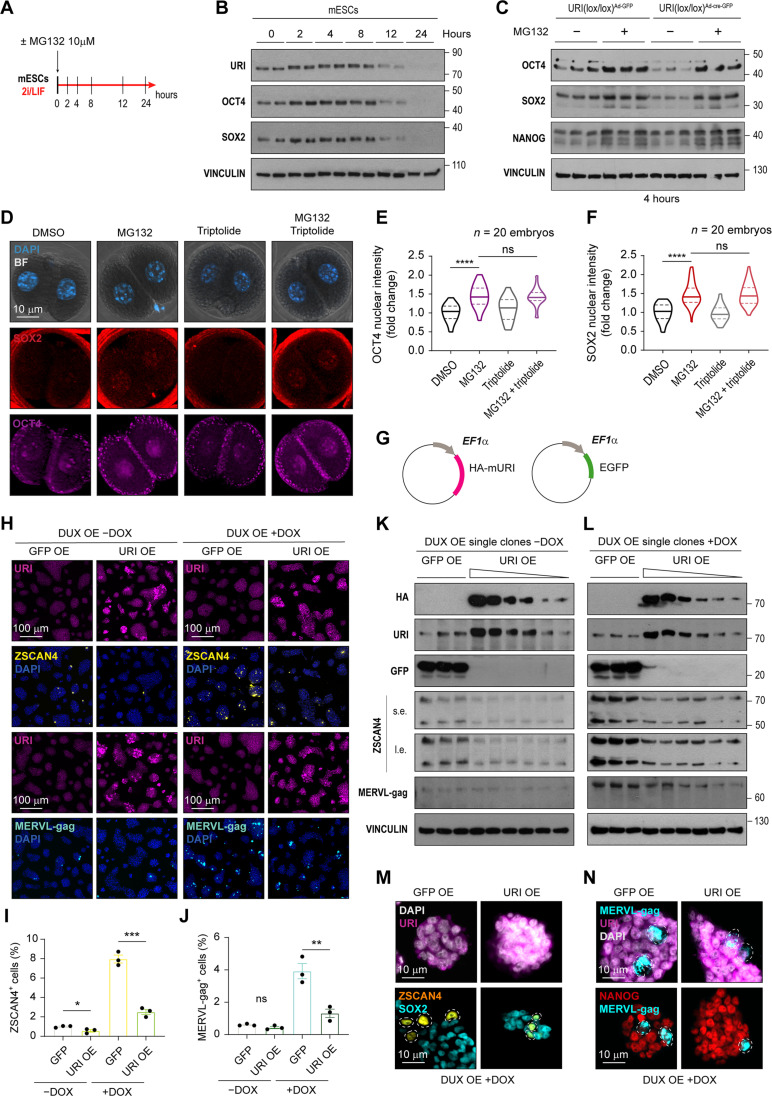
As URI depletion down-regulates the pluripotent factors at the protein but not the mRNA level, we further explored whether inhibition of the proteasome machinery was sufficient to reinstate the expression of pluripotent factors in URI-depleted mESCs. The results showed that the proteasome inhibitor MG132 increased the protein levels of URI and pluripotent factors over time in mESCs, before declining 12 hours after treatment (Fig. 7, A and B). Therefore, we treated URI-depleted mESCs with MG132 for 4 hours. Notably, inhibition of the proteasome reinstated the expression of OCT4, SOX2, and NANOG (Fig. 7C). MG132 treatment also restored the expression of core pluripotent factors in normal transitioning totipotent-like ZSCAN4+ or MERVL-gag+2C-like cells (fig. S6, A to E), which are known to have reduced expression of OCT4 and SOX2 (12), a hallmark of the totipotency state itself. In addition, in 2C-like mESCs stably transfected with the 2C-reporters Zscan4c-mEmerald or MERVL-LTR-tdTomato, treatment with 10 μM MG132 for 8 hours significantly decreased the population of plasmid labeled 2C-like cells (fig. S6, F to I) and up-regulated the core pluripotent factors (fig. S6, J and K). Together, these results suggest that the pluripotent factors are regulated at the protein stability level during the pluripotency-to-totipotency transition. Therefore, they might pose a certain impediment or barrier to the conversion into a totipotency-like state.
Fig. 7. URI is necessary to protect core pluripotent factors from proteasome degradation and counters the 2C-like state.
(A) Scheme of treatment of mESCs with proteasome inhibitor MG132. (B) WB of URI, OCT4 and SOX2 in mESCs treated with 10 μM MG132 at different time points. (C) WB of MG132-treated, URI-depleted mESCs infected with either AdV-EGFP or AdV-Cre-EGFP. (D) IF of OCT4 and SOX2 in 2C embryos treated with 10 μM MG132 and/or 2 μM triptolide for 8 hours. Images for DAPI channel are merged within BF. Scale bar, 10 μm. (E) Intensity of OCT4 in blastomere nucleus from (D). ANOVA with Tukey post hoc test is applied; ****P < 0.0001. (F) Intensity of SOX2 in blastomere nucleus from (D). ANOVA with Tukey post hoc test is applied; *P < 0.05. (G) Plasmid construct for constitutive and stable mURI or control EGFP overexpression in mESCs. (H) IF of URI, ZSCAN4, and MERVL-gag in the pool of clones in rescue experiments by overexpressing control EGFP or URI in combination with inducible DUX expression. Scale bars, 100 μm. (I and J) Abundance of ZSCAN4-positive (I) or MERVL-gag–positive (J) cells from (H). t test; *P < 0.05, **P < 0.01, and ***P < 0.001; ns, nonsignificant. (K and L) WB of single clones from EGFP or URI overexpression in inducible DUX-dependent 2C-like cells in absence (K) or presence (L) of doxycycline. Short exposure (s.e.) and long exposure (l.e) for ZSCAN4 protein is displayed. (M) IF of URI, ZSCAN4, and SOX2 in single clones of EGFP or URI overexpression in combination with DUX expression under doxycycline treatment. Scale bars, 10 μm. (N) IF of NANOG, ZSCAN4, MERVL-gag, and SOX2 in single clones of EGFP or URI overexpression in combination with DUX expression under doxycycline treatment. Scale bars, 10 μm.
The Dux gene, encoding a double homeobox transcription factor (DUX), is a key driver of the totipotent-like state in mESCs and plays an important role during zygotic genome activation in murine 2C embryos (17). MG132-treated DUX overexpressing 2C-like cells displayed increased levels of OCT4 and SOX2 pluripotent factors (fig. S6, L to Q). Expression of OCT4 and SOX2 was also significantly increased in 2C embryos after treatment with 10 μM of MG132 (Fig. 7, D to F). An important observation was that OCT4 and SOX2 protein stabilization was maternally dependent, as transcription inhibition by 2 μM triptolide did not abolish proteasome inhibition effect (Fig. 7, D to F).
Building on the observation that proteasome inhibition restores pluripotent factor expression in 2C embryos and 2C-like cells, we investigated whether URI overexpression could similarly facilitate the stabilization of OCT4 and SOX2 factors. Exogenous URI expression was sufficient to strikingly block the conversion of pluripotent cells into 2C-like cells, both under normal conditions and through DUX overexpression induction (Fig. 7, G to L). In addition, induced totipotent-like cells under URI overexpression showed positive markers for pluripotent and totipotent factor ZSCAN4 but not for MERVL-gag (Fig. 7, M and N). Together, these data suggest that URI is necessary to maintain the stability of the core pluripotent factors during the totipotent-to-pluripotent transitions in mESCs and embryos (fig. S6R).
MERVL controls pluripotent factor levels during totipotency
URI protects OCT4 and SOX2 from proteasome degradation and opposes the 2C-like state and MERVL expression. However, it is intriguing to note that maternally inherited URI persists in the zygote and 2C embryo, despite proteasome-mediated degradation of pluripotent core factors in totipotent embryos. This suggests the presence of additional regulatory mechanisms at play that work in tandem with URI to ensure the proper expression of OCT4 and SOX2 during early embryonic development.
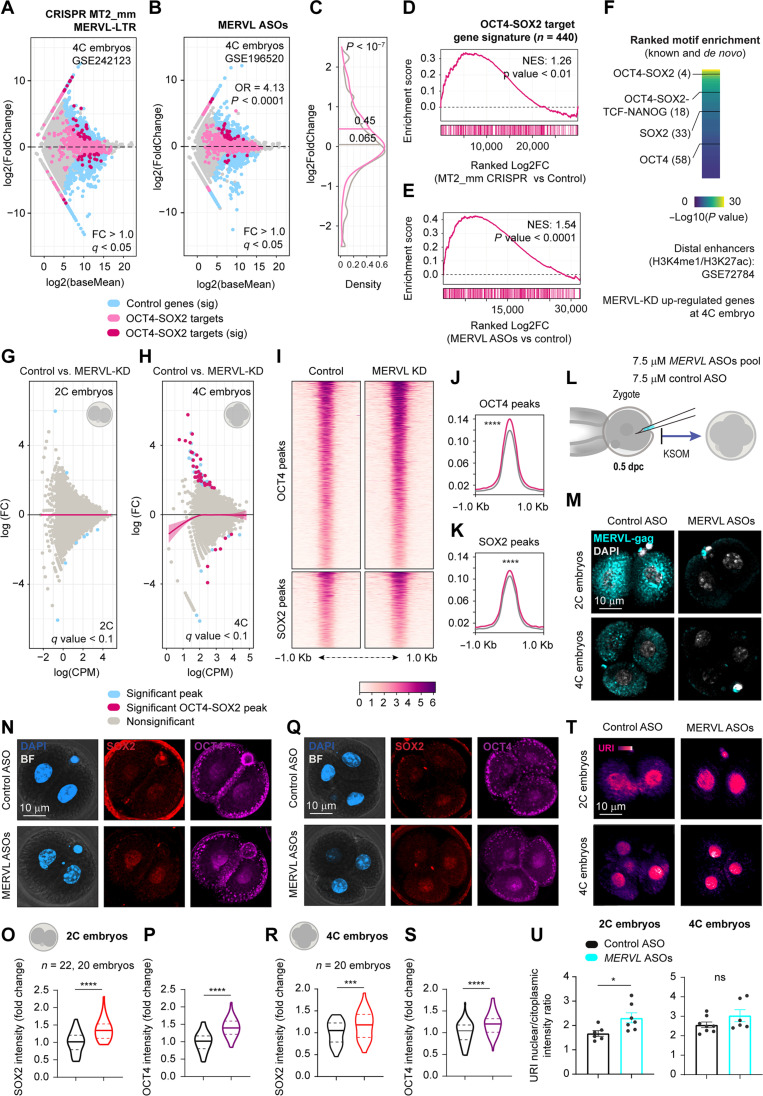
MERVL represents around 3% of the transcripts specifically in the totipotent embryo (12), and its depletion causes early embryo lethality with defects in pluripotency, mainly in the TE specification, via unknown mechanism (22, 23). Therefore, we decided to analyze the transcriptome of promoter MERVL-LTR depleted (57) and MERVL knockdown embryos (23) from a publicly available dataset. We showed that up-regulated genes at 4C stage displayed a tendency to be enriched with OCT4 and SOX2 target genes (Fig. 8, A to E). Accordingly, motif enrichment analysis of proximal and distal enhancers of up-regulated genes in MERVL knockdown embryos, displayed significant binding motifs for OCT4 and SOX2 in combination with others important lineage-segregation involved transcription factors (such as Krüppel-like factor family) (Fig. 8F). In addition, ATAC-seq assay revealed high chromatin accessibility in those pluripotency factor binding regions in MERVL-depleted embryos (Fig. 8, G to K), most likely due to an increase in the protein levels of OCT4 and SOX2 in the 2C and 4C stages. To confirm this hypothesis, we microinjected early zygote embryos with ASOs against MERVL as previously reported (23). Knockdown MERVL resulted in OCT4 and SOX2 up-regulation at both 2C and 4C stages, as shown by immunofluorescence (Fig. 8, O to T). Concurring this up-regulation, URI expression levels became more prominent in the nucleous (Fig. 8, U and V). These findings suggest that URI and MERVL could work together to ensure proper protein stability of pluripotent factors and orchestrate the delicate balance of pluripotency during early embryonic development.
Fig. 8. MERVL controls pluripotent core factors levels.
(A and B) Differential gene expression of MERVL-LTR knockout (A) and MERVL knockdown (KD) (B) 4C embryos. Fisher’s exact was calculated for general and OCT4-SOX2 target genes. ****P < 0.0001. (C) Density plot of gene expression distribution from (B). t test; ****P < 0.0001. (D and E) Gene enrichment analysis for OCT4 and SOX2 target genes from A (D) and B (E). (F) Motif enrichment in proximal and distal enhancers of up-regulated genes from (B). (G and H) MA plot for differentially accessible regions from ATAC-seq in 2C (G) and 4C (H) embryos. Lines are loess fits to each distribution with 95% confidence intervals. (I) Heatmap for normalized ATAC-seq reads from MERVL KD 4C embryos for OCT4 and SOX2 binding regions. Peaks are scaled within 1-kb window. (J and K) Density plots of coverage for OCT4 (J) and SOX2 (K) from (I). Wilcoxon rank-sum test (Benjamini-Hochberg correction); ****q < 0.0001. (L) Scheme of zygote microinjection for targeting MERVL mRNA. (M) IF of MERVL-gag in 2C embryos from (L). Scale bar, 10 μm. (N) IF of OCT4 and SOX2 in 2C embryos from (L). Scale bar, 10 μm. (O and P) Intensity of SOX2 (O) or OCT4 (P) per blastomere nucleous from (N). t test; ****P < 0.0001. (Q) IF of OCT4 and SOX2 in 4C embryos from (L). Scale bar, 10 μm. (R and S) Intensity of SOX2 (R) or OCT4 (S) per blastomere nucleous from (Q). t test; **P < 0.01; ns, nonsignificant. (T) IF of URI in microinjected 4C embryos from (L). Scale bar, 10 μm. (V) URI nuclear/cytoplasmic intensity in 2C (left) or 4C (right) embryos from (T). t test; *P < 0.05. Total number of embryos is referred in each panel. Repository accession number for sequencing dataset analysis are indicated in respective panel and compiled in table S1. Gene signature lists are arranged in table S2.
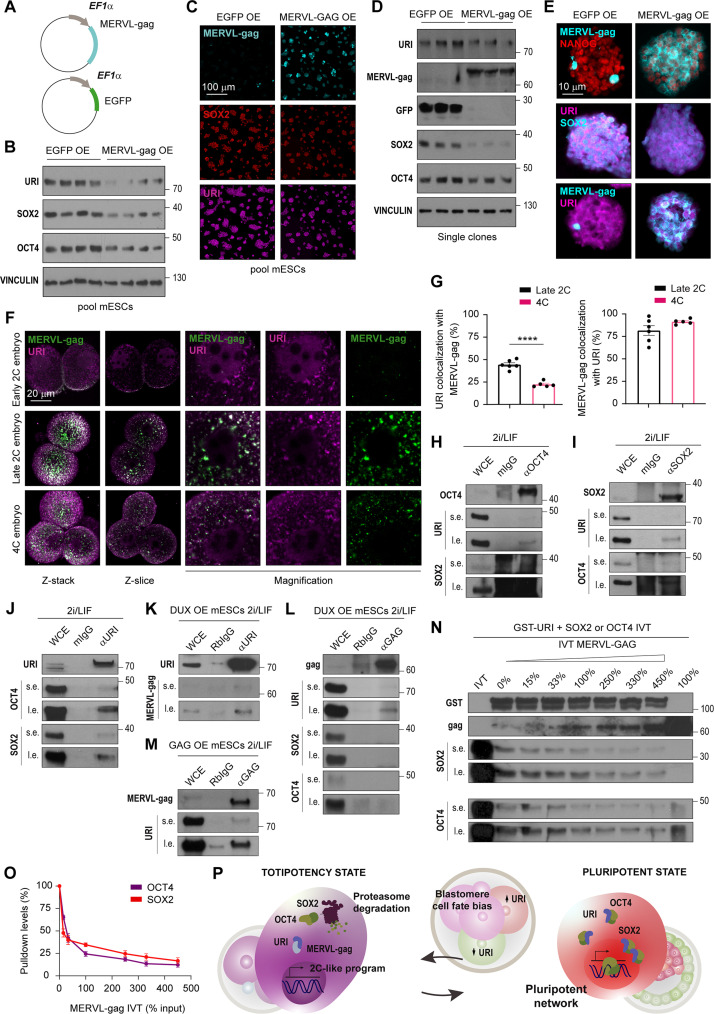
MERVL-gag blocks the binding of URI to OCT4 and SOX2
The MERVL open reading frame 1 protein MERVL-gag is present in almost 600 full copies in the murine genome. It is specifically present in the 2C embryo and 2C-like cells (12, 23). However, its function within the host cell remains unknown. Overexpression of MERVL-gag in pluripotent ESCs reduced levels of URI, OCT4, and SOX2 proteins (Fig. 9, A to E). This suggests that ERV proteins could potentially play an unexplored role in regulating the stability of essential cell lineage transcriptional factors. Coimmunofluorescence analysis revealed that MERVL-gag expression colocalized with URI in the totipotent late 2C embryo. However, at 4C stage, coinciding with a drop in MERVL-gag levels, URI expression underwent lower colocalization than at 2C stage, but its level was still increased in both cytoplasm and nucleus (Fig. 9, F and G). Therefore, the negative correlation between URI and MERVL-gag expressions, along with their implication in regulating pluripotent factor stability, indicates a potential competition between URI and MERVL-gag.
Fig. 9. MERVL-gag blocks URI binding to OCT4 and SOX2.
(A) Plasmid construct for constitutive and stable EGFP or MERVL-gag overexpression in mESCs. (B) WB of pool of clones from EGFP or MERVL-gag overexpression in mESCs. (C) IF of MERVL-gag, SOX2, and URI in the pool of clones from MERVL-gag overexpressing mESCs. Scale bar, 100 μm. (D) WB of single clones from EGFP or MERVL-gag overexpression in mESCs. (E) IF of MERVL-gag, NANOG, SOX2, and URI in single clones from EGFP or MERVL-gag overexpression in mESCs. Scale bars, 10 μm. (F) IF of MERVL-gag and URI in totipotent early 2C, late 2C, and 4C embryo. Projection, single Z-slice and magnifications for each channel are depicted. Scale bar, 20 μm. (G) Colocalization levels of URI with MERVL-gag (left) and vice versa (right) from (F). t test; ****P < 0.0001. (H to J) Coimmunoprecipitation assays of OCT4 (H), SOX2 (I), and URI (J) in mESCs cultured in presence of 2i/LIF. Short (s.e.) and long (l.e.) exposure are shown. (K to M) Coimmunoprecipitation assays of URI (K) and MERVL-gag in DUX-induced 2C-like cells (L) or MERVL-gag overexpressing cells (M). s.e. and l.e. are shown. (N) Competition pulldowns of IVT SOX2 or OCT4 in combination with increasing IVT MERVL-gag using GST-URI. IVT MERVL-gag quantity is annotated as percentage of loaded IVT inputs. s.e. and l.e. exposures are shown. (O) Pulldown levels of SOX2 and OCT4 from (N). (P) Working model. A 4% of the input was loaded as WCE in coimmunoprecipitation experiments.
Next, we investigated whether URI and MERVL-gag directly binds the core pluripotent factors to regulate their stability. Notably, IP assays conducted in mESCs cultured with 2i/LIF showed that URI, OCT4, and SOX2 were reciprocally coimmunoprecipitated (Fig. 9, H to J). The addition of 2i/LIF, which mimics the naïve pluripotent state, increased the levels of URI and pluripotent factors (fig. S7, A to C), with URI showing the longest protein turnover and half-lifetime in mESCs (fig. S7, D to F). LIF, representing the minimal condition necessary to preserve pluripotency in vitro, or MEKi alone were sufficient to sustain the protein expression of URI in mESCs following differentiation and inhibitor withdrawal, respectively (fig. S7, G to J). Inhibition of MEK significantly increased the interaction between URI and OCT4/SOX2, as shown by coimmunoprecipitation assays (fig. S7, K to M). Therefore, increased URI expression may be sufficient to reinforce pluripotency core factor stability.
Notably, reciprocal coimmunoprecipitation in totipotent-like cells revealed that MERVL-gag interacted with URI but not with OCT4 and SOX2 (Fig. 9, K to M). To further understand the link between URI, MERVL-gag, and pluripotent factors, we checked for their direct interactions by pulling down in vitro translated (IVT) proteins with purified glutathione S-transferase (GST)–fused proteins in reciprocal experiments. Clearly, OCT4 and SOX2 were bound to URI (fig. S8, A to F), possibly via several binding sites for OCT4 and SOX2 with preference for the regions flanking the RPB5 domain (fig. S8, G to M). However, MERVL-gag was found to be specifically bound to URI, but not to OCT4 and SOX2 (fig. S9, A and B). Since MERVL-gag regulates pluripotent factor stability, we hypothesized that it might block URI from its binding to OCT4 and SOX2, causing their degradation. Notably, MERVL-gag displaced the interaction between GST-URI and IVT pluripotent factors in a dose-dependent manner (Fig. 9, N and O). Last, we found that URI interacts with MERVL-gag in a highly conserved region when compared to its human counterpart, a reconstituted gag protein from HERVL sequences, specifically expressed in human 8C embryos and totipotent-like cells (fig. S9, C to F) (58, 59). Together, these results indicate that URI directly binds to OCT4 and SOX2, serving as a hub that safeguards them against proteasome-mediated degradation. The presence of MERVL-gag disrupts the URI complex with OCT4 and SOX2, consequently leading to their degradation through the proteasome pathway, promoting or reinforcing the totipotent state (Fig. 9P).
DISCUSSION
Here, we help elucidate partial mechanisms of lineage segregation and cell potency transitions in the very early embryonic development. Developmental potency and cell lineage contribution in the early embryo are heterogeneously biased as early as the 2C-4C stages due to the slow and stable binding of OCT4 and SOX2 factors to chromatin, resulting in a high contribution to the pluripotent compartment (8, 9). The widespread presence of ERV sequences in the mammal genome plays a crucial role in establishing and controlling the totipotency transcriptome (12, 17, 23). In addition, ERVs potentially function as chromatin organizers of the early murine embryo (13). However, their specific roles remain so far obscure.
It is demonstrated that embryo lethality following the loss of MERVL expression is attributed to the transcription process of the transcripts itself, although the underlying mechanism remains unknown (23). However, we demonstrate in this study here that the encoded protein MERVL-gag, which is specifically expressed in 2C embryos and 2C-like cells, also plays an important role and impedes the binding of URI to pluripotent factors. This disruption leads to the degradation of OCT4 and SOX2 degradation via the proteasome pathway. Hence, URI serves as a crucial guardian of OCT4 and SOX2, ensuring their integrity during both the onset pluripotency onset and maintenance of pluripotency.
Active transcription of MERVL transcripts has been shown to hijack enhancer machinery, driving a change in the transcriptome towards totipotency-like (13). In a similar way, the encoded MERVL-gag protein could target URI and consequently destabilize OCT4 and SOX2 to avoid pluripotent superenhancer configuration during early embryo transitions. Depletion of URI is sufficient to impair the full transition from totipotent-to-pluripotent embryo, and its expression coincides with pluripotency bias in mouse blastomeres. The interaction between URI and OCT4/SOX2 is controlled by pluripotent naïve conditions in vitro. In addition, the levels of URI may be transcriptionally regulated by a superenhancer region, which is essential during cell state transitions (36). These revelations might underscore the essential interplay between pluripotent factors and URI within the core network of pluripotency.
We also show that the loss of URI leads to embryo arrest at the 2C stage and possibly promotes a stable totipotent-like state in mESCs, potentially resulting in early embryonic lethality. Hence, the attenuation or suppression of URI expression emerges as a promising avenue to explore for cultivating more stable ESCs endowed with augmented developmental capabilities. Such engineered ESCs could find diverse applications including regenerative medicine, disease modeling, and the creation of artificial embryos.
In sum, the transcriptional burst of ERV during zygotic genome activation could control pluripotent factor stability to give enough time to the embryo to adjust and coordinate the smooth transition from totipotency to pluripotency and cell lineage specification during embryo development. ERV activation could constitute an additional heterogenous source of developmental potency bias in the blastomere, playing a key in cell lineage segregation in the early embryo. Thus, ERVs have evolved symbiotically with host cells to finely modulate cell potency transitions and ensure the timely progression and cell lineage specification during early embryo development.
MATERIALS AND METHODS
Mouse models
Mouse models are referenced in table S3. To study the contribution of zygotic Uri, zygotic-knockout (Uri Z-KO) embryos were obtained by crossing CAG-Cre mice, carrying a Cre recombinase under the control of the hybrid cytomegalovirus (CMV)–enhancer–chicken β-actin (CAG) promoter with conditional Uri knockout (Uri lox) mouse. To assess the role of Uri in the pluripotent compartment, conditional Uri knockout (Uri lox) males were crossed with the sex-determining region Y-box 2 (SOX2) promoter–driven Cre expressing female mice to generate 3.5 days postconception (dpc) Uri ICM-KO mouse embryos. For specific generation of maternal (M-KO) or zygotic-maternal Uri knockout (MZ-KO) embryos, conditional Uri KO mice were crossed with the zona pellucida 3 (ZP3) promoter–driven Cre expressing mice. URI was depleted in mESCs via the expression of an inducible Cre recombinase that is under the control of a ubiquitous human ubiquitin C (hUBC) promoter and which specifically deletes the loxP-flanked stop cassette in the Uri locus. All lines were backcrossed over at least seven generations and maintained in a pure C57BL6/J background. Embryo microinjection and staining experiments were carried out using hybrid F1 offspring females from mated C57BL6/J females with CBA males.
Mouse conditions and housing
Mouse experiments were hosted at the animal facility of the Spanish National Cancer Research Centre (CNIO). Mice were housed in specific pathogen–free stabulated conditions. Animals were maintained within 12-hour light/dark cycle between 8:00 a.m. and 8:00 p.m. in a 22° ± 1°C temperature-controlled room in individually ventilated units. Access to water and food was provided ad libitum. Animal experimentation was approved by the CNIO-ISCIII (Instituto de Salud Carlos III) Ethics Committee and Community of Madrid and performed in accordance with the guidelines for ethical conduct in experimental animal care and use of the European Union.
Embryo dissection
Embryos at preimplantation stages were recovered at different time points between 0.5 and 2.5 dpc from mated pregnant female mouse oviducts. In addition, early and late blastocyst 3.0 to 4.0 dpc were recovered by flushing uterine horns with M2 medium under a stereomicroscope. Eventually, some female mice were superovulated prior to mating. The 5- to 6-week-old female mice were injected intraperitoneally (i.p.) between 1:00 and 3:00 p.m. with one single dose of 5 IU (100 μl) of pregnant mare serum gonadotropin. Following 46 to 48 hours, mice were receiving via i.p. 5 IU (100 μl) human chorionic gonadotropin (hCG) and placed in a mating with fertile stud males. Embryos were recovered at the indicated timepoints post hCG injection: 20 hours for 1C, 48 hours for 2C, 58 hours for 4C, 72 hours for 8C, 80 hours for morula, and 98 hours for blastocyst embryos.
Peri-implanted embryos from 4.5 to 5.5 dpc extraction were performed by dissecting uterine horns in dissection media [DMEM/F12, 25 mM HEPES (pH 7.0), and 5% fetal bovine serum (FBS)]. Then, anti-mesometrium side was cut and implantation sites were pinched off and exposed. Then, embryo was scratching off with a pipette or forceps and handled with a glass capillary by mouth pipetting. Postimplanted embryo pickup was accomplished by extracting deciduae across the uterus after peeling off muscle layers. Embryo exposure and extraction were done by opening and snipping deciduae off through the antimesometrium side. Then, embryos were pulled off and collected. Yolk sacs were used for genotyping procedure.
Ex vivo embryo culturing
Zygotes were recovered at 0.5 dpc after vaginal plug visualization from mated pregnant female mice. Briefly, the cumulus oocyte mass complexes were extracted from an incision in the ampulla region below the infundibulum of the oviducts under a stereomicroscope in M2 medium and then treated with hyaluronidase type IV-S (2 mg ml−1) for 1 to 2 min for zona denudation. Zygotes were washed several times in M2 medium and placed in final M16 or KSOM medium drops inside a CO2 tissue culture incubator.
Embryos were cultured in M16 or KSOM medium media drops with penicillin-streptomycin (100 U ml−1) and covered with mineral paraffin oil under 5% CO2 at 37°C. Embryos were followed up daily until blastocyst-hatching stage or time of death. Subsequently, they were used for immunostaining or genotyping procedures.
Mouse and embryo genotyping
For DNA extraction, finger DNA from mouse pups was extracted by overnight incubation in lysis buffer containing 50 mM tris (pH 8.0), 100 mM NaCl, 100 mM EDTA, 1% SDS, and proteinase K (70 μg μl−1), following a saturated salt (2.5 M NaCl) with 0.7 volumes of ice-cold isopropanol precipitation. Sequences for primers are listed in table S4. Universal polymerase chain reaction (PCR) program used was as follows: initial 5-min denaturation step at 94°C, then 35 cycles of 94°C for 30 s, annealing 60°C for 45 s, and extension at 72°C for 90 s followed by a last extension step for additional 5 min.
For embryo genotyping, individual pre- and peri-implanted embryos (up to 6.5-dpc gastrula) were placed in 20 μl of embryo lysis buffer [50 mM tris (pH 8.5), 1 mM EDTA, 0.5% Tween 20, and proteinase K (15 μg ml−1)] and incubated at 55°C for 30 min. Last, proteinase K was inactivated by incubating samples at 95°C for 10 min prior to PCR. PCR was performed with 2- to 5-μl sample in a high cycle number (45 to 65 cycles) depending of the embryo stage. For 7.5-dpc embryos and beyond were genotyped by dissecting and treating yolk sac for DNA extraction.
Mouse ES cell culture
ES cells were cultured over γ-irradiated inactivated feeder cells (in-house made) on gelatin-coated plates during ES cell derivation and up to passage numbers 5 to 6. Subsequently, cells were maintained in 0.1% gelatin-coated plates with high-glucose DMEM supplemented with 15% knockout serum replacement, LIF (1000 U ml−1), 1% nonessential amino acids, penicillin-streptomycin (100 U ml−1), 2.5 mM l-glutamine and 50 μM β-mercaptoethanol. To promote naïve state, 2is (1 μM PD0325901 and 3 μM CHIR99021) were added to the medium (named 2i/LIF medium). For CreERT2 recombinase activation in the conditional URI-KO cells, 1 μM 4-hydroxytamoxifen (4-OHT) was used for a total of 12 to 24 hours. Rosette-like or in vitro lumenogenesis assay described ahead was carried out following previous work (52). Cells were changed and adapted to N2B27 medium (DMEM/F12 supplemented with N2 and B27) with 7.5% bovine serum albumin (BSA) in the presence of 2i for several days and passages as further described below. In addition, doxycycline (1 μg ml−1) was added for tetracycline inducible overexpressing (tetO-rtTA) systems in cells and 10 μM MG132 in 2i/LIF medium at assay-indicated times was used for protein stability assessment by proteasome inhibition.
For clonal selection after plasmid electroporation described below, puromycin (0.25 μg ml−1), neomycin (200 μg ml−1; G418), hygromycin B (350 μg ml−1), or blasticidin S (5 μg ml−1) was used, respectively, for a total of 5 to 7 days.
Cells were routinely splited by trypsinization using 0.25% trypsin-EDTA every 2 to 3 days and seeded with splitting ratios from 1:2 to 1:6. Media was refreshed daily or every other day. All performed assays used murine ESC lineages with a total number of passages between 8 and 30 to 35 since they were derived and established. Cultures were also routinely tested for mycoplasma by using a PCR test kit.
Feeder generation
Primary mouse embryonic fibroblasts (MEFs) were obtained from embryo dissection at 13.5 dpc from wild-type C57/BL6J mice. Embryos were dissected from harvested uterine horns on ice-cold phosphate-buffered saline (PBS)/10% FBS using pine-pointed forceps and separated from yolk sac and placenta. Fetuses were beheaded and liver and heart were teared out. Carcass were washed in ice-cold PBS and minced thoroughly with a scalpel blade and placed in 0.25% trypsin-EDTA for 10 min at 37°C. Disaggregated cells were next resuspended in high-glucose DMEM [supplemented with 10% of FBS, penicillin-streptomycin (100 U ml−1), 1% nonessential amino acids, 2.5 mM l-glutamine, and 1 mM sodium pyruvate] in 0.1% gelatin-coated plates. Fresh medium was added 16 hours later, when most of the cells were attached. Second or third passages of MEFs were trypsinized and pooled. Cell suspensions were γ-irradiated for a total dose of 24 grays (Gy) (with a Cs137 isotope source and a dose rate of 1.56 Gy min−1) and frozen. Then, before experimental assays, γ-irradiated inactivated MEFs were seeded at a density of 25,000 to 30,000 cells cm−2 in 0.1% gelatin-coated plates and cultured for 24 to 48 hours.
Zygote microinjection
Hybrid 5- to 6-week-old females (C57BL/6.CBA.F1) were superovulated as described in previous section. Hormone-treated females were then immediately crossed with fertile C57BL/6J stud males and next day embryos were recovered. Subsequently, zygotes were intracytoplasm microinjected with 10 or 5 μM Uri antisense oligonucleotides (ASOs) with sequence 5′-CATCTTCTGCAAATCT-3′ (ASO1) and 5′-ATTTAGCCGTGAATAG-3′ (ASO2) or nontargeting control ASO with sequence 5′-AACACGTCTATACGC-3′ diluted in microinjection buffer [10 mM tris (pH 8.0) and 0.1 mM EDTA]. Targeting the ERV MERVL was performed using previously described ASOs sequences by male pronucleus microinjection (23). Microinjection was performed in M2 medium under mineral oil using a microscope equipped with differential interference contrast (Nomarski) optics, an electronic microinjector, and micromanipulators. Additional controls for noninjected and buffer-injected zygotes were also performed. Microinjected and control zygotes were cultured in KSOM medium followed up and used for immunostaining techniques.
Chimeric embryo generation
Cultured ES cells were trypsinized to a single-cell suspension and collected in ES cell medium without 2i. Meanwhile 8C embryos were harvested from superovulated Hsd:ICR (CD-1) donor females 72 hours after hCG injection (2.5 dpc) by flushing dissected maternal oviducts with 0.5 ml of M2 medium and then transferred to KSOM culture medium in droplets covered with mineral oil and placed into a tissue culture CO2 incubator. Individual ES cells (10 to 12) were microinjected into each single recipient 8C embryo under a microscope using a manual pneumatic microinjector. After, chimeric embryos were cultured in KSOM medium for additional 36 to 48 hours and nuclear stained with Hoechst dye for better visualization of cell allocation at blastocyst stage by confocal imaging.
Subsequently, embryos were washed, fixed, and immunostained. Imaging of the labeled cells in microinjected embryos was done before and after fixation using confocal system.
Immunofluorescence in embryos
Normal and chimeric embryos at preimplantation were handled by mouth pipetting. After washing in PBS, embryos were fixed in PBS/4% PFA at room temperature (RT) for 20 min with gentle shaking and permeabilized with PBS/0.3% Triton X-100/0.2% Tween 20 at RT for 20 min in four-well multidishes. Nonspecific epitopes in the embryos were blocked in blocking solution PBS/1% BSA/0.2% Tween 20 at RT for 1 hour. Detection of CARM1 nuclear speckles was performed as previously described by fixing embryos in 100% methanol at −20°C for 20 min and blocked overnight in PBS/3% BSA/0.2% Tween 20 (10). Primary antibodies referenced in table S3 were diluted at indicated concentration in blocking solution, and embryos were incubated for 1 to 2 hours at RT or overnight at 4°C. Subsequently, embryos were washed in PBS/0.2% Tween 20 and incubated with diluted secondary antibodies in blocking solution containing 4′,6-diamidino-2-phenylindole (DAPI; 1 μg ml−1) at RT for 1 hour. Next, the embryos were transferred to PBS/0.5% polyvinylpyrrolidone (PVP)/ 0.2% Tween 20 microdroplets in a μ-slide chambered coverslip or a glass bottom 35-mm dish covered with mineral oil for confocal imaging.
Generation of mESCs
All ES cell lines described in this work were in-house made from different mouse models with pure C57/BL6J background as described in table S3. Murine ES cells were obtained from Uri conditional knockout (Uri lox) mice and then treated with adenoviruses carrying either EGFP alone (AdV-EGFP), or together with Cre recombinase (AdV-Cre-EGFP). In addition, Uri knockout (Uri lox) mice were mated with a transgenic CreERT2-expressing mice to generate a second URI-KO ES cell line. All derived cell lines were genotyped following sodium chloride, isopropanol/ethanol precipitation, and extraction protocol as indicated above.
Technical procedure for ES cell line derivation was followed accordingly to published protocol (60). Briefly, 6-week-old female and 8-week-old male from indicated lines (C57/BL6J background) were mated. Next morning, vaginal plug was checked for assessing 0.5 dpc. After 3.5 dpc, early blastocysts were recovered by flushing uterine horns with M2 medium. For zona pellucida removal, embryos were treated with homemade acidic Tyrode’s solution [25 mM HEPES (pH 2.5), 120 mM NaCl, 5 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 0.2% PVP, and 0.1% glucose] and washed extensively in M2 medium.
Blastocysts were pipetted in gelatin-coated feeder-seeded individual p48 wells with 2- to 4-hour preconditioned 2i/LIF medium (see below). Embryo disaggregation from the ICM outgrowths was done in 0.25% trypsin-EDTA for 10 min at 37°C with gentle pipetting up and down. Colonies were expanded and growth up to five to six passages in feeder layers and then in 0.1% gelatin coating for genotyping and experimental procedures.
Adenovirus production, titration, and cell infection
Human adenovirus type 5 (ΔE1/E3) encoding Cre recombinase and EGFP under two independent CMV immediate early promoters or just EGFP were scale produced and purified by cesium chloride (CsCl) method. Briefly, permissive 293A cells with stable integrated copy of the E1 gene were infected with cited adenovirus with a multiply of infection (MOI) of 0.1. After 3 to 4 days, cells were harvested, pelleted down, and lysed in buffer [0.1 M tris (pH 8.0), 0.5% sodium deoxycholate, 1 mM MgCl2, and DNAse I (5 mg ml−1)] for 60 min at RT with intermittent gentle shaking. Viral lysate supernatant was loaded in a CsCl gradient density column (phase 1, 1.25 g cm−3; phase 2, 1.35 g cm−3; and phase 3, 1.5 g cm−3) and ultracentrifuged 30,000 rcf at 10°C for 20 hours. Viral band was aspirated in small volume and dialyzed within 10,000 molecular weight cutoff cassette in 10 mM tris (pH 8.0) for 24 hours with intermittent buffer changes. Viruses were aliquoted in dialysis buffer supplemented with 10% glycerol. Viral titration was made by median tissue culture infectious dose method, taking advantages of the EGFP labeling. Murine ES cell lines infection was done in suspension after seeding with 200 MOI dose. After 6 to 12 hours, media was freshly replaced.
Plasmids
Plasmids used in this study are listed and referenced among materials and reagents in table S3. For chimera microinjection and cell tracking, ES cells were transfected by electroporation with pSBbi-GN. Totipotent-like cell labeling was done with 2C::tdTomato reporter for MERVL-LTR expression or alternatively checking Zscan4c activation by pZscan4-mEmerald. Alternatively, specific URI depletion in totipotent-like cells was accomplished generating both MERVL-LTR-tdTomato-T2A-NLS-Cre and Zscan4-Emerald-T2A-NLS-Cre plasmids as previously reported (12, 56) by incorporating a posterior in-frame T2A-NLS-Cre sequence. The 2C-like state was induced by using DUX tetracycline-inducible overexpression system in mESCs with tetO-FLAG-mDUX with neomycin plasmid (61). Plasmids for overexpressing URI and MERVL-gag were generated in this study. Briefly, Uri complimentary DNA (cDNA; NCBI gene ID:19777) (UniProt accession: Q3TLD5) and MERVL-gag (GenBank accession: Y12713) protein encoding sequence (UniProt accession: V9H130) were codon optimized and cloned into pcDNA3.1 backbone with a EF1α promoter substituting the CMV sequence.
In vitro protein translation assays were carried out using pcDNA3.3-SOX2 and pcDNA3.3-OCT4 plasmid for human isoforms. For bacterial protein expression, pGEX4T-1-OCT4 was acquired. Moreover, to generate pGEX4T-1-SOX2, human Sox2 cDNA was cloned from previous cited pcDNA3.3-SOX2 and inserted into Nhe I–EcoR I restriction enzyme target sequence in pGEX4T-1-OCT4 plasmid. The same strategy was followed for cloning MERVL-gag into a pGEX4T-1 construct.
Plasmid electroporation
Cells were collected by trypsinization and subsequently washed, quantified, and resuspended for a concentration of 1 million cells per 100 μl of reduced serum media containing 5 μg of linearized vector. Samples were placed into 2-mm gap electroporation cuvettes and transfected using NEPA21 electroporator.
Sample impedance was assessed in a 30- to 50-ohm range and parameters were set up as follows: poring pulse, 125 V voltage; 5 ms pulse length and 5 ms pulse interval; two pulses, 10% decay rate and + polarity; transfer pulse, 20 V voltage; 50 ms pulse length and 50 ms pulse interval; and five number of pulses, 40% decay rate and ± polarity.
After electroporation, cells were plated in 2i/LIF medium. The medium was replaced after 8 to 16 hours. Positive cell clones were selected with respective antibiotics, as described in previous sections, starting 36 to 48 hours postelectroporation for a total of 5 to 7 days.
mESC spheroid formation
To mimic morphological changes of the EPI during implantation (polarization and lumenogenesis) with mouse ES cells, we followed previously published protocols (52, 62). Briefly, cover glasses on plates were covered and coated with growth factor–reduced Matrigel. ES cells were plated at a density of 20,000 cells cm−2. Once cells were well attached, medium was changed to 5% Matrigel diluted in N2B27. After 24 to 48 hours, postseeding samples were used for immunostaining. Primary antibodies are compiled in table S3.
Immunofluorescence in mESCs
ESCs were plated onto 0.1% gelatin-coated 24-well plates with glass covers on it for 24 to 48 hours, fixed in PBS/2% PFA for 20 to 30 min, permeabilized in 0.5% Triton X-100 in PBS for 20 min, and blocked in PBS/2.5% BSA for 1 hour at RT. Primary antibody incubations were performed with overnight at 4°C or 1 to 2 hours at RT incubation in blocking solution using specified dilutions listed in table S3. Then, cover glasses were washed trice in PBS and incubated in their respective fluorescently conjugated secondary antibodies in blocking solution containing DAPI (1 μg ml−1) for 1 hour at RT. Slides were mounted in aqueous mounting media [0.1 M tris (pH 8.5), 25% glycerol, Mowiol 4-88 (0.1 g ml−1)] for imaging acquisition.
Quantitative real-time PCR
For quantitative real-time PCR (qRT-PCR) analysis, total RNA was extracted from cell lysis using TRIzol following manufacturer’s instructions. First-strand cDNA was generated from up to 2 μg of total RNA using Moloney murine leukemia virus reverse transcriptase and random hexamer priming. qPCR was performed using SYBR green master mix in 96-well dishes in triplicate and repeated with at least three or more biological replicates (n ≥ 3). Relative gene expression levels were quantified by delta-delta Ct (ΔΔCt) method using both gapdh (glyceraldehyde-3-phosphate dehydrogenase) and actb (β-actin) housekeeping control genes and normalized to respective control condition in fold change. Primer sequences are listed in the table S4.
Immunoblotting
Whole-cell extracts (WCEs) were prepared by lysing ES cells in RIPA plus buffer [50 mM tris (pH 8.0), 150 mM NaCl, 2 mM MgCl2, 2 mM CaCl2, 0.5% sodium deoxycholate, 10% glycerol, 1% NP-40, and 0.1% SDS] supplemented with 1 mM sodium ortovanadate (NaOV), 1 mM sodium fluoride (NaF), aprotinin (5 mg ml−1), and 500 μM phenylmethylsulfonyl fluoride (PMSF).
Lysates were quantified by Bradford method, prepared, and boiled in Laemmli buffer [50 mM tris (pH 6.8), 2% SDS, 10% glycerol, and 0.01% bromophenol blue] with 5% 2-mercaptoetanol or 100 mM dithiothreitol (DTT). A total of 20 μg of protein was loaded in acrylamide gels. Then, gels were electrophoresed at 200 V for 60 min and ice-cold transferred to nitrocellulose membranes at 100 V for 120 min. Membranes were then blocked in 5% nonfat dry milk in TBS-T and blotted in assay-indicated primary antibodies compiled in table S3.
After several washes, horseradish peroxidase (HRP)–conjugated secondary antibodies were incubated for 1 hour at RT. Blots were developed using homemade ECL detection system [50 mM tris (pH 8.5), 2.5 mM luminol, 400 μM p-coumaric acid, and final 0.015% H2O2]. Western blot (WB) analysis was performed at least with triplicate biological replicates (n ≥ 3) and was repeated in different technical replicates.
Coimmunoprecipitation
Murine ES cells were seeded in 60-mm plates per IP condition, after 2 to 3 days cells were lysed in 2.1-ml ice-cold IP buffer [50 mM tris (pH 8.0), 100 to 250 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% NP-40, 7.5% glycerol, 1 mM NaOV, 1 mM, NaF, 500 μM PMSF, and aproteinin (10 μg ml−1)]. After 5-min incubation, the resulting supernatant was passed through an 18-gauge needle and clarified by centrifugation. WCE was transferred to a low protein-binding tubes and immunoprecipitated using primary and isotype control antibodies added to a final 1 μg ml−1 concentration in 1-ml WCE and incubated for 1 to 2 hours at 4°C in rotation. Then, 25 μl of dynabeads slurry, either protein A or G dynabeads, was washed in lysis buffer prior addition and incubated for 1 hour at 4°C in rotation. Beads were precipitated during 1 min at 1500 rcf and washed trice in lysis buffer. Last, antibodies were eluted from beads in 30 μl of 2× Laemmli buffer [100 mM tris (pH 6.8), 4% SDS, 20% glycerol and 0.02% bromophenol blue, and 200 mM DTT] and boiled at 95°C for 4 min before immunoblotting. A total of 4% input was loaded as WCE control.
Protein expression and purification
Genetically engineered BL21 Escherichia coli bacteria [CodonPlus (DE3)-RIPL] strain was expanded for generating chemically induced competent bacteria by calcium chloride method. Bacteria were then transformed with non–codon optimized GST fusion-protein plasmids (pGEX4T-1 backbone) for full-length URI and fragments, MERVL-gag, OCT4, and SOX2 as indicated in the compiled table S3. Bacteria were cultured in LB culture medium until 0.3 to 0.4 optical density (OD; 600-nm absorbance). Then, isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to a final 400 μM concentration and growth overnight (for 16 hours) at 30°C. For MERVL-gag, OCT4, and SOX2 expression, 0.6 to 0.7 OD bacteria cultures were incubated with 1 mM IPTG for 6 hours at 22°C. Bacterial cultures were centrifuged at 4,000 rcf at 4°C for 15 min. Pellets were resuspendend in lysis buffer [50 mM tris (pH 8.0), 100 mM NaCl2, 2.5 mM MgCl2, 1% NP-40, 2 mM EGTA, 7.5% glycerol, 1 mM PMSF, lysozyme (100 μg ml−1), and DNAse I (10 μg ml−1)] and sonicated thrice for 1 min at 70% amplitude with 1 min pause in between.
Lysates were clarified by centrifugation at 6,000 rcf at 4°C for 15 min. Next, supernatants were incubated with 150 to 200 μl of glutathione sepharose beads slurry previously washed in PBS several times and incubated in lysis buffer for 1 hour on ice. Pellets were stored at 4°C up to few days. Sample check for quality protein purification was done by coomassie staining. Briefly, after resolving, gel was prefixed (50% methanol (MeOH), 10% acetic acid (HOAc) for 10 min at RT, and stained with Coomassie staining solution (0.5% brillant blue G-250, 50% MeOH, and 10% HOAc) for 5 min. Subsequently, gel was destained in washing solution (40% MeOH and 10% HOAc) for 1 hour and several passages in ddH2O with overnight incubation.
In vitro protein binding assay
IVT proteins were synthetized by using reticulocyte lysate system within a final volume of 50 μl and following manufacturer’s instructions and as previously reported (26). Interaction assay was made by incubating 3 to 10 μl of IVT protein with 10 to 30 μl of GST-fused protein pulldowns in IP buffer [50 mM tris (pH 8.0), 100 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% NP-40, and 7.5% glycerol] or PBS/0.1 Tween 20 for 1 hour at 4°C and rotation. Pulldown beads were washed four times in 1 ml of IP buffer or PBS and eluted in 30 μl of 2× Laemmli buffer [100 mM tris (pH 6.8), 4% SDS, 20% glycerol, 0.02% bromophenol blue, and 200 mM DTT] for immunoblotting analysis. A total 100% input was loaded as IVT control.
Tissue immunostaining and histology
Uterine horns tissues were dissected from 4.5 to 5.0 dpc from pregnant female mice previously mated with a URI heterozygous KO male and disposed in formalin-fixed, paraffin-embedded tissue blocks with horizontal mesometrium-antimesometrium axis. In addition, ovaries from Uri flox crossed Zp3-Cre female mice were also processed in blocks. Tissue sections with 3 μm thickness were deparaffinized, rehydrated, and antigen retrieved with 1 M sodium citrate buffer (pH 6.5). Then, endogenous peroxidase activity was blocked by 3% H2O2 incubation for 15 min, lately permeabilized in PBS/0.5% Triton X-100 for 30 min and blocked with PBS/5% BSA for 1 hour at RT. Primary antibody incubation was performed at 4°C overnight. Antibody references and dilutions are compiled in table S3. Sections were subsequently washed and revealed using Vectastain ABC-HRP kit with 3-diaminobenzidinetetracloride chromogen and hematoxylin counterstained.
RNA-seq database analysis
For RNA new-generation sequencing analysis, sequence read files were imported or downloaded with sra-tools into Galaxy platform from respective and assay-indicated accession numbers reported in table S1. Fastq files were scanned with fastQC tool. Then, reads were quality trimmed using trimmomatic –SLIDINGWINDOW: 4:20 and adaptor removed by cutadapt --paired if required. Subsequently, reads were mapped to GRCm38/mm10 running Hisat2 with default parameters. Transcript assembly and quantification were done with stringtie generating both total counts and relative abundance estimation files. For transposable elements mapping was performed with Hisat2 with restricted unique alignments –k 1. Transposable element repeats annotation for quantification was built by filtering the RepeatMasker track downloaded from UCSC. Quantification of repeats was done with featureCounts (subread) by gene and family ID clustering. Differential gene expression analysis was performed using DESeq2, besides single-cell heatmaps were generated using pheatmap function in previously normalized counts by variance stabilizing transformation (DESeq2::vst).
GSEAs were carried out in single samples with ssGSEA code package in R. The averages of normalized enrichment score (NES) were calculated for replicates inside the same condition and paired with the average of normalized Uri gene counts [fragments per kilobase million (FPKM)] for correlation analysis. GSEAs were also performed by calculating Pearson’s correlation coefficients of Uri with the rest of expressed genes using psych package in R. Correlation coefficient ranking was used to score the signature enrichment, then plotted with clusterProfiler and enrichplot packages. Last, gene set for molecular signature of the 2C-like cells was obtained from a previous study (63). The generation of potential OCT4 and SOX2 target gene list was performed compiling set of genes from previous works as previously described (7) and with additional cross-overlap with public databases: ENCODE Consortium, ChEA transcription factor targets dataset, and Enrichr. Additional signatures for mouse EPI and TE compartment and highly variable OCT4-SOX2 target genes were compiled in this study. Software and algorithms used in this study are described in table S3. Gene signatures are compiled and specified in table S2.
Motif enrichment analysis in proximal and distal enhancers of up-regulated genes from MERVL knockdown microinjected embryos was carried out following two different approaches. First, annotated proximal genes to broad enhancer peaks were intersected with the list of up-regulated genes. Second, 50-kb upstream regions of all up-regulated genes were annotated and intersected with selected enhancer regions. Promoter and gene body peaks were excluded. Filtered region list in bed format was submittted to –findMotifsGenome.pl –size given via HOMER software. The enhancer regions were identified by overlapping regions of called H3K4me1 and K3K27ac ChIP-seq peaks in murine ES cells (see table S1).
Gene dispersion variability analysis and normalization
Technical noise in gene expression counts from different assays was accounted for batching effect with the sva package in R. Normalization for gene counts was applied by estimating size factors on endogenous genes with DESeq2 (DESeq2::estimateSizeFactorsForMatrix) and then dividing read counts by appropriate size factor. In addition, gene count normalization was reported in FPKM from RNA-seq processing as explained above.
Identification of highly variable genes was carried out as previously described (33) by setting the squared coefficient of variation (CV2) across gene expression as a function of the mean normalized counts. Gene expression among blastomeres was first regressed out to reduce inter-embryo variation by building a linear model following previous published formula (7). Briefly, gene expression in a cell is related to the intercept of the mean expression of the gene, the intra-embryo average and the fit residual. Then, model was used to predict corrected gene expression values. Data were winsorized for most outlier cells if indicated, plotted, and exported to Adobe Illustrator. Hierarchical cluster analysis was done by sample distance with the obtained highly variable gene list and using hclust–ward function (stats package). In addition, high variable genes were intercepted with a list for possible OCT4 and SOX2 targets (64) as previously reported done for murine 4C embryos (7). Software and algorithms used in this study are described in table S3.
ChIP-seq, ATAC-seq, and DNAse-seq database analysis
Raw data samples were gathered and quality analyzed as previously defined above. This time, trim galore was used to remove nextera or additional adapters and poor basecall qualities in ATAC-seq or ChIP-seq datasets. Sequence reads aligning by Bowtie2 --dovetail if paired and --very-sensitive options were executed. For ATAC-seq analysis in MERVL KD embryos, sample replicates were subsampled to similar unique read numbers using calculated downsampling factor –s (samtools view) by estimating library complexity (using ATACseqQC package), as previously described (65). Duplicate reads elimination was done by Markduplicates (Picard) --true. Differential accessibility analysis was performed by calculating naïve overlaps between replicates and pools. Plots and binomial models were computed with csaw and edgeR packages with MACS2 called regions and BAM files applying a loess-based normalization method, based on previous pipeline (65).
Bigwig files for reads coverage were generated via bamCoverage (deeptools) with a bin size of 20 bases and using determined –scaleFactor computed by –calcNormFactors (TMM method from edgeR package) to correct signal-to-noise ratio between conditions. For nonconditional ChIP-seq analysis and track visualization, output bam files from replicates were merged using SAMtools for better single plot view and normalization was performed via reads per kilobase per million normalization. Files were visualized and plotted with pyGenomeTracks and exported to Adobe Illustrator as vector images.
Peaks for OCT4 and SOX2 genomic binding regions were assessed and called using MACS2, default settings for building the shifting model a minimum q value cutoff detection of 0.01 was used. Intersecting and excluding peaks of OCT4 and SOX2 were processed with beedtools. In addition, signal values were calculated with computeMatrix (deepTools) function with peak region centering with 1000 bp upstream and downstream distance. Last, it was visualized with plotHeatmap (deepTools). Software and algorithms used in this study are described in table S3.
Image analysis and quantifications
Confocal multichannel image acquisition was done by using Leica SP5 WLL or Leica SP8 microscopes equipped with a PL APO 63×/1.20 W Corr/0.17 CS objective for nuclear resolution or PL APO 20×/0.70 IMM Corr CS objective for whole-embryo resolution. Confocal imaging in embryos was performed with a depths range of 30 to 40 μm and with a 1024 pixel resolution. Additional to glycerol or water-immersion objectives for signal attenuation decrease, later correction was applied by Imaris software.
Immunofluorescence quantification in confocal assays was done by using the average signal of the Z-stacked slides in different compartment, cell, or embryo as specified.
Speckles quantification was performed by using Imaris spot model tool and setting up 0.5 μm size and intensity exclusion parameters. To quantify immunostainings, the brightness contrast and attenuation correction of the images were adjusted using Fiji/ImageJ, Adobe Photoshop, or Imaris for confocal imaging and Z-stacking. Additional postprocessing and quantifications were also done using Fiji-ImageJ. Software and algorithms used in this study are described in table S3.
Statistical analysis
All statistical test analyses were performed with Graphpad Prism, except genome-wide data analyses which were performed in Galaxy and R/Bioconductor. Details of individual tests are mentioned within each figure legend, including biological or experimental replicate number (n) and the reported error either as SEM or 95% confidence intervals. All statistics are *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, nonsignificant (ns), or nondetected (nd). Reproducibility was confirmed by at least three independent experiments. Software and algorithms used in this study are described in table S3.
Acknowledgments
We thank all mouse providers as described in Materials and Methods. We also thank the CNIO Animal Facility for mouse maintenance. We acknowledge M. Ko (Keio University, Systems Medicine, Japan) for providing the pZscan4-Emerald reporter plasmid.
Funding: S.d.l.R. was supported by a fellowship from the Comunidad de Madrid and by funds from the Severo Ochoa-CNIO. This work was funded by grants to N.D. supported by the State Research Agency (AEI; 10.13039/501100011033) from the Spanish Ministry of Science and Innovation (RTI2018-094834-B-I00 and PID2021-122695OB-I00), also including the iDIFFER network of Excellence (RED2022-134792-T), cofunded by European Regional Development Fund (ERDF), by the Comunidad Autónoma de Madrid (S2017/BMD-3817), and by the Asociación Española Contra el Cáncer (AECC) (PRYGN211184DJOU). This work was developed at the CNIO funded by the Health Institute Carlos III (ISCIII) and the Spanish Ministry of Science and Innovation.
Author contributions: S.d.l.R. designed and performed the experiments and analyzed all the data. S.d.l.R. analyzed all the bioinformatics data. M.d.M.R. performed some experiments. P.V. and S.O. performed microinjection in vivo and chimera embryo assay. S.d.l.R and N.D. developed the project and wrote the manuscript. N.D. conceived the project and secured funding.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data are available in the main text or Materials and Methods. Materials are available upon request to N.D. and the sharing of materials described in this work will be subject to standard material transfer agreements.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Tables S1, S3 and S4
Legend for table S2
Other Supplementary Material for this manuscript includes the following:
Table S2
REFERENCES AND NOTES
- 1.Gou L. T., Lim D. H., Ma W., Aubol B. E., Hao Y., Wang X., Zhao J., Liang Z., Shao C., Zhang X., Meng F., Li H., Zhang X., Xu R., Li D., Rosenfeld M. G., Mellon P. L., Adams J. A., Liu M. F., Fu X. D., Initiation of parental genome reprogramming in fertilized oocyte by splicing kinase SRPK1-catalyzed protamine phosphorylation. Cell 180, 1212–1227.e14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collombet S., Ranisavljevic N., Nagano T., Varnai C., Shisode T., Leung W., Piolot T., Galupa R., Borensztein M., Servant N., Fraser P., Ancelin K., Heard E., Parental-to-embryo switch of chromosome organization in early embryogenesis. Nature 580, 142–146 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Borsos M., Perricone S. M., Schauer T., Pontabry J., de Luca K. L., de Vries S. S., Ruiz-Morales E. R., Torres-Padilla M. E., Kind J., Genome-lamina interactions are established de novo in the early mouse embryo. Nature 569, 729–733 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu M., Zernicka-Goetz M., Principles of self-organization of the mammalian embryo. Cell 183, 1467–1478 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casser E., Israel S., Witten A., Schulte K., Schlatt S., Nordhoff V., Boiani M., Totipotency segregates between the sister blastomeres of two-cell stage mouse embryos. Sci. Rep. 7, 8299 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morris S. A., Guo Y., Zernicka-Goetz M., Developmental plasticity is bound by pluripotency and the Fgf and Wnt signaling pathways. Cell Rep. 2, 756–765 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goolam M., Scialdone A., Graham S. J. L., Macaulay I. C., Jedrusik A., Hupalowska A., Voet T., Marioni J. C., Zernicka-Goetz M., Heterogeneity in Oct4 and Sox2 targets biases cell fate in 4-cell mouse embryos. Cell 165, 61–74 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plachta N., Bollenbach T., Pease S., Fraser S. E., Pantazis P., Oct4 kinetics predict cell lineage patterning in the early mammalian embryo. Nat. Cell Biol. 13, 117–123 (2011). [DOI] [PubMed] [Google Scholar]
- 9.White M. D., Angiolini J. F., Alvarez Y. D., Kaur G., Zhao Z. W., Mocskos E., Bruno L., Bissiere S., Levi V., Plachta N., Long-lived binding of Sox2 to DNA predicts cell fate in the four-cell mouse embryo. Cell 165, 75–87 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Hupalowska A., Jedrusik A., Zhu M., Bedford M. T., Glover D. M., Zernicka-Goetz M., CARM1 and paraspeckles regulate pre-implantation mouse embryo development. Cell 175, 1902–1916.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Torres-Padilla M. E., Parfitt D. E., Kouzarides T., Zernicka-Goetz M., Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature 445, 214–218 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macfarlan T. S., Gifford W. D., Driscoll S., Lettieri K., Rowe H. M., Bonanomi D., Firth A., Singer O., Trono D., Pfaff S. L., Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487, 57–63 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asimi V., Sampath Kumar A., Niskanen H., Riemenschneider C., Hetzel S., Naderi J., Fasching N., Popitsch N., du M., Kretzmer H., Smith Z. D., Weigert R., Walther M., Mamde S., Meierhofer D., Wittler L., Buschow R., Timmermann B., Cisse I. I., Ameres S. L., Meissner A., Hnisz D., Hijacking of transcriptional condensates by endogenous retroviruses. Nat. Genet. 54, 1238–1247 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Iaco A., Planet E., Coluccio A., Verp S., Duc J., Trono D., DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet. 49, 941–945 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ribet D., Louvet-Vallée S., Harper F., de Parseval N., Dewannieux M., Heidmann O., Pierron G., Maro B., Heidmann T., Murine endogenous retrovirus MuERV-L is the progenitor of the "orphan" epsilon viruslike particles of the early mouse embryo. J. Virol. 82, 1622–1625 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hendrickson P. G., Doráis J. A., Grow E. J., Whiddon J. L., Lim J. W., Wike C. L., Weaver B. D., Pflueger C., Emery B. R., Wilcox A. L., Nix D. A., Peterson C. M., Tapscott S. J., Carrell D. T., Cairns B. R., Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet. 49, 925–934 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang F., Huang X., Zang R., Chen J., Fidalgo M., Sanchez-Priego C., Yang J., Caichen A., Ma F., Macfarlan T., Wang H., Gao S., Zhou H., Wang J., DUX-miR-344-ZMYM2-mediated activation of MERVL LTRs induces a totipotent 2C-like state. Cell Stem Cell 26, 234–250.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ancelin K., Syx L., Borensztein M., Ranisavljevic N., Vassilev I., Briseño-Roa L., Liu T., Metzger E., Servant N., Barillot E., Chen C. J., Schüle R., Heard E., Maternal LSD1/KDM1A is an essential regulator of chromatin and transcription landscapes during zygotic genome activation. eLife 5, e08851 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dodge J. E., Kang Y. K., Beppu H., Lei H., Li E., Histone H3-K9 methyltransferase ESET is essential for early development. Mol. Cell. Biol. 24, 2478–2486 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J., Hevi S., Kurash J. K., Lei H., Gay F., Bajko J., Su H., Sun W., Chang H., Xu G., Gaudet F., Li E., Chen T., The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 41, 125–129 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Cammas F., Mark M., Dollé P., Dierich A., Chambon P., Losson R., Mice lacking the transcriptional corepressor TIF1beta are defective in early postimplantation development. Development 127, 2955–2963 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Huang Y., Kim J. K., do D. V., Lee C., Penfold C. A., Zylicz J. J., Marioni J. C., Hackett J. A., Surani M. A., Stella modulates transcriptional and endogenous retrovirus programs during maternal-to-zygotic transition. eLife 6, e22345 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakashita A., Kitano T., Ishizu H., Guo Y., Masuda H., Ariura M., Murano K., Siomi H., Transcription of MERVL retrotransposons is required for preimplantation embryo development. Nat. Genet. 55, 484–495 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gstaiger M., Luke B., Hess D., Oakeley E. J., Wirbelauer C., Blondel M., Vigneron M., Peter M., Krek W., Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science 302, 1208–1212 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Chaves-Perez A., Thompson S., Djouder N., Roles and functions of the unconventional prefoldin URI. Adv. Exp. Med. Biol. 1106, 95–108 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Djouder N., Metzler S. C., Schmidt A., Wirbelauer C., Gstaiger M., Aebersold R., Hess D., Krek W., S6K1-mediated disassembly of mitochondrial URI/PP1gamma complexes activates a negative feedback program that counters S6K1 survival signaling. Mol. Cell 28, 28–40 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Herranz-Montoya I., Park S., Djouder N., A comprehensive analysis of prefoldins and their implication in cancer. iScience 24, 103273 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tummala K. S., Gomes A. L., Yilmaz M., Graña O., Bakiri L., Ruppen I., Ximénez-Embún P., Sheshappanavar V., Rodriguez-Justo M., Pisano D. G., Wagner E. F., Djouder N., Inhibition of de novo NAD+ synthesis by oncogenic URI causes liver tumorigenesis through DNA damage. Cancer Cell 26, 826–839 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Buren S., Gomes A. L., Teijeiro A., Fawal M.-A., Yilmaz M., Tummala K. S., Perez M., Rodriguez-Justo M., Campos-Olivas R., Megías D., Djouder N., Regulation of OGT by URI in response to glucose confers c-MYC-dependent survival mechanisms. Cancer Cell 30, 290–307 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Chaves-Perez A., Yilmaz M., Perna C., de la Rosa S., Djouder N., URI is required to maintain intestinal architecture during ionizing radiation. Science 364, eaaq1165 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Lundin V. F., Srayko M., Hyman A. A., Leroux M. R., Efficient chaperone-mediated tubulin biogenesis is essential for cell division and cell migration in C. elegans. Dev. Biol. 313, 320–334 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Delgehyr N., Wieland U., Rangone H., Pinson X., Mao G., Dzhindzhev N. S., McLean D., Riparbelli M. G., Llamazares S., Callaini G., Gonzalez C., Glover D. M., Drosophila Mgr, a Prefoldin subunit cooperating with von Hippel Lindau to regulate tubulin stability. Proc. Natl. Acad. Sci. U.S.A. 109, 5729–5734 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brennecke P., Anders S., Kim J. K., Kołodziejczyk A. A., Zhang X., Proserpio V., Baying B., Benes V., Teichmann S. A., Marioni J. C., Heisler M. G., Accounting for technical noise in single-cell RNA-seq experiments. Nat. Methods 10, 1093–1095 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Piotrowska-Nitsche K., Perea-Gomez A., Haraguchi S., Zernicka-Goetz M., Four-cell stage mouse blastomeres have different developmental properties. Development 132, 479–490 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Lim H. Y. G., Alvarez Y. D., Gasnier M., Wang Y., Tetlak P., Bissiere S., Wang H., Biro M., Plachta N., Keratins are asymmetrically inherited fate determinants in the mammalian embryo. Nature 585, 404–409 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Whyte W. A., Orlando D. A., Hnisz D., Abraham B. J., Lin C. Y., Kagey M. H., Rahl P. B., Lee T. I., Young R. A., Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belteki G., Haigh J., Kabacs N., Haigh K., Sison K., Costantini F., Whitsett J., Quaggin S. E., Nagy A., Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Res. 33, e51 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayashi S., Lewis P., Pevny L., McMahon A. P., Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech. Dev. 119(Suppl 1), S97–S101 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Hayashi S., Tenzen T., McMahon A. P., Maternal inheritance of Cre activity in a Sox2Cre deleter strain. Genesis 37, 51–53 (2003). [DOI] [PubMed] [Google Scholar]
- 40.de Vries W. N., Binns L. T., Fancher K. S., Dean J., Moore R., Kemler R., Knowles B. B., Expression of Cre recombinase in mouse oocytes: A means to study maternal effect genes. Genesis 26, 110–112 (2000). [PubMed] [Google Scholar]
- 41.Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Schöler H., Smith A., Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95, 379–391 (1998). [DOI] [PubMed] [Google Scholar]
- 42.Avilion A. A., Nicolis S. K., Pevny L. H., Perez L., Vivian N., Lovell-Badge R., Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 17, 126–140 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Foygel K., Choi B., Jun S., Leong D. E., Lee A., Wong C. C., Zuo E., Eckart M., Reijo Pera R. A., Wong W. H., Yao M. W. M., A novel and critical role for Oct4 as a regulator of the maternal-embryonic transition. PLOS One 3, e4109 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan M. H., Au K. F., Leong D. E., Foygel K., Wong W. H., Yao M. W. M., An Oct4-Sall4-Nanog network controls developmental progression in the pre-implantation mouse embryo. Mol. Syst. Biol. 9, 632 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keramari M., Razavi J., Ingman K. A., Patsch C., Edenhofer F., Ward C. M., Kimber S. J., Sox2 is essential for formation of trophectoderm in the preimplantation embryo. PLOS One 5, e13952 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu G., Han D., Gong Y., Sebastiano V., Gentile L., Singhal N., Adachi K., Fischedick G., Ortmeier C., Sinn M., Radstaak M., Tomilin A., Schöler H. R., Establishment of totipotency does not depend on Oct4A. Nat. Cell Biol. 15, 1089–1097 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evans M. J., Kaufman M. H., Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156 (1981). [DOI] [PubMed] [Google Scholar]
- 48.Martin G. R., Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. U.S.A. 78, 7634–7638 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ying Q. L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A., The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruzankina Y., Pinzon-Guzman C., Asare A., Ong T., Pontano L., Cotsarelis G., Zediak V. P., Velez M., Bhandoola A., Brown E. J., Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell 1, 113–126 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bedzhov I., Zernicka-Goetz M., Self-organizing properties of mouse pluripotent cells initiate morphogenesis upon implantation. Cell 156, 1032–1044 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shahbazi M. N., Scialdone A., Skorupska N., Weberling A., Recher G., Zhu M., Jedrusik A., Devito L. G., Noli L., Macaulay I. C., Buecker C., Khalaf Y., Ilic D., Voet T., Marioni J. C., Zernicka-Goetz M., Pluripotent state transitions coordinate morphogenesis in mouse and human embryos. Nature 552, 239–243 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gaume X., Torres-Padilla M. E., Regulation of reprogramming and cellular plasticity through histone exchange and histone variant incorporation. Cold Spring Harb. Symp. Quant. Biol. 80, 165–175 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Lu F., Zhang Y., Cell totipotency: Molecular features, induction, and maintenance. Natl. Sci. Rev. 2, 217–225 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Niwa H., Toyooka Y., Shimosato D., Strumpf D., Takahashi K., Yagi R., Rossant J., Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123, 917–929 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Zalzman M., Falco G., Sharova L. V., Nishiyama A., Thomas M., Lee S. L., Stagg C. A., Hoang H. G., Yang H. T., Indig F. E., Wersto R. P., Ko M. S. H., Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature 464, 858–863 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.J. Yang, L. Cook, Z. Chen, Systematic perturbation of thousands of retroviral LTRs in mouse embryos. bioRxiv 2023.09.19.558531 (2023). 10.1101/2023.09.19.558531. [DOI]
- 58.Taubenschmid-Stowers J., Rostovskaya M., Santos F., Ljung S., Argelaguet R., Krueger F., Nichols J., Reik W., 8C-like cells capture the human zygotic genome activation program in vitro. Cell Stem Cell 29, 449–459.e6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goke J., Lu X., Chan Y.-S., Ng H.-H., Ly L.-H., Sachs F., Szczerbinska I., Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell 16, 135–141 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Bryja V., Bonilla S., Arenas E., Derivation of mouse embryonic stem cells. Nat. Protoc. 1, 2082–2087 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Fu X., Wu X., Djekidel M. N., Zhang Y., Myc and Dnmt1 impede the pluripotent to totipotent state transition in embryonic stem cells. Nat. Cell Biol. 21, 835–844 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee G. Y., Kenny P. A., Lee E. H., Bissell M. J., Three-dimensional culture models of normal and malignant breast epithelial cells. Nat. Methods 4, 359–365 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eckersley-Maslin M. A., Svensson V., Krueger C., Stubbs T. M., Giehr P., Krueger F., Miragaia R. J., Kyriakopoulos C., Berrens R. V., Milagre I., Walter J., Teichmann S. A., Reik W., MERVL/Zscan4 network activation results in transient genome-wide DNA demethylation of mESCs. Cell Rep. 17, 179–192 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sharov A. A., Masui S., Sharova L. V., Piao Y., Aiba K., Matoba R., Xin L., Niwa H., Ko M. S. H., Identification of Pou5f1, Sox2, and Nanog downstream target genes with statistical confidence by applying a novel algorithm to time course microarray and genome-wide chromatin immunoprecipitation data. BMC Genomics 9, 269 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reske J. J., Wilson M. R., Chandler R. L., ATAC-seq normalization method can significantly affect differential accessibility analysis and interpretation. Epigenetics Chromatin 13, 22 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Tables S1, S3 and S4
Legend for table S2
Table S2