Abstract
Background & Aims
Bulevirtide (BLV) was approved for the treatment of compensated chronic hepatitis D virus (HDV) infection in Europe in 2020. However, research into the effects of the entry inhibitor BLV on HDV-host dynamics is in its infancy.
Methods
Eighteen patients with HDV under nucleos(t)ide analogue treatment for hepatitis B, with compensated cirrhosis and clinically significant portal hypertension, received BLV 2 mg/day. HDV RNA, alanine aminotransferase (ALT), and hepatitis B surface antigen (HBsAg) were measured at baseline, weeks 4, 8 and every 8 weeks thereafter. A mathematical model was developed to account for HDV, HBsAg and ALT dynamics during BLV treatment.
Results
Median baseline HDV RNA, HBsAg, and ALT were 4.9 log IU/ml [IQR: 4.4-5.8], 3.7 log IU/ml [IQR: 3.4-3.9] and 106 U/L [IQR: 81-142], respectively. During therapy, patients fit into four main HDV kinetic patterns: monophasic (n = 2), biphasic (n = 10), flat-partial response (n = 4), and non-responder (n = 2). ALT normalization was achieved in 14 (78%) patients at a median of 8 weeks (range: 4-16). HBsAg remained at pre-treatment levels. Assuming that BLV completely (∼100%) blocks HDV entry, modeling indicated that two HDV-infected cell populations exist: fast HDV clearing (median t1/2 = 13 days) and slow HDV clearing (median t1/2 = 44 days), where the slow HDV-clearing population consisted of ∼1% of total HDV-infected cells, which could explain why most patients exhibited a non-monophasic pattern of HDV decline. Moreover, modeling explained ALT normalization without a change in HBsAg based on a non-cytolytic loss of HDV from infected cells, resulting in HDV-free HBsAg-producing cells that release ALT upon death at a substantially lower rate compared to HDV-infected cells.
Conclusion
The entry inhibitor BLV provides a unique opportunity to understand HDV, HBsAg, ALT, and host dynamics.
Impact and implications
Mathematical modeling of hepatitis D virus (HDV) treatment with the entry inhibitor bulevirtide (BLV) provides a novel window into the dynamics of HDV RNA and alanine aminotransferase. Kinetic data from patients treated with BLV monotherapy can be explained by hepatocyte populations with different basal HDV clearance rates and non-cytolytic clearance of infected cells. While further studies are needed to test and refine the kinetic characterization described here, this study provides a new perspective on viral dynamics, which could inform evolving treatment strategies for HDV.
Keywords: HDV RNA, Bulevirtide, mathematical modeling, HBsAg, ALT
Graphical abstract
Highlights
-
•
Bulevirtide (BLV)’s mechanism of action provides a unique opportunity to understand the dynamics of HDV and HBV infection.
-
•
Assuming ∼100% blockage of HDV entry by BLV, mathematical modeling suggested that there are two populations of HDV-infected liver cells.
-
•
Modeling explained how ALT levels can normalize without a change in HBsAg levels under BLV.
Introduction
Hepatitis D virus (HDV) is an infectious subviral agent that can only propagate in people infected with hepatitis B virus (HBV), which supplies the necessary envelope proteins needed to assemble infectious HDV progeny virions. HDV was recognized as a distinct agent in 19771,2 and consists of a negative-sense, single-stranded, circular RNA genome. HDV is a serious clinical concern because ∼9-19 million persons worldwide are chronically infected with HDV, i.e. about 5% of those with chronic HBV,3,4 and chronic hepatitis D (CHD) has been linked to a much more accelerated course of liver disease compared to chronic mono-infection with HBV5 making it the most severe form of chronic viral hepatitis in humans.6 Nucleos(t)ide analogues (NUCs) for HBV are not effective against CHD.7 Sustained suppression of HDV with pegylated interferon-α (IFN) is very low (∼25%) and relapse rates are high even with 5 years of treatment.8,9 The reason for such low HDV cure rates is not known, in part because there is little information on the HDV-HBV-host dynamics.
Bulevirtide (BLV) at the dose of 2 mg/day was recently approved for the treatment of compensated CHD in Europe.10 BLV blocks the binding site of the human sodium taurocholate co-transporting polypeptide on the HBV envelope, thereby inhibiting HDV/HBV entry into hepatocytes.11 However, clinical data collected thus far12,13 do not provide clear guidance on the optimal use of BLV treatment (alone or in combination with IFN) for CHD, including duration of therapy. Thus, there is an urgent need to understand HDV-host dynamics under BLV treatment and to develop optimal response-guided therapy.
Very recently, we reported high effectiveness (78% virological response; 83% biochemical response; 67% combined response) and a good safety profile of BLV 2 mg/day monotherapy in 18 patients with difficult-to treat HDV genotype-1 who had compensated cirrhosis and clinically significant portal hypertension (CSPH), with or without hepatocellular carcinoma (HCC).14 The current study is the first to characterize and mathematically model HDV RNA, hepatitis B surface antigen (HBsAg) and alanine aminotransferase (ALT) kinetics under BLV monotherapy in patients with compensated cirrhosis and CSPH.
Materials and methods
Patient population
Eighteen HBeAg-negative patients with HDV-related compensated cirrhosis and CSPH were included in this study at the outpatient Liver Clinic of the Hepatology Division of the Foundation IRCCS Ca Granda Ospedale Maggiore Policlinico as recently described.14 Patients were all Caucasian, with a mean age of 51 ± 14 years old and 67% were male. They had a mean BMI of 24 and a mean initial ALT of 110 ± 48 U/L (Table S1). Participants self-administered subcutaneous BLV 2 mg/day monotherapy starting in December 2020. All patients were receiving tenofovir disoproxil fumarate or entecavir treatment for HBV. CHD was defined as HDV RNA positivity for more than 6 months. Cirrhosis and CSPH were defined in these patients as recently reported.14 Baseline characteristics are provided in Table S1.
BLV treatment was approved on a case-by-case basis by the Italian Medicines Agency (AIFA) and provided in the context of the 5% AIFA fund program. Informed consent was obtained from all 18 participants, according to the Helsinki Declaration. The study was approved by the local IRB (Comitato Etico Area 2 Milano).
Follow-up and measurements
All kinetic data were collected at treatment baseline, week 4, 8, 16, 24 and 48. HDV RNA was quantified by RoboGene® HDV RNA quantification 2.0 (Aj-Roboscreen, Jena, Germany; lower limit of quantification [LLoQ] 6 IU/ml). HBsAg was quantified by Elecsys HBsAg II quantitative assay on the Cobas®e801 Analyzer (Roche Diagnostics Gmbh, Mannheim, Germany; LLoQ 0.05 IU/ml). HBV DNA was quantified by Cobas® HBV Test on the Cobas® 4800 System (Roche Diagnostics, Germany; LLoQ 10 IU/ml). Kinetic data at weeks 24 and 48 are provided in Table S2.
Characterization of HDV kinetic patterns
The viral decline patterns were categorized into three main groups by empirical analysis, as recently done to characterize hepatitis E virus and HDV under antiviral treatments.15,16 Each identified slope (or phase) was calculated by linear regression using scipy.stats package. We defined a viral decline as monophasic if only one phase was present or two viral decline slopes were identified but differed by less than a factor of 2. A biphasic (BP) decline pattern was defined in cases where the first phase rate of decline was at least 2-fold faster than the second phase rate of decline. A flat-partial response (FPR) was defined in individuals who had a first phase viral decline followed by an extremely slow second phase that was extremely slow or flat, i.e. slope was not significantly different from zero. A viral breakthrough was defined as a ≥1 log IU/ml increase from nadir viral load. ALT normalization was defined as the achievement of values ≤41 U/L for females and ≤59 U/L for males.
Mathematical model
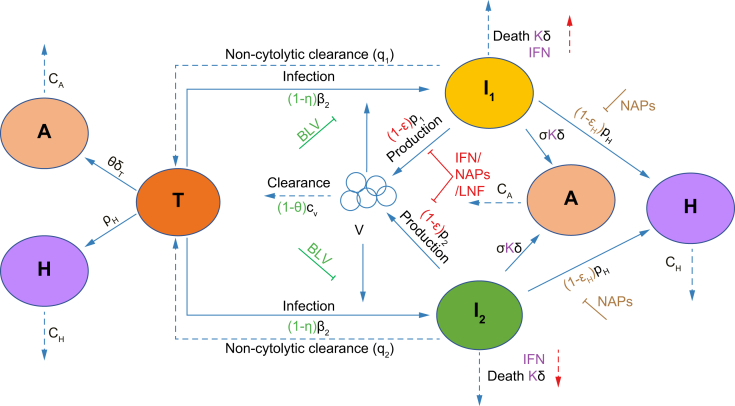
We further developed our previously published mathematical models of HDV infection and treatment[17], [18], [19] to describe HDV kinetics under treatment with BLV. The most important modifications of the previous models (Eq. 1 and Fig. 1) were the addition of two distinct infected cell populations and an estimate of ALT dynamics. In addition, we extended our previous models18,19 to explicitly describe the dynamics of HBsAg produced not only from HDV-infected cells but also from HDV-free target cells. For simplicity, since the model simulates BLV treatment in chronically HDV/HBV-coinfected patients, we ignored the dynamics of HBV-negative cells and/or HDV-monoinfected cells.[20], [21], [22], [23] The new model used here accounts for key features of the empirical data including a predominance of biphasic HDV RNA decline patterns and stability of HBsAg levels with declining HDV RNA and ALT levels. Mathematical modeling of viral kinetics predicts a monophasic viral decline under antiviral treatment that blocks viral infection.24 Modeling approaches with more than one HDV-infected cell population with differing life spans were considered to explain biphasic patterns of decline under ∼100% entry blockade with BLV. Non-cytolytic clearance of HDV from HBsAg-producing infected cells was hypothesized (supported, in part, by recent experimental findings25) to explain the stability of HBsAg during HDV RNA declines. Differing rates of ALT release upon death of HDV-infected and HDV-free (target) cells were hypothesized.
Fig. 1.
Schematic of the model.
Our mathematical model (Eq. 1) incorporates the viral dynamics of HBV-positive HDV-naïve target cells (T), two types of HDV-infected cells (I1 and I2), HDV virions (V), ALT (A), and HBsAg (H). Target cells become infected and convert to I1 at rate β1 and to I2 at rate β2. Virions are cleared at rate cV. I1 and I2 under non-cytolytic clearance to become target cells at rates q1 and q2, and die at rate δ. Infected cells produce virions at rate p. Target cells are produced at rate pT and die at rate δT. ALT is produced by target cell death at rate φ and by infected cell death at rate σ. ALT is cleared from blood at rate cA. HBsAg is produced by target cells and HDV-infected cells at rate pH and cleared from blood at rate cH. BLV blocks infection with efficacy η and may reduce viral clearance with efficacy θ as shown with the green symbols. Blocking of HDV production by IFN, LNF or NAPs is shown using red symbols (parameter ε). Secondary effects of IFN in increasing cell death are reflected by κ >1 and NAPs efficacy in blocking HBsAg production is shown with parameter εH. ALT, alanine aminotransferase; BLV, bulevirtide; HBsAg, hepatitis surface antigen; HBV, hepatitis B virus; HDV, hepatitis D virus; IFN, interferon-α/λ; LNF, lonafarnib; NAPs, nucleic acid polymers.
The model accounts for target liver cells, T; the serum HBsAg concentration, H; fast HDV-clearing infected cell population, I1; slow HDV-clearing infected cell population, I2; ALT concentration, A; and the serum HDV viral load concentration, V.
| (Eq. 1) |
where target cells, T, (already HBsAg-infected) are infected by HDV virions, V, and become fast HDV-clearing infected cells (I1) at rate β1, and slow HDV-clearing infected cells (I2) at rate β2. Target cells are also produced at rate pT and die at rate δT. Fast HDV-clearing infected cells (I1) undergo non-cytolytic clearance at rate q1 and slow HDV-clearing infected cells (I2) undergo non-cytolytic clearance at rate q2. Both I1 and I2 are assumed to die at rate constant δ. HDV virions (V) are produced by fast clearing infected cells at rate p1 and by slow clearing infected cells at rate p2 and are cleared at rate cV. The theoretical secondary effect of BLV in reducing HDV clearance17 is set at θ. Finally, HBsAg (H) is produced by all cells (I1, I2, and T) at rate pH (regardless of if produced from integrated HBV DNA or covalently closed circular DNA) and cleared at rate cH. ALT (A) is produced at rates σ and φ from the death of HDV-infected cells and HDV-free target cells, respectively, and is cleared in blood at rate cA. To account for the contribution of ALT from HBsAg-HDV-negative hepatocytes that are not accounted for in the model and for ALT produced by extrahepatic tissues, a fixed rate of ALT production from these sources, sx was included.
The efficacy of BLV in blocking HDV infection is represented by η, while the theoretical secondary effect of BLV in reducing HDV clearance17 has efficacy θ. Other anti-HDV treatments may block HDV RNA production, which is denoted by parameter ε, block production of HBsAg with efficacy εH19 and increase the death rate of HDV-infected cells (I1 and I2) with κ >1.
Model parameter estimations and initial conditions
We assume a pre-treatment steady state in HDV RNA viremia based on previously reported evidence of minor fluctuations over periods of weeks to months in the absence of treatment.26 The pre-treatment ratio of the concentration of fast HDV-clearing infected cells, I10, to slow HDV-clearing infected cells, I20, was defined as , where Iratio is the ratio of I1 and I2. We further set the following relations for the initial pre-treatment steady-state conditions:
Since we observe no decline in HBsAg, we assume similar target cell production and death rates, i.e., pT = δT = 0.001 day-1, respectively, in line with previous studies that estimated the HDV-infected cell death rate.27,28 HDV RNA, HBsAg and ALT clearance rates were set to day-1, day-1, and day-1 based on previous estimates.19,29,30 To reduce uncertainty and avoid identifiability issues, the following parameters were fixed to virions-1 day-1, virions cell-1 day-1, day-1, and sx=20 IU/ml/day.31 Pre-treatment ALT (A0) was set per each patient’s measured value (Table 1). As was done recently17 we assumed that BLV blocks HDV infection and might also slow HDV clearance which is reflected in the parameter , which we set to , with no effect on blocking of HBsAg (εH = 0) or HDV RNA (ε = 0) production or increase of HDV-infected-cell death (κ = 1).
Table 1.
Parameter fit values for individual patients
| Patient # | Viral kinetic pattern | V0 | H0 | A0 | q1 (t1/2) | q2 (t1/2) | I2_frac (%) | σδ/φ δT | HBsAg cells& (%) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | BP | 5.1 | 3.7 | 151 | 0.080 (8.6) | 0.014 (47.5) | 5.3 | 734 | 4.7 |
| 2∗∗ | BP | 5.3 | 3.9 | 172 | 0.042 (16.4) | 0.021 (33.7) | 3.9 | 56 | 4.2 |
| 3 | FPR | 5.7 | 3.7 | 107 | 0.045 (15.4) | 0.000 (-) | 0.6 | 41 | 7.7 |
| 4 | FPR+D | 3.3 | 3.7 | 79 | 0.116 (6.0) | 0.000 (-) | 2.2 | 2625 | 4.9 |
| 5 | BP | 4.7 | 3.9 | 99 | 0.181 (3.8) | 0.037 (18.6) | 6.5 | 369 | 7.8 |
| 6† | BP+B | 4.4 | 4.1 | 84 | 0.209 (3.3) | 0.014 (48.1) | 0.15 | 796 | 9.2 |
| 8 | MP | 5.5 | 4.1 | 49 | 0.032 (21.7) | 6.96 (0.1) | 0.1∗ | 4.5 | 3.1 |
| 10 | MP | 3.2 | 3.5 | 51 | 0.025 (27.4) | 15.2 (-) | 0.1∗ | 1130 | 4.6 |
| 11 | BP | 4.3 | 4.0 | 113 | 0.232 (3.0) | 0.027 (25.2) | 2.6 | 517 | 9.9 |
| 12 | BP | 4.5 | 2.6 | 155 | 0.045 (15.3) | 0.017 (40.1) | 7.1 | 756 | 2.2 |
| 13 | FPR | 3.9 | 3.0 | 86 | 0.065 (10.6) | 0.000 (-) | 3.1 | 2375 | 2.8 |
| 14 | BP | 6.1 | 4.3 | 222 | 0.035 (19.6) | 0.004 (197.3) | 0.3 | 11 | 1.5 |
| 15 | FPR | 6.2 | 3.9 | 105 | 0.042 (16.6) | 0.000 (-) | 1.5 | 12 | 1.7 |
| 16 | BP | 5.9 | 3.4 | 112 | 0.047 (14.6) | 0.009 (79.4) | 0.8 | 13 | 1.0 |
| 17∗∗∗ | BP+B | 3.7 | 3.8 | 32 | 0.095 (7.3) | 0.006 (121.9) | 1.9 | 40 | 4.1 |
| 18 | BP | 4.3 | 2.6 | 145 | 0.069 (10.1) | 0.019 (37.4) | 2.0 | 1564 | 3.1 |
| Med. | — | 4.6 | 3.8 | 106 | 0.056 (12.6) | 0.014 (47.5) | 2.0 | 443 | 4.8 |
We show the values of the baseline initial estimates of V0 (HDV RNA viral load) and H0 (HBsAg) for each patient, and the set initial value of ALT, A0. We also show the rates of clearance of fast HDV-clearing and slow HDV-clearing infected cells, along with their associated half-lives in parenthesis. We convert the Iratio to the fraction of cells that are slow HDV clearing. Finally, although only σ is explicitly fit, we report the more clinically meaningful ratio σδ/φδT, representing the excess ALT secretion from HDV-infected cells compared to uninfected cells. Fixed parameters were β1 = 1e-8, p2 = p1 =10, cH = 0.53, η = 1, θ = 0.1, cV = 0.42, cA = 2.5, ε = 0, εH = 0, κ = 1 and I20, T0, β2, pH and φ were set by steady state. &HBsAg cells represent the percentage of cells actively producing HBsAg in the model (i.e., T, I1, and I2) of total HBsAg-free hepatocytes (estimated at 107 cells/ml) in the liver, which are not modeled.
ALT, alanine aminotransferase; BP, biphasic; BP+B, biphasic followed by breakthrough; FPR, flat-partial response; FPR+D; flat-partial response followed by decline; HBsAg, hepatitis surface antigen; HDV, hepatitis D virus; MP, monophasic.
For these patients I2_frac was fixed because the second phase was not observed.
Fit only through week 40 due to later increase.
Fit only through week 32 due to later increase.
Fit only through week 16 due to increase.
The remaining parameters q1, q2, Iratio, and σ along with the pre-treatment HDV RNA (V0), and HBsAg (H0) were estimated by simultaneously fitting the model with measured HDV RNA, HBsAg, and ALT kinetic data for each patient.
All measured data points up to 48 weeks were included in the fit unless the patient reached HDV RNA under the LloQ (<6 log IU/ml) in which case the first point that was under the LLoQ was included, or if the patient experienced a rebound (Patients 6, 17) or sudden decrease (Patient 4) the last point up until the change in trend was included. Every included data point for HDV RNA, HBsAg and ALT had equal weight in the fitting based on minimizing least-squares. The error function was such that errors were fit on the log-transformed values of HDV RNA and HBsAg, whereas ALT was fit on a linear scale. To compensate for the fact that ALT differences would have a larger effect on our error, we overweighted errors in HDV RNA to have 100 times as much impact as errors in ALT or HBsAg. It is worth noting that because we cannot detect the respective compartments T, I1 and I2, we use a simplified fitting procedure, which can lead to bias. We used Python 3.7 and Scipy Version 1.0 to estimate the parameter values.
Statistical analysis
The Kruskal-Wallis test was used to compare how baseline characteristics correlated with HDV kinetic patterns. The Pearson Correlation test or t test was used to compare how estimated model parameters correlated with baseline characteristics. All tests were 2-sided and used a significance level of 0.05. Slopes were defined as flat (not different from zero) if the p value of linear regression was >0.05. The Bonferroni correction was applied in order to counteract the multiple testing problem.
Results
Characterization of HDV, HBsAg, and ALT kinetics under BLV monotherapy
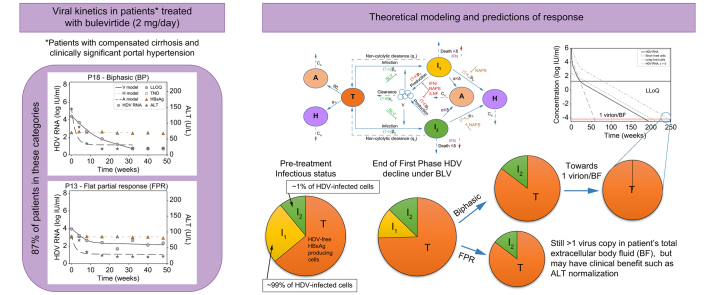
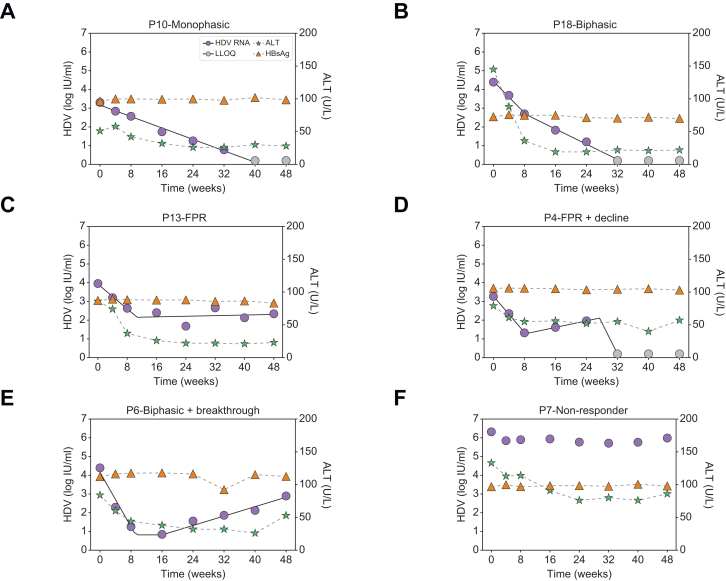
Median baseline HDV RNA, HBsAg, and ALT were 4.9 log IU/ml [IQR 4.4-5.8], 3.7 log IU/ml [IQR:3.4-3.9], and 106 U/L [IQR 81-142], respectively. HBV DNA was suppressed by NUCs, with 72% of patients having undetectable HBV DNA and median HBV DNA levels of 15 (range 14-22) IU/ml in the remaining patients. During 48 weeks of therapy, patients displayed several distinct HDV RNA kinetic patterns (Figs 2 and S1). Two patients had a monophasic decline in HDV RNA (Fig. 2A); eight patients experienced a biphasic decline (Fig. 2B); three had a FPR (Fig. 2C); one patient experienced a FPR followed by decline (Fig. 2D), while another two patients experienced a biphasic decline followed by a viral breakthrough (Fig. 2E), and two patients were non-responders (Fig. 2F).
Fig. 2.
HDV kinetic patterns under BLV monotherapy in patients with compensated cirrhosis and clinically significant portal hypertension.
(A) Monophasic, i.e. a single phase of viral decline (n = 2), (B) biphasic, viral decline consisting of a first phase decline that was 2-fold faster than the second phase decline (n = 8), (C) FPR, defined as a first phase viral decline followed by a plateau that was not significantly (p >0.05) different from slope=0 (n = 3), (D) FPR followed by decline (n = 1), (E) Biphasic followed by breakthrough (n = 2), and (F) non-responder, defined as having <∼1.0 log IU/ml decline from baseline (n = 2). Solid lines for HDV reflect modelling using piecewise linear regression and dashed lines between ALT markers are shown only to highlight the trajectory. ALT, alanine aminotransferase; BLV, bulevirtide; FPR, flat-partial response; HDV, hepatitis D virus.
The majority of patients (16/18) had no significant variation in HBsAg, i.e. <0.25 log over the course of treatment (Fig. 2). ALT normalization was achieved in 16 (89%) patients at a median of 12 weeks (range: 0 to 48 weeks) from initiation of BLV therapy (Figs 2 and S2). Overall, no significant associations were found between HDV RNA kinetic patterns and baseline characteristics (Table S1). Likewise, no significant associations were found between kinetic patterns and the presence of detectable HBV DNA or the time to ALT normalization.
Modeling suggests two populations of HDV-infected liver cells: A fast HDV-clearing and a slow HDV-clearing population
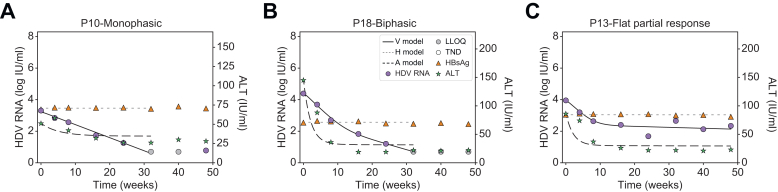
Modeling approaches considered differing numbers of HDV-infected cell populations and rates of non-cytolytic HDV clearance to transition from HDV-infected cells to HDV-free cells. A model with two cell populations – fast HDV-clearing and slow HDV-clearing cells (Eq. 1) – best explained the rapid first phase decline and the slower or flat second phase decline and calibrated well with each patient’s measured HDV RNA, HBsAg and ALT kinetics (Fig. 3 and Fig. S2). The median pre-treatment levels of HDV RNA and HBsAg were estimated as V0=4.6 log IU/ml and H0=3.8 log IU/ml, respectively (Table 1). The rapid first phase HDV RNA decline corresponded to a median half-life of 12.6 days for fast HDV-clearing cells. Slow HDV-clearing cells had a median half-life of 43.8 days across the 10 patients in whom a non-zero second HDV phase was observed. Fast HDV-clearing cells made up a median of 98.0% [92.9-99.9%] of all HDV-infected cells.
Fig. 3.
Representative model fits with patients experiencing monophasic, biphasic and flat-partial response in HDV.
We see that the viral load, V, shows excellent agreement with the measured data. We further see that the biphasic decline in viral load is explained by a larger cell population I1, which experiences fast clearance, together with a smaller cell population I2, experiencing slower clearance. Likewise, ALT and HBsAg are well described by our model. Model fit values are in Table 1. Fitting curves of all patients are shown in Fig. S2. ALT, alanine aminotransferase; BLV, bulevirtide; HBsAg, hepatitis surface antigen; HDV, hepatitis D virus; LLoQ, lower limit of quantification; TND, target not detected.
Modeling evaluated differing rates of ALT release upon death between HDV-infected cells and HDV-free (target) cells to understand ALT level declines. Based on the observed ALT kinetic data, the final model predicted that upon cell death, HDV-infected cells release substantially more ALT (443 times) compared to HDV-free (target) cells (Table 1). Overall, no significant associations were found among estimated model parameters (Table 1) and baseline characteristics (Table S1).
Implications for future response-guided therapy for HDV with BLV-based therapies
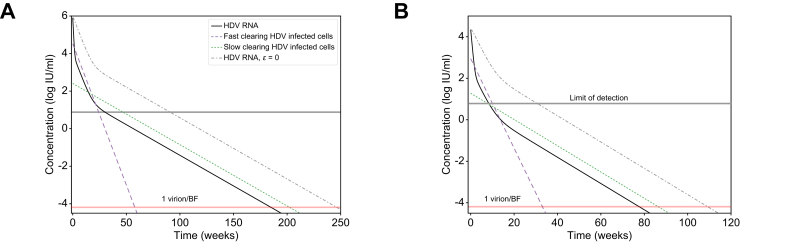
To further explore implications of the predicted two HDV-infected cell populations, we simulated the model (Eq. 1) under a theoretical combination therapy with BLV and anti-HDV drug(s) that block viral production with efficacy ε (e.g., interferon-α,18 interferon-λ,32 lonafarnib29 or nucleic acid polymers19). In Fig. 4 we examined such dual therapy in silico according to the estimated model parameters of patients 16 and 18 under BLV (Table 1) and considered the hypothetical effect of combination therapy with BLV blocking ∼100% HDV infection and potent drugs that block ∼99% of HDV production as previously estimated.18,19,29,32 Under dual therapy, HDV viral load was predicted to decline via three phases (compared to two phases under BLV alone, Fig. 4 dashed-dotted lines). The phases include a first rapid viral decline reflecting the half-life of HDV in blood due to the treatment that blocks viral production (ε = 99%), followed by a second slower phase that reflects the lifespan of fast HDV-clearing cells, followed by a final third phase that reflects the life span of slow HDV-clearing cells (Fig. 4). The in silico data suggest that the third phase might not always be evident from measurements of viral load in a clinical setting. One constraint on whether the third phase will be observable clinically is whether it begins before the viral load declines to below the limit of HDV RNA assay detection, 6 IU/ml for the Robogene 2.0 assay. For example, in Fig. 4B, the third phase would not be observed clinically since it only begins after the patient’s viral load is below the LLoQ. This could impact predictions about the duration of BLV-based therapy needed to reach <1 virus copy in the entire extracellular body fluid, reminiscent of the notion of predicting the duration of hepatitis C virus treatment with direct-acting antivirals needed to reach <1 virus copy in a patient’s total extracellular body fluid (BF).33 For example, in Fig. 4B a continuation of the second phase would lead to an estimated time to reach <1 virus copy/BF of around 40 weeks, while the less steep slope of the third phase leads to an estimated time of 80 weeks. Furthermore, even in Fig. 4A, where the third phase begins before the HDV viral load is below the LLoQ, it could still be missed clinically if sampling is not frequent enough to capture the third phase decline, again potentially leading to underestimation of the time to reach <1 virus/BF. In both theoretical patients, dual therapy shortened the time to reach <1 virus/BF by ∼48 weeks compared to BLV monotherapy (Fig. 4).
Fig. 4.
Model predictions of HDV decline under BLV monotherapy or in combination with anti-HDV drugs that block viral production.
Model prediction to reach <1 virus copy in representative patients’ total extracellular BF under combination therapy with blocking of ∼100% of HDV entry by BLV monotherapy (dashed-dotted black lines) or in combination with drug(s) that block ε = 99% of HDV production (solid black lines, blue and green dashed lines). The HDV RNA curve for ε = 0 is also shown for reference. Limit of HDV detection in the blood is shown as the upper horizontal line. 1 virus/BF is defined as 1 virus per patients’ total extracellular BF of 15 L (lower horizontal line). Model parameters in (A) and (B) were set to reflect the P16 and P18 parameter estimates shown in Table 1. For simplicity, additional treatment effects of nucleic acid polymer in blocking HBsAg production (εH) and interferon-α or interferon-λ in inducing death rate of HDV-infected cells (κ) in the model (Eq. 1 and Fig. 1) were not simulated, i.e. the parameters were set εH = 0 and κ = 1. BF, body fluid; BLV, bulevirtide; HDV, hepatitis D virus.
Discussion
We recently reported that that BLV 2 mg/day monotherapy administered for 48 weeks in 18 patients with HDV-related compensated cirrhosis and CSPH was safe and led to a virological response (i.e., ≥2 log IU/ml decline in HDV RNA from baseline) in 16 patients (89%). In the current study, we analyzed in detail the HDV RNA, HBsAg and ALT kinetics in these 16 patients with a virological response and found that only two patients (13%) experienced a monophasic HDV RNA decline. The other 14 (87%) patients experienced a first phase rapid viral decline that was followed by a slower (biphasic) or extremely slow (FPR) second phase, of whom two patients experienced viral breakthrough. The kinetic description provides the first comprehensive picture of HDV RNA, ALT and HBsAg under BLV monotherapy in patients with cirrhosis and CSPH.
Since BLV is an entry inhibitor,11 which is unlikely to block viral production, it provides a unique opportunity to understand HDV-host dynamics. Mathematical modeling with or without hepatocyte proliferation predicts a monophasic viral decline under drugs that block viral infection, reflecting attrition of productively HDV-infected cells.17,24,32,34 To explain why 87% of patients experienced two viral kinetic phases (BP or FPR), we hypothesized the existence of two HDV-infected populations. HDV-infected hepatocytes with a rapid HDV clearance rate (non-cytolytic) could explain the rapid first phase HDV decline. In the case of FPR, a flat second phase suggests a population of infected cells with a very slow HDV clearance rate. Alternatively, the flat second phase viral load could be partially explained (not explored in silico herein) by (i) death of slow HDV-clearing cells that are being replaced by proliferation of bystander slow HDV-clearing cells, and/or (ii) the existence of a subpopulation of HDV-susceptible cells that are resistant to BLV treatment, though are still able to be infected with HDV despite maintaining a lower steady state viral load compared to pre-treatment levels, and/or (iii) suboptimal BLV dosage or drug distribution in the liver. Scenario (i) is unlikely since death of infected cells would be counter to the ALT normalization observed in the patients exhibiting FPR kinetics. Scenario (ii) could be partially explained if HDV cell-free infection is blocked by BLV while cell-to-cell spread is not. However, in that case, we would not expect a flat second phase to be maintained for several weeks without further HDV cell-to-cell spread in the liver (resulting in a breakthrough increase in HDV RNA) unless cell-to-cell spread is restricted in the liver. Scenario (iii) is possible since none of the reported preliminary results in three patients with CHD treated with 10 mg of BLV indicated a FPR,17 although unlike in the current study, these patients did not have CSPH. Moreover, we cannot completely exclude that missed doses of BLV could have contributed to suboptimal BLV exposure, although patients reported adherence to therapy. Interestingly, a phase III study35 of BLV for chronic HDV showed a higher proportion of patients achieving undetectable HDV RNA at different time points with 10 mg compared to 2 mg, providing support for scenario (iii). Future studies could explore the effect of BLV dose increases in FPR cases, however, data regarding the safety and effectiveness of 10 mg in patients with advanced cirrhosis (and CSPH) are currently limited.
In our model we assumed two infected cell populations that both have similar HDV RNA production rates, viz. p1=p2=10. However we cannot rule out that the two different cell populations actually have different HDV production rates reminiscent of in vitro studies.36,37 Due to identifiability issues, differences in production rate cannot be determined based on the available data. Likewise, these in vitro studies might suggest the existence of different hepatocyte phenotypes. It is possible that the two cell types we propose here are actually different phases of infection within hepatocytes displaying different phenotypes. Future experimental and theoretical efforts could examine whether the two populations hypothesized here are actually distinct cell types or different phenotypes of the same cells.
The current model explains not only the HDV RNA kinetic patterns but also the observed ALT normalization and stable HBsAg levels during BLV treatment. To account for the latter, we assumed non-cytolytic HDV clearance from infected cells such that these cells still produced HBsAg concomitantly to the dramatic decline in HDV RNA levels in serum. This HDV loss from infected cells is reminiscent of the strong intrahepatic HDV declines in HDV RNA and HDAg+ cells that were recently reported in patients treated with 2 mg/day of BLV for 48 weeks.25 Furthermore, stable HBsAg (in parallel to declines in HDV RNA) could not be explained by hepatocyte proliferation unless only HBsAg-producing cells that are not HDV-infected proliferate, or HDV-infected cells proliferate in a way that the daughter cells are not HDV positive. The latter scenario is unlikely based on in vitro studies that demonstrated that HDV propagates to both cells during cell division.38 Likewise, we believe that the other scenarios are less likely than non-cytolytic clearance, however experimental confirmation is needed. To account for ALT normalization, the modelling results suggest that once HDV-infected cells become HDV-free under BLV, they release ALT into circulation upon their death at a lower rate compared to the ALT released by the death of HDV-infected cells. This prediction needs to be experimentally examined as well.
Reminiscent of the notion of modeling-based response-guided therapy for hepatitis C virus kinetics, where predictions were made for the duration of direct-acting-agent therapy to reach <1 virus copy in a patient’s total extracellular BF (i.e., sustained virologic response or cure),33 we recently predicted the time to reach <1 copy of HDV RNA with biphasic kinetics in three patients who initiated treatment with 10 mg/day of BLV.17 All three patients achieved HDV RNA target not detected (TND) within 29-43 weeks post initiation of BLV treatment. Modeling HDV RNA kinetics retrospectively suggested that a patient who had viral rebound (from HDV RNA TND) after stopping 52 weeks of 10 mg/day BLV did not reach <1 virion/BF at the end of therapy. In addition, modeling predicted17 that the other two patients who were treated for 144 weeks could have already reached <1 virion/BF. Indeed one of these two patients was recently reported to remain HDV RNA undetectable 72 weeks after BLV discontinuation, suggesting HDV viral cure.39 In the current study, only one patient (Patient 4) reached HDV RNA TND by week 48 after having had a transient viral increase between 8 and 24 weeks before HDV RNA became TND. Because the current model was not designed to explain spontaneous transient viral increase (i.e., not related to compliance), it was not possible to model the time to reach <1 virion/BF. Thus, the limited 48-week treatment duration does not allow us to predict the duration needed to reach <1 virion/BF in patients who achieved HDV RNA TND. While the current treatment does not seem capable of lowering HDV levels to <1 virion/BF in patients with FPR kinetics, ALT normalization and sustained lower HDV RNA levels (compared to baseline) could potentially provide clinical benefit, at least while patients remain on therapy.
The combination of 2 mg/day of BLV with IFN led to higher rates of HDV RNA TND during 48 weeks of therapy compared to BLV alone.40,41 Model simulations helped to explain the greater efficacy of BLV+IFN treatment compared to BLV alone (Fig. 4), by assuming that the mode of action of IFN is to block HDV production as previously predicted in treated patients.18 Whether IFN’s additional modes of action, such as the recent in vitro findings of suppression of cell division-mediated HDV spread,38,42 have a significant effect on HDV RNA kinetics in vivo is not known. Two patients experienced viral breakthrough after having a ∼3 log IU/ml decline in viral load compared to pre-treatment levels. Since non-adherence to BLV is unlikely (though cannot be completely ruled out) this breakthrough could reflect the development of BLV resistance and/or cell division-mediated HDV spread. While recent studies suggested that BLV resistance in non-responders is unlikely,42,43 further research is needed to identify the reason(s) for viral breakthrough.
In the current model (Eq. 1) we assume no new cell infections occur after BLV treatment initiation. Exploring the effect of suboptimal efficacy of BLV in blocking infection (e.g., due to NTCP turnover), would require further theoretical efforts. For example, the current model predicts a biphasic viral decline pattern under BLV efficacy of η = 80% in the same in silico patient (Fig. S3). Notably for much lower values of η = 50%, the decline appears monophasic since the infection of new cells maintains the dominance of fast HDV-clearing cells over slow HDV-clearing cells and thus the second phase is not observed. Likewise, declines in BLV efficacy over time could explain the observed biphasic declines and FPR even without two infected cell populations. In the supplementary information we explored the case where blocking of infection by BLV drops from η = 1 to η = 0.5, 0.1 or 0.0. We found that a drop to η = 0.5 could explain a biphasic decline and a drop to η = 0.1 could explain a FPR (Fig. S4). However, such a dramatic decline in effectiveness from 100% blocking of infection down to only 10% blocking seems unlikely. Other models may be explored as well. For example, we previously introduced the notion of a critical drug efficacy, such that if overall drug efficacy (εtot) is higher than the critical drug efficacy (εc) then viral levels will continually decline on therapy, while if εtot < εc, then viral loads will initially decline but will ultimately stabilize at a new set point, as seen in FPR. As previously shown, using the idea of critical drug efficacy and including hepatocyte proliferation in a viral kinetic model allows for prediction of complex viral decay profiles.32,34,44 Lastly, additional models could consider ways of incorporating cell-to-cell HDV spread e.g., via cell division-mediated spread, in addition to cell-free spread that is partially or entirely blocked by BLV.
Our current framework has limitations. One simplification that we chose to make is to leave HBsAg-negative hepatocytes and HDV-monoinfected hepatocytes out of our model that focuses on chronic infection. While during acute HBV infection the majority of the liver becomes infected and undergoes non-cytolytic clearance,23 in the setting of chronic HBV/HDV infection in the current study, the number of HDV-producing and HBsAg-producing infected cells is likely more stable. Furthermore, even if HBsAg-infected cells comprise a small proportion of the total liver (<∼10%), they are responsible for the majority of HBV/HDV dynamics, and thus we did not include non-HBsAg-producing hepatocytes.45 Notably, however, to account for the contribution of ALT by non-HBsAg-producing hepatocytes (and extrahepatic tissue sources) that are not included in the model, we added a fixed rate (sx). Patients in the current study were HBeAg negative. Previous studies indicated that only a small fraction (<10%) of total hepatocytes (estimated at ∼107 cells/ml46,47) are HBsAg-producing cells45 in HBeAg-negative cases. Therefore, the total concentration of HBsAg-producing cells (infected or not infected with HDV) in the model was kept at approximately ∼105-106 cells/ml (Table 1). Incorporating HBV-negative hepatocytes along with HDV-monoinfected hepatocytes would require longitudinal intrahepatic experimental measurements during BLV treatment to distinguish HDV-monoinfected hepatocytes (that do not secret HDV) from HDV-producing hepatocytes, as well as significantly more complex modelling efforts, as previously shown.20,21
In conclusion, this is the first report analyzing and modeling HDV RNA, HBsAg and ALT kinetics measured under BLV 2 mg/day monotherapy administrated for 48 weeks in patients with HDV-related compensated cirrhosis and CSPH. Mathematical modeling suggests that there are two populations of HDV-infected liver cells: a fast HDV-clearing and a slow HDV-clearing population. Modeling explained how ALT levels can normalize without a change in HBsAg levels under BLV by assuming non-cytolytic clearance of HDV from infected hepatocytes that remain HBsAg-producing cells and release ALT upon death at a reduced rate compared to liver cells infected with HDV. The findings provide an initial step toward developing response-guided treatment strategies for HDV. The model developed here provides a starting point to optimize the use of new agents for the treatment of HDV. Further studies are needed to test and refine the viral kinetic patterns described here, their prevalence among HDV-infected populations, as well as the model structure and parameter values.
Financial support
This work was supported in part by a grant from “Ricerca Corrente RC2021/105-01”, Italian Ministry of Health, and by U.S. NIH grants R01AI144112 and R01AI146917.
Authors’ contributions
Concept and design: Harel Dahari, Pietro Lampertico. Kinetic analysis and in-silico modeling: Louis Shekhtman, Harel Dahari. Data collection: Elisabetta Degasperi, Maria Paola Anolli, Sara Colonia Uceda Renteria, Dana Sambarino, Marta Borghi, Riccardo Perbellini, Floriana Facchetti, Ferruccio Ceriotti. Writing of the article: Louis Shekhtman, Scott J Cotler, Elisabetta Degasperi Harel Dahari, Pietro Lampertico. All authors approved the final version of the manuscript.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, upon request, to any qualified researcher.
Conflicts of interest
Elisabetta Degasperi: Advisory Board: AbbVie; Speaking and teaching: Gilead, MSD, AbbVie. Pietro Lampertico: Advisor and speaker bureau for BMS, Roche, Gilead Sciences, GSK, MSD, Abbvie, Janssen, Arrowhead, Alnylam, Eiger, MYR Pharma, Antios, Aligos, VIR. Other authors have nothing to disclose. The other authors declare no conflicts of interest that pertain to this work.
Please refer to the accompanying ICMJE disclosure forms for further details.
Footnotes
Author names in bold designate shared co-first authorship
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2023.100966.
Contributor Information
Pietro Lampertico, Email: pietro.lampertico@unimi.it.
Harel Dahari, Email: hdahari@luc.edu.
Supplementary data
The following are the supplementary data to this article:
:
References
- 1.Rizzetto M., Canese M.G., Arico S., et al. Immunofluorescence detection of new antigen-antibody system (delta/anti-delta) associated to hepatitis B virus in liver and in serum of HBsAg carriers. Gut. 1977;18:997–1003. doi: 10.1136/gut.18.12.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rizzetto M., Hoyer B., Canese M.G., et al. Delta Agent: association of delta antigen with hepatitis B surface antigen and RNA in serum of delta-infected chimpanzees. Proc Natl Acad Sci U S A. 1980;77:6124–6128. doi: 10.1073/pnas.77.10.6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stockdale A.J., Kreuels B., Henrion M.Y.R., et al. The global prevalence of hepatitis D virus infection: systematic review and meta-analysis. J Hepatol. 2020;73:523–532. doi: 10.1016/j.jhep.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen H.Y., Shen D.T., Ji D.Z., et al. Prevalence and burden of hepatitis D virus infection in the global population: a systematic review and meta-analysis. Gut. 2019;68:512–521. doi: 10.1136/gutjnl-2018-316601. [DOI] [PubMed] [Google Scholar]
- 5.Wedemeyer H., Manns M.P. Epidemiology, pathogenesis and management of hepatitis D: update and challenges ahead. Nat Rev Gastroenterol Hepatol. 2010;7:31–40. doi: 10.1038/nrgastro.2009.205. [DOI] [PubMed] [Google Scholar]
- 6.Rizzetto M., Hamid S., Negro F. The changing context of hepatitis D. J Hepatol. 2021;74:1200–1211. doi: 10.1016/j.jhep.2021.01.014. [DOI] [PubMed] [Google Scholar]
- 7.Wedemeyer H., Yurdaydin C., Hardtke S., et al. Peginterferon alfa-2a plus tenofovir disoproxil fumarate for hepatitis D (HIDIT-II): a randomised, placebo controlled, phase 2 trial. Lancet Infect Dis. 2019;19:275–286. doi: 10.1016/S1473-3099(18)30663-7. [DOI] [PubMed] [Google Scholar]
- 8.Yurdaydin C., Abbas Z., Buti M., et al. Treating chronic hepatitis delta: the need for surrogate markers of treatment efficacy. J Hepatol. 2019;70:1008–1015. doi: 10.1016/j.jhep.2018.12.022. [DOI] [PubMed] [Google Scholar]
- 9.Heller T., Rotman Y., Koh C., et al. Long-term therapy of chronic delta hepatitis with peginterferon alfa. Aliment Pharmacol Ther. 2014;40:93–104. doi: 10.1111/apt.12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang C., Syed Y.Y. Bulevirtide: first approval. Drugs. 2020;80:1601–1605. doi: 10.1007/s40265-020-01400-1. [DOI] [PubMed] [Google Scholar]
- 11.Urban S., Bartenschlager R., Kubitz R., et al. Strategies to inhibit entry of HBV and HDV into hepatocytes. Gastroenterology. 2014;147:48–64. doi: 10.1053/j.gastro.2014.04.030. [DOI] [PubMed] [Google Scholar]
- 12.Degasperi E., Anolli M.P., Lampertico P. Bulevirtide for patients with compensated chronic hepatitis delta: a review. Liver Int. 2022;43:80–86. doi: 10.1111/liv.15389. [DOI] [PubMed] [Google Scholar]
- 13.Jachs M., Schwarz C., Panzer M., et al. Response-guided long-term treatment of chronic hepatitis D patients with bulevirtide-results of a "real world" study. Aliment Pharmacol Ther. 2022;56:144–154. doi: 10.1111/apt.16945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Degasperi E., Anolli M.P., Uceda Renteria S.C., et al. Bulevirtide monotherapy for 48 Weeks in hdv patients with compensated cirrhosis and clinically significant portal hypertension. J Hepatol. 2022;77(6):1525–1531. doi: 10.1016/j.jhep.2022.07.016. [DOI] [PubMed] [Google Scholar]
- 15.Lhomme S., DebRoy S., Kamar N., et al. Plasma hepatitis E virus kinetics in solid organ transplant patients receiving ribavirin. Viruses. 2019;11 doi: 10.3390/v11070630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Etzion O., Hamid S., Lurie Y., et al. Treatment of chronic hepatitis d with peginterferon lambda - the phase 2 LIMT-1 clinical trial. Hepatology. 2023;77(6):2093–2103. doi: 10.1097/HEP.0000000000000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shekhtman L., Cotler S.J., Ploss A., et al. Mathematical modeling suggests that entry-inhibitor bulevirtide may interfere with hepatitis D virus clearance from circulation. J Hepatol. 2022;76:1229–1231. doi: 10.1016/j.jhep.2021.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guedj J., Rotman Y., Cotler S.J., et al. Understanding early serum hepatitis D virus and hepatitis B surface antigen kinetics during pegylated interferon-alpha therapy via mathematical modeling. Hepatology. 2014;60:1902–1910. doi: 10.1002/hep.27357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shekhtman L., Cotler S.J., Hershkovich L., et al. Modelling hepatitis D virus RNA and HBsAg dynamics during nucleic acid polymer monotherapy suggest rapid turnover of HBsAg. Sci Rep. 2020;10:7837. doi: 10.1038/s41598-020-64122-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar U., Uchida T., Hailegiorgis A., et al. Understanding HDV and HBV dynamics during acute co-infection in humanized uPA/SCID chimeric mice using an agent-based modeling appraoch. J Hepatol. 2018;68:S783. [Google Scholar]
- 21.Packer A., Forde J., Hews S., et al. Mathematical models of the interrelated dynamics of hepatitis D and B. Math Biosci. 2014;247:38–46. doi: 10.1016/j.mbs.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 22.de Sousa B.C., Cunha C. Development of mathematical models for the analysis of hepatitis delta virus viral dynamics. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guidotti L.G., Rochford R., Chung J., et al. Viral clearance without destruction of infected cells during acute HBV infection. Science. 1999;284:825–829. doi: 10.1126/science.284.5415.825. [DOI] [PubMed] [Google Scholar]
- 24.Neumann A.U., Lam N.P., Dahari H., et al. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science. 1998;282:103–107. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 25.Allweiss L., Volmari A., Ladiges Y., et al. Strong intrahepatic decline of Hepatitis D virus RNA and antigen after 48 weeks of treatment with bulevirtide in chronic HBV/HDV co-infected patients: interim results from a multicenter, openlabel, randomized phase 3 clinical trial (MYR301) Hepatology. 2021;74:S148A. [Google Scholar]
- 26.Mederacke I., Bremer B., Heidrich B., et al. Establishment of a novel quantitative hepatitis D virus (HDV) RNA assay using the Cobas TaqMan platform to study HDV RNA kinetics. J Clin Microbiol. 2010;48:2022–2029. doi: 10.1128/JCM.00084-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kadelka S., Dahari H., Ciupe S.M. Understanding the antiviral effects of RNAi-based therapy in HBeAg-positive chronic hepatitis B infection. Sci Rep. 2021;11:200. doi: 10.1038/s41598-020-80594-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dahari H., Shudo E., Ribeiro R.M., et al. Modeling complex decay profiles of hepatitis B virus during antiviral therapy. Hepatology. 2009;49:32–38. doi: 10.1002/hep.22586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koh C., Canini L., Dahari H., et al. Oral prenylation inhibition with lonafarnib in chronic hepatitis D infection: a proof-of-concept randomised, double-blind, placebo-controlled phase 2A trial. Lancet Infect Dis. 2015;15:1167–1174. doi: 10.1016/S1473-3099(15)00074-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giannini E.G., Testa R., Savarino V. Liver enzyme alteration: a guide for clinicians. CMAJ. 2005;172:367–379. doi: 10.1503/cmaj.1040752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ribeiro R.M., Layden-Almer J., Powers K.A., et al. Dynamics of alanine aminotransferase during hepatitis C virus treatment. Hepatology. 2003;38:509–517. doi: 10.1053/jhep.2003.50344. [DOI] [PubMed] [Google Scholar]
- 32.Cardozo-Ojeda E.F., Duehern S., Cotler S.J., et al. Mathematical modeling of HDV RNA, ALT and HBsAg kinetics suggests a dual mode of action of pegyinterferon lambda: the LIMT-1 study. Hepatology. 2023;78(S1):S579–S580. [Google Scholar]
- 33.Etzion O., Dahari H., Yardeni D., et al. Response guided therapy for reducing duration of direct acting antivirals in chronic hepatitis C infected patients: a Pilot study. Sci Rep. 2020;10 doi: 10.1038/s41598-020-74568-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dahari H., Shudo E., Ribeiro R.M., et al. Modeling complex decay profiles of hepatitis B virus during antiviral therapy. Hepatology. 2009;49:32–38. doi: 10.1002/hep.22586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lampertico P., Aleman S., Blank A., et al. Efficacy of Bulevirtide as monotherapy for chronic hepatitis D (CHD): week 48 results from an integrated analysis. Hepatology. 2022;76(Suppl. 1):S228. [Google Scholar]
- 36.Ni Y., Zhang Z., Engelskircher L., et al. Generation and characterization of a stable cell line persistently replicating and secreting the human hepatitis delta virus. Scientific Rep. 2019;9 doi: 10.1038/s41598-019-46493-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lempp F.A., Schlund F., Rieble L., et al. Recapitulation of HDV infection in a fully permissive hepatoma cell line allows efficient drug evaluation. Nat Commun. 2019;10:2265. doi: 10.1038/s41467-019-10211-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z., Ni Y., Lempp F.A., et al. Hepatitis D virus-induced interferon response and administered interferons control cell division-mediated virus spread. J Hepatol. 2022;77:957–966. doi: 10.1016/j.jhep.2022.05.023. [DOI] [PubMed] [Google Scholar]
- 39.Anolli M.P., Degasperi E., Allweiss L., et al. Cure of hepatitis Delta infection following 3 years of bulevirtide monotherapy in a patient with compenstated advanced cirrhosis. Hepatology. 2022;76:S217–S218. [Google Scholar]
- 40.de Lédinghen V., Metivier S., Bardou-Jacquet E., et al. Treatment with Bulevirtide in patients with chronic HBV/HDV coinfection. Safety and efficacy at month 18 in real-world settings. J Hepatol. 2022;77(S1):S840. [Google Scholar]
- 41.Fontaine H., Fougerou-Leurent C., Gordien E., et al. Real life study of bulevirtide in chronic hepatitis delta: preliminary results of the ANRS HD EP01 BuleDelta prospective cohort. J Hepatol. 2022;77(S1):S72. [Google Scholar]
- 42.Mateo R., Xu S., Shornikov A., et al. Broad-spectrum activity of bulevirtide against clinical isolates of HDV and recombinant pan-genotypic combinations of HBV/HDV. JHEP Rep. 2023 doi: 10.1016/j.jhepr.2023.100893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hollnberger J., Liu Y., Xu S., et al. No virologic resistance to bulevirtide monotherapy detected in patients through 24 weeks treatment in phase II and III clinical trials for chronic hepatitis delta. J Hepatol. 2023;79(3):657–665. doi: 10.1016/j.jhep.2023.04.027. [DOI] [PubMed] [Google Scholar]
- 44.Dahari H., Ribeiro R.M., Perelson A.S. Triphasic decline of hepatitis C virus RNA during antiviral therapy. Hepatology. 2007;46:16–21. doi: 10.1002/hep.21657. [DOI] [PubMed] [Google Scholar]
- 45.Werle–Lapostolle B., Bowden S., Locarnini S., et al. Persistence of cccDNA during the natural history of chronic hepatitis B and decline during adefovir dipivoxil therapy. Gastroenterology. 2004;126:1750–1758. doi: 10.1053/j.gastro.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 46.Mackay I.R. Hepatoimmunology: a perspective. Immunol Cel Biol. 2002;80:36–44. doi: 10.1046/j.1440-1711.2002.01063.x. [DOI] [PubMed] [Google Scholar]
- 47.Dahari H., Layden–Almer J.E., Kallwitz E., et al. A mathematical model of hepatitis C virus dynamics in patients with high baseline viral loads or advanced liver disease. Gastroenterology. 2009;136:1402–1409. doi: 10.1053/j.gastro.2008.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
:
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, upon request, to any qualified researcher.