ABSTRACT
Multidrug efflux is one of the major mechanisms of antibiotic resistance identified in clinical isolates of the human pathogen Acinetobacter baumannii. The multiple antibiotic resistance in this species is often enabled by the overproduction of the tripartite efflux pump AdeABC. In this pump, AdeB is the inner membrane transporter from the resistance-nodulation-division (RND) superfamily of proteins, which is responsible for the recognition and efflux of multiple structurally unrelated compounds. Like other RND transporters, AdeB is a trimeric protein with ligand-binding sites located in the large periplasmic domains. Previous structural studies, however, highlighted the uniqueness of AdeB interactions with ligands. Up to three ligand molecules were bound to one protomer of AdeB, mapping its substrate translocation path. In this study, we introduced single and double substitutions in the identified ligand-binding sites of AdeB. Our results show that the mechanism of substrate translocation by AdeB is different from that of other characterized RND transporters and that the functional interactions between the sites are nonadditive. We identified AdeB mutants with both the loss and the gain of antibiotic susceptibility phenotypes, as well as AdeB mutations making A. baumannii cells overproducing such pump variants even more susceptible to multiple antibiotics than efflux-deficient cells.
IMPORTANCE
Multidrug efflux pumps of the resistance-nodulation-division superfamily of proteins are important contributors to various aspects of bacterial physiology and antibiotic resistance. Studies of the best-characterized model transporter AcrB from Escherichia coli suggested that these transporters operate by a functional rotation mechanism in which various substrates bind to at least two different binding sites. This study suggests that the mechanism of AdeB is distinct and that the binding sites in this transporter are functionally linked.
KEYWORDS: Acinetobacter baumannii, multidrug efflux, antibiotic resistance
INTRODUCTION
Acinetobacter baumannii is currently classified by the Infectious Disease Society of America and the Center for Disease Control as one of the six most threatening multi-drug-resistant organisms in hospitals worldwide (1, 2). It is estimated that A. baumannii is the culprit behind up to 10% of hospital-acquired infections. As many as 43% of A. baumannii clinical isolates have developed resistance to at least three different classes of antibiotics (3). Mechanisms of resistance include changes in outer membrane proteins, drug target modification, activity of enzymes such as β-lactamases, and the expression of efflux pumps, particularly those of the resistance-nodulation-division (RND) superfamily of transporters, which are recognized as major contributors to multidrug resistance in other Gram-negative pathogens (4 – 6). AdeABC is one of the major A. baumannii multidrug efflux pumps, which are frequently overproduced in clinical isolates and are the major antibiotic resistance determinant (7, 8). This pump is composed of three subunits, of which AdeB is the transporter belonging to the RND superfamily of proteins, AdeA is a periplasmic membrane fusion protein, and AdeC is an outer membrane factor protein. In this complex, AdeB is responsible for the recognition and active efflux of structurally diverse drugs, including aminoglycoside antibiotics (9, 10), whereas AdeA and AdeC stimulate AdeB and create a protein conduit for the efflux of drugs across the outer membrane (9).
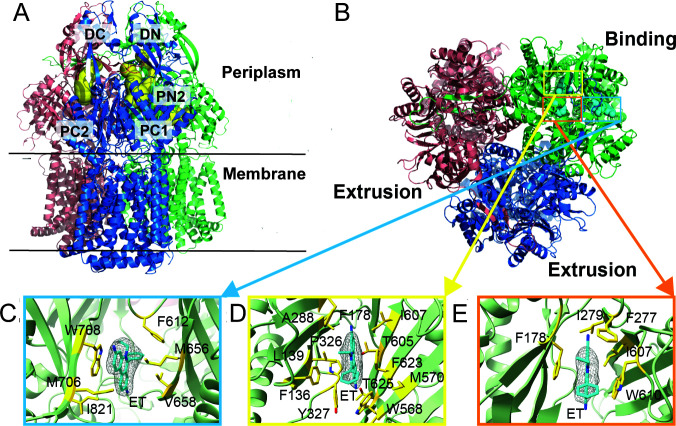
The structural studies showed that AdeB shares certain features with other RND transporters, such as the presence of large periplasmic domains for capturing drugs from the periplasm (Fig. 1A). On the other hand, AdeB also possesses unique properties. A characteristic feature of AdeB and homologous RND transporters is their trimeric structure, which is believed to be critical for their proposed molecular mechanism (11 – 13). In a model RND transporter such as AcrB from Escherichia coli, each protomer is thought to cycle sequentially through at least three conformations, which enable the access, binding, and extrusion of drugs (14). Until recently, this conformational rotation, which is not a physical rotation, was thought to occur in an asymmetric manner so that each protomer of the trimer is distinct and assumes only one of the three states at a time. The major outcome of such asymmetry is that a ligand molecule can bind either an “access” or a “binding” protomer but not the “extrusion” protomer (11, 15). In contrast, the AdeB structure was strikingly different (2). In the apo-state, all three protomers of AdeB were symmetric and assumed the extrusion conformation, whereas in the ligand-bound state, one of the protomers adopted a binding state with three molecules of ethidium (Et) bound to it at different locations, likely representing different snapshots along the substrate extrusion pathway (Fig. 1B through E) (2). Thus, the AdeB transporter could differ from a model AcrB in its substrate binding and extrusion pathways.
Fig 1.
Structure and ligand-binding sites of AdeB. (A) Ribbon diagram of the structure of an Et-bound AdeB viewed in the membrane plan. (B) Top view of the AdeB trimer depicting three bound Et ligands located at the binding protomer. The three bound Et ligands are depicted as cyan spheres. (C to E) The Et binding sites at the entrance (C), distal (D), and hydrophobic patch (E) sites. The electron microscopy densities of bound Et ligands are shown as gray meshes. The bound Et ligands are represented as cyan sticks. Residues that are involved in Et binding are represented as yellow sticks. The secondary structural elements of the binding protomer are depicted as green ribbons. Modified and reproduced from reference (2).
The structural analyses suggested that a drug molecule entering the periplasmic cleft between subdomains PC1 and PC2 of AdeB will presumably be guided by the flexible loop (F-loop, residues 661–670) to bind at the proximal multidrug binding site (PBP) (16). This drug will then pass through the gate loop (G-loop, residues 609–618) and reach the distal multidrug binding site (DBP) for extrusion. The three bound Et molecules were found in three distinct locations of the binding protomer of AdeB and appear to line the elongated channel formed by the protomer (2). The first bound Et is situated at the entrance of the periplasmic cleft of AdeB, where the entrance residues M656, V658, M706, W708, and I821 are responsible for providing hydrophobic and aromatic interactions to anchor bound Et (Fig. 1C). The second Et molecule is located at the distal drug binding site approximately 20 Å above the membrane surface, with multiplere sidues contributing to the binding of this bound Et (Fig. 1D). The third bound Et is observed to entangle in the hydrophobic patch of the distal binding site, with residues F178, F277, I279, I607, and W610 interacting with this Et (Fig. 1E). When analyzed in E. coli, a direct comparison of functional residues in AdeB and AcrB showed that substitutions in the analogous residues in the two pumps lead to different outcomes (17). Therefore, the AdeB mechanism appears to be different from that of AcrB.
In this study, we targeted by mutagenesis the key residues implicated in the binding of Et and its translocation by AdeB. The constructed mutants were either integrated onto the chromosome of the efflux-deficient AbΔ3 (IL119) cells or expressed from the plasmid in AbΔ3-Pore (IL139) cells. We unexpectedly found that the F178C substitution in the distal binding pocket leads to either antibiotic hypersusceptibility or hyposusceptibility phenotypes, depending on the substrate. To determine whether the three Et binding sites interact with each other, the F178C substitution was combined with substitutions in the same hydrophobic patch as well as with substitutions in the cleft and interphase binding sites. Even the substitutions that did not have any effect on the translocation of substrates on their own were detrimental to the ability of AdeB to protect cells against different antibiotics, whereas the expression of AdeB with the double F178C/F277C and F178C/W610C substitutions dropped the antibiotic susceptibility of the transporter below an efflux-deficient level. Our results suggest that the three binding sites of AdeB closely communicate and changes in one of the sites have a long-range effect on the substrate translocation path.
RESULTS
Single amino acid substitutions in AdeB binding sites generate both the loss- and gain-of-function phenotypes
We constructed 10 mutated AdeB variants containing single amino acid substitutions located along the Et translocation pathway (Table 1; Fig. 1; Fig. S1). We targeted (i) E89, F178, F277, W568, and W610 from the DBP and its hydrophobic patch; (ii) I663 from the conserved flexible loop connecting the cleft entrance to the PBP; (iii) D664 and E665 from the PBP; (iv) W708 located at the entrance of the periplasmic cleft; and (v) N932 involved in the proton relay network in the transmembrane domain. The mutations were introduced into a pTJ1 plasmid carrying adeABC operon from the multidrug-resistant A. baumannii AYE strain (10). Plasmids carrying mutated adeB and genes encoding its AdeA and AdeC subunits were introduced and integrated onto the chromosomes of AbΔ3 cells. In these cells, the adeABC variants were expressed under arabinose-inducible promoters from both the chromosome and the plasmid (10).
TABLE 1.
Minimal inhibitory (MIC) and half-maximal inhibitory (IC50) concentrations of antibacterial agents in A. baumannii Ab∆3 cells carrying an empty vector or plasmids producing AdeABC with indicated AdeB variants
| AdeB | Site a | Et | Gentamicin | Zeocin | Azithromycin | Erythromycin | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC | IC50 | MIC | IC50 | MIC | IC50 | MIC | IC50 | MIC | IC50 | ||
| – b | 4–8 | 3.14 ± 0.78 | 8–16 | 4.35 ± 0.62 | 0.5–1 | 0.45 ± 0.14 | 0.64 | 0.16 ± 0.08 | 1.25–2.5 | 0.63 ± 0.02 | |
| WT c | 32–64 | 21.47 ± 2.31 | 32 | 15.16 ± 2.47 | 16–32 | 7.67 ± 1.32 | 10–20 | 5.39 ± 0.66 | 10–20 | 5.00 ± 0.23 | |
| F178C | DBP patch | 16–32 | 8.64 ± 1.35 | 64–128 | 44.33 ± 8.57 | >256 | >256 | 10 | 4.6 ± 0.37 | 10 | 2.96 ± 0.41 |
| F277C | DBP patch | 16 | 9.61 ± 0.27 | 16–32 | 6.91 ± 0.47 | 32 | 6.51 ± 2.24 | 10–20 | 4.86 ± 0.83 | 10 | 4.24 ± 1.83 |
| W610C | DBP patch | 32–64 | 19.85 ± 2.7 | 8–16 | 2.86 ± 0.61 | 16 | 1.66 ± 0.39 | 10–20 | 5.59 ± 0.38 | 10 | 1.07 ± 0.34 |
| E89A | DBP | 32–64 | 18.87 ± 1.68 | 8 | 4.23 ± 0.26 | 32 | 6.86 ± 0.82 | 20 | 7.53 ± 0.04 | 20 | 8.36 ± 0.8 |
| W568C | DBP | 32 | 10.89 ± 1.19 | 16 | 7.62 ± 0.53 | 32 | 7.5 ± 0.82 | 20 | 5.5 ± 0.29 | 20 | 4.19 ± 0.13 |
| I663C | PBP F-loop | 32–64 | 19.96 ± 2.77 | 8–16 | 2.89 ± 0.64 | 8–16 | 1.78 ± 0.37 | 10–20 | 8.15 ± 0.34 | 20 | 2.11 ± 0.36 |
| D664C | PBP F-loop | 16–32 | 8.42 ± 1.16 | 16 | 7.63 ± 1.54 | 4–8 | 1.2 ± 0.4 | 2.5–5 | 1.47 ± 0.26 | 5 | 2.55 ± 0.26 |
| E665A | PBP F-loop | 16–32 | 13.19 ± 3 | 16 | 8.35 ± 1.55 | 8 | 3.29 ± 0.74 | 10 | 2.53 ± 0.19 | 20 | 2.04 ± 0.45 |
| W708C | PBP | 32 | 16.47 ± 2.87 | 8–16 | 7.81 ± 1.38 | 8–16 | 2.82 ± 0.58 | 10 | 4.34 ± 0.03 | 20 | 2.51 ± 0.72 |
| N932C | TMD | 32 | 16.93 ± 4.07 | 16 | 9.27 ± 0.94 | 4 | 1.22 ± 0.15 | 5.0–10 | 2.43 ± 0.01 | 10 | 1.35 ± 0.43 |
DBP, distal binding pocket; PBP, proximal binding pocket; TMD, transmembrane domain. Values indicating the hypersusceptibility phenotype are underlined. Values indicating hyposusceptibility are shown in bold. IC50 values are averages of three independent experiments with two technical replicates. Errors are standard deviations (n = 6).
–: empty vector.
WT: wild type
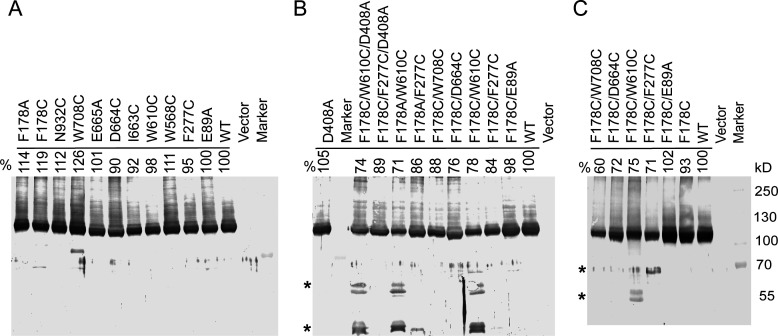
Immunoblotting analysis showed that all mutated AdeB proteins were produced at similar levels (Fig. 2A). However, cells producing AdeB mutants varied in their susceptibilities to five antibacterials: Et, gentamicin, zeocin, azithromycin, and erythromycin, the known substrates of the AdeABC efflux pump (10). As seen from the minimal inhibitory concentration (MIC) (Table 1), all constructed AdeB mutants retained at least partial activity. The N932C substitution reduced the MICs only of zeocin and two macrolides, suggesting that the role of this residue in proton-translocation is dispensable. Except for the F178C and W568C variants, all other AdeB mutants were defective in protecting cells against at least one of the tested antibiotics. The substitutions in the F-loop of the PBP negatively impacted the largest number of substrates, with the D664C variant being the least active against the macrolides azithromycin and erythromycin among all mutants. The substitutions in the DBP and the hydrophobic patch had antibiotic-specific effects. In particular, the E89A mutation reduced only the MIC of gentamicin; F277C reduced the MIC of Et only; and W610C reduced the MICs of both gentamicin and zeocin.
Fig 2.
Expression levels of the constructed AdeB variants. (A) Single substitution mutants produced in AbΔ3 cells. (B) AdeB variants with double and triple substitutions and with D408A substitution. (C) AdeB variants carrying the constructed F178C variants produced in AbΔ3-Pore cells. All panels: membrane fractions isolated from the cells producing the indicated AdeB variants were resolved by 8% SDS-PAGE, transferred into polyvinylidene fluoride membranes, and immunoblotted with the polyclonal antiAdeB antibody. The densities of the bands corresponding to the whole-length proteins were measured and expressed as a percent of the expression of the wild type (WT) protein. *, proteolytic fragments reacting with an antiAdeB polyclonal antibody.
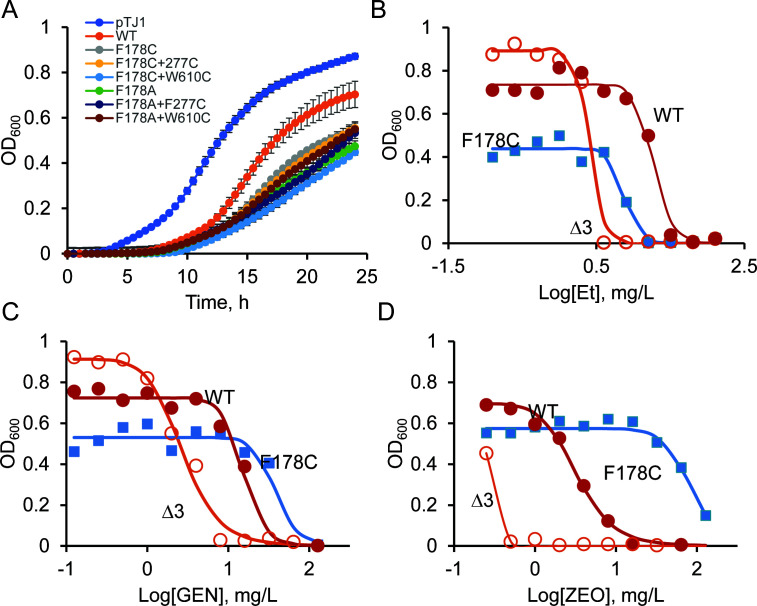
Among the substitutions, cells producing AdeB with F178C showed a rare phenotype. Although this mutation reduced the effectiveness of Et efflux, as predicted from the structural analyses, it also enhanced efflux of gentamicin and zeocin, as seen from increased MICs in cells producing the AdeB F178C variant (Fig. 1 to 3). For zeocin, the MIC increased by more than 16-fold in cells with AdeBF178C, and the effect for gentamicin was more modest, with a two- to fourfold increase in MIC. Overproduction of AdeABC is known to negatively affect A. baumannii growth physiology (10, 18). The introduction of the F178C substitution further reduced the growth rate of A. baumannii cells overproducing AdeABC pump (Fig. 3A). However, the antibiotic susceptibility phenotype of this and other mutants was independent of their growth rates (Table 1).
Fig 3.
Growth and antibiotic susceptibility of A. baumannii cells overproducing the indicated variants. (A) Growth curves of AdeBF178C/A and their derivatives grown in Luria-Bertani (LB) medium at 37°C with aeration (an average of two independent experiments with three technical repeats). (B-D) Growth inhibition curves of AbWT, AbΔ3, and AbΔ3 overproducing AdeB F178C variants in the presence of increasing concentrations of Et (B), gentamicin (C), and zeocin (D). The indicated strains were grown in 96-well plates in LB medium supplemented with doubling concentrations of the indicated antibiotics. OD600 values in each well were determined after 24 h of incubation at 37°C and plotted as a function of antibiotic concentration in the well.
Thus, the identified binding sites of AdeB are indeed important for the efflux of diverse substrates, and various substrates interact with specific amino acid residues in the binding pockets of AdeB. The replacement of the bulky aromatic F178 was beneficial for select substrates of AdeB. The importance of binding sites varies between the substrates, with substitutions in both the DBP and F-loop affecting the MICs of Et, gentamicin, and zeocin. In contrast, the activities of the macrolides azithromycin and erythromycin are only affected by PBP and the F-loop residues.
Nonadditive interactions between substrate binding sites in AdeB
To analyze putative interactions between the ligand-binding sites of AdeB, we introduced into the F178C variant the E89A, F277C, and W610C substitutions in the DBP and its hydrophobic patch, as well as the D664C and W708C substitutions in the F-loop and PBP, respectively. If the sites are independent from each other, we expect to find additive changes in MICs from combining the substitutions. In contrast, interactions between the sites will lead to nonadditive effects. All constructed double-substituted variants were produced in AbΔ3 cells at levels similar to those of the wild type (WT) (Fig. 2B). However, the F178C/W610C variant produced additional stable degradation products, suggesting that this combination affects the overall structure of the transporter.
We found that all combinations of substitutions abrogated the gain-of-function hyperresistant phenotype of the F178C variant (Table 2). This result suggests that the binding sites interact with each other even across the distance between the PBP and DBP. All double-substituted AdeB variants were more functionally compromised than their respective single substitution parents and the WT AdeB. Surprisingly, cells producing the F178C/F277C and F178C/W610C AdeB variants became up to fourfold more susceptible to all tested compounds than the efflux-deficient cells carrying an empty vector (the null control). This hypersusceptibility affected gentamicin and zeocin as well, against which the F178C variant was hyperresistant. The three residues, F178, F277, and W610, are located next to each other and form a hydrophobic patch in the DBP (Fig. 1).
TABLE 2.
MIC and IC50 values of antibacterial agents in A. baumannii Ab∆3 cells carrying an empty vector or plasmids producing the double-substituted AdeB F178C variants
| AdeB | Et | Gentamicin | Zeocin | Azithromycin | Erythromycin | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| MIC | IC50 | MIC | IC50 | MIC | IC50 | MIC | IC50 | MIC | IC50 | |
| – b | 4–8 | 3.14 ± 0.78 | 8.0–16 | 4.35 ± 0.62 | 0.5–1 | 0.45 ± 0.14 | 0.64 | 0.16 ± 0.08 | 1.25–2.5 | 0.63 ± 0.02 |
| WT | 32–64 | 21.47 ± 2.31 | 32 | 15.16 ± 2.47 | 16–32 | 7.67 ± 1.32 | 10–20 | 5.39 ± 0.66 | 10–20 | 5.00 ± 0.23 |
| F178C | 16–32 | 8.64 ± 1.35 | 64–128 a | 44.33 ± 8.57 | >256 | >256 | 10 | 4.6 ± 0.37 | 10 | 2.96 ± 0.41 |
| F178C/E89A | 16 | 3.62 ± 0.11 | 8.0–16 | 5.67 ± 0.56 | 16–32 | 3.97 ± 1.5 | 5 | 0.16 ± 0.004 | 5 | 0.55 ± 0.007 |
| F178C/F277C | 2–4 | 1.71 ± 0.12 | 4.0–8 | 2.93 ± 0.14 | 0.25 | 0.08 ± 0.001 | 0.64 | 0.03 ± 0.002 | 0.63–1.25 | 0.14 ± 0.03 |
| F178C/W610C | 1–2 | 0.39 ± 0.1 | 4.0–8 | 2.43 ± 0.13 | 0.25 | 0.09 ± 0.003 | 0.16–0.32 | 0.03 ± 0.007 | 0.63 | 0.2 ± 0.04 |
| F178C/D664C | 8 | 1.02 ± 0.32 | 8 | 4.31 ± 0.07 | 2.0–4 | 0.67 ± 0.11 | 1.25 | 0.2 ± 0.005 | 1.25 | 0.19 ± 0.007 |
| F178C/W708C | 8 | 2.36 ± 0.53 | 4.0–8 | 3.01 ± 0.78 | 8.0–16 | 1.6 ± 0.56 | 2.5 | 0.35 ± 0.04 | 2.5 | 0.42 ± 0.07 |
Values indicating the hypersusceptibility phenotype and the gain-of-function are underlined and shown in bold, respectively.
–: empty vector
These results suggest that during translocation, the ligand-binding sites constitute a single ligand translocation pathway, modifications of which in any of the sites affect the downstream interactions with the ligands. The hydrophobic patch is critical for both positive and negative interactions with ligands.
Double-substituted AdeB variants do not increase influx across the outer membrane
Like other RND transporters, AdeABC transports its substrates across the outer membrane, and its activity is synergistic with the low permeability barrier of the outer membrane. To analyze the contribution of the outer membrane to the observed gain- and loss-of-function phenotypes, we next transformed the F178C and its double-substituted derivatives into AbΔ3-Pore cells with the controllable permeability barrier of the outer membrane. In these cells, AdeABC variants were produced from the plasmid, and the addition of arabinose leads to both the expression of AdeABC and the hyperporination of the outer membrane. The latter increases the permeation of antibiotics and other substrates across the outer membrane permeability barrier (19). In addition, the levels of AdeB expression are lower in these cells because the protein is expressed only from the plasmid (10).
Measurements of MICs and half-maximal inhibitory concentrations (IC50s) of antibiotics in AbΔ3-Pore cells showed that AdeBWT provides four- to eightfold lower protection against all tested antibiotics (Table 3). The MICs of antibiotics in AbΔ3-Pore cells producing F178C variants were at least twofold lower than those of the WT variants, including the MICs against gentamicin and zeocin. Thus, the permeability barrier of the outer membrane and the amounts of AdeB both contribute to the hyposusceptibility phenotype of F178C seen in AbΔ3 cells.
TABLE 3.
MIC and IC50 values of antibacterial agents in A. baumannii AbΔ3-Pore cells carrying an empty vector or plasmids producing the indicated AdeB variants a
| AdeB | Et | Gentamicin | Zeocin | Azithromycin | Erythromycin | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| MIC | IC50 | MIC | IC50 | MIC | IC50 | MIC | IC50 | MIC | IC50 | |
| – b | 2–4 | 1.47 ± 0.38 | 4–8 | 3.59 ± 0.83 | 0.25–0.5 | 0.19 ± 0.11 | 0.16–0.32 | 0.16 ± 0.06 | 0.63–1.25 | 0.47 ± 0.06 |
| WT | 8–16 | 3.72 ± 0.85 | 16–32 | 8.72 ± 1.28 | 4–8 | 2.5 ± 0.11 | 2.5–5 | 0.96 ± 0.52 | 5.0–10 | 1.91 ± 0.42 |
| F178C | 2–4 | 2.47 ± 0.47 | 8–16 | 7.25 0.93 | 8 | 3.44 ± 1.42 | 1.25–2.5 | 0.63 ± 0.2 | 2.5–5 | 0.99 ± 0.42 |
| F178C + E89A | 4–8 | 0.91 ± 0.09 | 8 | 2.72 ± 0.21 | 8–16 | 5.01 ± 1.29 | 2.5 | 1.24 ± 0.21 | 2.5 | 1.18 ± 0.11 |
| F178C + F277C | 0.5 | 0.29 ± 0.01 | 4–8 | 1.26 ± 0.08 | 0.125–0.25 | 0.13 ± 0.01 | 0.32 | 0.07 ± 0.01 | 1.25 | 0.37 ± 0.09 |
| F178C + W610C | 1–2 | 0.50 ± 0.15 | 2–4 | 1.89 ± 0.98 | 0.064–0.125 | 0.085 ± 0.06 | 0.16–0.32 | 0.09 ± 0.01 | 0.63 | 0.08 ± 0.01 |
| F178C + D664C | 2–4 | 1.26 ± 0.66 | 8 | 1.35 ± 0.03 | 2 | 0.81 ± 0.27 | 0.64 | 0.27 ± 0.03 | 1.25 | 0.21 ± 0.07 |
| F178C + W708C | 2 | 0.58 ± 0.02 | 8 | 1.36 ± 0.24 | 8 | 3.07 ± 0.32 | 2.5 | 0.93 ± 0.22 | 2.5 | 1.03 ± 0.25 |
Values indicating the hypersusceptibility phenotype are underlined.
–: empty vector
Cells producing F178C/F277C and F178C/W610C AdeB variants remained more susceptible to antibiotics than the efflux-deficient control against Et, gentamicin, and zeocin but lost their hypersusceptibility phenotypes against azithromycin and erythromycin (Table 3). Interestingly, single substitutions in the DBP of AdeB did not have a notable effect against these antibiotics, suggesting that macrolides mostly bypass the DBP site and its hydrophobic patch. Thus, increased influx across the outer membrane is the major reason for the drop in MICs of macrolides against AbΔ3 cells producing F178C/F277C and F178C/W610C. In contrast, additional mechanisms are involved in the hypersusceptibility of these variants to Et, gentamicin, and zeocin.
Structural changes in AdeB but not its transport activity are responsible for the hypersusceptibility phenotypes of double-substituted F178C/F277C and F178C/W610C variants
In the AdeB structure, all three residues (F178, F277, and W610) are in proximity to each other (Fig. 1E). The analyses of the AdeB structure show that in the binding protomer, the Cys residues in these positions would be too far apart to form disulfide bridges (approx. 9–10 Å between γ-S atoms). However, in the extrusion or access protomer (based on the PDB: 7 KGI structure (2)), the distance on the F277C-F178C pair is approximately 5 Å, and the F178C-W610C would have approx. 6 Å distance between the γ-S atoms. Hence, it is likely that these residues, when substituted with cysteines, form disulfide bonds in extrusion and access conformations, which in turn could prevent interactions of substrates with this site. Alternatively, disulfide bonding could stabilize an alternative transitory AdeB conformation, which is different from the stable conformations seen in cryoEM structures and contributes to the hypersusceptibility phenotype. To test these possibilities, F178 residue was substituted for Ala, and the F178A variant was combined with F277C and W610C substitutions. We found that the F178A variant remained hyposusceptible against zeocin, although to a lesser degree (Table 4) and lost its mild hyposusceptibility against gentamicin. Since both F178C and F178A variants are produced in similar amounts (Fig. 2A and B), we conclude that the thiol side chain of Cys contributes positively to the hyposusceptibility phenotype.
TABLE 4.
MIC and IC50 values of antibacterial agents in A. baumannii Ab∆3 cells carrying an empty vector or plasmids producing AdeABC with indicated AdeB substitutions
| AdeB variants | Et | Gentamicin | Zeocin | Azithromycin | Erythromycin | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| MIC | IC50 | MIC | IC50 | MIC | IC50 | MIC | IC50 | MIC | IC50 | |
| – b | 4–8 | 3.14 ± 0.78 | 8.0–16 | 4.35 ± 0.62 | 1 | 0.45 ± 0.14 | 0.64 | 0.16 ± 0.08 | 1.25–2.5 | 0.63 ± 0.02 |
| WT | 32–64 | 21.47 ± 2.31 | 32 | 15.16 ± 2.47 | 16–32 | 7.67 ± 1.32 | 10–20 | 5.39 ± 0.66 | 10–20 | 5.00 ± 0.23 |
| F178C | 16–32 | 8.64 ± 1.35 | 64–128 a | 44.33 ± 8.57 | >256 | >256 | 10 | 4.6 ± 0.37 | 10 | 2.96 ± 0.41 |
| F178A | 8 | 3.42 ± 0.17 | 32–64 | 16.91 ± 2.87 | 64–128 | 36.16 ± 6.82 | 5 | 1.35 ± 0.46 | 5 | 0.75 ± 0.06 |
| F178A + F277C | 4–8 | 1.91 ± 0.82 | 32 | 15.62 ± 0.73 | 64–128 | 46.51 ± 3.24 | 2.5–5 | 1.88 ± 0.73 | 10 | 2.14 ± 0.43 |
| F178A + W610C | 1–2 | 0.49 ± 0.11 | 4 | 1.67 ± 0.33 | 0.5–1 | 0.35 ± 0.08 | 0.16–0.32 | 0.12 ± 0.01 | 1.25 | 0.32 ± 0.08 |
| D408A | 2 | 0.9 ± 0.35 | 4–8 | 1.95 ± 0.53 | 0.25–0.5 | 0.1 ± 0.02 | 0.32 | 0.17 ± 0.05 | 1.25 | 0.23 ± 0.09 |
| D408A + F178C+ F277C |
0.5–1 | 0.20 ± 0.1 | 4 | 1.06 ± 0.18 | 0.5–1 | 0.33 ± 0.03 | 0.32 | 0.09 ± 0.002 | 1.25 | 0.29 ± 0.05 |
| D408A + F178C+ W610C |
1 | 0.39 ± 0.03 | 2.0–4 | 1.86 ± 0.05 | 0.5 | 0.13 ± 0.001 | 0.32 | 0.08 ± 0.004 | 0.64 | 0.18 ± 0.04 |
Values indicating the hypersusceptibility phenotype and the gain-of-function are underlined and shown in bold, respectively.
–: empty vector
We also found that the F178A/F277C variant lost its hypersusceptibility phenotype against all antibiotics and regained hyposusceptibility against zeocin. This result suggested that indeed, the disulfide bond between F178C and F277C was responsible for hypersusceptibility. In contrast, cells producing the double-substituted F178A/W610C variant remained hypersusceptible to Et and gentamicin but had the null phenotype against zeocin, azithromycin, and erythromycin. Thus, the mechanism of hypersusceptibility of the double-substituted F178/W610 AdeB is more complex and likely arises due to both the disulfide bonding in the DP and additional mechanisms.
To determine whether the proton transport activity of AdeB contributes to the hypersusceptibility of double-substituted variants, we introduced the D408A substitution into the proton translocation center of AdeB. This residue is highly conserved in RND transporters and is directly involved in proton transport (20). Indeed, the AdeB D408A variant was found to have a null phenotype and was as susceptible to antibiotics as efflux-deficient AbΔ3 cells carrying an empty plasmid. The introduction of the D408A substitution into the F178C/F277C and F178C/W610C variants did not rescue the hypersusceptibility against Et, gentamicin, and zeocin. However, it was dominant in the F178C/F277C background against both azithromycin and erythromycin and against azithromycin in the F178C/W610C background. Thus, the proton transfer activity of AdeB contributes to hypersusceptibility against macrolides but not other tested antibiotics.
Taken together, our results suggest that disulfide bonding in the hydrophobic patch of AdeB traps the transporter in a conformation leading to the hypersusceptibility phenotype in AbΔ3. However, the mechanism of hypersusceptibility is specific to antibiotics. Macrolides can apparently bypass the DBP, and the hypersusceptibility is induced by the increased uptake of these antibiotics across the outer membrane due to the presence of the compromised AdeABC. In contrast, the hypersusceptibility against Et, gentamicin, and zeocin is caused by two different mechanisms. On one hand, disulfide bonding between F178C and F277C prevents binding to and/or translocation of these antibiotics through the DBP. On the other hand, inactivation of the proton transport does not prevent hypersusceptibility in F178C/277C as well as in F178C/W610C background. Thus, both variants are trapped in conformations that either increase the uptake or deplete the efflux of Et, gentamicin, and zeocin across the inner membrane.
DISCUSSION
In Gram-negative bacteria, RND efflux pumps share a three-component architecture, enabling efflux across the outer membrane. Most of the mechanistic insights into how active efflux occurs in these bacteria have been accumulated for model transporters such as AcrB from E. coli (14) and its close homolog MexB from Pseudomonas aeruginosa (21, 22). These transporters are thought to contain two binding pockets, the PBP and DBP, separated by the interface comprising the G-loop, with each pocket accessible for binding in one of the two conformers, access and binding, respectively. The proposed mechanism involves a functional rotation of each protomer so that ligands can occupy either PBP or DBP but not both sites simultaneously in the same protomer. Furthermore, the conformational transitions in two adjacent protomers are coupled to each other, so that in the active transporter, each protomer assumes one of the three possible states. Recent structural studies suggested that the mechanism of AdeB is different and that this transporter contains a ligand pathway enabling ligand binding in three different positions within the same protomer (2). This study provides functional validation of this mechanism and shows that the translocation pathways through AdeB differ between its ligands.
AdeABC is typically expressed in multidrug-resistant clinical isolates due to mutations in the regulatory regions (9, 23). The substrate specificity of this transporter is quite distinct from that of AcrB and MexB (10). It is particularly effective against cationic antibacterials such as Et, aminoglycoside antibiotics, and zeocin. AdeB is also efficient in the protection of cells against azithromycin, which is not a substrate of the AdeIJK pump constitutively expressed in A. baumannii. Site-directed mutagenesis of AdeB residues interacting with Et according to the cryo-electron microscopy structure showed that gentamicin and, to some extent, zeocin share the same translocation pathway with Et, whereas the translocation of macrolides is different. The latter are likely to bypass the DBP and not interact with its hydrophobic patch. The presence of two different pathways can be seen from MICs measured in cells producing AdeB with single substitutions in five different regions of PBP and DBP (Table 1). The mutations in DBP and in the patch had no effect on susceptibility against macrolides, but such substitutions reduced the activity of AdeB against Et, gentamicin, and zeocin. In contrast, mutations in PBP reduced the activity of AdeB against zeocin and macrolides. These results agree with previous structural analyses of AcrB that showed that erythromycin and other large antibiotics interact primarily with the PBP and are likely to bypass the DBP during translocation (24, 25). Zeocin is the largest ligand of AdeB with a molecular weight (MW) of 1,428 g/mol, followed by azithromycin with an MW of 749 g/mol and erythromycin with an MW of 733 g/mol. Although cells overproducing F178C/F277C and F178C/W610C variants were hypersusceptible to these antibiotics, this hypersusceptibility was largely due to the known synergy of efflux of these antibiotics with the permeability barrier of the outer membrane of A. baumannii (19, 26).
Both Et and gentamicin are notably smaller, with MW values of 394 and 478 g/mol, respectively, and their primary translocation site is the DBP. Interestingly, gentamicin and zeocin, both containing glycosidic moieties, share certain similarities that also make them distinct from Et. The F178C substitution makes their efflux by AdeB more efficient; in contrast, the same substitution reduces AdeB activity against Et (Fig. 3), suggesting that reduced hydrophobicity of the DBP or removal of the phenyl ring is beneficial for gentamicin and zeocin binding. Indeed, blocking this binding by introducing the disulfide F178C-F277C bond completely reversed the beneficial properties of F178C and even dropped the MICs of this and other antibiotics below the efflux null controls (Table 2). Apparently, the interactions of ligands in the hydrophobic patch have a long-range effect on the DBP and PBP because compromising interactions of ligands with E89 or D664 also abrogate the beneficial properties of the F178C variant. This finding suggests that the translocation pathway is an integer of interactions with multiple residues, and each ligand follows its own specific pathway.
It is also clear that the hydrophobic patch is not only a ligand-binding site, but it also plays an important role in the conformational dynamics of AdeB. The double substitutions of F178C and either F277C or W610C trap AdeB in a conformation that is more vulnerable to intrinsic proteolysis, as seen from anti-AdeB immunoblotting (Fig. 2B and C). This conformation is not caused by disulfide bonding between the residues because the same proteolysis is seen in the double mutants with the F178A residue. Also, the hypersusceptibility of cells producing either one of the F178C/F277C or F178C/W610C double-substituted AdeB variants cannot be reversed by inactivation of the proton transfer activity of AdeB (D408A mutant) or by hyperporination of the outer membrane. We conclude that hypersusceptibility is not caused by an increased concentration of the ligands in the periplasm but is due to the accumulation of the antibiotics in the cytoplasm. Such changes in the accumulation could be due to the increased influx of antibiotics into the cytoplasm or their reduced efflux from the cytoplasm. The conformationally trapped F178C/F277C and F178C/W610C variants could contribute to both by enabling the leakage of antibiotics through their transmembrane domain and thus increasing their influx, or by enabling the leakage of protons and thus reducing the overall proton motive force and reducing the activities of single-component efflux pumps acting across the inner membrane (27). The overproduction of the WT AdeABC is somewhat toxic to the cells, as seen from the reduced growth rate of A. baumannii cells (Fig. 3A) and changes in their morphology (10, 18). Since the expression of F178C variants is even more toxic than that of the WT (Fig. 3A), it is likely that the overproduction of this conformationally restrained AdeB depletes the proton motive force acting across the inner membrane and negatively affects all secondary transport reactions in the cells.
In conclusion, the function of AdeB is affected by substitutions in a manner different from previously reported results for the model RND transporters such as AcrB and MexB. There are at least two substrate translocation paths in AdeB, with one utilized by large antibiotics such as macrolides and another by cations such as Et and gentamicin. The amino acid residues involved in interactions with substrates in distant locations communicate with each other in a nonadditive manner. Our results suggest that trapping a specific conformation of AdeB could lead to an effective inhibition of the pump and even to hypersusceptibility to certain antibiotics. It is possible that binding of certain inhibitors in the hydrophobic patch of AdeB could mimic the conformations of the F178C/F277C and F178C/W610C variants.
MATERIALS AND METHODS
Bacterial strains and site-directed mutagenesis
Strains and plasmids used in this study are shown in Table 5. All substitutions in the adeB gene were constructed by the QuickChange XL Site-Directed Mutagenesis kit (Agilent) using pIL129 (pTJ1-AdeABC) as the template. Primers used in the mutagenesis are described in Table S1. Introduced mutations and the lack of undesired mutations were verified by DNA sequencing (Oklahoma Medical Research Foundation). These plasmids were inserted into AbΔ3 and AbΔ3-Pore strains as described previously (10).
TABLE 5.
Strains and plasmids used in this study
| Strain | Relevant genotype | Source |
|---|---|---|
| ATCC17978 (AbWT) | Drug-susceptible wild type | ATCC |
| JWW30 | ATCC 17978, spontaneous variant resistant to 100 mg/L streptomycin | (19) |
| IL119 (AbΔ3) | A. baumannii JWW30 ΔadeABΔadeFGHΔadeIJK | (19) |
| IL139 (AbΔ3-Pore) | A.baumannii IL119 attTn7::mini-Tn7T-Kanr-araC-P BAD -FhuA | (10) |
| IL122 | A.baumannii IL119 attTn7::miniTn7T-Tpr-araC-P BAD -MCS carrying pTJ1 | (19) |
| IL161 | A.baumannii IL139 attTn7::miniTn7T- Kanr -araC-P BAD -FhuA carrying pTJ1 | This study |
| IL140 | A. baumannii IL119 attTn7::mini-Tn7T-Tpr-araC-PBAD-adeABC carrying pTJ1-adeABC | (10) |
| IL146 | A.baumannii IL139 attTn7::mini-Tn7T-Kanr-araC-P BAD -FhuA carrying pTJ1-adeABC | (10) |
| C43(DE3) | F– ompThsdSB (rB− mB−) gal dcm (DE3) | (28) |
| Plasmids | ||
| pTJ1 | pUC18T-mini-Tn7T-Tp-araC-P BAD -MCS, Ampr, Tpr | (29) |
| pIL129 (pTJ1-adeABC) | pUC18T-mini-Tn7T-Tp-araC-P BAD-adeABC, Ampr, Tpr | (10) |
| pET-21 a (+) | T7lac, T7-Tag (N), His-Tag (C), Ampr | Novagen |
| pIL151 (pET21- AdeB) | AdeB inserted between NdeI and XhoI; Ampr | This study |
Drug-susceptibility testing
The susceptibility of AdeB mutant strains was determined in 96-well microtiter plates at 37°C in Luria-Bertani (LB) medium supplemented with 1% L-arabinose. MICs were analyzed using a twofold broth dilution method (19). Cell growth was determined visually, and the optical density (OD) at λ = 600 nm of the culture was measured by a microplate reader (Tecan Spark 10M) after incubation of the microtiter plates at 37°C for 24 h. The cell density values were plotted as a function of antibiotic concentrations in wells and then fitted into a Hill equation to determine the IC50 (30). All experiments have been done 2–3 times with technical duplicates. IC50 results were averaged, and standard deviations were calculated.
Production of polyclonal antiAdeB antibody
DNA fragments of the adeB gene were amplified by PCR using the genomic DNA of Ab WT (JWW30) as a template and cloned into pET21a with NdeI and XhoI as restriction sites to generate pET21-AdeB expressing the efflux transporter under the control of the isopropyl β-d-1-thiogalactopyranoside (IPTG)-inducible promoter. To purify AdeB, C43-AdeB cells were cultured in LB containing ampicillin and induced with 0.1 mM IPTG for an additional 5 h at an OD600 of ∼0.3. Protein purification was done as described before (31). Purified AdeB was separated by electrophoresis on an 8% SDS-polyacrylamide gel and visualized by Coomassie Brilliant Blue, cut out of the gel, and around 3 mg of protein was sent in phosphate buffered saline (PBS) buffer to ThermoFisherScientific for polyclonal antiAdeB antibody production in rabbits.
Protein expression and analyses
SDS-polyacrylamide gels and quantitative immunoblotting analyses were used to analyze the protein profiles and expression levels of mutated AdeB proteins in AbΔ3 and AbΔ3-Pore strains. Membrane fractions were isolated after 5 h of induction from A. baumannii cells by ultracentrifugation as described before (32). Proteins were normalized to the same total protein concentration, analyzed by 8% SDS-PAGE, and transferred to PVDF membranes for immunoblotting with primary polyclonal antiAdeB antibodies (ThermoFisher Scientific) and a secondary alkaline phosphatase-conjugated antirabbit immunoglobulin antibody (Sigma). The 5-bromo4-chloro-3-indolylphosphate and nitroblue tetrazolium substrates were used to visualize the bands.
ACKNOWLEDGMENTS
This work was supported by NIAID/NIH grant number R01AI052293 to H.I.Z.
Contributor Information
Helen I. Zgurskaya, Email: elenaz@ou.edu.
Michael Y. Galperin, NCBI, NLM, National Institutes of Health, Bethesda, Maryland, USA
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jb.00217-23.
Structure of the ligand-bound AdeB protomer. Three Et molecules bound to AdeB are shown as cyan spheres. The inset shows the mutated amino acid residues E89, W708, I663, D664, and E665.
Primers used in this study.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Tamma PD, Aitken SL, Bonomo RA, Mathers AJ, van Duin D, Clancy CJ. 2023. Infectious diseases society of America antimicrobial-resistant treatment guidance: gram-negative bacterial infections. Clin Infect Dis:ciad428. doi: 10.1093/cid/ciad428 [DOI] [PubMed] [Google Scholar]
- 2. Morgan CE, Glaza P, Leus IV, Trinh A, Su C-C, Cui M, Zgurskaya HI, Yu EW, Projan SJ. 2021. Cryoelectron microscopy structures of AdeB illuminate mechanisms of simultaneous binding and exporting of substrates. mBio 12:e03690-20. doi: 10.1128/mBio.03690-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Prevention CfDCa . 2022. COVID-19: U.S impact on antimicrobial resistance, special report 2022. https://www.cdc.gov/drugresistance/covid19.html.
- 4. Kyriakidis I, Vasileiou E, Pana ZD, Tragiannidis A. 2021. Acinetobacter baumannii antibiotic resistance mechanisms. Pathogens 10:373. doi: 10.3390/pathogens10030373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kornelsen V, Kumar A. 2021. Update on multidrug resistance efflux pumps in Acinetobacter spp. Antimicrob Agents Chemother 65:e0051421. doi: 10.1128/AAC.00514-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vrancianu CO, Gheorghe I, Czobor IB, Chifiriuc MC. 2020. Antibiotic resistance profiles, molecular mechanisms and innovative treatment strategies of Acinetobacter baumannii. Microorganisms 8:935. doi: 10.3390/microorganisms8060935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sun JR, Chan MC, Chang TY, Wang WY, Chiueh TS. 2010. Overexpression of the adeB gene in clinical isolates of tigecycline-nonsusceptible Acinetobacter baumannii without insertion mutations in adeRS. Antimicrob Agents Chemother 54:4934–4938. doi: 10.1128/AAC.00414-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Higgins PG, Wisplinghoff H, Stefanik D, Seifert H. 2004. Selection of topoisomerase mutations and overexpression of adeB mRNA transcripts during an outbreak of Acinetobacter baumannii. J Antimicrob Chemother 54:821–823. doi: 10.1093/jac/dkh427 [DOI] [PubMed] [Google Scholar]
- 9. Yoon EJ, Courvalin P, Grillot-Courvalin C. 2013. RND-type efflux pumps in multidrug-resistant clinical isolates of Acinetobacter baumannii: major role for AdeABc overexpression and AdeRS mutations. Antimicrob Agents Chemother 57:2989–2995. doi: 10.1128/AAC.02556-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Leus IV, Weeks JW, Bonifay V, Smith L, Richardson S, Zgurskaya HI, Silhavy TJ. 2018. Substrate specificities and efflux efficiencies of RND efflux pumps of Acinetobacter baumannii. J Bacteriol 200. doi: 10.1128/JB.00049-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morgan CE, Zhang Z, Bonomo RA, Yu EW. 2021. An analysis of the novel fluorocycline TP-6076 bound to both the ribosome and multidrug efflux pump AdeJ from Acinetobacter baumannii. mBio 13:e0373221. doi: 10.1128/mbio.03732-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shi Y, Hua X, Xu Q, Yang Y, Zhang L, He J, Mu X, Hu L, Leptihn S, Yu Y. 2020. Mechanism of eravacycline resistance in Acinetobacter baumannii mediated by a deletion mutation in the sensor kinase adeS, leading to elevated expression of the efflux pump AdeABC. Infect Genet Evol 80:104185. doi: 10.1016/j.meegid.2020.104185 [DOI] [PubMed] [Google Scholar]
- 13. Su CC, Morgan CE, Kambakam S, Rajavel M, Scott H, Huang W, Emerson CC, Taylor DJ, Stewart PL, Bonomo RA, Yu EW. 2019. Cryo-electron microscopy structure of an Acinetobacter baumannii multidrug efflux pump. mBio 10:e01295-19. doi: 10.1128/mBio.01295-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kobylka J, Kuth MS, Müller RT, Geertsma ER, Pos KM. 2020. AcrB: a mean, keen, drug efflux machine. Ann N Y Acad Sci 1459:38–68. doi: 10.1111/nyas.14239 [DOI] [PubMed] [Google Scholar]
- 15. Zhang Z, Morgan CE, Bonomo RA, Yu EW. 2021. Cryo-EM determination of eravacycline-bound structures of the ribosome and the multidrug efflux pump AdeJ of Acinetobacter baumannii. mBio 12:e0103121. doi: 10.1128/mBio.01031-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eicher T, Cha H, Seeger MA, Brandstätter L, El-Delik J, Bohnert JA, Kern WV, Verrey F, Grütter MG, Diederichs K, Pos KM. 2012. Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc Natl Acad Sci U S A 109:5687–5692. doi: 10.1073/pnas.1114944109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ornik-Cha A, Wilhelm J, Kobylka J, Sjuts H, Vargiu AV, Malloci G, Reitz J, Seybert A, Frangakis AS, Pos KM. 2021. Structural and functional analysis of the promiscuous AcrB and AdeB efflux pumps suggests different drug binding mechanisms. Nat Commun 12:6919. doi: 10.1038/s41467-021-27146-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yoon EJ, Balloy V, Fiette L, Chignard M, Courvalin P, Grillot-Courvalin C. 2016. Contribution of the Ade resistance-nodulation-cell division-type efflux pumps to fitness and pathogenesis of Acinetobacter baumannii. mBio 7:e00697-16. doi: 10.1128/mBio.00697-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krishnamoorthy G, Leus IV, Weeks JW, Wolloscheck D, Rybenkov VV, Zgurskaya HI, Bonomo RA. 2017. Synergy between active efflux and outer membrane diffusion defines rules of antibiotic permeation into gram-negative bacteria. mBio 8:e01172-17. doi: 10.1128/mBio.01172-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Krishnamoorthy G, Tikhonova EB, Zgurskaya HI. Role of the amino acid residues lining the vestibule region in Mexb of Pseudomonas Aeruginosa in substrate specificity.
- 21. Glavier M, Puvanendran D, Salvador D, Decossas M, Phan G, Garnier C, Frezza E, Cece Q, Schoehn G, Picard M, Taveau JC, Daury L, Broutin I, Lambert O. 2020. Antibiotic export by MexB multidrug efflux transporter is allosterically controlled by a MexA-OprM chaperone-like complex. Nat Commun 11:4948. doi: 10.1038/s41467-020-18770-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sennhauser G, Bukowska MA, Briand C, Grütter MG. 2009. Crystal structure of the multidrug exporter MexB from Pseudomonas aeruginosa. J Mol Biol 389:134–145. doi: 10.1016/j.jmb.2009.04.001 [DOI] [PubMed] [Google Scholar]
- 23. Ruzin A, Immermann FW, Bradford PA. 2010. RT-PCR and statistical analyses of adeABC expression in clinical isolates of Acinetobacter calcoaceticus-Acinetobacter baumannii complex. Microb Drug Resist 16:87–89. doi: 10.1089/mdr.2009.0131 [DOI] [PubMed] [Google Scholar]
- 24. Zwama M, Yamasaki S, Nakashima R, Sakurai K, Nishino K, Yamaguchi A. 2018. Multiple entry pathways within the efflux transporter AcrB contribute to multidrug recognition. Nat Commun 9:124. doi: 10.1038/s41467-017-02493-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tam HK, Foong WE, Oswald C, Herrmann A, Zeng H, Pos KM. 2021. Allosteric drug transport mechanism of multidrug transporter AcrB. Nat Commun 12:3889. doi: 10.1038/s41467-021-24151-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leus IV, Adamiak J, Chandar B, Bonifay V, Zhao S, Walker SS, Squadroni B, Balibar CJ, Kinarivala N, Standke LC, Voss HU, Tan DS, Rybenkov VV, Zgurskaya HI. 2023. Functional diversity of gram-negative permeability barriers reflected in antibacterial activities and intracellular accumulation of antibiotics. Antimicrob Agents Chemother 67:e0137722. doi: 10.1128/aac.01377-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Saha P, Sikdar S, Krishnamoorthy G, Zgurskaya HI, Rybenkov VV. 2020. Drug permeation against efflux by two transporters. ACS Infect Dis 6:747–758. doi: 10.1021/acsinfecdis.9b00510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miroux B, Walker JE. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol 260:289–298. doi: 10.1006/jmbi.1996.0399 [DOI] [PubMed] [Google Scholar]
- 29. Damron FH, McKenney ES, Barbier M, Liechti GW, Schweizer HP, Goldberg JB. 2013. Construction of mobilizable mini-Tn7 vectors for bioluminescent detection of gram-negative bacteria and single-copy promoter lux reporter analysis. Appl Environ Microbiol 79:4149–4153. doi: 10.1128/AEM.00640-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhao H, Petrushenko ZM, Walker JK, Baudry J, Zgurskaya HI, Rybenkov VV. 2018. Small molecule condensin inhibitors. ACS Infect Dis 4:1737–1745. doi: 10.1021/acsinfecdis.8b00222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fabre L, Ntreh AT, Yazidi A, Leus IV, Weeks JW, Bhattacharyya S, Ruickoldt J, Rouiller I, Zgurskaya HI, Sygusch J. 2021. A "drug sweeping" state of the TriABC triclosan efflux pump from Pseudomonas aeruginosa. Structure 29:261–274. doi: 10.1016/j.str.2020.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Krishnamoorthy G, Wolloscheck D, Weeks JW, Croft C, Rybenkov VV, Zgurskaya HI. 2016. Breaking the permeability barrier of Escherichia coli by controlled hyperporination of the outer membrane. Antimicrob Agents Chemother 60:7372–7381. doi: 10.1128/AAC.01882-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Structure of the ligand-bound AdeB protomer. Three Et molecules bound to AdeB are shown as cyan spheres. The inset shows the mutated amino acid residues E89, W708, I663, D664, and E665.
Primers used in this study.