Abstract
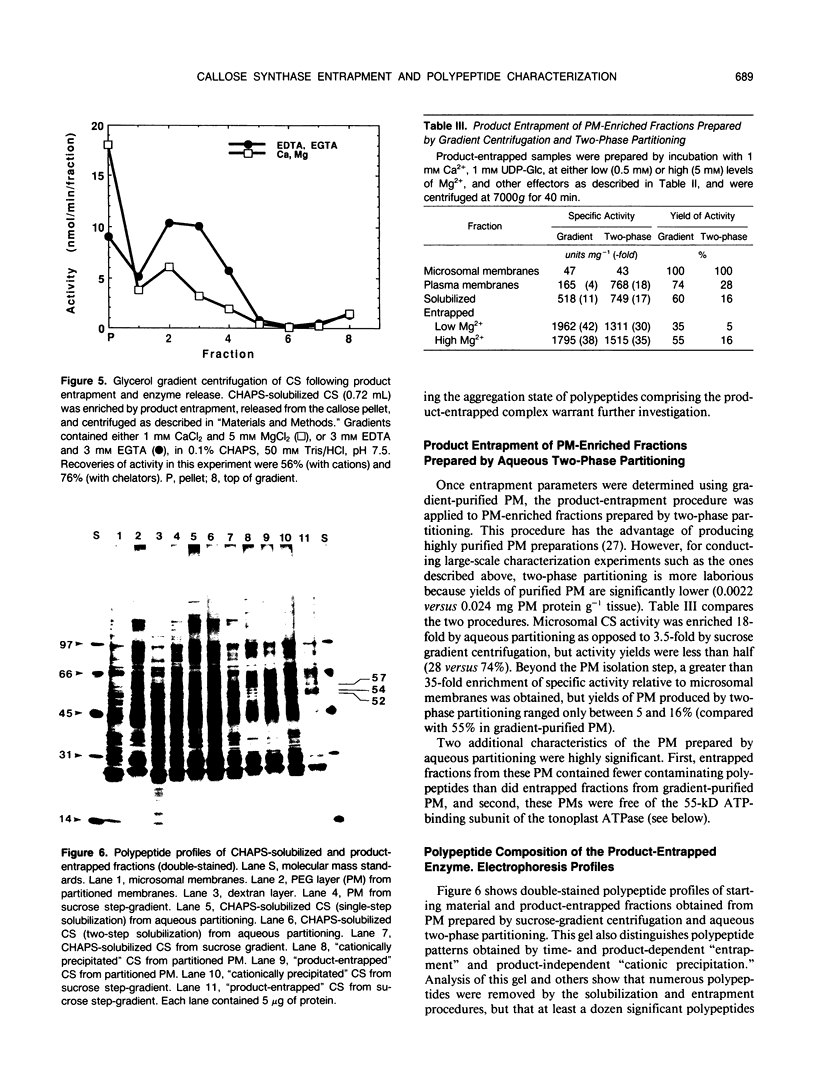
Rapid enrichment of CHAPS-solubilized UDP-glucose:(1,3)-β-glucan (callose) synthase from storage tissue of red beet (Beta vulgaris L.) is obtained when the preparation is incubated with an enzyme assay mixture, then centrifuged and the enzyme released from the callose pellet with a buffer containing EDTA and CHAPS (20-fold purification relative to microsomes). When centrifuged at high speed (80,000g), the enzyme can also be pelleted in the absence of substrate (UDP-Glc) or synthesis of callose, due to nonspecific aggregation of proteins caused by excess cations and insufficient detergent in the assay buffer. True time-dependent and substrate-dependent product-entrapment of callose synthase is obtained by low-speed centrifugation (7,000-11,000g) of enzyme incubated in reaction mixtures containing low levels of cations (0.5 millimolar Mg2+, 1 millimolar Ca2+) and sufficient detergent (0.02% digitonin, 0.12% CHAPS), together with cellobiose, buffer, and UDP-Glc. Entrapment conditions, therefore, are a compromise between preventing nonspecific precipitation of proteins and permitting sufficient enzyme activity for callose synthesis. Further enrichment of the enzyme released from the callose pellet was not obtained by rate-zonal glycerol gradient centrifugation, although its sedimentation rate was greatly enhanced by inclusion of divalent cations in the gradient. Preparations were markedly cleaner when product-entrapment was conducted on enzyme solubilized from plasma membranes isolated by aqueous two-phase partitioning rather than by gradient centrifugation. Product-entrapped preparations consistently contained polypeptides or groups of closely-migrating polypeptides at molecular masses of 92, 83, 70, 57, 43, 35, 31/29, and 27 kilodaltons. This polypeptide profile is in accordance with the findings of other callose synthase enrichment studies using a variety of tissue sources, and is consistent with the existence of a multi-subunit enzyme complex.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Biggs M. S., Harriman R. W., Handa A. K. Changes in Gene Expression during Tomato Fruit Ripening. Plant Physiol. 1986 Jun;81(2):395–403. doi: 10.1104/pp.81.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Bulone V., Girard V., Fèvre M. Separation and Partial Purification of 1,3-beta-Glucan and 1,4-beta-Glucan Synthases from Saprolegnia. Plant Physiol. 1990 Dec;94(4):1748–1755. doi: 10.1104/pp.94.4.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmer D. P., Solomon M., Read S. M. Direct Photolabeling with [P]UDP-Glucose for Identification of a Subunit of Cotton Fiber Callose Synthase. Plant Physiol. 1991 Feb;95(2):556–563. doi: 10.1104/pp.95.2.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhugga K. S., Ray P. M. A 55 kDa plasma membrane-associated polypeptide is involved in beta-1,3-glucan synthase activity in pea tissue. FEBS Lett. 1991 Jan 28;278(2):283–286. doi: 10.1016/0014-5793(91)80136-q. [DOI] [PubMed] [Google Scholar]
- Dhugga K. S., Ray P. M. Isoelectric Focusing of Plant Plasma Membrane Proteins : Further Evidence that a 55 Kilodalton Polypeptide Is Associated with beta-1,3-Glucan Synthase Activity from Pea. Plant Physiol. 1991 Apr;95(4):1302–1305. doi: 10.1104/pp.95.4.1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiberger L. L., Wasserman B. P. Partial Purification of Digitonin-Solubilized beta-Glucan Synthase from Red Beet Root. Plant Physiol. 1987 Apr;83(4):982–987. doi: 10.1104/pp.83.4.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost D. J., Read S. M., Drake R. R., Haley B. E., Wasserman B. P. Identification of the UDP-glucose-binding polypeptide of callose synthase from Beta vulgaris L. by photoaffinity labeling with 5-azido-UDP-glucose. J Biol Chem. 1990 Feb 5;265(4):2162–2167. [PubMed] [Google Scholar]
- Hayashi T., Read S. M., Bussell J., Thelen M., Lin F. C., Brown R. M., Delmer D. P. UDP-Glucose: (1-->3)-beta-Glucan Synthases from Mung Bean and Cotton: Differential Effects of Ca and Mg on Enzyme Properties and on Macromolecular Structure of the Glucan Product. Plant Physiol. 1987 Apr;83(4):1054–1062. doi: 10.1104/pp.83.4.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M. S., Elango N., Mattia E., Au-Young J., Robbins P. W., Cabib E. Isolation of chitin synthetase from Saccharomyces cerevisiae. Purification of an enzyme by entrapment in the reaction product. J Biol Chem. 1984 Dec 10;259(23):14966–14972. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lin F. C., Brown R. M., Jr, Drake R. R., Jr, Haley B. E. Identification of the uridine 5'-diphosphoglucose (UDP-Glc) binding subunit of cellulose synthase in Acetobacter xylinum using the photoaffinity probe 5-azido-UDP-Glc. J Biol Chem. 1990 Mar 25;265(9):4782–4784. [PubMed] [Google Scholar]
- Morré D. J., Morré D. M. Preparation of mammalian plasma membranes by aqueous two-phase partition. Biotechniques. 1989 Oct;7(9):946-8, 950-4, 956-8. [PubMed] [Google Scholar]
- Parry R. V., Turner J. C., Rea P. A. High purity preparations of higher plant vacuolar H+-ATPase reveal additional subunits. Revised subunit composition. J Biol Chem. 1989 Nov 25;264(33):20025–20032. [PubMed] [Google Scholar]
- Porzio M. A., Pearson A. M. Improved resolution of myofibrillar proteins with sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Biochim Biophys Acta. 1977 Jan 25;490(1):27–34. doi: 10.1016/0005-2795(77)90102-7. [DOI] [PubMed] [Google Scholar]
- Read S. M., Delmer D. P. Inhibition of Mung Bean UDP-Glucose: (1-->3)-beta-Glucan Synthase by UDP-Pyridoxal: Evidence for an Active-Site Amino Group. Plant Physiol. 1987 Dec;85(4):1008–1015. doi: 10.1104/pp.85.4.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan M. E., Rodis P., Wasserman B. P. CHAPS Solubilization and Functional Reconstitution of beta-Glucan Synthase from Red Beet Root (Beta vulgaris L.) Storage Tissue. Plant Physiol. 1987 Oct;85(2):516–522. doi: 10.1104/pp.85.2.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan M. E., Wasserman B. P. Susceptibility of UDP-Glucose:(1,3)-beta-Glucan Synthase to Inactivation by Phospholipases and Trypsin. Plant Physiol. 1989 Apr;89(4):1341–1344. doi: 10.1104/pp.89.4.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman B. P., McCarthy K. J. Regulation of Plasma Membrane beta-Glucan Synthase from Red Beet Root by Phospholipids : Reactivation of Triton X-100 Extracted Glucan Synthase by Phospholipids. Plant Physiol. 1986 Oct;82(2):396–400. doi: 10.1104/pp.82.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widell S., Lundborg T., Larsson C. Plasma membranes from oats prepared by partition in an aqueous polymer two-phase system : on the use of light-induced cytochrome B reduction as a marker for the plasma membrane. Plant Physiol. 1982 Nov;70(5):1429–1435. doi: 10.1104/pp.70.5.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]