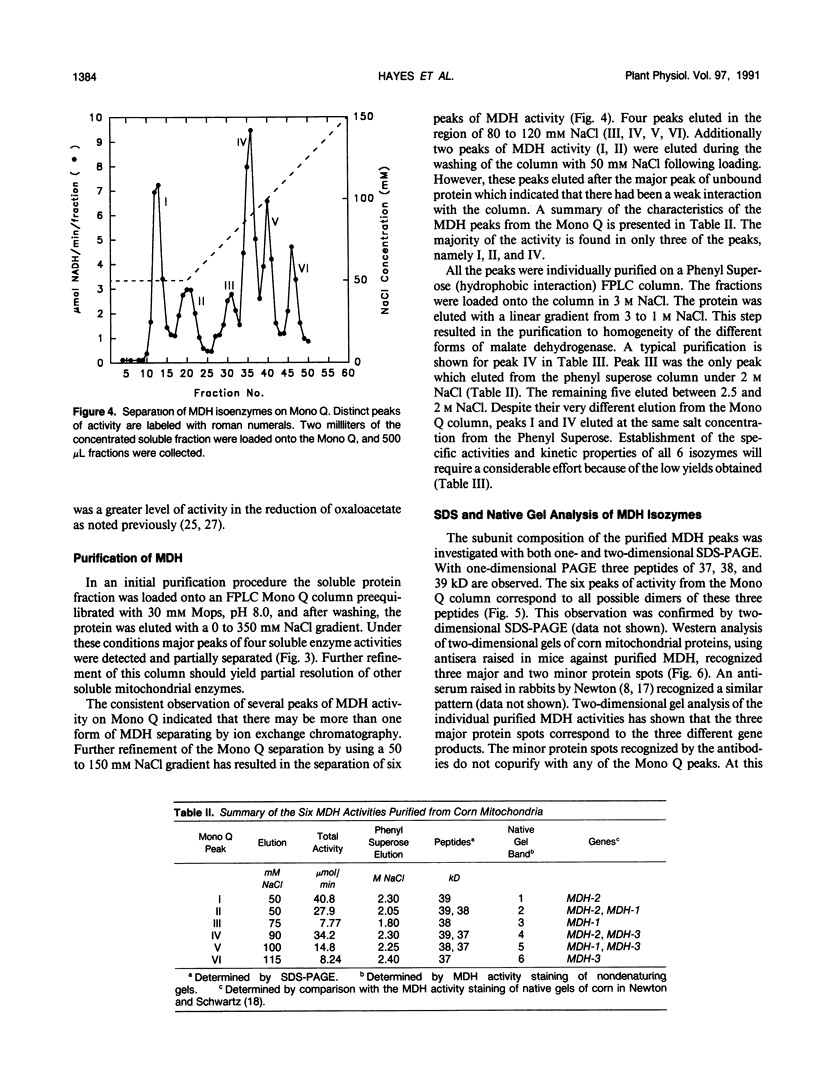
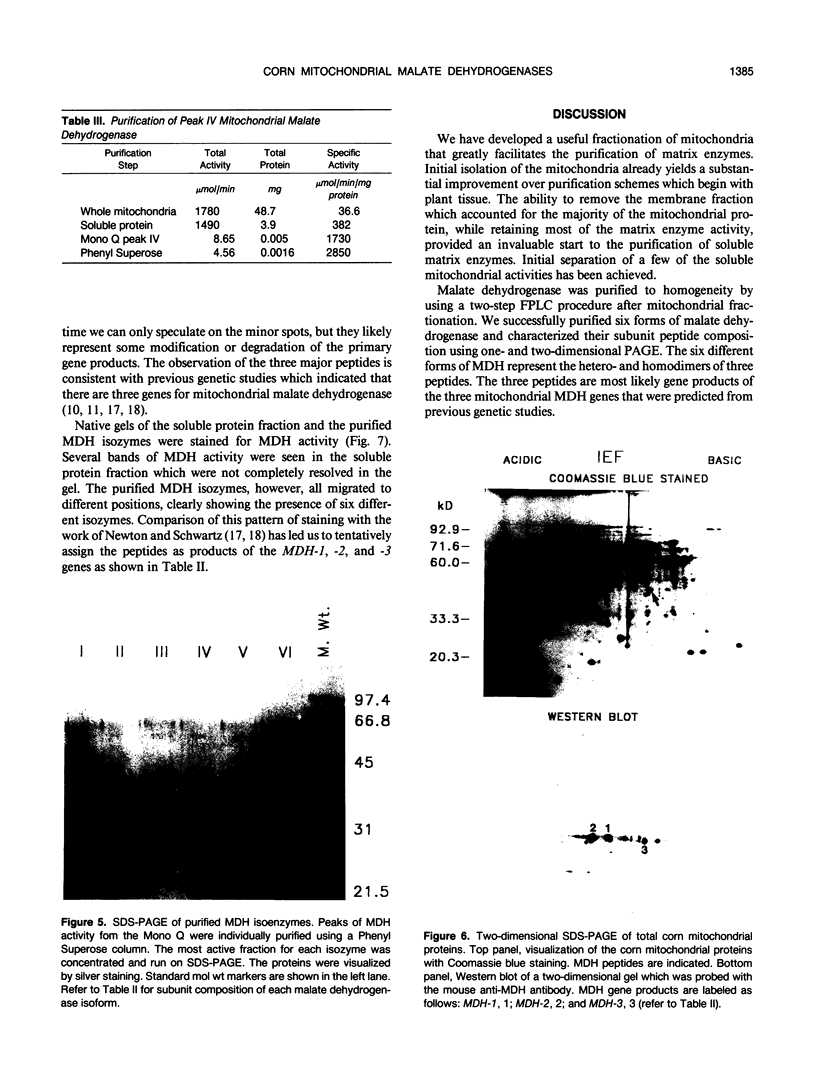
Abstract
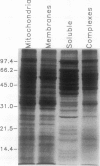
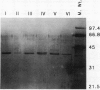
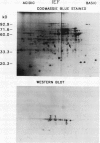
A method to fractionate corn (Zea mays L. B73) mitochondria into soluble proteins, high molecular weight soluble proteins, and membrane proteins was developed. These fractions were analyzed by both sodium dodecyl sulfate-polyacrylamide gel electrophoresis and assays of mitochondrial enzyme activities. The Krebs cycle enzymes were enriched in the soluble fraction. Malate dehydrogenase has been purified from the soluble fraction by a two-step fast protein liquid chromatography method. Six different malate dehydrogenase peaks were obtained from the Mono Q column. These peaks were individually purified using a Phenyl Superose column. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of the purified peaks showed that three of the isoenzymes consisted of different homodimers (I, III, VI) and three were different heterodimers (II, IV, V). Apparent molecular masses of the three different monomer subunits were 37, 38, and 39 kilodaltons. Nondenaturing gel analysis of the malate dehydrogenase peaks showed that each Mono Q peak contained a band of malate dehydrogenase activity with different mobility. These observations are consistent with three nuclear genes encoding corn mitochondrial malate dehydrogenase. Polyclonal antibodies raised against purified malate dehydrogenase were used to identify the gene products using Western blots of two-dimensional gels.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Douce R., Mannella C. A., Bonner W. D., Jr The external NADH dehydrogenases of intact plant mitochondria. Biochim Biophys Acta. 1973 Jan 18;292(1):105–116. doi: 10.1016/0005-2728(73)90255-7. [DOI] [PubMed] [Google Scholar]
- Elthon T. E., McIntosh L. Identification of the alternative terminal oxidase of higher plant mitochondria. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8399–8403. doi: 10.1073/pnas.84.23.8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietl C., Lehnerer M., Olsen O. Mitochondrial malate dehydrogenase from watermelon: sequence of cDNA clones and primary structure of the higher-plant precursor protein. Plant Mol Biol. 1990 Jun;14(6):1019–1030. doi: 10.1007/BF00019398. [DOI] [PubMed] [Google Scholar]
- Goodman M. M., Newton K. J., Stuber C. W. Malate dehydrogenase: viability of cytosolic nulls and lethality of mitochondrial nulls in maize. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1783–1785. doi: 10.1073/pnas.78.3.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M. M., Stuber C. W., Lee C. N., Johnson F. M. Genetic control of malate dehydrogenase isozymes in maize. Genetics. 1980 Jan;94(1):153–168. doi: 10.1093/genetics/94.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larson E., Howlett B., Jagendorf A. Artificial reductant enhancement of the Lowry method for protein determination. Anal Biochem. 1986 Jun;155(2):243–248. doi: 10.1016/0003-2697(86)90432-x. [DOI] [PubMed] [Google Scholar]
- Merril C. R., Goldman D., Van Keuren M. L. Gel protein stains: silver stain. Methods Enzymol. 1984;104:441–447. doi: 10.1016/s0076-6879(84)04111-2. [DOI] [PubMed] [Google Scholar]
- Newton K. J., Schwartz D. Genetic basis of the major malate dehydrogenase isozymes in maize. Genetics. 1980 Jun;95(2):425–442. doi: 10.1093/genetics/95.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwitzguebel J. P., Siegenthaler P. A. Purification of peroxisomes and mitochondria from spinach leaf by percoll gradient centrifugation. Plant Physiol. 1984 Jul;75(3):670–674. doi: 10.1104/pp.75.3.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger E. A., Hand J. M., Cashmore A. R., Vasconcelos A. C. Isolation of a cDNA encoding mitochondrial citrate synthase from Arabidopsis thaliana. Plant Mol Biol. 1989 Oct;13(4):411–418. doi: 10.1007/BF00015553. [DOI] [PubMed] [Google Scholar]
- Walk R. A., Hock B. Separation of malate dehydrogenase isoenzymes by affinity chromatography on 5'-AMP-Sepharose. Eur J Biochem. 1976 Dec;71(1):25–32. doi: 10.1111/j.1432-1033.1976.tb11085.x. [DOI] [PubMed] [Google Scholar]
- Yang N. S., Scandalios J. G. Purification and biochemical properties of genetically defined malate dehydrogenase in maize. Arch Biochem Biophys. 1974 Apr 2;161(2):335–353. doi: 10.1016/0003-9861(74)90314-2. [DOI] [PubMed] [Google Scholar]