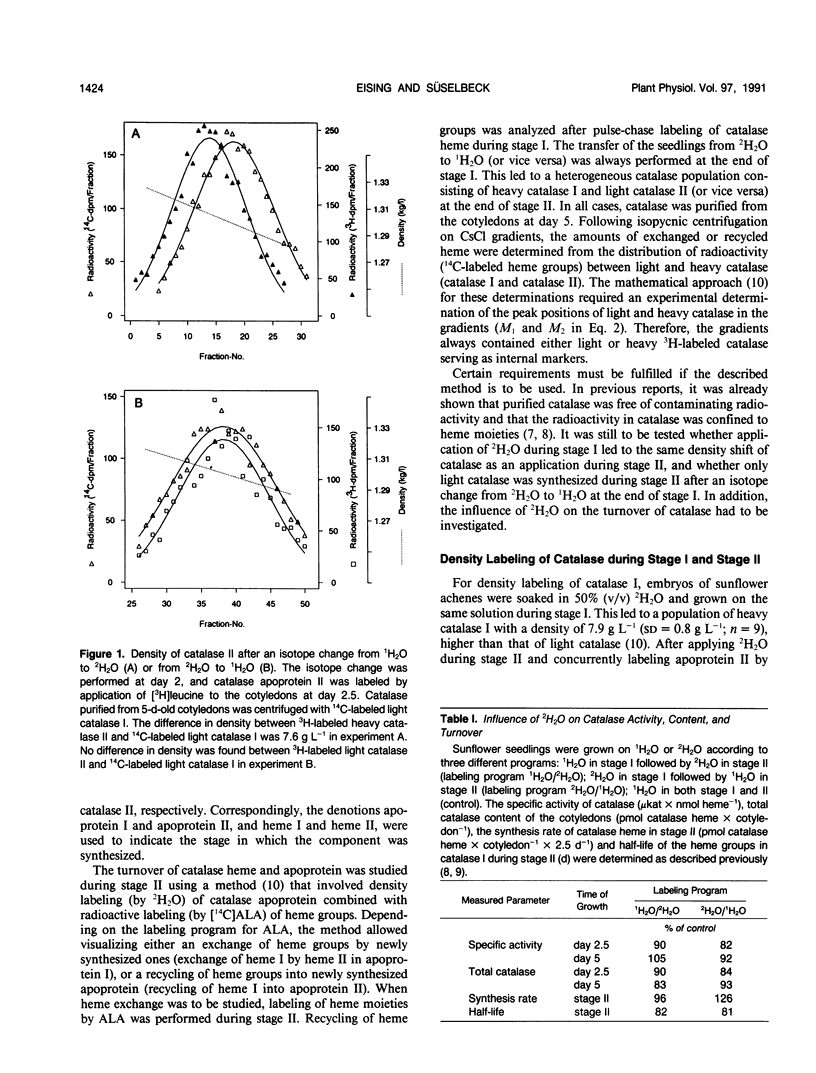
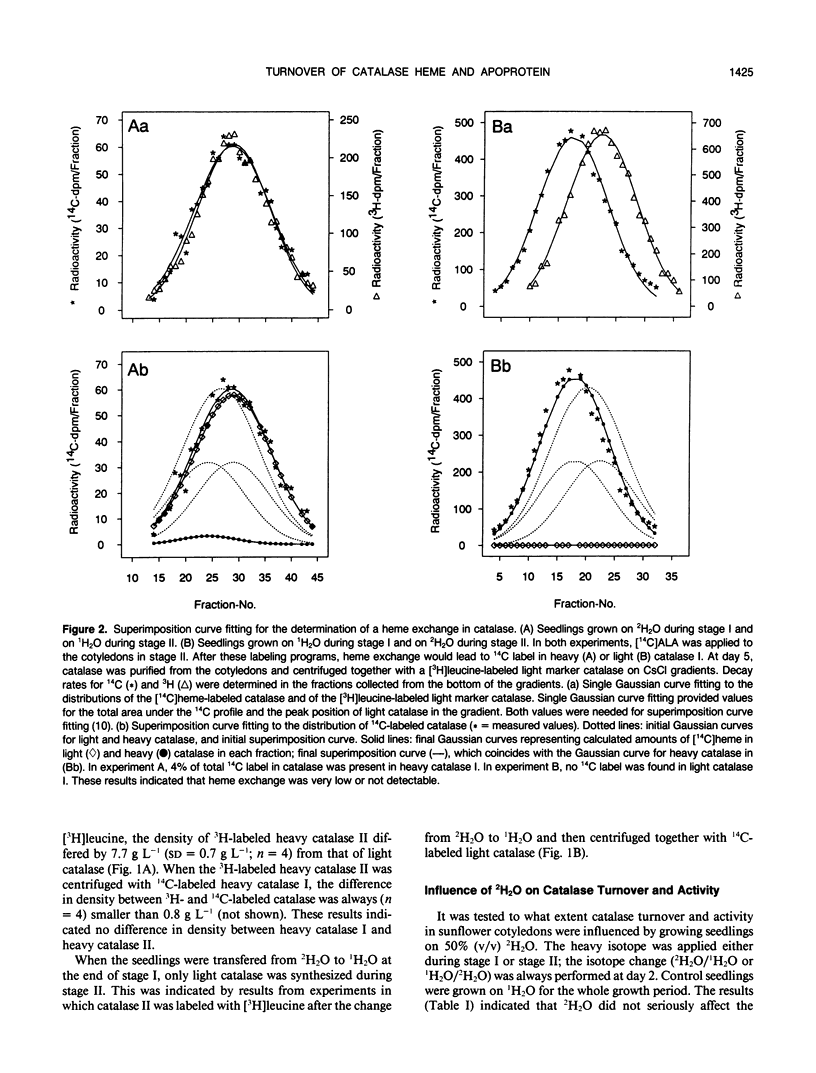
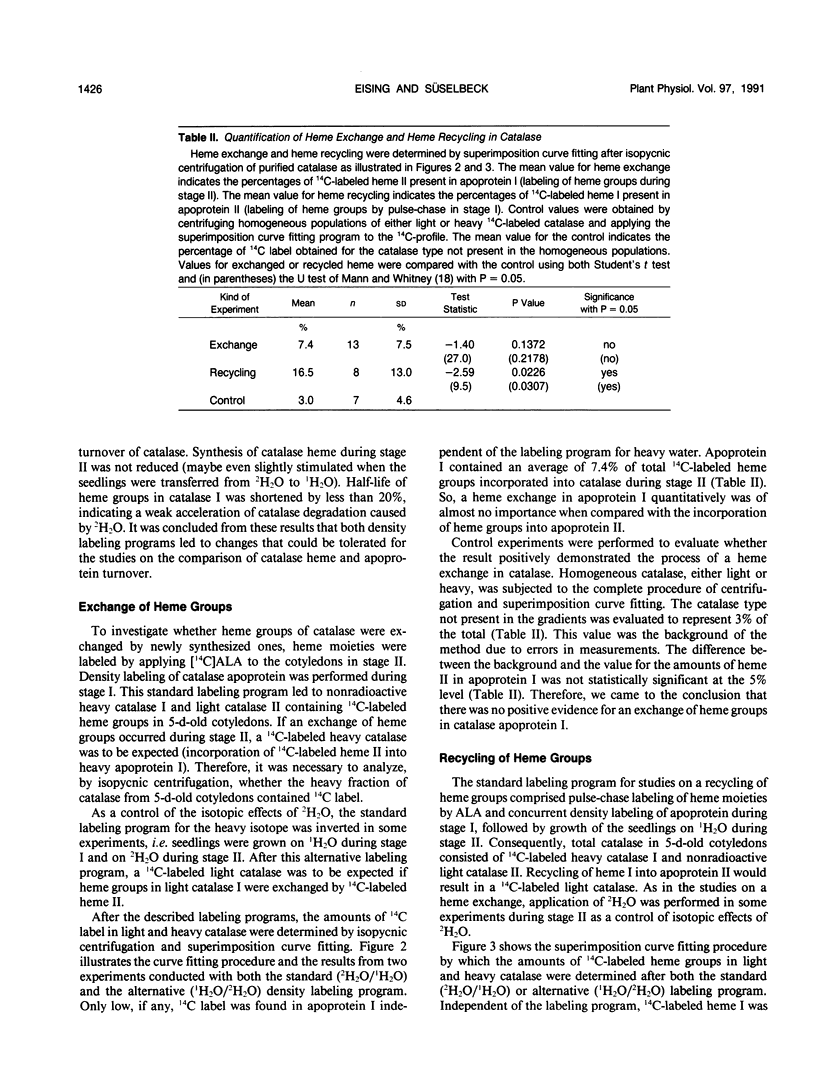
Abstract
The turnover of catalase apoprotein and catalase heme was studied in cotyledons of sunflower (Helianthus annuus L.) seedlings by density labeling of apoprotein and radioactive labeling of heme moieties. The heavy isotope (50% 2H2O) and the radioactive isotope ([14C]5-aminolevulinic acid) were applied either during growth in the dark (day 0-2.5) or in the light (day 2.5 and 5). Following isopycnic centrifugation of catalase purified from cotyledons of 5-day-old seedlings, superimposition curve fitting was used to determine the amounts of radioactive heme moieties in native and density-labeled catalase. Data from these determinations indicated that turnover of catalase heme and apoprotein essentially was coordinate. Only small amounts of heme groups were recycled into newly synthesized apoprotein during growth in the light, and no evidence was found for an exchange of heme groups in apoprotein moieties. It followed from these observations that degradation of catalase apoprotein was slightly faster than that of catalase heme. A degradation constant for catalase apoprotein of 0.263 per day was determined from the data on heme recycling and the degradation constant of catalase heme determined previously to be 0.205 per day (R Eising, B Gerhardt [1987] Plant Physiol 84: 225-232).
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Behrends W., Birkhan R., Kindl H. Transition form of microbodies. Overlapping of two sets of marker proteins during the rearrangement of glyoxysomes into leaf peroxisomes. Biol Chem Hoppe Seyler. 1990 Jan;371(1):85–94. doi: 10.1515/bchm3.1990.371.1.85. [DOI] [PubMed] [Google Scholar]
- Bock K. W., Siekevitz P. Turnover of heme and protein moieties of rat liver microsomal cytochrome b5. Biochem Biophys Res Commun. 1970 Oct 23;41(2):374–380. doi: 10.1016/0006-291x(70)90514-0. [DOI] [PubMed] [Google Scholar]
- Davies D. D., Humphrey T. J. Amino Acid recycling in relation to protein turnover. Plant Physiol. 1978 Jan;61(1):54–58. doi: 10.1104/pp.61.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druyan R., Jakovcic S., Rabinowitz M. Studies of cytochrome synthesis in rat liver. Biochem J. 1973 Jun;134(2):377–385. doi: 10.1042/bj1340377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eising R., Gerhardt B. Catalase Degradation in Sunflower Cotyledons during Peroxisome Transition from Glyoxysomal to Leaf Peroxisomal Function. Plant Physiol. 1987 Jun;84(2):225–232. doi: 10.1104/pp.84.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eising R., Gerhardt B. Catalase Synthesis and Turnover during Peroxisome Transition in the Cotyledons of Helianthus annuus L. Plant Physiol. 1989 Mar;89(3):1000–1005. doi: 10.1104/pp.89.3.1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzisket U., Gerhardt B. Synthesis of Isocitrate Lyase in Sunflower Cotyledons during the Transition in Cotyledonary Microbody Function. Plant Physiol. 1980 Jun;65(6):1081–1084. doi: 10.1104/pp.65.6.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarow P. B., Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- Lazarow P. B., de Duve C. The synthesis and turnover of rat liver of rat liver peroxisomes. IV. Biochemical pathway of catalase synthesis. J Cell Biol. 1973 Nov;59(2 Pt 1):491–506. doi: 10.1083/jcb.59.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarow P. B., de Duve C. The synthesis and turnover of rat liver peroxisomes. V. Intracellular pathway of catalase synthesis. J Cell Biol. 1973 Nov;59(2 Pt 1):507–524. doi: 10.1083/jcb.59.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson A., Thomas P. E., Ryan D. E., Levin W. The in vivo turnover of rat liver microsomal epoxide hydrolase and both the apoprotein and heme moieties of specific cytochrome P-450 isozymes. Arch Biochem Biophys. 1983 Aug;225(1):216–236. doi: 10.1016/0003-9861(83)90025-5. [DOI] [PubMed] [Google Scholar]
- Poole B., Leighton F., De Duve C. The synthesis and turnover of rat liver peroxisomes. II. Turnover of peroxisome proteins. J Cell Biol. 1969 May;41(2):536–546. doi: 10.1083/jcb.41.2.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole B. The kinetics of disappearance of labeled leucine from the free leucine pool of rat liver and its effect on the apparent turnover of catalase and other hepatic proteins. J Biol Chem. 1971 Nov;246(21):6587–6591. [PubMed] [Google Scholar]
- Schneider H. A. Enzymic capacities for chlorophyll biosynthesis. Activation and de novo synthesis of enzymes. Z Naturforsch C. 1976 Jan-Feb;31(1-2):55–63. doi: 10.1515/znc-1976-1-213. [DOI] [PubMed] [Google Scholar]
- Sugita Y., Tobe T., Sakamoto T., Higashi T. Immature precursor catalase in subcellular fractions of rat liver. J Biochem. 1982 Aug;92(2):509–515. doi: 10.1093/oxfordjournals.jbchem.a133958. [DOI] [PubMed] [Google Scholar]
- Thomas J., Weinstein J. D. Measurement of heme efflux and heme content in isolated developing chloroplasts. Plant Physiol. 1990 Nov;94(3):1414–1423. doi: 10.1104/pp.94.3.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trelease R. N., Becker W. M., Gruber P. J., Newcomb E. H. Microbodies (Glyoxysomes and Peroxisomes) in Cucumber Cotyledons: Correlative Biochemical and Ultrastructural Study in Light- and Dark-grown Seedlings. Plant Physiol. 1971 Oct;48(4):461–475. doi: 10.1104/pp.48.4.461. [DOI] [PMC free article] [PubMed] [Google Scholar]


