ABSTRACT
Macroautophagy/autophagy has been recognized as a central antiviral defense mechanism in plant, which involves complex interactions between viral proteins and host factors. Rhabdoviruses are single-stranded RNA viruses, and the infection causes serious harm to public health, livestock, and crop production. However, little is known about the role of autophagy in the defense against rhabdovirus infection by plant. In this work, we showed that Rice stripe mosaic cytorhabdovirus(RSMV) activated autophagy in plants and that autophagy served as an indispensable defense mechanism during RSMV infection. We identified RSMV glycoprotein as an autophagy inducer that interacted with OsSnRK1B and promoted the kinase activity of OsSnRK1B on OsATG6b. RSMV glycoprotein was toxic to rice cells and its targeted degradation by OsATG6b-mediated autophagy was essential to restrict the viral titer in plants. Importantly, SnRK1-glycoprotein and ATG6-glycoprotein interactions were well-conserved between several other rhabdoviruses and plants. Together, our data support a model that SnRK1 senses rhabdovirus glycoprotein for autophagy initiation, while ATG6 mediates targeted degradation of viral glycoprotein. This conserved mechanism ensures compatible infection by limiting the toxicity of viral glycoprotein and restricting the infection of rhabdoviruses.
Abbreviations: AMPK: adenosine 5‘-monophosphate (AMP)-activated protein kinase; ANOVA: analysis of variance; ATG: autophagy related; AZD: AZD8055; BiFC: bimolecular fluorescence complementation; BYSMV: barley yellow striate mosaic virus; Co-IP: co-immunoprecipitation; ConA: concanamycin A; CTD: C-terminal domain; DEX: dexamethasone; DMSO: dimethyl sulfoxide; G: glycoprotein; GFP: green fluorescent protein; MD: middle domain; MDC: monodansylcadaverine; NTD: N-terminal domain; OE: over expression; Os: Oryza sativa; PBS: phosphate-buffered saline; PtdIns3K: class III phosphatidylinositol-3-kinase; qRT-PCR: quantitative real-time reverse-transcription PCR; RFP: red fluorescent protein; RSMV: rice stripe mosaic virus; RSV: rice stripe virus; SGS3: suppressor of gene silencing 3; SnRK1: sucrose nonfermenting1-related protein kinase1; SYNV: sonchus yellow net virus; TEM: transmission electron microscopy; TM: transmembrane region; TOR: target of rapamycin; TRV: tobacco rattle virus; TYMaV: tomato yellow mottle-associated virus; VSV: vesicular stomatitis virus; WT: wild type; Y2H: yeast two-hybrid; YFP: yellow fluorescent protein.
KEYWORDS: ATG6, glycoprotein, rhabdovirus, RSMV, SnRK1
Introduction
Autophagy is a highly conserved cellular process in eukaryotes that removes damaged organelles or undesirable intracellular materials under stress conditions or during specific developmental processes [1,2]. Three major types of autophagy occur in eukaryotic cells, including macroautophagy, microautopahgy, and chaperone-mediated autophagy [3–5]. Macroautophagy (hereafter referred to as autophagy) is initiated in the cytoplasm through the formation of double-membraned autophagosomes, followed by the fusion of autophagosome with lysosomes for degradation or recycling. The execution of autophagy depends on a variety of membrane-associated factors called AuTophaGy-related (ATG) proteins [6,7]. About 40 ATGs have been identified from yeast by genetic screening, approximately half of them are indispensable for autophagosome formation, and the orthologs were subsequently identified in animals and plants [8,9].
The molecular machinery involved in autophagy has been characterized over the past two decades. When cells are challenged by nutrient-starved conditions, abiotic stresses or pathogen infection, the activation of an intricate cascade of kinases, including SnRK1/Snf1/AMPK (Sucrose Nonfermenting1-Related Protein Kinase1) and Target Of Rapamycin (TOR) kinase, converges on ATG1 kinase and its regulatory subunit ATG13 to modify their activities through phosphorylation. The activated ATG1 kinase initiates the formation of a complex called an autophagy precursor to trigger autophagy [10]. Meanwhile, SnRK1 can also stimulates the activity of the class III phosphatidylinositol-3-kinase (PtdIns3K) complex by phosphorylating ATG6/BECN1, which involves in the autophagic membranes remodeling [11,12]. Subsequently, the ubiquitin-like protein ATG8 interacts with several ATG proteins such as ATG1, ATG6/BECN1, and ATG7, which is central for autophagosome formation and recruitment of selective cargos [13,14]. Mature autophagosomes containing dysfunctional organelles and other components eventually fuse with lysosome for degradation [10].
Accumulating evidence has demonstrated that autophagy acts as a defense mechanism against diverse intracellular pathogens in metazoan and plant systems [15,16]. Although autophagy seems to play both antiviral and proviral roles, compelling evidence suggests that autophagy participates in innate and adaptive immune responses to eliminate pathogenic viruses [17,18]. For example, in Drosophila, vesicular stomatitis virus (VSV) infection induces autophagy, and the inhibition of autophagy by silencing ATGs leads to increased virus replication and fly mortality [19]. In humans, autophagy also restricts the infection of several viruses, like influenza A virus, human immunodeficiency virus, and herpes simplex virus [20–22]. In plants, a pioneering work reported that autophagy pathway is required for N gene-mediated resistance to tobacco mosaic virus (TMV) [23]. Recently, autophagy has been recognized as an important component of plant innate immunity against multiple viruses [24–32]. In the meanwhile, plant viruses have evolved strategies to harness host autophagy for their own benefits, including the protection of viral protein degradation by suppressing autophagy [24,28,33–35], the promotion of autophagic degradation of the RNA silencing components in plant cell [36–38], and even induction and utilization of autophagy in its vector insects to promote propagation and infection [39–43].
Rhabdoviruses forms large family in the order Mononegavirales, which are characterized by a negative-sense RNA genome of 11–16 kb [44]. This family has a broad host range including vertebrates, invertebrates, and plants, and some members cause serious harm to public health, livestock, and crop production [44–46]. The genome of rhabdoviruses encodes at least five canonical proteins in a conserved order: nucleocapsid protein (N), phosphoprotein (P), matrix protein (M), glycoprotein (G) and large polymerase protein (L). In addition, some members have two or more accessory genes frequently located between N – P, P – M, and/or G – L gene loci [45,47]. Among these viral genes, G gives the glycosylated spikes that extend through the viral membrane to the surface of the particles [45], and a few reports showed that some glycoproteins of animal rhabdoviruses, such as VSV and viral hemorrhagic septicemia virus (VHSV), are involved in autophagy induction [19,48]. Unfortunately, the glycoproteins encoded by plant rhabdoviruses are not well understood.
Rice stripe mosaic cytorhabdovirus (RSMV) is a recently reported cytoplasmic rhabdovirus constraining rice production [49]. RSMV is transmitted by Recilia dorsalis leafhoppers in a persistent-propagative manner [50], and the infected plants exhibit distinct symptoms, including yellow stripes, mosaic patterns, and twisted leaf tips [49,51]. The anti-genome of RSMV comprises seven open reading frames, encoding the canonical N-P-M-G-L proteins and two accessory proteins [49]. At present, it is largely unknown how rice defends against RSMV infection and how RSMV counter-defends to invade successfully [52].
In this study, we elucidated a conserved mechanism that rhabdovirus-encoded glycoprotein induced host autophagy by interacting with SnRK1 and promotes its kinase activity on ATG6. Moreover, glycoprotein of rhabdoviruses can be recognized by ATG6, which serves as a bridge to link glycoproteins to key autophagosome protein ATG8 for degradation. Importantly, we showed that the hyperaccumulation of RSMV glycoprotein was toxic for plant cells, and its targeted degradation by autophagy was essential to restrict the viral titer in plants. We conclude that autophagy is not only a defense strategy to restrict RSMV infection, but may also function to limit glycoprotein accumulation for the compatible RSMV infection.
Results
RSMV infection activates autophagy
Autophagy was recently shown to play an important role in defense against plant viruses [16,18]. To address whether RSMV infection regulates host autophagy, we first examined the expression of ATG genes in RSMV-infected rice plants using quantitative real-time reverse-transcription PCR (qRT-PCR). The results revealed that nine ATG genes (OsATG1a, OsATG5, OsATG6b, OsATG7, OsATG8a, OsATG8b, OsATG9a, OsATG13a, and OsATG18c) were significantly upregulated in rice plants upon RSMV infection (Figure 1A), suggesting that RSMV infection may activate host autophagy.
Figure 1.
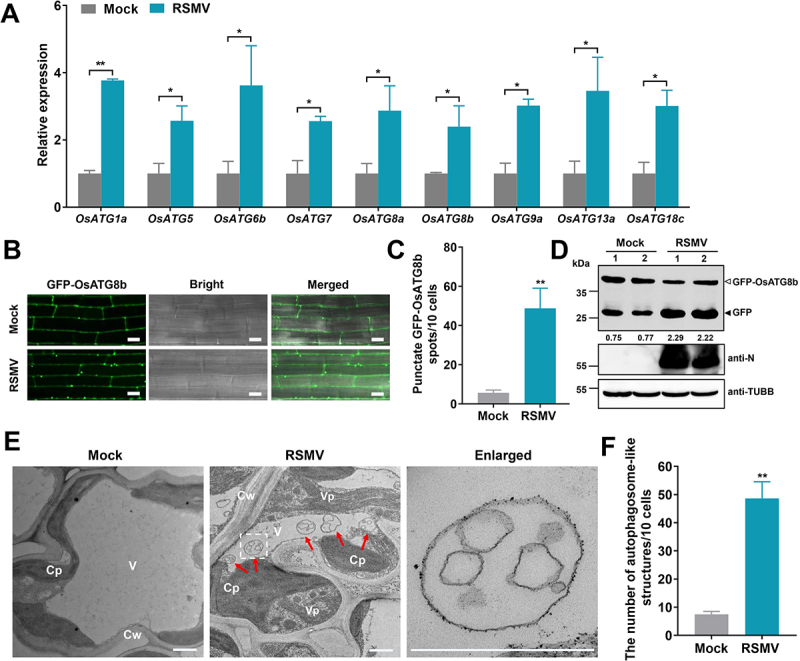
RSMV infection activates autophagy in rice plants. (A) Effects of RSMV infection on the expression of autophagy components in WT plants. Total RNAs were extracted from mock-infected or RSMV-infected rice leaves at 30 dpi. Values represent the mean relative to the mock-treated plants (n = 3 biological replicates) and were normalized with OsEf1α as an internal reference. Student’s t test was used for analyses (*P<.05). (B) relative autophagic activity revealed by GFP-OsATG8b transgenic rice plants infected by RSMV at 30 dpi. Autophagic bodies are revealed as GFP-positive puncta in the root cells. Bars: 10 μm. (C) the average number of GFP-OsATG8b spots in different samples in (B). Experiments were repeated six times and 60 cells in total were counted for the puncta in each treatment. Values represent the mean spots ± SD per 10 cells. Student’s t test was used for analyses (**P<.01). (D) accumulation of free GFP released from GFP-OsATG8b reporter upon RSMV infection. GFP-OsATG8b transgenic rice plants were RSMV-infected or mock-infected, and rice leaves were extracted at 30 dpi and then subjected to immunoblot analysis. The closed arrowhead indicates free GFP, and the open arrowhead indicates GFP-OsATG8b. The numbers below the bands represent the intensity ratio of free GFP verse GFP-OsATG8b. (E) Representative TEM images from the leaves of rice plants infected with mock or RSMV at 30 dpi. Obvious autophagic structures (red arrows) were observed in RSMV-infected samples and the corresponding region in the white box in the middle panel is magnified in the right panel. Cw cell wall, Cp chloroplast, V vacuole, Vp viroplasm. Bars: 1 μm. (F) the average number of typical double-membrane autophagosomes in different samples in (E). Experiments were repeated six times and 60 cells in total were counted for typical autophagic structures in each treatment. Values represent the mean number of autophagosomes ± SD per 10 cells. Student’s t test was used for analyses (**P<.01).
We then used monodansylcadaverine (MDC), an acidotropic dye widely used for autophagic structures detection in mammals and plants [53], as a probe to characterize autophagosomes. AZD8055, a well-known autophagy activator[54], induced significantly more autophagic structures in the root cells (Fig. S1A-B). Since RSMV could infect rice roots and accumulate virus titer to a level similar to that in leaves and stems (Fig. S1C), we monitored the MDC-positive punctated formation in the roots. The MDC-positive autophagic structures were frequently observed in the root cells of RSMV-infected rice plants but rarely in those of mock-infected plants (Fig. S1D-E).
Next, the transgenic rice plants expressing green fluorescent protein (GFP)-tagged Oryza sativa ATG8b (GFP-OsATG8b) were employed to monitor autophagy during RSMV infection. The GFP-positive punctated formation in the root cells represents pre-autophagosomal or autophagosomal structures. Compared to mock-infected plants, the number of punctate structures was increased in RSMV-infected plants by more than six folds (Figure 1B,C). We further evaluated autophagic flux that assesses autophagic transport to vacuoles by measuring the release of free GFP from the GFP-OsATG8b reporter. Free GFP signals readily accumulated when the GFP-OsATG8b transgenic plants were treated by AZD8055, indicating the effectiveness of the autophagic flux indicator (Fig. S1F). In RSMV-infected GFP-OsATG8b plants, the accumulation of free GFP was significantly higher than in mock-infected plants (Figure 1D), suggesting that RSMV infection triggers strong autophagic response in rice plants. In addition, Transmission electron microscopy (TEM) analysis demonstrated that RSMV infection induced an approximate six-fold increase of autophagosome-like double-membrane structures in the cytoplasm (Figure 1E,F). Taken together, these data indicate that RSMV infection strongly induces autophagy in rice plants.
RSMV glycoprotein induces autophagy
We next attempt to identify the viral protein responsible for host autophagy induction. Seven open reading frames (ORFs) of RSMV (Figure 2A) were expressed individually in Nicotiana benthamiana cells with GFP-tagged OsATG8c (GFP-OsATG8c). In the tobacco epidermal cells expressing RSMV glycoprotein, the number of GFP-positive puncta was significantly increased compared with that in the cells expressing other RSMV proteins or GUS protein (Figure 2B,C). Similar results were found in tobacco mesophyll cells (Fig. S2A-B). Furthermore, the vectors expressing MYC tag-fused proteins of RSMV were transfected into the protoplasts of GFP-OsATG8b rice plants to assess autophagic flux. The data showed that RSMV glycoprotein induced accumulation of free GFP signals was significantly higher than that of mock-transfected or other RSMV proteins expressing protoplasts (Figure 2D). These results suggest glycoprotein acts as an autophagic inducer during RSMV infection.
Figure 2.
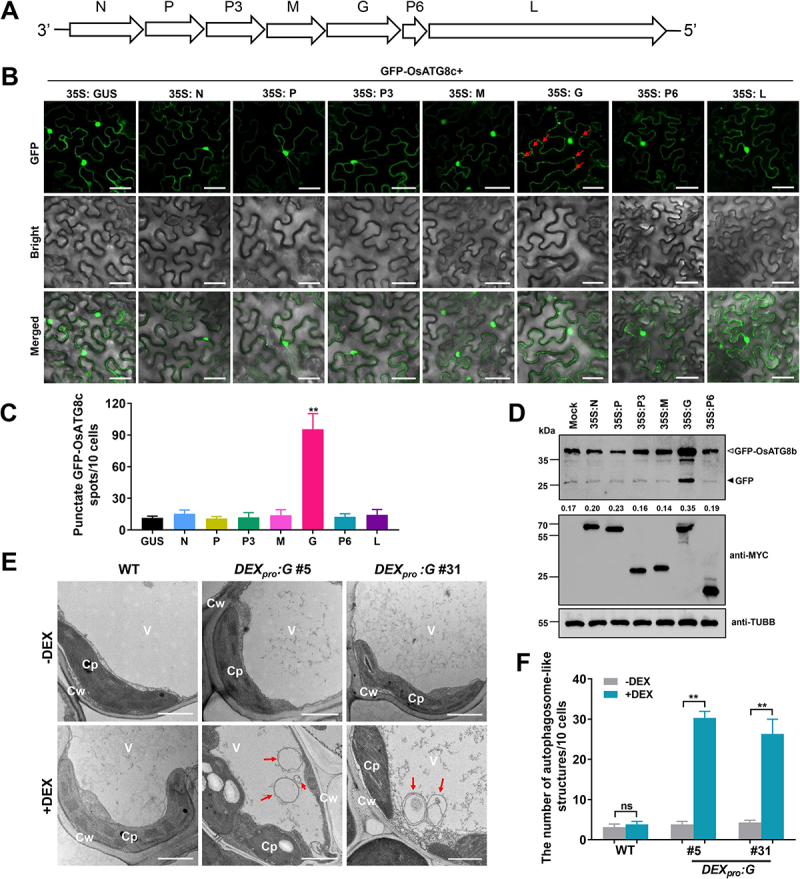
RSMV encoded glycoprotein induced autophagy in plant cells. (A) Schematic representation of the RSMV anti-genome and its encoded proteins. (B) relative autophagic activity revealed by GFP-OsATG8c in N. benthamiana leaves expressing different RSMV encoded proteins. Autophagic bodies are revealed as GFP-positive puncta in the epidermal cells and are indicated by red arrows. Each RSMV encoded proteins were driven by 35S promoter and GUS was used as a negative control. N nucleocapsid protein, P phosphoprotein, M matrix protein, G glycoprotein, L large polymerase protein. Bars: 50 μm. (C) the average number of GFP-OsATG8c spots in different samples in (B). Experiments were repeated six times and 60 cells in total were counted for the puncta in each treatment. Values represent the mean spots ± SD per 10 cells. One-way ANOVA followed by multiple-comparisons Tukey’s test was used for analyses (*P<.01). (D) accumulation of free GFP released from GFP-OsATG8b reporter upon RSMV proteins expression. Protoplasts generated from GFP-OsATG8b transgenic plants were transfected with plasmids expressing MYC tag-fused RSMV proteins, total protein was extracted after 12 h and then subjected to immunoblot analysis. The closed arrowhead indicates free GFP, and the open arrowhead indicates GFP-OsATG8b. The numbers below the bands represent the intensity ratio of free GFP verse GFP-OsATG8b. (E) Representative TEM images from the leaves of WT and DEXpro:G transgenic rice plants treated with or without DEX. Obvious autophagic structures (red arrows) were observed in the samples of DEX-treated DEXpro:G plants. Cw cell wall, Cp chloroplast, V vacuole. Bars: 1 μm. (F) the number of typical double-membrane autophagosomes in different samples in (E). Experiments were repeated six times and 60 cells in total were counted for typical autophagic structures in each treatment. Values represent the mean number of autophagosomes ± SD per 10 cells. Two-way ANOVA followed by multiple-comparisons Tukey’s test was used for analyses (**P<.01).
To further confirm that RSMV glycoprotein induces autophagy in rice plants, we attempted to generate transgenic rice plants constitutively expressing RSMV glycoprotein (35S:G). Unfortunately, no transgenic positive lines were obtained after four independent trials. We speculated that the constitutive expression of RSMV glycoprotein is lethal to rice callus (Fig. S3A). We then employed a dexamethasone (DEX)-inducible vector system [55,56] to express the glycoprotein fused with a Flag-4*MYC tag in the N-terminus (DEXpro:G). The glycoprotein can be ectopically controlled by application of DEX that does not occur in plants. After transformation, 32 T0 plants survived after antibiotic marker selection (Fig. S3B), and several lines displayed significant induction of glycoprotein transcript when their leaves were treated with 50 μM of DEX for 12 h (Fig. S3C). The DEXpro:G plants did not display growth defective phenotypes without DEX induction. However, intriguingly, DEX treatment during seedling stage resulted in rapid plant death within 12 h (Fig. S3D), while after four-leaf stage DEX treatment became unlethal, and the DEX-induced glycoprotein expression could last more than one day (Fig. S3E). Two homozygous DEXpro:G lines (#5 and #31) were generated and used for further experiments.
MDC staining showed that the formation of autophagic structures was induced by DEX application in DEXpro:G plants, but not in WT plants (Fig. S2C-D). qRT-PCR analysis confirmed the upregulation of ATG genes upon DEX induction in DEXpro:G plants, except for OsATG13a in DEXpro:G #31 (Fig. S2E). Further, TEM images revealed that DEX-induction caused greater than six-fold increase of double-membrane structures in the cytoplasm of DEXpro:G rice cells (Figure 2E,F). These results suggest that the presence of RSMV glycoprotein induces autophagy in plant cells.
Next, we employed immunoelectron microscopy to examine whether glycoprotein or RSMV particles were wrapped in autophagosomes. The gold labeled glycoprotein and RSMV particles were found to be accumulated in the double-membrane structures typical of autophagosomes (Fig. S4), suggesting that RSMV glycoprotein and the viral particles are targeted by autophagy.
Autophagy plays an antiviral role during RSMV Infection
To determine whether autophagy alters RSMV infection, we evaluated the viral symptom severity in osatg7 mutant defective in the core autophagy machinery. The WT and osatg7 plants were inoculated with RSMV at four-leaf stage, and 15 days post-inoculation (dpi) osatg7 plants showed severer phenotypes including stunting and mosaic patterns when compared to the WT plants (Figure 3A). The defective phenotype in osatg7 mutant was accompanied by the increased accumulation of RSMV RNA and proteins at 15 dpi (Figure 3B,C). Intriguingly, RSMV-infected osatg7 plants all perished before 30 dpi (Figure 3A), which may be due to the excessive accumulation of RSMV or glycoprotein.
Figure 3.
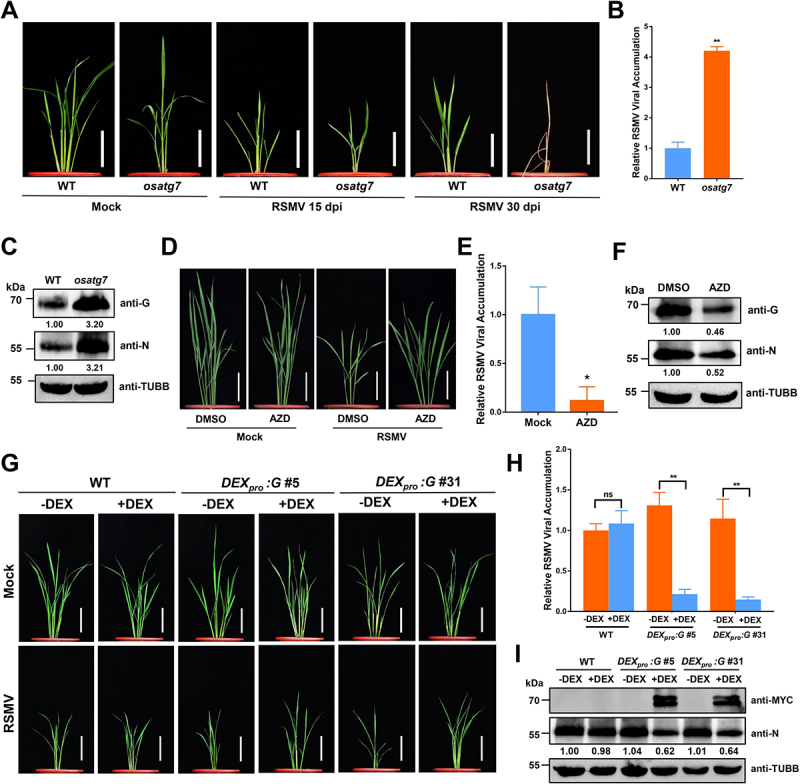
Autophagy plays an antiviral role during RSMV infection. (A) the symptoms of RSMV infection in osatg7 mutant and WT plants. The four-leaf stage plants were inoculated by viruliferous leafhopper and the photographs were taken at 15 and 30 dpi. Bars: 10 cm. (B) qRT-PCR analysis of RSMV viral RNA accumulation in osatg7 mutant and WT plants. Total RNAs were extracted from the RSMV infected leaves at 15 dpi. Values represent the mean relative to the WT plants (n = 3 biological replicates) and were normalized with OsEf1α as an internal reference. Student’s t test was used for analyses (*P<.01). (C) Western blot analysis of RSMV viral proteins accumulation in osatg7 mutant and WT plants. Total proteins were extracted from the RSMV infected leaves at 15 dpi, and were detected by specific antibodies against RSMV glycoprotein (anti-G) and nucleocapsid protein (anti-N). The relative protein band intensity was normalized against that of TUBB/tubulin. (D) the symptoms of RSMV infection in rice plants treated with DMSO or AZD. The plants were inoculated by viruliferous leafhopper and 10 μM AZD or DMSO as solvent control were treated for 3 h each by root soaking at 1, 4, 7, and 10 dpi. The photographs were taken at 30 dpi. Bars: 10 cm. (E) qRT-PCR analysis of RSMV viral RNA accumulation in rice plants treated with DMSO or AZD. Total RNAs were extracted from the RSMV infected leaves at 15 dpi. Values represent the mean relative to the WT plants (n = 3 biological replicates) and were normalized with OsEf1α as an internal reference. Student’s t test was used for analyses (**P<.05). (F) Western blot analysis of RSMV viral proteins accumulation in rice plants treated with DMSO or AZD. Total proteins were extracted from the RSMV infected leaves at 15 dpi, and were detected by specific antibodies against RSMV glycoprotein (anti-G) and nucleocapsid protein (anti-N). The relative protein band intensity was normalized against that of TUBB/tubulin. (G) the symptoms of RSMV infection in WT and DEXpro:G transgenic rice plants treated with or without DEX. The plants were inoculated by viruliferous leafhopper and then 50 μM DEX or solvent control was sprayed on the leaf surface at 1 dpi. The photographs were taken at 30 dpi. Bars: 10 cm. (H) qRT-PCR analysis of RSMV viral RNA accumulation in WT and DEXpro:G transgenic rice plants treated with or without DEX. Total RNAs were extracted from the RSMV infected leaves at 15 dpi. Values represent the mean relative to the WT plants (n = 3 biological replicates) and were normalized with OsEf1α as an internal reference. Two-way ANOVA followed by multiple-comparisons Tukey’s test was used for analyses (**P < 0.01). ns, not significant. (I) Western blot analysis of RSMV viral proteins accumulation in WT and DEXpro:G transgenic rice plants treated with or without DEX. Total proteins were extracted from the RSMV infected leaves at 15 dpi, and were detected by specific antibodies against MYC-tag (transgenic expressed glycoprotein fused with MYC tag) and RSMV nucleocapsid protein (anti-N). The relative protein band intensity was normalized against that of TUBB/tubulin.
Furthermore, AZD8055 was used to induce autophagy in rice plants. AZD8055 treatments attenuated the phenotype in RSMV-infected plants, (Figure 3D), and viral RNA and protein levels were also significantly decreased upon AZD8055 treatment at 15 dpi (Figure 3E,F). Together, these results indicate that autophagy functions as an antiviral defense response against RSMV.
Since RSMV-encoded glycoprotein induces autophagy in plant cells and activated autophagy limits viral infection, we speculate that RSMV glycoprotein may induce autophagy to fine-tune the viral load during infection. To test this hypothesis, we inoculated the DEXpro:G plants with RSMV, and the plants were treated with DEX to induce the glycoprotein overexpression. The symptoms caused by RSMV infection were ameliorated in the DEX-treated DEXpro:G plants (Figure 3G). In concordance, lower levels of viral RNAs and nucleocapsid proteins were detected in the DEX-treated DEXpro:G plants compared to controls (Figure 3H,I). Therefore, these data imply that glycoprotein-induced autophagy may function to limit the viral titer during RSMV infection and reduce the overall viral stress in host plant.
Glycoprotein of RSMV interacts with OsSnRK1B to modulate antiviral defense
To study the mechanism underlying the activation of autophagy by RSMV glycoprotein, we screened the host factors of RSMV glycoprotein through Y2H assay, and the rice OsSnRK1B protein was found to potentially interact with RSMV glycoprotein (Figure 4A), whose orthologs in yeast, metazoans, and other plants have been identified as positive regulators of autophagy57,58. The OsSnRK1B-glycorprotein interaction was further confirmed in vitro and in vivo by protein affinity-isolation assay (Figure 4B) and bimolecular fluorescence complementation (BiFC) assay (Figure 4C). In addition, the rice genome contains three SnRK1 members, designated as OsSnRK1A, OsSnRK1B, and OsSnRK1C (Fig. S5A), and the Y2H and BiFC results showed that RSMV glycoprotein only interacted with OsSnRK1B (Figure 4A and Fig. S5B). The expression of OsSnRK1B was significantly induced by RSMV infection (Fig. S5C), as well as the expression of glycoprotein in rice plants (Fig. S5D).
Figure 4.
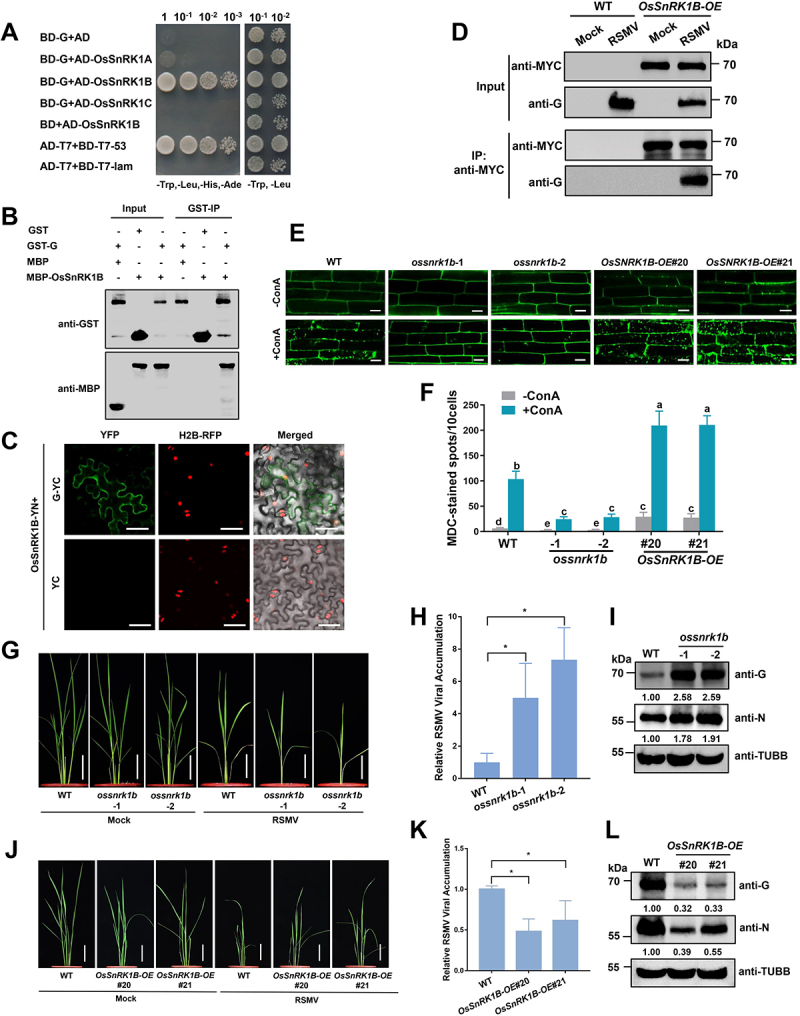
RSMV glycoprotein interacts with OsSnRK1B to modulate antiviral defense. (A) Y2H assay showing the interaction between OsSnRK1B and RSMV glycoprotein. Yeast strain Y2HGold cells co-transformed with the indicated plasmids were cultured separately on the SD-Trp-Leu-His-Ade and SD-Trp-Leu selection medium. (B) GST affinity-isolation assay showing the interaction between OsSnRK1B and RSMV glycoprotein in vitro. Purified MBP-OsSnRK1B or MBP was incubated with GST-G. After being immunoprecipitated with glutathione-Sepharose beads, the proteins were detected by protein gel blot analysis with anti-MBP or anti-GST antibodies. (C) BiFC analysis of the interaction between OsSnRK1B and RSMV glycoprotein. OsSnRK1B-YN was coexpressed with G-YC or YC in RFP-H2B transgenic N. benthamiana leaves. YFP signals were visualized by confocal microscopy. The nuclei showed red fluorescence. Bars: 50 μm. (D) co-IP analysis of the interaction between OsSnRK1B and RSMV glycoprotein. WT and OsSnRK1B-OE plants were mock or RSMV inoculated and were harvested at 15 dpi. Total proteins were immunoprecipitated with anti-MYC beads. Input and IP proteins were analyzed by protein gel blot analysis with anti-MYC and anti-G antibodies. (E) autophagic activity revealed by MDC-staining in the root cells of WT, ossnrk1b, and OsSnrk1B-OE plants pre-treated with or without concanamycin a (ConA). Bars: 10 μm. (F) the average number of MDC-stained spots in different samples in (E). Experiments were repeated six times and 60 cells in total were counted for the puncta in each treatment. Values represent the mean spots ± SD per 10 cells. Two-way ANOVA followed by multiple-comparisons Tukey’s test was used for analyses (different letters above the bars indicate significant differences between treatments at P < 0.05). (G and J) the symptoms of RSMV infection in WT and ossnrk1b (G) or OsSnrk1B-OE (J) plants. The plants were inoculated by viruliferous leafhopper and the photographs were taken at 30 dpi. Bars: 10 cm. (H and K) qRT-PCR analysis of RSMV viral RNA accumulation in WT and ossnrk1b (H) or OsSnrk1B-OE (K) plants. Total RNAs were extracted from the RSMV infected leaves at 15 dpi. Values represent the mean relative to the WT plants (n = 3 biological replicates) and were normalized with OsEf1α as an internal reference. Student’s t test was used for analyses (*P<.05). (I and L) Western blot analysis of RSMV viral proteins accumulation in WT and ossnrk1b (I) or OsSnRK1B-OE (L) plants. Total proteins were extracted from the RSMV infected leaves at 15 dpi, and were detected by specific antibodies against RSMV glycoprotein (anti-G) and nucleocapsid protein (anti-N). The relative protein band intensity was normalized against that of TUBB/tubulin.
We then generated OsSnRK1B overexpression transgenic lines (OsSnRK1B-OE) with a MYC tag fused to its C terminus (Fig. S5E). Co-immunoprecipitation (co-IP) assay showed that RSMV glycoprotein could be precipitated with OsSnRK1B-MYC upon RSMV infection (Figure 4D), confirming the presence of the OsSnRK1B-glycoprotein complex in planta.
To clarify the role of OsSnRK1B in autophagy, we generated ossnrk1b-1 and ossnrk1b-2 mutant rice lines by CRISPR-Cas9 (Fig. S5F). MDC staining was performed to monitor the basal autophagy level in ossnrk1b mutants and OsSnRK1B-OE plants. The two lines of OsSnRK1B-OE plants showed enhanced formation of autophagic structures compared to WT and ossnrk1b mutants (Figure 4E,F). The plants were then treated with concanamycin A (ConA), which facilitates the accumulation of autophagic bodies in vacuoles [59]. We observed reduced number of autophagic bodies in ossnrk1b mutants compared to WT plants, while in OsSnRK1B-OE plants there was enhanced formation of autophagic bodies (Figure 4E,F). These results suggest that OsSnRK1B positively regulates autophagy in rice plants.
Then we inoculated the WT, ossnrk1b mutant, and OsSnRK1B-OE transgenic seedlings with RSMV to investigate the role of OsSnRK1B in antiviral defense. The ossnrk1b mutants showed severer dwarf and mosaic symptoms after RSMV infection compared to WT plants (Figure 4G), while the OsSnRK1B-OE transgenic plants showed less growth inhibition (Figure 4J). These phenotypic changes were concomitant with higher levels of viral RNA and proteins in ossnrk1b mutants (Figure 4H,I), and reduced levels of viral RNA and nucleocapsid protein in the OsSnRK1B-OE plants (Figure 4K,L). We therefore conclude that OsSnRK1B is a host factor interacts with RSMV glycoprotein for antiviral defense.
Next we investigate the role of OsSnRK1B in RSMV glycoprotein induced autophagy. Unlike WT plants, we found that the expression levels of most ATG genes remained unchanged in RSMV-infected mutants compared to the mock infected mutants, except OsATG6 in ossnrk1b-2 and OsATG9a in ossnrk1b-1 which were still slightly induced (Figure 5A). Furthermore, MDC staining revealed an impaired formation of autophagic structures in ossnrk1b mutants upon RSMV infection (Figure 5B,C). These data suggest that RSMV-induced autophagy in rice plants is dependent on OsSnRK1B.
Figure 5.
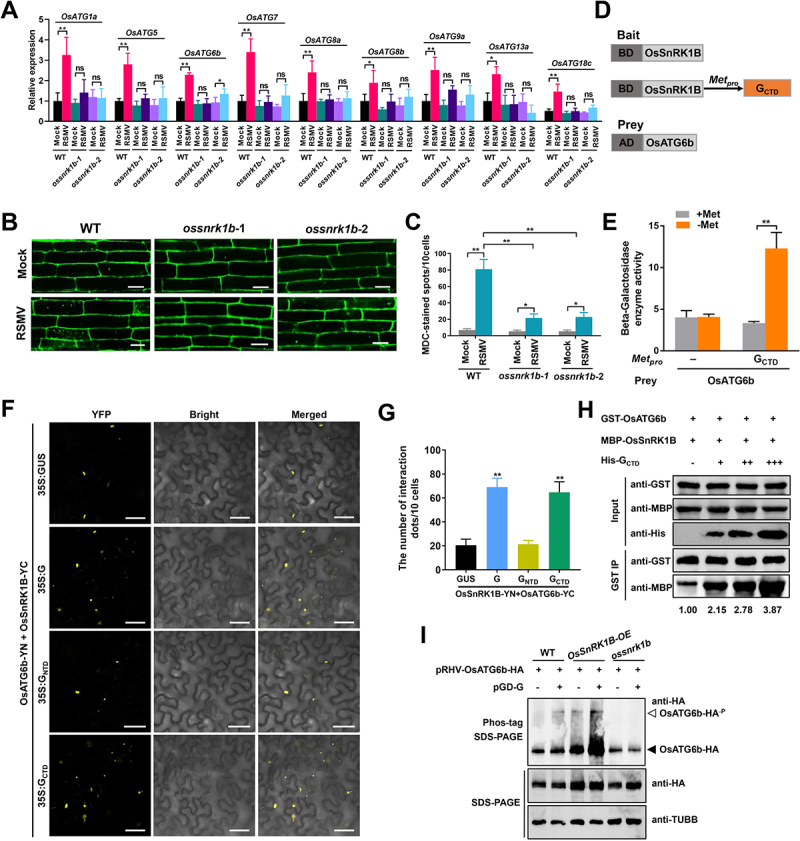
RSMV glycoprotein enhances OsSnRK1B interacting with OsATG6b. (A) Effects of RSMV infection on the expression of autophagy components in WT and ossnrk1b plants. Total RNAs were extracted from mock-infected or RSMV-infected rice leaves at 30 dpi. Values represent the mean relative to the mock-treated plants (n = 3 biological replicates) and were normalized with OsEf1α as an internal reference. Student’s t test was used for analyses (*P<.05, **P<.01). ns, not significant. (B) autophagic activity revealed by MDC-staining in the root cells of mock-infected or RSMV-infected WT and ossnrk1b plants. Bars: 10 μm. (C) the average number of MDC-stained spots in different samples in (B). Experiments were repeated six times and 60 cells in total were counted for the puncta in each treatment. Values represent the mean spots ± SD per 10 cells. Student’s t test was used for analyses (**P<.05, **P<.01). (D) Schematic representations of the bait and the prey constructs used in the yeast three-hybrid assays. OsSnRK1B was cloned into the pBridge vector and used as the bait. The CTD of RSMV glycoprotein was driven by the methionine-repressible Met25 promoter. OsATG6b was cloned into the pGADT7 vector as the prey. (E) yeast cultures containing the bait and prey constructs were grown on the SD/-Trp/-Leu medium for 3 days followed by in the liquid SD/-Trp/-Leu/-His/-Met medium with shaking. The β-Galactosidase enzyme activity was determined in the presence or the absence of RSMV glycoprotein. In the presence of 20 mM methionine, the expression of glycoprotein was inhibited. Values represent the mean activity ± SD. Student’s t test was used for analyses (**P<.05). (F) BiFC analysis of the interaction between OsSnRK1B and OsATG6b and the influence caused by full-length or truncated RSMV glycoprotein. A. tumefaciens strain EHA105 cultures carrying different combinations of constructs were infiltrated into N. benthamiana leaves, and YFP signals were visualized by confocal microscopy. Bars: 50 μm. (G) the average number of YFP spots in different samples in (F). Infiltration experiments were repeated six times and 60 cells in total were counted for the puncta. Values represent the mean spots ± SD per 10 cells. Student’s t test was used for analyses (**P<.01). (H) Dose-dependent affinity-isolation assays to assess RSMV glycoprotein influence on the OsSnRK1B-OsATG6b interaction in vitro. Equal amounts (3 ug) of purified GST-OsATG6b and MBP-OsSnRK1B proteins were mixed and incubated with increasing amounts of His-GCTD (0, 2, 4, or 8 μg). MBP-OsSnRK1B proteins were used to affinity isolate with GST-OsATG6b, and further detected by immunoblot. The relative protein band intensity MBP-OsSnRK1B was normalized against that of GST-OsATG6b. (I) RSMV glycoprotein promote the phosphorylation of OsATG6b in protoplasts. The pRHV-OsATG6b-HA plasmid was transfected with or without pGD-G plasmid into the protoplasts prepared from WT, OsSnRK1B-OE, or ossnrk1b plants for 14 h, and then the total protein was separated on Phos-tag and SDS-PAGE gels for immunoblot with anti-HA antibody. Immunoblot detection of TUBB/tubulin was used as loading controls. The closed arrowhead indicates the unphosphorylated OsATG6b-HA, and the open arrowhead indicates the phosphorylated OsATG6b-HA.
C-terminal cytoplasmic domain of RSMV glycoprotein interacts with OsSnRK1B to induce autophagy
Since OsSnRK1B plays an indispensable role in autophagy induction upon RSMV infection, we decided to map the interaction domains between RSMV glycoprotein and OsSnRK1B. OsSnRK1B has an N-terminal kinase domain (NTD), a C-terminal kinase associate domain (CTD), and an intermediate ubiquitin-associate domain (MD) (Fig. S6A). RSMV glycoprotein comprises a large N-terminal ectodomain (NTD), a short C-terminal cytoplasmic domain (CTD), and a transmembrane region (TM) in the middle (Fig. S6B). We generated different deletion mutants of OsSnRK1B and glycoprotein (i.e., NTD, MD, CTD of OsSnRK1B, and NTD, TM, CTD of RSMV glycoprotein) and analyzed their interactions through Y2H assays. The results showed that glycoprotein and OsSnRK1B interacted with each other through their CTD (Fig. S6C-D). Subcellular localization assay showed that full-length glycoprotein located in the cytoplasm while the CTD located in both cytoplasm and nucleus (Fig. S6E). Using GFP-OsATG8c as an autophagosome marker, we confirmed that RSMV glycoprotein CTD was able to induce autophagosome formation (Fig. S6F), and the induction efficiency was similar to the full-length RSMV glycoprotein (Fig. S6G). These results indicate that the 33 amino acids long CTD of RSMV glycoprotein can effectively induce autophagy through interacting with OsSnRK1B.
RSMV glycoprotein enhances the kinase activity of OsSnRK1B on OsAtg6b
SnRK1 is known as an energy-sensing protein kinase that activates autophagy by phosphorylating substrate proteins when cells are under stresses[58,60]. We screened the interacting proteins of OsSnRK1B and found that OsATG6b, but not OsAT6a and OsATG6c interacted with OsSnRK1B (Fig. S7A). The interaction was further validated by BiFC (Fig. S7B) and split luciferase assays (Fig. S7C). Subcellular localization analysis showed that OsSnRK1B was co-localized with OsATG6b, and RSMV glycoprotein also co-localized with these two host proteins (Fig. S7D).
We then examined whether RSMV glycoprotein affects the interaction between OsSnRK1B-OsATG6b using yeast three-hybrid assays. OsATG6b and OsSnRK1B were used as the prey and bait, respectively, and the CTD of RSMV glycoprotein was expressed via a methionine repressible Met25 promoter (Figure 5D). The presence of RSMV glycoprotein CTD promoted the interaction between OsSnRK1B and OsATG6b (Figure 5E). The interaction between OsSnRK1B and OsATG6b (the formation of interacting punctuate) was further confirmed by BiFC assay (Figure 5F), and the number of punctate structures were significantly increased when co-expressed with RSMV glycoprotein (Figure 5G). In addition, the CTD, but not the NTD, of RSMV glycoprotein was sufficient to enhance the interaction between OsSnRK1B and OsATG6b in BiFC assay (Figure 5F,G). Similar results were obtained by split luciferase assay, the luminescence intensity was significantly increased when co-expressed with RSMV glycoprotein (Fig. S7E-F).
Then dose-dependent affinity-isolation assays were used to assess whether glycoprotein regulated OsSnRK1B-OsATG6b interaction in vitro. The same amounts of GST-OsATG6b and MBP–OsSnRK1B were mixed with different concentration of His–GCTD in a GST affinity-isolation assay. The amounts of MBP-OsSnRK1B bound to GST-OsATG6b increased with escalating levels of His–GCTD (Figure 5H). Together, these data suggest that RSMV glycoprotein promotes the binding of OsSnRK1B with OsATG6b.
We speculated based on the Arabidopsis KIN10-ATG6 model [11] that OsATG6b is an OsSnRK1B substrate and the glycoprotein of RSMV could promote the OsATG6b phosphorylation to induce autophagy. To examine this hypothesis, we expressed OsATG6b with or without RSMV glycoprotein in protoplasts as an in vivo phosphorylation assay. An upshift in mobility during Phos-tag SDS-PAGE was observed for OsATG6b when co-expressed with RSMV glycoprotein in WT protoplasts, which was not detected when OsATG6b expressed alone. This upshift was more obvious in similarly treated protoplasts derived from OsSnRK1B-OE plants, while no such upshift in OsATG6b mobility was found in ossnrk1b protoplasts (Figure 5I), implying that RSMV glycoprotein promoted the OsSnRK1B catalyzed phosphorylation of OsATG6b.
OsAtg6b targets the glycoprotein of RSMV to restricts viral infection
ATG6 serves as a core component of the class III PtdIns3K complex and has been implicated in viral infection in many species [25,61]. Our results showed that RSMV infection and glycoprotein expression induced OsATG6b upregulation in rice plants (Figure 1A and Fig. S2E). In the screening of OsATG6b-interacting proteins during RSMV infection, we found that OsATG6b could interact with the glycoprotein of RSMV though Y2H assay (Figure 6A). The rice genome contains three OsATG6 members (OsATG6a, OsATG6b, and OsATG6c) (Fig. S8A), and RSMV glycoprotein could only interact with OsATG6b (Figure 6A), and this specific interaction was confirmed by in vivo BiFC assay (Figure 6B and Fig. S8B). Subcellular localization analysis showed that OsATG6b-YFP was distributed to a few bright puncta in the cytoplasm (Fig. S8C), consistent with the observations in Arabidopsis AtATG6/VPS30 and N. benthamiana NbBeclin1 [25,62]. CFP-fused glycoprotein (G-CFP) was co-localized with OsATG6b-YFP in the punctate structures in the cytoplasm (Fig. S8C), and their interaction was further validated by in vitro affinity-isolation assay (Figure 6C).
Figure 6.
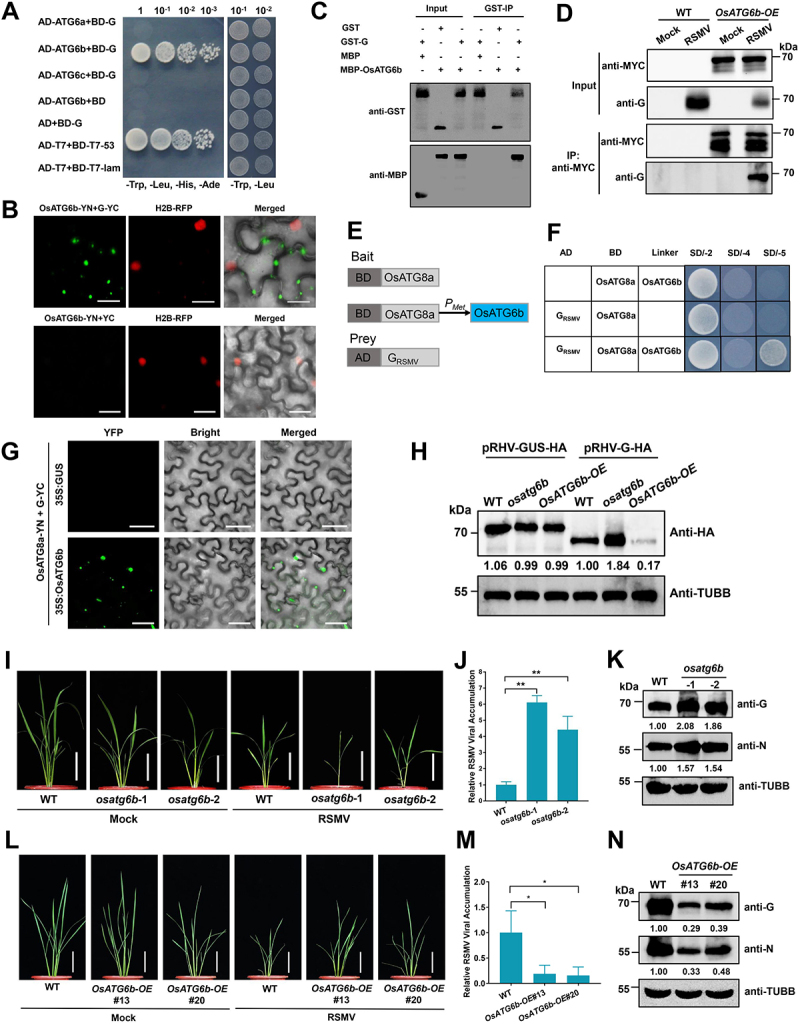
OsATG6b targets RSMV glycoprotein and restricts RSMV infection. (A) Y2H assay showing the interaction between OsATG6b and RSMV glycoprotein. Yeast strain Y2HGold cells co-transformed with the indicated plasmids were cultured separately on the SD-Trp-Leu-His-Ade and SD-Trp-Leu selection medium. (B) BiFC analysis of the interaction between OsATG6b and RSMV glycoprotein. OsATG6b-YN was coexpressed with G-YC or YC in RFP-H2B transgenic N. benthamiana leaves. YFP signals were visualized by confocal microscopy. The nuclei showed red fluorescence. Bars: 20 μm. (C) GST affinity-isolation assay showing the interaction between OsATG6b and RSMV glycoprotein in vitro. Purified MBP-OsATG6b or MBP was incubated with GST-G. After being immunoprecipitated with glutathione-Sepharose beads, the proteins were detected by immunoblot with anti-MBP or anti-GST antibodies. (D) co-IP analysis of the interaction between OsATG6b and RSMV glycoprotein. WT and OsATG6b-OE plants were mock or RSMV inoculated and were harvested at 15 dpi. Total proteins were immunoprecipitated with anti-MYC beads. Input and IP proteins were analyzed by immunoblot with anti-MYC and anti-G antibodies. (E) Schematic representations of the bait and the prey constructs used in the yeast three-hybrid assays. OsATG8a was cloned into the pBridge vector and used as the bait. The expression of OsATG6b was driven by the methionine-repressible Met25 promoter. RSMV glycoprotein was cloned into the pGADT7 vector as the prey. (F) yeast cells co-transformed with the indicated plasmids were cultured separately on the SD-Trp-Leu, SD-Trp-Leu-His-Ade, and SD-Trp-Leu-His-Ade-Met selection medium. (G) BiFC analysis of the interaction between OsATG8a and RSMV glycoprotein with the OsATG6b as a bridge. A. tumefaciens strain EHA105 cultures carrying different combinations of constructs were infiltrated into N. benthamiana leaves, and YFP signals were visualized by confocal microscopy. Bars: 50 μm. (H) transient expressed RSMV glycoprotein accumulation in the protoplasts generated from WT, osatg6b, or OsATG6b-OE. A pRHV vector expressing HA tagged glycoprotein or GUS were transfected in the protoplasts for 12 h, and total proteins were extracted for western blot analysis. The relative protein band intensity was normalized against that of TUBB/tubulin. (I and L) the symptoms of RSMV infection in WT and osatg6b (I) or OsAtg6b-OE (L) plants. The plants were inoculated by viruliferous leafhopper and the photographs were taken at 30 dpi. Bars: 10 cm. (J and M) qRT-PCR analysis of RSMV viral RNA accumulation in WT and osatg6b (J) or OsAtg6b-OE (M) plants. Total RNAs were extracted from the RSMV infected leaves at 15 dpi. Values represent the mean relative to the WT plants (n = 3 biological replicates) and were normalized with OsEf1α as an internal reference. Student’s t test was used for analyses (*P<.05). (K and N) Western blot analysis of RSMV viral proteins accumulation in WT and osatg6b (K) or OsAtg6b-OE (N) plants. Total proteins were extracted from the RSMV infected leaves at 15 dpi, and were detected by specific antibodies against RSMV glycoprotein (anti-G) and nucleocapsid protein (anti-N). The relative protein band intensity was normalized against that of TUBB/tubulin.
To further investigate the role of OsATG6b in RSMV infection, we generated OsATG6b overexpression transgenic lines (OsATG6b-OE) with a MYC tag fused to its C-terminus (Fig. S8D), and also osatg6b mutants by CRISPR-Cas9 (Fig. S8E). MDC staining showed that autophagy level was enhanced in OsATG6b-OE plants but reduced in osatg6b mutants (Fig. S8F-G). After the inoculation of RSMV on OsATG6b-OE plants, Co-IP assay showed the co-precipitation of OsATG6b-MYC and RSMV glycoprotein (Figure 6D), confirming their interaction in rice upon RSMV infection. In addition, OsATG6b has a coil-coil structure in the middle region (MD), and a NTD and a CTD (Fig. S8H). Y2H assays revealed that RSMV glycoprotein and OsATG6b interacted with each other through their NTD domains (Fig. S8I-J).
For selective autophagy, the binding of receptor proteins are required to adaptors to facilitate the docking of autophagy substrates to the autophagosomes [13]. We demonstrate that OsATG6b binds to OsATG8a, a member of the autophagy adaptor ATG8 family protein, through Y2H assay (Fig. S8K). Then we attempted to investigate whether OsATG6b serves as a scaffold to link RSMV glycoprotein to the autophagy adaptors OsATG8a, by examining the impact of OsATG6b on the interaction between RSMV glycoprotein and OsATG8a using yeast three-hybrid assays. RSMV glycoprotein and OsATG8a were used as the prey and bait, respectively, and OsATG6b was expressed via a methionine-repressible Met25 promoter (Figure 6E). The results showed that RSMV glycoprotein and OsATG8a did not interact with each other in the absence of OsATG6b, while they showed a strong interaction when OsATG6b was co-expressed (Figure 6F). These results were confirmed by in vivo BiFC assay, which demonstrated that RSMV glycoprotein and OsATG8a formed punctate structures in the cytoplasm only when OsATG6b was co-expressed (Figure 6G).
To test the direct effect of OsATG6b on the degradation of RSMV glycoprotein, we expressed RSMV glycoprotein or GUS protein as control in protoplasts generated from WT, osatg6b, or OsATG6b-OE. The results showed that GUS protein accumulated to equal amount in different protoplasts, while the glycoprotein accumulated more in osatg6b, but less in OsATG6b-OE protoplasts, comparing to the protoplasts generated from WT rice (Figure 6H). This data indicate that OsATG6b is required for glycoprotein degradation.
To investigate the role of OsATG6b in rice antiviral defense, two osatg6b mutant lines together with two lines of OsATG6b-OE (#13 and #20) were used to examine rice plant phenotype upon RSMV infection. Compared to the WT plants, osatg6b mutants showed very severe disease symptoms after RSMV infection (Figure 6I), while the OsATG6b-OE transgenic plants showed milder disease symptoms (Figure 6L). These phenotypic responses were consistent with the increased levels of RSMV RNA and nucleocapsid protein in the osatg6b mutants (Figure 6J,K), and the reduced levels in OsATG6b-OE plants (Figure 6M,N). Together, our data indicate that OsATG6b is required for glycoprotein degradation by autophagy and the restriction of RSMV infection.
Rhabdovirus glycoproteins induce autophagy as a conserved antiviral defense
To investigate whether other rhabdovirus-encoded glycoproteins induce autophagy through interacting with SnRK1 proteins, we cloned the glycoproteins from three other rhabdoviruses, barley yellow striate mosaic cytorhabdovirus (BYSMV), tomato yellow mottle-associated cytorhabdovirus (TYMaV), and sonchus yellow net nucleorhabdovirus (SYNV), identified as GBYSMV, GTYMaV and GSYNV, respectively (Fig. S9A). The canonical glycoproteins have a signal peptide, a large ectodomain, a transmembrane domain, and a cytoplasmic domain. GBYSMV does not have the transmembrane domain and cytoplasmic domain, and GTYMaV does not have the signal peptide but possesses two transmembrane domains (Fig. S9B).
We first determined the interaction between these glycoproteins and NbSnRK1, the OsSnRK1 ortholog of their host N. benthamiana. The Y2H assay results showed that GRSMV, GTYMaV, and GSYNV could interact with NbSnRK1, but GBYSMV failed to show interaction (Figure 7A). These results were further confirmed by BiFC assay (Figure 7B). Thus, we next investigated whether these glycoproteins can induce autophagy by using YFP-NbATG8a as an autophagosome marker in N. benthamiana leaves. Except for GBYSMV, all other glycoproteins were able to dramatically induce autophagy in tobacco epidermal cells (Figure 7C,D). Similar results were also found in tobacco mesophyll cells (Fig. S9C-D).
Figure 7.
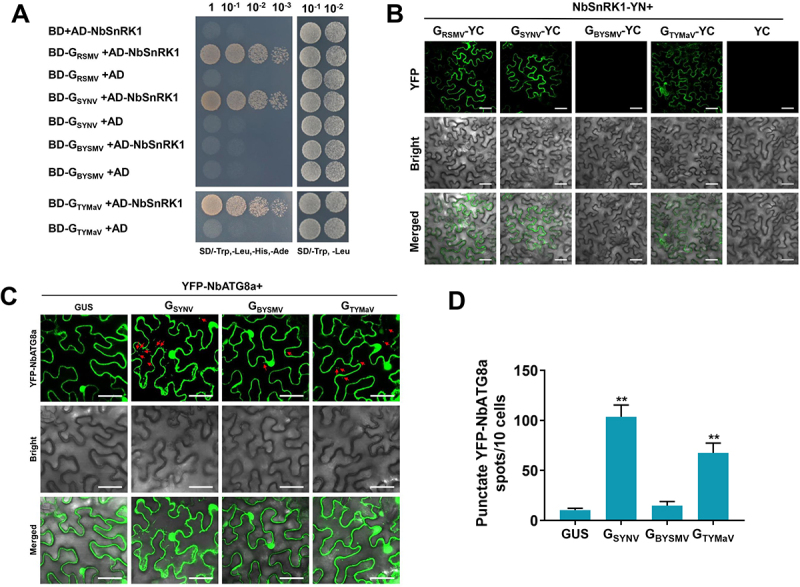
Rhabdoviruses induce host autophagy in a conserved way. (A) Y2H assay showing the interaction between NbSnRK1 and various glycoproteins. Yeast strain Y2HGold cells co-transformed with the indicated plasmids were cultured separately on the SD-Trp-Leu-His-Ade and SD-Trp-Leu selection medium. (B) BiFC analysis of the interaction between NbSnRK1 and various glycoproteins. NbSnRK1B-YN was coexpressed with glycoproteins-YC or YC in N. benthamiana leaves. YFP signals were visualized by confocal microscopy. Bars: 50 μm. (C) relative autophagic activity revealed by YFP-NbATG8a in N. benthamiana leaves expressing glycoproteins from different rhabdoviruses. Autophagic bodies are revealed as YFP-positive puncta in the epidermal cells. GUS was used as a negative control. Bars: 50 μm. (D) the average number of YFP-NbATG8a spots in different samples in (C). Experiments were repeated six times and 60 cells in total were counted for the puncta in each treatment. Values represent the mean spots ± SD per 10 cells. One-way ANOVA followed by multiple-comparisons Tukey’s test was used for analyses (*P<.01).
We further investigated whether these glycoproteins can be targeted by the ATG6 orthologs of N. benthamiana, NbBeclin1. Y2H assay and BiFC assay showed that all these glycoproteins interacted with NbBeclin1 (Figure 8A,B).
Figure 8.
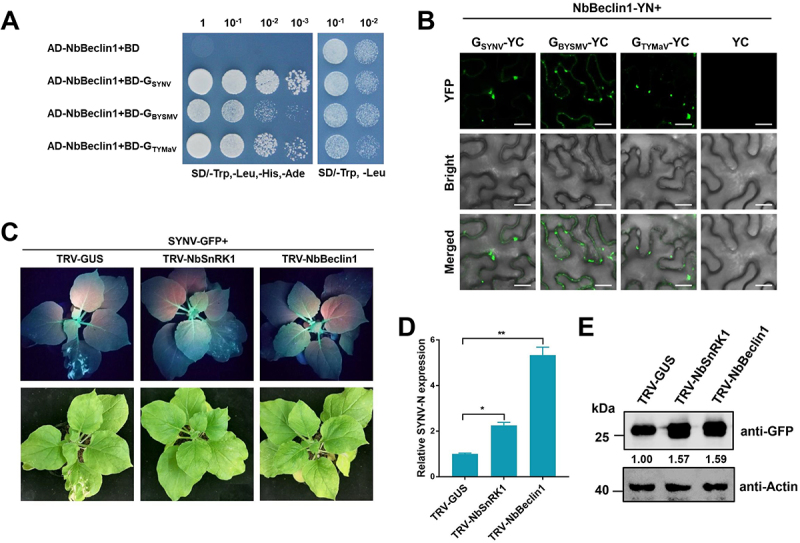
Rhabdoviruses are restricted by host autophagy in a conserved way. (A) Y2H assay showing the interaction between NbBeclin1 and various glycoproteins. Yeast strain Y2HGold cells co-transformed with the indicated plasmids were cultured separately on the SD-Trp-Leu-His-Ade and SD-Trp-Leu selection medium. (B) BiFC analysis of the interaction between NbBeclin1 and various glycoproteins. NbBeclin1-YN was coexpressed with glycoproteins-YC or YC in N. benthamiana leaves. YFP signals were visualized by confocal microscopy. Bars: 20 μm. (C) silencing of NbSnRK1 or NbBeclin1 promotes SYNV infection in N. benthamiana. GFP fluorescence and viral symptoms in plants pre-inoculated with TRV1 together with TRV2-GUS (control), TRV2-NbSnRK1, or TRV2-NbBeclin1 for 7 days and then infected by SYNV-GFP. Plants were photographed under UV light and regular light at 10 dpi. (D) qRT-PCR analysis of SYNV viral RNA accumulation in GUS-, NbSnRK1-, and NbBeclin1-silenced plants. Total RNAs were extracted from the SYNV-GFP infected leaves at 10 dpi. Values represent the mean relative to the GUS-silenced plants (n = 3 biological replicates) and were normalized with NbPP2A as an internal reference. Student’s t test was used for analyses (*P<.05, **P<.01). (E) Western blot analysis of the GFP protein accumulation in GUS-, NbSnRK1-, and NbBeclin1-silenced plants. Total proteins were extracted from the RSMV-GFP infected leaves at 15 dpi, and were detected by anti-GFP antibody. The relative protein band intensity was normalized against that of Actin.
To evaluate the role of autophagy in the infections of other rhabdovirus, the TRV VIGS vector was used to silence NbSnRK1 and NbBeclin1 (Fig. S9E-F), and then the silenced plants were challenged with SYNV-GFP [63]. Compared to the GUS-silenced plants, the silencing of NbSnRK1 and NbBeclin1 caused enhanced susceptibility to SYNV with more severe symptoms and stronger GFP signal (Figure 8C). Consistently, the viral RNA and protein levels of SYNV-GFP were significantly increased in the NbSnRK1- and NbBeclin1-silenced plants (Figure 8D,E). Therefore, we concluded that different rhabdovirus glycoproteins can induce autophagy as a conserved antiviral defense in plants.
Discussion
Viruses represent the simplest organisms composed of nucleic acids and proteins. To combat viral infection, plants have evolved several defense mechanisms targeting viral nucleic acid and viral proteins. Virus infection induces host RNA silencing to restrict the replication of viral nucleic acids [64], and the activation of autophagy can selectively degrade viral proteins as a defense line of host immunity [16,18]. Emerging evidence has demonstrated that plant cargo receptors can selectively interact with viral proteins to promote their autophagic degradation. For DNA viruses, Arabidopsis NBR1 was reported to target the degradation of capsid protein of cauliflower mosaic virus (CaMV) and restrict the establishment of viral infection [24]. ATG8 homologs from tobacco and Arabidopsis were reported to mediate the degradation of virulence factor βC1 or replication initiator protein C1 of several geminiviruses [26,29]. For RNA viruses, N. benthamiana Beclin1 (ATG6 homolog) was shown to target RNA-dependent RNA polymerase (RdRp) of several positive-sense RNA viruses [25]. Very recently, a novel tobacco and rice autophagosomal cargo receptor identified as a regulator of RNA-silencing suppressor protein p3 in rice stripe virus (RSV), by which the plants antagonize the infection of RSV31. In this study, we showed that RSMV-encoded glycoprotein is targeted by a rice ATG6 protein and delivered to autophagosomes for degradation (Figure 9). ATG6/Beclin1 is a core component of the class III PtdIns3K complex, and ATG6 has diversified to three homologs in rice plants, suggesting that OsATG6s has flexible functions and diverse protein partners (i.e., rice has seven OsATG8 homologs). Here, we showed that OsATG6b, but not OsATG6a or OsATG6c, specifically interacts with RSMV glycoprotein, suggesting that the expansion of ATG6s may have contribute to the diversification of selective autophagy pathways in rice plants. Since rhabdovirus encoded glycoprotein is essential for virion assembly and host cell entry [45], its autophagic degradation will inevitably restrict the viral infection and assembly.
Figure 9.
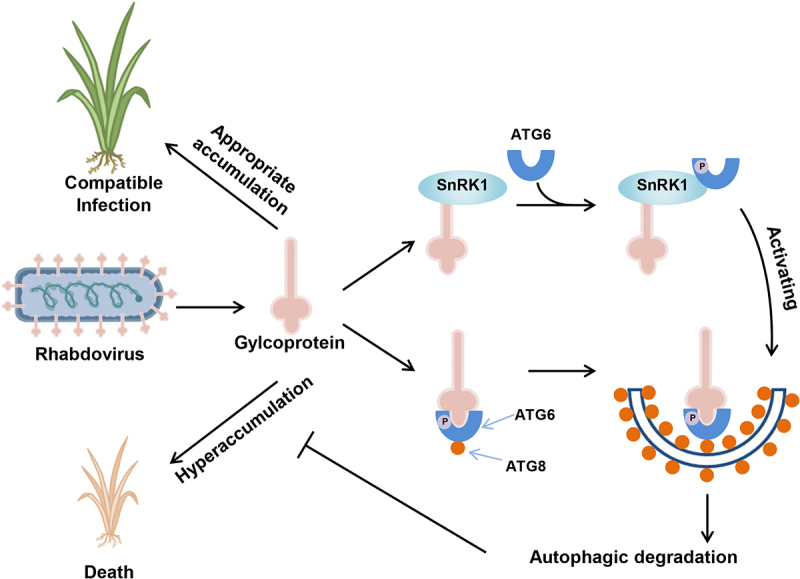
A proposed model illustrates how rhabdovirus encoded glycoprotein induces and harnesses host antiviral autophagy for maintaining its compatible infection. RSMV-encoded glycoprotein triggers host autophagy by interacting with OsSnRK1B and promotes its kinase activity on ATG6, and the glycoprotein can be recognized by ATG6, which serves as a bridge to link glycoproteins to key autophagosome protein ATG8 for degradation. The induction of autophagy functions as an antiviral mechanism against RSMV in rice plants. Moreover, the hyperaccumulation of RSMV glycoprotein is toxic for plant cells, so its targeted degradation by autophagy was essential to restrict the viral titer in plants.
On the other hand, plant viruses can also hijack autophagy to degrade host defense components for their own benefits, particularly for the viral suppressor of RNA silencing (VSR). For example, polerovirus-encoded P0 was reported to mediate autophagic degradation of AGO1 [37], and turnip mosaic virus (TuMV)-encoded VPg and tomato yellow leaf curl China virus (TYLCCNV)-encoded βC1 were found to mediate autophagic degradation of SGS3 [36,38]. These VSRs attack key components of host RNAi-based antiviral defense machinery via autophagy to establish the viral infection. In addition, RSV-encoded movement protein was reported to induce autophagic degradation of host remorin protein and overcome remorin-mediated inhibition of viral movement [65]. Very interestingly, we found that the hyperaccumulation of RSMV glycoprotein is toxic to rice plants (Fig. S3D), and the autophagy deficient mutant osatg7 were all perished after RSMV infection (Figure 3A). Although the underlying mechanism needs further analysis, existing evidence suggests that glycoprotein-induced autophagy seems to maintain appropriate accumulation level of glycoprotein to mitigate the viral toxicity to the plants, which balances the host survival and the proliferation of infected virus (Figure 9).
As described above, many viruses have been shown to regulate host autophagy. However, few viral effectors have been characterized regarding how they trigger or suppress autophagy. Apart from being the target of host autophagy, cotton leaf curl Multan virus (CLCuMuV)-encoded βC1 was found to activate autophagy by disrupting GAPCs – ATG3 interactions through its binding to GAPCs [27]. Moreover, barley stripe mosaic virus (BSMV)-encoded γb and tomato leaf curl Yunnan virus (TLCYnV)-encoded C2 subverts autophagy by disrupting the ATG7-ATG8 interaction through its binding to ATG7 [28,34]. In our study, we showed that RSMV-encoded glycoprotein and other rhabdovirus-encoded glycoproteins can interact with host SnRK1, a positive regulator of autophagy [57,58]. SnRK1 is a highly conserved energy sensor and is activated under energy deprivation, abiotic stresses, or pathogen infection [66]. In Arabidopsis, a subunit of SnRK1, named KIN10, was reported as a positive regulator of plant autophagy and showed that it acts by affecting the phosphorylation of ATG1 [67]. In addition, it was reported that the PtdIns3K complex is also activated via the phosphorylation of the PtdIns3K subunit ATG6 by the active KIN10, which bypasses the requirement of the ATG1 kinase complex for autophagy initiation [11]. We further demonstrated that RSMV glycoprotein promotes the kinase activity of OsSnRK1B on OsATG6b to trigger autophagy (Figure 9).
In summary, our study unveils a critical role of autophagy-induction by rice rhabdovirus-encoded glycoprotein, which seems to be conserved in other rhabdovirus-encoded glycoproteins. RSMV glycoprotein triggers autophagy by promoting SnRK1-dependent autophagy initiation, and the interaction with ATG6 mediates its autophagic degradation. The induction of autophagy functions as an antiviral mechanism against RSMV in rice plants. On the other hand, since the constitutive expression of RSMV glycoprotein is lethal, its degradation by autophagy reduces the toxicity to host plant and allows the viral replication at a suitable level (Figure 9). Thus, autophagy induction by RSMV glycoprotein restricts the viral infection to balance the host survival and viral propagation. These findings provide the insights into antiviral crop breeding based on autophagy regulation and the balance of viral glycoprotein accumulation.
Materials and methods
Plant materials and growth conditions
Rice (Oryza sativa subsp. japonica) cv. Zhonghua 11 (ZH11) was used in this study, include all the mutants and transgenic lines. OsSnRK1B-OE, OsATG6b-OE and DEXpro:G transgenic plants were generated in this study (as described below). The ossnrk1b and osatg6b mutant lines were generated using the CRISPR-Cas9 technology at the Biogle Genome Editing Center (Jiangsu, China). All the transgenic and mutant rice lines were generated from the ZH11 background. Rice plants were grown inside a greenhouse at 28–32°C and 60 ± 5% relative humidity with a 14-h-light/10-h-dark photoperiod. N. benthamiana plants were grown in growth chambers maintained at 25°C with a 16-h-light/8-h-dark dark photoperiod.
Virus, insect, and virus inoculation
RSMV was maintained in individual rice plant grown inside an insect-proof greenhouse. Leafhoppers (Recilia dorsalis) were reared and propagated on the RSMV-infected rice plants till viruliferous. Viruliferous leafhoppers were used to inoculate RSMV to rice seedlings. The presence of RSMV in RSMV-infected rice plants or viruliferous leafhoppers was confirmed by RT-PCR or RT-RAA-CRISPR-Cas12a visual detection as previously described [49,68].
SYNV-GFP infected tobacco leaves were a gift from Prof. Zhenghe Li (Zhejiang University) [63]. 0.5 g leaves infected with virus were ground in a chilled mortar containing 1 mL of freshly prepared inoculation buffer (0.5% sodium sulfite and 2% Celite [Sangon Biotech, A600547]) at 4°C. Two leaves of N. benthamiana plants were gently rubbed by hand with the leaf extracts. The mechanically inoculated plants were confirmed by RT-PCR or GFP expression under a UV lamp.
Plasmid construction
Primers used in this study are listed in Table S1. All the genes from rice and tobacco were amplified using the cDNA derived from corresponding plants as template. The coding sequences of each gene from RSMV were amplified using the cDNA of RSMV-infected rice plants as template. The coding sequence of glycoproteins from SYNV, TYMaV, and BYSMV were cloned from the plasmids containing corresponding fragment which were gifts from other labs [63,69].
For subcellular localization and BiFC assays, the corresponding genes were cloned individually into the pENTR/D-TOPO vector (ThermoFisher Scientific, K240020), and transferred into destination vectors using a Gateway LR reaction kit as instructed (ThermoFisher Scientific, 11791020).
For stable transgenic line generation, the coding sequences of OsSnRK1B and OsATG6b were cloned and inserted into the BamHI and NotI sites of the pRHVcMyc vector [70] using the In-Fusion cloning kit as instructed (Vazyme Biotech, C112–02). The coding sequence of each gene from RSMV was cloned into the pENTR/D-TOPO vector (ThermoFisher Scientific, K240020) and then transferred into the pBA-Flag-Myc4 vector [71] using a Gateway LR reaction kit as instructed (ThermoFisher Scientific, 11791020).
For transient expression in protoplasts, the coding sequence of OsATG6b was cloned and inserted into the BamHI and NotI sites of the pRHVcHA vector70 to obtain pRHV-OsATG6b-HA. The coding sequence of RSMV glycoprotein was cloned and inserted into the BamHI and SalI sites of the pGD vector [72] to obtain pGD-G. The cloning used the In-Fusion cloning kit (Vazyme Biotech, C112–02).
For inducible transgenic line generation, RSMV glycoprotein coding sequence was fused with a Flag-4*MYC tag on its N terminus and inserted into the XhoI and SpeI sites of the glucocorticoid-inducible vector [55,56] using the In-Fusion cloning kit (Vazyme Biotech, C112–02).
For the yeast two-hybrid (Y2H) assay, the glycoprotein coding sequences from different viruses were amplified and inserted into the pGBKT7-BD bait vector (Clontech, 630443). The autophagy-related protein coding sequences were amplified and cloned individually into the pGADT7-AD prey vector (Clontech, 630442) using the In-Fusion cloning kit (Vazyme Biotech, C112–02). For yeast three-hybrid (Y3H) assay, the coding sequence of OsSnRK1B or OsATG8a was cloned into the pBridge vector (Clontech, 630404) to express a DNA binding domain fusion protein. The coding sequence of RSMV glycoprotein or OsATG6b was amplified and cloned into the pBridge vector with a Met25 promoter.
For the in vitro affinity-isolation assays, the RSMV glycoprotein coding sequence was cloned into the pGEX4T1 vector, the OsSnRK1B and OsATG6b coding sequences were cloned into the pMBP28 vector [73], and the CTD of RSMV glycoprotein coding sequence was cloned into the pET28a vector (Novagene, 69864) using the In-Fusion cloning kit (Vazyme Biotech, C112–02).
For the VIGS assay, a partial fragment of NbSnRK1 was cloned into the pTRV2 vector [74] using the In-Fusion cloning kit (Vazyme Biotech, C112–02). The pTRV1, pTRV2-GUS, and pTRV2-Beclin1 were obtained from Prof. Fangfang Li (Institute of Plant Protection, Chinese Academy of Agricultural Sciences) [25].
RNA isolation and quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted from plant samples using the Total RNA Extraction Reagent (Vazyme Biotech, R401–01). For comparing the virus titer in roots, leaves, and stems of rice plants, semi-quantitative RT-PCR analysis was performed. Total RNAs extracted from different tissues of virus infected rice plants were adjusted to equal concentration and detected for RSMV by using HiScript II One Step RT-PCR Kit (Vazyme Biotech, P612–01) for 25 cycles, and OsEF1α was used as an internal control. For quantification of the virus titer or gene expression level in different samples, cDNA was synthesized using the isolated total RNA, oligo(dT), and a reverse transcriptase (Takara, RR037A). Quantitative PCR reactions were carried out on a CFX96 TouchTM real-time PCR detection system (Bio-Rad, Hercules, CA, USA) using the SYBR Premix Ex TaqTM II kit (Takara, DRR820A). Three independent biological replicates were performed, each in technical triplicates. The expression level of the OsEF1α in rice and NbPP2A in tobacco was used as an internal control, and the relative expression levels were calculated by the 2−ΔΔC(t) method [75]. Primers used for qRT-PCR are listed in Table S1.
Western blot and protein quantification analysis
Proteins were extracted from plant samples using RIPA Lysis buffer (CWBIO, CW0885M) containing Complete Mini protease inhibitor cocktail (Roche, 05056489001) and then separated in SDS-PAGE gels through electrophoresis. The PageRuler Prestained Protein Ladder (ThermoFisher Scientific, 26617) was used to show protein size. The separated protein bands were transferred to PVDF membranes (Millipore, IPVH00010), and detected using specific antibodies. The detection signal was then visualized using the Immobilon Western Chemiluminescent HRP Substrate as instructed (Millipore, WBKLS0500). Coomassie brilliant blue staining of Rubisco served as the loading control. The monoclonal antibody against nucleocapsid protein of RSMV was kindly provided by Prof. Jianxiang Wu (Zhejiang University, China), and the polyclonal antibody against RSMV glycoprotein (purified the 20–482 amino acids of the protein fused with His-tag) was produced by Abmart (Shanghai, China). The anti-GST (M20007S), anti-His (M30111S), anti-MYC (M20002S), anti-HA (M20003S), anti-Actin (M20009S), and anti-TUBB/tubulin (M30109S) antibodies were purchased from Abmart, and the anti-MBP (E8032S) antibody was purchased from New England Biolabs. For protein band quantification analyses, the signal intensity was quantified using the ImageJ software (http://rsb.info.nih.gov/ij/). The band intensity of a given protein was normalized against that of TUBB/tubulin in rice plants and Actin in tobacco plants.
Chemical treatment
For autophagosome observation, the rice roots were stained with 50 μM monodansylcadaverine (MDC; Sigma-Aldrich, 30432) in phosphate-buffered saline (PBS; Sangon Biotech, B540626), pH 7.4 for 15 min, followed by 3 washes with PBS. Roots were observed using a Zeiss LSM 780 confocal laser microscope with the excitation and emission wavelengths 405 nm and 430–480 nm, respectively.
For autophagy activation treatment, 20-day-old rice seedlings were first inoculated by RSMV-carrying leafhoppers and grown in nutrient solutions. At 1, 4, 7, and 10 dpi, the inoculated seedlings were transferred to nutrient solutions supplemented with 10 μM AZD8055 (Selleckchem, S1555) or DMSO as solvent control for 3 h and then transferred back to normal growth conditions. At 15 dpi, the accumulation of RSMV in the seedlings was examined by qRT-PCR and Western blot.
To visualize autophagic bodies, the roots were cut from rice plants and washed with sterile water, and then soaked in 10 mM MES-NaOH (pH 5.5) solutions, in the presence or the absence of 1 μM concanamycin A (Sigma-Aldrich, C9705), and incubated at room temperature for 8 h in darkness.
To induce the expression of DEXpro:G transgenic plants, a solution containing 50 μM dexamethasone (Sigma-Aldrich, D4902) in 0.01% Tween-20 was sprayed on the rice leaves. The treated plants were used for RSMV inoculation or TEM observation.
Yeast two-hybrid and three-hybrid assays
For the yeast two-hybrid (Y2H) assay, different combinations of pGBK and pGAD plasmids were transformed into yeast strain Y2HGold cells (Clontech, 630498). The transformants were cultivated on the SD/-Leu/-Trp (SD-L-T) medium and then on the SD/-Leu/-Trp-His-Ade (SD-L-T-H-A) selection medium to determine the protein-protein interaction. Yeast cells were photographed at 3 days post incubation at 30°C. All the experiments were repeated three times with similar results.
For the yeast three-hybrid (Y3H) assay, the pBridge and pGAD plasmids were co-transformed into yeast strain AH109 cells. The transformants were cultivated on the SD-L-T medium and then overnight in the liquid SD-L-T medium till the liquid became turbid. The yeast cultures were individually diluted (1:100; v:v) in the liquid SD/-Leu/-Trp-His-Met (SD-L-T-H-M) medium with or without 20 mM Met. Two days after incubation with shaking at 30°C, the β-Galactosidase enzymatic activity of each sample was determined as instructed (Solarbio Life Sciences, BC2585). Dilutions of suspended yeast were plated on SD-L-T-H-A and SD/-Leu/-Trp-His-Ade-Met (SD-L-T-H-A-M) solid media. Yeast cells were observed 3 days after incubation at 30°C. All the experiments were repeated at least three times with similar results.
In vitro affinity-isolation assay
The GST- and MBP-tagged proteins were expressed in E. coli and purified using glutathione Sepharose 4B beads (GE Healthcare, 17075601) and amylose resin (New England Biolabs, E8021S) as instructed by the manufacturer, respectively. A total amount of 3 μg purified GST-G was mixed with 6 μg purified MBP-OsSnRK1B, MBP-OsATG6b or MBP, and then incubated with 20 μL glutathione Sepharose 4B beads for 2 h at 4°C in the binding buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.25% Triton X-100 [Sigma-Aldrich, T8787]). After six washes with the binding buffer, the resin-bound proteins were released by 5 min boiling and the protein samples were detected by immunoblot assay using anti-GST and anti-MBP antibody.
For investigation of the interactions among three proteins, 3 μg purified GST-OsATG6b was mixed with 3 μg purified MBP-OsSnRK1B plus 0, 2, 4 or 8 μg purified His-GCTD, and then incubated with 20 μL glutathione sepharose 4B beads for 2 h at 4°C in 1× PBS. After six washes with 1× PBS, the resin-bound proteins were released by 5 min boiling and detected by immunoblot assay using anti-GST, anti-MBP and anti-His antibody.
Co-immunoprecipitation (co-IP) assay
Immunoprecipitation was performed using protein extracts from OsSnRK1B-OE, OsATG6b-OE, or WT plants with RSMV or mock infection. At 30 dpi, the rice leaves were ground in liquid nitrogen and the protein samples were extracted using IP buffer (40 mM Tris-HCl, pH 6.8, 150 mM NaCl, 5 mM MgCl2, 2 mM EDTA, 5 mM DTT, 2% glycerol, 0.1% Triton X-100, and protease inhibitor cocktail) from 5 g samples. After incubation at 4°C for 10 min, the mixtures were centrifuged at 1200 g for 10 min, and the supernatant was then incubated with 3 μL anti-MYC antibody (Abmart) for 2 h at 4°C, followed by incubation with 20 μL Protein A+G Agarose (Beyotime Technology, P2055). The immunoprecipitates were then washed four times with 1×PBS and resuspended in 20 μL 2×SDS-PAGE loading buffer (500 mM Tris-HCl, pH = 6.8, 50% glycerol, 10% SDS, 2% β-mercaptoethanol, and 1% bromophenol blue). Subsequently, the protein samples were boiled at 95°C for 10 min and separated on a 10% SDS-PAGE gel for western blotting analysis.
BiFC assay
Mixed A. tumefaciens strain EHA105 cultures carrying different combinations of expression vectors were individually infiltrated into N. benthamiana leaves. After 48–72 h, YFP fluorescence in the infiltrated leaf tissues was examined under a confocal laser microscopy (Zeiss LSM 780, Germany). The excitation wavelength of YFP was set at 514 nm and the emission wavelength was set at 520 to 580 nm.
Subcellular localization assay
To investigate the subcellular localization patterns of target proteins, plasmids were transiently expressed in N. benthamiana leaves via agro-infiltration. RFP-H2B expressed in the transgenic N. benthamiana leaves was used as a nuclear marker. Images were captured under a Zeiss LSM 780 confocal laser microscope. The excitation and emission wavelengths were set at 405 nm and 454–504 nm for CFP, 488 and 500–530 nm for GFP, and 543 and 588–641 nm for RFP, respectively.
The phosphorylation assay in protoplasts
Rice protoplasts were isolated from 10-day-old rice seedlings and transfected as described previously [76]. The pRHV-OsATG6b-HA plasmid with or without pGD-G plasmid was transiently expressed in protoplasts prepared from WT, OsSnRK1B-OE, or ossnrk1b plants for 14 h and then the total protein was extracted with RIPA Lysis Buffer (CWBIO, CW0885M) containing Halt Protease and Phosphatase Inhibitor Cocktail (ThermoFisher Scientific, 78442). The samples were separated on a 10–12% polyacrylamide gel containing 50 μM Phos-tag acrylamide (Wako Pure Chemicals, 198–17981). Immunoblot analysis was performed according to the instructions (Wako Pure Chemicals) and detected with the anti-HA antibody (Abmart, M20003S). Immunoblot analysis were also performed after protein separation on SDS-PAGE with anti-HA and anti-TUBB/tubulin.
Transmission electron microscope (TEM) observation
RSMV-infected and mock-infected rice plants, or DEX-treated or non-treated DEXpro:G and WT rice leaves were collected and cut into small pieces (3 mm × 1 mm). The sample tissues were fixed in 4% glutaraldehyde and 2% paraformaldehyde (in 100 mM phosphate buffer, pH 7.0). The samples were then post-fixed in OsO4, dehydrated in ethanol, and then embedded in Spurr resin(GE Healthcare, 14300) as instructed by the manufacturer (SPI-EM, Division of Structure Probe, Inc.). Ultrathin sections (~80 nm) were cut from the embedded tissues using a Leica UC7 Ultramicrotome and were collected on 3-mm copper slot grids, and then stained with uranyl acetate and lead citrate solution. The stained sections were examined and imaged under a Thermo Fisher Talos L120C transmission electron microscope set at 120 kV accelerating voltage.
Immuno-electron microscope observation for RSMV glycoprotein was performed as previously described [77]. The RSMV-infected and mock-infected rice plants were cut into small pieces, fixed with 0.1% glutaraldyhyde and 4% paraformaldehyde (in 100 mM phosphate buffer, pH 7.0) for 2 h and stored in 4% paraformaldehyde at 4°C overnight, and post-fixed in 0.5% OsO4 at 4°C for 1 h. After dehydration in serially diluted ethanol solutions, the fixed rice tissues were embedded in LR White Resin (GE Healthcare, 14381-UC). Ultra-thin sections (~80 nm) were cut from the embedded stem tissues using a Leica UC7 Ultramicrotomeand and the sections were mounted on formal supported nickel single slot grids. Sections were incubated in 100 mM phosphate buffer for 15 min, and then in a blocking buffer (1% BSA [Sigma-Aldrich, V900933] in 100 mM phosphate buffer, pH 7.0) for 15 min. The sections were then incubated in RSMV glycoprotein specific polyclonal antibody solution (1:100, v:v) for 1 h at room temperature, followed by incubation with 10 nm gold-conjugated goat-anti-rabbit IgG secondary antibody (Sigma-Aldrich, G7402) solution (1:100, v:v) for 1 h. The staining was performed with uranyl acetate and lead citrate, and the images were taken under the same transmission electron microscope.
Statistical analyses
Differences were analyzed using a one-way or two-way ANOVA followed by multiple-comparisons Tukey’s test. A p-value ≤0.05 was considered statistically significant. All analyses were performed using SPSS version 20 (SPSS, Inc. Chicago, Illinois, USA).
Supplementary Material
Acknowledgements
We thank Prof. Jianxiang Wu (Zhejiang University, Hangzhou, China) for providing RSMV nucleocapsid protein specific antibody; Prof. Zhenghe Li (Zhejiang University, Hangzhou, China) for providing the SYNV-GFP and the plasmids containing the glycoprotein coding sequences of SYNV and TYMaV; Prof. Xianbing Wang (China Agricultural University, Beijing, China) for providing the plasmids containing the glycoprotein coding sequence of BYSMV; Prof. Fangfang Li (Institute of Plant Protection, Chinese Academy of Agricultural Sciences) for providing the constructed pTRV2 and YFP-NbATG8a plasmids. No conflict of interest declared.
Correction Statement
This article has been corrected with minor changes. These changes do not impact the academic content of the article.
Funding Statement
This work was supported by the grants from the National Natural Science Foundation of China (32072388, 32222071), Guangdong Special Branch Plan for Young Talent with Scientific and Technological Innovation (2019TQ05N158), Natural Science Foundation of Guangdong Province (2022A1515010770), the Postdoctoral Science Foundation of China (2021M691083), Science and Technology Base and Talent Special Project of Guangxi Province (GuikeAD22035012), and Guangdong Provincial Innovation Team for General Key Technologies in Modern Agricultural Industry (2019KJ133).
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary Material
Supplemental data for this article can be accessed here
References
- [1].Liu Y, Bassham DC.. Autophagy: Pathways for Self-Eating in plant cells. Annu Rev Plant Biol. 2012;63:215–237. doi: 10.1146/annurev-arplant-042811-105441 [DOI] [PubMed] [Google Scholar]
- [2].Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nat Cell Biol. 2013;15(7):713–720. doi: 10.1038/ncb2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Massey A, Kiffin R, Cuervo AM. Pathophysiology of chaperone-mediated autophagy. Int J Biochem Cell Biol. 2004;36(12):2420–2434. doi: 10.1016/j.biocel.2004.04.010 [DOI] [PubMed] [Google Scholar]
- [4].Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118(1):7–18. doi: 10.1242/jcs.01620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].van Doorn WG, Papini A. Ultrastructure of autophagy in plant cells. Autophagy. 2013;9:1922–1936. doi: 10.4161/auto.26275 [DOI] [PubMed] [Google Scholar]
- [6].Khaminets A, Behl C, Dikic I. Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol. 2016;26(1):6–16. doi: 10.1016/j.tcb.2015.08.010 [DOI] [PubMed] [Google Scholar]
- [7].Mizushima N, Levine B, Cuervo AM, et al. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–1075. doi: 10.1038/nature06639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- [9].Wen X, Klionsky DJ. An overview of macroautophagy in yeast. J Mol Biol. 2016;428(9):1681–1699. doi: 10.1016/j.jmb.2016.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Marshall RS, Vierstra RD. Autophagy: The Master of Bulk and selective recycling. Annu Rev Plant Biol. 2018;69:173–208. doi: 10.1146/annurev-arplant-042817-040606 [DOI] [PubMed] [Google Scholar]
- [11].Huang X, Zheng C, Liu F, et al. Genetic analyses of the Arabidopsis ATG1 kinase complex Reveal both kinase-dependent and independent autophagic Routes during fixed-Carbon Starvation. Plant Cell. 2019;31(12):2973–2995. doi: 10.1105/tpc.19.00066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kim J, Kim Young C, Fang C, et al. Differential regulation of Distinct Vps34 Complexes by AMPK in nutrient stress and autophagy. Cell. 2013;152:290–303. doi: 10.1016/j.cell.2012.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Johansen T, Lamark T. Selective autophagy: ATG8 Family proteins, LIR Motifs and cargo Receptors. J Mol Biol. 2020;432:80–103. doi: 10.1016/j.jmb.2019.07.016 [DOI] [PubMed] [Google Scholar]
- [14].Soto-Burgos J, Zhuang X, Jiang L, et al. Dynamics of autophagosome formation plant physiology. Plant Physiol. 2017;176:219–229. doi: 10.1104/pp.17.01236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gomes Ligia C, Dikic I. Autophagy in Antimicrobial immunity. Molecular Cell. 2014;54:224–233. doi: 10.1016/j.molcel.2014.03.009 [DOI] [PubMed] [Google Scholar]
- [16].Yang M, Ismayil A, Liu Y. Autophagy in plant-virus interactions. Annu Rev Virol. 2020;7:403–419. doi: 10.1146/annurev-virology-010220-054709 [DOI] [PubMed] [Google Scholar]
- [17].Clavel M, Michaeli S, Genschik P. Autophagy: A Double-Edged Sword to Fight plant viruses. Trends Plant Sci. 2017;22(8):646–648. doi: 10.1016/j.tplants.2017.06.007 [DOI] [PubMed] [Google Scholar]
- [18].Huang X, Chen S, Yang X, et al. Friend or Enemy: A Dual role of autophagy in plant virus infection. Front Microbiol. 2020;11:11. doi: 10.3389/fmicb.2020.00736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Shelly S, Lukinova N, Bambina S, et al. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity. 2009;30(4):588–598. doi: 10.1016/j.immuni.2009.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sagnier S, Daussy CF, Borel S, et al. Autophagy restricts HIV-1 infection by selectively Degrading Tat in CD4+ T Lymphocytes. J Virol. 2015;89(1):615–625. doi: 10.1128/JVI.02174-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].O’Connell D, Liang C. Autophagy interaction with herpes simplex virus type-1 infection. Autophagy. 2016;12(3):451–459. doi: 10.1080/15548627.2016.1139262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gannagé M, Dormann D, Albrecht R, et al. Matrix protein 2 of Influenza a virus Blocks autophagosome Fusion with Lysosomes. Cell Host Microbe. 2009;6(4):367–380. doi: 10.1016/j.chom.2009.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Liu Y, Schiff M, Czymmek K, et al. Autophagy regulates Programmed Cell Death during the plant innate Immune response. Cell. 2005;121(4):567–577. doi: 10.1016/j.cell.2005.03.007 [DOI] [PubMed] [Google Scholar]
- [24].Hafrén A, Macia J-L, Love AJ, et al. Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc Natl Acad Sci, USA. 2017;114(10):E2026–E35. doi: 10.1073/pnas.1610687114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Li F, Zhang C, Li Y, et al. Beclin1 restricts RNA virus infection in plants through suppression and degradation of the viral polymerase. Nat Commun. 2018;9(1):1268. doi: 10.1038/s41467-018-03658-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Li F, Zhang M, Zhang C, et al. Nuclear autophagy degrades a geminivirus nuclear protein to restrict viral infection in solanaceous plants. New Phytol. 2020;225(4):1746–1761. doi: 10.1111/nph.16268 [DOI] [PubMed] [Google Scholar]
- [27].Ismayil A, Yang M, Haxim Y, et al. Cotton leaf curl Multan virus βC1 protein induces autophagy by disrupting the interaction of autophagy-related protein 3 with Glyceraldehyde-3-phosphate Dehydrogenases. Plant Cell. 2020;32:1124–1135. doi: 10.1105/tpc.19.00759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yang M, Zhang Y, Xie X, et al. Barley stripe mosaic virus γb protein Subverts autophagy to promote viral infection by disrupting the ATG7-ATG8 interaction. Plant Cell. 2018;30(7):1582–1595. doi: 10.1105/tpc.18.00122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Haxim Y, Ismayil A, Jia Q, et al. Autophagy functions as an antiviral mechanism against geminiviruses in plants. Elife. 2017;6:e23897. doi: 10.7554/eLife.23897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Nakahara KS, Masuta C, Yamada S, et al. Tobacco calmodulin-like protein provides secondary defense by binding to and directing degradation of virus RNA silencing suppressors. Proc Natl Acad Sci, USA. 2012;109(25):10113–10118. doi: 10.1073/pnas.1201628109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Jiang L, Lu Y, Zheng X, et al. The plant protein NbP3IP directs degradation of rice stripe virus p3 silencing suppressor protein to limit virus infection through interaction with the autophagy-related protein NbATG8. New Phytol. 2021;229(2):1036–1051. doi: 10.1111/nph.16917 [DOI] [PubMed] [Google Scholar]
- [32].Shukla A, Hoffmann G, Kushwaha NK, et al. Salicylic acid and the viral virulence factor 2b regulate the divergent roles of autophagy during cucumber mosaic virus infection. Autophagy. 2022;18:1–13. doi: 10.1080/15548627.2021.1987674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hafrén A, Üstün S, Hochmuth A, et al. Turnip mosaic virus Counteracts selective autophagy of the viral silencing suppressor HCpro. Plant Physiol. 2018;176(1):649–662. doi: 10.1104/pp.17.01198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Cao B, Ge L, Zhang M, et al. Geminiviral C2 proteins inhibit active autophagy to facilitate virus infection by impairing the interaction of ATG7 and ATG8. J Integr Plant Biol. 2023;65:1328–1343. doi: 10.1111/jipb.13452 [DOI] [PubMed] [Google Scholar]
- [35].Yang M, Wang Y, Li D, et al. Plant virus infection disrupts vacuolar acidification and autophagic degradation for the effective infection. Autophagy. 2022;18:1–2. doi: 10.1080/15548627.2022.2027194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Cheng X, Wang A, Simon AE. The Potyvirus silencing suppressor protein VPg mediates degradation of SGS3 via Ubiquitination and autophagy Pathways. J Virol. 2017;91(1):e01478–16. doi: 10.1128/JVI.01478-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Derrien B, Baumberger N, Schepetilnikov M, et al. Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc Natl Acad Sci, USA. 2012;109(39):15942–15946. doi: 10.1073/pnas.1209487109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Li F, Zhao N, Li Z, et al. A calmodulin-like protein suppresses RNA silencing and promotes geminivirus infection by degrading SGS3 via the autophagy pathway in Nicotiana benthamiana. PLOS Pathogens. 2017;13(2):e1006213. doi: 10.1371/journal.ppat.1006213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wang H, Zhang J, Liu H, et al. A plant virus hijacks phosphatidylinositol-3,5-bisphosphate to escape autophagic degradation in its insect vector. Autophagy. 2022;19:1–16. doi: 10.1080/15548627.2022.2116676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wang Q, Lu L, Zeng M, et al. Rice black-streaked dwarf virus P10 promotes phosphorylation of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) to induce autophagy in Laodelphax striatellus. Autophagy. 2022;18:1–20. doi: 10.1080/15548627.2021.1954773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chen Q, Jia D, Ren J, et al. VDAC1 balances mitophagy and apoptosis in leafhopper upon arbovirus infection. Autophagy. 2022;19:null–null. doi: 10.1080/15548627.2022.2150001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Chen Q, Zhang Y, Yang H, et al. GAPDH mediates plant reovirus-induced incomplete autophagy for persistent viral infection in leafhopper vector. Autophagy. 2022;19:1–14. doi: 10.1080/15548627.2022.2115830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Liang Q, Wan J, Liu H, et al. A plant nonenveloped double-stranded RNA virus activates and co-opts BNIP3-mediated mitophagy to promote persistent infection in its insect vector. Autophagy. 2022;19:null–null. doi: 10.1080/15548627.2022.2091904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Dietzgen RG, Kondo H, Goodin MM, et al. The family Rhabdoviridae: mono- and bipartite negative-sense RNA viruses with diverse genome organization and common evolutionary origins. Virus res. 2017;227:158–170. doi: 10.1016/j.virusres.2016.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Jackson AO, Dietzgen RG, Goodin MM, et al. Biology of plant rhabdoviruses. Annu Rev Phytopathol. 2005;43:623–660. doi: 10.1146/annurev.phyto.43.011205.141136 [DOI] [PubMed] [Google Scholar]
- [46].Kuzmin IV, Novella IS, Dietzgen RG, et al. The rhabdoviruses: Biodiversity, phylogenetics, and evolution. Infect Genet Evol. 2009;9:541–553. doi: 10.1016/j.meegid.2009.02.005 [DOI] [PubMed] [Google Scholar]
- [47].Ammar E-D, Tsai C-W, Whitfield AE, et al. Cellular and Molecular Aspects of rhabdovirus interactions with insect and plant Hosts. Annu Rev Entomol. 2009;54:447–468. doi: 10.1146/annurev.ento.54.110807.090454 [DOI] [PubMed] [Google Scholar]
- [48].García-Valtanen P, MdM O-V, Martínez-López A, et al. Autophagy-inducing peptides from mammalian VSV and fish VHSV rhabdoviral G glycoproteins (G) as models for the development of new therapeutic molecules. Autophagy. 2014;10:1666–1680. doi: 10.4161/auto.29557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Yang X, Huang J, Liu C, et al. Rice stripe mosaic virus, a Novel cytorhabdovirus Infecting rice via leafhopper transmission. Front Microbiol. 2017;7. doi: 10.3389/fmicb.2016.02140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Yang X, Zhang T, Chen B, et al. Transmission Biology of rice stripe mosaic virus by an Efficient insect vector Recilia dorsalis (Hemiptera: Cicadellidae). Front Microbiol. 2017;8. doi: 10.3389/fmicb.2017.02457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Chen S, Li W, Huang X, et al. Symptoms and yield loss caused by rice stripe mosaic virus. Virol J. 2019;16(1):145. doi: 10.1186/s12985-019-1240-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Wang Z, Chen B, Zhang T, et al. Rice stripe mosaic disease: Characteristics and control Strategies. Front Microbiol. 2021;12:12. doi: 10.3389/fmicb.2021.715223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Contento AL, Xiong Y, Bassham DC. Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J. 2005;42:598–608. doi: 10.1111/j.1365-313X.2005.02396.x [DOI] [PubMed] [Google Scholar]
- [54].Chresta CM, Davies BR, Hickson I, et al. AZD8055 is a Potent, selective, and Orally Bioavailable ATP-Competitive Mammalian target of Rapamycin kinase inhibitor with In vitro and In vivo Antitumor activity. Cancer Res. 2010;70:288–298. doi: 10.1158/0008-5472.CAN-09-1751 [DOI] [PubMed] [Google Scholar]
- [55].Aoyama T, Chua N-H. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997;11(3):605–612. doi: 10.1046/j.1365-313X.1997.11030605.x [DOI] [PubMed] [Google Scholar]
- [56].Koo AJK, Gao X, Daniel Jones A, et al. A rapid wound signal activates the systemic synthesis of bioactive jasmonates in Arabidopsis. Plant J. 2009;59(6):974–986. doi: 10.1111/j.1365-313X.2009.03924.x [DOI] [PubMed] [Google Scholar]
- [57].Wang Z, Wilson WA, Fujino MA, et al. Antagonistic controls of autophagy and Glycogen accumulation by Snf1p, the yeast Homolog of AMP-Activated protein kinase, and the Cyclin-dependent kinase Pho85p. Mol Cell Biol. 2001;21(17):5742–5752. doi: 10.1128/MCB.21.17.5742-5752.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Soto-Burgos J, Bassham DC, Otegui M. SnRK1 activates autophagy via the TOR signaling pathway in Arabidopsis thaliana. PLoS One. 2017;12(8):e0182591. doi: 10.1371/journal.pone.0182591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Yoshimoto K, Hanaoka H, Sato S, et al. Processing of ATG8s, ubiquitin-like proteins, and their Deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 2004;16(11):2967–2983. doi: 10.1105/tpc.104.025395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Kim J, Kundu M, Viollet B, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–141. doi: 10.1038/ncb2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Li F, Vierstra RD. Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 2012;17(9):526–537. doi: 10.1016/j.tplants.2012.05.006 [DOI] [PubMed] [Google Scholar]
- [62].Fujiki Y, Yoshimoto K, Ohsumi Y. An Arabidopsis Homolog of yeast ATG6/VPS30 is essential for Pollen Germination. Plant Physiol. 2007;143(3):1132–1139. doi: 10.1104/pp.106.093864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Wang Q, Ma X, Qian S, et al. Rescue of a plant negative-Strand RNA virus from cloned cDNA: Insights into Enveloped plant virus movement and Morphogenesis. PLOS Pathogens. 2015;11(10):e1005223. doi: 10.1371/journal.ppat.1005223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Ding S-W. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10(9):632–644. doi: 10.1038/nri2824 [DOI] [PubMed] [Google Scholar]
- [65].Fu S, Xu Y, Li C, et al. Rice stripe virus Interferes with S-acylation of remorin and induces its autophagic degradation to facilitate virus infection. Mol Plant. 2017;11:269–287. doi: 10.1016/j.molp.2017.11.011 [DOI] [PubMed] [Google Scholar]
- [66].Polge C, Thomas M. SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci. 2007;12(1):20–28. doi: 10.1016/j.tplants.2006.11.005 [DOI] [PubMed] [Google Scholar]
- [67].Chen L, Su Z-Z, Huang L, et al. The AMP-Activated protein kinase KIN10 is involved in the regulation of autophagy in Arabidopsis. Front Plant Sci. 2017;8. doi: 10.3389/fpls.2017.01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Wang J, Huang X, Chen S, et al. On-site and visual detection of sorghum mosaic virus and rice stripe mosaic virus based on reverse transcription-recombinase-aided amplification and CRISPR/Cas12a. Frontiers In Genome Ed. 2023;5. doi: 10.3389/fgeed.2023.1124794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Gao Q, Xu W-Y, Yan T, et al. Rescue of a plant cytorhabdovirus as versatile expression platforms for planthopper and cereal genomic studies. New Phytol. 2019;223:2120–2133. doi: 10.1111/nph.15889 [DOI] [PubMed] [Google Scholar]
- [70].He F, Zhang F, Sun W, et al. A versatile vector Toolkit for Functional analysis of rice genes. Rice. 2018;11(1):27. doi: 10.1186/s12284-018-0220-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Zhu H, Hu F, Wang R, et al. Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell. 2011;145(2):242–256. doi: 10.1016/j.cell.2011.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Goodin MM, Dietzgen RG, Schichnes D, et al. pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 2002;31(3):375–383. doi: 10.1046/j.1365-313X.2002.01360.x [DOI] [PubMed] [Google Scholar]
- [73].Cheng X, Wang X, Wu J, et al. βC1 encoded by tomato yellow leaf curl China betasatellite forms multimeric complexes in vitro and in vivo. Virology. 2011;409(2):156–162. doi: 10.1016/j.virol.2010.10.007 [DOI] [PubMed] [Google Scholar]
- [74].Ratcliff F, Martin-Hernandez AM, Baulcombe DC. Technical advance: tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 2001;25:237–245. doi: 10.1046/j.0960-7412.2000.00942.x [DOI] [PubMed] [Google Scholar]
- [75].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- [76].Zhang Y, Su J, Duan S, et al. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods. 2011;7(1):30. doi: 10.1186/1746-4811-7-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Zhao Y, Cao X, Zhong W, et al. A viral protein orchestrates rice ethylene signaling to coordinate viral infection and insect vector-mediated transmission. Mol Plant. 2022;15(4):689–705. doi: 10.1016/j.molp.2022.01.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


