Abstract
Recapitulating inherent heterogeneity and complex microarchitectures within confined print volumes for developing implantable constructs that could maintain their structure in vivo has remained challenging. Here, we present a combinational multimaterial and embedded bioprinting approach to fabricate complex tissue constructs that can be implanted postprinting and retain their three-dimensional (3D) shape in vivo. The microfluidics-based single nozzle printhead with computer-controlled pneumatic pressure valves enables laminar flow-based voxelation of up to seven individual bioinks with rapid switching between various bioinks that can solve alignment issues generated during switching multiple nozzles. To improve the spatial organization of various bioinks, printing fidelity with the z-direction, and printing speed, self-healing and biodegradable colloidal gels as support baths are introduced to build complex geometries. Furthermore, the colloidal gels provide suitable microenvironments like native extracellular matrices (ECMs) for achieving cell growths and fast host cell invasion via interconnected microporous networks in vitro and in vivo. Multicompartment microfibers (i.e., solid, core–shell, or donut shape), composed of two different bioink fractions with various lengths or their intravolume space filled by two, four, and six bioink fractions, are successfully printed in the ECM-like support bath. We also print various acellular complex geometries such as pyramids, spirals, and perfusable branched/linear vessels. Successful fabrication of vascularized liver and skeletal muscle tissue constructs show albumin secretion and bundled muscle mimic fibers, respectively. The interconnected microporous networks of colloidal gels result in maintaining printed complex geometries while enabling rapid cell infiltration, in vivo.
Keywords: compartmentalized bioprinting, 3D bioprinting, multimaterial extrusion bioprinting, vascular scaffolds, colloidal hydrogels
Graphical Abstract
1. INTRODUCTION
Implantable three-dimensional (3D) tissue constructs have been proposed as an alternative approach to reestablish the structure and function of injured or lost tissues and eventually address the issue of donor shortage in tissue transplantation. As a versatile additive manufacturing technique, bioprinting has been used to fabricate biomimetic tissue implants with different cellular and material compositions that capture the spatial heterogeneity of native biologic systems within a confined volume.1–4 However, the weak nature of cellular printed implants is likely to deform and destroy their free form and 3D complex architectures, post-implantation, due to mechanical constraints from surrounding tissues or physical stress during body movement. So far, relatively simple printed constructs (i.e., aligned or stacked cellular microfibrous constructs) have been implanted into animal defect models for tissue regeneration. However, these simple printed implants are inadequate to assess the structural advantages of 3D printing technologies for regenerative medicine compared with conventional scaffolds such as cellularized and micropatterned microporous hydrogels. Therefore, the printing of implantable constructs with complex architectures within an engineered extracellular matrix (ECM), which maintains structural functionality (i.e., cellular alignments or perfusable microchannel networks in printed constructs) and provides microenvironments like native ECMs for dynamic cellular behaviors surrounding printed constructs, is an attractive alternative to solve this major issue.
In previous studies, multicompartmental microfluidic devices have been developed to fabricate multimaterial components.5–7 These studies focus on fabricating single units (i.e., microfibers or microparticles) with different materials that have the potential to mimic the spatial heterogeneity of native biologic components. However, this microfluidics technology poses challenges to building the desired 3D complex constructs composed of multicompartmental single units, which are usually collected in the solution phase. Furthermore, the microfluidic devices are not as effective as the continuous extrusion methods such as bioprinting systems to build, in a continuous manner, multilayered and bigger constructs using a high concentration of cell-laden biomaterials with high viscosity while maintaining high cellular viability. Recent innovations in embedded bioprinting technology have made it possible to introduce support systems that can be retained in 3D complex architectures to provide a degree of functionality during the printing process, albeit with some limitations. So far, the roles of the supporting baths have been mainly used to improve printing fidelity and decrease the printing time, so they were usually removed after printing. The 3D-printed complex architectures obtained as such could be used for regenerative medicine, but such a route still has the potential to damage or deform the scaffold during or post-implantation.8,9 Consequently, the supporting baths, which are mainly composed of self-healing prepolymer solutions or microparticles, have not yet been well optimized to mimic ECMs in a way that allows for maintaining complex architectures and can be implantable with 3D-printed constructs without inhibiting host cell migration and vessel formation that are essential for tissue integration and regeneration in vivo.10,11
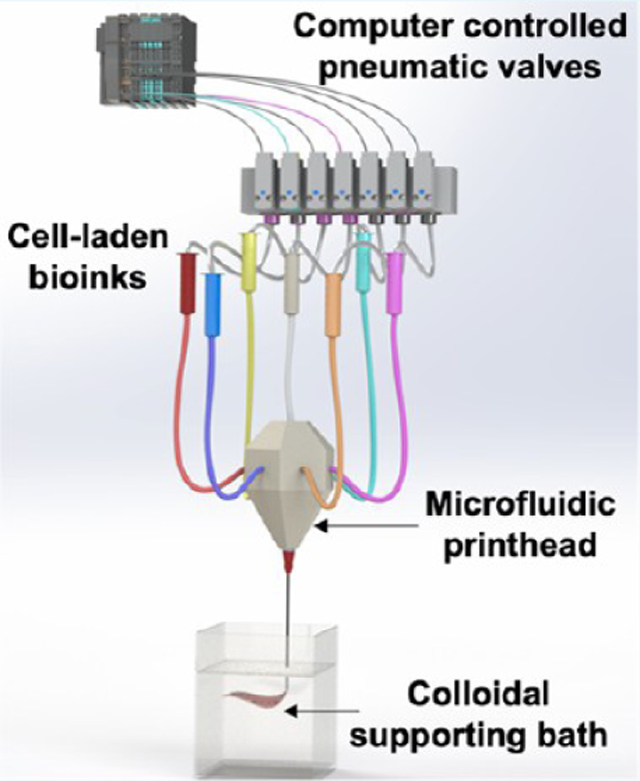
Here, we report on the development of a Microfluidic Multimaterial Manufacturing (M3) platform assisted by ECM-like colloidal gel-based support baths. Collectively, these offer innovations in terms of extrusion of compartmentalized and heterogeneous cellularized microfibrous bioinks within a defined volumetric space (compartmental) via a single microfluidic nozzle and support baths, which can support printed constructs and enhance their integration with the host while maintaining their structural architecture and hence achieve better outcomes by acting as ECM for the constructs in vivo.9,11–16 Additionally, for a multimaterial application, multinozzle systems have been used, which can affect printing outcomes owing to switching and alignment of different nozzles for different materials.1,3,17 As a proof of concept, switching of up to seven different bioinks is demonstrated via ink reservoirs connected to the microfluidic printhead. Deposition of inks is controlled by fine extrusion volume precision for a dynamic spatial control using computer-controlled pneumatic pumps. Although similar valve-based systems have been proposed in the past, these systems were limited by the number of different inks that can be extruded at once.18 Similarly, partially sacrificial suspension systems have been suggested. However, these materials have been used to fabricate the types of tissues that do not offer much heterogeneity in their composition or tissues that do not exhibit structure–function dependency.19,20 We report on the extrusion and subsequent printing of free form 3D constructs in a colloidal gel that acts as a support bath consisting of sub-10 μm particles. The advanced supporting bath platform presented in this study can not only extrude compartmentalized single units continuously in a desirable geometry and then cross-link the printed thick and large-scale constructs but also print with inks that vary in their viscosities and constitution. Additionally, the extrusions followed by cross-linking occur in a colloidal material, allowing us to form complex 3D architectures in the z-direction with soft, weak, and non-freestanding bioinks. The colloidal supporting bath can also help to prevent damage to these 3D-printed soft and weak complex architectures during the implantation process. The porosity of the colloidal gels eventually allows for host cell invasion, leading to better implant integration without damage to the complex architectures of 3D-printed implants (Scheme 1). The introduction of such gelatin-based colloidal gels offers the possibility of directly culturing or implanting the printed construct without the intermediary washing step. The interconnected microporous networks of the colloidal gels allow for a rapid infiltration of circulating cells around the printed constructs from the host tissue,21,22 leading to tissue integration while maintaining the correct 3D architecture of the printed constructs in vivo. Combined with the advantages of the multicompartmental microfluidic device and extrusion-based embedded bioprinting technology, we expect synergistic outcomes to create complex 3D architectures composed of compartmentalized single units continuously and in the future evaluate the regenerative ability of architecture advantages of 3D-printed implants.
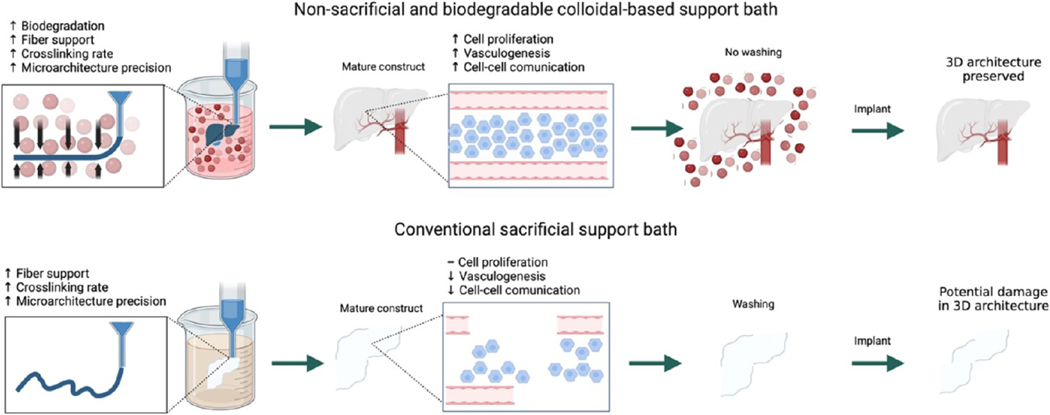
Scheme 1. Combination of Microfluidic Multimaterial Manufacturing (M3) Platform Assisted by ECM-Like Colloidal Gel-Based Support Bathsa.
a The single nozzle can dispense different bioinks of various viscosities without a backpressure buildup. The extruded bioinks are cross-linked in a support bath consisting of colloidal gels (CGs). The CGs degrade after implantation, maintaining complex 3D architectures and facilitating host cell migration, increased proliferation, and enhanced cell–cell communication, better than the corresponding contemporary sacrificial support baths.
2. MATERIALS AND METHODS
2.1. Materials.
Low-viscosity sodium salt of alginic acid of brown algae origin, porcine gelatin (type A, 300 bloom), methacrylic anhydride (MW 154.16 g/mol), 2-hydroxy-1-[4-(2-hydroxyethoxy) phenyl]-2-methyl-1-propanone (Irgacure 2959), agarose bioreagent for molecular biology, wide range/standard 3:1, HEPES buffer (N-(2-hydroxyethyl)piperazine-N′-(2-ethanesulfonic acid)), paraformaldehyde, N-isopropylacrylamide, acrylamide, cellulose nanofibrils, bovine serum albumin (BSA), goat serum, ethylenediaminetetraacetic acid (EDTA), and calcium chloride (CaCl2) were purchased from Sigma-Aldrich (St. Louis, MO) and used as received unless otherwise reported. Latex beads, carboxylate-modified polystyrene, and fluorescent green and red beads were purchased from Millipore Sigma. Primary antibodies for VE-cadherin (ab33168), MyHC (ab1242015), F-actin (ab205), and DAPI (ab228549) were purchased from Abcam (Cambridge, U.K.) and used according to the manufacturer’s protocol. PECAM-1 (CD31) and monoclonal antibodies for macrophages (CD86, CD206) were purchased from Developmental Studies Hybridoma Bank (DSHB; Iowa City, Iowa) and used according to their protocol.
2.2. 3D Bioprinter Design.
A delta 3D printer (Overlord Pro, Dreammaker) was adapted with a 3D-printed plastic holder, in that the position of the extruder was replaced with the 3D SLA printed printhead. The printhead was positioned along with reservoir tubing on this holder. Laser-cut poly(methyl methacrylate) (PMMA) holders for the syringes were positioned on the arms of the printer, and the reservoir syringes were snapped into them. A one-way pressure check valve with a silicone diaphragm was positioned in the path from the syringe reservoir to the single nozzle printhead, followed by a 23G metal connector that was connected to the tubing from the printhead. An array of plastic tubing was connected from a Wago system into 3D-printed plugs feeding into the reservoir syringes. A nitrogen gas tank was hooked onto the Wago system as a pressure source. The tank pressure was controlled manually with a pressure regulator. Pneumatic pressure valves and printhead movements were controlled via a MATLAB code through a computer. The Wago valves controlled the opening and closing of each channel, allowing just the bioink of a specific syringe or combinations of syringes to flow out.
2.3. Multimaterial Single Nozzle Printhead.
A diamond-shaped printhead with six side inlets placed at a 60° angle from neighboring inlets and one inlet from the top and an outlet from the bottom of the plane was designed using Solidworks software and printed in the Form 3 stereolithography 3D printer using clear resin (Formlabs). The printhead was postprocessed using the Formlabs washing equipment with isopropanol and then dried. 27G needles were inserted in the printhead holes. Afterward, nitrogen gas was flowed through the holes to clean any uncured material. Fifteen millimeter long metal connectors with an inner diameter of 250 μm were inserted in each of the inlets of the printhead and fixed using epoxy glue. A 23G needle was inserted in the outlet at the bottom end of the printhead and also fixed with epoxy glue. To characterize printed filament widths as a function of applied pressure and printing speed, the nozzle diameter was kept constant.
2.4. Simulations for Fluid Flow in the Printhead.
The fluid simulations were performed using ANSYS Fluent 2020. The material mixing structure was modeled and then transformed into a mesh capable of interacting with the simulated fluid mechanics, using defined and named inlet and outlet substructures to indicate the direction of fluid pressure and flow. Each of the seven material dispensers was defined as inlets, allowing the simulations to assume reasonable pressure, disallowing backflow through unused inlets. Each of the (up to) seven combinable fluid materials was given properties akin to the real-life material counterparts to accurately estimate the flow properties and shared shape of different material flow combinations. Simulated data were computed for the initial flow until a steady-state flow and then rendered using particles to imitate a steady material flow. Computation and rendering were performed using an Intel i9–10850k processor running at 4.8 GHz.
2.5. Agarose Colloidal Gel (ACG) and Gelatin Colloidal Gel (GCG) Preparation.
For ACG preparation, 2% (w/v) of agarose was dissolved in 100 mL of deionized (DI) water at 100 °C until the solution came to a boil. Once agarose dissolved, the magnetic stirrer was removed, the volume was brought back to 100 mL, and transferred to 4 °C. Another solution containing 60 mM CaCl2 in 100 mL of DI water was prepared and kept at 4 °C as well. Once the agarose formed a gel, the CaCl2 solution was added to the agarose and the solution was transferred to −20 °C for 20 min. The agarose–CaCl2 solution was placed in a blender and blended for 40 s in a pulse mode with 1 s OFF and 1 s ON intervals. ACG was poured into 50 mL of Falcon tubes and centrifuged for 5 min at 3000 rpm and then kept at 4 °C until further use. For bioprinting, high glucose DMEM culture medium with 10% FBS and 1% PS was used to wash ACG. Twenty-five milliliters of ACG was added to a Falcon tube along with 25 mL of prepared DMEM medium and centrifuged at 3000 rpm for 20 min. The supernatant was removed, and another washing step was repeated, centrifuging at 3000 rpm for 10 min.
For GCG preparation, 6.25% (w/v) of PEG300 solution was made by mixing 10 g of PEG300 in 160 mL of DI water while stirring at 50 °C until PEG300 was fully dissolved. Five grams of gelatin:GelMA (in different ratios) was separately dissolved in 100 mL of DI water at 50 °C to make a 5% (w/v) solution. A third solution consisting of 4.5% (w/v) NaCl (1.8 g) was made in 40 mL of DI water solution. Once ready, all three solutions were mixed together, and an additional 200 mL of PBS was added to this solution and equally distributed in 25 mL volumes per 50 mL falcon tubes and left on ice for 25 min. Twenty-five milliliters of ice-cold DI water were added to these tubes, and the solutions were centrifuged at 3700g for 15 min at 4 °C. The supernatant was removed, and the pellet was washed with 50 mL of cold DI water containing 2.5 mL of 1 M CaCl2. The resulting GCG was used without any further treatment.
2.6. Gelatin Methacryloyl (GelMA) Preparation.
Gelatin methacryloyl (GelMA) was synthesized following a previously published protocol.23 10% (w/v) of powdered type A gelatin (Sigma-Aldrich, 300 bloom from porcine skin) was dissolved in Dulbecco’s phosphate-buffered saline (DPBS, Gibco) while stirring at 60 °C. Methacrylic anhydride (Sigma-Aldrich) at 5% or 8% (v/v) was gradually added followed by constant stirring to the gelatin solution, representing a medium or high degree of methacryloyl substitution, respectively.24 After an hour of stirring at 60 °C and 500 rpm, two volumes of prewarmed (60 °C) DPBS were added to this solution. Dialysis of this solution was carried out for 1 week using 12–14 kDa cutoff membranes (Thermo Fisher Scientific; Waltham, MA) against DI water. The solution was filtered and lyophilized after freezing at −80 °C. GelMA as a white porous foam was obtained and left at room temperature until further use.
2.7. Bioink Preparation.
GelMA–alginate bioink was prepared by first mixing 6% of alginate in 25 mM HEPES buffer (pH 7.4) in a glass vial, and maintaining this solution at 60 °C for 1 h. A second solution containing 8% of GelMA (high methacrylation) and 1% of photoinitiator (PI) in 25 mM HEPES buffer (pH 7.4) was dissolved at 80 °C for 15 min. GelMA solution was added to the alginate solution and was left for mixing for 15 min. Green and red latex fluorescent beads at a concentration of 1% (v/v) were added to different glass vials to create green and red alginate–GelMA bioinks, respectively. The bioink was stored at 4 °C. The final concentrations were 3% of alginate, 4% of GelMA, and 0.5% of photoinitiator (PI). Cellulose nanofiber–GelMA–N-isopropylacrylamide (CNF– GelMA–NiPAM)–alginate bioink for vessel bioprinting was prepared by dissolving 2.5% of GelMA (high), 0.625% of gelatin, and 0.5% of PI in DPBS at 80 °C for 15 min. Once dissolved, 3% of alginate was added and the solution was left for mixing at 60 °C for 20 min until the alginate completely dissolved. This was followed by the addition of 12.45% of NiPAM and 0.07% of acrylamide, and the solution was kept at 60 °C for 15 min. The magnetic stirrer was removed from the vial, and 1% of CNF was added. The solution was kept at 80 °C for 5 min intervals along with vortex mixing for a total time of 15 min. Fluorescent nanobeads were added to acellular constructs to test different printing patterns.
2.8. Printing Codes and Printing Process.
The G-codes for the 3D-printed constructs were generated following two strategies: first, for the liver-like construct, a .stl file was downloaded from a free licensed website, Thingiverse, and loaded into Solidworks. The 3D model was then resized and exported as a .stl file. This was then converted to GCODE using CURA Software, and a code was developed in Python to change the coordinates from absolute to relative. A file with relative coordinates was made using the .gcode extension and used for printing. All other printed structures were designed in Solidworks and converted to .stl files for processing in the software Slic3r to obtain the path planning and extrusion movements.
Printing of 3D structures started by selecting the relevant file on the MATLAB console. Ten milliliters of syringe reservoirs were filled with bioinks, and the pressured N2 gas tubing was connected to the barrel flange. Air was opened manually for each syringe using the Wago system to ensure the correct flow of the bioinks. A poly-(dimethylsiloxane) (PDMS) cube was positioned at the center of the support bath container (5 cm × 5 cm × 10 cm clear plastic container purchased online) to hold up the embedded 3D prints. To minimize the effect of temperature on CGs, they were maintained at 4 °C before the printing, followed by keeping the CG containing bath container on an ice brick during the whole printing process. The bath contained approximately 200–250 mL of CGs over a depth of 5–7 cm, while printing was maintained to take place at the bottom with a thickness of 1 cm of the CG support bath to minimize the temperature effects. The cold ACG was added to the support bath container using a 25 mL pipette. The N2 gas tank was opened, and the GCODE and the Wago system code were then started to initiate the printing. The positioning of the bioinks on the syringes and the Wago system code for the valves was preplanned to achieve the desired multimaterial fiber configuration in the cross section and the outer structure. Once the construct was finished, it was photo-cross-linked using an 800 mW UV light source (Omnicure S2000, Excelitas Technologies) and allowed to cross-link for 100 s (20 s on lateral sides and 20 s from the top of the bath). The construct was removed carefully from the box using a spatula and placed in DPBS to avoid drying out. Our 3D-printed structures exhibited excellent postprinting stability and shape memory when resuspended in a solution.
2.9. Perfusion through Bioprinted Vessels.
After 3D bioprinting of hollow vessels, the constructs were removed carefully with stainless steel tweezers and washed with PBS. A platform was prepared on a Petri dish, where the vessel was placed in the center. Plastic tubing with 1 mm of the inner tube diameter was connected to the inlet and outlet of the vessel supported over a small slice of PDMS and pasted on the edges of the cylinder with a Loctite Ultra Gel Control Super Glue. A 1 mL syringe containing red food dye in DPBS was connected to the inlet plastic tube and flowed slowly. DPBS was passed through the vessel without any leakage for 1 min of a continuous, manual hand-controlled flow.
2.10. Mechanical and In Vitro Biological Characterizations.
Compressive strength testing of these hydrogels was performed on an Instron model 5542 mechanical tester installed with a 10 N load cell. To reach the equilibrium swelling state, hydrogels were incubated for 24 h at 37 °C in DPBS prior to mechanical testing. The samples were removed right before the mechanical characterization to ensure that the equilibrium swelling state was maintained. Prior to the tests, a digital caliper was used to measure the dimensions of the samples. With the strain rate set at 1 mm/min, the slope of the stress–strain curve in the linear kinetic region corresponding to 0–10% strain was used to calculate compressive moduli. DMEM with high glucose (Gibco, Thermo Fisher Scientific; Waltham, MA) containing 10% fetal bovine serum (FBS; Gibco, Thermo Fisher Scientific; Waltham, MA) was used to culture immortalized mouse myoblast, C2C12 (ATCC; Manassas, VA) cells, and endothelial growth medium (EGM; Lonza; Basel, Switzerland) was used for human umbilical vein endothelial cells (HUVECs; Lonza; Basel, Switzerland) before encapsulating them in 5 mm × 2 mm hydrogel discs of a 7.5% medium methacrylation bulk GelMA or GCG with different ratios of gelatin and GelMA. To assess the cell spreading/proliferation of HUVECs in hydrogels, cells were incubated with Prestoblue (Thermo Fisher Scientific; Waltham, MA) and used according to the manufacturer’s protocol. Cell proliferation was reported as the absorption at 570 nm. Cell viability was determined by staining C2C12 cells within the hydrogels using Live/Dead assay kit (calcein AM (0.5 μL/mL) and ethidium homodimer-1 (EthD-1, 2 μL/mL; Invitrogen, Thermo Fisher Scientific; Waltham, MA) and quantified according to the manufacturer’s instructions. The cells were then incubated at 37 °C for 45 min, followed by three DPBS washings to thoroughly remove the staining solutions. Subsequently, images were captured immediately under an inverted fluorescence microscope (Nikon; Minato City, Tokyo, Japan). Fiji software was used to count the number of living (green fluorescence) and dead cells (red fluorescence). Cell viability was calculated as a ratio of living cell numbers/total number of cells.
2.11. Bioprinting of Liver-Like and Muscle Fiber Mimic Tissues.
Immortal hepatocellular carcinoma, HepG2, cells (ATCC; Manassas, VA) were cultured using DMEM high glucose medium (Gibco, Thermo Fisher Scientific; Waltham, MA) containing 10% FBS, 1% L-glutamine, and 1% PS for over 1 week. A cell-laden bioink was created by encapsulating 5 mL of HepG2 cells at a concentration of 6 × 106 cells/mL and mixing with 5 mL of 6% alginate and 14% GelMA bioink. The final concentrations for cells were 3 × 106 cells/mL in an alginate/GelMA blend bioink with 3% alginate, 7% GelMA (w/v), and 0.5% PI. The 3D bioprinting area, the 3D bioprinter, and all of the materials used to print were sterilized with ethanol and UV light before the printing process. Cell-laden bioink was placed into the syringe reservoirs, and the printing process followed was identical for the acellular bioprints except when using CGs with DMEM media. The printing speed was maintained at 800 mm/min, and the pumping pressure was kept at 3 psi. To guarantee complete cross-linking of the constructs, the constructs with the GCG baths were removed postprinting and photo-cross-linked on each side with an ultra-violet (UV) light source (Omnicure S2000, Excelitas Technologies) at 800 mW for 20 s. Post-bioprinting and cross-linking, the bioprinted liver-like construct was then transferred to a Petri dish with culture medium using a spatula. The culture medium was changed three times on the day the liver-like construct was bioprinted to prevent contamination. The 3D-bioprinted liver-like construct was cultured for 21 days, and the media was collected for the albumin ELISA test at different time points.
For the muscle fiber mimic print, C2C12 (ATCC; Manassas, VA) cells were cultured using DMEM high glucose medium containing 10% of FBS. HUVECs (Lonza; Basel, Switzerland) were cultured in endothelial growth medium (EGM; Lonza; Basel, Switzerland) for over 2 weeks. A cell-laden bioink was prepared by encapsulating 5 mL of C2C12 cells at a concentration of 30 × 106 cells/mL and mixing with 5 mL of the alginate/GelMA (6:14% w/v) blend bioink for a final concentration of 3:7% (w/v) and 15 × 106 cells/mL for the blend bioink and C2C12 cells, respectively. A second cell-laden bioink was prepared in a similar fashion, albeit with HUVECs at a final concentration of 5 × 106 cells/mL. PI (0.5%) was added in both the bioinks for photo-cross-linking.
After sanitizing the area, the C2C12 bioink was placed into the central syringe reservoir and HUVEC bioink was filled in the lateral six syringe reservoirs of the printhead to create a core–shell print geometry with C2C12 cells forming the core and HUVECs forming the outer shell of the fibrils. A printing speed of 1000 mm/min and a pneumatic pumping pressure of 10 psi were employed. Afterward, the construct was photo-cross-linked in a similar manner as that for the liver-like construct print. The bioprinted muscle fiber mimic was then transferred to a Petri dish with culture medium using a spatula. To increase HUVEC attachment in the blended bioink, ethylenediaminetetraacetic acid (EDTA) was used as a chelation agent to remove alginate and enhance cell proliferation, as previously described.25 After 1 hour of culture, the printed muscle fiber mimic was treated with 20 mM of ethylenediaminetetraacetic acid (EDTA) solution in DPBS for 5 min and then returned to the culture medium. The culture medium was changed three times on the first day of the culture to prevent contamination. The muscle fiber mimic construct was cultured for 2 weeks, and then differentiation medium containing high glucose DMEM supplemented with 2% of horse serum (Gibco, Thermo Fisher Scientific; Waltham, MA) was used for the rest of the study duration.
2.12. 3D Image Reconstruction.
Histology images were converted to .tiff format with the Lossless Lempel–Ziv–Welch (LZW) CMYK compression, trimmed to a consistent aspect ratio, and transformed into 500 × 500 pixel images. The .tiff images were further reduced to 8-bits and stacked into a single stacked .tiff file. The images were transformed by setting tilt angles followed by image alignment, creating an axis correlation map. The hydrogel mass in different corresponding slices had to be aligned so that it represented the actual structure in the 3D space. This was followed by tilt alignment. The tilt defines the angles of bending on the z-axis. In the final step, the 3D reconstruction was achieved using a simultaneous iterative reconstruction technique (SIRT) algorithm.
2.13. Albumin ELISA.
Albumin levels in the media collected at different time points from the liver-like construct were measured employing a commercial human albumin ELISA kit (Invitrogen) and used according to the manufacturer’s protocol.
2.14. Immunostaining.
After 2 weeks of culture, the differentiation medium consisting of high glucose DMEM and 2% of horse serum was used for the rest of the days of cell culture. After a week of differentiation, the printed muscle mimic fibers were fixed in 4% paraformaldehyde (PFA). Blocking buffer was prepared by diluting 2.5 mL of goat serum and 50 μL of Triton X-100 in 22.45 mL of DPBS. The samples were incubated with blocking buffer for 2 h at room temperature (RT). Afterward, different samples were incubated with different antibodies for VE-cadherin (10 μg/mL), CD31 (2–5 μg/mL), or myosin heavy chain (MHC; 1 μg/mL) in blocking buffer overnight at 4 °C. The next day’s samples were washed three times with washing buffer for 5 min at RT. Additionally, for morphogenesis and microvascularization, the fibers were stained for F-actin and CD31 markers, respectively. Secondary antibodies were added in a dilution of 1:200 and incubated at RT for 2 h. The samples were washed with DPBS three times for 5 min at RT. F-actin was added at a dilution of 1:40 in DPBS and incubated for 2 h. Samples were washed with DPBS 3X for 5 min at RT and counterstained with DAPI at 2 μg/mL in DPBS for 15 min at RT. The samples were washed again three times with DPBS and imaged.
2.15. In Vivo Studies.
To evaluate the host cell infiltration capabilities of GCG in vivo, 5 mm × 5 mm × 5 mm cuboid CG hydrogels were fabricated and implanted on the backs of 12 week old Sprague Dawley male rats (Charles River Laboratories; Wilmington, MA). Animal procedures were approved by the Institutional Animal Use and Care Committee (IACUC) of Harvard Medical School under Protocol number: 2017N000114. In spontaneously breathing rats, anesthesia was induced and maintained with isoflurane, incisions were made on the backs of the rats, and cuboid hydrogels were implanted after making four deep pockets along the sides of the four limbs. Cuboid GelMA blocks of the same size were used as controls. Implanted blocks were surgically extracted after 1 or 3 weeks and paraffin-embedded before undergoing analysis for immune response, hydrogel degradation, and cellular invasion via histology. Cylindrical-shaped GCG hydrogels with a spiral structure printed inside them using the M3 system were implanted in a similar way. One week old explants were analyzed for the integration of the printed cylindrical geometry. For histology, sections were stained with DAPI for cell quantification, as were the vascular network formations elucidated by staining for CD31 and the immune response determined by staining for CD206 and CD86, respectively. H&E staining was used to evaluate sample integration and degradation. 8 mm wide × 3 mm thick cylindrical GCG and bulk hydrogel implants were subcutaneously implanted in the dorsal back pockets of rats. Implants were retrieved after 1 or 3 weeks post implantation and studied for biocompatibility, biodegradation, and host cell invasion.
2.16. Statistical Analyses.
All of the experiments were performed in triplicate. Graphpad Prism (version 8) was used to perform statistical analyses, and data were presented as the mean ± standard error of the mean (s.e.m.). Student’s t-tests and nonparametric Kruskal–Wallis test analysis were used to determine statistical significance. Asterisk signs represent *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 levels of significance.
3. RESULTS AND DISCUSSION
3.1. M3 Bioprinter.
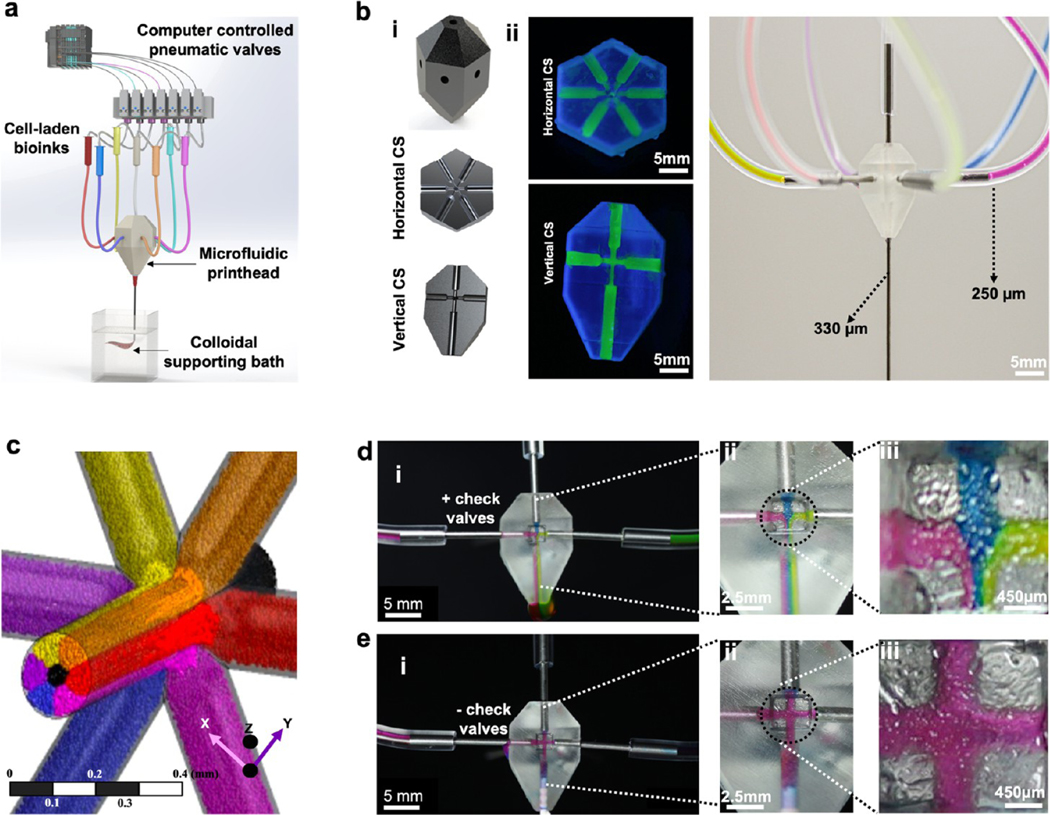
The M3 printing platform consists of a modified 3D printer and a single nozzle printhead that can route up to seven inks. The inks extrude in individual or simultaneous modes in a support bath, which facilitates complex geometries on a single microfiber (Figure 1a,b). Pneumatic pressure valves are advantageous over conventional extrusion-based printing systems in terms of feeding speed and switching for different inks after optimizing nozzle geometry, printing pressure, and ink rheology.26 However, the microfluidic printhead with pneumatic pressure system for flowing bioinks can cause backflow buildup, leading to intermixing of bioinks and undermining printing output. The introduction of check valves prevents the intermixing of different inks in the printhead (Figure 1c–e; mixing of inks in the presence and absence of check valves in Supporting Figure S1 and Supporting Movie S1). Studies for pressure valve gating predicted that up to seven different inks can be dispensed in tightly controlled volumes without any backflow buildup (Figure 1d; effect of check valves on ink mixing, Supporting Movies S2–S4). Microextrusion of ink volumes via pressure check valves and the printhead movements were achieved by a programmable automated Wago control system. Individual inks and a combination thereof could be switched at a frequency of 1–10 Hz without affecting the printing resolution (speed of material switching during printing, Supporting Movie S5).
Figure 1.
M3 bioprinter and characterization of the fluid flow of bioinks in the printhead. (a) Schematic of the M3 bioprinting system showing major components in the form of computer-controlled WAGO system pneumatic valves, microfluidic single nozzle printhead, and colloidal gel support bath where inks extrude and cross-link in the form of 3D bioprints. (b-i) Computer-aided design (CAD) of the printhead showing horizontal and vertical sections, (b-ii) an actual printhead, and (b-iii) an assembly showing the connected single nozzle seven-ink printhead. (c) Flow simulation of seven inks. Six of the bioinks enter the printhead from the sides, with a seventh ink coming from an out-of-the-plane inlet and all the inks extrude from a single outlet nozzle. (d, e) Cross-sectional view of the printhead to confirm the effect of flow patterns of three inks (red, blue, green) on the backflow and intermixing of different bioinks in (d i–iii), the presence and (e i–iii), absence of check valves.
3.2. Characterization of Colloidal Gels as Supporting Baths and Bioink.
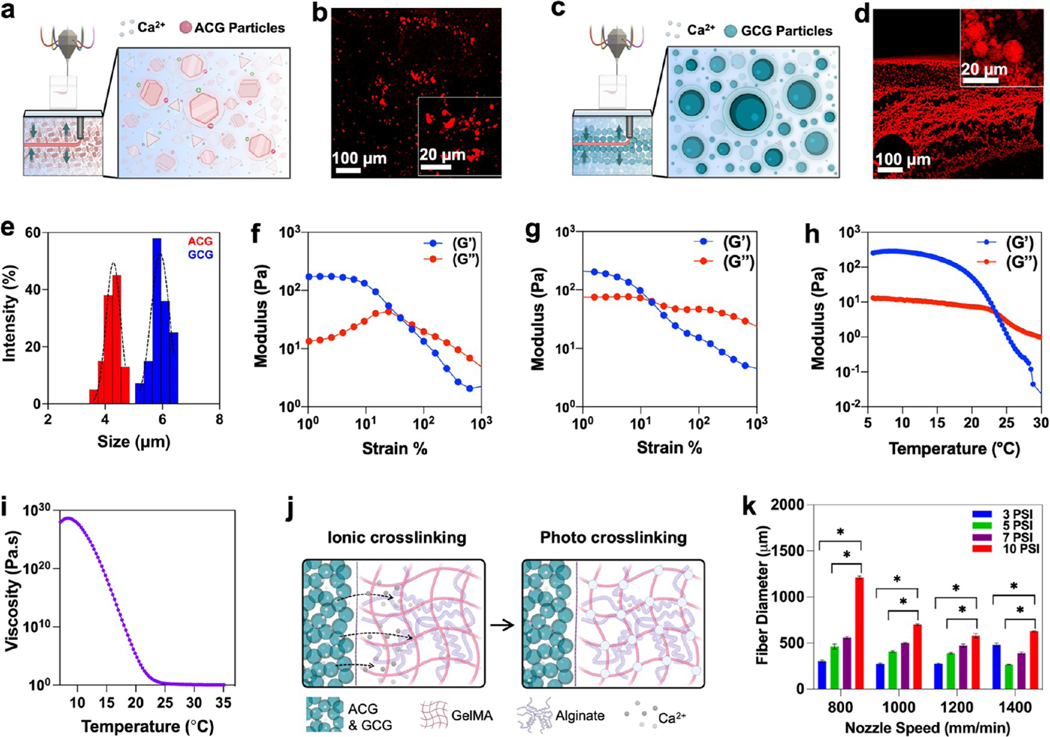
Agarose colloidal gel (ACG) and gelatin-based colloidal gel (GCG) are introduced as embedded supporting baths for free form bioprinting of blended bioinks made of alginate and gelatin methacryloyl (GelMA). The ACG is made by mixing a pregelled agarose in a precooled calcium chloride (CaCl2) solution and blending on a pulse mode,27 resulting in the production of polygon-shaped microparticles measuring 3.6 to 4.7 μm (Figure 2a–e). GCG is synthesized after a modification of previously published protocols,10,28 forming a coacervate of gelatin microparticles in the size range of 5.2–6.4 μm with poly(ethylene oxide) (PEO) in the presence of CaCl2 (Figure 2c–e).
Figure 2.
Characterization of CGs as a supporting bath and bioink. (a–d) Schematics showing self-healing and various molecular interactions such as ionic and hydrophobic interactions, and hydrogen bonding in (a) agarose colloidal gel (ACG) particles and (c) gelatin colloidal gel (GCG) particles. Fluorescent images of rhodamine incubated (b) ACG and (d) GCG. (e) Dynamic light scattering (DLS) size analysis of ACG and GCG particles. (f, g) Change in storage (G′) and loss moduli (G″) of the ACG and GCGs when stress is applied at a rate of 1–100–S, respectively. (h) Storage (G′) and loss moduli (G″) change in the 3% alginate–7% GelMA blend bioinks as a factor of temperature, suggesting a sol–gel transition. (i) Complex viscosity of the 3% alginate–7% GelMA blend bioink as a function of temperature. (j) Schematics showing two stages of cross-linking of alginate–GelMA blend bioinks; ionic cross-linking was achieved for alginate groups using Ca2+ ions and photo-cross-linking was achieved for the GelMA component of the bioink using excitation of photoinitiators. (k) Fiber diameter of the 3% alginate–7% GelMA blend bioink in GCG as a function of applied pressure and nozzle speed (n = 3; *p < 0.05).
Alginate and GelMA blend bioinks have been extensively used because they are inert and reveal a net neutral charge that is good for the chemical stability and biocompatibility of bioprinted cell-laden structures.25,29–32 As a bioink, alginate has been used as a shear-thinning biomaterial33 and can later be either removed via a brief treatment with EDTA25 or retained to tune the stiffness and degradation desired for the construct. To find a balance between cross-linking of the printed construct and promotion of cell survival and subsequent proliferation, we increased the GelMA concentration to 7% in cell-laden bioinks, while alginate content remained unchanged at 3%. It is pertinent to mention here that for seamless bioprinting, a balance has to be achieved between the self-healing time of the CG support bath and coagulation of the bioinks. The following major considerations were made before the printing. The concentration of CaCl2 in the support bath should not affect the cell viability, the extrusion rates and the nozzle speed should be in a range that does not affect the self-healing of the support bath, and the temperature of the support bath should not affect the viscosity of the extruded ink. Taking these factors into consideration, the CaCl2 concentration was maintained at ≤15 mM, the ink reservoirs were maintained at 25 °C, and CG support baths were maintained at 4 °C for the overall thermal stability of the CGs as well as the printed structures. Rheological investigations were performed on the CGs and the bioink. Both the colloidal gels (CGs) show thixotropic behavior under mechanical stress applied at a constant rate (Figure 2f,g). We observed that at low strain values (no nozzle movement, no bioprinting), storage moduli (G′) are higher than loss moduli (G″), demonstrating a solid-like elastic behavior of the CGs. When the nozzle extrudes ink at high pressure in the bath, the CGs show sol–gel transition. This is an interesting phenomenon observed in materials that show “self-healing” property. Interestingly, the sol–gel transition occurs earlier in GCGs as compared to that in ACGs. This is in line with the published literature on agarose and gelatin hydrogels.34,35 Also, the loss factor is greater in ACGs compared to that in GCGs. Applying strain at a shear rate of 1–100–S, the variation in Young’s modulus between the CG support baths and the bioinks ensured that the inks maintained their proper geometry and structure. Both the CGs showed a similar response in their change in storage (G′) and loss (G″) moduli under similar stress and temporal conditions. For printing, the ink reservoirs were kept warm (25 °C) to maintain a continuous flow during ink extrusion, as seen from a dip in viscosity beyond a temperature of around 14 °C (Figure 2h,i). Desired crosslinked shapes were attained via quick in situ ionic cross-linking with Ca2+ ions in the CGs for alginate followed by photo-crosslinking of the GelMA component with UV light (Figure 2j). The extruded fiber diameter was significantly affected by the applied pressure and the speed of the nozzle in the CG support bath (Figure 2k). In contrast, except for the high pressure (~10 psi) and low nozzle speed (~800 mm/min) printing conditions, all nozzle speeds used had a negligible effect on the overall diameter of the printed fibers. This observation was consistent in both the CGs and is important because it endows a flexibility to control the thickness of the printed microfibers by simply adjusting the pressure applied to the reservoir, irrespective of the nozzle speed. The 800–1400 mm/min print speeds achieved by the M3 printing platform, in comparison to the multimaterial extrusion-based bioprinters described in the literature, are significantly fast (comparison of extrusion printing speeds of different bioprinters; Supporting Table S1).
3.3. Linear and Compartmentalized Printing.
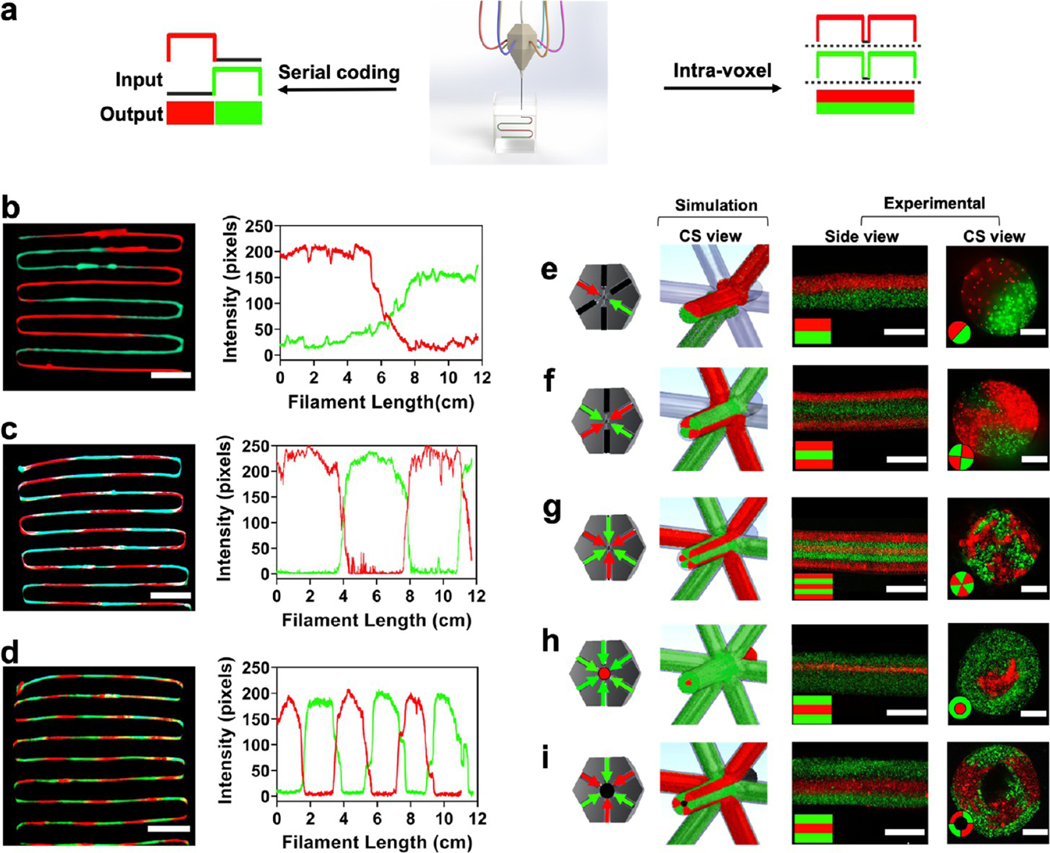
Next, we printed microfibrous filaments and shapes in linear serial coding and intravolumetric compartmentalized geometry (Figure 3a–d). In the linear geometry, all of the ink combinations could be extruded as individual fibers with different inks interspaced from each other controlled by the frequency with which the check valves for pneumatic pressure could be switched to open (on or 1) or close (off or 0). Printing serpentine linear filaments with inks consisting of two contrasting fluorescent bead colors, red and green, we observed negligible backflow or intermixing of the bioinks (Figure 3b–d). Intensity profiles as a function of the length of individual-colored filaments at different switching frequencies were observed, so that each individual color ink returned an intensity profile with pixel intensities following a periodic function. Interestingly, individual color comparisons, when plotted on top of each other, almost perfectly complemented with switching curves, reflecting a good complimentary match for the profiles of the two colors. For material switches, the 90–10% transition for the inks at a print speed of 1400 mm/min was measured at approximately 6, 4.5, and 1.8 cm, representing a switch time of 2.6, 1.9, and 0.78 s, respectively, for the filaments in Figure 3b–d. These values correspond to switching frequencies of 2.6, 3.9, and 5.2 Hz, respectively. An interesting inference from this observation is that the M3 bioprinter could achieve multimaterial bioprinting speeds that are faster than the conventional multimaterial bioprinters.3,8 Indeed, the current setup can be adapted for multiplexing for parallel multimaterial bioprinting by the addition of multiple nozzles and thus bridge the gap for the printing of complex bioprinted structures.
Figure 3.
Linear and compartmentalized printing. (a) Schematics showing linear and compartmentalized ink switching abilities of the M3 bioprinter using the single nozzle printhead. (b–d) Extrusions showing prints and intensity profiles for the red to green switch in large (~6 cm), medium (~3 cm), and short (~1.5 cm) linear switching modes, respectively. Scale bars: (b)–(d) = 1.5 cm. (e–i) Compartmentalized printing of different inks for different shapes showing channels open/close for the prints followed by cross-sectional simulation snapshot, experimental side-view, and cross-sectional view. The left panel shows a planar cross section of the printhead with different arrows showing the flow of different color inks. Black lines are for blocked/no-color channels. Simulation patterns on the right show the seven incoming inks (six planar and one in the z-axis) and the extruded output. Scale bars: (e–i) side view = 500 μm, cross-sectional view = 400 μm.
To demonstrate compartmentalized printing capability, we created a broad range of intricate patterns with the M3 bioprinter (Figure 3e–i). Using two colors, red and green, we simulated and printed structures that could be programmed to extrude in different volumes and for different time durations, creating patterned microfibrous scaffolds with distinct ink (color) separation. From a side view, the cylindrical filaments can be seen displaying different ink fractions with distinct color separations interspaced within the same confined volume. The respective cross sections showed the intravolume space filled with two, four, or six colors periodically interspersed and filling the desired regions. We next printed a core–shell arrangement representing a “donut-shaped” microfibrous pattern and a hollow donut. To be able to print these complex patterns, GelMA consisting of fluorescent green was used as the out-of-plane 4th ink from the top of the nozzle to print a “solid donut”. The three red fluorescent inks were fed in plane into the nozzle. After UV cross-linking, the printed structure had a red core of cross-linked GelMA in the middle in a green shell, giving it a shape of a solid donut. Instead of GelMA, 3% gelatin was used as the out-of-plane ink to print a hollow donut. After UV cross-linking the construct, gelatin could be washed away, creating a hollow donut shape. The right panels of Figure 3e–i show simulation snapshots of the respective ink arrangements extruded from the M3 nozzles (simulation of inks flowing in the printhead; Supporting Movies S6–S10). We successfully printed unconventional 3D shapes that are otherwise complex to be printed using conventional extrusion approaches (a two-ink hollow donut, insect eye, knots, and serpentine filaments; Supporting Figure S2). Among the printed constructs, all of the constituent microfibers maintained a well-defined cylindrical shape, with all of the ink components cohesively sticking to each other for a smooth and properly cross-linked structure. The switching of different inks showed a precise control of ink selection while maintaining printing resolution over a wide range of extruded volumes, both in linear and compartmentalized printing modes. This is an improvement over the conventional multimaterial bioprinters, as one can bioprint different tissues or organs where the contrast in tissue anatomy is large, despite the variation occurring over shorter depths such as in bone and muscle, muscle and nerve, or liver tissue and blood vessel pairs.3,8,36,37
3.4. Printing Complex Shapes and Hollow Vessels.
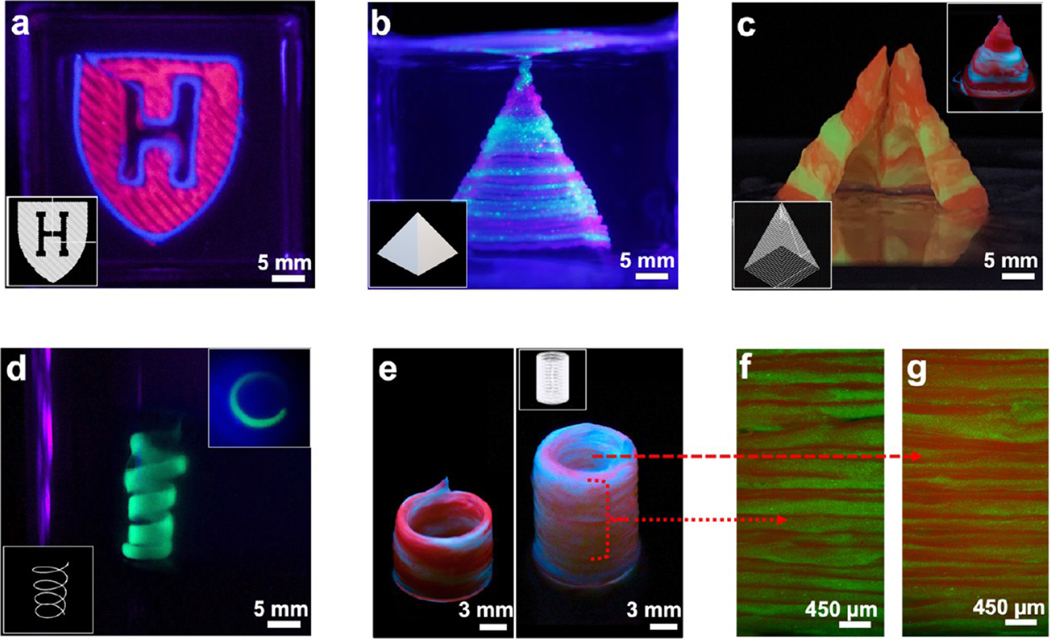
We further demonstrated the printing capability of the M3 bioprinter to fabricate sophisticated hierarchical 3D patterns in centimeter-sized 3D constructs. By spiking the bioinks with distinct fluorophores, the “Harvard University” logo was printed (Figure 4a), which showcases M3’s ability to print different shapes ranging from straight and inclined lines to curved boundaries, all within the same printed motif. To this end, complex shapes such as a spiral, as well as solid and hollow pyramids, were designed and printed (Figure 4b–d). While a spiral of 5 mm diameter could be printed using a single ink without losing the structural integrity (inset, Figure 4b), bigger constructs with different inks could be printed rapidly (~200 μL/min considering a 500 μm width of the printed filament and printing speed of 1000 mm/min) with a very distinct horizontal transition of the inks. With a fiber diameter of ~500 μm, printing 40–50 layers to make a larger construct was readily achieved. While this may be 10 times slower than a recently developed 3D printer that can switch between epoxy and silicone inks in 16–20 ms,38 in comparison to many fast-extruding multimaterial bioprinters, the M3 system is much faster. We were also able to print a straight-standing, macro-object showcasing an oblique ink transition and a hollow cylinder shape constituting two inks interspaced in a serpentine fashion all along the height of the construct (Figure 4e).38 We also fabricated a hollow cylinder shape where the inks could be switched at a single filament thickness. The spatial organization of the layer-by-layer deposition of microfibrous filaments was maintained on both the outer and the inner sidewalls of the cylinder (Figure 4f,g).
Figure 4.
Images of acellular prints of different shapes and geometries printed with M3 bioprinter. (a) Harvard Logo. (b) Spiral (inset: top view). (c, d) Solid and hollow pyramids (inset d: uncut hollow). (e) Hollow cylindrical tube with a serial ink change in a spiral (left) and horizontal (right) switching for red and green inks. (f, g) Transverse section fluorescence microscopy images of the outer and the inner walls of the right cylinder in panel e, respectively, showing that the resolution is maintained at a single fiber level at both the inside and outside of the printed construct.
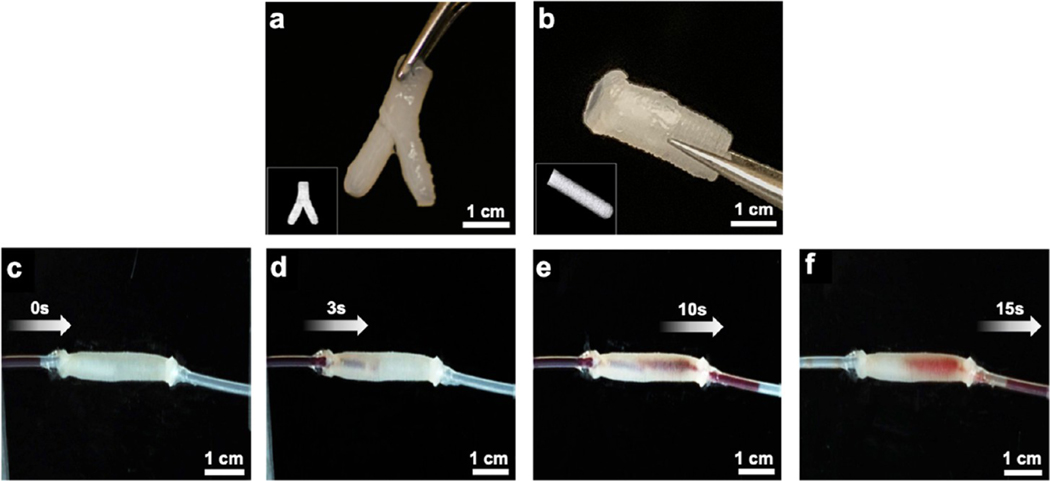
It is pertinent to mention here that adding various additives in the blend bioinks such as nanomaterials or cells at a high concentration could otherwise significantly change the rheological properties of bioinks such as viscosity and gelation and affect printability. However, due to the coagulation and self-healing function of the CG support bath, we were able to print complex 3D architectures without decreasing the printing ability and fidelity. To evaluate another feasibility of the M3 printer, we printed perfusable vessels using mechanically robust bioinks, which can potentially perform anastomosis with host blood vessels. To perform this, a nanocellulose component-dispersed alginate/GelMA blend bioink was used to fabricate perfusable vessels, which offers robustness to withstand fluid flow without any further treatment postprinting. Using a nanocomposite bioink consisting of CNF, GelMA, alginate, and NiPAM, we were able to print bifurcated Y-shaped and linear vessels (Figure 5a,b; bifurcated channel, Supporting Figure S3). Increasing the concentration of CNF in the nanocomposite bioinks significantly increased the overall mechanical properties such as elastic modulus, toughness, and elongation that are considered suitable mechanical properties of materials to make perfusable blood vessels (mechanical and rheological properties of CNF, NiPAM, and GelMA composite bioinks; Supporting Figure S4). The ink with a higher storage modulus (G′) value compared to the loss modulus (G″) usually provides good shape fidelity for the printed construct. The printed vessels were robust and easy to handle and, after being connected to plastic tubes, allowed the perfusion of biological fluid mimics without any leakage (Figure 5c–f; perfusion across a linear vessel, Supporting Movie S11). Notably, these printed channel constructs can directly be used for perfusion; they represent an interesting new candidate approach to potentially treat various vascular diseases.
Figure 5.
Images of acellular prints of different shapes and geometries printed with M3 bioprinter. (a) Printed branched and (b) linear vessels made of a composite bioink. (c–f) Snapshots of fluid perfusion across a printed linear vessel.
3.5. Bioprinting Cellularized 3D Tissue Constructs.
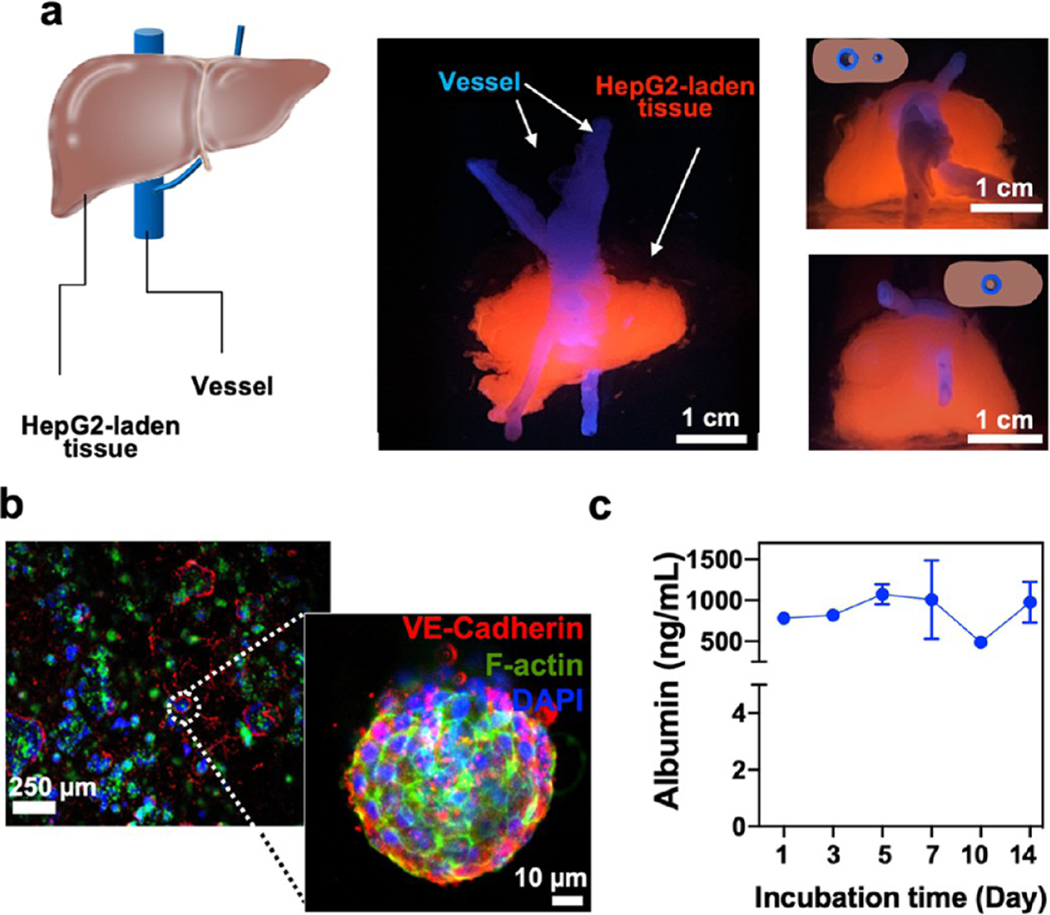
Exploiting the multicellular and multimaterial functionality of the M3 bioprinting platform, structural print form heterogeneity, and spatial arrangement, we next printed various living tissues. The combinatorial bioprinting approach introduced here allows us to extrude and cross-link different biomaterials varying in viscosity, such as alginate: GelMA or CNF–GelMA–NiPAM-based bioinks with different ratios of the constituent biomaterials, together with a high concentration of cells (>10 million cells/mL). It is challenging to print high cellladen microfibers with these types of bioinks using conventional extrusion-based printing system, owing to their high viscosity. In particular, we chose tissues that involve complex geometry, e.g., solid tissue with microvessels like liver (Figure 6) and different cellular structures aligned in a complex core–shell geometry, such as a muscle fiber mimic (Figure 7). We printed a liver-like construct (~1 cm thick) comprising of numerous bioprinted layers with an integrated perfusable channel, mimicking the hepatic vessel system. Although the vascularized liver-like construct has been reported recently,39 we show bioprinting a liver-like construct by first printing the vessel-like constructs (CNF–GelMA–NiPAM)–alginate followed by printing the hepatocellular carcinoma cell (HepG2)-laden alginate/GelMA blend bioink to create a liver-like construct.
Figure 6.
Cellular prints of liver-like and muscle-like 3D tissues. (a) Schematic and printed liver-like constructs. Front view (middle panel) shows liver-like constructs composed of vessels and HepG2-laden printed tissues. Top (top right) and bottom (bottom right) views show the perfusable channel passing through the length of the construct. (b) Fluorescent images of the printed liver-like construct showing staining for the proliferation of HepG2 cells via staining for VE-cadherin, F-actin, and DAPI after 21 days of culture. High-magnification microphotograph of the formed cellular spheroids within the printed 3D liver-like constructs. c, Albumin levels from the liver-like construct as determined by ELISA over 2 weeks of culture (n = 3).
Figure 7.
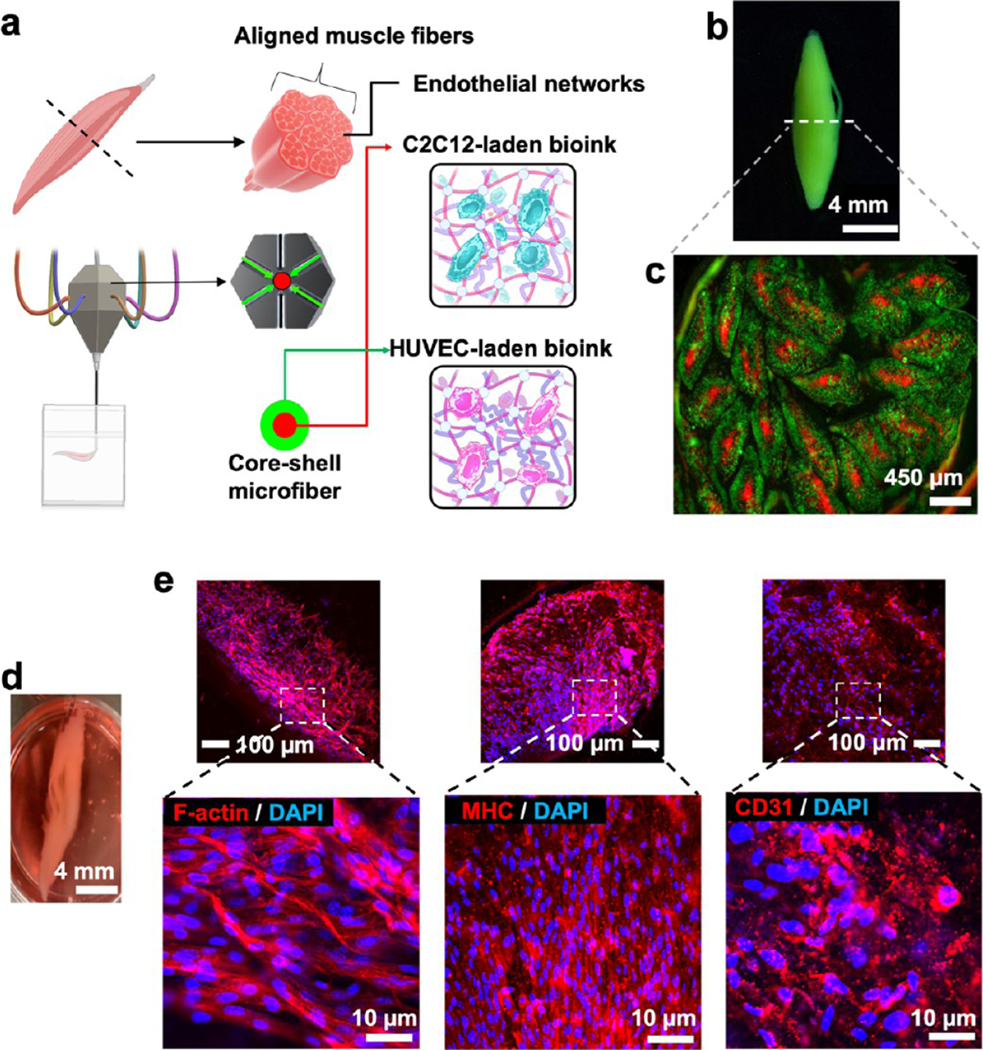
Cellular prints of muscle fiber mimic 3D tissues. (a–c) Schematics and printed construct of a muscle fiber mimic tissue with a core–shell geometry containing two cell types. The core contains C2C12 myocytes, and shell contains HUVECs. (d) Photograph and fluorescent microphotographs of the muscle fiber mimic after 21 days of culture. e, Immunofluorescent images show the differentiation of C2C12 cells into proliferating muscle cells, as depicted by f-actin and myosin heavy chain (MHC) staining. HUVECs exhibited a tendency to make microvasculature within different individual fibers in the muscle fiber mimic tissue, as depicted by CD31 staining.
Using conventional embedded bioprinting or multimaterial bioprinting approaches, it is a challenge to print two structurally and compositionally different structures simultaneously such as hollow microfibers and cell-laden scaffolds. Using our combinatorial bioprinting platform, we successfully printed a perfusable vessel component surrounded by a liver cell-laden bioink without changing or switching any type of nozzles. It is pertinent to mention that the biofabrication was independent of predefined microfluidic channels. The cellular tissue was comprised of immortal hepatocellular carcinoma cells (HepG2) (Figure 6a; bioprinted constructs of liver-like construct, Supporting Figure S5a–c; and bioprinting of liver-like construct, Supporting Movie S12) in the alginate/GelMA blend bioink. Immunohistochemical analysis revealed that the encapsulated HepG2 cells in the printed construct autonomously assembled into 3D spheroids throughout the printed construct after 21 days of culture. F-actin and VE-cadherin signals suggested the integrity of cellular junctions and spherical morphology, as expected of HepG2 spheroids (Figure 6b). The printed liver-like constructs demonstrated consistent performance in terms of albumin release for 14 days. The printed construct secreted approximately 44 μg of albumin/106 cells (Figure 6c) that correlated to the published literature on 2D- and 3D-cultured HepG2 samples.40,41 Together, these results demonstrated that the printed liver-like constructs could stably and continuously function over a prolonged period of time.
Next, we set out to fabricate skeletal muscle fiber mimic constructs that have a distinct architecture defined by myofiber bundles that run parallel to each other and are layered by muscle connective tissues (i.e., fasciculus) along with vessels and motor neurons to form a skeletal muscle fiber construct (Figure 7a). To replicate this well-defined structural heterogeneity, we printed the muscle fiber mimic within a defined compartmentalized space using two cell types: C2C12 myoblasts and human umbilical vein endothelial cells (HUVECs) (Figure 7b, bioprinted constructs of muscle fiber mimic, Supporting Figure S5d–f, CD31 immunostaining of bioprinted muscle fiber mimic scaffold, Supporting Figure S6, and bioprinting of a skeletal muscle fiber mimic construct, Supporting Movie S13). Spindle-shaped muscle fiber mimic with a thickness of 0.5 cm at its widest girth and 5 cm long, consisting of 23 fibrils (diameter: ~450 μm), was printed in a core–shell geometry and in nine layers. C2C12 myoblasts in the 3% alginate/7% GelMA blend bioink formed the core, and HUVECs in the blended bioink constituted the shell in the fiber. It is pertinent to mention that, compared to the liver-like construct, more than five times the number of cells were used in the muscle fiber mimic printing. These respective cell densities for C2C12 and HUVECs were chosen based on the previous literature.42,43 It is possible that with increased cell density, the viscosity of the bioink and viability of the encapsulated cells might be affected.44,45 However, we were able to extrude the bioinks with good viability, as demonstrated by the in vitro functionality of the encapsulated cells for over 3 weeks. This could be ascribed to the better proliferation opportunity that GCG presents compared to bulk gel, where the diffusion limit of the metabolites may not cater to the increased number of cells compared to GCG. Although recently muscle mimics have been bioprinted, the postprinting process involves the removal of sacrificial biomaterials,42 a time-consuming step that is bypassed by the M3 system. The printed cells in the construct showed a high expression of myosin heavy chain (MHC) (Figure 7f) and CD31 signals, implying the initiation or formation of microvasculature, resulting in the attainment of compartmentalized, aligned, and matured skeletal muscle fiber mimic tissues, as discussed previously in the literature.46
3.6. Biological Characterization of GCG and Stability of Printed Structures.
After introducing the printing capability of the M3 bioprinter for creating acellular and cellular 3D complex structures, we further studied whether the CGs provide suitable microenvironments like ECMs for achieving cell growth and fast host cell invasion via interconnected microporous networks in vitro and in vivo. To evaluate cellular behaviors in the GCG support gels, gelatin and GelMA were mixed in different ratios, such as 1:3 (25:75 GCG), 2:2 (50:50 GCG), and 0:1 (0:100 GCG), and then cross-linked by UV light to make stable hydrogels in biological conditions. In our previous study,47 bulk GelMA prepolymer solution as a thermal healing supporting bath was used as an ECM-mimic for fabricating neurospheroid brain-like constructs. Here, we emphasize the ECM-mimic function of the GCG support baths compared with the bulk GelMA hydrogel. The GCGs obtained showed interconnected networks composed of colloidal microgels, which showed significantly different morphologies compared with the bulk GelMA hydrogel (Figure 8a). Nevertheless, these GCGs with different ratios of gelatin and GelMA possessed similar or slightly higher compressive modulus than bulk GelMA hydrogel (Figure 8b and compressive moduli of GCGs with various ratios, Supporting Figure S7). Although the compressive moduli were not significantly different, a higher GelMA content ensured the fabrication of stable hydrogels that formed interconnected colloidal networks in the GCG hydrogel. Hence, the 2575 GCG hydrogel was preferred for further biological characterization. HUVECs and C2C12 were encapsulated in both 2575 GCG and bulk GelMA hydrogels (Figure 8c). Encapsulated HUVECs showed a better proliferation and formed worm-like cellular morphology when encapsulated in 2575 GCG compared to bulk GelMA hydrogel (Figure 8d,e). Similarly, C2C12 showed similar results suggesting that encapsulated cells had better cell viability and spreading in the 2575 GCGs compared with GelMA hydrogel (Figure 8f,g). Overall, we believe that the GCG support bath with microporous structures can provide a suitable microenvironment for improved cell spreading and proliferation while maintaining biomimetic mechanical strengths.
Figure 8.
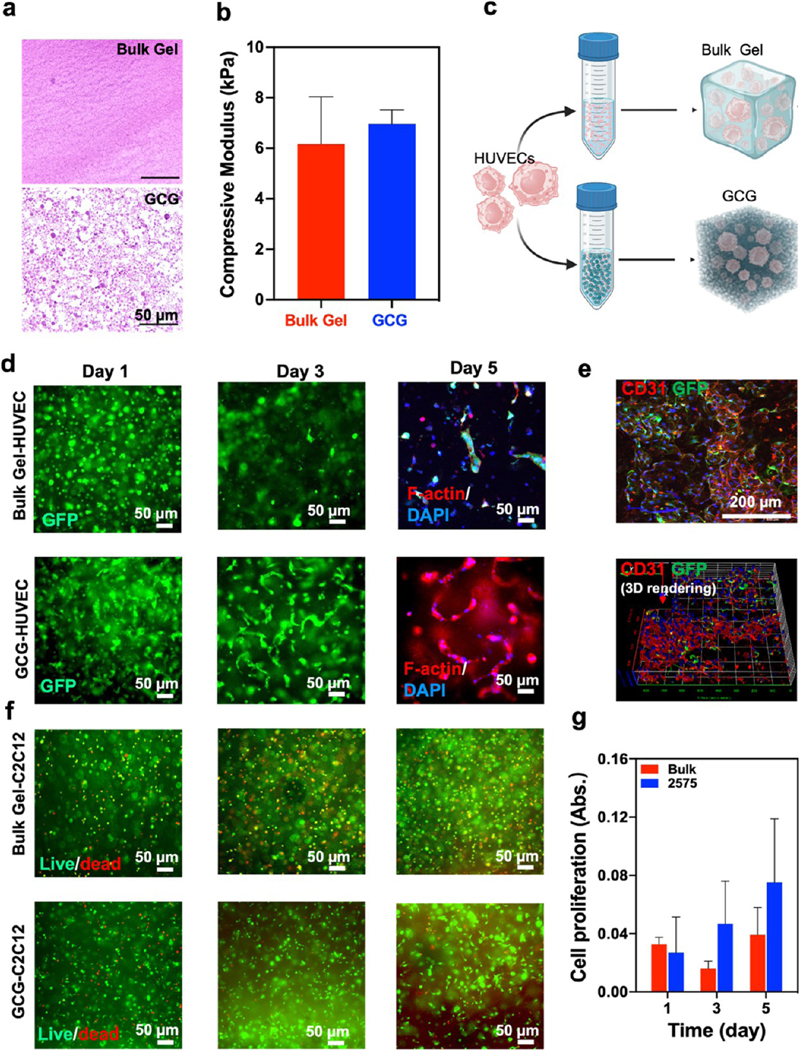
Mechanical properties and cellular behavior of encapsulated cells in the GCG in vitro. (a) H&E staining images of 2575 GCG and bulk GelMA hydrogels after UV cross-linking. (b) Compressive modulus of bulk GelMA and 2575 GCG hydrogels. (c) Schematic showing HUVEC encapsulation in the 2575 GCG and the bulk GelMA hydrogels. (d) Fluorescent microphotographs of encapsulated GFP-HUVECs in both 2575 GCG and the bulk GelMA hydrogels and showed F-actin/DAPI staining images at day 5. (e) Confocal and 3D reconstruction of Z-stack images of encapsulated GFP-HUVECs in the 2575 GCG hydrogel stained with CD31 and DAPI. The HUVECs exhibit a worm-like morphology in the 2575 GCG hydrogels and good cell spreading over a 2 week culture. (f) Live/dead staining images of encapsulated C2C12 in the bulk GelMA and 2575 GCG hydrogels. (g) Prestoblue assay to analyze the proliferation of encapsulated C2C12 in both 2575 GCG and the bulk GelMA hydrogels (n = 3).
We performed a subcutaneous in vivo biocompatibility, biodegradation, and host cell invasion study in the 2575 GCG hydrogel and compared the results with the bulk GelMA hydrogel (bulk gel) (Figure 9a). The GCG with its interconnected microporous nature showed better cell invasion and biodegradation than the bulk GelMA gel (Figure 9b–h). Of note, GCG was fully resorbed after 3 weeks, while the bulk GelMA gel still had some residual left (Figure 9c–e). Similarly, the GCG demonstrated a rapid cell invasion and endovascularization as compared to the traditional bulk GelMA hydrogel (Figure 9f). H&E staining together with immunostaining for CD86/CD206 and CD31 reveals the cells being primarily fibroblasts, immune cells, and endothelial cells, respectively (Figure 9g,h). CD86 is predominantly expressed by dendritic cells, macrophages, and memory B-cells and is associated with self-regulation and cell–cell association.48,49 CD206, on the other hand, is expressed by M2 macrophages and specific lymphatic or endothelial cells playing an important role in immune homeostasis.50 Advantageously, GCG hydrogels could therefore offer improved vascular integration via their interconnected microporous networks, maintaining complex architectures in the GCG scaffold, like native ECMs, and thus offer considerably better remodeling and retention rate than those in a bulk gel by the host animal.
Figure 9.
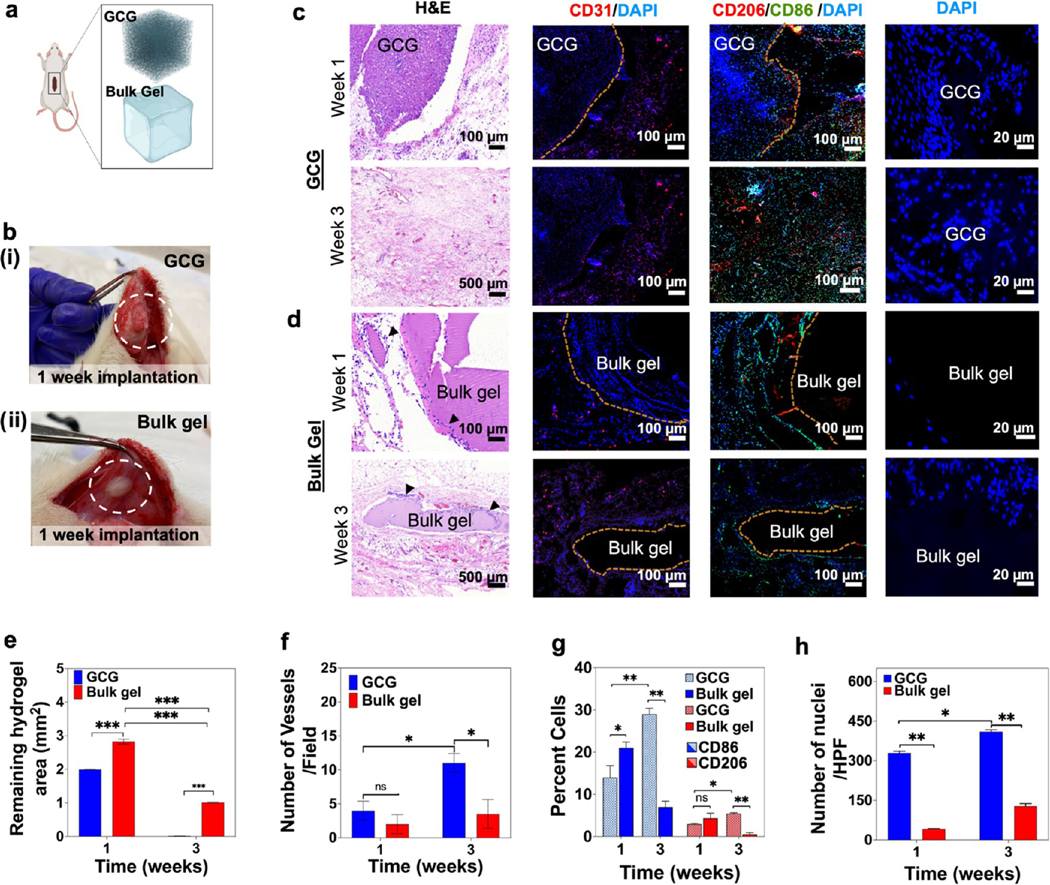
Biological characterization of a bioprinted GCG in vivo. (a) Schematics for the subcutaneous implantation of both the hydrogels. (b) GCG hydrogel (i), and bulk GelMA hydrogel (ii) explants 1 week post implantation. (c) GCG and (d) bulk GelMA hydrogel explants at 1 and 3 weeks were analyzed for cell invasion and degradation behavior (H&E), neovascularization (CD31), and macrophage induction (CD206 and CD86) along with DAPI staining. Black arrow heads in panel d (H&E) indicate host cells around the bulk GelMA hydrogel. (e–h) Semiquantification data for (e) degradation of the GCG and bulk hydrogels, (f) number of vessels, (g) percent of CD206- and CD86-stained macrophages, and (h) number of nuclei for host cell infiltration within the GCG and bulk hydrogels. (n = 3; ***p < 0.001, **p < 0.01, *p < 0.05.)
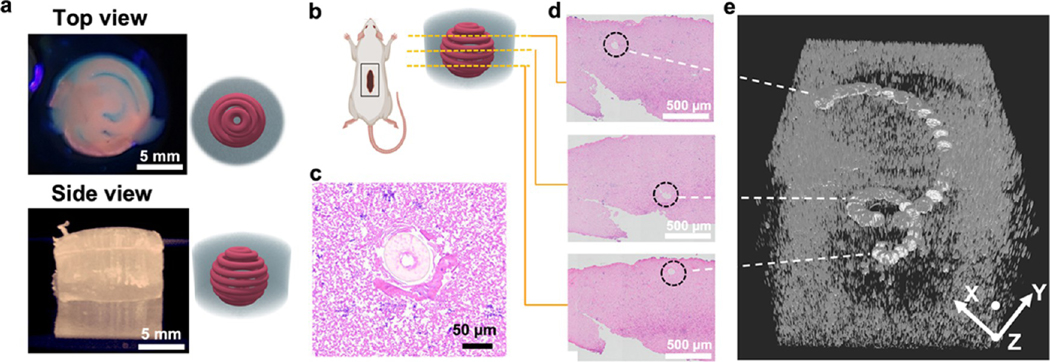
There is an intricate and intimate link between the functionality of living tissues and their highly specialized architecture. The development of bioprinting strategies that generate stable constructs and maintain structural identity to yield a healthy and mature functional tissue in vivo has remained a challenge in regenerative medicine.51 To evaluate the proof of concept of the stability of 3D-printed constructs provided by the GCG, a cylindrical GCG scaffold with a printed spiral construct using alginate/GelMA blend bioink was implanted in a rat subcutaneous model (Figure 10a,b). This structure was chosen for its simplicity in printing a 3D fibrous shape with ease across a compartmentalized space of the GCG. The printed construct-laden GCG was explanted 7 days post-implantation and analyzed histologically to investigate the structural integrity of the printed scaffold (Figure 10b). Various cross-sectional explant slices (Figure 10c,d) were stitched together using SIRT to reveal compartmental information across the z-axis (Figure 10e, reconstruction slices of the 3D volumetric rendition of the in vivo explant, Supporting Figure S8, and volumetric reconstruction of the bioprinted spiral 7 days post implantation, Supporting Movie S14). Clearly, the implanted construct in GCG is robust enough to retain its 3D fibrous shape 7 days post implantation while host cells can be seen invading near the printed fiber (Figure 10e). This important aspect is desirable for a few reasons, such as the ability it gives through GCG support baths to directly implant the printed scaffold, the retention of its morphological structure even under dynamic mechanical conditions in vivo, and the procurement of good host cell infiltration initiating neovascularization. It is an important step forward in terms of artificial tissues that need to maintain a certain geometry as an implant for proper function. Therefore, this concept might be helpful to maintain the function of the 3D-printed skeletal muscle implants such as maintaining myofiber alignment and the vessel networks in implants to retain their perfusable function or complex structure without any collapse or damage compared to the implanted “naked” 3D-printed constructs.
Figure 10.
Biological characterization of a bioprinted GCG in vivo. (a) Printed acellular structure (spiral) in the GCG supporting hydrogel. (b) 3D construct in the GCG supporting bath subcutaneously implanted in the back pocket of a rat for 1 week. (c, d) H&E-stained horizontal histological slices of the explanted printed construct. The representative histology slices presented are from different places along the thickness of the explant, showing the printed structure occupying different positions in space owing to its spiral architecture. (e) 3D volume mesh of the explant slices rendered by the SIRT volumetric algorithm revealing a spiral structure of the explanted printed construct. White dotted lines show the respective volume space in the mesh that is contributed by the histology slices on the left.
4. CONCLUSIONS
In conclusion, we believe that the M3 platform has the potential for bioprinting functional artificial tissue constructs and engineered organs that could be directly implanted, postprinting, and can maintain structural functionality while allowing expedited integration with the host. Coupled with a digitally tunable pneumatic pressure switching system operated at frequencies of up to 10 Hz enables microextrusion of all of the bioink components at printing speeds of >1000 mm/min, without compromising the printing resolution in a tightly controlled volume space. The freedom to choose different inks with different viscosities via fast switching of volumes at approximately 200 ms opens up possibilities of printing complex and uncommon shapes with varied types of bioinks, which has eluded conventional extrusion-based bioprinters. While the performance of cellularized printed constructs under in vitro conditions was excellent, further studies for optimization and with different tissue types are needed before such a technology can become mainstream. More importantly, there is a need to test the engineered tissues and organs printed with this platform in animals. Although, in our present study, we can use up to seven inks, we believe the printing process can be multiplexed by aligning several of these single nozzle printheads and expediting a printing process for bigger and more complex tissues and organs. Additionally, though the printer was exclusively used for bioinks, its abilities can be exploited for other non-biology inks for various applications. The bioprinter is being optimized for other tissue types to focus on alleviating the demand for tissues in treating different medical conditions.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health ( R 0 1 A R 0 7 4 2 3 4, R 2 1 E B 0 2 6 8 2 4, R 0 1 A R 0 7 3 8 2 2, R01AR077132), AHA Innovative Project Award (19IPLOI34660079), the Gillian Reny Stepping Strong Center for Trauma Innovation, and the Brigham Research Institute Innovation Evergreen Fund (IEF) at Brigham and Women’s Hospital. M.C.L. was supported by Basic Science Research Program through the National Research Foundation of Korea(NRF) funded by the Ministry of Education(NRF-2021R1A6A3A14039720). T.K. acknowledges funding from a Rubicon award (019.183EN.017) by the Dutch Research Council (NWO).
Footnotes
The authors declare the following competing financial interest(s): A provisional patent resulting from the work has been filed by the Brigham and Women’s Hospital (filed November 3, 2022).
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsami.2c12585.
Pressure valve gating (Movie S1) (MP4)
Printing in absence of check valves (Movie S2) (MP4)
Printing in presence of check valves and fast switching (Movie S3) (MP4)
Choice of bioink mixing (Movie S4) (MP4)
Multimaterial bioprinting at high speed (Movie S5) (MP4)
Simulation of 2 inks mixing and extrusion (Movie S6) (MP4)
Simulation of 4 inks mixing and extrusion (Movie S7) (MP4)
Simulation of 6 inks mixing and extrusion (Movie S8) (MP4)
Simulation of 7 inks mixing for core-shell geometry (Movie S9) (MP4)
Simulation of 7 inks mixing for donut shaped geometry (Movie S10) (MP4)
Perfusion across a linear vessel (Movie S11) (MP4) Bioprinting of liver-like construct (Movie S12) (MP4)
Bioprinting of a skeletal muscle fiber mimic construct (Movie S13) (MP4)
Volumetric reconstruction of the bioprinted spiral 7 days post implantation (Movie S14) (MP4)
Photographs of different components of the M3 bioprinter, different printed shapes, mechanical and rheological characterizations of CNF–NiPAM–GelMA composite bioink, bioprints of liver-like and skeletal muscle fiber mimic constructs, and movies on ink flow simulations, ink perfusion through a linear bioprinted vessel, bioprinting of liver-like and muscle fiber mimic constructs, and volumetric reconstruction of a bioprinted spiral 7 days post implantation (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acsami.2c12585
Contributor Information
Shabir Hassan, Division of Engineering in Medicine, Department of Medicine, Harvard Medical School, and Brigham and Women’s Hospital, Cambridge, Massachusetts 02139, United States; Department of Biology, Main Campus, Khalifa University, Abu Dhabi 127788, United Arab Emirates.
Eduardo Gomez-Reyes, Division of Engineering in Medicine, Department of Medicine, Harvard Medical School, and Brigham and Women’s Hospital, Cambridge, Massachusetts 02139, United States; Tecnológico de Monterrey at Monterrey, Monterrey, Nuevo León CP 64849, Mexico Eduardo.
Eduardo Enciso-Martinez, Division of Engineering in Medicine, Department of Medicine, Harvard Medical School, and Brigham and Women’s Hospital, Cambridge, Massachusetts 02139, United States; Tecnológico de Monterrey at Monterrey, Monterrey, Nuevo León CP 64849, Mexico.
Kun Shi, Division of Engineering in Medicine, Department of Medicine, Harvard Medical School, and Brigham and Women’s Hospital, Cambridge, Massachusetts 02139, United States; State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University and Collaborative Innovation Center of Biotherapy, Chengdu 610041, P. R. China.
Jorge Gonzalez Campos, Division of Engineering in Medicine, Department of Medicine, Harvard Medical School, and Brigham and Women’s Hospital, Cambridge, Massachusetts 02139, United States; Tecnológico de Monterrey at Monterrey, Monterrey, Nuevo León CP 64849, Mexico.
Oscar Yael Perez Soria, Division of Engineering in Medicine, Department of Medicine, Harvard Medical School, and Brigham and Women’s Hospital, Cambridge, Massachusetts 02139, United States; Tecnológico de Monterrey at Monterrey, Monterrey, Nuevo León CP 64849, Mexico.
Eder Luna-Cerón, Division of Engineering in Medicine, Department of Medicine, Harvard Medical School, and Brigham and Women’s Hospital, Cambridge, Massachusetts 02139, United States; Tecnológico de Monterrey at Monterrey, Monterrey, Nuevo León CP 64849, Mexico.
Myung Chul Lee, Division of Engineering in Medicine, Department of Medicine, Harvard Medical School, and Brigham and Women’s Hospital, Cambridge, Massachusetts 02139, United States Isaac.
Isaac Garcia-Reyes, Division of Engineering in Medicine, Department of Medicine, Harvard Medical School, and Brigham and Women’s Hospital, Cambridge, Massachusetts 02139, United States; Tecnológico de Monterrey at Monterrey, Monterrey, Nuevo León CP 64849, Mexico.
Joshua Steakelum, Division of Engineering in Medicine, Department of Medicine, Harvard Medical School, and Brigham and Women’s Hospital, Cambridge, Massachusetts 02139, United States; Department of Electrical and Computer Engineering, University of Massachusetts, Dartmouth, Massachusetts 02747, United States.
Haziq Jeelani, Institute of Electrical and Electronics Engineers (IEEE), New York 10016, United States.
Luis Enrique García-Rivera, Division of Engineering in Medicine, Department of Medicine, Harvard Medical School, and Brigham and Women’s Hospital, Cambridge, Massachusetts 02139, United States; Tecnológico de Monterrey at Monterrey, Monterrey, Nuevo León CP 64849, Mexico.
Minsung Cho, AltrixBio inc., Cambridge, Massachusetts 02139, United States.
Stephanie Sanchez Cortes, Division of Engineering in Medicine, Department of Medicine, Harvard Medical School, and Brigham and Women’s Hospital, Cambridge, Massachusetts 02139, United States; Tecnológico de Monterrey at Monterrey, Monterrey, Nuevo León CP 64849, Mexico.
Tom Kamperman, Division of Engineering in Medicine, Department of Medicine, Harvard Medical School, and Brigham and Women’s Hospital, Cambridge, Massachusetts 02139, United States; Department of Developmental Bioengineering, Faculty of Science and Technology, TechMed Centre, University Twente, Enschede 7522 NB, Netherlands.
Haihang Wang, Division of Engineering in Medicine, Department of Medicine, Harvard Medical School, and Brigham and Women’s Hospital, Cambridge, Massachusetts 02139, United States.
Jeroen Leijten, Department of Developmental Bioengineering, Faculty of Science and Technology, TechMed Centre, University Twente, Enschede 7522 NB, Netherlands.
Lance Fiondella, Department of Electrical and Computer Engineering, University of Massachusetts, Dartmouth, Massachusetts 02747, United States.
Su Ryon Shin, Division of Engineering in Medicine, Department of Medicine, Harvard Medical School, and Brigham and Women’s Hospital, Cambridge, Massachusetts 02139, United States.
REFERENCES
- (1).Ruiz-Cantu L; Gleadall A; Faris C; Segal J; Shakesheff K; Yang J. Multi-material 3D bioprinting of porous constructs for cartilage regeneration. Mater. Sci. Eng., C 2020, 109, No. 110578. [DOI] [PubMed] [Google Scholar]
- (2).Miri AK; Nieto D; Iglesias L; Goodarzi Hosseinabadi H; Maharjan S; Ruiz-Esparza GU; Khoshakhlagh P; Manbachi A; Dokmeci MR; Chen S; Shin SR; Zhang YS; Khademhosseini A. Microfluidics-Enabled Multimaterial Maskless Stereolithographic Bioprinting. Adv. Mater 2018, 30, No. 1800242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Rocca M; Fragasso A; Liu W; Heinrich MA; Zhang YS Embedded Multimaterial Extrusion Bioprinting. SLAS Technol. 2018, 23, 154–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Grigoryan B; Sazer DW; Avila A; Albritton JL; Padhye A; Ta AH; Greenfield PT; Gibbons DL; Miller JS Development, characterization, and applications of multi-material stereolithography bioprinting. Sci. Rep 2021, 11, No. 3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Cheng Y; Zheng F; Lu J; Shang L; Xie Z; Zhao Y; Chen Y; Gu Z. Bioinspired Multicompartmental Microfibers from Microfluidics. Adv. Mater 2014, 26, 5184–5190. [DOI] [PubMed] [Google Scholar]
- (6).Wu Z; Zheng Y; Lin L; Mao S; Li Z; Lin J-M Controllable Synthesis of Multicompartmental Particles Using 3D Microfluidics. Angew. Chem., Int. Ed 2020, 59, 2225–2229. [DOI] [PubMed] [Google Scholar]
- (7).Yu Y; Fu F; Shang L; Cheng Y; Gu Z; Zhao Y. Bioinspired Helical Microfibers from Microfluidics. Adv. Mater 2017, 29, No. 1605765. [DOI] [PubMed] [Google Scholar]
- (8).Liu W; Zhang YS; Heinrich MA; De Ferrari F; Jang HL; Bakht SM; Alvarez MM; Yang J; Li Y-C; Trujillo-de Santiago G; Miri AK; Zhu K; Khoshakhlagh P; Prakash G; Cheng H; Guan X; Zhong Z; Ju J; Zhu GH; Jin X; Shin SR; Dokmeci MR; Khademhosseini A. Rapid Continuous Multimaterial Extrusion Bioprinting. Adv. Mater 2017, 29, No. 1604630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Highley CB; Rodell CB; Burdick JA Direct 3D Printing of Shear-Thinning Hydrogels into Self-Healing Hydrogels. Adv. Mater 2015, 27, 5075–5079. [DOI] [PubMed] [Google Scholar]
- (10).Hinton TJ; Jallerat Q; Palchesko RN; Park JH; Grodzicki MS; Shue H-J; Ramadan MH; Hudson AR; Feinberg AW Three-dimensional printing of complex biological structures by freeform reversible embedding of suspended hydrogels. Sci. Adv 2015, 1, No. e1500758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Lee A; Hudson AR; Shiwarski DJ; Tashman JW; Hinton TJ; Yerneni S; Bliley JM; Campbell PG; Feinberg AW 3D bioprinting of collagen to rebuild components of the human heart. Science 2019, 365, 482–487. [DOI] [PubMed] [Google Scholar]
- (12).Song KH; Highley CB; Rouff A; Burdick JA Complex 3D-Printed Microchannels within Cell-Degradable Hydrogels. Adv. Funct. Mater 2018, 28, No. 1801331. [Google Scholar]
- (13).Bhattacharjee T; Zehnder SM; Rowe KG; Jain S; Nixon RM; Sawyer WG; Angelini TE Writing in the granular gel medium. Sci. Adv 2015, 1, No. e1500655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Kolesky DB; Homan KA; Skylar-Scott MA; Lewis JA Three-dimensional bioprinting of thick vascularized tissues. Proc. Natl. Acad. Sci. U.S.A 2016, 113, 3179–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Skylar-Scott MA; Uzel SGM; Nam LL; Ahrens JH; Truby RL; Damaraju S; Lewis JA Biomanufacturing of organ-specific tissues with high cellular density and embedded vascular channels. Sci. Adv 2019, 5, No. eaaw2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Noor N; Shapira A; Edri R; Gal I; Wertheim L; Dvir T. 3D Printing of Personalized Thick and Perfusable Cardiac Patches and Hearts. Adv. Sci 2019, 6, No. 1900344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).McCormack A; Highley CB; Leslie NR; Melchels FPW 3D Printing in Suspension Baths: Keeping the Promises of Bioprinting Afloat. Trends Biotechnol. 2020, 38, 584–593. [DOI] [PubMed] [Google Scholar]
- (18).Wang H; Guo K; Zhang L; Zhu H; Li S; Li S; Gao F; Liu X; Gu Q; Liu L; Zheng X. Valve-based consecutive bioprinting method for multimaterial tissue-like constructs with controllable interfaces. Biofabrication 2021, 13, No. 035001. [DOI] [PubMed] [Google Scholar]
- (19).Romanazzo S; Molley TG; Nemec S; Lin K; Sheikh R; Gooding JJ; Wan B; Li Q; Kilian KA; Roohani I. Synthetic Bone-Like Structures Through Omnidirectional Ceramic Bioprinting in Cell Suspensions. Adv. Funct. Mater 2021, 31, No. 2008216. [Google Scholar]
- (20).Jeon O; Lee YB; Jeong H; Lee SJ; Wells D; Alsberg E. Individual cell-only bioink and photocurable supporting medium for 3D printing and generation of engineered tissues with complex geometries. Mater. Horiz 2019, 6, 1625–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Griffin DR; Weaver WM; Scumpia PO; Di Carlo D; Segura T. Accelerated wound healing by injectable microporous gel scaffolds assembled from annealed building blocks. Nat. Mater 2015, 14, 737–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Ying G; Jiang N; Parra C; Tang G; Zhang J; Wang H; Chen S; Huang NP; Xie J; Zhang YS Bioprinted Injectable Hierarchically Porous Gelatin Methacryloyl Hydrogel Constructs with Shape-Memory Properties. Adv. Funct. Mater 2020, 30, No. 2003740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Zhang YS; Davoudi F; Walch P; Manbachi A; Luo X; Dell’Erba V; Miri AK; Albadawi H; Arneri A; Li X; Wang X; Dokmeci MR; Khademhosseini A; Oklu R. Bioprinted Thrombosis-on-a-Chip. Lab Chip 2016, 16, 4097–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Yue K; Trujillo-de Santiago G; Alvarez MM; Tamayol A; Annabi N; Khademhosseini A. Synthesis, properties, and biomedical applications of gelatin methacryloyl (GelMA) hydrogels. Biomaterials 2015, 73, 254–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Jia W; Gungor-Ozkerim PS; Zhang YS; Yue K; Zhu K; Liu W; Pi Q; Byambaa B; Dokmeci MR; Shin SR; Khademhosseini A. Direct 3D bioprinting of perfusable vascular constructs using a blend bioink. Biomaterials 2016, 106, 58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Hardin JO; Ober TJ; Valentine AD; Lewis JA Microfluidic Printheads for Multimaterial 3D Printing of Viscoelastic Inks. Adv. Mater 2015, 27, 3279–3284. [DOI] [PubMed] [Google Scholar]
- (27).Mirdamadi E. Agarose Slurry as a Support Medium for Bioprinting and Culturing Freestanding Cell-Laden Hydrogel Constructs. 3D Print. Addit. Manuf 2019, 6, 158–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Burgess DJ; Carless JE Manufacture of gelatin/gelatin coacervate microcapsules. Int. J. Pharm 1985, 27, 61–70. [Google Scholar]
- (29).Li Z; Huang S; Liu Y; Yao B; Hu T; Shi H; Xie J; Fu X. Tuning Alginate-Gelatin Bioink Properties by Varying Solvent and Their Impact on Stem Cell Behavior. Sci. Rep 2018, 8, No. 8020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Othman SA; Soon CF; Ma NL; Tee KS; Lim GP; Morsin M; Ahmad MK; Abdulmaged AI; Cheong SC Alginate-gelatin bioink for bioprinting of hela spheroids in alginate-gelatin hexagon shaped scaffolds. Polym. Bull 2021, 78, 6115–6135. [Google Scholar]
- (31).Neves MI; Moroni L; Barrias CC Modulating Alginate Hydrogels for Improved Biological Performance as Cellular 3D Microenvironments. Front. Bioeng. Biotechnol 2020, 8, No. 665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Babu RJ; Annaji M; Alsaqr A; Arnold RD Animal-Based Materials in the Formulation of Nanocarriers for Anticancer Therapeutics. In Polymeric Nanoparticles as a Promising Tool for Anti-cancer Therapeutics; Kesharwani P; Paknikar KM; Gajbhiye V, Eds.; Academic Press: 2019; Chapter 15, pp 319–341. [Google Scholar]
- (33).Chen Y; Xiong X; Liu X; Cui R; Wang C; Zhao G; Zhi W; Lu M; Duan K; Weng J; Qu S; Ge J. 3D Bioprinting of shear-thinning hybrid bioinks with excellent bioactivity derived from gellan/alginate and thixotropic magnesium phosphate-based gels. J. Mater. Chem. B 2020, 8, 5500–5514. [DOI] [PubMed] [Google Scholar]
- (34).Djabourov M. Gels. In NMR and MRI of Gels; The Royal Society of Chemistry, 2020; Chapter 1, pp 1–44. [Google Scholar]
- (35).Shiinoki Y; Yano T. Viscoelastic behavior of an agar gelatin mixture gel as a function of its composition. Food Hydrocolloids 1986, 1, 153–161. [Google Scholar]
- (36).Wang M; Li W; Mille LS; Ching T; Luo Z; Tang G; Garciamendez CE; Lesha A; Hashimoto M; Zhang YS Digital Light Processing Based Bioprinting with Composable Gradients. Adv. Mater 2022, 34, No. e2107038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Li W; Wang M; Mille LS; Robledo Lara JA; Huerta V; Uribe Velazquez T; Cheng F; Li H; Gong J; Ching T; Murphy CA; Lesha A; Hassan S; Woodfield TBF; Lim KS; Zhang YS A Smartphone-Enabled Portable Digital Light Processing 3D Printer. Adv. Mater 2021, 33, No. e2102153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Skylar-Scott MA; Mueller J; Visser CW; Lewis JA Voxelated soft matter via multimaterial multinozzle 3D printing. Nature 2019, 575, 330–335. [DOI] [PubMed] [Google Scholar]
- (39).Liu X; Wang X; Zhang L; Sun L; Wang H; Zhao H; Zhang Z; Liu W; Huang Y; Ji S; Zhang J; Li K; Song B; Li C; Zhang H; Li S; Wang S; Zheng X; Gu Q. 3D Liver Tissue Model with Branched Vascular Networks by Multimaterial Bioprinting. Adv. Healthcare Mater 2021, 10, No. e2101405. [DOI] [PubMed] [Google Scholar]
- (40).Schepers A; Li C; Chhabra A; Seney BT; Bhatia S. Engineering a perfusable 3D human liver platform from iPS cells. Lab Chip 2016, 16, 2644–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Bhise NS; Manoharan V; Massa S; Tamayol A; Ghaderi M; Miscuglio M; Lang Q; Shrike Zhang Y; Shin SR; Calzone G; Annabi N; Shupe TD; Bishop CE; Atala A; Dokmeci MR; Khademhosseini A, A liver-on-a-chip platform with bioprinted hepatic spheroids. Biofabrication 2016, 8, No. 014101. [DOI] [PubMed] [Google Scholar]
- (42).Kim JH; Seol Y-J; Ko IK; Kang H-W; Lee YK; Yoo JJ; Atala A; Lee SJ 3D Bioprinted Human Skeletal Muscle Constructs for Muscle Function Restoration. Sci. Rep 2018, 8, No. 12307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Wang WY; Lin D; Jarman EH; Polacheck WJ; Baker BM Functional angiogenesis requires microenvironmental cues balancing endothelial cell migration and proliferation. Lab Chip 2020, 20, 1153–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Gillispie G; Prim P; Copus J; Fisher J; Mikos AG; Yoo JJ; Atala A; Lee SJ Assessment methodologies for extrusion-based bioink printability. Biofabrication 2020, 12, 022003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Cidonio G; Glinka M; Dawson JI; Oreffo ROC The cell in the ink: Improving biofabrication by printing stem cells for skeletal regenerative medicine. Biomaterials 2019, 209, 10–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Liu L; Shi G-P CD31: beyond a marker for endothelial cells. Cardiovasc. Res 2012, 94, 3–5. [DOI] [PubMed] [Google Scholar]
- (47).Hirano M; Huang Y; Vela Jarquin D; De la Garza Hernández RL; Jodat YA; Luna Cerón E; García-Rivera LE; Shin SR 3D bioprinted human iPSC-derived somatosensory constructs with functional and highly purified sensory neuron networks. Biofabrication 2021, 13, No. 035046. [DOI] [PubMed] [Google Scholar]
- (48).Lenschow DJ; Su GH; Zuckerman LA; Nabavi N; Jellis CL; Gray GS; Miller J; Bluestone JA Expression and functional significance of an additional ligand for CTLA-4. Proc. Natl. Acad. Sci. U.S.A 1993, 90, 11054–11058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Ohue Y; Nishikawa H. Regulatory T (Treg) cells in cancer: Can Treg cells be a new therapeutic target? Cancer Sci. 2019, 110, 2080–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Azad AK; Rajaram MVS; Schlesinger LS Exploitation of the Macrophage Mannose Receptor (CD206) in Infectious Disease Diagnostics and Therapeutics. J. Cytol. Mol. Biol 2014, 1, No. 1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Levato R; Jungst T; Scheuring RG; Blunk T; Groll J; Malda J. From Shape to Function: The Next Step in Bioprinting. Adv. Mater 2020, 32, No. 1906423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.