Significance
This study uncovered a molecular mechanism of prokineticin receptor 1 (PROKR1) in skeletal muscle development. Activation of PROKR1 was found to stimulate exercise-responsive genes, particularly NR4A2 (nuclear receptor subfamily 4 group A member 2), through Gs-mediated cAMP response element-binding protein (CREB) phosphorylation. This activation led to the enhancement of oxidative muscle fibers, resulting in improved metabolic activity and mitochondrial function in mouse and human myotubes. Conversely, mice lacking Prokr1 displayed unfavorable metabolic traits, such as decreased lean mass, impaired glucose tolerance, and reduced energy expenditure. These findings highlight the significance of the PROKR1–CREB–NR4A2 axis in augmenting oxidative muscle fiber composition and overall metabolic function, and offer promising prospects for treating metabolic disorders and muscular diseases.
Keywords: PROKR1, NR4A2, oxidative muscle fiber
Abstract
Metabolic disorders are characterized by an imbalance in muscle fiber composition, and a potential therapeutic approach involves increasing the proportion of oxidative muscle fibers. Prokineticin receptor 1 (PROKR1) is a G protein–coupled receptor that plays a role in various metabolic functions, but its specific involvement in oxidative fiber specification is not fully understood. Here, we investigated the functions of PROKR1 in muscle development to address metabolic disorders and muscular diseases. A meta-analysis revealed that the activation of PROKR1 upregulated exercise-responsive genes, particularly nuclear receptor subfamily 4 group A member 2 (NR4A2). Further investigations using ChIP-PCR, luciferase assays, and pharmacological interventions demonstrated that PROKR1 signaling enhanced NR4A2 expression by Gs-mediated phosphorylation of cyclic adenosine monophosphate (cAMP) response element-binding protein (CREB) in both mouse and human myotubes. Genetic and pharmacological interventions showed that the PROKR1–NR4A2 axis promotes the specification of oxidative muscle fibers in both myocytes by promoting mitochondrial biogenesis and metabolic function. Prokr1-deficient mice displayed unfavorable metabolic phenotypes, such as lower lean mass, enlarged muscle fibers, impaired glucose, and insulin tolerance. These mice also exhibited reduced energy expenditure and exercise performance. The deletion of Prokr1 resulted in decreased oxidative muscle fiber composition and reduced activity in the Prokr1–CREB–Nr4a2 pathway, which were restored by AAV-mediated Prokr1 rescue. In summary, our findings highlight the activation of the PROKR1–CREB–NR4A2 axis as a mechanism for increasing the oxidative muscle fiber composition, which positively impacts overall metabolic function. This study lays an important scientific foundation for the development of effective muscular-metabolic therapeutics with unique mechanisms of action.
Muscular disorders are medical conditions that affect muscles in the body, and they can be caused by various factors, including genetics, infections, aging, and metabolic disorders (1). Cachexia, sarcopenia, and metabolic myopathies are some examples of muscular disorders that can lead to symptoms such as muscle weakness, wasting, low blood sugar levels, high levels of lactic acid in the blood, and metabolic acidosis (2, 3). Current treatments for muscular disorders aim to manage the symptoms and slow the progression of the condition. However, there are currently no disease-modifying therapeutics that can halt or reverse the progression of most muscular disorders.
Metabolic disorders, such as type 2 diabetes and obesity, are characterized by reduced oxidative muscle fibers and increased glycolytic fibers, causing insulin resistance and poor glucose tolerance (4–6). While there are currently treatments to alleviate symptoms, the prevalence of metabolic disorders continues to grow rapidly (7). The COVID-19 pandemic has highlighted the importance of addressing underlying diseases. Approximately 30% of patients who die of COVID-19 have diabetes. Additionally, overweight and obese patients are also reported to be infected with the virus and have a poor prognosis (8, 9). To reduce the prevalence of metabolic disorders and increase resistance to future infections, one breakthrough approach could be to increase the oxidative muscle fiber composition in muscle.
Prokineticin receptor 1 (Prokr1) is a G protein–coupled receptor (GPCR) that plays a critical role in various metabolic functions, primarily in peripheral tissues, such as fat, blood vessels, and the heart (10–12). Activation of Prokr1 through natural ligands, such as prokineticin 2 (PROK2), has been shown to inhibit adipocyte proliferation and differentiation and is downregulated in adipose tissue from mice with diet-induced obesity. Prokr1 also stimulates capillary development in endothelial cells, insulin-stimulated glucose uptake, and cardiomyocytic ventricular activity by promoting angiogenesis. Additionally, Prokr1 activates the phosphoinositide 3-kinase/Ak strain transforming/Protein kinase B (PI3K/AKT) signaling pathway to increase Glut4 translocation and improve insulin resistance in skeletal muscle cells (13). Despite these findings, the precise functions of Prokr1 in skeletal muscle physiology, particularly in relation to its role in oxidative muscle fiber specification, remain unclear.
In this study, we investigated the PROKR1–CREB–NR4A2 (cAMP response element-binding protein–nuclear receptor subfamily 4 group A member 2) axis and its effect on oxidative muscle fiber specification. The PROKR1 signaling pathway induced NR4A2 via CREB phosphorylation, resulting in the acquisition of oxidative muscle fiber properties. It also revealed that the loss-of-function of Prokr1 worsened the metabolic phenotype of the mice by reducing energy expenditure and exercise performance. These mice also had a shift in muscle fiber composition to glycolytic fibers by inactivating the Prokr1 signaling activity, which was restored by AAV-mediated Prokr1 rescue. These findings highlight the crucial role of the PROKR1–CREB–NR4A2 pathway in regulating oxidative muscle fiber composition and may lead to the development of effective therapeutic strategies for metabolic disorders.
Results
PROKR1 is Associated with Exercise-Responsive NR4A2 Gene Expression.
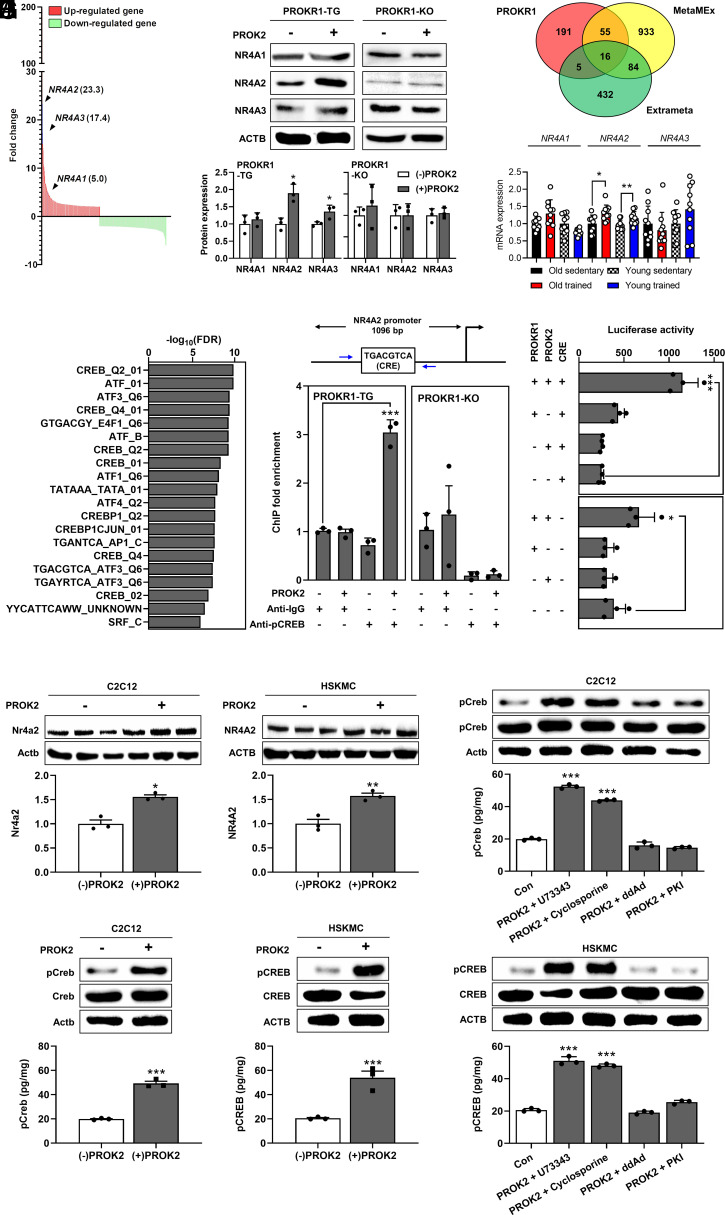
In a previous study, we identified 269 genes that were upregulated and 309 genes that were downregulated by PROKR1 activation (13). Among the up-regulated genes, nuclear receptor subfamily 4 group A members 1, 2, and 3 (NR4A1, NR4A2, and NR4A3) were identified (Fig. 1A and Dataset S1). These genes are well known as exercise-responsive transcription factors (TFs). To verify the PROKR1-dependent regulation of the NR4A family genes, their mRNA and protein levels were measured before and after PROK2-induced PROKR1 activation in HEK293T cells overexpressing or deficient in PROKR1. In PROKR1-overexpressing cells, the basal mRNA expression level of NR4A2 was markedly higher than that of NR4A1 and NR4A3, and PROK2 treatment significantly increased the mRNA level of NR4A2 by more than 9.5-fold (P < 0.001). In contrast, PROK2 treatment did not alter the mRNA expression levels of NR4A family genes in PROKR1-deficient cells (SI Appendix, Fig. S1). In line with the mRNA levels, PROK2 treatment significantly increased the protein expression levels of NR4A2 and NR4A3 by 1.9- and 1.4-fold, respectively, in PROKR1-overexpressing cells (P < 0.05); however, PROK2 treatment did not alter the protein levels of NR4A family genes in PROKR1-deficient cells (Fig. 1B).
Fig. 1.
Up-regulation of NR4A2 expression by PROKR1 signaling in muscle cells. (A) PROKR1-induced differentially expressed genes (DEGs). The X axis indicates each gene and the Y axis fold change for (+) PROK2 over (−) PROK2. (B) Western blotting of NR4A family genes (n = 3). (C) Meta-analysis of exercise-responsive gene expression. The red circle indicates PROKR1-induced up-regulated genes, yellow for acute aerobic exercise response genes from MetaMEx, and green for acute exercise response genes from Extrameta. (D) Age-independent mRNA expression of NR4A2 (n = 10). (E) TF binding motif analysis. The X axis indicates –log10(FDR); the Y axis indicates ranks the FDR values lowest to highest. (F) ChIP-PCR analysis of the pCREB binding site in HEK293T cells. The NR4A2 gene structure and CRE sequence are shown in the upper panel of the figure. The blue arrow indicates the primer binding site for ChIP-PCR. TG means PROKR1 overexpression, KO means PROKR1 deficiency (n = 4). Fold enrichment is relative to anti-IgG control. (G) CRE-dependent luciferase activity in HEK293T was measured at 24 h post-transfection and expressed as the fold change (n = 4). In the Left panel of the figure, PROKR1+/− indicates PROKR1 overexpression or knock-out, PROK2+/− for PROK2 treatment or no treatment, and CRE+/− for the presence or absence of CRE, respectively. (H) Western blotting of the PROKR1-induced NR4A2 gene in differentiated C2C12 and HSKMC cells (n = 3). (I) Western blotting and ELISA of PROKR1-induced CREB phosphorylation in differentiated C2C12 and HSKMC cells (n = 3). (J) Western blotting and ELISA of Gs-biased CREB phosphorylation with U7 (U73343, PLCβ inhibitor, 10 μM), CsA (Cyclosporine A, calcineurin inhibitor, 1.5 μM), ddAd (adenylate cyclase inhibitors, 10 μM), and PKI (PKC inhibitor, 10 μM) in differentiated C2C12 and HSKMC cells (n =3). Values are mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, unpaired two-tailed Student t test (B, D, H, and I). *P < 0.05, ***P < 0.001, one-way ANOVA followed by Dunnett’s post-hoc analysis (F, G, and J).
Next, to verify whether the PROKR1-induced up-regulated genes are exercise-responsive genes, a meta-analysis was conducted using human exercise-responsive gene sets. From MetaMEx (14) and Extrameta (15), 1,088 and 537 acute (aerobic) exercise-responsive genes in skeletal muscle tissue were collected, respectively. These genes were compared to the genes upregulated by PROKR1. There were 16 genes that were found to be common between the groups, and three members of the NR4A family were among these common genes (Fig. 1C and Dataset S2). We collected the exercise-responsive gene set according to age to determine which gene of the three members of the NR4A family was a true exercise-responsive gene independent of other confounding factors. The expression levels of NR4A2 were significantly increased before and after exercise in both younger and older healthy subjects (P < 0.05). In contrast, there was no change in NR4A1 gene expression in response to exercise, and exercise-induced up-regulation of NR4A3 was observed only in young healthy subjects (Fig. 1D). Based on these findings, it appears that NR4A2 may be associated with PROKR1, which prompted us to verify whether PROKR1 is an upstream regulator of NR4A2.
PROKR1 Induces Transcriptional Activation of NR4A2 through CREB Phosphorylation.
To determine whether NR4A2 is an effector of the PROKR1 signaling pathway, we conducted TF binding motif analysis using PROKR1-induced up-regulated genes to determine which TF regulates NR4A2 expression. Of the top 20 significantly enriched TF binding motifs (q < 3.55 × 10−7), 15 were observed as CREB and activating TF binding motifs, and most of the corresponding gene sets contained NR4A2 (Fig. 1E and Dataset S3). Chromatin immunoprecipitation (ChIP)-PCR showed that cells overexpressing PROKR1 were highly enriched with phosphorylated CREB (pCREB) by 4.7-fold in the promoter of the NR4A2 gene after PROK2 treatment (P < 0.001). However, in PROKR1-deficient cells treated with PROK2, no enrichment of pCREB was observed (Fig. 1F). In PROKR1-overexpressing cells transfected with a luciferase reporter harboring the CRE-containing promoter region of the NR4A2 gene, luciferase activity was significantly enhanced by 2.7-fold in response to PROK2 treatment (P < 0.001). However, cells transfected with the luciferase reporter into which the CRE-deleted promoter region of the NR4A2 gene was inserted did not exhibit enhanced luciferase activity, regardless of PROKR1 expression or PROK2 treatment (Fig. 1G). Therefore, these results showed that the expression of NR4A2 was regulated in a CREB-dependent manner activated by PROKR1.
It is crucial to determine whether PROKR1 phosphorylates CREB and controls the expression of NR4A2 in muscle cells to determine the role of PROKR1 as an exercise-mimicking factor. We differentiated mouse (C2C12) and human myoblasts (HSKMC) into myotubes and observed the expression levels of NR4A2 by PROK2 treatment. In both species of myotubes, PROK2 significantly increased the protein level by 1.6-fold (P < 0.05) (Fig. 1H). The phosphorylation level of CREB by PROK2 was significantly increased more than 2.5-fold in both myotubes (P < 0.001) (Fig. 1I). Furthermore, the pCREB level was increased transiently at 5 min of PROK2 treatment and then returned to the basal level, and NR4A2 expression was increased from 4 to 8 h of PROK2 treatment (SI Appendix, Fig. S2). As a result, it was established in muscle cells that the regulation of NR4A2 expression is dependent on CREB that has been activated by PROKR1. We then performed pharmacological intervention of Gs or Gq signaling molecules to determine which G protein in myotubes is involved in CREB phosphorylation by PROKR1. The increase in pCREB levels induced by PROK2 treatment in mouse and human myotubes was significantly inhibited by treatment with adenylyl cyclase (ddAd) and Protein kinase A (PKA) (PKI) inhibitors (P < 0.001). These inhibitors are known to act as downstream effectors of the Gs signaling pathway. In contrast, the pCREB level was not affected by treatment with PLCβ (U73343) or calcineurin (CsA) inhibitors, which are downstream effectors of the Gq signaling pathway (Fig. 1J). These findings provide strong evidence for the conclusion that PROKR1-mediated activation of CREB is predominantly biased toward the Gs signaling pathway. These results suggest that PROKR1 regulates NR4A2 expression in a Gs-PKA-CREB-dependent manner in mouse and human myotubes. In a subsequent study, the substantial effect of NR4A2 regulated by PROKR1 on the myotube phenotype was evaluated.
Skeletal Muscle Cells Acquired Oxidative Muscle Fiber Properties by PROKR1-Induced NR4A2.
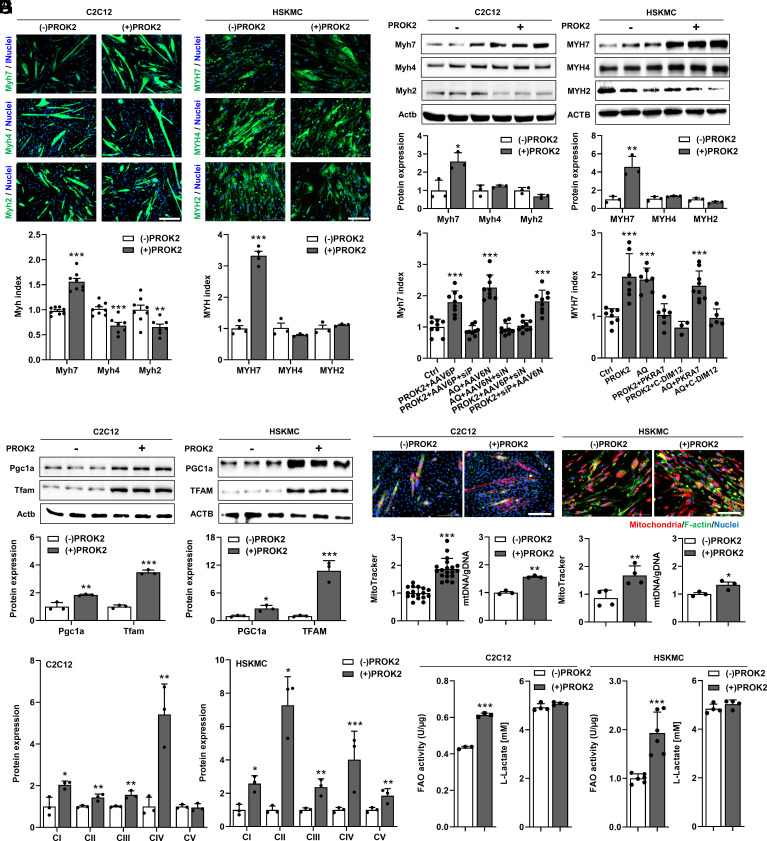
Activation of PROKR1 through PROK2 treatment induced oxidative myotube-specific differentiation of mouse and human myocytes. Immunocytochemistry demonstrated a significant increase in myosin heavy chain 7 (MYH7), an oxidative muscle marker, in both PROK2-treated myotubes (Fig. 2A). These results were supported by western blotting. The protein level of MYH7 was increased by 2.6-fold (P < 0.05) and 4.6-fold (P < 0.01) in mouse and human myotubes, respectively, while the expression of the glycolytic muscle markers MYH2 and MYH4 was not changed (Fig. 2B). Increased expression of oxidative muscle fiber marker genes by PROK2 was consistently observed at the mRNA level (SI Appendix, Fig. S3). These results suggest that PROKR1 plays a role in oxidative muscle fiber specification.
Fig. 2.
PROKR1-induced oxidative muscle fiber specification. (A) Immunocytochemistry of oxidative muscle fiber markers and glycolytic muscle fiber marker. The green fluorescence indicates MYH7, MYH4, and MYH2 in differentiated C2C12 (n = 8) and HSKMC cells (n = 4). Nuclei in all groups are stained in blue with DAPI. (Scale bar is 200 μm.) (B) Western blotting of oxidative muscle fiber and glycolytic muscle fiber marker in differentiated C2C12 and HSKMC cells (n = 3). (C) Genetic intervention of the Prokr1–Nr4a2 axis with AAV6P (AAV6-PROKR1), siP (siRNA-PROKR1), AAV6N (AAV6-NR4A2), and siN (siRNA-NR4A2) in differentiated C2C12 cells (n = 9). (D) Chemical intervention of the PROKR1–NR4A2 axis with AQ (Amodiaquine, NR4A2 agonist, 10 μM), PKRA7 (PROKR1 antagonist, 2 μM), and C-DIM12 (NR4A2 antagonist, 10 μM) in differentiated HSKMC cells (n = 3 to 9). (E) Western blotting of mitochondria biogenesis factors in differentiated C2C12 and HSKMC cells (n = 3). (F) Quantification of mitochondria after staining with Phallodin-488 (300 nM, green), MitoTracker (200 nM, red), nuclei in all groups are stained in blue with DAPI (n = 17 to 19). Mitochondrial DNA (mtDNA) was carried out using nuclear DNA (gDNA) as a standard through qPCR (n = 3) in differentiated C2C12 and HSKMC cells. (Scale bar is 200 μm.) (G) Western blotting of oxidative phosphorylation complexes in differentiated C2C12 and HSKMC cells (n = 3). (H) Metabolic shift in myofibers by assessing fatty acid oxidation (FAO) activity and L-lactate in differentiated C2C12 and HSKMC cells (n = 3 to 4). Values are mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, unpaired two-tailed Student t test (A, B, E, F, G, and H). ***P < 0.001, one-way ANOVA followed by Bonferroni’s post-hoc analysis (C and D). Source data are provided as a Source Data file.
We then asked about the role of NR4A2 in PROKR1-induced oxidative muscle fiber specification. To this end, Prokr1 and Nr4a2 expression was genetically interrupted. AAV6 was used as an effective adeno-associated virus (AAV) serotype in mouse myotubes (SI Appendix, Fig. S4), and the knockdown condition was optimized by combinatorial treatment with siRNAs (SI Appendix, Fig. S5). The results showed that Prokr1 overexpression increased Myh7 protein levels in mouse myotubes. However, knocking down Prokr1 with siRNAs prevented this increase. Similarly, increasing Nr4a2 expression using AAV6 led to a significant increase in Myh7, which was prevented by knocking down Nr4a2 with siRNAs. In addition, when Nr4a2 was knocked down in Prokr1-overexpressing myotubes, Myh7 was not increased. In contrast, overexpression of Nr4a2 in myotubes in which Prokr1 was knocked down increased Myh7 (Fig. 2C). Since genetic intervention in combination induced excessive cytotoxicity, the same study could not be performed in human myotubes. Instead, we conducted a pharmacological intervention study on PROKR1–NR4A2 with a similar scheme. In this paired study, we used PROK2 as a PROKR1 agonist, PKRA7 as a PROKR1 antagonist, amodiaquine (AQ) as an NR4A2 activator, and C-DIM12 as an NR4A2 inhibitor. Our results showed that PROK2 treatment increased MYH7 protein levels in human myotubes. However, simultaneous treatment with PROK2 and PKRA7 did not increase MYH7 expression. Similarly, AQ treatment increased MYH7 protein levels, but AQ and C-DIM12 did not increase MYH7 expression. Furthermore, PROK2 and C-DIM12 did not increase MYH7 expression. Conversely, PKRA7 and AQ increased MYH7 levels (Fig. 2D). These genetic and pharmacological intervention studies indicate that the PROKR1–NR4A2 axis makes a substantial contribution to oxidative muscle fiber specification.
Another important characteristic of oxidative muscle fibers is the content and metabolic functionality of the mitochondria. In PROKR1-induced mouse and human myotubes, the expression of PGC1a and TFAM, which promote mitochondrial biogenesis, was significantly enhanced at both the protein and mRNA levels (Fig. 2E and SI Appendix, Fig. S6). As a consequence of these changes, PROKR1-induced myotubes had quantitatively higher amounts of mitochondria (Fig. 2F). The expression levels of mitochondrial proteins involved in oxidative phosphorylation were also significantly increased in PROKR1-induced myotubes (Fig. 2G and SI Appendix, Fig. S7). These changes led to an enhancement in the metabolic function of mitochondria, as evidenced by the increased ability of these fibers to oxidize fatty acids (Fig. 2H). Taken together, these results demonstrated that PROKR1 acts as an upstream regulator of NR4A2 in muscle and that NR4A2 induced by PROKR1 enables differentiated myotubes to acquire oxidative muscle fiber phenotypes.
Prokr1 Deletion Worsened the Metabolic Phenotype in Mice.
To investigate the genetic effect of Prokr1 on living organisms, we generated mice with different Prokr1 genotypes using CRISPR/Cas9 (SI Appendix, Fig. S8). Mice were fed a high-fat diet (HFD) at 6 wk of age for 15 wk. HFD-fed mice had increased body weight from 2 wk after feeding compared to normal chow-fed mice, and the body weight of HFD-fed Prokr1 homozygotes was significantly increased compared to wild-type animals from 11 wk of age (P < 0.001) (SI Appendix, Fig. S9). There was no difference in food consumption among these groups (SI Appendix, Fig. S10); thus, the body weight changes were considered to be caused by the Prokr1 genotype. These genetic effects were more prominent in HFD-fed male mice than in females. Therefore, subsequent phenotypic analyses were conducted focusing on HFD-fed male mice.
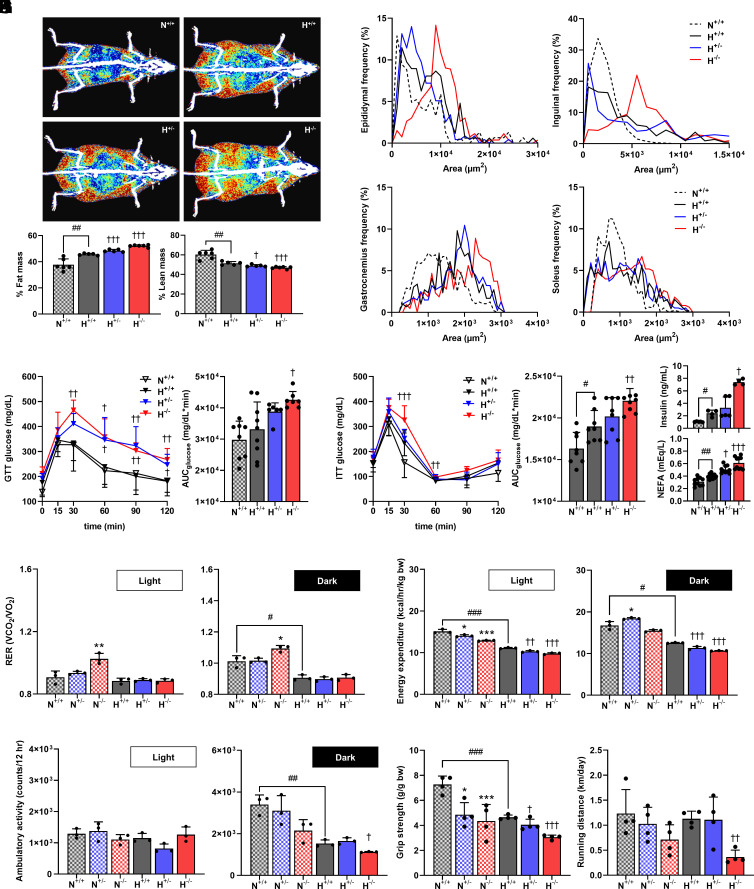
Body composition analysis showed that male mice fed the HFD developed more fat mass and less lean mass, which were inversely related to the Prokr1 gene dosage. In particular, Prokr1 homozygotes showed 6.8% higher fat mass (P < 0.001) and 4.7% lower lean mass (P < 0.05) than HFD-fed wild-type mice (Fig. 3A). Necropsy findings were consistent with the body composition analysis. The weights of epididymal and inguinal adipose tissues were increased inversely with the Prokr1 gene dosage, and the weights of gastrocnemius and soleus muscle tissues were reduced in parallel with the Prokr1 gene dosage (SI Appendix, Fig. S11). The size of epididymal adipocytes in HFD-fed wild-type mice was predominantly distributed at 2,000 μm2, but in Prokr1 heterozygotes and homozygotes, it was 4,000 μm2 and 9,000 μm2, respectively (Fig. 3B and SI Appendix, Fig. S12). The expansion of adipocyte size is a typical histological feature of obesity, and this change in size was similarly observed in inguinal adipose tissue. The size of gastrocnemius muscle fibers was significantly larger in Prokr1 heterozygotes and homozygotes, with sizes of 2,000 μm2 and 2,300 μm2, respectively, compared to the 1,800 μm2 in HFD-fed wild-type mice (Fig. 3C and SI Appendix, Fig. S13). Larger fiber size is a typical histologic feature of fast-twitch, glycolytic myofibers, and these hypertrophic changes were similarly observed in the soleus muscle tissue. HFD-fed mice showed fewer capillaries in their gastrocnemius muscle, and deleting Prokr1 further reduced capillary numbers. Similar findings were observed in the soleus muscle (SI Appendix, Fig. S14). Moreover, Prokr1 deletion significantly decreased total muscle fiber count (SI Appendix, Fig. S15).
Fig. 3.
Effect of Prokr1 genotype on metabolic phenotype, energy expenditure, and exercise in mice. (A) Representative DEXA (dual-energy X-ray absorptiometry) images showing the distribution of body fat and muscle percentage (n = 6). (B) Frequency distribution of adipocyte sizes from the hematoxylin and eosin (H&E) sections (n = 6). (C) Frequency distribution of myofiber sizes from the H&E sections (n = 6). (D) Glucose tolerance (n = 6 to 7). (E) Insulin tolerance (n = 8 to 9). The area under the curve (AUC) was calculated and shown as bar graphs in the Right panels. (F) Blood chemistry including fasting plasma insulin (n = 4 to 5) and fasting plasma NEFA (n = 10). (G) Averages of RER in light and dark cycles were calculated and are shown as bar graphs in the down panels (n = 3). (H) Averages of EE in light and dark cycles were calculated and are shown as bar graphs in the down panels (n = 3). (I) Averages of ambulatory activity in light and dark cycles was calculated and shown as bar graphs (n = 3). (J) Forelimb grip strengths normalized against lean body (n = 4). (K) Running distance measured using volunteer wheel running (n = 4). N means normal chow, H for high-fat diet, +/+ for Prokr1 wild-type mice, +/− Prokr1 heterozygous knock-out mice, −/− for Prokr1 homozygous knock-out mice. Values are mean ± SD. #P < 0.05, ##P < 0.01 vs. N+/+, #P < 0.05, ##P < 0.01 vs. H+/−, unpaired two-tailed Student t test between two groups (G–K). *P < 0.05, **P < 0.01, ***P < 0.001 vs. H+/+, one-way ANOVA followed by Bonferroni’s post-hoc analysis (A and D–K). Source data are provided as a Source Data file.
The Prokr1 genotype affected glucose metabolism in mice. Glucose tolerance in HFD-fed mice was impaired by Prokr1 gene dosage, and the glucose area under the curve in Prokr1 homozygotes was significantly increased by 22.1% compared with HFD-fed wild-type mice (P < 0.05) (Fig. 3D). These mice also had insulin insensitivity in a Prokr1 gene dosage–dependent manner (P < 0.01) (Fig. 3E). Blood chemistry showed that the circulating insulin and non-esterified fatty acids (NEFA) levels were elevated in mice depending on the Prokr1 gene dosage. Compared to HFD-fed wild-type mice with insulin concentrations of 3.3 ng/mL and NEFA concentrations of 0.4 mEq/L, Prokr1 homozygotes had significantly higher insulin and NEFA concentrations of 6.3 ng/mL (P < 0.01) and 0.6 mEq/L (P < 0.001), respectively (Fig. 3F). These results suggest that Prokr1 gene dosage may affect various metabolic parameters, including body composition changes. In particular, the quantitative and qualitative changes in muscle tissue caused by Prokr1 gene reduction prompted us to investigate the energy consumption and exercise performance of the mice.
Prokr1 Deletion Decreased Energy Expenditure and Exercise Performance in Mice.
We measured respiratory quotients to assess which fuel is metabolized as the main energy source depending on the genotype of Prokr1. Animals fed normal chow exhibited typical diurnal oscillatory respiratory exchange ratios (RERs). The range of the RER changed in correlation with the Prokr1 gene dosage. In particular, Prokr1 homozygotes showed an RER of 1.0 or more in both light and dark phases, which was significantly higher than that for the HFD-fed wild-type animals (P < 0.01). However, the RER of HFD-fed animals remained relatively constant at approximately 0.9 throughout the day, and no effect of the Prokr1 genotype was observed (Fig. 3G and SI Appendix, Fig. S16). These results suggest that Prokr1-deficient mice metabolize carbohydrates as their main energy source under normal diet conditions. Energy expenditure was more strongly influenced by the Prokr1 genotype. When fed normal chow in the light phase, Prokr1 heterozygotes and homozygotes showed energy expenditure rates of 14.0 and 12.9 kcal/h/kg bw (P < 0.05), respectively, both of which were significantly lower than that of wild-type animals (15.1 kcal/h/kg). A similar effect of the Prokr1 genotype was observed in HFD-fed animals, with energy expenditure rates of Prokr1 heterozygotes and homozygotes at 10.3 kcal/h/kg (P < 0.05) and 9.8 kcal/h/kg (P < 0.01), respectively, both of which were significantly lower than that of wild-type animals (11 kcal/h/kg). The Prokr1 gene dosage–dependent reduction in energy expenditure was not affected by diurnal oscillations (Fig. 3H and SI Appendix, Fig. S16).
The Prokr1 genotype affected exercise performance in mice. Regardless of the diet type, the ambulatory activity of animals during the dark phase was found to decrease in a Prokr1 gene dosage–dependent manner. For instance, Prokr1 homozygotes fed normal chow exhibited a significant reduction in ambulatory activity, with counts at 2,149/12 h, compared to the 3,398/12 h for wild-type animals (P < 0.05). Similarly, HFD-fed Prokr1 homozygotes also exhibited a significantly lower level of ambulatory activity, with counts at 1,126/12 h, compared to the 1,531/12 h for wild-type animals (P < 0.05) (Fig. 3I). Additionally, grip strength was found to decrease according to the Prokr1 genotype, as Prokr1 homozygotes exhibited lower grip strength than wild-type animals (P < 0.01) (Fig. 3J). Furthermore, the running distance of mice was significantly affected by the Prokr1 genotype. For instance, HFD-fed Prokr1 homozygotes ran a distance of 0.4 km/d. This was significantly shorter than that for wild-type animals, which was 1.1 km/d (P < 0.05) (Fig. 3K). In Prokr1 knock-out mice, there were no significant differences in heart structure and function compared to the wild-type mice. Diastolic blood pressure was lower but within the normal range, indicating no cardiac dysfunction (SI Appendix, Fig. S17). These findings suggest that Prokr1 plays a critical role in the regulation of energy metabolism and exercise performance in mice and that its effects may be mediated through changes in skeletal muscle properties.
Prokr1 Deletion Reduced Oxidative Muscle Fiber Composition in Mice.
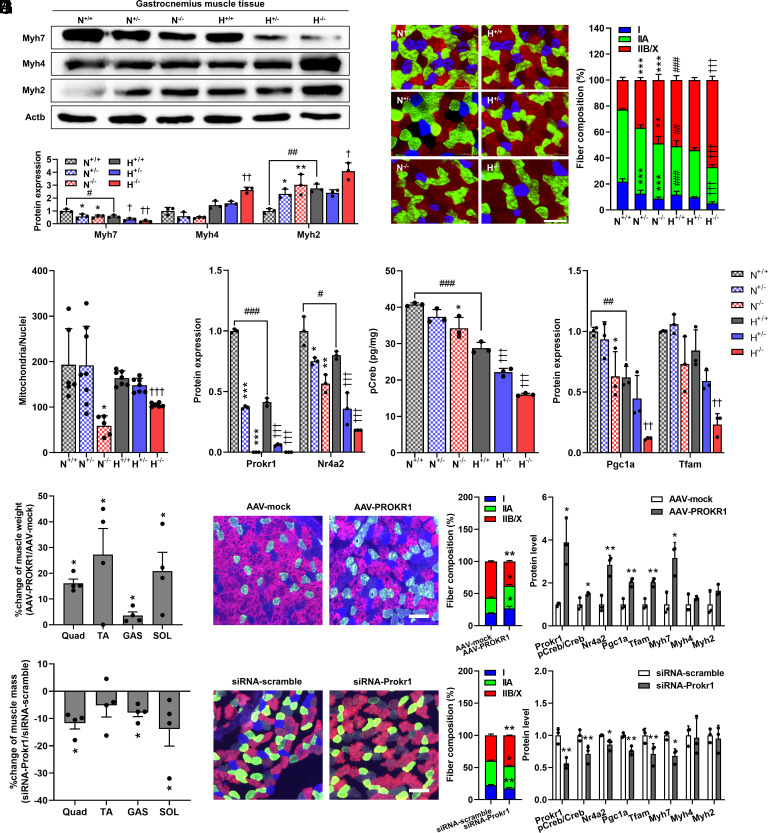
The effect of the Prokr1 genotype on muscle fiber composition was investigated. Western blotting revealed that the Prokr1 genotype had a significant effect on muscle fiber composition. In both the gastrocnemius and soleus muscles, there was a decrease in Myh7 protein levels and an increase in Myh4 and Myh2, which was inversely proportional to the Prokr1 gene dosage (Fig. 4A and SI Appendix, Fig. S18A). The effect of the Prokr1 genotype was more pronounced in HFD-fed animals. For instance, in the gastrocnemius muscle of HFD-fed Prokr1 homozygotes, there was a 2.9-fold decrease in Myh7 protein levels (P < 0.01), and Myh4 and Myh2 levels increased more than 1.5-fold compared to wild-type animals (P < 0.01). Similarly, in the soleus muscle of HFD-fed Prokr1 homozygotes, there was an 8.7-fold decrease in Myh7 protein levels (P < 0.01), and Myh4 and Myh2 levels increased more than 1.8-fold when compared to wild-type animals. These results were supported by histological analysis of muscle tissues. In the gastrocnemius muscle of normal chow-fed Prokr1 homozygotes, there was a significant decrease of 12.9% (P < 0.001) in the proportion of MyHC-I-positive muscle fibers and a significant increase of 26.1% (P < 0.001) in MyHC-IIB-positive muscle fibers compared to the wild-type animals (Fig. 4B). Similarly, in HFD-fed animals, there was a significant decrease of 6.8% (P < 0.001) in the proportion of MyHC-I-positive muscle fibers in Prokr1 homozygotes and a significant increase of 16.1% (P < 0.001) in MyHC-IIB-positive muscle fibers. In the soleus muscle of HFD-fed animals, a significant decrease of 7.8% (P < 0.001) in MyHC-I-positive muscle fibers was observed in Prokr1 homozygotes, while MyHC-IIB-positive muscle fibers increased by 15.2% (P < 0.001) compared to the wild-type animals (SI Appendix, Fig. S18B). Furthermore, the analysis of the fiber size by fiber type also demonstrated that the deletion of Prokr1 significantly increased the size of type II fibers (revised SI Appendix, Fig. S19). Overall, the results showed that the genotype of Prokr1 was highly correlated with changes in muscle fiber composition, with a Prokr1 gene dose–dependent decrease in oxidative muscle fibers and increase in glycolytic muscle fibers, particularly under HFD conditions.
Fig. 4.
Effect of Prokr1 on muscle fiber composition and signaling activity in the gastrocnemius muscle of male mice. (A) Western blotting and quantification of muscle fiber markers (n = 3). (B) Immunofluorescence staining and quantification of MyHC-I (blue), MyHC-IIA (green), and MyHC-IIB/X (red/black) proteins (n = 5). (Scale bar is 100 μm.) (C) MitoTracker staining and quantification of mitochondrial mass (n = 5 to 7). (D) Western blotting and quantification of Prokr1 signaling proteins (n = 3). Values are mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 vs. N+/+, †P < 0.05, ††P < 0.01, †††P < 0.001 vs. H+/+, one-way ANOVA followed by Dunnett’s post-hoc analysis, #P < 0.05, ##P < 0.01, ###P < 0.001 vs. N+/+, unpaired two-tailed Student t test. (E) Muscle weight changes after Prokr1 recovery (n = 4). Quad stands for quadriceps, TA for tibialis anterior, GAS for gastrocnemius, and SOL for soleus muscle. (F) Immunofluorescence staining and composition of MyHC-I (blue), MyHC-IIA (green), or MyHC-IIB/X (red/black)-positive fibers (n = 4). (Scale bar is 100 μm.) (G) Quantification of Prokr1 signaling proteins and muscle fiber markers (n = 3). (H) Muscle weight changes after Prokr1 knock-down (n = 4). Quad stands for quadriceps, TA for tibialis anterior, GAS for gastrocnemius, SOL for soleus muscle. (I) Immunofluorescence staining and quantification of MyHC-I (blue), MyHC-IIA (green), and MyHC-IIB/X (red/black) proteins (n = 4). (Scale bar is 100 μm.) (J) Quantification of Prokr1 signaling proteins and muscle fiber markers (n = 3). Values are mean ± SD. *P < 0.05, **P < 0.01 vs. AAV-mock or siRNA-scramble controls, unpaired two-tailed Student t test.
Mitochondrial amount was significantly reduced in a Prokr1 gene dosage–dependent manner, as evidenced by mitochondria-specific staining (SI Appendix, Fig. S20). In the gastrocnemius muscle, a 3.3-fold reduction was observed in normal chow-fed Prokr1 homozygotes compared to wild-type animals (P < 0.01). In HFD-fed Prokr1 homozygotes, a 1.6-fold reduction was also observed (P < 0.01) (Fig. 4C). Similarly, in the soleus muscle, Prokr1 homozygotes exhibited a more than 3.3-fold reduction compared to wild-type animals, regardless of diet type (P < 0.001) (SI Appendix, Fig. S18C). A decrease in mitochondrial amount according to the Prokr1 genotype was accompanied by a decrease in the protein levels of the mitochondrial respiratory chain complex (SI Appendix, Figs. S21and S22). These findings suggest that the Prokr1 genotype plays an essential role in determining muscle fiber composition and mitochondrial content. In particular, the deletion of Prokr1 appears to shift oxidative muscle fibers to glycolytic muscle fibers.
Prokr1–CREB–Nr4a2 Pathway Inactivation Reduces Oxidative Muscle Fiber Composition.
We investigated whether the Prokr1-dependent reduction in oxidative muscle fiber composition was influenced by Prokr1 signaling activity. We measured the expression levels of Prokr1 signaling proteins, including Prokr1, pCreb, and Nr4a2, as well as the downstream targets Pgc1a and Tfam, which induce muscle fiber composition shifts. Western blotting was used to analyze the expression levels of these proteins, and we found that their expression levels in both the soleus and gastrocnemius muscles reduced as Prokr1 gene dosage increased.
In the gastrocnemius muscle, we observed a more pronounced Prokr1 genotype–dependent decrease in the expression of signal proteins under HFD conditions. Specifically, the expression level of Nr4a2 in HFD-fed Prokr1 homozygotes was significantly reduced by 4.44-fold (P < 0.001) compared to that in wild-type animals. We also observed a significant 1.8-fold decrease in the level of pCreb in these animals (P < 0.001). Moreover, the expression levels of Pgc1a and Tfam were significantly reduced by 5.2- and 3.7-fold, respectively, in HFD-fed Prokr1 homozygotes (P < 0.01) (Fig. 4D and SI Appendix, Fig. S23). In the soleus muscle tissues of HFD-fed animals, the expression of Nr4a2 decreased inversely with increasing Prokr1 gene dosage. Specifically, Prokr1 homozygotes had a significant 2.6-fold decrease in Nr4a2 expression (P < 0.01) compared to wild-type animals. The levels of pCreb also decreased in a Prokr1 genotype–dependent manner, and this decrease was independent of diet type. Prokr1 homozygotes had a significant reduction of 1.3-fold in the normal chow condition and 2.2-fold in the HFD condition (P < 0.001). In addition, the expression levels of Pgc1a and Tfam were significantly reduced by 4.6-fold and 2.8-fold, respectively (P < 0.01), in HFD-fed animals (SI Appendix, Figs. S18D and S23).
To validate that Prokr1 plays a critical role in shifts in muscle fiber composition, we introduced AAV to restore Prokr1 in the leg muscles of Prokr1-deficient mice and siRNA to induce Prokr1 knock-down in wild-type mice. One week after injecting AAV-PROKR1, all leg muscle weights of mice were significantly increased by as little as 3.6% (gastrocnemius) and as much as 28.3% (tibialis anterior) compared to AAV-mock controls (P < 0.05) (Fig. 4E). AAV-PROKR1 significantly increased type I muscle fiber composition (P < 0.05) and decreased type II fibers in the gastrocnemius muscles of Prokr1-deficient mice (Fig. 4F). Furthermore, AAV-PROKR1 significantly increased the expression of Prokr1, Creb phosphorylation, and Nr4a2 (P < 0.05). It also increased the expression of mitochondria biogenesis factors such as Pgc1a and Tfam, with a concomitant increase in the expression of Myh7 (Fig. 4G). These increases in type I muscle fiber composition and Prokr1 signaling activity were observed in both quadriceps, tibialis anterior, and soleus muscles, too (SI Appendix, Fig. S24). We also found that pCreb was significantly enriched at the Nr4a2 CRE site in Prokr1-rescued quadriceps, tibialis anterior, gastrocnemius, and soleus muscle tissues compared to the AAV-mock controls (P < 0.01) (SI Appendix, Fig. S25). In contrast, all muscle weights in the legs of siRNA-Prokr1 injected mice were reduced by a minimum of 5.2% (tibialis anterior) and a maximum of 13.8% (soleus) compared to siRNA-scramble controls (P < 0.05) (Fig. 4H). siRNA-Prokr1 also significantly reduced type I muscle fiber composition (P < 0.01) and increased type II muscle fiber (Fig. 4I). Furthermore, all Prokr1 signaling molecules were significantly reduced (P < 0.05), and the expression of Pgc1a, Tfam, and Myh7 were also down-regulated (Fig. 4J). Overall, these findings suggest that the Prokr1 genotype induces a shift in muscle fiber composition toward oxidative fibers through Prokr1 signaling activity, which regulates the expression of key downstream targets involved in this process.
Discussion
In this study, we made important observations regarding the impact of PROKR1 on oxidative muscle fiber formation. These results demonstrated that the gain-of-function of PROKR1 promotes oxidative muscle fiber-specific myogenesis, whereas the loss-of-function of PROKR1 inhibits their formation. During myogenesis, the process by which myoblasts develop into mature myocytes and subsequently fuse to form multinucleated myofibers, there is a precise orchestration of signaling pathways and TFs. This intricate regulation ensures the proper development and function of muscles (16). Myocytes exhibit an intriguing characteristic known as myocyte plasticity, enabling them to selectively differentiate into oxidative or glycolytic fibers. This flexibility allows muscle fibers to adapt and modify their metabolic and contractile properties in response to diverse stimuli, such as exercise, hormonal signals, and environmental factors (17). For instance, prolonged periods of inactivity or disuse, such as immobilization or extended bed rest, can prompt a transition from oxidative to glycolytic fibers. This shift results in a faster, less oxidative, and more glycolytic phenotype in the affected muscle fibers. Conversely, endurance exercise training can facilitate the transformation of glycolytic fibers into oxidative fibers, leading to a slower, more oxidative phenotype (18). Multiple interventions have been identified that can regulate muscle fiber composition, highlighting the importance of myocyte plasticity. Activation of Sirt6 (19), overexpression of PGC1a (20), and treatment with narciclasine (21), and betaine (22) all demonstrate the potential to promote or maintain oxidative muscle fibers. These interventions emphasize the ability of myocytes to adapt and change their fiber-type composition. Here, we have identified PROKR1 as a key determinant of muscle fiber type. PROKR1 has been shown to enable differentiated myotubes to acquire an oxidative muscle fiber phenotype in mice and human myocytes. Additionally, the deletion of Prokr1 appears to shift oxidative muscle fibers to glycolytic muscle fibers in Prokr1-deficient mice. These results highlight the significance of PROKR1 in regulating muscle fiber specification.
Exercise induces physiological adaptations and remodeling of skeletal muscle, which exhibits remarkable plasticity of myocytes in response to stimuli and produces changes in metabolic capacity and morphology (23). Skeletal muscle adaptations through this plasticity include changes in fiber type composition, mitochondrial biogenesis, angiogenesis, and regulation of energy homeostasis. This suggests that the formation of oxidative muscle fibers through exercise may eventually be beneficial to skeletal muscle, playing an important role in preventing or delaying metabolic diseases, including insulin resistance and obesity, that affect overall health (24). The molecular mechanisms underlying these adaptations include changes in protein content, enzyme activity, and activation or inhibition of specific signaling pathways that regulate transcription and translation. TFs, such as NRFs, regulate DNA binding activity and gene activity in response to exercise, and these TF-mediated changes in gene activities promote exercise-induced structural remodeling of muscle and long-term improvements in metabolic function. These intrinsic adaptations in skeletal muscle maximize mitochondrial respiratory capacity and contractile function, ultimately enhancing exercise performance, maintaining homeostasis, and improving resistance to fatigue during future exercise challenges (23). NR4As, a subgroup of NRFs consisting of three members (NR4A1, NR4A2, and NR4A3), are immediately and transiently induced by a wide range of stimuli in various tissues and cells. These stimuli include various receptors, such as GPCRs and tyrosine kinase receptors, as well as mechanical stress and exercise (25). Interestingly, we observed in this study that activation of PROKR1 increased the expression of NR4A2 and selectively enhanced oxidative muscle fiber formation. This suggests that increased PROKR1 signaling activity may have exercise-mimicking effects and provides a mechanism of action for the generation of effective exercise-mimetic drugs.
Regulation of nuclear receptors by GPCRs is an important mechanism of cellular signaling in many physiological processes. For example, the androgen receptor is regulated by the luteinizing hormone receptor through PKA-mediated phosphorylation (26). Similarly, the estrogen receptor (ER) is regulated by the G protein–coupled ER through the PI3K/AKT pathway (27). Another example involves the peroxisome proliferator–activated receptor family; this family is regulated by GPCRs, such as the free fatty acid receptor family, through calcium/calmodulin-dependent protein kinase–mediated phosphorylation (28). These intricate pathways possess significant implications for metabolic and hormonal signaling, presenting potential opportunities for therapeutic strategies targeting hormonal and metabolic dysregulation. In line with these findings, our results suggest that the activation of PROKR1 stimulates the Gs signaling pathway, leading to an increase in the phosphorylation of CREB. This upregulation of pCREB is significantly enriched in the regulatory region of the NR4A2 gene, underscoring its critical role in NR4A2 expression regulation. The NR4A2 gene is known to regulate PGC1a, a key regulator of mitochondrial biogenesis and function (29). NR4A2 and PGC1a work together to regulate downstream target genes at the transcriptional level through protein–protein interactions (30, 31). Notably, PGC1a interacts with various TFs to activate mitochondrial genes, making it an important regulator of muscle development and oxidative muscle fiber specification (32–34). Therefore, the PROKR1-NR4A2-PGC1a pathway assumes a crucial role in modulating mitochondrial biogenesis and promoting oxidative muscle fiber specification, thereby presenting potential therapeutic avenues for mitigating musculoskeletal diseases associated with mitochondrial dysfunction.
Due to the limited availability of clinical data on the loss-of-function of PROKR1, it is currently challenging to extrapolate the implications of our findings to the clinic. However, a previous study on circulating PROK2 levels and associated human diseases provided valuable insight. Specifically, the elevated level of serum PROK2 is strongly associated with increased body mass and blood lipid, glucose, and uric acid levels in patients at high risk for cardiovascular disease and their susceptibility to metabolic syndrome. In addition, the recently discovered endogenous PROKR1-specific antagonist melanocortin receptor accessory protein 2 (MRAP2) has a significant effect on anthropometric traits in humans, depending on its genotype (35). The correlation between blood levels of PROK2, genetic variants of MRAP2, and metabolic phenotypes is an interesting finding that indicates a causal relationship between PROKR1 and metabolic diseases.
In this study, we did not observe the effects of the gain-of-function of PROKR1 on metabolic phenotypes in animals. Instead, genetic and pharmacological enhancement of the PROKR1–NR4A2 axis induced rodent and human myocytes into myotubes characterized by oxidative muscle fibers. Furthermore, the results of the AAV-mediated Prokr1 rescue study in Prokr1 knock-out mice demonstrated that Prokr1 could play an important role in oxidative muscle fiber specification and raise the possibility that activation of Prokr1 may contribute to the improvement of muscle-based metabolic phenotypes. It is controversial whether the metabolic effects of PROKR1 observed in this study originate in muscle or adipose tissues. Recent studies have shown that skeletal muscle plays an important role in interorgan communication through the secretion of myokines and metabolites. These muscle-derived signals can communicate with various organs, including the liver, adipose tissue, and brain, and influence their metabolic functions (36–39). Based on our findings, it is possible that the observed systemic effects of PROKR1 are a result of the effects of skeletal muscle-derived signaling on adipose tissue. The improvement in muscle mass and physiological function through PROKR1 and the subsequent reduction in fat mass may represent a mechanism of action for the development of therapeutics for metabolic diseases, including obesity. Based on the results presented in this study, further study of the interaction between muscle and adipose tissue is worth pursuing.
In summary, activation of the PROKR1–CREB–NR4A2 axis has been shown to improve the composition and functionality of oxidative muscle fibers in skeletal muscle, resembling the effects of exercise mimetics. This leads to a favorable impact on the overall metabolic function of the body. Therefore, this study is expected to provide an important scientific foundation for the development of effective exercise mimetics in the future.
Materials and Methods
A detailed protocol of TF binding motif analysis can be found in SI Appendix, Supplementary Materials and Methods. For in vitro experiments, information on reagents and cells used, the detailed protocols of Chromatin Immunoprecipitation, PCR, Luciferase Reporter Assay, Western Blotting, Enzyme-linked Immunosorbent Assay, Immunocytochemistry, siRNA Transfection and AAV Transduction, MitoTracker Red Staining, Fatty Acid Oxidation, and Lactate Measurement are described in SI Appendix, Supplementary Materials and Methods. The animal study protocol was approved by the Institutional Animal Care and Use Committees (IACUC) at Seoul National University under the reference number SNU-220504-2-1. Detailed information on mouse strain, genetic background, sources, and breeding strategies, and protocols of Prokr1 Knock-out Mouse Generation, Dual-Energy X-ray Absorptiometry, Glucose and Insulin Tolerance Tests, Non-Esterified Fatty Acid Measurement, Indirect Calorimetry, Grip Strength, Hematoxylin and Eosin Staining, Immunohistochemistry, and Prokr 1 Rescue Experiment can be found in SI Appendix, Supplementary Materials and Methods. Detailed information on the oligonucleotides used can be found in Dataset S4.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Acknowledgments
This research was supported by Basic Science Research Program through the National Research Foundation of Korea funded by Ministry of Science and ICT (2021R1A2C1006926).
Author contributions
J.M. and J.P. designed research; J.M. and J.H.P. performed research; S.C.Y. contributed new reagents/analytic tools; J.M., J.H.P., and J.P. analyzed data; and J.M. and J.P. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All data, code, and materials used in this study are described in the manuscript and are available in Datasets S1–S4.
Supporting Information
References
- 1.Collins K. H., et al. , Obesity, metabolic syndrome, and musculoskeletal disease: Common inflammatory pathways suggest a central role for loss of muscle integrity. Front. Physiol. 9, 112 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowen T. S., Schuler G., Adams V., Skeletal muscle wasting in cachexia and sarcopenia: Molecular pathophysiology and impact of exercise training. J. Cachexia Sarcopenia Muscle 6, 197–207 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Webster J. M., Kempen L., Hardy R. S., Langen R. C. J., Inflammation and skeletal muscle wasting during cachexia. Front. Physiol. 11, 597675 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanner C. J., et al. , Muscle fiber type is associated with obesity and weight loss. Am. J. Physiol. Endocrinol. Metab. 282, E1191–E1196 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Albers P. H., et al. , Human muscle fiber type-specific insulin signaling: Impact of obesity and type 2 diabetes. Diabetes 64, 485–497 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Stuart C. A., et al. , Slow-twitch fiber proportion in skeletal muscle correlates with insulin responsiveness. J. Clin. Endocrinol. Metab 98, 2027–2036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tahrani A. A., Barnett A. H., Bailey C. J., Pharmacology and therapeutic implications of current drugs for type 2 diabetes mellitus. Nat. Rev. Endocrinol. 12, 566–592 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Guan W. J., et al. , Clinical characteristics of Coronavirus disease 2019 in China. N. Engl. J. Med. 382, 1708–1720 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simonnet A., et al. , High prevalence of obesity in severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) requiring invasive mechanical ventilation. Obesity (Silver Spring) 28, 1195–1199 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dormishian M., et al. , Prokineticin receptor-1 is a new regulator of endothelial insulin uptake and capillary formation to control insulin sensitivity and cardiovascular and kidney functions. J. Am. Heart Assoc. 2, e000411 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szatkowski C., et al. , Prokineticin receptor 1 as a novel suppressor of preadipocyte proliferation and differentiation to control obesity. PLoS One 8, e81175 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Urayama K., et al. , The prokineticin receptor-1 (GPR73) promotes cardiomyocyte survival and angiogenesis. FASEB J. 21, 2980–2993 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Mok J., et al. , Prokineticin receptor 1 ameliorates insulin resistance in skeletal muscle. FASEB J. 35, e21179 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Pillon N. J., et al. , Transcriptomic profiling of skeletal muscle adaptations to exercise and inactivity. Nat. Commun. 11, 470 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amar D., et al. , Time trajectories in the transcriptomic response to exercise - a meta-analysis. Nat. Commun. 12, 3471 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Talbot J., Maves L., Skeletal muscle fiber type: Using insights from muscle developmental biology to dissect targets for susceptibility and resistance to muscle disease. Wiley Interdiscip. Rev. Dev. Biol. 5, 518–534 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams R. S., Annex B. H., Plasticity of myocytes and capillaries: A possible coordinating role for VEGF. Circ. Res. 95, 7–8 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Memme J. M., Slavin M., Moradi N., Hood D. A., Mitochondrial bioenergetics and turnover during chronic muscle disuse. Int. J. Mol. Sci. 22, 5179 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song M. Y., et al. , Sirt6 reprograms myofibers to oxidative type through CREB-dependent Sox6 suppression. Nat. Commun. 13, 1808 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schuler M., et al. , PGC1alpha expression is controlled in skeletal muscles by PPARbeta, whose ablation results in fiber-type switching, obesity, and type 2 diabetes. Cell Metab. 4, 407–414 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Julien S. G., et al. , Narciclasine attenuates diet-induced obesity by promoting oxidative metabolism in skeletal muscle. PLoS Biol. 15, e1002597 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du J., et al. , The regulation of skeletal muscle fiber-type composition by betaine is associated with NFATc1/MyoD. J. Mol. Med. (Berl) 96, 685–700 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Egan B., Zierath J. R., Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 17, 162–184 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Guan Y., Yan Z., Molecular mechanisms of exercise and healthspan. Cells 11, 872 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pearen M. A., Muscat G. E., Minireview: Nuclear hormone receptor 4A signaling: Implications for metabolic disease. Mol. Endocrinol. 24, 1891–1903 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiong S., et al. , Effects of luteinizing hormone receptor signaling in prostate cancer cells. Prostate 75, 141–150 (2015). [DOI] [PubMed] [Google Scholar]
- 27.DeLeon C., Wang D. Q., Arnatt C. K., G protein-coupled estrogen receptor, GPER1, Offers a novel target for the treatment of digestive diseases. Front. Endocrinol. (Lausanne) 11, 578536 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hara T., Kimura I., Inoue D., Ichimura A., Hirasawa A., Free fatty acid receptors and their role in regulation of energy metabolism. Rev. Physiol. Biochem. Pharmacol. 164, 77–116 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Nervina J. M., Magyar C. E., Pirih F. Q., Tetradis S., PGC-1alpha is induced by parathyroid hormone and coactivates Nurr1-mediated promoter activity in osteoblasts. Bone 39, 1018–1025 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Holla V. R., Wu H., Shi Q., Menter D. G., DuBois R. N., Nuclear orphan receptor NR4A2 modulates fatty acid oxidation pathways in colorectal cancer. J. Biol. Chem. 286, 30003–30009 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang L., et al. , Structural basis of binding of homodimers of the nuclear receptor NR4A2 to selective Nur-responsive DNA elements. J. Biol. Chem. 294, 19795–19803 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Handschin C., Spiegelman B. M., The role of exercise and PGC1alpha in inflammation and chronic disease. Nature 454, 463–469 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taherzadeh-Fard E., et al. , PGC-1alpha downstream transcription factors NRF-1 and TFAM are genetic modifiers of Huntington disease. Mol. Neurodegener. 6, 32 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Halling J. F., Pilegaard H., PGC-1alpha-mediated regulation of mitochondrial function and physiological implications. Appl. Physiol. Nutr. Metab. 45, 927–936 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Chaly A. L., Srisai D., Gardner E. E., Sebag J. A., The melanocortin receptor accessory protein 2 promotes food intake through inhibition of the prokineticin receptor-1. elife 5, e12397 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li V. L., et al. , An exercise-inducible metabolite that suppresses feeding and obesity. Nature 606, 785–790 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mittenbuhler M. J., et al. , Isolation of extracellular fluids reveals novel secreted bioactive proteins from muscle and fat tissues. Cell Metab. 35, 535–549.e7 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuramoto K., Liang H., Hong J. H., He C., Exercise-activated hepatic autophagy via the FN1-alpha5beta1 integrin pathway drives metabolic benefits of exercise. Cell Metab. 35, 620–632.e5 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scisciola L., et al. , Sarcopenia and cognitive function: Role of myokines in muscle brain cross-talk. Life (Basel) 11, 173 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Data Availability Statement
All data, code, and materials used in this study are described in the manuscript and are available in Datasets S1–S4.