Abstract
Human T‐cell leukemia virus type 1 (HTLV‐1) establishes chronic infection in humans and induces a T‐cell malignancy called adult T‐cell leukemia‐lymphoma (ATL) and several inflammatory diseases such as HTLV‐1‐associated myelopathy/tropical spastic paraparesis (HAM/TSP). Persistent HTLV‐1 infection is established under the pressure of host immunity, and therefore the immune response against HTLV‐1 is thought to reflect the status of the disease it causes. Indeed, it is known that cellular immunity against viral antigens is suppressed in ATL patients compared to HAM/TSP patients. In this study, we show that profiling the humoral immunity to several HTLV‐1 antigens, such as Gag, Env, and Tax, and measuring proviral load are useful tools for classifying disease status and predicting disease development. Using targeted sequencing, we found that several carriers whom this profiling method predicted to be at high risk for developing ATL indeed harbored driver mutations of ATL. The clonality of HTLV‐1‐infected cells in those carriers was still polyclonal; it is consistent with an early stage of leukemogenesis. Furthermore, this study revealed significance of anti‐Gag proteins to predict high risk group in HTLV‐1 carriers. Consistent with this finding, anti‐Gag cytotoxic T lymphocytes (CTLs) were increased in patients who received hematopoietic stem cell transplantation and achieved remission state, indicating the significance of anti‐Gag CTLs for disease control. Our findings suggest that our strategy that combines anti‐HTLV‐1 antibodies and proviral load may be useful for prediction of the development of HTLV‐1‐associated diseases.
Keywords: ATL, gag, HAM/TSP, HTLV‐1, tax
Combined analysis of anti‐HTLV‐1 antibodies and proviral load can efficiently divide ATL and HAM cases into different groups. This strategy may be useful for prediction of the development of HTLV‐1‐associated diseases.
1. INTRODUCTION
Viruses that cause chronic infections possess strategies to propagate in vivo despite the pressure of host immunity. An imbalance between these viral persistence strategies and the host immune response can lead to the onset of various diseases. Human T‐cell leukemia virus type 1 (HTLV‐1) was the first human pathogenic retrovirus to be discovered. 1 It causes adult T‐cell leukemia‐lymphoma (ATL) and an inflammatory disease of the spinal cord, HTLV‐1‐associated myelopathy/tropical spastic paraparesis (HAM/TSP). 2 , 3 HTLV‐1 mainly infects CD4+ T cells in vivo and propagates by two modes in vivo: de novo infection and clonal expansion. 4 The former mechanism involves cell‐to‐cell transmission of the viral particles, while the latter increases the number of cells of each infected clone. After infection with HTLV‐1, the clonal proliferation of infected cells is limited by host immunity. However, HTLV‐1 converts the immunophenotype of infected cells to regulatory T cell (Treg) like, which helps HTLV‐1 infected cells escape host immune surveillance through expression of immunosuppressive molecules. 5 , 6 , 7 For infected people in the carrier state, the conflict between the virus and host immune system shapes the diversity of infected cells and the immune reactions of each individual. In a small subset of carriers, the balance is disrupted at some point, and HTLV‐1‐associated diseases develop; 2.6%–6% of carriers develop ATL, and approximately 0.25% of carriers in the Japanese population and approximately 1.9% of carriers in the Caribbean population develop HAM/TSP. 8 , 9
The host immune response to HTLV‐1 differs among the associated diseases. HTLV‐1 encodes structural genes, such as gag, pol, and env, in addition to regulatory genes (tax and rex) and accessory genes (p12, p13, p30, and HBZ), which have various functions in infected cells. 10 Among them, Tax is recognized as a major target of cytotoxic T lymphocytes (CTLs) because of its high immunogenicity. 11 , 12 Tax is a potent transactivator and promotes viral replication by activation of the viral promoter, the 5′ long terminal repeat (5′‐LTR). 13 , 14 However, Tax expression is faint or inactivated by genetic/epigenetic mechanisms in ATL cells, 15 , 16 , 17 , 18 , 19 suggesting that escape from host immunity is closely associated with the development of ATL. In contrast, HAM/TSP cases show such strong immune responses to HTLV‐1 that infiltration of infected cells into the central nervous system (CNS) causes chronic inflammation. 20 In addition to cellular immunity, humoral immunity against HTLV‐1 also differs between patients with different HTLV‐1‐associated diseases. While HAM/TSP patients have high levels of antibodies against HTLV‐1 antigens in their blood and CSF, 21 , 22 ATL patients have low levels of antibody to Tax. 23 Having lower levels of antibody to Tax was suggested to be a risk factor for ATL. 24 These observations suggest that immunological parameters reflect the disease status induced by HTLV‐1 infection. However, no clinically useful method to pinpoint disease status using antibody levels to each of the HTLV‐1 antigens has been established until this one.
Previous studies proposed that high proviral load (PVL) 25 and clonality of infected cells 26 are possible risk factors for ATL; however, these criteria are insufficient to reliably identify groups at high risk for ATL or HAM/TSP. In this study, we employed the luciferase immunoprecipitation system (LIPS) assay, an efficient method to quantify the antibodies to HTLV‐1 antigens, and evaluated the usefulness of antibody levels for predicting the development of ATL and HAM/TSP. Multivariate analysis using the titers of several antiviral antibodies and PVL was able to efficiently divide ATL and HAM/TSP cases into different groups, and this strategy also enabled us to identify carriers at high risk for ATL. In addition, Gag‐specific CTLs were also observed in HTLV‐1‐infected individuals, indicating that Gag protein is immunogenic. These findings suggest that the evaluation of anti‐Gag immunity is useful for the prediction of HTLV‐1‐associated diseases.
2. MATERIALS AND METHODS
Details of clinical samples, protocols of LIPS assay, PVL quantification, next‐generation sequencing, ELISPOT, experimental procedures for detection of Gag in ATL cells, and multivariate analysis are described in Appendix S1 (SI Text).
3. RESULTS
3.1. Antibody responses to human T‐cell leukemia virus type 1 proteins
The initial purpose of this study was to find a way to identify HTLV‐1 carriers at high risk for the development of associated diseases using simple biomarkers. It is thought that the immune responses to viral antigens are suppressed in ATL patients but activated in HAM/TSP patients. Therefore, some immunological biomarkers may be useful to evaluate the risk of those diseases. Previously, we and another group reported that the LIPS assay was a sensitive and easy method to detect antibodies to HTLV‐1 antigens. 27 , 28 In this study, we modified the assay and screened patients for antibodies against each of several viral proteins to identify parameters suitable for predicting disease progression. First, the presence of antibodies to three Gag proteins (p15 nucleocapsid, p19 matrix, and p24 capsid), envelope (Env), Tax, Rex, p12, p13, p30, and HBZ was evaluated using a relatively small number of samples from HTLV‐1 carriers and non‐infected subjects (Figure S1). We found that antibody responses to Gag proteins (p15, p19, p24), Env, and Tax were higher in HTLV‐1‐infected individuals than uninfected controls, whereas no increase was detected in the luminometer units (LU) for any of the other antigens in infected subjects compared with uninfected ones, probably due to low titers of antibody to these antigens. Thus, we focused our remaining studies on antibodies to Gag proteins, Env, and Tax, and determined the antibody titers to them in 263 carriers, 56 HAM/TSP patients, and 25 ATL patients (Table S1).
Different patterns of antibody responses were observed in each clinical subgroup. HAM/TSP patients had significantly higher antibody levels to all antigens except for Env than carriers (Figure 1A). In contrast, ATL patients had significantly lower antibodies to Env than carriers. Interestingly, antibodies to p19 and p24 were significantly higher in ATL patients than those in carriers, indicating that low antibody responses to Tax and Env are not simply due to the immunodeficiency induced by ATL. To track the changes in antibody responses over time in each individual, serial blood samples from seven carrier cases were analyzed by LIPS. Although the samples were collected at different times depending on the cases, the levels of antibodies to each antigen were stable and almost unchanged in all cases (Table 1).
FIGURE 1.
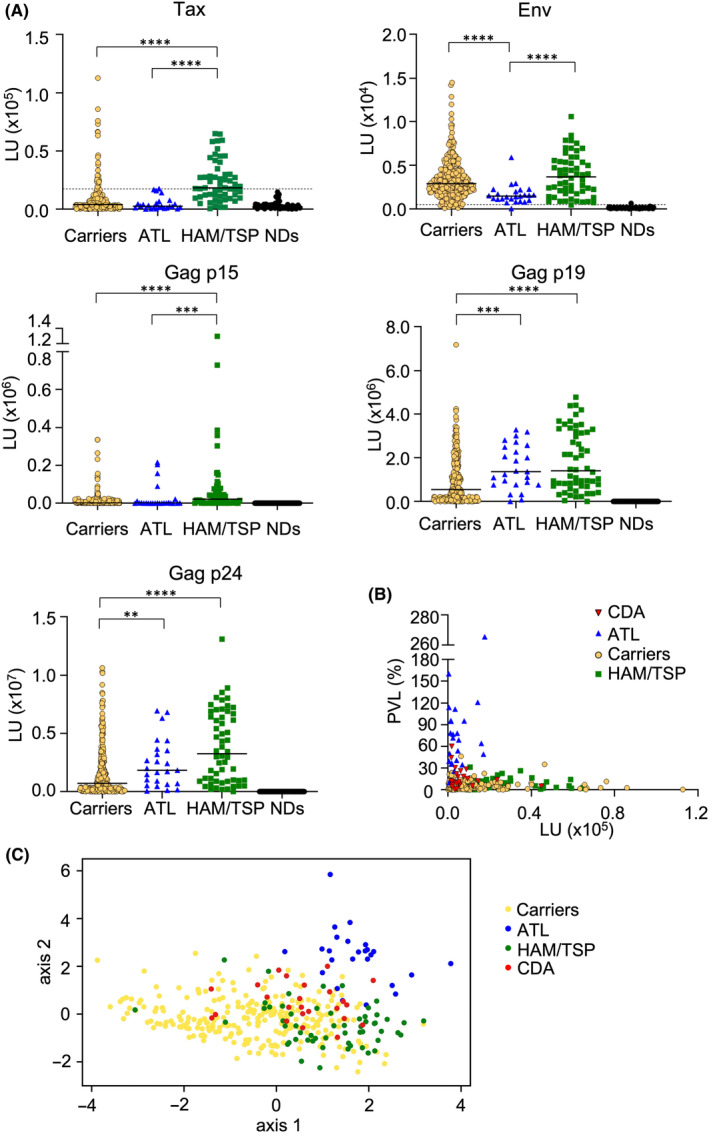
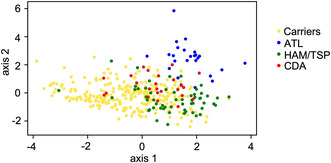
Antibody responses analyzed by luciferase immunoprecipitation system (LIPS) in human T‐cell leukemia virus type 1 (HTLV‐1) associated diseases. (A) The horizontal bar represents the median. The dotted lines are cutoff values calculated from the mean value of seronegative donors plus five standard deviations (SDs), as reported previously. 28 SDs of seronegative donors (NDs) were 2887, 75.63, 88.88, 455.6, and 1071 for Tax, Env, p15, p19, and p24, respectively. **P < 0.01, ***P < 0.005, ****P < 0.0001. (B) Tax antibody and proviral load (PVL) in carriers who developed ATL (CDA), ATL patients, HTLV‐1 carriers, and HTLV‐1‐associated myelopathy/tropical spastic paraparesis (HAM/TSP) patients. (C) PLS analysis using PVL and antibody responses to Tax, Env, and Gag. The scale of the PLS plot is a non‐dimensional quantity since horizontal and vertical axes represent integrated characteristics regarding symptoms.
TABLE 1.
Long‐term stability of antibody responses in carriers.
| Name and date | Tax (LU) | Env (LU) | Gag p15 (LU) | Gag p19 (LU) | Gag p24 (LU) | PVL (%) |
|---|---|---|---|---|---|---|
| Carrier1‐21 | 6447 | 2692 | 439 | 1,034,116 | 1,298,869 | 15.1 |
| Carrier1‐9 | 6137 | 2413 | 537 | 1,120,098 | 1,210,868 | 12.4 |
| Carrier1‐0 | 6347 | 2857 | 569 | 1,131,881 | 1,246,688 | 9.0 |
| Carrier2‐25 | 35,716 | 8497 | 706 | 2,565,449 | 5,518,157 | 22.5 |
| Carrier2‐13 | 33,634 | 7137 | 548 | 2,462,570 | 5,595,651 | 13.1 |
| Carrier2‐0 | 32,831 | 7784 | 810 | 2,267,793 | 5,167,180 | 9.5 |
| Carrier3‐22 | 18,805 | 1912 | 883 | 1,927,998 | 2,481,554 | 8.1 |
| Carrier3‐0 | 18,001 | 2030 | 771 | 2,255,472 | 2,298,831 | 4.1 |
| Carrier4‐11 | 53,202 | 7659 | 85,483 | 2,552,629 | 7,234,075 | 35.1 |
| Carrier4‐6 | 53,263 | 7718 | 95,916 | 2,608,591 | 7,623,197 | 58.0 |
| Carrier4‐0 | 53,338 | 7455 | 97,878 | 2,623,954 | 7,362,403 | 66.5 |
| Carrier5‐12 | 1833 | 3984 | 6315 | 273,786 | 521,893 | 15.0 |
| Carrier5‐0 | 1803 | 3869 | 6059 | 272,932 | 473,080 | 9.1 |
| Carrier6‐4 | 2766 | 769 | 321 | 326,841 | 4,209,567 | 9.5 |
| Carrier6‐0 | 3016 | 856 | 245 | 236,647 | 4,110,562 | 6.9 |
| Carrier7‐6 | 502 | 473 | 191 | 291,397 | 540,532 | 10.4 |
| Carrier7‐2 | 464 | 594 | 385 | 240,556 | 441,688 | 6.3 |
| Carrier7‐0 | 511 | 678 | 465 | 227,542 | 532,624 | 10.7 |
| Carrier8‐29 | 501 | 1147 | 58 | 575,385 | 2,842,144 | 15.2 |
| Carrier8‐2 | 463 | 1159 | 77 | 584,600 | 2,615,260 | 8.4 |
| ATL8‐0 | 491 | 994 | 63 | 596,678 | 2,405,165 | 6.9 |
Note: Date: 0 represents the date when the last sample was taken, and other numbers represent the number of months before the last sample was taken.
Case 8 developed ATL at time 0 after 29‐month observation.
Next, we evaluated the correlation between those antibodies and PVL (Figure S2A‐C, Table S2). In carriers, there were significant positive correlations between PVL and all antibodies (Figure S2A). In contrast, this correlation was not observed in ATL patients or HAM/TSP patients (Figure S2B,C), although it is possible that the sample sizes of these groups are too small to reveal a slight correlation. When the data from all subjects were combined, we saw that ATL and HAM/TSP were roughly divided into different populations, whereas carriers were widely distributed (Figure 1B; Figure S2D), indicating that carriers were heterogeneous. Some carriers had similar signatures to ATL or HAM/TSP. Samples from 25 carriers who later developed ATL (CDA) were additionally analyzed (Table S3). Antibody responses to viral antigens were also varied (red triangles in Figure 1B; Figure S2D).
3.2. Multivariate analysis for identifying high‐risk carriers for ATL
To develop more sensitive methods to identify high‐risk carriers, we analyzed these data by combining all the parameters. Partial least square (PLS) analysis was performed to determine how well the measured antibodies and PVL reflected the diagnosis. PLS is a variant of principle component analysis (PCA) used in medicine. PCA is a popular dimension reduction method that extracts a few useful interpretations from the whole array of measurements (antibody titers against Tax, Env, and Gag proteins and PVL) by minimizing the loss of information. PLS extends PCA by integrating diagnostic information such as HAM/TSP and ATL, enabling consideration of the association between the measurements and diagnostic information. Each diagnosis category (carrier, ATL, HAM/TSP, or CDA) formed a cluster on the plot (Figure 1C). Among several dimension reduction methods, we found that PLS performed better in cluster samples than other popular methods (as described in the Materials and Methods section). The following considerations may explain why only PLS, and not other popular methods, can achieve good clustering. First, a conventional PCA method can often face multicollinearity: highly correlated explanatory variables make it difficult to identify the explicit contribution of explanatory variables which are highly correlated. Multicollinearity is likely to occur in a clinical setting since selected biomarkers such as Gag‐p19 and Gag‐p24 often overlap biological characteristics. In several applications, PLS has been demonstrated to avoid the multicollinearity problem for analyzing metabolome datasets to which multicollinearity is inherent. 29 , 30 Second, PLS can utilize diagnostic information as a supervised machine learning method. This could be the reason why PLS outperforms nonlinear dimensionality reduction methods such as tSNE and uMAP. Using PLS analysis, HAM/TSP and ATL samples were well clustered in a two‐dimensional plane, respectively (Figure 1C). As expected, carrier samples overlap with HAM/TSP and ATL clusters, and CDA samples were located around the boundary between ATL and carriers.
We assumed carriers whose samples were located close to the ATL cluster on the two‐dimensional plane to be carriers at high risk for ATL. More specifically, the ranking of high‐risk carriers was calculated based on the distance from the centroid of CDA samples (Table S4). Based on this ranking, carriers showing antibody titers and PVLs similar to CDA were estimated to be at high risk. To verify whether these carriers were indeed at elevated risk of developing ATL, we tried to detect genetic mutations of driver genes in these individuals. Samples from eight asymptomatic carriers were analyzed; seven cases were close to the CDA cluster, and one was located in the HAM/TSP cluster in the PLS plot (Figure 2A). HTLV‐1‐infected cells (CD4+ CADM1+) were purified by cell sorting, and targeted sequencing was carried out. Five of the seven cases had somatic mutations associated with hematologic malignancies, including previously known driver mutations for ATL, such as TP73, CCR4, GPR183, and ARID2 (Figure 2A, Table 2; Table S5). 31 In contrast, no mutations were detected in two cases with lower rankings for ATL, even though one of them, KC‐30, had the highest PVL among the carriers analyzed. The median variant allele frequency of all analyzed cases was as low as 1.9%, and the clonality of infected cells in all cases was polyclonal (Figure 2A; Figure S3), suggesting that our prediction strategy has the potential to uncover carriers who are in a preleukemic stage.
FIGURE 2.
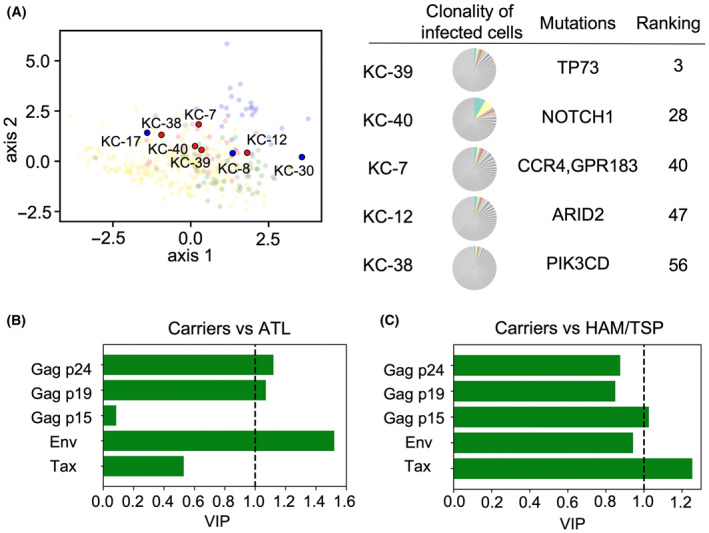
Identification of predictive markers for the development of ATL or human T‐cell leukemia virus type 1‐associated myelopathy/tropical spastic paraparesis (HAM/TSP). (A) Eight carriers were analyzed for somatic mutations of driver genes and clonality of infected cells. Red dots represent cases with at least one detected mutation, and blue dots represent cases with no detected mutation. Colored portions of pie charts represent the top 10 clones of each case. (B,C) VIP scores for prediction of ATL (B) and HAM/TSP (C). The VIP score for each variable represents the mathematical degree of contribution of that variable to the disease type (ATL or HAM/TSP). To determine whether a variable (PVL/antibody titer) is important or not based on the VIP score, a conventional criterion (threshold equal to 1) was employed (See Appendix S1 for details). Note that the VIP score represents the relative importance of an antibody's contribution to the assignment of a sample to a cluster.
TABLE 2.
Somatic mutations in carriers.
| Sample | Ranking | Gene | Type of mutation | VAF | PVL (%) | Tax | Env | Gag p15 | Gag p19 | Gag p24 |
|---|---|---|---|---|---|---|---|---|---|---|
| KC‐39 | 3 | TP73 | Nonsynonymous SNV | 0.017 | 9.5 | 2969 | 4041 | 551 | 403,674 | 3,397,985 |
| KC‐40 | 28 | NOTCH1 | Nonsynonymous SNV | 0.127 | 10.4 | 25,400 | 2001 | 1072 | 193,880 | 60,452 |
| KC‐8 | 36 | ND | 19.9 | 1649 | 7409 | 4386 | 2,312,792 | 4,837,946 | ||
| KC‐7 | 40 | CCR4 | Stopgain | 0.02 | 15.1 | 2484 | 1102 | 516 | 569,784 | 332,024 |
| GPR183 | Frameshift deletion | 0.012 | ||||||||
| KC‐12 | 47 | ARID2 | Stopgain | 0.032 | 22.5 | 17,410 | 5097 | 1540 | 2,868,154 | 3,644,769 |
| KC‐38 | 56 | PIK3CD | Nonsynonymous SNV | 0.01 | 8.9 | 248 | 2966 | 760 | 123,526 | 238,358 |
| KC‐17 | 98 | ND | 7.7 | 498 | 2313 | 419 | 104,451 | 25,882 | ||
| KC‐30 | 139 | ND | 66.5 | 52,423 | 6094 | 153,713 | 2,523,416 | 6,783,579 |
Abbreviation: ND, no mutation detected; PVL, proviral load; SNV, single nucleotide variant; VAF, variant allele frequency.
3.3. Significance of elevated anti‐Gag p19 and p24 antibodies in ATL cases
As expected, the titers of anti‐Tax and anti‐Env antibodies were lower in ATL cases compared with those of HAM/TSP patients and carriers. Unexpectedly, the titers of anti‐Gag p19 and p24 antibodies of ATL cases were higher than those of carriers, although a finding that there was no significant difference in anti‐Gag antibodies between ATL and HAM/TSP was consistent with previous reports. 32 To confirm which antibodies are important for discriminating ATL cases from carriers, we employed the variable importance in projection (VIP) scores derived from the PLS analysis. VIP scores identify antibody types that significantly contribute to the discrimination between the disease types (i.e., carrier, ATL, HAM/TSP, or CDA). A VIP score greater than one indicates an antibody type that makes an important contribution to the discrimination between disease types. (A more detailed explanation of VIP scores is given in Appendix S1: Materials and Methods.) Using VIP scores as convenient indicators for ranking antibody types, we found that high antibody titers against Gag p19, p24, and Env were important markers of ATL (Figure 2B). In contrast, elevated antibody titers to Tax and p15 appeared to be markers for HAM/TSP (Figure 2C). These findings suggest that antibody titers to Gag antigens are useful in predicting HTLV‐1‐related diseases.
3.4. Expression of Gag antigen in ATL cells
It is intriguing that the antibodies to Gag antigens have such an impact on the prediction of HTLV‐1‐associated diseases. In particular, we wondered why ATL patients had higher antibody responses against Gag p19 and p24 than carriers, even though responses to Tax and Env were lower in ATL cases (Figure 1). From those observations, we hypothesized that Gag antigens are expressed in ATL cells, even if Tax is not expressed. To detect gag transcripts, we synthesized cDNA using a gag‐specific primer and carried out quantitative RT‐PCR. As expected, gag mRNA was detected in all ATL cell lines, including Tax‐negative cell lines, although its expression level was quite low (Figure 3A). Since ED and ATL‐55 T cells have genetic mutations in the tax gene and TL‐Om1 has lost Tax expression due to DNA methylation of the 5′LTR, Tax is not expressed in these cell lines. Therefore, the gag mRNA we found in these cell lines is transcribed in a Tax‐independent manner. The gag gene transcript was also detected in fresh ATL cells (Figure 3B). There are no deletions in the provirus or mutations in the tax gene in these cases (Table S6). DNA methylation was observed in ATL‐1 and ATL‐3, consistent with the result that tax expression was repressed in those cases (Figure 3B). We also visualized gag mRNA using RNA fluorescence in situ hybridization (RNA‐FISH) in tax‐defective cells (Figure 3C,D). Expression of Gag protein in ED was confirmed by proximity ligation assay (Figure 3E,F). It is reported that the titer of antibody against the well‐known cancer‐testis antigen NY‐ESO‐1 is correlated with the expression level of the antigen's mRNA. 33 Thus, transcription of gag in leukemic cells regardless of Tax status is a possible reason for the presence of anti‐Gag antibodies in ATL cases.
FIGURE 3.
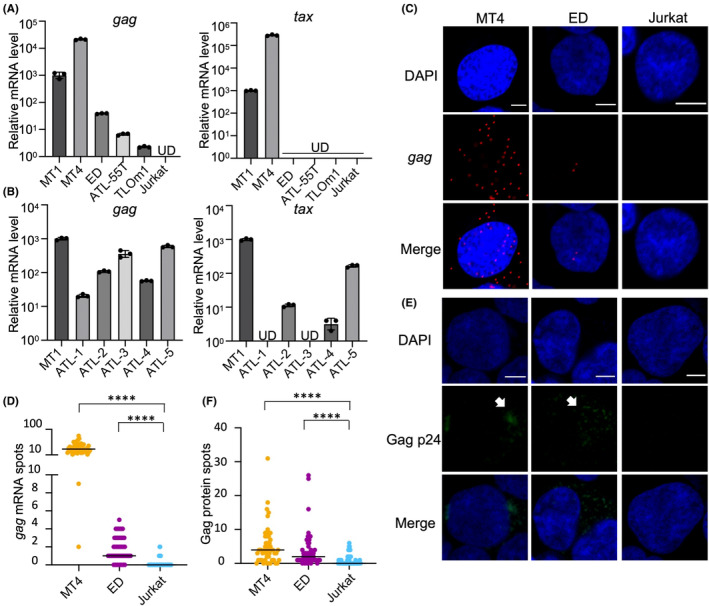
Gag expression in ATL cells. (A) RT‐qPCR of tax and gag in human T‐cell leukemia virus type 1 (HTLV‐1) infected cell lines and Jurkat cells. For quantification of gag mRNA, a no reverse transcriptase negative control was made, and gag mRNA was not detected. UD, undetected. (B) RT‐qPCR of tax and gag in primary ATL cells. All samples were obtained from ATL patients in which more than 80% of PBMCs were CD4+CADM1+, and they were confirmed to be monoclonal by inverse PCR. The amount of 18S rRNA in each sample was quantified and used as a reference to calculate the relative expression levels of tax and gag with the ddCt method. (C) RNA‐FISH of HTLV‐1 infected cell lines and Jurkat cells (scale bar, 5 μm). (D) gag mRNA spots per cell detected by RNA‐FISH in HTLV‐1‐infected cell lines and Jurkat cells. ****p < 0.0001. (E) Duolink PLA of HTLV‐1 infected cell lines and Jurkat cells (scale bar, 5 μm). Arrows indicate the signals of p24 protein. (F) Gag protein spots per cell detected by Duolink PLA in HTLV‐1 infected cell lines and Jurkat cells. ****p < 0.0001.
3.5. Cellular immunity against Gag in human T‐cell leukemia virus type 1‐infected individuals
Since HTLV‐1 propagates mainly by causing the proliferation of infected cells, cellular immunity is more critical than humoral immunity in regulating the number of infected cells. Based on the existence of anti‐Gag antibodies in ATL cases, we speculated that Gag‐specific CTLs may have a significant impact on the development of ATL. To evaluate the number and activity of CTLs to Gag antigens, we conducted ELISPOT assays using samples from patients with various clinical statuses including ATL patients who received hematopoietic stem cell transplantation (HSCT; Table S7). As shown in Figure 4, IFN‐γ‐producing cells were observed after stimulation with peptides from Gag antigens, indicating the presence of functional CTLs to those antigens. While the number of IFN‐γ‐producing cells was generally low in ATL patients and carriers, IFN‐γ‐producing cells were more common in some cases of HAM/TSP and, intriguingly, in several ATL patients who received HSCT. The two HSCT patients who had low numbers of IFN‐γ‐producing cells (HSCT‐3 and HSCT‐5) suffered clinical relapse after transplantation. These results suggest that when HSCT is successful in treating ATL, HSCT is associated with the rise of functional CTLs against Gag antigens.
FIGURE 4.
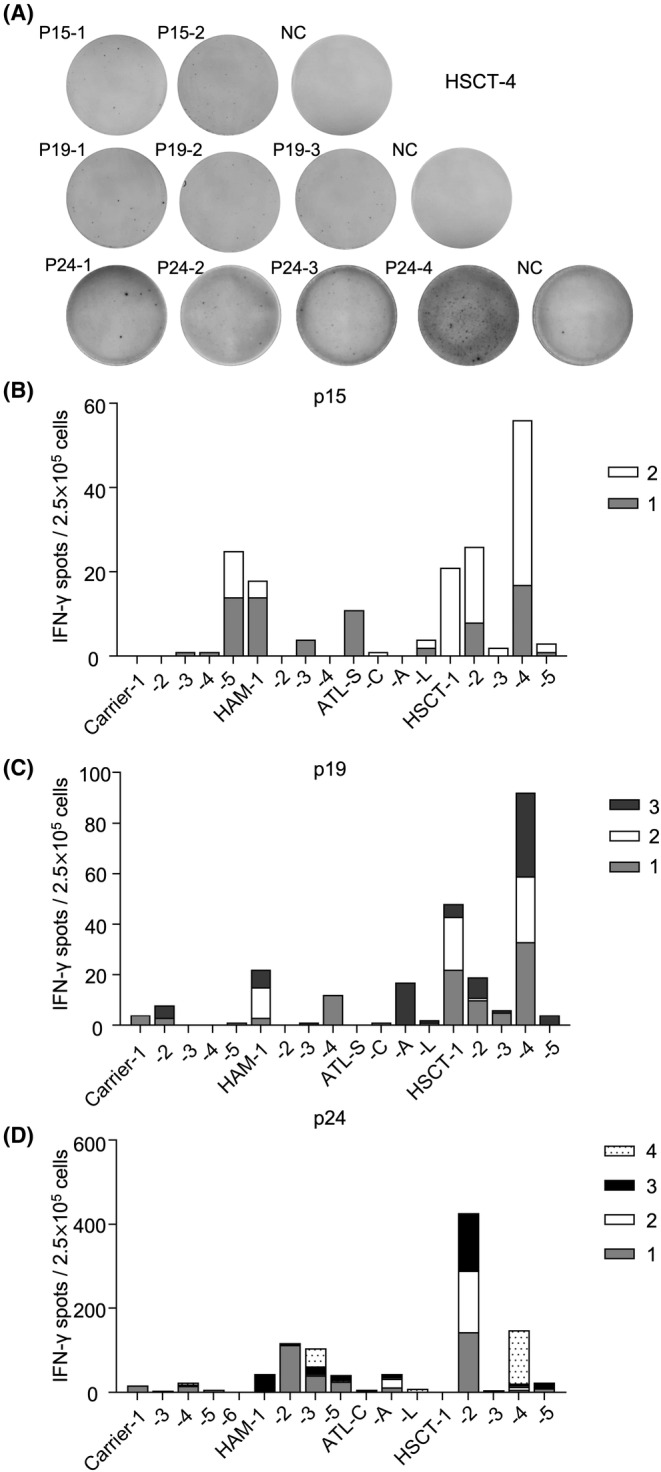
IFN‐γ ELISPOT assay for Gag. (A) Interferon‐gamma (IFN‐γ) spots of patient HSCT‐4 after stimulation with Gag antigen peptide pools. (B–D) The proportion of IFN‐γ spots in patients with various human T‐cell leukemia virus type 1 (HTLV‐1)‐associated diseases. Cells were stimulated with (B) p15 pooled peptides (1, 2), (C) p19 pooled peptides (1–3), and (D) p24 pooled peptides (1–4). The number of spots reported represents raw data minus the negative control (NC), which was stimulated without peptides. ATL‐A, acute type; ATL‐C, ATL chronic type; ATL‐L, lymphoma type; ATL‐S, smoldering type.
4. DISCUSSION
ATL shows a dismal prognosis regardless of intensive chemotherapy. Therefore, the identification of a high‐risk group among carriers is critical for successful treatment. ATL cells contain genetic changes in various pathways that are important for leukemogenesis. 31 Among these mutations, some of them are detected in the early stage of leukemogenesis. 34 , 35 However, in clinical practice, a method to identify high‐risk carriers is required without analyses of genetic changes. Here, we show that disease status can be classified by the multivariate analysis using PVL and antibodies to multiple viral antigens, such as Tax, Env, and Gag proteins. Our strategy is useful for identifying HTLV‐1 carriers at high risk for ATL at a very early stage of the disease since several driver mutations were found in the samples from individuals who are still in a polyclonal state. A previous study reported that all carriers who developed ATL had detectable mutations, while most carriers with high PVL but no detectable mutations did not develop ATL during a median of 10 years follow‐up, 34 suggesting that it is difficult to identify high‐risk cases by PVL alone. A more recent study showed that in evaluating the abundance of each HTLV‐1‐infected clone, the so‐called oligoclonality index is useful to identify a subset of HTLV‐1 carriers with high PVL at increased risk of developing ATL. 36 In the carrier state, HTLV‐1‐infected clones survive for years, and a clone that acquires malignant characteristics preferentially expands, resulting in the development of ATL. 37 Possibly our prediction method will be able to identify carriers with oncogenic mutations before the expansion of pre‐malignant clones. However, the number of the ATL cases analyzed is still small in this study. A larger number of cases and longer‐term follow‐up of the subjects will be needed to verify the usefulness of our strategy and to set the thresholds that define the risk of future disease development.
Among the antibodies to the viral antigens we analyzed, anti‐Gag antibodies were shown to be useful in predicting the development of HTLV‐1‐associated diseases. Gag is initially translated as a polyprotein and processed into three mature proteins: p19 (matrix), p24 (capsid), and p15 (nucleocapsid). Previous studies using the LIPS assay demonstrated that the titer of the antibody to full‐length Gag protein was not different between asymptomatic carriers, ATL, and HAM/TSP patients. 32 , 38 In this study, we intended to analyze the immune reaction against each of the mature Gag antigens and found that all antigens evoke both humoral and cellular immunities, which might be useful for the prediction of the disease statuses. It is noteworthy that the amount of anti‐Gag p15 was found to be an important parameter for predicting the onset of HAM/TSP. Gag precursor polyprotein is processed into each component by the viral protease. For efficient viral replication, ribosomal frameshift in the sequence of the p15‐encoding region is required to produce the Gag‐Pol polyprotein encoding the Gag proteins and viral enzymes such as the protease, reverse transcriptase, RNaseH, and integrase. 39 In our LIPS assay, the mature form of p15, which is generated by ribosomal frameshift and processing with the viral protease, was utilized as an antigen. Therefore, a high amount of anti‐p15 antibody in HAM/TSP patients might reflect the higher replication rate of the virus in those patients than in subjects with other disease statuses such as asymptomatic carriers or ATL. In sum, antibodies to these antigens appear to be promising markers for the prediction of both ATL and HAM/TSP, although more samples and different cohorts are still needed to verify this finding.
Human T‐cell leukemia virus type 1 replication is generally suppressed in vivo. Since Tax is critical for viral expression, 40 , 41 it has been thought that all viral transcripts encoded in the plus strand of the provirus are also suppressed in the absence of Tax. Tax is transiently induced in ATL cells by cellular stresses, and it functions to suppress apoptosis and promote viral replication. 42 In this study, through the profiling of antibodies against multiple viral antigens, we unexpectedly identified the expression of Gag in ATL cells. Gag proteins, which are translated from unspliced viral RNA, were observed even in Tax‐deficient ATL cells, indicating that the gene can be transcribed in a Tax‐independent manner. A previous study showed that, after ex vivo culture of ATL cells, induction of unspliced viral RNA was delayed but longer lasting than tax/rex expression, 43 suggesting that there are alternate mechanisms to regulate the expression of gag in addition to activation by Tax. Host transcription factors might be involved in this basal transcription. A recent study revealed that Yin Yang 1 (YY1) binds to viral RNA, but not DNA, and activates HTLV‐1 transcription. 44 The mechanism that triggers the initiation of viral transcription in a Tax‐independent manner is not yet understood. It is also important to consider the virological significance of Tax‐independent viral gene expression in ATL cells. It has been reported that the cis‐acting regulatory sequence (CRS) in unspliced viral RNA is pivotal in its transportation to the cytoplasm and stabilization by Rex. 45 The presence of low levels of unspliced transcripts might be an idling mechanism that allows viral replication to accelerate quickly in response to some stimulation such as cytotoxic stress.
Gag might also be a novel target for immunotherapy. Since the persistence of HTLV‐1 in vivo depends on the proliferation of infected cells rather than the replication of viral particles, cellular immunity plays an important role in regulating the abundance of the virus. Tax is recognized as the main target of CTLs, and a Tax‐targeting immunotherapy has been developed. 46 In this study, the ELISPOT assay showed that functional Gag‐specific CTLs are present in patients who respond well to HSCT. Cellular immunity against viral antigens is generally suppressed in ATL patients, and the graft versus ATL effect is thought to be important for long‐term remission after HSCT, 47 suggesting that anti‐Gag CTLs, in addition to anti‐Tax CTLs, have suppressive effects on the development and recurrence of ATL.
This study demonstrates the potential of anti‐Gag immunities as the predictive markers for HTLV‐1‐associated diseases. Evaluating the humoral immunity to Gag appears to be useful for predicting the onset of ATL and HAM/TSP, and Gag proteins have unforeseen, unexplored potential as novel targets for immunotherapy. It is hoped that further validation will lead to clinical application.
AUTHOR CONTRIBUTIONS
Asami Yamada: Conceptualization; data curation; formal analysis; methodology; writing – original draft. Jun‐ichirou Yasunaga: Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; resources; supervision; writing – original draft. Lihan Liang: Data curation; formal analysis. Wenyi Zhang: Data curation; formal analysis. Junya Sunagawa: Data curation; formal analysis; methodology; writing – original draft. Shinji Nakaoka: Data curation; formal analysis; methodology; supervision; writing – original draft. Shingo Iwami: Methodology; supervision. Yasunori Kogure: Data curation; formal analysis; methodology; writing – original draft. Yuta Ito: Data curation; formal analysis. Keisuke Kataoka: Methodology; supervision. Masanori Nakagawa: Resources. Masako Iwanaga: Resources. Atae Utsunomiya: Resources. Ki‐Ryang Koh: Resources. Toshiki Watanabe: Resources. Kisato Nosaka: Conceptualization; resources; supervision. Masao Matsuoka: Conceptualization; funding acquisition; investigation; supervision; writing – review and editing.
FUNDING INFORMATION
This work was supported by the Project for Cancer Research and Therapeutic Evolution (P‐CREATE) (21cm0106306h0006 to J.Y. and M.M.) and the Research Program on Emerging and Re‐emerging Infectious Diseases (21fk0108088h0003 to J.Y. and M.M.) from the Japan Agency for Medical Research and Development (AMED), KAKENHI (20H03514 to J.Y.; 19H03689 to M.M.) from the JSPS, the Japan Science and Technology Agency‐Mirai Program (JPMJMI18G1 to J.Y.), and the Platform of Supporting Cohort Study and Biospecimen Analysis (CoBiA) (16H06277 to T.W.) from the MEXT. This study was also supported in part by the JSPS Core‐to‐Core Program A, Advanced Research Networks.
CONFLICT OF INTEREST STATEMENT
Keisuke Kataoka and Masao Matsuoka are editorial board members, and the other authors have no conflicts of interest.
ETHICS STATEMENT
Approval of the research protocol by an Institutional Reviewer Board: Our experiments were approved by the Institutional Ethics Committee of Kumamoto University (accession numbers: G489, G499, and E2214).
Informed Consent: All clinical samples were collected after written informed consent was obtained in accordance with the Declaration of Helsinki.
Registry and the Registration No. of the study/trial: N/A.
Animal Studies: N/A.
Supporting information
Figures S1‐S3
Tables S1‐S7
Appendix S1
ACKNOWLEDGMENTS
The authors would like to thank Dr. Kaoru Uchimaru for providing samples, Drs. Kosuke Toyoda, Takafumi Shichijo, and Mikiko Izaki for helpful discussion, Mrs. Miho Matsumoto for helping with the experiments, Dr. Rika A Furuta and Dr. Naoya Shinohara for technical advice, and Dr. Linda Kingsbury for proofreading.
Yamada A, Yasunaga J‐i, Liang L, et al. Anti‐HTLV‐1 immunity combined with proviral load as predictive biomarkers for adult T‐cell leukemia‐lymphoma. Cancer Sci. 2024;115:310‐320. doi: 10.1111/cas.15997
REFERENCES
- 1. Poiesz BJ, Ruscetti FW, Gazdar AF, Bunn PA, Minna JD, Gallo RC. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T‐cell lymphoma. Proc Natl Acad Sci USA. 1980;77:7415‐7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Willems L, Hasegawa H, Accolla R, et al. Reducing the global burden of HTLV‐1 infection: an agenda for research and action. Antiviral Res. 2017;137:41‐48. [DOI] [PubMed] [Google Scholar]
- 3. Martin F, Tagaya Y, Gallo R. Time to eradicate HTLV‐1: an open letter to WHO. Lancet. 2018;391:1893‐1894. [DOI] [PubMed] [Google Scholar]
- 4. Bangham CR, Cook LB, Melamed A. HTLV‐1 clonality in adult T‐cell leukaemia and non‐malignant HTLV‐1 infection. Semin Cancer Biol. 2014;26:89‐98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Karube K, Ohshima K, Tsuchiya T, et al. Expression of FoxP3, a key molecule in CD4CD25 regulatory T cells, in adult T‐cell leukaemia/lymphoma cells. Br J Haematol. 2004;126:81‐84. [DOI] [PubMed] [Google Scholar]
- 6. Satou Y, Utsunomiya A, Tanabe J, Nakagawa M, Nosaka K, Matsuoka M. HTLV‐1 modulates the frequency and phenotype of FoxP3+CD4+T cells in virus‐infected individuals. Retrovirology. 2012;9:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sugiyama D, Nishikawa H, Maeda Y, et al. Anti‐CCR4 mAb selectively depletes effector‐type FoxP3+CD4+ regulatory T cells, evoking antitumor immune responses in humans. Proc Natl Acad Sci U S A. 2013;110:17945‐17950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Penova M, Kawaguchi S, Yasunaga JI, et al. Genome wide association study of HTLV‐1‐associated myelopathy/tropical spastic paraparesis in the Japanese population. Proc Natl Acad Sci US A. 2021;118:e2004199118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Iwanaga M. Epidemiology of HTLV‐1 infection and ATL in Japan: an update. Front Microbiol. 2020;11:1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Matsuoka M, Jeang KT. Human T‐cell leukaemia virus type 1 (HTLV‐1) infectivity and cellular transformation. Nat Rev Cancer. 2007;7:270‐280. [DOI] [PubMed] [Google Scholar]
- 11. Jacobson S, Shida H, McFarlin DE, Fauci AS, Koenig S. Circulating CD8+ cytotoxic T lymphocytes specific for HTLV‐I pX in patients with HTLV‐I associated neurological disease. Nature. 1990;348:245‐248. [DOI] [PubMed] [Google Scholar]
- 12. Kannagi M, Harada S, Maruyama I, et al. Predominant recognition of human T cell leukemia virus type I (HTLV‐I) pX gene products by human CD8+ cytotoxic T cells directed against HTLV‐I‐infected cells. Int Immunol. 1991;3:761‐767. [DOI] [PubMed] [Google Scholar]
- 13. Cann AJ, Rosenblatt JD, Wachsman W, Shah NP, Chen IS. Identification of the gene responsible for human T‐cell leukaemia virus transcriptional regulation. Nature. 1985;318:571‐574. [DOI] [PubMed] [Google Scholar]
- 14. Felber BK, Paskalis H, Kleinman‐Ewing C, Wong‐Staal F, Pavlakis GN. The pX protein of HTLV‐I is a transcriptional activator of its long terminal repeats. Science. 1985;229:675‐679. [DOI] [PubMed] [Google Scholar]
- 15. Furukawa Y, Kubota R, Tara M, Izumo S, Osame M. Existence of escape mutant in HTLV‐I tax during the development of adult T‐cell leukemia. Blood. 2001;97:987‐993. [DOI] [PubMed] [Google Scholar]
- 16. Takeda S, Maeda M, Morikawa S, et al. Genetic and epigenetic inactivation of tax gene in adult T‐cell leukemia cells. Int J Cancer. 2004;109:559‐567. [DOI] [PubMed] [Google Scholar]
- 17. Koiwa T, Hamano‐Usami A, Ishida T, et al. 5′‐long terminal repeat‐selective CpG methylation of latent human T‐cell leukemia virus type 1 provirus in vitro and in vivo. J Virol. 2002;76:9389‐9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Taniguchi Y, Nosaka K, Yasunaga J, et al. Silencing of human T‐cell leukemia virus type I gene transcription by epigenetic mechanisms. Retrovirology. 2005;2:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tamiya S, Matsuoka M, Etoh K, et al. Two types of defective human T‐lymphotropic virus type I provirus in adult T‐cell leukemia. Blood. 1996;88:3065‐3073. [PubMed] [Google Scholar]
- 20. Enose‐Akahata Y, Vellucci A, Jacobson S. Role of HTLV‐1 tax and HBZ in the pathogenesis of HAM/TSP. Front Microbiol. 2017;8:2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gessain A, Barin F, Vernant JC, et al. Antibodies to human T‐lymphotropic virus type‐I in patients with tropical spastic paraparesis. Lancet. 1985;2:407‐410. [DOI] [PubMed] [Google Scholar]
- 22. Osame M, Usuku K, Izumo S, et al. HTLV‐I associated myelopathy, a new clinical entity. Lancet. 1986;1:1031‐1032. [DOI] [PubMed] [Google Scholar]
- 23. Yokota T, Cho MJ, Tachibana N, et al. The prevalence of antibody to p42 of HTLV‐I among ATLL patients in comparison with healthy carriers in Japan. Int J Cancer. 1989;43:970‐974. [DOI] [PubMed] [Google Scholar]
- 24. Hisada M, Okayama A, Shioiri S, Spiegelman DL, Stuver SO, Mueller NE. Risk factors for adult T‐cell leukemia among carriers of human T‐lymphotropic virus type I. Blood. 1998;92:3557‐3561. [PubMed] [Google Scholar]
- 25. Iwanaga M, Watanabe T, Utsunomiya A, et al. Human T‐cell leukemia virus type I (HTLV‐1) proviral load and disease progression in asymptomatic HTLV‐1 carriers: a nationwide prospective study in Japan. Blood. 2010;116:1211‐1219. [DOI] [PubMed] [Google Scholar]
- 26. Imaizumi Y, Iwanaga M, Tsukasaki K, Hata T, Tomonaga M, Ikeda S. Natural course of HTLV‐1 carriers with monoclonal proliferation of T lymphocytes ("pre‐ATL") in a 20‐year follow‐up study. Blood. 2005;105:903‐904. [DOI] [PubMed] [Google Scholar]
- 27. Enose‐Akahata Y, Abrams A, Johnson KR, Maloney EM, Jacobson S. Quantitative differences in HTLV‐I antibody responses: classification and relative risk assessment for asymptomatic carriers and ATL and HAM/TSP patients from Jamaica. Blood. 2012;119:2829‐2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Furuta RA, Ma G, Matsuoka M, Otani S, Matsukura H, Hirayama F. Reevaluation of confirmatory tests for human T‐cell leukemia virus type 1 using a luciferase immunoprecipitation system in blood donors. Transfusion. 2015;55:880‐889. [DOI] [PubMed] [Google Scholar]
- 29. Chong I‐G, Jun C‐H. Performance of some variable selection methods when multicollinearity is present. Chemom Intel Lab Syst. 2005;78:103‐112. [Google Scholar]
- 30. Farahani HA, Rahiminezhad A, Same L. A comparison of partial least squares (PLS) and ordinary least squares (OLS) regressions in predicting of couples mental health based on their communicational patterns. Procedia Soc Behav Sci. 2010;5:1459‐1463. [Google Scholar]
- 31. Kataoka K, Nagata Y, Kitanaka A, et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet. 2015;47:1304‐1315. [DOI] [PubMed] [Google Scholar]
- 32. Burbelo PD, Meoli E, Leahy HP, et al. Anti‐HTLV antibody profiling reveals an antibody signature for HTLV‐I‐associated myelopathy/tropical spastic paraparesis (HAM/TSP). Retrovirology. 2008;5:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fujiwara S, Wada H, Kawada J, et al. NY‐ESO‐1 antibody as a novel tumour marker of gastric cancer. Br J Cancer. 2013;108:1119‐1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rowan AG, Dillon R, Witkover A, et al. Evolution of retrovirus‐infected premalignant T‐cell clones prior to adult T‐cell leukemia/lymphoma diagnosis. Blood. 2020;135:2023‐2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yamagishi M, Kubokawa M, Kuze Y, et al. Chronological genome and single‐cell transcriptome integration characterizes the evolutionary process of adult T cell leukemia‐lymphoma. Nat Commun. 2021;12:4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wolf SN, Haddow J, Greiller C, Taylor GP, Cook LBM, Rowan AG. Quantification of T cell clonality in human T cell leukaemia virus type‐1 carriers can detect the development of adult T cell leukaemia early. Blood Cancer J. 2021;11:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Etoh K, Tamiya S, Yamaguchi K, et al. Persistent clonal proliferation of human T‐lymphotropic virus type I‐infected cells in vivo. Cancer Res. 1997;57:4862‐4867. [PubMed] [Google Scholar]
- 38. Enose‐Akahata Y, Abrams A, Massoud R, et al. Humoral immune response to HTLV‐1 basic leucine zipper factor (HBZ) in HTLV‐1‐infected individuals. Retrovirology. 2013;10:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nam SH, Kidokoro M, Shida H, Hatanaka M. Processing of gag precursor polyprotein of human T‐cell leukemia virus type I by virus‐encoded protease. J Virol. 1988;62:3718‐3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen IS, Slamon DJ, Rosenblatt JD, Shah NP, Quan SG, Wachsman W. The x gene is essential for HTLV replication. Science. 1985;229:54‐58. [DOI] [PubMed] [Google Scholar]
- 41. Inoue J, Yoshida M, Seiki M. Transcriptional (p40x) and post‐transcriptional (p27x‐III) regulators are required for the expression and replication of human T‐cell leukemia virus type I genes. Proc Natl Acad Sci USA. 1987;84:3653‐3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mahgoub M, Yasunaga JI, Iwami S, et al. Sporadic on/off switching of HTLV‐1 tax expression is crucial to maintain the whole population of virus‐induced leukemic cells. Proc Natl Acad Sci USA. 2018;115:E1269‐e1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rende F, Cavallari I, Corradin A, et al. Kinetics and intracellular compartmentalization of HTLV‐1 gene expression: nuclear retention of HBZ mRNAs. Blood. 2011;117:4855‐4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang GZ, Goff SP. Yin Yang 1 is a potent activator of human T lymphotropic virus type 1 LTR‐driven gene expression via RNA binding. Proc Natl Acad Sci USA. 2020;117:18701‐18710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cavallari I, Rende F, Bona MK, et al. Expression of alternatively spliced human T‐cell leukemia virus type 1 mRNAs is influenced by mitosis and by a novel cis‐acting regulatory sequence. J Virol. 2016;90:1486‐1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Suehiro Y, Hasegawa A, Iino T, et al. Clinical outcomes of a novel therapeutic vaccine with tax peptide‐pulsed dendritic cells for adult T cell leukaemia/lymphoma in a pilot study. Br J Haematol. 2015;169:356‐367. [DOI] [PubMed] [Google Scholar]
- 47. Kannagi M, Harashima N, Kurihara K, et al. Tumor immunity against adult T‐cell leukemia. Cancer Sci. 2005;96:249‐255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1‐S3
Tables S1‐S7
Appendix S1


