Abstract
DectiSomes are anti‐infective drug‐loaded liposomes targeted to pathogenic cells by pathogen receptors including the Dectins. We have previously used C‐type lectin (CTL) pathogen receptors Dectin‐1, Dectin‐2, and DC‐SIGN to target DectiSomes to the extracellular oligoglycans surrounding diverse pathogenic fungi and kill them. Dectin‐3 (also known as MCL, CLEC4D) is a CTL pathogen receptor whose known cognate ligands are partly distinct from other CTLs. We expressed and purified a truncated Dectin‐3 polypeptide (DEC3) comprised of its carbohydrate recognition domain and stalk region. We prepared amphotericin B (AmB)‐loaded pegylated liposomes (AmB‐LLs) and coated them with this isoform of Dectin‐3 (DEC3‐AmB‐LLs), and we prepared control liposomes coated with bovine serum albumin (BSA‐AmB‐LLs). DEC3‐AmB‐LLs bound to the exopolysaccharide matrices of Candida albicans, Rhizopus delemar (formerly known as R. oryzae), and Cryptococcus neoformans from one to several orders of magnitude more strongly than untargeted AmB‐LLs or BSA‐AmB‐LLs. The data from our quantitative fluorescent binding assays were standardized using a CellProfiler program, AreaPipe, that was developed for this purpose. Consistent with enhanced binding, DEC3‐AmB‐LLs inhibited and/or killed C. albicans and R. delemar more efficiently than control liposomes and significantly reduced the effective dose of AmB. In conclusion, Dectin‐3 targeting has the potential to advance our goal of building pan‐antifungal DectiSomes.
Keywords: amphotericin B, C‐type lectins, DectiSomes, fungal pathogens, targeted liposomes
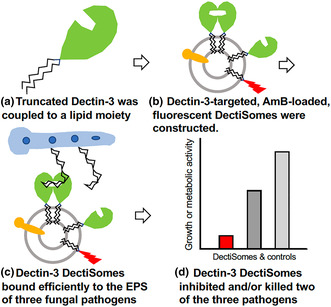
The carbohydrate recognition domain and stalk region of the C‐type lectin pathogen receptor Dectin‐3 (MCL, CLEC4D) were coupled to a lipid carrier (a) and, via its lipid moiety, inserted into an Amphotericin B‐loaded, red fluorescent liposome, producing a DectiSome (b). Dectin‐3 DectiSomes bound to the exopolysaccharide associated with three human fungal pathogens (c). Dectin‐3 DectiSomes significantly inhibited the growth and/or killed two of the pathogens in vitro and lowered the effective dose of Amphotericin B (d).
1. INTRODUCTION
In 2017, it was estimated that globally, there were approximately 750,000 cases of candidiasis, 900,000 cases of mucormycosis, and 220,000 cases of cryptococcosis (Bongomin et al., 2017). These three invasive fungal infections (IFIs) account for more than half of IFI‐associated deaths annually (Banerjee et al., 2021; Bongomin et al., 2017; Brown et al., 2012; Low & Rotstein, 2011; Varshney et al., 2017). Death rates are very high even following antifungal drug treatment, and in many cases of mucormycosis, the surgical removal of infected tissue is necessary. Candida albicans, Rhizopus delemar (R. oryzae), and Cryptococcus neoformans are the most common causative pathogens, respectively, and are the test species in the following study.
Our goal has been to design targeted pan‐antifungal liposomes, DectiSomes, which greatly improve the delivery and performance of antifungal drugs. DectiSomes are defined as liposomes loaded with an anti‐infective drug and targeted to pathogenic cells via a pathogen receptor protein such as a C‐type lectin (CTL). DectiSomes have several distinct advantages over more classical antibody‐targeted immunoliposomes, including a wider range of cognate ligands, lower cost, and greater avidity (Meagher et al., 2021, 2023). We have previously demonstrated the potential of three CTLs, Dectin‐1 (CLEC7A), Dectin‐2 (CLEC6A), and DC‐SIGN (CD209), to efficiently target Amphotericin B‐loaded DectiSomes to four human fungal pathogens: C. albicans, R. delemar, C. neoformans, and Aspergillus fumigatus (Ambati et al., 2022; Ambati, Ellis, et al., 2019, 2021; Ambati, Ferarro, et al., 2019; Ambati, Pham, et al., 2021; Choudhury et al., 2022; Meagher et al., 2021). One or more of the CTL‐targeted DectiSomes dramatically lowered the effective dose of Amphotericin B (AmB) when these pathogens were grown in vitro. DectiSomes were also shown to be effective at reducing fungal burden and/or improving mouse survival in murine models of aspergillosis and candidiasis (Ambati et al., 2022; Ambati, Ellis, et al., 2021; Ambati, Pham, et al., 2021).
We hoped to enhance the pan‐antifungal performance of DectiSomes by employing Dectin‐3 (MCL, Macrophage C‐type Lectin, CLEC4D) (Flornes et al., 2004; Goyal et al., 2018; Wilson et al., 2015). Dectin‐3 is expressed by several lymphoid cell types, particularly dendritic cells and macrophages. It is less well studied than the other CTLs. Dectin‐3 was initially reported to recognize the Ascomycete Candida spp. and the Basidiomycete Cryptococcus spp. (Goyal et al., 2018; Hole et al., 2016; Huang et al., 2018), and shortly thereafter the Ascomycetes Paracoccidioides brasiliensis (Preite et al., 2018) and Pneumocystis carinii (Kottom et al., 2019). When expressed in the membrane of lymphoid cell types, dimers of Dectin‐3's extracellular carbohydrate recognition domain (CRD) bind to fungal glycans. Then its intracellular immunoreceptor tyrosine‐based activation motif (ITAM) signals a fungal infection. Similar to Dectin‐2, Dectin‐3's CRD binds yeast alpha‐mannans. Evidence for Dectin‐3 recognition of a more expanded set of ligands comes primarily from analysis of artificial ligands (Decote‐Ricardo et al., 2019; Fonseca et al., 2009; Huang et al., 2018; Marr et al., 2004; Martinez & Casadevall, 2007). Its potential to recognize diverse novel fungal glycan and lipoglycan ligands was the rationale for building Dectin‐3‐targeted DectiSomes.
We cloned and expressed a truncated isoform of murine Dectin‐3 containing its extracellular CRD and stalk region (DEC3) and used it to coat Amphotericin B‐loaded liposomes to make the DectiSome DEC3‐AmB‐LL. We compared the ability of DEC3‐AmB‐LLs to bind, inhibit, and/or kill three highly evolutionarily divergent and morphologically distinct fungal pathogens—C. albicans (Ascomycete), R. delemar (Mucormycete, Zygomycete), and C. neoformans (Basidiomycete)—as compared to untargeted control liposomes.
2. RESULTS
2.1. Preparation of Dectin‐3‐targeted DectiSomes
We prepared Amphotericin B‐loaded pegylated liposomes (AmB‐LLs) with 11 to 12 moles percent AmB relative to 100 moles percent liposomal lipid, wherein AmB is intercalated into the liposomal membrane (Ambati, Ferarro, et al., 2019). They have a similar AmB concentration to the analogous commercial FDA‐approved un‐pegylated liposomal drug AmBisome (U.S. Food and Drug Administration, 1997). We cloned the N‐terminal portion of the sequence encoding the CRD and stalk region of murine Dectin‐3, which was codon optimized for E. coli expression (hereafter referred to as truncated Dectin‐3 or DEC3, Figure SF1). For this liposomal presentation, we omitted the transmembrane and intracellular ITAM signaling domains of Dectin‐3. The E. coli‐produced DEC3 polypeptide was affinity purified (Figure SF2a,b) and coupled to a pegylated lipid to make DEC3‐PEG‐DSPE. The DEC3‐PEG‐DSPE polypeptide was inserted via its DSPE lipid moiety into the AmB‐LLs at 1 mole percent to make the DectiSome DEC3‐AmB‐LL by methods we have described previously for other DectiSomes (Ambati, Ellis, et al., 2019; Ambati, Ferarro, et al., 2019; Ambati, Pham, et al., 2021). The construct allows the CRD and stalk region of DEC3 to float freely in the liposomal membrane and to form functional homodimers and multimers (Meagher et al., 2021, 2023). Bovine serum albumin (BSA) was also lipid‐modified and inserted via its DSPE moiety, but at 0.3 moles percent, accounting for its 3‐fold higher molecular weight relative to our truncated Dectin‐3 polypeptide (Ambati, Ferarro, et al., 2019). AmB‐LLs and BSA‐AmB‐LLs served as negative, untargeted liposome controls for non‐specific binding and killing. It was assumed that the protein coating of BSA‐AmB‐LLs would interfere with the untargeted interaction of AmB‐LLs with fungal cells (LaMastro et al., 2023). All three types of liposomes were fluorescently tagged with 2 moles percent Rhodamine‐B‐DHPE loaded into the liposomal membrane.
2.2. Qualitative and quantitative assessment of DectiSome binding to three fungal pathogens
C. albicans, R. delemar, and C. neoformans were grown in vitro, treated with rhodamine B‐labeled DEC3‐AmB‐LLs, BSA‐AmB‐LLs, or AmB‐LLs, washed, and then photographed using fluorescence microscopy. Under the in vitro growth conditions used, C. albicans cells exhibited both yeast and hyphal morphologies; R. delemar grew with a hyphal morphology; and C. neoformans was in the yeast form. These experiments allowed the qualitative assessment of liposome binding to various fungal cells of different morphotypes. The areas of rhodamine B red fluorescence were measured from multiple photographs to quantify these binding data. We began assessing fluorescent liposome binding area data by manually processing each image through ImageJ as done previously (Ambati, Ellis, et al., 2019; Ambati, Ferarro, et al., 2019; Ambati, Pham, et al., 2021; Choudhury et al., 2022). To reduce the labor involved in quantifying fluorescent liposome binding data and standardize the process, we created a CellProfiler pipeline, AreaPipe, that automates quantitative image analysis (Figure SF3).
2.3. C. albicans
Dendritic cell Dectin‐3 and purified epitope‐tagged Dectin‐3 polypeptides are reported to recognize C. albicans, at least in part, via their binding to extracellular oligo‐mannans (Goyal et al., 2018; Wang et al., 2016; Zhu et al., 2013). Because Dectin‐3's interactions with C. albicans are the best characterized for any fungal pathogen, we began our analysis of DEC3‐AmB‐LLs with this species. In our first experiment, C. albicans yeast cells were grown on microscope chamber slides for only 1.5 h in hyphal growth‐inducing media, wherein they just reached the yeast–hyphal transition stage of development. Fixed cells were stained with DEC3‐AmB‐LLs and control liposomes and viewed top‐down at high magnification (60×) using combined epifluorescence and phase contrast microscopy (Figure 1). DEC3‐AmB‐LLs bound efficiently to the exopolysaccharide (EPS) matrices surrounding all yeast–hyphal stage cells (Figure 1a), while binding by control BSA‐AmB‐LLs and AmB‐LLs was not detected (Figure 1b,c). Five pixels were substituted for the zero values of liposome binding in some control images, which was equivalent to the smallest numbers we had detected out of the 5 × 106 total pixel area in some experiments. The area of DEC3‐AmB‐LL binding was approximately 9,700‐fold larger than for BSA‐AmB‐LLs or AmB‐LLs (p < 0.0001, Figure 1d). The data are presented in a log10 scale scatter bar plot to reveal the dispersion of individual data points over a large dynamic range that would have been obscured in a linear plot.
FIGURE 1.
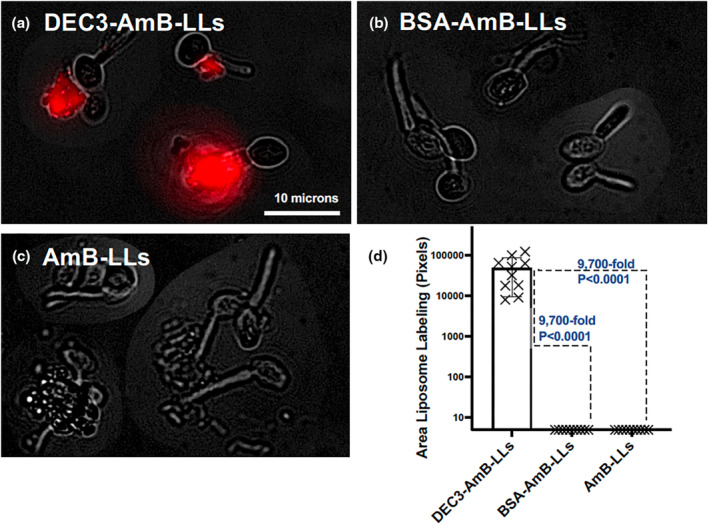
DEC3‐AmB‐LLs bound efficiently to Candida albicans during the yeast–hyphal transition stage of development. (a–c) respectively, show representative photographic images of red fluorescent DEC3‐AmB‐LLs, BSA‐AmB‐LLs, and AmB‐LLs binding to C. albicans in the yeast–hyphal transition stage. After plating in media that stimulated hyphal development, yeast cells were grown for 1.5 h to reach this stage. The scale bar corresponds to 10 μm. Images were acquired at 60× magnification and equivalently cropped to enlarge cells for presentation. Composite images were prepared because the cells were widely dispersed in each 60× field. (d) The relative area of red fluorescent liposome binding (log10) was quantified using AreaPipe and is shown in scatter bar plots. N = 10 for each bar. Standard errors from the mean and p values are indicated.
In our next experiment, C. albicans was grown on plastic microtiter plates until they produced mature hyphal colonies a few hundred microns in diameter, fixed, treated with liposomes, and viewed bottom‐up using combined epifluorescence and bright field microscopy (Figure 2). DEC3‐AmB‐LLs bound strongly (Figure 2a). Most of the binding appeared to be to patches of hyphal‐associated EPS at the periphery of the colonies, where hyphae had been growing most rapidly, and less binding was associated with older cells at the center of colonies. Control liposomes bound rarely (Figure 2b,c). The area of DEC3‐AmB‐LL binding was 1,150‐fold larger (PMW < 0.0001) than for BSA‐AmB‐LLs and 43‐fold larger (PMW < 0.0001) than for AmB‐LLs (Figure 2d). We speculate that the stronger binding of AmB‐LLs relative to BSA‐AmB‐LLs may be attributed to non‐specific binding of the liposomal membrane with hydrophobic components on the cells, such as cellular membranes (LaMastro et al., 2023), wherein a coating of BSA interfered with this interaction. Biological replicates of these two experiments with C. albicans showed similar results (Figure SF4a,b, respectively).
FIGURE 2.
DEC3‐AmB‐LLs bound efficiently to Candida albicans hyphae. (a–c) respectively, show representative photographic images of red fluorescent DEC3‐AmB‐LLs, BSA‐AmB‐LLs, and AmB‐LLs binding to 15‐hr‐old hyphal colonies. The scale bar indicates the size of the hyphae photographed at 20× magnification. (d) The relative area of red fluorescent liposome binding (log10) was quantified using AreaPipe and is shown in a scatter bar plot. N = 10 for each bar. Standard errors from the mean and PMW values are indicated.
2.4. R. delemar
Dectin‐3 has not previously been reported to recognize Mucormycetes such as R. delemar. Because R. delemar does not stick efficiently to plastic or glass growth substrates (Ibrahim et al., 2005), we cultured the cells on agar media in petri dishes, where we have shown that they stick efficiently (Choudhury et al., 2022). Agar plugs were removed, fixed, washed, stained with rhodamine B‐labeled DEC3‐AmB‐LLs, AmB‐LLs, and BSA‐AmB‐LLs, and in some cases also with calcofluor white (CW) for chitin, and washed extensively to remove unbound liposomes and CW (Choudhury et al., 2022). Stained cells were viewed top‐down by epifluorescence and, in some experiments, also by phase contrast.
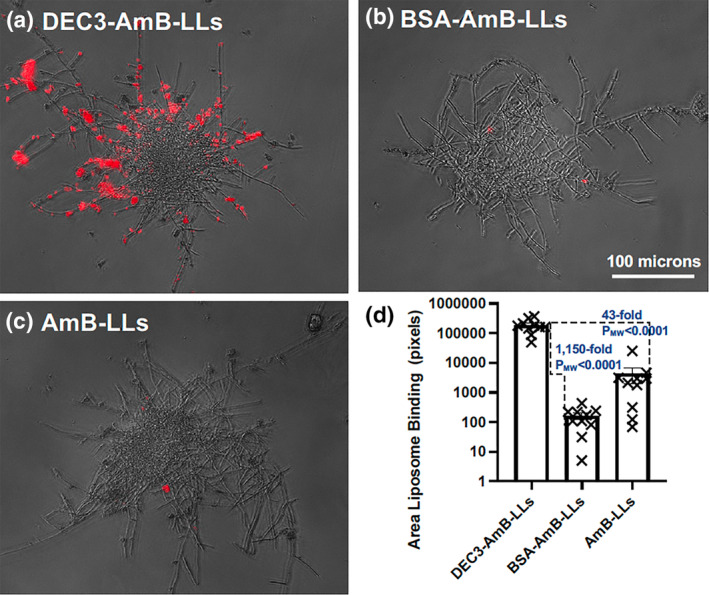
R. delemar cells were grown for 6 h to the germling stage or 15 h to the mature hyphal stage. We found that DEC3‐AmB‐LLs bound strongly to fixed swollen sporangiospores and germ tubes of germling stage cells (Figure 3a). By contrast, binding by the control liposomes was rarely observed (Figure 3b,c). DEC3‐AmB‐LL binding was heavily concentrated in the EPS deposits (yellow arrows). There was some staining that appeared to be tightly associated with the cell wall (white arrows), but it was not possible to distinguish binding to the cell wall itself from EPS that was closely allied with the cell wall. The patchy nature of the staining along the cell wall may be accounted for either by the deposition of small amounts of EPS proximal to the cell wall or by small regions in the cell wall containing cognate ligands of Dectin‐3 that are exposed, thus enabling DEC3‐AmB‐LL staining. The area of red fluorescent DEC3‐AmB‐LL binding was 164‐fold larger (PMW = 0.0022) than that for BSA‐AmB‐LLs and 188‐fold larger (PMW = 0.0022) than that for AmB‐LLs (Figure 3d). We did not observe preferential binding by AmB‐LLs relative to BSA‐AmB‐LLs.
FIGURE 3.
DEC3‐AmB‐LLs bound efficiently to Rhizopus delemar germlings. (a–c), respectively, show representative photographic images of red fluorescent DEC3‐AmB‐LLs, BSA‐AmB‐LLs, and AmB‐LLs binding to CW‐stained germinating sporangiospores and germling germ tubes. Sporangiospores were germinated on an agar surface for 6 h. The scale bar indicates the size of the germlings photographed at 20× magnification. The images were cropped to show enlarged germlings. (d) The relative area of red fluorescent liposome binding (log10) was quantified using AreaPipe and is shown in a scatter bar plot. N = 6 for each bar. Standard errors from the mean and PMW values are indicated.
When fixed hyphal colonies of R. delemar were examined (Figure 4), we again observed that DEC3‐AmB‐LLs bound to cell‐associated EPS deposits (Figure 4a), which are more obvious in a higher magnification image (Figure 4b). Control liposomes seldom bound (Figure 4c,d). The area of DEC3‐AmB‐LL binding was 614‐fold larger (PMW < 0.0001) than for BSA‐AmB‐LLs and 39‐fold larger (PMW < 0.0001) than for AmB‐LLs (Figure 4e). Consistent with the data for C. albicans, the AmB‐LLs showed higher levels of non‐specific binding than the BSA‐AmB‐LLs.
FIGURE 4.
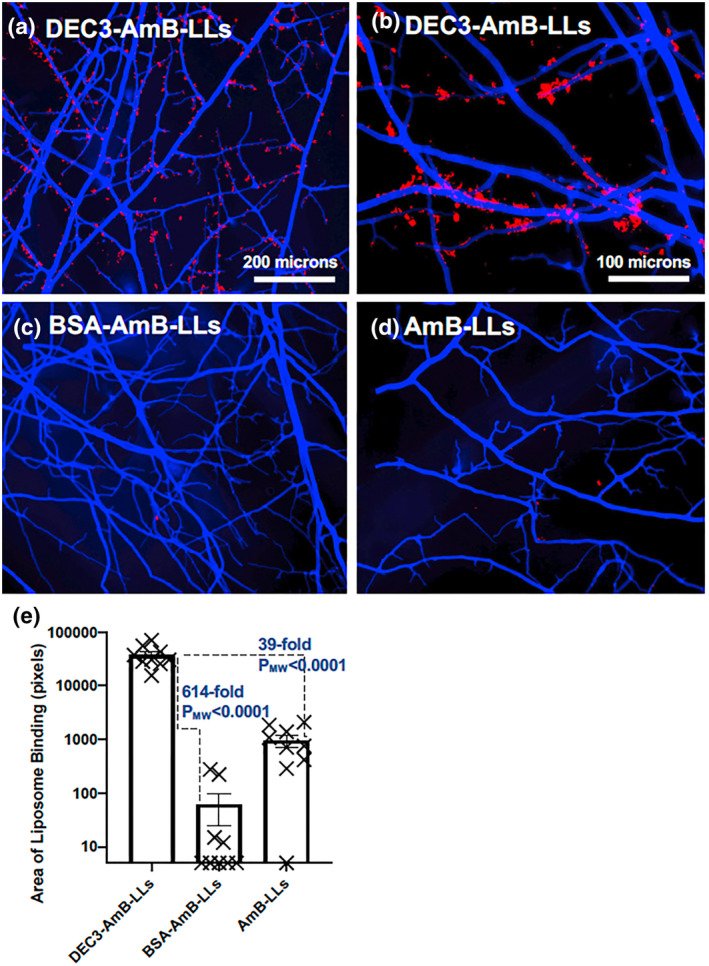
DEC3‐AmB‐LLs bound efficiently to fixed Rhizopus delemar hyphae. (a–d) respectively, show representative photographic images of red fluorescent DEC3‐AmB‐LLs, BSA‐AmB‐LLs, and AmB‐LLs binding to CW‐stained 15‐hr‐old hyphae grown on an agar surface photographed at 10×, and (b) shows DEC3‐AmB‐LLs photographed at 20× magnification. The scale bars indicate the size of the hyphae photographed at 10× (a, c, and d) or 20× magnification (b). (d) The relative area of red fluorescent liposome binding at 10X (log10) was quantified using AreaPipe and is shown in a scatter bar plot. N = 9 for each bar. Standard errors from the mean and PMW values are indicated.
When live mature hyphal colonies were examined (Figure 5a–d), we observed that DEC3‐AmB‐LLs bound to hyphal‐associated EPS deposits in a pattern similar to fixed hyphae (Figure 5a) and control liposomes seldom bound (Figure 5b,c). The relative area of DEC3‐AmB‐LL binding was less dramatic than for fixed cells. The area of DEC3‐AmB‐LL binding was 21‐fold larger (PMW = 0.0012) than for BSA‐AmB‐LLs and 73‐fold larger (PMW = 0.0006) than for AmB‐LLs. Perhaps the dynamic nature and turnover of cognate ligand sites in the EPS of live cells turn over DectiSome binding during the binding and washing steps, resulting in the weaker binding observed in live cells relative to fixed cells. Biological replicates of these three R. delemar liposome binding experiments showed similar results (Figure SF4c–e).
FIGURE 5.
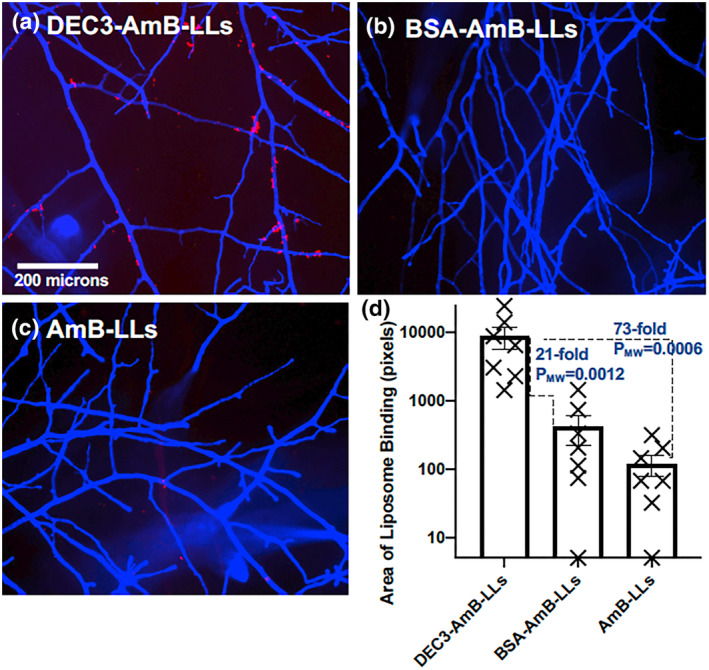
DEC3‐AmB‐LLs bound efficiently to live Rhizopus delemar hyphae. (a–c) respectively, show representative photographic images of red fluorescent DEC3‐AmB‐LLs, BSA‐AmB‐LLs, and AmB‐LLs binding to live CW‐stained hyphae. The scale bar indicates the size of the hyphae, which were photographed at 10× magnification. (d) The relative area of red fluorescent liposome binding (log10) was quantified using AreaPipe and is shown in a scatter bar plot. N = 7 for each bar. Standard errors from the mean and PMW values are indicated.
2.5. C. neoformans
Dendritic cell Dectin‐3 was previously reported to recognize C. neoformans (Goyal et al., 2018; Hole et al., 2016). Therefore, we anticipated that DEC3‐AmB‐LLs would bind to this species. Colonies of C. neoformans were grown on minimal agar plates for 6 or 18 h. Agar plugs were removed, fixed, washed, stained with rhodamine B‐labeled DEC3‐AmB‐LLs, AmB‐LLs, BSA‐AmB‐LLs, and CW, and washed extensively. Colonies were observed using top‐down epifluorescence (Figure 6).
FIGURE 6.
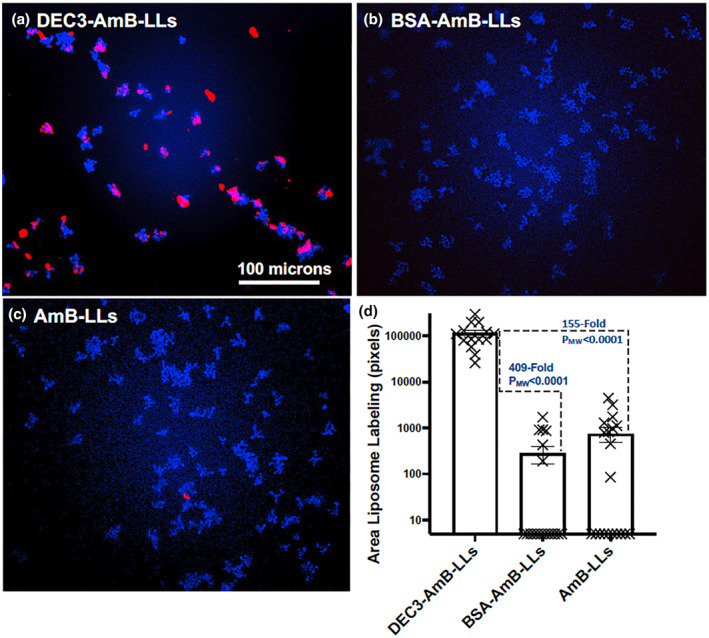
DEC3‐AmB‐LLs bound efficiently to the EPS matrices of small Cryptococcus neoformans colonies. (a–c) respectively, show representative photographic images of red fluorescent DEC3‐AmB‐LLs, BSA‐AmB‐LLs, and AmB‐LLs binding to CW‐stained 6‐hr‐old colonies of C. neoformans grown on an agar surface. The scale bar indicates the size of the colonies photographed at 20× magnification. (d) The relative area of red fluorescent liposome binding (log10) was quantified using AreaPipe and is shown in a scatter bar plot. N = 14 for each bar. Standard errors from the mean and PMW values are indicated.
Six‐hr‐old C. neoformans colonies were composed of only three cells to two dozen cells (Figure 6). DEC3‐AmB‐LLs bound strongly to the EPS matrices associated with almost every colony (Figure 6a), even those composed of only a few cells, while binding by control liposomes was rarely observed (Figure 6b,c). DEC3‐AmB‐LLs bound to a 409‐fold larger area (PMW < 0.0001) than BSA‐AmB‐LLs and a 155‐fold larger area (PMW < 0.0001) than AmB‐LLs (Figure 6d).
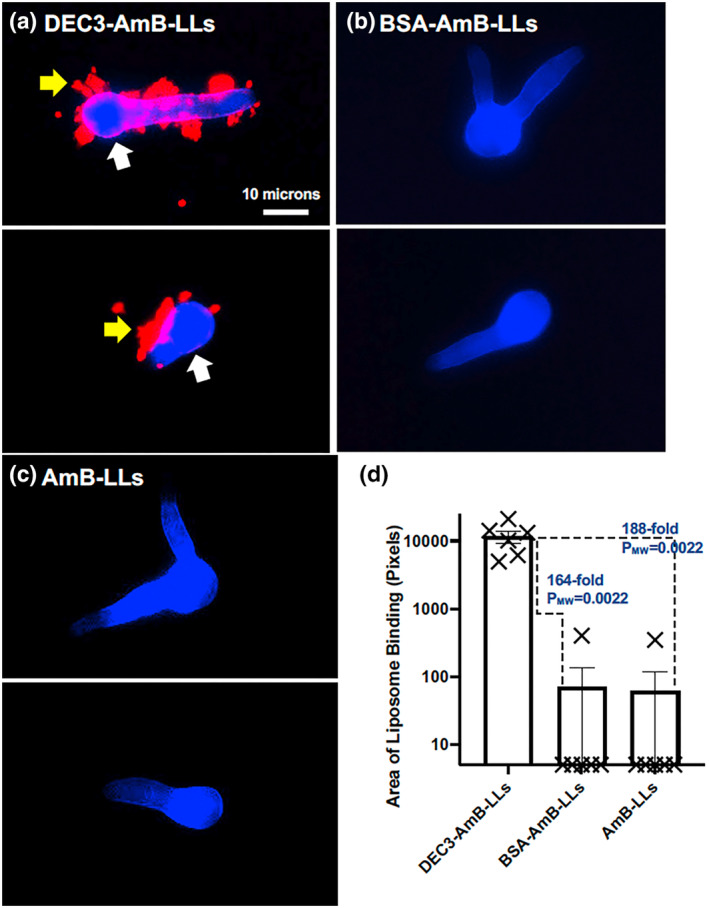
Eighteen‐hr‐old C. neoformans colonies were approximately 200 to 300 microns in diameter (Figure 7) and composed of thousands of cells. DEC3‐AmB‐LLs bound to numerous patches both within and at the boundary of mature colonies (Figure 7a). Some binding appeared to be specific to the EPS matrix at the periphery of the colonies (yellow arrows), while the binding that completely surrounded some small groups of cells might be specific to the cell capsules or capsule‐associated EPS of these spherical cells (white arrows). Binding by BSA‐AmB‐LLs and AmB‐LLs was seldom detected (Figure 7b,c), but is indicated for AmB‐LL binding by thin white arrows (Figure 7c). DEC3‐AmB‐LLs bound to a 6,160‐fold larger area (PMW < 0.0001) than BSA‐AmB‐LLs and to a 4,160‐fold larger area (PMW < 0.0001) than AmB‐LLs (Figure 7d). Biological replicates of these two C. neoformans liposome binding experiments produced similar results (Figure SF4f,g). To confirm that DEC3‐AmB‐LLs exhibit specific binding to fungal glycans, we tested the ability of various oligoglycans to inhibit binding to 16‐hr‐old C. neoformans colonies and found modest inhibition by oligomannans as expected and by glucuronic acid (Figure SF5) as recently reported in an independent study (Huang et al., 2018). The inhibition by laminarin shown in this experiment was unexpected and inconsistent among experimental replicates. Importantly, neither DEC3‐AmB‐LLs nor control liposomes BSA‐AmB‐LLs and AmB‐LLs bound detectably to human HEK293T cells, as shown in Figure SF6.
FIGURE 7.
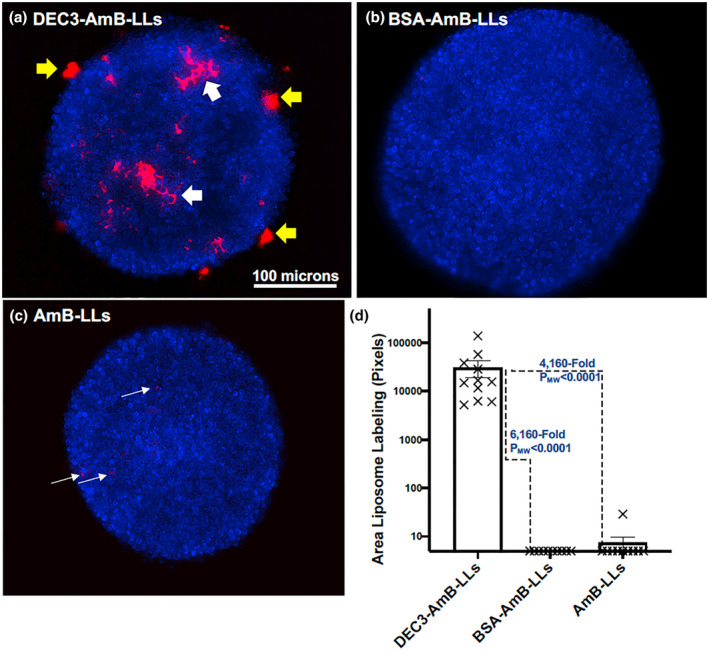
DEC3‐AmB‐LLs bound efficiently to large Cryptococcus neoformans colonies. (a–c) respectively, show representative photographic images of red fluorescent DEC3‐AmB‐LLs, BSA‐AmB‐LLs, and AmB‐LLs binding to CW‐stained 18‐hr‐old colonies of C. neoformans grown on an agar surface. Yellow arrows indicate DEC3‐AmB‐LL binding to EPS deposits, and large white arrows indicate binding that surrounds spherical yeast cells within the colonies. The scale bar indicates the size of the colonies photographed at 20× magnification. The uncommon red fluorescence of control liposome binding (white thin arrows in c) had to be greatly enhanced relative to that for DEC3‐AmB‐LLs to make it visible for presentation. (d) The relative area of red fluorescent liposome binding (log10) was quantified using AreaPipe and is shown in a scatter bar plot. N = 11 for each bar. Standard errors from the mean and PMW values are indicated.
2.6. Binding by Dectin‐3 protein
The exterior portion of the cell wall of C. albicans is rich in variously crosslinked oligomannans and mannoproteins (Garcia‐Rubio et al., 2020; Gow et al., 2017; Schiavone et al., 2014). The cell walls of Mucorales species contain small amounts of variously crosslinked mannans (Lecointe et al., 2019). The exterior portion of the C. neoformans capsule is rich in glucuronoxylomannans (GXMs), which are a known ligand of Dectin‐3 (Garcia‐Rubio et al., 2020, Gow et al., 2017). Therefore, we expected Dectin‐3‐targeted liposomes to bind to the cell wall or capsule of all three species. Yet, we did not observe DEC3‐AmB‐LLs to be unambiguously bound to their cell walls or capsules. We considered the possibility that the 100‐nanometer diameter of DEC3‐AmB‐LLs was simply too large to physically access the cognate ligands of Dectin‐3 within the highly crosslinked glycan matrices of the cell walls and/or capsules. We estimate the rotational diameter of the 23 kDa MW truncated Dectin‐3 polypeptide (DEC3) to be only a few nanometers. To test this hypothesis of size limitation for liposome binding, we directly conjugated rhodamine B (MW 0.48 kDa) to the Dectin‐3 polypeptide to make DEC3‐Rhod. DEC3‐Rhod bound to the EPS matrices surrounding the various developmental stages of all three species and has comparable spatial distributions to those of DEC3‐AmB‐LLs (Figure SF7a–e). The localization of both DEC3‐AmB‐LLs and DEC3‐Rhod staining varied widely from C. albicans colony to colony, as revealed by the distribution of data points in scatter bar plots, but no significant difference in binding was observed between the two reagents. As with the liposomal reagents, the DEC3‐Rhod protein reagent did not exhibit clear, unambiguous binding to the cell walls or capsules. Thus, the binding pattern of DEC3‐AmB‐LLs does not appear to have been dramatically altered by the physical size of liposomes.
2.7. Quantitative assessment of the inhibition and/or killing of three fungal pathogens
C. albicans, R. delemar, and C. neoformans were grown in vitro and treated with the DEC3‐AmB‐LLs, BSA‐AmB‐LLs, and AmB‐LLs using different temporal regimens, after which growth, viability, and/or metabolic activity were quantified.
2.8. C. albicans
An overnight culture of C. albicans yeast cells was suspended into RPMI +10% FBS hyphal‐inducing media and aliquoted into 96‐well microtiter plates. Shortly after hyphal development was initiated (1.5 h, 37°C; see morphology in Figure 1), cells were treated with the DEC3‐AmB‐LLs, BSA‐AmB‐LLs, or AmB‐LLs at the indicated AmB concentrations. Seventeen hours later, residual metabolic activity was measured using the CellTiter‐Blue (CTB) reagent (resazurin). Only live cells with an intact plasma membrane and a functional mitochondrial electron transport chain can reduce resazurin to the fluorescent product resorufin. The plate was incubated for approximately 60 min at 37°C, and the pink fluorescence of resorufin was quantified. The data are presented in a log10 scatter bar plot in Figure 8a. Cells receiving DEC3‐AmB‐LLs delivering 0.3, 0.2, and 0.1 μM AmB showed respectively, 24‐fold (PMW = 0.0003), 127‐fold (PMW = 0.0002), and 25‐fold (PMW = 0.0002) lower metabolic activity than the corresponding BSA‐AmB‐LL‐treated controls. Statistically significant reductions in metabolic activity were also observed relative to the AmB‐LL‐treated controls. Control liposomes delivering 0.3 μM AmB began to have strong inhibitory activity, thus limiting the resolution of differences at this and higher concentrations of AmB. A biological replicate produced similar results (Figure SF8a).
FIGURE 8.
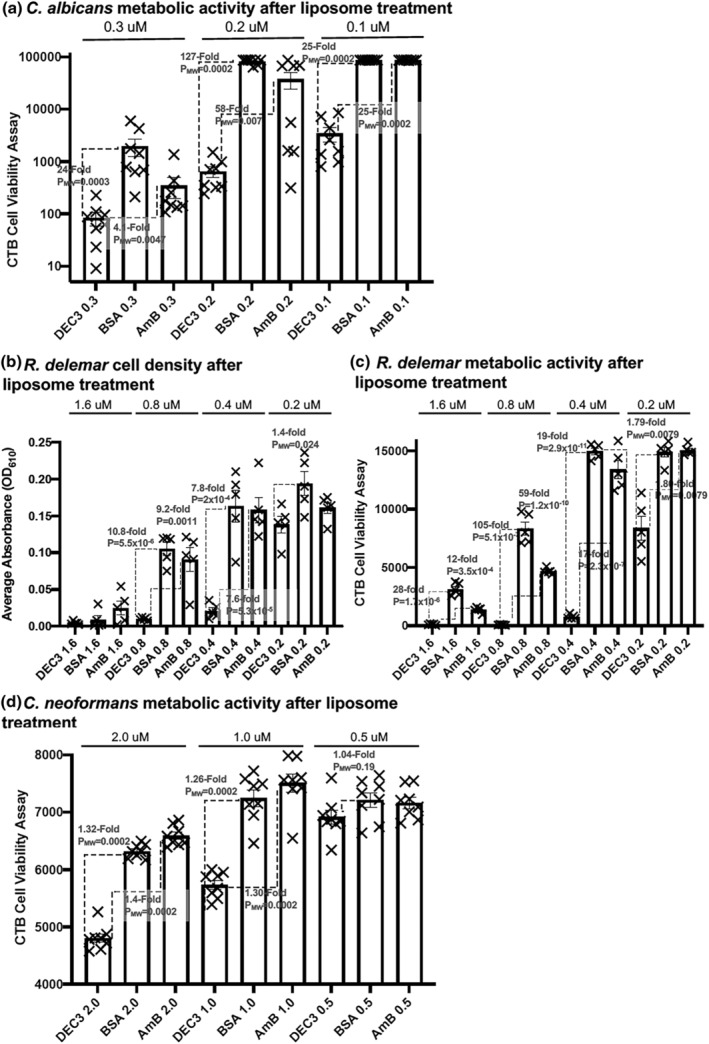
DEC3‐AmB‐LLs were efficient at inhibiting and/or killing two of the three fungal pathogens examined. Candida albicans, Rhizopus delemar, and Cryptococcus neoformans were grown in 96‐well microtiter plates and treated with DEC3‐AmB‐LLs, AmB‐LLs, and BSA‐AmB‐LLs, delivering the indicated micromolar concentrations of AmB. Growth inhibition and/or killing were assayed as reductions in cell density measured at A610 or reductions in viable cell metabolic activity by measuring the electrochemical reduction of the redox dye resazurin in CellTiter‐Blue reagent to fluorescent resorufin after liposome treatment. (a) Residual metabolic activity of C. albicans. (b) Cell density of R. delemar. (c) Residual metabolic activity of R. delemar. (d) Residual metabolic activity of C. neoformans. Cell density and metabolic activity are shown in scatter bar plots. N = 5 to 8 for each bar. Standard errors from the mean and p or PMW values are indicated.
2.9. R. delemar
R. delemar sporangiospores were plated in RPMI +0.165 M MOPS (pH 7) media in 96‐well microtiter plates. DEC3‐AmB‐LLs, BSA‐AmB‐LLs, and AmB‐LLs delivering different concentrations of AmB were immediately added to the respective wells, and the plates were incubated for 24 h at 37°C with shaking at 120 rpm. Cell density was measured as O.D. at A610. Cells receiving DEC3‐AmB‐LLs delivering 0.8, 0.4, and 0.2 μM AmB were 10.8‐fold (p = 5.5 × 10−6), 7.8‐fold (p = 2 × 10−4) and 1.4‐fold (p = 0.024) less dense than the respective BSA‐AmB‐LL‐treated controls, and the 0.8 and 0.4 μM samples were similarly less dense than the respective AmB‐LL‐treated controls (Figure 8b). After the cell density measurements were taken, CTB reagent was added to each well, and the plate was incubated at 37°C for an additional two hours. The pink fluorescence in each well was quantified. Cells receiving DEC3‐AmB‐LLs delivering 1.6, 0.8, 0.4, and 0.2 μM AmB had 28‐fold (p = 1.7 × 10−6), 105‐fold (p = 5.1 × 10−7), 19‐fold (p = 2.9 × 10−11), and 1.79‐fold (PMW = 0.0079) lower metabolic activity than the respective BSA‐AmB‐LL‐treated cells (Figure 8c). DEC3‐AmB‐LLs‐treated cells had similarly reduced levels of metabolic activity relative to the AmB‐LL‐treated controls. Biological replicates produced comparable results (Figure SF8b,c).
2.10. C. neoformans
C. neoformans yeast cells grown overnight in YPD were plated at 10,000 cells per well in 96‐well microtiter plates in RPMI + 0.165 M MOPS (pH 7) at 30°C. After a few hours of incubation in YPD, when the cells had formed small colonies equivalent to those shown in Figure 6, they were treated with liposomes delivering the indicated concentrations of AmB, and the plates were incubated for 1 h at 30°C with shaking at 50 rpm. The media with unbound liposomes was removed, CTB reagent in RPMI + MOPS media was added to each well, and the plate was incubated for an extended period at 30°C. Cells receiving DEC3‐AmB‐LLs delivering 2.0 and 1.0 μM AmB showed only 1.32‐fold (PMW = 0.0002) and 1.26‐fold (PMW = 0.0002) lower metabolic activity than the respective BSA‐AmB‐LL‐treated controls (Figure 8d). There were similar small reductions in CTB activity relative to the AmB‐LL‐treated controls. Because there was small variability in the results among eight individual samples, these results were highly statistically significant, but the fold improvements in inhibition by DEC3‐AmB‐LLs were clearly marginal. A biological replicate produced similar results for the comparison at 2.0 μM AmB but not for 1.0 or 0.5 μM (Figure SF8d). Variations in this CTB assay design, such as longer drug treatments or not removing unbound liposomes, did not improve the relative efficacy of DEC3‐AmB‐LLs. Attempts to measure the efficacy of DEC3‐AmB‐LLs at reducing cell viability by measuring reductions in cell density in microtiter plate assays, or reductions in CFUs in liquid suspension cultures, or increases in propidium iodide staining of dead cells all failed to give significant results. Apparently, these techniques were not sensitive or reproducible enough to detect the small changes in cell viability revealed by CTB assays.
3. DISCUSSION
We have shown that Dectin‐3's CRD targets DEC3‐AmB‐LLs specifically and efficiently to the yeast–hyphal transition stage and mature hyphal stage of the Ascomycete C. albicans, to the germling and mature hyphal stages of the Mucormycete R. delemar, and to small and large colonies of the Basidiomycete C. neoformans. We've added R. delemar to the list of fungal species recognized by Dectin‐3's CRD and suggest the possibility that there are other target pathogens for Dectin‐3 yet to be discovered. These three fungal pathogens diverged from common ancestral species several hundred million years ago and, hence, represent some of the extreme ancient diversity within the fungal kingdom (Kuramae et al., 2006; Spatafora et al., 2017; Taylor & Berbee, 2006). Fluorescent DectiSomes have tremendous power as probes for the novel binding of CTLs, such as Dectin‐3, because each liposome has a few thousand rhodamine B molecules and each has more than a thousand DEC3 molecules, thus creating avidity for target ligands. DEC3‐AmB‐LLs bound to the EPS matrices of these species, and there was little, if any, binding to their cell walls and/or capsules. The much smaller DEC3‐Rhod protein reagent also did not bind unambiguously to cell walls and/or capsules. Fungal pathogens are thought to produce EPS to adhere to host tissues and protect themselves from host immune defenses. EPS shed into host serum could decoy immune responses away from fungal cells. Dectin‐3 may have evolved its ability to bind to the EPS associated with and shed from diverse pathogens in order to aid in the complex host innate immune responses to infection. Our results presented here on Dectin‐3‐targeted DectiSomes binding to and killing C. albicans and R. delemar compare reasonably well with our previous publications on Dectin‐1‐, Dectin‐2‐, and DC‐SIGN‐targeted DectiSomes (Meagher et al., 2023).
Many CTLs form homodimers that form their glycan ligand binding sites at the junction of the two CRDs (Cummings & McEver, 2022). In our previous studies, we constructed liposomes containing the truncated forms of Dectin‐1, Dectin‐2, and DC‐SIGN alone, as we have done here with truncated Dectin‐3. In each case, this resulted in the efficient targeting of antifungal liposomes to fungal pathogens, and this design must have enabled homodimers to form functional ligand binding sites. Each showed dramatically improved antifungal efficacy over untargeted liposomal AmB toward one or more fungal pathogens, including Aspergillus fumigatus, C. albicans, C. neoformans, and R. delemar grown in vitro (Ambati, Ellis, et al., 2019; Ambati, Ferarro, et al., 2019; Ambati, Pham, et al., 2021; Choudhury et al., 2022) and A. fumigatus and C. albicans in mouse models of pulmonary aspergillosis and invasive candidiasis (Ambati et al., 2022; Ambati, Ellis, et al., 2021). However, these and several other CTLs are co‐expressed in mammalian dendritic cells and macrophages (Ariizumi et al., 2000; Hole et al., 2016; Kitai et al., 2021; Sun et al., 2013; Yoshikawa et al., 2021; Zhu et al., 2013) and hence have the potential to act cooperatively to enhance signaling. Dectin‐2 and Dectin‐3 appear to be on the same membrane rafts in bone marrow‐derived macrophages or when co‐transfected into monocyte/macrophage‐like RAW264.7 cells. This view of their linked behavior stems from results showing that when either binds a cognate ligand or is treated with either Dectin‐specific antibody, the two CTLs are co‐endocytosed (Zhu et al., 2013). The study used bimolecular fluorescence complementation of Dectin‐2 and Dectin‐3 yellow fluorescent protein fusions in HEK293 cells to show that the co‐expressed proteins physically interact on cell surfaces to form heterodimers that have enhanced and perhaps novel ligand binding and signaling properties. The implication is that physically connected heteromeric CRDs of the two CTLs are formed and have novel properties of ligand recognition. By contrast, when co‐transfected into embryonic kidney 293 T cells, Dectin‐2 did not appear to form heteromeric complexes with Dectin‐3 (Blankson et al., 2022). Thus, it is possible that other factors are required for their linked behavior. Now that we have shown that Dectin‐3 functions efficiently when floating on a liposomal surface, we can construct liposomes co‐presenting Dectin‐3 and Dectin‐2 and test their interaction in a cell‐free system. If liposomal co‐presentation of Dectin‐2/Dectin‐3 enhances and expands their ligand binding properties, this should make a more effective pan‐antifungal delivery vehicle than either Dectin alone.
The superfamily of CTL genes contains more than 30 members and several subgroups based on the sequences of their CRDs and their functional structures and signaling domains when positioned in the cell membrane (Drickamer & Taylor, 2015). Among the fifteen members of structural group 2 CTLs, Dectin‐3, Dectin‐2, and DCSIGN (CD209) all recognize oligomannans. This is a well‐represented class of glycans that form part of the cell walls and EPS matrices of most fungal pathogens (Briard et al., 2021; Drickamer & Taylor, 2015; Gow et al., 2017; Goyal et al., 2018). However, our previous studies showed that Dectin‐2‐ and DCSIGN‐targeted liposomes stained fungal EPS matrices with little, if any, staining of fungal cell walls or capsules (Ambati, Ellis, et al., 2019; Ambati, Pham, et al., 2021; Meagher et al., 2023). In these previous studies, as herein, microscope observations showed that binding was primarily localized to the material closely associated with the cells, but there was minimal binding tightly associated with the cell wall. We have inferred that this material is EPS, which is also known to contain cognate ligands of various CTLs. At the outset, we suspected that Dectin‐3 would recognize distinct fungal oligoglycans in both cell walls and EPS matrices and recognize diverse fungal pathogens. First, Dectin‐3 has a relatively distinct and divergent CRD amino acid sequence when compared to the other two Dectins. Second, lymphoid cell Dectin‐3 recognizes glucuronoxylomannan (GXM), which is characterized as a major glycan component of C. neoformans capsules and a lesser component of their biofilms (Albuquerque et al., 2014; Huang et al., 2018). GXM is not a single compound. It is composed of a heterogenous mixture of polysaccharides that vary widely in their molecular weight from less than 10 kDa to 300 kDa (Albuquerque et al., 2014) and are often crosslinked to other glycans, thus generating different GXM variants. We previously stained in vitro‐grown C. neoformans cells with the best‐characterized anti‐GXM antibody 18B7 and found robust staining of GXM within the capsule of most, but not all, individual cells and the EPS matrix associated with some cells (Ambati, Ellis, et al., 2019). Dectin‐2 liposomal staining only partially overlapped with that of the GXM antibody. Using a variety of monoclonal antibodies to GXM, including 18B7, it was shown that GXM encompasses many different immuno‐epitopes that are heterogeneously distributed in the C. neoformans capsule and EPS (Albuquerque et al., 2014). Because Dectin‐3 is reported to specifically recognize GXM, and GXM is a major glycan component of C. neoformans, we anticipated the potential for highly preferential binding to C. neoformans, and in particular its capsule, over binding to our other two test species. But instead, we only observed staining of the EPS matrix in small colonies and sporadic staining of the EPS surrounding some cells in older colonies. While it is possible that our DEC3‐AmB‐LL reagent recognized a very small subset of C. neoformans GXMs in the cell capsule, proving this would require extensive immunochemical and/or biochemical analyses. It is also possible that lymphoid cell Dectin‐3 recognition of GXM requires cooperation with other CTLs, and Dectin‐3 itself does not itself bind GXM. Our study does not attempt to identify the particular glycans or glycolipids recognized by Dectin‐3, and our results do suggest there is still much to be learned about Dectin‐3's range of cognate ligands.
Based on the two to three orders of magnitude higher levels of DEC3‐AmB‐LL binding to C. albicans, R. delemar, and C. neoformans relative to control liposome binding, we anticipated orders of magnitude more efficient inhibition and/or killing of all three species. When delivering the low concentrations of AmB of 0.1–1.6 μM, we showed that DEC3‐AmB‐LLs reduced the viable cell metabolic activity of C. albicans and R. delemar one to two orders of magnitude more efficiently than untargeted AmB‐loaded liposomal controls. Under the conditions of our assays, DEC3‐AmB‐LLs lowered the effective dose of liposomal AmB for inhibition and/or killing of these two species by at least two orders of magnitude. By contrast, and based on various assays, DEC3‐AmB‐LLs were much less effective at inhibiting or killing C. neoformans. This was unexpected based on DEC3‐AmB‐LL's efficient binding to this species. We cannot explain the lack of strong antifungal efficacy against C. neoformans.
As to the mechanism by which AmB‐loaded DectiSomes, and DEC3‐AmB‐LLs in particular, increase inhibition and killing of fungal pathogens relative to untargeted control liposomes, AmB‐LLs, our analog of AmBisome®, we propose the following. While it is known that AmBisome® liposomes pass through the cell wall and plasma membrane (Walker et al., 2018), it is reasonable to assume this transport process is relatively inefficient because the 60 to 90 nanometer diameter size of liposomes should impede movement through the cell wall and membrane. While a small fraction of the untargeted liposomes will make it through the cell wall and plasma membrane, most will diffuse far away from the cells or be washed away, and the AmB from these liposomes will be at much lower concentrations. By contrast, when AmB‐loaded DectiSomes are bound to the EPS, their close proximity to fungal cells increases the concentration of AmB diffusing from DectiSomes to fungal cells. The dimensions of AmB are measured in 10's of Angstroms, and it is known to penetrate into the cell membrane. The argument that AmB from DectiSomes will diffuse more rapidly to fungal cells than untargeted liposomal AmB is based on the Fokker–Planck equation showing that the rate of diffusion proceeds as the inverse square of the distance (Fokker, 1914; Planck, 1917). In the case of DectiSomes bound to the ligands in the EPS proximal to the fungal cells, there should be a more rapid and efficient diffusion of AmB to cells than by most of the untargeted liposomal AmB, which is more distant from fungal cells, and the rate of diffusion to fungal cells will be much slower. Additionally, the fact that various DectiSomes bind primarily to the EPS offers a big advantage to their antifungal activity. Depending upon their glycan compositions, the sheer size of the EPS should generate more available cognate ligand binding sites than the cell wall. Hence, EPS‐bound DectiSomes should provide a larger reservoir of AmB than if they were bound only to the cell wall.
While the goal is to develop DectiSomes as powerful pan‐antifungal therapeutics, this and our other recent studies leave many important issues unresolved (Meagher et al., 2023). We have only just begun to test DectiSomes in mouse models of invasive fungal disease (Ambati et al., 2022; Ambati, Ellis, et al., 2021; Ambati, Pham, et al., 2021). Extrapolating from in vitro binding and killing to in vivo studies is complicated by the fact that the glycan and lipoglycan ligands produced by fungi are dependent upon their growth environment. Dectins such as Dectin‐3 have the advantage over antibody‐targeted reagents in that a unique host Dectin sequence should have low immunogenicity relative to variable antibody sequences that often produce host‐specific immunogenic responses. However, Dectin‐1 and Dectin‐3 recognize endogenous ligands that discriminate self from non‐self (Mori et al., 2017; Saijo & Iwakura, 2011), and as more is known about Dectin‐3, it may also be shown to recognize host ligands. Thus, there is the potential that CTL‐targeted reagents could have toxicity issues. Our ongoing research explores a wide variety of topics concerning the efficacy of DectiSomes as drug delivery agents.
4. CONCLUSIONS
We presented statistically well‐supported data showing that Dectin‐3‐coated AmB‐loaded liposomes, DEC3‐AmB‐LLs, were an order of magnitude or more effective at binding to C. albicans, R. delemar, and C. neoformans than either of our control liposomes, AmB‐LLs and BSA‐AmB‐LLs. DEC3‐AmB‐LLs bound primarily to their EPS matrices and not to their cell walls or cell capsules. DEC3‐AmB‐LLs were similarly effective at inhibiting or killing C. albicans and R. delemar, but much less effective against C. neoformans. Our positive data showing the inhibition of hyphal stages of two highly divergent pathogens suggest that it would be worthwhile to explore the efficacy of DEC3‐AmB‐LLs in mouse models of candidiasis and mucormycosis and investigate the potential of Dectin‐3 to act synergistically and perhaps form heterodimers with Dectin‐2 on the same liposomes. The CellProfiler pipeline AreaPipe was introduced to automate and simplify the capture and quantification of fluorescent area data within TIFF or JPEG images, replacing the tedious, time‐consuming manual processing of area data in ImageJ.
5. EXPERIMENTAL PROCEDURES
5.1. Fungal strains and growth conditions
C. albicans strain SC5314 (Gillum et al., 1984) was grown in YPD (1% yeast extract, 2% peptone, 2% dextrose, ThermoFisher Cat#212750, #21677, Fisher Sci. Cat#D16‐10, respectively) liquid culture starting from a single colony on an agar plate and grown at 37°C overnight with vigorous shaking. Yeast cells were washed once into RPMI lacking phenol red dye (Sigma‐Aldrich, Cat# R8755) + 10% Fetal Bovine Serum (FBS) (GIBCO Cat#10437–028) adjusted to pH 7.5, plated at 20,000 cells/well in 1 mL and 35,000 cells/per well in 0.1 mL, and grown at 37°C to the germling or hyphal stage in wells of plastic 24‐well or 96‐well polystyrene microtiter plates or on poly‐L‐lysine‐coated glass microscope slides inside a glass chamber. When indicated, cells were fixed in 3.7% formalin (J.T. Baker, #2106–01) diluted in PBS for 1 h and washed three times in PBS before staining. The design of the reusable glass chambers and assembled chamber slides for growing cells is shown in Figure SF9a,b.
Sporangiospore stocks of R. delemar strain 99–880 (f.k.a. R. oryzae and R. arrhizus, ATCC MYA‐4621) were prepared as described recently (Choudhury et al., 2022) and stored at 4°C, where they remained greater than 90% viable for more than a month. Sporangiospores were germinated and grown to germling or hyphal stages on 1.5% agar plates made with RPMI +0.165 M MOPS (3‐(N‐morpholino) propane sulfonic acid; Sigma‐Aldrich, Cat# M1254) (Andrianaki et al., 2018) adjusted to pH 7 and incubated at 37°C (Choudhury et al., 2022). Agar plugs were removed and processed for top‐down microscopic imaging (Choudhury et al., 2022). For quantitative inhibition and killing experiments, cells were grown in liquid RPMI + MOPS media diluted to 800 cells per 90 μL per well in 96‐well microtiter plates. Because R. delemar does not stick well to microtiter plates but does stick efficiently to micropipette tips, reagents were added but never removed during growth and inhibition assays.
C. neoformans clinical isolate H99‐alpha (Montone, 2009) was grown at 30°C in liquid YPD (1% yeast extract, 2% peptone, 2% dextrose) with vigorous shaking overnight. Cells prepared for microscopy were then plated on RPMI + MOPS agar plates and incubated at 30°C, and agar plugs were later removed. Cells for viability assays were plated at 10,000 cells per 90 μL per well in YPD in 96‐well microtiter plates, grown for 4.5 h, and treated with liposomes, after which the media was removed and replaced with RPMI + MOPS media containing CTB reagent at the manufacturer's recommended concentration.
5.2. Production of Dectin‐3
Annotated Dectin‐3‐derived DNA and protein sequences are given in Figure SF1. The 219 amino acid (a.a.)‐long sequence of the native murine Dectin‐3 protein (NP_034949.3) was obtained from the National Center for Biotechnology Information (Figure SF1a). The sequence of the E. coli codon‐optimized construct of murine Dectin‐3 CRD and stalk region truncated and modified for cloning into pET‐45B and lipidation is shown in Figure SF1b and was deposited at the U.S. National Center for Biotechnology Information (BankIT, HK‐DEC3; accession number OQ366170). The sequence encodes a 199 a.a.‐long modified truncated Dectin‐3 polypeptide, DEC3, beginning at its N‐terminus with a vector‐encoded N‐terminal (His)6 affinity tag, an added flexible GlySerGly spacer, two lysine (K) residues, and another flexible GlySerGly spacer, followed by the 176 a.a.‐long N‐terminal end of murine Dectin‐3 (Figure SF1c). The E. coli strain BL21 containing the murine Dectin‐3‐pET45B plasmid was grown in 1 L of Luria broth at 37°C overnight without IPTG induction (Figure SF2) or with IPTG induction at O.D. 0.7 A600, followed by 4 more hours of growth. DEC3 was extracted from cell pellets using a 6 M guanidine hydrochloride (GuHCl) buffer and purified on nickel affinity resin in this buffer as described previously (Ambati, Ellis, et al., 2019; Ambati, Ferarro, et al., 2019). After affinity purification, the protein was approximately 80% pure (Figure SF2b). In various preparations, five to ten milligrams of affinity‐purified protein were typically recovered per liter of Luria broth culture.
5.3. Production of Dectin‐3‐coated drug‐loaded liposomes (DEC3‐AmB‐LLs)
AmB‐loaded liposomes, AmB‐LLs, were prepared as described previously, except that the drug loading was performed at 37°C instead of 60°C (Ambati, Ferarro, et al., 2019). Samples of Dectin‐3 were coupled to DSPE‐PEG‐3400‐NHS (Nanosoft Polymers, 1544–3400) in the same 6 M GuHCl buffer with the pH adjusted to 8.3 with triethanolamine and purified over Bio‐Gel P‐6 acrylamide molecular exclusion resin in a 1 M Arginine crowding buffer pH adjusted to 7.3 (Bio‐Rad Cat#150–0740) as we described previously for other CTLs (Ambati, Ellis, et al., 2019; Ambati, Ferarro, et al., 2019; Ambati, Pham, et al., 2021). DSPE‐PEG‐BSA was prepared from BSA (Sigma, Cat#A‐8022) by the same protocol, except that the coupling to DSPE‐PEG‐NHS was performed in 0.1 M pH 8.3 carbonate buffer, and it was subsequently desalted by P‐6 gel exclusion chromatography in 0.1 M sodium phosphate buffered saline pH 7.4 (Corning, Cat#21‐031‐CV). Modified Dectin‐3 and BSA were loaded into AmB‐LLs at 1 and 0.33 moles percent, respectively, relative to moles of liposomal lipid via their DSPE lipid moiety as described previously (Ambati, Ellis, et al., 2019, Ambati, Ferarro, et al., 2019). Typical preparations produced 800 μL of protein‐coated liposomes with 1.1 mg of modified Dectin‐3 or BSA (~1.4 mg/mL) and AmB at 750 μM (0.69 mg/mL). For each new batch of AmB‐loaded liposomes, the volumes of DEC3‐AmB‐LLs, BSA‐AmB‐LLs, and AmB‐LLs were adjusted to have the same concentration of AmB, typically 750 μM, to simplify dilutions of the three samples in subsequent experiments.
5.4. Liposome dilutions for binding and inhibition studies
For all liposome‐binding experiments, DEC3‐AmB‐LLs were diluted such that the Dectin‐3 protein was at a concentration of 1 μg/100 μL (1:100 w/v) in respective growth media. AmB‐LLs and BSA‐AmB‐LLs were diluted equivalently. For binding inhibition experiments, DEC3‐AmB‐LLs (1:100 w/v) were preincubated for 30 min with the following oligoglycans at a concentration of 10 mg/mL: yeast mannans extracted via alkaline extraction (Sigma Cat# M7504) or detergent extraction (Sigma Cat# M3640), laminarin (Sigma Cat# L9634), glucuronic acid (Sigma Cat# G5269), or the liposome dilution buffer as a control. The oligoglycan/DectiSome mixes were then added to pre‐blocked C. neoformans plugs. For killing studies, liposomes were diluted into growth media just before use, such that they delivered AmB final concentrations of 0.1 to 2 μM.
5.5. Liposome binding to a human cell line
The human embryonic kidney epithelial cell line HEK293T was obtained from the American Type Culture Collection (ATCC #CRL‐3216). Cells were grown on 24‐well plastic microtiter plates in RPMI media plus 10% fetal bovine serum at 37°C with 5% CO2. Cells were washed with PBS, fixed for 30 min with 4% freshly prepared formalin, and washed thrice more with PBS. Cells were stained with rhodamine‐tagged DEC3‐AmB‐LLs, BSA‐AmB‐LLs, and AmB‐LLs as described for staining fungal cells. Images were taken bottom up on an inverted revolving microscope at 10× magnification, combining the visible light channel and the fluorescent Texas Red channel.
5.6. CellProfiler analysis of fluorescent liposome staining
All microscopy was performed on an ECHO Revolve microscope (model #RVSF1000/Revolve R4). Red fluorescent liposome staining was captured in the TRITC channel, CW chitin staining in the DAPI channel, visible light images using the Ph1 phase condenser, and combined images as Revolve's zoverlay.jpg files. As we began this project, we manually captured the area of fluorescent staining image by image in ImageJ. The direct output from ImageJ presents fluorescence as a fraction of the image area, as we presented in our previous publications of DectiSomes and as shown in several supplemental figures herein (SF4C through SF4G). A CellProfiler (v4.2.4)‐based pipeline, AreaPipe (v. 5), was developed to analyze photographic images (e.g., jpg, tiff) for the area of fluorescent reagent binding (e.g., fluorescent liposomes) to cells (Carpenter et al., 2006). AreaPipe (AreaPipe_V5.cppipe) was deposited at the CellProfiler repository of published pipelines at the Broad Institute (https://cellprofiler.org/published‐pipelines). In short, the program eliminated the manual quantification of fluorescent imaging data. Figure SF3 shows the original rhodamine red fluorescent jpeg image of DEC3‐AmB‐LLs binding to a C. albicans hyphal colony (Figure SF3a) and compares the areas of fluorescence captured from the image using the manual analysis method in ImageJ (Figure SF3b) as done previously (Ambati et al., 2022; Ambati, Ellis, et al., 2019, 2021; Ambati, Ferarro, et al., 2019; Ambati, Pham, et al., 2021; Choudhury et al., 2022) to that obtained using AreaPipe (Figure SF3c). When using the manual method, each red fluorescent image (e.g., Figure SF3a) jpeg or tiff was (1) loaded into ImageJ, (2) converted to an 8‐bit black and white image under Image > Type, and (3) adjusted using threshold settings to capture the appropriate red liposome binding area under Image > Adjust > Threshold (30/255) > Apply. (4) These data were placed in a file using the Analyze>Measure functions, and (5) the ImageJ data were copied and pasted into an Excel file for subsequent quantitative analysis. To use AreaPipe, (1) CellProfiler was opened, (2) the jpeg or tiff images to be analyzed were pasted into the indicated window, (3) AreaPipe (AreaPipe_V5.cppipe) was dragged into the indicated window, and the operator answered “yes” to “load pipeline?”; (5) two separate folders were designated for the output data files “SaveImages” and “ExportToSpreadsheet”; (6) Analyze Images was selected; and (7) the quantitative area output data was automatically loaded into a .csv file and the jpeg images of the segmented areas into a folder (e.g., Figure SF3c). Some effort was devoted to ensuring that AreaPipe accurately captured even trace images with only a few pixels of liposomal fluorescence or could report zero pixels of fluorescence, which was not uncommon among images of control liposome‐treated samples. Typically, AreaPipe processed 30 jpeg images in 3 min using a standard laptop, which in our experience was at least 20‐times faster than the manual processing in ImageJ and avoided the excessive eye strain associated with the manual method. The only difference in the output is that the manual ImageJ method records a fraction of the fluorescent pixel area captured relative to the entire image area, while AreaPipe reports the actual number of fluorescent pixels captured from an image out of the total pixel area in images. In order to allow these data to be plotted on a log scale, when zero pixels were recorded, we substituted a value of 5 pixels, and with the manual method, when zero area was recorded, 0.0001 was substituted for the area, which was close to the lowest values recorded experimentally.
5.7. Quantitative inhibition and killing studies
Up to four types of assays were used in attempts to quantify DectiSome inhibition, killing, and/or survival after treatment with liposomal AmB. (1) Cell density was recorded at A610 nanometers in 96‐well microtiter plates (BioTeK Synergy HT microplate reader). (2) Colony‐forming units (CFUs) of C. neoformans were counted after plating dilutions of cells on YPD and incubating at 30°C. (3) Propidium iodide staining of dead cells followed our previous published protocol (Ambati, Ellis, et al., 2019). (4) CellTiter‐Blue reagent (CTB, resazurin, Promega Cat#G8081) was used to measure the reduction in live cell metabolic redox activity, with 20 μL of reagent being added to 100 μL of cells in 96‐well microtiter plates. After CTB was added, C. albicans, R. delemar, and C. neoformans were respectively incubated for approximately 1 h at 37°C, 2 h at 37°C, and 18 to 24 h at 30°C before the fluorescent product resorufin was measured at Ex485/Em590 (BioTek Synergy HT). We have not seen fluorescent CTB assays of C. neoformans reported previously, but an XTT colorimetric redox assay had been used previously (Martinez & Casadevall, 2007). Fluorescent CTB assays have a much larger dynamic range than colorimetric XTT assays. Long incubation times (e.g., 24 h) with CTB's resazurin dye are commonly used to measure the viability of Mycobacterium spp. following drug treatments (Amin et al., 2009; Franzblau et al., 1998).
5.8. Data management
Data were recorded and managed in Excel (v. 16.69). The CellProfiler AreaPipe output data were in the .csv file format, which is compatible with Excel. Scatter bar plots were prepared, and standard errors from the mean were estimated in GraphPad Prism 9 (v. 9.5.0). Most of the data were normally distributed, so the Student's two‐tailed t‐test was used to estimate p values (T.TEST in Excel). In cases where the data for at least one sample in a comparison appeared to be non‐parametric in their distribution, p values were estimated using the Mann–Whitney U test (Mann & Whitney, 1947) in Prism 9 and were indicated as PMW values.
AUTHOR CONTRIBUTIONS
Richard B. Meagher: Conceptualization; investigation; funding acquisition; writing – original draft; methodology; validation; writing – review and editing; visualization; project administration; formal analysis; data curation; supervision; resources. Quanita J. Choudhury: Conceptualization; investigation; writing – original draft; methodology; validation; writing – review and editing; visualization; data curation; formal analysis. Suresh Ambati: Conceptualization; investigation; methodology; visualization; validation; writing – review and editing; formal analysis; data curation. Collin D. Link: Investigation; methodology; validation; writing – review and editing; software; visualization. Xiaorong Lin: Conceptualization; funding acquisition; writing – review and editing; project administration; resources. Zachary A. Lewis: Conceptualization; funding acquisition; writing – review and editing; software; methodology; resources; supervision; project administration; validation.
CONFLICT OF INTEREST STATEMENT
UGARF has submitted patents to the United States Patent and Trademark Office. UGARF had no role in the design, execution, interpretation, or writing of this study. The authors declare that they have no conflicts of interest related to this study.
ETHICS STATEMENT
No animals were used in this research.
Supporting information
Supplemental Fig. SF1.
ACKNOWLEDGMENTS
We would like to thank the technical team from the University of Georgia Franklin College of Arts and Science Office of Information Technology for helping us maintain the computer hardware and software used to obtain, manage, process, and publish various data. We wish to thank Matthew Whittaker and Professor Eileen Kennedy of UGA's Pharmaceutical and Biomedical Sciences Department for supplying the HEK293T cells for staining and Annalee Picket in UGA's Center for Applied Isotope Studies' Glass Blowing Shop for preparing the glass microscope slide chambers. The work was funded by the University of Georgia Research Foundation, Inc. (UGARF to S.A. and R.B.M.) and the National Institute of Allergy and Infectious Diseases (R21AI144498 and R21AI148890 to R.B.M. and Z.A.L., and R01AI162989 to R.B.M., S.A., Z.A.L., and X.L.). The funders had no role in study design, data collection, interpretation, or the decision to submit the work for publication and are not responsible for the content of this article.
Choudhury, Q.J. , Ambati, S. , Link, C.D. , Lin, X. , Lewis, Z.A. & Meagher, R.B. (2023) Dectin‐3‐targeted antifungal liposomes efficiently bind and kill diverse fungal pathogens. Molecular Microbiology, 120, 723–739. Available from: 10.1111/mmi.15174
Quanita J. Choudhury and Suresh Ambati are co‐first authors.
Contributor Information
Zachary A. Lewis, Email: zlewis@uga.edu.
Richard B. Meagher, Email: meagher@uga.edu.
DATA AVAILABILITY STATEMENT
All new data that were presented and/or discussed within this publication and its supplemental data section are included herein, and any previously published data that were discussed were appropriately cited. The AreaPipe pipeline for fluorescent area analysis will be uploaded at the Broad Institute's site for CellProfiler programs and made public once this study is accepted for publication as per the Broad Institute's suggestion for the time of release.
REFERENCES
- Albuquerque, P.C. , Fonseca, F.L. , Dutra, F.F. , Bozza, M.T. , Frases, S. , Casadevall, A. et al. (2014) Cryptococcus neoformans glucuronoxylomannan fractions of different molecular masses are functionally distinct. Future Microbiology, 9, 147–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambati, S. , Ellis, E.C. , Lin, J. , Lin, X. , Lewis, Z.A. & Meagher, R.B. (2019) Dectin‐2‐targeted antifungal liposomes exhibit enhanced efficacy. mSphere, 4, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambati, S. , Ellis, E.C. , Pham, T. , Lewis, Z.A. , Lin, X. & Meagher, R.B. (2021) Antifungal liposomes directed by Dectin‐2 offer a promising therapeutic option for pulmonary aspergillosis. mBio, 12, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambati, S. , Ferarro, A.R. , Kang, S.E. , Lin, J. , Lin, X. , Momany, M. et al. (2019) Dectin‐1‐targeted antifungal liposomes exhibit enhanced efficacy. mSphere, 4, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambati, S. , Pham, T. , Lewis, Z.A. , Lin, X. & Meagher, R.B. (2021) DC‐SIGN targets amphotericin B‐loaded liposomes to diverse pathogenic fungi. Fungal Biology and Biotechnology, 8, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambati, S. , Pham, T. , Lewis, Z.A. , Lin, X. & Meagher, R.B. (2022) DectiSomes‐ glycan targeting of liposomal amphotericin B improves the treatment of disseminated candidiasis. Antimicrobial Agents and Chemotherapy, 66, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin, A.G. , Angala, S.K. , Chatterjee, D. & Crick, D.C. (2009) Rapid screening of inhibitors of mycobacterium tuberculosis growth using tetrazolium salts. Methods in Molecular Biology (Clifton, N.J.), 465, 187–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrianaki, A.M. , Kyrmizi, I. , Thanopoulou, K. , Baldin, C. , Drakos, E. , Soliman, S.S.M. et al. (2018) Iron restriction inside macrophages regulates pulmonary host defense against Rhizopus species. Nature Communications, 9, 3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi, K. , Shen, G.L. , Shikano, S. , Ritter, R., 3rd , Zukas, P. , Edelbaum, D. et al. (2000) Cloning of a second dendritic cell‐associated C‐type lectin (dectin‐2) and its alternatively spliced isoforms. The Journal of Biological Chemistry, 275, 11957–11963. [DOI] [PubMed] [Google Scholar]
- Banerjee, S. , Denning, D.W. & Chakrabarti, A. (2021) One health aspects & priority roadmap for fungal diseases: a mini‐review. The Indian Journal of Medical Research, 153, 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankson, V. , Lobato‐Pascual, A. , Saether, P.C. , Fossum, S. , Dissen, E. & Daws, M.R. (2022) Human macrophage C‐type lectin forms a heteromeric receptor complex with Mincle but not Dectin‐2. Scandinavian Journal of Immunology, 95, e13149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongomin, F. , Gago, S. , Oladele, R.O. & Denning, D.W. (2017) Global and multi‐national prevalence of fungal diseases‐estimate precision. Journal of Fungi (Basel), 3, 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briard, B. , Fontaine, T. , Kanneganti, T.D. , Gow, N.A.R. & Papon, N. (2021) Fungal cell wall components modulate our immune system. Cell Surface, 7, 100067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, G.D. , Denning, D.W. , Gow, N.A. , Levitz, S.M. , Netea, M.G. & White, T.C. (2012) Hidden killers: human fungal infections. Science Translational Medicine, 4, 165rv13. [DOI] [PubMed] [Google Scholar]
- Carpenter, A.E. , Jones, T.R. , Lamprecht, M.R. , Clarke, C. , Kang, I.H. , Friman, O. et al. (2006) CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biology, 7, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury, Q.J. , Ambati, S. , Lewis, Z.A. & Meagher, R.B. (2022) Targeted delivery of antifungal liposomes to Rhizopus delemar . Journal of Fungi (Basel), 8, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings, R.D. & McEver, R.P. (2022) Chapter 31. C‐type lectins. In: Essentials of glycobiology, 4th edition. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory PRess. [Google Scholar]
- Decote‐Ricardo, D. , LaRocque‐de‐Freitas, I.F. , Rocha, J.D.B. , Nascimento, D.O. , Nunes, M.P. , Morrot, A. et al. (2019) Immunomodulatory role of capsular polysaccharides constituents of Cryptococcus neoformans . Frontiers in Medicine, 6, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drickamer, K. & Taylor, M.E. (2015) Recent insights into structures and functions of C‐type lectins in the immune system. Current Opinion in Structural Biology, 34, 26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flornes, L.M. , Bryceson, Y.T. , Spurkland, A. , Lorentzen, J.C. , Dissen, E. & Fossum, S. (2004) Identification of lectin‐like receptors expressed by antigen presenting cells and neutrophils and their mapping to a novel gene complex. Immunogenetics, 56, 506–517. [DOI] [PubMed] [Google Scholar]
- Fokker, A.D. (1914) Die mittlere Energie rotierender elektrischer Dipole im Strahlungsfeld. Annalen der Physik, 348, 810–820. [Google Scholar]
- Fonseca, F.L. , Nimrichter, L. , Cordero, R.J. , Frases, S. , Rodrigues, J. , Goldman, D.L. et al. (2009) Role for chitin and chitooligomers in the capsular architecture of Cryptococcus neoformans . Eukaryotic Cell, 8, 1543–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzblau, S.G. , Witzig, R.S. , McLaughlin, J.C. , Torres, P. , Madico, G. , Hernandez, A. et al. (1998) Rapid, low‐technology MIC determination with clinical mycobacterium tuberculosis isolates by using the microplate Alamar blue assay. Journal of Clinical Microbiology, 36, 362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Rubio, R. , de Oliveira, H.C. , Rivera, J. & Trevijano‐Contador, N. (2020) The fungal cell wall: Candida, Cryptococcus, and Aspergillus species. Frontiers in Microbiology, 10, 2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillum, A.M. , Tsay, E.Y. & Kirsch, D.R. (1984) Isolation of the Candida albicans gene for orotidine‐5′‐phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Molecular & General Genetics, 198, 179–182. [DOI] [PubMed] [Google Scholar]
- Gow, N.A.R. , Latge, J.P. & Munro, C.A. (2017) The fungal cell wall: structure, biosynthesis, and function. Microbiology Spectrum, 5, 1‐25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal, S. , Castrillon‐Betancur, J.C. , Klaile, E. & Slevogt, H. (2018) The interaction of human pathogenic fungi with C‐type lectin receptors. Frontiers in Immunology, 9, 1261–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hole, C.R. , Leopold Wager, C.M. , Mendiola, A.S. , Wozniak, K.L. , Campuzano, A. , Lin, X. et al. (2016) Antifungal activity of plasmacytoid dendritic cells against Cryptococcus neoformans In vitro requires expression of Dectin‐3 (CLEC4D) and reactive oxygen species. Infection and Immunity, 84, 2493–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H.R. , Li, F. , Han, H. , Xu, X. , Li, N. , Wang, S. et al. (2018) Dectin‐3 recognizes glucuronoxylomannan of Cryptococcus neoformans serotype AD and Cryptococcus gattii serotype B to initiate host defense against cryptococcosis. Frontiers in Immunology, 9, 1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim, A.S. , Spellberg, B. , Avanessian, V. , Fu, Y. & Edwards, J.E., Jr. (2005) Rhizopus oryzae adheres to, is phagocytosed by, and damages endothelial cells in vitro. Infection and Immunity, 73, 778–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitai, Y. , Sato, K. , Tanno, D. , Yuan, X. , Umeki, A. , Kasamatsu, J. et al. (2021) Role of Dectin‐2 in the phagocytosis of Cryptococcus neoformans by dendritic cells. Infection and Immunity, 89, e0033021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottom, T.J. , Hebrink, D.M. , Monteiro, J.T. , Lepenies, B. , Carmona, E.M. , Wuethrich, M. et al. (2019) Myeloid C‐type lectin receptors that recognize fungal mannans interact with Pneumocystis organisms and major surface glycoprotein. Journal of Medical Microbiology, 68, 1649–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramae, E.E. , Robert, V. , Snel, B. , Weiss, M. & Boekhout, T. (2006) Phylogenomics reveal a robust fungal tree of life. FEMS Yeast Research, 6, 1213–1220. [DOI] [PubMed] [Google Scholar]
- LaMastro, V. , Campbell, K.M. , Gonzalez, P. , Meng‐Saccoccio, T. & Shukla, A. (2023) Antifungal liposomes: lipid saturation and cholesterol concentration impact interaction with fungal and mammalian cells. Journal of Biomedical Materials Research. Part A, 111, 644–659. [DOI] [PubMed] [Google Scholar]
- Lecointe, K. , Cornu, M. , Leroy, J. , Coulon, P. & Sendid, B. (2019) Polysaccharides cell wall architecture of Mucorales. Frontiers in Microbiology, 10, 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low, C.Y. & Rotstein, C. (2011) Emerging fungal infections in immunocompromised patients. F1000 medicine reports, 3, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann, H. & Whitney, D. (1947) On a test of whether one of two random variables is stochastically larger than the other. The Annals of Mathematical Statistics, 18, 50–60. [Google Scholar]
- Marr, K.A. , Balajee, S.A. , McLaughlin, L. , Tabouret, M. , Bentsen, C. & Walsh, T.J. (2004) Detection of galactomannan antigenemia by enzyme immunoassay for the diagnosis of invasive aspergillosis: variables that affect performance. The Journal of Infectious Diseases, 190, 641–649. [DOI] [PubMed] [Google Scholar]
- Martinez, L.R. & Casadevall, A. (2007) Cryptococcus neoformans biofilm formation depends on surface support and carbon source and reduces fungal cell susceptibility to heat, cold, and UV light. Applied and Environmental Microbiology, 73, 4592–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher, R. , Lewis, Z. , Ambati, S. & Lin, X. (2021) Aiming for a bull's‐eye: targeting antifungals to fungi with dectin‐decorated liposomes. PLoS Pathogens, 17, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher, R. , Lewis, Z. , Ambati, S. & Lin, X. (2023) DectiSomes: C‐type lectin receptor‐targeted liposomes as Pan‐antifungal drugs. Advanced Drug Delivery Reviews, 196, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montone, K.T. (2009) Regulating the T‐cell immune response toward the H99 strain of Cryptococcus neoformans . The American Journal of Pathology, 175, 2255–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, D. , Shibata, K. & Yamasaki, S. (2017) C‐type lectin receptor Dectin‐2 binds to an endogenous protein beta‐glucuronidase on dendritic cells. PLoS One, 12, e0169562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planck, M. (1917) Über einen Satz der statistischen Dynamik und seine Erweiterung in der Quantentheorie. Sitzungsberichte der Preussischen Akademie der Wissenschaften zu Berlin, 24, 324–341. [Google Scholar]
- Preite, N.W. , Feriotti, C. , Souza de Lima, D. , da Silva, B.B. , Condino‐Neto, A. , Pontillo, A. et al. (2018) The Syk‐Coupled C‐type lectin receptors Dectin‐2 and Dectin‐3 are involved in Paracoccidioides brasiliensis recognition by human plasmacytoid dendritic cells. Frontiers in Immunology, 9, 464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo, S. & Iwakura, Y. (2011) Dectin‐1 and Dectin‐2 in innate immunity against fungi. International Immunology, 23, 467–472. [DOI] [PubMed] [Google Scholar]
- Schiavone, M. , Vax, A. , Formosa, C. , Martin‐Yken, H. , Dague, E. & Francois, J.M. (2014) A combined chemical and enzymatic method to determine quantitatively the polysaccharide components in the cell wall of yeasts. FEMS Yeast Research, 14, 933–947. [DOI] [PubMed] [Google Scholar]
- Spatafora, J.W. , Aime, M.C. , Grigoriev, I.V. , Martin, F. , Stajich, J.E. & Blackwell, M. (2017) The fungal tree of life: from molecular systematics to genome‐scale phylogenies. Microbiology Spectrum, 5, 1‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H. , Xu, X.Y. , Shao, H.T. , Su, X. , Wu, X.D. , Wang, Q. et al. (2013) Dectin‐2 is predominately macrophage restricted and exhibits conspicuous expression during Aspergillus fumigatus invasion in human lung. Cellular Immunology, 284, 60–67. [DOI] [PubMed] [Google Scholar]
- Taylor, J.W. & Berbee, M.L. (2006) Dating divergences in the fungal tree of life: review and new analyses. Mycologia, 98, 838–849. [DOI] [PubMed] [Google Scholar]
- U.S. Food and Drug Administration . (1997) Ambisome (amphotericin B). Silver Spring, MD: U.S. Food and Drug Admistration, 050740. [Google Scholar]
- Varshney, A. , Scott, L.J. , Welch, R.P. , Erdos, M.R. , Chines, P.S. , Narisu, N. et al. (2017) Genetic regulatory signatures underlying islet gene expression and type 2 diabetes. Proceedings of the National Academy of Sciences of the United States of America, 114, 2301–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, L. , Sood, P. , Lenardon, M.D. , Milne, G. , Olson, J. , Jensen, G. et al. (2018) The viscoelastic properties of the fungal Cell Wall allow traffic of AmBisome as intact liposome vesicles. mBio, 9, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, T. , Pan, D. , Zhou, Z. , You, Y. , Jiang, C. , Zhao, X. et al. (2016) Dectin‐3 deficiency promotes colitis development due to impaired antifungal innate immune responses in the gut. PLoS Pathogens, 12, e1005662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, G.J. , Marakalala, M.J. , Hoving, J.C. , van Laarhoven, A. , Drummond, R.A. , Kerscher, B. et al. (2015) The C‐type lectin receptor CLECSF8/CLEC4D is a key component of anti‐mycobacterial immunity. Cell Host & Microbe, 17, 252–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa, M. , Yamada, S. , Sugamata, M. , Kanauchi, O. & Morita, Y. (2021) Dectin‐2 mediates phagocytosis of Lactobacillus paracasei KW3110 and IL‐10 production by macrophages. Scientific Reports, 11, 17737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, L.L. , Zhao, X.Q. , Jiang, C. , You, Y. , Chen, X.P. , Jiang, Y.Y. et al. (2013) C‐type lectin receptors Dectin‐3 and Dectin‐2 form a heterodimeric pattern‐recognition receptor for host defense against fungal infection. Immunity, 39, 324–334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. SF1.
Data Availability Statement
All new data that were presented and/or discussed within this publication and its supplemental data section are included herein, and any previously published data that were discussed were appropriately cited. The AreaPipe pipeline for fluorescent area analysis will be uploaded at the Broad Institute's site for CellProfiler programs and made public once this study is accepted for publication as per the Broad Institute's suggestion for the time of release.


