Abstract
In this study, the microbial ecology of three naturally fermented sausages produced in northeast Italy was studied by culture-dependent and -independent methods. By plating analysis, the predominance of lactic acid bacteria populations was pointed out, as well as the importance of coagulase-negative cocci. Also in the case of one fermentation, the fecal enterocci reached significant counts, highlighting their contribution to the particular transformation process. Yeast counts were higher than the detection limit (>100 CFU/g) in only one fermented sausage. Analysis of the denaturing gradient gel electrophoresis (DGGE) patterns and sequencing of the bands allowed profiling of the microbial populations present in the sausages during fermentation. The bacterial ecology was mainly characterized by the stable presence of Lactobacillus curvatus and Lactobacillus sakei, but Lactobacillus paracasei was also repeatedly detected. An important piece of evidence was the presence of Lactococcus garvieae, which clearly contributed in two fermentations. Several species of Staphylococcus were also detected. Regarding other bacterial groups, Bacillus sp., Ruminococcus sp., and Macrococcus caseolyticus were also identified at the beginning of the transformations. In addition, yeast species belonging to Debaryomyces hansenii, several Candida species, and Willopsis saturnus were observed in the DGGE gels. Finally, cluster analysis of the bacterial and yeast DGGE profiles highlighted the uniqueness of the fermentation processes studied.
Fermentation is one of the oldest technologies used to preserve food for long periods, and it is a process in which microbes, meat, and technology converge. Besides the valuable protein and fat nutrients, flavor compounds are abundant and can vary, depending on raw materials, starter cultures and processing conditions. Optimization and standardization of both organoleptic and safety characteristics require detailed knowledge of the relative contribution of these three factors (33).
The microbial flora that are established and that participate in the phenomena of fermentation and ripening of sausages originate from the environment, the equipment, and the raw materials used in the manufacture of the products. It is not an easy task to determine the contribution of the different bacterial groups in the reactions associated with ripening since the microbial flora are diverse and the effects of the different ingredients must be taken into account (18).
The first studies on the ecology of fermented sausages date back to 1970 (28). From then on, several investigations established two groups of microorganisms as being the main organisms responsible for the transformations involved during fermentation and ripening of sausages. Lactic acid bacteria (LAB), in particular Lactobacillus spp., and gram-positive coagulase-negative cocci (CNC), specifically Staphylococcus and Kocuria spp., are considered technologically fundamental.
LAB are responsible for lactic acid production, for the “tangy” flavor of sausages, and for the small amounts of acetic acid, ethanol, acetoin, carbon dioxide, and pyruvic acid that are produced during fermentation, depending on the starter applied, the carbohydrate substrate, and the sources of meat proteins and additives (2). Some LAB strains are also able to produce antimicrobial compounds (bacteriocins), thereby enchancing the safety of fermented sausages (20), a process often termed bioprotection or biopreservation (19, 47).
CNC participate in the development and stability of a desirable red color through nitrate reductase activity that leads to the formation of nitrosomyoglobin. Furthermore, nitrate reduction produces nitrite that can limit lipid oxidation (48). Moreover, various aromatic substances and organic acids are released from their protease and lipase activity (34).
Early in the 1990s, Muyzer et al. developed a culture-independent method that has the potential to study the microbial flora quickly and economically, termed denaturing gradient gel electrophoresis (DGGE) (35). This approach has the advantage of directly profiling microbial populations present in specific ecosystems by separating PCR products that have originated with universal primers, on the basis of the melting domain of the DNA molecules. The trend to move toward methods that avoid the use of selective cultivation and isolation of bacteria from natural samples is justified, considering the biases related to traditional culture-dependent methods. As a matter of fact, different authors described tremendous differences between isolated and naturally occurring species present in various habitats (17, 22, 23, 36).
DGGE has been widely used to study the ecology of fermented foods (3, 5, 6, 8, 30, 32, 39, 49) and to profile pathogens directly in food samples (7), and its use in food microbiology was recently reviewed (14).
In this paper, we describe the application of culture-dependent and -independent methods to study the ecology of fermented sausages produced in three different plants in northeast Italy. Sausages were collected at determined time points and subjected to traditional microbiological analysis. DNA was extracted directly from the samples and amplified by PCR, using universal bacterial and yeast primers. DGGE analysis and sequencing of the resulted bands allowed fingerprinting of the microbial populations present in the three fermentations. Moreover, cluster analysis of the bacterial and yeast DGGE profiles was carried out to determine their similarity.
MATERIALS AND METHODS
Fermented sausage technology and sampling procedures.
Three local meat factories in northeast Italy were selected to carry out the study. They are called plants C, L, and U in this study. For the production of the fermented sausages, traditional techniques were employed without the use of starter cultures. Plant C was characterized by extended maturation of the sausages that lasted 120 days, while plant U produced sausages with fast maturation (28 days). Plant L sold the final product after 45 days of ripening. The following ingredients were common to all three fermentations: pork meat (60 kg), lard (40 kg), sodium chloride (2.5 kg), nitrite and nitrate (200 ppm), and black pepper (70 g). For fermentation C, no sugars were added, while for fermentations L and U, 1.5 and 2.5 kg of sugars, respectively, were used. The mixture was used to stuff natural casings, and the procedure resulted in fresh sausages that were 25 cm long and 5 cm in diameter. Ripening parameters were different for the three plants considered. Plant C performed maturations at low temperatures (below 10°C) and relative humidity (RH) between 60 and 90% for the first 60 days, followed by a 60-day period with temperatures of 14°C and RH of 65 to 85%. For plants L and U, the first stage consisted of 2 days of drying with an RH of 85% at 22°C. The temperature was then decreased to 12 and 14°C, for plants L and U, respectively, at a rate of 2°C per day with an RH between 60 and 90%. Ripening was then carried out for the rest of the period at the temperatures reached in storerooms with 65 to 85% RH. Triplicate samples of the sausages were used for microbiological and molecular analyses. For plant C, samplings were performed at 0, 3, 10, 20, 30, 60, 90, and 120 days, whereas for plant L, samplings were performed at days 0, 3, 10, 20, 30, and 45. Sausages from plant U, characterized by fast ripening, were examined at 0, 3, 5, 7, 14, and 28 days.
pH measurements.
Potentiometric measurements of pH were carried out with a pin electrode of a pH meter (Radiometer Copenhagen pH M82, Cecchinato, Italy) inserted directly into the sample. Three independent measurements were done on each sample. Means and standard deviations were calculated.
Microbiological analysis.
The samples were subjected to a microbiological analysis to monitor the changes in the populations responsible for ripening of fermented sausages, as well as their hygienic quality. Twenty-five grams of each sample was transferred into a sterile stomacher bag, 225 ml of saline-peptone water (8 g of NaCl per liter, 1 g of bacteriological peptone per liter; Oxoid) was added, and the mixture was treated for 1.5 min in a stomacher machine (PBI, Milan, Italy). Further decimal dilutions were made, and the following analyses were carried out on duplicate agar plates: (i) total aerobic mesophilic flora on Gelysate agar (Oxoid) incubated for 48 to 72 h at 30°C, (ii) LAB on MRS agar (Oxoid) incubated with a double layer at 30°C for 48 h, (iii) CNC on mannitol salt agar (Oxoid) incubated at 30°C for 48 h, (iv) total enterobacteria and Escherichia coli on Coli-ID medium (Bio-Merieux, Marcy d'Etoile, France) incubated with a double layer at 37°C for 24 to 48 h, (v) fecal enterococci on kanamycin-esculin agar (Oxoid) incubated at 42°C for 24 h, (vi) Staphylococcus aureus on Baird Parker medium (Oxoid) with added egg yolk tellurite emulsion (Oxoid) incubated at 37°C for 24 to 48 h, and (vii) yeasts and molds on malt extract agar (Oxoid) supplemented with tetracycline (1 mg/ml; Sigma, Milan, Italy) incubated at 25°C for 48 to 72 h. For Listeria monocytogenes (24) and Salmonella spp. (25), the International Organization for Standardization ISO/DIS methods were performed. After counting, means and standard deviations were calculated.
Direct extraction of DNA from sausages.
At each sampling point, 10-g samples, in triplicates, were homogenized in a stomacher bag with 20 ml of saline-peptone water for 1 min. After each preparation had settled for 1 min, 1 ml of supernatant was transferred into a screw-cap tube containing 0.3 g of glass beads with a diameter of 0.5 mm and centrifuged at 4°C for 10 min at 14,000 × g. The resulting pellet was treated with 1 ml of petrol ether-hexane (1:1; Sigma) for 10 min at room temperature to extract lipids. A second centrifugation was performed, as described above, and the pellet was resuspended in 150 μl of proteinase K buffer (50 mM Tris-HCl, 10 mM EDTA, pH 7.5, 0.5% [wt/vol] sodium dodecyl sulfate [SDS]). Twenty-five microliters of proteinase K (25 mg/ml; Sigma) was added, and treatment at 65°C for 1 h was performed. After this step, 150 μl of 2× breaking buffer (4% Triton X-100 [vol/vol], 2% [wt/vol] SDS, 200 mM NaCl, 20 mM Tris, pH 8, 2 mM EDTA, pH 8) was mixed in the tubes, and 300 μl of phenol-chloroform-isoamyl alcohol (25:24:1, pH 6.7) (Sigma) was added. Then, three 30-s treatments at the maximum speed, with an interval of 10 s, were performed in a bead beader (Fast Prep; Bio 101, Vista, Calif.). The tubes were centrifuged at 12,000 × g at 4°C for 10 min, and the aqueous phase was collected and mixed with 1 ml of ice-cold absolute ethanol. The DNA was precipitated at 14,000 × g at 4°C for 10 min, and the pellets were dried under vacuum at room temperature. Fifty microliters of sterile water was added, and a 30-min incubation at 45°C facilitated the nucleic acid solubilization. One microliter of DNase-free RNase (Roche Diagnostics, Milan, Italy) was added to digest the RNA by incubation at 37°C for 1 h.
PCR.
The primers used in this study are described in Table 1. Amplifications were carried out in a final volume of 25 μl, containing 2 μl (50 ng total) of template DNA, 10 mM Tris HCl, pH 8.3, 50 mM KCl, 0.2 mM deoxynucleoside triphosphates (dNTPs), 1.25 U of Taq polymerase (Applied Biosystem, Milan, Italy), and 0.2 μM each primer. For primers P1 and P2, 1.5 mM MgCl2 was employed, whereas for primers NL1 and LS2, a concentration of 2 mM was used. Amplifications were carried out in a PTC-220 DNA Engine Dyad MJ Research thermalcycler (Celbio, Milan, Italy). The amplification cycle for primers P1 and P2 was characterized by an initial touchdown step in which the annealing temperature was lowered from 60 to 52°C in intervals of 2°C every 2 cycles, and 20 additional cycles were done with annealing at 50°C. Denaturation was performed at 95°C for 1 min, and extension was performed at 72°C for 1 min 30 s. For primers NL1 and LS2, reactions were run for 30 cycles of denaturation at 95°C for 60 s, annealing at 52°C for 45 s, and extension at 72°C for 60 s. For both amplification cycles, an initial denaturation at 95°C for 5 min and a final extension at 72°C for 7 min were carried out as well. Five microliters of each PCR mixture was analyzed by electrophoresis in a 0.5× Tris-borate-EDTA (TBE) agarose gel.
TABLE 1.
PCR primers used in this study
DGGE analysis.
The Dcode Universal Mutation Detection system (Bio-Rad, Hercules, Calif.) was used for DGGE analysis. Electrophoreses were performed in an 0.8-mm-thick polyacrylamide gel (8% [wt/vol] acrylamide-bisacrylamide at 37.5:1), using a denaturant gradient increasing in the direction of the electrophoretic run. For PCR products obtained with primers P1 and P2, a denaturant gradient from 40 to 60% (100% corresponded to 7 M urea and 40% [wt/vol] formamide) was used. Amplicons produced by primers NL1 and LS2 were analyzed in a denaturant gradient from 30 to 60%. Electrophoretic runs were carried out at a constant temperature of 60°C in 1.25× Tris-acetate-EDTA (TAE) for 3.5 h at 130 V for the P1 and P2 primers and for 4 h at 120 V for the NL1 and LS2 primers. After the electrophoresis, gels were stained for 20 min in 1.25× TAE containing 1× SYBR green (final concentration; Molecular Probes, Eugene, Oreg.) and visualized under UV light. Images were digitally captured and analyzed with the BioImaging System GeneGenius (SynGene, Cambridge, United Kingdom) for the recognition of the bands present. Comparisons of DGGE fingerprints obtained from the different sausages were performed with the pattern analysis software package Gel Compare, version 4.1 (Applied Maths, Kortrijk, Belgium). Calculation of similarity in the profiles of bands was based on Pearson product-moment correlation coefficient. Dendrograms were obtained by means of the unweighted pair group method using arithmetic average (UPGMA) clustering algorithm (50). DGGE analyses were performed at least twice. Normalization of the gels was performed by using band ladders. The bacterial ladder was the result of amplification of Lactobacilus casei DSM 20011 by primers P1 and P2, as described by Cocolin et al. (6), which was subsequently analyzed by DGGE. It was selected for its characteristic of giving a multiband pattern. The yeast ladder was a mixture of Hanseniaspora uvarum DBVPG 6717, Kluyveromyces thermotolerans DBVPG 6480, Saccharomyces cerevisiae DBVPG 6173, and Brettanomyces bruxellensins DBVPG 6706, amplified with primers NL1 and LS2, as reported by Cocolin et al. (5). To allow normalization before cluster analysis, ladders were loaded in the first and last wells of each gel run.
Sequencing of DGGE bands and sequence analysis.
Blocks of polyacrylamide gels containing selected DGGE bands were punched with sterile pipette tips. The blocks were then transferred in 50 μl of sterile water, and the DNA of the bands was allowed to diffuse overnight at 4°C. Two microliters of water, containing the eluted DNA, was used for the reamplification, and the PCR products, generated with the GC-clamped primer, were checked by DGGE with sausage amplified DNA as a control. Only products migrating as a single band and at the same position with respect to the control were amplified with the primer without the GC clamp, cloned in pGEM-T Easy vector (Promega, Milan, Italy), and sequenced by a commercial facility (MWG Biotech, Ebersberg, Germany). Searches in the GenBank database were performed with the BLAST program (1) to determine the closest known relatives of the partial 16S rRNA sequence obtained.
RESULTS
Traditional microbiological analysis.
The results of the plate counts obtained from the three fermentations monitored in this study are reported in Table 2. Both the microbial population changes during fermentation and the hygienic quality of the sausages were monitored. Fermentation C was characterized by a slow increase in the counts of almost all of the groups of microorganisms observed. Only the CNC showed a significant increase from 103 to 104 to 105 CFU/g from zero to 3 days, as well as yeasts that increased from 102 to 103 to 104 CFU/g. The total aerobic count and counts of LAB, fecal enterococci, total enterobacteria, and E. coli did not increase in the first 3 days. Significant growth was observed at 10 days, when almost all of the microbial groups showed an increase from 2 to 4 log10 CFU/g. For CNC and total enterobacteria, a slightly slower increase was noticed and counts moved from 104 to 105 to 105 to 106 CFU/g and from 102 to 103 to 103 to 104 CFU/g, respectively. At 10 days, the E. coli count was <10 CFU/g, and it remained below that limit. The rest of the transformation process was characterized by a slight increase in the counts of total bacteria and the establishment of the LAB population. LAB increased from 105 to 106 CFU/g at 10 days to 107 to 108 CFU/g at 20 days, counts that remained stable until the end of the process. CNC and yeasts showed a decrease during the central period of the fermentation, with an increase in the counts at the last sampling point, reaching 105 to 106 and 106 to 107 CFU/g, respectively. The same trend was observed for the fecal enterococci too, whereas total enterobacteria were undetectable starting from day 90 of the fermentation. Molds were below 100 CFU/g throughout the period observed, and S. aureus, L. monocytogenes, and Salmonella spp. were absent in 25 g of product in the first 20 days of fermentation.
TABLE 2.
Microbial dynamics, as determined by plating, of the fermentations monitored in this study
| Microbiological analysis parameter | Result (log10 CFU/g) for fermentation at day givena | |||||||
|---|---|---|---|---|---|---|---|---|
| Fermentation C | 0 | 3 | 10 | 20 | 30 | 60 | 90 | 120 |
| Total aerobic count | 4.46 (0.27) | 4.49 (0.04) | 7.03 (0.88) | 6.48 (0.41) | 6.97 (0.56) | 7.24 (0.26) | 7.11 (0.03) | 7.02 (0.02) |
| CNC | 3.23 (0.10) | 4.70 (0.63) | 5.37 (0.80) | 5.72 (0.16) | 5.09 (0.12) | 3.58 (0.16) | 4.30 (0.12) | 5.20 (0.21) |
| LAB | 3.17 (0.93) | 3.74 (0.07) | 6.37 (0.69) | 7.83 (0.02) | 7.58 (0.12) | 7.28 (0.50) | 7.87 (0.51) | 8.78 (0.21) |
| Yeasts | 2.00 (0.00) | 3.12 (0.46) | 5.36 (0.02) | 5.90 (0.14) | 4.98 (0.25) | 2.50 (0.08) | 3.12 (0.09) | 6.73 (0.05) |
| Molds | <100 (NA) | <100 (NA) | <100 (NA) | <100 (NA) | <100 (NA) | <100 (NA) | <100 (NA) | <100 (NA) |
| Fecal enterococci | 2.15 (0.27) | 2.40 (0.17) | 5.33 (0.33) | 7.18 (0.11) | 6.76 (0.07) | 6.81 (0.03) | 3.65 (0.15) | 4.12 (0.20) |
| Total enterobacteria | 1.73 (0.16) | 2.16 (0.43) | 3.20 (0.42) | 2.98 (0.58) | 1.97 (0.09) | 1.98 (0.02) | <10 (NA) | <10 (NA) |
| E. coli | 1.25 (0.24) | 1.43 (0.22) | <10 (NA) | <10 (NA) | <10 (NA) | <10 (NA) | <10 (NA) | <10 (NA) |
| S. aureus | <100 (NA) | <100 (NA) | <100 (NA) | <100 (NA) | <100 (NA) | <100 (NA) | <100 (NA) | <100 (NA) |
| L. monocytogenes | A | A | A | A | NP | NP | NP | NP |
| Salmonella spp. | A | A | A | A | NP | NP | NP | NP |
| Fermentation L | 0 | 3 | 10 | 20 | 30 | 45 | ||
| Total aerobic count | 4.74 (0.44) | 3.59 (0.36) | 8.03 (0.56) | 7.68 (0.14) | 8.55 (0.30) | 4.19 (0.23) | ||
| CNC | 3.11 (0.18) | 3.65 (0.15) | 6.83 (0.07) | 6.12 (0.71) | 5.86 (0.06) | 4.63 (0.20) | ||
| LAB | 5.29 (0.18) | 5.97 (0.27) | 7.81 (0.21) | 7.94 (0.12) | 7.93 (0.16) | 8.34 (0.14) | ||
| Yeasts | 3.55 (0.80) | 2.93 (0.05) | <100 (NA) | <100 (NA) | <100 (NA) | <100 (NA) | ||
| Molds | <100 (NA) | <100 (NA) | <100 (NA) | <100 (NA) | <100 (NA) | <100 (NA) | ||
| Fecal enterococci | 3.18 (0.12) | 4.47 (0.77) | 5.95 (0.15) | 4.95 (0.96) | 4.00 (0.00) | 5.47 (0.47) | ||
| Total enterobacteria | 1.94 (0.15) | 3.59 (0.36) | 4.29 (0.21) | 3.80 (0.66) | 4.28 (0.21) | 2.56 (0.09) | ||
| E. coli | 1.72 (0.63) | <10 (NA) | <10 (NA) | <10 (NA) | <10 (NA) | <10 (NA) | ||
| S. aureus | <100 (NA) | <100 (NA) | <100 (NA) | <100 (NA) | <100 (NA) | <100 (NA) | ||
| L. monocytogenes | A | A | A | A | NP | NP | ||
| Salmonella spp. | A | A | A | A | NP | NP | ||
| Fermentation U | 0 | 3 | 5 | 7 | 14 | 28 | ||
| Total aerobic count | 5.09 (0.20) | 5.41 (0.22) | 5.88 (0.19) | 5.33 (0.34) | 6.90 (0.56) | 9.11 (0.47) | ||
| CNC | 3.66 (0.10) | 3.80 (0.63) | 5.67 (0.57) | 4.62 (0.69) | 5.54 (0.06) | 5.01 (0.12) | ||
| LAB | 4.20 (0.17) | 8.24 (0.01) | 7.81 (0.10) | 8.21 (0.03) | 8.28 (0.16) | 8.45 (0.06) | ||
| Yeasts | 3.33 (0.06) | <100 (NA) | <100 (NA) | <100 (NA) | 3.53 (0.68) | 2.32 (0.28) | ||
| Molds | 2.59 (0.26) | <100 (NA) | <100 (NA) | <100 (NA) | <100 (NA) | <100 (NA) | ||
| Fecal enterococci | 3.73 (0.38) | 5.60 (0.00) | 5.83 (0.22) | 6.02 (0.16) | 6.08 (0.17) | 6.28 (0.32) | ||
| Total enterobacteria | 2.38 (0.06) | 4.94 (0.21) | 4.88 (0.38) | 2.18 (0.15) | 4.70 (0.41) | 4.66 (0.50) | ||
| E. coli | <10 (NA) | <10 (NA) | <10 (NA) | <10 (NA) | <10 (NA) | <10 (NA) | ||
| S. aureus | <100 (NA) | <100 (NA) | <100 (NA) | <100 (NA) | <100 (NA) | <100 (NA) | ||
| L. monocytogenes | A | A | A | A | A | NP | ||
| Salmonella spp. | A | A | A | A | A | NP | ||
Results are shown as the mean with standard deviation in parentheses. NA, not applicable; NP, not performed; A, absent in 25 g of product.
Fermentation L showed high counts of LAB populations right from the beginning of the fermentation. Already at day zero, the counts were 105 to 106 CFU/g, and the number rapidly increased to 107 to 108 CFU/g at 10 days. From then, the counts remained stable, with a slight increase at the end of the period, reaching 108 to 109 CFU/g at day 45. In terms of total bacterial count, CNC and fecal enterococci were characterized by a significant increase in the counts from 3 to 10 days, which were stable until the end of the fermentation. Yeasts were detected only at days zero and 3, while molds were always <100 CFU/g throughout the period monitored. E. coli was counted only at day zero, becoming undetectable from day 3. S. aureus, L. monocytogenes, and Salmonella spp. were not detected in the sausages produced in plant L.
Fermentation U was characterized by very fast growth of LAB populations: in fact, from the initial count of 104 CFU/g, they increased to 108 to 109 CFU/g already at 3 days. These counts were stable until the 28th day of fermentation. The total aerobic count was determined to be 105 CFU/g at day zero, which did not change significantly until the last day of sampling. As a matter of fact, at 28 days, it reached 109 CFU/g, which represented the highest count between the three fermentations monitored. CNC increased significantly between the 3rd and 5th days of transformation and remained stable after that point. As described for LAB, fecal enterococci showed a fast increase from zero to 3 days. They reached counts of 105 to 106 CFU/g at that moment, and from then on, a slight increase until the end of the fermentation was observed. The counts for yeasts were 103 to 104 CFU/g at day zero, and after a 2-week period, in which they were not detected, at 14 days, the yeast count was determined to be 103 to 104 CFU/g, decreasing to 102 to 103 CFU/g at the last day of sampling. Molds were only detected at day zero. Total enterobacteria grew appreciably between zero and 3 days of fermentation. The value of 104 to 105 CFU/g remained stable, with the exception of day 7, when the enterobacteria decreased to 102 to 103 CFU/g. E. coli, S. aureus, L. monocytogenes, and Salmonella were not detected.
pH measurements.
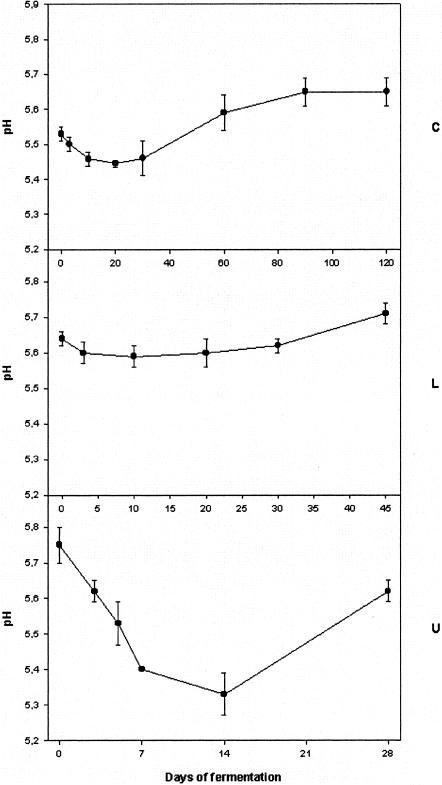
The results of the pH measurements are shown in Fig. 1. Fermentation C was characterized by an initial pH of 5.52 that decreased during the first 20 days of fermentation. From day 30, it started to increase, reaching a final value of about 5.65. For fermentation L, no significant decrease was observed at the beginning of the fermentation: from 5.62, pH decreased to 5.60 during the first 20 days. Only at the end of the transformation did it increase, reaching values of about 5.70. A different picture was obtained from fermentation U, in which a steep drop in the pH values was registered from day zero to day 14. From an initial pH of 5.75, it reached 5.32, which increased to 5.62, the final pH value of the sausages produced in plant U.
FIG. 1.
pH trends of the fermentations monitored in this study. Letters (C, L, and U) on the plots indicate the fermentation code.
DGGE profiles.
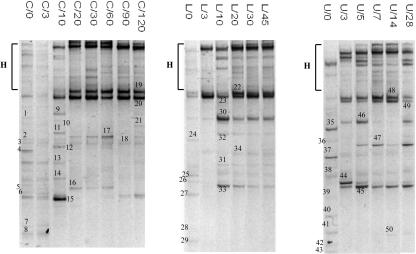
The bacterial and yeast DGGE profiles of the three fermentations are shown in Fig. 2 and 3, respectively, and the results of the band sequencing are reported in Tables 3 and 4. No differences were detected in the DGGE profiles when replicates obtained at the same sampling point were analyzed (data not shown). To make sure that no biases were introduced by the cloning step on PCR-amplified bands, representative clones were selected and subjected to PCR-DGGE analysis with sausage amplified DNA as a control. We always obtained a comigration between the original band cut from the DGGE gel and the band amplified from the clones, thereby confirming the validity of the approach used. Direct cloning of the cut bands could not be used because it failed in the case of faint bands (data not shown). Concerning the bacterial ecology as determined by DGGE, fermentation C was characterized by a complex picture during the first 10 days. Members of the LAB group (L. sakei and Lactobacillus curvatus), species of Staphylococcus (S. xylosus, S. sciuri or S. pulvereri, and S. equorum or S. succinus), Macrococcus caseolyticus, and Bacillus sp. were identified. Moreover, one unidentified bacterium, with the highest homology in the BLAST search to bacterium Ellin5218, was detected. This picture did not change significantly at 3 days, and only at day 10 did new bands show up in the DGGE gels. Different signals belonged to S. xylosus (Fig. 2, bands 9, 11, 12, and 14), while the remaining signals were identified as S. equorum or S. succinus. Interestingly, band 10 (Fig. 2) was determined to be Lactococcus garvieae. From day 20, the ecology of sausages produced in plant C was dominated by LAB members. L. sakei, L. curvatus, and Lactobacillus paracasei were the main populations detected from days 20 to 60. After 90 days of ripening, only L. paracasei was not detected anymore, and S. equorum or S. succinus showed up again in the gels. This situation remained stable until day 120. However a new band (Fig. 2, band 21) belonging to Brochothrix thermosphacta appeared at the last sampling point. Fermentation L was characterized, from day zero, by the presence of the specific doublet for L. curvatus and L. sakei, placed in the upper part of the gel, which was always present throughout the fermentation. At day zero, S. sciuri or S. pulvereri, L. paracasei, S. intermedius, Bacillus sp., and Ruminococcus sp. were detected. Moreover, a faint band identified as Lactococcus garvieae was present as well. This species was detected as well at all of the following sampling points. From day 10, the picture of the ecology did not change; in fact, the DGGE patterns were characterized by the same bands belonging to L. sakei, L. curvatus, Lactococcus garvieae, and S. equorum or S. succinus. Regarding fermentation U, a rich DGGE profile was obtained at day zero. B. thermosphacta, L. curvatus, S. equorum or S. succinus, S. sciuri or S. pulvereri, M. caseolyticus, and Bacillus pumilis were identified. This situation dramatically changed at 3 days, where L. sakei, L. curvatus, Lactococcus garvieae, L. paracasei, and S. equorum or S. succinus took over the fermentation process. The ecology showed slight changes as the transformation proceeded. A band belonging to S. xylosus (Fig. 2, band 47) appeared in the gel, and it was detected until day 28, while L. paracasei disappeared after day 5. At the last sampling points of fermentation U, M. caseolyticus and Lactococcus garvieae were detected again. In all three fermentations, different bands were visible in the upper part of the DGGE gels. They were all determined to be heteroduplexed by sequencing (data not shown).
FIG. 2.
Bacterial DGGE profiles. Lane designations indicate the fermentation and the day of sampling. The gels shown are not normalized. Bands indicated by numbers were excised and after reamplification subjected to sequencing. H, heteroduplex bands.
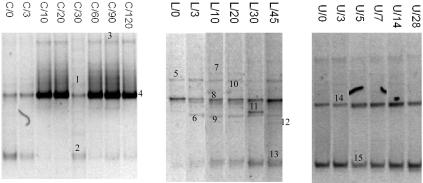
FIG. 3.
Yeast DGGE profiles. Lane designations indicate the fermentation and the time of sampling. The gels shown are not normalized. Bands indicated by numbers were excised and after reamplification subjected to sequencing.
TABLE 3.
Sequencing results from the bands cut from the bacterial DGGE gels
| Band(s)a | Size (bp) | Closest relative | % Identity | Sourceb |
|---|---|---|---|---|
| 1 | 113 | Lactobacillus sakei | 99.1 | AB124845 |
| 2 | 91 | Staphylococcus xylosus | 98.9 | AY126259 |
| 3, 18, 24, 36 | 113 | Lactobacillus curvatus | 99.6 | AY204894 |
| 4 | 75 | Bacterium Ellin5218 | 100 | AY234569 |
| 5, 25, 38 | 91 | Staphylococcus sciuri or S. pulvereric | 98.9 | AY126231 or AY126216 |
| 6 | 91 | Staphylococcus equorum or S. succinus | 98.2 | AF527483 or AY126240 |
| 7, 41 | 93 | Macrococcus caseolyticus | 96.7 | AY126157 |
| 8 | 91 | Bacillus sp. | 98.8 | AY461682 |
| 9 | 91 | Staphylococcus xylosus | 98.6 | AY126259 |
| 10, 30, 46 | 93 | Lactococcus garvieae | 99.6 | AY438044 |
| 11 | 91 | Staphylococcus xylosus | 97.9 | AY126259 |
| 12, 47 | 91 | Staphylococcus xylosus | 98.9 | AY126259 |
| 13, 37 | 91 | Staphylococcus equorum or S. succinusc | 98.7 | AF527483 or AY126240 |
| 14 | 91 | Staphylococcus xylosus | 99.0 | AY126259 |
| 15, 33, 45 | 91 | Staphylococcus equorum or S. succinus | 98.9 | AF527483 or AY126240 |
| 16, 26, 44 | 111 | Lactobacillus paracasei | 99.0 | AY204894 |
| 17, 32 | 111 | Lactobacillus curvatus | 99.1 | AY204894 |
| 19, 22, 48 | 111 | Lactobacillus curvatus | 97.2 | AY204894 |
| 20, 23, 49 | 113 | Lactobacillus sakei | 99.5 | AB124845 |
| 21, 35 | 91 | Brochothrix thermosphacta | 98.9 | M58798 |
| 27, 39 | 93 | Staphylococcus intermedius | 100 | D83369 |
| 28 | 91 | Bacillus sp. | 98.8 | AY461682 |
| 29 | 106 | Ruminococcus sp. | 100 | AY442822 |
| 31 | 114 | Lactobacillus sakei | 98.2 | AB124845 |
| 34 | 93 | Lactococcus garvieae | 98.9 | AY438044 |
| 41, 50 | 93 | Macrococcus caseolyticus | 98.9 | AY126157 |
| 42 | 95 | Macrococcus caseolyticus | 97.7 | AY126157 |
| 43 | 91 | Bacillus pumilis | 98.8 | X60637 |
Bands are numbered as indicated on the DGGE gels shown in Fig. 2.
Accession number of the sequence of the closest relative found by BLAST search.
The V1 regions were 100% identical, therefore avoiding identification.
TABLE 4.
Sequencing results of the bands cut from the yeast-DGGE gels
| Band(s)a | Size (bp) | Closest relative | % Identity | Sourceb |
|---|---|---|---|---|
| 1 | 241 | Candida tropicalis | 97.5 | AF267497 |
| 2, 13, 15 | 246 | Mayaca fluviatilis | 86.5 | AF293855 |
| 3 | 249 | Debaryomyces hansenii | 99.3 | AF485980 |
| 4 | 249 | Debaryomyces hansenii | 99.3 | AF485980 |
| 5 | 242 | Candida krisii | 100 | AJ539355 |
| 6 | 248 | Willopsis saturnus | 99.6 | AJ507804 |
| 7, 10 | 247 | Aerobasidium pullulans | 100 | AY185811 |
| 8 | 248 | Willopsis saturnus | 100 | AJ507804 |
| 9 | 248 | Candida fermentati | 100 | AY187283 |
| 10 | 248 | Willopsis saturnus | 99.8 | AJ507804 |
| 12 | 203 | Entyloma dahliae | 100 | AY272033 |
| 14 | 242 | Candida sake or C. austromarinac | 98.9 | AJ549822 or U62310 |
Bands are numbered as indicated on the DGGE gels shown in Fig. 3.
Accession number of the sequence of the closest relative found by BLAST search.
The D1-D2 loop regions were 100% identical, therefore avoiding identification.
The yeast ecology, as presented in Fig. 3, was considerably more simple than the bacterial ecology. In fermentation C, the main yeast detected was Debaryomyces hansenii, which was present right from the beginning of the fermentation. From day 10, its band was very intense until the end, and only at 30 days of fermentation did it decrease in intensity, allowing the detection of Candida tropicalis (Fig. 3, band 1). During fermentation C, another band was stably present in the DGGE gels. Band 2, absent only at days 10 and 20, was identified as Mayaca fluviatilis, although the identity retrieved from the GenBank database was too low to guarantee a correct identification. Fermentation L presented the highest number of bands within the fermentations monitored. Candida krisii and Willopsis saturnus were present from day zero, and Aerobasidium pullulans and Candida fermentati were detected at 3 and 30 days of fermentation and at days 10 and 20, respectively. In the last sampling point, a faint Entyloma dahliae band was detected. As described for fermentation C, also in this case, a band identified as M. fluviatilis was present. For fermentation U, the yeast ecology result was the simplest. Only two bands, corresponding to Candida sakei or Candida austomarina and M. fluviatilis were present throughout the fermentation.
Cluster analysis of the DGGE profiles.
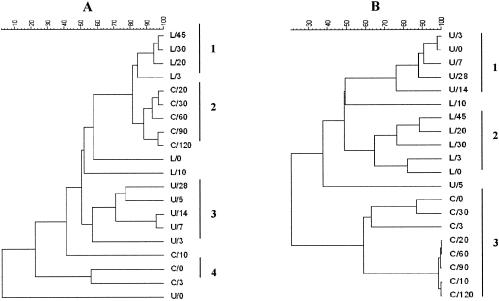
The results of the cluster analysis of the bacterial and yeast DGGE profiles of the three fermentations are reported in Fig. 4A and B, respectively. As shown, the bacterial ecology was determined to be fermentation specific, since samples coming from the same fermentation, collected at different time points, were almost always grouped in the same cluster. Fermentations L and C were more similar to each other than to fermentation U. As a matter of fact, two clusters grouping at four time points of fermentation L and five time points of fermentation C had a similarity of about 80% (Fig. 4A, clusters 1 and 2). The samples from fermentation U were more diverse than those from fermentations C and L, showing a similarity of about 60% (Fig. 4A, cluster 3). Samples taken from the first days of fermentation (days zero and 3 and in fermentation C also at day 10) formed single clusters, apart from samples taken at days zero and 3 during fermentation C, which were grouped in one cluster with a similarity of 60%. In Fig. 4B, the cluster analysis of the yeast DGGE is presented. In this case, a fermentation-specific distribution of the samples was obtained, creating clusters containing only samples from the same fermentation (Fig. 4B, clusters 1, 2, and 3). Already with a coefficient of discrimination of about 50%, the three fermentations could be separated, while only a sample analyzed at day 10 from fermentation L and a sample analyzed at day 5 from fermentation U were found to be highly diverse from the rest and did not belong to any cluster.
FIG. 4.
Cluster analysis of the profiles obtained by bacterial (A) and yeast (B) DGGE analysis. Dendrograms were obtained with the UPGMA clustering algorithm. Samples are indicated by the fermentation letter and the day of analysis. Identified clusters are indicated by numerals to the right of each panel.
DISCUSSION
Fermented sausages are a group of traditional products with great diversity in production methods and organoleptic characteristics between different countries and different regions of the same country. In Europe they have a long history, originating from Mediterranean countries during Roman times (28). Extensive studies have been performed on the characterization of traditional sausages produced in Greece, Italy, and Spain (4, 9, 10, 15, 16, 21, 31, 38, 41-44). A wide variety of microorganisms have been isolated from sausage fermentations by traditional methods. They belong to LAB and Staphylococcus and Kocuria spp. (12, 21). Among LAB, L. sakei, L. curvatus, and Lactobacillus plantarum are the species most frequently isolated in acid-fermented meat products (2). However, in some cases, the contribution of enterococci seems to be important (13). S. xylosus, S. carnosus, and S. saprophyticus are the CNC representatives involved in the fermentation of naturally fermented sausages (2, 9, 11, 29, 37).
The study of the ecology of the naturally fermented sausages has been carried out so far by traditional microbiological methods, based on plate counts, isolation, and biochemical identification. This approach has been repeatedly criticized because only easily culturable microorganisms can be detected, while members that need elective enrichments or that are in a particular physiological condition (in a sublethal or injured state) are lost. Only in a few cases and in the last 5 years has the development of molecular techniques been taken into account and exploited to study the microbial compositions of fermented sausages. Of particular interest are the direct culture-independent methods that are able to profile microbial populations avoiding the biases described above. In 2001, we published a paper on the development of a direct PCR-DGGE analysis to profile bacterial populations in Italian fermented sausages (6). The main evidence found was the great impact of LAB populations, which were stably present at both the DNA and RNA levels throughout the fermentation, possibly accounting for important transformations and determination of the organoleptic profile of these products. Concerning the CNC, after the first days of fermentation, only S. xylosus was detected.
In this paper, we wanted to compare the ecologies of three traditional fermented sausages produced in three plants of the northeastern part of Italy, as determined by traditional and molecular methods. The plants were placed in different locations of the Friuli-Venezia-Giulia region, and they were selected based on the different technologies used for the production and considering the fact that no use of starter cultures takes place. Plant C produces sausages with long maturation times (120 days), and fermentation is performed at low temperatures. In plant L, the sausages are sold after 45 days, while in plant U, fermentations end just after 28 days. The final products of the three plants have different organoleptic profiles. While sausages from plant U are characterized by an acid taste, the ones obtained from plant C are more aromatic. The products from plant L are slightly acid, with an appreciable aroma. To study the ecology of the naturally fermented sausages considered, a multiphasic approach was used, characterized by a simultaneous use of culture-dependent and -independent methods. Samples were analyzed on agar plates, and counts were determined after incubation. DNA was directly extracted from the sausages, amplified with universal bacterial and yeast primers, and after DGGE analysis of the amplicons obtained as well as sequencing of the bands present in the gels, the profiling of the ecology was revealed. Moreover, the fingerprints obtained by DGGE were subjected to cluster analysis to understand how different the three fermentations were.
The only significant difference in the microbial dynamics as determined by plating analysis was the diverse growth of the LAB in three sausages studied. These differences in the LAB trend become clear if the recipes used for the production and the fermentation conditions are taken into account. Fermentation U was characterized by the highest addition of fermentable sugars (about 2.5%) and a temperature of fermentation of 22°C, thereby allowing fast growth of the LAB populations right from the beginning. In fermentation L, the same temperature conditions were used, but most probably the sugar content (1.5%) did not support fast growth as described for fermentation U. In contrast, the slow growth of LAB in fermentation C must be the result of the low temperatures used in the fermentation process and the absence of added sugars. Nevertheless, for all of the fermentations, LAB reached about the same final number at the last sampling point (108 to 109 CFU/g). The LAB dynamics directly determined the pH values of the sausages studied. In fact, fermentation U was characterized by the highest jump in pH values at the onset of the fermentation (Fig. 1), while the pH drop for fermentations C and L was smoother and followed the increase in the LAB counts. It is interesting that in all of the fermentations considered in the study, the fecal enterococci reached a value of at least 106 CFU/g during fermentation, becoming an important population that possibly influences the final organoleptic characteristics of the product. Regarding yeasts, only for fermentation C was a significant increase observed. Concerning the safety aspect of the sausages studied, S. aureus, L. monocytogenes, and Salmonella spp. were never detected. Moreover, E. coli was only counted in the very first days of fermentations C and L. This information points out that good manufacturing practices are implemented and followed in the three plants considered in this research.
It is widely recognized that the main sources of microorganisms in a food fermentation, if starters are not employed, are the ingredients used (in the case of sausage fermentation, meat, salt, and spices) and the production environment. Thus, we can conclude that differences in the microbial species isolated in the three fermentations are due to the diverse distributions of these microbial species in the ingredients and the production environments of the three plants. The species naturally present are subject to a selection process that takes place during fermentation and are eventually defining the dynamics of the fermentations and the final characteristics of the products. In this study, only species able to grow at relatively low temperatures (22 to 12°C for fermentations L and U and 14 to 10°C for fermentation C) and salt resistant were able to colonize the product. Mainly LAB and CNC were responsible for the transformations, but contributions from yeasts and enterococci should be considered as well. The microbial ecologies described in this study are in agreement with the results previously described by other authors. LAB are mainly responsible for the acidification that occurs during the first days of fermentation, while CNC and yeasts participate in the proteolysis and lipolysis processes that correlate with the final characteristics of the product.
When the sausages were subjected to direct analysis by PCR-DGGE, important information became available. A general consideration is that the main differences detected in the ecology were not represented by the species of microorganisms identified by band sequencing but by their relative distribution between the fermentations. Considering the dynamic changes highlighted by the DGGE profiles, fermentation C presented wider biodiversity than fermentations L and U. In these two cases, already at day 3, intense bands were visible, underlining the predominance of specific microbial populations represented by L. sakei, L. curvatus, Lactococcus garvieae, and S. equorum or S. succinus. In fermentation C, complex DGGE profiles were observed until day 20, when species of LAB took over the fermentation. As reported in Tables 2 and 3, several DGGE bands belonged to the same bacterial or yeast species, confirming the presence of multiple copies of the rRNA genes, as previously described also by other authors (6, 30, 39). The bacterial ecology (Fig. 2 and Table 3) was mainly characterized by the strong presence of LAB populations. In all three fermentations, a stable signal from the beginning of the period studied was visible for L. curvatus and L. sakei, which remained constant throughout the transformation. Fermentation C was characterized by strong signal only after day 10, while for fermentations L and U, the signal was already appreciable after 3 days. This fact can be explained by considering once more the technological parameters used. In fact, in fermentation C, no sugars were added and low temperatures (<10°C) were used in the first part of the fermentation. Contribution by L. paracasei was found in fermentation C, where its specific band was visible from day 20 to day 60. Another LAB representative, Lactococcus garvieae, was determined to have an important impact on fermentations L and U. Lactococcus garvieae has been described as a common fish pathogen (40), but it is also frequently isolated from dairy products. Strains of this species are able to produce antimicrobial substances (51). Another important contribution to bacterial ecology was given by Staphylococcus species. S. equorum or S. succinus was present in all of the transformations considered. S. xylosus was mainly found in fermentation U; its specific band was present from day 5. It was not revealed in fermentation L, while in fermentation C, multiple bands belonging to S. xylosus were recognized only at day 10. As previously described (6), the main biodiversity is found at the beginning of the fermentation. As the ripening proceeds, the better adapted populations (LAB and CNC) outnumber the rest of the microflora. This dominance explains the less complex DGGE profile at the end of the fermentation. Bacillus spp., Ruminococcus sp., S. intermedius, and M. caseolyticus were detected at day zero. However, a band belonging to M. caseolyticus was present at the end of fermentation too for plant U. No L. plantarum, an LAB commonly associated with fermented meat products, was detected in the fermentations considered in the study, thereby confirming the results already reported in a previous study (6), in which dead L. plantarum cells were detected only at day zero.
The yeast ecology results are characterized by members of the genera Candida, Debaryomyces, and Willopsis. Fermentation C was the only one that showed a specific band for D. hansenii, described as proteolytic yeast during fermented sausage production (45), right from the beginning of the fermentation, and it was not detected in fermentations L and U. C. krisii and W. saturnus for fermentation L and C. sake or C. austromarina for fermentation U were the main species detected. It should be pointed out that a common band was detected in all of the fermentations monitored. After sequencing, the band was identified as M. fluviatilis, but the low identity retrieved (86.5%) suggests that the band cut from the gel belongs to a yeast species not yet identified or to a yeast member for which the 26S rRNA gene (D1-D2 loop) has not been deposited yet in GenBank. It should be underlined that growth of yeasts was observed only in fermentation C, where a species highly adapted to fermented sausages was identified. For fermentations L and U, no significant increase was detected. The species found in these two cases were present in the ingredients used for production, but they did not contribute to the fermentation.
Cluster analysis of the DGGE profiles highlighted how the three fermentations shared different level of similarities, when bacterial and yeast ecologies were considered. From the point of view of bacterial profiles, the results for fermentations L and C were very similar (>80%), while fermentation U was more distinct. Samples analyzed in the early stages of fermentations were more diverse, and they formed single clusters. Concerning the yeast ecology, the fermentations considered here showed a specific yeast pattern. On the basis of these results, the finding can be sustained that both bacterial and yeast ecologies were characteristic of each fermentation, thereby allowing their differentiation.
The application of culture-dependent and -independent methods highlighted the stable presence of the LAB members and the contribution to the bacterial ecology of Staphylococcus species. It was also observed that LAB DGGE band intensities did not correlate with LAB concentrations obtained by plating on MRS agar. Surprisingly, no bands belonging to fecal enterococci were detected in the DGGE gels, although their counts were above the detection limit of the method, determined to be 103 to 104 CFU/g when the predominant populations are above 108 CFU/g (6), in all of the fermentations studied. This evidence can be explained by considering masking effects by DNA belonging to the major populations present, but most probably is the result of poor amplification of Enterococcus spp. obtained by the protocol applied. Efforts to increase the intensity of the PCR amplicons produced from enterococcal DNA were carried out. Different primers, MgCl2 concentrations, and annealing temperatures were tested without success (data not shown). We admit that the use of V1 region does not allow the study of enterococci by the DGGE analysis protocol described here. A possible solution to the problem could be the use of different primers targeting other regions of the 16S rRNA gene. However, a previous study described that only the V1 region allows DGGE differentiation between LAB and CNC, without band comigration (6). In the case of yeast ecology, also when plate counts were below the detection limit, yeasts species were detected by DGGE.
The application of molecular methods allowed identification of strains belonging to Lactococcus garvieae and M. caseolyticus that are commonly isolated from dairy products. This is particularly important in the case of Lactococcus garvieae, since it was isolated in all three fermentations and was stably present throughout fermentation L, thereby deserving consideration from the point of view of its contribution to the final product. Finally, the possibility of applying cluster analysis to the DGGE profiles underlined how the three fermentations could be considered different processes from the point of view of the microbial ecology. Distinct clusters, containing samples from one specific fermentation only, were identified for both bacterial and yeast DGGE gels. In this study, the diverse technological procedures and ingredients used for the production resulted in distinct microbial ecologies that were mainly characterized by a different relative distribution of the same species within each fermentation, rather than the presence of different microorganisms. In our opinion, it is essential to underline, once more, how the use of multiphasic approaches to study food fermentations improves our understanding of these processes. Plating and DGGE analysis should not be considered separately; in fact, the combination of the results obtained allows us to carefully profile microbial dynamics. The methods described here allow a detailed depiction of the fermentation process, representing important tools to use to study the microbial ecology of sausages during transformation. They permit the identification of specific populations which can be selected for their particular characteristics and subsequently be used as starter cultures to improve the sensory profile and sanitary quality.
Acknowledgments
This study was supported by the Ministry of University, Rome, Italy, action PRIN (ex 40%).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Shaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aymerich, T., B. Martin, M. Garriga, and M. Hugas. 2003. Microbial quality and direct PCR identification of lactic acid bacteria and nonpathogenic staphylococci from artisanal low-acid sausages. Appl. Environ. Microbiol. 69:4583-4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.ben Omar, N., and F. Ampe. 2000. Microbial community dynamics during production of the Mexican fermented maize dough pozol. Appl. Environ. Microbiol. 66:3664-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cantoni, C., M. R. Molnar, P. Renon, and G. Gioletti. 1967. Lipolytic micrococci in pork fat. J. Appl. Bacteriol. 30:190-196. [DOI] [PubMed] [Google Scholar]
- 5.Cocolin, L., L. F. Bisson, and D. A. Mills. 2000. Direct profiling of the yeast dynamics in wine fermentations. FEMS Microbiol. Lett. 189:81-87. [DOI] [PubMed] [Google Scholar]
- 6.Cocolin, L., M. Manzano, C. Cantoni, and G. Comi. 2001. Denaturing gradient gel electrophoresis analysis of the 16S rRNA gene V1 region to monitor dynamic changes in the bacterial population during fermentation of Italian sausages. Appl. Environ. Microbiol. 67:5113-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cocolin, L., K. Rantsiou, L. Iacumin, C. Cantoni, and G. Comi. 2002. Direct identification in food samples of Listeria spp. and Listeria monocytogenes by molecular methods. Appl. Environ. Microbiol. 68:6273-6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cocolin, L., K. Rantsiou, L. Iacumin, R. Urso, C. Cantoni, and G. Comi. 2004. Study of the ecology of fresh sausages and characterization of populations of lactic acid bacteria by molecular methods. Appl. Environ. Microbiol. 70:1883-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Comi, G., B. Citterio, M. Manzano, and C. Cantoni. 1992. Evaluation and characterisation of Micrococcaceae strains in Italian dry fermented sausages. Fleischwirtschaft. 72:1679-1683. [Google Scholar]
- 10.Coppola, R., B. Giagnacovo, M. Iorizzo, and L. Grazia. 1997. Characterization of lactobacilli involved in the ripening of sopressata molisana, a typical southern Italian fermented sausage. Food Microbiol. 15:347-353. [Google Scholar]
- 11.Coppola, R., M. Iorizzo, R. Saotta, E. Sorrentino, and L. Grazia. 1997. Characterization of micrococci and staphylococci isolated from soppressata molisana, a Southern Italy fermented sausage. Food Microbiol. 14:47-53. [Google Scholar]
- 12.del Carmen de la Rosa, M., M. R. Mohino, M. Mohino, and M. A. Mosso. 1990. Characteristics of micrococci and staphylococci isolated from semi-preserved meat products. Food Microbiol. 7:207-215. [Google Scholar]
- 13.Dellapina, G., D. Blanco, E. Pancini, S. Barbuti, and M. Campanini. 1994. Microbiological evolution in Italian Felino, Milan and Hungarian-style salami. Ind. Conserve 69:85-90. [Google Scholar]
- 14.Ercolini, D. 2004. PCR-DGGE fingerprinting: novel strategies for detection of microbes in food. J. Microbiol. Methods 56:297-314. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Varona, M., E. M. Santos, I. Jaime, and J. Rovira. 2000. Characterization of Micrococcaceae isolated from different varieties of chorizo. Int. J. Food Microbiol. 54:189-195. [DOI] [PubMed] [Google Scholar]
- 16.Garriga, M., M. Hugas, P. Gou, M. T. Aymerich, J. Arnau, and J. M. Monfort. 1996. Technological and sensorial evaluation of Lactobacillus strains as starter cultures in fermented sausages. Int. J. Food Microbiol. 32:173-183. [DOI] [PubMed] [Google Scholar]
- 17.Head, I. M., J. R. Saunders, and R. W. Pickup. 1998. Microbial evolution, diversity and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb. Ecol. 35:1-21. [DOI] [PubMed] [Google Scholar]
- 18.Hierro, E., L. de la Hoz, and J. A. Ordonez. 1997. Contribution of microbial and meat endogenous enzymes to the lipolysis of dry fermented sausages. J. Agric. Food Chem. 45:2989-2995. [DOI] [PubMed] [Google Scholar]
- 19.Holzapfel, R. W., R. Geisen, and U. Schillinger. 1995. Biological preservation of foods with reference to protective cultures, bacteriocins and food-grade enzymes. Int. J. Food Microbiol. 24:343-362. [DOI] [PubMed] [Google Scholar]
- 20.Hugas, M. 1998. Bacteriocinogenic lactic acid bacteria for the biopreservation of meat and meat products. Meat Sci. 49:S139-S150. [PubMed] [Google Scholar]
- 21.Hugas, M., M. Garriga, T. Aymerich, and J. M. Monfort. 1993. Biochemical characterization of lactobacilli from dry fermented sausages. Int. J. Food Microbiol. 18:107-113. [DOI] [PubMed] [Google Scholar]
- 22.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hugenholtz, P., and N. R. Pace. 1996. Identifying microbial diversity in the natural environment: a molecular phylogenetic approach. Trends Biotechnol. 14:90-97. [DOI] [PubMed] [Google Scholar]
- 24.International Organization for Standardization. 1990. Microbiology—general guidance on methods for the detection of Listeria monocytogenes. Draft international standard ISO/DIS 11290. International Organization for Standardization, Geneva, Switzerland.
- 25.International Organization for Standardization. 1991. Microbiology—general guidance on methods for the detection of Salmonella. Draft international standard ISO/DIS 6579. International Organization for Standardization, Geneva, Switzerland.
- 26.Klijn, N., A. H. Weerkamp, and W. M. de Vos. 1991. Identification of mesophilic lactic acid bacteria by using polymerase chain reaction-amplified variable regions of 16S rRNA and specific DNA probes. Appl. Environ. Microbiol. 57:3390-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurtzman, C. P., and C. J. Robnett. 1998. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Leeuwenhoek 72:331-371. [DOI] [PubMed] [Google Scholar]
- 28.Lucke, F. K. 1974. Fermented sausages, p. 41-49. In B. J. B. Wood (ed.), Microbiology of fermented foods. Applied Science Publishers, London, England.
- 29.Mauriello, G., A. Casaburi, G. Blaiotta, and F. Villani. 2004. Isolation and technological properties of coagulase negative staphylococci from fermented sausages of Southern Italy. Meat Sci. 67:149-158. [DOI] [PubMed] [Google Scholar]
- 30.Meroth, C. B., J. Walter, C. Hertel, M. J. Brandt, and W. P. Hammes. 2003. Monitoring the bacterial population dynamics in sourdough fermentation process by using PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 69:475-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Metaxopoulos, J., J. Samelis, and M. Papadelli. 2001. Technological and microbiological evaluation of traditional processes as modified for the industrial manufacturing of dry fermented sausages in Greece. Ital. J. Food Sci. 13:3-18. [Google Scholar]
- 32.Mills, D. A., E. A. Johannsen, and L. Cocolin. 2002. Yeast diversity and persistence in botrytis-affected wine fermentation. Appl. Environ. Microbiol. 68:4884-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molly, K., D. Demeyer, G. Johansson, M. Raemaekers, M. Ghistelinck, and I. Geenen. 1997. The importance of meat enzymes in ripening and flavour generation in dry fermented sausages. First results of a European project. Food Chem. 59:539-545. [Google Scholar]
- 34.Montel, M. C., J. Reitz, R. Talon, J. L. Berdague, and S. Rousset. 1996. Biochemical activities of Micrococcaceae and their effects on the aromatic profiles and odours of dry sausages model. Food Microbiol. 13:489-499. [Google Scholar]
- 35.Muyzer, G., E. C. De Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-740. [DOI] [PubMed] [Google Scholar]
- 37.Papamanoli, E., P. Kotzekidou, N. Tzanetakis, and E. Litopoulou-Tzanetaki. 2002. Characterization of Micrococcaceae isolated from dry fermented sausages. Food Microbiol. 19:441-449. [DOI] [PubMed] [Google Scholar]
- 38.Parente, E., S. Griego, and M. A. Crudele. 2001. Phenotypic diversity of lactic acid bacteria isolated from fermented sausages produced in Basilicata (Southern Italy). J. Appl. Microbiol. 90:943-952. [DOI] [PubMed] [Google Scholar]
- 39.Randazzo, C. L., S. Torriani, A. D. L. Akkermans, W. M. de Vos, and E. E. Vaughan. 2002. Diversity, dynamics, and activity of bacterial communities during production of an artisanal Sicilian cheese as evaluated by 16S rRNA analysis. Appl. Environ. Microbiol. 68:1882-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ravelo, C., B. Magariños, S. López-Romalde, A. E. Toranzo, and J. L. Romalde. 2003. Molecular fingerprinting of fish-pathogenic Lactococcus garvieae strains by random amplified polymorphic DNA analysis. J. Clin. Microbiol. 41:751-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodriguez, M., F. Nunez, J. J. Cordoba, C. Sanabria, E. Bermudez, and M. A. Asensio. 1994. Characterization of Staphylococcus and Micrococcus spp. isolated from Iberian ham throughout the ripening process. Int. J. Food Microbiol. 24:329-335. [DOI] [PubMed] [Google Scholar]
- 42.Samelis, J., F. Maurogenakis, and J. Metaxopoulos. 1994. Characterisation of lactic acid bacteria isolated from naturally fermented Greek dry salami. Int. J. Food Microbiol. 23:179-196. [DOI] [PubMed] [Google Scholar]
- 43.Samelis, J., J. Metaxopoulos, M. Vlassi, and A. Pappa. 1998. Stability and safety of traditional Greek salami—a microbiological ecology study. Int. J. Food Microbiol. 44:69-82. [DOI] [PubMed] [Google Scholar]
- 44.Santos, E. M., C. Gonzalez-Fernandez, I. Jaime, and J. Rovira. 1998. Comparative study of lactic acid bacteria house flora isolated in different varieties of “chorizo”. Int. J. Food Microbiol. 39:123-128. [DOI] [PubMed] [Google Scholar]
- 45.Santos, N. N., R. C. Santos-Mendosa, Y. Sanz, T. Bolumar, M.-C. Aristoy, and F. Toldrà. 2001. Hydrolysis of pork muscle sarcoplasmatic proteins by Debaryomyces hansenii. Int. J. Food Microbiol. 68:199-206. [DOI] [PubMed] [Google Scholar]
- 46.Sheffield, V. C., D. R. Cox, L. S. Lerman, and R. M. Myers. 1989. Attachment of a 40-base pairs G+C rich sequence (GC clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc. Natl. Acad. Sci. USA 86:297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stiles, M. E. 1996. Biopreservation by lactic acid bacteria. Antonie Leeuwenhoek 70:331-345. [DOI] [PubMed] [Google Scholar]
- 48.Talon, R., D. Walter, S. Chartier, C. Barriere, and M. C. Montel. 1999. Effect of nitrate and incubation conditions on the production of catalase and nitrate reductase by staphylococci. Int. J. Food Microbiol. 52:47-56. [DOI] [PubMed] [Google Scholar]
- 49.van Beek, S., and F. G. Priest. 2002. Evolution of the lactic acid bacterial community during malt whisky fermentation: a polyphasic study. Appl. Environ. Microbiol. 68:297-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vauterin, L., and P. Vauterin. 1992. Computer-aided objective comparison of electrophoretic patterns for grouping and identification of microorganisms. Eur. Microbiol. 1:37-41. [Google Scholar]
- 51.Villani, F., M. Aponte, G. Blaiotta, G. Mauriello, O. Pepe, and G. Moschetti. 2001. Detection and characterization of a bacteriocin, garviecin L1-5, produced by Lactococcus garvieae isolated from raw cow's milk. J. Appl. Microbiol. 90:430-439. [DOI] [PubMed] [Google Scholar]