Abstract
Extracellular signal-regulated protein kinase (ERK, or mitogen-activated protein kinase [MAPK]) regulatory cascades in fungi turn on transcription factors that control developmental processes, stress responses, and cell wall integrity. CEK1 encodes a Candida albicans MAPK homolog (Cek1p), isolated by its ability to interfere with the Saccharomyces cerevisiae MAPK mating pathway. C. albicans cells with a deletion of the CEK1 gene are defective in shifting from a unicellular budding colonial growth mode to an agar-invasive hyphal growth mode when nutrients become limiting on solid medium with mannitol as a carbon source or on glucose when nitrogen is severely limited. The same phenotype is seen in C. albicans mutants in which the homologs (CST20, HST7, and CPH1) of the S. cerevisiae STE20, STE7, and STE12 genes are disrupted. In S. cerevisiae, the products of these genes function as part of a MAPK cascade required for mating and invasiveness of haploid cells and for pseudohyphal development of diploid cells. Epistasis studies revealed that the C. albicans CST20, HST7, CEK1, and CPH1 gene products lie in an equivalent, canonical, MAPK cascade. While Cek1p acts as part of the MAPK cascade involved in starvation-specific hyphal development, it may also play independent roles in C. albicans. In contrast to disruptions of the HST7 and CPH1 genes, disruption of the CEK1 gene adversely affects the growth of serum-induced mycelial colonies and attenuates virulence in a mouse model for systemic candidiasis.
Candida albicans, an opportunistic fungal pathogen, is the major causative agent of thrush and other forms of candidiasis. Diploid C. albicans alternates between a yeast form and mycelial and pseudomycelial forms but does not have a sexual cycle. Physiological temperatures, pH, and serum can promote the emergence of true hyphae from yeast cells in vitro, yet both these forms, as well as pseudohyphae, may be found in infected tissues (for a review, see reference 32). The roles of these different morphologies in pathogenesis have been controversial, but recently, hyphal differentiation has been found to be linked to systemic virulence (22, 26) and the ability of C. albicans cells to evade macrophages (26). Filamentous forms are also better than yeast forms at invading epithelial cells (7) and agar surfaces in vitro (5, 12, 34). This may be the result of both the mechanical advantages of hyphal forms in the penetration of solid substrates (11) and the production of hypha-specific hydrolytic enzymes such as some of the secreted aspartyl proteinases which also appear to contribute to virulence (14, 40).
Baker’s yeast, Saccharomyces cerevisiae, is also able to switch to a pseudohyphal growth mode, as a possible nutrient foraging mechanism (10). S. cerevisiae diploids, when exposed to severe nitrogen limitation, start to grow as chains of attached elongated cells which invade solid surfaces, rather than growing as individual budding cells. In response to nutrient starvation, S. cerevisiae haploid cells also start to grow invasively, but in a more random direction than diploid cells (35). Nutritional limitation of C. albicans (24) triggers the development of true parallel-sided hyphae as well as pseudohyphae; the latter are distinguished by conspicuous constrictions present at pseudohyphal cell-cell junctions (31). Nutrient starvation is a potential environmental signal for C. albicans in different microenvironments including the spleen and liver (41), but serum is the best inducer of the true parallel-sided hyphal form (12).
In both S. cerevisiae and C. albicans, environmental changes induce filamentous growth through at least two parallel signal transduction pathways (17, 26). Cells with homozygous deletions of both of two independently regulated putative transcriptional activators (called Ste12p and Phd1p in S. cerevisiae and Cph1p and Efg1p in C. albicans) are locked in the yeast phase and cannot grow filamentously (26), but cells with deletions of either of these transcriptional regulators demonstrate variable defects in filament development (24, 26, 42). In addition, C. albicans cph1 efg1 double null mutants are avirulent, but mutants with a deletion of either gene alone are virulent (26).
Little is known about the regulation of Phd1p in S. cerevisiae, but a regulatory kinase cascade which activates the Ste12p transcription factor functions not only during pseudohyphal development but also during the mating response of haploid cells (25, 35). These sequentially acting kinases from S. cerevisiae have been studied as a paradigm for eukaryotic mitogen-activated protein kinase (MAPK) modules which transduce signals from the cell surface to the nucleus for a wide variety of responses in eukaryotic cells (20, 37). In response to mating pheromone, the MAPK Fus3p is activated on tyrosine and threonine residues by the MAPK kinase (MAPKK, or MEK) Ste7p. Ste7p is activated by the serine/threonine MAPKK kinase (MAPKKK) Ste11p, which may itself be activated by the serine/threonine MAPKKK kinase (MAPKKKK) Ste20p. All of these MAPK cascade components, with the replacement of Fus3p with the homologous MAPK Kss1p, are also involved in activating the Ste12p transcription factor to drive filamentous growth (4, 25, 28, 35). While most of the MAPK cascade components are shared for the different developmental pathways, process-specific factors help to guide the specificity of response (27).
C. albicans homologs of these S. cerevisiae MAPK cascade elements include Cst20p (an Ste20p homolog) (16, 19), Hst7p (an Ste7p homolog) (3, 16, 19), Cek1p (a homolog of the Fus3p and Kss1p MAPKs) (5, 44), Cph1p (Ste12p transcription factor homolog) (24, 29), and also a MAPK phosphatase, Cpp1p (5). Cpp1p is a homolog of the S. cerevisiae MAPK phosphatase, Msg5p, which plays minor roles in the regulation of Fus3p (6, 46), and possibly the Mpk1p (Slt2p) MAPK (43) involved in cell wall integrity, bud emergence, and polarized cell growth (13, 45). Cst20p, Hst7p, and Cph1p are required for temperature-induced C. albicans hyphal formation under certain nutritional conditions on solid surfaces in vitro (16, 19, 24), and Cpp1p is a repressor of hyphal differentiation at ambient temperatures (5).
Here we show that the Cek1p MAPK, which was isolated by its ability to interfere with S. cerevisiae pheromone-induced cell cycle arrest when expressed on a high-copy-number plasmid (44), is part of the signal transducing machinery that coordinates hypha formation in C. albicans. Our studies suggest that Cst20p, Hst7p, Cek1p, and Cph1p act sequentially in the order of a canonical MAPK cascade to induce the C. albicans yeast-to-hypha transition in response to nutritional starvation. We also show that Cek1p fulfills an additional function during the growth of serum-induced mycelial colonies and that it is required for full pathogenicity during experimental systemic candidiasis.
MATERIALS AND METHODS
DNA manipulations and analysis.
DNA manipulations were performed by standard procedures (39). Southern blot analysis was performed with a nonradioactive labeling and detection kit (Boehringer Mannheim, Indianapolis, Ind.) according to the manufacturer’s recommendations.
Plasmid constructions.
For gene disruptions, pMO3 was created in several steps. First, a 5.2-kb XbaI/KpnI genomic DNA fragment containing the CEK1 gene was excised from plasmid M161p11 (44) and ligated to XbaI/KpnI-digested Bluescript KS(+) vector (Stratagene) to create pMO1. Oligodeoxynucleotide primers OT1 (5′GAAGATCTTATTCCATATGCTCCTTCTCC3′) and OT2 (5′GAAGATCTGTAAACATGTGTGAAGAATAG3′) were then used to delete a 1.2-kb region of the CEK1 gene containing most of the open reading frame from pMO1 by divergent PCR while BglII sites were added (underlined). This fragment was digested with BglII and self-ligated to create pMO2. pMO2 was digested with BglII and joined to a 4-kb BamHI/BglII hisG-URA3-hisG fragment from the plasmid p5921 (8) to create pMO3. Plasmids pCCa3 and pCCa4 were constructed by isolating the 5.2-kb XbaI/KpnI insert from pMO1 and joining this fragment to XbaI/KpnI-digested pBS-cURA3 (19) [pBluescript KS(+) containing the C. albicans URA3 gene between the NotI and XbaI polylinker—kindly provided by J. Douglas, I. D. Broadbent, and A. J. P. Brown, University of Aberdeen] for pCCa3 or the vector pVEC which contains URA3 as a selectable marker and a C. albicans autonomously replicating sequence (kindly provided by C. Nombela, University of Madrid, and B. Magee, University of Minnesota) for pCCa4.
Construction of pYPB1-ADHpt-HST7, containing HST7 driven by the ADH1 promoter, has been previously described (19). To construct plasmid pLJ19, carrying CPH1 under the control of the ADH1 promoter, the coding region of CPH1 flanked by BamHI sites with the 5′ oligonucleotide CGCGGATCCACTCTTTCGCCATGTCAATTAC and the 3′ oligonucleotide CGCGGATCCTATTCATCTATGTTTGTGACTG was amplified with plasmid pKB83.1 containing the CPH1 gene as a template (provided by K. Clark). The fragment was inserted into the BglII site of the plasmid YPB1-ADH1pl containing the ADH1 promoter (1), C. albicans URA3 as a selectable marker, and an autonomously replicating sequence (kindly provided by G. Bertram, I. D. Broadbent, P. J. F. Feldman, and A. J. P. Brown) (see reference 19).
Chromosomal disruption of the CEK1 gene.
A sequential gene disruption strategy (8) was used to replace both chromosomal copies of the CEK1 gene with an 8-kb KpnI/NotI exogenously provided DNA fragment, from the plasmid pMO3, in which most (1.2 kb) of the 1.6-kb open reading frame of CEK1 was replaced by a 4.0-kb fragment containing the selectable marker URA3 flanked by hisG direct repeats. Spheroplasts from the Ura3 auxotrophic strain, CAI4, were transformed (18) with the exogenously provided fragment. Mutations were verified by Southern analysis with a 3.2-kb KpnI/SacI fragment from pMO1 containing the CEK1 gene as a probe. Of 10 transformants analyzed, 9 had a disruption of one allele and 1 appeared to have a disruption of both alleles of the CEK1 gene. Two transformants (including CK43A [Table 1]) with a disruption of one allele of CEK1 (CEK1/cek1Δ::hisG-URA3-hisG) were chosen. Loss of the URA3 gene by recombination between repeats (leaving behind one copy of the hisG gene) was selected for on medium containing 5-fluoroorotic acid and uridine as described elsewhere (8) to create CEK1/cek1Δ::hisG strains (Table 1). These steps were repeated to obtain independent transformants with disruptions in both alleles of CEK1. CK43B-16 and CK43B-4 (cek1Δ::hisG-URA3-hisG/ cek1Δ::hisG [Table 1]) were chosen for further analysis.
TABLE 1.
Yeast strains used in this study
| C. albicans strain | Relevant genotype | Source or reference |
|---|---|---|
| SC5314 | Clinical isolate; Ura+ parent of CAI4 | 9 |
| CAI4 | ura3/ura3 | 8 |
| CK43A | ura3/ura3 CEK1/cek1Δ::hisG-URA3-hisG | This study |
| CK43AL | ura3/ura3 CEK1/cek1Δ::hisG | This study |
| CK43A-RI | ura3/ura3 CEK1/cek1Δ::hisG::CEK1-URA3 | This study |
| CK43B-16 | ura3/ura3 cek1Δ::hisG-URA3-hisG/cek1Δ::hisG | This study |
| CK43B-16L | ura3/ura3 cek1Δ::hisG/cek1Δ::hisG | This study |
| CK43B-RI | ura3/ura3 cek1Δ::hisG/cek1Δ::hisG::CEK1-URA3 | This study |
| CK43B-4 | ura3/ura3 cek1Δ::hisG-URA3-hisG/cek1Δ::hisG | This study |
| CK43B-4L | ura3/ura3 cek1Δ::hisG/cek1Δ::hisG | This study |
| CP29-1-7 | ura3/ura3 cpp1Δ::hisG-URA3-hisG/cpp1Δ::hisG | 5 |
| CP29-1-7L4 | ura3/ura3 cpp1Δ::hisG/cpp1Δ::hisG | 5 |
| CP29-1-7CK14 | ura3/ura3 cpp1Δ::hisG/cpp1Δ::hisG; cek1Δ::hisG-URA3-hisG/cek1Δ::hisG-URA3-hisG | 5 |
| CDH22 | ura3/ura3 cst20Δ::hisG-URA3-hisG/cst20Δ::hisG | 19 |
| CDH9 | ura3/ura3 hst7Δ::hisG-URA3-hisG/hst7Δ::hisG | 19 |
| JKC19 | ura3/ura3 cph1Δ::hisG-URA3-hisG/cph1Δ::hisG | 16 |
| CDH25 | ura3/ura3 cst20Δ::hisG/cst20Δ::hisG | 19 |
| CDH12 | ura3/ura3 hst7Δ::hisG/hst7Δ::hisG | 19 |
| CDH72 | ura3/ura3 cph1Δ::hisG/cph1Δ::hisG | 19 |
To target reintegration of CEK1 into the genome, the C. albicans reintegration plasmid containing the CEK1 gene and flanking sequences, pCCa3, was linearized with PstI and transformed into Ura− C. albicans containing the double or single deletion of the CEK1 gene. Strains were also transformed with the expression plasmid pCCa4.
Candida strains and growth conditions.
All strains are listed in Table 1. Standard growth of the yeast form in liquid culture was performed in YPD medium at 30°C (38). To induce germ tube formation in liquid culture, cells were diluted 10-fold from overnight cultures grown in Lee’s medium (23) into fresh Lee’s medium or from YPD into 10% fetal bovine serum (Intergen Inc., Purchase, N.Y.) and incubated for 3 h at 37°C.
To induce hyphal development on solid medium, budding C. albicans cells were grown overnight at 30°C with vigorous shaking in YPD medium and washed once with sterile water prior to being plated. Petri plates were divided into six sectors, and 5 to 25 cells per sector were then incubated for the indicated times at 30 or 37°C on different media. Solid Spider medium contains 1% nutrient broth, 0.2% K2HPO4, 1.4% agar, and 1% either glucose or mannitol (24). Lee M medium is a modification of the synthetic medium described by Lee et al. (23) with the substitution of mannitol for glucose. Synthetic low-ammonium–dextrose (SLAD) medium, containing 50 μM ammonium sulfate as sole nitrogen source, and synthetic ammonium-dextrose (SAD), containing 50 mM ammonium sulfate, were prepared as described elsewhere by omitting l-histidine from SLAHD and SAHD (10); the pH of these media was 4.0.
For serum plates, 10% fetal bovine serum was added to 1.4% agar at 50°C after autoclaving. Statistical analysis of the differences between mean mycelial colony diameters was performed with Student’s t test. Colony diameters obtained from 10 colonies of each strain grown on the same agar plate 18 h after incubation were measured in Adobe Photoshop (Adobe Systems, Inc.) after scanning photographs taken with a 2× objective. For comparative purposes, estimated mycelial colony growth rates were calculated from the mean colony diameters. To approximate growth rates, we assumed a direct relationship between mycelial colony diameter and growth rate (μ). We plotted the growth rates, obtained by counting hyphal tips for the wild-type SC5314 and the cpp1 null mutant (5), versus the mean mycelial colony diameters and used this as a standard curve to obtain growth rates for the other strains presented in this study.
Photomicroscopy of colonies and invasive growth was performed with a Nikon TMS inverted microscope, and plates were photographed with Kodak TMAX film.
For epistasis analysis, Ura3− derivatives of strains containing deletions of CST20 (CDH25), HST7 (CDH12), CPH1 (CDH72), and CEK1 (CK43B-16L) were transformed with plasmids YPB1-ADHpt, pYPB1-ADHpt-HST7, and pLJ19. The CPP1 null mutant strain (CP29-1-7L) was transformed with plasmids YPB1-ADHpt and pLJ19. These were plated on SLAD solid medium for the times indicated and observed by microscopy.
Diagnosis of pseudohyphal and true hyphal forms.
Filaments were classified as pseudohyphal if a constriction was apparent at junctions between two compartments (31). True hyphae were diagnosed if filaments were composed of parallel-sided compartments with no constrictions at compartment junctions and also by the presence of septa perpendicular to cell walls. To visualize these traits, photographs taken at a magnification of ×40 were scanned into the computer and images were magnified in Adobe Photoshop (Adobe Systems, Inc.). Filament compartments were traced, and the angles between walls and septa were examined.
Virulence studies.
Inbred female BALB/c mice were obtained from Charles River Breeding Laboratories (Sulzfeld, Germany) and used for infection at 8 to 10 weeks of age. C. albicans in vivo virulence testing was performed as described previously (5). Briefly, strains were grown to stationary phase in YPD. Cells were then harvested, washed, and adjusted to the desired density in phosphate-buffered saline and were injected intravenously into the tail vein in a final volume of 200 μl. Statistical analysis of the differences in survival between paired groups was performed with the Mantel-Haenszel log rank test with the GraphPad Prism 2.01 software package.
RESULTS
Disruption of CEK1 causes defects in the shift from a unicellular colonial mode of growth to an invasive hyphal mode of growth.
Cek1p belongs to the ERK family of MAP kinases and is closely related to the S. cerevisiae kinases Kss1 and Fus3 (44). Although CEK1 overexpression does not reestablish mating in a fus3 kss1 strain, disruption of the kinase domain of CEK1 blocks its ability to interfere with pheromone arrest (44). Because of previous reports of the roles of C. albicans CST20, HST7, and CPH1 genes in C. albicans hyphal development on solid medium containing mannitol as a carbon source (16, 19, 24), we disrupted CEK1 in order to explore whether Cek1p assists in hyphal development.
Null mutations of one or both alleles of CEK1 were made by sequential gene disruption and confirmed by Southern blot analysis (Fig. 1). The development of hyphae emanating from colonies grown for 4 days at 30°C on Lee’s (Fig. 2) or Spider (data not shown) medium containing mannitol as a carbon source was partially blocked when one allele of CEK1 was deleted from cells (cek1 heterozygous mutants) and was completely blocked when both CEK1 alleles were deleted (cek1 homozygous null mutants). Strains carrying null mutations of the CST20, HST7, and CPH1 genes were compared in parallel as controls and gave similar results (data not shown). The strain with the homozygous null mutation of the CEK1 gene regained the ability to make hyphae when wild-type CEK1 was reintroduced by the CEK1 expression plasmid pCCa4 (Fig. 2), and partial ability was regained by reintegration of the CEK1 gene at its chromosomal locus (data not shown). Like strains with homozygous CST20, HST7, and CPH1 null mutations, strains with homozygous CEK1 null mutations formed hyphae on solidified Lee’s or Spider medium containing glucose or on serum medium and in all hypha-inducing liquid media investigated (data not shown). These results indicated that Cek1p, like Cst20p, Hst7p, and Cph1p, is required for agar-invasive hypha formation from mature colonies under some conditions and suggested that they may act in the same regulatory cascade.
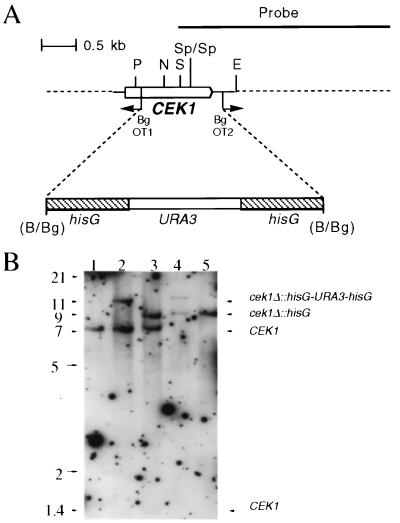
FIG. 1.
(A) Deletion of CEK1 in C. albicans; restriction map of and deletion strategy for the CEK1 gene. PCR with the divergent oligodeoxynucleotides OT1 and OT2 was used to delete a 1.2-kb fragment of the CEK1 gene. A 4.0-kb hisG-URA3-hisG cassette was then inserted. Restriction sites are as follows: B, BamHI; Bg, BglII; P, PstI; S, SacI; N, NsiI; E, EcoRI; Sp, SpeI. (B) Southern blot analysis of CEK1 disruptions with a 3.2-kb KpnI-SacI fragment containing the CEK1 gene as a probe. Genomic DNA samples from the following strains were digested with SpeI (absent from the hisG-URA3-hisG cassette): CEK1/CEK1 (CAI4) (lane 1), CEK1/cek1Δ::hisG-URA3-hisG (CK43A) (lane 2), CEK1/cek1Δ::hisG (CK43AL) (lane 3), cek1Δ::hisG-URA3-hisG/cek1Δ::hisG (CK43B-16) (lane 4), and cek1Δ::hisG/cek1Δ::hisG (CK43B-16L) (lane 5). Strains CK43B-4 and CK43B-4L gave results identical to the example shown here. Hybridization of a very small part (between the SacI and SpeI sites) of the probe to a 1.4-kb SpeI fragment, present only in the wild-type CEK1 gene, was barely detectable in these Southern blots and was not used for diagnostic purposes. The figure was assembled with Adobe Photoshop 3.0. Numbers on the left indicate sizes in kilobases.
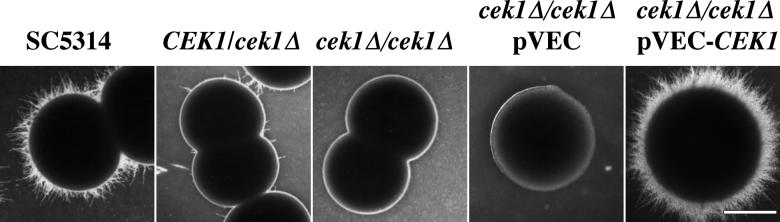
FIG. 2.
Growth of heterozygous and homozygous CEK1 deletion mutants on medium containing mannitol as a carbon source. Shown are uracil prototrophic strains: SC5314 (parental strain), CEK1/cek1Δ (CK43A), cek1Δ/cek1Δ (CK43B-16, identical to CK43B-4), cek1Δ/cek1Δ pVEC (plasmid vector pVEC in CK43B-4L), and cek1Δ/cek1Δ pVEC-CEK1 (plasmid pCCa4 containing the CEK1 gene in CK43B-4L). Cells were grown for 4 days at 30°C. Lee M medium (shown) and Spider medium gave similar results. Bar = 1.5 mm. The figure was assembled with Adobe Photoshop 4.0.
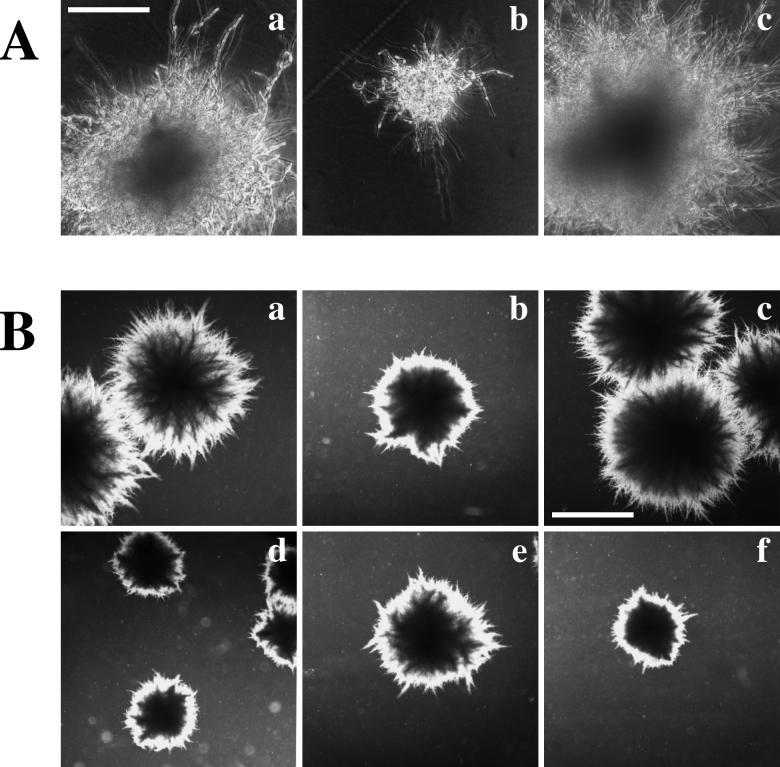
Because we found that agar-invasive growth of C. albicans hyphae was stimulated by severe nitrogen starvation (50 μM ammonium sulfate) on solid medium (SLAD) at a pH of 4.0 (Fig. 3), the condition that also stimulates pseudohyphal development in S. cerevisiae (10), we decided to explore whether the C. albicans MAPK cascade deletion mutants affected this process. Strains with deletions of both alleles of CST20, CPH1, HST7, or CEK1 were all defective in true hyphal outgrowth on solid SLAD at 30°C and made mainly large agar-invasive pseudohypha-type structures with constrictions between cells (Fig. 3). Strains with deletions of both alleles of the CPP1 gene, which encodes a tyrosine phosphatase that blocks hyphal development potentially by acting on the Cek1p MAPK (5), also developed parallel-sided hyphae on SLAD (Fig. 3). C. albicans wild type and null mutants grew only in the yeast form when the defined solid medium used was supplemented with 50 mM ammonium sulfate (SAD medium) as a nitrogen source (data not shown). In liquid culture, however, neither SLAD nor SAD medium (at 37°C) promoted C. albicans hyphal development (data not shown), suggesting that nitrogen limitation and contact with a solid surface, or an additional factor associated with colonial growth, act together to promote hyphal formation.
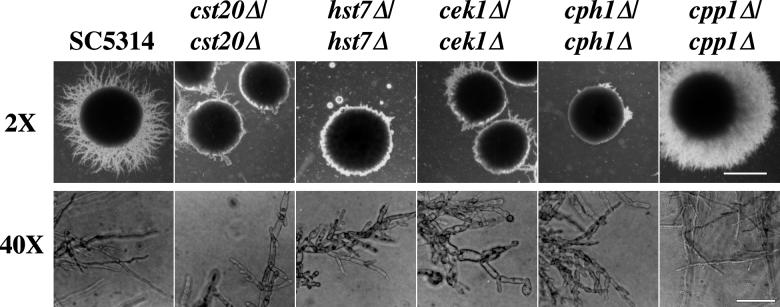
FIG. 3.
Defective formation of parallel-sided hyphae from uracil prototrophic MAPK cascade null mutants grown on low-ammonia–dextrose medium (SLAD). Shown are SC5314 (wild-type parent), cst20Δ/cst20Δ (CDH22), hst7Δ/hst7Δ (CDH9), cek1Δ/cek1Δ (CK43B-16), cph1Δ/cph1Δ (JKC19), and cpp1Δ/cpp1Δ (CP29-1-7). Cells were grown for 7 days at 30°C. (2× objective, bar = 1.5 mm; 40× objective, bar = 90 μm.) Constrictions around septa were examined. Parallel-sided hyphae with no constrictions at septa are seen emanating from the borders of SC5314 and cpp1Δ/cpp1Δ colonies. Pseudohyphal filaments of cst20Δ/cst20Δ, hst7Δ/hst7Δ, cek1Δ/cek1Δ, and cph1Δ/cph1Δ strains are aberrant in shape and have constrictions between cells. Noninvasive cells from colonies scraped off plates were composed entirely of blastospores (data not shown). The figure was assembled with Adobe Photoshop 4.0.
Suppression of the cek1 null mutant hyphal defect by CPH1 under the control of the strong ADH1 promoter but not by ADH1-driven HST7 during nitrogen limitation.
HST7 linked to the ADH1 promoter (1) on the plasmid pYPB-ADHpt-HST7 has been reported to be expressed 180-fold above wild-type HST7 levels (19). We examined whether HST7 and CPH1 under the control of the ADH1 promoter were able to overcome the hyphal growth defects of strains with homozygous deletions of the different MAPK cascade components. ADH-HST7 (pYPB-ADHpt-HST7) and ADH-CPH1 (pLJ19) bypassed the phenotypes of mutations in the C. albicans genes that would be expected to act earlier in the pathway by analogy with the S. cerevisiae pheromone response pathway. Colonies of cst20, hst7, cek1, and cph1 null mutant strains, containing ADH-CPH1, developed large zones of radial agar-penetrating hyphae, unlike colonies of null mutants containing the ADH1 vector (YPB1-ADHpt) alone (Fig. 4). The extensive mycelial networks seen were composed mainly of parallel-sided hyphae (data not shown) resembling those seen in Fig. 3 for SC5314. ADH-HST7 restored the ability of cst20 null mutant colonies to make parallel-sided hyphae (data not shown), resulting in more extensive agar-invasive radial hyphal growth (Fig. 4) than that for null mutants containing the vector alone, which produced pseudohyphae as shown in Fig. 3 (also see reference 19). The ADH-HST7 plasmid also restored radial hyphal growth to the hst7 null mutant, but not to the cek1 null or cph1 null strain, as assessed by examining the zones of radial mycelial growth extending beyond colony borders (Fig. 4) for several transformants.
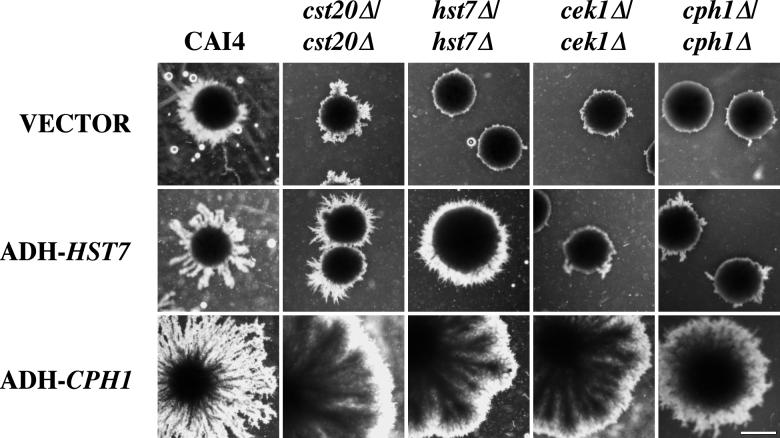
FIG. 4.
Complementation analysis of null mutants grown in SLAD medium. Shown are the uracil auxotrophic strains CAI4 (parent), cst20Δ/cst20Δ (CDH25), hst7Δ/hst7Δ (CDH12), cek1Δ/cek1Δ (CK43B-16L), and cph1Δ/cph1Δ (CDH72) transformed with pADH (YPB1-ADHpl), pADH-HST7 (pYPB1-ADHpt-HST7), or pADH-CPH1 (pLJ19). Cells were grown for 10 days at 30°C. As described above, filaments were examined for constrictions and for the radial penetration of filamentous growth beyond colony borders. The zones of filamentous growth surrounding pADH vector-transformed cst20/cst20 null mutants were composed of constricted pseudohyphae (as seen above), as were the less extensive filaments emanating from pADH vector-transformed hst7Δ/hst7Δ, cek1Δ/cek1Δ, and cph1Δ/cph1Δ null mutants. Bar = 1.5 mm. The figure was assembled with Adobe Photoshop 4.0.
In addition, hyperhyphal phenotypes were observed for CAI4 transformants containing the ADH-HST7 and ADH-CPH1 plasmids. This is consistent with the ability of HST7 and CPH1 to hyperactivate pseudohyphal growth when expressed in S. cerevisiae (3, 24). The abundant hyphal growth of strains containing the ADH-CPH1 plasmid resembled that seen for the cpp1 phosphatase null mutant, and the presence of ADH-CPH1 in the cpp1 null mutant strain did not enhance the hyper-radial-hyphal phenotype of cpp1 null mutants on SLAD (data not shown).
Disruption of CEK1 causes a mycelial colony radial growth defect on serum.
C. albicans cells plated on serum and incubated at 37°C display growth that is considered “truly mycelial” (12). All individual cells plated on solid medium containing agar and 10% serum make germ tubes within hours, and the growth kinetics of the entirely mycelial colony which penetrates and assimilates the agar has been well characterized elsewhere (12). Like strains with mutations of both alleles of the CST20, CPH1, and HST7 genes (16, 19, 24), cek1 homozygous null mutants make morphologically normal hyphae on serum (Fig. 5A), in contrast to strains with null mutations of EFG1, which make only pseudohyphae (26, 42), and cph1 efg1 double null mutants, which make only yeast cells (26). These results suggest that the MAPK cascade is at least partially active on serum. Despite the normal appearance of hyphae on serum, we observed significant differences (P < 0.05) in colony diameters and thus also in growth rate (μ) by paired comparisons of the wild-type (SC5314; μ = 0.12 h−1) and the heterozygous (CEK1/cek1; μ = 0.092 h−1) and homozygous (cek1/cek1; μ = 0.063 h−1) null strains (Table 2 and Fig. 5). The growth defect of the heterozygous mutants was reversed by two independent transformants containing reintegrations of the CEK1 gene (Fig. 5 and data not shown), and that of the homozygous cek1 null mutant was reversed by transformants containing the CEK1 gene on the plasmid pCCa4 (Fig. 5A).
FIG. 5.
Growth of invasive mycelial colonies on solidified serum. (A) Cells grown for 24 h on serum at 37°C (magnification, ×20; bar = 0.75 mm). Shown are uracil prototrophic strains SC5314 (parental strain) (a), cek1Δ/cek1Δ (CK43B-4) (b), and cek1Δ/cek1Δ (CK43B-4L) (c) transformed with the pVEC-CEK1 plasmid pCCa4. (B) Cells grown for 3 days on serum at 37°C (magnification, ×2; bar = 3 mm). Shown are uracil prototrophic strains SC5314 (parental strain) (a), CEK1/cek1Δ (CK43A) (b), CEK1/cek1Δ::CEK1-URA3 (CK43A-RI) (c), cek1Δ/cek1Δ (CK43B-4) (d), cek1Δ/cek1Δ cpp1Δ/cpp1Δ (CP29-1-7CK14) (e), and cpp1Δ/cpp1Δ (CP29-1-7) (f). The figure was assembled with Adobe Photoshop 4.0. Panels A and B are from independent experiments.
TABLE 2.
Mean mycelial colony diameters and estimated growth rates (μ) of strains grown on serum agar for 18 ha
| Ura3 prototrophic strain | Mean diam (mm) (n = 10) | SEM | Estimated growth rate (h−1) |
|---|---|---|---|
| SC5314 parent | 0.41 | 0.024 | 0.12 |
| CEK1/cek1 (CK43A) | 0.30 | 0.017 | 0.092 |
| cek1/cek1 (CK43B-4) | 0.19 | 0.025 | 0.063 |
| cpp1/cpp1 (CP29-1-7) | 0.18 | 0.011 | 0.062 |
| cpp1/cpp1 cek1/cek1 (CP29-1-7CK14) | 0.34 | 0.022 | 0.10 |
See Materials and Methods for details. Results presented here are for an experiment different from that for which the results are presented in Fig. 5.
Intriguingly, a mutant containing a double deletion of the CPP1 MAPK phosphatase and CEK1 together significantly (P < 0.05) suppresses defects in radial hyphal growth on serum of the independent cek1 and cpp1 null mutants (Fig. 5B; Table 2) (5). The estimated growth rate of the double mutant was 0.10 h−1, compared to similar rates of 0.062 and 0.063 h−1 for the individual cpp1 (5) and cek1 null mutants, respectively. Previously, we reported that the cek1 cpp1 double null mutant resembled the wild-type parent when grown on serum (5); however, further analysis demonstrated that the double null mutant has a minor but significant (P < 0.05) reduction in growth rate as assessed by differences in colony diameters (Table 2). Previous reports (16, 19) describe no influence of the cst20 or hst7 null mutant on hyphal growth on serum agar.
Virulence studies.
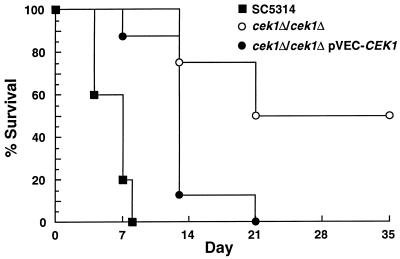
Mice were injected intravenously with the following uracil prototrophic strains: the parental strain (SC5314), cek1 null mutants (cek1Δ/cek1Δ), or null mutants containing the plasmid pCCa4 (cek1Δ/cek1Δ pVEC-CEK1). pCCa4 contains the CEK1 gene on the replicative vector pVEC, and pCCa4 suppressed the in vitro phenotypes of the cek1 null mutant (Fig. 2 and 5). Doubling times of the yeast form of these strains grown in YPD medium at 30°C were essentially the same. Two days after infection, mice injected with strains containing a wild-type copy of the CEK1 gene (cek1Δ/cek1Δ pVEC-CEK1) or the parental CEK1/CEK1 strain (SC5314) showed signs of severe systemic disease, including weight loss, while the cek1 null mutant-infected animals behaved normally. As illustrated in Fig. 6, infection with cek1 null mutants resulted in 50% mortality, while 100% mortality was observed when mice were infected with the control strains (cek1Δ/cek1Δ pVEC-CEK1 and SC5314) for the observed period of 35 days. These differences were significant (P < 0.05). Similar results were obtained with the null mutant strains CK43B-4 and CK43B-16 as well as with an additional independent null mutant.
FIG. 6.
Survival curves for BALB/c mice injected intravenously with 5 × 105 cells of the uracil prototrophic strains SC5314 (n = 5 mice; clinical isolate), cek1Δ/cek1Δ (CK43B-4; n = 8 mice), and cek1Δ/cek1Δ pVEC-CEK1 (CK43B-4L transformed with the plasmid pCCa4; n = 8 mice). Results were confirmed in two independent experiments. The cek1 null mutant strain CK43B-16 and another independent null mutant gave similar results (data not shown). Mortality data were quantified on days of sampling.
DISCUSSION
C. albicans harbors both structural and functional homologs of a starvation-activated MAPK cascade required for filamentation of the budding yeast S. cerevisiae. Our results show that the Cek1p MAPK plays a positive role in C. albicans hyphal development in response to nutritional starvation. We found that homozygous cek1Δ/cek1Δ null mutants, and to a lesser extent CEK1/cek1Δ heterozygous mutants, are defective in agar-invasive growth of hyphae in response to nutritional limitation, as are cells containing heterozygous and homozygous deletions of the C. albicans CST20, HST7, and CPH1 genes (16, 19, 24). Although Cek1p appears to function in this MAPK cascade, it also plays a role in modulating radial hyphal growth on serum and cells from which CEK1 has been deleted are less virulent when injected into mice, suggesting that Cek1p may be required for additional pathways in C. albicans.
Like pseudohyphal formation in S. cerevisiae, the development of agar-invasive parallel-sided Candida hyphae can be triggered from stationary colonies of yeast cells grown on medium severely limited for nitrogen. In S. cerevisiae, the starvation message is relayed to the MAPKKKK Ste20p via the GTP binding proteins Ras2p and Cdc42p (21, 30, 33) and two 14-3-3 homologs (36). We found that null mutation of MAPK cascade elements did not block C. albicans pseudohyphal development but did block the formation of parallel-sided hyphae instead. S. cerevisiae pseudohyphal formation is suppressed by deletion of elements of its cognate MAPK cascade, but a few pseudohyphae are still observable (26). In both organisms, two pathways involving the transcription factors Ste12p (Cph1p) and Phd1p (Efg1p) function together and independently to promote filament development (26, 42), whereas commitment to a polarized hyphal growth mode requires the action of the kinase CaCla4p (22). In C. albicans, the nature and intensity of stimuli combined with the differential activities of these signal transduction pathways may help determine whether cells undergo pseudohyphal or true hyphal differentiation.
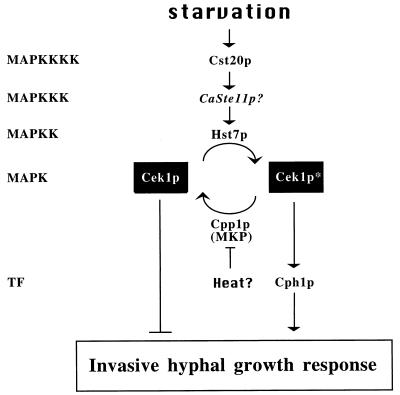
The similar phenotypes of strains with null mutations of the CEK1, CST20, HST7, and CPH1 genes under starvation conditions suggest that they function as part of the same regulatory cascade. We found that HST7 and CPH1 when expressed from the strong ADH1 promoter could complement deletions of CST20 and HST7, whereas expression of CPH1 but not of HST7 from the ADH1 promoter could complement deletions of CEK1 and CPH1. These epistatic relationships suggest that this filamentation MAPK cascade functions in an order typical of other MAPK cascades (Fig. 7) (see also reference 19). The Cek1p MAPK is thus required for the filamentation MAPK cascade in C. albicans and probably holds functions analogous to the Kss1p MAPK of S. cerevisiae (4, 28). Negative regulation by MAPK phosphatases can shut down or control the intensity of MAPK signaling (15, 46) (Fig. 7). Deletion of the CEK1 gene completely suppresses the extensive invasive hyphal growth that occurs at room temperature when the MAPK phosphatase gene CPP1 is removed from cells (5), suggesting that Cpp1p inhibits the hyphal-stimulating roles of Cek1p (Fig. 7). Higher temperatures probably lift the repression of hyphal development mediated by the Cpp1p MAPK phosphatase (5). Indeed, central control of hyphal formation through repression mechanisms may be a theme in C. albicans, since a strain with a deletion of the TUP1 gene encoding a general transcriptional repressor grew exclusively as filaments (2).
FIG. 7.
Model of the C. albicans MAPK cascade for the yeast-to-hypha transition. CaSte11p is an undescribed putative homolog of the S. cerevisiae Ste11p MAPKKK whose role has not yet been examined for this pathway. TF, transcription factor. The phosphorylated form of Cek1p is designated by an asterisk.
Like strains with null mutations of the CST20, CPH1, and HST7 genes (16, 19, 24, 26), serum-induced cek1 mutants grow as invasive hyphal colonies on solidified serum medium. Despite the normal hyphal development of null mutants of the MAPK cascade-regulated transcription factor Cph1p on serum, Cph1p nevertheless contributes to the development of hyphae on serum (26). A strain with deletion mutations of the two filamentation transcription factor genes CPH1 and EFG1 makes only single cells on solid serum medium, whereas efg1 null mutants make pseudohyphae (26, 42). This suggests that the MAPK cascade plays a role during serum-induced hyphal growth, which is usually masked by Efg1p. We found, however, that the zones of hyphal penetration of both heterozygous and homozygous cek1 null mutants were reduced on serum. We had previously suggested that a similar mycelial colony growth rate defect of strains with null mutations of the MAPK phosphatase gene CPP1 on serum was the result of detrimentally high levels of hypha-stimulating cellular activities because deletion of CEK1 suppressed the phenotype of the cpp1 null mutants (5). If Cek1p were the sole MAPK contributing to hyphal growth on serum, we would expect strains with double mutations of CEK1 and the MAPK phosphatase gene CPP1 to display the same phenotype as cek1Δ/cek1Δ single mutants, but the phenotype of the double null mutant is more similar to that of the wild type (5) (although they still have a minor defect in radial hyphal growth) than to those of the individual null mutants. A second MAPK, inappropriately active or hyperactive when the phosphatase is absent, could be partially compensating for loss of CEK1; this unknown MAPK could also, in its inactive form, inhibit hyphal growth when the phosphatase is present. Such a picture is plausible in view of the complexities of the roles of MAPKs during filamentous growth in S. cerevisiae (4, 28).
A role for the hyphal form of C. albicans in virulence is strongly supported by reports that a mutant carrying a deletion of the CaCLA4 gene, a regulator of polarized growth of the fungal germ tube into a true hyphal compartment, is completely avirulent (22), as are cph1 efg1 double null mutants which are locked in the yeast form (26). On the other hand, individual cph1, efg1 (26), and hst7 (19) null mutants are as pathogenic or almost as pathogenic as parental strains, suggesting that the starvation-induced MAPK pathway is not required alone for systemic virulence but can contribute to virulence. The cst20 (19) and cek1 null mutants are different. Injection of these strains into mice allows them to live longer on average than does injection of the parental strain, suggesting that they have overlapping or distinct pleiotropic functions in cells which affect virulence. Because cek1 cpp1 double null mutants (unpublished results) suppress both the attenuated virulence and the serum-induced mycelial colony growth defect common to single cek1 and cpp1 null mutants, it is possible that these phenotypes are related. Future analysis of these mutants should lead to further insights into the functions of the wild-type regulatory enzymes deleted from these strains and may unmask novel genes and cellular functions involved in the pathogenicity of this important fungal pathogen.
ACKNOWLEDGMENTS
We thank W. Fonzi, J. Kohler, and K. Clark for strains and plasmids and the Candida news group and colleagues at the ISHAM conference for discussions and collaborations. We thank J. Thorner for discussions and an in-press manuscript and C. Makris for discussions, Lyne Johnson for creating pLJ19, and the McGill Image Center for photography. We thank André Migneault for helping with figures.
C.C. was supported by a Medical Research Council of Canada (MRCC) postdoctoral fellowship. S.M. is a scholar of the MRCC. K.S. was supported by a grant of the Deutsche Forschungsgemeinschaft Schr 450/2-1.
Footnotes
National Research Council publication no. 41418.
REFERENCES
- 1.Bertram G, Swoboda R K, Gooday G W, Gow N A, Brown A J. Structure and regulation of the Candida albicans ADH1 gene encoding an immunogenic alcohol dehydrogenase. Yeast. 1996;12:115–127. doi: 10.1002/(sici)1097-0061(199602)12:2<115::aid-yea889>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 2.Braun B R, Johnson A D. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science. 1997;277:105–109. doi: 10.1126/science.277.5322.105. [DOI] [PubMed] [Google Scholar]
- 3.Clark K L, Feldmann P J, Dignard D, Larocque R, Brown A J, Lee M G, Thomas D Y, Whiteway M. Constitutive activation of the Saccharomyces cerevisiae mating response pathway by a MAP kinase kinase from Candida albicans. Mol Gen Genet. 1995;249:609–621. doi: 10.1007/BF00418030. [DOI] [PubMed] [Google Scholar]
- 4.Cook J G, Bardwell L, Thorner J. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature. 1997;390:85–88. doi: 10.1038/36355. [DOI] [PubMed] [Google Scholar]
- 5.Csank C, Makris C, Meloche S, Schröppel K, Röllinghoff M, Dignard D, Thomas D Y, Whiteway M. Derepressed hyphal growth and reduced virulence in a VH1 family-related protein phosphatase mutant of the human pathogen Candida albicans. Mol Biol Cell. 1997;8:2539–2551. doi: 10.1091/mbc.8.12.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doi K, Gartner A, Ammerer G, Errede B, Shinkawa H, Sugimoto K, Matsumoto K. MSG5, a novel protein phosphatase, promotes adaptation to pheromone response in S. cerevisiae. EMBO J. 1994;13:61–70. doi: 10.1002/j.1460-2075.1994.tb06235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fidel P, Jr, Sobel J D. The role of cell-mediated immunity in candidiasis. Trends Microbiol. 1994;2:202–206. doi: 10.1016/0966-842x(94)90112-i. [DOI] [PubMed] [Google Scholar]
- 8.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gillum A M, Tsay E Y H, Kirsch D R. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 10.Gimeno C J, Ljungdahl P O, Styles C A, Fink G R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 11.Gow N A. Growth and guidance of the fungal hypha. Microbiology. 1994;140:3193–3205. doi: 10.1099/13500872-140-12-3193. [DOI] [PubMed] [Google Scholar]
- 12.Gow N A, Gooday G W. Growth kinetics and morphology of colonies of the filamentous form of Candida albicans. J Gen Microbiol. 1982;128:2187–2194. doi: 10.1099/00221287-128-9-2187. [DOI] [PubMed] [Google Scholar]
- 13.Gray J V, Ogas J P, Kamada Y, Stone M, Levin D E, Herskowitz I. A role for the Pkc1 MAP kinase pathway of Saccharomyces cerevisiae in bud emergence and identification of a putative upstream regulator. EMBO J. 1997;16:4924–4937. doi: 10.1093/emboj/16.16.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hube B, Sanglard D, Odds F C, Hess D, Monod M, Schafer W, Brown A J, Gow N A. Disruption of each of the secreted aspartyl proteinase genes SAP1, SAP2, and SAP3 of Candida albicans attenuates virulence. Infect Immun. 1997;65:3529–3538. doi: 10.1128/iai.65.9.3529-3538.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keyse S M. An emerging family of dual specificity MAP kinase phosphatases. Biochim Biophys Acta. 1995;1265:152–160. doi: 10.1016/0167-4889(94)00211-v. [DOI] [PubMed] [Google Scholar]
- 16.Kohler J R, Fink G R. Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development. Proc Natl Acad Sci USA. 1996;93:13223–13228. doi: 10.1073/pnas.93.23.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kubler K, Mösch H-U, Rupp S, Lisanti M P. Gpa2p, a G-protein subunit, regulates growth and pseudohyphal development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J Biol Chem. 1997;272:20321–20323. doi: 10.1074/jbc.272.33.20321. [DOI] [PubMed] [Google Scholar]
- 18.Kurtz M B, Cortelyou M W, Kirsch D R. Integrative transformation of Candida albicans, using a cloned Candida ADE2 gene. Mol Cell Biol. 1986;6:142–149. doi: 10.1128/mcb.6.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leberer E, Harcus D, Broadbent I D, Clark K L, Dignard D, Ziegelbauer K, Schmidt A, Gow N A, Brown A J, Thomas D Y. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc Natl Acad Sci USA. 1996;93:13217–13222. doi: 10.1073/pnas.93.23.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leberer E, Thomas D Y, Whiteway M. Pheromone signalling and polarized morphogenesis in yeast. Curr Opin Genet Dev. 1997;7:59–66. doi: 10.1016/s0959-437x(97)80110-4. [DOI] [PubMed] [Google Scholar]
- 21.Leberer E, Wu C, Leeuw T, Fourest-Lieuvin A, Segall J E, Thomas D Y. Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J. 1997;16:83–97. doi: 10.1093/emboj/16.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leberer E, Ziegelbauer K, Schmidt A, Harcus D, Dignard D, Ash J, Johnson L, Thomas D Y. Virulence and hyphal formation of Candida albicans require the Ste20p-like protein kinase CaCla4p. Curr Biol. 1997;7:539–546. doi: 10.1016/s0960-9822(06)00252-1. [DOI] [PubMed] [Google Scholar]
- 23.Lee K L, Buckley H R, Campbell C C. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia. 1975;13:148–153. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- 24.Liu H, Kohler J, Fink G R. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 25.Liu H, Styles C A, Fink G R. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- 26.Lo H-J, Kohler J R, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink G R. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 27.Madhani H D, Fink G R. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- 28.Madhani H D, Styles C A, Fink G R. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 29.Malathi K, Ganesan K, Datta A. Identification of a putative transcription factor in Candida albicans that can complement the mating defect of Saccharomyces cerevisiae ste12 mutants. J Biol Chem. 1994;269:22945–22951. [PubMed] [Google Scholar]
- 30.Mosch H U, Roberts R L, Fink G R. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Odds F C. Candida and candidosis: a review and bibliography. 2nd ed. London, United Kingdom: Bailliere-Tindall; 1988. [Google Scholar]
- 32.Odds F C. Pathogenesis of Candida infections. J Am Acad Dermatol. 1994;31:S2–S5. doi: 10.1016/s0190-9622(08)81257-1. [DOI] [PubMed] [Google Scholar]
- 33.Peter M, Neiman A M, Park H O, van Lohuizen M, Herskowitz I. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 1996;15:7046–7059. [PMC free article] [PubMed] [Google Scholar]
- 34.Radford D R, Challacombe S J, Walter J D. A scanning electronmicroscopy investigation of the structure of colonies of different morphologies produced by phenotypic switching of Candida albicans. J Med Microbiol. 1994;40:416–423. doi: 10.1099/00222615-40-6-416. [DOI] [PubMed] [Google Scholar]
- 35.Roberts R L, Fink G R. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- 36.Roberts R L, Mosch H U, Fink G R. 14-3-3 proteins are essential for RAS/MAPK cascade signaling during pseudohyphal development in S. cerevisiae. Cell. 1997;89:1055–1065. doi: 10.1016/s0092-8674(00)80293-7. [DOI] [PubMed] [Google Scholar]
- 37.Robinson M J, Cobb M H. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 38.Rose M D, Winston F, Heiter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Sanglard D, Hube B, Monod M, Odds F C, Gow N A. A triple deletion of the secreted aspartyl proteinase genes SAP4, SAP5, and SAP6 of Candida albicans causes attenuated virulence. Infect Immun. 1997;65:3539–3546. doi: 10.1128/iai.65.9.3539-3546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Slauch J, Taylor R, Maloy S. Survival in a cruel world: how Vibrio cholerae and Salmonella respond to an unwilling host. Genes Dev. 1997;11:1761–1774. doi: 10.1101/gad.11.14.1761. [DOI] [PubMed] [Google Scholar]
- 42.Stoldt V R, Sonneborn A, Leuker C E, Ernst J F. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 1997;16:1982–1991. doi: 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watanabe Y, Irie K, Matsumoto K. Yeast RLM1 encodes a serum response factor-like protein that may function downstream of the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol Cell Biol. 1995;15:5740–5749. doi: 10.1128/mcb.15.10.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whiteway M, Dignard D, Thomas D Y. Dominant negative selection of heterologous genes: isolation of Candida albicans genes that interfere with Saccharomyces cerevisiae mating factor-induced cell cycle arrest. Proc Natl Acad Sci USA. 1992;89:9410–9414. doi: 10.1073/pnas.89.20.9410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zarzov P, Mazzoni C, Mann C. The SLT2(MPK1) MAP kinase is activated during periods of polarized cell growth in yeast. EMBO J. 1996;15:83–91. [PMC free article] [PubMed] [Google Scholar]
- 46.Zhan X L, Deschenes R J, Guan K L. Differential regulation of FUS3 MAP kinase by tyrosine-specific phosphatases PTP2/PTP3 and dual-specificity phosphatase MSG5 in Saccharomyces cerevisiae. Genes Dev. 1997;11:1690–1702. doi: 10.1101/gad.11.13.1690. [DOI] [PubMed] [Google Scholar]