Abstract
Chronic inflammation is crucial in developing insulin resistance and type 2 diabetes. Previous studies have shown that a leaf extract of Eucalyptus tereticornis, with ursolic acid (UA), oleanolic acid (OA), and ursolic acid lactone (UAL) as the main molecules (78 %) mixed with unknown minor metabolites (22 %), provided superior anti-inflammatory, hypoglycemic, and hypolipidemic effects than reconstituted triterpenoid mixtures in macrophage cell lines and a pre-diabetic mouse model. Further identification of the molecular mechanisms of action of this mixture of triterpenes is required. This study aims to analyse the RNA expression profiles of mouse and human macrophage cell lines treated with the natural extract and its components. Activated macrophage cell lines were treated with the natural extract, UA, OA, UAL or a triterpene mixture (M1). RNA was extracted and sequenced using the DNBseq platform and the EnrichR software to perform gene enrichment analysis using the Gene Ontology database, Kyoto Encyclopedia of Genes and Genomes, and Reactome. To conduct clustering analysis, we standardised the normalised counts of each gene and applied k-means clustering. The combination of molecules in the natural extract has an additive or synergic effect that affects the expression of up-regulated genes by macrophage activation. Triterpenes (M1) regulated 76 % of human and 68 % of mouse genes, while uncharacterised minority molecules could regulate 24 % of human and 32 % of mouse genes. The extract inhibited the expression of many cytokines (IL6, IL1, OSM), chemokines (CXCL3), inflammatory mediators (MMP8 and MMP13) and the JAK-STAT signalling pathway in both models. The natural extract has a more powerful immunomodulatory effect than the triterpene mixture, increasing the number of genes regulated in mouse and human models. Our study shows that Eucalyptus tereticornis extract is a promising option for breaking the link between inflammation and insulin resistance.
Keywords: Natural extract, Triterpenes, Immune system, Transcriptome, Inflammation, Macrophage
Graphical abstract
1. Introduction
Medicinal plants produce thousands of secondary metabolites to adapt to the environment and defend against environmental factors [1]. Traditional medicine has used these metabolites for centuries [2]. Eucalyptus (Eucalyptus spp.) is a genus of the Myrtaceae family, with about 900 species and subspecies. Different parts of this plant are rich in phytochemicals, and extracts from the leaves and fruits of this tree exhibit antioxidant, antidiabetic, antitumor, antibacterial, and antifungal properties [3,4]. Eucalyptus tereticornis Sm (Eu) is used in traditional medicine to treat diabetes mellitus [5]. The most common type of diabetes is type 2 diabetes (T2D), and the leading risk factor for T2D is obesity. Obesity leads to chronic, systemic inflammation and can lead to insulin resistance, b-cell dysfunction, and ultimately T2D. The recruitment, accumulation, and activation of pro-inflammatory macrophages in hypertrophic adipose tissue leads to the increase of production of pro-inflammatory adipokines such as TNF-α and IL-6 that is shown to induce insulin resistance in different tissues [[6], [7], [8], [9]].For this reason, new therapeutic options for managing diabetes mellitus and its complications target inflammation. An extract from Eu leaves, named OBE100, possesses hypoglycemic, hypolipidemic, antioxidant and anti-inflammatory activities [[10], [11], [12], [13]]. OBE100 contains three pentacyclic triterpenoids as main molecules: 47.6 % ursolic acid (UA), 14.1 % oleanolic acid (OA), and 16.3 % ursolic acid lactone (UAL) [11]. Triterpenoids are biomolecules composed of six isoprene units, and the most common and widely distributed are the pentacyclic triterpenoids [14]. These molecules have various biological properties, including antioxidant and anti-inflammatory activities. It has been shown that UA, OA and UAL reduce oxidative stress and inflammation, blocking the NF-kB activation, inhibiting the expression of proinflammatory genes or reducing the production of reactive oxygen species (ROS) [[15], [16], [17], [18]].
We have previously shown that OBE100 significantly decreases the expression of IL-6, TNFα, IL-1β and MCP-1 in both mouse and human macrophages [12] and confirmed that this Eu extract improves the inflammation process caused by a high-fat diet reducing the expression of proinflammatory cytokines, such as MCP-1, TNF-α, IL-1, and IL-6 in mice adipose tissue [13]. Interestingly, UA, OA, and UAL mixed with unknown minor metabolites in OBE100 extract present superior anti-inflammatory, hypoglycemic, and hypolipidemic effects than reconstituted triterpenoid mixtures [11]. Transcriptome sequencing (RNA-Seq) is a high-sensitivity technique that can be used to characterise the changes in gene expression and identify novel transcriptome features and cell culture is an essential technique to expand molecular knowledge.
The overall aim of this study was to use genome-wide mRNA-Seq analysis to compare and study the changes in gene expression profiles caused by triterpenoids and OBE100 in mouse and human macrophage cells to expand knowledge on how they modulate gene expression to regulate the inflammation pathways, and unravel the mechanistic details of the anti-inflammatory activities of these compounds comparing the reproducibility of their biological effects using two different cellular models.
2. Material and methods
2.1. Triterpenoid samples preparation
Eucalyptus tereticornis (Eu) leaves were collected in the district of Mariangola, Cesar, Valledupar - Colombia, in 2011. A specimen was deposited in the Herbarium of the University of Antioquia (# 178511). OBE100 was extracted as previously described [11]. Briefly, the dried leaves of Eu were extracted by a liquid-liquid separation with hexane: methanol: water 4:3:1 (v/v). The organic phase was collected and vacuum-filtered. The precipitate formed, named OBE100, was collected, stored, and submitted to an HPLC/MS analysis. The major compounds of OBE100 were identified as previously described [12]. We used UA, OA, and UAL as individual treatments and to prepare M1, an artificial mixture with the same concentrations of triterpenoids present in OBE100. UA and OA were acquired commercially from Sigma-Aldrich (St. Louis, Mo, USA). UAL is not available commercially, and it was obtained from the Eu as previously described [12].
2.2. Cell culture and treatments
J774A.1 (TIB-67™) mouse monocyte; macrophage cells and U-937 (CRL-1593.2) are human monocytic (pre-macrophage line) were purchased from ATCC (Manassas, VA, USA). Cells were cultured and maintained at 37 °C and 5 % CO2. J774A.1 cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) with 10 % fetal bovine serum (FBS), 1 % penicillin/streptomycin (P/S) (Sigma- Aldrich, St. Louis, MO, USA), and 5.5 mM glucose (Growth medium – GM1). The U-937 cells were cultured in RPMI culture medium with 10 % FBS, 1 % P/S, and 11 mM glucose (Growth medium – GM2). At 80 % confluence, J774A.1 macrophage cells were incubated in GM1 medium containing 1 mg/ml LPS and 20 ng/ml IFNγ for 24 h. In the last 6 h of activation, different treatments were added [19].
. U937 monocyte cells were differentiated into macrophages by adding 100 nM phorbol myristate acetate (PMA) to the growth medium for 72 h. Once differentiated, the cells were washed with PBS and activated with LPS at 100 ng/ml and 20 ng/ml INFγ in GM2 for 24 h [20]. In the last 6 h of activation, different treatments were added. RNA was extracted and collected at the end of treatments. Crude extract OBE100, M1, UA, OA, and UAL were reconstituted in dimethyl sulfoxide (DMSO) at 25 mg/ml (stock solution). The final concentration of the compounds in the culture medium was OBE100 100 μg/ml, M1 78 μg/ml (UA + OA + UAL mix), UA 47.7 μg/ml, OA 14 μg/ml, and UAL 16.3 μg/ml. The concentration used for the three different triterpenes, both in the mixture called M1 and individually, were those determined in the fraction called OBE100. Three independent experiments were performed, each in triplicate. We used two controls, cells with no activation and activated cells (AC).
2.3. RNA-Seq analysis
According to the manufacturer's instructions, total RNA was extracted from cells with the RNeasy kit (QIAGEN, Valencia, CA, USA). RNA quality control, library construction, and RNA sequencing using the DNBseq platform were performed by BGI Americas Corporation (Cambridge, MA, USA). mRNA libraries were sequenced with a sequencing depth of at least 20 million reads per sample, and data were analysed as described previously [21]. Genes were considered differentially expressed if the absolute value of log2 fold change ≥1 and the FDR ≤0.05. Human-mouse orthologous genes were extracted from the Mouse Genome Informatics (MGI) database [22]. The Pearson correlation in log2-fold changes between orthologous genes was calculated in R.
2.4. Gene enrichment analysis
The present study utilized the EnrichR software (https://maayanlab.cloud/Enrichr/) to conduct gene enrichment analysis, employing databases such as Gene Ontology, Kyoto Encyclopedia of Genes and Genomes (KEGG), and Reactome [23]. Enrichment outcomes were determined based on a combined score exceeding 15, calculated by EnrichR using P value (Fisher's exact test) and Z score (correction to the test) [23]. Subsequently, the enrichment results were presented as the percentage of genes enriched within each ontology category [23].
2.5. Clustering analysis of gene expression profiles
A clustering analysis approach to examine the RNAseq expression data was used. To conduct the analysis, we standardised the normalised counts of each gene and applied k-means clustering, utilising Euclidean distance as a metric with random initialisation and 10,000 executions. In this study, k-means clustering was employed as a method to categorize gene expression profiles as was implemented before [24]. The analysis was conducted using the R programming language (version 4.3.2) and specifically the kmeans function from the “stats” package. Prior to clustering, the normalised gene counts were subjected to scaling using z-score. To determine the optimal number of clusters (K), elbow method were applied to discern the intrinsic structure of the gene expression dataset. Subsequently, the kmeans function was utilized, taking the scaled gene expression data matrix, the specified number of clusters, euclidean distance as a metric and 10000 executions as input parameters. The algorithm iteratively assigned genes to clusters based on their expression profiles and updated cluster centroids until convergence. Cluster plots were employed to represent the categorized gene expression profiles.
2.6. Transcription factor motif analysis
Cytoscape app iRegulon was used to detect de transcription factor motifs overrepresented in the promoter of the differentially expressed genes [25].Gene promoters were considered between nucleotides − 300 and +50 relatives to the Transcription Start Site. The motif similarity was set as 0.001. Significance was tested using the normalised enrichment score greater than 5, and the transcription factors that passed the filter were used to construct the regulatory network visualised by Cytoscape [26].
2.7. Statistical analysis
The data were presented as the geometric mean plus or minus the standard deviation. GraphPad Prisma software v 6.07 (GraphPad Software, Inc., United States) was used for statistical analysis. A Student's t-test was utilized for all statistical analyses not specified elsewhere when comparing groups. Statistical significance was achieved when the p-value was less than 0.05.
3. Results
3.1. Activated mouse and human macrophage cell lines show a similar gene expression pattern
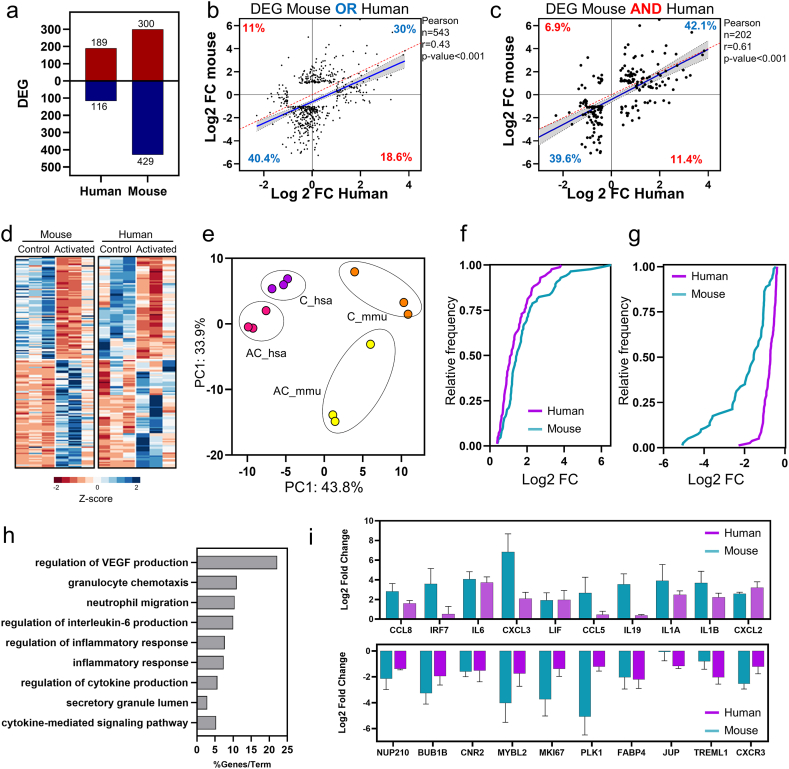
RNA-Seq data sets were obtained to analyse macrophage response genes to LPS and IFN- γ (Activated Control-AC) stimulation that was common in humans and mice. For each species, the AC is compared to the control (C), and the differentially expressed genes (DEG) are obtained (Fig. 1a). Three hundred-five differentially expressed genes for human-activated macrophages were found, of which 189 were up-regulated and 116 down-regulated (Supplementary Table 1). However, 729 differentially expressed genes for activated mouse macrophages were found; 300 were up-regulated, and 429 were down-regulated (Supplementary Table 1).
Fig. 1.
Concordance in expression profiles in activated human and mouse macrophages. (a) Bar plot representing the total number of up-regulated and downregulated genes for human and mouse-activated macrophages (b) Pearson correlation plot of the expression of orthologous genes that are DEGs in at least one species (conditional OR). (c) Pearson correlation plot of the expression of orthologous genes DEGs in both species (conditional AND). Each dot represents an orthologous gene for the correlation plots, and its expression is quantified as the log2 fold change. The blue line represents the linear regression, and the grey-shaded area represents the 95 % confidence interval. The red line represents perfect identity. (d) Heat map of Z-score normalised orthologue DEGs by unsupervised hierarchical clustering analysis. (e) Principal component analysis of orthologue DEGS genes using the FPKM. Each principal component (PC1 and PC2) shows the variance percentage. Cumulative distribution plots of the log2 fold change for a mouse (blue line) and human (purple) for up- (f) and (g) down-regulated genes. (h) Gene ontology analysis of common orthologue DEGs in human and mouse activated macrophages. Each horizontal black bar represents the ontology term fold enrichment compared to the total number of genes in each term. (i) Bar plots of orthologue DEGs in activated human and mouse macrophages. These genes were selected based on previous reports as crucial genes in macrophage activation. C: Control group, AC: Activated control. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The two lists of DEGs were combined (Union set/OR) and filtered to include only mouse-human orthologous genes (Supplementary Table 2). For this list, 543 orthologous genes differentially expressed in at least the mouse or human groups were tested for correlation (Fig. 1b). This analysis shows that 70.4 % of the genes coincide in the direction of regulation, 30 % positively regulated, and 40.4 % negatively regulated in both species (Fig. 1b). However, despite being a significant correlation, the correlation was only r = 0.43. To have common activation response genes in the two species, these latter were filtered to orthologous genes that were differentially expressed in mice AND humans (intersection set/AND). (Fig. 1c). For this set of genes, a strong correlation r = 0.61 was found, and of the 202 genes analysed, 165 (81.7 %) presented the same direction of change between the two species (42.1 % up-regulated and 39.6 % downregulated).
A heatmap was used to explore further the expression profile of the 165 orthologous genes in humans and mice (Fig. 1d). The heatmap shows that the 165 genes overlap significantly, and their expression profile is equal in response to macrophage activation between the two species. Additionally, we transformed the data to Fragments Per Kilobase of transcript per Million mapped reads (FPKM) to plot the genes in the same principal component analysis (PCA) (Fig. 1e). PCA showed high heterogeneity and a more significant difference between control and activated macrophages in mice. Nevertheless, for humans, the heterogeneity was less, suggesting that although the 165 orthologous genes share a similar profile, their magnitude of expression is different. To explore this possibility, we analysed the cumulative distributions of orthologous genes (Fig. 1f and g). For up-regulation, 25 % of the genes in the mouse model have a log2 fold change (FC) greater than 2.3, while for the human, it was greater than 1.8 (Fig. 1f). The same occurs for down-regulated genes, where for the mouse, 25 % of the genes have a log2 FC lower than −2.6, while for humans, the same fraction has a log2 FC lower than −1.1 (Fig. 1g).
Enrichment analysis was performed to identify in which biological processes the 165 orthologous genes were involved. The analysis showed nine gene ontologies, including genes related to VEGF regulation, granulocyte chemotaxis, neutrophil migration, IL-6 production, and inflammatory processes (Fig. 1h). Up-regulated genes for humans and mice are related to an inflammatory state mediated by cytokines and chemokines such as CCL8, IL6, LIF, CCL5, IL1A, IL1b, and CXCL2 (Fig. 1i), a typical profile of macrophages treated with LPS. Genes that control the cell cycle were identified among the down-regulated genes, such as NUP210, BUB1B, MYBL2, MKI67, and PLK1 (Fig. 1j). The cell cycle gene profile may be associated with IFN- γ being used in the activation treatment, which is known to arrest the cell cycle. These analyses illustrate that the 165 human-mouse orthologous genes show a similar activation profile. Their biological activity is related to macrophage activation with LPS and IFN- γ, making them good candidates for exploring the modulatory effect of plant extracts in these two model species.
3.2. OBE100 extract diminished the gene expression program of activated macrophages
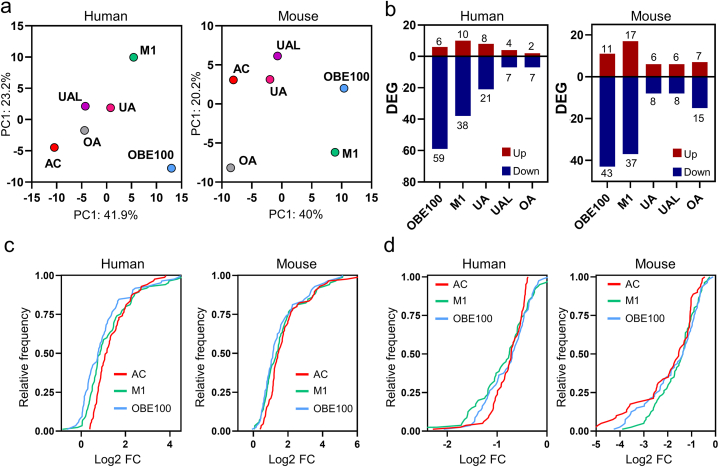
We utilized a PCA to evaluate the impact of OBE100 extract, an artificial mixture of triterpenes (M1), and individual triterpenes (UA, OA, and UAL) on the expression of 165 LPS and IFN-γ response genes in macrophages (Fig. 2a). The PCA analysis for the two species reveals three distinct patterns: 1) the treatment with OBE100 displays the most significant difference compared to activated macrophages; 2) the treatments with individual triterpenes are closer to the activated control, and 3) when combined (M1), they move further away from the activated control.
Fig. 2.
OBE100 treatment resulted in a reduction of the gene expression program associated with activated macrophages. (a) Principal component analysis (PCA) for human and mouse models of five treatments (AC: activated control, UA: Ursolic acid, UAL: Ursolic acid lactone, OA: Oleanolic acid, M1: triterpenes mix and OBE100) based on 165 common orthologue genes. (b) Bar plot displaying the cumulative count of up-regulated and down-regulated genes for each treatment condition. (c,d) Cumulative distribution plots of log2 fold change for OBE100 (light blue line) and M1 (green line) treatments compared to activated control (red line) for both up-regulated (c) and downregulated (d) genes. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To investigate whether the observed differences in the PCA were due to significant changes in gene expression, we calculated gene expression levels for each treatment concerning activated macrophages. DEG was defined as those with an absolute log2 fold change ≥0.38 or FDR <0.05 (Fig. 2b). Our analysis demonstrated that most DEG was downregulated across all treatments and in human and mouse models. Notably, the OBE100 treatment had the highest number of DEG (human: 59 and 6, mouse: 43 and 11, down and up-regulated, respectively). As the PCA shows, the treatment with M1 presents a more significant number of DEG than the treatments with individual molecules.
We performed cumulative frequency analysis to investigate further the impact of OBE100 and M1 treatments on gene expression levels (Fig. 2c and d). Our results show that the cumulative frequency distribution of up-regulated genes is shifted to the left of the activated control group in both humans and mice following OBE100 and M1 treatments, indicating a significant decrease in the fold change of DEG (Fig. 2c). Moreover, for down-regulated genes, the distribution of the accumulated frequency for OBE100 and M1 treatments was shifted to the right, approaching a zero log 2 FC, indicating that the treatments must reduce the effect of activation by LPS and IFN-γ (Fig. 2d). These results suggest that OBE100 and M1 treatments may attenuate the effect of LPS and IFN-γ activation on gene expression.
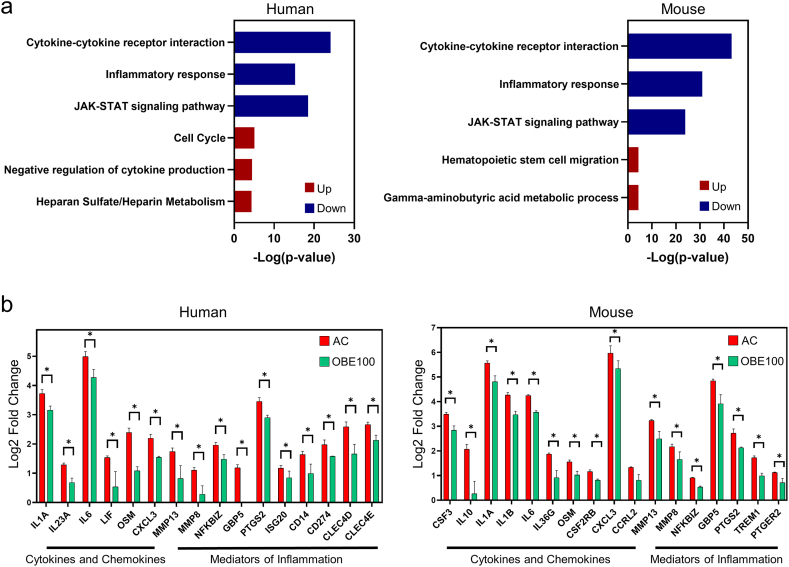
3.3. OBE100 modulates the inflammatory program in activated macrophages
To help elucidate the underlying biological mechanisms regulating OBE100 treatment, we conducted a comprehensive gene ontology (GO) analysis. Our findings indicate that OBE100 down-regulates the expression of genes associated with cytokine signalling and the inflammatory response, affecting the expression of the JAK-STAT signalling pathway (Fig. 3a). Specifically, our results reveal that activated macrophages treated with OBE100 exhibit a decreased expression of cytokines and chemokines, including IL1A, IL6, OSM, and CXCL3, in both human and murine models (Fig. 3b). Nevertheless, we also observed specific differences between the human and mouse models. For instance, the regulation of LIF, a member of the IL-6 family, and IL23A were unique to the human model, whereas CSF3, IL10, IL1B, and IL36G were more prominently regulated in mice (Fig. 3b). Furthermore, OBE100 treatment was associated with a reduction in several inflammation mediators, including metalloproteinases (MMP8 and MMP13), NF-KB transcription factor (NFKBIZ), a regulator of Inflammasomes (GBP5), and prostaglandin synthesis (PTGS2) (Fig. 3b). Notably, the human model showed the downregulation of membrane proteins that regulate inflammation, such as CD14, CD274 (also known as PD-L1), and type C lectin receptors (ClEC4D and ClEC4E) (Fig. 3b). These results show that OBE100 has an immunomodulatory effect on activated macrophages by reducing the expression of cytokines, chemokines, and inflammatory mediators.
Fig. 3.
The Modulation of the Inflammatory Program in Activated Macrophages by OBE. (a) Gene ontology enrichment analysis of DEG modulated by OBE in human and mouse macrophages. The coloured bars represent the percentage of genes present in the data set compared to the total number of genes in each term (% Genes/Term). The down-regulated and up-regulated ontologies are shown in blue and red, respectively. (b) Bar plots of normalised differentially expressed genes with a critical function on inflammation: Cytokines, chemokines, and inflammatory mediators. *p ≤ 0.05 compared with AC. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.4. The OBE100 extract has a more powerful immunomodulatory effect than the triterpene mixture
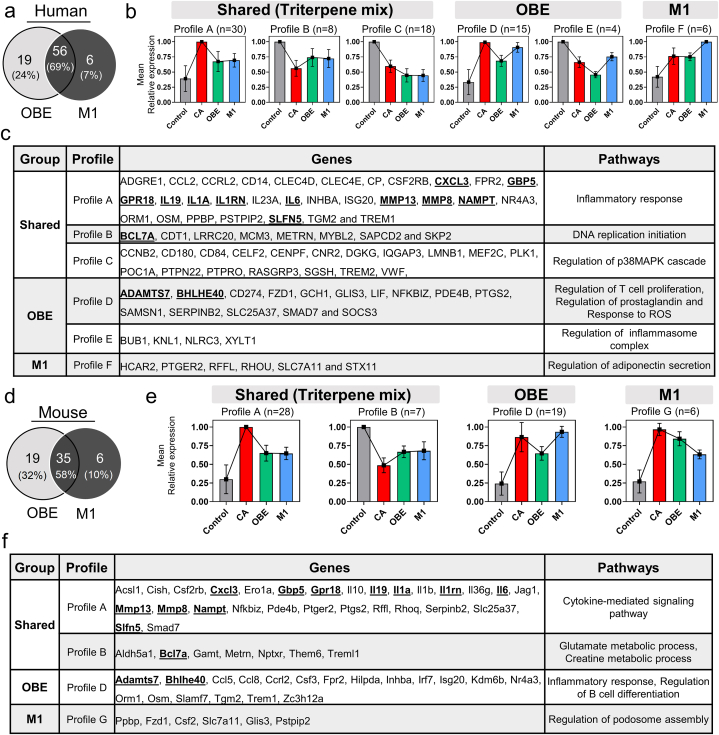
The immunomodulatory effect of the OBE100 extract on macrophages has been reported [11], but the molecules underlying this effect are not yet fully understood. It is unclear whether the main effect is due to the mixture of triterpenes (UA, UAL, and OA), the uncharacterised fraction in the OBE100 extract, or both. To identify which genes were regulated by the triterpenes and which were dependent on the uncharacterised fraction, a k-means clustering to categorize gene expression profiles was run. Genes expressed similarly in the two treatments are the genes regulated by the triterpene mixture. Genes only modulated by the OBE100 treatment are likely to be genes regulated explicitly by the uncharacterised fraction. Our analysis showed that OBE100 triterpenes regulated 69 % of human genes and 58 % of mouse genes, while uncharacterised molecules could regulate 24 % of human genes and 32 % of mouse genes (Fig. 4a and d, respectively).
Fig. 4.
The Immunomodulatory Effect of OBE Extract and its Relation to three major triterpene compounds. Venn Diagrams illustrating the modulation of DEGs by OBE and M1 Treatment in human (a) and mouse (d) macrophages. DEGs were categorized into seven profiles using k-means based on their transcript level patterns across the four human (b) and mouse (e) macrophage samples. A table presents the genes belonging to each profile, along with the Gene Ontology enrichment analysis of the eight profiles for human (c) and mouse (f) macrophages. Notably, the genes underlined indicate the shared genes between profiles in human and mouse models.
In order to gain a more comprehensive understanding of the differences in gene expression, the k-means clusters were divided into seven distinct profiles denoted as A through G (Fig. 4b and e). Specifically, three profiles were identified for the shared genes, A through C, regulated by the OBE100 triterpenes.
Profile A included positively regulated genes, such as CXCL3, GBP5, GPR18, IL19, IL1A, IL1RN, IL6, MMP8, MMP13, NAMPT and SLFN5 among others, during activation with LPS and IFN (Fig. 4c and f). However, expression of these genes was reduced following treatments with OBE100 and the M1, and their biological function in humans and mice is linked to inflammation. Profile B included genes negatively regulated during activation and whose expression was induced by treatments approaching basal levels (Fig. 4b and e). For this group, we found genes in humans that were associated with DNA replication and the cell cycle, such as BCL7A, CDT1, MCM3, MYBL2, SAPCD2, and SKP2, whereas genes in mice were linked to glutamate metabolism and creatine, such as Aldh5a1, Gamt, and Them6 (Fig. 4c and f). Lastly, profile C was only identified in humans and represented a group of down-regulated genes during activation and further reduced by treatments. This profile was associated with genes that regulated the p38MAPK cascade, such as PTPN22 and TREM2.
Moreover, we have identified two gene expression profiles, D and E, that could be regulated by the minority fraction (Fig. 4b and e); these are modulated by treatment with OBE100 but not by M1. Genes belonging to profile D were up-regulated in activated macrophages and diminished their expression with the treatment. Genes belonging to D profiles regulate immune system processes, such as inflammation (in humans: LIF, ADAMTS7, NFKBIZ, SOCS3; in mice: Ccl5, Ccl8, Ccrl2, Csf3, Adamts7 Irf7, and Isg20), the proliferation of T and B cells (SMAD7, CD274), prostaglandins (PTGS2) and reactive oxygen species (GCH1) (Fig. 4c and f). In contrast, profile E consists of only four genes, which were down-regulated during macrophage activation, and their expression is further reduced upon treatment with OBE100. Notably, the NLRC3 gene in profile E negatively regulates the inflammasome (Fig. 4c).
Finally, the analysis reveals the F and G profiles comprising specific genes exclusively modulated with the mixture of triterpenes treatment. Based on this, it can be inferred that the mix of triterpenes leads to the modulation of a particular set of genes, but the effect was concealed when the minority fraction was present (Fig. 4b and e). Interestingly, the F profile was unique for humans and the G profile for mice. Profile F exhibits genes up-regulated in the activation group but with a potentiated expression in the mix of triterpenes, such as the HCAR2 gene that regulates adiponectin secretion (Fig. 4b andc). On the other hand, the G profile was opposite to the F profile, given that the genes were up-regulated during activation and decreased their expression with the mix of triterpenes (Fig. 4e). For example, we have the CSF2 gene, which regulates the ensemble of Podosomes (Fig. 4f).
3.5. OBE100-modulated genes are associated with a set of crucial transcriptional factors
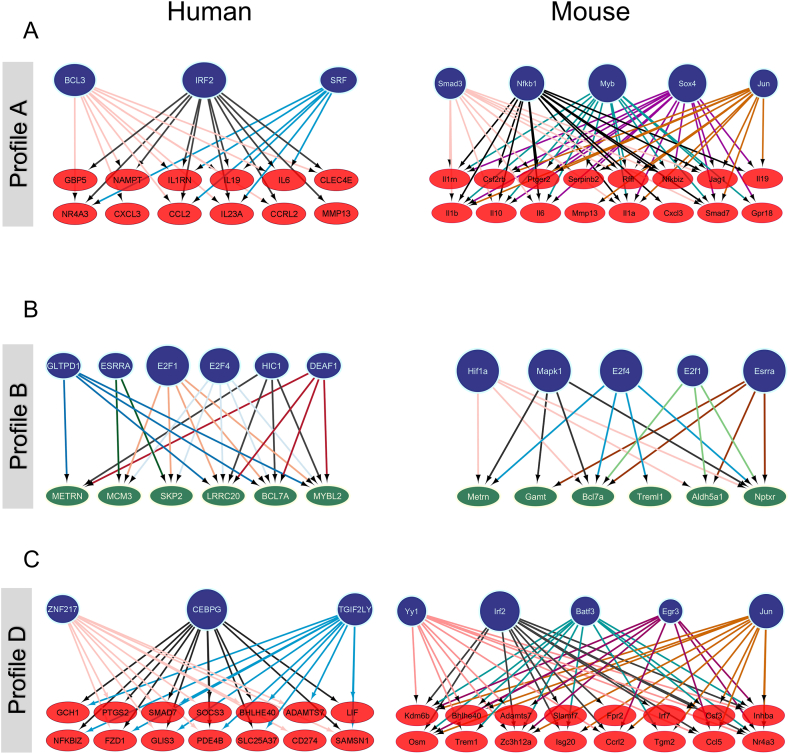
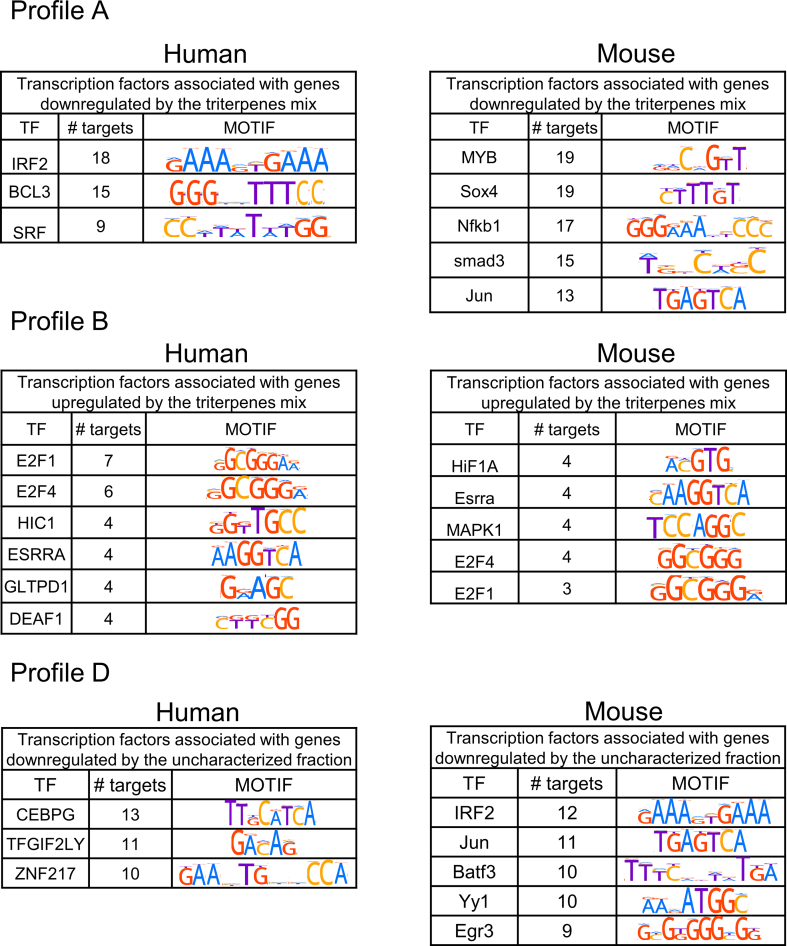
One of the molecular mechanisms by which the triterpene mixture (M1) or its associated uncharacterised fraction may modulate gene expression profiles is through transcriptional control. To investigate this potential mechanism, we assessed the enrichment of transcription factor (TF) motifs in the promoter sequences of DEGs dependent on the triterpene mixture (profiles A and B as depicted in Fig. 4) and those dependent on the uncharacterised fraction (profile D as shown in Fig. 4).
In the network representations, blue nodes denote transcription factors, while green and red nodes represent promoters of upregulated and downregulated genes, respectively. Our analyses revealed the identification of 12 TFs for humans and 15 for mice that regulate 32 and 38 genes, respectively (Fig. 5(A–C) and Fig. S1). Specifically, in profile A, the identified TFs differ between humans and mice, yet both are associated with inflammation genes. In humans, the transcription factors BCL3 and IRF2, known regulators of inflammation, were found, each governing 18 and 15 genes, respectively. In mice, transcription factors such as NFKb1 and Jun, recognized for regulating inflammatory genes, were identified. Additionally, TFs MYB, SOX4, and SMAD3, acknowledged for their involvement in various immune system processes, including ECM differentiation, migration, and remodeling, were also identified.
Fig. 5.
Association of OBE100-modulated genes with key transcription factors. The regulatory network illustrates the transcription factors and their target genes associated with each expression profile (A, B, and C) in both models. Blue circles represent transcription factors, while up-regulated and down-regulated genes are depicted by green and red ellipses, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
In profile B, TFs from the E2F family (E2F1 and E2f4) and estrogen-related receptor alpha (ESRRA) were identified in both models (Fig. 5B and Fig. S1). Particularly, E2F family factors, known for regulating cell cycle genes such as MCM3, skp2, and mybl2, were identified. TFs hif1a and mapk1a were found in mice, with the latter acting as a transcriptional repressor. Similarly, HIC1, another transcriptional repressor, was identified for humans. Analyzing the expression profile D (Fig. 5C and Fig. S1), the CEBPG TF stood out for humans, regulating inflammatory genes such as LIF and NFKBIZ. Notably, in mice, TFs IRF2, BATF3, and JUN were found, forming heterotrimers to regulate immune cell fate. Finally, TFs early growth response genes 3 (Egr3) and YY1 were identified, known for their role in maintaining immune homeostasis [[27], [28], [29], [30], [31], [32], [33]].Discussion.
Obesity is a global epidemic associated with several comorbidities, including insulin resistance, type 2 diabetes, cardiovascular diseases, and cancer. The mechanisms linking obesity with these diseases are not totally understood, but inflammation plays a crucial role. Obese people increase the accumulation of macrophages in tissues such as adipose tissue, skeletal muscle, liver, gut, pancreatic islet, and brain. These macrophages switch phenotypes from M0-or M2-like to M1-like, increasing the expression of proinflammatory cytokines and leading to metabolic dysfunctions and diseases linked to obesity [34,35]. Thus, targeting inflammation is a noteworthy approach to treating metabolic alterations associated with obesity. Previous works have shown that OBE100, an Eu extract, has hypoglycemic, hypolipidemic, antioxidant, and anti-inflammatory activities [[10], [11], [12], [13]]. This work analysed how the extract compounds modulate gene expression in mouse and human macrophages. Using these two cell lines can make it possible to validate the biological reproducibility of the results obtained. We used LPS and INF-γ to activate in vitro mouse and human macrophages. It has been described that the macrophage response to this in vitro classically activation is similar to the in vivo M1 macrophage signature [36]. After the activation, despite the heterogeneity shown in mice (Fig. 1e), we found that the expression profile of 165 genes changes in the same direction between the two species (Fig. 1c). As previously described, genes involved in immunoregulatory and inflammatory processes are up-regulated, and LPS and INF-γ treatment led to an overexpression of genes that code for proteins produced by activated monocytes, neutrophils, and expressed at sites of inflammation (Fig. 1i) [37,38]. Gene ontology analysis confirmed this result (Fig. 1h). Another effect of activating macrophages with LPS and INF-γ is the inhibition of cell proliferation [39,40]. In this work, LPS and INF-γ treatment downregulated genes involved in mitosis and cell cycle progression (e.g., PLK1, BUB1B, MYBL2, MKI67) (Fig. 1i). Thus, gene modification patterns followed previously described in activating these cells with LPS and INF-γ.
Triterpenoids are well-studied natural molecules found in plants. OBE100 extract has UA, OA, and UAL as the main molecules. These three triterpenoids have been shown to present antioxidant and anti-inflammatory effects [12,16,41,42]. In this study, the treatment of activated macrophages with the individual triterpenes changed the cells' expression pattern, and this change was increased when we mixed them (M1). Interestingly, combining these molecules with the minor components of the natural extract increased these changes in the expression pattern (Fig. 2a). This data was confirmed with the DEG analysis; Out of 165 orthologous genes, OBE100 was the treatment that altered the expression of the most significant number of genes in both human and mouse cells (Fig. 2b). The cumulative frequency analysis showed that M1 and OBE100 inhibited up-regulated gene expression by macrophage activation (Fig. 2c). However, the combination of molecules in the natural extract has an additive or synergic effect that affects, to the greatest extent, the expression of genes in these cells. Since these genes are involved in inflammatory processes, this may explain why OBE100 provides a superior anti-inflammatory effect, as previously described [11]. To reduce complexity, we analysed our data by examining GO. This analysis highlighted OBE100 inhibited gene expression associated with an inflammatory response in both human and murine cells (Fig. 3a), consistent with the protective effects of OBE100 [10]. Fig. 3b shows that the natural extract reduces gene expression in acute and chronic inflammatory responses. OBE100 inhibits the expression of NFKBIZ, OSM, GBP5, IL1A, and IL6 genes in both human and mouse cells. NFKBIZ, a member of the ankyrin-repeat family and induced by lipopolysaccharide (LPS), OSM, a member of the IL-6 family of cytokines, and GBP5, a member of the dynamin superfamily of GTPases, are involved in the induction of inflammatory genes, including IL6 and IL1A [[43], [44], [45], [46]]. IL6 encodes a cytokine that functions in inflammation and the maturation of B cells, and IL1A mediates the activation of NF-kappa B and the three MAPK pathways p38, p42/p44, and JNK pathways [47,48]. The IL-1 superfamily members are responsible for strong inflammatory responses with potential damaging effects, involved in the development of T2D [49].OBE100 also suppresses CXCL3 expression in both models. This chemokine is involved in inflammation and chemoattraction for neutrophils [50]. Interestingly, CXCL3 also has a role in adipocyte differentiation, and CXCL3 and its receptor, CXCR2, are highly expressed in mature adipocytes. CXCL3 induces adipogenesis in an autocrine and paracrine manner, and inhibition of this gene is associated with inhibiting adipogenic differentiation [51]. This latter is according to the effect observed in adipocytes after OBE100 treatment [11]. The extract also affects MMP expression, downregulating MMP8 and MMP13 gene expression in human and mouse models (Fig. 3b). It has been shown that MMPs are involved in the progression of various inflammatory conditions by regulating the proinflammatory cytokines TNF-α and IL-1β, and MMP8 expression and is associated with obesity, insulin resistance and T2D [[52], [53], [54]]. OBE100 also regulated the expression of genes differently in human and mouse cells. In any case, this differential expression enhances the anti-inflammatory effect in both models. LIF, IL23A, CD14, CD274, CLEC4D, and CLEC4E are significantly downregulated in human cells. All these genes involve inflammatory processes [36,55]. Interestingly, LPS induces inflammation through CD14 [56], and CD274 is up-regulated in adipose tissue immune cells in obese patients [57]. In the same way, OBE100 enhanced the anti-inflammatory effect in mouse cells, inhibiting the expression of genes associated with inflammation, such as IL1B, IL36G, CSF3, CCRL2, TREM1, and CSF2RB [36,51,[58], [59], [60], [61]].
The high content of triterpenes present in the natural extract may explain these biological activities. Terpenes such as UA, OA, and UAL present anti-inflammatory activities and inhibit the expression of inflammatory genes [62]. However, we have previously shown that OBE100 provides superior anti-inflammatory effects than the reconstituted triterpenoid mixture [11]. We analysed the differences in gene expression between the natural extract and the mix of terpenoids without the minor fraction to help understand this enhanced effect of OBE100. After a gene expression profile categorisation, we found that triterpenes regulate 76 % of genes in human cells and 68 % of genes in mouse cells, while 24 % more of genes in humans and 32 % of genes in mice were regulated by the effect of the unknown fraction alone or in combination with the terpenoids (Fig. 4a and d). We distinguished different expression profiles. Three of them, A, B, and C, are similar to OBE100 and M1 (Fig. 4b and e), so we may conclude that UA, OA, and UAL regulate the expression of the genes in these profiles. Profiles A and C include genes downregulated by OBE100 and M1 treatments (Fig. 4c and f). These genes play an essential role in regulating inflammation. Profile B collects genes up-regulated by OBE100 and M1, such as CDT1, MCM3, MYBL2, and SKP2. These genes involve DNA replication and cell cycle progression [[61], [62], [63]].
Interestingly, LPS and IFN-γ inhibit cell proliferation, and triterpenes may up-regulate genes involved in cell cycle control to help suppress macrophage activation. However, it has been shown that the pharmacological properties of these molecules in cancer prevention are attributed to inhibiting the cell cycle and inducing cellular apoptosis [63]. In our experiment, triterpenes doses used have no toxicity; it can be hypothesised that UA, OA, and UAL at lower concentrations may regulate the activated macrophage cell cycle differently. Profiles D and E (Fig. 4b and e) are exclusive of OBE100 treatment, and in both profiles, downregulated gene expression may be due to the minority fraction of the natural extract. We cannot conclude if this effect is produced by the molecules in this fraction or by combining the molecules with triterpenoids. The inhibited genes are involved in the inflammation process, such as CD274, LIF NFKBIZ, CCRL2, TREM1, and OSM (Fig. 4c and f). This result may explain the OBE100 superior anti-inflammatory effect observed and agrees with traditional medicine when whole plants or mixtures of plants are used rather than isolated compounds.
Common TFs control some of the genes regulated under these different expression profiles. Analyses were performed to identify which TFs were associated with regulating differentially expressed genes that the triterpene treatments could directly or indirectly regulate. Fig. 5a shows that genes inhibited by terpenes (OBE100 and M1 – Profile A) are controlled by TFs associated with inflammation. The TFs that regulate the most significant number of genes are BCL3, IRF2, MYB, SOX4, and NFKB1 (Fig. S1). NFKB1 regulates inflammatory responses, inducing the expression of proinflammatory genes; BCL3, a member of the IκB family of NF-κB inhibitors, also regulates the activity of the NF-κB pathway.
Moreover, BCL3 inhibition regulates lipid metabolism, improving hepatic steatosis and adipose tissue hypertrophy. SOX4 regulates inflammation and is also associated with the NF-κB pathway. IRF2 is an essential regulator of the proinflammatory response and may be a target to treat diseases like obesity with inflammatory disorders [[27], [28], [29], [30]]. The analysis of genes regulated by the minority fraction of OBE100 (Profile D) revealed that TFs that regulate the most significant number of genes are CEBPG, BATF3, and Yy1. C/EBPs are a family of transcription factors involved in differentiation, cellular proliferation, and inflammation processes, and Yy1 depletion is associated with a reduction of the cellular inflammatory response [64]. Our findings suggest that treating triterpenes and their minor fraction could simultaneously modulate TFs regulating the immune system. The networks also indicate a combined action of TFs on target gene promoters, suggesting that they may be acting cooperatively or in sequential steps in regulating gene expression.
4. Conclusion
This study provides evidence that an extract of Eu (OBE100) has anti-inflammatory activity through its ability to downregulate proinflammatory gene expression. UA, OA, and UAL are the main molecules that regulate the expression of inflammatory genes. However, unknown minor metabolites in the extract provide a synergistic or additive superior anti-inflammatory effect. Further studies are required to identify the minor compounds of the extract. OBE100 is a promising extract for diabetes treatment that could break the link between inflammation and insulin resistance.
Funding
This work was supported by Minciencias, Grant No. 111577756880, and Universidad de Antioquia, Grant No. 2017-16968. The funders played no role in the study design, data collection, analysis, publication decision, or manuscript preparation.
Data availability statement
The datasets generated for this study are in the Gene Expression Omnibus (GEO) DataSets (https://www.ncbi.nlm.nih.gov/gds) under the accession number GSE250471.
CRediT authorship contribution statement
Alejandro Mejia-Garcia: Writing – review & editing, Methodology, Investigation, Formal analysis, Data curation. Geysson Javier Fernandez: Writing – review & editing, Writing – original draft, Methodology, Investigation, Formal analysis, Data curation. Luis Fernando Echeverri: Writing – review & editing, Resources, Methodology. Norman Balcazar: Writing – review & editing, Writing – original draft, Supervision, Resources, Project administration, Funding acquisition, Formal analysis, Conceptualization. Sergio Acin: Writing – review & editing, Writing – original draft, Supervision, Resources, Project administration, Methodology, Funding acquisition, Formal analysis, Conceptualization.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Sergio Acin reports financial support was provided by Colombia Ministry of Science Technology and Innovation. Sergio Acin reports a relationship with University of Antioquia that includes: employment. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e24382.
Contributor Information
Norman Balcazar, Email: norman.balcazar@udea.edu.co.
Sergio Acin, Email: s.acin@udea.edu.co.
Appendix A. Supplementary data
The following are the supplementary data to this article:
figs1.
References
- 1.Yang L., Wen K.S., Ruan X., et al. Response of plant secondary metabolites to environmental factors. Molecules. 2018;23:1–26. doi: 10.3390/molecules23040762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jain Chitra, Khatana Shivani, Vijayvergia Rekha. Bioactivity of secondary metabolites of various plants: a review. Int J Pharm Sci Res. 2019;10:494–504. doi: 10.13040/IJPSR.0975-8232.10(2).494-04. [DOI] [Google Scholar]
- 3.Surbhi Kumar A., Singh S., et al. Eucalyptus: phytochemical composition, extraction methods and food and medicinal applications. Adv Tradit Med. 2021 doi: 10.1007/s13596-021-00582-7. [DOI] [Google Scholar]
- 4.Chandorkar N., Tambe S., Amin P., et al. A systematic and comprehensive review on current understanding of the pharmacological actions, molecular mechanisms, and clinical implications of the genus Eucalyptus. Phytomedicine. 2021;1 doi: 10.1016/j.phyplu.2021.100089. [DOI] [Google Scholar]
- 5.Villaseñor I.M., Lamadrid M.R.A. Comparative anti-hyperglycemic potentials of medicinal plants. J. Ethnopharmacol. 2006;104:129–131. doi: 10.1016/j.jep.2005.08.067. [DOI] [PubMed] [Google Scholar]
- 6.Tsalamandris S., Antonopoulos A.S., Oikonomou E., et al. Risk factors and cardiovascular disease prevention the role of inflammation in diabetes : current Concepts and future perspectives. Eur Cardiol Rev. 2019;14:50–59. doi: 10.15420/ecr.2018.33.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rohm T.V., Meier D.T., Olefsky J.M., et al. Inflammation in obesity, diabetes, and related disorders. Immunity. 2022;55:31–55. doi: 10.1016/j.immuni.2021.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oguntibeju O.O. Type 2 diabetes mellitus, oxidative stress and inflammation: examining the links. Int J Physiol Pathophysiol Pharmacol. 2019;11:45–63. [PMC free article] [PubMed] [Google Scholar]
- 9.Kwon H., Pessin J.E. Adipokines mediate inflammation and insulin resistance. Front. Endocrinol. 2013;4:1–13. doi: 10.3389/fendo.2013.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Acín S., Muñoz D.L., Guillen A., et al. Triterpene-enriched fractions from Eucalyptus tereticornis ameliorate metabolic alterations in a mouse model of diet-induced obesity. J. Ethnopharmacol. 2021 doi: 10.1016/j.jep.2020.113298. [DOI] [PubMed] [Google Scholar]
- 11.Betancur L.I., Muñoz D.L., Guillen A., et al. Major triterpenoids from eucalyptus tereticornis have enhanced beneficial effects in cellular models when mixed with minor compounds present in raw extract. An. Acad. Bras. Cienc. 2021;93:1–15. doi: 10.1590/0001-3765202120201351. [DOI] [PubMed] [Google Scholar]
- 12.Ceballos S., Guilí En A., Lorena D., et al. Immunometabolic regulation by triterpenes of Eucalyptus tereticornis in adipose tissue cell line models. Immunometabolic regulation by triter- penes of Eucalyptus tereticornis in adipose tissue cell line models. Phytomedicine Phytomedicine. 2018 doi: 10.1016/j.phymed.2018.03.059. [DOI] [PubMed] [Google Scholar]
- 13.Guillén A., Granados S., Rivas K.E., et al. Antihyperglycemic activity of Eucalyptus tereticornis in insulin-resistant cells and a nutritional model of diabetic mice. Adv Pharmacol Sci. 2015;2015 doi: 10.1155/2015/418673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen H.N., Ullevig S.L., Short J.D., et al. Ursolic acid and related analogues: triterpenoids with broad health benefits. Antioxidants. 2021;10:1–24. doi: 10.3390/antiox10081161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castellano J.M., Ramos-Romero S., Perona J.S. Oleanolic acid: extraction, characterization and biological activity. Nutrients. 2022;14 doi: 10.3390/nu14030623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balcazar N., Betancur L.I., Muñoz D.L., et al. Ursolic acid lactone obtained from eucalyptus tereticornis increases glucose uptake and reduces inflammatory activity and intracellular neutral fat: an in vitro study. Molecules. 2021;26 doi: 10.3390/molecules26082282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hyun M.K., Kim D.H., Park C.H., et al. Protective mechanisms of loquat leaf extract and ursolic acid against diabetic pro-inflammation. J. Mol. Med. 2022;100:1455–1464. doi: 10.1007/s00109-022-02243-x. [DOI] [PubMed] [Google Scholar]
- 18.Luan M., Wang H., Wang J., et al. Advances in anti-inflammatory activity, mechanism and therapeutic application of ursolic acid. Mini-Rev. Med. Chem. 2022;22:422–436. doi: 10.2174/1389557521666210913113522. [DOI] [PubMed] [Google Scholar]
- 19.Hwang J., Zheng M., Wiraja C., et al. Reprogramming of macrophages with macrophage cell membrane-derived nanoghosts. Nanoscale Adv. 2020;2:5254–5262. doi: 10.1039/d0na00572j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Córdova-Dávalos L.E., Cervantes-García D., Ballona-Alba M.F., et al. Protective effect of glycomacropeptide on the inflammatory response of U937 macrophages. Foods. 2023;12 doi: 10.3390/foods12071528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ribeiro M.A., Estill M.S., Fernandez G.J., et al. Integrative transcriptome and microRNome analysis identifies dysregulated pathways in human Sertoli cells exposed to TCDD. Toxicology. 2018;409:112–118. doi: 10.1016/j.tox.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 22.Ringwald M., Richardson J.E., Baldarelli R.M., et al. Mouse genome Informatics (MGI): latest news from MGD and GXD. Mamm. Genome. 2022;33:4–18. doi: 10.1007/s00335-021-09921-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Edward Y., Tan Christopher M., Yan Kou, Duan Qiaonan, Zichen Wang G.V.M., Ma’ayan1 N.R.C. Avi. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013;14 doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez G.J., Ramírez-Mejía J.M., Castillo J.A., et al. Vitamin D modulates expression of antimicrobial peptides and proinflammatory cytokines to restrict Zika virus infection in macrophages. Int. Immunopharm. 2023:119. doi: 10.1016/j.intimp.2023.110232. [DOI] [PubMed] [Google Scholar]
- 25.Janky R., Verfaillie A., Imrichová H., et al. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput. Biol. 2014;10 doi: 10.1371/journal.pcbi.1003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paul Shannon 1, Andrew Markiel 1, Owen Ozier, Nitin S 2, Baliga 1, Jonathan T., Wang 2, Daniel Ramage 2, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:426. doi: 10.1101/gr.1239303.metabolite. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang S., Gao J., Liu S., et al. Transcription coactivator bcl3 acts as a potential regulator of lipid metabolism through the effects on inflammation. J. Inflamm. Res. 2021;14:4915–4926. doi: 10.2147/JIR.S327858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui H., Banerjee S., Guo S., et al. IFN regulatory factor 2 inhibits expression of glycolytic genes and lipopolysaccharide-induced proinflammatory responses in macrophages. J. Immunol. 2018;200:3218–3230. doi: 10.4049/jimmunol.1701571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu T., Zhang L., Joo D., et al. NF-κB signaling in inflammation. Signal Transduct. Targeted Ther. 2017;2 doi: 10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei H., Gu Q. SOX4 promotes high-glucose-induced inflammation and angiogenesis of retinal endothelial cells by activating NF-$κ$B signaling pathway. Open Life Sci. 2022;17:393–400. doi: 10.1515/biol-2022-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Itoh Y., Saitoh M., Miyazawa K. Smad3-STAT3 crosstalk in pathophysiological contexts. Acta Biochim. Biophys. Sin. 2018;50:82–90. doi: 10.1093/abbs/gmx118. [DOI] [PubMed] [Google Scholar]
- 32.Kawai T., Autieri M.V., Scalia R. Adipose tissue inflammation and metabolic dysfunction in obesity. Am. J. Physiol. Cell Physiol. 2021;320:C375–C391. doi: 10.1152/ajpcell.00379.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orecchioni M., Ghosheh Y., Pramod A.B., et al. Macrophage polarization: different gene signatures in M1(Lps+) vs. Classically and M2(LPS-) vs. Alternatively activated macrophages. Front. Immunol. 2019;10:1–14. doi: 10.3389/fimmu.2019.01084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Held T.K., Weihua X., Yuan L., et al. Gamma interferon augments macrophage activation by lipopolysaccharide by two distinct mechanisms, at the signal transduction level and via an autocrine mechanism involving tumor necrosis factor alpha and interleukin-1. Infect. Immun. 1999;67:206–212. doi: 10.1128/iai.67.1.206-212.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su X., Yu Y., Zhong Y., et al. Interferon-γ regulates cellular metabolism and mRNA translation to potentiate macrophage activation. Nat. Immunol. 2015;16:838–849. doi: 10.1038/ni.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xaus J., Cardó M., Valledor A.F., et al. Interferon γ induces the expression of p21(waf-1) and arrests macrophage cell cycle, preventing induction of apoptosis. Immunity. 1999;11:103–113. doi: 10.1016/S1074-7613(00)80085-0. [DOI] [PubMed] [Google Scholar]
- 37.Xaus J., Comalada M., Valledor A.F., et al. Molecular mechanisms involved in macrophage survival, proliferation, activation or apoptosis. Immunobiology. 2001;204:543–550. doi: 10.1078/0171-2985-00091. [DOI] [PubMed] [Google Scholar]
- 38.Checker R., Sandur S.K., Sharma D., et al. Potent anti-inflammatory activity of ursolic acid, a triterpenoid antioxidant, is mediated through suppression of NF-??B, AP-1 and NF-AT. PLoS One. 2012;7 doi: 10.1371/journal.pone.0031318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee W., Yang E.J., Ku S.K., et al. Anti-inflammatory effects of oleanolic acid on LPS-induced inflammation in vitro and in vivo. Inflammation. 2013;36:94–102. doi: 10.1007/s10753-012-9523-9. [DOI] [PubMed] [Google Scholar]
- 40.Soysa R., Bean J.C., Wu X., et al. Early-Derived murine macrophages temporarily renounce tissue identity during acute systemic inflammation. J. Immunol. 2021;207:569–576. doi: 10.4049/jimmunol.2001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Slowikowski K., Nguyen H.N., Noss E.H., et al. CUX1 and IκBζ (NFKBIZ) mediate the synergistic inflammatory response to TNF and IL-17A in stromal fibroblasts. Proc. Natl. Acad. Sci. U.S.A. 2020;117:5532–5541. doi: 10.1073/pnas.1912702117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones S.A., Jenkins B.J. Recent insights into targeting the IL-6 cytokine family in inflammatory diseases and cancer. Nat. Rev. Immunol. 2018;18:773–789. doi: 10.1038/s41577-018-0066-7. [DOI] [PubMed] [Google Scholar]
- 43.Wu T., Han C., Shelhamer J.H. Involvement of p38 and p42/44 MAP kinases and protein kinase C in the interferon-γ and interleukin-1α-induced phosphorylation of 85-kDa cytosolic phospholipase A2 in primary human bronchial epithelial cells. Cytokine. 2004;25:11–20. doi: 10.1016/j.cyto.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 44.Rincon M. Interleukin-6: from an inflammatory marker to a target for inflammatory diseases. Trends Immunol. 2012;33:571–577. doi: 10.1016/j.it.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 45.Zhou L., Zhao H., Zhao H., et al. GBP5 exacerbates rosacea-like skin inflammation by skewing macrophage polarization towards M1 phenotype through the NF-$κ$B signalling pathway. J. Eur. Acad. Dermatol. Venereol. 2023;37:796–809. doi: 10.1111/jdv.18725. [DOI] [PubMed] [Google Scholar]
- 46.Metzemaekers M., Gouwy M., Proost P. Neutrophil chemoattractant receptors in health and disease: double-edged swords. Cell. Mol. Immunol. 2020;17:433–450. doi: 10.1038/s41423-020-0412-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kusuyama J., Komorizono A., Bandow K., et al. CXCL3 positively regulates adipogenic differentiation. J. Lipid Res. 2016;57:1806–1820. doi: 10.1194/jlr.M067207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lint P Van, Libert C. Chemokine and cytokine processing by matrix metalloproteinases and its effect on leukocyte migration and inflammation. 2007;82:1375–1381. doi: 10.1189/jlb.0607338. [DOI] [PubMed] [Google Scholar]
- 49.Behzadi P., Sameer A.S., Nissar S., et al. The interleukin-1 (IL-1) superfamily cytokines and their single nucleotide polymorphisms (SNPs) J Immunol Res. 2022:2022. doi: 10.1155/2022/2054431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winkelmann A., Lauhio A., Rissanen A., et al. Association of MMP-8 with obesity , smoking and insulin resistance. 2016;46:757–765. doi: 10.1111/eci.12649. [DOI] [PubMed] [Google Scholar]
- 51.Dambuza I.M., Brown G.D. vols. 21–27. 2015. (ScienceDirect C-type Lectins in Immunity : Recent Developments). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee H.S., Kim W.J. The role of matrix metalloproteinase in inflammation with a focus on infectious diseases. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms231810546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rietschel E.T.H., Schletter J., Weidemann B., et al. vol. 4. 1998. (Lipopolysaccharide and Peptidoglycan : CD14-dependent Bacterial Inducers of Inflammation P Opol Y Sacchari D Es). [DOI] [PubMed] [Google Scholar]
- 54.Tysoe PDL1 reduces adipose inflammation in obesity. Nat. Rev. Endocrinol. 2022;18:334. doi: 10.1038/s41574-022-00668-5. [DOI] [PubMed] [Google Scholar]
- 55.Yuan Z.C., Xu W.D., Liu X.Y., et al. Biology of il-36 signaling and its role in systemic inflammatory diseases. Front. Immunol. 2019;10 doi: 10.3389/fimmu.2019.02532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cancello R., Cle K. 2006. Is Obesity an Inflammatory Illness ? Role of Low-Grade Inflammation and Macrophage Infiltration in Human White Adipose Tissue; pp. 1141–1147. [DOI] [PubMed] [Google Scholar]
- 57.Tessarz A.S., Cerwenka A. The TREM-1/DAP12 pathway. 2008;116:111–116. doi: 10.1016/j.imlet.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 58.Hornero R.A., Dyczko A., Hamad I., et al. 2022. pp. 1–16. (Dissecting the Role of CSF2RB Expression in Human Regulatory T Cells). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schioppa T., Sozio F., Barbazza I., et al. Molecular Basis for CCRL2 Regulation of Leukocyte Migration. 2020;8:1–7. doi: 10.3389/fcell.2020.615031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Del Prado-Audelo M.L., Cortés H., Caballero-Florán I.H., et al. Therapeutic applications of terpenes on inflammatory diseases. Front. Pharmacol. 2021;12:1–7. doi: 10.3389/fphar.2021.704197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Assoian R.K., Yung Y. A reciprocal relationship between Rb and Skp2: implications for restriction point control, signal transduction to the cell cycle and cancer. Cell Cycle. 2008;7:24–27. doi: 10.4161/cc.7.1.5232. [DOI] [PubMed] [Google Scholar]
- 62.Musa J., Aynaud M.M., Mirabeau O., et al. MYBL2 (B-Myb): a central regulator of cell proliferation, cell survival and differentiation involved in tumorigenesis. Cell Death Dis. 2017;8:1–9. doi: 10.1038/CDDIS.2017.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li S., Kuo H.C.D., Yin R., et al. Epigenetics/epigenomics of triterpenoids in cancer prevention and in health. Biochem. Pharmacol. 2020;175 doi: 10.1016/j.bcp.2020.113890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qian Z., Zhao J. Silencing YY1 alleviates ox-LDL-induced inflammation and lipid accumulation in macrophages through regulation of PCSK9/LDLR signaling. J. Microbiol. Biotechnol. 2022;32:1406–1415. doi: 10.4014/jmb.2207.07011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study are in the Gene Expression Omnibus (GEO) DataSets (https://www.ncbi.nlm.nih.gov/gds) under the accession number GSE250471.