Abstract
Individuals with schizophrenia frequently experience co-occurring substance use, including tobacco smoking and heavy cannabis use, and substance use disorders. There is interest in understanding the extent to which these relationships are causal, and to what extent shared genetic factors play a role. We explored the relationships between schizophrenia (Scz), cannabis use disorder (CanUD), and ever-regular tobacco smoking (Smk) using the largest available genome-wide studies of these phenotypes in individuals of African and European ancestries. All three phenotypes were positively genetically correlated (rgs = 0.17 – 0.62). Causal inference analyses suggested the presence of horizontal pleiotropy, but evidence for bidirectional causal relationships was also found between all three phenotypes even after correcting for horizontal pleiotropy. We identified 439 pleiotropic loci in the European ancestry data, 150 of which were novel (i.e., not genome-wide significant in the original studies). Of these pleiotropic loci, 202 had lead variants which showed convergent effects (i.e., same direction of effect) on Scz, CanUD, and Smk. Genetic variants convergent across all three phenotypes showed strong genetic correlations with risk-taking, executive function, and several mental health conditions. Our results suggest that both horizontal pleiotropy and causal mechanisms may play a role in the relationship between CanUD, Smk, and Scz, but longitudinal, prospective studies are needed to confirm a causal relationship.
Introduction
Schizophrenia (Scz) is a psychiatric condition with an estimated twin-based heritability of around 80%1,2. Substance use disorders (SUDs) are highly prevalent in individuals with Scz3. Of these co-occurring SUDs, the role of cannabis use as a risk factor for Scz and first episode psychosis onset remains a classical “chicken or egg” problem in psychiatry4.
Some studies have suggested a causal, dose- and age-dependent effect of cannabis use on risk for onset of Scz and other forms of psychosis5–7. However, cannabis use and cannabis use disorder (CanUD) are heritable8 (twin heritability ~50%), and an alternative hypothesis is that shared genetic pathways underlie liability to Scz and cannabis use phenotypes9,10. Genetic correlations from genome-wide association studies (GWAS) have provided support for some genetic commonality (e.g., SNP-rg (Scz, cannabis use) = 0.2511, SNP-rg (Scz, CanUD) = 0.3712). A recent study identified 27 and 21 genome-wide significant loci contributing to the shared genetic etiology between Scz and cannabis use and CanUD, respectively13. However, the identification of shared loci was largely driven by genome-wide significant loci in the Scz GWAS, due to the relative difference in discovery power between the Scz and cannabis GWASs. Furthermore, these prior studies have largely been performed in samples predominantly of European ancestry, limiting the generalizability of these findings.
Horizontal pleiotropy (i.e., genetic variants independently contributing to both CanUD and Scz) and vertical pleiotropy (i.e., shared genetic associations via a causal path) are not mutually exclusive; both mechanisms may play a role in the co-occurrence of CanUD and Scz. Genetically informed studies of CanUD and Scz have reached mixed conclusions, with no single direction of causality receiving overwhelming support14,15. Several Mendelian Randomization analyses have suggested greater support for Scz causing cannabis use and CanUD than the opposite direction11,16, while the most recent GWAS of CanUD found a bidirectional causal association between Scz and CanUD17.
Few prior genetic studies have attempted to disentangle how nicotine/tobacco use genetics impacts the genetic relationship between Scz and CanUD. Approximately 72% of those with Scz report daily tobacco smoking (while this same report estimated 43% were regular cannabis users18), and there is evidence that individuals who smoke tobacco daily are at increased risk of psychosis19, an earlier age of onset of first psychotic episode19, and the development of schizophrenia20. The prevalence of tobacco use, whether as tobacco cigarettes or consumed with cannabis in certain preparations (e.g., blunts, where tobacco is removed from a cigar and replaced with cannabis, or spliffs, where cannabis and tobacco are rolled together), is also high in individuals with CanUD21,22. Prior studies have reported genetic correlations of tobacco smoking with CanUD (SNP-rg = 0.6117) and Scz (SNP-rg = 0.1423). Despite this, few epidemiologic studies have taken potential genetic sharing into account when reporting evidence for causal relationships between tobacco, cannabis, and Scz6,7. In turn, few genomic studies of cannabis and Scz have considered the role of tobacco13, despite the frequent co-occurrence of tobacco and cannabis use, especially in Europe24. In a prior study, we found that genetic liability for CanUD was positively associated with genetic liability for Scz even when accounting for the genetic components of cannabis ever-use, tobacco smoking, and nicotine dependence10. Another study found a causal effect of genetic liability to cannabis use on risk for schizophrenia, and this association was unchanged when accounting for tobacco smoking15. Thus, the genetic association between cannabis and Scz appears to be independent of tobacco use genetics to some extent, although the relatively low power of prior CanUD GWAS meant limited conclusions could be drawn from these earlier studies.
Given the significant genetic correlations between CanUD, tobacco smoking, and Scz, the increasing pace of cannabis legalization with emerging increases in CanUD incidence25, parallel increases in the popularity of nicotine vaping26, and the consequent potential impact on the course of Scz in those with heavy cannabis and tobacco use27–31, we investigated the evidence for causal relationships and horizontal pleiotropy between CanUD, tobacco smoking, and Scz. We used the largest genome-wide summary statistics available for Scz32 (European ancestry N = 161,405; African ancestry N = 15,846), CanUD17 (European ancestry N = 886,025; African ancestry N = 120,208), and ever-regularly smoking tobacco33 (Smk; European ancestry N = 805,431; African ancestry N = 24,278) in samples whose genetic ancestry is most similar to those historically from Europe (henceforth referred to as “European ancestry”) and samples whose genetic ancestry is most similar to those historically from Africa (henceforth referred to as “African ancestry”) to identify and characterize pleiotropic signals and conduct causal inference analyses. We focused on CanUD and Smk (as opposed to cannabis ever-use, or cigarettes per day) as CanUD was the cannabis phenotype with the largest genetic correlation with Scz, there was no available GWAS of cannabis consumption or heaviness of use, and current GWAS of nicotine dependence (relying on the Fagerström Test for Nicotine Dependence34 (FTND)) have been relatively under-powered compared to Smk.
Results
Genome-wide genetic correlations
Schizophrenia (Scz), Cannabis Use Disorder (CanUD), and ever-smoking tobacco regularly (Smk) were significantly genetically correlated in the European ancestry data (Table S1). The magnitude of the genetic correlation between Scz and CanUD (rg = 0.37, SE = 0.02, p = 2.97e–60) was statistically greater (pdiff = 6.5e–18) than the correlation between Scz and Smk (rg = 0.17, SE = 0.02, p = 6.88e-20) or between Scz and a measure of nicotine dependence more similar to CanUD, the FTND (rg = 0.22, SE = 0.04, p = 1.56e-7; pdiff = 0.002). This suggests that our choice of ever-regular smoking, rather than the FTND, as a measure of tobacco use was not the reason for the lower genetic correlation.
In the African ancestry data, the largest genetic correlation was between Scz and CanUD (rg = 0.61, SE = 0.14, p = 1.41e-5; Table S1). While the genetic correlation between Scz and Smk (rg = 0.34, SE = 0.15, p = 0.03) was of greater magnitude than in the European ancestry data, this estimate was not significantly different from zero after accounting for multiple testing, due to the much larger standard error.
Causal inference analyses
Using CAUSE35, a method that accounts for both correlated and uncorrelated horizontal pleiotropic effects, we found evidence for bidirectional causal relationships between all three phenotypes in the European ancestry data (Figure 1a, Table S2).
Figure 1.
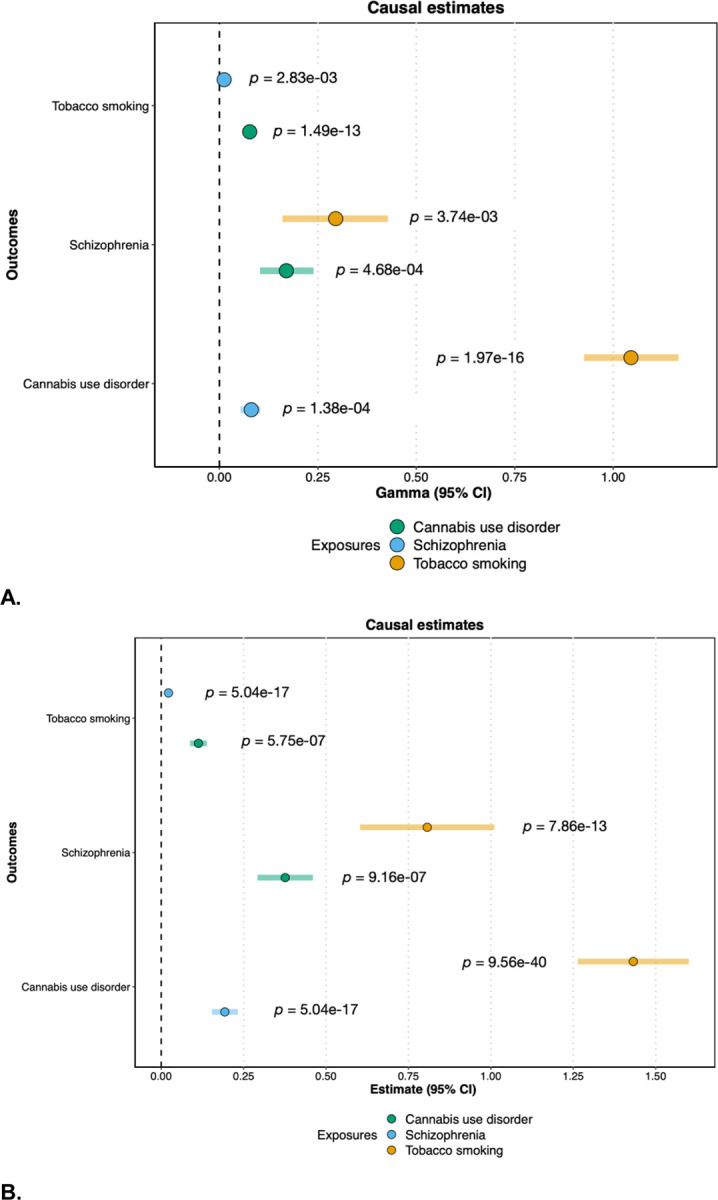
Panel A: Causal estimates (gamma) and 95% confidence intervals from CAUSE. Panel B: Causal estimates (beta) and 95% confidence intervals from MR-PRESSO. “Exposure” phenotypes are indicated by the color, while “Outcome” phenotypes are listed on the y-axis.
To explicitly test for the presence of horizontal pleiotropy, and to ensure our results were not isolated to a specific method of causal inference, we also performed Mendelian Randomization Pleiotropy RESidual Sum and Outlier (MR-PRESSO36) analyses in the European ancestry data. The MR-PRESSO global test for horizontal pleiotropy was significant for each pairwise test, and we found significant bidirectional causal effects between all three traits after the removal of outliers for horizontal pleiotropy (Figure 1b), consistent with the results from CAUSE. Results from other MR methods were generally consistent, with the same direction of effect (Table S3), although the more conservative MR-Egger test37,38 only showed a statistically significant causal effect of Smk on CanUD.
Cross-trait loci: European ancestry
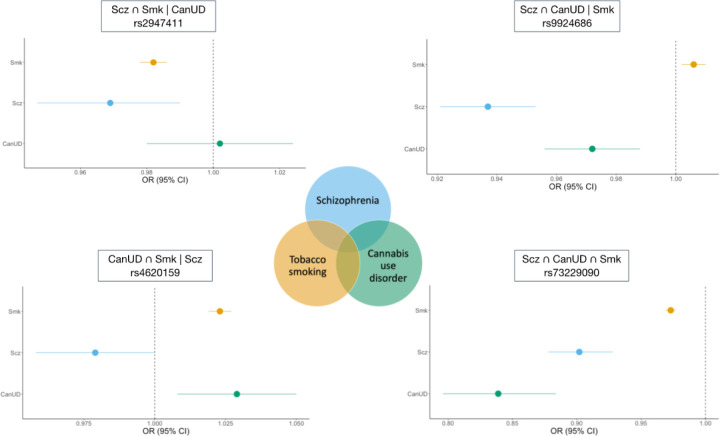
In consideration of the significant genetic correlations and evidence for horizontal pleiotropy from MR-PRESSO, we used ‘Association analysis based on SubSETs’ (ASSET39) to combine the GWAS summary data for CanUD, Smk and Scz (separately by ancestry), using the two-tailed meta-analysis approach. Unlike traditional meta-analysis approaches, ASSET accounts for SNPs with significant effects on multiple disorders even if the effects on the traits are in opposite directions. Following Lam et al.40, we use the following notation for each subset: ∩ represents variant subsets with the same directions of effect (+ or −), and | represents variant subsets whose effects are divergent across the different phenotypes. We therefore defined four subsets: (1) Scz ∩ CanUD ∩ Smk (i.e., a subset with convergent effects across all 3 traits); (2) Scz ∩ CanUD | Smk (i.e., a subset of variants with convergent effects for Scz and CanUD, but divergent effects for Smk); (3) Scz ∩ Smk | CanUD; and (4) CanUD ∩ Smk | Scz.
In total, we identified 439 pleiotropic genomic risk loci (i.e., loci where the lead SNP has an effect on all three phenotypes). Of these, 150 loci were novel (i.e., not genome-wide significant in any of the original GWAS; see Table S4 and Table S5), with 127 of these loci having lead SNP p-values ≤1e-5 in at least one of the original GWAS, and the remaining 23 having p-values ≤1.4e-4.
For the subset of SNPs with convergent effects across all 3 traits (Scz ∩ CanUD ∩ Smk) in the European ancestry samples, we identified 202 genomic risk loci with 259 lead SNPs (Table S6). The strongest locus was on chromosome 8, with the top lead SNP being rs73229090 (chr8:27442127, p = 1.5e-62; Figure 2), located in an intron of the non-coding gene GULOP, replicating previous associations with each trait (e.g.,41–43). This SNP is also an expression quantitative trait locus (eQTL) for EPHX2 in B cells, tibial artery, esophagus, and cultured fibroblast cells, CHRNA2 in the cerebellum, and CCDC25 in the nucleus accumbens.
Figure 2. Example forest plots from the ASSET European ancestry cross-disorder metaanalysis of CanUD, Smk, and Scz.
The lower right panel shows lead SNP (rs73229090) in Scz ∩ CanUD ∩ Smk subset. The upper right panel shows SNP (rs9924686) in Scz ∩ CanUD | Smk subset. The upper left panel shows SNP (rs2947411) in Scz ∩ Smk | CanUD subset. The lower left panel shows SNP (rs4620159) in CanUD ∩ Smk | Scz subset.
The Scz ∩ CanUD | Smk subset of SNPs revealed 37 genomic risk loci with 37 lead SNPs (Table S7). The top association was on chromosome 16, with lead SNP rs9924686 (chr16:30003076, p = 3.3e-15) within a locus previously implicated by Scz GWAS41. This SNP, located in the 3’ untranslated region of the serine/threonine-protein kinase gene TAOK2, has a CADD score of 18.16, suggesting deleteriousness, and a RegulomeDB score of 1f (eQTL + transcription factor binding/DNase peak), suggesting that this SNP is likely to affect transcription factor binding and linked to expression of a gene target. Furthermore, rs9924686 is an eQTL for several genes, including genes associated with metabolic and immunological traits44,45 (YPEL3 and INO80E in adipose tissue and several brain tissues) and alcohol intake45,46 (PPP4C and MVP in cultured cell fibroblasts).
We identified 46 genomic risk loci with 48 lead SNPs for the Scz ∩ Smk | CanUD subset (Table S8). Chromosome 2 had the strongest signal in this subset, with intergenic lead SNP rs2947411 (chr2:614168, p = 3.6e-19) that replicates previous associations with Smk43. This SNP was an eQTL for only one gene (SH3YL1 in whole blood).
There were 114 genomic risk loci and 143 lead SNPs for the CanUD ∩ Smk | Scz subset (Table S9). The strongest meta-analytic effect was at lead SNP rs4620159 on chromosome 6 (chr6:111744735, p = 1.8e-28); this locus was previously associated with Smk and CanUD47,48. The lead SNP is an intronic variant in REV3L, a gene previously associated with smoking and several metabolic traits23,45.
Cross-trait loci: African ancestry
No associations passed the genome-wide significance threshold (alpha = 5e–8) in the ASSET analysis of the African ancestry data. However, the 14,001 pleiotropic SNPs that were genome-wide significant in the European ancestry data showed smaller p-values than expected by chance in the African ancestry data (i.e., the distribution of p-values was significantly left-skewed, with a Kolmogorov-Smirnov goodness-of-fit test indicating significant (p < 2e-16) divergence from a distribution of 14,001 randomly sampled SNP p-values). This suggests that with larger sample sizes, future analyses might identify similar loci across both the European and African ancestry datasets.
Cross-ancestry meta-analysis
We performed a sample size-weighted cross-ancestry meta-analysis of the ancestry-specific one-sided meta-analysis results from ASSET. Unlike the ancestry-specific two-tailed meta-analyses described above, the one-sided meta-analysis in ASSET is more like a traditional meta-analysis, resulting in one effect size per SNP regardless of whether the SNP shows divergent directions of effect across traits. The cross-ancestry meta-analysis of CanUD, Smk, and Scz resulted in 448 genome-wide significant risk loci (Table S10).
Genetic associations with other phenotypes
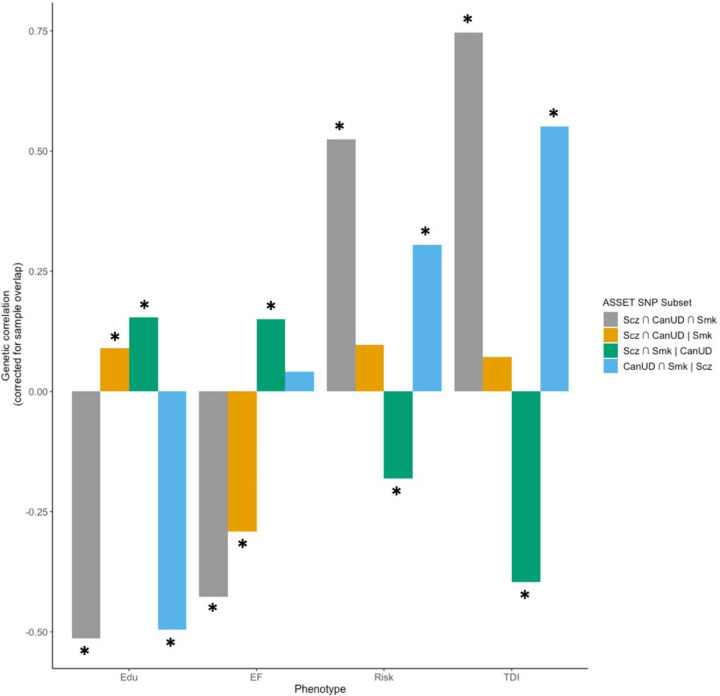
After defining SNP subsets using ASSET, we used GNOVA49 to estimate genetic correlations between the SNP subsets and educational attainment50 (Edu), executive function51, risk-taking46, and Townsend deprivation index (TDI; a regional measure of deprivation in the UK) in the European ancestry data (Figure 3, Table S11). Edu has previously been shown to be positively correlated with a subset of variants contributing to Scz risk40, despite negative genetic correlations between Scz and cognitive function52, and we expected that related socioeconomic status (i.e., TDI), executive function, and risk-taking phenotypes might be differentially associated with SNP subsets. For all subsets, the effect estimate was aligned with the direction of effect for CanUD.
Figure 3. Estimated genetic correlations between educational attainment (Edu), executive function (EF), risk-taking (Risk), and Townsend deprivation index (TDI) and SNP subsets from ASSET.
Asterisks (*) represent genetic correlations that are statistically significant after Bonferroni correction for 16 tests (p < 0.003).
The Scz ∩ CanUD ∩ Smk subset (i.e., variants with the same direction of effect on all three phenotypes) showed the strongest genetic correlations with all traits tested except Edu, where the Scz ∩ CanUD ∩ Smk and CanUD ∩ Smk | Scz subsets showed similar magnitudes of genetic correlation. For Edu, risk-taking, and TDI, the Scz ∩ CanUD ∩ Smk and CanUD ∩ Smk | Scz subsets showed the same direction of genetic correlation, while the Scz ∩ Smk | CanUD subset showed correlations in the opposite direction. In other words, genetic variants with the same direction of effect on CanUD and Smk, regardless of the direction of effect on Scz, showed similar negative genetic correlations with Edu, and positive genetic correlations with risk-taking and TDI, while genetic variants with the same direction of effect on Scz and Smk but not CanUD showed correlations in the opposite direction. Notably, the Scz ∩ CanUD ∩ Smk and Scz ∩ CanUD | Smk subsets were negatively genetically correlated with executive function, while the Scz ∩ Smk | CanUD subset was positively correlated and the CanUD ∩ Smk | Scz subset was not significantly correlated, suggesting a pivotal role of the intersection of CanUD and Scz, regardless of Smk, on executive functioning.
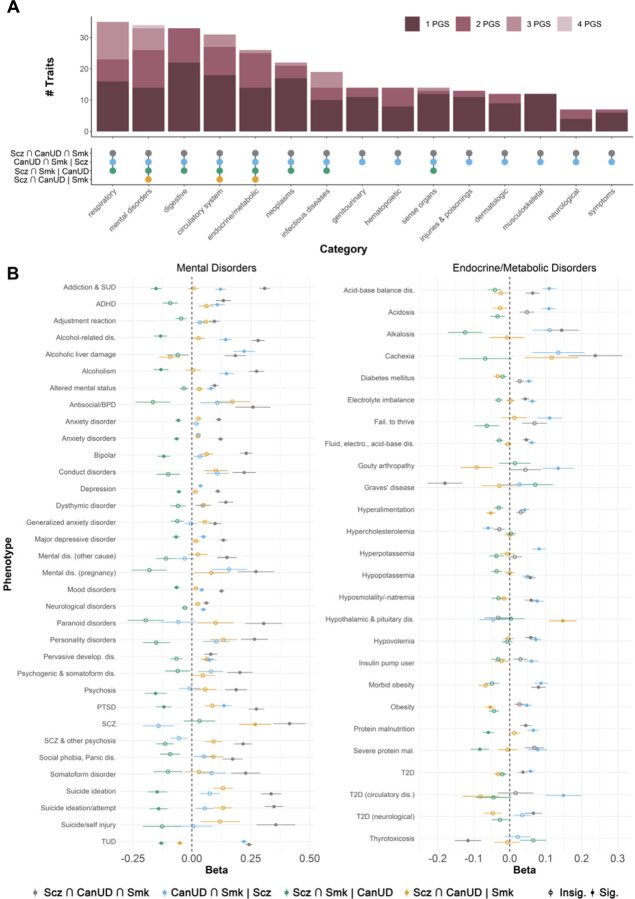
We also created polygenic scores (PGS) from each SNP subset in the European ancestry data and tested their associations with a range of health-related phenotypes in the BioVU biobank. In line with the genetic correlations in GNOVA, the PGS for the convergent subset of SNPS (Scz ∩ CanUD ∩ Smk) showed the strongest associations overall with most subsets of traits (Figure 4A), especially suicide attempt, psychosis, PTSD, conduct disorders, antisocial/borderline personality disorder, bipolar disorder, and alcohol-related disorders, among other psychiatric phenotypes (Figure 4B). Exceptions to this pattern included metabolic and endocrine phenotypes (Figure 4B), for which the PGS for the CanUD ∩ Smk | Scz subset had the greatest magnitude of associations with many of these traits, including acidosis, adult failure to thrive, type 2 diabetes, and hyperkalemia.
Figure 4. Associations between polygenic scores for SNP subsets from ASSET and health-related phenotypes in the BioVU biobank.
Panel A: Upset plot showing the number of phenotypes within different categories associated with one or more PGS. Panel B: Forest plots showing associations between the four ASSET SNP subset PGSs and mental disorders (left panel) and endocrine/metabolic traits (right panel) in BioVU.
Partitioned genetic covariance analysis
When stratified by broad tissue type, the genetic covariance between CanUD and Scz was significantly enriched for brain tissues in the European ancestry data (ρ = 0.029, p = 8.3e-4), while the genetic covariance between Smk and Scz was not significantly enriched for any tissue category (Figure S1, Table S12).
Discussion
The nature of the relationship between cannabis use and schizophrenia is a compelling and fiercely debated question in psychiatry, one that is complicated by the possibility of shared genetic factors and the frequent co-occurrence with tobacco smoking. There are major public health implications associated with a causal effect of cannabis use on schizophrenia risk, so a resolution of this question is important. Here, we describe the largest genome-wide, crossancestry and cross-disorder analyses of cannabis use disorder (CanUD), tobacco smoking (Smk), and schizophrenia (Scz) to date.
Our analyses revealed three key findings. First, CanUD and Smk are both genetically correlated with Scz, and this was consistent in both the European and African ancestry datasets. However, CanUD and Scz showed a greater degree of genetic overlap than Smk and Scz. Second, causal inference analyses suggested evidence of bidirectional causality for genetic liability to Scz, CanUD, and Smk, albeit in the presence of horizontal pleiotropy. Third, genomic loci that comprise the intersection between CanUD and Scz are associated with other mental health conditions and executive functioning.
In causal inference analyses that accounted for both correlated and uncorrelated forms of horizontal pleiotropy, we saw evidence for bidirectional causal relationships between all three phenotypes. We found evidence of horizontal pleiotropy for all trait pairs through the MR-PRESSO global test, but again found significant bidirectional causal estimates even after removing outlier SNPs for horizontal pleiotropy. Collectively, these results support causal links between CanUD, Smk, and Scz, although it is worth noting that the MR-Egger test did not support any causal relationships except for genetic liability for Smk causing CanUD. Convergent evidence from additional sources (especially longitudinal, prospective cohort studies) are needed53, especially in light of conflicting results from epidemiological studies5,6,54 and the limitations (and assumptions) associated with genetic methods of causal inference55.
Over 200 loci had convergent genome-wide significant effects on CanUD, Smk and Scz. The strongest convergent locus was on chromosome 8, with the lead SNP being a brain eQTL for EPHX2, CHRNA2, and CCDC25. While CHRNA2, a nicotinic cholinergic receptor (nAChR), seems an intuitive finding for Smk, this locus was most strongly associated with Scz and CanUD (Figure 2), and the top lead variant in all recent CanUD GWASs has mapped to this locus12,17,42. The role of cholinergic disturbance in positive56 symptoms and cognitive symptoms57 of Scz raise the potential for use of nChR agonists for treatment of comorbid Scz, CanUD and Smk58. EPHX2 encodes soluble epoxide hydrolase (sEH), the overexpression of which has been implicated in Scz59 and other diseases with a neuroinflammatory component (e.g., Alzheimer’s Disease). There is evidence for synergy between sEH and fatty acid amide hydrolase (FAAH60), which metabolizes endogenous cannabinoids and the inhibition of which is being evaluated for the treatment of pain. Given the emerging and paradoxical role of CanUD and a proinflammatory state61, the role of EPHX2 at the intersection of these disorders is intriguing.
Variants previously implicated in metabolic phenotypes emerged from the Scz ∩ CanUD | Smk subset. For instance, the lead SNP rs9924686 in TAOK2 was negatively associated with CanUD and Scz but not Smk and has been implicated in numerous prior GWAS of metabolic traits62,63. Further, while PGS derived from this subset were associated with both psychiatric and metabolic traits, the direction of association differed between this subset and the polygenic score of the fully convergent subset for metabolic but not psychiatric phenotypes (Figure 4).
Genetic predisposition for executive functioning was negatively correlated with the subset of fully convergent variants (Scz ∩ CanUD ∩ Smk), as well as those in the Scz ∩ CanUD | Smk subset, but positively associated with the other two subsets (i.e., where effects diverged for either CanUD or Scz), suggesting that only variation shared by CanUD and Scz related to lower executive functioning. Executive functioning deficits are a defining feature of Scz52 and a broad range of substance use disorders51,64,65, consistent with our findings. Executive functioning deficits have also been implicated in a broader range of mental health conditions51, which also aligns with our observation that variants influencing CanUD and ScZ, regardless of their effects of Smk, appear to index serious psychiatric comorbidity. Notably, while executive functioning is related to educational attainment51, the pattern of associations between convergent subsets of variants and educational attainment appeared to be quite different – for instance, any subset where CanUD and Smk diverged was associated with greater educational attainment, suggesting that educational attainment was more closely related to convergent signals for CanUD and Smk, rather than Scz. Thus, our study implicates the genetic liability to lower executive functioning as a common mechanism undergirding CanUD and Scz.
The genetic correlation between CanUD and Scz (rg = 0.37, SE = 0.02) was significantly greater (pdiff = 6.5e-18) than that between Smk and Scz (rg = 0.17, SE = 0.02) in the European ancestry data (with a similar but non-significant pattern in the African ancestry data: rg(CanUD, Scz) = 0.61, SE = 0.14 vs. rg(Smk, Scz) = 0.34, SE = 0.15). This suggests a greater proportion of shared genetic effects for CanUD and Scz than for Smk and Scz. When we partitioned the genetic covariance between phenotype pairs (Scz and CanUD, and Scz and Smk) into broad tissue types, the genetic covariance of CanUD and Scz was enriched for brain tissue, while the genetic covariance between Smk and Scz was not significantly enriched in any tissue category. These results are consistent with an overall pattern of findings in our study: the degree of genetic overlap, and the extent to which it is enriched in meaningful biological categories, is greater for CanUD and Scz than for Smk and Scz.
Our analyses of African ancestry data increase the generalizability of our findings. However, the smaller sample size of the individual African ancestry GWASs and limited available data for follow-up analyses (e.g., annotation files for partitioned genetic covariance analyses) constrained the extent to which we were able to accomplish our goal of equitable analyses. The genetic correlation between CanUD and Scz was substantially larger in the African ancestry data (rg = 0.610, SE = 0.140, p = 1.41e-5) than in the European ancestry data (rg = 0.373, SE = 0.023, p = 2.97e-60), albeit with a much larger standard error, suggesting that with increasing sample size, there could be considerable opportunity to identify pleiotropic loci.
Several other limitations applied to our study. First, while early age of cannabis initiation and use of high-potency cannabis have been suggested as risk factors for Scz, we did not have data available on potency and did not include age at first use in our analyses, as the only available GWAS for this phenotype was relatively underpowered and had non-significant SNP-heritability66. Similarly, we were unaware of any GWAS of cannabis consumption (i.e., heaviness or frequency of use). Another limitation is that the individual GWAS likely contain comorbid cases (e.g., a SCZ case with co-occurring CanUD), and this could artificially inflate our estimates of genetic correlations. Furthermore, cross-trait assortative mating has been shown to bias genetic correlations (e.g., between alcohol use disorders and schizophrenia)67, although the extent to which this could be affecting estimates of correlation between Scz, CanUD, and Smk specifically has not yet been quantified. Finally, there may be some sample overlap among the different GWAS (especially for CanUD and Smk), which could have inflated our MR results (but not CAUSE, which accounts for sample overlap).
Overall, our results add to the body of literature suggesting that both Smk and CanUD may be important predisposing factors as well as sequela of Scz. We demonstrate that the relationship between Smk, CanUD, and Scz may be due to both correlated genetic and reciprocal causal effects. Further, we identify executive functioning as a potential phenotype that links genetic liability for CanUD and Scz. While cigarette use is generally decreasing68, nicotine exposure through vaping is increasing26,69 and cannabis legalization and use are becoming more widespread worldwide70. As substance use policies and modes of use continue to change, it is important to carefully monitor epidemiologic trends in mental health conditions, especially schizophrenia and other psychotic disorders, and consider targeted interventions that may benefit individuals with heavy cannabis and tobacco use.
Methods
Genome-wide summary statistics
We used summary statistics from the largest available GWAS of each trait: Scz, CanUD and tobacco smoking.
Schizophrenia (Scz): We used data from the most recent Psychiatric Genomics Consortium (PGC) Schizophrenia genome-wide association study (GWAS) metaanalysis of individuals of European ancestry (N = 161,405; Ncases = 67,390)32. We also analyzed summary statistics from a GWAS meta-analysis of schizophrenia in African ancestry individuals (N = 15,846; Ncases = 7509), from the Cooperative Studies Program (CSP) #572 and the Genomic Psychiatry Cohort71.
Cannabis use disorder (CanUD): We used data from Levey et al.’s recent GWAS meta-analysis of cannabis use disorder17, which combined data from the Million Veteran Program, the Psychiatric Genomics Consortium, the Lundbeck Foundation Initiative for Integrative Psychiatric Research, and deCODE Genetics (European ancestry N = 886,025; Ncases = 42,281; African ancestry N = 120,208; Ncases = 19,065).
Ever-smoked tobacco regularly (Smk): We used summary statistics from the GWAS & Sequencing Consortium of Alcohol and Nicotine use (GSCAN) GWAS of self-reported ever/never regular cigarette smoking (European ancestry N = 805,431; Never = 393,707; African ancestry N = 24,278; Ncases = 9,916)72. We used the publicly-available set of summary statistics, which does not include data from 23andMe; the sample sizes reported here reflect that exclusion. This phenotype was measured in a variety of ways in different cohorts (e.g., “Have you smoked over 100 cigarettes over the course of your life?”, “Have you ever smoked every day for at least a month?”, “Have you ever smoked regularly?”). We selected this phenotype over others reflecting smoking quantity (cigarettes per day) or dependence because it had the largest sample size and the most genome-wide significant loci of any tobacco-related GWAS and shows considerable overlap with other nicotine use traits; thus, it seems likely that the genetics of Smk would be inclusive of most genetic factors related to tobacco involvement.
We also used genome-wide summary statistics for educational attainment, executive function, risk-taking, and the Townsend Deprivation Index (TDI):
Educational attainment: We used data from a GWAS of educational attainment from Lee et al.50 (2018) in individuals of European ancestry (N = 766,345).
Executive function: We used summary statistics from a GWAS of executive function in the European ancestry subset of the UK Biobank by Hatoum et al.51 (2023; N = 427,037).
Risk-taking: We used data from a GWAS by Linnér et al.46 (2019) of a single item that queried whether someone was a risk-taker. This GWAS was a meta-analysis of the UK Biobank and 10 replication cohorts (N = 466,571).
TDI: We used summary statistics from the Neale Lab GWAS (https://www.nealelab.is/uk-biobank) of the Townsend Deprivation Index (a measure of material deprivation in a region, incorporating data on unemployment, non-car-owning households, non-home-owning households, and household overcrowding) in the European ancestry subset of the UK Biobank (N = 336,798).
Genome-wide genetic correlation analyses
We used linkage disequilibrium score regression73,74 (LDSC) to estimate pairwise genome-wide genetic correlations (rg) between Scz, Smk, and CanUD. For the European ancestry summary statistics, we used pre-computed LD scores from the 1000 Genomes Phase 3 European reference panel (available from the LDSC website). For the African ancestry summary statistics, we used pre-computed LD scores from the PanUKBB African ancestry sample (available from https://pan.ukbb.broadinstitute.org/downloads).
We further tested whether genetic correlations were significantly different from each other using a block-jackknife method74,75. The block-jackknife method is a resampling approach, where the difference between resampling genetic correlations is used to calculate a jackknife standard error. From this standard error a Z-statistic is estimated and used in a two-tailed Z-test to determine if the difference between two genetic correlations is significantly different from zero (i.e., H0: rg(Scz, Smk) - rg(Scz, CanUD) = 0).
Causal inference analyses
We tested for causal relationships between Scz, CanUD, and Smk using CAUSE35. Compared to traditional Mendelian Randomization methods, CAUSE has the advantage of accounting for correlated horizontal pleiotropic effects (i.e., a genetic instrument is associated with a confounder which is related to both the exposure and the outcome) as well as uncorrelated horizontal pleiotropy. CAUSE uses a less stringent p-value threshold (p < 1e–3) to incorporate data from more variants across the genome. CAUSE constructs two nested models: a sharing model and causal model. Both models allow for horizontal pleiotropic effects; however, only the causal model includes a causal effect parameter (gamma). CAUSE compares the sharing and causal models to each other, to determine which model best fits the data, by estimating the difference in the expected log pointwise posterior density (ΔELPD). CAUSE then computes a z-score from the ΔELPD that can be compared to a normal distribution to obtain a one-sided p-value, which corresponds to a test of the null hypothesis that the sharing model fits the data at least as well as the causal model. Significant p-values therefore indicate the presence of a causal effect, after accounting for pleiotropy.
We performed additional causal inference analyses, including Mendelian Randomization Pleiotropy RESidual Sum and Outlier (MR-PRESSO36) analyses to test for horizontal pleiotropy and causal relationships among Scz, CanUD, and Smk. We performed these analyses using the TwoSampleMR R package76,77. We required SNP instruments to have p-value < 5e-8 and performed LD-based clumping. We report the results from the MR-PRESSO global test for horizontal pleiotropy, MR-PRESSO test for causality after removing outliers for horizontal pleiotropy, MR-Egger, weighted median, inverse variance weighted, simple mode, and weighted mode tests for causality, heterogeneity tests for the inverse variance weighted and MR-Egger tests, and the MR-Egger pleiotropy test (Table S3).
We only performed these analyses using the European ancestry summary statistics, because the African ancestry summary statistics were relatively under-powered for a causal inference analysis, particularly the Scz summary statistics.
Cross-disorder genome-wide association study meta-analysis
We used ‘Association analysis based on SubSETs’ (ASSET39) to combine the GWAS summary data for CanUD, Smk and Scz (separately by ancestry), using the two-tailed meta-analysis approach to obtain a single cross-disorder association statistic. Unlike traditional meta-analysis approaches, ASSET takes into account SNPs with significant effects on multiple disorders even if the effects on the traits are in opposite directions. We used the LDSC genetic covariance intercept to approximate the degree of sample overlap amongst the studies and included it in the ASSET covariance matrix. Default parameters were applied using the ‘h.traits’ function.
We then separated the ASSET results into subsets. Following Lam et al.40, we use the following notation for each subset: ∩ represents variant subsets with the same directions of effect (+ or −), and | represents variant subsets whose effect sizes are in the opposite direction of those for one versus the other two traits. We defined four subsets: (1) Scz ∩ CanUD ∩ Smk (i.e., a subset with convergent effects across all 3 traits); (2) Scz ∩ CanUD | Smk (i.e., a subset of variants with convergent effects for Scz and CanUD, but divergent effects for Smk); (3) Scz ∩ Smk | CanUD; and (4) CanUD ∩ Smk | Scz.
For each subset, we used FUMA v1.6.178 for annotation and identification of genomewide significant risk loci and independent lead SNPs. We used the matching ancestry subset of the 1000 Genomes Project Phase 379 reference panel for clumping and annotation of SNPs (e.g., the African ancestry reference panel for our African ancestry cross-disorder summary statistics). We used the default parameters for FUMA, with “independent SNPs” defined as those with p < 5e-8 and independent of each other with LD r2 < 0.6, and “lead SNPs” as independent SNPs which are strictly independent at a more stringent LD r2 < 0.1. Genomic risk loci (defined by LD blocks of independent SNPs) that were 250 kb or closer were merged into a single locus.
To perform a cross-ancestry meta-analysis, we used the ancestry-specific one-sided meta-analysis results from ASSET. Unlike the two-tailed approach described above, the one-sided meta-analysis in ASSET is more akin to a traditional meta-analysis and results in one effect size per SNP, regardless of whether the SNP shows divergent directions of effect across traits. We combined the ancestry-specific ASSET results using a sample-size weighted meta-analysis scheme in METAL80. As before, we uploaded METAL results to FUMA for clumping and annotation, using the 1000 Genomes Project Phase 379 all ancestries reference panel.
Identification of novel loci
To determine whether the ASSET meta-analysis revealed any novel loci in the European ancestry data that were not genome-wide significant in the original GWAS (CanUD, Smk, Scz), we used the LDLink package81 in R to identify all LD proxy SNPs (r2>0.6) for each of the 439 lead pleiotropic SNPs. We then merged these results with the summary statistics for the original CanUD, Smk, and Scz GWASs to determine whether the locus had been identified as genome-wide significant in any of the original GWASs.
Genetic correlations with other relevant phenotypes
After defining SNP subsets using ASSET, we used GeNetic cOVariance Analyzer (GNOVA49) to estimate genetic covariances (ρg) and correlations (rg) between the SNP subsets and relevant phenotypes in the European ancestry data. For all subsets, the effect estimate was aligned with the direction of effect for CanUD, for ease of interpretation. It was unclear how best to weight the estimate for each subset; following the example of Lam et al.40, we used the largest absolute effect size from the three phenotypes as SNP weights in each subset (flipping the sign of the estimate as necessary, to align with the direction of effect for CanUD).
Polygenic scores of ASSET-derived SNP subsets and associations in BioVU
We created polygenic scores for each ASSET-derived SNP subset in the European ancestry subset of the BioVU biobank (N=72,225)82,83. We fitted a logistical regression model to each of 1,338 case/control phenotypes (“phecodes”) to estimate the odds of diagnosis given each PGS. Models were adjusted for sex, median age of the longitudinal electronic health records, and the first 10 PCs. Analyses were conducted using the PheWAS version 0.99.5–2 R package. Phecodes were excluded from the analysis if they did not have at least two International Disease Classification codes mapping to a PheWAS disease category (Phecode Map 1.2; https://phewascatalog.org/phecodes) and had less than 100 cases. The disease phenotypes included 145 circulatory system, 123 genitourinary, 118 endocrine/metabolic, 125 digestive, 118 neoplasms, 91 musculoskeletal, 85 sense organs, 73 injuries & poisonings, 68 dermatological, 76 respiratory, 69 neurological, 64 mental disorders, 42 infectious diseases, 42 hematopoietic, 34 congenital anomalies, 34 symptoms, and 31 pregnancy complications. The phenome-wide significance threshold was set at a Bonferroni-adjusted threshold of p ≤ 3.62e-5.
Partitioned genetic covariance analyses
We used GNOVA49 to partition the genetic covariance (ρg) between CanUD, Smk, and Scz into salient annotation categories. These included tissue-specific functionality (GenoSkyline annotations, which are tissue-specific functional regions defined by integrating high-throughput epigenetic annotations). GNOVA is robust to potential sample overlap between summary statistics. We applied Bonferroni correction for multiple testing across all 3 trait pairs (CanUD ~ Smk, CanUD ~ Scz, and Smk ~ Scz) and 7 tissue types tested (e.g., we corrected for 3 × 7 = 21 tests, for an α = 0.002.) We only performed these analyses using the European ancestry summary stats, as the annotation data was derived using European ancestry samples.
Supplementary Material
Funding and competing interests
ECJ received support from K01DA051759. DAAB received funding from K99AA030808. JYK received support from a Canada Research Chair in Translational Neuropsychopharmacology (CIHR). HHAT is funded by a Canadian Institutes of Health Research Postdoctoral Fellowship. SSR was supported by T32IR5226 and DP1DA054394. DFL received support from 1IK2BX005058-01A2. JG received support from R01DA058862. J.G. is a holder of US patent 10,900,082 titled: ‘Genotype-guided dosing of opioid agonists’, issued 26 January 2021. J.G. is paid for their editorial work on the journal Complex Psychiatry.
Funding Statement
ECJ received support from K01DA051759. DAAB received funding from K99AA030808. JYK received support from a Canada Research Chair in Translational Neuropsychopharmacology (CIHR). HHAT is funded by a Canadian Institutes of Health Research Postdoctoral Fellowship. SSR was supported by T32IR5226 and DP1DA054394. DFL received support from 1IK2BX005058-01A2. JG received support from R01DA058862. J.G. is a holder of US patent 10,900,082 titled: ‘Genotype-guided dosing of opioid agonists’, issued 26 January 2021. J.G. is paid for their editorial work on the journal Complex Psychiatry.
References
- 1.Polderman T. J. C. et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet 47, 702–709 (2015). [DOI] [PubMed] [Google Scholar]
- 2.PF S., KS K. & MC N. Schizophrenia as a complex trait: Evidence from a meta-analysis of twin studies. Arch Gen Psychiatry 60, 1187–1192 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Volkow N. D. Substance use disorders in schizophrenia—clinical implications of comorbidity. Schizophrenia bulletin vol. 35 469–472 Preprint at (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gage S. H., Hickman M. & Zammit S. Association between cannabis and psychosis: epidemiologic evidence. Biol Psychiatry 79, 549–556 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Di Forti M. et al. Proportion of patients in south London with first-episode psychosis attributable to use of high potency cannabis: a case-control study. Lancet Psychiatry 2, 233–238 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Compton W. et al. Association between cannabis use disorder and schizophrenia stronger in young males than in females. Psychol Med 1–7 (2023) doi:DOI: 10.1017/S0033291723000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andréasson S., Engström A., Allebeck P. & Rydberg U. CANNABIS AND SCHIZOPHRENIA A Longitudinal Study of Swedish Conscripts. The Lancet 330, 1483–1486 (1987). [DOI] [PubMed] [Google Scholar]
- 8.Deak J. D. & Johnson E. C. Genetics of substance use disorders: a review. Psychol Med 1–12 (2021) doi:DOI: 10.1017/S0033291721000969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reginsson G. W. et al. Polygenic risk scores for schizophrenia and bipolar disorder associate with addiction. Addiction Biology (2017) doi: 10.1111/adb.12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson E. C. et al. The relationship between cannabis and schizophrenia: a genetically informed perspective. Addiction (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pasman J. A. et al. GWAS of lifetime cannabis use reveals new risk loci, genetic overlap with psychiatric traits, and a causal influence of schizophrenia. Nat Neurosci 21, 1161–1170 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson E. C. et al. A large-scale genome-wide association study meta-analysis of cannabis use disorder. Lancet Psychiatry (2020) doi: 10.1016/S2215-0366(20)30339-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng W. et al. The relationship between cannabis use, schizophrenia, and bipolar disorder: a genetically informed study. Lancet Psychiatry 10, 441–451 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillespie N. A. & Kendler K. S. Use of genetically informed methods to clarify the nature of the association between cannabis use and risk for schizophrenia. JAMA Psychiatry 78, 467–468 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Vaucher J. et al. Cannabis use and risk of schizophrenia: a Mendelian randomization study. Mol Psychiatry 23, 1287 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gage S. H. et al. Assessing causality in associations between cannabis use and schizophrenia risk: a two-sample Mendelian randomization study. Psychol Med 47, 971–980 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levey D. F. et al. Multi-ancestry genome-wide association study of cannabis use disorder yields insight into disease biology and public health implications. Nat Genet 55, 2094–2103 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartz S. M. et al. Comorbidity of severe psychotic disorders with measures of substance use. JAMA Psychiatry 71, 248 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gurillo P., Jauhar S., Murray R. M. & MacCabe J. H. Does tobacco use cause psychosis? Systematic review and meta-analysis. Lancet Psychiatry 2, 718–725 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kendler K. S., Lönn S. L., Sundquist J. & Sundquist K. Smoking and schizophrenia in population cohorts of Swedish women and men: a prospective co-relative control study. American Journal of Psychiatry 172, 1092–1100 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agrawal A., Budney A. J. & Lynskey M. T. The co‐occurring use and misuse of cannabis and tobacco: a review. Addiction 107, 1221–1233 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agrawal A. & Lynskey M. T. Tobacco and cannabis co-occurrence: Does route of administration matter? Drug Alcohol Depend 99, 240–247 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu M. et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet 51, 237–244 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hindocha C., Freeman T. P., Ferris J. A., Lynskey M. T. & Winstock A. R. No smoke without tobacco: a global overview of cannabis and tobacco routes of administration and their association with intention to quit. Front Psychiatry 7, 104 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cerdá M. et al. Association Between Recreational Marijuana Legalization in the United States and Changes in Marijuana Use and Cannabis Use Disorder From 2008 to 2016. JAMA Psychiatry 77, 165–171 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miech R. et al. Trends in use and perceptions of nicotine vaping among US youth from 2017 to 2020. JAMA Pediatr 175, 185–190 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oluwoye O. et al. Impact of tobacco, alcohol and cannabis use on treatment outcomes among patients experiencing first episode psychosis: Data from the national RAISE-ETP study. Early Interv Psychiatry 13, 142–146 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iasevoli F., Balletta R., Gilardi V., Giordano S. & De Bartolomeis A. Tobacco smoking in treatment-resistant schizophrenia patients is associated with impaired cognitive functioning, more severe negative symptoms, and poorer social adjustment. Neuropsychiatr Dis Treat 1113–1120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang C.-Y. et al. Cigarette smoking in patients with schizophrenia in China: prospective, multicentre study. Australian & New Zealand Journal of Psychiatry 44, 456–462 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Dickerson F. et al. Cigarette smoking among persons with schizophrenia or bipolar disorder in routine clinical settings, 1999–2011. Psychiatric services 64, 44–50 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Foti D. J., Kotov R., Guey L. T. & Bromet E. J. Cannabis use and the course of schizophrenia: 10-year follow-up after first hospitalization. American Journal of Psychiatry 167, 987–993 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trubetskoy V. et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 604, 502–508 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saunders G. R. B. et al. Genetic diversity fuels gene discovery for tobacco and alcohol use. Nature 612, 720–724 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quach B. C. et al. Expanding the genetic architecture of nicotine dependence and its shared genetics with multiple traits. Nat Commun 11, 5562 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morrison J., Knoblauch N., Marcus J. H., Stephens M. & He X. Mendelian randomization accounting for correlated and uncorrelated pleiotropic effects using genome-wide summary statistics. Nat Genet 52, 740–747 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verbanck M., Chen C.-Y., Neale B. & Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet 50, 693–698 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bowden J., Davey Smith G. & Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 44, 512–525 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bowden J. et al. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: the role of the I 2 statistic. Int J Epidemiol 45, 1961–1974 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhattacharjee S. et al. A Subset-Based Approach Improves Power and Interpretation for the Combined Analysis of Genetic Association Studies of Heterogeneous Traits. The American Journal of Human Genetics 90, 821–835 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lam M. et al. Pleiotropic Meta-Analysis of Cognition, Education, and Schizophrenia Differentiates Roles of Early Neurodevelopmental and Adult Synaptic Pathways. The American Journal of Human Genetics 105, 334–350 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Consortium S. W. G. of the P. G. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Demontis D. et al. Genome-wide association study implicates CHRNA2 in cannabis use disorder. Nat Neurosci 1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Erzurumluoglu A. M. et al. Meta-analysis of up to 622,409 individuals identifies 40 novel smoking behaviour associated genetic loci. Mol Psychiatry 25, 2392–2409 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Astle W. J. et al. The Allelic Landscape of Human Blood Cell Trait Variation and Links to Common Complex Disease. Cell 167, 1415–1429.e19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watanabe K. et al. A global overview of pleiotropy and genetic architecture in complex traits. Nat Genet (2019) doi: 10.1038/s41588-019-0481-0. [DOI] [PubMed] [Google Scholar]
- 46.Linnér R. K. et al. Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat Genet 51, 245 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu H. et al. Identifying genetic loci and phenomic associations of substance use traits: A multi-trait analysis of GWAS (MTAG) study. Addiction 118, 1942–1952 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pasman J. A. et al. Genetic Risk for Smoking: Disentangling Interplay Between Genes and Socioeconomic Status. Behav Genet 52, 92–107 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu Q. et al. A Powerful Approach to Estimating Annotation-Stratified Genetic Covariance via GWAS Summary Statistics. The American Journal of Human Genetics 101, 939–964 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee J. J. et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet 50, 1112–1121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hatoum A. S. et al. Genome-wide association study shows that executive functioning is influenced by GABAergic processes and is a neurocognitive genetic correlate of psychiatric disorders. Biol Psychiatry 93, 59–70 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song J. et al. The impact of educational attainment, intelligence and intellectual disability on schizophrenia: a Swedish population-based register and genetic study. Mol Psychiatry 27, 2439–2447 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Munafò M. R. & Davey Smith G. Robust research needs many lines of evidence. Nature 553, 399–401 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Elser H. et al. State Cannabis Legalization and Psychosis-Related Health Care Utilization. JAMA Netw Open 6, e2252689–e2252689 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davies N. M., Holmes M. V & Smith G. D. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. bmj 362, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caton M., Ochoa E. L. M. & Barrantes F. J. The role of nicotinic cholinergic neurotransmission in delusional thinking. NPJ Schizophr 6, 16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.D’Souza M. S. & Markou A. Schizophrenia and tobacco smoking comorbidity: nAChR agonists in the treatment of schizophrenia-associated cognitive deficits. Neuropharmacology 62, 1564–1573 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lieberman J. A., Javitch J. A. & Moore H. Cholinergic Agonists as Novel Treatments for Schizophrenia: The Promise of Rational Drug Development for Psychiatry. American Journal of Psychiatry 165, 931–936 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Ma M. et al. Key role of soluble epoxide hydrolase in the neurodevelopmental disorders of offspring after maternal immune activation. Proceedings of the National Academy of Sciences 116, 7083–7088 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kodani S. D. et al. Design and Potency of Dual Soluble Epoxide Hydrolase/Fatty Acid Amide Hydrolase Inhibitors. ACS Omega 3, 14076–14086 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wei T.-T. et al. Cannabinoid receptor 1 antagonist genistein attenuates marijuanainduced vascular inflammation. Cell 185, 1676–1693.e23 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pulit S. L. et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum Mol Genet 28, 166–174 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yengo L. et al. Meta-analysis of genome-wide association studies for height and body mass index in ~700000 individuals of European ancestry. Hum Mol Genet 27, 3641–3649 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koob G. F. & Volkow N. D. Neurobiology of addiction: a neurocircuitry analysis. The Lancet Psychiatry vol. 3 760–773 Preprint at 10.1016/S2215-0366(16)00104-8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hatoum A. S. et al. The addiction risk factor: a unitary genetic vulnerability characterizes substance use disorders and their associations with common correlates. Neuropsychopharmacology 47, 1739–1745 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Minică C. C. et al. Genome-wide association meta-analysis of age at first cannabis use. Addiction 113, 2073–2086 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Border R. et al. Cross-trait assortative mating is widespread and inflates genetic correlation estimates. Science (1979) 378, 754–761 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meza R., Jimenez-Mendoza E. & Levy D. T. Trends in tobacco use among adolescents by grade, sex, and race, 1991–2019. JAMA Netw Open 3, e2027465–e2027465 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tattan-Birch H., Brown J., Shahab L. & Jackson S. E. Trends in use of e-cigarette device types and heated tobacco products from 2016 to 2020 in England. Sci Rep 11, 13203 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hall W. et al. Public health implications of legalising the production and sale of cannabis for medicinal and recreational use. The Lancet 394, 1580–1590 (2019). [DOI] [PubMed] [Google Scholar]
- 71.Bigdeli T. B. et al. Contributions of common genetic variants to risk of schizophrenia among individuals of African and Latino ancestry. Mol Psychiatry 1–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saunders G. R. B. et al. Genetic diversity fuels gene discovery for tobacco and alcohol use. Nature 612, 720–724 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bulik-Sullivan B. K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bulik-Sullivan B. K. et al. An atlas of genetic correlations across human diseases and traits. Nat Genet 47, 1236 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Coleman J. R. I. et al. Genome-wide gene-environment analyses of major depressive disorder and reported lifetime traumatic experiences in UK Biobank. Mol Psychiatry 25, 1430–1446 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hemani G. et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 7, e34408 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hemani G., Tilling K. & Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet 13, e1007081-(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Watanabe K., Taskesen E., van Bochoven A. & Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun 8, 1826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Consortium T. 1000 G. P. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Willer C. J., Li Y. & Abecasis G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Myers T. A., Chanock S. J. & Machiela M. J. LDlinkR: An R Package for Rapidly Calculating Linkage Disequilibrium Statistics in Diverse Populations. Front Genet 11, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dennis J. et al. Genetic risk for major depressive disorder and loneliness in sex-specific associations with coronary artery disease. Mol Psychiatry 1–11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dennis J. K. et al. Clinical laboratory test-wide association scan of polygenic scores identifies biomarkers of complex disease. Genome Med 13, 6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.